Abstract
Lichens are the result of a symbiotic interaction between fungi (mycobionts) and algae (phycobionts). Aside from mycobionts, lichen thalli can also contain non-lichenised fungal species, such as lichenicolous and endolichenic fungi. For this study, three surveys were conducted in China’s Yunnan Province and Inner Mongolia Autonomous Region between 2017 and 2020. Several samples of four lichen species were collected during these surveys: Candelariafibrosa, Flavoparmeliacaperata, Flavopuncteliaflaventior and Ramalinasinensis. Six isolates of Coniochaeta were recovered from these four lichen species. The phylogenetic and morphological analyses revealed that two of these isolates were previously identified species, Coniochaetavelutinosa and C.acaciae. Those remaining were from potentially unknown species. We used molecular and morphological data to describe these previously-unknown species as Coniochaetafibrosaesp. nov., C.mongoliaesp. nov. and C.sinensissp. nov. The findings of this study significantly improve our understanding of the variety and habitat preferences of Coniochaeta in China and globally.
Keywords: Coniochaetaceae , lichens, molecular phylogeny, Mongolia, Yunnan Province
Introduction
Lichens are a symbiotic relationship between heterotrophic fungi and algae (including cyanobacteria) that are usually referred to as mycobiont and phycobiont, respectively (Nash and Thomas 2008; Tripathi and Joshi 2019). Lichens exhibit a diversity of colours, thallus morphology and fruiting bodies (Ahmadjian 1993). Lichens have a limited fossil record, yet recent molecular-clock analyses suggested their being at least 250 million years old (Nelsen et al. 2020). Apart from the mycobionts, a lichen thallus can also house non-lichenised fungal species, such as lichenicolous and endolichenic fungi. The former utilise lichens as their hosts (Lawrey and Diederich 2003), whereas the latter behave similar to ‘endophytes’ (Arnold et al. 2009; Suryanarayanan and Thirunavukkarasu 2017). Various species of Coniochaeta are examples of endolichenic fungi (Zhang et al. 2016; Harrington et al. 2019).
Coniochaeta is a genus of pleomorphic yeasts belonging to the Coniochaetales (Ascomycota) with global distribution (García et al. 2006; Damm et al. 2010; Raja et al. 2012; Vazquez-Campos et al. 2014; Nasr et al. 2018; Harrington et al. 2019). This genus has distinct asexual and sexual states in its life cycle. Previously, the genus Lecythophora was erected to include asexual states of Coniochaeta (Weber 2002). After the dual nomenclature of pleomorphic fungi was discontinued (Hawksworth 2011), following the principle of priority, these genera were reclassified under Coniochaeta (Khan et al. 2013; Réblová et al. 2016).
The sexual state of Coniochaeta is characterised by dark brown to black ascomata with setae. These ascomata can either be pyriform ostiolate or globose non-ostiolate. Asci are thin-walled, producing single-celled, smooth ascospores with an elongated embryo crack (García et al. 2006; Asgari et al. 2007). In contrast, the asexual state of Coniochaeta has distinctive pink salmon to dark brown colonies producing phialidic conidiogenous cells (Checa 1988; Damm et al. 2010; Khan et al. 2013). Coniochaeta has been isolated from various substrates, such as butter, faeces, wood, soil, uranium wastewater, plants and lichens (Weber 2002; García et al. 2006; Vazquez-Campos et al. 2014; Harrington et al. 2019). Some Coniochaeta species are also known to be human and animal pathogens (Hoog et al. 2000; Perdomo et al. 2013; Troy et al. 2013).
Several Coniochaeta species have been isolated from Asia (Kamiya et al. 1995; García et al. 2006; Asgari et al. 2007). Previously, three undescribed Coniochaeta species were identified from China growing on plant litters and herbivore faeces, but none associated with liches (Chang and Wang 2011; Hyde et al. 2020). In this study, six isolates of Coniochaeta species were recovered from four lichen species collected from the Yunnan Province and the Inner Mongolia Autonomous Region of China. Analyses of molecular and morphological data indicated these six isolates represented five species of Coniochaeta. Amongst these were two previously-described taxa, C.velutinosa and C.acaciae, whereas the remaining three were undescribed. Here, we describe these species as Coniochaetamongoliae sp. nov., C.sinensis sp. nov. and C.fibrosae sp. nov. This study substantially augments our current knowledge on the diversity and host range of Coniochaeta and endolichenic fungi from China.
Materials and methods
Collection of lichen samples
Between 2017 and 2020, three surveys were conducted in the Yunnan Province and Inner Mongolia Autonomous Region of China. During these surveys, multiple samples of four lichens species were collected. Samples of Flavoparmeliacaperata (2017), Flavopuncteliaflaventior (2017) and Candelariafibrosa (2020) were collected from the Yunnan Province, whereas Ramalinasinensis was collected from the Inner Mongolia Autonomous Region in 2019. During their transit, all lichen samples were stored separately in paper bags.
Isolation of fungi from lichen thalli
All lichen samples were repeatedly rinsed with tap water followed by deionised water. Using a Leica Zoom 2000 stereomicroscope, the upper cortex was scraped off with a sterile blade. The medullary layer was carefully dissected and rinsed using sterile deionised water. Thereafter, these medullary tissues were placed on to 2% potato dextrose agar (PDA) plates, amended with 0.05% streptomycin. All Petri plates were incubated for 14 days at 25 °C. Hyphal tips of mycelia emerging from the medullary tissues were sub-cultured on to fresh PDA plates.
Ex-holotype cultures of undescribed fungal species, described in this study, were deposited in the China General Microbiological Culture Collection Center (CGMCC), Beijing, China. The holotype specimens were deposited in the culture collection of the Institute of Microbiology (HMAS), Beijing, China (Accession numbers are listed in Table. 1).
Table 1.
GenBank accession numbers Coniochaeta species used for the phylogenetic analyses. T = ex-type isolates.
| Taxa | Strain | HMAS | GenBank accession number | ||
|---|---|---|---|---|---|
| LSU | ITS | ||||
| Coniochaeta acaciae | MFLUCC 17-2298T | MG062737 | MG062735 | ||
| C. acaciae | CX37 | MW750757 | MW750761 | ||
| C. africana | CBS:120868T | NG_066150 | NR_137725 | ||
| C. angustispora | CBS:144.70 | MH871308 | MH859528 | ||
| C. arenariae | MFLUCC 18-0405T | MN017893 | - | ||
| C. baysunika | MFLUCC 17-0830T | MG828996 | MG828880 | ||
| C. boothii | CBS:381.74T | AJ875226 | NR_159776 | ||
| C. cateniformis | UTHSC 01-1644T | HE610329 | NR_111517 | ||
| C. cephalothecoides | L821 | KY064030 | KY064029 | ||
| C. coluteae | MFLUCC 17-2299T | MG137252 | MG137251 | ||
| C. cruciata | FMR 7409 | AJ875222 | - | ||
| C. cymbiformispora | NBRC 32199 | LC146726 | LC146726 | ||
| C. cipronana | CBS:144016T | - | NR_157478 | ||
| C. decumbens | CBS:153.42T | NG_067257 | NR_144912 | ||
| C. dendrobiicola | DLCCR7 | MK225603 | MK225602 | ||
| C. discoidea | CBS:158.80T | NG_064120 | NR_159779 | ||
| C. discospora | CBS:168.58 | MH869278 | MH857740 | ||
| C. ellipsoidea | CBS:137.68T | MH870804 | MH859091 | ||
| C. endophytica | AEA 9094T | EF420069 | EF420005 | ||
| C. euphorbiae | CBS:139768 = 1001T | - | KP941076 | ||
| C. extramundana | CBS:247.77T | MH872828 | MH861057 | ||
| C. fasciculata | CBS:205.38T | FR691988 | NR_154770 | ||
| C. fibrosae | CGMCC3.20304T | 350271 | MW750758 | MW750760 | |
| C. fibrosae | CX04D1 | MW750755 | MW750756 | ||
| C. fodinicola | FRL = CBS:136963T | KF857172 | JQ904603 | ||
| C. gigantospora | ILLS:60816T | JN684909 | JN684909 | ||
| C. hansenii | CBS:885.68 | AJ875223 | - | ||
| C. hoffmannii | CBS:245.38T | AF353599 | NR_167688 | ||
| C. iranica | CBS:139767 = 0806T | - | KP941078 | ||
| C. krabiensis | MFLU 16-1230T | MN017892 | - | ||
| C. leucoplaca | CBS:486.73 | MH872465 | - | ||
| C. ligniaria | 98.1105 | AF353585 | - | ||
| C. lignicola | CBS:267.33T | NG_067344 | NR_111520 | ||
| C. luteorubra | UTHSC 01-20T | HE610328 | HE610330 | ||
| C. luteoviridis | CBS:206.38T | NG_067348 | NR_154769 | ||
| C. malacotricha | F2106 | AF353589 | - | ||
| C. marina | MFLUCC 18-0408T | MK458765 | MK458764 | ||
| C. mutabilis | CBS:157.44T | NG_042382 | NR_111519 | ||
| C. navarrae | LTA3 = CBS:141016T | KU762326 | KU762326 | ||
| C. nepalica | NBRC 30584T | LC146727 | LC146727 | ||
| C. ornata | FMR7415T | AJ875228 | - | ||
| C. ostrea | CBS:507.70T | NG_064080 | NR_159772 | ||
| C. polymorpha | CBS:132722T | HE863327 | NR_121473 | ||
| C. polysperma | CBS:669.77T | MH872868 | MH861109 | ||
| C. prunicola | CBS:120875T | GQ154602 | GQ154540 | ||
| C. pulveracea | CAB683 | GQ351559 | - | ||
| C. punctulata | CBS:159.80 | MH873024 | MH861254 | ||
| C. mongoliae | CGMCC3.20250T | 350270 | MW077646 | MW077645 | |
| C. rhopalochaeta | CBS:109872T | GQ351561 | - | ||
| C. rosae | TASM:6127T | NG_066204 | NR_157509 | ||
| C. savoryi | CBS:725.74T | MH872627 | MH860890 | ||
| C. simbalensis | NFCCI:4236T | MG917738 | NR_164024 | ||
| C. sinensis | CGMCC3.20306T | 350269 | MW422265 | MW422269 | |
| C. sordaria | CBS:492.73 | MH878380 | - | ||
| C. subcorticalis | CBS:551.75 | AF353593 | - | ||
| C. taeniospora | LTA = CBS:141014T | KU762324 | KU762324 | ||
| C. tetraspora | CBS:139.68 | MH870806 | MH859093 | ||
| C. velutina | CBS:981.68 | MH870991 | MH859264 | ||
| C. velutinosa | Co29 | GU553330 | GU553327 | ||
| C. velutinosa | CGMCC3.20249 | MW346687 | MW298866 | ||
| C. verticillata | CBS:816.71T | AJ875232 | NR_159774 | ||
| C. vineae | KUMCC 17-0322T | - | NR_168225 | ||
| C. canina | UTHSC 11-2460 | NG_042720 | NR_120211 | ||
| Zanclospora jonesii | MFLUCC15-1015T | NG_067549 | KY212753 | ||
| Paragaeumannomyces garethjonesii | MFLUCC 15-1012T | NG_059017 | KY212751 | ||
Morphology and growth studies
Colony morphologies of ex-holotypes, representing four potentially new fungal species, were described from eight-day-old cultures growing at 25 °C. A Leica DM6 compound microscope attached to a Zeiss Axio Imager Z2 camera was used for measuring and photographing microscopic morphological characters. A minimum of 50 conidia and conidiogenous cells per isolate were measured using the software ImageJ (Rasband 1997; Schneider et al. 2012).
For the growth study, ex-holotype isolates were sub-cultured on to PDA and incubated for five days at 25 °C. Thereafter, 5 mm diam. agar plugs were placed at the centre of 90 mm Petri dishes. Three replicates per ex-type isolate were incubated at 5, 10, 15, 20, 25, 30 and 35 °C (± 0.5 °C). The colony diameter of each isolate was measured daily up to the eighth day.
DNA extraction, PCR amplification and sequencing
For all undescribed fungal species, eight-day-old cultures growing at 25 °C were used for the extraction of total genomic DNA using PrepManTM Ultra Sample Preparation Reagent (Applied Biosystems, California, USA), following the manufacturer’s instructions. The complete internal transcribed spacers (ITS) and the partial 28S nuclear ribosomal large subunit rRNA gene (LSU) were amplified using the primer pairs ITS1/ITS4 (White et al. 1990) and LR0R/LR5 (Vilgalys and Hester 1990; White et al. 1990), respectively.
Each 25 μl of PCR reaction included 10.5 μl of PCR grade water, 12.5 μl of 1–5TM 2× High-Fidelity Master Mix (buffer, MgCl2, dNTPs and Taq; Tsingke Co., China), 0.5 μl each of forward and reverse primers and 1 μl DNA template. For both gene regions, PCR amplifications were conducted with an initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 sec, 56 °C for 1 min, 72 °C for 1 min; final extension at 72 °C for 10 min. Positive amplifications were verified using agarose gel electrophoresis.
All the PCR products were sequenced by QingDao MDBio Biotech Co., Ltd., China. The resulting sequences were assembled using Geneious v.10.2.2 (Biomatters, Auckland, New Zealand). Preliminary identification of the sequences was undertaken using the BLAST algorithm (Altschul et al. 1990) available through the NCBI GenBank. All the sequences, generated in this study, were deposited at GenBank (Table 1).
Phylogenetic analyses
For the purpose of phylogenetic analyses, we constructed three separate datasets. These are as follows: a) ITS, b) LSU and c) ITS + LSU. Each dataset included sequences generated in this study and those retrieved from the NCBI GenBank. Where available, ex-type sequences of previously-known Coniochaeta species were added to the datasets. For all three datasets, Paragaeumannomycesgarethjonesii and Zanclosporajonesii were selected as the outgroup taxa (Table 1). All datasets were aligned using MAFFT v. 7 (Katoh and Standley 2013); thereafter, manually adjusted if needed using MEGA v.7 (Kumar et al. 2016). All aligned sequence datasets were deposited to TreeBase (Acc. No 28404).
Software for Maximum Likelihood (ML) and Bayesian Inference (BI) phylogenetic analysis was accessed through the CIPRES Science Gateway platform (Miller et al. 2010). jModeltest 2.2 (Nylander et al. 2008) was used for selecting appropriate substitution models. ML analyses were done using RAxML v. 8.2.4 (Stamatakis 2006; Stamatakis et al. 2008) using the GTR substitution model and 1000 bootstrap replicates. BI analyses were undertaken using MrBayes v.3.2 (Ronquist et al. 2012). Four MCMC chains were run from a random starting tree for five million generations and trees were sampled every 100th generation. A quarter of the sampled trees were discarded during burn-in. The remaining trees were used for constructing consensus trees. The resulting ML and BI trees were viewed with FigTree v.1.4 (Rambaut 2009).
Results
Isolation
In this study, four lichen species were collected from Yunnan Province and the Inner Mongolia Autonomous Region in 2017, 2019 and 2020. A total of six isolates of Coniochaeta were recovered from these four lichen species. These are CX03C1 and CX04D1 from Candelariafibrosa, 8004b from Flavoparmeliacaperata, CS-04 and CS-09 from Ramalinasinensis and CX37 from Flavopuncteliaflaventior.
Preliminary identification of these isolates, using the BLAST algorithm, indicated isolates 8004b and CX37 were known Coniochaeta species, C.velutinosa and C.acaciae, respectively, whereas, CX03C1, CX04D1, CS-04 and CS-09 were potentially undescribed species.
Phylogenetic analyses
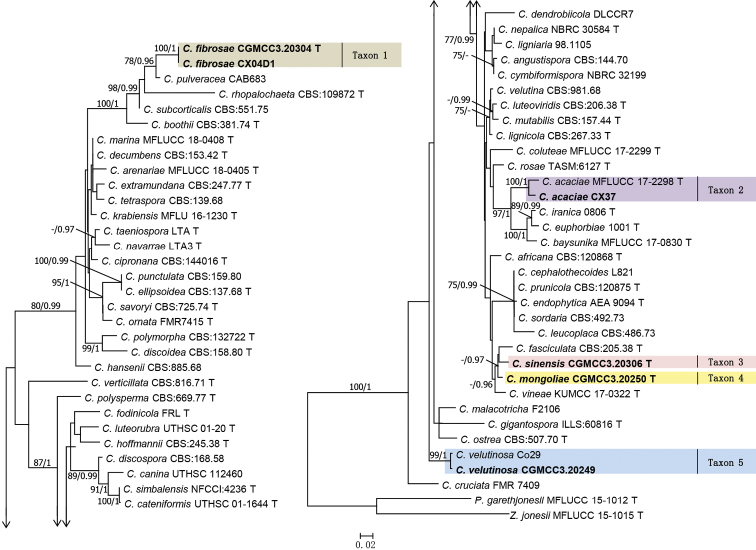
Both single gene and concatenated datasets were used for phylogenetic analyses using ML and BI approaches. The single gene dataset for ITS included 53 taxa, whereas the LSU had 61 taxa. The concatenated dataset included 65 taxa and 1489 characters including gaps (ITS: 1–655; LSU: 656–1489). Individual gene trees for Coniochaeta species had similar topologies and were congruent with the tree generated using the concatenated dataset when taxon sampling overlapped. Bootstrap values < 75% and posterior probability < 0.95 were considered unreliable (Fig. 1, Suppl. material 1 and Suppl. material 2).
Figure 1.
Maximum Likelihood tree constructed using ITS+LSU dataset. Bootstrap support values ≥ 75% and posterior probabilities ≥ 0.95 are indicated above the nodes as ML / PP. The isolates obtained in this study are shown in bold. T = ex-type isolates.
In the phylogenetic trees, constructed using the concatenated dataset, isolates CX03C1 and CX04D1 formed a monophyletic clade (Taxon 1) and sister to C.pulveracea (Fig. 1). Even though, in the phylogenetic trees using a single gene, isolates of Taxon 1 emerged as a monophyletic clade, yet the sister taxon varied. For ITS, C.boothii was found sister to Taxon 1, whereas for LSU, it was C.pulveracea (Suppl. material 1: Fig. S1 and Suppl. material 2: S2).
In the tree constructed using the concatenated dataset, isolate CX37 (Taxon 2) formed a monophyletic clade with C.acaciae with high statistical support. Similar topologies were also observed in the ITS and LSU trees.
The phylogenetic position of isolates CS-04 (Taxon 3) and CS-09 (Taxon 4) substantially varied across the phylogenetic trees. In the trees using the concatenated dataset, isolates CS-04 (Taxon 3) and CS-09 (Taxon 4) nested within a clade that included C.fasciculata and C.vineae (Fig. 1). In ITS gene trees, isolates CS-04 and CS-09 nested within a clade that included C.coluteae, C.fasciculata and C.vineae (Suppl. material 1). In the LSU trees, isolate CS-04 (Taxon 3) grouped with a clade that included C.leucoplaca, C.cephalothecoides, C.endophytica, C.prunicola and C.sordaria (Suppl. material 2), whereas, isolate CS-09 formed a monophyletic clade with C.mutabilis (Suppl. material 2). Irrespective of the trees, the statistical support for all the groups was unreliable.
Irrespective of the datasets and phylogenetic approaches, isolate 8004b (Taxon 5) was grouped with C.velutinosa (Asgari and Zare 2006) with high statistical support (Fig. 1, Suppl. material 1 and Suppl. material 2).
Taxonomy
Coniochaeta fibrosae
H. L. Si & Y. M. Su sp. nov.
6DC014B1-5414-520D-B337-698C4393AC2C
839390
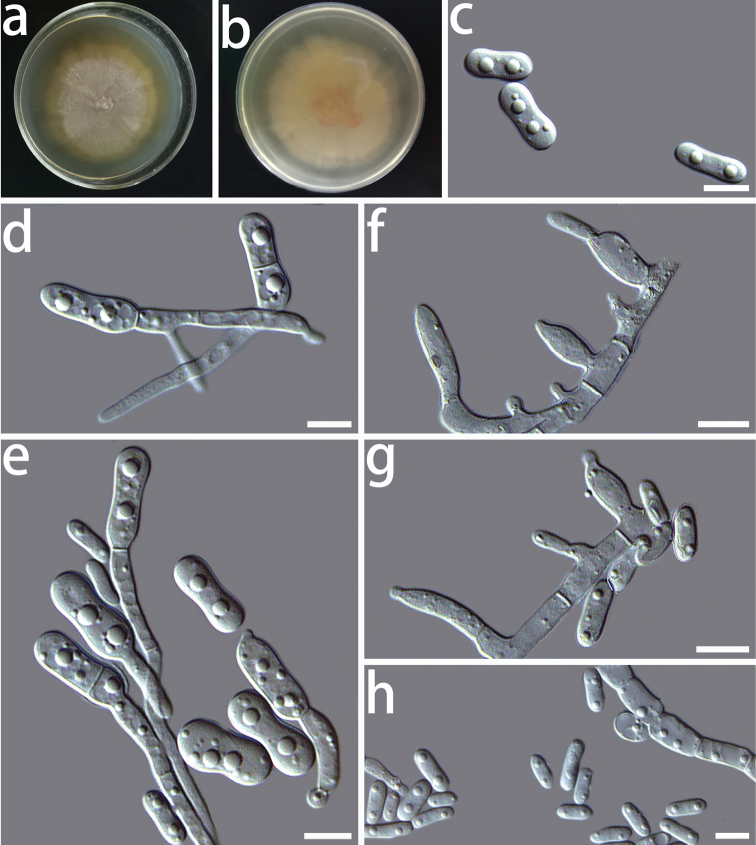
Figure 2.
Morphological characters of Coniochaetafibrosae sp. nov. (HMAS 350271) a, b cultures on PDA from the surface and reverse c swollen conidia d, e swollen conidia germinate hyphae f, g conidiogenous cells h conidia. Scale bars: 10 μm.
Holotype.
China, Yunnan Province: Tiesuo township, 26°32'71"N, 100°57'3"E, ca. 2120 m elev., isolated from Candelariafibrosa, 13 Nov 2020, H. L. Si, CX03C1 (HMAS 350271, holotype), ex-type culture CGMCC3.20304.
Etymology.
The name relates to the lichen Candelariafibrosa and both isolates of this fungus were isolated from its medulla.
Description.
Colony on PDA after 8 d, hyphae hyaline, multi-guttulate, septate, smooth-walled; conidiophores short; conidiogenous cells hyaline, phialidic or oval in shape, single or in clusters on short lateral branches, measuring 2.9–7.2 × 1.8–3.7 μm (x̄= 4.7 × 2.6 μm, n = 50) (Fig. 2f, g); two types of conidia were observed, swollen conidia were hyaline, one-celled, dumb-bell-shaped, with hyphae emerging from both ends (Fig. 2d, e), measuring 7.6–16.5 × 2.3–4.1 μm (x̄ = 9.7 × 3.1 μm, n = 50) (Fig. 2c), oblong conidia were hyaline, one-celled, often oblong to ellipsoidal in shape, measuring 3.4–6.8 × 1.4–2.7 μm (x̄ = 4.7 × 1.8 μm, n = 50) (Fig. 2h). Chlamydospores absent. Sexual morph unknown.
Culture characteristics.
The optimal temperature for growth was 25 °C on PDA. No growth was detected at 5 and 35 °C. Colonies on PDA after 8 d at 25 °C were white, circular, margin entire, flat, dense, partially immersed in the medium and sticky protuberance at the centre of the colony.
Additional specimen examined.
China, Yunnan Province: Tiesuo township, 26°32'71"N, 100°57'3"E, ca. 2120 m elev., isolated from on Candelariafibrosa, 13 Nov 2020, H. L. Si, CX04D1.
Notes.
In the phylogenetic analyses, both isolates of C.fibrosae sp. nov. formed a monophyletic clade, but the sister taxon differed between datasets. These sibling species were either C.boothii (ITS) or C.pulveracea (LSU and concatenated). Both of these sibling species were described, based on their sexual state and chlamydospores (Manoharachary and Ramarao 1973; Romero et al. 1999; García et al. 2006). However, we did not find sexual reproductive structures in our species. As a result, we were unable to compare the morphology of these species.
Coniochaeta sinensis
H. L. Si & Y. M. Su sp. nov.
4E57B146-2AA6-53BF-8D09-25FAD27BF614
839388
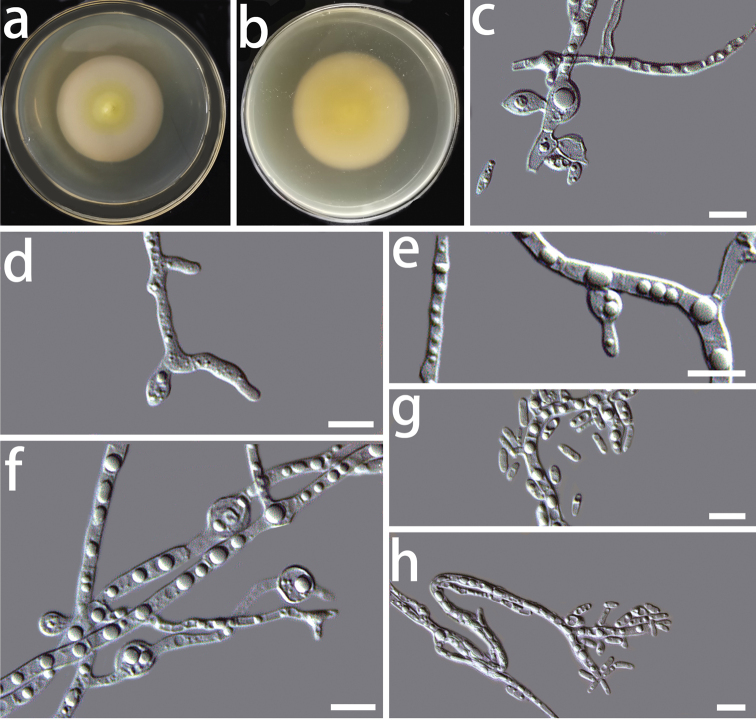
Figure 3.
Morphological characters of Coniochaetasinensis sp. nov. (HMAS 350269) a, b cultures on PDA from the surface and reverse c, d conidiogenous cells e conidiogenous cell that is producing conidia f chlamydospores g, h conidia. Scale bars: 10 μm.
Holotype.
China, the Inner Mongolia Autonomous Region: Chifeng City, 44°13'46"N, 118°44'57"E, ca. 1500 m elev., isolated from the medulla of Ramalinasinensis, 11 Oct 2019, H. L. Si, CS-04 (HMAS 350269, holotype), ex-type culture CGMCC3.20306.
Etymology.
The name relates to the lichen Ramalinasinensis, as a single isolate of this fungus was obtained from the medulla of this lichen.
Description.
Colony on PDA after 8 d, hyphae hyaline, multi-guttulate, septate, smooth-walled, often hyphal strands consolidating to form bundles, conidiophores short or absent; conidiogenous cells hyaline, phialidic or oval in shape, single or in clusters on short lateral branches, measuring 2.8–7.1 × 1.1–3.7 μm (x̄ = 4.2 × 2.3 μm, n = 50) (Fig. 3c–e); conidia hyaline, one-celled, often oblong to ellipsoidal in shape, measuring 2.5–4.6 × 0.7–2.1 μm (x̄ = 3.3 × 1.2 μm, n = 50) (Fig. 3g, h); chlamydospore solitary or in short chains, hyaline, thick-walled, elongate ellipsoidal or almost globose in shape, measuring 3.7–6.6 × 2.5–5.4 μm (x̄ = 4.8 × 3.7 μm, n = 50) (Fig. 3f). Sexual morph unknown.
Culture characteristics.
The optimal temperature for growth is 30 °C. No growth was detected at 5 and 35 °C. Colonies on PDA after 8 d at 30 °C were yellow in the centre and white around the edges, circular, margin entire, flat, dense, partially immersed in the medium, the centre of the colony slightly bulging.
Notes.
Coniochaetasinensis sp. nov. clusters with C.vineae, C.fasciculata and C.mongoliae sp. nov. in our phylogenetic tree, constructed using the concatenated dataset, but the statistical support was insignificant. Amongst these species, C.vineae is only known in its sexual morph (Hyde et al. 2020). There are, however, significant morphological differences amongst C.sinensis sp. nov., C.fasciculata and C.mongoliae sp. nov. These are (1) the shapes and sizes of conidiogenous cells, (2) the shapes and sizes of conidia and (3) the shapes and sizes of chlamydospores. When compared to C.mongoliae sp. nov., C.sinensis sp. nov. has smaller conidiogenous cells and conidia. The conidia of C.sinensis sp. nov. are significantly smaller than those of C.fasciculata (Beyma 1939). Aside from that, the chlamydospores of C.sinensis sp. nov. are longer than those of C.mongoliae sp. nov.
Coniochaeta mongoliae
H. L. Si & Y. M. Su sp. nov.
F4350B40-E623-595F-9E93-A90FC25D35DD
839389
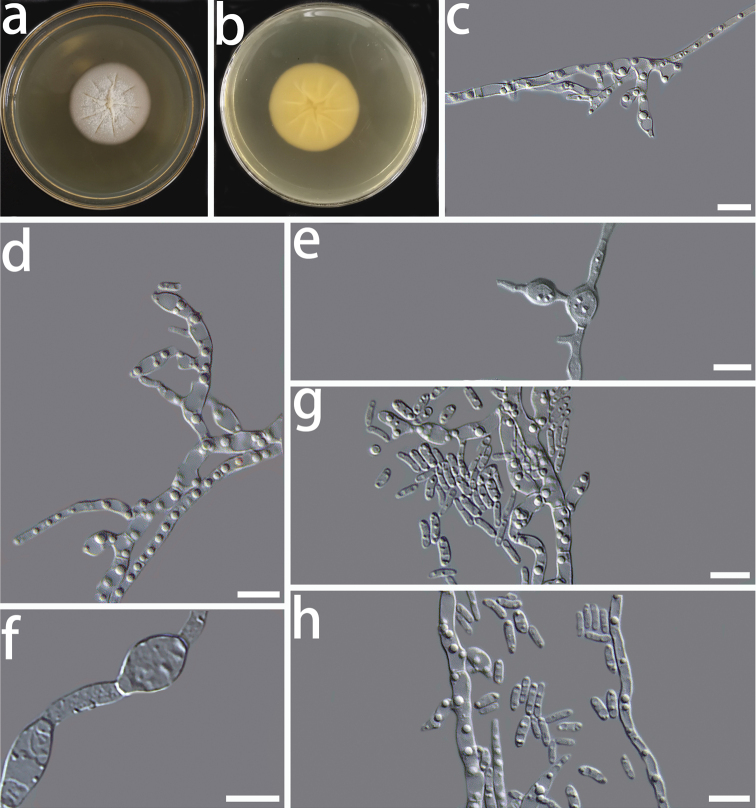
Figure 4.
Morphological characters of Coniochaetamongoliae sp. nov. (HMAS 350270) a, b cultures on PDA from the surface and reverse, c, d conidiogenous cells e, f chlamydospores g, h conidia. Scale bars: 10 μm.
Holotype.
China, the Inner Mongolia Autonomous Region, Chifeng City, 44°13'46"N, 118°44'57"E, ca. 1500 m elev., isolated from the medulla of Ramalinasinensis, 11 Oct 2019, H. L. Si, CS-09 (HMAS 350270, holotype), ex-type living culture, CGMCC 3.20250.
Etymology.
The lichen was collected in the Inner Mongolia Autonomous Region, thus the name.
Description.
Colony on PDA after 8 d, hyphae hyaline, multi-guttulate, septate, smooth-walled, often with hyphal strands consolidating to form bundles; conidiophores short or absent; conidiogenous cells hyaline, flask or acicular in shape, measuring 3.3–12.5 × 1.6–5.1 μm (x̅ = 6.6 × 2.9 μm, n = 50) (Fig. 4c, d); conidia hyaline, smooth-walled, ellipsoidal, 3.3–8.4 × 0.6–1.9 μm (x̅ = 4.8 × 1.3 μm, n = 50) (Fig. 4g and h); chlamydospore solitary or in short chains, hyaline, thick-walled, elongate ellipsoidal or almost globose in shape, measuring 2.7–6.7 × 2.6–5.4 μm (x̅ = 4.6 × 3.8 μm, n = 50) (Fig. 4e, f). Sexual morph unknown.
Culture characteristics.
The optimal temperature for growth is 25 °C. No growth was detected at 5 °C and 35 °C. Colonies on PDA after 8 d at 25 °C were white to light pink in colour, circular, flat, dense, partially immersed in the medium, the centre of the colony is rough, forming radial grooves.
Notes.
In the phylogenetic tree using the concatenated dataset, Coniochaetamongoliae sp. nov. clustered in a clade that included C.sinensis sp. nov., C.vineae and C.fasciculata, but with low statistical support. Moreover, these four species have substantial morphological differences (for details, see the notes for C.sinensis sp. nov.).
Discussion
In the present study, Candelariafibrosa, Flavoparmeliacaperata, Flavopuncteliaflaventior and Ramalinasinensis were collected from the Yunnan and Inner Mongolia Regions of China between 2017 and 2020. We isolated six Coniochaeta isolates from these lichens, which we classified into five species. Two of these were previously-described species, while the other three were unknown. Here, we describe these three previously-unknown species as C.fibrosae sp. nov., C.sinensis sp. nov. and C.mongoliae sp. nov.
The majority of species in the genus Coniochaeta are saprophytes or pathogens of plants and humans, while many others have an unknown ecological function (Harrington et al. 2019). Species of Coniochaeta are frequently isolated from asymptomatic tissues of woody plants and lichens throughout temperate and northern North America (Del Olmo-Ruiz 2012). Some of these species were found exclusively on plants or lichens, such as C.endophytica and C.hoffmannii, respectively (Zhang et al. 2016; Harrington et al. 2019) or on both, such as Coniochaeta sp. Clade 9 (Del Olmo-Ruiz 2012). The two previously-described species recovered in this study (C.acaciae and C.velutinosa) were also isolated from barley leaves in Iran (Asgari and Zare 2006) and dead Acacia species branches in Uzbekistan (Samarakoon et al. 2018). This demonstrates Coniochaeta’s ability to thrive in a variety of habitats, yet their ecological role in all these environments is still largely unknown.
The lack of sequences for protein-coding gene regions is one of the pitfalls in identifying taxa in the genus Coniochaeta. For the majority of species, only ITS and LSU sequences are currently available. Sequences for the largest subunit of RNA polymerase II (rpb1), the second-largest subunit of RNA polymerase II (rpb2), translation elongation factor 1-alpha (tef1) or β-tubulin gene (tub2) were only used in a few studies involving a limited number of species (Spatafora et al. 2006; Voglmayr et al. 2016; Samarakoon et al. 2018; Harrington et al. 2019). Due to the paucity of sequences, we could not include those gene regions in this study. Moreover, even after repeated attempts, we could not amplify the rpb1, rpb2 and tub2 gene regions for the species isolated in this study. Consequently, there is an urgent need for primers that can successfully amplify protein-coding genes from a wide variety of taxa in order to demystify the taxonomy for this genus.
In this study, we identified the isolate CX37 as Coniochaetaacaciae. This is because, in the phylogenies using both concatenated and single-gene datasets, isolate CX37 and ex-type sequences of C.acaciae grouped into a monophyletic clade. However, pair-wise comparison of gene regions showed there were at least 15 bps (ITS) and 6 bps (LSU) differences between CX37 and ex-type sequences of C.acaciae (Samarakoon et al. 2018). Moreover, following the protocol suggested by Damm et al. (2010) and Harrington et al. (2019), we could not induce ascomata formation in the isolate CX37. This hindered us from comparing the sexual structures of this species. In the future, the discovery of more isolates of C.acaciae will allow us to clarify the taxonomy of this species.
In the present study, through repeated sampling, we isolated five Coniochaeta species associated with four lichen species in China. Amongst these, three were previously-undescribed species. Data emerging from this study substantially augmented our current knowledge on the diversity and host range of this genus in China and globally. However, our surveys were exclusively conducted in two Provinces in China. Currently, more surveys should be conducted in various ecoregions of China to catalogue the diversity of Coniochaeta and various other endolichenic fungi.
Supplementary Material
Acknowledgements
This study was funded by The National Natural Science Foundation of China (Grant# 31600100). Thanks to Prof. Li-Song Wang from Kunming Institute of Botany, Chinese Academy of Sciences and Prof. Zun-Tian Zhao from Shandong Normal University for their assistance with the identification of lichens.
Citation
Si H-L, Su Y-M, Zheng X-X, Ding M-Y, Bose T, Chang R-L (2021) Phylogenetic and morphological analyses of Coniochaeta isolates recovered from Inner Mongolia and Yunnan revealed three new endolichenic fungal species. MycoKeys 83: 105–121. https://doi.org/10.3897/mycokeys.83.71140
Supplementary materials
Figure S1. ML tree generated from ITS sequence data
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hong-Li Si, Yue-Min Su, Xiao-Xiao Zheng, Meng-Yao Ding, Tanay Bose, Run-Lei Chang
Data type
Pdf. file
Explanation note
Maximum Likelihood tree constructed using ITS dataset. Bootstrap support values ≥ 75% and posterior probabilities ≥ 0.95 are indicated above the nodes as ML / PP. The isolates obtained in this study are shown in bold. T = ex-type isolates.
Figure S2. ML tree generated from LSU sequence data
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hong-Li Si, Yue-Min Su, Xiao-Xiao Zheng, Meng-Yao Ding, Tanay Bose, Run-Lei Chang
Data type
Pdf. file
Explanation note
Maximum Likelihood tree constructed using LSU dataset. Bootstrap support values ≥ 75% and posterior probabilities ≥ 0.95 are indicated above the nodes as ML / PP. The isolates obtained in this study are shown in bold. T = ex-type isolates.
References
- Ahmadjian V. (1993) The lichen symbiosis. John Wiley & Sons Inc, 250 pp.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. Jorunal of Molecular Biology 215: 403–410. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Arnold AE, Jolanta Miadlikowska, K. Lindsay Higgins, Snehal D. Sarvate, Paul Gugger, Amanda Way, Valérie Hofstetter, Frank Kauff, Lutzoni F. (2009) A phylogenetic estimation of trophic transition networks for Ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? Systematic Biology 58: 283–297. 10.1093/sysbio/syp001 [DOI] [PubMed]
- Asgari B, Zare R. (2006) Two new Coniochaeta species from Iran. Nova Hedwigia Band 82: 227–236. 10.1127/0029-5035/2006/0082-0227 [DOI] [Google Scholar]
- Asgari B, Zare R, Gams W. (2007) Coniochaetaershadii, a new species from Iran, and a key to well-documented Coniochaeta species. Nova Hedwigia 84: 175–187. 10.1127/0029-5035/2007/0084-0175 [DOI] [Google Scholar]
- Beyma FH van. (1939) Beschreibung einiger neuer Pilzarten aus dem Centraalbureau voor Schimmelcultures Baarn (Holland). V. Mitteilung. Zentralblatt für Bakteriologie und Parasitenkunde, Abteilung 2. 99: 381–394. [Google Scholar]
- Chang Jong-How, Wang Yei-Zeng. (2011) Taxonomy of Coniochaetaleucoplaca and C.velutina: morphological and molecular studies based on LSU rDNA of isolates from Taiwan. Nova Hedwigia 92: 57–67. 10.1127/0029-5035/2011/0092-0057 [DOI] [Google Scholar]
- Checa J. (1988) The genus Coniochaeta (Sacc.) Cooke (Coniochaetaceae, Ascomycotina) in Spain. Crypt Mycol 9(1): 1–34. [Google Scholar]
- Damm U, Fourie PH, Crous PW. (2010) Coniochaeta (Lecythophora), Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia - Molecular Phylogeny and Evolution of Fungi 24: 60–80. 10.3767/003158510X500705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Olmo-Ruiz M. (2012) Diversity, distributions, and host affiliations of fungal endophytes associated with seedless vascular plants. PhD Thesis, University of Arizona, Arizona.
- García D, Stchigel AM, Cano J, Calduch M, Hawksworth DL, Guarro J. (2006) Molecular phylogeny of Coniochaetales. Mycological Research 110: 1271–1289. 10.1016/j.mycres.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Harrington AH, Olmo-Ruiz M, U’Ren JM, Garcia K, Arnold AE. (2019) Coniochaetaendophytica sp. nov., a foliar endophyte associated with healthy photosynthetic tissue of Platycladus orientalis (Cupressaceae). Plant and Fungal Systematics 64: 65–79. 10.2478/pfs-2019-0008 [DOI] [Google Scholar]
- Hawksworth DL. (2011) A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. IMA Fungus 2: 155–162. 10.3897/mycokeys.1.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog Gd, Guarro J, Gene J, Figueras M. (2000) Atlas of clinical fungi 2nd ed. Baard-Delft: Centraalbureau vor Schimmelcultures/Universitat Rovira Virgili.
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Jones EBG, Liu N-G, Abeywickrama PD, Mapook A, Wei D, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao D-F, Li J, Samarakoon MC, Chaiwan N, Lin C-G, Phutthacharoen K, Zhang S-N, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang H-B, Yang J, Zeng M, Huanraluek N, Liu J-K, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang S-K, Thiyagaraja V, Lu Y-Z, Jayawardena RS, Dong W, Yang E-F, Singh SK, Singh SM, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Alvarenga RLM, Gibertoni TB, Pfliegler WP, Horváth E, Imre A, Alves AL, da Silva Santos AC, Tiago PV, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu J, Sheng J. (2020) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Kamiya S, Uchiyama S, Udagawa SI. (1995) Two new species of Coniochaeta with a cephalothecoid peridium wall. Mycoscience 36: 377–383. 10.1007/BF02268619 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, Gené J, Ahmad S, Cano J, Al-Sweih N, Joseph L, Chandy R, Guarro J. (2013) Coniochaetapolymorpha, a new species from endotracheal aspirate of a preterm neonate, and transfer of Lecythophora species to Coniochaeta. Antonie van Leeuwenhoek 104: 243–252. 10.1007/s10482-013-9943-z [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolutionl 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrey JD, Diederich P. (2003) Lichenicolous fungi: interactions, evolution, and biodiversity. Bryologist 106: 80–120. 10.1093/molbev/msw054 [DOI] [Google Scholar]
- Manoharachary C, Ramarao P. (1973) Thielaviaboothii sp. nov. from pond mud. Transactions of the British Mycological Society 61: 196–198. 10.1016/S0007-1536(73)80105-6 [DOI] [Google Scholar]
- Miller MA, Pfeiffer WT, Schwartz T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE), 2010, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Nash, Thomas H. (2008) Lichen biology. University Press, Cambridge, 498 pp. 10.1017/CBO9780511790478 [DOI] [Google Scholar]
- Nasr S, Bien S, Soudi MR, Alimadadi N, Fazeli SS, Damm U. (2018) Novel Collophorina and Coniochaeta species from Euphorbiapolycaulis , an endemic plant in Iran. Mycological Progress 17: 755–771. 10.1007/s11557-018-1382-9 [DOI] [Google Scholar]
- Nelsen MP, Lücking R, Boyce CK, Lumbsch HT, Ree RH. (2020) No support for the emergence of lichens prior to the evolution of vascular plants. Geobiology 18: 3–13. 10.1111/gbi.12369 [DOI] [PubMed] [Google Scholar]
- Nylander J, Wilgenbusch JC, Dan LW, Swofford DL. (2008) AWTY (Are We There Yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24: 581–583. 10.1093/bioinformatics/btm388 [DOI] [PubMed] [Google Scholar]
- Perdomo H, Garcia D, Gene J, Cano J, Sutton DA, Summerbell R, Guarro J. (2013) Phialemoniopsis, a new genus of Sordariomycetes, and new species of Phialemonium and Lecythophora. Mycologia 105: 398–421. 10.3852/12-137 [DOI] [PubMed] [Google Scholar]
- Raja HA, Fournier J, Shearer CA, Miller AN. (2012) Freshwater ascomycetes: Coniochaetagigantospora sp. nov. based on morphological and molecular data. Mycoscience 53: 373–380. 10.1007/s10267-012-0181-4 [DOI] [Google Scholar]
- Rambaut A. (2009) FigTree v1. 3.1. http://tree bio ed ac uk/software/figtree/
- Rasband W. (1997) ImageJ, US National Institutes of Health, Bethesda, Maryland, USA. http://imagej.nih.gov/ij
- Réblová M, Miller AN, Rossman AY, Seifert KA, Crous PW, Hawksworth DL, Abdel-Wahab MA, Cannon PF, Daranagama DA, De Beer ZW, Huang S-K, Hyde KD, Jayawardena R, Jaklitsch W, Jones EBG, Ju Y-M, Judith C, Maharachchikumbura SSN, Pang K-L, Petrini LE, Raja HA, Romero AI, Shearer C, Senanayake IC, Voglmayr H, Weir BS, Wijayawarden NN. (2016) Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7: 131–153. 10.5598/imafungus.2016.07.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero AI, Carmaran CC, Lorenzo LE. (1999) A new species of Coniochaeta with a key to the species known in Argentina. Mycological Research 103: 689–695. 10.1017/S0953756298007564 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon MC, Gafforov Y, Liu N, Maharachchikumbura S, Bhat JD, Liu JK, Promputtha I, Hyde KD. (2018) Combined multi-gene backbone tree for the genus Coniochaeta with two new species from Uzbekistan. Phytotaxa 336: 43–58. 10.11646/phytotaxa.336.1.3 [DOI] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, Hesse C, O’Rourke B, Serdani M, Spotts R, Lutzoni F, Hofstetter V, Miadlikowska J. (2006) A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. 10.1080/15572536.2006.11832630 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Suryanarayanan TS, Thirunavukkarasu N. (2017) Endolichenic fungi: the lesser known fungal associates of lichens. Mycology 8: 189–196. 10.1080/21501203.2017.1352048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi M, Joshi Y. (2019) What are lichenized fungi? In: Endolichenic Fungi: Present and Future Trends. Springer, Singapore, 1–26. 10.1007/978-981-13-7268-1_1 [DOI]
- Troy GC, Panciera DL, Phillip PJ, Sutton DA, Josepa G, Cano JF, Josep G, Thompson EH, Wickes BL. (2013) Mixed infection caused by Lecythophoracanina sp. nov. and Plectosphaerellacucumerina in a German shepherd dog. Medical mycology: official publication of the International Society for Human and Animal Mycology 51(5): 455–460. 10.3109/13693786.2012.754998 [DOI] [Google Scholar]
- Vazquez-Campos X, Kinsela A, Waite T, Collins R, Neilan B. (2014) Fodinomycesuranophilus gen. nov. sp. nov. and Coniochaetafodinicola sp. nov., two uranium mine-inhabiting Ascomycota fungi from northern Australia. Mycologia 106: 1073–1089. 10.3852/14-013 [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr, Hermann, Friebes, Gernot, Jaklitsch, Gernot M, Garcia, Susana (2016) Lopadostomataeniosporum revisited and a new species of Coniochaeta. Sydowia 68: 87–97. 10.1016/j.simyco.2017.05.001 [DOI] [Google Scholar]
- Weber E. (2002) The Lecythophora-Coniochaeta complex: I. Morphological studies on Lecythophora species isolated from Piceaabies. Nova Hedwigia 74: 159–185. 10.1127/0029-5035/2002/0074-0159 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols.Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Zhang T, Wei X-L, Wei Y-Z, Liu H-Y, Yu L-Y. (2016) Diversity and distribution of cultured endolichenic fungi in the Ny-Ålesund Region, Svalbard (High Arctic). Extremophiles 20: 461–470. 10.1007/s00792-016-0836-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. ML tree generated from ITS sequence data
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hong-Li Si, Yue-Min Su, Xiao-Xiao Zheng, Meng-Yao Ding, Tanay Bose, Run-Lei Chang
Data type
Pdf. file
Explanation note
Maximum Likelihood tree constructed using ITS dataset. Bootstrap support values ≥ 75% and posterior probabilities ≥ 0.95 are indicated above the nodes as ML / PP. The isolates obtained in this study are shown in bold. T = ex-type isolates.
Figure S2. ML tree generated from LSU sequence data
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hong-Li Si, Yue-Min Su, Xiao-Xiao Zheng, Meng-Yao Ding, Tanay Bose, Run-Lei Chang
Data type
Pdf. file
Explanation note
Maximum Likelihood tree constructed using LSU dataset. Bootstrap support values ≥ 75% and posterior probabilities ≥ 0.95 are indicated above the nodes as ML / PP. The isolates obtained in this study are shown in bold. T = ex-type isolates.