Abstract
Neural stem cells (NSCs) undergo massive molecular and cellular changes during neuronal differentiation. These include mitochondria and metabolism remodelling, which were thought to be mostly permissive cues, but recent work indicates that they are causally linked to neurogenesis. Striking remodelling of mitochondria occurs right after mitosis of NSCs, which influences the postmitotic daughter cells towards self-renewal or differentiation. The transitioning to neuronal fate requires metabolic rewiring including increased oxidative phosphorylation activity, which drives transcriptional and epigenetic effects to influence cell fate. Mitochondria metabolic pathways also contribute in an essential way to the regulation of NSC proliferation and self-renewal. The influence of mitochondria and metabolism on neurogenesis is conserved from fly to human systems, but also displays striking differences linked to cell context or species. These new findings have important implications for our understanding of neurodevelopmental diseases and possibly human brain evolution.
Highlights
-
•
Mitochondrial dynamics after mitosis controls neuronal fate commitment.
-
•
Developmental metabolic shifts drive neural stem cell amplification and differentiation.
-
•
Metabolic intermediates impact neurogenesis through post-translational modifications.
-
•
Loss of tricarboxylic acid cycle–associated genes impairs neurogenesis in the embryonic and adult brain.
-
•
Species differences in mitochondria dynamics and function could be linked to the evolution of neurogenesis.
Introduction
Neurogenesis is a key event of neural development, by which neural stem/progenitor cells (NSPCs) stop self-renewing and dividing, to convert to postmitotic neurons. The delicate balance between self-renewal and differentiation of NSPCs is controlled by intrinsic and extrinsic cues, in particular transcription factors and morphogen signals (reviewed in [1,2]). However, recent evidence has uncovered the key influence of mechanisms once considered to be merely permissive: cell metabolism and mitochondria dynamics.
Mitochondria play essential roles in energy production, calcium homeostasis and cell signalling. These organelles constitute a network that is constantly remodelled through a process of fission or fusion or mitochondria dynamics [3]. Mitochondria dynamics have been associated with cell fate decisions in various systems [4]. It can influence stem cell renewal and differentiation in part through the direct or indirect production of metabolites that can modulate developmental pathways, in particular through post-translational modifications (PTMs), such as histone acetylation and methylation [5,6]. The impact of mitochondria on neuronal development and function has long been illustrated by the major neurological consequences of mitochondrial diseases (mostly caused by mutations in mitochondria-related genes, the most exemplative disease being Leigh syndrome), including motor and sensory hypofunction, epilepsy, autism spectrum behaviours and dementia [7], but their implication in early neural development and neurogenesis had remained mostly unexplored until recently.
Mitochondria dynamics during neurogenesis
A first hint of the implication of mitochondria dynamics in neurogenesis was provided by the morphological inspection of mitochondria in various types of NSPCs and neurons, in particular in the mouse developing cerebral cortex [8,9]. This revealed that radial glial cells (RGC), the major neural stem cells (NSCs) in the developing brain, display mostly fused mitochondria, whereas intermediate progenitor cells, which constitute transit amplifying cells already committed to neuronal fate, displayed fragmented mitochondria [8,9]. Surprisingly, newly born neurons were found to display even smaller mitochondria, suggesting a high level of mitochondria fission during the neurogenic transition, which is then followed by a gradual increase in mitochondria size during neuronal maturation [9]. Moreover, disruption of genes that promote mitochondria fusion, such as Opa1 (optic atrophy 1) and MFN1/2 (mitofusin 1 and 2), leads to decrease of RGC self-renewal and increased neuronal differentiation, whereas conversely the disruption of Drp1 (dynamin-related protein 1), the main effector of fission, leads to increased self-renewal and decreased neurogenesis [8,9].
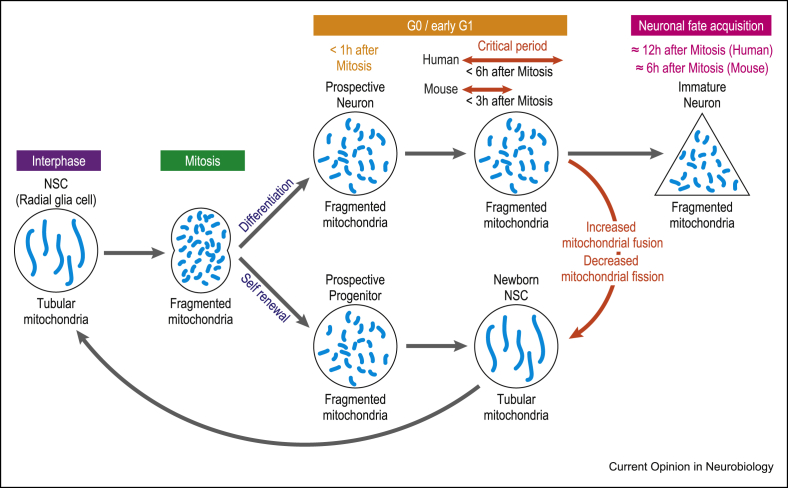
Mitochondria dynamics is closely linked to the cell cycle, with increased fusion during G1/S phases followed by fission during G2 and mitosis [10], which allows for partitioning of mitochondria between the daughter cells. How then can mitochondria dynamics be articulated with the progression of neurogenesis, which is by definition a postmitotic event? This was addressed with a novel in vitro tracking method to follow mitochondrial dynamics of cortical NSPCs throughout self-renewal and neuronal differentiation, from cell division to fate acquisition [9]. This confirmed that mitochondrial fission occurred during NSPC mitosis, as expected, but it also revealed a dichotomic behaviour of the daughter cells: those destined to remain NSPCs displayed high levels of mitochondria fusion, whereas prospective neuronal cells maintained higher levels of mitochondrial fission (Figure 1). Moreover, the acute induction of mitochondria fusion shortly after mitosis (using fusion activator or fission inhibitor molecules) dramatically altered the balance of cell fate, leading most NSPCs to undergo self-renewal instead of neuronal differentiation (Figure 1). The impact of mitochondria dynamics after NSPC mitosis was also assessed in vivo, using the FlashTag method to label mitotic cortical progenitors [11], confirming the positive impact of mitochondrial fusion on NSPC self-renewal [9]. These results demonstrate that mitochondria dynamics have a profound influence on cortical neurogenesis. In addition, the timing of the events indicates that fate commitment is not irreversibly determined in postmitotic daughter cells but that there is a postmitotic critical period of plasticity of fate commitment. This finding extends the classical view of neuronal fate decision that is thought to occur before mitosis as early as G1 in the mother cell [12].
Figure 1.
Mitochondria dynamics regulate neurogenesis during a postmitotic critical period. The mouse cortical NSC/RGC has tubular mitochondria during interphase and undergoes mitochondrial fission during mitosis. After mitosis, both daughter cells display fragmented mitochondria. However, the cells that undergo mitochondrial fusion in the next hours will remain NSCs, whereas those that keep fragmented mitochondria will become neurons. During this critical period, which is double in human NSCs compared with mouse NSCs, the fate of the cells can be changed by manipulating mitochondria dynamics.
Could these findings be conserved beyond mouse corticogenesis? Human cortical progenitors display increased self-renewal potential that enables expansion of progenitors and ultimately increased neuronal output, which underlies the increase in cortical size in the human lineage [1]. The same tracking system of mitochondria and cell fate was applied to human cortical progenitors derived from pluripotent stem cells, revealing conserved relationships between mitochondria dynamics and neurogenesis [9]. However, mitochondria dynamics was found to be able to influence cell fate for a much longer, doubled period after cell division, when compared with mouse cortical progenitors (Figure 1). The prolonged critical period of postmitotic fate plasticity in human cortical progenitors could be in line with their increased self-renewal potential, although this remains to be tested further [9].
Overall, these data strongly suggest that mitochondria remodelling plays a crucial instructive role in neurogenesis, raising the question of the downstream mechanisms. Mitochondria fission and fusion are tightly linked to mitochondria activity, in part through the regulation of mitochondria cristae in which oxidative phosphorylation (OXPHOS) molecular effectors and the electron transport chain (ETC) are concentrated [3]. Indeed OXPHOS is essential for proper neurogenesis, whether in NSCs themselves or during the neurogenesis transition, as explained in the following.
Glycolysis to OXPHOS metabolic shifts during neurogenesis
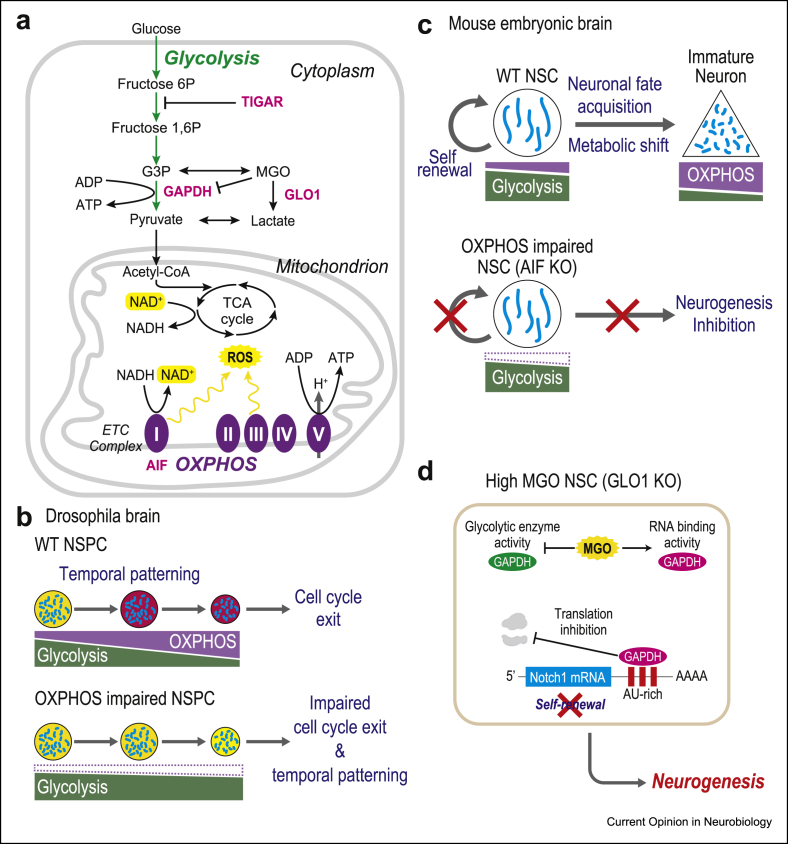
Glycolysis in the cytosol and OXPHOS in the mitochondria are the two main processes that generate ATP within cells (reviewed in [13]) (Figure 2a). The glycolytic pathway leads to pyruvate generation, which can be converted into lactate. Cytoplasmic pyruvate can also enter mitochondria to feed the tricarboxylic acid (TCA) cycle, which generates the fuels for OXPHOS to generate ATP via the ETC. Glycolysis generates much less ATP than OXPHOS does, but its kinetics is overall faster, and it feeds in most biosynthetic pathways to support cell growth and proliferation [13]. Therefore, highly proliferative cells tend to rely mostly on glycolysis, even in the presence of high oxygen levels, a process known as aerobic glycolysis, once thought to be a landmark property of cancer cells [13]. On the other hand, mitochondria respiration through OXPHOS is the most efficient way for the cell to generate ATP, which is therefore heavily used in energy-demanding differentiated cells such as neurons. Indeed, metabolic analyses of NSPCs and their neuronal progeny in many systems and species have shown that NSPCs rely predominantly on glycolysis over OXPHOS, whereas differentiated neurons exhibit higher mitochondrial respiration [8,14, 15, 16, 17, 18, 19]. But do these pathways play a role in the neurogenic transition? A first direct implication of glycolysis–OXPHOS metabolic balance in neurogenesis was provided by genetic disruption of mitochondrial OXPHOS-related genes in the Drosophila brain: OXPHOS genes were shown to be required at the end of neurogenesis to reduce the size of NSPCs and induce their cell cycle exit towards neuronal fate acquisition [14] (Figure 2). These data indicate that glycolysis to OXPHOS metabolic shift is a likely cause and not a mere consequence of neuronal differentiation. On the other hand, the disruption of OXPHOS genes in fly neuroblasts has also consequences in NSPCs themselves, including reduced proliferation rates and altered temporal patterning, that is, temporal changes in their identity, which also perturb the transition to cell cycle exit [20]. Consistent with these findings, genetic disruption in the mouse apoptosis-inducing factor (AIF), a mitochondrial protein essential for OXPHOS, results in impaired self-renewal capacity of cortical NSPCs together with impaired cell cycle exit and neuronal differentiation, ultimately leading to microcephaly [21]. The impact of mitochondria on neurogenesis has also been studied in the context of adult neurogenesis in the mouse hippocampus [22]. In this case, mitochondria activity is required for the amplification and differentiation of neurogenic precursors, but notably, the NSC mitochondria display a mostly fragmented shape, and progressive fusion takes place in neurogenic precursors and differentiated neurons [22]. This suggests that the relationships between mitochondria morphology, dynamics and function depend on the cell context, which should be investigated further. Finally, the requirement of OXPHOS for human neurogenesis was explored recently in the context of mitochondrial disease modelling, using pluripotent stem cell–derived NSCs bearing pathogenic mutations in the mitochondria EC assembly gene SURF1 (causative for Leigh syndrome), which displayed reduced neuronal fate acquisition and maturation [23].
Figure 2.
Glycolysis–OXPHOS metabolic shift and neurogenesis.(a) A simplified scheme of glycolysis and OXPHOS pathways. Genes involved in neurogenesis are highlighted in pink and purple. (b) In the Drosophila brain, NSPCs display a gradual metabolic shift from glycolysis to OXPHOS, which is required for temporal changes in cell identity, culminating in cell cycle exit. (c) During mouse corticogenesis, a glycolysis to OXPHOS shift occurs during the conversion of mouse NSCs into immature neurons. OXPHOS inhibition (AIF KO) leads to impaired self-renewal and neurogenesis. (d) Mechanisms of action of glycolysis metabolite MGO. MGO directly inhibits the glycolytic activity of GAPDH, while promoting its binding to the mRNA of Notch1, thereby inhibiting its translation and downstream self-renewal pathways.
Collectively, these data indicate that OXPHOS is required at multiple steps of neurogenesis, from NSC self-renewal to neuronal fate acquisition. How about glycolysis? Disruption of glycolysis does not appear to affect neurogenesis in fly neuroblasts [20], but recent evidence suggests that active regulation of glycolysis can participate in mouse cortical neurogenesis (Figure 2) [24, 25, 26∗∗]. The TP53-inducible glycolysis and apoptosis regulator (TIGAR), an endogenous inhibitor of glycolysis, is upregulated in neurons, whereas its disruption leads to decreased mouse cortical neurogenesis in vitro [25]. Moreover, a specific glycolysis metabolite, methylglyoxal (MGO), can influence NSPC self-renewal while regulating glycolytic rates [26] (Figure 2). Increased levels of MGO lead to decreased self-renewal capacity of NSPCs in the developing mouse cortex [26,27]. MGO acts by binding to the key glycolysis enzyme GAPDH, which diverts GAPDH from its enzymatic activity towards another function, as an RNA-binding protein [28]. GAPDH was found to bind the mRNA of Notch1, a crucial regulator of NSPC self-renewal, leading to Notch1 decreased translation and thereby increased neurogenesis [26]. These data suggest that glycolytic metabolites such as MGO can coordinate metabolism with neurogenesis, through direct post-translational interactions between key effectors of glycolysis and developmental pathways. The modulation of PTM by specific metabolites is indeed an emerging theme linking metabolism with cell fate control: an increasing number of metabolites have been found to serve as rate-limiting substrates for PTM, in particular for histones but also other proteins, or modulators of PTM effector enzymes such as sirtuin deacetylases [5,29]. As we shall see further in the following, this is likely to be the case for neurogenesis as well.
Links between mitochondria OXPHOS and neurogenesis: ROS and sirtuins
The data abovementioned implicate mitochondria-dependent OXPHOS to promote neurogenesis, but what could be the underlying mechanisms? Cellular reactive oxygen species (ROS), which are produced not only by mitochondrial OXPHOS via ETC activity but also by Nox enzymes at the plasma membrane, can have a physiological impact on cell behaviour and fate [30]. ROSs were initially found to increase NSPC self-renewal and proliferation [31], but more recent data indicate pleiotropic, context-dependent effects. In adult mouse neurogenesis, high levels of ROS have been associated with NSPC quiescence [32], whereas in the embryonic cortex, ROS increase during the conversion from NSPCs to neurons [8]. In this case, ROS appears to act through one of its physiological targets, the transcription factor NRF2, leading to upregulation of the BOTCH gene, a Notch inhibitor, thereby favouring the transition towards neuronal fate [8]. ROS production during neurogenesis has been reported upstream or downstream of various other classes of transcription factors [33, 34, 35, 36], providing additional levels of complexity between ROS levels and transcriptional regulation during neurogenesis, which remain to be explored.
Another main consequence of OXPHOS activity is the change in REDOX (reduction–oxidation) balance, in particular through the pair formed by the oxidised and reduced form of NAD (NAD+ and NADH). NAD+/NADH ratios are influenced by glycolysis–OXPHOS balance and have been implicated in many aspects of neuronal fate acquisition and differentiation, mostly through the activity of the NAD+-dependent deacetylase sirtuin family [37]. In particular, sirtuin-1 (Sirt1) is required for cell fate determination of NSPCs during embryonic development [38, 39, 40]. In mouse cortical development, Sirt1 is recruited to selective DNA targets by the transcription repressor factor Bcl6 and induces selective epigenetic repression of neighbouring genes by histone deacetylation [39]. Interestingly, BCL6 acts by repressing key genes of the Notch, Wnt, SHH and FGF pathways, all of which promote NSPC self-renewal and proliferation: their repression by the BCL6/Sirt1complex thus leads to cell cycle exit and neuronal differentiation [40].
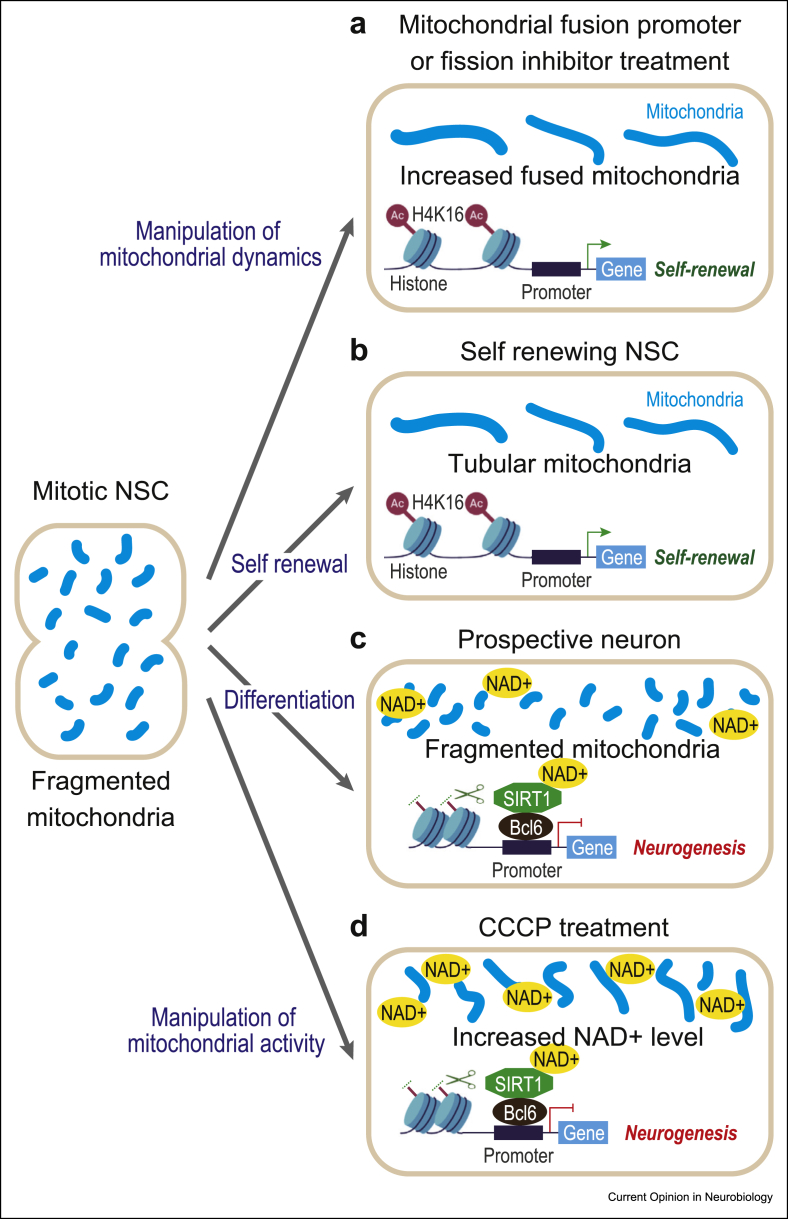
Recently, the relationship between Sirt1 activity and mitochondrial function/dynamics was assessed during neuronal fate determination [9] (Figure 3). This revealed that the pharmacological increase of the ETC activity using Carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a proton ionophore , which is expected to lead to increased NAD+/NADH ratios [41], could promote cortical neurogenesis in a Sirt1-dependent manner. Conversely, the induction of NSPC self-renewal induced by mitochondria fusion (Figure 1) could be blocked by sirtuin activation. Finally, manipulation of either sirtuin activity or mitochondria fusion, during the neurogenic conversion, could lead to acute changes in acetylation patterns of histone H4 lysine 16 (H4K16), a main target of Sirt1/Bcl6 (Figure 3). Collectively, these data suggest that mitochondrial influence on neuronal fate determination involves the redox-sensitive Sirt1 activity, which could lead to chromatin remodelling, favouring neuronal fate acquisition. However, it remains to be determined how the changes in mitochondrial morphology actually lead to increased mitochondrial NAD+ levels and how this can then affect Sirt1 activity in the nucleus [42]. Moreover, it will be important to understand how the observed histone acetylation changes actually lead to selective transcriptional responses required for neurogenesis and to explore whether other mitochondria-derived metabolites can drive PTM of histones or other relevant proteins [5,6]. Finally, it will be interesting to study modulation of sirtuins by mitochondrial activity in other cell fate conversions in which OXPHOS is implicated, such as direct neuronal reprogramming and neural tumorigenesis [43,44].
Figure 3.
The model of the links between mitochondrial dynamics and sirtuin signalling during neurogenesis. After mitosis, NSCs that display high levels of mitochondria fusion (following fusion activation/fission inhibition (a) or physiologically (b)) will remain NSCs, through the action of activated genes that promote self-renewal. On the other hand, the cells that maintain high levels of fission will convert into neurons (c), in part through the action of NAD+-dependent sirtuins that deacetylate histones, thereby maintaining self-renewal genes in a repressed mode. The same impact on neuronal fate acquisition and histone remodelling can be obtained with treatment of the cells with the OXPHOS activator CCCP (d), even with mitochondria in a fused state.
Beyond and between glycolysis and OXPHOS: TCA and lipid metabolism in neurogenesis
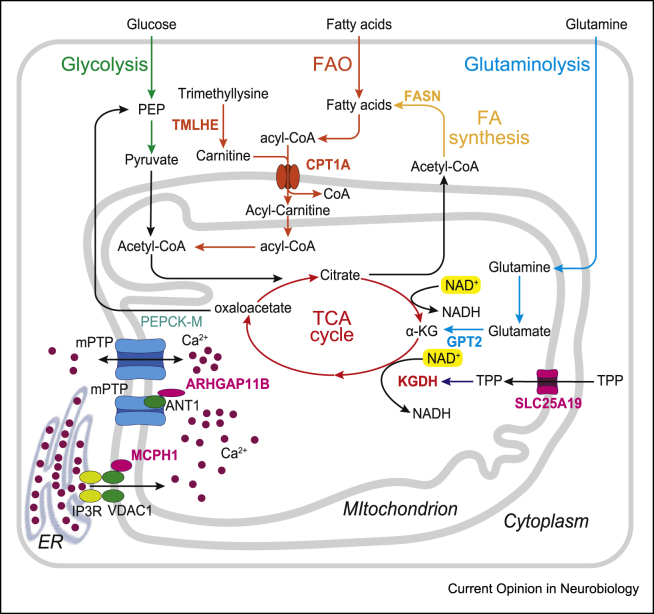
Although these data illustrate the key influence of OXPHOS on neurogenesis, other mitochondria-dependent metabolic functions have been implicated, in particular in link with the TCA cycle and lipid metabolism (Figure 4).
Figure 4.
Mitochondrial metabolic networks and associated genes that contribute to neurogenesis. The major mitochondria-dependent metabolic pathways are coloured: glycolysis (green), TCA cycle (red), glutaminolysis (light blue), FA synthesis (yellow), FA β-oxidation (FAO, orange). The microcephaly gene-encoded protein MCPH1 interacts with mitochondrial voltage-dependent anion channel 1 (VDAC1), which together with IP3 receptor (IP3R) controls calcium influx from endoplasmic reticulum (ER) into mitochondria. ARHBGA11B binds to ANT1, an mPTP component, and inhibits mPTP channel function. FAO and FA synthesis genes (TMLHE, CPT1A and FASN) and TCA-linked genes (GPT2, SLC25A19 and KGDH) are required for mouse and human cortical neurogenesis. mPTP, mitochondrial permeability transition pore.
Primary microcephaly (MCPH) is a genetic neurodevelopmental disorder (NDD) characterised by highly reduced brain size, in particular of the cerebral cortex [45]. One of the causative genes, microcephalin 1 (MCPH1), was previously involved in NSPC proliferation and survival through the regulation of chromatin segregation during mitosis [45]. Recently, however, the MCPH1-encoded protein was detected at the level of mitochondria outer membranes [46]. MCPH1 was found to interact with and activate mitochondrial calcium channels linking the endoplasmic reticulum (ER) to mitochondria (Figure 4). Deletion of MCPH1 in mouse cortical NSPCs leads to a reduced intramitochondrial calcium level, which can affect the activity of TCA cycle key enzymes including α-ketoglutarate dehydrogenase (KGDH) [47], which is required for neurogenesis in fly neuroblasts [14]. MCPH1 deficiency could also be linked to decreased expression of mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M or PCK2). PCK2 can promote glycolysis and glutaminolysis in tumour growth through pyruvate regeneration, to feed acetyl-CoA to the TCA cycle [48]. These results point to potential roles of MCPH1 in neurogenesis through regulation of the mitochondrial calcium level and metabolic activity. Interestingly, several gene mutations associated with other forms of microcephaly have been linked to the TCA cycle, in particular at the level of glutaminolysis (Figure 4). These include SLC25A19 (solute carrier family 25 member 19), the mitochondrial thiamine pyrophosphate transporter [49], the deletion of which leads to early neural defects in the mouse and to a reduction in activity of KGDH through a depletion of mitochondrial thiamine pyrophosphate [50], KGDH itself [51] and glutamate pyruvate transaminase 2 (GPT2) [52]. Finally, ARHGAP11B (Rho GTPase-activating protein 11B), a human-specific gene that drives proliferation of cortical NSPCs, was also localised to mitochondria [53], where its gain of function can lead to blockade of the mitochondrial permeability transition pore (mPTP), thereby leading to increased calcium levels. The expansion of mouse cortical NSPCs induced by ARHGAP11B overexpression could be blocked by pharmacological inhibition of glutaminolysis, which could also reduce the number of proliferative NSPCs in ex vivo cultures of the human fetal cortex [53]. It is intriguing that both MCPH1 and ARHGAP11B appear to control mitochondrial calcium levels, albeit though different mechanisms. It will be interesting to test further the impact of mitochondrial calcium on neurogenesis, as it can influence cell fate in cardiomyogenesis through Notch pathway modulation [54].
Overall, these studies highlight the importance of the TCA cycle and related pathways in embryonic cortical neurogenesis across species. It will be important to determine whether these effects contribute to NSPC proliferation through the generation of ATP and TCA-dependent anabolism, through the feeding of OXPHOS, or whether they control NSPC biology in a more instructive fashion through TCA-derived signalling metabolites, such as α-KG that could promote histone H3 demethylation, which is required for neurogenesis [55,56].
Finally, another important aspect of mitochondria function related to neurogenesis is fatty acids (FAs) metabolism (Figure 4). In mammalian cells, FAs are obtained either by direct exogenous uptake or by de novo synthesis from acetyl-CoA in the cytoplasm, in this latter case catalysed by fatty acid synthase (FASN). A first direct implication of FA metabolism in neuronal fate determination was shown in the context of mouse adult neurogenic niches, where proliferating NSCs show high FASN activity, and its genetic disruption leads to reduced proliferation and neurogenesis [57]. Conversely, a gain-of-function mutation in the same gene (FASN-R1819W, leading to enhanced FASN activity) leads to intellectual disability [58], whereas mutant mice bearing the same mutation also display reduced adult hippocampal neurogenesis [59]. Interestingly, although FASN-R1819W–mutant mice show seemingly normal embryonic cortical development, human cerebral organoids carrying the patient's mutation show reduced proliferation of NSPCs [59]. These results suggest that the impact of FA metabolism is cell type–specific, but could also be linked to species differences.
On the other hand, FAs are converted back to acetyl-CoA via mitochondrial β-oxidation (FAO), which is also implicated in adult and embryonic neurogenesis [60,61] (Figure 4). In adult neurogenesis, the key FAO enzyme, carnitine palmitoyltransferase 1a (Cpt1a), is required for the maintenance of quiescence of NSCs [60]. In the mouse embryonic cortex, downregulation of FAO by knockdown of Cpt1a, or of trimethyllysine hydroxylase, epsilon (TMLHE, involved in FAO through carnitine biosynthesis), leads to decreased self-renewal and apoptosis of cortical NSPCs [60,61]. These data indicate the importance of FA metabolism in neurogenesis, although as for TCA, it will be important to determine the underlying downstream mechanisms, permissive and/or instructive.
Conclusions and perspectives
Mitochondria and related metabolism have emerged as a crucial source of cues required for proper neurogenesis, from NSC self-renewal to neuronal fate commitment, but many open questions are still standing. Among these, it remains unclear how mitochondria dynamics and function are actually linked to one another during neurogenesis, and by which upstream mechanisms they are controlled, in synergy with developmental signals. Conversely, it will be important to find out which downstream mechanisms are involved in each effect of mitochondria on neurogenesis, whether through permissive regulation of metabolite production or through more instructive signalling modes, such as metabolism-driven PTMs.
On the other hand, the finding that mitochondria dynamics–dependent neuronal fate acquisition displays striking timing differences between mouse and human species suggests that metabolism could be causally linked to species-specific properties relevant to brain evolution, perhaps in link with recently uncovered species differences in protein turnover related to developmental timing in other systems [62,63].
Finally, although mitochondrial defects have long been thought to strike preferentially the brain because of its high energy needs, the new findings reviewed here invite to revisit the mechanisms of mitochondria-linked NDD, exploring early neural defects as well as direct links with known neurodevelopmental pathways.
Conflict of interest statement
Nothing declared.
Acknowledgements
The authors wish to apologise to the authors whose work could not be discussed because of space constraints. Work from the P.V. laboratory described here was funded by the European Research Council (ERC Adv Grant), the Fondation Roger de Spoelberch, the Belgian FWO and FRS/FNRS, the AXA Research Fund, the Belgian Queen Elizabeth Foundation and the Fondation ULB. R.I. was a postdoctoral fellow of the Belgian FRS/FNRS.
This review comes from a themed issue on Molecular Neuroscience
Edited by Frank Bradke and Yukiko Goda
References
- 1.Bonnefont J., Vanderhaeghen P. Neuronal fate acquisition and specification: time for a change. Curr Opin Neurobiol. 2021 doi: 10.1016/j.conb.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villalba A., Götz M., Borrell V. Current topics in developmental biology. Academic Press Inc.; 2020. The regulation of cortical neurogenesis; pp. 1–66. [DOI] [PubMed] [Google Scholar]
- 3.Pernas L., Scorrano L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol. 2016;78:505–531. doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarty R.P., Chandel N.S. Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell. 2021;28:394–408. doi: 10.1016/j.stem.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarazona O.A., Pourquié O. Exploring the influence of cell metabolism on cell fate through protein post-translational modifications. Dev Cell. 2020;54:282–292. doi: 10.1016/j.devcel.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diehl K.L., Muir T.W. Chromatin as a key consumer in the metabolite economy. Nat Chem Biol. 2020;16:620–629. doi: 10.1038/s41589-020-0517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falk M.J. Neurodevelopmental manifestations of mitochondrial disease. J Dev Behav Pediatr. 2010;31:610–621. doi: 10.1097/DBP.0b013e3181ef42c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khacho M., Clark A., Svoboda D.S., Azzi J., MacLaurin J.G., Meghaizel C., Sesaki H., Lagace D.C., Germain M., Harper M.E. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]; Khacho and coll. uncover the impact of mitochondria dynamics on mouse neurogenesis, and its potential links with mitochondria-derived reactive oxygen species.
- Iwata R., Casimir P., Vanderhaeghen P. Mitochondrial dynamics in postmitotic cells regulate neurogenesis. Science. 2020;862:858–862. doi: 10.1126/science.aba9760. (80- ) [DOI] [PubMed] [Google Scholar]; Iwata and coll. demonstrate that mitochondria dynamics act after the last division of neural stem/progenitor cells to influence neurogenesis, during a critical period of fate plasticity that is doubled in human vs. mouse NSC. They link the impact of mitochondria dynamics on cell fate to Sirtuin activity and its activity on chromatin remodeling.
- 10.Mitra K., Wunder C., Roysam B., Lin G., Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Telley L., Govindan S., Prados J., Stevant I., Nef S., Dermitzakis E., Dayer A., Jabaudon D. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science. 2016 doi: 10.1126/science.aad8361. (80- ) [DOI] [PubMed] [Google Scholar]
- 12.Dehay C., Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 13.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 14.Homem C.C.F.F., Steinmann V., Burkard T.R., Jais A., Esterbauer H., Knoblich J.A. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014;158:874–888. doi: 10.1016/j.cell.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Agathocleous M., Love N.K., Randlett O., Harris J.J., Liu J., Murray A.J., Harris W.A. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14:859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agostini M., Romeo F., Inoue S., Niklison-Chirou M.V., Elia A.J., Dinsdale D., Morone N., Knight R.A., Mak T.W., Melino G. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 2016;23:1502–1514. doi: 10.1038/cdd.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Brien L.C., Keeney P.M., Bennett J.P. Differentiation of human neural stem cells into motor neurons stimulates mitochondrial biogenesis and decreases glycolytic flux. Stem Cells Dev. 2015;24:1984–1994. doi: 10.1089/scd.2015.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F.H., Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:1–25. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz C., Lesimple P., Bukowiecki R., Zink A., Inak G., Mlody B., Singh M., Semtner M., Mah N., Auré K. Human iPSC-derived neural progenitors are an effective drug discovery model for neurological mtDNA disorders. Cell Stem Cell. 2017;20:659–674.e9. doi: 10.1016/j.stem.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Van Den Ameele J., Brand A.H. Neural stem cell temporal patterning and brain tumour growth rely on oxidative phosphorylation. Elife. 2019;8:1–20. doi: 10.7554/eLife.47887. [DOI] [PMC free article] [PubMed] [Google Scholar]; van den Ameele and Brand demonstrate the requirement of mitochondria OXPHOS activity on NSC self-renewal as well as temporal patterning of fate transitions.
- 21.Khacho M., Clark A., Svoboda D.S., MacLaurin J.G., Lagace D.C., Park D.S., Slack R.S. Mitochondrial dysfunction underlies cognitive defects as a result of neural stem cell depletion and impaired neurogenesis. Hum Mol Genet. 2017;26:3327–3341. doi: 10.1093/hmg/ddx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckervordersandforth R., Ebert B., Schäffner I., Moss J., Fiebig C., Shin J., Moore D.L., Ghosh L., Trinchero M.F., Stockburger C. Role of mitochondrial metabolism in the control of early lineage progression and aging phenotypes in adult hippocampal neurogenesis. Neuron. 2017;93:560–573.e6. doi: 10.1016/j.neuron.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inak G., Rybak-wolf A., Lisowski P., Pentimalli T.M., Jüttner R., Gla P., Uppal K., Bottani E., Brunetti D., Secker C. Defective metabolic programming impairs early neuronal morphogenesis in neural cultures and an organoid model of Leigh syndrome. Nat Commun. 2021;12:1–22. doi: 10.1038/s41467-021-22117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lange C., Turrero Garcia M., Decimo I., Bifari F., Eelen G., Quaegebeur A., Boon R., Zhao H., Boeckx B., Chang J. Relief of hypoxia by angiogenesis promotes neural stem cell differentiation by targeting glycolysis. EMBO J. 2016;35:924–941. doi: 10.15252/embj.201592372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou W., Zhao T., Du J., Ji G., Li X., Ji S., Tian W., Wang X., Hao A. TIGAR promotes neural stem cell differentiation through acetyl-CoA-mediated histone acetylation. Cell Death Dis. 2019;10 doi: 10.1038/s41419-019-1434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D.C., Harvey E.M., Suraj R., Erickson S.L., Mohammad L., Ren M., Liu H., He G., Kaplan D.R., Ellis J. Methylglyoxal couples metabolic and translational control of Notch signalling in mammalian neural stem cells. Nat Commun. 2020;11 doi: 10.1038/s41467-020-15941-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rodrigues et al. describe how a specific glycolysis metabolite, Methylglyoxal, can modulate neurogenesis through a functional switch of GAPDH, from a glycolytic enzyme to an RNA binding protein that regulates the Notch pathway.
- 27.Yang G., Cancino G.I., Zahr S.K., Guskjolen A., Voronova A., Gallagher D., Frankland P.W., Kaplan D.R., Miller F.D. A glo1-methylglyoxal pathway that is perturbed in maternal diabetes regulates embryonic and adult neural stem cell pools in murine offspring. Cell Rep. 2016;17:1022–1036. doi: 10.1016/j.celrep.2016.09.067. [DOI] [PubMed] [Google Scholar]
- 28.Chang C.H., Curtis J.D., Maggi L.B., Faubert B., Villarino A.V., O'Sullivan D., Huang S.C.C., Van Der Windt G.J.W., Blagih J., Qiu J. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid M.A., Dai Z., Locasale J.W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol. 2017;19:1298–1306. doi: 10.1038/ncb3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmström K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 31.Le Belle J.E., Orozco N.M., Paucar A.A., Saxe J.P., Mottahedeh J., Pyle A.D., Wu H., Kornblum H.I. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8:59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adusumilli V.S., Walker T.L., Overall R.W., Klatt G.M., Zeidan S.A., Zocher S., Kirova D.G., Ntitsias K., Fischer T.J., Sykes A.M. ROS dynamics delineate functional states of hippocampal neural stem cells and link to their activity-dependent exit from quiescence. Cell Stem Cell. 2020;28:1–15. doi: 10.1016/j.stem.2020.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo H., Lyssiotis C.A., Zhang Y., Ying H., Asara J.M., Cantley L.C., Paik J.H. FoxO3 coordinates metabolic pathways to maintain redox balance in neural stem cells. EMBO J. 2013;32:2589–2602. doi: 10.1038/emboj.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poché R.A., Zhang M., Rueda E.M., Tong X., McElwee M.L., Wong L., Hsu C.W., Dejosez M., Burns A.R., Fox D.A. RONIN is an essential transcriptional regulator of genes required for mitochondrial function in the developing retina. Cell Rep. 2016;14:1684–1697. doi: 10.1016/j.celrep.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chui A., Zhang Q., Dai Q., Shi S.H. Oxidative stress regulates progenitor behavior and cortical neurogenesis. Development. 2020;147 doi: 10.1242/dev.184150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue M., Iwai R., Tabata H., Konno D., Komabayashi-Suzuki M., Watanabe C., Iwanari H., Mochizuki Y., Hamakubo T., Matsuzaki F. Prdm16 is crucial for progression of the multipolar phase during neural differentiation of the developing neocortex. Development. 2017;144:385–399. doi: 10.1242/dev.136382. [DOI] [PubMed] [Google Scholar]
- 37.Lautrup S., Sinclair D.A., Mattson M.P., Fang E.F. NAD+ in brain aging and neurodegenerative disorders. Cell Metab. 2019;30:630–655. doi: 10.1016/j.cmet.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prozorovski T., Schulze-Topphoff U., Glumm R., Baumgart J., Schröter F., Ninnemann O., Siegert E., Bendix I., Brüstle O., Nitsch R. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 39.Tiberi L., Van Den Ameele J., Dimidschstein J., Piccirilli J., Gall D., Herpoel A., Bilheu A., Bonnefont J., Iacovino M., Kyba M. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat Neurosci. 2012;15:1627–1635. doi: 10.1038/nn.3264. [DOI] [PubMed] [Google Scholar]
- 40.Bonnefont J., Tiberi L., van den Ameele J., Potier D., Gaber Z.B.B.Z.B., Lin X., Bilheu A., Herpoel A., Velez Bravo F.D.F.D.D., Guillemot F. Cortical neurogenesis requires bcl6-mediated transcriptional repression of multiple self-renewal-promoting extrinsic pathways. Neuron. 2019;103:1096–1108 e4. doi: 10.1016/j.neuron.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patgiri A., Skinner O.S., Miyazaki Y., Schleifer G., Marutani E., Shah H., Sharma R., Goodman R.P., To T.L., Robert Bao X. An engineered enzyme that targets circulating lactate to alleviate intracellular NADH:NAD+ imbalance. Nat Biotechnol. 2020;38:309–313. doi: 10.1038/s41587-019-0377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boon R., Silveira G.G., Mostoslavsky R. Nuclear metabolism and the regulation of the epigenome. Nat Metab. 2020;2:1190–1203. doi: 10.1038/s42255-020-00285-4. [DOI] [PubMed] [Google Scholar]
- 43.Russo G.L., Sonsalla G., Natarajan P., Breunig C.T., Bulli G., Merl-Pham J., Schmitt S., Giehrl-Schwab J., Giesert F., Jastroch M. CRISPR-mediated induction of neuron-enriched mitochondrial proteins boosts direct glia-to-neuron conversion. Cell Stem Cell. 2020 doi: 10.1016/j.stem.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonnay F., Veloso A., Steinmann V., Köcher T., Abdusselamoglu M.D., Bajaj S., Rivelles E., Landskron L., Esterbauer H., Zinzen R.P. Oxidative metabolism drives immortalization of neural stem cells during tumorigenesis. Cell. 2020;182:1490–1507.e19. doi: 10.1016/j.cell.2020.07.039. [DOI] [PubMed] [Google Scholar]
- 45.Jayaraman D., Bae B. Il, Walsh C.A. The genetics of primary microcephaly. Annu Rev Genom Hum Genet. 2018;19:177–200. doi: 10.1146/annurev-genom-083117-021441. [DOI] [PubMed] [Google Scholar]
- Journiac N., Gilabert-Juan J., Cipriani S., Benit P., Liu X., Jacquier S., Faivre V., Delahaye-Duriez A., Csaba Z., Hourcade T. Cell metabolic Alterations due to Mcph1 mutation in microcephaly. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.03.070. [DOI] [PubMed] [Google Scholar]; Journiac and coll. discover that MCPH1, a gene mutated in human microcephaly previously associated with mitotic regulation, encodes a protein found in mitochondria, where it regulates several aspects of metabolism possibly linked to neurogenesis.
- 47.McCormack J.G., Halestrap A.P., Denton R.M. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev. 1990;70:391–425. doi: 10.1152/physrev.1990.70.2.391. [DOI] [PubMed] [Google Scholar]
- 48.Montal E.D., Dewi R., Bhalla K., Ou L., Hwang B.J., Ropell A.E., Gordon C., Liu W.J., DeBerardinis R.J., Sudderth J. PEPCK coordinates the regulation of central carbon metabolism to promote cancer cell growth. Mol Cell. 2015;60:571–583. doi: 10.1016/j.molcel.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenberg M.J., Agarwala R., Bouffard G., Davis J., Fiermonte G., Hilliard M.S., Koch T., Kalikin L.M., Makalowska I., Morton D.H. Mutant deoxynucleotide carrier is associated with congenital microcephaly. Nat Genet. 2002;32:175–179. doi: 10.1038/ng948. [DOI] [PubMed] [Google Scholar]
- 50.Lindhurst M.J., Fiermonte G., Song S., Struys E., De Leonardis F., Schwartzberg P.L., Chen A., Castegna A., Verhoeven N., Mathews C.K. Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc Natl Acad Sci U S A. 2006;103:15927–15932. doi: 10.1073/pnas.0607661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon W.H., Sandoval H., Nagarkar-Jaiswal S., Jaiswal M., Yamamoto S., Haelterman N.A., Putluri N., Putluri V., Sreekumar A., Tos T. Loss of nardilysin, a mitochondrial Co-chaperone for α-ketoglutarate dehydrogenase, promotes mTORC1 activation and neurodegeneration. Neuron. 2017;93:115–131. doi: 10.1016/j.neuron.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hengel H., Keimer R., Deigendesch W., Rieß A., Marzouqa H., Zaidan J., Bauer P., Schöls L. GPT2 mutations cause developmental encephalopathy with microcephaly and features of complicated hereditary spastic paraplegia. Clin Genet. 2018;94:356–361. doi: 10.1111/cge.13390. [DOI] [PubMed] [Google Scholar]
- 53.Namba T., Pinson A., Wimberger P., Chinopoulos C., Huttner W.B. Human-specific ARHGAP11B acts in mitochondria to expand neocortical progenitors by glutaminolysis article acts in mitochondria to expand neocortical progenitors by glutaminolysis. Neuron. 2020 doi: 10.1016/j.neuron.2019.11.027. [DOI] [PubMed] [Google Scholar]
- 54.Kasahara A., Cipolat S., Chen Y., Dorn G.W., Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and notch signaling. Science. 2013 doi: 10.1126/science.1241359. (80- ) [DOI] [PubMed] [Google Scholar]
- 55.Shan Y., Zhang Y., Zhao Y., Wang T., Zhang J., Yao J., Ma N., Liang Z., Huang W., Huang K. JMJD3 and UTX determine fidelity and lineage specification of human neural progenitor cells. Nat Commun. 2020;11:1–16. doi: 10.1038/s41467-019-14028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X., Xu B., Mulvey B., Evans M., Jordan S., Wang Y.D., Pagala V., Peng J., Fan Y., Patel A. Differentiation of human pluripotent stem cells into neurons or cortical organoids requires transcriptional co-regulation by UTX and 53BP1. Nat Neurosci. 2019;22:362–373. doi: 10.1038/s41593-018-0328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knobloch M., Braun S.M.G., Zurkirchen L., Von Schoultz C., Zamboni N., Araúzo-Bravo M.J., Kovacs W.J., Karalay Ö., Suter U., MacHado R.A.C. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 2013;493:226–230. doi: 10.1038/nature11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Bowers M., Liang T., Gonzalez-Bohorquez D., Zocher S., Jaeger B.N., Kovacs W.J., Röhrl C., Cramb K.M.L., Winterer J., Kruse M. FASN-dependent lipid metabolism links neurogenic stem/progenitor cell activity to learning and memory deficits. Cell Stem Cell. 2020;27:98–109. doi: 10.1016/j.stem.2020.04.002. e11. [DOI] [PubMed] [Google Scholar]; Bowers et al. describe the mechanisms by which specific mutations in Fasn, the Fatty acid synthase gene, affects several aspects of human embryonic neurogenesis.
- 60.Knobloch M., Pilz G.A., Ghesquière B., Kovacs W.J., Wegleiter T., Moore D.L., Hruzova M., Zamboni N., Carmeliet P., Jessberger S. A fatty acid oxidation-dependent metabolic shift regulates adult neural stem cell activity. Cell Rep. 2017;20:2144–2155. doi: 10.1016/j.celrep.2017.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie Z., Jones A., Deeney J.T., Hur S.K., Bankaitis V.A. Inborn errors of long-chain fatty acid β-oxidation link neural stem cell self-renewal to autism. Cell Rep. 2016;14:991–999. doi: 10.1016/j.celrep.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuda M., Hayashi H., Garcia-Ojalvo J., Yoshioka-Kobayashi K., Kageyama R., Yamanaka Y., Ikeya M., Toguchida J., Alev C., Ebisuya M. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science. 2020;1455:1450–1455. doi: 10.1126/science.aba7668. (80- ) [DOI] [PubMed] [Google Scholar]
- 63.Rayon T., Stamataki D., Perez-Carrasco R., Garcia-Perez L., Barrington C., Melchionda M., Exelby K., Lazaro J., Tybulewicz V.L.J., Fisher E.M.C. Species-specific pace of development is associated with differences in protein stability. Science. 2020 doi: 10.1126/SCIENCE.ABA7667. (80- ) [DOI] [PMC free article] [PubMed] [Google Scholar]