ABSTRACT
The survival of viruses depends on their ability to resist host defenses and, of all animal virus families, the poxviruses have the most antidefense genes. Orthopoxviruses (ORPV), a genus within the subfamily Chordopoxvirinae, infect diverse mammals and include one of the most devastating human pathogens, the now eradicated smallpox virus. ORPV encode ∼200 genes, of which roughly half are directly involved in virus genome replication and expression as well as virion morphogenesis. The remaining ∼100 “accessory” genes are responsible for virus-host interactions, particularly counter-defense of innate immunity. Complete sequences are currently available for several hundred ORPV genomes isolated from a variety of mammalian hosts, providing a rich resource for comparative genomics and reconstruction of ORPV evolution. To identify the provenance and evolutionary trends of the ORPV accessory genes, we constructed clusters including the orthologs of these genes from all chordopoxviruses. Most of the accessory genes were captured in three major waves early in chordopoxvirus evolution, prior to the divergence of ORPV and the sister genus Centapoxvirus from their common ancestor. The capture of these genes from the host was followed by extensive gene duplication, yielding several paralogous gene families. In addition, nine genes were gained during the evolution of ORPV themselves. In contrast, nearly every accessory gene was lost, some on multiple, independent occasions in numerous lineages of ORPV, so that no ORPV retains them all. A variety of functional interactions could be inferred from examination of pairs of ORPV accessory genes that were either often or rarely lost concurrently.
KEYWORDS: Gene gain, Gene loss, Host range, Poxvirus, Poxvirus evolution, Virus evolution, Virus phylogeny, Adaptive mutations, Innate immunity
INTRODUCTION
Orthopoxviruses (ORPV) comprise a genus within the subfamily Chordopoxvirinae of the family Poxviridae (1). ORPV infect diverse mammalian hosts and include smallpox (variola) virus (VARV), the causative agent of one of the most devastating albeit now eradicated diseases of humans, other pathogens, such as monkeypox virus (MPXV), as well as cowpox virus (CPXV) and vaccinia virus (VACV), the first described virus vaccines (1–3). Given the health relevance of the ORPV, their recent development as recombinant vaccines for heterologous diseases and as oncolytic viruses, and their utility as laboratory models for studying virus molecular biology and virus-host interactions, hundreds of ORPV genomes have been sequenced. These double-stranded (ds) DNA genomes are from 170 to 210 kb (kb) in length and encode about 200 proteins (4, 5).
The localization of genes with different functions in the genomes of ORPV is highly non-random. The central part of the genome, of about 100 kb, is occupied by genes that are shared with other poxviruses (and in many cases, with other large dsDNA viruses in the phylum Nucleocytoviricota (6), also known as Nucleocytoplasmic Large DNA Viruses, or NCLDV) and encode about 100 proteins involved, primarily, in virus genome replication and expression as well as virion morphogenesis (7). The peripheral regions of the ORPV genomes include inverted terminal repeats of 0.1 to 12 kb and contain primarily genes encoding proteins involved in various facets of virus-host interaction and define the virus host range (8). More specifically, most of these “accessory” ORPV proteins counteract host antivirus responses including innate immunity and programmed cell death (1, 9–13). The accessory genes of the ORPV appear to have been derived from the hosts at diverse stages of evolution, with the exception of several very short putative genes (4) that might have originated de novo from non-coding sequences. Some of the accessory proteins retain high sequence similarity with their cellular homologs and likely ancestors, whereas others are highly diverged such that identification of the specific ancestry can be difficult or even (presently) unfeasible (14, 15).
The accessory ORPV genes include several families of diverged paralogs, most of which encode proteins containing widespread protein-protein interaction domains involved in various intracellular and extracellular signaling pathways. Such domains found in multiple ORPV proteins include the BTB (BR-C, ttk and bab) domain, also known as the POZ (Poxvirus and Zinc finger) domain, Kelch repeats (16, 17), Ankyrin (ANK) repeats, a diverged variant of the F-box known as PRANC (Poxvirus protein Repeat of ANkyrin C-terminal) domain (18), the Bcl-2 domain (19), and a distinct, apparently, chordopoxvirus-specific chemokine-binding PIE (Poxvirus Immune Evasion) domain (20). A notable feature of two paralogous families of ORPV proteins is the fusion of two distinct domains, ANK-PRANC in one case and BTB-Kelch in another. The proteins of both these families are adaptors that mediate the exploitation of the host ubiquitin-proteasome by poxviruses (21–23). Most likely, these gene families evolved by gene duplication during the evolution of the poxviruses, after the initial capture of the respective family founder from the host.
Comparative genome analysis of ORPV has shown that these viruses have captured very few genes after their divergence from the common ancestor with other chordopoxviruses. In contrast, numerous accessory genes were inactivated by frameshift mutations or deleted in different ORPV lineages. In particular, and perhaps counterintuitively, variola virus strains, including the most virulent ones, have lost many accessory genes which correlates with their narrow (exclusively human) host range (4). Dissection of the patterns of evolution, and in particular, the trends in the lineage-specific loss of the accessory genes is crucial for understanding the ORPV biology, including the evolution of host range, evasion of the host immunity, pathogenicity and virulence. Furthermore, poxviruses are unique among animal viruses in terms of the diversity of proteins that affect various signaling pathways involved in the host antivirus defense (8). Thus, characterization of the mechanisms and interactions of the accessory proteins of ORPV has the potential to elucidate not only the virus-host interactions and coevolution, but also the functions of different components and the cross talk among these pathways themselves.
Here, we sought to obtain a comprehensive picture of the evolution of the ORPV accessory genes. To this end, we constructed Clusters of Orthologous Genes (COGs) for all ORPV with complete genome sequences, and amended these with orthologs of ORPV genes from other chordopoxviruses. We also reannotated the ORPV accessory genes taking into account the current published information and predicting the functions of several previously uncharacterized genes. Based on these analyses, a unified, formal nomenclature of ORPV genes (OPG) is proposed. We then reconstructed the history of gains and losses for all accessory genes and investigated the emerging evolutionary patterns and their functional implications.
RESULTS
Clusters of orthologs and functional reannotation of ORPV genes.
For the purpose of reannotation of the ORPV accessory genes and reconstruction of their genome evolution, we first assembled the complete set of COGs (24–26) for the 235 available unique complete genome sequences of ORPV, starting with the previously described set of 214 genes (4) and using the clustering procedure described under Materials and Methods. In this work, we were interested in the evolution of the accessory genes of ORPV, whereas accessory genes present exclusively in other chordopoxvirus genomes were outside of the scope of the study. However, because ORPV share many of the accessory genes with some of the chordopoxviruses of other genera, it was necessary to tally the representation of these ORPV genes across all chordopoxviruses in order to map their capture to specific stages of evolution. To this end, position-specific scoring matrices (PSSMs) constructed from multiple alignments of the ORPV accessory proteins were mapped to 100 selected genomes of well-characterized viruses representing all genera of chordopoxviruses. PSI-BLAST searches were carried out against the annotated protein complements, and additionally, against six-frame genome translations, in order to identify proteins that might have been missed during previous genome annotations. We also translated in 6 frames all the ORPV intergenic regions longer than 100 bp and performed clustering of the resulting putative protein sequences by similarity as well as PSI-BLAST and HHPRED searches to identify potential homologs. However, these procedures failed to identify any additional OPGs.
Among the 214 OPGs of the Old World ORPV, 109 were represented in all ORPV and thus were operationally defined as core genes, whereas the remaining ones could not be identified or were lost due to deletions, frameshifts or nonsense mutations in various subsets of the ORPV and so were operationally defined as accessory genes. It should be noted that, under the automated procedure we implemented for calling gene presence, a gene was considered present if the encoded protein comprised more than 80% of the mean length of the protein encoded by the respective OPG. This hard cut-off was based on the results of the previous analysis of the ORPV genomes (4) but missed at least three special cases of gene disruption that nevertheless yield functional proteins. For two of such cases, additional OPGs (OPG010a and OPG188a) were introduced to accommodate distinct forms of ORPV proteins encoded by the same genes as OPG010 and OPG188, respectively, bringing the total OPG count to 216. The third case is OPG065 (E3L), for which we retained a single OPG because of the relatively small size of the eliminated portion of the gene. These special cases in OPG evolution are discussed below in the respective section.
Two ORFs encoding uncharacterized, very small proteins (OPG024 and OPG207), both predicted to be intrinsically disordered and missing in many ORPV genomes, are dubious as protein-coding genes, but were kept as OPGs for consistency (4). Additionally, four genes were shared by AKPV and/or New World ORPV with centapoxviruses, but were missing (apparently, lost) in the Old World ORPV (denoted X1-4 in Table S1). It should be noted that the definitions of the core and accessory genes adopted here are based solely on the patterns of gene loss and do not perfectly correspond to the functional divide between the genes involved in virus replication, expression and morphogenesis, versus the genes implicated in virus-host interactions (Table 1). Thus, some of the core genes, for example, OPG027 (C7L) and OPG159 (A31R; see details below), encode proteins that target host defense pathways, whereas a few accessory genes, for example, OPG178 (A48R, thymidylate kinase) encode proteins involved in virus genome replication.
TABLE 1.
ORPV genes implicated in virus-host interaction (103 genes)
| OPG | Protein domain architecture, localization, function | Cop |
|---|---|---|
| 1 | PIE domain, chemokine binding protein | C23L* |
| 2 | TNF-α receptor homolog and PIE domain, chemokine binding protein, CrmB | C22L* |
| 3 | ANK and PRANC domains, binds to the NF-κB p105 subunit and to SCP1 | C19L-C21L* |
| 4 | ANK and PRANC domains | C17L-C18L* |
| 5 | Bcl-2 domain, host range factor | C16L* |
| 6 | Bcl-2 domain | |
| 7 | PIE domain | |
| 8 | BTB domain, no kelch domain | |
| 9 | ANK and PRANC domains | |
| 10 | C-type lectin domain, putative decoy ligand for NK cell inhibitory receptor | |
| 10a | Type II transmembrane, TAP inhibitor, inhibits peptide loading on MHC I and antigen presentation | |
| 11 | BTB and Kelch domains | |
| 12 | PIE domain, SCP-2 | |
| 13 | TNF receptor vCD30 homolog, binds CD153, prevents CD30/CD153 interaction | |
| 14 | ANK and PRANC domains, binds to cullin 2 and inhibits NF-κB activation | |
| 15 | ANK and PRANC domains | |
| 16 | MHC class I homolog (OMCP), secreted, blocks NKG2D receptor | |
| 17 | ANK and PRANC domains | |
| 18 | Paralog of OPG27, VACV C7 protein | |
| 19 | Epidermal growth factor homolog, EGFR-dependent MAPK/ERK1/2 activation | C11R |
| 20 | N-term prolyl hydroxylase fold, C-term IL-1 receptor antagonist, antagonizes DNA-PK by binding Ku | C10L |
| 21 | KilA-N and RING domains, E3 ubiquitin ligase, blocks apoptosis, virulence factor | |
| 22 | IL-18 binding protein homolog, secreted, blocks IL-18 receptor binding site | |
| 23 | ANK and PRANC domains, host range, CP77 | |
| 24 | Intrinsically disordered | |
| 25 | ANK and PRANC domains, targets IFITs for degradation, IFN resistance | C9L |
| 26 | PIE domain, secreted | C8L |
| 27 | Host range, inhibits type I IFN production and signaling, paralog of OPG18 | C7L |
| 28 | Transmembrane domain | |
| 29 | Bcl-2 domain, inhibits type I IFN production and signaling, targets HDAC5 for degradation | C6L |
| 30 | BTB domain without Kelch domain | C5L |
| 31 | Inactive prolyl hydroxylase, induction of hypoxic response, inhibits NF-kB signaling, binds Ku | C4L |
| 32 | Complement control protein-like (CCP), prevents complement activation, secreted | C3L |
| 33 | BTB and Kelch domains, inhibits inflammation, reduces immunopathology | C2L |
| 34 | Bcl-2 domain | C1L |
| 35 | Bcl-2 domain, inhibits NF-kB activation and apoptosis | N1L |
| 36 | Bcl-2 domain, IRF3 activation inhibitor, α-amanitin target | N2L |
| 37 | ANK domain (no PRANC domain), inhibits intrinsic apoptosis at the level of the apoptosome | M1L |
| 38 | PIE domain, blocks SD28-mediated T cell activation, secreted | M2L |
| 39 | ANK domain, no PRANC domain, NF-kB inhibitor, host range | K1L |
| 41 | Mimic of eIF2α, pseudo-substrate for PKR , PKR inhibitor, IFN-resistance | K3L |
| 42 | Phospholipase-D homolog | K4L |
| 43 | Monoglyceride lipase homolog | K5L, K6L |
| 44 | Bcl-2 domain, binds DDX3, blocks IFN-β promoter induction | K7R |
| 45 | Bcl-2 domain, localized to mitochondria, blocks mitochondrial apoptosis | F1L |
| 47 | BTB and Kelch domains, contributes to virulence, larger lesion size | F3L |
| 49 | Immunoglobulin domain, type I membrane protein, affects plaque morphology | F5L |
| 50 | No detectable domains | F6L |
| 51 | No detectable domains | F7L |
| 52 | No detectable domains, non-essential for morphogenesis and actin rearrangement in VACV | F8L |
| 55 | RhoA-mDia signaling inhibitor, promotes cell motility | F11L |
| 60 | Predicted all-β domain, highly conserved in most chordopoxviruses | F15L |
| 61 | Non-functional serine recombinase | F16L |
| 65 | Z-DNA binding domain, dsRNA-binding, PKR inhibitor, IFN resistance | E3L |
| 67 | 2 DNA-binding BEN domains, cGAS and IFN type 1 inhibitor, virulence factor | E5R |
| 74 | C-terminal TM, activates of ERK1/2 signaling pathway, increases virulence | O1L |
| 91 | NLPc/P60 superfamily protein, predicted hydrolase, non-essential in cell culture | G6R |
| 159 | US22 family, SUKH superfamily, predicted dsRNA binding | A31R |
| 163 | MHC class II antigen presentation inhibitor, virulence factor | A35R |
| 165 | Inactive prolyl hydroxylase, induction of hypoxic response?, paralog of C10, C4 proteins | A37R |
| 166 | No detectable domains, uncharacterized | |
| 167 | CD47 homolog, integral membrane protein, promotes Ca2+ influx into infected cells | A38L |
| 168 | Semaphorin homolog, secreted, blocks semaphorin cell surface receptor | A39R |
| 169 | Lectin homolog, type II TM, cell surface | A40R |
| 170 | PIE domain, secreted chemokine binding protein, interferes with chemokine-GAG interaction | A41L |
| 171 | Profilin homolog, interacts with A-type inclusion protein and tropomyosin | A42R |
| 172 | Type I membrane glycoprotein, localizes to Golgi and plasma membranes | A43R |
| 173 | Inhibitor of host translation initiation, suppresses innate and adaptive immunity | A43.5R |
| 174 | Hydroxysteroid dehydrogenase, inflammatory response inhibitor | A44L |
| 176 | Bcl-2 domain, inhibits TLR- and IL-1β mediated NF-kB activation | A46R |
| 177 | Gasdermin homolog, pyroptosis inhibitor? Immunoprevalent | A47L |
| 179 | Bcl-2 domain, disordered 17 amino acid N-terminus mimics IκBα, inhibits NF-kB activation | A49R |
| 181 | Distant homolog of OPG202, VACV B17 protein | A51R |
| 182 | Bcl-2 domain, NF-kB pathway inhibitor, blocks TLR signaling | A52R |
| 183 | TNF-receptor homolog, CrmC, secreted, prevents TNF binding to cell receptor | A53R |
| 184 | BTB and Kelch domains, strong interaction with Cullin 3, could sequester Cullin 3 | A55R |
| 188 | Poxin-Schlafen fusion protein, blocks DNA sensing and prevents IFN induction | B2R-B3R |
| 188a | Poxin, 2’-3’-cGAMP nuclease, blocks DNA sensing and IFN induction | B2 |
| 189 | ANK and PRANC domains, affects ECTV spread in mouse organs | B4R |
| 191 | HEAT/ARM repeats, C-terminal transmembrane domain | B6R |
| 192 | PIE domain, ER resident, SCP-3, influences lesion size in mice | B7R |
| 193 | IFN-γ receptor mimic, secreted, sequesters IFN-γ | B8R |
| 194 | Surface glycoprotein type 1 TM domain, absent in CPXV Gri and BRI, present in CPXV GER91-3 | |
| 195 | PIE, ER localized, retains MHC I in ER, prevents antigen presentation | B9R |
| 196 | BTB and Kelch domains | B10R |
| 197 | Disordered | B11R |
| 198 | Serine/threonine kinase homolog, nonfunctional | B12R |
| 199 | Serpin (SPI2), CrmA, caspase-1 and 8 inhibitor, apoptosis (pyroptosis) inhibitor | B13R, B14R |
| 200 | Bcl-2, NF-kB signaling inhibitor, prevents phosphorylation of IkBα, regulates MAPK/AP-1 activation | B15R |
| 201 | 3 immunoglobulin domains, IL-1 receptor homolog, blocks IL-1β, prevents fever in mice | B16R |
| 202 | Distant homolog of OPG181, VACV A51 protein | B17L |
| 203 | ANK and PRANC domains, binds to SCF ubiquitin ligase complex | B18R |
| 204 | 3 immunoglobulin domains, IFN type-1 decoy receptor, secreted, attaches to GAGs, inhibits IFN | B19R |
| 205 | ANK and PRANC domains | B20R |
| 206 | BTB and Kelch domains interacts with C3RL complex | |
| 207 | No detectable domains, uncharacterized | C11.5R |
| 208 | Serpin (SPI1), host range factor, apoptosis inhibitor | C12L |
| 209 | PIE domain, SCP-1 | C13L, C14L |
| 210 | Surface glycoprotein, type I transmembrane domain, T cell response suppressor, virulence factor | |
| 211 | ANK and PRANC domains | C15L/B21R |
| 212 | TNF receptor homolog and PIE domain, CrmD | |
| 213 | TNF receptor homolog, secreted, CrmE, blocks TNF binding to cell receptors | |
| 214 | N-methyl D-aspartate receptor homolog, Golgi localized, vGAAP, suppresses apoptosis |
Genes that are present on both ends of the Cop genome in internal terminal repeat.Abbreviations: Cop, Copenhagen; IFN, interferon; TM, transmembrane; EV, enveloped virus.
Functional annotation of the ORPV genes. (A) OPG number. (B) Accession number for OPGs of CPXV-Gri; when the corresponding gene is absent in CPXV-Gri, the accession number of CPXV-Ger91-3, CPXV-Bri or VACV-Cop is indicated. (C) Protein lengths (amino acid number) in CPXV-Gri; when the corresponding gene is absent in CPXV-Gri, protein length is indicated for CPXV Ger91-3, CPXV-Bri or VACV-Cop. (D) State of the gene in CPXV Gri genome. (E) Domain architecture and/or function. (F) Gene capture in the evolution of chordopoxviruses. Node numbers correspond to the phylogenetic tree of chordopoxviruses in Fig. 2. A, ancestral gene present in the common ancestor of all chordopoxviruses; Node 2, prior to divergence of Crocodylidpoxviruses from the rest of the Chordopoxviruses (41 genes); Node 3, prior to divergence of Avipoxviruses and mammalian poxviruses (43 genes); Node 4, prior to divergence of Molluscipox (5 genes); Node 5 prior to divergence of Parapox (7 genes); Node 6 prior to divergence of Squirrel pox UK (3 genes); Node 7 Prior to divergence of Pteropox (node 1 genes); Node 8 prior to divergence of Squirrel pox Berlin (5 genes); Node 9 prior to divergence of Cetacean (1 gene); Node 10 Prior to divergence of leporipoxviruses (30 genes); Node 11, prior to divergence of Centapoxviruses (22 genes); Node 12, prior to divergence of New World ORPV (6 genes); Node 15 prior to divergence of Ectromelia-Abatino-CPXV 5 (2 genes); Node 16 prior to divergence of major cowpox branch CPXV 4 within ORPV (1 gene); (G, I, K) State of the gene in VACV Copenhagen, VACV WR, and CPXV Brighton, respectively. X, ORF sequence can’t be identified; D, large deletion representing 50+ bp or >1/3 reference length; F, frameshift(s); N, in-frame nonsense mutation(s). (H, J, L) Gene names in VACV Copenhagen, VACV WR and CPXV Brighton, respectively. Abbreviations: EV, enveloped virion; IV, immature virion; MV, mature virion; WV, wrapped virion; TM, transmembrane; Cop, Copenhagen VACV nomenclature; EFC, entry fusion complex; VMAP, virion membrane assembly protein. Download Table S1, XLSX file, 0.03 MB (35.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We propose the OPGs as the unified nomenclature for ORPV genes, from OPG001 at the left end of the genome to OPG214 at the right end (Table S1). Each OPG corresponds to a set of orthologous genes that are conserved in a subset of the ORPV so that orthologs in different viruses share the same name. The previous nomenclatures based on the position of the ORPV genes on restriction endonuclease fragments are obsolete, although we still indicate the VACV Copenhagen gene names for their historical familiarity (27). Further, unlike previous nomenclatures of ORPV genes, the OPGs included only genes that are intact in at least one ORPV and hence predicted to be functional, to the exclusion of all pseudogenes.
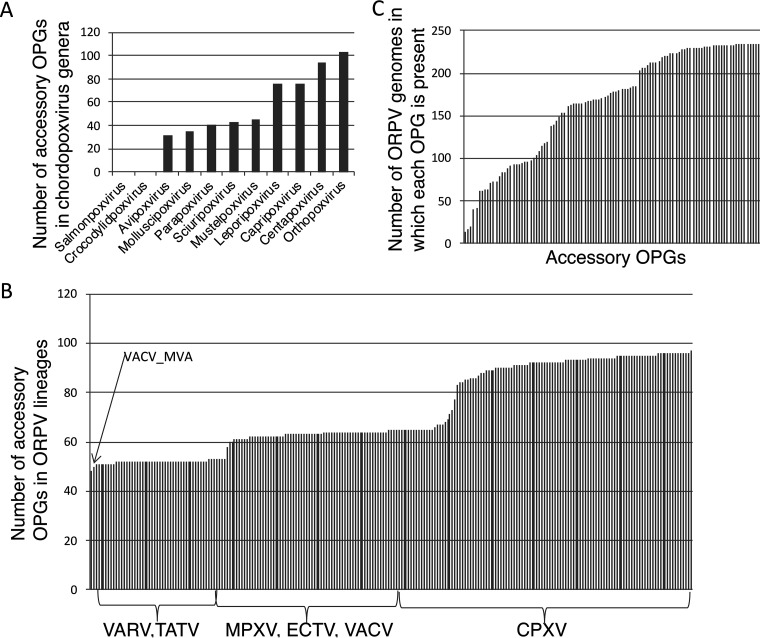
Most of the core ORPV genes are widely represented in other chordopoxviruses although many are missing in the deepest branching genus, the Salmonpoxviruses (28) (Table S2). In contrast, the representation of ORPV accessory genes in other chordopoxvirus genera varies from the near complete absence in salmonpoxvirus and crocodylidpoxviruses to the near complete representation in the Centapoxvirus genus (Fig. 1A), depending on the evolutionary distance from ORPV (see below). The ORPV themselves also notably differ in the representation of the accessory genes, from about half of the full set (53-55 genes) in VARV to about 60 genes in MPXV, VACV and ECTV to the near full complement in some of the CPXV strains (Fig. 1B and Table S3A). An orthogonal perspective on the accessory genes shows a broad range of representation of the OPGs among the ORPV, from the presence in only a small minority of the genomes to near ubiquity (Fig. 1C). The 214 OPGs represent the pangenome (29, 30) of the Old World ORPV, that is, the union of all genes identified in these viruses, with the exception of AKPV that has an additional gene. There is no ‘mother of ORPV’ genome that would contain all the 214 OPGs, but many CPXV strains lack only a few genes, and one strain, CPXV _RatKre08_2|KC813505, has 213 genes. The location of the core and accessory genes closely follows the ‘core in the center’ rule, with very few accessory genes breaking the central block of the core genes (Tables S1 and S3).
FIG 1.
Distribution of ORPV accessory genes among chordopoxviruses. (A) Number of ORPV accessory genes (OPG) represented in each chordopoxvirus genus. (B) Distribution of accessory genes among the major lineages of ORPV. (C) Representation of accessory genes in ORPV genomes. Each bar represents one of the 106 accessory OPGs.
Orthologs of the OPGs in other chordopoxviruses. Presence of an intact OPG is denoted by 1 and absence by 0. Download Table S2, XLSX file, 0.3 MB (272.7KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representation of Orthopoxvirus genes (OPG) in ORPV genomes. (A) Sheet 1: binary pattern of OPG presence-absence in ORPV genomes. Gene presence is shown by 1 and gene absence by 0. (B) Sheet 2: breakdown of the OPG inactivation events. Abbreviations: D, decaying gene (within gene deletion); F, frameshift; N, in-frame stop codon; X, absent gene (large deletion). Download Table S3, XLSX file, 0.3 MB (349.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We revised and updated the functional annotations of all accessory ORPV genes through a combination of literature review and database searches for functionally characterized homologs and conserved domains using the PSI-BLAST and HHPred methods, in cases where the available information was insufficient. As a result, we were able to infer the domain organizations and potential functions of three previously uncharacterized genes (OPG159, OPG165, OPG177) that are described immediately below, whereas 10 genes remained completely uncharacterized. The accessory OPG and their known domain architectures and/or functions are described in Table 1 and, in further detail, in Table S1.
Domain architectures and predicted functions of previously uncharacterized ORPV genes.
While reannotating the accessory genes of the ORPV, we were able to infer the domain organizations and potential functions of three previously uncharacterized genes. The OPG159 (A31R) gene, which is conserved in all chordopoxviruses except for the deepest branch (the salmonpoxviruses), encodes a protein that is similar to the N-terminal domains of the US22 family of the SUKH superfamily (31). This connection was not detectable with PSI-BLAST, whereas the HHPred search yielded a moderate similarity to profile of the US22 family of herpesvirus proteins (probability of 43.45%). However, the fact that A31 and the US22 family proteins aligned from the N termini of both proteins, and then, almost throughout the length of OPG159, and the close similarity of the predicted secondary structures (Fig. S1A), suggest that this match reflects genuine homology. Indeed, in an HHPred search initiated with the fowlpox virus OPG159 ortholog, the US22 family came up with a higher score of 63.76. The SUKH domain forms a distinct α−β structural fold with 6 conserved β-strand and 4 conserved α-helices, and all these elements are readily predictable in OPG159 and its homologs from other chordopoxviruses (Fig. S1A). Crocodylidpoxviruses and avipoxviruses encode additional, paralogous US22 domain proteins (Fig. S1A), as previously reported (31). Apparently, this paralog was lost during the subsequent evolution of chordopoxviruses.
Multiple sequence alignments of previously uncharacterized ORPV proteins. (A) OPG159 (A31), a US22 family member; (B) OPG165 (A37), an inactive prolyl hydroxylase homolog, putative elicitor of hypoxic response. The residues that are predicted to bind the Fe2+ ion are marked with red rectangles. (C) OPG181 (A47), gasdermin homolog, a predicted apoptosis inhibitor. Secondary structure elements extracted from crystal structures or predicted with Ali2D are indicated (red, α-helix; blue, β-strand). Download FIG S1, PPT file, 0.2 MB (161.4KB, ppt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The SUKH domains are widespread in bacteria, eukaryotes and RNA viruses, and appear to be involved in a broad range of defense functions, often, in interaction with various nucleases (31). The US22 family of the SUKH domains is represented in many herpesviruses (hence the name of the family, after the US22 gene of betaherpesviruses), which typically encode multiple paralogs of SUKH domain proteins, and some iridoviruses and adenoviruses. Two of the SUKH domain proteins of human cytomegalovirus, pTRS1 and pIRS1, have been shown to bind dsRNA via the US22 domain (a non-canonical dsRNA-binding domain) and inhibit the antivirus protein kinase R (PKR) (32–34). However, PKR inhibition by these herpesvirus proteins does not require the US22 domain or dsRNA binding (35). Inhibition of PKR is one of the primary effects of poxviruses on the host innate immunity and one of the principal host range determinants (36, 37). ORPV and some of the other chordopoxviruses encode two previously characterized PKR inhibitors, K3 (OPG041), a eIF2α mimic (38), and E3 (OPG065), a dsRNA-binding protein (39). The possibility exists that OPG159 is a third PKR inhibitor, albeit functioning via a mechanism distinct from that of the cytomegalovirus pTRS1 and pIRS1, that could back-up E3 and K3, conceivably, adding robustness to the virus antidefense response. However, it has been shown that a double E3L, K3L deletion mutant of VACV can replicate only in PKR or RNase L knockout cell lines (36, 40), suggesting that OPG159 is insufficient to prevent the inhibitory activity of PKR and is more likely to inhibit a different dsRNA-dependent defense activity. OPG159 might have been acquired by an ancestral chordopoxvirus from a herpesvirus, the only case of such apparent gene transfer between these two unrelated groups of large animal DNA viruses.
The OPG165 (A37R) represented in the ORPV and some other chordopoxviruses encodes a protein homologous to prolyl hydroxylases (HHPred probability 90.24% with prolyl-4 hydroxylase from Chlamydomonas reinhardtii, PDB 2JIG) (Fig. S1B). The double-stranded β-helix fold of prolyl hydroxylases (41) has been previously detected in ORPV proteins OPG020 (C10L) and OPG031 (C4L) (42) that are highly similar to each other, but only distantly related to OPG165 (HHPred probability 87.29%). Furthermore, the similarity of the OPG031 and OPG020 proteins to prolyl hydroxylases is far more pronounced than that of OPG165. Examination of the multiple alignments of these 3 chordopoxvirus proteins containing the prolyl hydroxylase domain shows that the residues coordinating the iron cation required for prolyl hydroxylase activity are missing in all three poxvirus proteins (Fig. S1B), which implies that this domain was exapted for non-enzymatic functions. It has been shown that OPG020 protein induces the hypoxic response in VACV-infected cells by binding the N-terminal domain of the human oxygen sensor, prolyl-hydroxylase domain containing protein 2 (PHD2), and preventing it from hydroxylating a proline residue in the transcription factor HIF-1a that activates cellular genes involved in the hypoxic response (42). Apparently, the N-terminal domain of OPG020 acts as a structural mimic of the oligomerization domain of PHD2 but instead of an enzymatically active complex yields an inactive one, that is, inhibits PHD2 via a dominant negative mechanism. It appears likely that OPG165 functions in a similar manner, recapitulating the theme of partially redundant anti-immune mechanisms in ORPV. Given the disruption of the catalytic site in all ORPV prolyl hydroxylase homologs, it appears that, notwithstanding the high divergence of OPG165, they represent a family of paralogs that evolved via two duplications after initial capture by an ancestral virus.
The OPG177 (A47L) encodes a protein homologous to the C-terminal domain of gasdermins, a distinct α-helical bundle (100% HHPred probability with human gasdermin D, 5NH1). Gasdermins are key effectors of pyroptosis, a lytic, proinflammatory cell death pathway, and are involved also in other programmed cell death pathways (43). The N-terminal domains of gasdermins oligomerize and form membrane pores that cause membrane leakage, release of proinflammatory molecules and eventual cell death (43, 44). However, under normal conditions, this activity of gasdermins is inhibited by the C-terminal repressor domain (45). Under stress, cleavage of gasdermins by proteases, such as caspase 1, leads to unregulated pore formation and induces pyroptosis or other forms of cell death. The ORPV, thus, encode only the repressor domain that could either inhibit the activity of the cleaved form of gasdermin in trans or the cleavage itself. Notably, centapoxviruses, the chordopoxvirus genus that is the sister group to ORPV, as well as New World ORPV, encode an additional paralog of OPG177 (OPGX2, Table S1 and Fig. S1C), suggesting a complex evolutionary history of this gene (see discussion below).
Phylogeny and diversity of ORPV.
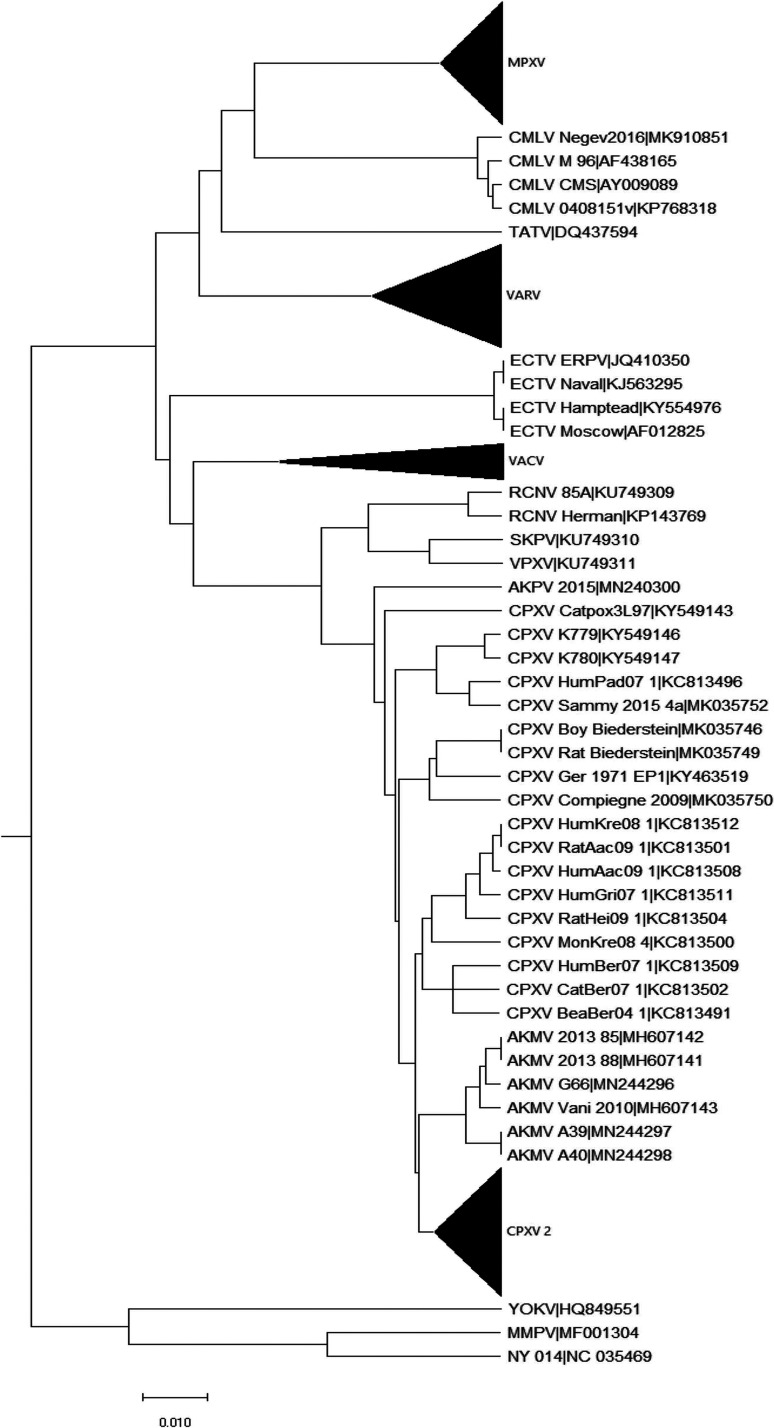
A phylogenetic tree of 235 ORPV genomes combined with 3 genomes of centapoxviruses was constructed from a multiple alignment of the nucleotide sequences of the central portions representing about 118 kb of the genomes, from OPG048 (F4L) to OPG160 (A32L), using centapoxvirus sequences as the outgroup (Fig. S2). The resulting tree topology is largely compatible with the topologies of the trees previously published for smaller collections of ORPV genomes (4, 46–48). As expected, the deepest split among the ORPV is between the New World (RCNV, SKPV, VPXV) and Old World ORPV. The AKPV and AKMV are the deepest branches in the Old World clade, and the rest of the tree is a pool of CPXV strains from which several tight groups of viruses originate, including ECTV, VARV, CMLV-TATV, VACV and MPXV. As noted previously (48), CPXV is not a monophyletic group: there are five distinct clades, among which CPXV1 (splitting into CPXV1.1 and CPXV1.2) belongs in the same branch with VACV, CPXV2 is the sister group of VARV-CMLV-TATV, whereas CPXV3 and 4 are deeper branches, and CPXV5 is the sister group of ECTV-ORPV Abatino (OPVA, Fig. S2).
Complete phylogenetic tree of ORPV. Phylogenetic tree was constructed from a multiple alignment of the central portions of 235 ORPV genomes combined with genomes of three centapoxviruses. Node numbers are shown. Download FIG S2, PDF file, 0.9 MB (951.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We estimated the sequence divergence within the tight clusters of ORPV genomes (Table S4). Members of the CPXV3 and CPXV4 clades have diversified substantially suggesting thousands of years of evolution. In contrast, the mean divergence among the VARV strains was only 0.36%, after excluding the oldest sequenced strains that come from 11th century Vikings (43) and the 17th century (44). This level of divergence apparently reflects approximately 400 years of VARV evolution. The diversity among the MPXV strains was even less, suggestive of recent diversification of this virus. In contrast, the diversity of VACV strains was relatively high (more than a 1% mean divergence), which could reflect fast evolution during vaccine propagation in domesticated animals and cell culture.
Evolutionary divergence within the major clades of ORPV. The mean distances between the genomes in the respective clades are given as percentage nucleotide difference. The main diagonal shows the within-clade distances between the strains across the root of the clade. Download Table S4, DOCX file, 0.01 MB (14.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The four dominant families of paralogous accessory genes.
The accessory genes sets of ORPV are dominated by four families of paralogous genes encoding proteins implicated in the modulation of various pathways of the host immune response and programmed cell death (Table 1, Table S1): 1) 11 PIE domain proteins, five of which also contain the Tumor Necrosis Factor (TNF) receptor domain at the N-terminus, 2) 12 Bcl-2 fold proteins, 3) 7 Kelch repeat proteins also containing the BTB domain at the N-terminus, 4) 14 ANK repeat proteins, most of which contain the additional PRANC domain, a distinct variant of the F-box. Altogether, these four paralogous families include 45 genes, that is, nearly half of the ORPV accessory genes. As discussed below, apart from the ORPV, all four major families of paralogs are represented in many other chordopoxviruses, suggesting that the ancestors of each family were captured by chordopoxviruses at early stages of their evolution.
For the PIE domain family, there are no detectable homologs outside poxviruses, suggesting that this all-beta fold protein evolved by rearrangement of one of the host β-barrel domains, which obliterated the signatures of ancestry (20). For the other paralogous families, there are numerous homologs in diverse organisms, but the exact ancestry is difficult to pinpoint due to the low sequence similarity between poxvirus and cellular proteins. Most of the paralogous family members are significantly more similar to each other than they are to any non-poxvirus homologs (8). Notably, in the process of the ORPV COGs construction, most of the members of each of the four families accurately resolved into individual COGs, indicating that they represent distinct evolutionary lineages, although some manual intervention was required as indicated under Materials and Methods.
The genes encoding members of the four families are scattered over both ends of the ORPV genomes, including several tandem blocks, such as the block of three Bcl-2 genes, OPG34-36 (Table 1). The phylogenetic trees of each family (Fig. S3A-D) are well resolved, with most branches supported by high bootstrap values, and reflect the history of serial duplications that could be mapped to different stages in the evolution of chordopoxviruses as discussed below. Notably, however, there is not a single case of a pair of paralogous genes that are adjacent in the genome being sister groups in the phylogenetic trees of the respective families (Fig. S3A-D). Thus, if the original duplications occurred in tandem, the poxvirus genomes were subsequently extensively rearranged. The proximity of some paralogous genes, e.g., OPG34-36, likely resulted from secondary translocations.
Phylogenetic trees of the four major families of paralogous genes in ORPV. (A) PIE-domain family. (B) Bcl-2 family. (C) BTB-Kelch family (Only Kelch domain was used for the tree construction). (D) Ankyrin-PRANC family FIG S3, PPT file, 0.1 MB (123.4KB, ppt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reconstruction of gains and losses in the evolution of ORPV accessory genes.
We further sought to reconstruct the history of the acquisition (gain) and loss of the ORPV accessory genes during the evolution of chordopoxviruses, in order to uncover potential functionally relevant patterns and connections between genes. Given that capture of a gene from an external source by evolving viruses is a rare event, we assumed that each OPG was gained only once during chordopoxvirus evolution but could have been lost on multiple, independent occasions. Under this assumption (known as the Dollo law in zoology), the gain and loss events can be deterministically (given the phylogenetic tree topology) reconstructed using the Dollo parsimony approach (49, 50). An early study has reported a reconstruction of gene gain and loss for 20 poxvirus genomes using a similar approach (51). The vastly expanded collection of chordopoxvirus genomes provides for a more precise mapping of the evolutionary events on the phylogenetic tree. A cautionary note is that the Dollo assumption for some genes might be violated (that is, some genes potentially could be regained) as a result of recombination among chordopoxviruses themselves. However, we did not address this possibility (with the exception of one specific case discussed below).
We applied Dollo parsimony (see Materials and Methods for details) to the patterns of OPG presence-absence in virus genomes (Table S3A) to obtain independent gain and loss reconstructions of the OPGs. Given that most of the accessory OPGs are shared with different groups of other chordopoxviruses, the reconstructions were performed on 100 genomes that were selected to represent the diversity of chordopoxviruses. The phylogenetic tree of these 100 viruses was constructed from a concatenated multiple alignment of 35 conserved proteins and rooted with the Salmon Gill poxvirus, the deepest known branch of chordopoxviruses. To investigate the patterns of gene loss and possible additional gain among ORPV, a separate reconstruction was performed on the tree of 238 ORPV and centapoxvirus genomes described in the preceding section. The inferred patterns of gene gain and loss are described in the following sections.
Waves of gene gain during the evolution of chordopoxviruses.
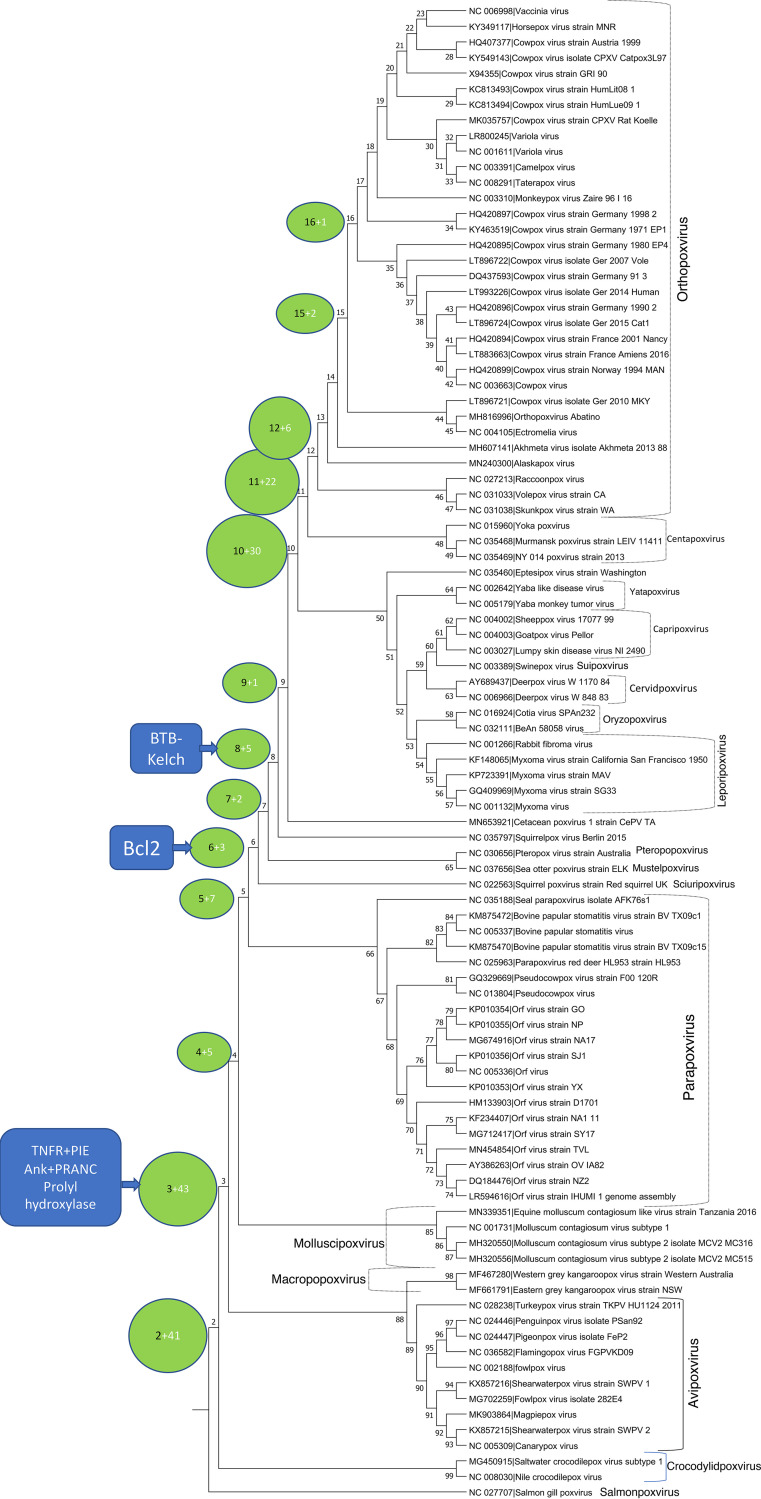
According to our reconstruction, there were four major waves of gene capture during the evolution of chordopoxviruses (Fig. 2, Table S1, column F and Table S5A). The first wave occurred following the split of the genus Salmonpoxvirus from the common ancestor with the rest of the chordopoxviruses. At this time, 41 genes were inferred to have been captured, all representatives of the core set of ORPV genes. As noticed previously, salmon gill poxvirus lacks most of the genes involved in membrane biogenesis in other chordopoxviruses (28), so all these genes were acquired en route to the common ancestor of the tetrapod-infecting chordopoxviruses. As noticed above, however, the core set of ORPV genes also includes some genes implicated in virus-host interactions. The only gene in that category acquired in this early stage of chordopoxvirus evolution is OPG159 (A31R), a member of the US22 family and a putative inhibitor of a dsRNA-dependent host defense activity, that most likely was captured by the ancestral chordopoxvirus from a herpesvirus (see above).
FIG 2.
Gain of ORPV genes at different stages of evolution of chordopoxviruses. Each circle associated with a tree branch shows the node number from which the branch emits (see Table S5A) and the number of genes inferred to have been gained at the branch. The callouts (blue boxes) indicate the inferred points of entry of the ancestors of the major families of paralogous accessory genes into the evolving poxvirus genomes. The chordopoxvirus genera are indicated to the right of the tree.
Reconstruction of gene gains and losses in the evolution of chordopoxviruses and ORPV. (A) Sheet 1: gain and loss of OPG in the evolution of chordopoxviruses. Gene gain (1) and loss (-1) events shown for individual virus strains (tips of the tree) and internal tree nodes denoted by numbers as in Fig. 2. (B) Sheet 2: gain and loss of OPG in the evolution of ORPV. Gene gain (1) and loss (-1) events shown for individual virus strains (tips of the tree) and internal tree nodes denoted by numbers as in Fig. S2. Download Table S5, XLSX file, 0.4 MB (440.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The second major wave of gene acquisition maps to the divergence of crocodilepox viruses from the largest chordopoxvirus branch that unites viruses infecting birds and mammals. By that point in chordopoxvirus evolution, the set of core genes was largely complete, and the 43 genes inferred to have been acquired at this juncture comprise about half of the accessory genes (Fig. 2, Table S5A). Importantly, this is the point of entrance into poxvirus genomes of three of the dominant chordopoxvirus gene families: TNFR-PIE fusion proteins, ANK-PRANC fusion proteins and the prolyl hydroxylase domain-containing proteins. Among the 12 OPGs that consist of ANK-PRANC proteins, 11 were inferred to have been acquired at this stage, suggestive of fast expansion of this family via a series of gene duplications after the capture of the founder of the family from the host. In addition, two genes encoding ANK repeats without a PRANC domain appear at this point, likely, via duplication followed by truncation. For the other two families, the founders were the only representatives at this stage, indicating that duplication occurred later in chordopoxvirus evolution.
The third wave of gene proliferation corresponds to the divergence of the ORPV and their sister group, centapoxviruses, from the common ancestor with leporipoxviruses and capripoxviruses. In contrast to the previous wave, this stage of evolution primarily involved duplication of previously acquired accessory genes, in particular, the Bcl-2 and BTB-Kelch proteins that were captured between the first and second waves of gene gain (Fig. 2 and Table S5A).
The fourth, final wave of gene gain occurred prior to the divergence of ORPV from the common ancestor with centapoxviruses (Fig. 2 and Table S5A). This stage involved further duplication of the genes in the BTB-Kelch and Bcl-2 families as well as capture of several genes from the hosts such as, for example, guanylate kinase (OPG186, A57R) and an inactivated protein kinase (OPG198, B12R). The genes apparently gained during the evolution of the ORPV are discussed in the next section.
As a necessary cautionary note, it should be indicated that the reconstruction of the history of gene gain by duplication in paralogous families, even if largely compatible with the gene phylogenies for these families, is more error-prone than the inferences for the rest of the genes because of the possibility that lineage-specific duplications are mistaken for ancient ones by the COG approach. In particular, late occurrence of at least some of the duplications in the paralogous families has been suggested previously (8). Thus, the counts of ancient gene gains by duplications given here should be considered the upper bound.
Gene gain in ORPV.
Only nine genes were gained throughout the evolution of ORPV (Fig. 2, Table S1, column F and Table S5B). Six new genes were inferred to have been gained at the base of ORPV, including additional members of the large families of paralogs, namely, PIE (OPG192), Bcl-2 (OPG029), and BTB domain without Kelch (OPG030). OPG192 seems to have evolved by duplication of the pre-existing OPG026 (Fig. S3A), followed by relocation of one of the paralogs in the genome. In contrast, the specific origins of OPG029 and OPG030 are difficult to pinpoint, apparently, because of the high rates of evolution of these genes resulting in long branches in the phylogenetic trees of the respective families (Fig. S3B,C). OPG028 is a special case, a gene encoding a tiny membrane protein that might have emerged de novo, from a non-coding sequence. Subsequently, this gene seems to have been lost in only one VARV strain, suggestive of an important role in ORPV reproduction. Of note is the array of three co-located new genes, OPG028-030, an apparent hot spot of ORPV gene birth. OPG058 (F14L), which is conserved in all ORPV, apparently evolved by duplication of OPG156 (A30L), a small virion phosphoprotein. The conservation of both paralogs in all ORPV suggests that they perform distinct, essential functions. Finally, OPG188 is another special case, where a new domain was added to an ancestral poxvirus protein as discussed below.
Three additional genes were gained later in ORPV evolution. OPG207 (C11.5L) encodes a tiny protein without any distinct features, the origin of which maps to the branch separating AKMV from the rest of the Old World ORPV and that apparently was lost on 19 independent occasions during the subsequent ORPV evolution (Table S5B). This gene should be considered questionable until the production of the protein is experimentally validated. OPG010a is another special case, where a protein with a new function was derived from a preexisting protein as discussed in the next section.
The latest gained gene in ORPV is OPG214, N-methyl d-aspartate receptor homolog, an apoptosis inhibitor (52), that was acquired before the divergence of CPXV4 from the rest of ORPV but was subsequently lost on 19 independent occasions (Table S5B). This is the only gene that was acquired by ORPV from the hosts as supported by the high amino acid sequence identity (about 75%) with mammalian homologs. Nearly identical 21 bp sequences flanking the open reading frame are indicative of a target site duplication, consistent with gene capture resulting from LINE-1 mediated retrotransposition of the mRNA of the ancestral host gene (53). Conceivably, this protein plays an accessory role in apoptosis (pyroptosis) inhibition that becomes irrelevant in many ORPV.
Loss of genes during the evolution of ORPV.
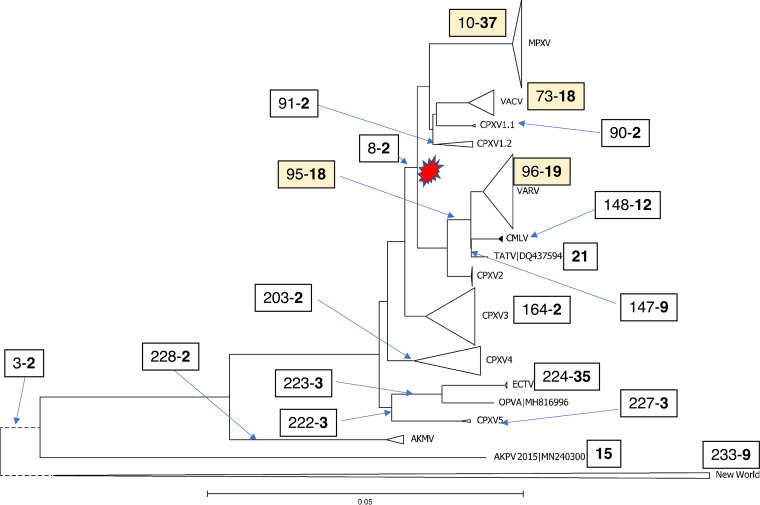
The distribution of the gene losses across the phylogenetic tree of the ORPV is highly non-uniform (Fig. 3). The most extensive gene loss, 37 genes, was mapped to the branch that leads to the common ancestor of the MPXV, followed by the loss of 35 genes at the base of the ECTV branch. The stem of VARV is associated with the loss of 19 genes, and 18 more genes were lost on the path to the common ancestor of CMLV-TATV and VARV. Thus, altogether, the emergence of VARV was accompanied by the loss of as many genes as the advent of MPXV, followed by the loss of 12 additional genes within the VARV clade, after the deepest split between the Viking strain and the rest of the strains (Table S5B). At the base of the VACV branch, 18 genes were lost, followed by substantial additional losses in different groups of VACV strains, for example, 18 genes in the ancestor of the Tashkent strains. Apart from these deep losses, some individual ORPV strains have lost multiple genes as well. Striking examples include the VACV strain MVA, which was passaged more than 500 times in chicken embryo fibroblasts, and IHD-W1, each losing 18 genes.
FIG 3.
Phylogenetic tree of ORPV and gene loss in ORPV evolution. The large branches are collapsed and shown with triangles. Each box associated with a tree branch shows the node number from which the branch emits (see Fig. S2) and the number of genes inferred to have been lost at the branch (numbers in bold). The four colored boxes show major clades with many genes lost at the stem. For branches without associated boxes, no or one gain was inferred. The red shape shows the high-loss clade of ORPV.
Several pairs of sister branches of ORPV including closely related viruses showed sharply contrasting amounts and rates of gene loss (Fig. 3). The strongest case in point is the ECTV-OPVA branch, where the ancestor of ECTV strains lost 35 genes whereas OPVA lost none. Similarly, 19 genes were lost at the base of the VACV, in contrast, to only two genes lost in CPXV1 (Fig. 3). These contrasting patterns of gene loss presumably reflect drastic changes in the virus biology following the divergence of the respective clades. The nature of the change is clear in the case of VACV because of multiple passages in domesticated animals for vaccine production or in cell cultures, but far less obvious in the case of ECTV although a switch to a narrow host range is a clear possibility. Except for the early branching ECTV clade, all groups of ORPV that underwent extensive genes loss belong within the same major clade in the phylogenetic tree that splits from CPXV3 (Fig. 3). However, only two genes were lost at the stem of this clade, indicating that massive gene loss occurred in parallel, independently, at the bases of the MPXV, VACV and VARV branches within the ‘high-loss’ clade of ORPV.
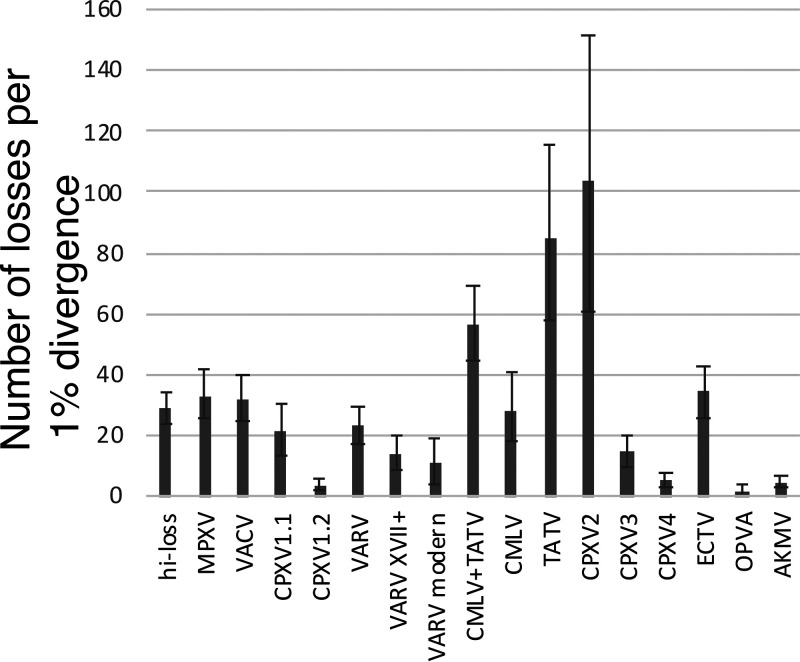
The rates of accessory gene loss for each clade were compared to the percent divergence of the nucleotide sequence in the central conserved core region of the genome, which serves as a proxy for time, under the molecular clock assumption (Fig. 4). Thus, the number of genes lost in a clade (for example, at the base of the VARV branch and in all its descendants) per 1% divergence of the nucleotide sequences in the central region of the genome, shows that the groups in the high-loss branch (Fig. 3) as well as ECTV lose genes at similar rates of 20 to 35 (Fig. 4). Interestingly, when the loss rate for the VARV branch was estimated without the deepest Viking branch, and then, without the 17th century branch, a lower estimate of 10 was obtained for the modern strains, suggesting that gene loss in VARV had been slowing down after the initial acceleration. The deep branches of ORPV, CPXV4 and AKPV had low loss rates, whereas the loss rate in CPXV3 was close to that in the modern strains of VARV. Remarkably, however, CPXV2, the sister clade of the VARV-CMLV-TATV branch, shows an extremely high gene loss rate of about 100, and TATV had only a slightly lower rate of about 80. These estimates indicate that the evolution of ORPV involved several major accelerations of gene loss of roughly the same magnitude, with the notable exceptions of the dramatic accelerations in CPXV2 and TATV.
FIG 4.
Estimated rates of gene loss in the major clades of ORPV. The plot shows the loss rate in the stem of the respective clade and each of the branches within the clade. The gene loss rates were estimated as the number of genes lost per 1% nucleotide distance from the common ancestor of the respective group. Whiskers show the 90% confidence interval (5th and 95th percentile of the loss rates across 1000 bootstrap samples of genes).
The trends of gene loss are reflected in the topology of the gene content evolution tree of ORPV that was constructed by comparing the patterns of gene presence-absence (54) and shows substantial differences in topology from the phylogenetic tree built from the sequence alignment of the core genes (Fig. 5). Unlike in the sequence-based phylogeny, in the gene presence-absence tree, all CPXV strains that lost few genes form a tight cluster that also includes the AKMV strains and is joined by New World ORPV, and then, by VACV that underwent a moderate gene loss at the onset of their evolution. Virus groups that underwent extensive gene loss, namely, MPXV, VARV, TATV and CMLV, form another major cluster (Fig. 5). This tree topology reflects the substantial overlap in the sets of genes that were independently lost in these four groups of ORPV as discussed below. In this tree, CPXV2, the group of strains with an anomalously high gene loss rate, forms a deep branch in the CPXV clade, indicating that its gene loss pattern was distinct from that in the rest of the high-loss groups.
FIG 5.
Gene content evolution tree of ORPV derived from the patterns of gene presence-absence. The Unweighted Pair Group Method with Arithmetic mean (UPGMA) dendrogram was constructed from the matrix of distances between OPG presence-absence patterns, and therefore, reflects the overlap between the sets of lost genes (Table S3A).
Genomic landscape of gene losses in ORPV.
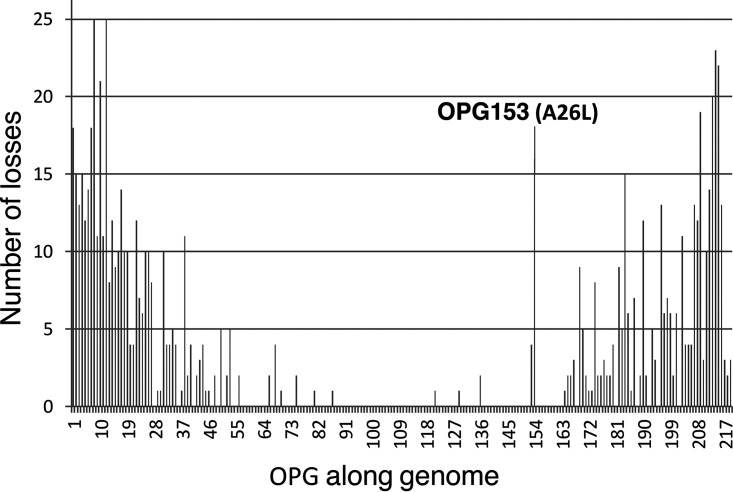
By definition, the great majority of the genes lost by ORPV are accessory genes located at both ends of the genomes (operationally defined as the 50 genes at each end). Among these genes, 88 comprising 44 from each end were lost at least once during ORPV evolution, and 81 genes were lost at least twice, independently. Moreover, many genes were lost on multiple, independent occasions, up to 25 times (Fig. 6 and Table S5B). The frequency of gene loss notably drops from each of the ends toward the middle of the genome.
FIG 6.
Distribution of inferred gene losses along the ORPV genomes. Each bar shows the number of times a given OPG was lost during the evolution of the ORPV. OPG153 (A26L), a gene with an anomalously high loss rate for its position away from the genome end, is indicated.
The accessory genes were often lost in blocks (Tables S3A and S5B). Strikingly, the same block of 10 genes, located near the left end of the genome, was lost in the three major branches of ORPV, namely, MPXV, VARV, and VACV, and in the latter two groups, this block extends to 13 genes. The sole exception within these three groups is horsepox virus (HSPV), which belongs inside the VACV branch (Fig. S2), but retains three of the genes in the 10 gene block (Table S3A). The 10 gene block consists, primarily, of genes encoding members of the large accessory protein families (see above), including three paralogous ANK-PRANC proteins, two Bcl-2 proteins, two proteins containing the BTB domain (one with Kelch domain and another without), and one protein containing the PIE domain (Table 1). In the case of this and other, shorter loss-prone gene blocks, the respective genes have been entirely deleted (the genes at the border of the deleted gene blocks are typically present but are disrupted by frameshifts or within gene deletions). However, the exact borders of the deletions do not coincide in different groups of ORPV, and furthermore, the presence of genes in the middle of the block in the HSPV genome implies that, at least, in VACV, the loss of this gene block involved more than one deletion. Similarly, two or even more deletions in this gene block occurred in ECTV and in many strains of CPXV (Table S3A). Another region of the genome that is particularly prone to deletions is the 13 gene block near the right end, from OPG177 to OPG188a (Tables S3A and S5B).
There were only a few losses in the central part of the genome that deviated from this trend (Tables S3A and S5B). In particular, OPG041 (K3L) was lost in MPXV and ECTV, and MPXV also lost OPG067 (E5R). The loss of OPG041, one of the two OPGs that inhibit PKR via distinct mechanisms (see above), appears to reflect the experimentally demonstrated lower level of dsRNA production in cells infected with MPXV (55) and ECTV (56), compared with VACV, which apparently makes the former viruses less sensitive to the dsRNA-dependent inhibition of virus reproduction mediated by PKR (57). Notably, in addition to the inactivation of OPG041, MPXV encodes an N-terminally truncated form of the E3 protein (OPG065) lacking the Z-DNA-binding domain (8) as discussed below, in the section on unusual cases of gene disruption in ORPV.
The specific functions of OPG067 are not known, but the presence of two DNA-binding BEN (BANP, E5R and Nac1) domains (58) in this protein suggests the possibility of involvement in a different but, perhaps, related anti-immune mechanism that is likely to involve recognition of cytoplasmic DNA by host defense factors.
The common ancestor of the VARV strains, with the exception of the deepest branch, lost the OPG069 (E7R) gene that encodes a component of the enveloped virions (59). The common ancestor of TATV and CMLV as well as the MVA strain of VACV lost OPG074 (O1L) that has been shown to activate the ERK kinase, which is required for optimal reproduction of other VACV strains in cell culture (60). Several additional genes from the middle of the genome were found to be lost from individual strains (Table S3A) although, in these cases, it is harder to rule out a sequencing error.
Concurrent and exclusive gene loss and potential interactions among ORPV genes.
We sought to explore potential functional connections among ORPV genes by detecting patterns of concurrent gene loss (groups of genes that are lost together more often than expected) or, conversely, exclusive gene loss (groups of genes that are lost together unexpectedly rarely). In many previous studies, concurrent gene loss patterns have been used to predict functionally linked genes, for example, those encoding proteins that form a complex or otherwise cooperate within the same functional pathway (61, 62). Conversely, patterns of exclusive (complementary) gene loss can point to functional redundancy, or in other words, to alternative genes responsible for the same essential biological function. For example, genes with complementary loss patterns could affect the same pathway, perhaps, at different stages and via different mechanisms as in the case of OPG041 and OPC065 discussed above. Such genes are likely to behave as synthetic lethal, that is, pairs of genes that cause lethality only when both are inactivated. Although requiring much caution, due to the complexity of interactions among genes, analysis of patterns of gene loss can have considerable predictive power (61–63).
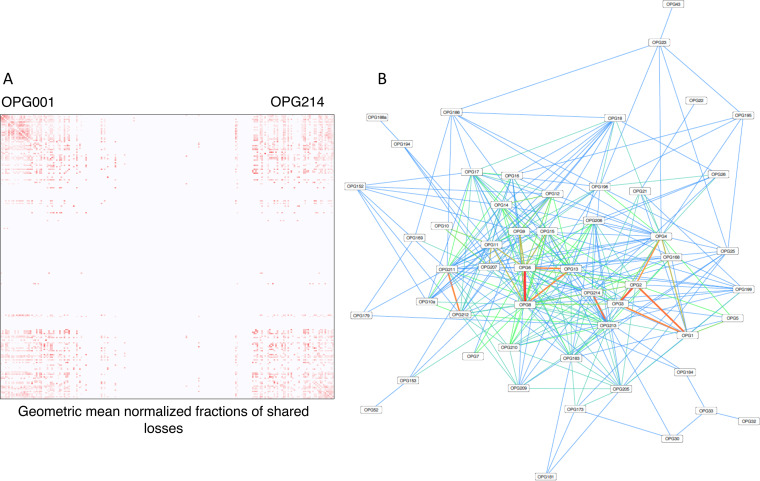
The patterns of concurrent gene loss can be represented in the form of a heat map (Fig. 7A) or a network (Fig. 7B) (see Table S6 for complete information). Predictably, high density of concurrent losses of OPG was observed at both ends of the genomes, whereas closer to the middle of the genome the density of concurrent losses dropped precipitously (Fig. 7A). At least in part, this trend could be attributed simply to the high deletion rate near the genome termini. However, the pattern persists, even though the concomitant loss frequency was normalized by the geometric mean of the individual gene losses (Table S6), suggesting that the strongest connections might reflect functional associations. For example, the most strongly connected cluster of three concomitantly lost genes in the concurrent gene loss network (Fig. 7B) included OPG006 (a Bcl-2 protein), OPG008 (a BTB domain protein) and OPG013 (a TNF receptor homolog) that potentially could affect different steps of the same, perhaps, species-specific host pathway. There are, also, multiple associations among genes located closer to the central part of the genome (Table S6). A case in point is the gene pair OPG032 (C3L, secreted complement control factor) and OPG033 (C2L, a BTB-Kelch protein): each of the four times OPG032 was lost during the ORPV evolution, the loss occurred jointly with OPG033 (which was only once lost separately). This concurrent loss of the two genes could not be explained simply by the high probability of joint deletion because OPG032 was never lost together with its other neighbor OPG031, and OPG033 was only once lost together with OPG034. Thus, a functional interpretation of the excessive joint loss of OPG032 and OPG033 appears to be more likely than a mechanistic one.
FIG 7.
Concurrent gene loss in ORPV. (A) Heatmap of gene losses. The all against all map of OPG is shown. The intensity of the red color is proportional to the frequency of the concurrent loss of the respective OPG. (B) Network of gene losses. Shown is a sub-network of the complete network of concurrent gene loses that includes 53 genes, with edges connecting genes with at least 3 independent concurrent losses. The color goes from blue (3 shared losses) to red (13 shared losses). The network was visualized using Cytoscape v.3.8.2.
Concurrent and exclusive gene losses in ORPV. Sheet 1: number of concurrent losses for all pairs of OPG. The intensity of the red highlighting is proportional to the frequency of losses. Sheet 2: node degrees (the number of nodes connected with the given node) of the OPGs in the complete network of concurrent losses. Sheet 3: number of concurrent losses for all pairs of OPG normalized by the geometric mean of the number of individual losses for the respective OPG. Sheet 4: number of genomes where both genes are absent for all pairs of 102 OPGs with at least one absence in the Old World ORPV. Sheet 5: number of genomes where both genes are absent for all pairs of 102 OPGs with at least one absence in the Old World ORPV, normalized by the geometric mean of the number of Old World ORPV genomes where this gene is absent for the respective OPGs Table S6, XLSX file, 0.4 MB (413.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Perhaps, the most notable is the case of OPG074 (O1L), a non-essential membrane protein encoded in the central part of the ORPV genomes. This gene was lost twice during the ORPV evolution, in the stem branch of CMLV-TATV and, independently, in VACV strain MVA. On both occasions, OPG074 was lost jointly with OPG034 (C1L, a Bcl-2 domain protein) and OPG039 (K1L, an ANK repeat protein). In this case, given the low incidence of gene loss in the middle of the genome, involvement of the three genes in the same functional pathway appears likely.
The network of concurrent losses of ORPV genes (Fig. 7B and Table S6) is dense, with many genes having a high node degree, that is, having been lost concurrently with numerous other genes, up to 84 connections (Fig. S4A,B and Table S6). The network displays a near uniform node degree distribution (Fig. S4A) and a high clustering coefficient (Fig. S4B), which reflects the existence of many tightly connected groups of genes. These features differentiate this network from both random networks and scale-free networks that are common in biology (64) and seem to suggest numerous interactions among the OPG that are distributed across multiple, overlapping gene modules. Although many of these connections are likely to be spurious, this high connectedness of the network suggests multiple, complex functional interactions among the accessory genes of ORPV. Interestingly, both most highly linked genes, OPG213 and OPG183 (A53R), encode TNF receptor homologs, suggesting a central role of these genes in the interactions of ORPV with the host immune responses.
Parameters of the network of concurrent loss of ORPV genes. (A) Node degree distribution for the complete network of the concurrent gene losses. The distribution is shown for the complete network of concurrent losses where edges connect genes that were concomitantly lost at least once. P.d.f. probability density function. (B) Clustering coefficient distribution for the complete network of the concurrent gene losses. p.d.f. probability density function FIG S4, PPT file, 0.4 MB (385.4KB, ppt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A notable case of repeated loss of the same gene is OPG153 (A26L) that was lost on 18 independent occasions during ORPV evolution. This is an anomalously high rate of loss for a gene located so far from the end of the genome: among the topmost frequently lost ORPV genes, A26L is the only one located more than 50 genes away from the end (Fig. 6 and Table S1). The A26 protein is contained in mature ORPV virions and contributes to the binding of MV to plasma membrane and to the suppression of virus-cell fusion (65), and in some ORPV, also is involved in occluding mature virions in dense inclusions (66). Evolutionary experiments in cell culture have shown that an A26L disruption mutant came to fixation in the virus population after multiple passages of a VACV mutant with a defect in the A8R gene encoding a transcription factor (67). Our present analysis shows that A26L was lost concurrently with a variety of ORPV genes, suggesting that analogous compensatory phenomena occurred repeatedly during the ORPV evolution.
To summarize, although the available data is insufficient to make compelling predictions of functionally linked ORPV genes, many tentative inferences amenable to experimental validation can be made.
We next explored the pairs and larger clusters of genes that were often lost individually but never or infrequently together (Table S6). The most striking such pair consists of OPG005 (C16L, Bcl-2 protein) and OPG037 (M1L, ANK protein without the PRANC domain, apoptosis inhibitor) that have been lost 12 and 11 times, respectively, during ORPV evolution, but are never missing together. In this case, it can be predicted with a degree of confidence that these proteins affect the same host apoptotic pathway and form a synthetic lethal pair. Furthermore, OPG005 is never missing together with OPG204 (B19R, decoy interferon receptor). OPG019 (C11R, epidermal growth factor homolog) is never missing together with several genes, including the frequently lost OPG001 and OPG003 (PIE domain and Ank-PRANC domain proteins, respectively), again suggesting functional redundancy. Overall, as in the case of concurrently lost genes, the exclusive loss or absence of genes in ORPV seems to have limited predictive power but several instances of likely functional redundancy can be inferred.
Gene loss in the four major families of paralogous ORPV genes.
We examined in greater detail the patterns of gene loss within the four largest families of paralogous genes of ORPV, in relation to the major branches of ORPV (Table 2). None of the ORPV has lost any of the families completely. Conversely, each of the members of each family was lost at least once during ORPV evolution, except for two Bcl-2 proteins, OPG35 (N1L) and OPG200 (B15R). There were notable differences in the extents of gene loss across the four families of paralogs and the major lineages of ORPV (Table 2). Thus, MPXV lost 6 of the 8 BTB-Kelch proteins, whereas VACV and ECTV each lost only one. Overall, the most extensive loss was observed among the ANK-PRANC proteins, whereas Bcl-2 proteins and PIE proteins were lost rarely, in comparison. An exception is the loss of nearly half of the Bcl-2 family in ECTV (Table 2). The genes within each family are lost non-uniformly, following the general trend of preferential gene loss at genome ends. Thus, among the BTB-Kelch genes OPG008 and OPG011 located near the left end of the genome are the most extensively lost genes, each on 25 occasions, whereas OPG047 (F3L), which is closer to the middle of the genome, was lost only twice. Similarly, among the Ank-PRANC genes, the near-terminal OPG211 was lost on 20 occasions, whereas OPG189 (B4R) only twice. This signal is even more prominent among the Bcl-2 family genes. Thus, the near-terminal OPG5 and OPG6 were lost 12 and 14 times, respectively, whereas OPG035 (N1L) was never lost (as mentioned above), and the two adjacent Bcl-2 genes located toward the middle of the genome, OPG44 (K7R) and OPG45 (F1L), were lost only once each (table S5B). Taken together, the pattern of gene losses in the families of paralogous ORPV genes suggest that, although most of these genes individually can be dispensable depending on the biology of specific viruses, there seems to be an essential basic function of each family in suppression of host immunity that is essential for ORPV reproduction.
TABLE 2.
Loss of the genes of the four dominant large families of paralogs in the major clades of ORPV
| ORPV branch | Total lost genes | BTB-Kelch (8) | Ank-PRANC (14) | Bcl-2 (12) | PIE (11) |
|---|---|---|---|---|---|
| MPXV | 37 | 6 | 5 | 3 | 1 |
| VARV | 37 | 6 | 7 | 2 | 2 |
| VACV | 18 | 1 | 6 | 1 | 2 |
| ECTV | 35 | 1 | 7 | 6 | 3 |
The total number of members in each paralogous family is indicated in parentheses.
Progression of ORPV genes en route to elimination.
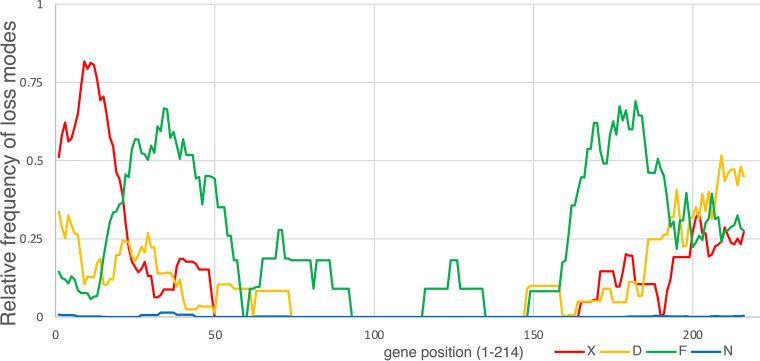
We were further interested in the more mechanistic details of gene loss during the evolution of ORPV. Among the total of 2502 accessory genes that were found to be inactivated in the 235 ORPV genomes, 795 were disrupted by frameshift mutations, 947 contain within gene deletions, only one contained an in-frame stop codon resulting from a nonsense mutation, and 759 were (nearly) completely deleted, in many cases, as parts of larger deletions (Table S3B). The different types of gene inactivation showed distinct distributions over the length of ORPV genomes such that large deletions occur primarily near both genome termini, whereas genes located deeper into the genomes are mostly inactivated by frameshifts; within-genes deletions are less common and occur more uniformly along the genome (Fig. 8). Notably, the distributions of all events displayed a high degree of symmetry with respect to the middle of the genome.
FIG 8.
Distribution of different gene disruption events along the ORPV genomes. The distributions of relative frequencies of 4 types of gene disruption events along the ORPV genomes are shown. The events are color-coded as shown beneath the plot: D, decaying (deletion); F, frameshift; N, in-frame stop nonsense mutation; X, complete absence (large deletion). Relative frequencies of disruption events in each gene were averaged in an 11-gene sliding windows.
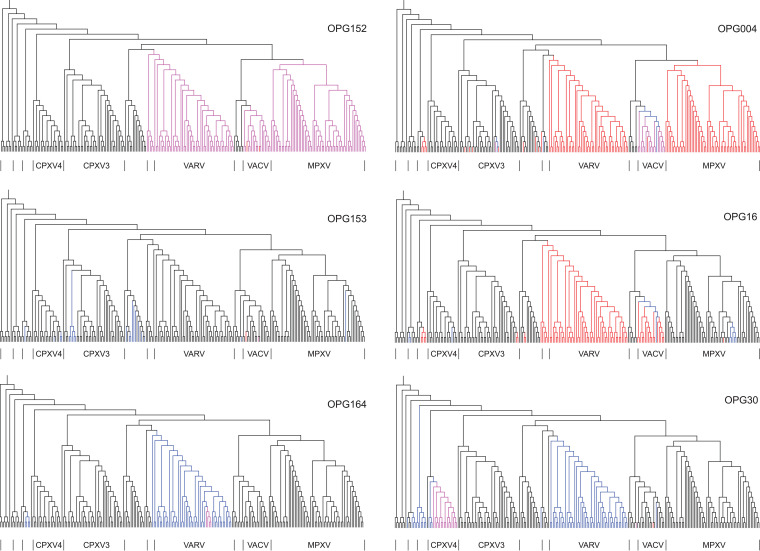
By mapping gene inactivation events onto the phylogenetic tree of ORPV, the paths of gene elimination could be traced. We performed such an analysis for six selected accessory genes that showed distinct histories depending on their genomic positions (Fig. 9). Thus, OPG004, located near the left end of the genome, was lost as a result of independent large deletions in the MPXV and VARV branches (as already noted above), whereas within the VACV branch as well as CPXV3, frameshifts were followed by either within-gene or larger deletions. OPG016 was eliminated by large deletions in VARV and some CPXV branches, whereas in VACV, such deletions are traceable to a founding frameshift. In the case of OPG030, closer to the central part of the genome, frameshifts independently occurred in VARV and ECMV, and the resulting pseudogenes persisted in all viruses of the respective groups; in contrast, in CPXV4, a frameshift was followed by a within gene deletion. OPG152 and OPG153, two adjacent genes located close in the middle portion of the genome, where gene disruption is overall rare, display contrasting patterns of gene loss. OPG152 was disrupted by independent within gene deletions in MPXV, VARV and VACV, whereas OPG153 was inactivated by multiple independent frameshifts (as discussed above), of which several occurred in the same position within the gene, in a stretch of 7 A bases that is likely to trigger DNA polymerase slippage. Finally, for OPG164, a frameshift at the base of the VARV clade was followed by a within gene deletion in one branch.
FIG 9.
Routes of gene disruption in ORPV. The gene disruption events for 6 OPGs are mapped onto the phylogenetic tree of the ORPV (Fig. S2). Magenta, decaying (within gene deletion); blue, frameshift; red, missing (large deletion).
Overall, the routes of gene elimination in ORPV are likely to be determined by multiple factors. These include genomic position, whereby deletions appear to preferentially occur near the ends, the presence of frameshift or deletion prone sequences within the respective genes, and possibly, additional effects of selection.
Unusual fates of disrupted genes in ORPV evolution.
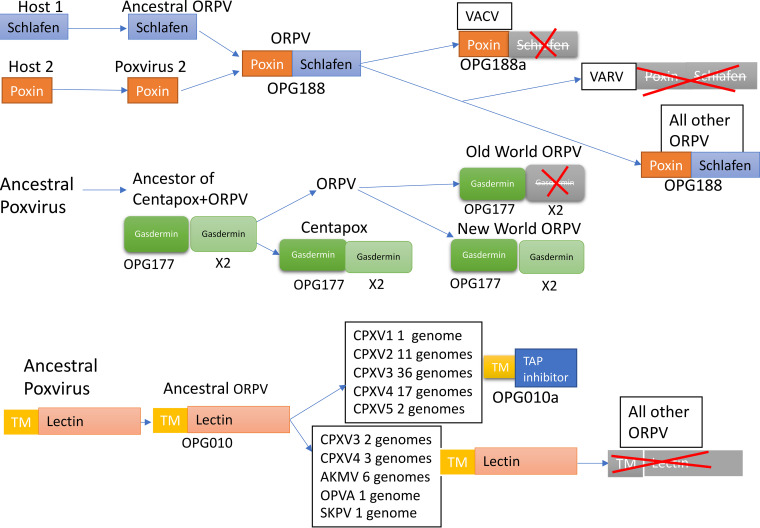
One notable example is the recruitment of a frameshift variant of an ancestral gene for a new function. The OPG010 protein consists of about 160 amino acids and contains an N-terminal transmembrane domain followed by a C-type lectin domain. It is conserved in most chordopoxviruses and was apparently acquired prior to the divergence of mammalian poxviruses and avipoxviruses from their common ancestor (Table S1 and S5A). This gene, however, is disrupted in many ORPV (Fig. 10 and Table S3A). Remarkably, one of the disrupted variants, 69 amino acids in length and retaining the transmembrane domain, acquired a new C-terminal sequence, derived from an alternative, originally non-coding reading frame in the lectin-coding region, and gained a distinct function, as an ER-located TAP inhibitor, that was fixed in evolution (68). This functional truncated variant is found in many strains from all five branches of CPXV (Fig. 10), and remarkably, the nucleotide sequences encoding the short protein are (nearly) identical in all these genomes (with only one or two substitutions in some of the strains). The only plausible explanation for this highly unusual case of evolution of a new functional gene in ORPV involves broad dissemination across the diversity of CPXV via homologous recombination following the initial frameshift mutation. The other ORPV, including the major high-loss branches, MPXV, VARV and VACV, the OPG010 gene was disrupted to yield fragments of variable lengths that originated from different parts of the protein, are not conserved even among closely related strains and thus apparently are non-functional (Fig. 10). In the course of the ORPV gene reannotation, we opted to construct two OPGs for this unusual case: one for the full-length protein containing the lectin domain (OPG010) and the other one for the functional truncated variant (OPG010a). Because most of the accessory genes were truncated through frameshifts in different lineages of ORPV, we searched for possible cases of evolutionary conservation of truncated proteins, especially, those containing signal peptides, but failed to identify convincing evidence of this route of evolution for any other genes. Thus, so far, OPG010a remains the only clear case of a disrupted gene escaping pseudogenization through exaptation.
FIG 10.
Unique evolutionary scenarios for disrupted genes in ORPV. The inferred evolutionary events are shown for three OPGs with unusual histories. OPG188: fusion of Poxin with Schlafen followed by disruption of Schlafen, leaving the intact Poxin, or disruption of the entire fused gene. OPG177, Gasdermin homolog, duplication in branch leading to the common ancestor of ORPV and Centapoxviruses followed by the loss of one paralog in Old World ORPV. Poxvirus X denotes the common ancestor of Centapoxviruses and ORPV. Shapes with pale gray coloring and red cross denotes gene disruption. OPG010, OPG010a: exaptation of a disrupted remnant of an endoplsmic reticulum protein containing a lectin domain for a function as a TAP inhibitor (OPG10a); OPG10a evolved convergently in several groups of ORPV (see the text).
Another example of an unusual fate among the OPG involves fusions and fissions of Poxin and Schlafen. Most of the CPXV and MPXV strains encode a protein of about 500 amino acids, OPG188, which is a fusion of the Poxin, a cGAMP-specific nuclease that blocks the STING-dependent interferon pathway (69, 70), and the RNA-binding protein Schlafen (71). Poxin is apparently ancestral in poxviruses, being represented in some entomopoxviruses, whereas Schlafen was acquired by the ancestor of ORPV and fused with poxin (Fig. 10). Subsequently, the fused gene OPG188 was disrupted in the common ancestor of the CPXV1 and VACV lineages, recreating the active Poxin and generating a C-terminal fragment that is apparently non-functional; furthermore, in VARV, the OPG188 gene was disrupted completely (Fig. 10). A full size, stand-alone Schlafen protein was not detected in any poxvirus genomes. For the ORPV gene reannotation, we once again created two different OPGs, OPG188 for the fusion protein and OPG188a for the solo Poxin. Poxin and Schlafen, in all likelihood, function in concert in the fusion protein, although the specific role of this protein, and what Schlafen adds to the Poxin activity, remains unclear. To our knowledge, the evolutionary history of Poxin-Schlafen is unique in that disruption of the fusion protein recreates the original, active state of one of the fusion components.
The well-characterized case of OPG065 (E3L) (8) is, in principle, similar to the case of OPG188/188a. This gene encoding a protein containing an N-terminal Z-DNA-binding domain (Zα) and a C-terminal dsRNA-binding domain (dsRBD) was acquired prior to the divergence of parapoxviruses from the rest of the mammalian chordpoxviruses (Table S1 and Ref. 8). The dsRBD domain is necessary and sufficient for the inhibiton of PKR, whereas the specific function of the Zα domain remains unclear although disruption of this domain in VACV leads to severely reduced virulence in model systems which correlates with the reduction of Z-DNA binding (72–74). The upstream portion of OPG065 is disrupted in leporipoxviruses and, independently, in MPXV and Volepox virus, inactivating the Zα domain, whereas the dsRBD remains intact and active (8) (Table S3B). Thus, similar to the case of OPG188, multiple independent disruptions of OPG065 eliminate one domain and a distinct function of a multdomain, multifunctional ORPV protein without affecting a second domain with its distict function. The disruption of OPG065 is thought to affect the virulence and host range of the respective chordopoxviruses (8).
Gasdermins have a complex history in ORPV. As discussed above, OPG177 (A47L) is a homolog of the C-terminal portion of gasdermins and a likely pyroptosis inhibitor. This gene was captured from the host prior to the divergence of leporipoxviruses and capripoxviruses from the rest of mammalian poxviruses (Table S1 and S5A) and was subsequently duplicated prior to the divergence of centapoxviruses and ORPV. Both centapoxviruses and the New World ORPV encode two gasdermin paralogs but the Old World ORPV have lost one of the copies (OPGX2 in Table S1 and Fig. 10), followed by the loss of the remaining one (OPG177) in MPXV and CMLV (Fig. 10).
DISCUSSION
We initiated this work with the goal of elucidating consistent trends in ORPV evolution and harnessing comparative genomics for predicting functions of uncharacterized genes likely involved in potential virus-host interactions. Even before pursuing these goals directly, our first task was to develop an evolutionary classification of ORPV genes and the corresponding uniform nomenclature, and reannotating the accessory genes combining literature searches with sequence analysis using sensitive computational methods. Notwithstanding the obvious drawback of introducing a new, unfamiliar nomenclature, we believe this to be an essential step because of the advantage of using the same name for orthologous genes in all ORPV. Inconsistent naming of orthologs in different viruses substantially complicates comparative genomic studies. We included the commonly used nomenclature of the VACV Copenhagen genes along with the OPGs. This practice will probably continue in further ORPV studies, but the introduction of the unified nomenclature appears indispensable. We suggest that the OPGs, which entail a convenient, uniform numbering that immediately conveys the gene position in the virus genome, accommodate all Old World ORPV genes and exclude pseudogenes, become the standard tool for annotation of ORPV genomes and the study of ORPV evolution. The current reannotation of the accessory OPG highlights the remarkable multifunctionality of many of these proteins: even those accessory proteins that, unlike those in the ANK-PRANC and BTB-Kelch families, encompass only a single domain typically have been shown to affect multiple host immune pathways (Table 1 and Table S1). To this end, we employed the well-established COG approach (24–26). All 214 OPGs accurately resolved into COGs, with minimal manual intervention to disambiguate the assignment of paralogous genes.
The OPGs are divisible into two classes of nearly equal size: about 100 genes in the central part of the genome that encode proteins involved in viral housekeeping functions (virion structure, replication, transcription) and are mostly essential for virus reproduction in cell culture, and about 50 genes from each end that encode accessory proteins involved in virus-host interactions. There are a few exceptions to this partitioning whereby several genes affecting host immune mechanisms are located close to the middle of the genome, and conversely, a few genes involved in replication are close to the ends. Furthermore, some of the essential, housekeeping genes possess additional functions in the abrogation of host responses to infection. A striking case in point is OPG062 (F17R), which is a major DNA-binding protein in VACV virion cores (75, 76), but additionally, has been shown to suppress the host interferon response by dysregulating mTOR kinase (77, 78). Another notable example is OPG106 (H1L), a dual specificity phosphatase that plays an essential role in virion morphogenesis by dephosphorylating virion proteins, and additionally, inhibits the STAT1-dependent interferon pathway (79, 80). These exceptions notwithstanding, the evolutionary regimes of the central part and the periphery of the ORPV genome are dramatically different. The genes in the central region, except for several lineage-specific losses, all persisted throughout the evolution of chordopoxviruses infecting tetrapods. In contrast, the peripheral regions are highly dynamic and were completely reshaped during the same evolutionary span. Gain and loss of the accessory genes of ORPV occurred on different evolutionary scales. Most of the accessory genes apparently accrued in the last common ancestor of ORPV as a result of three waves of gene capture. On the short scale, evolution of the ORPV was dominated by gene loss, which accompanied the emergence of each major ORPV branch, and in the MPXV and VARV branches, involved up to 40% of the accessory genes. Notably, the rate of gene loss in ORPV showed a steep gradient from the ends toward the central part of the genome. Whether the cause of the decreasing rate of gene loss from the periphery to the center of the ORPV genomes is mechanistic or adaptive, is hard to ascertain. Deletions might occur more frequently near the ends of the genome via recombination between gene copies during the replication of head-to-head concatemers (81, 82). However, it is also conceivable that selection favors genome rearrangements that cluster less essential genes and push genes with more important functions toward the central part of the genome. These two explanations of the observed trend of gene loss are not mutually exclusive.
In this work, we did not systematically investigate recombination between ORPV genomes that could be an important factor in the evolution given the high level of sequence similarity allowing homologous recombination. Indeed, likely recombination events between ORPV and also between ORPV and centapoxviruses have been reported in several previous studies (46, 83–85). The remarkable case of OPG010a, a novel gene that appears to have disseminated among CPXV strains via multiple recombination events, underlines the role of recombination in ORPV evolution that remains to be comprehensively characterized by phylogenomic analysis.
A notable feature of the accessory gene repertoires of the ORPV is the high prevalence of paralogous gene families including the striking proliferation of ANK-PRANC and Bcl-2 proteins. Acquisition of new genes by poxviruses requires complete reverse transcription of the respective host mRNAs followed by illegitimate recombination with the virus genome. The feasibility of this scenario has been demonstrated experimentally (53), but incorporation and subsequent fixation of a functional gene is likely to be extremely rare. Therefore, in the course of their adaptive evolution, poxviruses take the maximum advantage of each acquired gene via serial duplication and functional diversification. According to our reconstruction, the expansion of paralogous families via duplication typically occurred shortly after poxviruses captured the founding members of the respective families, especially, in the case of the ANK-PRANC family. Amplification of accessory genes under selective pressure in evolutionary experiment, dubbed the evolutionary accordion (86, 87), suggests that gene duplications are relatively frequent in poxviruses, in accord with the evolutionary reconstruction. The differences between the specific roles of paralogs are poorly understood but involve both distinct specificities and (partial) functional redundancy (8). As we observe here, none of the large families of paralogs is lost completely in any orthopoxvirus, suggesting that the basic activity of each class of domains, such as, for example, the role of ANK-PRANC protein in the modulation of the ubiquitin-proteasome network, is essential for virus reproduction, regardless of the host range.
The default explanation for the massive gene loss at the bases of the major branches of ORPV (MPXV, VARV, VACV, ECTV) seems to be a ratchet of gene elimination upon transition from a broad to a narrow host range. Conceivably, when a gene(s) required for virus reproduction in a broad range of hosts is accidentally inactivated, the virus becomes locked within a single host, and the ratchet kicks off, such that fixation of further gene deletions is driven by selection for replication speed. However, it is also conceivable that subsets of the accessory gene are rendered dispensable by biological factors other than the host range, such as for example, a generally slow reproduction rate.
An alternative or additional interpretation of the massive gene loss as direct adaptation cannot be dismissed, namely, that inactivation of certain genes is actually advantageous for a virus under given conditions. In evolutionary experiments with VACV in cell culture, inactivation of either of the OPG153 (A26L), OPG091 (G6R) and OPG141 (A14.5L) genes, which are all mature virion components, led to a substantial increase in the fitness of a mutant defective in the function of OPG134 (A8R), a transcription factor (67). Inactivation of any of these genes in the wild type VACV did not enhance virus fitness. Here, we observed that A26L was frequently lost, jointly with a variety of other genes, during the evolution of ORPV, whereas G6R and A14.5L were fully conserved. These findings emphasize the complex, non-obvious functional relationships among ORPV genes that substantially depend on the specific biological context.
The largely uniform estimated rate of gene loss along with major deviations in some lineages of ORPV (Fig. 4) might be best compatible with a combination of neutral and adaptive factors underlying the observed patterns.
Most of the genes that have been lost in the ORPV were initially disrupted by frameshifts, and these mechanistic causes of frequent frameshift mutations occurring during ORPV replication might exist. For example, in the frequently disrupted OPG153, frameshifts occurred on a run of 7 T on multiple occasions (67). Although detailed analysis of the frameshifts is beyond the scope of this work, given that the ORPV genomes are AT-rich, the presence of runs of T in the coding sequences of many genes is likely to substantially increase the frequency of frameshift mutations that become the first step en route to gene loss.
One of the principal goals at the inception of this work was to infer functional interactions among ORPV genes through patterns of concurrent and exclusive loss. The analysis of these patterns did lead to predictions that might merit experimental follow-up, but these are few and relatively weak. There seem to be multiple, complementary reasons behind the limited success of this effort. First, the interactions among the ORPV genes appear to be highly complex, because of which many patterns are not readily interpretable. The high density and clustering of the network of concurrent gene losses in ORPV (Fig. 8) implies multiple functionalities of most of the accessory genes and the existence of multiple, overlapping modules affecting the even more complex host defense pathways. Second, many of the connections in that network are likely to reflect generic rather than specific functional links whereby, upon changes in the host range or virus-host interactions in general, many genes involved in the suppression of diverse host defense pathways become dispensable simultaneously, obscuring specific functional interactions. Third, although there are many ORPV genomes now available, the data is not as rich as it might seem because the major groups, such as MPXV, VARV, and VACV, are clusters of numerous, closely related genomes.
Although the apparent multifunctionality of most of the accessory proteins of the ORPV (1) complicates analysis of the gene loss patterns among sets of genes involved in the inhibition of the same defense pathway, a clear trend is noticeable. The genes affecting the same pathway appear to form echeloned anti-immune systems such that, depending on the host range and other biological factors, some but not all components can become redundant and hence dispensable. A straightforward example are the three genes eliciting hypoxic response (OPG020, 031, 165): one or two of these genes were lost in multiple ORPV, but at least one gene was always retained. Further accumulation of diverse ORPV genome sequences can be expected to unravel the full potential of comparative genomic approaches for deciphering the networks of virus-host interactions.
MATERIALS AND METHODS
Gene complements of chordopoxviruses, ORPV and centapoxviruses, construction of clusters of orthologous ORPV genes and reannotation of the accessory gene functions.
Complete chordopoxvirus genomes, available as of September 2020, were downloaded from GenBank (88). A non-redundant set (‘100 chordopoxvirus genomes’) was constructed by comparing all genomes using MEGABLAST (89), clustering genomes using BLASTCLUST (90), with the coverage threshold of 98% and identity threshold of 98%, and selecting one representative per cluster. Proteins that are encoded in the genomes but are not annotated in GenBank were predicted using PRODIGAL (91). ORPV and centapoxvirus genomes from the non-redundant set were used to produce clusters of OPG as follows. Predicted proteins of the 34 ORPV and 3 centapoxvirus genomes were pooled and clustered using BLASTCLUST with a 50% coverage threshold and a 50% identity threshold; out-paralogs, that is, paralogs resulting from duplications preceding the emergence of the ORPV (92), were separated manually on the basis of their conserved positions in the ORPV genome. Protein sequences within each cluster were aligned using MUSCLE (93). Position-specific scoring matrices (PSSM) derived from the alignments were used as queries in a PSI-BLAST search (94) against the set of 238 high-quality ORPV and centapoxvirus genomes; top hits were collected into new clusters and re-aligned. This procedure produced 216 clusters of OPG that represent the ORPV pangenome.
The 216 OPGs were mapped to the 100 chordopoxvirus genomes in the following manner: Position-specific scoring matrices (PSSM) derived from the OPG alignments were used as queries in a PSI-BLAST search (94). OPGs with short reference sequences (<100 aa) were searched against the six-frame translations of the genomic DNA; OPGs with longer reference sequences were searched against the GenBank-annotated proteins encoded in these genomes. Hits covering at least 75% of the query were registered as gene presence.
A different, more accurate procedure was applied for the analysis of the ORPV and centapoxvirus genomes. Alignment consensus was obtained for each OPG to determine the majority-supported protein boundaries and used further as the reference protein sequence for each of the 216 OPGs. PSSM derived from the OPG alignments were used as queries in a PSI-BLAST search (94) against the six-frame translations of the set of 238 ORPV and centapoxvirus genomes. The footprints produced by these searches were examined, and those passing the following criteria, modified from those employed previously for ORPV genome analysis (4), were considered to indicate the presence of a functional gene: (i) no more than 50 bp from the 5′-side of the footprint are missing; (ii) the footprint covers at least 80% of the OPG consensus length; (iii) the footprint contains no stop codons except within 50 bp from either end. The phyletic pattern was further examined and, where required, corrected case by case, to ensure agreement with the published data on the presence or absence of the respective gene. Orthopoxvirus pangenome genes were ordered and numbered according to their consensus location in the set of 235 ORPV genomes.
The initial functional annotation of each OPG was taken from previously published ORPV genome analyses and surveys (1, 4, 5). In order to update and enhance the annotation, all sequences of accessory proteins were searched against the NCBI non-redundant protein sequence database using PSI-BLAST (94) and against the PDB, PFAM and CDD databases using HHPred (95). The putative homologs identified in these searches were assessed for validity and functional relevance case by case. Protein secondary structure was predicted with Ali2D (96).
Phylogenetic analysis.
Protein sequences were aligned using MUSCLE (93); sites containing more than 50% of gap characters and with homogeneity below 0.1 (97) were removed. Phylogenetic trees were constructed using FastTree with the WAG evolutionary model and gamma-distributed site rates (98).
The phylogenetic tree for the selected representative set of chordopoxviruses was constructed using concatenated alignments of 35 proteins conserved across chordopoxviruses, with alignment filtering and Fasttree as described above. The following OPGs were included: 053, 063, 066, 071, 072, 084, 089, 091, 094, 095, 102, 104, 105, 106, 107, 109, 111, 113, 116, 117, 124, 125, 126, 127, 129, 133, 136, 137, 143, 145, 147, 148, 151, 155, 160. The tree was rooted with the salmon gill poxvirus as the outgroup.
The phylogenetic tree for the ORPV and centapoxvirus genomes was constructed as follows. The highly conserved regions of the genomes, spanning the segment between OPG048 and OPG160 were extracted from all 238 genomes and aligned using MAFFT (99). All sites containing gaps were removed from the alignment. A maximum likelihood phylogenetic tree was reconstructed using IQ-tree with the GTR+F+R5 evolutionary model, selected by built-in ModelFinder (100). The tree was rooted between ORPV and centapoxviruses.
Similarities between the presence-absence patterns in a pair of genomes were calculated as the number of shared orthologs divided by the geometric mean of the gene complements. Similarities were converted to distances using the d = -ln(s) transformation. UPGMA dendrogram was constructed from the matrix of pairwise genome distances.
Reconstruction of gene gain and loss.
The history of gene gains and losses in the set of 100 chordopoxvirus genomes and 238 genomes of ORPV and centapoxviruses was reconstructed, under the Dollo parsimony principle, using the DOLLOP program of the PHYLIP package (101). To elucidate the specific modes of gene loss, including frameshifts, in-frame nonsense mutations, and partial or complete gene deletions, a reference sequence was selected for each of the 216 OPGs that gave the highest-scoring TBLASTN (94) match in a search against the 238 genomes of ORPV and centapoxviruses, covering 100% of the protein query and containing no stop codons. Reference ORFs were used as queries in a BLASTN search against the 231 Old World ORPV genome, the best match in each genome was identified, and the nucleotide sequences were extracted and aligned together with the corresponding reference sequence using MAFFT (99). Alignments that involved genes marked as absent in a given genome were classified into the following categories: missing if no BLASTN hits with an e-value better than 10−5, at least 50% identity, and with at least 200 bp matching the query, are found; decaying if the alignment with the reference ORF was found to be missing more than 33% of the length or more than 50 bp relative to the reference ORF; frameshifted if the alignment satisfied the coverage criteria, but contained at least one frameshift; and nonsense if the gene sequence was not frameshifted, but contained in-frame stop codons.
Clade-specific gene loss rate was calculated by dividing the number of independent losses within a clade (including the branch at the base of the clade) by the total tree branch length of the clade (measured in substitutions per site). Confidence intervals were obtained by calculating the numbers of losses within 1000 bootstrap samples of 216 genes and discarding top and bottom 5% of the results.
Data availability.
All the genome sequences used for this analysis were from GenBank. The alignments of 35 conserved chordopoxvirus proteins used for constructing the phylogenetic tree shown in Fig. 2 and the alignment of the central regions of the ORPV genomes used for the tree shown in Fig. S2 are available at ftp://ftp.ncbi.nih.gov/pub/yutinn/Poxvir_2021/.
ACKNOWLEDGMENTS
Research was supported by the Intramural Research Program of the National Library of Medicine, NIH (N.Y., Y.I.W., E.V.K.) and by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH (T.G.S., B.M.).
ADDENDUM IN PROOF
After this article was accepted for publication, it came to our attention that the homology of OPG177 (VACV A47L) with gasdermins has been independently identified by Mirzakhanyan and Gershon (102).
Footnotes
This article is a direct contribution from Bernard Moss, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Stefan Rothenburg, University of California, Davis, School of Medicine, and Yan Xiang, University of Texas Health Science Center at San Antonio.
Citation Senkevich TG, Yutin N, Wolf YI, Koonin EV, Moss B. 2021. Ancient gene capture and recent gene loss shape the evolution of orthopoxvirus-host interaction genes. mBio 12:e01495-21. https://doi.org/10.1128/mBio.01495-21.
Contributor Information
Bernard Moss, Email: bmoss@nih.gov.
Blossom Damania, University of North Carolina, Chapel Hill.
REFERENCES
- 1.Moss D, Smith GL. Poxviridae: The Viruses and Their Replication. In: Fields Virology. Vol 2, 7 ed: LWW; 2021. [Google Scholar]
- 2.Fenner F, Wittek R, Dumbell KR. 1988. The Otrhopxviruses. Academic Press, Boca Raton, FL. [Google Scholar]
- 3.Meyer H, Ehmann R, Smith GL. 2020. Smallpox in the Post-Eradication Era. Viruses 12:138. doi: 10.3390/v12020138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrickson RC, Wang C, Hatcher EL, Lefkowitz EJ. 2010. Orthopoxvirus genome evolution: the role of gene loss. Viruses 2:1933–1967. doi: 10.3390/v2091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu SL, Upton C. 2019. Bioinformatics for Analysis of Poxvirus Genomes. Methods Mol Biol 2023:29–62. doi: 10.1007/978-1-4939-9593-6_2. [DOI] [PubMed] [Google Scholar]
- 6.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini FM, Kuhn JH. 2020. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol Mol Biol Rev 84. doi: 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. 2003. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol 77:7590–7600. doi: 10.1128/jvi.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bratke KA, McLysaght A, Rothenburg S. 2013. A survey of host range genes in poxvirus genomes. Infect Genet Evol 14:406–425. doi: 10.1016/j.meegid.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith GL, Talbot-Cooper C, Lu Y. 2018. How Does Vaccinia Virus Interfere With Interferon? Adv Virus Res 100:355–378. doi: 10.1016/bs.aivir.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Smith GL, Benfield CTO, Maluquer de Motes C, Mazzon M, Ember SWJ, Ferguson BJ, Sumner RP. 2013. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol 94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 11.Suraweera CD, Hinds MG, Kvansakul M. 2020. Poxviral Strategies to Overcome Host Cell Apoptosis. Pathogens 10:6. doi: 10.3390/pathogens10010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols DB, De Martini W, Cottrell J. 2017. Poxviruses Utilize Multiple Strategies to Inhibit Apoptosis. Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothenburg S, Brennan G. 2020. Species-Specific Host-Virus Interactions: Implications for Viral Host Range and Virulence. Trends Microbiol 28:46–56. doi: 10.1016/j.tim.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odom MR, Hendrickson RC, Lefkowitz EJ. 2009. Poxvirus protein evolution: family wide assessment of possible horizontal gene transfer events. Virus Res 144:233–249. doi: 10.1016/j.virusres.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes AL. 2002. Origin and evolution of viral interleukin-10 and other DNA virus genes with vertebrate homologues. J Mol Evol 54:90–101. doi: 10.1007/s00239-001-0021-1. [DOI] [PubMed] [Google Scholar]
- 16.Prag S, Adams JC. 2003. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics 4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi X, et al. 2019. Kelch-like proteins: Physiological functions and relationships with diseases. Pharmacol Res 148:104404. doi: 10.1016/j.phrs.2019.104404. [DOI] [PubMed] [Google Scholar]
- 18.Herbert MH, Squire CJ, Mercer AA. 2015. Poxviral ankyrin proteins. Viruses 7:709–738. doi: 10.3390/v7020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez JM, Esteban M. 2010. A poxvirus Bcl-2-like gene family involved in regulation of host immune response: sequence similarity and evolutionary history. Virol J 7:59. doi: 10.1186/1743-422X-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson CA, Epperson ML, Singh S, Elliott JI, Fremont DH. 2015. Structural Conservation and Functional Diversity of the Poxvirus Immune Evasion (PIE) Domain Superfamily. Viruses 7:4878–4898. doi: 10.3390/v7092848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barry M, et al. 2010. Poxvirus exploitation of the ubiquitin-proteasome system. Viruses 2:2356–2380. doi: 10.3390/v2102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Villa NY, McFadden G. 2009. Interplay between poxviruses and the cellular ubiquitin/ubiquitin-like pathways. FEBS Lett 583:607–614. doi: 10.1016/j.febslet.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 23.Shchelkunov SN. 2010. Interaction of orthopoxviruses with the cellular ubiquitin-ligase system. Virus Genes 41:309–318. doi: 10.1007/s11262-010-0519-y. [DOI] [PubMed] [Google Scholar]
- 24.Tatusov RL, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 26.Galperin MY, Kristensen DM, Makarova KS, Wolf YI, Koonin EV. 2019. Microbial genome analysis: the COG approach. Brief Bioinform 20:1063–1070. doi: 10.1093/bib/bbx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goebel SJ, Johnson GP, Perkus ME, Davis SW, Winslow JP, Paoletti E. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247–266. 517–263. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 28.Gjessing MC, et al. 2015. Salmon Gill Poxvirus, the Deepest Representative of the Chordopoxvirinae. J Virol 89:9348–9367. doi: 10.1128/JVI.01174-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vernikos G, Medini D, Riley DR, Tettelin H. 2015. Ten years of pan-genome analyses. Curr Opin Microbiol 23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 30.Koonin EV, Makarova KS, Wolf YI. 2021. Evolution of Microbial Genomics: Conceptual Shifts over a Quarter Century. Trends Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Iyer LM, Aravind L. 2011. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res 39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bierle CJ, Schleiss MR, Geballe AP. 2012. Antagonism of the protein kinase R pathway by the guinea pig cytomegalovirus US22-family gene gp145. Virology 433:157–166. doi: 10.1016/j.virol.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valchanova RS, Picard-Maureau M, Budt M, Brune W. 2006. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J Virol 80:10181–10190. doi: 10.1128/JVI.00908-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hakki M, Geballe AP. 2005. Double-stranded RNA binding by human cytomegalovirus pTRS1. J Virol 79:7311–7318. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent HA, Ziehr B, Moorman NJ. 2017. Mechanism of Protein Kinase R Inhibition by Human Cytomegalovirus pTRS1. J Virol 91. doi: 10.1128/JVI.01574-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao J, Varga J, Deschambault Y. 2020. Poxvirus encoded eIF2alpha homolog, K3 family proteins, is a key determinant of poxvirus host species specificity. Virology 541:101–112. doi: 10.1016/j.virol.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Park C, Peng C, Brennan G, Rothenburg S. 2019. Species-specific inhibition of antiviral protein kinase R by capripoxviruses and vaccinia virus. Ann N Y Acad Sci 1438:18–29. doi: 10.1111/nyas.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dar AC, Sicheri F. 2002. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell 10:295–305. doi: 10.1016/s1097-2765(02)00590-7. [DOI] [PubMed] [Google Scholar]
- 39.Langland JO, Jacobs BL. 2004. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology 324:419–429. doi: 10.1016/j.virol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 40.Vipat S, Brennan G, Park C, Haller SL, Rothenburg S. 2020. Rapid, Seamless Generation of Recombinant Poxviruses using Host Range and Visual Selection. J Vis Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longbotham JE, et al. 2015. Structure and Mechanism of a Viral Collagen Prolyl Hydroxylase. Biochemistry 54:6093–6105. doi: 10.1021/acs.biochem.5b00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazzon M, et al. 2013. A mechanism for induction of a hypoxic response by vaccinia virus. Proc Natl Acad Sci U S A 110:12444–12449. doi: 10.1073/pnas.1302140110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang L, Lu C, Zheng G, Burgering BM. 2020. Emerging insights on the role of gasdermins in infection and inflammatory diseases. Clin Transl Immunology 9:e1186. doi: 10.1002/cti2.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flores-Romero H, Ros U, Garcia-Saez AJ. 2020. Pore formation in regulated cell death. EMBO J 39:e105753. doi: 10.15252/embj.2020105753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broz P, Pelegrin P, Shao F. 2020. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol 20:143–157. doi: 10.1038/s41577-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 46.Gigante CM, et al. 2019. Genome of Alaskapox Virus, A Novel Orthopoxvirus Isolated from Alaska. Viruses 11. doi: 10.3390/v11080708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruber C, Giombini E, Selleri M, Tausch S, Andrusch A, Tyshaieva A, Cardeti G, Lorenzetti R, De Marco L, Carletti F, Nitsche A, Capobianchi M, Ippolito G, Autorino G, Castilletti C. 2018. Whole Genome Characterization of Orthopoxvirus (OPV) Abatino, a Zoonotic Virus Representing a Putative Novel Clade of Old World Orthopoxviruses. Viruses 10:546. doi: 10.3390/v10100546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mauldin M, Antwerpen M, Emerson G, Li Y, Zoeller G, Carroll D, Meyer H. 2017. Cowpox virus: What's in a Name? Viruses 9:101. doi: 10.3390/v9050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Csuros M, Rogozin IB, Koonin EV. 2011. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput Biol 7:e1002150. doi: 10.1371/journal.pcbi.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alekseyenko AV, Lee CJ, Suchard MA. 2008. Wagner and Dollo: a stochastic duet by composing two parsimonious solos. Syst Biol 57:772–784. doi: 10.1080/10635150802434394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLysaght A, Baldi PF, Gaut BS. 2003. Extensive gene gain associated with adaptive evolution of poxviruses. Proc Natl Acad Sci U S A 100:15655–15660. doi: 10.1073/pnas.2136653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veyer DL, Carrara G, Maluquer de Motes C, Smith GL. 2017. Vaccinia virus evasion of regulated cell death. Immunol Lett 186:68–80. doi: 10.1016/j.imlet.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 53.Fixsen SM, et al. 2020. Poxviruses capture host genes by LINE-1 retrotransposition. https://wwwbiorxivorg/content/101101/20201026356402v1. [DOI] [PMC free article] [PubMed]
- 54.Wolf YI, Rogozin IB, Grishin NV, Koonin EV. 2002. Genome trees and the tree of life. Trends Genet 18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- 55.Arndt WD, et al. 2016. Monkeypox virus induces the synthesis of less dsRNA than vaccinia virus, and is more resistant to the anti-poxvirus drug, IBT, than vaccinia virus. Virology 497:125–135. doi: 10.1016/j.virol.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frey TR, et al. 2017. Ectromelia virus accumulates less double-stranded RNA compared to vaccinia virus in BS-C-1 cells. Virology 509:98–111. doi: 10.1016/j.virol.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park C, et al. 2021. Orthopoxvirus K3 orthologs show virus- and host-specific inhibition of the antiviral protein kinase PKR. PLoS Pathog 17:e1009183. doi: 10.1371/journal.ppat.1009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khan A, et al. 2015. BEND3 represses rDNA transcription by stabilizing a NoRC component via USP21 deubiquitinase. Proc Natl Acad Sci U S A 112:8338–8343. doi: 10.1073/pnas.1424705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin KH, Franke CA, Hruby DE. 1999. Novel acylation of poxvirus A-type inclusion proteins. Virus Res 60:147–157. doi: 10.1016/s0168-1702(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 60.Schweneker M, et al. 2012. The vaccinia virus O1 protein is required for sustained activation of extracellular signal-regulated kinase 1/2 and promotes viral virulence. J Virol 86:2323–2336. doi: 10.1128/JVI.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galperin MY, Koonin EV. 2000. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol 18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- 62.Aravind L, Watanabe H, Lipman DJ, Koonin EV. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci U S A 97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marcotte EM. 2000. Computational genetics: finding protein function by nonhomology methods. Curr Opin Struct Biol 10:359–365. doi: 10.1016/s0959-440x(00)00097-x. [DOI] [PubMed] [Google Scholar]
- 64.Barabasi AL, Oltvai ZN. 2004. Network biology: understanding the cell's functional organization. Nat Rev Genet 5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 65.Chiu WL, Lin CL, Yang MH, Tzou DL, Chang W. 2007. Vaccinia virus 4c (A26L) protein on intracellular mature virus binds to the extracellular cellular matrix laminin. J Virol 81:2149–2157. doi: 10.1128/JVI.02302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howard AR, Weisberg AS, Moss B. 2010. Congregation of orthopoxvirus virions in cytoplasmic A-type inclusions is mediated by interactions of a bridging protein (A26p) with a matrix protein (ATIp) and a virion membrane-associated protein (A27p). J Virol 84:7592–7602. doi: 10.1128/JVI.00704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Senkevich TG, Zhivkoplias EK, Weisberg AS, Moss B. 2020. Inactivation of Genes by Frameshift Mutations Provides Rapid Adaptation of an Attenuated Vaccinia Virus. J Virol 94. doi: 10.1128/JVI.01053-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luteijn RD, Hoelen H, Kruse E, van Leeuwen WF, Grootens J, Horst D, Koorengevel M, Drijfhout JW, Kremmer E, Früh K, Neefjes JJ, Killian A, Lebbink RJ, Ressing ME, Wiertz EJHJ. 2014. Cowpox virus protein CPXV012 eludes CTLs by blocking ATP binding to TAP. Ji 193:1578–1589. doi: 10.4049/jimmunol.1400964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eaglesham JB, Pan Y, Kupper TS, Kranzusch PJ. 2019. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature 566:259–263. doi: 10.1038/s41586-019-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eaglesham JB, McCarty KL, Kranzusch PJ. 2020. Structures of diverse poxin cGAMP nucleases reveal a widespread role for cGAS-STING evasion in host-pathogen conflict. Elife 9. doi: 10.7554/eLife.59753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bustos O, et al. 2009. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene 447:1–11. doi: 10.1016/j.gene.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brandt T, et al. 2005. The N-terminal domain of the vaccinia virus E3L-protein is required for neurovirulence, but not induction of a protective immune response. Virology 333:263–270. doi: 10.1016/j.virol.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 73.Brandt TA, Jacobs BL. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J Virol 75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim YG, et al. 2003. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci U S A 100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedersen K, et al. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J Virol 74:3525–3536. doi: 10.1128/jvi.74.8.3525-3536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmidt FI, et al. 2013. Vaccinia virus entry is followed by core activation and proteasome-mediated release of the immunomodulatory effector VH1 from lateral bodies. Cell Rep 4:464–476. doi: 10.1016/j.celrep.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 77.Meade N, King M, Munger J, Walsh D. 2019. mTOR Dysregulation by Vaccinia Virus F17 Controls Multiple Processes with Varying Roles in Infection. J Virol 93. doi: 10.1128/JVI.00784-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meade N, et al. 2018. Poxviruses Evade Cytosolic Sensing through Disruption of an mTORC1-mTORC2 Regulatory Circuit. Cell 174:1143–1157 e1117. doi: 10.1016/j.cell.2018.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Derrien M, Punjabi A, Khanna M, Grubisha O, Traktman P. 1999. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J Virol 73:7287–7296. doi: 10.1128/JVI.73.9.7287-7296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mann BA, et al. 2008. Vaccinia virus blocks Stat1-dependent and Stat1-independent gene expression induced by type I and type II interferons. J Interferon Cytokine Res 28:367–380. doi: 10.1089/jir.2007.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Willer DO, Yao XD, Mann MJ, Evans DH. 2000. In vitro concatemer formation catalyzed by vaccinia virus DNA polymerase. Virology 278:562–569. doi: 10.1006/viro.2000.0686. [DOI] [PubMed] [Google Scholar]
- 82.Senkevich TG, et al. 2015. Mapping vaccinia virus DNA replication origins at nucleotide level by deep sequencing. Proc Natl Acad Sci U S A 112:10908–10913. doi: 10.1073/pnas.1514809112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao J, Gigante C, Khmaladze E, Liu P, Tang S, Wilkins K, Zhao K, Davidson W, Nakazawa Y, Maghlakelidze G, Geleishvili M, Kokhreidze M, Carroll D, Emerson G, Li Y. 2018. Genome Sequences of Akhmeta Virus, an Early Divergent Old World Orthopoxvirus. Viruses 10:252. doi: 10.3390/v10050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franke A, Pfaff F, Jenckel M, Hoffmann B, Höper D, Antwerpen M, Meyer H, Beer M, Hoffmann D. 2017. Classification of Cowpox Viruses into Several Distinct Clades and Identification of a Novel Lineage. Viruses 9:142. doi: 10.3390/v9060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qin L, Favis N, Famulski J, Evans DH. 2015. Evolution of and evolutionary relationships between extant vaccinia virus strains. J Virol 89:1809–1824. doi: 10.1128/JVI.02797-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elde NC, et al. 2012. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brennan G, Kitzman JO, Rothenburg S, Shendure J, Geballe AP. 2014. Adaptive gene amplification as an intermediate step in the expansion of virus host range. PLoS Pathog 10:e1004002. doi: 10.1371/journal.ppat.1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benson DA, et al. 2018. GenBank. Nucleic Acids Res 46:D41–D47. doi: 10.1093/nar/gkx1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 90.Kuznetsov A, Bollin CJ. 2021. NCBI Genome Workbench: Desktop Software for Comparative Genomics, Visualization, and GenBank Data Submission. Methods Mol Biol 2231:261–295. doi: 10.1007/978-1-0716-1036-7_16. [DOI] [PubMed] [Google Scholar]
- 91.Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koonin EV. 2005. Orthologs, Paralogs and Evolutionary Genomics. Annu Rev Genet in press. [DOI] [PubMed] [Google Scholar]
- 93.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 96.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 97.Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. 2008. The deep archaeal roots of eukaryotes. Mol Biol Evol 25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Felsenstein J. 1996. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol 266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 102.Mirzakhanyan Y, Gershon PD. 2020. Structure-based deep mining reveals first-time annotations for 46 percent of the dark annotation space of the 9,671-member superproteome of the nucleocytoplasmic large DNA viruses. J Virol 94:e00854-20. doi: 10.1128/JVI.00854-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional annotation of the ORPV genes. (A) OPG number. (B) Accession number for OPGs of CPXV-Gri; when the corresponding gene is absent in CPXV-Gri, the accession number of CPXV-Ger91-3, CPXV-Bri or VACV-Cop is indicated. (C) Protein lengths (amino acid number) in CPXV-Gri; when the corresponding gene is absent in CPXV-Gri, protein length is indicated for CPXV Ger91-3, CPXV-Bri or VACV-Cop. (D) State of the gene in CPXV Gri genome. (E) Domain architecture and/or function. (F) Gene capture in the evolution of chordopoxviruses. Node numbers correspond to the phylogenetic tree of chordopoxviruses in Fig. 2. A, ancestral gene present in the common ancestor of all chordopoxviruses; Node 2, prior to divergence of Crocodylidpoxviruses from the rest of the Chordopoxviruses (41 genes); Node 3, prior to divergence of Avipoxviruses and mammalian poxviruses (43 genes); Node 4, prior to divergence of Molluscipox (5 genes); Node 5 prior to divergence of Parapox (7 genes); Node 6 prior to divergence of Squirrel pox UK (3 genes); Node 7 Prior to divergence of Pteropox (node 1 genes); Node 8 prior to divergence of Squirrel pox Berlin (5 genes); Node 9 prior to divergence of Cetacean (1 gene); Node 10 Prior to divergence of leporipoxviruses (30 genes); Node 11, prior to divergence of Centapoxviruses (22 genes); Node 12, prior to divergence of New World ORPV (6 genes); Node 15 prior to divergence of Ectromelia-Abatino-CPXV 5 (2 genes); Node 16 prior to divergence of major cowpox branch CPXV 4 within ORPV (1 gene); (G, I, K) State of the gene in VACV Copenhagen, VACV WR, and CPXV Brighton, respectively. X, ORF sequence can’t be identified; D, large deletion representing 50+ bp or >1/3 reference length; F, frameshift(s); N, in-frame nonsense mutation(s). (H, J, L) Gene names in VACV Copenhagen, VACV WR and CPXV Brighton, respectively. Abbreviations: EV, enveloped virion; IV, immature virion; MV, mature virion; WV, wrapped virion; TM, transmembrane; Cop, Copenhagen VACV nomenclature; EFC, entry fusion complex; VMAP, virion membrane assembly protein. Download Table S1, XLSX file, 0.03 MB (35.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Orthologs of the OPGs in other chordopoxviruses. Presence of an intact OPG is denoted by 1 and absence by 0. Download Table S2, XLSX file, 0.3 MB (272.7KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representation of Orthopoxvirus genes (OPG) in ORPV genomes. (A) Sheet 1: binary pattern of OPG presence-absence in ORPV genomes. Gene presence is shown by 1 and gene absence by 0. (B) Sheet 2: breakdown of the OPG inactivation events. Abbreviations: D, decaying gene (within gene deletion); F, frameshift; N, in-frame stop codon; X, absent gene (large deletion). Download Table S3, XLSX file, 0.3 MB (349.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Multiple sequence alignments of previously uncharacterized ORPV proteins. (A) OPG159 (A31), a US22 family member; (B) OPG165 (A37), an inactive prolyl hydroxylase homolog, putative elicitor of hypoxic response. The residues that are predicted to bind the Fe2+ ion are marked with red rectangles. (C) OPG181 (A47), gasdermin homolog, a predicted apoptosis inhibitor. Secondary structure elements extracted from crystal structures or predicted with Ali2D are indicated (red, α-helix; blue, β-strand). Download FIG S1, PPT file, 0.2 MB (161.4KB, ppt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Complete phylogenetic tree of ORPV. Phylogenetic tree was constructed from a multiple alignment of the central portions of 235 ORPV genomes combined with genomes of three centapoxviruses. Node numbers are shown. Download FIG S2, PDF file, 0.9 MB (951.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Evolutionary divergence within the major clades of ORPV. The mean distances between the genomes in the respective clades are given as percentage nucleotide difference. The main diagonal shows the within-clade distances between the strains across the root of the clade. Download Table S4, DOCX file, 0.01 MB (14.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Phylogenetic trees of the four major families of paralogous genes in ORPV. (A) PIE-domain family. (B) Bcl-2 family. (C) BTB-Kelch family (Only Kelch domain was used for the tree construction). (D) Ankyrin-PRANC family FIG S3, PPT file, 0.1 MB (123.4KB, ppt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reconstruction of gene gains and losses in the evolution of chordopoxviruses and ORPV. (A) Sheet 1: gain and loss of OPG in the evolution of chordopoxviruses. Gene gain (1) and loss (-1) events shown for individual virus strains (tips of the tree) and internal tree nodes denoted by numbers as in Fig. 2. (B) Sheet 2: gain and loss of OPG in the evolution of ORPV. Gene gain (1) and loss (-1) events shown for individual virus strains (tips of the tree) and internal tree nodes denoted by numbers as in Fig. S2. Download Table S5, XLSX file, 0.4 MB (440.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Concurrent and exclusive gene losses in ORPV. Sheet 1: number of concurrent losses for all pairs of OPG. The intensity of the red highlighting is proportional to the frequency of losses. Sheet 2: node degrees (the number of nodes connected with the given node) of the OPGs in the complete network of concurrent losses. Sheet 3: number of concurrent losses for all pairs of OPG normalized by the geometric mean of the number of individual losses for the respective OPG. Sheet 4: number of genomes where both genes are absent for all pairs of 102 OPGs with at least one absence in the Old World ORPV. Sheet 5: number of genomes where both genes are absent for all pairs of 102 OPGs with at least one absence in the Old World ORPV, normalized by the geometric mean of the number of Old World ORPV genomes where this gene is absent for the respective OPGs Table S6, XLSX file, 0.4 MB (413.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Parameters of the network of concurrent loss of ORPV genes. (A) Node degree distribution for the complete network of the concurrent gene losses. The distribution is shown for the complete network of concurrent losses where edges connect genes that were concomitantly lost at least once. P.d.f. probability density function. (B) Clustering coefficient distribution for the complete network of the concurrent gene losses. p.d.f. probability density function FIG S4, PPT file, 0.4 MB (385.4KB, ppt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
All the genome sequences used for this analysis were from GenBank. The alignments of 35 conserved chordopoxvirus proteins used for constructing the phylogenetic tree shown in Fig. 2 and the alignment of the central regions of the ORPV genomes used for the tree shown in Fig. S2 are available at ftp://ftp.ncbi.nih.gov/pub/yutinn/Poxvir_2021/.