Abstract
The endogenous circadian period of animals and humans is typically very close to 24 hours. Individuals with much longer circadian periods have been observed, however, and in the case of humans, these deviations have health implications. Previously, we observed a line of Drosophila with a very long average period of 31.3 hours for locomotor activity behavior. Preliminary mapping indicated that the long period did not map to known canonical clock genes but instead mapped to multiple chromosomes. Using RNA-Seq we surveyed the whole transcriptome of fly heads from this line across time and compared it to a wild-type control. A three-way generalized linear model revealed that approximately two-thirds of the genes were expressed differentially among the two genotypes, while only one quarter of the genes varied across time. Using these results we applied algorithms to search for genes that oscillated over 24 hours, identifying genes not previously known to cycle. We identified 166 differentially expressed genes that overlapped with a previous GWAS of circadian behavior, strongly implicating them in the long period phenotype. We tested mutations in 45 of these genes for their effect on the circadian period. Mutations in Alk, alph, CG10089, CG42540, CG6034, Kairos (CG6123), CG8768, klg, Lar, sick, and tinc had significant effects on the circadian period, with seven of these mutations increasing the circadian period of locomotor activity behavior. Genetic rescue of mutant Kairos restored the circadian period to wild-type levels, suggesting it has a critical role in determining period length in constant darkness.
Keywords: long circadian period, Drosophila melanogaster, RNA-Seq, systems genetics, circadian rhythms
Introduction
Circadian rhythms are ubiquitous, exhibited at the molecular, biochemical, physiological, behavioral, and population levels across all organisms (Merrow et al., 2005; Huang et al., 2011). These biological processes typically fluctuate over 24 hours, and are under partial genetic control (Emery et al., 1995; Hofstetter et al., 1995; Harbison et al., 2019; Keenan et al., 2020). Both naturally-occurring and chemically-induced EMS mutations in canonical clock genes can produce free-running locomotor period lengths significantly deviant from the characteristic 24 hour circadian rhythm (Konopka and Benzer, 1971; Rothenfluh et al., 2000; Tauber et al., 2007). For instance, a missense point mutation in the Per-Arnt-Sim domain encoding region of the canonical clock gene period (per) lengthened the circadian period to 28.6 hours in constant darkness (Konopka and Benzer, 1971; Hardin et al., 1990). Period lengthening mutants of timeless (tim) have also been identified using forward genetic screens, with periods ranging from 26–28 hours (Rothenfluh et al., 2000). Thus, mutations in a single canonical clock gene can substantially increase the circadian period.
In Drosophila melanogaster, circadian rhythms require oscillations in the transcripts of canonical clock genes; accordingly, the transcripts of per and tim oscillate over a 24-hour cycle (Merrow et al., 2005; Huang et al., 2011). A previous study showed an inverse relationship between per transcript levels and the behavioral periodicity- i.e., flies with low levels of per transcript levels displayed longer periods and those with higher per abundance exhibited shorter periodicities in their locomotor activity (Baylies et al., 1987). Thus, transcript levels of canonical genes play an important role in determining/influencing the length of circadian rhythms in Drosophila.
We recently observed flies with a very long circadian period in the process of conducting a Genome-Wide Association Study (GWAS) of circadian behavior in the Drosophila Genetic Reference Panel (DGRP) (Harbison et al., 2019). DGRP_892 is a wild-derived inbred line of the DGRP with an average circadian period of 31.3 hours (Harbison et al., 2019). Unlike the known canonical clock polymorphisms producing a long period, preliminary mapping suggested that the long period phenotype mapped to multiple regions in the genome (Harbison et al., 2019). Importantly, DGRP_892 was removed from the genome-wide association analysis as the long period phenotype was an outlier. The remaining lines of the DGRP were used to compute the genotype-phenotype associations. The GWAS identified 584 SNPs tagging 268 genes associated with circadian period and rhythmicity index (Harbison et al., 2019).
In principle, the genes contributing to the long period phenotype in DGRP_892 could be among the 268 identified in the GWAS. However, picking the most promising genes from a large list of potential candidates remains one of the challenges of functional genomics. One way to prioritize candidate genes for further study is to choose those genes repeatedly identified among disparate data analyses. This systems genetics approach has been applied previously to identify gene networks affecting sleep, sleep deprivation, and recovery sleep in mice, where the results of quantitative trait locus (QTL) mapping, expression QTL mapping, predicted annotation effects, differential gene expression analyses, and quantitative trait transcript mapping were combined to prioritize genes (Diessler et al., 2018). Transcriptional expression quantitative trait locus mapping data were also used to refine large genomic QTL for sleep in flies (Smith and Macdonald, 2020). Thus, combinatorial systems genetics approaches can assist in the prioritization of important candidate genes for complex traits.
With this approach in mind, we surveyed the transcriptome of whole heads in DGRP_892 over time and compared it to the transcriptome of a line with a 24-hour circadian period. We identified transcripts differentially expressed among genotype, time, sex, and their interactions. We then compared the transcriptional data to the previous GWAS (Harbison et al., 2019). We found that 62% of the genes identified by the GWAS also had genes differentially expressed between the two genotypes. In addition, using RAIN and JTK_CYCLE, we identified transcripts with putatively aberrant cycling in DGRP_892. These combined analyses suggested 166 candidate genes that might affect circadian period.
Of the 166 candidate genes, 45 had Minos insertion lines available for testing. We assessed the circadian period of these Minos insertions and confirmed 11 genes with significant differences in circadian period as compared to the isogenic control. These genes may therefore contribute to the long period in DGRP_892. In addition, we rescued one of the long-period phenotypes by precise excision of a Minos element in CG6123, a gene we have renamed Kairos. In this way we demonstrate that the relatively common variants we identified in the DGRP have altered regulation of gene expression in DGRP_892 that may explain its long circadian period.
Materials and Methods
Drosophila stocks
We investigated gene expression in DGRP_892, a line from the Drosophila Genetic Reference Panel (Mackay et al., 2012). We previously demonstrated that DGRP_892 has a very long circadian period; on average, the circadian period was 31.3 hours in these flies (Harbison et al., 2019). We used w1118; Canton-S B (CSB) as a control line; these flies have an average circadian period of 23.9 hours (Harbison et al., 2019).
To confirm the genes that we identified, we tested circadian behavior in Minos element mutants targeting 45 candidate genes. These transposable element mutations are readily available from the Bloomington, IN stock center (BDSC, Indiana University) and have isogenic control lines (Bellen et al., 2011). Ten Minos elements targeted genes with expression that was predicted to cycle and overlapped previous GWAS results (Harbison et al., 2019). We tested an additional 35 candidate genes that were differentially expressed between the DGRP_892 and CSB genotypes and overlapped the previous GWAS results (Harbison et al., 2019). Table S1 lists the mutant lines tested.
Sample collection for RNA-Seq
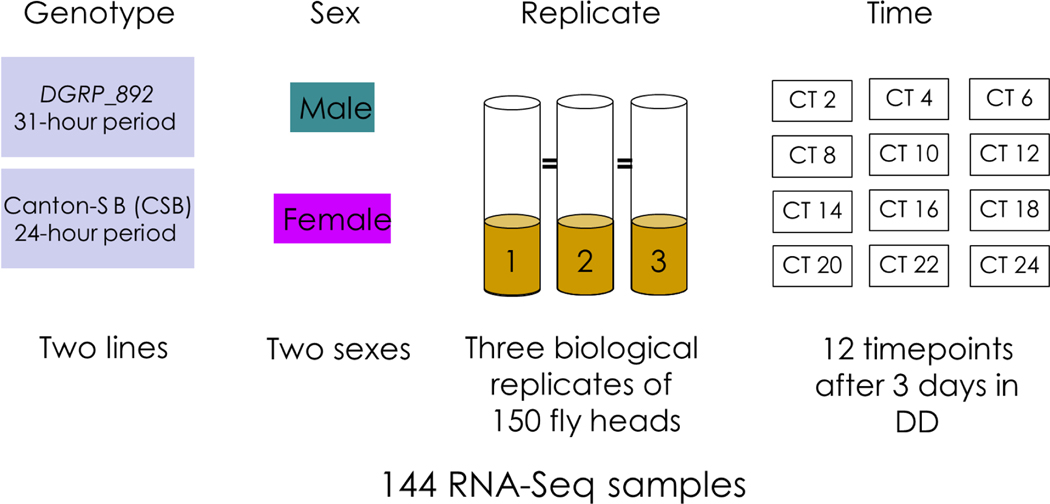
Flies were maintained under standard culture (cornmeal-sucrose-agar medium, 25°C, 60–75% relative humidity) and lighting conditions (12-hour light: dark cycle) (LD). Five virgin males and five females of were used to set up w1118; Canton-S B and DGRP_892 cultures in an LD incubator. Soon after eclosion, 24 separate-sex vials with 150 virgin males and 150 virgin females per vial were collected for each genotype. One set of flies was entrained in an incubator with a 12:12 LD cycle (6:00 AM ‘lights on’; 6:00 PM ‘lights off’) and another set was entrained in an incubator with a reverse LD cycle (6:00 PM ‘lights on’ and 6:00 AM ‘lights off’) for 3 days. Both groups of flies were then transferred to a common incubator in constant darkness (DD). On the 3rd day of DD, flies were flash-frozen on dry ice every 2 hours starting at circadian time (CT02/CT12) and maintained at −80°C until further processing. Flies were decapitated on dry ice for RNA extraction. We replicated this procedure three times to produce 144 RNA samples (3 biological replicates, 2 sexes, 2 genotypes and 12 time points).
RNA extraction
Samples were randomized and extracted in groups of 12. For each RNA sample, 150 heads were placed in Omni-ruptor tubes (Omni International, Kennesaw, GA) with 4 RNase/DNase-free metal beads (Omni International, Kennesaw, GA) and homogenized in 1 ml TRIzol (Thermo Fisher Scientific, Grand Island, NY) using an Omni-bead ruptor with the following settings: Speed, 5.65 m/s; Time, 20 sec; Duration, 30 sec; Cycles, 2. Homogenates were incubated at room temperature for 5 minutes. 200 μl of chloroform (Mallinckrodt Baker, Center Valley, PA) were added to each tube; samples were then mixed by shaking for 15 seconds, incubated at room temperature for 3 minutes, and centrifuged at 4°C, 12,000 x g for 15 minutes. The 500-μl aqueous phase was transferred to new tubes and RNA was further purified using the PureLink RNA Mini Kit (Life Technologies, Carlsbad, CA) with on-column DNase treatment, according to the manufacturer’s instructions. Purified total RNA was eluted in 60 μl of water and stored at −80°C. RNA was quantified by NanoDrop 8000 (Thermo Fisher Scientific, Grand Island, NY) and quality verified by BioAnalyzer 2100 using the Agilent RNA 6000 Pico Kit (Agilent Technologies, Santa Clara, CA).
RNA library preparation and sequencing
Prior to poly(A) RNA selection, 0.02 μl ERCC Spike-In Control Mix 1 (Life Technologies, Carlsbad, CA) was added to 1 μg total RNA. Poly(A)-selected stranded mRNA libraries were constructed from 1 μg total RNA using Illumina TruSeq Stranded mRNA Sample Prep Kits (Illumina, San Diego, CA) per the manufacturer’s instructions with the following exception: PCR amplification was performed for 10 cycles rather than 15 to minimize the risk of over-amplification. Unique barcode adapters were applied to each library; dual indices were used. Libraries were pooled for sequencing.
The pooled libraries were sequenced on multiple lanes of a HiSeq4000 using version 4 chemistry to achieve a minimum of 12 million 76-bp read pairs. The sequences were processed using RTA version 1.18.64 and CASAVA 1.8.2 (Illumina, San Diego, CA).
Alignment
Quality Control (QC) of FASTQ files were assessed using FastQC toolkit (v0.11.5) using default parameters (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). The paired-end reads were aligned against the Drosophila melanogaster reference genome (dmel.r6.16) (dos Santos et al., 2015) using STAR (v2.5.3a) alignment software with the following parameters “--twopassMode basic --sjdbOverhang 75” (Dobin et al., 2013). sjdbOverhang is the parameter for the length of the donor/acceptor sequence on each side of the junctions. Gene level read counts were produced by featureCounts (v1.5.2) (Liao et al., 2013; Liao et al., 2014) using only uniquely mapped reads with setting “-p -s 2” (paired-end, reverse- stranded). We used the trimmed mean of M-values (TMM) normalization method to normalize raw read counts (Robinson and Oshlack, 2010).
Sequence Analysis
We computed the distribution of TMM normalized read counts mapping to either genic or intergenic regions. We used the 95th percentile of that distribution to detect the threshold for removing low count genes, which was 4.67 log2(TMM normalized counts +1) (Lin et al., 2016b). Genes with read counts smaller than the threshold for all samples were discarded from further analysis. Using the ERCC spike-in reads, we noted a bias in the data according to the date of the sequencing run and confirmed it in the sequence data using Principal Component Analysis (Supplemental Figure S1). We accounted for this bias with an additional parameter in our models (see below). We performed a likelihood ratio test using DESeq2 (nbinomLRT) to identify differentially expressed genes (Love et al., 2014). We made the following comparisons for each gene i.
To identify genes with significant differential gene expression across timepoints, we compared the full model
| (1) |
to the reduced model
| (2) |
where G is genotype (DGRP_892 or Canton-S B), S is sex, T is time, and B is batch effect due to sequencing date. We identified genes with significant differential expression across genotype and sex in the same manner. We identified significant two-way interactions among these terms (i.e., G×S, G×T, and S×T) by comparing the full model
| (3) |
to the reduced model given by equation (1);
| (4) |
to the reduced model given by equation (3); and
| (5) |
to the reduced model given by equation (4). Finally, we identified significant three-way interactions (G×S×T) by comparing the full model
to the reduced model given by equation (5). Adjusted P values were calculated using the software default, which uses the Benjamini-Hochberg method to determine false discovery rates (Benjamini and Hochberg, 1995). Our threshold was an adjusted P value of 0.05 or less.
Genes with significant differential expression were further classified for potential gene-gene interactions using Modulated Modularity Clustering (MMC) (Stone and Ayroles, 2009). MMC calculates the correlation of gene expression between any two gene pairs. The magnitude of the correlation is presumed to reflect the level of interaction among gene pairs. MMC applies an iterative graphical method to cluster correlated genes into highly interconnected groups. The MMC algorithm determines the number of highly interconnected clusters (or ‘modules’) rather than relying on a user-selected number of clusters. The correlations are re-ordered and graphed with the most highly interconnected groups of genes presented at the top left, while the least interconnected genes are graphed at the bottom right.
We further analyzed genes having significant differential expression among model terms containing time (i.e., T, G×T, and G×T×S) for possible circadian cycling using the JTK_Cycle and RAIN packages (Hughes et al., 2010; Thaben and Westermark, 2014). First, we removed the batch effect in our dataset from TMM-normalized reads using the ComBat function in the sva R package (Leek and Storey, 2007): (ComBat(data,batch=date,mod=”geno+sex+time”). Using RAIN, DGRP_892 and CSB were analyzed separately by sex with the following settings, rain(t(data),deltat=2,period=24,nr.series=3,period.delta=0,peak.border=c(0.35,0.65). In JTK_Cycle, we used the following settings:(jtkdist(12,30 jtk.init(10:12,2)) for 12 samples, 3 replicates, min_period: 20, max_period: 24. These settings enabled us to search for rhythmic gene expression over a 24-period. We repeated this analysis but adjusted the parameters in JTK_Cycle and RAIN to search for gene expression that was potentially cycling at longer periods, up to 32 hours. The Benjamini-Hochberg procedure was used to control the false discovery rate.
Creation of Minos precise excision line
We followed the protocol for Minos element excision as previously reported to excise the insertion in CG6123 (Metaxakis et al., 2005). For the generation of excision events in the germ line of flies with single Minos insertions, flies homozygous for a Mi[ET1] transposon insertion on the X chromosome (Mi[ET1]CG6123[MB02356]) were crossed with flies carrying helper chromosome w[1118]; sna[Sco]/SM6a, P[w[+mC]=hsILMiT]2.4 (F1). Two days after setting up the crosses, the parent flies were removed and larvae in the vials were heat-shocked for one hour per day for the next three days in a 37°C water bath. Transformed progeny were screened for the presence of red mosaic GFP eyes, indicating a partially transposed Minos element. Individual transformed males were backcrossed to females from the original Minos insertion line, CG6123[MB02356] (F2). The female progeny of this cross were heterozygous flies containing one copy of the GFP-positive Minos insertion and one copy of the GFP-negative putative excision chromosome, while GFP-negative males could be selected for directly. These progeny were crossed, and GFP-negative progeny were selected to create homozygous excision lines. DNA was extracted from the cultures to confirm precise excisions via PCR amplification and sequencing. We analyzed the Mi[ET1] excision event by PCR using a set of oligos flanking the original insertion site. The oligo sequences used were: Forward Primer1-TTGGTTGCGGAGTAAGTACG; Reverse Primer1-TGCGAATACAGATGCGGATA, Forward primer2 CATGGCCTTTGAACAGGATT; Reverse primer2- TGCGAATACAGATGCGGATA, MI.3.Forward primer- ATGATAGTAAATCACATTACG; MI.3.Reverse primer- CAATAATTTAATAATTTCCC, MI.5.Forward primer- CAAAAGCAACTAATGTAACGG; MI.5.Reverse primer-TTGCTCTTCTTGAGATTAAGGTA.
Validation of candidate genes and precise excision line
Flies were maintained under standard culture (cornmeal-sucrose-agar medium, 25°C, 60–75% relative humidity) and lighting conditions (12-h light: dark cycle (LD). Five virgin male and five virgin females were used to set up cultures in an LD incubator. Virgin flies were collected and maintained in sex-separate vials for 3 days at LD. Individual flies were then transferred to DAM2 monitors (Trikinetics, Waltham, MA). Flies were placed into DAM monitor tubes with 5% sucrose, 2% agar food. Flies were monitored for 4 days in LD followed by 14 days in DD to assess their entrainment and free-running behavior respectively. Sixteen flies per sex per line were tested. We calculated circadian period using the ClockLab program (Actimetrics, Wilmette, IL). We analyzed the data using the ANOVA model Y = μ + G + S + G × S + ε, where G is genotype and S is sex. For the precise excision line, we compared the circadian period of the excision line (CG6123MB*), the heterozygote (CG6123MB*/MB02356), the homozygous mutant line (CG6123MB02356), and the w1118 control with a post-hoc Tukey test. We retested Minos and precise excision lines having significant genotypic effects, and analyzed the data with the ANOVA model Y = μ + G + S + R + G×S + G×R + R×S + G×S×R + ε, where G and S are as defined above and R is replicate.
Data availability
All RNA-Seq data from this study are available from the National Center for Biotechnology Information (NCBI) GEO under the accession number GSE155018, including raw read counts and batch-adjusted TMM-normalized read counts for each gene per genotype/replicate/sex. All supplemental figures and tables are available at figshare under the following link: https://figshare.com/s/637491852db02ea31527.
Results
Previously, we identified a line of the Drosophila Genetic Reference Panel (DGRP) with an unusually long circadian period of 31.3 hours (Harbison et al., 2019). We collected and sequenced RNA from fly heads of this line, DGRP_892, and compared it to a line with a 24-hour circadian period, CSB. We collected the RNA at 2-hour intervals over a 24-hour period in order to identify cycling transcripts. Fig. 1 shows the experimental setup: three replicate biological samples were collected according to genotype, timepoint, and sex. We found 11,474 transcripts with detectable expression in fly heads. Using a generalized linear model, we identified genes that were differentially expressed by Genotype, Time, and Sex, and their interactions. Table 1 lists the numbers and percentages of differentially expressed genes according to the experimental factor. Normalized read counts for each gene and additional details can be found in Table S2. In total, we found 10,119 genes with differential expression (FDR adjusted P value < 0.05). Increasing the stringency of the FDR adjusted P value to 0.01 or 0.001 (Table S3) had little effect on the numbers of differentially expressed genes among Genotype or Sex but did affect model terms containing the environmental factor of Time, as might be anticipated from previous work (Lin et al., 2016b).
Figure 1.
Experimental setup.
Table 1.
Numbers of differentially expressed genes by factor in the generalized linear model. Percentages of differentially expressed genes are based on the 11,474 genes with detected expression in each sample.
| Factor | No. differentially expressed genes (%) |
|---|---|
|
| |
| Genotype (G) | 7,685 (66.9) |
| Time (T) | 2,692 (23.4) |
| Sex (S) | 7,658 (66.7) |
| G × T | 27 (0.23) |
| G × S | 1,953 (17.0) |
| S × T | 84 (0.73) |
| G × T × S | 1 (0.01) |
Differential expression between DGRP_892 and CSB
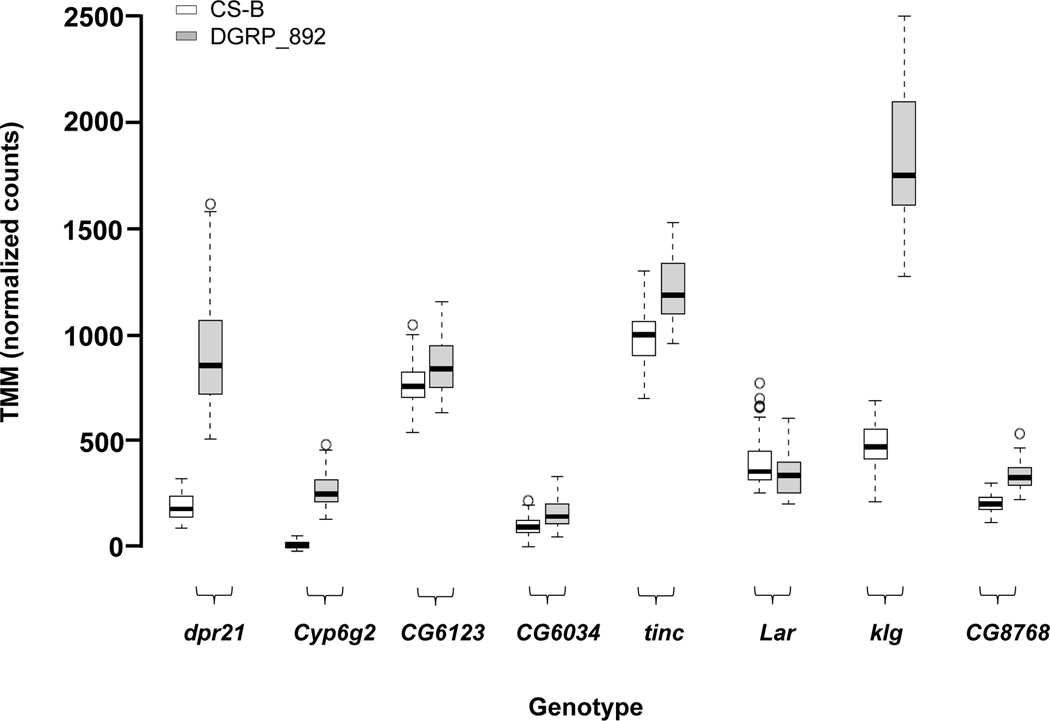
Many of the 11,474 detectable genes were differentially expressed between DGRP_892 and CSB: 66.9% (7,685) genes were differentially expressed by Genotype, similar to other studies which observed that 66%−72% of the detected genes had differential gene expression among genotypes of the DGRP (Lin et al., 2016a; Everett et al., 2020). Some examples of genes with differential expression between the two genotypes are shown in Fig. 2A. Slightly less than half (47.9%) of the transcripts had higher gene expression in DGRP_892 compared to CSB. The vast majority of these genes (94.8%) were protein coding genes, with the remaining classified as non-protein coding (Table S2). We used Modulated Modularity Clustering (MMC) to cluster all 7,685 significantly differentially expressed genes into modules (Stone and Ayroles, 2009) (Table S4). This procedure clustered genes with highly correlated expression into 15 large modules (Fig. 2B). These modules predict co-regulated networks of gene expression (Harbison et al., 2009). Gene Ontology (GO) analyses (Yu et al., 2012) identified significant enrichment for many different terms in each module (Table S5). For example, Module 2 was enriched for genes involved in circadian rhythms and sleep, such as Cul3, Hk, jet, Pka-R2, Rdl, Sh, and sgg (Fig. 2B; Table S5) (Park et al., 2000; Martinek et al., 2001; Cirelli et al., 2005; Koh et al., 2006; Bushey et al., 2007; Parisky et al., 2008; Stavropoulos and Young, 2011). Module 2 contains a large group of genes (725 total) potentially interacting with these circadian rhythm/sleep genes. Thus, most of the genes with detectable expression in the head were differentially expressed among genotypes, and some of these genes included known circadian and sleep genes.
Figure 2.
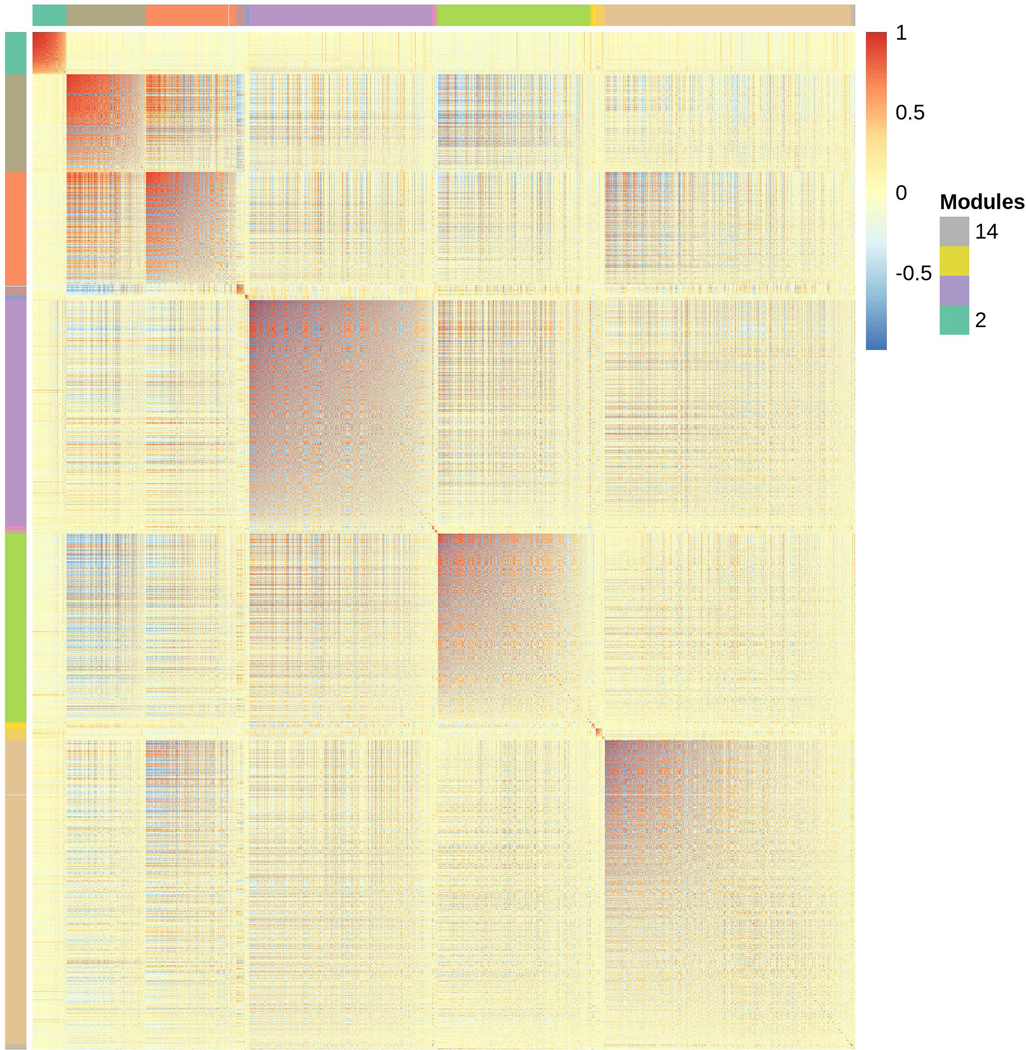
Genes differentially expressed between DGRP_892 and CSB. (a) Representative boxplots of TMM-normalized gene expression for dpr21, Cyp6g2, CG6123, CG6034, tinc, Lar, klg, and CG8768. (b) Plot of modules identified using Modulated Modularity Clustering. Each square in the plot represents the correlation between two genes; all 7,685 genes are shown in the plot. The red-white-blue scale bar ranges high positive correlation (1.0, red) to high negative correlation (−1.0, blue). Colored bars demarcate the boundary of each module. Clusters of genes (modules) range from the most highly interconnected (upper left) to the least interconnected (lower right).
We found that 166 of the 7,685 differentially expressed genes overlapped with 268 previously identified in the genome-wide association study of circadian behavior, an enrichment over what would be expected by chance (Table S6; P < 0.0001, hypergeometric test). Interestingly, only 34 of the 166 genes were also significant for the main effect of Time or the Genotype × Time interaction, raising the possibility that some of the effects on circadian period occur in a time-independent fashion. We therefore chose candidate genes from the overlap of the two studies for further testing (see Verification of Candidate Genes below).
Differential gene expression across time
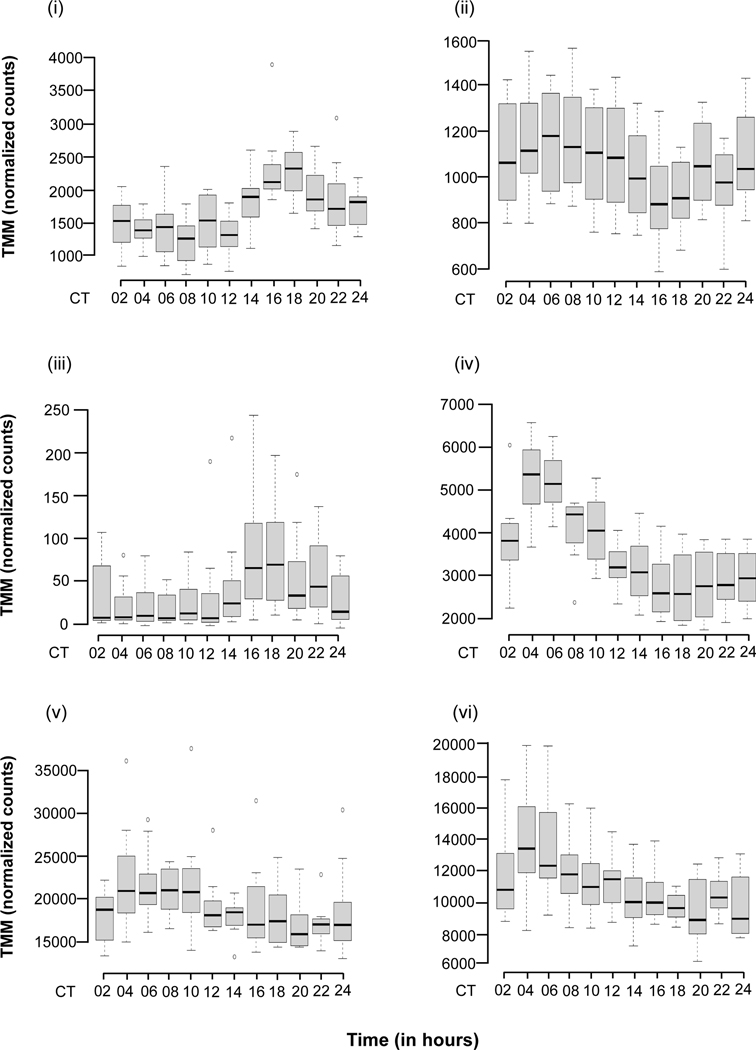
Roughly one quarter of the genes with detectable expression also varied significantly by time point across 24 hours (Table 1; Table S2). Some representative examples of changes in gene expression over time can be seen in Fig. 3A, including the canonical circadian clock genes per and Clk, whose transcripts are known to cycle over 24 hours (Hardin et al., 1990; Hardin, 2004). We used MMC to cluster the gene expression of all 2,692 genes. MMC analysis clustered the correlation among each gene pair into 8 modules, including one very large module with 1,637 genes, Module 4 (Fig. 3B; Table S7). Only Module 4 was enriched for GO terms (Table S8). Genes that vary significantly with time may be cycling in a circadian fashion, or they may simply have differential gene expression at a single time point. We therefore used these 2,692 genes in a further analysis to determine whether the time-specific differences in gene expression could be attributed to cycling behavior (see JTK_CYCLE and RAIN analyses below).
Figure 3.
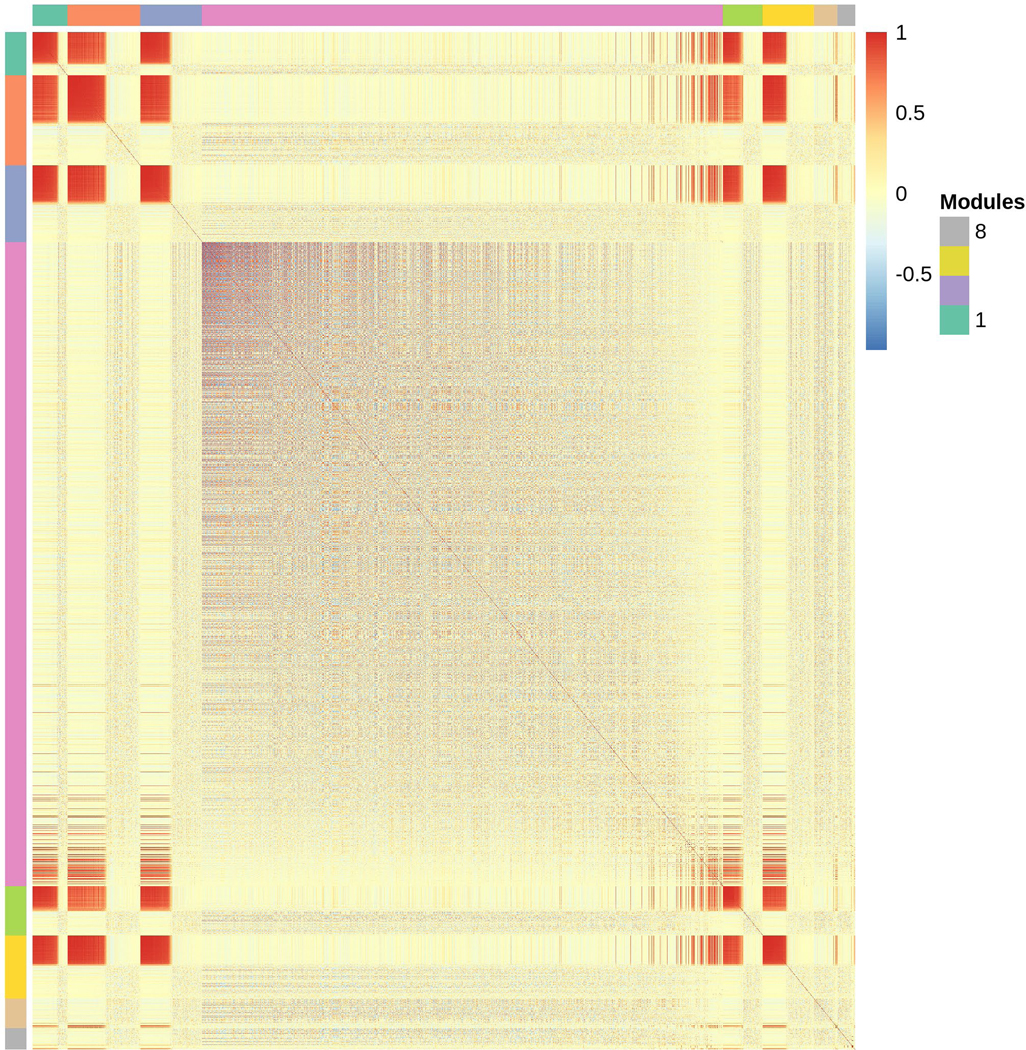
Genes differentially expressed with time. (a) Representative boxplots of TMM-normalized gene expression for (i) per, (ii) Clk, (iii) dy, (iv) gol, (v) inaF-B, and (vi) Slob are plotted against time point. The box plots represent the distribution of TMM-normalized gene expression values for both genotypes and sexes combined. CT denotes circadian time in constant darkness. (b) Plot of modules identified using Modulated Modularity Clustering. Each square in the plot represents the correlation between two genes; all 2,692 genes are shown in the plot. The red-white-blue scale bar ranges from high positive correlation (1.0, red) to high negative correlation (−1.0, blue). Colored bars demarcate the boundary of each module. Clusters of genes (modules) range from the most highly interconnected (upper left) to the least interconnected (lower right).
Significant Genotype × Time differential gene expression
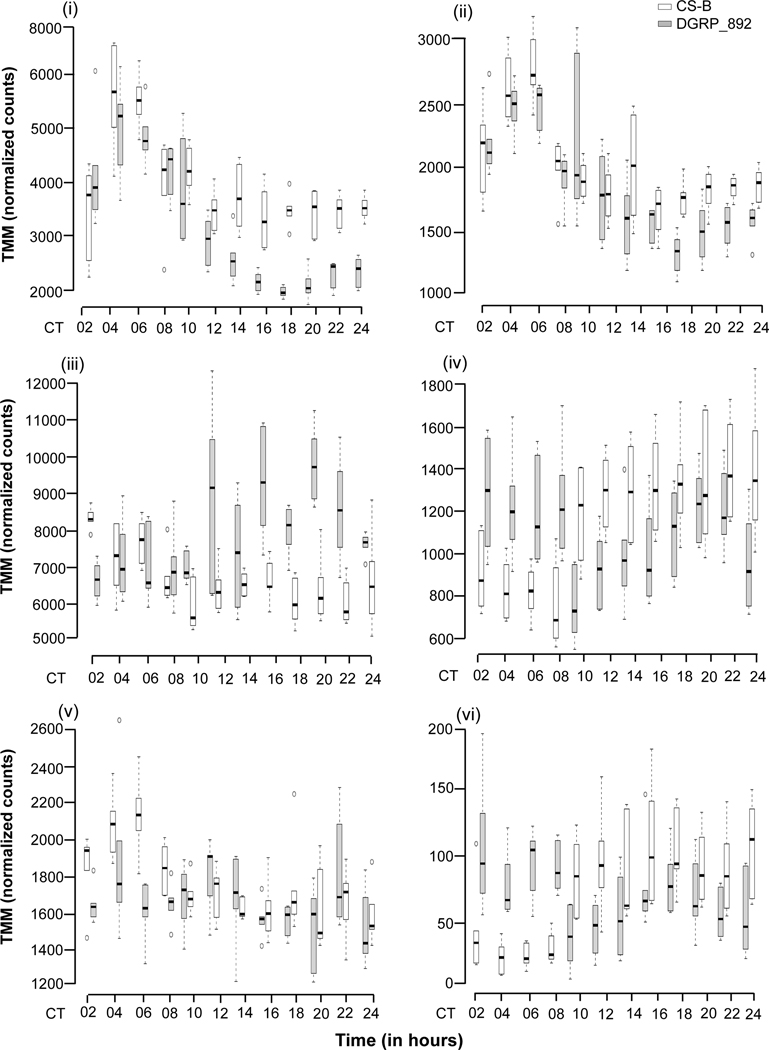
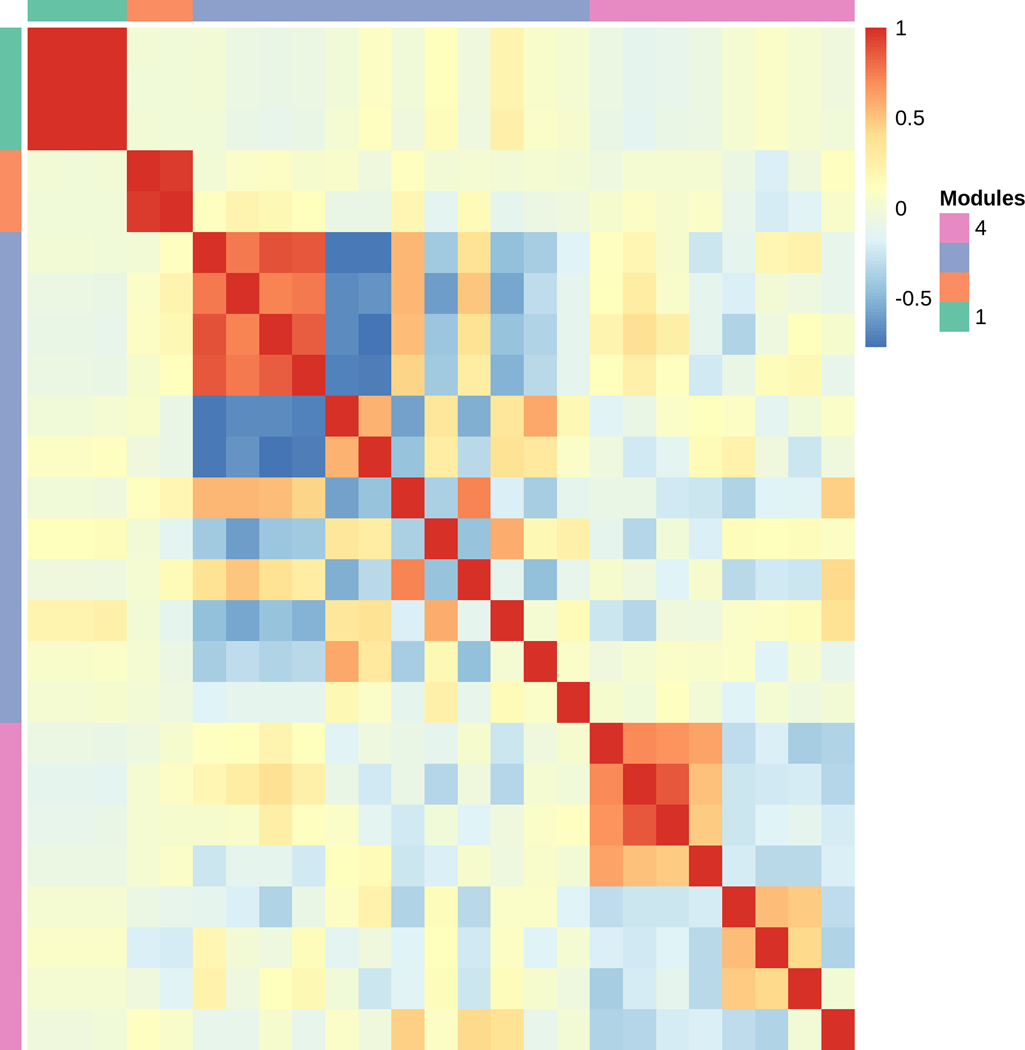
In addition to genes differentially expressed by the main effects of Genotype and Time, we wanted to know whether any of the genes were differentially expressed by the interaction of Genotype and Time. We hypothesized that if significant Genotype × Time interactions in gene expression existed, it would suggest potential differences in cycling behavior that might underlie the long circadian period. Only 27 genes had significant differential gene expression for this interaction term (Table S2). per was the only canonical clock gene with a significant Genotype × Time interaction. Some representative genes have been plotted in Fig. 4A. Using MMC, the pairwise correlation of gene expression among these 27 genes clustered into four modules (Table S9). Due to the small number of genes significant for Genotype × Time, we did not observe much GO term enrichment in the modules, with the exception of Module 1 (Table S10). This module consisted of three heat shock proteins, Hsp23, Hsp26, and Hsp27, which were associated with the GO terms protein folding/refolding and response to temperature and heat shock stress. As with the genes that were differentially expressed by Time, significant Genotype × Time terms might represent circadian cycling. These genes were also used in the analysis to identify cycling genes outlined below.
Figure 4.
Genes with a significant Genotype × Time interaction. (a) Representative boxplots of TMM-normalized gene expression for (i) gol, (ii) Usp8, (iii) Rh5, (iv) Cyp6a21, (v) Dnr1, and (vi) CG33965 are plotted against time point for DGRP_892 (gray boxes) and CSB (white boxes). CT denotes circadian time in constant darkness. (b) Plot of modules identified using Modulated Modularity Clustering. Each square in the plot represents the correlation between two genes; all 27 genes are shown in the plot. The red-white-blue scale bar shows ranges from high positive correlation (1.0, red) to high negative correlation (−1.0, blue). Colored bars demarcate the boundary of each module. Clusters of genes (modules) range from the most highly interconnected (upper left) to the least interconnected (lower right).
Sex-specific differential gene expression
Although both sexes had an elongated circadian period, a 50.4-minute difference was observed between male and female flies of DGRP_892, with males having a longer circadian period than females (Harbison et al., 2019). Thus, sex-specific differences in gene expression may contribute differentially to the long period. Accordingly, we sequenced RNA from males and females separately. As anticipated from many previous experiments using Drosophila (Jin et al., 2001; Arbeitman et al., 2002; Parisi et al., 2003; Ranz et al., 2003; Harbison et al., 2005; Wayne et al., 2007; Zhang et al., 2007; Ayroles et al., 2009; Huylmans and Parsch, 2014; Huang et al., 2015; Lin et al., 2016a; Everett et al., 2020), most of the genome (66.7%) was differentially expressed between male and female heads (Table 1; Table S2). We observed an enrichment of sex-specific differential expression on the X chromosome. The number of differentially expressed genes on the X chromosome, 1,290, is 9.95% greater than would be expected by chance, while the number of differentially expressed genes on Chromosome 2L, 1,422, is 7.62% lower than expected by chance (Chi-squared test, P = 0.001). MMC analysis clustered these genes into 7 large modules with many significant GO terms (Tables S11 and S12). Similarly, 1,953 genes were differentially expressed by the Genotype × Sex interaction term, and 84 by the Sex × Time interaction term (Table 1). MMC clustering identified 13 modules for Genotype × Sex and 4 modules for Sex × Time (Tables S13 and S14, respectively) and corresponding GO terms (Tables S15 and S16, respectively). Only one gene was significant for the three-way interaction of Genotype × Time × Sex, Btk29A. Sex-specific differences in gene expression were ubiquitous in fly heads and may therefore contribute to sex-specific differences in the long circadian period.
JTK_CYCLE and RAIN analysis to identify cycling gene expression
As mentioned previously, differences in gene expression across time may be due to circadian cycling, or they may be due to a difference in gene expression at a single time point (Hsu and Harmer, 2012). We used more robust analyses to determine whether the genes differentially expressed by Time or Genotype × Time were in fact cycling. We used JTK_CYCLE and RAIN analysis to predict whether these genes were cycling with a 24-hour period (Hughes et al., 2010; Thaben and Westermark, 2014). We performed the analysis for each genotype and sex separately. Using JTK_CYCLE, 343 unique genes were predicted to cycle with 24-hour periodicity (adjusted P < 0.05). RAIN analysis predicted 1,286 unique genes would cycle with 24-hour periodicity (adjusted P < 0.05), and all of the genes identified by JTK_CYCLE were also predicted by RAIN (Table S17). Cycling transcripts were easier to detect in female heads than in males: only 34.4% of the transcripts predicted to cycle came from the male data. As expected, most of the putatively cycling transcripts were identified from CSB, the line with the 24-hour period; only 14.3% of the transcripts predicted to cycle came from DGRP_892 data. Overall, 132 genes were predicted to cycle in both DGRP_892 and CSB; 1,102 were predicted to cycle in CSB only, and 52 were predicted to cycle in DGRP_892 only.
Canonical clock genes were predicted to cycle using these analyses. For CSB, per was the only canonical clock gene predicted to cycle in both sexes using JTK_Cycle, while Clk and vri were predicted to cycle in CSB in one sex only. None of the canonical clock genes were predicted to cycle in DGRP_892 by JTK_Cycle. When the less restrictive RAIN algorithm was used, however, canonical clock genes per, tim, and Clk were predicted to cycle in both genotypes, while Pdp1 and vri were predicted to cycle in CSB only.
Additionally, we searched for genes putatively cycling at longer period lengths, from 26 to 32 hours. Only one gene, gol, was predicted to cycle over a 28-hour period in DGRP_892 using JTK_Cycle, and it was specifically predicted to cycle in females. Interestingly, gol was also predicted to cycle in both sexes of CSB with a 24-hour period. We found that 24 of the 1,286 genes predicted to cycle by RAIN overlapped with the 268 identified in the previous GWAS, including gol (Harbison et al., 2019). We subjected a subset of these genes to further verification and testing as outlined below.
Verification of candidate genes
As mentioned previously, 166 genes significant for the main effect of Genotype overlapped with the GWAS. Among these 166 genes, 24 genes were predicted to cycle from the JTK_CYCLE/RAIN analysis, and 39 are expressed in clock neurons (Kula-Eversole et al., 2010; Nagoshi et al., 2010; Abruzzi et al., 2017) (Table S18). We identified 45 Minos insertion mutant lines available from these 166 genes to test for their effects on circadian period. Ten of these genes were predicted to cycle in CSB and not DGRP_892. The remaining 35 lines came from the overlap of Genotype/GWAS. Initially, 17 mutants exhibited significant effects on circadian period. We repeated the phenotypic measures and found that 11 mutant alleles had significant effects on circadian period that were confirmed by re-test: Alk, alph, CG10089, CG42540, CG6034, CG6123, CG8768, klg, Lar, sick, and tinc (Bonferroni corrected P value = 0.001; Table S1). Interestingly, the effect of the mutation in Alk was largely confined to females, while the effect of CG10089 was predominantly due to a small shift in male behavior (Table S1). In addition, CG6123 was reported to have an effect on circadian period previously (Harbison et al., 2019). Minos insertions in CG6123, klg, and tinc had the greatest increases in circadian period over the control, increasing period by 1.63, 1.70, and 0.97 hours on average in both males and females, respectively (Table S1). Thus, we identified genes altering circadian period, including some mutants that produced large increases in period when tested.
Genetic rescue of CG6123
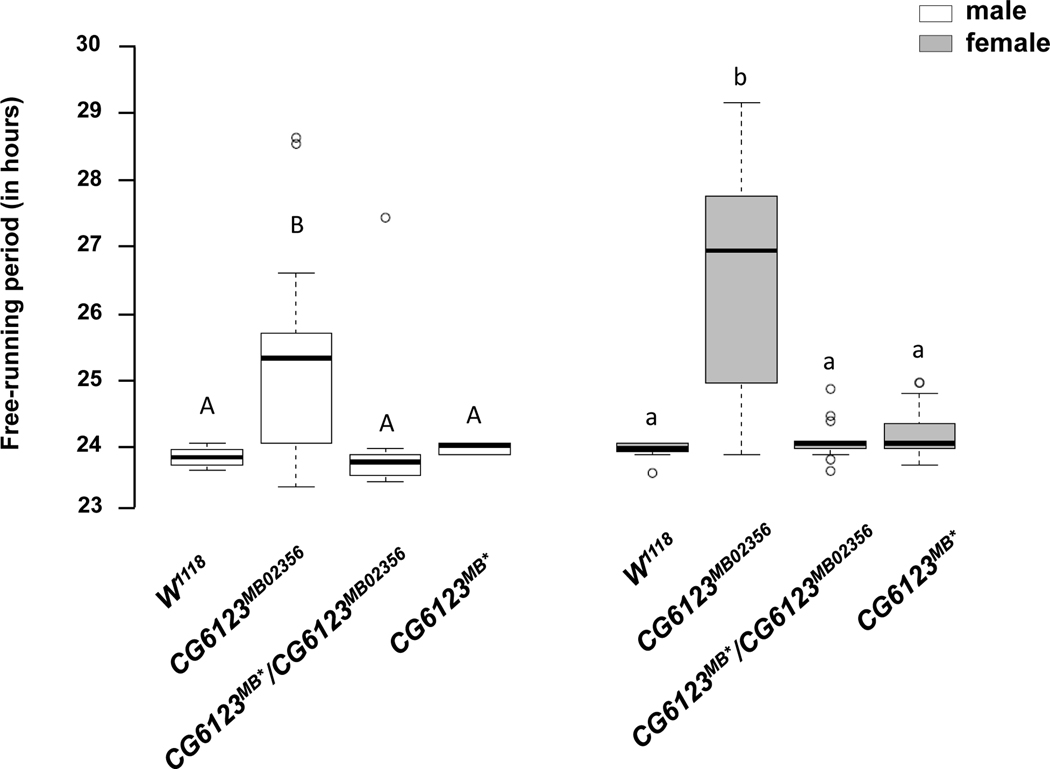
We generated a precise excision of the Minos element in CG6123 (see Materials and Methods). We verified the excision through PCR and sequencing analysis (Fig. S2, S3, and S4). We measured the locomotor activity rhythms and compared the circadian period of the precise excision line to the original homozygous CG6123 Minos element line, a heterozygous cross between the CG6123 Minos element line and the excision line, and the w1118 control line. As Fig. 5 shows, both the homozygous excision line and the heterozygous cross were indistinguishable from the w1118 control line. This demonstrates genetic rescue of the long period phenotype, and the effects of the rescued allele are dominant to the mutant allele. We have re-named CG6123 Kairos, a Greek term which refers to an appropriate or suitable moment for action.
Figure 5.
Genetic rescue of the Minos element in Kairos (CG6123). Free-running period of the w1118 isogenic control, homozygous CG6123MB02356 mutant, CG6123MB02356/ CG6123MB* (heterozygote) and the homozygous CG6123MB* precise excision under constant dark conditions. White boxes indicate males and gray boxes indicate females. N=16 in each case except for CG6123MB* females where N=15. Letters above the boxplots indicate significant differences by post-hoc Tukey test (P < 0.05).
Discussion
In the present study, we have surveyed the transcriptome of a wild-type Drosophila line--DGRP_892, which exhibits an extremely long period in its locomotor activity behavior in constant darkness. Our approach to the experimental design followed the “Guidelines for genome-scale analysis of biological rhythms”(Hughes et al., 2017) and included 1) sample collection every two hours; 2) biological replicates (there are six samples for every timepoint for each genotype); 3) greater than ten million reads per sample for flies (14,638,141 on average per sample for our data); 4) pooling of five or more individuals (we used 150 heads per sample); 5) samples collected in constant darkness and under constant temperature; and 6) samples from both sexes were collected. The only exception to the guidelines in our experiment was the collection of samples over two circadian cycles (i.e., 48 hours); instead, we collected samples over 24 hours only. As the guidelines state, this lack of duplicate timepoints could potentially lead to false negatives when attempting to identify cycling genes (Hughes et al., 2017). per was the only canonical clock gene with a significant Genotype × Time interaction term. per was also the only canonical clock gene predicted to cycle for both sexes in CSB using JTK_Cycle, while Clk and vri were predicted to cycle for one sex only. However, all of the canonical clock genes (per, tim, Pdp1, Clk, and vri) were predicted to cycle in CSB using the less stringent algorithm in RAIN. In addition, cycling transcripts are more easily detected when clock neurons are assayed separately as opposed to extracting RNA from the entire head (Kula-Eversole et al., 2010). These results suggest that clock gene expression measured using RNA-Seq techniques is relatively noisy and cycling transcripts may evade detection unless additional timepoints are sampled, more permissive algorithms are used, or smaller subsets of tissue are sampled.
Fewer oscillating transcripts were present in DGRP_892 than CSB. Canonical clock genes did not appear to cycle in DGRP_892 when the data were analyzed using JTK_Cycle; however, per, tim, and Clk were predicted to cycle with 24-hour periodicity when the more permissive RAIN algorithm was used. These contradictory findings suggest two distinct possibilities. The first is that per, tim, and Clk are cycling in 24 hours, and the gene(s) contributing to the long circadian period act downstream of the molecular clock. The second possibility is that per, tim, and Clk are not cycling in DGRP_892, and that the identification of these genes as cycling by RAIN is a false positive result. If the genes are not cycling in DGRP_892, then the potential exists for at least one of the genes contributing to the long circadian period to interact directly with the molecular clock. We previously measured gene expression in constant darkness in per, tim, and Pdp1 in DGRP_892 using RT-PCR and analyzed the data using JTK_Cycle; none of the genes cycled (Harbison et al., 2019), consistent with the second possibility.
We found transcripts that have not been shown to cycle in previous studies (Hughes et al., 2012; Rodriguez et al., 2013). We compared the total number of putative oscillating genes across previous expression studies, and found that 10 to 35 genes overlap (Supplementary Table S19) (Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Ceriani et al., 2002; Lin et al., 2002; Ueda et al., 2002; Keegan et al., 2007). The relatively low overlap of cycling genes between these previous studies and ours could have several origins, including 1) the genotypes assayed; 2) the platform used to assess gene expression (RNA-Seq versus microarray); 3) the number of replicated samples; 4) the number of timepoints assayed; and 5) the algorithms used to identify cycling genes.
Approximately two-thirds of all detected transcripts (out of 11,474) were differentially expressed between genotypes while only 27 were differentially expressed for the Genotype x Time interaction, which suggests that at least some of the effects of gene expression on circadian period may be due to time-independent gene activity. Combining GWAS (Harbison et al., 2019) and RNA-seq analysis, we were able to narrow down candidate genes that might be critical for regulating circadian rhythms of the locomotor activity behavior. Further testing confirmed 11 genes affecting circadian period for the locomotor activity rhythms. Genetic rescue revealed that Kairos (CG6123) is necessary for regulating free-running locomotor activity behavior. Our RNA-seq data also revealed that Kairos does not cycle in constant darkness. The molecular function of Kairos is unknown. It is conserved among Diptera and other insects, but does not have a known mammalian ortholog (Hu et al., 2011). It is highly enriched in the ventral lateral neurons (LNvs) (Abruzzi et al., 2017), particularly the small ventral lateral neurons (sLNvs) (Kula-Eversole et al., 2010), suggesting a potential role as an input molecule to the core clock. Further experiments will determine whether it interacts directly with canonical circadian clock genes.
In addition to Kairos, we also verified that Alk, alph, CG10089, CG42540, CG6034, CG8768, klg, Lar, sick, and tinc alter the period lengths of rest: activity behavior. The Minos insertion mutants of klg in particular displayed a comparatively longer period than the rest. klg encodes a cell-adhesion molecule expressed in both neurons and glia and is required for long-term memory formation (Matsuno et al., 2015). klg expression has been localized to the sLNvs (Kula-Eversole et al., 2010). klg regulates the transcriptional activator repo in glial cells (Matsuno et al., 2015). It would be interesting to investigate the role of klg in the regulation of circadian rhythms given the importance of glial cells in rhythmic behavior (Ng et al., 2011).
The candidate genes sick, tinc, CG10089, CG42540, CG6034, and CG8768 have not been previously reported for their role in circadian regulation in flies, nor have their mammalian homologs. sick is expressed in dorsal neurons (DN1) as well as large and small LNvs (Kula-Eversole et al., 2010; Abruzzi et al., 2017). sick encodes a cytoskeleton protein and is highly expressed in F-actin rich axons of mushroom bodies (Abe et al., 2014). Accordingly, sick mutants exhibit axonal growth defects in the mushroom bodies (Abe et al., 2014). Interestingly, knockdown of sick in the dopaminergic neurons enhances olfactory memory, which suggests that the normal function of sick is to act as a memory suppressor gene (Walkinshaw et al., 2015). tinc is highly enriched in central and peripheral nervous system and encodes a transmembrane protein, which is involved in development of the compound eyes in Drosophila melanogaster (Hirota et al., 2005). CG10089 is expressed in both large and small LNvs (Kula-Eversole et al., 2010). Electronic annotation suggests that CG10089 has protein tyrosine/serine/threonine phosphatase activity, but little else is known about this gene (dos Santos et al., 2015). Likewise, CG42540 has an unknown function but exhibits some homology to human stomatin; CG6034 is predicted to have oxidoreductase activity; and CG8768 is predicted to have catalytic activity (dos Santos et al., 2015). Further investigation is needed to elucidate the role of these genes in modifying the circadian period of locomotor activity behavior.
Several of the genes we verified have been noted in previous circadian and sleep studies. Lar is gene implicated in Drosophila rhythmic behavior in constant darkness (Agrawal and Hardin, 2016) and in environmental sensitivity to waking activity (Harbison et al., 2013). Lar encodes a receptor protein tyrosine phosphatase leukocyte-antigen-related protein, and Lar mutants lack the dorsal projections of core clock neurons, which renders flies completely arrhythmic in constant darkness (Agrawal and Hardin, 2016). The arrhythmic phenotype of Lar mutants was largely attributed to impairment of PDF accumulation in the core clock neurons instead of effects on the core clock gene expression levels (Agrawal and Hardin, 2016). These results are in contrast with our results as the Minos insertion mutants of Lar that we tested are completely rhythmic. One possibility is that the Lar mutants used in our study act as a hypomorph rather than a deletion as the Minos insertion is in an intron. Other possibilities may include differences in the genetic background, the external environment, and their interaction, which are known to influence expressivity and penetrance differentially (Chandler et al., 2017). Alk encodes Anaplastic lymphoma kinase, which is involved in sleep regulation and memory formation (Bai and Sehgal, 2015). Mutants of Alk have increased sleep when its expression levels are reduced in the mushroom bodies (Bai and Sehgal, 2015). Interestingly, previous work found that Alk does not affect locomotor circadian behavior, though it is known to cycle in dorsal lateral neurons (LNds) (Bai and Sehgal, 2015; Abruzzi et al., 2017). However, only males were tested for circadian behavior (Bai and Sehgal, 2015). Our test, which examined both male and female mutants of Alk, revealed an effect on circadian period predominantly in females, who had a 43-minute decrease in circadian period compared to the control (Table S1). alph has been shown to cycle in constant darkness in an earlier microarray study, but did not cycle in LD (Ueda et al., 2002). We did not observe significant cycling for alph. alph encodes a Serine-Threonine phosphatase involved in RAS/MAPK signaling (Baril and Therrien, 2006). Thus, several of the genes we identified have known roles in circadian rhythms and sleep.
Systems genetics approaches combine the results of large-scale measures of the genome, transcriptome, proteome, etc., to identify the most promising candidate genes influencing a complex trait (Civelek and Lusis, 2014). Our combinatorial analysis of transcriptomic data together with GWAS results enabled us to identify genes that may underlie the long period phenotype seen in DGRP_892. Of the 11 mutants with significant effects on circadian period, none precisely phenocopy the 31-hour circadian period of DGRP_892. This strongly indicates that multiple genes are interacting to produce the long period, a notion supported by work reporting the interaction of naturally-occurring variants to affect rhythmic behavior in flies (Peschel et al., 2006). Using CRISPR, it is possible to perturb up to four genes simultaneously with a single polycistronic construct (Port and Bullock, 2016). The application of CRISPR-Cas9 tools will enable us to perturb multiple combinations of genes in DGRP_892 in order to understand how putative genetic interactions produce the long circadian period. It is also possible that some of the candidate genes may have effects that are developmental in origin. Perturbation of candidate genes using conditional GAL4 systems (Osterwalder et al., 2001; McGuire et al., 2003) will reveal any temporal dependency. Genes can be placed into their proper genomic context by verifying the transcriptional changes among associated genes common to a given module. For example, perturbations made to Alk in the DGRP_892 background should lead to a corresponding transcriptional response in the circadian/sleep genes highly correlated with Alk from the same module (Harbison et al., 2009). In this way, the contributing effectors producing the long period can be placed among the known circadian and sleep network.
Supplementary Material
Supplemental Fig S1: Principal component analysis reveals a bias in the data according to the date of the sequencing run.
Supplemental Fig S2: Agarose gel showing genotyping strategy used for determining the presence of Minos insertion in the CG6123 genomic region as well as its precise excision in the rescued line. Yellow and red denote the oligos used for PCR amplification flanking the Minos insertion whereas green and purple arrows denote the oligos used for checking the presence of the Minos insertion in the original CG6123MB02356 mutant.
Supplemental Fig S3: Top panel shows the presence of the Minos insertion in the CG6123 genomic region. Black flanking arrows denote the oligos used for Sanger based sequencing. Lower panel shows the snapshot of Flybase snapshot of nucleotide blast of W1118 control line. The transparent red box denotes the location of the Minos insertion in the original mutant of CG6123.
Supplemental Fig S4: Top panel shows the presence of the Minos insertion in the CG6123 genomic region. Black flanking arrows denote the oligos used for Sanger based sequencing. Lower panel shows the snapshot of Flybase snapshot of nucleotide blast of rescued CG6123 Minos insertion line. The transparent red box denotes the location of the original Minos insertion.
Supplemental Table S1. Mutant test results. For each gene the table lists the Minos genotype, Bloomington stock number, and isogenic control line. The number of flies tested and period length is given for females and males separately and for sexes combined. Significance is indicated for those lines having a Bonferroni-corrected P value of 0.0011 or less for females and males separately, and sexes combined. An asterisk indicates that the gene was significant upon retest. Genes predicted to cycle are indicated, as are those with significant differences by Genotype (DGRP_892 versus Canton-S B).
Supplemental Table S2. RNA-Seq analysis results. For each of the 11,474 genes for which expression was detected, the table lists feature type (protein coding, non-protein coding, etc.), chromosome arm location, gene symbol, mean TMM-normalized read counts for each genotype/sex/time point/replicate, and adjusted P-value for each factor analyzed in the experiment. Rep, replicate.
Supplemental Table S3. Numbers of differentially expressed genes at different false-discovery rates. For each factor in the generalized linear model, the table shows the numbers of genes that would be called significantly differentially expressed.
Supplemental Table S4. Modulated Modularity Clustering results for genes with significant differential expression among genotypes. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S5. Gene Ontology (GO) enrichment for genes with significant differential expression by genotype. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S6. Overlap between genes identified in the GWAS of circadian rhythms (Harbison et al., 2019) and those with significant differential expression by genotype. The table lists overlapping genes by Flybase ID and Gene Symbol.
Supplemental Table S7. Modulated Modularity Clustering results for genes with significant differential expression across timepoints. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S8. Gene Ontology (GO) enrichment for genes with significant differential expression among timepoints. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S9. Modulated Modularity Clustering results for genes with significant Genotype × Time differential expression. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S10. Gene Ontology (GO) enrichment for genes with significant Genotype × Time differential expression. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S11. Modulated Modularity Clustering results for genes with significant differential expression among sexes. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S12. Gene Ontology (GO) enrichment for genes with significant differential expression by sex. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S13. Modulated Modularity Clustering results for genes with significant Genotype × Sex differential expression. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S14. Modulated Modularity Clustering results for genes with significant Time × Sex differential expression. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S15. Gene Ontology (GO) enrichment for genes with significant Genotype × Sex differential expression. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S16. Gene Ontology (GO) enrichment for genes with significant Time × Sex differential expression. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S17. RAIN/JTK_Cycle results. For each gene, the table lists chromosome, Flybase ID, Gene symbol, predicted period, predicted phase, the adjusted P value from the RAIN analysis, and the adjusted P value from the JTK_Cycle analysis.
Supplemental Table S18. Genes expressed in clock neurons. The 166 genes identified in this study were compared to three studies examining gene expression in clock neurons. The overlap is indicated in the table. The genes indicated in bold had mutations with significant effects on circadian period in this study. DN1, dorsal neurons; LNd, dorsal lateral neurons; LNv, ventral lateral neurons; lLNv, large ventral lateral neurons; sLNv, small ventral lateral neurons; clock, includes all clock neurons. Genes expressed at a particular timepoint are indicated. Please see the text for additional information and references.
Supplemental Table S19: Putative cycling genes common across studies. The table lists genes identified as cycling by RAIN by Flybase ID, Gene Symbol, Line, and Sex, and indicates genes found in previous studies with an ‘x’.
Acknowledgements:
We thank the National Intramural Sequencing Center Consortium for the RNA sequences. This work used the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the Intramural Research Program of the NIH, the National Heart Lung and Blood Institute.
Footnotes
Declaration of Conflicting Interests: The authors declare that there is no conflict of interest.
References
- Abe T, Yamazaki D, Murakami S, Hiroi M, Nitta Y, Maeyama Y, and Tabata T (2014) The NAV2 homolog Sickie regulates F-actin-mediated axonal growth in Drosophila mushroom body neurons via the non-canonical Rac-Cofilin pathway. Development 141:4716–4728. [DOI] [PubMed] [Google Scholar]
- Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, Shafer O, and Rosbash M (2017) RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS genetics 13:e1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal P, and Hardin PE (2016) The Drosophila Receptor Protein Tyrosine Phosphatase LAR Is Required for Development of Circadian Pacemaker Neuron Processes That Support Rhythmic Activity in Constant Darkness But Not during Light/Dark Cycles. J Neurosci 36:3860–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, and White KP (2002) Gene expression during the life cycle of Drosophila melanogaster. Science 297:2270–2275. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RR, and Mackay TF (2009) Systems genetics of complex traits in Drosophila melanogaster. Nature genetics 41:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, and Sehgal A (2015) Anaplastic Lymphoma Kinase Acts in the Drosophila Mushroom Body to Negatively Regulate Sleep. PLoS Genet 11:e1005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril C, and Therrien M (2006) Alphabet, a Ser/Thr phosphatase of the protein phosphatase 2C family, negatively regulates RAS/MAPK signaling in Drosophila. Dev Biol 294:232–245. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bargiello TA, Jackson FR, and Young MW (1987) Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature 326:390–392. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, Hoskins RA, and Spradling AC (2011) The Drosophila gene disruption project: Progress using transposons with distinctive site specificities. Genetics 188:731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc B 57:289–300. [Google Scholar]
- Bushey D, Huber R, Tononi G, and Cirelli C (2007) Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci 27:5384–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, and Kay SA (2002) Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci 22:9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler CH, Chari S, Kowalski A, Choi L, Tack D, DeNieu M, Pitchers W, Sonnenschein A, Marvin L, Hummel K, Marier C, Victory A, Porter C, Mammel A, Holms J, Sivaratnam G, and Dworkin I (2017) How well do you know your mutation? Complex effects of genetic background on expressivity, complementation, and ordering of allelic effects. PLoS Genet 13:e1007075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, and Tononi G (2005) Reduced sleep in Drosophila Shaker mutants. Nature 434:1087–1092. [DOI] [PubMed] [Google Scholar]
- Civelek M, and Lusis AJ (2014) Systems genetics approaches to understand complex traits. Nat Rev Genet 15:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, and Young MW (2001) Circadian regulation of gene expression systems in the Drosophila head. Neuron 32:657–671. [DOI] [PubMed] [Google Scholar]
- Diessler S, Jan M, Emmenegger Y, Guex N, Middleton B, Skene DJ, Ibberson M, Burdet F, Gotz L, Pagni M, Sankar M, Liechti R, Hor CN, Xenarios I, and Franken P (2018) A systems genetics resource and analysis of sleep regulation in the mouse. PLoS Biol 16:e2005750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, and Consortium tF (2015) FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res 43:D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery PT, Birley AJ, and Morgan E (1995) Investigation of natural genetic variation in the circadian system of Drosophila melanogaster. II. Biometrical analyses of locomotor activity rhythms recorded in constant darkness. Chronobiol Int 12:77–86. [DOI] [PubMed] [Google Scholar]
- Everett LJ, Huang W, Zhou S, Carbone MA, Lyman RF, Arya GH, Geisz MS, Ma J, Morgante F, St Armour G, Turlapati L, Anholt RRH, and Mackay TFC (2020) Gene expression networks in the Drosophila Genetic Reference Panel. Genome research 30:485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Carbone MA, Ayroles JF, Stone EA, Lyman RF, and Mackay TF (2009) Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nature genetics 41:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Chang S, Kamdar KP, and Mackay TF (2005) Quantitative genomics of starvation stress resistance in Drosophila. Genome biology 6:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Kumar S, Huang W, McCoy LJ, Smith KR, and Mackay TFC (2019) Genome-Wide Association Study of Circadian Behavior in Drosophila melanogaster. Behav Genet 49:60–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, McCoy LJ, and Mackay TF (2013) Genome-wide association study of sleep in Drosophila melanogaster. BMC genomics 14:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE (2004) Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms 19:348–360. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, and Rosbash M (1990) Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343:536–540. [DOI] [PubMed] [Google Scholar]
- Hirota Y, Sawamoto K, Takahashi K, Ueda R, and Okano H (2005) The transmembrane protein, Tincar, is involved in the development of the compound eye in Drosophila melanogaster. Dev Genes Evol 215:90–96. [DOI] [PubMed] [Google Scholar]
- Hofstetter JR, Mayeda AR, Possidente B, and Nurnberger JI Jr. (1995) Quantitative trait loci (QTL) for circadian rhythms of locomotor activity in mice. Behav Genet 25:545–556. [DOI] [PubMed] [Google Scholar]
- Hsu PY, and Harmer SL (2012) Circadian phase has profound effects on differential expression analysis. PLoS One 7:e49853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, and Mohr SE (2011) An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Carbone MA, Magwire MM, Peiffer J, Lyman RF, Stone EA, Anholt RR, and Mackay TF (2015) Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc Natl Acad Sci U S A:E 6010–E6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ramsey KM, Marcheva B, and Bass J (2011) Circadian rhythms, sleep, and metabolism. J Clin Invest 121:2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Abruzzi KC, Allada R, Anafi R, Arpat AB, Asher G, Baldi P, de Bekker C, Bell-Pedersen D, Blau J, Brown S, Ceriani MF, Chen Z, Chiu JC, Cox J, Crowell AM, DeBruyne JP, Dijk DJ, DiTacchio L, Doyle FJ, Duffield GE, Dunlap JC, Eckel-Mahan K, Esser KA, FitzGerald GA, Forger DB, Francey LJ, Fu YH, Gachon F, Gatfield D, de Goede P, Golden SS, Green C, Harer J, Harmer S, Haspel J, Hastings MH, Herzel H, Herzog ED, Hoffmann C, Hong C, Hughey JJ, Hurley JM, de la Iglesia HO, Johnson C, Kay SA, Koike N, Kornacker K, Kramer A, Lamia K, Leise T, Lewis SA, Li J, Li X, Liu AC, Loros JJ, Martino TA, Menet JS, Merrow M, Millar AJ, Mockler T, Naef F, Nagoshi E, Nitabach MN, Olmedo M, Nusinow DA, Ptacek LJ, Rand D, Reddy AB, Robles MS, Roenneberg T, Rosbash M, Ruben MD, Rund SSC, Sancar A, Sassone-Corsi P, Sehgal A, Sherrill-Mix S, Skene DJ, Storch KF, Takahashi JS, Ueda HR, Wang H, Weitz C, Westermark PO, Wijnen H, Xu Y, Wu G, Yoo SH, Young M, Zhang EE, Zielinski T, and Hogenesch JB (2017) Guidelines for Genome-Scale Analysis of Biological Rhythms. J Biol Rhythms 32:380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Grant GR, Paquin C, Qian J, and Nitabach MN (2012) Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res 22:1266–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, and Kornacker K (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25:372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huylmans AK, and Parsch J (2014) Population- and sex-biased gene expression in the excretion organs of Drosophila melanogaster. G3 4:2307–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, and Gibson G (2001) The contributions of sex, genotype, and age to transcriptional variance in Drosophila melanogaster. Nature genetics 29:389–395. [DOI] [PubMed] [Google Scholar]
- Keegan KP, Pradhan S, Wang JP, and Allada R (2007) Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol 3:e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan BT, Galante RJ, Lian J, Simecek P, Gatti DM, Zhang L, Lim DC, Svenson KL, Churchill GA, and Pack AI (2020) High-throughput sleep phenotyping produces robust and heritable traits in Diversity Outbred mice and their founder strains. Sleep 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Zheng X, and Sehgal A (2006) JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312:1809–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, and Benzer S (1971) Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A 68:2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, and Rosbash M (2010) Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci U S A 107:13497–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, and Storey JD (2007) Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS genetics 3:1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. [DOI] [PubMed] [Google Scholar]
- Lin Y, Chen ZX, Oliver B, and Harbison ST (2016a) Microenvironmental Gene Expression Plasticity Among Individual Drosophila melanogaster. G3 (Bethesda) 6:4197–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Golovnina K, Chen Z-X, Lee HN, Serrano Negron YL, Sultana H, Oliver B, and Harbison ST (2016b) Comparison of normalization and differential expression analyses using RNA-Seq data from 726 individual Drosophila melanogaster. BMC genomics 17:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, and Taghert PH (2002) Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A 99:9562–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RR, Barron M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu LL, Qu C, Ramia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu YQ, Yamamoto A, Zhu Y, Bergman CM, Thornton KR, Mittelman D, and Gibbs RA (2012) The Drosophila melanogaster Genetic Reference Panel. Nature 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, and Young MW (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105:769–779. [DOI] [PubMed] [Google Scholar]
- Matsuno M, Horiuchi J, Yuasa Y, Ofusa K, Miyashita T, Masuda T, and Saitoe M (2015) Long-term memory formation in Drosophila requires training-dependent glial transcription. J Neurosci 35:5557–5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, and Rosbash M (2001) Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107:567–578. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, and Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302:1765–1768. [DOI] [PubMed] [Google Scholar]
- Merrow M, Spoelstra K, and Roenneberg T (2005) The circadian cycle: daily rhythms from behaviour to genes. EMBO Rep 6:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Oehler S, Klinakis A, and Savakis C (2005) Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, and Rosbash M (2010) Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci 13:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Tangredi MM, and Jackson FR (2011) Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr Biol 21:625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, and Kesheshian H (2001) A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A 98:12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, and Oliver B (2003) Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299:697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, and Griffith LC (2008) PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Sedore SA, Cronmiller C, and Hirsh J (2000) Type II cAMP-dependent protein kinase-deficient Drosophila are viable but show developmental, circadian, and drug response phenotypes. J Biol Chem 275:20588–20596. [DOI] [PubMed] [Google Scholar]
- Peschel N, Veleri S, and Stanewsky R (2006) Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila’s circadian clock. Proc Natl Acad Sci U S A 103:17313–17318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, and Bullock SL (2016) Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods 13:852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, and Hartl DL (2003) Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300:1742–1745. [DOI] [PubMed] [Google Scholar]
- Robinson MD, and Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome biology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Tang CH, Khodor YL, Vodala S, Menet JS, and Rosbash M (2013) Nascent-Seq analysis of Drosophila cycling gene expression. Proc Natl Acad Sci U S A 110:E275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Abodeely M, Price JL, and Young MW (2000) Isolation and analysis of six timeless alleles that cause short- or long-period circadian rhythms in Drosophila. Genetics 156:665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BR, and Macdonald SJ (2020) Dissecting the Genetic Basis of Variation in Drosophila Sleep Using a Multiparental QTL Mapping Resource. Genes (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N, and Young MW (2011) insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72:964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA, and Ayroles JF (2009) Modulated Modularity Clustering as an exploratory tool for functional genomic inference. PLoS genetics 5:e1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, Daga A, Selmin A, Monger K, Benna C, Rosato E, Kyriacou CP, and Costa R (2007) Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science 316:1895–1898. [DOI] [PubMed] [Google Scholar]
- Thaben PF, and Westermark PO (2014) Detecting rhythms in time series with RAIN. J Biol Rhythms 29:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, and Hashimoto S (2002) Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem 277:14048–14052. [DOI] [PubMed] [Google Scholar]
- Walkinshaw E, Gai Y, Farkas C, Richter D, Nicholas E, Keleman K, and Davis RL (2015) Identification of genes that promote or inhibit olfactory memory formation in Drosophila. Genetics 199:1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne ML, Telonis-Scott M, Bono LM, Harshman LG, Kopp A, Nuzhdin SV, and McIntyre LM (2007) Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proc Natl Acad Sci U S A 104:18577–18582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, and Oliver B (2007) Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature 450:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig S1: Principal component analysis reveals a bias in the data according to the date of the sequencing run.
Supplemental Fig S2: Agarose gel showing genotyping strategy used for determining the presence of Minos insertion in the CG6123 genomic region as well as its precise excision in the rescued line. Yellow and red denote the oligos used for PCR amplification flanking the Minos insertion whereas green and purple arrows denote the oligos used for checking the presence of the Minos insertion in the original CG6123MB02356 mutant.
Supplemental Fig S3: Top panel shows the presence of the Minos insertion in the CG6123 genomic region. Black flanking arrows denote the oligos used for Sanger based sequencing. Lower panel shows the snapshot of Flybase snapshot of nucleotide blast of W1118 control line. The transparent red box denotes the location of the Minos insertion in the original mutant of CG6123.
Supplemental Fig S4: Top panel shows the presence of the Minos insertion in the CG6123 genomic region. Black flanking arrows denote the oligos used for Sanger based sequencing. Lower panel shows the snapshot of Flybase snapshot of nucleotide blast of rescued CG6123 Minos insertion line. The transparent red box denotes the location of the original Minos insertion.
Supplemental Table S1. Mutant test results. For each gene the table lists the Minos genotype, Bloomington stock number, and isogenic control line. The number of flies tested and period length is given for females and males separately and for sexes combined. Significance is indicated for those lines having a Bonferroni-corrected P value of 0.0011 or less for females and males separately, and sexes combined. An asterisk indicates that the gene was significant upon retest. Genes predicted to cycle are indicated, as are those with significant differences by Genotype (DGRP_892 versus Canton-S B).
Supplemental Table S2. RNA-Seq analysis results. For each of the 11,474 genes for which expression was detected, the table lists feature type (protein coding, non-protein coding, etc.), chromosome arm location, gene symbol, mean TMM-normalized read counts for each genotype/sex/time point/replicate, and adjusted P-value for each factor analyzed in the experiment. Rep, replicate.
Supplemental Table S3. Numbers of differentially expressed genes at different false-discovery rates. For each factor in the generalized linear model, the table shows the numbers of genes that would be called significantly differentially expressed.
Supplemental Table S4. Modulated Modularity Clustering results for genes with significant differential expression among genotypes. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S5. Gene Ontology (GO) enrichment for genes with significant differential expression by genotype. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S6. Overlap between genes identified in the GWAS of circadian rhythms (Harbison et al., 2019) and those with significant differential expression by genotype. The table lists overlapping genes by Flybase ID and Gene Symbol.
Supplemental Table S7. Modulated Modularity Clustering results for genes with significant differential expression across timepoints. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S8. Gene Ontology (GO) enrichment for genes with significant differential expression among timepoints. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S9. Modulated Modularity Clustering results for genes with significant Genotype × Time differential expression. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S10. Gene Ontology (GO) enrichment for genes with significant Genotype × Time differential expression. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S11. Modulated Modularity Clustering results for genes with significant differential expression among sexes. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S12. Gene Ontology (GO) enrichment for genes with significant differential expression by sex. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S13. Modulated Modularity Clustering results for genes with significant Genotype × Sex differential expression. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S14. Modulated Modularity Clustering results for genes with significant Time × Sex differential expression. For each differentially expressed gene, the table lists the gene symbol, module, average degree (connectivity) for the module, and degree (connectivity) of the gene.
Supplemental Table S15. Gene Ontology (GO) enrichment for genes with significant Genotype × Sex differential expression. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S16. Gene Ontology (GO) enrichment for genes with significant Time × Sex differential expression. The table lists GO classification (Biological Process, Molecular Function, or Cellular Component); the GO Term ID; the GO term description; enrichment P value, adjusted P value, list of genes enriched by gene symbol, and module number.
Supplemental Table S17. RAIN/JTK_Cycle results. For each gene, the table lists chromosome, Flybase ID, Gene symbol, predicted period, predicted phase, the adjusted P value from the RAIN analysis, and the adjusted P value from the JTK_Cycle analysis.
Supplemental Table S18. Genes expressed in clock neurons. The 166 genes identified in this study were compared to three studies examining gene expression in clock neurons. The overlap is indicated in the table. The genes indicated in bold had mutations with significant effects on circadian period in this study. DN1, dorsal neurons; LNd, dorsal lateral neurons; LNv, ventral lateral neurons; lLNv, large ventral lateral neurons; sLNv, small ventral lateral neurons; clock, includes all clock neurons. Genes expressed at a particular timepoint are indicated. Please see the text for additional information and references.
Supplemental Table S19: Putative cycling genes common across studies. The table lists genes identified as cycling by RAIN by Flybase ID, Gene Symbol, Line, and Sex, and indicates genes found in previous studies with an ‘x’.
Data Availability Statement
All RNA-Seq data from this study are available from the National Center for Biotechnology Information (NCBI) GEO under the accession number GSE155018, including raw read counts and batch-adjusted TMM-normalized read counts for each gene per genotype/replicate/sex. All supplemental figures and tables are available at figshare under the following link: https://figshare.com/s/637491852db02ea31527.