Abstract
In Saccharomyces cerevisiae, the Wee1 family kinase Swe1p is normally stable during G1 and S phases but is unstable during G2 and M phases due to ubiquitination and subsequent degradation. However, perturbations of the actin cytoskeleton lead to a stabilization and accumulation of Swe1p. This response constitutes part of a morphogenesis checkpoint that couples cell cycle progression to proper bud formation, but the basis for the regulation of Swe1p degradation by the morphogenesis checkpoint remains unknown. Previous studies have identified a protein kinase, Hsl1p, and a phylogenetically conserved protein of unknown function, Hsl7p, as putative negative regulators of Swe1p. We report here that Hsl1p and Hsl7p act in concert to target Swe1p for degradation. Both proteins are required for Swe1p degradation during the unperturbed cell cycle, and excess Hsl1p accelerates Swe1p degradation in the G2-M phase. Hsl1p accumulates periodically during the cell cycle and promotes the periodic phosphorylation of Hsl7p. Hsl7p can be detected in a complex with Swe1p in cell lysates, and the overexpression of Hsl7p or Hsl1p produces an effective override of the G2 arrest imposed by the morphogenesis checkpoint. These findings suggest that Hsl1p and Hsl7p interact directly with Swe1p to promote its recognition by the ubiquitination complex, leading ultimately to its destruction.
Entry into mitosis is triggered by the activation of Cdc2-type cyclin-dependent kinases (Cdc28p in Saccharomyces cerevisiae) by mitotic B-type cyclins (35, 37). Cyclin B-Cdc2 complexes can accumulate in an inactive form if the Cdc2 subunit is phosphorylated on a critical tyrosine residue (amino acid 19 in Cdc28p) and, in some cells, also on the adjacent threonine residue (10, 35). Checkpoint controls that regulate entry into mitosis utilize this inhibitory phosphorylation to restrain activation of Cdc2 until key cell cycle events have been completed (38, 42). Cdc2 tyrosine phosphorylation is catalyzed by Wee1-related kinases (Swe1p in S. cerevisiae), and dephosphorylation is catalyzed by Cdc25-related phosphatases (Mih1p in S. cerevisiae) (7, 10, 45). It is therefore of great interest to elucidate the regulatory pathways that control the activity of the Wee1 family and Cdc25 family enzymes.
In S. cerevisiae, Cdc28p Tyr19 phosphorylation is induced by the morphogenesis checkpoint, which helps to coordinate the nuclear cycle with the process of bud development (22, 33). For example, several environmental insults, including rapid changes in ambient temperature or osmolarity, trigger a temporary disruption of actin polarity, causing delays in bud formation. The morphogenesis checkpoint responds by delaying mitosis so that cells do not undergo nuclear division before a bud has been constructed, thus preventing the formation of binucleate cells (22, 33). The cell cycle delay induced by the morphogenesis checkpoint requires Swe1p (49). Swe1p abundance varies during the cell cycle as a result of regulated transcription and degradation. SWE1 transcription is periodic, with a peak in late G1 (24, 29, 49) phase, and Swe1p is stable early in the cell cycle but becomes unstable during G2 and M phases as a consequence of Cdc28p activation by the B-type cyclins Clb1p to Clb4p (48). Thus, Swe1p accumulates during late G1 and S phases and is degraded during G2-M in the unperturbed cell cycle. However, Swe1p is stabilized in response to perturbations of actin organization, and the resulting persistence or continued accumulation of the protein (possibly in conjunction with changes in its activity and/or localization) leads to G2 arrest (48).
Two putative upstream regulators of Swe1p, Hsl1p and Hsl7p, were discovered serendipitously during a genetic screen for mutations displaying synthetic lethality with a deletion of the amino terminus of histone H3 (HSL [histone synthetic lethal]) (29). Although the basis for the genetic interaction with histones was not clarified, the data suggested that Hsl1p and Hsl7p act in some manner to lower the level of Swe1p activity. In particular, it was found that hsl1 and hsl7 mutants displayed a G2 delay that was eliminated upon deletion of SWE1 (29). However, many mutants with defects in cell morphogenesis display similar Swe1p-dependent G2 delays, produced by the morphogenesis checkpoint in response to the mutant defect (33). It is therefore important to determine whether Hsl1p and Hsl7p indeed act directly on Swe1p or whether they simply cause a morphogenesis defect that activates the checkpoint response.
The sequence of Hsl7p has not yet suggested possible biochemical activities for this protein, but homology searches have identified close relatives in several other eukaryotes, including Schizosaccharomyces pombe and humans (16, 29). In contrast, support for the hypothesis that Hsl1p is a direct negative regulator of Swe1p has come from the similarity between the kinase domain of Hsl1p and that of Nim1, an S. pombe protein that has been shown to directly phosphorylate and inhibit Wee1 (9, 40, 55). In addition, HSL1 (also called NIK1) was isolated independently in a screen for S. cerevisiae genes that could serve as multicopy suppressors of a temperature-sensitive cdc2 mutant in S. pombe (54). This circumstantial evidence suggests that Hsl1p may also act by directly phosphorylating and inhibiting Swe1p. Homology searches have revealed that S. cerevisiae contains two other kinases, Gin4p and Kcc4p, that are approximately as similar to Nim1 as is Hsl1p (4, 25). Recently, it was suggested that these kinases play a redundant role in Swe1p regulation (4). However, none of these kinases has yet been shown to regulate Swe1p directly.
In this report, we provide evidence that both Hsl1p and Hsl7p are bona fide negative regulators of Swe1p that appear to function interdependently in a pathway that targets Swe1p for degradation. During a checkpoint response, Hsl1p and Hsl7p do not target Swe1p for degradation, suggesting that the checkpoint mechanism may stabilize Swe1p by inhibiting Hsl1p or Hsl7p function.
MATERIALS AND METHODS
Yeast strains and plasmids.
Standard genetic and molecular biology methods (30, 46) were used to generate all strains and plasmids used in this study, except as indicated below. The yeast strains used are listed in Table 1. Plasmids containing the swe1ΔLEU2 (7), mih1ΔLEU2 (45), GAL:SWE1myc:URA3 (33), CDC28Y19F:TRP1 (48), hsl1ΔURA3 (29), and hsl7ΔURA3 (29) alleles have been described previously; appropriate fragments were introduced into yeast strains by direct transformation and confirmed by diagnostic PCR (26) and phenotypic tests.
TABLE 1.
S. cerevisiae strains used in this study
| Straina | Relevant genotype |
|---|---|
| JMY1469 | a SWE1myc:HIS2 |
| JMY1503 | a SWE1myc:HIS2 hsl1ΔURA3 |
| JMY1505 | a SWE1myc:HIS2 hsl7ΔURA3 |
| JMY1507 | a SWE1myc:HIS2 hsl1ΔURA3 hsl7ΔURA3 |
| JMY1470 | a SWE1myc:HIS2 SWE1myc:TRP1 |
| JMY1477 | a SWE1myc:HIS2 SWE1myc:TRP1 hsl1ΔURA3 |
| JMY1475 | a SWE1myc:HIS2 SWE1myc:TRP1 hsl7ΔURA3 |
| JMY1479 | a SWE1myc:HIS2 SWE1myc:TRP1 hsl1ΔURA3 hsl7ΔURA3 |
| JMY1569 | a/α swe1ΔLEU2/SWE1 mih1ΔLEU2/MIH1 hsl1ΔURA3/HSL1 |
| JMY1570 | a/α swe1ΔLEU2/SWE1 mih1ΔLEU2/MIH1 hsl7ΔURA3/HSL7 |
| JMY1571 | a/α mih1ΔTRP1/MIH1 hsl1ΔURA3/HSL1 GAL:HSL7:LEU2/HSL7 |
| JMY1572 | a/α mih1ΔTRP1/MIH1 hsl7ΔURA3/HSL7 GAL:HSL1:LEU2/HSL1 |
| DLY657 | a cdc24-1 bar1 |
| DLY690 | a cdc24-1 swe1ΔLEU2 bar1 |
| JMY1284 | a GAL:HSL7:LEU2 cdc24-1 bar1 |
| JMY1495 | a GAL:HSL1:LEU2:HSL1:TRP1 cdc24-1 bar1 |
| JMY1472 | a SWE1myc:HIS2 cdc24-1 bar1 |
| JMY1494 | a SWE1myc:HIS2 SWE1myc:TRP1 cdc24-1 bar1 |
| JMY1493 | a SWE1myc:HIS2 SWE1myc:TRP1(3X) cdc24-1 bar1 |
| JMY1300 | a hsl1ΔURA3 cdc24-1 bar1 |
| JMY1301 | a hsl7ΔURA3 cdc24-1 bar1 |
| JMY1500 | a HSL1myc:URA3 bar1 |
| M-1295* | a HSL7-3HA:kan |
| M-1505* | a GAL:SWE1myc:URA3 |
| M-1537* | a GAL:SWE1myc:URA3 HSL7-3HA:kan |
| JMY1521* | a GAL:SWE1myc:URA3 HSL7-3HA:kan bar1ΔTRP1 |
| JMY1539* | a GAL:SWE1myc:URA3 HSL7-3HA:kan hsl1ΔTRP1 |
| DLY1 | a bar1 |
| RSY342 | a GAL:SWE1myc:URA3 CDC28Y19F:TRP1 |
| RSY361 | a GAL:SWE1myc:URA3 hsl1ΔURA3 CDC28Y19F:TRP1 bar1 |
| RSY356 | a GAL:SWE1myc:URA3 hsl7ΔURA3 CDC28Y19F:TRP1 bar1 |
| RSY366 | a GAL:SWE1myc:URA3 GAL:HSL1:LEU2 CDC28Y19F:TRP1 bar1 |
| RSY370 | a GAL:SWE1myc:URA3 GAL:HSL7:LEU2 CDC28Y19F:TRP1 bar1 |
To create the SWE1myc:HIS2 allele, a 2.6-kb EcoRI/BamHI fragment containing the COOH terminus of SWE1 tagged with one hemagglutinin (HA) epitope, 12 myc epitopes, and downstream sequences was isolated from pRS306-GAL:SWE1myc (33) and ligated into the EcoRI/BglII sites of vector YIpGAP2 (49). Digestion of this plasmid with KpnI targets integration to the SWE1 locus, creating a full-length SWE1myc allele tagged with HIS2 adjacent to a 3′ truncated SWE1. To create the SWE1myc:TRP1 allele, a 4.0-kb PstI/BamHI fragment containing the SWE1 gene and flanking sequences was removed from plasmid pJM1024 (isolated from a YCp50 genomic library [44]) and ligated into the corresponding sites of vector YIplac204 (14). A 2.0-kb ClaI/BamHI fragment from pRS306-GAL:SWE1myc (33) that carries the COOH terminus of SWE1 tagged as described above was inserted in place of the corresponding untagged fragment in the YIplac204-SWE1 plasmid to create a full-length myc-tagged SWE1 expressed from the SWE1 promoter. This plasmid was digested with EcoRV to target integration to the TRP1 locus. In addition to transformants containing a single copy of the SWE1myc:TRP1 allele, one transformant contained three copies integrated at TRP1 as determined by Southern blot analysis. This allele is referred to as SWE1myc:TRP1(3X).
The GAL:HSL1:LEU2 and GAL:HSL7:LEU2 alleles were constructed by similar strategies. In each case, the 5′ end of the gene was amplified from genomic DNA by PCR. A BamHI site was incorporated into each primer with the 5′ site just upstream of the start codon. The primers used were 5′-TTATTGGATCCACACGACATGACTGGTCAC-3′ and 5′-GTTTATTAGGATCCTCTAATGCTGCCATGCCG-3′ (HSL1) and 5′-GGTTCAGGATCCATATGCATAGCAACG-3′ and 5′-CATACGAAGGATCCCTGGTTCTTGGCAAAGC-3′ (HSL7). The PCR products (0.8 kb for HSL1 and 0.7 kb for HSL7) were cut with BamHI and ligated into the corresponding site of vector YIpG2 (13, 53), which placed the fragments downstream of the GAL1 promoter. The YIpG2-HSL1 plasmid was targeted to integrate at the HSL1 locus by digestion with XbaI; this created a full-length GAL:HSL1:LEU2 allele adjacent to a 3′ truncated HSL1. The YIpG2-HSL7 plasmid was targeted to integrate at the HSL7 locus by digestion with NruI; this created a full-length GAL:HSL7:LEU2 allele adjacent to a 3′ truncated HSL7. To create the GAL:HSL1:LEU2:HSL1:TRP1 allele, a 7.3-kb BamHI/SacI fragment containing HSL1 and surrounding genomic sequence was isolated from plasmid pNE30 (11) and ligated into the corresponding sites of the vector pRS304 (50). The resulting plasmid was digested with StuI to target integration to the GAL:HSL1:LEU2 locus, creating a strain that contains GAL-regulated HSL1 adjacent to wild-type HSL1 under its own promoter.
The HSL7-3HA:kan allele was constructed as described by Longtine et al. (27). To create the HSL1myc:URA3 allele, a 0.65-kb fragment corresponding to the 3′ end of HSL1 and including the last coding base of the HSL1 open reading frame was amplified by PCR with primers (5′-CTCTAGAATCTAAAAAAGTAGGTGGGGG-3′ and 5′-CGTCGACTGAACGTCCGGCATTTCGAATTAC-3′) that placed an XbaI site upstream of the fragment and a SalI site downstream. This PCR product was digested with XbaI and SalI and inserted into XbaI/SalI-digested pRS306-GAL:SWE1myc (33), thus replacing the entire SWE1 open reading frame and upstream sequences with the COOH-terminal HSL1 fragment. This created an in-frame fusion of the 3′ end of HSL1 with the myc tag in the plasmid. The resulting plasmid was targeted to integrate at the HSL1 locus by digestion with EcoRI, thus creating the HSL1myc:URA3 allele adjacent to a 5′ deleted HSL1.
The mih1ΔTRP1, bar1ΔTRP1, and hsl1ΔTRP1 alleles were constructed by using the PCR disruption method (5, 28) and plasmid pRS304 (50) as a template. The PCR products were transformed directly into yeast to delete all or nearly all of the open reading frames of interest. The PCR primers used were: 5′-TGGA CAAACCAGGATTGAAGTCAGCGAGGGTGAAGAAACCGCGCGTTTC GGTGATGAC-3′ and 5′-AATAACGATCTTCTTGCGGGCCTGGGTAAATCTTCTCGGTTTTCCTGATGCGGTATTTTCTCCT-3′ for MIH1, 5′-CCATT ACTGCTTTAACAAACGATGGCACTGGTCACTTAGAGCGCGTTTCGG TGATGAC-3′ and 5′-ACACTGCCCGAATTTGCCATAGTCGAGGATAATTCTAATTTAGTTTCCTGATGCGGTATTTTCTCCT-3′ for BAR1, and 5′- TCAAATAGGTTGGATATCCATCATACTACTTGCTACTAATGCGCGTT TCGGTGATGAC-3′ and 5′-GAATTTATGAACGTCCGGCATTTCGAATTACTCTCTCCACTTTCCTGATGCGGTATTTTCTCCT-3′ for HSL1. All disruptions were confirmed by diagnostic PCR (26).
Media, growth conditions, and cell synchrony.
Strains were grown in YEPD (1% yeast extract, 2% Bacto Peptone, 2% dextrose, and 0.01% adenine), YEPS (YEPD but with 2% sucrose instead of dextrose), or YEPG (YEPD but with 2% galactose instead of dextrose) medium. For α-factor arrest-release experiments, exponentially growing cells (2 × 106 to 5 × 106 cells/ml) were incubated with 20 to 25 ng of α-factor (custom synthesized by Research Genetics, Huntsville, Ala.) per ml for 2 to 3 h, harvested by centrifugation, and resuspended in a fresh medium to release the α-factor-induced cell cycle block. bar1 strains were used in all such experiments, and microscopic examination confirmed that >90% of the arrested cells were unbudded. Cells were arrested in G2/M by incubation with 15 μg of nocodazole (Sigma, St. Louis, Mo.; stored as a 10-mg/ml stock solution in dimethylsulfoxide at −20°C) per ml for 3 to 4 h (18). Microscopic examination confirmed that >80% of the treated cells had large buds, indicative of G2/M arrest.
Fluorescence staining and microscopy.
To visualize nuclear DNA, cells were fixed in 70% ethanol for >1 h, harvested by centrifugation, and resuspended in 0.2 μg of DAPI (4′6-diamidino-2-phenylindole; Sigma). Cells were viewed on an Axioskop apparatus (Zeiss, Thornwood, N.Y.) equipped with epifluorescence and differential interference contrast optics. Images were captured by using a cooled model charge-coupled device (CCD) camera (Princeton Instruments, Princeton, N.J.). Microscopic images of whole yeast microcolonies were captured similarly.
Preparation of lysates, immunoprecipitation, immunoblotting, and phosphatase treatment.
Yeast cells were washed with ice-cold H2O and harvested by centrifugation. Cell pellets were stored frozen at −80°C. Lysates were made by resuspending the pellets in a lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% NP-40, 1 mM sodium pyrophosphate, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and 2 μg each of pepstatin A and leupeptin (Sigma) per ml and vortexing with acid-washed glass beads. Lysates were clarified by centrifugation for 8 min at 14,000 rpm in an Eppendorf Microfuge, and the protein concentration was determined by using the Bio-Rad (Hercules, Calif.) protein assay.
For electrophoresis and immunoblotting, 20 μg of total protein per gel lane were mixed with hot (95°C) 2× sample loading buffer (final concentrations, 62.5 mM Tris-HCl [pH 6.8], 1% sodium dodecyl sulfate [SDS], 25% glycerol, 355 mM β-mercaptoethanol, 0.01% bromophenol blue) and incubated at 95°C for 5 min prior to electrophoresis on SDS–6 or 8% polyacrylamide gels. Proteins were then transferred electrophoretically to nitrocellulose membranes (Schleicher and Schuell, Keene, N.H.) and stained with anti-myc (9E10; Santa Cruz Biotechnology, Santa Cruz, Calif.) or anti-HA (12CA5; Boehringer Mannheim, Indianapolis, Ind.) antibody. Before staining, membranes were blocked with 5% nonfat dry milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBS-Tween). Primary antibodies were used at a 1:1,000 dilution in PBS-Tween containing 1% nonfat dry milk. The secondary antibody (horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G; Jackson Immunoresearch Laboratories, West Grove, Pa.) was used at a 1:2,500 dilution in the same solution. Incubations were carried out for 1 h each and separated by three washes with PBS-Tween. Blots were developed with Renaissance Western Blot Chemiluminescence Reagent Plus (NEN Life Sciences Products, Boston, Mass.).
For immunoprecipitation, 200 μg of lysate was incubated for 1 h with 1 μl of antibody and then for a further 1 h with 30 μl of a 50% slurry of protein A-Sepharose (Sigma) at 4°C with gentle rocking. Beads were washed three times with lysis buffer (see above) without protease inhibitors and then heated in 1× sample loading buffer, and proteins were separated and immunoblotted as described above.
For phosphatase treatment of Hsl7p-HA, anti-HA immunoprecipitate from 400 μg of lysate was washed twice with lysis buffer (see above) and twice with a solution containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM sodium pyrophosphate, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS and then divided into three equal aliquots. These aliquots were resuspended in 40 mM PIPES (piperazine-N,N′-bis[2-ethanesulfonic acid]) (pH 6.0) containing 1 mM dithiothreitol, 7.5 mM phenylmethylsufonyl fluoride, 37.5 μg of aprotinin (Sigma) per ml, and 25 μg each of benzamidine (Sigma), leupeptin, and pepstatin A per ml. Type II potato acid phosphatase (0.14 U) (Sigma) was added to two of the samples, and all samples were incubated at 30°C for 30 min. Sodium orthovanadate (10 mM) was added to inhibit phosphatase activity in one of the samples.
Pulse-chase analysis of Swe1p-myc stability.
GAL:SWE1myc:URA3 cells were grown in YEPS at 30°C and induced to express Swe1p-myc by the addition of 2% galactose for 10 min or 3 h. Cells were then harvested by centrifugation, resuspended at a density of 108 cells/ml in a labeling medium (6.7 g of yeast nitrogen base without methionine and cysteine [Bio 101, Vista, Calif.] per liter, 2% sucrose, and 2% galactose, plus 0.25 mCi of Trans35S-Label [ICN Pharmaceuticals, Costa Mesa, Calif.] per ml [0.183 mM]) and incubated for a further 10 min to label newly synthesized proteins with [35S]methionine and cysteine. Labeled cells were collected by filtration, washed with a prewarmed medium, and resuspended at a density of 3 × 107 cells/ml in fresh YEPD or YEPG supplemented with 3 mM methionine and 0.5% Casamino Acids to prevent further labeling. Incubation was continued, and aliquots of cells were diluted into ice-cold 10 mM NaN3, harvested by centrifugation, washed with ice-cold 10 mM NaN3, and frozen at −80°C. For some experiments, the protocol was modified as follows: α-factor (50-ng/ml final concentration) or nocodazole (15-μg/ml final concentration) was added 1 h prior to the addition of galactose, and subsequent incubations were performed in media containing the same concentration of α-factor or nocodazole.
For analysis, cell pellets were lysed as described above, and Swe1p-myc was immunoprecipitated by using anti-myc antibody (see above) from samples containing 3 μCi of radioactive label. Immunoprecipitates were washed three times with a lysis buffer, heated for 5 min in 1× sample loading buffer, and separated in SDS–8% polyacrylamide gels. Dried gels were exposed to a Molecular Dynamics (Sunnyvale, Calif.) storage phosphor screen for 24 to 48 h, scanned on a Molecular Dynamics model 445 SI PhosphorImager, and analyzed with ImageQuant, version 1.2 software.
RESULTS
Negative regulation of Swe1p in unperturbed cells by a pathway involving both Hsl1p and Hsl7p.
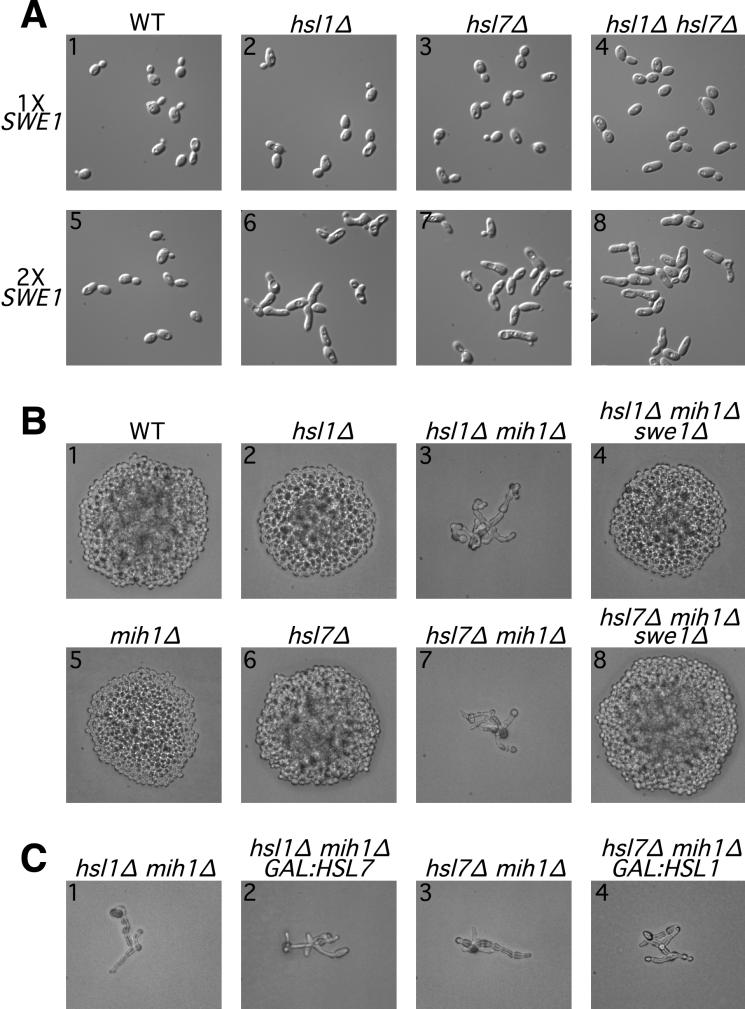
Swe1p-mediated inhibition of Cdc28p leads to a G2 delay during which bud growth continues primarily at the bud tip, resulting in a distinctive elongated-bud morphology (7, 23). The finding that hsl1 and hsl7 mutants exhibited a SWE1-dependent elongated-bud phenotype originally suggested that Hsl1p and Hsl7p might be negative regulators of Swe1p (29). However, the mutant phenotypes are quite variable depending on the strain background and growth conditions (54; see below), raising the question of how generally this conclusion might apply. For example, in our strain background, the deletion of HSL1 or HSL7 did not cause a pronounced phenotype in cells growing exponentially on our standard medium (Fig. 1A, panels 1 to 3). However, when the populations approached the stationary phase, a fraction of the hsl1Δ and hsl7Δ cells displayed elongated buds; this effect was not seen if SWE1 was also deleted (data not shown). In addition, doubling the copy number of SWE1, a manipulation that has little effect on otherwise wild-type cells (Fig. 1A, panel 5), caused a pronounced elongated-bud phenotype in hsl1Δ or hsl7Δ cells even during exponential growth (Fig. 1A, panels 6 and 7).
FIG. 1.
Negative regulation of Swe1p in unperturbed cells by a pathway involving both Hsl1p and Hsl7p. (A) G2 delay (resulting in bud elongation) when SWE1 copy number is doubled in the absence of Hsl1p, Hsl7p, or both. Wild-type (WT) (JMY1469), hsl1Δ (JMY1503), hsl7Δ (JMY1505), and hsl1Δ hsl7Δ (JMY1507) strains and related strains containing an extra copy of SWE1 (JMY1470, JMY1477, JMY1475, and JMY1479) were observed by using differential interference contrast optics during exponential growth (5 × 106 cells/ml) in YEPD medium. (B and C) Genetic interactions among SWE1, HSL1, HSL7, and MIH1. (B) Diploid strains JMY1569 (upper row) and JMY1570 (lower row) were sporulated to generate haploid segregants with the indicated genotypes (confirmed by replica plating and analysis of marker genes). Following tetrad dissection, spores were allowed to grow on YEPD medium for 2 days before the resulting microcolonies were photographed (C). Diploid strains JMY1571 (panels 1 and 2) and JMY1572 (panels 3 and 4) were sporulated, and spores of the indicated genotypes were grown on YEPG medium to induce overexpression of the GAL-regulated genes.
The phosphatase Mih1p antagonizes Swe1p activity by reversing the Swe1p-catalyzed phosphorylation of Cdc28p (45). In the course of other studies, we observed that the steady-state levels of Mih1p declined as populations approached the stationary phase (32), perhaps explaining why hsl1Δ or hsl7Δ strains would show a Swe1p-dependent G2 delay only at high cell densities (see above). To investigate further the interplay among Swe1p, Mih1p, and Hsl1p-Hsl7p, we constructed double-mutant and triple-mutant strains. Although the deletion of MIH1 (Fig. 1B, panel 5), like the deletion of HSL1 or HSL7 (Fig. 1B, panels 2 and 6), has little effect in otherwise wild-type cells, both hsl1Δ mih1Δ and hsl7Δ mih1Δ double mutants were inviable and produced extremely elongated buds, suggestive of G2 arrest (Fig. 1B, panels 3 and 7). The deletion of SWE1 restored normal growth to these strains (Fig. 1B, panels 4 and 8), confirming that the G2 arrest was a result of Swe1p activity.
Taken together, these data suggest that Hsl1p and Hsl7p indeed function generally as negative regulators of Swe1p. This negative regulation appears to play a minor role in unperturbed cells unless the activity of Swe1p is artificially increased or the activity of Mih1p is decreased to the point that it cannot effectively antagonize the action of the unregulated Swe1p.
To ask if Hsl1p and Hsl7p function in the same or separate pathways for regulation of Swe1p, we constructed hsl1Δ hsl7Δ double-mutant strains. Like the hsl1Δ and hsl7Δ single mutants, a double mutant that was otherwise wild type showed no conspicuous abnormalities during exponential growth (Fig. 1A, panel 4). Upon approach to the stationary phase (data not shown) or when the SWE1 copy number was doubled (Fig. 1A, panel 8), the double mutant displayed elongated buds, but this phenotype did not appear more severe than those of the single mutants. This panel of strains provides a very sensitive readout of Swe1p activity, because doubling the SWE1 dose has a large effect. Thus, the absence of an additive or synergistic effect in the double mutant implies that it has no more active Swe1p than the single mutants, suggesting that Hsl1p and Hsl7p act in the same pathway to inhibit Swe1p.
We attempted to order the actions of Hsl1p and Hsl7p in this pathway by testing whether the overexpression of either gene could compensate for the loss of the other. However, no such rescue was observed (Fig. 1C), suggesting that neither Hsl1p nor Hsl7p can effectively down-regulate Swe1p on its own and hence that these proteins play interdependent roles in the same step of Swe1p control.
Function of Hsl1p and Hsl7p during the morphogenesis checkpoint response.
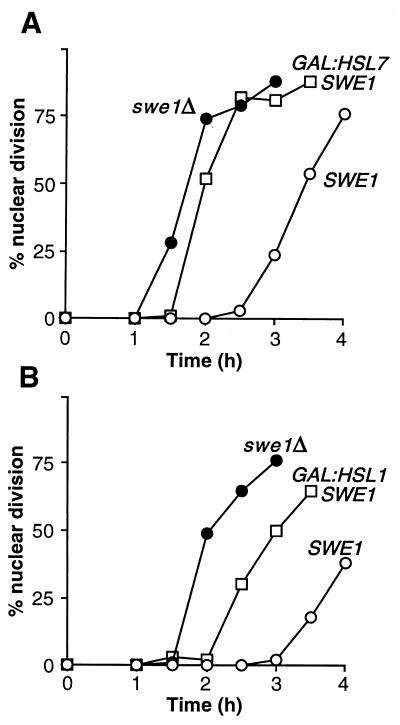
If Hsl1p and Hsl7p really act directly as negative regulators of Swe1p, then excess Hsl1p or Hsl7p might cause an inappropriate inhibition of Swe1p that could override the G2 delay imposed by the morphogenesis checkpoint. To test this possibility, we generated strains that expressed HSL1 or HSL7 under the control of the regulatable GAL1 promoter and also harbored a temperature-sensitive cdc24 mutation. At a restrictive temperature, the cdc24 mutant is unable to polarize actin and consequently exhibits a prolonged Swe1p-dependent G2 delay (1, 22, 52) (Fig. 2A). However, when Hsl7p was overexpressed by growing the GAL:HSL7 strain on galactose, the G2 delay was virtually eliminated, and the cells traversed mitosis with kinetics similar to those of cells that lacked Swe1p altogether (Fig. 2A). The corresponding experiment for Hsl1p was more complicated because the constitutive overexpression of Hsl1p caused a Swe1p-independent growth defect associated with severe morphological aberrations (data not shown). However, when this problem was circumvented by growing the GAL:HSL1 cells on galactose for only a short time, it was clear that the overexpression of Hsl1p could also override the checkpoint-induced G2 delay of cdc24 cells (Fig. 2B). It is not clear whether the less complete override of the checkpoint in this experiment reflects intrinsic differences in the abilities of Hsl1p and Hsl7p to inhibit Swe1p or simply a lesser degree of overexpression in the Hsl1p experiment. Nevertheless, these data suggest strongly that both Hsl1p and Hsl7p are bona fide negative regulators of Swe1p.
FIG. 2.
Override of the morphogenesis checkpoint by overexpression of HSL1 or HSL7. (A) Strains DLY657 (cdc24-1 SWE1) (○), DLY690 (cdc24-1 swe1Δ) (●), and JMY1284 (cdc24-1 SWE1 GAL:HSL7:LEU2) (□) were grown overnight at 24°C (permissive temperature) in YEPG to induce the GAL promoter, synchronized in G1 phase with α-factor, and released into fresh YEPG at 37°C (restrictive temperature), where actin polarization and bud formation did not occur. At 30-min intervals, cells were fixed and stained to monitor the kinetics of nuclear division; 200 cells were scored in each sample. (B) Strains DLY657 (○), DLY690 (●), and JMY1495 (cdc24-1 SWE1 GAL:HSL1:LEU2) (□) were grown at 24°C in YEPS (noninducing nonrepressing medium for the GAL promoter) and arrested in G1 phase with α-factor. Galactose was then added to induce the GAL promoter, and 1 h later the cells were released into fresh YEPG at 37°C and monitored as described above.
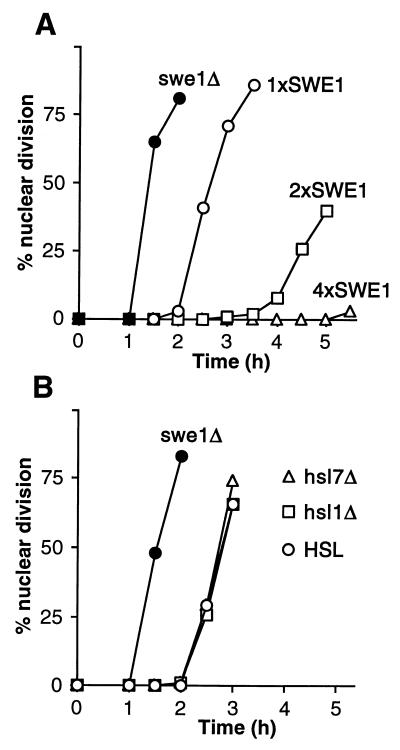
To investigate whether Hsl1p and Hsl7p normally play a role in the morphogenesis checkpoint response, we examined this response in cells with HSL1 or HSL7 deleted. Under the conditions used, the duration of the G2 delay is very sensitive to SWE1 gene dosage (49) (Fig. 3A). Nonetheless, the deletion of HSL1 or HSL7 did not produce a detectable lengthening of the G2 delay in the cdc24 mutant (Fig. 3B). The simplest interpretation of this result is that Hsl1p and Hsl7p are already turned off when the checkpoint response is induced, so that deleting the genes produces no additional increase in Swe1p activity (or, thus, in G2 delay) under these conditions.
FIG. 3.
Equivalent checkpoint delays in Hsl+ and Hsl− cells. (A) Strains JMY1472 (cdc24-1 SWE1) (○), DLY690 (cdc24-1 swe1Δ) (●), JMY1494 (cdc24-1 2xSWE1) (□), and JMY1493 (cdc24-1 4xSWE1) (▵) were grown at 24°C (permissive temperature), synchronized in G1 phase with α-factor, and released at 37°C (restrictive temperature), where actin polarization and bud formation did not occur. At 30-min intervals, cells were fixed and stained to monitor the kinetics of nuclear division; 200 cells were scored in each sample. (B) Strains DLY657 (cdc24-1 SWE1) (○), DLY690 (●), JMY1300 (cdc24-1 SWE1 hsl1Δ) (□), and JMY1301 (cdc24-1 SWE1 hsl7Δ) (▵) were synchronized and analyzed as described for panel A.
Periodic accumulation of Hsl1p during the cell cycle.
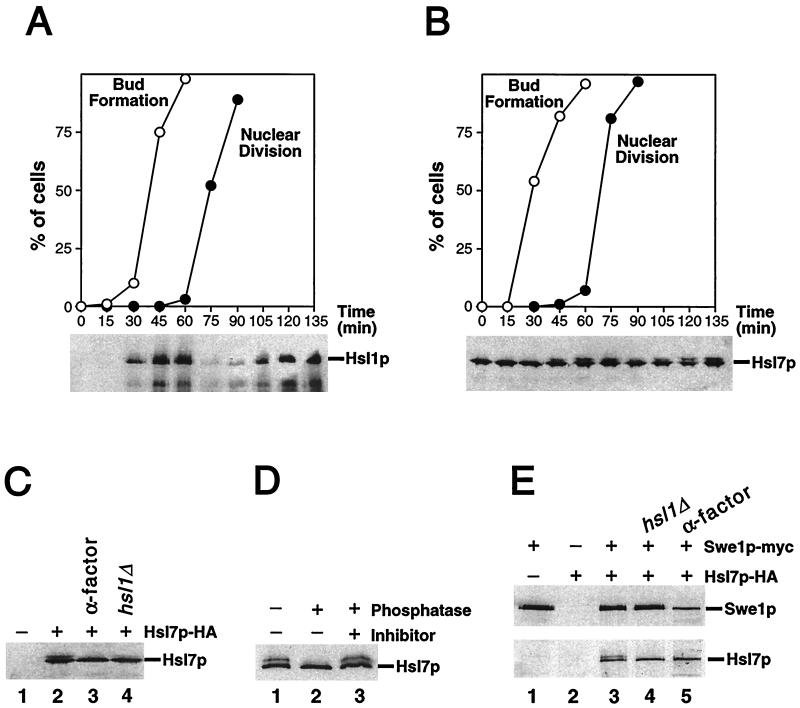
To examine the behavior of Hsl1p and Hsl7p during the cell cycle, we generated strains expressing epitope-tagged versions of these proteins. The Hsl1p-myc and Hsl7p-HA proteins (expressed under the control of their own promoters at their normal genomic loci) were fully functional by the criteria that HSL1myc:URA3 mih1ΔLEU2 and HSL7-3HA:kan mih1ΔLEU2 strains were viable and had normal cell morphology (data not shown). In synchronized cells, Hsl1p-myc was absent in G1, accumulated during S phase to a peak in G2/M, and disappeared coincident with nuclear division (Fig. 4A). This pattern of protein accumulation is consistent with the previously described pattern of HSL1 mRNA accumulation, which is periodic with a peak in late G1 (54).
FIG. 4.
Characterization of Hsl1p and Hsl7p. (A) Periodic accumulation of Hsl1p during the cell cycle. Wild-type cells expressing Hsl1p-myc (strain JMY1500) were grown in YEPD, synchronized in G1 phase with α-factor, and released into a fresh medium. Cells were harvested at the indicated times, and separate aliquots were lysed to detect Hsl1p or fixed to monitor bud formation and nuclear division. Hsl1p-myc was immunoprecipitated from lysates containing 200 μg of total protein, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblotted with anti-myc antibody. (B to D) Hsl1p-dependent phosphorylation of Hsl7p during the cell cycle. (B) Wild-type cells expressing Hsl7p-HA (strain JMY1521) were synchronized as described above. Lysates containing 20 μg of total protein were separated by SDS-PAGE, and Hsl7p-HA was detected by immunoblotting with anti-HA antibody. (C) Lysates were prepared from an Hsl7p-HA-expressing strain (JMY1521) that had been arrested in G1 phase with α-factor (lane 3) and from cells of strains expressing or lacking Hsl7p-HA, Swe1p-myc (GAL regulated), and Hsl1p as indicated (lane 1, M-1505; lane 2, M-1537; and lane 4, JMY1539) that had been arrested in G2 phase by growth for 3 h after galactose was added to induce overexpression of Swe1p-myc. Proteins were separated by SDS-PAGE and Hsl7p-HA was detected by immunoblotting with anti-HA antibody. (D) Anti-HA immunoprecipitates were prepared from lysate of a strain (M-1537) expressing Hsl7p-HA and divided into three equal aliquots that were subjected to a mock phosphatase treatment (lane 1), treatment with potato acid phosphatase (lane 2), or treatment with phosphatase together with the phosphatase inhibitor sodium orthovanadate (lane 3). Proteins were separated by SDS-PAGE, and Hsl7p-HA was detected by immunoblotting with anti-HA antibody. (E) Coimmunoprecipitation of Hsl7p-HA with Swe1p-myc. Lysates were prepared from strains expressing Hsl7p-HA, Swe1p-myc (GAL regulated), and/or Hsl1p, as indicated (lane 1, M-1505; lane 2, M-1295; lane 3, M-1537; and lane 4, JMY1539), that had been arrested in G2 phase as described for panel C. Lysate was also prepared from a strain expressing both tagged proteins that had been arrested in G1 phase with α-factor (JMY1521 [lane 5]). Anti-myc immunoprecipitates were prepared from samples containing 200 μg of total protein, separated by SDS-PAGE, and immunoblotted with anti-myc (upper blot) or anti-HA (lower blot) antibody.
Periodic Hsl1p-dependent phosphorylation of Hsl7p.
In contrast to Hsl1p, Hsl7p was present at approximately constant levels throughout the cell cycle (Fig. 4B). However, a fraction of the Hsl7p protein was modified in a cell cycle-dependent manner, as indicated by the periodic appearance of a more slowly migrating species (Fig. 4B). This species was also apparent in cells that had been arrested in G2 by overexpression of Swe1p (Fig. 4C, lane 2), suggesting that Clb1p-4p–Cdc28p activity was not required for Hsl7p modification. Because the appearance of the modified Hsl7p protein was correlated with the peak in Hsl1p abundance during the cell cycle, we tested whether the modification was Hsl1p dependent. Indeed, the modified Hsl7p protein was not detectable in an hsl1Δ strain (Fig. 4C, lane 4). The modified Hsl7p protein disappeared following phosphatase treatment (Fig. 4D), indicating that the modification was phosphorylation. Thus, Hsl1p promotes the periodic phosphorylation of Hsl7p. It is not yet clear whether this effect is direct or indirect.
Hsl1p-independent association of Hsl7p with Swe1p.
To determine if the negative regulation of Swe1p by Hsl7p reflects a physical interaction, we tested for coimmunoprecipitation of these proteins. Indeed, when immunoprecipitates were prepared with anti-myc antibodies from cells expressing both Swe1p-myc and Hsl7p-HA, the latter protein was readily detected by immunoblotting (Fig. 4E, lane 3). In control experiments with cells expressing just one of the tagged proteins, no Hsl7p-HA was detected (Fig. 4E, lanes 1 and 2). Both phosphorylated and unphosphorylated forms of Hsl7p were coimmunoprecipitated with Swe1p (Fig. 4E, lane 3), and the association did not depend on Hsl1p (Fig. 4E, lane 4) and was detectable in cells arrested in G1 by α-factor (Fig. 4E, lane 5). These data are consistent with the hypothesis that Hsl7p is a direct regulator of Swe1p.
Dependence of Swe1p degradation on Hsl1p and Hsl7p during the unperturbed cell cycle.
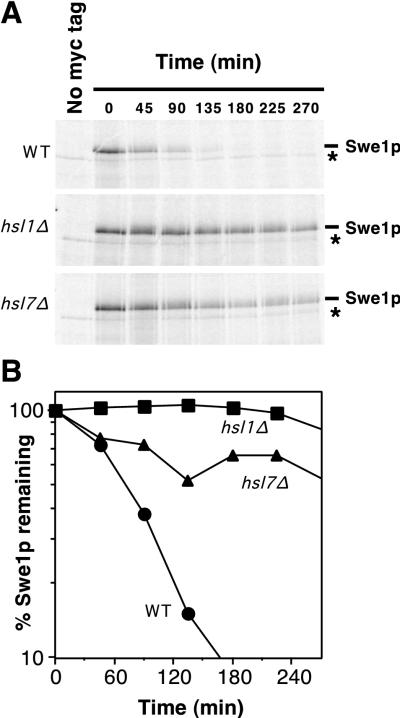
Swe1p is normally stabilized in response to activation of the morphogenesis checkpoint (see the introduction). If the checkpoint acts (at least in part) by down-regulation of Hsl1p and/or Hsl7p, then the deletion of HSL1 or HSL7 should also result in Swe1p stabilization. Indeed, pulse-chase experiments (Fig. 5) showed that Swe1p was dramatically stabilized in both hsl1Δ and hsl7Δ strains, relative to the wild type, suggesting that Hsl1p and Hsl7p are required to target Swe1p for degradation.
FIG. 5.
Stabilization of Swe1p in hsl1Δ and hsl7Δ strains. (A) CDC28Y19F GAL:SWE1myc strains RSY342 (wild type [WT]) (HSL1 HSL7) (top), RSY361 (hsl1Δ HSL7) (middle), and RSY356 (HSL1 hsl7Δ) (bottom) were grown in YEPS and induced to express Swe1p-myc by 10 min of growth in the presence of galactose. The cells were harvested, pulse labeled with [35S]methionine and cysteine for 10 min, harvested again, and resuspended in fresh YEPD (to repress the GAL promoter) containing nonradioactive methionine and cysteine. The amounts of 35S-labeled Swe1p-myc were determined at intervals by immunoprecipitation and SDS-PAGE. Cells of a strain (DLY1) not expressing Swe1p-myc were pulse labeled and processed as described above, providing a control shown in the left-hand lane of each gel. The asterisk indicates a labeled band that is present in cells lacking Swe1p-myc (left lanes) and binds to the protein A beads used for immunoprecipitation. (B) The radioactive signals from the gels shown in panel A were quantitated with a phosphorimager. These experiments were performed with CDC28Y19F strains to avoid potential complications arising from the dependence of Swe1p degradation on Cdc28p activity (48); i.e., if the Swe1p produced during the pulse substantially inhibited Cdc28p, an artifactual stabilization of Swe1p might be observed during the chase period. However, Cdc28pY19F, which lacks the Swe1p phosphorylation site, is largely resistant to inhibition by Swe1p. We confirmed that cell proliferation indeed continued through the pulse-chase protocol in all strains (data not shown).
Cell cycle-specific acceleration of Swe1p degradation by overexpression of Hsl1p.
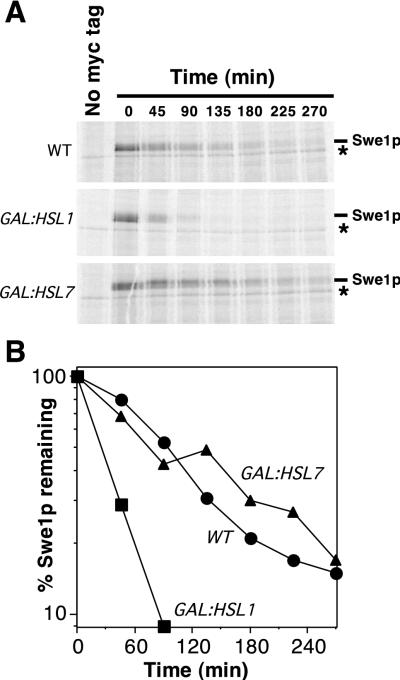
If Hsl1p or Hsl7p is rate limiting for Swe1p degradation, then the overexpression of one or both of these proteins might accelerate Swe1p degradation. To test this possibility, we performed pulse chase experiments in strains that simultaneously overexpressed Swe1p and Hsl1p or Swe1p and Hsl7p. It was observed previously that the overexpression of Swe1p in such experiments slows its degradation, presumably by saturating the capacity of a limiting component involved in Swe1p degradation (48) (also compare WT in Fig. 6 with WT in Fig. 5, in which Swe1p was not overexpressed). Strikingly, the overexpression of Hsl1p (but not of Hsl7p) accelerated Swe1p degradation (Fig. 6), suggesting that Hsl1p levels are rate limiting for Swe1p degradation, at least under conditions of overexpression.
FIG. 6.
Acceleration of Swe1p degradation by overexpression of Hsl1p. (A) CDC28Y19F GAL:SWE1myc strains RSY342 (WT), RSY366 (GAL:HSL1), and RSY370 (GAL:HSL7) were grown in YEPS and induced to overexpress the GAL-regulated genes by addition of galactose for 3 h. The cells were harvested, pulse labeled with [35S]methionine and cysteine for 10 min, harvested again, and resuspended in fresh YEPG containing nonradioactive methionine and cysteine. The amount of 35S-labeled Swe1p-myc was determined by immunoprecipitation and SDS-PAGE. The asterisk indicates a labeled band that is present in cells lacking Swe1p-myc (left lanes) and binds to the protein A beads used for immunoprecipitation. (B) The radioactive signals from the gels shown in panel A were quantitated with a phosphorimager.
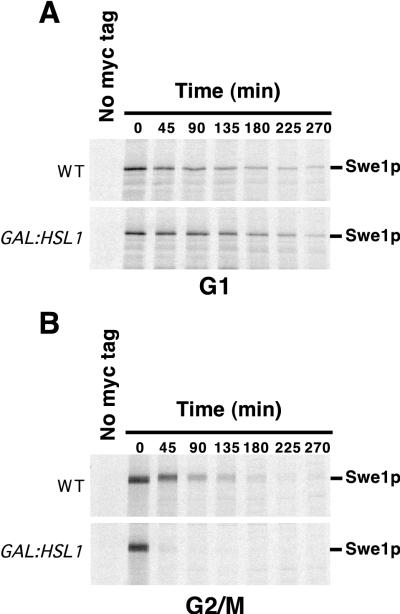
Swe1p is normally stable during G1 and unstable during G2/M (48). Given the data described above, it seemed possible that the stability of Swe1p in G1 cells might be due simply to the absence of Hsl1p. The acceleration of Swe1p degradation upon overproduction of Hsl1p (Fig. 6) might reflect either more efficient degradation during G2/M, inappropriate degradation during G1, or both. To distinguish among these possibilities, we repeated the experiment whose results are shown in Fig. 6 with cells synchronized in G1 with α-factor or in G2/M with nocodazole. As shown in Fig. 7A, Swe1p was stable in G1 cells even when Hsl1p was overexpressed. In contrast, excess Hsl1p promoted more rapid Swe1p degradation in the G2/M-arrested cells (Fig. 7B). Thus, Hsl1p appears to be rate limiting for degradation of overproduced Swe1p, but only at later stages of the cell cycle, suggesting the existence of a cell cycle-regulated step in Swe1p degradation in addition to the periodic accumulation of Hsl1p.
FIG. 7.
Cell cycle specificity of the acceleration of Swe1p degradation by overexpression of Hsl1p. CDC28Y19F GAL:SWE1myc strains RSY342 (WT) and RSY366 (GAL:HSL1) were grown in YEPS and induced to overexpress the GAL-regulated genes by addition of galactose. Just after galactose addition, the culture was split, and α-factor (50 ng/ml) was added to one set (A) while nocodazole (15 μg/ml) was added to the other (B). After incubation for 4 h, the cells were harvested, pulse labeled with [35S]methionine and cysteine for 10 min, harvested again, and resuspended in fresh YEPG containing nonradioactive methionine and cysteine. The labeling and chase media also contained α-factor or nocodazole to maintain the cell cycle arrest throughout. The amount of 35S-labeled Swe1p-myc remaining was determined by immunoprecipitation and SDS-PAGE.
DISCUSSION
Down-regulation of Swe1p by Hsl1p and Hsl7p.
It was observed previously that hsl1 and hsl7 mutants display a Swe1p-dependent G2 delay, suggesting that Hsl1p and Hsl7p act as negative regulators of Swe1p (29). However, many mutants defective for aspects of cell morphogenesis also display Swe1p-dependent G2 delays (22, 33), so that it was not clear whether Hsl1p and Hsl7p were involved primarily in morphogenesis or acted more directly on Swe1p. Our finding that the overexpression of Hsl1p or Hsl7p can override a morphogenesis checkpoint-induced G2 delay provides a strong argument that these proteins function directly in down-regulating Swe1p. This down-regulation appears to depend, at least in part, on targeting Swe1p for degradation. The stability of Swe1p normally varies during the cell cycle: it is moderately stable during G1 and becomes quite unstable in G2/M (48). Our data indicate that both Hsl1p and Hsl7p are required for the rapid degradation of Swe1p and that Hsl1p is rate limiting for Swe1p degradation, at least under conditions of Swe1p overexpression. In addition, the genetic data indicate that Hsl1p and Hsl7p act in a single pathway to down-regulate Swe1p and that neither one can effectively down-regulate Swe1p in the absence of the other.
It has been shown previously that Swe1p degradation involves its ubiquitination by a complex, called SCFMet30, that contains the F box protein Met30p and the ubiquitin-conjugating enzyme Cdc34p (19). Detailed analyses of the ubiquitination of the Cdc28p inhibitor Sic1p by a similar complex, SCFCdc4, have revealed that phosphorylation of Sic1p is a necessary prelude to its ubiquitination and subsequent degradation (3, 12, 31, 51). Swe1p, like Sic1p, becomes hyperphosphorylated prior to its degradation (48). Thus, it is plausible that a part of this hyperphosphorylation is due to Hsl1p (in conjunction with Hsl7p) and that this phosphorylation targets Swe1p for recognition and ubiquitination by SCFMet30. Swe1p degradation also requires Clb-Cdc28p activity (48). Our data indicate that Hsl1p overexpression accelerates Swe1p degradation during G2/M but is unable to do so during G1. Taken together, the data suggest that Clb-Cdc28p activity is required either to activate Hsl1p-Hsl7p or to collaborate with Hsl1p-Hsl7p in targeting Swe1p for degradation (Fig. 8A).
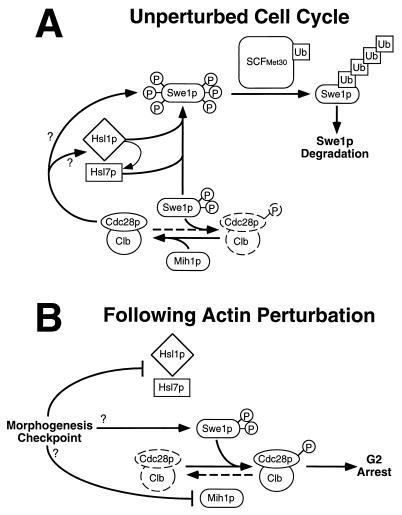
FIG. 8.
Model for control of the S. cerevisiae cell cycle by the morphogenesis checkpoint. During the unperturbed cell cycle (A), Hsl1p and Hsl7p promote Swe1p hyperphosphorylation (P) leading to recognition by SCFMet30, which catalyzes polyubiquitination (Ub), resulting in the subsequent degradation of Swe1p. Clb-Cdc28p complexes also contribute to Swe1p degradation, acting either through Hsl1p-Hsl7p or separately on Swe1p. Although Hsl7p can bind to Swe1p in the absence of Hsl1p, the fate of the complex may be regulated by Hsl1p-mediated phosphorylation of Hsl7p. The net effect of these interactions is to promote Swe1p degradation in G2/M phase, which promotes the activation of Clb-Cdc28p complexes and hence the unimpeded progression of cells through mitosis. (Note, however, that the activity of Mih1p appears normally to be high enough to keep Clb-Cdc28p largely in the active state even if Swe1p degradation does not occur on schedule.) Following perturbation of the actin cytoskeleton (B), the morphogenesis checkpoint inhibits Hsl1p-Hsl7p, thus preventing Swe1p degradation. However, Swe1p stabilization alone is insufficient to promote G2 arrest, and other checkpoint-responsive pathways must also act to regulate Swe1p and/or Mih1p, so that the balance of their activities is tilted in favor of the phosphorylation and inhibition of Cdc28p, leading to G2 arrest.
The conclusion that Hsl1p is a negative regulator of Swe1p was anticipated because the Hsl1p kinase domain (although not the large noncatalytic domain) is closely related to that of S. pombe Nim1, which is a negative regulator of Wee1. However, the finding that Hsl1p targets Swe1p for degradation was surprising, because Nim1 has been shown to phosphorylate Wee1 directly, inhibiting its kinase activity (9, 40, 55). We do not yet known whether Hsl1p can phosphorylate Swe1p directly and/or inhibit Swe1p kinase activity. Similarly, it is not known whether Nim1 influences Wee1 degradation in S. pombe, and it is possible that the phosphorylation of Swe1p by Hsl1p and of Wee1 by Nim1 functions both to inhibit their kinase activity and to target them for degradation. However, it is also possible that Hsl1p phosphorylates other substrates that are important for Swe1p degradation. One candidate is Hsl7p, which is in constant abundance through the cell cycle but is phosphorylated in an Hsl1p- and cell cycle-dependent manner; the cell cycle dependence may reflect the periodic accumulation of Hsl1p.
In addition to Hsl1p, there are two other Nim1-related kinases in S. cerevisiae, Gin4p and Ycl024Wp/Kcc4p (2, 4, 17, 25, 29, 39, 54). All of these kinases have diverged from Nim1 to similar extents (4, 17, 25), suggesting that they might play a redundant role in the down-regulation of Swe1p. However, Hsl7p phosphorylation did not occur, and Swe1p was completely stabilized in hsl1Δ mutants even though Gin4p and Kcc4p were present. These data suggest that Hsl1p (together with Hsl7p) plays a unique role in Swe1p regulation that is not shared with Gin4p or Kcc4p. The hypothesis that the Nim1-related kinases play distinct roles in S. cerevisiae is supported by the finding that Gin4p, but not Hsl1p or Kcc4p, is important for proper septin organization (25, 27).
Hsl7p is also important for targeting Swe1p for degradation. Hsl7p is a member of a protein family that is highly conserved across species but has no known biochemical activity or informative sequence motifs. Coimmunoprecipitation experiments indicate that Hsl7p is physically associated with Swe1p and that this association does not require the phosphorylation of Hsl7p or the presence of Hsl1p. Furthermore, the Hsl7p-Swe1p association also occurs in G1-arrested cells, in which Swe1p is not hyperphosphorylated (48). These data suggest that the Swe1p-Hsl7p interaction might be a very early step in the targeting of Swe1p for degradation, preceding the accumulation of Hsl1p, the phosphorylation of Hsl7p, and the hyperphosphorylation of Swe1p (Fig. 8A).
In contrast to the apparent role of Hsl7p as a negative regulator of Swe1p, a recent study with S. pombe failed to identify a role for the Hsl7p homolog, Skb1, in regulating Wee1 (15). Instead, genetic interactions between skb1, wee1, and cdc25 mutations suggested that Skb1, like Wee1, acts to delay entry into mitosis (15). It is not clear how to reconcile the seemingly opposite roles suggested for Hsl7p and Skb1, but it seems possible that the genetic interactions observed in S. pombe reflect an effect of skb1 mutations in perturbing morphogenesis (or other processes) rather than a direct effect of Skb1 in cell cycle control.
Hsl1p, Hsl7p, and the morphogenesis checkpoint.
Although Hsl1p and Hsl7p down-regulate Swe1p in unperturbed cells, hsl1Δ and hsl7Δ mutations appear to have no effect on Swe1p function in cdc24 mutant cells undergoing a checkpoint response. This suggests either that Hsl1p and/or Hsl7p is itself down-regulated under checkpoint-inducing conditions (the model we prefer [Fig. 8B]) or that Swe1p is somehow protected from the action of Hsl1p and Hsl7p under these conditions.
We and others have shown that Hsl1p and Hsl7p, together with a fraction of cellular Swe1p, are localized to the mother-bud neck in a septin-dependent manner (4, 27, 36, 47). In addition, Barral et al. (4) made the intriguing observation that Hsl1p kinase activity (assayed by autophosphorylation in vitro) declined in septin mutants, suggesting that Hsl1p activity is dependent upon its proper localization. Because cdc24 mutants fail to assemble a septin ring (20, 41), this suggests that Hsl1p would be inactive in these mutants, thus providing a mechanism for the proposed down-regulation of Hsl1p-Hsl7p by the morphogenesis checkpoint.
However, the model that the morphogenesis checkpoint-induced G2 delay is due simply to Hsl1p delocalization in response to septin defects (4) is inconsistent with much of the available data. First, the checkpoint override observed in a cdc24 mutant upon overexpression of Hsl1p or Hsl7p indicates that these proteins are able to function in the absence of assembled septins or neck structures, at least when present in excess. Second, Swe1p-dependent G2 delays are triggered by several conditions (e.g., osmotic shock or treatment with latrunculin A in wild-type cells; tpm1Δ mutations) that affect the actin cytoskeleton but do not appear to affect septin organization or the mother-bud neck (33). Indeed, treatment with latrunculin A did not displace Hsl1p or Hsl7p from the neck (27). These conditions all cause Swe1p stabilization (48), suggesting that Hsl1p and Hsl7p are no longer effective in targeting Swe1p for degradation. It seems possible that the ability of Hsl1p and Hsl7p to down-regulate Swe1p can itself be down-regulated by more than one mechanism, but further research will be needed to test this hypothesis and to elucidate the pathway(s) involved.
Whatever the detailed mechanism(s) responsible for the checkpoint-induced stabilization of Swe1p, the data presented here also demonstrate that relieving the down-regulation of Swe1p by Hsl1p and Hsl7p is not sufficient to explain the checkpoint-induced G2 delay. In our strain background, the deletion of HSL1 or HSL7 did not induce a detectable G2 delay in otherwise wild-type cells during exponential growth, indicating that the G2 delay caused by the morphogenesis checkpoint must involve additional pathways. Such pathways could include an increase in Swe1p specific activity, a change in Swe1p localization, or an inhibition of Mih1p, the phosphatase that counteracts Swe1p-mediated phosphorylation of Cdc28p (Fig. 8B). The last mechanism certainly has the potential to combine very effectively with Hsl1p-Hsl7p down-regulation, as hsl1Δ mih1Δ and hsl7Δ mih1Δ cells undergo a lethal Swe1p-dependent G2 arrest.
Conclusions.
We report here that Hsl1p and Hsl7p play a direct role in targeting Swe1p for degradation, and we suggest that down-regulation of the Hsl1p-Hsl7p pathway plays a role in the morphogenesis checkpoint response. Homologs of Hsl1p and Hsl7p have been identified in many species (8, 15, 21, 29). In S. cerevisiae, the control of Swe1p degradation is linked to the morphogenesis checkpoint. However, Wee1 degradation in Xenopus is regulated by the DNA replication checkpoint (34). It may be that a conserved degradation control pathway has been linked to different checkpoint sensors in different cells.
ACKNOWLEDGMENTS
We thank Y. Barral, D. Kellogg, A. Myers, M. Snyder, and J. Thorner for communicating results prior to publication. We thank Sally Kornbluth, Robin Wharton, John York, and Jake Harrison for critical reading of the manuscript, and the members of the Lew and Pringle labs for stimulating interactions.
J.N.M. and M.S.L. were supported by NIH postdoctoral fellowships GM18455 and GM15766, respectively. This work was supported by NIH grant GM31006 to J.R.P. and by NIH grant GM53050 and funds from the Searle Scholars Program/The Chicago Community Trust to D.J.L.
REFERENCES
- 1.Adams A E M, Pringle J R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman R, Kellogg D. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 4.Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi E, Pringle J R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Booher R N, Deshaies R J, Kirschner M W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chervitz S A, Aravind L, Sherlock G, Ball C A, Koonin E V, Dwight S S, Harris M A, Dolinski K, Mohr S, Smith T, Weng S, Cherry J M, Botstein D. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman T R, Tang Z, Dunphy W G. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 1993;72:919–929. doi: 10.1016/0092-8674(93)90580-j. [DOI] [PubMed] [Google Scholar]
- 10.Dunphy W G. The decision to enter mitosis. Trends Cell Biol. 1994;4:202–207. doi: 10.1016/0962-8924(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 11.Edgington N P, Blacketer M J, Bierwagen T A, Myers A M. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol Cell Biol. 1999;19:1369–1380. doi: 10.1128/mcb.19.2.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 13.Ghiara J B, Richardson H E, Sugimoto K, Henze M, Lew D J, Wittenberg C, Reed S I. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991;65:163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- 14.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 15.Gilbreth M, Yang P, Bartholomeusz G, Pimental R A, Kansra S, Gadiraju R, Marcus S. Negative regulation of mitosis in fission yeast by the shk1 interacting protein skb1 and its human homolog, Skb1Hs. Proc Natl Acad Sci USA. 1998;95:14781–14786. doi: 10.1073/pnas.95.25.14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb M H, Marcus S. The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc Natl Acad Sci USA. 1996;93:13802–13807. doi: 10.1073/pnas.93.24.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter T, Plowman G D. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs C W, Adams A E M, Szaniszlo P J, Pringle J R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser P, Sia R A L, Bardes E G S, Lew D J, Reed S I. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, H. B., B. K. Haarer, and J. R. Pringle. Unpublished results.
- 21.Krapivinsky G, Pu W, Wickman K, Krapivinsky L, Clapham D E. pICln binds to a mammalian homolog of a yeast protein involved in regulation of cell morphology. J Biol Chem. 1998;273:10811–10814. doi: 10.1074/jbc.273.18.10811. [DOI] [PubMed] [Google Scholar]
- 22.Lew D J, Reed S I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim H H, Goh P-Y, Surana U. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longtine M S, Fares H, Pringle J R. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtine M S, McKenzie III A, DeMarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Longtine, M. S., C. L. Theesfeld, J. N. McMillan, E. Weaver, J. R. Pringle, and D. J. Lew. Unpublished data. [DOI] [PMC free article] [PubMed]
- 28.Lorenz M C, Muir R S, Lim E, McElver J, Weber S C, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 29.Ma X-J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 31.Mathias N, Johnson S L, Winey M, Adams A E M, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMillan, J. N. Personal communication.
- 33.McMillan J N, Sia R A L, Lew D J. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael W M, Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282:1886–1889. doi: 10.1126/science.282.5395.1886. . (Erratum, 283:35, 1999.) [DOI] [PubMed] [Google Scholar]
- 35.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 36.Myers, A. 1999. Personal communication.
- 37.Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 38.Ohi R, Gould K L. Regulating the onset of mitosis. Curr Opin Cell Biol. 1999;11:267–273. doi: 10.1016/s0955-0674(99)80036-2. [DOI] [PubMed] [Google Scholar]
- 39.Okuzaki D, Tanaka S, Kanazawa H, Nojima H. Gin4 of S. cerevisiae is a bud neck protein that interacts with the Cdc28 complex. Genes Cells. 1997;2:753–770. doi: 10.1046/j.1365-2443.1997.1590358.x. [DOI] [PubMed] [Google Scholar]
- 40.Parker L L, Walter S A, Young P G, Piwnica-Worms H. Phosphorylation and inactivation of the mitotic inhibitor Wee1 by the nim1/cdr1 kinase. Nature. 1993;363:736–738. doi: 10.1038/363736a0. [DOI] [PubMed] [Google Scholar]
- 41.Pringle J R, Bi E, Harkins H A, Zahner J E, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- 42.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson H E, Wittenberg C, Cross F R, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 44.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 45.Russell P, Moreno S, Reed S I. Conservation of mitotic controls in fission and budding yeast. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- 46.Sherman F, Fink G, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 47.Shulewitz M J, Inouye C J, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sia R A L, Bardes E S G, Lew D J. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 1998;17:6678–6688. doi: 10.1093/emboj/17.22.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sia R A L, Herald H A, Lew D J. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 52.Sloat B F, Adams A E M, Pringle J R. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stueland C S, Lew D J, Cismowski M J, Reed S I. Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3744–3755. doi: 10.1128/mcb.13.6.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka S, Nojima H. Nik1: a Nim1-like protein kinase of S. cerevisiae interacts with the Cdc28 complex and regulates cell cycle progression. Genes Cells. 1996;1:905–921. doi: 10.1046/j.1365-2443.1996.d01-213.x. [DOI] [PubMed] [Google Scholar]
- 55.Wu L, Russell P. Nim1 kinase promotes mitosis by inactivating Wee1 tyrosine kinase. Nature. 1993;363:738–741. doi: 10.1038/363738a0. [DOI] [PubMed] [Google Scholar]