Abstract
CRISPR-Cas9 technologies have dramatically increased the ease of targeting DNA sequences in the genomes of living systems. Fusion of chromatin-modifying domains to the nuclease-deactivated dCas9 has enabled targeted epigenome editing in both cultured cells and animal models. However, delivering large dCas9 fusion proteins to target cells and tissues is an obstacle to widespread adoption of these tools for in vivo studies. Here we describe the generation and characterization of two conditional transgenic mouse lines for epigenome editing, Rosa26:LSL-dCas9-p300 for gene activation and Rosa26:LSL-dCas9-KRAB for gene repression. By targeting gRNAs to transcriptional start sites or distal enhancer elements, we demonstrate regulation of target genes and corresponding changes to epigenetic states and downstream phenotypes in the brain and liver in vivo, and in T cells and fibroblasts ex vivo. These mouse lines are convenient and valuable tools for facile, temporally controlled, and tissue-restricted epigenome editing and manipulation of gene expression in vivo.
Epigenome editing with CRISPR-Cas9 systems has become a widespread approach for interrogating fundamental aspects of biological processes through targeted regulation of genes and the non-coding genome.1 Fusions of the nuclease-deactivated Cas9 (dCas9) to transcriptional activator or repressor domains enables targeted manipulation of gene expression. Initial work characterizing dCas9 fused to the tetramer of the VP16 acidic activation peptide (dCas9VP64) laid the framework for the development of a wide range of next-generation transcriptional activators, including SAM, VPR, p300core, Tet1CD, and others.2-9 In addition, dCas9 fusions that can repress gene expression have been characterized to function through direct or indirect chromatin modification using domains and enzymes such as KRAB, MECP2, and DNMT3a.2, 10, 11 In particular, we and others have shown that dCas9p300 and dCas9KRAB can be targeted to enhancers or near transcriptional start sites, leading to targeted histone acetylation or methylation and subsequent changes in gene expression.3, 12 These epigenome modifying dCas9-fusion proteins have been used for studies of gene regulation,13-15 directed cell differentiation,16-20 therapeutic gene modulation,21, 22 and high-throughput screening of putative gene regulatory elements.23-26
Although the majority of studies using CRISPR-based epigenome editing tools have focused on ex vivo cell culture systems in which delivery challenges are readily addressable, there are several examples of the powerful utility of targeted gene activation and repression in vivo. The smaller dCas9 from Staphylococcus aureus27 was incorporated into a dCas9KRAB fusion protein and delivered with a gRNA expression cassette via adeno-associated virus (AAV) for targeted gene repression in the mouse liver.21 Short “dead” gRNAs were used to activate gene expression in the liver and muscle of the Cas9 transgenic mouse28 using dual AAV delivery of these gRNAs with an MPH activator module.29 A dCas9-SunTag transgenic mouse has been used to activate gene expression in the midbrain30 and liver31 when a transcriptional activation domain was provided in trans. Another study used a constitutively expressed dCas9VP64 knock-in to activate expression of the Sim1 gene and rescue obesity resulting from Sim1 haploinsufficiency when crossed to mice transgenic for gRNA expression cassettes.22 While most previous studies focused on gene activation, one recent study generated a tetracycline-inducible dCas9KRAB mouse line. Hematopoietic stem cells (HSCs) from this line were then engineered ex vivo to assess the impact of five transcription factors on hematopoietic lineage determination after transplantation into a host animal.32 Plasmids encoding either dCas9VP64 or dCas9KRAB have also been transfected into the mouse brain to interrogate the role of epigenetic regulation in addiction behaviors.13
These experiments highlight the potential for studies of gene regulation using CRISPR-based epigenome editing. Accordingly, we sought to generate widely applicable transgenic mouse lines to readily perform temporally controlled and tissue-restricted epigenome editing for both gene activation and repression with simple addition of the gRNA. There are several potential advantages to using Cre-inducible expression of dCas9-based epigenome editors from transgenic mouse lines compared to viral delivery. First, several epigenome editors, such as dCas9 fusions to the acetyltransferase p300, are too large for viral vectors. Second, a single genomic copy ensures a more uniform expression level between cells and animals. Third, the use of a genetically encoded Cre recombinase adds tissue or cell type specificity when a corresponding promoter that can be packaged into viral vectors is unavailable. Fourth, it is possible to genetically encode both the gRNA and the Cre recombinase to alter gene expression or program epigenetic states in cell types that are not readily transduced by viral vectors. Fifth, the transgenic expression of dCas9-based epigenome editors may overcome challenges associated with immune system recognition of the bacterial Cas9 protein.33-37 Here we characterize and demonstrate the utility of two new mouse lines, Rosa26-LSL-dCas9-p300core (dCas9p300) and Rosa26-LSL-dCas9-KRAB (dCas9KRAB), for in vivo epigenome editing and modulation of gene expression.
Generation and Characterization of the Mouse Lines
We generated dCas9 epigenome editor mice by inserting a Cre-inducible cassette into the Rosa26 locus using traditional homologous recombination in mouse embryonic stem cells (Figure 1A). The inserted transgene consists of a CAG promoter followed by a loxP-stop-loxP (LSL) cassette, followed by the cDNA encoding the dCas9 fusion protein containing a FLAG epitope. This allows for inducible expression of the dCas9 fusion protein in response to Cre recombinase activity that results in removal of the stop signal between the loxP sites. To evaluate the inducibility of the dCas9p300 and dCas9KRAB lines, we treated the mice with AAV9:CMV.Cre and found Cre-dependent expression in spleen, skeletal muscle, liver, pancreas, and heart (Figure S1 and S9B).
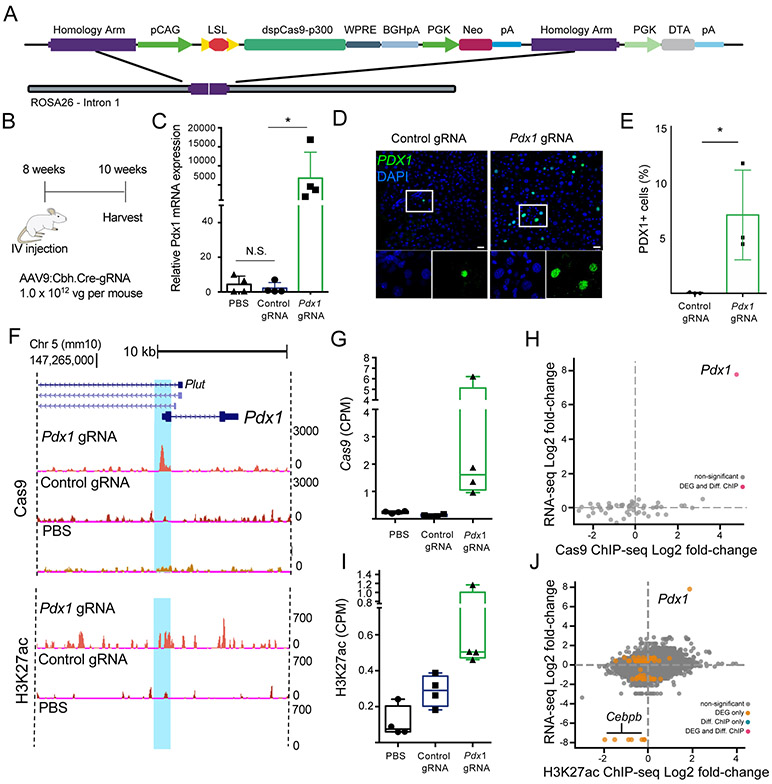
Figure 1: AAV-based gRNA and Cre recombinase delivery to Rosa26:LSL-dCas9p300 mice activates Pdx1 gene expression and catalyzes targeted histone acetylation.
A) Schematic of Rosa26:LSL-dCas9p300 (dCas9p300) knock-in locus. B) Experimental design for in vivo Pdx1 activation. C) Pdx1 mRNA quantification 8 weeks post-injection in liver tissue lysates isolated from mice injected with phosphate-buffered saline (PBS), or AAV9 encoding Cre and either a control non-targeting or Pdx1-targeting gRNA (n=4 per group, Kruskal-Wallis one-way ANOVA with Dunnett’s post-hoc, *p=0.0132). D) PDX1 immunostaining of liver tissue sections at 14 days post-injection of mice treated with control and Pdx1-targeted gRNAs. Scale bar = 50 μm E) Quantification of PDX1+ nuclei in control gRNA and Pdx1 gRNA treated animals (7.0% vs 0.03%, p=0.019, students t-test, n = 3 animals, with 3 images counted per animal). F) Representative browser tracks of dCas9 and H2K27ac ChIP-Seq data from treated livers at 2 weeks post-treatment (replicates are presented in Figures S3 and S4). G) dCas9 ChIP-seq quantification of sequencing counts (CPM = counts per million) in the gRNA target region of Pdx1 in samples from mice treated with Pdx1-targeted gRNA, control gRNA, or PBS (n = 4, one-way ANOVA with Dunnett’s post-hoc, p=0.059). H) RNA-seq and Pdx1 ChIP-seq analyses showing the relationship between changes in gene expression and occupancy of Cas9 genome-wide. Log2(fold-change) was calculated using the ratio of read counts between samples treated with the Pdx1-targeted gRNA relative to control non-targeting gRNA in dCas9p300 cells (n = 4, FDR < 0.05). I) H3K27ac ChIP-seq quantification of sequencing counts within a 1 kb window centered on the gRNA target site near the TSS of Pdx1 in samples from mice treated with the Pdx1-targeted gRNA, control gRNA, or PBS (n = 4, two-tailed student’s t-test, p = 0.07). J) RNA-seq and H3K27ac ChIP-seq analysis showing the relationship between changes in gene expression and genome-wide H3K27 acetylation for samples from mice treated with the Pdx1-targeted gRNA and control gRNA, (n = 4, FDR < 0.05, DEG = differentially expressed gene, orange dot; Diff. ChIP = differentially enriched ChIP-seq signal, blue dot; DEG and Diff. ChIP = differentially expressed gene and ChIP-seq enrichment, red dot). All bar-plot error bars represent standard deviation, all boxplots are drawn from 25th to 75th percentile with the horizontal bar at the mean and whiskers extending to the minima and maxima
Targeted Gene Activation in the Liver
To demonstrate activation of gene expression with dCas9p300 in cells harboring a single genomic insertion of the dCas9p300 expression cassette, we isolated primary fibroblasts from the gastrocnemius and tibialis anterior hindlimb muscles of the dCas9p300 mouse to test gRNAs in cell culture. We tested four gRNAs targeting the transcriptional start site (TSS) of Pdx1 (pancreatic and duodenal homeobox 1) (Figure S2A). Pdx1 was originally selected as a target due to its ability to induce liver cells into a pancreatic beta cell-like phenotype, including glucose-responsive insulin expression.38 Cells were co-transduced with lentiviral vectors encoding Cre and a single gRNA expression cassette and cultured for 4 days, at which point RNA was isolated for gene expression analysis by qRT-PCR. Two of the four gRNAs significantly increased Pdx1 mRNA levels compared to a control gRNA targeting Myod. For all future experiments, we continued with gRNA #4 as it showed the greatest activation of Pdx1 mRNA expression at ~150-fold increase relative to controls (Figure S2B).
This Pdx1-targeting gRNA was cloned into an AAV vector containing a ubiquitous CBh promoter driving Cre (AAV9:Cbh.Cre-Pdx1.gRNA), which we injected into 8 week old mice and collected liver tissue two weeks later to assess Pdx1 mRNA expression (Figure 1B). The Pdx1 mRNA levels were increased several thousand-fold compared to treatment with a corresponding AAV vector containing a non-targeting control gRNA (Figure 1C). We confirmed an increase in PDX1 protein levels in tissue sections of the treated livers as compared to a non-targeting gRNA control as measured by immunofluorescence staining (Figure 1D). In the animals treated with Pdx1 gRNA, 7.0% of nuclei were PDX1+, compared to 0.03% of cells in livers treated with the control gRNA (Figures 1E). However, we did not detect an increase in insulin mRNA by qRT-PCR or protein by immunostaining in any treated animals. This result is supported by previous studies showing that hyperactive PDX1-VP16 overexpression was necessary to achieve robust insulin induction.39
After demonstrating robust target gene activation, we characterized the specificity of gene regulation in the dCas9p300 mice. While the p300 acetyltransferase has many well-characterized targets involved in gene regulation, previous studies showed deposition of acetylation of lysine 27 on histone subunit 3 (H3K27ac) in regions of dCas9p300 binding that is concomitant with an increase in target gene mRNA levels.3, 14, 19, 40-43 To assess the genome-wide specificity of both dCas9 binding and targeted acetylation, we performed chromatin precipitation and sequencing (ChIP-seq) using antibodies against dCas9 and H3K27ac. Significant levels of specific binding of dCas9p300 to the targeted region of Pdx1 (Figures 1F-G, S3) and increased H3K27ac in the surrounding regions were readily detectable (Figures 1F, S4). Genome-wide comparisons showed that the only statistically significant differential dCas9 binding site between the Pdx1-targeted treatment and non-targeting gRNA controls was at the Pdx1 gRNA target site (Figure 1H). Genome-wide, H3K27ac levels were not significantly different between the treatment and gRNA control (Figure S5A). However, we observed a 2-fold increase in H3K27ac at the Pdx1 gRNA target binding site as compared to the non-targeting control gRNA (Figure 1I). When comparing mice injected with AAV encoding Cre and the Pdx1-targeted gRNA to control mice injected with saline, we found widespread differential H3K27ac levels which appears to be attributable to gRNA-independent consequences of overexpression of the constitutively active p300 acetyltransferase catalytic domain (Figure S5D).
To assess the specificity of changes in gene expression downstream of targeted histone acetylation, we performed RNA sequencing (RNA-seq) on mRNA harvested from treated mouse livers. When compared to treatment with Cre and a non-targeting gRNA, the Pdx1-targeted gRNA led to increases in Pdx1 expression that were the greatest change in gene expression transcriptome-wide (Figure 1H, 1J, S5G, S6A). This is consistent with previous levels of specificity reported for dCas9p300 in cultured cells.3 However, when comparing Pdx1-targeting gRNA or control non-targeting gRNA to saline-injected controls, we observed widespread gRNA-independent changes in gene expression as a result of dCas9p300 expression (Figure S6B-C). These results support gRNA-mediated specific epigenome editing and changes to target gene expression in vivo in this dCas9p300 mouse, but also underscore the need for proper controls to account for gRNA-independent effects when overexpressing constitutively active catalytic domains of epigenome editors.
Targeted Gene Activation in the Brain
To test gene activation in differentiated neurons with a single genomic insertion of the dCas9p300 expression cassette, we first sought to regulate gene expression in cultured primary neurons. Previously, we showed that gRNA-mediated recruitment of dCas9VP64 overexpressed from a lentiviral vector is sufficient to drive expression of the mature neuronal NMDA receptor subunit Grin2c in developing cerebellar granule neurons (CGNs).15 Here, to determine whether the Cre-inducible dCas9p300 transgene is similarly effective for activating gene expression, we cultured CGNs heterozygous for the dCas9p300 transgene and then either induced dCas9p300 expression by lentiviral delivery of Cre or over-expressed dCas9VP64 from a lentiviral vector for comparison. Recruitment of dCas9p300 to the Grin2c promoter activated Grin2c expression to a similar degree as dCas9VP64 recruitment (Figure S7A). This activation was specific for Grin2c and not due to accelerated neuronal maturation because the expression of another developmentally upregulated gene, Wnt7a, was not different in any of the conditions (Figure S7B). In addition, these data suggest that the dCas9p300 transgene is at least as effective as lentiviral dCas9VP64 for inducing expression of developmental genes in cultured primary neurons.
To determine the ability of transgenic dCas9p300 to regulate gene expression in neurons in vivo, we measured induction of neuronal activity-dependent genes in the hippocampus, a brain region that is important for spatial learning and memory. Fos transcription is rapidly and robustly induced by the physiological changes in neuronal firing that follow sensory experience. The Fos gene is flanked by five enhancer elements that regulate its stimulus-dependent transcription. We previously showed that expression of dCas9p300 from a transfected plasmid and its recruitment to enhancer 2 (Enh2) is sufficient to increase Fos mRNA and protein expression levels in cultured neurons.14 To recruit dCas9p300 to Enh2 in vivo, we delivered an AAV vector encoding a gRNA expression cassette, Cre recombinase under the control of the neuron-specific Syn1 promoter, and GFP to track transduced cells, by stereotactic injection into the dorsal hippocampus of dCas9p300 heterozygous mice (Figure 2A, S8A). For each mouse, one side of the brain was injected with a gRNA targeting Fos Enh2 and the other side was injected with a control non-targeting gRNA against LacZ as an in-animal control. Both sides of the hippocampus showed similar GFP expression and immunofluorescence staining showed robust detection of Cas9 in neurons, confirming that the Cre virus was inducing conditional dCas9p300 expression (Figure S8B). We allowed one cohort of the mice to explore a set of novel objects in the open field, which is a stimulus that induces expression of Fos in the hippocampus.14 As expected, immunostaining for FOS protein in the dentate gyrus region of the hippocampus showed sparse cells expressing high levels of induced FOS. However, there was a significantly greater number of high FOS-positive cells on the side of each brain that expressed the Fos Enh2-targeting gRNA compared with the side expressing the LacZ control gRNA (Figure 2B-C). Importantly, quantification of FOS protein levels across all of the transduced cells showed that the distribution of FOS was significantly increased in the cells expressing the Fos Enh2-targeting gRNA compared to those expressing the LacZ-targeting gRNA (Figure S8C), consistent with dCas9p300-dependent regulation of Fos gene expression in a cell-autonomous manner.
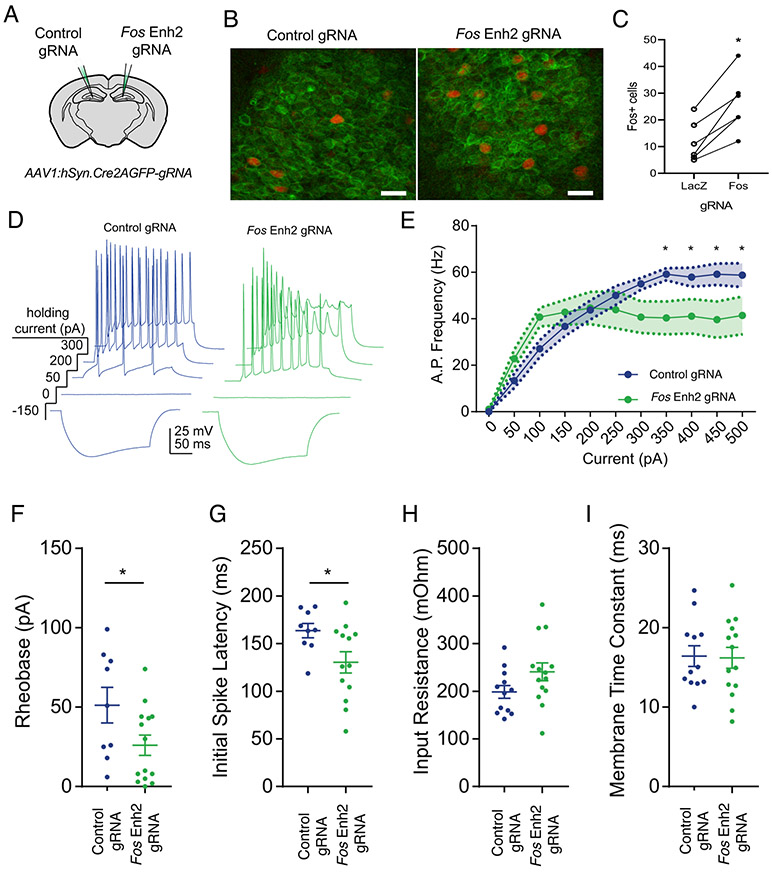
Figure 2: Epigenomic enhancement of Fos in vivo increases excitability in CA1 neurons.
A) Contralateral AAV injection strategy for comparison of targeting and non-targeting control gRNAs. AAV:Syn1-Cre.gRNA was injected into in the hippocampus of dCas9p300 mice. A gRNA targeting LacZ was used as control for the gRNA targeting Fos enh2. B) Immunoflourescence imaging of neurons in the dendate gyrus region of the hippocampus after transduction with AAV containing LacZ control-gRNA or Fos Enh2-gRNA and stimulation with novel objects. (AAV-gRNA+ neurons (green), FOS+ neurons (red). scale bar = 20 μm). C) Quantification of those FOS+ neurons in (B). The lines connect measurements from the two sides of the same mouse. Counts of FOS+ cells in tissue slices from n=6 paired ROIs per condition from 3 animals. p = 0.002 by student’s paired two-sided t-test. D) Representative current clamp traces from acute hippocampal slices. Virally-expressed GFP was used to identify CA1 neurons for recording. E) Summary of input/output. For current F(10,240) = 31.12, p < 0.0001, virus F(1,24) = 1.05, p = 0.32 and current x virus interaction F(10,240) = 4.64, p < 0.0001. For Fos Enh2 vs LacZ control at 350 pA p = 0.016, at 400 pA p = 0.031, at 450 pA p = 0.012, and at 500 pA p = 0.026 (two-way repeated measures ANOVA with post-hoc Fisher’s LSD test; * denotes p < 0.05.) F) Rheobase, minimal current to spike. G) Latency to first spike (p = 0.018). H) Input resistance (p = 0.092). I) Membrane time constant (p = 0.091). n=12 LacZ, n=14 Fos Enh2, each from 2 animals (For F-I, two-tailed students t-test; horizontal bars show mean; error bars show SEM).
A key application of using transgenic dCas9p300 mice to regulate neuronal gene expression in vivo is the ability to determine the consequences of epigenome editing on the function of physiologically relevant neuronal circuits in the intact brain. To determine whether dCas9p300-driven increases in FOS protein levels were sufficient to change neuronal physiology, we cut acute slices of hippocampus from a second cohort of dCas9p300 mice and performed current clamp recordings from virally transduced CA1 neurons (Figure S8D-E). Neurons expressing either the Fos Enh2 gRNA or the LacZ gRNA fired action potentials in response to progressive current steps (Figure 2D). However, the action potential stimulus-response curves were significantly different between the two treatments. The maximum firing rate was reduced for the neurons expressing the Fos Enh2 gRNA compared with neurons expressing the LacZ control gRNA (Figure 2E). However, the rheobase, which reports the input current at which neurons begin to fire spikes, and the initial spike latency were both significantly reduced in neurons expressing the Fos Enh2 gRNA compared to LacZ gRNA controls (Figure 2F and G). These data indicate increased excitability of the Fos Enh2 edited neurons. Importantly, the input resistance and membrane time constants between conditions were unchanged (Figure 2H and I). This suggests that the differences in action potential firing were due to changes in active, rather than passive, membrane properties. Taken together, these data show that dCas9p300-mediated epigenome editing of a single Fos enhancer in the adult brain in vivo induces changes in FOS protein levels that are sufficient to modulate neuronal physiology.
Targeted Gene Repression in the Liver
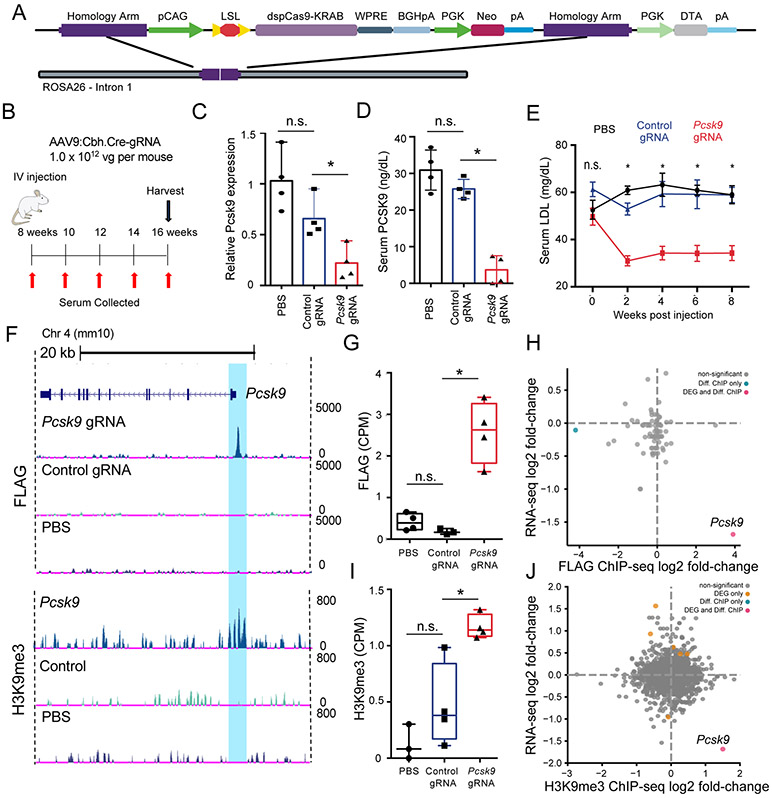
For characterization of the dCas9KRAB mice, we chose to target Pcsk9 since loss of PCSK9 protein is known to reduce serum levels of LDL cholesterol.44 In a previous study, AAV co-delivery of the smaller dCas9 from S. aureus fused to the KRAB domain and a Pcsk9-targeted gRNA repressed Pcsk9 in the liver, resulting in reduced serum LDL cholesterol levels.21 We delivered AAV9 encoding Cre and an S. pyogenes gRNA targeting the same sequence in the Pcsk9 promoter by tail vein injection to 8-week old dCas9KRAB mice (Figure 3A), as well as control mice injected with saline or a corresponding vector containing a non-targeting control gRNA (Figure 3B). At eight weeks post-injection, we harvested the mouse livers and assessed Pcsk9 mRNA levels. We found that Pcsk9 mRNA levels were reduced ~70% when dCas9KRAB was targeted to the promoter of Pcsk9 compared to the non-targeting gRNA and saline controls (Figure 3C). We also measured serum PCSK9 levels at 4 weeks post-injection and found a ~90% reduction in samples treated with the Pcsk9-targeted gRNA as compared to controls (Figure 3D). Additionally, serum was collected every two weeks during the eight-week experiment to assess LDL cholesterol levels (Figure 3B). There was a ~45% reduction in serum LDL cholesterol levels at each time point from 2 to 8 weeks (Figure 3E). Upon Pcsk9 repression, LDL receptor protein levels increase due to lack of receptor degradation,45 which was confirmed by western blot in three of the four Pcsk9-targeted animals (Figure S9A).
Figure 3: AAV-based gRNA and Cre recombinase delivery to Rosa26:LSL-dCas9KRAB mice represses Pcsk9 and catalyzes targeted histone methylation.
A) Schematic of Rosa26:LSL-dCas9KRAB (dCas9KRAB) knock-in locus. B) Schematic of the experimental design to test Pcsk9 repression in the dCas9KRAB mice. C) Pcsk9 mRNA quantification 8 weeks post injection in liver tissue lysates isolated from mice injected with phosphate-buffered saline (PBS), or AAV9 encoding Cre and either a control non-targeting or Pcsk9-targeting gRNA (n = 4 per group, one-way ANOVA with Dunnett’s post-hoc, *p = 0.0021) D) PCSK9 serum protein levels at 4 weeks post injection of PBS or AAV9 encoding Cre and either a control non-targeting or Pcsk9-targeting gRNA (n = 4 per group, one-way ANOVA with Dunnett’s post-hoc, * p=0.0001). E) LDL cholesterol levels in the serum at 8 weeks following administration of PBS, or AAV9:Cbh.Cre containing either a control non-targeting or Pcsk9-targeting gRNA (N=4 per group, two-way ANOVA with Tukey’s, * denotes p<0.0001). F) Representative browser tracks of dCas9KRAB (FLAG epitope) and H3K9me3 ChIP-Seq data from liver samples at 8 weeks post treatment (replicates are presented in Figures S10 and S11). Highlighted region corresponds to the area surrounding the gRNA target site in the Pcsk9 promoter. G) dCas9-FLAG ChIP-seq quantification of sequencing counts (CPM = counts per million) in the Pcsk9 promoter in samples treated with Pcsk9-targeted gRNA (n = 4), control non-targeting gRNA (n = 3), or PBS (n = 4) (one-way ANOVA with Dunnett’s post-hoc, p < 0.05.). H) Log2(fold-change) of RNA-seq and FLAG ChIP-seq signal comparing read counts in peaks between samples treated with Pcsk9-targeted gRNA (n = 4) and control non-targeting gRNA (n = 3) showing the relationship between gene expression and genome-wide dCas9KRAB binding. (Diff. ChIP = differentially enriched ChIP-seq signal, blue; DEG and Diff. ChIP = differentially expressed gene and ChIP-seq enrichment, red; FDR < 0.05. I) H3K9me3 ChIP-seq quantification of sequencing counts (CPM = counts per million) in the Pcsk9 promoter in samples treated with Pcsk9-targeted gRNA (n = 4), control non-targeting gRNA (n = 4), or PBS (n = 4) (one-way ANOVA with Dunnett’s post-hoc, p < 0.05.). J) Log2(fold-change) of RNA-seq and H3K9me3 ChIP-seq signal comparing read counts in peaks for samples treated with the Pcsk9-targeting gRNA (n = 4) and control non-targeting gRNA (n = 4) (DEG = differentially expressed gene, orange; Diff. ChIP = differentially enriched ChIP-seq signal, blue; DEG and Diff. ChIP = differentially expressed gene and ChIP-seq enrichment, red; FDR < 0.05). All bar-plot error bars represent standard deviation, all boxplots are drawn from 25th to 75th percentile with the horizontal bar at the mean and whiskers extending to the minima and maxima
To assess the specificity of gene regulation by dCas9KRAB in these transgenic mice, we performed RNA-seq and ChIP-seq on liver tissue samples from dCas9KRAB heterozygous mice injected with AAV encoding Cre and the Pcsk9-targeted gRNA, AAV encoding Cre and the control non-targeting gRNA, or saline controls. For RNA-seq, we performed comparisons of treatment with the Pcsk9-targeted gRNA to both the control gRNA and saline controls. In both cases, Pcsk9 was one of the most strongly downregulated genes, with only 15 and 6 other genes, respectively, significantly changed transcriptome-wide (FDR <0.01, Figure S6D-F, Supplementary Data). ChIP-seq was performed using an anti-FLAG antibody for dCas9 binding specificity and a H3K9me3 antibody to detect histone methylation as a result of dCas9KRAB-mediated recruitment of methyltransferases (Figure 3F-J, S10, and S11). Specific and significantly enriched dCas9 binding to the gRNA target site upstream of Pcsk9 was readily detected (Figure 3F-G). Additionally, significant increases in H3K9me3 deposition in the target region of the Pcsk9-targeted gRNA were evident compared to the non-targeting control gRNA or saline controls (Figure 3F,I). The dCas9KRAB binding was also highly specific genome-wide, as the Pcsk9 gRNA target site was one of only two significantly differential peaks between samples treated with the Pcsk9 gRNA and the control gRNA in the dCas9 ChIP (Figure 3H) and the only differential peak in the H3K9me3 ChIP (Figure 3J).
Targeted Gene Activation and Repression in Primary Immune Cells
To further demonstrate the versatility of the dCas9p300 and dCas9KRAB mice, we tested modulation of expression of the master regulator transcription factor Foxp3 in CD4+ T cells. Foxp3 is essential for the development of a specialized arm of regulatory CD4+ T cells (Tregs) that has a critical role in the attenuation and maintenance of the immune response to self and foreign pathogens.46-49 Activation of Foxp3 expression and the generation of Tregs with immunosuppressive properties is of interest for cellular immunotherapies. Prior work has shown that lentiviral delivery of dCas9p300 targeted to the promoter of Foxp3 was sufficient to increase FOXP3 protein levels.50
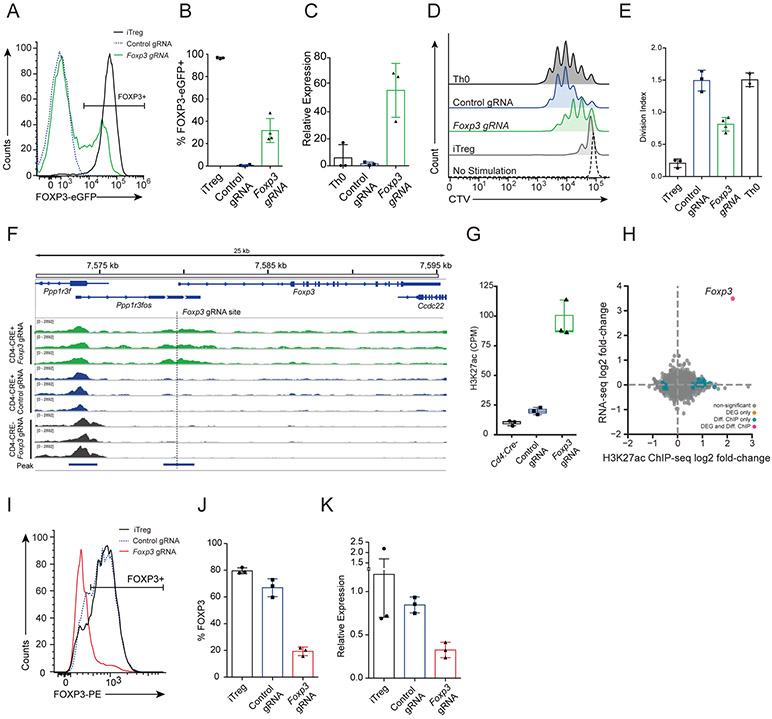
We activated Foxp3 in T cells from the dCas9p300 mice using a Foxp3-eGFP; CD4-Cre; Rosa26-LSL-dCas9p300 cross to conditionally express dCas9p300 in all CD4+ T cells. Spleen and lymph nodes were isolated from these mice, dissociated, and CD4+/CD25−/CD44lo/CD62Lhi naïve T (Tn) cells were purified by fluorescence activated cell sorting (FACS) (Figure S12A and S12B,C). Tn cells were cultured in TCR-stimulating conditions (Th0) consisting of plate bound α-CD3, α-CD28, the blocking antibodies α-IL4 and α-IFNg, and IL-2 prior to their transduction with retroviral supernatant containing a gRNA expression cassette. We first tested a panel of 12 gRNAs targeting the Foxp3 promoter and identified four gRNAs immediately upstream of the TSS which had an activating effect (Figure S2B, D-E). We selected Foxp3-gRNA-5 and a non-targeting control gRNA for all downstream experiments. Following retroviral delivery of this Foxp3-targeting gRNA, Foxp3 activation was detected in ~30% of transduced cells as measured by FOXP3-eGFP signal via flow cytometry (Figure 4A-B). We verified the gene activation by qRT-PCR and found that the recruitment of dCas9p300 to the Foxp3 promoter led to a ~50-fold increase of Foxp3 expression relative to the control gRNA or untreated Th0 T cells (Figure 4C). Using RNA-seq to assay gene expression in Foxp3-gRNA treated Th0 cells, we found that relative to Th0 cells treated with the control gRNA, FoxP3 was the only differentially expressed gene genome-wide (Figure 4H, S13).
Figure 4: Epigenome editing in T cells for activation and repression of Foxp3.
A) Flow cytometry analysis of FOXP3-eGFP expression in CD4+ T cells purified from Rosa26:LSL-dCas9p300;CD4:Cre;Foxp3:eGFP mice cultured in vitro under Th0 polarization conditions (rIL-2) after transduction with retrovirus encoding the cell surface marker Thy1.1, for tracking transduced cells, and either the Foxp3-gRNA (green) or control-gRNA (blue). Cells cultured in iTreg polarization conditions (rIL-2, hTGFβ1) were included as a positive control for Foxp3 expression (black). B) Summary data depicting the percentage of Thy1.1+ cells that were FOXP3-EGFP+ (p<0.0001; one-way ANOVA with Dunnett’s post-hoc, iTreg n = 3, control gRNA and Foxp3-gRNA n = 4). C) qRT-PCR measurement of Foxp3 mRNA levels in Th0 cells compared to cells treated with Foxp3-targeting gRNA (green), control non-targeting gRNA (blue), or no virus (black) (p = 0.0190, one-way ANOVA with Dunnett’s post-hoc, n = 3 per condition). D) Flow cytometry histograms showing proliferation of Cell Trace Violet (CTV)-labelled CD4+/FOXP3-eGFP− conventional T cells (Tconv) after 72 hrs of in vitro co-culture with aCD3/aCD28 dynabeads and either FOXP3-eGFP+ iTreg cells (grey, n = 3), Th0 cells (black, n = 2), FACS-purified Thy1.1+/FOXP3-eGFP+ cells treated with Foxp3-gRNA (green, n = 3), or Thy1.1+ cells treated with control non-targeting gRNA (blue, n = 3). Tconv cells with no aCD3/aCD28 and rIL-7 served as a no activation control (dashed). E) Suppressive capacity of Tregs summarized as division index of Tconv (p < 0.0001 one-way ANOVA with Tukey’s post-hoc. F) Browser track of H3K27ac ChIP-seq read counts at the Foxp3 locus in Th0 cells from a Rosa26:LSL-dCas9p300 mouse that was either Cd4:Cre+ or Cd4:Cre−. Cells were treated with retrovirus containing Foxp3-targeting or control non-targeting gRNA and Thy1.1 reporter. ChIP-seq was performed 72hrs after transduction on sort-purified Thy1.1+ cells (red = Cd4:Cre+ Foxp3-gRNA; yellow = Cd4:Cre+ control-gRNA; blue = Cd4:Cre− Foxp3-gRNA). G) Summary of H3K27ac ChIP-seq reads counts per million within the MACS2-called peak that intersects the Foxp3-gRNA target site for each genotype and gRNA treatment (green = Foxp3-gRNA treated Cd4:Cre+ Th0, blue = control-gRNA treated Cd4:Cre+ Th0; black = Foxp3-gRNA treated Cd4:Cre− Th0, n = 3 per condition). H) Scatter plot depicting log2(fold-change) of gene expression and H3K27ac enrichment when comparing read counts from Cd4:Cre+ Rosa26:LSL-dCas9p300 Th0 cells treated with Foxp3-gRNA to control-gRNA (FDR < 0.01; DEG = differentially expressed gene, orange dot; Diff. ChIP = differentially enriched ChIP-seq signal, blue dot; DEG and Diff. ChIP = differentially expressed gene and ChIP-seq enrichment, red dot) I) Flow cytometry analysis of FOXP3 expression in CD4+ T cells purified from Rosa26:LSL-dCas9KRAB mice cultured in vitro under iTreg polarization conditions and transduced with retrovirus encoding the indicated gRNAs. J) Summary data showing the percentage of Thy1.1+ cells that were FOXP3-EGFP+ for each gRNA treatment (p < 0.0001, one-way ANOVA with Dunnett’s post-hoc, n = 3 per condition). K) qRT-PCR measurement of Foxp3 mRNA levels in iTreg cells treated without virus (black), control-gRNA (blue), or Foxp3-targeting gRNA (red) (p < 0.0135, one-way ANOVA with Dunnett’s post-hoc, n = 3 per condition). All bar-plot error bars represent standard deviation, all boxplots are drawn from 25th to 75th percentile with the horizontal bar at the mean and whiskers extending to the minima and maxima.
To quantify changes in genome-wide histone acetylation made by dCas9p300, we performed ChIP-seq for H3K27ac in Th0 cells from Cd4-Cre+ dCas9p300+ mice transduced with Foxp3-targeting or control gRNAs. We identified a significant enrichment in H3K27ac at the gRNA target site and only 25 other H3K27ac peak regions were detected as differentially acetylated genome-wide (Figure 4F-H, Figure S5B, Supplementary Data). Together, the RNA-seq and ChIP-seq analyses highlight the specificity and efficacy of targeting dCas9p300 to one location of the genome (Figure 4H). However, when we compared RNA-seq and ChIP-seq in Th0 cells treated with the Foxp3-gRNA from mice with or without Cd4-Cre, we observed numerous differences in gene expression and H3K27 acetylation (Figure S5H, Supplementary Data). When comparing RNA-seq data from Cre− vs Cre+ cells, the same set of genes were found to be differentially expressed regardless of whether cells were treated with Foxp3-targeting or control gRNA, further supporting a guide-independent effect of active dCas9p300 expression on the baseline epigenetic state in this mouse strain (Figure S13). This effect is specific to the dCas9p300 mice and is not observed in dCas9KRAB mice when comparing liver samples from mice treated with either Pcsk9-targeting gRNA or saline (Figure S5F,I). These results reiterate the need for experimental controls that include an active dCas9p300 for comparisons to an accurate baseline epigenetic state.
To test the capacity of FOXP3+ Th0 cells generated by targeted epigenome editing to function as Tregs, we used an in vitro suppression assay to test their ability to limit proliferation of activated CD4+ T cells in co-culture. The cells treated with Foxp3-targeting gRNA showed enhanced suppression relative to untreated cells or cells treated with control gRNA controls, showing suppressive activity similar to the positive control iTregs generated in vitro by established protocols (Figure 4D and E). To verify that the suppression activity was mediated by the gRNA-treated cells, we performed the suppression assay with serial dilutions of the number of Foxp3-induced cells in co-culture and found a dose-dependent suppressive effect (Figure S12F and G).
To demonstrate gene repression in another cell type from the dCas9KRAB mice ex vivo, we isolated naïve CD4+ T cells from the lymph nodes and spleen of Cd4:Cre/dCas9KRAB mice and generated FOXP3+ iTregs in vitro via TCR activation and treatment with IL-2 and TGFβ1. iTregs were transduced with retrovirus containing either the Foxp3-targeting or non-targeting gRNAs. We achieved ~70% reduction in Foxp3 mRNA levels in cells with dCas9KRAB targeted to the Foxp3 promoter as compared to untreated iTregs or those treated with the control gRNA (Figure 4I). In addition, we assessed FOXP3 protein levels by immunostaining and found a significant reduction in the percentage of FOXP3+ cells when transduced with the Foxp3-targeting gRNA as compared to controls (Figure 4J-K). These results confirm our ability to repress genes in multiple tissues and cell types of the dCas9KRAB mouse line both in vivo and ex vivo.
Discussion
Perturbation of gene expression has long been a powerful tool for uncovering the functions of genes. Recent technological advances have enabled not only the study of gene function, but also the mechanism by which the genes themselves are regulated. Using dCas9 fused to epigenome modifiers has proven to be a productive strategy to uncover unknown gene function, to map regions of the non-coding genome, and to validate potential therapeutic applications using CRISPR-based epigenome editors in vivo. Here we describe the generation and characterization of Cre-inducible dCas9p300 and dCas9KRAB transgenic mouse lines for targeted activation or repression of promoters and non-coding regulatory elements in vivo or in primary cells ex vivo, including in the liver, T cells, fibroblasts, and neurons. We demonstrate induction of dCas9 epigenome editor expression using viral or transgenic Cre delivery combined with viral gRNA delivery. The targeted gene activation or repression induced both changes in mRNA transcript levels and protein changes that elicit downstream phenotypes. Concomitant with expression changes, we show targeted deposition of histone marks subsequent to the recruitment of dCas9-based epigenetic effectors at both promoters and enhancers.
An unbiased, comprehensive, genome-wide analyses of epigenome editing specificity by ChIP-seq and RNA-seq showed both a high level of precision in DNA-targeting and interesting differences between the dCas9p300 and dCas9KRAB editors. The dCas9p300 editor was highly precise and robust in binding to its genomic target (Figure 1H), which led to corresponding specificity in catalyzing H3K27ac (Figure 1F-J, 4F-H) and activating target gene expression (Figure S5A, B) when compared to a control non-targeting gRNA. However, when compared to saline injection or Cd4:Cre− control samples, in which Cre is not present and the dCas9p300 would not be induced, there were widespread genome-wide changes to H3K27ac and gene expression (Figure S5G, H). Interestingly, these changes did not appear to result in any adverse phenotype. Since the off-target changes were similar between the targeting and non-targeting gRNA in T cells (Figure S13), we interpret the changes to be gRNA-independent and related to the expression of the constitutively active core catalytic domain of the p300 acetyltransferase that may alter the genome-wide epigenetic baseline of these cells. This is consistent with previous observations that dCas9 fused to catalytic enzymes such as DNA methyltransferases can exhibit gRNA-independent changes,51 and underscores the importance of using proper gRNA controls to determine the consequences of modulating gene expression through epigenome editing. In contrast to dCas9p300, which has inherent non-specific catalytic activity, dCas9KRAB serves as a scaffold to recruit other enzymatic histone modifiers. Accordingly, target changes to gene expression and epigenetic state were highly specific when comparing to either the non-targeting gRNA or saline controls (Figure S5C, F, I), which was consistent with the high level of specificity of DNA-targeting (Figure 3H) and histone modifications (Figure 3J).
Following the development of CRISPR-Cas9 for genome editing in human cells,52-56 a Cre-inducible Cas9 transgenic mouse was quickly generated and widely distributed for in vivo studies of gene function genetics of disease.28 This mouse line has been essential to numerous important breakthroughs including in vivo dissection of gene function57 studies of gene regulatory networks,58 and in vivo genetic screens.59 We expect these mice to be similarly powerful for enabling in vivo studies of endogenous gene knockdown, activation, and perturbation of epigenetic states. Accordingly, the mice have been deposited for distribution via The Jackson Laboratory (Stock No: 033065 (dCas9p300) and 033066 (dCas9KRAB)).
Materials and Methods
Generation of Rosa26:LSL-dCas9-p300 and Rosa26:LSL-dCas9-KRAB transgenic lines
All experiments involving animals were conducted with strict adherence to the guidelines for the care and use of laboratory animals of the National Institute of Health (NIH). All experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Duke University.
To generate mouse lines for conditional expression of these dCas9 fusion proteins, we used a modified pAi9 targeting vector. The pAi9 vector targets the Rosa26 locus and contains the following features: 5’ Rosa homology arm, CAG promoter, a loxP flanked triple polyadenylation signal (pA) stop cassette (lox-stop-lox; LSL), a codon optimized dCas9-p300 cassette3 or dCas9-KRAB12 with either a 1X-FLAG or 3X-FLAG respectively, a woodchuck hepatitis posttranscriptional regulatory element (WPRE), a bovine growth hormone pA, a PGK-Neo-pA selection cassette, and the 3’ homology arm. This modified pAi9 targeting vector was electroporated into hybrid G4 B6N/129S6 ES cells and targeting of the ROSA locus was confirmed by PCR and sequencing. Positive clones were expanded and injected into the 8-cell morulae of ICR mice. Chimeric mice were then mated to establish the transgenic line. Mice are genotyped using the following primers: Forward Primer: GCAGCCTCTGTTCCACATACAC, Reverse Primer: TAAGCCTGCCCAGAAGACTC, Second F primer: AAAGTCGCTCTGAGTTGTTAT. The following PCR conditions are used for genotyping PCR: 95°C for 5 min, 35 cycles of: 95°C for 30 seconds, 57°C for 45 seconds, 72°C for 1 minutes, 72°C for 3 minutes, and 12°C Hold. Expected product sizes are as follows: WT band is 235bp; Knock-In is 162bp. All mice used in experiments were dCas9p300 or dCas9KRAB heterozygous animals.
The new mouse lines are available via The Jackson Laboratory: Stock No: 033065 ∣ Rosa26-LSL-dCas9-p300; Stock No: 033066 ∣ Rosa26-LSL-dCas9-KRAB.
All mice were housed in pathogen free barrier conditions with the exception of those mice used for stereotaxic injections. Mice were kept in ventilated racks at a temperature of 22°C ± 1°C and humidity of 30-70% with food and water ad libitum. Mice used for liver and brain targeting experiments experienced a 12/12h light/dark cycle, whereas mice used for T cell experiments experienced a 14/10h light/dark cycle. All mice were used between 8-12 weeks old and not selected based on specific sex, with the exception of those adult males used only for sterotaxic injection experiments.
Isolation and culture of primary dCas9p300 cells
Fibroblasts were isolated from gastrocnemius and tibialis anterior of hindlimbs of 6 week old dCas9p300 heterozygous animals using a modified version of a protocol by Springer et al60. Instead of proceeding with myoblast isolation, fibroblasts were selected through selective trypsinization of the myoblasts out of the culture using 0.25% Trypsin. Fibroblasts were then grown in DMEM +10% FBS.
Cerebellar granule neurons (CGNs) from male and female postnatal day 7 (P7) dCas9p300 heterozygous pups were cultured following our published protocols15. Briefly, the cerebellar cortex was removed and dissociated with papain, granule neuron progenitors were purified by centrifugation through a Percoll gradient, and neurons were plated on poly-D-lysine coated plates in neurobasal media with B27 supplements (Invitrogen), 1% FBS, and pen-strep. On day 1 of in vitro culture (DIV1), neurons were transduced with lentiviruses. All neurons received either 2 gRNAs targeting the Grin2c promoter (Site 2-2. Site 2-3 from Frank et al.15 or a control non-targeting gRNA complementary to a sequence in the LacZ gene14. gRNAs were delivered in a FUGW-based U6 chimeric gRNA expression vector co-expressing GFP. In addition to the gRNA virus, one set of cells received a lentivirus expressing the Cre recombinase under the control of the hSyn1 promoter (Addgene 86641). This induced expression of the dCas9p300 transgene in the neurons. Other neurons received lentivirus expressing dCas9VP64 with the gRNAs to compare the functions of the two transgenes in neurons from the same cultures5. Viruses were titered on HEK293T cells before use and delivered at a MOI of 10.
AAV cloning and production
An AAV backbone containing a gRNA cloning site, pCBh.Cre-WPRE-hGHpA was obtained from Addgene (plasmid #60229). Two oligonucleotides per gRNA were purchased, annealed, and cloned into the SapI cloning site. Prior to AAV production, ITRs were verified by SmaI digest. pCre-Pdx1.gRNA and pCre-control.gRNA were used to generate high titer AAV9. Viral supernatants at the indicated titers were dialyzed w/350 mM NaCl and 5% D-sorbitol in PBS. For neurons, gRNAs for LacZ or Fos Enh2 sequences were cloned in the Sap1 site of an AAV backbone (Addgene #60231) with hSyn-Cre-2A-GFP. High titer AAVs for intracranial delivery were produced in serotype AAV2/1 by the Duke University Viral Vector Core Facility.
Lentiviral cloning and production
Lentiviral production was performed using standard procedures in HEK293T (ATCC CRL-11268) cells using second generation lentiviral vectors VSVg and d8.961. Lentiviral preps for fibroblast transduction were purified using Lenti-XTM (Clonetech, Cat#631232) to a 20X concentration. Fibroblasts were transduced at 1X concentration in the absence of polybrene for 24 hours, media was then changed and fibroblasts were returned to growth media. For neurons, viruses were purified by ultracentrifugation and resuspended at a titer of ~5x105 infectious particles per μL of PBS. Neurons were transduced in a solution of DMEM + 0.5 μg/mL polybrene for 6 hrs, then neurons were washed and returned to conditioned medium for 5 days to allow for viral gene expression.
Retroviral cloning and production
Retrovirus was produced in PLAT-E cells (provided by Dan Littman) following standard procedures62. Briefly, individual gRNA were cloned into the BbsI site of a Mouse Stem Cell Virus (MSCV) backbone that contained a gRNA expression cassette controlled by the mU6 promoter and a Thy1.1 reporter controlled by an hPGK promoter. Cloned vectors were transfected into PLAT-E cells using Lipofectamine 3000 (Thermo, Cat#L3000008) and culture media was replaced after 16 hrs. Virus-containing media was collected 48 hrs or 72 hrs after transfection, spun at 800 x g for 10 minutes and the viral supernatant was snap frozen in liquid nitrogen until the transduction of T cells.
All sequences for gRNA protospacers can be found in Supplementary Table 1.
RNA isolation and qRT-PCR
RNA was isolated from fibroblasts using a Qiagen RNAeasy (Cat#74136). cDNA synthesis was performed using Superscript III with Vilo buffer. For Pdx1 quantification, Taqman probes (Pdx1 (4453320 (Mm00435565_m1) and Gapdh (4448489 (Mm99999915_g1) were used along with Perfecta qPCR Fastmix II (Cat #95118-012). For Pcsk9 quantification, primers and conditions from a previously published report were used21. Neurons were harvested for RNA extraction (Qiagen RNAeasy kit with on-column DNAse treatment), oligo(dT) and random primers primed for cDNA synthesis with iScript™ cDNA Synthesis Kit (BioRad Cat#1708891), and run for SYBR qPCR on a Quantstudio 3 realtime PCR system (Thermo). All gene expression values were normalized to Gapdh in the same well to control for sample handling. Sequences for qPCR primers can be found in Supplemental Table 1. T cells were collected in Cells-to-CT 2-Step Taqman kit (Thermo, Cat#A35377) following the manufacturer’s instructions. Foxp3 quantification was performed using Taqman probes Foxp3 (4331182 (Mm00475162_m1)) and Gapdh (4448489 (Mm99999915_g1)).
RNA-seq and differential expression analysis
For liver tissue, RNA was isolated using the Qiagen RNA plus universal Kit (Cat#73404). RNA-seq was performed after Total RNA was polyA tail-enriched, rRNA-depleted, and libraries prepped with a NEBNext Ultra II kit (E7645L). For T cells, transduced cells were FACS-purified using an SH800 and fluorescent antibody targeting Thy1.1. Cell pellets were processed using Qiagen RNeasy RNA extraction kit. RNA-seq libraries were built from rRNA-depleted mRNA using the Illumina TruSeq mRNA library prep kit (Cat#20020595). All RNA-seq samples were first validated for consistent quality using FastQC v0.11.2 (Babraham Institute). Raw reads were trimmed to remove adapters and bases with average quality score (Q) (Phred33) of < 20 using a 4bp sliding window (SLIDINGWINDOW:4:20) with Trimmomatic v0.3263. Trimmed reads were subsequently aligned to the primary assembly of the GRCm38 mouse genome using STAR v2.4.1a64 removing alignments containing non-canonical splice junctions (--outFilterIntronMotifs RemoveNoncanonical).Aligned reads were assigned to genes in the GENCODE vM13 comprehensive gene annotation65 using the featureCounts command in the subread package with default settings (v1.4.6-p466). Differential expression analysis was performed using DESeq2 (v1.22.067) running on R (v3.5.1). Briefly, raw counts were imported and filtered to remove genes with low or no expression, keeping genes having two or more counts per million (CPMs) in two or more samples. Filtered counts were then normalized using the DESeq function which uses estimated size factors to account for library size as well as gene and global dispersion. In order to find significant differentially expressed genes, the nbinomWaldTest was used to test the coefficients in the fitted Negative Binomial GLM using the previously calculated size factors and dispersion estimates. Genes having a Benjamini-Hochberg false discovery rate (FDR) less than 0.05 were considered significant (unless otherwise indicated). Log2(fold-change) values were shrunk towards zero using the adaptive shrinkage estimator from the ‘ashr’ R package68. For estimating transcript abundance, Transcript Per Million (TPMs) were computed using the rsem-calculate-expression function in the RSEM v1.2.21 package69. Data were visualized using pandas v0.23.3 and seaborn v0.9.0 packages in Python 2.7.11.
Protein isolation and western blot
Protein from tissue was isolated using a Biomasher II mortar and pestle tubes in conjunction with RIPA or TGH buffer. Protein was quantified using BCA assay. Novex NuPage 4-12% Bis-Tris gels were run for 45 minutes in MES running buffer and transferred onto nitrocellulose membranes for an hour at 4 °C in towbin’s buffer with 20% MeOH. Membranes were blocked for non-specific binding overnight in 5% milk in TBS-T. 1° Antibodies were incubated in 5% milk in TBS-T and used at the following manner: Anti-Cas9 (EnCor Biotechnology, Cat#MCA-3F, 1:2000) at room temp for 2-3 hours, Anti-Gapdh (Cell Signaling, Cat#2118L, 1:5000), Anti-LDLR (Abcam, Cat#ab52818, 1:1000), and anti-Actin (EMD Millipore, Cat#MAB1501, 1:5000) overnight at 4°C. After washing, secondary antibodies (Anti-rabbit HRP (Sigma, Cat#A6154, 1:5000) and Anti-Mouse HRP (Santa Cruz, Cat#sc-2005, 1:5000) were incubated in 3% BSA in TBS-T. Membranes were visualized using Clarity western ECL substrate (Cat#1705061) from BioRad and imaged using a BioRad ChemiDoc XRS+. For neurons, cells were lysed directly in 1X RIPA buffer and blots were incubated with goat anti-mouse 680 (Biotium, Cat#20253, 1:5000). Fluorescent immunoreactivity was imaged on a LICOR Odyssey and analyzed with LiCor Image Studio v4.0.
Stereotaxic injection of gRNA AAVs in mouse hippocampus
AAV containing GFP and a gRNA targeting either LacZ (control) or Fos Enh2 (experimental treatment) were stereotaxically injected into each hemisphere of the dorsal hippocampus of adult male dCas9p300 heterozygous mice such that each mouse had one control side and one experimental side (stereotaxic coordinates AP: -2.3, ML +/-1.8, DV: -1.8). 3 wks following transduction, mice were either used for evaluation of Fos expression (n=3 mice) or for electrophysiology (n=2 mice).
Histological staining
For PDX1 staining, livers were fixed overnight with 4% PFA and 12 μm-thick cryostat sections were processed for immunofluorescence. Slides were washed in PBS (Phosphate-buffered saline), boiled in citrate buffer (10 mM citric acid, 0.05% Tween-20, pH 6.0) for 25 min at 95 °C and were then kept for 20 minutes at room temperature (antigen retrieval). Slides were permeabilized with PBS containing 0.2% Triton for 20 minutes, washed with PBS and blocked with 3% BSA + 0.1% Tween in PBS for 1 hour. Primary antibody (Abcam, Cat#ab47267, 1:100) was incubated overnight at 4°C in blocking reagent. Sections were then washed and incubated with specific secondary antibody coupled to Alexa-Fluor-488 (Life Technologies, Cat#A27034, 1:200) and DAPI. Confocal images were acquired with a Zeiss LSM 880 microscope.
For Fos staining in hippocampal sections, after 3 weeks following viral injection, mice were placed in an open field and allowed to explore three novel objects for a period of 2 hours. Mice were then perfused with 4% paraformaldehyde and brains were coronally sectioned on a freezing microtome for immunostaining. Primary antibodies used were rabbit anti-c-Fos (Calbiochem, Cat#PC38, 1:1000) or rabbit anti-Cas9 (EnCor Biotechnology, Cat#RPCA-CAS9-Sp, 1:1000). These were detected by anti-rabbit Cy3 (1:500). Z-stack images through the dentate gyrus were obtained using a Leica SP8 upright confocal with a 40X objective plus additional digital zoom. For each animal, the hemisphere infected with gRNA targeting Fos Enh2 was compared to the control hemisphere expressing the control LacZ gRNA. To count high Fos-expressing cells, z-stacks were converted to sum projections and thresholded using ImageJ. These counts were compared using a paired t-test. To determine whether there were changes in Fos intensity across the population of GFP-positive cells, ROIs were created for each GFP-positive cell within a given region while blind to Fos expression. Fluorescence intensity of the Fos channel was then measured for each of these cells, creating a distribution of Fos intensities. The Fos signal for each cell was normalized to the average control hemisphere Fos value for a given animal. The resulting distributions were then compared using a Kolmogorov-Smirnoff test.
ChIP-Sequencing and analysis
Livers were pulverized and fixed as described by Savic et al.70 Following fixation, livers were rocked at 4°C in 10 ml Farnham Lysis Buffer for 10 minutes followed by centrifugation. Pellets were resuspended in 4 mL RIPA buffer. 1 mL RIPA and 1 mL RIPA containing resuspended liver was placed into a 15 mL tube with 800 mg of Diagenode sonication beads (Diagenode, C01020031). This tube was sonicated for 30 cycles of 30s on and 30s off using a Bioruptor Pico (Diagenode) at 4°C. For CD4+ T cells, 2 million cells were fixed with 1% paraformaldehyde at room temperature for 10 minutes, quenched with 0.125M glycine and washed twice with PBS. Fixed cells were lysed with 1mL of Farnhyme Lysis Buffer for 5 minutes on ice followed by centrifugation at 800 x g for 5 minutes at 4°C. Cell pellets were snap frozen before being resuspended in 100μL of RIPA buffer. Cells were then sonicated for 10 cycles of 30s on 30s off using a Bioruptor Pico as above. ChIP for all samples was performed using 5 μg of either Cas9 (Diagenode C15200229-100), Flag-M2 (Sigma, F1804), H3K27ac (Abcam, ab4729), or H3K9me3 (Abcam, ab8898) antibodies. Following ChIP, ChIP-seq libraries were prepared using a Kapa Hyper Prep kit (Roche 07962312001), and sequenced using 50bp single end reads on a Illumina HiSeq4000 or 25bp paired end reads on an Illumina NextSeq 500. For analysis, adapter sequences were removed from the raw reads using Trimmomatic v0.32.63 Reads were aligned to the primary assembly of the GRCm38 mouse genome using Bowtie v1.0.071, reporting the best alignment with up to 2 mismatches (parameters --best --strata -v 2). Duplicates were marked using Picard MarkDuplicates v1.130 (http://broadinstitute.github.io/picard/), while low mappability or blacklisted regions identified by the ENCODE project were filtered out from the final BAM files. Signal files were generated with deeptools bamCoverage (v3.0.172) ignoring duplicates, extending reads 200bp and applying RPKM normalization. Using the sequenced input controls, binding regions were identified using the callpeak function in MACS2 v2.1.1.2016030973. All MACS2 peaks were annotated with the TSS of the nearest gene using Gencode vM19 basic annotation data. For the differential binding analysis, first, a union peakset was computed merging individual peak calls using bedtools2 v2.25.074. Then, reads in peaks were estimated using featureCounts from the subread package v1.4.6-p466. The difference in binding was assessed with DESeq2 v1.22.067 using the nbinomWaldTest to test coefficients in the fitted Negative Binomial GLM. Data were visualized using pandas v0.23.3 and seaborn v0.9.0 packages in Python 2.7.11 or R v3.5.1.
ChIP-qPCR and analysis
For cerebellar granule neurons, chromatin immunoprecipitation was performed following the protocol of EZ-ChIP (Millipore, 17-371). Briefly, cells were lysed by SDS Lysis Buffer and sonicated for 1.5 hrs (Diagenode Bioruptor) at 4°C on the high setting with 30 seconds on/off interval. 20 μl Dynabeads Protein G (ThermoFisher, 10003D) was pre-incubated with 2 μg antibodies in 1XPBS buffer for 4 to 6 hours at 4°C. Mouse anti-Cas9 (Encor MCA-3F9 and Diagenode C15200229-100) antibodies were used. Cell lysates were then incubated overnight with antibody-bead complexes at 4°C. Subsequently, the beads were washed with Low Salt Immune Complex Wash Buffer, High Salt Immune Complex Wash Buffer, LiCl Immune Complex Wash Buffer, and TE Buffer. Bound protein/DNA complexes were eluted by ChIP elution buffer and then reversed the crosslinks. Samples were treated with RNase A and Proteinase K for post-immunoprecipitation and then the DNA was purified using QIAquick PCR Purification Kit (Qiagen, 28104). ChIP primers sequences can be found in Supplemental Table 1. ChIP for Grin2c was normalized to ChIP for Gapdh in the same sample to control for concentration and sample handling.
Hippocampal slice electrophysiological recordings
3 wks after viral injections into dCas9p300 heterozygous mice, mice were anesthetized with isofluorane, and transcardially perfused with ice-cold NMDG artificial cerebrospinal fluid (NMDG-ACSF; containing 92 mM NMDG, 2.5 mM KCl, 1.2 mM NaH2PO4, 30 mM NaHCO3, 20 mM HEPES, 2 mM glucose, 5 mM sodium ascorbate, 2 mM thiourea, 3 mM sodium pyruvate, 10 mM MgSO4, 0.5 mM CaCl2) that was bubbled with 5% CO2/95% O2. The brain was extracted and sectioned into 300 μm thick sagittal slices using a vibratome (VT-1000S, Leica Microsystems) in ice-cold oxygenated NMDG-ACSF. Coronal sections including the dorsal hippocampus were bubbled in the same solution at 37 °C for 8 min, and transferred to oxygenated modified-HEPES ACSF at room temperature (20–25 °C; 92 mM NaCl, 2.5 mM KCl, 1.2 mM NaH2PO4, 30 mM NaHCO3, 20 mM HEPES, 2 mM glucose, 5 mM sodium ascorbate, 2 mM thiourea, 3 mM sodium pyruvate, 2 mM MgSO4, 2 mM CaCl2) for at least 1 h before recording. Recordings were performed in a submerged chamber, superfused with continuously bubbled ACSF (125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 25 mM NaHCO3, 25 mM glucose, 2 mM CaCl2, 1 mM MgCl2, 2 mM sodium pyruvate) at near-physiological temperature (34 ± 1 °C). For whole-cell current-clamp recordings, the electrodes (4–6 MΩ) were filled with an intracellular solution containing 130 mM K-methylsulfonate, 5 mM NaCl, 10 mM HEPES, 15 mM EGTA, 12 mM phosphocreatine, 3 mM Mg-ATP, 0.2 mM Mg-GTP, 0.05 AlexaFlour 594 cadaverine, and 1% biotin. Current clamp responses were recorded with a Multiclamp 700B amplifier (filtered at 2-4 kHz) and digitized at 10 kHz (Digidata 1440). Series resistance was always <20 MΩ and was compensated at 80%-95%. Virally transduced pyramidal cells expressing GFP were visualized by infrared differential interference contrast and fluorescence video microscopy (Olympus; CoolLED) with a 40x objective. Data were collected and analyzed off-line using AxographX v1.7.6 and Neuromatic v3.0 packages (Think Random) in Igor Pro 6.22A software (WaveMetrics). Resting membrane potential was measured while cells were held at 0 pA. The action potential frequency was measured during holding currents of 0 to 500 pA. Rheobase was measured as the minimal amount of current needed to depolarize the cell membrane. Initial spike latency was defined as the time from the onset of current holding change to the first measurable deflection of the potential from baseline. Membrane time constant was defined as the time it took the cell’s voltage level to decay to resting state after delivering a 200ms step of -150pA. Input resistance was measured using “ΔV/pA =mΩ” for each current step and averaging across all current steps for individual neurons. All measurements were averaged across neurons. Finally, recorded neurons were labeled with biotin delivered through the patch pipette, and post-hoc confocal imaging of fixed slices confirmed GFP expression in the recorded neurons.
Novel object exploration and hippocampal expression of Fos
AAV vectors containing GFP and gRNA targeting either LacZ (control) or Fos Enh2 were injected into either hemisphere of the dorsal hippocampus of adult male dCas9p300 mice such that each mouse had one control hemisphere and one experimental hemisphere (stereotaxic coordinates AP: -2.3, ML +/-1.8, DV: -1.8). Three weeks following AAV injection, mice were placed in an open field and allowed to explore three novel objects for a period of 2 hours. Mice were then perfused with 4% paraformaldehyde and brains were coronally sectioned on a freezing microtome for immunostaining. Primary antibodies used were rabbit anti-c-Fos (CalBiochem, Cat#PC38, 1:1000) and rabbit anti-Cre recombinase (Cell Signaling #15036, 1:1000). These were detected by anti-rabbit Cy3 (1:500). Z-stack images through the dentate gyrus were obtained using a Leica SP8 upright confocal with a 40X objective plus additional digital zoom. For each animal, the hemisphere infected with gRNA targeting Fos Enh2 was compared to the control hemisphere expressing the control LacZ gRNA. To count high Fos-expressing cells, z-stacks were converted to sum projections and thresholded using ImageJ v1.46j (National Institutes of Health). These counts were compared using a paired t-test. To determine whether there were changes in Fos intensity across the population of GFP-positive cells, ROIs were created for each GFP-positive cell within a given region while blind to Fos expression. Fluorescence intensity of the Fos channel was then measured for each of these cells, creating a distribution of Fos intensities. The Fos signal for each cell was normalized to the average control hemisphere Fos value for a given animal. The resulting distributions were then compared using a Kolmogorov-Smirnoff test.
Purification of CD4 T cells for in vitro cultures
Rosa26-LSL-dCas9-p300 or Rosa26-LSL-dCas9-KRAB mice were backcrossed with a B6-Foxp3-eGFP mouse line (Jax stock# 006772)75, 76 for six generations and then crossed with the B6-CD4:Cre mouse line (Jax stock# 022071)77 to generate heterozygous Cd4:Cre+/dCas9-KRAB+/Foxp3-eGFP+ or Cd4:Cre+/dCas9-p300+/Foxp3-eGFP+ experimental mice. Spleens and lymph nodes were harvested, dissociated, and processed with ammonium chloride potassium (ACK) lysis buffer and passed through the Magnisort mouse CD4 T cell enrichment kit (Thermo, Cat#8804-6821-74). Lymphocytes were surface-stained with antibodies for the purification of nT cells (CD4+CD25−CD62LhiCD44lo) through FACS using either a Beckman Culture Astiros (CD4-FITC 1:400; CD25-eFluor450 1:300; CD62L-APC 1:500; CD44-PE 1:500, and fixable viability dye e780 1:1000) or SONY SH800 (CD4-PECy7 1:500; CD25-PE 1:500; CD44-FITC 1:400; CD62L-PEcy5 1:500; Live/Dead Red 1:1000) instrument to achieve ≥98% purity. For ChIP-seq, Tn cells were purified using Magnisort mouse Naïve CD4+ Tcell enrichment kit (Thermo, Cat#8804-6824-74) rather than sorting in order to increase starting material. Antibody information: CD4 (eBiosciences, Cat#25-0042-82), CD25 (eBiosciences, Cat#12-0251-83), CD44 (eBiosciences, Cat#11-0441-85), CD62L (eBiosciences, Cat#15-0621-82).
In vitro culture conditions for CD4 T cell subset polarization
All T cell polarizations were performed using Olympus tissue-culture plastic that was coated with anti-Hamster IgG (20ng/μL, MP Biomedical, Cat#856984) at 37°C for at least 4 hours prior. Tn cells were seeded at ~250k cells/mL. Th0 activating conditions contained hamster-anti-CD3e (0.25 μg/mL), hamster-anti-CD28 (1 μg/mL), blocking antibodies anti-IL4 (2μg/mL, eBiosciences, Cat#16-7041-85) and anti-IFNg (2μg/mL, eBiosciences Cat#16-7311-85), and mIL-2 (10 ng/mL, Peprotech Cat#212-12). iTreg polarizing conditions contained the above and was supplemented with hTGFb1 (5 ng/mL, eBiosciences Cat#14-8348-62).
T cell retroviral transduction and in vitro culture
Tn cells were isolated and purified from the spleen and lymph nodes of Cd4:Cre+/dCas9-p300+/Foxp3-eGFP+ or Cd4:Cre+/dCas9-KRAB+ mice via FACS as described above and cultured in Th0 activating conditions at 37°C and 5% CO2. Between 20-24 hours later, the activated T cells were transduced via spinfection with viral supernatant containing 6.66 ng/mL polybrene and Mouse Stem Cell Virus (MSCV) encoding a Foxp3 promoter-targeting gRNA, or a non-targeting control gRNA. Viral supernatant was replaced with Th0 or iTreg media and cells were cultured for 7 or 3 days for p300 and KRAB experiments, respectively.
Purification of p300-induced Treg effector cells for co-culture
At 7 days after viral transduction, Treg effector cells were stained with fixable viability dye e780 (eBioscience Cat#65-0865-18) and Thy1.1 (CD90.1-PE, StemCell, Cat#60024PE, 1:400). Viable transduced cells expressing FOXP3-eGFP were FACS purified (Thy1.1+/FOXP3-eGFP+) and resuspended at 500,000 cells/mL in RPMI.
Purification of p300-induced Treg effector T cells for ChIP and RNA-seq
Tn cells were activated for 20-24 hours in Th0 conditions as above before transduction with Foxp3 or control gRNA. At three days post transduction, 100K or 2M viable, Thy1.1+ transduced Tcells were FACS purified for RNA-seq or ChIP-seq, respectively.
Purification of conventional T cells and suppression assay
Spleens and lymphnodes from Foxp3-eGFP mice (JAX stock# 006769) were processed as above and the recovered splenocytes were stained for viability (Live/Dead red; Thermo, Cat#L34971, 1:1000) and CD4-PEcy7 (1:500). CD4+FOXP3−eGFP− live cells were purified using a SONY SH800 FACS machine. Cells were washed with 30 mL of PBS and stained with 5 μM Cell Trace Violet (CTV; Thermo, Cat#C34557) following manufacturer’s instructions. Stained Tconv cells were diluted to 500,000 cells/mL in RPMI and co-cultured with 25,000 Treg cells per well for 72 hours with 25,000 aCD3/aCD28 Dynabeads (Thermo, Cat#11161). Cells were then removed from the magnetic beads, stained for viability with e780, and processed for flow cytometric analysis.
FOXP3 protein staining and flow cytometry analysis
For dCas9KRAB-based experiments, in vitro cultured, transduced T cells were stained with Thy1.1-PerCP-Cy5.5 (eBioscience, Cat#45-0900-82, 1:300) and Fixable Viability Dye eFluor780 (1:1000) prior to fixation and permeabilization with FOXP3 Transcription Factor Staining Buffer Kit (eBioscience, Cat#00-5523-00) following manufacturer’s protocol. Cells were stained intracellularly with Foxp3-PE (eBioscience, Cat#12-5773-82, 1:200) and data was collected with a BD FACSCanto II cytometer using FACSDiva v8.0.3. All flow cytometry was analyzed using FlowJo v10.6.2
Statistics and Reproducibility
No statistical method was used to predetermine samples size. Sample sizes were chosen to be consistent with other published reports of dCas9-based activators and repressors used in vivo21, 28, 78. In each of these three studies, n = 3 or 4 was used in the majority of assays measuring changes in gene expression. For high-throughput sequencing based assays, sample sizes were chosen to be consistent with ENCODE standards79, 80. All statistical analysis was conducted using Prism v6 or v9 (GraphPad) with the exception of Supplementary Figure 2 which was conducted using R v3.5.1. Sequencing data were only excluded for failing to pass pre-existing criteria for technical quality. RT-PCR data were excluded only if our control for RNA harvesting and sample processing showed abnormally low yield, defined as more than 2 standard deviations from the mean of the sample set. Electrophysiology data were excluded only if the measurement of basal membrane properties suggested that the cell was dying. Mice for all experiments were selected randomly with respect to parent of origin, age, and sex with the exception of mice for stereotaxic injections for which only adult male animals were used. Sample collection and analysis for liver-targeted experiments were performed blind. Animals were injected, numbered, and housed mixed in cages and samples were collected based on animal number only. Sample collection and analysis for T cell, neuron, and brain experiments were not performed blind.
Data availability
Raw sequencing files are available from the NCBI Gene Expression Omnibus via SuperSeries accession GSE146848.
Code availability
Processing and analysis code will be made available upon request.
Supplementary Material
Acknowledgements:
We thank Jay Deng and Brendan Ryu for assistance with viral injections and Kevin Franks for his support in performing the electrophysiology recordings. This work has been supported by the Allen Distinguished Investigator Program through The Paul G. Allen Frontiers Group to C.A.G., Translating Duke Health Initiative to K.D.P., Open Philanthropy to C.A.G., National Institutes of Health (NIH) grants R33DA041878 to A.E.W. and C.A.G., R01DA036865 to C.A.G., U01AI146356 to C.A.G., UM1HG009428 to T.E.R., M.C., and C.A.G., UG3AR075336 to A.A., and R01GM115474 to M.C., National Science Foundation (NSF) grant EFMA-1830957 to C.A.G., Defense Advanced Research Projects Agency (DARPA) grant HR0011-19-2-0008 to C.A.G., a Pfizer-NCBiotech Distinguished Postdoctoral Fellowship in Gene Therapy to J.C.B., and a Swiss National Science Foundation Postdoctoral Fellowship to V.C.
Footnotes
Conflict of Interest Statement
CAG, IBH, and TER have filed patent applications related to CRISPR technologies for genome engineering. CAG is an advisor to Tune Therapeutics, Sarepta Therapeutics, Levo Therapeutics, and Iveric Bio, and a co-founder of Tune Therapeutics, Element Genomics, and Locus Biosciences. AA is a co-founder and advisor to StrideBio and TorqueBio. TER is a co-founder of Element Genomics. MPG is a co-founder and employee of Tune Therapeutics. All other authors declare no conflicts of interest.
References
- 1.Thakore PI, Black JB, Hilton IB & Gersbach CA Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods 13, 127–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilton IB et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33, 510–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeder ML et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10, 977–979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Pinera P et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10, 973–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavez A et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS & Vale RD A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu XS et al. Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–247 e217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stepper P et al. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res 45, 1703–1713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeo NC et al. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat Methods 15, 611–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thakore PI et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter MD et al. Nr4a1 suppresses cocaine-induced behavior via epigenetic regulation of homeostatic target genes. Nat Commun 11, 504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen LF et al. Enhancer Histone Acetylation Modulates Transcriptional Bursting Dynamics of Neuronal Activity-Inducible Genes. Cell Rep 26, 1174–1188 e1175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank CL et al. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci 18, 647–656 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black JB et al. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell 19, 406–414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y et al. CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell 23, 758–771 e758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weltner J et al. Human pluripotent reprogramming with CRISPR activators. Nat Commun 9, 2643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu P, Chen M, Liu Y, Qi LS & Ding S CRISPR-Based Chromatin Remodeling of the Endogenous Oct4 or Sox2 Locus Enables Reprogramming to Pluripotency. Cell Stem Cell 22, 252–261 e254 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty S et al. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Reports 3, 940–947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thakore PI et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat Commun 9, 1674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matharu N et al. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klann TS et al. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fulco CP et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fulco CP et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat Genet 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simeonov DR et al. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549, 111–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platt RJ et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao HK et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507 e1415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci 21, 440–446 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Wangensteen KJ et al. Combinatorial genetics in liver repopulation and carcinogenesis with a in vivo CRISPR activation platform. Hepatology 68, 663–676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K et al. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat Commun 11, 485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chew WL et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods 13, 868–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson CE et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li A et al. AAV-CRISPR Gene Editing Is Negated by Pre-existing Immunity to Cas9. Mol Ther (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno AM et al. Immune-orthogonal orthologues of AAV capsids and of Cas9 circumvent the immune response to the administration of gene therapy. Nat Biomed Eng 3, 806–816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferdosi SR et al. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat Commun 10, 1842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferber S et al. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med 6, 568–572 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Tang DQ et al. Reprogramming liver-stem WB cells into functional insulin-producing cells by persistent expression of Pdx1− and Pdx1-VP16 mediated by lentiviral vectors. Lab Invest 86, 83–93 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T et al. Chemically Controlled Epigenome Editing through an Inducible dCas9 System. J Am Chem Soc 139, 11337–11340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat Commun 9, 2949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang AS et al. The Histone Chaperone FACT Induces Cas9 Multi-turnover Behavior and Modifies Genome Manipulation in Human Cells. Mol Cell 79, 221–233 e225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo L et al. A combination strategy targeting enhancer plasticity exerts synergistic lethality against BETi-resistant leukemia cells. Nat Commun 11, 740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abifadel M et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 34, 154–156 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Zhang DW et al. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem 282, 18602–18612 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Brunkow ME et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 27, 68–73 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Fontenot JD, Gavin MA & Rudensky AY Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Hori S, Nomura T & Sakaguchi S Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Khattri R, Cox T, Yasayko SA & Ramsdell F An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol 4, 337–342 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Okada M, Kanamori M, Someya K, Nakatsukasa H & Yoshimura A Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells. Epigenetics Chromatin 10, 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galonska C et al. Genome-wide tracking of dCas9-methyltransferase footprints. Nat Commun 9, 597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jinek M et al. RNA-programmed genome editing in human cells. Elife 2, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho SW, Kim S, Kim JM & Kim JS Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 230–232 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Swiech L et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33, 102–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parnas O et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell 162, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Springer ML, Rando TA & Blau HM Gene delivery to muscle. Curr Protoc Hum Genet Chapter 13, Unit13 14 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Salmon P & Trono D Production and titration of lentiviral vectors. Curr Protoc Neurosci Chapter 4, Unit 4 21 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Morita S, Kojima T & Kitamura T Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7, 1063–1066 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harrow J et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao Y, Smyth GK & Shi W The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stephens M False discovery rates: a new deal. Biostatistics 18, 275–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Savic D, Gertz J, Jain P, Cooper GM & Myers RM Mapping genome-wide transcription factor binding sites in frozen tissues. Epigenetics Chromatin 6, 30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramirez F, Dundar F, Diehl S, Gruning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haribhai D et al. Regulatory T cells dynamically control the primary immune response to foreign antigen. J Immunol 178, 2961–2972 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Lin W et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol 8, 359–368 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Lee PP et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001). [DOI] [PubMed] [Google Scholar]
- 78.Liao HK et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507.e1415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Landt SG et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome research 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conesa A et al. A survey of best practices for RNA-seq data analysis. Genome Biol 17, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing files are available from the NCBI Gene Expression Omnibus via SuperSeries accession GSE146848.