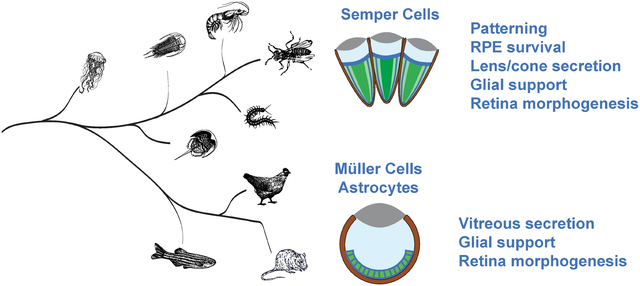
Abstract
The arthropod compound eye represents one of two major eye types in the animal kingdom and has served as an essential experimental paradigm for defining fundamental mechanisms underlying sensory organ formation, function, and maintenance. One of the most distinguishing features of the compound eye is the highly regular array of lens facets that define individual eye (ommatidial) units. These lens facets are produced by a deeply conserved quartet of cuticle-secreting cells, called Semper cells (SCs). Also widely known as cone cells, SCs were originally identified for their secretion of the dioptric system, i.e. the corneal lens and underlying crystalline cones. Additionally, SCs are now known to execute a diversity of patterning and glial functions in compound eye development and maintenance. Here, we present an integrated account of our current knowledge of SC multifunctionality in the Drosophila compound eye, highlighting emerging gene regulatory modules that may drive the diverse roles for these cells. Drawing comparisons with other deeply conserved retinal glia in the vertebrate single lens eye, this discussion speaks to glial cell origins and opens new avenues for understanding sensory system support programs.
Keywords: glia, IRM proteins, Pax2, PaxB, Prospero, Prox1, SoxB, Müller glia, eye evolution, lens, animal vision, crystalline cone
Graphical Abstract
Introduction
Compound eyes are a signature trait and the primary visual sense organ of arthropods, animals unparalleled for their prolific diversification into aquatic, terrestrial, and aerial life environments (Feller et al., 2020; Koenig and Gross, 2020). The defining architecture of compound eyes is the highly organized grid of individual eye units, called ommatidia. Each ommatidium comprises a central light-focusing dioptric structure (a corneal lens facet and crystalline cone) and an underlying light-detecting neural retina (rhabdomeric photoreceptors [PRs]). This light-sensing core is optically separated from neighboring ommatidia by light-insulating pigment cells (interommatidial pigment cells or the retinal pigmented epithelium [RPE]). Ommatidial design is thus not unlike that of the single lens eye in vertebrates, in which a neural retina lies behind a dioptric system, surrounded by an RPE (Fig.1).
Figure 1: Structural comparison of arthropod compound eye and vertebrate single lens eye.
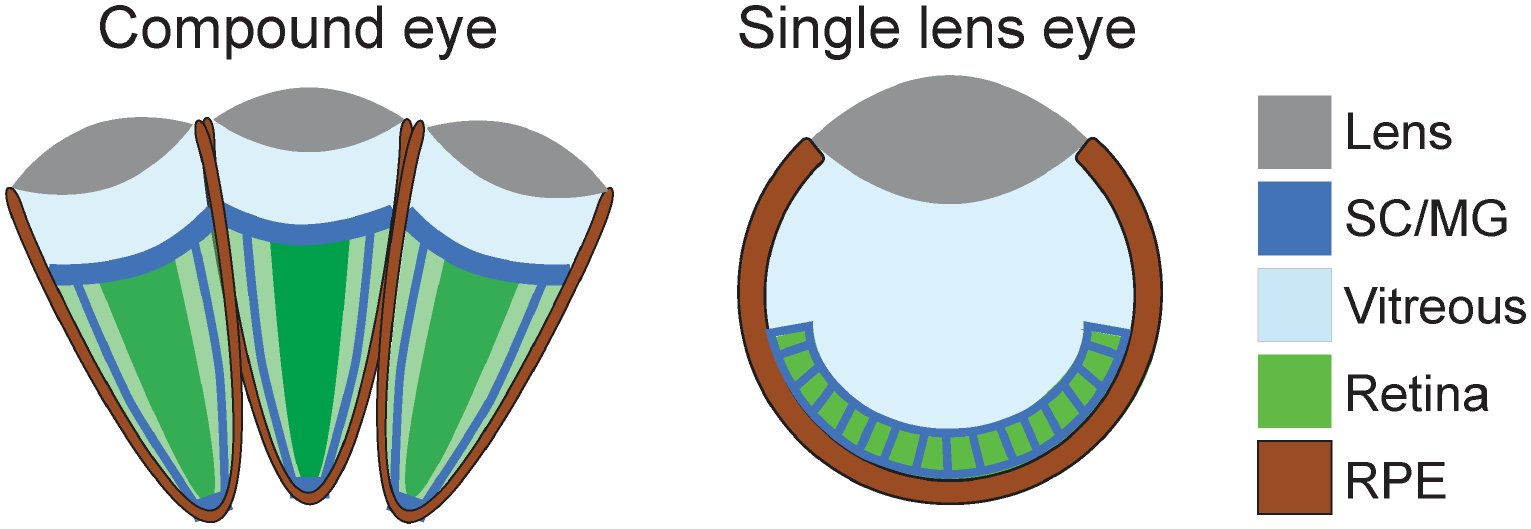
SC/MG represents radial glia-like support cells, Semper cells (SCs) in arthropod eyes and Müller glia (MG) in vertebrate eyes. RPE = retinal pigment epithelium.
Cellular, physiological, and genetic studies of the insect compound eye have led to the discovery of a wide range of deeply conserved neurobiological processes (Bellen et al., 2010). Besides finding similarities with the vertebrate camera eye in retinal neuron development and function, for example, a growing body of research has uncovered common support/glial roles for the RPE on PR health in both vertebrates and invertebrates (Charlton-Perkins and Cook, 2010; Cook et al., 2011; Hartenstein and Reh, 2002; Sparrow et al., 2010; Strauss, 2005; Viets et al., 2016). Studies of the RPE in honeybees, for instance, were foundational for understanding the lactate shuttle of energy support and ion homeostasis between neurons and glia (Coles, 1989; Tsacopoulos and Magistretti, 1996). In Drosophila, studies on white, the first genetic mutation identified, provided insights into the role of ABC transporters in neuroprotection (Borycz et al., 2008; Ferreiro et al., 2018; W. S. Kim et al., 2008; Lee and Montell, 2004; Morgan, 1910). More recent studies have uncovered roles of the fly RPE in the visual cycle, neurotransmitter storage, and lipid metabolism (Chaturvedi et al., 2014; Han et al., 2017; Liu et al., 2017, 2015; Wang et al., 2012, 2010). Thus, glial-like support mechanisms of pigment cells are well-documented in both insect compound eyes and vertebrate single lens eyes (Bharti et al., 2006; Klettner and Dithmar, 2020; Sparrow et al., 2010; Strauss, 2005).
Here, we focus on a second type of canonical support cell in arthropod ommatidia that is widely recognized for its roles in the formation of the corneal lens and the underlying crystalline cone. We highlight how these cells also contribute to ocular organization, development, and visual system maintenance, which can now be dissected for more general roles in sensory system assembly and support using the power of fly genetics.
Semper’s cells: Discovery and phylogenetic origin
The dioptric system of the compound eye captivated some of the very first microscopists and physiologists (Bernhard et al., 1972; Exner, 1989; Nilsson, 1989). Defining the cellular nature of this light-focusing system arose from studies of ommatidial development in a variety of arthropods, eventually published by Edouard Claparède in 1860 (Claparède, 1860). In this work, he credits the German zoologist Carl Semper for sharing his preliminary findings on these cells in both insect and crustacean eyes, hence naming them Semper’s cells (SCs) (Claparède, 1860). Claparède and Semper established that, in addition to being the sole source of the prominent vitreous-like compartment called the crystalline cone (Nilsson and Kelber, 2007), the SCs also frequently contribute the major part of the overlying cuticular corneal lens facet (Claparède, 1860). Reflecting their ubiquitous role in crystalline cone production, SCs have also become widely known as cone cells (Cagan and Ready, 1989; Nilsson and Kelber, 2007). However, given their broader functionality and the unfortunate terminological match with the cone PRs of the vertebrate eye, we have returned to their first namesake: SCs (Charlton-Perkins et al., 2017).
The vast number of comparative ultrastructural studies in the second half of the 20th century confirmed that a quartet of SCs is present in the ommatidia of most insect and crustacean species (the arthropod clade Pancrustacea). The presence of four SCs is so characteristic of Pancrustacea compound eyes that the term “Tetraconata” has been introduced as an equivalent name for this group (Dohle, 2001; Richter, 2002; Schwentner et al., 2018). Recent studies of fossil samples revealed crystalline cone structures in the elaborate compound eyes of the extinct trilobites (Schoenemann et al., 2017; Schoenemann and Clarkson, 2020; Scholtz et al., 2019), pointing to an even deeper conserved existence of SCs. SCs have also been described in centipedes (Myriapoda) and horseshoe crabs (an ancient lineage of Chelicerates), suggesting their origin dates minimally back to the dawn of arthropod emergence, i.e. ~500 million years ago (Fahrenbach, 1968; Nilsson, 1989). This positions SCs as originating near the separation of vertebrates and invertebrates, when visual function became more demanding as predator-prey interactions started to emerge (Parker, 2004).
Semper cell specification: A neural fate decision
SC development has been most thoroughly analyzed in the Drosophila compound eye (Cagan and Ready, 1989; Kumar, 2012; Morrison et al., 2018; Quan et al., 2012; Tomlinson, 1985; Tomlinson et al., 2011; Tomlinson and Ready, 1987; Treisman, 2013; Tsachaki and Sprecher, 2012; Voas and Rebay, 2004; Wolff and Ready, 1993). These efforts were propelled by the discovery that one of the first behavioral mutants isolated is caused by misspecification of the UV-sensitive R7 PR to that of a SC (Banerjee et al., 1987; Benzer, 1967; Harris et al., 1976; Tomlinson and Ready, 1986). Today, the choice to be or not to be a SC represents a classic paradigm for understanding cell fate decisions.
Briefly, the events leading up to this decisive step first involve the expansion of eyeless (ey)/Pax6-positive eye progenitors by cell proliferation through the majority of postembryonic (larval) development (Kumar, 2011). This expansion phase comes to an end when a front of cell differentiation forms just prior to pupation, stopping progenitor proliferation and initiating cell type specification (Wolff and Ready, 1991). The ensuing phase of retinogenesis involves progressive ommatidial cluster formation through two rounds of cell cycle exit. The progenitors that exit first (early progenitors) give rise to 5 PRs (R8–2/5–3/4), while later-exiting progenitors that undergo one more round of cell division (late progenitors) generate the last 3 PRs (R1/6–7) and 4 SCs (Kumar, 2012; Wolff and Ready, 1993) (Fig.2). After pupation, remaining precursors differentiate into three pigment cell populations or are eliminated through apoptosis (Brachmann and Cagan, 2003) (Fig.3).
Figure 2: Cellular events during Drosophila late larval eye development.
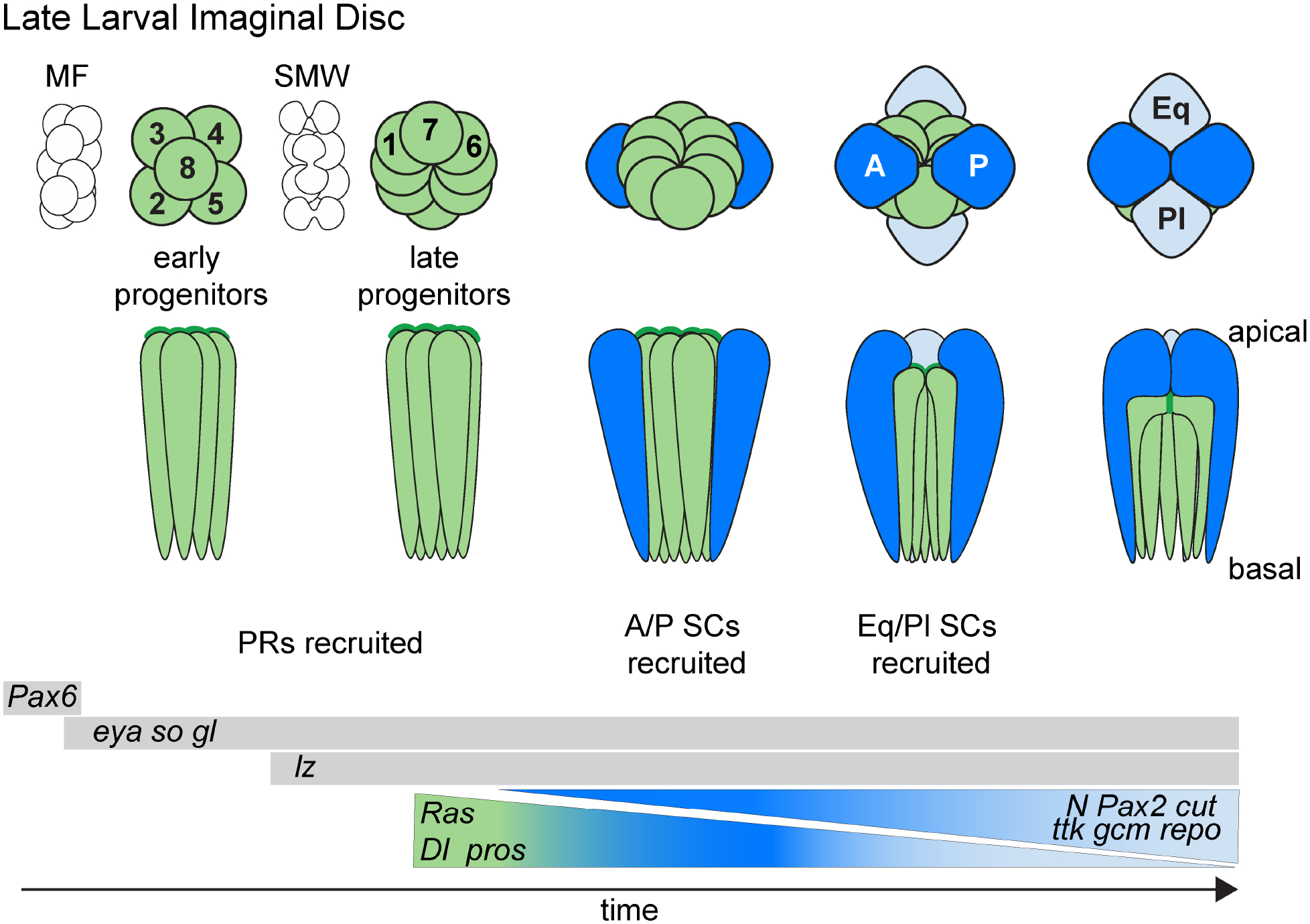
Green = retinal neurons (photoreceptors), dark blue = anterior (A) and posterior (P) Semper cells (SCs), light blue = equatorial (Eq) and Polar (Pl) SCs. MF = morphogenetic furrow, a moving front that stops progenitor cell proliferation in G1 and initiates a first wave of neurogenesis of 5 neurons/ommatidia (PRs R8, 2/5 and 3/4). An additional round of proliferation at the second mitotic wave (SMW) provides sufficient progenitors for building the remainder of the eye. Key transcription factors are listed as markers for proliferating ocular progenitors (eyeless [ey]), early retinal progenitors (eyes absent [eya], sine oculis [so] and glass [gl]), and with additional lozenge [Lz] expression in late progenitors. Pax2, tramtrack (ttk), cut, glial cells missing (gcm) and reversed polarity (repo) are pro-glial in SCs. prospero (pros) marks the last PR (R7) and the SC progenitors. R7 fate is promoted by a pros/Ras positive feedback loop, whereas SC fate is promoted by a Pax2/Notch positive feedback loop. Anterior to the left.
Figure 3: Cellular events during early pupal stages of Drosophila eye development.
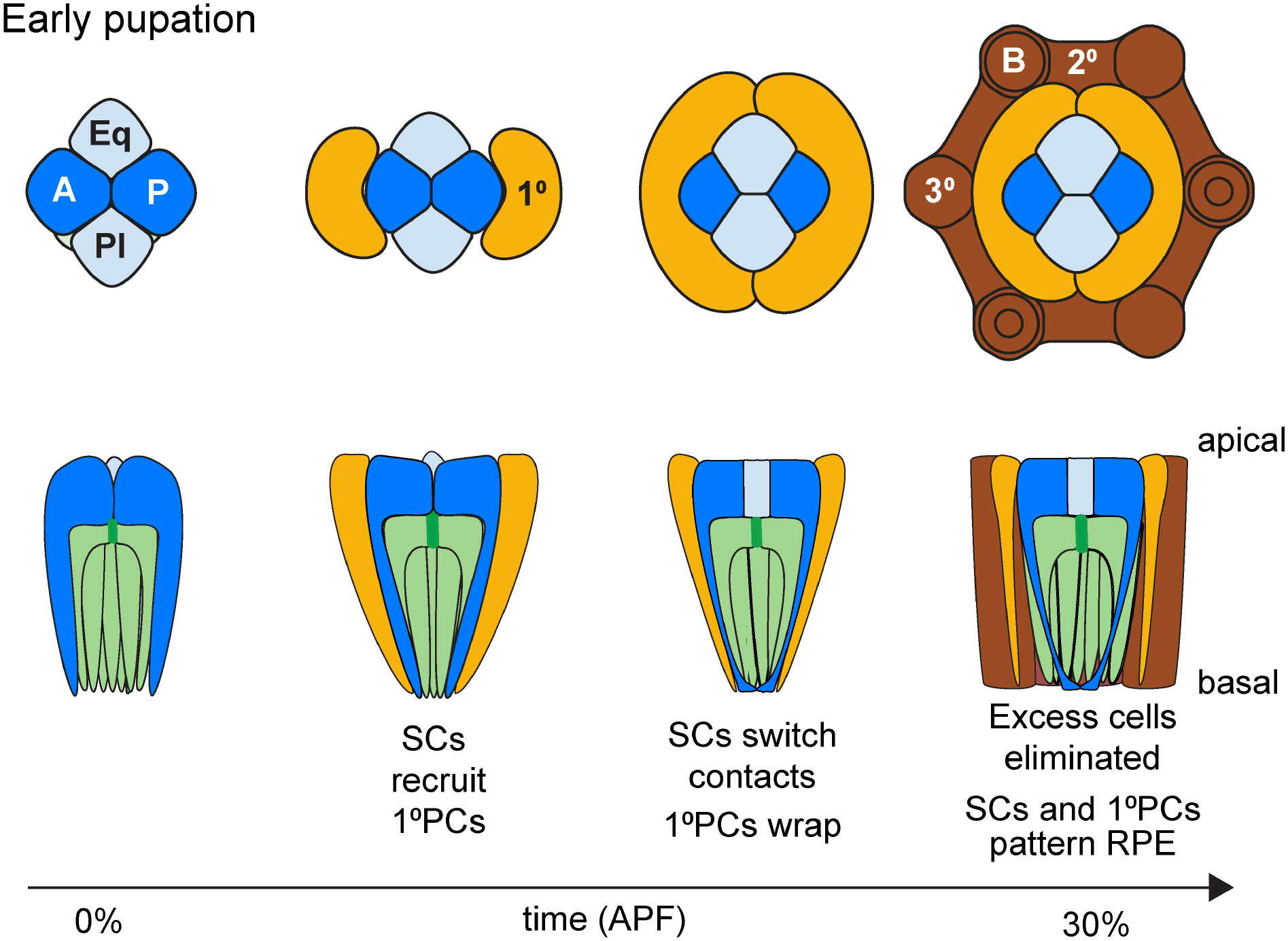
Color code as in Figure 1 for retina (green) and Semper cells (blue). Light orange = primary pigment cells (PPCs), dark brown = interommatidial cells (IOCs). IOCs include the highly pigmented elongated secondary (2°) and hexagonal tertiary (3°) cells (= i, RPE). Interommatidial bristles indicated with circles.
At the transcriptional level, both early and late retinal progenitors express the transcription factor-encoding genes sine oculis, eyes absent, and glass, with the late progenitors also expressing the Runt homology protein-encoding gene lozenge (Flores et al., 1998; Morrison et al., 2018; Pignoni et al., 1997; Tanaka-Matakatsu and Du, 2008; Yan et al., 2003; Zhang et al., 2006) (Fig. 2). From here, the different ommatidial cell types arise through a combinatorial transcriptional code and synergistic integration with differential Ras and Notch signaling levels (Charlton-Perkins et al., 2011b; Frankfort and Mardon, 2002; Hayashi et al., 2008; Nagaraj and Banerjee, 2003; Tomlinson et al., 2011; Voas and Rebay, 2004; Xu et al., 2000; Yan et al., 2003). The pivotal decision to develop as an R7 (the last retinal neuron to be specified) vs a SC (an accessory/glial fate) is made in a subset of late progenitors, called the R7 equivalence group. This group of 5 cells shares the expression of the proneural transcription factor-encoding gene prospero (pros) (Choksi et al., 2006; Doe et al., 1991; Kauffmann et al., 1996; Vaessin et al., 1991). Localized activation of Sevenless/Ras/MAPK signaling promotes R7 fate in a single R7 equivalence group member contacting the R8 PR, while the remaining four members acquire the SC default fate through high Notch and low Ras signaling (Flores et al., 2000; Lai and Rubin, 2001; Raabe, 2000; Simon, 1994; Tomlinson and Ready, 1987; Zipursky and Rubin, 1994) (Fig. 3). Combined, fly retinogenesis follows a developmental theme common to other parts of the fly and vertebrate nervous system where pools of “early” and “late” progenitors give rise to all-neuronal vs neuronal-glial cell types, respectively, with high Notch signaling driving glial fate (Cepko, 1993; Cepko et al., 1996; Dorsky et al., 1995; Flores et al., 2000; Gaiano and Fishell, 2002; Hojo et al., 2000; Lai and Rubin, 2001; MacDonald et al., 2015; Satow et al., 2001; Stiemke and Hollyfield, 1995; Vetter and Moore, 2001; Young, 1985).
Consistent with a glial specification paradigm for SC fate, transcription factors generally known to control glial cell fate are also expressed in early SCs. This includes the fly-specific factors glial cell missing (gcm), reversed polarity (repo), and tramtrack (ttk), as well as more deeply conserved gliogenic factors like pros/Prox1 and Pax2 (Badenhorst, 2001; Charlton-Perkins et al., 2011b, 2017; Choksi et al., 2006; Fu and Noll, 1997; Kato et al., 2015; Mavromatakis and Tomlinson, 2016; Shi and Noll, 2009; Soukkarieh et al., 2007; Wallace et al., 2016). Interestingly, pros/Prox1 and Pax2 cooperate in SCs to both control early glial fate decisions as well as later glial functions (Charlton-Perkins et al., 2011b, 2017)(Charlton-Perkins and Cook, in preparation). Given that these factors are each common to other sensory system glia, including the vertebrate retina intrinsically-generated radial Müller glia (MG) (Charlton-Perkins et al., 2011b, 2017; Cid et al., 2010; Freeman and Doe, 2001; Kavaler et al., 1999; Stanke et al., 2010), pros/Prox1 and Pax2 may be a part of a conserved genetic module that imposes sensory glial fate.
Semper cell subtypes
Despite the apparent common roles for the full quartet of SCs in the mature eye, early studies of retinal development in Drosophila revealed that not all four SCs are created equal. Instead, the “primary” anterior and posterior (A/P) SCs form before the “accessory” equatorial and polar (Eq/Pl) SCs (Tomlinson, 1985; Tomlinson and Ready, 1987) (Fig. 2). At the molecular level, these SC subtypes transiently differ in expression levels of cell fate determinants. Pax2 and its downstream target cut show higher expression in A/P SCs relative to pros, while pros is more strongly expressed in the Eq/Pl SCs (Charlton-Perkins et al., 2011b). SC subtypes continue to show gene expression differences at least through mid-pupation. For example, A/P SCs consistently express higher levels of Delta (Dl), which is a Ras/MAPK-induced Notch ligand. Additionally, reporters for Ras/MAPK signaling exhibit SC subtype-selective gene expression patterns (Parks et al., 1995; Tsuda et al., 2002; Wildonger et al., 2005). Consistent with SCs experiencing different signaling environments, the formation of only two SCs is often observed when the EGFR, Notch or Wingless (Wg)/Wnt signaling pathways are perturbed (Bhattacharya and Baker, 2009; Cordero and Cagan, 2010; Matsuo et al., 1997; Pickup et al., 2009; Rebay and Rubin, 1995; Treier et al., 1995). Intriguing comparative evidence for deeper evolutionary conservation of SC subtypes is provided by the shrimp-like marine crustacean Meganyctiphanes norvegica (Hallberg and Nilsson, 1983). In this species, SCs have diversified to the effect that the A/P SCs produce the lens while the Eq/Pl SC maintain contact with the PRs. The complete separation of lens secretion and PR support is intriguing with respect to the possibility that separable genetic and functional modules exist in SCs. Such subfunctionalization is rare, however, suggesting strong developmental constraints for maintaining the A/P and Eq/Pl subfate organization. Indeed, as described next, the SC subfates do play distinct roles in the following phase of ommatidial patterning.
Semper cell-directed patterning of the ommatidial array
Once SC recruitment is complete by early pupation, the differentiating SCs adopt active roles in the patterning of the remainder of the ommatidial unit. First, the apical portions of the SCs migrate over the top of the PRs, covering the surface of the retina (Fig. 2B). At this point, the A/P SCs are in physical contact with each other, pushing the Eq/Pl SC pair to the outside of the cluster (Cagan and Ready, 1989). In this configuration, the A/P SCs recruit the pair of primary pigment cells (PPCs) through Dl-driven Notch activation in neighboring progenitors (Nagaraj and Banerjee, 2007; Parks et al., 1995). After PPC recruitment, the Eq/Pl SCs surfaces migrate above those of the A/P SCs, establish contact with each other, and push the A/P SC pair apart (Cagan and Ready, 1989). From here, the SCs and PPCs join forces to promote the organization and patterning of the interommatidial pigment cells, i.e. the RPE (Bao and Cagan, 2005; Miller and Cagan, 1998) (Fig.3).
SCs drive their own mirror symmetry and steer their concentric arrangement with the PPCs and RPE through differential deployment of two families of cell adhesion proteins: Cadherins and Nephrin/Neph1-related Irre-C Recognition Module (IRM) proteins (Bao et al., 2010; Bao and Cagan, 2005; Fischbach et al., 2009; Grillo-Hill and Wolff, 2009; Grzeschik and Knust, 2005; Hayashi and Carthew, 2004). Both sets of proteins are concentrated at adherens junctions that form near the apical surfaces of cells (Harris and Tepass, 2010; Tepass and Harris, 2007). While all cells in the eye express E-cadherin, N-cadherin is restricted to the SC-SC interfaces during early ommatidial patterning. This selective enrichment in SCs helps to stabilize the stereotypic “soap bubble” organization of the SC quartet (Hayashi and Carthew, 2004). In addition, N-cadherin controls the SC-PPC-RPE concentric organization by driving MyoIIdependent contractility differences across the surface of the eye (Blackie et al., 2020; Chan et al., 2017; Hayashi and Carthew, 2004).
In addition to these Cadherin-mediated functions, Nephrin and Neph1-like IRM-driven adhesion differences are likewise responsible for correct ommatidial patterning (Bao et al., 2010; Bao and Cagan, 2005; Grzeschik and Knust, 2005; Reiter et al., 1996). The Nephrin and Neph1-like subfamilies of adhesion factors show differential distribution in the eye. The Nephrin paralog pair, Hibris and Sticks-N-Stones, is expressed in SCs and PPCs while the Neph1 paralogs Roughest/irregular chiasm (Irre) and Kin of Irre (Kirre) are restricted to the interommatidial cells (IOCs) (Bao et al., 2010; Fischbach et al., 2009). Homophilic interactions between the Nephrin IRMs ensure SC pair switching and PPC wrapping of the SC quartet by dictating maximal contact among SCs and PPCs. This process also effectively separates the apical surfaces of SCs from surrounding interommatidial precursor cells (Bao and Cagan, 2005; Grillo-Hill and Wolff, 2009).
Whereas Nephrin-associated homophilic interactions drive central ommatidial organization, stronger heterophilic interactions between the Nephrins and Neph1-like molecules maximize PPC-RPE contacts, ultimately leading to the efficient separation of individual ommatidia (Bao et al., 2010; Bao and Cagan, 2005). Importantly, SC-dependent signals are essential for directing this interplay of Nephrin-Neph1 interactions. For instance, the same Dl-Notch activation that promotes PPC recruitment also ensures robust Nephrin expression in these cells, leading to the robust accumulation of Neph1 at the PPC-IOC interface (Bao, 2014). In this way, SCs drive an inside-out signaling relay that influences cell adhesion molecule expression two cell diameters away. This process, combined with Myo-II mediated biomechanical forces, pulls individual ommatidia together (Blackie et al., 2020; Chan et al., 2017; Johnson et al., 2011; Larson et al., 2010), bringing the morphology of the differentiating eye one step closer to its mature architecture.
Not much is known regarding the molecular pathways controlling the differential expression of the adhesion molecules needed to drive ommatidial organization. However, it is worth noting that in the radial glia of the zebrafish retina, i.e. the MG, the Nephrin and Neph1 IRM proteins Nphs1 and Kirre1a ensure that the highly ordered tiling of the retina is properly established (Charlton-Perkins et al., 2019). Further notable is that, in both fish and flies, Pax2 lies upstream of IRM expression to control similar patterning processes (Charlton-Perkins et al., 2019) (Charlton-Perkins and Cook, in preparation). This raises the intriguing possibility that a deeply conserved Pax2-IRM regulatory module controls retinal subunit organization.
Semper cells as RPE survival mediators
Besides regulating the concentric organization and patterning of SCs, PPCs and RPE, SCs further assist in the final shaping of the Drosophila eye by influencing programmed cell death. In this context, SCs have been implicated in at least two peaks of apoptosis that unfold during pupal compound eye development (Baker, 2001; Baker and Yu, 2001; Brachmann and Cagan, 2003; Cordero et al., 2004; Lim and Tomlinson, 2006; Lin et al., 2004). First, in the main portion of the eye, SCs provide EGFR-dependent survival signals to IOCs. These signals allow IOCs to avert Notch-dependent pruning during the final stages of RPE patterning, leading to the appropriate separation of individual ommatidia (Miller and Cagan, 1998; Monserrate and Brachmann, 2007; Querenet et al., 2015). Second, at the periphery of the eye, SCs promote survival of the RPE, but in addition, induce death of non-RPE cells (Kumar et al., 2015). This striking dual property is driven by Wg/Wnt signaling from the head capsule tissue surrounding the ocular field. In particular, Wg from the head capsule promotes restricted wg expression in SCs located at the periphery of the eye, which in turn leads to the localized activation of Snail transcription factor expression in the SCs (Kumar et al., 2015). These peripheral SCs promote RPE survival, while in parallel, remove remaining ommatidial cells (including the SCs themselves) through apoptosis (Kumar et al., 2015). As a result, incomplete ommatidial units are eliminated at the edge of the eye and the retina becomes optically isolated from the rest of the head by a thick band of RPE cells.
Besides these pupal functions, SCs contribute to IOC progenitor survival in the late larval retina. Here again, studies on Pax2 provide critical insights, as mutations in this SC fate determinant cause widespread cell death in imaginal discs, producing adult eyes largely lacking an RPE (Fu and Noll, 1997; Siddall et al., 2003). Notably, similar precocious cell death is observed in animals lacking sufficient EGFR activation (Baker and Yu, 2001). As EGFR contributes to Pax2 expression, these results raise the possibility that Pax2 is essential for this signal (Flores et al., 2000). Alternatively, or in addition, because Pax2 can suppress Dl expression (Charlton-Perkins et al., 2011b), young Pax2-positive SC progenitors could delay Notch-dependent apoptotic signals by blocking the expression of its ligand. Of note, Pax2 plays pro-survival roles in other developmental systems, including the vertebrate eye (Bosze et al., 2021; Bouchard et al., 2002; Burton et al., 2004; Park et al., 2006; Viringipurampeer et al., 2012). Therefore, the fly eye provides a valuable model for dissecting a deeply conserved Pax2-associated cell survival regulatory module (Park et al., 2006).
Facilitating photoreceptor morphogenesis
The above discussion highlights the role of SCs in patterning the non-neuronal cell types in the fly eye. In addition, SCs help build the retina’s neuronal compartment and provide structural support to the PRs. The first SC-dependent role in retinal morphogenesis occurs soon after the start of pupation. At this time, the apical surface of the SCs migrate above the PRs. As the SCs and PRs are attached through zonula adherens (ZAs), this migration leads to the inward rotation of the PRs apical membrane 90 degrees (Longley and Ready 1995). This reorientation of the PR apical surfaces is a critical process that ultimately allows the light-gathering rhabdomeres to extend in the distal-proximal direction to reach from the surface to the base of the retina (Longley and Ready 1995) (Fig. 4). Eventually, the SCs form an electron-dense “rhabdomere cap” that binds the SCs to the PRs via integrin-rich desmosome-like structures (Longley and Ready, 1995) (Fig. 5).
Figure 4: Final stages of Drosophila eye development.
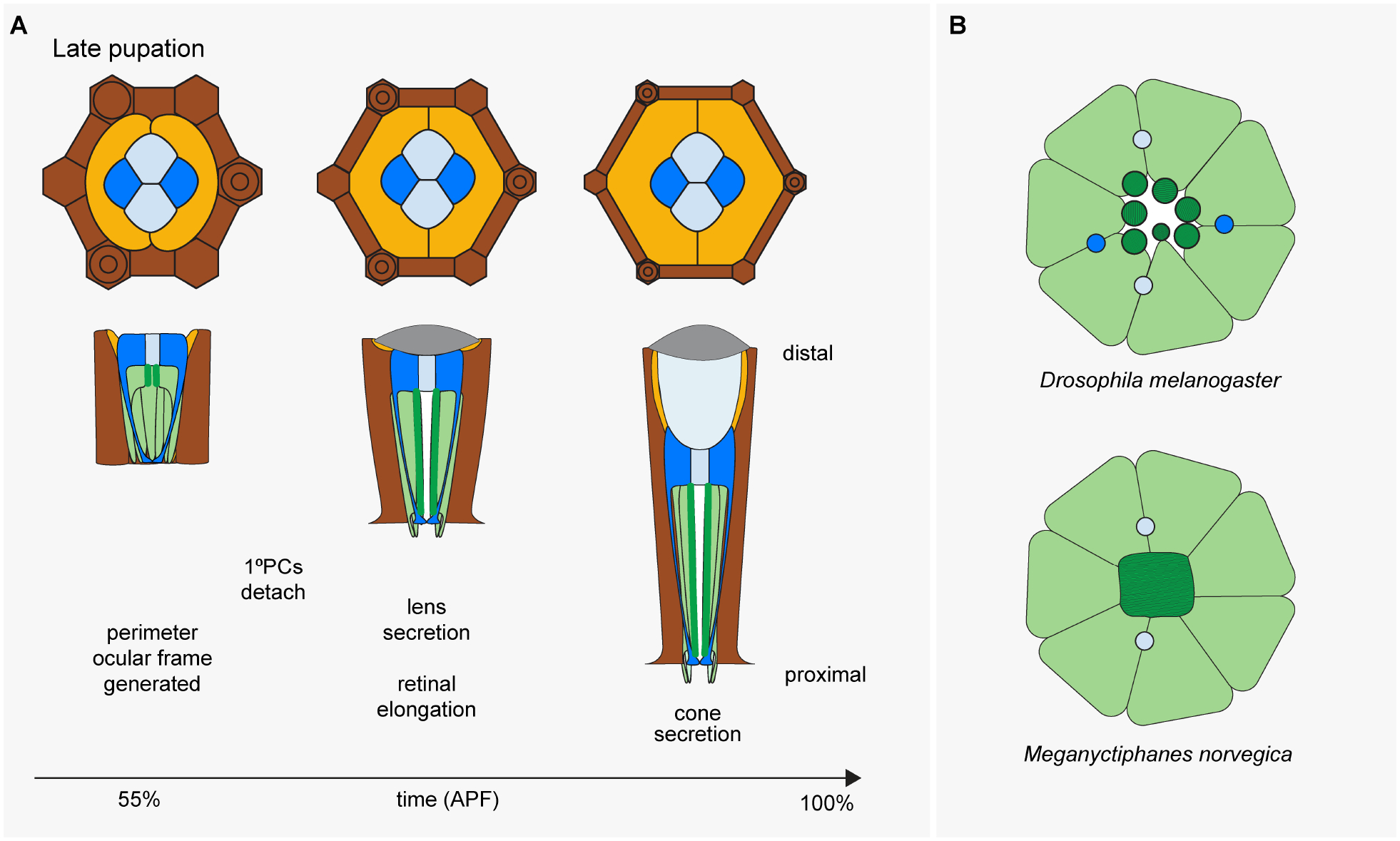
(A) Same color code as Figure 3. (B) SC-based interretinular fiber locations in the adult eyes of Drosophila melanogaster (open rhabdomeres) and Meganyctiphanes norvegica (fused rhabdome). Rhabdomeres are indicated in dark green, while photoreceptor cell bodies are in light green.
Figure 5: Junctions present in mature Semper cells.
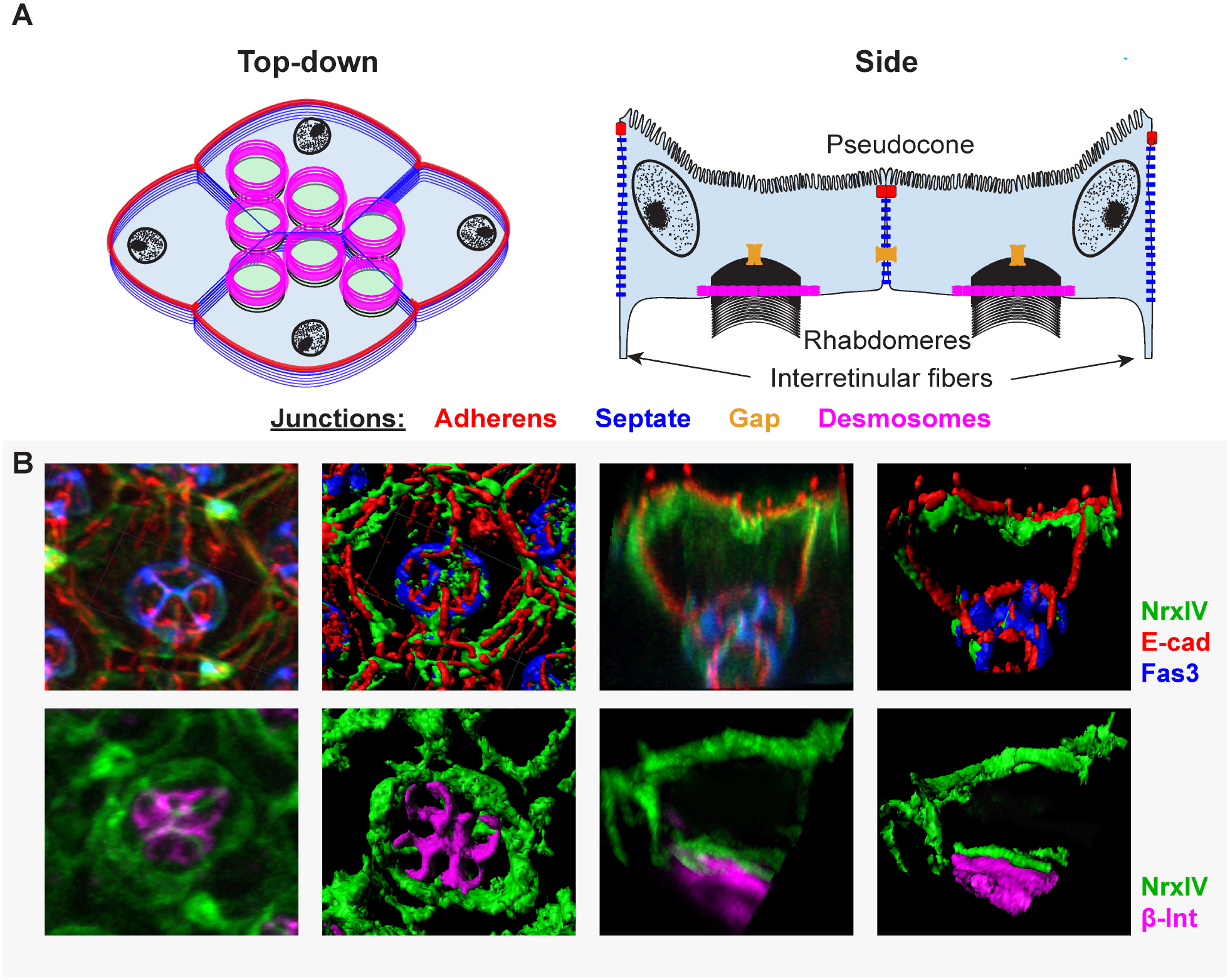
(A) Top down and side view representations of different junctional complexes found in SCs. These include adherens junctions (zonula adherens, red), septate junctions (ladder-like junctions found beneath zonula adherens, blue), and gap junctions (yellow). Confocal images represent individual ommatidia stained for: the septate junction markers Neurexin IV (NrxIV, green) and Fasciclin 3 (Fas3, blue) and the adherens junction marker E-cadherin (E-cad, red) (top) or NrxIV (green) and hemi-desmosome marker β-integrin (β-int, magenta) (bottom).
Just as SCs cover the surface of the PRs apically, they also form a plate below the PRs basally. Plate formation proceeds in a pairwise fashion. The A/P SCs initially insert their endfeet into the center of the ommatidial core at ~14% pupation, while the Eq/Pl SCs insert their endfeet at ~37% pupation. Together, the quartet of SC feet form a plate at the base of the retina and connect to the retinal floor through integrins. Once the PRs are oriented and encapsulated at either end, their actin-rich apical membranes begin to expand and grow proximally until reaching the SC-generated floor, where they attach with basal ZAs. The rhabdomeres then continue expanding and deepening through the entire length of the retina in two additional growth spurts at ~55 and 75% pupation (Ready, 2002). While much of rhabdomere growth is likely intrinsic to PRs, evidence suggests SCs also contribute to this process. For example, Pax2 mutant eyes have abnormally shaped and shortened PR rhabdomeres, suggesting that the SC-PR interface is necessary for proper rhabdomere elongation (Charlton-Perkins et al., 2017; Fu and Noll, 1997).
Stretching to connect the apical cap and the basal plate of the SCs are thin processes, called interretinular fibers. One per SC, these fibers run immediately adjacent to the PRs’ zonula adherens (ZA) junctions (Müller et al., 2003; Waddington and Perry, 1963) The presence and positioning of the SC fibers are remarkably conserved across Pancrustacea, highlighting their likely importance in supporting the integrity of the retina(Fischer et al., 2000, 2019; Horridge, 1966; Melzer et al., 2000, 1997; Menzel, 1972). Interestingly, at least in some arthropod eyes, microtubules are present in these SC extensions, raising the possibility that vesicular transport occurs between the distal SC soma to the endfeet (Burton and Stockhammer, 1969; Chen et al., 2019; Horridge and Giddings, 1971; Meyer- Rochow, 1977) (U. Wolfrum, personal communication). In this respect, it is worth noting that, while other cells in the fly eye have their pigment granules distributed through the cell body, the ommochrome pigment granules found in SCs are specifically located in the SC feet but absent in their soma at the top of the eye (Wolff and Ready, 1993). This asymmetric distribution thus allows light to pass through apically, and perhaps prevents light from penetrating into the brain at the base (Tomlinson, 2012; Waddington and Perry, 1963).
Drosophila PRs generate an extracellular matrix filled lumen, called the inter-rhabdomeric space (Husain et al., 2006; Nie et al., 2014; Zelhof et al., 2006). Because the SCs form a basket around the PR cluster, this lumen is encapsulated by the SCs. Thus, once mature, the SCs contact 3 three separate receptacles: the pseudocone, the inter-rhabdomeric space, and the brain-retina-barrier. The latter has been shown to involve a third important intercellular connection particularly abundant among SCs: septate junctions (SJs) (Banerjee et al., 2008). These junctions form just basal to ZAs in insects and utilize proteins present in junctions associated with barriers and paracellular transport in vertebrates, such as tight junctions (TJs) and the axo-glial nodes of Ranvier (Banerjee et al., 2006; Faivre-Sarrailh, 2020). Support for similar barrier functions in the fly eye comes from genetic analysis of Neurexin-IV (NrxIV)/dCaspr, a deeply conserved factor required for SJ/TJ formation. NrxIV protein is present in the basal feet of the SCs and RPE cells where it controls the fly brain-retina-barrier (Banerjee et al., 2008). Interestingly, while NrxIV is present in both RPE and SCs proximally, it is specifically enriched in SCs distally (Banerjee et al., 2008). Another SJ-associated protein, Fasciclin 3, is likewise enriched distally in SCs (Charlton-Perkins et al., 2017). It thus appears that SC-SC septate junctions are unique, perhaps to form strong selective barriers between the vitreous-like pseudocone, the inter-rhabdomeric space and the brain-retina-barrier (Fig. 5).
Secreting the corneal lens
Lens formation and composition:
As all cell specification and patterning events are complete by mid-pupation, the developing eye transitions to terminal differentiation events (Cagan and Ready, 1989; Ready, 2002; Waddington and Perry, 1960). At this point, i.e. by about 50% pupation, the SCs, together with primary and secondary pigment cells, begin to secrete the biconvex corneal lens (Cagan and Ready, 1989). This laminated cuticular structure is largely finalized by 75% of pupal development, although some protein incorporation has been observed in newly eclosed adults (Yoon et al., 1997). Drosocrystallin and Retinin represent the major cuticular constituents of the lens and are expressed by both the SCs and PPCs (Charlton-Perkins et al., 2017; Janssens and Gehring, 1999; E. Kim et al., 2008; Komori et al., 1992; Kryuchkov et al., 2020; Stahl et al., 2017; Xu et al., 2004). There are also two lower abundance cuticular proteins Cpr72Ec and Cpr66D, that are primarily expressed in the interommatidial pigment cells (Kryuchkov et al., 2020; Stahl et al., 2017). Such cell type-specific expression levels presumably set up the differential gradients needed to generate the curved lens structure and achieve appropriate refractive properties.
Corneal lens surface formation:
Another important aspect of the corneal lens is its outer coating, a lawn of tiny protuberances known as corneal nipples. A bioengineering inspiration for surface coatings on solar cells and optic lenses (Brunner et al., 2012; Dewan et al., 2012), these nanostructures are anti-reflective and thus enhance light transmittance. Recent studies revealed that Drosophila corneal nipple formation requires retinin and the arachidonic acid-specific lysophospholipid acyltransferase-encoding gene farjavit (Kryuchkov et al., 2020). Importantly, the failure of corneal nipple formation is considered to be the cause of the glossy or glassy eye phenotypes seen in a variety of Drosophila mutants (Bernhard et al., 1963; Kryuchkov et al., 2011). This includes the eye-specific allele of Pax2, spapol (named for its sparkling, polished eye appearance), thus tying Pax2 to corneal lens surface formation.
Conservation of Pax-Sox mediated crystallin production:
As the most abundant corneal lens protein in Drosophila, Drosocrystallin has been most thoroughly studied (Komori et al., 1992; Stahl et al., 2017). Like other crystallins in animal lenses, Drosocrystallin is of critical functional importance outside of the lens, including in the peritrophic membrane of the gut, revealing broader requirements for this structural protein (Charlton-Perkins et al., 2011a; Dziedzic et al., 2009; Janssens and Gehring, 1999; Kozmik et al., 2003; Kuraishi et al., 2011; Piatigorsky et al., 1988; Vopalensky and Kozmik, 2009). Molecularly, an enhancer that recapitulates Drosocrystallin’s expression in the eye is activated by Pax2, further exemplifying the pervasive role of this factor in SC biology (Dziedzic et al., 2009; Janssens and Gehring, 1999). Remarkably, Pax2 not only activates a fly crystallin gene enhancer, but can also substitute for another Pax gene, Pax6, to activate an enhancer of the chicken gamma-crystallin gene DC5, when the latter is introduced into the fly (Blanco et al., 2005; Kamachi et al., 2001; Kozmik, 2005). Furthermore, just as Pax6 cooperates with Sox2 to activate the DC5 enhancer in their authentic host, i.e. the chicken, Pax2 cooperates with Drosophila SoxN to regulate the enhancer in flies (Blanco et al., 2005; Kamachi et al., 2001; Kozmik, 2005). These findings raise the possibility of a deeply conserved Pax-Sox regulatory module in charge of lens production.
The interchangeability of SoxN and Sox2 is consistent with them both being members of the SoxB1 subgroup of the SRY-related HMG-box transcription factor gene family (Guth and Wegner, 2008). Similarly, the participation of Pax6 and Pax2 in bilaterian crystallin gene regulation is explained by shared ancestry and functional conservation. Both factors are phylogenetically related to the PaxB factors in the Cnidaria (Suga et al. 2010; Kozmik et al. 2003), an ancient metazoan phylum that diversified in parallel to the emergence of the Bilateria. In this ancient group, lens-forming eyes originated multiple times (Picciani et al., 2018). Functional homology of cnidarian PaxB with fly Pax2 has been demonstrated by studies which revealed that the PaxB homologs of two distantly related cnidarian species, the box jellyfish Tripedalia cystophora (Cubozoa) and the hydrozoan Cladonema radiatum rescue the eye-specific Pax2 spapol mutant phenotype (Kozmik et al., 2003; Suga et al., 2010). Remarkably, with respect to crystallin regulation, T. cystophora PaxB was found to activate the promoters of three crystallin genes of the same species (Kozmik et al., 2003)). Thus, the transcriptional control of crystallin genes by PaxB factors appears to constitute a deeply conserved aspect of animal eye development. The intriguing prediction that SoxB is likewise involved in cnidarian crystallin gene regulation remains to be tested.
Secreting the pseudocone
Following lens secretion, the SCs switch to secreting the pseudocone, the extracellular vitreous-like substance that represents the dipteran-type of crystalline cone. In Drosophila, this secretory switch happens at 75% pupation. As the pseudocone gains in volume, the SCs are pushed away from the hard cuticular lens, with the major portion of their cell bodies being pressed onto the PRs’ tops, and their nuclei being forced aside, thus minimizing light interference (Ready, 2002). While crystalline cone formation is the most characteristic role for SCs, very little is known about its content, even in the well-studied Drosophila model. The same holds for how the correct focal distance between the corneal lens and the PRs is achieved to establish what is known as emmetropia. Recent work in a range of arthropod eyes revealed that, unlike the single lens eye types in vertebrates and squids, arthropods do not seem to require visual input to reach emmetropia, suggesting that an intrinsic system is in place for this process (Owens et al., 2020; Sivak and Sivak, 2019). As for the pseudocone contents, the only known protein marker identified thus far is an 85 kDa glycoprotein recognized by the now unavailable monoclonal antibody 3G6 (Edwards and Meyer, 1990). Originally isolated for its specificity for neuropil glia in crickets, the epitope recognized by this 3G6 is concentrated in crystalline cones in insects, crustacea, and even myriapods (Edwards and Meyer, 1990; Meyer et al., 1987). While preliminary, these data indicate the existence of a deeply conserved molecular component produced by both nervous system glia and SCs.
More recently, the amino acid B-alanine was also shown to be enriched in the Drosophila pseudocone (Borycz et al., 2012; Han et al., 2017; Xu et al., 2015). B-alanine is a substrate for both energy production and the recycling of the fly PR neurotransmitter histamine. Knockdown of the B-alanine synthesis enzyme pyd3 in SCs resulted in weaker neuronal activity in neighboring PRs, but showed no changes in neurotransmission (Charlton-Perkins et al., 2017). These observations hint at the possibility that the pseudocone not only contributes to the focusing of the eye, but also stores metabolic substrates, not unlike other fluid-filled tissues in the vertebrate nervous system (e.g., ventricles, vitreous).
Semper cell-mediated homeostatic support in the adult retina
Until recently, our understanding of SC function was confined to their role in supporting ommatidial development and structure. However, SC-targeted transcriptomic and genetic approaches have uncovered an additional glial support role for these cells in the adult retina via deeply conserved effector genes (Charlton-Perkins et al., 2017). Glia maintain nervous system function by providing structural and physiological support. Structurally, the SCs radially encapsulate the rhabdomeres and provide a Pax2-dependent scaffold for rhabdomere elongation as alluded to above (Charlton-Perkins et al., 2017). As with many radial-like glia, including retinal MG, the mechanics of this glial structural support requires further exploration (MacDonald et al., 2015).
Physiologically, SCs participate in at least two glial support roles: energy maintenance and ion homeostasis. In highly active areas of the nervous system like the retina, the exceptional energy demand is met by neuron-glia metabolic coupling (Bittern et al., 2020; Country, 2017; Magistretti, 2004). In the most simplistic model of this interaction, glia maintain glycolysis utilizing glycogen or glucose reserves to generate citric acid cycle substrates (Tsacopoulos et al., 1994; Tsacopoulos and Magistretti, 1996; Volkenhoff et al., 2015). These resources in turn are transported or diffuse to neighboring neurons for mitochondrial ATP production (Rittschof and Schirmeier, 2018). Consistent with similar coupling in other insect eyes, ultrastructural studies of the honeybee retina uncovered an abundance of glycogen reserves in SCs (Perrelet 1970). SCs also appear to use similar metabolic effectors as vertebrate glia for providing nutrients to their intimately linked PRs (Hurley et al., 2015; Reichenbach and Bringmann, 2013; Rittschof and Schirmeier, 2018; Vohra and Kolko, 2018). This is suggested by findings in genetic studies that Drosophila SCs require the glucose transporter Glut1 and lactate dehydrogenase, dLdh, to help sustain neighboring PR function (Charlton-Perkins et al., 2017).
Another interesting function of SCs is glutamate recycling. Glutamate is the primary neurotransmitter in the vertebrate retina, while the fly retina uses histamine (Hardie, 1987; Sarthy, 1991). However, highly active neurons release glutamate and ammonium as metabolic byproducts that are recycled by neighboring glia to either alanine (for reuse) or glutamine (as storage for future energy demands) (Rittschof and Schirmeier, 2018; Tsacopoulos et al., 1997, 1994). The glutamate-sodium symporters EAAT1 and EAAT2 as well as the glutamate-ammonia ligase glutamine synthase (GS2) are strongly expressed in SCs and, in the case of EAAT1, similarly help to sustain PR function (Charlton-Perkins et al., 2017). In light of this, SCs have become an attractive model to study glutamate recycling in the context of glial energy production rather than neurotransmission (Chaturvedi et al., 2016; Han et al., 2017; Xu et al., 2015).
Potassium ion homeostasis is another essential support function for glia to maintain the membrane potentials of highly active neurons (Beckner, 2020). Throughout the nervous system, including the retina, this support relies on a complex potassium-ion buffering system consisting of glia-expressed inward rectifying potassium (Irk, or Kir) channels and an interconnected glial Connexin network of gap junctions (Beckner, 2020; Reichenbach and Bringmann, 2013). Potassium ions that are released during neuronal activity have an immediate impact on membrane potential; however, uptake of these by interconnected glia through Irk channels offsets this change, thereby stabilizing neural activity (Beckner, 2020). While the Drosophila SCs express two Irks (Irk1 and Irk2), knockdown experiments indicate that Irk2 is specifically critical in SC-mediated support of PR activity (Charlton-Perkins et al., 2017). SCs are also uniquely enriched with the Connexin-related innexin gene family member innexin 6 (Stebbings et al., 2002), raising the possibility that SCs may use a similar gap junction network for maintaining ion homeostasis in the compound eye.
The SCs’ ability to provide metabolic and homeostatic glial support is a salient finding that invites further studies to identify additional functions they might perform. For instance, SCs are enriched with the single Na/K-ATPase alpha subunit (Atpα) and two of its beta subunits (Nrv2 and Nrv3), each of which are required for PR activity (Charlton-Perkins et al., 2017). Na/KATPases (Na/K pumps) are broadly expressed in animal cells, executing a diverse set of functions ranging from transport of water, ions, neurotransmitters, amino acids, to that of metabolites. Moreover, Na/K pumps are a critical component of several cellular junctions (Clausen et al., 2017; Genova and Fehon, 2003; Rajasekaran et al., 2001; Vagin et al., 2006). We therefore expect further metabolic and structural roles of SCs to emerge from future studies.
A cooperative Pax2-Prox module for retinal gliogenic potential
The above discussion highlights a number of deeply conserved elements that ensure the proper organization (e.g. Nephrins) and maintenance (e.g. Glut1, Kir channels, and EAAT1) of retinas in the two major animal eye types. This raises the question as to whether evolutionarily more deeply seated factors contribute to retinal glia support roles. Candidates for these are the transcriptional regulators pros and Pax2, which not only cooperate during early SC specification, as mentioned earlier, but also in later functions. For example, Pax2 provides photoreceptor structural support, whereas pros provides homeostatic support (Charlton-Perkins et al., 2017).
A closer look reveals some parallels of Pax2 functions between fly and vertebrate eyes. In flies, Pax2 ensures the restriction of retinal neurons to the retina, participates in ommatidial organization, and drives glial fate (Charlton-Perkins et al., 2011b, 2017; Siddall et al., 2003). In vertebrates, Pax2 is expressed in the optic stalk where it works to close around the axons of the optic nerve, form the boundary between the neural retina and glial optic stalk, and promote glial fate in the optic stalk (Bosze et al., 2021; Schwarz et al., 2000; Soukkarieh et al., 2007; Torres et al., 1996). Thus, in both cases, Pax2 controls the organization of the ocular structure and glial formation. While most genetic studies on vertebrate Pax2 have focused on its role in optic nerve glia, Pax2 is also expressed in other retinal glia. In animals lacking intra-retinal vasculature (e.g. fish and chick), Pax2 is expressed in MG, whereas in vascularized retinas (e.g. mammals), Pax2 expression is instead expressed in the intra-retinal astrocytes (Stanke et al., 2010). While the cause for this apparent switch has not specifically been addressed, it is notable that this change in Pax2 expression correlates with the glia most involved in the blood-retina-barrier. Given Pax2’s association with other neural tissue barriers (Bosze et al., 2021; Rhinn and Brand, 2001), it seems promising to further explore the role of Pax2 in its boundary-forming processes using the fly eye as a model. In addition, it is worth noting that the other vertebrate Pax2-related factors, Pax5 and Pax8, are transcriptomically enriched in mammalian MG, raising the possibility that radial glia-enriched expression has subfunctionalized to other PaxB orthologs during evolution (Hoang et al., 2020; Nelson et al., 2011; Roesch et al., 2008; Uhlén et al., 2015) (https://www.proteinatlas.org/ENSG00000196092-PAX5/celltype; https://www.proteinatlas.org/ENSG00000125618-PAX8/celltype). We predict that comparative transcriptomics for PaxB-related targets in various model systems will unearth deeply conserved Pax-mediated gene regulatory modules.
The possible conservation of pros functions in retinal gliogenesis is less clear. In Drosophila, pros is initially weakly expressed in a subpopulation of retinal precursors with neuro-glial potential. Pros then accumulates and maintains strong expression in the last neuron to form, i.e., R7, where it controls later terminal differentiation events for this UV-sensitive PR (Cook et al., 2003; Kauffmann et al., 1996; Morey et al., 2008). Weaker, more dynamic expression is observed in the SCs, where its presence is likewise important for terminal differentiation events (Cook et al., 2003; Kauffmann et al., 1996). For example, the knockdown of pros in SCs leads to reduced PR activity and premature retinal degeneration in adult flies (Charlton-Perkins et al., 2017). This gliogenic role is in line with studies of pros in the developing Drosophila nervous system where, in addition to its well-established role in neural progenitor cell cycle exit, pros initiates the expression of several glia-associated gene products and aids astrocyte differentiation (Choksi et al., 2006; Peco et al., 2016).
In mice, Prox1 is mostly known for its role in neural progenitor cell cycle exit and neuronal differentiation. For example, in the mouse retina, Prox1 affects early progenitor cell cycle exit, participates in horizontal cell fate decisions, and later in bipolar cell differentiation (Dyer et al., 2003). However, Prox1 expression is also detected in subsets of vertebrate glial cells, including radial glia, astrocytes, and oligodendrocytes (Bunk et al., 2016; Kato et al., 2015; Lavado and Oliver, 2007). In addition, weak expression of Prox1 has been reported in both fish and mammalian MG (Charlton-Perkins et al., 2019; Cid et al., 2010), raising the possibility that Prox1 plays later glial roles not yet tested in the vertebrate eye. Remarkably, the pros homolog in the nematode Caenorhabditis elegans also directs glial function, revealing the importance of pros for gliogenesis even in a highly reduced sensory system (Wallace et al., 2016).
As a whole, these findings suggest that pros/Prox1 may represent an ancient player in providing a “neural” environment during early development, and homeostatic glial function in more mature cell types, while Pax2 plays important roles in promoting glial fate and providing structural support for the developing retina.
Evolution and conservation of retinal support cells
As this review underscores, SCs control a wide range of functions: patterning, cell survival, lens and pseudocone secretion, retinal morphogenesis, and PR-directed glial support. Of note, the recently reported expression of a specialized opsin in both SCs and pigment cells of butterfly compound eyes gives reason to expect even further roles yet to be uncovered (Macias-Muñoz et al., 2019). Besides cementing the broad significance of SCs for retinal development and function, all of this begs the question of how this multifunctional cell type originated. As highlighted throughout the review, comparative genetic data indicate four potentially deeply conserved candidate modules representing defined SC functions: a PaxB-SoxB1 module related to crystallin gene expression, a Pax2-pros module controlling gliogenesis and glial support functions, a Pax2-IRM module directing cellular patterning events and a separate Pax2 module regulating cell survival (Fig. 6). The strikingly pervasive involvement of Pax2 in these modules is consistent with its deep associations with sensory system formation (Fritzsch et al., 2005; Vopalensky and Kozmik, 2009).
Figure 6: Hypothesized gene regulatory modules utilized by retinal support cells.
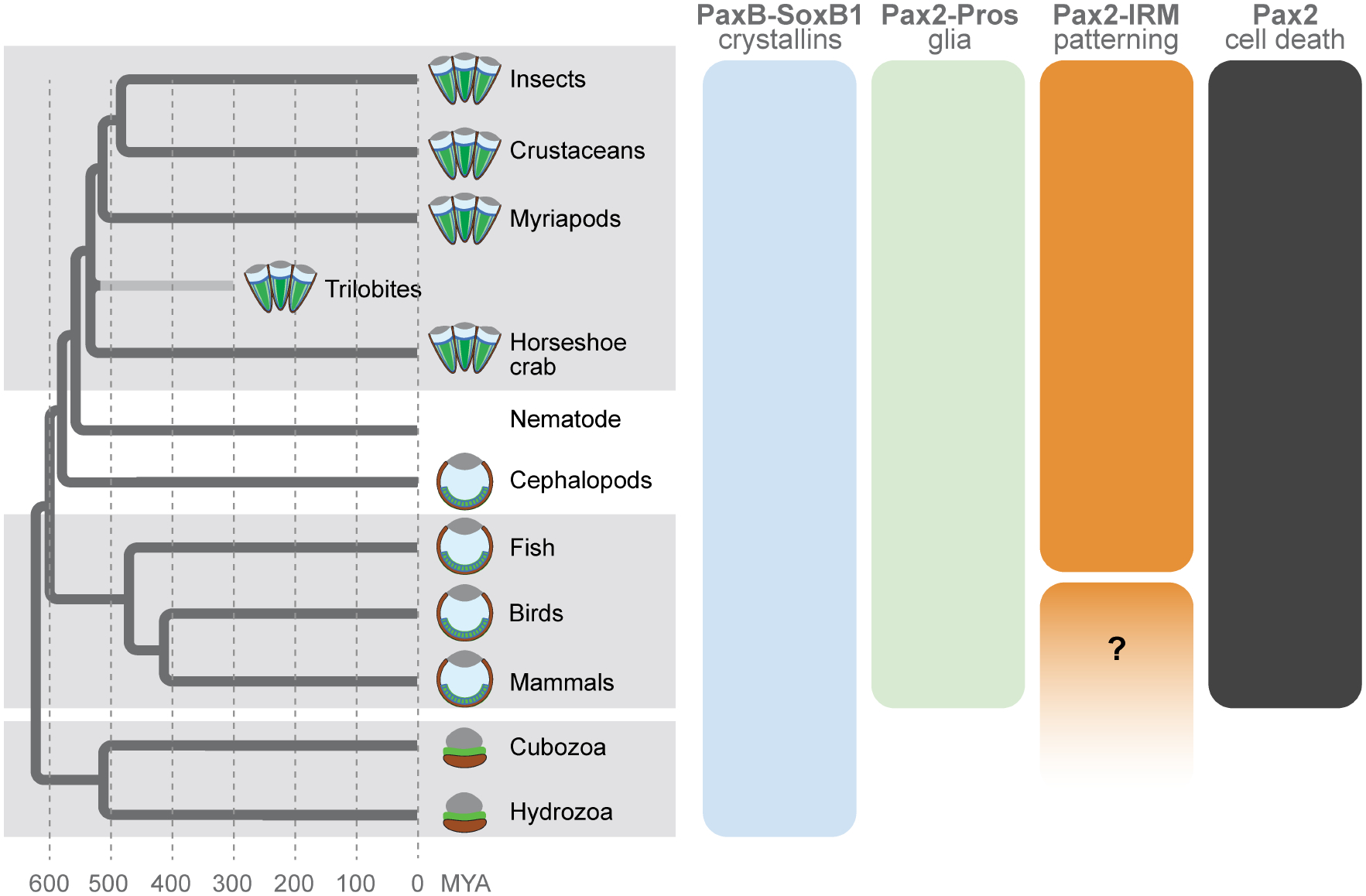
Phylogenetic tree depicts the relationships between taxa and species explored in the review with arthropods represented by five major lineages: insects, crustaceans, myriapods, the extinct trilobites, and chelicerates. The latter are represented by its only member with a compound eye, the horseshoe crab Limulus polyphemus. Schematics at branch tips depict generalized versions of the major eye types, ranging from compound eyes (arthropods) to independently evolved camera eyes of squids (cephalopods) and vertebrates (e.g. fish, birds, and mammals), as well as the independently derived eye types in the cnidarian species like Tripedalia cystophora (Cubozoa) and Cladonema radiatum (Hydrozoa). Bars to the right depict hypothesized ancient gene regulatory modules utilized in diverse visual organs as discussed in the text.
Compelling support for the modular nature of SC functions at the genetic level is provided by the earlier mentioned crustacean M. norvegica, in which only the A/P SCs are lens-secreting, while only the Eq/Pl SCs reach fibres to the retinal floor (Hallberg and Nilsson, 1983). In this case, it is tempting to speculate that the putative PaxB-SoxB1 lens production module has become A/P SC-specific, while the Pax2-pros PR support module may be partitioned into the Eq/Pl SCs. While such a possibility is intriguing, the existence of these candidate modules requires further scrutiny, by necessity in a broader sample of species. Their possible existence, however, speaks to a scenario in which SCs originated by co-option of conserved kernel-like gene regulatory modules that drive distinct cellular functions (Arendt et al., 2016). The same scenario, of course, may apply to MG, a vertebrate counterpart to arthropod SCs with respect to retinal homeostatic and structural support. While both cell types can be traced back over hundreds of millions of years to the early evolution of Bilateria (Fig. 6), pinpointing the existence and nature of SC/MG-like precursor cells represents a greater challenge. Recent morphological and immunohistochemical studies in a variety of protostomes provided evidence for a secretory radial glial cell type in simpler sensory systems, raising the possibility that such a cell type did emerge at the beginning of bilaterian evolution (Hartline, 2011; Helm et al., 2017; Losada-Perez, 2018; Rey et al., 2020). The shared genetic modules of SCs and MG could thus be explained through inheritance from a common visual system-specific primordial radial glia.
Concluding remarks
The important question of the existence of primordial radial glia should be addressed by a new generation of studies of glial support sources in animal visual organs. While mammalian camera lens eyes and arthropod compound eyes represent key reference points, similarities in the structures, functions, and genetic programs of different eye types in the animal tree of life highlight the potential for expanding cellular and molecular studies of deeply conserved gliogenic programs. This includes species with no apparent accessory cells except pigment cells, as the latter are part of the PR support system in both vertebrates and invertebrates. The most critical information on the origin of SC/MG can be expected to emerge from the camera eyes of squids (Cephalopoda) (Fig. 6). The retinas of these sophisticated visual organs are populated by pigmented radial glial-like support cells that extend proximo-distal fibres (Koenig et al., 2016; Saibil, 1990; Tomarev et al., 1997) The time seems ripe to elucidate the transcriptomic cell signatures in this third type of high resolution animal eye (Nilsson, 2013), given the success of temporal cross-species developmental transcriptomic studies. Such studies have already started to provide new insights into the existence of conserved regulatory modules responsible for cell type-specific development, structure, and function in the retina (Charlton-Perkins et al., 2019, 2017; Hoang et al., 2020). Thus, cultivating this area of retinal biology will give rise to new and exciting discoveries that will likely elucidate conserved accessory/glial cell type fates and functions in animal vision.
Highlights.
Drosophila Semper cells, also known as cone cells, are highly multifunctional
Semper cells direct the differentiation of all three tissue types in the compound eye
Semper cells serve homeostatic support roles in the adult compound eye
Semper cells share many characteristics with vertebrate retinal Muller glia
Conserved gene regulatory modules are associated with specific Semper cell functions
ACKNOWLEDGEMENTS
We thank Bob Johnston for the opportunity to contribute this review, as well as Marla Spain, Elke Buschbeck, and two anonymous reviewers for their helpful comments on the manuscript. We also thank Uwe Wolfrum for sharing his unpublished findings on SC-associated cytoskeletal elements in the owlfly Ascalaphus. This work was supported by NIH-EY031526 and NSF-IOS1856241 (TAC), an unrestricted grant from Research to Prevent Blindness (Wayne State University Department of Ophthalmological, Visual and Anatomical Sciences), and Wellcome Trust and MSCA-IF-2015-707668 Marie Curie IF (MCP).
Abbreviations:
- SC
Semper cells
- PR
Photoreceptors
- MG
Müller glia
- PPC
Primary pigment cell
- IOC
Interommatidial cell
- RPE
Retinal pigment epithelium
- IRM
Irre-C Recognition Module
- pros
prospero
- Dl
Delta
- ZA
zonula adherens
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Arendt D, Musser JM, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, Wagner GP, 2016. The origin and evolution of cell types. Nat. Rev. Genet 17, 744–757. [DOI] [PubMed] [Google Scholar]
- Badenhorst P, 2001. Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development 128, 4093–4101. [DOI] [PubMed] [Google Scholar]
- Baker NE, 2001. Cell proliferation, survival, and death in the Drosophila eye. Seminars in Cell & Developmental Biology. 10.1006/scdb.2001.0274 [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY, 2001. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell 104, 699–708. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Bainton RJ, Mayer N, Beckstead R, Bhat MA, 2008. Septate junctions are required for ommatidial integrity and blood–eye barrier function in Drosophila. Dev. Biol 317, 585–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Sousa AD, Bhat MA, 2006. Organization and Function of Septate Junctions: An Evolutionary Perspective. Cell Biochemistry and Biophysics. 10.1385/cbb:46:1:65 [DOI] [PubMed] [Google Scholar]
- Banerjee U, Renfranz PJ, Pollock JA, Benzer S, 1987. Molecular characterization and expression of sevenless, a gene involved in neuronal pattern formation in the Drosophila eye. Cell 49, 281–291. [DOI] [PubMed] [Google Scholar]
- Bao S, 2014. Notch controls cell adhesion in the Drosophila eye. PLoS Genet. 10, e1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Cagan R, 2005. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev. Cell 8, 925–935. [DOI] [PubMed] [Google Scholar]
- Bao S, Fischbach K-F, Corbin V, Cagan RL, 2010. Preferential adhesion maintains separation of ommatidia in the Drosophila eye. Dev. Biol 344, 948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckner ME, 2020. A roadmap for potassium buffering/dispersion via the glial network of the CNS. Neurochem. Int 136, 104727. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Tong C, Tsuda H, 2010. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci 11, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S, 1967. BEHAVIORAL MUTANTS OF Drosophila ISOLATED BY COUNTERCURRENT DISTRIBUTION. Proc. Natl. Acad. Sci. U. S. A 58, 1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard CG, Gemne G, Seitz G, 1972. Optical Properties of the Compound Eye, in: Abramov I, O’Bryan P, Bernhard CG, Fuortes MGF, Gemne G, Gouras P, Hartline HK, Kropf A, Levick WR, Levinson JZ, Mauzerall D, Ratliff F, Seitz G, Sickel W, Stell WK, Tomita T, Trujillo-Cenoz O, Westheimer G, Cohen AI, Fuortes MGF (Eds.), Physiology of Photoreceptor Organs. Springer Berlin Heidelberg, Berlin, Heidelberg, pp. 357–379. [Google Scholar]
- Bernhard CG, Miller WH, Móller AR, 1963. Function of the Corneal Nipples in the Compound Eyes of Insects. Acta Physiologica Scandinavica. 10.1111/j.1748-1716.1963.tb02661.x [DOI] [PubMed] [Google Scholar]
- Bharti K, Nguyen M-TT, Skuntz S, Bertuzzi S, Arnheiter H, 2006. The other pigment cell: specification and development of the pigmented epithelium of the vertebrate eye. Pigment Cell Res. 19, 380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE, 2009. The HLH protein Extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Dev. Biol 327, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittern J, Pogodalla N, Ohm H, Brüser L, Kottmeier R, Schirmeier S, Klämbt C, 2020. Neuron–glia interaction in the Drosophila nervous system. Developmental Neurobiology. 10.1002/dneu.22737 [DOI] [PubMed] [Google Scholar]
- Blackie L, Walther RF, Staddon MF, Banerjee S, Pichaud F, 2020. Cell-type-specific mechanical response and myosin dynamics during retinal lens development in Drosophila. Mol. Biol. Cell 31, 1355–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J, Girard F, Kamachi Y, Kondoh H, Gehring WJ, 2005. Functional analysis of the chicken delta1-crystallin enhancer activity in Drosophila reveals remarkable evolutionary conservation between chicken and fly. Development 132, 1895–1905. [DOI] [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Edwards TN, Boulianne GL, Meinertzhagen IA, 2012. The metabolism of histamine in the Drosophila optic lobe involves an ommatidial pathway: - alanine recycles through the retina. Journal of Experimental Biology. 10.1242/jeb.060699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Kubów A, Lloyd V, Meinertzhagen IA, 2008. Drosophila ABC transporter mutants white, brown and scarlet have altered contents and distribution of biogenic amines in the brain. J. Exp. Biol 211, 3454–3466. [DOI] [PubMed] [Google Scholar]
- Bosze B, Suarez-Navarro J, Soofi A, Lauderdale JD, Dressler GR, Brown NL, 2021. Multiple roles for Pax2 in the embryonic mouse eye. Dev. Biol 472, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Mandler M, Neubüser A, Busslinger M, 2002. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 16, 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Cagan RL, 2003. Patterning the fly eye: the role of apoptosis. Trends Genet. 19, 91–96. [DOI] [PubMed] [Google Scholar]
- Brunner R, Sandfuchs O, Pacholski C, Morhard C, Spatz J, 2012. Lessons from nature: biomimetic subwavelength structures for high-performance optics. Laser Photonics Rev. 6, 641–659. [Google Scholar]
- Bunk EC, Ertaylan G, Ortega F, Pavlou MA, Gonzalez Cano L, Stergiopoulos A, Safaiyan S, Völs S, van Cann M, Politis PK, Simons M, Berninger B, Del Sol A, Schwamborn JC, 2016. Prox1 Is Required for Oligodendrocyte Cell Identity in Adult Neural Stem Cells of the Subventricular Zone. Stem Cells 34, 2115–2129. [DOI] [PubMed] [Google Scholar]
- Burton PR, Stockhammer KA, 1969. Electron microscopic studies of the compound eye of the toadbug,Gelastocoris oculatus. Journal of Morphology. 10.1002/jmor.1051270208 [DOI] [Google Scholar]
- Burton Q, Cole LK, Mulheisen M, Chang W, Wu DK, 2004. The role of Pax2 in mouse inner ear development. Dev. Biol 272, 161–175. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF, 1989. The emergence of order in the Drosophila pupal retina. Dev. Biol 136, 346–362. [DOI] [PubMed] [Google Scholar]
- Cepko CL, 1993. Chapter 1 Retinal cell fate determination. Progress in Retinal Research. 10.1016/0278-4327(93)90002-b [DOI] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D, 1996. Cell fate determination in the vertebrate retina. Proc. Natl. Acad. Sci. U. S. A 93, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EH, Chavadimane Shivakumar P, Clément R, Laugier E, Lenne P-F, 2017. Patterned cortical tension mediated by N-cadherin controls cell geometric order in the Drosophila eye. Elife 6. 10.7554/eLife.22796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Almeida AD, MacDonald RB, Harris WA, 2019. Genetic control of cellular morphogenesis in Müller glia. Glia. 10.1101/392902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins MA, Sendler ED, Buschbeck EK, Cook TA, 2017. Multifunctional glial support by Semper cells in the Drosophila retina. PLoS Genet. 13, e1006782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Brown NL, Cook TA, 2011a. The lens in focus: a comparison of lens development in Drosophila and vertebrates. Mol. Genet. Genomics 286, 189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Cook TA, 2010. Building a fly eye: terminal differentiation events of the retina, corneal lens, and pigmented epithelia. Curr. Top. Dev. Biol 93, 129–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Whitaker SL, Fei Y, Xie B, Li-Kroeger D, Gebelein B, Cook T, 2011b. Prospero and Pax2 combinatorially control neural cell fate decisions by modulating Ras- and Notch-dependent signaling. Neural Dev. 6, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R, Luan Z, Guo P, Li H-S, 2016. Drosophila Vision Depends on Carcinine Uptake by an Organic Cation Transporter. Cell Rep. 14, 2076–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R, Reddig K, Li H-S, 2014. Long-distance mechanism of neurotransmitter recycling mediated by glial network facilitates visual function in Drosophila. Proc. Natl. Acad. Sci. U. S. A 111, 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q-X, Chen Y-W, Li W-L, 2019. Ultrastructural comparison of the compound eyes of the Asian corn borer Ostrinia furnacalis (Lepidoptera: Crambidae) under light/dark adaptation. Arthropod Struct. Dev 53, 100901. [DOI] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH, 2006. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11, 775–789. [DOI] [PubMed] [Google Scholar]
- Cid E, Santos-Ledo A, Parrilla-Monge M, Lillo C, Arévalo R, Lara JM, Aijón J, Velasco A, 2010. Prox1 expression in rod precursors and Müller cells. Exp. Eye Res 90, 267–276. [DOI] [PubMed] [Google Scholar]
- Claparède R-É, 1860. Zur Morphologie der zusammengesetzten Augen bei den Arthropoden. Wilhelm Engelmann. [Google Scholar]
- Clausen MV, Hilbers F, Poulsen H, 2017. The Structure and Function of the Na,K-ATPase Isoforms in Health and Disease. Front. Physiol 8, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles JA, 1989. Functions of glial cells in the retina of the honeybee drone. Glia 2, 1–9. [DOI] [PubMed] [Google Scholar]
- Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C, 2003. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev. Cell 4, 853–864. [DOI] [PubMed] [Google Scholar]
- Cook T, Zelhof A, Mishra M, Nie J, 2011. 800 facets of retinal degeneration. Prog. Mol. Biol. Transl. Sci 100, 331–368. [DOI] [PubMed] [Google Scholar]
- Cordero JB, Cagan RL, 2010. Canonical wingless signaling regulates cone cell specification in the Drosophila retina. Dev. Dyn 239, 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero J, Jassim O, Bao S, Cagan R, 2004. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech. Dev 121, 1523–1530. [DOI] [PubMed] [Google Scholar]
- Country MW, 2017. Retinal metabolism: A comparative look at energetics in the retina. Brain Research. 10.1016/j.brainres.2017.07.025 [DOI] [PubMed] [Google Scholar]
- Dewan R, Fischer S, Meyer-Rochow VB, Özdemir Y, Hamraz S, Knipp D, 2012. Studying nanostructured nipple arrays of moth eye facets helps to design better thin film solar cells. Bioinspir. Biomim 7, 016003. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP, 1991. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, 451–464. [DOI] [PubMed] [Google Scholar]
- Dohle W, 2001. Are the insects terrestrial crustaceans? A discussion of some new facts and arguments and the proposal of the proper name’Tetraconata’for the monophyletic unit Crustacea+ Hexapoda, in: Annales de La Société Entomologique de France. pp. 85–103. [Google Scholar]
- Dorsky RI, Rapaport DH, Harris WA, 1995. Xotch inhibits cell differentiation in the Xenopus retina. Neuron 14, 487–496. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G, 2003. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat. Genet 34, 53–58. [DOI] [PubMed] [Google Scholar]
- Dziedzic K, Heaphy J, Prescott H, Kavaler J, 2009. The transcription factor D-Pax2 regulates crystallin production during eye development in Drosophila melanogaster. Dev. Dyn 238, 2530–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Meyer MR, 1990. Conservation of antigen 3G6: a crystalline cone constituent in the compound eye of arthropods. J. Neurobiol 21, 441–452. [DOI] [PubMed] [Google Scholar]
- Exner S, 1989. The physiology of the compound eyes of insects and crustaceans. Springer-Verlag GmbH & Co. KG. [Google Scholar]
- Fahrenbach WH, 1968. The morphology of the eyes of Limulus. Zeitschrift für Zellforschung und Mikroskopische Anatomie 93, 451–483. [DOI] [PubMed] [Google Scholar]
- Faivre-Sarrailh C, 2020. Molecular organization and function of vertebrate septate-like junctions. Biochimica et Biophysica Acta (BBA) - Biomembranes. 10.1016/j.bbamem.2020.183211 [DOI] [PubMed] [Google Scholar]
- Feller KD, Sharkey CR, McDuffee-Altekruse A, Bracken-Grissom HD, Lord NP, Porter ML, Schweikert LE, 2020. Surf and turf vision: Patterns and predictors of visual acuity in compound eye evolution. Arthropod Struct. Dev 101002. [DOI] [PubMed] [Google Scholar]
- Ferreiro MJ, Pérez C, Marchesano M, Ruiz S, Caputi A, Aguilera P, Barrio R, Cantera R, 2018. Drosophila melanogaster White Mutant w1118 Undergo Retinal Degeneration. Front. Neurosci 11, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach K-F, Linneweber GA, Andlauer TFM, Hertenstein A, Bonengel B, Chaudhary K, 2009. The irre cell recognition module (IRM) proteins. J. Neurogenet 23, 48–67. [DOI] [PubMed] [Google Scholar]
- Fischer C, Mahner M, Wachmann E, 2000. The rhabdom structure in the ommatidia of the Heteroptera (Insecta), and its phylogenetic significance. Zoomorphology 120, 1–13. [Google Scholar]
- Fischer S, Lu Z, Meinertzhagen IA, 2019. Three-dimensional ultrastructural organization of the ommatidium of the minute parasitoid wasp Trichogramma evanescens. Arthropod Struct. Dev 48, 35–48. [DOI] [PubMed] [Google Scholar]
- Flores GV, Daga A, Kalhor HR, Banerjee U, 1998. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development 125, 3681–3687. [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U, 2000. Combinatorial signaling in the specification of unique cell fates. Cell 103, 75–85. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Mardon G, 2002. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development 129, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Doe CQ, 2001. Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in the Drosophila CNS. Development 128, 4103–4112. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Piatigorsky J, Tessmar-Raible K, Jékely G, Guy K, Raible F, Wittbrodt J, Arendt D, 2005. Ancestry of Photic and Mechanic Sensation? Science 308, 1113–1114. [DOI] [PubMed] [Google Scholar]
- Fu W, Noll M, 1997. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 11, 2066–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiano N, Fishell G, 2002. The role of notch in promoting glial and neural stem cell fates. Annu. Rev. Neurosci 25, 471–490. [DOI] [PubMed] [Google Scholar]
- Genova JL, Fehon RG, 2003. Neuroglian, Gliotactin, and the Na+/K+ ATPase are essential for septate junction function in Drosophila. J. Cell Biol 161, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo-Hill BK, Wolff T, 2009. Dynamic cell shapes and contacts in the developingDrosophilaretina are regulated by the Ig cell adhesion protein hibris. Developmental Dynamics. 10.1002/dvdy.21981 [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Knust E, 2005. IrreC/rst-mediated cell sorting during Drosophila pupal eye development depends on proper localisation of DE-cadherin. Development 132, 2035–2045. [DOI] [PubMed] [Google Scholar]
- Guth SIE, Wegner M, 2008. Having it both ways: Sox protein function between conservation and innovation. Cell. Mol. Life Sci 65, 3000–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg E, Nilsson D-E, 1983. The euphausiid compound eye — a morphological reinvestigation (Crustacea: Euphausiacea). Zoomorphology 103, 59–66. [Google Scholar]
- Han Y, Xiong L, Xu Y, Tian T, Wang T, 2017. The β-alanine transporter BalaT is required for visual neurotransmission in. Elife 6. 10.7554/eLife.29146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC, 1987. Is histamine a neurotransmitter in insect photoreceptors? J. Comp. Physiol. A 161, 201–213. [DOI] [PubMed] [Google Scholar]
- Harris TJC, Tepass U, 2010. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol 11, 502–514. [DOI] [PubMed] [Google Scholar]
- Harris WA, Stark WS, Walker JA, 1976. Genetic dissection of the photoreceptor system in the compound eye of Drosophila melanogaster. J. Physiol 256, 415–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Reh TA, 2002. Homologies between vertebrate and invertebrate eyes. Results Probl. Cell Differ. 37, 219–255. [DOI] [PubMed] [Google Scholar]
- Hartline DK, 2011. The evolutionary origins of glia. Glia 59, 1215–1236. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Carthew RW, 2004. Surface mechanics mediate pattern formation in the developing retina. Nature 431, 647–652. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Xu C, Carthew RW, 2008. Cell-type-specific transcription of prospero is controlled by combinatorial signaling in the Drosophila eye. Development 135, 2787–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm C, Karl A, Beckers P, Kaul-Strehlow S, Ulbricht E, Kourtesis I, Kuhrt H, Hausen H, Bartolomaeus T, Reichenbach A, Bleidorn C, 2017. Early evolution of radial glial cells in Bilateria. Proc. Biol. Sci 284. 10.1098/rspb.2017.0743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T, Wang J, Boyd P, Wang F, Santiago C, Jiang L, Yoo S, Lahne M, Todd LJ, Jia M, Saez C, Keuthan C, Palazzo I, Squires N, Campbell WA, Rajaii F, Parayil T, Trinh V, Kim DW, Wang G, Campbell LJ, Ash J, Fischer AJ, Hyde DR, Qian J, Blackshaw S, 2020. Gene regulatory networks controlling vertebrate retinal regeneration. Science 370. 10.1126/science.abb8598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R, 2000. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development 127, 2515–2522. [DOI] [PubMed] [Google Scholar]
- Horridge GA, 1966. The retina of the locust, in: The Functional Organization of the Compound Eye: Proceedings of the International Symposium. Pergamon Press. [Google Scholar]
- Horridge GA, Giddings C, 1971. The ommatidium of the termite mastotermes darwiniensis. Tissue Cell 3, 463–476. [DOI] [PubMed] [Google Scholar]
- Hurley JB, Lindsay KJ, Du J, 2015. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J. Neurosci. Res 93, 1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain N, Pellikka M, Hong H, Klimentova T, Choe K-M, Clandinin TR, Tepass U, 2006. The agrin/perlecan-related protein eyes shut is essential for epithelial lumen formation in the Drosophila retina. Dev. Cell 11, 483–493. [DOI] [PubMed] [Google Scholar]
- Janssens H, Gehring WJ, 1999. Isolation and characterization of drosocrystallin, a lens crystallin gene of Drosophila melanogaster. Dev. Biol 207, 204–214. [DOI] [PubMed] [Google Scholar]
- Johnson RI, Sedgwick A, D’Souza-Schorey C, Cagan RL, 2011. Role for a Cindr–Arf6 axis in patterning emerging epithelia. MBoC 22, 4513–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H, 2001. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 15, 1272–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Konno D, Berry M, Matsuzaki F, Logan A, Hidalgo A, 2015. Prox1 Inhibits Proliferation and Is Required for Differentiation of the Oligodendrocyte Cell Lineage in the Mouse. PLoS One 10, e0145334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann RC, Li S, Gallagher PA, Zhang J, Carthew RW, 1996. Ras1 signaling and transcriptional competence in the R7 cell of Drosophila. Genes Dev. 10, 2167–2178. [DOI] [PubMed] [Google Scholar]
- Kavaler J, Fu W, Duan H, Noll M, Posakony JW, 1999. An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development 126, 2261–2272. [DOI] [PubMed] [Google Scholar]
- Kim E, Choi Y, Lee S, Seo Y, Yoon J, Baek K, 2008. Characterization of the Drosophila melanogaster retinin gene encoding a cornea-specific protein. Insect Mol. Biol 17, 537–543. [DOI] [PubMed] [Google Scholar]
- Kim WS, Weickert CS, Garner B, 2008. Role of ATP-binding cassette transporters in brain lipid transport and neurological disease. J. Neurochem 104, 1145–1166. [DOI] [PubMed] [Google Scholar]
- Klettner AK, Dithmar S, 2020. Retinal Pigment Epithelium in Health and Disease. Springer Nature. [Google Scholar]
- Koenig KM, Gross JM, 2020. Evolution and development of complex eyes: a celebration of diversity. Development 147. 10.1242/dev.182923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig KM, Sun P, Meyer E, Gross JM, 2016. Eye development and photoreceptor differentiation in the cephalopod Doryteuthis pealeii. Development 143, 3168–3181. [DOI] [PubMed] [Google Scholar]
- Komori N, Usukura J, Matsumoto H, 1992. Drosocrystallin, a major 52 kDa glycoprotein of the Drosophila melanogaster corneal lens. Purification, biochemical characterization, and subcellular localization. J. Cell Sci 102 (Pt 2), 191–201. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, 2005. Pax genes in eye development and evolution. Curr. Opin. Genet. Dev 15, 430–438. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Daube M, Frei E, Norman B, Kos L, Dishaw LJ, Noll M, Piatigorsky J, 2003. Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev. Cell 5, 773–785. [DOI] [PubMed] [Google Scholar]
- Kryuchkov M, Bilousov O, Lehmann J, Fiebig M, Katanaev VL, 2020. Reverse and forward engineering of Drosophila corneal nanocoatings. Nature 585, 383–389. [DOI] [PubMed] [Google Scholar]
- Kryuchkov M, Katanaev VL, Enin GA, Sergeev A, Timchenko AA, Serdyuk IN, 2011. Analysis of Micro- and Nano-Structures of the Corneal Surface of Drosophila and Its Mutants by Atomic Force Microscopy and Optical Diffraction. PLoS ONE. 10.1371/journal.pone.0022237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, 2012. Building an ommatidium one cell at a time. Dev. Dyn 241, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, 2011. My what big eyes you have: How the Drosophila retina grows. Developmental Neurobiology. 10.1002/dneu.20921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SR, Patel H, Tomlinson A, 2015. Wingless mediated apoptosis: How cone cells direct the death of peripheral ommatidia in the developing Drosophila eye. Dev. Biol 407, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraishi T, Binggeli O, Opota O, Buchon N, Lemaitre B, 2011. Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A 108, 15966–15971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Rubin GM, 2001. Neuralized is essential for a subset of Notch pathway-dependent cell fate decisions during Drosophila eye development. Proc. Natl. Acad. Sci. U. S. A 98, 5637–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Johnson RI, Swat M, Cordero JB, Glazier JA, Cagan RL, 2010. Computer simulation of cellular patterning within the Drosophila pupal eye. PLoS Comput. Biol 6, e1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavado A, Oliver G, 2007. Prox1 expression patterns in the developing and adult murine brain. Dev. Dyn 236, 518–524. [DOI] [PubMed] [Google Scholar]
- Lee S-J, Montell C, 2004. Suppression of constant-light-induced blindness but not retinal degeneration by inhibition of the rhodopsin degradation pathway. Curr. Biol 14, 2076–2085. [DOI] [PubMed] [Google Scholar]
- Lim H-Y, Tomlinson A, 2006. Organization of the peripheral fly eye: the roles of Snail family transcription factors in peripheral retinal apoptosis. Development 133, 3529–3537. [DOI] [PubMed] [Google Scholar]
- Lin HV, Rogulja A, Cadigan KM, 2004. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development 131, 2409–2418. [DOI] [PubMed] [Google Scholar]
- Liu L, MacKenzie KR, Putluri N, Maletić-Savatić M, Bellen HJ, 2017. The Glia-Neuron Lactate Shuttle and Elevated ROS Promote Lipid Synthesis in Neurons and Lipid Droplet Accumulation in Glia via APOE/D. Cell Metab. 26, 719–737.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, Li Z, Hui J, Graham BH, Quintana A, Bellen HJ, 2015. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley RL Jr, Ready DF, 1995. Integrins and the development of three-dimensional structure in the Drosophila compound eye. Dev. Biol 171, 415–433. [DOI] [PubMed] [Google Scholar]
- Losada-Perez M, 2018. Glia: from “just glue” to essential players in complex nervous systems: a comparative view from flies to mammals. Journal of Neurogenetics. 10.1080/01677063.2018.1464568 [DOI] [PubMed] [Google Scholar]
- MacDonald RB, Randlett O, Oswald J, Yoshimatsu T, Franze K, Harris WA, 2015. Müller glia provide essential tensile strength to the developing retina. J. Cell Biol 210, 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias-Muñoz A, Rangel Olguin AG, Briscoe AD, 2019. Evolution of phototransduction genes in Lepidoptera. Genome Biol. Evol 11, 2107–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistretti PJ, 2004. Brain Energy Metabolism. From Molecules to Networks. 10.1016/b978-012148660-0/50004-4 [DOI] [Google Scholar]
- Matsuo T, Takahashi K, Kondo S, Kaibuchi K, Yamamoto D, 1997. Regulation of cone cell formation by Canoe and Ras in the developing Drosophila eye. Development 124, 2671–2680. [DOI] [PubMed] [Google Scholar]
- Mavromatakis YE, Tomlinson A, 2016. R7 Photoreceptor Specification in the Developing Drosophila Eye: The Role of the Transcription Factor Deadpan. PLoS Genet. 12, e1006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer RR, Diersch R, Nicastro D, Smola U, 1997. Compound Eye Evolution: Highly Conserved Retinula and Cone Cell Patterns Indicate a Common Origin of the Insect and Crustacean Ommatidium. Naturwissenschaften 84, 542–544. [Google Scholar]
- Melzer RR, Michalke C, Smola U, 2000. Walking on insect paths? Early ommatidial development in the compound eye of the ancestral crustacean, Triops cancriformis. Naturwissenschaften 87, 308–311. [DOI] [PubMed] [Google Scholar]
- Menzel R, 1972. Feinstruktur des Komplexauges der Roten Waldameise, Formica polyctena (Hymenoptera, Formicidae). Zeitschrift für Zellforschung und Mikroskopische Anatomie 127, 356–373. [PubMed] [Google Scholar]
- Meyer MR, Reddy GR, Edwards JS, 1987. Immunological probes reveal spatial and developmental diversity in insect neuroglia. The Journal of Neuroscience. 10.1523/jneurosci.07-02-00512.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer- Rochow VB, 1977. Structure and possible function of the unusual compound eye of Sericesthis geminata (Coleoptera: Scarabaeidae). N. Z. J. Zool 4, 21–34. [Google Scholar]
- Miller DT, Cagan RL, 1998. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development 125, 2327–2335. [DOI] [PubMed] [Google Scholar]
- Monserrate JP, Brachmann CB, 2007. Identification of the death zone: a spatially restricted region for programmed cell death that sculpts the fly eye. Cell Death Differ. 14, 209–217. [DOI] [PubMed] [Google Scholar]
- Morey M, Yee SK, Herman T, Nern A, Blanco E, Zipursky SL, 2008. Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature 456, 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH, 1910. SEX LIMITED INHERITANCE IN DROSOPHILA. Science 32, 120–122. [DOI] [PubMed] [Google Scholar]
- Morrison CA, Chen H, Cook T, Brown S, Treisman JE, 2018. Glass promotes the differentiation of neuronal and non-neuronal cell types in the Drosophila eye. PLoS Genet. 14, e1007173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CHG, Rosenberg J, Richter S, Benno Meyer-Rochow V, 2003. The compound eye of Scutigera coleoptrata (Linnaeus, 1758) (Chilopoda: Notostigmophora): an ultrastructural reinvestigation that adds support to the Mandibulata concept. Zoomorphology 122, 191–209. [Google Scholar]
- Nagaraj R, Banerjee U, 2007. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development 134, 825–831. [DOI] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U, 2003. Reiterative and Concurrent Use of EGFR and Notch Signaling during Drosophila Eye Development. Handbook of Cell Signaling. 10.1016/b978-012124546-7/50619-7 [DOI] [Google Scholar]
- Nelson BR, Ueki Y, Reardon S, Karl MO, Georgi S, Hartman BH, Lamba DA, Reh TA, 2011. Genome-wide analysis of Müller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS One 6, e22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, Mahato S, Zelhof AC, 2014. The actomyosin machinery is required for Drosophila retinal lumen formation. PLoS Genet. 10, e1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D-E, 2013. Eye evolution and its functional basis. Vis. Neurosci 30, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D-E, 1989. Optics and Evolution of the Compound Eye, in: Facets of Vision. Springer; Berlin Heidelberg, pp. 30–73. [Google Scholar]
- Nilsson D-E, Kelber A, 2007. A functional analysis of compound eye evolution. Arthropod Struct. Dev 36, 373–385. [DOI] [PubMed] [Google Scholar]
- Owens M, Giordullo I, Buschbeck EK, 2020. Establishment of correctly focused eyes may not require visual input in arthropods. J. Exp. Biol 223. 10.1242/jeb.216192 [DOI] [PubMed] [Google Scholar]
- Park D, Jia H, Rajakumar V, Chamberlin HM, 2006. Pax2/5/8 proteins promote cell survival in C. elegans. Development 133, 4193–4202. [DOI] [PubMed] [Google Scholar]
- Parker A, 2004. In The Blink Of An Eye: How Vision Sparked The Big Bang Of Evolution. Basic Books. [Google Scholar]
- Parks AL, Turner FR, Muskavitch MA, 1995. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech. Dev 50, 201–216. [DOI] [PubMed] [Google Scholar]
- Peco E, Davla S, Camp D, M Stacey S, Landgraf M, van Meyel DJ, 2016. Drosophila astrocytes cover specific territories of the CNS neuropil and are instructed to differentiate by Prospero, a key effector of Notch. Development 143, 1170–1181. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J, O’Brien WE, Norman BL, Kalumuck K, Wistow GJ, Borras T, Nickerson JM, Wawrousek EF, 1988. Gene sharing by delta-crystallin and argininosuccinate lyase. Proc. Natl. Acad. Sci. U. S. A 85, 3479–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciani N, Kerlin JR, Sierra N, Swafford AJM, Ramirez MD, Roberts NG, Cannon JT, Daly M, Oakley TH, 2018. Prolific origination of eyes in Cnidaria with co-option of non-visual opsins. Curr. Biol 28, 2413–2419.e4. [DOI] [PubMed] [Google Scholar]
- Pickup AT, Ming L, Lipshitz HD, 2009. Hindsight modulates Delta expression during Drosophila cone cell induction. Development 136, 975–982. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL, 1997. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91, 881–891. [DOI] [PubMed] [Google Scholar]
- Quan X-J, Ramaekers A, Hassan BA, 2012. Transcriptional control of cell fate specification: lessons from the fly retina. Curr. Top. Dev. Biol 98, 259–276. [DOI] [PubMed] [Google Scholar]
- Querenet M, Goubard V, Chatelain G, Davoust N, Mollereau B, 2015. Spen is required for pigment cell survival during pupal development in Drosophila. Dev. Biol 402, 208–215. [DOI] [PubMed] [Google Scholar]
- Raabe T, 2000. The sevenless signaling pathway: variations of a common theme. Biochim. Biophys. Acta 1496, 151–163. [DOI] [PubMed] [Google Scholar]
- Rajasekaran SA, Palmer LG, Moon SY, Peralta Soler A, Apodaca GL, Harper JF, Zheng Y, Rajasekaran AK, 2001. Na,K-ATPase activity is required for formation of tight junctions, desmosomes, and induction of polarity in epithelial cells. Mol. Biol. Cell 12, 3717–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready DF, 2002. Drosophila compound eye morphogenesis: blind mechanical engineers? Results Probl. Cell Differ 37, 191–204. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM, 1995. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81, 857–866. [DOI] [PubMed] [Google Scholar]
- Reichenbach A, Bringmann A, 2013. New functions of Müller cells. Glia 61, 651–678. [DOI] [PubMed] [Google Scholar]
- Reiter C, Schimansky T, Nie Z, Fischbach KF, 1996. Reorganization of membrane contacts prior to apoptosis in the Drosophila retina: the role of the IrreC-rst protein. Development 122, 1931–1940. [DOI] [PubMed] [Google Scholar]
- Rey S, Zalc B, Klämbt C, 2020. Evolution of glial wrapping: A new hypothesis. Dev. Neurobiol 10.1002/dneu.22739 [DOI] [PubMed] [Google Scholar]
- Rhinn M, Brand M, 2001. The midbrain–hindbrain boundary organizer. Curr. Opin. Neurobiol 11, 34–42. [DOI] [PubMed] [Google Scholar]
- Richter S, 2002. The Tetraconata concept: hexapod-crustacean relationships and the phylogeny of Crustacea. Org. Divers. Evol 2, 217–237. [Google Scholar]
- Rittschof CC, Schirmeier S, 2018. Insect models of central nervous system energy metabolism and its links to behavior. Glia 66, 1160–1175. [DOI] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL, 2008. The transcriptome of retinal Müller glial cells. J. Comp. Neurol 509, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil HR, 1990. Structure and Function of the Squid Eye, in: Gilbert DL, Adelman WJ, Arnold JM (Eds.), Squid as Experimental Animals. Springer US, Boston, MA, pp. 371–397. [Google Scholar]
- Sarthy PV, 1991. Histamine: a neurotransmitter candidate for Drosophila photoreceptors. J. Neurochem 57, 1757–1768. [DOI] [PubMed] [Google Scholar]
- Satow T, Bae SK, Inoue T, Inoue C, Miyoshi G, Tomita K, Bessho Y, Hashimoto N, Kageyama R, 2001. The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J. Neurosci 21, 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenemann B, Clarkson ENK, 2020. Insights into a 429-million-year-old compound eye. Scientific Reports. 10.1038/s41598-020-69219-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenemann B, Pärnaste H, Clarkson ENK, 2017. Structure and function of a compound eye, more than half a billion years old. Proc. Natl. Acad. Sci. U. S. A 114, 13489–13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtz G, Staude A, Dunlop JA, 2019. Trilobite compound eyes with crystalline cones and rhabdoms show mandibulate affinities. Nature Communications. 10.1038/s41467-019-10459-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Cecconi F, Bernier G, Andrejewski N, 2000. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. [DOI] [PubMed]
- Schwentner M, Richter S, Rogers DC, Giribet G, 2018. Tetraconatan phylogeny with special focus on Malacostraca and Branchiopoda: highlighting the strength of taxon-specific matrices in phylogenomics. Proc. Biol. Sci 285. 10.1098/rspb.2018.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Noll M, 2009. Determination of cell fates in the R7 equivalence group of the Drosophila eye by the concerted regulation of D-Pax2 and TTK88. Dev. Biol 331, 68–77. [DOI] [PubMed] [Google Scholar]
- Siddall NA, Behan KJ, Crew JR, Cheung TL, Fair JA, Batterham P, Pollock JA, 2003. Mutations in lozenge and D-Pax2 invoke ectopic patterned cell death in the developing Drosophila eye using distinct mechanisms. Dev. Genes Evol 213, 107–119. [DOI] [PubMed] [Google Scholar]
- Simon MA, 1994. Signal Transduction during the Development of the Drosophila R7 Photoreceptor. Developmental Biology. 10.1006/dbio.1994.1327 [DOI] [PubMed] [Google Scholar]
- Sivak JG, Sivak JM, 2019. Conserved characteristics of ocular refractive development – Did the eye evolve once? Experimental Eye Research. 10.1016/j.exer.2018.05.007 [DOI] [PubMed] [Google Scholar]
- Soukkarieh C, Agius E, Soula C, Cochard P, 2007. Pax2 regulates neuronal-glial cell fate choice in the embryonic optic nerve. Dev. Biol 303, 800–813. [DOI] [PubMed] [Google Scholar]
- Sparrow RJ, Hicks D, Hamel PC, 2010. The retinal pigment epithelium in health and disease. Curr. Mol. Med 10, 802–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl AL, Charlton-Perkins M, Buschbeck EK, Cook TA, 2017. The cuticular nature of corneal lenses in Drosophila melanogaster. Dev. Genes Evol 227, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke J, Moose HE, El-Hodiri HM, Fischer AJ, 2010. Comparative study of Pax2 expression in glial cells in the retina and optic nerve of birds and mammals. J. Comp. Neurol 518, 2316–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbings LA, Todman MG, Phillips R, Greer CE, Tam J, Phelan P, Jacobs K, Bacon JP, Davies JA, 2002. Gap junctions in Drosophila: developmental expression of the entire innexin gene family. Mech. Dev 113, 197–205. [DOI] [PubMed] [Google Scholar]
- Stiemke MM, Hollyfield JG, 1995. Cell birthdays in Xenopus laevis retina. Differentiation 58, 189–193. [DOI] [PubMed] [Google Scholar]
- Strauss O, 2005. The retinal pigment epithelium in visual function. Physiol. Rev 85, 845–881. [DOI] [PubMed] [Google Scholar]
- Suga H, Tschopp P, Graziussi DF, Stierwald M, Schmid V, Gehring WJ, 2010. Flexibly deployed Pax genes in eye development at the early evolution of animals demonstrated by studies on a hydrozoan jellyfish. Proc. Natl. Acad. Sci. U. S. A 107, 14263–14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Du W, 2008. Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Developmental Biology. 10.1016/j.ydbio.2007.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Harris KP, 2007. Adherens junctions in Drosophila retinal morphogenesis. Trends Cell Biol. 17, 26–35. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Callaerts P, Kos L, Zinovieva R, Halder G, Gehring W, Piatigorsky J, 1997. Squid Pax-6 and eye development. Proc. Natl. Acad. Sci. U. S. A 94, 2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, 2012. The origin of the Drosophila subretinal pigment layer. J. Comp. Neurol 520, 2676–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, 1985. The cellular dynamics of pattern formation in the eye of Drosophila. J. Embryol. Exp. Morphol 89, 313–331. [PubMed] [Google Scholar]
- Tomlinson A, Mavromatakis YE, Struhl G, 2011. Three distinct roles for notch in Drosophila R7 photoreceptor specification. PLoS Biol. 9, e1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF, 1987. Cell fate in the Drosophila ommatidium. Dev. Biol 123, 264–275. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF, 1986. Sevenless: a cell-specific homeotic mutation of the Drosophila eye. Science 231, 400–402. [DOI] [PubMed] [Google Scholar]
- Torres M, Gómez-Pardo E, Gruss P, 1996. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development 122, 3381–3391. [DOI] [PubMed] [Google Scholar]
- Treier M, Bohmann D, Mlodzik M, 1995. JUN cooperates with the ETS domain protein pointed to induce photoreceptor R7 fate in the Drosophila eye. Cell 83, 753–760. [DOI] [PubMed] [Google Scholar]
- Treisman JE, 2013. Retinal differentiation in Drosophila. Wiley Interdiscip. Rev. Dev. Biol 2, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsachaki M, Sprecher SG, 2012. Genetic and developmental mechanisms underlying the formation of the Drosophila compound eye. Dev. Dyn 241, 40–56. [DOI] [PubMed] [Google Scholar]
- Tsacopoulos M, Magistretti PJ, 1996. Metabolic coupling between glia and neurons. J. Neurosci 16, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsacopoulos M, Poitry-Yamate CL, Poitry S, 1997. Ammonium and glutamate released by neurons are signals regulating the nutritive function of a glial cell. J. Neurosci 17, 2383–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsacopoulos M, Veuthey AL, Saravelos SG, Perrottet P, Tsoupras G, 1994. Glial cells transform glucose to alanine, which fuels the neurons in the honeybee retina. J. Neurosci 14, 1339–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipursky SL, Banerjee U, 2002. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell 110, 625–637. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F, 2015. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN, 1991. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell 67, 941–953. [DOI] [PubMed] [Google Scholar]
- Vagin O, Tokhtaeva E, Sachs G, 2006. The role of the beta1 subunit of the Na,K-ATPase and its glycosylation in cell-cell adhesion. J. Biol. Chem 281, 39573–39587. [DOI] [PubMed] [Google Scholar]
- Vetter ML, Moore KB, 2001. Becoming glial in the neural retina. Dev. Dyn 221, 146–153. [DOI] [PubMed] [Google Scholar]
- Viets K, Eldred K, Johnston RJ Jr, 2016. Mechanisms of Photoreceptor Patterning in Vertebrates and Invertebrates. Trends Genet. 32, 638–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viringipurampeer IA, Ferreira T, DeMaria S, Yoon JJ, Shan X, Moosajee M, Gregory-Evans K, Ngai J, Gregory-Evans CY, 2012. Pax2 regulates a fadd-dependent molecular switch that drives tissue fusion during eye development. Hum. Mol. Genet 21, 2357–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voas MG, Rebay I, 2004. Signal integration during development: insights from the Drosophila eye. Dev. Dyn 229, 162–175. [DOI] [PubMed] [Google Scholar]
- Vohra R, Kolko M, 2018. Neuroprotection of the inner retina: Müller cells and lactate. Neural Regeneration Res. 13, 1741–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkenhoff A, Weiler A, Letzel M, Stehling M, Klämbt C, Schirmeier S, 2015. Glial Glycolysis Is Essential for Neuronal Survival in Drosophila. Cell Metab. 22, 437–447. [DOI] [PubMed] [Google Scholar]
- Vopalensky P, Kozmik Z, 2009. Eye evolution: common use and independent recruitment of genetic components. Philos. Trans. R. Soc. Lond. B Biol. Sci 364, 2819–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH, Perry MM, 1963. Inter-retinular fibres in the eyes of Drosophila. Journal of Insect Physiology. 10.1016/0022-1910(63)90058-1 [DOI] [Google Scholar]
- Waddington CH, Perry MM, 1960. The ultra-structure of the developing eye of Drosophila. Proceedings of the Royal Society of London. Series B. Biological Sciences 153, 155–178. [Google Scholar]
- Wallace SW, Singhvi A, Liang Y, Lu Y, Shaham S, 2016. PROS-1/Prospero Is a Major Regulator of the Glia-Specific Secretome Controlling Sensory-Neuron Shape and Function in C. elegans. Cell Rep. 15, 550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang T, Jiao Y, von Lintig J, Montell C, 2010. Requirement for an enzymatic visual cycle in Drosophila. Curr. Biol 20, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang T, Ni JD, von Lintig J, Montell C, 2012. The Drosophila visual cycle and de novo chromophore synthesis depends on rdhB. J. Neurosci 32, 3485–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildonger J, Sosinsky A, Honig B, Mann RS, 2005. Lozenge directly activates argos and klumpfuss to regulate programmed cell death. Genes Dev. 19, 1034–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Ready DF, 1993. Pattern formation in the Drosophila retina. The development of Drosophila melanogaster 2, 1277–1325. [Google Scholar]
- Wolff T, Ready DF, 1991. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113, 841–850. [DOI] [PubMed] [Google Scholar]
- Xu C, Kauffmann RC, Zhang J, Kladny S, Carthew RW, 2000. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103, 87–97. [DOI] [PubMed] [Google Scholar]
- Xu H, Lee S-J, Suzuki E, Dugan KD, Stoddard A, Li H-S, Chodosh LA, Montell C, 2004. A lysosomal tetraspanin associated with retinal degeneration identified via a genomewide screen. EMBO J. 23, 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, An F, Borycz JA, Borycz J, Meinertzhagen IA, Wang T, 2015. Histamine Recycling Is Mediated by CarT, a Carcinine Transporter in Drosophila Photoreceptors. PLoS Genet. 11, e1005764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Canon J, Banerjee U, 2003. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev. Biol 263, 323–329. [DOI] [PubMed] [Google Scholar]
- Yoon CS, Hirosawa K, Suzuki E, 1997. Corneal lens secretion in newly emerged Drosophila melanogaster examined by electron microscope autoradiography. J. Electron Microsc 46, 243–246. [DOI] [PubMed] [Google Scholar]
- Young RW, 1985. Cell differentiation in the retina of the mouse. Anat. Rec 212, 199–205. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Hardy RW, Becker A, Zuker CS, 2006. Transforming the architecture of compound eyes. Nature 443, 696–699. [DOI] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F, 2006. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133, 4881–4889. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Rubin GM, 1994. Determination of neuronal cell fate: lessons from the R7 neuron of Drosophila. Annu. Rev. Neurosci 17, 373–397. [DOI] [PubMed] [Google Scholar]


