Abstract
The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) is an enormous, 470-kDa protein serine/threonine kinase that has homology with members of the phosphatidylinositol (PI) 3-kinase superfamily. This protein contributes to the repair of DNA double-strand breaks (DSBs) by assembling broken ends of DNA molecules in combination with the DNA-binding factors Ku70 and Ku80. It may also serve as a molecular scaffold for recruiting DNA repair factors to DNA strand breaks. This study attempts to better define the role of protein kinase activity in the repair of DNA DSBs. We constructed a contiguous 14-kb human DNA-PKcs cDNA and demonstrated that it can complement the DNA DSB repair defects of two mutant cell lines known to be deficient in DNA-PKcs (M059J and V3). We then created deletion and site-directed mutations within the conserved PI 3-kinase domain of the DNA-PKcs gene to test the importance of protein kinase activity for DSB rejoining. These DNA-PKcs mutant constructs are able to express the protein but fail to complement the DNA DSB or V(D)J recombination defects of DNA-PKcs mutant cells. These results indicate that the protein kinase activity of DNA-PKcs is essential for the rejoining of DNA DSBs in mammalian cells. We have also determined a model structure for the DNA-PKcs kinase domain based on comparisons to the crystallographic structure of a cyclic AMP-dependent protein kinase. This structure gives some insight into which amino acid residues are crucial for the kinase activity in DNA-PKcs.
The DNA-dependent protein kinase (DNA-PK) is an enzyme consisting of a 470-kDa catalytic subunit (DNA-PKcs) and a heterodimeric regulatory complex called Ku, which is composed of 70 (Ku70)- and 86 (Ku80)-kDa subunits (16, 21). Detailed characterization of several ionizing-radiation-sensitive rodent cell lines, including that of scid mice, has demonstrated that the DNA-PK complex is involved in the repair of DNA double-strand breaks (DSBs) induced by ionizing radiation as well as in the rejoining of V(D)J recombination intermediates (for reviews, see references 1, 22, 23, and 46). Although the genetic requirement for DNA-PK in DSB repair is well documented, the precise role of this enzyme during repair processes is not known.
In vitro experiments have demonstrated that the stable Ku protein heterodimer, consisting of the 70- and 86-kDa subunits (Ku70/80), can bind to free DNA ends in a sequence-independent manner (3, 17). The Ku70/80-DNA complex can then stabilize the association of the 470-kDa DNA-PKcs to form the DNA-PK holoenzyme (16, 40). These observations prompted the hypothesis that DNA-PK functions in DNA repair by phosphorylating protein substrates that colocalize with it on the ends of broken DNA.
The human DNA-PKcs gene transcribes a 12,228-bp open reading frame that encodes a polypeptide consisting of 4,127 amino acids, with a predicted molecular mass of 470 kDa (5, 11, 21). The carboxy-terminal end of this protein consists of 380 amino acid residues with high homology to the carboxy-terminal catalytic domains of proteins that fall into the phosphatidylinositol (PI) 3-kinase superfamily.
Although a reasonable working hypothesis would suggest that this kinase domain is vital to the DNA repair function of DNA-PKcs, analysis of the mutational defects in cell lines known to be altered in the DNA-PKcs gene do not provide unequivocal proof that the kinase domain is essential for DNA repair. Murine scid cells, for example, contain a nonsense mutation at Tyr-4046 that causes truncation of the C-terminal end by 83 amino acids (2, 21). This fragment, however, does not contain elements of the serine/threonine protein kinase domain. Equine scid cells have a frameshift mutation at amino acid 3155 which causes a loss of 25% of the DNA-PKcs peptide (37). This deleted quarter of the protein does contain the PI 3-kinase domain but also carries other sites that may be essential, including the probable Ku70 binding site (24). And finally, the murine SX9 cell line has a point mutation (Leu to Pro at amino acid 3191) at a site outside the kinase domain (15).
The enormous size of the DNA-PK complex has led others to suggest that, in addition to protein kinase activity, DNA-PK may also serve as a molecular scaffold that recruits DNA repair factors to DNA DSBs (22, 23). Mutations that disrupt this hypothetical scaffolding would also influence DNA DSB repair.
As an initial step toward dissecting the functional domains of this protein, we have assembled a functional, full-length, 13.5-kb DNA-PKcs cDNA and developed recombinant protein expression systems for a variety of mammalian cells. This system was used to determine the importance of the serine/threonine protein kinase activity during DSB repair by creating both deletion and site-directed mutations within the conserved PI 3-kinase domain of DNA-PKcs.
MATERIALS AND METHODS
Library screening of DNA-PKcs cDNA.
Human DNA-PKcs cDNA clones were obtained by screening a human pEBS7 library or a human pREP4 cDNA library (30, 39). Hybridization probes used for screening the libraries were eight PCR-amplified fragments (PDP) that were created from almost entire regions of DNA-PKcs cDNA, based on the HSU47077 sequence in the GenBank database (accession no. U47077). Primer sets used for PCR were DP-10/DP-15 for PDP-1, DP-14/DP-19 for PDP-2, DP-18/DP-5 for PDP-6, DP-27/DP-15 for PDP-9, DP-32/DP-33 for PDP-10, DP-4/DP-5 for PDP-11, DP-1/DP-44 for PDP-12, and DP-45/DP-42 for PDP-13. Individual primer sequences are as follows: DP-1, TTCAGTGCCAAGAGATCTTCCTTC; DP-4, TAGACGTGATGTATTCTCGCCTTCC; DP-5, TAACAAGCTTGGCTAAGAAGAGACG; DP-10, TTCCAGAGATTTCGGTTTGCTTG; DP-14, TGAATCTGAAGACCACCGTGCTT; DP-15, GATCCACGTAGTCTTTGTATGTG; DP-18, AGCAGGAGAAGAGTCCAGTAAAC; DP-19, TCGTAGTGAACGGAAATTATTAT; DP-27, GCACGCGCGGGAGCGGGACTC; DP-32, TGGTGAAGGGTGGCGAGGACCTG; DP-33, TAACTTGCAGCACTTGTAAATGC; DP-42, ACTCAGCTCTTGACTGTAATGAA; DP-44, ATCTGCAGTCTGTGTCAGTGTGA; DP-45, AAGTACTGTTCTCACTCCGATGT. One hundred nanograms of plasmid DNA purified from whole pEBS7 or pREP4 cDNA library was used as a template for PCR. PCRs were carried out in 20-μl volumes that contained 0.5 U of Taq polymerase (Perkin-Elmer), 1 μM dNTP, 1 μM primers, and PCR buffer (purchased from Perkin-Elmer). XL-PCR kits (catalog no. N808-0187; Perkin-Elmer) were used to amplify PDP-14 with pEBS7 cDNA library DNA as a template and primers DP-46 (TCCCTGCTGACATTTATTGACA) and DP-47 (NheI) (CTGCTAGCGGGGTAAGCTTCTCCTCTATTT). DNA sequencing reactions were performed by the dye terminator method with Taq-FS DNA polymerase sequencing kits (Applied Biosystems) and an Applied Biosystems model 373 DNA sequencer.
Site-directed mutagenesis.
Site-directed mutagenesis was performed with a QuikChange site-directed mutagenesis kit (catalog no. 200518; Stratagene). A 1.5-kb fragment of the DNA-PKcs cDNA was liberated by digestion with PmlI and KpnI restriction enzymes and subcloned into the pT7Blue plasmid vector (Novagen). Primers used for site-directed mutagenesis are as follows: MQ-1, CTGGATCCTCGGGATTGGAAACAGACATCTGAAC; MQ-2, GTTCAGATGTCTGTTTCCAATCCCGAGGATCCAG; MQ-3, CCCTTTCCTGGTGAGGGGTGGCGAGGACCTGCGG; MQ-4, CCGCAGGTCCTCGCCACCCCTCACCAGGAAAGGG (underlined nucleotides are mutation sites). The MQ-1 and MQ-2 primers introduce the D3921N mutation, and the MQ-3 and MQ-4 primers introduce K3752R. Introduction of each mutation was confirmed by DNA sequence analysis, as described above. During the mutagenesis protocol, a secondary mutation was accidentally introduced into the same domain, resulting in a clone that has two mutations, L3750R and K3752R.
After the introduction of the directed mutations, the PmlI-KpnI fragments were isolated and inserted into the pPGDP-8 cDNA expression vector. During this process, a single clone lost a base within the restriction site of PmlI, CACGTG, causing a frame shift at the position of amino acid 3715 that resulted in truncation of the protein after 10 amino acids and loss of the entire PI 3-kinase domain.
Cell culture and transfection.
The DNA-PKcs expression constructs were tested for V(D)J recombination, DNA DSB repair, and radiation survival after transfection into the following cell lines: human glioma cell lines M059K (wild type) and M059J (DNA-PKcs mutant) (29) and Chinese hamster ovary (CHO) cell lines AA8 (wild type) and V3 (DNA-PKcs mutant) (31). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2 in air by using alpha-MEM medium supplemented with 10% fetal calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml.
Transfection of the DNA-PKcs expression plasmid was performed with a calcium phosphate transfection system (catalog no. 18306-019; Gibco-BRL). For each 106 cells in a 100-mm tissue culture dish, 10 μg of the DNA-PKcs expression vector and 10 μg of the pSV2neo or pPur plasmid were transfected. Forty-eight hours after each transfection, cells were replated with the appropriate selection medium containing either 400 μg of G418 per ml or 0.5 μg of puromycin per ml. After 7 to 21 days of selection, individual colonies were isolated and further cultured.
Radiation survival assays.
Survival curves for each cell line were obtained by measuring the colony-forming abilities of irradiated cell populations. Three hundred cells were plated on 60-mm plastic petri dishes and irradiated with 137Cs γ rays at 2 h after plating at a rate of 2.2 Gy/min to achieve a cumulative dose of 1, 2, 3, or 5 Gy. After 7 to 14 days, cells were fixed and stained with 1% crystal violet in a 70% ethanol solution, colonies containing more than 20 cells were scored, and the mean value for triplicate culture dishes was determined. Cell survival was normalized to plating efficiency of untreated controls for each cell type.
Protein extract preparation, Western blotting, and in vitro protein kinase assays.
Whole-cell extracts were prepared as described previously (32). Protein concentrations of extracts were determined by Bradford analysis using bovine serum albumin as a standard. Western blot analysis of DNA-PKcs was performed as described previously (32) with the DNA-PKcs antibody [42-26] (10). As the loading control, anti-c-Abl monoclonal antibody [24-11] (1:500 dilution) (catalog no. sc-23; Santa Cruz Biotechnology, Inc.) and anti-Ku80 monoclonal antibody (a gift from Ning-Hsing Yeh) (1:1,000 dilution) were used.
DNA-PK activity was measured using recombinant replication protein A (RPA) as a substrate (32). To enrich for DNA-PK in cell extracts, immunoprecipitation reactions were performed with protein A-Sepharose CL-4B (catalog no. 17-0780-01; Pharmacia) beads conjugated with anti-human DNA-PKcs monoclonal antibody [25-4] (10) and 800 μg of whole-cell extracts. Immunoprecipitation reaction mixtures were incubated for 5 h at 4°C, washed four times with 1 ml of TM buffer (50 mM Tris-Cl [pH 7.9]–12.5 mM MgCl2–1 mM EDTA–20% glycerol) containing 100 mM KCl and two times with TM buffer containing 50 mM KCl. The precipitates were then incubated for 60 min at 30°C with 12.5 μM ATP containing 5 μCi of [γ-32P]ATP (1 Ci = 37 GBq), 100 ng of sonicated salmon sperm DNA, and 2.5 μg of recombinant RPA in TM buffer with 100 mM KCl. Protein kinase reactions were terminated by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and denatured proteins were resolved in an SDS–10% PAGE gel. Phosphorylated RPA was visualized by autoradiography.
In vitro V(D)J recombination assays.
V(D)J recombination assays were performed with plasmid substrates (pJH200 and pJH290) and RAG1 and RAG2 expression vectors, as described previously (8, 9). For each experiment, 12 μg of RAG1 vector, 10 μg of RAG2 vector, and 5 μg of pJH200 (or pJH290) substrate were transfected for each 100-mm dish of cultured cells. DNA substrates were recovered 48 h after transfection, and V(D)J recombination events were scored in a quantitative bacterial transformation assay. The number of rearranged plasmid molecules was determined as the number of bacterial ampicillin- plus chloramphenicol-resistant (Ampr + Cmr) colonies. Recombination frequency (R) is calculated as percentage of the number of Ampr + Cmr colonies from the number of Ampr colonies. For each DNA sample, the number of bacterial colonies is a mean of values obtained from two agar plates. Two independent transfections of each cell line with each V(D)J recombination substrate were performed to determine V(D)J recombination potentials.
DNA DSB repair assay.
DNA DSB repair activity following exposure to ionizing radiation was measured by two different methods: (i) rejoining kinetics, plotted as a function of time course after irradiation; and (ii) measure of residual DNA DSB lesions following exposure and recovery to three doses (0, 20, and 40 Gy) of 137Cs γ rays. Exposures consisted of a dose rate of 4 Gy/min on ice. Immediately following irradiation, the cold medium was replaced with medium that had been warmed to 37°C and the cells were placed in a 37°C tissue culture incubator for 4 h to allow for DNA DSB repair. The cells were then trypsinized on ice, washed, suspended in agarose plugs, lysed, and electrophoresed. Residual DNA DSB lesions were determined by CHEF pulsed-field gel electrophoresis combined with a storage phosphorimaging system (38). Rejoined lesions were defined as the fraction of DNA that had regained sizes large enough to prevent migration during electrophoresis (DNA retained).
Molecular modeling of the DNA-PKcs kinase domain.
An initial model of the DNA-PKcs kinase domain was built on the basis of the crystallographic structure of the cyclic AMP (cAMP)-dependent protein kinase (cAPK) catalytic subunit (48). The sequence of the ATP binding pocket of the human DNA-dependent protein kinase (HSU47077 from GenBank) was aligned with that of the catalytic subunit of cAPK (1APM from Protein Data Bank [PDB] [33a]) by using the program ALIGN from GeneStream (15a).
We modeled the structure of the ATP binding pocket of the human DNA-PK (amino acids [aa] 3795 to 3858 and 3912 to 3949) on the crystal structure of the catalytic subunit of cAPK (1APM). The structure of ATP in the complex was modeled by using the structure of ANP (adenylyl imidodiphosphate), taken from a different crystal structure of the catalytic subunit of cAPK (1CDK from PDB). The two crystal structures (1CDK and 1APM) are very similar, with the root-mean-square difference between the two sets of C-alpha atoms being 0.3 Å. Structures of the insertions and deletions were modeled with a loop modeling algorithm developed at Los Alamos National Laboratory (43). With the modified main-chain structure (including insertions and deletions), side-chain atoms were added by using an in-house software program. Special attention was paid to ensure that no close van der Waals contact exists between each of the side chains and the remainder of the molecule. The modeled structure was then subjected to a full atomic energy minimization by using AMBER (12).
RESULTS
Expression of DNA-PKcs in human and rodent DNA-PKcs mutant cell lines.
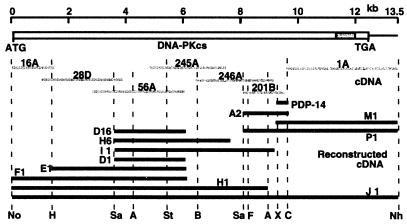
DNA-PKcs cDNA clones were identified by screening two plasmid cDNA libraries, pEBS7 (30) and pREP4 (39), with probes created by PCR amplification of eight fragments (PDP) of the DNA-PKcs gene. Seven cDNA fragments (1A, 16A, 28D, 56A, 245A, 246A, and 201B) that covered the entire DNA-PKcs cDNA region were identified (Fig. 1). A small (23-bp) gap between the 201B and 1A cDNA clones was closed by PCR amplification of a 400-bp cDNA (PDP-14) with high-fidelity rTth DNA polymerase.
FIG. 1.
Schematic presentation of the human DNA-PKcs cDNA and the process used in assembly of cDNA fragments. The full-length DNA-PKcs cDNA with the PI 3-kinase domain is shown just below the DNA size scale. The gray lines represent seven cDNAs identified from libraries and one PCR fragment, which were used for reconstruction of the full-length DNA-PKcs cDNA. The solid lines in the bottom groups represent assembled cDNAs that were cloned into plasmid vectors. The restriction enzyme sites (N, NotI; H, HindIII; Sa, SalI; A, AvrII; St, StuI; B, BglII; F, FseI; X, XbaI; C, ClaI; N, NheI) shown at the bottom of figure were used for assembling the full-length cDNA.
A full-length contiguous human DNA-PKcs cDNA clone was created from three individual clones (pKDP-F1, pBDP-I1, and pBDP-P1) that contained overlapping cDNA sequences that covered the entire DNA-PKcs coding sequence (Fig. 1). These three fragments were then assembled to create the full-length DNA-PKcs cDNA clone (pKDP-J1). Since the full-length clone was created in a vector backbone (pBluescript II KS; Stratagene) that did not contain promoter elements to direct expression in mammalian cells, three different mammalian promoter regulatory sequences were then introduced into the pKDP-J1 construct to direct expression in mammalian cells. These constructs are pPGDP-8, which utilizes the mouse phosphoglycerate kinase promoter; pCMDP-6, which utilizes a cytomegalovirus promoter; and pCHDP-6, which utilizes the same cytomegalovirus promoter but is additionally fused with a six-His polypeptide at the N terminus.
The capacity of these three full-length DNA-PKcs cDNAs to complement endogenous DNA-PKcs mutations was tested by transfecting these constructs into two DNA-PKcs mutant cell lines, the human glioma cell line M059J (29) and the CHO cell line V3 (31). Either plasmid pPur (Clontech) or plasmid pSV2neo was cotransfected along with the constructs to allow selection for drug resistance to puromycin or neomycin, respectively. Individual drug-resistant colonies were isolated for each clone, and these clones were examined for expression of recombinant DNA-PKcs protein.
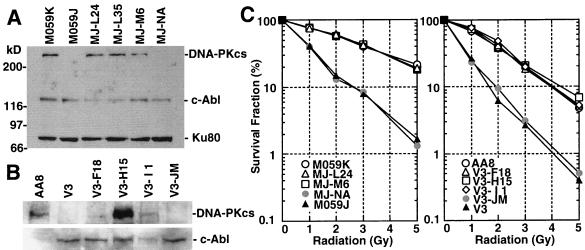
Expression of the recombinant DNA-PKcs proteins was analyzed by Western blotting with monoclonal DNA-PKcs antibody [42-26] (10). Expression of recombinant DNA-PKcs protein was readily detected in the human glioma M059J-derived cell clones MJ-L24, MJ-L35, and MJ-M6, which were transfected with pCHDP-6, pCHDP-6, and pCMDP-6, respectively (Fig. 2A). Expression of DNA-PKcs in CHO clones V3-F18, V3-H15, and V3-I1 (transfected with pPGDP-8, pCHDP-6, and pCMDP-6, respectively) was significantly lower than in human cells and required loading 10 times more protein extract (80 μg) and longer chemiluminescence detection exposure times (Fig. 2B).
FIG. 2.
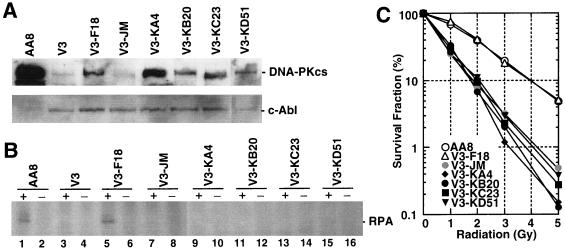
(A) Western blot analysis of recombinant DNA-PKcs protein expression. Eight micrograms each of protein extract from the human glioma cell line M059K (wild-type); M059J (DNA-PKcs mutant); MJ-L24, MJ-L35, and MJ-M6 (M059J cells transfected with the full-length DNA-PKcs cDNA expression vector); and MJ-NA (M059J cells transfected with the control plasmid pPur) were resolved by SDS-PAGE, and DNA-PKcs expression was detected by Western blot analysis. As a loading control, c-Abl and Ku80 expression was also examined. (B) For the CHO-derived cell line, AA8 (wild type); V3 (DNA-PKcs mutant); V3-F18, V3-H15, and V3-l1 (intact DNA-PKcs cDNA-transfected V3 clones); and V3-JM (control transfection V3 clone), 80 μg of each protein extract was used for Western blot analysis. The c-Abl loading control was also examined. (C) Complementation of radiation sensitivity by recombinant human DNA-PKcs. Human glioma cell lines (left) and CHO V3 cell lines (right) were assayed for radiation sensitivity.
Individual cell clones from each of the different recombinant transfections were selected and analyzed for radiation survival following acute dose irradiation (Fig. 2C). The human M059J-derived cells, MJ-L24 and MJ-M6, showed resistance levels similar to wild-type M059K cells (29). The CHO V3-derived cell lines V3-F18, V3-H15, and V3-I1 also recovered radioresistance to levels resembling parental wild-type AA8 cells. Control cell lines for each experiment, MJ-NA and V3-JM, which were created by transfecting each of the cell lines with the antibiotic resistance plasmids pPur or pSV2neo, respectively, showed no increased radioresistance.
Effects of PI 3-kinase domain mutations on DNA DSB rejoining.
Site-directed mutations were introduced within the conserved PI 3-kinase domain of the DNA-PKcs cDNA to determine whether this domain is required for DNA protein kinase activity and DNA DSB rejoining activity. Additional information about the conserved kinase domain can be found in Fig. 5A and in “Molecular modeling of the DNA-PKcs kinase domain,” below. Two residues that are conserved between serine/threonine protein kinase and PI 3-kinase domains were site mutated as described earlier. In subdomain II of serine/threonine protein kinases, the conserved lysine FLVKGGEDLRO (aa 3749 to 3759) was mutated to arginine (K3752R). This residue is believed to be critical for ATP binding within the kinase active site (19, 25). A second mutation in subdomain VIb of serine/threonine protein kinases was created within the highly conserved motif DRHLNN (aa 3921 to 3926). The conserved aspartic acid at position 3921, which is thought to function in catalysis, was mutated to asparagine (D3921N). Three independent clones that contain mutations within these kinase domain sites were produced: pPKM-A1 (K3752R), pPKM-B1 (L3750R and K3752R), and pPKM-C2 (D3921N). In addition to these clones containing point substitutions, a frameshift (pPKM-C1) was also created to examine the effects of a truncated DNA-PKcs where the entire PI 3-kinase domain is missing.
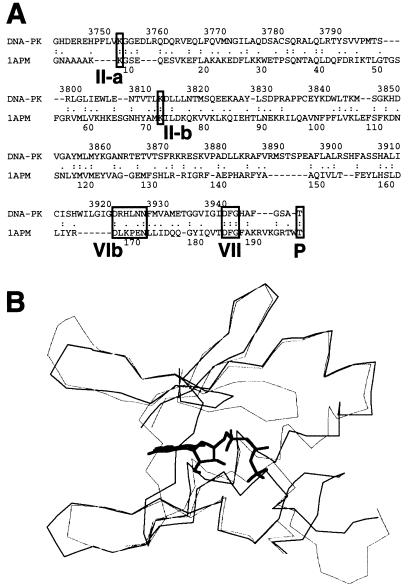
FIG. 5.
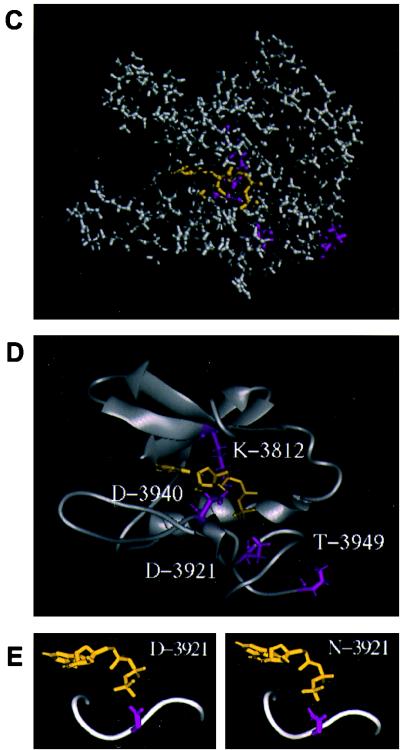
Three-dimensional modeling of the DNA-PKcs PI 3-kinase domain. (A) The sequence of the ATP binding pocket of the human DNA-PKcs was aligned with that of the catalytic subunit of cAPK (1APM). Each open box shows the serine/threonine protein kinase subdomain II homology domain within the PI 3-kinase superfamily (II-a), subdomain II from 1APM (II-b), and subdomain VIb and VII homology regions within DNA-PKcs and 1APM (VIb, VII). The phosphorylation site of threonine near the catalytic site is also shown as an open box (P). (B) Superimposed image of the ATP binding pocket of the modeled DNA-PKcs structure and the crystal structure of the 1APM active site. The thick black line in the center is ANP, the thin black line is the modeled structure of the DNA-PKcs, and the thin gray line is the crystal structure of 1APM. (C) Energy-minimized structure of the ATP binding pocket of the human DNA-PKcs. The ATP binding pocket is drawn in light gray, with ANP drawn in yellow and three conserved residues (K-3812, D-3921, and D-3940) drawn in magenta. (D) The backbone fold image of the binding pocket of the DNA-PKcs is shown together with ATP, the two conserved residues (K-3812 and D-3940) that are important for ATP binding, and another conserved residue, D-3921, that contributes to catalytic activity. The glycosylated T-3949, which may contribute to control kinase activity by phosphorylation, is also shown. (E) Structure model of the mutated residue D3921N of kinase subdomain VIb. The catalytic aspartate D-3921 is oriented with the carboxyl group facing the γ-phosphate of ATP (left). In the mutant molecule, N-3921 has an amino group pointing toward the γ-phosphate of ATP (right).
Each of the four independent kinase domain mutant cDNAs (pPKM-A1, pPKM-B1, pPKM-C1, and pPKM-C2) was transfected into V3 cell lines by the protocols described earlier. One clone that expressed DNA-PKcs protein at a level equal to or greater than the recombinant wild-type clone V3-F18 was chosen from each of the transfected cell pools (Fig. 3A). Expression of DNA-PKcs in the parental AA8 strain was greater than the human recombinant wild-type DNA-PKcs detected in clone V3-F18. Very low, residual DNA-PKcs expression was observed in the mutant V3 cell line and the vector transfection control line V3-JM. The cell lines V3-KA4 (K3752R), V3-KB20 (L3750R and K3752R), V3-KC23 (frameshift mutant), and V3-KD51 (D3921N) all expressed DNA-PKcs at levels comparable to the recombinant wild-type V3-F18 cells. As expected, the truncated mutant protein V3-KC23 had a faster gel mobility.
FIG. 3.
(A) Expression analysis of DNA-PKcs PI 3-kinase domain mutants in CHO V3 cells by Western blotting. AA8, wild type; V3, DNA-PKcs mutant; V3-F18, intact DNA-PKcs-transfected V3 cell line; V3-JM, transfection control V3 cell line; V3-KA4, kinase domain II mutant; V3-KB20, domain II mutant; V3-KC23, frameshift mutant which makes the truncated protein; V3-KD51, domain VIb mutant. As a loading control, c-Abl expression was also examined. (B) DNA-activated protein kinase activity of wild-type and mutant DNA-PKcs-expressing cells. DNA-PKcs was immunoprecipitated from whole-cell extracts. Protein kinase activity was analyzed in the absence or presence of added RPA, as indicated. The position of the 32-kDa subunit of recombinant human RPA is indicated. Phosphorylated RPA signals are observed in lane 1 (AA8) and lane 5 (V3-F18). (C) Radiation sensitivity of wild-type and mutant DNA-PKcs-expressing V3 cell lines. V3 cells expressing intact human DNA-PKcs (V3-F18) and each of the V3 cell lines expressing the four kinase domain mutant DNA-PKcs proteins were assayed for radiation sensitivity. Radiation sensitivity of the control pSV2neo transfectant V3-JM is also shown.
DNA-PK activity of the recombinant wild-type DNA-PKcs clone and each point mutant clone was then measured with recombinant RPA as a substrate (Fig. 3B). In wild-type rodent cells, the DNA-PKcs, Ku70, and Ku80 components of DNA-PK appear to be present at much lower levels than those found in primate cells (1, 22). In addition, it has been shown previously that kinase activity of the DNA-PKcs is much lower in rodent cells than in primate cells (13). Because recombinant wild-type DNA-PKcs expression in deficient rodent cells is even lower than normal expression in AA8 cells (rodent wild type), a conventional DNA-PKcs kinase assay could not detect the kinase activities of recombinant proteins. Therefore, we developed a protein kinase assay based on immunoprecipitation of the DNA-PKcs with the human anti-DNA-PKcs antibody [25-4] (10) rather than utilizing the less sensitive DNA-agarose bead method (14). This sensitive, qualitative assay clearly demonstrates positive RPA phosphorylation signals from extracts of wild-type AA8 and V3-F18 cells and also reflects the lack of RPA phosphorylation activity in extracts from all of the point mutants, V3, and V3 transfection controls (V3-JM). These data indicate that appropriate mutations within the PI 3-kinase domain will abolish the protein serine/threonine kinase activity of the DNA-PKcs.
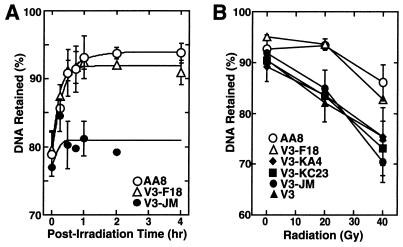
The DNA-PKcs kinase activity appears to be essential for mammalian cells to maintain a wild-type phenotype following exposure to ionizing radiation. In both cell survival (Fig. 3) and DNA DSB repair (Fig. 4) assays, none of the four cells expressing the individual kinase mutant proteins could restore V3 cell lines to the radiation-resistant phenotype of cells harboring a wild-type DNA-PKcs gene. Pulsed-field gel electrophoresis of radiation-treated cells indicated that both DNA DSB rejoining kinetics and analysis of residual lesions from V3-F18 cells closely followed those of the parental AA8 cells (Fig. 4A). Conversely, V3-JM cells showed little DSB rejoining during the 4-h postirradiation recovery period required to drive DSB rejoining to completion (Fig. 4A). The site-directed mutant constructs and radiation-sensitive lines (V3, V3-KC23, V3-KA4, and V3-JM) all showed a substantially greater fraction of unrejoined DNA DSBs at both 20 and 40 Gy compared to data obtained with AA8 and V3-F18 cells (Fig. 4B).
FIG. 4.
DNA DSB repair capacities of wild-type and mutant DNA-PKcs-expressing cell lines. DNA DSB repair is expressed as the percentage of DNA retained in the agarose plug after pulsed-field gel electrophoresis (PFGE) analysis of irradiated cell cultures. (A) Time courses of DNA DSB repair activities for AA8 (wild type), V3-F18 (intact DNA-PKcs-transfected V3 cell line), and V3-JM (control pSV2neo-transfected V3 cell line). (B) Comparisons of DNA DSB repair capacities of wild-type and DNA-PKcs mutant cell lines at 4 h postirradiation with 20- and 40-Gy doses. V3-KA4, kinase domain II mutant; V3-KC23, frameshift mutant (truncated DNA-PKcs); V3, DNA-PKcs mutant; V3-JM, control pSV2neo transfectant.
DNA DSB repair is also an important component of V(D)J recombination rejoining (4, 23, 28, 41), and the ability of kinase domain mutant constructs to participate in either coding or recombination signal sequence (RSS) joint formation is illustrated in Table 1. Recombinant wild-type V3-F18 cells are 200-fold-more proficient at coding joint formation than radiation-sensitive V3-JM cells. The three kinase cDNA mutants are also deficient in coding joint formation, showing activity that is similar to those of V3-JM cells.
TABLE 1.
V(D)J recombination assay of V3 cells with intact or mutated recombinant DNA-PKcs
| Cell line | No. of colonies
|
Ratio (%)a | Ratio avg ± SD | |
|---|---|---|---|---|
| Ampr | Ampr + Camr | |||
| Coding joining (pJH290) | ||||
| NIH 3T3 | 2,690,000 | 16,700 | 0.62 | 0.85 ± 0.33 |
| 7,540,000 | 81,600 | 1.08 | ||
| AA8 | 204,000 | 15,700 | 7.70 | 7.96 ± 0.36 |
| 26,800 | 2,200 | 8.21 | ||
| V3-F18 | 973,200 | 17,870 | 1.84 | 2.82 ± 1.39 |
| 54,200 | 2,060 | 3.80 | ||
| V3-JM | 595,000 | 62 | 0.010 | 0.014 ± 0.006 |
| 183,000 | 33 | 0.018 | ||
| V3-KA4 | 906,000 | 145 | 0.016 | 0.014 ± 0.003 |
| 533,300 | 64 | 0.012 | ||
| V3-KC23 | 1,140,000 | 80 | 0.007 | 0.009 ± 0.003 |
| 54,500 | 6 | 0.011 | ||
| V3-KD51 | 23,100 | 6 | 0.026 | 0.026 |
| RSS joining (pJH200) | ||||
| NIH 3T3 | 4,215,000 | 165,400 | 3.92 | 3.92 |
| AA8 | 61,500 | 4,590 | 7.46 | 7.28 ± 0.25 |
| 48,600 | 3,450 | 7.10 | ||
| V3-F18 | 49,700 | 1,590 | 3.20 | 3.55 ± 0.50 |
| 85,100 | 3,320 | 3.90 | ||
| V3-JM | 57,500 | 184 | 0.32 | 0.46 ± 0.20 |
| 96,700 | 580 | 0.60 | ||
| V3-KA4 | 76,500 | 260 | 0.34 | 0.54 ± 0.28 |
| 171,600 | 1,270 | 0.74 | ||
| V3-KC23 | 863,000 | 4,400 | 0.51 | 0.69 ± 0.25 |
| 247,000 | 2,150 | 0.87 | ||
| V3-KD51 | 12,500 | 25 | 0.20 | 0.20 |
Derived from the formula [(Ampr + Camr)/Ampr] × 100.
Conversely, DNA-PKcs-deficient cells have been reported to be only weakly impaired in RSS joint formation (4, 23, 28, 41). Our results indicate that the CHO V3-JM cell line produces RSS joint formation that is significantly lower (15-fold) than that found in wild-type cells. This activity could be restored by intact DNA-PKcs cDNA but not by the kinase-inactivating mutant constructs.
Molecular modeling of the DNA-PKcs kinase domain.
The disruptive nature of the mutations we created can be better understood by examining the PI 3-kinase domain from a model structure that was built by using the crystallographic structure of the cAPK catalytic subunit as a guide (48). Alignment of the protein sequence of the ATP binding pocket of the human DNA-PKcs (HSU47077 from GenBank) with that of the catalytic subunit of cAPK (Fig. 5A) showed the two sequences to be highly homologous, with 22% identity and 50% similarity. The structure of the ATP binding pocket of the human DNA-PKcs (aa 3795 to 3858 and 3912 to 3949) was then modeled by using the crystal structure of the catalytic subunit of cAPK (1APM from PDB) (33a) and the structure of the ANP co-crystal structure of the catalytic subunit of cAPK (1CDK from PDB) (7). Our modeled structure of DNA-PKcs superimposes tightly with the crystal structure of cAPK (Fig. 5B). The energy-minimized structure of the ATP binding pocket of the human DNA-PKcs complexed with ANP is shown in Fig. 5C, and its main-chain fold image is shown in Fig. 5D. ANP is drawn in yellow, and three important residues (Lys-3812, Asp-3921, and Asp-3940) are drawn in magenta. Lys-3752, which we initially thought would function in ATP binding, is localized outside of the ATP binding pocket (this residue is not shown in Fig. 5C and D). The conserved residue D-3921, which is important for the catalytic reaction, and glycosylated T-3949, a potential autophosphorylation site, are also shown in Fig. 5D.
The structure of the mutated residue D3921N of kinase subdomain VIb is modeled with an in-house software program. The catalytic aspartate D-3921 is oriented with the carboxyl group facing the γ-phosphate of ATP (Fig. 5E, left). This arrangement is suitable for the kinase molecule to phosphorylate its substrate (25). In the mutant molecule, N-3921 has an amino group pointing toward the γ-phosphate of ATP (Fig. 5E, right).
DISCUSSION
The role of DNA-PK in the repair of DNA DSBs in vivo is not known, but it is thought to function by initiating a protein phosphorylation cascade that is activated by its association with the ends of broken DNA. The induction of DNA-PK-mediated protein phosphorylation may facilitate DNA DSB rejoining directly by stimulating the catalytic activity of other DNA repair factors that colocalize to damaged DNA ends. Intrinsic to this model is the necessity for DNA-PK to function as a protein kinase. If this model is accurate, mutations that abolish the protein kinase activity of DNA-PK should disrupt this putative DNA strand break signaling cascade and inactivate DNA DSB rejoining pathways. In this report, we provide evidence that supports this model by directly demonstrating that the protein kinase activity from DNA-PKcs is essential for the repair of ionizing radiation-induced DNA DSB and the rejoining of V(D)J intermediates.
The kinase domain of the DNA-PKcs is highly conserved between other members of the PI 3-kinase superfamily (for reviews, see references 1, 27, and 45). Other members of this family also function in cellular pathways that intersect with DNA damage responses. The ATM gene, which is defective in patients with ataxia telangiectasia, is required for the DNA damage-induced checkpoints in the G1, S, and G2 phases of the cell cycle and for normal repair of chromosomal DNA damage (35). The Drosophila melanogaster mei-41 gene product is functionally similar to ATM, as are the Saccharomyces cerevisiae MEC1 (ESR1) and the Schizosaccharomyces pombe rad3 gene products (20, 26, 36, 44).
Three regions of homology within the PI 3-kinase domain are shared by the members of the DNA-PKcs subgroup (1, 19, 25): first, a conserved region containing protein sequences corresponding to subdomain II of the serine/threonine protein kinases; second, a region that includes protein sequences corresponding to subdomains VIb and VII; and third, a motif found at the extreme carboxy terminus that is found only in the DNA-PKcs subgroup. In the scid mouse, this domain of the DNA-PKcs protein is missing (2, 5). We show here that mutations that remove the conserved lysine in subdomain II (K3752R or L3750R + K3752R) or the conserved aspartic acid in subdomain VIb (D3921N) disrupt the protein kinase activity of the DNA-PKcs. In addition, each of these mutant DNA-PKcs proteins could not function in the repair of ionizing radiation-induced DNA DSBs or in V(D)J recombination.
Using information obtained from the crystal structure of cAPK, we generated a structure model for the PI 3-kinase domain of the DNA-PKcs. The conserved serine/threonine protein kinase subdomains VIb and VII of these two proteins were readily aligned, and the DNA-PKcs structure could be superimposed over the cAPK crystal structure. Asp-3912 in subdomain VIb and the DFG motif in subdomain VII are well conserved between these two proteins. Based on the position of Lys-3752 within the primary sequence of DNA-PKcs, it was thought that this residue may interact with the α- and β-phosphates of ATP (1, 19, 25). However, superimposed structures show that this lysine residue may reside outside of the ATP binding pocket and therefore may not directly interact with ATP. However, our analysis indicates that Lys-3752 is important for kinase activity, and it is therefore likely that this amino acid has an alternative role in maintaining the structure of the active site. Interestingly, Lys-3812, which is unique to the DNA-PKcs, aligns remarkably well with the ATP-binding lysine of cAPK in the modeled structure. Site-directed mutagenesis of this lysine residue may help to support this notion.
In cAPK, Thr-197 is a phosphorylation site that is important in modulating the catalytic flux of cAPK (25). In addition, the cAPK peptide inhibitor PKI [5-24] appears to localize in the vicinity of Thr-197 (48). The modeling comparison indicates that Thr-3949 of DNA-PKcs is localized in the corresponding space and could serve as a phosphorylation site. Autophosphorylation of Thr-3949 may have a role in self-inhibition of DNA-PKcs kinase activity.
The contribution of DNA-PKcs to the coding and RSS joint formation of V(D)J recombination has been somewhat ambiguous because scid mice exhibit a leaky phenotype for T- and B-cell development (6). Previous studies indicate that scid cells have relatively normal RSS joint formation rates but that this process may be somewhat error prone (4, 28, 41). In contrast, our results indicate that RSS joint formation in DNA-PKcs mutant cells is also impaired but with less magnitude than coding joint formation. Because there is about 10% residual RSS joint formation activity in DNA-PKcs mutant cells, based on our results, and almost no activity in Ku70 or Ku80 mutant cells (18, 41), we can speculate that there is an alternative pathway for RSS joint repair which is DNA-PKcs independent and Ku70/80 dependent. On the other hand, coding joint formation is almost entirely DNA-PKcs dependent, suggesting that DNA-PKcs is essential in the resolution of hairpin structure intermediates common in coding joint formation (34, 42).
Reconstructed human DNA-PKcs cDNA fully complements the radiation sensitivity and DNA DSB repair-deficient phenotypes of V3 cells, but the level of coding and RSS joint formation is only partially restored when V3-F18 activity is compared to those of the wild-type AA8 cells. Several possibilities could explain these differences. One may be that V(D)J recombination activities between DNA-PKcs proteins from different species may not be able to fully complement each other, i.e., human DNA-PKcs may not be able to fully complement Chinese hamster function. Alternatively, the human DNA-PKcs V(D)J recombination activity may be expressed at suboptimal levels in hamster V3-F18 cells. Inherent differences between the AA8 and V3 cell lines may have also caused differences in the base levels of V(D)J recombination activity between these two cell lines, since V3 has been maintained as a separate cell line for a considerable period of time. In addition, inconsistencies between coding and RSS joint formation between the hamster-derived AA8 cells and V3-F18 cells may not be significant if one considers the differences in coding and RSS joint formation between wild-type NIH 3T3 mouse cells and AA8 hamster cells.
Our complementation levels are significantly higher than previous results with chromosome transfer or YAC fusion experiments. The latter constructs only partially restored the mutant phenotype (4, 33, 47). The plausible explanation for this is that these introduced chromosomes or YAC DNAs are relatively unstable and are easily lost during propagation, resulting in subpopulations of cells exhibiting the uncomplemented phenotype. Our reconstructed cDNA provides an excellent complementation system for analyzing other components of DNA-PKcs function.
ACKNOWLEDGMENTS
We thank P. Pardington, D. Manter, and Y. Zheng for technical assistance; R. Legerski for the pEBS7 cDNA library; M. Buchwald for the pREP4 cDNA library; J. H. J. Petrini for plasmid pPGKneo; T. H. Carter for anti-DNA-PKcs antibodies [25-4] and [42-26]; N.-H. Yeh for anti-Ku80 monoclonal antibody; and R. T. Okinaka for assistance in editing of the manuscript. Also, we thank C. W. Anderson for a suggestion about DNA-PKcs PI 3-kinase domain mutagenesis.
This work was supported by U.S. Department of Energy and NIH grants CA50519 to D.J.C. and CA06294 to M.D.S.
REFERENCES
- 1.Anderson C W, Carter T H. The DNA-activated protein kinase—DNA-PK. Curr Top Microbiol Immunol. 1996;217:91–111. doi: 10.1007/978-3-642-50140-1_7. [DOI] [PubMed] [Google Scholar]
- 2.Araki R, Fujimori A, Hamatani K, Mita K, Saito T, Mori M, Fukumura R, Morimyo M, Muto M, Itoh M, Tatsumi K, Abe M. Nonsense mutation at Tyr-4046 in the DNA-dependent protein kinase catalytic subunit of severe combined immune deficiency mice. Proc Natl Acad Sci USA. 1997;94:2438–2443. doi: 10.1073/pnas.94.6.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blier P R, Griffith A J, Craft J, Hardin J A. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 4.Blunt T, Finnie N J, Taccioli G E, Smith G C, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 5.Blunt T, Gell D, Fox M, Taccioli G E, Lehmann A R, Jackson S P, Jeggo P A. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc Natl Acad Sci USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosma M J, Carroll A M. The SCID mouse mutant: definition, characterization, and potential uses. Annu Rev Immunol. 1991;9:323–350. doi: 10.1146/annurev.iy.09.040191.001543. [DOI] [PubMed] [Google Scholar]
- 7.Bossemeyer D, Engh R A, Kinzel V, Ponstingl H, Huber R. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24) EMBO J. 1993;12:849–859. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boubnov N V, Hall K T, Wills Z, Lee S E, He D M, Benjamin D M, Pulaski C R, Band H, Reeves W, Hendrickson E A, et al. Complementation of the ionizing radiation sensitivity, DNA end binding, and V(D)J recombination defects of double-strand break repair mutants by the p86 Ku autoantigen. Proc Natl Acad Sci USA. 1995;92:890–894. doi: 10.1073/pnas.92.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boubnov N V, Wills Z P, Weaver D T. V(D)J recombination coding junction formation without DNA homology: processing of coding termini. Mol Cell Biol. 1993;13:6957–6968. doi: 10.1128/mcb.13.11.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connelly M A, Zhang H, Kieleczawa J, Anderson C W. Alternate splice-site utilization in the gene for the catalytic subunit of the DNA-activated protein kinase, DNA-PKcs. Gene. 1996;175:271–273. doi: 10.1016/0378-1119(96)00135-7. [DOI] [PubMed] [Google Scholar]
- 12.Cornell W D, Cieplak P, Bayly C I, Could I P, Merz K M J, Ferguson D M, Spellmeyer D C, Fox T, Caldwell J W, Kollman P A. A second generation force-field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 13.Finnie N J, Gottlieb T M, Blunt T, Jeggo P A, Jackson S P. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fried L M, Koumenis C, Peterson S R, Green S L, van Zijl P, Allalunis-Turner J, Chen D J, Fishel R, Giaccia A J, Brown J M, Kirchgessner C U. The DNA damage response in DNA-dependent protein kinase-deficient SCID mouse cells: replication protein A hyperphosphorylation and p53 induction. Proc Natl Acad Sci USA. 1996;93:13825–13830. doi: 10.1073/pnas.93.24.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukumura R, Araki R, Fujimori A, Mori M, Saito T, Watanabe F, Sarashi M, Itsukaichi H, Eguchi-Kasai K, Sato K, Tatsumi K, Abe M. Murine cell line SX9 bearing a mutation in the dna-pkcs gene exhibits aberrant V(D)J recombination not only in the coding joint but also in the signal joint. J Biol Chem. 1998;273:13058–13064. doi: 10.1074/jbc.273.21.13058. [DOI] [PubMed] [Google Scholar]
- 15a.GeneStream. 1999. [Online.] Institut de Génétique Humaine, Montpellier, France. http: //www2.igh.cnrs.fr/bin/align-guess.cgi. [17 March 1999, last date accessed.]
- 16.Gottlieb T M, Jackson S P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 17.Griffith A J, Blier P R, Mimori T, Hardin J A. Ku polypeptides synthesized in vitro assemble into complexes which recognize ends of double-stranded DNA. J Biol Chem. 1992;267:331–338. [PubMed] [Google Scholar]
- 18.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanks S K, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 20.Hari K L, Santerre A, Sekelsky J J, McKim K S, Boyd J B, Hawley R S. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- 21.Hartley K O, Gell D, Smith G C, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 22.Jackson S P. DNA-dependent protein kinase. Int J Biochem Cell Biol. 1997;29:935–938. doi: 10.1016/s1357-2725(97)00006-x. [DOI] [PubMed] [Google Scholar]
- 23.Jeggo P A. DNA-PK: at the cross-roads of biochemistry and genetics. Mutat Res. 1997;384:1–14. doi: 10.1016/s0921-8777(97)00009-8. [DOI] [PubMed] [Google Scholar]
- 24.Jin S, Kharbanda S, Mayer B, Kufe D, Weaver D T. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J Biol Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 25.Johnson L N, Noble M E, Owen D J. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 26.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keith C T, Schreiber S L. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 28.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 29.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day R S R, Barron G M, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 30.Peterson C, Legerski R. High-frequency transformation of human repair-deficient cell lines by an Epstein-Barr virus-based cDNA expression vector. Gene. 1991;107:279–284. doi: 10.1016/0378-1119(91)90328-9. [DOI] [PubMed] [Google Scholar]
- 31.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson S R, Stackhouse M, Waltman M J, Chen F, Sato K, Chen D J. Characterization of two DNA double-stranded break repair-deficient cell lines that express inactive DNA-dependent protein kinase catalytic subunits. J Biol Chem. 1997;272:10227–10231. doi: 10.1074/jbc.272.15.10227. [DOI] [PubMed] [Google Scholar]
- 33.Priestley A, Beamish H J, Gell D, Amatucci A G, Muhlmann-Diaz M C, Singleton B K, Smith G C, Blunt T, Schalkwyk L C, Bedford J S, Jackson S P, Jeggo P A, Taccioli G E. Molecular and biochemical characterisation of DNA-dependent protein kinase-defective rodent mutant irs-20. Nucleic Acids Res. 1998;26:1965–1973. doi: 10.1093/nar/26.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Protein Data Bank. 26 January 1999, revision date. [Online.] Brookhaven National Laboratory. http: //www.pdb.bnl.gov. [17 March 1999, last date accessed.]
- 34.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 35.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 36.Seaton B L, Yucel J, Sunnerhagen P, Subramani S. Isolation and characterization of the Schizosaccharomyces pombe rad3 gene, involved in the DNA damage and DNA synthesis checkpoints. Gene. 1992;119:83–89. doi: 10.1016/0378-1119(92)90069-2. [DOI] [PubMed] [Google Scholar]
- 37.Shin E K, Perryman L E, Meek K. A kinase-negative mutation of DNA-PK(CS) in equine SCID results in defective coding and signal joint formation. J Immunol. 1997;158:3565–3569. [PubMed] [Google Scholar]
- 38.Story M D, Mendoza E A, Meyn R E, Tofilon P J. Pulsed-field gel electrophoretic analysis of DNA double-strand breaks in mammalian cells using photostimulable storage phosphor imaging. Int J Radiat Biol. 1994;65:523–528. doi: 10.1080/09553009414550611. [DOI] [PubMed] [Google Scholar]
- 39.Strathdee C A, Gavish H, Shannon W R, Buchwald M. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature. 1992;356:763–767. doi: 10.1038/356763a0. . (Erratum, 358:434.) [DOI] [PubMed] [Google Scholar]
- 40.Suwa A, Hirakata M, Takeda Y, Jesch S A, Mimori T, Hardin J A. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc Natl Acad Sci USA. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 42.Thompson C B. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 43.Tung C S. A computational approach to modeling nucleic acid hairpin structures. Biophys J. 1997;72:876–885. doi: 10.1016/s0006-3495(97)78722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 45.Zakian V A. ATM-related genes: what do they tell us about functions of the human gene? Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 46.Zdzienicka M Z. Mammalian mutants defective in the response to ionizing radiation-induced DNA damage. Mutat Res. 1995;336:203–213. doi: 10.1016/0921-8777(95)00003-3. [DOI] [PubMed] [Google Scholar]
- 47.Zdzienicka M Z, Jongmans W, Oshimura M, Priestley A, Whitmore G F, Jeggo P A. Complementation analysis of the murine scid cell line. Radiat Res. 1995;143:238–244. [PubMed] [Google Scholar]
- 48.Zheng J, Knighton D R, ten Eyck L F, Karlsson R, Xuong N, Taylor S S, Sowadski J M. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32:2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]