Summary
B cell follicles (BCFs) in lymph nodes (LNs) are generally exempt of CD8+ T and NK cells. African green monkeys (AGMs), a natural host of simian immunodeficiency virus (SIV), display NK cell-mediated viral control in BCF. NK cell migration into BCF in chronically SIVagm-infected AGM is associated with CXCR5+ NK cells. We aimed to identify the mechanism leading to CXCR5 expression on NK cells. We show that CXCR5+ NK cells in LN were induced following SIVagm infection. CXCR5+ NK cells accumulated preferentially in BCF with proliferating B cells. Autologous NK-B cell co-cultures in transwell chambers induced CXCR5+ NK cells. Transcriptome analysis of CXCR5+ NK cells revealed expression of bcl6 and IL6R. IL-6 induced CXCR5 on AGM and human NK cells. IL6 mRNA was detected in LN at higher levels during SIVagm than SIVmac infection and often produced by plasma cells. Our study reveals a mechanism of B cell-dependent NK cell regulation.
Subject areas: immunology, virology
Graphical abstract
Highlights
-
•
IL-6 can induce CXCR5 on NK cells
-
•
CXCR5+ NK cells expressed high levels of bcl6 and IL6R
-
•
More IL-6+ plasmablast/plasma cells in lymph nodes in SIVagm than SIVmac infection
-
•
B cells participate in the regulation of NK cell migration into BCF
Immunology; Virology
Introduction
Natural killer (NK) cells promote rapid effector response contributing to the control of viral infections and defense against tumors (Choucair et al., 2019; van Erp et al., 2019; Vivier et al., 2011). Depending on the strength and combination of multiple possible inhibitory and activating signals, NK cells can be either activated or inhibited (Choucair et al., 2019; van Erp et al., 2019). The modulation of this equilibrium between activating or inhibitory receptors is used in clinical studies to improve NK cell effector activities (Di Vito et al., 2019; Khan et al., 2020). In humans, NK cells are usually divided into two major subsets (CD56bright and CD56dim) that differ according to anatomical localizations, phenotypes, and functions. It has become evident that the spectrum of human NK cell diversity is broader than originally appreciated (Strauss-Albee et al., 2015; Strauss-Albee and Blish, 2016) and that NK cells have the capacity to adapt and differentiate into long-lived memory cells (Flórez-Álvarez et al., 2018; Pahl et al., 2018; Paust et al., 2017). Moreover, NK cell diversity is influenced by age, history of infection, and the tissue micro-environment (Freud et al., 2017; Strauss-Albee et al., 2015; Strauss-Albee and Blish, 2016). Chronic viral infection can sustainably modulate NK cells toward improved or impaired function (Hammer et al., 2018). The presence of impaired NK cells in HIV infection was reported early on (Giavedoni et al., 2000; Mavilio et al., 2003). Similar observations have been made in the nonhuman primate (NHP) model for HIV, i.e. the macaque infected with SIVmac (Alter et al., 2005; Costanzo et al., 2018; Giavedoni et al., 2000; Huot et al., 2018; LaBonte et al., 2006; Maria et al., 2003; Schafer et al., 2015). NK cell modulation therapies are therefore being investigated in the context of HIV cure research (Flórez-Álvarez et al., 2018; Peppa, 2019; Saez-Cirion et al., 2014). Assays to improve NK cell-mediated viral control include NK cell-stimulation via IFN-α, TLR agonists, IL-21 or IL-15 superagonist associated with antiretroviral (ARV) treatment (Chehimi et al., 2010; Davenport et al., 2019; Ellis-Connell et al., 2018; Garrido et al., 2018; Harper et al., 2021; Jost et al., 2014; Papasavvas et al., 2019; Schlaepfer and Speck, 2008; van Hall et al., 2019; Vibholm et al., 2017). However, studies on NK cells in tissues during HIV and/or SIVmac infections are scarce, despite the fact that NK cells in blood and tissues show clear phenotypical and functional differences and that the major viral reservoirs reside in tissues. During ARV treatment, the virus persists in cellular and anatomical reservoirs, such as in memory CD4+ T cells of the mucosa, in PD-1+ and CTLA4+ CD4+ T cells in the T zone of lymph nodes (LNs) and particularly in follicular helper T (TFH) cells of the B cell follicles (BCFs), thereby potentially perturbating B cell responses (Banga et al., 2016; Fletcher et al., 2014; Fukazawa et al., 2015; Kuo and Lichterfeld, 2018; McGary et al., 2017; Vanhamel et al., n.d.). To progress on NK cell-targeting therapies for cancers and chronic viral infections, one must better understand NK cell trafficking and differentiation in tissues.
In LN, NK cells are predominantly located in the medulla and paracortex at steady state (Bajénoff et al., 2006; Ferlazzo and Carrega, 2012). Upon HIV-1 or SIVmac infection, NK cells in LN might decrease in numbers and display a reduced migratory capacity (Huot et al., 2017; Luteijn et al., 2011). African nonhuman primates, such as sooty mangabeys or African green monkeys (AGMs), have most likely been natural hosts for SIV for over one million years (Raehtz et al., 2016; Svardal et al., 2017). Once these monkeys are infected with SIV they do not progress to AIDS despite persistent high viremia (Huot et al., 2018; Paiardini et al., 2009). The tissue viral distribution in these species is different as compared with pathogenic HIV-1/SIVmac infections. Indeed, while a robust viral production in the intestine has been reported in both non-pathogenic and pathogenic infections, only natural hosts exhibit a strong control of SIV in secondary lymphoid tissues (SLTs) with a lack of virus in TFH cells (Brenchley et al., 2012; Diop et al., 2000; Estes, 2013; Gueye et al., 2004; Huot et al., 2018; Jacquelin et al., 2014; Martinot et al., 2013; Zeng et al., 2011). We previously showed that NK cells are responsible for this control in AGM (Huot et al., 2017). The capacity to migrate into BCF during chronic SIVagm infections was linked to high numbers of CXCR5+ NK cells in SLTs (Huot et al., 2017, 2021).
It is of major importance to understand how NK cells acquire the capacity to migrate into BCF and hence to upregulate CXCR5. This is of importance not only in the context of HIV/SIV infections but also more broadly as NK cells contribute in regulating humoral responses (Bradley et al., 2018; Gao et al., 2005; Rydyznski et al., 2018). Previous studies have for example shown that human NK cells can stimulate autologous resting B cells to synthesize Ig or modulate antibody responses through TFH cell killing (Blanca et al., 2001). NK cells capable to migrate into BCF display potentially enhanced potential to modulate humoral responses. The regulation of CXCR5 expression has previously been studied on B cells and TFH cells. Pre-TFH cells derive from CXCR5-CD4+T cells after stimulation by dendritic cells (DC) and soluble factors (IL-6/IL-21/IL-27), the latter inducing transcription factors including B-cell lymphoma 6 (Bcl-6). Bcl-6 represses transcription factors involved in lineage differentiation (T-bet, GATA3, RORγC) and favors expression of IL-21 and CXCR5 (Kroenke et al., 2012). These pre-TFH cells co-express CXCR5 and CCR7, which leads to their migration into the interfollicular zone where they encounter B cells. Pre-TFH-B cell interactions trigger activation (CD40L/CD40, MHC-II/TCR-CD4, CD80-CD86/CD28) and auto/paracrine (IL-6/IL-21/IL-27) signalization, allowing pre-TFH cells (CCR7+CXCR5+) to switch to TFH cells with a CCR7-CXCR5+ phenotype capable entering into BCF (Chavele et al., 2015; Eivazi et al., 2016; Wu et al., 2018).
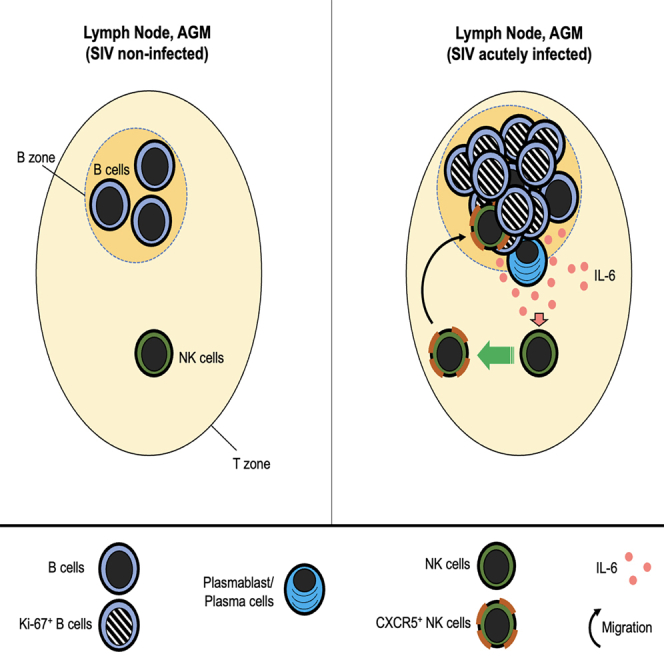
While the process of CXCR5 induction on TFH and B cells has been well studied, knowledge about the mechanism of CXCR5 induction on NK cells is lacking. Herein, we studied CXCR5 induction on NK cells by taking advantage of the AGM/SIVagm model. We show that CXCR5+ NK cells accumulate preferentially in BCF with high numbers of proliferating B cells. We explored the NK-B cell cross talk and observed a proximity between NK and B cells inside and around BCF in acute SIVagm infection. Autologous co-cultures of NK with B cells in transwell chambers induced CXCR5 on NK cells. Transcriptome analysis of CXCR5+ NK cells revealed high expression of Bcl6 and Interleukine-6 receptor (IL6R). IL-6 mRNA was detected to high levels in LN during SIVagm acute infection. IL-6 induced in a dose-dependent manner CXCR5 on NK cells from AGM in vitro and also up-regulated CXCR5 on NK cells from humans. IL-6 was produced by plasma cells in the LN of chronically SIVagm-infected AGMs. Our study reveals a B-NK cell cross talk impact and identifies IL-6 as a major factor contributing in the induction of CXCR5+ NK cells.
Results
CXCR5+ NK cells accumulate early after SIVagm infection in activated B cell follicles
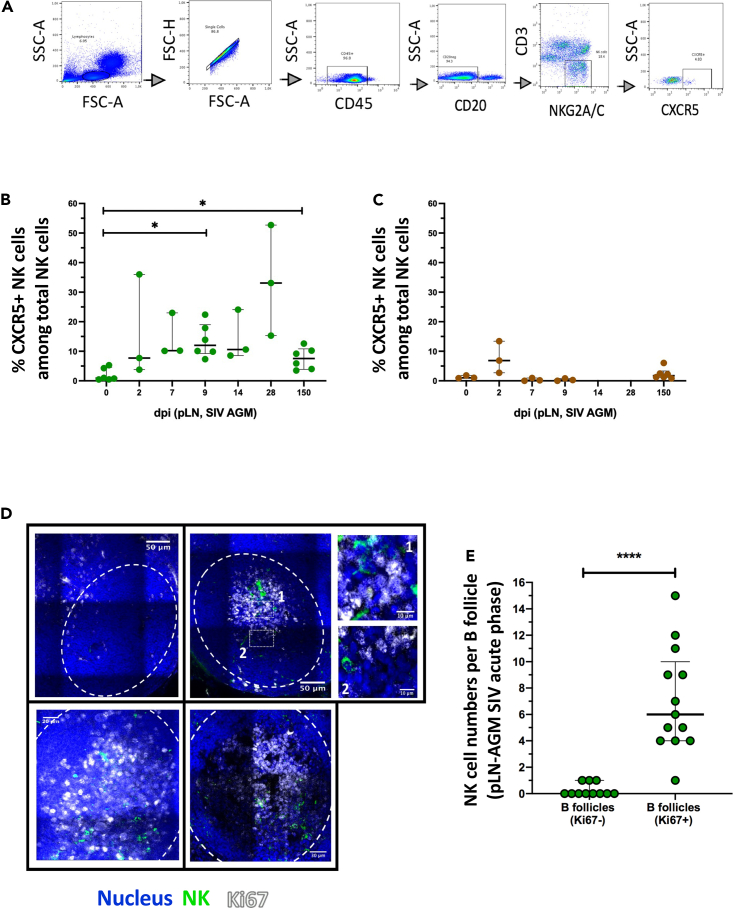
We previously showed that CXCR5+ NK cells were elevated in peripheral LN (pLN) of chronically SIVagm-infected AGM in contrast to SIVmac-infected macaques (Huot et al., 2017, Huot et al., 2021). To further decipher the kinetics of appearance of CXCR5+ NK cells after SIVagm infection, we followed CXCR5+ NK cells in pLN at days 0, 2, 7, 9, 14, 28 and 150 post-infection (p.i.). NK cells were gated as previously described (Huot et al., 2017) (Figures 1A and S1A). Before SIV infection, the median percentage of CXCR5+ NK cells in LN was 0.88% (Figure 1B). After SIVagm infection, the median percentage of CXCR5+ NK cells in the pLN was ranging from 7.69% to 33.1% between days 2 and 28 p.i. and was 7.5% at day 150 p.i (Figure 1B), thus similar to levels previously reported for chronic SIVagm infection (Huot et al., 2017). There was a strong inter-individual variability between the animals. However, the levels of CXCR5+ NK cells clearly increased in response to SIVagm infection. In contrast, there was no sign of such an increase of CXCR5+ NK cells in pLN of chronically SIVmac-infected macaques, although we cannot exclude an increase at day 2 p.i (Figures 1C and S1B).
Figure 1.
Temporal and spatial dynamics of CXCR5+ NK cells in peripheral lymph nodes during primary SIVagm infection
(A) Gating strategy of CXCR5+ NK cells in pLN. NK cells were gated as CD45+CD3−CD20−NKG2A/C+CXCR5+ cells as previously reported (Huot et al., 2017; Mavilio et al., 2005, p. 80). A representative image is shown. The example corresponds to pLN cells from an African Green Monkey infected by SIVagm at day 9 post-infection.
(B) Kinetic of CXCR5+ NK cell percentages in pLN during SIVagm infection, dpi = days post-infection. Each green circle indicates an individual animal (depending on the time point, n = 3 or 6 AGM); p value = ∗ <0.05, Wilcoxon test.
(C) Kinetic of CXCR5+ NK cell percentages during SIVmac infection. Each brown square indicates an individual animal (n = 3 or 6 MAC).
(D) Spatial distribution of NK cells and proliferating B cells in pLN during acute SIVagm infection (day 9 - 14 p.i.). The white dashed line delineates the B cell follicle (BCF). pLN from 5 individual AGMs were analyzed. The large images show representative BCFs for 4 distinct animals. White numbered squares (1 & 2) show zoomed areas.
(E) NK cell counts in B cell follicles in presence or absence of proliferating B cells as indicated by Ki-67 staining. The pLN were analyzed at day 9-14 p.i. Each green circle indicates one BCF (BCF n = 24), (n = 5 AGM; p value = ∗∗∗∗ <0.0001, Mann-Whitney test). Each dot bar indicates median values, and error bars indicate interquartile ranges.
To comprehend the spatial distribution of NK cells in LN during SIVagm infection, we analyzed the CCR7 expression on CXCR5+ NK cells (Figure S1C). The results suggest that between day 0 and day 2 p.i. the dynamics of CCR7+ and CCR7- CXCR5+ NK cells were comparable, but starting from day 7 p.i., the levels of CCR7−CXCR5+ NK cells appeared to be higher than the CCR7+CXCR5+ NK cells (Figure S1D). However, the number of animals analyzed was too low to conclude.
In chronic SIVagm infection, NK cells were found either around or inside BCF (Huot et al., 2017). We wondered whether the distinct anatomical location of NK cells was related to the activation state of the BCF (Phan et al., 2009). We stained pLN collected during acute SIVagm infection with Ki-67 to identify proliferating B cells. NK cells localized predominantly inside those BCF harboring high numbers of Ki-67 + cells (Figures 1D and 1E). Altogether these data show that CXCR5+ NK cells are induced rapidly after SIVagm infection and that migration into BCF occurs when B cells in BCF are strongly proliferating.
Induction of CXCR5+ NK cells by soluble factor(s) produced in autologous NK-B cell co-cultures
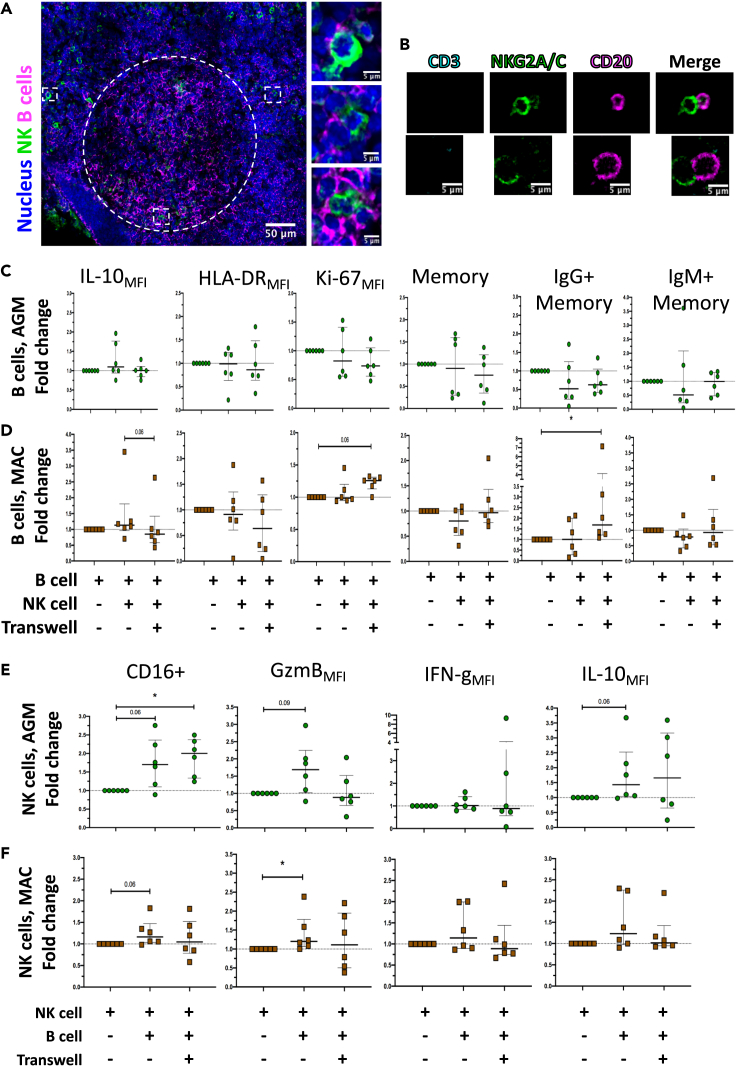
In order to identify the causative link between the presence of NK cells and activated B cells, we investigated the cross talk between these two cell types. A pre-requisite for a cross-talk is proximity. We therefore analyzed the location of NK and B cells in situ inside the pLN of SIVagm-infected AGM. We observed multiple zones of possible direct contact between NK and B cells in the interfollicular areas around BCF and inside the BCF (Figure 2A). Then, to examine further NK-B cell interactions, autologous NK and B cells were isolated from uninfected AGMs and co-cultured. Microscopy analyses were realized after 1 day of co-culture. Multiple contacts between B and NK cell membranes were observed in the co-cultures (Figures 2B and S1E). In order to investigate if NK and B cell contacts affect the phenotype of NK and/or B cells, we performed co-cultures of autologous NK and B cells from uninfected AGMs and macaques and analyzed them by flow cytometry. The cells were cultured for 6 days as follows: (i) NK cells alone; (ii) B cells alone, (iii) NK cells co-cultured with autologous B cells, and (iv) NK cells co-cultured with autologous B cells separated by transwell chambers for preventing direct contacts. We first analyzed the phenotype of B cells in the co-cultures. We did not observe any significant changes with the AGM cells (Figure 2C). In the macaque cell cultures, IgG + B memory cell increased in the transwell condition (Figure 2D). We then analyzed the NK cell phenotype. The frequency of CD16+ and Granzyme B + NK cells increased in the co-cultures with B cells of AGMs and/or macaques (Figures 2E and 2F). We then evaluated CXCR5+ NK cells (Figures 3A and 3B). The frequency of CXCR5+ NK cells was increased in co-cultures with B cells as compared to NK cells alone, both for AGM and macaques (Figures 3C and 3D). Strikingly, CXCR5+ NK cell levels were significantly increased only in the transwell condition, but not in cultures allowing contacts (Figures 3C and 3D). We cannot exclude that by studying more animals, an increase would also be seen for co-cultures in contact. However, the increases of CXCR5+ NK cells appeared higher in the transwells when compared with NK-B cell cultures with contact (p = 0.03 for AGM, p = 0.06 for MAC). These results indicated the presence of one or more soluble factor(s) produced during the co-culture of NK and B cells that induced CXCR5 expression on NK cells.
Figure 2.
Contact and mutual impact between NK and B cells
(A) Co-staining of B cells and NK cells in pLN during primary SIVagm infection. pLN were analyzed at day 9 p.i. A representative section showing a BCF and the surrounding interfollicular area is shown. The white dashed line delineates the BCF. White squares show zoomed areas, (n = 3 AGM).
(B) Analysis of potential contacts between NK and B cells isolated from PBMC of non-infected AGM after 1 day of co-culture. (n = 3 AGM). Two representative examples for each marker are shown (top and bottom, from the same culture). The CD3 labeling is used here as a control and is absent on either NK or B cells.
(C) Analysis of the phenotype of B cells after co-culture with autologous NK cells or co-culture with autologous NK cells separated by a transwell membrane. B and NK cells were isolated from PBMC of uninfected AGMs. The fold change in B cell frequency of a given phenotype among total B cells compared to B cells cultured alone is shown after 6 days of co-culture (n = 6 AGM).
(D) Analysis of the phenotype of B cells after 6 days of co-culture with autologous NK cells, separated or not by a transwell membrane. Cells were isolated from PBMC of uninfected MAC (n = 6 MAC). Fold-change compared with B cells cultured alone (p value = ∗ <0.05, trends (p = 0.06) are also indicated, Wilcoxon test).
(E) Analysis of the phenotype of NK cells co-cultured with B cells or co-cultured with B cells separated by a transwell membrane. NK and B cells were isolated from PBMC of uninfected AGMs. The fold change in NK cell frequency of a given phenotype among total NK cells compared with NK cells cultured alone is shown after 6 days of co-culture (n = 6 AGM) (p value = ∗ <0.05, trends (p = 0.06 and p = 0.09) are also indicated, Wilcoxon test).
(F) Analysis of the phenotype of NK cells after 6 days of co-culture with autologous B cells, separated or not by a transwell membrane. Cells were isolated from PBMC of uninfected MAC (n = 6 MAC). Fold-change compared with NK cells cultured alone (p value = ∗ <0.05, trends (p = 0.06) are also indicated, Wilcoxon test). Each dot bar indicates median values, and error bars indicate interquartile ranges.
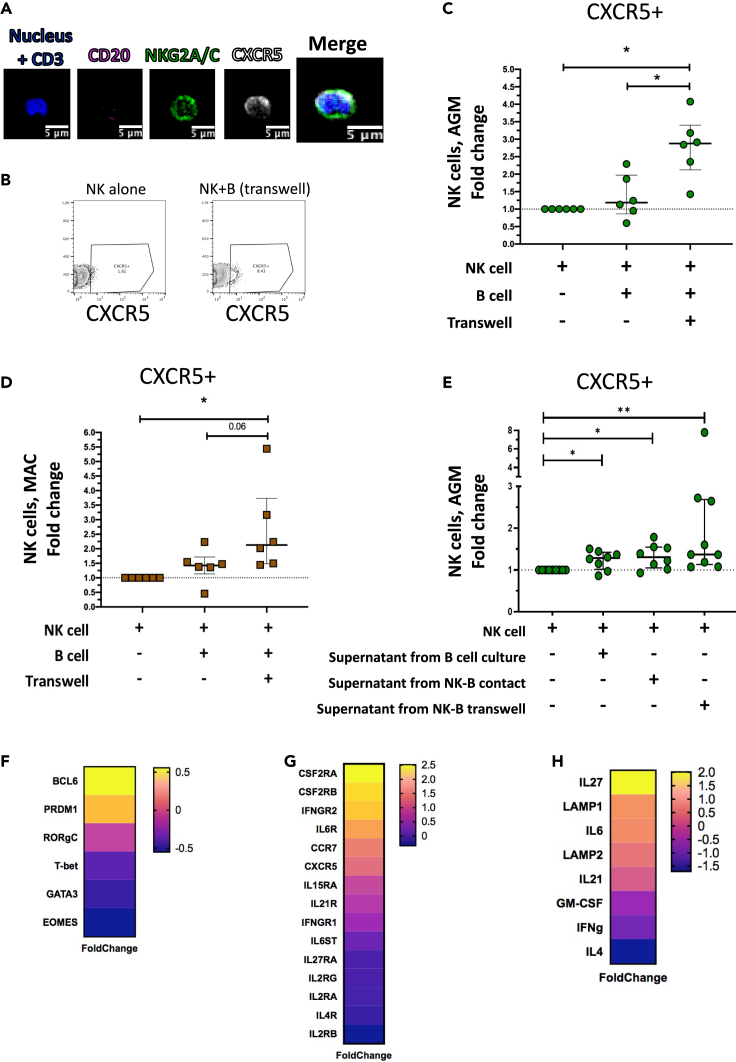
Figure 3.
CXCR5+ NK cell induction and gene expression profile
(A) Staining of NK cells co-cultured with autologous B cells separated by a transwell membrane illustrating CXCR5 expression on NK cells. The CD3 labeling is used here as a negative control.
(B) Dot plot representative of CXCR5+ NK cells from uninfected AGM after 6 days of culture: NK cells cultured alone and NK cells co-cultured with autologous B cells separated by a transwell membrane.
(C) The fold change of CXCR5+ NK cell frequencies among total NK cells compared with the frequency of CXCR5+ NK cells in NK cells cultured alone is shown after 6 days of co-culture. B and NK cells were isolated from PBMC of 6 non-infected AGMs (p value = ∗ <0.05, Wilcoxon test).
(D) The fold change of CXCR5+ NK cell frequencies among total NK cells compared with the frequency of CXCR5+ NK cells in NK cells cultured alone is shown after 6 days of co-culture. B and NK cells were isolated from PBMC of 6 non-infected MAC (p value = ∗ <0.05, trends (p = 0.06) are also indicated, Wilcoxon test).
(E) The fold change of CXCR5+ NK cell frequencies cultured 2 days with supernatants collected after 6 days of B cells cultured alone, of B and NK cell co-cultures and of B and NK cells cultured separated by a transwell membrane. Cells were isolated from blood of uninfected AGM (p value = ∗ <0.05 and ∗∗ <0.01, Wilcoxon test). Each green dot indicates an individual animal (Table S5).
(F–H) Transcriptomic profile of CXCR5+ NK cells isolated from pLN of chronically SIVagm-infected AGM. The gene expression profiles of CXCR5+ NK cells were compared with that of the predominant NK cell subpopulation in pLN, i.e. NKG2a/chighCD16- NK cells (n = 3 AGM). Each row represents the Log2 (Foldchange) of an individual gene as indicated. (F) Expression profiles of genes coding for transcription factors known to be associated with the CXCR5 pathway on TFH and B cells. (G) Expression profiles of genes coding for cellular cytokine and chemokine receptors. (G) Expression profiles of genes coding for soluble factors known to be associated with the CXCR5 pathway on TFH and B cells. Each dot bar indicates median values, and error bars indicate interquartile ranges.
To address this, we collected the supernatants from the cultures and added them to the culture of NK cells from SIV-negative AGM for two days. The supernatants collected from NK and B cell co-cultures in transwell chambers were sufficient to induce CXCR5 expression on NK cells (Figure 3E). The supernatants of NK and B cells cultured together and the supernatant of B cells cultured alone did also increase the levels of CXCR5+ NK cells but to a lesser extent than the supernatants from the co-cultures in transwells (Figure 3E). Altogether, these data show that one or more soluble factor(s) produced during NK-B cell co-cultures induced CXCR5 expression on NK cells.
CXCR5+ NK cells expressed high levels of IL-6 receptor
To identify this(ese) soluble factor(s), we analyzed the signaling pathways active in CXCR5+ NK cells from pLN of SIVagm-infected AGM. To this end, we used RNAseq to map the transcriptional profile of the CXCR5+ NK cell subpopulation compared with the major NK cell population present in LN: CD45 + CD3-NKG2AhighCD16- NK cells (Figure S2C) (Huot et al., 2021). We analyzed the expression profiles of genes coding for transcription factors, cytokines and cytokine-receptors (Figures 3F–3H; Table S4). Among the transcription factors, Bcl-6 was highly expressed in CXCR5+ NK cells (Figure 3F), similar to what is known for the CXCR5-expressing TFH lineage (Liu et al., 2012). The cytokine-receptor coding genes showing high levels of expression in CXCR5+ NK cells were the colony stimulating factor 2 receptor alpha subunit (CSF2RA/B) coding for the GM-CSF receptor, IFNGR2, IL6R, IL-15RA and IL21R (Figure 3G). CXCR5+ NK cells expressed high levels of IL-27, IL-6 and LAMP1 (CD107a) and low levels of IFN-g and IL-4 (Figure 3H). Previous studies have identified IL-6, IL-21 and IL-27 as major soluble factors responsible for the upregulation of CXCR5 on CD4+ T cells (Chavele et al., 2015; de Wit et al., 2015; Eivazi et al., 2016; Ma et al., 2012; Moukambi et al., 2019; Wu et al., 2018). Of note, the IL-6 receptor was expressed more strongly on CXCR5+ NK cells than other receptors such as IL-27R (Figure 3G), suggesting that IL-6 might be one of the factors involved in CXCR5 upregulation on NK cells.
IL-6 induced CXCR5 expression on NK cells in vitro
We next evaluated whether IL-6 was produced in our culture system. Intracellular IL-6 production was measured in the in vitro cultured B cells. A median of 24.5% B cells were IL-6 positive (Figure 4A). To define the level of IL-6 production ex vivo, we measured intracellular IL-6 in B cells from blood of uninfected AGM. Median levels of 2.5% IL-6+ B cells were observed ex vivo (Figure 4A). This indicates a strong enrichment of IL-6 production by B cells in our culture condition.
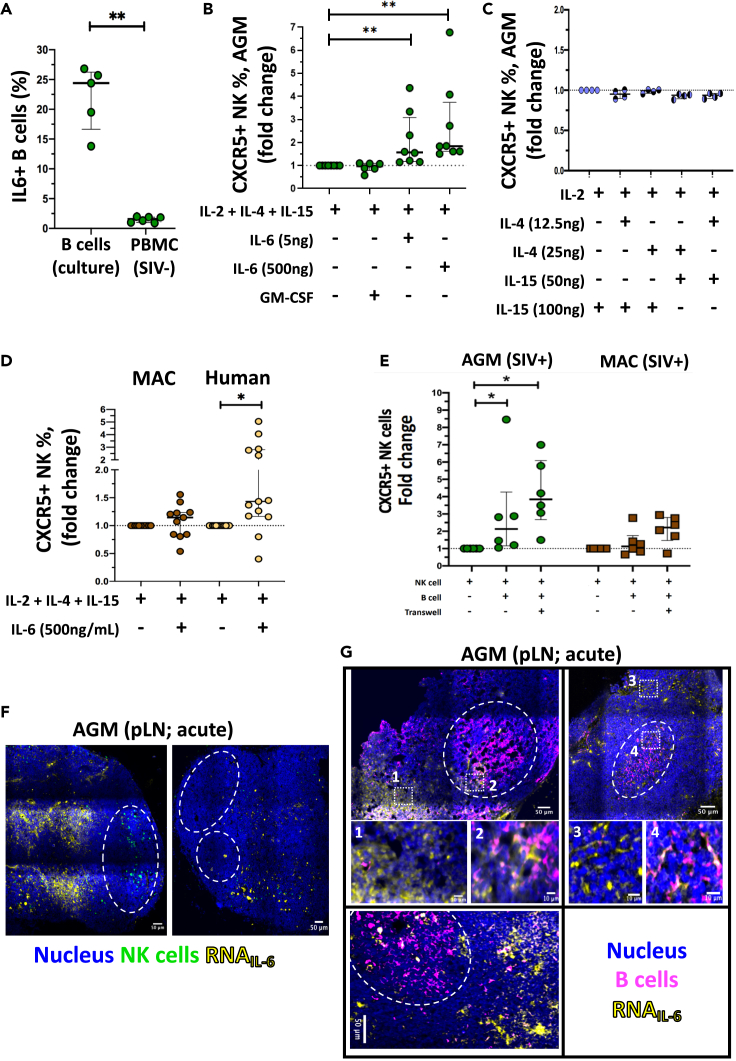
Figure 4.
IL-6 a soluble factor inducer of CXCR5 expression on NK cells in vitro
(A) Percentage of IL-6+ B cells isolated from blood of uninfected AGM after 6 days in culture or ex vivo in PBMC of uninfected and chronically SIVagm-infected AGM. Each green circle indicates an individual animal (n = 3–6 AGM), (p value = ∗∗ <0.01, Wilcoxon test).
(B) Impact of IL-6 on CXCR5+NK cell frequencies. NK cells were isolated from PBMC of uninfected AGMs and cultured for 2 days with distinct doses of IL-6 (5 or 500ng/mL) or with GM-CSF (250ng/mL). The fold change of CXCR5+ NK cell frequencies as compared to NK cells cultured without IL-6 or GM-CSF is shown. Each green circle indicates an individual animal (n = 5–7 AGM), (p value = ∗∗ <0.01, Wilcoxon test).
(C) Evaluation if the cytokines in the medium used for the cell cultures impacted on CXCR5+ NK cell frequencies. Cells were isolated from PBMC of uninfected AGM (n = 4 AGM) and cultured for 6 days with IL-2 (100 UI/mL), IL-4 (12.5ng/mL), IL-4 (25ng/mL), IL-15 (50ng/mL) or IL-15 (100ng/mL). Fold-change of CXCR5+ NK cell percentages compared to NK cells cultured alone.
(D) Impact of IL-6 on NK cells from macaques and humans. NK cells were isolated from PBMC of uninfected animals and healthy donors and cultured for 2 days with IL-6. The fold change of CXCR5+ NK cell frequencies as compared to NK cells cultured without IL-6 is shown (n = 11 MAC, n = 13 healthy donors), (p value = ∗ <0.05, Wilcoxon test).
(E) The fold change of CXCR5+ NK cell frequencies among total NK cells compared to the frequency of CXCR5+ NK cells in NK cells cultured alone is shown after 6 days of co-culture. B and NK cells were isolated from PBMC of 6 chronically SIV-infected AGMs (green) and 6 MACs (brown). (p value = ∗ <0.05, Wilcoxon test).
(F and G) IL-6 mRNA production in pLN during acute SIVagm infection (day 9 p.i.), (n = 6 AGM). IL-6 mRNA was co-stained with antibodies labeling (F) NK cells and (G) B cells. White squares (numbered 1–4) show zoomed regions. BCF are indicated by a dashed white line. Representative examples are shown. Each dot bar indicates median values, and error bars indicate interquartile ranges.
To study the impact of IL-6 on CXCR5 expression on NK cells, we cultured blood NK cells of uninfected AGM in the presence of IL-6. IL-6 induced an increase of CXCR5+ NK cell levels (Figure 4B). In contrast, GM-CSF, which similarly to IL-6 is known to be also produced by B cells (Cyster and Allen, 2019; Oleinika et al., 2019) and whose receptor was highly expressed on CXCR5+ NK cells, did not induce any change of the CXCR5+ NK cell frequencies (Figure 4B). Noteworthy, we also did not observe any impact of the cytokines used in the culture medium (IL-2, IL-4, IL-15) alone or combined on the CXCR5 expression on NK cells (Figure 4C).
We next investigated the IL-6 effect on NK cells from macaques and humans. We observed that human but not macaque NK cells upregulated CXCR5 expression in presence of IL-6 (Figure 4D). The fold increase of human CXCR5+ NK cells was comparable to that of AGM cells (Figures 4B and 4D). These different responses to IL-6 stimulation according to the species could not be explained by differences in IL6R macaque sequence compared with human and AGM IL6R sequences, the latter being closely related to the macaque IL6R (Figure S2D).
We searched for potential differences in the cultures with contact and in transwell condition. The levels of B and NK cells were the same between the two conditions, and importantly, the IL-6 production by B cells was not different between the co-culture with contact compared with the transwell condition (Figure S2E and F).
We then analyzed if there are differences before and after SIV infection. NK cells isolated during chronic SIVagm infection were more susceptible CXCR5 induction than NK cells isolated in chronic SIVmac infection, suggesting a loss of susceptibility to induction of CXCR5 expression on the NK cells in the SIVmac-infected macaques (Figure 4E). Altogether, we show that IL-6 in vitro is capable to induce CXCR5 on AGM and human NK cells.
IL-6 expression in pLN during acute SIVagm infection in vivo
We addressed the question of whether IL-6 could be relevant in vivo during SIVagm infection for inducing CXCR5+ NK cells. We therefore evaluated whether IL-6 could be locally produced in pLN using in situ microscopy analyses. We analyzed IL-6 mRNA expression in pLN during SIVagm and SIVmac infection (Figures 4F, 4G, 5A, and S3A). The IL-6 mRNA levels were higher in SIVagm than SIVmac acute infection (Figure 5B).
Figure 5.
IL-6 and CXCR5+ NK cells in chronic phase of SIV infection
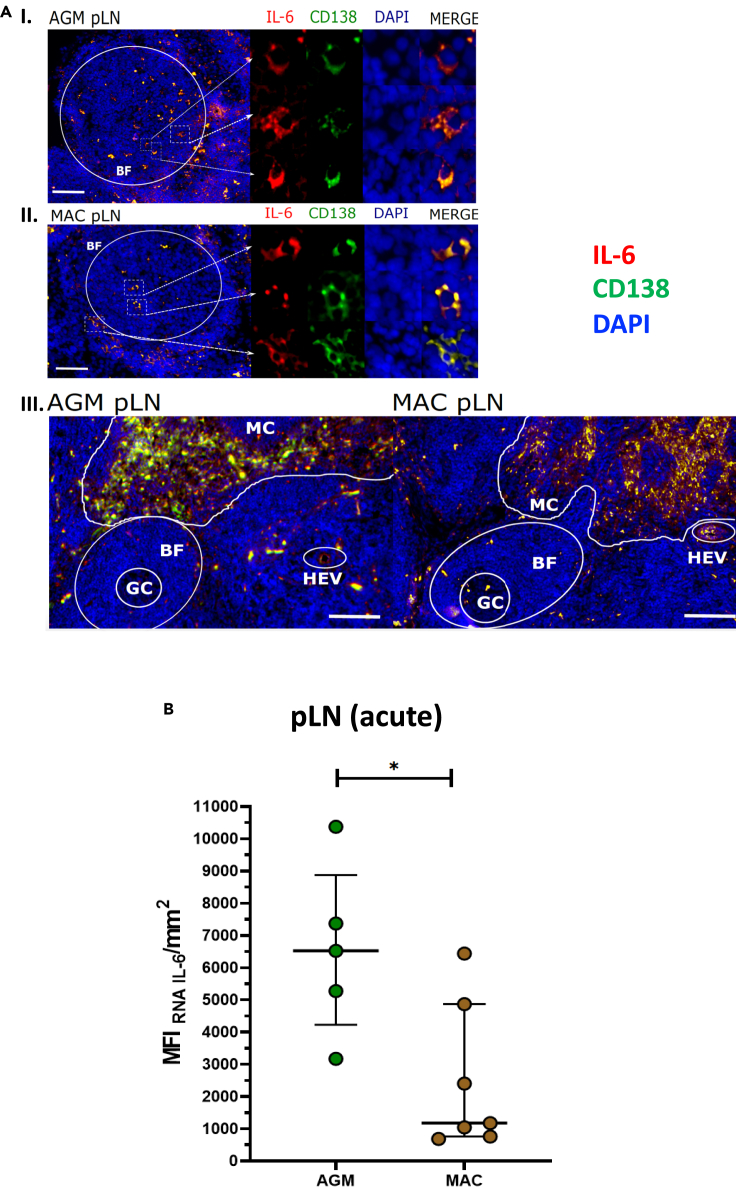
(A) IL-6 mRNA production by plasma cells identification (CD138) in pLN during chronic SIVagm and SIVmac infections (around day 200 p.i.), (n = 4 AGM and n = 4 MAC). Germinal Center (GC), B Follicle (BF), High Endothelial Venules (HEV) and Medulla Cords (MC) were identified by anatomical structure. The scale bar on image I and image II represent 90μm. The scale bar on image III represents 250μm.
(B) Quantification of IL-6 mRNA signal in pLN SIV acute infection from AGM and MAC. The pLN were analyzed at day 9-14 p.i. (n = 6 AGM, n = 7 MAC; p value = ∗ <0.05, Mann-Whitney test). Each dot bar indicates median values, and error bar indicate interquartile ranges.
To study the location of IL-6 production and if it is produced in vicinity of NK cells, we co-stained NK cells in pLN of acutely infected AGM (Figure 4F) macaques (Figure S3A). Cells expressing IL-6 were observed both in the BCF and T cell zone. In the interfollicular area of the T zone, foci of IL-6 mRNA expression were also detected, in particular in the AGM LNs (Figures 4F, 4G, and 5A).
We then addressed the question which cells expressed the IL-6. By analyzing LN cells from acutely infected AGM and MAC ex vivo using flow cytometry, we observed that T cells, CD20 + B cells, and CD3−CD20- cells produced IL-6 (Figure S3B). Using imaging analyses, we detected in LN during chronic SIVagm and SIVmac infection IL-6 expression in CD20+ B cells within the BCF (Figure 4G). In the medulla and around BCF of LN from SIVagm-infected AGM, IL-6 was produced by many plasma cells (Figure 5A). At the opposite, only few plasma cells expressed IL-6 in macaque LNs (Figure 5A). Altogether, IL-6 expression was higher in AGM than macaques during acute SIV infection (Figure 5B). IL-6 was expressed by distinct cells in several areas of the LN and often by plasma cells during chronic SIVagm infection.
Discussion
In this study, we investigated aspects of the B-NK cell cross-talk as well as the regulation of CXCR5 expression on NK cells. Several studies have already demonstrated that NK cells can directly impact humoral responses. It is therefore of key importance to better understand the regulation of NK migration into BCF. To contribute to this milestone, we performed functional, transcriptomic and imaging assays to study the NK-B cell cross talk in primate models and studied environmental factors participating to the induction of CXCR5 expression on NK cell during SIV infection.
We identified a soluble factor, IL-6, that by its own was capable of inducing CXCR5 on NK cells from AGM and humans. IL-6 is known to be increased in HIV-1 and SIVmac infections in plasma of untreated individuals (Petrovas et al., 2012), in contrast to SIVagm-infected AGM, known to exhibit similar IL-6 plasma concentration than uninfected animals (Campillo-Gimenez et al., 2010; Favre et al., 2009; Jacquelin et al., 2014). The lower levels of IL-6 in blood of SIVagm-infected AGMs compared with SIVmac infection therefore seems a paradox with regard to the higher levels of CXCR5+ NK cells in SIVagm compared with SIVmac infection. However, CXCR5+ NK cells have been detected in LN but not in blood or other tissues of SIVagm-infected AGM (Huot et al., 2017). IL-6 production is known to be increased by classical activation of B cells and/or by adding SIV on B cells (Epeldegui et al., 2010; Teleshova et al., 2006; Urashima et al., 1996). We show both in vitro and in vivo in pLN, that B cells expressed IL-6. We observed foci of IL-6 expression by plasmablasts/plasma cells in the interfollicular zones during acute SIVagm infection. It is thus possible that IL-6 produced by plasma cells up-regulate CXCR5 on NK cells allowing them to enter into the BCFs. It is not mutually exclusive that IL-6 production in the pLN during SIV infection is also due to other cells, such as macrophages, DC and T cells, which can be a major source of IL-6 in LN (Chavele et al., 2015; Hope et al., 1995). DC are well-known to cross-talk with NK cells (Fernandez et al., 1999; Walzer et al., 2005) and to participate in the induction of CXCR5+ pre-TFH cells (Eivazi et al., 2016; Rao, 2018; Suh, 2014). It is possible that plasma cells and DC in the interfollicular region and B cells in the BCF are the major sources of IL-6 in the pLN during SIVagm infection. The higher levels of IL-6 in LN during SIVagm compared to SIVmac infection might be explained by a lack of DC or plasma cell depletion in SIVagm compared to SIVmac infection. Interestingly, plasma cell/plasmablast frequency was significantly lower in the people living with HIV under ART compared with healthy individuals (Planchais et al., 2018). These data suggest that plasma cells/plasmablasts can have an important indirect role in LN homeostatic regulation during HIV/SIV infection.
We observed in CXCR5+ NK cells an upregulation of mRNA coding for several cytokine receptors, in particular GM-CSF receptors (CSF2RA/B), IFNGR and IL6R as compared with the main NK cell population in pLN of SIVagm-infected animals. GM-CSF did not induce CXCR5 expression on NK cells. In contrast, IL-6 induced CXCR5 expression on NK cells. We have previously shown that NK cells increased in numbers in the BCF in response to SIVagm infection and accumulated preferentially inside those BCF where follicular dendritic cells presented high levels of membrane-bound IL-15 (Huot et al., 2017). Membrane-bound IL-15 could concomitantly enhance B cell proliferation (Park et al., 2004) and augment the survival of the induced CXCR5+ NK cells in the germinal centers.
The co-cultures of B cells with autologous NK cells from macaques induced CXCR5+ NK cells in transwell condition, which indicates that these cells can potentially be induced in macaques. However, in line with previous data on macaques in chronic stage of SIVmac infection (Huot et al., 2017), there was no detectable increase of CXCR5+ NK cells in pLN during SIVmac infection. The absence of heightened levels of CXCR5+ NK cells in macaques might be explained by the need of local high concentrations of IL-6 as observed in LN during SIVagm infection and which were lacking in acute SIVmac infection. It could also be that the type of cells producing IL-6 and/or the location of the IL-6 producing cells are more important than the overall IL-6 concentrations in the pLN. Indeed, the anatomical distribution of IL-6 producing cells in pLN was distinct between SIVagm and SIVmac infections, in particular in the interfollicular zones. Moreover, multiple differences have been described between SIVmac and SIVagm infections regarding the inflammatory environment and/or maturation state of myeloid cells (Jacquelin et al., 2014; Shaffer et al., 2002; Wijewardana et al., 2010). It is possible that in addition to plasma cells, more DC producing IL-6 were present in LN in the acutely infected AGM than in the acutely infected MAC. Alternatively, NK cells from SIV-infected macaques might become less susceptible to IL-6 for other reasons such as a distinct differentiation stage in response to SIV infection, lower expression of IL6R and/or low levels of STAT3 (Theurich et al., 2017).
The observation that CXCR5+ NK cells were induced with supernatants from NK-B cell co-cultures but less with supernatants from B cells cultured alone provides evidence for a cross talk between NK and B cells. The impact of the cross talk on CXCR5+ NK cell levels might be explained by the presence of eventual co-factors secreted by NK cells and/or B cells after co-culture, and/or a higher susceptibility of NK cells to IL-6 (such as for example IL6R expression) after co-culture with B cells in transwell conditions. Of note, CXCR5+ NK cells were induced in the transwell chambers but not or to a lower extent in the co-cultures allowing contacts between NK and B cells. This indicates that the contact between NK cells and B cells was not necessary to induce CXCR5 and demonstrates CXCR5 upregulation on NK cells by soluble co-factors. Among the other cytokines known to participate in the induction of CXCR5 on CD4+T cells are IL-21 and IL-27 (Chavele et al., 2015; Eivazi et al., 2016; Moukambi et al., 2019). It is possible that these cytokines or other soluble factors play a role in CXCR5 upregulation in NK cells in vivo. The fact that contact was not necessary also demonstrated that CXCR5 on the NK cells was not acquired by trogocytosis. Surprisingly, the contact could inhibit CXCR5+ NK cell induction. Although there was a trend for NK cells to upregulate Granzyme B in the cultures in contact with B cells but not in the transwell chambers, it is unlikely that the NK cells killed the IL-6 producing B cells in the cultures with contact since the levels of total B cells and of IL-6 producing B cells remained the same. Moreover, the supernatant from cells in contact induced the same level of CXCR5+ NK cells than the supernatant from B cells cultured alone. Also, the IL-6+ B cell frequency was similar in co-cultures with or without contact. It is thus rather possible that NK cells cultured in transwells were more susceptible for IL-6, either through distinct levels of IL6R and/or distinct expression of signaling molecules. These hypotheses need to be addressed in future studies.
The analysis of transcription factors revealed that Bcl-6 was upregulated in the CXCR5+ NK cells in vivo, while the expressions of RORC, t-bet, gata-3, and EOMES were downregulated when compared with the predominant NK cell population in pLN of SIVagm-infected animals. This observation is in line with the literature regarding Bcl-6 and its capacity of this transcriptional factor to upregulate CXCR5, PD1, IL-21R, and IL6R expression and to repress the other transcriptional factors and CCR7 in TFH cells (Yu and Vinuesa, 2010; Xu et al., 2019). We also observed a lower expression of IL-2 receptors (IL2RA/B/G). These data are reminiscent of the IL-6-mediated decrease of IL-2 receptor expression on the surface of germinal center-TFH cells (Papillion et al., 2019). Altogether, our data emphasize on various major similarities between NK and TFH cells in the transcriptional profiles that may lead to CXCR5 expression. Surprisingly, the expression of PRDM1 was also upregulated in the CXCR5+ NK cells. PRDM1 encodes B lymphocyte-induced maturation protein-1 (Blimp1) (Shaffer et al., 2002). Blimp1 is an antagonist of Bcl-6 in B and T lymphocytes and thus the concomitant up-regulation might be surprising. However, it is known that in NK cells, the interaction of Blimp1 with Bcl-6 is distinct from B and T lymphocytes (Kallies et al., 2011). In human NK cells, Blimp1 is associated with Granzyme B expression and represses IFN-γ, TNF-α, and TNF-β production (Kallies et al., 2011). This profile is in line with the phenotype of the NK cells observed after co-culture.
Knowledge on the mechanism regulating CXCR5 expression in NK cells was scarce. This study contributes to elucidate CXCR5 induction on NK cells. These insights can be of interest for the efforts aiming at directing NK cells into BCF during HIV infections. They might also be of general relevance for other infections. Abnormally elevated levels of IL-6, for instance, are produced in other infections such as SARS-Cov2 infections and in some chronic inflammatory diseases (Costela-Ruiz et al., 2020). IL-6 is also produced in response to vaccinations (Vanden Bush et al., 2009). Depending on where this IL-6 is produced, it might influence the CXCR5+ NK cell levels. Little attention has been given so far to CXCR5+ NK cells and to NK cells in LN in general (Garcia et al., 2012; Theurich et al., 2017; Yang et al., 2019),. More knowledge is needed on these cells in lymphoid tissues in distinct settings and on their potential influence on B cell responses. Indeed, B cell responses are among many other factors also under the influence of NK cells during immunization strategies (Cook et al., 2015; Farsakoglu et al., 2019). NK cells have the potential to kill TFH and B cells and thereby strongly influence humoral responses (Blanca et al., 2001). Conversely, activated B cells could influence NK cell responses. We show here for instance an increase of CD16 + NK cells after co-culture with B cells and the preferential presence of NK cells in BCF containing high levels of proliferating B cells during SIVagm infection.
In conclusion, we deliver here key elements to understand how CXCR5+ NK cells are induced. We provide evidence for a cross talk between NK cells and B cells and importantly describe an interaction between IL-6 producing cells and NK cells. Our results thus open an unexplored research field regarding the regulation of NK cells by IL-6. CXCR5+ NK cells might play multiple beneficial (i.e. viral control) and harmful roles (i.e. reducing B cell responses) depending on the context (Bradley et al., 2018). Our approach thus is relevant regarding HIV cure, and moreover has a more general interest in immunology, including the treatment and prevention strategies of other infections and non-communicable inflammatory diseases.
Limitation of the study
In this study, we have analyzed CXCR5 induction on NK cell in SIVagm infection. Here, we observed in vitro transwell condition co-culture NK-B cell can induce CXCR5 on NK cells. However, we did not investigate the mechanism of CXCR5 induction in NK cells by the activation of B cell. Also, as discussed in the article, other cytokines (as IL-21 or IL27) are known to participate at the CXCR5 induction in CD4+ T cell. Yet, we did not study the effect of cytokine mixes on NK cells to induce CXCR5 at their surface. To further verify the mechanism of IL-6, it will be important to understand the NK signaling in NK cells and their susceptibility to IL-6 according to their species origin (AGM, MAC or Human).
Another interesting future challenge could be the study of the myeloid impact of IL-6 production in LN during SIVagm and SIVmac infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD45 | BD | 558411 |
| Anti-CD3 | BD | 560770 |
| Anti-CD16 | Beckman | A33098 |
| Anti-NKG2A | Beckman | PNA60797 |
| Anti-CD20 | BD | 555624 |
| Anti-Granzyme B | BD | 561151 |
| Anti-CXCR5 | MABTech | 11-9185-42 |
| Anti-PD-1 | Biolegend | 329922 |
| Anti-CD4 | BD | 560811 |
| Anti-MHC-E | Biolegend | 342603 |
| Anti-HLA-DR | OZYME | 307626 |
| Anti-IFNg | BD | 554702 |
| Anti-IL-10 | Biolegend | 501420 |
| Anti-IL6 | BD | 559331 |
| Anti-CD8 | Miltenyi | BW135/80 |
| Anti-CD3 | BD | 560770 |
| Anti-CD20 | BD | 560631 |
| Anti-IgD | Southern Biotech | 2030-07 |
| Anti-CD27 | BD | 560609 |
| Anti-CD107a | BD | 555802 |
| Anti-IgG | BD | 555786 |
| Anti-IgM | BD | 562031 |
| Anti-Ki67 | BD | 556027 |
| Anti-CD14 | BD | 557154 |
| Anti-PE microbeads | Miltenyi | 130-048-801 |
| Anti-CD20 microbeads | Miltenyi | 130-091-105 |
| Critical commercial assays | ||
| Human NK cell isolation kit | Stemcell | 17955 |
| RNeasy Mini Kit | Qiagen | 205113 |
| High-Sensitivity DNA Assay Kit | Invitrogen | Q33120 |
| Recombinant proteins | ||
| Human IL-2 | Miltenyi | 130-097-746 |
| Human IL-4 | Miltenyi | 130-093-917 |
| Human IL-15 | Miltenyi | 130-095-760 |
| Human IL-6 | Miltenyi | 130-093-931 |
| Human GM-CSF | Miltenyi | 130-093-862 |
| Experimental models: organisms/strains | ||
| Wild-type SIVagm.sab92018 | Diop OM et al., J. Virol., 2000 (https://10.1128/jvi.74.16.7538-7547.2000) | N/A |
| SIVmac251 isolate | Huot et al. Nat. Med. 2017 (https://doi.org/10.1038/nm.4421) | N/A |
| Oligonucleotides | ||
| Primers for IL-6, see STAR Methods | ACD | https://acdbio.com |
| Deposited data | ||
| RNA sequencing raw data | This paper and Huot et al. Nat. Med. 2021 (https://doi.org/10.1038/s41467-021-21402-1) | GSE140600 |
| Software and algorithms | ||
| Fiji (ImageJ) | Open Source | https://imagej.net/software/fiji/ |
| Prism | GraphPad | https://www.graphpad.com |
| CLC Sequence Viewer 7 | Open Source | https://clc-sequence-viewer.software.informer.com |
| BD FACSDiva | BD | https://www.bdbiosciences.com |
| FlowJo | BD | https://www.flowjo.com |
| Other | ||
| Monkey IL6 genome references (ENSMFAG00000000229, ENSCSAG00000013925) | Ensembl | https://www.ensembl.org |
| Monkey IL6R genome references (ENSCSAG00000001948, ENSMFAT00000012800, ENSMFAT00000012809, ENSMFAT00000012806, ENST00000344086, ENST00000368485, ENST00000476006, ENST00000512471, ENST00000622330) | Ensembl | https://www.ensembl.org |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Michaela Müller-Trutwin (mmuller@pasteur.fr).
Materials availability
Materials generated in this study will be made available on reasonable requests with a completed Materials Transfer Agreement. The authors declare that all other data supporting the findings of this study are available within the article and its supplemental information files or are available from the authors upon request.
Experimental model and subject details
Ethics statement and animal studies
Twenty AGMs (Caribbean Chlorocebus sabaeus) and twenty-eight cynomolgus macaques (Macaca fascicularis [MAC]) were studied. Nine of the twenty AGM were uninfected and thirteen AGM were exposed intravenously with the wild-type isolate SIVagm.sab92018 as previously reported (Diop et al., 2000; Huot et al., 2017). The MAC were imported from Mauritian AAALAC certified breeding centers. Seventeen of these MAC were uninfected and eleven MAC were exposed intravenously with the SIVmac251 isolate as previously reported (Huot et al., 2017) (Table S1).
The AGM and MAC were housed in IDMIT infrastructure facilities (CEA, Fontenay-aux-Roses, France) under animal facility authorization #D92-032-02 (Prefecture des Hauts de Seine, France) and in compliance with European Directive 2010/63/EU, the French regulations and the Standards for Human Care and Use of Laboratory Animals, of the Office for Laboratory Animal Welfare (OLAW, assurance number #A5826-01, US). The study was approved by the institutional ethical committee “Comité d’Ethique en Expérimentation Animale du Commissariat à l’Energie Atomique et aux Energies Alternatives” (CEtEA #44) under statement numbers A13-005, A15-035, A16-016, A17-059 and authorized by the “Research, Innovation and Education Ministry” under registration numbers 01114.02, APAFIS#2453-2015102713323361, APAFIS#4442-2016030818243239 and APAFIS# 11236-2017091214402801. The animals used here had been purchased for other programs and samples were shared with our study according to the 3R rule. Some results of these other programs have already been published (Huot et al, 2017, 2020; Palgen et al., 2020).
Uninfected animals were housed in groups and infected animals were housed in adjoining individual cages allowing social interactions, and maintained under controlled conditions with respect to humidity, temperature, and light (12 h light/12 h dark cycles). Water was available ad libitum. Animals were monitored and fed once or twice daily commercial monkey chow and fruit by trained personnel. Environmental enrichment was provided whenever possible including toys, novel foodstuffs, and music under the supervision of the CEA Animal Welfare Body. Experimental procedures (animal handling, viral inoculations, and samplings) were conducted after sedation with ketamine hydrochloride (10 mg/kg) or tiletamine (4 mg/kg) and zolazepam (4 mg/kg). For tissues collected at necropsy: animals were sedated with ketamine chlorhydrate (10 mg/kg) or tiletamine (4 mg/kg) and zolazepam (4 mg/kg) then humanely euthanized by intravenous injection of 180 mg/kg sodium pentobarbital. The sample size varied between three and eleven monkeys per group depending on the type of experiment.
Human blood collection and isolation of PBMC
Human blood samples from healthy donors were obtained from the French Blood Bank (Etablissement Français du Sang, EFS) in agreement with national and European legislation (C CPSL UNT, number 15/EFS/023). The samples were anonymous and sex and gender are unknown. The sample size is indicated in the figures. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation (Eurobio, CMSMSL01-01) (Table S6).
Processing of blood and tissues samples from NHP
PBMC were isolated as described above and the PBMCs either used fresh or frozen in FBS (Eurobio, CVFSVF00-01) with 10% of DMSO (Sigma, D2650-100ML) for later use.
After biopsy, the peripheral lymph nodes were transported to the laboratory and kept during the transport for 2–3 h at 4°C in RPMI medium. The pLN were then cut into pieces, grinded with a gentleMACS Dissociator (Miltenyi, 130-093-235) in the same RPMI medium. The pLN cell suspension was centrifugated at 1500g for 5 min to pellet the cells. The cells were used fresh or were frozen in FBS with 10% of DMSO and stored in liquid nitrogen until further use. The supernatants were immediately filtered at 70μm (Clearline, 141379C), centrifugated (1000g for 20 min) to clean from remaining cell debris and then stored at −80°C.
Isolation of NK cells and B cells from NHP
PBMCs were saturated for non-specific sites with FcR blocking reagent (Miltenyi, 130-059-901). NHP NK and B cells respectively express NKG2A/C and CD20 (Brocca-Cofano et al., 2017; Huot et al., 2017). To isolate NK cells, cells were incubated with NKG2A/C antibodies (Z199-PE) for 20 min at 4°C. Using anti-PE microbeads (Miltenyi, 130-048-801), NKG2A/C+ cells were selected through magnetic columns (Miltenyi, 130-042-401). To isolate B cells from PBMCs, we used anti-CD20 microbeads (Miltenyi, 130-091-105). Cells were passed through magnetic columns (Miltenyi, 130-042-401) to recover B cells.
Isolation of human NK cells
NK cells were isolated from PBMC by RoboSep (Stemcell) using the human NK cell isolation kit (17955, Stemcell).
Method details
Culture of NK cells and B cells
Co-cultures were realized between autologous NK and B cells (ratio 1:5) in 96 well plates and 96 transwell plates (Life Sciences, 3381). Cells were cultured in medium containing IL-2 (Miltenyi, 130-097-746; 100IU), IL-4 (Miltenyi, 130-093-917; 25ng/mL), IL-15 (Miltenyi, 130-095-760; 100ng/mL), IL-6 (Miltenyi, 130-093-931; 5 or 500ng/mL) and GM-CSF (Miltenyi, 130-093-862; 250ng/mL). The supernatants were collected after 2 or 6 days of culture and used after centrifugation.
Tissue inclusion preparation
To conserve frozen tissues for ulterior image analyses, tissue pieces or small biopsies were incubated in PFA 4% (AlfaAesar, J61899) overnight. After several washes, the tissues were immersed into sucrose (Sigma, S0389-500G) 10% at 4°C during 72h. Then the incubation medium was changed to sucrose 20%, and after 72h to sucrose 30%. Tissues were then snap-frozen in O.C.T. (Tissue-Tek, 4583). Tissues were conserved at −80°C or in liquid nitrogen. All tissues in O.C.T were cut at 10μm using a cryostat (LEICA CM 3050S) and put on slides (Thermo Scientific, J3800AMNZ). These slides were stored at −20°C.
Flow cytometry staining
Cells were thawed at room temperature. For the NK and B cell panels (Table S2), we saturated non-specific sites by incubation with PBS containing 4% of NHP sera during 20 min. We centrifuged at 1500 x g for 5 min. Then we added the antibodies to the samples (1 million of cells) and homogenized. We incubated the mix for 30 min at 4°C. We added 5 mL of PBS, centrifuged at 1500 x g for 5 min and discarded the supernatant. We permeabilized the cells by Cytofix solution (BD, 554,655) for 20 min at 4°C. Five mL of PermWash (X1) solution (BD, 51-2091KZ) was added and we centrifuged at 1900 x g for 5 min. Antibodies were added on samples and homogenized followed by incubation for 20 min at 4°C. We added 5mL of PermWash (X1) solution and centrifuged at 1900 x g for 5 min. We homogenized the pellet in PFA 4%. The fluorescence staining was observed and captured by a flow cytometer (BD, LSR II) and software (BD, Diva). Images were analyzed with FlowJo.
Immunofluoresce and in situ hybridization
For immunofluorescent staining of the tissue sections, the epitope revelation protocol was realized with PBS incubation at room temperature (RT) for 15 min. Then, the slides were incubated in methanol (Fisher Chemical, M/3950/15) at −20°C during 2 h (h) and subsequently in formaldehyde 2% at RT (Sigma, F8775-500mL) for 15 min. The non-specific sites were saturated with BSA 4% (Sigma, A6003-25G) during 2h at RT followed by a washing step for 1h with PBS. The primary antibodies (Table S3; 1:200) were added overnight at 4°C. Then the slides were washed for 1h in PBS. Secondary antibodies (Table S3; 1:200) and DAPI (Table S3; 1:1000) were added for 1h at 4°C. After a washing step (1h in PBS) the mounting medium was added (Invitrogen, 00-4958-02). We finally dropped off the cover (Fisher Scientific, 15,165,452 – Dutscher, 100038N) on the slide with mounting medium.
For the fluorescent in situ hybridization realized to measure IL-6 mRNA expression in LN, we followed the RNAscope protocol with IL-6 probe utilization (Advanced Cell Diagnostics Europe, 804,341-C2) and ACD HybEZ Hybridization system (Advanced Cell Diagnostics Europe, 310,013). The Macaca fascicularis (ENSMFAG00000000229) is known to present three transcripts (splice variants), while for the IL-6 gene from Chlorocebus sabaeus (ENSCSAG00000013925) one transcript is published. We analyzed the sequences by alignment on CLC Sequence Viewer. We identified a large region with 99% homology between all sequences and selected this region for constructing the probe. To exclude gDNA, we selected primers that hybridize on two exons. Based on this, the ACD company produced an IL-6 probe (300 nucleotides) to be used with RNAscope technology. Given the length and the homology of the probe for the 2 species, a distinct binding of the probe to macaque or AGM IL-6 RNA is not supported. The reference sequence used for IL-6 probe was CACTTGCCTGGTGAAAATCATCACTGGTCTTTTGGAGTTTGAGGTATACCTAGAGTACCTCCAGAACAGGTTTGAGAGTAGTGAGGAGCAAGCCAGAGCTGTGCAAATGAGTACAAAAGTCCTGATCCAGTTCCTGCAGAAAAAGGCAAAGAATCTAGATGCAATAACCACCCCTGAACCAACCACAAATGCCAGCCTGCTGACGAAGCTGCAGGCACAGAACCAGTGGCTGCAGGACATGACGACGCATCTCATCCTGCGCAGCTTTAAGGAGTTCCTGCAGTCCAGCCTGAGGGCTCTTCGGCAAATGT. Nucleotides underline represent differences between AGM and MAC IL-6 cDNA sequences from Ensembl databases (ENSCSAG00000013925, ENSMFAG00000000229). The antibodies used for stainings were used as described above. The fluorescence staining was captured in all assays by a spinning-disk (Yokagawa, CellVoyager CV1000, x20). Images were analyzed by ImageJ (Fiji).
Immunostaining of in vitro cultured cells was realized using coated wells with poly-L-lysine (P8920-100ML) for 1h at 37°C. NK and B cells were cultured one day before coating. Antibodies were added to the coated cells (Table S3). The fluorescence staining was captured in all assays by a confocal microscope (Zeiss, LSM700, x40). Images were analyzed by ImageJ (Fiji).
Cell sorting of NK cell subsets for RNAseq
Eight-color panels were used to phenotype, surface stain and sort NK cells. Total pLN cells were thawed in 20% FBS-containing media supplemented with benzonase nuclease. Cells were washed and stained with Aqua Live/Dead stain (Molecular Probes). Cells were washed and the sites saturated using normal mouse IgG (Caltag). pLN cells were surface stained for CD45 (D058-1283, BD), CD3 (SP34.2, BD), CD8 (BW135/80, Miltenyi), CD16 (3G8, Beckman Coulter, Inc.), NKG2a/c (Z199, Beckman Coulter, Inc.), CD20 (2H7, Biolegend), CD14 (M5E2, BD) and CXCR5 (MU5UBEE, MABTech). The antibodies are listed in Table S2. Post-staining, cells were washed, filtered and sorted on a FACS ARIA II (BD). Cells were directly collected in an RLT lysis buffer (Qiagen) that contained TCEP (tris(2-carboxyethyl) phosphine). The purity of the cells was >97%.
RNA extraction, library preparation and sequencing
RNA was isolated from the sorted NK cells using the RNeasy Mini Kit (205113, Qiagen). RNA integrity was verified with the Agilent Bioanalyzer. DNase-treated RNA was submitted to library preparation using the Truseq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, CA), according to manufacturer's instructions. An initial poly(A) RNA isolation step (included in the Illumina protocol) was performed on 10 ng of total RNA to keep only the polyadenylated RNA fraction and remove the ribosomal RNA. A step of fragmentation was then performed on the enriched fraction, by divalent ions at high temperature. The fragmented RNA samples were randomly primed for reverse transcription followed by second-strand synthesis to create double-stranded cDNA fragments. No end repair step was necessary. An adenine was added to the 3′-end and specific Illumina adapters were ligated. Ligation products were submitted to PCR amplification. The obtained oriented libraries were controlled by Bioanalyzer DNA1000 Chips (Agilent, # 5067-1504) and quantified by spectrofluorimetry (Quant-iT High-Sensitivity DNA Assay Kit, #Q33120, Invitrogen). Sequencing was performed on the Illumina Hiseq2500 platform to generate single-end 100 bp reads bearing strand specificity.
Phylogenetic analysis
Sequence alignment was operated by CLC Sequence Viewer 7 software. References of IL6R genes were obtained from cDNA Ensembl database. The genes correspond to IL6R_AGM (ENSCSAG00000001948), IL6R_MAC-X1 (ENSMFAT00000012800), IL6R_MAC-X2 (ENSMFAT00000012809), IL6R_MAC-X3 (ENSMFAT00000012806), IL6R_Hum-201 (ENST00000344086), IL6R_Hum-202 (ENST00000368485), IL6R_Hum-203 (ENST00000476006), IL6R_Hum-206 (ENST00000512471), and IL6R_Hum-208 (ENST00000622330). Phylogenetic analysis was carried out by the CLC Sequence Viewer 7 software. Neighbor Joining was the tree construction method used and the nucleotide distance measure is in Jukes-Cantor.
Quantification and statistical analysis
Bioinformatic analysis of the RNA sequence data
Bioinformatic analyses were performed using the RNA-seq pipeline from Sequana (Cokelaer et al., 2017). Reads were cleaned of adapter sequences, and low-quality sequences were removed using cutadapt version 1.11 (Martin, 2011). Only sequences ≥25 nucleotides (nt) in length were considered for further analysis. STAR version 2.5.0a, with default parameters, was used for alignment on the reference genome (Chlorocebus sabaeus, from Ensembl release 90). Genes were counted using feature counts version 1.4.6-p3 (Liao et al., 2014) from Subreads package (parameters: -t gene, -g ID and -s 1). Data were analyzed using R version 3.4.3 and the Bioconductor package DESeq2 version 1.18.1 (Love et al., 2014). Normalization and dispersion estimation were performed with DESeq2, using the default parameters, and statistical tests for differential expression were performed by applying the independent filtering algorithm. A generalized linear model, including the monkey identifier as a blocking factor, was used to test for the differential expression between the biological conditions. For each pairwise comparison, raw p Values were adjusted for multiple testing according to the Benjamini and Hochberg (BH) procedure (Benjamini and Hochberg, 1995). Each list was used to query the Kyoto Encyclopedia of genes and Genomes (KEGG), GO-biological function database and Wiki pathways. Genes with an adjusted p Value < 0.05 were considered differentially expressed (Table S4). Analyses and vizualisation of gene expressions by heat maps were performed using Prism (GraphPad, La Jolla, CA)
Image quantification analyses
To quantify the number of NK cells per B follicle in pLN from acute SIVagm infection, we first defined the BCF according its circular structure and density and/or CD20 + expressing cells. Then we counted the number of NK cells in Ki-67- and Ki-67 + BCF. The potential difference was analyzed using a Mann-Whitney test. To quantify MFI of RNAIL-6/mm2, we acquired the signal on calibrated image with exactly the same parameters (20% excitation, 200ms exposition, 20% gain) on CellVoyager (Yokagawa, CV1000) at x20 for each pLN. In ImageJ software (Fiji), the signal was collected via MFI measured on pLN section and MFI value according to a specific area was converted in mm2 (Campillo-Gimenez et al., 2010; Hirao et al., 2014; Raehtz et al., 2020).
Statistical analysis
In order to illustrate specific gene sets, we draw heatmaps based on variance-stabilizing transformed counts. For the other data, a Wilcoxon matched-pairs signed rank test was used with subsequent Bonferroni correction to evaluate whether there was a statistically significant difference in the level of one given marker at a given time point post-infection when compared to the baseline medians (day 0). Baseline medians in blood consisted of 3–6 pre-infection values per animal. All statistical computations were performed using Prism (GraphPad, La Jolla, CA). The statistical details of experiments can be found in the figures and their legends.
Acknowledgments
We are grateful for the excellent contributions from the veterinarians and staff at the IDMIT Center (Benoit Delache, Jean-Marie Helies, Raphaël Ho Tsong Fang, Delphine Desjardins and Nastasia Dimant). We also thank all the people from the Illumina Hiseq 2500 platform at the Institut Pasteur and especially Caroline Proux and Jean-Yves Coppee. NH was supported by the Fondation Beytout and Institut Pasteur. PR was recipient of a PhD fellowship from the University Paris Diderot, Sorbonne Paris Cité and also supported by the NIH (R01AI143457) and Institut Pasteur. CaP was recipient of a Roux-Cantarini Postdoctoral Fellowship. CyP was supported by the ERC-2013-StG 337146 program. We would like to acknowledge grant support from ANRS, the Fondation J. Beytout, MSD Avenir, the Fondation Les Ailes and from the NIH (R01AI143457) to MMT. IDMIT infrastructure is supported by the “Program Investissements d’Avenir” (PIA), managed by the ANR under reference ANR-11-INBS-0008 and by the PIA grant ANR-10-EQPX-02-01. We equally acknowledge the Investments for Future grant ANR-10–INSB–04 to support the UtechS Photonic BioImaging (Imagopole) and C2RT facilities at Institut Pasteur.
Author contributions
PR, NH, BJ, ML and CaP performed the experiments. PR, NH, BJ, CyP, HM and MMT designed the experiments. VC, NDB and BJ coordinated the animal studies. PR, NH, BJ, HM, RLG and MMT analyzed the data. MMT obtained the funding. PR and MMT wrote the manuscript and all co-authors reviewed it.
Declaration of interests
The authors declare no competing interests.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103109.
Supplemental information
The gene expression profiles of CXCR5+ NK cells were compared to that of the predominant NK cell subpopulation in pLN, i.e. NKG2a/chighCD16- NK cells (n = 3 AGM).
Data and code availability
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All raw data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alter G., Teigen N., Davis B.T., Addo M.M., Suscovich T.J., Waring M.T., Streeck H., Johnston M.N., Staller K.D., Zaman M.T. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood. 2005;106:3366–3369. doi: 10.1182/blood-2005-03-1100. [DOI] [PubMed] [Google Scholar]
- Bajénoff M., Breart B., Huang A.Y.C., Qi H., Cazareth J., Braud V.M., Germain R.N., Glaichenhaus N. Natural killer cell behavior in lymph nodes revealed by static and real-time imaging. J. Exp. Med. 2006;203:619–631. doi: 10.1084/jem.20051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga R., Procopio F.A., Noto A., Pollakis G., Cavassini M., Ohmiti K., Corpataux J.-M., de Leval L., Pantaleo G., Perreau M. PD-1 + and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med. 2016;22:754–761. doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Blanca I.R., Bere E.W., Young H.A., Ortaldo J.R. Human B cell activation by autologous NK cells is regulated by CD40-CD40 ligand interaction: role of memory B cells and CD5+ B cells. J. Immunol. 2001;167:6132–6139. doi: 10.4049/jimmunol.167.11.6132. [DOI] [PubMed] [Google Scholar]
- Bradley T., Peppa D., Pedroza-Pacheco I., Li D., Cain D.W., Henao R., Venkat V., Hora B., Chen Y., Vandergrift N.A. RAB11FIP5 expression and altered natural killer cell function are associated with induction of HIV broadly neutralizing antibody responses. Cell. 2018;175:387–399.e17. doi: 10.1016/j.cell.2018.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J.M., Vinton C., Tabb B., Hao X.P., Connick E., Paiardini M., Lifson J.D., Silvestri G., Estes J.D. Differential infection patterns of CD4+ T cells and lymphoid tissue viral burden distinguish progressive and nonprogressive lentiviral infections. Blood. 2012;120:4172–4181. doi: 10.1182/blood-2012-06-437608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocca-Cofano E., Kuhrt D., Siewe B., Xu C., Haret-Richter G.S., Craigo J., Labranche C., Montefiori D.C., Landay A., Apetrei C., Pandrea I. Pathogenic correlates of simian immunodeficiency virus-associated B cell dysfunction. J. Virol. 2017;91 doi: 10.1128/JVI.01051-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillo-Gimenez L., Cumont M.-C., Fay M., Kared H., Monceaux V., Diop O., Müller-Trutwin M., Hurtrel B., Lévy Y., Zaunders J. AIDS progression is associated with the emergence of IL-17–producing cells early after simian immunodeficiency virus infection. J. Immunol. 2010;184:984–992. doi: 10.4049/jimmunol.0902316. [DOI] [PubMed] [Google Scholar]
- Chavele K.-M., Merry E., Ehrenstein M.R. Cutting edge: circulating plasmablasts induce the differentiation of human T follicular helper cells via IL-6 production. J. Immunol. 2015 doi: 10.4049/jimmunol.1401190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehimi J., Papasavvas E., Tomescu C., Gekonge B., Abdulhaqq S., Raymond A., Hancock A., Vinekar K., Carty C., Reynolds G. Inability of plasmacytoid dendritic cells to directly lyse HIV-infected autologous CD4+ T cells despite induction of tumor necrosis factor-related apoptosis-inducing ligand. J. Virol. 2010;84:2762–2773. doi: 10.1128/JVI.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choucair K., Duff J.R., Cassidy C.S., Albrethsen M.T., Kelso J.D., Lenhard A., Staats H., Patel R., Brunicardi F.C., Dworkin L., Nemunaitis J. Natural killer cells: a review of biology, therapeutic potential and challenges in treatment of solid tumors. Future Oncol. 2019;15:3053–3069. doi: 10.2217/fon-2019-0116. [DOI] [PubMed] [Google Scholar]
- Cokelaer T., Desvillechabrol D., Legendre R., Cardon M. “Sequana”: a set of snakemake NGS pipelines. J. Open Source Softw. 2017;2:352. doi: 10.21105/joss.00352. [DOI] [Google Scholar]
- Cook K.D., Kline H.C., Whitmire J.K. NK cells inhibit humoral immunity by reducing the abundance of CD4+ T follicular helper cells during a chronic virus infection. J. Leukoc. Biol. 2015;98:153–162. doi: 10.1189/jlb.4HI1214-594R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M.C., Kim D., Creegan M., Lal K.G., Ake J.A., Currier J.R., Streeck H., Robb M.L., Michael N.L., Bolton D.L. Transcriptomic signatures of NK cells suggest impaired responsiveness in HIV-1 infection and increased activity post-vaccination. Nat. Commun. 2018;9:1212. doi: 10.1038/s41467-018-03618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster J.G., Allen C.D.C. B cell responses: cell interaction dynamics and decisions. Cell. 2019;177:524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport M.P., Khoury D.S., Cromer D., Lewin S.R., Kelleher A.D., Kent S.J. Functional cure of HIV: the scale of the challenge. Nat. Rev. Immunol. 2019;19:45–54. doi: 10.1038/s41577-018-0085-4. [DOI] [PubMed] [Google Scholar]
- de Wit J., Jorritsma T., Makuch M., Remmerswaal E.B.M., Klaasse Bos H., Souwer Y., Neefjes J., ten Berge I.J.M., van Ham S.M. Human B cells promote T-cell plasticity to optimize antibody response by inducing coexpression of T(H)1/T(FH) signatures. J. Allergy Clin. Immunol. 2015;135:1053–1060. doi: 10.1016/j.jaci.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Di Vito C., Mikulak J., Zaghi E., Pesce S., Marcenaro E., Mavilio D. NK cells to cure cancer. Semin. Immunol. 2019;41:101272. doi: 10.1016/j.smim.2019.03.004. [DOI] [PubMed] [Google Scholar]
- Diop O., Gueye A., Dias-Tavares M., Kornfeld C., Faye A., Ave P., Huerre M., Corbet S., Barre-Sinoussi F., Müller-Trutwin M. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in african green monkeys. J. Virol. 2000;74:7538–7547. doi: 10.1128/JVI.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eivazi S., Bagheri S., Hashemzadeh M.S., Ghalavand M., Qamsari E.S., Dorostkar R., Yasemi M. Development of T follicular helper cells and their role in disease and immune system. Biomed. Pharmacother. 2016;84:1668–1678. doi: 10.1016/j.biopha.2016.10.083. [DOI] [PubMed] [Google Scholar]
- Ellis-Connell A.L., Balgeman A.J., Zarbock K.R., Barry G., Weiler A., Egan J.O., Jeng E.K., Friedrich T., Miller J.S., Haase A.T. ALT-803 transiently reduces simian immunodeficiency virus replication in the absence of antiretroviral treatment. J. Virol. 2018;92 doi: 10.1128/JVI.01748-17. e01748–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epeldegui M., Thapa D.R., Cruz J.D.L., Kitchen S., Zack J.A., Martínez-Maza O. CD40 ligand (CD154) incorporated into HIV virions induces activation-induced cytidine deaminase (AID) expression in human B lymphocytes. PLoS One. 2010;5:e11448. doi: 10.1371/journal.pone.0011448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes J.D. Pathobiology of HIV/SIV-associated changes in secondary lymphoid tissues. Immunol. Rev. 2013;254:65–77. doi: 10.1111/imr.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsakoglu Y., Palomino-Segura M., Latino I., Zanaga S., Chatziandreou N., Pizzagalli D.U., Rinaldi A., Bolis M., Sallusto F., Stein J.V., Gonzalez S.F. Influenza vaccination induces NK-Cell-Mediated type-II IFN response that regulates humoral immunity in an IL-6-dependent manner. Cell Rep. 2019;26:2307–2315.e5. doi: 10.1016/j.celrep.2019.01.104. [DOI] [PubMed] [Google Scholar]
- Favre D., Lederer S., Kanwar B., Ma Z.-M., Proll S., Kasakow Z., Mold J., Swainson L., Barbour J.D., Baskin C.R. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog. 2009;5:e1000295. doi: 10.1371/journal.ppat.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlazzo G., Carrega P. Natural killer cell distribution and trafficking in human tissues. Front. Immunol. 2012;3 doi: 10.3389/fimmu.2012.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez N.C., Lozier A., Flament C., Ricciardi-Castagnoli P., Bellet D., Suter M., Perricaudet M., Tursz T., Maraskovsky E., Zitvogel L. Dendritic cells directly trigger NK cell functions: cross-talk relevant in innate anti-tumor immune responses in vivo. Nat. Med. 1999;5:405–411. doi: 10.1038/7403. [DOI] [PubMed] [Google Scholar]
- Fletcher C.V., Staskus K., Wietgrefe S.W., Rothenberger M., Reilly C., Chipman J.G., Beilman G.J., Khoruts A., Thorkelson A., Schmidt T.E. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. U S A. 2014 doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flórez-Álvarez L., Hernandez J.C., Zapata W. NK cells in HIV-1 infection: from basic science to vaccine strategies. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freud A.G., Mundy-Bosse B.L., Yu J., Caligiuri M.A. The broad spectrum of human natural killer cell diversity. Immunity. 2017;47:820–833. doi: 10.1016/j.immuni.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y., Lum R., Okoye A.A., Park H., Matsuda K., Bae J.Y., Hagen S.I., Shoemaker R., Deleage C., Lucero C. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat. Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., Dang T., Dunnick W.A., Collins J.T., Blazar B.R., Yuan D. Receptors and counterreceptors involved in NK-B cell interactions. J. Immunol. 2005;174:4113–4119. doi: 10.4049/jimmunol.174.7.4113. [DOI] [PubMed] [Google Scholar]
- Garcia Z., Lemaître F., van Rooijen N., Albert M.L., Levy Y., Schwartz O., Bousso P. Subcapsular sinus macrophages promote NK cell accumulation and activation in response to lymph-borne viral particles. Blood. 2012;120:4744–4750. doi: 10.1182/blood-2012-02-408179. [DOI] [PubMed] [Google Scholar]
- Garrido C., Abad-Fernandez M., Tuyishime M., Pollara J.J., Ferrari G., Soriano-Sarabia N., Margolis D.M. Interleukin-15-Stimulated natural killer cells clear HIV-1-Infected cells following latency reversal ex vivo. J. Virol. 2018;92 doi: 10.1128/JVI.00235-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavedoni L.D., Velasquillo M.C., Parodi L.M., Hubbard G.B., Hodara V.L. Cytokine expression, natural killer cell activation, and phenotypic changes in lymphoid cells from rhesus macaques during acute infection with pathogenic simian immunodeficiency virus. J. Virol. 2000;74:1648–1657. doi: 10.1128/JVI.74.4.1648-1657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueye A., Diop O.M., Ploquin M.J.Y., Kornfeld C., Faye A., Cumont M.-C., Hurtrel B., Barré-Sinoussi F., Müller-Trutwin M.C. Viral load in tissues during the early and chronic phase of non-pathogenic SIVagm infection. J. Med. Primatol. 2004;33:83–97. doi: 10.1111/j.1600-0684.2004.00057.x. [DOI] [PubMed] [Google Scholar]
- Hammer Q., Rückert T., Borst E.M., Dunst J., Haubner A., Durek P., Heinrich F., Gasparoni G., Babic M., Tomic A. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 2018;19:453–463. doi: 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]
- Harper J., Huot N., Micci L., Tharp G., King C., Rascle P., Shenvi N., Wang H., Galardi C., Upadhyay A.A. IL-21 and IFNα therapy rescues terminally differentiated NK cells and limits SIV reservoir in ART-treated macaques. Nat. Commun. 2021;12:2866. doi: 10.1038/s41467-021-23189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao L.A., Grishina I., Bourry O., Hu W.K., Somrit M., Sankaran-Walters S., Gaulke C.A., Fenton A.N., Li J.A., Crawford R.W. Early mucosal sensing of SIV infection by paneth cells induces IL-1β production and initiates gut epithelial disruption. PLoS Pathog. 2014;10:e1004311. doi: 10.1371/journal.ppat.1004311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope J.C., Cumberbatch M., Fielding I., Dearman R.J., Kimber I., Hopkins S.J. Identification of dendritic cells as a major source of interleukin-6 in draining lymph nodes following skin sensitization of mice. Immunology. 1995;86:441–447. [PMC free article] [PubMed] [Google Scholar]
- Huot N., Bosinger S.E., Paiardini M., Reeves R.K., Müller-Trutwin M. Lymph node cellular and viral dynamics in natural hosts and impact for HIV cure strategies. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot N., Jacquelin B., Garcia-Tellez T., Rascle P., Ploquin M.J., Madec Y., Reeves R.K., Derreudre-Bosquet N., Müller-Trutwin M. Natural killer cells migrate into and control simian immunodeficiency virus replication in lymph node follicles in African green monkeys. Nat. Med. 2017;23:1277–1286. doi: 10.1038/nm.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot N., Rascle P., Petitdemange C., Contreras V., Palgen J.-L., Stahl-Hennig C., Le Grand R., Beignon A.-S., Jacquelin B., Müller-Trutwin M. Non-human primate determinants of NK cells in tissues at steady-state and during SIV infection. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.02134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot N., Rascle P., Petitdemange C., Contreras V., Stürzel C.M., Baquero E., Harper J.L., Passaes C., Legendre R., Varet H. SIV-induced terminally differentiated adaptive NK cells in lymph nodes associated with enhanced MHC-E restricted activity. Nat. Commun. 2021;12:1282. doi: 10.1038/s41467-021-21402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquelin B., Petitjean G., Kunkel D., Liovat A.-S., Jochems S.P., Rogers K.A., Ploquin M.J., Madec Y., Barré-Sinoussi F., Dereuddre-Bosquet N. Innate immune responses and rapid control of inflammation in african green monkeys treated or not with interferon-alpha during primary SIVagm infection. PLoS Pathog. 2014;10:e1004241. doi: 10.1371/journal.ppat.1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost S., Tomezsko P.J., Rands K., Toth I., Lichterfeld M., Gandhi R.T., Altfeld M. CD4+ T-cell help enhances NK cell function following therapeutic HIV-1 vaccination. J. Virol. 2014;88:8349–8354. doi: 10.1128/JVI.00924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Carotta S., Huntington N.D., Bernard N.J., Tarlinton D.M., Smyth M.J., Nutt S.L. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood. 2011;117:1869–1879. doi: 10.1182/blood-2010-08-303123. [DOI] [PubMed] [Google Scholar]
- Khan M., Arooj S., Wang H. NK cell-based immune checkpoint inhibition. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke M.A., Eto D., Locci M., Cho M., Davidson T., Haddad E.K., Crotty S. Bcl6 and maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H.-H., Lichterfeld M. Recent progress in understanding HIV reservoirs. Curr. Opin. HIV AIDS. 2018;13:137–142. doi: 10.1097/COH.0000000000000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonte M.L., McKay P.F., Letvin N.L. Evidence of NK cell dysfunction in SIV-infected rhesus monkeys: impairment of cytokine secretion and NKG2C/C2 expression. Eur. J. Immunol. 2006;36:2424–2433. doi: 10.1002/eji.200635901. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu X., Yan X., Zhong B., Nurieva R.I., Wang A., Wang X., Martin-Orozco N., Wang Y., Chang S.H., Esplugues E. Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med. 2012;209:1841–1852. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn R., Sciaranghella G., Lunzen J.V., Nolting A., Dugast A.-S., Ghebremichael M.S., Altfeld M., Alter G. Early viral replication in lymph nodes provides HIV with a means by which to escape NK-cell-mediated control. Eur. J. Immunol. 2011;41:2729–2740. doi: 10.1002/eji.201040886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.S., Deenick E.K., Batten M., Tangye S.G. The origins, function, and regulation of T follicular helper cells. J. Exp. Med. 2012;209:1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria A.D., Fogli M., Costa P., Murdaca G., Puppo F., Mavilio D., Moretta A., Moretta L. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur. J. Immunol. 2003;33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Martinot A.J., Meythaler M., Pozzi L.-A., Boisvert K.D., Knight H., Walsh D., Westmoreland S., Anderson D.C., Kaur A., O’Neil S.P. Acute SIV infection in sooty mangabey monkeys is characterized by rapid virus clearance from lymph nodes and absence of productive infection in germinal centers. PLoS One. 2013;8:e57785. doi: 10.1371/journal.pone.0057785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio D., Benjamin J., Daucher M., Lombardo G., Kottilil S., Planta M.A., Marcenaro E., Bottino C., Moretta L., Moretta A., Fauci A.S. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc. Natl. Acad. Sci U S A. 2003;100:15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio D., Benjamin J., Kim D., Lombardo G., Daucher M., Kinter A., Nies-Kraske E., Marcenaro E., Moretta A., Fauci A.S. Identification of NKG2A and NKp80 as specific natural killer cell markers in rhesus and pigtailed monkeys. Blood. 2005;106:1718–1725. doi: 10.1182/blood-2004-12-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary C.S., Deleage C., Harper J., Micci L., Ribeiro S.P., Paganini S., Kuri-Cervantes L., Benne C., Ryan E.S., Balderas R. CTLA-4+PD-1- memory CD4+ T cells critically contribute to viral persistence in antiretroviral therapy-suppressed, SIV-infected rhesus macaques. Immunity. 2017;47:776–788.e5. doi: 10.1016/j.immuni.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moukambi F., Rabezanahary H., Fortier Y., Rodrigues V., Clain J., Benmadid-Laktout G., Zghidi-Abouzid O., Soundaramourty C., Laforge M., Estaquier J. Mucosal T follicular helper cells in SIV-infected rhesus macaques: contributing role of IL-27. Mucosal Immunol. 2019;12:1038–1054. doi: 10.1038/s41385-019-0174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleinika K., Mauri C., Salama A.D. Effector and regulatory B cells in immune-mediated kidney disease. Nat. Rev. Nephrol. 2019;15:11–26. doi: 10.1038/s41581-018-0074-7. [DOI] [PubMed] [Google Scholar]
- Pahl J.H.W., Cerwenka A., Ni J. Memory-like NK cells: remembering a previous activation by cytokines and NK cell receptors. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini M., Pandrea I., Apetrei C., Silvestri G. Lessons learned from the natural hosts of HIV-related viruses. Annu. Rev. Med. 2009;60:485–495. doi: 10.1146/annurev.med.60.041807.123753. [DOI] [PubMed] [Google Scholar]
- Palgen J.-L., Tchitchek N., Rodriguez-Pozo A., Jouhault Q., Abdelhouahab H., Dereuddre-Bosquet N., Contreras V., Martinon F., Cosma A., Lévy Y. Innate and secondary humoral responses are improved by increasing the time between MVA vaccine immunizations. NPJ Vaccin. 2020;5:24. doi: 10.1038/s41541-020-0175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasavvas E., Azzoni L., Kossenkov A.V., Dawany N., Morales K.H., Fair M., Ross B.N., Lynn K., Mackiewicz A., Mounzer K. NK response correlates with HIV decrease in pegylated IFN-α2a–Treated ART-suppressed subjects. J. Immunol. 2019 doi: 10.4049/jimmunol.1801511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillion A., Powell M.D., Chisolm D.A., Bachus H., Fuller M.J., Weinmann A.S., Villarino A., O’Shea J.J., León B., Oestreich K.J., Ballesteros-Tato A. Inhibition of IL-2 responsiveness by IL-6 is required for the generation of GC-TFH cells. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.-S., Yoon S.-O., Armitage R.J., Choi Y.S. Follicular dendritic cells produce IL-15 that enhances germinal center B cell proliferation in membrane-bound form. J. Immunol. 2004;173:6676–6683. doi: 10.4049/jimmunol.173.11.6676. [DOI] [PubMed] [Google Scholar]
- Paust S., Blish C.A., Reeves R.K. Redefining memory: building the case for adaptive NK cells. J. Virol. 2017;91 doi: 10.1128/JVI.00169-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppa D. Entering a new era of harnessing natural killer cell responses in HIV infection. EBioMedicine. 2019;44:26–27. doi: 10.1016/j.ebiom.2019.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C., Yamamoto T., Gerner M.Y., Boswell K.L., Wloka K., Smith E.C., Ambrozak D.R., Sandler N.G., Timmer K.J., Sun X. CD4 T follicular helper cell dynamics during SIV infection. J. Clin. Invest. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Gray E.E., Cyster J.G. The microanatomy of B cell activation. Curr. Opin. Immunol. 2009;21:258–265. doi: 10.1016/j.coi.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais C., Hocqueloux L., Ibanez C., Gallien S., Copie C., Surenaud M., Kök A., Lorin V., Fusaro M., Delfau-Larue M.-H. Early antiretroviral therapy preserves functional follicular helper T and HIV-specific B cells in the gut mucosa of HIV-1–Infected individuals. J. Immunol. 2018 doi: 10.4049/jimmunol.1701615. [DOI] [PubMed] [Google Scholar]
- Raehtz K., Pandrea I., Apetrei C. The well-tempered SIV infection: pathogenesis of SIV infection in natural hosts in the wild, with emphasis on virus transmission and early events post-infection that may contribute to protection from disease progression. Infect. Genet. Evol. 2016;46:308–323. doi: 10.1016/j.meegid.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raehtz K.D., Barrenäs F., Xu C., Busman-Sahay K., Valentine A., Law L., Ma D., Policicchio B.B., Wijewardana V., Brocca-Cofano E. African green monkeys avoid SIV disease progression by preventing intestinal dysfunction and maintaining mucosal barrier integrity. PLOS Pathog. 2020;16:e1008333. doi: 10.1371/journal.ppat.1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao D.A. T cells that help B cells in chronically inflamed tissues. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski C.E., Cranert S.A., Zhou J.Q., Xu H., Kleinstein S.H., Singh H., Waggoner S.N. Affinity maturation is impaired by natural killer cell suppression of germinal centers. Cell Rep. 2018;24:3367–3373.e4. doi: 10.1016/j.celrep.2018.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Cirion A., Jacquelin B., Barré-Sinoussi F., Müller-Trutwin M. Immune responses during spontaneous control of HIV and AIDS: what is the hope for a cure? Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130436. doi: 10.1098/rstb.2013.0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer J.L., Li H., Evans T.I., Estes J.D., Reeves R.K. Accumulation of cytotoxic CD16+ NK cells in simian immunodeficiency virus-infected lymph nodes associated with in situ differentiation and functional anergy. J. Virol. 2015;89:6887–6894. doi: 10.1128/JVI.00660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer E., Speck R.F. Anti-HIV activity mediated by natural killer and CD8+ cells after toll-like receptor 7/8 triggering. PLoS One. 2008;3:e1999. doi: 10.1371/journal.pone.0001999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Lin K.-I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K., Staudt L.M. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/S1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Strauss-Albee D.M., Blish C.A. Human NK cell diversity in viral infection: ramifications of ramification. Front. Immunol. 2016;7 doi: 10.3389/fimmu.2016.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss-Albee D.M., Fukuyama J., Liang E.C., Yao Y., Jarrell J.A., Drake A.L., Kinuthia J., Montgomery R.R., John-Stewart G., Holmes S., Blish C.A. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci. Transl. Med. 2015;7:297ra115. doi: 10.1126/scitranslmed.aac5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh W.-K. Life of T Follicular helper cells. Mol. Cells. 2014;38:195–201. doi: 10.14348/molcells.2015.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svardal H., Jasinska A.J., Apetrei C., Coppola G., Huang Y., Schmitt C.A., Jacquelin B., Ramensky V., Müller-Trutwin M., Antonio M. Ancient hybridization and strong adaptation to viruses across African vervet monkey populations. Nat. Genet. 2017;49:1705–1713. doi: 10.1038/ng.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleshova N., Kenney J., Williams V., Nest G.V., Marshall J., Lifson J.D., Sivin I., Dufour J., Bohm R., Gettie A., Pope M. CpG-C ISS-ODN activation of blood-derived B cells from healthy and chronic immunodeficiency virus-infected macaques. J. Leukoc. Biol. 2006;79:257–267. doi: 10.1189/jlb.0205084. [DOI] [PubMed] [Google Scholar]
- Theurich S., Tsaousidou E., Hanssen R., Lempradl A.M., Mauer J., Timper K., Schilbach K., Folz-Donahue K., Heilinger C., Sexl V. IL-6/Stat3-Dependent induction of a distinct, obesity-associated NK cell subpopulation deteriorates energy and glucose homeostasis. Cell Metab. 2017;26:171–184.e6. doi: 10.1016/j.cmet.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Urashima M., Chauhan D., Hatziyanni M., Ogata A., Hollenbaugh D., Aruffo A., Anderson K.C. CD40 ligand triggers interleukin-6 mediated B cell differentiation. Leuk. Res. 1996;20:507–515. doi: 10.1016/0145-2126(95)00098-4. [DOI] [PubMed] [Google Scholar]
- van Erp E.A., van Kampen M.R., van Kasteren P.B., de Wit J. Viral infection of human natural killer cells. Viruses. 2019;11:243. doi: 10.3390/v11030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hall T., André P., Horowitz A., Ruan D.F., Borst L., Zerbib R., Narni-Mancinelli E., van der Burg S.H., Vivier E. Monalizumab: inhibiting the novel immune checkpoint NKG2A. J. Immunother. Cancer. 2019;7:263. doi: 10.1186/s40425-019-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Bush T.J., Buchta C.M., Claudio J., Bishop G.A. Cutting Edge: importance of IL-6 and cooperation between innate and adaptive immune receptors in cellular vaccination with B lymphocytes. J. Immunol. 2009;183:4833–4837. doi: 10.4049/jimmunol.0900968. [DOI] [PubMed] [Google Scholar]
- Vanhamel J., Bruggemans A., Debyser Z. Establishment of latent HIV-1 reservoirs: what do we really know? J. Virus Erad. 2019;5:3–9. [PMC free article] [PubMed] [Google Scholar]
- Vibholm L., Schleimann M.H., Højen J.F., Benfield T., Offersen R., Rasmussen K., Olesen R., Dige A., Agnholt J., Grau J. Short-course toll-like receptor 9 agonist treatment impacts innate immunity and plasma viremia in individuals with human immunodeficiency virus infection. Clin. Infect. Dis. 2017;64:1686–1695. doi: 10.1093/cid/cix201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E., Raulet D.H., Moretta A., Caligiuri M.A., Zitvogel L., Lanier L.L., Yokoyama W.M., Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T., Dalod M., Vivier E., Zitvogel L. Natural killer cell-dendritic cell crosstalk in the initiation of immune responses. Expert Opin. Biol. Ther. 2005;5:S49–S59. doi: 10.1517/14712598.5.1.s49. [DOI] [PubMed] [Google Scholar]
- Wijewardana V., Soloff A.C., Liu X., Brown K.N., Barratt-Boyes S.M. Early myeloid dendritic cell dysregulation is predictive of disease progression in simian immunodeficiency virus infection. PLoS Pathog. 2010;6:e1001235. doi: 10.1371/journal.ppat.1001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Deng Y., Zhao M., Zhang J., Zheng M., Chen G., Li L., He Z., Lu Q. Molecular control of follicular helper T cell development and differentiation. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Zhao X., Wang X., Feng H., Gou M., Jin W., Wang X., Liu X., Dong C. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 2019;51:826–839.e5. doi: 10.1016/j.immuni.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Yang C.-L., Zhang P., Liu R.-T., Zhang N., Zhang M., Li H., Du T., Li X.-L., Dou Y.-C., Duan R.-S. CXCR5-negative natural killer cells ameliorate experimental autoimmune myasthenia gravis by suppressing follicular helper T cells. J. Neuroinflammation. 2019;16:282. doi: 10.1186/s12974-019-1687-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Vinuesa C.G. The elusive identity of T follicular helper cells. Trends Immunol. 2010;31:377–383. doi: 10.1016/j.it.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Zeng M., Smith A.J., Wietgrefe S.W., Southern P.J., Schacker T.W., Reilly C.S., Estes J.D., Burton G.F., Silvestri G., Lifson J.D. Cumulative mechanisms of lymphoid tissue fibrosis and T cell depletion in HIV-1 and SIV infections. J. Clin. Invest. 2011;121:998–1008. doi: 10.1172/JCI45157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The gene expression profiles of CXCR5+ NK cells were compared to that of the predominant NK cell subpopulation in pLN, i.e. NKG2a/chighCD16- NK cells (n = 3 AGM).
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All raw data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.