Abstract
Recent advances in cancer immunotherapy have completely revolutionized cancer treatment strategies. Nonetheless, the increasing incidence of immune-related adverse events (irAEs) is now limiting the overall benefits of these treatments. irAEs are well-recognized side effects of some of the most effective cancer immunotherapy agents, including antibody blockade of the cytotoxic T-lymphocyte-associated protein 4 and programmed death protein 1/programmed-death ligand 1 pathways. To develop an action plan on the key elements needed to unravel and understand the key mechanisms driving irAEs, the Society for Immunotherapy for Cancer and the American Association for Cancer Research partnered to bring together research and clinical experts in cancer immunotherapy, autoimmunity, immune regulation, genetics and informatics who are investigating irAEs using animal models, clinical data and patient specimens to discuss current strategies and identify the critical next steps needed to create breakthroughs in our understanding of these toxicities. The genetic and environmental risk factors, immune cell subsets and other key immunological mediators and the unique clinical presentations of irAEs across the different organ systems were the foundation for identifying key opportunities and future directions described in this report. These include the pressing need for significantly improved preclinical model systems, broader collection of biospecimens with standardized collection and clinical annotation made available for research and integration of electronic health record and multiomic data with harmonized and standardized methods, definitions and terminologies to further our understanding of irAE pathogenesis. Based on these needs, this report makes a set of recommendations to advance our understanding of irAE mechanisms, which will be crucial to prevent their occurrence and improve their treatment.
Keywords: autoimmunity, immunotherapy, translational medical research
State of the field
Immune checkpoint blockade (ICB) has revolutionized the treatment of cancer, leading immunotherapy to become the fourth pillar of cancer therapeutics.1 ICB agents are monoclonal antibodies that block key negative regulators of T cell function, such as cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed death protein 1 (PD-1) and programmed-death ligand 1 (PD-L1),2 resulting in enhanced antitumor responses. CTLA-4, the first coinhibitory receptor discovered3 4 and the target for the first approved ICB immunotherapy,5 6 plays a critical role in downregulating T cell activation and maintaining immune homeostasis through cell-intrinsic signaling mechanisms and cell-extrinsic competition with CD28 for ligands.7 CTLA-4 blockade can alter and expand the repertoire of tumor-reactive or autoreactive T cells.7 Similarly, the PD-1:PD-L1/L2 pathway also inhibits T cell responses and maintains immune tolerance and homeostasis by reducing T cell receptor (TCR)/CD28 signaling, cytokine production and target cell lysis.8 Both CTLA-4 and PD-1 are upregulated following T cell activation, but CTLA-4 and its ligands are more abundant in secondary lymphoid organs while PD-1 ligand distribution across a diverse set of hematopoietic and non-hematopoietic cells suggests more impact in peripheral tissues.7 Of note, loss of CTLA-4 or PD-1/PD-L1 can accelerate autoimmunity.7
More than 10% of advanced cancer patients achieve true benefit from ICB,9 but the majority (66%–72%) will also experience some form of immune toxicity from these therapies, termed immune-related adverse events (irAEs).10–12 These irAEs can affect all organ systems (eg, skin, musculoskeletal, gastrointestinal, neurologic, cardiovascular, hepatic, endocrine) with a range of severity and often mirror aspects of known primary autoimmune diseases (eg, rheumatoid arthritis, inflammatory bowel disease and other inflammatory conditions), though there are also clear distinctions, and investigations are underway to understand the differences.13 Not surprisingly, irAEs are treated with many of the same therapeutic interventions as primary autoimmune diseases, including glucocorticosteroids and biological agents that are immunosuppressive or anti-inflammatory.14
Immuno-oncology remains a rapidly developing field and it is estimated that there are many more coinhibitory pathways beyond CTLA-4 and PD-1 that could be valuable therapeutic targets. In fact, the future of cancer immunotherapy likely lies in combination therapy to increase antitumor responses,15 16 which will likely also drive the development of novel irAEs and the need for new strategies to identify and mitigate these immune toxicities while maintaining effective antitumor immunity. Since treatment for irAEs may inhibit ICB-induced antitumor responses, uncovering the pathways that differentiate irAEs from antitumor immunity is key to understanding the underlying biology and identifying treatments that reduce adverse events while preserving the anticancer immune response.
Taken together, this suggests a critical need to invest in translational and basic research efforts that could further our understanding of the spectrum of irAE disease presentation and associated mechanisms, and lead to the development of (1) rapid diagnostic tools, (2) biomarkers to identify patients at risk for developing irAEs, and (3) better targeted therapies to manage irAEs without impacting immunotherapy efficacy. In particular, there is a pressing need to identify precision treatment solutions that go beyond withholding ICBs and/or giving glucocorticosteroids and reflect the specific cancer and immunotoxicities experienced by the patient to optimize both cancer and irAE outcomes.17 18 Development of such targeted therapies will require studying irAEs directly in patients’ tissue and blood samples, as most irAE presentations have yet to be faithfully recapitulated in animal models.19 20
The Society for Immunotherapy of Cancer and the American Association for Cancer Research held a joint workshop in Houston, TX in March 2020. The workshop, ‘The Cancer Biology Underlying Immunotherapy-Induced Autoimmunity’, discussed the biological mechanisms of irAEs to ICB and was coorganized by Lisa H. Butterfield, PhD, Elizabeth M. Jaffee, MD, and Arlene H. Sharpe, MD, PhD. The wide spectrum of irAE clinical presentations (eg, colitis, pancreatitis, hepatitis, myocarditis, neurotoxicities, thyroiditis, ICB-induced insulin dependent diabetes mellitus and lymphocytic hypophysitis) was discussed in detail during the meeting. As these clinical presentations have been thoroughly reviewed recently,21–28 this report focuses on summarizing the critical challenges and opportunities posed by immunotherapy-induced irAEs and strategies that should be the focus of future investigations that were identified during the meeting.
Critical questions for understanding and treating Immunotoxicities
The future for cancer patient care will entail the development of precision strategies empowering effective immunotherapy treatment for cancer that take into consideration individual intrinsic and extrinsic risk factors for developing irAEs and combine new and existing therapeutic regimens. To increase the success of the next generation of immunotherapy strategies while reducing the negative impact of irAEs on long-term outcomes for patients with cancer, the workshop participants identified several critical questions to be addressed:
What mechanisms drive the correlation of irAEs with antitumor responses? Can the two be delineated and uncoupled?
What is the impact of ICB cessation or immunosuppression in response to irAE development?
What are the intrinsic and extrinsic risk factors for irAEs?
Can we use clinical data and models to improve preclinical models of irAEs following ICB to support improved mechanistic understanding of irAEs?
How can we harmonize clinical metadata to empower irAE clinical model development?
What types of translational research efforts are needed to unravel irAE biological drivers?
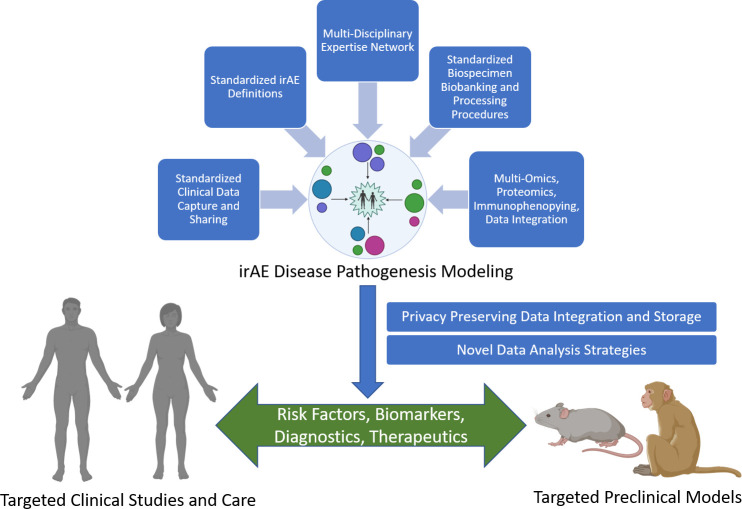
To answer these questions and better understand, prevent and treat irAEs following ICB therapy, it is essential to develop representative animal models for preclinical studies, collect tissue samples and real-world clinical care data for clinical model development and establish large-scale collaborative working groups to develop and standardize novel data collection, translational research and analysis strategies. The next section frames the challenges and opportunities in the field raised by these questions and strategies to address them. Figure 1 outlines our overall proposed framework for tackling these questions.
Figure 1.
Future directions for irAE research. Effective clinical modeling of irAEs will require the integration of a variety of data, including: (1) standardized definitions of irAEs and tools to identify them, (2) clinical data from electronic health records that reflect the critical components of irAEs and cancer descriptions and outcomes that have been standardized using common data models to support data sharing, (3) collaborative biospecimen biobanking programs with standardized operating procedures for tissue collection, processing and storage, and (4) high-quality, high-throughput multiomics, proteomics and immunophenotyping data integration strategies that provide mechanistic information. The overall clinical modeling of irAE disease pathogenesis efforts will require multidisciplinary clinicians and scientists and the development of modeling, management and analysis strategies and data collection from a large number of organizations to represent the wide range of irAEs, particularly rare irAEs. Clinical modeling will also require data storage technologies that support integration of clinical and mechanistic data while preserving participant privacy and novel machine learning strategies to derive insights about risk factors and biomarkers of disease that can lead to new diagnostics and therapeutics for clinical care as well as support refinement and development of more effective preclinical models. irAE, immune-related adverse event.
Challenge: decoupling irAES from antitumor responses
A subset of irAE presentations following ICB treatment is associated with positive tumor outcomes. For example, the appearance of vitiligo and hypothyroidism, two relatively low severity irAEs, are associated with effective antitumor responses and are thought to indicate the activation of on-target, off-tumor immune activity.29–37 However, many irAEs (eg, ICB-induced diabetes and myocarditis) are much more severe, and improving clinical management of immunotoxicity remains a critical unmet need, as is an enhanced understanding of how irAE treatment strategies impact ICB efficacy. For instance, the first line of treatment for irAEs is often corticosteroids.14 While the impact of low-dose corticosteroids is still debated, glucocorticosteroids present at the time of treatment initiation and high-dose corticosteroids may mitigate ICB therapy38 even though they may be mandatory for management of life-threatening severe toxicities.14 Preclinical work examining the mechanisms underlying both tumor immunity and irAEs following ICB therapy, together with a deeper understanding of patient responses to ICB therapy and biomarkers of the potential for irAE development, will be essential for developing targeted immunotherapy that maximizes the anticancer response while minimizing immunotoxicity.
Challenge: defining intrinsic and extrinsic mediators of irAEs
A central challenge for optimizing immunotherapy is understanding the intrinsic and extrinsic mediators of irAE development and host-directed immune activation, which are key to ultimately distinguishing the biology associated with antitumor immune responses from irAEs. Like many complex trait presentations such as autoimmune diseases, irAE pathogenesis may result from multifactorial parameters, involving gene (intrinsic)-by-environment (extrinsic) interactions. Yet, little is known about the interplay of intrinsic and extrinsic drivers of irAEs, in part because of the lack of large, well-annotated cohorts of patients spanning many healthcare institutions, needed to properly power studies of the genetic and environmental drivers across the range of rare and more common irAEs and different cancer types (see the Opportunities section). However, the field of irAE biology can start deriving potential insights from ongoing studies of autoimmune diseases, cancer and rare immunodeficiency disorders.
Genetic loci are some of the key intrinsic risk factors impacting autoimmunity, with HLA alleles being the strongest known factor for susceptibility. Numerous studies have shown that carriers of certain HLA alleles are at higher risk of developing specific autoimmune diseases.39 In addition, HLA alleles have also been associated with melanoma risk and response to certain anticancer therapies, such as high-dose interleukin-2.40–46 For example, HLA-Cw*06, which has been associated with better relapse-free and overall survival, has also been correlated with development of psoriasis.47 Interestingly, a recent report has also demonstrated that polygenic risk for skin autoimmunity is associated with longer overall survival in bladder cancer treated with anti-PD-L1.48 Lessons learned from genetic association studies in autoimmune diseases and cancer clearly suggest that mapping susceptibility loci associated with the range of irAEs could provide important biological insights into the manifestation of irAEs.
Beyond deriving biology from complex trait genetics, additional insights can be derived from studying rare monogenic traits. For example, uniallelic germline mutations in checkpoint molecules themselves have been implicated in irAE-like immune dysfunction, particularly CTLA4.49 50 Similar to CTLA4-/- mouse models, humans with heterozygous novel and pathological changes in CTLA4 DNA sequence exhibit lymphocytic organ infiltration and signs of autoimmunity.51 Heterozygous mutations in CTLA4 result in diminished CTLA-4 expression and function, leading to overt immune dysregulation with diverse autoimmune presentations, including type 1 diabetes and lymphoproliferative disorders. CTLA4 haploinsufficiency can predispose to lung and gut diseases, with histological evidence confirming extensive target tissue infiltration by lymphocytes. Bone marrow failure and infiltrative brain lesions have also been described in this setting.52 With the caveat of variable penetrance and a wide spectrum of expressivity, increased predisposition to lymphoma, non-lymphoid malignancies and hypogammaglobulinemia are also within the clinical spectrum. Haploinsufficiency of CTLA4 is also linked to reduced Treg inhibitory function and hyperproliferation of effector cells with a robust alteration of T and B cell homeostasis. Interestingly, treatment of CTLA4 insufficiency has been successful in limited clinical settings. Abatacept, a fusion protein of the CTLA-4 extracellular domain with an antibody Fc region, has been used in combination with corticosteroids and mammalian target of rapamycin (mTOR) inhibitors to treat CNS lesions, lung and gut disease presentations, as well as autoimmune cytopenia in CTLA4-deficient patients.52–55
Given the similarities between some clinical presentations of irAEs and the phenotypes observed with these rare monogenic immunodeficiencies, studying the disease evolution and treatment of patients with such intrinsic predispositions to disrupted immune homeostasis could offer another window of opportunity to further our understanding and support development of treatments for irAEs in patients undergoing CTLA-4 blockade for cancer. In addition, lessons from studying and treating these rare immunodeficiency conditions have the potential to inform treatment solutions for irAE presentations. Such efforts will ultimately require many independent groups to assemble very large datasets of tissue samples and well-controlled and annotated clinical variables (such as heterogeneity of prior treatments), given the breadth and depth of human variability, cancer variability and the general rarity of most irAEs.
Extrinsic triggers of autoimmune diseases are poorly understood but may derive from the infectome (some infections are protective, others are triggers),56 microbiome (increase of proinflammatory or reduction of anti-inflammatory bacteria)57 58 or exposome. Given the similarities in clinical presentations between certain irAEs and autoimmune diseases, it is very likely that several similar extrinsic triggers also contribute to irAE pathogenesis, which should be the focus of future efforts that will involve assembling large well-annotated patient cohorts to tackle such questions.
Of note, cancer often involves the creation of neoantigens in the tumor that have varying auto-antigenicity59; thus, antitumor responses may increase the likelihood of autoimmunity and therefore of irAEs and mortality—similar to the immune response to pathogens that triggers autoimmunity. In some instances, autoimmunity may be associated with cancer; for example, Lambert-Eaton myasthenic syndrome is associated with antitumor immunity against small-cell lung carcinoma, while systemic lupus erythematosus has been associated with development of certain cancers.60 61 Pre-existing chronic inflammation may also create an environment that promotes evasion of immune surveillance by cancer that can be reversed in some cases by immunotherapy.62 While we currently have a limited understanding of the relationship between cancer and autoimmunity and the role of extrinsic factors in driving pathogenesis, some mouse models may enable examination of this relationship and help shed light on specific mechanisms driving irAEs.63 64 For example, ICB exacerbates autoimmunity in non-obese diabetic (NOD) mice and PD-1/CTLA-4 inhibition has tissue-dependent impacts.
Challenge: preclinical model development
Translational biomedical research centers on the concept of ‘bench to bedside and back to the bench’, necessitating preclinical modeling designed to best mirror the clinical paradigm. The complexity of cancer immune responses highlights the need for ‘in vivo veritas’, especially given the increasing clinical use of ICB to treat a large range of cancer types and the diverse array of irAEs limiting ICB antitumor efficacy. The inbred laboratory mouse is the bedrock for both discovery and assessing efficacy of immunotherapeutic approaches in cancer as well as autoimmunity. This is due to low cost, ease of access and biological reproducibility due to optimal control of all variables and availability of reagents. However, even considering the not-insignificant species immune disparities, critical questions revolve around how representative the commonly used tumor models are to study cancer biology, as these models often involve subcutaneous implantation of extensively cultured and rapidly growing established mouse tumor cell lines into very young (age-equivalent to teenage human) healthy inbred mice housed under specific-pathogen-free (SPF) conditions. Due to rapid tumor growth and/or use of therapeutics not of mouse origin, therapeutic interventions are given for extremely short times and off-target effects other than cytokine-storm phenomena are rarely observed. The lack of fully murine reagents also represents a significant problem as it results in neutralizing responses to the xenogeneic proteins by the immunocompetent host, limiting the long-term application of therapy which is often used clinically and may be important for the development of irAEs in the mouse models. This claim is further supported by the paucity of reports demonstrating the phenocopy of irAEs observed in patients in conventional mouse tumor models treated with ICB. However, there is appreciable literature on the impact of ICB in different mouse autoimmune models, indicating these may be valuable resources to combine with tumor models to tease out mechanisms and pathways that could then be linked with clinical presentations.65 Finally, incorporating human modifying elements (eg, age, biological sex, body mass index and exposure to pathogens) in murine tumor immunotherapy models is of critical importance, given reports that both age and obesity can markedly impact irAE and cancer outcomes, both positively and negatively.66 67 In addition, the microbiome associated with different mouse vendors is also important for ICB responses, highlighting the constraints of using SPF mice as well as potentially affecting model reproducibility with different laboratories.68 All of these aforementioned variables likely impact autoimmune mouse models as well.
Xenogeneic models, including patient-derived xenograft and humanized mouse models, while useful in assessing direct human tumor cell responses, have limitations when used to model complex ICB responses and irAEs.69 For instance, extensive species differences exist with regard to the effects of cytokines and adhesion molecules on the host or immune cells.70–72 Additionally, human immune cells have varying capacities to survive or engraft in a murine host, and the impact of human immune cells placed or developed in the xenogeneic murine environment is poorly characterized. Ex vivo organoid models are attractive given they add the complexity of cellular interactions not seen with tumor cell lines. Nonetheless, the challenge of reproducing biologically meaningful immunological interactions due to complex culture conditions that impact both tumor and immune cell component survival and function creates significant limitations to the use of such model systems, which is further complicated with ICB assessment. Finally, both xenogeneic modeling and ex vivo models also can have significant contrivances if using immune cells allogeneic to the tumor. Thus, while these models appear attractive for ICB and irAE modeling, the numerous immunological deviations inherent with them significantly limit their extrapolation potential and more work is needed to assess in which experimental conditions these models could be most helpful for modeling.
Large animal models, such as client-owned canine patients with spontaneous cancers or non-human primates, offer clear advantages with regard to being outbred and, in the case of canine models, having spontaneous cancers that can be treated with immunotherapy. However, these models are severely hampered by the (1) extreme cost, (2) recipient heterogeneity, being outbred with comorbidities, (3) extremely limited number of patients available for research purposes, (4) limited access to tissues, (5) lack of suitable reagents (particularly with ICB) compatible with large animals, and (6) limited occurrence of irAEs with the immunotherapies assessed thus far in these models. Thus, beyond directly studying human tissue specimens, syngeneic mouse tumor models in conjunction with commonly used autoimmune mouse models remain, for now, the best means to mechanistically study irAEs associated with the use of ICB. Nevertheless, it remains imperative that the community develops a deeper understanding of the limitations of the commonly used mouse tumor models and funding opportunities are needed to foster the development of more appropriate model systems.
In preclinical modeling, little attention outside of overt toxicity of an approach is given, with the emphasis in most studies being centered on the antitumor response that often needs to occur rapidly. Given the tremendous cost and challenges associated with using aged mice (eg, a 22-month-old mouse can cost as much as US$500 and obtaining sufficient aged mice from the National Institute on Aging (NIA) for therapy studies is predicated on having NIA funding) or in generating diet-induced obese mice of both genders in therapy studies (particularly germane given the well-described impact of biological sex on autoimmunity and recent reports suggesting an impact of gender in ICB67), there is a significant need to support development and availability of these resources to the cancer research community. There is also a need to develop and assess immunotherapy reagents (antibodies, cytokines, etc) that are of mouse origin to more properly mirror the clinical paradigm, allowing long-term application that could lead to irAE manifestation. Furthermore, using more indolent tumors or genetically engineered mouse models would offer more reflective immune-tumor cell dynamics and therapy, but the challenge remains the prohibitive cost associated with using such models for therapy studies and assessing long-term treatment impacts which are often not viewed as sufficiently innovative or mechanistic to warrant funding. Understanding the pathways involved in driving and sustaining ICB-associated irAEs observed in the clinic and linking them with preclinical models are vital for next steps. This will require large investments from public and private sectors given the lack of representative models and the costs associated with what are viewed as studying negative events. Such investments are urgently needed to ultimately overcome the irAE impediments on immunotherapy treatment efficacy.
Opportunity: harmonizing clinical metadata and clinical model development
Challenges with current preclinical models and the need for deeper understanding of the human immune response to ICB present an opportunity to flip the research axis and translate clinical experience to bench-testable hypotheses. While clinical trial data provide a foundation for this work, trials generally involve the collection of a narrow set of study-focused clinical data and biospecimen samples over a specific window of time from a highly selected patient population that may not be representative of the broader population of people who will receive therapy.73–76 Thus, we need a ‘bedside-to-bench’ approach for clinical modeling of the development of irAEs that includes a more representative patient sample and leverages a broad array of clinical and laboratory data, and to translate these models into the preclinical space to explore biological mechanisms of irAE development in more detail.
The adoption of electronic health record (EHR) technology in the USA77 parallels the rise of FDA-approved ICB strategies for cancer treatment.1 As of 2015, when many of the anti-PD-1/L1 antibodies were first approved for use, over 95% of hospitals accepting Medicare78 and 80% of ambulatory care settings79 had adopted EHRs to document clinical care. Thus, almost all patients receiving ICB have had their treatment course documented in an EHR. EHR data, particularly for cancer patients who have high contact with healthcare systems, contains a rich source of longitudinal clinical observations, including those prior to the development of cancer, during treatment and post-therapy, as well as highly rigorous laboratory test results.
To capture irAEs in sufficient numbers that reflect the full range of presentations across a wide range of cancers, it is necessary to integrate data from a large number of healthcare organizations. Given that the underlying data structures of many EHR systems (even systems provided by the same vendor) are not always the same, integration and alignment of clinical data requires the development of ‘common data models’ (CDMs)—or shared data languages—for data alignment and exchange. With the expansion of EHR technology, a number of networks built on CDMs have been established. The Patient-Centered Outcomes Research Network (PCORnet),80 81 built on the PCORnet CDM,82 the Accrual to Clinical Trials Network,83 built on the Informatics for Integrating Biology and the Bedside data model84 and the Observational Health Data Sciences and Informatics (ODHSI) Network,85 built on the Observational Medical Outcomes Partnership CDM,86 represent three large-scale clinical data networks including healthcare organizations across the country that have agreed to map their local data to a CDM to support research. While some organizations may participate in more than one network, mapping local data to a CDM and participating in a data-sharing network can be costly. Thus, the Department of Health and Human Services (HHS) Office of the Assistant Secretary for Planning and Evaluation has engaged critical federal agencies within HHS, including the Food and Drug Administration, National Cancer Institute, National Library of Medicine and the Office of the National Coordinator for Health Information Technology to participate in the CDM Harmonization project to support data translation between CDMs and cross-institution sharing.87
The large-scale CDMs are early in their lifespans and development has focused on the most commonly used clinical concepts, such as patient demographics and vitals, general diagnoses, medications ordered, laboratory results and procedures performed. More complex concepts, particularly those that are domain-specific or primarily documented in the free-text clinical notes, are often unrepresented. Thus, to support irAE research, it is necessary to create and integrate specialized terminologies (such as the International Classification of Diseases for Oncology) to describe concepts critical to oncology, immunotherapy and irAEs—such as cancer morphology, stage and disease progression and immunological milieu—into these CDMs. Also critical is to have standardized definitions of individual irAEs such that they can be documented using the established clinical terminologies or consistently inferred from standard clinical data. Since the most complex clinical concepts related to understanding cancer care and irAE development are often only present in clinical notes, it will also be necessary to develop natural text mining tools that can rapidly and accurately convert clinical text in medical records into machine-interpretable concepts that can then be shared across organizations engaged in irAE research.
Large bodies of clinical data without tissue-based insights limit many investigations to the realm of correlation and association. For bedside-based mechanistic studies aiming to elucidate the underlying biology of irAEs, clinical information must be linked to tissue samples. Cancer patients, in addition to having significant clinical documentation, also have frequent laboratory tests that are an opportunity for minimally invasive collection of supplementary biospecimens that can be used to explore the physiological mechanisms of irAEs. Similar to the development of clinical data sharing networks, significant activities are underway nationally to develop strategies for integrating primary research data derived from biospecimens and biobanks associated with clinical data. The Electronic Medical Records and Genomics (eMERGE) Network,88 89 funded by the National Human Genome Research Institute, is a national consortium of academic medical centers that have integrated their local biospecimen-derived genomic data with their EHR data to perform genomic studies based on disease-specific cohorts identified through medical record information. The All of Us Research Program,90 building on the successful strategies of eMERGE, is collecting data and biospecimens from over 1 million people, in large part by leveraging health care-providing organizations to recruit participants who will share their medical records as well as tissue samples.
Opportunity: expanding clinical and translational models with biobank data
To understand and study the immune mechanisms of irAEs, access to patients’ blood and tissue specimens are key. Collection of fresh biospecimens is challenging, particularly in a way that supports asking functional immune questions as opposed to molecular questions that can be asked of fixed cells. To obtain reproducible data, biospecimens collected under standardized conditions are needed, as are assays that are conducted in a way that conforms to the spirit of CLIA rules (accurate, precise, reproducible).91 92 Humans can have high variability in immune cell numbers and activity, but biomarkers pointing toward mechanisms can still be identified, for example, target tissue-associated autoantibodies, like GAD-65 (glutamic acid decarboxylase) for type 1 diabetes mellitus (T1DM). Functional tests can yield greater insights than more descriptive phenotypic readouts.93–95 The Autoimmune Events Resulting from Systemic Modulation by Immunotherapy (AEROSMITH) Biobank is in development by the Parker Institute for Cancer Immunotherapy, JDRF and the Helmsley Trust. This biorepository will include blood and stool specimens from >1000 patients with cancer pretreatment and on-treatment receiving standard of care ICB to identify biomarkers of irAEs.96 The collection has been developed specifically to interrogate mechanisms using genetic, molecular, cellular and soluble protein data generated from blood combined with clinical assays and medical record data. This biorepository and other similar tissue banks will provide an important resource for future research questions.
In addition to a call to action to develop consensus definitions with comprehensive nomenclature for irAEs, a call to standardize biospecimen collection across institutions is critical. While samples biobanked for ‘omics’ analyses, such as those collected by the All of Us Research Program,90 will be important for cancer immunotherapy and irAE research to help define biomarkers and predictors of immunotherapy response and toxicity, ultimately biospecimens collected throughout the course of cancer therapy and at the time of the irAE will be critical to better define mechanisms and identify drivers of irAEs that could be therapeutically targeted.
While early irAE translational work focused on biospecimens collected during a very specific window of time in the midst of ICB efficacy trials, it was not standard practice to collect tissue/blood at time of the development or resolution of irAEs. We need to develop and implement standardized biospecimen biobanking procedures to enable the collection and sharing of a range of biospecimens (eg, irAE-affected tissue biopsies, blood, plasma, serum, stool and body fluids such as urine and cerebral spinal fluid). Integrating specimen collection within routine clinical care with the patient’s appropriate informed consent is preferred.97 This practice supports specimen collection prior to ICB treatment and at the time of irAE diagnosis before the initiation of immunosuppression treatment to mitigate irAEs, in addition to reducing the need for higher risk ‘research-only’ biopsies. Moreover, the establishment of rapid autopsy programs represents another important source of biospecimens, especially for organ systems that are challenging to biopsy (eg, brain, pancreas, heart). It also offers an opportunity to sample multiple tissues from the same individual simultaneously, enabling study of the immune response across different organs and tumors that ultimately led to the patient’s demise, and provides a glimpse of the level of baseline inflammation in any particular organ after ICB.
Opportunity: tailored clinical data collection and translational efforts
Effective clinical sample collection and biobanking requires an effective and reproducible framework. The Massachusetts General Hospital has pioneered implementation of an immunotoxicity service that maximizes patient care, clinical sample collection and translational research efforts.98 99 This service has developed and implemented a framework for understanding irAEs and ultimately providing better care for patients with cancer, that is based on six critical elements, including: (1) creating an infrastructure and expert knowledge for rapid identification of patients presenting with irAEs; (2) aligning experts across divisions of medical specialties to develop best practices for clinical care and optimal disease phenotyping; (3) developing a platform for oncologists, medicine subspecialists and scientists to connect regularly; (4) establishing infrastructure for sample collection in clinical settings and identifying champions to spearhead sample collection and standardized biobanking; (5) collecting the highest yield tissue samples at clinically relevant time points; and (6) accessing and leveraging the most optimal and cutting-edge technologies and big data integrative analysis frameworks to test biological hypotheses. These six key elements are further illustrated below.
First, the Severe Immunotherapy Complication (SIC) service comprises over 51 clinicians and researchers across 6 Departments (i.e., medicine, dermatology, neurology, pathology, radiology, ophthalmology) and 10 Divisions of Medicine (i.e., hematology/oncology, gastroenterology, pulmonology, cardiology, palliative care, allergy/immunology, infectious disease, endocrinology, rheumatology, nephrology), who work together to study and deeply characterize phenotypically these irAE clinical presentations,100–107 with the ultimate goal of developing more optimal treatment solutions for patients with cancer. Second, the MGH SIC service organizes a bi-weekly multidisciplinary irAE clinical case conference, which serves as a platform for clinician subspecialists, oncologists and scientists to actively connect and brainstorm about new innovative approaches and solutions. Third, by leveraging the SIC Service for irAE patient identification, the MGH team has collected biospecimens from over 300 patients across a range of irAEs prior to any administration of immunosuppressive agents to treat the irAEs. Fourth, they also collect samples serially to dynamically map key cellular and secreted factors tracking with irAEs’ evolution and resolution. Finally, the team has optimized and customized different protocols to process blood, serum and tissue specimens to enable a range of downstream immunophenotyping, functional experiments, microscopy, secreted factor analyses and molecular profiling, including single-cell RNA sequencing with paired TCR and BCR sequencing of tissue and paired blood samples to map culprit cell populations and signaling pathways involved in driving and sustaining irAEs. Integration of these different types of measurements with accurate clinical data and phenotyping will enable identification of key drivers of irAE pathogenesis to support the development of new diagnostic tools and therapeutic targets. In addition, such integrative bench-to-bedside translational efforts will ultimately allow leveraging the results from big-data integrative analyses to inform the design and target selection of the next generation of clinical trials aiming at mitigating irAEs while maintaining antitumor immunity.
Access to tissue where the irAE and inflammation unfold is key to understanding pathogenesis, while analysis of paired blood collected together with the irAE tissue enables mapping and identifying potential circulating biomarkers. Ultimately, well-clinically annotated patient phenotypes and access to irAE tissue, coupled with the use of cutting-edge ‘multiomics’, proteomics, immunophenotyping and microscopy strategies, will allow for identification of underlying irAE mechanisms driving pathogenesis, along with therapeutic target candidates that could lead to further study in preclinical and human models. Partnerships between clinical teams, translational researchers that are using systems biology and ‘multiomics’ strategies and bioinformaticians, such as those pioneered at the Massachusetts General Hospital,98 will provide lessons learned to optimize sample collection, leverage translational research and analysis frameworks, and sharing of biospecimens and patient metadata for clinical and irAE mechanism research.
Opportunity: multidisciplinary and multiorganization collaboration
Beyond local institutional multidisciplinary efforts,97 large national biobanking and translational efforts are also essential, particularly for rare cancers and irAEs. Similar to the CDMs needed for effectively sharing, aligning and integrating clinical data across institutions, the development of standardized operating procedures for sample collection from each organ subtype is required,108 and specific cryopreservation approaches are needed for a range of downstream measurements. These gold-standard procedures should be consensus driven and disseminated publicly so that consortia of universities, academic medical centers and community hospitals can bank biospecimens paired with key clinical data in a uniform way and pool resources to empower larger and more comprehensive translational research efforts, which are essential to study some of the rarest (eg, T1DM) and most fatal irAEs (eg, myocarditis with myasthenia).
There are already numerous ongoing national and international biobanking efforts, such as the aforementioned ‘AEROSMITH’ project,96 the Predictive Markers of Immune-related Adverse Events in Patients Treated with Immune Stimulatory Drugs study109 and the National Cancer Institute (NCI)-Alliance A151804,110 a multi-institutional study open at sites across the National Clinical Trial Network that has established a national biorepository to advance studies of irAEs. This alliance is committed to making biospecimens and data available to investigators following future submission and approval of proposals. While the logistics of standardizing sample collection across multiple sites will remain more challenging than single institutional efforts (such as for analyzing freshly collected tissue specimens from irAE lesions), these multi-institutional efforts are critical to empower the discovery and validation of the next generation of irAE predictors and biomarkers that promise to significantly impact the care of patients with cancer treated with ICB. These important biobanking efforts, clinical annotation and in-depth biospecimen studies using cutting-edge experimental and analysis frameworks are promising strategies for solving the underpinnings of irAEs. However, these endeavors remain costly and there is a pressing need for more funding to support collaborative efforts and sustain new and ongoing efforts.
Currently, primary research data from clinically derived biospecimens lives outside the clinical data space. Informatics technologies that ensure accurate linkage between datasets while protecting patient privacy will be critical to derive insights from the clinical and research data, and to connect these data with other valuable sources of information—such as healthcare claims data, data from wearable devices, environmental exposure data and patient reported outcomes.111 112 In addition, data storage and management solutions that support rapid input, processing and validation of large scale multi-dimensional data and that are extensible to include new data and concepts, such as the Parker Institute for Cancer Immunotherapy’s CANcer Data and Evidence Library platform113 and the National Heart Lung and Blood Institute’s BioData Catalyst114 that provides access to the Trans-Omics for Precision Medicine Program,115 will be essential for supporting rapid and replicable data analysis. Finally, deriving insights from data of this complexity will require intersection of the data with modern machine learning strategies that can leverage large scale, multi-dimensional datasets and provide interpretable results that can be further investigated by primary and translational research teams to improve preclinical models of irAEs, help design the next generation of clinical trials and enhance precision medicine strategies for clinical care.
Conclusions
Taken together, there are three core areas that will be essential to drive irAE research forward:
Developing strategies to integrate and share clinical and research data sets at a national scale to support irAE surveillance and development of biomarkers for irAEs.
Integrating biobanks with detailed clinical data and the development of biospecimen collection standard operating procedures supporting minimally invasive and disruptive collection of biospecimens (tissue, blood, body fluid) during routine care and at time of autopsy.
Leveraging insights from clinical and high-throughput ‘multi-omics’ data to develop preclinical models that more accurately model irAEs following immunotherapy and to help guide and prioritize the therapeutic target candidates to be assessed in the next generation of clinical trials.
The development of robust models to understand the etiology and progression of irAEs following cancer immunotherapy based on real-world clinical information integrated with primary research data will require creation of novel large-scale data resources, analysis strategies and effective interpretation of results. These investigations will require a multidisciplinary team of informaticists, data scientists, clinical subspecialists, oncologists, basic science and translational researchers and patients, along with multiorganizational collaborations that span healthcare systems, federal research agencies, commercial partners, academic researchers and patient advocacy groups. These partnerships, which will require substantial funding, will enable effective strategies for collection and sharing of clinical data and biospecimens, development of novel analysis strategies and implementation of critical findings into clinical practice.
Acknowledgments
The authors acknowledge SITC and AACR staff for their contributions including Emily B. Shepard, PhD, (SITC) for medical writing and editorial support, SITC staff Lianne Wiggins and Tara Withington and AACR staff Lisa Haubein, PhD, Dan Erkes, PhD, and Sarah Martin, PhD for project and workshop management and assistance. Additionally, the authors wish to thank both SITC and AACR for supporting the manuscript development. The full list of participants in the 'SITC-AACR Workshop: The Cancer Biology Underlying Immunotherapy-Induced Autoimmunity' follows. Lisa H. Butterfield, PhD – Parker Institute for Cancer Immunotherapy and University of California, San Francisco – Organizer. Elizabeth M. Jaffee, MD – Sidney Kimmel Cancer Center, Johns Hopkins University – Organizer. Arlene H. Sharpe, MD, PhD – Harvard Medical School – Organizer. Robert A. Anders, MD, PhD – Johns Hopkins University. Nicholas L. Bayless, PhD – Parker Institute for Cancer Immunotherapy. Jeffrey A. Bluestone, PhD – University of California, San Francisco and Sonoma Biotherapeutics. Laura Boos, MD – Royal Marsden Hospital. Katarzyna Bourcier, PhD – National Cancer Institute. Samantha Bucktrout, PhD – Parker Institute for Cancer Immunotherapy. Michael Dougan, MD, PhD – Massachusetts General Hospital. Robert J. Fontana, MD – University of Michigan. Helen Gogas, PhD – National and Kapodistrian University of Athens, Greece. Ellen Gravallese, MD – Brigham and Women’s Hospital. Aida Habtezion, MD, MSc – Stanford University. David A. Hafler, MD, FANA – Yale University. Toby Hecht, PhD – National Cancer Institute. Christian A. Koch, MD, PhD – Fox Chase Cancer Center. Vijay K. Kuchroo, DVM, PhD – Brigham and Women’s Hospital. Le Min, MD, PhD – Brigham and Women’s Hospital. William J. Murphy, PhD – University of California, Davis School of Medicine. Katherine L. Nathanson, MD – University of Pennsylvania. Jill O’Donnell-Tormey, PhD – Cancer Research Institute. Hideho Okada, MD, PhD – University of California, San Francisco. Virginia Pascual, MD – Weill Cornell Medicine. Bart O. Roep, PhD – City of Hope. Annette L. Rothermel, PhD – National Institute of Allergy and Infectious Diseases. Bianco D. Santomasso, MD, PhD – Memorial Sloan Kettering Cancer Center. Cheryl Selinsky, PhD – Parker Institute for Cancer Immunotherapy. Elad Sharon, MD, MPH – National Cancer Institute. Sara Siebel Marr, MS – Parker Institute for Cancer Immunotherapy. Mark Stewart, PhD – Friends of Cancer Research. Ryan J. Sullivan, MD – Massachusetts General Hospital Cancer Center. Samra Turajlic, PhD, MRCP – The Francis Crick Institute. Gulbu Uzel, MD – National Institute for Allergies and Infectious Diseases. Alexandra-Chloé Villani, PhD – Massachusetts General Hospital, Harvard Medical School, and the Broad Institute. Angela Vincent, MBBS, MSc – University of Oxford. Theresa Walunas, PhD – Northwestern University Feinberg School of Medicine. Jennifer A. Wargo, MD, MMSc – University of Texas MD Anderson Cancer Center. Figure partially developed using BioRender.com.
Footnotes
Twitter: @NickBayless, @villanilab, @TheresaWalunas
Contributors: WJM, A-CV and TLW serve as corresponding authors for this manuscript. Questions regarding the preclinical modeling aspects of the manuscript can be addressed to WJM. Questions regarding biobanking strategies, developing effective multidisciplinary teams and frameworks bridging between clinic and lab, translational research endeavors aiming to map the underpinnings of irAEs and autoimmunity, the use of cutting-edge technologies and computational integrative frameworks to infer predictors and drivers of irAE can be addressed to A-CV. Questions regarding clinical informatics or the use of electronic health record information can be addressed to TLW. All other authors equally drafted content and provided critical review during manuscript development process and are therefore listed in alphabetical order. All authors have read and approved the final version of this manuscript.
Funding: This manuscript was funded by the Society for Immunotherapy of Cancer.
Competing interests: JAB is a cofounder, CEO and a Board member of Sonoma Biotherapeutics (with salary and ownership interests). He is a cofounder of Celsius Therapeutics with ownership interests; a member of the Board of Directors of Gilead and Provention Bio with compensation and ownership interests, and the Parker Institute for Cancer Immunotherapy; and is a member of the scientific advisory boards of Arcus Biosciences, Solid Biosciences, Rheos Medicines and Vir Biotechnology. LHB has received consulting fees from Calidi, Takeda, Western Oncolytics, Khloris, Pyxis, Cytomix, Roche-Genentech, DC Prime and RAPT in the last 24 months. EMJ is a paid consultant for Adaptive Biotech, CSTONE, Achilles, DragonFly, Candel Therapeutics and Genocea. She receives funding from Lustgarten Foundation and Bristol Myer Squibb. She is the Chief Medical Advisor for Lustgarten and SAB advisor to the Parker Institute for Cancer Immunotherapy (PICI) and for the C3 Cancer Institute. She is a founding member of Abmeta. AHS receives royalty from Pfizer; has IP rights with Roche, Merck, Bristol Myers Squibb, EMD-Serono, Boehringer-Ingelheim, AstraZeneca, Dako and Novartis; receives consulting fees from Surface Oncology, Elstar, SQZ Biotechnologies, Selecta, Elpiscience, Monopteros, Bicara, GlaxoSmithKline and Janssen advisory boards, the Stand Up to Cancer Grant Catalyst Executive Advisory Committee and Review Panel, the Bloomberg Kimmel Institute for Cancer Immunotherapy at Johns Hopkins EAB, the Human Oncology and Pathogenesis Program at Memorial Sloan Kettering Cancer Center EAB, and the Massachusetts General Hospital Cancer Center EAB; has contracted research with Roche, Merck, AbbVie and Quark Ventures; has partner consulting fees from Roche, Bristol Myers Squibb, Xios and Origimed; and has partner ownership interest in Nextpoint, Triursus and Xios. All other authors have nothing to disclose. SITC staff (EBS) has nothing to disclose.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not required.
References
- 1.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350–5. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol 2018;18:153–67. 10.1038/nri.2017.108 [DOI] [PubMed] [Google Scholar]
- 3.Walunas TL, Lenschow DJ, Bakker CY, et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1994;1:405–13. 10.1016/1074-7613(94)90071-x [DOI] [PubMed] [Google Scholar]
- 4.Tivol EA, Borriello F, Schweitzer AN, et al. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 1995;3:541–7. 10.1016/1074-7613(95)90125-6 [DOI] [PubMed] [Google Scholar]
- 5.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996;271:1734–6. 10.1126/science.271.5256.1734 [DOI] [PubMed] [Google Scholar]
- 6.Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchbinder EI, Desai A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am J Clin Oncol 2016;39:98–106. 10.1097/COC.0000000000000239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keir ME, Butte MJ, Freeman GJ, et al. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 2008;26:677–704. 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haslam A, Gill J, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for immune checkpoint inhibitor drugs. JAMA Netw Open 2020;3:e200423-e.. 10.1001/jamanetworkopen.2020.0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun L, Zhang L, Yu J, et al. Clinical efficacy and safety of anti-PD-1/PD-L1 inhibitors for the treatment of advanced or metastatic cancer: a systematic review and meta-analysis. Sci Rep 2020;10:2083. 10.1038/s41598-020-58674-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robert C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat Commun 2020;11:3801. 10.1038/s41467-020-17670-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 13.Postow MA, Sidlow R, Hellmann MD. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med 2018;378:158–68. 10.1056/NEJMra1703481 [DOI] [PubMed] [Google Scholar]
- 14.Brahmer JR, Abu-Sbeih H, Ascierto PA, et al. Society for Immunotherapy of Cancer (SITC) clinical practice guideline on immune checkpoint inhibitor-related adverse events. J Immunother Cancer 2021;9. 10.1136/jitc-2021-002435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ott PA, Hodi FS, Kaufman HL, et al. Combination immunotherapy: a road map. J Immunother Cancer 2017;5:16. 10.1186/s40425-017-0218-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalil DN, Smith EL, Brentjens RJ, et al. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol 2016;13:394. 10.1038/nrclinonc.2016.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drakaki A, Dhillon PK, Wakelee H, et al. Association of baseline systemic corticosteroid use with overall survival and time to next treatment in patients receiving immune checkpoint inhibitor therapy in real-world US oncology practice for advanced non-small cell lung cancer, melanoma, or urothelial carcinoma. Oncoimmunology 2020;9:1824645. 10.1080/2162402X.2020.1824645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrelli F, Signorelli D, Ghidini M, et al. Association of steroids use with survival in patients treated with immune checkpoint inhibitors: a systematic review and meta-analysis. Cancers 2020;12. 10.3390/cancers12030546. [Epub ahead of print: 27 02 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Elsas A, Hurwitz AA, Allison JP. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J Exp Med 1999;190:355–66. 10.1084/jem.190.3.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curran MA, Montalvo W, Yagita H, et al. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A 2010;107:4275–80. 10.1073/pnas.0915174107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perdigoto AL, Kluger H, Herold KC. Adverse events induced by immune checkpoint inhibitors. Curr Opin Immunol 2021;69:29–38. 10.1016/j.coi.2021.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright JJ, Powers AC, Johnson DB. Endocrine toxicities of immune checkpoint inhibitors. Nat Rev Endocrinol 2021;17:389–99. 10.1038/s41574-021-00484-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cappelli LC, Bingham CO. Expert perspective: immune checkpoint inhibitors and rheumatologic complications. Arthritis Rheumatol 2021;73:553–65. 10.1002/art.41587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison RA, Tummala S, de Groot J. Neurologic toxicities of cancer immunotherapies: a review. Curr Neurol Neurosci Rep 2020;20:27. 10.1007/s11910-020-01038-2 [DOI] [PubMed] [Google Scholar]
- 25.Haugh AM, Probasco JC, Johnson DB. Neurologic complications of immune checkpoint inhibitors. Expert Opin Drug Saf 2020;19:479–88. 10.1080/14740338.2020.1738382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dougan M. Gastrointestinal and hepatic complications of immunotherapy: current management and future perspectives. Curr Gastroenterol Rep 2020;22:15. 10.1007/s11894-020-0752-z [DOI] [PubMed] [Google Scholar]
- 27.Bellaguarda E, Hanauer S. Checkpoint inhibitor-induced colitis. Am J Gastroenterol 2020;115:202–10. 10.14309/ajg.0000000000000497 [DOI] [PubMed] [Google Scholar]
- 28.Quandt Z, Young A, Anderson M. Immune checkpoint inhibitor diabetes mellitus: a novel form of autoimmune diabetes. Clin Exp Immunol 2020;200:131–40. 10.1111/cei.13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atkins MB, Mier JW, Parkinson DR, et al. Hypothyroidism after treatment with interleukin-2 and lymphokine-activated killer cells. N Engl J Med 1988;318:1557–63. 10.1056/NEJM198806163182401 [DOI] [PubMed] [Google Scholar]
- 30.Weijl NI, Van der Harst D, Brand A, et al. Hypothyroidism during immunotherapy with interleukin-2 is associated with antithyroid antibodies and response to treatment. J Clin Oncol 1993;11:1376–83. 10.1200/JCO.1993.11.7.1376 [DOI] [PubMed] [Google Scholar]
- 31.Scalzo S, Gengaro A, Boccoli G, et al. Primary hypothyroidism associated with interleukin-2 and interferon alpha-2 therapy of melanoma and renal carcinoma. Eur J Cancer 1990;26:1152–6. 10.1016/0277-5379(90)90275-x [DOI] [PubMed] [Google Scholar]
- 32.Krouse RS, Royal RE, Heywood G, et al. Thyroid dysfunction in 281 patients with metastatic melanoma or renal carcinoma treated with interleukin-2 alone. J Immunother Emphasis Tumor Immunol 1995;18:272–8. 10.1097/00002371-199511000-00008 [DOI] [PubMed] [Google Scholar]
- 33.Phan GQ, Attia P, Steinberg SM, et al. Factors associated with response to high-dose interleukin-2 in patients with metastatic melanoma. J Clin Oncol 2001;19:3477–82. 10.1200/JCO.2001.19.15.3477 [DOI] [PubMed] [Google Scholar]
- 34.Becker JC, Winkler B, Klingert S, et al. Antiphospholipid syndrome associated with immunotherapy for patients with melanoma. Cancer 1994;73:1621–4. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg SA, White DE. Vitiligo in patients with melanoma: normal tissue antigens can be targets for cancer immunotherapy. J Immunother Emphasis Tumor Immunol 1996;19:81–4. [PubMed] [Google Scholar]
- 36.Nordlund JJ, Kirkwood JM, Forget BM, et al. Vitiligo in patients with metastatic melanoma: a good prognostic sign. J Am Acad Dermatol 1983;9:689–96. 10.1016/s0190-9622(83)70182-9 [DOI] [PubMed] [Google Scholar]
- 37.Bystryn JC, Rigel D, Friedman RJ, et al. Prognostic significance of hypopigmentation in malignant melanoma. Arch Dermatol 1987;123:1053–5. [PubMed] [Google Scholar]
- 38.Faje AT, Lawrence D, Flaherty K, et al. High-dose glucocorticoids for the treatment of ipilimumab-induced hypophysitis is associated with reduced survival in patients with melanoma. Cancer 2018;124:3706–14. 10.1002/cncr.31629 [DOI] [PubMed] [Google Scholar]
- 39.Klein J, Sato A. The HLA system. Second of two parts. N Engl J Med 2000;343:782–6. 10.1056/NEJM200009143431106 [DOI] [PubMed] [Google Scholar]
- 40.Rubin JT, Day R, Duquesnoy R, et al. HLA-DQ1 is associated with clinical response and survival of patients with melanoma who are treated with interleukin-2. Ther Immunol 1995;2:1–6. [PubMed] [Google Scholar]
- 41.Marincola FM, Shamamian P, Rivoltini L, et al. HLA associations in the antitumor response against malignant melanoma. J Immunother Emphasis Tumor Immunol 1995;18:242–52. 10.1097/00002371-199511000-00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marincola FM, Venzon D, White D, et al. HLA association with response and toxicity in melanoma patients treated with interleukin 2-based immunotherapy. Cancer Res 1992;52:6561–6. [PubMed] [Google Scholar]
- 43.Scheibenbogen C, Keilholz U, Mytilineos J, et al. HLA class I alleles and responsiveness of melanoma to immunotherapy with interferon-alpha (IFN-alpha) and interleukin-2 (IL-2). Melanoma Res 1994;4:191–4. 10.1097/00008390-199406000-00008 [DOI] [PubMed] [Google Scholar]
- 44.Lee JE, Reveille JD, Ross MI, et al. HLA-DQB1*0301 association with increased cutaneous melanoma risk. Int J Cancer 1994;59:510–3. 10.1002/ijc.2910590413 [DOI] [PubMed] [Google Scholar]
- 45.Lee JE, Lu M, Mansfield PF, et al. Malignant melanoma: relationship of the human leukocyte antigen class II gene DQB1*0301 to disease recurrence in American Joint Committee on Cancer stage I or II. Cancer 1996;78:758–63. [DOI] [PubMed] [Google Scholar]
- 46.Lee JE, Abdalla J, Porter GA, et al. Presence of the human leukocyte antigen class II gene DRB1*1101 predicts interferon gamma levels and disease recurrence in melanoma patients. Ann Surg Oncol 2002;9:587–93. 10.1007/BF02573896 [DOI] [PubMed] [Google Scholar]
- 47.Gogas H, Kirkwood JM, Falk CS, et al. Correlation of molecular human leukocyte antigen typing and outcome in high-risk melanoma patients receiving adjuvant interferon. Cancer 2010;116:4326–33. 10.1002/cncr.25211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan Z, Di Nucci F, Kwan A, et al. Polygenic risk for skin autoimmunity impacts immune checkpoint blockade in bladder cancer. Proc Natl Acad Sci U S A 2020;117:12288–94. 10.1073/pnas.1922867117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuehn HS, Ouyang W, Lo B, et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science 2014;345:1623–7. 10.1126/science.1255904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Notarangelo LD, Uzel G, Rao VK. Primary immunodeficiencies: novel genes and unusual presentations. Hematology Am Soc Hematol Educ Program 2019;2019:443–8. 10.1182/hematology.2019000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farmer JR, Uzel G. Mapping out autoimmunity control in primary immune regulatory disorders. J Allergy Clin Immunol Pract 2021;9:653–9. 10.1016/j.jaip.2020.12.024 [DOI] [PubMed] [Google Scholar]
- 52.Schindler MK, Pittaluga S, Enose-Akahata Y, et al. Haploinsufficiency of immune checkpoint receptor CTLA4 induces a distinct neuroinflammatory disorder. J Clin Invest 2020;130:5551–61. 10.1172/JCI135947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uzel G, Karanovic D, Su H, et al. Management of cytopenias in CTLA4 haploinsufficiency using abatacept and sirolimus. Blood 2018;132:2409. 10.1182/blood-2018-99-120185 [DOI] [Google Scholar]
- 54.Lee S, Moon JS, Lee C-R, et al. Abatacept alleviates severe autoimmune symptoms in a patient carrying a de novo variant in CTLA-4. J Allergy Clin Immunol 2016;137:327–30. 10.1016/j.jaci.2015.08.036 [DOI] [PubMed] [Google Scholar]
- 55.Jamee M, Hosseinzadeh S, Sharifinejad N, et al. Comprehensive comparison between 222 CTLA-4 haploinsufficiency and 212 LRBA deficiency patients: a systematic review. Clin Exp Immunol 2021;205:28–43. 10.1111/cei.13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ercolini AM, Miller SD. The role of infections in autoimmune disease. Clin Exp Immunol 2009;155:1–15. 10.1111/j.1365-2249.2008.03834.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 2016;7:10391. 10.1038/ncomms10391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gopalakrishnan V, Helmink BA, Spencer CN, et al. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 2018;33:570–80. 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiaro J, Kasanen HHE, Whalley T, et al. Viral molecular mimicry influences the antitumor immune response in murine and human melanoma. Cancer Immunol Res 2021. 10.1158/2326-6066.CIR-20-0814. [Epub ahead of print: 08 Jun 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Titulaer MJ, Maddison P, Sont JK, et al. Clinical Dutch-English Lambert-Eaton myasthenic syndrome (LEMS) tumor association prediction score accurately predicts small-cell lung cancer in the LEMS. J Clin Oncol 2011;29:902–8. 10.1200/JCO.2010.32.0440 [DOI] [PubMed] [Google Scholar]
- 61.Song L, Wang Y, Zhang J, et al. The risks of cancer development in systemic lupus erythematosus (SLE) patients: a systematic review and meta-analysis. Arthritis Res Ther 2018;20:270. 10.1186/s13075-018-1760-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura K, Smyth MJ. Targeting cancer-related inflammation in the era of immunotherapy. Immunol Cell Biol 2017;95:325–32. 10.1038/icb.2016.126 [DOI] [PubMed] [Google Scholar]
- 63.Anderson MS, Bluestone JA. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol 2005;23:447–85. 10.1146/annurev.immunol.23.021704.115643 [DOI] [PubMed] [Google Scholar]
- 64.Young A, Quandt Z, Bluestone JA. The balancing act between cancer immunity and autoimmunity in response to immunotherapy. Cancer Immunol Res 2018;6:1445–52. 10.1158/2326-6066.CIR-18-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Blake SJ, Smyth MJ, et al. Improved mouse models to assess tumour immunity and irAEs after combination cancer immunotherapies. Clin Transl Immunology 2014;3:e22-e. 10.1038/cti.2014.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, Aguilar EG, Luna JI, et al. Paradoxical effects of obesity on T cell function during tumor progression and PD-1 checkpoint blockade. Nat Med 2019;25:141–51. 10.1038/s41591-018-0221-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McQuade JL, Daniel CR, Hess KR, et al. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol 2018;19:310–22. 10.1016/S1470-2045(18)30078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084–9. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shultz LD, Keck J, Burzenski L, et al. Humanized mouse models of immunological diseases and precision medicine. Mamm Genome 2019;30:123–42. 10.1007/s00335-019-09796-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mestas J, Hughes CCW, Mice O. Of mice and not men: differences between mouse and human immunology. J Immunol 2004;172:2731–8. 10.4049/jimmunol.172.5.2731 [DOI] [PubMed] [Google Scholar]
- 71.Scheerlinck JP. Functional and structural comparison of cytokines in different species. Vet Immunol Immunopathol 1999;72:39–44. 10.1016/s0165-2427(99)00115-4 [DOI] [PubMed] [Google Scholar]
- 72.Pulendran B, Davis MM. The science and medicine of human immunology. Science 2020;369:eaay4014. 10.1126/science.aay4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rothwell PM. External validity of randomised controlled trials: "to whom do the results of this trial apply?". Lancet 2005;365:82–93. 10.1016/S0140-6736(04)17670-8 [DOI] [PubMed] [Google Scholar]
- 74.Grant SR, Lin TA, Miller AB, et al. Racial and ethnic disparities among participants in US-based phase 3 randomized cancer clinical trials. JNCI Cancer Spectr 2020;4:pkaa060. 10.1093/jncics/pkaa060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen MS, Lara PN, Dang JHT, et al. Twenty years post-NIH Revitalization act: enhancing minority participation in clinical trials (EMPaCT): laying the groundwork for improving minority clinical trial accrual. Cancer 2014;120:1091–6. 10.1002/cncr.28575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bleyer A, Budd T, Montello M. Adolescents and young adults with cancer. Cancer 2006;107:1645–55. 10.1002/cncr.22102 [DOI] [PubMed] [Google Scholar]
- 77.Blumenthal D. Launching HITECH. N Engl J Med 2010;362:382–5. 10.1056/NEJMp0912825 [DOI] [PubMed] [Google Scholar]
- 78.Henry J, Pylypchuk Y, Searcy T. ONC data brief, no. 352016. Adoption of electronic health record systems among U.S. non-federal acute care hospitals: 2008-2015. Washington, DC: Office of the National Coordinator for Health Information Technology; 2016. [Google Scholar]
- 79.. 2019. Office-based physician health IT adoption and use. National Electronic Health Records Survey.
- 80.Fleurence RL, Curtis LH, Califf RM, et al. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc 2014;21:578–82. 10.1136/amiajnl-2014-002747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Forrest CB, McTigue KM, Hernandez AF, et al. PCORnet® 2020: current state, accomplishments, and future directions. J Clin Epidemiol 2021;129:60–7. 10.1016/j.jclinepi.2020.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.PCORnet . Common data model, 2021. Available: https://pcornet.org/data/
- 83.Visweswaran S, Becich MJ, D'Itri VS, et al. Accrual to Clinical Trials (ACT): a Clinical and Translational Science Award Consortium network. JAMIA Open 2018;1:147–52. 10.1093/jamiaopen/ooy033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17:124–30. 10.1136/jamia.2009.000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 86.Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc 2012;19:54–60. 10.1136/amiajnl-2011-000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.. Harmonization of various common data models and open standards for evidence generation; 2020. https://aspe.hhs.gov/sites/default/files/private/pdf/259016/CDMH-Final-Report-14August2020.pdf
- 88.Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 2013;15:761–71. 10.1038/gim.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4:13. 10.1186/1755-8794-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.All of Us Research Program Investigators, Denny JC, Rutter JL, et al. The "All of Us" research program. N Engl J Med 2019;381:668–76. 10.1056/NEJMsr1809937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dobbin KK, Cesano A, Alvarez J, et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume II - clinical validation and regulatory considerations. J Immunother Cancer 2016;4:77. 10.1186/s40425-016-0179-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Masucci GV, Cesano A, Hawtin R, et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume I - pre-analytical and analytical validation. J Immunother Cancer 2016;4:76. 10.1186/s40425-016-0178-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuan J, Hegde PS, Clynes R, et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J Immunother Cancer 2016;4:3. 10.1186/s40425-016-0107-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stroncek DF, Butterfield LH, Cannarile MA, et al. Systematic evaluation of immune regulation and modulation. J Immunother Cancer 2017;5:21. 10.1186/s40425-017-0223-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gnjatic S, Bronte V, Brunet LR, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer 2017;5:44. 10.1186/s40425-017-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parker Institute for Cancer Immunotherapy . Autoimmunity and cancer immunotherapy. Available: https://www.parkerici.org/research-project/autoimmunity-and-cancer-immunotherapy/
- 97.Johnson DB, Reynolds KL, Sullivan RJ, et al. Immune checkpoint inhibitor toxicities: systems-based approaches to improve patient care and research. Lancet Oncol 2020;21:e398–404. 10.1016/S1470-2045(20)30107-8 [DOI] [PubMed] [Google Scholar]
- 98.Massachusetts General Hospital . Severe immunotherapy complications service. Available: https://www.massgeneral.org/cancer-center/treatments-and-services/severe-immunotherapy-complications
- 99.Leyre Z, Molina G, Mooradian M. Effect of a multidisciplinary severe immunotherapy complications service on outcomes for patients receiving immune checkpoint inhibitor therapy for cancer with cancer undergoing ICI therapy. J Immunother Cancer 2021. (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang L, Zlotoff DA, Awadalla M, et al. Major adverse cardiovascular events and the timing and dose of corticosteroids in immune checkpoint inhibitor-associated myocarditis. Circulation 2020;141:2031–4. 10.1161/CIRCULATIONAHA.119.044703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Molina GE, Reynolds KL, Chen ST. Diagnostic and therapeutic differences between immune checkpoint inhibitor-induced and idiopathic bullous pemphigoid: a cross-sectional study. Br J Dermatol 2020;183:1126–8. 10.1111/bjd.19313 [DOI] [PubMed] [Google Scholar]
- 102.Reynolds KL, Guidon AC. Diagnosis and management of immune checkpoint inhibitor-associated neurologic toxicity: illustrative case and review of the literature. Oncologist 2019;24:435–43. 10.1634/theoncologist.2018-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leaf RK, Ferreri C, Rangachari D, et al. Clinical and laboratory features of autoimmune hemolytic anemia associated with immune checkpoint inhibitors. Am J Hematol 2019;94:563–74. 10.1002/ajh.25448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Faje A, Reynolds K, Zubiri L, et al. Hypophysitis secondary to nivolumab and pembrolizumab is a clinical entity distinct from ipilimumab-associated hypophysitis. Eur J Endocrinol 2019;181:211–9. 10.1530/EJE-19-0238 [DOI] [PubMed] [Google Scholar]
- 105.Mooradian MJ, Nasrallah M, Gainor JF, et al. Musculoskeletal rheumatic complications of immune checkpoint inhibitor therapy: a single center experience. Semin Arthritis Rheum 2019;48:1127–32. 10.1016/j.semarthrit.2018.10.012 [DOI] [PubMed] [Google Scholar]
- 106.Sise ME, Seethapathy H, Reynolds KL. Diagnosis and management of immune checkpoint inhibitor-associated renal toxicity: illustrative case and review. Oncologist 2019;24:735–42. 10.1634/theoncologist.2018-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Durbin SM, Mooradian MJ, Fintelmann FJ, et al. Diagnostic utility of CT for suspected immune checkpoint inhibitor enterocolitis. J Immunother Cancer 2020;8:e001329. 10.1136/jitc-2020-001329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gastman B, Agarwal PK, Berger A, et al. Defining best practices for tissue procurement in immuno-oncology clinical trials: consensus statement from the Society for Immunotherapy of Cancer Surgery Committee. J Immunother Cancer 2020;8:e001583. 10.1136/jitc-2020-001583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.U.S. National Library of Medicine . Predictive markers of immune-related adverse events in patients treated with immune stimulatory drugs (PREMIS) ClinicalTrials.gov, 2019. Available: https://clinicaltrials.gov/ct2/show/NCT03984318
- 110.Kozono DE, Sharon E, Le-Rademacher J, et al. Alliance A151804: establishment of a national biorepository to advance studies of immune-related adverse events. Journal of Clinical Oncology 2020;38:TPS3154. 10.1200/JCO.2020.38.15_suppl.TPS3154 [DOI] [Google Scholar]
- 111.Kho AN, Cashy JP, Jackson KL, et al. Design and implementation of a privacy preserving electronic health record linkage tool in Chicago. J Am Med Inform Assoc 2015;22:1072–80. 10.1093/jamia/ocv038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Randall SM, Ferrante AM, Boyd JH, et al. Privacy-preserving record linkage on large real world datasets. J Biomed Inform 2014;50:205–12. 10.1016/j.jbi.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 113.Parker Institute for Cancer Immunotherapy . CANDEL: a first-of-its-kind data analysis engine for scientific discovery, 2020. Available: https://www.parkerici.org/research-project/candel-data-analysis-platform
- 114.National Heart, Lung, and Blood Institute . BioData catalyst, 2020. Available: https://biodatacatalyst.nhlbi.nih.gov/
- 115.NHLBI Trans-Omics for Precision Medicine . About TOPMed: National Heart, Lung and Blood Institute, 2020. Available: https://www.nhlbiwgs.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- . 2019. Office-based physician health IT adoption and use. National Electronic Health Records Survey.