Abstract
New treatments are required for advanced prostate cancer; however, there are fewer preclinical models of prostate cancer than other common tumor types to test candidate therapeutics. One opportunity to increase the scope of preclinical studies is to grow tissue from patient-derived xenografts (PDXs) as organoid cultures. Here we report a scalable pipeline for automated seeding, treatment and an analysis of the drug responses of prostate cancer organoids. We established organoid cultures from 5 PDXs with diverse phenotypes of prostate cancer, including castrate-sensitive and castrate-resistant disease, as well as adenocarcinoma and neuroendocrine pathology. We robotically embedded organoids in Matrigel in 384-well plates and monitored growth via brightfield microscopy before treatment with poly ADP-ribose polymerase inhibitors or a compound library. Independent readouts including metabolic activity and live-cell imaging–based features provided robust measures of organoid growth and complementary ways of assessing drug efficacy. Single organoid analyses enabled in-depth assessment of morphological differences between patients and within organoid populations and revealed that larger organoids had more striking changes in morphology and composition after drug treatment. By increasing the scale and scope of organoid experiments, this automated assay complements other patient-derived models and will expedite preclinical testing of new treatments for prostate cancer.
Keywords: organoids, prostate cancer, patient-derived xenograft, PARP inhibitor, high-content imaging
Introduction
Patients with advanced prostate cancer invariably develop resistance to all currently available systemic treatments. Initially, patients receive androgen deprivation therapy (ADT), sometimes in combination with other androgen receptor (AR)–directed treatments or taxane-based chemotherapy.1–5 When ADT fails, patients develop castration-resistant prostate cancer (CRPC), and they may receive further AR-directed treatments or chemotherapies. However, disease control is limited to several months on average. A small subset of patients with genomic alterations in DNA damage repair genes can also be treated with poly ADP-ribose polymerase (PARP) inhibitors.6,7 Unfortunately, various mechanisms of resistance emerge in CRPC, including enrichment of genomic alterations associated with poor prognosis, aberrations that maintain the activation of the AR pathway, and phenotypic transformation into AR-null and neuroendocrine phenotypes.8,9 New therapies are required for these diverse forms of CRPC; however, preclinical development is often hampered by a shortage of patient-derived models and the limited ability to deploy them in drug screening.10
Two of the most common ways in which human prostate cancer tissue is used in preclinical research is as patient-derived xenografts (PDXs) and organoids. Serially transplantable PDXs are tumors that are continually transplanted in immunocompromised mice.11 Organoids are three-dimensional (3D) cultures of prostate cancer cells embedded in the extracellular matrix, often Matrigel.12–14 It is challenging to establish PDXs and organoids from fresh prostate cancer tissue, with lower success rates than other common tumors.13,15,16 Nevertheless, once these models are established, they provide renewable sources of tumor cells. Therefore, PDXs and organoids can be integrated in preclinical studies by implanting organoids into mice to establish xenografts15,17 or by digesting PDXs and growing them as organoids.18–22 The corresponding organoids consistently maintain histological, transcriptional, and genomic features of the parental PDXs.17,19,22,23 This is important because organoids and PDXs are complementary models with distinct advantages and disadvantages for preclinical testing.11,24
Organoid cultures are more complex than two-dimensional (2D) cultures of cells lines and therefore may more accurately reflect some aspects of patient tumors.25 For example, prostate cancer organoids represent the histopathological features of the original patient tumors or PDXs, including cytology and expression of phenotypic biomarkers.13,17,19,26 The promise of organoid experiments is their scalability, making them particularly useful for screening large numbers of different compounds or comprehensive dose responses of individual compounds, which would not be feasible with in vivo PDX experiments. Indeed, previous studies have used prostate cancer organoids, either from patient or PDX tissues, to test drug libraries or specific therapeutics.17,19,27
However, significant work remains to establish stricter quality controls to standardize phenotypic analyses and make 3D models more reproducible.28 Furthermore, previous studies have predominantly used whole-well and endpoint readouts, such as PrestoBlue or CellTiter-Glo (CTG) activity, to measure organoid viability. Endpoint readouts are often destructive of patient-derived material and make it more difficult and labor intensive to assess growth kinetics. In addition, there are limited options for assessing organoid morphology and therefore no ability to compare the variability of the organoid population in each well. In contrast, a cell-level and/or temporal-spatial analysis would exploit more of the purported advantages and physiologically informative content of multicellular 3D models. High-content imaging preserves both population-specific and spatial information and is highly amenable to multiplexing. This enables extraction of more information per well for models that tend to be more labor intensive and costly to manufacture than their 2D counterparts.29
We have developed a novel quantitative high-throughput imaging assay that harnesses the scalability of organoid cultures. Using this approach, we can capture the complexity of the growth of different patient-derived organoids over time, measure multiple whole-well metrics and single-organoid readouts, and show how the morphology and composition of discrete subpopulations of organoids can differ in response to drug treatment.
Materials and Methods
Patient-Derived Xenografts
Patient samples were acquired with informed, written consent according to human ethics approvals from Monash Health (RES-20-0000-103C and RES-20-0000-107C) and the Peter MacCallum Cancer Centre (15/98, 97_27). Prostate cancer tissue was obtained from radical prostatectomy samples of patients with treatment-naïve prostate cancer (224R, 287R, and 305R) or from rapid autopsy samples of a patient with CRPC who consented to the CASCADE rapid autopsy program (201A).30
Serially transplantable PDXs were established by the Melbourne Urological Research Alliance (MURAL), as previously described.18,31 All animal procedures were conducted in accordance with Monash University Animal ethics approvals (MARP 2018/087). Briefly, patient tissues were initially grafted under the renal capsule of 6- to 8-week-old male NOD-SCID interleukin 2–receptor gamma chain knockout (NSG; RRID:IMSR_JAX:005557) mice supplemented with testosterone implants (Monash Animal Research Laboratories, Monash Breeding Colony approval number MMCA 209/25BC and 15160).32 Once grafts reached 1000 mm3, they were harvested and implanted into new mice. After three to five generations, grafts were transferred to the subcutaneous site of host mice. PDXs 224R, 305R, 201.1A, and 201.2A were also regrafted into precastrated NSG mice, to mimic the low circulating androgen levels of men on ADT (designated 224R-Cx, 305R-Cx, 201.1A-Cx, and 201.2A-Cx).
The phenotype of PDXs was routinely monitored using immunohistochemistry for markers of human adenocarcinoma and neuroendocrine prostate cancer (CK8/18, AR, PSA, PSMA, chromogranin A, synaptophysin, CD56). Using staining for CD45, we confirmed that the PDXs did not contain lymphoma. The identity of PDXs was routinely authenticated by profiling short tandem repeats (GenePrint 10 System, Promega, Madison, WI) at the Australian Genome Research Facility, Melbourne. The patient-derived models used in this are available from the Melbourne Urological Research Alliance on request.
Data from targeted DNA sequencing of PDXs was extracted from the NCBI Sequence Read Archive BioProject PRJNA675382. We examined alterations in 12 DNA damage repair genes used in multiple clinical trials of PARP inhibitors in prostate cancer (ATM, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK2, FANCA, PALB2, RAD51B, RAD51C, RAD51D).33–36 The following thresholds were used for copy number alterations: (gain >log2[2.8/2]; amplification >log2[8/2]; loss <log2[1.2/2]; deep deletion <log2[0.2/2]). Variants were included if they were known or likely to be pathogenic, based on high IMPACT scores with the Ensembl Variant Effect Predictor (VEP) v90.37 All variants were heterozygous, with allele frequencies less than 0.5.
Organoid Cultures and High-Throughput Seeding
Organoid cultures were cultured from PDX tissues using previously described methods (Fig. 1A).13,18,19 Briefly, PDX tissue was finely minced with scalpels and digested for 40 min at 37 °C in RPMI-1640 containing 0.2 mg/mL DNase I (Roche, Basel, Switzerland) and 0.65 U/mL Liberase TM (Roche). Digested samples were centrifuged, filtered through 100 µM cell strainers, treated with Red Blood Cell Lysis Buffer (Sigma-Aldrich, St. Louis, MO), and washed with RPMI containing 10% fetal bovine serum, 10 U/mL penicillin, and 10 mg/mL streptomycin. Cells were seeded in growth factor–reduced, phenol-red free, IdEV-free Matrigel (catalog No. 356231, lot No. 0020004, concentration 9.4 mg/mL) at a density of 1 × 105 cells per 40 µL of Matrigel in 24-well plates. Organoids were cultured in ENR media (201.1A-Cx, 201.2A-Cx, and 287R) or ENR-2 media (224R-Cx and 305R-Cx) as previously described.12,19 ENR media were advanced DMEM/F12 (Thermo Fisher, Waltham, MA) containing 50 ng/mL epithelial growth factor (Sigma-Aldrich), 5% (vol/vol) R-spondin 1 (Monash Biomedicine Discovery Institute Organoid Program), 10% (vol/vol) noggin (Monash Biomedicine Discovery Institute Organoid Program), 10 ng/mL fibroblast growth factor 10 (VWR International, Queensland, Australia), 5 ng/mL fibroblast growth factor 2 (PeproTech, Rocky Hill, NJ), 1 nM DHT, 10 mM nicotinamide (Sigma-Aldrich), 0.5 µM A83-01 (Sigma-Aldrich), 10 µM SB202190 (Sigma-Aldrich), 1 × B27 additive (Thermo Fisher), 1.25 mM N-acetyl-L-cysteine (Sigma-Aldrich), 2 mM glutamax (Thermo Fisher), and 1 µM prostaglandin (Tocris Bioscience, Bristol, UK). ENR-2 media had the same formulation but without the addition of 1.25 mM N-acetylcysteine (Sigma-Aldrich) and 10 µM SB202190 (Sigma-Aldrich). During organoid establishment, 10 µM Y-27632 dihydrochloride (Selleck Chemicals, Houston, TX) was also added to the culture medium.
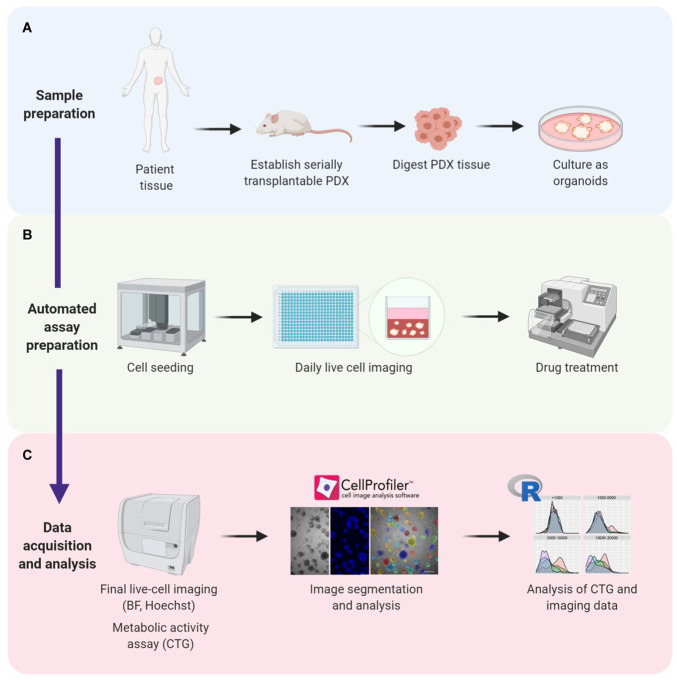
Figure 1.
Establishing a high-throughput assay for automated seeding, treatment, and analysis of prostate cancer organoids. (A) Prostate cancer tissue is acquired from patients and used to establish serially transplantable patient-derived xenografts (PDXs). PDX tissue is then digested and cultured in Matrigel as organoids. (B) Organoids are robotically seeded in 384-well plates and monitored at different intervals using live-cell brightfield microscopy. Drug treatment started at 8 d in culture and concluded at day 21. (C) After treatment, live-cell imaging followed by endpoint CTG measurements were performed. Microscopy images were segmented and quantified using CellProfiler software. CTG and imaging data were analyzed using R software. BF = brightfield microscopy; CTG = CellTiter-Glo; PDX = patient-derived xenograft.
Organoids were passaged via dissociation with TrypLE (Sigma-Aldrich) for 10 min at 37 °C once before automated seeding. Cells (3 × 103 per well) were dispensed in 384-well plates at 10 µL of 35% to 80% Matrigel using the JanusG3 liquid handling robot with a cooled stage (PerkinElmer, Waltham, MA; Fig. 1B). Plates were pulse-centrifuged to 500 rpm after dispensing to disperse the Matrigel across the whole well and to remove bubbles. Plates were left at 37 °C in the incubator for 10 min to set the Matrigel, then ENR or ENR-2 organoid media were added (35 µL), and the plates were incubated in a LiCONiCs STX220 high-throughput incubator (LiCONiC, Lichtenstein).
Drug Compound Preparation and Treatment
Talazoparib and carboplatin (Selleck Chemicals) were used as proof of concept for measuring changes in organoid growth and morphology in response to treatment. The stock solutions (1 mM talazoparib in 100% DMSO, 1 mM carboplatin in water + 0.1% Triton X) were pipetted directly into a T8 cartridge of a Tecan D300e drug printer (Tecan Trading AG, Männedorf, Switzerland) and automatically dispensed to the wells at final concentrations of 500, 100, 50, and 10 nM (five replicates each) or 0.05% DMSO as vehicle control (20 replicates for 201.1A-Cx, 224R, and 287R; 15 replicates for 201.2A-Cx and 305R-Cx; Suppl. Fig. S1, with representative plate maps). The drug compound titrations were added to the wells in the 384-well plates on day 8 after seeding and then every 2 to 3 d until day 21 (Fig. 1B).
The drug screen consisted of 42 compounds, provided in 384-well plates by Compounds Australia (Nathan QLD, Australia), at three concentrations of compound stock solution in 100% DMSO (0.1, 1, and 10 µM) in technical duplicates on each plate. Six positive controls and negative controls (0.2% DMSO) were included in the outer wells in multiple replicate wells (Suppl. Fig. S1, with representative plate maps). The drug stock plates were then hydrated with organoid media and the compounds added to two biological replicate cell plates per organoid type using the Sciclone ALH3000 robot (PerkinElmer) on day 8 after seeding and then every 2 to 3 d until day 20.
Imaging and Endpoint Readouts
Live organoid cultures were imaged daily via brightfield microscopy using the Cytation5 Cell Imaging Multi-Mode Reader (BioTek, Winooski, VT) at 2.5× magnification (one field/well, maximum projection of a stack of three z-heights) to establish growth curves. After drug treatment for 12 d, nuclei were stained for 30 min at room temperature with Hoechst 33342 (10 mg/mL) at 1:1000 (Thermo Fisher), and brightfield and fluorescent images were taken on live cells (Cytation5, 2.5× magnification, one field/well, maximum projection of a stack of three z-heights). Organoid segmentation, based on brightfield images, was performed using CellProfiler software (Broad Institute of MIT and Harvard, version 3.0.0).38 After identification of organoids as primary objects, we added a filter step to exclude everything with an area <160 px2 from further analysis. These objects are usually smaller than single cells and are therefore most likely cell debris. For subsequent quantification of organoid morphology, including area, radius, and eccentricity, as well as texture and intensity features, we used well-level averages, except for single-organoid analyses.
Supplementary Figure S2 briefly explains the most relevant imaging features used for organoid analyses (http://cellprofiler-manual.s3.amazonaws.com/CellProfiler-3.0.0/modules/measurement.html).
Whole-population metabolic activity was assessed in the same wells using the CTG Luminescent Cell Viability Assay (Promega). For this, we added 20 µL of undiluted CTG reagent directly onto the 35 µL of media in each well using the automated dispenser (BioTek), sealed the plate, and vigorously shook it on an orbital shaker at room temperature for 20 min. After that, the plates were pulse spun at 1000 rpm, and luminescence intensity was measured at the default gain of 135 on the Cytation5 plate reader.
Data Processing and Quality Controls
Data were imported into and processed in the R Studio statistical environment (v1.2.1335)39 using the tidyverse package (v1.3.0).40
As part of quality control, the 15 to 20 vehicle control wells were analyzed for outliers using the find_HDoutliers function for the detection of anomalies in high-dimensional data.41 Next, heat maps were generated based on raw organoid area values at day 8 using the dplyr (v1.0.0) and platetools (v0.1.2) packages to visually confirm consistent distribution of average organoid areas across all plates before drug treatment. We looked for suspicious patterns, such as consistently lower values in the outside wells of the plate (potential edge effects) or two adjacent rows having dramatically lower/higher values than other rows (potential liquid-handling issue during organoid seeding). Additional quality control measures included the calculation of the mean, standard deviation (SD), and coefficient of variation (CV) for the main readouts (Table 1). Ideally, untreated controls should have CVs of less than 15%, and replicate wells treated with toxic compounds should have CVs of less than 24%. Finally, the Z′ factor metric (Eq. (1)) was calculated to assess the degree of overlap between the mean of the DMSO vehicle controls (n = 15–20) and the mean of each talazoparib concentration (500, 100, 50, and 10 nM, n = 5 per plate) to judge whether the specific cellular feature was suitable as a statistically robust assay readout.
Table 1.
Explanation of Imaging Features.
| Imaging Feature | Explanation |
|---|---|
| Area | Number of pixels in the region (region defined as organoid) |
| MeanRadius | Mean distance of any pixel in the object to the closest pixel outside of the object |
| Eccentricity | Ratio of the distance between the foci of the ellipse and its major axis length (the value is between 0 and 1: 0 is a circle, whereas 1 is a line) |
| FormFactor | Calculated as 4 × π × area/perimeter2: 1 is a perfectly circular object |
| MeanIntensity | Mean of pixel intensity values in the objects across the well |
| Texture | Spatial distribution of intensities within each object; more precisely, the fraction of total stain in the center of the objects |
| Texture_Variance | Measures intensity variations in gray-scale images. An object or entire image without much texture has a smooth appearance; an object or image with a lot of texture will appear rough and show a wide variety of pixel intensities |
| (1) |
The constant factor 3 in the definition of the Z′ factor (Eq. (1)) is motivated by the normal distribution, for which more than 99% of values occur within 3 SD of the mean. To account for potentially larger variability in 3D high-throughput screens, we also calculated all Z′ factors with only 2 SD, which still captured more than 95% of the values.
The Z′ factor values were categorized as follows: Z′ = 0.5 to 1, excellent (0.5 is equivalent to a separation of 12 SDs between DMSOmean and PosCtrlmean), 0 to 0.5 is acceptable, and <0 is unacceptable (too much overlap between positive and negative controls).42
Compound Screen Analysis
To ensure that all features were the same scale for correlation calculations and phenotypic profiles, we calculated robust Z-scores from the raw values using the following equation (Eq. (2)).
| (2) |
The robust Z-score is the number of median absolute deviations by which the raw value of a sample is above or below the median value of the controls per plate for this particular readout. This method does not assume a normal Gaussian distribution to the underlying data and is more robust in dealing with outliers or otherwise nonsymmetric distributions.43 Raw values above the median have positive Z-scores, whereas those below the median have negative Z-scores.
To calculate and visualize the correlation of the 13 readouts (12 imaging features and CTG), we used the stats (v3.6.1) and the corrplot (v0.84) packages in R. We used data from 201.1A-Cx organoids, including all screened compounds (42 library drugs and 6 control compounds). Technical duplicates on each plate were averaged, but biological replicates were not, resulting in 288 conditions in the final correlation data set. We used the default Pearson correlation method to compute correlation coefficients.
Single Organoid Data Visualization
Single organoid distributions were visualized using the tidyverse (ggplot) and plotly (v4.9.2.1) package, generating histograms (geom_histogram) and violin plots (geom_quasirandom). Correlation plots to compare the histogram distributions of different PDX models or drug dose responses were plotted using the corrplot (v0.84) package based on Pearson’s chi-squared test residuals. Residuals were colored by association, with positive residuals in blue, specifying an attraction between the corresponding row (area bin) and column (cell type/drug concentration) variables. Negative residuals were shown in red, implying a negative association between the corresponding row and column variables.
Statistical Analysis
All statistical analyses, with the exception of Pearson’s chi-squared test of histogram distributions, were conducted using GraphPad Prism 7 software (GraphPad Software Inc, La Jolla, CA), with statistical significance set at p < 0.05. All statistical tests were two tailed and are listed in the corresponding figure legend and supplementary tables. Data are expressed as mean ± SEM.
Results
Establishing a High-Throughput Assay That Supports the Growth of Prostate Cancer Organoids
PDXs and organoids are complementary models with different capabilities for preclinical testing. To integrate these models, we developed a method to grow PDX tissues as organoids and measure their responses to drug treatment (Fig. 1A–C). The samples were prepared by collecting fresh patient tumor tissues, grafting them into immunocompromised mice to establish serially transplantable PDXs, digesting the PDX tissues, and seeding the cells in Matrigel using previously described media for prostate cancer organoids12,13,19 (Fig. 1A). Once the organoids were established, they were reseeded in Matrigel in 384-well plates with a Janus G3 robot with a cooled stage (Fig. 1B). The organoids were grown for 8 d to develop larger structures and then treated with drug for 12 d. Regular live-cell imaging with brightfield microscopy was used to monitor the morphology of organoids before, during, and after treatment (Fig. 1B, C). On day 21, organoids were stained with Hoechst and imaged with fluorescence microscopy. Metabolic activity was measured using CTG. Images of brightfield and fluorescence microscopy were analyzed with CellProfiler software, followed by further statistical analysis with customized R software tools (Fig. 1C; Suppl. Fig. S2).38,39
To examine the performance of this assay, we used five different PDXs from the Melbourne Urological Research Alliance (Fig. 2A). We selected tumors that exhibit active growth as organoids when they are manually seeded in Matrigel (personal communication, Drs. Natalie Lister and Birunthi Niranjan, Monash University). These PDXs represent diverse phenotypes of prostate cancer. They were established from three treatment-naïve, high-risk primary tumors (224R-Cx, 287R, 305R-Cx) and two different therapy-resistant metastases (201.1A-Cx, 201.2A-Cx) from a patient who failed multiple treatments and consented to the CASCADE rapid autopsy program18, 30. This set of PDXs includes castrate-sensitive adenocarcinoma (287R), neuroendocrine prostate cancer (224R-Cx, 305R-Cx), and metastatic CRPC with adenocarcinoma (201.1-Cx) or double-negative (201.2-Cx) pathology (Fig. 2A). The castrate-sensitive PDX (287R) is propagated in testosterone-supplemented mice, whereas the four castration-resistant tumors are grown in castrated mice. None of the patients were treated with PARP inhibitors in the clinic before their tumor tissue was collected (Fig. 2A).
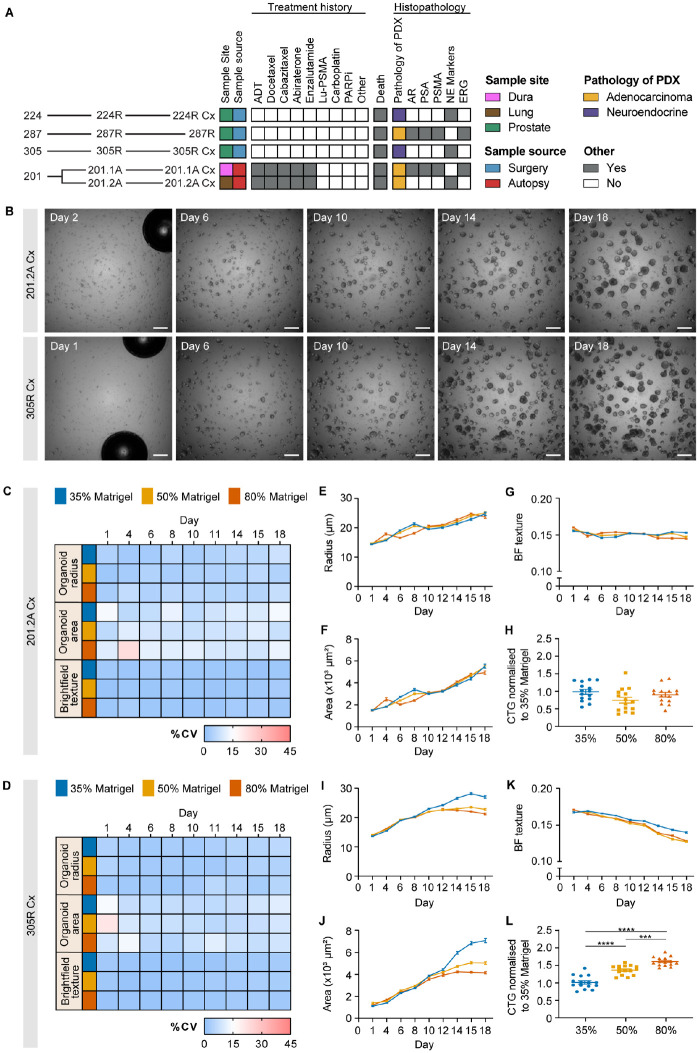
Figure 2.
Identifying robust endpoints for measuring drug responses of prostate cancer organoids. (A) Heat map summarizing features of prostate cancer PDXs established from five specimens obtained from four patients. The data include the site of the original tumor, the source of the tissue, patient treatment history before the tissue was obtained, follow-up (death), and the histopathology of the PDXs. (B) Representative images of 201.2A-Cx and 305R-Cx organoids treated with vehicle control on days 1/2, 6, 10, 14, and 18. Scale bars = 500 µm. (C, D) Heat maps of the percentage coefficient of variation for (C) 201.2A-Cx and (D) 305R-Cx organoids across 18 d for organoid radius, area, and brightfield texture in 35% (blue), 50% (yellow), and 80% Matrigel (orange). (E–G) Growth curves of 201.2A-Cx organoids in different Matrigel concentrations over 18 d for (E) radius, (F) area, and (G) brightfield texture. (H) Metabolic activity (normalized to 35% Matrigel), as measured by CTG, in 201.2A-Cx organoids grown in different concentrations of Matrigel; one-way analysis of variance with Tukey’s post hoc test. (I–K) Growth curves of 305R-Cx organoids over 18 d for (I) radius, (J) area, and (K) brightfield texture. (L) Metabolic activity (normalized to 35% Matrigel), as measured by CTG, in 305R-Cx organoids grown in different concentrations of Matrigel. The radius, area, and brightfield texture values represent the well-level average from each well, expressed as the mean across n = 15–20 replicate wells. Error bars represent SEM. ***p < 0.001; ****p < 0.0001; one-way analysis of variance with Tukey’s post hoc test. CV = coefficient of variation, CTG = CellTiter-Glo; PDX = patient-derived xenograft.
To examine whether there was consistent growth of prostate cancer organoids after automated seeding, we embedded cells from each PDX in Matrigel and monitored their features over time using brightfield microscopy. We used three different concentrations of Matrigel (35%, 50%, and 80%) to determine whether it affected the size and growth rate of the organoids (see plate layout in Suppl. Fig. S1A). As shown for 201.2-Cx and 305R-Cx (Fig. 2B), there were numerous small clusters of cells after seeding, which developed into large multicellular organoids over 18 d. To compare organoid growth between wells, we examined the %CV of the well average organoid area, radius, and brightfield texture measured using CellProfiler (Fig. 2C, D; Suppl. Fig. S3A–C). Overall, there was a low CV across replicate wells, typically less than 15%, for the different models, time points, Matrigel concentrations, and measurements of organoid growth. Most variation was observed during the first 1 to 3 d after seeding, which may be due to small bubbles in the media or Matrigel that could impair imaging of the whole well at early time points (Fig. 2B, day 1/2).
We also used well averages of organoid radius, area, and brightfield texture to compare the growth of organoids over time in different concentrations of Matrigel (Fig. 2E–K; Suppl. Fig. S3D–O; Suppl. Tables S1 and S2). In all concentrations of Matrigel, the mean radius and area of organoids from all models increased over time, confirming their active growth after automated seeding (Fig. 2E, F, I, J; Suppl. Fig. S3D, E, H, I, L, M; Suppl. Table S1). However, the magnitude of growth differed across the models: 305R-Cx organoids grew significantly larger in lower Matrigel concentrations (Fig. 2I, J) but 201.2-Cx organoids did not (Fig. 2E, F; Suppl. Table S2). Conversely, 201.1A-Cx, 224R-Cx, and 287R organoids were all smaller in lower concentrations of Matrigel (Suppl. Fig. S3D, E, H, I, L, M; Suppl. Table S2). Brightfield texture, a quantitative measure of the intensity distribution within structures, had no consistent association with different Matrigel concentrations (Fig. 2G, K; Suppl. Fig. S3F, J, N; Suppl. Table S2). Instead, brightfield texture scores decreased over time in all models across all concentrations of Matrigel, except for 201.2-Cx organoids in 35% Matrigel. This trend likely reflects the change from small, compact structures to larger, more variable structures as the organoids grow over time.
A major difference between the Matrigel concentrations was the distribution of structures across the focal plane. In lower concentrations of Matrigel (35% and 50%), organoids settled into a more consistent low focal plane than in 80%, expediting imaging because a smaller z-stack was required to capture the whole population in each well (Suppl. Fig. S4). One outlier was 201.1A-Cx, where cells grew on the bottom of plates over time when seeded in 35% and, to a lesser extent, 50% Matrigel. Finally, to further examine the growth of prostate cancer organoids in different concentrations of Matrigel, we used CTG to measure the total metabolic activity of each well after 21 d of culture (Fig. 2H, L; Suppl. Fig. S3G, K, O). The average CTG activity was significantly greater in 201.1A-Cx and 305R-Cx organoids grown in higher concentrations of Matrigel. This was despite the smaller radius and area of 305R-Cx organoids in higher concentrations of Matrigel (Fig. 2L), suggesting that these smaller structures had more viable cells for this particular tumor. There was no consistent association between CTG activity and Matrigel concentration for 201.2-Cx, 224R-Cx, and 287R organoids. Thus, no particular Matrigel concentration was superior for the growth of all models measured across all endpoints. Automated seeding successfully supported the growth of all five models in each concentration of Matrigel. For subsequent experiments, we used 50% Matrigel to reduce cost and ensure robust organoid growth of structures at a consistent focal plane for efficient imaging. Overall, we recommend testing different concentrations of Matrigel for new organoid cultures, especially if they are challenging to grow. If cells grow on the bottom of wells, this can be prevented by increasing the Matrigel concentration or perhaps coating wells with a basement layer of 100% Matrigel.
Robust Endpoints for Measuring Drug Responses of Prostate Cancer Organoids
To assess the performance of the assay for preclinical testing and the usefulness of different endpoints, we treated organoids from each tumor with a candidate drug. We used talazoparib, a potent PARP inhibitor that is currently in a phase 3 registration trial for metastatic CRPC (NCT03395197) and approved for patients with HER2-negative breast cancer with germline BRCA1/2 mutations.34,44,45 The dose range (10–500 nM) was based on previous experiments with prostate cells lines, with 500 nM representing a high dose.46 In vivo, 224R-Cx regresses with talazoparib treatment, whereas the other tumors have only a partial (PDX-305R-Cx) or no response (personal communication. Drs. Laura Porter and Ashlee Clark, Monash University). Consistent with this, all talazoparib doses significantly reduced CTG activity of 224R-Cx organoids compared with the vehicle control (Fig. 3A; Suppl. Table S3). The decrease in viability of 224-Cx organoids, treated with increasing doses of talazoparib, was supported by brightfield microscopy and Hoechst staining (Fig. 3B). The 201.2A-Cx and 287R organoids were not affected by talazoparib, whereas 201.1A-Cx and 305R-Cx organoids responded only at the highest dose (Fig. 3A; Suppl. Table S3). Thus, the in vitro organoid data reflected the relative in vivo sensitivity of the PDXs to talazoparib, with 224R-Cx organoids being highly sensitive to talazoparib, whereas the others had minimal responses.
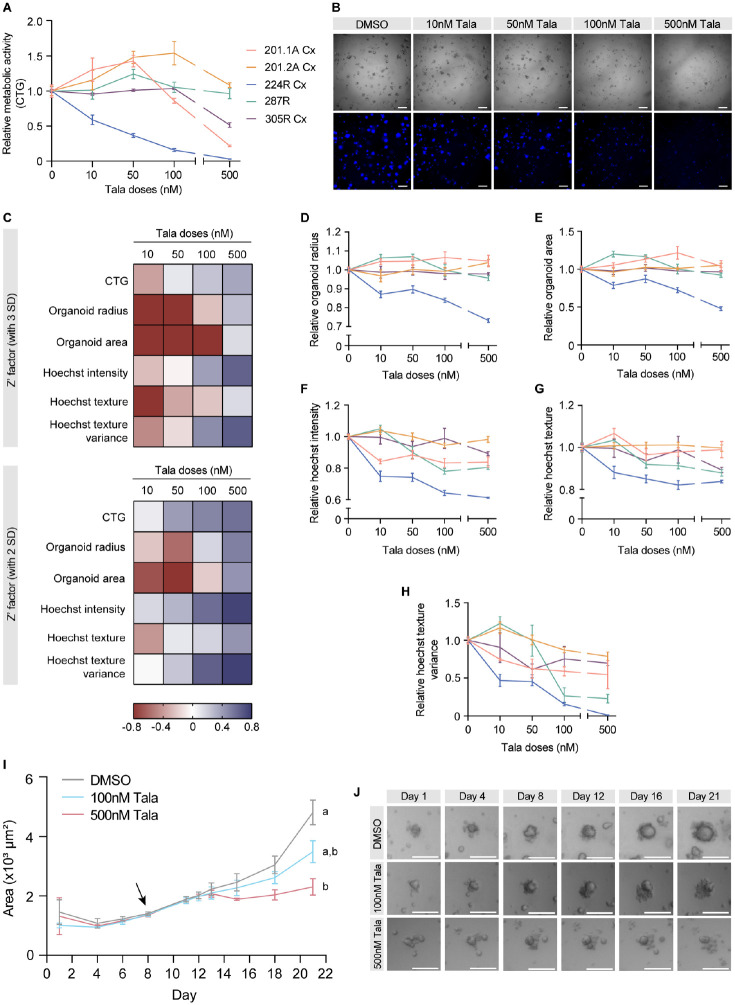
Figure 3.
Comparison of talazoparib responses of organoids from diverse phenotypes of prostate cancer. (A) Relative metabolic activity, as measured by CTG, in 201.1A-Cx (red), 201.2A-Cx (yellow), 224R-Cx (blue), 287R (green), and 305R-Cx (purple) organoids in increasing doses of talazoparib (n = 5). (B) Representative images of brightfield microscopy and Hoechst staining of 224R-Cx organoids treated for 12 d with vehicle or increasing doses of talazoparib. Scale bars = 500 µm. (C) Heat map summarizing the Z′ factor for each readout (CTG, organoid radius, area, Hoechst intensity, texture, and texture variance) in increasing doses of talazoparib. Z′ factors were calculated by using either three (top) or two (bottom) standard deviations in the formula. (D–H) Dose-response curves of organoids generated from measurements of (D) radius, (E) area, (F) Hoechst intensity, (G) texture, and (H) texture variance. Dose-response curves for each organoid treated with talazoparib were normalized to their respective negative control (DMSO). (I) Graph showing the mean area of 224R-Cx organoids treated with DMSO, 100 nM talazoparib, or 500 nM talazoparib over 21 d. The arrow indicates the first day of treatment (day 8). Data are mean ± SEM. ap < 0.001, statistical analysis of change in organoid area across time within each treatment group; bp < 0.001, statistical analysis of organoid area within each treatment group compared with vehicle control; two-way analysis of variance with Dunnett’s post hoc test. (J) Representative images of a single organoid treated with DMSO, 100 nM talazoparib, or 500 nM talazoparib over 21 d. Scale bars = 250 µm. Imaging measurements represent well-level averages, and all data are mean ± SEM. CTG = CellTiter-Glo; SD = standard deviation; Tala = talazoparib.
We also examined whether the responses of tumors to talazoparib were associated with genomic defects of genes in the DNA damage repair pathway, as this predicts the sensitivity to PARP inhibitors in patients.47 None of the tumors have germline mutations, high allele frequency somatic mutations, or deep deletions of genes in the homologous recombination DNA damage repair pathway, indicating that the mechanism underlying the sensitivity of 224R-Cx requires further investigation (Suppl. Fig. 5A). The limited sensitivity to talazoparib of 201.1-Cx, which has a heterozygous BRCA2 mutation, and of 305R-Cx, which has copy number loss of BRCA2, is consistent with the lower response rate of prostate cancers with monoallelic versus biallelic alterations in BRCA2 to PARP inhibitors.6
To assess the performance of the different assay readouts in discriminating between the positive control (talazoparib) and the vehicle control, we calculated the Z′ factor using CTG data from 224R-Cx organoids. The Z′ factor was within the acceptable range of 0 to 0.5 for talazoparib doses of 50 nM and greater (Fig. 3C). Given the expected higher variability in screening 3D organoid cultures, we also calculated Z′ factors based on a lower threshold (2 SDs instead of 3) and found that now all four doses of talazoparib resulted in a Z′ factor greater than zero (Fig. 3C). In addition to CTG, we analyzed brightfield and Hoechst-stained cells at the end of treatment and compared the Z′ factor for a range of imaging-based features (Suppl. Fig. S2) to identify the most robust readouts for further analyses. Overall, five readouts had acceptable Z′ factor scores calculated using at least 2 SD (organoid radius and area based on brightfield images and Hoechst intensity, texture, and texture variance based on fluorescence images; Fig. 3C).
Consistent with the sensitivity of 224R-Cx organoids to talazoparib, their average radius and area decreased across all talazoparib doses (Fig. 3D, E). After talazoparib treatment, 224R-Cx organoids also became less dense, based on decreased Hoechst intensity (Fig. 3F), and less uniform in composition, based on decreased Hoechst texture (Fig. 3G). Furthermore, Hoechst texture variance, a measure of intensity variations between objects within an image, also decreased with increasing doses of talazoparib (Fig. 3H), suggesting consistent changes in the composition of organoids within treated wells. Compared with 224R-Cx organoids, there was minimal change in the dose-response curves with each readout for the other tumors, especially at talazoparib doses up to 100 nM (Fig. 3D–H; Suppl. Table S3). Collectively, these data demonstrate that in addition to measuring metabolic activity with CTG, imaging-based measures of organoid size and composition are robust and valuable readouts of organoid growth.
We found that live-cell brightfield imaging was extremely useful in quantifying structure growth and responses to drug treatment over time. We measured organoid area over the course of the assay to compare the responses of 224R-Cx and 287R organoids to talazoparib. Intriguingly, after initiating treatment on day 8, the average area of 224R-Cx organoids in the vehicle and treatment groups started to diverge only 7 d later, on day 15 of treatment (Fig. 3I, J). By 10 d posttreatment, the 224R-Cx organoids treated with talazoparib were significantly smaller in area than those treated with the vehicle control. In contrast, 287R organoids were unresponsive to talazoparib and increased in area at a similar rate in both control and treatment groups (Suppl. Fig. S5C–D).
In addition to talazoparib, we treated 224R-Cx organoids with carboplatin (500 nM), a platinum chemotherapy, for 21 d (Suppl. Fig. S5B) to show that live-cell measurements of organoid area also detected the effects of a different compound. This highlights the importance of monitoring organoids at several time points to generate a temporal measure of response and ensure that any drug-induced changes are not overlooked.
Using Robust Endpoints for a Compound Screen with Organoids
After identifying robust readouts, based on talazoparib treatment, we applied the high-throughput imaging assay to a pilot compound screen. The screen included 42 compounds from a custom drug library and 6 controls, all at three doses, across three different organoids (201.1A-Cx, 201.2A-Cx, and 287R; see the screen plate layout in Suppl. Fig. S1B). To identify potential hits, we used CTG and Hoechst texture variance as readouts. Both are sensitive endpoints (Fig. 3C), and there was no major correlation between them based on Pearson correlation coefficients (Fig. 4A). This suggested that the combination of both endpoints would yield additional, nonredundant information on the effect of the drugs. The average fold changes in CTG and Hoechst texture variance for each compound (across two technical and two biological plate replicates) were compared in scatter plots for each dose (0.1, 1, or 10 µM; Fig. 4B). The number of compounds in the red zone (with <0.5-fold change to control for both readouts) increased with higher doses as expected.
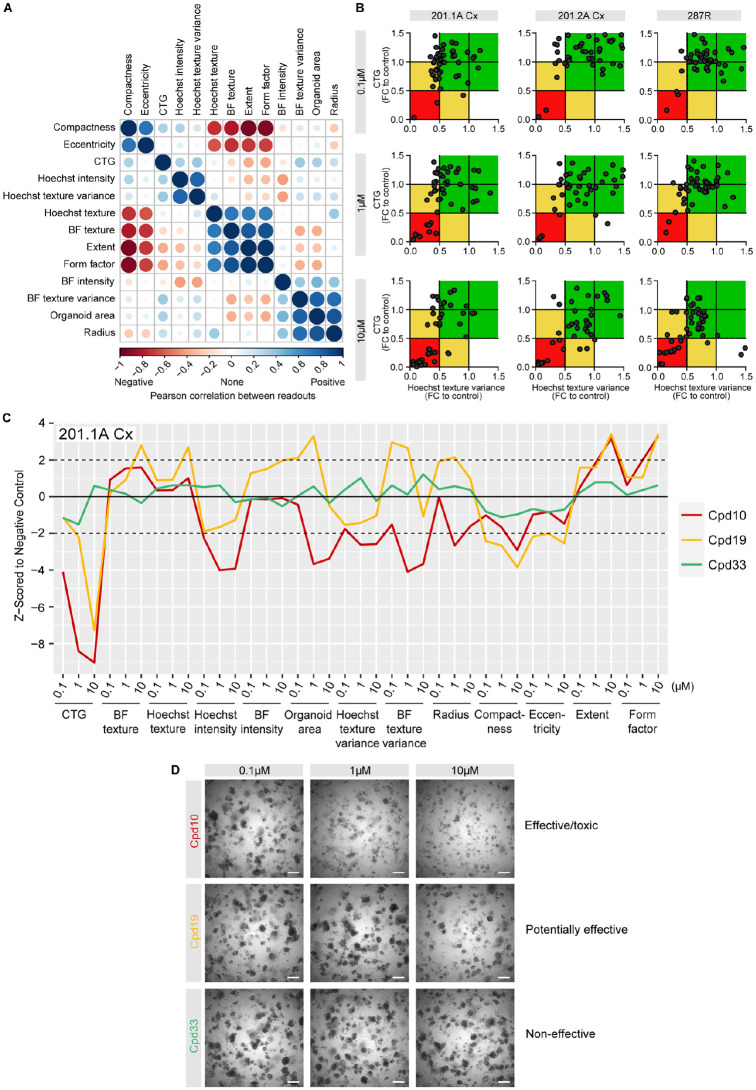
Figure 4.
Multiparametric analysis of compound screen with organoids. (A) Graphical matrix of positive (blue) and negative (red) Pearson correlation coefficients for 13 readouts (CTG and 12 features from the brightfield and Hoechst images) with 201.1A-Cx, 201.2A-Cx, and 287R organoids across all three doses of a compound library. The size of a circle is proportional to the magnitude of the correlation. (B) Scatter plot showing values for CTG (y-axis) and Hoechst texture variance (x-axis) in 201.A-Cx, 201.2A-Cx, and 287R organoids treated with 0.1, 1, and 10 µM of 48 compounds. The red zone includes compounds with <0.5-fold change to control for both readouts. The yellow zone includes compounds with <0.5-fold change to control for only one of the readouts, whereas the green zone includes compounds with >0.5-fold change to control for both readouts. (C) Graph of the phenotypic profiles of 201.1A-Cx organoids based on robust Z-scores from the 13 readouts in response to three doses of Cpd10 (red; effective/toxic), Cpd19 (yellow; potentially effective), and Cpd33 (green; noneffective). (D) Representative images of 201.1A-Cx organoids treated with 0.1, 1, and 10 µM Cpd10, Cpd19, and Cpd33 over 12 d. Scale bars = 500 µm. CTG = CellTiter-Glo, BF = brightfield texture; FC = fold change.
Across the three organoids, only one of the toxic positive control compounds was a hit at 0.1 µM. At the 1 µM dose, in addition to the toxic positive control, compound 10 (Cpd10) was a hit for all three organoids. At the highest dose of 10 µM, seven compounds were consistently in the red zone, which we classified as “toxic/effective” (Fig. 4B).
To further compare CTG with high-throughput imaging readouts, we used the robust Z-scores of 13 readouts to plot the profile of 201.1A-Cx organoids in response to all three doses of three compounds (Fig. 4C). Cpd10 represents an “effective/toxic” hit, with both 1 and 10 µM in the red zone. It induced a marked reduction in CTG, Hoechst intensity, Hoechst texture variance, and area, even at the lowest dose of 0.1 µM (Fig. 4C). Brightfield images confirmed the clear toxicity of 1 and 10 µM doses of Cpd10 (Fig. 4D). Conversely, Cpd33 was classified as a “noneffective” compound (green zone), as it did not significantly change any imaging feature or CTG as compared with control (Fig. 4C) or alter organoid morphology (Fig. 4D). Finally, Cpd19 was classified as a “potentially effective” compound (yellow zone), because the CTG fold change was below the cutoff but the Hoechst texture variance fold change was not. Cpd19 would have been classified as highly toxic based on CTG alone. Interestingly, the additional imaging features showed that Cpd19 induced a very different phenotypic profile from Cpd10, with a strong reduction in CTG but an increase in area and texture features (Fig. 4C). Altogether, these data show that the assay can be used to screen organoids of prostate cancer with compound libraries and stratify hits based on multiple readouts, which is not feasible with a single feature.
Brightfield Microscopy and Hoechst Staining Capture Inter- and Intratumor Heterogeneity in Organoid Morphology and Composition
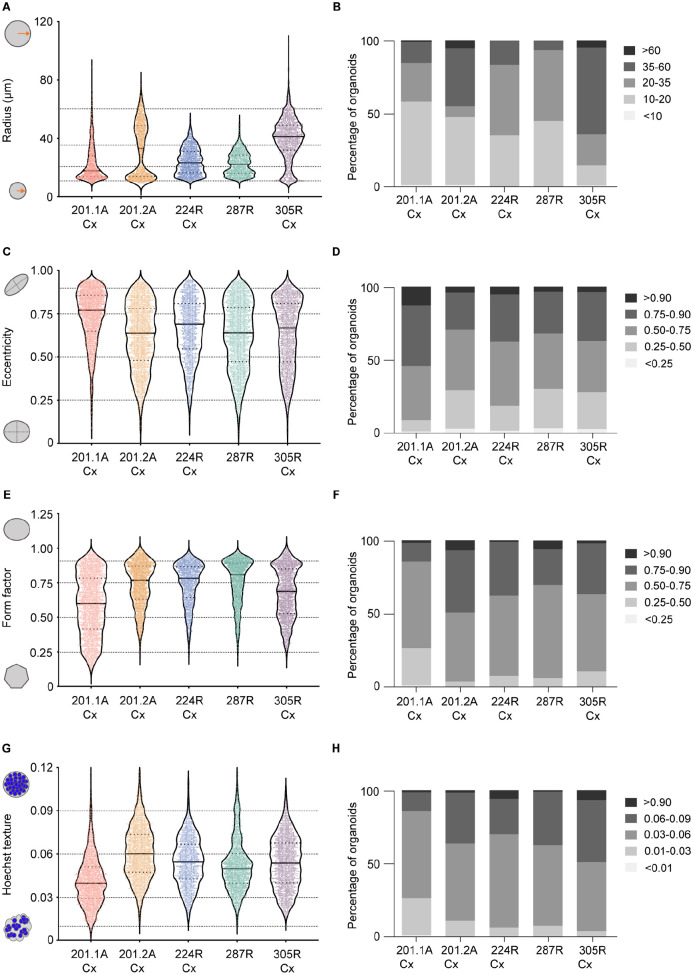
To examine the heterogeneity within and between organoid cultures, we measured the morphology and composition of individual untreated organoids from different tumors based on their radius, eccentricity, form factor, and Hoechst texture (Fig. 5A–H). The variation in these readouts between single organoids was evident from violin plots of each culture (Fig. 5A, C, E, G). To enable quantitative comparison of the distribution of single organoids, we segregated the values for each parameter into five discrete bins (Fig. 5A, C, E, G). Stacked plots showed that the proportion of organoids within each bin varied, highlighting the heterogeneity of organoid cultures (Fig. 5B, D, F, H). Therefore, we used Pearson’s chi-squared tests to determine whether there were significantly different numbers of organoids in each bin (Suppl. Fig. S6). Organoid radius was the most sensitive feature to distinguish between the tumor models and showed higher variability between tumors. Both 201.2A-Cx and 305R-Cx organoids had a higher proportion of large radius structures (>35 µm), whereas 201.1A-Cx, 224R-Cx, and 287R organoids were often smaller (<35 µm; Fig. 5B; Suppl. Fig. S6A). Despite this, organoids from four of the five models had similar morphologies based on eccentricity and form factor. These measures of circularity were evenly distributed with a slight trend toward a rounder shape (Fig. 5D F,; Suppl. Fig. S6B, C). The 201.1A-Cx organoids were an exception, being more elliptical based on the distribution of eccentricity and form factor. There were also more irregular structures within 201.1A-Cx cultures, resulting in the lowest overall Hoechst texture of the five models (Fig. 5H; Suppl. Fig. S6D). This may be due to some cells growing on the bottom of plates instead of forming compact, round organoids. Collectively, these data highlight the heterogeneity in the size and shape of prostate cancer organoids, even within cultures of each tumor. Given this complexity, it may be informative to evaluate the effects of drug treatments in defined subpopulations of organoids in addition to the average changes across each well.
Figure 5.
Brightfield microscopy and Hoechst staining readouts capture inter- and intrapatient heterogeneity in organoid morphology and composition. The distribution of organoid radius (A, B), eccentricity (C, D), form factor (E, F), and Hoechst texture (G, H) is shown for untreated organoids from five PDXs (n = 15 wells per tumor). In the violin plots (A, C, E, G), each dot denotes an individual organoid. For each specific parameter, organoids from each PDX were segregated into five discrete bins. Stacked bar graphs (B, D, F, H) show the percentage of organoids in each bin for each organoid model. PDX = patient-derived xenograft.
Single-Organoid Analyses Show Talazoparib Alters the Morphology and Composition of PARP Inhibitor–Sensitive Organoids
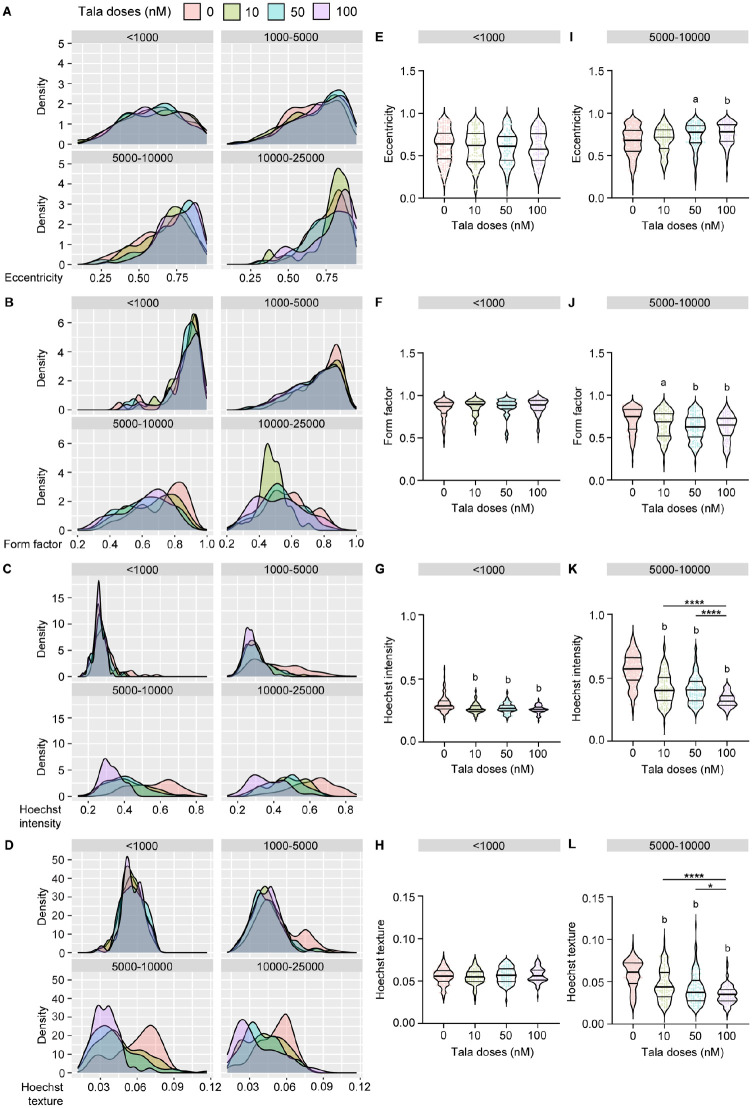
To examine the response of 224R-Cx to talazoparib at the single-structure level, we divided organoids into four bins based on their area, stratifying them as <1000 µm2, 1000 to 5000 µm2, 5000 to 10,000 µm2, and 10,000 to 25,000 µm2. We excluded the 10,000 to 25,000 µm2 bin from further analysis to ensure that any large artifacts or poorly segmented organoids would not bias the outcome. We also excluded the high 500 nM dose because of significant toxicity in the 224R-Cx model. Next, we evaluated changes in the distribution of eccentricity (Fig. 6A), form factor (Fig. 6B), Hoechst intensity (Fig. 6C), and texture (Fig. 6D) in each bin after talazoparib treatment. In the small organoids (<1000 µm2), there was little variation in the distribution of eccentricity (Fig. 6E), form factor (Fig. 6F), and Hoechst texture (Fig. 6H) after treatment. Only Hoechst intensity (Fig. 6G) differed from the vehicle control. Thus, the small organoids maintained a rounded morphology after treatment, but there was a decrease in their cellular density.
Figure 6.
Single organoid analyses show that talazoparib alters the morphology and composition of PARP inhibitor–sensitive organoids. Untreated and treated 224R-Cx organoids were divided into four groups based on their area: <1000 µm2, 1000 to 5000 µm2, 5000 to 10,000 µm2, and 10,000 to 25,000 µm2. (A–D) Density plots showing the change in distribution of (A) eccentricity, (B) form factor, (C) Hoechst intensity, and (D) texture values in organoids after treatment compared with vehicle control in each group. (E–H) Violin plots of the (E) eccentricity, (F) form factor, (G) Hoechst intensity, and (H) texture values in organoids after treatment compared with vehicle control in <1000 µm2 organoids. (I–L) Violin plots for the (I) eccentricity, (J) form factor, (K) Hoechst intensity, and (L) texture values in organoids after treatment compared with vehicle control in 5000 to 10,000 µm2 organoids. Each dot denotes an individual organoid from n = 15 (DMSO) or n = 5 wells (talazoparib). ap < 0.01; bp < 0.0001, statistical analysis of organoid measurements compared with vehicle control. *p < 0.05, ****P < 0.0001; one-way analysis of variance with post hoc Tukey’s test. PARP = poly ADP-ribose polymerase; tala = talazoparib.
As organoids increased in area, there were greater changes in the distribution of each endpoint across all doses of talazoparib. Focusing on organoids in the 5000 to 10,000 µm2 bin, we showed there was a significant difference in eccentricity and form factor after talazoparib treatment (Fig. 6I, J). Hoechst intensity and texture also significantly decreased in the larger organoids (Fig. 6K, L). This effect was significantly different between each dose of talazoparib, indicating a dose-dependent decrease in the uniformity and density organoids with treatment. Collectively, these in-depth analyses of organoids based on their size show that more striking changes in morphology and composition occurred in larger structures. Thus, imaging-based readouts of individual organoids can extend the findings from whole well-level measurements of cell viability.
Discussion
For preclinical testing, organoids have the potential to maintain the scalability of 2D cell lines, while adding the clinical relevance of PDXs. In practice, however, this remains challenging, particularly for tumor types that are difficult to grow, such as prostate cancer. In this study, we developed an automated, high-throughput assay that enables the growth of organoids grown from prostate cancer PDXs, with low well-to-well variance over time. In addition, we identified several robust endpoints to quantify the responses of organoids to drug treatment, including CTG cell viability and 3D size and morphology captured using brightfield microscopy and Hoechst staining. These readouts can be integrated to identify hits in compound screens. Beyond these well-level readouts, we demonstrated that high-content imaging can be used to monitor differences in organoid growth over time and to divide cultures into specific subpopulations for single-organoid measurements. Therefore, this assay provides a high-throughput approach to grow, treat, and analyze organoids in depth.
Using 3D organoids from PDXs for high-throughput screening presents a unique set of challenges. Recognizing those challenges, and then addressing them by selecting the appropriate automation, instrumentation, analysis software, readouts, optimization steps, and controls is vital for successfully implementing 3D image–based drug screening.
Automation of 3D organoid imaging pipelines with liquid handling and robotics has been established by other research groups to improve the imaging efficiency with multiple patient samples and large drug compound libraries.48,49 However, in those assays, organoids were layered on top of the gel, rather than fully embedded within the 3D extracellular matrices. Here, we used the JanusG3 liquid handling robot (PerkinElmer) for automatic dispensing of a complete organoid/Matrigel suspension—sitting in a cold block—into multiwell plates. This overcame the challenges of temperature control and gel viscosity and enabled us to test multiple concentrations of Matrigel in our routine assay development pipeline to quickly optimize the growth conditions for prostate cancer organoids.
We decided to capture all readouts—brightfield and fluorescence images as well as CTG luminescence—on the Cytation5 Cell Imaging Multi-Mode Reader (BioTek) at a very low magnification of 2.5×. We selected a variety of readouts with the goal of extracting as much information from the 3D cultures as possible. In addition, all image and data analyses were performed with open-access software tools and freely available pipelines. This affordable, all-in-one instrument and open-access software will be more accessible to a wide variety of laboratories compared with advanced confocal or high-resolution microscopes with subscription-based analysis tools. Alternative approaches for analyzing 3D cultures include the AMIDA (Automated Morphometric Image Data Analysis) package, which has been used with prostate cancer cell lines.50
Patient organoids can have different growth rates and drug responses depending on their genetic and environmental features.51 Although previous studies analyzed the growth rate of different organoids, they have often relied on fluorescent labeling techniques with dyes or lentiviral constructs.28 Here, we monitored the organoids over 3 wk with label-free live-cell imaging with brightfield microscopy, generating growth curves and kinetic data to monitor drug treatment efficiently.
It is critical to consider interpatient, intrapatient, and intratumor variability when screening for new treatments.52 One approach has been to generate multiple cancer organoid cultures from each patient cancer specimen and screen them separately.53 In our image analyses, we segmented every organoid in each well individually to obtain a single-organoid data set. This allowed us to compare the morphology of organoid populations from five tumors with each other. More importantly, we could also deeply analyze subsets of organoids within each culture and show that talazoparib caused more measureable changes in organoid morphology and composition in larger structures.
As the capability for growing and analyzing organoids increases, there will be greater consideration of how best to integrate them with other patient-derived models in preclinical research. Here, we grew organoids from PDXs, circumventing the need for fresh patient samples and creating an opportunity to use the same tumors for in vitro and in vivo experiments in future studies. Regardless of the source of organoids and PDXs, and whether they come from the same or different patients, they are complementary models for measuring drug responses.24,54 A limitation is that it continues to be difficult to establish PDXs and organoids from prostate cancer with existing protocols, and not all PDXs grow as efficiently in vitro.18,19 In our hands, approximately 90% of PDXs form organoids, and 20% of those can be passaged for at least five generations. Nevertheless, when organoids are grown successfully, they are useful for testing drug treatments across broad dose responses or screening compound libraries and carefully quantifying well-level and single-organoid changes in growth and morphology. In comparison, PDX experiments are low throughput but are invaluable for measuring changes in tumor volume over time in a complex host environment where pharmacokinetics and pharmacodynamics can affect treatment responses. Thus, the advantages of each model address the disadvantages of the other.24
The flexibility and scalability of automated organoid experiments will enable several future applications of this approach. One important application is compound screening, and here we showed that CTG and imaging endpoints provide complementary, but not redundant, measures of organoid responses to a drug library. Independent readouts can be integrated to prioritize compound hits for further testing and to compare how different compounds alter the viability, morphology, and composition of organoids. For example, we identified two compounds that both reduced CTG activity and elicited distinct changes in organoid morphology. Another application of the organoid assay is to assess drug synergy of combination therapies.15,55–57 In the future, using this methodology to analyze co-cultures of organoids and other cell types would address a common criticism of tumor “organoids”: that they usually do not contain cell from the tumor microenvironment. It would also provide scope for testing a greater range of treatments, including immunotherapies.
In conclusion, this study demonstrates the ability to automate preclinical testing with prostate cancer organoids with diverse phenotypes. We have shown that using a combination of plate-reader and imaging-based parameters can generate robust readouts. Our approach can be used to quantify changes in the growth of heterogeneous 3D cultures to candidate drugs or compound libraries and across whole wells or specific subpopulations of organoids. Coupling this approach with other patient-derived models will expand the capacity and rigor of preclinical testing of new treatments before they enter clinical trials.
Supplemental Material
Supplemental material, sj-pdf-1-jbx-10.1177_24725552211020668 for High-Throughput Imaging Assay for Drug Screening of 3D Prostate Cancer Organoids by Nicholas Choo, Susanne Ramm, Jennii Luu, Jean M. Winter, Luke A. Selth, Amy R. Dwyer, Mark Frydenberg, Jeremy Grummet, Shahneen Sandhu, Theresa E. Hickey, Wayne D. Tilley, Renea A. Taylor, Gail P. Risbridger, Mitchell G. Lawrence and Kaylene J. Simpson in SLAS Discovery
Supplemental material, sj-pdf-2-jbx-10.1177_24725552211020668 for High-Throughput Imaging Assay for Drug Screening of 3D Prostate Cancer Organoids by Nicholas Choo, Susanne Ramm, Jennii Luu, Jean M. Winter, Luke A. Selth, Amy R. Dwyer, Mark Frydenberg, Jeremy Grummet, Shahneen Sandhu, Theresa E. Hickey, Wayne D. Tilley, Renea A. Taylor, Gail P. Risbridger, Mitchell G. Lawrence and Kaylene J. Simpson in SLAS Discovery
Acknowledgments
We acknowledge the people of the Kulin and Wurundjeri Nations, on whose land these studies were done. We pay our respects to their Elders, past and present. We thank the patients and families who generously supported this research by consenting to provide tissue. We thank the members of the Melbourne Urological Research Alliance, Melissa Papargiris, and Jenna Kraska for providing PDXs; Kathryn Alsop, Lisa Devereux, Heather Thorne, and the CASCADE rapid autopsy program; Wallace Crellin and James McPherson for invaluable advice; David Goode, Andrew Bakshi, and Shivakumar Keerthikumar for targeted sequencing data; Melissa Bullock, Ashlee Clark, Natalie Lister, Samantha O’Dea, Birunthi Niranjan, and Hong Wang for laboratory assistance and technical advice; Monash University Animal Research Laboratories; and the Monash Biomedicine Discovery Institute Organoid Program. We thank Compounds Australia at Griffith University for provision of compounds and logistics management.
Footnotes
Supplemental material is available online with this article.
Author Contributions: M.G.L. and K.J.S. had full access to all the data in the study and take full responsibility for the integrity of the data and the accuracy of the data analysis. N.C., S.R., L.A.S., T.E.H., W.D.T., R.A.T., G.P.R., M.G.L., K.J.S., study concept and design; N.C., S.R., J.L., acquisition of data; N.C., S.R., J.M.W., W.D.T., R.A.T., G.P.R., M.G.L., K.J.S., analysis and interpretation of data; N.C., S.R., M.G.L., K.J.S., drafting of manuscript; N.C., S.R., statistical analysis; L.A.S., T.E.H., W.D.T., R.A.T., G.P.R., M.G.L., K.J.S., obtaining funding; J.M.W., A.R.D., M.F., J.G., S.S., administrative, technical, or material support; W.D.T., R.A.T., G.P.R., M.G.L., K.J.S., supervision; all authors, critical revision of the manuscript for important intellectual content.
Declaration of Conflicting Interests: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: G. Risbridger, R. Taylor, and M. Lawrence, provision of reagents for laboratory research (Zenith Epigenetics) and other research collaborations (Pfizer, Astellas). J. Grummet, honoraria from BK Ultrasound, Biobot, Mundipharma; travel: Astellas; owner of MRI PRO Pty Ltd., an online training platform. S. Sandhu, grant funding to institution from AstraZeneca, Novartis, Amgen, Pfizer, Genentech, Merck Sharp and Dohme, Merck Serono; honoraria donated to institution from Merck Sharp and Dohme, Astra Zeneca, and Bristol Myer Squibb. All other authors have no conflicts of interest to declare.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Health and Medical Research Council, Australia (fellowship to G.P.R. 1102752, project grants 1138242 and 1156570; ideas grant 1186647 to W.D.T.); the Department of Health and Human Services acting through the Victorian Cancer Agency (fellowships to M.G.L. MCRF18017, R.A.T. MCRF15023, CAPTIV Program); Movember & the National Breast Cancer Foundation (NBCF; Collaboration Initiative Grant to T.E.H, G.P.R, L.A.S., and W.D.T.); the US Department of Defense through the Prostate Cancer Research Program (G.P.R. W81XWH1810349; opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense); the CASS Foundation (medical science grant 7139 to M.G.L.); the Movember Foundation (Global Action Plan 1); the EJ Whitten Foundation; the Peter and Lyndy White Foundation; TissuPath Pathology; The Hospital Research Foundation (ID 2018-06-Strategic-R; T.E.H., L.A.S., W.D.T.); NBCF (fellowship IIRS-19-009 to T.E.H.); Cancer Council SA Beat Cancer Project (Early Career Cancer Research Fellowship to J.M.W.); the University of Adelaide (W.D.T.); and Flinders Health and Medical Research Institute, Flinders University (L.A.S.). L.A.S. is supported by principal cancer research fellowships awarded by the Cancer Council’s Beat Cancer project on behalf of its donors, the state government through the Department of Health, and the Australian government through the Medical Research Future Fund. The Victorian Centre for Functional Genomics (K.J.S.) is funded by the Australian Cancer Research Foundation (ACRF), Phenomics Australia (PA) through funding from the Australian Government’s National Collaborative Research Infrastructure Strategy (NCRIS) program, the Peter MacCallum Cancer Centre Foundation, and the University of Melbourne Research Collaborative Infrastructure Program (MCRIP).
ORCID iD: Kaylene J. Simpson  https://orcid.org/0000-0001-9136-1781
https://orcid.org/0000-0001-9136-1781
References
- 1.James N. D., Sydes M. R., Clarke N. W., et al. Addition of Docetaxel, Zoledronic Acid, or Both to First-Line Long-Term Hormone Therapy in Prostate Cancer (STAMPEDE): Survival Results from an Adaptive, Multiarm, Multistage, Platform Randomised Controlled Trial. Lancet 2016, 387, 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sweeney C. J., Chen Y.-H., Carducci M., et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. N. Engl. J. Med. 2015, 373, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis I. D., Martin A. J., Stockler M. R., et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N. Engl. J. Med. 2019, 381, 121–131. [DOI] [PubMed] [Google Scholar]
- 4.Chi K. N., Agarwal N., Bjartell A., et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2019, 381, 13–24. [DOI] [PubMed] [Google Scholar]
- 5.Fizazi K., Tran N., Fein L., et al. Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2017, 377, 352–360. [DOI] [PubMed] [Google Scholar]
- 6.Abida W., Patnaik A., Campbell D., et al. Rucaparib in Men with Metastatic Castration-Resistant Prostate Cancer Harboring a BRCA1 or BRCA2 Gene Alteration. J. Clin. Oncol. 2020, 38, 3763–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bono J., Mateo J., Fizazi K., et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [DOI] [PubMed] [Google Scholar]
- 8.Coutinho I., Day T. K., Tilley W. D., et al. Androgen Receptor Signaling in Castration-Resistant Prostate Cancer: A Lesson in Persistence. Endocr. Relat. Cancer 2016, 23, T179–T197. [DOI] [PubMed] [Google Scholar]
- 9.Davies A., Zoubeidi A., Selth L. A.The Epigenetic and Transcriptional Landscape of Neuroendocrine Prostate Cancer. Endocr. Relat. Cancer 2020, 27, R35–R50. [DOI] [PubMed] [Google Scholar]
- 10.Obinata D., Lawrence M. G., Takayama K., et al. Recent Discoveries in the Androgen Receptor Pathway in Castration-Resistant Prostate Cancer. Front. Oncol. 2020, 10, 581515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Risbridger G. P., Lawrence M. G., Taylor R. A.PDX: Moving beyond Drug Screening to Versatile Models for Research Discovery. J. Endocr. Soc. 2020, 4, bvaa132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drost J., Karthaus W. R., Gao D., et al. Organoid Culture Systems for Prostate Epithelial and Cancer Tissue. Nat. Protoc. 2016, 11, 347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao D., Vela I., Sboner A., et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014, 159, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chua C. W., Shibata M., Lei M., et al. Single Luminal Epithelial Progenitors Can Generate Prostate Organoids in Culture. Nat. Cell Biol. 2014, 16, 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauli C., Hopkins B. D., Prandi D., et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risbridger G. P., Lawrence M. G.Towards Best Practice in Establishing Patient-Derived Xenografts. In Patient-Derived Xenograft Models of Human Cancer; Wang Y., Lin D., Gout P. W., Eds.; Humana Press, 2017. [Google Scholar]
- 17.Puca L., Bareja R., Prandi D., et al. Patient Derived Organoids to Model Rare Prostate Cancer Phenotypes. Nat. Commun. 2018, 9, 2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence M. G., Obinata D., Sandhu S., et al. Patient-Derived Models of Abiraterone and Enzalutamide-Resistant Prostate Cancer Reveal Sensitivity to Ribosome-Directed Therapy. Eur. Urol. 2018, 74, 562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beshiri M. L., Tice C. M., Tran C., et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin. Cancer Res. 2018, 24, 4332–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong E. L., Martinez M., Yang J., et al. Hydrogel-Based 3D Model of Patient-Derived Prostate Xenograft Tumors Suitable for Drug Screening. Mol. Pharm. 2014, 11, 2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young S. R., Saar M., Santos J., et al. Establishment and Serial Passage of Cell Cultures Derived from LuCaP Xenografts. Prostate 2013, 73, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saar M., Zhao H., Nolley R., et al. Spheroid Culture of LuCaP 147 as an Authentic Preclinical Model of Prostate Cancer Subtype with SPOP Mutation and Hypermutator Phenotype. Cancer Lett. 2014, 351, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes R. C., Toubia J., Townley S., et al. Post-Transcriptional Gene Regulation by MicroRNA-194 Promotes Neuroendocrine Transdifferentiation in Prostate Cancer. Cell Rep. 2021, 34, 108585. [DOI] [PubMed] [Google Scholar]
- 24.Gleave A. M., Ci X., Lin D., et al. A Synopsis of Prostate Organoid Methodologies, Applications, and Limitations. Prostate 2020, 80, 518–526. [DOI] [PubMed] [Google Scholar]
- 25.Horvath P., Aulner N., Bickle M., et al. Screening out Irrelevant Cell-Based Models of Disease. Nat. Rev. Drug Discov. 2016, 15, 751–769. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence M. G., Porter L. H., Clouston D., et al. Knowing What’s Growing: Why Ductal and Intraductal Prostate Cancer Matter. Sci. Transl. Med. 2020, 12, eaaz0152. [DOI] [PubMed] [Google Scholar]
- 27.Jansson K. H., Tucker J. B., Stahl L. E., et al. High-Throughput Screens Identify HSP90 Inhibitors as Potent Therapeutics That Target Inter-Related Growth and Survival Pathways in Advanced Prostate Cancer. Sci. Rep. 2018, 8, 17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S., Choung S., Sun R. X., et al. Comparison of Cell and Organoid-Level Analysis of Patient-Derived 3D Organoids to Evaluate Tumor Cell Growth Dynamics and Drug Response. SLAS Discov. 2020, 25, 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wardwell-Swanson J., Suzuki M., Dowell K. G., et al. A Framework for Optimizing High-Content Imaging of 3D Models for Drug Discovery. SLAS Discov. 2020, 25, 709–722. [DOI] [PubMed] [Google Scholar]
- 30.Alsop K., Thorne H., Sandhu S., et al. A Community-Based Model of Rapid Autopsy in End-Stage Cancer Patients. Nat. Biotechnol. 2016, 34, 1010–1014. [DOI] [PubMed] [Google Scholar]
- 31.Porter L. H., Lawrence M. G., Wang H., et al. Establishing a Cryopreservation Protocol for Patient-Derived Xenografts of Prostate Cancer. Prostate 2019, 79, 1326–1337. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence M. G., Taylor R. A., Toivanen R., et al. A Preclinical Xenograft Model of Prostate Cancer Using Human Tumors. Nat. Protoc. 2013, 8, 836–848. [DOI] [PubMed] [Google Scholar]
- 33.Abida W., Patnaik A., Campbell D., et al. Rucaparib in Men with Metastatic Castration-Resistant Prostate Cancer Harboring a BRCA1 or BRCA1 Alteration. J. Clin. Oncol. 2020, 38, 3763–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Bono J. S., Higano C. S., Saad F., et al. TALAPRO-1: An Open-Label, Response Rate Phase II Study of Talazoparib (TALA) in Men with DNA Damage Repair (DDR) Defects and Metastatic Castration-Resistant Prostate Cancer (mCRPC) Who Previously Received Taxane-Based Chemotherapy (CT) and Progressed on Greater Than or Equal to One Novel Hormonal Therapy (NHT). J. Clin. Oncol. 2019, 37, TPS342–TPS342. [Google Scholar]
- 35.de Bono J., Mateo J., Fizazi K., et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [DOI] [PubMed] [Google Scholar]
- 36.Smith M. R., Sandhu S. K., Kelly W. K., et al. Phase II Study of Niraparib in Patients with Metastatic Castration-Resistant Prostate Cancer (mCRPC) and Biallelic DNA-Repair Gene Defects (DRD): Preliminary Results of GALAHAD. J. Clin. Oncol. 2019, 37, 202–202.30523719 [Google Scholar]
- 37.McLaren W., Gil L., Hunt S. E., et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McQuin C., Goodman A., Chernyshev V., et al. CellProfiler 3.0: Next-Generation Image Processing for Biology. PLoS Biol. 2018, 16, e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, 2013. [Google Scholar]
- 40.Wickham H., Averick M., Bryan J., et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4. [Google Scholar]
- 41.Wilkinson L.Visualizing Big Data Outliers through Distributed Aggregation. IEEE Trans. Vis. Comput. Graph. 2018, 24, 256–266. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J. H., Chung T. D., Oldenburg K. R.A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- 43.Bray M. A., Carpenter A., Imaging Platform B. I. o. M. a. H., et al. Assay Guidance Manual. Eli Lilly & Co, 2004. [Google Scholar]
- 44.Agarwal N., Shore N. D., Dunshee C., et al. TALAPRO-2: A Two-Part, Placebo-Controlled Phase III Study of Talazoparib (TALA) with Enzalutamide (ENZA) in Metastatic Castration-Resistant Prostate Cancer (mCRPC). J. Clin. Oncol. 2019, 37, TPS337–TPS337. [Google Scholar]
- 45.Litton J. K., Rugo H. S., Ettl J., et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murai J., Huang S.-y. N., Renaud A., et al. Stereospecific PARP Trapping by BMN 673 and Comparison with Olaparib and Rucaparib. Mol. Cancer Ther. 2014, 13, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mateo J., Carreira S., Sandhu S., et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czerniecki S. M., Cruz N. M., Harder J. L., et al. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 2018, 22, 929–940.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francies H. E., Barthorpe A., McLaren-Douglas A., et al. Drug Sensitivity Assays of Human Cancer Organoid Cultures. Methods Mol. Biol. 2019, 1576, 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Härmä V., Schukov H. P., Happonen A., et al. Quantification of Dynamic Morphological Drug Responses in 3D Organotypic Cell Cultures by Automated Image Analysis. PLoS One 2014, 9, e96426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlachogiannis G., Hedayat S., Vatsiou A., et al. Patient-Derived Organoids Model Treatment Response of Metastatic Gastrointestinal Cancers. Science 2018, 359, 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Witte C. J., Espejo Valle-Inclan J., Hami N., et al. Patient-Derived Ovarian Cancer Organoids Mimic Clinical Response and Exhibit Heterogeneous Inter- and Intrapatient Drug Responses. Cell Rep. 2020, 31, 107762. [DOI] [PubMed] [Google Scholar]
- 53.Li L., Knutsdottir H., Hui K., et al. Human Primary Liver Cancer Organoids Reveal Intratumor and Interpatient Drug Response Heterogeneity. JCI Insight 2019, 4, e121490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Risbridger G. P., Toivanen R., Taylor R. A.Preclinical Models of Prostate Cancer: Patient-Derived Xenografts, Organoids, and Other Explant Models. Cold Spring Harb. Perspect. Med. 2018, 8, a030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuster B., Junkin M., Kashaf S. S., et al. Automated Microfluidic Platform for Dynamic and Combinatorial Drug Screening of Tumor Organoids. Nat. Commun. 2020, 11, 5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ceder S., Eriksson S. E., Cheteh E. H., et al. A Thiol-Bound Drug Reservoir Enhances APR-246-Induced Mutant p53 Tumor Cell Death. EMBO Mol. Med. 2021, 13, e10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Behrenbruch C., Foroutan M., Lind P., et al. Targeting of TP53-Independent Cell Cycle Checkpoints Overcomes FOLFOX Resistance in Metastatic Colorectal Cancer. bioRxiv 2021, 2021.02.04.429849. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jbx-10.1177_24725552211020668 for High-Throughput Imaging Assay for Drug Screening of 3D Prostate Cancer Organoids by Nicholas Choo, Susanne Ramm, Jennii Luu, Jean M. Winter, Luke A. Selth, Amy R. Dwyer, Mark Frydenberg, Jeremy Grummet, Shahneen Sandhu, Theresa E. Hickey, Wayne D. Tilley, Renea A. Taylor, Gail P. Risbridger, Mitchell G. Lawrence and Kaylene J. Simpson in SLAS Discovery
Supplemental material, sj-pdf-2-jbx-10.1177_24725552211020668 for High-Throughput Imaging Assay for Drug Screening of 3D Prostate Cancer Organoids by Nicholas Choo, Susanne Ramm, Jennii Luu, Jean M. Winter, Luke A. Selth, Amy R. Dwyer, Mark Frydenberg, Jeremy Grummet, Shahneen Sandhu, Theresa E. Hickey, Wayne D. Tilley, Renea A. Taylor, Gail P. Risbridger, Mitchell G. Lawrence and Kaylene J. Simpson in SLAS Discovery