Abstract
Coexpression of the human TATA-binding protein (TBP)-associated factor 28 (hTAFII28) with the altered-specificity mutant TBP spm3 synergistically enhances transcriptional activation by the activation function 2 of the nuclear receptors (NRs) for estrogen and vitamin D3 from a reporter plasmid containing a TGTA element in mammalian cells. This synergy is abolished by mutation of specific amino acids in the α2-helix of the histone fold in the conserved C-terminal region of hTAFII28. Critical amino acids are found on both the exposed hydrophilic face of this helix and the hydrophobic interface with TAFII18. This α-helix of hTAFII28 therefore mediates multiple interactions required for coactivator activity. We further show that mutation of specific residues in the H1′ α-helix of TBP either reduces or increases interactions with hTAFII28. The mutations which reduce interactions with hTAFII28 do not affect functional synergy, whereas the TBP mutation which increases interaction with hTAFII28 is defective in its ability to synergistically enhance activation by NRs. However, this TBP mutant supports activation by other activators and is thus specifically defective for its ability to synergize with hTAFII28.
The RNA polymerase II transcription factor TFIID is a multiprotein complex composed of the TATA-binding protein (TBP) and a series of TBP-associated factors (TAFIIs) (3, 7). Not only are TAFIIs components of transcription factor TFIID, but distinct subsets of TAFIIs are also components of the SAGA, PCAF, and TFTC complexes (13, 14, 21, 31, 40). For human TFIID (hTFIID), cDNAs for 11 hTAFIIs have been characterized (10, 16, 18, 24, 25).
Genetic and biochemical experiments show that some TAFIIs are important for promoter recognition and expression of a subset of promoters (15, 38, 39), while others are more generally required for transcription in Saccharomyces cerevisiae (2, 26, 27, 29). An increasing body of results also shows that hTAFII28, hTAFII135, and hTAFII105 can act as specific transcriptional coactivators in mammalian cells. Expression of hTAFII135 specifically potentiates activation by the ligand-dependent activation function 2 (AF-2) of the nuclear receptors (NRs) for all-trans retinoic acid (retinoic acid receptor), thyroid hormone (thyroid hormone receptor), and vitamin D3 (vitamin D3 receptor [VDR]) (24). Distinct domains of hTAFII135 interact specifically with Sp1, CREB, and E1A, and coexpression of the TAFII135 domains with which these activators interact has a dominant negative effect on their activity (23, 28, 33, 36).
Similar experiments have shown that hTAFII105 interacts specifically with the p65 subunit of NF-κB and that TAFII105 expression strongly potentiates activation by NF-κB in mammalian cells (42). Coexpression of hTAFII28 and/or TBP strongly potentiates activation by the viral Tax protein, and Tax interacts directly with hTAFII28 and TBP to form a ternary complex (8).
Expression of hTAFII28 also potentiates ligand-dependent activation by the AF-2s of many NRs, the most dramatic effects being seen with the receptors for 9-cis retinoic acid (retinoid X receptor), estrogen (estrogen receptor [ER]), and the VDR (22). Deletion analysis showed that coactivator activity required amino acids 150 to 179 in the C-terminal domain of hTAFII28. Subsequent determination of the three-dimensional structure of the hTAFII28/hTAFII18 heterodimer at 2.6-Å resolution by X-ray crystallography indicated that these two proteins interact via a histone fold motif present in the C-terminal domain of hTAFII28 and in the central region of hTAFII18 (4). Amino acids 150 to 179 required for coactivator activity form the amphipathic α2-helix of the hTAFII28 histone fold. In the hTAFII28/hTAFII18 heterodimer, residues on the hydrophobic face of the α2-helix make intermolecular contacts with hTAFII18, while the residues on the mainly hydrophilic solvent-exposed face are available for mediating interactions with other proteins.
Although the ability of hTAFII28 to act as a transcriptional coactivator did not require direct interactions with the NRs, it apparently required interactions with TBP. hTAFII28 interacts directly with TBP both in vitro and in transfected mammalian cells (22, 25). This interaction requires amino acids 150 to 179 of hTAFII28, since deletion of this region dramatically reduced interactions with TBP. However, as this region is also required for interaction with hTAFII18, the possible roles of these different interactions in hTAFII28 coactivator activity could not be determined.
We have used the structural information to better characterize the amino acids required for hTAFII28 coactivator activity. We show that coexpression of hTAFII28 with the altered-specificity mutant TBP spm3 results in a synergistic enhancement of NR AF-2-activated transcription from a reporter plasmid with a mutated TGTA element. Mutation of several residues on the solvent-exposed surface and one residue on the hydrophobic surface of the α2-helix of the hTAFII28 histone fold abolishes this synergy. The amino acids on the solvent-exposed surface are also required for hTAFII28 to interact with coexpressed TBP.
We further show that mutations in the α-helix H1′ of TBP affect interactions between TAFII28 and TBP. Several of these TBP mutations strongly reduce interaction with hTAFII28, while one mutation results in increased interaction. Surprisingly, however, TBP mutations which reduce interactions with hTAFII28 do not impair the functional synergy. In contrast, no synergy is observed with the TBP mutant which shows increased interaction with hTAFII28, although this mutant does support activation by other activators.
MATERIALS AND METHODS
Construction of recombinant plasmids.
Mutations in hTAFII28 were generated by PCR amplification with the appropriate oligonucleotides and cloning of the resulting fragments in expression vector pXJ41 (41). Mutations in TBP were constructed in the same way in the TBP spm3 background and cloned in expression vector pSG5. The previously described E271R and L275R mutants (a kind gift from A. Berk) were recloned into the pSG5 expression vector. All plasmids were verified by automated DNA sequencing, and further details of constructions are available on request. The G4-NR, G4-VP16, and G4-AP2 expression vectors are also as described previously (22, 25).
Transfection of Cos cells and immunoprecipitations.
Cos cells were transfected by the calcium phosphate coprecipitation technique, and immunoprecipitations were performed as previously described (22, 25). Forty-eight hours following transfection, the cells were harvested by three cycles of freezing-thawing in buffer A (50 mM Tris-HCl [pH 7.9], 20% glycerol, 1 mM dithiothreitol, and 0.1% Nonidet P-40) containing 0.5 M KCl. The expression of the transfected proteins was verified on Western blots. For immunoprecipitations, cell extracts were incubated for 1 h at 4°C with 1 to 2 μg of the indicated monoclonal antibodies (MAbs), after which time 50 μl of protein G-Sepharose was added and incubation was continued for another 2 h. The protein G-Sepharose was then washed four times for 10 min each at room temperature with buffer A containing 1.0 M KCl and once with buffer A containing 0.1 M KCl. The resin was resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, boiled for 5 min, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The bound proteins were detected on Western blots with the indicated antibodies with an ECL kit (Amersham). For functional assays, in addition to the expression vectors described for each figure, all transfections contained 2 μg of pXJ-LacZ as internal standard for luciferase assays, 5 μg of the TGTA-Fos-Luc reporter, and pBSK− DNA as carrier. Transfections were performed in dextran-charcoal-treated medium, and ligands were added [50 nM all-trans-retinoic acid, 100 nM 1,25(OH)2D3, and 15 nM E2] at the same time as the DNA-calcium phosphate coprecipitate. Cells were harvested 48 h after transfection, and β-galactosidase and luciferase assays were performed by standard procedures. In all cases, similar results (±10%) were obtained in at least three independent transfections, and the results of typical experiments are shown in the figures.
Antibody preparation.
MAbs against hTAFII28 (15TA and 1C9), TBP (3G3), and the B10 epitope of the ER were as previously described (1, 5, 18, 25).
RESULTS
Amino acids in the hTAFII28 histone fold required for enhancement of NR activity in transfected mammalian cells.
Coexpression of hTAFII28 potentiates ligand-dependent transcriptional activation by the ER and VDR AF-2s in transfected Cos cells (22, 24). To better understand the molecular mechanism of this effect, we tested the ability of hTAFII28 to potentiate activation by the VDR and ER AF-2s in the presence of the altered-specificity mutant TBP spm3 (35) with a promoter with a TGTA rather than a TATA element.
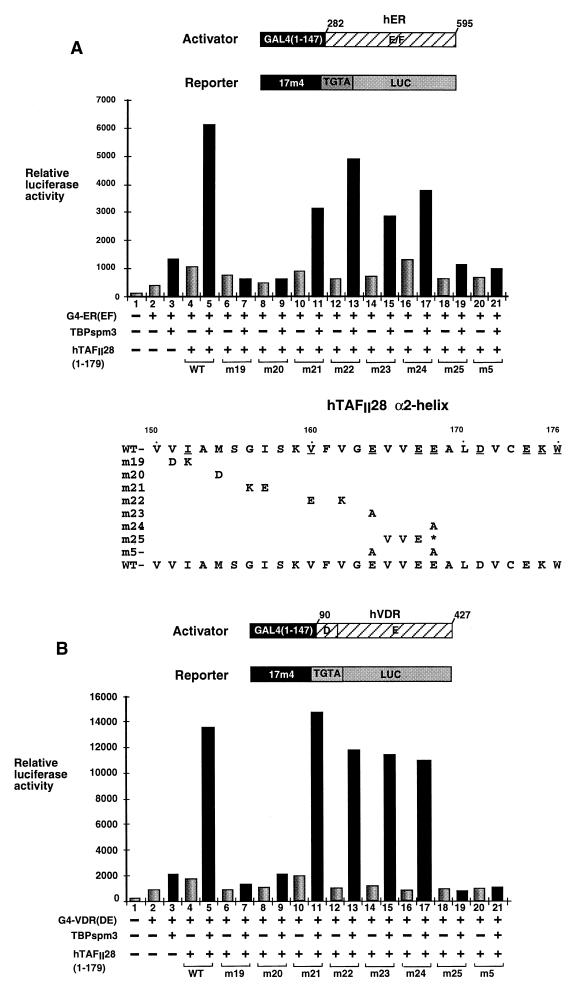
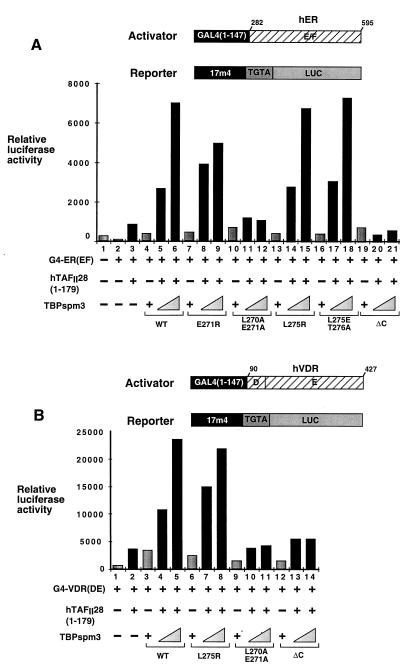
Expression of a chimera containing the DNA binding domain of the yeast activator GAL4 and the ligand binding domains (containing the AF-2) of the hER [G4-ER(EF)] or the hVDR [G4-VDR(DE)] led to only a small increase in the activity of a cotransfected luciferase reporter gene driven by a G4-responsive promoter with a TGTA element (6) (see Materials and Methods) (Fig. 1A and B, lanes 1 and 2). Expression of either TBP spm3 or TAFII28(1–179) significantly increased activation by G4-ER(EF) or G4-VDR(DE) (Fig. 1A and B, lanes 3 and 4), while coexpression of both led to a much stronger synergistic activation (lanes 3 to 5). All of the effects described were ligand dependent (data not shown). Thus, coexpressed TBP and TAFII28 synergistically enhance activation by the AF-2s of these NRs.
FIG. 1.
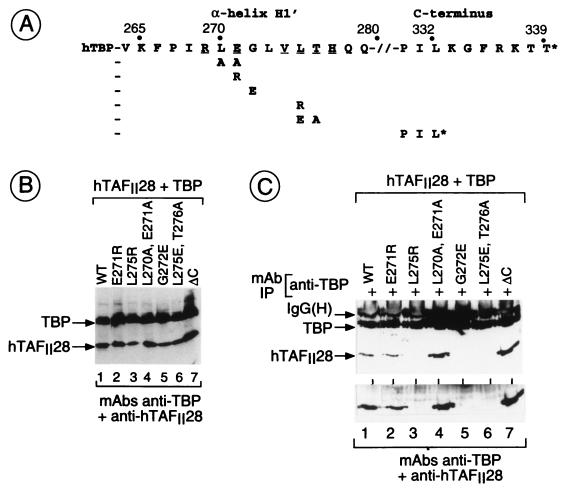
Functional analysis of hTAFII28 mutants. (A and B) Graphic representation of luciferase assays (arbitrary units). Cells were transfected with the vectors shown below each lane. Transfections contained 250 ng of the G4-ER or G4-VDR expression vectors, 1 μg of the TBP expression vector, 4 μg of the hTAFII28 expression vectors, 5 μg of the luciferase reporter vector, and 2 μg of the pXJ-β-galactosidase reporter as an internal control. All transfections contained the appropriate ligands. The structures of the G4-NR activator plasmids and the luciferase reporter are schematized. The numbers represent the amino acid coordinates in the natural receptors. The mutants used in panels A and B are shown below the graph in panel A. (C) Transfections were as described for panels A and B. The mutants used are schematized below the graph. (D) Transfections contained 2 (lane 9) or 4 (lanes 5, 6, and 10) μg of the hTAFII18 expression vector as indicated along with the amounts of the other expression vectors described above. WT, wild type.
We have previously shown that mutation of three of the exposed glutamic acid residues (E164P, E167P, and E168R) in the hTAFII28 α2-helix abolishes hTAFII28 coactivator activity (22). However, since two of these substitutions introduced prolines which would disrupt the α-helical structure, this did not permit identification of precise amino acids within the α2-helix involved in activation.
To better define the hTAFII28 amino acids required for coactivator activity, residues located on the hydrophobic and/or hydrophilic faces of the α2-helix were mutated either individually or in groups. Hydrophobic residues were replaced by charged residues, and charged residues, predominantly E and D, were replaced by alanine (see Materials and Methods) (Fig. 1A and B and 2A).
The ability of hTAFII28 mutants with substitutions in the α2-helix to synergistically enhance activation by the ER or VDR AF-2s in the presence of TBP spm3 was evaluated. Close to wild-type activity was seen when mutant m21 (G156K-I157E) or m22 (V160E-V162K) was coexpressed with TBP spm3 (Fig. 1A and B, lanes 11 and 13). Similarly, the double substitutions in m8 (E167A-D171A) and m10 (C173K-W176A) or the triple mutation m11 (G163E-L170K-E174A) also did not significantly affect the ability of hTAFII28 to synergize with TBP spm3 (Fig. 1C, lanes 12 to 17). In contrast, coexpression of mutants m19 (V151D-I152K), m20 (M154D), and m25, in which the C-terminal portion of the helix is deleted, along with TBP spm3 did not result in the strong increase in activation seen with wild-type TAFII28 with either activator (Fig. 1A and B, lanes 5, 7, 9, and 19). These mutations have therefore completely abrogated the synergy with TBP.
The single substitutions in m23 (E164A) and m24 (E168A) had only a minor effect on activation by G4-ER and no significant effect on activation by G4-VDR (Fig. 1A and B, lanes 14 to 17). The corresponding double mutant m5, in which both residues were mutated (E164A-E168A), was, however, completely inactive (Fig. 1A and B, lanes 20 and 21, and 1C, lanes 8 and 9). Similarly, mutant m6, in which K175 is additionally mutated, was also inactive while mutation m4 (K175A alone) had no effect (Fig. 1C, lanes 10 and 11 and 6 and 7). Therefore, synergy between hTAFII28 and TBP is abolished by mutation (m5) of two residues (E164 and E168) on the solvent-exposed surface and residue M154 (m20) on the hydrophobic surface. Synergy is also abolished by m19, in which residues on both surfaces are mutated.
The above results show that TAFII28 is functionally limiting in the Cos cells, since the increase in its intracellular concentration brought about during transfection increases activation by the NR AF-2s. This would imply that its heterodimeric partner is not functionally limiting for these activators, since hTAFII18 was not overexpressed in these experiments. To test this idea, hTAFII18 was coexpressed either in the absence or in the presence of both TBP spm3 and hTAFII28.
As described above, coexpression of hTAFII28 and TBP spm3 strongly increased activation by the G4-VDR chimera (Fig. 1D, lanes 1, 3, 7, and 8). In contrast, coexpression of hTAFII18 did not potentiate VDR AF-2 activity either in the presence or in the absence of TBP spm3 (lanes 5 and 6). Similarly, coexpression of hTAFII18 did not further potentiate the activation seen in the presence of hTAFII28 and TBP spm3 (lanes 9 and 10). Thus, in contrast to TAFII28, TAFII18 is not functionally limiting, since its overexpression does not potentiate activation by the VDR AF-2.
Residues in the α2-helix of the hTAFII28 histone fold required for interaction with TBP.
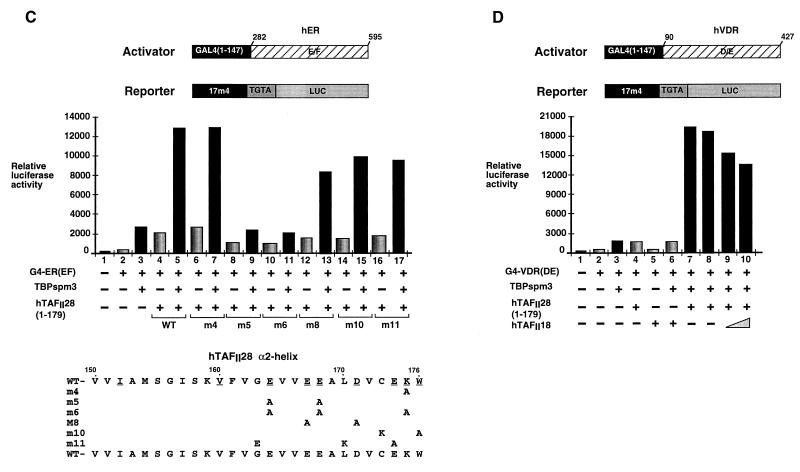
To determine whether the above mutations in hTAFII28 were defective in their interactions with TBP and/or hTAFII18, they were coexpressed in Cos cells, where we have previously shown interactions between these overexpressed proteins (18, 22). In this way, interaction assays are performed under the same conditions as those for the functional assays, thus facilitating a comparison of the two.
The mutant TAFII28 proteins were expressed in Cos cells along with TBP or hTAFII18, and the recombinant proteins were detected in the transfected cell extracts by immunoblotting with the appropriate MAbs (see Materials and Methods). All of the mutant hTAFII28 proteins were expressed at comparable levels (Fig. 2B and data not shown). The complexes formed between the overexpressed proteins were then precipitated with MAbs against TBP or TAFII28.
FIG. 2.
hTAFII28-TBP and hTAFII28-hTAFII18 interactions. (A) The sequence of the hTAFII28 α2-helix is shown. The numbers indicate the amino acid coordinates. Amino acids exposed in the hTAFII28/hTAFII18 heterodimer are underlined. The amino acid substitutions in each mutant are shown below the wild-type sequence. M25 contains a truncated α2-helix. (B) Coexpression of TBP and hTAFII28 mutants. Extracts from cells transfected with the vectors shown above each lane were analyzed by immunoblotting with the MAbs shown below the panel. The locations of TBP and the TAFII28 mutants are indicated. (C and D) Coimmunoprecipitation of hTAFII28 mutants with TBP. Extracts from cells transfected with the vectors shown above each lane were immunoprecipitated with the anti-TBP MAb 3G3 or the anti-hTAFII28 antibody 15TA as indicated above the panel, and the immunoprecipitated proteins were analyzed on immunoblots with the antibodies indicated below the panel. The positions of TBP and hTAFII28 are indicated along with the heavy chain of the antibody used in the immunoprecipitations revealed by the peroxidase-conjugated secondary antibody used in the immunoblots. (E) Coprecipitation of hTAFII28 mutants with B10-hTAFII18. The layout is as described for panels C and D. Lane 1 shows the precipitation of extracts from cells transfected with only B10-hTAFII18. IgG(H), immunoglobulin G heavy chain; IP, immunoprecipitation; WT, wild type.
As previously shown, hTAFII28(1–179) can be coprecipitated by the anti-TBP MAb 3G3 following coexpression of both proteins in Cos cells (Fig. 2C, lane 1). Compared to hTAFII28(1–179) wild type, interaction with TBP was not affected by hTAFII28 mutations m20, which abolishes coactivator activity, m21, and m22 (Fig. 2C, lanes 3 and 4, and data not shown, summarized in Fig. 3). Similarly, the double mutations m8 and m10 or the triple mutation m11 had no effect on interaction with TBP (summarized in Fig. 3).
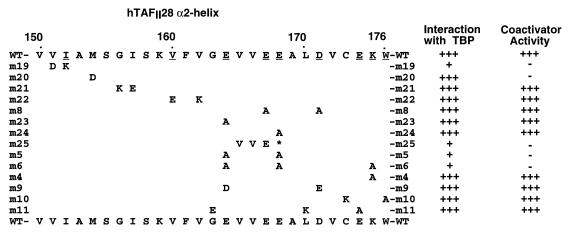
FIG. 3.
The effects of mutations in hTAFII28 on physical and functional interactions with TBP are summarized. WT, wild type.
In contrast, coprecipitation with TBP was strongly reduced by mutations m19 and m25 (Fig. 2C, lanes 2 and 5). Mutants m23 and m24 interacted with TBP (Fig. 2C, lanes 6 and 7, and 2D, lanes 1 to 4), whereas with m5, in which both residues were mutated, strongly reduced interaction was observed (Fig. 2D, lanes 5 and 6). These results show that hTAFII28 residues V151 and/or I152, E164, and E168 play critical roles in the interaction with TBP.
The ability of these mutations to affect heterodimerization with hTAFII18 was also determined by coexpression of the mutants with a B10 epitope-tagged derivative of hTAFII18 and immunoprecipitation with the anti-B10 MAb (22, 25). When compared with the wild-type proteins (Fig. 2E, lane 2), comparable coprecipitation of all the mutants was observed (lanes 3 to 7 and data not shown). Therefore, while some of the above hTAFII28 mutations significantly diminished interactions with TBP, they did not abolish interactions between coexpressed hTAFII28 and hTAFII18. This is true even for alleles, for example, m20 and m22, harboring radical mutations on the hydrophobic face of the α2-helix. Similarly, none of the mutations affected the coprecipitation of hTAFII28 with hTAFII55 or hTAFII135 (data not shown).
Amino acids in α-helix H1′ of TBP required for interaction with hTAFII28.
The histone fold region of hTAFII28 shows marked sequence homology with the C-terminal region of the SAGA and PCAF subunit SPT3 (4). SPT3 in yeast plays a role in transcription from a subset of promoters with nonconsensus TATA elements (9). Genetic experiments have shown a reciprocal allele-specific suppression of mutants in the putative α2-helix of yeast SPT3 (ySPT3) (E240K) and the H1′ helix of yTBP (allele spt15-2; G174E) (9, 11, 20). This suggests that these two helices mediate the functional and perhaps also physical interactions between the respective proteins. The homology with SPT3 suggested to us that the H1′ helix of hTBP may also be involved in physical and functional interactions with hTAFII28. Furthermore, mutation F237V in the yTBP C terminus is an intragenic suppressor of the G174E mutation in the H1′ helix, and yTBP mutation K239E is an extragenic suppressor of the E240K mutation in the putative α2-helix of ySPT3 (11). This suggests that residues at the C terminus of the TBP H2′ α-helix may also contribute to SPT3-TBP interactions in accordance with the fact that this region and the H1′ helix are in close proximity in the folded TBP molecule (17, 30).
To test the possible contribution of both of these regions in interactions with hTAFII28, TBP spm3 derivatives with additional substitutions in the H1′ helix or deletion of the C-terminal region (Materials and Methods and Fig. 4A) were coexpressed along with wild-type hTAFII28 in Cos cells. Each TBP mutant accumulated to similar levels in the transfected Cos cells when coexpressed along with hTAFII28 (Fig. 4B). The coexpressed proteins were then immunoprecipitated with MAb 3G3 directed against the extreme N terminus of TBP (19).
FIG. 4.
Mutations in TBP which affect interactions with hTAFII28. (A) The sequences of the H1′ and C-terminal regions of hTBP are shown along with the amino acid coordinates. The amino acid substitutions are shown below the wild-type sequence. The asterisk indicates the stop codon in the C-terminal deletion. (B) Coexpression of TBP mutants and hTAFII28. The layout is as described for Fig. 1B. (C) Coprecipitation of hTAFII28 with TBP mutants. The layout is as described for Fig. 1C. The lower panel shows a longer exposure of the region of the gel containing hTAFII28. IgG(H), immunoglobulin G heavy chain; IP, immunoprecipitation; WT, wild type.
Compared with TBP spm3, interaction with hTAFII28 was not affected by the mutation E271R in the N-terminal portion of H1′ (Fig. 4C, lanes 1 and 2). In contrast, interaction with hTAFII28 was strongly reduced by mutations in the C-terminal portion of this helix, L275R and L275E-T276A (lanes 3 and 6). Interaction with TAFII28 was also strongly reduced by the mutation G272E (lane 5), equivalent to mutation G174E of allele spt15-21 in yTBP. Interestingly, interaction of hTAFII28 with mutant L270A-E271A and with the ΔC mutant was actually increased (Fig. 4C, lanes 1, 4, and 7). Therefore, specific residues in the TBP H1′ helix are required for efficient interactions with hTAFII28.
A mutation in the H1′ helix of TBP which specifically impairs synergy with hTAFII28.
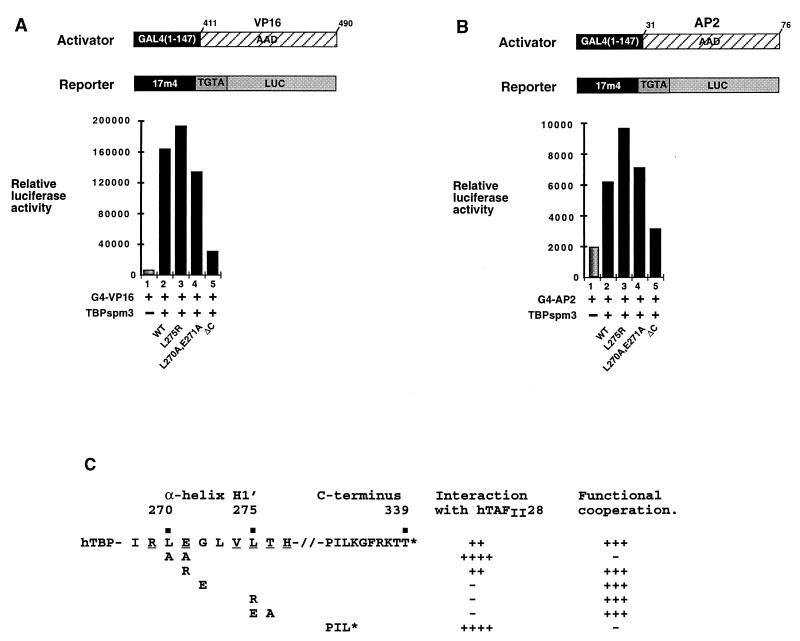
Mutations in α-helix H1′ of TBP do not affect interaction with any of the identified basal factors and have only a minor effect on the ability of TBP to support activation by the VP16 and E1A activation domains in transfected cells (6). We tested the ability of several of the above mutants in this helix to support activation by the G4-NR chimeras. As described above, coexpression of hTAFII28 and TBP spm3 synergistically enhanced activation by the ER AF-2 (Fig. 5A, lanes 3 to 6) TBP spm3 mutants E271R, G272E, L275R, and L275E-T276A also synergized with hTAFII28 to enhance activation by the ER AF-2 (lanes 7 to 9 and 13 to 18 and data not shown, summarized in Fig. 6C). In contrast, expression of mutant L270A-E271A did not enhance activation by the ER AF-2 activity in the presence of hTAFII28 (lanes 10 to 12). Furthermore, no synergy was seen with this TBP mutant in titration experiments over a 20-fold range of the corresponding expression vectors (0.5 to 10 μg), nor in the presence of coexpressed hTAFII18 (data not shown). Similarly, the ΔC mutant also failed to synergize with coexpressed hTAFII28 (lanes 19 to 21).
FIG. 5.
Functional analysis of TBP mutants. The layout is as described for Fig. 2. Transfections contained 1 or 2 μg of the TBP expression vectors along with 4 μg of the hTAFII28 expression vector as indicated. WT, wild type.
FIG. 6.
Effect of TBP mutations on activation by VP16 and AP2. (A and B) Transfections contained 1 μg of the TBP expression vectors and 100 ng of the G4-VP16 or G4-AP2 expression vector. (C) The effects of mutations in TBP on physical and functional interactions with hTAFII28 are summarized. WT, wild type. Exposed residues are underlined.
Wild-type TBP spm3 and mutant L275R also synergized with hTAFII28 to enhance activation by the VDR AF-2 (Fig. 5B, lanes 3 and 4 and 6 to 8). However, as observed with the ER AF-2, mutants L270A-E271A and ΔC did not synergize with hTAFII28 to enhance activation by the VDR AF-2 (lanes 9 to 14). Similar results were obtained with a G4-retinoic acid receptor chimera (data not shown).
The ability of these mutants to support activation by unrelated activators G4-VP16 and G4-AP2, whose activities are not enhanced by coexpression of hTAFII28 (reference 22 and data not shown), was also tested. Comparable activities were seen with wild-type TBP spm3, L270A-E271A, and L275R, whereas the activity of mutant ΔC was severely reduced (Fig. 6A and B). These results show that mutant L270A-E271A conserves its intrinsic ability to mediate activation but is specifically unable to functionally cooperate with hTAFII28, while mutant ΔC is generally defective in its ability to mediate activation.
DISCUSSION
The α2-helix of the hTAFII28 histone fold plays a critical role in functional interactions with TBP.
We have identified specific amino acids in the hTAFII28 α2-helix which are required for synergy with TBP in mammalian cells. Simultaneous mutation of amino acids E164 and E168, which are close together on the solvent-exposed surface of the α2-helix (Fig. 7), abolishes functional cooperation with TBP. This same mutation also results in a loss of interaction with TBP. Therefore, the hydrophilic face of the α2-helix, which is not involved in intermolecular interactions with hTAFII18, mediates interaction with other proteins, one being TBP.
FIG. 7.
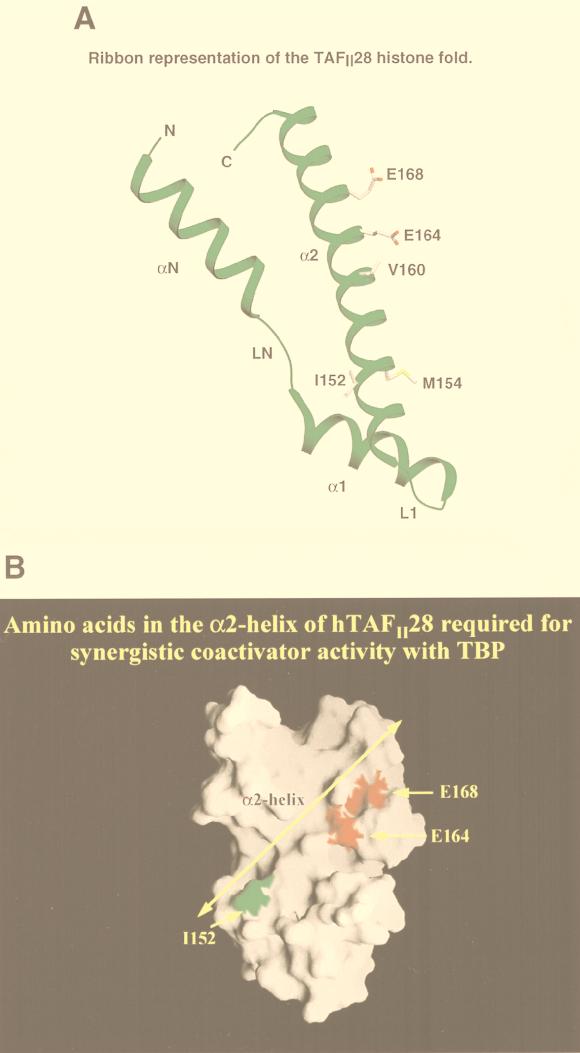
(A) A ribbon representation of the hTAFII28 histone fold. The locations of the αN-, α1-, and α2-helices are shown along with the LN and L1 loops. For the sake of clarity, the α3-helix has been deleted. The α2-helix has been aligned so that the hydrophobic interface with hTAFII18 is oriented to the rear, while the exposed residues are oriented toward the viewer. The side chains of several pertinent amino acids are depicted. (B) Surface contour map of the hTAFII28/hTAFII18 heterodimer. The locations of the hTAFII28 α2-helix and amino acids I152, E164, and E168 are indicated. M154 was also colored but is invisible on the surface, showing that it is completely buried in the interface with hTAFII18.
Functional synergy and interaction with TBP are also abolished by mutation of V151 and I152. Of these two amino acids, only I152 is on the exposed surface (Fig. 7). As none of the other mutations on the hydrophobic face of the α2-helix abolish interaction with TBP, it is probable that it is mutation of I152 which is responsible for the loss of interaction with TBP. While I152 is not adjacent to E164 and E168, it is nevertheless part of an epitope on the exposed face of the α2-helix involved in interactions with TBP. Furthermore, by analogy with the results obtained with E164 and E168, it is probable that it is the mutation of I152 which also causes the loss of synergy with TBP.
The m19 and m5 mutants discussed above, which have lost the ability to synergize with TBP, have changes in amino acids on the exposed surface of the α2-helix. However, synergy is also lost upon mutation of M154 (m20) on the hydrophobic surface. Since M154 is not on the exposed surface of the α2-helix (Fig. 7A), it is unlikely that it contributes to physical interactions with TBP, and indeed, coprecipitation of mutant m20 with TBP is comparable to that of the wild type.
As M154 is on the hydrophobic interface with TAFII18, this rather suggests that heterodimerization with TAFII18 is involved in coactivator activity. However, m20 does not abolish heterodimerization with hTAFII18. Indeed, mutation of several other residues (I157 and V162), which must surely disrupt the hydrophobic interactions between the hTAFII28 and hTAFII18 α2-helices, does not abolish heterodimer formation and has no effect on synergy. Therefore, the tight hTAFII28-hTAFII18 interaction, involving not only the two α2-helices but also the strong interface formed by the hTAFII28 αN- and hTAFII18 α1-helices (4), cannot be totally disrupted by mutation of only one or two amino acids in the α2-helix. Such mutations are nevertheless likely to significantly affect heterodimer stability or conformation. Our results show that the perturbations introduced by mutation of I157 and V162 do not affect synergy, whereas the changes in structure and/or stability induced by mutation of M154 are not compatible with function. Therefore, although it is not clear why M154 is particularly sensitive to mutation, the result obtained with this mutant indicates that interaction with hTAFII18 is required for coactivator activity.
Mutation of exposed residues of the αN- and α1-helices of the histone fold had no effect on synergy with TBP (18a), further highlighting the unique and critical role played by the α2-helix in this process. All together, our results would suggest that this reflects the ability of this helix to interact with factors essential for NR AF-2 activity via the exposed face and a requirement for heterodimerization with TAFII18.
Possible molecular mechanisms underlying the coactivator activity of hTAFII28.
Our results show that specific residues in the H1′ α-helix of TBP are required for interactions with hTAFII28. Interaction with hTAFII28 was severely reduced by mutation of G272 and L275. Mutation E271R had no effect on interaction, while mutation L270A-E271A and deletion of the C-terminal domain of TBP led to increased interaction with hTAFII28. The effect of these mutations was specific to hTAFII28, since they did not affect, either positively or negatively, interactions with coexpressed hTAFII18 or hTAFII20 (18a).
While mutations in hTAFII28 which affect interaction with TBP also affect synergy, reciprocal TBP mutations which clearly diminish interactions with hTAFII28 do not abolish the synergy between these two proteins. This synergy is, however, abolished by the L270A-E271A mutation in TBP which increases interaction with hTAFII28. This does not reflect a general loss of TBP function, since this mutant supports activation by VP16 and AP2. This mutant therefore exhibits a selective defect in its ability to synergize with hTAFII28, which correlates with the increased interaction.
How do TAFII28 and TBP synergize to enhance activation by NRs? Our results would be consistent with the incorporation of hTAFII28 and TBP spm3 into a TFIID complex comprising endogenous TAFIIs. Indeed, we and others have previously shown that a fraction of transfected TBP spm3 and hTAFII28 (and indeed other TAFIIs) do associate with the endogenous cellular TFIID (12, 22, 32). Transfected hTAFII28 may be assembled into TFIID complexes via TAF-TAF rather than TAF-TBP interactions, explaining why the mutations in TBP do not abolish functional cooperation. Heterodimerization with TAFII18 is an obvious alternative interaction allowing incorporation of TAFII28 into a TFIID complex.
In our experiments, incorporation of hTAFII28 and TBP spm3 into endogenous TFIID complexes does not necessarily arise by the modification of existing TFIID complexes. In the course of the 48 h of the experiment, several cell divisions take place and de novo TFIID complexes are assembled. These new complexes are assembled under conditions where the intracellular concentration of TAFII28 and/or TBP spm3 is much higher than normal. Such conditions would of course favor the incorporation of these proteins during assembly of TFIID complexes. The net result would be to increase the concentration of TFIID complexes containing TAFII28 and TBP spm3.
Further evidence that association of hTAFII28 with endogenous TFIID is required for activation comes from the observation that expression of hTAFII28 leads to enhanced activation by the NR AF-2s even in the absence of TBP spm3 (we have previously made similar observations with TAFII135; for discussion, see reference 24). This is in keeping with our observation that expression of hTAFII28 alone suffices to increase activation with a reporter with a TATA element (22). The lower increase seen here with the TGTA-containing reporter reflects the low affinity of the endogenous TBP for this element, which is only partially compensated by the expression of TAFII28. These results show that hTAFII28 expression facilitates NR AF-2-dependent activation via the endogenous TFIID, an observation most readily explained by the incorporation of hTAFII28 into endogenous TFIID.
Taken altogether, our results would suggest that the synergistic activation observed here requires the formation of TFIID complexes with TBP spm3, which allows efficient recognition of the TGTA element, and hTAFII28, which facilitates the efficient use of the complexes containing TBP spm3 by the NR AF-2s.
The TFIID complexes comprising hTAFII28 and/or TBP spm3 may efficiently mediate NR AF-2 activity due to interactions between hTAFII28 and factors required for NR AF-2 activity. Further evidence for this comes from consideration of the effect of mutations in hTAFII28. Since the hTAFII28 mutations abrogate coactivator activity and this is not simply a consequence of loss of interaction with TBP, it is probable that these mutations also affect interactions with another protein(s) which is indispensable for transcriptional activation.
The phenotype of the TBP mutant L270A-E271A is particularly interesting in this respect. As discussed above, the hTAFII28 α2-helix may interact with a protein other than TBP required for transcriptional activity. It is therefore possible that the increased interaction with this TBP mutant impairs these other hTAFII28 interactions, resulting in a loss of coactivator activity. While we cannot exclude more complex scenarios, altogether our observations would be consistent with a model in which hTAFII28 coactivator activity involves dynamic hTAFII28-TBP interactions which have to be dissociated to allow hTAFII28 to subsequently interact with other proteins required for transcriptional enhancement.
One surprising observation was that deletion of the C-terminal residues of the TBP H2′ helix led to a loss of function. This TBP mutant not only was defective in its ability to synergize with hTAFII28 but was generally defective, since it did not efficiently mediate activation by the VP16 activation domain. These C-terminal residues are not known to be required for interaction with other components of the preinitiation complex (6, 37), and the mutations of Y329 and K337 had only mild effects on activated transcription in mammalian cells (6). Nevertheless, two mutations in the equivalent region of yTBP (E236P and F237D) led to defects in activated transcription but did not affect interaction with the TATA element (34). The F237D mutation led to a general loss of TBP interactions with other basal transcription factors, possibly due to an altered conformation. While the molecular basis for the loss of function seen with C-terminal deletion in hTBP is not clear, our results show that this region is necessary for TBP function in mammalian cells.
ACKNOWLEDGMENTS
We thank A. Berk for expression vectors and reporter plasmids; P. Chambon for support; L. Perletti and M. Vigneron for critical comments; S. Vicaire and D. Stephane for DNA sequencing; Y. Lutz and the MAb facility; the staff of cell culture and oligonucleotide facilities; B. Boulay, J. M. Lafontaine, R. Buchert, and C. Werlé for illustrations; and Roussel-Uclaf for providing 1,25(OH)2D3.
G.M. and A.-C.L. were supported by fellowships from the Ligue Nationale contre le Cancer and the Association pour la Recherche contre le Cancer. This work was supported by grants from the CNRS, the INSERM, the Hôpital Universitaire de Strasbourg, the Ministère de la Recherche et de la Technologie, the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer, and the Human Frontier Science Programme.
REFERENCES
- 1.Ali S, Lutz Y, Bellocq J P, Chenard-Neu M P, Rouyer N, Metzger D. Production and characterization of monoclonal antibodies recognising defined regions of the human oestrogen receptor. Hybridoma. 1993;12:391–405. doi: 10.1089/hyb.1993.12.391. [DOI] [PubMed] [Google Scholar]
- 2.Apone L M, Virbasius C A, Holstege F C, Wang J, Young R A, Green M R. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- 3.Bell B, Tora L. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp Cell Res. 1999;246:11–19. doi: 10.1006/excr.1998.4294. [DOI] [PubMed] [Google Scholar]
- 4.Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne A C, Davidson I, Moras D. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- 5.Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly J M, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant G O, Martel L S, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 7.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 8.Caron C, Mengus G, Dubrowskaya V, Roisin A, Davidson I, Jalinot P. Human TAFII28 interacts with the human T cell leukemia virus type I Tax transactivator and promotes its transcriptional activity. Proc Natl Acad Sci USA. 1997;94:3662–3667. doi: 10.1073/pnas.94.8.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collart M A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubrovskaya V, Lavigne A C, Davidson I, Acker J, Staub A, Tora L. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 1996;15:3702–3712. [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:131–131. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 12.Farmer G, Colgan J, Nakatani Y, Manley J L, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol Cell Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, Workman J L. A subset of TAFs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 14.Grant P A, Sterner D E, Duggan L J, Workman J L, Berger S L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 15.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 16.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 17.Kim J L, Burley S K. 1.9 A resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nat Struct Biol. 1994;1:638–653. doi: 10.1038/nsb0994-638. [DOI] [PubMed] [Google Scholar]
- 18.Lavigne A C, Mengus G, May M, Dubrovskaya V, Tora L, Chambon P, Davidson I. Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J Biol Chem. 1996;271:19774–19780. doi: 10.1074/jbc.271.33.19774. [DOI] [PubMed] [Google Scholar]
- 18a.Lavigne, A.-C., Y.-G. Gangloff, L. Carré, G. Mengus, C. Birck, O. Poch, C. Romier, D. Moras, and I. Davidson. Unpublished data. [DOI] [PMC free article] [PubMed]
- 19.Lescure A, Lutz Y, Eberhard D, Jacq X, Krol A, Grummt I, Davidson I, Chambon P, Tora L. The N-terminal domain of the human TATA-binding protein plays a role in transcription from TATA-containing RNA polymerase II and III promoters. EMBO J. 1994;13:1166–1175. doi: 10.1002/j.1460-2075.1994.tb06366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez E, Kundu T K, Fu J, Roeder R G. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 22.May M, Mengus G, Lavigne A C, Chambon P, Davidson I. Human TAFII28 promotes transcriptional stimulation by activation function 2 of the retinoid X receptors. EMBO J. 1996;15:3093–3104. [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzarelli J M, Mengus G, Davidson I, Ricciardi R P. The transactivation domain of adenovirus E1A interacts with the C terminus of human TAFII135. J Virol. 1997;71:7978–7983. doi: 10.1128/jvi.71.10.7978-7983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mengus G, May M, Carre L, Chambon P, Davidson I. Human TAFII135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 25.Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. Cloning and characterization of hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J. 1995;14:1520–1531. doi: 10.1002/j.1460-2075.1995.tb07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel B, Komarnitsky P, Buratowski S. Histone-like TAFs are essential for transcription in vivo. Mol Cell. 1998;2:663–673. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- 27.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 29.Natarajan K, Jackson B M, Rhee E, Hinnebusch A G. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- 30.Nikolov D B, Burley S K. 2.1 A resolution refined structure of a TATA box-binding protein (TBP) Nat Struct Biol. 1994;1:621–637. doi: 10.1038/nsb0994-621. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 32.Sadovsky Y, Webb P, Lopez G, Baxter J D, Fitzpatrick P M, Gizang-Ginsberg E, Cavailles V, Parker M G, Kushner P J. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol Cell Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saluja D, Vassallo M F, Tanese N. Distinct subdomains of human TAFII30 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stargell L A, Struhl K. A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol Cell Biol. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strubin M, Struhl K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 36.Tanese N, Saluja D, Vassallo M F, Chen J L, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Sun X, Reinberg D, Ebright R H. Protein-protein interactions in eukaryotic transcription initiation: structure of the preinitiation complex. Proc Natl Acad Sci USA. 1996;93:1119–1124. doi: 10.1073/pnas.93.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 39.Walker S S, Shen W C, Reese J C, Apone L M, Green M R. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 40.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 41.Xiao J H, Davidson I, Matthes H, Garnier J M, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 42.Yamit-Hezi A, Dikstein R. TAFII105 mediates activation of anti-apoptotic genes by NF-kappaB. EMBO J. 1998;17:5161–5169. doi: 10.1093/emboj/17.17.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]