Abstract
The Caenorhabditis elegans SKN-1 protein binds DNA through a basic region like those of bZIP proteins and through a flexible amino-terminal arm segment similar to those with which numerous helix-turn-helix proteins bind to bases in the minor groove. A recent X-ray crystallographic structure suggests that the SKN-1 amino-terminal arm provides only nonspecific DNA binding. In this study, however, we demonstrate that this segment mediates recognition of an AT-rich element that is part of the preferred SKN-1 binding site and thereby significantly increases the sequence specificity with which SKN-1 binds DNA. Mutagenesis experiments show that multiple amino acid residues within the arm are involved in binding. These residues provide binding affinity through distinct but partially redundant interactions and enhance specificity by discriminating against alternate sites. The AT-rich element minor groove is important for binding of the arm, which appears to affect DNA conformation in this region. This conformational effect does not seem to involve DNA bending, however, because the arm does not appear to affect a modest DNA bend that is induced by SKN-1. The data illustrate an example of how a small, flexible protein segment can make an important contribution to DNA binding specificity through multiple interactions and mechanisms.
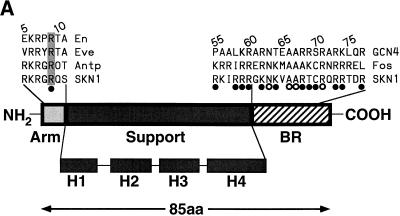
Maternally expressed SKN-1 protein is required for cell fate specification during the earliest embryonic stages in Caenorhabditis elegans (3, 4). SKN-1 is a transcription factor that binds DNA as a monomer (2). Its unusual DNA binding domain (the Skn domain) (Fig. 1A) includes a carboxyl-terminal basic region (BR) like those of dimeric basic-leucine zipper (bZIP) proteins and a flexible amino-terminal arm like those with which homeodomains and other helix-turn-helix proteins bind in the minor groove (2). The consensus-preferred SKN-1 binding site, as determined by in vitro selection, consists of an AT-rich element (A/TA/TT) immediately 5′ of the sequence G/ATCAT (2), which corresponds to the GTCAT half-site recognized by many bZIP protein BRs (2, 32). Although individual BR peptides do not bind DNA stably as monomers, the purified Skn domain binds to a preferred site with a dissociation constant (Kd) in the range of 10−9 M (8). The Skn domain BR is stabilized on the G/ATCAT sequence primarily by the adjacent α-helical support segment (Fig. 1A and B), which is unrelated to either a ZIP segment or a helix-turn-helix motif and forms a globule that leaves the BR exposed in the major groove (8, 26, 32). The Skn domain amino-terminal arm, which is almost identical in sequence to that of the antennapedia homeodomain (Fig. 1A), is also important for binding affinity and has been proposed to mediate specific recognition of this AT-rich element (2, 8).
FIG. 1.
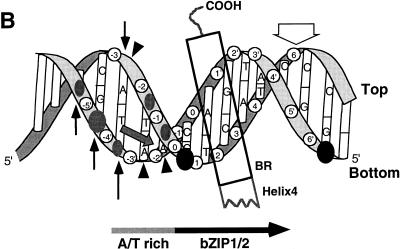
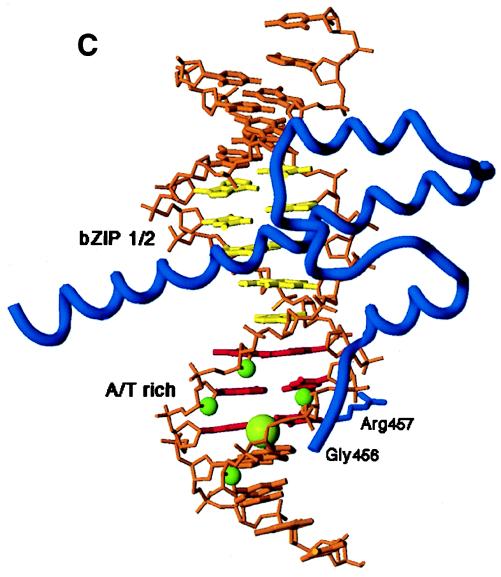
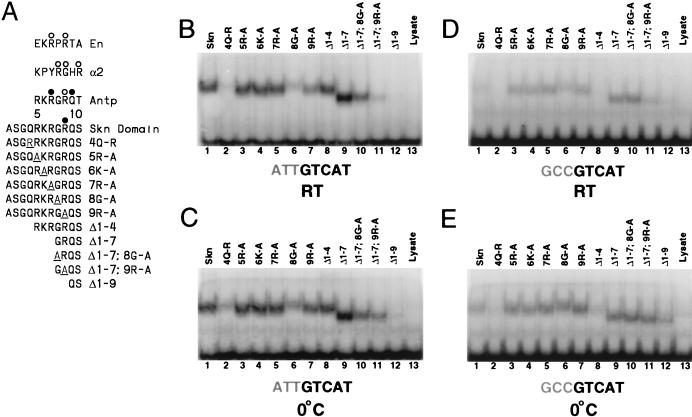
Skn domain and its interactions with DNA. (A) Diagram of the Skn domain, which consists of the carboxyl-terminal 85 amino acids (aa) of SKN-1 (residues 449 to 533) and is numbered as described in reference 2. The sequences of the amino-terminal arm and BR are compared with the corresponding sequences from the indicated homeodomain and bZIP proteins, respectively. The four support segment α-helices identified by nuclear magnetic resonance and X-ray crystallography (8, 26, 32) are labeled H1 to H4. The arm and BR residues that are designated by open and filled circles contact DNA bases and the backbone, respectively, in the Skn domain structure determined by X-ray crystallography (32). Arg 9 in the amino-terminal arm (SKN-1 residue 457) is shaded because it is highly conserved in homeodomains, where it is designated position 5 (34). Sequences are from references 8 and 34. (B) Results of biochemical analyses of Skn domain DNA binding. The BR α-helix extends into the major groove directly from support segment helix 4. Black circles indicate positions of maximum hydroxyl radical protection by both the Skn domain and Δ1–9. Shaded ovals indicate where protection by the Skn domain is greater than by Δ1–9, with the size of the oval indicating the extent of difference (8). Black vertical arrows indicate where prior hydroxyl radical cleavage enhances Skn domain binding (8). Black arrowheads indicate positions at which adenine methylation binding interference is greater for the Skn domain (Fig. 4). The small shaded arrow in the minor groove indicates the approximate direction that corresponds to the amino terminus of a homeodomain arm, relative to the location of the major groove recognition helix. The wide downward-pointing arrow indicates the approximate position at which the major groove corresponds to the direction in which the Skn domain and Δ1–9 bend DNA. (C) Structure of the Skn domain bound to DNA (32). The Skn domain backbone is shown in blue, along with the Arg 9 (Arg 457) side chain. Other side chains are not shown because they are not positioned close to the AT-rich region (32). The DNA backbone is orange, the GTCAT base pairs are yellow, and the AT-rich region bases are red. Green spheres indicate the sugar residues protected from hydroxyl radical attack specifically by the amino-terminal arm (Fig. 1B). Residues that are located amino terminal to Gly 8 (Gly 456) are disordered and not visible in the structure (32).
However, the recent crystal structure of an Skn domain-DNA complex does not support the idea that the amino-terminal arm contributes to binding sequence specificity (32). This structure (Fig. 1C) was determined with a preferred SKN-1 DNA binding site (2) and an Skn domain derivative in which a tag of six histidine (His) residues was attached immediately amino terminal to the arm at position 2 (Fig. 1A) (32). The crystal structure is generally consistent with mutagenesis, footprinting, and nuclear magnetic resonance spectroscopic data with respect to how the support segment stabilizes the BR on DNA (32). However, this structure does not suggest a mechanism for how the amino-terminal arm might mediate any base sequence preferences. It indicates that the amino terminus of the arm points away from the support segment (Fig. 1C) (32) in an orientation opposite to that of most helix-turn-helix protein arm segments (Fig. 1B), which usually lie within the minor groove and contact bases and the adjacent DNA backbone (1, 10, 14, 17–19, 23, 43, 44). In this structure, the Skn domain arm does not interact with the DNA minor groove and appears to make only a single direct contact with the backbone (Fig. 1C) (32). Based upon these findings, it has been concluded that the similarity of the SKN-1 amino-terminal arm segment to homeodomain arms (Fig. 1A) derives simply from a clustering of basic residues, that this segment contributes only nonspecific DNA binding affinity, and that the SKN-1 binding site consists only of G/ATCAT (32).
For multiple reasons, it is important to elucidate how the amino-terminal arm contributes to SKN-1 DNA binding. It is of interest to determine whether the arm increases the number of bases specified by SKN-1, because we have only a limited understanding of which target genes SKN-1 might regulate in vivo (49). In addition, homeodomain arm segments can be bound by protein cofactors (41, 43, 48) and are critical for functional specificity that is not directly attributable to DNA binding (11, 22, 24, 47), suggesting that the Skn domain arm might be involved in similar interactions. Finally, it is of importance to identify general principles by which such small peptide segments can contribute to DNA binding affinity and specificity.
In this study, we demonstrate that the Skn domain amino-terminal arm increases binding specificity significantly by conferring a sequence preference for the AT-rich element. Multiple amino acid residues within the arm appear to interact with the DNA but are not individually essential for binding affinity, suggesting conformational flexibility. A conserved arginine (Arg 9) (Fig. 1A and C) appears to be the most important of the basic residues within the arm, but each of them contributes specificity by destabilizing binding to nonpreferred sites. A glycine that is interspersed among these basic residues (Gly 8) influences interaction of the entire arm with the DNA (Fig. 1A and C) and may contact DNA directly. Binding of the arm appears to depend upon the AT-rich region minor groove and to influence DNA conformation. This conformational effect does not seem to involve DNA bending, however, because binding of the arm does not substantially influence a modest DNA bend (<10°) that is mediated by the BR and support segment. These experiments demonstrate that the Skn domain amino-terminal arm, like those of helix-turn-helix proteins, is an important DNA specificity segment and that it establishes these sequence preferences through multiple interactions. The results of these experiments also suggest a model for how this small protein segment binds to the DNA.
MATERIALS AND METHODS
Protein expression and DNA binding assays.
The Skn domain protein and a mutant protein that lacks the arm (amino acids 1 to 9) of the Skn domain (Δ1–9) were expressed in Escherichia coli, purified, and quantitated by spectroscopic and amino acid analysis as described in reference 8. Expression and purification of the glutathione S-transferase–SKN-1 protein were described previously (2). Kds were determined at room temperature by electrophoretic mobility shift assay (EMSA) under conditions of protein excess as described previously (8), except that only siliconized tips and tubes were used. The Kd with which the Skn domain binds its preferred site had been measured as 1 (±0.5) × 10−9 M (8), but under these latter conditions this Kd was approximately 3 × 10−10 M.
Deletion mutations were produced by PCR and confirmed by DNA sequencing. Substitution mutations were constructed by the circular-mutagenesis change reaction (45). Expression and quantitation of in vitro-translated proteins were performed essentially as described in reference 8 with a combined transcription-translation kit (Promega). Each protein was quantitated multiple times by 35S-labeled translation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The variability among these translations generally fell within a range of 1.5-fold. EMSAs were performed as described previously (8), with poly(dI-dC) present at 0.0125 μg/μl in each sample. Each protein was tested as described in the legend to Fig. 2 with at least two different preparations of unlabeled protein to ensure reproducibility.
FIG. 2.
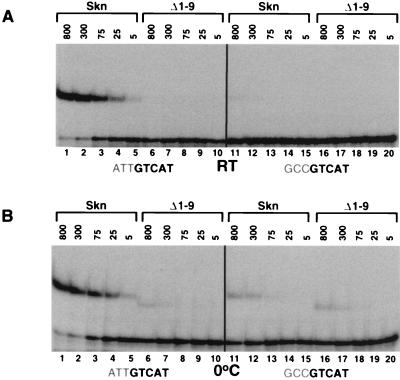
Involvement of the Skn domain amino-terminal arm in DNA binding affinity and specificity. (A) Binding of the Skn domain and the Δ1–9 Skn domain mutant (2) to DNA, assayed by EMSA at room temperature (RT). The ATTGTCAT preferred site assayed is the 22-bp oligonucleotide SK1 (2). The GCCGTCAT mutant site (−3,−2,−1 GC [2]) is identical to SK1 except for these three base pairs. Protein concentrations are indicated above the gel (in 10−12 molar units), and specific DNA is present at 5 × 10−12 M. (B) Binding of the Skn domain and the Δ1–9 mutant to the SK1 and −3,−2,−1 GC sites at 0°C, assayed as described for panel A.
The DNA probe and procedures used in the methylation interference assay have been described previously (2). To ensure that the observed effects were reproducible and derived from specific binding, each experiment was performed multiple times under conditions at which an EMSA (not shown) demonstrated that 10 to 50% of the input DNA was in the bound fraction and that only a single protein molecule was bound.
Circular-permutation assay.
Probes for the circular-permutation assay were constructed from a Bluescript plasmid containing an in vitro-selected SKN-1 binding sequence that includes the ATTGTCAT preferred binding site (2). Three different sets of PCR primers were used to generate probes A, B, and C, which were each 160 bp in length. In probe A, the SKN-1 binding site was 12 bp from the 5′ end; in probe B, it was 77 bp from the 5′ end; and in probe C, it was 10 bp from the 3′ end. The probes were purified and end labeled with 32P by polynucleotide kinase. The EMSA was performed on 6% polyacrylamide gels as described in reference 2, with the Skn domain protein, Δ1–9, and full-length SKN-1 present at concentrations of 1, 30, and 50 nM, respectively.
Ligation-mediated cyclization kinetics assay.
The cyclization kinetics assay was performed essentially as described by Kahn and Crothers (15). Ligation substrates were constructed with plasmids (31 and 37 pBluescript II KS 11T15F) (15, 27) that were gifts of David Fisher. Each contained a Max protein binding site located 31 or 37 bp from a sequence of six regularly spaced poly(dA) tracts, which induces an intrinsic bend. The Max binding sites were replaced by a single preferred SKN-1 binding site, which was placed at different positions by PCR. These plasmids were used to generate the cyclization probes (SK32 to SK41) listed in Table 1, as described previously (27).
TABLE 1.
Summary of cyclization kinetics analysesa
| Probe | Ratio of Jbound to Jfree
|
|
|---|---|---|
| Skn domain | Δ1–9 | |
| SK32 | 1 | 1 |
| SK34 | 0.6 | 0.7 |
| SK36 | 0.5 | 0.5 |
| SK38 | 1 | 1 |
| SK40 | 2.0 | 2.3 |
| SK41 | 1.5 | 1.8 |
The value J is derived from the relative ratio of circularization to bimolecular ligation and is proportional to the slopes of the plots in Fig. 8D and E (15). The difference in J that results from protein binding is obtained from the ratio of the respective slopes and is indicative of bending angle (12, 13, 15). The J ratios shown were determined by averaging results of multiple representative experiments such as those shown in Fig. 8B to E. In these experiments, the final value for ln(total DNA/linear DNA) ranged between 0.7 and 1.6, as shown in Fig. 8B and C. The number in each probe name corresponds to the distance in base pairs from the link between SKN-1 binding site positions −1 and −2 (Fig. 1B) to the center of the nearest dA tract.
To establish protein concentrations that were ideal for complete and specific binding to these cyclization probes, EMSAs were performed under the conditions that were used for ligation (see below). Nonspecific binding was assayed by performing side-by-side EMSAs with minicircle substrate identical to the substrate used in the above-described assays except that it lacked an SKN-1 binding site (27). The Skn domain concentrations chosen for the ligation experiments varied between 10 and 25 nM (not corrected for activity), depending upon the protein preparation. However, under these conditions, the levels of specific and nonspecific binding by Δ1–9 differed by only 1 order of magnitude at room temperature, making it advantageous to perform these experiments at 0°C, which stabilized specific binding (not shown). The Δ1–9 concentrations then chosen for ligation experiments ranged between 50 and 125 nM (not corrected for activity), also depending upon the protein stock. Under these conditions, a nonspecific probe was not bound appreciably by either the Skn domain or Δ1–9 and the bulk of specific probe was bound by a single protein molecule (not shown).
Cyclization kinetics assays were performed at a DNA concentration of 3 × 10−11 M, as described previously (27), except that 0.1% Nonidet P-40 was added. Experiments involving Δ1–9 were performed at 0°C, and those involving the Skn domain were performed at room temperature, but the latter results were confirmed at 0°C (not shown). Each ligation reaction mixture contained DNA at a concentration of 32 pM. Reaction mixtures were incubated for 30 min, and then cyclizations were initiated by the addition of 50 to 1,000 U of ligase. At 10 different time points, 16 μl of the reaction mixture was removed and the ligation was halted by the addition of 8 μl of the proteinase K mixture described in reference 15. By varying the amount of ligase added, these experiments were performed over times ranging between 4 and 21 h. Reaction mixtures were heated and then analyzed in a 6% native gel as described previously (15). Dried gels were analyzed by PhosphorImaging (Molecular Dynamics).
RESULTS
The amino-terminal arm specifies the AT-rich element.
To determine whether the AT-rich element that was selected in vitro (2) is important for high-affinity binding, we assayed binding of the Skn domain to an oligonucleotide (SK1) (Fig. 2A and B, lanes 1 to 5) that matches the preferred consensus (2) and to an otherwise identical site in which the AT-rich element had been changed to GCC (−3,−2,−1 GCC) (Fig. 2A and B, lanes 11 to 15). The purified Skn domain (8) bound to SK1 with an approximately fivefold higher affinity than it bound to the −3,−2,−1 GCC site (not shown), as indicated by titration in an EMSA in which the Kd was estimated from the protein concentration that binds 50% of the input DNA (6).
We also tested binding of the Skn domain to these sites by EMSA titration of in vitro-translated protein, performed at a low concentration (5 × 10−12 M) of specific DNA to obtain a semiquantitative measurement of binding. These conditions may be more similar to those of a cellular environment than are those of assays of binding by a purified protein, because both reticulocyte lysate proteins and poly(dI-dC) competitor are present. In this assay, the in vitro-translated Skn domain appeared to bind the SK1 site with a moderately higher affinity than was measured previously for the purified Skn domain (Fig. 2A, lanes 1 to 5) (see Materials and Methods). Significantly, a >100-fold-higher concentration of the Skn domain protein was required to bind −3,−2,−1 GCC at a level comparable to that at which it bound the preferred site, SK1 (Fig. 2A, lanes 5 and 11), demonstrating that the AT-rich element is an important part of the high-affinity consensus SKN-1 binding site.
To determine whether the amino-terminal arm segment is involved in this sequence specificity, we assayed binding to these sites of an Skn domain mutant that lacks the arm (Δ1–9) (Fig. 1A) (2). In an assay performed with purified proteins at room temperature, the Skn domain protein bound SK1 at a fivefold higher affinity than did Δ1–9 (8). However, binding by in vitro-translated Δ1–9 was detectable only at 0°C (Fig. 2A and B, lanes 6). In the latter assay, the concentrations of the Skn domain protein and Δ1–9 that gave similar levels of SK1 binding differed by about 2 orders of magnitude (Fig. 2B, lanes 5 and 6). In contrast, these proteins bound at comparable levels to the −3,−2,−1 GCC mutant site (Fig. 2B, lanes 11 to 20), and Δ1–9 bound as well to this site as to SK1 (Fig. 2B, lanes 6 to 10 and 16 to 20), demonstrating that the specificity of the Skn domain for the AT-rich element is mediated by the amino-terminal arm.
Multiple Skn domain amino-terminal arm residues contribute to DNA binding affinity and specificity.
In most homeodomain amino-terminal arms, except for those in yeast proteins such as α2 (Fig. 3A), the Arg residue that corresponds to Skn domain Arg 9 is conserved and lies carboxyl terminal to a residue that varies according to protein class but is often Gly or Pro (Fig. 1A and 3A) (34). The more distal arm residues (positions 5 to 7) are usually basic but are less conserved (Fig. 1A and 3A) (34). To investigate how these individual residues contribute to Skn domain DNA binding, we created mutations in the arm that included amino-terminal deletions and alanine substitutions to replace individual side chains with a methyl group (Fig. 3A) and tested their abilities to bind to the SK1 and −3,−2,−1 GCC sites. In the EMSA, a low temperature noticeably stabilizes binding that involves either the Δ1–9 mutant protein or the −3,−2,−1 GCC mutant DNA sequence (Fig. 2A and B, lanes 5 to 20) but not specific binding of the Skn domain to the optimal sequence, SK1, as indicated by comparison of the bound and free DNA fractions (Fig. 2A and B, lanes 1 to 5). Binding by various other mutant proteins and DNA sequences that we have analyzed was also relatively enhanced at low temperature (see below), presumably because these mutant protein-DNA complexes are less stable. We therefore tested these mutants for binding at both room temperature and 0°C to allow their binding to be compared stringently with optimal Skn domain-DNA binding and still permit analysis of weak mutants such as Δ1–9.
FIG. 3.
Mutational analysis of the Skn domain amino-terminal arm. (A) Mutations constructed within the arm, which is compared with homeodomain arm segments. In each of these proteins, the amino-terminal arm was altered as shown, but the remainder of the Skn domain (Fig. 1) was intact. Each sequence is preceded by an initiation methionine. Open and closed circles indicate contacts with bases and the backbone, respectively, that were revealed by structural studies (1, 17, 23, 32). (B) Binding of the indicated proteins (described in panel A) to the SK1 oligonucleotide site (Fig. 2). Equal concentrations (25 pM) of these in vitro-translated proteins were assayed for DNA binding at room temperature (RT) by EMSA. Only the preferred sequence is shown below the gel, with the AT-rich region being shaded. (C) EMSA carried out as described for panel B but performed at 0°C. (D) EMSA of the indicated proteins for binding to the −3,−2,−1 GC mutant site (Fig. 2) at room temperature. (E) EMSA carried out as described for panel D but performed at 0°C.
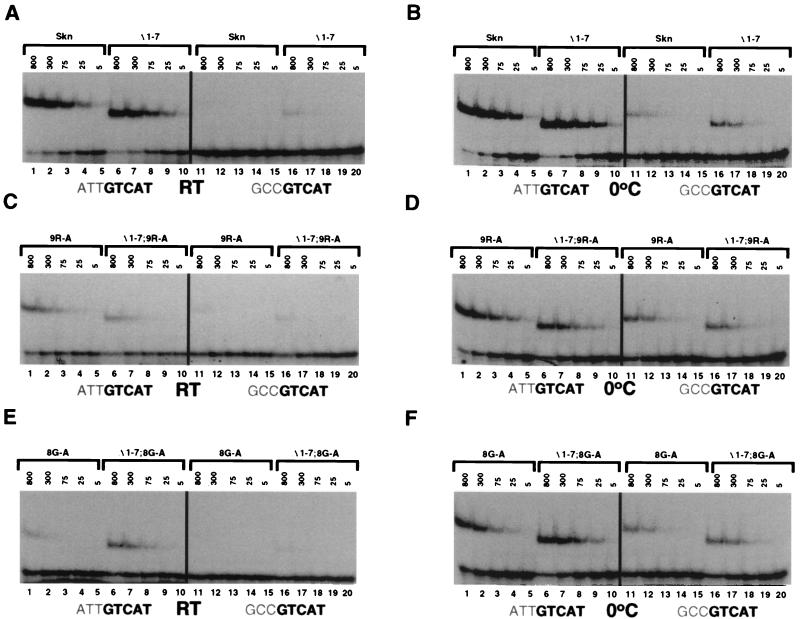
The deletion mutants Δ1–4 and Δ1–7 (Fig. 3A) both bound well to SK1 (Fig. 3B, lanes 8 and 9), and an EMSA titration indicated that Δ1–7 and the Skn domain bound to this site with very similar affinities (Fig. 4A, lanes 1 to 10), indicating that the more distal (amino-terminal) residues in the arm (positions 1 to 7) (Fig. 3A) are not required for binding affinity. In contrast, the dramatic difference in levels of binding between Δ1–9 and Δ1–7 (Fig. 2A and 4A, lanes 6 to 10, and 3B and C, lanes 9 and 12) suggests that residues 8 and 9 are particularly important. This finding is consistent with the Skn domain crystal structure, in which Arg 9 appears to contact the DNA directly (Fig. 1C) (32).
FIG. 4.
EMSA titration of DNA binding by Skn domain amino-terminal arm mutants. (A) Binding of the Skn domain and Δ1–7 proteins (Fig. 3A) to the indicated sites, assayed at room temperature (RT) as described for Fig. 2. Protein concentrations are indicated above the gel (in 10−12 molar units), and specific DNA was present at 5 × 10−12 M. (B) EMSA carried out as described for panel A but performed at 0°C. (C) Binding of the 9R-A and Δ1–7 9R-A proteins (Fig. 3A) to the indicated sites, assayed as described for panel A. (D) EMSA carried out as described for panel C but performed at 0°C. (E) Binding of the 8G-A and Δ1–7 8G-A proteins (Fig. 3A) to the indicated DNA sequences, assayed as described for panel A. (F) EMSA carried out as described for panel E but performed at 0°C.
Binding to the preferred SK1 site was not affected by substitution of Ala for basic residues 5, 6, and 7 (Fig. 3A; Fig. 3B and C, lanes 1 and 3 to 5) but, surprisingly, was only partially impaired by substitution of Ala for Arg 9 (Fig. 3A; Fig. 3B and C, lanes 1 and 7). The latter finding was confirmed by an EMSA titration experiment (Fig. 4A to D, lanes 1 to 5), and it suggested that interaction of the Skn domain arm with DNA may be more complicated than was indicated by the crystal structure (Fig. 1C) (32). Consistent with this idea, although deletion of residues 1 to 7 did not decrease the affinity of the Skn domain for SK1 (Fig. 3B and C, lanes 1 and 9, and 4A and B, lanes 1 to 10), this deletion modestly impaired binding in the context of the Arg 9-to-Ala substitution (Fig. 3B and C, lanes 7 and 11, and 4C and D, lanes 1 to 10), suggesting that the more distal basic residues can contribute affinity. Given these results, it appears likely that multiple basic residues in the Skn domain arm can interact with DNA but that they are partially redundant for binding. In addition, replacement of Gln 4 with Arg (4Q-R) inhibited binding (Fig. 3B and C, lanes 2), suggesting that this even more amino-terminal residue may also be located close to the DNA.
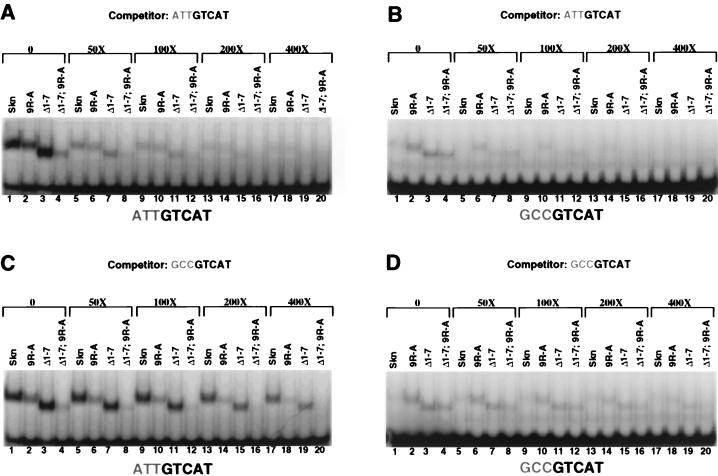
To evaluate how individual residues in the arm contribute to sequence specificity, we assayed binding of these mutants to the −3,−2,−1 GCC site (Fig. 3D and E). Although the Arg 9-to-Ala substitution (Fig. 3A) decreased binding of the Skn domain to SK1 (see above), surprisingly, this mutation modestly increased its affinity for −3,−2,−1 GCC (Fig. 3B to E, lanes 1 and 7, and 4A to D, lanes 1 to 5 and 11 to 15). Supporting this idea, in a binding site competition assay, the SK1 site more effectively competed binding by the Skn domain than by the 9R-A mutant protein, but the −3,−2,−1 GC site more effectively competed binding by the 9R-A mutant than by the Skn domain (Fig. 5A and C, lanes 1, 2, 13, and 14). Surprisingly, replacement of individual distal basic residues (positions 5 to 7) (Fig. 3A) with Ala comparably enhanced binding of the Skn domain to the −3,−2,−1 GCC site (Fig. 3B to E, lanes 1 and 3 to 5). Consistent with this finding, the Δ1–4 mutant retained the complete AT-rich element preference (Fig. 3B to E, lane 8), but the Δ1–7 mutant bound with higher affinity than the Skn domain to the −3,−2,−1 GCC site (Fig. 3D and E, lanes 1 and 9; 4A and B, lanes 1 to 20; and 5C, lanes 1, 3, 17, and 19). Simultaneous removal of these distal basic residues (positions 1 to 7) and replacement of Arg 9 with alanine further diminished the relative preference for the AT-rich element (Figs. 3B to E, lane 11, and 4C, lanes 6 to 10 and 16 to 20), as confirmed by binding site competition analysis (Fig. 5, lanes 4 and 8). However, the Δ1–7 9R-A mutation did not impair binding as severely as did the Δ1–9 deletion (Fig. 3B to E, lanes 11 and 12), a difference that may involve additional DNA contacts by the mutant arm (see below) or a stabilizing effect on the overall Skn domain fold (35). Together, these experiments indicate that although individual basic residues in the arm are not required for binding affinity, each enhances specificity by diminishing binding to the GC-rich mutant site.
FIG. 5.
Binding site competition analysis of Skn domain amino-terminal arm mutants. (A) Binding of the indicated in vitro-translated proteins (Fig. 3A) to the labeled SK1 site at room temperature, assayed as described in the legend to Fig. 3B. Protein and labeled DNA concentrations were 45 pM and 1 nM, respectively. Unlabeled SK1 competitor was added to binding reaction mixtures at the indicated ratios of competitor to labeled probe. (B) EMSA of binding to the labeled −3,−2,−1 GC mutant site, performed as described for panel A, with unlabeled SK1 competitor added as indicated. (C) EMSA of binding to the SK1 site, with unlabeled −3,−2,−1 GC competitor added as indicated. (D) EMSA of binding to the −3,−2,−1 GC site, with unlabeled −3,−2,−1 GC competitor added as indicated.
Mutagenesis experiments also indicated that Gly 8 (Fig. 3A) is important for DNA binding. The Gly 8-to-Ala mutation significantly impaired binding of the Skn domain to SK1, thereby diminishing the AT-rich preference (Fig. 3B to E, lane 6, and 4, B, E, and F, lanes 1 to 5 and 11 to 15). In the context of the Δ1–7 mutant, however, this substitution decreased specific binding affinity to a lesser extent (Fig. 3B to E, lanes 6, 9, and 10, and 4A, B, E, and F, lanes 1 to 10), indicating that the distal arm residues (positions 1 to 7) destabilize binding when Gly 8 is replaced with Ala. The last observation is in sharp contrast to the finding that these distal residues stabilize binding by the 9R-A mutant (see above). These experiments suggest that the Gly 8-to-Ala substitution may destabilize Skn domain DNA binding by impairing the interaction of the entire arm with DNA, a model that may explain why the DNA-bound 8G-A Skn domain mutant migrates at a relatively decreased mobility (Fig. 3B to E, lanes 1 and 6), but they are also consistent with the possibility that Gly 8 contacts DNA directly.
Binding of the amino-terminal arm at the AT-rich region.
In the Skn domain crystal structure, the amino-terminal arm does not interact with bases or appear to influence DNA conformation at the AT-rich region (Fig. 1C) (32). Instead, this structure indicates that the Arg 9 amino group is located approximately 3.7 and 4.7 Å (Protein Data Bank; 1SKN) from the bottom-strand phosphates at −3 and −4, respectively (Fig. 1B and C) and that salt bridges may occur between the arm and DNA positions further from the GTCAT consensus (32). The Skn domain amino-terminal arm protects DNA from hydroxyl radical cleavage around −4 on the bottom strand and to a lesser extent at −1 and −2 on the top strand (Fig. 1B and C) (8), suggesting that the arm lies close to the backbone, or the minor groove, at and distal to the AT-rich region. The crystal structure is consistent with the protection detected at −4 but does not readily explain the remaining protection. In addition, discrimination by the Skn domain against GCC at positions −3 through −1 is mostly relieved by replacement of these G · C pairs with inosine (I) · C pairs (2). Inosine lacks the guanine amino group that protrudes into the minor groove; therefore I · C base pairs are indistinguishable from A · T base pairs in this case. The last finding indicates that the AT-rich region minor groove is important for Skn domain binding.
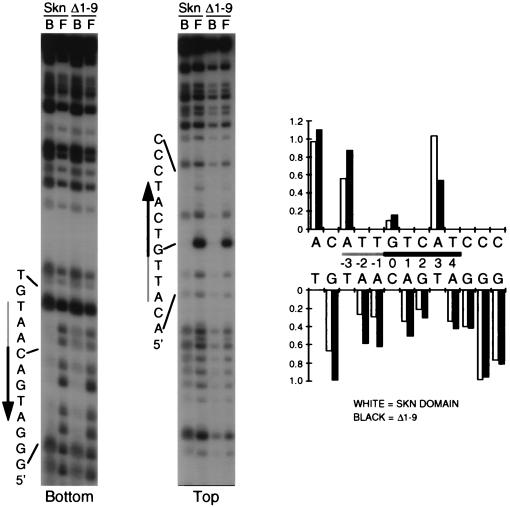
To investigate further how the Skn domain arm might specify the AT-rich region, we studied how N3 methylation of adenine in the minor groove interferes with DNA binding by the Skn domain protein and the Δ1–9 mutant. The methylation interference pattern for the Skn domain (Fig. 6) was not appreciably different from that of full-length SKN-1 (2). Adenine methylation within the AT-rich region, especially at positions −2 and −1 on the bottom strand, had a greater relative inhibitory effect on DNA binding by the Skn domain than that by Δ1–9 (Fig. 1B and 6; compare the relative levels of interference at −2, −1, and +4 on the bottom strand). This is similar to how, at these positions, the presence of guanine NH2 groups in the minor groove interfered with binding by the Skn domain but not by Δ1–9 (see above; Fig. 2B). In contrast, adenine methylation at −5 and positions further from the bZIP half site did not impair Skn domain binding (Fig. 6). These findings further support the model that the AT-rich region minor groove is specifically involved in binding of the amino-terminal arm.
FIG. 6.
Methylation interference with Skn domain DNA binding. An end-labeled and partially methylated SKN-1 binding site was bound by the indicated purified protein (Skn domain or Δ1–9) and then bound (lanes B) and free (lanes F) DNA fractions were separated by EMSA. After cleavage, these DNA samples were run on sequencing gels, which were analyzed with a PhosphorImager. A representative experiment is shown. Shaded and black arrows alongside the gel indicate the AT-rich region and bZIP half-site, respectively, with the arrow pointing away from the center of the complete bZIP binding site. Top and bottom indicate the DNA strands shown between the graphs on the right. Band intensities at each position were converted to ratios of bound-to-free DNA fractions, which were normalized to 1 at positions at which no binding occurred, and are represented in graphs to the right. Site positions are numbered so that 0 corresponds to the center of a bZIP dimer site.
DNA bending associated with SKN-1 binding.
The importance of the AT-rich region minor groove may potentially reflect direct binding by the SKN-1 amino-terminal arm, or alternatively, this sequence may more readily accommodate an indirect conformational effect. Consistent with the latter model, prior hydroxyl radical cleavage next to the AT-rich region enhanced binding of the Skn domain but not of Δ1–9 (Fig. 1B) (8). This observation indicates that binding of the arm involves an energy cost which is decreased by breaking the DNA backbone at these positions, suggesting that the arm normally stabilizes a less favorable DNA conformation. Since A · T base pairs allow bending towards the minor groove, it is possible that the arm specifies the AT-rich region because it can more readily bend or otherwise distort the DNA through this particular arrangement of base pairs.
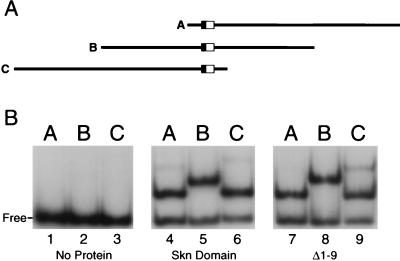
We used the circular-permutation assay (46) to test whether the Skn domain protein or the Δ1–9 protein promotes DNA bending. If a protein bends DNA, the electrophoretic mobility of the protein-DNA complex is influenced by the position of the binding site along a linear DNA fragment. If the binding site is in the middle of the fragment (probe B) (Fig. 7A), bending will induce a more distorted shape than if the site is at either end (probes A and C) (Fig. 7A), resulting in a relatively slower gel mobility. In the absence of added protein, probes A, B, and C migrated with indistinguishable mobilities (Fig. 7B, lanes 1 to 3). In contrast, complexes of either the Skn domain, Δ1–9, or full-length SKN-1 with probe B (Fig. 7A) migrated with comparably decreased mobilities (Fig. 7B, lanes 4 to 9, and data not shown). These findings suggest that full-length SKN-1, the Skn domain, and Δ1–9 proteins may each bend DNA, but they should be interpreted conservatively because the shapes of certain DNA binding domains can cause similar gel mobility anomalies (27, 36, 37).
FIG. 7.
Circular-permutation analysis of Skn domain DNA binding. (A) Diagrams of probes A, B, and C. In each probe the SKN-1 binding site is located at a different position relative to the ends. The AT-rich region and bZIP half-site within the SKN-1 binding site are indicated by a filled and open box, respectively. (B) EMSA in which the purified proteins designated below the gel were bound to probes A, B, and C, as indicated above the gel.
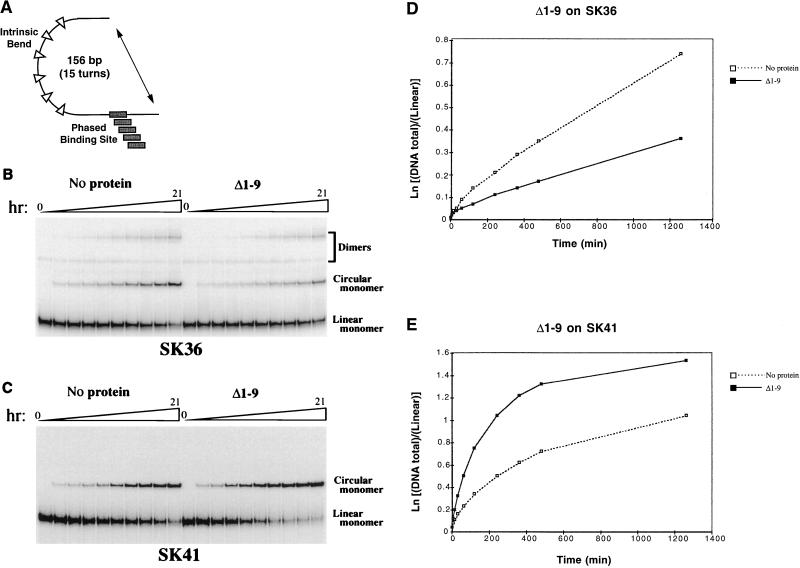
As an independent bending assay, we performed cyclization kinetics (15), which measures whether protein binding affects the rate at which DNA that contains a fixed bend can form a ligated minicircle (Fig. 8A). Cyclization is enhanced if a protein bends DNA in the same direction as that of the fixed bend and is inhibited if the DNA is bent in the opposite direction (15) or held straight (27). The relative kinetics of circular versus bimolecular ligation can be quantitated as the J factor (Table 1), which is proportional to the bending angle (15). This assay also reveals the approximate direction of bending, because in these substrates the site is phased around a helical turn of DNA relative to the fixed bend (Fig. 8A).
FIG. 8.
Cyclization kinetics analysis of DNA bending by SKN-1. (A) Cartoon (not drawn to scale) of minicircle ligation probes in which the SKN-1 binding site is phased along a turn of the DNA helix relative to a fixed bend. (B) Effects of Δ1–9 binding on ligation of the SK36 probe. The products of a representative ligation experiment were analyzed by electrophoresis. These products can include monomeric circles and dimeric species, as shown. The proportion of dimers varied among these experiments but was never greater than that apparent here and was usually undetectable, as in panel C. The time course of ligation is depicted above the gel and represented in the graph in panel D. Comparable results were obtained with shorter time courses (not shown). (C) Effects of Δ1–9 binding on ligation of the SK41 probe, in which the SKN-1 binding site is located 5 bp further from the fixed bend than in SK36. A representative ligation experiment was analyzed by electrophoresis. (D and E) Plots of the results of the ligation experiments shown in panels B and C, respectively.
Binding of either Δ1–9 or the Skn domain protein inhibited cyclization of the SK34 and SK36 substrates (Fig. 8B and D; Table 1) but stimulated SK40 and SK41 cyclization (Fig. 8C and E; Table 1). These effects correlated with phasing of the SKN-1 binding site around a DNA helical turn (10.5 bp), so that opposite effects were observed on opposite sides of the helix (Table 1), suggesting that these proteins bend DNA in similar directions. Consistent with this idea, neither protein affected cyclization of SK32 and SK38 (Table 1), in which the sites are located at intermediate positions. The J ratios derived from binding of either of these proteins to SK34 or SK36 differed from those associated with SK40 or SK41 binding by a factor of approximately 3 (Table 1), suggesting that they each induce a modest DNA bend (5 to 10°) (12, 13). The positions of these SKN-1 binding sites relative to the fixed bend indicate that both proteins bend DNA approximately towards the major groove at position +6 within the SKN-1 binding site (Fig. 1B). The magnitude and direction of this DNA bend are consistent with the 7° bend that occurs in the Skn domain-DNA crystal structure (32). These experiments suggest that the amino-terminal arm does not significantly influence DNA bending but leave open the possibility that AT-rich sequence specificity derives from a different type of conformational effect.
Sequence preferences of the amino-terminal arm.
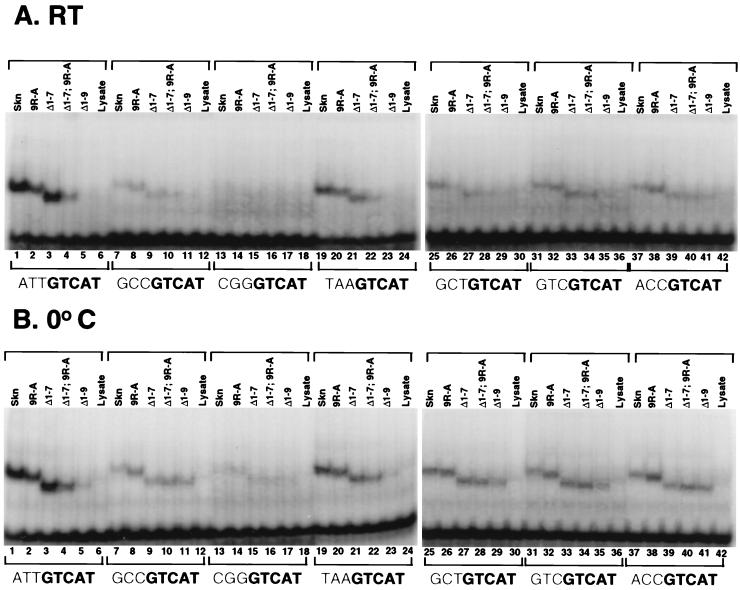
To investigate further how the amino-terminal arm might specify base recognition within the AT-rich region, we assayed how mutations within the arm affected binding of the Skn domain to a panel of binding site mutants. The Skn domain and various mutant derivatives bound at higher affinities to the −3,−2,−1 GCC mutant site than to a site in which ATT was replaced with CGG (Fig. 9, lanes 7 to 17), indicating that its sequence specificity in this region may be even greater than was indicated by the experiments described above. The difference in levels of binding between these two mutant sites does not derive entirely from the amino-terminal arm, however, because the Δ1–9 mutant also bound at lower affinity to the −3,−2,−1 CGG site (Fig. 9, lanes 11 and 17). Binding by either the Skn domain or Δ1–7 was reduced by replacement of ATT with TAA (Fig. 9, lanes 1, 3, 19, and 21), indicating that the particular arrangement of these A · T base pairs is important for binding. To investigate the contribution of individual A · T base pairs, we back-substituted the corresponding SK1 residue for each residue within the −3,−2,−1 GCC site. Each of these substitutions enhanced binding by the Skn domain (Fig. 9, lanes 7, 25, 31, and 37), suggesting that each base pair normally contributes to binding. In addition, the Δ1–7 mutant bound comparably to each back-substituted site, indicating that no single A · T base pair is sufficient to restore high-level binding by this truncated arm (Fig. 9, lanes 3, 27, 33, and 39). This last finding is consistent with the model suggesting that binding of the arm involves a conformational effect that depends upon each of these positions. However, some residues within the arm may still interact specifically with particular positions within the AT-rich element, as was suggested by the finding that the 9R-A mutant binds at higher affinity to the −3,−2,−1 ACC and −3,−2,−1 GTC sites than to the −3,−2,−1 GCT site (Fig. 9, lanes 26, 32, and 38).
FIG. 9.
Binding of the Skn domain to mutant DNA sequences. Shown are results of EMSAs at room temperature (RT) (A) and at 0°C (B) of the indicated proteins (Fig. 3A) to determine binding to the SK1 preferred site (ATTGTCAT) or to the various mutant sites shown, which differ from SK1 only at the indicated bases. DNA and specific protein concentrations were 1 nM and 50 pM, respectively.
DISCUSSION
Multiple amino-terminal arm residues contribute to SKN-1 DNA binding specificity.
We have demonstrated that an AT-rich element adjacent to the bZIP half-site is important for high-affinity DNA binding by the Skn domain and that this sequence specificity is mediated almost exclusively by the amino-terminal arm (Fig. 2), although the remainder of the Skn domain may have a minor influence (Fig. 9, lanes 5, 11, and 17). Each base pair within the AT-rich element contributes to this specificity, and variations within this sequence can result in a range of binding affinities (Fig. 9). In light of previous experiments, which have indicated that the preferences of the Skn domain and full-length SKN-1 for the AT-rich element are comparable (2), our findings suggest that this sequence element is likely to be relevant to SKN-1 function in vivo.
Site-directed mutagenesis experiments showed that individual residues within the Skn domain amino-terminal arm make distinct contributions to DNA binding affinity and specificity, particularly at room temperature (Fig. 3 and 4). Replacement of Arg 9 decreases binding affinity, especially when the distal arm residues (positions 1 to 7) are deleted, and diminishes the AT-rich preference further by enhancing binding to the −3,−2,−1 GCC site (Fig. 3B to E, lanes 1, 7, 9, and 11; Fig. 4 and 5). The Skn domain structure predicts that Arg 9 is likely to be important, because it suggests that this residue contacts DNA directly (Fig. 1C) (32). However, replacement of Gly 8 within the Skn domain even more markedly impairs binding (Fig. 3B to E, lanes 1, 6, and 7; Fig. 4), even though this residue does not contact DNA in the crystal structure (Fig. 1C) (32). Gly 8 is less critical for binding affinity when residues 1 to 7 are removed (Fig. 3B to E, lanes 1, 6, and 10, and 4E and F), indicating that it might provide the rotational flexibility required for proper positioning of these residues. In addition, contacts between glycine residues and nucleic acids have been observed (10, 23, 31), suggesting that Gly 8 may also interact directly with the DNA. The distal basic residues in the arm (positions 5 to 7) (Fig. 3A) are disordered in the Skn domain structure (Fig. 1C) (32) and not required for binding affinity (Fig. 3B, lane 9; Fig. 4), but individual Ala substitutions at these positions each diminish the AT-rich preference (Fig. 3B to E, lanes 3 to 5). In addition, residues 1 to 7 stabilize binding by the 9R-A mutant (Fig. 3B to E, lanes 7 and 11; Fig. 4 and 5), indicating that they can make otherwise redundant contributions to binding affinity. These last observations suggest that residues 5 to 7 can contribute to DNA binding, presumably through heterogeneous or unstable interactions. The distinct functions of these different Skn domain arm residues indicate that this segment does not represent simply a random collection of basic residues. However, the functional significance of its similarity to homeodomain arms (Fig. 1A) remains to be determined.
The finding that alanine substitution for any basic residue in the Skn domain arm enhances binding to the −3,−2,−1 GC mutant site (Fig. 3B to E) suggests that these residues interact with DNA more readily if the AT-rich element is present and destabilize the protein-DNA complex if they are not bound to the DNA. This effect may contribute to destabilization of 8G-A DNA binding by residues 1 to 7 (Fig. 3B to E, lanes 6 and 10) and may involve not only steric incompatibility but also their highly basic charge. For example, basic DNA binding regions related to the Skn domain BR have an intrinsic α-helical character (20, 33, 42) but form an α-helix only upon DNA binding (see reference 8), presumably because they require some neutralization of positive charges. Similarly, the SKN-1–DNA complex may potentially be destabilized if the amino-terminal arm cannot properly engage a nonpreferred site and if its basic charges are not at least partially neutralized. This mechanism might have contributed to destabilization of binding by introduction of an Arg residue at position 4 (4Q-R) (Fig. 3B to E, lanes 2), at which an initiation methionine is tolerated (Δ1–4) (Fig. 3B and D, lanes 8).
The DNA sequence specificity of protein-DNA binding is generally understood to involve hydrogen bonding and van der Waals interactions that contribute affinity, and numerous examples of how individual amino acid residues can bind to particular bases have been described (29, 39). Specificity can also be profoundly influenced by intrinsic DNA structure (30) and by how well the protein and DNA adapt to each other (38). Our findings suggest an additional mechanism, in which residues that do not necessarily contribute affinity can enhance binding specificity by inhibiting interactions with nonpreferred DNA sequences. Presumably, such residues destabilize the overall interaction if they are not properly engaged in the binding surface. This mechanism of energetic exclusion can narrow the field of potentially favorable binding sites that are specified on the basis of affinity alone.
Interaction of the SKN-1 amino-terminal arm with DNA.
Our experiments indicate that models for how the Skn domain amino-terminal arm interacts with DNA must account for the AT-rich sequence specificity mediated by the arm and for the involvement of multiple amino acid residues within the arm in binding. The models must also explain how binding of the arm apparently affects DNA conformation, which was suggested by the observation that prior hydroxyl radical cleavage adjacent to the AT-rich element enhanced binding by the Skn domain but not by Δ1–9 (Fig. 1B) (8). Supporting the idea that specification of the AT-rich element may involve indirect mechanisms, these cleavage experiments did not identify individual bases within this region that are specifically required for binding of the arm (8), and each individual base pair made a comparably modest contribution to binding by the Δ1–7 mutant, in which the arm is truncated (Fig. 9A and B, lanes 9, 27, 33, and 39). Models for binding of the amino-terminal arm to DNA should also account for the importance of the AT-rich element minor groove, which was indicated by hydroxyl radical footprinting, the I · C substitution experiments, and the methylation interference assay (2) (Fig. 1B and 6). Finally, the locations of the arm and the adjacent helix 1 (Fig. 1A) in the Skn domain crystal structure (Fig. 1C) (32) provide an additional caveat, because they indicate that a major conformational adaptation would be required if the arm is oriented analogously to homeodomain amino-terminal arms (Fig. 1B) and lies deeply in the AT-rich element minor groove.
In light of the propensity of A · T base pairs to bend toward the minor groove, one plausible model by which the AT-rich element minor groove might be important for a conformational effect is that it allows the arm to promote a DNA bend or kink. However, our experiments suggest that the Skn domain arm does not promote DNA bending but also that the Skn domain BR and support segment induce a modest DNA bend that should increase the surface area along which SKN-1 interacts with DNA (32). Various assays have indicated that some related bZIP proteins bend DNA (16, 21, 30, 40), but this issue has remained controversial (12, 28, 36, 37). However, our evidence comes from both gel-based (circular-permutation) and solution (circularization kinetics) assays and is in general agreement with crystallographic data (32). An effect of DNA bending or conformation on SKN-1 binding may be important because transcription factors generally function within multiprotein complexes, the composition of which can be influenced by subtle intermolecular interactions and effects on DNA conformation (7). In addition, SKN-1 activity in vivo is modulated by the POP-1 protein, a high-mobility-group protein of the TCF (TCF/LEF) family (25), members of which are both Wnt signalling targets and “architectural” proteins that bend DNA (5, 9).
A model that is consistent with the experimental evidence is that the amino-terminal arm interacts with the DNA backbone around position −4 and that it lies above or across the minor groove, as suggested by the hydroxyl radical protection footprint (Fig. 1B and C). In doing so, it induces a localized conformational effect that is favored by the AT-rich element minor groove (29, 39) and makes multiple interactions with the DNA, including some that are unstable. These interactions help discriminate against G · C base pairs in this region, and some may directly involve bases in the distal portion of the AT-rich element. The data also do not rule out a more extensive direct interaction with the AT-rich element minor groove. However, that model would require the arm to be oriented more similarly to those of homeodomains (Fig. 1B) and is more difficult to reconcile with the Skn domain crystal structure (Fig. 1C) (32). Further structural investigations will be necessary to determine exactly how the arm specifies the AT-rich element as it interacts with DNA. However, our experiments illustrate how mutagenesis and biochemical analyses can complement and extend structural studies and how short polypeptide segments that are relatively unstructured can make important contributions to binding specificity.
ACKNOWLEDGMENTS
We thank Tom Ellenberger, David Fisher, Phil Auron, and members of the Blackwell lab for helpful discussions and critically reading the manuscript, and we thank Karen Kim for help with sequencing. For protein purification we thank Dara Gilbert, Jim Cheung, and Tom Ellenberger, whom we also thank for computer graphics.
This work was supported by a grant from the NIH (GM50900) to T.K.B., who is a Searle Scholar.
REFERENCES
- 1.Billeter M, Qian Y Q, Otting G, Muller M, Gehring W, Wuthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993;234:1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- 2.Blackwell T K, Bowerman B, Priess J, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- 3.Bowerman B, Draper B W, Mello C, Priess J. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–452. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- 4.Bowerman B, Eaton B A, Priess J R. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- 5.Cadigan K M, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 6.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 7.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 8.Carroll A S, Gilbert D E, Liu X, Cheung J W, Michnowicz J E, Wagner G, Ellenberger T E, Blackwell T K. SKN-1 domain folding and basic region monomer stabilization upon DNA binding. Genes Dev. 1997;11:2227–2238. doi: 10.1101/gad.11.17.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clevers H, van de Wetering M. TCF/LEF factor earn their wings. Trends Genet. 1997;13:485–489. doi: 10.1016/s0168-9525(97)01305-x. [DOI] [PubMed] [Google Scholar]
- 10.Feng J-A, Johnson R C, Dickerson R E. Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions. Science. 1994;263:348–355. doi: 10.1126/science.8278807. [DOI] [PubMed] [Google Scholar]
- 11.Furukubo-Tokunaga K, Flister S, Gehring W J. Functional specificity of the Antennapedia homeodomain. Proc Natl Acad Sci USA. 1993;90:6360–6364. doi: 10.1073/pnas.90.13.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagerman P J. Do basic region-leucine zipper proteins bend their DNA targets … does it matter? Proc Natl Acad Sci USA. 1996;93:9993–9996. doi: 10.1073/pnas.93.19.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hockings S C, Kahn J D, Crothers D M. Characterization of the ATF/CREB site and its complex with GCN4. Proc Natl Acad Sci USA. 1998;95:1410–1415. doi: 10.1073/pnas.95.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson E M, Li P, Leon-del-Rio A, Rosenfeld M G, Aggarwal A K. Structure of Pit-1 POU domain bound to DNA as a dimer:unexpected arrangement and flexibility. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 15.Kahn J D, Crothers D M. Protein-induced bending and DNA cyclization. Proc Natl Acad Sci USA. 1992;89:6343–6347. doi: 10.1073/pnas.89.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerppola T K, Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991;66:317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- 17.Kissinger C R, Liu B, Martin-Blanco E, Kornberg T B, Pabo C. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–580. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 18.Klemm J D, Rould M A, Aurora R, Herr W, Pabo C O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 19.Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 20.Krebs D, Dahmani B, El Antri S, Monnot M, Convert O, Mauffret O, Troalen F, Fermandjian S. The basic subdomain of the c-Jun oncoprotein: a joint CD, Fourier-transform infrared and NMR study. Eur J Biochem. 1995;231:370–380. doi: 10.1111/j.1432-1033.1995.tb20709.x. [DOI] [PubMed] [Google Scholar]
- 21.Leonard D A, Rajaram N, Kerppola T K. Structural basis of DNA bending and oriented heterodimer binding by the basic leucine zipper domains of Fos and Jun. Proc Natl Acad Sci USA. 1997;94:4913–4918. doi: 10.1073/pnas.94.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, He X, Gerrero M R, Mok M, Aggarwal A, Rosenfeld M G. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes Dev. 1993;7:2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Stark M R, Johnson A D, Wolberger C. Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- 24.Lin L, McGinnis W. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev. 1992;6:1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- 25.Lin R, Thompson S, Priess J R. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 26.Lo M-C, Ha S, Pelczer I, Pal S, Walker S. The solution structure of the DNA binding domain of Skn-1. Proc Natl Acad Sci USA. 1998;95:8455–8460. doi: 10.1073/pnas.95.15.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick R J X, Badalian T, Fisher D E. The leucine zipper may induce electrophoretic mobility anomalies without DNA bending. Proc Natl Acad Sci USA. 1996;93:14434–14439. doi: 10.1073/pnas.93.25.14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGill G, Fisher D E. DNA bending and the curious case of Fos/Jun. Chem Biol. 1998;5:R29–R38. doi: 10.1016/s1074-5521(98)90139-9. [DOI] [PubMed] [Google Scholar]
- 29.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 30.Paolella D N, Palmer C R, Schepartz A. DNA targets for certain bZIP proteins distinguished by an intrinsic bend. Science. 1994;264:1130–1133. doi: 10.1126/science.8178171. [DOI] [PubMed] [Google Scholar]
- 31.Puglisi J D, Chen L, Blanchard S, Frankel A D. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 32.Rupert P B, Daughdrill G W, Bowerman B, Matthews B W. The binding domain of Skn-1 in complex with DNA: a new DNA-binding motif. Nat Struct Biol. 1998;5:484–491. doi: 10.1038/nsb0698-484. [DOI] [PubMed] [Google Scholar]
- 33.Saudek V, Pasley H S, Gibson T, Gausepohl H, Frank R, Pastore A. Solution structure of the basic region from the transcriptional activator GCN4. Biochemistry. 1991;30:1310–1317. doi: 10.1021/bi00219a022. [DOI] [PubMed] [Google Scholar]
- 34.Scott M P, Tamkun J W, Hartzell G W I. The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 35.Shang Z, Issaac V E, Li H, Patel L, Catron K M, Curran T, Montelione G T, Abate C. Design of a “minimAl” homeodomain: the N-terminal arm modulates DNA binding affinity and stabilizes homeodomain structure. Proc Natl Acad Sci USA. 1994;91:8373–8377. doi: 10.1073/pnas.91.18.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sitlani A, Crothers D M. DNA-binding domains of Fos and Jun do not induce DNA curvature: an investigation with solution and gel methods. Proc Natl Acad Sci USA. 1998;95:1404–1409. doi: 10.1073/pnas.95.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sitlani A, Crothers D M. Fos and Jun do not bend the AP-1 recognition site. Proc Natl Acad Sci USA. 1996;93:3248–3252. doi: 10.1073/pnas.93.8.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spolar R S, Record M T., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 39.Steitz T A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990;23:205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- 40.Strauss-Soukup J K, Maher L J. DNA bending by GCN4 mutants bearing cationic residues. Biochemistry. 1997;36:10026–10032. doi: 10.1021/bi970215u. [DOI] [PubMed] [Google Scholar]
- 41.Treacy M N, Neilson L I, Turner E E, He X, Rosenfeld M G. Twin of I-POU: a two amino acid difference in the I-POU homeodomain distinguishes an activator from an inhibitor of transcription. Cell. 1992;68:491–505. doi: 10.1016/0092-8674(92)90186-g. [DOI] [PubMed] [Google Scholar]
- 42.Weiss M A. Thermal unfolding studies of a leucine zipper domain and its specific DNA complex: implications for scissors grip recognition. Biochemistry. 1990;29:8020–8024. doi: 10.1021/bi00487a004. [DOI] [PubMed] [Google Scholar]
- 43.Wilson D S, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 44.Wolberger C, Vershon A K, Liu B, Johnson A D, Pabo C O. Crystal structure of a MATα2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 45.Wright D A, Futcher B, Ghosh P, Geha R S. Association of human fas (CD95) with a ubiquitin-conjugating enzyme (UBC-FAP) J Biol Chem. 1996;271:31037–31043. doi: 10.1074/jbc.271.49.31037. [DOI] [PubMed] [Google Scholar]
- 46.Wu H-M, Crothers D M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 47.Zeng W, Andrew D J, Mathies L D, Horner M A, Scott M P. Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functional specificity. Development. 1993;118:339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Catron K M, Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc Natl Acad Sci USA. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu J, Hill R J, Heid P J, Fukuyama M, Sugimoto A, Priess J R, Rothman J H. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]