Abstract
Promoting residential cells, particularly endogenous neural stem and progenitor cells (NSPCs), for tissue regeneration represents a potential strategy for the treatment of spinal cord injury (SCI). However, adult NSPCs differentiate mainly into glial cells and contribute to glial scar formation at the site of injury. Gsx1 is known to regulate the generation of excitatory and inhibitory interneurons during embryonic development of the spinal cord. In this study, we show that lentivirus-mediated expression of Gsx1 increases the number of NSPCs in a mouse model of lateral hemisection SCI during the acute stage. Subsequently, Gsx1 expression increases the generation of glutamatergic and cholinergic interneurons and decreases the generation of GABAergic interneurons in the chronic stage of SCI. Importantly, Gsx1 reduces reactive astrogliosis and glial scar formation, promotes serotonin (5-HT) neuronal activity, and improves the locomotor function of the injured mice. Moreover, RNA sequencing (RNA-seq) analysis reveals that Gsx1-induced transcriptome regulation correlates with NSPC signaling, NSPC activation, neuronal differentiation, and inhibition of astrogliosis and scar formation. Collectively, our study provides molecular insights for Gsx1-mediated functional recovery and identifies the potential of Gsx1 gene therapy for injuries in the spinal cord and possibly other parts of the central nervous system.
Keywords: Gsx1, spinal cord injury, gene therapy, neurogenesis, glial scar, NSPC signaling, functional recovery
Graphical abstract
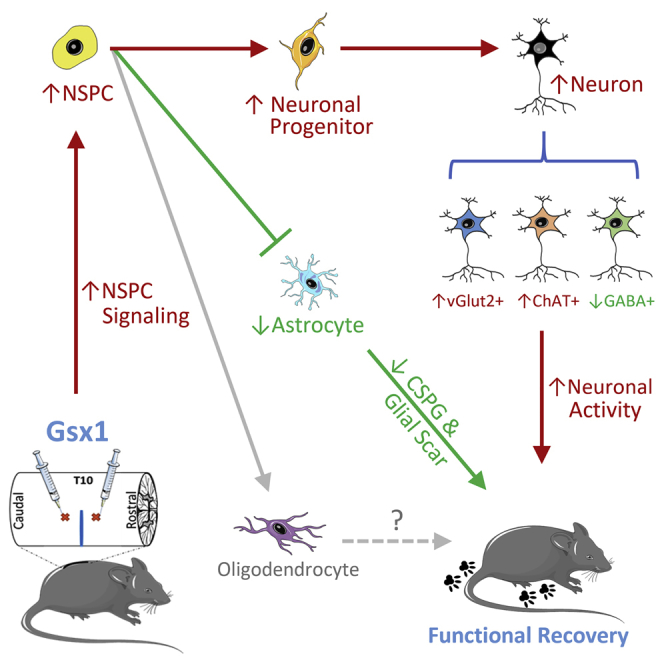
The authors show that lentivirus-mediated Gsx1 expression promotes locomotor functional recovery in a mouse model of spinal cord injury. Gsx1 increases the number of interneurons and neuronal activity, and it reduces glial scarring at the lesion site, demonstrating its therapeutic potentials for traumatic injuries in the central nervous system.
Introduction
To restore function after spinal cord injury (SCI), it is essential to repair and reconstruct the damaged local circuitry. Major hurdles in neural regeneration include a limited level of neurogenesis in the adult spinal cord and an inflammatory microenvironment that inhibits neurogenesis, axon regeneration, neuronal relay formation, and myelination at the injury site.1,2 Endogenous neural stem and progenitor cells (NSPCs) that reside around the central canal in the ependymal region provide a potential cell source for damage repair and regeneration.3, 4, 5 In general, it is thought that resident NSPCs have a limited contribution to cell replacement. Studies have shown that ependyma is not a major source of endogenous NSPCs nor does it provide neuroprotective astrocytes after SCI;6,7 however, several other studies implicate that ependymal cells are resident NSPCs and can contribute to axon remyelination and stimulate functional recovery.8,9 SCI is known to activate NSPCs and differentiate mainly into astrocytes and oligodendrocytes.5,10,11 Cell transplantation approaches using exogenous NSPCs have been tested; however, issues related to immunogenicity, integration, efficacy, and safety present significant challenges for the treatment of SCI.12 Studies have reported that residential glial cells can be reprogrammed into neurons by forced expression of neurogenic transcription factors, e.g., Sox2 or NeuroD1, in the injured brain and spinal cord, but they led to limited or no functional improvement.13 A more recent study has demonstrated that reprogramming of residential NG2 glial progenitors promotes adult neurogenesis and functional improvement following SCI.14 It has also been shown that astrocytes become reactive and produce chondroitin sulfate proteoglycans (CSPGs), which result in permanent functional deficits.15 Thus, the attenuation of glial scar formation represents a potential strategy to promote axonal regeneration and functional recovery.16, 17, 18 Furthermore, SCI induces over-inhibition by the GABAergic interneurons, rendering neurons spared from injury non-functional.19,20 Studies have demonstrated that by reducing the excitability of inhibitory interneurons or re-establishing the excitation-inhibition homeostasis, the dormant relay pathways can be reactivated after SCI and lead to improved locomotor function.21 Therefore, identifying an innovative method to promote neurogenesis, reduce glial scarring, and control excitation-inhibition homeostasis is thus fundamental to the development of an effective SCI therapy.
Many transcription factors play essential roles in neurogenesis during the embryonic development of the spinal cord.22 Among these factors, genomic screened homeobox 1 (Gsx1 or Gsh1) and NK6 homeobox 1 (Nkx6.1) are known to regulate the proliferation and differentiation of interneuron progenitors.23,24 Gsx1 and its homolog Gsx2 are expressed in NSPCs and control the choice between excitatory and inhibitory cell fates of the interneurons in the developing spinal cord.24, 25, 26 In addition, studies have shown that Gsx2 maintains progenitors of the lateral ganglionic eminence in an undifferentiated state, whereas Gsx1 promotes progenitor maturation and the acquisition of neuronal phenotypes, indicating that Gsx factors control the balance between proliferation and differentiation in the neuronal progenitor pool.26 Furthermore, our previous studies have established that Gsx1 and Nkx6.1 factors bind to a Notch1 enhancer and regulate the expression of Notch1 during the embryonic development of the brain and spinal cord.27,28 Gsx1 expression is typically low or nondetectable in the adult spinal cord.29 We thus hypothesize that reactivating Gsx1 expression in the adult injured spinal cord promotes neurogenesis and generation of interneurons important for the re-establishment of local neural circuits and functional recovery after injury.
In this study, a lentiviral vector was used to transduce Gsx1 to cells at the lesion site in a mouse model of SCI. Interestingly, we found that forced Gsx1 expression promotes cell proliferation and increases the number of NSPCs at the injury site during the acute stage of injury. In the chronic stage, Gsx1 increases the number of glutamatergic and cholinergic neurons and decreases the number of GABAergic interneurons. Importantly, Gsx1 expression attenuates glial scar formation and dramatically improves locomotor function in the injured mice. Transcriptome analysis by RNA sequencing (RNA-seq) reveals that Gsx1 induces signaling pathways associated with NSPCs (e.g., Notch and Wnt signaling pathways) and inhibits gene expression associated with reactive and scar-forming astrocytes. Taken together, our study provides molecular insights for Gsx1-mediated functional recovery after SCI and identifies the use of Gsx1 expression as a potential treatment for injuries in the spinal cord and possibly other parts of the central nervous system (CNS).
Results
Lentivirus-mediated expression of Gsx1 in a mouse model of SCI
Given the important role of Gsx1 during embryonic development of the spinal cord, we hypothesized that the upregulation of Gsx1 expression in the adult injured spinal cord promotes neural regeneration. We thus performed a lateral hemisection from the midline to the left side of the spinal cord at the thoracic (T) 9–10 level. This injury model was chosen for the following reasons: (1) it simulates an injury more likely to be seen clinically than complete transection and allows for comparison between injured and healthy tissue in the same animal; (2) it is suitable for the examination of locomotor function and recovery in different spinal tracts or to compare deficits in function of contralateral and ipsilateral lesions; (3) the model is also suitable for the investigation of gene therapy and nerve grafting, a potentially promising surgical treatment for SCI; and (4) hemisection results in a less severe injury than contusion or complete transection. The completeness and consistency of the lateral hemisection SCI was confirmed by the observation of paralysis in the left hindlimb. Immediately after the SCI, lentivirus (~1 μL/site of 1 × 108 transducing units [TU]/mL) encoding Gsx1 and a reporter red fluorescent protein (RFP) (lenti-Gsx1-RFP) were injected into the injured spinal cord, approximately 1 mm rostral and caudal to the injury site (Figure 1A). Lentivirus encoding only the RFP reporter (lenti-Ctrl-RFP) was used as a control. The cytomegalovirus (CMV) immediate-early enhancer and promoter were chosen to drive the expression of Gsx1 in the spinal cord. CMV is a strong promoter and is commonly used to drive gene expression in a variety of cells in the spinal cord, including stem cells.30,31 We assigned animals randomly into the following three groups (6–12 mice/group): (1) sham (exposing the spinal cord without injury); (2) injured mice with an injection of lenti-Ctrl-RFP (SCI+Ctrl); and (3) injured mice with an injection of lenti-Gsx1-RFP (SCI+Gsx1). We confirmed that the lentivirus-mediated Gsx1 expression in the spinal cord tissue at 3 days post-injury (DPI) and 7 DPI by immunohistochemistry and quantitative real-time PCR. Lentivirus injection significantly increased Gsx1 expression in virus-transduced cells, i.e., RFP+ cells (Figures S1A–S1D). In addition, we performed immunohistochemistry analysis on virally transduced cells and determined their neuronal versus glial identity using antibodies specific to neurons (NeuN, Figure S1F) and astrocytes (GFAP, Figure S1G) at 3 DPI. No significant differences in the percentages of co-labeled cells (RFP+/NeuN+ or RFP+/GFAP+) were detected between the SCI+Ctrl or SCI+Gsx1 groups (Figure S1H), indicating that the Gsx1 encoding virus does not preferentially infect the mature neuronal or astrocyte cell population.
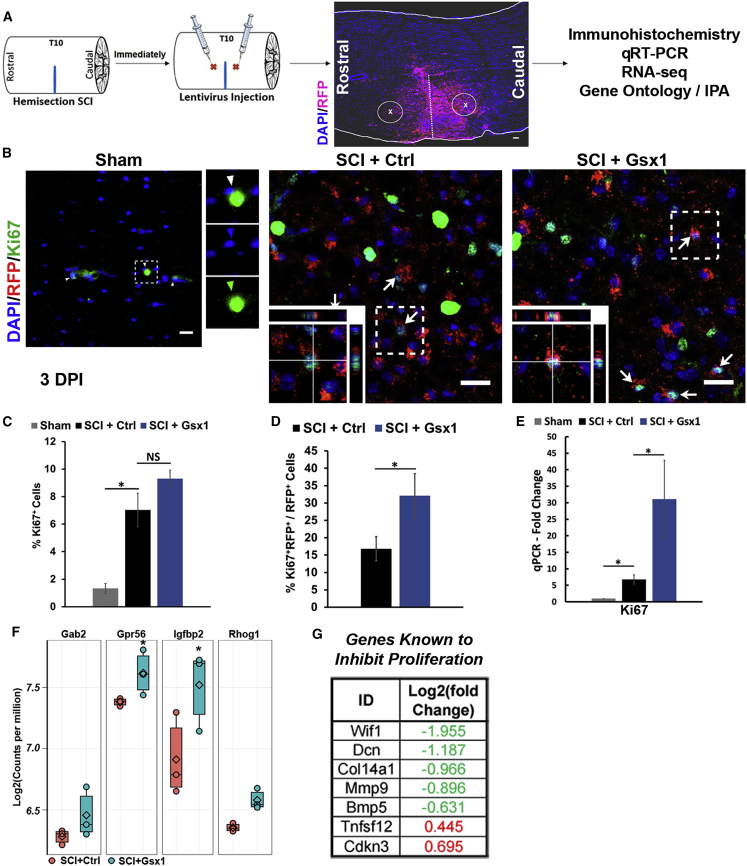
Figure 1.
Gsx1 expression promotes cell proliferation in the injured spinal cord
(A) Lateral hemisection SCI was performed on 8- to 12-week-old mice at the T9–T10 level immediately followed by the injection of lentivirus encoding Gsx1 along with RFP reporter (lenti-Gsx1-RFP). Lentivirus encoding only the reporter RFP was used as a control (lenti-Ctrl-RFP). Spinal cord tissues were analyzed by immunohistochemistry, RNA-seq, Ingenuity Pathway Analysis (IPA), and quantitative real-time PCR analysis. Scale bar, 100 μm. (B) Confocal images of sagittal sections of spinal cord tissue at 3 DPI show the expression of the viral reporter RFP and cell proliferation marker Ki67 (n = 3 for sham and n = 6 for SCI+Ctrl and SCI+Gsx1). Arrows indicate Ki67+/RFP+ co-labeled cells. Images in the bottom left corner show a higher magnification z stack view of the area denoted by a dashed white box. Scale bars, 20 μm. (C and D) Quantification of all Ki67+ cells and Ki67+/RFP+ cells. (E) Quantitative real-time PCR analysis of Ki67 mRNA expression at 3 DPI, normalized to the sham group; n = 4. Mean ± SEM. ∗p < 0.05 by Students’ t test (D) and one-way ANOVA followed by a Tukey post hoc test (C and E). (F) Box plot of genes known to promote cell proliferation (n = 3). Each dot represents the gene expression as log2(count per million) for one biological replicate sample. (G) List of differentially expressed genes that are known to inhibit cell proliferation between SCI+Gsx1 and SCI+Ctrl groups by RNA-seq analysis (n = 3).
Gsx1 promotes cell proliferation in the injured spinal cord
SCI is known to induce cell proliferation at the lesion site.32 To determine whether Gsx1 enhances injury-induced cell proliferation, we examined the expression of a cell proliferation marker Ki67 at 3 DPI by immunohistochemistry followed by confocal imaging analysis (Figure 1B). The RFP+ and Ki67+ cells were found to be located around the injection sites. The number of Ki67+ cells and DAPI+ cells was manually counted in the control and experimental groups, that is, sham (n = 3), SCI+Ctrl (n = 6), and SCI+Gsx1 (n = 6). We observed a significant increase in the percentage of Ki67+ cells in both of the injury groups that received a viral injection as compared to the sham mice, with the highest increase found in the SCI+Gsx1 group (Figure 1C). When the percentage of Ki67+/RFP+ co-labeled cells among RFP+ cells was calculated, this percentage was significantly higher in the SCI+Gsx1 group than in the SCI+Ctrl group at 3 DPI (Figure 1D). Furthermore, the increase in Ki67 mRNA expression was validated by quantitative real-time PCR. A significantly higher level of Ki67 mRNA was detected in the SCI+Gsx1 group as compared to the SCI+Ctrl and sham groups (~4-fold increase; Figure 1E).
The transcriptome analysis using RNA-seq was performed to identify genes and pathways induced by Gsx1 expression. We found 475, 1,447, and 3,946 differentially expressed genes (DEGs) between the SCI+Ctrl and SCI+Gsx1 groups at 3, 14, and 35 DPI, respectively (Figure S2). Gene enrichment analysis of the top 40 DEGs at 3 DPI using REVIGO33 revealed that Gsx1 upregulated developmental processes, e.g., cell proliferation and differentiation (Table S1; Figures S3A and S3D). REVIGO is a web application that summarizes lists of Gene Ontology (GO) terms by finding a representative subset of the terms using a simple clustering algorithm that relies on semantic similarity measures. Gsx1 expression upregulated genes known to promote cell proliferation (e.g., Gab2, Gpr56, Igfbp2, Rhog; Figure 1F) and downregulated genes known to inhibit cell proliferation (e.g., Wif1, Dcn, Mmp9; Figure 1G). To determine the effect of Gsx1 expression on cell proliferation in the uninjured spinal cord, we also perform viral injection in the sham groups. We noticed a significantly increased percentage of Ki67+/RFP+ co-labeled cells among RFP+ cells in the sham+Gsx1 group as compared to the sham+Ctrl group (Figure S4). These data suggest that Gsx1 expression enhances cell proliferation in the adult spinal cord without or with injury.
Gsx1 increases the number of NSPCs in the injured spinal cord
Our previous studies have shown that the reporter GFP labels NSPCs that preferentially differentiate into interneurons in Notch1CR2-GFP transgenic mice during embryonic development.27,28 Furthermore, the adult endogenous NSPCs exist in quiescent states under normal conditions and they become activated after injury.11,34 Thus, we used the Notch1CR2-GFP transgenic mouse model to examine the behavior of NSPCs in the injured spinal cord. We found that the transection injury significantly increased GFP+ cells at 7 and 14 DPI as compared to the sham group (n = 3 for each time point, Figures 2A–2D). In the sham group, only a few GFP+ cells were detected in the ependymal region around the central canal (Figure 2A), a region known to contain NSPCs in the spinal cord. In contrast, SCI significantly increased the number of GFP+ cells. These cells were found mainly in the dorsal half of the spinal cord at 3 and 7 DPI. By 14 DPI, an increasing number of GFP+ cells spread throughout the entire spinal cord (Figures 2B–2D). Injury-induced GFP+ cells were co-labeled with Notch1, Sox9, and Nkx6.1 at 3 DPI (Figure 2E), indicating that they were NSPCs. We next examined the effect of Gsx1 expression on GFP+ cells in Notch1CR2-GFP mice after SCI. Compared with the SCI+Ctrl group (~35% of GFP+ cells; n = 3), Gsx1 expression significantly increased the percentage of GFP+ cells among RFP+ cells at 7 DPI (~62%; n = 3, p = 0.0038; Figure 2F). These results suggest that Gsx1 expression enhanced NSPC activation and further increased the number of NSPCs in the injured spinal cord.
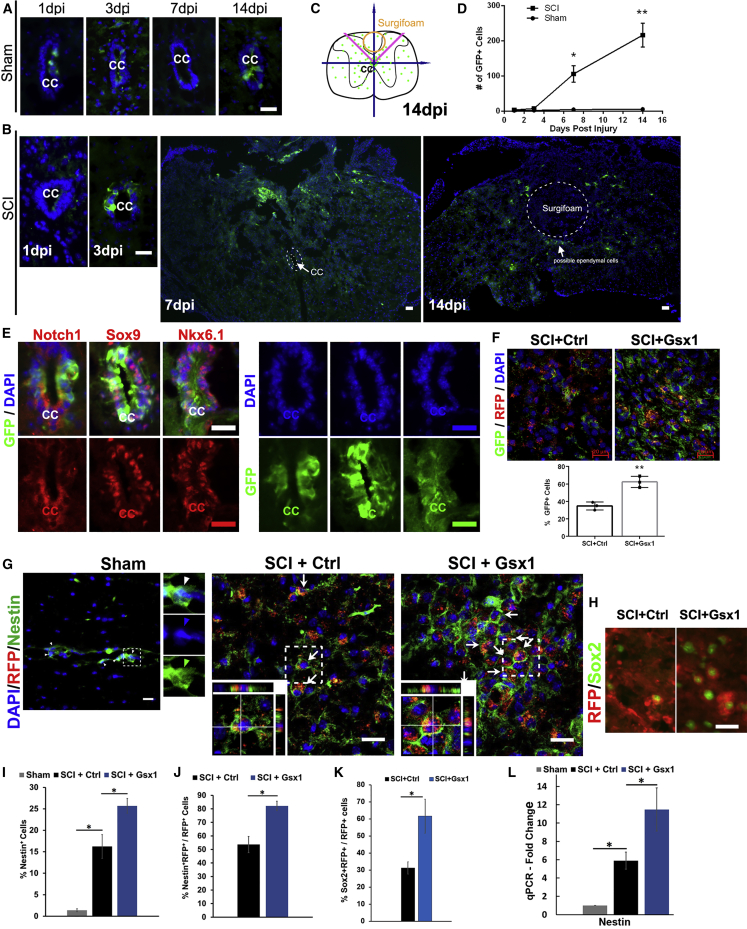
Figure 2.
Gsx1 increases the number of NSPCs after SCI
Full transection spinal cord injury (Tx) was performed on Notch1CR2-GFP transgenic mice at the T9–T10 level. (A) Fluorescence images of the cross-section through the central canal (CC) at the T9–10 level of the spinal cord from the sham groups show GFP+ cells at 1, 3, 7 and 14 DPI. Only a few GFP+ cells can be detected in the ependymal cell population lining the wall of the CC. (B) The location of GFP+ cells near the ependymal region at 1, 3, 7, and 14 DPI in the Tx animals. (C) A schematic shows the distribution of GFP+ cells. (D) Quantification of the number of GFP+ cells in the sham and SCI groups (n = 3 for all time points). (E) GFP+ ependymal cells co-labeled with Notch1, Sox9, and Nkx6.1. GFP, DAPI, and cell markers are also shown in separate channels. (F) Lentivirus injection was performed in transgenic animals with lateral hemisection SCI. Immunofluorescence analysis of the spinal cord sections shows that Gsx1 expression increased the percentage of GFP+ cells among virally transduced cells (RFP+) (n = 3). (G and H) Representative images of sagittal sections of spinal cord tissues show the expression of viral reporter RFP and the NSPC marker Nestin (G) and Sox2 (H) at 3 DPI (n = 3 for the sham group and n = 6 for the SCI+Ctrl and SCI+Gsx1). Arrows indicate Nestin+/RFP+ co-labeled cells. Images in the bottom left corner show a higher magnification z stack view of the area denoted by a dashed white box. (I–K) Quantification of all Nestin+ cells (I), Nestin+/RFP+ co-labeled cells (J), and Sox2+/RFP+ co-labeled cells (K). (L) Quantitative real-time PCR analysis shows Nestin mRNA expression at 3 DPI, normalized to the sham group; n = 4. Mean ± SEM. ∗p < 0.05 by a Students’ t test (F, J, and K) and one-way ANOVA followed by a Tukey post hoc test (D, I, and L). Scale bars, 50 μm (A–E) and 20 μm (F and G).
The effect of Gsx1 expression on NSPCs in the injured spinal cord was further examined in C57BL/6J mice with additional markers, Nestin and Sox2, at 3 DPI. Confocal imaging analysis identified Nestin+ (Figure 2G) and Sox2+ (Figure 2H) cells around the injury and injection sites. Compared to the sham group, a significantly increased percentage of Nestin+ and Sox2+ cells was found at the lesion site in the SCI groups (SCI+Ctrl and SCI+Gsx1), with the highest increase in the SCI+Gsx1 group (Figure 2I). A significantly higher percentage of Nestin+/RFP+ co-labeled cells among RFP+ cells was found in the SCI+Gsx1 group as compared to the SCI+Ctrl group (Figure 2J). Similarly, a significantly higher percentage of Sox2+/RFP+ cells among RFP+ cells was found in the SCI+Gsx1 group (Figure 2K). The quantitative real-time PCR analysis confirmed that Gsx1 expression significantly increased Nestin mRNA expression as compared to the SCI+Ctrl and sham groups (Figure 2L). To determine the effect of Gsx1 expression on NSPCs in the uninjured spinal cord, we also perform viral injection in the sham groups. We noticed a significantly increased percentage of Nestin+/RFP+ co-labeled cells among RFP+ cells in the sham+Gsx1 group as compared to the sham+Ctrl group (Figure S5), indicating that Gsx1 expression can also activate endogenous adult NSPCs without injury.
Gsx1 upregulates signaling pathways associated with NSPCs
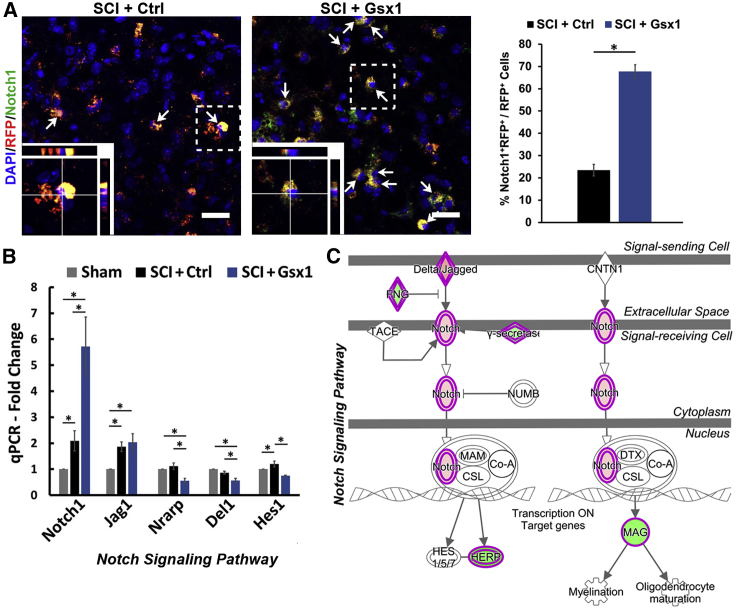
Many signaling pathways play essential roles in the maintenance of adult NSPCs.35, 36, 37, 38 We next explored the effect of Gsx1 expression on genes associated with NSPC signaling pathways. RNA-seq and Ingenuity Pathway Analysis (IPA) analysis revealed the upregulation of Notch, Nanog, and Wnt signaling pathways (see Tables S1, S2, and S3). We then performed immunohistochemistry analysis using the anti-Notch1 antibody. A significant increase in the number of Notch1+/RFP+ co-labeled cells among RFP+ cells was observed in the SCI+Gsx1 group as compared to the SCI+Ctrl group at 3 DPI (Figure 3A). A further increase in mRNA expression of Notch1 and Jag1 (a ligand for the Notch1 receptor) was found in the SCI+Gsx1 group as compared to the sham and SCI+Ctrl groups (Figure 3B). In contrast, Nrarp was downregulated in the SCI+Gsx1 group (Figure 3B). Nrarp is a negative regulator of the Notch signaling pathway that physically interacts with the Notch intracellular domain (NICD) and blocks Notch transcription.39,40 Furthermore, compared to the control treatment, Gsx1 expression decreased the mRNA levels of Del1 and Hes1 (Figure 3B), key components in Notch signaling. The RNA-seq and IPA revealed the upregulation of the Notch signaling pathway (Figure 3C), including increased expression of Hes7 and Rbpj (Figure S6A). The RNA-seq analysis also revealed the genes known to positively and negative regulate NSPCs (Figures S6B and S6C). In addition, there was an increased expression of genes associated with activation of the Nanog signaling pathway, e.g., Akt2, Map2k2, Pik3cd, and Pik3cg, and NSPC genes Rap2, Sox11, and Tyk2 at 3 DPI (Figure S6D). In contrast, Notch/Nanog signaling pathways were not detected at 35 DPI by RNA-seq and IPA (Figures S2 and S3). Furthermore, RNA-seq analysis and quantitative real-time PCR also revealed the upregulation of Wnt signaling pathways at 3, 14, and 35 DPI (Figure S7). These results support the notion that Gsx1 expression transiently activates/enhances signaling pathways involved in NSPCs, e.g., Notch, Nanog, and Wnt signaling pathways, after SCI.
Figure 3.
Gsx1 upregulates Notch signaling in the acute stage of SCI
(A) Representative confocal images of sagittal sections through the T9–10 spinal cord at 3 DPI show the expression of viral reporter RFP and NSPC marker Notch1 (n = 4 for SCI+Ctrl and SCI+Gsx1). Arrows indicate Notch1+/RFP+ co-labeled cells. Images in the bottom left corner show a higher magnification z stack view of the area denoted by a dashed white box. Scale bars, 20 μm. The quantification of Notch1+ cells among RFP+ cells is shown in a histogram on the right. (B) A histogram shows the quantitative real-time PCR analysis of the genes involved in the Notch signaling pathway (Notch1, Jag1, Nrarp, Del1, and Hes1). Mean ± SEM. ∗p < 0.05 by a Students’ t test (A) and one-way ANOVA followed by a Tukey post hoc test (B). (C) Diagram depicts the upregulated Notch signaling pathway by Gsx1 expression revealed by IPA.
Gsx1 induces specific types of interneurons in the injured spinal cord
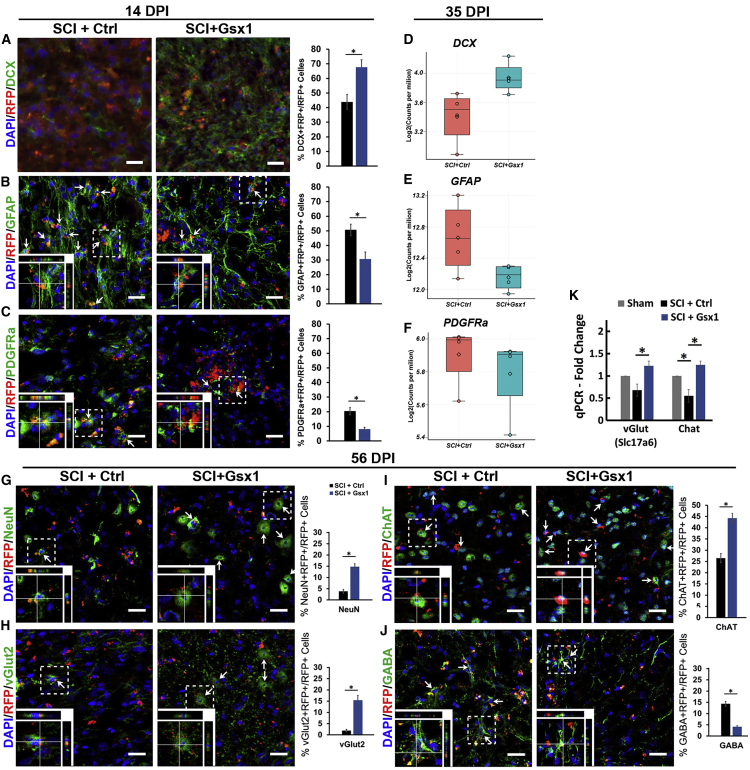
In the adult spinal cord, injury-activated NSPCs mostly generate astrocytes and oligodendrocytes.5,11 To investigate the role of Gsx1 in NSPC differentiation after SCI, we examined spinal cord tissues at 14 DPI with an early neuronal marker doublecortin (DCX), an astrocyte marker GFAP, and an oligodendrocyte progenitor marker PDGFRa in the SCI+Ctrl (n = 6) and SCI+Gsx1 (n = 6) groups. Compared to the control group (SCI+Ctrl), Gsx1 expression significantly increased the percentage of DCX+/RFP+ co-labeled cells (Figure 4A) and decreased the percentage of GFAP+/RFP+ (Figure 4B) and PDGFRa+/RFP+ (Figure 4C) co-labeled cells among RFP+ cells. There was no significant difference in the number of oligodendrocytes (Olig2+/RFP+ cells) between the SCI+Ctrl and SCI+Gsx1 groups (Figure S8). These results suggest that Gsx1 expression skewed NSPC differentiation toward the neuronal over the glial lineage during the chronic stage of SCI. The upregulation of DCX (Figure 4D) and the downregulation of GFAP (Figure 4E) and PDGFRa (Figure 4F) in the SCI+Gsx1 group at 35 DPI was confirmed by RNA-seq analysis. GO analysis of DEGs at 14 and 35 DPI revealed that enrichment of Gsx1 induced DEGs involved in CNS development, neurogenesis, cell differentiation, neuronal projection, and synapse organization (Table S4; Figures S2 and S3). The identity of virally infected cells was further examined at 56 DPI using a mature neuronal marker NeuN (Figure 4G), a glutamatergic interneuron marker vGlut2 (Figure 4H), a cholinergic neuronal marker ChAT (Figure 4I), and a GABAergic interneuron marker GABA (Figure 4J). Gsx1 expression significantly increased the percentage of NeuN+, ChAT+, and vGlut2+ cells and decreased the percentage of GABA+ cells among RFP+ cells (n = 4) as compared to the SCI+Ctrl group (n = 4). The quantitative real-time PCR analysis detected a significantly increased mRNA level of vGlut (or Slc17a6) and ChAT in the SCI+Gsx1 group (n = 4) as compared to the sham (n = 4) and SCI+Ctrl (n = 4) groups at 35 DPI (Figure 4K). These results indicate that Gsx1 expression preferentially increased the number of excitatory glutamatergic and cholinergic interneurons and decreased the number of inhibitory GABAergic interneurons in the injured spinal cord.
Figure 4.
Gsx1 affects the generation of specific types of interneurons in the injured spinal cord
(A–C) Confocal images of sagittal sections of spinal cord tissues at 14 DPI show the expression of the viral reporter RFP and early neuronal marker doublecortin (DCX) (A), astrocyte marker GFAP (B), and oligodendrocyte progenitor marker PDGFRa (C). Arrows indicate cell marker+/RFP+ co-labeled cells. Images in the bottom left corner show a higher magnification z stack view of the area denoted by a dashed white box. Scale bars, 20 μm. (D–F) Quantification of virally transduced cells co-labeled with DCX, GFAP, or PDGFRa (n = 3). Gene expression box plots of DCX (D), GFAP (E), and PDGFRa (F) at 35 DPI between SCI+Ctrl and SCI+Gsx1 groups. Each dot represents the gene expression as log2(count per million) for one biological replicate sample. Mean ± SEM. ∗p < 0.05 by Students’ t test. (G–J) Confocal images of sagittal sections of spinal cord tissues at 56 DPI show the expression of viral reporter RFP and a mature neuron marker NeuN (G), glutamatergic neuron marker vGlut2 (H), cholinergic neuron marker ChAT (I), and GABAergic neuron marker GABA (J) with quantification (n = 4). Images in the bottom left corner show a higher magnification z stack view of the area denoted by a dashed white box. Scale bars, 20 μm. (K) Quantitative real-time PCR analysis shows vGlut and Chat mRNA expression at 35 DPI, normalized to the sham group; n = 3. Mean ± SEM ∗p < 0.05 by one-way ANOVA followed by a Tukey post hoc test.
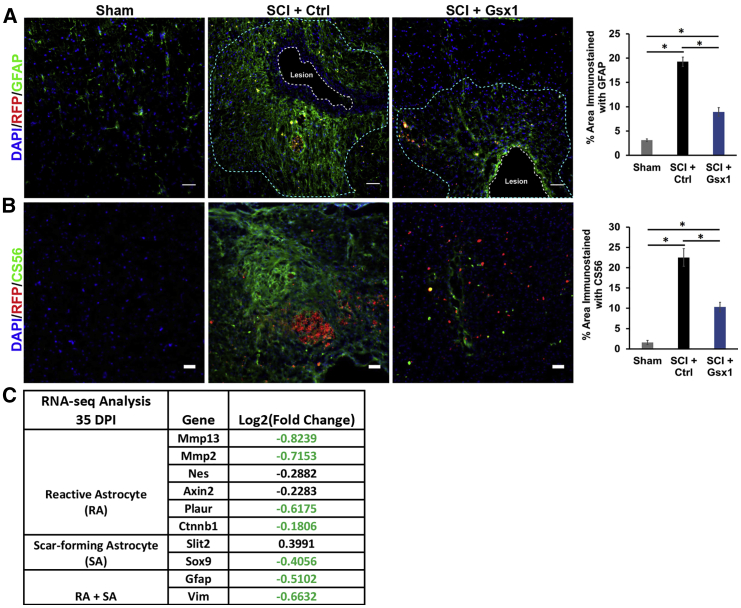
Gsx1 attenuates glial scar formation
As Gsx1 affected neural differentiation and reduced the number of glial cells, we next examined whether Gsx1 affects glial scar formation. Immunofluorescence analysis was performed on spinal cord tissues using GFAP as a marker for reactive astrocytes and CSPG as a marker for glial scar formation. Gsx1 expression significantly reduced the GFAP and CS56 immunostained area around the lesion site in the SCI+Gsx1 group as compared to the SCI+Ctrl group (Figures 5A and 5B). The RNA-seq analysis showed a downregulation of genes associated with reactive astrocytes (e.g., Mmp13, Mmp2, Nes, Axin2, Slit2, Plaur, and Ctnnb1), scar-forming astrocytes (e.g., Slit2 and Sox9), and both reactive and scar-forming astrocytes (e.g., GFAP and Vim)41 at 35 DPI (Figure 5C). In addition, we examined the effect of Gsx1 expression on glial scar formation in the uninjured spinal cord (sham group). Although the viral injection itself increased the GFAP immunostained area at the injection site (compared to the sham alone group), no significant difference was noticed in the immunostained area for GFAP or CS56 between the sham+Gsx1 and sham+Ctrl groups (Figure S9). These results suggest that Gsx1 represses reactive and scar-forming astrocytes and attenuates glial scar formation after SCI.
Figure 5.
Gsx1 attenuates astrogliosis and glial scar formation
(A and B) Representative fluorescence images of sagittal sections through the lesion site in the spinal cord at 56 DPI show the expression of viral reporter RFP, GFAP (A), and chondroitin sulfate proteoglycan (CSPG, stained with CS56) (B), and the quantification of the immunostained area with anti-GFAP and anti-CS56 around the injury site is shown on the right. Scale bars, 50 μm. n = 4 for sham and n = 6 for SCI+Ctrl and SCI+Gsx1 groups. Mean ± SEM. ∗p < 0.05 by one-way ANOVA followed by a Tukey post hoc test. (C) A partial list of genes associated with reactive astrocytes (RAs) and scar-forming astrocytes (SAs) identified by RNA-seq analysis. Fold change in green font indicates statistical significance.
In addition, we have performed cell culture experiments using the NE-4C cell line, a neural ectoderm-derived neural stem cell line42, 43, 44 with lenti-Gsx1 transduction. We observed that there was a significantly increased number of Map2+ neurons and reduced number of GFAP+ astrocytes in the cultured NE-4C cells 14 days after lenti-Gsx1 transduction as compared to the lenti-Ctrl group (Figure S10). This result further supports Gsx1 function in neural differentiation and is consistent with the known function of Gsx1 in regulating neural differentiation during embryonic spinal cord development.24
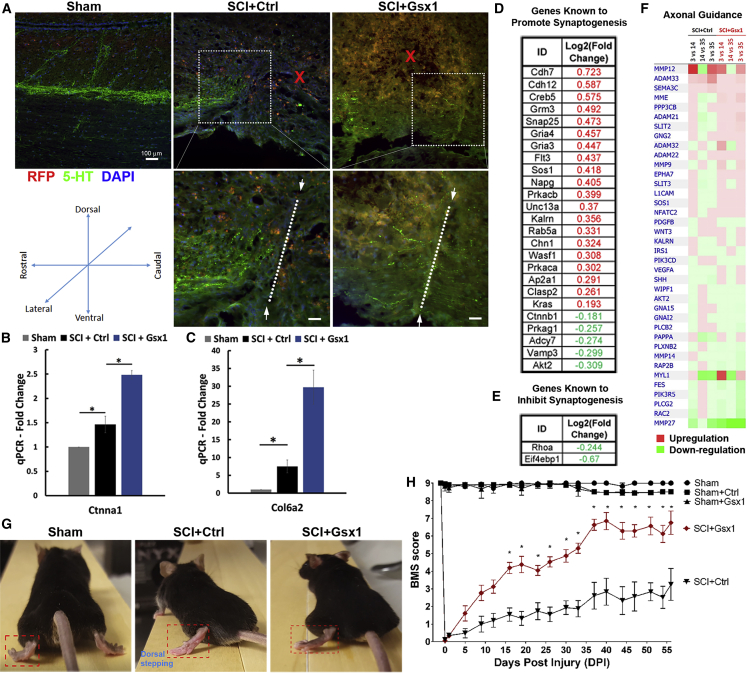
Gsx1 promotes 5-HT neuronal activity and improves locomotor function after SCI
Neurotransmission of serotonin (5-HT) in the spinal cord is required for modulating sensory, motor, and autonomic functions, and positive 5-HT staining is associated with neuronal activity.45 After SCI, 5-HT-positive axons caudal to the injury site degenerate, while rostral to the injury site they sprout.46,47 Thus, we examined the 5-HT immunoactivity in spinal cord tissues isolated at 35 DPI. In the sham group (n = 3), the 5-HT-labeled axons were detected continuously through the T9–T10 region of the spinal cord (Figure 6A). However, in the SCI+Ctrl group (n = 3), the 5-HT-labeled axons were detected rostrally but not caudally to the injury site (Figures 6A and S11; white dotted line indicates the hemisection site). In contrast, the 5-HT-labeled axons in the SCI+Gsx1 group (n = 3) extended caudally to the injury site (Figures 6A and S11). This result suggests that Gsx1 expression promotes axon sprouting and growth after SCI. To determine the molecular basis for Gsx1-induced axon sprouting/growth, we examined the expression of a selected set of genes involved in axon growth and synaptogenesis. Gsx1 expression (n = 4) significantly increased mRNA level of Ctnna1 and Col6a2 as compared to the SCI+Ctrl group (n = 4) at 35 DPI (Figures 6B and 6C) by quantitative real-time PCR analysis. The RNA-seq analysis and GO analysis revealed that Gsx1 expression led to an upregulation of genes known to promote synaptogenesis (Figure 6D) and a downregulation of genes known to inhibit synaptogenesis (Figure 6E). In addition, Gsx1 expression upregulated genes associated with axonal guidance pathways (Figure 6F). Finally, we assessed the effect of Gsx1 expression on locomotor behavior using an open-field locomotion test starting from the day before the injury (−1 DPI) to 56 DPI (8 weeks). For each mouse, a Basso Mouse Scale (BMS) score48 was assigned double-blindly by three observers. BMS scores range from 0 (complete paralysis and no ankle movement) to 9 (normal walking).48 All mice with a lateral hemisection SCI at the T9–10 level exhibited paralysis in the left hindlimb, while the sham, sham+Ctrl, and sham+Gsx1 animal groups displayed a normal locomotor behavior with a BMS score of ~9 from −1 to 56 DPI (Figures 6G and 6H). Mice with SCI (both SCI+Ctrl and SCI+Gsx1 groups) exhibited paralysis in the left hindlimb with a BMS score of 0 on the day of hemisection injury (0 DPI) (Figures 6G and 6H; Videos S1, S2, and S3), confirming lateral hemisection-induced SCI. For mice in the SCI+Ctrl group (n = 6), the BMS scores gradually improved to ~3 (dorsal stepping) by 56 DPI. In contrast, mice in the SCI+Gsx1 group (n = 12) demonstrated profoundly improved locomotor function with the BMS scores around 6–7 by 42 DPI and the scores maintained above 6 until the end of the tests (Figure 6H). The BMS scores for the SCI+Gsx1 group increased significantly higher than did those of the SCI+Ctrl group starting at 16 DPI (Figure 6H; ∗p < 0.05). These results demonstrated that Gsx1 expression significantly improved locomotor functional recovery after SCI.
Figure 6.
Gsx1 promotes 5-HT neuronal activity and improves locomotor function after SCI
(A) Sagittal sections of the spinal cord through the T9–T10 level show 5-HT staining at 35 DPI. The boxed region is shown in a higher magnification. The white dotted line indicates the hemisection site. “X” indicates sites of viral injection. (B and C) Quantitative real-time PCR analysis of differentially expressed genes (Ctnna1 and Col6a2) involved in axon guidance at 35 DPI (n = 4; two-way ANOVA analysis followed by a post hoc test). (D and E) Partial list of Gsx1-induced differentially expressed genes involved in synaptogenesis. (F) Heatmap shows genes involved in axonal guidance from RNA-seq analysis and IPA comparing among 3, 14, and 35 DPI groups (n ≥ 3). Mean ± SEM. ∗p < 0.05 by Student’s t test. Red color indicates gene upregulation and green indicates downregulation. (G and H) Representative images of walking posture at 56 DPI (G) and a plot of the BMS scores (H) of left hindlimb over 56 DPI (n = 6 for all data points and groups, except the SCI+Gsx1 group, for which n = 12). ∗p < 0.05 by a two-way repeated measures ANOVA followed by a Tukey post hoc test.
Discussion
Limited neurogenesis, neurite growth, increased reactive astrogliosis, and scar formation are the major hurdles for neural regeneration and functional recovery after SCI.1,2 In this study, we show that exogenous Gsx1 expression increases the number of NSPCs and the generation of specific subtypes of interneurons, i.e., vGlut2+ and ChAT+ interneurons, and 5-HT neuronal activity, and it reduces the number of inhibitory GABAergic neurons. Moreover, Gsx1 attenuates reactive astrogliosis and glial scar formation, and it promotes locomotor functional recovery in mice with SCI.
It has been shown that transcription factors, including Sox2, Oct4, Klf4, and NeuroD1, can reprogram the endogenous glial cells into neurons or NSPCs. However, these approaches largely led to limited or no functional recovery after SCI.14,49, 50, 51 The failure of reprogrammed neurons to elicit functional recovery may be attributed to the following reasons: (1) low reprogramming efficiency, which resulted in an insufficient number of functional neurons; (2) Oct4, Sox2, and NeuroD1 are general neurogenic factors, which may not be capable of inducing specific neuronal types required for recovering from SCI, e.g., Sox2-induced neurons resemble GABAergic interneurons50 (in fact, the induction of additional inhibitory interneurons might have caused a further imbalance of the excitation/inhibition homeostasis); and (3) functional recovery may require the generation of various specific cell types. In the current study, we have shown that Gsx1 promoted the generation of glutamatergic and cholinergic interneurons, and reduced the generation of GABAergic interneurons. These spinal interneurons have been demonstrated to be essential for transmitting both motor and sensory impulses.52,53 Furthermore, reducing the excitability of spinal cord inhibitory interneurons enhances functional recovery in mice with SCI.21 It is thus possible that Gsx1-induced functional recovery was partially due to the reduced inhibitory GABAergic interneurons and increased excitatory glutamatergic and cholinergic interneurons. This is also consistent with the established role of Gsx1 to control the generation of excitatory and inhibitory interneurons during embryonic development of the spinal cord.24 Thus, by affecting interneuron subpopulations, Gsx1 contributes to the restoration of the excitation/inhibition homeostasis in the injured spinal cord.
Previous studies have demonstrated that resident astrocytes can be reprogrammed into neurons using astrocyte-specific GFAP promoter, but a significant functional recovery has generally not been reported.49,50,54 Thus, we used a strong ubiquitous CMV promoter to drive Gsx1 expression and target all possible cell types in the injured spinal cord. The observation that Gsx1 expression led to increased cell proliferation (with an increased number of Ki67+/RFP+ cells) suggests that Gsx1 mainly affects the NSPC population. This role of Gsx1 in NSPCs was further demonstrated in the Notch1CR2-GFP transgenic SCI model, in which the reporter GFP-labeled cells are NSPCs and committed to become interneurons.27,28
Injury to the brain and spinal cord leads to NSPC activation.55 The observation that Gsx1 expression leads to a further increase in the number of GFP+ NSPCs during acute stage of injury implicates a role of Gsx1 in NSPC activation and proliferation. The finding of similar levels of Ki67+ cells and Nestin+ cells in the sham+Gsx1 and SCI+Ctrl groups (Figures S4 and S5) further confirms such a role of Gsx1. This is also supported by RNA-seq analysis that Gsx1-induced genes were involved in cell proliferation. Further pathway analysis reveals that Gsx1 upregulates NSPC signaling pathways, including Notch, Nanog, and Wnt signaling. Studies have established a role for Gsx1 in the regulation of Notch signaling for neuronal differentiation.27,28,56 Although our data strongly indicate that Gsx1 induces NSPC differentiation into functional interneurons, we cannot rule out the possibility that Gsx1 can reprogram residential glial cells into neurons or promote survival of the neurons at the lesion site. Future cell-lineage tracing experiments using a cell-specific promoter for targeted expression of Gsx1 in glial cells or NSPCs are needed to determine the origin of Gsx1-induced neurons.
SCI causes activation of microglia and astrocytes, which leads to reactive astrogliosis and glial scar formation.15,57,58 The glial scar is mostly composed of reactive astrocytes, non-neuronal cells (e.g., pericytes and meningeal cells), and proteoglycan-rich extracellular matrix (ECM).41,59,60 Activated astrocytes secrete CSPG, which constitutes the major component of the glial scar.61 Inhibition of CSPG represents an important therapeutic strategy for achieving functional recovery after SCI. The observation that Gsx1 reduces reactive astrogliosis and thus glial scar formation is well correlated with functional recovery after SCI; such a role for Gsx1 has not been reported. In fact, the adult NSPCs give rise to mainly astrocytes after CNS injury.62,63 Gsx1 expression significantly decreases the expression of genes associated with reactive astrocytes and scar-forming astrocytes. It is likely that Gsx1 induces the generation of neurons at the expense of the astrocyte lineage, and thus reduced astrogliosis leads to the attenuation of scar formation during the chronic stage of SCI. A recent study has shown that the overexpression of Oct4 and Klf4 led to reduced glial scar formation and improved motor function after SCI.51 Thus, neurogenic factors may generally have the potential to suppress astrogliosis and scar formation in the injured spinal cord. However, an effective therapeutic strategy for SCI may also require specific subtypes of spinal cord interneurons for maintaining excitatory/inhibitory homeostasis.
In summary, we have demonstrated that lentivirus-mediated Gsx1 expression in the injured spinal cord is sufficient to reduce glial scarring, increase neurogenesis of specific interneurons, and promote neuronal activity and locomotion function after SCI. These findings unveil Gsx1 gene therapy as a promising treatment for injuries to the spinal cord and perhaps other parts of the CNS.
Materials and methods
Animals
Young adult (8- to 12-week-old) mice (Notch1CR2-GFP transgenic27,28 and C57BL/6J, Jackson Laboratory, 000664) were used in this study. All of the proposed animal work was conducted under compliance with the Institutional Animal Care and Use Committee (IACUC) at Rutgers University. All animals were housed in an animal care facility with a 12-h light/12-h dark cycle. Mice under each experimental condition were assigned randomly with an equal number of male and female mice when possible.
Lentivirus production
Lentiviruses encoding Gsx1 and a reporter RFP (lenti-Gsx1-RFP) and control lentiviruses (encoding only the reporter RFP, lenti-Ctrl-RFP) (ABM, LV465366 and LV084) were generated by transfecting human embryonic kidney (HEK)293T cells with a mixture of target vector (lenti-Gsx1-RFP or lenti-Ctrl-RFP), envelope plasmids (pMD2.G/VSVG, Addgene, 12259), and third-generation packaging plasmids (pMDLg/pRRE, Addgene, 12251 and pRSV-Rev, Addgene, 12253). HEK293T cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% nonessential amino acids (MEM NEAAs 100×, Life Technology, 11140050), and 1% GlutaMAX I 100× (Life Technology 35050061). Transfection of the HEK293T cells was performed when the culture reached ~50%–60% confluency. Virus-containing supernatant was collected at day 2 and day 4 after transfection. Viruses were concentrated by precipitating the virus supernatant by the polyethylene glycol 6000 (PEG6000) method. Viral titer was determined by infecting HEK293T cells.
Hemisection SCI and lentivirus injection
For hemisection SCI and lentiviral injections, mice were first anesthetized with 5% isoflurane inhalation for 3–4 min and then maintained at 2.5% isoflurane for the remainder of the surgery. Next, the skin was disinfected with betadine scrub and 70% ethanol wipes. Laminectomy was performed around T9–T10 to expose the spinal cord. Next, local anesthesia (0.125% Marcaine [bupivacaine hydrochloride]) was applied and dorsal blood vessels were burned using a cauterizer. Then, a lateral cut was made to the left side of the spinal cord and the cut ends at the midline of the spinal cord for hemisection SCI. Immediately after the injury, ~1–2 μL of virus (1 × 108 TU/mL) was injected approximately 1 mm rostral and caudal to the lesion epicenter (~0.5 mm in depth). Injection was performed using a 10-μL Hamilton syringe with a 32G needle mounted on a micromanipulator and moved as little as a micrometer at a time. The virus was injected at approximately 1 μL/min and the needle was left in place for 2–3 min to allow diffusion and prevent leakage or backflow. Injection volume has an error of ±0.1 μL. For the sham animals, the skin and muscle were cut to expose the spinal cord, but no injury was introduced. Muscles were sutured, and the skin was stapled back together. Immediately after surgery, 1 mg/kg meloxicam, a pain killer, and 50 mg/kg cefazoline, an antibiotic, were administered subcutaneously.
Animals were divided into the following three groups (6–12 mice/group): (1) sham mice (exposed the spine without injury, sham); (2) SCI mice with an injection of lenti-control-RFP (SCI+Ctrl); and (3) SCI mice with an injection of lenti-Gsx1-RFP (SCI+Gsx1).
Behavioral/locomotor assessment
Locomotion of each animal was evaluated based on the BMS from an open-field test.48 The BMS scale ranges from 0 (completely paralyzed) to 9 (normal). The BMS score assessment was given after 2–3 min of observation per animal by three independent observers who were blinded to the type of treatment. The BMS assessment was performed once before the surgery and then twice a week for up to 56 DPI.
Tissue processing
Spinal cord tissues at 3, 7, 14, 35, and 56 DPI were harvested after intracardial perfusion with 1× phosphate-buffered saline (PBS) followed by 4% (w/v) paraformaldehyde (PFA), and then microsurgically dissected and fixed overnight (18–24 h) in 4% PFA on a rotor. Fixed spinal cord tissues were washed three times with 1× PBS for 30 min and then placed in 30% (w/v) sucrose overnight until tissue sank to the bottom. Next, tissues were cryopreserved by embedding in Tissue-Tek optimum cutting temperature (OCT) and stored at −80°C until needed. Sagittal or cross-sections (12-μm thickness) were generated using a cryostat (Thermo Scientific).
Immunohistochemistry
Immunostaining was performed following a previously established protocol with minor modifications.27 Briefly, sections were treated with cold methanol for 10 min at room temperature for fixation and antigen retrieval. All antibodies were diluted in blocking solution containing 0.05% Triton X-100, 2% donkey serum, 3% bovine serum albumin (BSA), and PBS (1×) (pH 7.4). Sections were incubated with primary antibodies (Table S5) overnight at 4°C and washed three times for 10 min with PBS, and then incubated with secondary antibodies (Table S5) for 1 h at room temperature and washed three times for 10 min with PBS. For nuclear staining, 4′,6-diamidino-2-phenylindole (DAPI; 200 ng/mL) was added and then samples were washed three times with PBS and sealed with Cytoseal 60 (Thermo Fisher Scientific, 8310-4).
Imaging and image analysis
At least five sections from each slide/animal were analyzed. Images were captured at the same exposure and threshold, and at the same intensity per condition using a Zeiss LSM 800 confocal microscope or Zeiss AxioVision imager A1. The automatic cell counter in ImageJ64 was used to count the total number of cells. Co-labeled cells with cell type-specific markers and viral marker RFP were counted manually using ImageJ in separate RGB channels and with the following stereological considerations: (1) systematic and random sampling; (2) calculation of total cell numbers instead of signal densities; (3) counting of cells, not cell profiles; and (4) specific staining to clearly identify the cells of interest.
RNA extraction and quality control
Spinal cord tissues of 3, 14, and 35 DPI (n ≥ 3 for each time point) were isolated and segments containing injured/injected parenchymal segments (spanning ~2–3 mm from each side of the lesion) were rapidly snap-frozen in liquid nitrogen. Total RNA was isolated from spinal cord tissues using the RNeasy Lipid Tissue mini kit (QIAGEN, 74804) following the manufacturer’s protocol. The concentration of the total RNA was determined using a Qubit RNA broad-range (BR) assay kit (Life Technologies), and quality of the total RNA was determined using the RNA 6000 Nano chip on the 2100 Bioanalyzer automated electrophoresis system (Agilent Technologies).
Library preparation and RNA-seq
Library preparation and RNA-seq were performed by Admera Health (South Plainfield, NJ, USA). Total RNA was used for library preparation of each sample, which was subsequently bar-coded and prepared according to the manufacturer’s instructions (Illumina). The libraries were prepared using an Illumina MiSeq paired-end kit and sequenced as paired-end, 2 × 150-bp on the Illumina MiSeq. The sequencing run was performed according to the manufacturer’s instructions and generated a total of 40 million reads per sample.
RNA-seq data analysis and pathway analysis
After a quality check of the raw fastq files using FastQC,65 all sequences were aligned to the mouse reference genome, mm10, with STAR version 2.0.66 The raw read counts were generated using HTSeq (version 0.6.0).67 DESeq268,69 (an R/Bioconductor package) was used to normalize the counts and call differential gene expression on a counts matrix generated by HTSeq. Differentially expressed transcripts/genes between Gsx1 expression and control groups were defined by statistical significance (p value) and biological relevance (fold change). Downstream pathway analysis was carried out using IPA (QIAGEN, Redwood City, CA, USA; https://digitalinsights.qiagen.com/products/ingenuity-pathway-analysis). Pathway identification was performed using IPA, which is built on the manually curated content of the QIAGEN Knowledge Base to help scientists understand the biological context of expression analysis experiments. Differentially expressed genes and their expression changes (log2 fold change) were used as input. Box plots of gene expression were generated from count matrix from the HTSeq using START70 and the edgeR71 algorithm. Each dot on the box plot represents one biological sample.
Quantitative real-time PCR analysis
Complementary DNA (cDNA) was synthesized from total RNA using the SuperScript III first-strand synthesis system (Invitrogen, 18080051) following the manufacturer’s protocol. Quantitative real-time PCR analysis was performed with Power SYBR Green PCR master mix and gene-specific primers (Table S6) using a StepOnePlus real-time PCR system (Applied Biosystems). GAPDH was used as a reference housekeeping gene. The Levak method was used to calculate the fold change by normalizing it to the sham.
NE4C cell culture
NE4C cells (ATCC CRL-2925) were maintained in Eagle’s minimum essential medium (EMEM) with l-glutamine (ATCC 30-2003) supplemented with 10% FBS, 2 mM GlutaMAX (Gibco GlutaMAX 100×), and 1% penicillin-streptomycin (pen-strep) at 37°C with 5% CO2. For cell passage, subconfluent cultures were detached using TrypLE Express (GIBCO) diluted 4-fold with 1X PBS and transferred into poly-l-lysine (PLL)-coated dishes. For neural differentiation, NE-4C cells were cultured in EMEM 5% FBS with 2 mM GlutaMAX and 1% pen-strep. Retinoic acid (RA) was used to induce neuron formation. RA treatment (10−7 M) was added in media every other day starting 1 day after plating NE-4C cells and continuing until day 8. RA was then removed and NE4C culture was continued until day 14. Cell cultures were fixed with 4% PFA in PBS at room temperature for 15 min. Cells were then washed three times with PBS before adding blocking buffer (0.05% Triton X-100, 2% donkey serum, and 3% BSA in PBS) for 1 h at room temperature. All antibodies were diluted in PBS. Cells were incubated with primary antibodies overnight at 4°C and washed three times for 5 min with PBS. Cells were incubated with secondary antibodies for 1 h 30 min at room temperature followed by washing three times for 5 min with PBS. Cell nuclei were stained with DAPI (200 ng/mL) and mounted to coverslips with Vectashield Plus antifade mounting medium (Vector Laboratories, H-1900),
Statistical analysis
All data were analyzed using GraphPad Prism version 6.0. Statistical significance between two conditions was calculated by a Student’s t test, and multi-group comparison was performed using a one-way ANOVA, followed by a Tukey post hoc test. For BMS behavior analysis, a two-way repeated measures ANOVA was performed. Data are presented as mean ± standard error of the mean (SEM). A p value of less than 0.05 was considered statistically significant.
Data availability
The raw RNA-seq gene expression data described in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO: GSE171441.
Acknowledgments
We thank members of the Cai lab for constructive comments and discussion, Drs. Kelvin Kwan and Rick Cohen for advice for lentivirus production and cell culture experiments, and Catherine Cai for video editing. This work was supported by the State of New Jersey Commission on Spinal Cord Research grant 15IRG006 and Rutgers TechAdvance Fund (to L.C.); a U.S. Department of Education GAANN Precision and Personalized Medicine Pre-Doctoral Training Fellowship (to M.P.); NIH Biotechnology Pre-Doctoral Training Fellowship T32GM008339 (to M.P. and J.A.); and New Jersey Commission on Spinal CordResearch Graduate Fellowship CSCR12FEL001 (to Y.L.). K.-B.L acknowledges partial financial support from the NSF (CHE-1429062) and NIH (R01 1R01DC016612).
Author contributions
L.C. and Y.L.L. conceived the idea. M.P., Y.L.L., and L.C. designed the experiments. M.P., Y.L., J.A., S.C.-P., R.S., S.L., Z.F., B.R., and F.E. performed the experiments. K.-B.L. assisted with qPCR analysis. M.P., Y.L.L., and L.C. analyzed the data and wrote the manuscript.
Declaration of interests
L.C., M.P., and Y.L.L. are listed as co-inventors in patent applications related to this work. L.C. is a founder of NeuroNovus Therapeutics Inc. and a member of the scientific advisory board. The remaining authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.04.027.
Supplemental information
References
- 1.Tran A.P., Warren P.M., Silver J. The biology of regeneration failure and success after spinal cord injury. Physiol. Rev. 2018;98:881–917. doi: 10.1152/physrev.00017.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sofroniew M.V. Dissecting spinal cord regeneration. Nature. 2018;557:343–350. doi: 10.1038/s41586-018-0068-4. [DOI] [PubMed] [Google Scholar]
- 3.Lacroix S., Hamilton L.K., Vaugeois A., Beaudoin S., Breault-Dugas C., Pineau I., Lévesque S.A., Grégoire C.A., Fernandes K.J. Central canal ependymal cells proliferate extensively in response to traumatic spinal cord injury but not demyelinating lesions. PLoS ONE. 2014;9:e85916. doi: 10.1371/journal.pone.0085916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonough A., Martínez-Cerdeño V. Endogenous proliferation after spinal cord injury in animal models. Stem Cells Int. 2012;2012:387513. doi: 10.1155/2012/387513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meletis K., Barnabé-Heider F., Carlén M., Evergren E., Tomilin N., Shupliakov O., Frisén J. Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol. 2008;6:e182. doi: 10.1371/journal.pbio.0060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren Y., Ao Y., O’Shea T.M., Burda J.E., Bernstein A.M., Brumm A.J., Muthusamy N., Ghashghaei H.T., Carmichael S.T., Cheng L., Sofroniew M.V. Ependymal cell contribution to scar formation after spinal cord injury is minimal, local and dependent on direct ependymal injury. Sci. Rep. 2017;7:41122. doi: 10.1038/srep41122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paniagua-Torija B., Norenberg M., Arevalo-Martin A., Carballosa-Gautam M.M., Campos-Martin Y., Molina-Holgado E., Garcia-Ovejero D. Cells in the adult human spinal cord ependymal region do not proliferate after injury. J. Pathol. 2018;246:415–421. doi: 10.1002/path.5151. [DOI] [PubMed] [Google Scholar]
- 8.Becker C.G., Becker T., Hugnot J.P. The spinal ependymal zone as a source of endogenous repair cells across vertebrates. Prog. Neurobiol. 2018;170:67–80. doi: 10.1016/j.pneurobio.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Llorens-Bobadilla E., Chell J.M., Le Merre P., Wu Y., Zamboni M., Bergenstråhle J., Stenudd M., Sopova E., Lundeberg J., Shupliakov O. A latent lineage potential in resident neural stem cells enables spinal cord repair. Science. 2020;370:eabb8795. doi: 10.1126/science.abb8795. [DOI] [PubMed] [Google Scholar]
- 10.Barnabé-Heider F., Frisén J. Stem cells for spinal cord repair. Cell Stem Cell. 2008;3:16–24. doi: 10.1016/j.stem.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Sabelström H., Stenudd M., Frisén J. Neural stem cells in the adult spinal cord. Exp. Neurol. 2014;260:44–49. doi: 10.1016/j.expneurol.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 12.Assinck P., Duncan G.J., Hilton B.J., Plemel J.R., Tetzlaff W. Cell transplantation therapy for spinal cord injury. Nat. Neurosci. 2017;20:637–647. doi: 10.1038/nn.4541. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Chen G. In vivo reprogramming for CNS repair: Regenerating neurons from endogenous glial cells. Neuron. 2016;91:728–738. doi: 10.1016/j.neuron.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai W., Wu W., Wang L.L., Ni H., Chen C., Yang J., Zang T., Zou Y., Xu X.M., Zhang C.L. In vivo reprogramming of NG2 glia enables adult neurogenesis and functional recovery following spinal cord injury. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.02.009. Published online February 25, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silver J., Miller J.H. Regeneration beyond the glial scar. Nat. Rev. Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 16.Cafferty W.B., Yang S.H., Duffy P.J., Li S., Strittmatter S.M. Functional axonal regeneration through astrocytic scar genetically modified to digest chondroitin sulfate proteoglycans. J. Neurosci. 2007;27:2176–2185. doi: 10.1523/JNEUROSCI.5176-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dias D.O., Kim H., Holl D., Werne Solnestam B., Lundeberg J., Carlén M., Göritz C., Frisén J. Reducing pericyte-derived scarring promotes recovery after spinal cord injury. Cell. 2018;173:153–165.e22. doi: 10.1016/j.cell.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin J.M., Fackelmeier B., Clemett C.A., Fong D.M., Mouravlev A., Young D., O’Carroll S.J. Astrocyte-selective AAV-ADAMTS4 gene therapy combined with hindlimb rehabilitation promotes functional recovery after spinal cord injury. Exp. Neurol. 2020;327:113232. doi: 10.1016/j.expneurol.2020.113232. [DOI] [PubMed] [Google Scholar]
- 19.Courtine G., Song B., Roy R.R., Zhong H., Herrmann J.E., Ao Y., Qi J., Edgerton V.R., Sofroniew M.V. Recovery of supraspinal control of stepping via indirect propriospinal relay connections after spinal cord injury. Nat. Med. 2008;14:69–74. doi: 10.1038/nm1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossignol S., Frigon A. Recovery of locomotion after spinal cord injury: Some facts and mechanisms. Annu. Rev. Neurosci. 2011;34:413–440. doi: 10.1146/annurev-neuro-061010-113746. [DOI] [PubMed] [Google Scholar]
- 21.Chen B., Li Y., Yu B., Zhang Z., Brommer B., Williams P.R., Liu Y., Hegarty S.V., Zhou S., Zhu J. Reactivation of dormant relay pathways in injured spinal cord by KCC2 manipulations. Cell. 2018;174:521–535.e13. doi: 10.1016/j.cell.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S.K., Pfaff S.L. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat. Neurosci. 2001;4(Suppl):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X., Chen Y., Zhu Q., Huang H., Teng P., Zheng K., Hu X., Xie B., Zhang Z., Sander M., Qiu M. Control of astrocyte progenitor specification, migration and maturation by Nkx6.1 homeodomain transcription factor. PLoS ONE. 2014;9:e109171. doi: 10.1371/journal.pone.0109171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuguchi R., Kriks S., Cordes R., Gossler A., Ma Q., Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat. Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- 25.Chapman H., Riesenberg A., Ehrman L.A., Kohli V., Nardini D., Nakafuku M., Campbell K., Waclaw R.R. Gsx transcription factors control neuronal versus glial specification in ventricular zone progenitors of the mouse lateral ganglionic eminence. Dev. Biol. 2018;442:115–126. doi: 10.1016/j.ydbio.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei Z., Wang B., Chen G., Nagao M., Nakafuku M., Campbell K. Homeobox genes Gsx1 and Gsx2 differentially regulate telencephalic progenitor maturation. Proc. Natl. Acad. Sci. USA. 2011;108:1675–1680. doi: 10.1073/pnas.1008824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Tzatzalos E., Kwan K.Y., Grumet M., Cai L. Transcriptional regulation of Notch1 expression by Nkx6.1 in neural stem/progenitor cells during ventral spinal cord development. Sci. Rep. 2016;6:38665. doi: 10.1038/srep38665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzatzalos E., Smith S.M., Doh S.T., Hao H., Li Y., Wu A., Grumet M., Cai L. A cis-element in the Notch1 locus is involved in the regulation of gene expression in interneuron progenitors. Dev. Biol. 2012;372:217–228. doi: 10.1016/j.ydbio.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen K., Deng S., Lu H., Zheng Y., Yang G., Kim D., Cao Q., Wu J.Q. RNA-seq characterization of spinal cord injury transcriptome in acute/subacute phases: A resource for understanding the pathology at the systems level. PLoS ONE. 2013;8:e72567. doi: 10.1371/journal.pone.0072567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieuwenhuis B., Haenzi B., Hilton S., Carnicer-Lombarte A., Hobo B., Verhaagen J., Fawcett J.W. Optimization of adeno-associated viral vector-mediated transduction of the corticospinal tract: comparison of four promoters. Gene Ther. Gene Ther. 2021;28:56–74. doi: 10.1038/s41434-020-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrow K.M., Perez-Campo F.M., Ward C.M. Use of the cytomegalovirus promoter for transient and stable transgene expression in mouse embryonic stem cells. Methods Mol. Biol. 2006;329:283–294. doi: 10.1385/1-59745-037-5:283. [DOI] [PubMed] [Google Scholar]
- 32.Zhao C., Deng W., Gage F.H. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 33.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S., Chen Z. Employing endogenous NSCs to promote recovery of spinal cord injury. Stem Cells Int. 2019;2019:1958631. doi: 10.1155/2019/1958631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imayoshi I., Sakamoto M., Yamaguchi M., Mori K., Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chavali M., Klingener M., Kokkosis A.G., Garkun Y., Felong S., Maffei A., Aguirre A. Non-canonical Wnt signaling regulates neural stem cell quiescence during homeostasis and after demyelination. Nat. Commun. 2018;9:36. doi: 10.1038/s41467-017-02440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bengoa-Vergniory N., Kypta R.M. Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell. Mol. Life Sci. 2015;72:4157–4172. doi: 10.1007/s00018-015-2028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wexler E.M., Paucer A., Kornblum H.I., Palmer T.D., Geschwind D.H. Endogenous Wnt signaling maintains neural progenitor cell potency. Stem Cells. 2009;27:1130–1141. doi: 10.1002/stem.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izon D.J., Aster J.C., He Y., Weng A., Karnell F.G., Patriub V., Xu L., Bakkour S., Rodriguez C., Allman D., Pear W.S. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16:231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 40.Pirot P., van Grunsven L.A., Marine J.C., Huylebroeck D., Bellefroid E.J. Direct regulation of the Nrarp gene promoter by the Notch signaling pathway. Biochem. Biophys. Res. Commun. 2004;322:526–534. doi: 10.1016/j.bbrc.2004.07.157. [DOI] [PubMed] [Google Scholar]
- 41.Hara M., Kobayakawa K., Ohkawa Y., Kumamaru H., Yokota K., Saito T., Kijima K., Yoshizaki S., Harimaya K., Nakashima Y., Okada S. Interaction of reactive astrocytes with type I collagen induces astrocytic scar formation through the integrin-N-cadherin pathway after spinal cord injury. Nat. Med. 2017;23:818–828. doi: 10.1038/nm.4354. [DOI] [PubMed] [Google Scholar]
- 42.Demeter K., Herberth B., Duda E., Domonkos A., Jaffredo T., Herman J.P., Madarász E. Fate of cloned embryonic neuroectodermal cells implanted into the adult, newborn and embryonic forebrain. Exp. Neurol. 2004;188:254–267. doi: 10.1016/j.expneurol.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 43.Schlett K., Madarász E. Retinoic acid induced neural differentiation in a neuroectodermal cell line immortalized by p53 deficiency. J. Neurosci. Res. 1997;47:405–415. [PubMed] [Google Scholar]
- 44.Varga B.V., Hádinger N., Gócza E., Dulberg V., Demeter K., Madarász E., Herberth B. Generation of diverse neuronal subtypes in cloned populations of stem-like cells. BMC Dev. Biol. 2008;8:89. doi: 10.1186/1471-213X-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrin F.E., Noristani H.N. Serotonergic mechanisms in spinal cord injury. Exp. Neurol. 2019;318:174–191. doi: 10.1016/j.expneurol.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Faden A.I., Gannon A., Basbaum A.I. Use of serotonin immunocytochemistry as a marker of injury severity after experimental spinal trauma in rats. Brain Res. 1988;450:94–100. doi: 10.1016/0006-8993(88)91548-x. [DOI] [PubMed] [Google Scholar]
- 47.Camand E., Morel M.P., Faissner A., Sotelo C., Dusart I. Long-term changes in the molecular composition of the glial scar and progressive increase of serotoninergic fibre sprouting after hemisection of the mouse spinal cord. Eur. J. Neurosci. 2004;20:1161–1176. doi: 10.1111/j.1460-9568.2004.03558.x. [DOI] [PubMed] [Google Scholar]
- 48.Basso D.M., Fisher L.C., Anderson A.J., Jakeman L.B., McTigue D.M., Popovich P.G. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J. Neurotrauma. 2006;23:635–659. doi: 10.1089/neu.2006.23.635. [DOI] [PubMed] [Google Scholar]
- 49.Chen W., Zhang B., Xu S., Lin R., Wang W. Lentivirus carrying the NeuroD1 gene promotes the conversion from glial cells into neurons in a spinal cord injury model. Brain Res. Bull. 2017;135:143–148. doi: 10.1016/j.brainresbull.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Su Z., Niu W., Liu M.L., Zou Y., Zhang C.L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang X., Wang C., Zhou X., Wang J., Xia K., Yang B., Gong Z., Ying L., Yu C., Shi K. Overexpression of the transcription factors OCT4 and KLF4 improves motor function after spinal cord injury. CNS Neurosci. Ther. 2020;26:940–951. doi: 10.1111/cns.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bannatyne B.A., Hao Z.Z., Dyer G.M.C., Watanabe M., Maxwell D.J., Berkowitz A. Neurotransmitters and motoneuron contacts of multifunctional and behaviorally specialized turtle spinal cord interneurons. J. Neurosci. 2020;40:2680–2694. doi: 10.1523/JNEUROSCI.2200-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paixão S., Loschek L., Gaitanos L., Alcalà Morales P., Goulding M., Klein R. Identification of spinal neurons contributing to the dorsal column projection mediating fine touch and corrective motor movements. Neuron. 2019;104:749–764.e6. doi: 10.1016/j.neuron.2019.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noristani H.N., Sabourin J.C., Boukhaddaoui H., Chan-Seng E., Gerber Y.N., Perrin F.E. Spinal cord injury induces astroglial conversion towards neuronal lineage. Mol. Neurodegener. 2016;11:68. doi: 10.1186/s13024-016-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson J., Patel M., Forenzo D., Ai X., Cai C., Wade Q., Risman R., Cai L. A novel mouse model for the study of endogenous neural stem and progenitor cells after traumatic brain injury. Exp. Neurol. 2020;325:113119. doi: 10.1016/j.expneurol.2019.113119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang B., Waclaw R.R., Allen Z.J., 2nd, Guillemot F., Campbell K. Ascl1 is a required downstream effector of Gsx gene function in the embryonic mouse telencephalon. Neural Dev. 2009;4:5. doi: 10.1186/1749-8104-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bovolenta P., Wandosell F., Nieto-Sampedro M. CNS glial scar tissue: a source of molecules which inhibit central neurite outgrowth. Prog. Brain Res. 1992;94:367–379. doi: 10.1016/s0079-6123(08)61765-3. [DOI] [PubMed] [Google Scholar]
- 58.Rolls A., Shechter R., Schwartz M. The bright side of the glial scar in CNS repair. Nat. Rev. Neurosci. 2009;10:235–241. doi: 10.1038/nrn2591. [DOI] [PubMed] [Google Scholar]
- 59.Göritz C., Dias D.O., Tomilin N., Barbacid M., Shupliakov O., Frisén J. A pericyte origin of spinal cord scar tissue. Science. 2011;333:238–242. doi: 10.1126/science.1203165. [DOI] [PubMed] [Google Scholar]
- 60.Silver J., Schwab M.E., Popovich P.G. Central nervous system regenerative failure: Role of oligodendrocytes, astrocytes, and microglia. Cold Spring Harb. Perspect. Biol. 2014;7:a020602. doi: 10.1101/cshperspect.a020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgenstern D.A., Asher R.A., Fawcett J.W. Chondroitin sulphate proteoglycans in the CNS injury response. Prog. Brain Res. 2002;137:313–332. doi: 10.1016/s0079-6123(02)37024-9. [DOI] [PubMed] [Google Scholar]
- 62.Faulkner J.R., Herrmann J.E., Woo M.J., Tansey K.E., Doan N.B., Sofroniew M.V. Reactive astrocytes protect tissue and preserve function after spinal cord injury. J. Neurosci. 2004;24:2143–2155. doi: 10.1523/JNEUROSCI.3547-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benner E.J., Luciano D., Jo R., Abdi K., Paez-Gonzalez P., Sheng H., Warner D.S., Liu C., Eroglu C., Kuo C.T. Protective astrogenesis from the SVZ niche after injury is controlled by Notch modulator Thbs4. Nature. 2013;497:369–373. doi: 10.1038/nature12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data.https://qubeshub.org/resources/fastqc [Google Scholar]
- 66.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nelson J.W., Sklenar J., Barnes A.P., Minnier J. The START App: A web-based RNAseq analysis and visualization resource. Bioinformatics. 2017;33:447–449. doi: 10.1093/bioinformatics/btw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq gene expression data described in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO: GSE171441.