Abstract
Background
Lung cancer claims more lives than any other cancer worldwide. Lung adenocarcinoma (LUAD) accounts for approximately 40% of all lung cancers. Members of the Transducin-like Enhancer of split (TLE) protein family repress transcription through multiple mechanisms; however, their prognostic value in LUAD is still unclear.
Methods
A dataset from The Cancer Genome Atlas was used to analyze the relationship between the expression of TLE family members and outcomes of LUAD. The expression of TLE family members in 59 normal and 513 tumor samples in the TCGA dataset was selected. For paired analysis, 57 normal and 57 tumor paired tissues were selected. Gene Ontology (GO) term and Reactome pathway enrichment analyses of the TLE family members were performed. Progression-free survival (PFS) and overall survival (OS) served as endpoints in this study. All statistical analyses were performed with R 3.6.0.
Results
The expression levels of TLE family proteins differed between 59 normal and 513 tumor samples. High TLE1 and low TLE2 levels were associated with poor progression-free and OS (all P<0.050). Multivariate analysis demonstrated that high TLE1 expression and low TLE2 expression were independent risk factors for a poor outcome in LUAD. Moreover, the combined expression of these two proteins was a good tool for prognostication.
Conclusions
High TLE1 expression and low TLE2 are independent adverse prognostic factors in LUAD and can serve as prognostic biomarkers.
Keywords: TLE family, lung adenocarcinoma (LUAD), prognosis
Introduction
Lung cancer takes more lives each year than any other cancer worldwide. In 2018 alone, there were 2.1 million new lung cancer cases (1). Lung adenocarcinoma (LUAD) is the most common type of lung cancer (2). The outcomes of patients with LUAD are capricious and unpredictable (3,4). Therefore, reliable and predictive biomarkers are needed for the identification and stratification of patients.
Members of the Transducin-like Enhancer of split (TLE) family of proteins are involved in many developmental processes, including lateral inhibition, segmentation, sex determination, eye development, and pancreatic development (5). TLE family proteins repress the transcription process through multiple mechanisms (6). Instead of binding to DNA directly, TLE proteins are recruited to the template by DNA-bound repressor proteins (7,8). Seven TLE homologs have been found in humans, namely TLE 1–7. Four TLE proteins (TLE 1–4) are full length, while another two homologous proteins, TLE5 and TLE6, are expressed in truncated forms (9,10). All TLE proteins interact with Tcf1 and Lef1. They are also enriched in the Wnt signaling pathway, suggesting their involvement in T-cell development and function (11-13). TLE corepressors predominantly partner with Tcf1/Lef1 to form CD8+ T-cell identity and are physiologically required for T-cell development (14).
TLE1 and TLE4 have been found to function as tumor suppressors in myeloid leukemia (15). Overexpression of TLE1 was also detected in a subset of aggressive and advanced lung cancers, and might regulate lung cancer aggressiveness (16). Yao et al. reported that TLE1 promoted epithelial-mesenchymal transition in A549 lung cancer cells by suppressing E-cadherin. In human lung cancer cells, TLE1 promotes tumorigenicity and inhibits anoikis through ZEB1-mediated E-cadherin repression (17,18). Furthermore, TLE4 interacts with PU.1 and Pax5 transcription factor in blood lineage cells, indicating that it plays a role in B-cell function (19).
Lee JH reported that immunohistochemical expression of TLE1 in invasive breast cancer and its association with clinicopathological parameters, prognosis, and intrinsic subtype (20). Hu S provides new insights and evidence that TLE2 functions as a tumor suppressor gene and prognostic marker in pancreatic ductal adenocarcinoma (21). However, these two paper focus on only one member’ function. The novel idea in this paper is to analysis the whole TLE family members, and then select two family members (TLE1 and TLE2) according to the statistic significance. Furthermore, we combined these two members as one group. So, the combined biomarkers could be more precise and effective. More research is needed to ascertain the prognostic value of TLEs in LUAD. Therefore, we conducted a study of the correlation between TLE protein expression and survival outcomes in LUAD. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/tlcr-21-582).
Methods
Patients
Gene expression data of LUAD tissues from The Cancer Genome Atlas (TCGA) were obtained via the Genomic Data Commons (GDC, available at: http://potal.gdc.cancer.gov/) data portal on November 6, 2019. The expression of TLE family members in 59 normal and 513 tumor samples in the TCGA dataset was quantified as fragments per kilobase of exon, per million reads mapped (FPKM). For paired analysis, 57 normal and 57 tumor paired tissues were selected. In accordance with the database policy, access to the de-identified linked dataset was obtained from TCGA.
Clinicopathological data for the patients in the TCGA dataset, including age, sex, pathologic stage, molecular data, and survival information, were also retrieved. Only patients with survival information and expression data available at that time point were included in the study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene Ontology (GO) term and Reactome pathway enrichment analyses of the TLE family members were performed. GO terms include biological processes (BPs), cellular components (CCs), and molecular functions (MFs). An adjusted P value <0.05 was considered to be statistically significant in both GO and Reactome pathway analysis.
Statistical analysis
Progression-free survival (PFS) and overall survival (OS) served as endpoints in this study. PFS refers to the length of time for which a patient lives with the disease without progression. OS is defined as the time from diagnosis to death from any cause or the last follow-up visit.
The Wilcoxon rank-sum test was employed for numerical data comparisons. Categorical data were compared using Fisher’s exact test. For survival analysis, the Kaplan-Meier method and log-rank test were applied. For the analysis of PFS and OS, univariate and multivariate Cox proportional hazard models were constructed, and 95% confidence intervals (95% CIs) were calculated. All statistical analyses were performed with R 3.6.0.
Results
The expression of TLEs in normal and LUAD tissue samples
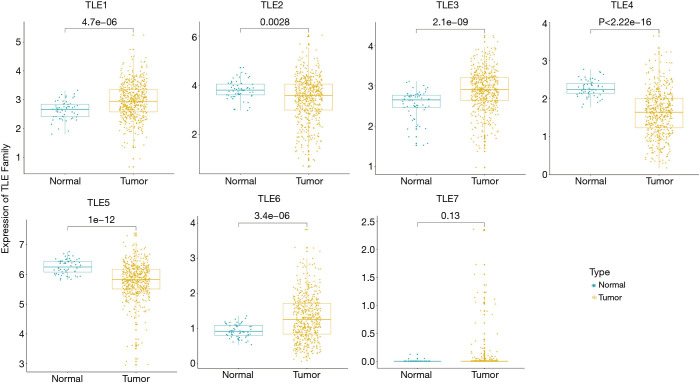
In this study, we examined the expression levels of TLEs in LUAD tissue samples in a TCGA dataset. We observed remarkable differences in the expression levels of TLEs in tumor samples compared with normal tissues (Figure 1). The same trends were also validated in the paired samples (Figure S1). The results suggested that the expression levels of TLEs might be related to LUAD progression.
Figure 1.
The expression levels of TLEs in normal and LUAD tissues in the TCGA dataset. The x-axis represents the sample type; the y-axis represents the TLE expression levels. The different expression levels of TLE 1–7 in normal (n=59) and tumor (n=513) tissues. TLE, Transducin-like Enhancer of split; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
The prognostic value of TLE expression levels in LUAD
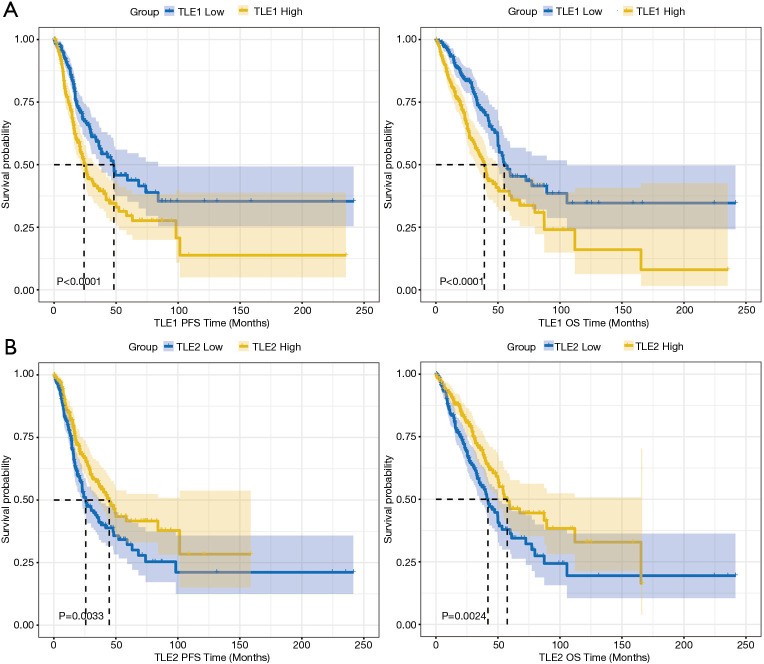
The patients in the TCGA dataset were divided into two groups according to the median expression level of each TLE family member. Table 1 shows the differences between the high and low expression groups. A high expression of TLE1 (Figure 2A) and a low expression of TLE2 were found to have an adverse affect on both PFS and OS (Figure 2B) (PFS: P<0.001, P=0.003; OS: P<0.0001, P=0.004, respectively).
Table 1. Comparison of PFS and OS between the high and low expression levels of TLE family in 513 LUAD samples of TCGA.
| Family members | PFS | OS | |||
|---|---|---|---|---|---|
| χ2 | P value | χ2 | P value | ||
| TLE1 (high vs. low) | 15.382 | <0.001 | 15.445 | <0.001 | |
| TLE2 (high vs. low) | 8.633 | 0.003 | 8.167 | 0.004 | |
| TLE3 (high vs. low) | 0.621 | 0.431 | 0.438 | 0.508 | |
| TLE4 (high vs. low) | 0.014 | 0.907 | 0.158 | 0.691 | |
| TLE5 (high vs. low) | 4.490 | 0.034 | 2.688 | 0.101 | |
| TLE6 (high vs. low) | 0.709 | 0.400 | 0.565 | 0.452 | |
| TLE7 (high vs. low) | 3.609 | 0.057 | 0.653 | 0.419 | |
PFS, progression-free survival; OS, overall survival; TLE, Transducin-like Enhancer of split; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
Figure 2.
The Kaplan-Meier curves of PFS and OS in the TCGA cohort. (A) Patients with high TLE1 expression had shorter PFS and OS than those with low TLE1 expression; (B) patients with high TLE2 expression had longer PFS and OS than those with low expression. PFS, progression-free survival; OS, overall survival; TCGA, The Cancer Genome Atlas; TLE, Transducin-like Enhancer of split.
The expression of TLE family members in LUAD
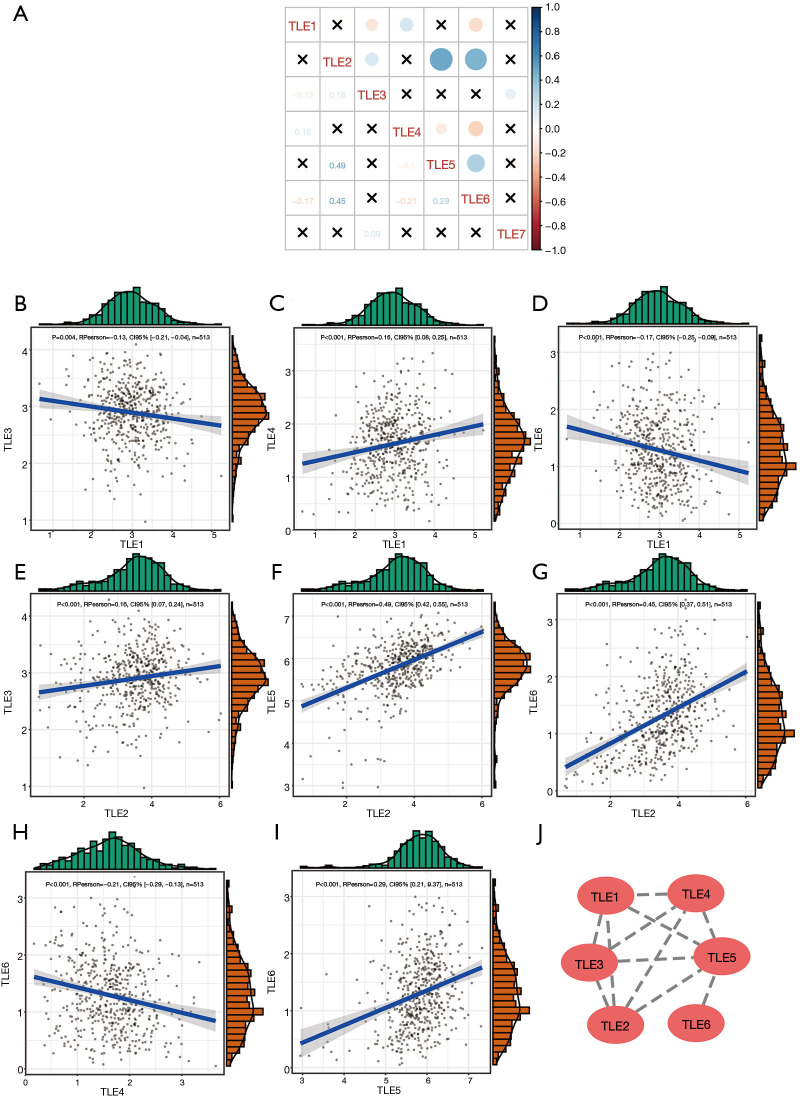
The correlations between the expression levels of TLE members in LUAD were analyzed. As shown in Figure 3, the expression levels of the TLE family members were interrelated (all |RPearson| >0.1, Figure 3A), especially those of TLE3/TLE1, TLE4/TLE1, TLE6/TLE1, TLE3/TLE2, TLE5/TLE2, TLE6/TLE2, TLE6/TLE4, and TLE6/TLE5 (all |RPearson| >0.1, Figure 3B,C,D,E,F,G,H,I). These results suggest that the TLEs might be transcriptionally co-regulated together. Previous research has shown that TLE1, TLE2, and TLE5 work as a complex to regulate DNA transcription. A protein-protein interaction (PPI) network was constructed for the TLE family members and revealed interactions between TLE 1–6 (Figure 3J).
Figure 3.
Correlations between the expression levels and PPI network of the TLE family members. (A) Co-expression heat map of TLE genes in TCGA; (B) co-expression relationship between the TLE3 and TLE1 genes in TCGA; (C) co-expression relationship between the TLE4 and TLE1 genes in TCGA; (D) co-expression relationship between the TLE6 and TLE1 genes in TCGA; (E) co-expression relationship between the TLE3 and TLE2 genes in TCGA; (F) co-expression relationship between the TLE5 and TLE2 genes in TCGA; (G) co-expression relationship between the TLE6 and TLE2 genes in TCGA; (H) co-expression relationship between the TLE6 and TLE4 genes in TCGA; (I) co-expression relationship between the TLE6 and TLE5 genes in TCGA; (J) the PPI network of the TLE family members. PPI, protein-protein interaction; TLE, Transducin-like Enhancer of split; TCGA, The Cancer Genome Atlas.
Clinical and molecular characteristics
The clinical and molecular characteristics of 513 LUAD patients in TCGA are shown in Table 2. Compared to those in the TLE1low group, patients in the TLE1high group were more likely to have received radiation therapy (P=0.021). Furthermore, high TLE1 expression showed a trend toward pathologic T1 stage (P=0.060). The TLE1high group also had a lower proportion of patients with RET (rearranged during transfection) mutation (48.25% vs. 51.95%, P=0.029).
Table 2. Patients’ characteristics of 513 LUAD patients in TCGA.
| Characteristics | TLE1 | TLE2 | |||||
|---|---|---|---|---|---|---|---|
| Low (n=256) | High (n=257) | P value | Low (n=256) | High (n=257) | P value | ||
| Age, mean (range) | 65.78 (33–88) | 64.41 (38–87) | 0.980 | 65.24 (33–88) | 64.96 (39–87) | 0.690 | |
| Gender, n (%) | 0.892 | 0.102 | |||||
| Female | 139 (54.3) | 137 (53.31) | 128 (50.00) | 148 (57.59) | |||
| Male | 117 (45.7) | 120 (46.69) | 128 (50.00) | 109 (42.41) | |||
| Race, n (%) | 0.906 | 0.226 | |||||
| White | 198 (86.09) | 199 (86.9) | 203 (88.65) | 194 (84.35) | |||
| Others | 32 (13.91) | 30 (13.1) | 26 (11.35) | 36 (15.65) | |||
| Stage, n (%) | 0.460 | 0.011 | |||||
| I | 135 (52.53) | 145 (56.64) | 158 (61.48) | 122 (47.66) | |||
| II | 61 (23.74) | 59 (23.05) | 48 (18.68) | 72 (28.12) | |||
| III | 47 (18.29) | 33 (12.89) | 39 (15.18) | 41 (16.02) | |||
| IV | 11 (4.28) | 14 (5.47) | 8 (3.11) | 17 (6.64) | |||
| Pathologic T, n (%) | 0.060 | 0.001 | |||||
| T1 | 73 (28.4) | 98 (38.28) | 104 (40.47) | 67 (26.17) | |||
| T2 | 146 (56.81) | 129 (50.39) | 132 (51.36) | 143 (55.86) | |||
| T3 | 24 (9.34) | 22 (8.59) | 16 (6.23) | 30 (11.72) | |||
| T4 | 11 (4.28) | 7 (2.73) | 4 (1.56) | 14 (5.47) | |||
| Pathologic N, n (%) | 0.386 | 0.191 | |||||
| N0 | 164 (63.81) | 171 (66.8) | 175 (68.09) | 160 (62.5) | |||
| N1 | 45 (17.51) | 49 (19.14) | 38 (14.79) | 56 (21.88) | |||
| N2 | 40 (15.56) | 29 (11.33) | 35 (13.62) | 34 (13.28) | |||
| N3 | 2 (0.78) | 0 (0) | 2 (0.78) | 0 (0) | |||
| NX | 5 (1.95) | 7 (2.73) | 6 (2.33) | 6 (2.34) | |||
| Pathologic M, n (%) | 0.140 | 0.212 | |||||
| M0 | 184 (71.6) | 158 (61.72) | 171 (66.54) | 171 (66.8) | |||
| M1 | 10 (3.89) | 14 (5.47) | 8 (3.11) | 16 (6.25) | |||
| MX | 61 (23.74) | 81 (31.64) | 74 (28.79) | 68 (26.56) | |||
| Pharmaceutical therapy, n (%) | 0.681 | 0.057 | |||||
| No | 36 (14.01) | 34 (13.28) | 33 (12.84) | 37 (14.45) | |||
| Yes | 35 (13.62) | 29 (11.33) | 24 (9.34) | 40 (15.62) | |||
| Radiation therapy, n (%) | 0.021 | 0.242 | |||||
| No | 32 (12.45) | 42 (16.41) | 32 (12.45) | 42 (16.41) | |||
| Yes | 41 (15.95) | 22 (8.59) | 28 (10.89) | 35 (13.67) | |||
| Surgery locoregional procedure, n (%) | 0.420 | 0.059 | |||||
| No | 54 (21.01) | 47 (18.36) | 40 (15.56) | 61 (23.83) | |||
| Yes | 6 (2.33) | 3 (1.17) | 5 (1.95) | 4 (1.56) | |||
| Surgery metastatic procedure, n (%) | 0.702 | 0.015 | |||||
| No | 44 (17.12) | 38 (14.84) | 31 (12.06) | 51 (19.92) | |||
| Yes | 12 (4.67) | 10 (3.91) | 8 (3.11) | 14 (5.47) | |||
| EGFR mutation, n (%) | 0.306 | 0.529 | |||||
| No | 104 (40.47) | 87 (33.98) | 102 (39.69) | 89 (34.77) | |||
| Yes | 40 (15.56) | 41 (16.02) | 39 (15.18) | 42 (16.41) | |||
| KRAS mutation, n (%) | 0.133 | 0.435 | |||||
| No | 135 (52.53) | 117 (45.7) | 131 (50.97) | 121 (47.27) | |||
| Yes | 34 (13.23) | 29 (11.33) | 34 (13.23) | 29 (11.33) | |||
| EML4 ALK, n (%) | 0.392 | 0.168 | |||||
| No | 111 (43.19) | 96 (37.5) | 112 (43.58) | 95 (37.11) | |||
| Yes | 17 (6.61) | 18 (7.03) | 20 (7.78) | 15 (5.86) | |||
| High NRAS, n (%) | 110 (42.97) | 147 (57.2) | 0.728 | 159 (62.11) | 98 (38.13) | 0.074 | |
| High ALK, n (%) | 131 (51.17) | 126 (49.03) | 0.197 | 119 (46.48) | 138 (53.7) | 0.153 | |
| High PIK3CA, n (%) | 110 (42.97) | 147 (57.2) | 0.128 | 138 (53.91) | 119 (46.3) | <0.001 | |
| High ROS1, n (%) | 140 (54.69) | 117 (45.53) | 0.300 | 107 (41.8) | 150 (58.37) | 0.003 | |
| High EGFR, n (%) | 126 (49.22) | 131 (50.97) | 0.576 | 118 (46.09) | 139 (54.09) | 0.287 | |
| High BRAF, n (%) | 128 (50) | 129 (50.19) | 0.245 | 116 (45.31) | 141 (54.86) | <0.001 | |
| High RET, n (%) | 133 (51.95) | 124 (48.25) | 0.029 | 119 (46.48) | 138 (53.7) | 0.714 | |
| High KRAS, n (%) | 116 (45.31) | 141 (54.86) | 0.546 | 137 (53.52) | 120 (46.69) | 0.011 | |
| High ERBB2, n (%) | 115 (44.92) | 142 (55.25) | 0.099 | 97 (37.89) | 160 (62.26) | 0.508 | |
LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
Compared to the TLE2low group, high TLE2 expression was correlated with stage (P=0.011) and pathologic T stage (P=0.001). Moreover, high TLE2 expression showed a trend toward more pharmaceutical therapy (P=0.057). In the TLE2high group, lower proportions of patients had PIK3CA mutation (46.3% vs. 53.91%, P<0.001) and KRAS mutation (46.69% vs. 53.52%, P=0.011), but a higher proportion of patients had BRAF mutation (54.86% vs. 45.31%, P<0.001), compared with the TLE2low group.
Univariate and multivariate analysis of PFS and OS
To further assess the prognostic significance of TLE1/2, the expression levels of TLE 1–7 (high vs. low), age (≥60 vs. <60 years), sex, and common genetic mutations (ALK, BRAF, EGFR, ERBB2, KRAS, NRAS, PIK3CA, RET, and ROS1) were enrolled in univariate and multivariate analysis (Table 3). The univariate analysis identified three independent risk factors for PFS. They were high expression levels of TLE1, TLE2, and TLE5 (P<0.001, P=0.004, and P=0.035). While, the univariate analysis identified five independent risk factors for OS. They were high expression levels of TLE1, TLE2, and TLE5, ALK mutation, and NRAS mutation (P<0.0001, P=0.003, P=0.024, P=0.036, and P=0.045). The multivariate analysis identified high TLE1 and TLE2 expression as independent risk factors for both PFS and OS (PFS: P<0.001, P=0.020; OS: P<0.0001, P=0.047, respectively).
Table 3. Univariate and multivariate cox regression analyses of PFS and OS in 559 multiple myeloma patients.
| Characteristics | Univariate cox regression | Multivariate cox regression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PFS | OS | PFS | OS | ||||||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | ||||
| AGE (≥60 vs. <60) | 0.97 (0.71–1.32) | 0.839 | 1.40 (1.03–1.89) | 0.029 | 0.89 (0.64–1.24) | 0.492 | 1.42 (1.04–1.95) | 0.027 | |||
| Gender | 1.05 (0.77–1.43) | 0.750 | 0.97 (0.72–1.32) | 0.860 | 1.22 (0.87–1.70) | 0.257 | 1.03 (0.75–1.43) | 0.842 | |||
| ALB | 0.72 (0.53–0.97) | 0.033 | 0.47 (0.35–0.64) | <0.001 | 0.82 (0.59–1.13) | 0.220 | 0.55 (0.40–0.76) | <0.001 | |||
| B2M | 1.72 (1.27–2.33) | <0.001 | 2.22 (1.64–3.01) | <0.001 | 1.13 (0.75–1.70) | 0.553 | 1.34 (0.89–2.00) | 0.157 | |||
| BMPC | 1.63 (1.18–2.27) | 0.003 | 1.82 (1.30–2.56) | 0.001 | 1.39 (0.95–2.03) | 0.089 | 1.45 (0.98–2.14) | 0.066 | |||
| CREAT | 1.36 (0.99–1.87) | 0.055 | 1.73 (1.28–2.35) | <0.001 | 1.07 (0.74–1.56) | 0.714 | 1.19 (0.83–1.70) | 0.344 | |||
| CRP | 0.97 (0.71–1.34) | 0.869 | 1.49 (1.10–2.01) | 0.011 | 0.85 (0.6–1.19) | 0.338 | 1.17 (0.84–1.62) | 0.361 | |||
| LDH | 2.58 (1.65–4.05) | <0.001 | 3.68 (2.53–5.37) | <0.001 | 2.62 (1.62–4.23) | <0.001 | 2.91 (1.93–4.40) | <0.001 | |||
| HGB | 0.54 (0.39–0.74) | <0.001 | 0.61 (0.45–0.84) | 0.002 | 0.70 (0.48–1.01) | 0.055 | 1.04 (0.73–1.49) | 0.819 | |||
| MRI | 1.26 (0.93–1.71) | 0.141 | 1.89 (1.38–2.60) | <0.001 | 1.61 (1.17–2.22) | 0.004 | 2.21 (1.58–3.08) | <0.001 | |||
| SUN1 (high vs. low) | 1.21 (0.89–1.64) | 0.235 | 1.21 (0.89–1.64) | 0.222 | 1.03 (0.74–1.43) | 0.853 | 1.08 (0.78–1.49) | 0.649 | |||
| SUN2 (high vs. low) | 1.40(1.03–1.90) | 0.029 | 1.42 (1.05–1.91) | 0.024 | 1.34 (0.97–1.84) | 0.074 | 1.52 (1.11–2.07) | 0.009 | |||
| SUN3 (high vs. low) | 0.90 (0.67–1.22) | 0.509 | 1.02 (0.75–1.37) | 0.920 | 0.90 (0.65–1.25) | 0.527 | 0.99 (0.72–1.37) | 0.968 | |||
| SPAG4 (high vs. low) | 0.60 (0.44–0.82) | 0.001 | 0.65 (0.48–0.88) | 0.006 | 0.69 (0.49–0.97) | 0.031 | 0.65 (0.47–0.91) | 0.012 | |||
| SUN5 (high vs. low) | 1.10 (0.81–1.50) | 0.525 | 1.12 (0.83–1.52) | 0.445 | 1.15 (0.83–1.59) | 0.416 | 1.15 (0.83–1.59) | 0.393 | |||
| CCND1 | 0.65 (0.48–0.88) | 0.006 | 0.74 (0.55–1.01) | 0.054 | 0.59 (0.42–0.83) | 0.003 | 0.88 (0.63–1.22) | 0.440 | |||
| CDK4 | 1.22 (0.90–1.66) | 0.197 | 1.36 (1.01–1.84) | 0.045 | 1.07 (0.76–1.50) | 0.689 | 1.15 (0.81–1.63) | 0.422 | |||
| CDK5 | 0.88 (0.65–1.19) | 0.396 | 1.09 (0.81–1.47) | 0.579 | 0.70 (0.49–1.00) | 0.050 | 0.82 (0.58–1.15) | 0.247 | |||
| FGFR3 | 0.90 (0.67–1.22) | 0.508 | 0.80 (0.60–1.09) | 0.154 | 0.84 (0.60–1.17) | 0.310 | 0.78 (0.57–1.08) | 0.136 | |||
| HK2 | 1.74 (1.28–2.37) | 0.000 | 1.65 (1.22–2.25) | 0.001 | 1.85 (1.33–2.57) | 0.000 | 1.61 (1.15–2.24) | 0.005 | |||
| LIG4 | 0.84 (0.62–1.14) | 0.268 | 0.85 (0.63–1.14) | 0.276 | 0.89 (0.65–1.22) | 0.454 | 0.94 (0.69–1.29) | 0.714 | |||
| TP53 | 1.06 (0.78–1.44) | 0.700 | 0.85 (0.63–1.15) | 0.291 | 1.05 (0.76–1.46) | 0.757 | 0.86 (0.62–1.20) | 0.382 | |||
PFS, progression-free survival; OS, overall survival; CR, complete remission; HR, hazard ratio; CI, confidence interval; ALB, albumin, 35 g/L; B2M, beta-2 microglobulin, mg/L; HGB, haemoglobin, g/dL.
The combined prognostic effect of TLE1 and TLE2 in LUAD
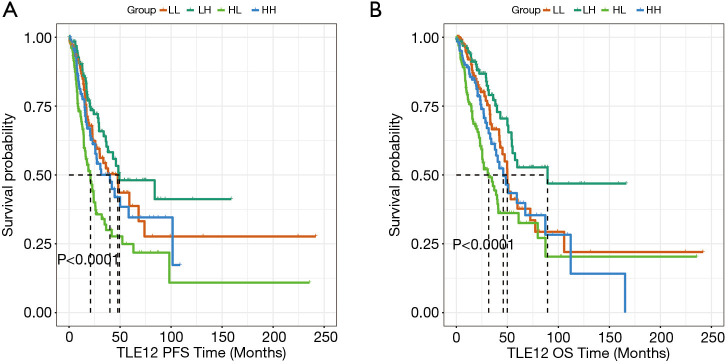
The effects of TLE1 and TLE2 on PFS and OS were subsequently examined. The results showed that the subgroup with both TLE1high and TLE2low had significantly shorter PFS and OS than the other subgroups (TLE1low/TLE2low, TLE1low/TLE2high, TLE1high/TLE2high) (P<0.0001 and P<0.0001, respectively, Figure 4).
Figure 4.
Kaplan-Meier curves of PFS and OS for patients in TCGA. (LL: TLE1low/TLE2low, LH: TLE1low/TLE2high, HL: TLE1high/TLE2low, HH: TLE1high/TLE2high). (A) TLE1high/TLE2low expression had shorter PFS; (B) TLE1high/TLE2low expression had shorter OS. PFS, progression-free survival; OS, overall survival; TLE, Transducin-like Enhancer of split.
GO and Reactome pathway enrichment analysis of the TLE family members
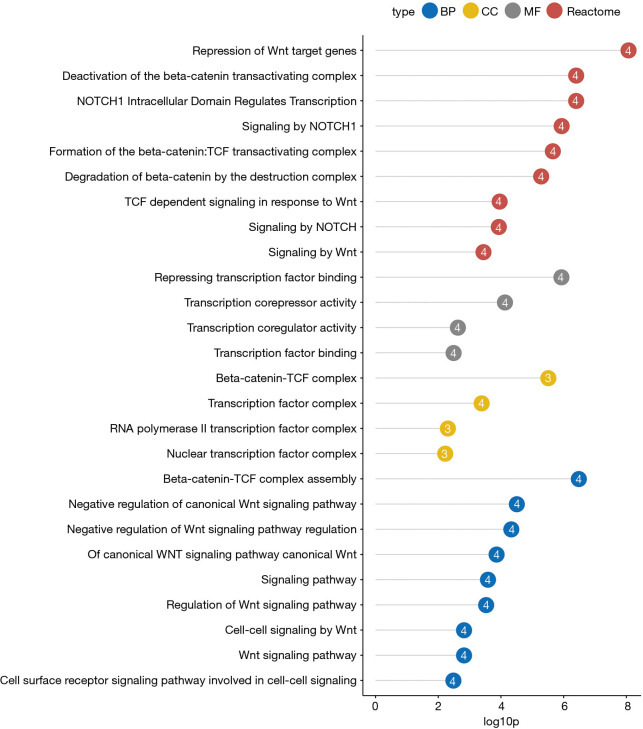
Reactome pathway enrichment analysis of the TLE family members showed that they are related to the repression of WNT target genes, TCF-dependent signaling in response to WNT, and deactivation of the beta-catenin transactivating complex (Figure 5). GO term enrichment analysis revealed that the TLE family members were related to repressing transcription factor binding, beta-catenin-TCF-complex, transcription factor complex, negative regulation of canonical WNT signaling pathway, and cell-cell signaling by WNT (Figure 5). A model illustrating the relationship between the TLE family members and Wnt pathway is shown in Figure S2.
Figure 5.
Gene Ontology term and Reactome pathway enrichment analyses of the TLE family members. TLE: Transducin-like Enhancer of split.
Discussion
This study found an increasing trend in TLE1 expression and a decreasing trend in TLE2 expression in LUAD. Furthermore, a high TLE1 level and a low TLE2 level were poor prognostic factors in patients with LUAD. Co-expression analysis confirmed that TLE1 and TLE2 were strongly correlated in LUAD. Based on multivariate analysis, TLE1 and TLE2 were also found to be independent risk factors for PFS and OS. Compared with the use of a single marker, the combined application of biomarkers seems to have higher value in the assessment of tumor prognosis.
The relationships between TLE family members and cancers have been studied recently. TLE1 has been shown to bind to Runx1, which is essential for the maintenance and generation of hematopoietic stem/progenitor cells (HSPCs) (22,23). TLE1 is related to chromatin through its interactions with the amino-terminal tail of histone H3 (24). The silencing function of the TLE1 gene is dependent on recruitment of the histone deacetylase (HDAC) protein. In a study by Chen et al., acetyl groups were removed from nearby DNA bound histones (25). TLE1 is highly expressed in diseased metaplastic and neoplastic transformed states as well as in proliferative epithelial tissues (26). TLE1 is also involved in the pathogenesis of cancers. For instance, it is abnormally expressed in various human cancers including synovial sarcoma (27), breast cancer (28), and lung cancer (16).
There were function differences of TLE proteins in different types of tumors. Seo et al. found that inhibition of TLE1 altered cancer cell proliferation and apoptosis through suppression of Bcl-2 expression (27). TLE1 is selectively over-expressed in invasive breast tumors compared to non-invasive ductal carcinoma in situ and normal mammary epithelial tissue (28). Allen et al. found that transgenic mice carrying the human TLE1 homolog developed LUAD. TLE1 was found to be overexpressed in approximately 11% of patients with lung squamous cell carcinomas and 20% of patients with LUADs. It is a putative lung-specific oncogene that positively regulates Bcl2 expression and ErbB1/ErbB2 signaling to promote cancer progression (16). Yao et al. found that the ZEB1/TLE1/E-cadherin transcriptional mechanism was a pathway that promoted the oncogenicity of lung cancer cells (18). TLE1 was also found to be an effector of epithelial-mesenchymal transition via transcriptional silencing of the epithelial marker E-cadherin in lung cancer cells (17). The TLE1 gene functions as a tumor suppressor in myeloid leukemia. Epigenetic inactivation of the TLE1 gene was shown to promote myeloid cell proliferation and survival (15). Elevated expression of TLE1 is frequently observed in glioblastoma, and is associated with poor OS (29). TLE1 expression is a good prognostic indicator in gastric cancer and that it exerts oncogenic effects (30). TLE1 may act as a tumor suppressor in hepatocellular carcinoma (31).
TLE1, which is a negative regulator of apoptosis, has been observed to inhibit low potassium-induced neuronal apoptosis (32), and to exhibit anti-neurogenic activity in mammalian forebrain development. Furthermore, Yao et al. reported that upregulation of TLE1 in transgenic mice inhibited neuronal development in vivo (33). In their study, Nuthall et al. observed that ectopic TLE1 expression in neural progenitor cells promoted their un-differentiation status with concomitant increased proliferation ability (34). In conjunction with Forkhead box protein G1 (FoxG1), TLE1 promoted survival in post-mitotic neurons in Zhang et al.’s study (32). It has also been observed to inhibit caspase-independent cell death induced by Bit1 (Bcl2-inhibitor of transcription 1) (35), and to positively regulate Bcl2 expression and ErbB1 and ErbB2 signaling (16). Furthermore, downregulation of TLE1 can result in excess inflammatory cytokine production (36). Also, Sonderegger et al. observed that exogenous TLE1 expression stimulated anchorage-independent growth in chicken embryo fibroblasts (37). These findings show that TLE1 play an important role in the BP.
TLE2 can form a complex with the replication and transcription activator (RTA) to access the cognate DNA sequence of the RTA-responsive element at different promoters and subsequently inhibit replication and transactivation (38). TLE2 and β-catenin are physically associated with NDRG1 to affect the Wnt pathway in esophageal cancer cells (39). TLE2 is also associated with the survival and progression of bladder cancer. A low expression of TLE2 was associated with shorter OS and PFS, whereas a high expression of TLE2 was associated with favorable OS. This was consistent with Wu et al.’s analysis of a cohort from the TCGA (40). In our study, the results showed that the subgroup with both TLE1high and TLE2low had significantly shorter PFS and OS than the other subgroups. High TLE1 expression and low TLE2 are independent adverse prognostic factors in LUAD and can serve as prognostic biomarkers.
Regarding TLE3, Qian et al. found that it was highly expressed in slow-proliferating and -migrating breast cancer cell lines like MCF-7; however, it was downregulated in more malignant stromal cells like ZR-75-30. These results revealed TLE3 as the possible key factor in breast cancer cell proliferation and migration (41).
The transcriptional pathways of the TLE family include Wnt, Notch, Pax2, and Runx2 (19). Cell fate determination during embryogenesis in adults is mediated by Wnt growth factors. Co-repressors of TLEs are known to contribute to the repression of Wnt targets in the absence of signaling; however, inactivation or displacement of TLE by Wnt signaling is still poorly understood. Ramakrishnan et al. reviewed the prevalence and molecular mechanisms of the Wnt transcription switch, and found that the regulation of gene expression by the Wnt pathway is complex (42). Fu et al. revealed that loss of Lats1/2 mobilized a previously unrecognized TLE/YAP/TAZ-Groucho interaction to suppress Wnt/TCF-mediated transcription, which led to Wnt-uncoupled progenitor expansion and intestinal stem cell depletion (43). Wnt acts by stabilizing cellular levels of the transcriptional coactivator beta-catenin, which forms complexes with sequence-specific DNA-binding Tcf/Lef transcription factors. Tcf/Lefs acts as transcriptional repressors by binding to Groucho/TLE proteins in the absence of nuclear β-catenin. Hanson et al. revealed a mechanism by which XIAP-mediated removal of Gro/TLE from TCF/Lef allows assembly of β-catenin-TCF/Lef complexes that initiate a Wnt-specific transcriptional program (44). Daniels et al. confirmed that β-catenin displaced Groucho/TLE from Tcf/Lef by binding to a previously unidentified second, low-affinity binding site on Lef-1 that includes sequences just N-terminal to the DNA-binding domain, and that overlaps the Groucho/TLE-binding site (45). Wu et al. found that the removal of O-GlcNAc was important for gene activation via Wnt-responsive promoters and demonstrated how Groucho/TLEs repress gene transcription (46).
In conclusion, we found that the expression levels of TLEs shows a significantly different trend with the progression of LUAD. A high expression TLE1 and a low expression of TLE2 were associated with a poor prognosis. Furthermore, TLE1 and TLE2 were found to be independent prognostic factors for patients with LUAD, and might also be potential therapeutic targets. Moreover, the combined use of TLE1 and TLE2 was better than the use of a single biomarker. However, further studies of the molecular mechanism of TLEs in LUAD are needed.
Supplementary
The article’s supplementary files as
Acknowledgments
Thank you Yue Wang and Jianyan Wen for providing professional writing services and materials.
Funding: This study is supported by the funding from the research and demonstration application of clinical diagnosis and treatment technology in Beijing capital city (Z191100006619008).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-582
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-582). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Travis WD. Pathology of lung cancer. Clin Chest Med 2011;32:669-92. 10.1016/j.ccm.2011.08.005 [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Yamada S, Wu Y, et al. Polypeptide N-acetylgalactosaminyltransferase-6 expression independently predicts poor overall survival in patients with lung adenocarcinoma after curative resection. Oncotarget 2016;7:54463-73. 10.18632/oncotarget.9810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenhalgh J, Dwan K, Boland A, et al. First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer. Cochrane Database Syst Rev 2016;(5):CD010383. 10.1002/14651858.CD010383.pub2 [DOI] [PubMed] [Google Scholar]
- 5.Agarwal M, Kumar P, Mathew SJ. The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life 2015;67:472-81. 10.1002/iub.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene 2000;249:1-16. 10.1016/S0378-1119(00)00161-X [DOI] [PubMed] [Google Scholar]
- 7.Jennings BH, Ish-Horowicz D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol 2008;9:205. 10.1186/gb-2008-9-1-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turki-Judeh W, Courey AJ. Groucho: a corepressor with instructive roles in development. Curr Top Dev Biol 2012;98:65-96. 10.1016/B978-0-12-386499-4.00003-3 [DOI] [PubMed] [Google Scholar]
- 9.Gasperowicz M, Otto F. Mammalian Groucho homologs: redundancy or specificity? J Cell Biochem 2005;95:670-87. 10.1002/jcb.20476 [DOI] [PubMed] [Google Scholar]
- 10.Buscarlet M, Stifani S. The 'Marx' of Groucho on development and disease. Trends Cell Biol 2007;17:353-61. 10.1016/j.tcb.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 11.Brantjes H, Roose J, van De Wetering M, et al. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res 2001;29:1410-9. 10.1093/nar/29.7.1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue HH, Zhao DM. Regulation of mature T cell responses by the Wnt signaling pathway. Ann N Y Acad Sci 2012;1247:16-33. 10.1111/j.1749-6632.2011.06302.x [DOI] [PubMed] [Google Scholar]
- 13.Steinke FC, Xue HH. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunol Res 2014;59:45-55. 10.1007/s12026-014-8545-9 [DOI] [PubMed] [Google Scholar]
- 14.Xing S, Shao P, Li F, et al. Tle corepressors are differentially partitioned to instruct CD8+ T cell lineage choice and identity. J Exp Med 2018;215:2211-26. 10.1084/jem.20171514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayyani F, Wang J, Yeh JR, et al. Loss of TLE1 and TLE4 from the del(9q) commonly deleted region in AML cooperates with AML1-ETO to affect myeloid cell proliferation and survival. Blood 2008;111:4338-47. 10.1182/blood-2007-07-103291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen T, van Tuyl M, Iyengar P, et al. Grg1 acts as a lung-specific oncogene in a transgenic mouse model. Cancer Res 2006;66:1294-301. 10.1158/0008-5472.CAN-05-1634 [DOI] [PubMed] [Google Scholar]
- 17.Yao X, Ireland SK, Pham T, et al. TLE1 promotes EMT in A549 lung cancer cells through suppression of E-cadherin. Biochem Biophys Res Commun 2014;455:277-84. 10.1016/j.bbrc.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao X, Pham T, Temple B, et al. TLE1 inhibits anoikis and promotes tumorigenicity in human lung cancer cells through ZEB1-mediated E-cadherin repression. Oncotarget 2017;8:72235-49. 10.18632/oncotarget.19703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberhard D, Jiménez G, Heavey B, et al. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J 2000;19:2292-303. 10.1093/emboj/19.10.2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Bae SB, Oh MH, et al. Clinicopathologic and Prognostic Significance of Transducin-Like Enhancer of Split 1 Protein Expression in Invasive Breast Cancer. J Breast Cancer 2017;20:45-53. 10.4048/jbc.2017.20.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu S, Chen Z, Gu J, et al. TLE2 is associated with favorable prognosis and regulates cell growth and gemcitabine sensitivity in pancreatic cancer. Ann Transl Med 2020;8:1017. 10.21037/atm-20-5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levanon D, Goldstein RE, Bernstein Y, et al. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci U S A 1998;95:11590-5. 10.1073/pnas.95.20.11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai X, Gao L, Teng L, et al. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell 2015;17:165-77. 10.1016/j.stem.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem 1997;272:26604-10. 10.1074/jbc.272.42.26604 [DOI] [PubMed] [Google Scholar]
- 25.Chen G, Fernandez J, Mische S, et al. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev 1999;13:2218-30. 10.1101/gad.13.17.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Dehni G, Purcell KJ, et al. Epithelial expression and chromosomal location of human TLE genes: implications for notch signaling and neoplasia. Genomics 1996;31:58-64. 10.1006/geno.1996.0009 [DOI] [PubMed] [Google Scholar]
- 27.Seo SW, Lee H, Lee HI, et al. The role of TLE1 in synovial sarcoma. J Orthop Res 2011;29:1131-6. 10.1002/jor.21318 [DOI] [PubMed] [Google Scholar]
- 28.Brunquell C, Biliran H, Jennings S, et al. TLE1 is an anoikis regulator and is downregulated by Bit1 in breast cancer cells. Mol Cancer Res 2012;10:1482-95. 10.1158/1541-7786.MCR-12-0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verginelli F, Perin A, Dali R, et al. Transcription factors FOXG1 and Groucho/TLE promote glioblastoma growth. Nat Commun 2013;4:2956. 10.1038/ncomms3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Son MW, Kim KJ, et al. Prognostic and Clinicopathological Significance of Transducer-Like Enhancer of Split 1 Expression in Gastric Cancer. J Gastric Cancer 2016;16:21-7. 10.5230/jgc.2016.16.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Masi A, Viganotti M, Antoccia A, et al. Characterization of HuH6, Hep3B, HepG2 and HLE liver cancer cell lines by WNT/β - catenin pathway, microRNA expression and protein expression profile. Cell Mol Biol (Noisy-le-grand) 2010;56 Suppl:OL1299-317. [PubMed] [Google Scholar]
- 32.Zhang X, Chen HM, Jaramillo E, et al. Histone deacetylase-related protein inhibits AES-mediated neuronal cell death by direct interaction. J Neurosci Res 2008;86:2423-31. 10.1002/jnr.21680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao J, Liu Y, Lo R, et al. Disrupted development of the cerebral hemispheres in transgenic mice expressing the mammalian Groucho homologue transducin-like-enhancer of split 1 in postmitotic neurons. Mech Dev 2000;93:105-15. 10.1016/S0925-4773(00)00278-1 [DOI] [PubMed] [Google Scholar]
- 34.Nuthall HN, Joachim K, Stifani S. Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol Cell Biol 2004;24:8395-407. 10.1128/MCB.24.19.8395-8407.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jan Y, Matter M, Pai JT, et al. A mitochondrial protein, Bit1, mediates apoptosis regulated by integrins and Groucho/TLE corepressors. Cell 2004;116:751-62. 10.1016/S0092-8674(04)00204-1 [DOI] [PubMed] [Google Scholar]
- 36.Ramasamy S, Saez B, Mukhopadhyay S, et al. Tle1 tumor suppressor negatively regulates inflammation in vivo and modulates NF-κB inflammatory pathway. Proc Natl Acad Sci U S A 2016;113:1871-6. 10.1073/pnas.1511380113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonderegger CK, Vogt PK. Binding of the corepressor TLE1 to Qin enhances Qin-mediated transformation of chicken embryo fibroblasts. Oncogene 2003;22:1749-57. 10.1038/sj.onc.1206308 [DOI] [PubMed] [Google Scholar]
- 38.He Z, Liu Y, Liang D, et al. Cellular corepressor TLE2 inhibits replication-and-transcription- activator-mediated transactivation and lytic reactivation of Kaposi's sarcoma-associated herpesvirus. J Virol 2010;84:2047-62. 10.1128/JVI.01984-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ai R, Sun Y, Guo Z, et al. NDRG1 overexpression promotes the progression of esophageal squamous cell carcinoma through modulating Wnt signaling pathway. Cancer Biol Ther 2016;17:943-54. 10.1080/15384047.2016.1210734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S, Nitschke K, Heinkele J, et al. ANLN and TLE2 in Muscle Invasive Bladder Cancer: A Functional and Clinical Evaluation Based on In Silico and In Vitro Data. Cancers (Basel) 2019;11:1840. 10.3390/cancers11121840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian TT. The effects of Tle3/Grg3 Expression on the proliferation and Migration of Human Breat Cancer cells. Hunan University 2014:1-68. [Google Scholar]
- 42.Ramakrishnan AB, Sinha A, Fan VB, et al. The Wnt Transcriptional Switch: TLE Removal or Inactivation? Bioessays 2018. doi: . 10.1002/bies.201700162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu V, Guan KL. Tales from the Cryptkeeper: New Roles for Lats1/2 in Wnt-driven Homeostasis. Cell Stem Cell 2020;26:612-4. 10.1016/j.stem.2020.04.011 [DOI] [PubMed] [Google Scholar]
- 44.Hanson AJ, Wallace HA, Freeman TJ, et al. XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Mol Cell 2012;45:619-28. 10.1016/j.molcel.2011.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 2005;12:364-71. 10.1038/nsmb912 [DOI] [PubMed] [Google Scholar]
- 46.Wu J, Bowe DB, Sadlonova A, et al. O-GlcNAc transferase is critical for transducin-like enhancer of split (TLE)-mediated repression of canonical Wnt signaling. J Biol Chem 2014;289:12168-76. 10.1074/jbc.M114.553859 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as