Abstract
The mitogen-activated protein kinase pathway is one of the most frequently altered pathways in cancer. It is involved in the control of cell proliferation, invasion, and metabolism, and can cause resistance to therapy. A number of aggressive malignancies, including melanoma, colon cancer, and glioma, are driven by a constitutively activating missense mutation (V600E) in the v-Raf murine sarcoma viral oncogene homolog B (BRAF) component of the pathway. Mitogen-activated protein kinase kinase (MEK) inhibition is initially effective in targeting these cancers, but reflexive activation of mammalian target of rapamycin (mTOR) signaling contributes to frequent therapy resistance. We have previously demonstrated that combination treatment with the MEK inhibitor trametinib and the dual mammalian target of rapamycin complex 1/2 inhibitor TAK228 improves survival and decreases vascularization in a BRAFV600E mutant glioma model. To elucidate the mechanism of action of this combination therapy and understand the ensuing tumor response, we performed comprehensive unbiased proteomic and phosphoproteomic characterization of BRAFV600E mutant glioma xenografts after short-course treatment with trametinib and TAK228. We identified 13,313 proteins and 30,928 localized phosphosites, of which 12,526 proteins and 17,444 phosphosites were quantified across all samples (data available via ProteomeXchange; identifier PXD022329). We identified distinct response signatures for each monotherapy and combination therapy and validated that combination treatment inhibited activation of the mitogen-activated protein kinase and mTOR pathways. Combination therapy also increased apoptotic signaling, suppressed angiogenesis signaling, and broadly suppressed the activity of the cyclin-dependent kinases. In response to combination therapy, both epidermal growth factor receptor and class 1 histone deacetylase proteins were activated. This study reports a detailed (phospho)proteomic analysis of the response of BRAFV600E mutant glioma to combined MEK and mTOR pathway inhibition and identifies new targets for the development of rational combination therapies for BRAF-driven tumors.
Keywords: glioma, trametinib, TAK228, sapanisertib, tandem MS, phosphoproteome, mouse models, apoptosis, angiogenesis, signal transduction, drug targets
Abbreviations: ACN, acetonitrile; BRAF, v-Raf murine sarcoma viral oncogene homolog B; CDK, cyclin-dependent kinase; EGFR, epidermal growth factor receptor; ERF, erythroblast transformation-specific 2 repressor factor; ESI, electrospray ionization; FAK, focal adhesion kinase; FDA, Food and Drug Administration; FDR, false discovery rate; HDAC, histone deacetylase; IMAC, immobilized metal affinity chromatography; KSEA, kinase–substrate enrichment analysis; MAPK, mitogen-activated protein kinase; MEK, mitogen-activated protein kinase kinase; mTOR, mammalian target of rapamycin; mTORC1/2, mammalian target of rapamycin complex 1/2; PCA, principal component analysis; PSM, peptide-to-spectrum match; PTK2, protein tyrosine kinase 2; RT, room temperature; TMT, tandem mass tag; VEGF, vascular endothelial growth factor
Graphical Abstract

Highlights
-
•
Large-scale proteomic and phosphoproteomic analysis of an in vivo BRAFV600E glioma model.
-
•
In-depth analysis of tumor and stromal cellular response to protein kinase inhibitors.
-
•
Insights into acquired resistance and alternative therapeutic approaches.
In Brief
BRAFV600E is a key oncogenic driver in glioma, melanoma, and colon cancer. These tumors escape mitogen-activated protein kinase pathway inhibition by upregulating mammalian target of rapamycin signaling. Using comprehensive unbiased proteomic and phosphoproteomic analysis of an in vivo BRAFV600E mutant glioma model treated with inhibitors of both these key pathways, we characterize the tumor and stromal response and suggest additional therapeutic targets for BRAF-driven cancers, including epidermal growth factor receptor and class 1 histone deacetylases.
Mutations in the mitogen-activated protein kinase (MAPK) pathway are among the most common in cancer (1). Components of the pathway, including the tumor suppressor neurofibromin 1, and the oncogenes RAS and RAF, regulate cell proliferation, invasion, metabolism, and resistance to therapy. In particular, constitutively activating mutation of the v-Raf murine sarcoma viral oncogene homolog B (BRAF) gene through missense mutation (V600E) is a key driver event in melanoma, thyroid cancer, lung cancer, colon cancer, and glioma (2, 3, 4, 5, 6, 7). The BRAFV600E mutation is commonly treated with inhibitors of the MAPK pathway, such as the mitogen-activated protein kinase kinase (MEK) inhibitor trametinib (7, 8, 9, 10). Activation of BRAF also signals to the mammalian target of rapamycin (mTOR) pathway, and many tumors that initially respond to MEK inhibition develop reliance on the mTOR pathway as an escape mechanism (11, 12). To prevent such an escape, we previously tested the combination of trametinib along with an mTOR inhibitor TAK228 (also known as sapanisertib) in MEK-activated glioma (13). TAK228 is a dual mammalian target of rapamycin complex 1/2 (mTORC1/2) kinase inhibitor that blocks both mTORC1 and mTORC2 activation, thereby eliminating most aspects of the mTOR pathway (14, 15, 16, 17). Trametinib is in clinical trials for MEK-activated glioma (NCT02124772). TAK228 penetrates the brain and inhibits the activation of mTORC1/2 in preclinical models of aggressive brain tumors (17). TAK228 is in early phase clinical trials for glioma (NCT02133183). Both MEK and mTOR can signal to promote cell growth, survival, alter cellular metabolism, and prevent apoptosis (18, 19). We therefore hypothesized that trametinib and TAK228 would combine to inhibit MEK-activated glioma cell growth (13). Trametinib and TAK228 synergized in MEK-activated glioma both in vitro and in vivo. Combination therapy decreased cell proliferation, induced apoptosis, and decreased tumor vascularity, suggesting multiple potential mechanisms of action. We found that the combination extended survival of mice bearing BRAFV600E mutant glioma xenografts more than monotherapy. To better understand how TAK228 and trametinib act as monotherapy and in combination, we treated established BRAFV600E mutant glioma tumors with a short course of each inhibitor alone and in combination and performed comprehensive proteomics and phosphoproteomics.
Experimental Procedures
Chemicals
Urea, sodium chloride, EDTA, bovine aprotinin, leupeptin (Roche), PMSF, sodium fluoride, Phosphatase Inhibitor Cocktail 2, and Phosphatase Inhibitor Cocktail 3, O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate, iodoacetamide, and ammonium formate were purchased from Sigma–Aldrich. Tris–HCl, 1,4-DTT, and all MS-grade solvents, including water, formic acid, trifluoroacetic acid, and acetonitrile (ACN), were purchased from Thermo Fisher Scientific. Lysyl endopeptidase (Lys-C) was purchased from Wako Chemicals, and trypsin protease was purchased from Promega.
Sample Generation and Preparation
The in vivo experiment utilized the patient-derived BT40 xenograft line, which was previously characterized as a pediatric glioma containing the BRAFV600E mutation (20, 21). BT40 only grows as serially passaged flank tumors in immunocompromised mice and will not form tumors intracranially or grow in cell culture. 1 × 106 BT40 tumor cells, in 50% Dulbecco's modified Eagle's medium/F12 and 50% Matrigel (Corning), were injected into the flanks of 4- to -6-week-old female NU/NU mice (Charles River). This animal experiment was approved by The Johns Hopkins University Institutional Animal Care and Use Committee. Treatment started after the flank tumors were visible and measurable. Mice were randomized into four groups of three mice with 1 to 2 flank tumors per mouse. Mice were treated by oral gavage with the MEK inhibitor trametinib (1.5 mg/kg per day for 5 days, Monday through Friday), the dual mTORC1/2 inhibitor TAK228 (1 mg/kg once on Friday), or the combination of these drugs. We administered the MEK inhibitor trametinib for 5 days because of the long half-life of this drug and the need to achieve steady state. In our prior work, we demonstrated robust inhibition of the MAPK pathway at this dosing schedule in vivo (13). We administered TAK228, which has a short half-life, as a single dose and harvested tumors 4 h later, which is when we find maximal inhibition of mTOR targets in vivo (13, 14, 17, 22). Control animals received vehicle solution accordingly. The mice were sacrificed 4 h after the last dose of drug, and the tumors were removed and flash frozen in liquid nitrogen. The five tumors from each of the aforementioned treatment condition were subsequently processed for proteomic analysis. Tissue lysis and sample preparation for global proteomic and phosphoproteomic characterization were performed as previously described (23, 24). Cryopulverized tissue was resuspended in lysis buffer (8 M urea, 75 mM sodium chloride, 50 mM Tris–HCl, pH 8.0, 1 mM EDTA, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF, 10 mM sodium fluoride, Phosphatase Inhibitor Cocktail 2 and Phosphatase Inhibitor Cocktail 3 [1:100 dilution], and 20 μM O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate). Lysates were clarified by centrifugation at 20,000g for 10 min at 4 °C. For each sample, 1 mg of protein lysate was subjected to reduction with 5 mM 1,4-DTT for 30 min at room temperature (RT), followed by alkylation with 10 mM iodoacetamide for 45 min at RT in the dark. Urea concentration was reduced <2 M using 50 mM Tris–HCl, pH 8.0. Samples were subjected to tandem digestion of Lys-C at a ratio of enzyme-to-substrate 1:50 for 2 h at RT followed by trypsin at a ratio of enzyme-to-substrate 1:50 overnight at RT. The generated peptides were acidified to a final concentration of 1% formic acid, subjected to cleanup using C-18 SepPak columns, and then dried. Peptides were resuspended in 50 mM Hepes, pH 8.5 buffer, and measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). For tandem mass tag (TMT) labeling, samples were assigned to a specific isobaric channel, and 300 μg of peptides from each individual sample was labeled with 10-plex TMT reagents (Thermo Fisher Scientific) following manufacturer's instructions. A pooled reference comprised of equal amounts of peptide material from each of the individual samples was generated, 300 μg of which was included in the individual TMT plexes. Following TMT labeling and reaction quenching, the individual samples were pooled and subjected to cleanup using C-18 SepPak columns and then dried.
Peptide Fractionation by Basic Reversed-phase Liquid Chromatography
The desalted TMT-labeled samples were reconstituted in a volume of 20 mM ammonium formate (pH 10) and 2% ACN and subjected to basic reversed-phase chromatography using 1220 LC System (Agilent) with solvent A (2% ACN, 5 mM ammonium formate, and pH 10) and a nonlinear gradient of solvent B (90% ACN, 5 mM ammonium formate, and pH 10) at 1 ml/min as follows: 0% solvent B (9 min), 6% solvent B (4 min), 6% to 28.5% solvent B (50 min), 28% to 34% solvent B (5.5 min), 34% to 60% solvent B (13 min), and then held at 60% solvent B for 8.5 min on a Agilent 4.6 mm × 250 mm RP Zorbax 300 A Extend-C18 column with 3.5 μm size beads (Agilent). Fractions were collected using an Analyst-FC fraction collector (Agilent) and concatenated as previously described (23, 24). In brief, fraction collection began 2 min after sample injection, with the first 12 fractions pooled and representative of the column flowthrough. The remaining collected fractions were concatenated into 24 fractions by combining four fractions that are 24 fractions apart (i.e., combining fractions 1, 25, 49, and 73; 2, 26, 50, and 74; and so on), resulting in a total 25 fractions. About 5% of each of the fractions was aliquoted for global proteomic analysis, dried down, and resuspended in 3% ACN, 0.1% formic acid prior to electrospray ionization (ESI)–LC–MS/MS analysis.
Enrichment of Phosphopeptides by Fe–Immobilized Metal Affinity Chromatography
The remaining 95% of the sample was utilized for phosphopeptide enrichment and was further concatenated, combining fraction that are 12 fractions apart (i.e., combining fractions 1, 13, and 2, 14; and so on), with the flow-through fraction excluded from concatenation. A total of 13 fractions were subjected to phosphopeptide enrichment using immobilized metal affinity chromatography (IMAC) as previously described (25). Nickel–nitrilotriacetic acid agarose beads (Qiagen) were utilized to prepare Fe3+–nitrilotriacetic acid agarose beads, and then peptides from the individual fractions reconstituted in 80% ACN/0.1% trifluoroacetic acid were incubated with 10 μl of the Fe3+–IMAC beads for 30 min. Samples were then spun down, and the supernatant containing unbound peptides was removed. The beads were washed twice and then loaded onto equilibrated C-18 Stage Tips with 80% can and 0.1% trifluoroacetic acid. Tips were rinsed twice with 1% formic acid, followed by sample elution off the Fe3+–IMAC beads and onto the C-18 State Tips with 70 μl of 500 mM dibasic potassium phosphate, pH 7.0 three times. C-18 Stage Tips were washed twice with 1% formic acid, followed by elution of the phosphopeptides from the C-18 Stage Tips with 50% ACN, 0.1% formic acid twice. Samples were dried down and resuspended in 3% ACN and 0.1% formic acid prior to ESI–LC–MS/MS analysis.
Global Proteome and Phosphoproteome ESI–LC–MS/MS Data Acquisition
Global proteome and phosphoproteome fractions were analyzed using the same instrumentation and methods. About ~1 μg of peptide was separated using Easy nLC 1200 UHPLC system (Thermo Fisher Scientific) on an in-house packed 20 cm × 75 μm diameter C18 column (1.9 μm Reprosil-Pur C18-AQ beads [Dr Maisch GmbH]; Picofrit 10 μm opening [New Objective]). The column was heated to 50 °C using a column heater (Phoenix-ST). The flow rate was 0.300 μl/min with 0.1% formic acid and 2% ACN in water (A) and 0.1% formic acid, 90% ACN (B). The peptides were separated with a 6 to 30% B gradient in 84 min and analyzed using the Thermo Fusion Lumos mass spectrometer (Thermo Fisher Scientific). The following were the parameters: MS1: resolution—60,000; mass range—350 to 1800 m/z; radiofrequency lens—30%, automatic gain control target—4.0e5; maximum injection time—50 ms; charge state include —2 to 6; dynamic exclusion—45 s; top 20 ions selected for MS2; MS2: resolution—50,000; scan range mode was set to auto normal, with first mass −110; high-energy collision dissociation activation energy—37; isolation width (m/z)—0.7, automatic gain control target—2.0e5; and maximum injection time—105 ms.
Data Processing
All LC–MS/MS files were analyzed by MS-PyCloud, a cloud-based proteomic pipeline developed in Johns Hopkins University to perform database search for spectrum assignments (26), using MS-GF+ in this study against a combined human and mouse RefSeq database (version 20160914; 37,405 entries) (27, 28). A decoy database was used to assess the false discovery rate (FDR) at peptide-to-spectrum match (PSM), peptide, and protein levels (29). Peptides were searched with two tryptic ends, allowing up to two missed cleavages. Search parameters included 20 ppm precursor tolerance and 0.06 Da fragment ion tolerance, static modification of carbamidomethylation at cysteine (+57.02146), TMT-label modification of N terminus and lysine (+229.16293) and variable modifications of oxidation at methionine (+15.99491) and phosphorylation at serine, threonine, and tyrosine (+79.96633). Filters used for global data analysis included one PSM per peptide and two peptides per protein, with a 1% FDR threshold at the protein level. Of note, modified and nonmodified versions of the same peptide species are not considered as distinct entries for our two peptides per protein criterion. Filters used for phosphoproteome data included one PSM per peptide and one peptide per protein, with a 1% FDR threshold at the peptide level. For peptide and protein quantitation, first PSMs are sorted by score (−log10[MS-GF+ SpectralEValue]) for each charge state, and then a threshold score at the specified PSM-level FDR is determined. All PSMs with a score above the threshold are retained and used for quantification and adjusted based on TMT reporter tag lot correction factors. Individual PSM intensities are summed, and the data are rolled up to peptide/protein/gene level. Data normalization entailed median normalization relative to the reference channel for each set at the PSM level, followed by log2 of the ratio relative to the reference channel. Data are then rolled up to the peptide/protein/gene level by taking the median at each step. An additional median step was included for this dataset that involved calculation of the median value for each sample/TMT channel, followed by determination of the median of the TMT channel median. The phosphosites were localized using the LuciPHOr2 software package (http://luciphor2.sourceforge.net) (30); the results were filtered to keep all localized PSMs with an estimated false localization rate of ≤10%. Phosphosites were normalized to the abundance of the respective protein (log2[abundance] phosphosite − log2[abundance] protein); for proteins with multiple phosphosites, each phosphosite was treated independently.
Protein and Phosphosite Species Assignment
For the proteome dataset, if all contributing PSMs were mouse, then the protein was considered mouse (stroma). This is also true for the phosphoproteome dataset, that is, if all contributing PSMs were mouse, then the phosphosite was considered mouse. Otherwise, if the contributing PSMs were either human or shared between human and mouse, then the protein or phosphosite was considered human (tumor) (31).
Druggable Protein Analysis
We compiled a list of druggable/targetable proteins using the Drug Gene Interaction Database, which we queried for Food and Drug Administration (FDA)–approved and non–FDA-approved inhibitors (32). The final list used for the analysis included 221 druggable proteins.
Experimental Design and Statistical Rationale
A total of five biological replicates for each experimental condition (vehicle, n = 5; trametinib, n = 5; TAK228, n = 5; and trametinib–TAK228 combination, n = 5) were investigated, based on the assumption that these genetically homogeneous tumors and mice would exhibit generally concordant responses to the same inhibitors administered in the same fashion to each mouse. No technical replicates were required. In our previous work on BT40 glioma treated with the same drug combination, we were able to see statistically significant differences in tumor volume (after 12 days of treatment) with a total sample size of 20 across our four experimental conditions (13). Based on that previous result, we felt confident we would see statistically significant changes in the global proteome and phosphoproteome after a shorter course (5 days) of treatment. In addition, a sample size of 20 tumors across four groups produces an “E” value of 16, which is adequate for statistically significant results (33). The control (vehicle) group served as a comparison reference for the other experimental conditions. Initial analysis of global proteomics and phosphoproteomics datasets in order to generate volcano plots and mean difference calculations was performed using Perseus (https://maxquant.net/perseus/) (34). To generate volcano plots for the human and mouse proteomes and phosphoproteomes within Perseus, an FDR of <0.01 was used, and the S0 parameter was chosen to target a mean difference of ~0.5 or greater (which is approximately equivalent to a fold change of 1.5). Correlation analysis (using Pearson correlation coefficient) was performed using Morpheus from the Broad Institute (https://software.broadinstitute.org/morpheus). Protein pathway analyses were performed using the Panther (35, 36) database. For all analyses, unless otherwise specified, an FDR <0.05 was used as the confidence threshold. Principal component analysis (PCA) was performed using the PCA function from the Scikit-learn Python package, which used a singular value decomposition to project the data to two-dimensional space (37). Volcano plots (of protein pathway data) were generated using Excel (Microsoft). Kinase activity was inferred from the (phospho)proteomic data using kinase–substrate enrichment analysis (KSEA) (38); for KSEA results, an FDR <0.05 was used as the confidence threshold. KSEA scores each kinase based on the relative hyperphosphorylation or dephosphorylation of the majority of its substrates, as identified from phosphosite-specific kinase–substrate databases; therefore, the z-score of a kinase is based on the collective phosphorylation status of it substrates. The negative or positive value of the z-score implies a decrease or increase in the kinase's overall activity relative to the control.
Results
Unbiased MS Proteomics and Phosphoproteomics Data Separate Tumors into Groups Based on Treatment With MEK Inhibitor, mTOR Inhibitor, or Combination Therapy
We used high-resolution MS to characterize changes in the proteome and phosphoproteome in a BRAFV600E mutant glioma xenograft model (BT40). Mice were randomized into four groups of three mice with 1 to 2 flank tumors per mouse (for a total number of 20 tumors, with five tumors per treatment group). These BT40 tumor-bearing mice were treated with the MEK inhibitor trametinib alone, the dual mTORC1/2 kinase inhibitor TAK228 alone, or a combination of trametinib and TAK228 for a short time course as described previously; control mice received vehicle solution. Five tumors per treatment condition were snap frozen upon removal and subsequently homogenized via cryopulverization and then processed for proteomic and phosphoproteomic analysis as previously described (23, 24). Proteomics and phosphoproteomics analyses identified a total of 13,313 proteins and 30,928 localized phosphosites, respectively, of which 12,526 proteins and 17,444 phosphosites were quantified across all samples; after normalizing localized phosphosites on the abundance of the respective proteins, there were 10,767 normalized localized human phosphosites and 3912 normalized localized mouse phosphosites (Fig. 1A and supplemental Tables S1–S7). PCA demonstrated that the four treatment groups are, overall, well separated and distinct from one another with regard to the human (tumor) proteome and phosphoproteome, as well as the mouse (stroma) proteome and phosphoproteome (Fig. 1, B and C and supplemental Fig. S1, A and B); this result was confirmed by Pearson correlation (supplemental Fig. S1, C–F). We note that one of the TAK228 samples (TAK228-3) separated from the other TAK228 samples and grouped with the vehicle samples. A potential reason for this separation is that the TAK228 treatment was a single oral gavage dose 4 h prior to sacrificing the animals; it is possible that the mouse did not receive the full dose (e.g., if there was an air bubble in the gavage syringe or if the mouse regurgitated some of the drug). Regardless, this sample was included in all the analyses given that mTOR signaling was appropriately inhibited in this sample, as evidenced by decreased p4E-BP1 (Fig. 2A). In order to determine statistically significantly altered proteins and phosphosites for subsequent analysis, volcano plots were generated (as described) for each treatment condition compared with vehicle for both the proteome and phosphoproteome and for both human and mouse (Fig. 1, D and E, supplemental Figs. S2 and S3, and supplemental Tables S8–S11).
Fig. 1.
Global proteomics and phosphoproteomics separated tumors into groups based on treatment with trametinib monotherapy, TAK228 monotherapy, or combination therapy.A, proteomic and phosphoproteomic metrics. Thirteen thousand three hundred thirteen proteins were identified, of which 12,526 were quantified across all 20 samples. Thirty thousand nine hundred twenty-eight localized phosphosites were identified, of which 17,444 were quantified across all 20 samples. If all contributing PSMs were mouse, then the protein or phosphosite was considered mouse (stroma). If contributing PSMs were either distinctly human or human and mouse, then the protein or phosphosite was considered human (tumor). For phosphosites, in parentheses are listed the total number of phosphosites (10,767 human and 3912 mouse) that could be normalized to their respective protein. Only normalized phosphosites were used for all subsequent analysis. Principal component analysis of human proteome (B) and phosphoproteome (C) shows distinct grouping of the four treatment groups (vehicle, trametinib, TAK228, and combination), with the exception that one TAK228 sample more closely grouped with the vehicle samples. This TAK228 sample was included in all analyses given that the mTOR signaling pathway was appropriately inhibited in this sample (Fig. 2A). Volcano plots of human proteome (D) and human phosphoproteome (E) of mean difference (combination—vehicle) versus −log(p). For these analyses, FDR cutoff was <0.01. FDR, false discovery rate; mTOR, mammalian target of rapamycin; PSM, peptide-to-spectrum match.
Fig. 2.
Trametinib–TAK228 combination therapy inhibited both the MAPK and mTOR pathways.A, heat map of log2(abundance) of MAPK and mTOR pathway phosphorylation sites. All sites included were statistically significantly decreased in combination as compared with vehicle based on prior volcano plot (Fig. 1E and supplemental Table S9). Diagram of MAPK and mTOR signaling pathways (B) with proteins whose phosphorylations are statistically significantly reduced in combination as compared with vehicle shaded blue. MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin.
Phosphoproteomic Analysis Demonstrates Broad MAPK and mTOR Pathway Inhibition After Combined Trametinib–TAK228 Therapy
Phosphoproteomic analysis confirmed statistically significant inhibition of a panel of MAPK signaling pathway targets by trametinib–TAK228 combination therapy (Fig. 2, A and B and supplemental Fig. S4A). Trametinib monotherapy suppressed BRAF-T401, MAPK1-Y187, and STMN1-S38 (supplemental Fig. S4A), whereas trametinib–TAK228 combination therapy in addition inhibited MAPK3-Y204, ARAF-S186, and other downstream MAPK pathway targets in addition to STMN-S38, including RP6SKA1-S26, and two phosphorylation sites on erythroblast transformation-specific domain–containing transcription factor erythroblast transformation-specific 2 repressor factor (ERF), ERF-S21 and ERF-T526. Of note, stathmin phosphorylation at S38 is an important regulator of synaptogenesis and has been characterized as a key regulator of gliomagenesis (39). Phosphoproteomic analysis further confirmed inhibition of the mTOR signaling pathway after TAK228 monotherapy and trametinib–TAK228 combination therapy (Fig. 2, A and B and supplemental Fig. S4B). TAK228 alone inhibited MTOR-S2481, 4E-BP1-T37T46, 4E-BP1-S65, 4E-BP1-T70, RICTOR-S1591, and AKT1S1-S183, and trametinib–TAK228 combination therapy in addition inhibited MTOR-S1261 and RICTOR-S1577. Consistent with the known compensatory activation of the mTOR pathway after MEK inhibition in BRAF-mutant tumors (11, 12), trametinib alone caused statistically significant increases in AKT1S1-S183, RICTOR-S1577, EIF4EBP1-T37T46, EIF4EBP1-S65, and EIF4EBP1-T70 (Fig. 2A and supplemental Fig. S4B).
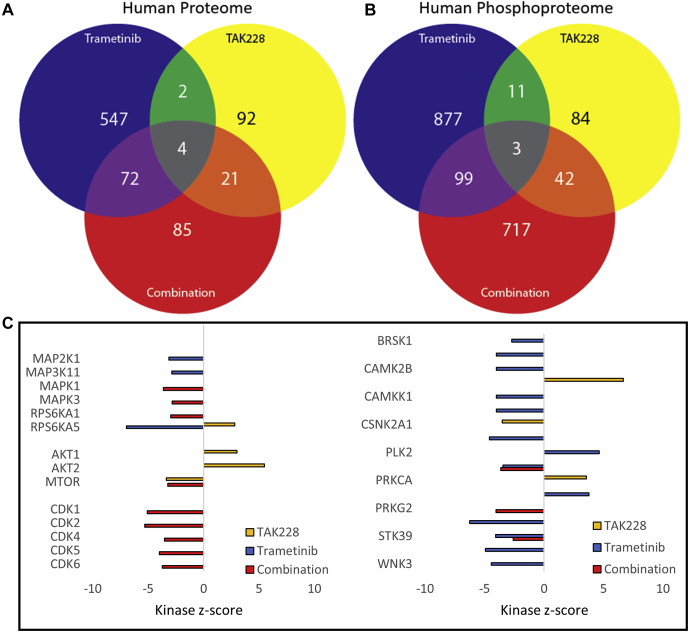
Global Proteomic–Phosphoproteomic Analysis Identifies Distinct Response Signatures Associated With Trametinib, TAK228, or Combination Therapy
A global analysis of the human proteome and phosphoproteome after trametinib monotherapy, TAK228 monotherapy, or trametinib–TAK228 combination therapy compared with vehicle revealed distinct treatment signatures, with statistically significant changes in the abundance of proteins and normalized localized phosphosites that are unique to each treatment condition and that are shared by two treatment conditions (trametinib and combination, TAK228 and combination, trametinib and TAK228); there are also a number of protein changes (four total) and phosphosite changes (three total) that are shared between monotherapies and combination therapy (Fig. 3, A and B, supplemental Fig. S5; and supplemental Tables S12–S15). Of the shared proteins and phosphosites, there were increases in tropomyosin beta chain (TPM2), transgelin actin cross-linking gelling protein (TAGLN), retrotransposon Gag-like protein 8C (RTL8C), and a phosphorylation site on DNA mismatch repair protein MSH3 (MSH3-S33), and there were decreases in capZ-interacting protein (RCSD1), a phosphorylation site on girdin (CCDC88A-S1652), and, interestingly, a phosphorylation site on tropomyosin beta chain (TPM2-S283) (supplemental Fig. S6). Five of these seven shared proteins and phosphosites, two increased and three decreased (TPM2, TPM2-S283, TAGLN, RCSD1, and CCDC88A-S1652), are involved in actin cytoskeleton dynamics, which play a role in diverse cancer cell processes, including mitosis, proliferation, invasion, and metastasis (40, 41, 42, 43).
Fig. 3.
Global proteomic–phosphoproteomic analysis identified distinct response signatures associated with trametinib, TAK228, or combination therapy. Venn diagrams showing statistically significant unique and shared protein (A) or phosphosite (B) changes in each treatment condition (compared with vehicle). C, kinase-substrate enrichment analysis (KSEA) kinase z-scores are represented in a bar graph showing inhibition of mTOR activity after TAK228 and combination treatment, inhibition of MAPK signaling after trametinib and combination treatment, and inhibition of cyclin-dependent kinases after combination treatment. Only kinases for which the FDR was <0.05 were included. FDR, false discovery rate; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin.
Combination Therapy Broadly Suppresses Cyclin-dependent Proteins
In order to investigate the impact of trametinib alone, TAK228 alone, and trametinib–TAK228 combination therapy on kinase activity within BRAFV600E mutant glioma xenografts, we utilized KSEA (38). We identified 1297 kinase–substrate relationships in the dataset (supplemental Table S16). We confirmed appropriate inhibition of mTOR by TAK228, with upstream compensatory activation of AKT1 and AKT2, which is a known feedback result of mTOR inhibition (44). We also confirmed inhibition of MAPK signaling by trametinib. In trametinib–TAK228 combination, both mTOR and MAPK are inhibited. There was also notable inhibition in the combination-treated tumors of cyclin-dependent kinases (CDK1, CDK2, CDK4, CDK5, and CDK6), proteins critical for cell cycle progression and tumor growth (Fig. 3C and supplemental Table S16).
Combination Therapy Broadly Suppresses Progrowth Signaling Pathways
Reactome protein pathway over representation analysis (using the Panther database) generated from statistically significantly altered proteins and normalized localized phosphosites in trametinib–TAK228 combination compared with vehicle plotted in a volcano plot of fold enrichment versus −log10(FDR) validated inhibition of the mTOR and MAPK pathways after trametinib–TAK228 combination therapy. In addition, this analysis demonstrated inhibition of vascular endothelial growth factor (VEGF) signaling after both trametinib monotherapy and trametinib–TAK228 combination therapy (Fig. 4, supplemental Table S17, and supplemental Fig. S7), a result we had previously confirmed by immunoblotting and histology of BT40 xenograft tumors (13). Furthermore, phosphoproteomic analysis confirmed inhibition of proliferation as measured by phosphorylated retinoblastoma protein (supplemental Fig. S8), a result we had previously seen by immunoblotting (13). Of note, extracellular matrix organization, a pathway that has previously been shown to be upregulated at baseline in BRAFV600E mutant glioma (39), was downregulated in response to trametinib–TAK228 combination therapy, despite being upregulated after trametinib or TAK228 monotherapy (supplemental Fig. S7). The majority of upregulated protein pathways after combination therapy were associated with mRNA and rRNA processing, suggesting that the tumors may be compensating for the suppression of translation as a result of mTOR and MAPK pathway inhibition (45) by upregulating both transcription (mRNA) and the translation machinery (rRNA). Many of the RNA pathways upregulated after combination therapy (rRNA processing, RNA polymerase II transcription termination, processing of capped intron-containing pre-mRNA, and mRNA 3′-end processing) were not similarly upregulated after either monotherapy (supplemental Fig. S7), suggesting that dual inhibition of both the MAPK and mTOR pathways is required to see these impacts on RNA processing pathways.
Fig. 4.
Trametinib–TAK228 combination therapy broadly inhibited progrowth signaling pathways, while activating pathways involved in RNA processing. Reactome protein pathways significantly altered (FDR <0.05) in an over-representation analysis (ORA) of statistically significantly altered proteins and phosphosites after trametinib–TAK228 combination therapy are represented in a scatter plot of fold enrichment versus −log10(FDR). FDR, false discovery rate.
Combination Therapy Increases Activation and Overall Levels of Proapoptotic Proteins
In addition to inhibition of progrowth pathways by trametinib–TAK228 combination therapy, proteomic and phosphoproteomic analysis demonstrated that dual inhibition of the MAPK and mTORC1/2 pathways increased expression of proapoptotic proteins and their activating phosphorylations (Fig. 5A). Trametinib monotherapy also increased expression of proapoptotic proteins and their phosphorylations, with specific overlap of GSN, DDX20-S505, and SCRIB-S1309 (supplemental Fig. S9A). Of note, the increase in proapoptotic signaling protein levels in the human proteome was not duplicated in the mouse proteome (supplemental Fig. S10A), indicating that the induction of apoptosis is specific to the glioma tumor, with the surrounding stroma being spared. This result confirmed our previous results that apoptosis was not induced in normal mouse tissues in response to 28 days of treatment with trametinib–TAK228 combination therapy (13).
Fig. 5.
Trametinib–TAK228 combination therapy activated apoptotic signaling and inhibited angiogenesis in BRAF-mutant glioma. Stacked bar graphs of log2(abundance) of proteins and phosphorylation sites demonstrating statistically significant (A) increases in human proteins and phosphosites involved in apoptosis in combination compared with vehicle in BRAF-mutant glioma, (B) decreases in human proteins and phosphosites involved in angiogenesis/VEGF signaling in BRAF-mutant glioma. FDR <0.01 for all data points included. BRAF, v-Raf murine sarcoma viral oncogene homolog B; VEGF, vascular endothelial growth factor.
Combination Therapy Suppresses Signals Promoting Vascularity of Tumors
We had previously shown that combination-treated tumors had decreased levels of VEGF and phosphorylated focal adhesion kinase (FAK, also known as protein tyrosine kinase 2 [PTK2]) by Western blot and decreased vascularity by histology (13). Proteomic and phosphoproteomic analysis from our short-course trametinib–TAK228 combination treatment of BT40 flank tumors validated these results, showing statistically significant reductions in proteins (including kinase insert domain receptor/VEGF receptor 2) and activating phosphorylations (including PTK2/FAK at PTK2–Y576) involved in angiogenesis signaling (Fig. 5B). Trametinib monotherapy also led to significant reductions in angiogenesis signaling (supplemental Fig. S9B and supplemental Table S17). Mouse proteins and phosphorylation sites involved in angiogenesis signaling were also significantly downregulated (supplemental Fig. S10B), indicating that the suppression of new vessel formation by combined trametinib–TAK228 therapy is a result of inhibition of VEGF/angiogenesis signaling in both the glioma tumor and the stroma.
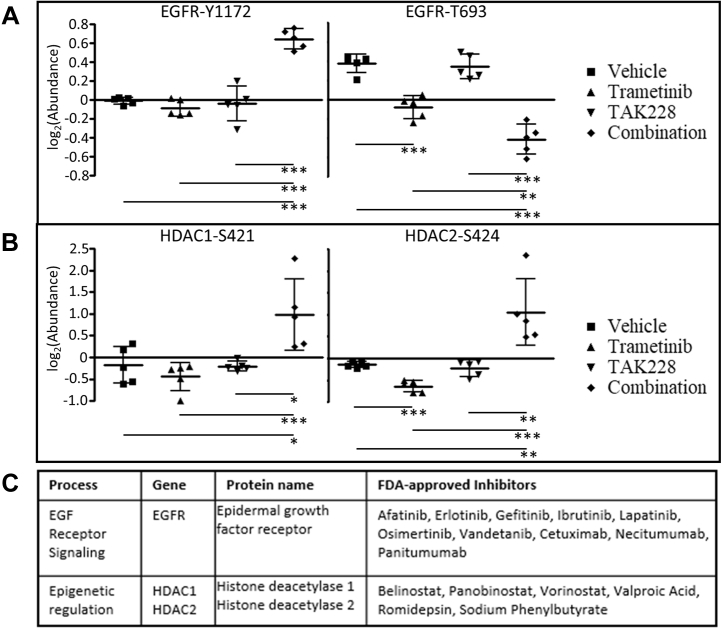
MEK/mTORC1/2 Inhibition Upregulates Epidermal Growth Factor
Having used detailed proteomic and phosphoproteomic analysis to characterize the cellular response of BRAFV600E mutant glioma to combined inhibition of the MAPK and mTOR pathways, and understanding that acquired resistance to such targeted therapies will invariably develop, we next sought to investigate our wealth of (phospho)proteomic data to identify potential new targets for future rational combination therapies. Analysis of protein and phosphosite expression corresponding to targets for existing drug therapies (supplemental Table S18) showed significant increases in a number of targetable proteins and activating phosphorylations (supplemental Tables S19–S21). One notable change was in two regulatory phosphorylations of the epidermal growth factor receptor (EGFR): an increase in the activating autophosphorylation site EGFR–Y1172 (46, 47, 48, 49), with a concomitant decrease in the inhibitory phosphorylation EGFR–T693 (50, 51, 52). Both these changes were present after combination therapy, whereas TAK228 monotherapy did not show any statistically significant change in either EGFR phosphorylation. Trametinib monotherapy only decreased the inhibitory T693 phosphorylation (Fig. 6A). Investigating further into the mechanism for this upregulation of EGFR, we identified an overall downregulation of the EGFR pathway in combination-treated tumors (Fig. 4 and supplemental Fig. S11). We interpret this upregulation of the cell surface receptor tyrosine kinase after combined trametinib–TAK228 therapy as a feedback activation in response to the downstream pathway inhibition. Feedback reactivation of EGFR after MAPK pathway inhibition is a well-characterized phenomenon in BRAFV600E mutant melanoma, so this result in a BRAFV600E mutant glioma reinforces this known biological feedback response (53, 54).
Fig. 6.
Druggable protein analysis after combination trametinib–TAK228 therapy identified a number of potentially targetable upregulated proteins, including epidermal growth factor receptor (EGFR) and class 1 HDACs. Aligned dot plots of log2(abundance) of listed phosphosites in each treatment condition, vehicle, trametinib monotherapy, TAK228 monotherapy, and trametinib–TAK228 combination therapy: (A) two EGFR phosphosites, one activating (EGFR-Y1172) and one inhibitory (EGFR-T693), showing that the level of EGFR-Y1172 was statistically significantly increased after combination therapy, and the level of the EGFR-T693 was statistically significantly decreased after trametinib monotherapy and combination therapy; (B) two class 1 HDAC-activating phosphosites, HDAC1-S421 and HDAC2-S424, showing that the levels of both of these activating phosphorylations were statistically significantly increased after combination therapy, but the level of HDAC-S424 was decreased after trametinib monotherapy. For A and B, ∗∗∗p < 0.001, ∗∗p between 0.001 and 0.01, ∗p between 0.01 and 0.05. C, the table listing FDA-approved inhibitors for EGFR and HDAC1/2. EGFR, epidermal growth factor receptor; FDA, Food and Drug Administration; HDAC, histone deacetylase.
Combination Therapy Upregulates Class 1 Histone Deacetylase Proteins
Another notable change was an increase in activating phosphorylations of both histone deacetylase 1 (HDAC1-S421) and histone deacetylase 2 (HDAC2-S424) after trametinib–TAK228 combination therapy (Fig. 6B). The latter was actually significantly decreased after trametinib monotherapy. Otherwise, there was no statistically significant change in these key regulatory sites with either trametinib or TAK228 monotherapy.
Based on this upregulation of EGFR and class 1 HDACs as a result of combined trametinib–TAK228 therapy in our BRAFV600E mutant glioma model, we can identify existing FDA-approved inhibitors that could potentially be added to MEK inhibition and mTOR inhibition to improve response and potentially preclude, or at least forestall, the development of resistance (Fig. 6C).
Discussion
Low-grade gliomas are the most common pediatric brain tumors (55). Activating mutations in the BRAF gene are one of the hallmarks of this disease. BRAF-activating mutations also occur in high-grade gliomas in children and adults, as well as in other cancer types, including melanoma, thyroid cancer, and colon cancer (2, 3, 4, 5, 6, 7). BRAF activation leads to increased signaling along the MAPK pathway as well as the mTOR pathway (56). Low-grade gliomas have activation of mTOR as well as MAPK (57). Treatment of low-grade gliomas with the mTOR inhibitor everolimus stabilizes disease in many patients and produces regressions in some patients (58, 59). Clinical trials are currently underway in pediatric low-grade glioma combining MEK and mTORC1-inhibiting rapalogs (NCT04485559). TAK228 is superior to rapalogs since it more fully inhibits the mTOR pathway; by dual targeting both mTORC1 and mTORC2, TAK288 obviates the reflexive feedback activation of mTORC2 that results from targeted mTORC1 inhibition with rapalogs like everolimus (13, 14, 15, 16, 17). An alternative treatment strategy in BRAFV600E mutant tumors has been to combine the BRAF-specific inhibitor dabrafenib with trametinib, which has been effective in treating melanoma (60). However, similar results have not been seen with this combination in BRAFV600E mutant colorectal cancer, indicating that such a vertical inhibition approach may not effectively treat all BRAF-mutant solid tumors (61). Given the active clinical investigation of combined MEK and mTOR inhibition in patients with pediatric low-grade glioma, we further investigated the impact of combined MEK and mTOR inhibition in a xenograft model of BRAFV600E mutant glioma. Our initial studies demonstrated that combination of MEK and mTOR inhibition in the BT40 BRAFV600E model decreased tumor growth and decreased tumor vascularity, suggesting an effect on both the tumor cells directly as well as on the mouse vasculature (13). In the current study, we sought to better understand the response of BRAF-activated tumors to short-term treatment with a MEK inhibitor, a dual mTORC1/2 kinase inhibitor, and combination therapy.
Our comprehensive proteomic and phosphoproteomic analysis demonstrated broad downregulation of the MAPK and mTOR pathways after combined trametinib–TAK228 treatment. We confirmed our prior findings (13) that the combination of trametinib and TAK228 induced apoptosis in the tumor while sparing the stroma. Combined therapy also suppressed endothelial cell growth and new blood vessel formation through downregulation of PTK2/FAK and VEGF signaling.
Interestingly, we found marked decreases in CDK activity by KSEA, including CDKs 1, 2, 4, 5, and 6. In a recent analysis of the proteogenomic landscape of pediatric brain tumors from the Clinical Proteomic Tumor Analysis Consortium, CDK1 and CDK2 were activated in more proliferative tumors in a range of histologic subtypes, including high-grade and low-grade gliomas; and CDK5 was upregulated in a subset of gliomas (39). CDKs are a target of novel cancer therapies that are being explored for aggressive gliomas as well as BRAF-activated melanoma (62). Herein, we have demonstrated that the combination of MEK and mTOR inhibition using trametinib and TAK228 effectively inhibit cell proliferation and CDKs in BRAFV600E mutant glioma.
The feedback upregulation we observed in EGFR in our BRAFV600E mutant glioma model after short-course combination of trametinib–TAK228 therapy has previously been reported in BRAFV600E mutant melanoma and colorectal cancer after BRAF or MEK inhibitor monotherapy; these findings have led to successful clinical trials of combination therapies with BRAF–MEK inhibitors and EGFR inhibitors in BRAFV600E mutant colorectal cancer (53, 54, 63). Whether the upregulation of EGFR after short-course treatment would contribute to the eventual development of resistance to combined MAPK- and mTOR-pathway inhibition in BRAF-mutant glioma would require investigation of the phosphoproteome after a longer course of treatment. The upregulation of HDACs implicates a role for addition of HDAC inhibitors to MEK inhibitors to improve response in BRAF-mutant glioma, a combination that is effective in RAS-mutant non–small cell lung cancer and pancreatic cancer and is currently being investigated in BRAF-mutant, RAS-mutant, and neurofibromin 1–mutant melanoma (64, 65, 66, 67). Furthermore, there is evidence that the combination of mTOR and HDAC inhibitors effectively target aggressive nervous system malignancies (68, 69), indicating that combining HDAC inhibition with MEK inhibition and/or mTOR inhibition to treat BRAF-mutant glioma is a compelling new avenue for investigation.
We investigated the proteome and phosphoproteome after short-term treatment with targeted pathway inhibitors in order to detect changes in signaling pathways that occur in treatment-naive tumors. In the future, we plan to compare the proteome and phosphoproteome of naive tumors with those that have developed resistance to the inhibitors after weeks of treatment. These analyses may provide further insights into the adaptive responses BRAF-mutant cancer cells use to escape precision therapies.
In vivo comprehensive proteomics and phosphoproteomics in well-defined human tumors in mice provides an opportunity to understand how cancer cells interact with normal stromal cells and adapt to therapeutic interventions. In this work, we confirmed and extended our findings in treating an aggressive BRAFV600E mutant human glioma model with MEK and mTORC1/2 inhibitors as monotherapy and in combination. Our findings highlight changes in tumor–stromal signaling affecting tumor vasculature, tumor cell cycle proteins, EGFR, and class 1 HDACs as a result of trametinib–TAK228 combination therapy. Insights gained from these studies will impact the development of additional rational combination therapies for aggressive BRAF-driven tumors.
Data Availability
All the MS proteomics data (both global proteome and phosphoproteome), including raw MS data, processed peak lists, and respective search results have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) (70) via the PRIDE (71) partner repository with the dataset identifier PXD022329.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
Funding and additional information
M. J. M. is funded in part by fellowship support from the St. Baldrick's Foundation, as well as a NextGen award from the Children's Cancer Foundation, a Hyundai Hope on Wheels Young Investigator Award, and a grant from the National Institutes of Health Loan Repayment Program (L40 CA242540). Additional funding is provided by the Imagine an Answer to Kids' Brain Cancer (E. H. R.), the Giant Food Pediatric Cancer Research Fund, and a National Cancer Institute core grant to the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center (CA006973). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
M. J. M., A. A., D. J. C., and E. H. R. conceptualization; M. J. M., A. A., L. C., T.-S. M. L., M. S., D. J. C., and E. H. R. methodology; H. S., T.-S. M. L., and M. S. software; M. J. M. and A. A. validation; M. J. M., H. S., and M. S., formal analysis; M. J. M., A. A., L. C., D. J. C., and E. H. R. investigation; H. Z., E. H. R., and C. G. E. resources; H. S., T.-S. M. L., M. S., D. J. C., and E. H. R. data curation; M. J. M., A. A., M. S., D. J. C., and E. H. R. writing—original draft; M. J. M., A. A., M. S., D. J. C., E. H. R., and J. A. R. writing—review and editing; M. J. M. and H. S. visualization; C. G. E., H. Z., D. J. C., and E. H. R. supervision; H. Z., D. J. C., and E. H. R. project administration; M. J. M. and E. H. R. funding acquisition.
Contributor Information
Micah J. Maxwell, Email: mmaxwel1@jhmi.edu.
Eric H. Raabe, Email: eraabe2@jhmi.edu.
Supplemental Data
References
- 1.Burotto M., Chiou V.L., Lee J.M., Kohn E.C. The MAPK pathway across different malignancies: A new perspective. Cancer. 2014;120:3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bond C.E., Whitehall V.L.J. How the BRAF V600E mutation defines a distinct subgroup of colorectal cancer: Molecular and clinical implications. Gastroenterol. Res. Pract. 2018;2018:9250757. doi: 10.1155/2018/9250757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dankner M., Rose A.A.N., Rajkumar S., Siegel P.M., Watson I.R. Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene. 2018;37:3183–3199. doi: 10.1038/s41388-018-0171-x. [DOI] [PubMed] [Google Scholar]
- 4.Nucera C., Lawler J., Parangi S. BRAF(V600E) and microenvironment in thyroid cancer: A functional link to drive cancer progression. Cancer Res. 2011;71:2417–2422. doi: 10.1158/0008-5472.CAN-10-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Leary C.G., Andelkovic V., Ladwa R., Pavlakis N., Zhou C., Hirsch F., Richard D., O'Byrne K. Targeting BRAF mutations in non-small cell lung cancer. Transl. Lung Cancer Res. 2019;8:1119–1124. doi: 10.21037/tlcr.2019.10.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olow A., Mueller S., Yang X., Hashizume R., Meyerowitz J., Weiss W., Resnick A.C., Waanders A.J., Stalpers L.J., Berger M.S., Gupta N., James C.D., Petritsch C.K., Haas-Kogan D.A. BRAF status in personalizing treatment approaches for pediatric gliomas. Clin. Cancer Res. 2016;22:5312–5321. doi: 10.1158/1078-0432.CCR-15-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreck K.C., Grossman S.A., Pratilas C.A. BRAF mutations and the utility of RAF and MEK inhibitors in primary brain tumors. Cancers (Basel) 2019;11:1262. doi: 10.3390/cancers11091262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty K.T., Infante J.R., Daud A., Gonzalez R., Kefford R.F., Sosman J., Hamid O., Schuchter L., Cebon J., Ibrahim N., Kudchadkar R., Burris H.A., 3rd, Falchook G., Algazi A., Lewis K. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondyli M., Larouche V., Saint-Martin C., Ellezam B., Pouliot L., Sinnett D., Legault G., Crevier L., Weil A., Farmer J.P., Jabado N., Perreault S. Trametinib for progressive pediatric low-grade gliomas. J. Neurooncol. 2018;140:435–444. doi: 10.1007/s11060-018-2971-9. [DOI] [PubMed] [Google Scholar]
- 10.Maraka S., Janku F. BRAF alterations in primary brain tumors. Discov. Med. 2018;26:51–60. [PubMed] [Google Scholar]
- 11.Conciatori F., Ciuffreda L., Bazzichetto C., Falcone I., Pilotto S., Bria E., Cognetti F., Milella M. mTOR cross-talk in cancer and potential for combination therapy. Cancers (Basel) 2018;10:23. doi: 10.3390/cancers10010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sievert A.J., Lang S.S., Boucher K.L., Madsen P.J., Slaunwhite E., Choudhari N., Kellet M., Storm P.B., Resnick A.C. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5957–5962. doi: 10.1073/pnas.1219232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold A., Yuan M., Price A., Harris L., Eberhart C.G., Raabe E.H. Synergistic activity of mTORC1/2 kinase and MEK inhibitors suppresses pediatric low-grade glioma tumorigenicity and vascularity. Neuro Oncol. 2020;22:563–574. doi: 10.1093/neuonc/noz230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghobrial I.M., Siegel D.S., Vij R., Berdeja J.G., Richardson P.G., Neuwirth R., Patel C.G., Zohren F., Wolf J.L. TAK-228 (formerly MLN0128), an investigational oral dual TORC1/2 inhibitor: A phase I dose escalation study in patients with relapsed or refractory multiple myeloma, non-Hodgkin lymphoma, or Waldenstrom's macroglobulinemia. Am. J. Hematol. 2016;91:400–405. doi: 10.1002/ajh.24300. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Prat A., Rodriguez-Vida A., Juanpere-Rodero N., Arpi O., Menendez S., Soria-Jimenez L., Martinez A., Iarchouk N., Rojo F., Albanell J., Brake R., Rovira A., Bellmunt J. Novel oral mTORC1/2 inhibitor TAK-228 has synergistic antitumor effects when combined with paclitaxel or PI3Kalpha inhibitor TAK-117 in preclinical bladder cancer models. Mol. Cancer Res. 2019;17:1931–1944. doi: 10.1158/1541-7786.MCR-18-0923. [DOI] [PubMed] [Google Scholar]
- 16.Miyahara H., Yadavilli S., Natsumeda M., Rubens J.A., Rodgers L., Kambhampati M., Taylor I.C., Kaur H., Asnaghi L., Eberhart C.G., Warren K.E., Nazarian J., Raabe E.H. The dual mTOR kinase inhibitor TAK228 inhibits tumorigenicity and enhances radiosensitization in diffuse intrinsic pontine glioma. Cancer Lett. 2017;400:110–116. doi: 10.1016/j.canlet.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubens J.A., Wang S.Z., Price A., Weingart M.F., Allen S.J., Orr B.A., Eberhart C.G., Raabe E.H. The TORC1/2 inhibitor TAK228 sensitizes atypical teratoid rhabdoid tumors to cisplatin-induced cytotoxicity. Neuro Oncol. 2017;19:1361–1371. doi: 10.1093/neuonc/nox067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang L., Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 19.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bid H.K., Kibler A., Phelps D.A., Manap S., Xiao L., Lin J., Capper D., Oswald D., Geier B., DeWire M., Smith P.D., Kurmasheva R.T., Mo X., Fernandez S., Houghton P.J. Development, characterization, and reversal of acquired resistance to the MEK1 inhibitor selumetinib (AZD6244) in an in vivo model of childhood astrocytoma. Clin. Cancer Res. 2013;19:6716–6729. doi: 10.1158/1078-0432.CCR-13-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolb E.A., Gorlick R., Houghton P.J., Morton C.L., Neale G., Keir S.T., Carol H., Lock R., Phelps D., Kang M.H., Reynolds C.P., Maris J.M., Billups C., Smith M.A. Initial testing (stage 1) of AZD6244 (ARRY-142886) by the pediatric preclinical testing program. Pediatr. Blood Cancer. 2010;55:668–677. doi: 10.1002/pbc.22576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore K.N., Bauer T.M., Falchook G.S., Chowdhury S., Patel C., Neuwirth R., Enke A., Zohren F., Patel M.R. Phase I study of the investigational oral mTORC1/2 inhibitor sapanisertib (TAK-228): Tolerability and food effects of a milled formulation in patients with advanced solid tumours. ESMO Open. 2018;3 doi: 10.1136/esmoopen-2017-000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark D.J., Dhanasekaran S.M., Petralia F., Pan J., Song X., Hu Y., da Veiga Leprevost F., Reva B., Lih T.M., Chang H.Y., Ma W., Huang C., Ricketts C.J., Chen L., Krek A. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell. 2019;179:964–983.e931. doi: 10.1016/j.cell.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mertins P., Tang L.C., Krug K., Clark D.J., Gritsenko M.A., Chen L., Clauser K.R., Clauss T.R., Shah P., Gillette M.A., Petyuk V.A., Thomas S.N., Mani D.R., Mundt F., Moore R.J. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc. 2018;13:1632–1661. doi: 10.1038/s41596-018-0006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mertins P., Qiao J.W., Patel J., Udeshi N.D., Clauser K.R., Mani D.R., Burgess M.W., Gillette M.A., Jaffe J.D., Carr S.A. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L., Zhang B., Schnaubelt M., Shah P., Aiyetan P., Chan D., Zhang H., Zhang Z. MS-PyCloud: An open-source, cloud computing-based pipeline for LC-MS/MS data analysis. bioRxiv. 2018 doi: 10.1101/320887. [preprint] [DOI] [Google Scholar]
- 27.Kim S., Gupta N., Pevzner P.A. Spectral probabilities and generating functions of tandem mass spectra: A strike against decoy databases. J. Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S., Pevzner P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 30.Fermin D., Avtonomov D., Choi H., Nesvizhskii A.I. LuciPHOr2: Site localization of generic post-translational modifications from tandem mass spectrometry data. Bioinformatics. 2015;31:1141–1143. doi: 10.1093/bioinformatics/btu788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang K.L., Li S., Mertins P., Cao S., Gunawardena H.P., Ruggles K.V., Mani D.R., Clauser K.R., Tanioka M., Usary J., Kavuri S.M., Xie L., Yoon C., Qiao J.W., Wrobel J. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat. Commun. 2017;8:14864. doi: 10.1038/ncomms14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotto K.C., Wagner A.H., Feng Y.Y., Kiwala S., Coffman A.C., Spies G., Wollam A., Spies N.C., Griffith O.L., Griffith M. DGIdb 3.0: A redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charan J., Kantharia N.D. How to calculate sample size in animal studies? J. Pharmacol. Pharmacother. 2013;4:303–306. doi: 10.4103/0976-500X.119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 35.Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas P.D., Kejariwal A., Guo N., Mi H., Campbell M.J., Muruganujan A., Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 38.Wiredja D.D., Koyuturk M., Chance M.R. The KSEA app: A web-based tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics. 2017;33:3489–3491. doi: 10.1093/bioinformatics/btx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petralia F., Tignor N., Reva B., Koptyra M., Chowdhury S., Rykunov D., Krek A., Ma W., Zhu Y., Ji J., Calinawan A., Whiteaker J.R., Colaprico A., Stathias V., Omelchenko T. Integrated proteogenomic characterization across major histological types of pediatric brain cancer. Cell. 2020;183:1962–1985.e1931. doi: 10.1016/j.cell.2020.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dvorakova M., Nenutil R., Bouchal P. Transgelins, cytoskeletal proteins implicated in different aspects of cancer development. Expert Rev. Proteomics. 2014;11:149–165. doi: 10.1586/14789450.2014.860358. [DOI] [PubMed] [Google Scholar]
- 41.Hernandez-Valladares M., Kim T., Kannan B., Tung A., Aguda A.H., Larsson M., Cooper J.A., Robinson R.C. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat. Struct. Mol. Biol. 2010;17:497–503. doi: 10.1038/nsmb.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sliwinska M., Robaszkiewicz K., Wasag P., Moraczewska J. Mutations Q93H and E97K in TPM2 disrupt Ca-dependent regulation of actin filaments. Int. J. Mol. Sci. 2021;22:4036. doi: 10.3390/ijms22084036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Enomoto A., Weng L., Mizutani Y., Abudureyimu S., Esaki N., Tsuyuki Y., Chen C., Mii S., Asai N., Haga H., Ishida S., Yokota K., Akiyama M., Takahashi M. Girdin/GIV regulates collective cancer cell migration by controlling cell adhesion and cytoskeletal organization. Cancer Sci. 2018;109:3643–3656. doi: 10.1111/cas.13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan X., Harkavy B., Shen N., Grohar P., Helman L.J. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene. 2007;26:1932–1940. doi: 10.1038/sj.onc.1209990. [DOI] [PubMed] [Google Scholar]
- 45.Roux P.P., Topisirovic I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorkin A., Helin K., Waters C.M., Carpenter G., Beguinot L. Multiple autophosphorylation sites of the epidermal growth factor receptor are essential for receptor kinase activity and internalization. Contrasting significance of tyrosine 992 in the native and truncated receptors. J. Biol. Chem. 1992;267:8672–8678. [PubMed] [Google Scholar]
- 47.Sorkin A., Waters C., Overholser K.A., Carpenter G. Multiple autophosphorylation site mutations of the epidermal growth factor receptor. Analysis of kinase activity and endocytosis. J. Biol. Chem. 1991;266:8355–8362. [PubMed] [Google Scholar]
- 48.Imami K., Sugiyama N., Imamura H., Wakabayashi M., Tomita M., Taniguchi M., Ueno T., Toi M., Ishihama Y. Temporal profiling of lapatinib-suppressed phosphorylation signals in EGFR/HER2 pathways. Mol. Cell. Proteomics. 2012;11:1741–1757. doi: 10.1074/mcp.M112.019919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun T., Aceto N., Meerbrey K.L., Kessler J.D., Zhou C., Migliaccio I., Nguyen D.X., Pavlova N.N., Botero M., Huang J., Bernardi R.J., Schmitt E., Hu G., Li M.Z., Dephoure N. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144:703–718. doi: 10.1016/j.cell.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mertins P., Yang F., Liu T., Mani D.R., Petyuk V.A., Gillette M.A., Clauser K.R., Qiao J.W., Gritsenko M.A., Moore R.J., Levine D.A., Townsend R., Erdmann-Gilmore P., Snider J.E., Davies S.R. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteomics. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winograd-Katz S.E., Levitzki A. Cisplatin induces PKB/Akt activation and p38(MAPK) phosphorylation of the EGF receptor. Oncogene. 2006;25:7381–7390. doi: 10.1038/sj.onc.1209737. [DOI] [PubMed] [Google Scholar]
- 52.Yi L., Shi T., Gritsenko M.A., X'Avia Chan C.Y., Fillmore T.L., Hess B.M., Swensen A.C., Liu T., Smith R.D., Wiley H.S., Qian W.J. Targeted quantification of phosphorylation dynamics in the context of EGFR-MAPK pathway. Anal. Chem. 2018;90:5256–5263. doi: 10.1021/acs.analchem.8b00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn A., Chatterjee A., Eccles M.R. The slow cycling phenotype: A growing problem for treatment resistance in melanoma. Mol. Cancer Ther. 2017;16:1002–1009. doi: 10.1158/1535-7163.MCT-16-0535. [DOI] [PubMed] [Google Scholar]
- 54.Dratkiewicz E., Simiczyjew A., Pietraszek-Gremplewicz K., Mazurkiewicz J., Nowak D. Characterization of melanoma cell lines resistant to vemurafenib and evaluation of their responsiveness to EGFR- and MET-inhibitor treatment. Int. J. Mol. Sci. 2019;21:113. doi: 10.3390/ijms21010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raabe E., Kieran M.W., Cohen K.J. New strategies in pediatric gliomas: Molecular advances in pediatric low-grade gliomas as a model. Clin. Cancer Res. 2013;19:4553–4558. doi: 10.1158/1078-0432.CCR-13-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prabowo A.S., Iyer A.M., Veersema T.J., Anink J.J., Schouten-van Meeteren A.Y., Spliet W.G., van Rijen P.C., Ferrier C.H., Capper D., Thom M., Aronica E. BRAF V600E mutation is associated with mTOR signaling activation in glioneuronal tumors. Brain Pathol. 2014;24:52–66. doi: 10.1111/bpa.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hutt-Cabezas M., Karajannis M.A., Zagzag D., Shah S., Horkayne-Szakaly I., Rushing E.J., Cameron J.D., Jain D., Eberhart C.G., Raabe E.H., Rodriguez F.J. Activation of mTORC1/mTORC2 signaling in pediatric low-grade glioma and pilocytic astrocytoma reveals mTOR as a therapeutic target. Neuro Oncol. 2013;15:1604–1614. doi: 10.1093/neuonc/not132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wahl M., Chang S.M., Phillips J.J., Molinaro A.M., Costello J.F., Mazor T., Alexandrescu S., Lupo J.M., Nelson S.J., Berger M., Prados M., Taylor J.W., Butowski N., Clarke J.L., Haas-Kogan D. Probing the phosphatidylinositol 3-kinase/mammalian target of rapamycin pathway in gliomas: A phase 2 study of everolimus for recurrent adult low-grade gliomas. Cancer. 2017;123:4631–4639. doi: 10.1002/cncr.30909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright K.D., Yao X., London W.B., Kao P.C., Gore L., Hunger S., Geyer R., Cohen K.J., Allen J.C., Katzenstein H.M., Smith A., Boklan J., Nazemi K., Trippett T., Karajannis M. A POETIC phase II study of continuous oral everolimus in recurrent, radiographically progressive pediatric low-grade glioma. Pediatr. Blood Cancer. 2021;68 doi: 10.1002/pbc.28787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robert C., Karaszewska B., Schachter J., Rutkowski P., Mackiewicz A., Stroiakovski D., Lichinitser M., Dummer R., Grange F., Mortier L., Chiarion-Sileni V., Drucis K., Krajsova I., Hauschild A., Lorigan P. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 61.Corcoran R.B., Atreya C.E., Falchook G.S., Kwak E.L., Ryan D.P., Bendell J.C., Hamid O., Messersmith W.A., Daud A., Kurzrock R., Pierobon M., Sun P., Cunningham E., Little S., Orford K. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J. Clin. Oncol. 2015;33:4023–4031. doi: 10.1200/JCO.2015.63.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schettini F., De Santo I., Rea C.G., De Placido P., Formisano L., Giuliano M., Arpino G., De Laurentiis M., Puglisi F., De Placido S., Del Mastro L. CDK 4/6 inhibitors as single agent in advanced solid tumors. Front. Oncol. 2018;8:608. doi: 10.3389/fonc.2018.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corcoran R.B., Andre T., Atreya C.E., Schellens J.H.M., Yoshino T., Bendell J.C., Hollebecque A., McRee A.J., Siena S., Middleton G., Muro K., Gordon M.S., Tabernero J., Yaeger R., O'Dwyer P.J. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov. 2018;8:428–443. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chao M.W., Chang L.H., Tu H.J., Chang C.D., Lai M.J., Chen Y.Y., Liou J.P., Teng C.M., Pan S.L. Combination treatment strategy for pancreatic cancer involving the novel HDAC inhibitor MPT0E028 with a MEK inhibitor beyond K-Ras status. Clin. Epigenetics. 2019;11:85. doi: 10.1186/s13148-019-0681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maertens O., Kuzmickas R., Manchester H.E., Emerson C.E., Gavin A.G., Guild C.J., Wong T.C., De Raedt T., Bowman-Colin C., Hatchi E., Garraway L.A., Flaherty K.T., Pathania S., Elledge S.J., Cichowski K. MAPK pathway suppression unmasks latent DNA repair defects and confers a chemical synthetic vulnerability in BRAF-, NRAS-, and NF1-mutant melanomas. Cancer Discov. 2019;9:526–545. doi: 10.1158/2159-8290.CD-18-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamada T., Amann J.M., Tanimoto A., Taniguchi H., Shukuya T., Timmers C., Yano S., Shilo K., Carbone D.P. Histone deacetylase inhibition enhances the antitumor activity of a MEK inhibitor in lung cancer cells harboring RAS mutations. Mol. Cancer Ther. 2018;17:17–25. doi: 10.1158/1535-7163.MCT-17-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeon M., Kim Y., Jung H.S., Jeoung D. Histone deacetylase inhibitors to overcome resistance to targeted and immuno therapy in metastatic melanoma. Front. Cell Dev. Biol. 2020;8:486. doi: 10.3389/fcell.2020.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malone C.F., Emerson C., Ingraham R., Barbosa W., Guerra S., Yoon H., Liu L.L., Michor F., Haigis M., Macleod K.F., Maertens O., Cichowski K. mTOR and HDAC inhibitors converge on the TXNIP/thioredoxin pathway to cause catastrophic oxidative stress and regression of RAS-driven tumors. Cancer Discov. 2017;7:1450–1463. doi: 10.1158/2159-8290.CD-17-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meng W., Wang B., Mao W., Wang J., Zhao Y., Li Q., Zhang C., Ma J. Enhanced efficacy of histone deacetylase inhibitor panobinostat combined with dual PI3K/mTOR inhibitor BEZ235 against glioblastoma. Nagoya J. Med. Sci. 2019;81:93–102. doi: 10.18999/nagjms.81.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deutsch E.W., Bandeira N., Sharma V., Perez-Riverol Y., Carver J.J., Kundu D.J., Garcia-Seisdedos D., Jarnuczak A.F., Hewapathirana S., Pullman B.S., Wertz J., Sun Z., Kawano S., Okuda S., Watanabe Y. The ProteomeXchange consortium in 2020: Enabling 'big data' approaches in proteomics. Nucleic Acids Res. 2020;48:D1145–D1152. doi: 10.1093/nar/gkz984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the MS proteomics data (both global proteome and phosphoproteome), including raw MS data, processed peak lists, and respective search results have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) (70) via the PRIDE (71) partner repository with the dataset identifier PXD022329.