Abstract
The meninges contain adaptive immune cells that provide immunosurveillance of the CNS. These cells are thought to derive from the systemic circulation. Through single cell analyses, confocal imaging, bone marrow chimeras and parabiosis experiments, we show that meningeal B cells derive locally from the calvaria, which harbors a bone marrow niche for hematopoiesis. B cells reach the meninges from the calvaria through specialized vascular connections. This calvaria-meningeal path of B cell development may provide the CNS with a constant supply of B cells educated by CNS antigens. Conversely, we show that a subset of antigen-experienced B cells that populate the meninges in aging mice are blood-borne. These results identify a private source for meningeal B cells which may help maintain immune privilege within the CNS.
One-sentence summary:
Meningeal B cells originate from the skull bone marrow, but blood-derived antigen-experienced B cells infiltrate the meninges during aging.
The central nervous system (CNS) is enveloped by the meninges, which harbor different immune cell types that provide constant surveillance at the CNS border (1-3). Although meningeal lymphocytes are thought to derive exclusively from the systemic circulation, recent findings question this premise. Vascular connections between the calvaria (the flat bones forming the top dome of the skull) and meninges have recently been described (4-6), and two multidimensional studies identified a cluster of developmentally immature B cells in the mouse CNS (7, 8). We hypothesized that these B cells may derive from calvarial hematopoiesis. Here, we show that meningeal B cells encompass multiple stages of B cell development, spanning pro-B to mature B cells. Using parabiotic mice and bone marrow (BM) chimeras with selective reconstitution of the skull BM, we demonstrate that most meningeal B cells originate from the calvaria. We envision that calvaria-derived B cells are locally educated by CNS-derived antigens to prevent the generation of immunoglobulins (Ig) with high affinity for CNS epitopes. Conversely, a population of age-associated B cells (ABCs) infiltrates the mouse meninges from the circulation during aging. This study sheds light on the origin and phenotypes of meningeal B cells in homeostasis and during aging, challenging the widely accepted idea that meningeal adaptive immunity originates exclusively from systemic circulation.
Results
Meningeal B cells are extravascular and can exit the CNS compartment through the dura lymphatics
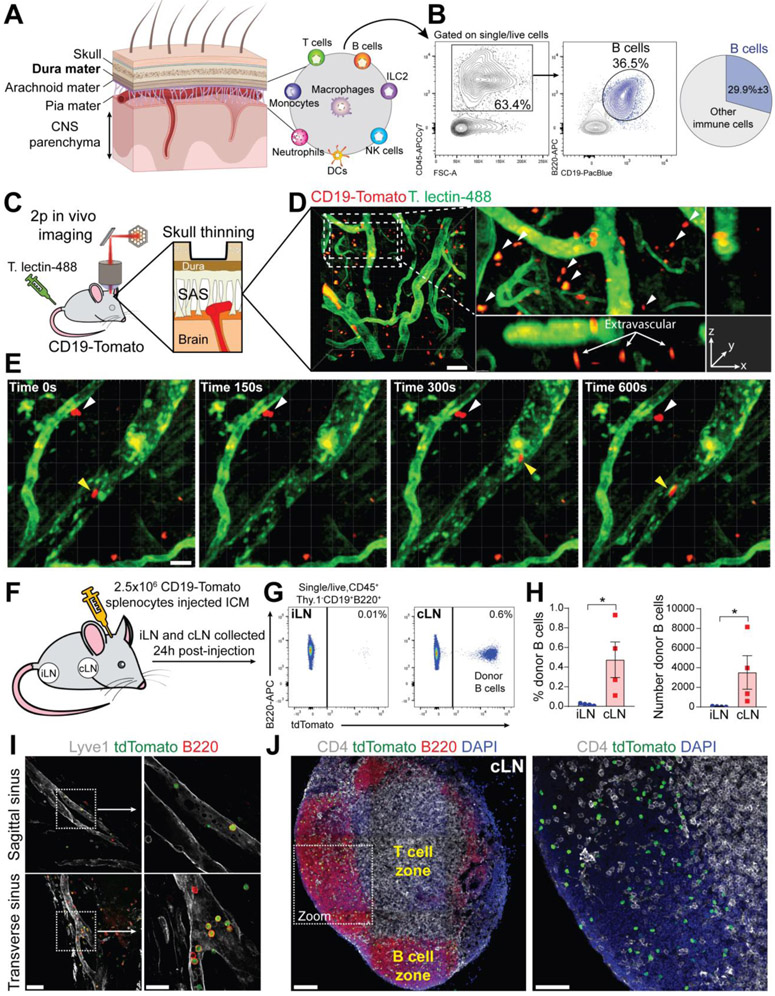
Meninges are formed by three membranes. The dura mater is the most outer layer attached onto the skull periosteum, whereas the two inner layers, the arachnoid and pia mater (leptomeninges), cover the brain cortex (Fig. 1A). This region is enriched of different immune cell types, and B cells represent about 30% of the total CD45+ cells in mouse meninges (Fig. 1B, S1A). Because the dura contains a relative abundance of immune cells and can be more easily dissected than the leptomeninges, we focused most of our subsequent studies on dural B cells. By flow cytometry, B cells were found in both the dura mater and the brain/leptomeninges samples (fig. S1B). The majority of meningeal B cells in young-adult mice were B2 type, while innate B cells (B1a and B1b) represented a minor population (fig. S1C). We surveyed B cells by confocal imaging in the brain and spinal cord of Cd19Cre:Rosa26-tdTomato mice (hereafter CD19-Tomato), which express tdTomato specifically in CD19+ cells (9). While no B cells were found in the brain parenchyma (fig. S1D), B cells were present in the leptomeninges along the brain surface (fig. S1E), indicating that brain B cells are extra-parenchymal. Two-photon in vivo imaging in the subdural space of the CD19-Tomato mice showed that most meningeal B cells were localized in the extravascular compartment (Fig. 1, C and D) and appeared relatively immobile as compared to intravascular B cells (Fig. 1E and Movie S1). The dura mater contains blood and lymphatic vessels (LV) along the sagittal and transverse sinuses (fig. S2A). These areas were particularly enriched in B cells, some of which were located within dura LV (fig. S2B). Dura lymphatics drain immune cells and molecules to the cervical lymph nodes (cLNs) (10, 11), suggesting that meningeal B cells may undergo a similar fate. To test this hypothesis, CD19-Tomato splenocytes were introduced in the CSF (cerebrospinal fluid) of wild-type mice by intracisterna magna (ICM) injection (Fig. 1F), and 24 hours later were found accumulating in the cLNs (Fig. 1, G and H). Donor-derived B cells appeared to be located in both the dura LV and cLN B cell zone (Fig. 1, I and J). Thus, LV may serve as a migratory route for B cells exiting the CNS compartment.
Fig. 1. B cells represent a main immune cell type in mouse meninges and are capable of trafficking through meningeal lymphatics.
(A) Cartoon representing the structural organization of the meninges. (B) Representative flow cytometry plot showing the proportion of B cells within the overall CD45+ population in mouse dura (average of n=4 mice, data generated from a single experiment). (C) Schematic depiction of the experimental approach to perform in vivo two-photon imaging in the subdural space of CD19-Tomato mice. (D) Representative two-photon image of extravascular B cells in CD19-Tomato mouse meninges (scale bar=50μm). (E) Two-photon time-lapse imaging in the meninges of a CD19-Tomato mouse. Intravascular B cell: yellow arrowhead; extravascular B cells: white arrowhead (scale bar=20μm). (F) Schematic depiction of the experimental approach (ICM: Intra-Cisterna Magna). (G) Flow cytometry analysis of donor (CD19-Tomato) derived B cells in inguinal lymph nodes (iLN) and cervical lymph nodes (cLN) 24h post-injection. (H) Frequency and absolute number of donor-derived B cells in iLN and cLN (mean ± SEM; n=4 mice; Mann Whitney U test *P<0.05; data generated from a single experiment). (I) Representative confocal image of donor (CD19-Tomato) B cells trafficking through the dura lymphatics 24h post-injection (low magnification image scale bar=50μm; high magnification image scale bar=20μm). (J) Representative confocal image of donor (CD19-Tomato) derived B cells in cLN 24h post-injection (low magnification image scale bar=100μm; high magnification image scale bar=50μm).
Meningeal B cells are phenotypically similar to bone marrow B cells
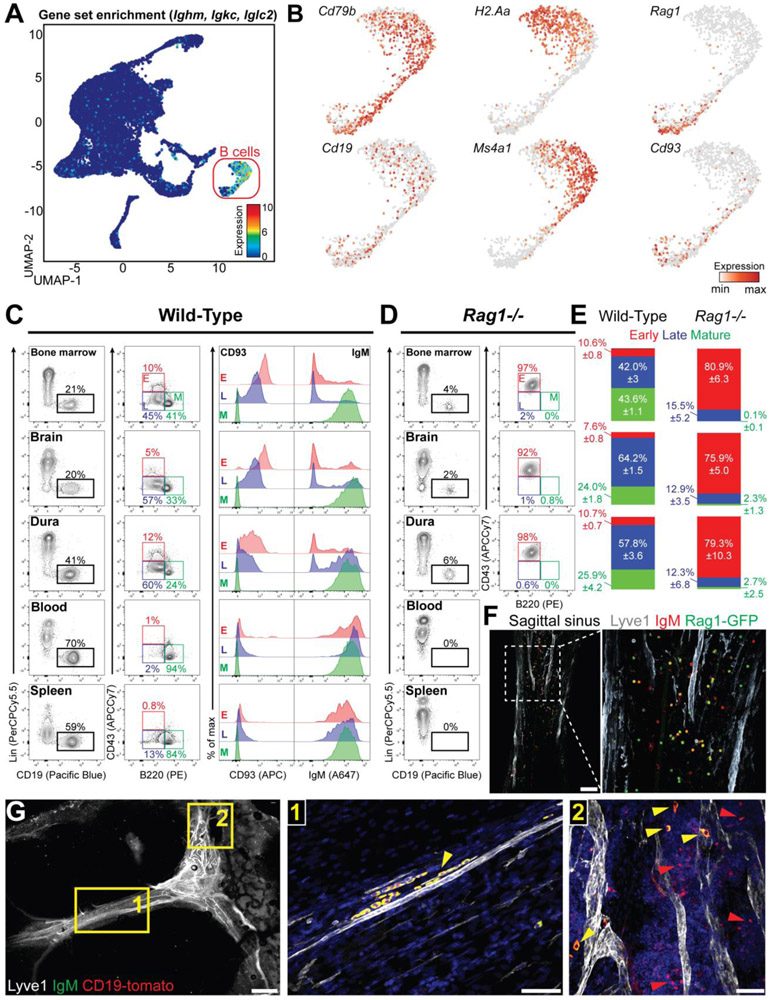
Although B cells represent a main immune population in the meninges, little is known about their composition under homeostasis. A previous scRNA-seq study on mouse brain immune cells identified two distinct B cell subsets, labeled as “mature” and “immature” (7). We re-clustered these transcriptomic data (GSE98969) and identified B cells based on the high levels of Ig transcripts (Fig. 2A). This population uniformly expressed pan-B cell signature genes (Cd79b and Cd19). However, mature B cell genes (H2-Aa and Ms4a1) and early B cell genes (Rag1 and Cd93) appeared unevenly distributed in this cluster (Fig. 2B), suggesting that B cells in the CNS encompass different stages of their development. Next, we compared the phenotype of meningeal B cells to that of B cells in the BM, blood, and spleen (Fig. 2C). Based on Hardy fraction (12, 13) early B cells were identified as CD19+B220loCD43hi (bona fide fraction B-C), late B cells as CD19+B220loCD43lo (bona fide fraction D-E), and mature B cells as CD19+B220hiCD43− (fraction F). BM and meningeal B cells appeared similarly distributed into these three subsets. In both compartments, early B cells were IgM−CD93+, mature B cells were IgM+CD93−, and late B cells had an intermediate phenotype. By contrast, most blood and spleen B cells exhibited a mature phenotype. We then repeated this analysis in Rag1−/− mice, which lack mature B cells (Fig. 2D), and found a population of B220loCD43hi B cells in the BM, brain, and dura, whereas the blood and spleen completely lacked B cells. Thus, the composition of meningeal B cells closely resembles the BM, both in wild-type and Rag1−/− mice (Fig. 2E). As a validation, we imaged the dura mater from Rag1-GFP knock-in mice (14) and found Rag1-expressing B cells, preferentially located along the sagittal sinus (Fig. 2F, S3A). We also performed confocal imaging of the dura mater from CD19-Tomato mice stained for IgM and found distinct subsets of IgM+ and IgM− B cells (Fig. 2G). Immunofluorescent imaging of CD93, Ki67, GL7 (markers of early B cells) and CD20 (marker of mature B cells) in the dura mater confirmed that early B cells are present in mouse meninges under homeostasis (fig. S3B). Lastly, we analyzed the expression of various markers differentially expressed across B cell development and confirmed that BM and meninges harbor overlapping B cell phenotypes (fig. S4).
Fig. 2. Mouse meninges harbor a heterogeneous B cell population that encompass multiple developmental stages.
(A) Uniform manifold approximation and projection (UMAP) of CD45+ brain cells from a deposited scRNA-seq dataset (GSE98969). Normalized expression for Ighm, Igkc and Iglc2 was color-coded for transcript counts. The cell cluster highly enriched for the gene set was inferred as B cells. (B) Enrichment for transcripts linked to different B cell maturation stages. Pan-B cell: Cd79b and Cd19; mature B cell: H2-Aa and Ms4a1; immature B cell: Rag1 and Cd93. (C) Flow cytometry analysis of bone marrow (BM), brain, dura, blood and spleen B cell in C57BL/6 mice. B cells were gated as CD45+CD19+ and lineage (CD3, CD11b, F4/80 and Gr-1) negative (left column) and were further divided into early (B220loCD43hi), late (B220loCD43lo), and mature (B220hiCD43−) subset (middle column). CD93 and IgM staining is shown for the three subsets (right column). (D) Flow cytometry analysis of Rag1−/− mice. B cells derived from BM, brain, and dura were gated as CD19+ and lineage (CD3, CD11b, F4/80 and Gr-1) negative (left column) and were further divided into early (B220loCD43hi), late (B220loCD43lo), and mature (B220hiCD43−) subsets (right column). (E) Quantification of B cell subsets in bone marrow, brain and dura of C57BL/6 and Rag1−/− mice (n=8 and 7 mice respectively; data generated from two independent experiments). (F) Representative confocal images of the dura mater from Rag1-GFP mice (scale bar=100μm). (G) Representative confocal image of IgM+ (yellow arrowhead) and IgM− B cells (red arrowhead) in CD19-Tomato mice (low magnification image scale bar=1mm; high magnification image scale bar=50μm).
Single-cell analyses resolve multiple developmental stages of dura B cells
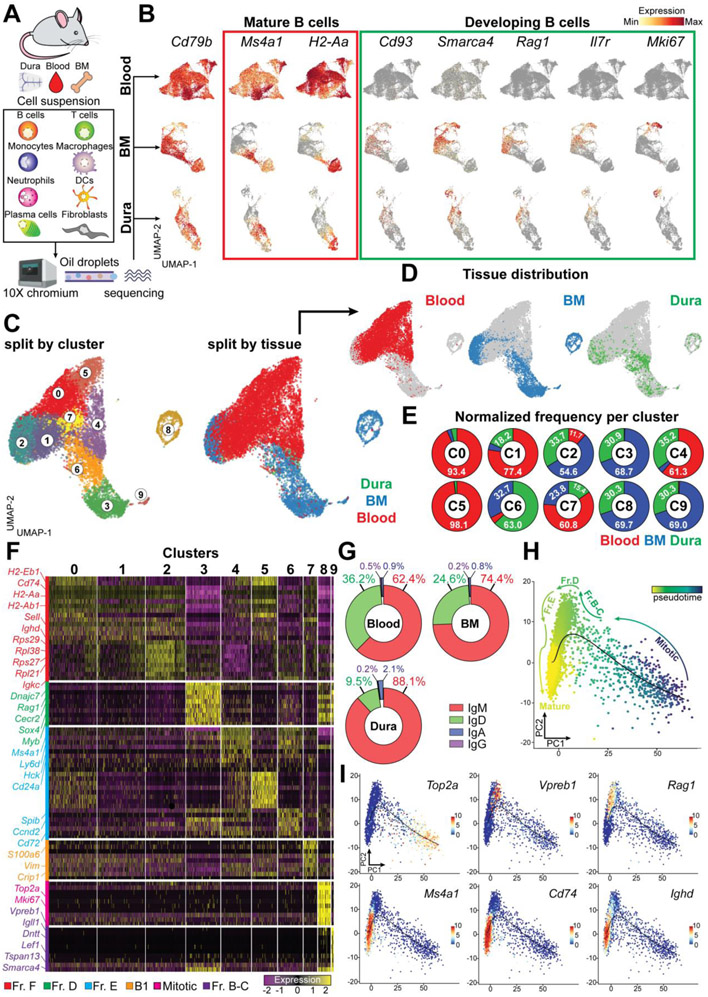
To gain in-depth insight into the diversity of meningeal B cells, we performed single-cell RNA sequencing (scRNA-seq) of cells isolated from dura mater, blood and BM. We analyzed unsorted cell suspensions to minimize experimental artifacts due to cell stress and to include both immune and non-immune cells (Fig. 3A). Cells that passed the quality control underwent unsupervised clustering and were displayed on the UMAP space (fig. S5, S6, S7). In the dura mater we resolved clusters of neutrophils (43.7%), B cells (26.3%), T cells (3.2% double negative, 2.8% CD8+ and 1.7% CD4+), monocytes (2.5% Ly6C+, 1.2% Ly6C−), NK cells (1.6%), macrophages (1.3%), mast cells (1.1%), plasmacytoid dendritic cells (pDCs) (1%), classical dendritic cells (cDCs) (0.9%), type-2 innate lymphoid cells (ILC2) (0.4%), and plasma cells (0.3%) (fig. S5). Dura fibroblasts (5.7%) were identified based on the expression Coll1a and Mgp, as recently reported (15). Focusing on the B cell compartment, we found multiple B cell clusters in all analyzed tissues. Indeed, all B cells in the blood exhibited a rather homogeneous expression of the mature markers Ms4a1 and H2-Aa, whereas the early B cell markers Cd93, Smarca4, Rag1, IL7R, and Mki67 were not detected (Fig 3B). The diversity in blood B cells was mostly driven by the expression of κ or λ light chains (fig. S7). By contrast, B cells in the BM and dura exhibited a non-overlapping enrichment for both mature and early markers (Fig 3B), confirming the presence of multiple stages of B cell development in these niches.
Fig. 3. scRNA-seq analysis reveals a similar transcriptomic pattern in dura and BM B cells.
(A) Schematic depiction of the experimental design related to scRNAseq. (B) Expression of featured genes denoting mature and developing B cells in blood, BM and dura. (C) UMAP of 13,281 B cells aggregated from blood, BM, and dura collected from three C57BL6 mice and colored by cluster (left) or tissue origin (middle) (data generated from a single experiment). (D) UMAP plots split by tissue showing the distribution of blood, BM, and dura B cell. E) Proportional contribution of the three tissues (blood, BM, and dura) to the ten B cell clusters. (F) Gene expression heatmap of the top 10 signature genes per cluster. (G) Frequency of C-region usage per tissue determined by scBCRseq. (H) Developmental trajectory displayed on PCA map and colored by slingshot pseudotime. (I) Enrichment of key transcripts differentially expressed throughout B cell maturation.
Next, we reclustered all B cells from the three tissues on a single UMAP space (Fig. 3C) and found that B cells from the BM and dura occupied overlapping territories, whereas most of the blood B cells clustered separately (Fig. 3D). This analysis yielded ten different clusters (C0-9). C0 and C5 were almost entirely (>90%) formed by blood B cells; C3, C6, C8 and C9 contained B cells from both BM and dura, but were largely depleted of blood B cells (<5%); C1 and C4 contained B cells from both blood and dura, but were depleted of BM B cells (<5%); and C2 and C7 contained B cells from all compartments (Fig. 3E). To determine the identity of each cluster we performed a differential gene expression analysis (cluster vs total; cutoff=log2FC>0.5) (Fig. 3F and Table S1), followed by enrichment analysis of Gene Ontology (GO) biological process (fig. S8, A and B). Based on these analyses, we determined that blood contained primarily mature naïve B cells (C0, C1, C2, C5; fraction F), a small subset of immature B cells (C4; fraction E) and B1b cells (C7). By contrast, BM and dura contained mature naïve B cells (C2; Fraction F), immature B cells (C6; fraction E), pre-B cells (C3; fraction D), pro-B cells (C9; fraction B-C), and mitotic B cells (C8). BCR-seq analysis showed that 88.1% of dura B cells with productive V(D)J rearrangement were IgM type, again confirming their early stage of development (Fig. 3G). Finally, we performed pseudotime analysis (Fig. 3H) and identified a developmental trajectory that originated from mitotic B cells, then traversed pro-B and pre-B intermediate stages, before eventually giving rise to mature B cells (Fig. 3I). To empirically test this model, we performed BrdU pulse-chase experiments focusing on BM and dura B cells (fig. S8C). At 24 hours post BrdU pulse, early B cells exhibited the maximal incorporation of BrdU. At 3 days, the percentage of BrdU+ early B cells was dramatically reduced, but it increased in both late and mature B cell populations. At 5 days, almost no BrdU+ B cells could be detected, presumably because BrdU-labeled cells underwent apoptosis, further differentiated, or were progressively diluted by newly generated B cells. Thus, the molecular signature of dura B cells highlights a stepwise developmental continuum, comprehensive of multiple intermediate phenotypes that are normally found in the BM, but not in the periphery.
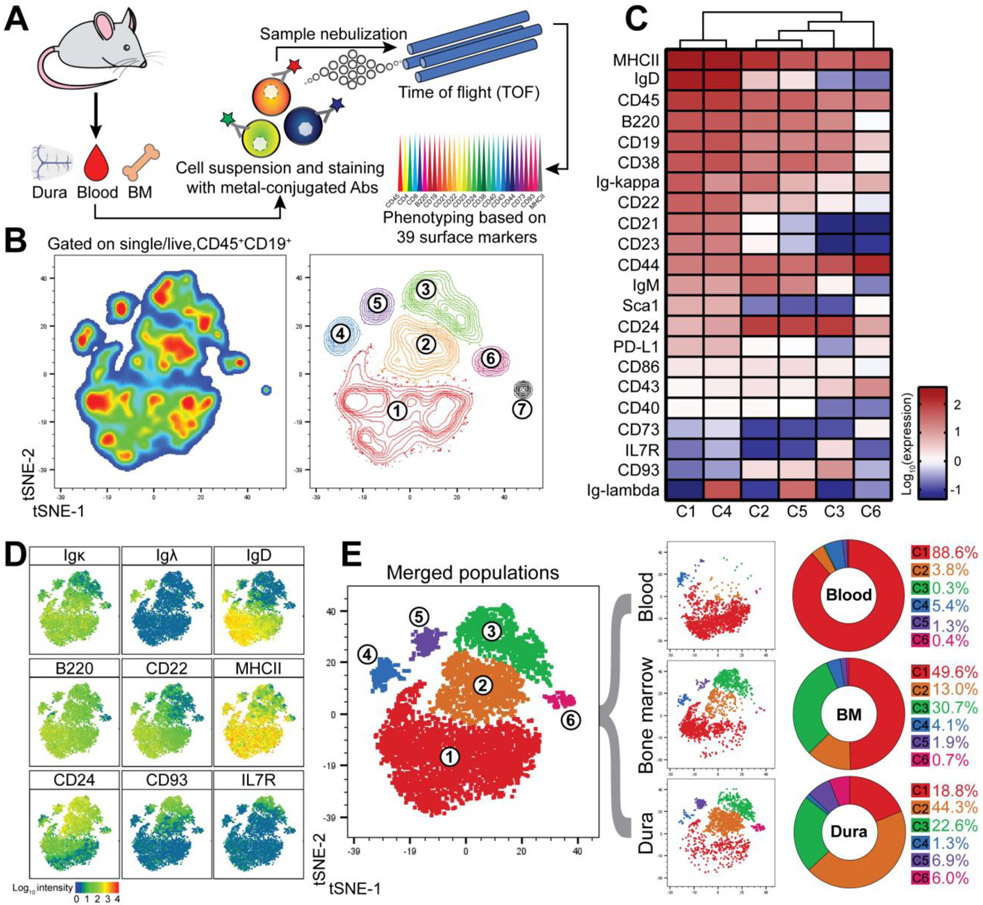
To validate the scRNA-seq data we performed a mass cytometry study of dura, blood, and BM B cells (Fig. 4A, S9). Unsupervised clustering of the CD45+CD19+ population using t-SNE identified seven distinct clusters (C1-7) (Fig. 4B and Table S2). We analyzed clusters C1-C6 in detail (Fig. 4, C and D), whereas C7 was excluded from further analysis because it contained only 17 cells. C1 and C4 exhibited a typical mature phenotype (MHC-II+IgM+IgD+), but differentially expressed κ or λ light chains. C2 and C5 had an immature B cell phenotype (IgDloCD22loCD21loCD23loCD24+) while κ or λ light chains were differentially expressed. C3 had an early B cell phenotype (IgD−CD21−CD23−IgMloCD24+CD43+IL7R+CD93+). C6 was negative or low for both naïve B cell markers and pre-B markers, but expressed CD73, CD43, PD-L1 and CD44, indicating an activated phenotype (16). Next, we determined the distribution of B cells from each compartment among the six clusters (Fig. 4E). Blood B cells were maximally enriched in C1 (88.6%), BM B cells were distributed among all clusters (C1=49.6%, C2=13%, C3=30.7%), and dura B cells were primarily concentrated in C2 (44.3%) with substantial fractions in C1 (18.8%) and C3 (22.6%). Blood and BM were equally represented in C4 (5.4% and 4.1% respectively), whereas the dura was more abundant than either blood or BM in C5 (6.9% vs 1.3% and 1.9%). Thus, mass cytometry analysis confirms that the dura mater contains a large proportion of developing B cells.
Fig. 4. Mass cytometry confirms developmental heterogeneity of dura B cells.
(A) Schematic depiction of the experimental design related to CyTOF analysis. (B) Unsupervised clustering by means of t-distributed stochastic neighbor embedding (t-SNE) of 6,000 live singlet CD45+CD19+ B cells obtained from concatenation of three tissues (blood, BM, and dura) collected from three C57BL6 mice (data generated from a single experiment). Cells are displayed by pseudocolor (left) and contour plots (right). (C) Heatmap showing the surface protein expression of selected B cell markers among the six B cell clusters (cluster 7 was arbitrarily excluded due to low cell number). (D) Staining enrichment of representative surface markers for mature and developing B cells. (E) Merged tSNE plot colored by cluster (left) and tSNE plot split by tissue (middle). Frequency of each cluster in blood, BM, and dura (right).
Meningeal B cells mostly derive from the calvaria
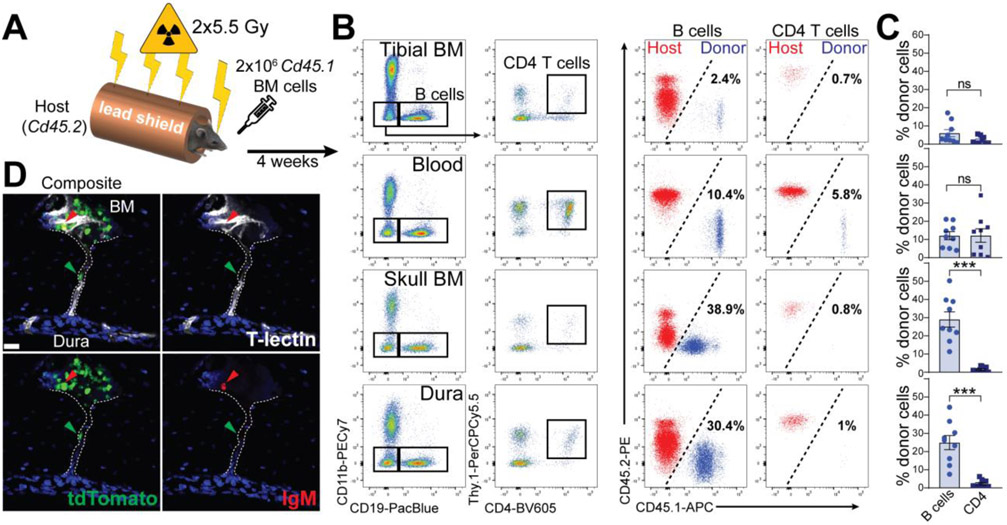
Because only a few immature B cells were present in the blood, it was unlikely that developing B cells in the dura originated from the systemic circulation. We hypothesized that dura B cells originated in the calvarial BM. Indeed, the caudal region of the skull harbored a large hematopoietic niche (fig. S10, A and B), containing a variety of immune cells, including IL7R+ early B cells and LSK stem cells (fig. S10C). EM and confocal imaging highlighted a complex cellular system in this area, containing both myeloid and lymphoid cells (Fig. S10, D and E). To test our hypothesis, we performed an atypical bone-marrow transplantation experiment. During irradiation, the mouse body was protected with a lead shield, leaving only the head fully exposed (Fig. 5A). Head-irradiated Cd45.2 mice received Cd45.1 BM cells and four weeks later the percentage of B cell chimerism was assessed in different tissues (Fig. 5, B and C). In both tibial BM and peripheral blood, only a negligible percentage of donor-derived B cells was found (6% and 12.1% respectively), whereas a large population of donor-derived B cells was present in skull BM and dura (29% and 24.9% respectively). By contrast, almost no donor-derived CD4+ T cells were found in these compartments. We performed the reversal experiment, using a Cd45.1 host, transplanted with Cd45.2 BM cells and obtained comparable results (fig. S11A). To determine whether circulating B cells may potentially contribute to the pool of meningeal B cells under homeostasis, we performed parabiosis experiment using wild-type and CD19-Tomato mice. After four weeks of shared circulation, the percentage of tdTomato-positive B cells was assessed in wild-type mice. A large population of parabiont-derived B cells was present in the blood and spleen (39.2% and 36.4% respectively), whereas a minor infiltration could be found in brain and dura (11.3% and 8.0% respectively) (fig. S11, B and C). Comparable results were obtained by adoptive transfer of CD19-Tomato splenocytes into wild-type recipient mice (fig. S11, D and E). Thus, most meningeal B cells appear to originate in the calvarial BM and not from the circulation.
Fig. 5. Skull BM chimeras demonstrate that dura B cells originate from the calvaria.
(A) Schematic depiction of the experimental design of calvaria BM transplantation. (B) Representative flow cytometry plots of B cells (gated on CD19+CD11b− cells) and CD4 T cells (gated on CD19−CD11b−Thy.1+CD4+ cells) from multiple compartments in recipient mice (left). Percentage of donor-derived (CD45.1+) and host-derived (CD45.2+) B and CD4 T cells are shown (right). (C) Frequency of donor-derived B cells and CD4 T cells per compartment (mean ± SEM; n=9 mice; Mann Whitney U test ***P<0.001; data generated from two independent experiments). (D) Representative confocal image of an IgM− B cell trafficking from the calvarial BM towards meninges through a skull vascular channel.
It has been recently shown that vascular channels form a direct communication between the calvaria and the meningeal space, thus allowing migration of immune cells independently of the systemic circulation (4, 6). We performed confocal imaging of skull cryosections after immunofluorescent staining for endothelial markers (tomato-lectin, CD34, and CD31). B cells were visualized by means of the CD19-Tomato reporter. In agreement with these studies, we found vascular channels through the inner skull bone (fig S12A). As a complementary technique, we performed X-ray tomography on intact skull specimens impregnated with radiopaque metals. The resulting 3D image displayed in z-series reveals the presence of radiolucent areas (seemingly vessels) running through the calvaria and opening at the base of the sagittal sinus (fig. S12b and Movie 2). Finally, we provide evidence of IgM-negative B cells trafficking through these channels, confirming that calvaria-derived B cells reach the meningeal compartment at an early stage of their development (Fig. 3D, S12, C and D).
Dura fibroblasts produce crucial factors for B cell development
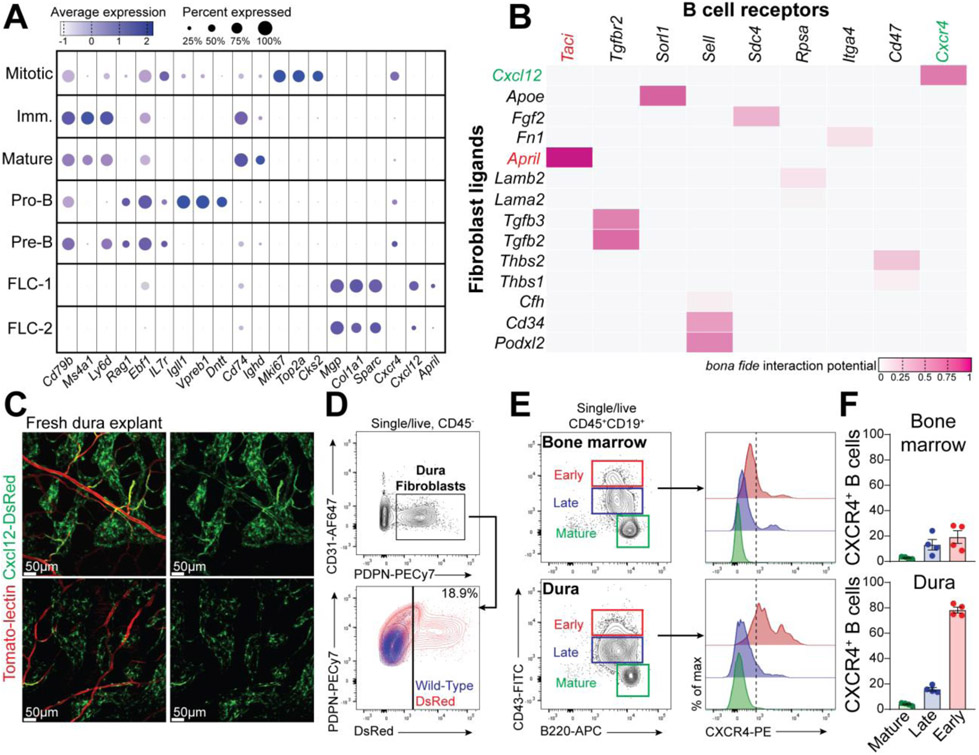
Notably, one of the top signature genes of the fibroblast-like cells (FLC) in dura was Cxcl12 (17), whereas dura early B cells express Cxcr4 (Fig. 6A). NicheNet analysis highlighted a network of molecular communications between FLC and dura B cells. Amongst the several ligands detected in dura fibroblasts, Cxcl12, April, Tgfb2, and Apoe were assigned to a high probability score (Fig. 6B). Expression of Cxcl12 in dural fibroblasts was confirmed by imaging and flow cytometry using Cxcl12-DsRed reporter mice (Fig. 6, C and D). Surface expression of CXCR4 in early B cells was also confirmed by flow cytometry (Fig. 6E). Interestingly, early B cells in dura exhibited a remarkable surface expression of CXCR4 (Fig. 6F), suggesting that calvaria-derived B cells may require this receptor for their homing in the meningeal compartment. Lastly, confocal imaging showed B cells in close contact to Cxcl12-expressing FLC in the sagittal sinus (fig. S13). The CXCL12-CXCR4 axis is crucial for B cells development, therefore we suggest that dura fibroblasts may support survival and differentiation of early B cells derived from the calvarial BM.
Fig. 6. Ligand-receptor interactions between CXCL12+ fibroblasts and CXCR4+ early B cells in the dura.
(A) Dot plot presenting the expression level of featured genes in fibroblasts and B cells at different maturation stages. (B) NicheNet analysis displaying the putative interaction score between fibroblasts-derived ligands and B cell receptors. (C) Representative confocal images of freshly prepared dura explant from a Cxcl12-DsRed reporter mouse (Tomato-lectin-488 injected iv. before perfusion). (D) FACS analysis of dura fibroblasts from Cxcl12-DsRed reporter mice (representative of three mice; data generated from a single experiment). (E) FACS analysis of CXCR4 expression in early, late, and mature B cells from dura and BM. (F) frequency of CXCR4+ B cells in the three B cells subsets from dura and BM (mean ± SEM; n=4 mice; data generated from a single experiment).
ABCs and plasma cells accumulate in mouse meninges during aging
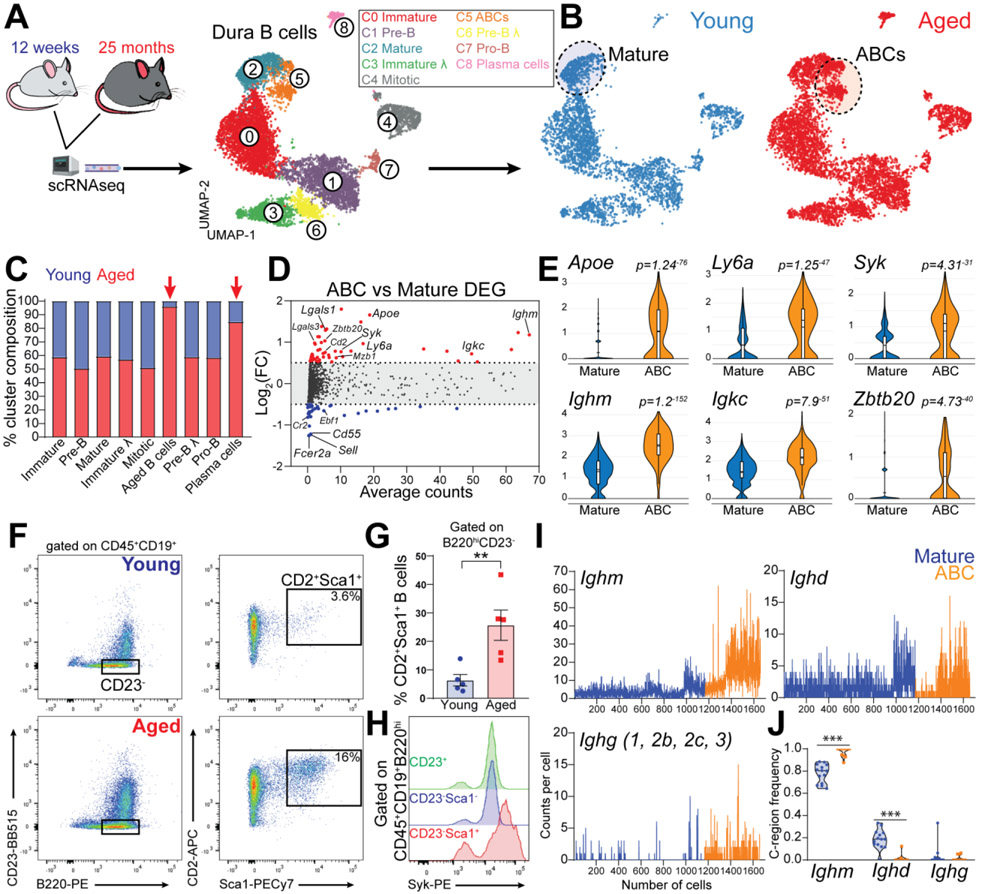
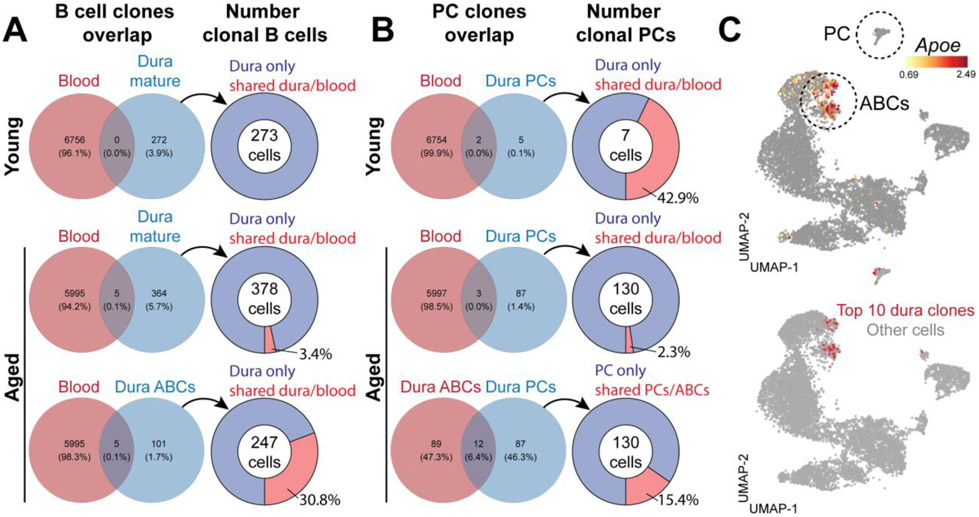
Lymphocytes exhibit clonal and phenotypical imbalance during aging (18, 19). Indeed, we found a dramatic increase in the total number of B cells in the dura of aged mice (fig. S14, A and B). We then investigated the impact of aging on meningeal B cells by comparing young (8-12-week-old) and aged (20-25-month-old) mice by scRNA-seq and scBCR-seq (Fig. 7A). An accumulation of clonal B cells was found in the aged dura (fig. S14C), as well as an increased clonal overlap with blood B cells (fig. S14D), suggesting infiltration from the periphery. All B cell clusters in dura were equally represented in young and aged mice, except for one cluster which was almost entirely (96.1%) derived from aged mice (Fig. 7, B and C) and was therefore annotated as “age associated B cells” (ABCs). Differential gene expression analysis between ABCs and mature B cells (log2FC>0.5; adj. P-value<0.01) revealed 105 differentially expressed genes (Table S3). The top upregulated transcripts included Apoe, Ly6a, Ighm, Igkc, Cd2, Lgals1, Zbtb20, and Syk, whereas the top downregulated transcripts contained Fcer2a, Cr2, Cd55, Sell, and Ebf1 (Fig. 7, D and E). Using flow cytometry, we confirmed a significant expansion of B220hiCD23−CD2+Sca1+ ABC in the dura of aged mice (Fig. 7, F and G). Consistent with the scRNA-seq data, this cell population also expressed increased levels Syk and ApoE protein (Fig. 7H, S14E). Furthermore, dura ABCs had increased numbers of Ighm heavy chain transcripts compared to mature B cells (Fig. 7I). scBCR-seq further confirmed increased Ighm, and reduced Ighd, C-region usage in ABCs (Fig. 7J). Additionally, ABCs exhibited greater similarity in their V gene usage profiles (fig. S14F), as well as accumulation of somatic mutations in the BCRs of cells expressing the Mu heavy chain (fig. S14G). Altogether, these data indicate that dura ABCs are antigen experienced B cells. Using these BCR data, we determined the percentage of B cells in clones that were shared between dura and blood (Fig. 8A). We found that dura mature B cells had no clonal overlap with blood in young mice, whereas a minor overlap could be observed in aged mice. By contrast, many dura ABCs were members of clones shared with blood (30.8%), suggesting that these cells had trafficked from the periphery.
Fig. 7. Age-associated B cells disseminate throughout the dura of aged mice.
(A) Schematic depiction of the experimental design of scRNAseq comparing young and aged mice. UMAP of 9,352 B cells aggregated from seven 12-week-old and seven 25-month-old C57BL6 female mice (data generated from two independent experiments). (B) Distribution of B cells from young and aged mice. (C) Contribution of young versus aged mice to each cluster. (D) Differential gene expression analysis of ABCs versus mature B cells. (E) Violin plots showing top up-regulated genes in ABCs compared to mature B cells. (F) Dura ABCs gated as the B220hiCD23−CD2+Sca1+ cells. (G) ABC population is significantly increased in the dura of aged mice compared to young mice (mean ± SEM; n=5 mice; unpaired Student’s t test **P<0.01; data generated from two independent experiments). (H) Flow cytometry histogram showing increased levels of Syk protein in dura ABCs (representative of three 18-month-old female mice; data generated from a single experiment). (I) Ig heavy chains transcript counts per cell in ABCs and mature B cells. (J) Frequency of heavy chains usage determined by BCRseq in ABCs and mature B cells (violin plot; n=11-14 mice per group; two-ways ANOVA and Bonferroni post-hoc test ***P<0.001).
Fig. 8. Age-associated B cells in the aged dura originate from the periphery.
(A) Venn diagrams representing the proportion of detected B cell clones shared between dura mature B cells and blood in young and aged mice, or between dura ABCs and blood. The connected pie chart represents the percentage of dura B cells (mature or ABCs) belonging to shared clonotypes. (B) Venn diagrams representing the proportion of detected plasma cell (PC) clones shared between dura and blood in young and aged mice, or between dura plasma cells and dura ABCs. The connected pie chart represents the percentage of dura plasma cells belonging to shared clonotypes. (C) UMAP showing the distribution of the ten most frequent clones in dura.
Next, we focused on dura plasma cells (fig. S15, A and B), which also appeared increased in the aged dura (Fig. 7C). Differential gene expression analysis revealed that the expression of Ig isotypes was markedly shifted between the two age groups. In young mice, dura plasma cells were predominantly IgA+, but became mostly IgM+ in aged mice (fig. S15C). By confocal imaging, we surveyed plasma cells located along the sagittal sinus (fig. S15D) and confirmed the massive increase of these cells in aged mice (fig. S15E), especially IgG and IgM types, while the number of IgA plasma cells was unchanged (fig. S14F). Lastly, we assessed the redundancy of plasma cell clones detected by scBCR-seq to gain insight into their origin (Fig. 8B). Although in the young dura only a few plasma cell clones could be detected, we found an important clonal overlap with blood, indicating that young plasma cells (mostly IgA) infiltrate the dura from the periphery, as recently demonstrated (20). By contrast, plasma cell clones in the aged dura (mostly IgM) exhibited a negligible overlap with the blood repertoire, indicating that most of these cells were not blood-derived. Of note, we found a clonal overlap (15.4%) between dura plasma cells and dura ABCs, suggesting that, in aged mice, some dura ABCs may locally undergo terminal differentiation into IgM secreting plasma cells. The top ten most expanded clones detected in the aged dura specifically mapped on both ABC and plasma cell clusters (Fig. 8C), supporting the possibility of a clonal relatedness between these two populations.
Discussion
Recent studies have shed light on the origin and phenotypic diversity of the myeloid cell landscape at the CNS borders (8, 21-24). Yet, little is known about B cells resident at CNS interfaces. Using complementary techniques, we demonstrated that meningeal B cells encompass multiple stages of the development. These B cells originate in calvaria and infiltrate the meninges via a network of channels uncoupled from the systemic circulation. Early B cells may complete their maturation within the meningeal compartment, wherein dura fibroblasts can provide critical molecules for B cell development, such as CXCL12 (25, 26). Using Nur77GFP mice, in which GFP expression reflects BCR engagement by self-antigens (27), we showed that transitional and mature meningeal B cells are equipped with a functional BCR, evinced by GFP expression in these populations (Fig. S16, A-C). Thus, calvaria may provide an early and prompt source of B cells that develop in the dura resulting in negative selection of B cells with high affinity for local self-epitopes. In support of this hypothesis, transgenic mice carrying the IgH chain of the mAb 8.18C5 specific for myelin oligodendrocyte glycoprotein (MOG) (28) showed a significant reduction of MOG-specific B cells in dura compared to tibial BM, suggesting that a negative selection may occur locally (Fig. S16, D-F). The calvaria was recently described as a source of meningeal neutrophils (4) and a companion paper shows that calvarial BM supplies the CNS with myeloid cells, both under homeostasis and inflammation (29). We further integrate these findings showing that meningeal B cells share a similar origin.
In aged mice, we identified a distinct population of meningeal B cells corresponding to ABC (30), which have been mostly described in the spleen (19, 31-33), but never in the CNS. scBCR-seq indicates that dura ABCs are antigen experienced B cells infiltrated from the systemic circulation. Additionally, aged dura features a dramatic expansion of IgM and, to a lower extent, IgG plasma cells. By contrast, meningeal IgA plasma cells, which derive from the gut (20, 34), appeared unaffected by aging. The infiltration of blood-borne ABC, as well as the accumulation of plasma cells in meninges, may endanger the immune privileged CNS environment during aging. Since B cells are key player in neuroinflammatory and autoimmune disorders (35, 36), these findings may help better understand the origin of self-reactive B cells in these pathologic conditions.
Materials and Methods
Animals
All mice used in this study were C57BL/6J housed under specific pathogen free conditions at Washington University School of Medicine animal facility. Homozygous Cd19Cre mice were purchased from the Jackson Laboratory (JAX stock #006785) and mated with homozygous Rosa26-STOP-tdTomato (Ai14) mice bred in house. Heterozygous Cxcl12DsRed mice were purchased from the Jackson Laboratory (JAX stock #022458) and mated with C57BL/6J mice bred in house. Nur77GFP mice were provided by Dr. P. Allen (Department of Pathology and Immunology, Washington University School of Medicine). IGHMOG mice were provided by Dr. G.F. Wu (Department of Neurology, Washington University in Saint Louis). All experiments involving laboratory animals were performed under the approval of the Institutional Animal Care and Use Committee at Washington University in St. Louis (protocol #19-0981). For flow cytometry and imaging experiments, both genders were used and equally distributed among the experimental groups. For CyTOF, scRNAseq and parabiosis experiments, females only were used to minimize biological variability. For studies on young-adult mice, 8-12-week-old mice were used. For studies on aged mice, 20-25-month-old mice were used. On the day of sacrifice, mice received a lethal dose of ketamine-xylazine injected i.p. After complete loss of the paw-pinch reflex, blood samples were collected by heart puncture. All other tissues analyzed in this study (dura, brain, spleen, bone marrow) were collected upon perfusion with 30 ml of ice-cold PBS. Parabiotic mice were prepared by the surgery core in the Hope Center for Neurological Disorders (Washington University, St. Louis, MO) as previously described (37).
Intravenous injection of CD19-Tomato splenocytes
Five-month-old CD19-Tomato mice were sacrificed with CO2 and spleens were immediately collected into ice-cold PBS. Splenocytes were mechanically extracted by mashing the spleen on a 70 μm strainer and collected in a 50 ml falcon tube. Samples were pelleted by centrifugation at 300g for 15 min at 4°C, followed by red blood cells lysis on ice for 2 min. Splenocytes were then washed in sterile PBS, pelleted by centrifugation at 300g for 5 minutes at 4°C, and resuspended in 1 ml of sterile PBS. Before counting, cells were filtered through a 35 μm sterile strainer. Three-month-old gender-matched C57BL6 mice were used as recipients. Mice were anesthetized with a dose of 80 mg per kilogram ketamine and 10 mg per kilogram xylazine administered by i.p. injection. After complete loss of the paw-pinch reflex, mice received a retro-orbital injection of 20×106 freshly prepared splenocytes from CD19-Tomato mice. Recipient mice were sacrificed on week later and blood, spleen, brain, and dura were analyzed by flow cytometry.
ICM injection of CD19-Tomato splenocytes
A single CD19-Tomato mouse was perfused under sterile conditions and spleen was immediately collected into ice-cold PBS. Splenocytes were mechanically extracted by mashing the spleen on a 70 μm strainer and collected in a 50 ml falcon tube. Samples were pelleted by centrifugation at 300g for 15 min at 4°C, followed by red blood cells lysis on ice for 2 min. Splenocytes were then washed in sterile PBS, pelleted by centrifugation at 300g for 5 min at 4°C, and resuspended at a concentration of 0.5×106 cells/μl. Three-month-old gender-matched C57BL6 mice were used as recipients. Intra cisterna magna (ICM) injection was performed as previously described (10). Under deep anesthesia, 2.5×106 freshly prepared CD19-Tomato splenocytes (in a volume of 5 μl) were administered ICM at a flow rate of 0.5 μl/min. Mice were sutured and monitored until complete recovery. Dura mater, cervical lymph nodes, and inguinal lymph nodes were collected 24 hours post-injection.
Sample preparation for flow cytometry or scRNAseq
Blood samples underwent red blood cells lysis at room temperature for 5 min. Hereafter, sample preparation was entirely carried out either on ice or at 4°C. No enzymatic digestion was used in this study. Dura mater was stripped off the inner skull surface using precision tweezers. Single cell suspension of brain, dura and calvarial bone marrow was generated via mechanical dissociation using a dounce homogenizer. Cells were then filtered through a 70 μm strainer and collected in a 50 ml conical centrifugation tube. Splenocytes were mechanically extracted by mashing the spleen on a 70 μm strainer and collected in a 50 ml falcon tube. Tibial bone marrow was extracted from open tibias into a 1.5 ml microcentrifuge tube by centrifugation at top-speed for 20 s using a bench centrifuge. Spleen and bone marrow samples underwent red blood cells lysis on ice for 2 min. Samples were pelleted by centrifugation at 300g for 15 min at 4°C. Dura pellets were resuspended in 5 ml of 75% isotonic percoll, overlaid with 3 ml of PBS. Stromal cells and debris were depleted by centrifugation at 1000g for 30 min at 4°C (acceleration 0, break 1). The interface percoll-PBS was collected for analysis. Brain pellets were resuspended in 5 ml of 30% isotonic percoll, overlaid with 2 ml of PBS. Myelin was depleted by centrifugation at 1000g for 30 min at 4°C (acceleration 0, break 1). The pellet was collected for analysis.
Flow cytometry analysis
Single cell suspensions were washed in PBS followed by live/dead staining (Zombie Aqua or Zombie UV, Biolegend) at 1:1000 dilution for 15 min on ice. Fc-receptor blockade was performed using CD16/32 blocking antibody (clone 93, Biolegend; or clone 197, made in house) incubated 10 min on ice. Surface staining was always performed between 30 min and 1 hour on ice (complete antibody list in Table S4). For staining of intracellular markers, fixation and permeabilization were performed using either the BD Cytofix/Cytoperm kit (BD Bioscience), or the Foxp3/Transcription Factor Staining kit (eBioscience), according to the product instructions. For ApoE intracellular staining, 1 mg/ml of purified anti-ApoE (clone HJ6.3, kindly provided by David Holtzman) was conjugated with AlexaFluor647 and fixed/permeabilized cells were stained overnight (1:100 dilution) at 4°C. For staining of IGHMOG BCR, B cells were incubated with 2 mg/ml biotin-conjugated recombinant MOG (1:100 dilution) for 30 min on ice prior to surface markers staining. Flow cytometry analysis was performed on BD X20, BD LSR Fortessa, or BD Canto-II (BD Bioscience). Raw data were analyzed with FlowJo v10.
Skull bone marrow transplantation
Twelve-week-old C57BL/6J (Cd45.2) mice were anesthetized with a dose of 80 mg per kilogram ketamine and 10 mg per kilogram xylazine administered by i.p. injection. Mice were inserted into a one-inch-thick lead shield (1 inch-thick Lead Vial Shield, 50 ml, Pinestar Technology) leaving only the head exposed. Each mouse received 11 Gy of gamma-irradiation, split into two doses 4 hours apart. Immediately after the second dose, mice were reconstituted with 2×106 Cd45.1 bone marrow cells injected i.v. Mice were left undisturbed in their home cage for the following four weeks. On the day of experiment, samples were processed as described above. Percentage of chimerism within different immune populations was determined by staining for CD45.2 and CD45.1.
Immunofluorescence staining
Brains, spinal cords, and skull caps were fixed in 4% paraformaldehyde (PFA) at 4°C overnight. Fixed specimens were then dehydrated in 30% sucrose solution for at least 48 hours and then cut into 60-100 μm-thick sections at the cryostat (Leica). Staining on free-floating sections was performed. Cryosections were blocked for 4 hours in 5% BSA solution with 0.5% Triton X-100. Primary antibody staining was performed in 1% BSA solution with 0.5% Triton X-100, for 48 hours at 4°C. Secondary staining with fluorochrome-conjugated antibodies was performed at room temperature for 2 hours (complete antibody list in Table S4). Immunostained sections were mounted on Superfrost glass slides (Fisher Scientific) and embedded in Prolong Glass anti-fade mounting media (Thermofisher). Lymph nodes were fixed and dehydrated as above and sliced into 20 μm-thick sections at the cryostat. Sections were directly mounted on the Superfrost glass slides and air dried. Blocking was performed for 1 hour (5% BSA, 0.5% Triton X-100), and antibody staining for 24 hours at 4°C (1% BSA, 0.5% Triton X-100). For whole mount preparation of dura mater, skull caps were fixed as above, and then decalcified in 0.5M pH 8.0 EDTA solution (Corning) for 48 hours at 4°C. Blocking and staining were performed as above. The whole skull was mounted on Superfrost glass slides and embedded in Fluoromount-G anti-fade mounting media (Southern Biotech). Stained samples (both cryosections and whole mount preparations) were covered with 1.5H high precision cover glass (Marienfeld Superior) and let dry overnight at room temperature before imaging.
Confocal imaging
Confocal imaging of dura mater whole mounts preparations and brain cryosections was performed using a Zeiss LSM880 airyscan inverted confocal microscope equipped with a 34-channel GaAsp (gallium arsenide phosphide) detector. Cryosections were imaged with a 40X/NA1.4 oil-immersion objective. Dura whole mounts were imaged with a 40X/NA1.2 water-immersion objective. Images were acquired at 2048×2048 pixel resolution, 20/50μm-thick z-stack z-step=1μm, line averaging=2, using ZEN Black (ZEISS Efficient Navigation) software (Zeiss). When needed, tile-scan mode was used to generate large filed-of-view images. Confocal imaging of leptomeninges was performed on fresh unfixed brains from young adult CD19Cre:Rosa26-TdTomato mice. Briefly, mice were anesthetized with a lethal dose of ketamine and xylazine administered by i.p. injection. After complete loss of the paw-pinch reflex, blood vessels were fluorescently labeled by retro-orbital injection of 100 μl of Tomato-Lectin DyLight-488 (Vector Laboratories, 1:1 dilution). Two minutes later, the mouse was decapitated, and the brain was immediately rinsed in ice-cold PBS and imaged using a Leica SP8 inverted confocal microscope. Images were acquired with a 25X/NA0.95 water-immersion objective, at 2048×2048 or 4096×4096 pixel resolution, 70 μm-thick z-stack, z-step=5μm, line and frame average=3, using Leica Application Suite, LAS X (Leica Microsystems). Maximal projections were rendered in Imaris V8.3 (Bitplane, Zurich, Switzerland).
Two-photon imaging
Young adult CD19Cre:Rosa26-TdTomato mice were anesthetized with a dose of 80 mg per kilogram ketamine and 10 mg per kilogram xylazine administered by i.p. injection. After complete loss of the paw-pinch reflex, fur on the head and neck was shaved and the skin on the parietal skull bone was surgically removed. Skull bone was thinned using an electrical micro drill and the head was fixed on a metal holder to minimize movements during the live imaging. Immediately before imaging, blood vessels were fluorescently labeled by retro-orbital injection of 100 μl of Tomato-Lectin DyLight-488 (Vector Laboratories, 1:1 dilution). Time-lapse in vivo imaging was performed with a Leica SP8 2-photon imaging system. DyLight-488 and Tdtomato were excited with Mai Tai Deepsee and Insight Deepsee lasers (Specta Physics, Santa Clara Ca, USA) optimally tuned to 920 nm and 1050 nm respectively. Emission spectrums were collected on ultrasensitive hybrid detectors as follows: Tdtomato >560 nm, DyLight-488 between 495 and 560nm, second harmonic <458nm. The subdural space was identified using the second harmonic generation and the blood vessels as territory landmarks. Time-lapse imaging was performed with a 25X/NA0.95 water-immersion objective, at 1024×1024 pixel resolution, z-stacks=40 μm, z-step=2.5 μm, acquisition speed=4 frames per min, imaging time=30 min, using Leica Application Suite, LAS X (Leica Microsystems). Maximal projection and video editing were performed with Imaris V8.3 (Bitplane, Zurich, Switzerland).
Mass Cytometry (CyTOF) analysis
Single cell suspension from dura mater, tibial bone marrow and blood were obtained as described above. Cells were resuspended in Cy-FACS buffer (PBS, Rockland; 0.1% BSA, Sigma A3059; 0.02% NaN3; 2mM EDTA). Fc-receptor blockade was performed using CD16/32 blocking antibody (clone 93, Biolegend, 1:100). Cell were stained for 1 hours on ice with the surface staining cocktail (complete antibody list in Table S4). Viability staining was performed by resuspending cells in 2.5 μM Cisplatin for 60 s. Cells were then fixed with 4% PFA for 30 min on ice followed by overnight DNA staining with Iridium-labeled intercalator (1:3000). Cells were acquired with the Mass Cytometry system CyTOF2 (Fluidigm) and analyzed using Cytobank. Immune cells were hierarchically gated as follows: nucleated cells, exclusion EQ calibration beads, live cells, single cells, CD45+. Dimensionality reduction analysis was performed on CD45+ cells using the viSNE toolX in Cytobank to apply the Barnes-Hut implementation of the t-SNE algorithm. Unsupervised clustering of the total CD45+ cells was performed using the following parameters: iterations = 2000; perplexity = 100; Theta = 0.5. This setup has been chosen as it demonstrated to produce the best separation of biologically meaningful populations in the three compartments. Next, CD45+CD19+ population from blood, BM and dura were further analyzed with FlowJo v10. B cells populations from independent replicates were aggregated and down-sampled to 2000 single/live B cells per compartment. Lastly, down-sampled files were concatenated and re-clustered by tSNE algorithm using default settings (iterations = 1000; perplexity = 20; Theta = 0.5).
BrdU staining for cell proliferation
Mice received two i.p. injections 6 hours apart of 5 mg BrdU (Sigma) dissolved in PBS. Mice were randomized and sacrificed at multiple timepoints post-injection (24 hours, 3 days, 5 days, 7 days). Samples were prepared as described above, followed by BrdU staining using the BrdU flow Kit (BD Pharmingen). Briefly, after surface staining, cells were fixed with fix/perm BD buffer for 20 min on ice, followed by 10 min nuclear permeabilization with BD perm-plus buffer, and a second fixation for 5 min at room temperature. Fixed/permeabilized cells were incubated with DNase-I (300 μg/ml) for 1 hours at 37°C, followed by intranuclear BrdU staining (Biolegend clone 3D4, 5 μl BrdU-PE per sample) for 30 min at room temperature. Flow cytometry analysis was performed on BD LSR Fortessa.
Transmission Electron Microscopy
After perfusion with ice-cold PBS, skull caps were removed and fixed with 4% PFA, 0.1% glutaraldehyde (Polysciences Inc., Warrington, PA) in phosphate-buffer 0.1M, pH 7.2 overnight at 4°C. Samples were decalcified in 0.5M pH 8.0 EDTA solution (Corning) for 48 hours at 4°C and fixed again in 2% PFA, 2.5% glutaraldehyde in 100 mM sodium cacodylate buffer, pH 7.2 for 2 hours at room temperature. Samples were washed in sodium cacodylate buffer at room temperature and post-fixed in 2% osmium tetroxide (Polysciences Inc.) for 2 hours at room temperature. Samples were then rinsed extensively in dH20 prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella Inc., Redding, CA) for 1 hour at room temperature. After several rinses in dH20, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella Inc.). Thick sections of 400 nm were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems Inc., Bannockburn, IL) and stained with 1% toluidine blue, 2% sodium borate. Brightfield images were acquired on a Zeiss AxioObserver D1 inverted microscope (Carl Zeiss Inc. Thrownwood, NY) equipped with an Axiocam 503 color camera. Areas of interest were identified, and images were acquired with 20X/NA0.5 and 40X/NA1.2 objectives. Ultrathin sections of 95 nm from the areas of interest were then collected on Cu grids, stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA Inc., Peabody, MA) equipped with an AMT 8 megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques, Woburn, MA).
X-ray tomography
Upon perfusion, mouse cranium was fixed in 4% PFA overnight at 4°C and stained with 10% Lugols iodine solution (38) at 21°C for 5 days prior to imaging. Following incubation, samples were embedded in 2% agarose, and imaged using a Zeiss Versa 520 (Carl Zeiss Microscopy, White Plains, NY) using either a 0.4X flat panel detector or a 4X objective at 80 kV. Final tomograms were visualized, and resultant images were generated in Zeiss XM3DViewer v.1.2.9. Z-projections were generated in ImageJ.
Single-cell RNA sequencing and single-cell BCR sequencing library preparation
For scRNAseq three 10-week-old and three 25-month-old female mice were used. Single-cell suspension from dura mater, tibial bone marrow and blood were obtained as described above. No sorting or enrichment was used as this experiment aimed to achieve a comprehensive transcriptomic representation of both immune and stromal cells. Samples were resuspended into low binding Eppendorf tube with PBS + 0.04% BSA at a final concentration of ~1000 cells/μl. Single cells gene-expression and V(D)J libraries were prepared by the McDonnell Genome Institute (MGI) at Washington University using the 5' Single Cell with V(D)J Enrichment Reagents Kit from 10X Genomics. Using the 10X Chromium VDJ + 5' Gene expression v.1 platform, up to 17,500 cells were partitioned into nanoliter Gel-bead-in-Emulsions (GEMs) droplets. Each GEM undergoes retro-transcriptase reaction to generate single-cell cDNA and receives a unique 12 nt cell barcode and unique molecular identifier (UMI). GEM cDNA was amplified for 11 cycles before being purified using SPRIselect beads. For scRNAseq, 10μl of purified cDNA was used to generate the library for RNA sequencing. Libraries were sequenced on a NovaSeq S4 (Illumina, 300 cycle kit) flow cell, targeting 50K read pairs/cell. For scBCRseq, 2μl of purified cDNA was used to generate the sequencing of the V(D)J region. Sequencing was performed using a NovaSeq S4 (Illumina, 300 cycle kit) flow cell, targeting 5K read pairs per cell.
Single-cell RNAseq analysis
Cell Ranger Software Suite (v3.1.0) from 10X Genomics was used for sample demultiplexing, barcode processing, and single-cell counting. Cell Ranger count was used to align samples to the reference genome GRCm38 (2.0.1), quantify reads, and filter reads and barcodes. Contamination by ambient RNA in droplets was normalized using SoupX (v1.4.5) (https://github.com/constantAmateur/SoupX). Furthermore, contaminating genes from erythrocytes and platelets (Tubb1, Gp1bb, Gp9, Clu, Nrgn, Ppbp, Pf4, Gng11, Hbb-b1, Hbb-b2, Hba-a1, Hba-a2) were filtered out from the dataset. The Seurat (v3.2.2) package in R was used for downstream analysis. For quality control, cells with mitochondrial content >20% were removed. Cells with low UMI and gene number per cell were filtered out. Cutoffs for UMI and gene number were determined on the basis of histograms showing cell density as a function of UMI per gene counts. For dura and bone marrow samples, cutoffs of >500 UMI and >250 genes were applied. For blood samples, cutoffs of >500 UMI and >1000 genes were applied. After filtering, a total of 98,378 cells remained with a median 3152 UMI and median 1264 genes per cell. Genes expressed in fewer than 10 cells were removed from the dataset.
Samples from different tissues were clustered separately. For each tissue, data were normalized using the SCTransform method regressed on mitochondrial gene percentage and integrated using FindIntegrationAnchors function and Canonical Correlation Analysis (CCA) (39, 40). Principle component analysis was performed, and the top 40 principle components were selected for dimensionality reduction using the Uniform Approximation and Projection (UMAP) algorithm. For identification of marker genes and differential expression analysis, we used the MAST algorithm implemented through the Seurat R package (41). Marker genes were identified by comparing each cluster against all other clusters using the FindConservedMarkers function with default settings (log-fold change threshold of 0.25 and >10% cells expressing the gene) and age as the grouping variable. Cell clusters from each tissue were annotated based on marker gene expression and clusters consisting of doublets were manually removed. For further analyses of B cell heterogeneity, B cell clusters were extracted and re-clustered. For re-clustering analyses, data were normalized using a scaling factor of 10,000 and mitochondrial read percentage was regressed with a negative binomial model. Principal component analysis was performed using the top 2,000 most variable genes and UMAP analysis was performed using the top 10 principle components. Clustering was performed using a resolution of 0.4. For data visualization and figure preparation of scRNA-seq data BBrowser version 2.7.5 was used (42).
Single-cell pseudotime trajectories were inferred using diffusion map algorithms implemented through the R package destiny (43). Normalized expression values were used as input for the generation of diffusion maps. Cells were ordered based on the first diffusion component. To further visualize B lineage differentiation, pseudotime was inferred using the slingshot R package (44). Gene Ontology analysis for biological processes enriched in different B cell populations was performed on the top 50 transcripts in each cluster using Metascape (https://metascape.org) (45). Receptor-ligand interactions were mined from the scRNA-seq data using the NicheNet algorithm (46). In brief, NicheNet analysis was performed using the marker genes for all clusters of dura B cell and dura fibroblast using default settings. After calculation of interaction scores between possible receptor-ligand combinations, pairs were filtered for those bona fide interactions that were documented in the literature and publicly available databases.
Single-cell BCR repertoire analysis
Sample demultiplexing and barcode processing was performed using the Cell Ranger Single-Cell Software Suite (10x Genomics). CellRanger-5.0.12 was used to align reads to the reference genome (vdj_GRCm38_alts_ensembl-5.0.0) and assemble BCRs. BCR sequencing data was processed using hte immcantation framework (immcantation.org) (47, 48). Germline V(D)J gene annotation was performed for all 10x Genomics BCR sequences using IgBLAST v1.16.0, with a curated set of Ig reference alleles specific for the C57BL/6 strain of Mus musculus (49). IgBLAST output was parsed using Change-O v0.4.6 (47). Additional quality control required sequences to be productively rearranged and have valid V and J gene annotations, consistent chain annotation (excluding sequences annotated with H chain V gene and L chain J gene), and a junction length that is a multiple of 3. Cells with exactly the same barcode and BCR sequences between different samples were excluded. Furthermore, only cells with exactly one H chain sequence paired with at least one L chain sequence were kept. After processing, there were 52,509 cells with paired H and L chains of which 19,447 had paired gene expression data.
B cell clonal lineages were inferred using hierarchical clustering with single linkage (50, 51). Cells were first partitioned based on common H and L chain V and J gene annotations and junction region lengths. Within each partition, cells whose H chain junction regions were within 0.07 normalized Hamming distance from each other were clustered as clones. This distance threshold was determined by manual inspection to identify the local minimum between the two modes of the bimodal distance-to-nearest distribution. The clones were further split based on the common L chain V and J gene.
Mutation frequency was calculated for cells by counting the number of nucleotide mismatches from the germline sequence in the H chain variable segment leading up to the CDR3. The calculation was performed using the calcObservedMutations function from SHazaM v1.0.2.
Statistical analysis
For flow cytometry and imaging data, graphs and statistics were produced using the Graph-Pad Prism 8 software package. Statistical difference between two groups was determined by either two-tailed unpaired Student’s t test (when both groups passed the Kolmogorov Smirnov normality test) or two-tailed Mann Whitney U test (when at least one group did not pass the Kolmogorov Smirnov normality test). When the effects of two independent variables were considered, the two-way analysis of variance (ANOVA) with Bonferroni post-hoc test was used. When more than two groups were compared, one-way analysis of variance (ANOVA) with Bonferroni post-hoc test was used. All statistical analyses display individual values for each biological replicate, mean value, and standard error of the mean (SEM). Statistical significance was set at p-value <0.05.
Supplementary Material
Acknowledgments:
We thank Dr. E. Lantelme for the excellent management of the flow cytometry core facility at the department of Pathology and Immunology (Washington University in St. Louis). We thank Dr. D. Bender and R. Lin from the Bursky Center for Human Immunology & Immunotherapy Programs (CHiiPs) for the precious support concerning sample preparation for CyTOF analysis and acquisition of CyTOF data. We thank B. Saunders, Dr. B. Zinselmeyer and Dr. G. Randolph for the assistance with the two-photon in-vivo imaging. We thank Y. Chen and Dr. D. Holtzman for providing the anti-ApoE antibody. We also thank Dr. L. Adamo for providing CD19-Tomato mice for preliminary experiments, Dr. P. Allen for providing Nur77GFP mice, Dr. A. Archambault for providing the IGHMOG mice, Dr. N. Sakaguchi for providing Rag1-GFP mice, and K. Ayasoufi for helping with the perfusion and shipment of Rag1-GFP samples. Lastly, we thank Dr. R. Pelanda, J. Andersson, F. Melchers, A. Pernis, P. Kendall and A. Ellebedy for the critical comments and helpful suggestions during the manuscript preparation.
Funding:
This research was supported in part by NIH R01AI104739 (S.H.K.) and the Gruber Foundation (M.W.). V.P was supported by F30 DK127540-01 and T32 DK 77653-28 NIH fellowship awards. R.S.C. was supported by FA-2020-01-IBD-1 fellowship award from the Lawrence C. Pakula, MD IBD Education & Innovation Fund.
Footnotes
Competing interests: All authors declare no conflict of interest.
Data and materials availability: scRNA-seq and scBCR-seq data available from NCBI GEO with accession number GSE173144. All other data and materials are available in the paper and supplementary materials.
References:
- 1.Alves de Lima K, Rustenhoven J, Kipnis J, Meningeal Immunity and Its Function in Maintenance of the Central Nervous System in Health and Disease. Annu. Rev. Immunol (2020), doi: 10.1146/annurev-immunol-102319-103410. [DOI] [PubMed] [Google Scholar]
- 2.Engelhardt B, Vajkoczy P, Weller RO, The movers and shapers in immune privilege of the CNS. Nat. Immunol (2017), , doi: 10.1038/ni.3666. [DOI] [PubMed] [Google Scholar]
- 3.Rua R, McGavern DB, Advances in Meningeal Immunity. Trends Mol. Med (2018), , doi: 10.1016/j.molmed.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herisson F, et al. , Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration. Nat. Neurosci (2018), doi: 10.1038/s41593-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai R, et al. , Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections. Nat. Neurosci (2019), doi: 10.1038/s41593-018-0301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao H, et al. , Leukaemia hijacks a neural mechanism to invade the central nervous system. Nature (2018), doi: 10.1038/s41586-018-0342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keren-Shaul H, et al. , A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Mundt S, Mrdjen D, Utz SG, Greter M, Schreiner B, Becher B, Conventional DCs sample and present myelin antigens in the healthy CNS and allow parenchymal T cell entry to initiate neuroinflammation. Sci. Immunol (2019), doi: 10.1126/sciimmunol.aau8380. [DOI] [PubMed] [Google Scholar]
- 9.Rickert RC, Roes J, Rajewsky K, B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. (1997), doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louveau A, et al. , CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat. Neurosci (2018), , doi: 10.1038/s41593-018-0227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn JH, et al. , Meningeal lymphatic vessels at the skull base drain cerebrospinal fluid. Nature (2019), doi: 10.1038/s41586-019-1419-5. [DOI] [PubMed] [Google Scholar]
- 12.Tung JW, Mrazek MD, Yang Y, Herzenberg LA, Herzenberg LA, Phenotypically distinct B cell development pathways map to the three B cell lineages in the mouse. Proc. Natl. Acad. Sci. U. S. A (2006), doi: 10.1073/pnas.0511305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K, Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med (1991), doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuwata N, Igarashi H, Ohmura T, Aizawa S, Sakaguchi N, Cutting edge: absence of expression of RAG1 in peritoneal B-1 cells detected by knocking into RAG1 locus with green fluorescent protein gene. J. Immunol (1999). [PubMed] [Google Scholar]
- 15.DeSisto J, et al. , Single-Cell Transcriptomic Analyses of the Developing Meninges Reveal Meningeal Fibroblast Diversity and Function. Dev. Cell (2020), doi: 10.1016/j.devcel.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schena F, et al. , Dependence of Immunoglobulin Class Switch Recombination in B Cells on Vesicular Release of ATP and CD73 Ectonucleotidase Activity. Cell Rep. (2013), doi: 10.1016/j.celrep.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Rustenhoven J, et al. , Functional characterization of the dural sinuses as a neuroimmune interface. Cell. 184, 1000–1016.e27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almanzar N, et al. , A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature (2020), doi: 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogilenko DA, et al. , Comprehensive Profiling of an Aging Immune System Reveals Clonal GZMK+ CD8+ T Cells as Conserved Hallmark of Inflammaging. Immunity (2021), doi: 10.1016/j.immuni.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Fitzpatrick Z, et al. , Gut-educated IgA plasma cells defend the meningeal venous sinuses. Nature (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldmann T, et al. , Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol 17, 797–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordão MJC, et al. , Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science (80-. ). (2019), doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 23.Van Hove H, et al. , A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci (2019), doi: 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 24.Mrdjen D, et al. , High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity. 48, 380–395.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Fistonich C, et al. , Cell circuits between B cell progenitors and IL-7+mesenchymal progenitor cells control B cell development. J. Exp. Med (2018), doi: 10.1084/JEM.20180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie Y, Waite J, Brewer F, Sunshine MJ, Littman DR, Zou YR, The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med (2004), doi: 10.1084/jem.20041185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zikherman J, Parameswaran R, Weiss A, Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 489, 160–164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litzenburger T, et al. , B lymphocytes producing demyelinating autoantibodies: Development and function in gene-targeted transgenic mice. J. Exp. Med 188, 169–180 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cugurra JKA, Mamuladze T, Rustenhoven J, Dykstra T, Beroshvili G, Greenberg ZJ, Baker W, Papadopoulos Z, Drieu A, Blackburn S, Kanamori M, Brioschi S, Herz J, Schuettpelz LG, Colonna M, Smirnov I, Skull and vertebral bone marrow are myeloid reservoirs for the meninges and CNS parenchyma. Science. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancro MP, Age-Associated B Cells. Annu. Rev. Immunol (2020), doi: 10.1146/annurev-immunol-092419-031130. [DOI] [PubMed] [Google Scholar]
- 31.Rubtsov AV, et al. , Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c+ B-cell population is important for the development of autoimmunity. Blood (2011), doi: 10.1182/blood-2011-01-331462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao Y, O’Neill P, Naradikian MS, Scholz JL, Cancro MP, A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood (2011), doi: 10.1182/blood-2011-01-330530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manni M, et al. , Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat. Immunol (2018), doi: 10.1038/s41590-018-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rojas OL, et al. , Recirculating Intestinal IgA-Producing Cells Regulate Neuroinflammation via IL-10. Cell (2019), doi: 10.1016/j.cell.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauser SL, et al. , B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N. Engl. J. Med (2008), doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 36.Hauser SL, et al. , Ofatumumab versus Teriflunomide in Multiple Sclerosis. N. Engl. J. Med (2020), doi: 10.1056/nejmoa1917246. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, et al. , TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med 213, 667–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metscher BD, MicroCT for developmental biology: A versatile tool for high-contrast 3D imaging at histological resolutions. Dev. Dyn (2009), doi: 10.1002/dvdy.21857. [DOI] [PubMed] [Google Scholar]
- 39.Stuart T, et al. , Comprehensive Integration of Single-Cell Data. Cell (2019), doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. (2019), doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finak G, et al. , MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. (2015), doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le T, et al. , BBrowser : Making single-cell data easily accessible (2020), doi: 10.1101/2020.12.11.414136. [DOI] [Google Scholar]
- 43.Angerer P, Haghverdi L, Büttner M, Theis FJ, Marr C, Buettner F, Destiny: Diffusion maps for large-scale single-cell data in R. Bioinformatics (2016), doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 44.Street K, et al. , Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics (2018), doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y, et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun (2019), doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Browaeys R, Saelens W, Saeys Y, NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH, Change-O: A toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics (2015), doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vander Heiden JA, et al. , pRESTO : a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. 30, 1930–1932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alsoussi WB, et al. , A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol 205, 915–922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta NT, Adams KD, Briggs AW, Timberlake SC, Vigneault F, Kleinstein SH, Hierarchical Clustering Can Identify B Cell Clones with High Confidence in Ig Repertoire Sequencing Data. J. Immunol 198, 2489–2499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou JQ, Kleinstein SH, Cutting Edge: Ig H Chains Are Sufficient to Determine Most B Cell Clonal Relationships. J. Immunol 203, 1687–1692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.