Abstract
Vitamin D receptor (VDR) executes the main biological functions of its ligand vitamin D. VDR/vitamin D plays critical roles in regulating host immunity, maintaining barrier functions, and shaping gut microbiome. Reduction of intestinal VDR has been reported in various diseases, including inflammatory diseases and colon cancer. However, it is always challenging to get biopsies to test the pathologic changes of VDR in intestine. In the current study, we reported a simple and sensitive quantitative PCR (qPCR) method to detect reduction of intestinal VDR using fecal samples. We validated this method in several experimental models, such as colitis, bacterial infection, and aging. We further correlated the qPCR data of VDR with the protein level of VDR in colon or serum 25 (OH)D3 in mice with different VDR status (VDR+/+, VDR+/-, and VDR−/−). Our data indicate that the qPCR method to test VDR using fecal samples could detect the expression level of intestinal VDR in various diseases. Our study highlights the feasibility, sensitivity, and simplicity of a molecular method to study the status of VDR as a biomarker.
Keywords: Aging, Biomarker, Correlation, Infection, Inflammation, Microbiome, Salmonella, Vitamin D deficiency
Background
Clinically, the serum 25(OH)D3 is used as a biomarker for the vitamin D status of human body. However, the biological most active vitamin D is 1, 25(OH)2D3, which binds to vitamin D receptor (VDR). The biological function of 1, 25(OH)2D3 is executed via VDR, a nuclear receptor. Vitamin D/VDR plays critical roles regulating host immunity, maintaining barrier functions, and shaping gut microbiome.1, 2, 3 A fundamental relationship among vitamin D/VDR, gut microbiota, and genetic factors is essential not only for intestinal homeostasis, but also for the development of chronic inflammation, such as inflammatory bowel diseases (IBD).2,4 This knowledge could be used to develop intestinal VDR as a clinical biomarker for identifying patients who might benefit from currently available interventions, as well as for the eventual development of novel strategies for the prevention and treatment of human IBD and other chronic diseases.5, 6, 7, 8 Reduction of intestinal VDR has been also reported in other diseases, such as colon cancer.9, 10, 11 However, it is always challenging to get biopsies to test the pathologic changes of VDR in intestine.
We hypothesize that the fecal samples contain sufficient shed epithelial cells expressing VDR, which allow the detection by qPCR. In the current study, we reported a simple qPCR method to measure the mRNA level of VDR, using fecal samples. We validated this method in several experimental models, such as colitis, chronic infection, and aging.
Materials and methods
Animals
VDR−/− mice on a C57BL/6 background were obtained by breeding heterozygous VDR+/− mice.12 VDRLoxP mice were originally reported by Dr. Geert Carmeliet.13 VDRΔIEC mice were obtained by crossing the VDRLoxP mice with villin-cre mice (Jackson Laboratory, 004586), as we previously reported.5 Mice were provided with water ad libitum and maintained in a 12-h dark/light cycle. The animal work was approved by the UIC Office of Animal Care.
Colon tissues and fecal samples
The intestinal tissues and fecal samples were obtained from specific-pathogen-free C57BL/6 mice12,14 or mouse models used in previous studies,15, 16, 17 including a chronic Salmonella infection model,18 a dextran sulfate sodium (DSS) treated colitis model.17 Samples of aging mice were from 20 to 24 week-old C57BL/6 mice, and the control mice were 7–8 week-old.
Immunohistochemistry (IHC)
The immunohistochemistry was performed on paraffin-embedded tissue sections (4 μm). After preparation of the slides, as described previously,19,20 antigen retrieval was achieved by incubation of the slides for 15 min in the hot preheating sodium citrate (pH 6.0) buffer, and 30 min of cooling at room temperature. Endogenous peroxidases were quenched by incubating the slides in 3% hydrogen peroxide for 10 min, followed by three rinses with HBSS, and incubation for 1 h in 3% BSA +1% goat serum in HBSS to reduce nonspecific background. Primary antibodies VDR (1:300, Santa Cruz, Cat.No.SC-13133) was applied for overnight in a cold room. After three rinses with HBSS, the slides were incubated in secondary antibody (1:100, Jackson ImmunoResearch Laboratories, Cat.No.115-065-174) for 1 h at room temperature. After washing with HBSS for 10 min, the slides were incubated with vectastain ABC reagent (Vector Laboratories, Cat.No. PK-6100) for 1 h. After washing with HBSS for 5 min, color development was achieved by applying peroxidase substrate kit (Vector Laboratories, Cat.No. SK-4800) for 2–5 min, depending on the primary antibody. The duration of peroxidase substrate incubation was determined through pilot experiments and was then held constant for all of the slides. After washing in distilled water, the sections were counterstained with hematoxylin (Leica, Cat.No.3801570), dehydrated through ethanol and xylene, and cover-slipped using a permount (Fisher Scientific, Cat.No.SP15-100).
Fecal sample collection
The tubes for fecal sample collection were autoclaved and then irradiated with ultraviolet light to destroy the contaminating environmental bacterial DNA. Fresh fecal pellets were collected by holding the mouse in one hand, during which the mouse can defecate directly in 1.5 mL tube held in the other hand. About 3–4 fresh fecal pellets were collected from each mouse. Alternatively, individual mice were put in sterile cages, and fresh fecal pellets were collected from the cages. Tubes containing fecal pellets were stored in −80 °C freezer.
RNA extraction from fecal sample
Approximately 100 mg of frozen fecal pellet was added to a sterile 1.5 mL tube containing 0.5 mL TRIzol Reagent (Thermo Fisher Scientific, Cat.No.15596018), and homogenized with a pestle for 2–3 min. After homogenization, the slurry was poured into 1.5 mL tubes, which were centrifuged at 12,000×g for 5 min at 4 °C. The supernatant from each tube was transferred carefully to new sterile 1.5 mL tubes. To each tube, 0.3 mL of TRIzol Reagent and 0.3 mL chloroform were added, the tubes were shaken vigorously for 30 s, incubated for 5 min at 4 °C, and centrifuged at 12,000×g for 15 min at 4 °C. The aqueous phase from each tube was removed carefully without contamination from the interface and transferred to a fresh 1.5 mL tube. An equal volume of 70% ethanol was added, and the tubes were vortexed vigorously for 30 s. The mixed solution (700 μL) was added to an RNeasy minispin column (Qiagen, Cat No.217004), and the columns were centrifuged at 10,000×g for 15 s at 4 °C. The remaining steps were performed according to the manufacturer's instructions. Total RNA concentrations were determined by ultraviolet spectrophotometry, and the RNA samples were stored at −80 °C.
RNA extraction from colonic tissue sample
Mouse colonic epithelial cells were collected by scraping the tissue from the colon of the mouse, including the proximal and distal regions. Total RNA was extracted from mouse epithelial cells using TRIzol reagent (Invitrogen, Grand Island, NY, USA) by following company's instructions. Per 5–10 × 106 of cells were lysed with 1 mL of TRIzol Reagent by repetitive pipetting, and then followed by 5-min incubation at room temperature to permit the complete dissociation of nucleoprotein complexes. Then 0.2 mL of chloroform per 1 mL of TRIzol Reagent was used. Samples were vortexed vigorously for 15 s followed by incubation of 2–3 min at room temperature. Samples were then subjected to centrifugation at no more than 12,000×g at 4 °C for 15 min. Upper aqueous phase was carefully transferred without disturbing the interphase into a new Eppendorf tube, and then precipitated using 0.5 mL of isopropyl alcohol per 1 mL of TRIzol Reagent for 10 min at room temperature. Pellet after 10-min centrifugation at 12,000×g at 4 °C was washed with at least 1 mL of 75% ethanol per 1 mL of TRIzol Reagent used for the initial homogenization. After the centrifugation of 7500×g for 5 min at 4 °C, RNA pellet was air-dried for 5–10 min, and 40 ul of RNase-free water was added for resuspension by pipetting. The solution can be incubated at 55–60 C° for 10 min if the pellet is hard to dissolve. Total RNA concentrations were determined by ultraviolet spectrophotometry, and the RNA samples were stored at −80 °C.
qPCR analysis
RNA reverse transcription was performed using the iScript cDNA synthesis kit (Bio-Rad, Cat.No.170889) according to the manufacturer's protocol. The RT cDNA reaction products were subjected to qPCR using Bio-Rad CFX 96 Real-time system and SYBR green supermix (Bio-Rad, Cat.No. 1725121) according to the manufacturer's protocol. All expression levels were normalized to β-actin levels of the same sample. Percent expression was calculated as the ratio of the normalized value of each sample to that of the corresponding control. All qPCR reactions were performed in triplicate. Optimal primer sequences were designed using Primer-BLAST or obtained from PrimerBank. The primer pairs used in this study were listed as follows: mouse β-actin, Forward 5′-TGTTACCAACTGGGACGACA-3′ and Reverse 5′-CTGGGTCATCTTTTCACGGT-3′; mouse VDRF, Forward 5′-GAATGTGCCTCGGATCTGTGG-3′ and Reverse 5′-ATGCGGCAATCTCCATTGAAG-3′ (Table 1).
Table 1.
Real-time PCR primers.
| Primers name | Sequence |
|---|---|
| mβ-actinF | 5′-TGTTACCAACTGGGACGACA-3′ |
| mβ-actinR | 5′-CTGGGTCATCTTTTCACGGT-3′ |
| mVDRF | 5′-GAATGTGCCTCGGATCTGTGG-3′ |
| mVDRR | 5′-ATGCGGCAATCTCCATTGAAG-3′ |
Western blot
Mouse colonic epithelial cells were collected by scraping the proximal and distal regions of colon tissues. The harvested cells were sonicated in lysis buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, pH 8.0, 1% Triton X-100) with 0.2 mM sodium ortho-vanadate, and protease inhibitor cocktail. Cultured cells were rinsed twice with ice-cold HBSS, lysed in protein loading buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol). After sonication, protein concentration was measured using the BioRad Reagent (BioRad, Cat.No. 500-0006). Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with primary antibodies: anti-Villin (Santa Cruz Biotechnology, SC-58897), and anti-VDR (Santa Cruz Biotechnology, SC-13133). Membranes probed with more than one antibody were stripped before re-probing. Visualization was performed by using ECL (Thermo Fisher Scientific, Cat.No. 32106).
Serum 25-hydroxyvitamin D3
25-Hydroxyvitamin D3 in mouse serum was measured using ELISA kit (Biomatik, Cat.No. EKL54383) according to the manufacturer's protocol. Mouse serum (50 μL per well, 1:10 dilution) was added to each well, then followed by 50 μL of detection reagent A and 2-h incubation at 37 °C. After wash, 100 μL of detection reagent B working solution was added to each well and incubated for 1 h at 37 °C. Following complete washing, 90 μL of Substrate solution was added to each well. After 15–25 min incubation at 37 °C, 50 μL of Stop Solution was added. Then the samples were ready for absorbance measurement at 450 nm.
Statistical analysis
All the data are expressed as the mean ± SD. All statistical tests were 2-sided. The P values < 0.05 were considered statistically significant. Student's t-test was used to compare the differences between two-groups samples. Pairwise correlation analyses and scatter plots were conducted among fold changes of VDR protein, serum vitamin D3, and VDR mRNA, using SAS version 9.4 (SAS Institute, Inc., Cary, NC, USA). Other statistical analyses were performed using GraphPad Prism 6 (GraphPad, Inc., San Diego, CA., USA).
Results and discussion
Establish a qPCR method to measure the VDR expression in feces
To establish a method to test VDR expression in feces, we first examined the total RNA extracted from mouse feces. Agarose gel electrophoresis in Fig. 1A showed that the RNA extracted from the fecal samples had high quality with the fragment length between 28S and 18S bands, suggesting the amount and quality of mammal RNAs were sufficient for the studies.
Figure 1.
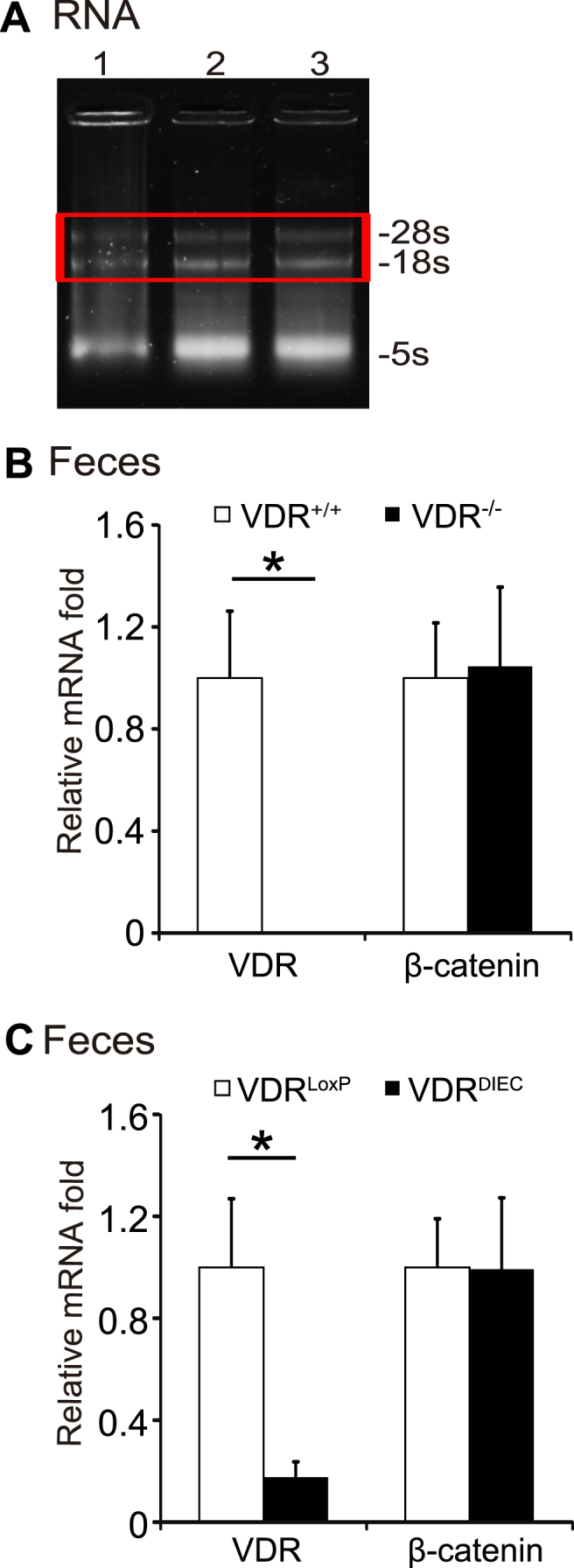
Establish a qPCR method to test VDR expression in fecal samples. (A) Agarose gel electrophoresis of total RNA extracted from fecal samples. The results showed that the RNA extracted from the fecal samples had high quality with the fragment length between 28S and 18S bands. Lanes 1–3: three different fecal samples. (B) RNA expression of VDR and β-catenin in fecal samples from VDR+/+ or VDR−/− mice. No VDR mRNA expression in fecal samples from VDR−/− mice, while β-catenin expression is normal in VDR−/− mice. Data are expressed as mean ± SD. n = 3, Student's t-test, ∗P < 0.05. (C) RNA expression of VDR and β-catenin in fecal samples from VDRLoxp or VDRΔIEC mice. Data are expressed as mean ± SD. n = 6, Student's t-test, ∗P < 0.05.
We then tested the mRNA expression of VDR in fecal samples from wild-type and VDR knockout (VDR−/−) mice. No VDR mRNA expression was detected in fecal samples from VDR−/− mice, whereas the β-catenin expression was detectable in both VDR+/+ and VDR−/− mice, suggesting the PCR method was specific to differentiate the VDR level in these mice (Fig. 1B).
We further examined the expression levels of VDR and β-catenin, using fecal samples from the intestinal epithelial VDR conditional knockout (VDRΔIEC) mice. Fecal samples from the VDRLoxp mice were used as a control group. We found significantly reduced VDR mRNA expression in VDRΔIEC mice, compared with the VDRLoxp mice (Fig. 1C). Because VDRΔIEC mice had specific VDR deletion in intestinal epithelial cells, feces had mixture of shed cells from other cell types. We could still detect some mRNA of VDR in feces of VDRΔIEC mice. However, we did not find the significant changes of β-catenin expression in feces of VDRΔIEC mice, compared with the VDRLoxp mice (Fig. 1C). Our data suggest that this established method is specific and sensitive.
Reduced VDR expression in a colitis mouse model
VDR expression is known to be down-regulated in colons of patients with IBD, in both UC and CD, compared to normal colons.5,21 In a DSS-induced colitis model, VDR was also down-regulated in colon (Fig. 2A). To validate the new method for VDR expression, we compared the expression profiles of Vdr gene from paired DSS-treated and control fecal samples. We were able to test the down-regulated VDR expression at the mRNA level in feces from the DSS-treated mice (Fig. 2B). We further found that VDR expression was also significantly down-regulated at the mRNA level in colon samples from DSS-treated mice (Fig. 2C). These observations in colon were consistent with the data using the fecal samples.
Figure 2.
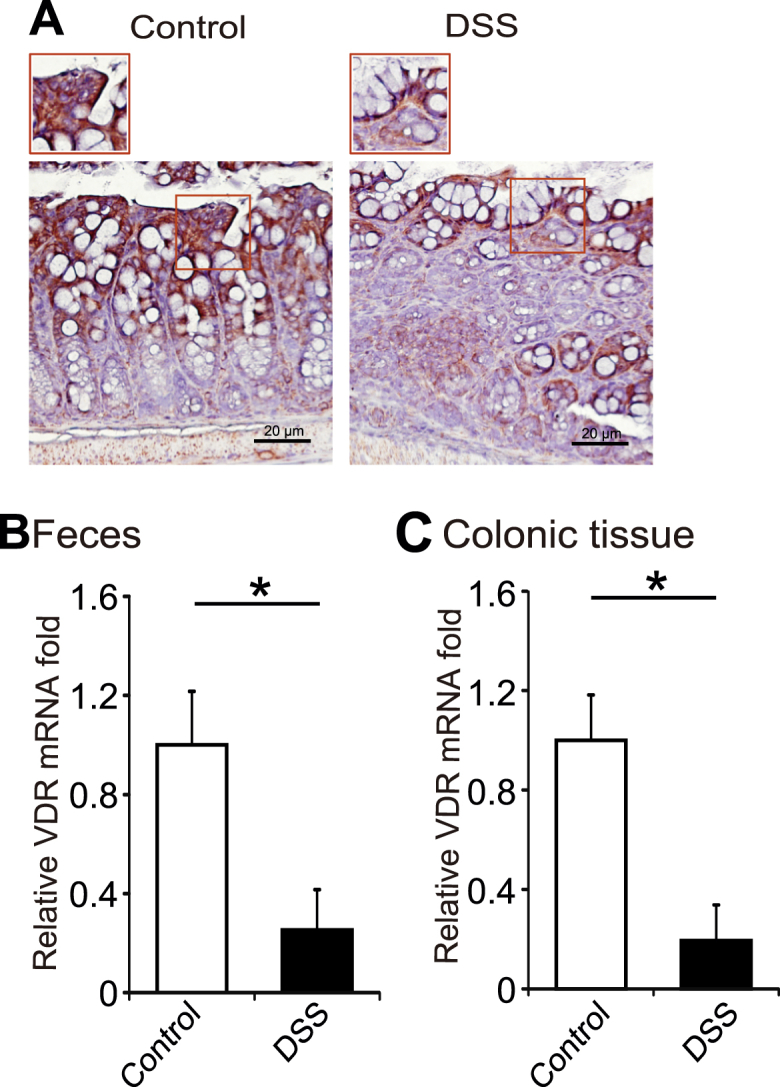
Reduced VDR expression in a colitis mouse model. (A) VDR staining in DSS mice model. Images were representative of experiments in triplicate; Normal, n = 6; DSS, n = 8. (B) Comparison of RT-PCR expression profiles of Vdr gene from paired DSS-treated and control fecal samples. VDR expression was down-regulated in DSS-induced IBD mice model. Data are expressed as mean ± SD. n = 6, Student's t-test, ∗P < 0.05. (C) Comparison of RT-PCR expression profiles of Vdr gene from paired DSS-treated and control intestinal samples. VDR expression was reduced in colon from DSS-treated mice. Data are expressed as mean ± SD. n = 6, Student's t-test, ∗P < 0.05.
Using qPCR method to test the level of VDR in a Salmonella infection model
Chronic Salmonella infection led to the reduction of VDR in colon.16,17 Here, we showed less VDR expression in the Salmonella treated colon 4 days post infection (Fig. 3A). Comparing qPCR expression profiles of Vdr gene from paired Salmonella-treated and control feces, we found that Vdr expression was significantly down-regulated in feces from Salmonella infected mice (Fig. 3B) (n = 6, Student's t-test, P < 0.05). VDR mRNA level was also down-regulated in intestinal tissue samples from Salmonella infected mice (Fig. 3C) (Data are expressed as mean ± SD. n = 6, Student's t-test, P < 0.05). Again, these observations in colon tissue were consistent with the data using the fecal samples.
Figure 3.
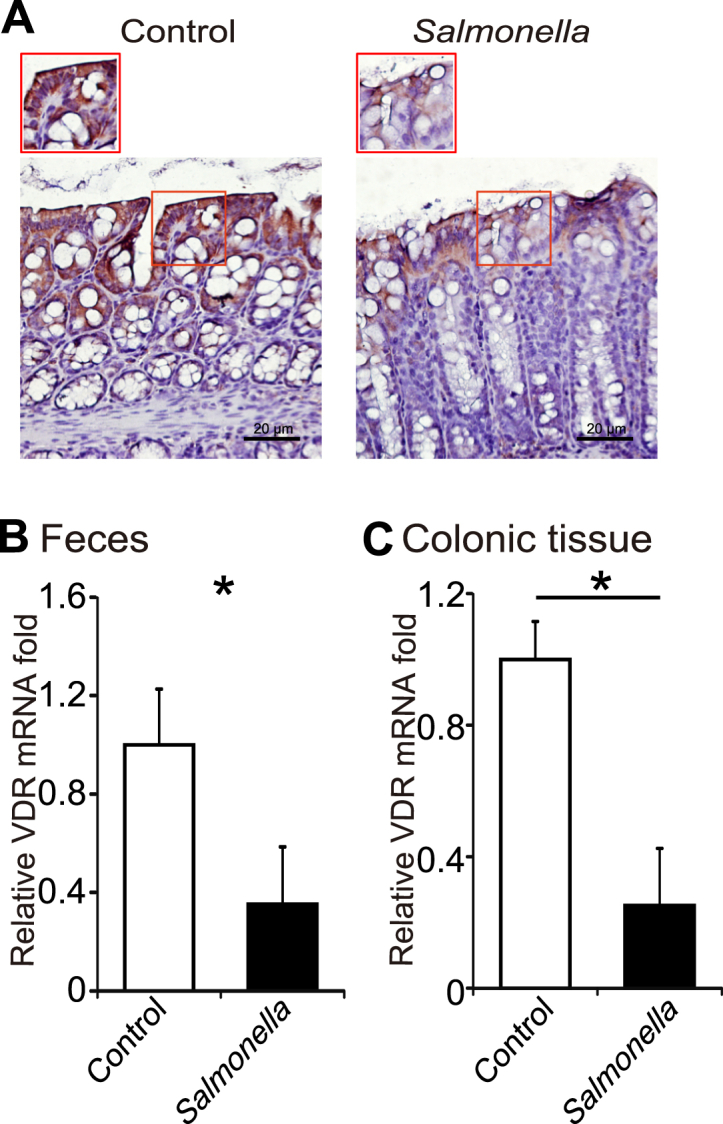
Using the qPCR method to test VDR in a Salmonella-colitis model. (A) VDR staining in colon of Salmonella-treatment mice. Images were representative of experiments in triplicate; Control, n = 6; Salmonella, n = 8. (B) Comparison of RT-PCR expression profiles of VDR gene from paired Salmonella-treated and control fecal samples. VDR expression was down-regulated in feces from chronic Salmonella infected mice (4 days post infection). Data are expressed as mean ± SD. n = 6, Student's t-test, ∗P < 0.05. (C) Comparison of qPCR expression profiles of Vdr gene from paired Salmonella-treated and control intestinal samples. VDR expression was down-regulated in Salmonella-treated mice intestinal samples. Data are expressed as mean ± SD. n = 6, Student's t-test, ∗P < 0.05.
VDR level in the young and aging mice
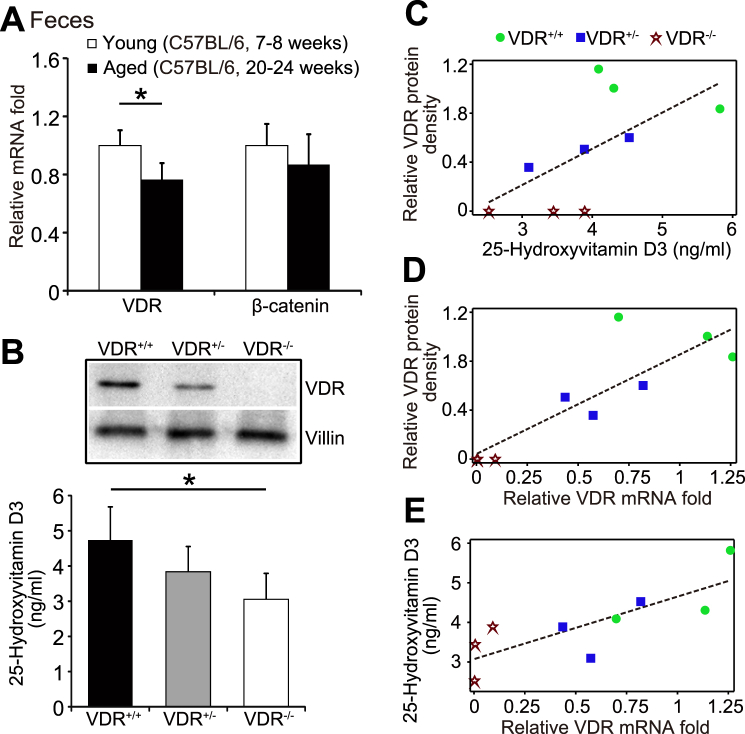
VDR expression was significantly reduced over aging process.22,23 We further examined the expression levels of VDR, using fecal samples from the young (C57BL/6, 7–8 weeks) and aged (C57BL/6, 20–24 weeks) mice. Compared with the younger mice, feces of aged mice significantly reduced the expression level of VDR mRNA. However, we did not find distinct changes of β-catenin expression between the young and aging mice (Fig. 4A) (Data are expressed as mean ± SD. n = 5, Student's t-test, P < 0.05).
Figure 4.
VDR in feces from young and aging mice and correlation analysis among serum 25-hydroxyvitamin D3, colonic VDR protein, and fecal VDR mRNA. (A) Comparison of VDR mRNA levels in fecal samples between young (C57BL/6, 7–8 week old) and aging (C57BL/6, 20–24 week old) mice. Data are expressed as mean ± SD. n = 5, Student's t-test, ∗P < 0.05. (B) Expression of colonic VDR protein and serum 25-hydroxyvitamin D3 in the VDR+/+, VDR+/−, and VDR−/− mice. Data are expressed as mean ± SD. n = 3–4, Student's t-test, ∗P < 0.05. (C) The correlation analysis of serum 25-hydroxyvitamin D3 with the level of intestinal VDR protein. The correlation analysis and scatter plot were performed between fold changes of VDR protein and vitamin D3. P = 0.0734, total n = 9 in VDR protein (n = 3 for VDR+/+, VDR+/−, and VDR−/−, respectively); total n = 10 in vitamin D3 (n = 3 for VDR+/+, VDR+/−, respectively and n = 4 for VDR−/−). (D) The correlation analysis of intestinal VDR protein and fecal VDR. The correlation analysis and scatter plot were performed. P = 0.0032, total n = 9 in VDR protein and total n = 9 for fecal VDR (n = 3 for VDR+/+, VDR+/−, and VDR−/−, respectively). (E) The correlation analysis of serum 25-hydroxyvitamin D3 and fecal VDR mRNA. The correlation analysis and scatter plot were performed. P = 0.0110, total n = 10 in vitamin D3 (n = 3 for VDR+/+, VDR+/−, respectively and n = 4 for VDR−/−) and total n = 9 for fecal VDR (n = 3 for VDR+/+, VDR+/−, and VDR−/−, respectively).
Positive correlation differentiated by the VDR status and serum 25-hydroxyvitamin D3
Expression of colonic VDR protein was dramatically reduced in the VDR+/− and VDR−/− mice. Compared with wild-type VDR+/+ mice, our Western blot data showed the reduced VDR protein in colon of VDR+/− mice and no-VDR protein in the whole-body VDR−/− mice (Fig. 4B Western blots of VDR protein). In the meantime, 25-hydroxyvitamin D3 levels in serum of VDR+/− mice and VDR−/− mice were significantly reduced, respectively, compared to the VDR+/+ mice (Fig. 4B ELISA data of 25-hydroxyvitamin D3). Thus, VDR expression in colon correlates with the serum level of 25-hydroxyvitamin D3 in mice.
To quantify and visualize the correlations of serum 25-hydroxyvitamin D3 level and intestinal VDR protein, we performed correlation analysis and scatter plot between fold changes of VDR protein and serum 25-hydroxyvitamin D3 (Fig. 4C). The results showed that serum 25-hydroxyvitamin D3 and intestinal VDR were positively associated with Pearson correlation coefficient (P-values) (0.86, P = 0.0734). In Fig. 4D, correlation analysis also showed a positive correlation of intestinal VDR and fecal VDR (0.79, P = 0.0032). Serum 25-hydroxyvitamin D3 and fecal VDR were also positively correlated (0.62, P = 0.0110), as shown in Fig. 4E.
In summary, we have established a simple qPCR method to detect reduction of VDR using fecal samples from different experimental models. The qPCR data from feces are consistent with the pathological IHC data and PCR data of VDR in colon. A positive correlation was differentiated by VDR status among serum 25-hydroxyvitamin D3, intestinal VDR protein, and fecal VDR mRNA.
We recognize the limit of current study. In the future study, we plan to examine the samples from patients with vitamin D deficiency and chronic diseases to further validate our method.
Conclusion
Our data indicate that the qPCR method to test VDR using fecal samples could detect the reduction of VDR at the mRNA level. We highlight the feasibility, sensitivity, and simplicity of a molecular method to determine the status of VDR as a biomarker in various diseases, such as colitis, bacterial infection, and aging.
Ethics approval and consent to participate
All animal studies were performed following ACC guidelines at the University of Illinois at Chicago (UIC), IL, USA.
Consent for publication
Not applicable.
Availability of data and material
Data and material will be available by request.
Authors contribution
Yong-guo Zhang performed qPCR, IHC, and detailed analysis, prepared for figures and draft, and analyzed the data. Yinglin Xia directed the project for the statistical analysis of data. Jun Sun obtained funds, designed the study, and directed the project. All authors contributed to the writing of the manuscript.
Conflict of Interests
The authors declare no conflict of interest.
Funding
We would like to acknowledge the support from UIC Cancer Center, the NIDDK grant R01DK105118 and R01DK114126 to Jun Sun.
Acknowledgements
We would like to thank Jason Xia for helping with proofreading.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Sun J. Dietary vitamin D, vitamin D receptor, and microbiome. Curr Opin Clin Nutr Metab Care. 2018;21(6):471–474. doi: 10.1097/MCO.0000000000000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shang M., Sun J. Vitamin D/VDR, probiotics, and gastrointestinal diseases. Curr Med Chem. 2017;24(9):876–887. doi: 10.2174/0929867323666161202150008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark A., Mach N. Role of vitamin D in the hygiene hypothesis: the interplay between vitamin D, vitamin D receptors, gut microbiota, and immune response. Front Immunol. 2016;7:e627. doi: 10.3389/fimmu.2016.00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen O.H., Hansen T.I., Gubatan J.M., Jensen K.B., Rejnmark L. Managing vitamin D deficiency in inflammatory bowel disease. Frontline Gastroenterol. 2019;10(4):394–400. doi: 10.1136/flgastro-2018-101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu S., Zhang Y.G., Lu R. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2015;64(7):1082–1094. doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakke D., Sun J. Ancient nuclear receptor VDR with new functions: microbiome and inflammation. Inflamm Bowel Dis. 2018;24(6):1149–1154. doi: 10.1093/ibd/izy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakke D., Chatterjee I., Agrawal A., Dai Y., Sun J. Regulation of microbiota by vitamin D receptor: a nuclear weapon in metabolic diseases. Nucl Receptor Res. 2018;5:e101377. doi: 10.11131/2018/101377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulten H.J., Bangash M., Karim S. Comprehensive molecular biomarker identification in breast cancer brain metastases. J Transl Med. 2017;15(1):e269. doi: 10.1186/s12967-017-1370-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meeker S., Seamons A., Maggio-Price L., Paik J. Protective links between vitamin D, inflammatory bowel disease and colon cancer. World J Gastroenterol. 2016;22(3):933–948. doi: 10.3748/wjg.v22.i3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon S.M., Shin E.A. Exploring vitamin D metabolism and function in cancer. Exp Mol Med. 2018;50(4):1–14. doi: 10.1038/s12276-018-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J. The role of vitamin D and vitamin D receptors in colon cancer. Clin Transl Gastroenterol. 2017;8(6):e103. doi: 10.1038/ctg.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y.G., Wu S., Lu R. Tight junction CLDN2 gene is a direct target of the vitamin D receptor. Sci Rep. 2015;5:e10642. doi: 10.1038/srep10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Cromphaut S.J., Dewerchin M., Hoenderop J.G. Duodenal calcium absorption in vitamin D receptor-knockout mice: functional and molecular aspects. Proc Natl Acad Sci U S A. 2001;98(23):13324–13329. doi: 10.1073/pnas.231474698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z., Zhang Y.G., Xia Y., Xu X., Jiao X., Sun J. Salmonella enteritidis effector AvrA stabilizes intestinal tight junctions via the JNK pathway. J Biol Chem. 2016;291(52):26837–26849. doi: 10.1074/jbc.M116.757393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R., Bosland M., Xia Y.L., Zhang Y.G., Kato I., Sun J. Presence of Salmonella AvrA in colorectal tumor and its precursor lesions in mouse intestine and human specimens. Oncotarget. 2017;8(33):55104–55115. doi: 10.18632/oncotarget.19052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S., Yoon S., Zhang Y.G. Vitamin D receptor pathway is required for probiotic protection in colitis. Am J Physiol Gastrointest Liver Physiol. 2015;309(5):G341–G349. doi: 10.1152/ajpgi.00105.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y.G., Lu R., Xia Y.L. Lack of vitamin D receptor leads to hyperfunction of claudin-2 in intestinal inflammatory responses. Inflamm Bowel Dis. 2019;25(1):97–110. doi: 10.1093/ibd/izy292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S., Lu R., Zhang Y.G., Sun J. Chronic Salmonella infected mouse model. J Vis Exp. 2010;39:e1947. doi: 10.3791/1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S., Liao A.P., Xia Y. Vitamin D receptor negatively regulates bacterial-stimulated NF-kappaB activity in intestine. Am J Pathol. 2010;177(2):686–697. doi: 10.2353/ajpath.2010.090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y.G., Zhu X., Lu R. Intestinal epithelial HMGB1 inhibits bacterial infection via STAT3 regulation of autophagy. Autophagy. 2019;15(11):1935–1953. doi: 10.1080/15548627.2019.1596485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W., Chen Y., Golan M.A. Intestinal epithelial vitamin D receptor signaling inhibits experimental colitis. J Clin Invest. 2013;123(9):3983–3996. doi: 10.1172/JCI65842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong T., Liu Z.H., Zhang H. Age-dependent expression of the vitamin D receptor and the protective effect of vitamin D receptor activation on H2O2-induced apoptosis in rat intervertebral disc cells. J Steroid Biochem Mol Biol. 2019;190:126–138. doi: 10.1016/j.jsbmb.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Campolina-Silva G.H., Maria B.T., Mahecha G.A.B., Oliveira C.A. Reduced vitamin D receptor (VDR) expression and plasma vitamin D levels are associated with aging-related prostate lesions. Prostate. 2018;78(7):532–546. doi: 10.1002/pros.23498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and material will be available by request.