Abstract
We have previously shown that replication of fission yeast chromosomes is initiated in distinct regions. Analyses of autonomous replicating sequences have suggested that regions required for replication are very different from those in budding yeast. Here, we present evidence that fission yeast replication origins are specifically associated with proteins that participate in initiation of replication. Most Orp1p, a putative subunit of the fission yeast origin recognition complex (ORC), was found to be associated with chromatin-enriched insoluble components throughout the cell cycle. In contrast, the minichromosome maintenance (Mcm) proteins, SpMcm2p and SpMcm6p, encoded by the nda1+/cdc19+ and mis5+ genes, respectively, were associated with chromatin DNA only during the G1 and S phases. Immunostaining of spread nuclei showed SpMcm6p to be localized at discrete foci on chromatin during the G1 and S phases. A chromatin immunoprecipitation assay demonstrated that Orp1p was preferentially localized at the ars2004 and ars3002 origins of the chromosome throughout the cell cycle, while SpMcm6p was associated with these origins only in the G1 and S phases. Both Orp1p and SpMcm6p were associated with a 1-kb region that contains elements required for autonomous replication of ars2004. The results suggest that the fission yeast ORC specifically interacts with chromosomal replication origins and that Mcm proteins are loaded onto the origins to play a role in initiation of replication.
Initiation of replication in eukaryotic cells is tightly regulated during the cell cycle. The regulation presumably involves interactions of specific proteins with specific DNA sequences in replication origins.
Accumulating evidence suggests that the structures of replication origins in eukaryotic species are very different from each other (15, 23). In budding yeast, Saccharomyces cerevisiae, certain chromosome fragments shorter than 200 bp have been shown to replicate autonomously (autonomously replicating sequences [ARSs]) (28, 56). All of the ARSs contain a match to an 11-bp ARS consensus sequence that is essential for replication (9, 41, 63). In contrast, replication origins in other eukaryotes have larger and more complex structures than those in budding yeast. In fission yeast, Shizosaccharomyces pombe, replication of chromosomes is initiated in specific regions that can be isolated as ARS fragments (10, 20, 50). Detailed analyses of fission yeast ARSs have shown that regions required for replication are several times larger and no equivalent short essential sequence exists (19, 42, 52). Instead, clustered adenine/thymine stretches are required for ARS activity (13, 19, 32, 51). On the other hand, for initiation of replication from a distinct site on mammalian chromosomes, multiple regions at distant locations have been shown to be required (2, 34). Thus, budding yeast replication origins might have a rather exceptional structure compared with replication origins in other eukaryotes.
An origin recognition complex (ORC) composed of six subunits has been identified as a protein complex that binds to the budding yeast ARSs (6). Analyses of temperature-sensitive orc2-1 and orc5-1 mutants have shown that the ORC has an essential role in initiation of replication of the yeast genome (38). Recently, homologues of ORC components have been identified in many eukaryotes (11, 22, 24, 37, 45, 54). Depletion of XOrc1p or XOrc2p from Xenopus egg extracts abolishes replication of sperm chromatin (11, 54). Analysis of an orp1-4 mutant of fission yeast showed that orp1+, a counterpart of ORC1, is required for cell cycle progression into the S phase (25). However, it was not elucidated whether the ORC in most eukaryotic species except budding yeast participates in chromosomal replication through its interaction with replication origins or not.
It is proposed that prereplicative complexes (Pre-RCs) assembled at budding yeast replication origins in the G1 phase are required for coupling of S phase with mitosis and for limiting of DNA replication to once per cell cycle (16). While the ORC remains at origins during most or all of the cell cycle, a Cdc6 protein that interacts with the ORC is required for Pre-RC formation (38). The minichromosome maintenance (Mcm) proteins, which are considered to be components of a replication licensing factor that limits replication to once in a cell cycle (8), are loaded onto chromatin after passage through mitosis in a Cdc6-dependent manner (18). Associations of budding yeast Orc1, Cdc6, and Mcm proteins with chromosomal replication origins have been demonstrated by precipitation of chromosome fragments with specific antibodies (chromatin immunoprecipitation [CHIP] assay) (4, 60).
The fission yeast mis5+ gene, originally isolated from a mutant defective in chromosome segregation, encodes the sixth Mcm family protein (SpMcm6) and is required for replication of the fission yeast genome (58). Six fission yeast Mcm proteins, including SpMcm6p and SpMcm2p, which is encoded by nda1+/cdc19+, exist as a heterohexamer complex in exponentially growing cells (1). Although interaction of SpMcm4p, which is encoded by cdc21+, with Orc1p and Cdc18p, a counterpart of Cdc6p, has been suggested (25), it is still unclear whether fission yeast Mcm proteins function at replication origins.
In this study, we focused on whether the fission yeast ORC and Mcm proteins specifically interact with chromosomal replication origins. The finding that Orp1p is associated with chromosomal replication origins during most or all of the cell cycle while SpMcm6p is loaded onto the origins in G1 phase, depending on the expression of Cdc18p, suggests that fission yeast replication origins are recognized by the ORC and a cell cycle-dependent protein complex containing Mcm proteins is specifically formed at the origins.
MATERIALS AND METHODS
Strains and media.
The S. pombe haploid strains used were 972 h− and HM123 h− leu1 (44) and the temperature-sensitive cdc10-129 (46) and cdc25-22 (21) mutant strains. TTY15 h− orp1::5xFLAG-orp1 carries a 5xFLAG tag at the amino terminus of Orp1p (59). OGC12 h− cdc10-129 leu1 orp1::5xFLAG-orp1 and OGC9 h− cdc25-22 orp1::5xFLAG-orp1 were made by standard genetic methods as described earlier (3). Fission yeast strains were cultured in the complete medium YPD (1% yeast extract, 2% polypeptone, 2% glucose) and the minimal medium EMM (43). Media containing 2% agar were used for plating. The cdc10-129 and cdc25-22 derivatives were grown at 25°C and shifted to 36°C for 4 h to arrest cells in the G1 phase and at the G2/M boundary, respectively. To arrest cells in the early S phase, they were cultured with hydroxyurea (HU), an inhibitor of ribonucleotide reductase, at a final concentration of 10 mM at 30°C for 3 h. Transformation was performed by electroporation (26). pREP81-cdc18+ carrying the cdc18+ gene cloned under the control of the thiamine-regulatable nmt1 promoter (5) was provided by P. Nurse. The nmt1 promoter was repressed by adding 5 μg of thiamine per ml to EMM.
Plasmid DNA was prepared from Escherichia coli DH5α transformants as described previously (47). E. coli BL21 (DE3) (57) was used for peptide production for immunization. E. coli strains were grown in LB broth (1% Bacto Tryptone, 0.5% Bacto Yeast Extract, 1% NaCl).
Preparation of antisera.
To express the 43- to 458-amino-acid region of SpMcm6p in E. coli, an XhoI linker (5′CCCTCGAGGG3′) was inserted into the Hpal site of the mis5+ gene (58) and the 1,245-bp XhoI fragment was inserted into the Xhol site of pET-3bx, which had been made by insertion of the XhoI linker into the end-filled BamHI site of pET-3b. For expression of the 202- to 527-amino-acid region of SpMcm2p, a BamHI linker (5′CCGGATCCGG3′) was inserted into the EcoRV site of the nda1+ gene cloned in pET-3 (49) and the 975-bp BamHI fragment was inserted into the BamHI site of pET-3c.
E. coli BL21 (DE3) transformants with pET derivatives grown at 37°C in LB broth containing 30 μg of ampicillin per ml and 60 μg of methicillin per ml were added to 1 mM isopropyl-β-d(−)-thiogalactopyranoside and cultured for 4 h for induction of peptides. The peptides recovered as insoluble inclusion bodies were purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroeluted, and concentrated with Centriprep concentrators (Amicon) for immunization. Rabbit antibodies against the SpMcm6 peptide were affinity-purified with a N-hydroxysuccinimide-activated column (Pharmacia) cross-linked with the peptide as recommended by the manufacturer. The affinity-purified antibody bound to a protein in the fission yeast extract with an apparent molecular mass of 110 kDa on SDS-PAGE. The difference from the value predicted from the amino acid sequence (95.5 kDa) might be due to acidic amino acid residues clustered in SpMcm6p, as reported for SpMcm2p (49).
Preparation of cell extracts and immunoblotting.
S. pombe haploid cells (3 × 108 cells) were resuspended in 0.3 ml of HB buffer (25 mM MOPS [pH 7.2], 15 mM MgCl2, 15 mM EGTA, 60 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 0.1 mM Na3VO4, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 20 μg of leupeptin per ml, 20 μg of aprotinin per ml) and disrupted with acid-washed glass beads using a bead beater (Biospec Products). Proteins in the extracts were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Immobilon; Millipore Corp.). The membranes were incubated for 1 h at room temperature in blocking solution (10 mM Na-PO4 [pH 7.5], 154 mM NaCl, 0.05% Tween 20, 1% bovine serum albumin [BSA], 0.3% skim milk) and then reacted with affinity-purified rabbit anti-SpMcm6, anti-SpMcm2 antibody or mouse anti-FLAG monoclonal antibody M2 (Kodak) at a 1:1,000 dilution in blocking solution for 1 h at room temperature. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G (IgG; Amersham) was used as the secondary antibody. Binding was visualized with the Amersham ECL system.
Preparation of chromatin-enriched fractions.
For separation of chromatin-enriched insoluble fractions from soluble proteins, the method for budding yeast (18) was used with some modifications. Fission yeast cells (5 × 108 cells) placed in ice-cold STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3) for 5 min were incubated at 37°C for 20 min in PEMS (100 mM PIPES [pH 6.9], 1 mM EGTA, 1 mM MgSO4, 1 M sorbitol) containing 1.2 mg of Lysing enzymes (Sigma) per ml and 0.4 mg of Zymolyase 20T (Seikagaku Corporation) per ml and washed with ice-cold 1 M sorbitol in 25 mM morpholinepropanesulfonic acid (MOPS; pH 7.2). Cells resuspended in HBS buffer (HB buffer supplemented with 0.4 M sorbitol) at a concentration of 109 cells/ml were lysed by addition of Triton X-100 at a final concentration of 0.5% for 5 min on ice, and the insoluble fraction was recovered by centrifugation for 15 min at 20,000 × g.
For digestion of cellular DNA with micrococcal nuclease (MNase), the insoluble fraction, washed and resuspended in 50 μl of digestion buffer (HBS containing 2.5 mM CaCl2 instead of EGTA), was incubated with 5 or 50 U of MNase for 2 min at 37°C. After centrifugation at 10,000 × g for 3 min, the pellet was subjected to another digestion as described above and the supernatant sum (100 μl) was used as the MNase supernatant fraction.
Immunostaining of whole cells.
Immunostaining of fission yeast cells was carried out as described previously (3). Briefly, cells fixed in 4% paraformaldehyde for 1 h at room temperature were incubated at 37°C in PEMS containing 1 mg of Lysing enzymes per ml and 0.3 mg of Zymolyase 20T per ml until approximately 10% of the cells lost their cell walls as observed under a microscope. The cells were permeabilized with 1% Triton X-100 in phosphate-buffered saline (PBS) and then transferred onto polylysine-coated glass slides. The slides were sequentially incubated at room temperature for 1 h in blocking solution containing 3% BSA and 0.1% skim milk in PBS, for 12 h with affinity-purified anti-SpMcm6 antibody, and then for 8 h with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG antibody (Amersham). The sample was stained with 0.2 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml in PBS for 5 min and mounted with antifade-containing Vectashield mounting medium (Vector Laboratories).
Preparation of spread nucleoids.
Immunostaining of spread nucleoids was performed as described earlier (7) with modifications. Cells incubated at 37°C in PEMS containing 1.2 mg of Novozyme 234 per ml and 0.4 mg of Zymolyase 20T per ml for 20 min until about 90% of the cells became spheroplasts were washed once and resuspended in MES/sorbitol buffer (0.1 M 2-[N-morpholino]ethanesulfonic acid [pH 6.5], 1 mM EDTA, 0.5 mM MgCl2, 1 M sorbitol) at a concentration of 5 × 108 cells/ml. This cell suspension (20 μl), placed on glass slides was mixed with 40 μl of 1% (wt/vol) paraformaldehyde in 3.4% sucrose, and then 80 μl of 0.1% Lipsol was immediately added. When about 80% of the spheroplasts had burst, 80 μl of 1% (wt/vol) paraformaldehyde in 3.4% sucrose was added and the cell suspension was spread over the glass slide surface and dried for 5 h.
Slides washed with 0.2% Photoflo (Kodak) were incubated for 15 min at room temperature in 3% BSA and 0.1% skim milk in TBS (20 mM Tris-HCl [pH 7.5], 0.15 M NaCl) and reacted with diluted anti-SpMcm6 antibody overnight at 4°C. After several washes with TBS, the samples were reacted with FITC-conjugated anti-rabbit IgG antibody under the same conditions used for the primary antibody, except that the incubation was for 2 h at room temperature in the dark. The samples were then stained with DAPI and mounted as described above.
Centrifugal elutriation.
Cells grown in YPD at 30°C to 107 cells/ml were pumped at 55 ml/min into a Beckman JE-6 elutriation rotor with a 10-ml chamber spinning at 2,500 rpm. Fractions (150 ml) were collected with an increase in the flow rate of 5 ml/min. The small-cell fractions, corresponding to approximately 10% of the total cell population, were concentrated by filtration and cultured in prewarmed YPD for progression through the cell cycle. Populations of septum-containing cells (septation index) were measured every 15 min to monitor the cell cycle.
CHIP.
CHIP was performed as described previously (55, 60), with some modifications. Fission yeast cells (5 × 108) fixed in 1% formaldehyde for 15 min at room temperature and then in 125 mM glycine for 5 min were incubated at 37°C in PEMS with 1.2 mg of Lysing enzymes per ml and 0.4 mg of Zymolyase 20T per ml until about 10% had lost their cell walls. After being washed twice with TBS and resuspended in 0.4 ml of lysis buffer (50 mM HEPES-KOH [pH 7.4], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml), the cells were disrupted with glass beads by using a bead beater for 1 min. The broken cells were sonicated four times for 15 s each until chromatin DNA was sheared into 500- to 700-bp fragments. Supernatant obtained by centrifugation at 12,000 × g for 10 min was used for immunoprecipitation with magnetic beads (Dynal) associated with mouse anti-FLAG or rabbit anti-SpMcm6 antibodies. Immunoprecipitates were washed and DNA was purified as described (55).
PCR amplification by ampli-Taq Gold (Perkin-Elmer) was performed in 30 μl of supplemented buffer with a 1/50 dilution of immunoprecipitated DNA or a 1/7,200 dilution of total DNA and a mixture of three sets of primers. The nucleotide sequences of the primers used were as follows: ars2004F, 5′-ATGGTAGATGGAGAAACGGG-3′; ars2004R, 5′-CACGGCATCTTTCTTCACGA-3′; ars3002F, 5′-TTGGCGCTAAACAATCTCTG-3′; ars3002R, 5′-TCCTTGTCGAACTCAATTGC-3′; nonARS1F, TCGAAGATCCTACCGCTTTC-3′; nonARS1R, 5′-GATTCACATAACCCGCTAGC-3′; nonARS2F, 5′-ATGTATAGCTGGAACGCCTG-3′; nonARS2R, 5′-TTCCTCAAATCACCCCACGT-3′. The concentrations of the ars2004, ars3002, nonARS1, and nonARS2 primers were 0.3, 0.4, 0.5, and 0.25 μM, respectively. The sequences of the primers within the ars2004 locus are as follows: ars2004-1F, 5′-AAAGAAGATTCGCGAGGCAC-3′; ars2004-1R, 5′-CAAGTTTATCCCCACTGATCC-3′; ars2004-2F, 5′-ATGGTAGATGGAGAAACGGG-3′; ars2004-2R, 5′-CACGGCATCTTTCTTCACGA-3′; ars2004-3F, 5′-CGCAGAAGTCCAACCTAAAA-3′; ars2004-3R, 5′-AATGGGAAAGGATGGACGGA-3′; ars2004-4F, 5′-GTGGTGGCAACTTTTGATGAATG-3′; ars2004-4R, 5′-CGATCGTTTTTGTTAGGGTGTG-3′. These primers were used at a concentration of 0.3 μM. An initial incubation for 9 min at 95°C to activate Taq polymerase was followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 53°C, and elongation for 2 min at 72°C and a final extension for 7 min at 72°C. PCR products were separated in 2.3% agarose gels and visualized with 0.5 μg of ethidium bromide per ml. The gel images obtained with a charge-coupled device camera (Epi-UV FA1100; Aisin Cosmos) were processed by using Photoshop (Adobe).
RESULTS
Association of fission yeast Orp1 and Mcm proteins with chromatin.
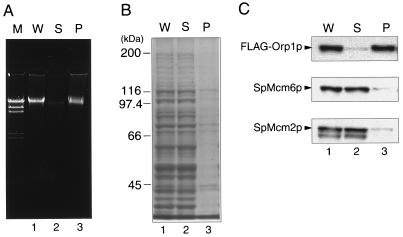
To detect possible association of the fission yeast ORC and Mcm proteins with chromatin, we constructed haploid strain TTY15 carrying 5xFLAG-tagged orp1 under the control of the native promoter in place of the orp1+ gene on the chromosome. Exponentially growing TTY15 cells were gently lysed after digestion of their cell walls, and the chromatin-enriched insoluble fraction was separated from soluble proteins by centrifugation as described previously (18). Chromosomal DNA was precipitated in the pellet (Fig. 1A), while most of the proteins were recovered in the soluble fraction (Fig. 1B). By Western blotting of both fractions with anti-FLAG antibody, most of the FLAG-Orp1p was detected in the insoluble fraction (Fig. 1C). These results showed that Orp1p was associated with insoluble cellular components, including chromatin DNA. In contrast, the results of immunoblotting with the anti-SpMcm6 and anti-SpMcm2 antibodies showed that these proteins existed mostly in the soluble protein fraction of exponentially growing cells (Fig. 1C).
FIG. 1.
Immunoblotting of Orp1 and Mcm proteins in soluble and insoluble fractions of fission yeast extract. Whole cell extract (W; lane 1), soluble supernatant (S; lane 2), and insoluble chromatin-enriched pellet (P; lane 3) were prepared from TTY15 cells carrying 5xFLAG-orp1 as described in Materials and Methods. (A) DNA was extracted with phenol, precipitated with ethanol, and analyzed by electrophoresis in a 0.8% agarose gel containing 0.5 μg of ethidium bromide per ml. HindIII-digested λ phage DNA was included as molecular size markers in lane M. (B) Proteins separated by SDS-PAGE were stained with Coomassie brilliant blue. Positions of some molecular size markers are shown on the left. (C) Proteins separated by SDS-PAGE were immunoblotted with anti-FLAG (top), anti-SpMcm6 (middle), and anti-SpMcm2 (bottom) antibodies. Positions of FLAG-Orp1p, SpMcm6p, and SpMcm2p are indicated. Bands migrating faster than SpMcm2p appear to be unrelated proteins, since their abundance did not increase with overproduction of SpMcm2p under the control of the nmt1 promoter (48).
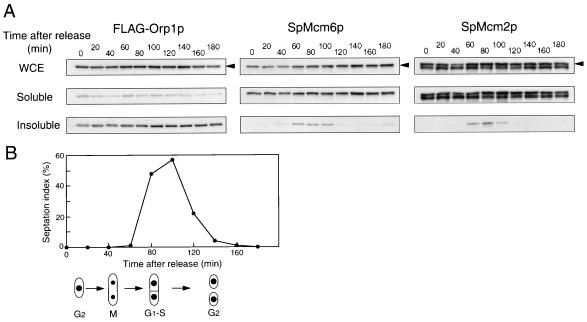
To examine whether SpMcm6p and SpMcm2p exhibited cell cycle-specific binding to chromatin, the temperature-sensitive cdc25-22 mutant cells carrying the FLAG-orp1+ gene were arrested at the G2/M boundary by incubation at the restrictive temperature and released into cell cycle progression at the permissive temperature. Progression through the cell cycle was monitored by assessing populations of septum-containing cells (septation index). Cytokinesis occurs much later than nuclear division in the fission yeast cell cycle, and septum-containing cells are in the G1 and S phases. Western blotting analysis with anti-FLAG antibody showed that the majority of FLAG-Orp1p existed in the insoluble fraction at all time points (Fig. 2A, left blots). In metaphase-arrested nda3-KM311 cells at the restrictive temperature, in G1-arrested cdc10-129 cells, or in wild-type cells arrested early in the S phase by addition of HU, which reduces cellular concentrations of deoxyribonucleotides, most of the FLAG-Orp1p was recovered in the insoluble fraction (data not shown). These results suggested that Orp1p is associated with chromatin or another insoluble component(s) throughout the cell cycle. In contrast, SpMcm6p was not detected in the insoluble fraction in cells arrested in G2/M (0 min) or within 20 to 40 min after release. However, approximately 20% of the SpMcm6p was detected in the insoluble fraction at 60 min and this persisted until the 100-min time point (Fig. 2A, middle blots). SpMcm2p was present in the insoluble fraction at the same time points as SpMcm6p (Fig. 2A, right blots). SpMcm6p and SpMcm2p appeared in the insoluble fraction as the septation index increased (Fig. 2B), suggesting that these proteins are associated with chromatin during the G1 and S phases.
FIG. 2.
Chromatin association of Orp1 and Mcm proteins in synchronized cells. OGC9 (h− cdc25-22 orp1::5xFLAG-orp1) cells were incubated at 36°C for 4 h to produce arrest at the G2/M boundary and released at the permissive temperature (25°C). (A) Every 20 min after release, whole cell extract (WCE), the soluble fraction, and the insoluble chromatin fraction were analyzed by immunoblotting with anti-FLAG, anti-SpMcm6, or anti-SpMcm2 antibodies. The amounts of proteins were adjusted by immunoblotting with anti-tubulin antibody. The arrowheads indicate the positions of FLAG-Orp1p, SpMcm6p, and SpMcm2p. (B) The septation index is shown along with schematic illustrations of corresponding phases of the cell cycle.
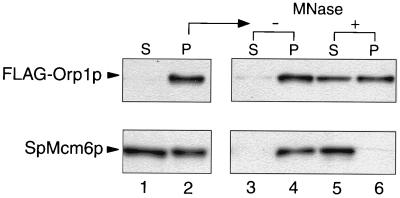
We next examined the effects of MNase digestion on the association of FLAG-Orp1p and SpMcm6p with the insoluble fraction. In cells arrested at the early S phase by addition of HU, almost all of the FLAG-Orp1p and about half of the SpMcm6p were associated with the insoluble fraction (Fig. 3, lanes 1 and 2). Association of more abundant SpMcm6p in the insoluble fraction of HU-arrested cells compared with that of synchronized cdc25-22 cells might be due to a difference between the populations of G1 and S-phase cells. Incubation without MNase did not cause dissociation of FLAG-Orp1p or SpMcm6p from the fraction (Fig. 3, lanes 3 and 4). Upon incubation with MNase, however, the SpMcm6p was completely released (Fig. 3, bottom, lanes 5 and 6). In contrast, only about half of the FLAG-Orp1p was released into the soluble fraction (Fig. 3, top, lanes 5 and 6), even with a 10-fold higher concentration of MNase (data not shown). The effects of MNase on the distribution of Orp1p were essentially the same when exponentially growing cells were examined (48). Thus, most of the Mcm6p present in the insoluble fraction was associated with chromatin DNA while some Orp1p molecules might be anchored to an insoluble component(s) other than chromatin DNA.
FIG. 3.
Dissociation of Orp1p and SpMcm6p from the insoluble fraction by MNase digestion. TTY15 cells were arrested by addition of 10 mM HU and incubation at 30°C for 3 h, and the soluble fraction (S; lane 1) and insoluble pellet (P; lane 2) were separated by centrifugation and analyzed by immunoblotting with anti-FLAG or anti-SpMcm6 antibody. The insoluble pellet was incubated in the absence (lanes 3 and 4) or presence (lanes 5 and 6) of 2 U of MNase and then recentrifuged.
Requirement of cdc18+ expression for association of SpMcm6p with chromatin.
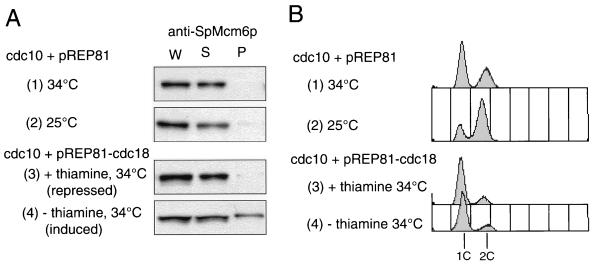
In fission yeast cells, G1- and S-specific expression of various genes is regulated by cdc10+ (40). In cdc10-129 mutant cells arrested in the G1 phase at the restrictive temperature, SpMcm6p was not present in the chromatin fraction (Fig. 4A, part 1), suggesting that its chromatin association depended on certain gene products regulated by cdc10+. It has been shown that expression of cdc18+, a counterpart of budding yeast CDC6, is regulated by cdc10+ (31). To examine whether Cdc18p is required for chromatin association of SpMcm6p, we used cdc10-129 cells transformed with pREP81-cdc18+ carrying the cdc18+ gene placed under the control of the nmt1 promoter, which is repressed in the presence of thiamine (5). By repressing the nmt1 promoter at the restrictive temperature, SpMcm6p was hardly detected in the chromatin fraction (Fig. 4A, part 3). In contrast, with constitutive expression of cdc18+ at the restrictive temperature, approximately one-third of the SpMcm6p was associated with chromatin (Fig. 4A, part 4). Flow cytometry analysis showed that most of the cdc10-129 cells expressing cdc18+ at the restrictive temperature contained 1C DNA content, suggesting that DNA replication did not occur under these conditions (Fig. 4B, part 4). Although the cdc18+ gene regulated by the nmt1 promoter has been shown to suppress the defect of the cdc10-129 mutation (31), cdc10-129 cells transformed with pREP81-cdc18+ grew very slowly at 34°C (data not shown). Some other factor(s) regulated by cdc10+ may not have been activated in G1-arrested cdc10-129 cells.
FIG. 4.
Effect of constitutive expression of Cdc18p on association of SpMcm6p with chromatin. OGC12 cells transformed with a pREP81 vector carrying the nmt1 promoter (parts 1 and 2) or pREP81-cdc18+ (parts 3 and 4) were cultured at 25°C in EMM without thiamine, and then portions of the culture were shifted to 34°C for 3 h in the presence (part 3) or absence (parts 1 and 4) of 5 μg of thiamine per ml. (A) Proteins in the whole cell extract (W), soluble fraction (S), and insoluble pellet (P) were separated by SDS-PAGE and immunoblotted with anti-SpMcm6 antibody. (B) DNA contents of cells were analyzed by FACScan. Positions of 1C and 2C DNA contents of one (1c) and two (2c) copies are indicated.
Cell cycle-dependent localization of SpMcm6p at discrete foci on spread nucleoids.
We examined the cellular localization of SpMcm6p in exponentially growing wild-type cells by immunostaining them with affinity-purified anti-SpMcm6 antibody. As shown in Fig. 5A, part b, SpMcm6-signals were located exclusively in the nuclei. The majority of exponentially growing fission yeast cells were in G2 phase. The nuclei in the binuclear cells (shown by the arrowheads in Fig. 5A, parts a and b), which were assumed to be in post-M to S phase, were also stained by the anti-SpMcm6 antibody. Furthermore, SpMcm6 signals were located in the nuclei of G1-arrested cdc10-129 mutant cells, G2/M-arrested cdc25-22 cells, and early-S-arrested wild-type cells cultured in the presence of HU (48). Nuclear localization of SpMcm6p throughout the cell cycle was thus indicated.
FIG. 5.
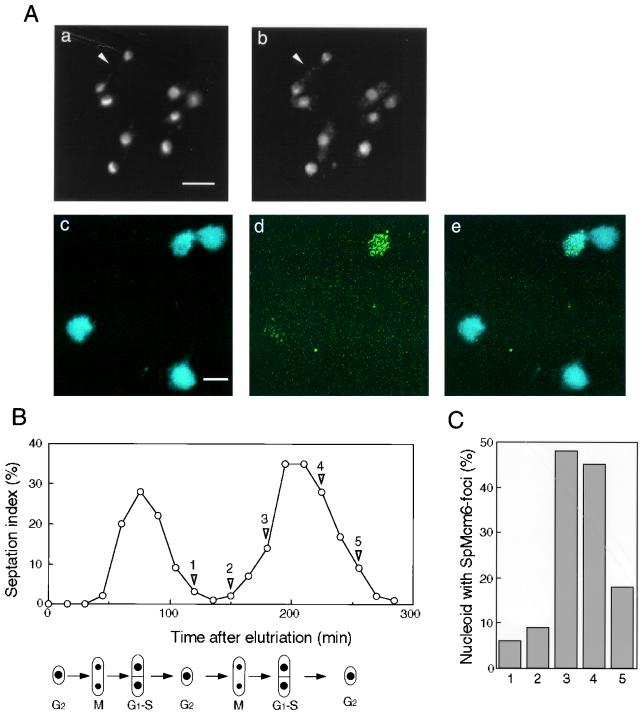
Subnuclear localization of SpMcm6p. (A) Exponentially growing strain 972 cells were fixed and stained with DAPI (a) or immunostained with affinity-purified anti-SpMcm6 antibody and FITC-conjugated anti-rabbit IgG (b). A binuclear cell in post-M to S phase is indicated with an arrowhead in parts a and b. Spread nucleoids prepared as described in Materials and Methods were stained with DAPI (c) or immunostained with anti-SpMcm6 antibody (d). Merged images of DAPI and immunostaining are shown in part e. Bars, 5 μm. (B) Small G2 phase cells collected by centrifugal elutriation were incubated in YPD at 30°C. The population of cells with septums (septation index) was measured every 15 min. (C) At the indicated time points (points 1 to 5, shown by arrowheads), spread nucleoids were immunostained with anti-SpMcm6 antibody. Although some foci partly overlapped, they were separable on enlarged images. The number of foci per nucleoid was determined for approximately 100 nucleoids, and percentages of cells containing more than five foci are shown.
To examine the subnuclear localization of SpMcm6p, we employed a method by which chromatin-associated proteins are retained on spread nuclear materials called nucleoids while soluble proteins are washed out (7, 35). DAPI staining of nucleoids of exponentially growing cells showed the chromosomal DNA to be spread over an area about five times larger than an intact nucleus (Fig. 5A, part c). SpMcm6 signals were not observed in most nucleoids by immunostaining with anti-SpMcm6 antibody. However, approximately 20% of the nucleoids contained SpMcm6p signals, which were observed as 5 to 50 discrete foci with various sizes and shapes (Fig. 5A, part d). The number of foci in a nucleoid was about 15, on average. The results showed that SpMcm6p was associated with distinct regions of the chromatin in a certain fraction of exponentially growing cells.
To examine whether SpMcm6 foci represent cell cycle-specific association of SpMcm6p with chromatin, cells synchronized by centrifugal elutriation were analyzed by immunostaining. As shown in Fig. 5B, the septation index increased during two periods, from 45 to 105 min and from 165 to 255 min. SpMcm6 foci were hardly observed on the nucleoids at time point 1 (120 min) or 2 (150 min), when most cells were in G2 phase (Fig. 5C). However, the population of SpMcm6 focus-containing nucleoids increased to about 50% at time points 3 (180 min) and 4 (225 min) while it was reduced at time point 5 (255 min). These results showed that SpMcm6 foci were observed in the G1 and S phases. Consistent with these results, SpMcm6 foci accumulated in cells arrested in the early S phase by addition of HU but were not observed in G2/M-arrested cdc25-22 cells or in G1-arrested cdc10-129 cells (48). From these results, we concluded that SpMcm6p was associated with discrete regions of chromatin in the G1 and S phases. We failed to localize FLAG-Orp1p on spread nucleoids due to weak immunostaining signals, although the protein was localized in the nuclei throughout the cell cycle (48).
Localization of Orp1p and SpMcm6p at chromosomal replication origins.
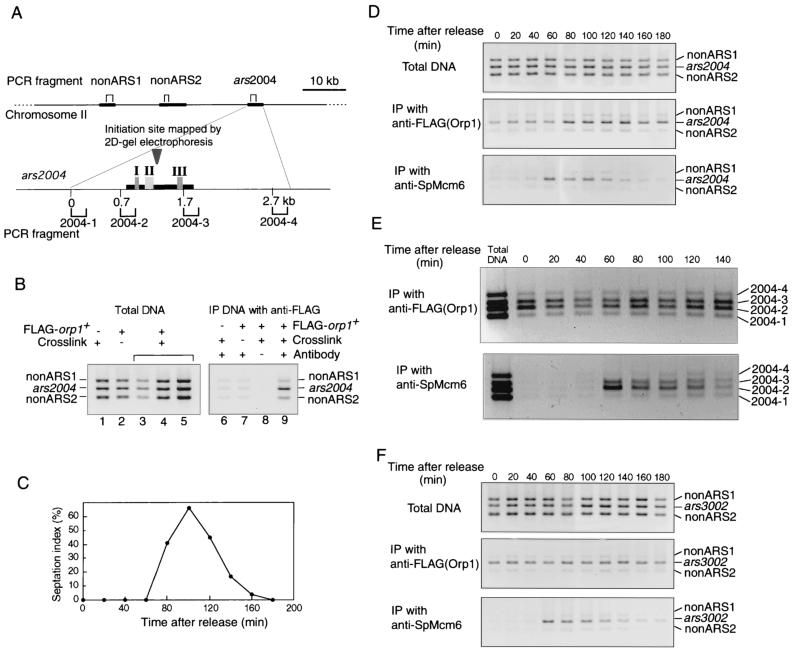
To examine whether Orp1p and SpMcm6p were specifically associated with chromosomal replication origins, we employed the CHIP assay (4, 55, 60). Exponentially growing cells carrying the FLAG-orp1 gene were cross-linked with formaldehyde, and chromosomal DNA was fragmented by sonication. The DNA fragments recovered by immunoprecipitation with anti-FLAG or anti-SpMcm6 antibody were subjected to PCR amplification with specific primers. We have previously shown that the ars2004 locus on chromosome II functions as a replication origin in almost every cell cycle (50). Three sets of primers, namely, ars2004 primers to amplify a 239-bp segment in the ars2004 region and two non-ARS primer sets to amplify 195- and 289-bp fragments approximately 18 and 30 kb, respectively, distant from the ars2004 locus (Fig. 6A), were used for PCR. As shown in Fig. 6B, three DNA fragments were amplified to similar extents from the total cellular DNA without immunoprecipitation (lanes 1 to 5). On the other hand, the ars2004 fragment was preferentially amplified from DNA immunoprecipitated with the anti-FLAG antibody (Fig. 6B, lane 9). No specific DNA fragment was amplified without the FLAG-orp1 gene, cross-linking, or the anti-FLAG antibody (Fig. 6B, lanes 6 to 8). These results demonstrated that Orp1p specifically bound to the chromosomal ars2004 locus.
FIG. 6.
CHIP assay with anti-FLAG and anti-SpMcm6 antibodies. (A) Positions of fragments examined by PCR are shown by brackets above and below the maps of chromosome II and ars2004, respectively. The replication initiation site in ars2004 mapped by two-dimensional gel electrophoresis (50) is shown by the inverted triangle. The 940-bp fragment (thick bar) of ars2004 is sufficient for ARS activity; and regions I, II, and III, which are required for ARS activity, are indicated (51). (B) PCR products amplified with three sets of primers from total cellular DNA without immunoprecipitation (lanes 1 to 5) or from immunoprecipitated DNA (lanes 6 to 9) were analyzed by agarose gel electrophoresis. The samples were prepared from strain 972 cells without FLAG-orp1 (lanes 1 and 6) or from strain TTY15 cells carrying FLAG-orp1 (lanes 2 to 5 and 7 to 9). Cells were either cross-linked with formaldehyde (lanes 1, 3 to 6, 8, and 9) or not cross-linked (lanes 2 and 7) before immunoprecipitation. Extracts were incubated with magnetic beads associated with anti-FLAG antibody (lanes 6, 7, and 9) or with the beads without an antibody (lane 8). Lanes 3 to 5 show the products obtained with a four-fold serial dilution of the template DNA, and lane 4 corresponds to the same amount of template obtained under the standard condition. (C) OGC9 cells were synchronized as described for Fig. 2. The septation index is shown as a percentage. Samples were collected every 20 min, and total cellular DNA without immunoprecipitation, immunoprecipitated with anti-FLAG antibody, or immunoprecipitated with anti-SpMcm6 antibody was used as the template for PCR amplification. PCR products obtained with three sets of primers, (ars2004, nonARS1, and nonARS2 [D]), with four sets of primers [2004-1, 2004-2, 2004-3, and 2004-4 [E]), and with three other sets of primers (ars3002, nonARS1, and nonARS2 [F]) are presented.
To examine the association of SpMcm6p with chromosomal replication origins by CHIP assay, cdc25-22 mutant cells carrying the FLAG-orp1 gene were arrested at the G2/M boundary and released at the permissive temperature. By immunoprecipitation with anti-FLAG antibody, the ars2004 fragment was preferentially amplified at all of the time points examined (Fig. 6D, middle gel), indicating that the Orp1 protein was bound to the ars2004 locus throughout the cell cycle. In contrast, by immunoprecipitation with anti-SpMcm6 antibody, no specific DNA was amplified from 0 to 40 min after release (Fig. 6D, bottom gel). However, the ars2004 fragment was preferentially amplified from 60 to 120 min and thereafter diminished. Two non-ARS fragments were not significantly amplified at any time points (Fig. 6D, bottom gel). These results showed that SpMcm6p was specifically associated with the ars2004 locus during a certain period of the cell cycle. Judging from the septation index, which increased to nearly 50% in the period from 80 to 120 min (Fig. 6C), SpMcm6p was associated with the ars2004 locus during the G1 and S phases.
We have shown that the central 940-bp fragment (ars2004M) of ars2004 is sufficient for ARS activity and contains three regions required for ARS activity (51). In order to examine whether Orp1p and SpMcm6p are associated with the ars2004M segment or with a broad region containing the segment, four primer sets separated by 0.6 to 1 kb within and outside ars2004M (Fig. 6A) were used for PCR amplification. By immunoprecipitation with anti-FLAG antibody, fragments 2 and 3 within ars2004M were preferentially amplified at all of the time points examined after synchronization (Fig. 6E, upper gel). In contrast, fragments 1 and 4 outside of ars2004M were not effectively amplified (Fig. 6E, upper gel). By immunoprecipitation with anti-SpMcm6, fragments 2 and 3 were preferentially amplified at 60 min and then gradually diminished (Fig. 6E, lower gel). These results showed that Orp1p and Mcm6p were associated with ars2004M but not with outside regions. The intensity of fragment 2 was significantly higher than that of fragment 3.
We next examined whether Orp1p and SpMcm6p were associated with another replication origin, ars3002, located downstream of the ura4+ gene (19, 20). From the DNA precipitated with anti-FLAG antibody, the ars3002 fragment was preferentially amplified at all of the time points examined after synchronization (Fig. 6F). By immunoprecipitation with anti-SpMcm6 antibody, the ars3002 fragment was amplified from 60 to 120 min (Fig. 6F). Thus, the results were almost the same as those obtained for ars2004. It can be concluded that the Orp1 protein is constantly bound to chromosomal replication origins, while SpMcm6p is preferentially associated with these origins during the G1 and S phases of the cell cycle.
DISCUSSION
In budding yeast replication origins, the 11-bp ARS consensus sequence is essential for interaction with the ORC. However, replication origins in other eukaryotic species, including fission yeast, do not appear to contain a short essential sequence (15, 23) and it has not been known whether the ORC is located at chromosomal replication origins. The present study demonstrated that a fission yeast ORC subunit and an Mcm protein are specifically localized at chromosomal replication origins. Orp1p is located at the ars2004 and ars3002 loci throughout the cell cycle, while SpMcm6p is associated with these origins only in the G1 and S phases. To our knowledge, this is the first indication of preferential localization of the ORC and Mcm proteins at the chromosomal replication origins in eukaryotic species except for budding yeast.
The CHIP assay finding that Orp1p was localized at ars2004 and ars3002 but not at non-ARS regions (Fig. 6) suggests that a certain sequence or DNA structure in the replication origins is recognized by the fission yeast ORC. Although there is no extensive sequence homology between these two ARS fragments, the regions required for the ARS activities are rich in adenines on a DNA strand (19, 51). Orp1p was preferentially located within a 1-kb region of ars2004M (Fig. 6) in which the replication initiation site had been mapped (50). ars2004M contains two adenine/thymine-rich regions, regions I and III, which are essential for ARS activity (51). Thus, an adenine-thymine-rich stretch is the primary candidate for the ORC-binding site. Segments 2 and 3, which are about 1 kb apart and proximal to regions I and III, respectively, were similarly amplified in the CHIP assay. The ORC would interact either with the unique region at the center of segments 2 and 3 or with multiple regions within ars2004M. Since the central region of ars2004M is not essential for ARS activity (51), it seems likely that ORCs interact with both regions I and III. Regions I and III can be functionally replaced with repeats of AAAT, but not AAT or AT, suggesting that various sequences with three or more consecutive adenines, rather than mere AT richness, are required for interaction with the ORC. These results suggest that the mechanism of interaction of the fission yeast ORC with replication origins might be different from that of the budding yeast ORC. Recently, fission yeast Orp4, a homologue of budding yeast Orc4, has been shown to contain AT-hook motifs that are involved in interaction with the minor groove of AT tracts in DNA (12). The N-terminal domain of Orp4p, with nine AT-hook motifs, specifically binds to the fission yeast ars1 fragment that contains adenine-thymine stretches. The DNA binding activity of Orp4p might participate in recognition of fission yeast replication origins. Further genetic and biochemical studies are required for better understanding of the nature of the fission yeast ORC.
The results of MNase digestion (Fig. 3) suggest that some Orp1p molecules are associated with insoluble nuclear components. We observed that chromosomal origin regions were not readily released from the insoluble fraction by sonication (48), suggesting that Orp1p in the insoluble fraction interacts with both chromatin and insoluble components. Interaction of the ORC with insoluble nuclear components might be involved in the localization of origins at certain nuclear structures. It has been reported that DNA replication in mammalian cells occurs in a distinct nuclear structure, called a replication factory, which is tightly attached to the nuclear matrix (27). On the other hand, the budding yeast ORC has been shown to be involved in cellular functions other than chromosome replication, such as silencing of mating type control (39) and mitotic chromatin condensation (17). The relationship of these functions to the subnuclear localization of the ORC remains to be elucidated.
In addition to Orp1p, SpMcm6p is preferentially located within an about 1-kb region of ars2004, as shown by the CHIP assay. SpMcm6p was associated with the ars2004 and ars3002 loci only during the G1 and S phases, at the same time as when it was associated with chromatin (compare Fig. 2 and 6). In a similar period of the cell cycle, SpMcm6p was detected as distinct foci on spread nucleoids (Fig. 5), suggesting that the foci contain replication origins associated with SpMcm6p. The number of SpMcm6 foci is, at most, 50, even in cells arrested early in S phase by addition of HU (48). Thus, the number of SpMcm6 foci per nucleoid is about 10 times smaller than the number of putative replication origins (50). Multiple replication origins that are associated with the ORC and Mcm proteins could be clustered in the foci. The clustering might facilitate effective activation and inactivation of a set of replication origins by cell cycle signals. Budding yeast Mcm7p has also been shown to be found as distinct foci on spread chromatin by immunostaining (60). In higher eukaryotic cells, however, the entire chromosome regions are immunostained with anti-Mcm antibodies (33, 36, 62). It remains to be solved whether higher eukaryotic Mcm proteins are located at specific regions of the chromosome or exist along the entire chromosome. Even in the former case, several thousand Mcm signals might not be separable as distinct foci due to possible overlapping.
For association of SpMcm6p with chromatin, Cdc18p, a homologue of Cdc6p, was required (Fig. 4). This is consistent with the requirement of Cdc6p, as well as the ORC, for chromatin loading of Mcm proteins in budding yeast and Xenopus egg extract (14, 18, 54). The ORC, Cdc6p, and Mcm proteins become sequentially associated with replication origins to form Pre-RCs (18). A certain temperature-sensitive mutation in orp1 impaired the association of Mcm6p with chromatin at the restrictive temperature (4a), suggesting dependency of chromatin-association of Mcm proteins on the ORC function. After loading of Mcm, however, the budding yeast and Xenopus ORCs and Cdc6p can be removed from chromatin without removing Mcm proteins, suggesting that Mcm proteins, once loaded, are associated with some other component (18, 29). The CHIP assay with anti-SpMcm6p showed that segment 2 was more efficiently amplified than segment 3 in ars2004M, while both segments were similarly amplified by immunoprecipitation with anti-FLAG antibody. The location of SpMcm6p within ars2004 might not be the same as that of the ORC. In human cells, the ORC and Mcm proteins are not colocalized within a 500-bp region of the chromosome (53).
Mcm proteins, components of Pre-RCs in budding yeast, are relocated at nonorigin regions with kinetics similar to those of DNA polymerase ε, which moves with replication forks (4). Although we have not observed significant association of SpMcm6p with non-ARS fragments (Fig. 6), transient localization of Mcm proteins in non-ARS regions might not be detected due to faster fork movement under our conditions. On the other hand, Mcm proteins might be involved in possible unwinding of replication origins, which results in association of replication protein A (RPA), a single-stranded DNA binding protein complex (61). Ishimi has shown that a hexamer complex composed of human Mcm4, -6, and -7 proteins exhibits weak DNA helicase activity (30). Possible Mcm protein DNA helicase activity could be responsible for origin unwinding, replication fork movement, or both. Since association and dissociation of Mcm proteins from replication origins could be key reactions in the regulation of replication, more-detailed analysis of molecular mechanisms of interaction of Mcm proteins with replication origins is necessary. Fission yeast replication origins composed of functional elements located far from each other might be suitable for analysis of the assembly of various protein factors during the process of initiation of replication.
ACKNOWLEDGMENTS
We thank D. Gilbert, T. Yonesaki, H. Araki, J. T. Lee, C. Obuse, and T. Tsurimoto for critical reading of the manuscript and helpful discussions; M. Yanagida, P. Nurse, and B. Stillman for providing yeast strains and plasmids; T. Tanaka for advice on chromatin immunoprecipitation analysis; and T. Usui for advice on preparation of spread nucleoids.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–479. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier R E K, Wahl G M, Epner E M. Participation of human β-globin locus controling region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 3.Alfa C, Fantes P, Hyams J, Mcleod M, Warbrick E. Experiments with fission yeast. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 4.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 4a.Asahara, T., and H. Masukata. Unpublished data.
- 5.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;15:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 6.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 7.Bishop D K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 8.Blow J J, Laskey R A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 9.Broach J R, Li Y Y, Feldman J, Jayaram M, Abraham J, Nasmyth K A, Hicks J B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harbor Symp Quant Biol. 1983;47:1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- 10.Caddle M S, Calos M P. Specific initiation at an origin of replication from Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1796–1805. doi: 10.1128/mcb.14.3.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter P B, Mueller P R, Dunphy W G. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 12.Chuang R-Y, Kelly T J. The fission yeast homologue of Orc4 binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clyne R K, Kelly T J. Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman T R, Carpenter P B, Dunphy W G. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 15.DePamphilis M L. Origin of DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 48–86. [Google Scholar]
- 16.Diffley J F, Cocker J H, Dowell S J, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 17.Dillin A, Rine J. Roles for ORC in M phase and S phase. Science. 1998;279:1733–1737. doi: 10.1126/science.279.5357.1733. [DOI] [PubMed] [Google Scholar]
- 18.Donovan S, Harwood J, Drury L S, Diffley J F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubey D D, Kim S M, Todorov I T, Huberman J A. Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- 20.Dubey D D, Zhu J, Carlson D L, Sharma K, Huberman J A. Three ARS elements contribute to the ura4 replication origin region in the fission yeast, Schizosaccharomyces pombe. EMBO J. 1994;13:3638–3647. doi: 10.1002/j.1460-2075.1994.tb06671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fantes P. Epistatic gene interactions in the control of division in fission yeast. Nature. 1979;279:428–430. doi: 10.1038/279428a0. [DOI] [PubMed] [Google Scholar]
- 22.Gavin K A, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert D M. Replication origins in yeast versus metazoa: separation of the haves and the have nots. Curr Opin Genet Dev. 1998;8:194–199. doi: 10.1016/s0959-437x(98)80141-x. [DOI] [PubMed] [Google Scholar]
- 24.Gossen M, Pak D T, Hansen S K, Acharya J K, Botchan M R. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 25.Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 26.Hood M T, Stachow C. Transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1990;18:688–692. doi: 10.1093/nar/18.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hozak P, Hassan A B, Jackson D A, Cook P R. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao C L, Carbon J. High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci USA. 1979;76:3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua X, Newport I. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 31.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 32.Kim S M, Huberman J A. Multiple orientation-dependent, synergistically interacting similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol Cell Biol. 1998;18:7294–7303. doi: 10.1128/mcb.18.12.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura H, Nozaki N, Sugimoto K. DNA polymerase alpha associated protein P1, a murine homolog of yeast MCM3, changes its intranuclear distribution during the DNA synthetic period. EMBO J. 1994;13:4311–4320. doi: 10.1002/j.1460-2075.1994.tb06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human β-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 35.Klein F, Laroche T, Cardenas M E, Hofmann J F, Schweizer D, Gasser S M. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krude T, Musahl C, Laskey R A, Knippers R. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J Cell Sci. 1996;109:309–318. doi: 10.1242/jcs.109.2.309. [DOI] [PubMed] [Google Scholar]
- 37.Leatherwood J, Lopez-Girona A, Russell P. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature. 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- 38.Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 39.Loo S, Fox C A, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowndes N F, McInerny C J, Johnson A L, Fantes P A, Johnston L H. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+ Nature. 1992;355:449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- 41.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 42.Maundrell K, Hutchison A, Shall S. Sequence analysis of ARS elements in fission yeast. EMBO J. 1988;7:2203–2209. doi: 10.1002/j.1460-2075.1988.tb03059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchison J M. Physiological and cytological methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970;4:131–165. [Google Scholar]
- 44.Miyake S, Okishio N, Samejima I, Hiraoka Y, Toda T, Saitoh I, Yanagida M. Fission yeast genes nda1+ and nda4+, mutations of which lead to S-phase block, chromatin alteration and Ca2+ suppression, are members of the CDC46/MCM2 family. Mol Biol Cell. 1993;4:1003–1015. doi: 10.1091/mbc.4.10.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muzi-Falconi M, Kelly T J. Orp1, a member of the Cdc18/Cdc6 family of S-phase regulators, is homologous to a component of the origin recognition complex. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 47.Obuse C, Okuno Y, Okazaki T, Masukata H. A replication-enhancing element with transcriptional silencer activity in autonomously replicating human chromosomal fragments. Mol Biol Cell. 1996;7:43–55. doi: 10.1091/mbc.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogawa, Y. Unpublished data.
- 49.Okishio N, Adachi Y, Yanagida M. Fission yeast Nda1 and Nda4, MCM homologs required for DNA replication, are constitutive nuclear proteins. J Cell Sci. 1996;109:319–326. doi: 10.1242/jcs.109.2.319. [DOI] [PubMed] [Google Scholar]
- 50.Okuno Y, Okazaki T, Masukata H. Identification of a predominant replication origin in fission yeast. Nucleic Acids Res. 1997;25:530–537. doi: 10.1093/nar/25.3.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okuno Y, Satoh H, Sekiguchi M, Masukata H. Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol Cell Biol. 1999;19:6699–6709. doi: 10.1128/mcb.19.10.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olsson T, Ekwall K, Ruusala T. The silent P mating type locus in fission yeast contains two autonomously replicating sequences. Nucleic Acids Res. 1993;21:855–861. doi: 10.1093/nar/21.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritzi M, Baack M, Musahl C, Romanowski P, Laskey R A, Knippers R. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J Biol Chem. 1998;273:24543–24549. doi: 10.1074/jbc.273.38.24543. [DOI] [PubMed] [Google Scholar]
- 54.Rowles A, Chong J P, Brown L, Howell M, Evan G I, Blow J J. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 55.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 56.Struhl K, Stinchcomb D T, Scherer S, Davis R W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takahashi, T. Unpublished data.
- 60.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Todorov I T, Attaran A, Kearsey S E. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van-Houten J V, Newlon C S. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol Cell Biol. 1990;10:3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]