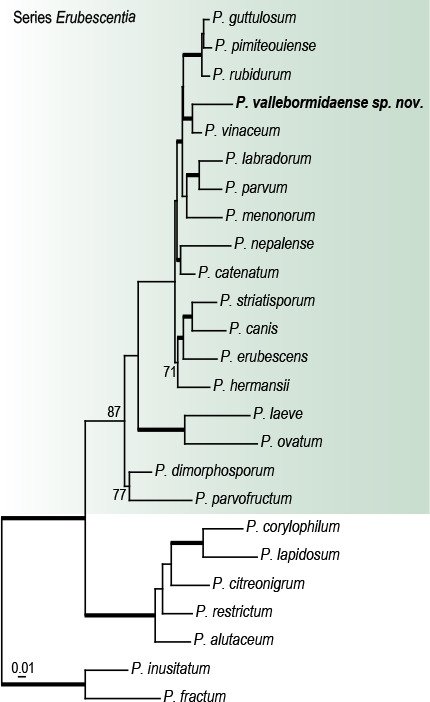
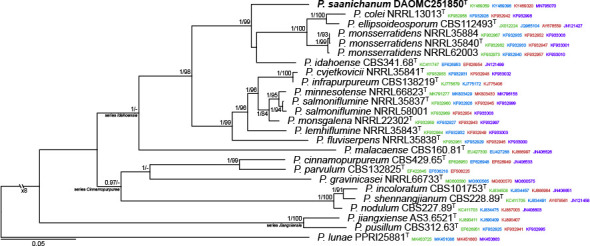
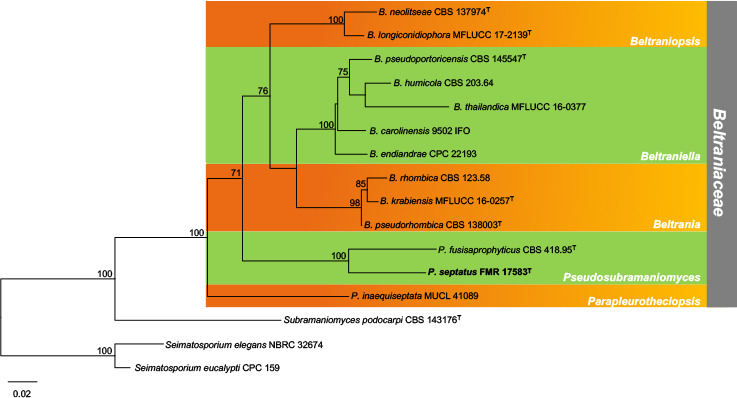
Abstract
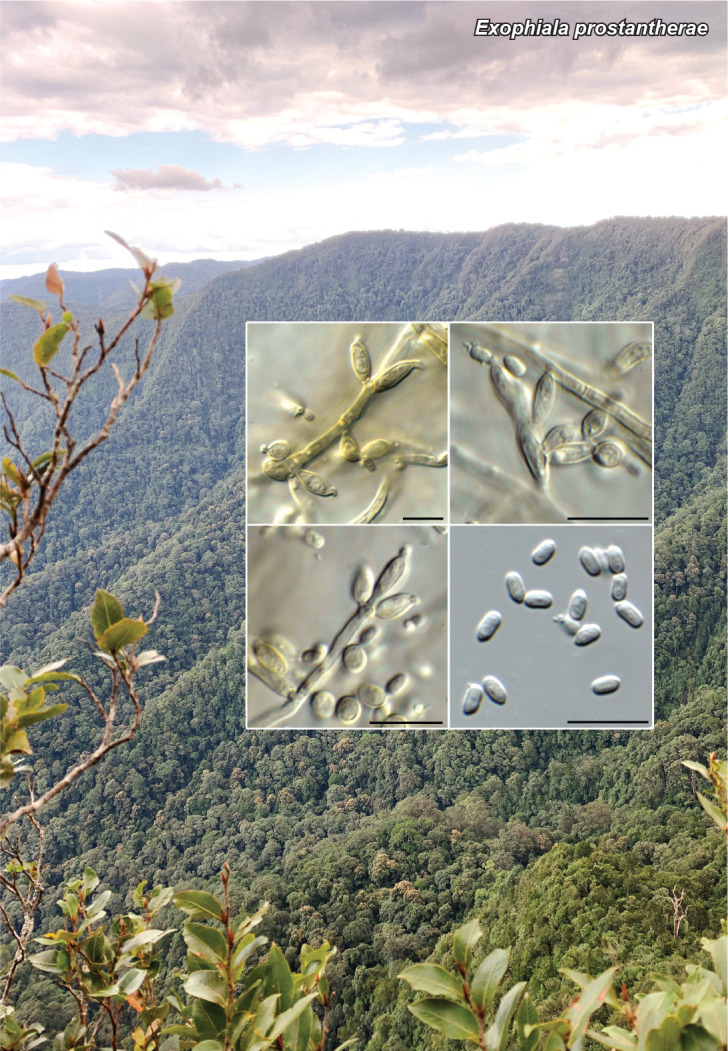
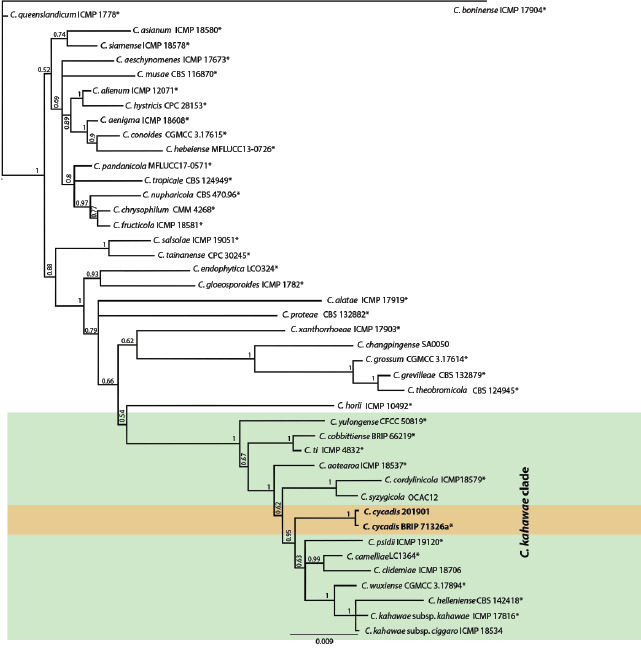
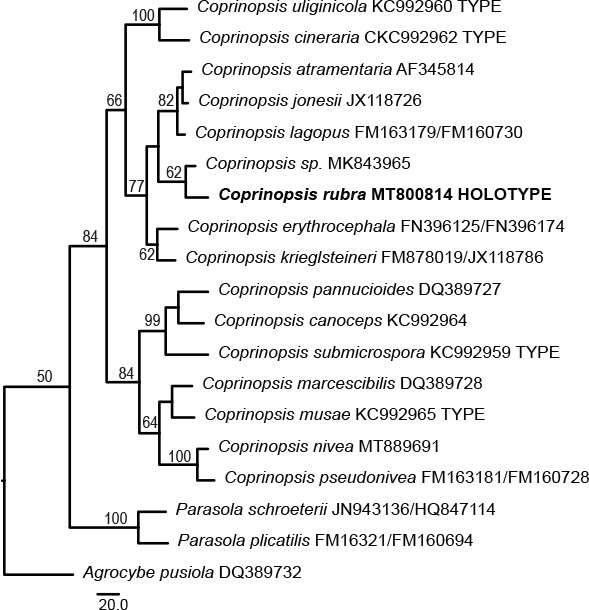
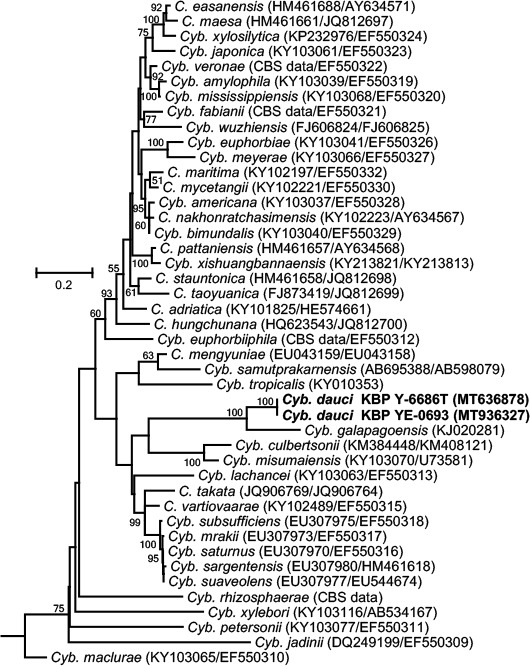
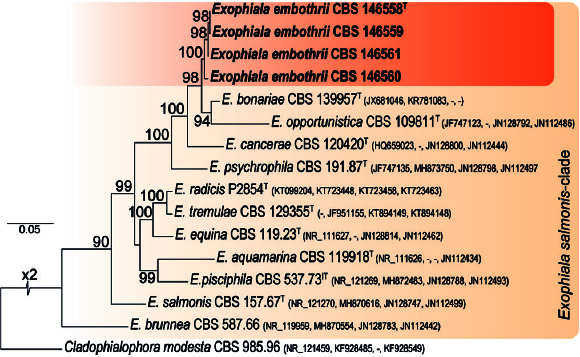
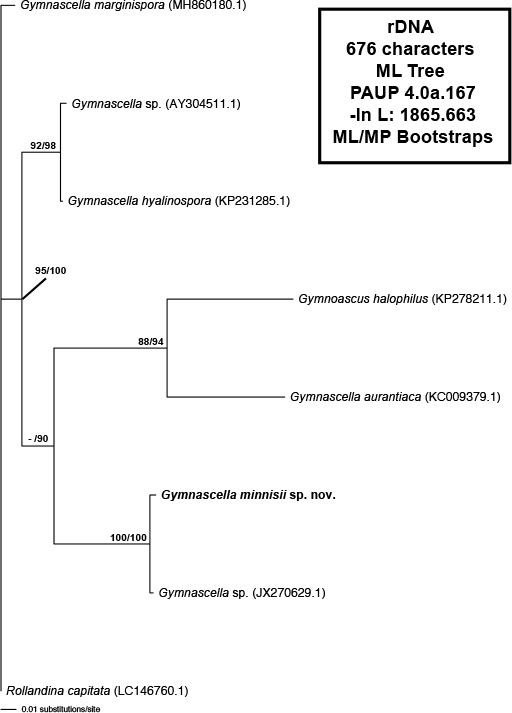
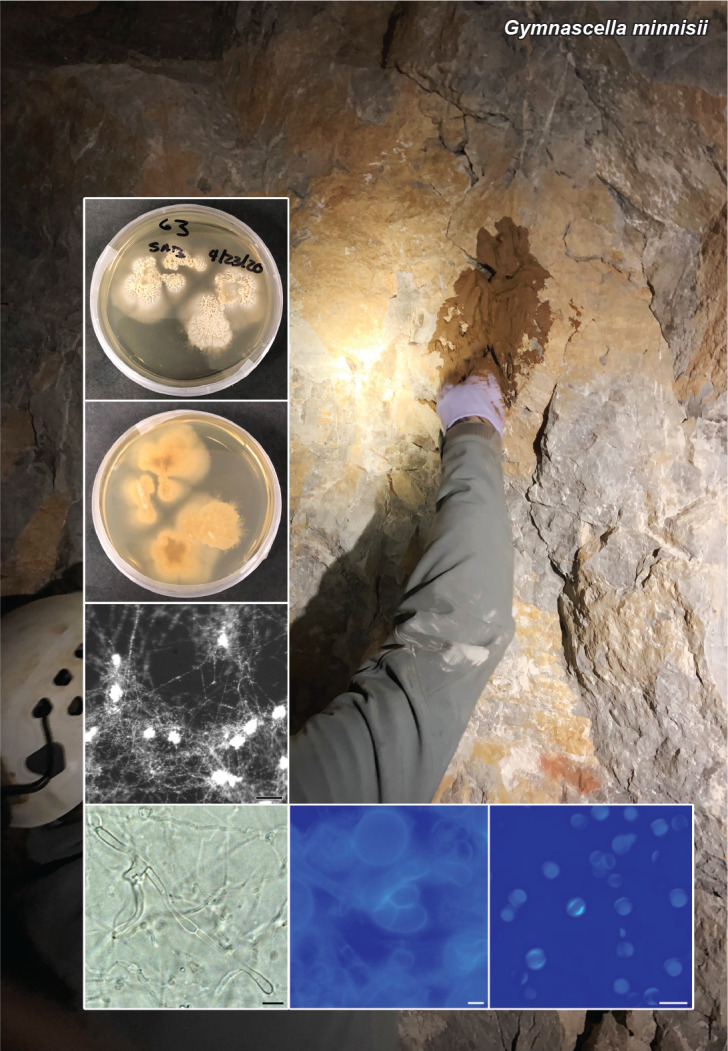
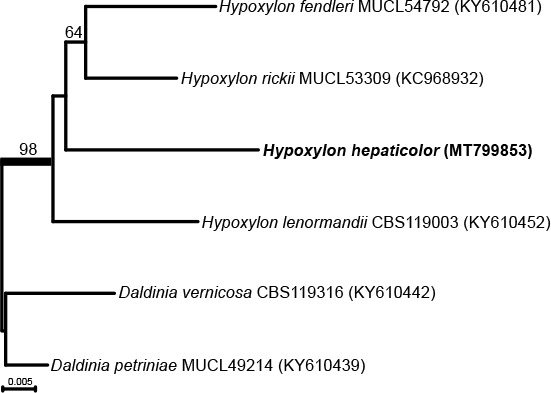
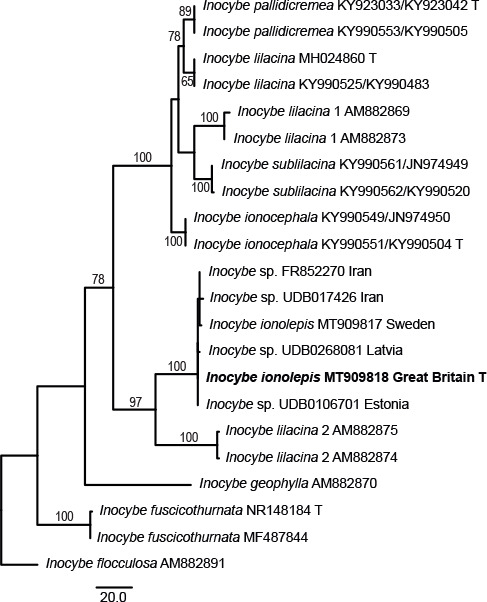
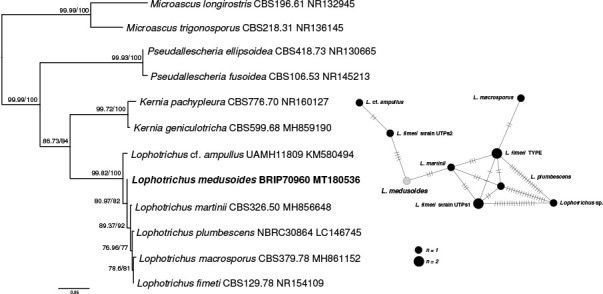
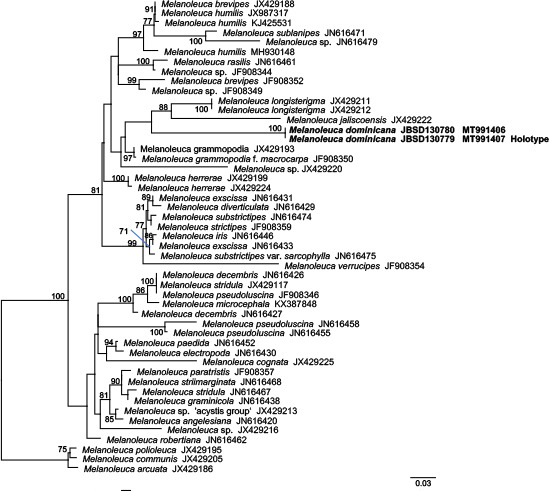
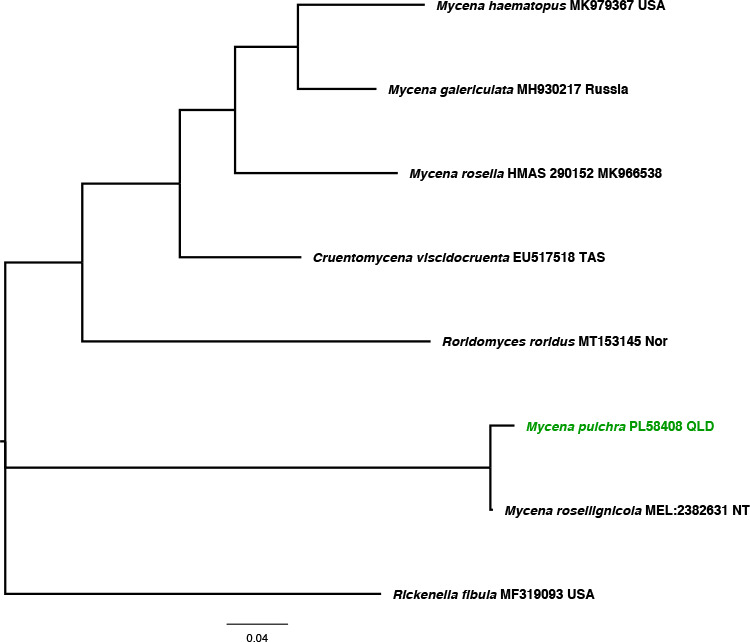
Novel species of fungi described in this study include those from various countries as follows: Australia, Austroboletus asper on soil, Cylindromonium alloxyli on leaves of Alloxylon pinnatum, Davidhawksworthia quintiniae on leaves of Quintinia sieberi, Exophiala prostantherae on leaves of Prostanthera sp., Lactifluus lactiglaucus on soil, Linteromyces quintiniae (incl. Linteromyces gen. nov.) on leaves of Quintinia sieberi, Lophotrichus medusoides from stem tissue of Citrus garrawayi, Mycena pulchra on soil, Neocalonectria tristaniopsidis (incl. Neocalonectria gen. nov.) and Xyladictyochaeta tristaniopsidis on leaves of Tristaniopsis collina, Parasarocladium tasmanniae on leaves of Tasmannia insipida, Phytophthora aquae-cooljarloo from pond water, Serendipita whamiae as endophyte from roots of Eriochilus cucullatus, Veloboletus limbatus (incl. Veloboletus gen. nov.) on soil. Austria, Cortinarius glaucoelotus on soil. Bulgaria, Suhomyces rilaensis from the gut of Bolitophagus interruptus found on a Polyporus sp. Canada, Cantharellus betularum among leaf litter of Betula, Penicillium saanichii from house dust. Chile, Circinella lampensis on soil, Exophiala embothrii from rhizosphere of Embothrium coccineum. China, Colletotrichum cycadis on leaves of Cycas revoluta. Croatia, Phialocephala melitaea on fallen branch of Pinus halepensis. Czech Republic, Geoglossum jirinae on soil, Pyrenochaetopsis rajhradensis from dead wood of Buxus sempervirens. Dominican Republic, Amanita domingensis on litter of deciduous wood, Melanoleuca dominicana on forest litter. France, Crinipellis nigrolamellata (Martinique) on leaves of Pisonia fragrans, Talaromyces pulveris from bore dust of Xestobium rufovillosum infesting floorboards. French Guiana, Hypoxylon hepaticolor on dead corticated branch. Great Britain, Inocybe ionolepis on soil. India, Cortinarius indopurpurascens among leaf litter of Quercus leucotrichophora. Iran, Pseudopyricularia javanii on infected leaves of Cyperus sp., Xenomonodictys iranica (incl. Xenomonodictys gen. nov.) on wood of Fagus orientalis. Italy, Penicillium vallebormidaense from compost. Namibia, Alternaria mirabibensis on plant litter, Curvularia moringae and Moringomyces phantasmae (incl. Moringomyces gen. nov.) on leaves and flowers of Moringa ovalifolia, Gobabebomyces vachelliae (incl. Gobabebomyces gen. nov.) on leaves of Vachellia erioloba, Preussia procaviae on dung of Procavia capensis. Pakistan, Russula shawarensis from soil on forest floor. Russia, Cyberlindnera dauci from Daucus carota. South Africa, Acremonium behniae on leaves of Behnia reticulata, Dothiora aloidendri and Hantamomyces aloidendri (incl. Hantamomyces gen. nov.) on leaves of Aloidendron dichotomum, Endoconidioma euphorbiae on leaves of Euphorbia mauritanica, Eucasphaeria proteae on leaves of Protea neriifolia, Exophiala mali from inner fruit tissue of Malus sp., Graminopassalora geissorhizae on leaves of Geissorhiza splendidissima, Neocamarosporium leipoldtiae on leaves of Leipoldtia schultzii, Neocladosporium osteospermi on leaf spots of Osteospermum moniliferum, Neometulocladosporiella seifertii on leaves of Combretum caffrum, Paramyrothecium pituitipietianum on stems of Grielum humifusum, Phytopythium paucipapillatum from roots of Vitis sp., Stemphylium carpobroti and Verrucocladosporium carpobroti on leaves of Carpobrotus quadrifolius, Suttonomyces cephalophylli on leaves of Cephalophyllum pilansii. Sweden, Coprinopsis rubra on cow dung, Elaphomyces nemoreus from deciduous woodlands. Spain, Polyscytalum pini-canariensis on needles of Pinus canariensis, Pseudosubramaniomyces septatus from stream sediment, Tuber lusitanicum on soil under Quercus suber. Thailand, Tolypocladium flavonigrum on Elaphomyces sp. USA, Chaetothyrina spondiadis on fruits of Spondias mombin, Gymnascella minnisii from bat guano, Juncomyces patwiniorum on culms of Juncus effusus, Moelleriella puertoricoensis on scale insect, Neodothiora populina (incl. Neodothiora gen. nov.) on stem cankers of Populus tremuloides, Pseudogymnoascus palmeri from cave sediment. Vietnam, Cyphellophora vietnamensis on leaf litter, Tylopilus subotsuensis on soil in montane evergreen broadleaf forest. Morphological and culture characteristics are supported by DNA barcodes.
Keywords: ITS nrDNA barcodes, LSU, new taxa, systematics
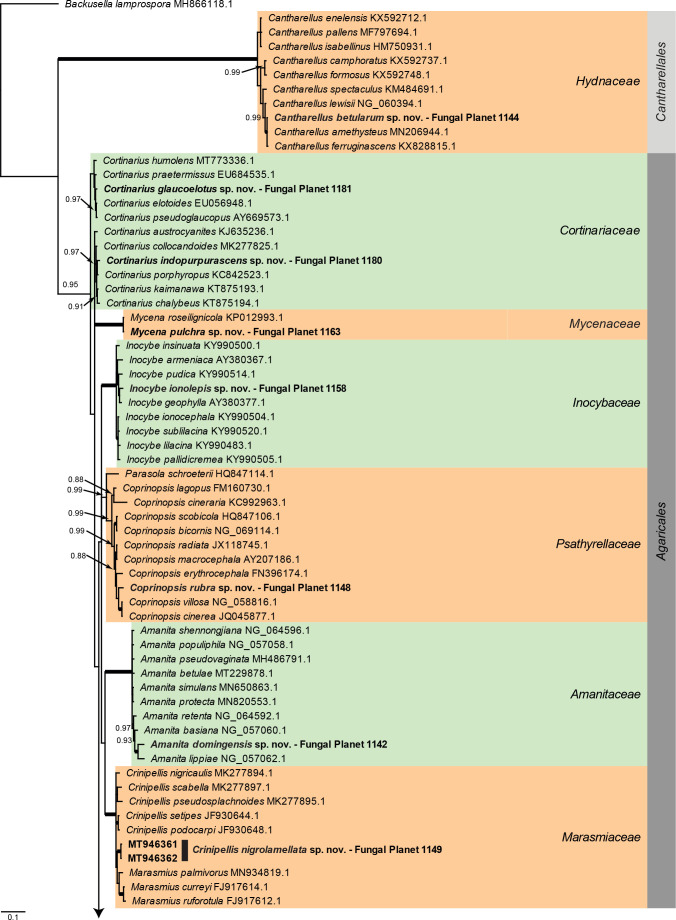
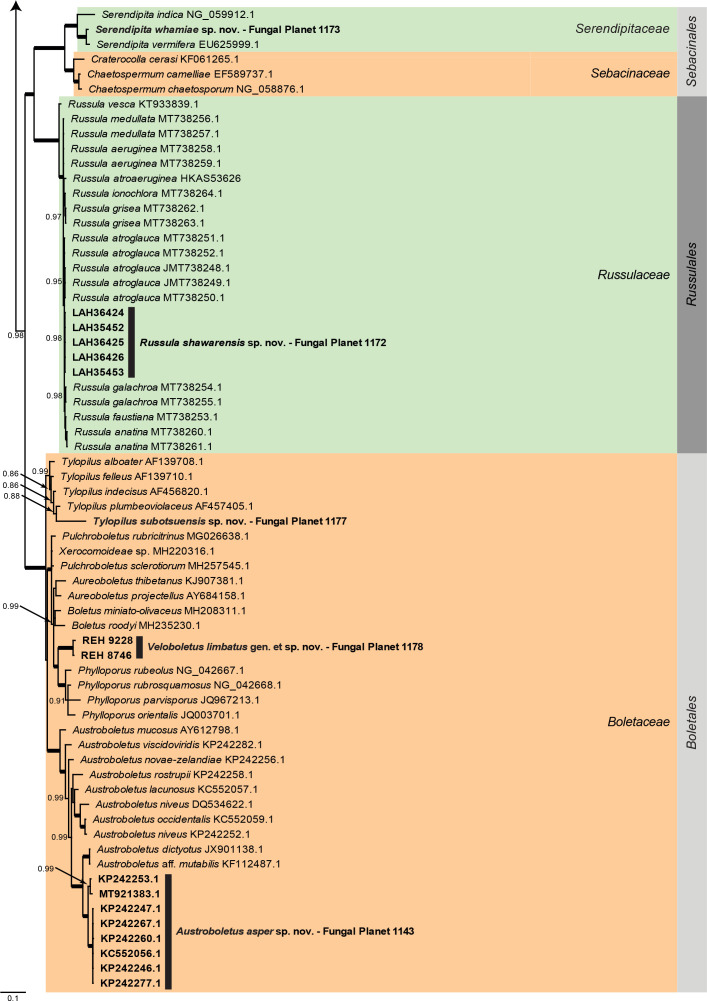
Overview Agaricomycetes phylogeny – part 1 .
Consensus phylogram (50 % majority rule) of 435 752 trees resulting from a Bayesian analysis of the LSU sequence alignment (130 sequences including outgroup; 979 aligned positions; 571 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Backusella lamprospora (GenBank MH866118.1) and the taxonomic novelties described in this study for which LSU sequence data were available are indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
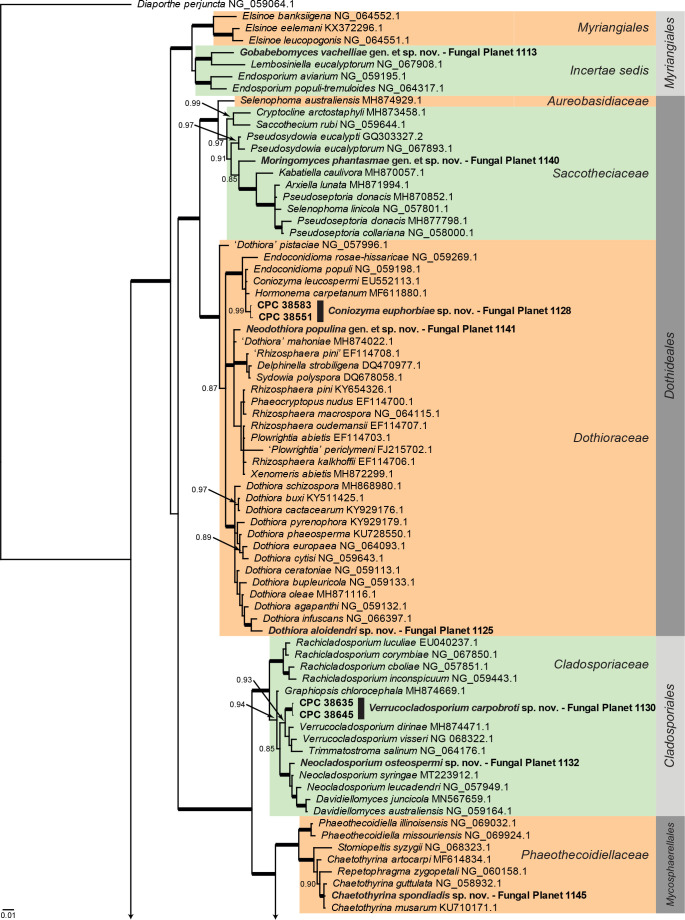
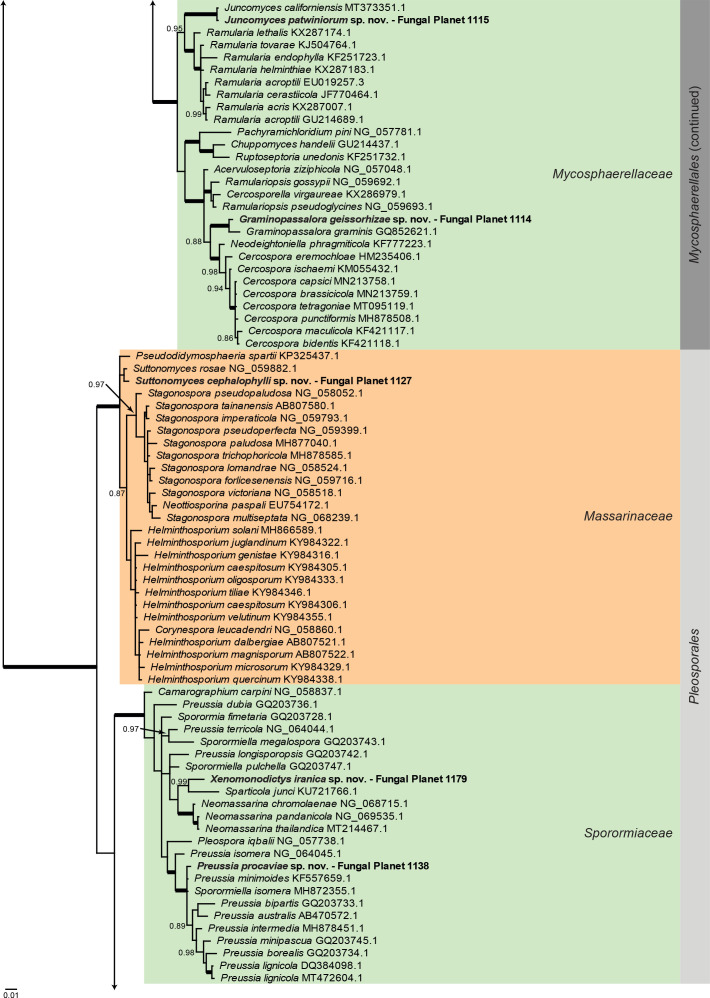
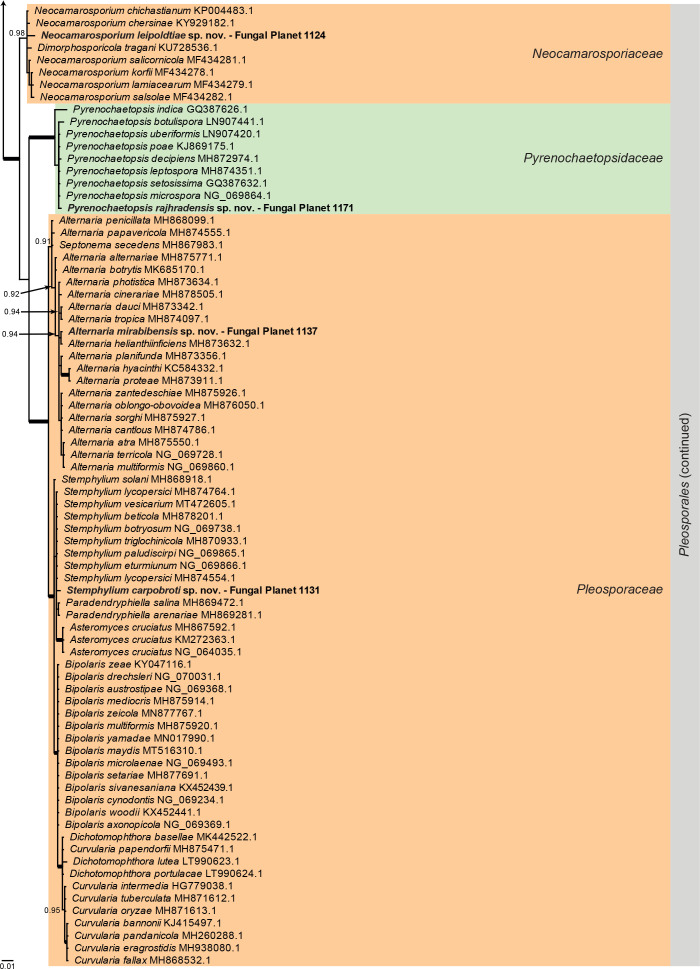
Overview Dothideomycetes phylogeny – part 1 .
Consensus phylogram (50 % majority rule) of 146 328 trees resulting from a Bayesian analysis of the LSU sequence alignment (233 sequences including outgroup; 824 aligned positions; 341 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Diaporthe perjuncta (GenBank NG_059064.1) and the taxonomic novelties described in this study for which LSU sequence data were available are indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
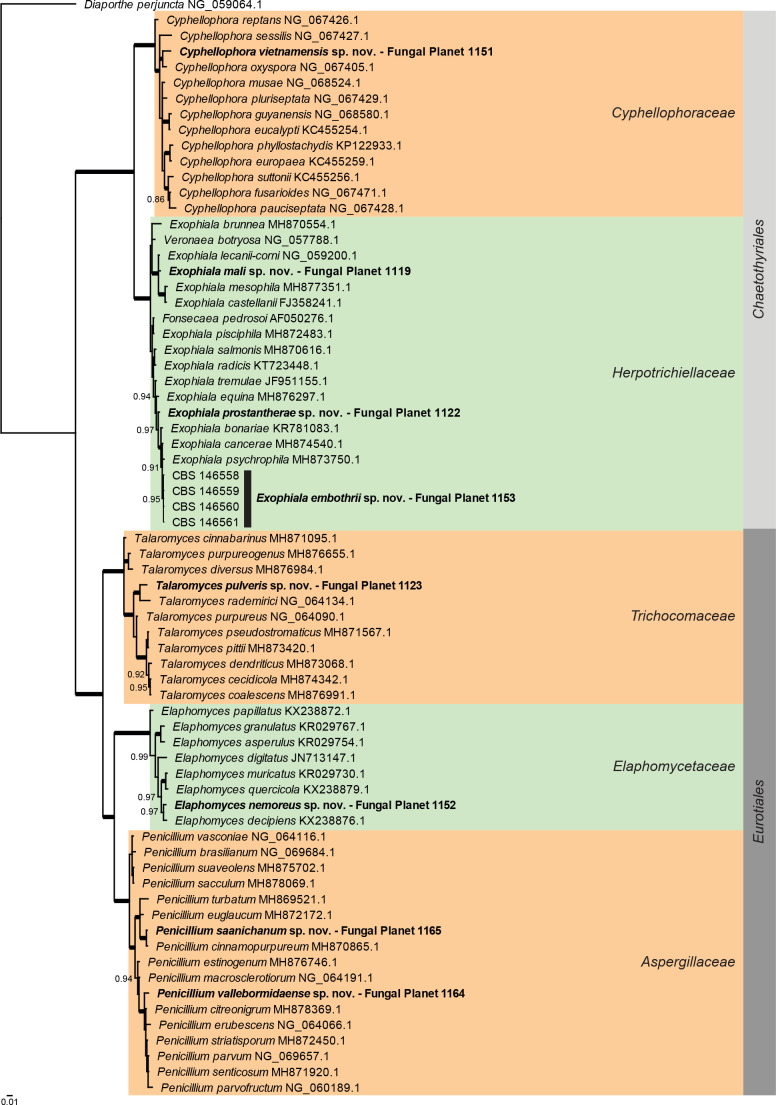
Overview Eurotiomycetes phylogeny .
Consensus phylogram (50 % majority rule) of 146 252 trees resulting from a Bayesian analysis of the LSU sequence alignment (70 sequences including outgroup; 838 aligned positions; 267 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Diaporthe perjuncta (GenBank NG_059064.1) and the taxonomic novelties described in this study for which LSU sequence data were available are indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
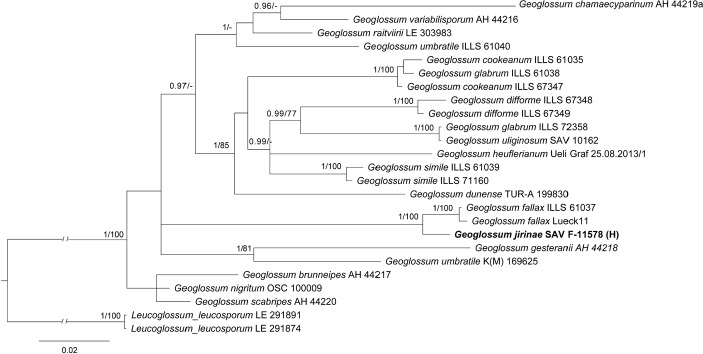
Overview Geoglossomycetes phylogeny .
Consensus phylogram (50 % majority rule) of 46 502 trees resulting from a Bayesian analysis of the LSU sequence alignment (18 sequences including outgroup; 930 aligned positions; 223 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. The family and order are indicated with coloured blocks to the right of the tree. GenBank accession or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Aspergillus niger (GenBank KC119204.1) and the taxonomic novelty described in this study for which LSU sequence data were available is indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
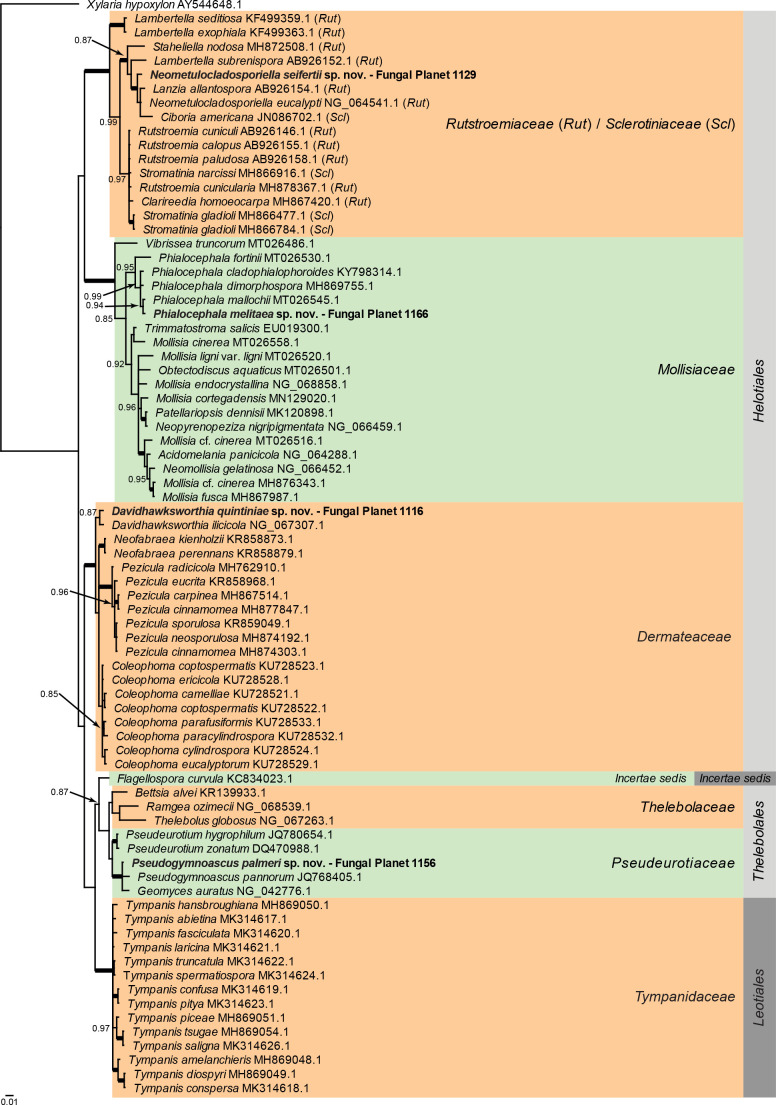
Overview Leotiomycetes phylogeny .
Consensus phylogram (50 % majority rule) of 222 002 trees resulting from a Bayesian analysis of the LSU sequence alignment (78 sequences including outgroup; 839 aligned positions; 258 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. Families and orders are indicated with coloured blocks to the right of the tree. Family assignment for Helotiales follows Johnston et al. (2019). GenBank accession or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Xylaria hypoxylon (GenBank AY544648.1) and the taxonomic novelties described in this study for which LSU sequence data were available are indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
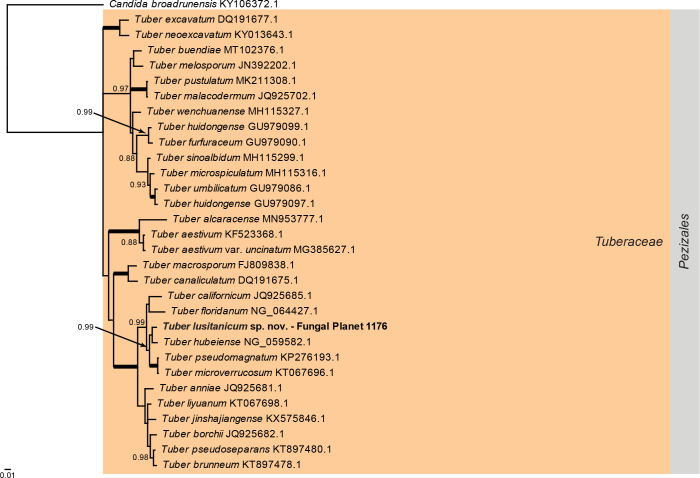
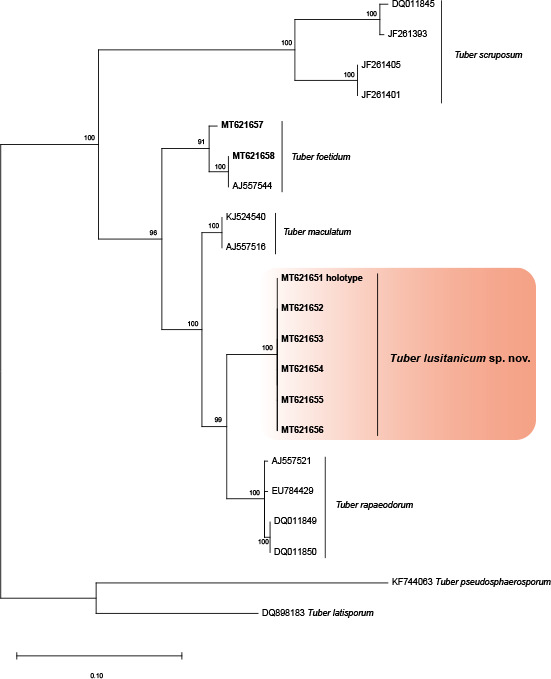
Overview Pezizomycetes phylogeny .
Consensus phylogram (50 % majority rule) of 52 502 trees resulting from a Bayesian analysis of the LSU sequence alignment (31 sequences including outgroup; 821 aligned positions; 204 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. The family and order are indicated with coloured blocks to the right of the tree. GenBank accession or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Candida broadrunensis (GenBank KY106372.1) and the taxonomic novelty described in this study for which LSU sequence data were available is indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
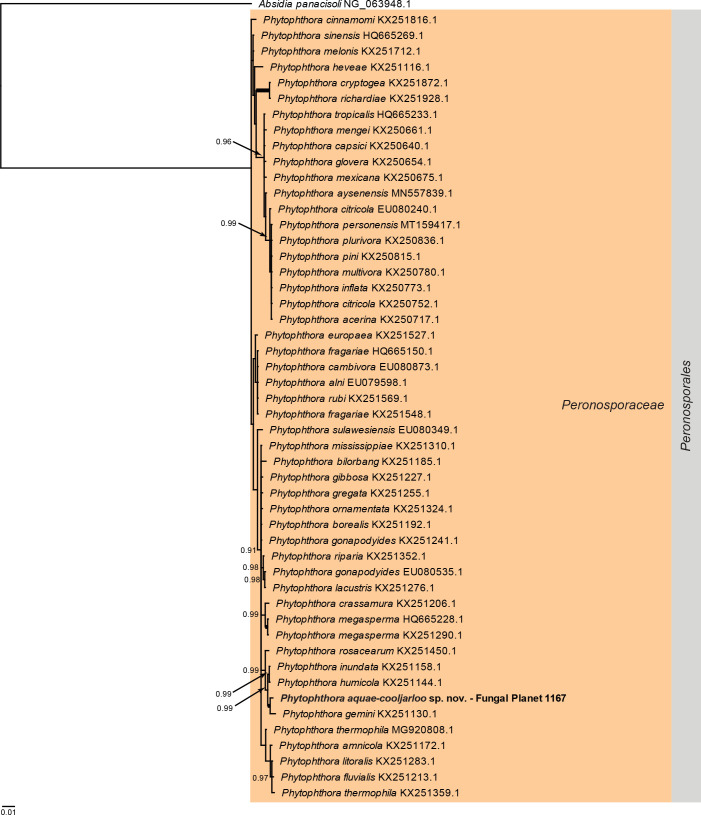
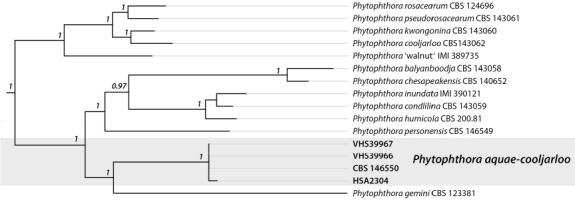
Overview Phytophthora phylogeny .
Consensus phylogram (50 % majority rule) of 1 260 002 trees resulting from a Bayesian analysis of the LSU sequence alignment (51 sequences including outgroup; 1 305 aligned positions; 130 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. The family and order are indicated with coloured blocks to the right of the tree. GenBank accession or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Absidia panacisoli (GenBank NG_063948.1) and the taxonomic novelty described in this study for which LSU sequence data were available is indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
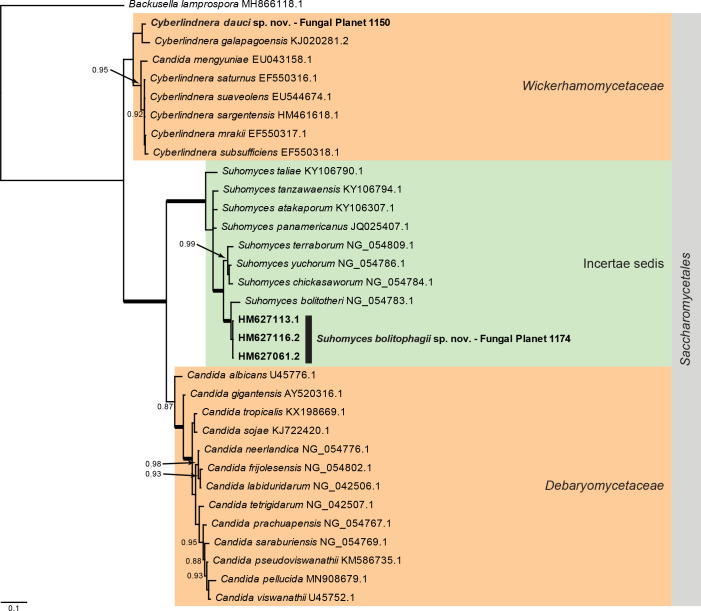
Overview Saccharomycetes phylogeny .
Consensus phylogram (50 % majority rule) of 69 002 trees resulting from a Bayesian analysis of the LSU sequence alignment (33 sequences including outgroup; 553 aligned positions; 198 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. The families and order are indicated with coloured blocks to the right of the tree. GenBank accession and/or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Backusella lamprospora (GenBank MH866118.1) and the taxonomic novelties described in this study for which LSU sequence data were available are indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
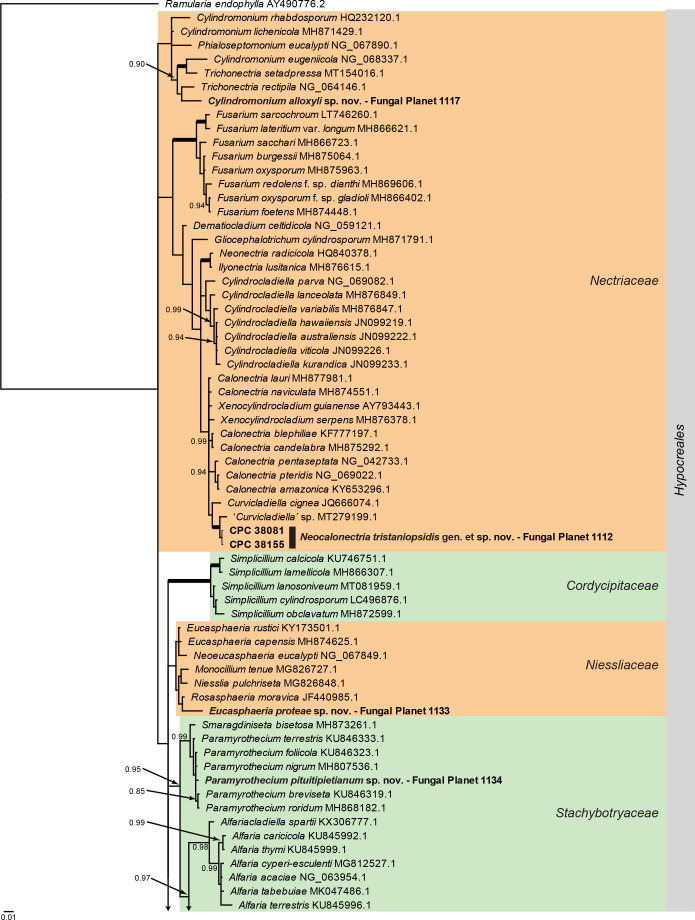
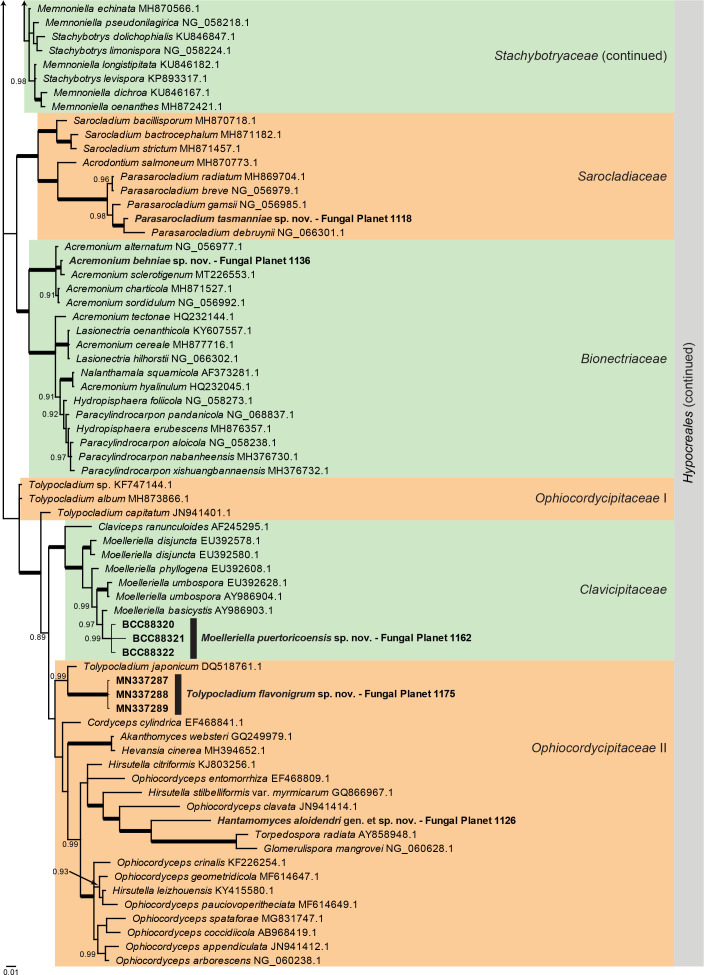
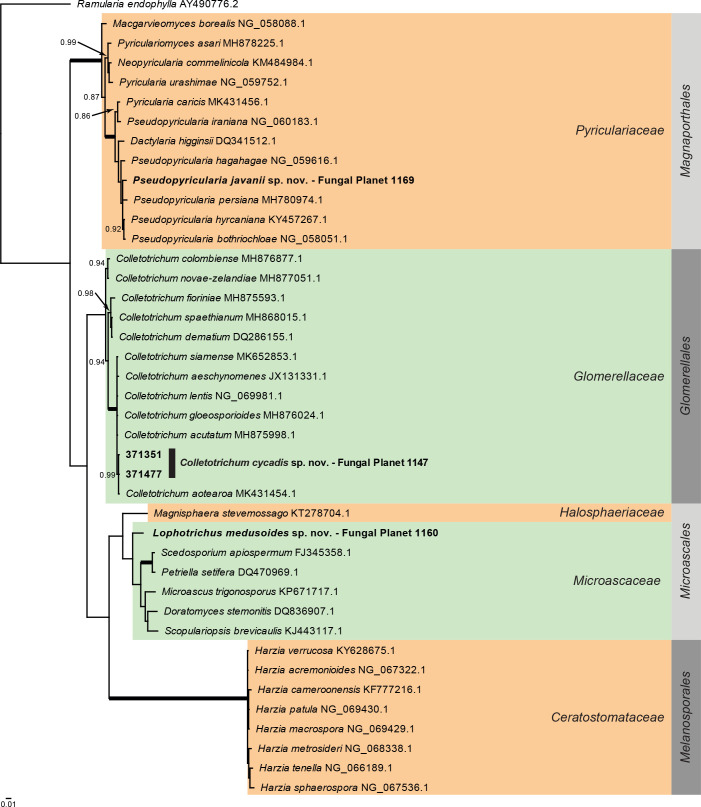
Overview Sordariomycetes ( Hypocreales ) phylogeny – part 1 .
Consensus phylogram (50 % majority rule) of 1 695 002 trees resulting from a Bayesian analysis of the LSU sequence alignment (135 sequences including outgroup; 812 aligned positions; 341 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. Families and the order are indicated with coloured blocks to the right of the tree. GenBank accession and/or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Ramularia endophylla (GenBank AY490776.2) and the taxonomic novelties described in this study for which LSU sequence data were available are indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
Overview Sordariomycetes (Other orders) phylogeny .
Consensus phylogram (50 % majority rule) of 60 752 trees resulting from a Bayesian analysis of the LSU sequence alignment (41 sequences including outgroup; 807 aligned positions; 247 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. Families and orders are indicated with coloured blocks to the right of the tree. GenBank accession and/or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Ramularia endophylla (GenBank AY490776.2) and the taxonomic novelties described in this study for which LSU sequence data were available are indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
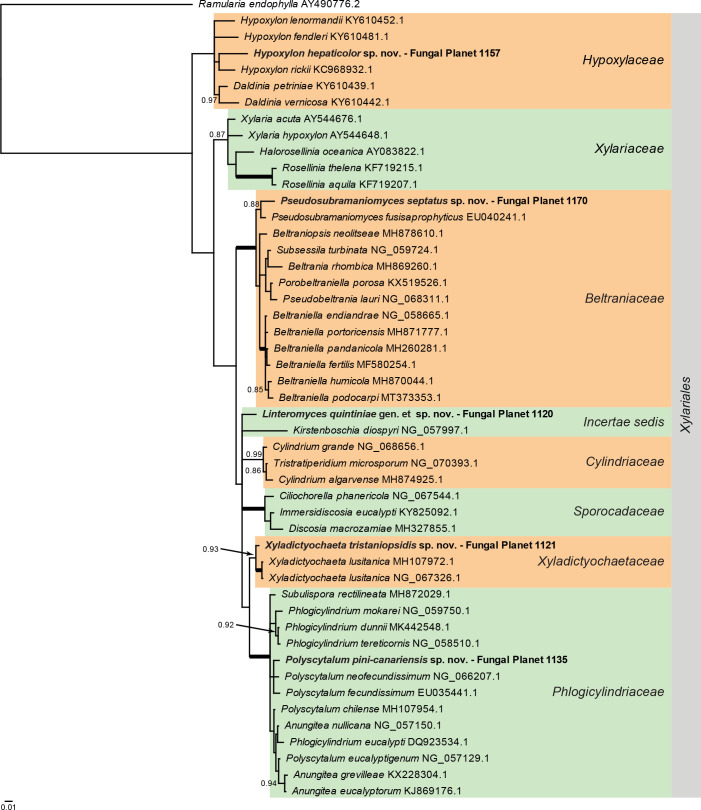
Overview Sordariomycetes ( Xylariales ) phylogeny .
Consensus phylogram (50 % majority rule) of 528 002 trees resulting from a Bayesian analysis of the LSU sequence alignment (49 sequences including outgroup; 815 aligned positions; 218 unique site patterns) using MrBayes v. 3.2.7a (Ronquist et al. 2012). Bayesian posterior probabilities (PP) > 0.84 are shown at the nodes and thickened lines represent nodes with PP = 1.00. The scale bar represents the expected changes per site. Families and the order are indicated with coloured blocks to the right of the tree. GenBank accession and/or Fungal Planet numbers are indicated behind the species names. The tree was rooted to Ramularia endophylla (GenBank AY490776.2) and the taxonomic novelties described in this study for which LSU sequence data were available are indicated in bold face. The alignment and tree were deposited in TreeBASE (Submission ID 27179).
Acknowledgements
Pedro Crous acknowledges Brett A. Summerell (Royal Botanic Gardens, Sydney, Australia) and Michael J. Wingfield (FABI, University of Pretoria, South Africa), for making several site photographs and field collections available for study. Jan Dijksterhuis is thanked for SEM photomicrographs of Neocalonectria tristaniopsis. Katrina Syme and co-authors thank the curation staff at BRI, MEL, PERTH for their help with loans and processing of collections. Funding for fieldwork and sequencing was provided by the Helen McLellan Fund (RBG Victoria). A. Vizzini thanks R. Berndt (Curator of Fungus Collections, Herbaria Z+ZT) for the loan of specimens. The 2015 collecting trips to Martinique directed by R. Courtecuisse were made possible through financial help from Communauté Territoriale de Martinique, Parc Naturel Régional de Martinique (PNRM) and French national Forestry Office (ONF). The study of Aleksey V. Kachalkin and colleagues was supported by the Russian Science Foundation (grant No. 19-74-10002). Isabel Iturrieta-González and colleagues were partially supported by the Spanish Ministerio de Economía, Industria y Competitividad (grant CGL2017-88094-P). Financial support was provided by the VEGA grant agency (project 2/0061/19) to Viktor Kučera and Marek Slovák. The studies of V. Antonín and H. Ševčíková were enabled by support provided to the Moravian Museum by the Ministry of Culture of the Czech Republic as part of its long-term conceptual development programme for research institutions (MK000094862). Abigail E. Rea-Ireland and colleagues acknowledge the National Fish & Wildlife Foundation, the Pennsylvania Game Commission, Greg Turner for material support on project, Lock Haven University, Temple University, Joseph Calabrese, Jacob Adam, Collin Wesley, Alden Mileto, and Alina Pislar, as well as Karen Hughes for allowing Abigail Rea-Ireland to finish this undergraduate research while starting graduate studies at the University of Tennessee, Knoxville. Jacques Fournier gratefully acknowledges the Parc Naturel Amazonien de Guyane for having initiated, funded and organized the field work in Saül in 2018 and 2019, in the context of the ABC inventory project during which the new species Hypoxylon hepaticolor was collected. Jed Calvert acknowledges support from the Maxim Foundation for travel, collection and help in the discovery of this taxon. The research of Cobus M. Visagie, Rafik Assabgui & Keith A. Seifert was supported by a grant from the Alfred P. Sloan Foundation Program (grant 2014-06-03) on the Microbiology of the Built Environment. Neven Matočec, Ivana Kušan, Ana Pošta, Zdenko Tkalčec, and Armin Mešić are grateful to the Croatian Science Foundation for their financial support under the project grant HRZZ-IP-2018-01-1736 (ForFungiDNA) and to Miro Pucar for his assistance during the fieldwork. Ana Pošta thanks Croatian Science Foundation for their support under the grant HRZZ-2018-09-7081. Shaun D. Langenhoven and colleagues are grateful to the South African Table Grape Industry, Winetech, the National Research Foundation (grant number: 99916) and Technology and Human Resources for Industry Programme (THRIP) for funding. The grant holders acknowledge that opinions, findings and conclusions or recommendations expressed in any publication generated by the NRF-supported research are that of the authors, and that the NRF accepts no liability whatsoever in this regard. The authors would like to thank Meagan van Dyk for help in formatting the article. This study of Daniel Torres-Garcia, Josepa Gené and Dania García was partially supported by the Spanish Ministerio de Economía, Industria y Competitividad (grant CGL2017-88094-P). Kanoksri Tasanathai and colleagues would like to thank Morakot Tanticharoen and Somvong Tragoonrung, Platform Technology Management Section, National Center for Genetic Engineering and Biotechnology (BIOTEC), Grant No. P19-50231 and CPMO Grant No. P11-00331 for their support of the program Biodiversity studies of entomopathogenic fungi in Thailand. Jean Lodge for her suggestions about the specimens collected in Puerto Rico. The study of Olga V. Morozova was carried out within the framework of a research project of the Komarov Botanical Institute RAS (AAAA-A19-119020890079-6) using equipment of its Core Facility Centre ‘Cell and Molecular Technologies in Plant Science’ with the financial support of Russian Foundation for Basic Research (project no. 20-04-00349). The study of Alina V. Alexandrova was supported by Moscow State University Grant for Leading Scientific Schools ‘Depository of the Living Systems’ in the framework of the MSU Development Program. Roy E. Halling acknowledges grants from the National Science Foundation (USA) DEB–0414665, DEB–1020421 and the National Geographic Society Committee for Research and Exploration in grant #8457–08. Logistical support from the Queensland Herbarium (BRI) aided field studies in Queensland. The Queensland Parks and Wildlife Service offered accommodation and orientation on Fraser Island. The staff and resources of the L.B. & D. Cullman Laboratory at the New York Botanical Garden aided in DNA extraction and amplifications. André De Kesel is thanked for providing insights regarding the morphological and developmental terminology of Clémençon (2012). Sandra Abell, Timothy Baroni, Teresa Lebel, Gregory Mueller, Todd Osmundson, and Klaus Querengasser are thanked for field assistance. Sushma Mandava kindly assisted early on in generating tef1 and LSU sequences and Pooja Singh and Olga Khmelnitsky are thanked for assistance in this project with preliminary molecular phylogenetic and morphological assessments. Dilnora Gouliamova and colleagues were supported by a grant from the Bulgarian Science Fund (KP-06-H31/19). The authors express their gratitude for Borislav Guéorguiev from National Museum of Natural History (Sofia, Bulgaria) for the identification of beetles. Asunción Morte is grateful to AEI/FEDER, UE (CGL2016-78946-R) and Fundación Séneca- Agencia de Ciencia y Tecnología de la Región de Murcia (20866/PI/18) for financial support. Patrick Leonard and John Dearnaley are grateful for the help and advice given by T. Lebel and F. Guard. Milan Spetik and colleagues acknowledge funding by an Internal Grant of Mendel University (IGA-ZF/2020-DP003). The study of Bálint Dima was partly supported by the ELTE Institutional Excellence Program supported by the National Research, Development and Innovation Office (NKFIH-1157-8/2019-DT) in Hungary. Kesiban Özdemir is thanked for sequencing help. Cortinarius glaucoelotus was sequenced within ABOL, subproject HRSFM University of Vienna, supported by the Austrian Federal Ministry of Education, Science and Research. Kamal C. Semwal and Vinod K. Bhatt are grateful to the Uttarakhand State Council for Science and Technology (UCoST), Dehradun, Uttarakhand, India for the financial support provided under project no. UCS&T/R&D/LS-1/12-13/4912. The research of Ellen Larsson and Mikael Jeppson was supported by the Swedish Taxonomy Initiative, SLU Artdatabanken (grant 2019.4.3-13).
REFERENCES
- Anisimova M, Manuel G., Dufayard J-F, et al. 2011. Survey of branch support methods demonstrates accuracy, power, and robustness of Fast Likelihood-based approximation schemes. Systematic Biology 60: 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonín V, Ryoo R, Ka K-H, et al. 2014. Three new species of Crinipellis and one new variety of Moniliophthora (Basidiomycota, Marasmiaceae) described from the Republic of Korea. Phytotaxa 170: 86–102. [Google Scholar]
- Arauzo S, Iglesias P.2014. La familia Geoglossaceae ss. str. en la península Ibérica y la Macaronesia. Errotari 11: 166–259. [Google Scholar]
- Attili-Angelis D, Duarte APM, Pagnocca FC, et al. 2014. Novel Phialophora species from leaf-cutting ants (tribe Attini). Fungal Diversity 65: 65–75. [Google Scholar]
- Australian Plant Pest Database, online database, (accessed 06 August 2020). https://appd.ala.org.au/appd-hub/search#tab_simpleSearch.
- Baral H-O. 1987. Lugol’s solution / IKI versus Melzer’s reagent: hemiamyloidity, a universal feature of the ascus wall. Mycotaxon 29: 399–450. [Google Scholar]
- Basiewicz M, Weiss M, Kogel K-H, et al. 2012. Molecular and phenotypic characterization of Sebacina vermifera strains associated with orchids, and the description of Pirformospora williamsii sp. nov. Fungal Biology 116: 204–213. [DOI] [PubMed] [Google Scholar]
- Baten MA, Mingzhu L, Motohashi K, et al. 2015. Two new species, Phytopythium iriomotense sp. nov. and P. aichiense sp. nov., isolated from river water and water purification sludge in Japan. Mycological Progress 14: 1–12. [Google Scholar]
- Bezerra JDP, Sandoval-Denis M, Paiva LM, et al. 2017. New endophytic Toxicocladosporium species from cacti in Brazil, and description of Neocladosporium gen. nov. IMA Fungus 8: 77–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon M.1984. Novitates – Combinaisons et taxons nouveaux. Documents Mycologiques 14 (53): 6. [Google Scholar]
- Borchsenius F.2009. FastGap 1.2. Department of Bio-sciences, Aarhus University, Denmark. http://www.aubot.dk/FastGap_home.htm. [Google Scholar]
- Brasier CM, Sanchez-Hernandez E, Kirk SA. 2003. Phytophthora inundata sp. nov., a part heterothallic pathogen of trees and shrubs in wet or flooded soils. Mycological Research 107: 477–484. [DOI] [PubMed] [Google Scholar]
- Braun U, Crous PW, Nakashima C.2015. Cercosporoid fungi (Mycosphaerellaceae) 3. Species on monocots (Poaceae, true grasses). IMA Fungus 6: 25–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U, Hill CF.2002. Some new micromycetes from New Zealand. Mycological Progress 1: 19–30. [Google Scholar]
- Burgess TI, Simamora A, White D, et al. 2018. New species from Phytophthora Clade 6a: evidence for recent radiation. Persoonia 41: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyck B.2000. Le genre Cantharellus en Europe. Bulletin de la Société Mycologique de France 116: 91–1378. [Google Scholar]
- Buyck B, Hofstetter V.2011. The contribution of tef-1 sequences for species delimitation in the Cantharellus cibarius complex in the southeastern USA. Fungal Diversity 49: 35–46. [Google Scholar]
- Ceruti A, Fontana A, Nosenzo C.2003. Le specie europee del genere Tuber. Una revision storica. Museo Regionale di Scienze Naturali, Torino. [Google Scholar]
- Chaverri P, Liu M, Hodge KT.2008. A monograph of the entomopathogenic genera Hypocrella, Moelleriella, and Samuelsia gen. nov. (Ascomycota, Hypocreales, Clavicipitaceae), and their aschersonia-like anamorphs in the Neotropics. Studies in Mycology 60: 1–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Bañuls A-L, et al. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clémençon H. 2012. Cytology and plectology of the hymenomycetes. 2nd ed. Cramer, Stuttgart. [Google Scholar]
- Corner EHJ.1994. Agarics in Malesia II Mycenoid. Beihefte zur Nova Hedwigia 109: 1–271. [Google Scholar]
- Crous PW.2002. Taxonomy and pathology of Cylindrocladium (Calonectria) and allied genera. APS Press. [Google Scholar]
- Crous PW, Braun U, Schubert K, et al. 2007a. Delimiting Cladosporium from morphologically similar genera. Studies in Mycology 58: 33–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Decock C, Schoch CL.2001. Xenocylindrocladium guianense and X. subverticillatum, two new species of hyphomycetes from plant debris in the tropics. Mycoscience 42: 559–566. [Google Scholar]
- Crous PW, Groenewald JZ.2016. They seldom occur alone. Fungal Biology 120: 1392–1415. [DOI] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ.2017. The genera of fungi – G 4: Camarosporium and Dothiora. IMA Fungus 8: 131–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG.2011. Fungal Planet description sheets: 69–91. Persoonia 26: 108–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Luangsa-ard JJ, Wingfield MJ, et al. 2018a. Fungal Planet description sheets: 785–867. Persoonia 41: 238–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Mohammed C, Glen M, et al. 2007b. Eucalyptus microfungi known from culture. 3. Eucasphaeria and Sympoventuria genera nova, and new species of Furcaspora, Harknessia, Heteroconium and Phacidiella. Fungal Diversity 25: 19–36. [Google Scholar]
- Crous PW, Schumacher RK, Akulov A, et al. 2019a. New and interesting fungi. 2. Fungal Systematics and Evolution 3: 57–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, et al. 2018b. New and interesting fungi. 1. Fungal Systematics and Evolution 1: 169–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. 2014. Fungal Planet description Sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Wingfield MJ, et al. 2012. Fungal Planet description sheets: 128–153. Persoonia 29: 146–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2017a. Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2018c. Fungal Planet description sheets: 716–784. Persoonia 40: 240–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI.2017b. Fungal Planet description sheets: 558–624. Persoonia 38: 240–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Cheewangkoon R, et al. 2019b. Foliar pathogens of eucalypts. Studies in Mycology 94: 125–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Chooi Y-H, et al. 2020a. Fungal Planet description sheets: 1042–1111. Persoonia 44: 301–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Lombard L, et al. 2019c. Fungal Planet description sheets: 951–1041. Persoonia 43: 223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, et al. 2020b. New and interesting fungi. 3. Fungal Systematics and Evolution 6: 157–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz ACR, Gusmão LFP, Castañeda-Ruíz RF. 2007. Conidial fungi from the semi-arid Caatinga biome of Brazil. Subramaniomyces pulcher sp. nov. and notes on Sporidesmium circinophorum. Mycotaxon 102: 25–32. [Google Scholar]
- Daranagama DA, Camporesi E, Liu XZ, et al. 2016. Tristratiperidium microsporum gen. et sp. nov. (Xylariales) on dead leaves of Arundo plinii. Mycological Progress 15: 8. [Google Scholar]
- Darriba D, Taaboada GL, Doallo R, et al. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Atri NS, Buyck B.2013. Three new species of Russula (Russulales) from Sikkim (India). Mycosphere 4: 722–732. [Google Scholar]
- Das K, Rossi W, Leonardi M, et al. 2018. Fungal Biodiversity Profiles 61–70. Cryptogamie, Mycologie 39: 381–418. [Google Scholar]
- Day MJ, Gibas CFC, Fujimura KE, et al. 2006. Monodictys arctica, a new hyphomycete from the roots of Saxifraga oppositifolia collected in the Canadian High Arctic. Mycotaxon 98: 261–272. [Google Scholar]
- De Beer ZW, Duong TA, Barnes I, et al. 2014. Redefining Ceratocystis and allied genera. Studies in Mycology 79: 187–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Vicente VA, Najafzadeh MJ, et al. 2011. Waterborne Exophiala species causing disease in cold-blooded animals. Persoonia 27: 46–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Weenink XO, Gerrits van den Ende AHG.1999. Taxonomy of the Phialophora verrucosa complex with the description of two new species. Studies in Mycology 43: 107–122. [Google Scholar]
- DeCock C, Hennebert GL, Crous PW.1997. Nectria serpens sp. nov. and its hyphomycetous anamorph Xenocylindrocladium gen. nov. Mycological Research 101: 786–790. [Google Scholar]
- Deighton FC.1967. Studies on Cercospora and allied genera. II. Passalora, Cercosporidium and some species of Fusicladium on Euphorbia. Mycological Papers 112: 1–80. [Google Scholar]
- Dennis RWG.1952. Lepiota and allied genera in Trinidad, British West Indies. Kew Bulletin 7: 459–500. [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research 36 (Web Server issue), W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EJ.1908. The Geoglossaceae of North America. Annales Mycologici 6: 387–477. [Google Scholar]
- Edgar RC.2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries EM.1836–1838. Epicrisis systematis mycologici seu synopsis Hymenomycetum. Uppsala, Sweden. [Google Scholar]
- Gams W.1971. Cephalosporium-artige Schimmelpilze (Hyphomycetes). Fischer, Stuttgart. [Google Scholar]
- Gams W, Holubová-Jechová V. 1976. Chloridium and some other dematiaceous hyphomycetes growing on decaying wood. Studies in Mycology 13: 1–99. [Google Scholar]
- Garnica S, Weiß M, Oertel B, et al. 2009. Phylogenetic relationships in Cortinarius, section Calochroi, inferred from nuclear DNA sequences. BMC Evolutionary Biology 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon LM, Rueda LJ, Celis AM, et al. 2019. Exophiala psychrophila: a new agent of chromoblastomycosis. Medical Mycology Case Reports 23: 31–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelardi M, Simonini G, Ercole E, et al. 2014a. Alessioporus and Pulchroboletus (Boletaceae, Boletineae), two novel genera for Xerocomus ichnusanus and X. roseoalbidus from the European Mediterranean basin: molecular and morphological evidence. Mycologia 106: 1168–1187. [DOI] [PubMed] [Google Scholar]
- Gelardi M, Vizzini A, Ercole E, et al. 2014b. New collection, iconography and molecular evidence for Tylopilus neofelleus (Boletaceae, Boletoideae) from southwestern China and the taxonomic status of T. plumbeoviolaceoides and T. microsporus. Mycoscience 56: 373–386. [Google Scholar]
- Gomes RR, Vicente VA, De Azevedo CMPS, et al. 2016. Molecular epidemiology of agents of human chromoblastomycosis in Brazil with the description of two novel species. PLoS Neglected Tropical Diseases 10 (11): e0005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarro J, Gene J, Stchigel AM, et al. 2012. Atlas of soil ascomycetes. CBS Biodiversity Series 10. Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands. [Google Scholar]
- Guevara-Suarez M, García D, Cano-Lira JF, et al. 2020. Species diversity in Penicillium and Talaromyces from herbivore dung, and the proposal of two new genera of penicillium-like fungi in Aspergillaceae. Fungal Systematics and Evolution 5: 39–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. 2010a. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, et al. 2010b. PHYML online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Research 33 (Web Server issue): W557–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakelier N.1967. Three new Swedish species of Geoglossum. Svensk Botanisk Tidskrift 61: 419–424. [Google Scholar]
- Heilmann-Clausen J, Verbeken A, Vesterholt J. 1998. The genus Lactarius. Fungi of Northern Europe Vol 2. [Google Scholar]
- Hernández-Restrepo M, Gené J, Castañeda-Ruiz RF, et al. 2017. Phylogeny of saprobic microfungi from Southern Europe. Studies in Mycology 86: 53–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera M, Bandala VL, Montoya L.2018. Cantharellus violaceovinosus, a new species from tropical Quercus forests in eastern Mexico. MycoKeys 32: 91–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesseltine CW, Fennell DI.1955. The genus Circinella. Mycologia 47: 193–212. [Google Scholar]
- Hongo T.1966. Notulae Mycologicae (5). Memoirs of the Faculty of Education, Shiga University. Natural Science (Otsu) 16: 57–62. [Google Scholar]
- Hongo T.1973. Enumeration of the Hygrophoraceae, Boletaceae and Strobilomycetaceae. Mycological reports from New Guinea and the Solomon Islands (16–21). Bulletin of the National Science Museum (Tokyo) 16: 537–557. [Google Scholar]
- Hosoya T, Huhtinen S.2002. Hyaloscyphaceae in Japan (7): Hyaloscypha albohyalina var. monodictys var. nov.. 43: 405–409. [Google Scholar]
- Houbraken J, Kocsubé S, Visagie CM, et al. 2020. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H.2012. Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Diversity 52: 75–98. [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, et al. 2008. NCBI BLAST: a better web interface. Nucleic Acids Research 36 (suppl_2): W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston PR, Quijada L, Smith CA, et al. 2019. A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD.2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerekes JF, Desjardin DE.2009. A monograph of the genera Crinipellis and Moniliophthora from Southeast Asia including a molecular phylogeny of the nrITS region. Fungal Diversity 37: 101–152. [Google Scholar]
- Kirk PM.1982. New or interesting microfungi IV. Dematiaceous hyphomycetes from Devon. Transactions of the British Mycological Society 78: 55–74. [Google Scholar]
- Kirk PM.1983. New or interesting microfungi IX. Dematiaceous hyphomycetes from Esher Common. Transactions of the British Mycological Society 80: 449–467. [Google Scholar]
- Klaubauf S, Tharreau D, Fournier E, et al. 2014. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Studies in Mycology 79: 85–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko WH, Ann PJ.1985. Phytophthora humicola, a new species from soil of a citrus orchard in Taiwan. Mycologia 77: 631–636. [Google Scholar]
- Kornerup A, Wanscher JH.1967. Methuen Handbook of Colour. Methuen & Co Ltd, London, England. [Google Scholar]
- Kornerup A, Wanscher JH.1978. Methuen Handbook of Colour, 3rd ed. Eyre Methuen, London. [Google Scholar]
- Kornerup A, Wanscher JH.1983. Methuen Handbook of Colour, 3rd ed. Eyre Methuen, Ltd., London. [Google Scholar]
- Kumar S, Stecher G, Li M, et al. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP.2001. Six new anamorphic ascomycetous yeasts near Candida tanzawaensis. FEMS Yeast Research 1: 177–185. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ, Blackwell M.2016. Description of Teunomyces gen. nov. for the Candida kruisii clade, Suhomyces gen. nov. for the Candida tanzawaensis clade and Suhomyces kilbournensis sp. nov. FEMS Yeast Research 16: 1–9. [DOI] [PubMed] [Google Scholar]
- Kušan I, Matočec N, Antonić O, et al. 2014. Biogeographical variability and re-description of an imperfectly known species Hamatocanthoscypha rotundispora (Helotiales, Hyaloscyphaceae). Phytotaxa 170: 1–12. [Google Scholar]
- Kušan I, Matočec N, Mešić A, et al. 2015. A new species of Thecotheus from Croatia with a key to the known species with apiculate spores. Sydowia 67: 51–63. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 4: 772–773. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D.2015. POPART: full-feature software for haplotype network construction. Methods in Ecology and Evolution 6: 1110–1116. [Google Scholar]
- Li GJ, Hyde KD, Zhao RL, et al. 2016. Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78: 1–237. [Google Scholar]
- Li GJ, Zhao Q, Zhao D, et al. 2013. Russula atroaeruginea and R. sichuanensis spp. nov.from southwest China. Mycotaxon 124: 173–188. [Google Scholar]
- Liu YJ, Whelen S, Hall BD.1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799e1808. [DOI] [PubMed] [Google Scholar]
- Lombard L, Houbraken J, Decock C, et al. 2016. Generic hyper-diversity in Stachybotriaceae. Persoonia 36: 156–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van der Merwe NA, Groenewald JZ, et al. 2015. Generic concepts in Nectriaceae. Studies in Mycology 80: 189–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch JM, Lindner DL, Gargas A, et al. 2013. A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white nose syndrome. Mycologia 105: 237–252. [DOI] [PubMed] [Google Scholar]
- Maas Geesteranus RA, Horak E. 1995. Mycena and related genera from Papua New Guinea and New Caledonia. Bibliotheca Mycologica 159: 143–329. [Google Scholar]
- Maciá-Vicente JG, Glynou K, Piepenbring M. 2016. A new species of Exophiala associated with roots. Mycological Progress 15: 18. [Google Scholar]
- Madrid H, Hernández-Restrepo M, Gené J, et al. 2016. New and interesting chaetothyrialean fungi from Spain. Mycological Progress 15: 1179–1201. [Google Scholar]
- Mains EB.1957. Species of Cordyceps parasitic on Elaphomyces. Bulletin of the Torrey Botanical Club 84: 243–251. [Google Scholar]
- Man in’t Veld WA, Rosendahl KC, Brouwer H, et al. 2011. Phytophthora gemini sp. nov., a new species isolated from the halophilic plant Zostera marina in the Netherlands. Fungal Biology 115: 724–732. [DOI] [PubMed] [Google Scholar]
- Man in’t Veld WA, Rosendahl KC, Van Rijswick PC, et al. 2019. Multiple Halophytophthora spp. and Phytophthora spp. including P. gemini, P. inundata and P. chesapeakensis sp. nov. isolated from the seagrass Zostera marina in the Northern hemisphere. European Journal of Plant Pathology 153: 341–57. [Google Scholar]
- Marin-Felix Y, Groenewald JZ, Cai L, et al. 2017a. Genera of phytopathogenic fungi: GOPHY 1. Studies in Mycology 86: 99–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Crous PW. 2020. Multi-locus phylogeny of the genus Curvularia and description of ten new species. Mycological Progress 19: 559–588. [Google Scholar]
- Marin-Felix Y, Senwanna C, Cheewangkoon R, et al. 2017b. New species and records of Bipolaris and Curvularia from Thailand. Mycosphere 8: 1556–1574. [Google Scholar]
- Marincowitz S, Crous PW, Groenewald JZ, et al. 2008. Microfungi occurring on Proteaceae in the fynbos. CBS Biodiversity Series 7: 1–166. [Google Scholar]
- Massimo NC, Nandi Devan MM, Arendt KR, et al. 2015. Fungal endophytes in aboveground tissues of desert plants: infrequent in culture, but highly diverse and distinctive symbionts. Microbial Ecology 70: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny PB, Swenie RA.2018. The Inocybe geophylla group in North America: a revision of the lilac species surrounding I. lilacina. Mycologia 110: 618–634. [DOI] [PubMed] [Google Scholar]
- Matsushima T.1971. Microfungi of the Solomon Islands and Papua-New Guinea. Published by the author, Kobe. [Google Scholar]
- McLeod A, Botha WJ, Meitz JC, et al. 2009. Biodiversity of Pythium species in South African agricultural systems. Mycological Research 113: 933–951. [DOI] [PubMed] [Google Scholar]
- Mello A, Vizzini A, Longato S, et al. 2000. Tuber borchii versus Tuber maculatum: neotype studies and DNA analyses. Mycologia 92: 326–331. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA: 1–8. [Google Scholar]
- Miller MA, Schwartz T, Pickett BE, et al. 2015. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evolutionary Bioinformatics 11: EBO-S21501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OK, Jr, Lodge DJ, Baroni TJ.2000. New and interesting ectomycorrhizal fungi from Puerto Rico, Mona, and Guana Islands. Mycologia 92: 558–570. [Google Scholar]
- Minh BQ, Nguyen MAT, Von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnis AM, Lindner DL.2013. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Mycology 117: 638–649. [DOI] [PubMed] [Google Scholar]
- Minter DW, Cannon PF.2015. Geoglossum cookeanum. IMI Descriptions of Fungi and Bacteria, No. 204, Sheet 2031.
- Molia A, Larsson E, Jeppson M, et al. 2020. Elaphomyces section Elaphomyces (Eurotiales, Ascomycota) – taxonomy and phylogeny of North European taxa, with the introduction of three new species. Fungal Systematics and Evolution 5: 283–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Jones G. 1975. Notes on Hyphomycetes. VIII. Lylea, a new genus. Mycotaxon 3: 129–132. [Google Scholar]
- Moser M.1961. ‘1960. ’. Die Gattung Phlegmacium (Schleimköpfe). Die Pilze Mitteleuropas, Band IV. Klinkhardt, Germany. [Google Scholar]
- Mouzouras R, Jones EBG.1985. Monodictys pelagica, the anamorph of Nereiospora cristata (Halosphaeriaceae). Canadian Journal of Botany 63: 2444–2447. [Google Scholar]
- Müller E, Von Arx JA. 1962. Die Gattungen der didymosporen Pyrenomyceten. Beiträge zur Kryptogamenflora der Schweiz 11: 1–922. [Google Scholar]
- Munsell. 1975. Munsell Soil Colour Charts. Macbeth, Baltimore. [Google Scholar]
- Munsell AH. Munsell Color (Firm). 2000. Munsell soil color charts. New Windsor, NY: Munsell Color. [Google Scholar]
- Nagy LG, Desjardin DE, Vágvölgyi C, et al. 2013. Phylogenetic analyses of Coprinopsis section Lanatuli and Atramentarii identify multiple species within morphologically defined taxa. Mycologia 105: 112–124. [DOI] [PubMed] [Google Scholar]
- Najafzadeh MJ, Vicente VA, Feng P, et al. 2018. Rapid identification of seven waterborne Exophiala species by RCA DNA padlock probes. Mycopathologia 183: 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase T, Itoh M, Takematsu A, et al. 1988. Candida tanzawaensis, a new species of yeast isolated from moss collected in Japan. Transactions of the Mycological Society of Japan 29: 331–338. [Google Scholar]
- Nannfeldt JA.1942. The Geoglossaceae of Sweden (with regard also to the surrounding countries). Arkiv för Botanik 30A: 1–67. [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, et al. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa J, Nakashima C.2020. Japanese species of Alternaria and their species boundaries based on host range. Fungal Systematics and Evolution 5: 197–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberwinkler F.1964. Intrahymeniale Heterobasidiomyceten: Fruchtkörperlose Sebacina-Sippen und ihre systematische Stellung. Nova Hedwigia 7: 489–499. [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M, et al. 2012. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 28: 1166–1167. [DOI] [PubMed] [Google Scholar]
- Paden JW.1971. Three new species of Eupenicillium from soil. Mycopathologia et Mycologia Applicata 43: 259–268. [DOI] [PubMed] [Google Scholar]
- Palmer JM, Kubatova A, Novakova A, et al. 2014. Molecular characterization of a heterothallic mating system in Pseudogymnoascus destructans, the fungus causing white-nose syndrome of bats. G3: Genes, Genomes, Genetics 4: 1755–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz A, Bellanger JM, Lavoise C, et al. 2017. The genus Elaphomyces (Ascomycota, Eurotiales): a ribosomal DNA-based phylogeny and revised systematics of European ‘deer truffles’. Persoonia 38: 197–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck CH.1985. Gymnascella. Mycotaxon 24: 68–93. [Google Scholar]
- Pegler DN.1983. Agaric flora of the Lesser Antilles. Kew Bulletin Additional Series 9: 1–668. [Google Scholar]
- Petch T.1906. Descriptions of new Ceylon fungi. Annals of the Royal Botanic Gardens Peradeniya 3: 1–10. [Google Scholar]
- Pitt J.1980. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press, London. [Google Scholar]
- Pordel A, Khodaparast SA, McKenzie EHC, et al. 2017. Two new species of Pseudopyricularia from Iran. Mycological Progress 16: 729–736. [Google Scholar]
- Raithelhuber J.1974. Hongos argentinos. Tomo I. Buenos Aires: Compañía Impresora Argentina. [Google Scholar]
- Rayner RW.1970. A Mycological Colour Chart. Commonwealth Mycological Institute, Kew and British Mycological Society. [Google Scholar]
- Réblová M, Hernández-Restrepo M, Fournier J, et al. 2020. New insights into the systematics of Bactrodesmium and its allies and introducing new genera, species and morphological patterns in the Pleurotheciales and Savoryellales (Sordariomycetes). Studies in Mycology 95: 415–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Untereiner WA, Réblová K. 2013. Novel Evolutionary lineages revealed in the Chaetothyriales (Fungi) based on multigene phylogenetic analyses and comparison of ITS secondary structure. PLoS One 8 (5): e63547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Buckley E.2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Riess K, Oberwinkler F, Bauer R, et al. 2014. Communities of endophytic Sebacinales associated with roots of herbaceous plants in agricultural and grassland ecosystems are dominated by Serendipita herbamans sp. nov. PLoS ONE 9: E94676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P.1993. Exidiopsis species from Devon, including the new segregate genera Ceratosebacina, Endoperplexa, Microsebacina, and Serendipita. Mycological Research 97: 467–478. [Google Scholar]
- Ronquist F, Huelsenbeck JP.2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biolology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryberg M, Nilsson RH, Kristiansson E, et al. 2008. Mining metadata from unidentified ITS sequences in GenBank: a case study in Inocybe (Basidiomycota). BMC Evolutionary Biology 8: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar G, Brandrud TE, Dima B, et al. 2014. Cortinarius untergattung Phlegmacium sektion Purpurascentes in Europa. Journal des JEC 16: 140–161. [Google Scholar]
- Samson RA.1972. Notes on Pseudogymnoascus, Gymnoascus and related genera. Acta Botanica Neerlandica 21: 517–527. [Google Scholar]
- Samson RA, Yilmaz N, Houbraken J, et al. 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology 70: 159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-García M, Cifuentes-Blanco J, Matheny PB. 2013. Revisión taxonómica del género Melanoleuca en México y descripción de especies nuevas. Revista Mexicana de Biodiversidad 84 (supplemento micologia): 111–127. [Google Scholar]
- Sandoval-Denis M, Gené J, Sutton DA, et al. 2016. Redefining Microascus, Scopulariopsis and allied genera. Persoonia: Molecular Phylogeny and Evolution of Fungi. 36: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnari M.1998. Monografia illustrate del genere Russula in Europa. Tromo Primo, Italy. [Google Scholar]
- Sarwar S, Aziz T, Hanif M, et al. 2019. Russula swatica: A new species of Russula based on molecular, light microscopy, and scanning electron microscopy analyses from Swat Valley of Khyber Pakhtunkhwa province of Pakistan. Microscopy Research and Technique 82: 1700–1705. [DOI] [PubMed] [Google Scholar]
- Saxena AS, Mukerji KG.1970. Fungi of Delhi XV. Lophotrichus indicus sp. nov. Acta Botanica Neerlandica 19: 722–726. [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, et al. 2011. The genera of Hyphomycetes. CBS Biodiversity Series no. 9: 1–997. Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. [Google Scholar]
- Silvestro D, Michalak I.2012. raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution 12: 335–337. [Google Scholar]
- Singer R.1942. A monographic study of the genera Crinipellis and Chaetocalathus. Lilloa 8: 441–534. [Google Scholar]
- Singer R.1955. Type Studies on Basidiomycetes. VIII. Sydowia 9: 367–431. [Google Scholar]
- Singer R, Digilio APL.1951. Pródromo de la Flora Agaricina Argentina. Lilloa 25: 5–461. [Google Scholar]
- Singtripop C, Hongsanan S, Li J, et al. 2016. Chaetothyrina mangiferae sp. nov., a new species of Chaetothyrina. Phytotaxa 255: 21–33. [Google Scholar]
- Soop K, Dima B, Cooper JA, et al. 2019. A phylogenetic approach to a global supraspecific taxonomy of Cortinarius (Agaricales) with an emphasis on the southern mycota. Persoonia 42: 261–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.2014. RAxML Version 8: A tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.2015. Using RAxML to infer phylogenies. Current Protocols in Bioinformatics 51: 6.14.1–6.14.14. [DOI] [PubMed] [Google Scholar]
- Stecher G, Tamura K, Kumar S.2020. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Molecular Biology and Evolution 37: 1237–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JA.1975. Fungi of Puerto Rico and the American Virgin Islands. Contribution of the Reed Herbarium 23: 1–743. [Google Scholar]
- Suh S-Q, McHugh JV, Blackwell M. 2004. Expansion of the Candida tanzawaensis yeast clade: 16 novel Candida species from basidiocarp-feeding beetles. International Journal of Systematic and Evolutionary Microbiology 54: 2409–2429. [DOI] [PubMed] [Google Scholar]
- Summerbell RC, Gueidan C, Guarro J, et al. 2018. The protean acremonium. A. sclerotigenum/egyptiacum: Revision, food contaminant, and human disease. Microorganisms 6: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL.2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0b10. Sunderland, Massachusetts: Sinauer Associates. [Google Scholar]
- Swofford DL.2003. PAUP* 4.0b10. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Hirayama K, Yonezawa H, et al. 2015. Revision of the Massarineae (Pleosporales, Dothideomycetes). Studies in Mycology 82: 75–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanney JB, Douglas B, Seifert KA.2016. Sexual and asexual states of some endophytic Phialocephala species of Picea. Mycologia 108: 255–280. [DOI] [PubMed] [Google Scholar]
- Tanney JB, Seifert KA.2020. Mollisiaceae: An overlooked lineage of diverse endophytes. Studies in Mycology 95: 293–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Crous PW.2001. Morphological variation and cultural characteristics of Coniothyrium leucospermi associated with leaf spots of Proteaceae. Mycoscience 42: 265–271. [Google Scholar]
- Thorn RG, Kim JI, Lebeuf R, et al. 2017. The golden chanterelles of Newfoundland and Labrador: a new species, a new record for North America, and a lost species rediscovered. Botany 95: 547–560. [Google Scholar]
- Trifinopoulos J, Nguyen L-T, Von Haeseler A, et al. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research: 44 (W1): W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneda A, Hambleton S, Currah RS.2004. Morphology and phylogenetic placement of Endoconidioma, a new endoconidial genus from trembling aspen. Mycologia 96: 1128–1135. [PubMed] [Google Scholar]
- Uljé CB. 2005. Coprinus. In: Noordeloos ME, Kuyper TW, Vellinga EC. (eds), Flora Agaricina Neerlandica 6: 22–109. Taylor & Francis, Boca Raton. [Google Scholar]
- Van der Plaats-Niterink AJ. 1981. Monograph of the genus Pythium. Studies in Mycology 21: 1–242. [Google Scholar]
- Varghese KI, Rao VG.1980. ‘1979. ’. Forest microfungi I. Subramaniomyces, a new genus of hyphomycetes. Kavaka 7: 83–85. [Google Scholar]
- Velmurugan S, Prasannakumar C, Manokaran S, et al. 2013. DNA barcodes for marine fungal identification and discovery. Fungal Ecology 6: 408–418. [Google Scholar]
- Videira SIR, Groenewald JZ, Nakashima C, et al. 2017. Mycosphaerellaceae – chaos or clarity? Studies in Mycology 87: 257–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, et al. 2014. Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D, Groenewald M, De Vries M, et al. 2019. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Hyde KD, Jeewon R, et al. 2017. Phylogenetic revision of Camarosporium (Pleosporineae, Dothideomycetes) and allied genera. Studies in Mycology 87: 207–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warcup JH, Talbot PHB.1967. Perfect states of Rhizoctonias associated with orchids. New Phytologist 66: 631–641. [Google Scholar]
- Watling R, Gregory NM.1986. Observations on the boletes of the Cooloola Sandmass, Queensland and notes on their distribution in Australia. Proceedings of the Royal Society of Queensland 97: 97–128. [Google Scholar]
- Weir BS, Johnston PR, Damm U.2012. The Colletotrichum gloeosporioides species complex. Studies in Mycology 73: 115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M, Waller F, Zuccaro A, et al. 2016. Sebacinales – one thousand and one interactions with land plants. New Phytologist 211: 20–40. [DOI] [PubMed] [Google Scholar]
- Wendt L, Sir EB, Kuhnert E, et al. 2018. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycological Progress 17: 115–154. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: A guide to methods and applications: 315–322. Academic Press, Inc., New York. [Google Scholar]
- Whitehead MR, Catullo RA, Ruibal M, et al. 2017. Evaluating multilocus Bayesian species delimitation for discovery of cryptic mycorrhizal diversity. Fungal Ecology 26: 74–84. [Google Scholar]
- Wijayawardene NN, Hyde KD, Bhat DJ, et al. 2015. Additions to brown spored coelomycetous taxa in Massarinae, Pleosporales: introducing Phragmocamarosporium gen. nov. and Suttonomyces gen. nov. Cryptogamie, Mycologie 36: 213–224. [Google Scholar]
- Woudenberg JHC, Hanse B, Van Leeuwen GCM, et al. 2017. Stemphylium revisited. Studies in Mycology 87: 77–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg JHC, Seidl MF, Groenewald JZ, et al. 2015. Alternaria section Alternaria: Species, formae speciales or pathotypes? Studies in Mycology 82: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Li YC, Zhu XT, et al. 2016. One hundred noteworthy boletes from China. Fungal Diversity 81: 25–188. [Google Scholar]
- Yilmaz N, Visagie CM, Houbraken J, et al. 2014. Polyphasic taxonomy of the genus Talaromyces. Studies in Mycology 78: 175–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Lu YF, Liu XJ, et al. 2017. Cyberlindnera xishuangbannaensis f.a., sp. nov., a yeast isolated from rotting wood. International Journal of Systematic and Evolutionary Microbiology 67: 5051–5055. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ.2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation, The University of Texas at Austen. [Google Scholar]