Abstract
Mitogen-activated protein kinases (MAPKs) are inactivated by dual-specificity and protein tyrosine phosphatases (PTPs) in yeasts. In Saccharomyces cerevisiae, two PTPs, Ptp2 and Ptp3, inactivate the MAPKs, Hog1 and Fus3, with different specificities. To further examine the functions and substrate specificities of Ptp2 and Ptp3, we tested whether they could inactivate a third MAPK, Mpk1, in the cell wall integrity pathway. In vivo and in vitro evidence indicates that both PTPs inactivate Mpk1, but Ptp2 is the more effective negative regulator. Multicopy expression of PTP2, but not PTP3, suppressed growth defects due to the MEK kinase mutation, BCK1-20, and the MEK mutation, MKK1-386, that hyperactivate this pathway. In addition, deletion of PTP2, but not PTP3, exacerbated growth defects due to MKK1-386. Other evidence supported a role for Ptp3 in this pathway. Expression of MKK1-386 was lethal in the ptp2Δ ptp3Δ strain but not in either single PTP deletion strain. In addition, the ptp2Δ ptp3Δ strain showed higher levels of heat stress-induced Mpk1-phosphotyrosine than the wild-type strain or strains lacking either PTP. The PTPs also showed differences in vitro. Ptp2 was more efficient than Ptp3 at binding and dephosphorylating Mpk1. Another factor that may contribute to the greater effectiveness of Ptp2 is its subcellular localization. Ptp2 is predominantly nuclear whereas Ptp3 is cytoplasmic, suggesting that active Mpk1 is present in the nucleus. Last, PTP2 but not PTP3 transcript increased in response to heat shock in a Mpk1-dependent manner, suggesting that Ptp2 acts in a negative feedback loop to inactivate Mpk1.
Mitogen-activated protein kinase (MAPK) pathways regulate growth, development, and the response to stress in eukaryotes (2, 11, 28, 30, 36, 77). These pathways regulate mitosis and differentiation in vertebrates, vulval development in Caenorhabditis elegans, and photoreceptor development in Drosophila melanogaster (42). In the yeast Saccharomyces cerevisiae, six different MAPK pathways regulate mating, osmosensing, pseudohyphal growth, invasive growth, spore formation, and cell wall biosynthesis (28, 44). The proper regulation of MAPK pathways is important since mutations that alter their activity cause defects in growth and development. Mutations that hyperactivate MAPK modules lead to cell transformation in vertebrates, developmental defects in C. elegans and Drosophila, and growth defects in S. cerevisiae (12, 30, 33, 45, 68, 71, 73, 77). Common to all MAPK pathways are MAPK modules comprising three sequentially acting kinases termed MEK kinase (MEKK), MEK, and MAPK (11). MEKK activates MEK, which in turn activates MAPK by phosphorylating a conserved threonine and tyrosine residue in the phosphorylation lip sequence (11, 42, 74).
Although the activation of MAPK pathways has been intensively investigated, the inactivation of these pathways is less well understood. Most is known about the inactivation of MAPKs. Since MAPKs require phosphorylation of both a Thr and a Tyr for full activity (11, 42), it is possible to inactivate them by dephosphorylating either phosphothreonine, phosphotyrosine (pY), or both residues. Dual-specificity phosphatases, capable of dephosphorylating both phosphothreonine and pY residues, inactivate MAPKs in vertebrates, Drosophila, and S. cerevisiae (17, 25, 41, 42, 47, 70). In vertebrates, there are at least nine dual-specificity phosphatases that inactivate the three MAPK family members, ERK, JNK/SAPK, and p38, with variable specificity (42). In Drosophila, a dual-specificity phosphatase, puckered, has been identified as a negative regulator of JNK in a developmental pathway (47). In the S. cerevisiae genome, six genes encode proteins similar to dual-specificity phosphatases. One of these, Msg5, has been shown to inactivate the MAPK, Fus3, in the pheromone response pathway (17, 76).
In yeasts, MAPKs have also been shown to be inactivated by a novel mechanism involving protein tyrosine phosphatases (PTPs) specific for pY (33, 50, 66, 73). The S. cerevisiae MAPKs, Hog1, in the osmotic stress-activated high-osmolarity glycerol (HOG) pathway, and Fus3, in the pheromone response pathway, are inactivated by two protein tyrosine phosphatases, Ptp2 and Ptp3 (33, 73, 76). These two PTPs contain a catalytic domain of ∼400 residues 57% similar to each other and significantly similar to vertebrate PTPs (5, 7, 9). Analysis of human PTP1B showed that the Cys residue in the conserved sequence, (I/V)HCXAGXXR(S/T)G, acts as a nucleophile in pY hydrolysis (26). Mutation of the corresponding residue in Ptp2 and Ptp3 inactivates them in vivo and in vitro (33, 73, 76). In Schizosaccharomyces pombe, two protein tyrosine phosphatases, Pyp1 and Pyp2, have been shown to inactivate a MAPK, Spc1/Sty1, in a stress response pathway analogous to the HOG pathway (14, 50, 66).
Protein phosphatases that regulate MAPKs show variable levels of specificity for their substrates. For example, the vertebrate dual-specificity phosphatase, MKP-3/PYST1 (23, 53, 56), is highly specific for ERK, while other dual-specificity phosphatases, MKP-1, MKP-2, and MKP-4, inactivate multiple MAPKs (3, 10, 42, 54, 70). Part of this specificity is due to noncatalytic domains that recognize their substrates through tight binding interactions (55). PTPs also show different specificities for MAPKs. For example, the S. cerevisiae Ptp2 is a more effective negative regulator of Hog1 than Ptp3 (33, 73). We have shown that Ptp2 binds Hog1 more effectively than Ptp3 (33), and this likely contributes to its greater ability to inactivate Hog1. In contrast, Ptp3 is a more effective negative regulator of Fus3 than Ptp2 (76), but the reason for this is unknown.
To further explore the functions of S. cerevisiae Ptp2 and Ptp3 in MAPK signaling and their different specificities for MAPKs, we examined whether these phosphatases could inactivate Mpk1, a MAPK in the cell wall integrity pathway (37). This pathway activates the biosynthesis of the yeast cell wall during vegetative growth, during mating, and in response to stresses such as heat and hypo-osmotic shock (6, 13, 22, 34, 40, 43, 46, 49, 52, 75). The activation of this pathway has been well characterized, but its inactivation is poorly understood. The transmembrane proteins Hcs77, Mid2, and Mtl1 signal to Rho1, protein kinase C, and a MAPK module comprising a MEKK called Bck1, redundant MEKs called Mkk1 and Mkk2, and a single MAPK, Mpk1 (22, 32, 35, 37, 38, 40, 58, 61). MSG5 was isolated as a multicopy suppressor of growth defects due to the hyperactive MEK mutant, MKK1-386; however, it has not been established whether it is a physiologically relevant regulator of Mpk1 (71).
In this work, we show that both Ptp2 and Ptp3 inactivate Mpk1; however, Ptp2 is a more potent negative regulator than Ptp3. The difference in their activities is likely explained by at least three factors. First, Ptp2 binds Mpk1 and dephosphorylates Mpk1-pY more effectively than Ptp3. Second, these PTPs show differences in subcellular localization; Ptp2 is predominantly nuclear, while Ptp3 is cytoplasmic. The nuclear localization of Ptp2 may account for its greater effectiveness, since activation of MAPKs often results in their translocation to the nucleus. Last, heat shock increased the level of PTP2 transcript in a Mpk1-dependent manner, suggesting that Ptp2 acts in a negative feedback loop to inactivate Mpk1. Thus, Ptp2 and Ptp3 are global regulators of MAPK signaling, inactivating the HOG, pheromone response, and cell wall integrity pathways but show differences in specificity toward the MAPKs in these pathways.
MATERIALS AND METHODS
Strains, media, and genetic techniques.
All strains were derived from the wild-type haploid strain BBY48 (MATα trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801) (1) unless otherwise noted. A strain bearing a deletion of the MPK1 gene was produced by transformation of BBY48 with an MPK1 deletion construct in plasmid pJL2, described below. This plasmid was digested with BamHI and EcoRI and transformed into BBY48, and His+ transformants were selected. Southern analysis identified transformants bearing the deletion allele at the MPK1 locus (63). Briefly, genomic DNA was digested with KpnI, and a Southern blot was probed with a 522-bp BamHI-SmaI fragment corresponding to the 5′ end of MPK1. A 5.2-kb fragment characteristic of the mpk1Δ::HIS3 allele integrated at the correct locus was detected in several transformants. Backcrossing mpk1Δ::HIS3 strains to the isogenic wild-type strain BBY45 (1), sporulation, and dissection resulted in a 2:2 segregation of His+:His− spore clones. All His+ spore clones were temperature sensitive for growth as expected for a mpk1Δ strain (37). Strains lacking MPK1 and HOG1, PTP2, PTP3, or PTP2 and PTP3 were produced by crossing the mpk1Δ strain to a hog1Δ (33), ptp2Δ (33), or ptp3Δ (33) strain. The resulting strains were CMY3 (MATα mpk1Δ::HIS3), CMY5 (MATα mpk1Δ::HIS3 hog1Δ::TRP1), CMY6 (MATα mpk1Δ::HIS3 ptp2Δ::HIS3), CMY7 (MATα mpk1Δ::HIS3 ptp3Δ::TRP1), and CMY8 (MATα mpk1Δ::HIS3 ptp2Δ::HIS3 ptp3Δ::TRP1).
Galactose-inducible yeast strains bearing deletions of PTP2, PTP3, or both PTP2 and PTP3 were produced as follows. A strain lacking PTP2, CMY1 (MATa ptp2Δ::HIS3 trp1-Δ63 ura3-52 his3-Δ200 leu2-3,112 lys2-801 GAL+) was produced by transformation of the wild-type diploid JD51 (MATa/MATα trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 lys2-801/lys2-801 GAL+) (16) with a PTP2 deletion construct (59), and ptp2Δ::HIS3 strains were identified as described previously (59). A strain lacking PTP3, CMY2 (MATα ptp3Δ::HIS3 trp1-Δ63 ura3-52 his3-Δ200 leu2-3,112 lys2-801 GAL+), was produced by transformation of JD51 (16) with a ptp3Δ::HIS3 allele in plasmid pCM1. Plasmid pHF1, designed for construction of ptp3Δ alleles (33), was digested with SmaI, and a blunt-ended HIS3 fragment from YEp6 (69) was ligated to produce pCM1. This plasmid was digested with EcoRI and BamHI and transformed into JD51, and His+ transformants were selected. A PstI digest of genomic DNA from the His+ transformants was probed with a 1-kb SmaI-EcoRI fragment corresponding to the 3′ end of the PTP3 gene to detect the 5.7-kb fragment indicative of ptp3Δ::HIS3 transformants (33). Sporulation and dissection of heterozygous ptp3Δ::HIS3/PTP3 diploids resulted in 2:2 segregation of His+:His− spore clones. Strain CMY23 (MATa ptp2Δ::HIS3 ptp3Δ::HIS3 trp1-Δ63 ura3-52 his3-Δ200 leu2-3,112 lys2-801 GAL+) was produced by mating CMY1 and CMY2, sporulating diploids, dissecting tetrads, and identifying His+ spore clones from nonparental ditype tetrads.
Yeast strains DL245 (EG123 BCK1-20) and EG123 (MATa leu2-3,112 ura3-52 trp1-1 his4 can1r) were kindly provided by D. Levin (Department of Biochemistry, Johns Hopkins University School of Public Health) (38). Glutathione S-transferase (GST) fusions to Ptp2 and Ptp3, described previously (33), were expressed in S. cerevisiae 334 (MATa leu2-3,112 gal1 reg1-501 ura3-52 pep4-3 prb1-112) (31). Media to culture yeast and bacteria were produced as described by Sherman et al. (65). Yeast transformations were performed as described by Dohmen et al. (15). Standard procedures for yeast manipulations (27) were used. Recombinant DNA manipulations were carried out in Escherichia coli MC1061 or DH5α (63).
Plasmids.
The PTP2-green fluorescent protein (GFP) and PTP3-GFP fusion proteins were constructed by using a 750-bp NotI GFP fragment from pSF-GP1, kindly provided by J. Hirsch (Columbia University). Plasmid pPTP2GFP, where GFP is fused to the carboxy terminus of Ptp2, was constructed by replacing the PTP2 stop codon with a NotI site, using PCR. The oligonucleotide containing the NotI site, 5′-TCTAGAGCGGCCGCAACAAGGTAACGCGTTC-3′, was paired with another oligonucleotide, 5′-GCGCTCGAGCAACAAGCACTAAGTCCCTAGCG-3′, 462 bp upstream of the PTP2 start codon. The resulting 2.7-kb PCR product was digested with NotI and XhoI and ligated into pSK (Stratagene) to generate pSKXNPTP2. Plasmid pSKXNPTP2 digested with NotI was ligated to the 750-bp NotI fragment of GFP, and the orientation of the GFP insert was determined by digestion with SpeI. The resulting plasmid, pSKPTP2-GFP, was digested with KpnI and SacI, and the ∼3.5-kb PTP2-GFP fragment was cloned into YEplac181, a plasmid containing a 2μm origin of replication and bearing the LEU2 gene (20), to produce pPTP2-GFP. The Ptp2-GFP fusion is functional since it suppressed lethality of a sln1Δ strain which hyperactivates the HOG pathway, as determined by the 5-fluoro-orotic acid assay described previously (33, 59). Briefly, functional PTP2 carried on a low-copy-number or multicopy plasmid allows an sln1Δ strain, IMY101 (MATa sln1Δ::HIS3 + pSLN1-URA3) to grow on media containing 5-fluoro-orotic acid (59). The pPTP2-GFP plasmid produced a single protein of expected size, ∼111 kDa, that cross-reacted with the monoclonal anti-GFP antibody (Clontech).
Plasmid pPTP3-GFP was constructed by replacing the PTP3 stop codon with a NotI site, using PCR. The oligonucleotide containing the NotI site, 5′-TCTAGAGCGGCCGCATTGTGGCAATTCTTTC-3′, was paired with the oligonucleotide 5′-CGGGATCCATGAAGGACAGTGTAGACTGC-3′ (start codon in bold). The resulting 2.8-kb fragment was digested with NotI and BamHI and ligated into pSK to generate pSKBNPTP3. Plasmid pSKBNPTP3 was digested with NotI and ligated to the 750-bp NotI GFP fragment. The orientation of the GFP insert was determined by digestion with BamHI. The ∼3.5-kb PTP3-GFP fragment was isolated from pSKPTP3-GFP by digestion with KpnI and SacI and cloned into Yeplac181 to produce p181PTP3-GFP. A 1.3-kb SphI fragment containing the endogenous PTP3 promoter and 105 bp of the PTP3 open reading frame isolated from pAF12 (33) was ligated into p181PTP3-GFP to produce pPTP3-GFP. This construct expresses a functional Ptp3-GFP fusion protein since it suppressed lethality of the sln1Δ strain, IMY101, as described above. Strains transformed with pPTP3-GFP expressed a protein of expected size, ∼131 kDa that cross-reacted with the monoclonal anti-GFP antibody (Clontech).
The PTP3 promoter was fused to lacZ by using the yeast integrating vector YIp357 (57), containing lacZ (American Tissue Type Collection). The promoter region, from −785 to the start codon, was amplified by pairing the primers 5′-CCCGGTACCGTTAGACCGCAAGCGAGTTTACC-3′ and 5′-CCCAAGCTTCATGTTCTATATGATAAGTAGATC-3′ (start codon in bold). The fragment was digested with KpnI and HindIII and ligated into the corresponding sites of YIp357. The resulting plasmid, YIp-PTP3Z, was linearized by digestion with SnaBI and transformed into the wild-type haploid, BBY48 (1). Integration of YIp-PTP3Z at the PTP3 locus was confirmed by Southern analysis (63).
To construct the mpk1Δ::HIS3 allele, PCR was used to produce a 522-bp fragment corresponding to the 5′ end of the gene having a BamHI site upstream of the start site and a SmaI site at the start codon. PCR was also used to generate a 340-bp fragment corresponding to the 3′ end of the gene having a SmaI site 1.39 kb downstream of the start site and an EcoRI site 205 bp downstream of the stop codon. Both fragments were ligated into pUC19 digested with BamHI and EcoRI to generate plasmid pJL1. This plasmid was digested with SmaI, and a 1.8-kb blunt-ended BamHI fragment of HIS3 (69) was ligated to produce pJL2, containing the mpk1Δ::HIS3 allele.
To construct plasmid p181-MPK1HA, a 2.2-kb KpnI-NarI fragment from pFL44-SLT2HA (75), kindly provided by M. Snyder (Yale University, Department of Biology), was ligated into YEplac181. Plasmid pNV7MKK1386 was a kind gift from K. Irie and K. Matsumoto (Nagoya University, Nagoya, Japan) (71). Plasmid p112PTP2 was constructed by isolating a 4-kb PvuII fragment from pHS6.7 (59) and ligating it into the corresponding sites of YEplac112 (20). Plasmid p112PTP3 was produced by isolating a 5.6-kb SalI fragment from pAF12 (33) and ligating it into the corresponding sites of YEplac112. Ptp2 and Ptp3 were expressed in E. coli by using the pRSETa vector (Invitrogen). The PTP2 fragment from pGST-PTP2 (33) was isolated as a 2.44-kb BamHI-HindIII fragment and cloned into pRSETa. The PTP3 fragment from pGST-PTP3 (33) was isolated as a 2.88-kb BamHI-SnaBI fragment and cloned into pRSETa digested with BamHI and PvuII.
The GST-Mpk1 fusion protein was produced by cloning MPK1 into the vector pEG(KT) (51). PCR was used to introduce a BamHI site just upstream of the MPK1 start codon and a second BamHI site 175 bp downstream of the MPK1 stop codon, using primers 5′-CGGGATCCATGGCTGATAAGATAGAGAGGC-3′ (start codon in bold) and 5′-CGGGATCCCCGCGGAGTACGATTAAGATAAGC-3′. The 1.63-kb PCR product was digested with BamHI and ligated to pEG(KT) to produce plasmid pGST-MPK1. Strains transformed with this plasmid expressed an 81-kDa protein that cross-reacted with anti-GST antibody (Pharmacia).
Yeast dilution assay.
To assess the ability of Ptp2 and Ptp3 to suppress growth defects due to the hyperactive MKK1 allele, MKK1-386, pNV7MKK1386 and the multicopy plasmids bearing PTPs were cotransformed into galactose-inducible yeast strains, wild-type strain JD52 (MATa trp1-Δ63 ura3-52 his3-Δ200 leu2-3,112 lys2-801 GAL+) (16) and PTP deletion strains CMY1 (ptp2Δ), CMY2 (ptp3Δ), and CMY23 (ptp2Δ ptp3Δ). The strains were cultured in synthetic medium lacking uracil and tryptophan for 48 h at 30°C and diluted to 3.5 U (A600), and fivefold serial dilutions were aliquoted into microtiter dishes. Dilutions were spotted by using a stainless steel pin replicator onto selective medium containing either glucose or galactose and lacking uracil and tryptophan. The plates were incubated at 30°C for 3 days.
Immunoblot analysis.
The level of Mpk1-pY was monitored by immunoblotting. Cultures of mpk1Δ, mpk1Δ ptp2Δ, mpk1Δ ptp3Δ, and mpk1Δ ptp2Δ ptp3Δ strains (in the BBY48 background) carrying p181-MPK1HA were grown to exponential phase at 25°C, heat shocked by the addition of an equal volume of medium at 53°C, and further incubated at 39°C. Cells from 10 ml of culture were harvested by centrifugation, resuspended in 60 μl of L1 buffer (29) (90 mM Na-HEPES [pH 7.5], 2% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol [DTT]), and heated at 100°C for 3 min. On ice, 60 μl of L2 buffer (29) (50 mM Na-HEPES [pH 7.5], 5 mM EDTA, 150 mM NaCl, 1% Triton X-100) with protease inhibitors (leupeptin, pepstatin A, antipain, aprotinin, and chymostatin [each at 20 μg/ml] and phenylmethylsulfonyl fluoride [PMSF; 35 μg/ml]) and protein phosphatase inhibitors (1 mM sodium orthovanadate and 40 mM β-glycerophosphate) was added. Protein samples, 100 μg for anti-pY blots and 10 μg for anti-hemagglutinin (HA) blots, were boiled in sample buffer, and Western analysis was performed as described previously (33). Mpk1-pY was detected by immunoblotting with anti-pY antibody PY-99 (Santa Cruz Biotechnology), and Mpk1-HA was detected with anti-HA antibody 12CA5 (BAbCo). The secondary antibody was rabbit anti-mouse alkaline phosphatase-conjugated antibody (Promega). Immunoreactivity was visualized with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium (Promega).
Coprecipitation assay.
Binding between mutant PTPs and Mpk1 was detected as follows. The catalytically inactive PTP mutants, Ptp2-C666S and Ptp3-C804A, were fused to the C terminus of GST under regulation of the GAL1-10 promoter in the vector pEG(KT). Plasmids pGST-PTP2-C666S (33), pGST-PTP3-C804A (33), and pEG(KT) were coexpressed with p181-MPK1HA in S. cerevisiae 334 (31). Yeast cells were grown at 30°C in synthetic medium lacking uracil and leucine and containing 2% galactose. Cells from 100 ml of culture grown at 30°C to ∼1 U (A600) were heat shocked by the addition of an equal volume of selective medium at 48°C and further incubated at 39°C for 25 min in prewarmed flasks. Cells were harvested by centrifugation and lysed by glass beading in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM MgCl2, 0.2% Triton X-100, 0.1% 2-mercaptoethanol, 50 mM β-glycerophosphate) with protease inhibitors (leupeptin, pepstatin A, antipain, aprotinin, and chymostatin [each at 20 μg/ml] and 35 μg of PMSF per ml). The lysates were centrifuged, and the resulting supernatant was incubated with 45 μl of a 1:1 slurry of glutathione-Sepharose beads (Pharmacia Biotech Inc.). The beads were washed three times with 600 μl of lysis buffer and then three times with lysis buffer containing 400 mM NaCl. The beads were boiled in sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Purification of Ptp2 and Ptp3 from E. coli.
His6-Ptp2 and His6-Ptp3 were expressed in E. coli BL21(DE3)pLysS (Novagen) by using an inducible T7-based expression system. Cells were grown in 2× YT medium to early log phase, 0.3 U (A600), cooled to 23°C for 1 h, induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside, and grown to 1.0 U (A600). To isolate His6-PTPs, cells were lysed in sonication buffer (20 mM Tris-HCl [pH 8], 100 mM NaCl), and the lysate was centrifuged at 17,000 × g for 15 min. The supernatant was incubated with 1.5 ml of Co2+ immobilized metal affinity resin (Talon; Clontech) for 4 h at 4°C. After binding, the beads were washed twice with 30 ml of sonication buffer. The resin was transferred to a column, washed with 3 bed volumes of 20 mM Tris-HCl (pH 8)–100 mM NaCl–10 mM imidazole, and eluted with 2 bed volumes of 20 mM Tris-HCl (pH 8)–100 mM NaCl–75 mM imidazole. Fractions containing His6-PTPs were identified by immunoblotting with anti-His6 antibody (BAbCo). Assays with the small-molecule phosphatase substrate p-nitrophenyl phosphate (PNPP) showed that PTPs had comparable but low activity. Incubation of 2.3 μg of PTPs with 20 mM PNPP in 50 mM imidazole (pH 7.5) and 1 mM DTT for 2.5 h at 30°C led to changes in A405 of 0.263 for Ptp2 and 0.163 for Ptp3.
Ptp2 dephosphorylation of Mpk1-pY in vitro.
GST-Mpk1 was isolated from the wild-type strain 334 (31), transformed with plasmid pGST-MPK1. The strain was grown to ∼1 U (A600) at 23°C in synthetic medium lacking uracil and containing 2% galactose. To activate GST-Mpk1, the culture was heat shocked by adding an equal volume of media heated to 55°C, which brought the final temperature to 39°C. After 1 h of incubation at 39°C, cells were harvested by centrifugation and ∼1.2 × 109 cells were lysed in 800 μl of buffer A (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 0.1% Triton X-100, 2.5 mM sodium orthovanadate, 1 mM DTT, 50 mM β-glycerophosphate, 35 μg of PMSF per ml, 20 μg each of aprotinin, leupeptin, pepstatin, chymostatin, and antipain per ml). The lysate was clarified by centrifugation and mixed with 70 μl of a 1:1 slurry of glutathione-Sepharose (Pharmacia) for 1 h at 4°C. The beads were washed twice with 1 ml of buffer A, twice with 1 ml of buffer B (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 2.5 mM sodium orthovanadate, 1 mM DTT, 50 mM β-glycerophosphate), twice with 500 μl of 1× phosphatase buffer (50 mM imidazole [pH 7.5], 3 mM DTT), and once with 300 μl of 2× phosphatase buffer. GST-Mpk1 bound to resin was resuspended in 2× phosphatase buffer and aliquoted into four 20-μl fractions, each containing ∼100 ng of GST-Mpk1. Aliquots of bound GST-Mpk1 were incubated at 30°C for 1 h with 0 to 1 μg of Ptp2, with or without sodium orthovanadate (15 mM). The reaction was terminated by adding 300 μl of ice-cold buffer B and boiling in sample buffer. The samples were analyzed by SDS-PAGE and immunoblotting. Half of the sample was immunoblotted with anti-pY antibody PY-99 (Santa Cruz Biotechnology), and the other half was immunoblotted with anti-GST antibody (Pharmacia).
RNA analysis.
Yeast strains grown in YPD at 30°C to 1 U (A600) were untreated or heat shocked by the addition of an equal volume of YPD prewarmed to 48°C. Heat-shocked cultures were further incubated for 2 h at 39°C in prewarmed flasks. Cells were harvested by centrifugation, and RNA was prepared by freezing in phenol and SDS (64). Total RNA, ∼40 μg, was electrophoresed in formaldehyde-containing agarose gels, and hybridization was performed by standard methods (63). The blot was hybridized with 32P-labeled probes to PTP2, PTP3, and TUB1. A 750-bp PstI fragment internal to the PTP2 open reading frame, a 1-kb ClaI fragment internal to the PTP3 open reading frame, and a 1.1-kb BglII-ClaI fragment from TUB1 were used to produce 32P-labeled probes (63). The blots were quantified by PhosphorImager (Molecular Dynamics) analysis.
Fluorescence microscopy.
Strains expressing GFP fusion proteins were grown to 1 U (A600) in synthetic medium lacking leucine to select for plasmids. To visualize the nucleus, cultures were diluted to 0.5 U (A600), 4′,6-diamidino-2-phenylindole (DAPI) was added to a concentration of 2.5 μg/ml, and the cultures were grown under low-light conditions for 30 min at 30°C (27). Cells were adsorbed to polylysine-coated slides for 10 min and covered with coverslips. Fluorescence images were captured by using a Zeiss Axioplan fluorescence microscope with a Photometrics Sensys digital charge-coupled device camera system. Images were processed by using IP-LAB Spectrum software.
RESULTS
Ptp2 and Ptp3 inactivate the cell wall integrity pathway in yeast.
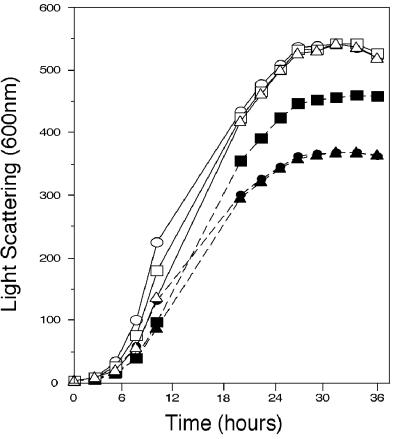
To test whether Ptp2 and Ptp3 could inactivate the cell wall integrity pathway, we determined whether overexpression of these phosphatases could alleviate growth defects due to mutations that hyperactivate this pathway. The MEKK mutation, BCK1-20, which leads to constitutive activation of this pathway, conferred a modest growth defect, where the doubling time in rich medium at 30°C was 110 min, compared to 90 min for the wild type (38). This growth defect is likely due to constitutive activation of the downstream MEKs, Mkk1 and Mkk2, and the MAPK, Mpk1 (32, 37). Multicopy plasmids bearing PTP2 or PTP3 were transformed into a strain expressing BCK1-20, and the growth of these strains was monitored in selective media at 30°C. Although only slight changes in doubling times were observed during exponential growth, overexpression of PTP2, but not PTP3, allowed BCK1-20 strains to reach a higher culture density after 24 h of growth (Fig. 1). Neither overexpression of PTP2 nor overexpression of PTP3 altered the growth of the wild type.
FIG. 1.
Ptp2 but not Ptp3 suppresses growth defects due to constitutive activation of the cell wall integrity pathway by the hyperactive MEKK mutation BCK1-20. DL245, a strain bearing a hyperactive MEKK allele, BCK1-20, and EG123, the isogenic wild-type strain, were transformed with a multicopy plasmid carrying PTP2 (p112PTP2), PTP3 (p112PTP3), or empty vector YEplac112. Cultures of these strains were diluted in selective medium and incubated at 30°C. The increase in optical density of the cultures at 600 nm was measured at the indicated times with a photoelectric colorimeter. Data shown are the average of at least two independent experiments. Strains: DL245/p112PTP2 (closed squares); DL245/p112PTP3 (closed triangles); DL245/YEplac112 (closed circles); EG123/YEplac112 (open circles); EG123/p112PTP3 (open triangles); EG123/p112PTP2 (open squares).
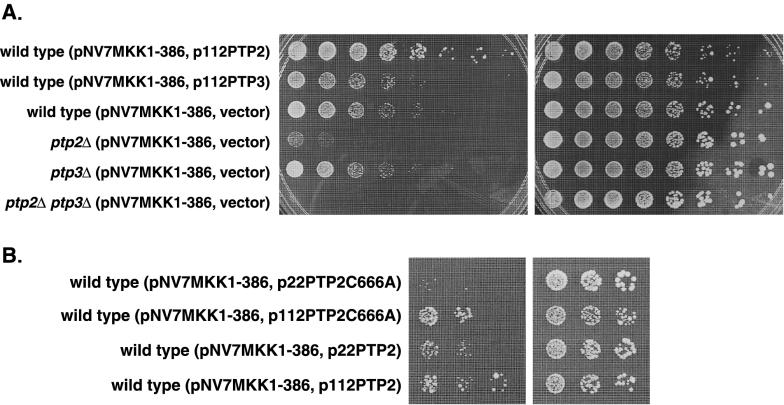
These results suggested that Ptp2 inactivates the redundant MEKs, Mkk1 and/or Mkk2, or the MAPK, Mpk1, in this pathway. To test whether Ptp2 acts downstream of the MEKs, we used the MKK1-386 mutation, which confers a growth defect due to constitutive activation of Mpk1 (71, 72, 74). The growth of wild-type cells expressing MKK1-386, under regulation of the GAL1 promoter, was ∼25-fold worse on galactose than on glucose (Fig. 2A). A 125-fold dilution of a culture with an optical density at 600 nm of 3.5 U grew on galactose-containing media, while the same culture diluted 3,125-fold grew on glucose containing media. Expression of PTP2 from a multicopy plasmid allowed cells expressing MKK1-386 to grow ∼25-fold better than cells overexpressing PTP3 or containing empty vector, suggesting that Ptp2 acts downstream of the MEK, Mkk1 (Fig. 2A). Ptp2 also suppressed the growth defect associated with MKK1-386 when expressed from a low-copy-number centromere-based plasmid, but not as well as multicopy expression (compare Fig. 2A, row 1, to Fig. 2B, row 3). Thus, Ptp2 most likely inactivates the cell wall integrity pathway by acting on the MAPK, Mpk1.
FIG. 2.
Growth defects associated with overexpression of a constitutively active MEK allele are altered by PTP expression level. (A) Overexpression of PTP2 suppressed the growth defect due to expression of the constitutively active MKK1 allele, MKK1-386, while strains lacking PTPs exacerbated these growth defects. Wild-type, ptp2Δ, ptp3Δ, and ptp2Δ ptp3Δ strains carrying pNV7MKK1386 and either YEplac112, p112PTP2, or p112PTP3 were grown in selective medium containing glucose. The cultures were normalized to 3.5 U (A600); fivefold serial dilutions were made, spotted onto synthetic medium containing galactose (left panel) or glucose (right panel), and incubated at 30°C. (B) The catalytic activity of Ptp2 is important for suppression of the growth defect due to MKK1-386. Wild-type JD52 carrying pNV7MKK1386 and either p22PTP2C666A, p112PTP2C666A, or p22PTP2 was grown in synthetic medium containing glucose, and growth was examined as for panel A.
To determine whether suppression required the phosphatase activity of Ptp2, a catalytically inactive mutant, Ptp2-C666S (33), was tested for suppression of the MKK1-386 defect. When expressed from a low-copy-number plasmid, Ptp2-C666S did not improve the growth of cells expressing MKK1-386, indicating that phosphatase activity is required for suppression (Fig. 2B, row 1). The mutant expressed from a multicopy plasmid, however, allowed the MKK1-386 strain to grow as well as the strain expressing wild-type PTP2 on a low-copy-number plasmid (Fig. 2B; compare rows 2 and 3). The ability of multicopy Ptp2-C666A to inactivate this pathway may be due to binding Mpk1 (see below). Although the mutant is catalytically inactive, binding and sequestration of Mpk1 may prevent activation of downstream targets. A similar result was observed with Ptp2-C666A and Hog1 (33).
Since overexpression of PTP2 suppressed growth defects due to hyperactivation of the cell wall integrity pathway, we tested whether deletion of PTP2 and/or PTP3 would further exacerbate growth defects associated with MKK1-386. A strain lacking PTP2 poorly tolerated MKK1-386 compared to the wild type (Fig. 2A). In contrast, a strain lacking PTP3 grew as well as the wild type in the presence of MKK1-386. Expression of MKK1-386 in a ptp2Δ ptp3Δ double mutant was lethal (Fig. 2A). Thus, both PTPs have a role in inactivation of this pathway and may act synergistically.
Ptp2 and Ptp3 regulate Mpk1 Tyr phosphorylation in vivo.
Overexpression of PTP2 suppressed growth defects due to constitutively active MEKK and MEK in this pathway. Thus, MAPK is the likely target of this phosphatase, since it requires phosphorylation of both a Thr and a Tyr residue in the phosphorylation lip sequence for activation (11, 42). Therefore, we examined whether Ptp2 and possibly Ptp3 could inactivate the pathway by dephosphorylating Mpk1-pY. To test this, the level of Mpk1-pY was monitored in wild-type and PTP deletion strains by using an anti-pY antibody, since it most directly assesses the role of PTPs. If the PTPs dephosphorylate Mpk1-pY, then PTP deletion strains should show higher levels of Mpk1-pY compared to the wild type after heat shock activation of Mpk1.
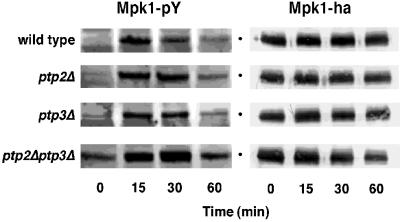
The level of Mpk1-pY was monitored in wild-type, ptp2Δ, ptp3Δ, and ptp2Δ ptp3Δ strains expressing epitope-tagged Mpk1, Mpk1-HA, during continuous exposure to heat stress at 39°C. In the wild type, the level of Mpk1-pY reached a maximum after 15 min of heat stress (Fig. 3). In the ptp2Δ and ptp3Δ strains, heat shock induction of Mpk1-pY was similar to that of the wild type at 15 min (Fig. 3). In contrast, the ptp2Δ strain consistently showed a slightly higher level of Mpk1-pY at the 30-min time point. The most obvious increase in the level of Mpk1-pY was observed in the ptp2Δ ptp3Δ strain, where a substantial increase in the basal (0 min) and heat shock-induced (15 min and beyond) levels of Mpk1-pY was seen. Since the total amount of Mpk1 protein did not increase during heat stress, as judged by immunoblotting with anti-HA antibody (Fig. 3), increased Mpk1-pY levels in PTP deletion mutants is due to their inability to dephosphorylate Mpk1. Similar results were obtained for untagged Mpk1 (data not shown). These results indicate that PTPs are required to maintain a low basal level of Mpk1-pY and to set an upper limit for Mpk1 activation in response to heat stress.
FIG. 3.
Ptp2 and Ptp3 regulate the level of Mpk1-pY in vivo. The level of Mpk1-pY was assessed in strains lacking either Ptp2, Ptp3, or both PTPs before and during continuous exposure to heat stress. Cultures were grown at 25°C, heat shocked at 39°C, and harvested at the indicated times. The samples were analyzed by SDS-PAGE and immunoblotting with anti-pY and anti-HA antibodies.
These data also suggest that PTPs are involved in adaptation, or inactivation of the pathway following heat stress, since the PTP deletion strains dephosphorylated Mpk1-pY more poorly than the wild type. In the wild type, the level of Mpk1-pY began decreasing by 30 min of heat stress, while the level of Mpk1-pY continued to increase at 30 min in the ptp2Δ and ptp2Δ ptp3Δ strains (Fig. 3). The ptp2Δ ptp3Δ strain had the most severe defect since Mpk1-pY levels remained high up to 60 min. The amount of Mpk1-pY at this time point is likely underestimated since the ptp2Δ ptp3Δ strain is temperature sensitive for growth at 37°C (60). This is reflected in the lower level of Mpk1-HA protein at the 60-min time point (Fig. 3). Therefore, both Ptp2 and to a lesser extent Ptp3 are required for adaptation to heat stress in this pathway.
PTPs physically associate with Mpk1.
If Mpk1 is a substrate for Ptp2 and Ptp3, it should be able to bind the PTPs. To test this, we used catalytically inactive PTPs since such mutants have been shown to bind their substrates with greater affinity than their wild-type counterparts (17, 66, 70, 73). Presumably, tighter binding occurs because the tyrosine-phosphorylated substrate is not hydrolyzed and as a result is not readily released from the enzyme (19, 70). Catalytically inactive Ptp2 (Ptp2-C666S) or Ptp3 (Ptp3-C804A) fused to GST was coexpressed with Mpk1-HA in yeast. Cells were heat shocked for 25 min at 39°C to activate Mpk1 and the GST-PTP proteins isolated on glutathione-Sepharose beads. Both GST–Ptp2-C666S and GST–Ptp3-C804A bound Mpk1-HA (Fig. 4). GST-PTP mutants also bound Mpk1 when cells were untreated, suggesting that Mpk1 phosphorylation is not required to bind PTPs (data not shown). Binding between PTPs and Mpk1 was specific since GST alone did not coprecipitate with Mpk1-HA (Fig. 4). In multiple experiments, the Ptp2 mutant bound more Mpk1-HA than the Ptp3 mutant. In addition, the interaction between GST–Ptp2-C666S and Mpk1 was more resistant to disruption by washing with buffer containing high concentrations of sodium chloride than the complex between Mpk1 and GST–Ptp3-C804A. These differences in binding are not due to differences in Mpk1-HA levels since they were the same in all strains (Fig. 4). That GST–Ptp2-C666S bound more Mpk1-HA than GST–Ptp3-C804A provides a possible biochemical mechanism for Ptp2 being a more effective negative regulator than Ptp3.
FIG. 4.
Ptp2 and Ptp3 bind Mpk1 in vitro. GST fusions to the catalytically inactive PTP mutants, GST-Ptp2C666S and GST-Ptp3C804A, or GST alone were coexpressed with Mpk1-HA in S. cerevisiae 334. After 25 min of heat stress at 39°C, cells were lysed by glass beading, and the GST-containing proteins were isolated by binding to glutathione-Sepharose. Following extensive washing, the bound proteins were analyzed by SDS-PAGE and immunoblotting with anti-GST and anti-HA antibodies. Data shown are representative of three independent experiments.
Ptp2 dephosphorylates Mpk1-pY in vitro.
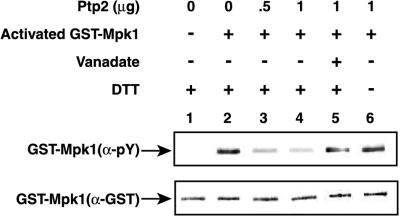
Since the results above suggested that Ptp2 and Ptp3 inactivate Mpk1 directly, we tested whether they could dephosphorylate Mpk1-pY in vitro. Ptp2 and Ptp3 tagged with six His repeats at the amino terminus were expressed in E. coli and isolated by immobilized metal affinity chromatography. The PTPs were incubated with activated GST-Mpk1 isolated from heat-shocked yeast. Treatment of GST–Mpk1-pY with Ptp2 resulted in dephosphorylation of the pY residue in Mpk1, as judged by immunoblotting with anti-pY antibody (Fig. 5). Dephosphorylation was due to PTP activity since orthovanadate, an inhibitor of PTPs and dual-specificity phosphatases (26, 70), prevented dephosphorylation of Mpk1-pY. Dephosphorylation required the presence of a reducing agent, DTT (Fig. 5) or 2-mercaptoethanol (data not shown), which likely prevents oxidation of Cys666 in Ptp2, which acts as a nucleophile during dephosphorylation (26, 33, 73). Compared to Ptp2, Ptp3 had significantly weaker PTP activity toward GST–Mpk1-pY. Ptp2 and Ptp3 had similar abilities to dephosphorylate the small-molecule substrate, PNPP (see Materials and Methods). Approximately 3.3 times more PNPP units of Ptp3 did not dephosphorylate Mpk1-pY as well as Ptp2 (data not shown). Thus, Ptp2 but not Ptp3 is an effective protein phosphatase for Mpk1-pY in vitro.
FIG. 5.
Ptp2 dephosphorylates Mpk1-pY in vitro. Inactive GST-Mpk1 isolated from yeast grown at 23°C (lane 1) and active GST-Mpk1 isolated from yeast heat shocked at 39°C (lanes 2 to 6) were incubated for 1 h at 30°C with buffer alone (lanes 1 and 2), Ptp2 (lanes 3 and 4), Ptp2 in the presence of vanadate (lane 5), and Ptp2 in buffer lacking DTT (lane 6). The samples were analyzed by SDS-PAGE and immunoblotting. Half of the sample was immunoblotted with anti-pY antibody, and the other half was immunoblotted with anti-GST antibody.
PTP2 transcript is elevated by heat shock in a Mpk1-dependent manner.
Activation of MAPK pathways often leads to an increase in phosphatase transcript levels (24, 33, 50) and is generally thought to be required for adaptation. Therefore, we tested whether the cell wall integrity pathway upregulates the expression of PTPs. Since it was previously shown that the PTP2 transcript is upregulated in response to heat stress (59), we tested whether PTP2 and possibly PTP3 transcripts might be regulated in a Mpk1-dependent manner. In addition, since Hog1 is required for upregulation of PTP2 and PTP3 transcripts in response to osmotic stress (33), we tested whether Hog1 might also be required to induce PTPs in response to heat stress. Northern analysis was performed to examine the steady-state levels of PTP2 and PTP3 transcripts before and after heat shock for 2 h at 39°C (Fig. 6). Heat shock elevated the PTP2 transcript ∼6-fold above the basal level in the wild-type and hog1Δ strains but not in the mpk1Δ or mpk1Δ hog1Δ strain. Thus, Mpk1, but not Hog1, is required to upregulate PTP2 expression in response to heat stress. No obvious increase in the level of PTP3 transcript was observed in response to heat stress. Since it was difficult to detect PTP3 by Northern analysis, a PTP3 promoter-lacZ fusion was constructed and integrated at the PTP3 locus. Heat shock did not alter the expression of PTP3::lacZ, as judged by assaying β-galactosidase activity. Therefore, heat shock activation of the cell wall integrity pathway upregulates PTP2 but not PTP3 expression.
FIG. 6.
PTP2 transcript increases in response to heat shock in a Mpk1-dependent manner. The effect of heat shock on the steady-state level of PTP2 transcript was examined in wild-type, mpk1Δ, hog1Δ, and mpk1Δ hog1Δ strains by Northern analysis. Total RNA was isolated from untreated cells grown at 30°C or cells heat shocked at 39°C for 2 h. The Northern blot was probed with 32P-labeled PTP2 and TUB1 fragments and visualized by PhosphorImager analysis.
Ptp2 and Ptp3 are localized in different subcellular compartments.
Ptp2 and Ptp3 show differences in the ability to inactivate the cell wall integrity pathway and other MAPK pathways. For example, Ptp2 is a more potent negative regulator of Mpk1 and Hog1 than Ptp3 (33, 73). In the pheromone response pathway, the opposite is true: Ptp3 has a greater influence than Ptp2 (76). We have shown that Ptp2 has greater activity toward Mpk1-pY in vitro than Ptp3. In vivo, an additional factor may be that Ptp2 and Ptp3 are localized in different compartments in the cell. This seems a reasonable hypothesis since Mpk1 is present in both the nucleus and the cytoplasm (34).
To examine Ptp2 and Ptp3 subcellular localization, PTPs were fused to GFP. The PTP-GFP fusions are functional, since they suppressed growth defects due to constitutive activation of the HOG (see Materials and Methods) and cell wall integrity pathways (data not shown). Ptp2-GFP is predominantly nuclear when expressed from a multicopy plasmid in a ptp2Δ strain (Fig. 7A). In contrast, Ptp3-GFP appears cytoplasmic and excluded from the nucleus when expressed from a multicopy plasmid in a ptp3Δ strain (Fig. 7B). The nuclear localization of Ptp2 may therefore contribute to its greater effectiveness on Mpk1. Heat stress did not change the localization of PTPs; Ptp2-GFP remained nuclear and Ptp3-GFP remained cytoplasmic. Deletion of MPK1 did not detectably alter the subcellular localization of either PTP-GFP fusion under standard growth conditions. Since PTPs also inactivate the HOG and pheromone response pathways, we also tested whether osmotic stress or treatment with α-factor would alter their localization. Neither signal affected PTP localization. Thus, similar to findings for other MAPK inactivating phosphatases, the subcellular localization of PTPs is not altered by signals that activate MAPKs.
FIG. 7.
Subcellular localization of Ptp2 and Ptp3. Exponentially growing cells of a ptp2Δ strain carrying pPTP2-GFP or a ptp3Δ strain carrying pPTP3-GFP were grown in the presence of DAPI (2.5 μg/ml) for 30 min under low-light conditions. GFP and DAPI were visualized by fluorescence microscopy.
DISCUSSION
Protein phosphatases can be nonspecific or specific regulators of MAPK signaling pathways. For example, the vertebrate dual-specificity phosphatases MKP-1 and MKP-2 are nonspecific regulators, inactivating ERK, JNK/SAPK, and p38 (3, 4, 10, 42, 70). Other dual-specificity phosphatases, such as PAC1, recognize a subset of MAPK substrates, ERK and p38 (10). One vertebrate dual-specificity phosphatase, MKP-3, is highly specific for ERK (23, 53, 55, 56). In S. cerevisiae, the protein tyrosine-specific phosphatases Ptp2 and Ptp3 show differences in the ability to inactivate MAPK pathways. Ptp2 is a more effective negative regulator of Hog1 than Ptp3, while Ptp3 is a more effective negative regulator of Fus3 than Ptp2 (33, 73, 76).
To further examine the substrate specificities of Ptp2 and Ptp3, we tested whether they could inactivate another MAPK in yeast called Mpk1 and, if so, whether they would differ in the ability to inactivate Mpk1. We predicted that Ptp3 would be a more potent negative regulator of Mpk1 than Ptp2, since the phosphorylation lip sequences in Mpk1 and Fus3 are nearly identical but different from that in Hog1 (11). Our data show that both PTPs inactivate Mpk1, demonstrating that Ptp2 and Ptp3 regulate multiple MAPK pathways in yeast. Contrary to our prediction, however, Ptp2 is a more effective negative regulator of Mpk1 than Ptp3.
Genetic and biochemical data show that Ptp2 inactivates the cell wall integrity pathway by inactivating Mpk1. Multicopy expression of PTP2 suppressed growth defects due to the MEKK mutation, BCK1-20, and the MEK mutation, MKK1-386, that hyperactivate this pathway. In addition, deletion of PTP2 exacerbated growth defects due to MKK1-386 (Fig. 2). These results suggested that Ptp2 acts downstream of Mkk1. Since the catalytically inactive Ptp2 mutant, Ptp2-C666S, bound Mpk1-pY, and wild-type Ptp2 dephosphorylated Mpk1-pY in vitro, Ptp2 likely inactivates this pathway by dephosphorylating Mpk1-pY in vivo. Consistent with this, the level of Mpk1-pY in response to heat stress was higher in the ptp2Δ strain than in the wild-type strain.
Ptp3 also inactivates this pathway but is a weaker negative regulator than Ptp2. For example, multicopy expression of PTP2 suppressed growth defects due to the hyperactive MEKK and MEK alleles, but multicopy expression of PTP3 did not. Genetic interactions were observed, however, when PTP3 was deleted together with PTP2. Expression of the hyperactive MEK allele, MKK1-386, was lethal in the ptp2Δ ptp3Δ strain but not in either single PTP deletion strain. Biochemical data also support a role for Ptp3 in this pathway. Heat stress induced Mpk1-pY to a higher level in the ptp2Δ ptp3Δ strain compared to either single ptp mutant or the wild type. Coprecipitation and activity assays also indicate that Ptp3 inactivates Mpk1 but not as efficiently as Ptp2. Ptp3-C804A coprecipitated with Mpk1-pY, but it bound less Mpk1-pY than mutant Ptp2. Furthermore, Ptp3 was less effective than Ptp2 at dephosphorylating Mpk1-pY in vitro. These results taken together show that Ptp3 is a weaker negative regulator of Mpk1 than Ptp2 but has an important role in this pathway.
The differential localization of PTPs may also explain why Ptp2 is a more effective negative regulator than Ptp3. Fluorescence localization of Ptp2-GFP indicates that it is predominantly nuclear whereas Ptp3-GFP is chiefly cytoplasmic. This difference in localization is significant because MAPKs are present in both the nucleus and the cytoplasm, with activation of MAPKs often leading to their nuclear accumulation (8, 39). MAPK activation, however, does not always result in their nuclear translocation. For example, a significant fraction of active ERK has been shown to remain cytoplasmic after activation (21, 62). Interestingly, Mpk1 is concentrated in the nucleus at 24°C and becomes more uniformly distributed between the cytoplasm and the nucleus after 30 min of heat stress at 39°C (34). Thus, active Mpk1 may be present in both compartments. We speculate that at least some active Mpk1 is nuclear and that this nuclear Mpk1 is responsible for growth defects. This would explain why altering the expression level of PTP2 has a greater effect on the activity of this pathway than altering the expression of PTP3 (Fig. 1 and 2). Since we cannot rule out the possibility that some Ptp2 is cytoplasmic, Ptp2 may have a greater effect on this pathway for other reasons. As described above, Ptp2 binds Mpk1 more effectively than Ptp3 and dephosphorylates Mpk1-pY more efficiently than Ptp3. The combination of these effects likely accounts for the greater activity of Ptp2 on this pathway.
A further implication of the differential localization of Ptp2 and Ptp3 is that Mpk1-pY freely distributes between the nucleus and the cytoplasm. For example, the ptp2Δ and ptp3Δ strains dephosphorylate Mpk1-pY nearly as well as the wild type, although the ptp2Δ ptp3Δ strain shows a severe defect. Since nuclear Ptp2 can compensate for cytoplasmic Ptp3 in the ptp3Δ strain, this would suggest that cytoplasmic Mpk1-pY translocates to the nucleus to be dephosphorylated by nuclear Ptp2. In the ptp2Δ strain, nuclear Mpk1-pY becomes available to cytoplasmic Ptp3 upon Mpk1-pY redistribution to the cytoplasm. Thus, contrary to current hypotheses, dephosphorylation of MAPK may not be required for its export from the nucleus.
Analysis of the level of Mpk1-pY during heat shock activation suggests three roles for Ptp2 and Ptp3. One role is in maintaining a low basal activity of Mpk1. In the absence of heat stress, a low level of Mpk1-pY was observed in wild-type, ptp2Δ, and ptp3Δ strains. The ptp2Δ ptp3Δ mutant, however, showed a significantly higher basal level of Mpk1-pY. These results are similar to those observed for Hog1 and Fus3. The ptp2Δ and ptp2Δ ptp3Δ strains had a high basal level of Hog1-pY (33, 73), and the ptp3Δ and ptp2Δ ptp3Δ strains had an increased basal level of Fus3-pY (76). Therefore, PTPs may prevent the activation of MAPKs in the absence of signal. A second role of PTPs, not seen in other MAPK pathways, is to set an upper limit on Mpk1 Tyr phosphorylation. Mpk1-pY reached a higher level in the single PTP deletion strains and in the ptp2Δ ptp3Δ strain compared to the wild type. Perhaps this mechanism prevents growth defects due to activation of a large fraction of Mpk1.
A third role for Ptp2 and Ptp3 is in adaptation, or inactivating Mpk1-pY during prolonged exposure to heat stress. Previously, it was reported that the level of Mpk1-pY remains high and shows no obvious decrease for up to 2 h of heat stress (75). In our strain background, however, heat stress induced Mpk1-pY to a maximum at ∼15 min but began decreasing by 30 min. The reasons for these differences are not known but may be due to strain-specific effects. Since the level of Mpk1-pY decreased in our strain background, the role of PTPs in adaptation could be examined. We find that the level of Mpk1-pY remained high in both ptp2Δ and ptp2Δ ptp3Δ strains compared to the wild type, consistent with a role for PTPs in adaptation. The residual dephosphorylation of Mpk1-pY observed in the ptp2Δ ptp3Δ strain may be due to Msg5, a dual-specificity phosphatase isolated as a multicopy suppressor of MKK1-386 growth defects (71). That Msg5 acts as a negative regulator in this pathway is further supported by genetic evidence. Although ptp2Δ and msg5Δ strains are not lethal when MKK1-386 is overexpressed, it is lethal in a ptp2Δ msg5Δ strain (48). Whether Msg5 dephosphorylates Mpk1 remains to be tested.
Transcriptional activation of PTP2 may also play a role in adaptation. In this work, we showed that the PTP2 transcript is upregulated by heat shock in a Mpk1-dependent manner, suggesting that an increase in the level of Ptp2 is required to inactivate Mpk1. Similarly, osmotic stress elevates PTP2 and PTP3 transcripts in a Hog1-dependent manner (33). In S. pombe, osmotic stress induces expression of the protein tyrosine phosphatase, pyp2+, and this is dependent on the MAPK, Spc1 (50). This regulation is not limited to PTPs since dual-specificity phosphatases have also been shown to be induced by MAPK pathways in S. cerevisiae and vertebrates (3, 17, 47). Upregulation of protein phosphatases may therefore be a universal mechanism for MAPK pathway adaptation.
In summary, Ptp2 and Ptp3 are global regulators of MAPK signaling pathways since they inactivate at least three MAPKs, Hog1, Fus3, and Mpk1. These phosphatases show distinct preferences for MAPKs, as has been observed for dual-specificity phosphatases in vertebrates (42). Ptp2 is a more effective negative regulator of Mpk1 and Hog1 than Ptp3 (33, 73). In contrast, Ptp3 is a more potent negative regulator of Fus3 than Ptp2 (76). These differences are likely due to at least two factors. First, PTPs that are better able to bind their substrates more efficiently dephosphorylate their substrates. Ptp2 binds more Mpk1 and Hog1 than Ptp3, and Ptp2 more efficiently dephosphorylates Mpk1 (Fig. 4) and Hog1 (67) in vitro. Based on these results, we would predict that Ptp3 is better able to bind and dephosphorylate Fus3 than Ptp2. Second, the localization of PTPs likely influences their ability to inactivate MAPKs. We propose that the nuclear localization of Ptp2 contributes to its greater ability to dephosphorylate Mpk1 and Hog1, because a fraction of active Mpk1 and Hog1 accumulates in the nucleus (18, 34, 48). Further examination of how PTP localization influences their ability to inactivate MAPKs and identification of regions within PTPs responsible for discrimination between MAPKs is under way.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation and National Institutes of Health training grant GM07135.
We thank David Levin, Jean Hirsch, Michael Snyder, Kenji Irie, Kunihiro Matsumoto, Xiao-Li Zhan, Kun-Liang Guan, Natalie Ahn, Paul Shapiro, and Stephanie Mohr for yeast strains and reagents; we thank Randall Bass, Tim Lewis, Janel Warmka, and Ann Whalen for helpful discussions and comments on the manuscript.
REFERENCES
- 1.Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 3.Brondello J M, Brunet A, Pouyssegur J, McKenzie F R. The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J Biol Chem. 1997;272:1368–1376. doi: 10.1074/jbc.272.2.1368. [DOI] [PubMed] [Google Scholar]
- 4.Brondello J M, McKenzie F R, Sun H, Tonks N K, Pouyssegur J. Constitutive MAP kinase phosphatase (MKP-1) expression blocks G1 specific gene transcription and S-phase entry in fibroblasts. Oncogene. 1995;10:1895–1904. [PubMed] [Google Scholar]
- 5.Brown-Shimer S, Johnson K A, Lawrence J B, Johnson C, Bruskin A, Green N R, Hill D E. Molecular cloning and chromosome mapping of the human gene encoding protein phosphotyrosyl phosphatase 1B. Proc Natl Acad Sci USA. 1990;87:5148–5152. doi: 10.1073/pnas.87.13.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buehrer B M, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charbonneau H, Tonks N K, Kumar S, Diltz C D, Harrylock M, Cool D E, Krebs E G, Fischer E H, Walsh K A. Human placenta protein-tyrosine-phosphatase: amino acid sequence and relationship to a family of receptor-like proteins. Proc Natl Acad Sci USA. 1989;86:5252–5256. doi: 10.1073/pnas.86.14.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R H, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chernoff J, Schievella A R, Jost C A, Erikson R L, Neel B G. Cloning of a cDNA for a major human protein-tyrosine-phosphatase. Proc Natl Acad Sci USA. 1990;87:2735–2739. doi: 10.1073/pnas.87.7.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu Y, Solski P A, Khosravi-Far R, Der C J, Kelly K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J Biol Chem. 1996;271:6497–6501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 11.Cobb M H, Goldsmith E J. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 12.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 13.Davenport K R, Sohaskey M, Kamada Y, Levin D E, Gustin M C. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 14.Degols G, Shiozaki K, Russell P. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:2870–2877. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dohmen R J, Strasser A W, Honer C B, Hollenberg C P. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991;7:691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- 16.Dohmen R J, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 17.Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrigno P, Posas F, Koepp D, Saito H, Silver P A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint A J, Tiganis T, Barford D, Tonks N K. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez F A, Seth A, Raden D L, Bowman D S, Fay F S, Davis R J. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling membrane and the nucleus. J Cell Biol. 1993;122:1089–1101. doi: 10.1083/jcb.122.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groom L A, Sneddon A A, Alessi D R, Dowd S, Keyse S M. Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 1996;15:3621–3632. [PMC free article] [PubMed] [Google Scholar]
- 24.Grumont R J, Rasko J E, Strasser A, Gerondakis S. Activation of the mitogen-activated protein kinase pathway induces transcription of the PAC-1 phosphatase gene. Mol Cell Biol. 1996;16:2913–2921. doi: 10.1128/mcb.16.6.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan K L, Butch E. Isolation and characterization of a novel dual specific phosphatase, HVH2, which selectively dephosphorylates the mitogen-activated protein kinase. J Biol Chem. 1995;270:7197–7203. doi: 10.1074/jbc.270.13.7197. [DOI] [PubMed] [Google Scholar]
- 26.Guan K L, Dixon J E. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. J Biol Chem. 1991;266:17026–17030. [PubMed] [Google Scholar]
- 27.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 28.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 29.Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 30.Horvitz H R, Sternberg P W. Multiple intercellular signaling systems control the development of the Caenorhabditis elegans vulva. Nature. 1991;351:535–541. doi: 10.1038/351535a0. [DOI] [PubMed] [Google Scholar]
- 31.Hovland P, Flick J, Johnston M, Sclafani R A. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 32.Irie K, Takase M, Lee K S, Levin D E, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacoby T, Flanagan H, Faykin A, Seto A G, Mattison C, Ota I. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J Biol Chem. 1997;272:17749–17755. doi: 10.1074/jbc.272.28.17749. [DOI] [PubMed] [Google Scholar]
- 34.Kamada Y, Jung U S, Piotrowski J, Levin D E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 35.Kamada Y, Qadota H, Python C P, Anraku Y, Ohya Y, Levin D E. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 36.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 37.Lee K S, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin D E. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee K S, Levin D E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenormand P, Sardet C, Pages G, L’Allemain G, Brunet A, Pouyssegur J. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levin D E, Fields F O, Kunisawa R, Bishop J M, Thorner J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62:213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- 41.Lewis T, Groom L A, Sneddon A A, Smythe C, Keyse S M. XCL100, an inducible nuclear MAP kinase phosphatase from Xenopus laevis: its role in MAP kinase inactivation in differentiated cells and its expression during early development. J Cell Sci. 1995;108:2885–2896. doi: 10.1242/jcs.108.8.2885. [DOI] [PubMed] [Google Scholar]
- 42.Lewis T S, Shapiro P S, Ahn N G. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 43.Madden K, Sheu Y J, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- 44.Madhani H D, Fink G R. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 45.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande Woude G F, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 46.Marini N J, Meldrum E, Buehrer B, Hubberstey A V, Stone D E, Traynor-Kaplan A, Reed S I. A pathway in the yeast cell division cycle linking protein kinase C (Pkc1) to activation of Cdc28 at START. EMBO J. 1996;15:3040–3052. [PMC free article] [PubMed] [Google Scholar]
- 47.Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky A M, Martinez-Arias A. Puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12:557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattison, C. P., and I. M. Ota. Unpublished data.
- 49.Mazzoni C, Zarov P, Rambourg A, Mann C. The SLT2 (MPK1) MAP kinase homolog is involved in polarized cell growth in Saccharomyces cerevisiae. J Cell Biol. 1993;123:1821–1833. doi: 10.1083/jcb.123.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millar J B, Buck V, Wilkinson M G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- 51.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 52.Mizunuma M, Hirata D, Miyahara K, Tsuchiya E, Miyakawa T. Role of calcineurin and Mpk1 in regulating the onset of mitosis in budding yeast. Nature. 1998;392:303–306. doi: 10.1038/32695. [DOI] [PubMed] [Google Scholar]
- 53.Muda M, Boschert U, Dickinson R, Martinou J C, Martinou I, Camps M, Schlegel W, Arkinstall S. MKP-3, a novel cytosolic protein-tyrosine phosphatase that exemplifies a new class of mitogen-activated protein kinase phosphatase. J Biol Chem. 1996;271:4319–4326. doi: 10.1074/jbc.271.8.4319. [DOI] [PubMed] [Google Scholar]
- 54.Muda M, Boschert U, Smith A, Antonsson B, Gillieron C, Chabert C, Camps M, Martinou I, Ashworth A, Arkinstall S. Molecular cloning and functional characterization of a novel mitogen-activated protein kinase phosphatase, MKP-4. J Biol Chem. 1997;272:5141–5151. doi: 10.1074/jbc.272.8.5141. [DOI] [PubMed] [Google Scholar]
- 55.Muda M, Theodosiou A, Gillieron C, Smith A, Chabert C, Camps M, Boschert U, Rodrigues N, Davies K, Ashworth A, Arkinstall S. The mitogen-activated protein kinase phosphatase-3 N-terminal noncatalytic region is responsible for tight substrate binding and enzymatic specificity. J Biol Chem. 1998;273:9323–9329. doi: 10.1074/jbc.273.15.9323. [DOI] [PubMed] [Google Scholar]
- 56.Muda M, Theodosiou A, Rodrigues N, Boschert U, Camps M, Gillieron C, Davies K, Ashworth A, Arkinstall S. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J Biol Chem. 1996;271:27205–27208. doi: 10.1074/jbc.271.44.27205. [DOI] [PubMed] [Google Scholar]
- 57.Myers A M, Tzagoloff A, Kinney D M, Lusty C J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 58.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ota I M, Varshavsky A. A gene encoding a putative tyrosine phosphatase suppresses lethality of an N-end rule-dependent mutant. Proc Natl Acad Sci USA. 1992;89:2355–2359. doi: 10.1073/pnas.89.6.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ota, I. M. Unpublished data.
- 61.Rajavel M, Philip B, Buehrer B M, Errede B, Levin D E. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3969–3976. doi: 10.1128/mcb.19.6.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reszka A A, Seger R, Kiltz C D, Krebs E G, Fischer E H. Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc Natl Acad Sci USA. 1995;92:8881–8885. doi: 10.1073/pnas.92.19.8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 64.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 66.Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- 67.Spencer, S., and I. M. Ota. Unpublished data.
- 68.Stevenson B J, Rhodes N, Errede B, Sprague G F., Jr Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 69.Struhl K, Stinchcomb D T, Scherer S, Davis R W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun H, Charles C H, Lau L F, Tonks N K. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 71.Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wurgler-Murphy S M, Maeda T, Witten E A, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yashar B, Irie K, Printen J A, Stevenson B J, Sprague G F, Jr, Matsumoto K, Errede B. Yeast MEK-dependent signal transduction: response thresholds and parameters affecting fidelity. Mol Cell Biol. 1995;15:6545–6553. doi: 10.1128/mcb.15.12.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zarzov P, Mazzoni C, Mann C. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]
- 76.Zhan X L, Deschenes R J, Guan K L. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
- 77.Zipursky S L, Rubin G M. Determination of neuronal cell fate: lessons from the R7 neuron of Drosophila. Annu Rev Neurosci. 1994;17:373–397. doi: 10.1146/annurev.ne.17.030194.002105. [DOI] [PubMed] [Google Scholar]