Abstract
Polycomb complexes have traditionally been prescribed roles as transcriptional repressors, though increasing evidence demonstrate they can also activate gene expression. However, the mechanisms underlying positive gene regulation mediated by Polycomb proteins are poorly understood. Here, we show that RING1B, a core component of Polycomb Repressive Complex 1, regulates enhancer–promoter interaction of the bona fide estrogen-activated GREB1 gene. Systematic characterization of RNA:DNA hybrid formation (R-loops), nascent transcription and RNA Pol II activity upon estrogen administration revealed a key role of RING1B in gene activation by regulating R-loop formation and RNA Pol II elongation. We also found that the estrogen receptor alpha (ERα) and RNA are both necessary for full RING1B recruitment to estrogen-activated genes. Notably, RING1B recruitment was mostly unaffected upon RNA Pol II depletion. Our findings delineate the functional interplay between RING1B, RNA and ERα to safeguard chromatin architecture perturbations required for estrogen-mediated gene regulation and highlight the crosstalk between steroid hormones and Polycomb proteins to regulate oncogenic programs.
INTRODUCTION
RING1B is a core component of Polycomb Repressive Complex 1 (PRC1) and mono-ubiquitinates histone H2A at lysine 119 (H2AK119ub1). Mechanistically, PRC1 halts RNA Polymerase II (RNA Pol II) elongation and modulates 3D chromatin architecture by compacting chromatin, leading to gene repression maintenance (1–3). Notably, PRC1 and RING1B can also be recruited to active enhancers and gene promoters (4). We recently showed that RING1B is a critical regulator of the estrogen receptor alpha (ERα) transcriptional regulatory circuit in ER+ breast cancer (5). However, the molecular mechanisms behind RING1B-mediated enhancer and gene activation remain a mystery. Indeed, the determinants underpinning the recruitment of Polycomb proteins to transcriptionally active genes and enhancers are largely unknown.
Estrogen (17-β estradiol, E2) binds to its receptor, estrogen receptor alpha (ERα) and regulates many biological processes such as bone growth, reproductive maturation and energy homeostasis (6). Moreover, liganded-ERα drives proliferation of ER+ breast cancer cells, which accounts for ∼80% of all breast cancer cases (7). Liganded-ERα activates transcription of E2-responsive genes primarily by binding to chromatin at sites containing consensus DNA sequences known as estrogen response elements (EREs) (8). At sites lacking EREs, ERα can indirectly bind chromatin by tethering to other transcription factors (TFs) (e.g. SP1, FOS and cJUN) (9). Pioneer TFs such as FOXA1, GATA3 and AP-2γ bind to their cognate DNA sequences in condensed chromatin and facilitate ERα chromatin binding (10). Liganded ERα recruits a large cohort of coactivators to coordinate changes to the local chromatin environment. These changes include histone modifications, enhancer activation and enhancer–promoter looping at active ERα target genes, ultimately leading to transcription via RNA Pol II (11). A plethora of studies characterized the coordinated physical and functional interactions between liganded ERα and its large cohort of coactivators. Detailed genome-wide examination of the E2 signaling pathway not only revealed multiple ERα-mediated mechanisms of gene regulation but also highlighted significant gaps in our mechanistic understanding of this key physiological and oncogenic pathway (12–14). Indeed, filling this gap in knowledge is crucial for developing the next generation of therapeutic approaches targeting ER+ breast cancer.
Here, we elucidated the role of RING1B in promoting ERα target gene expression and explored how RING1B is recruited to chromatin in response to estrogen. Circularized chromatin conformation capture followed by massively parallel sequencing (4C-seq) revealed RING1B as a key orchestrator of E2-induced enhancer–promoter looping. Additionally, our results show that RING1B is directly involved in the transcription of ERα target genes and that its depletion handicaps the formation of nascent transcripts as well as R-loops at key RING1B/ERα co-target genes. RNA-seq analysis coupled with genome-wide assessment of E2-induced RNA Pol II recruitment demonstrates that RING1B is a molecular sensor of E2 responsiveness. We also found that RING1B recruitment to chromatin is dependent on ERα and RNA. Interestingly, depletion of RNA Pol II did not abrogate the E2-induced chromatin recruitment of RING1B and ERα. Instead, binding of both was increased at promoters, demonstrating that despite the lack of transcription, increasing ERα recruitment was sufficient to increase binding of RING1B.
MATERIALS AND METHODS
Cell lines
T47D and MCF7 (ATCC HTB-133 and HTB-22) were maintained at 37°C with 5% CO2 and split every 3 days (T47D) or 4 days (MCF7) according to ATCC recommendations. Culture media were supplemented with 1× penicillin/streptomycin and 1× glutaMAX, and complete culture media for each cell line were as follows: T47D, RPMI 1640 with 10% FBS and insulin (10 μg/ml); MCF7, EMEM with 10% FBS and insulin (10 μg/ml). Prior to adding estrogen (10 nM, Sigma-Aldrich E2758), cells were maintained in media without phenol red and 10% charcoal:dextran stripped FBS (GeminiBio 100–119) for 72 h before treating with ethanol (vehicle) or E2. Cells were routinely tested to be free of mycoplasma infection.
Generation of stably transduced cells
HEK293T cells (ATCC CRL-3216) were maintained at 37°C with 5% CO2 in DMEM with 10% FBS supplemented with 1× penicillin/streptomycin and 1× glutaMAX. To produce shRNA lentiviruses, 2 × 106 HEK293T cells were plated on to a 10 cm2 plate for at least 8 h before transfection with calcium phosphate and 8 μg of pLKO-shRNA vectors (Control, Addgene 10879; RNF2, Sigma-Aldrich TRCN0000033697; ESR1 #1, Sigma-Aldrich TRCN0000010774; ESR1 #2, Sigma-Aldrich TRCN0000003301), 2 μg of pCMV-VSV-G and 6 μg of pCMV-dR8.91. The transfection media were removed following overnight incubation and replaced with 8.5 ml of HEK293T cell culture media for 48 h. The viral supernatant was then collected, passed through a 0.45 μM polyethersulfone filter and used to transduce T47D and MCF7 cells. Specifically, 5 × 106 T47D or MCF7 cells were plated on to a 15 cm2 plate. The next day, the media were removed, and the cells were incubated with a mixture of viral supernatant and cell culture media at a ratio of 50:50 in a total volume of 5 ml, with the addition of polybrene (1 μg/ml, Sigma-Aldrich TR-1003-G). After 16 h of incubation, viruses were removed and complete culture medium corresponding to each cell type was added for cell recovery. Cells were selected 24 h after recovery with puromycin (2 μg/ml, Biogems 5855822) and were maintained in selection. All experiments were performed within 2 weeks after transduction.
Western blotting
Cells were lysed in high-salt buffer (300 nM NaCl, 50 mM Tris-HCl pH 8, 10% glycerol, and 0.2% NP-40) supplemented with protease inhibitors (Sigma-Aldrich 04693132001) and sonicated 5 min at 4°C with a Bioruptor in 30 s on/off cycles. After centrifugation at 16 000 × g for 15 min, soluble material was quantified by Bradford assay (Bio-Rad 5000006). Western blotting was performed using standard protocols and imaged on an Odyssey CLx imaging system (Li-COR), and various exposures within the linear range were captured using Image Studio software. Images were rotated, resized and cropped using Adobe Photoshop CC 2019 and imported into Adobe Illustrator CC 2019 to be assembled into figures.
ATAC-seq library preparation and analysis
ATAC-seq experiments were performed as previously described (15). FASTQ data were processed with default parameters using the ATAC-seq/ENCODE pipeline from the Kundaje lab (https://github.com/kundajelab/atac_dnase_pipelines) and aligned to the hg19 genome. BigWig file output from the pipeline was visualized in the UCSC genome browser.
4C-seq library preparation
4C-seq was performed following Brouwer et al. (16). Specifically, 8–10 × 106 cells from each experimental condition were resuspended in 10 ml of culture media and 1 ml of 16% methanol-free formaldehyde (Thermo Fisher Scientific 28908) was added to the cell suspension for a final concentration of 1.45% to crosslink the cells at room temperature for 10 min. The crosslinking reaction was quenched with 575 μl of 2.5 M glycine (Sigma-Aldrich G8898). The cells were then centrifuged for 8 min at 340 × g at 4°C before being lysed 10–15 min on ice in 5 ml of prechilled lysis buffer (10 mM tris-HCl pH 8, 10 mM NaCl, 0.2% NP-40) supplemented with protease inhibitors (Sigma-Aldrich S8830). Nuclei from the lysed cells were pelleted by centrifuging at 650 × g at 4°C for 5 min, washed with 1× PBS, and aliquoted into 4–5 × 106 nuclei pellets before flash freezing and storing at -80°C. One aliquot of each experimental condition was resuspended in 0.5 ml of 1.2× buffer B (Thermo Fisher Scientific BB5) followed by incubation with 15 μl of 10% SDS then with 50 μl of 20% Triton X-100, each for 1 h at 37°C with gentle agitation at 900 rpm. A 10 μl aliquot was removed and stored at -20°C prior to chromatin digestion with 400U of CviQI (Thermo Fisher Scientific ER0211) overnight at 37°C and gentle agitation at 900 rpm. A 10 μl aliquot was removed prior to decrosslinking with 10 μl of proteinase K (NEB P8107S) and 10 mM tris-HCl pH 8 at 37°C for 1 h in a final volume of 100 μl. Digestion efficiency was determined by running 20 μl of the pre- and post-digestion samples on a 0.6% agarose gel. If a smear was observed, CviQI was heat inactivated by incubating at 65°C for 20 min. To finish generating the 3C library, the samples were transferred to a 50 ml tube for ligation overnight at 16°C with 6.125 ml of 1.15× T4 ligation buffer (NEB B0202S) and 100U of T4 DNA ligase (NEB M0202L). Next, samples were decrosslinked with 30 μl of proteinase K at 65°C for at least 4 h and treated with 30 μl of RNase A (10 mg/ml, Thermo Fisher Scientific AM9780) for 30 min at 37°C. After cooling the samples down to room temperature, 7 ml of phenol/chloroform/isoamyl alcohol (Thermo Fisher Scientific 15–593–049) was added and samples were shaken vigorously then centrifuged at 3200 × g for 15 min. The upper aqueous layer was transferred to a new 50 ml tube to which 7 ml of water, 1.5 ml of 3M sodium acetate (pH 5.4) and 35 ml of 100% ethanol were added. The samples were frozen at -80°C for 2 h then centrifuged at 3200 × g at 4°C for 45 min and washed with 10 ml of 70% ethanol. After air drying the pellets, they were resuspended in 150 μl of 10 mM Tris-HCl, pH 8. Ligation efficiency was determined by running 5 μl of 3C material on a 0.6% agarose gel, and a significant upward shift in the digested DNA should be observed. To generate the 4C library from the 3C material, the 3C library was digested with 50U of MboI (NEB R0147L) in a 500 μl reaction overnight. A 10 μl aliquot was removed from the reaction to determine digestion efficiency on a 1.5% agarose gel. If digestion was successful (the DNA appears as a smear of fragments <1000 bp), the reaction was heat inactivated for 20 min at 65°C. Next, samples were transferred to a 50 ml tube to which 1.4 ml of 10× T4 DNA ligase buffer, 200U of T4 DNA ligase, and water up to 14 ml were added before ligation at 16°C overnight. The samples were then precipitated with 14 μl of glycogen (Thermo Fisher Scientific R0561), 1.4 ml of 3M sodium acetate (pH 5.4), and 35 ml of 100% ethanol at -80°C for 2 h. The samples were pelleted at 3200 × g for 45 min and washed with 10 ml of 70% ethanol before air drying and resuspending in 150 μl of 10 mM Tris-HCl, pH 7.5. The DNA samples were purified using the QIAquick PCR Purification Kit (Qiagen 28106) following the manufacturer’s protocol, and the concentration was measured via nanodrop. To amplify the DNA template and generate the 4C library, 3 equivalent PCR reactions with 200 ng of input 4C material were ran simultaneously using the Expand Long Template PCR System (Sigma-Aldrich 11681842001) and 5 μl from each reaction was ran on a 1.5% agarose gel to verify the success of the reaction. All successful reactions were combined and purified using the High Pure PCR Product Purification Kit (Sigma-Aldrich 11732676001) and analyzed on an Agilent Tapestation using the D5000 ScreenTape, reagents, and ladder (Agilent 5067–5588, 5067–5589, 5067–5590, respectively). The 4C libraries were sequenced (75 bp, single-end) on an Illumina NovaSeq 6000 instrument for a total of 1 million reads per sample.
RT-qPCR and nascent transcript RT-qPCR
RNA was extracted from cell pellets with Trizol (Thermo Fisher Scientific 15–596–018) following the manufacturer’s protocol and the concentration measured using nanodrop. The qScript cDNA Synthesis Kit (VWR 101414–098) was used to convert 1 μg of RNA to cDNA for all mature transcript RT-qPCRs experiments performed. For nascent transcript RT-qPCR experiments, mock cDNA was synthesized in the same manner but without the addition of reverse transcriptase to ensure identify any genomic DNA contamination. To perform the qPCR, the cDNA from 1 μg of RNA was first diluted 1:10. A reaction master mix was then prepared consisting of 5 μl of SYBR Green Supermix (Bio-Rad 1725124), 2 μl water and 1 μl of primer mix (10 μM each of forward and reverse primer) per sample. Total volume of mastermix was calculated for two technical replicates per sample per gene queried. The RT-qPCR reaction was carried out in a 96-well plate (Bio-Rad MLL9601) on a Bio-rad CFX96 using 8 μl of mastermix and 2 μl of diluted cDNA. The thermocycling protocol adapted from Bio-rad is as follows: 1 cycle for 2 min at 95°C, 40 cycles of 15 s at 95°C followed by 30 s at 60°C, finishing with a melt curve cycle where the temperature is increased from 55°C to 95°C in 0.5°C increments for 30 s. Relative enrichment of each queried gene was measured against the RPLP0 gene.
DRIP-qPCR and DRIP-Seq library preparation
DNA:RNA immunoprecipitation was performed following Sanz et al. (17). Specifically, 5 × 106 cells per experimental condition were harvested and washed with 1× PBS. The cells were resuspended in 1.55 ml of TE buffer with 100 μl of 10% SDS and 5 μl of 20 mg/ml proteinase K (NEB P8107S) were added before incubation overnight at 37°C. DNA was extracted with 1.6 ml of phenol/chloroform/isoamyl alcohol (Thermo Fisher Scientific 15–593–049) in a 15 ml high-density Maxtract phase-lock gel tube (Qiagen 129065), centrifuged at 1500 × g for 10 min and precipitated with 4 ml of 100% ethanol and 160 μl of 3M sodium acetate (pH 5.4). The DNA was washed 3× with 80% ethanol, air dried and resuspended in 125 μl of TE buffer. Next, 100 μl of DNA was digested overnight at 37°C using an enzyme cocktail consisting of 30U each of EcoRI-HF (NEB R3101L), XbaI (NEB R0145L), BsrGI-HF (NEB R3575L), HindIII (NEB R0104L) and SspI (NEB R0132L) along with 1.5 μl of BSA (NEB B9000S), 15 μl of 10× Cutsmart buffer (NEB B7204S), 1.5 μl of spermidine (Sigma-Aldrich 05292) and water up to 150 μl. The DNA mixture should be drastically reduced in viscosity. To the digested DNA, 100 μl of water and 250 μl of phenol/chloroform/isoamyl alcohol were added and the mixture transferred to 2 ml phase-lock gel light tubes (VWR 10847–800) and centrifuged at 16 000 × g for 10 min followed by incubation for 1 h at -20°C with 1.5 μl of glycogen (Thermo Fisher Scientific R0561), 25 μl of 3M sodium acetate (pH 5.4) and 625 μl of 100% ethanol. DNA was precipitated by centrifuging at 16 000 × g for 35 min at 4°C and washed with ice cold 80% ethanol before air drying and resuspending in 50 μl of TE buffer. DNA concentration was measured using a nanodrop and 10 μg was digested with 4 μl of RNase H (NEB M0297L), 20 μl of RNase H buffer and water up to 200 μl for 4 h at 37°C to produce the RNase H treated negative control. For each sample, 8 μg of DNA was diluted in 500 μl of TE buffer and 50 μl was aliquoted at -20°C as input. R-loops were immunoprecipitated using 10 μl of S9.6 antibody (Sigma-Aldrich MABE1095) and 51 μl of 10× DRIP binding buffer (100 mM sodium phosphate, pH 7; 1.4M NaCl; 0.5% Triton X-100) in a 500 μl reaction for 14–16 h at 4°C. Protein G Dynabeads (Thermo Fisher Scientific 10004D) were washed 2× with 700 μl of 1× DRIP binding buffer and the immunoprecipitation reaction was added to 50 μl of beads for 2 h at 4°C to pull down the DNA. The beads were separated from the solution on a magnetic rack and washed 2× with 700 μl of 1× DRIP binding buffer. DNA was eluted from the beads with 300 μl of DRIP elution buffer (50 mM Tris-HCl, pH 8.0; 10 mM EDTA, pH 8.0; 0.5% SDS) and 7 μl of proteinase K was added to the samples, inputs, and negative controls then incubated at 55°C for 45 min. After incubation, the eluted DNA was extracted using a phase-lock gel tube with 300 μl of phenol/chloroform/isoamylalcohol centrifuged at 16 000 × g for 10 min. The aqueous layer was transferred to a new 1.5 ml tube and the DNA was precipitated with 1.8 μl of glycogen, 30 μl of 3 M sodium acetate (pH 5.4), and 750 μl of 100% ethanol. The precipitated DNA was washed 1× with 80% ethanol, air dried and resuspended in 50 μl of nuclease-free water. To perform DRIP-qPCR, the precipitated DNA, the RNase H-treated negative control, and input for each sample were diluted 1:4, followed by RT-qPCR as described. For DRIP-seq, the DNA samples, negative controls, and inputs were diluted to a total volume of 100 μl in TE buffer and transferred into Diagenode Bioruptor NGS 0.65 ml microtubes (Diagenode C30010011) then sonicated on a Bioruptor Pico Sonication Device (Diagenode B01060010) for 19 total 30 s on/off cycles (with a 10 min break after the first 10 cycles) to generate fragment sizes of around 250 bp. Sonication efficiency was verified on an Agilent Tapestation using High Sensitivity D1000 screentapes and reagents (Agilent Technologies 5067–5584 and 5067–5585, respectively). DRIP-seq libraries were prepared with at least 2 ng of DNA starting material using the NEBNext Ultra DNA library prep kit for Illumina (NEB E7370). Libraries were purified using Agencourt AMPure XP beads (Beckman Coulter A63881) and library fragment sizes were measured again then sequenced on an Illumina Novaseq 6000 instrument.
ChIP/rChIP and ChIP/rChIP-seq library preparation
ChIP experiments were performed as previously described (5). rChIP was performed by incorporating the methodology from Long et al. into our ChIP protocol (18). Specifically, 2 μg of RNase A (10mg/ml, Thermo Fisher Scientific AM9780) was added to the sample during the immunoprecipitation step. The immunoprecipitated DNA was used to either perform ChIP-qPCR or generate libraries using the NEBNext Ultra DNA library prep kit for Illumina (NEB E7370) as previously described then sequenced (75 bp, single-end, on an Illumina NovaSeq 6000 instrument.
LC-MS/MS
RING1B and IgG pull-downs were performed as a ChIP assay. Before mass spectrometry analysis the proteins were alkylated with 20 mM iodoacetamide in the dark for 30 min. Afterward, phosphoric acid was added to the sample at a final concentration of 1.2%. Samples were diluted in six volumes of binding buffer (90% methanol and 10 mM ammonium bicarbonate, pH 8.0). After gentle mixing, the protein solution was loaded to an S-trap filter (Protifi) and spun at 500 g for 30 s. The sample was washed twice with binding buffer. Finally, 1 μg of sequencing grade trypsin (Promega), diluted in 50 mM ammonium bicarbonate, was added into the S-trap filter and samples were digested at 37°C for 18 h. Peptides were eluted in three steps: (i) 40 μl of 50 mM ammonium bicarbonate, (ii) 40 μl of 0.1% TFA and (iii) 40 μl of 60% acetonitrile and 0.1% TFA. The peptide solution was pooled, spun at 1000 g for 30 s and dried in a vacuum centrifuge. Prior to mass spectrometry analysis, samples were desalted using a 96-well plate filter (Orochem) packed with 1 mg of Oasis HLB C-18 resin (Waters). Briefly, the samples were resuspended in 100 μl of 0.1% TFA and loaded onto the HLB resin, which was previously equilibrated using 100 μl of the same buffer. After washing with 100 μl of 0.1% TFA, the samples were eluted with a buffer containing 70 μl of 60% acetonitrile and 0.1% TFA and then dried in a vacuum centrifuge. Samples were then resuspended in 10 μl of 0.1% TFA and loaded onto a Dionex RSLC Ultimate 300 (Thermo Scientific), coupled online with an Orbitrap Fusion Lumos (Thermo Scientific). Chromatographic separation was performed with a two-column system, consisting of a C-18 trap cartridge (300 μm ID, 5 mm length) and a picofrit analytical column (75 μm ID, 25 cm length) packed in-house with reversed-phase Repro-Sil Pur C18-AQ 3 μm resin. Peptides were separated using a 120 min gradient from 4–30% buffer B (buffer A: 0.1% formic acid, buffer B: 80% acetonitrile + 0.1% formic acid) at a flow rate of 300 nl/min. The mass spectrometer was set to acquire spectra in a data-dependent acquisition (DDA) mode. Briefly, the full MS scan was set to 300–1200 m/z in the orbitrap with a resolution of 120 000 (at 200 m/z) and an AGC target of 5 × 105. MS/MS was performed in the ion trap using the top speed mode (2 s), an AGC target of 10e4 and an HCD collision energy of 35. Raw files were searched using Proteome Discoverer software (v2.4, Thermo Scientific) using SEQUEST search engine and the SwissProt human database (updated February 2020). The search for total proteome included variable modification of N-terminal acetylation, and fixed modification of carbamidomethyl cysteine. Trypsin was specified as the digestive enzyme with up to two missed cleavages allowed. Mass tolerance was set to 10 pm for precursor ions and 0.2 Da for product ions. Peptide and protein false discovery rate was set to 1%. Each analysis was performed with three biological replicates. Prior statistics, proteins were log2 transformed, normalized by the average value of each sample and missing values were imputed using a normal distribution two standard deviations lower than the mean. Statistical regulation was assessed using homoscedastic one-tail t-test (if P-value < 0.05). Data distribution was assumed to be normal but this was not formally tested.
DRIP/ChIP/rChIP-seq analysis
FASTQ data were processed using the ENCODE-DCC/chip-seq-pipeline2 pipeline from the Kundaje lab (https://github.com/ENCODE-DCC/chip-seq-pipeline2) with default parameters and aligned to the hg19 genome. Peaks with signal (fold enrichment over input generated from MACS2) > 4 and a q-value < 0.05 were used to downstream analysis. Homer annotatePeaks was used for peak annotation and gene assignment while findMotifsGenome was used for motif analysis. Bedtools v2.29.0 was used to determine peak overlaps and NGS Plot v2.63 was used to generate density plots and heatmaps. BigWig file output from the pipeline was visualized in the UCSC genome browser.
RNA-seq analysis
FASTQ data were processed with the LSF RNAseq Pipeline (https://github.com/diderote/LSF-RNAseq). Briefly, cutadapt v2.3 (–nextseq-trim = 20 -m 18) was used to remove low-quality reads. Expected gene counts were obtained using RSEM v1.3.0 and STAR v2.6.1a was used to align the reads to the human hg19 transcriptome (GENCODE V19 annotation). RUVseq v1.12.0 was used to adjust gene counts by removing unwanted variance using exogenous ERCC spike-in RNA. Differential expression was determined using DESeq2 v1.18.1 and R (version 3.4.1) with a q-value < 0.05 and an FC > 1.5 (Wald test). Heat maps were generated using variance stabilized gene counts from DESeq2. For GSEAs, the Wald statistic of each time point compared to hormone-deprived conditions was used as input for the Preranked module of GSEA v3.0 on Hallmark gene sets (-scoring_scheme weighted –nrom meandiv). Analysis of Figure 2C: The RNA-seq data were analyzed by the DESeq2 differential gene expression analysis pipeline, which normalized the transcript counts by median of ratios. Normalization via median of ratios considers sequencing depth as well as RNA composition, thereby optimizing comparisons between samples and for differential gene expression analyses. The dark blue and the white colors used for the color code in Figure 2C are scaled to the highest and the lowest normalized counts detected for each gene within all ten samples, respectively. For example, the highest level of FMN1 expression was detected in the 24 h WT rescue sample at normalized count of 6309.075 while the lowest was detected in the HD shRING1B sample at normalized count of 1077.12. Therefore, the dark blue is scaled to represent 6309.075 while the white is scaled to represent 1077.12.
Figure 2.
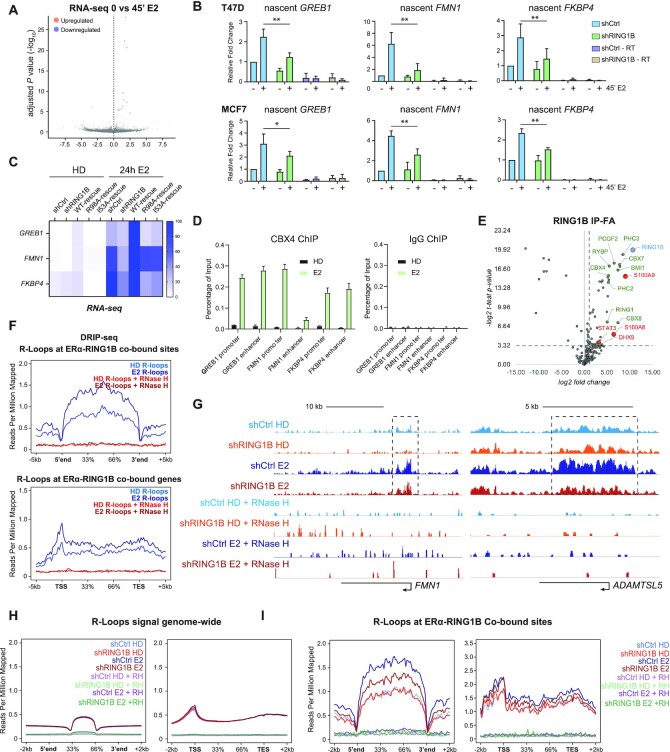
RING1B promotes R-loop formation at ERα target genes via direct participation in their transcription. (A) RNA-seq volcano plot depicting one significantly downregulated and 11 significantly upregulated genes (P-value < 0.05, FC > 1.5) in parental T47D cells before and after 4′ of E2 stimulation; n = 2. (B) RT-qPCR measuring levels of GREB1, FMN1 and FKBP4 nascent transcripts in T47D and MCF7 shCTR and shRING1B cells before and after 45′ of E2 stimulation. shCTR – RT and shRING1B – RT are RNA samples subjected to the cDNA conversion process in the absence of reverse transcriptase. Enrichment detected in these samples represent genomic DNA contamination. Error bars represent the standard deviation of three biological replicates. *P-value < 0.05, **P-value < 0.01, one-tailed paired t-test. (C) RNA-seq heatmap depicting expression of GREB1, FMN1, and FKBP4 in T47D shCTR, shRING1B, as well as shRING1B cells rescued with wildtype (WT), nucleosome-binding mutant (R98A) and a catalytic-dead mutant (I53A) RING1B; n = 2. (D) CBX4 and IgG, as a negative control, ChIP-qPCR of RING1B/ERα co-bound sites before and after 45 min of E2 administration; n = 2. (E) Endogenous RING1B immunoprecipitation with nuclear extracts after crosslinking with 1% FA for 10 min. Proteins bound to RING1B were identified by LC-MS/MS, and enrichment was calculated based on fold change over IgG enrichment and P-value < 0.05. IgG was used as a negative control. Experiments were performed in three biological replicates. Proteins labeled in green are PRC1 subunits. Proteins labeled in red are new RING1B-associated proteins. (F) Average DRIP-seq signals in parental T47D cells before and after 45′ of E2 stimulation at ERα and RING1B co-bound sites (top) and genes (bottom). No signal was detected in samples treated with RNase H, indicating that the signal observed was specific for R-loops. (G) T47D shCTR and shRING1B DRIP-seq signal before and after 45 min of E2 at the FMN1 and the ADAMTSL5 genes. (H) Average genome-wide DRIP-seq signal (left) and with respect to genes that contain R-loops (right). (I) Average DRIP-seq signal at all RING1B and ERα co-bound sites (left) and genes (right) that contain R-loops.
4C-seq analysis
FASTQ files were trimmed to a fixed length of 36 bps. The trimmed reads were analyzed using the 4Cseqpipe pipeline (http://compgenomics.weizmann.ac.il/tanay/?page_id=367) along default parameters to generate domainograms (19). The first step in the pipeline converted the trimmed reads in fastq files to raw files for extracting the appropriate sequences. The second step mapped the sequences from the step 1 to the fragmented hg19 genome. The last step generated the domainograms around the viewpoint of GREB1 promoter, centered around the following genomic coordinate: chr2:11,679,794–11,680,120.
Statistical analysis
Significance was determined by Student’s t test (one tailed, paired) and paired Mann–Whitney U test. Error bars in figures represent SD of at least two independent experiments. The ggbloxplot function of the ggpubr package (https://cran.r-project.org/web/packages/ggpubr/index.html) was used to generate boxplots for data visualization. All other bar graphs were plotted using Graphpad Prism Version 8.
RESULTS
RING1B coordinates estrogen-induced enhancer-promoter interactions
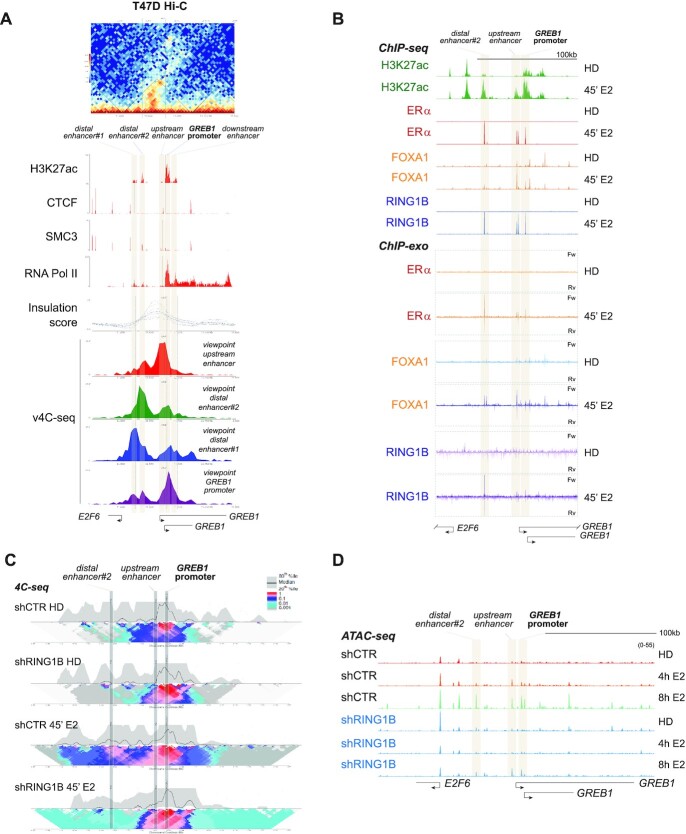
In pluripotent stem cells, PRC1 compacts chromatin which regulates distal and local chromatin architecture to maintain repression of developmental genes (3). However, in ER+ breast cancer cells, RING1B promotes E2-induced gene expression (5). It is unknown whether RING1B regulates dynamic chromatin architecture perturbations to activate gene transcription upon E2 stimulation. We recently showed that RING1B and ERα are co-recruited to enhancers and promoters following E2 stimulation. Therefore, we assessed whether RING1B was important for enhancer–promoter interactions of genes activated in this context. As a proof of principle, we investigated GREB1, a well-characterized ERα and RING1B target gene that is rapidly induced by E2. To characterize the 3D chromatin architecture around the GREB1 locus, we conducted a virtual 4C-seq (v4C-seq) using data from Hi-C experiments performed on T47D cells cultured in media containing physiological levels of E2 (Figure 1A, top) (20). Hi-C is often used to determine genome-wide chromatin organization while v4C-seq identifies potential interactions between one locus and regions elsewhere in the genome. Within the CTCF and SMC3 (cohesin complex) marked boundary of the topologically-associated domain (TAD) encompassing the GREB1 locus, we observed five putative regulatory regions based on H3K27ac ChIP-seq signal (Figure 1A, panel labeled ‘H3K27ac’). Centering the viewpoint around the GREB1 promoter (purple v4C-seq track), we detected potential interactions between the promoter and two distal enhancers (distal enhancer #1 and #2), an upstream enhancer and a downstream enhancer of the promoter (Figure 1A). To further strengthen this observation, we also used as a viewpoint the two distal enhancers (green and blue v4C-seq tracks) and the upstream enhancer (red v4C-seq track). Notably, all three potential regulatory sites likely also interact with the GREB1 promoter (Figure 1A).
Figure 1.
RING1B coordinates estrogen-induced 3D chromatin architectural changes. (A) Top: Hi-C heatmap of 3D chromatin interactions around the GREB1 promoter showing the formation of a distinct TAD. Middle: CTCF and SMC3 ChIP-seq tracks define the boundary of the TAD containing five sites enriched for H3K27ac. The TAD is further demarcated by the insulation score. RNA Pol II track demarcates the GREB1 TSS. Bottom: v4C-seq centered (i) around the upstream enhancer (red track), (ii) distal enhancer#2 (green track), (iii) distal enhancer#1 (blue track) and (iv) the GREB1 promoter (purple track) show extensive intra-TAD interactions around the GREB1 promoter. (B) H3K27ac, ERα, FOXA1 and RING1B ChIP-seq tracks as well as the ChIP-exo tracks of ERα, FOXA1, and RING1B before and after 45 min of E2 stimulation depicting colocalization of the second distal enhancer (site 2), the proximal enhancer (site 3) and the GREB1 promoter (site 4) with ERα, RING1B and FOXA1. (C) 4C-seq in T47D shCTR and shRING1B cells before (HD) and after 45 min of E2 stimulation, centering the viewpoint around the GREB1 promoter. The median and 20th and 80th percentiles of sliding 5-kb windows determine the main trend line. Color scale represents enrichment relative to the maximum attainable 12-kb median value. (D) T47D shCTR and shRING1B ATAC-seq tracks after 4 and 8h of E2 stimulation, n = 2.
Using our ChIP-seq and ChIP-exo data, we found that E2 induced the recruitment of RING1B and ERα to the second distal enhancer (site 2), the proximal enhancer (site 3), and the GREB1 promoter (site 4) (Figure 1B). To confirm these interactions and to determine the role of RING1B in enhancer–promoter interactions, we applied high-resolution 4C-seq to the GREB1 promoter. We used its promoter as a viewpoint for the assay. 4C-seq was performed in control (shCTR) and RING1B-depleted (shRING1B) cells that were hormone deprived (HD) and stimulated with E2 for 45 min (Figure 1C). E2 stimulation increased the interaction (shown as contact intensity, see figure caption) between the GREB1 promoter and the distal enhancer#2 in shCTR cells; however, RING1B depletion prevented these interactions (Figure 1C and Supplementary Figure S1A). Importantly, although 45 min of E2 induced enhancer–promoter interactions concomitant with RING1B and ERα co-recruitment to both the promoter and enhancers, the chromatin accessibility of these sites remained unaffected until after 4 h of E2 treatment (Figure 1D and Supplementary Figure S1B). Notably upon RING1B depletion, these sites remain inaccessible until after 8 h of E2 stimulation, at which point a modest increase in chromatin accessibility was observed (Figure 1D). Altogether, these results show that RING1B plays a major role in mediating E2-induced 3D chromatin architecture and accessibility at the GREB1 locus in ER+ breast cancer cells.
RING1B promotes R-loop formation at ERα target genes via direct participation in their transcription
We previously demonstrated that RING1B is crucial for global E2-induced expression of ERα target genes. However, it was unclear whether RING1B is directly involved in the transcription of ERα target genes. RNA-seq of T47D cells before and after 45 min of E2 stimulation revealed upregulation of only 11 genes (Figure 2A). Upregulation of bona fide E2-induced genes such as GREB1, FMN1, FKPB4 and TFF1 was not detected by RT-qPCR of total RNA (Supplementary Figure S2A) even though ERα, RING1B and RNA Pol II were recruited to their promoters as early as 20 min after E2 stimulation (Supplementary Figure S2B). These results suggest that although mature RNA was not detected, 45 min of estrogen stimulation initiates the transcription of ERα target genes. Therefore, we performed RT-qPCR using primer pairs spanning intron–exon junctions to determine the levels of nascent transcripts of the RING1B/ERα co-target genes, GREB1, FMN1 and FKBP4, before and after 45 min of E2. In both T47D and MCF7 ER+ breast cancer cell lines, RING1B depletion attenuated E2-induced transcriptional increase (Figure 2B and Supplementary Figure S2C) indicating that RING1B is directly involved in the transcription of ERα target genes in response to E2 stimulation. Importantly, expression of these genes was rescued upon ectopic expression of wild type (WT) RING1B. Expression of catalytically dead (RING1BI53A) and nucleosome-binding deficient (RING1BR98A) mutants in RING1B depleted cells only restored expression of FMN1 (Figure 2C). Specifically, within the 109 RING1B/ERα co-targeted genes at 45 min of E2 that are upregulated after 24 h of E2, 44 and 30 genes were upregulated in rescued cells expressing RING1BI53A and RING1BR98A, respectively. These results indicate that both the catalytic and the nucleosome-binding abilities of RING1B are important for proper E2-induced transcription of many ERα target genes such as GREB1 and FKBP4. To test if RING1B deposited mono-ubiquitination of histone H2A on lysine 119 (H2AK119ub1) at these sites, we performed H2AK119ub1 ChIP-qPCR before and after 45 min of E2 administration. We did not detect significant H2AK119ub1 enrichment either before or after E2 stimulation (Supplementary Figure S2D). Therefore, we conclude that RING1B may mono-ubiquitinate non-histone substrates.
We next asked whether RING1B was recruited to E2-responsive genes in a PRC1 dependent or independent fashion. We recently showed estrogen stimulation induced a strong recruitment of a canonical PRC1 (cPRC1) complex containing CBX4 and PCGF2 only after prolonged (24 h) estrogen administration. Because residency of PRC1 complexes is highly dynamic and only a small fraction (∼20%) of complexes are stably interacting with chromatin (21), we performed CBX4 ChIP assays from double crosslinked chromatin. We reasoned that double crosslinking would increase the likelihood of capturing CBX4 binding at RING1B/ERα co-sites upon 45 min of E2 administration. Results in Figure 2D show CBX4 occupancy was strongly detected under this experimental condition, indicating that a cPRC1 complex is recruited to E2-induced genes. To further strength this observation, we performed co-immunoprecipitations of endogenous RING1B-associated protein complexes from crosslinked nuclear extracts followed by Liquid Chromatography with tandem mass spectrometry (LC-MS/MS) (Figure 2E). Notably, after 45 min of E2, RING1B mainly associates with cPRC1 subunits (CBX4/7/8, PCGF2/4, PHC2/3) and only with one non-canonical PRC1 (ncPRC1) subunit (RYBP). Moreover, we also found association of RING1B with novel partners such as STAT3, S100A8/A9 and DHX9. These new RING1B-associated proteins functionally link cPRC1 to the estrogen pathway as the transcription factor STAT3 is recruited to ERα sites (22), and the calcium-binding proteins S100A8/A9 are transcriptional activators recruited to STAT3 and AP-1 binding sites in breast cancer cells (23). Interestingly, the RNase helicase DHX9 has been implicated in many cellular processes including DNA replication, transcription and genome stability, and more recently in R-loop formation (24).
We then asked whether RING1B was involved in R-loop formation. Gene transcription is highly regulated in many cellular contexts by R-loops (25). R-loops are DNA:RNA hybrid structures that form when the nascent transcript anneals with the template DNA strand, leaving the complementary strand exposed. R-loops formed directly downstream of promoters can promote gene expression by enhancing the deposition of active histone marks, while those formed at terminator sites facilitate RNA Pol II pause and release (26). Nevertheless, transcription is necessary for formation of R-loops that mediate higher-order chromatin architecture and are docking sites for chromatin-associated machinery such as Polycomb complexes (27). Recently, it has been shown that E2-induced R-loops in ER+ breast cancer cells exposed to E2 for 24 h (28). Whether acute administration of E2 induces R-loop formation and whether Polycomb proteins play a role in R-loop formation and resolution are not known. To address these questions, we performed DNA–RNA immunoprecipitation followed by massive parallel sequencing (DRIP-seq) using the R-loop-specific S9.6 antibody in parental T47D cells before and after 45 min of E2 stimulation (17). Notably, we found an increase in R-loop formation at ERα and RING1B co-bound sites as well as within the bodies of ERα and RING1B co-regulated genes. Importantly, RNAse H treatment eliminated the R-loop signal (Figure 2F). Moreover, we stably expressed a doxycycline (dox)-inducible Flag-tagged RNase H and observed a dramatic dose- and time-dependent R-loops resolution upon RNase H expression (Supplementary Figure S2E and F), further confirming that R-loops are formed after 45 min of E2 administration. We then performed DRIP-qPCR in shCTR and shRING1B T47D and MCF7 cells before and after 45 min of E2 stimulation at previously characterized ERα and RING1B co-bound genes. Notably, RING1B depletion attenuated the E2-induced formation of R-loops at these sites while having no impact on R-loop formation at RPL13A, a gene not targeted by ERα nor RING1B (28). Additionally, in agreement with a previous study, no R-loops were detected at a previously characterized negative control region ‘83/84’ (Supplementary Figure S2G) (28). RING1B depletion impaired E2-induced R-loop accumulation at the FMN1 and ADAMTSL5 loci, both of which are co-targeted by ERα and RING1B (Figure 2G). Notably, global levels of R-loops remained unaffected by E2 stimulation and RING1B depletion (Figure 2H). However, at sites and genes co-bound by both ERα and RING1B, E2-induced R-loop formation was abrogated by RING1B depletion (Figure 2I), while R-loop formation was not affected at genes regulated only by ERα but not RING1B (Supplementary Figure S2H). Altogether, our data indicate that RING1B affects E2-induced expression of ERα target genes and R-loop accumulation, specifically at RING1B/ERα co-targets.
RING1B genomic localization predicts whether ERα target genes are transcriptionally responsive to E2
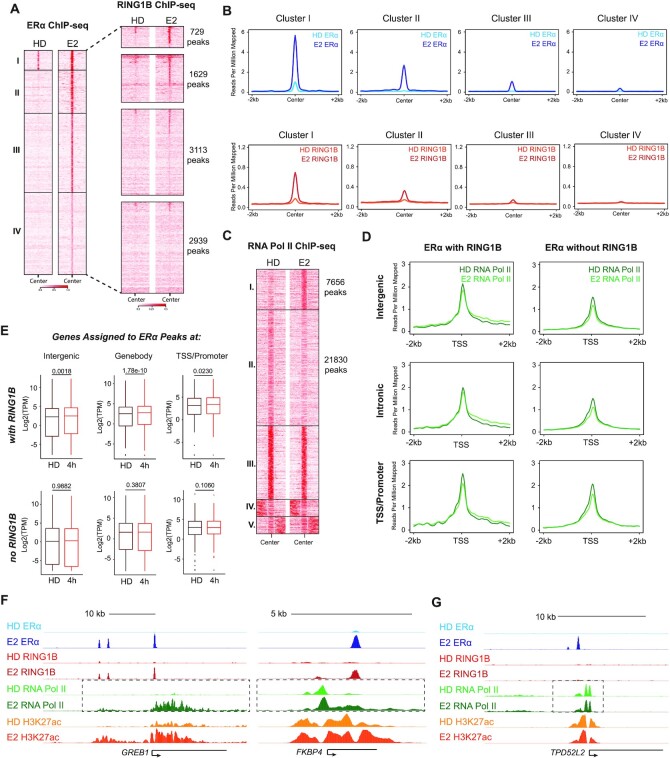
Given that RING1B directly participates in the E2-induced transcription of ERα target genes, we next sought to examine the behavior of RNA Pol II at these genes in response to E2 stimulation. We performed ChIP-seq of ERα and RING1B in parental T47D cells with two biological replicates before and after 45 min of E2, then performed a clustering analysis with combined replicates (Figure 3A and Supplementary Figure S3A,B). ERα recruitment was divided into four clusters with RING1B binding overlapping with sites from clusters I and II (Figure 3A). The strongest ERα and RING1B co-bound sites were found in cluster I followed by cluster II (Figure 3B), reinforcing the notion that RING1B is a key regulator of ERα-mediated gene regulation. Of note, of all the R-loops detected genome-wide, 249 sites contained both RING1B and ERα while 1,100 sites contained only ERα. About 92% of the R-loop sites containing both RING1B and ERα were found at cluster 1 ERα peaks, while only 41% of R-loop sites containing only ERα were found at cluster 1 ERα peaks.
Figure 3.
RING1B genomic localization predicts whether ERα target genes are transcriptionally responsive to E2. (A) ChIP-seq heatmap depicting genome wide ERα bound chromatin sites centered on peak summits before and after 45 min of E2 in parental T47D cells clustered by ERα occupancy (left). Heatmap of RING1B chromatin binding before and after 45 min of E2 organized by ERα clusters (right); n = 2. (B) Average signal of ERα (top) and RING1B (bottom) at sites within each of the 4 clusters before and after 45 min of E2. (C) ChIP-seq heatmap depicting genome-wide RNA Pol II chromatin binding before and after 45 min of E2, clustered by the levels of RNA Pol II occupancy. (D) Average RNA Pol II signal before and after 45 min of E2 with respect to the TSS of genes regulated by both ERα and RING1B (left) or by ERα alone (right) at intergenic regions (top), sites within a gene body (middle), or at promoter/TSS regions (bottom). (E) TPM values of genes regulated by both ERα and RING1B (top) or by ERα alone (bottom), before and after 4 h of E2 stimulation, from an intergenic site (left), within the gene body (middle), or at a promoter/TSS (right). (F and G) ERα, RING1B, RNA Pol II and H3K27ac signal before and after 45 min of E2 at the RING1B and ERα co-regulated genes GREB1 and FKBP4 (F) and at TPD52L2 (G), which is regulated by ERα alone.
Next, we performed ChIP-seq of total RNA Pol II in T47D cells before and after 45 min of E2 to determine its E2-induced occupancy dynamics with respect to ERα and RING1B. As expected, RNA Pol II binding accumulated (cluster I), decreased (cluster II) and remained unaffected by E2 stimulation at thousands of sites (Figure 3C). Notably, we only detected accumulation of RNA Pol II binding within the gene bodies at RING1B and ERα co-target genes (cluster I) (Figure 3D). Regardless of whether ERα was recruited to the intergenic regions, gene bodies (introns/exons) or promoter/TSS regions of genes, E2 induced higher levels of RNA Pol II occupancy within the body of the genes only if they were co-occupied by RING1B, indicating that the localization of RING1B to an ERα target gene is predictive of its transcriptional activity after 45 min of E2 stimulation (Figure 3D). Importantly, E2-induced recruitment of RNA Pol II was abolished in RING1B depleted cells (Supplementary Figure S3C). Genomic annotation of cluster I and II RNA Pol II sites, which exhibited changes in RNA Pol II binding upon E2 treatment, showed that these sites were primarily located within the promoters and gene bodies, with some located at intergenic regions (Supplementary Figure S3D). TF motif and gene ontology (GO) analyses (29) revealed, as expected, that cluster I genes were significantly enriched for EREs and the early estrogen response pathway, respectively (Supplementary Figure S3E,F), while cluster 2 genes did not reveal any significant enrichments (data not shown). Furthermore, E2 treatment induced significant upregulation of genes bound by both ERα and RING1B detectable as early as at 4 h while genes bound by ERα alone were not significantly upregulated (Figure 3E). Specifically, RNA Pol II elongation was observed at GREB1 and FKBP4 (RING1B/ERα co-targets) but not at genes lacking RING1B recruitment (TPD52L2 and LPAR1) (Figure 3F,G and Supplementary Figure S3G). Notably, 24 h of E2 administration induced activation (Fold change > 2, q-value < 0.05) of 835 genes in control cells, while only 224 of these genes was upregulated in RING1B depleted cells (Supplementary Figure S3H). Our results indicate that RING1B occupancy is a molecular sensor of E2 induced ERα target gene expression, further affirming RING1B as a key factor directly involved in the transcription of ERα target genes.
RING1B recruitment to chromatin is dependent on ERα and chromatin-associated RNA
Elucidating the molecular determinants targeting RING1B and Polycomb proteins to specific genomic locations is crucial to better understanding of how epigenetic factors determine cell fate, homeostasis and cellular response to stimuli. We previously found that after 45 min of E2 induction, RING1B is recruited to chromatin in a PRC1-independent fashion and it is required for full ERα recruitment to specific loci (5). Given that RING1B and ERα exhibit such extensive functional interaction and genomic co-localization, we reasoned that RING1B recruitment to chromatin may, at least partially, be dependent on ERα.
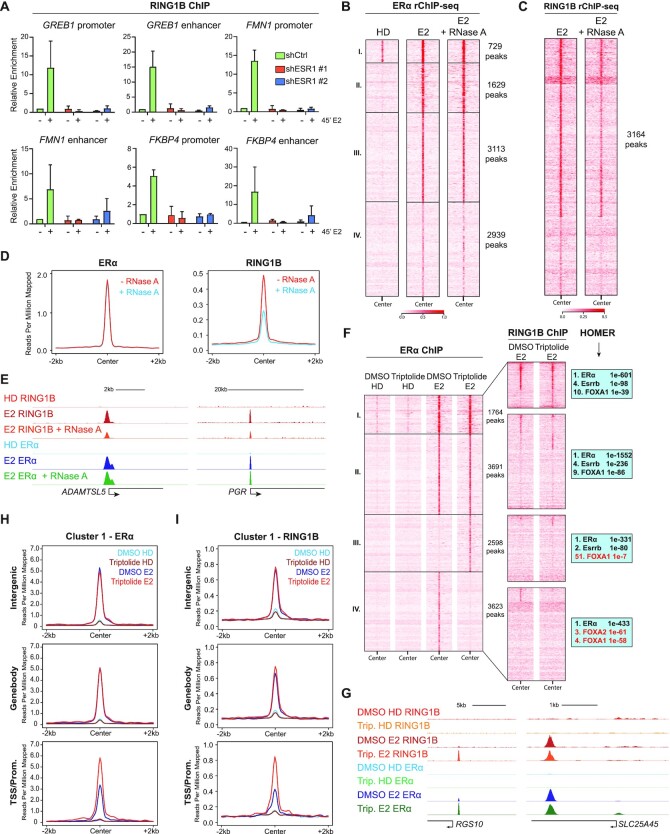
As expected, upon ERα depletion, E2 stimulation did not induce ERα-target gene expression (Supplementary Figure S4A,B). We then performed RING1B and ERα ChIP-qPCR at RING1B/ERα co-targets and found that upon ERα depletion, RING1B recruitment was abolished (Figure 4A and Supplementary Figure S4C). Our previous ChIP-exo studies revealed that RING1B is recruited to sites within 10 bp of EREs rather than to a discrete cognate sequence. Therefore, we assessed whether RING1B recruitment was also dependent on other factors that are not TFs such as single-stranded RNA molecules (mRNA and non-coding RNAs). Interestingly, a previous study showed that depletion of enhancer RNA (eRNAs) abrogated recruitment of ERα co-factors to enhancers (30). To this end, we performed two replicates of ERα and RING1B ChIP-seq with RNase A treatment (rChIP) (18) in which RNase A is added during the immunoprecipitation step to resolve protein-chromatin interactions that are dependent on chromatin-associated RNA. We found that treatment with RNase A did not affect the global stability of ERα chromatin binding (Figure 4B and Supplementary Figure S4D). However, RNase A treatment decreased the stability of RING1B at chromatin at all RING1B-bound sites ∼50% genome-wide (Figure 4C–E and Supplementary Figure S4D). These results were further confirmed by rChIP-qPCR (Supplementary Figure S4E,F). We then tested if R-loops were a determinant for RING1B stabilization at chromatin. To test this, we performed rChIP assays with RNase H treated chromatin. We found that RNase H did not affect RING1B-chromatin interaction, suggesting that R-loops are not required for stability of RING1B at chromatin (Supplementary Figure S4G). In line with the lack of evidence supporting direct binding of RING1B to RNA (31), electrophoresis mobility shift assays (EMSA) did not detect direct RING1B interaction with either RNA or R-loops (Supplementary Figure S4H). These results indicate that the association between RING1B and chromatin is partially dependent on chromatin-bound single-stranded RNA and ERα.
Figure 4.
RING1B recruitment to chromatin is dependent on ERα and chromatin-associated RNA. (A) RING1B ChIP-qPCR in shCTR and 2 shESR1 T47D cell lines before and after 45 min of E2 administration at six sites with known RING1B and ERα co-occupancy. (B and C) rChIP-seq heatmap depicting E2-induced ERα (B) and RING1B (C) chromatin binding at the previously described 4 clusters (Figure 3A); n = 2. (D) Average signal of ERα and RING1B chromatin binding after 45 min of E2 stimulation with or without RNase A treatment. (E) RING1B and ERα ChIP-seq signal after 45 min of E2 stimulation with or without RNase A treatment at ADAMTSL5 and PGR. (F) ERα (left) and RING1B (center) ChIP-seq heatmaps before and after 45 min of E2 treated with vehicle (DMSO) or 10 μM triptolide for 9 h clustered by ERα occupancy. HOMER motif analysis of the four clusters (right). Cluster III ERα sites contain very little FOXA1 motif enrichment. (G) RING1B and ERα ChIP-seq before and after 45 min of E2 in cells treated with DMSO or triptolide showing increased ERα and RING1B recruitment at the RGS10 promoter but little change at the SLC25A45 gene body. (H–I) Average signal of ERα (H) and RING1B (I) at cluster I, classified by genomic annotation, before and after 45 min of E2 in cells treated with DMSO or triptolide.
Transcriptional inhibition increases ERα and RING1B recruitment to promoters but not enhancers
Since depletion of chromatin-bound RNA decreased the association of RING1B with chromatin, we decided to deplete RNA transcripts with triptolide. Triptolide inhibits global transcription by inducing proteasome-dependent degradation of RNA Pol II (32). Treatment with 10 μM of triptolide for 9 h depleted RNA Pol II and inhibited the transcription of ERα target genes without affecting the expression of ERα and RING1B (Supplementary Figure S5A,B). ERα and RING1B ChIP-seq in triptolide and E2 treated cells revealed four distinct clusters of ERα binding sites (Figure 4F and Supplementary Figure S5C). Cluster I sites exhibited the greatest overlap with RING1B ChIP-seq followed by cluster II while almost no RING1B occupancy was observed in clusters III and IV (Figure 4F), but induced de novo ERα binding sites (cluster III) and decreased (cluster IV) E2-induced ERα binding. Given that cluster I overlapped with RING1B the most, genes assigned to this cluster exhibited the highest expression levels compared to genes assigned to other clusters (Supplementary Figure S5E). Interestingly, HOMER motif analysis (33) revealed that cluster III sites were the least enriched for FOXA1, a pioneer TF indispensable for the chromatin recruitment of ERα and RING1B (Figure 4F), a result we further confirmed by FOXA1 ChIP-seq (Supplementary Figure S5G). This observation suggests that RNA Pol II depletion may create a novel dependency on another pioneer TF for ERα. Importantly, this result further highlights RING1B’s dependency on ERα for targeting to chromatin. Clusters I and II were further annotated and classified into intergenic, gene body and promoter/TSS regions. On average, triptolide treatment did not affect E2-induced ERα recruitment to intergenic regions and gene bodies (Figure 4G, 4H and Supplementary Figure S5D, S5F). However, depletion of RNA Pol II increased E2-induced recruitment of ERα and RING1B to promoter/TSS sites (Figure 4G–I and Supplementary Figure S5D, S5F). Together, these results suggest that despite RING1B being dependent on RNA for binding to chromatin, inhibiting transcription of RNA via RNA Pol II depletion did not result in decreased RING1B binding upon an increase in ERα recruitment.
DISCUSSION
Here we show that RING1B regulates enhancer-promoter interaction of GREB1, a classic E2-responsive gene, concomitantly with promoting the transcription of ERα target genes. R-loops generated upon acute E2 stimulation are also dependent on RING1B. We found that RING1B is a molecular sensor for ERα recruitment and the E2 response. Moreover, the RING1B recruitment to chromatin is dependent on ERα and is partially regulated via chromatin-associated RNA. These results suggest a potential positive feedback loop between RING1B and ERα wherein RING1B promotes the transcription of ERα target genes. Interestingly, chromatin accessibility was dispensable for both the enhancer–promoter looping of GREB1 and gene activation following 45 min of E2 stimulation. In fact, chromatin accessibility at these sites was only observed after 4–8 h after E2 treatment. Taken together, these results show that an increase in chromatin accessibility (i) does not necessarily correlate with changes in enhancer–promoter interactions and (ii) is not required for a gene to be transcriptionally active. Indeed, we suggest that chromatin accessibility can be required to maintain gene activity rather than transcriptional initiation.
Canonical and non-canonical PRC1 complexes, defined by the presence or absence of CBX subunits respectively, can be recruited in a PRC2-dependent and independent manner (34). Recently, R-loops have also been found to recruit RING1B to a sub-set of repressed Polycomb target genes in stem cells (27). How RING1B is targeted to active enhancers and promoters is largely unknown. Here we show for the first time that a cPRC1 complex can be also recruited to sites prone to R-loop formation upon stimulation of gene transcription and that RING1B is indispensable for R-loop accumulation. Moreover, we recently showed that RING1B promoted: (i) the deposition of H3K27ac, (ii) eRNA transcription at RING1B/ERα co-target enhancers, (iii) full ERα engagement with the chromatin and (iv) expression of E2-responsive genes. As to how RING1B is recruited to chromatin, our studies provided several clues. First, we found that RING1B is dependent on FOXA1 as a pioneer factor, just like ERα. Upon FOXA1 depletion, RING1B was not recruited to chromatin. Moreover, RING1B chromatin binding was dependent on RNA. However, previously published studies, as well as our EMSA experiment, indicate that RING1B does not physically interact with RNA (31). Interestingly, macromolecules such as RNA are known to be included into phase-separated condensates with ribonucleoproteins through liquid–liquid phase separation (35,36), which occurs when two distinct phases form within a homogeneous mixture. Formation of these condensates, which some refer to as membrane-less organelles, are required for many biological processes, and dysregulated phase separation has been implicated in a variety of pathological processes (37). E2 is known to stimulate the phase separation of ERα (38). Furthermore, ERα can coordinate the formation of these phase-separated condensates at E2-responsive active enhancers, which are characterized by high levels of eRNA transcription upon E2 stimulation (30). Within these condensates are important ERα co-factors such as GATA3 and AP-2γ which mediate the activation of ERα-containing enhancers, reflected by an increase in eRNAs. Chemical disruption of condensate formation at these enhancers significantly decreased the recruitment of ERα co-factors to these enhancers as well as abrogated enhancer activation. eRNA also plays an indispensable role in promoting the formation of phase-separated transcriptional complexes at ERα target enhancers. The binding of ERα, however, is not affected in either of these scenarios since ERα binds directly to DNA. Importantly, the recruitment of ERα and cofactors to constitutively active enhancers is also not affected by disrupting phase separation, suggesting that phase-separated condensates only form upon E2 stimulation (30). These findings draw many similarities to ours. We demonstrated that not only RING1B chromatin association is dependent on RNA but also that ERα chromatin association is not affected by RNA resolution. In addition to this positive feedback loop between RING1B, ERα and RNA, RING1B is also recruited to ERα target enhancers to mediate their activation. This recruitment is specific to de novo enhancers that are formed upon E2 and not at pre-existing E2-independent enhancers, which parallels the finding that phase separated condensates are only formed at E2-responsive enhancers. Disruption of phase separation, and therefore, the recruitment of ERα coactivators, abrogates E2-induced gene transcription, which is highly similar to the attenuation we observed upon RING1B depletion.
A recent study tracking PRC1 occupancy on the chromatin revealed that the majority of RING1B and PRC1 are not stably bound (21). This result suggests a high degree of turnover in condensate components that is consistent with the observation that proteins within phase-separated condensates can diffuse in and out with ease (39), such as those at ERα target enhancers. It is plausible that RING1B is recruited to active, E2-responsive ERα target enhancers and genes by being included into phase-separated condensates, whose stability and formation is dependent on the presence of RNA. As the exact stoichiometry of RNA to protein is crucial for the formation of RNA-containing condensates, we theorize that RNA resolution with RNase A in our RING1B rChIP disrupted the integrity of the crosslinked RNA-based condensate structures on the chromatin (40). Since RING1B may be part of the crosslinked condensate structure, a proportion of chromatin-associated RING1B is thus released upon RNase A treatment.
Nevertheless, it is important to note that RNase A treatment reduced RING1B binding to chromatin by about 50%, therefore showing that RING1B depends on other ERα-related mechanisms for chromatin binding. In fact, upon depletion of RNA Pol II via triptolide treatment, RING1B binding increased at ERα-bound promoters, concomitant with an increase in ERα binding. Upon E2 stimulation, ERα is poly-ubiquitinated and targeted for proteasomal degradation (41). This mechanism is important for maintaining proper E2-responsive gene expression (42,43). Inhibiting proteasomal function, poly-ubiquitination or ERα transcriptional activity increases the amount of ERα in the cell (44). Transcriptional inhibition via the depletion of RNA Pol II, therefore, is likely to prevent the degradation of ERα upon E2 stimulation and lead to accumulation of ERα. Interestingly, we only observed an overall increase in ERα at promoters and not at purported enhancers located within intergenic regions and gene bodies. This suggests that proteasomal degradation may be specific to ERα involved in gene transcription and not in enhancer activation. Additionally, the increased RING1B binding upon RNA Pol II depletion suggests that ERα plays a more important role in recruiting RING1B to the chromatin than does RNA. So far, the depletion of ERα is the only known mechanism in ER+ breast cancer cells to block the recruitment of RING1B to the chromatin. However, it is unclear whether ERα acts solely as an estrogen sensor, as a recruitment scaffold, or both, for targeting RING1B to the chromatin. Further exploration is warranted to determine the exact mechanism underlying E2-induced and ERα-dependent recruitment of RING1B to chromatin.
DATA AVAILABILITY
All raw and processed ChIP–seq and RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE173860.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Ho Lam Chan for help in preparing the manuscript and to members of the Morey laboratory, Gloria Mas and Joyce M. Slingerland for discussions, as well as the Oncogenomics Core Facility, Cancer Modeling Shared Resource, and the Flow Cytometry Core Facility at the Sylvester Comprehensive Cancer Center. This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Yusheng Zhang, Sylvester Comprehensive Cancer Center, Biomedical Research Building, 1501 NW 10th Avenue, Miami, FL 33136, USA; Department of Human Genetics, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Tong Liu, Department of Computer Science, University of Miami, 1365 Memorial Drive, P.O. Box 248154, Coral Gables, FL 33124, USA.
Fenghua Yuan, Sylvester Comprehensive Cancer Center, Biomedical Research Building, 1501 NW 10th Avenue, Miami, FL 33136, USA; Department of Biochemistry and Molecular Biology, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Liliana Garcia-Martinez, Sylvester Comprehensive Cancer Center, Biomedical Research Building, 1501 NW 10th Avenue, Miami, FL 33136, USA; Department of Human Genetics, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Kyutae D Lee, Sylvester Comprehensive Cancer Center, Biomedical Research Building, 1501 NW 10th Avenue, Miami, FL 33136, USA; Department of Human Genetics, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Stephanie Stransky, Department of Biochemistry, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
Simone Sidoli, Department of Biochemistry, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
Ramiro E Verdun, Sylvester Comprehensive Cancer Center, Biomedical Research Building, 1501 NW 10th Avenue, Miami, FL 33136, USA; Division of Hematology, Department of Medicine, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Yanbin Zhang, Sylvester Comprehensive Cancer Center, Biomedical Research Building, 1501 NW 10th Avenue, Miami, FL 33136, USA; Department of Biochemistry and Molecular Biology, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Zheng Wang, Department of Computer Science, University of Miami, 1365 Memorial Drive, P.O. Box 248154, Coral Gables, FL 33124, USA.
Lluis Morey, Sylvester Comprehensive Cancer Center, Biomedical Research Building, 1501 NW 10th Avenue, Miami, FL 33136, USA; Department of Human Genetics, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Sylvester Comprehensive Cancer Center; Florida Health Bankhead-Coley Cancer Research Program [20B15]; V Foundation [DEC2020-009]; Lampert Breast Cancer Research Fund (to L.M.); Leukemia Research Foundation (Hollis Brownstein New Investigator Research Grant); AFAR (Sagol Network GerOmics award); Deerfield (Xseed award); NIH [1S10OD030286-01] (to S.S., S.D.); Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA240139. This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding for open access charge: Sylvester Comprehensive Cancer Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stock J.K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A.G., Pombo A.. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007; 9:1428–1435. [DOI] [PubMed] [Google Scholar]
- 2.Endoh M., Endo T.A., Endoh T., Isono K., Sharif J., Ohara O., Toyoda T., Ito T., Eskeland R., Bickmore W.A.et al.. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLos Genet. 2012; 8:e1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eskeland R., Leeb M., Grimes G.R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A.I., Wutz A.et al.. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell. 2010; 38:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan H.L., Beckedorff F., Zhang Y., Garcia-Huidobro J., Jiang H., Colaprico A., Bilbao D., Figueroa M.E., LaCava J., Shiekhattar R.et al.. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun. 2018; 9:3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Chan H.L., Garcia-Martinez L., Karl D.L., Weich N., Slingerland J.M., Verdun R.E., Morey L.. Estrogen induces dynamic ERα and RING1B recruitment to control gene and enhancer activities in luminal breast cancer. Sci. Adv. 2020; 6:eaaz7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mauvais-Jarvis F., Clegg D.J., Hevener A.L.. The role of estrogens in control of energy balance and glucose homeostasis. Endocr. Rev. 2013; 34:309–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.American Cancer Society Breast cancer facts and figures 2019–2020. 2019; American Cancer Society. [Google Scholar]
- 8.Klinge C.M.Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001; 29:2905–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjornstrom L., Sjoberg M.. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol. Endocrinol. 2005; 19:833–842. [DOI] [PubMed] [Google Scholar]
- 10.Zaret K.S., Carroll J.S.. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011; 25:2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Martinez L., Zhang Y., Nakata Y., Chan H.L., Morey L.. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021; 12:1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F.et al.. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006; 38:1289–1297. [DOI] [PubMed] [Google Scholar]
- 13.Stender J.D., Kim K., Charn T.H., Komm B., Chang K.C., Kraus W.L., Benner C., Glass C.K., Katzenellenbogen B.S.. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol. Cell. Biol. 2010; 30:3943–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swinstead E.E., Miranda T.B., Paakinaho V., Baek S., Goldstein I., Hawkins M., Karpova T.S., Ball D., Mazza D., Lavis L.D.et al.. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell. 2016; 165:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J.. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015; 109:21.29.21–21.29.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouwer R.W., van den Hout M.C., van I.W.F., Soler E., Stadhouders R.. Unbiased interrogation of 3D genome topology using chromosome conformation capture coupled to high-throughput sequencing (4C-Seq). Methods Mol. Biol. 2017; 1507:199–220. [DOI] [PubMed] [Google Scholar]
- 17.Sanz L.A., Chédin F.. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2019; 14:1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long Y., Hwang T., Gooding A.R., Goodrich K.J., Rinn J.L., Cech T.R.. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet. 2020; 52:931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Werken H.J., Landan G., Holwerda S.J., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Oz Y., Bouwman B.A.et al.. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods. 2012; 9:969–972. [DOI] [PubMed] [Google Scholar]
- 20.Wolff J., Rabbani L., Gilsbach R., Richard G., Manke T., Backofen R., Grüning B.A.. Galaxy HiCExplorer 3: a web server for reproducible Hi-C, capture Hi-C and single-cell Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 2020; 48:W177–W184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huseyin M.K., Klose R.J.. Live-cell single particle tracking of PRC1 reveals a highly dynamic system with low target site occupancy. Nat. Commun. 2021; 12:887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siersbæk R., Scabia V., Nagarajan S., Chernukhin I., Papachristou E.K., Broome R., Johnston S.J., Joosten S.E.P., Green A.R., Kumar S.et al.. IL6/STAT3 signaling hijacks estrogen receptor α enhancers to drive breast cancer metastasis. Cancer Cell. 2020; 38:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song R., Struhl K.. S100A8/S100A9 cytokine acts as a transcriptional coactivator during breast cellular transformation. Sci. Adv. 2021; 7:eabe5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakraborty P., Huang J.T.J., Hiom K.. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 2018; 9:4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niehrs C., Luke B.. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020; 21:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skourti-Stathaki K., Proudfoot N.J.. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014; 28:1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skourti-Stathaki K., Torlai Triglia E., Warburton M., Voigt P., Bird A., Pombo A.. R-loops enhance polycomb repression at a subset of developmental regulator genes. Mol. Cell. 2019; 73:930–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stork C.T., Bocek M., Crossley M.P., Sollier J., Sanz L.A., Chédin F., Swigut T., Cimprich K.A.. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife. 2016; 5:e17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A.. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair S.J., Yang L., Meluzzi D., Oh S., Yang F., Friedman M.J., Wang S., Suter T., Alshareedah I., Gamliel A.et al.. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 2019; 26:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S.et al.. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013; 152:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Lu J.J., He L., Yu Q.. Triptolide (TPL) inhibits global transcription by inducing proteasome-dependent degradation of RNA polymerase II (Pol II). PLoS One. 2011; 6:e23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G.. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017; 171:34–57. [DOI] [PubMed] [Google Scholar]
- 35.Alshareedah I., Moosa M.M., Raju M., Potoyan D.A., Banerjee P.R.. Phase transition of RNA-protein complexes into ordered hollow condensates. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:15650–15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A.. A phase separation model for transcriptional control. Cell. 2017; 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch Let al.. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018; 28:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Jove Navarro M., Kashida S., Chouaib R., Souquere S., Pierron G., Weil D., Gueroui Z. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat. Commun. 2019; 10:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W., Slingerland J.M.. Links between oestrogen receptor activation and proteolysis: relevance to hormone-regulated cancer therapy. Nat. Rev. Cancer. 2014; 14:26–38. [DOI] [PubMed] [Google Scholar]
- 42.Fan M., Nakshatri H., Nephew K.P.. Inhibiting proteasomal proteolysis sustains estrogen receptor-alpha activation. Mol. Endocrinol. 2004; 18:2603–2615. [DOI] [PubMed] [Google Scholar]
- 43.Powers G.L., Ellison-Zelski S.J., Casa A.J., Lee A.V., Alarid E.T.. Proteasome inhibition represses ERalpha gene expression in ER+ cells: a new link between proteasome activity and estrogen signaling in breast cancer. Oncogene. 2010; 29:1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wijayaratne A.L., McDonnell D.P.. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J. Biol. Chem. 2001; 276:35684–35692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed ChIP–seq and RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE173860.