Abstract
We have examined structural differences between the proto-oncogene c-Myb and the cyclic AMP-responsive factor CREB that underlie their constitutive or signal-dependent activation properties. Both proteins stimulate gene expression via activating regions that articulate with a shallow hydrophobic groove in the KIX domain of the coactivator CREB-binding protein (CBP). Three hydrophobic residues in c-Myb that are conserved in CREB function importantly in cellular gene activation and in complex formation with KIX. These hydrophobic residues are assembled on one face of an amphipathic helix in both proteins, and mutations that disrupt c-Myb or CREB helicity in this region block interaction of either factor with KIX. Binding of the helical c-Myb domain to KIX is accompanied by a substantial increase in entropy that compensates for the comparatively low enthalpy of complex formation. By contrast, binding of CREB to KIX entails a large entropy cost due to a random coil-to-helix transition in CREB that accompanies complex formation. These results indicate that the constitutive and inducible activation properties of c-Myb and CREB reflect secondary structural characteristics of their corresponding activating regions that influence the thermodynamics of formation of a complex with CBP.
Gene-specific factors regulate transcription via activating regions that interact with specific targets in the RNA polymerase II (Pol II) machinery. Indeed, the strength of an activating region appears to reflect its intrinsic affinity for one or more such target proteins (20). But, apart from a general abundance of certain amino acids such as proline, glutamine, or acidic residues, no consensus motif for interaction with any individual target has emerged.
Ideally, the identification of a motif that is conserved between activating regions might best be approached with transcription factors that associate with a common target in the Pol II apparatus. The Pol II-associated coactivator CREB-binding protein (CBP) and its paralog P300 (hereafter referred to as CBP/P300), for example, associate with a number of activators via several interaction domains. One of these, referred to as the KIX domain, has been shown to interact functionally with the cyclic AMP-responsive factor CREB (12) as well as with certain constitutive activators such as c-Myb (4), SREBP (11), Stat-1 (22), Tax (9), and cubitus interruptus (1). A three-helix structure that contains a shallow hydrophobic groove, the KIX domain is highly conserved in all species that express CBP/P300 homologues (16). The functional importance of this domain for cellular gene expression has been most extensively evaluated in the context of cyclic AMP and CREB signaling.
CREB triggers target gene expression, following its protein kinase A (PKA)-mediated phosphorylation at Ser133, via a kinase-inducible activation domain (KID) that binds to KIX (2, 12). Direct interactions between the Ser133 phosphate moiety and KIX account for about half of the free energy of formation of a complex between KID and KIX (13, 16). The Ser133 phosphate in KID forms a hydrogen bond with Tyr658 and a salt bridge with Lys662 in KIX, and mutagenesis of both Tyr658 and Lys662 reduces the affinity of phospho(Ser133)KID for KIX about 1,000-fold.
Allosteric contributions from the Ser133 phosphate group also figure prominently in this interaction. The phosphate moiety promotes formation of an amphipathic helix in KID, termed αB, in part by stabilizing the helix macrodipole and also by hydrogen bonding with the backbone Ser133 amide (16). Ser133 phosphorylation per se is not sufficient to induce the helical transition in αB, however; complex formation with KIX is also required. Residues on the hydrophobic face of helix αB stabilize the helical transition via contacts with a shallow hydrophobic groove in KIX.
The importance of KIX for target gene induction via numerous activators suggests a common mode of binding, perhaps via a conserved motif, but casual inspection of these activating regions does not reveal any extensive sequence similarity to support that notion. The ability of one domain in CBP to accommodate both constitutive and signal-dependent factors has prompted us to compare the mechanisms underlying both types of interactions, using c-Myb and CREB. Our results suggest that c-Myb- and CREB-activating regions actually have limited sequence similarities that reflect comparable surface interactions with the KIX domain; however, they have remarkable differences in secondary structure that potently affect the thermodynamics of complex formation. We propose that these structural differences in c-Myb and CREB form the basis for their constitutive and inducible activation properties.
MATERIALS AND METHODS
Plasmids.
Wild-type and mutant KIX cDNAs were constructed by PCR amplification and were cloned into a pGEX-LT vector. All KIX proteins contained amino acids (aa) 586 through 672 of the murine CBP. Rous sarcoma virus (RSV)-CREB M1 (S133A) and RSV-CREB Δ136-160 were previously described (6, 7). −411 CD13/APN Luc was previously described (18). 5×Gal-Luc (pGL-2/G5B) is described elsewhere (8). Gal-KIX was constructed by inserting a KIX cDNA fragment encoding CBP aa 553 through 679 into pM3 (17). Gal-Myb 186-325 was constructed by inserting a c-Myb cDNA fragment into pM2 (17). pCMX c-Myb was constructed by subcloning the full-length murine c-Myb cDNA from pRmb3Svneo into pCMX. RSV2×VP16 Myb vectors were developed by ligating two repeats of VP16 in tandem into the RSV expression vector pGR. A c-Myb PCR fragment (described below) was then inserted downstream of the VP16 sequences. Other plasmids were constructed by using standard cloning techniques.
Mutagenesis.
Murine c-Myb point mutants were generated by using Stratagene’s Quickchange protocol, and mutations were confirmed by DNA sequencing. Myb truncation mutants were made by PCR amplification by using internal primers to c-Myb.
GST pulldown and fluorescence polarization assays.
GST-KIX S/B fusion proteins were purified from Escherichia coli, and glutathione S-transferase (GST) pulldown assays with 35S-labeled methionine-labeled in vitro-transcribed and -translated c-Myb were performed as previously described (21). About 20 μg of GST-KIX S/B was used per reaction. Input levels of c-Myb (about 10,000 cpm) were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or by trichloroacetic acid precipitation and scintillation counting and were within ±15% of the level of wild-type c-Myb. Quantitation of c-Myb following SDS-PAGE was performed with a Molecular Dynamics Storm phosphorimager. For fluorescence anisotropy experiments, c-Myb (aa 276 through 315) and KID (aa 88 through 160) peptides were N-terminally treated with fluorescein, and fluorescence polarization values were obtained with wild-type and mutant KIX polypeptides by using a Pan Vera Beacon 2000 Variable-Temperature Fluorescence Polarization system (13). KIX proteins were added to samples containing fluorescein-treated (fl) Myb or KID peptides (final concentration for each, 5 to 16 nM), and binding assays were performed in a total volume of 150 μl at 22°C after a 90-s delay with 16-s integration.
Transient-transfection assays.
For the mammalian two-hybrid assays, COS-7 cells in 3.5-cm-diameter dishes were transfected with 0.5 μg of 5×Gal-Luc reporter plasmid, 0.5 μg of Gal-KIX S/B expression plasmid, and 1.5 μg of RSV2×VP16-Myb, RSV-Myb, or pGR (RSV) expression plasmid and 0.01 μg of pRL-SV40 (SV40, simian virus 40) (Promega) by the calcium phosphate method. Cells were harvested after approximately 23 h and assayed for luciferase derived from the reporter. For c-Myb activity assays, 293 embryonal kidney cells in 3.5-cm-diameter dishes were transfected with 0.5 μg of reporter plasmid, 0.5 μg of either pCMX-c-Myb, Gal-Myb 186-325, or Gal-Myb 290-315 expression vector, and 0.5 μg of RSV-β-Gal. After about 17 h cells were harvested and assayed for reporter plasmid-derived luciferase activity. −411 CD13/APN Luc reporter was used to test pCMX c-Myb activity, and 5×Gal-Luc reporter was used to test Gal-Myb activity. F9 cell transfection assays to test CREB activity were performed as previously described (3). Reporter gene-derived luciferase or chloramphenicol acetyltransferase (CAT) activity was normalized to an internal β-galactosidase (RSV-β-Gal), luciferase (RSV-Luc), or Renilla luciferase (pRL-SV40) control.
ITC.
Isothermal titration of c-Myb (aa 291 to 315) (2.0 mM) or pKID (aa 100 to 160) (0.3 mM) into KIX polypeptide (aa 586 to 672) (0.225 and 0.022 mM, respectively) was performed at 27°C in 50 mM Tris (pH 7.0)–50 mM NaCl. The experiments were carried out by using an MCS titration calorimeter from MicroCal, Inc. (Northampton, Mass.), with a 250-μl injection syringe during stirring at 4,000 rpm. An initial 1-μl injection was followed by 29 7-μl injections of pKID or 28 10-μl injections of Myb. Integration of the thermogram and subtraction of the blanks gave a binding isotherm that fitted best to a model of one-site interaction. The data were fitted by using the variables K (association constant), ΔH (enthalpy), and N (stoichiometric ratio), using the isothermal titration calorimetry (ITC) data analysis software in ORIGIN version 2.3 (MicroCal Software, Northampton, Mass.). ΔG and ΔS were then obtained by the following basic thermodynamic equations: ΔG° = −RTlnK = ΔH° − TΔS°. The minor deviations of the stoichiometric ratio N from the theoretical value of 1.0 (2% for pKID and 14% for Myb) were used to correct ΔH and K for concentration determination errors. The C values (C = K × [KIX] × N) indicated that binding-constant determination by ITC was very accurate for pKID (C = 60) and reasonably accurate for Myb (C = 10). Even an unreasonably large error in the determination of K would not change the sign of the calculated ΔS.
RESULTS
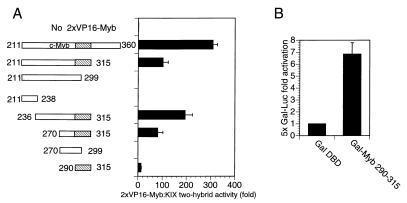
To determine whether CREB and c-Myb have a common KIX interaction motif, we performed mammalian two-hybrid studies using a series of nested fragments within the c-Myb activation domain. A c-Myb-VP16 fusion polypeptide containing residues in c-Myb that were previously shown to associate with CBP (aa 211 to 360) (4) potently stimulated Gal4 KIX activity on a cotransfected 5×Gal4 luciferase reporter plasmid in COS-7 cells (Fig. 1A). Shorter polypeptides containing residues 270 through 315 or 290 through 315 in c-Myb were also capable of associating with KIX in two-hybrid assays, but c-Myb constructs lacking those sequences were not (Fig. 1A). These experiments indicate that residues 290 through 315 in c-Myb encode a minimal KIX binding domain.
FIG. 1.
Characterization of a minimal transactivation domain in c-Myb that binds to the KIX domain of CBP. (A) Mammalian two-hybrid assays performed by using Gal4-KIX and c-Myb/VP16 expression vectors to examine interactions of c-Myb with CBP. COS-7 cells were transfected with 5×Gal4-Luc and pRL-SV40 Renilla luciferase reporters and with expression vectors for Gal-KIX and 2×VP16-Myb fragments. The hatched box represents the minimal KIX interaction domain of c-Myb encompassing aa 290 to 315. Inclusive amino acid endpoints of c-Myb fragments fused to tandem VP16 activation domains are indicated. Results are shown as fold activation (mean ± standard deviation, n = 2) over that by empty RSV expression vector (without VP16-Myb). c-Myb/VP16 polypeptides were expressed at comparable levels in transfected cells (data not shown). Inclusive endpoints of c-Myb are shown. (B) c-Myb (aa 290 to 315) stimulates target gene expression when fused to the Gal4 DNA binding domain (DBD). 293 cells were transfected with 5×Gal Luc and RSV-β-Gal reporter plasmids as well as vectors expressing Gal4 DBD or Gal-Myb (aa 290 to 315). Activity is reported as fold activation over that by Gal DBD (mean ± standard deviation, n = 3).
To evaluate the transcriptional activity of this region, we prepared a Gal4-cMyb construct containing c-Myb residues (aa 290 through 315) fused to the Gal4 DNA binding domain (aa 1 through 147). Following cotransfection with a 5×Gal4 luciferase reporter, the Gal-Myb (aa 290 through 315) polypeptide stimulated target gene activity in 293 cells sevenfold compared to the Gal4 DNA binding domain alone (Fig. 1B).
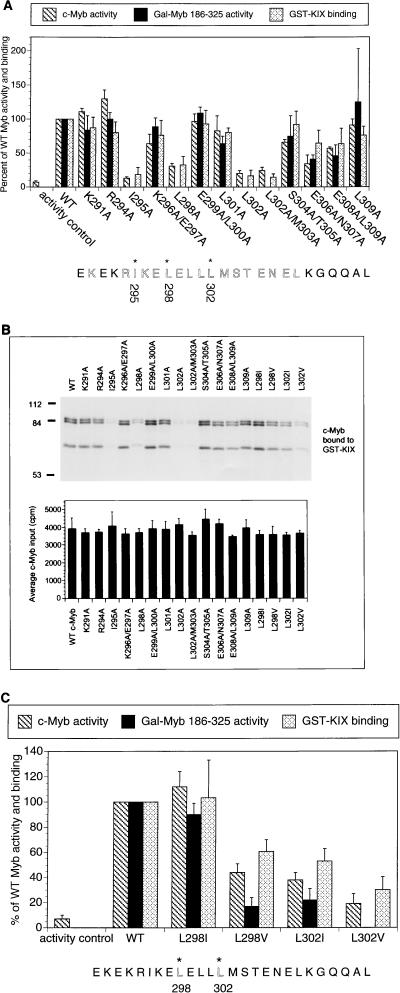
The ability of c-Myb (aa 290 through 315) to stimulate target gene activity and to bind to KIX prompted us to evaluate the functional importance of this region in the context of the full-length c-Myb protein by site-directed mutagenesis. Using a c-Myb-responsive reporter construct (CD13/APN), we identified three hydrophobic residues (Ile295, Leu298, and Leu302) that blocked target gene activation 70 to 90% when mutated individually to alanine (Fig. 2A). Alanine substitutions at basic or acidic residues within the same region (aa 290 to 315) had only minimal effects on c-Myb activity, however, suggesting that hydrophobic contacts with CBP are selectively required for target gene activation. The same three hydrophobic residues (Ile295, Leu298, and Leu302) also appear to function importantly in the context of a GAL4:c-Myb (186-325) polypeptide, arguing against nonspecific effects of these mutations on nuclear targeting or DNA binding activities of the full-length c-Myb protein (Fig. 2A). Indeed, mutant c-Myb polypeptides containing alanine substitutions at any of the three hydrophobic residues (Ile295, Leu298, and Leu302) were unable to interact efficiently with GST-KIX polypeptides in pulldown assays, indicating that target gene activation via these amino acids is CBP/P300 dependent (Fig. 2A and B).
FIG. 2.
Hydrophobic residues in c-Myb are necessary for formation of a complex with KIX and for target gene activation. (A) Alanine mutagenesis reveals that Ile295, Leu298, and Leu302 are necessary for c-Myb function. Shown are activities of mutant c-Myb polypeptides, either in the context of full-length c-Myb (c-Myb activity; hatched bars) or as a Gal-Myb (aa 186 to 325) fusion protein (Gal-Myb 186-325 activity; black bars). Full-length c-Myb activity was evaluated by using a CD13/APN luciferase reporter, whereas Gal4-Myb activity was assessed with a 5×Gal4-luciferase reporter in 293 cells. The activity controls are empty expression vector control for the c-Myb transactivation assays and Gal DNA binding domain expression vector for the Gal-Myb 186-325 activity assays. Activity is compared to that of wild-type (WT) c-Myb (set at 100%). In vitro binding assays were performed by using full-length c-Myb and GST-KIX polypeptides (GST-KIX binding; stippled bars). Binding of WT c-Myb to KIX is compared to those by mutant c-Myb polypeptides. Asterisks at the bottom indicate residues in c-Myb critical for transcriptional activity and for KIX binding. Results for transcriptional activity are presented as means ± standard deviations (n ≥ 2). The GST-KIX pulldown data are presented as means ± standard deviations (n = 3). Comparable expression of mutant c-Myb polypeptides, either in the context of the full-length protein or as GAL4-Myb chimeras, was verified by Western blot assay of transfected cells (data not shown). (B) Top, representative autoradiogram obtained following SDS-PAGE of 35S-labeled c-Myb proteins bound to GST-KIX resin. Bottom, average input (40% of total, expressed as counts per minute) for each c-Myb polypeptide. Data are expressed as means ± standard deviations (n = 2). Bands of 84 and 85 kDa represent full-length c-Myb polypeptides. Although the precise amino acid endpoints were not determined, the 65-kDa band represents a C-terminally truncated c-Myb polypeptide extending through the first 600 amino acids of c-Myb and containing the KIX binding domain (aa 295 to 315). (C) Leu302 in c-Myb forms specialized contacts with the KIX domain of CBP. The effects of conservative substitutions at Leu298 and Leu302 in c-Myb on target gene activation and complex formation with KIX are shown. Activities are shown as percentages of that for WT c-Myb (hatched bars) or Gal4 Myb 186-325 (black bars) polypeptides. Results from pulldown assays (see Fig. 2B) with GST-KIX resin are also shown (stippled bars).
To evaluate the specificity with which hydrophobic residues in c-Myb bind to KIX, we prepared mutant c-Myb polypeptides containing conservative substitutions at either Leu298 or Leu302. Mutagenesis of Leu298 to isoleucine had a minimal impact on target gene activation or formation of a complex with KIX in vitro, but a valine substitution at the same position inhibited binding of mutant c-Myb to KIX 40% and blocked reporter induction two- to fivefold (Fig. 2C). By contrast, mutation of Leu302 to either isoleucine or valine compromised both transactivation and KIX binding activities of c-Myb, suggesting that Leu302 forms specialized contacts with KIX (Fig. 2C). In this regard, the KIX binding domain of c-Myb has modest sequence similarity with the αB region of KID, particularly in the spacing of hydrophobic residues that are critical for association with KIX (Fig. 3). In this alignment, Leu302 of c-Myb should correspond to Leu141 in CREB, a critical residue that anchors the KID domain by projecting into a deep hydrophobic pocket in KIX (16).
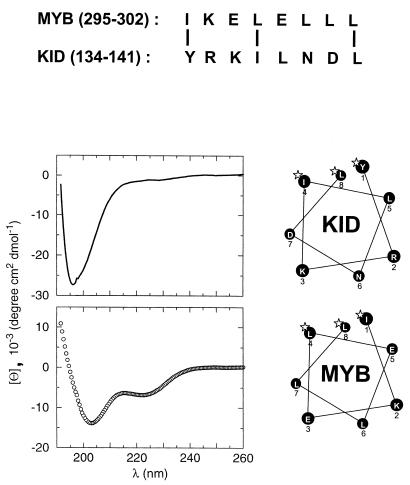
FIG. 3.
The KIX binding domain of c-Myb (aa 290 to 315) forms an amphipathic helix that has similarity with the αB region of KID. Top, sequence alignment of c-Myb (aa 295 to 302) and KID (aa 134 to 141). Bottom left, circular dichroism spectra of the αB region of KID (aa 128 to 147) (solid line, top graph) and c-Myb activating region (aa 290 to 315) (circles, bottom graph) recorded under identical conditions. The helical content of c-Myb is more than one order of magnitude greater than that of KID, as indicated by its θ220/θ230 ratio. Bottom right, helical wheel diagrams of KID (134-141) αB in the complexed state and c-Myb. Homologous residues essential for KIX binding are marked with asterisks.
The importance of a random coil-to-helix transition in the αB region of phospho(Ser133)KID for high-affinity interaction with KIX (13) prompted us to examine whether the KIX binding domain in c-Myb also folds into a helical structure. Circular dichroism spectroscopy revealed that the helical content of this domain (aa 290 to 315), estimated by its θ222-nm/θ203-nm ratio, was about 30% (Fig. 3). By contrast, the αB region in KID (aa 128 to 147) was only 1% helical, reflecting the random coil structure of this peptide in the unbound state (12) (Fig. 3). Helical wheel analysis of the c-Myb (aa 290 to 315)-activating region indicates that this peptide may form an amphipathic helix, with the three functionally important hydrophobic residues (Ile295, Leu298, and Leu302) clustering on one face and, like Tyr134, Ile137, and Leu141 in CREB, interacting with the hydrophobic groove in KIX (Fig. 3).
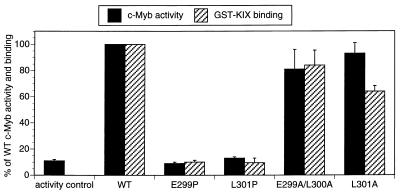
The ability of c-Myb to form an amphipathic helix led us to examine the importance of helical structure for complex formation and for target gene activation. We selected residues in c-Myb (Leu301 and Glu299) that do not appear to form surface contacts with KIX, because alanine substitutions at these positions have little effect on complex formation or target gene activation (Fig. 4). Consistent with the notion that the secondary structure of c-Myb is essential for its activation properties, mutagenesis of either Leu301 or Glu299 to proline, however, severely compromised target gene activation and disrupted KIX binding in vitro (Fig. 4).
FIG. 4.
Helical structure of the c-Myb activating region is critical for complex formation with KIX and for target gene activation. The bar graph shows KIX binding activity of full-length wild-type (WT) and mutant c-Myb polypeptides (stippled bars). Effects of mutations with the helix-sparing amino acid alanine and the helix-breaking amino acid proline on KIX binding activity are shown. Binding of 35S-labeled c-Myb polypeptides to GST-KIX resin was quantitated and expressed as the percentage of WT c-Myb binding activity. Transient luciferase assays were performed on 293 cells transfected with WT and mutant c-Myb polypeptides (hatched bars) containing alanine or proline substitutions at amino acid positions listed under each bar. Luciferase activities derived from the CD13/APN-Luc reporter gene were normalized to β-galactosidase activity derived from cotransfected RSV-β-Gal control plasmid and expressed as percentages of WT c-Myb activity. Results are shown as means ± standard deviations (n = 3). Mutant and WT c-Myb polypeptides were comparably expressed in transfected 293 cells (data not shown).
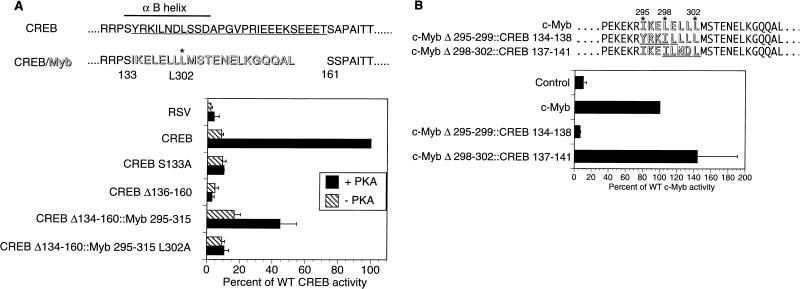
The presence of comparably positioned hydrophobic residues in both c-Myb (aa 290 to 315) and KID (αB) helices prompted us to evaluate whether these domains could functionally substitute for one another. In transient assays of F9 cells, wild-type CREB stimulated a CRE-CAT reporter plasmid 12-fold in response to PKA (7), and deletion of αB sequences in KID (aa 136 to 160) blocked reporter induction, demonstrating the importance of this region for CBP/P300 recruitment (Fig. 5A).
FIG. 5.
A putative α-helix in c-Myb is functionally similar to the αB region of CREB. (A) Insertion of a c-Myb fragment containing a putative α-helix rescues PKA inducibility from an inactive CREB polypeptide lacking the αB helix. F9 cells were transfected with CRE-CAT reporter and expression vectors for either CREB, CREB S133A (nonphosphorylatable), CREB Δ136-160 (lacking the αB helix), CREB Δ134-160::Myb 295-315, or CREB Δ134-160::Myb 295-315 L302A (containing the Leu302-to-Ala mutation in the Myb insert). Cotransfection with wild-type (WT) PKA (+ PKA) or catalytically inactive PKA (− PKA) expression vector is indicated. Activities are reported as means ± standard deviations (n ≥ 3) compared to that of WT CREB with PKA (100%). RSV is an empty expression vector control. CREB residues that form the αB helix are indicated by a thick line; residues deleted in the CREBΔ136-160 mutant are indicated by the thin underline; c-Myb residues (295 through 315) that were inserted into CREBΔ134-160 are shown in shadowed type (CREB/Myb). The positions of CREB aa S133 and S161 and c-Myb aa L302 are shown. (B) A portion of the CREB αB helix can substitute for the comparable region of the putative α-helix of c-Myb. Activity was tested on the c-Myb-responsive CD13/APN Luc reporter in 293 cells, along with cytomegalovirus expression vectors for either c-Myb, c-MybΔ295-299::CREB134-138, c-MybΔ298-302::CREB137-141, or an empty vector (Control). c-Myb (outlined type) sequences replaced by CREB residues (underlined and outlined type) are indicated. Activities are reported relative to WT c-Myb (100%) and expressed as means ± standard deviations (n ≥ 2).
Inserting c-Myb (295-315) in place of αB partially restored PKA inducibility in the context of full-length CREB protein (Fig. 5A). Reflecting the constitutive helical structure of Myb (aa 295 to 315), the basal activity of the CREB::Myb295-315 chimera was also two- to threefold higher than that of wild-type CREB (Fig. 5A). By contrast, substitution of a KIX interaction-defective c-Myb L302A mutant fragment for αB had no such effect, indicating that both constitutive and PKA-inducible activities of the CREB::Myb295-315 chimera are dependent on recruitment of CBP (Fig. 5A). In reciprocal exchange experiments, residues 137 to 141 in the αB region of CREB (ILNDL) were found to substitute for c-Myb residues 298 to 302 (LELLL) in the context of the full-length c-Myb protein (Fig. 5B). However, αB residues 134 to 138 (YRKIL) did not support c-Myb activity when inserted in place of aa 295 to 299 (IKELE) in c-Myb, potentially reflecting a negative determinant of helicity in this region of CREB (Fig. 5B).
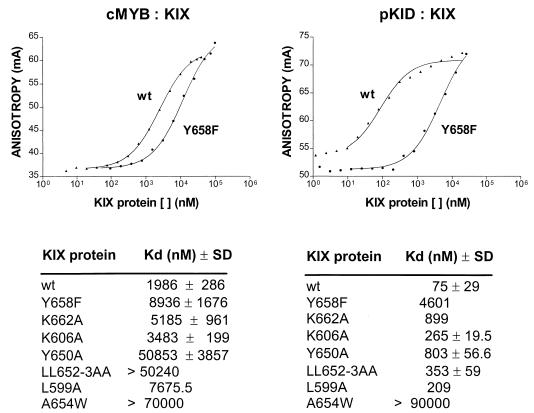
Structure-function studies showing that an amphipathic αB helix in KID articulates with a shallow hydrophobic groove in KIX (13, 16) led us to examine whether c-Myb binds to KIX by forming similar contacts. To evaluate the relative importance of surface residues in KIX for formation of a complex with c-Myb and CREB, we performed fluorescence anisotropy experiments. In equilibrium binding assays, an fl-Myb (aa 276 to 315) peptide associated with KIX with a predicted Kd of 2 ± 0.3 μM, whereas a mutant L302A fl-Myb did not bind detectably (Kd > 50 μM; data not shown) to KIX under the same conditions (Fig. 6). By contrast, the Kd for complex formation between fl-phospho(Ser133)KID (aa 88 to 160) and KIX was 75 ± 30 nM, suggesting that KID forms surface contacts with KIX additional to those formed by c-Myb.
FIG. 6.
Apparent affinity of wild-type (wt) and mutant KIX polypeptides for complex formation with fl-c-Myb (276-315) or KID (88-160) peptides, as indicated. Top, representative curve for binding of wt and Y658F mutant KIX polypeptides to each peptide. Bottom, table showing calculated Kd values for binding of wt and Y658F, K662A, K606A, Y650A, and LL652-3AA mutant KIX polypeptides. Results show means for measurements from three or more independent experiments with standard deviations. Total fluorescence intensity in each experiment remained constant with increasing concentrations of KIX protein.
On average, alanine substitution at hydrophobic residues (Leu652, Leu653, Lys606, and Tyr650) within the shallow groove of KIX lowered the Kd values for both CREB and c-Myb binding, although these effects were more drastic in the case of c-Myb (Fig. 6B). Tyr650 appeared to be the most important of the shallow groove residues for complex formation, reflecting its combined contribution to both shallow groove structures and deep pocket structures in KIX. Correspondingly, mutagenesis of Tyr650 in KIX to alanine reduced the apparent affinity about 10-fold for KID (Kd = 803 ± 57 nM) and about 25-fold for c-Myb (Kd = 51 ± 3.9 μM) (Fig. 6).
By contrast with these shallow groove contacts, residues in KIX that coordinate with the phosphate moiety in CREB did not appear to be essential for c-Myb binding. Notably, Tyr658 forms a hydrogen bond with the Ser133 phosphate group in CREB, and mutagenesis of this residue to phenylalanine lowered the apparent Kd for phospho(Ser133)KID 60-fold, from 75 nM to 4.6 μM (13) (Fig. 6). Mutagenesis of Tyr658 to Phe in KIX reduced c-Myb binding only 4.5-fold, however, indicating that hydrogen bond contributions from the Tyr658 hydroxyl group are unlikely to stabilize complex formation with c-Myb significantly.
Lys662 in KIX forms a salt bridge with the Ser133 phosphate, and substitution of Lys662 with alanine disrupted binding to phospho(Ser133)KID 12-fold (Kd = 0.9 μM) (13) (Fig. 6), but complex formation between fl-Myb and Lys662Ala KIX was reduced only 2.6-fold. Taken together, these results indicate that, whereas binding of KIX to KID requires both phosphate group interactions and shallow groove interactions, binding to c-Myb may depend primarily on hydrophobic contacts via the shallow groove in KIX (Fig. 6).
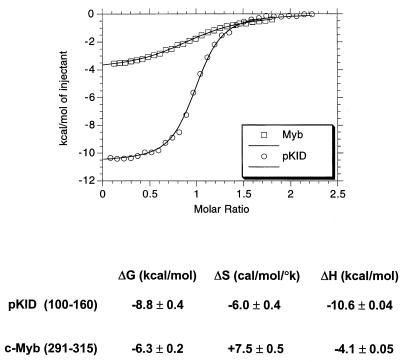
Differences in secondary structure between the unbound c-Myb and KID domains prompted us to compare the relative thermodynamics of formation of complexes with KIX. In agreement with Kd measurements obtained by fluorescence anisotropy, the ΔG for the KID/KIX complex evaluated by ITC was −8.8 kcal/mol, whereas it was −6.0 kcal/mol for the c-Myb/KIX complex (Fig. 7). Reflecting in part the random coil-to-helix transition in KID, binding of phospho(Ser133)KID to KIX entailed a substantial entropy penalty of −6 cal/mol/°K that offset the high enthalpy of complex formation (ΔH = −10.6 kcal/mol ± 0.5%).
FIG. 7.
Thermodynamics of complex formation with KIX differ for CREB and c-Myb. ITC results for KID(100-160):KIX and Myb(291-315):KIX complexes are shown. Top, graph shows heat (kilocalories per mole) generated with increasing ratio of c-Myb to KIX (squares) or phospho(Ser133)KID to KIX (circles). The midpoint of the ITC curve corresponds to the stoichiometric ratio. Bottom, data are calculated free energy (ΔG, expressed in kilocalories per mole), enthalpy (ΔH, expressed in kilocalories per mole), and entropy (ΔS, expressed in calories per mole per degree kelvin) components for each complex. The enthalpy was obtained from the fitting process, while the free energy and entropy were calculated as described in the Materials and Methods section. The Kd measured by fluorescence anisotropy method for the same phospho(Ser133)KID peptide agrees within the range of experimental error with that measured by ITC. Any apparent discrepancy between the results of the two methods can be attributed to the differences in solution conditions (temperature, pH, and ionic strength) and protein-peptide constructs used.
By contrast, binding of c-Myb (aa 291 to 315) to KIX was accompanied by a large positive change in entropy (ΔS = +7.5 cal/mol/°K) that compensated for the relatively low enthalpy of complex formation (ΔH = −4.1 kcal/mol ± 1%) (Fig. 7). Preliminary nuclear magnetic resonance (NMR) analysis indicates that the secondary structure of KIX remains unchanged in the free and bound states for both KID:KIX and c-Myb:KIX complexes (23). Thus, the entropy of complex formation, in both cases, reflects the positive contribution of removal of ordered water molecules from the hydrophobic surfaces offset by the negative contribution of conformational changes in the activation domains. The dramatic rearrangement of pKID was observed directly by NMR (16) and by circular dichroism (Fig. 3) as well as indirectly by ITC (Fig. 7). Taken together, these results indicate that formation of a complex between KID and KIX is enthalpy driven, whereas the c-Myb:KIX complex is dependent on both enthalpy and entropy components.
DISCUSSION
Activating regions have been proposed to interact with the transcriptional apparatus via simple interactions that do not require elements of secondary structure (15). Our results indicate, on the contrary, that the constitutive and inducible properties of certain activators in part reflect their intrinsic helical propensity.
Both CREB and c-Myb appear to stimulate transcription via surface contacts with hydrophobic residues lining the shallow groove in KIX. Fluorescence anisotropy and ITC experiments reveal that c-Myb binds to KIX with affinity nearly 50-fold lower than that of CREB, due in large part to the absence of hydrogen bond and salt bridge contributions from Tyr658 and Lys662 in KIX with the phosphate moiety. Indeed, binding of phospho(Ser133)CREB to mutant Y658F KIX is comparable to that of c-Myb, suggesting that the Ser133 phosphate interactions account, in large part, for affinity differences between c-Myb and CREB.
Why then is CREB unable to bind KIX in the unphosphorylated state? Circular dichroism and NMR studies reveal that KID assumes a random coil structure in the unbound state, and ITC experiments indicate that the helical transition in KID entails a substantial entropy cost that is not compensated by hydrophobic contacts with the shallow groove sufficiently to permit KIX binding. The ΔS for formation of a complex between KID and KIX thus effectively precludes recruitment of CBP to the promoter in unstimulated cells and ensures low levels of target gene expression in the basal state. Indeed, replacement of the αB region in KID with the helical c-Myb (aa 290 to 315) region stimulated basal reporter expression, demonstrating the importance of the structural transition in CREB for optimal transcriptional control.
The difference between KID and c-Myb in affinity for KIX (50-fold) is difficult to reconcile with their comparable activities in transient-transfection assays. However, this discrepancy may be explained, at least in part, by differing kinetic profiles for transcriptional activation via the two factors. Cyclic AMP stimulates CREB phosphorylation and target gene expression with burst-attenuation kinetics: CREB activity is maximal within 15 to 30 min of induction, but declines to baseline levels after 2 to 4 h, due to Ser133 dephosphorylation by the Ser/Thr phosphatase PP-1 (7a). By contrast, c-Myb stimulates target gene expression constitutively, and the cumulative effect of Myb activity may therefore be comparable to that of CREB.
Remarkably, the KIX interaction domain of c-Myb contains an LXXLL motif (aa 298 to 302) that has been shown to mediate binding of members of the SRC family of coactivators to the nuclear hormone receptors (5, 10, 19). Crystal structures of liganded SRC-1:PPARγ and GRIP-1:TR complexes reveal that the LXXLL motif folds into a helical structure that articulates with a shallow groove in the ligand binding domain of each type of receptor (5, 10). Although it is unclear whether c-Myb interacts functionally with certain nuclear receptors, the importance of this motif for KIX binding illustrates a potentially conserved mechanism by which unrelated activators recruit the transcriptional apparatus. In this regard, it should be of considerable interest to evaluate whether the presence of an LXXLL motif in c-Myb contributes to cross-coupling with nuclear receptor pathways (14), whereby certain nuclear receptors compete with CBP for binding to the activation domain of c-Myb.
ACKNOWLEDGMENTS
We thank Meghan Mitchell, Stephen Kelly, Xiaoying Wang, Mike Long, and Geli Gao for technical assistance. We also thank Steve Harrison and Chris Walsh for helpful suggestions and Gerry Zambetti and Jan van Deursen for comments on the manuscript.
This work was supported by NIH grants CA70909 (L.H.S.), RO1 CA76385 (P.K.B.), and R01GM37828 (M.M.), by National Cancer Institute Cancer Center Support (CORE) grant P30 CA21765, and by the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital.
D.P. and M.R. contributed equally to this work.
REFERENCES
- 1.Akimaru H, Chen Y, Dai P, Hou D, Nonaka M, Smolik S, Armstrong S, Goodman R, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 2.Arias J, Alberts A, Brindle P, Claret F, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–228. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 3.Brindle P, Linke S, Montminy M. Analysis of a PK-A dependent activator in CREB reveals a new role for the CREM family of repressors. Nature. 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 4.Dai P, Akimaru H, Tanaka Y, Hou D, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 5.Darimont B, Wagner R, Apriletti J, Stallcup M, Kushner P, Baxter J, Fletterick R, Yamamoto K. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez G A, Menzel P, Leonard J, Fischer W H, Montminy M R. Characterization of motifs which are critical for activity of the cyclic AMP-responsive transcription factor CREB. Mol Cell Biol. 1991;11:1306–1312. doi: 10.1128/mcb.11.3.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 7a.Hagiwara M, Alberts A, Brindle P, Meinkoth J, Feramisco J, Deng T, Karin M, Shenolikar S, Montminy M. Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell. 1992;70:105–113. doi: 10.1016/0092-8674(92)90537-m. [DOI] [PubMed] [Google Scholar]
- 8.Kasper L H, Brindle P K, Schnabel C A, Pritchard C E J, Cleary M L, van Deursen J M A. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwok R, Laurance M, Lundblad J, Goldman P, Shih H, Connor L, Marriott S, Goodman R. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 10.McInerney E, Rose D, Flynn S, Westin S, Mullen T, Krones A, Inostroza J, Torchia J, Nolte R, Assa-Munt N, Milburn M, Glass C, Rosenfeld M. Determinants of coactivator LXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliner J, Andresen J, Hansen S, Zhou S, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 12.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker D, Jhala U, Radhakrishnan I, Yaffe M, Reyes C, Shulman A, Cantley L, Wright P, Montminy M. Analysis of an activator: coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- 14.Pfitzner E, Kirfel J, Becker P, Rolke A, Schule R. Physical interaction between retinoic acid receptor and the oncoprotein Myb inhibits retinoic acid-dependent trans-activation. Proc Natl Acad Sci USA. 1998;95:5539–5544. doi: 10.1073/pnas.95.10.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan I, Perez-Alvarado G, Parker D, Dyson H J, Montminy M, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator-coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 17.Sadowski I, Bell B, Broad P, Hollis M. Gal4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro L. Myb and Ets proteins cooperate to transactivate an early myeloid gene. J Biol Chem. 1995;270:8763–8767. doi: 10.1074/jbc.270.15.8763. [DOI] [PubMed] [Google Scholar]
- 19.Torchia J, Rose D, Inostroza J, Kannei Y, Westin S, Glass C K, Rosenfield M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, Reece R, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, Shapiro L, Rivera M, Kumar A, Brindle P K. A role for CREB binding protein and P300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Vinkemeier U, Gu W, Chakravarti D, Horvath C, Darnell J. Two contact regions between Stat1 and CBP/P300 in interferon signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zor, T., and P. E. Wright. Unpublished observations.