Abstract
STUDY QUESTION
What are the long-term developmental, reproductive and genetic consequences of mitochondrial replacement therapy (MRT) in primates?
SUMMARY ANSWER
Longitudinal investigation of MRT rhesus macaques (Macaca mulatta) generated with donor mtDNA that is exceedingly distant from the original maternal counterpart suggest that their growth, general health and fertility is unremarkable and similar to controls.
WHAT IS KNOWN ALREADY
Mitochondrial gene mutations contribute to a diverse range of incurable human disorders. MRT via spindle transfer in oocytes was developed and proposed to prevent transmission of pathogenic mtDNA mutations from mothers to children.
STUDY DESIGN, SIZE, DURATION
The study provides longitudinal studies on general health, fertility as well as transmission and segregation of parental mtDNA haplotypes to various tissues and organs in five adult MRT rhesus macaques and their offspring.
PARTICIPANTS/MATERIALS, SETTING, METHODS
MRT was achieved by spindle transfer between metaphase II oocytes from genetically divergent rhesus macaque populations. After fertilization of oocytes with sperm, heteroplasmic zygotes contained an unequal mixture of three parental genomes, i.e. donor (≥97%), maternal (≤3%), and paternal (≤0.1%) mitochondrial (mt)DNA. MRT monkeys were grown to adulthood and their development and general health was regularly monitored. Reproductive fitness of male and female MRT macaques was evaluated by time-mated breeding and production of live offspring. The relative contribution of donor, maternal, and paternal mtDNA was measured by whole mitochondrial genome sequencing in all organs and tissues of MRT animals and their offspring.
MAIN RESULTS AND THE ROLE OF CHANCE
Both male and female MRT rhesus macaques containing unequal mixture of three parental genomes, i.e. donor (≥97%), maternal (≤3%), and paternal (≤0.1%) mtDNA reached healthy adulthood, were fertile and most animals stably maintained the initial ratio of parental mtDNA heteroplasmy and donor mtDNA was transmitted from females to offspring. However, in one monkey out of four analyzed, initially negligible maternal mtDNA heteroplasmy levels increased substantially up to 17% in selected internal tissues and organs. In addition, two monkeys showed paternal mtDNA contribution up to 33% in selected internal tissues and organs.
LARGE SCALE DATA
N/A.
LIMITATIONS, REASONS FOR CAUTION
Conclusions in this study were made on a relatively low number of MRT monkeys, and on only one F1 (first generation) female. In addition, monkey MRT involved two wildtype mtDNA haplotypes, but not disease-relevant variants. Clinical trials on children born after MRT will be required to fully determine safety and efficacy of MRT for humans.
WIDER IMPLICATIONS OF THE FINDINGS
Our data show that MRT is compatible with normal postnatal development including overall health and reproductive fitness in nonhuman primates without any detected adverse effects. ‘Mismatched’ donor mtDNA in MRT animals even from the genetically distant mtDNA haplotypes did not cause secondary mitochondrial dysfunction. However, carry-over maternal or paternal mtDNA contributions increased substantially in selected internal tissues / organs of some MRT animals implying the possibility of mtDNA mutation recurrence.
STUDY FUNDING/COMPETING INTEREST(S)
This work has been funded by the grants from the Burroughs Wellcome Fund, the National Institutes of Health (RO1AG062459 and P51 OD011092), National Research Foundation of Korea (2018R1D1A1B07043216) and Oregon Health & Science University institutional funds. The authors declare no competing interests.
Keywords: heteroplasmy, mtDNA segregation, mitochondrial replacement therapy (MRT), germline transmission, maternal mtDNA carryover, paternal mtDNA transmission
Introduction
Mitochondrial (mt)DNA encodes 13 proteins of the oxidative phosphorylation (OXPHOS) machinery, while the remaining components are provided by the nuclear genome (nDNA) (Shoubridge, 2001; Wallace, 2007). Neutral mtDNA sequence differences between individuals, observed as single-nucleotide polymorphisms (SNPs), form the basis for human haplotype variations (Wallace and Chalkia, 2013). In addition to neutral SNPs, pathogenic mtDNA variants have been identified that are associated with serious maternally inherited disorders including myopathies, neurodegenerative diseases and diabetes (Stewart and Chinnery, 2015; Russell et al., 2020). In the context of mitochondrial replacement therapy (MRT), the mutant mtDNA in a mother’s unfertilized oocyte can be removed and replaced with donor genomes by meiotic spindle transfer (Tachibana et al., 2009, 2010, 2013; Paull et al., 2013). Specifically, the entire cytoplasm, including the mitochondrial complement of the mother’s metaphase II-arrested oocyte, is replaced with donor cytoplasm and mtDNA derived from a healthy donor oocyte using spindle transfer. MRT can also be achieved after fertilization by pronuclear transfer (PNT) in zygotes (McGrath and Solter, 1983; Craven et al., 2010; Hyslop et al., 2016a, b). Since a randomly selected donor mtDNA haplotype is different than the original maternal mtDNA haplotype, it has been speculated that resulting ‘mismatch’ could lead to a secondary mitochondrial dysfunction in MRT children (Chinnery et al., 2014; Eyre-Walker, 2017). This is due to potential incompatibility between the nuclear genome of the mother and the new mitochondrial genome of the donor. For instance, reciprocal mtDNA replacement between two mouse subspecies with widely divergent mtDNA sequences can lead to high embryonic loss and stillborn rates (Ma et al., 2016). Mixture of two normal but different mouse mtDNAs can be genetically unstable and produce adverse physiological effects including altered behavior and cognition (Sharpley et al., 2012).
MRT does not eliminate entirely maternal mtDNA complement, leading to residual carryover of maternal haplotype, typically less than 3% for spindle transfer or less than 10% for PNT. In addition, a few copies of paternal mtDNA are also introduced during fertilization resulting in unequal heteroplasmy in a MRT zygote.
To investigate developmental consequences and vertical transmission and segregation of heteroplasmic mtDNA to somatic and germline lineages, five rhesus macaques (Macaca mulatta) (F1 (first generation), named Mito, Tracker, Spindler, Spindy and Crysta) were produced by MRT between 2009 and 2012 in our laboratory (Tachibana et al., 2009, 2013) employing parental mtDNA from geographically isolated sub-populations of Macaca mulatta from India and China. These two groups of macaques, isolated approximately 162,000 years ago (Hernandez et al., 2007), became genetically distinct exceeding those of other sub-populations and even those between different mammalian species (Smith, 2005). While these F1 MRT monkeys were generated using nuclear DNA from one sub-population and donor mtDNA from another, complete mtDNA sequence differences between parental genomes for each MRT monkey had not been yet analyzed.
Materials and methods
Experimental model and subject details
This study was performed with two groups of rhesus macaques (Macaca mulatta) from India and China housed at the Oregon National Primate Research Center (ONPRC). Five F1 MRT monkeys including four males (Mito, Tracker, Spindler and Spindy) and one female (Crysta) and three F2 (second generation) MRT offspring including two females (Nona and Mia) and one male (Mac) were included in the study. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the ONPRC of Oregon Health & Science University (OHSU).
Method details
Sperm collection and analyses
Prior to sperm collection, males were trained over a course of 6 weeks with positive reinforcement methods to sit in a chair by a trained behaviorist. At this point, two electrodes were applied to the monkey’s penis attached to a P-T Electronics Electro Ejaculator machine. Protocol for the machine was followed until a sample was collected. This sample was then analyzed by the ART Core at ONPRC for standard semen parameters including volume, color, count, motility, agglutination and morphology.
Time-mated breeding
At the onset of menses, daily blood draws were performed (normally females will ovulate on Day 12 of their menses cycle) to monitor P4 (progesterone) and E2 (estradiol) levels. When E2 reached 100 pg/ml pairing with a male ensued. Daily blood draws continued through ovulation when E2 level peaked and P4 increased. Animals remained paired for 1 day post-ovulation. At 2–4 weeks post-pairing, a prolonged rise in serum P4 and an E2 level of 100 pg/ml were taken as indicative of pregnancy that was further confirmed by ultrasound one month after pairing.
Controlled ovarian stimulation
Controlled ovarian stimulation (COS) was performed by the ART Core at ONPRC. At the onset of menses, 30 IUs of recombinant human FSH was administered twice daily for 6 consecutive days. On Days 7 and 8, 30 IUs of FSH and 30 IUs of recombinant human LH were co-injected twice per day. Approximately 0.1 mg/kg body weight of Antide was given when the estradiol level reached >200 pg/ml to block circulating GnRH, thereby preventing the endogenous LH surge. Approximately 36 h prior to follicle aspiration a single dose of 1100 IUs of hCG was injected to stimulate ovulation. Laparoscopic follicular aspiration and oocyte retrieval were performed by veterinary staff at ONPRC.
DNA extractions from oocytes and tissues
DNA from oocytes was extracted using Pico Pure DNA extraction kit (Applied BioSystems, Grand Island, NY, USA). DNA from all other tissues was extracted using Gentra Puregene DNA purification kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions.
mtDNA heteroplasmy quantification by amplification refractory mutation system (ARMs)-quantitative (q) PCR
Amplification refractory mutation system (ARMs)-quantitative (q) PCR (ARMs-qPCR) was used to measure mtDNA heteroplasmy as previously described (Ma et al., 2015). Individual primers and probes were designed specifically for each MRT monkey to detect unique SNP differences between donor and maternal mtDNA.
Whole mitochondrial genome sequencing
Whole mtDNA was amplified by a single PCR reaction using the primer set: rh2076-F: (AGACACTAGGAAAAAACCTTATAGAGAGAGT) and rh2075-R: (AAAGAGCTGTCCCTCTTTAGACTAGC) using TAKARA LA Taq polymerase (Takara Biotechnology, Shiga, Japan) under the following conditions: 94°C for 2 min followed by 30 cycles of 94°C for 20 s, 56°C for 1 min and 68°C for 16 min, with final extension at 72°C for 10 min. Concentration of PCR products was measured by the Qubit 2.0 Fluorometer (Invitrogen). Amplified PCR product was used for library preparation with the Nextera XT DNA Kit (Illumina, San Diego, CA, USA) following manufacturer’s instructions. Sequencing was performed on the Illumina MiSeq platform at the Molecular Technologies Support Core of ONPRC and/or at Asan Medical Center.
Sequence analysis by NextGENe
Mitochondrial genome sequencing (MiSeq) data were analyzed using NextGENe (Softgenetics) software. Sequence reads ranging from 200 to 300 base pair (bp) were quality filtered and processed using an algorithm similar to BLAT (BLAST-Like Alignment Tool). The sequence consolidation feature (condensation) was performed to reduce false-positive variants and produce sample consensus sequences and variant calls. Alignment without sequence condensation was used to calculate the percentage of the mitochondrial genome covered with an average depth of coverage of 1000. Quality FASTQ reads were inputted, further quality filtered and converted to FASTA format. Filtered reads were then aligned to a reference sequence specific to the sample’s pedigree, using a matching requirement of at least 12 bases or 90% similarity to the reference, rigorous alignment and removal of ambiguously mapped reads. Variants representing less than 2% total reads (pre-condensation) were excluded following a prior study (He et al., 2010). Presence of carry-over maternal or paternal mtDNA was called when >30% of SNP variants were detected in each sample. In Supplementary Tables SIV and SV, heteroplasmy values were calculated by NextGENe software as follows: base heteroplasmy (mutant allele frequency %) = mutant allele (forward + reverse)/total coverage of all alleles C, G, T and A (forward + reverse) × 100.
Genetic distance analysis for parental mtDNA
Maternal and donor consensus sequences for each MRT offspring were submitted to MITOS webserver (http://mitos.bioinf.uni-leipzig.de/index.py) for automatic annotation following default parameters. The MITOS pipeline uses a BLASTX homology search of all previously annotated NCBI (National Center for Biotechnology Information) rhesus macaque mitochondrial genomes applying an aggregation-reconciliation algorithm to determine protein-coding gene boundaries, and covariance models of sequence similarity and secondary structure to identify non-coding RNA. No manual annotation was performed. MITOS-annotated genes were aligned by Mesquite 3.2 (http://www.mesquiteproject.org). SNPs were counted manually in the nucleotide view option. Codon changes were determined by translating the sequence and manually counting amino acid differences. Heteroplasmic sites were excluded from informative SNP counts between donor and maternal mtDNA of MRT monkeys.
Statistical analysis
One-way ANOVA was used for the comparisons of sperm count and motility. A P-value less than 0.05 was considered statistically significant. No statistical methods were used to predetermine sample size.
Results
Sequence divergence between maternal and donor mtDNAs
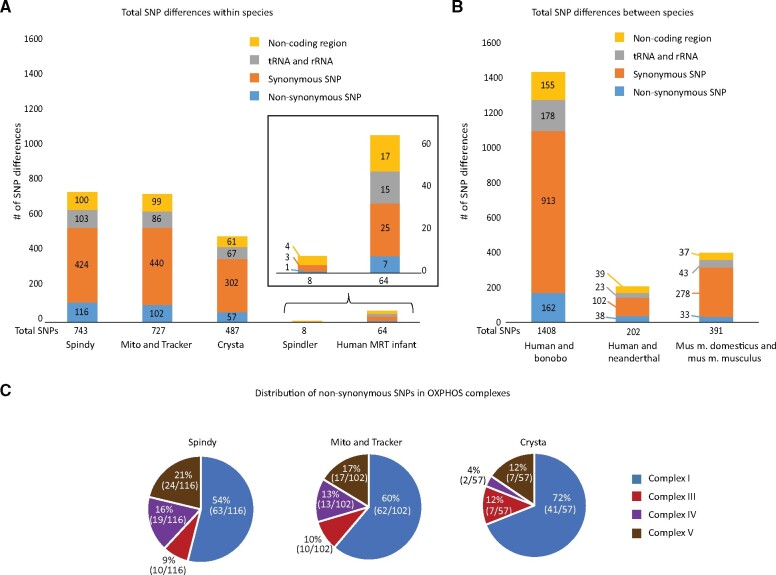
We sequenced entire mtDNA from maternal and donor blood samples for the F1 MRT offspring by next-generation sequencing using MiSeq platform. The NCBI rhesus macaque reference sequence (NC_005943.1) was used for gene boundary determination and informative SNP identification. Sequence analyses revealed that donor and maternal mtDNA in Spindler, both of Indian origin, varied in 8 homoplasmic SNP positions. While heteroplasmic SNPs were also present, only homoplasmic variants were considered for determining contributions of parental mtDNA (Supplementary Table SI). In contrast, donor/maternal mtDNA sequence differences in the other four animals produced by Indian/Chinese combinations ranged from 487 SNPs in Crysta to 743 SNPs in Spindy, representing an extreme degree of mtDNA sequence mismatch (Fig. 1A). By comparison, 64 SNPs (not including the mutation site) differentiated the maternal and donor mtDNA in a recent clinical MRT case resulting in the birth of a healthy baby (Zhang et al., 2017) (Fig. 1A). MtDNA sequence distance i.e. SNP differences, is expected to increase between sub-species and different species, for example, 391 SNPs were found between two subspecies of mice, 202 SNPs between humans and Neanderthals and 1408 SNPs between humans and bonobo (Fig. 1B).
Figure 1.
Comparisons of mitochondrial DNA (mtDNA) sequence differences. (A) Sequence differences as single-nucleotide polymorphisms (SNPs) between maternal and donor mtDNA for five mitochondrial replacement therapy (MRT) monkeys and for the human MRT infant. (B) mtDNA sequence differences in between species and distant mouse strains (PWD and C57BL/6). (C) Distribution of non-synonymous SNPs in oxidative phosphorylation (OXPHOS) complexes. Most changes resulting in a protein sequence alteration occur in complex I of the oxidative phosphorylation pathway.
In MRT monkeys, the greatest number of SNPs was found in the protein-coding genes, including non-synonymous substitutions resulting in amino acid change. The majority of non-synonymous SNPs (54–72%) were located in genes encoding the complex I protein. The remaining SNPs were distributed between the complexes III, IV and V (Fig. 1C). SNPs were also located in the RNA-coding genes and the non-coding control region (Fig. 1A). Thus, the diverse genetic background between these two sub-species of rhesus macaques is much greater than those between even the most distant human populations. These results challenge the claims that MRT macaques were generated from the ‘same troop’, arguing that this model is not representative of human MRT (Reinhardt et al., 2013).
Postnatal development of F1 MRT monkeys
While the birth of a child following MRT now has been reported (Zhang et al., 2017), questions remain regarding whether an experimental ‘mismatch’ between maternal and donor mtDNA may cause secondary mitochondrial dysfunction in children and impair their postnatal development. To address these critical issues, we have previously reported on the health of MRT macaque infants from birth to 3 years of age (Tachibana et al., 2013). Longitudinal follow-up evaluations for these macaques presented here include adult animals up to 9 years of age, corresponding to middle-aged humans (Simmons, 2016). Overall health fitness has been determined by periodic body weight measurements, routine veterinary examinations, hematological analyses and basic metabolic panels of blood samples, as well as routine testing for colony excluded viruses (SIV (Simian Immunodeficiency Virus), STLV (Simian T-cell Leukemia Virus), SRV (Simian Retrovirus) and Herpes B). Viral testing for the preceding viruses was uniformly negative for animals involved in this project. None of the animals experienced any serious physical illness. The mean body-weights (a non-invasive measure of growth) of MRT macaques were similar to the mean for a cohort of 20 control animals of same age (10 for each gender) (Supplementary Fig. S1A and B). The values for hemoglobin, red and white blood cell counts, mean corpuscular volume and hemoglobin concentration were also within normal ranges (Supplementary Table SII). Moreover, basic metabolic panels were also within the normal range (Supplementary Table SII). Furthermore, 4 adult MRT macaques (Tracker, Spindler, Spindy and Crysta) were subjected to in-depth histological evaluations of all major organs and tissues including respiratory, cardiovascular, hematopoietic, genitourinary, endocrine, musculoskeletal and central nervous systems, liver, pancreas and gastrointestinal tract. Morphological and histological evaluations revealed no significant differences from the control animals.
Reproductive fitness of MRT monkeys
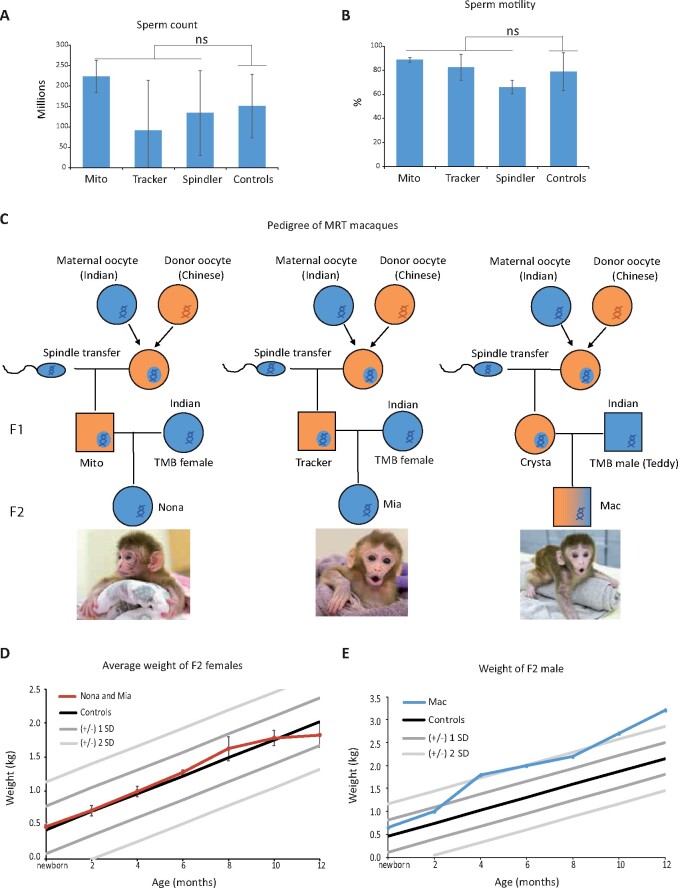
Reproductive health and specifically reduced male fertility of MRT offspring have been cautioned about based on studies of hybrid model animals (Clancy et al., 2011; Yee et al., 2013; Bhattacharyya et al., 2014). Therefore, we investigated reproductive fitness of male and female MRT macaques. Semen analysis was assessed in sexually mature adult males subjected to an IACUC approved sperm collection protocol. Two independent ejaculates from each of the three adult males were collected and analyzed for sperm volume, count, motility and sperm morphology. Control sperm samples were collected from five males of the same age. All measured semen parameters for MRT monkeys were normal and similar to controls (Fig. 2A and B; Supplementary Table SIII). Two MRT males, Mito and Tracker, were subsequently tested for natural fertility by pairing with adult females following a standard time-mated breeding (TMB) protocol. Mito established a pregnancy upon his second pairing, which resulted in a term pregnancy and birth of a healthy female infant (Nona, Fig. 2C). The first pairing of Tracker with a TMB female also resulted in a pregnancy ending in a miscarriage during the first trimester. No significant morphological abnormalities were noted after gross examination of aborted fetal tissues. However, tissue culture showed a positive reaction to alpha-hemolytic Streptococcus suggesting that bacterial infection may have caused placentitis and fetal loss. The second pairing of Tracker with another TMB female resulted in a term pregnancy and birth of a healthy female infant (Mia, Fig. 2C). The pregnancy rate of MRT males (3 pregnancies out of 4 parings, 75%) is comparable to the rate achieved with control males (29/54, 54%) (Supplementary Table SIII). The sole MRT female, Crysta, was paired with a TMB male (Teddy) resulting in pregnancy and birth by natural delivery of a healthy male infant (Mac, Fig. 2C).
Figure 2.
Reproductive fitness of MRT monkeys. (A) Sperm count for three mitochondrial replacement therapy (MRT) males (two ejaculates from each monkey) were analyzed and compared to five controls of similar age. (B) Sperm motility for the MRT males were analyzed and compared to five controls of similar age. Error bars in A and B represent the mean ± SD (n = 2 per monkey, biological replicates). ns denotes P ≥ 0.05. (C) Pedigree of MRT rhesus macaques. The Indian sub-population is represented with blue and the Chinese with orange color. (D) Comparison of weight change with age in MRT F2 females and age-matched controls. (E) Comparison of weight change with age in an MRT second generation (F2) male and age-matched controls. ns, non-significant.
Microsatellite parentage analysis by short tandem repeat assay confirmed that all three F2 infants were offspring of respective MRT parents (Supplementary Table SIII). Growth and development of these infants were also routinely examined and the growth curves of two females were within the normal ranges for rhesus macaques (Fig. 2D; Supplementary Table SIII) (Haertel et al., 2018). The male offspring Mac showed body weight within the SD in the first 8 months. However, subsequently, this animal gained more body weight than controls of the same age (Fig. 2E). We measured body weights of Mac’s father Teddy and Teddy’s other offspring (offspring 1), produced by natural breeding with non-MRT females. Their body weight curves fall within the SD as controls (Supplementary Fig. S1C). Since Mac was sacrificed at 1 year age, the cause of abnormal weight gain after 8 months remains unclear.
These results suggest normal fertility of adult MRT monkeys and imply that ‘mismatched’ donor mtDNA in these trans-mitochondrial macaques did not cause any detectable reproductive abnormalities.
Contributions of parental mtDNAs to the first generation MRT monkeys
During the MRT procedure, a small but measurable fraction of the maternal oocyte cytoplasm is inevitably co-transferred with the spindle-chromosomal complex leading to the presence of residual maternal mtDNA in MRT oocytes. We previously measured maternal and donor mtDNA copy numbers in MRT oocytes and estimated an average of 0.6% heteroplasmy for maternal mtDNA with a maximum of 3% that can be passed to MRT embryos and offspring (Tachibana et al., 2009, 2013; Lee et al., 2012; Kang et al., 2016). In clinical situations, this may result in the transmission of the mutant maternal mtDNA to children, albeit at low heteroplasmy levels. However, we and others recently reported that during in vitro passaging, some human embryonic stem cell (ESC) lines derived from MRT embryos showed a rapid increase of carry-over maternal mtDNA while donor mtDNA declined, often leading to complete reversal to homoplasmy for the maternal mtDNA (Kang et al., 2016; Yamada et al., 2016; Hyslop et al., 2016a,b). Thus far, this phenomenon was observed in vitro in ESCs but not in MRT offspring leading to questions whether the reversal is an in vitro ESC artifact or may take place in vivo in MRT children.
In addition to the low but detectable residual maternal mtDNA, sperm also introduces a few paternal mitochondria during fertilization resulting in the 3rd mtDNA complement in MRT zygotes. However, because of very low (3–5) mtDNA copy numbers per sperm versus (200 000–300 000) in oocytes, estimated sperm mtDNA heteroplasmy in zygotes or preimplantation embryos is extremely low and remains well below reliable detection levels by most existing methods (Santos et al., 2006; Wu et al., 2019).
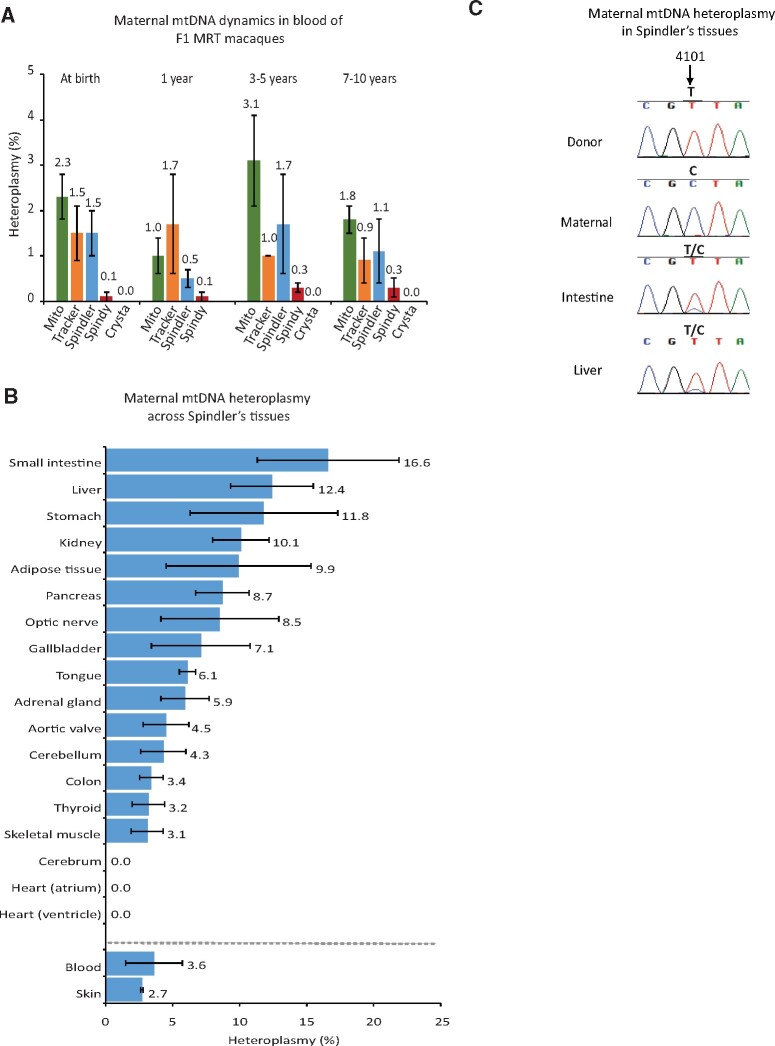
We initially investigated the presence and heteroplasmy dynamics of maternal and donor mtDNA in blood and skin of MRT macaques from birth to adults (10 years) using ARMs-qPCR enabling the measurement of heteroplasmy at ≤1% (Lee et al., 2012). All macaques carried predominantly donor mtDNA; however, maternal mtDNA was also detectable at low (≤3%) heteroplasmy levels in some blood and skin samples. This heteroplasmy did not change notably during regular samplings from infancy to adulthood (Fig. 3A, Supplementary Fig. S2A and Table SIV). Urine samples also showed the presence of maternal mtDNA at low heteroplasmy levels (Supplementary Fig. S2B and Table SIV).
Figure 3.
Detection of maternal carry-over mtDNA in MRT monkeys. (A) Maternal mtDNA heteroplasmy measured by amplification refractory mutation system (ARMs)-quantitative (q) PCR in blood of MRT monkeys at different age. (B) Heteroplasmy levels for carry-over maternal mtDNA across 20 different tissues in F1 mitochondrial replacement therapy (MRT) monkey (Spindler). The data in A and B represent the mean ± SD (n = 3, biological replicates). (C) Chromatograms of Sanger sequencing demonstrating maternal mtDNA heteroplasmy in Spindler’s intestine and liver tissues.
Previous studies reported tissue-specific segregation of heteroplasmic mtDNA in mice resulting in different heteroplasmy levels in various organs and tissues (Jenuth et al., 1997; Burgstaller et al., 2014). We reasoned that peripheral tissue samples may not fully represent heteroplasmy levels in internal tissues/organs. Therefore, we sampled all major organs and tissues from post-necropsy adult MRT monkeys and performed in-depth mtDNA analyses. In addition to ARMs-qPCR and Sanger, we also employed whole mitochondrial genome sequencing using the Illumina MiSeq platform. From the MiSeq consensus sequence for each monkey, only homoplasmic variants (SNPs) discriminating maternal from donor as well as paternal from donor mtDNA were identified and used for parentage determination.
Because of the Indian origin of the donor, maternal and paternal mtDNA for Spindler, we found only six unique informative SNP variants defining the maternal mtDNA and two unique SNPs for defining paternal mtDNA inheritance in this animal (Supplementary Table SIV). Compared to maternal mtDNA heteroplasmy levels measured in skin (2.7%) and blood (3.6%), 18 analyzed internal organs/tissues displayed a range of heteroplasmy levels from undetectable in cerebrum and heart to 10.1% in kidney, 11.8% in stomach, 12.4% in liver and 16.6% in small intestine (Fig. 3B and 3C; Supplementary Table SIV). Paternal unique mtDNA variants were undetectable by MiSeq and Sanger techniques.
For Crysta and Spindy, generated from Indian and Chinese macaques, 267 and 352 unique SNP variants respectively were available to differentiate maternal and donor mtDNA (Supplementary Table SIV). In these two animals, only traces of maternal mtDNA were observed in selected internal organs represented by only one or a few SNP variants. Unique paternal SNP variants were not present in any tested tissues/organs from these monkeys (Supplementary Table SIV).
Incidentally, Tracker’s maternal and paternal mtDNA consensus sequences appeared to be identical to each other but differed in 718 SNP variants from the donor mtDNA. Several internal organs/tissues, including skin, kidney heart, thymus, optic nerve and lung, displayed substantial heteroplasmy levels for maternal/paternal mtDNA variants ranging from 2.5% to 14.8%, while heteroplasmy was undetectable in blood, liver, spinal cord and urine samples (Supplementary Table SIV). Since no unique SNPs were available for differentiating maternal from paternal mtDNA, it remains unclear which of these parents contributed their mtDNA to the offspring.
Transmission of parental mtDNAs to the second generation MRT monkeys
We next investigated the transmission of donor, maternal and paternal mtDNA from the F1 MRT monkeys to their offspring using the same approach. As described above, Nona and Mia, offspring of F1 MRT males Mito and Tracker, were produced by natural breeding with TMB females (Fig. 2C). Since Mito or Tracker predominantly carried donor mtDNA of Chinese origin, they were bred with Indian females carrying mtDNA haplotypes distinct by 203 and 198 SNPs, respectively (Supplementary Table SV). MiSeq analysis of 27 sampled organs/tissues of Nona and Mia did not detect any paternal mtDNA variants and these monkeys carried exclusively maternal mtDNA (Supplementary Table SV).
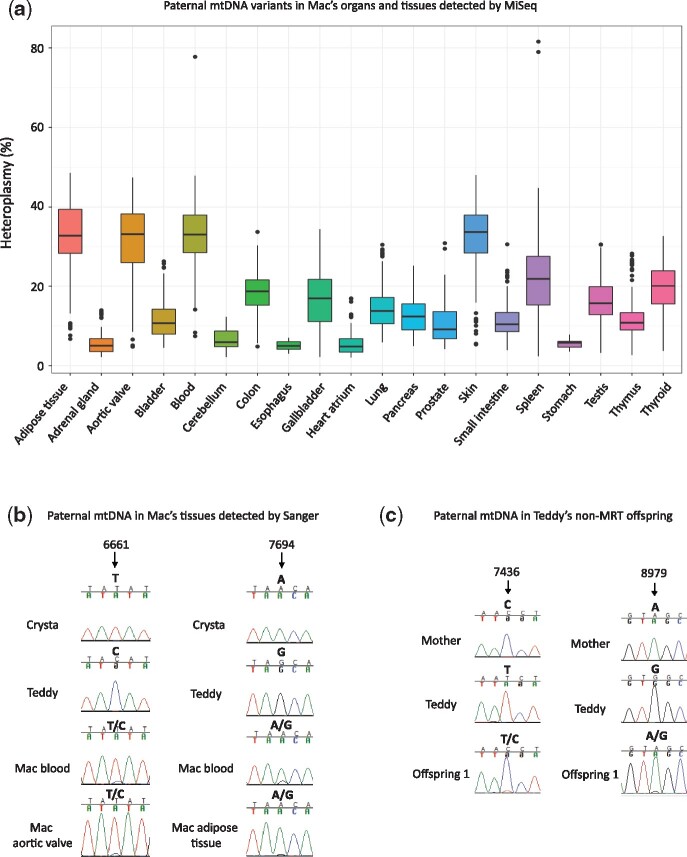
Similarly, F1 female MRT Crysta, carrying predominantly donor Chinese mtDNA, was paired with Indian male Teddy and gave birth to male infant Mac (Fig. 2C). A total of 458 unique SNP variants were different between Teddy’s and Crysta’s donor (major) mtDNA in addition to 265 unique SNPs discriminating Crysta’s donor mtDNA from Crysta’s maternal (carryover) mtDNA (Supplementary Table SV). Among 28 analyzed organs/tissues of Mac, 20 showed a mixture of variants that matched to both paternal and maternal (Crysta’s donor) mtDNA haplotypes, while the remaining eight organs carried exclusively maternal (Crysta’s donor) mtDNA (Supplementary Table SV). The mean heteroplasmy for the paternal mtDNA variants varied from 5% in esophagus to 33% in blood (Fig. 4A; Supplementary Table SV). To exclude the possibility of sample mix-up and/or contamination, MiSeq on Mac’s samples were independently repeated and verified by our collaborators from Asan Medical Center in South Korea. In addition, Sanger sequencing further corroborated the presence of paternal mtDNA in these tissues (Fig. 4B). MiSeq and Sanger analyses did not detect any of the unique variants matched to maternal carryover mtDNA in Mac’s tissues. However, since variants with heteroplasmy levels at 2% and less were excluded from analysis, we cannot rule out the possibility that paternal or maternal carryover mtDNA is present at low levels in negative organs/tissues.
Figure 4.
Paternal mtDNA transmission in rhesus macaques. (A) Detection of paternal mitochondrial DNA variants in tissues of Mac. Each box represents the interquartile range (25th to 75th percentile) of all heteroplasmic variants in each tissue, the dot represents outliers and the line within the box depicts the median. (B) Chromatograms of Sanger sequencing showing paternal mtDNA in Mac’s various tissues. (C) Sanger chromatograms showing paternal mtDNA heteroplasmy in one of Teddy’s non-MRT offspring.
Given that transmission of donor mtDNA to the next generation was studied just on one live offspring, we investigated mtDNA content in individual oocytes retrieved from Crysta (Supplementary Fig. S3A). A total of 57 mature oocytes were collected following a COS protocol and maternal and donor mtDNA levels were measured using ARMs-qPCR. Maternal mtDNA (‘carryover’) was detected at 1.6% heteroplasmy level in one oocyte and the remaining 56 oocytes carried exclusively donor mtDNA (Supplementary Fig. S3B). Independent MiSeq and Sanger sequencing corroborated ARMs-qPCR results (Supplementary Fig. S3C). These data demonstrate germline transmission of donor mtDNA from the MRT female macaque to her oocytes and live offspring, but maternal mtDNA may still persist at a low heteroplasmy level in some oocytes. These conclusions, while important, are limited by only one MRT female analyzed.
In addition, our results provide strong evidence that in rare cases a small amount of original paternal mtDNA in zygotes can be selectively amplified during development in offspring resulting in biparental inheritance in rhesus macaques. However, this transmission occurred asymmetrically in organs and tissues leading to varied heteroplasmy levels of paternal mtDNA in different tissues.
To determine whether Teddy’s paternal mtDNA transmission is secondary to nDNA/mtDNA ‘mismatch’ in Crysta, we performed MiSeq on blood DNA samples from 10 of Teddy’s other offspring (5 of each sex) produced by natural breeding with non-MRT females. Sequencing analysis revealed that one male offspring (offspring 1 in Supplementary Table SV) carried variants matched to the paternal mtDNA at 31% heteroplasmy. In the other 9 offspring, heteroplasmy for paternal mtDNA was not detected (Supplementary Table SV). We also corroborated the MiSeq results by Sanger sequencing (Fig. 4C). These results suggest that paternal mtDNA transmission could be specific to individual males but independent of MRT.
To investigate possible molecular mechanisms responsible for selective amplification of maternal or paternal mtDNA in some MRT rhesus macaques, we carried out in-depth mtDNA sequence analysis. Our previous study on human MRT embryos suggested the replication positively correlated with the number of guanosine (G) residues in the conserved sequence box II (CSBII) region (Supplementary Fig. S2C) (Kang et al., 2019). Rhesus macaques, similar to humans, carry conserved guanosine sequence structure CSBII sequences except for thymine (T) substitution of adenine (A) between the two guanosine clusters (Supplementary Fig. S2D). Analysis revealed that the CSBII sequences between the parental mtDNAs for Spindler, Mac and Tracker were identical (Supplementary Fig. S2D). Among the two MRT animals with tissue-specific rebound of maternal/paternal mtDNA, Tracker was produced with a combination of Chinese/Indian mtDNA that differed in 727 SNPs (Supplementary Table SI). In contrast, donor/maternal (Indian/Indian) mtDNA differences in Spindler were confined only to eight SNPs. Thus, it seems unlikely that mtDNA sequence differences alone contributed to the selective amplification of maternal or paternal mtDNA in MRT offspring.
Discussion
The risk of transmitting pathogenic mtDNA mutations from a mother’s oocytes to her children can be substantially reduced by MRT. Clinical applications of this form of germline gene therapy for families with maternally inherited mitochondrial disease have been considered in many countries. However, concerns were raised about potential incompatibility of pairing a randomly selected donor mtDNA with the maternal nuclear genome (Cristina Kenney et al., 2014; Ma et al., 2016). Since both mtDNA and nDNA co-encode OXPHOS proteins, their proper assembly and interactions are crucial for normal mitochondrial function (Schon et al., 2012). In addition, nDNA provides all the replication, transcription and translation machinery required for mtDNA encoded genes. These considerations led to calls to pre-select donor oocytes with mtDNA haplotypes that would match the patient’s maternal haplotype for MRT (Reinhardt et al., 2013). Results of the current longitudinal study on MRT monkeys generated with donor mtDNA that is exceedingly distant from the original maternal counterpart suggest that their growth, general health fitness and fertility is unremarkable and similar to controls. As noted above, the genetic distance between donor and maternal mtDNA for some MRT monkeys was much greater than those for the most distant human populations (Blanco et al., 2011; Burgstaller et al., 2014). Thus, it is reasonable to suggest that MRT with ‘unmatched’ donor mtDNA haplotype within a species should be safe and effective as long as these sequence variations are non-pathogenic.
A common problem associated with most MRT approaches, including spindle transfer, PNT and polar body transfer, is the incomplete replacement of the pathogenic maternal mtDNA (Tachibana et al., 2009; Craven et al., 2010; Paull et al., 2013; Wang et al., 2014; Kang et al., 2016; Ma et al., 2017). A small, residual amount of maternal mtDNA, known as carryover, remains and is passed to MRT offspring. Although the initial levels of maternal carryover mtDNA are low and generally may not cause mitochondrial disease, its persistence and possible expansion during embryo development is a possibility. This phenomenon was observed in some human ESCs derived from MRT embryos, suggesting selective replicative advantage of residual maternal mtDNA over donor mtDNA leading to a complete reversal to the maternal mtDNA (Kang et al., 2016; Yamada et al., 2016; Hyslop et al., 2016a,b). While these studies were modeled on in vitro grown ESCs, our investigation confirms maternal mtDNA levels may also increase in vivo during normal development and aging in MRT primates. We demonstrate that in some MRT monkeys, originally low heteroplasmy levels of the maternal mtDNA may increase 10-fold to 17%. It is important to note that in contrast to human ESC studies, we did not observe complete reversal to maternal mtDNA in our MRT monkeys. The increased heteroplasmy levels in MRT animals were still below the known mutation thresholds for causing mitochondrial disease. Another caveat is that maternal mtDNA increase in MRT monkeys was not uniform but occurred in selected internal tissues and organs. Therefore, routine testing of peripheral tissues such as blood, urine or skin may not reveal actual heteroplasmy levels in internal organs. While the precise mechanism and timing of when such genetic drift and segregation occurs remains unknown, our results imply that levels of the mutant, maternal mtDNA may increase randomly in selected organs and tissues of children born post-MRT. Therefore, it is vital to further optimize MRT procedures to lower or completely eliminate the carryover of maternal mtDNA.
These conclusions have implications for the clinical application of human MRT and patients would obviously need to be informed of potential risks. Furthermore, our results in nonhuman primate model of MRT underscore the need for clinical trials. MRT is also considered for infertility due to oocyte cytoplasmic deficiencies. In this case, both the maternal and donor mtDNA are non-pathogenic and the reversal could be less of a concern, as it would not be expected to cause classical mtDNA disease in resulting children. Therefore, it might be safer to test MRT on infertility patients. In addition, ongoing and future clinical applications of MRT along with additional research on animal models should provide safety evaluations for possible harmful secondary outcomes reflecting nuclear–mitochondrial incompatibility (Chinnery et al., 2014).
Since the discovery of mtDNA in 1963 (Nass and Nass, 1963a, b), it has been a common notion that inheritance of this cytoplasmic genome occurs in a strict maternal manner. Increasingly, evidence from experimental crosses of occasional paternal inheritance has also been reported in animals including Drosophila melanogaster (Kondo et al., 1990; Sherengul et al., 2006; Polovina et al., 2020), mouse hybrids of Mus musculus and Mus spretus (Gyllensten et al., 1991; Shitara et al., 1998), sheep (Zhao et al., 2004), periodical cicada (Fontaine et al., 2007), honeybee (Meusel and Moritz, 1993) and monkeys born by nuclear transfer (St John and Schatten, 2004). The first report of paternal transmission in humans came from a patient with mitochondrial myopathy harboring a paternally derived 2-bp mtDNA deletion in the MTND2 gene in muscle tissues (Schwartz and Vissing, 2002). Most recently, paternal mtDNA transmission was documented in three unrelated families, which further challenges the central dogma of strict maternal transmission of mtDNA to offspring (Luo et al., 2018). In the current study, we present strong evidence of paternal mtDNA transmission to offspring of one particular rhesus macaque male. Moreover, we show that paternal mtDNA contribution may substantially differ in heteroplasmy levels between various organs and tissues of the same animal ranging from undetectable to 33%.
Diverse molecular mechanisms underlying the phenomenon of strict maternal inheritance have been described. One explanation is the stochastic loss of paternal mtDNA in a zygote after fertilization due to dilution (Birky, 1983). In animals, a mature sperm carries just a few copies of mtDNA, in contrast, an oocyte contains 105 to 108 copies of mtDNA (Jansen and de Boer, 1998; Chiaratti et al., 2020). Therefore, the sperm mtDNA heteroplasmy in zygotes or preimplantation embryos is well below the detection levels by most existing methods (Santos et al., 2006; Wu et al., 2019). Another mechanism is selective elimination of sperm mitochondria in a zygote by the oocyte’s machinery. For instance, selective degradation of paternal mitochondria and their mtDNA through CPS-6 (endonuclease G)-mediated autophagy in Caenorhabditis elegans (Al Rawi et al., 2011; Sato and Sato, 2011), and elimination of paternal mtDNA through ubiquitin-proteasome machinery in monkeys (Sutovsky et al., 1999).
Our results demonstrate substantial levels of paternal mtDNA in some organs and tissues of offspring, likely due to selective replication of sperm mtDNA. While the underlying mechanisms remain unknown, previous human studies suggest that sequence polymorphism in CSBII region can confer preferential replicative advantage of specific mtDNA haplotypes (Agaronyan et al., 2015; Kang et al., 2019). Analysis of mtDNA sequences of monkeys did not find any sequence differences within the CSBII region suggesting that other regulatory regions or epigenetic mechanisms may be involved (Wolf et al., 2017). For example, TFAM (transcription factor A, mitochondrial) is a core transcription factor in human mitochondria and has two important functions of binding and bending mtDNA (Parisi and Clayton, 1991). The binding function suggests a direct relationship between TFAM and mtDNA copy number, while bending suggests a role in mtDNA replication initiation. 7S DNA is a short mtDNA strand, which is formed through premature termination of heavy-strand replication. The function of 7S DNA is still unknown, but it may serve as a primer for mtDNA replication under the strand-displacement mtDNA replication model (Nicholls and Minczuk, 2014). It is possible that only 7S-containing mtDNA are primed for replication. Further investigations are necessary to clarify the exact mechanisms of the paternal mtDNA leakage and maternal mtDNA reversal post-MRT.
Supplementary data
Supplementary data are available at Human Reproduction online.
Data availability
The MiSeq dataset analyzed and reported in this study is available from the SRA of NCBI with the accession number PRJNA630763.
Supplementary Material
Acknowledgements
The authors acknowledge the OHSU Institutional Animal Care and Use Committee (IACUC) at the ONPRC for providing oversight and guidance. We thank the Division of Comparative Medicine, Assisted Reproductive Technologies, Endocrine Technologies, Integrated Pathology and Molecular Technologies Cores at ONPRC for their services and support. We are indebted to UC Davis veterinary genetics laboratory for microsatellite genotyping.
Authors’ roles
H.M., P.A. and S.M. conceived the study, designed the experiments and supervised the project. H.M., C.V.D., A.K., Y.Li. and N.M.G. performed the necropsy tissue collection and DNA extractions. H.M., C.V., H.D. and R.T.H. performed mtDNA ARMs-qPCR analyses. H.D., H.M., A.K., E.K. and Y.L. performed MiSeq sequencing and H.D., A.M. and H.M. performed MiSeq Data analysis. H.S. performed routine examination and provided veterinary care and welfare oversight for all animals. C.R. performed sperm preparation and analysis, oocyte collection and hormone assays. T.H. performed TMB and ultrasounds. Y.Li., D.L., and A.M. performed PCR and Sanger sequencing. H.M., C.V. and S.M. wrote the paper.
Funding
The studies were supported by the grants from the Burroughs Wellcome Fund, the National Institutes of Health (NIH) (RO1AG062459 and P51 OD011092), National Research Foundation of Korea (2018R1D1A1B07043216) and Oregon Health & Science University institutional funds.
Conflict of interest
The authors declare no competing interests.
References
- Agaronyan K, Morozov YI, Anikin M, Temiakov D. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science 2015;347:548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 2011;334:1144–1147. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya T, Reifova R, Gregorova S, Simecek P, Gergelits V, Mistrik M, Martincova I, Pialek J, Forejt J. X Chromosome control of meiotic chromosome synapsis in mouse inter-subspecific hybrids. PLoS Genet 2014;10:e1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW Jr. Relaxed cellular controls and organelle heredity. Science 1983;222:468–475. [DOI] [PubMed] [Google Scholar]
- Blanco R, Mayordomo E, Montoya J, Ruiz-Pesini E. Rebooting the human mitochondrial phylogeny: an automated and scalable methodology with expert knowledge. BMC Bioinformatics 2011;12:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgstaller JP, Johnston IG, Jones NS, Albrechtova J, Kolbe T, Vogl C, Futschik A, Mayrhofer C, Klein D, Sabitzer S et al. MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Rep 2014;7:2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaratti MR, Macabelli CH, Augusto Neto JD, Grejo MP, Pandey AK, Perecin F, Collado MD. Maternal transmission of mitochondrial diseases. Genet Mol Biol 2020;43:e20190095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery PF, Craven L, Mitalipov S, Stewart JB, Herbert M, Turnbull DM. The challenges of mitochondrial replacement. PLoS Genet 2014;10:e1004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Hime GR, Shirras AD. Cytoplasmic male sterility in Drosophila melanogaster associated with a mitochondrial CYTB variant. Heredity (Edinb ) 2011;107:374–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven L, Tuppen HA, Greggains GD, Harbottle SJ, Murphy JL, Cree LM, Murdoch AP, Chinnery PF, Taylor RW, Lightowlers RN et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature 2010;465:82–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristina Kenney M, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, Tarek M, Caceres-del-Carpio J, Nesburn AB, Boyer DS et al. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum Mol Genet 2014;23:3537–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. Mitochondrial replacement therapy: are mito-nuclear interactions likely to be a problem? Genetics 2017;205:1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine KM, Cooley JR, Simon C. Evidence for paternal leakage in hybrid periodical cicadas (Hemiptera: Magicicada spp.). PLoS One 2007;2:e892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U, Wharton D, Josefsson A, Wilson AC. Paternal inheritance of mitochondrial DNA in mice. Nature 1991;352:255–257. [DOI] [PubMed] [Google Scholar]
- Haertel AJ, Prongay K, Gao L, Gottlieb DH, Park B. Standard growth and diarrhea-associated growth faltering in captive infant rhesus macaques (Macaca mulatta). Am J Primatol 2018;80:e22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 2010;464:610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science 2007;316:240–243. [DOI] [PubMed] [Google Scholar]
- Hyslop LA, Blakeley P, Craven L, Richardson J, Fogarty NM, Fragouli E, Lamb M, Wamaitha SE, Prathalingam N, Zhang Q et al. Corrigendum: towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature 2016. a;538:542–542. [DOI] [PubMed] [Google Scholar]
- Hyslop LA, Blakeley P, Craven L, Richardson J, Fogarty NM, Fragouli E, Lamb M, Wamaitha SE, Prathalingam N, Zhang Q et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature 2016. b;534:383–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP, de Boer K. The bottleneck: mitochondrial imperatives in oogenesis and ovarian follicular fate. Mol Cell Endocrinol 1998;145:81–88. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet 1997;16:93–95. [DOI] [PubMed] [Google Scholar]
- Kang E, Koski A, Amato P, Temiakov D, Mitalipov S. Reply to: Reversion after replacement of mitochondrial DNA. Nature 2019;574:E12–E13. [DOI] [PubMed] [Google Scholar]
- Kang E, Wu J, Gutierrez NM, Koski A, Tippner-Hedges R, Agaronyan K, Platero-Luengo A, Martinez-Redondo P, Ma H, Lee Y et al. Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature 2016;540:270–275. [DOI] [PubMed] [Google Scholar]
- Kondo R, Satta Y, Matsuura ET, Ishiwa H, Takahata N, Chigusa SI. Incomplete maternal transmission of mitochondrial DNA in Drosophila. Genetics 1990;126:657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Ma H, Juanes RC, Tachibana M, Sparman M, Woodward J, Ramsey C, Xu J, Kang EJ, Amato P et al. Rapid mitochondrial DNA segregation in primate preimplantation embryos precedes somatic and germline bottleneck. Cell Rep 2012;1:506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Valencia CA, Zhang J, Lee NC, Slone J, Gui B, Wang X, Li Z, Dell S, Brown J et al. Biparental inheritance of mitochondrial DNA in humans. Proc Natl Acad Sci USA 2018;115:13039–13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Folmes CDL, Wu J, Morey R, Mora-Castilla S, Ocampo A, Ma L, Poulton J, Wang X, Ahmed R et al. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature 2015;524:234–238. [DOI] [PubMed] [Google Scholar]
- Ma H, Marti Gutierrez N, Morey R, Van Dyken C, Kang E, Hayama T, Lee Y, Li Y, Tippner-Hedges R, Wolf DP et al. Incompatibility between nuclear and mitochondrial genomes contributes to an interspecies reproductive barrier. Cell Metab 2016;24:283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, O’Neil RC, Marti Gutierrez N, Hariharan M, Zhang ZZ, He Y, Cinnioglu C, Kayali R, Kang E, Lee Y et al. Functional human oocytes generated by transfer of polar body genomes. Cell Stem Cell 2017;20:112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Solter D. Nuclear transplantation in the mouse embryo by microsurgery and cell fusion. Science 1983;220:1300–1302. [DOI] [PubMed] [Google Scholar]
- Meusel MS, Moritz RF. Transfer of paternal mitochondrial DNA during fertilization of honeybee (Apis mellifera L.) eggs. Curr Genet 1993;24:539–543. [DOI] [PubMed] [Google Scholar]
- Nass MM, Nass S. Intramitochonfrial fibers with DNA characteristics. I. Fixation and electron staining reactions. J Cell Biol 1963. a;19:593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass S, Nass MM. Intramitochondrial fibers with DNA characteristics. II. Enzymatic and other hydrolytic treatments. J Cell Biol 1963. b;19:613–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls TJ, Minczuk M. In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol 2014;56:175–181. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 1991;252:965–969. [DOI] [PubMed] [Google Scholar]
- Paull D, Emmanuele V, Weiss KA, Treff N, Stewart L, Hua H, Zimmer M, Kahler DJ, Goland RS, Noggle SA et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature 2013;493:632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polovina ES, Parakatselaki ME, Ladoukakis ED. Paternal leakage of mitochondrial DNA and maternal inheritance of heteroplasmy in Drosophila hybrids. Sci Rep 2020;10:2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt K, Dowling DK, Morrow EH. Medicine. Mitochondrial replacement, evolution, and the clinic. Science 2013;341:1345–1346. [DOI] [PubMed] [Google Scholar]
- Russell OM, Gorman GS, Lightowlers RN, Turnbull DM. Mitochondrial diseases: hope for the future. Cell 2020;181:168–188. [DOI] [PubMed] [Google Scholar]
- Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril 2006;85:584–591. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 2011;334:1141–1144. [DOI] [PubMed] [Google Scholar]
- Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet 2012;13:878–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. N Engl J Med 2002;347:576–580. [DOI] [PubMed] [Google Scholar]
- Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M, Crimi M, Waymire K, Lin CS, Masubuchi S, Friend N, Koike M et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell 2012;151:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherengul W, Kondo R, Matsuura ET. Analysis of paternal transmission of mitochondrial DNA in Drosophila. Genes Genet Syst 2006;81:399–404. [DOI] [PubMed] [Google Scholar]
- Shitara H, Hayashi JI, Takahama S, Kaneda H, Yonekawa H. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 1998;148:851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge EA. Nuclear genetic defects of oxidative phosphorylation. Hum Mol Genet 2001;10:2277–2284. [DOI] [PubMed] [Google Scholar]
- Simmons HA. Age-associated pathology in rhesus macaques (Macaca mulatta). Vet Pathol 2016;53:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG. Genetic characterization of Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta). Comp Med 2005;55:227–230. [PubMed] [Google Scholar]
- St John JC, Schatten G. Paternal mitochondrial DNA transmission during nonhuman primate nuclear transfer. Genetics 2004;167:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet 2015;16:530–542. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature 1999;402:371–372. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Amato P, Sparman M, Woodward J, Sanchis DM, Ma H, Gutierrez NM, Tippner-Hedges R, Kang E, Lee HS et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature 2013;493:627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Mitalipov S. Chromosome transfer in mature oocytes. Nat Protoc 2010;5:1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, Woodward J, Li Y, Ramsey C, Kolotushkina O, Mitalipov S. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature 2009;461:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol 2013;5:a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem 2007;76:781–821. [DOI] [PubMed] [Google Scholar]
- Wang T, Sha H, Ji D, Zhang HL, Chen D, Cao Y, Zhu J. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell 2014;157:1591–1604. [DOI] [PubMed] [Google Scholar]
- Wolf DP, Hayama T, Mitalipov S. Mitochondrial genome inheritance and replacement in the human germline. EMBO J 2017;36:2177–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Huffman AM, Whitcomb BW, Josyula S, Labrie S, Tougias E, Rahil T, Sites CK, Pilsner JR. Sperm mitochondrial DNA measures and semen parameters among men undergoing fertility treatment. Reprod Biomed Online 2019;38:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Emmanuele V, Sanchez-Quintero MJ, Sun B, Lallos G, Paull D, Zimmer M, Pagett S, Prosser RW, Sauer MV et al. Genetic drift can compromise mitochondrial replacement by nuclear transfer in human oocytes. Cell Stem Cell 2016;18:749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee WK, Sutton KL, Dowling DK. In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr Biol 2013;23:R55–R56. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu H, Luo S, Lu Z, Chavez-Badiola A, Liu Z, Yang M, Merhi Z, Silber SJ, Munne S et al. Live birth derived from oocyte spindle transfer to prevent mitochondrial disease. Reprod Biomed Online 2017;34:361–368. [DOI] [PubMed] [Google Scholar]
- Zhao X, Li N, Guo W, Hu X, Liu Z, Gong G, Wang A, Feng J, Wu C. Further evidence for paternal inheritance of mitochondrial DNA in the sheep (Ovis aries). Heredity (Edinb ) 2004;93:399–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MiSeq dataset analyzed and reported in this study is available from the SRA of NCBI with the accession number PRJNA630763.