Abstract
The adoptive transfer of Chimeric Antigen Receptor (CAR) T-cells represents a breakthrough in clinical oncology, yet both between- and within-patient differences in autologously-derived T-cells are a major contributor to therapy failure. In order to interrogate the molecular determinants of clinical CAR T-cell persistence, we extensively characterized the pre-manufacture T-cells of 71 patients with B-cell malignancies on trial to receive anti-CD19 CAR T-cell therapy. We performed RNA-Seq on sorted T-cell subsets from all 71 patients, followed by paired CITE-Seq and single-cell ATAC-Seq on T-cells from 6 of these patients. We found that chronic interferon signaling regulated by IRF7 was associated with poor CAR T-cell persistence across T-cell subsets, and that the TCF7 regulon not only associates with the favorable naive T-cell state, but is maintained in effector T-cells among patients with long-term CAR T-cell persistence. These findings provide key insights into the underlying molecular determinants of clinical CAR T-cell function.
Introduction
Chimeric Antigen Receptor (CAR) T-cell therapy has been a breakthrough in cancer therapy, yet failure to achieve long-term CAR T-cell persistence remains a major barrier to sustained remission in many patients. While complete response rates in pediatric B-cell Acute Lymphoblastic Leukemia (B-ALL) are high, differences in apheresis T-cell products have been shown to play a critical role in determining the duration of anti-tumor CAR T-cell function (1). Recent studies have provided strong evidence that differences in apheresed T-cells are a major determinant in therapeutic failure of CAR T-cell therapy in adult malignancies such as Chronic Lymphocytic Leukemia (CLL) and Multiple Myeloma (MM) (2,3), suggesting that a deeper understanding of intrinsic T-cell function is essential for improving existing and future CAR T-cell therapies for both pediatric and adult cancers.
At the core of this challenge is the heterogeneous nature of autologous T-cells that form the starting material for CAR T-cell therapy. Most CAR T-cell therapy trials to date have engineered CAR T-cells from the bulk population of T-cells extracted from the patient, consisting of a mixture of CD4+ and CD8+ T-cells across naive, memory, and effector lineages. Clinical and preclinical studies have led to a growing understanding of a positive role of naive and memory T-cell subsets in CAR T-cell efficacy, suggesting that cellular stemness and memory formation are critical to maintaining long-term remission (4–6). These studies have provided foundational insight into the composition of favorable T-cell phenotypes, yet a more comprehensive understanding has been hindered by limited sample sizes, confounding by T-cell subtype, and lack of deeper molecular analyses.
In order to address these challenges, we developed a subtype-specific transcriptomic atlas of pre-manufacture T-cells from 71 patients on trial to receive anti-CD19 CAR T-cell therapy. A crucial component of our experimental design was to sort T-cell subsets prior to RNA-sequencing, allowing us to directly account for the confounding effect of T-cell subset composition and identify clinically associated molecular pathways that act within T-cell subsets. We developed a web server for visualization of genes and regulons from our transcriptional atlas, which can be accessed at https://tanlab4generegulation.shinyapps.io/Tcell_Atlas/. Based on our findings regarding interferon signaling and TCF7 expression in effector T-cell subtypes, we performed integrative CITE-Seq and single-cell ATAC-Seq on 6 of the patients in the study, allowing us to more deeply characterize the interplay and regulation of these clinically-associated biological pathways.
Results
Naive and early memory T-cell composition predicts clinical CAR T-cell persistence
We identified 71 children or young adults who were enrolled to receive anti-CD19 CAR T-cell therapy at the Children’s Hospital of Philadelphia (Supplementary Table S1). The mean age at enrollment was 11.7 (95% confidence interval [10.1–13.3]), and there was a balanced ratio of female and male patients (47.9%, [36.0–60.0] female). Seventy patients had relapsed or refractory B-ALL, and one was a young adult with Hodgkin’s lymphoma. CAR T-cell manufacture was successful in 65 of the 71 patients. The primary clinical endpoint for functional CAR T-cell persistence was duration of B-cell aplasia (BCA), consistent with prior studies which have established that BCA is a sensitive marker of functional anti-CD19 CAR T-cell persistence in pediatric populations (1,7). Long term CAR T-cell persistence was defined as BCA ≥ 6 months, whereas short-term or failed persistence was defined as BCA < 6 months. Duration of BCA was considered to be right-censored if the patient had ongoing BCA at the most recent follow-up, proceeded to bone marrow transplant, died of other causes, or had CD19− relapse with continued BCA. In an intention-to-treat manner, the median duration of CAR T-cell persistence among patients in our cohort was 11 months, similar to event-free survival durations reported in recent clinical trials in B-ALL (8).
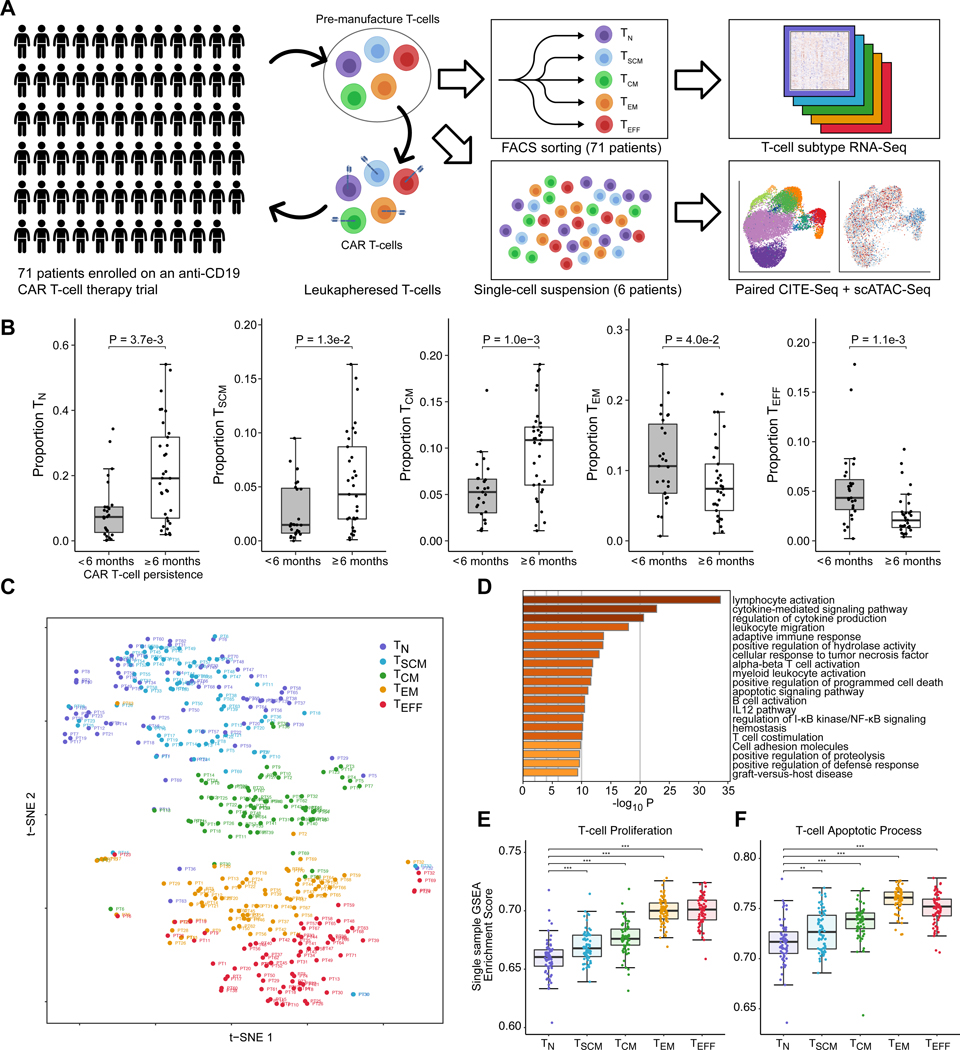
In order to assess the composition and molecular pathways of pre-manufacture T-cells, we curated a biobank of T-cells from these 71 patients (Fig. 1A). These T-cells were aliquoted at time of clinical leukapheresis, and represent the populations of the T-cells used for CAR T-cell manufacture. We used Fluorescence-Activated Cell Sorting (FACS) to sort T-cells into five T-cell subsets: Naive (TN), Stem Cell Memory (TSCM), Central Memory (TCM), Effector Memory (TEM), and Effector (TEFF) (Supplementary Fig. S1A). We performed RNA-Seq on the five sorted T-cell populations for each of the patients, producing an atlas of 355 transcriptomic profiles representing the T-cell composition across pediatric pre-manufacture T-cells.
Figure 1. A transcriptome atlas of pre-manufacture T-cells among children and young adults enrolled to receive anti-CD19 CAR T-cell therapy.
(A) T-cells from 71 patients were collected at time of clinical leukapheresis and sorted into five T-cell subsets: Naive (TN), Stem Cell Memory (TSCM), Central Memory (TCM), Effector Memory (TEM), and Effector (TEFF). All 355 T-cell populations underwent RNA-Sequencing. For 6 of these patients, paired CITE-Seq and single-cell ATAC-Seq was performed. (B) Association between proportion of TN, TSCM, TCM, TEM and TEFF at time of leukapheresis with long-term CAR T-cell persistence, assessed by duration of B-cell Aplasia (BCA). Pairwise statistical significance was assessed using the Wilcoxon rank-sum test, and multiple testing correction was performed using the Benjamini-Hochberg procedure. (C) t-distributed stochastic neighbor embedding (t-SNE) plot of transcriptome data, capturing the functional continuum from naive, memory, and effector T-cell lineages. (D) Enriched pathways among differentially expressed genes from comparison of T-cell subsets (ANOVA F-test FDR < 0.05, top 500 genes). (E) Single-sample Gene Set Enrichment Analysis scores of T-cell proliferation and (F) apoptotic pathways across T-cell subsets. Statistical significance for pairwise comparisons was performed with Welch’s t-test.
We found that higher proportions of TN, TSCM, and TCM were associated with clinical CAR T-cell persistence beyond 6 months (False Discovery Rate (FDR)-adjusted p = 3.7e-3, 1.3e-2, 1.0e-3), and lower proportions of TEM and TEFF were associated with clinical CAR T-cell persistence beyond 6 months (FDR-adjusted p = 4.0e-2, p = 1.1e-3) (Fig. 1B). Combined proportions of TN, TSCM, TCM, and TEM were associated with clinical CAR T-cell persistence, with greatest discrimination observed when combining TN, TSCM, and TCM proportions (Log-rank test p = 6.2e-3 to 1.4e-2, Supplementary Fig. S1B–C), suggesting that a general and robust trend for a role of naive and memory subsets as a prognostic marker for clinical CAR T-cell function.
We performed CIBERSORTx (9) analysis to estimate the relative proportion in CD4+ and CD8+ T-cells in each T-cell subset. In order to robustly estimate deconvolution proportions, we ran the algorithm using bulk and single-cell references, finding that estimated proportions were robust and broadly concordant with naive, memory, and activated T-cell phenotypes based on reference annotations (Supplementary Fig. S2A–B). We found that TN, TSCM, and TCM subsets were predominantly CD4+, whereas TEM and TEFF subsets were predominantly CD8+ (Supplementary Figure S2C). Using the deconvoluted data, we found that a greater proportion of CD8+ TN and TSCM cells was positively associated with clinical CAR T-cell persistence (FDR-adjusted p = 3.1e-2, 4.1e-2; Supplementary Fig. S2D). This trend was observed both in our deconvolution estimates and in differential expression of CD8 genes (Supplementary Fig. S2E). These data suggest a key role of naive T-cells, particularly in the CD8+ compartment, in contributing to CAR T-cell efficacy.
TEFF and TEM highly express genes for T-cell activation and proliferation, but persistence may be limited by increased apoptosis
Since the composition of naive, memory, and effector T-cell lineages was robustly associated with clinical CAR T-cell persistence, we next sought to investigate the biological pathways defining these T-cell subtypes among our patient cohort. Our bulk RNA-Seq data on sorted patient T-cells captured the functional continuum of T-cell differentiation, from TN, TSCM, TCM, TEM and TEFF subtypes (Fig. 1C). At a global transcriptomic level, between-subtype differences in T-cells were the dominant effect; T-cells of the same subtype clustered strongly between patients, whereas T-cells derived from individual patients generally separated by subtype. TN and TSCM shared similar transcriptomic profiles and were largely indistinguishable at the transcriptomic level, whereas TCM, TEM, and TEFF separated strongly.
Differential expression analysis with Linear Models for Microarray Data (Limma) (10,11) revealed 6,953 differentially expressed genes associated with T-cell type (FDR < 0.05, Supplementary Table S2). Pathway analysis of the differentially expressed genes revealed an enrichment in lymphocyte activation, cytokine signaling, and leukocyte migration pathways (p = 1.93e-34, 1.49e-23, 2.54e-21; Fig. 1D); single-sample Gene Set Enrichment Analysis (12) revealed that these pathways were enriched among the effector T-cell lineages (Supplementary Fig. S3A).
We found that expression of lymphocyte proliferation pathways increased along a gradient towards effector T-cells (p = 2.5e-15, Fig. 1D–E), associated with higher Ki-67 expression in TEM and TEFF cells (Supplementary Fig. 3A). Effector T-cells exhibited extensive metabolic reprogramming supportive of a functionally activated state. Expression of LDHA, GLUT1, and GAPDH was up-regulated among effector T-cells, indicating a metabolic shift to aerobic glycolysis; and pathways involving biosynthesis of protein, DNA, and lipid were enriched, suggesting biosynthetic support for cellular division (Supplementary Fig. S3B). Counteracting these pathways, we found that apoptotic pathways, including both intrinsic and extrinsic apoptotic signaling, were strongly enriched among genes expressed higher in the effector lineages (p = 2.6e-12, Fig. 1F, Supplementary Fig. S3C–D). Up-regulated expression of cytotoxic markers, cytokines, and inhibitory receptors among TEM and TEFF cells suggest that these subsets likely represent functional, endogenously activated T-cell phenotypes (Supplementary Fig. S4A–B). Together, these data suggest that while the more differentiated T-cells exhibited a proliferative phenotype at baseline, their proliferative ability may be inherently limited by apoptotic pathways that act as a barrier to the years-long cellular survival and proliferation necessary for long-term success of CAR T-cell therapy.
Network analysis reveals critical roles of TCF7 and LEF1 in maintaining naive and early memory T-cell states
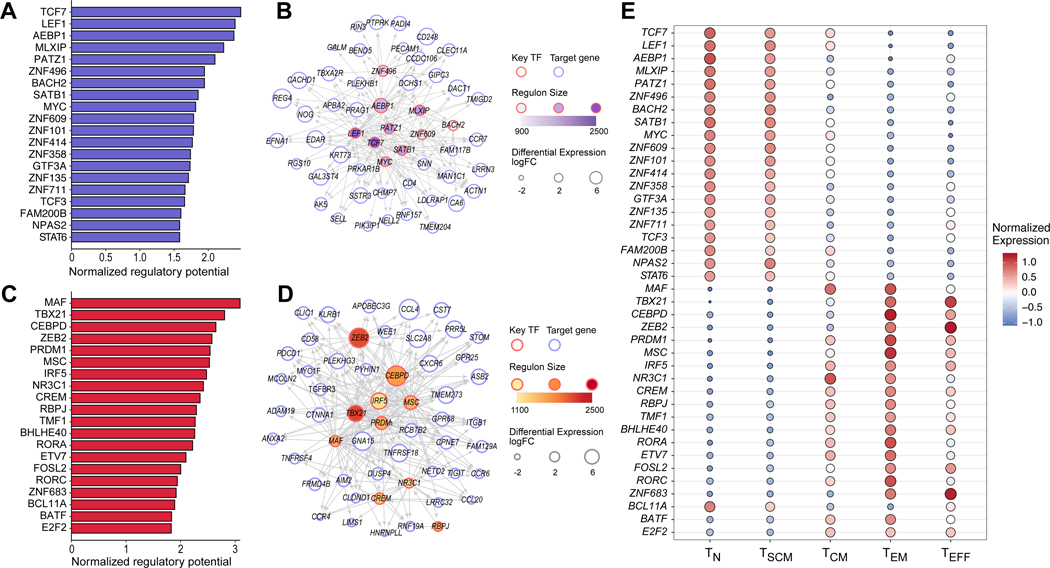
Next, we sought to investigate the transcriptional regulators that act to maintain the naive and early memory phenotypes associated with long-term CAR T-cell persistence. We sought to construct a robust transcriptional regulatory network (TRN) consisting of predicted interactions between transcription factors (TFs) and target genes. Benchmarking studies for TRN inference have demonstrated that no individual computational method performs optimally across data sets, and a consensus approach achieves superior and more robust performance (13). Following this principle, we applied top-performing gene regulatory inference algorithms (14–17) to our expression data to generate base networks, from which we constructed a consensus TRN using the Borda Count principle as previously described (13). In order to identify T-cell specific TF-gene interactions, we repeated these steps to construct a consensus TRN for non-T-cell immune populations from a compendium of 43 microarray datasets from Becht et al. (18), representing 708 samples, and defined a T-cell specific TRN as the network whose edges are present in the T-cell TRN but not present in the non-T-cell immune TRN from public data (see Methods). To validate this network approach, we considered a set of T-cell TFs involved in T-cell differentiation (19) as a benchmark, and assessed the node degree of these TFs in the T-cell specific network and the network constructed from non-T-cell public immune data. The node degree of these benchmark TFs was significantly higher in the T-cell specific network compared to the null distribution (Wilcoxon rank-sum p = 7.6e-5, Supplementary Fig. S5A), but not different from the null distribution in the non-T-cell immune network (p = 0.32), demonstrating the capacity of the constructed T-cell specific network to capture T-cell transcriptional regulatory biology.
We have previously described a method for identifying key TFs involved in regulating transcriptional fate, which we have recently applied to study hematopoiesis and type I diabetes (20–22). We extended this method to our T-cell specific network in order to identify key TFs acting to maintain naive and stem cell memory state, as well as TFs driving effector T-cell development. Among the TFs with the strongest predicted regulatory potential driving naive and early memory T-cell state were TCF7 and LEF1 (FDR < 0.05, Fig. 2A–B), which have been previously described as essential in early thymocyte development (23,24) and maintenance of T-cell memory function (19,25). Among the TFs with the strongest regulatory potential for effector T-cell states were TBX21 (T-bet), which has been associated with both effector CD8+ and CD4+ TH1 differentiation (19,26,27); PRDM1 (Blimp-1), which has been associated with differentiation of CD8+ cells and non-Tfh CD4+ cells (19,26,28); and ZEB2, a transcriptional repressor that has been recently shown to cooperate with T-bet to promote a terminal CD8+ T-cell differentiation program (29,30) (FDR < 0.05, Fig. 2C–D). These were supported with strong differential expression of these transcription factors across T-cell subtype (Fig. 2E, Supplementary Fig. S5B–F).
Figure 2. Transcriptional regulation of naive and effector T-cell states.
(A) Top 20 predicted transcription factors (TFs) associated with TN cells sorted by normalized regulatory potential (FDR < 0.05). (B) Transcriptional regulatory network of top 10 predicted TFs in (A) and top 50 differentially expressed genes (DEGs). (C) Top 20 predicted key TFs associated with TEFF cells (FDR < 0.05). (D) Transcriptional regulatory network of the top 10 predicted TFs in (C) and top 50 DEGs. (E) Differential expression of TFs predicted to regulate naive and effector T-cell states. Log-transformed gene expression values were normalized across the 355 T-cell samples using the z-score transformation.
Chronic interferon response associates with poor clinical CAR T-cell persistence
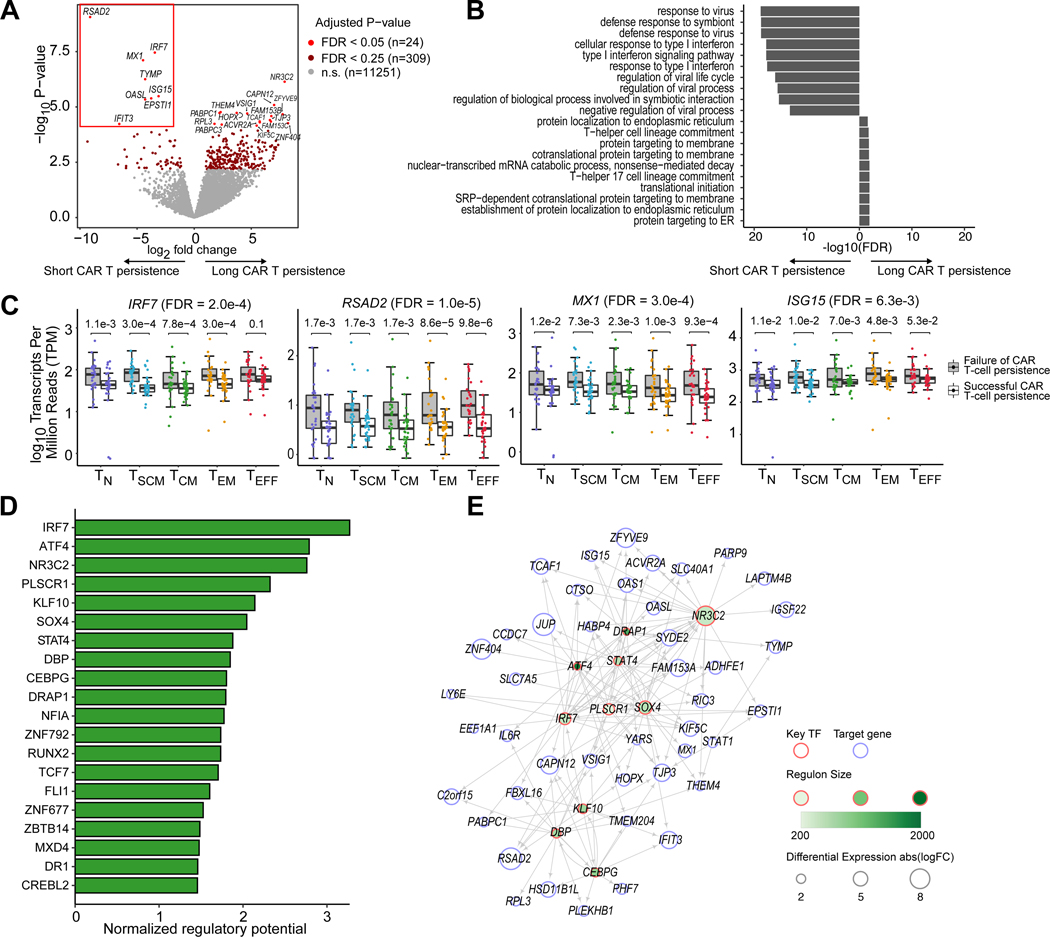
We designed a mixed-effects regression model to identify differentially expressed genes within T-cell subtypes associated with clinical CAR T-cell persistence (Supplementary Table S2; Methods). This approach allowed us to identify genes and pathways associated with clinical CAR T-cell persistence in a manner that accounts for the potential confounding effect of T-cell subtype composition (Fig. 3A). We performed pathway analysis on the differentially expressed genes, and found that type I interferon signaling response was the most significantly enriched pathway (Fig. 3B, p = 2.67e-16). The genes most differentially up-regulated among patients with poor CAR T-cell persistence included the type I interferon response genes RSAD2, IRF7, MX1, ISG15, OASL, and IFIT3 (FDR < 0.05; Fig. 3A). Interestingly, these genes were differentially expressed between patients even among the naive and memory T-cell subtypes (Fig. 3C).
Figure 3. Interferon signaling pathway genes are up-regulated across T-cell subsets among patients with poor CAR T-cell persistence.
(A) Volcano plot showing differentially expressed genes in patients with long-term (≥ 6 months) vs failed (< 6 months) CAR T-cell persistence across T-cell subsets. Significantly differentially expressed genes associated with failed CAR T-cell persistence (FDR < 0.05) are highlighted with a red box. (B) Top enriched pathways among up-regulated and down-regulated differentially expressed genes between patients with long-term vs failed CAR T-cell persistence. (C) Boxplots of example differentially expressed genes in the interferon response pathway. The overall ANOVA F-test FDR is shown next to the gene. FDRs for pairwise comparisons within each T-cell subtype are shown on top of each pair of boxplots. (D) Top 20 predicted key transcription factors (TFs) associated with the long-term vs failed persistence sorted by normalized regulatory potential (FDR < 0.05). (E) Transcriptional regulatory network of the top 10 predicted TFs in (D) and top 50 DEGs.
We extended our gene network inference methods to define Sregressed, a T-cell specific network generated from expression data regressed for the potentially confounding effect of T-cell subtype (see Methods). We applied our TF prioritization method to identify transcription factors associated with clinical CAR T-cell persistence. Our network highlighted IRF7 as the gene with the strongest predicted regulatory potential discriminating between clinical CAR T-cell persistence (Fig. 3D–E). Among other top hits were STAT4, which has also been associated with interferon response signaling and TH1 differentiation (27,31), and SOX4 which has been associated with T-cell differentiation (32).
Type I interferon response genes have been shown to play an important role in physiological T-cell activation (33); indeed, we found that the type I interferon response pathway was up-regulated in the TEM and TEFF (Supplementary Fig. S6A). We asked whether this pathway was up-regulated among patients with poor CAR T-cell persistence in the naive and early memory subsets. To assess the robustness of this pathway, we repeated our differential expression analysis on T-cell subsets that both included and excluded the TEM and TEFF populations. IRF7, RSAD2, MX1, and ISG15 were broadly differentially expressed among all T-cell subsets, and interferon genes were among the top genes associated with poor CAR T-cell persistence in comparisons including strictly naive and early memory T-cell subsets (Supplementary Fig. S6B). For example, in our mixed-effects interaction model, we identified 16 genes associated with clinical CAR T-cell persistence within the TN, TSCM, and TCM subtypes, seven of which were negatively associated with clinical CAR T-cell persistence: IRF7, RSAD2, SLC7A5, OASL, TYMP, MX1, and ISG15 (Supplementary Fig. S7A). We found that among these early memory T-cell subsets, IRF7 was the top-ranked transcription factor associated with poor clinical CAR T-cell persistence, suggesting that the interferon signaling response pathways were enriched across T-cell phenotypes, including the naive and early memory subsets (Supplementary Fig. S7B–D).
The TCF7 network in patients with long-term CAR T-cell persistence is maintained in effector T-cell lineages
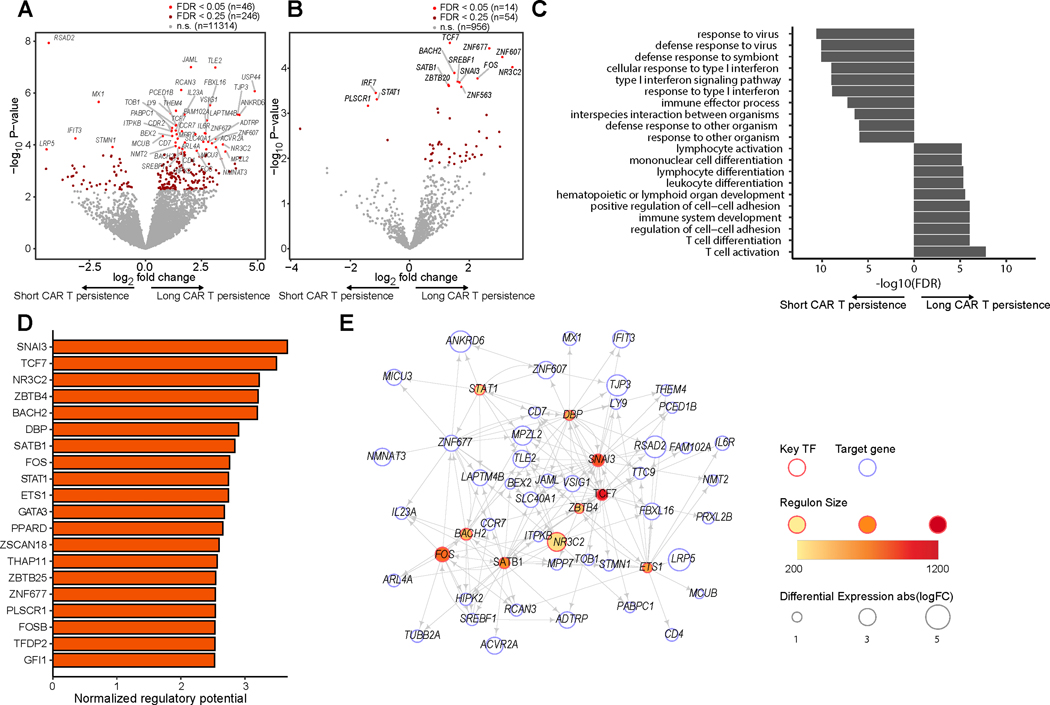
We next hypothesized that differential molecular pathways between patients with long and short CAR T-cell persistence may manifest in transcriptional differences in the effector T-cell phenotypes. We focused on the TEM and TEFF subsets, identifying genes (Fig. 4A) and transcription factors (Fig. 4B) differentially expressed between patients with short and long CAR T-cell persistence. We observed an enrichment of interferon signaling response pathways (p = 2.49e-8), and the most differentially expressed genes associated with poor CAR T-cell persistence were interferon response genes including RSAD2, MX1, and IFIT3 (Fig. 4A). Unlike our previous analyses where interferon response pathways dominated the differential expression analysis, the highest-ranked pathways for TEM and TEFF were those associated with T-cell activation and differentiation (p = 3.34e-13, Fig. 4C), suggesting that regulators of T-cell differentiation state may have an additional role in maintaining T-cell persistence within these differentiated subtypes.
Figure 4. Maintenance of the TCF7 network among effector T-cells associates with long CAR T-cell persistence.
(A) Volcano plot displaying differentially expressed genes in TEM and TEFF between patients with long-term (≥ 6 months) vs failed (< 6 months) CAR T-cell persistence. (B) Volcano plot displaying differentially expressed transcription factors in TEM and TEFF between patients with long-term (≥ 6 months) vs failed (< 6 months) CAR T-cell persistence. (C) Top enriched pathways among up-regulated and down-regulated differentially expressed genes between patients with long-term vs failed CAR T-cell persistence in TEM and TEFF. (D) Top 20 predicted TFs associated with long CAR T-cell persistence in TEM and TEFF (FDR < 0.05). (E) Transcriptional regulatory network of the top 10 predicted TFs in (D) and top 50 DEGs.
Narrowing our differential expression analysis to transcription factors in these effector phenotypes, we found that TCF7 was the most significantly up-regulated TF associated with long CAR T-cell persistence (FDR = 0.018, Fig. 4B). Network analysis revealed that TCF7 was among the top-ranked TFs associated with CAR T-cell persistence in TEM and TEFF; among the other top hits expressed at higher levels among patients with long CAR T-cell persistence were BACH2, FOS, and GATA3, which is associated with TH2 development; among top hits expressed at higher levels in patients with short CAR T-cell persistence was STAT1, which has been associated with TH1 development and interferon signaling (27,34). (Fig. 4D–E). Strictly within the TEFF subset, we found TCF7 was the most significantly up-regulated transcription factor associated with long CAR T-cell persistence, and T-cell activation remained the top-enriched pathway (Supplementary Fig. S8A–D). We repeated our network analysis using exclusively TEFF expression data, and found that TCF7 remained among the top 20 ranked TFs, along with GATA3 and BACH2 (Supplementary Fig. S8E–F).
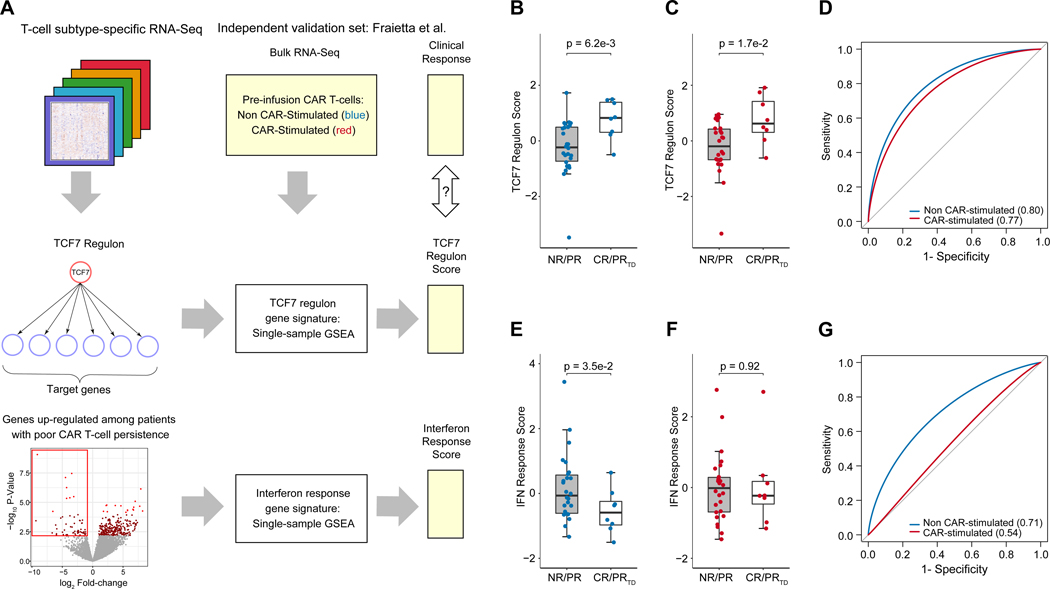
The TCF7 regulon and interferon response gene signatures are associated with clinical CAR T-cell therapy outcomes in an independent validation set
We next sought to validate transcriptional gene signatures representing the TCF7 regulon and interferon signaling response (Fig. 5A). During network construction from our complete transcriptome dataset involving all T-cell subtypes, TCF7 was the transcription factor with the greatest node connectivity, with 2,496 predicted targets, of which 1,487 had a positive expression correlation with TCF7. We defined the TCF7 regulon as the set of predicted target genes of TCF7 with a positive expression correlation, and defined the TCF7 regulon score as the single-sample GSEA enrichment score using the gene set composed of TCF7 and its regulon (Supplementary Table S3). Since this score was defined in a manner agnostic to T-cell type or clinical outcome labels, we first assessed whether this score discriminated between naive and effector T-cell types, as well as between clinical response groups among effector T-cells. Indeed, this TCF7 regulon score discriminated between T-cell subtypes, was significantly higher in TEFF among patients with long-term CAR T-cell persistence, and discriminated between these clinical outcome groups using leave-one-patient-out and leave-one-T-cell-type-out cross-validation (Supplementary Fig. S9A–D). Next, we sought to assess this score in an independent validation set. We assessed this score on the RNA-Seq dataset of Fraietta et al., which consisted of pre-infusion anti-CD19 CAR T-cells generated from adults with Chronic Lymphocytic Leukemia (CLL) (5) and is, to our knowledge, the largest clinically-annotated RNA-Seq dataset from patients receiving CAR T-cell therapy prior to our present study. The CAR T-cells of these 34 patients assessed either underwent mock stimulation (non-CAR stimulated group) or bead-based anti-CD19 CAR stimulation in vitro (CAR-stimulated group). This dataset had several differences from ours, since it was generated from engineered CAR T-cells, collected from adults with CLL, and was not sorted by T-cell subtype. Despite these differences, we sought to evaluate whether our gene signatures were robust enough to validate in this independent dataset. Indeed, we found that our TCF7 regulon score was positively associated with favorable clinical response in the non-CAR-stimulated samples (p = 0.0062, Fig. 5B–D), as well as in the CAR-stimulated group (p = 0.017). The discriminative ability of this gene signature even in the CAR-stimulated group suggests that the prognostic relevance of TCF7 regulon is maintained throughout CAR T-cell manufacture and stimulation.
Figure 5. Validation of TCF7 regulon and interferon response gene signature in an independent dataset.
(A) Workflow for development and validation of gene signatures representing the TCF7 regulon and interferon response pathways acting between and within T-cell subsets. (B-C) Evaluation of the TCF7 regulon score on the independent dataset of Fraietta et al. on both the unstimulated (blue) and CAR-stimulated (red) CAR T-cells from patients with chronic lymphoblastic leukemia (CLL). Each point represents a patient. Clinical response groups were as previously described in the independent validation set5, with unfavorable outcomes, NR = non-responder, PR = partial responder; and favorable outcomes PRTD = partial responder with highly active T-cell products, CR = complete remission. Statistical significance between clinical response groups was assessed with Welch’s t-test. (D) Receiver operating characteristic curve for the TCF7 regulon score. (E-F) Evaluation of the interferon response score on the independent dataset of Fraietta et al. on both the unstimulated (blue) and CAR-stimulated (red) CAR T-cells. Statistical significance was assessed with Welch’s t-test. (G) Receiver operating characteristic curve for the interferon response score.
Next, we developed a prognostic gene signature representing the interferon response observed across T-cell subtypes. We identified 55 genes significantly up-regulated among those patients with short CAR T-cell persistence, which was strongly enriched for type I interferon response pathways (Fig. 3A, C). We thus defined an interferon response score as the single-sample GSEA enrichment score based on these differentially expressed genes (Supplementary Table S3). This gene signature was associated with T-cell subtypes, and was up-regulated among the TCM, TEM, and TEFF subsets compared to TN (Supplementary Fig. S9E). We performed cross-validation based on a leave-one-patient-out and leave-one-T-cell-type-out approach, finding that this method for gene signature discovery robustly discriminated between patients with long-term and short-term CAR T-cell persistence across T-cell subtypes (Area Under Receptor Operating Characteristic Curve (AUROC) = 0.63–0.76, Supplementary Fig. S9F–G). We next assessed this gene signature on the independent validation set by Fraietta et al. (5) We found that for the non-CAR-stimulated group, our interferon response signature was significantly associated with poor clinical response (p = 0.035, AUROC = 0.71, Fig. 5E). However, for the CAR-stimulated group, this gene signature was not significantly prognostic (p = 0.92, AUROC = 0.54, Fig. 5F–G), suggesting that the T-cell activation pathways triggered by CAR stimulation may negate the prognostic significance of the interferon response signature.
Analysis of T-cell subset markers and TF expression in the manufactured CAR T-cell product
We asked to what extent the engineered CAR T-cells differ from pre-manufacture T-cells and retain an association with long-term clinical CAR T-cell persistence. We identified engineered CAR T-cells from 11 patients on trial to receive CAR T-cell therapy (study identifier: NCT01626495) who were not previously studied in our bulk RNA-Seq analysis: four had long-term B-cell aplasia greater or equal to 6 months, five had short-term B-cell aplasia less than six months, and two were not ultimately infused due to medical contraindications. We identified pre-manufacture T-cells from three of these patients. FACS analysis revealed increased protein expression of IRF7 in the CAR T-cell products compared to pre-manufacture T-cells across T-cell subsets (FDR = 0.001 to 0.062, Supplementary Fig. S9H), suggestive of interferon signaling induced during the CAR T manufacturing process. Among the post-manufacture CAR T-cell samples from the nine patients who received a CAR T-cell infusion, proportions of T-cell subsets defined by CD62L and CD45RO were not significantly associated with long-term CAR T-cell persistence (FDR = 0.948, Supplementary Fig. S9I). RNA expression assessed by RT-qPCR of transcription factors was assessed for association with clinical CAR T-cell persistence, with expression of TCF7 trending towards higher expression in patients with long CAR T persistence, IRF7 and TBX21 trending towards higher expression in patients with short CAR T persistence, and no clear trend for LEF1 and PRDM1 expression; these trends were not statistically significant (p=0.161–0.794; Supplementary Fig. S9J–K). The relatively small sample size likely contributed to the lack of statistical significance in these trends; power analysis suggested that 24, 22, and 17 patients per group would be required to achieve statistical significance at 0.05 with 80% power for expression of TCF7, IRF7, and TBX21 respectively. In addition, in vitro stimulation during CAR T manufacture may at least partially abrogate between-patient T-cell differences as reported in a previous study (1).
Integrative single-cell analysis demonstrates that the TCF7 regulon is not mutually exclusive to interferon response
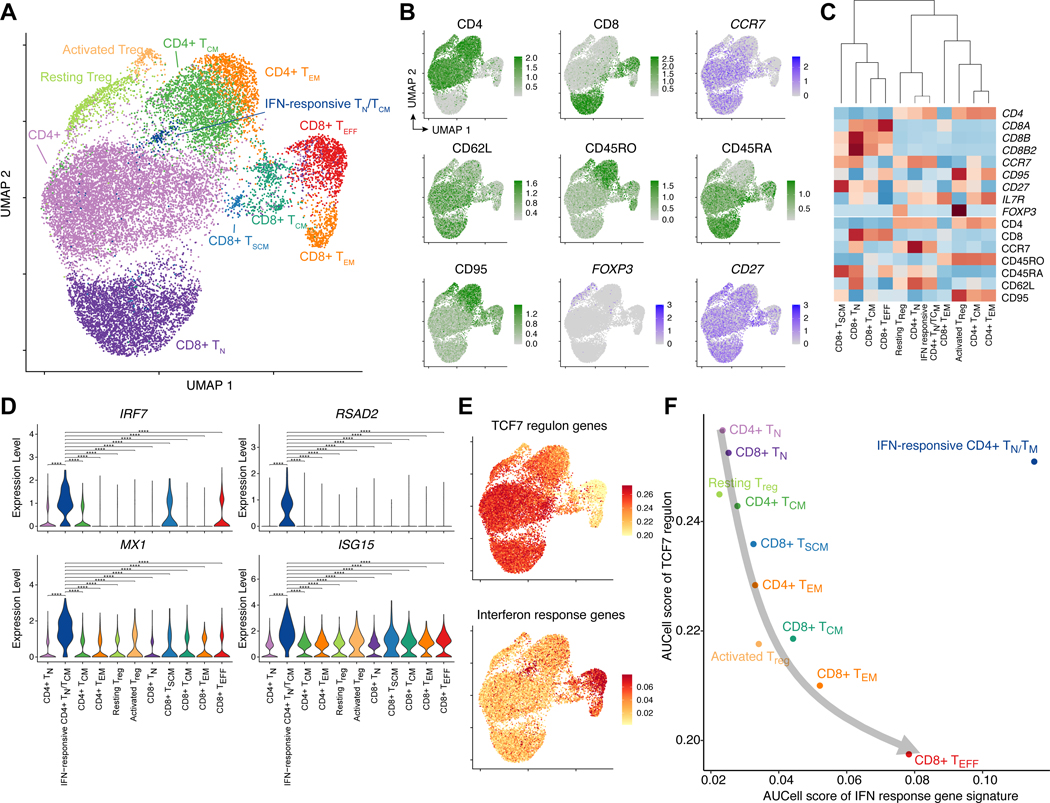
In order to more deeply understand the relationship between the TCF7 regulon and interferon response in pre-manufacture T-cells, we performed CITE-Seq (35) and single-cell ATAC-Seq (scATAC-Seq) on 6 of the 71 patients included in this study. These 6 patients were selected based on sample availability and representing a range of clinical CAR T-cell persistence outcomes, from failure at 2 months to persistence greater than 22 months. From our CITE-Seq data, we obtained a dual readout of cell surface protein expression and RNA quantification within the same cell; scATAC-Seq was performed in a separate aliquot of T-cells from the same patients. After filtering out low-quality cells, our single-cell data consisted of 17,750 and 19,673 cells from the CITE-Seq and scATAC-seq data respectively. We first focused on the CITE-Seq data, performing data integration, dimensionality reduction, and clustering to reveal a single-cell landscape of CD4+ and CD8+ T-cells (Methods, Fig. 6A–C, Supplementary Fig. S10A). The data broadly mixed by patient and separated by both RNA and protein markers, suggestive of good integration (Supplementary Fig. S10B–C). The scRNA-Seq data were initially clustered into 21 clusters based on global transcriptomic profiles, and clusters were subsequently merged based on selected RNA and protein markers (Fig. 6C). This led to the identification of 11 final T-cell subsets, including CD4+ and CD8+ naive, memory, and effector subsets, as well as resting and activated regulatory T-cells (Treg cells) highly expressing the transcription factor FOXP3. The proportion of naive and early memory T-cells ranged from 92.6% in Patient 51 with greater than 6 months of CAR T-cell persistence, to 72.7% in Patient 66 with failed CAR T-cell persistence at 3 months (Supplementary Fig. S10D–E).
Figure 6. CITE-Seq captures the heterogeneity of pre-manufacture T-cells and demonstrates that the TCF7 regulon and interferon response gene signatures are not mutually exclusive.
(A) UMAP projection of 17,750 CITE-Seq cells, colored by 11 T-cell clusters. (B) Protein and RNA expression of selected marker genes based on CITE-Seq antibody-derived tags (protein, green) and normalized RNA expression (blue). (C) Cluster dendrogram of marker gene expression based on RNA expression (top) and protein expression (bottom). Complete-linkage hierarchical clustering was performed on these selected marker genes. (D) Violin plots indicating the normalized RNA expression of interferon response genes for each T-cell cluster. Pairwise statistical significance was assessed using the Wilcoxon rank-sum test. ****: p < 0.0001; ***: p < 0.001; **: p < 0.01; *: 0.01, n.s.: p ≥ 0.05. (E) AUCell enrichment scores associated with the TCF7 regulon and interferon response gene signatures defined based on our bulk RNA-Seq analysis. (F) Association between TCF7 regulon and interferon response gene signatures across 11 T-cell clusters, demonstrating that these pathways are inversely related in normal T-cell differentiation (gray arrow) but not necessarily mutually exclusive, as the IFN-responsive CD4+ naive/memory cluster was concurrently enriched in both signature scores.
Intriguingly, we identified a cluster of CD4+ T-cells expressing naive and memory markers such as CCR7, CD62L, and CD45RA, but also strongly expressing interferon response genes such as IRF7, RSAD2, MX1, and ISG15 as well as STAT1, which has been shown to be involved in interferon signaling and TH1 development (27,34) (Fig. 6A, D, Supplementary Fig. S10F–G). This cluster did not express other TH1 TFs such as STAT4 or TBX21, and did not express characteristic TH2 or Treg TFs such as GATA3, STAT6, or FOXP3 (Supplementary Fig. S10G). This cluster was relatively rare (approximately 1% of total T-cells), and not significantly associated with CAR T-cell persistence in the n = 6 patients assessed (p = 0.40). However, we reasoned that this population may be helpful in understanding the relationship between the TCF7 regulon and interferon response pathway. We performed AUCell analysis (36) to define single-cell enrichment scores for the previously defined TCF7 regulon and interferon response gene signatures. We found that the TCF7 regulon score was enriched among naive and memory T-cells, whereas the interferon response gene signature was strongly enriched among activated T-cells, particularly CD8+ TEM and TEFF (Fig. 6E). These gene signatures shared a strong inverse relationship among the T-cell subtypes, with the notable exception of the interferon-responsive CD4+ naive/memory population, which was strongly enriched for both signatures (Fig. 6F). These data provide cellular-level support for our prior finding that interferon response plays a role not only in activated T-cell states, but in naive and memory states. Furthermore, this finding suggests that the TCF7 regulon and interferon response pathway are not mutually exclusive, and may impart independent effects of T-cell function and CAR T-cell efficacy.
Epigenetic regulation of TCF7 and its downstream targets remains partially active in effector T-cells
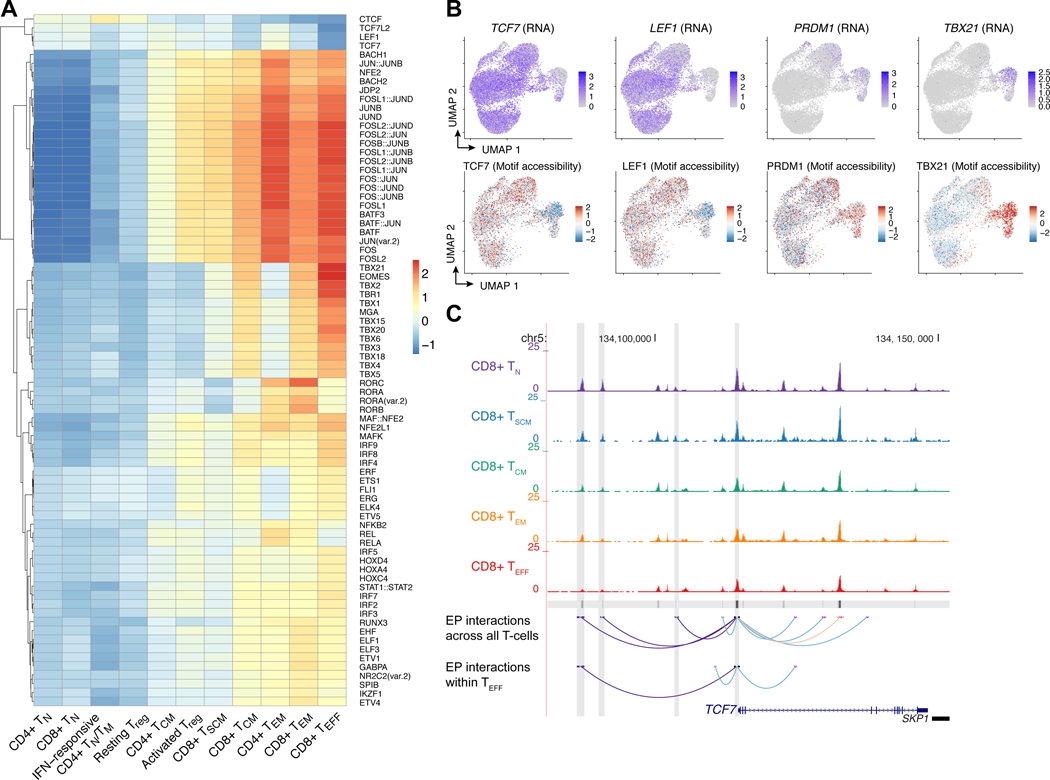
We investigated the epigenetic regulation of T-cell states and TCF7 function through integration of single-cell ATAC-Seq data with CITE-Seq data. We integrated patient scATAC-Seq data using Seurat and projected these data onto the UMAP defined by the CITE-Seq analysis. We assessed chromatin accessibility of TF motifs with chromVAR (37), finding that accessibility of the TCF7/LEF1 motif was strongly associated with naive and memory T-cell states, whereas accessibility of the PRDM1 (Blimp-1) motif was associated with effector memory and effector T-cells, and accessibility of the TBX21 motif was strongly associated with CD8+ TEM and TEFF (Fig. 7A–B). The strongest differentially accessible motif was the AP-1 motif, which was closed among naive T-cells and strongly opened among the TEM and TEFF subsets (Fig. 7A, Supplementary Fig. S11A).
Figure 7. Single-cell ATAC-Seq captures the epigenetic regulation of T-cell functional states and reveals that transcriptional regulation of TCF7 is partially maintained in CD8+ TEFF.
(A) Heatmap of chromVAR deviation scores associated with TF motifs across T-cell clusters. Shown are motifs that were significantly associated with T-cell cluster (ANOVA FDR < 0.05) and for which at least one T-cell subtype had an absolute deviation score greater than 0.90. (B) Normalized RNA expression of TCF7, LEF1, PRDM1, and TBX21, and associated chromVAR motif deviation scores. (C) Chromatin accessibility tracks for CD8+ TN, TSCM, TCM, TEM, and TEFF at the TCF7 locus. The bottom tracks show high-confidence enhancer-promoter (EP) interactions predicted based on chromatin co-accessibility (Cicero score > 0.25, light red), metacell-based regression (Bonferroni-adjusted p < 0.05, light blue), and EP interactions that were concordantly predicted with both methods (dark purple). EP interactions were predicted across all T-cells using the complete scATAC-Seq and CITE-Seq datasets, as well as on only the CD8+ TEFF cells.
While TCF7 expression and motif accessibility were diminished among effector T-cells (Fig. 7B), we asked whether its enhancer accessibility and downstream targets remained partially active, which could provide an explanation for the positive association of TCF7 with clinical CAR T-cell persistence observed in our bulk RNA-Seq analysis. We predicted enhancer-promoter interactions using two methods: chromatin co-accessibility from scATAC-Seq alone using Cicero, and correlation of chromatin accessibility with RNA expression using a metacell-based regression approach as previously described (38). Using stringent cutoffs of Cicero co-accessibility scores (>0.25) and metacell-based Bonferroni adjusted P-values (<0.05), we identified three upstream enhancers with strong evidence of interaction with the TCF7 promoter (Fig. 7C). We repeated this analysis with the single-cell data from CD8+ TEFF alone, for which gene expression as well as chromatin accessibility of TCF7 and its enhancers was diminished. Despite the use of this restricted subset of cells, we found that one of the enhancers retained both chromatin co-accessibility and association with TCF7 expression (co-accessibility score = 0.37, Bonferroni adjusted p = 4.3e-3), indicating that the epigenetic regulation of TCF7 remains partially active in this differentiated T-cell subset (Fig. 7C). Physical interactions between predicted enhancer-promoter interactions were validated using chromosome conformation capture in healthy donor T-cells, demonstrating that these predicted enhancers were strongest among the naive and early memory T-cell types and partially maintained across T-cell subtypes (Supplementary Fig. S11B–C; Supplementary Table S4).
Finally, we asked whether our data support the notion that TCF7 partially maintains its regulatory interaction with its downstream targets in effector T-cells. We considered the set of chromatin peaks containing the TCF7 motif, and the genes within the TCF7 regulon defined by our bulk RNA-Seq analysis. We first considered all T-cells in our scATAC-Seq data, finding that the chromatin co-accessibility scores associated with these TCF7 peak-promoter pairs were enriched compared to the null distribution of non-TCF7-related promoter-peak pairs (Supplementary Fig. S11D–E; Wilcoxon p = 9.01e-22). We repeated this analysis restricted to the scATAC-Seq data associated with the CD8+ TEFF data, asking whether the enrichment of TCF7 regulatory interactions is maintained. While the overall mean co-accessibility score was diminished within this restricted T-cell subset compared to the pan-T-cell analysis, we found that the co-accessibility scores for TCF7 peak-to-promoter pairs remained enriched compared to the null peak-to-promoter scores within the CD8+ TEFF subset alone (Supplementary Fig. S11E, p = 2.73e-3). Furthermore, the mean expression of TCF7 targets based on peak-to-promoter pairs within CD8+ TEFF was significantly greater than the background of non-TCF7 target genes (p = 3.07e-22, Supplementary Fig. S11F). These data support the notion that the regulatory interactions between TCF7 and its targets are partially maintained even among the CD8+ effector T-cells, providing a mechanistic explanation for our observation of TCF7 expression as a positive association with clinical CAR T-cell persistence in effector T-cells.
Discussion
To date, studies of the T-cell determinants of CAR T-cell persistence have been limited by modest clinical sample sizes and confounded by T-cell subtype composition. By performing subtype-specific transcriptomic analysis of 71 pediatric patients and integrative single-cell analysis of 6 patients, we developed, to our knowledge, the largest and most comprehensive molecular portrait of the landscape of autologously-derived T-cells in CAR T-cell therapy. Our approach allowed us to deeply characterize the composition and transcriptional regulation of favorable T-cell states, and importantly, to account for the confounding effect of T-cell subtype to identify TCF7 and interferon signaling as pathways associated with CAR T-cell persistence.
We found that higher proportions of naive and early memory T-cells were positively associated with long-term clinical CAR T-cell persistence. While TEM and TEFF subsets were highly enriched in proliferative and metabolically active pathways — phenotypes that may superficially appear to be beneficial for effective CAR T-cell function — we observed concomitant enrichment in intrinsic and extrinsic apoptotic signaling in these subsets, suggesting that programmed cell death ultimately hinders the contribution of these T-cells to long-term CAR T-cell persistence. Regulatory network analysis nominated TCF7 and LEF1 (in contrast to other TCF family members) as core transcription factors maintaining naive and memory T-cell states, and PRDM1 and TBX21 as associated with effector T-cell states; the epigenetic basis of these lineage factors was strongly supported by motif enrichment analysis in our scATAC-Seq analysis. Given the evidence of a favorable role of naive and early memory T-cells in other CAR constructs (39) and in intrinsic T-cell expansion potential (4), these transcriptional regulators are likely of general importance to other forms of CAR T-cell therapy.
The TCF7 gene, encoding for the Tcf1 transcription factor, has been shown to profoundly remodel the T-cell chromatin landscape, and knockout studies have provided evidence that it acts as a key initiating factor in early thymocyte development (23,40). Our single-cell RNA-Seq and chromatin accessibility suggest a profound shift in the transcriptional regulatory machinery, from TCF7 and LEF1 to PRDM1 and TBX21, with TBX21 expression and motif accessibility particularly notable among CD8+ cells. Whether one or two TFs represent pioneering TFs that drive the rest, or if these cell fate transitions represent a cooperative transcription factor circuit, remains unclear (19). Intriguingly, our data revealed several lines of evidence suggesting that TCF7 plays a role not only in maintaining the naive and early memory T-cell states, but also in maintaining a favorable phenotype in effector lineages. While TCF7 has been known to be highly expressed in naive and memory T-cell subsets, recent mouse models of chronic viral infection and solid tumors have implicated TCF7 in cell fate decisions in effector and exhausted phenotypes. For example, recent experimental studies have led to the discovery of PD1+Tcf1+Tim3- progenitor exhausted T-cells; TCF7 appears to be a critical regulator of maintaining stemness in exhausted T-cells, and may be responsible for the proliferative burst seen in anti-PD1 therapy (41–43). Our findings represent a key translation to a novel clinical domain, suggesting that TCF7 acts within effector T-cells to maintain a favorable T-cell phenotype not only in chronic viral infection and solid tumors, but in other forms of immunotherapy including adoptive T-cell immunotherapy.
The interferon response pathway has been shown to play a major role in T-cell function and dysfunction, and its role depends strongly on the context and duration of response. While interferon signaling stimulates T-cell responses on the short term, chronic interferon signaling has been shown in experimental models to drive T-cell dysfunction (44). IRF7, a key regulator of the type I interferon response (45), emerged in our data as a transcriptional regulator associated with poor CAR T-cell persistence across T-cell subsets. Our data suggest an immunosuppressive effect of chronic interferon signaling among pre-manufacture T-cells that contribute to failure of long-term CAR T-cell persistence. Interferon signaling was associated with poor CAR T-cell persistence even in naive and early memory T-cells, suggesting that this pathway is likely driven not by repeated cognate antigen binding as in T-cell exhaustion, but by alternative mechanisms such as circulating inflammatory factors. Mouse models of persistent LCMV infection have shown that blockage of type I interferon is associated with improved T-cell responses (33,46), highlighting the immunosuppressive effect of chronic interferon signaling. In contrast, Zhao et al. found that CAR stimulation strongly induces IRF7 expression, and that knockdown of IRF7 reduced the cytolytic potential of CAR T-cells (47). This suggests that during the acute phase of CAR-stimulated T-cell activation, IRF7 and the type I interferon network may be necessary for optimal activation and stimulation.
Understanding the T-cell determinants of successful CAR T-cell therapy is fundamental in improving clinical outcomes for pediatric B-cell malignancies, as well as in the development of adoptive T-cell therapies for other malignancies including solid tumors. By analyzing autologously-derived, pre-manufacture T-cells, our unique dataset provides valuable insights for generalizable T-cell mechanisms that are not confounded by specific perturbations in the CAR T-cell manufacturing process. Moreover, our analysis of sorted T-cell subsets and single-cell analyses enabled us to identify gene regulatory mechanisms that account for the potential confounding effect of T-cell subtype composition. Together, our data provide an expanded view of the molecular mechanisms associated with the long-term efficacy of clinical CAR T-cell therapy.
Materials and Methods
Patient identification and clinical annotation
Patients were identified by the clinical practices at the Children’s Hospital of Philadelphia Division of Oncology in Philadelphia, PA. Patients were enrolled onto Children’s Hospital of Philadelphia Institutional Review Board-approved clinical trials NCT01626495 and NCT02906371, with written informed consent obtained by patients or their guardians in accordance with the U.S. Common Rule. Pre-manufacture T-cells were acquired from the IRB-approved local institutional trial CHP-784.
For event-free survival analyses, we defined a CAR T-cell failure event as the clinical progression of leukemic blast cells or detection of circulating B-cells. Event-free survival was recorded as a censored data point if the patient had B-cell aplasia and proceeded to receive a bone marrow transplant, died of other causes such as cytokine release syndrome, had CD19- relapse, or had continued B-cell aplasia at the most recent follow-up.
T-cell enrichment and cell sorting
T-cells were enriched from apheresis samples by negative selection using EasySep™ Human T Cell Enrichment Kit (#19051; StemCell Technologies) according to the manufacturer’s instructions. Previous work (48,49) has demonstrated that CCR7, CD62L, CD45RO and CD95 can be used to differentiate the various T cell phenotypes by using the following expression patterns: naïve (TN) – CCR7+, CD62L+, CD45RO–, CD95–; stem central memory (TSCM) – CCR7+, CD62L+, CD45RO–, CD95+; central memory (TCM) – CCR7+, CD62L+, CD45RO+, CD95+; effector memory (TEM) – CCR7–, CD62L–, CD45RO+, CD95+; terminal effector (TEFF) – CCR7–, CD62L–, CD45RO–, CD95+. Briefly, cells were resuspended in fluorescence activated cell sorting (FACS) buffer (Ca++ & Mg++ free phosphate-buffered saline + 1% BSA), then incubated with CCR7 antibody for 20 min at 37°C, washed once, and then incubated with remaining antibody cocktails for 25 min at 4°C. Samples were then washed twice, and sorting was done using MoFlo® Astrios™ EQ High-Speed Cell Sorter (Beckman Coulter, Inc). The antibodies used for cell sorting described above were CCR7-FITC (#561271; BD Biosciences), CD95-PE (#556641; BD Biosciences), CD45RO-BV421 (#304224; BioLegend) and CD62L-PerCP/Cyanine5.5 (#304824; BioLegend).
Bulk RNA Sequencing
Cells were sorted into TRIzol-LS (Invitrogen), and total RNA was purified using RNeasy micro kit (Cat #: 74004, Qiagen) and analyzed for purity and integrity using RNA 6000 Pico Kit for Bioanalyzer 2100 (Cat #: 5067–1513, Agilent). Full-length cDNA was synthesized and amplified using SMART-Seq V4 Ultra Low Input RNA Kit (Cat #: 634891, Takara) from 1 ng total RNA per sample. Full-length cDNA was purified using SPRIselect beads (Cat #: B23318, Beckman Coulter). Sequencing libraries were prepared using Nextera XT DNA Library Prep Kit (Cat #: FC-131–1096) and 200 pg purified full-length cDNA per sample. Libraries were sequenced on an Illumina Hiseq 2500 in paired-end mode with the read length of 100 nt.
CITE-Seq
Sorted cells were blocked with Human TruStain FcX™ (BioLegend, Cat# 422301) and then stained with a TotalSeq-A antibody panel (See supplemental information). Stained cells were immediately processed using the 10x Genomics Chromium controller and the Chromium Single Cell 3’ reagent kits (V3). 3’ GEX libraries were constructed using 10x Genomics library preparation kit. ADT libraries were constructed using KAPA HiFi HotStart ReadyMix kit (Kapa Biosystems, Cat# KK2601). Library quality was checked using Agilent High Sensitivity DNA kit and Bioanalyzer 2100. Libraries were quantified using dsDNA High-Sensitivity (HS) assay kit (Invitrogen) on Qubit fluorometer and the qPCR-based KAPA quantification kit. Libraries were sequenced on an Illumina Nova-Seq 6000 with 28:8:0:87 paired-end format.
scATAC-Seq
Sorted cells were centrifuged at 300g for 5 min at 40C. 45uL of chilled lysis buffer was added to cell pellets and mixed by pipetting gently three times, and incubated 3 min on ice. After incubation, 50uL of pre-chilled wash buffer was added without mixing and centrifuged immediately at 300g for 5 min at 40C. 95uL supernatant was carefully discarded and 45uL pre-chilled diluted nuclei buffer (10x Genomics) was added without mixing and sample was centrifuged at 300g for 5 min at 40C. The nuclei pellet was then resuspended in 7uL pre-chilled diluted nuclei buffer and nuclei concentration was determined using a Countess II cell counter (Invitrogen). 7,000–20,000 nuclei were used for the transposition reaction in bulk, and then loaded to the 10x Genomics Chromium controller and processed with the Chromium Single Cell ATAC Reagent kit. Library quality was checked using Agilent High Sensitivity DNA kit and Bioanalyzer 2100. Libraries were quantified using dsDNA High-Sensitivity (HS) assay kit (Invitrogen) on Qubit fluorometer and the qPCR-based KAPA quantification kit. Libraries were sequenced on an Illumina Nova-Seq 6000 with 49:8:16:49 paired-end format.
Bulk and single-cell data have been submitted to dbGaP under Study Accession phs002323.v1.p1.
Additional methods are described in the Supplementary Methods.
Supplementary Material
Significance.
In order to improve clinical outcomes for CAR T-cell therapy, there is a need to understand the molecular determinants of CAR T-cell persistence. These data represent the largest clinically-annotated molecular atlas in CAR T-cell therapy to date, and significantly advance our understanding of the mechanisms underlying therapeutic efficacy.
Acknowledgements
We thank the Children’s Hospital of Philadelphia Flow Cytometry Core for their assistance with cell sorting, and the Research Information Services for providing computing support. This work was supported by National Institutes of Health of United States of America grants CA232361 (to D.M.B. and S.A.G) and CA233285 (to K.T.), a Doris Duke Charitable Foundation Clinical Scientist Development Award and a Stand Up To Cancer Innovative Research Grant, Grant Number SU2C-AACR-IRG 12–17 (to D.M.B.), a CIHR Doctoral Foreign Study Award #433117 (to G.M.C), a National Institutes of Health/National Child Health and Human Development award T32HD043021 and National Cancer Institute award K12CA076931 (to Y.Y.D), NIH Medical Scientist Training Program T32 GM07170 (to S.B.), and a 2020 Blavatnik Family Fellowship in Biomedical Research (to Q.Z.). Stand Up To Cancer (SU2C) is a division of the Entertainment Industry Foundation. The indicated SU2C research grant is administered by the American Association for Cancer Research, a scientific partner of SU2C.
S.A.G. is a paid consultant for Novartis, Vertex, CBMG, Adaptimmune, TCR2, Juno, Jazz, Eureka, Cellectis, Roche, GlaxoSmithKline, Cure Genetics, Humanigen, and Janssen/Johnson & Johnson, reports receiving commercial research grants from Kite, Servier, and Novartis, is listed as inventor on a patent for toxicity management for antitumor activity of CARs (WO 2014011984 A1; managed according to the University of Pennsylvania patent policy), and reports receiving other remuneration from McNaul Ebel. D.M.B. is now an employee of Tmunity Therapeutics, Inc.
Footnotes
No potential conflicts of interest were disclosed by the other authors.
References
- 1.Finney OC, Brakke HM, Rawlings-Rhea S, Hicks R, Doolittle D, Lopez M, et al. CD19 CAR T cell product and disease attributes predict leukemia remission durability. J Clin Invest. 2019;129:2123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N Engl J Med. 2017;377:2531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen AD, Garfall AL, Stadtmauer EA, Melenhorst JJ, Lacey SF, Lancaster E, et al. B cell maturation antigen–specific CAR T cells are clinically active in multiple myeloma. J Clin Invest. The American Society for Clinical Investigation; 2019;129:2210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh N, Perazzelli J, Grupp SA, Barrett DM. Early memory phenotypes drive T cell proliferation in patients with pediatric malignancies. Sci Transl Med. 2016;8:320ra3. [DOI] [PubMed] [Google Scholar]
- 5.Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. 2018;24:563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, Zhang M, Ramos CA, Durett A, Liu E, Dakhova O, et al. Closely related T-memory stem cells correlate with in vivo expansion of CAR.CD19-T cells and are preserved by IL-7 and IL-15. Blood. 2014;123:3750–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N Engl J Med. 2018;378:439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marbach D, Costello JC, Küffner R, Vega NM, Prill RJ, Camacho DM, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods [Internet]. PLoS ONE. 2010. page e12776. Available from: 10.1371/journal.pone.0012776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haury A-C, Mordelet F, Vera-Licona P, Vert J- P. TIGRESS: Trustful Inference of Gene REgulation using Stability Selection. BMC Syst Biol. 2012;6:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonneau R, Reiss DJ, Shannon P, Facciotti M, Hood L, Baliga NS, et al. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006;7:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao L, Tober J, Gao P, Chen C, Tan K, Speck NA. RUNX1 and the endothelial origin of blood. Exp Hematol. 2018;68:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao P, Uzun Y, He B, Salamati SE, Coffey JKM, Tsalikian E, et al. Risk variants disrupting enhancers of TH1 and TREG cells in type 1 diabetes. Proc Natl Acad Sci U S A. 2019;116:7581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao P, Chen C, Howell ED, Li Y, Tober J, Uzun Y, et al. Transcriptional regulatory network controlling the ontogeny of hematopoietic stem cells. Genes Dev. 2020;34:13–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu S, Zhou X, Steinke FC, Liu C, Chen S-C, Zagorodna O, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber BN, Chi AW-S, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Xue H-H. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J Immunol. 2012;189:2722–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Best JA, Blair DA, Knell J, Yang E, Mayya V, Doedens A, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013;14:404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nat Rev Immunol. Springer Science and Business Media LLC; 2012;12:799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG, et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. Elsevier BV; 2011;35:622–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med. 2015;212:2027–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med. 2015;212:2041–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, et al. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297:2063–6. [DOI] [PubMed] [Google Scholar]
- 32.Kuwahara M, Yamashita M, Shinoda K, Tofukuji S, Onodera A, Shinnakasu R, et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat Immunol. 2012;13:778–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KCF, Welch M, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson APS, et al. Functional crosstalk between type I and II interferon through the regulated expression of STAT1. PLoS Biol. Public Library of Science (PLoS); 2010;8:e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Q, Gao P, Tober J, Bennett L, Chen C, Uzun Y, et al. Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood. 2020;136:845–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sommermeyer D, Hudecek M, Kosasih PL, Gogishvili T, Maloney DG, Turtle CJ, et al. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia. 2016;30:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson JL, Georgakilas G, Petrovic J, Kurachi M, Cai S, Harly C, et al. Lineage-Determining Transcription Factor TCF-1 Initiates the Epigenetic Identity of T Cells. Immunity. 2018;48:243–257.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol. 2016;1:eaai8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minn AJ, Wherry EJ. Combination Cancer Therapies with Immune Checkpoint Blockade: Convergence on Interferon Signaling. Cell. 2016;165:272–5. [DOI] [PubMed] [Google Scholar]
- 45.Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12:399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Z, Condomines M, van der Stegen SJC, Perna F, Kloss CC, Gunset G, et al. Structural Design of Engineered Costimulation Determines Tumor Rejection Kinetics and Persistence of CAR T Cells. Cancer Cell. 2015;28:415–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das RK, Vernau L, Grupp SA, Barrett DM. Naïve T-cell Deficits at Diagnosis and after Chemotherapy Impair Cell Therapy Potential in Pediatric Cancers. Cancer Discovery. 2019;9:492–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.