Abstract
Motor stereotypies occurring in early-onset neuropsychiatric diseases are associated with dysregulated basal ganglia direct-pathway activity. Disruptions in network connectivity through impaired neuronal structure have been implicated in both rodents and humans. However, the neurobiological mechanisms leading to direct-pathway neuron disconnectivity in stereotypy remain poorly understood. We have a mouse line with Tropomyosin receptor kinase B (TrkB) receptor deletion from D1-expressing cells (D1-Cre-flTrkB) in which a subset of animals shows repetitive rotations and head tics with juvenile onset. Here we demonstrate these behaviors may be associated with abnormal direct-pathway activity by reducing rotations using chemogenetic inhibition of dorsal striatum D1-medium spiny neurons (D1-MSNs) in both juvenile and young adult mice. Taking advantage of phenotypical differences in animals with similar genotype, we then interrogated the D1-MSN specific translatome associated with repetitive behavior by using RNA-sequencing of ribosome-associated mRNA. Detailed translatome analysis followed by multiplexed gene expression assessment revealed profound alterations in neuronal projection and synaptic structure related genes in stereotypy mice. Examination of neuronal morphology demonstrated dendritic atrophy and dendritic spine loss in dorsal striatum D1-MSNs from mice with repetitive behavior. Together, our results uncover phenotype-specific molecular alterations in D1-MSNs that relate to morphological adaptations in mice displaying stereotypy behavior.
Introduction
Stereotypies are repetitive, rhythmic behaviors ranging from simple movement sequences to complex actions1 that are observed in multiple neuropsychiatric diseases associated with basal ganglia dysfunction1–3. Imbalance in basal ganglia output-pathways arising from dopamine 1 receptor (D1, direct-pathway) vs. dopamine 2 receptor (D2, indirect-pathway) containing striatal medium spiny neurons (MSNs, aka- spiny projection neurons) is implicated in the repetitive movements that characterize disorders including Tourette Syndrome (TS), Autism Spectrum Disorders (ASD), and Obsessive Compulsive Disorder (OCD)1, 3–6. Activation of striatal dopamine D1 receptors induces stereotypies in non-human animals7, including those with dopamine deficiency8 and hyperdopaminergic function9, implicating a dysregulated direct-pathway in stereotypy behavior. Similarly, TS and OCD patients display increased sensitivity of striatal dopamine receptors and downregulation of D1 receptors10, 11 likely contributing to striatal hyperactivity6, 12. Additionally, mutations in genes regulating synaptic function and dendritic growth occur in TS, OCD and ASD13–17 and have been confirmed by striatal transcriptome analyses in TS and OCD patients18, 19. Thus, impaired MSN morphology combined with disrupted physiological properties may underlie neuronal hyperactivity and/or dopamine hypersensitivity and ultimately altered direct pathway output8.
Typically, stereotypies emerge during childhood and/or adolescence3, 20, suggesting a neurodevelopmental basis21. Striatal MSN development requires cortical brain-derived neurotrophic factor (BDNF) 22, 23. In humans, impairment of BDNF-Tropomyosin receptor kinase B (TrkB) signaling is associated with stereotypy disorders24–26 and conditional knockout of the BDNF-TrkB signaling pathway in striatal circuits leads to motor dysfunction including hyperlocomotion and stereotypy in mice23, 27. However, corticostriatal BDNF-TrkB knockout in mice usually affects both direct and indirect pathway MSNs to a similar extent, and BDNF deletion drastically impairs MSN development and survival23, 27, 28. In these conditions, parsing out the contribution of the BDNF-TrkB signaling in each pathway is complex. Cell-type specific TrkB deletion from enkephalin-expressing cells (D2-MSNs) induces spontaneous hyperlocomotion in middle-aged mice, which resembles Huntington’s disease (HD)29 and HD mouse models show impaired TrkB signaling in D2-MSNs30. Surprisingly, while evidence suggests that BDNF-TrkB signaling in direct pathway neurons could have an important role in the emergence of stereotypy, to date no study has assessed the effect of TrkB deletion in dopamine D1-MSNs on stereotypy behavior.
We have a mouse line in which the TrkB receptor is deleted from D1 expressing cells (D1-Cre-flTrkB). Interestingly, a subset of the animals (14.4%) exhibit repetitive circling behavior and head tics. Chemogenetic inhibition of striatal D1-MSNs reduced repetitive circling in the stereotypy mice. Thus, we took advantage of this phenotypic difference to probe molecular alterations associated with stereotypy in striatal D1-MSNs. Cell-type specific RNA-sequencing of ribosome-associated mRNA identified translatome alterations of morphology-related genes and morphological analysis demonstrated D1-MSN dendritic atrophy in mice with stereotypy.
Materials and methods
Experimental subjects
D1-Cre-flTrkB mice were generated by breeding D1-Cre (Line FK150)31 on a C57Bl/6 background with flTrkB32 mice on a mixed C57Bl/6 x FVB/N background. D1-Cre-TrkBflox/+ 33 were crossed with TrkBflox/+ to obtain D1-Cre-TrkBflox/flox, D1-Cre-TrkBflox/+, and D1-Cre-TrkB+/+. For behavioral and neuronal morphology experiments, D1-Cre-TrkB+/+ (referred to as D1-Cre) and D1-Cre-flTrkBflox/flox (referred to as D1-Cre-flTrkB) were used. TrkB knockout from D1-positive cells resulted in full-length TrkB deletion as described previously32. For cell-type specific RNA-sequencing studies D1-Cre-flTrkB mice were bred with RiboTag (RT)+/+ mice (Rpl22tm1.1Psam/J)34 on a C57Bl/6 background to generate D1-Cre-RT+/− and D1-Cre-flTrkB-RT+/− mice. No differences were observed between sexes in circling behavior: both males (26/55; i.e. 47.3%) and females (29/55; i.e. 52.7%) (Supplementary Figure S1a) were used in behavioral and neuronal morphology experiments. Studies were conducted in accordance with guidelines set up by the IACUC at University of Maryland School of Medicine (UMSOM). Animals were given food and water ad libitum and housed in UMSOM facilities during the whole study. For all experiments animals were not randomly assigned to groups: assignment was based on genotype/phenotype.
Stereotypic behavior
Behavioral characterization was performed in 15 min weekly sessions between the age of 3 and 8 weeks in a 5L cylinder. Mice were first habituated to the cylinder for 15 min. In each session, rotations, head tics and grooming were video recorded and then analyzed. Rotations were defined as complete 360° turns35 while head tics were defined as uncontrolled head waving/shaking with increased neck rigidity occurring separately from any other motor behavior. Repetitive circling was the predominant stereotypic behavior and head tics co-occurred in every mouse in addition to rotations. The number of rearings was counted with video tracking software (CleverSys). For chemogenetic inhibition of D1-MSNs, a similar procedure was used. Briefly, mice were habituated to the cylinder before being injected with saline solution (i.p.;10mL/kg) 30 min prior to behavioral testing. On the next day, mice were injected with 0.5mg/kg clozapine-N-oxide (i.p.; LKT Laboratories) 30 min prior to behavioral testing. For pharmacological testing, following habituation mice were injected with vehicle solution (saline with 10% (2-hydroxypropyl)-beta-cyclodextrin s.c.; 5mL/kg) 15min prior to behavioral assessment. Mice received injections of the D1-antagonist Ecopipam (SCH-39166; 0.02mg/kg, s.c., Tocris) and the D2-antagonist Haloperidol (0.05mg/kg, s.c., Sigma Aldrich) in a randomized, counter-balanced way. Both drug tests had their own vehicle session and were separated by 3 days of washout.
Locomotor activity
For Baseline locomotion experiments, mice were recorded for 30min. For pharmacology experiments, mice were recorded during 15min sessions, 15min after receiving vehicle, Ecopipam or Haloperidol. The same vehicle session was used to compare the effect of both drugs. In both conditions, total distance was measured (CleverSys) as described in36.
Stereotaxic surgery
Under isoflurane anesthesia, the dorsal striatum (from Bregma, anterior/posterior: +0.6, medial/lateral: +/−1.8, dorsal/ventral: −3) of juvenile (PND18–20) D1-Cre-flTrkB mice was bilaterally injected with a Cre-inducible, double inverted open (DIO)-reading frame adeno-associated virus (AAV): AAV5-hSyn-DIO-hM4D(Gi)-mCherry (Addgene; #44362) or AAV5-Ef1a-DIO-Cherry (UNC vector core) for Designer Receptor Exclusively Activated by Designer Drugs (DREADDs) experiments and with AAV5-DIO-eYFP (UNC vector core) diluted to 1.5×1011 VP/mL for neuronal morphology37. A minimum of 15 days was allowed for viral expression.
Cell-type specific RNA sequencing and Bioinformatics
Immunoprecipitated polyribosomes were prepared from dorsal striatum of D1-Cre-RT and D1-Cre-flTrkB-RT mice according to our previous studies36–38. Dorsal striatum punches were collected from a single animal, homogenized and 800μL of the supernatant was incubated in HA-coupled magnetic beads (Invitrogen: 100.03D; Covance: MMS-101R) overnight at 4°C. Next, magnetic beads were washed in high salt buffer. After adding TRK lysis buffer, RNA was extracted with the RNeasy Micro kit (Qiagen: 74004). For RNA-sequencing, only samples with RNA integrity numbers > 8 were used. Samples were submitted in biological triplicates for RNA-sequencing at the UMSOM Institute for Genome Sciences (IGS) and processed as described previously39. Libraries were prepared from 90ng of RNA from each sample using the NEBNext Ultra kit (New England BioLabs). Samples were sequenced on an Illumina HiSeq 4000 with a 75 bp paired-end read. An average of 66–97 millions reads were obtained for each sample. Reads were aligned to the mouse genome (Mus_musculus.GRCm38) using TopHat40 (version 2.0.8; maximum number of mismatches = 2; segment length = 30; maximum multi-hits per read = 25; maximum intron length = 50000). The number of reads that aligned to the predicted coding regions were determined using HTSeq41. Significant differential expression (SDE) was assessed using DEseq. Post-analysis was conducted to avoid over-inflation of fold change. All the values are quantile normalized Transcripts Per Kilobase Million (TPM) and values lower than 10% of the dataset value were replaced with the 10th quantile. Only genes with cutoff >1.2 (min(group1)/max(group2)>1.2, or min(group2)/max(group1)>1.2) and the absolute value of Log Fold Change (LFC)≥ 1 in either of the pairwise comparisons were considered as differentially expressed (DE; 816 genes). All the DE genes in either of the pair-wise comparisons were uploaded into EXPANDER 2.042 for non-supervised clustering using CLICK algorithm43 to find genes with similar patterns of expression and TANGO for GO functional enrichment analysis. Transcriptional regulator network were obtain with iRegulon44 by analyzing genes for GO terms based on CLICK cluster separation. Interaction maps between transcription factors and target genes were made using Cytoscape software45 (v. 3.6.1). RNA-sequencing data are available through the Gene Expression Omnibus database (GEO accession number: GSE146274), as well as the gEAR Portal (https://umgear.org/p?s=afc1b9f8).
RiboTag validation and Gene expression
To validate the specific enrichment of D1-MSN associated genes (Drd1, Chrm4, Tac1), we synthetized 100ng of RiboTagged cDNA (see above for detailed procedure) and non-precipitated RNA (input) using the iScript cDNA synthesis kit (Bio-Rad). cDNA was preamplified using TaqMan assay46. FAM probes for genes of interest (20X TaqMan Gene Expression Assay, Applied Biosystems; see Supplementary Table 1 for primer information) were multiplexed and diluted to a final concentration of 0.2X and amplified for 14 cycles using a PCR thermal cycler (C1000 Touch, Biorad). After amplification, samples were diluted at a 1:10 concentration with TE buffer. qRT-PCR was run with TaqMan Gene expression Master Mix (2X, Applied Biosystems) on a CFX-384 Touch (Bio-Rad) and quantification of mRNA changes was performed using the ΔΔCT method47. For mRNA analysis in dorsal striatum tissue, RNA was extracted using the MicroElute kit (Omega) and cDNA was synthetized from 400ng of RNA with the iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed as described above (See Supplementary Table 1 for primer sequences).
RNA-sequencing data were confirmed in a separate set of RiboTag-processed samples. Following RNA isolation, cDNA was synthetized (iScript cDNA; Bio-Rad). 10ng of cDNA was amplified using the Low Input Kit (NanoString technologies) before being processed with nCounter Master kit (NanoString Technologies) by UMSOM IGS on a custom-made gene expression Code set (see Supplementary Table 1 for primer sequences). Data were analyzed with nSolver Analysis software as in48.
Immunostaining
Mice were perfused with 0.1M PBS and 4% paraformaldehyde and brains were post-fixed for 24h For DREADDs virus validation, 40μm sections were blocked in 3% normal donkey serum and 0.3% Triton X-100 in PBS for 30 min, then incubated in rabbit anti-ds-Red (1:1500; Clontech: #632496) overnight at room temperature. Slices were incubated for 2h in anti-rabbit-Cy3 (1:1000, Jackson Immuno; #111–166-003). Following PBS wash, slices were mounted with Vectashield containing DAPI, and imaged on a confocal microscope. For D1-MSN morphology, 100 μm slices were stained as described in37, 49 using chicken anti-GFP (1:500; Aves Lab, #GFP-1020) and Anti-Chicken Alexa 488 (1:500; Jackson Immuno, #111–545-144). Slices were mounted with Vectashield mounting media.
Neuronal morphology
Sections containing dorsal striatum were sampled from bregma AP: 1.2–0.6mm and Z-stack images were acquired on a confocal microscope (Olympus FV500) at 0.6μm increments using a 40x objective. As described previously36, 37, D1-MSNs were 3D reconstructed using Imaris 8.3 software (Bitplane, Oxford Instruments). 2D image from single MSN were generated for sholl analysis. Concentric ring intersections were determined using the ImageJ sholl analysis plugin50 at 10 μm increments from soma. Spine images were acquired from secondary dendrites with 0.2μm increments Z-stacks using a 60x objective with 2x digital magnification and quantified with Neuron Studio software51. For all morphological analyses, 3–4 cells were averaged per animal. Morphology was blindly assessed.
Statistical analysis
Graphpad Prism 6.0 software was used for statistical analysis. Normality was assessed with Bartlett’s test. ANOVAs and Kruskal-Wallis tests were run for normal and non-normal data respectively. Tukey, Sidak and Dunn’s post-hoc tests were used. Samples were excluded if not detected (molecular analysis) or if they failed Grubbs’ outlier test. Sample sizes were determined from previous studies using mouse behavior, cell-type specific RNA isolation and neuronal morphology36–38. In figure legends: *p≤0.05, **p<0.01, ***p<0.001. All graphs represent mean ± s.e.m. Individual values are plotted to report that variation and variance is similar between groups that are compared. See captions of supplementary figures for supplementary data statistics.
Results
A subset of D1-Cre-flTrkB mice exhibit D1-MSN related repetitive behavior
While the majority of D1-Cre-flTrkB mice showed no stereotypy (further referred to as D1-Cre-flTrkB-NS), behavioral characterization revealed that a subset of both male and female D1-Cre-flTrkB mice (further referred to as D1-Cre-flTrkB-S) exhibited stereotypic behavior (55/381; 14.4%; Supplementary Figure S1a, Supplementary Video 1). D1-Cre-flTrkB-S displayed significantly more rotational behavior: 2-Way RM-ANOVA: Group: F(2;29)=21.42; p<0.0001; Tukey post-hoc: p<0.05 when compared to D1-Cre and D1-Cre-flTrkB-NS from the ages 3 to 8 weeks (Figure 1a). Both clockwise and counter-clockwise circling was observed. D1-Cre-flTrkB-S was the only group to exhibit head tics during that same time period: 2-Way RM-ANOVA: Group: F(2;29)=63.83; p<0.0001; Tukey post-hoc: p<0.01 (Figure 1b). Although both repetitive circling and head tics were present in every stereotypy mouse, high interindividual variability was observed, and no correlation was found between the rotations and the head tics (Supplementary Figure S1k). No differences in rearing behavior were observed between groups (Supplementary Figure S1b) and although knockout mice from both phenotypes exhibited lower grooming time at 3 and 4 weeks, this difference did not persist at later ages (Supplementary Figure S1c). Overall, D1-Cre-flTrkB-S mice displayed increased behavioral activation and locomotion in an open field at both 4 and 8 weeks (Supplementary Figure S1d). This further suggests that circling behavior was unrelated to the test environment. Next we sought to test if ecopipam52 and haloperidol53, two drugs used in stereotypy disorders, could affect repetitive behaviors in our young adult mice. For both drugs, we used a dose that reduced locomotion in D1-Cre-flTrkB-S mice to levels comparable to D1-Cre and D1-Cre-flTrkB-NS mice without eliciting akinesia (Supplementary Figure S1e). Both drugs efficiently reduced circling behavior without altering normal motor behavior in D1-Cre and D1-Cre-flTrkB-NS mice: −50.1% for ecopipam, 2-Way RM-ANOVA: Group x Drug: F(2;12)=11.49; p<0.002, Tukey post-hoc: p<0.001 (Figure 1c) and −64.4% for haloperidol, 2-Way RM-ANOVA: Group: F(2;12)=53.95; p<0.0001, Tukey post-hoc: p<0.001 (Figure 1d). However, head tics were not altered by ecopipam or haloperidol (Supplementary Figure S1f). Balanced activation between direct and indirect pathways controls movement54. The effectiveness of low doses of dopamine antagonists on stereotypy implicates increased activation of the direct pathway in D1-Cre-flTrkB-S mice. To further evaluate this, we infused a Cre-dependent hM4D(Gi) inhibitory DREADDs in the dorsal striatum to transiently inhibit D1-MSN synaptic transmission55 (Figure 1e; Supplementary Figure S1g). Inhibition of D1-positive cells drastically decrease locomotor activity56 but low concentrations of clozapine-N-oxide (CNO) can only partially decrease synaptic transmission in intact neuronal circuits55. Our hypothesis was that if a partial reduction in synaptic transmission could reduce circling behavior without affecting normal movement, this would reveal impaired D1-MSN activity. We used a 0.5mg/kg (i.p.) CNO dose that had no effect on locomotion in an open field (Supplementary Figure S1h). Further, this dose was insufficient to impact rotational behavior in the absence of the hM4D(Gi) receptor (Supplementary Figure S1i). In humans, stereotypic behaviors appear during childhood and persist through adolescence3, 57. We thus tested, in two separate cohorts, the effect of D1-MSN inhibition at both juvenile (4 weeks) and young adult (8 weeks) ages. D1-Cre-flTrkB-S mice receiving CNO displayed a significant 27% decrease in circling behavior relative to pre-CNO vehicle injection at 4 weeks: 2-Way RM-ANOVA: Group x Treatment: F(2,21)=4.87; p=0.02; Sidak post-hoc: p<0.01 (Figure 1f) and a significant 31% decrease relative to pre-CNO vehicle injection at 8 weeks: 2-Way RM-ANOVA: Group x Treatment: F(2,20)=8.71; p=0.002; Sidak post-hoc: p<0.001 (Figure 1g). However, CNO had no effect on head tics at both 4 and 8 weeks (Supplementary Figure S1j). In both D1-Cre and D1-Cre-flTrkB-NS mice, D1-MSN inhibition had no effect on rotations further confirming that our inhibition protocol had no impact on normal motor behavior (Figure 1f, g).
Figure 1:
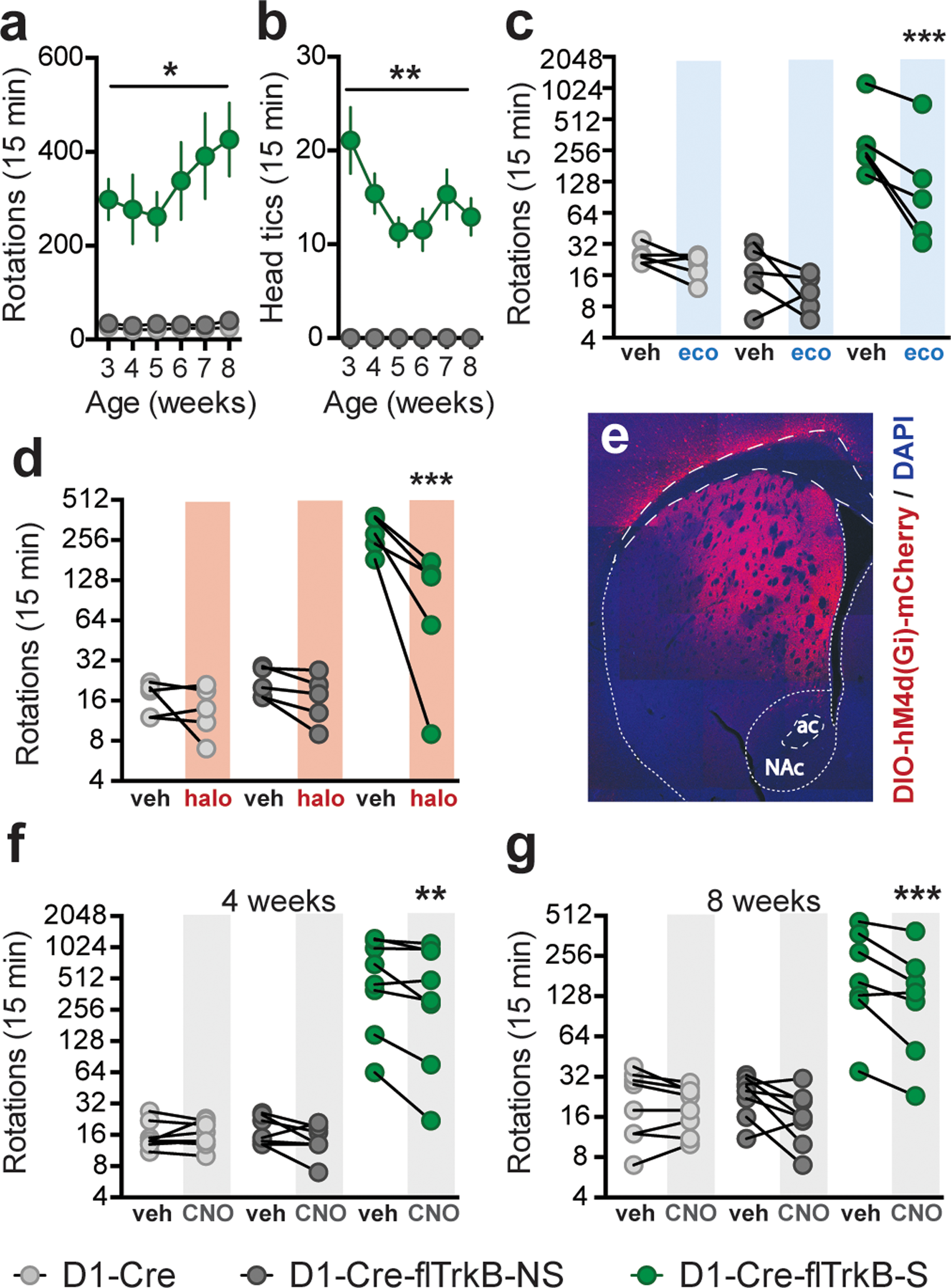
Behavioral and pharmacological characterization of D1-Cre-flTrkB mice: a. Rotational behavior in mice displaying D1-Cre-flTrkB-S, D1-Cre-flTrkB-NS, and D1-Cre control mice from juvenile to young adult ages (*p<0.05); b. Only D1-Cre-flTrkB-S mice display head tics between the age of 3 and 8 weeks (**p<0.01); (a-b) n= 13 D1-Cre-flTrkB-S, 7 D1-Cre-flTrkB-NS, and 12 D1-Cre; c. Rotational behavior following vehicle (veh) and 0.02 mg/kg (sc.) ecopipam (SCH-39166) in 8 week-old mice (***p<0.001); d. Rotational behavior following vehicle (veh) and 0.05 mg/kg (sc.) haloperidol in 8 week-old mice (***p<0.001); (c-d) n=5 in each group; e. Representative image of AAV-DIO-hM4d(Gi)-mCherry virus injection in the dorsal striatum (ac= anterior commissure, NAc= nucleus accumbens); f. Rotational behavior following vehicle (veh) and 0.5 mg/kg (ip.) clozapine-N-oxide (CNO) in 4 week-old mice (**p<0.01), n=8 in each group; g. Rotational behavior following vehicle and 0.5 mg/kg (ip.) CNO in 8 week-old mice (***p<0.001), n= 8, 8, 7 respectively. For ‘Rotations’, please note the scale of the y-axis.
Molecular profiling of striatal D1-MSNs
First, using the RiboTag procedure we demonstrate significant enrichment of D1-MSN-related mRNAs in immunoprecipitated samples relative to input from dorsal striatum (Supplementary Figure S2a), consistent with our previous studies in ventral striatum and other studies in dorsal striatum 36–38, 47, 58–60. Since individual differences in stereotypy behavior were observed between D1-Cre-flTrkB-S vs. D1-Cre-flTrkB-NS mice and stereotypy was partially reduced by chemogenetic inhibition of striatal D1-MSNs, we then probed molecular adaptations in the D1-MSNs. We used D1-Cre-flTrkB- RiboTag (RT)-S, -NS, and D1-Cre-RT control mice, at age 4 weeks when stereotypy behavior is observed and reduced by chemogenetic inhibition of striatal D1-MSNs. Ribosome-associated mRNA was isolated from dorsal striatum D1-MSNs from each group followed by translatome profiling (Figure 2a). Using RNA-sequencing, we identified a total of 14,764 genes expressed in our samples (Supplementary Figure S2b). The full RNA-profiling dataset is available in Supplementary Table 2.
Figure 2:
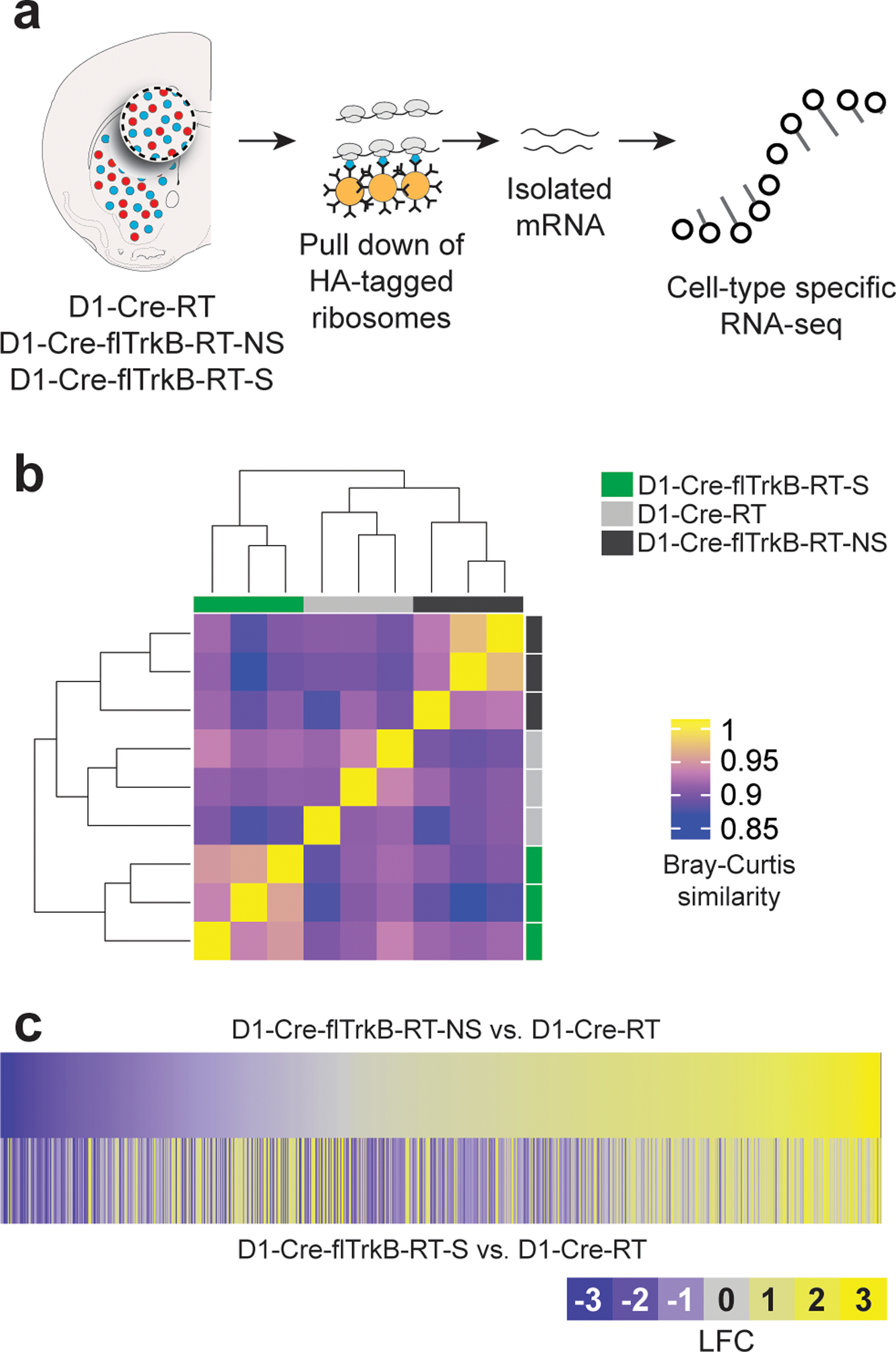
D1-MSNs of mice with stereotypy display distinct translatome profiles: a. Schematic of the RiboTag isolation of D1-MSN mRNA followed by cell-type specific RNA-sequencing; b. Heat map and dendrogram for unbiased hierarchical clustering (scale= Bray-Curtis similarity values); c. Heatmaps of the 816 expressed genes displaying differential gene expression for D1-Cre-flTrkB-RT-NS and D1-Cre-flTrkB-RT-S mice compared to D1-Cre-RT (LFC= Log fold change).
Interestingly, the overall translatome profile of D1-MSNs in D1-Cre-flTrkB-RT-NS mice was more similar to D1-Cre-RT than to D1-Cre-flTrkB-RT-S mice (Figure 2b). However, differences in gene expression between D1-Cre-flTrkB-RT-NS and D1-Cre-flTrkB-RT-S emerged (Supplementary Figure S2b), which may reflect that repetitive behaviors lead to different impact on gene expression level. Overall, we found 816 genes with significantly different expression (Figure 2c; Supplementary Table 3). Based on non-supervised clustering algorithm (CLICK algorithm), all the differentially expressed genes were grouped to 4 clusters (overall average homogeneity=0.807; overall average separation=−0.336): 1: lower expression in D1-Cre-flTrkB-RT-S mice (338 genes); 2: higher expression in D1-Cre-flTrkB-RT-S (245 genes); 3: lower expression in D1-Cre-RT (122 genes); 4: higher expression in D1-Cre-RT (111 genes) (Figure 3a; Supplementary Table 3). Ontology (GO) analysis was further conducted of these 4 clusters. Cluster 1 presented the most GO terms (i.e. 124) compared to Cluster 2 (53 terms), Cluster 3 (9 terms) and Cluster 4 (36 terms). Clusters showing changes occurring in both D1-Cre-flTrkB-RT-NS and D1-Cre-flTrkB-RT-S were related to ‘Cell surface receptor signaling pathway’ (Cluster 3) or ‘Regulation of signal transduction’ (Cluster 4), ‘Regulation of kinase activity’ (Cluster 4), ‘Negative regulation of MAPK cascade’ (Cluster 4), ‘Receptor binding’ (Cluster 4) (Supplementary Figure S2c, d). Among the 124 enriched GO categories in Cluster 1 (decreased expression in D1-Cre-flTrkB-RT-S), 43 were related to neuronal, dendritic and synaptic development as well as receptor and/or transporter activity and signaling. Of particular interest, the GO term ‘Regulation of signal transduction’ (GO:0009966; 74 genes; 22% frequency in set; p=2.42 E-14), ‘Cell projection organization’ (GO:0030030; 34 genes; 10% frequency in set; p=6.62 E-9), ‘Cell projection morphogenesis’ (GO:0048858; 27 genes; 8% frequency in set; p=6.10 E-13), ‘Regulation of neuron projection development’ (GO:0010975; 23 genes; 6.8% frequency in set; p=3.02 E-8) and ‘Regulation of synapse structure or activity’ (GO:0050803; 12 genes; 3.6% frequency in set; p=9.82 E-8) were significantly changed in D1-Cre-flTrkB-RT-S mice (Figure 3b; Supplementary Figure S3a). In Cluster 2 (increased expression in D1-Cre-flTrkB-RT-S), one GO term with significant enrichment particularly reflected the phenotype present in our stereotypy mice: ‘Regulation of locomotion’ (GO:004001227; 29 genes; 12% frequency in set; p=1.78 E-12; Figure 3b; Supplementary Figure S3a). Detailed analysis of gene expression for ‘Cell projection morphogenesis’ (GO:0048858) revealed that, out of 27 genes showing differential expression in our dataset, 11 were significantly different in D1-Cre-flTrkB-RT-S compared to D1-Cre-RT and 25 compared to D1-Cre-flTrkB-RT-NS (Figure 3c; Supplementary Table 3). Similarly for the other GO terms identified here, D1-Cre-flTrkB-RT-S showed significant differences for 82 to 94% of genes from Cluster 1 and for 62% of genes from Cluster 2 when compared to D1-Cre-flTrkB-RT-NS (Supplementary Figure S2c; Supplementary Table 3). Together, the GO analysis suggested that D1-MSNs from mice with stereotypy display altered gene expression for molecules in dendritic and synaptic pathways.
Figure 3:
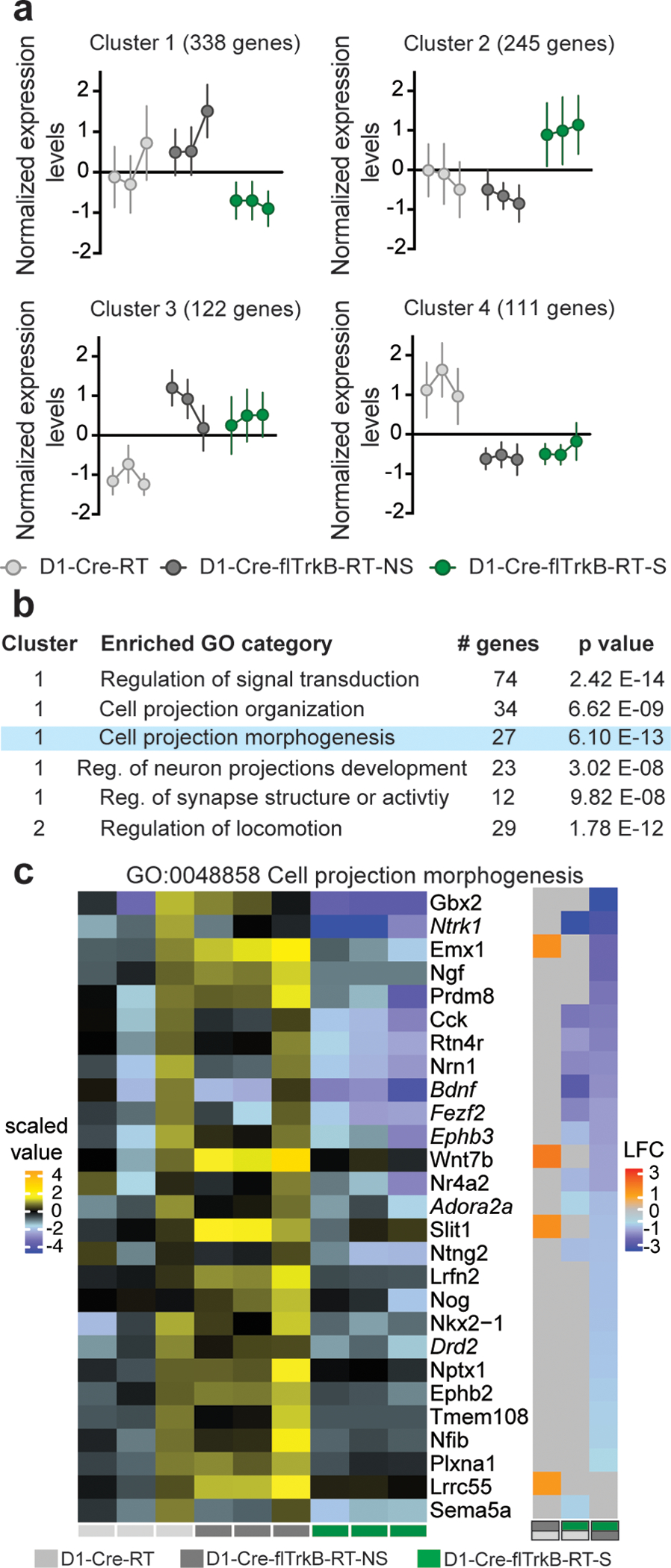
Gene ontology analysis reveals alterations in morphology-related genes in D1-MSNs of mice with stereotypy: a. Non-supervised clustering algorithm (CLICK algorithm) identified 4 clusters based on gene expression: 1: low expression (338 genes) and 2: high expression (245 genes) in mice with stereotypy, 3: low expression (122 genes) and 4: high expression (111 genes) in control mice; b. List of the top GO terms related to neuronal morphology and motor function. Blue highlight denotes the GO term detailed in the following panel; c. Left: Heat map for the biological replicates for the GO term “Cell projection morphogenesis”, Right: Pairwise comparisons for the corresponding genes in the 3 conditions: D1-Cre-flTrkB-RT-S showed decreased expression for 25 genes when compared to D1-Cre-flTrkB-RT-NS and 11 genes when compared to D1-Cre-RT mice (LFC= Log fold change). Light grey (D1-Cre-RT), dark grey (D1-Cre-flTrkB-RT-NS) and green (D1-Cre-flTrkB-RT-S) labels below the heat maps indicate groups and group comparisons. Genes in italic were selected for multiplexed mRNA measurements (see Figure 4).
To further identify whether transcriptional regulons are also impacted by stereotypy, we conducted transcriptional regulator network analysis using iRegulon44. Three of the GO terms we identified appeared to share a common transcriptional regulator. The transcription factor Nr2f1 shares a binding motif with: 5 genes (42%) of the ‘Regulation of synapse structure or activity’ term: motif similarity: FDR<8.136 E-4; 22 genes (30%) of the ‘Regulation of signal transduction’ term: motif similarity: FDR<2.502 E-4; 19 genes (66%) of the ‘Regulation of locomotion’ term: motif similarity: FDR<1.122 E-4 (Figure 4a). Based on this list as well as genes involved in ‘Cell projection morphogenesis’ (Figure 3c), we selected several target genes and measured multiplexed mRNA levels in D1-MSNs at both 4 and 8 weeks using NanoString. The expression of a subset of these genes was not detected at 4 weeks but reached detection threshold at 8 weeks. Additionally, many genes showed trends of altered expression at 4 weeks and reached significance at 8 weeks (Figure 4b). To account for this evolution in gene expression, we reported both significant (p≤0.05) and trending (p≤0.08) results. At 4 weeks, the expression of genes such as Macf1, Mme, Ndnf, S1pr1 and Strn was altered in D1-Cre-flTrkB-RT-S mice (Figure 4b, Supplementary Table 4). Alterations in gene expression continued at 8 weeks for over 60% of the genes from our list in D1-Cre-flTrkB-RT-S mice. Overall, most genes showed decreased levels, which is consistent with Cluster 1 profile, and genes associated with ‘Regulation of locomotion’ were upregulated following Cluster 2 profile (Figure 4b; Supplementary Table 4). Interestingly the expression of Nr2f1, which regulates the expression of numerous genes in our list, was significantly reduced in D1-Cre-flTrkB-RT-S mice at 8 weeks. Finally at both time points, the expression of Ntrk2 (TrkB) was drastically decreased in both D1-Cre-flTrkB-RT-NS and D1-Cre-flTrkB-RT-S mice confirming both the effective knockout of TrkB in our mice as well as the cell-type specificity of our RiboTag samples (Figure 4b; Supplementary Table 4). Of note, we also observed distinct patterns of expression of D2-MSN enriched genes: Adora2a, Drd2 and Penk mRNA were upregulated in non-stereotypy mice and down-regulated in stereotypy mice at 8 weeks (Figure 4b; Supplementary Table 4). Penk was also reduced in stereotypy mice at 4 weeks. When assessed in total dorsal striatum tissue, Adora2a and Drd2 were increased in both non-stereotypy and/or stereotypy mice. Penk levels were unchanged in total tissue (Supplementary Figure S3b). While D1-MSN enriched genes did not display distinct patterns of expression, we observed significant changes in two of these genes: Chrm4 was upregulated in D1-Cre-flTrkB-S at 4 weeks and Pdyn was upregulated in D1-Cre-fltrkB-NS at 8 weeks in D1-MSNs using RiboTag tissue (Figure 4b; Supplementary Table 4).
Figure 4.
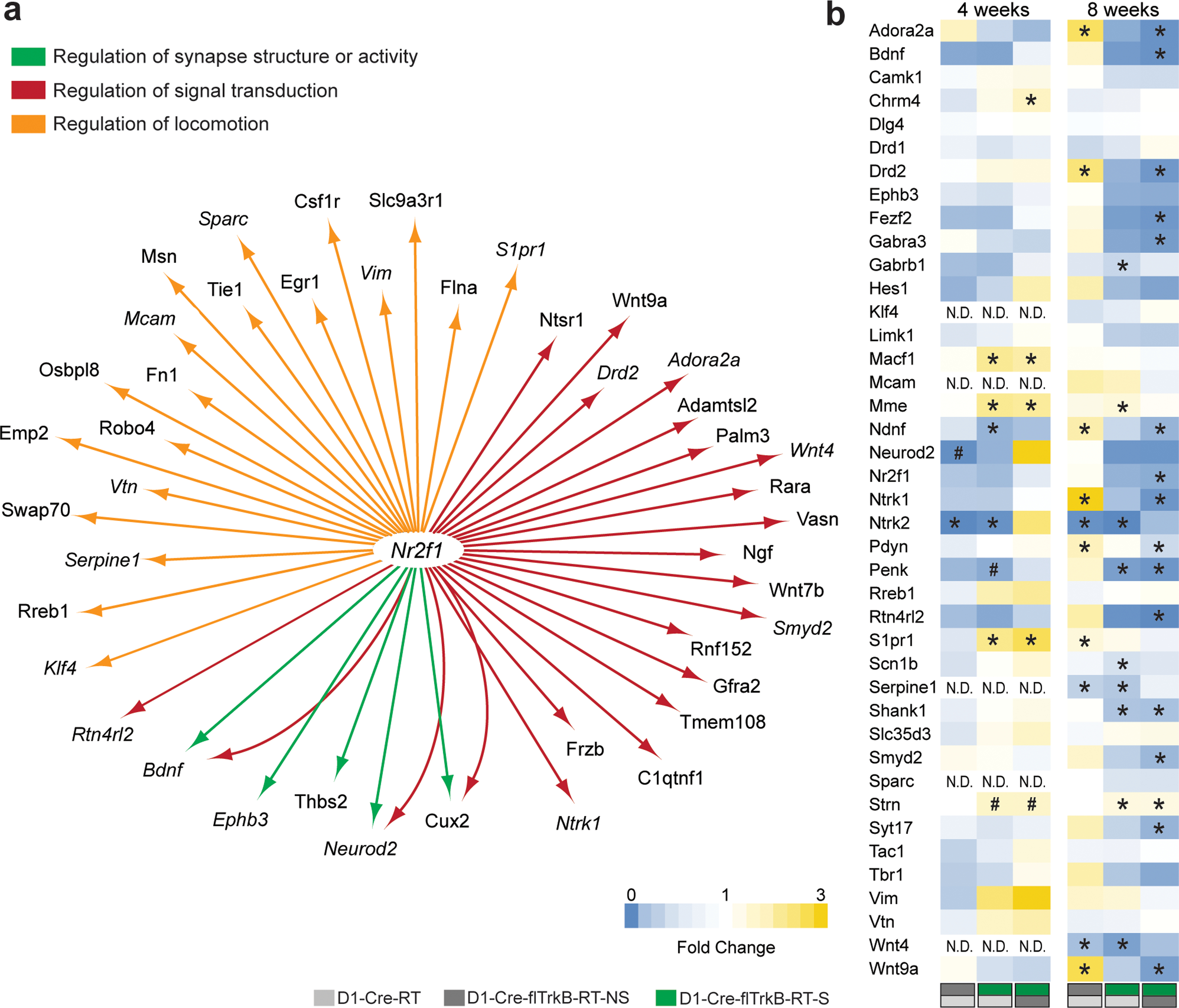
Multiplexed mRNA analysis of D1-MSNs. a. Transcriptional regulator network analysis shows that Nr2f1 is a common regulator for multiple morphology-related genes. Genes in italic were selected for multiplexed mRNA measurements; b. Multiplexed mRNA analysis (NanoString) on both juvenile and young adult mice (# p≤0.08, *p≤0.05). Light grey (D1-Cre-RT), dark grey (D1-Cre-flTrkB-RT-NS) and green (D1-Cre-flTrkB-RT-S) labels below the heat maps indicate groups and group comparisons.
Stereotypy mice exhibit D1-MSN atrophy
The translatome analyses pointed toward morphological alterations in D1-MSNs of mice with stereotypy behavior. To assess whether these impairments were present in early stages, we sparsely labeled dorsal striatum D1-MSNs with AAV-DIO-eYFP and assessed different morphological parameters at both 4 and 8 weeks (Figure 5a,h). At 4 weeks, the number of branch points was not significantly altered: 1-way ANOVA: F(2;12)=1.53; p=0.26 (Figure 5b). Sholl analysis revealed however decreased D1-MSN complexity: 2-way ANOVA Group: F(2;252)=25.11; p<0.0001; Tukey post-hoc: p<0.05 (Figure 5c) and a reduced dendritic length: 1-way ANOVA: F(2;12)= 4.65; p= 0.03; Tukey post-hoc: p<0.05, in D1-Cre-flTrkB-S mice only (Figure 5d). However, soma volume was unchanged: 1-way ANOVA: F(2;12)=0.70; p=0.52, suggesting that morphological alterations were not due to general neuronal reduction (Figure 5e). Since the GO analysis suggested altered synaptic structure, we measured dendritic spines in all groups (Figure 5f). Spine density was significantly reduced in D1-Cre-flTrkB-S mice: 1-way ANOVA: F(2;12)=8.42; p=0.005; Tukey post-hoc: p≤0.05 (Figure 5g). Stereotypy mice displayed reduced thin spine density compared to the other groups (Supplementary Figure S4a).
Figure 5:
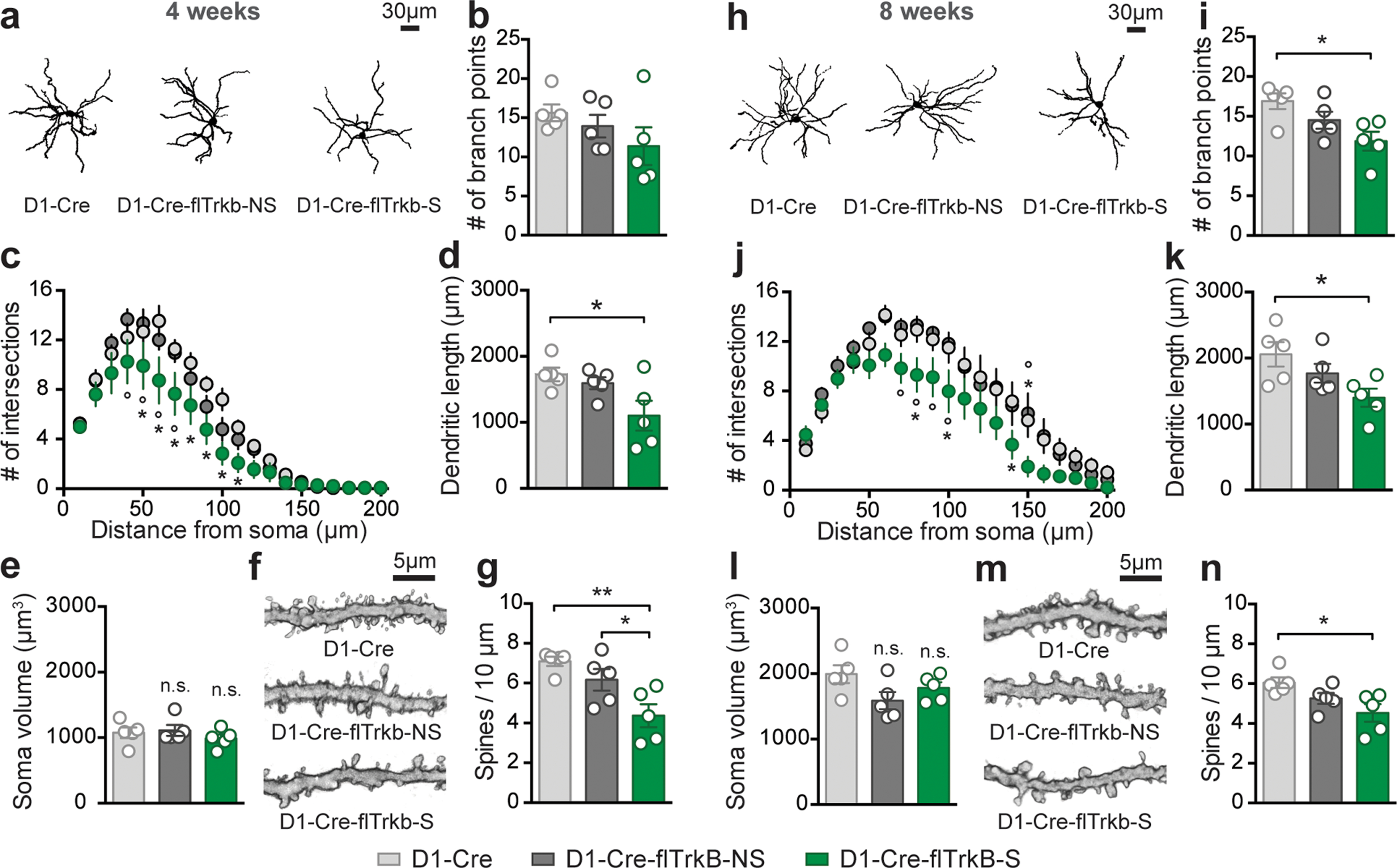
Mice with stereotypy display D1-MSN dendritic atrophy and reduced dendritic spines at both 4 and 8 weeks: a. Representative images of dorsal striatum D1-MSN from each group at 4 weeks; b. Number of branch points is not significantly changed at 4 weeks; c. Decreased neuronal complexity (Sholl analysis, *p<0.05 from D1-Cre and °p<0.05 from D1-Cre-flTrkB-NS); d. Dendritic length is reduced in D1-Cre-flTrkB-RT-S mice (*p<0.05); e. Soma volume is unchanged in both D1-Cre-flTrkB-NS and D1-Cre-flTrkB-S mice; f. Representative images of D1-MSN dendritic spines from each group at 4 weeks; g. Decreased spine density in D1-Cre-flTrkB-S mice (*p≤0.05). h. Representative images of dorsal striatum D1-MSN from each group at 8 weeks; i. Number of branch points is significantly reduced in Cre-flTrkB-RT-S mice (*p<0.05); j. Decreased neuronal complexity (Sholl analysis, *p<0.05 from D1-Cre and °p<0.05 from D1-Cre-flTrkB-NS); k. Dendritic length is reduced in D1-Cre-flTrkB-RT-S mice (*p<0.05); l. Soma volume is unchanged in both D1-Cre-flTrkB-NS and D1-Cre-flTrkB-S mice; m. Representative images of D1-MSN dendritic spines from each group at 8 weeks; n. Decreased spine density in D1-Cre-flTrkB-S mice (*p≤0.05). For all graphs, n= 5 mice/group, 3–4 cells/mouse.
At 8 weeks, we observed significantly decreased number of branch points: 1-way ANOVA: F(2;12)=5.31; p=0.02; Tukey post-hoc: p<0.05 (Figure 5i) and reduction in D1-MSN complexity in D1-Cre-flTrkB-S mice: 2-way ANOVA Group: F(2;252)=29.65; p<0.0001; Tukey post-hoc: p<0.05 (Figure 5j). Similarly, D1-Cre-flTrkB-S mice have significantly shorter dendrites: 1-way ANOVA: F(2;12)=4.48; p=0.03; Tukey post-hoc: p<0.05 (Figure 5k). Again, soma volume was unchanged: 1-way ANOVA: F(2;12)=2.83; p=0.1, confirming morphological alterations were not due to general neuronal reduction (Figure 5l). Spine density was significantly reduced in D1-Cre-flTrkB-S mice: 1-way ANOVA: F(2;12)=5.07; p=0.03; Tukey post-hoc: p<0.05 (Figure 5m,n). Thin spine density in both D1-Cre-flTrkB-NS and D1-Cre-flTrkB-S was decreased compared to D1-Cre controls (Supplementary Figure S4b). Thus as indicated by the GO analysis, we confirmed that D1-Cre-flTrkB-S mice displayed altered D1-MSN morphology.
Discussion
In this study we showed that TrkB knockout from D1-expressing cells results in stereotypic rotational behavior and head tics in a subset of mice. Dopamine antagonists used in patients with stereotypy reduced repetitive circling and chemogenetic inhibition of striatal D1-MSNs supports a role for these neurons in mediating the stereotypy behavior. By comparing the cell-type specific translatome in mice with a similar genotype but different phenotype, we showed that D1-MSNs from mice with stereotypy displayed alterations in ribosome-associated mRNAs for gene networks including those regulating neuronal morphology and locomotion. Finally, we observed dendritic atrophy and reduced dendritic spines in D1-MSNs of D1-Cre-flTrkB-S mice, which is consistent with alterations in neuronal morphology related gene networks.
Stereotypies are associated with increased direct motor pathway activity and treatments inhibiting direct pathway activity or disinhibiting indirect pathway activity are routinely used in patients1, 3, 8, 9, 52. Here, both ecopipam and haloperidol reduced circling behavior at doses that did not affect normal locomotion. This implicates alterations in basal ganglia motor pathways in the mice displaying stereotypy. Moreover in humans, impairments in BDNF-TrkB signaling are associated with several stereotypy disorders24–26. We found that only a subset of mice lacking the TrkB receptor in direct-pathway neurons exhibit repetitive circling behavior. This suggests that TrkB deletion was necessary but not sufficient to support repetitive behavior. To provide further insight into the molecular differences between both phenotypes, we interrogated the translatome of dorsal striatum D1-MSNs in juvenile mice using cell type-specific RNA-sequencing. We focused our analysis on this time point because stereotypies appear during childhood and/or adolescence in humans3, 20. First, unbiased hierarchical clustering performed on the entire dataset revealed that similarities in gene expression pattern were not shared by mice with similar genotype but rather by similar phenotype. This suggested that although D1-MSNs of D1-Cre-flTrkB mice undeniably share similarities in gene expression related to TrkB knockout such as kinase activity or receptors signaling pathways (see Cluster 3 and 4 of Figure 3a and Supplementary Figure S2c, d), transcriptional alterations in D1-MSNs associated with repetitive behavior were greater than the effect of gene deletion alone. This was supported by the non-supervised clustering analysis (CLICK algorithm), revealing that 71% of the post-filtrated genes with significant changes belonged to the mice with stereotypy.
To obtain better insight into the alterations present in D1-MSNs of mice with stereotypy, we conducted a gene ontology analysis on down- or up-regulated genes in mice with repetitive behavior. Interestingly, 35% of the down-regulated genes were related to dendritic and synaptic development as well as receptor and/or transporter activity and signaling. Detailed analysis of genes belonging to these GO categories allowed us to generate a list of genes that can potentially support cellular adaptations underlying repetitive behavior as well as a common transcriptional regulator. Multiplexed mRNA analysis of 4-week-old animals confirmed key genes coding for cytoskeleton, neurite outgrowth, spines or synapse formation such as Macf161, Mme62, Ndnf63, Strn64 or S1pr165 had altered expression in mice with repetitive behavior. These genes have fundamental roles throughout brain development. Impaired expression, observed here at juvenile age, could support altered D1-MSN functioning leading to repetitive behaviors. While some transcriptional changes were already present at 4 weeks, more reached statistical significance at 8 weeks. This might reflect neural development processes in mice during which neurotransmission, synaptic function and neuronal networks undergo maturation and pruning stages between PND 25 and PND 49 to reach a stable, fully formed, adult brain at PND >6066. Additionally, the establishment of transcriptional differences at 8 weeks in mice with stereotypy could be due to a gradual decreased expression of Nr2f1 (COUP-TF1), which was significantly decreased at 8 weeks. Nr2f1 was identified as a potential common transcriptional regulator for multiple genes in our dataset and is involved in neurite outgrowth67. Impaired transcriptional regulation could thus have a broad impact on neuronal morphology. Altogether, our mRNA data confirmed a wide range of transcriptional alterations in stereotypy mice and suggested that differential expression of morphology-related genes persist until young-adult stages.
Based on these translatome findings, we evaluated D1-MSN morphology in both juvenile and young adult mice. In mice with stereotypy, we observed lower dendritic complexity and dendritic length at 4 weeks. This suggests impaired neuronal morphology occurs early in development. In young adults, these alterations became more apparent. Again, this is consistent with our gene analysis in which important gene changes were seen at 4 weeks but most of the changes were significant at 8 weeks. Striatal neurons integrate cortical and subcortical information to provide error detection monitoring in order to generate harmonious motor behaviors6, 68. D1-MSN dendritic atrophy could result in altered functional connectivity and aberrant signal processing supporting abnormal behavior69–71. This is further supported by the reduced expression of the spine-related genes Shank1 and Strn, both associated with impaired circuitry in ASD64, 72 as well as reduced thin spine density at both 4 and 8 weeks. Reduced excitatory input can induce changes in intrinsic excitability at the postsynaptic level to maintain homeostasis in circuit activity73. Our group has recently described this mechanism for stress-induced dendritic atrophy in ventral striatal D1-MSNs37, 38, 74. Further, our previous studies demonstrate reduced inhibition of ventral striatal D1-MSNs in D1-Cre-flTrkB mice75. In mice with stereotypy, reduced dendritic complexity could thus lead to a reduction in inhibitory synapses or a homeostatic increase in excitability in dorsal striatal D1-MSNs. This would favor an increased up-state in which D1-MSN activation would be triggered by stimuli that are insufficient to activate these neurons in controls or non-stereotypy animals. This is consistent with our chemogenetic inhibition of D1-MSNs, at a low CNO dose, reducing circling behavior in stereotypy mice only. This is also in line with a decrease in the voltage-gated sodium channel subunit gene, Scn1b, in D1-MSNs of stereotypy mice, since loss of function of this molecule can lead to the development of hyperexcitability in the brain76. It is however difficult to untangle whether increased D1-MSN excitability results from homeostatic scaling or abnormal neuronal development. Future experiments comparing D1-MSN excitability in both mice with and without stereotypy to D1-Cre control mice, at different ages, will be important to better characterize these mechanisms and how they emerge.
Along with alterations in morphology and locomotor related molecules we observed changes in D2-MSN enriched genes: Adora2a, Drd2, and Penk. mRNA for these genes was significantly upregulated in D1-MSNs of non-stereotypy mice and downregulated in stereotypy mice at 8 weeks. Penk was also significantly reduced in stereotypy mice at 4 weeks. This could reflect a greater proportion of MSNs expressing both D1 and D2 enriched molecules in non-stereotypy mice. Both Drd2 and Adora2a were upregulated in total dorsal striatum tissue of both stereotypy and non-stereotypy mice at 8 weeks, which could be a consequence of TrkB knockout from D1-MSNs. Indeed, BDNF released into the striatum would potentially be more readily available to bind to TrkB receptors on D2-MSNs, resulting in increased D2- and A2A-receptor expression77, 78. A previous study showed that enhancing Drd2 expression in striatum leads to increased presence of D1-MSN bridging collaterals to the globus pallidus externus (GPe) and enhanced inhibition of GPe neurons79. A similar mechanism could be occurring here with enhanced Drd2 in D1-MSNs of non-stereotypy mice. Given the morphological and physiological differences between both MSN subtypes80, 81, this could contribute to stabilized activity between the direct and indirect pathways in these mice. While patterns of expression in groups of D1-MSN enriched genes was not observed between stereotypy and non-stereotypy mice we did observe an increase of the striosome D1-MSN enriched gene, Pdyn- the precursor for the dynorphin peptide82, in D1-MSNs of non-stereotypy mice at 8 weeks. Further, Pdyn was reduced in D1-MSNs of stereotypy mice when compared to non-stereotypy mice. In contrast mRNA for the matrix D1-MSN enriched gene, Chrm4 83 - the gene for acetylcholine muscarinic receptor 4, is enhanced in D1-MSNs of stereotypy mice at 4 weeks. The differential regulation of Pdyn vs. Chrm4 could reflect an imbalance in striosome vs. matrix function, which is implicated in stereotypy behavior2. Additionally, the responsiveness of stereotypy mice to the low dose of CNO, acting on the DREADD hM4D(Gi), derived from Chrm4, could be a consequence of enhanced sensitivity of M4 signaling that might occur through upregulation of endogenous Chrm4.
Finally, head tics were not impacted by chemogenetic inhibition or dopamine receptor antagonism. This suggests that these movements are independent from striatal dopaminergic function. For instance, studies in TS have highlighted impairments in GABAergic function in different regions of the basal ganglia84. Reduced GABAA binding in the thalamus is associated with tics and thalamic deep brain stimulation showed efficacy in reducing tic severity in TS patients84, 85. Future experiments exploring impairments in different structures of the basal ganglia will be needed to better understand the underpinning of head tics in our mice.
In summary, our study reveals deletion of TrkB in D1 expressing neurons leads to individual differences in repetitive behavior, that are associated with altered D1-MSN translatome and dendritic morphology. These cellular alterations potentially disrupt D1-MSN output activity to promote repetitive behavior. How the differences in D1-MSN translatomes between D1-Cre-flTrkB mice with and without stereotypy emerge remains unclear and could involve epigenetic changes associated with impaired BDNF-TrkB signaling25 that may either aggravate or compensate for the effect of TrkB knockout. Additionally, factors such as prenatal stress can affect neurodevelopment and have been associated with ASD or hyperactivity and altered striatal dopamine receptor expression86, 87. Insight into the distinct molecular and cellular adaptations underlying stereotypy or lack of stereotypy could uncover neural mechanisms of repetitive behavior relevant to many disorders such as TS, OCD and ASD. Further, understanding mechanisms that prevent these behaviors, which may be occurring in non-stereotypy mice, has relevance for combating stereotypy behavior in brain disorders.
Supplementary Material
Acknowledgments
This work was funded by Tourette Syndrome Association and NIH R01DA038613 (MKL), R01DC013817 (RH) and Association Française du Syndrôme de Gilles de la Tourette (ME). The authors would like to thank Sunayana Mitra, Maggie S Mattern, Victoria Rhodes, Katherine Duarte and Heather Brewer-Scotti for their technical help.
Footnotes
The authors declare that they have no conflict of interest.
References
- 1.Peter Z, Oliphant ME, Fernandez TV. Motor Stereotypies: A Pathophysiological Review. Frontiers in neuroscience 2017; 11: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canales JJ, Graybiel AM. A measure of striatal function predicts motor stereotypy. Nature neuroscience 2000; 3(4): 377–383. [DOI] [PubMed] [Google Scholar]
- 3.Albin RL, Mink JW. Recent advances in Tourette syndrome research. Trends in neurosciences 2006; 29(3): 175–182. [DOI] [PubMed] [Google Scholar]
- 4.Saka E, Graybiel AM. Pathophysiology of Tourette’s syndrome: striatal pathways revisited. Brain & development 2003; 25Suppl 1: S15–19. [DOI] [PubMed] [Google Scholar]
- 5.Maia TV, Frank MJ. From reinforcement learning models to psychiatric and neurological disorders. Nature neuroscience 2011; 14(2): 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guehl D, Benazzouz A, Aouizerate B, Cuny E, Rotge JY, Rougier A et al. Neuronal correlates of obsessions in the caudate nucleus. Biological psychiatry 2008; 63(6): 557–562. [DOI] [PubMed] [Google Scholar]
- 7.Berridge KC, Aldridge JW. Super-stereotypy I: enhancement of a complex movement sequence by systemic dopamine D1 agonists. Synapse 2000; 37(3): 194–204. [DOI] [PubMed] [Google Scholar]
- 8.Chartoff EH, Marck BT, Matsumoto AM, Dorsa DM, Palmiter RD. Induction of stereotypy in dopamine-deficient mice requires striatal D1 receptor activation. Proceedings of the National Academy of Sciences of the United States of America 2001; 98(18): 10451–10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Kim H, Kim JE, Park JY, Choi J, Lee JE et al. Excessive D1 Dopamine Receptor Activation in the Dorsal Striatum Promotes Autistic-Like Behaviors. Molecular neurobiology 2018; 55(7): 5658–5671. [DOI] [PubMed] [Google Scholar]
- 10.Singer HS, Butler IJ, Tune LE, Seifert WE Jr., Coyle JT. Dopaminergic dsyfunction in Tourette syndrome. Annals of neurology 1982; 12(4): 361–366. [DOI] [PubMed] [Google Scholar]
- 11.Olver JS, O’Keefe G, Jones GR, Burrows GD, Tochon-Danguy HJ, Ackermann U et al. Dopamine D1 receptor binding in the striatum of patients with obsessive-compulsive disorder. Journal of affective disorders 2009; 114(1–3): 321–326. [DOI] [PubMed] [Google Scholar]
- 12.Visser-Vandewalle V DBS in tourette syndrome: rationale, current status and future prospects. Acta neurochirurgica Supplement 2007; 97(Pt 2): 215–222. [DOI] [PubMed] [Google Scholar]
- 13.Zuchner S, Wendland JR, Ashley-Koch AE, Collins AL, Tran-Viet KN, Quinn K et al. Multiple rare SAPAP3 missense variants in trichotillomania and OCD. Molecular psychiatry 2009; 14(1): 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nature genetics 2007; 39(1): 25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 2005; 310(5746): 317–320. [DOI] [PubMed] [Google Scholar]
- 16.Corbit VL, Manning EE, Gittis AH, Ahmari SE. Strengthened inputs from secondary motor cortex to striatum in a mouse model of compulsive behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK et al. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 2014; 158(1): 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennington JB, Coppola G, Kataoka-Sasaki Y, Fernandez TV, Palejev D, Li Y et al. Transcriptome Analysis of the Human Striatum in Tourette Syndrome. Biological psychiatry 2016; 79(5): 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisboa BCG, Oliveira KC, Tahira AC, Barbosa AR, Feltrin AS, Gouveia G et al. Initial findings of striatum tripartite model in OCD brain samples based on transcriptome analysis. Scientific reports 2019; 9(1): 3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter AS, Pollock RA. Obsessive compulsive disorder in childhood. Current opinion in pediatrics 2000; 12(4): 325–330. [DOI] [PubMed] [Google Scholar]
- 21.American-Psychiatry-Association. Diagnostic and Statistical Manual of Mental Disorders, 5th edition. American Psychiatry Publishing: Arlington, VA, 2013. [Google Scholar]
- 22.Altar CA, Cai N, Bliven T, Juhasz M, Conner JM, Acheson AL et al. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature 1997; 389(6653): 856–860. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Yui D, Luikart BW, McKay RM, Li Y, Rubenstein JL et al. Conditional ablation of brain-derived neurotrophic factor-TrkB signaling impairs striatal neuron development. Proceedings of the National Academy of Sciences of the United States of America 2012; 109(38): 15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alonso P, Gratacos M, Menchon JM, Saiz-Ruiz J, Segalas C, Baca-Garcia E et al. Extensive genotyping of the BDNF and NTRK2 genes define protective haplotypes against obsessive-compulsive disorder. Biological psychiatry 2008; 63(6): 619–628. [DOI] [PubMed] [Google Scholar]
- 25.Liu DY, Shen XM, Yuan FF, Guo OY, Zhong Y, Chen JG et al. The Physiology of BDNF and Its Relationship with ADHD. Molecular neurobiology 2015; 52(3): 1467–1476. [DOI] [PubMed] [Google Scholar]
- 26.Liu S, Cui J, Niu Z, Yi M, Zhang X, Che F et al. Do obsessive-compulsive disorder and Tourette syndrome share a common susceptibility gene? An association study of the BDNF Val66Met polymorphism in the Chinese Han population. The world journal of biological psychiatry : the official journal of the World Federation of Societies of Biological Psychiatry 2015; 16(8): 602–609. [DOI] [PubMed] [Google Scholar]
- 27.Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. The Journal of neuroscience : the official journal of the Society for Neuroscience 2004; 24(17): 4250–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baydyuk M, Xu B. BDNF signaling and survival of striatal neurons. Frontiers in cellular neuroscience 2014; 8: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besusso D, Geibel M, Kramer D, Schneider T, Pendolino V, Picconi B et al. BDNF-TrkB signaling in striatopallidal neurons controls inhibition of locomotor behavior. Nature communications 2013; 4: 2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plotkin JL, Day M, Peterson JD, Xie Z, Kress GJ, Rafalovich I et al. Impaired TrkB receptor signaling underlies corticostriatal dysfunction in Huntington’s disease. Neuron 2014; 83(1): 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. The Journal of neuroscience : the official journal of the Society for Neuroscience 2007; 27(37): 9817–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luikart BW, Nef S, Shipman T, Parada LF. In vivo role of truncated trkb receptors during sensory ganglion neurogenesis. Neuroscience 2003; 117(4): 847–858. [DOI] [PubMed] [Google Scholar]
- 33.Lobo MK, Covington HE 3rd, Chaudhury D, Friedman AK, Sun H, Damez-Werno D et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science 2010; 330(6002): 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America 2009; 106(33): 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engeln M, Bastide MF, Toulme E, Dehay B, Bourdenx M, Doudnikoff E et al. Selective Inactivation of Striatal FosB/DeltaFosB-Expressing Neurons Alleviates L-DOPA-Induced Dyskinesia. Biological psychiatry 2016; 79(5): 354–361. [DOI] [PubMed] [Google Scholar]
- 36.Chandra R, Engeln M, Schiefer C, Patton MH, Martin JA, Werner CT et al. Drp1 Mitochondrial Fission in D1 Neurons Mediates Behavioral and Cellular Plasticity during Early Cocaine Abstinence. Neuron 2017; 96(6): 1327–1341 e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox ME, Chandra R, Menken MS, Larkin EJ, Nam H, Engeln M et al. Dendritic remodeling of D1 neurons by RhoA/Rho-kinase mediates depression-like behavior. Molecular psychiatry 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francis TC, Chandra R, Gaynor A, Konkalmatt P, Metzbower SR, Evans B et al. Molecular basis of dendritic atrophy and activity in stress susceptibility. Molecular psychiatry 2017; 22(11): 1512–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matern MS, Beirl A, Ogawa Y, Song Y, Paladugu N, Kindt KS et al. Transcriptomic Profiling of Zebrafish Hair Cells Using RiboTag. Frontiers in cell and developmental biology 2018; 6: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 2013; 14(4): R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology 2010; 11(10): R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R et al. EXPANDER--an integrative program suite for microarray data analysis. BMC bioinformatics 2005; 6: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharan R, Shamir R. CLICK: a clustering algorithm with applications to gene expression analysis. Proceedings International Conference on Intelligent Systems for Molecular Biology 2000; 8: 307–316. [PubMed] [Google Scholar]
- 44.Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS computational biology 2014; 10(7): e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 2003; 13(11): 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu QR, Rubio FJ, Bossert JM, Marchant NJ, Fanous S, Hou X et al. Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from a single rat dorsal striatum using fluorescence-activated cell sorting (FACS). Journal of neurochemistry 2014; 128(1): 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandra R, Francis TC, Konkalmatt P, Amgalan A, Gancarz AM, Dietz DM et al. Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. The Journal of neuroscience : the official journal of the Society for Neuroscience 2015; 35(20): 7927–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chessum L, Matern MS, Kelly MC, Johnson SL, Ogawa Y, Milon B et al. Helios is a key transcriptional regulator of outer hair cell maturation. Nature 2018; 563(7733): 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engeln M, Mitra S, Chandra R, Gyawali U, Fox ME, Dietz DM et al. Sex specific role for Egr3 in nucleus accumbens D2-medium spiny neurons following long term abstinence from cocaine self-administration. Biological psychiatry 2019; 10.1016/j.biopsych.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ et al. Neuronal morphometry directly from bitmap images. Nature methods 2014; 11(10): 982–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PloS one 2008; 3(4): e1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert DL, Budman CL, Singer HS, Kurlan R, Chipkin RE. A D1 receptor antagonist, ecopipam, for treatment of tics in Tourette syndrome. Clinical neuropharmacology 2014; 37(1): 26–30. [DOI] [PubMed] [Google Scholar]
- 53.Maia TV, Conceicao VA. Dopaminergic Disturbances in Tourette Syndrome: An Integrative Account. Biological psychiatry 2018; 84(5): 332–344. [DOI] [PubMed] [Google Scholar]
- 54.Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, Deisseroth K et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature 2010; 466(7306): 622–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stachniak TJ, Ghosh A, Sternson SM. Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus-->midbrain pathway for feeding behavior. Neuron 2014; 82(4): 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez JL, Bonaventura J, Lesniak W, Mathews WB, Sysa-Shah P, Rodriguez LA et al. Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science 2017; 357(6350): 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abramowitz JS, Taylor S, McKay D. Obsessive-compulsive disorder. Lancet 2009; 374(9688): 491–499. [DOI] [PubMed] [Google Scholar]
- 58.Chandra R, Engeln M, Francis TC, Konkalmatt P, Patel D, Lobo MK. A Role for Peroxisome Proliferator-Activated Receptor Gamma Coactivator-1alpha in Nucleus Accumbens Neuron Subtypes in Cocaine Action. Biological psychiatry 2017; 81(7): 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 2008; 135(4): 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nature neuroscience 2006; 9(3): 443–452. [DOI] [PubMed] [Google Scholar]
- 61.Ka M, Kim WY. Microtubule-Actin Crosslinking Factor 1 Is Required for Dendritic Arborization and Axon Outgrowth in the Developing Brain. Molecular neurobiology 2016; 53(9): 6018–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higuchi Y, Hashiguchi A, Yuan J, Yoshimura A, Mitsui J, Ishiura H et al. Mutations in MME cause an autosomal-recessive Charcot-Marie-Tooth disease type 2. Annals of neurology 2016; 79(4): 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuang XL, Zhao XM, Xu HF, Shi YY, Deng JB, Sun GT. Spatio-temporal expression of a novel neuron-derived neurotrophic factor (NDNF) in mouse brains during development. BMC neuroscience 2010; 11: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen YK, Chen CY, Hu HT, Hsueh YP. CTTNBP2, but not CTTNBP2NL, regulates dendritic spinogenesis and synaptic distribution of the striatin-PP2A complex. Molecular biology of the cell 2012; 23(22): 4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toman RE, Payne SG, Watterson KR, Maceyka M, Lee NH, Milstien S et al. Differential transactivation of sphingosine-1-phosphate receptors modulates NGF-induced neurite extension. The Journal of cell biology 2004; 166(3): 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Progress in neurobiology 2013; 106–107: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Armentano M, Filosa A, Andolfi G, Studer M. COUP-TFI is required for the formation of commissural projections in the forebrain by regulating axonal growth. Development 2006; 133(21): 4151–4162. [DOI] [PubMed] [Google Scholar]
- 68.Ahmari SE, Spellman T, Douglass NL, Kheirbek MA, Simpson HB, Deisseroth K et al. Repeated cortico-striatal stimulation generates persistent OCD-like behavior. Science 2013; 340(6137): 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan S, van den Heuvel OA, Cath DC, de Wit SJ, Vriend C, Veltman DJ et al. Altered Functional Connectivity in Resting State Networks in Tourette’s Disorder. Frontiers in human neuroscience 2018; 12: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Worbe Y, Marrakchi-Kacem L, Lecomte S, Valabregue R, Poupon F, Guevara P et al. Altered structural connectivity of cortico-striato-pallido-thalamic networks in Gilles de la Tourette syndrome. Brain : a journal of neurology 2015; 138(Pt 2): 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hao H, Chen C, Mao W, Xia W, Yi Z, Zhao P et al. Alterations in resting-state local functional connectivity in obsessive-compulsive disorder. Journal of affective disorders 2019; 245: 113–119. [DOI] [PubMed] [Google Scholar]
- 72.Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S et al. SHANK1 Deletions in Males with Autism Spectrum Disorder. American journal of human genetics 2012; 90(5): 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell 2008; 135(3): 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Francis TC, Gaynor A, Chandra R, Fox ME, Lobo MK. The Selective RhoA Inhibitor Rhosin Promotes Stress Resiliency Through Enhancing D1-Medium Spiny Neuron Plasticity and Reducing Hyperexcitability. Biological psychiatry 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koo JW, Lobo MK, Chaudhury D, Labonte B, Friedman A, Heller E et al. Loss of BDNF signaling in D1R-expressing NAc neurons enhances morphine reward by reducing GABA inhibition. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 2014; 39(11): 2646–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brackenbury WJ, Yuan Y, O’Malley HA, Parent JM, Isom LL. Abnormal neuronal patterning occurs during early postnatal brain development of Scn1b-null mice and precedes hyperexcitability. Proceedings of the National Academy of Sciences of the United States of America 2013; 110(3): 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guillin O, Diaz J, Carroll P, Griffon N, Schwartz JC, Sokoloff P. BDNF controls dopamine D3 receptor expression and triggers behavioural sensitization. Nature 2001; 411(6833): 86–89. [DOI] [PubMed] [Google Scholar]
- 78.Lee FS, Chao MV. Activation of Trk neurotrophin receptors in the absence of neurotrophins. Proceedings of the National Academy of Sciences of the United States of America 2001; 98(6): 3555–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cazorla M, de Carvalho FD, Chohan MO, Shegda M, Chuhma N, Rayport S et al. Dopamine D2 receptors regulate the anatomical and functional balance of basal ganglia circuitry. Neuron 2014; 81(1): 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gertler TS, Chan CS, Surmeier DJ. Dichotomous anatomical properties of adult striatal medium spiny neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 2008; 28(43): 10814–10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma YY, Cepeda C, Chatta P, Franklin L, Evans CJ, Levine MS. Regional and cell-type-specific effects of DAMGO on striatal D1 and D2 dopamine receptor-expressing medium-sized spiny neurons. ASN neuro 2012; 4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization in the basal ganglia. Annual review of neuroscience 1992; 15: 285–320. [DOI] [PubMed] [Google Scholar]
- 83.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 2003; 425(6961): 917–925. [DOI] [PubMed] [Google Scholar]
- 84.Lerner A, Bagic A, Simmons JM, Mari Z, Bonne O, Xu B et al. Widespread abnormality of the gamma-aminobutyric acid-ergic system in Tourette syndrome. Brain : a journal of neurology 2012; 135(Pt 6): 1926–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Porta M, Brambilla A, Cavanna AE, Servello D, Sassi M, Rickards H et al. Thalamic deep brain stimulation for treatment-refractory Tourette syndrome: two-year outcome. Neurology 2009; 73(17): 1375–1380. [DOI] [PubMed] [Google Scholar]
- 86.Bronson SL, Bale TL. Prenatal stress-induced increases in placental inflammation and offspring hyperactivity are male-specific and ameliorated by maternal antiinflammatory treatment. Endocrinology 2014; 155(7): 2635–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan JC, Nugent BM, Bale TL. Parental Advisory: Maternal and Paternal Stress Can Impact Offspring Neurodevelopment. Biological psychiatry 2018; 83(10): 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


