Abstract
Independent but closely spaced DNA binding sites for Stat3 and c-Jun are required for maximal enhancer function in a number of genes, including the gene encoding the interleukin-6 (IL-6)-induced acute-phase response protein, α2-macroglobulin. In addition, a physical interaction of Stat3 with c-Jun, based on yeast two-hybrid interaction experiments, has been reported. Here we confirm the existence of an interaction between Stat3 and c-Jun both in vitro, with recombinant proteins, and in vivo, during transient transfection. Using fragments of both proteins, we mapped the interactive sites to the C-terminal region of c-Jun and to two regions in Stat3, within the coiled-coil domain and in a portion of the DNA binding domain distant from DNA contact sites. In transient-transfection experiments with the α2-macroglobulin enhancer, Stat3 and c-Jun cooperated to yield maximal enhancer function. Point mutations of Stat3 within the interacting domains blocked both physical interaction of Stat3 with c-Jun and their cooperation in IL-6-induced transcription directed by the α2-macroglobulin enhancer. While the amino acid sequences and the three-dimensional structures of Stat3 and Stat1 cores are very similar, fragments of Stat1 failed to bind c-Jun in vitro. Although Stat1 binds in vitro to the gamma interferon gene response (GAS) element in the α2-macroglobulin enhancer, Stat1 did not stimulate transcription, nor did Stat1 and c-Jun cooperate in driving transcription controlled by the α2-macroglobulin enhancer.
Clustered specific DNA binding sites for an array of activating transcription factors, plus proteins that bend DNA to facilitate contact between bound proteins, have been documented for a number of vertebrate genes (15, 21, 25, 37). These composite structures have been called enhanceosomes (8). The T-cell receptor alpha (15) and beta-interferon (IFN-β) (25) enhanceosomes, which are assembled in response to dimerization of the T-cell receptor or double-stranded RNA, respectively, have been most thoroughly and profitably explored. Two classes of genes that are very likely dependent on enhanceosome assembly have received a great deal of attention: genes expressed in a tissue-specific manner that acquire multiple binding proteins during development, and genes that are acutely activated by an external stimulus. The latter structures hold appeal for study because they can be examined in cultured cells, in which induced synchronous changes occur in all of the cells under observation, potentially allowing the acute assembly and disassembly of proteins in an enhanceosome to be revealed.
The STAT family of transcription factors is activated by the attachment of polypeptide ligands to specific cell surface receptors and, after tyrosine phosphorylation, dimerization, and translocation to the nucleus, can participate within minutes in gene activation (11). It seems likely that STAT molecules bind DNA regions where preenhanceosome structures exist (26, 27) and that the arrival of an activated STAT dimer(s) is the key to forming an active enhanceosome (27). Such a possibility is suggested by experiments showing closely spaced binding sites for STATs and other proteins in the response elements of a number of genes (17, 24, 27, 41). Furthermore, DNase and permanganate treatment of cell nuclei revealed proteins bound at or near Stat1 sites before polypeptide treatment. This was followed by detection of STAT molecules binding close to the same DNA regions after induction (26).
One intensively studied set of physiologically important genes that are transcriptionally induced in the liver are the acute-phase response proteins, whose levels increase in the wake of bacterial infections and other toxic assaults. Interleukin-6 (IL-6) stimulation of hepatocytes, via the activation of Stat3, is thought to be the main trigger for inducing the acute-phase genes (18). One of the best-studied enhancers of acute-phase response genes is the α2-macroglobulin enhancer (20) (reviewed in reference 18), a DNA fragment 100 bases long with binding sites for both Stat3 (also called a GAS site) and for AP-1, which includes members of the Fos, Jun, and activating transcription factor (ATF) families of transcription factors. Extracts from liver nuclei of IL-6-treated animals or transformed hepatocytes (hepatoma cells) in culture indicated induction of binding to this region. Since Stat3 and c-Jun interacted in yeast two-hybrid assays and cooperated in maximizing the transcription responses of reporter genes containing the ∼100-bp enhancer (30, 31), it seemed likely that this genomic region would form a STAT-dependent enhanceosome. The experiments presented here were designed to explore this possibility and to uncover the physical basis of c-Jun–Stat3 cooperation. We report evidence, in vitro and in vivo, for an interaction between a region within c-Jun and specific sites within Stat3. Mutations in the proposed contact residues of Stat3 both reduce c-Jun–Stat3 protein interaction and disrupt the cooperation between these two proteins that is required for maximal IL-6-dependent gene activation driven by the α2-macroglobulin enhancer.
MATERIALS AND METHODS
Cell culture and antibodies.
Human HepG2 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 15% fetal bovine serum (HyClone). Human 293T cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Anti-Stat3 serum and anti-Stat1 serum were produced in rabbits as previously described (32, 33, 44, 45) and diluted 1:1,000 for Western blotting or 1:10 for supershifting DNA-protein complexes in electrophoretic mobility shift assays (EMSAs). Monoclonal c-Jun antibody (Santa Cruz) was diluted 1:500 for Western blotting. Anti-phospho-Stat3 (Tyr 705) antibody (New England Biolabs) was used at a 1:5,000 dilution and anti-phospho-Stat3 (Ser 727) antibody (New England Biolabs) was used at a 1:1,000 dilution for Western blotting. Anti-FLAG monoclonal antibody (Kodak/IBI) was used at a 1:1,000 dilution for Western blotting and at a 1:10 dilution for supershifting DNA-protein complexes. Human IL-6 was purchased from Boehringer Mannheim and was used at a concentration of 5 ng/ml. The recombinant soluble form of the human IL-6 receptor was purchased from R&D Systems and was used at a concentration of 5 ng/ml. IFN-γ was a gift from Amgen Inc. and was used at 5 ng/ml for 30 min.
Plasmid constructions.
Glutathione S-transferase (GST) fusion constructs containing various Stat3 fragments were generated by PCR using primers containing 5′ BamHI sites and 3′ NotI sites. Amplified products were digested with appropriate enzymes and cloned into pGEX-5X-1 (Pharmacia). Construction of the expression vector pRcCMV (Invitrogen), containing Stat1 and Stat3, was as previously described (39). The expression vector for c-Jun, pRSV-Jun, was a gift from Daniel Besser (The Rockefeller University). The luciferase reporter plasmid was constructed by releasing the α2-macroglobulin promoter fragment from α2-macroglobulin-TK-CAT-WT (a gift from Daniel Nathans, Johns Hopkins University School of Medicine) (30) and inserting it into vector pTATA (a gift from Daniel Besser), which has the TATA box of the thymidine kinase gene. The luciferase reporter gene containing three Ly6E sites (3xLy6E) was previously described (39). pCMVβ-gal was purchased from Invitrogen.
GST fusion protein association assay.
Preparation of GST fusion proteins was carried out by induction of Escherichia coli containing the fusion vector at 30°C with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Following lysis by sonication, GST proteins were purified on glutathione-Sepharose beads (Pharmacia) and washed extensively with phosphate-buffered saline. For in vitro translation of proteins, full-length c-Jun cDNA was used for program-coupled transcription and translation reactions (TNT; Promega) in the presence of 35S-labeled methionine (DuPont/NEN) according to the manufacturer’s directions. GST protein association assays with translation products or HepG2 extracts were carried as previously described (43). After being washed, the resulting complexes were eluted in sodium dodecyl sulfate (SDS) gel loading buffer and separated by SDS–10% polyacrylamide gel electrophoresis (PAGE).
Transfection experiments.
Transient transfections were performed in 24-well plates with 2.5 × 105 cells per well by the calcium phosphate method as instructed by the manufacturer (GIBCO/BRL). The total amount of DNA transfected was brought up to 2 μg per well by addition of sonicated salmon sperm DNA. Twenty-four hours after transfection, cells were treated with either IL-6 or IFN-γ for 6 h or left untreated. Luciferase assays were performed according to the manufacturer’s directions (Promega), and β-galactosidase (β-gal) assays were done as previously described (2). All results shown in the figures are luciferase activities normalized against the internal-control β-gal activity. Each assay was performed in triplicate, in a single experiment, and repeated in three different experiments with similar results.
Cell extracts and immunoblotting.
Whole-cell lysates and nuclear extracts were prepared as described previously (35). Immunoprecipitation and Western blotting were carried out by standard methods (2).
Site-directed mutagenesis.
The QuickChange site-directed mutagenesis method (Promega) was used to introduce mutations into Stat3. Primer 5′ CACCCAACAGCCGCCGTAGCAACAGAGAAGCAGVAGATG 3′ was used to create the V137A mutant, 5′ GCCGTAGTGACAGAGAAGGCACAGATGTTGGAGCAGCAT 3′ was used to create the Q141A mutant, 5′ GCCGTAGTGACAGAGAAGCAGCAGATGGCAGAGCAGCATCTTCAGGATGTC 3′ was used to create the L144A mutant, 5′ ATGTTGGAGCAGCATGCTCAGGATGTCCGGAAGC 3′ was used to create the L148A mutant, 5′ GCAGCATCTTCAGGATGCACGGAAGCGAGTGCAGG 3′ was used to create the V151A mutant, and 5′ CAACTCAGGAAATTTGACCAGCAACGCGACTGCCGTGGCAAACTGGACACCAGTCTTG 3′ was used to create the TKR mutant. (Underlined residues are codon mutations.)
EMSA.
Nuclear extracts (∼2 to 3 μg of protein) from IL-6-treated 293T cells transfected with FLAG-tagged Stat3 constructs were incubated with 1 ng of 32P-labeled M67 probe (38) for 20 min at room temperature. Nuclear extracts (2 to 3 μg) from HepG2 cells, either untreated and treated with either IL-6 or IFN-γ, were incubated with 32P-labeled α2MGAS probe, which contains the GAS element (underlined) in the α2-macroglobulin enhancer (5′ AATCCTTCTGGGAATTC 3′). The protein-DNA complexes were analyzed by EMSA as previously described (13).
RESULTS
Stat3 and Stat1 interact with c-Jun in vivo.
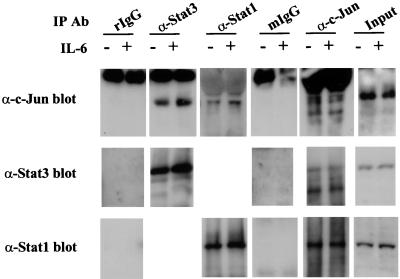
In preliminary experiments using yeast two-hybrid assays, we attempted to detect interactions of Stat1 and -3 with c-Jun. Weak interactions of c-Jun with amino-terminal portions of Stat3, but not of Stat1, were observed (data not shown). Low-dose IL-6 treatment of cells favors activation of Stat3, and IL-6 treatment at higher doses also leads to activation of Stat1 (29, 45). Therefore, we next tested whether coimmunoprecipitation of c-Jun with either Stat1 or Stat3 could be observed when using nuclear extracts from IL-6-treated and untreated HepG2 cells. In both treated and untreated cell extracts, both Stat1 and -3 could be coprecipitated by c-Jun antibody; STAT antibodies also precipitated c-Jun, while control antibodies did not coimmunoprecipitate c-Jun, Stat1, or Stat3 (Fig. 1). Although no definitive conclusions about STAT–c-Jun affinities can be drawn from such experiments or from the earlier yeast two-hybrid results (30), we were encouraged to search for sites of protein-protein interactions between STATs and c-Jun. Since we had earlier demonstrated an interaction between an IFN regulatory factor (IRF) family protein, p48, and Stat1 to occur in the region between 150 and 200 amino acids from the N terminus (in the coiled-coil region of the Stat structure), we anticipated that this region might also contain binding sites for other nuclear proteins (19).
FIG. 1.
Stat1 and Stat3 interact with c-Jun in vivo. Nuclear extracts (300 μg) from IL-6-treated (+) or untreated (−) HepG2 cells were immunoprecipitated (IP) with the antibodies (Ab) indicated at the top, and the immunoprecipitates were then subjected to SDS–10% PAGE followed by Western blotting with the antibodies indicated on the left. Rabbit immunoglobulin (rIgG) and mouse immunoglobulin (mIgG) (Santa Cruz) were used as controls for the Stat1 and -3 or c-Jun immunoprecipitations, respectively. α-c-Jun, anti-c-Jun; α-Stat3, anti-Stat3; αStat1, anti-Stat1.
Mapping the c-Jun–Stat binding domains.
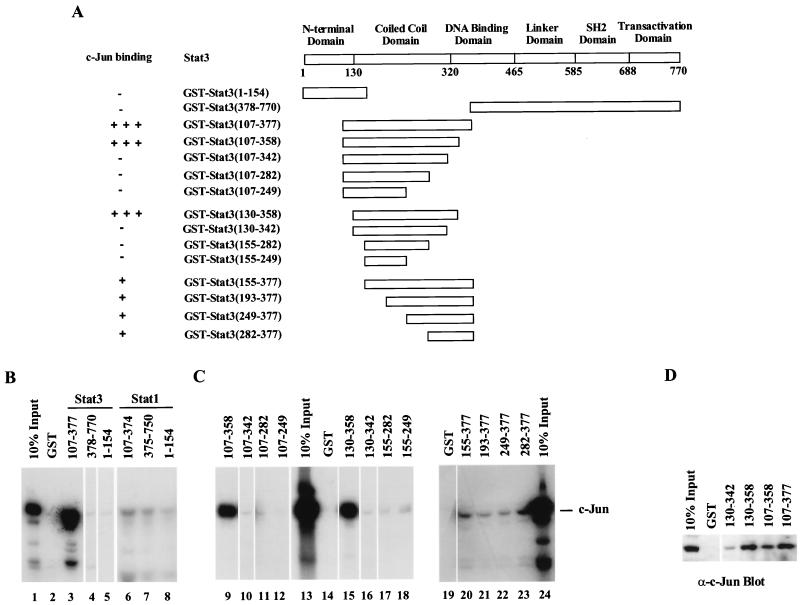
The domain boundaries of Stat1 and -3 in Fig. 2A are marked according to a recent crystallographic study of the Stat3β core dimer on DNA (4). These domains are virtually identical in both Stat3 (4) and Stat1 (9), for which we had the crystallographic coordinates. In order to define domains of Stat1 or -3 that potentially interact with c-Jun, GST fusion proteins containing three different regions of Stat3 (amino acids 1 to 154, 107 to 377, and 378 to 770) and of Stat1 (amino acids 1 to 154, 107 to 374, and 375 to 750) were prepared and coupled to Sepharose beads. Full-length 35S-labeled c-Jun produced by in vitro translation was incubated with the different sections of STATs, and the bound proteins were analyzed by gel electrophoresis and autoradiography (approximately equal amounts of GST fusion proteins were used in each fragment assay) (Fig. 2B). The GST-Stat3(107–377) fusion protein interacted strongly with c-Jun (Fig. 2B, lane 3), while the NH2-terminal [GST-Stat3(1–154)] and COOH-terminal [GST-Stat3(378–770)] Stat3 fusion fragments bound very little c-Jun (Fig. 2B, lanes 4 and 5). Residues 107 to 377 of Stat3 include the entire coiled-coil domain evident in the crystal structure and 57 amino acid residues of the DNA binding domain. In contrast, no fragment of Stat1 tested bound strongly to c-Jun in several attempts with this assay, although weak interactions were observed (Fig. 2B, lanes 6 to 8). These very clear results contrast with the coimmunoprecipitation experiments of Fig. 1. Perhaps the Stat1(107–374) fragment does not fold correctly, such that interaction sites are not presented, or some additional protein is required for Stat1–c-Jun interaction. At any rate, we have not pursued further any potential Stat1–c-Jun physical interaction.
FIG. 2.
Mapping of the regions in Stat1 and -3 that interact with in vitro-translated c-Jun by GST pull-down assays. (A) A schematic diagram of the structural domains of Stat3, and a summary of the interactions between c-Jun and various GST-Stat3 fusion fragments. −, no binding evident; +, weak binding exhibited; +++, strong binding demonstrated. (B) c-Jun interacts with GST-Stat3(107–377). (C) Mapping of the minimal c-Jun-interactive region in Stat3. Equivalent amounts of each GST-Stat3 fusion protein attached to glutathione-Sepharose beads were incubated with in vitro-translated full-length c-Jun labeled with [35S]methionine. The bound proteins were analyzed by SDS–10% PAGE and radiography. (D) Endogenous c-Jun interacts with Stat3-GST fusion proteins. HepG2 cell extracts were incubated with GST-Stat3 fusion proteins bound to glutathione-Sepharose beads. The precipitates were analyzed by SDS–10% PAGE and blotted with an anti-c-Jun antibody (α-c-Jun).
Further deletions from either or both ends of the Stat3(107–377) segment were generated, and GST fusion proteins were prepared to map the minimal region of Stat3 required for the observed in vitro c-Jun binding (Fig. 2A and C). Equivalent amounts of the different bead-bound GST fusion protein were again incubated with in vitro-translated full-length c-Jun. Residues 130 to 358 of Stat3 were essential and sufficient for c-Jun binding (Fig. 2C, lane 15). Deletion of N-terminal residues up to residue 154 decreased c-Jun binding (lane 20), and deletion of C-terminal residues 343 to 358 abolished c-Jun binding (lane 16). Thus, these two regions were candidates for areas that contain residues involved in c-Jun binding.
To determine whether the Stat3 fusion proteins could bind endogenous c-Jun from HepG2 whole-cell extracts, three interacting Stat3-GST fusion fragments were incubated with HepG2 cell extracts. The protein was eluted from the Stat3-bound beads, separated by SDS-PAGE, and immunoblotted with c-Jun antibody (Fig. 2D). Consistent with the results obtained with in vitro-synthesized c-Jun, the negative control, GST-Stat3(130–342), showed very weak c-Jun binding, but three other Stat3 fragments [GST-Stat3(130–358), GST-Stat3(107–358), and GST-Stat3(107–377)] all reacted strongly with the c-Jun in the cell extracts.
Stat3-interactive region in c-Jun lies within residues 105 to 334.
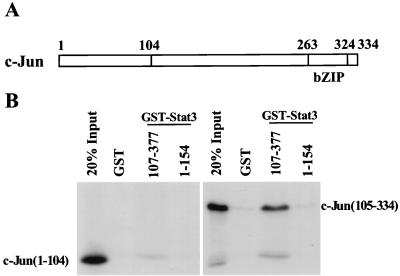
To define the Stat3 binding segment of c-Jun, the N-terminal region containing residues 1 to 104 and the C-terminal region containing residues 105 to 334 of c-Jun were labeled with 35S by in vitro translation. These labeled products were incubated with the GST-Stat3 fragments containing either residues 107 to 377 or 1 to 154. While the N-terminal region of c-Jun did not bind to GST-Stat3(107–377), the C-terminal region of c-Jun was bound strongly to GST-Stat3(107–377) (Fig. 3B). The C-terminal segment of c-Jun contains the bZIP region of c-Jun(263–324) that, in association with c-Fos and DNA, was studied crystallographically (16). Since the residue 263 to 324 region of c-Jun engages in dimerization and DNA binding, it is tempting to speculate that the residue 105 to 263 region of c-Jun contains amino acids that might contact Stat3 when the two proteins are bound simultaneously to DNA.
FIG. 3.
Mapping of the Stat3-interactive region in c-Jun by GST pull-down assays. (A) Schematic diagram of the structural domains of c-Jun. The fragments of c-Jun that were in vitro translated were residues 1 to 104 and 105 to 334. (B) The fragment containing residues 105 to 334 of c-Jun is sufficient to bind to GST-Stat3(107–377). bZIP, basic leucine zipper.
Site-directed mutagenesis in two regions of Stat3.
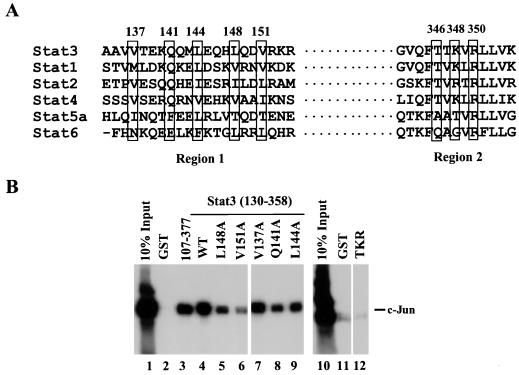
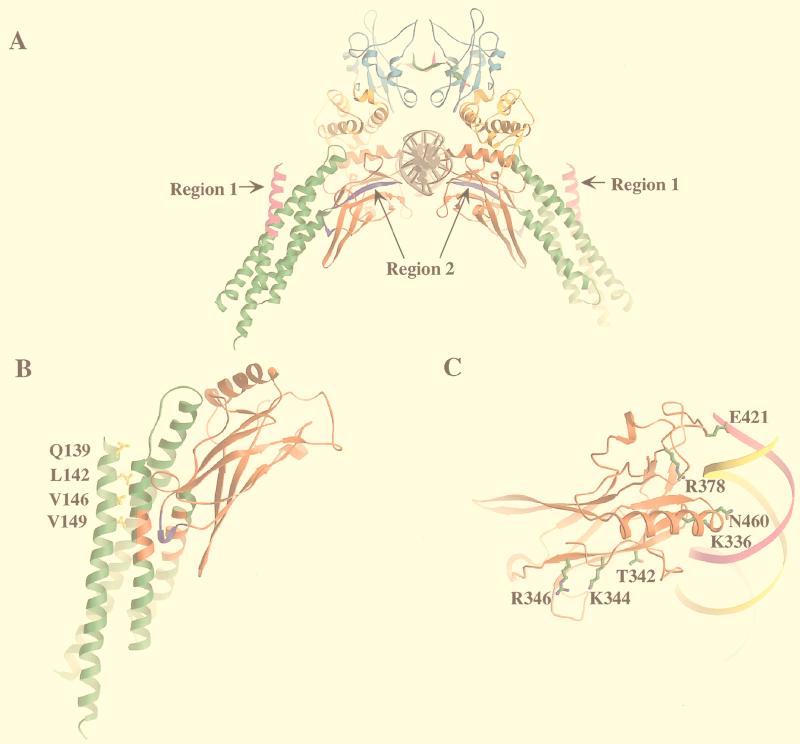
In order to identify specific residues of Stat3 that might be important for Stat3–c-Jun interaction, and guided by the deletion results showing that Stat3 residues between 130 and 154 (region 1) and 342 to 358 (region 2) are important in Stat3–c-Jun interaction (Fig. 2A), site-directed mutagenesis was performed in these two regions. Sequence alignment of seven mammalian STAT proteins revealed five conserved residues in region 1 (Fig. 4A). Each of the conserved residues was changed to alanine (Fig. 5B). Region 2 lies toward the NH2-terminal end of the structural domain that contains DNA contact residues; three conserved residues that do not make close contact with DNA were all changed to alanine (Fig. 4A and 5C).
FIG. 4.
Site-directed mutagenesis in regions 1 and 2 of the Stat3 molecule. (A) Sequence alignment of STAT proteins in regions 1 and 2. The five boxed residues in Stat3 were changed to alanine individually. The three boxed residues in region 2 were changed to alanines simultaneously. (B) Three Stat3 mutants showed decreased c-Jun binding. Mutants L148A and V151A (lanes 5 and 6) demonstrated weaker c-Jun binding. Mutant TKR (lane 12) in region 2 lost the c-Jun binding. WT, wild-type GST-Stat3(130–358).
FIG. 5.
Ribbon diagrams of regions 1 and 2, where site-directed mutagenesis was performed, and the corresponding mutated residues in the Stat1 molecule. (A) Two c-Jun-interactive regions in Stat3 are shown in a ribbon diagram of the Stat1 core dimer on DNA. Region 1 is shown in magenta, and region 2 is shown in purple. The coiled-coil domain is shown in green, the DNA binding domain is in red, the linker domain is in orange, and the SH2 domain is in cyan. The tail segments are shown in green and in magenta. (B) Four corresponding mutated residues in region 1 of Stat3 are shown in a ribbon diagram of the coiled-coil domain (green) and DNA binding domain (red) of the Stat1 monomer. M135 in Stat1, the residue corresponding to V137 in Stat3, is not included in the ribbon diagram. (C) Three corresponding mutated residues in region 2 of Stat3 are shown in a ribbon diagram of the DNA binding domain of the Stat1 monomer with DNA.
Stat3 cDNAs encoding region 131 to 358 with the corresponding mutations were expressed as GST fusion proteins and tested for their binding ability to labeled c-Jun. Two region 1 mutants, L148A and V151A, demonstrated a weaker binding of c-Jun (Fig. 4B, lanes 5 and 6). The triple mutation (T346K348R350) in region 2 virtually abolished c-Jun binding (Fig. 4B, lane 12). Thus, it appeared that residues within the coiled-coil domain as well as within the first three β-strands of the DNA binding domain of Stat3 may be involved in the Stat3–c-Jun interaction. To evaluate the functional importance of the c-Jun–Stat3 interactions indicated by these experiments, we employed a transient-transfection analysis (Fig. 6). We included Stat1 in these experiments both to see whether it could supplant Stat3 and as a closely related control protein.
FIG. 6.
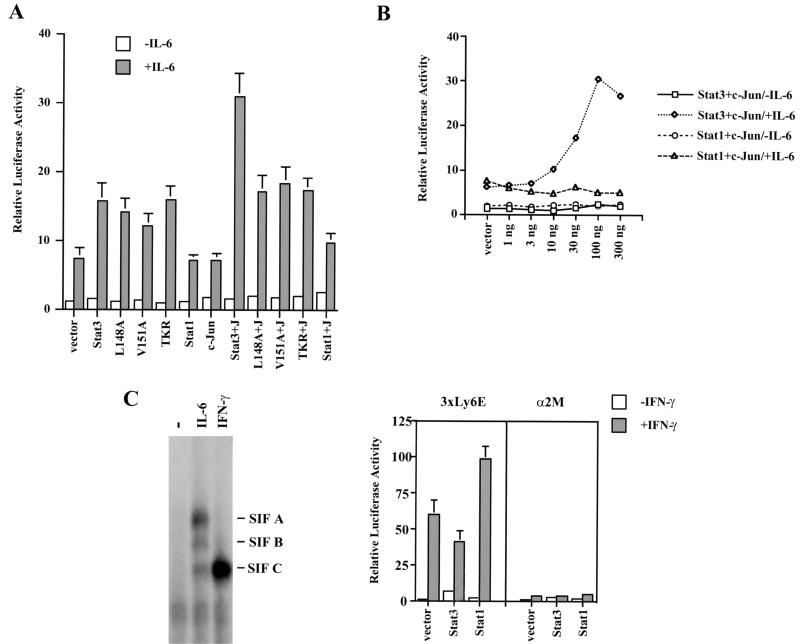
Requirement of Stat3–c-Jun interaction for maximal activation of an IL-6-inducible α2-macroglobulin reporter gene containing both Stat3 and AP-1 binding sites. (A) Cotransfection of wild-type Stat3 and c-Jun boosted the IL-6-dependent response, while Stat1 and three noninteractive Stat3 mutants were ineffective with c-Jun at increasing the IL-6-dependent response. HepG2 cells were transfected with 0.5 μg of luciferase reporter, 0.2 μg of CMVβgal, 50 ng of Stat3, or 50 ng of c-Jun. Twenty-four hours after transfection, cells were left untreated (−) or were treated with 5 ng of IL-6 per ml for 6 h (+) prior to being harvested for luciferase and β-gal assays. Results shown are the means ± standard deviations of data from three experiments. The luciferase activity was normalized against the internal-control β-gal activity and calculated relative to the activity of cells transfected with the vector plasmid pRcCMV. (B) Stat1 was ineffective at cooperating with c-Jun to activate the IL-6-induced transcriptional response. HepG2 cells were cotransfected with 0.5 μg of α2-macroglobulin luciferase reporter, 50 ng of c-Jun, and increasing amounts of either Stat3 or Stat1 as indicated. (C) Stat1 is functionally active on IFN-γ treatment in HepG2 cells. (Left panel) EMSA with 32P-labeled α2MGAS probe. IL-6 treatment led to the activation of Stat1 and Stat3 while IFN-γ treatment led to the activation of Stat1 in HepG2 cells. −, no cytokines added. SIF A, Stat3 homodimer; SIF B, Stat3-Stat1 heterodimer; SIF C, Stat1 homodimer. (Right panel) IFN-γ induced activation of Stat1 with the reporter gene 3xLy6E, but not with the α2-macroglobulin reporter gene (α2M).
Stat3 and c-Jun cooperatively activate an IL-6-inducible α2-macroglobulin reporter gene containing both STAT and c-Jun binding sites.
The DNA segment from the α2-macroglobulin gene (nucleotides −189 to −95) contains a STAT binding site (a GAS element, identified by the TTN5AA motif) and an AP-1 binding site, and both sites are required for maximal IL-6-induced transcription (18, 20, 30). This DNA segment was therefore used as the enhancer of a luciferase reporter gene construct. HepG2 cells express endogenous Stat3, Stat1, and c-Jun, and cells transfected with the reporter gene construct by itself exhibited an approximately sevenfold higher transcriptional response when induced by IL-6 (Fig. 6A, gray bar of vector lane) than when uninduced (Fig. 6A, white bar of vector lane). Thus, supplemental effects of wild-type proteins or interfering effects of mutants must be distinguished from this rather high background. Transfection of the reporter gene and the expression vector for wild-type Stat3 boosted the IL-6-dependent response to about 15-fold higher than that of the uninduced vector alone. Transfection of the c-Jun vector did not increase the level of IL-6-induced transcription. Simultaneous transfection of the vectors for wild-type Stat3 and c-Jun led to an IL-6-dependent response of the reporter gene of approximately 30-fold higher than that of the uninduced vector alone (Fig. 6A, lane Stat3+J). These results, plus the earlier work from other labs showing a requirement for binding sites for each type of factor, is the basis for concluding that there may be a physical interaction between Stat3 and c-Jun in stimulating transcription.
The above results with wild-type Stat3 provided a basis for comparing the functions of mutant Stat3 molecules. All three mutants tested (L148A, V151A, and TKR) by themselves, without extra c-Jun, improved the IL-6-dependent response to almost the same extent as did wild-type Stat3, implying that the mutations did not affect the protein in some drastic or undefined manner (Fig. 6A). However, none of the mutants provided appreciable cooperation in the presence of extra c-Jun. These results support the conclusion that the mutations in regions 1 and 2 of Stat3 (Fig. 4 and 5) block cooperation between Stat3 and c-Jun.
Transient transfection was used to examine more thoroughly the effects of Stat1 on transcription driven by the α2-macroglobulin enhancer. There was no stimulation of transcription of the reporter gene by Stat1 compared to that achieved with the vector alone (Fig. 6A, Stat1 lane), in contrast to the situation in which extra Stat3 was added. Stat1, along with c-Jun, also was ineffective in boosting the IL-6-dependent response (Fig. 6A, Stat1+J lane). Even at high concentrations the Stat1 expression vector failed to cooperate with c-Jun to stimulate transcription (Fig. 6B), whereas increasing the Stat3 concentration together with addition of extra c-Jun progressively supplemented the IL-6 response to a maximum of about fourfold above background (Fig. 6B). We did observe, however, as has been repeatedly reported, that IL-6 at 5 ng/ml, the concentration used in these experiments, activated both Stat1 and Stat3 as DNA binding proteins (Fig. 6C, left panel). The same experiment was also performed with IL-6 at 10 ng/ml, with a consequent stronger induction of Stat1 DNA binding activity. Again, however, there was no evidence of a supplemental transcriptional stimulation by Stat1 (data not shown).
We next questioned whether the α2-macroglobulin promoter would respond to Stat1 if that molecule were stimulated by IFN-γ. In spite of a very strong STAT DNA binding activity, IFN-γ did not activate the α2-macroglobulin enhancer. Moreover, regardless of whether extra Stat1 or Stat3 was supplied (Fig. 6C, right panel), IFN-γ did not activate transcription driven by the α2-macroglobulin promoter. Functional activation by IFN-γ of endogenous and supplemental Stat1 in HepG2 cells did, however, activate the known Stat1- or Stat3-sensitive synthetic promoter, Ly6E (Fig. 6C, right panel), which contains not one but rather three Stat binding sites. This reporter gene, long known to respond to IFN-γ (11, 39), was stimulated about 50-fold by endogenous protein (Stat1), and this response was doubled by additional Stat1 expression. Hence, there is no doubt that Stat1 can be activated in HepG2 cells, but it does not participate in activating transcription driven by the α2-macroglobulin enhancer.
The noninteractive Stat3 mutants can bind DNA and activate noncooperative IL-6-induced transcription.
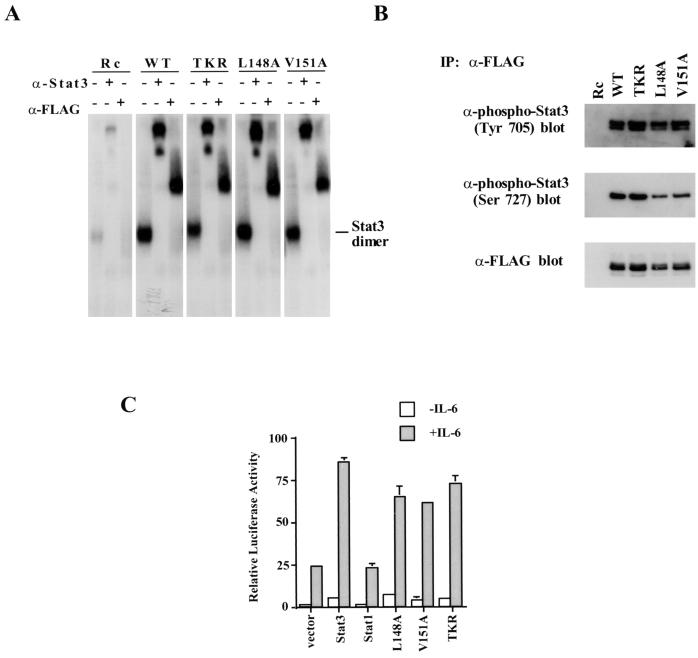
The coiled-coil and DNA binding region Stat3 mutants failed to cooperate with c-Jun, but we tested whether these proteins individually retained the ability to stimulate IL-6-driven transcription. First, the DNA binding abilities of the Stat3 mutants, compared with that of the wild-type protein, were examined by overexpression of proteins in 293T cells, since these cells are known to have relatively low levels of endogenous Stat3 and Stat1 proteins. Cells expressing either wild-type Stat3 or a Stat3 mutant were treated with IL-6 and IL-6 soluble receptor for 30 min, and nuclear extracts were prepared. All three of the Stat3 mutants exhibited DNA-binding abilities indistinguishable from that of wild-type Stat3 in a standard EMSA using a 32P-labeled M67 probe (Fig. 7A). Antibody-mediated supershifting experiments proved that the complexes were specific. The overexpressed proteins were tagged with the FLAG epitope, and both anti-FLAG and anti-Stat3 antibodies retarded the complexes (Stat1 antibody had no effect on these complexes [data not shown]). In addition, both wild-type and mutant proteins were phosphorylated on tyrosine and serine residues, as tested by Western blotting with anti-phospho-Stat3 (Tyr 705) and anti-phospho-Stat3 (Ser 727) antibodies (Fig. 7B). The IL-6-dependent transcriptional activity of three Stat3 mutants was also evaluated in transient-transfection assays using the reporter gene containing three copies of Ly6E sites (3xLy6E), which has been shown to be dependent on Stat3 for IL-6-activated transcription in HepG2 cells (34). All of the proteins were capable of driving transcription of this reporter gene (Fig. 7C), indicating successful activation, dimerization, nuclear translocation, DNA binding, and communication with the basal RNA polymerase II machinery. For all purposes other than c-Jun binding, these proteins are indistinguishable from the wild-type protein.
FIG. 7.
The noninteractive Stat3 mutants can bind DNA and activate IL-6-dependent transcription. (A) The DNA binding abilities of three noninteractive Stat3 mutants were examined by gel mobility shift analysis with 32P-labeled M67 probe. 293T cells were transiently transfected with either wild-type (WT) Stat3 or mutant Stat3 cDNAs treated with IL-6 at a concentration of 5 ng/ml and recombinant human IL-6 soluble receptor at a concentration of 5 ng/ml for 30 min. Nuclear extracts were prepared from these cells, and 3 mg of extract was used in each EMSA. Rc, pRcCMV; α-Stat3, anti-Stat3; α-FLAG, anti-FLAG. (B) Phosphorylation on tyrosine and serine residues of the three Stat3 mutants was indistinguishable from that of wild-type Stat3. Nuclear extracts (75 μ) from transfected 293T cells were immunoprecipitated (IP) with anti-FLAG antibody, and the immunoprecipitates were then subjected to SDS–7% PAGE followed by Western blotting with the antibodies indicated. (C) The IL-6-dependent transcriptional activities of three Stat3 mutants were examined by using the 3xLy6E luciferase reporter. −, no IL-6 added; +, IL-6 present.
DISCUSSION
Transcriptional activation of mammalian genes is now universally regarded as requiring the cooperative effect of many proteins (8, 28). In this work, we employed the now widely used approach of locating required protein-protein interactions between two cooperating transcription factors by in vitro association of domains of each protein. We were successful with GST fusion fragments of Stat3, but not those of Stat1, in locating a segment from residues ∼130 to 358 of Stat3 that bound to the COOH half of c-Jun. Transfection experiments showed that mutations which prevent the protein-protein interaction also prevent cooperative transcriptional activation driven by a promoter containing binding sites for both c-Jun and Stat3. Thus, these experiments are encouraging in terms of eventually determining the importance of Stat3–c-Jun interactions in enhanceosomes that may be dependent on the arrival of a STAT in the nucleus. c-Jun is thought to be a constitutive nuclear molecule, but any STAT necessary for enhanceosome function, together with c-Jun, would require activation in the cytoplasm, translocation into the nucleus, and DNA binding.
From the present experiments, we cannot conclude that other Stat3–c-Jun, or in fact Stat1–c-Jun, interactions do not occur. Moreover, while we found mutations that block both Stat3–c-Jun in vitro interaction and transcriptional cooperation, we cannot state unequivocally that these regions contact each other. Only structural analysis can provide proof of that. The mutations described in this work could, of course, point to regions of contact between c-Jun and Stat3. Interactions within the coiled-coil (L148 and V151) would, from the crystal structure, seem logical, since this region presents extensive surfaces for interaction. However, the finding of a potential contact site between c-Jun and Stat3 within the DNA binding domain was somewhat of a surprise. The Stat DNA binding domain is fairly large compared to other such domains and presents obvious surfaces away from the single surface that interacts with DNA. So the opportunity to interact with additional molecules that may be bound to DNA certainly exists for the DNA binding domain of the STATs. The two regions of Stat3 that interact with c-Jun are reasonably close together (about 20 Å) (Fig. 5A) in the three-dimensional structure, so that binding by a ∼30-kDa domain of c-Jun between those two regions of the STAT protein does not seem unreasonable.
The specificity of the in vitro protein interaction between Stat3 and c-Jun and the ability of Stat3, but not Stat1, to stimulate transcription in the context of the α2-macroglobulin enhancer are noteworthy. The sequence similarities between Stat1 and Stat3 in the two regions where the Stat3–c-Jun contacts occur are strong, and indeed in coprecipitation experiments (Fig. 1) some interaction between Stat1 and c-Jun seemed to occur. However, when protein fragments were used, very little interaction between Stat1 and c-Jun was detectable, especially compared to the strong Stat3 fragment interaction. In addition, transfection experiments using the α2-macroglobulin promoter showed that Stat1 is unable to activate transcription from this promoter either alone or with c-Jun, in contrast to Stat3. Thus, Stat3 and Stat1 are functionally differentiated and Stat3 is not replaceable by Stat1 in gene activation driven by the α2-macroglobulin enhancer.
Several recently published studies suggest that Stat3–c-Jun interaction may occur in other enhanceosomes. For example, ciliary neurotrophic factor activation of the vasoactive intestinal peptide gene (12, 23) involves Stat3 activation. A 180-bp cytokine response element of the vasoactive intestinal peptide promoter includes a Stat3 and the AP-1 site, which are both required for the ciliary neurotrophic factor-induced gene activation (36). A second example involves the matrix metalloproteinases, a group of proteins directly involved in extracellular-matrix breakdown (40). The induced expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases requires an oncostatin M-responsive element containing an AP-1 site and a STAT site, both of which are necessary to achieve maximal induction (22). Finally, in what is perhaps the most intensively studied of all polypeptide-induced promoters, the c-fos promoter, there are Stat3 and AP-1 sites, both of which are required for regulated expression of c-fos in animals from a chromosomal site and in fibroblast cells derived from transgenic animals (27). Thus, it appears that it might be fruitful to pursue the c-Jun–Stat3 interaction as a likely example of a frequently used interaction in enhanceosome formation. We make a final tentative suggestion, although a more thorough evaluation of STAT-activated genes could invalidate it. c-jun is a proto-oncogene, and either overexpression of this gene or increased stability of the resultant protein can result in cellular transformation (1, 3, 5). Constitutively active Stat3 is being increasingly recognized as a cooperating partner in cellular tranformation (6, 14, 42), while Stat1 induced by IFN-α and IFN-γ operates to restrain cellular proliferation (7, 10). Thus, the Stat3–c-Jun interaction could be important for transformation. This will be tested in conjunction with v-src, for which maximal transformation depends on Stat3 (6).
ACKNOWLEDGMENTS
We thank Lois Cousseau for preparing the manuscript; Jacqueline F. Bromberg, Daniel Besser, and Jillian J. Zhang for scientific discussions; and Yanxiang Zhao, David Jeruzalmi, and Xiaomin Chen for preparing the ribbon diagram of the Stat1 molecule.
This work was supported by NIH grants AI32489 and AI34420 to J.E.D. X.Z. is supported by NIH training grant CA09673. M.H.W. is supported by a Cancer Research Institute postdoctoral fellowship.
REFERENCES
- 1.Alani R, Brown P, Binétruy B, Dosaka H, Rosenberg R K, Angel P, Karin M, Birrer M J. The transactivating domain of the c-Jun proto-oncoprotein is required for cotransformation of rat embryo cells. Mol Cell Biol. 1991;11:6286–6295. doi: 10.1128/mcb.11.12.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Baichwal V R, Tjian R. Control of c-Jun activity by interaction of a cell-specific inhibitor with regulatory domain delta: differences between v- and c-Jun. Cell. 1990;63:815–825. doi: 10.1016/0092-8674(90)90147-7. [DOI] [PubMed] [Google Scholar]
- 4.Becker S, Groner B, Muller C W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 5.Bohmann D, Tjian R. Biochemical analysis of transcriptional activation by Jun: differential activity of c- and v-Jun. Cell. 1989;59:709–717. doi: 10.1016/0092-8674(89)90017-2. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg J F, Horvath C M, Besser D, Lathem W W, Darnell J E., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both IFN-α and IFN-γ. Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 10.Chin Y E, Kitagawa M, Su W C, You Z H, Iwamoto Y, Fu X Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 11.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 12.Fann M J, Patterson P H. A novel approach to screen for cytokine effects on neuronal gene expression. J Neurochem. 1993;61:1349–1355. doi: 10.1111/j.1471-4159.1993.tb13628.x. [DOI] [PubMed] [Google Scholar]
- 13.Fried M, Crothers D M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia R, Yu C L, Hudnall A, Catlett R, Nelson K L, Smithgall T, Fujita D J, Ethier S P, Jove R. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 15.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 16.Glover J N M, Harrison S C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos–c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 17.Guyer N B, Severns C W, Wong P, Feghali C A, Wright T M. IFN-γ induces a p91/Stat1α-related transcription factor with distinct activation and binding properties. J Immunol. 1995;155:3472–3480. [PubMed] [Google Scholar]
- 18.Heinrich P C, Horn F, Graeve L, Dittrich E, Kerr I, Muller-Newen G, Grotzinger J, Wollmer A. Interleukin-6 and related cytokines: effect on the acute phase reaction. Z Ernaehrwiss. 1998;37:43–49. [PubMed] [Google Scholar]
- 19.Horvath C M, Stark G R, Kerr I M, Darnell J E., Jr Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 complex. Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T, Tanahashi H, Misumi Y, Sakaki Y. Nuclear factors interacting with an interleukin-6 responsive element of rat alpha 2-macroglobulin gene. Nucleic Acids Res. 1989;17:9425–9435. doi: 10.1093/nar/17.22.9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 22.Korzus E, Nagase H, Rydell R, Travis J. The mitogen-activated protein kinase and JAK-STAT signaling pathways are required for an oncostatin M-responsive element-mediated activation of matrix metalloproteinase 1 gene expression. J Biol Chem. 1997;272:1188–1196. doi: 10.1074/jbc.272.2.1188. [DOI] [PubMed] [Google Scholar]
- 23.Lewis S E, Rao M S, Symes A J, Dauer W T, Fink J S, Landis S C, Hyman S E. Coordinate regulation of choline acetyltransferase, tyrosine hydroxylase, and neuropeptide mRNAs by ciliary neurotrophic factor and leukemia inhibitory factor in cultured sympathetic neurons. J Neurochem. 1994;63:429–438. doi: 10.1046/j.1471-4159.1994.63020429.x. [DOI] [PubMed] [Google Scholar]
- 24.Look D C, Pelletier M R, Holtzman M J. Selective interaction of a subset of interferon-gamma response element-binding proteins with the intercellular adhesion molecule-1 (ICAM-1) gene promoter controls the pattern of expression on epithelial cells. J Biol Chem. 1994;269:8952–8958. [PubMed] [Google Scholar]
- 25.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 26.Mirkovitch J, Decker T, Darnell J E., Jr Interferon induction of gene transcription analyzed by in vivo footprinting. Mol Cell Biol. 1992;12:1–9. doi: 10.1128/mcb.12.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson L M, Kerppola T K, Vendrell M, Luk D, Smeyne R J, Bocchiaro C, Morgan J I, Curran T. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 28.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1997;21:327–335. [PubMed] [Google Scholar]
- 29.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 30.Schaefer T S, Sanders L K, Nathans D. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc Natl Acad Sci USA. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaefer T S, Sanders L K, Park O K, Nathans D. Functional differences between Stat3α and Stat3β. Mol Cell Biol. 1997;17:5307–5316. doi: 10.1128/mcb.17.9.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindler C, Fu X-Y, Improta T, Aebersold R, Darnell J E., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91 and 84 kDa ISGF-3 proteins that are activated by interferon-α. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindler C, Shuai K, Prezioso V R, Darnell J E., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–815. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 34.Sengupta T K, Talbot E S, Scherle P A, Ivashkiv L. Rapid inhibition of interleukin-6 signaling and Stat3 activation mediated by mitogen-activated protein kinases. Proc Natl Acad Sci USA. 1998;95:11107–11112. doi: 10.1073/pnas.95.19.11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shuai K, Schindler C, Prezioso V R, Darnell J E., Jr Activation of transcription by IFN-γ: tyrosine phosphorylation of a 91 kD DNA binding protein. Science. 1992;259:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 36.Symes A, Lewis S, Corpus L, Rajan P, Human S E, Fink J S. STAT proteins participate in the regulation of the vasoactive intestinal peptide gene by the ciliary neurotrophic factor family of cytokines. Mol Endocrinol. 1994;8:1750–1763. doi: 10.1210/mend.8.12.7708062. [DOI] [PubMed] [Google Scholar]
- 37.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 38.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription of Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 40.Werb Z, Alexander C M, Adler R R. Expression and function of matrix metalloproteinase in development. Matrix Suppl. 1992;1:337–343. [PubMed] [Google Scholar]
- 41.Xu X A, Sun Y L, Hoey T. Cooperative DNA binding and sequence selective recognition conferred by the Stat amino terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 42.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong Z, Wen Z, Darnell J E., Jr Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc Natl Acad Sci USA. 1994;91:4806–4810. doi: 10.1073/pnas.91.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong Z, Wen Z, Darnell J E., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]