ABSTRACT
The ability to inducibly repress gene expression is critical to the study of organisms, like Chlamydia, with reduced genomes in which the majority of genes are likely to be essential. We recently described the feasibility of a CRISPR interference (CRISPRi) system to inducibly repress gene expression in Chlamydia trachomatis. However, the initial system suffered from some drawbacks, primarily leaky expression of the anhydrotetracycline (aTc)-inducible dCas9 ortholog and plasmid instability, which prevented population-wide studies (e.g., transcript analyses) of the effects of knockdown. Here, we describe various modifications to the original system that have allowed us to measure gene expression changes within a transformed population of C. trachomatis serovar L2. These modifications include (i) a change in the vector backbone, (ii) the introduction of a weaker ribosome binding site driving dCas9 translation, and (iii) the addition of a degradation tag to dCas9 itself. With these changes, we demonstrate the ability to inducibly repress a target gene sequence, as measured by the absence of protein by immunofluorescence analysis and by decreased transcript levels. Importantly, the expression of dCas9 alone (i.e., without a guide RNA [gRNA]) had minimal impact on chlamydial growth or development. We also describe complementation of the knockdown effect by introducing a transcriptional fusion of the target gene 3′ to dCas9. Finally, we demonstrate the functionality of a second CRISPRi system based on a dCas12 system that expands the number of potential chromosomal targets. These tools should provide the ability to study essential gene function in Chlamydia.
KEYWORDS: Chlamydia, CRISPR interference, inducible repression, gene expression, CRISPRi
INTRODUCTION
Chlamydia spp. are obligate intracellular bacteria. In evolving to this niche, these unique bacteria have significantly reduced their genome sizes and contents. Chlamydia trachomatis, the leading cause of bacterial sexually transmitted infections as well as of preventable infectious blindness (1–3), encodes approximately 900 open reading frames (ORFs) in roughly 1 Mbp (4). A complicating factor in the study of Chlamydia is the developmental cycle these bacteria utilize to alternate between functional and morphologic forms, namely the infectious but nondividing elementary body (EB) and the noninfectious but dividing reticulate body (RB) (5). The entirety of the developmental cycle occurs within a pathogen-specified parasitic organelle termed the inclusion (6). Given its limited gene repertoire, the majority of ORFs are likely essential to the organism, which further complicates studies of Chlamydia. Indeed, if we assume that the core chlamydial genome is essential, then over 500 ORFs (>55%) would fall within this category (7). Essential genes in Chlamydia are not only those required to replicate and divide (e.g., rpoB, which encodes an RNA polymerase subunit), in a classical sense, but those that are required to transition between developmental forms for successful completion of the developmental cycle. Examples of the latter class might include components of the type III secretion system that are necessary to secrete effectors that mediate entry into the host cell and establishment of the inclusion (8, 9) or the histone-rich proteins that are needed to compact the chromosome in the EB (10). Thus, a gene in Chlamydia, which, when disrupted, results in the inability to increase EB output, may be considered essential for this organism, at least when studied in cell culture. The subset of genes that may be essential for in vivo growth could be larger, as the growth conditions in cell culture are likely more permissive than in more relevant infection models (11). To date, less than 10% of ORFs have been disrupted in C. trachomatis using newly developed genetic tools (12), including group II intron insertion (13), allelic exchange (14), and transposon mutagenesis (15).
Stable transformation of Chlamydia is a relatively recent development (12), and new genetic tools continue to be deployed. We previously reported on the feasibility of using CRISPR interference (CRISPRi) to inducibly knock down gene expression in C. trachomatis serovar L2 (16). Such a tool would represent a significant advance in our ability to study essential gene function in this obligate intracellular bacterium. CRISPRi relies on (i) the inducible expression of a catalytically inactive Cas9 in combination with (ii) the constitutive expression of a guide RNA (gRNA) that recognizes a complementary chromosomal sequence next to a protospacer-adjacent motif (PAM) sequence. In Escherichia coli, these components are typically encoded on two plasmids (17), but for Chlamydia, we created a single-plasmid system (16). We were unable to transform with a single vector encoding Streptococcus pyogenes dCas9 plus a gRNA, but did have success with Staphylococcus aureus dCas9 with a constitutively expressed gRNA. Nonetheless, we noted two key issues with the system that required improvement before widespread adoption of this technology (16). First, we observed issues related to plasmid stability during generation of the transformant, which resulted in a mixed population of bacteria that precluded population-wide analyses (e.g., transcript studies). Second, we noted leaky expression of the dCas9 ortholog that could result in target inhibition in the absence of induction.
Here, we describe improvements to the CRISPRi system for Chlamydia that should make it suitable for widespread use to inducibly knock down target gene function. To overcome prior limitations, we made three key alterations to the system. First, we moved the components to a different plasmid backbone—this eliminated the plasmid stability issues observed when propagating transformants. Second, we modified the ribosome binding site (RBS) that drives dCas9 translation initiation to reduce its efficiency. Third, we appended a C-terminal degradation tag to dCas9 that renders the protein unstable. These latter two modifications reduced issues related to leaky expression. These changes allowed us to generate a stable population of transformants that could be analyzed for population-wide gene expression changes. Finally, we also describe here the implementation of a second CRISPRi system based on a dCas12 ortholog that utilizes a different PAM sequence. These two systems will allow for broad targeting of the chlamydial genome and for analysis of essential gene function in a straightforward and easy approach.
RESULTS
Development of a modified plasmid for CRISPRi in Chlamydia.
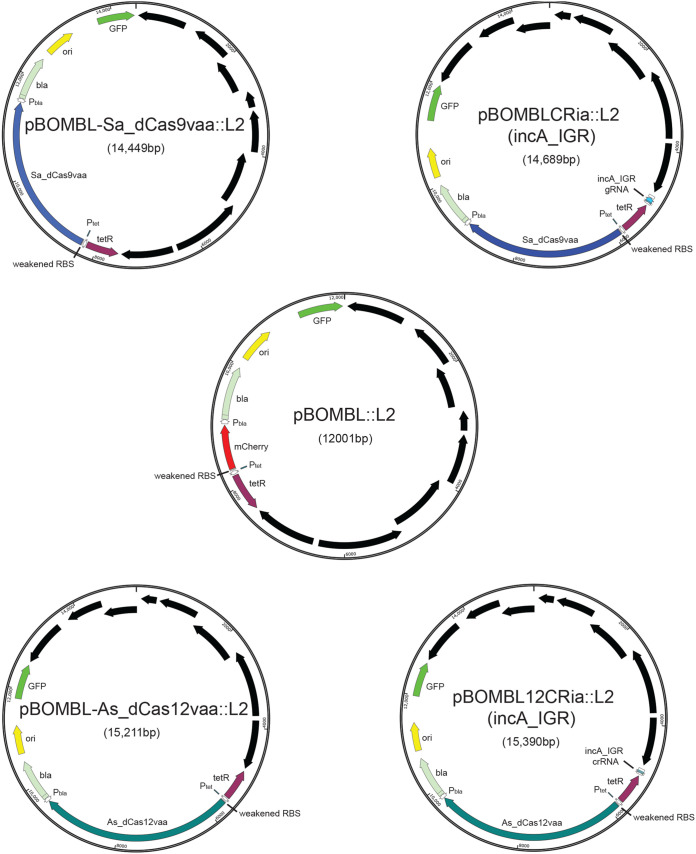
Given the noted issues with plasmid stability in our original implementation of a dCas9-based CRISPRi system in Chlamydia (16), our first approach to address this was to move the constituents to the pBOMB4-Tet plasmid (18), creating pBOMBCRi::L2. This was an empirical decision designed to determine if general plasmid backbone characteristics were responsible for the observed plasmid instability in the original CRISPRi design. We again targeted the incA gene due to its dispensability, so that any leaky expression would be detectable (using an antibody against IncA) and not detrimental to the propagation of the transformant (19). Although we readily obtained a pure population of transformants, thereby eliminating the plasmid stability issue, we noticed extensive leaky expression of the dCas9 that blocked incA expression in the absence of induction (data not shown). Therefore, we included two further modifications in an attempt to reduce or eliminate leaky expression of the dCas9. First, we modified the ribosome binding site in the pBOMB4-Tet::L2 vector to reduce its efficiency. In doing so, we created the vector pBOMBL::L2 (Fig. 1). Second, we appended a degradation tag (val-ala-ala; VAA), derived from the SsrA tagging system (20, 21), to the dCas9 ortholog that would target it for degradation by the chlamydial ClpXP protease system (22). This resulted in the plasmid pBOMBL-Sa_dCas9vaa::L2 [also referred to as pBOMBLCRia::L2 (e.v.)] (Fig. 1). We then inserted a constitutively expressed gRNA targeting the template strand of the intergenic region upstream of incA into this new vector to create pBOMBLCRia::L2 (incA_IGR) (Fig. 1). This vector was transformed into C. trachomatis L2 and further characterized as described below.
FIG 1.
Illustration of plasmids used in the current study and their indicated sizes in bp. Relevant genes and gene regulatory factors are indicated, as well as the overall size of the vector. Black arrows represent the chlamydial plasmid open reading frames (ORFs) that are part of the shuttle vector. RBS, ribosome binding site; GFP, green fluorescent protein. Plasmid maps were generated using SnapGene Viewer.
Potential targets for CRISPRi are limited by the PAM sequence of the dCas ortholog being used. The Staphylococcus aureus dCas9 uses the purine-rich PAM sequence NNGRRT (23). Chlamydia trachomatis is an AT-rich organism (4), and thus the presence of the G within the NNGRRT sequence potentially limits the chromosomal targets. By searching for these sequences in the C. trachomatis L2/434/Bu chromosome, we found 32,523 sites of all combinations of NNGRRT (highlighted in Table S2 in the supplemental material). By developing a second CRISPRi system utilizing a dCas ortholog that recognizes an AT-rich PAM sequence, the total number and location of potential interference sites would be greatly expanded. Therefore, we inserted the Acidoaminococcus dCas12 ortholog into the aforementioned pBOMBL vector and appended the VAA degradation tag to create the plasmid pBOMBL-As_ddCpf1vaa::L2 (also referred to as pBOMBL12CRia::L2 [e.v.]). This dCas12 ortholog recognizes the PAM sequence TTTV (V = not T) (24), present in 46,789 sites in the C. trachomatis L2/434/Bu chromosome (highlighted in Table S3 in the supplemental material). Insertion of the constitutively expressed CRISPR RNA (crRNA) designed to target the nontemplate strand of the upstream intergenic region of incA resulted in pBOMBL12CRia::L2 (incA_IGR). The pBOMBL12CRia::L2 construct, with or without the incA_IGR crRNA, was transformed into C. trachomatis L2.
Expression of the dCas9 in combination with the guide RNA targeting incA blocks expression of IncA.
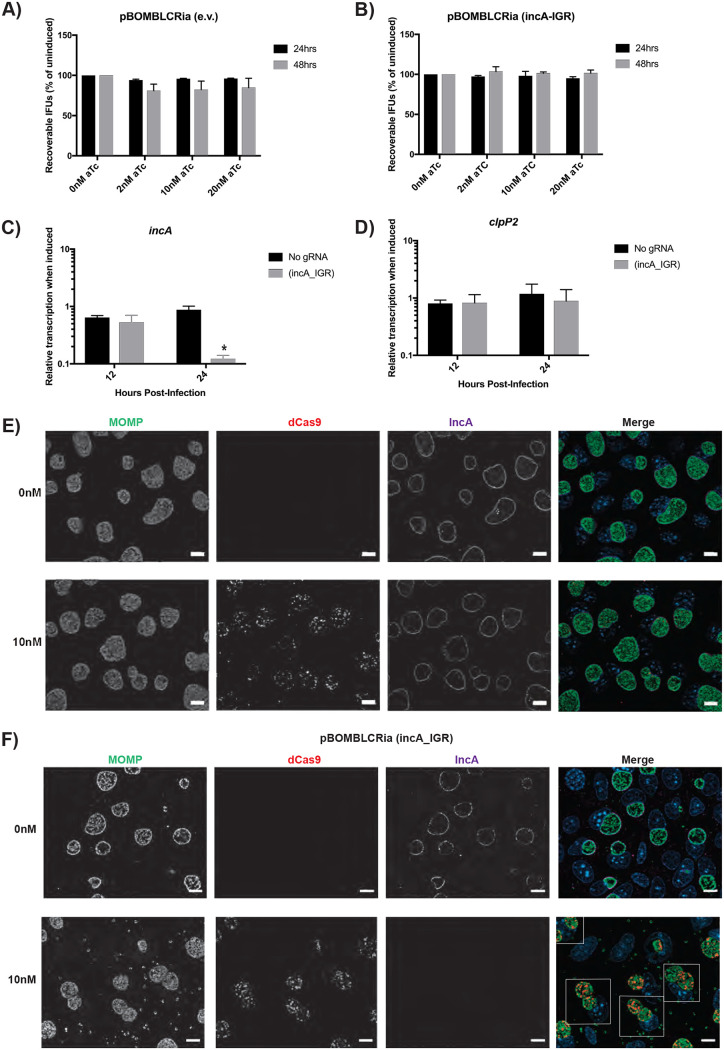
We previously noted that expression of the dCas9 in chlamydiae did not impact inclusion or bacterial morphology (16). However, to directly quantify the effect of dCas9 expression on bacterial growth, we performed an inclusion-forming unit (IFU) assay, which measures the production of infectious progeny (i.e., EBs) from an infected cell culture. Cells were infected with the transformants, and dCas9 expression was induced or not at 4 h postinfection (hpi) with 0, 2, 10, or 20 nM anhydrotetracycline (aTc). Samples were collected at 24 and 48 hpi and subsequently titrated onto fresh cell monolayers to quantify IFUs. Importantly, the pBOMB plasmids encode green fluorescent protein (GFP) to visualize transformants. Therefore, when quantifying IFUs, we compared the GFP-positive (GFP+) inclusion counts to the total inclusion count (i.e., GFP+ and GFP− inclusions) to assess plasmid stability after dCas9 induction. IncA is nonessential and should have limited impact on chlamydial growth (19). After inducing dCas9 expression in each transformant, we observed no biologically or statistically significant effect on IFU production at any time point assessed and irrespective of the concentration of aTc used (Fig. 2A and B). In addition, when comparing the growth of the wild-type L2 strain to transformant strains (including the empty vector strain lacking any CRISPRi components), we observed each of the uninduced transformants grew to approximately 80% of the level of the wild type (see Fig. S1A in the supplemental material), likely reflecting the added metabolic burden of replicating and expressing the shuttle vector components (e.g., beta-lactamase, Tet repressor, etc.) in the transformants. These data indicate that dCas9 expression is not toxic to Chlamydia and that, in the presence of the gRNA targeting the incA intergenic region, this too has no deleterious effect on chlamydial growth.
FIG 2.
dCas9 induction effectively represses IncA expression without off-target growth defects. (A and B) McCoy cells were infected with dCas9 transformants with or without an incA-targeting guide RNA (gRNA). Transformants were induced at 4 hours postinfection (hpi) with the various concentrations of aTc and allowed to proceed for 24 hpi, at which time the cells were harvested. The elementary bodies (EBs) were infected onto a fresh monolayer of cells, and this secondary infection was allowed to proceed for 24 h, at which time the inclusion-forming units (IFUs) were counted. No changes between the induced and uninduced control were significant. (C and D) Transformants were induced at 8 hpi using 10 nM aTc. RNA and DNA were isolated at 12 and 24 hpi for quantitative PCR (qPCR). Quantified cDNA was normalized to gDNA and is expressed as induced relative to uninduced samples. Student’s t test was used to compare induced to uninduced samples following log10 transformation, *, P < 0.05. No differences in the clpP2 transcripts were significant. (E and F) Transformants were induced at 4 hpi with 10 nM aTc and allowed to proceed until 24 hpi. Cells were then fixed and stained using primary antibodies to major outer membrane protein (MOMP), ddCpf1 (dCas12), and IncA. Boxes outline cells containing multiple inclusions. All images were acquired on an Axio Imager.Z2 microscope with ApoTome.2 at ×100 magnification. Bars, 10 μm.
We performed reverse transcription-quantitative PCR (RT-qPCR) analysis to test the specific knockdown of incA transcription in the transformant pBOMBLCRia::L2 (incA_IGR) in comparison with the no-gRNA strain pBOMBLCRia::L2 (e.v.). Briefly, McCoy cells were infected with transformants and induced or not with 10 nM aTc at 8 hpi. At 12 hpi and 24 hpi, RNA and DNA samples were collected. The DNA samples were processed and analyzed by qPCR to quantify the genomic C. trachomatis DNA levels. RNA samples were processed and analyzed by RT-qPCR to detect transcript levels of incA and three additional genes, euo (25), clpP2 (26), and omcB (27), representing early, middle, and late stages of development, respectively. RT-qPCR results were then normalized to the genomic DNA levels (28). Results obtained indicated that dCas9 induction in the pBOMBLCRia::L2 (e.v.) showed no effect on any of the genes analyzed (Fig. 2C and D; see also Fig. S2A and B in the supplemental material). However, for the pBOMBLCRia::L2 (incA_IGR) strain, inducing dCas9 expression resulted in a significant decrease only in incA transcripts at 24 hpi (84% reduction in incA transcript levels; Fig. 2C). This, therefore, supports the hypothesis that dCas9 induction in this improved CRISPRi method results in specific knockdown of the targeted gene.
The simplest way to determine the efficacy of our modified CRISPRi system was to assess localization of IncA on the inclusion membrane after inducing knockdown (16, 19). McCoy cells were infected with transformants carrying either the pBOMBLCRia::L2 (incA_IGR) plasmid, encoding the full CRISPRi system, or the pBOMBLCRia::L2 (e.v.) plasmid, which encodes no gRNA expression cassette. The expression of the dCas9 was induced or not at 8 hpi by the addition of 2 nM or 10 nM concentrations of aTc. Infected cells were fixed with 100% methanol at 16 h postinfection (hpi), 24 hpi, and 48 hpi and processed for immunofluorescence for IncA expression, dCas9 expression, and a marker for chlamydiae (major outer membrane protein [MOMP]). As observed in Fig. 2E and F, dCas9 was clearly present within the chlamydiae at 24 hpi, as expected (see also Fig. S3 in the supplemental material for additional time points). Furthermore, IncA expression was clearly localized to the inclusion membrane in the pBOMBLCRia::L2 (e.v.) transformant, thus indicating that the expression of the dCas9 without the gRNA has no impact on IncA expression. For the pBOMBLCRia::L2 (incA_IGR) transformant, the expression of dCas9 in combination with the gRNA targeting the intergenic region upstream of incA blocked the expression of IncA (Fig. 2E and F and Fig. S3). Importantly, the lack of IncA was associated with multiple inclusions per cell, which is the expected phenotype for an IncA-null strain (see boxed cells in Fig. 2F) (13, 19). The combination of these data indicates (i) the successful implementation of the dCas9-based CRISPRi system to knock down expression of a target gene and (ii) the ability to perform population-wide assessments of the impact of knockdown on said gene.
A CRISPRi system based on dCas12 is also capable of inducibly repressing incA gene expression in Chlamydia.
Given the potentially restrictive NNGRRT PAM sequence of the S. aureus dCas9 in the AT-rich C. trachomatis, we sought to evaluate the efficacy of a second dCas ortholog, the Acidoaminococcus dCas12, that uses a TTTV PAM sequence (24). As for the dCas9 system, we investigated the possibility of growth defects being caused by dCas12 expression by performing IFU assays to quantify the production of infectious progeny generated throughout the developmental cycle, RT-qPCR to monitor transcriptional changes, and immunofluorescence assays (IFA) to monitor organism morphology and visualize IncA (Fig. 3).
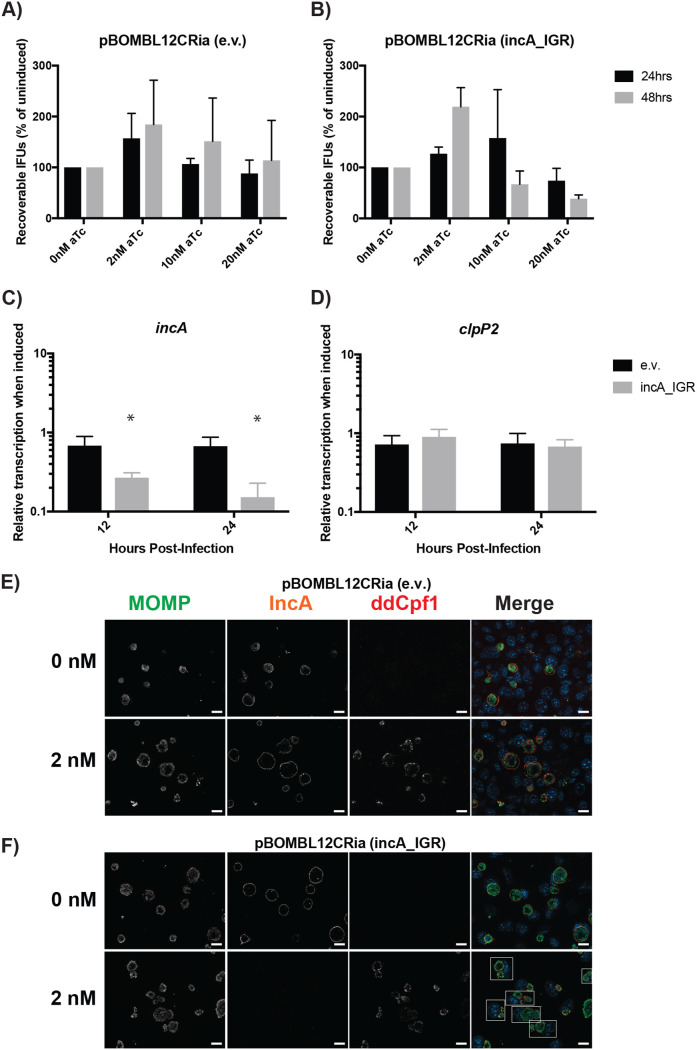
FIG 3.
dCas12 induction effectively represses IncA expression without off-target growth defects. McCoy cells were infected with dCas12 transformants with or without an incA targeting crRNA. (A and B) Transformants were induced at 4 hpi with the designated concentration of aTc and allowed to proceed. Cells were harvested at 24 hpi, and EBs were allowed to infect a fresh monolayer. The secondary infection was allowed to proceed for 24 h before IFUs were enumerated. No changes between the induced cells and the uninduced control were significant. (C and D) Transformants were induced at 8 hpi using 2 nM aTc. RNA and DNA were isolated at 12 and 24 hpi for qPCR. Quantified cDNA was normalized to gDNA and is expressed as induced relative to uninduced. Student’s t test was used to compare induced to uninduced samples following log10 transformation, *, P < 0.05. No differences in the clpP2 transcripts were significant. (E and F) Transformants were induced at 4 hpi with 2 nM aTc and allowed to proceed until 24 hpi. Cells were then fixed and stained using primary antibodies to major outer membrane protein (MOMP), ddCpf1 (dCas12), and IncA. Boxes outline cells containing multiple inclusions. All images were acquired on an Axio Imager.Z2 microscope with ApoTome.2 at ×100 magnification. Bars, 5 μm.
Briefly, McCoy cells were infected with pBOMBL12CRia::L2, with or without the incA_IGR crRNA. Samples were induced with 0, 2, 10, or 20 nM aTc at 4 hpi. At 24 and 48 hpi, samples were fixed for IFA or harvested and used to infect a second monolayer of McCoy cells. The secondary infection progressed for 24 h before IFUs were quantified and enumerated. Induction of dCas12, regardless of the presence of a crRNA, did not have a deleterious effect on chlamydial growth (Fig. 3A and B). However, IFU results and organism morphology became increasingly variable when inducing with 10 nM aTc or greater in these transformants. All further experiments were thus performed using 2 nM aTc to eliminate the possibility that this variability could impact results. As noted above for the dCas9 transformants, these strains grew to at least 70% of the wild-type L2 strain (Fig. S1B).
RT-qPCR was performed to validate the absence of deleterious effects associated with dCas12 expression and to determine incA transcript levels. Cells were infected with transformants and induced or not with 2 nM aTc at 8 hpi. DNA and RNA samples were collected at 12 and 24 hpi. DNA samples were processed and analyzed via qPCR to quantify C. trachomatis gDNA levels. RNA was analyzed using RT-qPCR to detect transcript levels of incA, euo, clpP2, and omcB, as noted above, and normalized to genomic DNA levels. Notably, induction of the transformant lacking a crRNA had no effect on any of the genes analyzed (Fig. 3C and D and Fig. S2C and D). However, induction of the incA_IGR targeting transformant resulted in a significant reduction only in incA transcripts at both 12 (73% decreased) and 24 hpi (85% decreased; Fig. 3C).
To determine whether the reduction in incA transcripts resulted in the loss of IncA at the inclusion membrane, we performed an IFA assay as noted above for the dCas9 system. As for the dCas9 CRISPRi, our IFA results demonstrated a loss of IncA only when pBOMBL12CRia::L2 (incA_IGR) is induced (Fig. 3E and F; see also Fig. S4 in the supplemental material). In addition, the lack of IncA was associated with multiple inclusions per cell, as noted for the dCas9 experiments in Fig. 2 (see boxed cells in Fig. 3F). Taken together, these data support the efficacy of dCas12 as an alternative dCas ortholog to be utilized in CRISPRi gene repression, expanding the possible targets for repression due to the alternative PAM sequence recognized.
Removal of anhydrotetracycline leads to loss of the dCas ortholog staining.
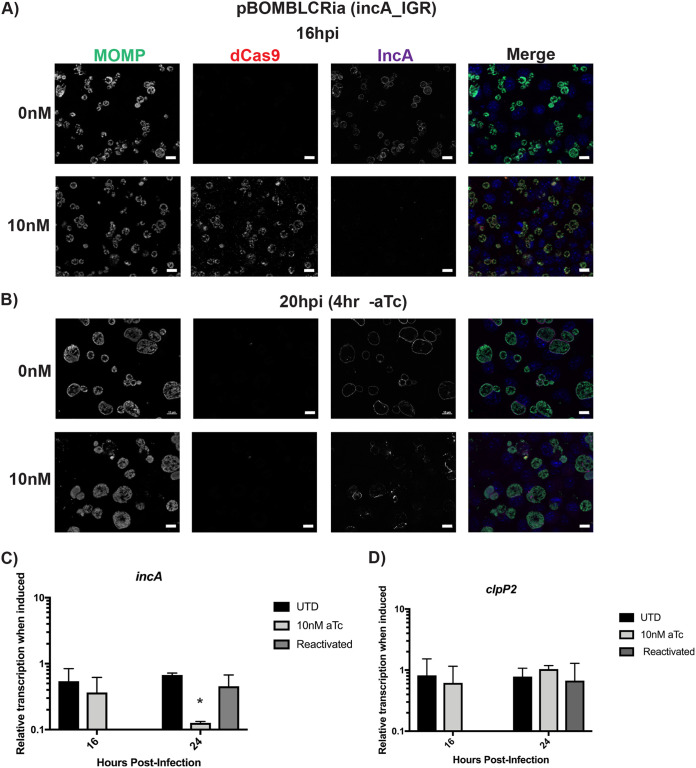
To expand the potential utility of the CRISPRi system, the inhibition of gene expression should ideally be reversible. One modification we made to the CRISPRi system to reduce leaky expression was to append a degradation tag to the dCas ortholog. This tag allows it to be recognized by the ClpXP protease system for degradation (22). Therefore, we next wanted to determine whether removal of the aTc inducer would lead to loss of dCas9/12 labeling by IFA. Briefly, infected cells were induced at 8 hpi for dCas9/12 expression by the addition of 10 nM (dCas9) or 2 nM (dCas12) aTc. At 16 hpi, the medium was aspirated and washed 2 times with 1× Dulbecco’s phosphate-buffered saline (DPBS). New medium lacking aTc was then added to the cells, and cells were subsequently fixed at 20 hpi (for dCas9) or 36 hpi (for dCas12). The samples were processed for immunofluorescence for dCas9/12, IncA, and MOMP. We observed that the removal of aTc led to a rapid loss in dCas9 staining within 4 h (Fig. 4A and B). However, restoration of IncA labeling of the inclusion membrane was less efficient and more heterogeneous (Fig. 4A and B). For dCas12, similar observations were noted at 36 hpi (20 h postreactivation; see Fig. S5 in the supplemental material). It should be noted that, owing to the developmental cycle of Chlamydia, incA transcription declines as secondary differentiation progresses, and loss of IncA does not stop developmental cycle progression (19). When we measured incA transcripts 8 h after removing aTc from the dCas9 cultures, we observed a recovery of incA transcript levels but no effect on the unrelated clpP2 transcript levels (Fig. 4C and D).
FIG 4.
Removal of aTc leads to the loss of dCas9. McCoy cells were infected with dCas9 transformants with an incA-targeting gRNA. Transformants were induced at 4 hpi with 10 nM aTc and allowed to proceed until 16 hpi (A). At 16 hpi a subset of the samples was washed with Dulbecco’s phosphate-buffered saline (DPBS) and fixed with 100% MeOH (B). For the next subset of samples, the medium was aspirated and the cells washed with DPBS. Fresh medium without any aTc was added, and the infection was allowed to proceed for an additional 4 h and then fixed with 100% MeOH. All fixed samples were stained using primary antibodies to major outer membrane protein (MOMP), IncA, and dCas9. All images were acquired on an Axio Imager.Z2 microscope with ApoTome.2 at ×100 magnification. Bars, 10 μm. (C and D). Transcript levels for incA and clpP2 were measured at the indicated times from the indicated culture conditions as described in the legend of Fig. 2. Student’s t test was used to compare induced to uninduced samples following log10 transformation, *, P < 0.05.
Expression of incA_FLAG transcriptionally fused to dCas9 restores IncA expression.
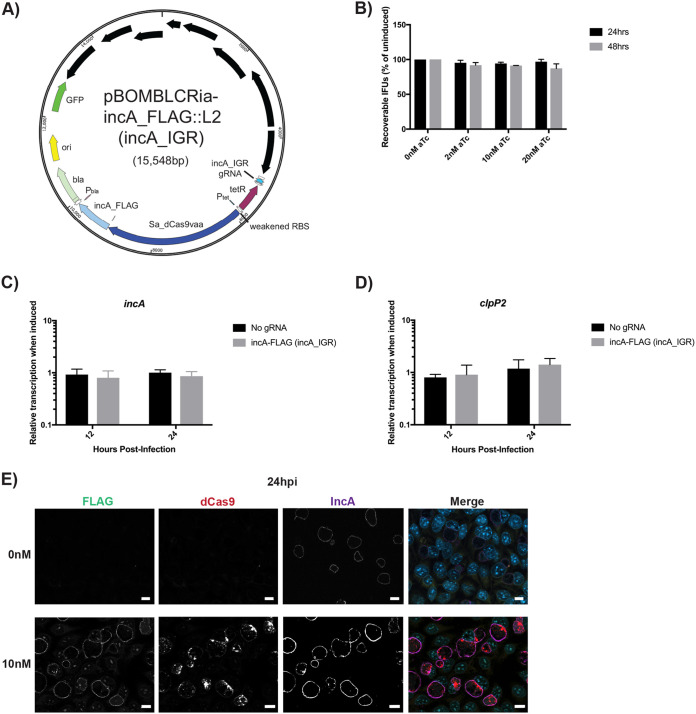
Molecular Koch’s postulates require that a given phenotype be investigated by disrupting, and then replacing, a gene hypothesized to control said phenotype (29). In the case of an inducible CRISPRi knockdown system, fulfilling molecular Koch’s postulates would require expressing an additional copy of the target gene, ideally in an inducible manner, during dCas9/12 induction, without its being targeted by the gRNA or crRNA. To accomplish this, we created a transcriptional fusion of dCas9 with incA_FLAG and created the plasmid pBOMBLCRia-incA_FLAG (incA_IGR) (Fig. 5A). The incA_FLAG allele is resistant to the knockdown effect because the gRNA targets the chromosomal intergenic region 5′ to incA. Thus, the plasmid copy will be transcribed and translated independently of the incA promoter. This plasmid was subsequently transformed into C. trachomatis L2.
FIG 5.
Induction of dCas9 transcriptionally fused to incA_FLAG functionally complements IncA expression at the inclusion membrane. (A) Illustration of the plasmid map for pBOMBLCRia-incA_FLAG::L2 (incA_IGR). The plasmid map was generated using SnapGene Viewer. (B) Cells were infected with the transformant, and expression of the Sa_dCas9 and incA_FLAG was induced at 4 hpi with the indicated concentrations of aTc. At 24 hpi, the cells were harvested. EBs were infected onto a fresh monolayer of cells, and this secondary infection was allowed to proceed for 24 h, at which time the IFUs were counted. (C and D) Cells were infected with the transformant, and expression of the Sa_dCas9 and incA_FLAG was induced at 8 hpi using 10 nM aTc. RNA and DNA were isolated at 12 and 24 hpi for qPCR. Quantified cDNA was normalized to gDNA and is expressed as induced relative to uninduced samples. Student’s t test was used to compare induced to uninduced samples following Log10 transformation, *, P < 0.05. (E) Cells were infected with the transformant, and expression of the Sa_dCas9 and incA_FLAG was induced at 4 hpi with 10 nM aTc and allowed to proceed until 24 hpi. Cells were then fixed and stained using primary antibodies to FLAG, dCas9, and IncA. All images were acquired on an Axio Imager.Z2 microscope with ApoTome.2 at ×100 magnification. Bars, 10 μm.
To test whether there was any effect of dCas9 and IncA_FLAG expression on bacterial growth, we measured the production of infectious progeny (i.e., EBs) from an infected cell culture by an IFU assay. Cells were infected with the transformants, and various concentrations of aTc were used to induce dCas9 and incA_FLAG expression at 4 hpi. At the designated time points (i.e., 24 and 48 hpi), samples were collected and subsequently titrated onto fresh cell monolayers, and the infection was allowed to proceed for a further 24 h. The number of inclusions were quantified from this secondary infection. We observed that there was no effect on IFU production at any time point assessed, irrespective of the concentration of aTc used (Fig. 5B and Fig. S1A). We then performed RT-qPCR to measure transcriptional changes as described above. The cells were infected with pBOMBLCRia-incA_FLAG (incA_IGR) transformants and induced or not with 10 nM aTc at 8 hpi. DNA and RNA samples were collected at 12 and 24 hpi. Transcript levels of incA, clpP2, euo, and omcB were normalized to genomic DNA levels. Results indicated that the induction of the transformant restored incA transcript levels and had no effect on any of the other genes analyzed (Fig. 5C and D; see also Fig. S6 in the supplemental material). To test for IncA expression during dCas9 induction, we infected cells and induced the expression of dCas9 and incA_FLAG at 8 hpi by the addition of 10 nM aTc. The infection was allowed to proceed until 24 hpi, at which time point the cells were fixed and processed for immunofluorescence for IncA, dCas9, and FLAG expression. We observed that IncA was clearly present within both induced and uninduced samples whereas FLAG staining was visible in the induced cultures only (Fig. 5E and Fig. S6). Furthermore, we did not observe multiple inclusions per cell, as was apparent in the knockdown strains (Fig. 2 and 3), thus highlighting that the phenotype of incA knockdown was restored by complementation. We conclude from these data that inducibly knocked down gene expression can be complemented by expressing an additional copy of the target gene as a transcriptional fusion with dCas9.
DISCUSSION
In evolving to its obligate intracellular niche, Chlamydia has significantly reduced its chromosomal gene content. For example, the human pathogen C. trachomatis encodes roughly 900 open reading frames (4). Given its limited gene repertoire, its obligate dependence on a host cell, and its unique developmental cycle, it is likely that many genes in Chlamydia will prove to be essential. Indeed, many of these essential genes may confer to Chlamydia its unique traits. Traditional approaches to study gene function in model bacterial organisms rely on generating a knockout, conditional or otherwise, and then complementing the phenotype by expressing the gene under investigation elsewhere on the chromosome or on a plasmid. Although recent advances in Chlamydia genetic manipulations have allowed for the generation of targeted knockouts of nonessential genes (7, 8), there is not yet a facile means for generating a conditional knockout for essential genes. This has hindered our ability to explore chlamydial biology more directly.
To circumvent this limitation, we previously reported on the feasibility of adapting CRISPR interference to inducibly knockdown gene expression in C. trachomatis (16). In the current study, we demonstrate the efficacy of two different CRISPRi systems to inducibly repress gene expression. We show that both systems, based on dCas9 and dCas12 orthologs, are capable of reducing transcript levels of the target gene with a concurrent reduction in its protein levels as assessed by immunofluorescence analysis. Interestingly, the dCas12 knockdown effect was detectable at 12 hpi (4 h postinduction), which was not the case for the dCas9 knockdown. However, each system is targeting a different sequence and strand; therefore, it is difficult to directly compare efficiencies of knockdown in this context. Whether dCas12 is more rapid in its effect will be revealed as more targets are assessed. Notably, both systems reduced transcripts more than 80% at the later time point. Importantly, no negative impact on chlamydial growth was associated with expressing the dCas9 or dCas12 protein when induced with concentrations of aTc that also blocked target gene expression.
We further extended the utility of the CRISPRi system in two ways. The first modification we implemented was to append a degradation tag to render the dCas ortholog unstable. This allowed for its rapid disappearance after removing the aTc inducer. This should allow, in cases where chlamydial developmental cycle progression is halted by inhibiting a specific gene’s expression, the ability to “reactivate” gene expression and monitor recovery of growth. Given that incA is a dispensable gene and that its transcription peaks during the logarithmic RB growth phase (19, 30), we were unable to successfully recover high levels of IncA on the inclusion membrane, since its transcription is already declining during the period when we “reactivated” its expression. These data suggest that expression and secretion of Inc proteins are tightly regulated such that, even with recovery of transcript levels, IncA was not efficiently detected on the inclusion membrane when its gene transcription was delayed due to CRISPR interference. What these data indicate about type III secretion function more generally requires further investigation. Importantly, our data demonstrate the loss of the dCas ortholog after removal of the aTc inducer, thus indicating the feasibility of restoring transcription levels and, potentially protein levels, in cases where the developmental cycle is halted or delayed.
The second modification we implemented was to functionally complement knockdown by coexpressing the target gene (incA) as a transcriptional fusion with the dCas9 ortholog. The gRNA target sequence for incA that we used is within its upstream 5′ intergenic region, and thus there was no risk that the CRISPRi system would target the plasmid copy. However, we envision scenarios where a coding sequence is directly targeted by a gRNA or a crRNA and, in these instances, complementation should be feasible by making synonymous changes within the plasmid copy to prevent gRNA/crRNA binding. Indeed, we have successfully implemented this approach in the study of a novel bacterial morphology determinant that is the second gene within an operon (34).
Our demonstration of the efficacy of two different dCas orthologs to inducibly repress gene expression in Chlamydia expands the utility of the CRISPRi approach in this obligate intracellular pathogen. This is potentially important given the constraints in PAM sequence recognition by a given dCas ortholog. In the case of the S. aureus dCas9, its PAM sequence is NNGRRT (23). When designing gRNA sequences, we have observed instances where a suitable PAM is not present on the nontemplate strand in the intergenic region or the 5′ coding region of a target ORF (data not shown). The use of the Acidoaminococcus dCas12, with its PAM sequence of TTTV (24), should circumvent this limitation given the greater instances of this PAM in the chromosome. In total, both dCas systems target almost 80,000 sites (see Table S4 in the supplemental material). Given that the size of the C. trachomatis chromosome is ∼1 Mbp and those of the gRNA or crRNA are 20 or 21 bp, a simplistic calculation would indicate a maximum 1.5× coverage of the entire genome. Anecdotally, we have been able to find a target sequence covered by either the dCas9 or dCas12 system in every ORF or upstream intergenic region we have analyzed (>50 of 895 ORFs). In principle, and as described in the original CRISPRi study in E. coli (16), targeting of either strand within the promoter region is favored for transcriptional knockdown, whereas targeting of the nontemplate strand in the coding region is necessary for the same effect. Our vectors were constructed to test the former strategy (see also reference 16), but we have successfully used the latter to target the second gene within an operon (Brockett, Lee, et al., unpublished data).
In eukaryotic systems, the use of CRISPR to generate knockouts comes with the potential for off-target cutting events, as only a single cut on a given chromosome is required to generate a knockout (31). Such off-target effects for CRISPR interference are less concerning, due to the necessity of having the dCas ortholog remain bound to the chromosomal target sequence at all times with high enough affinity to block transcription (32). In Chlamydia, off-target effects of CRISPRi are further mitigated by its small genome size. In addition, most off-target effects have gRNA binding sites with at most 3-bp mismatches (31). For the dCas9 system, the incA-targeting gRNA showed a maximum homology of 14 of 21 bp to a sequence in recB but without a nearby PAM sequence. For the dCas12 system, the incA-targeting crRNA displayed a maximum homology of 11 of 20 bp to multiple sequences but with no PAM sequence near these chromosomal sites. In designing gRNAs or crRNAs, we have yet to encounter homology greater than the 14 of 21 bp we described for the incA-targeting gRNA. Therefore, the probability of blocking transcription elsewhere on the chromosome is extremely low, since this would require at least a 7-bp mismatch and an appropriate PAM in the off-target sequence.
Potential limitations of using CRISPRi could arise in Chlamydia if targeting an entire operon, yet our demonstration that a target ORF could be complemented as a transcriptional fusion with the dCas9 suggests a possible means to address this. For example, in a three-gene operon, XYZ, if the entire operon is knocked down, then gene X could be investigated by complementing back YZ. Similarly, gene Y could be investigated by complementing back XZ. As CRISPRi is widely implemented in the field, the success of such strategies should become apparent. Another potential limitation is the degree to which the dCas ortholog blocks transcription. We have observed transcript levels reduced by 80 to 90%; thus, for highly transcribed genes, this may not be sufficient to reveal a phenotypic effect. Worth noting, however, is that this level of knockdown in eukaryotes using small interfering RNA (siRNA) is sufficient to ablate protein expression in a system where transcription and translation are not coupled as occurs in prokaryotes (33). Finally, the lack of antibodies against a target protein could hamper direct measurement of a phenotype and may require monitoring chlamydial microbiology by transcriptome or proteome assessments. This is one reason we chose to investigate IncA as a target, since we possess an antibody that recognizes it and it has a well-characterized phenotype when knocked out (i.e., lack of inclusion fusion [18]). In spite of these potential limitations, CRISPRi should add a powerful tool to our ability to dissect gene function in Chlamydia.
In sum, we have demonstrated the application of CRISPRi to inducibly knock down gene expression in Chlamydia. This approach should allow for the study of essential gene function in this unique bacterium and will greatly accelerate our understanding of chlamydial biology. We recently demonstrated such utility in the study of the essential clpP2X operon (22).
MATERIALS AND METHODS
Plasmid construction.
All primer sequences and plasmid descriptions can be found in Table S1 in the supplemental material.
The original pBOMB4-Tet::L2 plasmid (a kind gift of T. Hackstadt, Rocky Mountain Labs, NIH [18]) was modified to reduce its ribosome binding efficiency by first digesting with AatII and PstI and treating with alkaline phosphatase (FastDigest enzymes; Fermentas, Thermo Fisher, Grand Island, NY). PCR products encoding tetR with its promoter and mCherry with overlapping segments encoding a reduced ribosome binding efficiency site were amplified from pBOMB4-Tet::L2 using Phusion DNA polymerase (New England Biolabs, Ipswich, MA) and purified using a Qiagen (Germantown, MD) PCR purification kit according to the manufacturers’ guidelines. The digested pBOMB4 plasmid was mixed with the PCR products in an NEBuilder HiFi DNA assembly (NEB) reaction following the manufacturer’s guidelines and transformed into chemically competent E. coli 10-β (NEB) using standard techniques. The resulting colonies were screened for the correct plasmid, pBOMBL::L2, which was isolated by miniprep (Qiagen) and verified by restriction digestion and sequencing across the promoter region and ribosome binding site.
The dCas9 gene from Staphylococcus aureus was PCR-amplified with Phusion DNA polymerase using the plasmid pX603-AAV-CMV::NLS-dSaCas9(D10A,N580A)-NLS-3xHA-bGHpA (a gift of F. Zhang; Addgene plasmid 61594 [23]) as the template. The purified PCR product was inserted into the EagI- and KpnI-digested plasmid pBOMBL::L2, which was also treated with alkaline phosphatase, using NEBuilder HiFi DNA assembly kit. The product was transformed into chemically competent E. coli 10-β using standard techniques. The resulting colonies were screened for the correct plasmid, pBOMBL-Sa_dCas9vaa::L2 [also referred to as pBOMBLCRia::L2 (e.v.)], as described above. The gRNA cassette targeting the 5′ region upstream of incA was synthesized by Integrated DNA Technologies (IDT, Coralville, IA) as a synthetic double-stranded DNA (gBlock), the sequence of which is listed in the supplemental material. The gRNA gBlock was used directly in an NEBuilder HiFi DNA assembly reaction with BamHI-digested and alkaline phosphatase-treated pBOMBL-Sa_dCas9vaa::L2. The resulting plasmid, pBOMBLCRia::L2 (incA_IGR), was transformed into competent E. coli 10-β, isolated, and confirmed as described above.
The dCas12 (ddAsCpf1) gene from Acidaminococcus was PCR amplified with Phusion DNA polymerase using the plasmid pXX55-ddAsCpf1 (a kind gift of J. Wang [24]) as the template. The PCR product was inserted into EagI- and KpnI-digested plasmid pBOMBL::L2 as described above to create the plasmid pBOMBL-ddCpf1vaa::L2 [also referred to as pBOMBL12CRia::L2 (e.v.)]. The crRNA cassette targeting the 5′ region upstream of incA was synthesized by IDT and directly inserted into BamHI-digested pBOMBL-ddCpf1vaa::L2 as noted above for the dCas9 system to create the plasmid pBOMBL12CRia::L2 (incA_IGR).
To complement the incA knockdown in the dCas9 system, incA was PCR amplified from C. trachomatis L2 genomic DNA. The reverse primer added a 3′ FLAG tag to the product and contained overlapping regions with Sa_dCas9vaa, which was itself amplified to encode overlapping regions with incA and an internal ribosome binding site for a transcriptional fusion product. The PCR products were inserted into EagI- and KpnI-digested plasmid pBOMBL::L2 as noted above to create the plasmid pBOMBLCRia-incA_FLAG::L2 (incA_IGR).
Organism and cell culture.
The plasmidless (−pL2) strain of C. trachomatis serovar L2 (a kind gift of I. Clarke) was propagated in McCoy cells as described elsewhere for use in transformations (12). The murine fibroblast cell line McCoy (a kind gift of H. Caldwell, NIH) and the human cervical epithelial HeLa cell line were routinely cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco/Thermo Fisher) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 10 μg/ml gentamicin (Gibco/Thermo Fisher) and incubated at 37°C with 5% CO2. Bacterial and eukaryotic cell cultures are routinely tested for Mycoplasma contamination using LookOut Mycoplasma PCR detection kit (Millipore-Sigma, St. Louis, MO) according to the manufacturer’s instructions.
Chlamydial transformation.
Transformations were performed as described elsewhere using demethylated plasmids prepared from the dam− dcm− strain of E. coli (NEB) (12). Briefly, 2 μg of demethylated plasmid was added to 2.5 × 106 C. trachomatis serovar L2 -pL2 in Tris-CaCl2 buffer and incubated at room temperature for 30 min (14). McCoy cells plated the day before at 106 per well in a 6-well plate were subsequently infected with the transformation mix (one well per transformation). Penicillin (1 U/ml) and cycloheximide (1 μg/ml) (MilliporeSigma) were added at 8 h postinfection (hpi). Infected cells were harvested at 48 hpi, centrifuged at 17,000 × g for 30 min at 4°C, and resuspended in 1 ml of Hanks’ balanced salt solution (HBSS; Gibco). The suspension was centrifuged for 5 min at 400 × g at 4°C, and the supernatant was added to 1 ml HBSS to infect a new monolayer of McCoy cells. This process of infecting and harvesting infected cells was repeated until wild-type inclusions were visible and the penicillin-resistant bacteria were isolated.
Inclusion forming unit assay.
McCoy cells were routinely cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco) at 37°C with 5% CO2 supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT). All infected and uninfected cell cultures were incubated at these conditions. Cells were plated in 24‐well culture plates at a density of 1 × 105 cells per well. Glass coverslips were placed in one well subset of the 24-well culture plate for immunofluorescence microscopy. Cells were infected with transformed C. trachomatis L2 strains, i.e., pBOMBL::L2, pBOMBL-Sa_dCas9vaa::L2, pBOMBLCRia::L2 (incA_IGR), pBOMBLCRia-incA_FLAG (incA_IGR), pBOMBL-ddCpf1vaa::L2, and pBOMBL12CRia::L2 (incA_IGR) at a multiplicity of infection of 1. The cells were centrifuged at 400 × g for 15 min in the Eppendorf 5810R centrifuge and cultured at 37°C with 5% CO2. Samples were induced with increasing concentrations of aTc, i.e., 0 nM, 2 nM, 10 nM, and 20 nM at 4 h postinfection and harvested at 24 hpi and 48 hpi. At the designated time points, cell lysates, which contain elementary bodies (EBs), were collected from infected McCoy cell cultures by scraping cells into 2-sucrose-phosphate (2SP) solution. The resulting mixture was frozen at −80°C and used to infect a fresh McCoy cell monolayer in a series of 10-fold dilutions. The secondary infection was allowed to progress for 24 h prior to fixation with 4% paraformaldehyde. Labeling for immunofluorescence microscopy of the organisms was done using primary goat anti-MOMP antibody and a secondary donkey anti-goat antibody labeled with Alexa Fluor 594. Titers were enumerated by calculating the total number of inclusions per field based on counts from 15 fields of view for each triplicate well, giving a total of 45 fields of view per experiment. Three independent replicates were performed, and the totals for each experiment were averaged. Results were normalized as a percentage of the uninduced samples for each time point. Student’s two-tailed t test to compare the induced samples to the uninduced samples was performed using the averages of each biological replicate. No statistical differences (i.e., P > 0.05) were observed for any IFU experiments using Student’s t test.
Indirect immunofluorescence microscopy.
Cells were cultured on glass coverslips in 24-well tissue culture plates and infected with C. trachomatis L2 strains at a multiplicity of infection (MOI) of 2. At 8 hpi, the samples were induced with various concentrations of aTc, i.e., 0 nM, 2 nM, 10 nM, and 20 nM. At the designated time points the samples were then washed three times with DPBS and fixed with 100% methanol for 1 min. For a subset of the samples, at 16 hpi, the DMEM medium was aspirated and washed three times with DPBS and new DMEM added without aTc and cultured until the designated time points for fixation. Organisms were stained using a primary mouse antibody specific to C. trachomatis major outer membrane protein (MOMP), primary sheep antibody specific to IncA, primary rabbit antibody specific to Sa-dCas9 (Abcam, Cambridge, MA). DAPI (4′,6-diamidino-2-phenylindole) was added to visualize DNA. For the pBOMBLCRia-incA_FLAG::L2 (incA_IGR) strain, a primary mouse antibody specific for FLAG (Millipore-Sigma) was also used in conjunction with a secondary donkey anti-mouse antibody conjugated to Alexa Fluor 488 (Jackson Laboratory). The samples were observed with a Zeiss Imager.Z2 equipped with an Apotome.2 using a 100× lens objective.
Transcriptional analyses.
RNA extraction was performed on infected cell monolayers using TRIzol (Invitrogen/Thermo Fisher). DNA contamination was removed by using Turbo DNAfree (Ambion/Thermo Fisher) according to manufacturer’s instructions. DNA-free RNA was converted to cDNA using random nonamers (New England Biolabs, Ipswich, MA) and SuperScript III RT (Invitrogen/Thermo Fisher) per the manufacturer’s instructions. Reaction end products were diluted 10-fold with molecular biology grade water and stored at −80°C. Equal volumes of cDNA reaction mixture were used with SYBR green master mix (Applied Biosystems) and quantified by QuantStudio 3 (Applied Biosystems/Thermo Fisher) using the standard amplification cycle with a melting curve analysis. Results were compared to a standard curve generated against purified C. trachomatis L2 genomic DNA. Replicate wells were harvested for DNA extraction using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). Chlamydial genomes were quantified by using equal total DNA quantities in qPCR with a groEL1 primer set. Resulting genomic values were used to normalize respective transcript data (27, 28). RT-qPCR results were normalized for efficiency with typical results demonstrating an r2 of >0.995 and efficiencies greater than 90%. Student’s t test was used to compare the normalized transcript levels of induced and uninduced samples following log10 transformation. Data with an asterisk (*) indicate a P value of <0.05.
ACKNOWLEDGMENTS
We acknowledge E. Rucks (UNMC), T. Hackstadt (RML/NIAID), H. Caldwell (NIH), I. Clarke (University of Southampton), and J. Wang (Shanghai Institutes for Biological Sciences) for reagents.
S.P.O. and colleagues were supported by funding from NIAID (grant 1R21AI141933-01) at the National Institutes of Health and a CAREER award (1810599) from the National Science Foundation.
Footnotes
Supplemental material is available online only.
Contributor Information
Scot P. Ouellette, Email: scot.ouellette@unmc.edu.
Craig R. Roy, Yale University School of Medicine
REFERENCES
- 1.Satterwhite C, Gottlieb S, Romaguera R, Bolan G, Burstein G, Schuler C, Popovic T. 2011. CDC Grand Rounds: Chlamydia prevention: challenges and strategies for reducing disease burden and sequelae. MMWR Morb Mortal Wkly Rep 60:370–373. [PubMed] [Google Scholar]
- 2.Anonymous. 2019. U.S. STDs break record. Science 366:287–288. [Google Scholar]
- 3.Mabey DCW, Solomon AW, Foster A. 2003. Trachoma. Lancet 362:223–229. 10.1016/S0140-6736(03)13914-1. [DOI] [PubMed] [Google Scholar]
- 4.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 5.Abdel-Rahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Moore ER, Ouellette SP. 2014. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front Cell Infect Microbiol 4:157. 10.3389/fcimb.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigalova OM, Chaplin AV, Bochkareva OO, Shelyakin PV, Filaretov VA, Akkuratov EE, Burskaia V, Gelfand MS. 2019. Chlamydia pan-genomic analysis reveals balance between host adaptation and selective pressure to genome reduction. BMC Genomics 20:710. 10.1186/s12864-019-6059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf K, Betts HJ, Chellas-Géry B, Hower S, Linton CN, Fields KA. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol Microbiol 61:1543–1555. 10.1111/j.1365-2958.2006.05347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stone CB, Bulir DC, Emdin CA, Pirie RM, Porfilio EA, Slootstra JW, Mahony JB. 2011. Chlamydia pneumoniae CdsL regulates CdsN ATPase activity, and disruption with a peptide mimetic precents bacterial invasion. Front Microbiol 2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackstadt T, Baehr W, Ying Y. 1991. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc Natl Acad Sci U S A 88:3937–3941. 10.1073/pnas.88.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nogueira AT, Braun KM, Carabeo RA. 2017. Characterization of the growth of Chlamydia trachomatis in in vitro-generated stratified epithelium. Front Cell Infect Microbiol 7:438. 10.3389/fcimb.2017.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CM, Fisher DJ. 2013. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One 8:e83989. 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller KE, Wolf K, Fields KA. 2016. Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. mBio 7:e01817-15. 10.1128/mBio.01817-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, LaBrie SD, Carrell SJ, Suchland RJ, Dimond ZE, Kwong F, Rockey DD, Hefty PS, Hybiske K. 2019. Development of transposon mutagenesis for Chlamydia muridarum. J Bacteriol 201:e00366-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouellette SP. 2018. Feasibility of a conditional knockout system for Chlamydia based on CRISPR interference. Front Cell Infect Microbiol 8:59. 10.3389/fcimb.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suchland RJ, Rockey DD, Bannantine JP, Stamm WE. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect Immun 68:360–367. 10.1128/iai.68.1.360-367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn JM, Neher SB, Kim Y-I, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11:671–683. 10.1016/S1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 21.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823:15–28. 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood NA, Blocker AM, Seleem MA, Conda-Sheridan M, Fisher DJ, Ouellette SP. 2020. The ClpX and ClpP2 orthologs of Chlamydia trachomatis perform discrete and essential functions in organism growth and development. mBio 11:e02016-20. 10.1128/mBio.02016-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. 2015. Crystal structure of Staphylococcus aureus Cas9. Cell 162:1113–1126. 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Wang J, Cheng Q, Zheng X, Zhao G, Wang J. 2017. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov 3:17018. 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wichlan DG, Hatch TP. 1993. Identification of an early-stage gene of Chlamydia psittaci. J Bacteriol 175:2936–2942. 10.1128/jb.175.10.2936-2942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood NA, Chung KY, Blocker AM, Rodrigues de Almeida N, Conda-Sheridan M, Fisher DJ, Ouellette SP. 2018. Initial characterization of the two ClpP paralogs of Chlamydia trachomatis suggests unique functionality for each. J Bacteriol 201:e00635-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouellette SP, Hatch TP, AbdelRahman YM, Rose LA, Belland RJ, Byrne GI. 2006. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol Microbiol 62:1387–1401. 10.1111/j.1365-2958.2006.05465.x. [DOI] [PubMed] [Google Scholar]
- 28.Ouellette SP, AbdelRahman YM, Belland RJ, Byrne GI. 2005. The Chlamydia pneumoniae type III-secretion related lcrH gene clusters are developmentally expressed operons. J Bacteriol 187:7853–7856. 10.1128/JB.187.22.7853-7856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falkow S. 1988. Molecular Koch’s postulates applied to microbial pathogenicity. Rev Infect Dis 10:S274–276. 10.1093/cid/10.Supplement_2.S274. [DOI] [PubMed] [Google Scholar]
- 30.Gauliard E, Ouellette SP, Rueden KJ, Ladant D. 2015. Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis. Front Cell Infect Microbiol 5:13. 10.3389/fcimb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konerman S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. 2013. DNA targeting specifity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832. 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Y, Shen W, Zhang J, Yang B, Liu Y-N, Qi H, Yu X, Lu S-Y, Chen Y, Xu Y-Z, Li Y, Gage FH, Mi S, Yao J. 2018. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat Neurosci 21:447–454. 10.1038/s41593-018-0077-5. [DOI] [PubMed] [Google Scholar]
- 33.Holmes K, Williams CM, Chapman EA, Cross MJ. 2010. Detection of siRNA induced mRNA silencing by RT-qPCR: considerations for experimental design. BMC Res Notes 3:53. 10.1186/1756-0500-3-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brockett MR, Lee J, Cox JV, Liechti GW, Ouellette SP. 2021. A dynamic, ring-forming bactofilin critical for maintaining cell size in the obligate intracellular bacterium Chlamydia trachomatis. Infect Immun. 10.1128/IAI.00203-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download IAI.00108-21-s0001.pdf, PDF file, 124 KB (123.6KB, pdf)
Table S2. Download IAI.00108-21-s0002.pdf, PDF file, 2.85 MB (2.9MB, pdf)
Table S3. Download IAI.00108-21-s0003.pdf, PDF file, 3.40 MB (3.4MB, pdf)
Table S4. Download IAI.00108-21-s0004.pdf, PDF file, 4.35 MB (4.4MB, pdf)
Fig. S1 to S6. Download IAI.00108-21-s0005.pdf, PDF file, 1.15 MB (1.2MB, pdf)