Abstract
Eukaryotic genomes contain domesticated genes from integrating viruses and mobile genetic elements. Among these are homologs of the capsid protein (known as Gag) of long terminal repeat (LTR) retrotransposons and retroviruses. We identify several mammalian Gag homologs that form virus-like particles (VLPs) and one LTR retrotransposon homolog, PEG10, that preferentially binds and facilitates vesicular secretion of its own mRNA. We show the mRNA cargo of PEG10 can be reprogrammed by flanking genes of interest with Peg10’s untranslated regions. Taking advantage of this reprogrammability, we develop Selective Endogenous eNcapsidation for cellular Delivery (SEND) by engineering both mouse and human PEG10 to package, secrete, and deliver specific RNAs. Together, these results demonstrate SEND as a modular platform suited for development as an efficient therapeutic delivery modality.
One-Sentence Summary:
The LTR retrotransposon-derived Gag protein PEG10 can be harnessed to facilitate efficient and specific intercellular delivery of cargo mRNAs in mammalian cells.
More than 8% of the human genome is composed of sequences derived from LTR retroelements, including retroviruses, that have integrated into mammalian genomes throughout evolution (1–5). Retroviruses and retrotransposons have many common mechanistic features including the core structural gene (known as gag), however, while retrotransposons replicate intracellularly, the acquisition of the envelope (env) gene by retroviruses has enabled intercellular replication (6). Most endogenous retroelements have lost their original functions, but some of their genes have been recruited for diverse roles in normal mammalian physiology. For example, the fusogenic syncytins evolved from retroviral env proteins (7). The gag homolog, Arc, which forms capsids and has been reported to transfer its own mRNA (8–10), is involved in memory consolidation and regulates inflammation in the skin (11, 12). Another gag homolog, the LTR retrotransposon-derived protein PEG10, which has been reported to bind RNA and also forms capsids (13), is involved in mammalian placenta formation (14, 15). These examples raise the possibility that additional retroelement-derived proteins encoded in the mammalian genome transfer specific nucleic acids, providing a potentially programmable mechanism for intercellular communication.
Computational survey of mammalian capsid-forming gag homologs
To identify genes with the potential to transfer specific nucleic acids, we focused on homologs of gag containing the core capsid (CA) domain, which protects the genome of both retrotransposons and exogenous retroviruses (16, 17). Previous genome analyses identified many endogenous gag homologs in mammalian genomes (18), and experimental efforts have validated the ability of some of these proteins including MmArc and MmPEG10 to form capsid-like particles that are secreted within extracellular vesicles (EVs) (10, 13). To ensure a complete list of candidates, we searched the human and mouse genomes for gag homologs. This search identified 48 gag-derived genes in the human genome and 102 gag homologs in the mouse genome; for 19 human genes, an orthologous relationship between human and mouse was readily traced (in several cases, with additional mouse paralogs), whereas the remaining ones appeared to be species-specific (Table S1 and S2).
Canonical genomes of both LTR retrotransposons and retroviruses encode a long polyprotein consisting of several conserved domains: The matrix (MA), CA, and nucleocapsid (NC) form the gag subdomain and are responsible for membrane attachment, capsid formation, and genome binding, respectively. The pol subdomain contains the protease (PRO), responsible for cleaving the polyprotein, the reverse transcriptase (RT), which converts retroelement RNA into DNA, and the integrase (IN) domain, which integrates the genome into that of the host. Some families of endogenous gag-containing proteins, such as the PNMA family, contain only the CA and NC subdomains of gag, whereas others, such as RTL1 and PEG10 (also known as RTL2 or Mart2), additionally include subdomains of pol, namely a PRO domain and a predicted RT-like domain (Fig. 1A, Table S1). Phylogenetic analysis of Peg10 and its homologs supports the origin of this gene from LTR retrotransposons (fig. S1, A and B).
Fig. 1. Identification of mammalian retroelement derived Gag homologs that form capsids and are secreted.
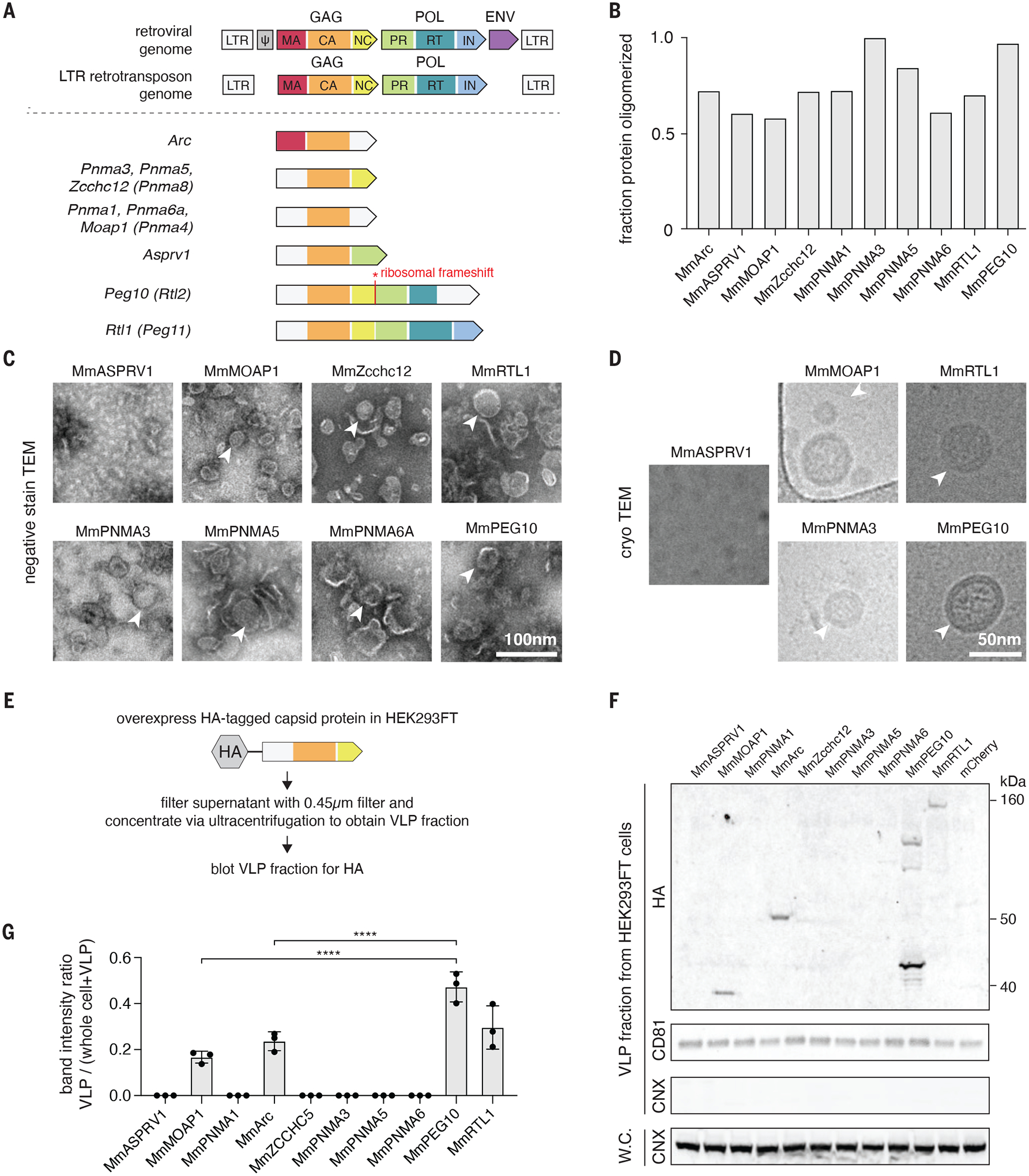
A. Domain architectures of selected Capsid (CA)-containing mammalian Gag homologs compared to that of typical retrovirus and LTR retrotransposons. Each group of Gag homologs contains a distinct combination of predicted CA, Nucleocapsid (NC), Protease (PR), and Reverse Transcriptase (RT) domains. LTR, long terminal repeat; MA, matrix; IN, integrase.
B. Fraction of the total bacterially-produced protein that forms oligomers (>600 kD), as determined by size exclusion chromatography.
C. Representative negative stain transmission electron micrographs (TEM) of the Mus musculus (Mm) orthologues of the CA-domain containing proteins. Scale bar, 100 nm.
D. Representative electron micrographs using cryogenic electron microscopy (cryoTEM) of a selected subset of the identified CA-domain containing proteins. Scale bar, 50 nm.
E. Method for detecting extracellular forms of CA-domain containing homologs.
F. Representative blots of CA-domain containing proteins in the cell-free fraction. CD81 was used as loading control for the ultracentrifuged cell-free fraction. Whole cell (W.C.) and VLP fraction blots for the endoplasmic reticulum marker CALNEXIN (CNX) ensure equal loading of whole cell protein and the purity of cell-free VLP fraction.
G. Quantification of extracellular CA-domain containing proteins (as in F) based on n=3 replicates.
Among these genes, Arc is the most well studied. Drosophila Arc1 (darc1) is a gag homolog which contains the MA, CA, and NC domains. It has been shown to form capsids, bind its own mRNA, and transfer it from motor neurons to muscles at the neuromuscular junction (9). darc1 mRNA binding is dependent on its own 3’ untranslated region (UTR), and fusion of this sequence to heterologous mRNAs can initiate their export and transfer as well. Mus musculus Arc (MmArc), by contrast, contains only the CA domain and has also been shown to form capsids and transfers its own mRNA across synapses (10), but thus far this transfer has not been shown to be specific to Arc mRNA, likely due to the lack of a NC domain.
To narrow down the scope of our analysis, we focused on CA-domain containing proteins that are conserved between human and mouse and have detectable levels of mRNA in adult human tissues, reasoning that such proteins were most likely to have been co-opted for important physiological roles in mammals (fig. S2). We produced mouse versions of the selected CA-containing proteins in E. coli and found that a number of these formed higher molecular weight oligomers identified by size exclusion (Fig. 1B, and fig. S3A), as previously noted for some of these proteins, such as MmArc (10). Electron microscopy of these aggregated proteins showed that MmMOAP1, MmZCCHC12, MmRTL1, MmPNMA3, MmPNMA5, MmPNMA6a, and MmPEG10 self-assemble into capsid-like particles, many of which appear spherical (Fig. 1, C and D, and fig. S3, B and C).
MmPEG10 binds and secrets its own mRNA
To determine if these proteins are secreted within an EV, we overexpressed an epitope tagged mouse ortholog of each CA-containing gene in HEK293FT cells and harvested both the whole cell lysate and the virus-like particle (VLP) fraction by clarification and ultracentrifugation of the culture media (Fig. 1E). We found that MmMOAP1, MmArc, MmPEG10, and MmRTL1 were all present in the VLP fraction (Fig. 1F, and fig. S4A), but MmPEG10 was the most abundant protein in the VLP fraction (Fig. 1G). Additionally, endogenous MmPEG10, but not MmMOAP1 or MmRTL1, was readily detectable in cell-free adult mouse serum (fig. S4B).
We next tested whether any of the capsid-like particles formed by Gag homologs contained specific mRNAs using RNA sequencing. To avoid the possibility of transfected Gag homolog expression plasmids contributing to high background signal during sequencing, we used CRISPR activation (19) to induce expression of endogenous genes in mouse N2a cells (Fig. 2A and S5A). We performed mRNA sequencing on whole cell lysate and the VLP fraction (following nuclease treatment to remove any residual, unencapsidated RNA) to identify RNA species in the VLP fraction. We found MmPeg10 transcriptional activation led to accumulation of significant amounts of full length MmPeg10 mRNA transcripts in the VLP fraction (Fig. 2, B and C). Previous work on MmPEG10 demonstrated that it binds a number of mRNAs inside of trophoblast stem cells including itself (13), however, here we further show that MmPEG10 binds and secretes its own mRNA into the VLP fraction. An important caveat of this experiment is that some of these proteins, particularly MmArc, are subject to regulation at the level of translation, so lack of enrichment in the VLP fraction could be due to low protein expression (20).
Fig 2. MmPEG10 protein and mRNA is secreted in vesicles by cells in vitro.
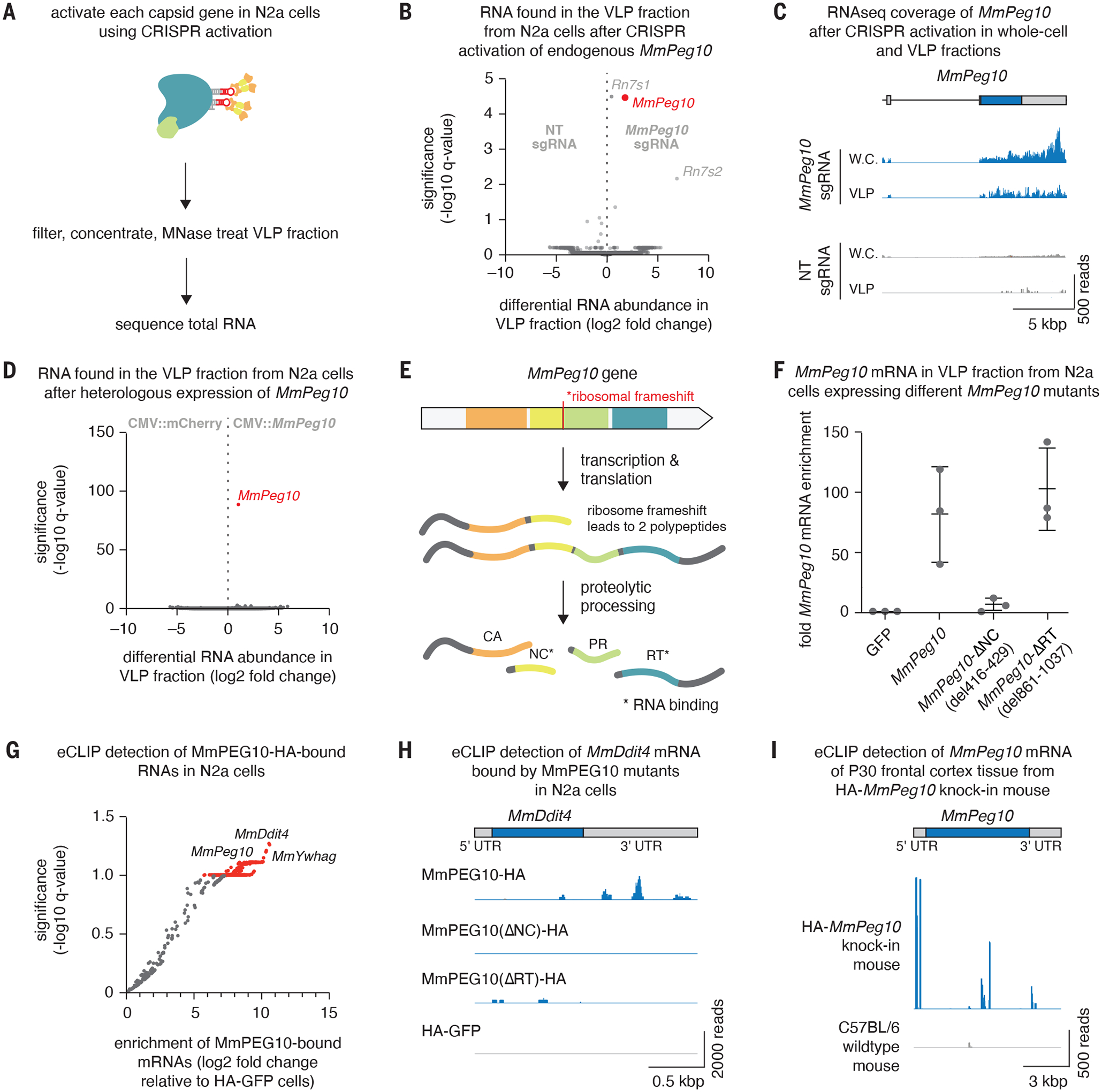
A. Method for identifying nucleic acids that are secreted in the VLP fraction upon gene activation of CA-domain containing proteins.
B. Differential RNA abundance and significance in the VLP fraction from N2a cells after CRISPR activation of endogenous MmPeg10.
C. Alignment of sequencing reads showing sequencing coverage of the MmPeg10 mRNA from (B).
D. Differential RNA abundance and significance in the VLP fraction from N2a cells after heterologous transfection of MmPeg10. n=3 replicates.
E. The four domains of MmPEG10 are translated into two isoforms. These are self-processed by the PEG10 protease into separate domains, of which the NC and RT bind RNA.
F. Fold enrichment of MmPeg10 mRNA compared to GFP in the VLP fraction from N2a cells transfected with wildtype MmPeg10 or deletions of the predicted nucleocapsid (ΔNC) and reverse transcriptase (ΔRT) domains.
G. Log2 fold change and significance of bound RNAs from eCLIP data comparing HA-GFP to WT MmPEG10-HA.
H. Representative sequencing alignment histogram of the MmDdit4 locus generated from eCLIP of N2a cells transfected with wildtype or mutant MmPeg10.
I. Representative sequencing alignment histogram of the MmPeg10 locus generated from eCLIP data of n = 3 HA-PEG10 and n = 3 untagged animals.
To confirm our observation for MmPeg10, we transiently transfected overexpression plasmids of UTR-flanked MmPeg10 into N2a cells and found only enrichment for MmPeg10 mRNA in the VLP fraction (Fig. 2D) under this overexpression condition. PEG10 contains two putative nucleic acid binding domains, namely the NC and RT, which are released from the polypeptide upon PEG10 self processing (21) (Fig. 2E, Supplementary Text 1, fig. S5, B–D). We generated deletions of these domains and found that mRNA export depends on the MmPEG10 NC, as loss of the nucleic acid-binding zinc finger CCHC motif (residues 416–429) from the MmPEG10 NC substantially reduced export of its mRNA (Fig. 2F).
To better understand the roles of the nucleic acid binding domains of MmPEG10 in RNA binding, we performed eCLIP in N2a cells after transient transfection with HA-tagged MmPeg10 as well as the NC and RT mutants (fig. S6, A and B). Compared to the control, MmPEG10 strongly bound a number of mRNAs in N2a cells including its own mRNA (Fig. 2G). Importantly, both the NC and the RT domains are required for the binding of these mRNAs by MmPEG10 (Fig. 2H and fig. S6C). To confirm MmPEG10’s cellular role in an in vivo context, we generated knock-in mice carrying an N-terminal HA-tag on the endogenous MmPEG10 protein (fig. S6D). Expression of MmPeg10 in cortical neurons has been demonstrated previously (fig. S6E) (22). Endogenous MmPEG10 was also found to bind its own mRNA as well as other transcripts abundant in neurons (fig. S6, F and G); in contrast to previous datasets, we detected strong MmPEG10 binding in the 5’ UTR as well as some additional binding near the boundary between the nucleocapsid and protease coding sequences and in the beginning of the 3’ UTR (Fig. 2I) (13).
MmPEG10’s binding of mRNA has been reported to increase the cellular abundance of target transcripts (13). To confirm this role of MmPEG10 in its native context in vivo, we perturbed MmPeg10 gene expression in the postnatal mouse brain and assessed expression changes of MmPEG10 bound transcripts (Supplementary Text 2). We found that the mRNAs of 49 genes downregulated in the brain upon MmPeg10 knockout are bound to MmPEG10 in the age-matched mouse brain (fig. S7F), suggesting that one of the functions of MmPEG10 is to bind and stabilize mRNAs with fundamental roles in neurodevelopment.
Pseudotyped PEG10 VLPs can deliver engineered cargo mRNAs bearing RNA packaging signals from PEG10 UTRs
To reprogram MmPEG10 to bind and package heterologous RNA, we tested whether a cargo mRNA consisting of both the 5’ and 3’ UTR of MmPeg10 flanking a gene of interest would be efficiently packaged, exported, delivered, and translated in recipient cells (Fig. 3A). This UTR grafting approach has been demonstrated for the Ty3 retroelement and darc1 (9, 23). We first used a Cre-loxP system, a highly sensitive system for tracking RNA exchange that has been used previously with exosomes in vivo (24). We flanked the Cre recombinase coding sequence with the MmPeg10 UTRs and co-transfected it with MmPeg10 with and without a fusogen, the vesicular stomatitis virus envelope protein (VSVg) (Fig. 3A). We found that MmPEG10 VLPs pseudotyped with VSVg are secreted within EVs that mediate transfer of Cre mRNA, not protein, into target loxP-GFP reporter N2a cells in a VSVg- and MmPeg10 UTR-dependent manner (Fig. 3B, C, D, and S8, Supplementary Text 3). This result suggests that addition of the Peg10 UTRs enables functional intercellular transfer of an mRNA via VLPs and that these VLPs require a fusogenic protein for cell entry.
Fig 3. Flanking mRNA with MmPeg10 5’ and 3’ UTRs enables functional intercellular transfer of mRNA into a target cell.
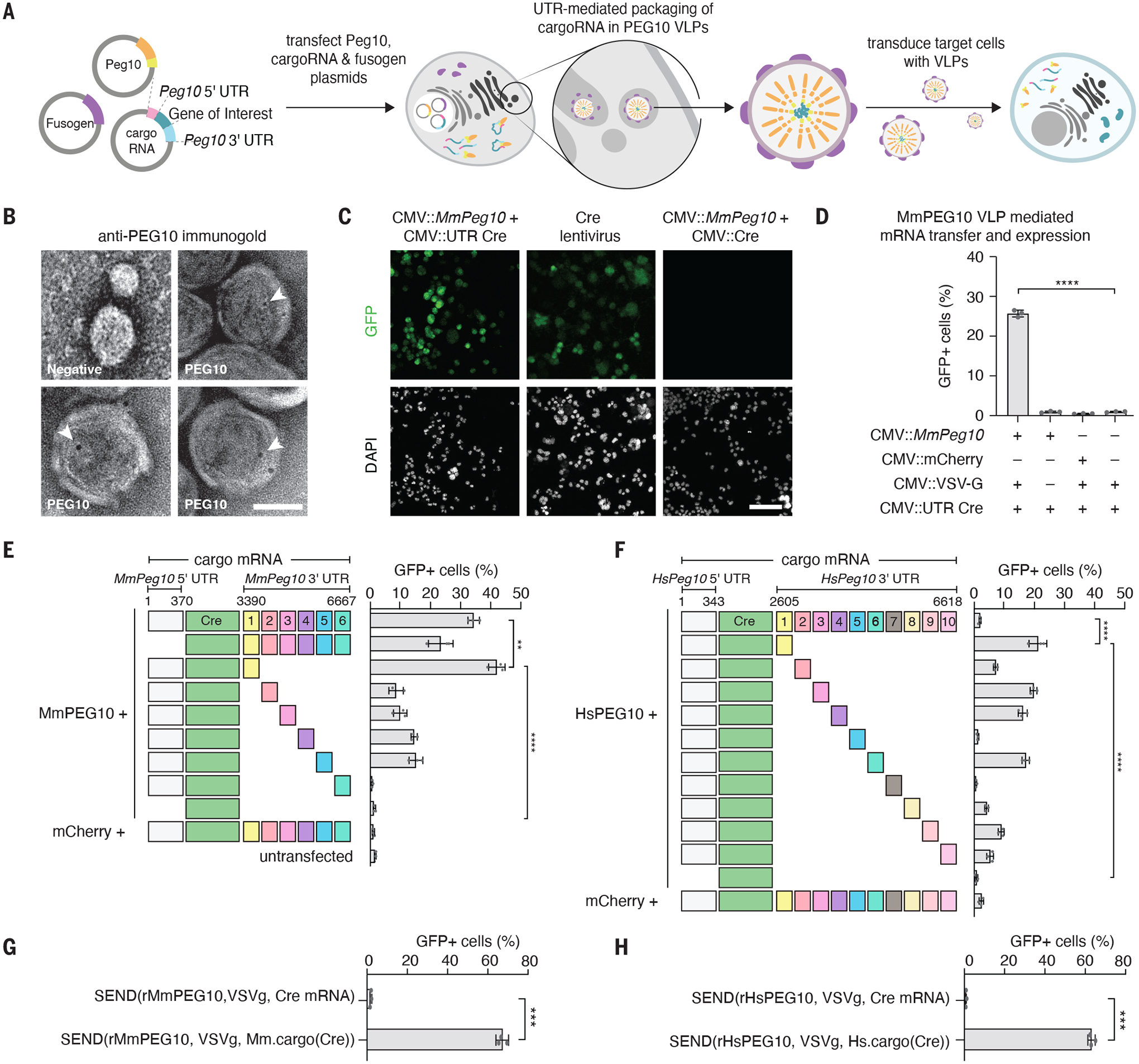
A. Schematic showing reprogramming MmPEG10 for functional delivery of a cargo RNA flanked with the MmPeg10 5′ and 3′ UTRs (hereafter, “cargo(RNA)”)
B epresentative TEM micrographs of VLP fraction immunogold labeled for MmPEG10. Text labels indicate transfection of cells with MmPeg10 or mock (negative). Arrowheads indicate gold labeling. Scale bar, 50 nm.
C. Representative images of loxP-GFP N2a cells treated with VSVg pseudotyped MmPEG10 VLPs produced by transfecting Mm.cargo(Cre) or Cre mRNA, and a lentivirus encoding Cre. Scale bar, 100 um.
D. Functional transfer of RNA into loxP-GFP N2a cells mediated by VSVg pseudotyped MmPEG10 VLPs. Data quantified by flow cytometry 72 hours after VLP addition, n = 3 replicates.
E. Functional transfer of RNA into loxP-GFP N2a cells mediated by VSVg pseudotyped VLPs produced with MmPeg10 or mCherry and Mm.cargo(Cre) constructs encoding tiles of the MmPeg10 3’UTR. Data quantified by flow cytometry 72 hours after VLP addition, n = 3 replicates.
F. Functional transfer of RNA into loxP-GFP N2a cells mediated by VSVg pseudotyped VLPs produced with HsPEG1010 or mCherry and Hs.cargo(Cre) constructs encoding tiles of the HsPeg10 3’UTR. Data quantified by flow cytometry 72 hours after VLP addition, n = 3 replicates.
G. Functional transfer of RNA into loxP-GFP N2a cells mediated by VSVg pseudotyped VLPs produced with rMmPeg10 and Mm.cargo(Cre) or Cre mRNA. Data quantified by flow cytometry 72 hours after VLP addition, n = 3 replicates.
H. Functional transfer of RNA into loxP-GFP N2a cells mediated by VSVg pseudotyped VLPs produced with rHsPeg10 and Hs.cargo(Cre) or Cre mRNA. Data quantified by flow cytometry 72 hours after VLP addition, n = 3 replicates. For all panels *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, One-Way ANOVA.
We next asked if there is a minimal UTR packaging signal for mediating efficient packaging and functional transfer. The 3’ UTR of MmPeg10 is approximately 4-kB long, but eCLIP indicates only portions of the 3’ UTR are bound by MmPEG10 (Fig. 2I). We created constructs encoding the MmPeg10 5’ UTR, Cre, and 500-bp segments of the MmPeg10 3’ UTR. We found that the proximal 500 bp of the MmPeg10 3’ UTR are sufficient for efficient functional transfer of Cre mRNA into target reporter cells (Fig. 3E). Importantly, no efficient functional mRNA transfer was observed for non-UTR flanked Cre or for Cre without the proximal 500 bp of the 3’ UTR. Moving forward, we refer to RNA cargo flanked by the MmPeg10 5’ UTR and the proximal 500 bp of the 3’ UTR as Mm.cargo(RNA), where “(RNA)” specifies the cargo being flanked (e.g., Mm.cargo(Cre)).
Like the mouse ortholog, HsPEG10 is an abundantly secreted protein in the VLP fraction (fig. S10A). Using the same approach we employed with MmPeg10, we identified that the 5’ UTR and the first 500 bp of the HsPEG10 3’ UTR are sufficient to mediate functional transfer of Cre mRNA, hereafter denoted as Hs.cargo(RNA) (Fig. 3F). Interestingly, these functional regions of the UTRs are highly conserved across mammals (fig. S10B). Similar to its mouse ortholog, the human system is specific, requiring HsPEG10 UTR sequences for functional mRNA transfer whereas non-flanked Cre produced only minimal reporter cell activity (Fig. 3F).
To further boost the packaging of a cargo(RNA) by PEG10, we explored the impact of removing any additional PEG10 cis binding elements within the MmPeg10/HsPEG10 coding sequence. For both human and mouse, transfer was increased as a result of recoding the sequence between the NC and the PRO domains, which corresponds to the MmPEG10-bound region in the eCLIP experiments (Fig. 2I, Supplementary Text 4).
Combining these optimizations, we produced VLPs with the recoded mouse and human PEG10 (rMmPEG10 or rHsPEG10), VSVg, and the optimized cargo(RNA) containing the first 500 bp of the 3’ UTR; we refer to this system as Selective Endogenous eNcapsidation for cellular Delivery (SEND). With SEND we detected a substantial (up to 60%) increase in functional transfer of cargo(Cre) into N2a cells for both human and mouse PEG10 (Fig. 3, G and H). Furthermore, we show that VLPs produced with rMmPEG10 can mediate the functional transfer of H2B-mCherry (fig. S12, A and B). Comparison of SEND to previously developed delivery vectors showed that SEND is 4–5 fold less potent than an integrating lentiviral vector as assayed by digital droplet PCR and functional titration (fig. S12, B, C, D, and E). However, given that SEND delivers mRNA rather than integrating an overexpression cassette, we expect it to perform competitively against other mRNA delivery vehicles.
PEG10 is a modular platform for RNA delivery
To generate a fully endogenous SEND system, we tested whether VSVg can be replaced with an endogenous fusogenic transmembrane protein. Given the overlapping tissue expression of MmPeg10/HsPEG10 and syncytin genes (Supplementary Text 5), we tested the feasibility of pseudotyping the mouse SEND system with MmSYNA, or MmSYNB compared to pseudotyping with VSVg. Pseudotyped particles were incubated with tail-tip fibroblasts from loxP-tdTomato reporter mice, a cell type which we have found amenable to transduction by these fusogens. Based on previous reports, we added the transduction enhancer vectofusin-1 to the supernatant for MmSYNA and MmSYNB particles to enhance in vitro transduction (25). In these primary cells, both VSVg and MmSYNA enabled SEND-mediated functional transfer of Mm.cargo(Cre) while MmSYNB did not (Fig. 4A and B). Again, this packaging was highly specific, as only UTR flanked mRNA (i.e., Mm.cargo(Cre)) was functionally transferred. Together with MmSYNA, SEND can be configured as a fully endogenous system for functional gene transfer.
Fig. 4. SEND is a modular system capable of delivering gene editing tools into human and mouse cells.
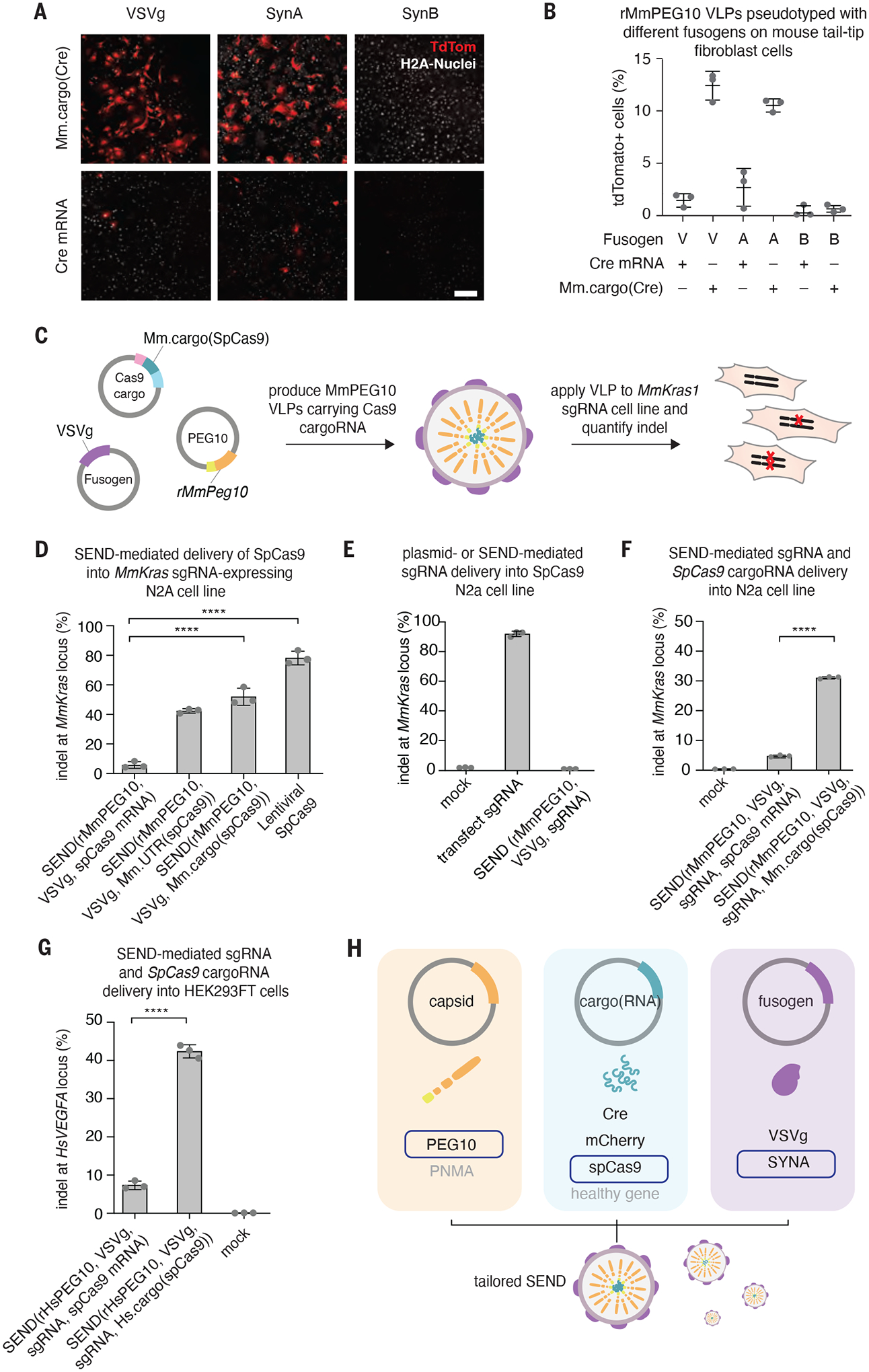
A. Representative images demonstrating functional transfer of Mm.cargo(Cre) or Cre mRNA in rMmPEG10 VLPs pseudotyped with VSVg (V), MmSYNA (A), or MmSYNB (B) in Ai9 (loxP-tdTomato) tail tip fibroblasts. Scale bar, 200 μm.
B ercent of tdTomato positive cells out of the total number of H2A stained nuclei from high content imaging of n=3 replicates of (A).
C. Schematic representing the retooling of SEND for genome engineering.
D. Indels at the MmKras locus in MmKras1-sgRNA-N2a cells treated with SEND (VSVg pseudotyped rMmPEG10 VLPs) containing either SpCas9 mRNA, Mm.UTR(SpCas9), or Mm.cargo(SpCas9) and a lentivirus encoding SpCas9. Indels quantified by NGS 72 hours after VLP or lentivirus addition, n=3 replicates.
E. Indels at the mouse MmKras locus in a constitutively expressing SpCas9 N2a cell line either transfected with a plasmid carrying the MmKras sgRNA or treated with SEND (rMmPEG10, VSVg, MmKras sgRNA). Indels quantified by NGS after 72 hours, n=3 replicates.
F. Indels at the MmKras locus in N2a cells treated with SEND (VSVg pseudotyped rMmPeg10 SEND VLPs) containing either SpCas9 mRNA or Mm.cargo(SpCas9) and sgRNA. Indels quantified by NGS 72 hours after VLP addition, n=3 replicates.
G. Indels at the HsVEGFA locus in HEK293FT cells treated with SEND (VSVg pseudotyped rHsPEG10 VLPs) containing either SpCas9 mRNA or Hs.cargo(SpCas9) and an unmodified sgRNA. Indels determined by NGS 72 hours after VLP addition, n=3 replicates.
H. SEND is a modular delivery platform combining an endogenous Gag homolog, cargo mRNA, and fusogen, which can be tailored for specific contexts.
Supported by our understanding of the minimal requirements for PEG10-mediated mRNA delivery (i.e., UTRs and an endogenous fusogen), we could begin to probe the endogenous role of MmPEG10-mediated MmPeg10 RNA delivery in neurons. Functional transfer of MmSYNA pseudotyped VLPs carrying the native PEG10 transcript into primary mouse cortical neurons led to upregulation of a number of genes involved in neurodevelopment (Supplementary Text 6). This finding reinforces the notion that one role of endogenous MmPeg10 delivery is binding and stabilizing specific mRNA transcripts in recipient cells. RNA sequencing of N2a cells receiving Mm.cargo(Peg10) revealed substantial gene expression changes upon MmPeg10 delivery that were largely abrogated with PEG10 mediated Mm.cargo(Cre) delivery (Supplementary Text 7). This suggests transferring a reprogrammed cargo does not have the same impact on recipient cells as transferring MmPeg10 and indicates that MmPeg10 transcript delivery rather than the delivery of MmPEG10 protein is responsible for the observed gene expression changes. It remains unclear whether MmPEG10 VLPs are natively pseudotyped by the endogenous fusogen MmSYNA to enable cellular uptake of PEG10 VLPs in the central nervous system.
To further characterize the modularity of the components of this system, we tested different cargoRNAs. Using the same pipeline developed for cargo(Cre), we tested whether SEND could mediate the functional transfer of a large ~5 kb Mm.cargo(SpCas9) into N2a cell lines constitutively expressing an sgRNA against MmKras (Fig. 4C). SEND was able to functionally transfer SpCas9, leading to ~60% indels in recipient cells (Fig. 4D); similar to the results with Cre, SEND is specific and only able to efficiently functionally transfer SpCas9 flanked by either the full length or optimized Peg10 UTR sequences.
To create an all-in-one vector for delivery of sgRNA and SpCas9, we first tested if an sgRNA can be efficiently delivered by SEND. We independently packaged an sgRNA targeting Kras into rMmPeg10 VLPs by co-expressing rMmPeg10 with VSVg and a U6 driven sgRNA and incubated them with Cas9 expressing N2a cells; we saw very little activity even though direct transfection of the guide showed robust indel formation (Fig. 4E). We found, however, that co-packaging the guide alongside Mm.cargo(SpCas9) by co-expressing Mm.cargo(SpCas9) with a U6 driven sgRNA on a separate plasmid was sufficient to mediate 30% indels (Fig. 4F). To determine the reproducibility of this genome editing approach, we repeated this co-packaging strategy with the human SEND system and were able to generate ~40% indels in HEK293FT cells at the HsVEGFA locus (Fig. 4G).
The development of SEND from an endogenous retroelement complements existing delivery approaches using lipid nanoparticles (26), VLPs derived from bona fide retroviruses (27–29), and active mRNA loading approaches in EVs (30, 31). Moreover, SEND may have reduced immunogenicity compared to currently available viral vectors (32) due to its use of endogenous human proteins. Supporting this is gene expression data from the developing human thymus which demonstrates that HsPEG10 is highly expressed compared to other CA-containing genes in the thymic epithelium (33) (fig. S16), which is responsible for T cell tolerance induction. As a modular, fully endogenous system, SEND has the potential to be extended into a minimally immunogenic delivery platform that can be repeatedly dosed, greatly expanding the applications for nucleic acid therapy.
Supplementary Material
Acknowledgments:
We thank D.S. Yun for electron microscopy assistance, A. Koller for mass spectrometry assistance, Lin Wu and the Harvard GMF for the generation of transgenic animals, A. Tang for illustration assistance, and the entire Zhang laboratory for support and advice.
Funding:
Simons Center for the Social Brain at MIT (MS)
National Institutes of Health grant 1R01-HG009761 (FZ)
National Institutes of Health grant 1DP1-HL141201 (FZ)
Howard Hughes Medical Institute (FZ)
Open Philanthropy Project (FZ)
Harold G. and Leila Mathers Foundation (FZ)
Edward Mallinckrodt, Jr. Foundation (FZ)
Poitras Center for Psychiatric Disorders Research at MIT (FZ)
Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT (FZ)
Yang-Tan Center for Molecular Therapeutics at MIT (FZ)
Phillips family (FZ)
R. Metcalfe (FZ)
J. and P. Poitras (FZ)
Footnotes
Competing interests: M.S., B.L., and F.Z. are co-inventors on a U.S. provisional patent application filed by the Broad Institute related to this work. F.Z. is a cofounder of Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, and Sherlock Biosciences.
Data and materials availability:
Expression plasmids are available from Addgene under a uniform biological material transfer agreement. Additional information available via the Zhang Lab website (https://zhanglab.bio). Next generation sequencing data generated are available from NCBI SRA with accession number PRJNA743280. All other data are available in the paper and supplementary materials.
References
- 1.Goodier JL, Kazazian HH Jr, Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 135, 23–35 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Smit AF, Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev 9, 657–663 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Patel MR, Emerman M, Malik HS, Paleovirology - ghosts and gifts of viruses past. Curr. Opin. Virol 1, 304–309 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guio L, González J, New Insights on the Evolution of Genome Content: Population Dynamics of Transposable Elements in Flies and Humans. Methods Mol. Biol 1910, 505–530 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Feschotte C, Gilbert C, Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet 13, 283–296 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Kim FJ, Battini J-L, Manel N, Sitbon M, Emergence of vertebrate retroviruses and envelope capture. Virology. 318, 183–191 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T, Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U. S. A 106, 12127–12132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myrum C, Baumann A, Bustad HJ, Flydal MI, Mariaule V, Alvira S, Cuéllar J, Haavik J, Soulé J, Valpuesta JM, Márquez JA, Martinez A, Bramham CR, Arc is a flexible modular protein capable of reversible self-oligomerization. Biochem. J 468, 145–158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T, Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell. 172, 262–274.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, Yoder N, Belnap DM, Erlendsson S, Morado DR, Briggs JAG, Feschotte C, Shepherd JD, The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell. 173, 275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erica Korb SF, Arc in synaptic plasticity: from gene to behavior. Trends Neurosci. 34, 591 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barragan-Iglesias P, De La Pena JB, Lou TF, Loerch S, Intercellular Arc signaling regulates vasodilation (2020) (available at https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3684856). [DOI] [PMC free article] [PubMed]
- 13.Abed M, Verschueren E, Budayeva H, Liu P, Kirkpatrick DS, Reja R, Kummerfeld SK, Webster JD, Gierke S, Reichelt M, Anderson KR, Newman RJ, Roose-Girma M, Modrusan Z, Pektas H, Maltepe E, Newton K, Dixit VM, The Gag protein PEG10 binds to RNA and regulates trophoblast stem cell lineage specification. PLoS One. 14, e0214110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, Kohda T, Ogura A, Yokoyama M, Kaneko-Ishino T, Ishino F, Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet 38, 101–106 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Henke C, Strissel PL, Schubert M-T, Mitchell M, Stolt CC, Faschingbauer F, Beckmann MW, Strick R, Selective expression of sense and antisense transcripts of the sushi-ichi-related retrotransposon--derived family during mouse placentogenesis. Retrovirology. 12, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krupovic M, Koonin EV, Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. U. S. A 114, E2401–E2410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodonova SO, Prinz S, Bilanchone V, Sandmeyer S, Briggs JAG, Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses. Proc. Natl. Acad. Sci. U. S. A 116, 10048–10057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campillos M, Doerks T, Shah PK, Bork P, Computational characterization of multiple Gag-like human proteins. Trends Genet. 22, 585–589 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F, Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace CS, Lyford GL, Worley PF, Steward O, Differential intracellular sorting of immediate early gene mRNAs depends on signals in the mRNA sequence. J. Neurosci 18, 26–35 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golda M, Mótyán JA, Mahdi M, Tőzsér J, Functional Study of the Retrotransposon-Derived Human PEG10 Protease. Int. J. Mol. Sci 21 (2020), doi: 10.3390/ijms21072424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, Goeva A, Nemesh J, Kamitaki N, Brumbaugh S, Kulp D, McCarroll SA, Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell. 174 (2018), pp. 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clemens K, Bilanchone V, Beliakova-Bethell N, Sequence requirements for localization and packaging of Ty3 retroelement RNA. Virus Res. (2013) (available at https://www.sciencedirect.com/science/article/pii/S0168170212003784). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridder K, Sevko A, Heide J, Dams M, Rupp A-K, Macas J, Starmann J, Tjwa M, Plate KH, Sültmann H, Altevogt P, Umansky V, Momma S, Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology. 4, e1008371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coquin Y, Ferrand M, Seye A, Menu L, Galy A, Syncytins enable novel possibilities to transduce human or mouse primary B cells and to achieve well-tolerated in vivo gene transfer. Cold Spring Harbor Laboratory (2019), p. 816223. [Google Scholar]
- 26.Kowalski PS, Rudra A, Miao L, Anderson DG, Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther 27, 710–728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mock U, Riecken K, Berdien B, Qasim W, Chan E, Cathomen T, Fehse B, Novel lentiviral vectors with mutated reverse transcriptase for mRNA delivery of TALE nucleases. Sci. Rep 4, 6409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaczmarczyk SJ, Sitaraman K, Young HA, Hughes SH, Chatterjee DK, Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. U. S. A 108, 16998–17003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangeot PE, Risson V, Fusil F, Marnef A, Laurent E, Blin J, Mournetas V, Massouridès E, Sohier TJM, Corbin A, Aubé F, Teixeira M, Pinset C, Schaeffer L, Legube G, Cosset F-L, Verhoeyen E, Ohlmann T, Ricci EP, Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun 10, 45 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kojima R, Bojar D, Rizzi G, Hamri GC-E, El-Baba MD, Saxena P, Ausländer S, Tan KR, Fussenegger M, Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Park\inson’s disease treatment. Nat. Commun 9, 1305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hung ME, Leonard JN, A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J Extracell Vesicles. 5, 31027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirley JL, de Jong YP, Terhorst C, Herzog RW, Immune Responses to Viral Gene Therapy Vectors. Mol. Ther 28, 709–722 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J-E, Botting RA, Domínguez Conde C, Popescu D-M, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, Wilbrey-Clark A, Roberts K, Kedlian VR, Ferdinand JR, He X, Webb S, Maunder D, Vandamme N, Mahbubani KT, Polanski K, Mamanova L, Bolt L, Crossland D, de Rita F, Fuller A, Filby A, Reynolds G, Dixon D, Saeb-Parsy K, Lisgo S, Henderson D, Vento-Tormo R, Bayraktar OA, Barker RA, Meyer KB, Saeys Y, Bonfanti P, Behjati S, Clatworthy MR, Taghon T, Haniffa M, Teichmann SA, A cell atlas of human thymic development defines T cell repertoire formation. Science. 367 (2020), doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, Ishino F, A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 73, 232–237 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Volff J, Körting C, Schartl M, Ty3/Gypsy retrotransposon fossils in mammalian genomes: did they evolve into new cellular functions? Mol. Biol. Evol 18, 266–270 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Shigemoto K, Brennan J, Walls E, Watson CJ, Stott D, Rigby PW, Reith AD, Identification and characterisation of a developmentally regulated mammalian gene that utilises -1 programmed ribosomal frameshifting. Nucleic Acids Res. 29, 4079–4088 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park J-E, Stephenson E, Polański K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA, Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, Yamaoka Y, Nakamura Y, Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 63, 3043–3048 (2003). [PubMed] [Google Scholar]
- 39.Liang J, Liu N, Xin H, Knockdown long non-coding RNA PEG10 inhibits proliferation, migration and invasion of glioma cell line U251 by regulating miR-506. Gen. Physiol. Biophys 38, 295–304 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Kawai Y, Imada K, Akamatsu S, Zhang F, Seiler R, Hayashi T, Leong J, Beraldi E, Saxena N, Kretschmer A, Oo HZ, Contreras-Sanz A, Matsuyama H, Lin D, Fazli L, Collins CC, Wyatt AW, Black PC, Gleave ME, Paternally Expressed Gene 10 (PEG10) Promotes Growth, Invasion, and Survival of Bladder Cancer. Mol. Cancer Ther 19, 2210–2220 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Kim S, Thaper D, Bidnur S, Toren P, Akamatsu S, Bishop JL, Colins C, Vahid S, Zoubeidi A, PEG10 is associated with treatment-induced neuroendocrine prostate cancer. J. Mol. Endocrinol 63, 39–49 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F, CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M, Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U. S. A 93, 14440–14445 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Käll L, Storey JD, Noble WS, Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics. 24, i42–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nesvizhskii AI, Keller A, Kolker E, Aebersold R, A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 75, 4646–4658 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V, A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Challis RC, Kumar SR, Chan KY, Challis C, Jang MJ, Rajendran PS, Tompkins JD, Shivkumar K, Deverman BE, Gradinaru V, Widespread and targeted gene expression by systemic AAV vectors: Production, purification, and administration. Cold Spring Harbor Laboratory (2018), p. 246405. [Google Scholar]
- 48.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, Weitz DA, Rozenblatt-Rosen O, Zhang F, Regev A, Massively parallel single-nucleus RNA-seq with DroNc-seq. Nature Methods. 14 (2017), pp. 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrews S, FastQC: A quality control tool for high throughput sequence data. Online publication (2010).
- 51.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L, Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Connelly JP, Pruett-Miller SM, CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing. Scientific Reports. 9 (2019), doi: 10.1038/s41598-019-40896-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB, Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nature Protocols. 9 (2014), pp. 263–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Nostrand EL, Nguyen TB, Gelboin-Burkhart C, Wang R, Blue SM, Pratt GA, Louie AL, Yeo GW, Robust, Cost-Effective Profiling of RNA Binding Protein Targets with Single-end Enhanced Crosslinking and Immunoprecipitation (seCLIP). Methods Mol. Biol 1648, 177–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Expression plasmids are available from Addgene under a uniform biological material transfer agreement. Additional information available via the Zhang Lab website (https://zhanglab.bio). Next generation sequencing data generated are available from NCBI SRA with accession number PRJNA743280. All other data are available in the paper and supplementary materials.


