Significance
We show that Bouncer’s homolog in mammals, SPACA4, is required for efficient fertilization in mice. In contrast to fish, in which Bouncer is required for female fertility, SPACA4 is expressed exclusively in the sperm and is required for male fertility. SPACA4 and Bouncer present an intriguing example of homologous proteins that both play key roles in reproduction yet diverged in terms of gene expression pattern and mode of action. Overall, our work identifies SPACA4 as an important sperm protein necessary for zona pellucida penetration during mammalian fertilization. Since human SPACA4 is also expressed exclusively in sperm, we anticipate that our findings in mice will have relevance to human biology.
Keywords: fertilization, sperm–egg interaction, zona pellucida, mouse
Abstract
Fertilization is the fundamental process that initiates the development of a new individual in all sexually reproducing species. Despite its importance, our understanding of the molecular players that govern mammalian sperm–egg interaction is incomplete, partly because many of the essential factors found in nonmammalian species do not have obvious mammalian homologs. We have recently identified the lymphocyte antigen-6 (Ly6)/urokinase-type plasminogen activator receptor (uPAR) protein Bouncer as an essential fertilization factor in zebrafish [S. Herberg, K. R. Gert, A. Schleiffer, A. Pauli, Science 361, 1029–1033 (2018)]. Here, we show that Bouncer’s homolog in mammals, Sperm Acrosome Associated 4 (SPACA4), is also required for efficient fertilization in mice. In contrast to fish, in which Bouncer is expressed specifically in the egg, SPACA4 is expressed exclusively in the sperm. Male knockout mice are severely subfertile, and sperm lacking SPACA4 fail to fertilize wild-type eggs in vitro. Interestingly, removal of the zona pellucida rescues the fertilization defect of Spaca4-deficient sperm in vitro, indicating that SPACA4 is not required for the interaction of sperm and the oolemma but rather of sperm and the zona pellucida. Our work identifies SPACA4 as an important sperm protein necessary for zona pellucida penetration during mammalian fertilization.
Fertilization is the fundamental process by which two gametes, the sperm and the egg, fuse to form a single cell, which gives rise to a new organism. Despite being essential for all sexually reproducing organisms, the molecular mechanisms that mediate sperm–egg interaction remain poorly understood. One important step toward gaining mechanistic insights into fertilization is the identification of molecules that can mediate sperm–egg interaction. Several proteins that are specifically required for gamete interaction in mammals have been identified with the help of genetic mouse models (reviewed in refs. 1–3). Required proteins on the sperm are the transmembrane proteins IZUMO1 (4, 5), SPACA6 (6, 7), TMEM95 (7–9), FIMP (10), DCST1/2 (11, 12), and the secreted protein SOF1 (8). The factors required on the egg are the tetraspanin protein CD9 (13–15) as well as the glycosylphosphatidylinositol (GPI)-anchored protein JUNO (16). The only known interacting protein pair is IZUMO1 and JUNO (16–20), which is known to mediate adhesion between sperm and egg (4, 5, 16, 19). How the other known mammalian fertilization factors enable sperm–egg binding and/or fusion remains unclear and is subject to active research.
In addition to sperm or egg surface proteins, genetic analyses also revealed the importance of proteins of the mammalian egg coat, called zona pellucida (ZP), in fertilization. ZP proteins were shown to be required for the initial step of sperm binding to the ZP as well as the subsequent block to polyspermy, which is induced by the first sperm entering the egg (21–25). While IZUMO1, CD9, SPACA6, DCST1/2, and ZP proteins have homologs in nonmammalian vertebrates, phylogenetic analyses by us and others suggest that the other known essential mammalian fertilization factors JUNO, TMEM95, FIMP, and SOF1 lack clear nonmammalian homologs (SI Appendix, Fig. S1) (9, 26). In line with this observation, rapid protein evolution and divergence have been noted as general hallmarks of proteins involved in reproduction (27).
This evolutionary divergence limits the direct transfer of knowledge gained from studies in invertebrates (28–31) and plants (reviewed in ref. 32) to fertilization in vertebrates. For example, the mechanistically best-understood fertilization proteins are lysin and Verl from the marine mollusk abalone (reviewed in ref. 33). Lysin is a sperm-expressed and highly abundant secreted protein, whereas Verl is an egg-coat protein that shows structural homology to mammalian ZP2 (34). Species-specific binding of lysin to Verl causes nonenzymatic disruption of the abalone egg coat, thereby allowing conspecific sperm to fertilize the egg (34–37). However, lysin has no known homolog in vertebrates, leaving it open whether a similar mechanism might contribute to mammalian sperm passage through the ZP.
Apart from the lack of clear homologs, identification of further factors required for sperm–egg interaction in vertebrates has also been hampered by the almost exclusive focus on mammalian fertilization. Mammalian fertilization occurs internally, which poses additional experimental challenges due to the low number of eggs and inaccessibility of gametes. Moreover, possible functional redundancies among fertilization factors have limited the informative value of single-gene knockout studies in mice. Accordingly, many genes that had been implicated as potential fertilization factors based on in vitro studies were later shown to be dispensable for fertilization in vivo (38–40). Other gamete-specific proteins that might play a role in fertilization have not yet been analyzed for their function in vivo, leaving their roles during mammalian fertilization unclear.
One of these gamete-specific proteins is SPACA4 (Sperm Acrosome Associated 4; also called SAMP14 [Sperm Acrosomal Membrane Associated 14]), which was initially identified by mass spectrometry in a screen for membrane-bound human sperm proteins (41). SPACA4 is particularly interesting for three reasons: First, its fish homolog Bouncer was recently shown to be essential for sperm–egg membrane interaction in zebrafish (42). Secondly, while Bouncer is expressed exclusively in the egg in fish and frogs, its closest mammalian homolog SPACA4 is expressed exclusively in the testis (41, 42). Thirdly, the incubation of human sperm with SPACA4-specific antibodies was shown to decrease the binding and fusion of sperm with ZP-free hamster eggs in vitro (41). Taken together, these observations point toward an important function of SPACA4 for mammalian fertilization.
Here, we investigate the functional relevance of SPACA4 in mammals by analyzing the phenotypic consequence of genetic loss of SPACA4 in mice.
Results
Murine SPACA4 Is Expressed in the Testis and Localizes to the Inner Sperm Membrane.
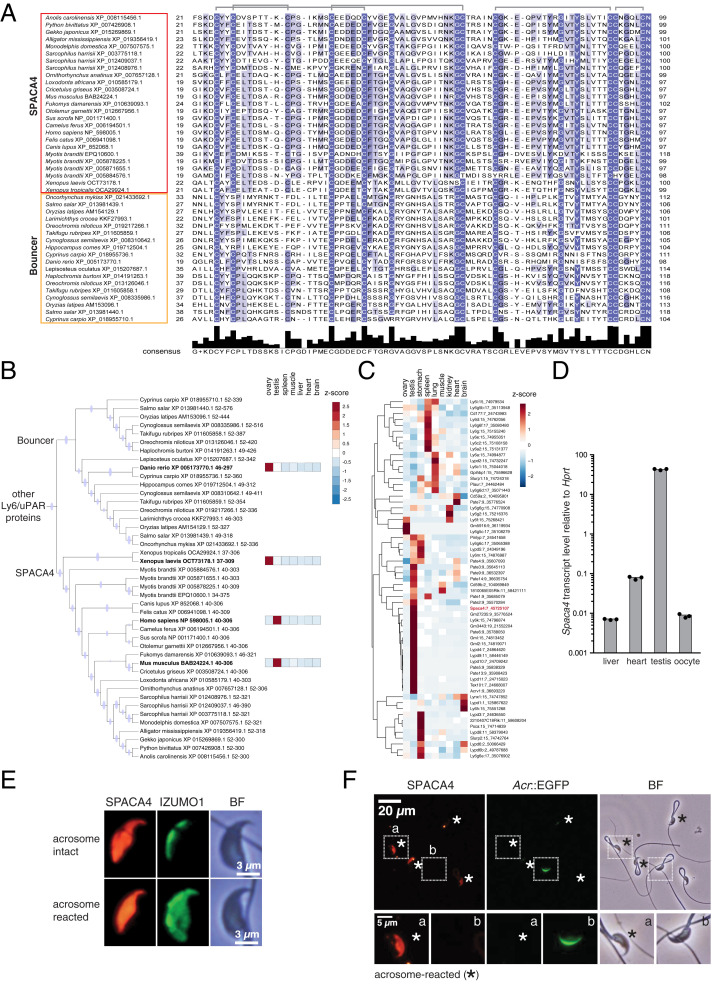
Bouncer was recently discovered as an essential fertilization factor in zebrafish that is attached to the egg surface via a GPI anchor and enables sperm binding to the egg membrane (42). Evolutionary analysis revealed that Bouncer has a mammalian homolog, SPACA4 (Fig. 1 A and B and SI Appendix, Fig. S2A) (42), which raised the immediate question whether SPACA4 might also be important for mammalian reproduction. Bouncer and SPACA4 belong to the large lymphocyte antigen-6 (Ly6)/urokinase-type plasminogen activator receptor (uPAR) protein family, which is characterized by a conserved 60 to 80 amino acid protein domain containing 8 to 10 cysteines that adopt a characteristic three-finger fold (43). Most Ly6/uPAR-type genes occur in clusters in the mouse genome (SI Appendix, Fig. S2B), consistent with their origin by gene duplication (43), and are expressed in diverse tissues in mice and humans (Fig. 1C and SI Appendix, Fig. S2C). While mammalian Spaca4 is not the only Ly6/uPAR-type gene expressed specifically in the male germline (testis) (Fig. 1C and SI Appendix, Fig. S2C), it stands out for having homologs in fish (bouncer) and amphibians (Spaca4) that are also germline specifically expressed yet in the opposite sex (ovary) (Fig. 1B and SI Appendix, Fig. S2A).
Fig. 1.
SPACA4 is expressed in murine sperm. (A) Protein sequence alignment of the mature domain of SPACA4/Bouncer protein family members. Shown is the mature three-finger domain of SPACA4 proteins (red box) and Bouncer proteins (orange box) lacking the N-terminal signal peptide and the carboxyl-terminal GPI-anchoring sequence. Amino acid divergence among different species is indicated based on the percent amino acid identity (blue shading); predicted cysteine bridges (gray) are shown above the alignment. Adapted from ref. 42. (B) Mammalian SPACA4 is the homolog for fish Bouncer and is expressed specifically in testis. Part of a maximum-likelihood phylogenetic tree based on Ly6/uPAR protein sequence alignments across vertebrates showing SPACA4 and Bouncer (refer to SI Appendix, Fig. S2A for the full tree; adapted from ref. 42). Branches supported by ultrafast bootstrap values (≥95%) are marked with a blue dot. Z-scores of averaged gene expression values across adult tissues are shown on the right for Danio rerio (11), Xenopus laevis (75), Mus musculus (73), and Homo sapiens (www.gtexportal.org). While Bouncer/Spaca4 mRNAs are expressed in oocytes in fish and frogs, mammalian Spaca4 mRNAs are expressed in testis. (C) Mouse Ly6/uPAR genes show diverse expression patterns across adult tissues. The heatmap is color coded based on z-scores of the normalized gene expression values (average of the square root) of RNA-Seq data from murine adult tissues (73). The clustering and dendrogram (Left) are based on expression scores. Spaca4 is highlighted in red. Numbers behind gene names indicate chromosome location in mice. (D) Mouse Spaca4 mRNA is expressed in the male germline. RT-qPCR from murine cDNA from different adult tissues reveals enrichment of Spaca4 specifically in mouse testis. Primers amplifying Hypoxanthineguanine phosphoribosyl transferase (Hprt) were used as a control. (E) Mouse SPACA4 protein localizes to the sperm head. Immunostaining of SPACA4 (red) and IZUMO1 (green) under permeabilizing conditions detects SPACA4 and IZUMO1 in the sperm head of wild-type B6D2F1 mice. In contrast to IZUMO1 that relocalizes after the acrosome reaction, SPACA4 does not change its localization. BF, brightfield image. (Scale bar, 3 µm.) (F) Murine SPACA4 localizes to the inner sperm membrane. Immunofluorescence staining of SPACA4 (red) in mouse spermatozoa under nonpermeabilizing conditions. Spermatozoa were derived from transgenic mice expressing EGFP under the control of the Acrosin promoter (65), labeling sperm with intact acrosomes (green). AR spermatozoa are highlighted by an asterisk (loss of green label). Boxed areas are shown below at higher magnification (a: AR sperm; b: spermatozoa with intact acrosome). SPACA4 is detected in AR spermatozoa (a) but not in spermatozoa with intact acrosomes (b). [Scale bar, 20 µm (Top) and 5 µm (Bottom).]
To confirm that Spaca4 was indeed expressed in male but not female gametes in mice, we analyzed the expression level of Spaca4 messenger RNA (mRNA) in different tissues using RT-qPCR. Murine Spaca4 was detected specifically in the testis and was enriched 100- to 1,000-fold compared to other tissues (Fig. 1D and SI Appendix, Fig. S2D), which agrees with published RNA-sequencing (RNA-Seq) data from mice (Fig. 1 B and C and SI Appendix, Fig. S2C) and with the reported testis-specific expression in humans (41) (SI Appendix, Fig. S2C). Analysis of published single-cell RNA-Seq data from murine spermatogenesis revealed a peak of Spaca4 mRNA expression in round spermatids, which resembles the expression of Izumo1 mRNA in timing and magnitude (SI Appendix, Fig. S3) (44). Using anti-mouse SPACA4 antibodies, SPACA4 was found to localize to the sperm head (Fig. 1E) and was readily detected on nonpermeabilized acrosome-reacted (AR), but not on acrosome-intact, live sperm (Fig. 1F). This expression pattern is consistent with the reported acrosomal membrane localization of human SPACA4 (41).
Male Mice Lacking SPACA4 Are Subfertile.
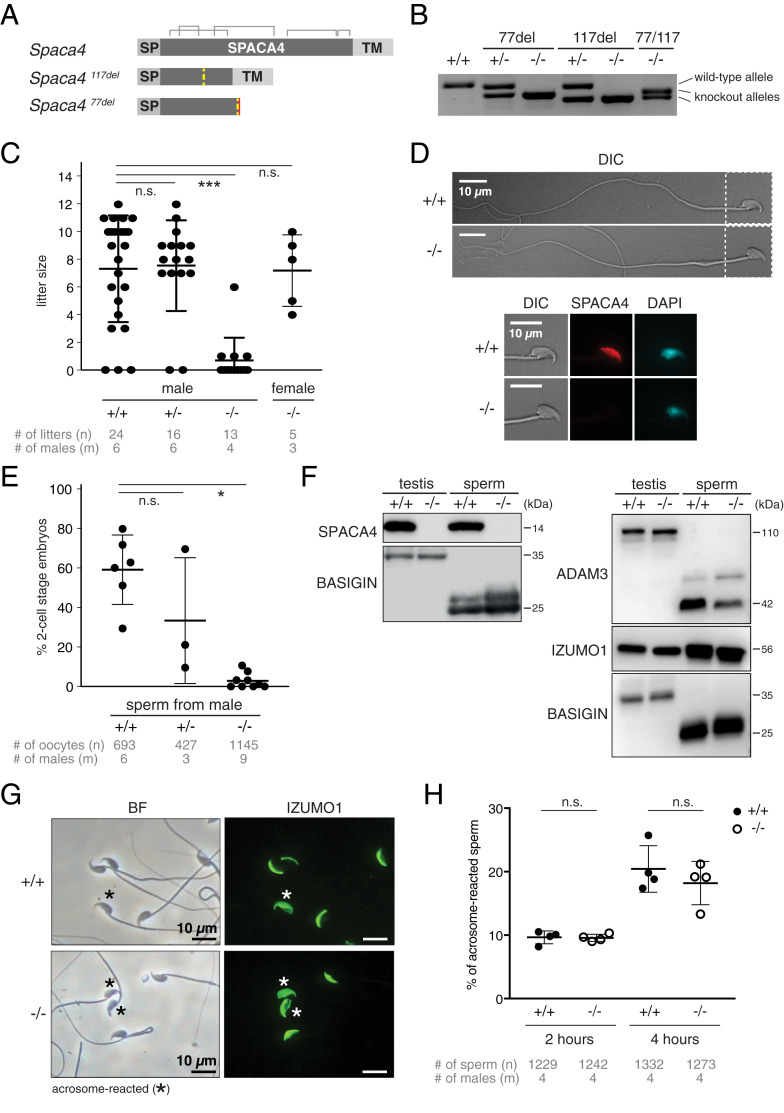
To investigate the function of mammalian SPACA4 in vivo, we generated Spaca4 knockout mice by CRISPR-Cas9–mediated targeted mutagenesis. We recovered two mutant alleles in the C57BL/6J background. The first one, in the following called Spaca477del, contains a 77-nt deletion leading to a frameshift after amino acid 42 (Fig. 2 A and B and SI Appendix, Fig. S4A). The second allele, in the following called Spaca4117del, contains a 117-nt in-frame deletion, which removes half (39 amino acids) of the mature SPACA4 protein, is therefore unable to fold into the characteristic three-finger conformation, and is thus also predicted to result in a full knockout mutation (Fig. 2 A and B and SI Appendix, Fig. S4A). A third, independent Spaca4 knockout mouse (Spaca4tm1Osb) was generated in the B6D2 background by replacing the whole exon of the Spaca4 gene with a neomycin resistance cassette using homologous recombination (SI Appendix, Fig. S5 A–C). Spaca4 knockout mice appeared indistinguishable from wild-type and heterozygous littermates, revealing that SPACA4 is dispensable for somatic development in mice.
Fig. 2.
SPACA4 is required for efficient fertilization in male mice. (A and B) Overview of the C57BL/6J-Spaca4 knockout alleles generated by CRISPR-Cas9–mediated targeted mutagenesis. One allele contains a 117-nt in-frame deletion after amino acid 42. The other allele contains a 77-nt out-of-frame deletion after amino acid 47. (A) Schematic of the wild-type and knockout alleles. Yellow dashed lines indicate the site of the deletions. Predicted disulfide bridges are indicated in gray. SP, signal peptide; TM, transmembrane region. (B) Genotyping of Spaca4 knockout mice. Spaca4 PCR products are separated based on their sizes on an agarose gel. (C) Spaca4 knockout male mice are subfertile. Litter sizes of C57BL/6J-Spaca4 wild-type (+/+), heterozygous (+/−), and transheterozygous (−/−) males caged with B6129F1 wild-type females or B6129F1 wild-type males caged with transheterozygous (−/−) females. Successful mating was confirmed by plug checks. Data are means ± SD. ***P < 0.0001 (Kruskal–Wallis test with Dunn multiple-comparisons test); n.s., not significant. n = number of litters; m = number of male mice tested. (D) Sperm morphology is normal in the absence of SPACA4 protein. DIC, differential interference contrast image. The sperm heads (boxed areas) are shown below at higher magnification. Immunostaining of sperm detects SPACA4 protein (red) under permeabilizing conditions in the head of sperm from wild-type (+/+) mice but not in sperm from Spaca4 knockout (−/−) mice. DAPI (cyan) staining labels the sperm nucleus. (Scale bars, 10 µm.) (E) Sperm from Spaca4 knockout mice has a severely reduced ability to fertilize wild-type oocytes. IVF of oocytes from C57BL/6J wild-type females using sperm from C57BL/6J-Spaca4 wild-type (+/+), heterozygous (+/−), or transheterozygous (−/−) males. Plotted is the percentage of two-cell stage embryos as a measure of successful fertilization. Data are means ± SD. *P = 0.014 (Kruskal–Wallis test with Dunn multiple-comparisons test); n.s., not significant. n = total number of oocytes; m = number of males tested. (F) IZUMO1 and ADAM3 proteins are expressed and processed normally in sperm and testis from Spaca4 knockout mice. Western blot analysis showing the absence of SPACA4 protein in testes and spermatozoa of Spaca4 knockout mice (Left) and expression and processing of ADAM3 and IZUMO1 proteins similar to wild-type controls. Testes and spermatozoa of wild-type mice, as well as BASIGIN protein, which undergoes proteolytic cleavage during sperm maturation (7), are used as control. (G) IZUMO1 localization is normal in Spaca4 knockout mice. Immunostaining of sperm under permeabilizing conditions shows normal localization of IZUMO1 (green) in acrosome-intact wild-type (+/+) and SPACA4-deficient (−/−) sperm and normal relocalization of IZUMO1 in AR sperm (asterisk) in both genotypes. BF, brightfield image. (Scale bars, 10 µm.) (H) Quantification of the percentage of AR sperm 2 or 4 h after incubation of capacitated sperm. Acrosome reaction was assessed by immunostaining for IZUMO1. Data are means ± SD; n.s., not significant. n = total number of sperm; m = number of males tested.
In line with SPACA4’s sperm-specific expression in mammals, we found that SPACA4 is necessary for male fertility. The litter size of transheterozygous (Spaca4117del/77del) as well as homozygous mutant male mice was significantly lower than the litter sizes of wild-type or heterozygous male mice (P < 0.0001) (Fig. 2C and SI Appendix, Figs. S4B and S5D). In contrast, female fertility was not affected by the Spaca4 mutation (Fig. 2C). The observed defect in male fertility was not due to an inability of Spaca4 knockout males to mate, as verified by the presence of vaginal plugs.
The severely reduced fertility of males lacking SPACA4 could have multiple reasons, including reduced sperm count, immotility of the sperm, or a defect in gamete interaction. Sperm morphology and numbers were similar in wild-type and knockout mice, and sperm derived from knockout males was motile (Fig. 2D and SI Appendix, Fig. S4 C–E), suggesting that SPACA4 is required during sperm–egg interaction. To test this hypothesis, we performed in vitro fertilization (IVF) experiments, which revealed that Spaca4 mutant sperm was severely compromised in its ability to fertilize wild-type oocytes in vitro: Spermatozoa from transheterozygous (Spaca4117del/77del) male mice resulted in a severely reduced average fertilization rate of 2.9% (two-cell stage embryos), whereas the average fertilization rates using spermatozoa from wild-type or heterozygous (Spaca4117del/+ or Spaca477del/+) mice were at 59.4% or 33.4%, respectively (Fig. 2E). Similar defects in IVF were observed in Spaca4tm1Osb mutant sperm (SI Appendix, Fig. S5E).
The inability of SPACA4-deficient sperm to efficiently fertilize wild-type oocytes could be due to multiple reasons, including 1) a failure to passage the uterotubal junction, a defect which has been shown to correlate with the lack of the presence and/or posttranslational processing of ADAM3 (45, 46), 2) loss of expression and/or improper localization of IZUMO1, or 3) a defect in undergoing the acrosome reaction. However, Western blotting for SPACA4, ADAM3, and IZUMO1 and immunofluorescence staining for IZUMO1 revealed that ADAM3 protein was processed into the shorter protein isoform in knockout sperm (Fig. 2F) and that sperm of Spaca4 knockout mice showed normal rates of acrosome reactions as well as normal expression and relocalization of IZUMO1 upon acrosome reaction (Fig. 2 G and H). Even though ADAM3 processing was slightly reduced in Spaca4 mutant sperm compared to wild-type sperm, this decrease cannot account for the severe fertilization defect in Spaca4 mutants given that much less of processed ADAM3 is required to rescue infertility mutants (47). Overall, we conclude that SPACA4 is an important, albeit not absolutely essential, protein required at the step of mammalian gamete interaction.
SPACA4 Is Required for Efficient Penetration of the ZP.
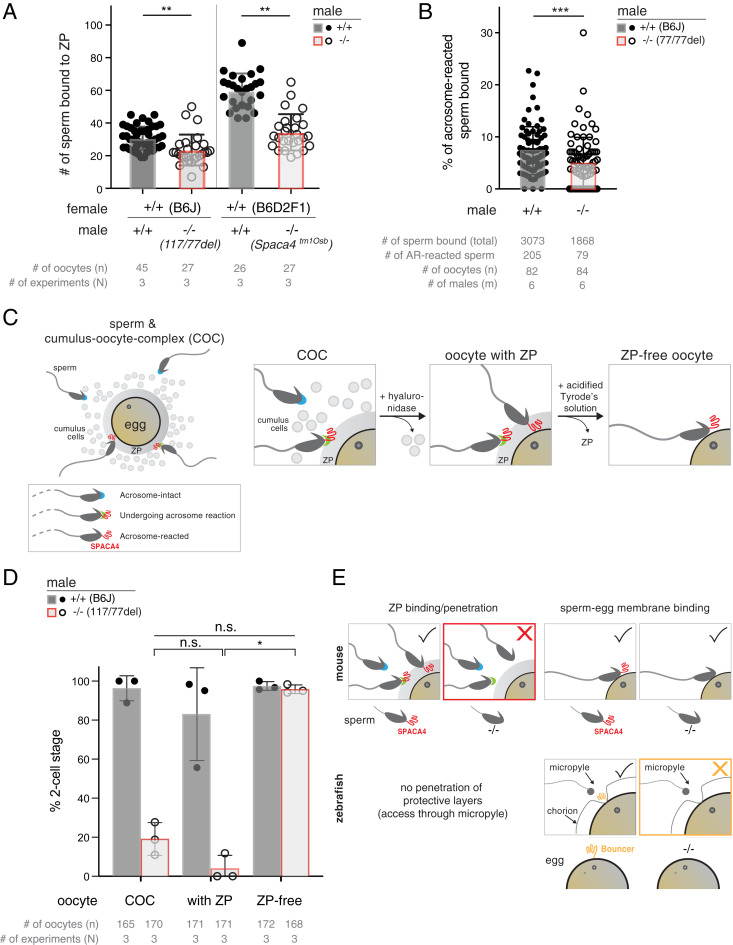
The marked IVF defect of sperm derived from Spaca4−/− males showed that SPACA4 is necessary for sperm to efficiently interact with the egg. In mammals, this interaction occurs in two steps: Sperm first needs to bind and penetrate the ZP before binding to the egg membrane (oolemma), which enables sperm–egg fusion. To determine at which stage of fertilization SPACA4-deficient sperm is impaired, we quantified the number of spermatozoa bound to the ZP 30 min after insemination. We found that in both Spaca4−/− mutant strains, fewer mutant spermatozoa bound the ZP of wild-type oocytes compared to control wild-type sperm (23.5 ± 9.5 in the case of Spaca4117/77 versus 30.7 ± 7.0 for wild-type [B6J] [P < 0.001]; 34.3 ± 11.1 in the case of Spaca4tm1Osb versus 59.7 ± 10.7 for wild-type [B6D2F1] [P < 0.001]) (Fig. 3A). A closer analysis of the observed defect in ZP binding revealed that both acrosome-intact and AR sperm from Spaca4 knockout males showed decreased binding to wild-type oocytes (4.5% AR sperm bound per oocyte [0.9 ± 0.8 AR sperm out of 22.2 ± 8.7 total sperm bound per oocyte] in the case of Spaca477/77 versus 7.4% AR sperm bound per oocyte [2.5 ± 1.2 AR sperm out of 37.5 ± 11.7 total sperm bound per oocyte] in the case of wild-type [B6J] [P < 0.001]) (Fig. 3B and SI Appendix, Fig. S6 A and B). Analysis of sperm motility showed slightly reduced motility of mutant sperm (SI Appendix, Fig. S6C). However, given that the difference in motility between the two different wild-type strains (B6J and B6D2F1) was larger than between the mutant and background-matched wild-type spermatozoa and that sperm motility of B6D2 knockout sperm was higher than that of wild-type B6J sperm (SI Appendix, Fig. S6C), we conclude that the small decrease in motility cannot explain the defect in fertilizing ability of Spaca4 knockout spermatozoa. Instead, the main defect of Spaca4 mutant spermatozoa appears in sperm binding.
Fig. 3.
SPACA4 enables sperm to bind to and penetrate the ZP. (A) SPACA4 is required for efficient binding of sperm to the ZP. Sperm of the indicated genotypes was incubated for 30 min with hyaluronidase-treated oocytes (with intact ZP) of matching genetic backgrounds. Plotted is the number of sperm bound to ZP-containing oocytes. Data are means ± SD. P values (**P < 0.01) are by Student’s t test. n = total number of oocytes; N = number of replicates. (B) Quantification of the percentage of AR sperm derived from wild-type (+/+) and Spaca477nt-del/77nt-del (−/−) males bound to ZP-containing wild-type oocytes. Data are means ± SD. The P value (***P < 0.001) is by Student’s t test. Numbers of total sperm bound, AR sperm, oocytes (n) and males (m) tested are indicated (SI Appendix, Fig. S6 A and B). (C) Schematic of sperm bound to the COC and experimental treatments used to remove the cumulus cells (by treatment of COCs with hyaluronidase) and the ZP (by treatment of cumulus-free oocytes with acidified Tyrode’s solution) from the COCs. Acrosome-intact sperm, blue cap; sperm undergoing the acrosome reaction, green cap, and SPACA4 (red) getting exposed; AR sperm, no cap, and SPACA4 (red) exposed. (D) SPACA4 is required for ZP penetration but not for oolemma binding and fusion. IVF performed with COCs from superovulated C57BL/6J females, cumulus cell–free oocytes (oocyte with ZP), and ZP-free oocytes with sperm from either wild-type C57BL/6J males or age-matched Spaca4117nt-del/77nt-del males. Plotted is the percentage of two-cell stage embryos as a measure of successful fertilization. Data are means ± SD. P values (*P < 0.05; n.s., not significant) are by a Kruskal–Wallis test with Dunn multiple-comparisons test. n = total number of oocytes; N = number of replicates. (E) Comparison of SPACA4’s function in murine fertilization versus Bouncer’s role in fish fertilization. (Top) In the mouse, sperm-expressed SPACA4 (red) is exposed in AR sperm (green cap and no cap) and is required for efficient ZP penetration (Left) but not for sperm–egg membrane binding (Right). Acrosome-intact sperm, blue cap. (Bottom) In zebrafish, sperm can access the egg membrane via the preformed funnel, the so-called micropyle, in the chorion. Egg-expressed Bouncer (orange) is required for sperm–egg membrane binding in zebrafish (42). The tick mark (yes) or cross (no) at the top right in each box indicates whether this step of fertilization can occur in the wild-type or mutant condition.
The reduced binding of SPACA4-deficient sperm to the ZP prompted us to ask at which step of sperm–oocyte complex interaction SPACA4 was required, since mammalian sperm first needs to penetrate the protective layers that surround the oocyte, namely, the cumulus layer and the ZP (Fig. 3C). To test whether removal of the cumulus cells and/or the ZP can rescue the fertility defect of SPACA4-deficient sperm in IVF, wild-type oocytes were treated with hyaluronidase and acidified Tyrode’s solution to remove the cumulus layer and ZP, respectively (48) (Fig. 3C). Fertilization by wild-type sperm was not significantly affected by the different treatments (Fig. 3D). Similarly, removal of the cumulus cells did not rescue the low fertilization rate of SPACA4-deficient sperm (18.8% cumulus–oocytle complexes [COCs] versus 4.7% [cumulus cells removed] [P = 0.884]; Fig. 3D). However, removal of the ZP enabled SPACA4-deficient sperm to fertilize zona-free oocytes at a similarly high rate (95.8%) as wild-type sperm (97.1%) (P value for mutant sperm fertilizing oocytes with ZP versus ZP-free = 0.033) (Fig. 3D). We therefore conclude that murine SPACA4 is required for the sperm’s ability to efficiently bind to and traverse the ZP (Fig. 3E).
Discussion
Here, we reveal that SPACA4, the mammalian homolog of fish Bouncer (Fig. 1 A and B and SI Appendix, Fig. S2A) (42), is required for normal male fertility in mice (Fig. 2 and SI Appendix, Fig. S5). We find that SPACA4 is expressed in the sperm head and gets exposed by the acrosome reaction (Figs. 1 E and F and 2D), which enables sperm to efficiently bind to and penetrate the ZP (Fig. 3). SPACA4, therefore, differs from the proposed function of other known sperm fertility factors required for sperm–egg interaction in mice (SPACA6, IZUMO1, SOF1, FIMP, TMEM95, and DCST1/2), which are required at the later step of sperm–egg membrane binding and/or fusion (4–12). Thus, our work provides genetic evidence for an important function of SPACA4 for the step of ZP binding in mice. Together with previous SPACA4-antibody–blocking experiments with human sperm and the conserved expression pattern of SPACA4 in human sperm (41) (Fig. 1B and SI Appendix, Fig. S2 A and C), our results in mice could have direct relevance for male fertility in humans. Although both our results presented here from genetic knockout studies in mice, as well as the antibody-blocking experiments with human sperm (41), are consistent with human SPACA4 playing an important role in human reproduction, the precise function of human SPACA4 is still unclear. While our studies in mice demonstrate a role for murine SPACA4 in ZP binding, but not in the subsequent step of sperm–egg membrane interaction, the results reported by Shetty et al. for human SPACA4 suggested a role in sperm–egg membrane interaction but did not assess ZP binding (41). The difference in the reported function of SPACA4 between human and mice could be due to technical reasons [e.g., due to known difficulties in biochemical blocking studies to uncover the real physiological function of proteins, as has been observed, for example, in the case of Acrosin (49, 50), Fertilin (51, 52), and CD46 (53, 54) (reviewed in refs. 40 and 55)]. In this case, human SPACA4 may function at the step of ZP binding, as we report here for murine SPACA4. Alternatively, it could reflect a real biological difference between mouse and human SPACA4’s function. Future work will be needed to explore a possible link between Spaca4 mutations in humans and subfertility in men presenting normal sperm count, morphology, and motility.
SPACA4 and Bouncer are not the only Ly6/uPAR proteins that have been linked to vertebrate reproduction. Several other members of this large gene family show a testis-restricted expression pattern in mammals (Fig. 1C and SI Appendix, Fig. S2C), some of which (Tex101, Ly6k, Lypd4, Pate4, and the Pate gene cluster) have been confirmed in genetic knockout studies to be required for male fertility in mice (56–58). In light of these known male-specific requirements for Ly6/uPAR proteins in mammals, SPACA4 and Bouncer present an interesting example of homologous proteins that diverged in terms of gene expression pattern and mode of action. Our phylogenetic sequence analysis shows that Bouncer and SPACA4 are the closest homologs among all other Ly6/uPAR family members (Fig. 1B and SI Appendix, Fig. S2A), yet they have opposing germline-specific expression patterns that broadly correlate with external (fish, amphibians; expressed in the egg) versus internal (mammals, reptiles; expressed in the sperm) fertilization (Fig. 1B and SI Appendix, Fig. S2A). We currently do not know how this different gene expression pattern arose or whether it evolved as a consequence of the different fertilization modes. One possibility is that an ancestral Ly6/uPAR protein might have been expressed in both male and female gonads and that sex-specific loss of expression occurred either by chance in different lineages or in response to a functional benefit of the restricted expression of SPACA4/Bouncer to the male or female germline. Acquisition of a restricted expression domain from an initially broader expression pattern has been proposed for other members of the Ly6/uPAR protein family, namely, for snake toxins that evolved a venom gland–restricted expression pattern (59).
The difference in expression pattern also extends to a difference in mode of action, at least in the case of the two example model organisms zebrafish (42) and mice (this work): While Bouncer in fish is required in the egg for sperm binding to the oolemma (42), the results presented here reveal that SPACA4 is dispensable for sperm binding to the oolemma in mice and is instead required for the preceding process of ZP penetration and/or binding. In this regard, it is interesting to note that mammalian and fish gametes differ in key aspects: Mammalian sperm has an acrosome, a specialized vesicle in the sperm head that must undergo exocytosis to expose important membrane-localized fertility factors (5, 60). This so-called acrosome reaction is important for successful ZP penetration and fertilization. Moreover, mammalian sperm needs to first bind to and penetrate the outer coat before gaining access to the egg membrane. Fish sperm, on the other hand, lacks an acrosome and has direct access to the oolemma through the micropyle, a preformed funnel in the outer protective layer of the fish egg. One can therefore speculate that acquisition of a sperm-specific expression of SPACA4 in mammals was beneficial to allow sperm to pass the additional outer barrier. In zebrafish, current data suggests that Bouncer acts in a unilateral manner by interacting with a still-unknown factor on the opposing gamete, since successful sperm entry requires compatibility between a species’ Bouncer and the sperm (42). Whether a similar mode of action (e.g., a SPACA4 interacting protein expressed in the oocyte and a possible involvement in species-specificity of sperm–egg interaction) also applies to mammals is currently unclear. Given the divergence in function of Ly6/uPAR proteins (43, 61), it is possible that SPACA4 acquired a different function from Bouncer [e.g., by acting in cis through interacting with other sperm-expressed membrane proteins or by interacting with molecules in the extracellular matrix surrounding the oocyte as suggested for its interaction with plasminogen (62)]. Moreover, one can speculate that for species with internal fertilization, a selection step determining gamete compatibility at the stage of sperm–egg interaction might be less important, since mating partner selection alone can ensure that only the selected partner’s sperm will be available for fertilization. This is not the case for species performing external fertilization who cannot guarantee by premating choice that only conspecific sperm reaches the egg (28, 63) and whose oocyte-specific expression of Bouncer could contribute to postcopulation female mate choice (also called cryptic female mate choice) (64). Thus, sperm-expressed proteins like SPACA4 could promote the efficiency of fertilization, while oocyte-expressed proteins like Bouncer could support the selection of conspecific sperm. Future experiments will be required to elucidate how SPACA4 promotes fertilization in mammals and to what extent the function and mechanism differ between SPACA4 and Bouncer.
Overall, our study on SPACA4/Bouncer highlights an interesting example of a vertebrate-specific sperm–egg interaction protein that evolved a different gene expression pattern, mode of action, and, possibly, function in fish versus mammals.
Materials and Methods
Mouse Lines and Husbandry.
All mouse experiments were conducted according to Austrian and European guidelines for animal research and approved by local Austrian authorities or by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University (Biken-AP-H30-01). Mice were maintained under a 10-/14-h light/dark cycle (IMP) or under a 12-h light/dark cycle (Osaka University). Wild-type mice were purchased from CLEA Japan and Japan SLC (Osaka University). The mouse strain Tg(Acr-EGFP)1Osb has been reported before (65).
Generation of Spaca4 Knockout Mice.
The mouse Spaca4 gene consists of a single exon and maps to chromosome 7. Two strategies were used to generate Spaca4 knockout mice: 1) CRISPR-Cas9–based gene targeting (IMP) and 2) replacement of the Spaca4-encoding exon with a neomycin selection cassette via gene targeting of embryonic stem (ES) cells (Osaka University).
CRISPR-Cas9–based gene targeting (resultant Spaca4 alleles: C57BL/6J-Spaca4117del and C57BL/6J-Spaca477del).
The Spaca4 knockout mice were generated using CRISPR-Cas9–based gene targeting. Two guide RNAs (gRNAs) targeting the coding region of Spaca4 were generated according to published protocols (66) by oligo annealing followed by T7 polymerase–driven in vitro transcription (gene-specific targeting oligo: SPACA4_gRNA1 and SPACA4_gRNA2; common tracer oligo: SI Appendix, Table S1). For gRNA injections, zygotes were isolated from superovulated donor female (C57BL/6J) mice on the day of the coagulation plug (= E0.5). To remove cumulus cells, zygotes were incubated in hyaluronidase solution (∼0.3 mg/mL). The injection mix (50 ng/µL Cas9 mRNA [Sigma-Aldrich] and 50 ng/µL SPACA4_gRNA1 and SPACA4_gRNA2) was injected into the cytoplasm of the zygotes. Injected zygotes were incubated for at least 15 min at 37 °C and 5% CO2. Surviving zygotes were transferred into the oviducts of pseudopregnant recipient females. The resulting pups were genotyped using primers SPACA4_gt_F and SPACA4_gt_R (SI Appendix, Table S1).
Gene targeting in ES cells (resultant Spaca4 allele: C57BL/6N-Spaca4tm1Osb).
The targeting vector was constructed using pNT1.1 (https://www.ncbi.nlm.nih.gov/nuccore/JN935771). A 1.9-kb NotI-XhoI short arm fragment and a 5.1-kb PacI-MfeI long arm fragment were obtained by PCR amplification using Bacterial Artificial Chromosome (BAC) DNA (RP24-343L2) as a template. The primers used were Spaca4_targeting-s_F and Spaca4_targeting-s_R for the short arm; Spaca4_targeting-l_F and Spaca4_targeting-l_R for the long arm (SI Appendix, Table S1). The targeting construct was linearized with ClaI and electroporated into EGR-G101 [C57BL/6N-Tg(CAG/Acr-EGFP)] ES cells (67). Potentially targeted ES cell clones were separated by positive/negative selection with G418 and ganciclovir. Correct targeting of the Spaca4 allele in ES cell clones and germ-line transmission were determined by PCR. Screening primers used were Spaca4_screening+gt#781 and Spaca4_screening+gt#5081 for the short arm; Spaca4_screening+gt#5173 and Spaca4_screening+gt#678 for the long arm (SI Appendix, Table S1). The mutant ES clones were injected into eight-cell stage ICR embryos, and chimeric blastocysts were transferred into the uterine horns of pseudopregnant ICR females the next day. To confirm germ-line transmission, chimeric males were mated with B6D2F1 females. Offspring from heterozygous intercrosses were genotyped by PCR. The genotyping primers used were Spaca4_gt#5269, Spaca4_gt#5298, and Spaca4_screening+gt#781 (SI Appendix, Table S1). Two bands, a 0.3-kb band as the wild-type allele and a 0.5-kb band as the knockout allele, were amplified by PCR.
Genotyping: Extraction of gDNA from Mouse Ear Clips and Genotyping PCR.
Mice were genotyped at weaning age (around 3 wk) using ear clips. gDNA was extracted from the ear clips using one of the following three alternative protocols. According to one protocol, ear clips were lysed by incubation in 25 µL QuickExtract Solution (QE09050, Lucigen) at 65 °C and shaking at 600 rpm for 30 min. According to a second protocol, ear clips were lysed in 100 µL lysis buffer (0.1 M NaCl, 0.5% sodium dodecyl sulfate (SDS), 10 mM Tris pH 8.0, 0.25 mM ethylenediaminetetraacetic acid (EDTA), 2 µg/µL proteinase K) at 55 °C for 3 h. To precipitate the cellular debris, 60 µL NaCl was added and the sample was centrifuged at 21,000 × g for 10 min at 4 °C. The supernatant was centrifuged a second time (21,000 × g, 10 min at 4 °C). To precipitate the DNA, 160 µL cold 100% ethanol was added, and the sample was centrifuged again (21,000 × g, 10 min at 4 °C). The pellet was washed a second time using 250 µL 75% ethanol (centrifugation at 21,000 × g, 5 min at 4 °C). The cleaned DNA pellet was dried at 37 °C for 5 to 15 min and solubilized in 50 µL ddH2O. According to a third protocol, a commercial lysis buffer was used to extract the gDNA from ear clippings (DirectPCR Lysis Reagent [Mouse Tail], ViagenBiotech). A total of 200 µL commercial lysis buffer and 1 µL 100 mg/mL proteinase K were added to each ear clip and incubated at 55 °C under shaking (600 rpm) overnight. For heat inactivation, the samples were heated to 85 °C for 45 min under vigorous shaking (800 rpm).
To amplify the Spaca4 coding region, standard Taq Polyermase or Q5 Hot Start polymerase (New England Biolabs) was used with primers SPACA4_gt_F and SPACA4_gt_R (SI Appendix, Table S1) according to the manufacturer’s protocol. The size of the PCR product was analyzed on a 2% agarose gel.
Identification of the Nature of the SPACA477del and SPACA4117del Mutations.
To analyze the nature of the Spaca4 mutations, the PCR products from the genotyping of the first outcross (generation F1) were cloned into the cloning vectors provided by the StrataClone PCR cloning kit (Stratagene). The cloning was performed according to the manufacturer’s protocol. Of the resulting bacterial colonies, 96 colonies were picked and sequenced using the primer SPACA4_gt_F (SI Appendix, Table S1). Two mice, one with a 77-nt and the other one with a 117-nt deletion in the coding sequence of Spaca4, were selected and used for further in- and outcrossing. Both deletions were also confirmed by sequencing the PCR products from the genotyping of the next generation (F2).
Wild-type Spaca4 Open Reading Frame (ORF) (the bold sequence is deleted in the mutant Spaca477del; the underlined sequence is deleted in the mutant Spaca4117del): Atggtccttggctggccactgcttctggtgttggttctttgcccaggtgtgacaggcatcaaggactgcgtcttctgtgagctgactgactctgctcggtgccctggcacacacatgcgctgtggggatgacgaagattgcttcacaggccacggagtagcccagggtgtggggcccatcatcaacaaaggctgcgtgcactccaccagctgtggccgcgaggaacccatcagctacatgggcctcacatacagtctcaccaccacctgctgttctggccacctttgcaataagggcactggcctttccacaggggctaccagcctgtcactgggtctgcagctgctcctgggcctgttgctgctgcttcaatactggctgtga.
Wild-type SPACA4 protein (underlined sequence, signal peptide; the bold sequence is deleted in the mutant Spaca477del; the underlined italicized sequence is deleted in the mutant Spaca4117del): MVLGWPLLLVLVLCPGVTGIKDCVFCELTDSARCPGTHMRCGDDEDCFTGHGVAQGVGPIINKGCVHSTSCGREEPISYMGLTYSLTTTCCSGHLCNKGTGLSTGATSLSLGLQLLLGLLLLLQYWL*.
Spaca477del ORF is as follows: Atggtccttggctggccactgcttctggtgttggttctttgcccaggtgtgacaggcatcaaggactgcgtcttctgtgagctgactgactctgctcggtgccctggcacacacatgcgctgtggggatgacgaagattgcttaggaacccatcagctacatgggcctcacatacagtctcaccaccacctgctgttctggccacctttgcaataa.
SPACA477del protein (the bold sequence does not exist in the wild-type, as it is a consequence of the out-of-frame deletion) is as follows:
MVLGWPLLLVLVLCPGVTGIKDCVFCELTDSARCPGTHMRCGDDEDCLGTHQLHGPHIQSHHHLLFWPPLQ*.
Spaca4117del ORF is as follows: Atggtccttggctggccactgcttctggtgttggttctttgcccaggtgtgacaggcatcaaggactgcgtcttctgtgagctgactgactctgctcggtgccctggcacacacatgcgctgtggcctcacatacagtctcaccaccacctgctgttctggccacctttgcaataagggcactggcctttccacaggggctaccagcctgtcactgggtctgcagctgctcctgggcctgttgctgctgcttcaatactggctgtga.
SPACA4117del protein is as follows: MVLGWPLLLVLVLCPGVTGIKDCVFCELTDSARCPGTHMRCGLTYSLTTTCCSGHLCNKGTGLSTGATSLSLGLQLLLGLLLLLQYWL*.
RT-PCR and RT-qPCR of Spaca4.
Mouse complementary DNA (cDNA) was prepared from multiple adult tissues of ICR mice and from testes of Spaca4 knockout mice. Total RNA was reverse-transcribed into cDNA using a SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). The amplification conditions for PCR were 2 min at 50 °C and 30 s at 95 °C, followed by 39 cycles of 95 °C for 15 s and 60 °C for 1 min (+ plate read) (RT-qPCR: Fig. 1C) or 1 min at 94 °C, followed by 30 cycles of 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 30 s, with a final 7-min extension at 72 °C (RT-PCR: SI Appendix, Fig. S2D), using primers targeting Spaca4 (Spaca4_qPCR_F2 and Spaca4_qPCR_R2) and Hprt (HPRT_qPCR_F and HPRT_qPCR_R) (Fig. 1C) or Spaca4 (Spaca4_qPCR_F1 and Spaca4_qPCR_R1) and Gapdh (Gapdh_RT_F and Gapdh_RT_R) (SI Appendix, Fig. S2D).
In Vivo Fertility Assays.
Fertility assays in mice were performed according to two alternative methods: In the case of CRISPR-generated C57BL/6J-Spaca4 alleles, C57BL/6J-Spaca4 wild-type, heterozygous, transheterozygous (Spaca4117del/77del), or homozygous (Spaca4117del/117del or Spaca477del/77del) mutant male or female mice were caged with 2- to 4-mo-old B6129F1 wild-type mice in the evening. Females were checked for plugs every morning and separated from the males as soon as a plug could be observed. The number of pups for each female was counted within a week of birth. In the case of the mutant male mice, this procedure was repeated at least once before the mutant mice were kept caged with a B6129F1 female for 3 to 10 wk after the initial plug.
In the case of C57BL/6N-Spaca4tm1Osb mutants, sexually mature wild-type, heterozygous or homozygous mutant male mice were caged with 2-mo-old B6D2F1 for several months, and the number of pups in each cage was counted within a week of birth. Average litter sizes are presented as the number of total pups born divided by the number of litters for each genotype.
IVF Assays.
Before IVF, female mice were superovulated by injection of CARD HyperOva (KYD-010-EX, Cosmo Bio Co) approximately 63 h before and human chorionic gonadotropin (hCG, Chorulon) 14 to 16 h before harvesting the oocytes. IVF was performed using Toyoda, Yokoyama, Hoshi (TYH) medium, CARD MEDIUM (KYD-003-EX, Cosmo Bio Co), and CARD FERTIUP Preincubation Medium (KYD-002-EX, Cosmo Bio Co) according to the manufacturer’s protocol. Sperm was prepared from the cauda epididymides and capacitated in CARD FERTIUP medium for 1 h or TYH medium for 2 h. Oocytes from superovulated female mice (C57BL/6J) were introduced into a drop of CARD MEDIUM or TYH medium. To prepare cumulus- or zona-free oocytes (Fig. 3C), COCs were collected in M2 medium (MR-015P-5D, Merck) and treated with 300 µg/mL hyaluronidase (H3884, Sigma-Aldrich) until the cumulus cells were removed, and cumulus cell-free oocytes were washed in M2. For ZP removal, the cumulus cell–free oocytes were moved to a droplet of acidified Tyrode’s solution (T1788, Sigma-Aldrich) for a few seconds, then washed with M2 and finally transferred into CARD MEDIUM. Afterward, the preincubated sperm was added to the differently treated oocytes for fertilization. Sperm and eggs were incubated at 37 °C and 5% CO2 and washed 3 h after incubation. Fertilization rates were recorded by counting the number of two-cell stage embryos on the next day.
Sperm Number and Motility Analyses.
The number (SI Appendix, Fig. S4D) and overall motility (motile, progressive motile; SI Appendix, Fig. S4E) of sperm were measured using the computer-assisted sperm analysis system CEROS II animal (Hamilton Thorne) according to the manufacturer’s protocol. In brief, 1 µL sperm was diluted 1:200 in Dulbecco's phosphate-buffered saline (DPBS, MR-006C, Merck), and then overall motility was assessed on a CEROS II.
To quantify sperm motility under IVF conditions (SI Appendix, Fig. S6C), cauda epididymal spermatozoa were squeezed out and then dispersed in TYH (for sperm motility and IVF). After incubation of 10 and 120 min in TYH, sperm motility patterns were examined using the CEROS II sperm analysis system (58, 68).
Assessment of Sperm Binding to the ZP.
The sperm ZP-binding assay was performed as described previously (69). In brief, cumulus cells of oocytes were removed by treatment with bovine testicular hyaluronidase (175 U/mL; Sigma-Aldrich) for 5 min. In the TYH medium, cumulus-free oocytes were mixed and incubated for 2 h with spermatozoa of the indicated genotypes and fixed with 0.25% glutaraldehyde for 30 min. The bound spermatozoa were observed with an Olympus IX73 microscope (Olympus) (Fig. 3A) or an IX-70 fluorescent microscope (Olympus) (Fig. 3B and SI Appendix, Fig. S6 A and B). IZUMO1 was used as a marker of the acrosome reaction in spermatozoa bound to the ZP. After fixation, the oocytes were incubated with rat anti-IZUMO1 antibody (KS64-125) for 30 min, followed by incubation with goat anti-rat immunoglobulin G (IgG) Alexa Fluor 488 for 30 min. The AR spermatozoa were observed with a BZ-X710 fluorescent microscope (Keyence) (Fig. 3B and SI Appendix, Fig. S6 A and B).
Generation of Anti-Mouse SPACA4 Monoclonal Antibodies.
Monoclonal antibody production was performed as described previously (69). Briefly, cauda epididymal sperm were collected from the ICR mouse strain, and sperm heads and tails were separated by mild sonication on ice. The sample was layered on a discontinuous sucrose gradient. Sperm heads were in the pellet after ultracentrifugation. The pellet fraction was used as antigen. Three female Wistar rats were immunized using the sperm head solution. Spleen cells were harvested after the third immunization and fused with mouse myeloma cells (P3U1). The hybridomas were screened by an enzyme-linked immunosorbent assay, and the positive clones were grown. Their supernatants were used for immunostaining and immunoblot analyses.
Immunostaining of Mouse Sperm.
Immunostaining of mouse sperm was performed as described previously (70). Briefly, all samples were mounted on glass slides and dried. After washing with phosphate-buffered saline (PBS), slides were blocked with 10% Normal Goat Serum/PBS (B6J) or 10% Newborn Calf Serum (NBCS)/PBS (B6D2F1) for 1 h and incubated with primary antibodies [rat monoclonal anti-mouse SPACA4 (KS139-281) (71), rat monoclonal anti-mouse IZUMO1 (KS64-125) (71, 72), and rabbit anti-mouse IZUMO1 (4)] in blocking buffer at 4 °C overnight. After washing with PBS + 0.1% Tween 20 (PBS-T) (B6J) or 10% NBCS/PBS-T (B6D2F1), the slides were incubated with secondary antibodies (goat anti-rat IgG Alexa Fluor 488 [A11006], goat anti-rat IgG Alexa Fluor 546 [A11081], and goat anti-rabbit IgG Alexa Fluor 488 [A11008], all purchased from Thermo Fisher Scientific) in blocking buffer for 1 h. After washing with PBS containing 0.05% Tween-20, the slides were observed under a fluorescence microscope (B6J: Axio Imager.Z2, Zeiss; B6D2F1: IX70, Olympus). The mouse strain Tg(Acr-EGFP)1Osb was used to identify spermatozoa with unreacted acrosomes (65).
Western Blotting.
Immunoblot analysis from mouse sperm was performed as described previously (69). Briefly, sperm samples were collected from the cauda epididymis and vas deferens. These samples were homogenized in lysis buffer containing 1% Triton X-100 and 1% protease inhibitors (Nacalai Tesque), centrifuged, and the supernatants were collected. Protein lysates were resolved by SDS/polyacrylamide gel electrophoresis (PAGE) under reducing condition and transferred to polyvinylidene fluoride (PVDF) membranes (Merck Millipore). After blocking, blots were incubated with primary antibodies [rat monoclonal anti-mouse SPACA4 (KS139-281) (71), mouse monoclonal anti-ADAM3 sc-365288 (Santa Cruz Biotechnology), rat monoclonal anti-IZUMO1 KS64-125 (71, 72), goat polyclonal anti-BASIGIN/EMMPRIN sc-9757, or mouse monoclonal anti-BASIGIN/EMMPRIN sc-46700 (Santa Cruz Biotechnology)] overnight at 4 °C and then incubated with secondary antibodies conjugated with horseradish-peroxidase (HRP) (HRP-conjugated goat anti-rat IgGs [112-035-167] and HRP-conjugated goat anti-mouse IgGs [115-036-062] [Jackson ImmunoResearch Laboratories]). The detection was performed using an enhanced chemiluminescence (ECL) plus Western blotting detection kit (GE Healthcare) and Chemi-Lumi One Super (Nacalai Tesque).
Protein Sequence Alignments of Fertility Factors.
Protein sequences of human CD9 (P21926), IZUMO1 (Q8IYV9), SPACA6 (W5XKT8), JUNO/IZUMO1R (A6ND01), TMEM95 (Q3KNT9), FIMP (Q96LL3), and mouse LLCFC/SOF1 (Q9D9P8) were downloaded from uniprot (https://www.uniprot.org/) and searched via the National Center for Biotechnology Information (NCBI) protein BLAST blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) for homologous protein sequences in Mus musculus (taxid:10090) or Homo sapiens (taxid:9606), reptiles (taxid:8459), Xenopus (taxid:8353), fugu (taxid:31032), Danio rerio (taxid:7955), platypus (taxid:9258), and armadillo (taxid:9359). If no homologous protein sequence was found, the search was extended to translated nucleotide sequences using tblastn. Protein sequence alignments were generated with clustal omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) using default parameters and plotted with JalView (https://www.jalview.org/).
Bioinformatic Analyses of Ly6/uPAR Genes.
Our analysis of the mouse Ly6/uPAR family considered 66 mouse genes annotated in the Ensembl release 79 of GRCm38. The Ensembl release was chosen to match the mouse expression data (73). The set of genes was selected based on sequence similarity to the mouse Ly6/uPAR family published in ref. 43 and domain similarity searches with Ly6/uPAR domain Hidden Markov Models (HMMs) against the Ensembl proteome (42). For defining the set of human orthologs of the mouse Ly6/uPAR genes, mouse gene symbols were used to query against Diopt version 8.0 (74), and hits were only considered with Best.Score, Best.Score.Reverse, and Rank “high.” Mouse expression data were obtained from ref. 73. Human expression data were obtained from GTEx version 8 (www.gtexportal.org; GTEx_Analysis_2017-06-05_v8_RNASeQCv1.1.9_gene_median_tpm). Xenopus laevis expression data were derived from the NCBI Gene Expression Omnibus (GEO) entry GSE73419 (75). Danio rerio expression data were derived from GEO entries GSE111882 (testis, ovary, and mature oocytes) (42), GSE147112 (oogenesis and mature oocytes) (76), and GSE171906 (adult tissues) (11). Heatmaps were used to illustrate the relative expression levels of genes across tissues. To this end, published library size and length-corrected expression values (transcript per million mapped reads (TPM)/fragments per kilo base per million mapped reads (FPKM)) were obtained; after square-root–based variance stabilizing transformation, the obtained values were centered and scaled per gene (z-score).
Statistical Analysis.
Statistical analysis was performed with the GraphPad Prism 7 software. Statistical tests are detailed in each figure legend. Differences were considered significant at *P < 0.05, **P < 0.01, and ***P < 0.001; n.s., not significant). Error bars represent SD. Figure legends indicate the number of n values and number of experiments for each analysis.
Supplementary Material
Acknowledgments
We thank the animal facility personnel at the Research Institute of Molecular Pathology (IMP)/Institute of Molecular Biotechnology (IMBA), in particular Jasmin Ecker and Lukas Laister, and Naoko Nagasawa at Osaka University/National Cerebral and Cardiovascular Center for excellent care of animals and for help with mouse husbandry; J.R. Wojciechowski and L. Haas from the Obenauf laboratory, A. Fedl from the Busslinger laboratory, D. Keays and I. Baptista from the Keays laboratory, and E. Chatzidaki and J. Gassler from the Tachibana laboratory for technical assistance and advice with mouse work; K. Aumayer and her team at the biooptics facility at the Vienna Biocenter for support with microscopy; and the entire A.P. laboratory, particularly Krista Gert and Victoria Deneke, for valuable discussions and comments on the project and manuscript. This work was supported on the Austrian side (A.P. laboratory) by the IMP, Boehringer Ingelheim, the Austrian Academy of Sciences, the Austrian Research Promotion Agency (FFG) (Headquarter Grant FFG-852936), the Austrian Science Fund (FWF) START program (Y 1031-B28) to A.P., the Human Frontier Science Program (HFSP) Career Development Award (CDA00066/2015) and the HFSP Young Investigator Award to A.P., European Molecular Biology Organization (EMBO) Young Investigator Programme (YIP) funds to A.P., a doctoral (DOC) Fellowship from the Austrian Academy of Sciences to S.H., and a Boehringer Ingelheim Fonds PhD fellowship to A.B. This work was supported on the Japanese side (M.I. laboratory and Y.F. laboratory) by Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science KAKENHI Grants JP15H05573, JP16KK0180, JP20KK0155, JP21K19198, and JP21H02397 to Y.F. and JP19H05750, JP21H04753, and JP21H05033 to M.I.; Japan Agency for Medical Research and Development Grant JP21gm5010001 to M.I.; Takeda Science Foundation Grants to Y.F. and M.I.; a Mochida Memorial Foundation for Medical and Pharmaceutical research grant to Y.F.; the Sumitomo Foundation Grant for Basic Science Research Projects to Y.F.; a Senri Life Science Foundation grant to Y.F.; the Intramural Research Fund (Grants 21-2-6, 30-2-5, and 31-6-3) for Cardiovascular Diseases of the National Cerebral and Cardiovascular Center to Y.F.; Eunice Kennedy Shriver National Institute of Child Health and Human Development Grants R01HD088412 and P01HD087157 to M.I.; and the Bill & Melinda Gates Foundation (Grant INV-001902 to M.I.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108777118/-/DCSupplemental.
Data Availability
Previously published data were used for this work that are available on the publicly accessible database GEO [Danio rerio expression data were derived from GEO entries GSE111882 (testis, ovary, and mature oocytes) (42), GSE147112 (oogenesis and mature oocytes) (76), and GSE171906 (adult tissues) (11)]. The Spaca4 knockout mice, C57BL/6N-Spaca4tm1Osb, were deposited into the RIKEN BioResource Center (https://mus.brc.riken.jp/en/) and are available to the scientific community. All data are included in the article and/or SI Appendix, and are freely available to the scientific community.
References
- 1.Evans J. P., Sperm-egg interaction. Annu. Rev. Physiol. 74, 477–502 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Krauchunas A. R., Marcello M. R., Singson A., The molecular complexity of fertilization: Introducing the concept of a fertilization synapse. Mol. Reprod. Dev. 83, 376–386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okabe M., Sperm-egg interaction and fertilization: Past, present, and future. Biol. Reprod. 99, 134–146 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Inoue N., Ikawa M., Isotani A., Okabe M., The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434, 234–238 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Satouh Y., Inoue N., Ikawa M., Okabe M., Visualization of the moment of mouse sperm-egg fusion and dynamic localization of IZUMO1. J. Cell Sci. 125, 4985–4990 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Lorenzetti D., et al., A transgenic insertion on mouse chromosome 17 inactivates a novel immunoglobulin superfamily gene potentially involved in sperm-egg fusion. Mamm. Genome 25, 141–148 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Noda T., et al., Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm-oocyte fusion in mice. Proc. Natl. Acad. Sci. U.S.A. 117, 11493–11502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Fuertes B., et al., Subfertility in bulls carrying a nonsense mutation in transmembrane protein 95 is due to failure to interact with the oocyte vestments. Biol. Reprod. 97, 50–60 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Pausch H., et al., A nonsense mutation in TMEM95 encoding a nondescript transmembrane protein causes idiopathic male subfertility in cattle. PLoS Genet. 10, e1004044 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujihara Y., et al., Spermatozoa lacking fertilization influencing membrane protein (FIMP) fail to fuse with oocytes in mice. Proc. Natl. Acad. Sci. U.S.A. 117, 9393–9400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noda T., et al., Sperm membrane proteins DCST1 and DCST2 are required for the sperm-egg fusion process in mice and fish. bioRxiv [Preprint] (2021) 10.1101/2021.04.18.440256 (Accessed 7 May 2021). [DOI]
- 12.Inoue N., Hagihara Y., Wada I., Evolutionarily conserved sperm factors, DCST1 and DCST2, are required for gamete fusion. eLife 10, 1–3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaji K., et al., The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat. Genet. 24, 279–282 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Miyado K., et al., Requirement of CD9 on the egg plasma membrane for fertilization. Science 287, 321–324 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Le Naour F., Rubinstein E., Jasmin C., Prenant M., Boucheix C., Severely reduced female fertility in CD9-deficient mice. Science 287, 319–321 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Bianchi E., Doe B., Goulding D., Wright G. J., Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aydin H., Sultana A., Li S., Thavalingam A., Lee J. E., Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature 534, 562–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohto U., et al., Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization. Nature 534, 566–569 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kato K., et al., Structural and functional insights into IZUMO1 recognition by JUNO in mammalian fertilization. Nat. Commun. 7, 12198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han L., et al., Divergent evolution of vitamin B9 binding underlies Juno-mediated adhesion of mammalian gametes. Curr. Biol. 26, R100–R101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rankin T. L., et al., Human ZP3 restores fertility in Zp3 null mice without affecting order-specific sperm binding. Development 125, 2415–2424 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Rankin T., et al., Mice homozygous for an insertional mutation in the Zp3 gene lack a zona pellucida and are infertile. Development 122, 2903–2910 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Gahlay G., Gauthier L., Baibakov B., Epifano O., Dean J., Gamete recognition in mice depends on the cleavage status of an egg’s zona pellucida protein. Science 329, 216–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avella M. A., Baibakov B., Dean J., A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. J. Cell Biol. 205, 801–809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avella M. A., Xiong B., Dean J., The molecular basis of gamete recognition in mice and humans. Mol. Hum. Reprod. 19, 279–289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grayson P., Izumo1 and Juno: The evolutionary origins and coevolution of essential sperm-egg binding partners. R. Soc. Open Sci. 2, 150296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanson W. J., Vacquier V. D., The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Levitan D. R., Do sperm really compete and do eggs ever have a choice? Adult distribution and gamete mixing influence sexual selection, sexual conflict, and the evolution of gamete recognition proteins in the sea. Am. Nat. 191, 88–105 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Kosman E. T., Levitan D. R., Sperm competition and the evolution of gametic compatibility in externally fertilizing taxa. Mol. Hum. Reprod. 20, 1190–1197 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Evans J. P., Sherman C. D. H., Sexual selection and the evolution of egg-sperm interactions in broadcast-spawning invertebrates. Biol. Bull. 224, 166–183 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Palumbi S. R., Speciation and the evolution of gamete recognition genes: Pattern and process. Heredity 102, 66–76 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Dresselhaus T., Sprunck S., Wessel G. M., Fertilization mechanisms in flowering plants. Curr. Biol. 26, R125–R139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gert K. R., Pauli A., Species-Specific Mechanisms during Fertilization (Elsevier Inc., ed. 1, 2019). [DOI] [PubMed] [Google Scholar]
- 34.Raj I., et al., Structural basis of egg coat-sperm recognition at fertilization. Cell 169, 1315–1326.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyon J. D., Vacquier V. D.. Regions mediating species-specific recognition of the abalone egg vitelline envelope. Dev. Biol. 159, 151–159 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Swanson W. J., Vacquier V. D., The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc. Natl. Acad. Sci. U.S.A. 94, 6724–6729 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis C. A., Talbot C. F., Vacquier V. D., A protein from abalone sperm dissolves the egg vitelline layer by a nonenzymatic mechanism. Dev. Biol. 92, 227–239 (1982). [DOI] [PubMed] [Google Scholar]
- 38.Okabe M., Beware of memes in the interpretation of your results—Lessons from gene-disrupted mice in fertilization research. FEBS Lett. 592, 2673–2679 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Park S., et al., CRISPR/Cas9-mediated genome-edited mice reveal 10 testis-enriched genes are dispensable for male fecundity. Biol. Reprod. 103, 195–204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikawa M., et al., Fertilization: A sperm’s journey to and interaction with the oocyte. J. Clin. Invest. 120, 984–994 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shetty J., et al., SAMP14, a novel, acrosomal membrane-associated, glycosylphosphatidylinositol-anchored member of the Ly-6/urokinase-type plasminogen activator receptor superfamily with a role in sperm-egg interaction. J. Biol. Chem. 278, 30506–30515 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Herberg S., Gert K. R., Schleiffer A., Pauli A., The Ly6/uPAR protein Bouncer is necessary and sufficient for species-specific fertilization. Science 361, 1029–1033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loughner C. L., et al., Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum. Genomics 10, 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst C., Eling N., Martinez-Jimenez C. P., Marioni J. C., Odom D. T., Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun. 10, 1251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujihara Y., Miyata H., Ikawa M., Factors controlling sperm migration through the oviduct revealed by gene-modified mouse models. Exp. Anim. 67, 91–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiyozumi D., et al., NELL2-mediated lumicrine signaling through OVCH2 is required for male fertility. Science 1135, 1132–1135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujihara Y., Oji A., Kojima-Kita K., Larasati T., Ikawa M., Co-expression of sperm membrane proteins CMTM2A and CMTM2B is essential for ADAM3 localization and male fertility in mice. J. Cell Sci. 131, 1–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behringer R., Gertsenstein M., Kristina Nagy A. N., Manipulating the Mouse Embryo (Cold Spring Harbor Laboratory Press, 2014). [Google Scholar]
- 49.Salonen I., Kallajoki M., Monoclonal antibody against human sperm acrosome inhibits sperm penetration of zona-free hamster eggs. Int. J. Androl. 10, 731–739 (1987). [DOI] [PubMed] [Google Scholar]
- 50.Baba T., Azuma S., Kashiwabara S., Toyoda Y., Sperm from mice carrying a targeted mutation of the acrosin gene can penetrate the oocyte zona pellucida and effect fertilization. J. Biol. Chem. 269, 31845–31849 (1994). [PubMed] [Google Scholar]
- 51.Blobel C. P., et al., A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature 356, 248–252 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Cho C., et al., Fertilization defects in sperm from mice lacking fertilin β. Science 281, 1857–1859 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Okabe M., et al., Homology of an acrosome-reacted sperm-specific antigen to CD46. J. Pharmacobiodyn. 15, 455–459 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Inoue N., et al., Disruption of mouse CD46 causes an accelerated spontaneous acrosome reaction in sperm. Mol. Cell. Biol. 23, 2614–2622 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okabe M., The cell biology of mammalian fertilization. Development 140, 4471–4479 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Fujihara Y., et al., Identification of multiple male reproductive tract-specific proteins that regulate sperm migration through the oviduct in mice. Proc. Natl. Acad. Sci. U.S.A. 116, 18498–18506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujihara Y., Okabe M., Ikawa M., GPI-anchored protein complex, LY6K/TEX101, is required for sperm migration into the oviduct and male fertility in mice. Biol. Reprod. 90, 60 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Noda T., et al., Seminal vesicle secretory protein 7, PATE4, is not required for sperm function but for copulatory plug formation to ensure fecundity. Biol. Reprod. 100, 1035–1045 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hargreaves A. D., Swain M. T., Hegarty M. J., Logan D. W., Mulley J. F., Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins. Genome Biol. Evol. 6, 2088–2095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirohashi N., Yanagimachi R., Sperm acrosome reaction: Its site and role in fertilization. Biol. Reprod. 99, 127–133 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Vasilyeva N. A., Loktyushov E. V., Bychkov M. L., Shenkarev Z. O., Lyukmanova E. N., Three finger proteins from the Ly6 / uPAR family: Functional diversity within one structural motif. Biochemistry (Mosc.) 82, 1702–1715 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Ferrer M. J. S., Xu W., Shetty J., Herr J., Oko R., Plasminogen improves mouse IVF by interactions with inner acrosomal membrane-bound MMP2 and SAMP14. Biol. Reprod. 94, 88 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Levitan D. R., Ferrell D. L., Selection on gamete recognition proteins depends on sex, density, and genotype frequency. Science 312, 267–270 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Firman R. C., Gasparini C., Manier M. K., Pizzari T., Postmating female control: 20 years of cryptic female choice. Trends Ecol. Evol. 32, 368–382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakanishi T., et al., Real-time observation of acrosomal dispersal from mouse sperm using GFP as a marker protein. FEBS Lett. 449, 277–283 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Gagnon J. A., et al., Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9, e98186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujihara Y., Kaseda K., Inoue N., Ikawa M., Okabe M., Production of mouse pups from germline transmission-failed knockout chimeras. Transgenic Res. 22, 195–200 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Goodson S. G., Zhang Z., Tsuruta J. K., Wang W., O’Brien D. A., Classification of mouse sperm motility patterns using an automated multiclass support vector machines model. Biol. Reprod. 84, 1207–1215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamaguchi R., Yamagata K., Ikawa M., Moss S. B., Okabe M., Aberrant distribution of ADAM3 in sperm from both angiotensin-converting enzyme (Ace)- and calmegin (Clgn)-deficient mice. Biol. Reprod. 75, 760–766 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Yamaguchi R., et al., Cd52, known as a major maturation-associated sperm membrane antigen secreted from the epididymis, is not required for fertilization in the mouse. Genes Cells 13, 851–861 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Fujihara Y., et al., Expression of TEX101, regulated by ACE, is essential for the production of fertile mouse spermatozoa. Proc. Natl. Acad. Sci. U.S.A. 110, 8111–8116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ikawa M., et al., Calsperin is a testis-specific chaperone required for sperm fertility. J. Biol. Chem. 286, 5639–5646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li B., et al., A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA-seq. Sci. Rep. 7, 4200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu Y., et al., An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Session A. M., et al., Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cabrera-Quio L. E., Schleiffer A., Mechtler K., Pauli A., Zebrafish Ski7 tunes RNA levels during the oocyte-to-embryo transition. PLoS Genet. 17, e1009390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published data were used for this work that are available on the publicly accessible database GEO [Danio rerio expression data were derived from GEO entries GSE111882 (testis, ovary, and mature oocytes) (42), GSE147112 (oogenesis and mature oocytes) (76), and GSE171906 (adult tissues) (11)]. The Spaca4 knockout mice, C57BL/6N-Spaca4tm1Osb, were deposited into the RIKEN BioResource Center (https://mus.brc.riken.jp/en/) and are available to the scientific community. All data are included in the article and/or SI Appendix, and are freely available to the scientific community.