Abstract
Three Cbfa motifs are strategically positioned in the bone-specific rat osteocalcin (rOC) promoter. Sites A and B flank the vitamin D response element in the distal promoter and sites B and C flank a positioned nucleosome in the proximal promoter. The functional significance of each Cbfa element was addressed by mutating individual or multiple Cbfa sites within the context of the −1.1-kb rOC promoter fused to a chloramphenicol acetyltransferase reporter gene. Promoter activity was assayed following transient transfection and after stable genomic integration in ROS 17/2.8 osteoblastic cell lines. We show that all three Cbfa sites are required for maximal basal expression of the rOC promoter. However, the distal sites A and B each contribute significantly more (P < 0.001) to promoter activity than site C. In a genomic context, sites A and B can largely compensate for a mutation at the proximal site C, and paired mutations involving site A (mAB or mAC) result in a far greater loss of activity than the mBC mutation. Strikingly, mutation of the three Cbfa sites leads to abrogation of responsiveness to vitamin D. Vitamin D-enhanced activity is also not observed when sites A and B are mutated. Significantly, related to these losses in transcriptional activity, mutation of the three Cbfa sites results in altered chromatin structure as reflected by loss of DNase I-hypersensitive sites at the vitamin D response element and over the proximal tissue-specific basal promoter. These findings strongly support a multifunctional role for Cbfa factors in regulating gene expression, not only as simple transcriptional transactivators but also by facilitating modifications in promoter architecture and chromatin organization.
Bone tissue-restricted expression of the osteocalcin (OC) gene during development of the osteoblast phenotype requires a multiplicity of transactivating factors. Among the key regulators of OC expression are transcription factors that play essential roles in embryonic formation of the skeleton and osteoblast differentiation. These include the Cbfa (core binding factors α)/AML (acute myelogenous leukemia) family of runt homology domain (rhd) DNA binding proteins (reviewed in reference 33), the Msx and Dlx homeodomain proteins (28, 51, 63), AP-1 proteins (41), and steroid hormone receptors (reviewed in reference 40).
The Cbfa/AML family of transcriptional activators are critical factors for the development of hematopoietic and skeletal tissues. Each of three known genes, Cbfa1 (human AML-3/mouse Pebp2a [hAML-3/mPebp2a]), Cbfa2 (hAML-1/mPebp2b), and Cbfa3 (hAML-2/mPebp2c), encodes several mRNA splice variants (1, 37, 56). The tissue-specific transcriptional properties of the Cbfa proteins are in part accounted for by their selective representation in distinct cellular phenotypes. Cbfa2/AML-1 primarily regulates expression of genes related to the development of thymus and hematopoietic tissues, and a null mutant of this gene results in embryonic lethality due to the absence of definitive hematopoiesis. Several isoforms of Cbfa1 have been described; one is expressed in hematopoietic tissues (71), and another is highly expressed in osteoblast lineage cells of bone (6, 14, 58, 60, 66) and in hypertrophic chondrocytes (34). Ablation of the Cbfa1 gene in mice reveals the importance of this factor in development of the skeleton with a consequent absence of mineralized connective tissues (34, 48, 49). The Cbfa class of rhd proteins was initially identified in bone as an osseous-cell-specific DNA binding complex extracted from the nuclear matrix (9, 43). Subsequently, Cbfa proteins were shown to regulate tissue-specific expression of the OC promoter (5, 13). Although overexpression of the Cbfa1/AML-3, Cbfa2/AML-1, or Cbfa3/AML-2 factors in nonosseous cells can confer expression of the bone-specific OC gene (5), the DNA binding activity present in mature osteoblasts consists primarily of the Cbfa1 gene product (6, 14).
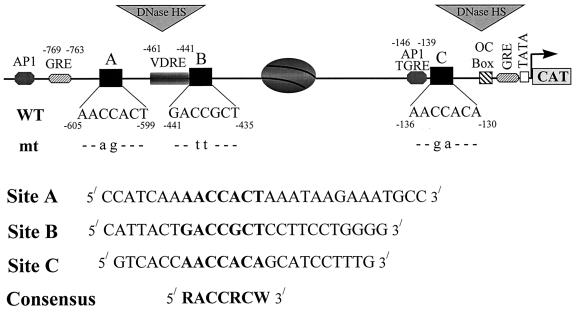
The bone-specific rat OC promoter contains three recognition sites for Cbfa interactions (sites A, B, and C [43]). Notably, all three motifs bind a similar osteoblast-specific DNA binding complex, first designated NMP-2 (9, 43). While only one Cbfa site fused to a minimal OC promoter is sufficient to confer enhancer activity in osseous and nonosseous cells (5), the presence and positioning of multiple Cbfa sites suggest that spatial organization of the native OC promoter may be important for interaction of Cbfa proteins with other OC promoter regulatory factors. For example, transcription of the rat and human OC genes is strongly influenced by 1,25-dihydroxyvitamin D. Cbfa sites A and B flank the vitamin D response element which mediates 3- to 10-fold enhancer activity of the rat and human promoters. A third Cbfa site, C (also designated OSE2 in mice [13]), is located in the proximal promoter (nucleotides [nt] −136 to −130). A nucleosome is positioned between Cbfa sites B and C in the transcriptionally active rat OC promoter (46). Because Cbfa factors associate with the nuclear matrix (57) and can recruit other factors into complexes (3, 29, 36, 50, 56), Cbfa binding sites may impose structural constraints on the OC promoter to facilitate interaction and activities of the proximal and distal regulatory elements (e.g., the vitamin D response element [VDRE] and TATA domains). The function of the Cbfa motifs within the context of the native rat OC promoter has yet to be examined.
In these studies, we establish that all three Cbfa elements within the rat OC promoter contribute to basal transcriptional activity, suggesting a functional interaction among the three sites. In contrast to recent studies of the mouse OC promoter (17), the distal site A (nt −605 to −599) in the rat OC promoter contributes far more to Cbfa-dependent promoter activity than the proximal site C/OSE2. Furthermore, the presence of the upstream Cbfa sites is critical for vitamin D induction of OC promoter activity. Mutation of all three Cbfa sites results in altered chromatin organization, as reflected by loss of DNase I-hypersensitive sites in the OC promoter. These findings suggest that transcriptional activity of the OC gene, which requires interactions of proximal and distal regulatory elements, may be facilitated through spatial constraints of the promoter imposed by the binding of the nuclear-matrix-associated Cbfa factors to critically positioned recognition sequences.
MATERIALS AND METHODS
Plasmid constructs.
Site-directed mutagenesis was performed to incorporate 2-nt substitutions into the core binding motif (ACC) of each individual Cbfa site (RACCRCW) in the 1.1-kb rat OC promoter fragment (Fig. 1). Mutations were generated by a PCR-based approach (4) with the following synthetic oligonucleotides (Integrated DNA Technologies, Inc., Coralville, Iowa): Site A, 5′ CCATCAAAAagACTAAATAAGAAATGCC 3′; Site B, 5′ CATTACTGAttGCTCCTTCCTGGGG 3′; and Site C, 5′ GTCACCAAgaACAGCATCCTTTG 3′. The mutations (indicated by lowercase letters) within the Cbfa consensus motif (underlined) were selected such that no other DNA-protein binding sites would be generated. Plasmids bearing mutations in two of the three Cbfa sites were generated by replacing the wild-type (WT) sequence with the mutant by restriction digestion of the −1.1-kb ratOC promoter. The mABC plasmid bearing mutations in all three Cbfa sites was obtained by digestion of mAB with BglII-HindIII to release the WT site C, which was replaced with the mutant site C plasmid fragment.
FIG. 1.
Wild-type and mutated Cbfa motifs in the rat OC gene promoter. The positions and nucleotide numbers of the three Cbfa sites (A, B, and C) relative to the VDRE, the glucocorticoid response element (GRE), a TGFβ-responsive AP-1 site, and two primary transcriptional elements requisite for basal transcription, the OC box and the TATA box, are indicated. The positioned nucleosome and DNase I-hypersensitive sites that are present when the gene is transcribed are also indicated. The Cbfa core recognition sequence at each site is indicated in boldface, with mutant nucleotides (mt) designated below. The lower panel shows the WT oligonucleotide probes, used in gel mobility shift assays, containing the site A, B, and C Cbfa motifs within the context of flanking sequences of the rat OC promoter.
Transient transfection and CAT reporter assays.
Ros 17/2.8 cells were plated at a density of 8 × 104/well in six-well plates and transfected 24 h later with 3 μg of either WT or mutant plasmids and 100 ng of Rous sarcoma virus luciferase. The total amount of exogenous DNA was maintained at 5 μg/well with salmon sperm DNA. The cells were transfected with DNA in the presence of 50 μg of DEAE-dextran (Pharmacia, Piscataway, N.J.)/ml and 50 μg of chloroquine (Sigma, St. Louis, Mo.)/ml and incubated at 37°C for 2.5 h with occasional swirling. The transfection mix was aspirated, and the cells were shocked for 90 s with 10% glycerol in serum-free F12 medium, washed twice with phosphate-buffered saline (PBS), and then incubated at 37°C in F12 medium supplemented with 5% fetal calf serum (Gibco Life Technology, Grand Island, N.Y.) for 24 to 48 h. The cells were washed twice with ice-cold PBS and lysed with 300 μl of reporter lysis buffer (Promega Corp., Madison, Wis.) at room temperature for 30 min. The cell lysates were collected and stored at −70°C or used immediately for chloramphenicol acetyltransferase (CAT) assays as described previously (18). Luciferase activity was determined in the same lysate with luciferase assay reagents from Promega Corp. Luminescence was quantitated with a Monolite TM 2010 instrument (Analytical Luminescence Laboratory, San Diego, Calif.).
Construction of stable cell lines.
Ros 17/2.8 cells with genomically integrated WT and Cbfa mutant constructs were generated by the calcium phosphate method. For each construct, four 100-mm-diameter plates were transfected with 15 μg of the −1.1-kb OC-CAT plasmid and 5 μg of pCEP-4 (Invitrogen, San Diego, Calif.) encoding the hygromycin B phosphotransferase gene. The cells were harvested at 95% confluency and replated for selection in medium containing 55 U of hygromycin B (Calbiochem, La Jolla, Calif.)/ml based upon preliminary killing curves. Resistant colonies (60 to 75) from each plate were pooled and propagated as polyclonal cell lines. Each pool was expanded until 20 × 108 cells were available for preparation of frozen stocks. The cells were routinely maintained in medium containing hygromycin B for measuring CAT activity and responsiveness to steroid hormones and growth factors. Stable cells were plated at 2 × 105/well in a six-well plate and treated with 10−8 M 1,25-dihydroxyvitamin D3. The cells were washed twice with PBS and lysed by adding 300 μl of reporter lysis buffer (Promega Corp.) at room temperature for 30 min. CAT activity assays were performed as described above.
Nuclear extracts and electrophoretic mobility shift assay.
Nuclear extracts from Ros 17/2.8 cells were prepared as described previously (12) with 0.45 M KCl for extraction. For the electrophoretic mobility shift assay, 10 mM HEPES (pH 7.9), 0.2 mM EDTA, and 100 mM KCl were combined with 10 μl of a DNA mixture containing 20 fmol of probe DNA and 1 μg of poly(dI-dC) · (dI-dC) as a nonspecific competitor and incubated at room temperature for 30 min. For competition experiments, unlabeled double-stranded oligonucleotide (240 to 2,000 fmol) was added to the binding reaction mixture with the other components. Samples were loaded without tracking dye onto a 4% acrylamide–bisacrylamide (30:1) gel in 0.5× Tris borate–EDTA. Electrophoresis was performed for 2 to 2.5 h at 200 V. The gels were dried and subjected to autoradiography.
Studies of DNase I-hypersensitive sites.
DNase I digestion analysis was performed according to the indirect end-labeling method (65). ROS 17/2.8 cells were plated at a density of 106 per 100-mm-diameter plate, and nuclei were isolated on day 9 by Dounce homogenization (loose pestle) in 8 volumes of RSB buffer (10 mM Tris HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2) with 0.5% (vol/vol) NP-40. To evaluate cell lysis, an aliquot of nuclei was stained with 0.4% Trypan blue 1:1 (vol/vol).
The nuclear suspension was diluted by adding an equal volume of RSB buffer, and nuclei were collected by centrifugation. The pelleted nuclei were resuspended in RSB buffer, and the DNA concentration was estimated by absorption at 260 nm. Aliquots of 20 A260 units were digested with increasing concentrations of DNase I (0 to 5 U) (Worthington Biochemicals, Freehold, N.J.) in a 1-ml final volume for 10 min at room temperature. The reaction was stopped by adding EDTA, sodium dodecyl sulfate, RNase I (Promega), and proteinase K (Fisher Biotech, Fairlawn, N.J.) to final concentrations of 25 mM, 0.5% (vol/vol), 1 U/ml, and 200 μg/ml, respectively, and incubated at 37°C overnight. The samples were extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and twice with chloroform-isoamyl alcohol (24:1). Nucleic acids were precipitated with 2.5 volumes of ethanol at −70°C for at least 4 h and then resuspended in 25 mM Tris, pH 7.8. DNA was digested with BamHI or XbaI (4 U/μg of DNA) to release a 4.3-kb fragment from the OC gene (45) or a 2.78-kb fragment from the chromosomally integrated pOCZCAT fusion gene, respectively (19). The digested DNA was extracted with phenol-chloroform, precipitated with ethanol, and resuspended in 25 mM Tris, pH 7.8. DNA samples (10 μg) were electrophoresed in a 1.2% agarose gel (Bio-Rad, Hercules, Calif.) and then transferred to a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Arlington Heights, Ill.) according to the manufacturer’s instructions.
Hybridization probes were prepared by restriction digestion of pOC 3.4 (38), containing the rat OC gene and flanking sequences, with XbaI-BamHI and digestion of pOCZCAT with XbaI-NcoI. The probes were labeled by the random-primer method with [α-32P]dCTP and the Prime-It II kit (Stratagene, La Jolla, Calif.). Hybridization was carried out at 65°C with 1 ng of probe (specific activity, 109 cpm/μg) per 10-cm2 membrane. The blots were analyzed by autoradiography or by using a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Statistical representation and analysis of the data.
The data are displayed as box-and-whisker plots (25), where the length of the bar represents the range of observations between the first and third quartile (i.e., the interquartile range [IQR]). The length of the whiskers represents the interval between the first or third quartile and the most extreme observation that does not meet the definition for an outlier, i.e., a value more than 1 1/2 IQRs from either the first or third quartile, depending upon whether the observation is above or below the median.
The distributional characteristics of promoter activities were evaluated graphically by using histograms and the Kolmogorov-Smirnov one-sample test for normality (53). If data were not normally distributed, monotonic transformations were applied (i.e., natural logarithms) to achieve normality. The significance of Cbfa sites to promoter activity was evaluated by analysis of variance for mixed models with restricted estimation by maximum likelihood (42).
RESULTS
Full basal activity of the OC promoter requires the distal Cbfa sites.
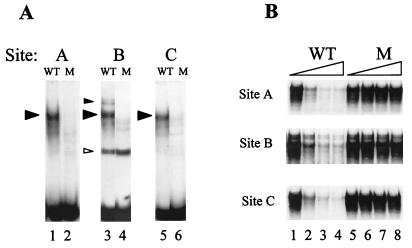
A single Cbfa element is sufficient to confer tissue-specific transactivation of the proximal OC promoter (5, 14). To address the contributions of the multiple Cbfa sites in the native rat OC promoter to OC transcription, we carried out site-directed mutagenesis of each of the three Cbfa elements, designated sites A, B, and C (Fig. 1). Initially, we established 2-nt substitution mutations in the core of each OC Cbfa site which abrogate bone-specific Cbfa binding activity (Fig. 2). The appropriate WT and mutant oligonucleotide sequences for each site (shown in Materials and Methods) were examined in gel mobility shift assays with nuclear extracts from ROS 17/2.8 rat osteosarcoma cell lines. These extracts contain abundant levels of Cbfa1, which forms an osteoblast-specific complex that can be supershifted by Cbfa1-specific antibody (5, 6). Figure 2A demonstrates that each mutation results in loss of the Cbfa binding complex. Because competition for binding to the WT sequence is not observed in the presence of 80-fold excess (1 nM) of mutant oligonucleotide representing site A, B, or C (Fig. 2B, lanes 5 to 8), the mutations have completely abrogated Cbfa binding.
FIG. 2.
Mutations of the three rat OC Cbfa sites result in loss of Cbfa binding. (A) Formation of the osteoblast-specific complex from nuclear extracts of ROS 17/2.8 cells compared with oligonucleotides containing WT sequences representing Cbfa site A (lane 1), site B (lane 3), and site C (lane 5) and mutated (M) Cbfa sequences of sites A (lane 2), B (lane 4), and C (lane 6) in gel mobility shift assays. Cbfa complexes are indicated by solid arrowheads. A nonspecific complex (site B) is indicated by an open arrowhead. (B) Site A, site B, and site C show corresponding competition assays for each Cbfa site with the WT sequence as probe with increasing amounts (0, 20, 60, and 80×) of either WT oligonucleotide (lanes 1 to 4, respectively) or Cbfa site-mutated oligonucleotides (lanes 5 to 8, respectively) as the competitor.
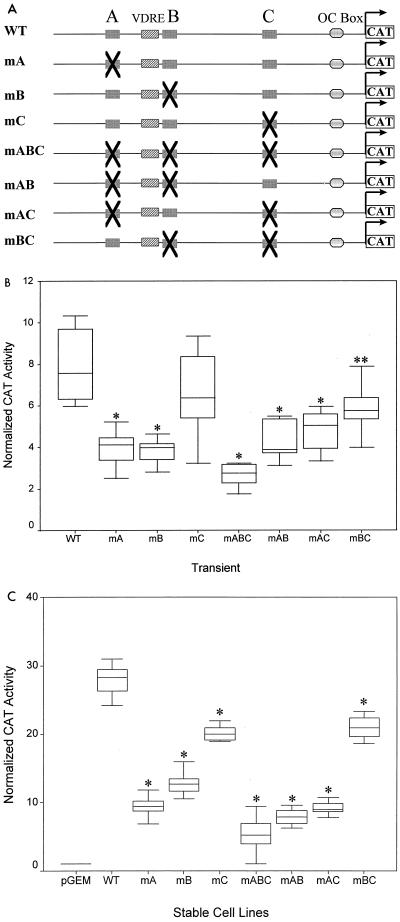
The locations of the Cbfa sites within the rat OC promoter (43) suggest different activities, while promoter deletion analysis (5, 27) indicates redundant function for the multiple Cbfa sites. The functionality of each site was therefore tested following transient transfection of the WT and mutant OC promoter (−1.1-kb) constructs into ROS 17/2.8 cells (Fig. 3B and D). Activity of the OC WT promoter was compared to activity of promoters having single or multiple Cbfa site mutations. While mutation of each of the sites reduced promoter activity compared to that of the control, mutation of the proximal site C had the least effect (83% of WT; P value of 0.206 is nonsignificant). Mutation of sites A and B independently or of the two sites AB and AC reduced promoter activity to approximately 50% of that of the WT, but high statistical significance of the effect was found in these transient assays only for mAB (P < 0.001). Transcriptional activity of the Cbfa three-site mutation, mABC, was decreased to 40% of the control level (P < 0.001).
FIG. 3.
Requirement for multiple Cbfa sites for maximal transcriptional activity of the rat OC gene promoter. (A) Rat OC 5′ sequences (−1.1 kb) containing either single or multiple site mutations are schematically illustrated. Single-site mutations are designated mA, mB, or mC; two-site mutations are designated mAB, mAC, or mBC; and mABC is the triple mutation. (B) Following transient transfections in ROS 17/2.8 cells, normalized CAT reporter activity of the WT promoter and promoters with Cbfa site mutations are compared. Reporter activity was assayed 24 h after transfection of ROS 17/2.8 cells. The [14C]CAT activity was quantitated by a Betascope analyzer (Betagen, Waltham, Mass.) and normalized to that of luciferase. (Aliquots of the lysate were assayed for luciferase activity to normalize CAT activity). Each bar represents the LS mean ± standard error of the mean (SEM) (n = 12). (C) Activity of WT and Cbfa site-mutated promoters stably integrated into ROS 17/2.8 cells. Each group represents promoter activity in four independent cell lines, with each cell line assayed in four separate experiments in triplicate. The cells were harvested 3 days after being plated as the cells reached monolayer confluency for quantitation of CAT activity (normalized to total protein of the cell lysate). Each bar represents the LS mean ± SEM (n = 18). The pGEM control represents a promoterless CAT-containing stable cell line. Single asterisks, statistically less than WT, P < 0.001; double asterisks, P < 0.01.
To determine the contribution of the Cbfa sites to OC gene promoter transcription within a genomic context, we established a series of ROS 17/2.8 cell lines which contain stably integrated OC-CAT reporter gene constructs with single, paired, or triple Cbfa mutations in the −1.1-kb rat OC promoter (Fig. 3A). As described in Materials and Methods, we examined four independent pools of cell lines for each construct to compensate for positional effects on promoter activity. Due to site-of-integration effects and copy number, basal activity for each cell pool varied among the cell lines over a twofold range (data not shown). Figure 3C shows that mutation of each of the individual Cbfa sites resulted in significantly decreased promoter activity, with reductions in transcription greater than those observed following transient transfection (Fig. 3B). Again, mutation of the proximal site C minimally affected transcription; even as a stable integrant in osteoblasts (P < 0.05). Mutations involving site A had a more pronounced effect in decreasing promoter activity in the stable cell lines (from 20 to 30% of that of the control; P < 0.001). This conclusion is further supported by the finding that transcription of mBC is only reduced to 80% of that of the control, indicating that site A contributes a significant level of Cbfa-dependent activity to the OC promoter (Fig. 3C). Because all possible mutation groups were examined and sample sizes within a group were large, the statistical significance of the contributions of all Cbfa sites to OC promoter activity could be compared (Table 1). On the basis of whether each of the sites is present or absent in a construct, the effect of site A or B, but not C, was always significant. Furthermore, two-way analysis of variance of the least-squares mean of each group revealed a significant interaction between sites A and B. There is a two- to threefold-greater effect contributed by site A when site B is present and by site B when site A is present. Taken together, these findings indicate that all of the Cbfa sites in the rat OC promoter contribute to basal transcription and that the distal sites A and B play predominant roles in supporting promoter activity within a chromosomal context.
TABLE 1.
Contribution of distal Cbfa sites A and B to promoter activity
| Cbfa site(s) |
P values for activity tested against all groups in:
|
|
|---|---|---|
| Transient assays | Stable cell lines | |
| A | 0.0001 | 0.0001 |
| B | 0.0001 | 0.0001 |
| C | NSa | 0.0001 |
| Interaction betweenb: | ||
| A and B | 0.0013 | 0.0001 |
| A and C | NS | 0.0062 |
| B and C | NS | NS |
| A, B, and C | 0.0001 | 0.0001 |
NS, not significant (i.e., >0.05).
The interactions between the indicated sites were tested for significance. When sites A and B or A, B, and C are present together in transient assays or stable cells and sites A and C are present together in a genomic context, the presence of the other site(s) influences the contribution to transcriptional activity of the promoter.
Functional compensatory activity of the three Cbfa sites in OC transcription.
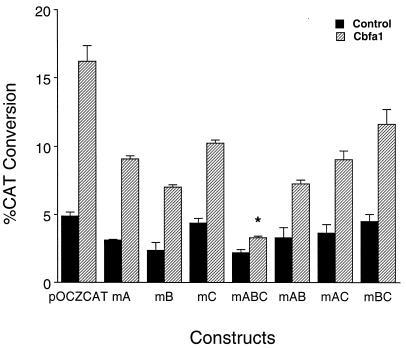
The mutagenesis studies suggest that the distal sites A and B provide a significant level of functional activity and that the proximal site C does not contribute equally to basal promoter activity. To further assess the involvement of each of the three Cbfa sites in transcription of the rat OC gene, we examined the consequences of forced expression of Cbfa1 on activity of WT and mutant promoter constructs in nonosseous cells, which do not express OC. In HeLa cells, for example, OC promoter activity is very low, and transcription factors necessary for bone tissue-specific expression are unlikely to be present. Cbfa forced expression in HeLa cells results in detectable WT OC promoter activity (3.3-fold induction [Fig. 4]); as expected, mutation of all three Cbfa sites dramatically reduces Cbfa-dependent OC promoter activity. In contrast, mutation of the single site A, B, or C resulted in induction of OC promoter activity to approximately the same extent, but somewhat less than WT. Together, these findings suggest that any one of the three sites can support induction of the OC promoter when cellular levels of Cbfa1 are available. However, compensatory effects are partial, indicating that all three sites are necessary for maximal promoter activity. The level of induction by forced Cbfa expression for each of the mutations compared to the WT promoter in ROS 17/2.8 cells is similar to that in HeLa cells (data not shown). Thus, these findings indicate that Cbfa-dependent induction of promoter activity at each Cbfa site is similar, suggesting functional compensatory activity. Notably, the mutational analyses support a selective contribution of the distal sites versus the proximal site C to basal promoter activity (Fig. 3).
FIG. 4.
Cbfa1-mediated transactivation of OC promoters containing mutated Cbfa sites in HeLa cells. Cbfa1 expression plasmid (0.5 μg) and indicated OC-CAT plasmids (2.5 μg) were cotransfected into HeLa cells (n = 9 for each sample) and assayed 36 h following transfection. The CAT activity was quantitated by direct counting with a Betascope analyzer (Betagen). CAT activity was calculated as percent conversion and normalized for luciferase values used as internal controls. A statistically significant difference (P < 0.001) of the mABC group from WT is indicated by an asterisk.
Vitamin D enhancement of OC transcription is dependent on Cbfa1 regulatory elements.
Our mutational studies suggest that the two distal Cbfa sites which flank the VDRE are essential to basal expression of the OC gene (Fig. 3C). The VDRE is a strong enhancer of OC transcription, and vitamin D responsiveness necessitates interaction of the VDR-RXR heterodimer complex at the distal VDRE (nt −461 to −441) with TFIIB and other general transcription factors at the proximal TATA binding element (nt −32 to −29). We therefore addressed the potential contribution of Cbfa sites in mediating vitamin D responsiveness within the context of chromosomal integration of the OC promoter in stable cell lines. The importance of examining vitamin D responsiveness as influenced by the Cbfa motifs within a chromosomal context is supported by our previous studies demonstrating the presence of a positioned nucleosome between Cbfa sites B and C on the OC promoter and increased DNase I hypersensitivity in response to vitamin D (45, 46).
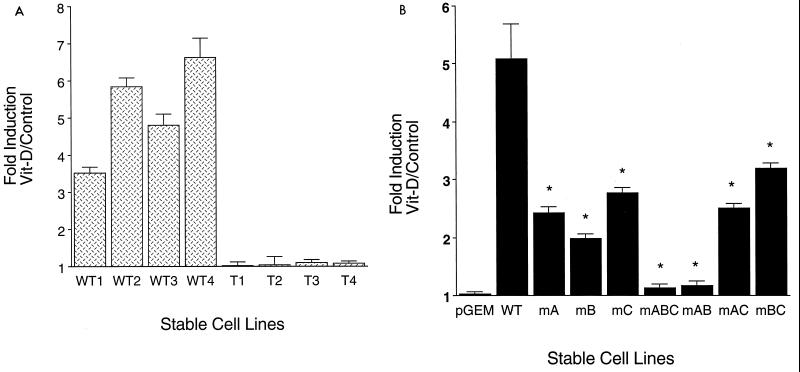
Expression of the stably integrated WT promoter was enhanced by vitamin D from four- to sevenfold in the four stable cell line pools, independent of basal promoter activity (Fig. 5A). In striking contrast, the four stable cell lines in which all three Cbfa sites of the integrated OC promoter were mutated (mABC cell lines, designated T1 to T4 [Fig. 5A]) exhibited nearly complete inhibition of vitamin D stimulation of transcription. This abrogation of vitamin D responsiveness of the mABC promoter was also observed following transient transfection of the WT and mABC promoters into ROS 17/2.8 cells. In three independent experiments (total n = 18), the mean fold induction of the WT OC promoter by 10−8 M 1,25(OH)2D3 in 24 h is 7.4 (± 0.2 standard deviation [SD]) but only 1.6 (± 0.3 SD) for the mABC construct.
FIG. 5.
Cbfa sites regulate vitamin D-mediated transcription of the OC gene. (A) Loss of vitamin D (Vit-D) responsiveness of the rat OC promoter with three mutated Cbfa sites (mABC). Four independent ROS 17/2.8 cell lines with stably integrated WT promoters (WT1, WT2, WT3, and WT4) or with the triple Cbfa site mutation (mABC) (T1, T2, T3, and T4) were treated 3 days after being plated for 24 h with 10−8 M 1,25(OH)2D3 and assayed for CAT activity normalized to total protein in the cell lysate. Each bar represents the mean value of three determinations. (B) Distal Cbfa sites A and B in the OC promoter are required for vitamin D-induced transcriptional activity. ROS 17/2.8 cell lines containing stably integrated WT and the indicated Cbfa mutant promoter-CAT reporter constructs were examined for responsiveness to vitamin D (10−8 M 1,25(OH)2D3; 24 h). pGEM control is a promoterless-CAT stable cell line. The effect of vitamin D (vitamin D-treated/control untreated cells) is reported as fold induction. Each bar represents pooled data from three or four separate cell lines, each assayed in triplicate in two to four independent experiments. Asterisk, P < 0.001 (statistical significance of mutant cell line versus WT). The error bars indicate SD.
We then proceeded to examine the contribution of each Cbfa site to vitamin D-dependent promoter activity. Mutation of any one of the single Cbfa sites (mA, mB, or mC) decreased the vitamin D response from 5-fold induction (WT) to 2.5- and 2.8-fold induction for mutant sites A and C, respectively, and 1.9-fold stimulation with mutant site B (mB) (Fig. 5B). When vitamin D responsiveness of the paired mutations was examined (Fig. 5B), the contribution of the distal sites to functional activity of the VDRE was further defined. Notably, the mAB two-site mutation nearly eliminated vitamin D stimulatory activity, similar to the effect of the triple mutant. In contrast, two-site mutations involving the proximal site C (mAC and mBC) had less effect, reducing vitamin D responsiveness to 2.5- and 3-fold induction, respectively, similar to the consequences of the single site C mutation. Thus, these results confirm a critical role for sites A and B, not only in basal expression of the OC promoter but also in vitamin D regulation of OC promoter activity. These findings suggest that the Cbfa sites support structural organization of the OC promoter that is permissive for interaction of the distal VDRE and proximal TATA binding factors required for enhancer activity of the VDR-RXR complex (21, 23, 57).
Cbfa binding factors contribute to chromatin organization of the OC promoter.
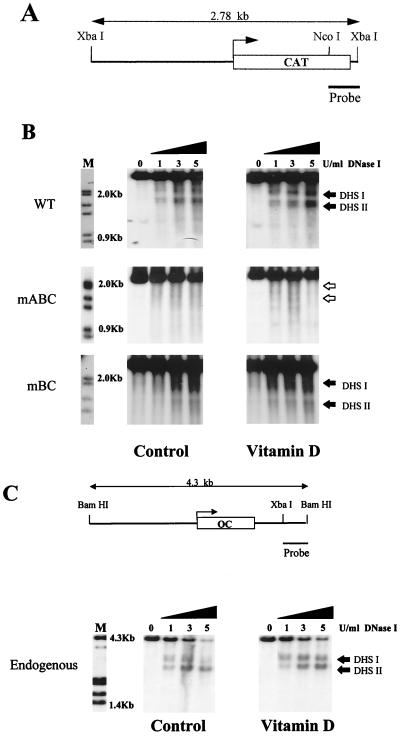
Previous studies carried out by our laboratory have shown the presence of DNase I-hypersensitive sites in the native OC promoter as well as in the transgene of stable cell lines with an integrated OC promoter-CAT gene (pOCZCAT) (45–47). These promoters exhibit two DNase I-hypersensitive sites, designated DHS I in the basal promoter region and DHS II in the distal promoter encompassing the VDRE. The nuclease accessibility of both regions is functionally related to the extent to which the OC gene or the transgene (pOCZCAT) is expressed. Based on these findings and the observation that the triple Cbfa mutant (mABC) does not respond to vitamin D, we examined the effect of this mutation on the nuclease hypersensitivity of the OC promoter. Figure 6 shows the consequences of DNase I digestion of nuclei from the ROS 17/2.8 cell line carrying the wild-type −1.1-kb rat OC promoter transgene. Two DNase I-hypersensitive sites are present both in the control cells and in cells treated with vitamin D (Fig. 6B). An increase in DNase I hypersensitivity is observed following vitamin D treatment. In contrast, DNase I-hypersensitive sites are not detected in nuclei from either untreated or vitamin D-treated cells carrying the mABC OC promoter-CAT transgene (Fig. 6B). The specificity of the DNase I-hypersensitive sites was confirmed by reprobing the blot with a −0.4-kb fragment (XbaI-BanI) that detected the vector backbone (data not shown).
FIG. 6.
DNase I-hypersensitive profile of WT and mutated OC promoters. (A) Diagrammatic illustration of the OC promoter-CAT transgene showing the region used as a probe. (B) Nuclei were isolated from untreated (left-hand panels) and vitamin D-treated [10−8 M 1,25(OH)2D3 for 24 h) (right-hand panels) ROS 17/2.8 stable cell lines having the 1.1-kb rat WT, mABC, or mBC OC promoter. Nuclei were incubated with increasing amounts of DNase I (from 0 to 5 U per 20 optical density at 260 μm units of nuclei) for 10 min at room temperature. The DNase I concentrations (U/ml) are designated above the lanes of the Southern blot. The purified DNA was digested with XbaI to detect the transgene. (C) DNase I hypersensitivity of the endogenous OC promoter from the mABC stable cell line. The BamHI-XbaI fragment of the OC gene used as a probe is shown above. All samples (10 μg) were fractionated electrophoretically in a 1.2% agarose gel, and the blots were hybridized with the corresponding probe. Lane M, markers from λ DNA digested with HindIII and EcoRI. The two DNase I-hypersensitive sites (DHS I and DHS II) are indicated by solid arrowheads.
To ensure that the complete loss of DNase I hypersensitivity in mABC is not due to a nonspecific alteration of the promoter, we examined the DNase I profile of mutant BC, which retains significant basal activity and vitamin D responsiveness (Fig. 3C and 5B). Interestingly, we observed DNase I hypersensitivity in the distal domain (DHS II), which encompasses the VDRE. Thus, there is a correlation between vitamin D-enhanced transcription and the level of DNase I hypersensitivity. The competency of the mBC cell line to respond to vitamin D suggests that the ability of the VDR receptor complex to interact with its specific binding sequence in the OC promoter remains intact. Significantly, in mBC, the proximal DHS I which resides over the mutated site C is very weak. Therefore, modifications in DNase I hypersensitivity are linked to mutation of Cbfa sites. Together, these results demonstrate that the chromatin structure of the OC promoter requires the integrity of the Cbfa sites.
The specificity of the contribution of Cbfa elements to chromatin structure is further demonstrated by the DNase I hypersensitivity of the native gene in the mABC cell line (Fig. 6C). The endogenous OC gene in the mABC mutant cell line retains basal DNase I hypersensitivity, which increases upon treatment with vitamin D. Similar responsiveness of the endogenous OC gene to DNase I was observed in the cell line carrying the WT transgene (data not shown). A schematic illustration summarizing these modifications in DNase I hypersensitivity of the WT and mABC OC promoters is presented in Fig. 7. These studies provide compelling evidence that Cbfa factors are determinants of chromatin organization that supports transcriptional activity of the OC promoter.
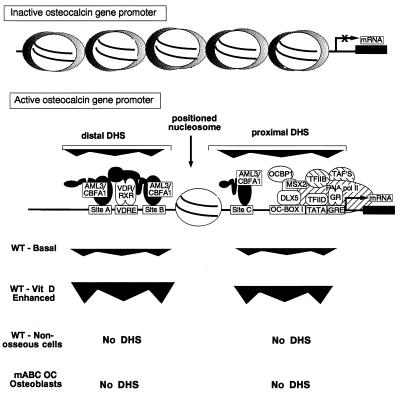
FIG. 7.
Illustration of modifications in chromatin organization of the rat OC promoter. The top line diagrams random distribution of nucleosomes across −1.7 kb of the rat OC promoter in cells that do not express OC, as established by micrococcal nuclease digestion (46). The second line diagrams the actively transcribed OC promoter, showing the span of two DNase I-hypersensitive (DHS) sites together with regulatory elements in the proximal and distal DHS sites. The extent of DNase I hypersensitivity is compared for basal OC expression (WT-basal) and vitamin D-treated (WT–vitamin D-enhanced) osteoblasts (ROS 17/2.8) cells. No DHS, undetectable DNase I hypersensitivity in nonosseous cells or undetectable mABC OC promoter in osteoblasts.
DISCUSSION
The OC gene promoter provides a blueprint for defining factors that regulate osteoblast-specific expression of the gene. These factors contribute to the complexity of molecular mechanisms associated with development of the osteoblast phenotype. We and others (6, 7, 28, 61–63) have shown by deletion analysis of the rat OC promoter from −1.7 kbp to −108 bp that a significant loss of transcriptional activity occurs when the proximal Cbfa site C is not present. This observation suggests either that there is functional redundancy of the Cbfa sites or that the distal recognition sites do not contribute to promoter activity. To understand the contribution of Cbfa factors to regulation of OC gene transcription, it was necessary to address the function of the multiple Cbfa sites in the native OC promoter.
Our studies have established the importance of the distal Cbfa sites A and B to activity of the rat OC promoter in a genomic context. We used a series of cell lines with stable integrants of the −1.1-kb OC promoter or with site-specific mutations of each Cbfa sequence alone, as well as combinatorial mutations of two or all three Cbfa sites. While transient-transfection assays have greatly expanded our knowledge of transcriptional mechanisms regulating gene expression, such transfected promoter-reporter constructs may not completely reflect the regulation of endogenous genes. In stable integrants of the OC promoter in ROS 17/2.8 cells, mutation of either site A or B results in a significant loss of promoter activity. Mutation of the proximal site C (OSE-2 [13]) alone has the least effect in modifying bone-specific basal activity of the OC promoter, suggesting strong compensatory activity by the distal Cbfa sites. However, when we carried out Cbfa expression studies to confirm the importance of each Cbfa site, the results demonstrated that all three sites are necessary for maximal basal promoter activity. The more pronounced effects of the Cbfa site mutations observed when the OC promoter is stably integrated into the genome of ROS 17/2.8 cells suggest that the chromatin context strongly influences activity of the promoter.
Our results indicate that regulatory elements other than Cbfa sites contribute to OC gene transcription. We find that in transient assays, 40% of wild-type OC promoter activity remains when Cbfa sites A, B, and C are mutated (mABC [Fig. 3B]), while 20% of activity is retained by the mABC mutant in stable cell lines (Fig. 3C). This residual promoter activity of mABC is consistent with results from promoter deletion analysis of the rat OC gene (26, 27) and reflects the contribution of the highly conserved OC box I (−99 to −76) regulatory element, which is also necessary for tissue-specific basal expression.
The major products of the various Cbfa/AML genes have several shared domains that contribute to transcriptional regulation of tissue-specific genes; the Cbfa factors may function as architectural proteins that serve to assemble macromolecular complexes involved in gene regulation. These structurally and functionally homologous segments include the conserved DNA binding rhd and transcriptional activation and suppression domains (24, 35, 44), as well as subcellular targeting signals (29, 69). The promoter-organizing functions of Cbfa factors may involve Cbfa-interacting proteins, including Cbfβ (5, 6, 32, 56), ALY (10), and Groucho/TLE (3, 22, 36, 60). Interestingly, Groucho/TLE proteins have been shown to contact the N terminus of histone H3 (15, 50). The Cbfa class of transcription factors has also been shown to associate with the nuclear matrix (29, 68, 69), the structural scaffold of the nucleus, through a 31-amino-acid nuclear matrix targeting sequence (NMTS) in the C terminus of full-length Cbfa isoforms (reference 69 and unpublished data). The NMTS directs Cbfa factors to transcriptionally active subnuclear sites (68), similar to the NMTS-dependent targeting of the rat glucocorticoid receptor (59). Together, the multiple protein-protein interaction domains of Cbfa factors may operate by a promoter architectural mechanism to functionally support physiologically regulated expression of the tissue-specific OC gene.
We have shown that mutation of the Cbfa sites results in a striking loss of responsiveness of the rat OC promoter to vitamin D and other physiological mediators of osteoblast differentiation, including glucocorticoids and TGFβ (preliminary data [30]). These signalling molecules regulate OC transcription through, respectively, VDR-RXR, GR, and AP-1 factors that act at non-Cbfa elements. Furthermore, mutation of the three Cbfa sites results in complete loss of DNase I hypersensitivity and the dynamic vitamin D-dependent modifications in chromatin structure which are essential for normal activity of the OC promoter. Indeed, a similar absence of DNase I hypersensitivity is observed in the silent endogenous OC gene within nonosseous cells (Fig. 7). Therefore, the competency of this promoter to undergo chromatin remodeling for maximal transcriptional responsiveness involves key contributions of Cbfa factors. Our results are consistent with the concept that Cbfa factors contribute to a promoter conformation that mediates accessibility or recruitment of factors to DNA regulatory elements.
Steroid hormone-dependent transcriptional activation is known to involve a modification in chromatin organization. In the rat OC gene, we have established that binding of the vitamin D-liganded VDR-RXR complex to the VDRE in the distal promoter induces architectural changes in chromatin that facilitate requisite interactions with the proximal basal promoter complex (21, 72). Glucocorticoid regulation of the mouse mammary tumor virus promoter (8, 16, 55, 64, 67) involves the GR-mediated conversion of a repressive chromatin state to an open configuration allowing NF1 and Oct1 access to their binding sites. Subsequently, activation of transcription occurs through interactions of GR with the TFIID basal complex (2, 54). The results presented here suggest that interaction of the OC promoter with the nuclear matrix-associated Cbfa1 factor is an essential step for steroid hormone-dependent activity of the OC promoter.
The significance of the three Cbfa sites in the rat OC promoter, two of which flank the VDRE, in contributing to maximal expression and physiologic responsiveness of the gene is highlighted by the opposing effects of vitamin D on the mouse OC promoter (11, 39, 70). Vitamin D does not mediate enhanced activity of the mouse OC promoter (70), and in fact, the mouse OC VDRE exhibits weak downregulation by the hormone (39). Consistent with this finding, the mouse VDRE sequence is not flanked by two functional Cbfa sites as occurs in the rat OC promoter. In the mouse OC promoter, the distal Cbfa site resides upstream of the VDRE in a position similar to that in the rat promoter (nt −608 to −602), but site B (−441 to −435 in the rat), which is critically involved in vitamin D regulation of the rat promoter, is not present in the mouse promoter (17). Mutational analysis of the mouse Cbfa sites has established that the distal site contributes far less to transcription than the proximal Cbfa/OSE2 site (17). Clearly, our studies demonstrate that the distal Cbfa A and B sites are important for basal activity of the rat OC promoter and essential for vitamin D responsiveness. The subtle differences between mouse and rat in the organization of Cbfa motifs may be necessary to regulate OC expression and responsiveness to physiologic mediators of bone formation and turnover at different levels, depending on the species. Thus, caution must be exercised when generalizing conclusions with respect to regulation of OC promoters from different species.
Many tissue-specific genes contain multiple Cbfa sites which are strategically positioned relative to other cis-acting elements (20, 31, 52). This heterogeneity in promoter organization of Cbfa-dependent genes, together with a series of context-dependent activation domains in the C termini of Cbfa factors, suggests an inherent difficulty in predicting the transcriptional effect of a given Cbfa site. This molecular complexity provides the necessary versatility to accommodate the different biological functions of the broad spectrum of Cbfa-regulated genes. Our studies provide the first evidence that Cbfa1 factors in osteoblasts regulate bone tissue-specific transcription not only through their DNA binding activities but also as nuclear-matrix-associated factors that mediate chromatin organization and facilitate transcriptional activity by association with other transactivating factors.
ACKNOWLEDGMENTS
We thank Judy Rask for editorial assistance.
This work was supported by grants from the National Institutes of Health (AR39588, AR45689, DE12528, and TW00990).
REFERENCES
- 1.Ahn M Y, Bae S C, Maruyama M, Ito Y. Comparison of the human genomic structure of the runt domain-encoding PEBP2/CBFα gene family. Gene. 1996;168:279–280. doi: 10.1016/0378-1119(95)00751-2. [DOI] [PubMed] [Google Scholar]
- 2.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 3.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 5.Banerjee C, Hiebert S W, Stein J L, Lian J B, Stein G S. An AML-1 consensus sequence binds an osteoblast-specific complex and transcriptionally activates the osteocalcin gene. Proc Natl Acad Sci USA. 1996;93:4968–4973. doi: 10.1073/pnas.93.10.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee C, McCabe L R, Choi J-Y, Hiebert S W, Stein J L, Stein G S, Lian J B. Runt homology domain proteins in osteoblast differentiation: AML-3/CBFA1 is a major component of a bone specific complex. J Cell Biochem. 1997;66:1–8. doi: 10.1002/(sici)1097-4644(19970701)66:1<1::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee C, Stein J L, van Wijnen A J, Frenkel B, Lian J B, Stein G S. Transforming growth factor-beta 1 responsiveness of the rat osteocalcin gene is mediated by an activator protein-1 binding site. Endocrinology. 1996;137:1991–2000. doi: 10.1210/endo.137.5.8612540. [DOI] [PubMed] [Google Scholar]
- 8.Beato M. Chromatin structure and the regulation of gene expression: remodeling at the MMTV promoter. J Mol Med. 1996;74:711–724. doi: 10.1007/s001090050076. [DOI] [PubMed] [Google Scholar]
- 9.Bidwell J P, van Wijnen A J, Fey E G, Dworetzky S, Penman S, Stein J L, Lian J B, Stein G S. Osteocalcin gene promoter-binding factors are tissue-specific nuclear matrix components. Proc Natl Acad Sci USA. 1993;90:3162–3166. doi: 10.1073/pnas.90.8.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 11.Clemens T L, Tang H, Maeda S, Kesterson R A, DeMayo F, Pike J W, Gundberg C M. Analysis of osteocalcin expression in transgenic mice reveals a species difference in vitamin D regulation of mouse and human osteocalcin genes. J Bone Miner Res. 1997;12:1570–1576. doi: 10.1359/jbmr.1997.12.10.1570. [DOI] [PubMed] [Google Scholar]
- 12.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducy P, Zhang R, Geoffroy V, Ridall A L, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 15.Fisher A L, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 16.Fragoso G, Pennie W D, John S, Hager G L. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol Cell Biol. 1998;18:3633–3644. doi: 10.1128/mcb.18.6.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frendo J L, Xiao G, Fuchs S, Franceschi R T, Karsenty G, Ducy P. Functional hierarchy between two OSE2 elements in the control of osteocalcin gene expression in vivo. J Biol Chem. 1998;273:30509–30516. doi: 10.1074/jbc.273.46.30509. [DOI] [PubMed] [Google Scholar]
- 18.Frenkel B, Mijnes J, Aronow M A, Zambetti G, Banerjee C, Stein J L, Lian J B, Stein G S. Position and orientation-selective silencer in protein-coding sequences of the rat osteocalcin gene. Biochemistry. 1993;32:13636–13643. doi: 10.1021/bi00212a031. [DOI] [PubMed] [Google Scholar]
- 19.Frenkel B, Montecino M, Green J, Aslam F, Desai R, Banerjee C, Stein J L, Lian J B, Stein G S. Basal and vitamin D-responsive activity of the rat osteocalcin promoter in stably transfected osteosarcoma cells: requirement of upstream sequences for control by the proximal regulatory domain. Endocrinology. 1996;137:1080–1088. doi: 10.1210/endo.137.3.8603577. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y H, Shinki T, Yuasa T, Kataoka-Enomoto H, Komori T, Suda T, Yamaguchi A. Potential role of cbfa1, an essential transcriptional factor for osteoblast differentiation, in osteoclastogenesis: regulation of mRNA expression of osteoclast differentiation factor (ODF) Biochem Biophys Res Commun. 1998;252:697–702. doi: 10.1006/bbrc.1998.9643. [DOI] [PubMed] [Google Scholar]
- 21.Guo B, Aslam F, van Wijnen A J, Roberts S G E, Frenkel B, Green M, DeLuca H, Lian J B, Stein G S, Stein J L. YY1 regulates VDR/RXR mediated transactivation of the vitamin D responsive osteocalcin gene. Proc Natl Acad Sci USA. 1997;94:121–126. doi: 10.1073/pnas.94.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo B, Javed A, Green J, Stein J L, Lian J B, van Wijnen A J, Stein G S. Groucho/TLE proteins associate with the nuclear matrix and repress Cbfa/AML mediated transactivation on osteocalcin promoter. Bone. 1998;23:S184. . (Abstract.) [Google Scholar]
- 23.Haussler M R, Haussler C A, Jurutka P W, Thompson P D, Hsieh J C, Remus L S, Selznick S H, Whitfield G K. The vitamin D hormone and its nuclear receptor: molecular actions and disease states. J Endocrinol. 1997;154:S57–S73. [PubMed] [Google Scholar]
- 24.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussell M F, Gilliland D G, Lenny N, Meyers S. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoaglin D, Mosteller F, Tukey J. Understanding robust and exploratory data analysis. New York, N.Y: John Wiley & Sons; 1983. [Google Scholar]
- 26.Hoffmann, H., J. Green, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. Expression screening of factors binding to the osteocalcin bone-specific promoter element OC Box I: isolation of a novel osteoblast differentiation-specific factor. J. Cell. Biochem., in press. [DOI] [PubMed]
- 27.Hoffmann H M, Beumer T L, Rahman S, McCabe L R, Banerjee C, Aslam F, Tiro J A, van Wijnen A J, Stein J L, Stein G S, Lian J B. Bone tissue-specific transcription of the osteocalcin gene: role of an activator osteoblast-specific complex and suppressor hox proteins that bind the OC box. J Cell Biochem. 1996;61:310–324. doi: 10.1002/(sici)1097-4644(19960501)61:2<310::aid-jcb14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann H M, Catron K M, van Wijnen A J, McCabe L R, Lian J B, Stein G S, Stein J L. Transcriptional control of the tissue-specific, developmentally regulated osteocalcin gene requires a binding motif for the Msx family of homeodomain proteins. Proc Natl Acad Sci USA. 1994;91:12887–12891. doi: 10.1073/pnas.91.26.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito Y. The runt protein and its companion PEBP2: a close link between this transcription factor and AML. Leukemia. 1997;11(Suppl. 3):279–280. [PubMed] [Google Scholar]
- 30.Javed A, Gutierrez S, van Wijnen A J, Stein G S, Stein J L, Lian J B. Cbfa binding sites are required for hormonal responsiveness in the osteocalcin promoter. Bone. 1998;23:S196. . (Abstract.) [Google Scholar]
- 31.Ji C, Casinghino S, Chang D J, Chen Y, Javed A, Ito Y, Hiebert S W, Lian J B, Stein G S, McCarthy T L, Centrella M. CBFa(AML/PEBP2)-related elements in the TGF-beta type I receptor promoter and expression with osteoblast differentiation. J Cell Biochem. 1998;69:353–363. [PubMed] [Google Scholar]
- 32.Kanno T, Kanno Y, Chen L F, Ogawa E, Kim W Y, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor alpha subunit revealed in the presence of the beta subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komori T. Cbfa1, a transcription factor for osteoblast differentiation and bone formation. J Bone Miner Metab. 1998;16:1–4. [Google Scholar]
- 34.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y-H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 35.Lenny N, Meyers S, Hiebert S. Functional domains of the t(8;21) fusion protein, AML-1/ETO. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 36.Levanon D, Goldstein R E, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 38.Lian J, Stewart C, Puchacz E, Mackowiak S, Shalhoub V, Collart D, Zambetti G, Stein G. Structure of the rat osteocalcin gene and regulation of vitamin D-dependent expression. Proc Natl Acad Sci USA. 1989;86:1143–1147. doi: 10.1073/pnas.86.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lian J B, Shalhoub V, Aslam F, Frenkel B, Green J, Hamrah M, Stein G S, Stein J L. Species-specific glucocorticoid and 1,25-dihydroxyvitamin D responsiveness in mouse MC3T3-E1 osteoblasts: dexamethasone inhibits osteoblast differentiation and vitamin D downregulates osteocalcin gene expression. Endocrinology. 1997;138:2117–2127. doi: 10.1210/endo.138.5.5117. [DOI] [PubMed] [Google Scholar]
- 40.Lian J B, Stein G S, Stein J L, van Wijnen A J. Regulated expression of the bone-specific osteocalcin gene by vitamins and hormones. In: Litwack G, editor. Vitamins and hormones. San Diego, Calif: Academic Press; 1998. pp. 443–509. [DOI] [PubMed] [Google Scholar]
- 41.McCabe L R, Banerjee C, Kundu R, Harrison R J, Dobner P R, Stein J L, Lian J B, Stein G S. Developmental expression and activities of specific fos and jun proteins are functionally related to osteoblast maturation: role of fra-2 and jun D during differentiation. Endocrinology. 1996;137:4398–4408. doi: 10.1210/endo.137.10.8828501. [DOI] [PubMed] [Google Scholar]
- 42.McLean R A, Sanders W L, Stroup W W. A unified approach to mixed linear models. Am Stat. 1991;45:54–64. [Google Scholar]
- 43.Merriman H L, van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 44.Meyers S, Lenny N, Sun W-H, Hiebert S W. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 45.Montecino M, Frenkel B, Lian J, Stein J, Stein G. Requirement of distal and proximal promoter sequences for chromatin organization of the osteocalcin gene in bone-derived cells. J Cell Biochem. 1996;63:221–228. doi: 10.1002/(SICI)1097-4644(19961101)63:2%3C221::AID-JCB9%3E3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 46.Montecino M, Lian J, Stein G, Stein J. Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry. 1996;35:5093–5102. doi: 10.1021/bi952489s. [DOI] [PubMed] [Google Scholar]
- 47.Montecino M, Pockwinse S, Lian J, Stein G, Stein J. DNase I hypersensitive sites in promoter elements associated with basal and vitamin D dependent transcription of the bone-specific osteocalcin gene. Biochemistry. 1994;33:348–353. doi: 10.1021/bi00167a045. [DOI] [PubMed] [Google Scholar]
- 48.Mundlos S, Otto F, Mundlos C, Mulliken J B, Aylsworth A S, Albright S, Lindhout D, Cole W G, Henn W, Knoll J H M, Owen M J, Mertelsmann R, Zabel B U, Olsen B R. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 49.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp G W H, Beddington R S P, Mundlos S, Olsen B R, Selby P B, Owen M J. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 50.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 51.Ryoo H-M, Hoffmann H M, Beumer T L, Frenkel B, Towler D A, Stein G S, Stein J L, van Wijnen A J, Lian J B. Stage-specific expression of Dlx-5 during osteoblast differentiation: involvement in regulation of osteocalcin gene expression. Mol Endocrinol. 1997;11:1681–1694. doi: 10.1210/mend.11.11.0011. [DOI] [PubMed] [Google Scholar]
- 52.Selvamurugan N, Chou W Y, Pearman A T, Pulumati M R, Partridge N C. Parathyroid hormone regulates the rat collagenase-3 promoter in osteoblastic cells through the cooperative interaction of the activator protein-1 site and the runt domain binding sequence. J Biol Chem. 1998;273:10647–10657. doi: 10.1074/jbc.273.17.10647. [DOI] [PubMed] [Google Scholar]
- 53.Siegel S. Nonparametric statistics for the behavioral sciences. New York, N.Y: McGraw-Hill Book Company; 1956. [Google Scholar]
- 54.Smith C L, Hager G L. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J Biol Chem. 1997;272:27493–27496. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 55.Smith C L, Htun H, Wolford R G, Hager G L. Differential activity of progesterone and glucocorticoid receptors on mouse mammary tumor virus templates differing in chromatin structure. J Biol Chem. 1997;272:14227–14235. doi: 10.1074/jbc.272.22.14227. [DOI] [PubMed] [Google Scholar]
- 56.Speck N A, Stacy T. A new transcription factor family associated with human leukemias. Crit Rev Eukaryot Gene Expr. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 57.Stein G S, Lian J B, van Wijnen A J, Stein J L. The osteocalcin gene: a model for multiple parameters of skeletal-specific transcriptional control. Mol Biol Rep. 1997;24:185–196. doi: 10.1023/a:1006803615430. [DOI] [PubMed] [Google Scholar]
- 58.Stewart M, Terry A, Hu M, O’Hara M, Blyth K, Baxter E, Cameron E, Onions D E, Neil J C. Proviral insertions induce the expression of bone-specific isoforms of PEBP2alphaA (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci USA. 1997;94:8646–8651. doi: 10.1073/pnas.94.16.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Y, Getzenberg R H, Vietmeier B N, Stallcup M R, Eggert M, Renkawitz R, DeFranco D B. The DNA-binding and τ2 transactivation domains of the rat glucocorticoid receptor constitute a nuclear matrix targeting signal. Mol Endocrinol. 1998;12:1420–1431. doi: 10.1210/mend.12.9.0169. [DOI] [PubMed] [Google Scholar]
- 60.Thirunavukkarasu K, Mahajan M, McLarren K W, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbf beta. Mol Cell Biol. 1998;18:4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Towler D A, Bennett C D, Rodan G A. Activity of the rat osteocalcin basal promoter in osteoblastic cells is dependent upon homeodomain and CP1 binding motifs. Mol Endocrinol. 1994;8:614–624. doi: 10.1210/mend.8.5.7914673. [DOI] [PubMed] [Google Scholar]
- 62.Towler D A, Rodan G A. Identification of a rat osteocalcin promoter 3′,5′-cyclic adenosine monophosphate response region containing two PuGGTCA steroid hormone receptor binding motifs. Endocrinology. 1995;136:1089–1096. doi: 10.1210/endo.136.3.7867563. [DOI] [PubMed] [Google Scholar]
- 63.Towler D A, Rutledge S J, Rodan G A. Msx-2/Hox 8.1: a transcriptional regulator of the rat osteocalcin promoter. Mol Endocrinol. 1994;8:1484–1493. doi: 10.1210/mend.8.11.7877617. [DOI] [PubMed] [Google Scholar]
- 64.Truss M, Bartsch J, Mows C, Chavez S, Beato M. Chromatin structure of the MMTV promoter and its changes during hormonal induction. Cell Mol Neurobiol. 1996;16:85–101. doi: 10.1007/BF02088169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 66.Xiao Z S, Thomas R, Hinson T K, Quarles L D. Genomic structure and isoform expression of the mouse, rat and human Cbfa1/Osf2 transcription factor. Gene. 1998;214:187–197. doi: 10.1016/s0378-1119(98)00227-3. [DOI] [PubMed] [Google Scholar]
- 67.Zaret K S, Yamamoto K R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984;38:29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
- 68.Zeng C, McNeil S, Pockwinse S, Nickerson J A, Shopland L, Lawrence J B, Penman S, Hiebert S W, Lian J B, van Wijnen A J, Stein J L, Stein G S. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, Hiebert S W. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang R, Ducy P, Karsenty G. 1,25-dihydroxyvitamin D3 inhibits osteocalcin expression in mouse through an indirect mechanism. J Biol Chem. 1997;272:110–116. doi: 10.1074/jbc.272.1.110. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y W, Bae S C, Takahashi E, Ito Y. The cDNA cloning of the transcripts of human PEBP2alphaA/CBFA1 mapped to 6p12.3-p21.1, the locus for cleidocranial dysplasia. Oncogene. 1997;15:367–371. doi: 10.1038/sj.onc.1201352. [DOI] [PubMed] [Google Scholar]
- 72.Zhou H, Manji S S, Findlay D M, Martin T J, Heath J K, Ng K W. Novel action of retinoic acid. Stabilization of newly synthesized alkaline phosphatase transcripts. J Biol Chem. 1994;269:22433–22439. [PubMed] [Google Scholar]