Abstract
The Ste20/p21-activated kinase homolog Shk1 is essential for viability and required for normal morphology, mating, and cell cycle control in the fission yeast Schizosaccharomyces pombe. Shk1 is regulated by the p21 G protein Cdc42, which has been shown to form a complex with the SH3 domain protein Scd2 (also called Ral3). In this study, we investigated whether Scd2 plays a role in regulating Shk1 function. We found that recombinant Scd2 and Shk1 interact directly in vitro and that they interact in vivo, as determined by the two-hybrid assay and genetic analyses in fission yeast. The second of two N-terminal SH3 domains of Scd2 is both necessary and sufficient for interaction with Shk1. While full-length Scd2 interacted with only the R1 N-terminal regulatory subdomain of Shk1, a C-terminal deletion mutant of Scd2 interacted with both the R1 and R3 subdomains of Shk1, suggesting that the non-SH3 C-terminal domain of Scd2 may be involved in defining specificity in SH3 binding domain recognition. Overexpression of Scd2 stimulated the autophosphorylation activity of wild-type Shk1 in fission yeast but, consistent with results of genetic analyses, did not stimulate the activity of a Shk1 protein lacking the R1 subdomain. Results of additional two-hybrid experiments suggest that Scd2 may stimulate Shk1 catalytic function, at least in part, by positively modulating protein-protein interaction between Cdc42 and Shk1. We propose that Scd2 functions as an organizing center, or scaffold, for the Cdc42 complex in fission yeast and that it acts in concert with Cdc42 to positively regulate Shk1 function.
p21-activated kinases (PAKs) have been remarkably conserved through evolution, with homologs identified in eukaryotes ranging from yeasts to mammals (34). PAKs have diverse functions in eukaryotic organisms, including roles in regulation of cytoskeletal organization and cellular morphology (21, 24, 27, 35), growth factor-induced signaling pathways (14, 19, 24, 28, 43), mitosis and meiosis (11, 12, 16, 39), and apoptosis (31). PAKs are direct binding targets for the related p21 G proteins Cdc42 and Rac, but they do not bind to Rho, Ras, or other small G proteins (34). Cdc42 and Rac bind in a GTP-dependent fashion to a highly conserved sequence, referred to as the CRIB (Cdc42 and Rac interactive binding) domain, found in the N-terminal regulatory domains of all characterized PAKs (6). In most cases, PAK catalytic activity can be stimulated by Cdc42 and Rac in vitro (34). However, the mammalian Pak4 kinase cannot be stimulated by Cdc42 and Rac, even though it binds to both proteins (1), and a PAK identified from Acanthamoeba can be stimulated by Cdc42 and Rac only in the presence of acidic lipids (5). These findings, combined with the diverse cellular functions attributed to PAKs, lead to the hypothesis that there may be additional regulators for PAKs besides Cdc42 and Rac.
Most known PAKs contain proline rich sequences within their N-terminal regulatory domains that could serve as potential docking sites for SH3 domain proteins involved in PAK regulation. Indeed, it has been demonstrated that mutations of potential SH3 binding sites in Pak1/α-Pak reduce the efficiency with which it triggers cytoskeletal changes when overexpressed in mammalian cells (13, 35). In addition, it has been shown in mammalian cells that one of the SH3 domains in the adapter protein Nck interacts with Pak1 (4, 35), and the SH3 domain in α- and β-PIX (also called Cool-1 and Cool-2), two closely related Cdc42/Rac guanine nucleotide exchange factors (GEFs), interacts with both Pak1/α-Pak and Pak3/β-Pak (3, 23). Overexpression of either Nck or PIX leads to activation of PAKs in vivo (4, 10, 23, 35), and recombinant PIX protein can activate PAK immunoprecipitated from mammalian cells in vitro (10). Nck and PIX have been proposed to function primarily in the recruitment of PAKs to specific cellular locales (i.e., focal complexes by PIX and growth factor-receptor tyrosine kinase complexes by Nck) where PAKs can be subsequently activated by Cdc42 and Rac. A budding yeast PAK, Ste20, has also been shown to form a complex with an SH3 domain protein, Bem1, although it has not yet been demonstrated whether the two proteins interact directly or whether Bem1 plays any role in regulating Ste20 catalytic function (20).
We have been studying the function and regulation of PAKs in the fission yeast Schizosaccharomyces pombe, which possesses two known PAKs, Shk1 (also called Pak1 and Orb2) and Shk2 (also called Pak2) (24, 27, 33, 39, 42). Shk1 is essential for viability of fission yeast cells and is required for normal cell morphology and cytoskeletal regulation, efficient mating response, and proper cell cycle regulation (16, 24, 27, 39, 42). The second known fission yeast PAK, Shk2, is dispensable for normal growth, morphology, and mating and appears to be largely redundant in function with Shk1 (33, 42). The N-terminal regulatory domain of Shk1 can be divided into three subdomains, which we have designated R1 (amino acid residues 1 to 146), R2 (amino acid residues 147 to 203), and R3 (amino acid residues 204 to 380) (Fig. 1). The R2 subdomain contains the CRIB domain. Neither the R1 nor R3 subdomain of Shk1 exhibits significant homology with the corresponding domains of other known PAKs. A variety of genetic data indicate that Shk1 is a key effector for Cdc42 in fission yeast. These data include the observations that the terminal phenotypes resulting from shk1 and cdc42 null mutations are similar (growth arrest as small, round cells) (24, 26, 27), that overexpression of dominant inhibitory alleles of the cdc42 and shk1 genes causes similar defects in morphology and mating (24, 27), and that gain of Shk1 function can partially suppress the mating defect of fission yeast cells expressing a dominant inhibitory mutant allele of cdc42 (24). Furthermore, Tu and Wigler recently used the two-hybrid assay to provide evidence that Cdc42 may activate Shk1 in vivo by blocking autoinhibitory association of the Shk1 regulatory and catalytic domains (37). We previously described a second potential regulator of Shk1, Skb1, which interacts with the R3 subdomain of Shk1 (17). Skb1 functions as a mitotic inhibitor in fission yeast, and this function is dependent on Shk1 (16). Genetic data suggest that Skb1, like Cdc42, functions to positively modulate Shk1 function in vivo. Although genetic data strongly suggest that Cdc42 and Skb1 positively regulate Shk1 function in vivo, we have been unable to detect direct stimulation of Shk1 catalytic function by either purified Skb1 or Cdc42 protein in vitro (27a).
FIG. 1.
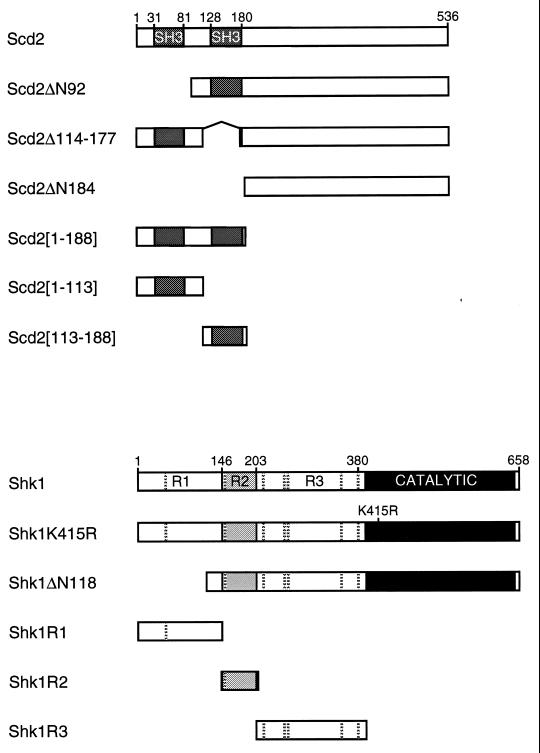
Schematic representation of various forms of Scd2 and Shk1 proteins used in this study. For two-hybrid experiments, scd2 coding sequences were fused to the GBD-encoding sequence in plasmid pHP5, while shk1 coding sequences were fused to the GAD-encoding sequence in plasmid pGADGH (Materials and Methods). Hatched horizontal bars shown in Shk1 proteins indicate the positions of PxxP motifs that could potentially serve as SH3 binding sites. The R2 subdomain of Shk1 contains the Cdc42 binding domain.
In a previous study (7), we demonstrated that Cdc42 forms a quaternary protein complex containing the Ras proto-oncoprotein homolog Ras1, the presumptive Cdc42 GEF Scd1 (also called Ral1 [15]), and Scd2 (also called Ral3 [15]). Scd2 possesses two N-terminally positioned SH3 domains (Fig. 1) and is both structurally and functionally related to the budding yeast protein Bem1 (7). Since Shk1 has several potential SH3 docking sites in its N-terminal regulatory domain (Fig. 1), we conducted a study to determine whether Scd2 and Shk1 proteins interact and, if so, whether and how Scd2 might affect the catalytic function of Shk1. In this report, we present in vitro and in vivo evidence for SH3 domain-dependent interaction between Scd2 and Shk1 and show that Scd2 stimulates Shk1 catalytic function in vivo. We also provide evidence that Scd2 positively modulates the interaction between Cdc42 and Shk1 in vivo. These and additional results described herein suggest that Scd2 is a direct regulator of Shk1 in fission yeast.
MATERIALS AND METHODS
Yeast strains and manipulations.
The S. pombe strain used was CHP428 (h+ ade6-210 his7-366 leu1-32 ura4-D18) (42). The Saccharomyces cerevisiae two-hybrid reporter strains used were SFY526 (MATa ade2-201 his3-200 leu2-3,112 lys2-801 trp1-901 ura3-52 canR gla4-542 gal80-538, URA3::GAL1UAS-GAL1TATA-lacZ) (Clontech) and L40 (MATa ade2 his3 leu2 trp1 LYS2::lexA-HIS3 URA3::lexA-lacZ) (40). Yeast cells were transformed by the lithium acetate procedure (2). S. pombe cultures were grown on either YEA (2% yeast extract, peptone, 2% dextrose, 75 mg of adenine per liter) or synthetic minimal medium (EMM) with appropriate supplements (2). S. cerevisiae cultures were grown in dropout medium with supplements (30). Freshly transformed cells were used in those studies in which full length Shk1 and Scd2 were overexpressed in fission yeast because the levels of expression of these proteins decreased dramatically as cultures aged.
Plasmids.
The backbone two-hybrid plasmids were pGADGH (for expression of Gal4 transcriptional activation domain [GAD] fusions) and pHP5 and pGBT9 (for expression of Gal4 DNA binding domain [GBD] fusions) (42). Plasmids pGADShk1, pGADShk1ΔR1, pGADShk1R3, pGADScd1[40-872], pGADRas1, pGADCdc42, pGBDScd2, pLBDShk1 GBDByr2, pAAUCMShk1, and pAAUCMShk1ΔN118 have been described elsewhere (17, 24, 42). pGBDShk1, for expression of GBD-Shk1, was constructed by cloning the shk1 protein coding sequence from pLBDShk1 into pHP5. PCRs were used to amplify the Shk1 R1 and R2 subdomain-encoding as well as the Shk1ΔN118-encoding sequences for cloning into pGADGH to produce pGADShk1R1, pGADShk1R2, and pGADShk1ΔN118. The Shk1 R3 subdomain-encoding sequence from pGADGHShk1R3 was cloned into pAAUCM (24) for expression of c-Myc epitope-tagged Shk1R3 (CMShk1R3) in fission yeast. pTrcHisShk1FL was constructed by cloning a BamHI/SalI fragment encoding Shk1 from pLBDShk1 into pTrcHisB (Invitrogen). pTrcHisH-RasG12V has been described elsewhere (36). pREP1Scd2 and pTrcHisScd2 were constructed by cloning a BamHI fragment of Scd2 (7) into the corresponding sites of pREP1 (25) and pTrcHisB, respectively. pAAUCMShk1R1 was constructed by cloning a BamHI/KpnI fragment encoding Shk1R1 from pGADGHShk1R1 into pAAUCM. pAAScd2 has been described previously (7) and was used to express Scd2 from a third vector in the two-hybrid host strain SFY526. Scd2ΔN92 and Scd2ΔN184 coding sequences were amplified by PCR and cloned into pART1CM (7), for expression of c-Myc epitope-tagged Scd2ΔN92 (CMScd2ΔN92), and into pGADGH and pHP5 to create pART1CMScd2ΔN92 and pGBDScd2ΔN92, respectively. The Scd2ΔN92 coding sequence was also cloned into pTrcHisB and pRP259 (7), which express cloned genes as polyhistidine (His6) and glutathione S-transferase (GST) fusion proteins, respectively, to create pTrcHisScd2ΔN92, and pGSTScd2ΔN92. Similarly, we created pART1CMScd2ΔN184, pGADScd2ΔN184, pGBDScd2ΔN184, pTrcHisScd2ΔN184, and pGSTScd2ΔN184. The DNA sequences encoding the second SH3 domain of Scd2 (residues 128 to 180 [Fig. 1]) are flanked by SacI (5′) and NheI (3′) sites. To delete the second SH3 domain from Scd2 to create Scd2Δ114-177, we exchanged the original SacI/NheI fragment with a new PCR-amplified fragment, scd2Δ114-177, in which the SacI site was placed after the coding region for the second SH3 domain. The PCR-amplified Scd2Δ114-177 encoding sequence was cloned into pHP5 and pART1CM to create pGBDScd2Δ114-177 and pART1CMScd2Δ114-177. To construct pGBDScd21-113, a novel stop codon was introduced at the SacI site in pGBDScd2Δ114-177 by an amber linker (CTAGTCTAGACTAG; New England Biolabs). The coding sequence for Scd21-188 was created by PCR. This fragment was cloned into pHP5 to create pGBDScd21-188. To construct pTrcHisScd2113-188, pGBDScd21-188 was digested with SacI and PstI, and the resulting fragment was cloned into pTrcHisB. To generate pGBDScd2113-188, a BamHI fragment capable of encoding Scd2113-188 was isolated from pTrcHisScd2113-188 and cloned into pHP5. A PstI/EcoRI fragment containing the coding sequence for c-Myc-Byr1 (41) was cloned into pTrcHisC to create pTrcHisCMBYR1. The sequences of all PCR products were confirmed by sequencing.
β-Galactosidase assay.
The filter assay for testing two-hybrid interactions was performed as described previously (38). The liquid assay for β-galactosidase activity was performed as described elsewhere (30); β-galactosidase activity (Miller units) was calculated by using the following formula: (A420 × 1.7)/(0.0045 × protein concentration [mg/ml] × extract volume [ml] × time [min]).
Quantitative S. pombe mating Assays.
Mating assays were performed essentially as described previously (18). Briefly, transformants were patched on EMM plates and incubated for 3 days at 30°C. The percentage of mating (number of asci/total number of cells) was then determined by microscopic analysis. Values represent the average mating detected for at least three independent transformants.
Far-Western analysis.
Plasmids expressing GST and His6-tagged proteins were transformed into Escherichia coli BL21(DE3)pLysS. The lysis buffer for various forms of GST-tagged Scd2 was phosphate-buffered saline plus 1 mM phenylmethylsulfonyl fluoride. Triton X-100 was added to the lysate to 1%. The final crude lysate was incubated on ice for 30 min and then cleared by centrifugation. Purified proteins were resuspended in phosphate-buffered saline to a final concentration of 1 μg/ml. The lysis buffer for His6-Shk1 and Scd1ΔN was the 1× binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]; Novagen). The protocol for far-Western analysis is essentially as described elsewhere (8). Approximately 1 μg each of purified His6-tagged protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane, and then probed with 10 μg of purified GST-tagged Scd2 proteins (4°C, 1 h). The presence of GST-tagged proteins was detected by Western blotting using antibody specific for GST (Sigma).
Immune complex kinase assays.
c-Myc epitope-tagged proteins were immunoprecipitated from yeast lysates by the method described previously except that 1% NP-40 was included in the lysis buffer (1). Immunoprecipitates were washed three times with lysis buffer and once with kinase buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgCl2, 1 mM MnCl2). During kinase buffer wash, samples were divided in two. One set was resuspended in 25 μl of kinase buffer containing 10 μCi of [γ-32P]ATP (6,000 Ci/mmol), 20 μM ATP, and 2.5 μg of myelin basic protein (Sigma). The other set was processed the same way as the kinase reaction but without [γ-32P]ATP. Kinase reactions were terminated after 20 min at 30°C and handled as described previously (42).
Kinase assays using bacterially expressed recombinant proteins.
BL-21(DE3)pLysS cells were transformed with pTrcHisScd2 and grown in 2×-YT at 37°C. Subsequently, they were induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) for 2 h after reaching an A600 of ≈0.6. The lysis buffer (5 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 7.9]) contains a protease inhibitor cocktail of 1 mM phenylmethylsulfonyl fluoride, 10 μM pepstatin A, leupeptin (50 μg/ml), 36 E-64 (36 μg/ml), and aprotinin (10 μg/ml), which was used in all buffers for protein purification. After clarification for 50 min at 30,000 × g at 4°C, the lysate was bound batchwise with the nickel resin (Invitrogen) for 1.5 h at 4°C. The resin was washed with wash buffer (60 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 7.9]) at 4°C. His6-Scd2 was eluted batchwise with 1 M imidazole in wash buffer at 4°C. Purified His6-Scd2 was concentrated and exchanged into 50 mM Tris-HCl (pH 8.0) in a Centriprep-5 (Millipore) as instructed by manufacturer. Purified His6-Scd2 protein was stored in 10% glycerol with 0.1% Triton X-100 and 10 mM reduced glutathione at −80°C.
pTrcHisShk1 and pTrcHisH-RasG12V were transformed into BL21(DE3)pLYSS cells, and cultures were grown as described above except that they were incubated at 30°C. The lysis buffers were 50 mM NaF–10 mM Na3VO4–10 mM C3H7O6PNa2–137 mM NaCl–50 mM Tris-HCl (pH 7.5) for pTrcHisShk1-transformed cells and 10 mM MgCl2–150 mM NaCl–50 mM Tris-HCl (pH 7.5) for pTrcHisH-RasG12V cells. Lysates were incubated for 1 h at 4°C in the presence of 10% glycerol and 0.5% dodecyl-β-d-maltoside (Calbiochem) for His6-H-RasG12V or in the presence of 1% NP-40 and 10% glycerol for His6-Shk1. After centrifugation for 60 min at 30,000 × g at 4°C, the clarified lysate was incubated overnight with nickel resin. The resin was washed with 20 mM imidazole–300 mM NaCl–50 mM Tris-HCl (pH 7.0)–2 mM MgCl2 for His6-H-RasG12V or 50 mM Tris-HCl (pH 7.0)–300 mM NaCl–20 mM imidazole–50 mM NaF–10 mM Na3 VO4–10 mM C3H7O6PNa2 for His6-Shk1. His6-H-RasGV12 and His6-Shk1 proteins were eluted batchwise, using wash buffers containing 500 mM imidazole. Both His6-H-RasG12V and His6-Shk1 were concentrated and exchanged with 20 mM HEPES (pH 7.5)–100 mM NaCl–2 mM MgCl2–1 mM dithiothreitol as described above. Purified His6-H-RasG12V and His6-Shk1 proteins were stored in 15% glycerol at −80°C.
For kinase assays, approximately 80 ng of His6-Shk1 was mixed with test proteins as indicated in a total of 25 μl of kinase buffer (50 mM HEPES [pH 7.4], 10 mM MgCl2, 2 mM MnCl2, 1 mM dithiothreitol, 0.05% Triton X-100, 20 μM ATP, 10 μCi of [γ-32P]ATP (6,000 Ci/mmol]) and incubated at 30°C for 20 min. Reactions were terminated as described above. Proteins were resolved by SDS-PAGE on a 4 to 15% gradient gel and processed as described above.
RESULTS
Scd2 binds directly to Shk1 in vitro.
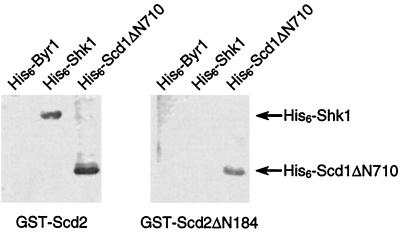
A far-Western assay was performed to address whether Shk1 and Scd2 can interact directly in vitro. Scd2 and Shk1 proteins were expressed as recombinant GST- and His6-tagged fusion proteins, respectively, in bacterial cells. Purified recombinant His6-tagged proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and incubated with either GST-Scd2 or GST fused to a truncated Scd2 protein lacking the two SH3 domains (GST-Scd2ΔN184) (Fig. 1). As shown in Fig. 2, GST-Scd2 was capable of binding to immobilized His6-Shk1 but not to His6-Byr1, which was used as a negative control. GST-Scd2ΔN184, by contrast, was not capable of binding to His6-Shk1 but was capable of binding a positive control, His6-Scd1ΔN710, which contains the C terminus of Scd1 that has been shown to bind directly to Scd2 (7). These data demonstrate that Scd2 binds directly to Shk1 in vitro and suggest the possibility that this interaction is dependent on at least one of the SH3 domains of Scd2.
FIG. 2.
Scd2 binds to Shk1 in vitro. Purified recombinant His6-Byr1 (0.5 μg; lane 1), His6-Shk1 (1 μg; lane 2), and His6-Scd1ΔN710 (1 μg; lane 3) proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were then subjected to far-Western analysis (Materials and Methods) by incubating with 10 μg of either GST-Scd2 (left) or GST-Scd2ΔN184 (right). Membrane bound GST-Scd2 proteins were detected by immunoblotting with anti-GST antibody. The positions of His6-Shk1 and His6-ScdΔN710 are indicated by the arrows.
Mapping the binding sites between Scd2 and Shk1 by using the yeast two-hybrid system.
We used the yeast two-hybrid system to map the domains of Scd2 and Shk1 that are required for the two proteins to interact. Full-length Scd2 and a series of Scd2 truncation and deletion mutant proteins were fused to the GBD, while a series of Shk1 and Shk1 truncation mutant proteins were fused to the GAD (Fig. 1). The results of two-hybrid interactions tested between these GBD-Scd2 and GAD-Shk1 fusion proteins are summarized in Table 1. Consistent with the far-Western data described above, full-length Shk1 protein (GAD-Shk1) interacted with full-length Scd2 protein (GBD-Scd2) but not with the Scd2 mutant protein lacking the two SH3 domains (GBD-Scd2ΔN184). We also determined that Scd2 did not interact detectably in the two-hybrid assay with the second known fission yeast PAK Shk2 (33, 42), with the budding yeast PAKs Cla4 (9) and Ste20 (19, 29), or with mammalian Pak1/α-Pak (22) (data not shown). These results demonstrate that the Scd2-Shk1 interaction is highly specific in nature.
TABLE 1.
Pairwise interactions between wild-type and mutant forms of Scd2 and Shk1 tested in the yeast two-hybrid assay
| GAD fusion protein | β-Galactosidase activity with indicated GBD fusion proteina
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Scd2 | Scd2ΔN92 | Scd2Δ 114-177 | Scd2ΔN184 | Scd2 [1-188] | Scd2 [1-113] | Scd2 [113-188] | Cdc42 | Byr2 | |
| Shk1 | + (20.26) | + (30.55) | − (0.11) | − (0.11) | + (57.26) | − (0.10) | + (89.22) | + | − |
| Shk1ΔN118 | − (0.29) | − | − | − | + | − | + (12.32) | + | − |
| Shk1R1 | + (27.20) | + | − | − | + | − | + (44.70) | − | ND |
| Shk1R2 | − (0.22) | − | − | − | − | − | − (0.10) | + | ND |
| Shk1R3 | − (0.18) | − | − | − | + | − | + (3.05) | − | ND |
| Scd1[40-872] | + (109.00) | + | + | + | − | − | − (0.11) | ND | − |
| Ras1 | − (0.17) | − (0.15) | − (0.15) | − (0.10) | − (0.14) | − (0.09) | − (0.12) | − | + |
+ and − indicate whether β-galactosidase activity was detected between pairs of GBD and GAD fusion proteins in the β-galactosidase filter assay (see Materials and Methods). At least eight independent transformants were tested for each determination made by the filter assay. Values in parentheses indicate β-galactosidase activity detected by the quantitative liquid β-galactosidase assay (see Materials and Methods). Values are expressed as Miller units and represent the average activity assayed for two independent yeast transformants. ND, not determined.
We next sought to define the domain of Shk1 with which Scd2 interacts. Shk1 has potential SH3 docking sites (PxxP motifs) in each of its three N-terminal regulatory subdomains (R1, R2, and R3 [Fig. 1]). We found that GBD-Scd2 interacted with a GAD fusion of the Shk1 R1 subdomain (GAD-Shk1R1) but not with an N-terminally truncated Shk1 protein lacking most of the R1 subdomain (GAD-Shk1ΔN118), including the first PxxP motif, or with either the Shk1 R2 or R3 subdomains (GAD-Shk1R2 or GAD-Shk1R3, respectively). We conclude that the full-length Scd2 protein interacts specifically with the R1 subdomain of Shk1.
We next defined the structural determinants of Scd2 required for it to interact with Shk1. A GBD fusion of an Scd2 deletion mutant lacking the first SH3 domain (GBD-Scd2ΔN93) retained the ability to interact with GAD-Shk1. Furthermore, like full-length Scd2, Scd2ΔN93 interacted specifically with both full-length Shk1 and the R1 subdomain of Shk1 but not with either the Shk1 R2 or R3 subdomain or with Shk1ΔN118. By contrast, an Scd2 mutant protein lacking the second SH3 domain (GBD-Scd2Δ114-177) was not capable of interacting with full-length Shk1 or with any of the various deletion mutants of Shk1, even though it interacted with the positive control GAD-Scd1[40-872]. These results demonstrate that the second SH3 domain of Scd2 is required for its interaction with Shk1.
To investigate whether the second SH3 domain alone is sufficient to interact with the R1 subdomain of Shk1, we constructed GBD fusions of mutant Scd2 proteins containing either the first (Scd2[1-113]) or second (Scd2[113-188]) SH3 domain or both (Scd2[1-188]) SH3 domains of Scd2 and tested for interactions with various forms of GAD-Shk1. Scd2[1-113] showed no detectable interaction with full-length Shk1 or with any of the Shk1 subdomains tested. By contrast, both Scd2[1-188] and Scd2[113-188] interacted not only with full-length Shk1 and the Shk1 R1 subdomain but also with the Shk1 R3 subdomain. Scd2[1-188] and Scd2[113-188] did not interact with the Shk1 R2 subdomain or with either Scd1[40-872] or Ras1, which were used as negative controls. These intriguing results suggest that while the second SH3 domain of Scd2 is both necessary and sufficient for binding to Shk1, the sequence C-terminal of the SH3 domains may be required for specificity in SH3 binding domain recognition.
Specific interaction between Scd2 and the R1 subdomain of Shk1 in fission yeast.
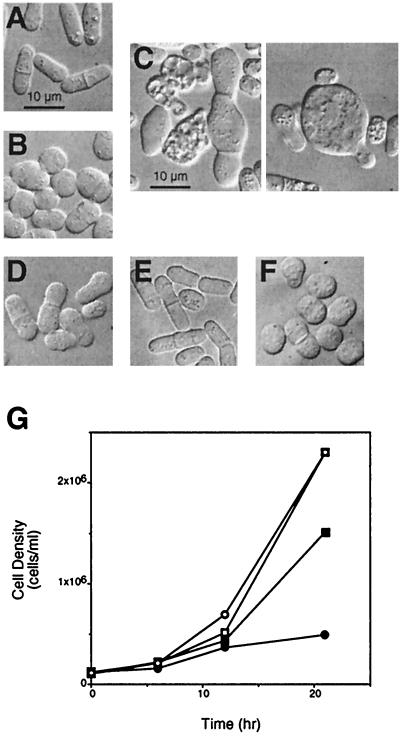
Wild-type fission yeast cells have a rod-like morphology (Fig. 3A), whereas mutants defective for scd2 and shk1 are spheroidal in appearance (7, 24, 27). Overexpression of Scd2 also causes fission yeast cells to become spheroidal, similar to loss of function of Scd2 (7) (Fig. 3B). We tested whether overexpression of the R1 subdomain of Shk1 might block the effects of Scd2 overexpression on cell morphology. Interestingly, we found that overexpression of the Shk1 R1 subdomain alone caused fission yeast cells to exhibit significant and varied morphological abnormalities (Fig. 3C) and to become significantly impaired for growth (Fig. 3G). In contrast to cells that overexpressed either Scd2 or Shk1R1 alone, cells that overexpressed both Scd2 and Shk1 R1 were much more similar to wild-type cells in both morphology (Fig. 3D) and rate of growth (Fig. 3G). These results demonstrate that overexpression of the Shk1 R1 domain partially counteracts the effects of Scd2 overexpression and vice versa. Fission yeast cells that overexpressed the Shk1 R3 domain, which does not interact detectably with full-length Scd2 in the two-hybrid system but contains five potential SH3 binding sites, were similar to wild-type cells in morphology (Fig. 3E) and exhibited no obvious growth defects (data not shown). Furthermore, cells that overexpressed both Shk1R3 and Scd2 were morphologically indistinguishable from cells that overexpressed only Scd2 (Fig. 3F). Thus, the specific interaction between the Shk1 R1 domain and Scd2, as determined by the two-hybrid system, is retained in fission yeast.
FIG. 3.
Genetic evidence for interaction between Scd2 and the Shk1 R1 subdomain in fission yeast. (A to F) Photomicrographs of wild-type CHP428 cells (Materials and Methods) cotransformed with control plasmids pREP1 and pAAUCM (A), pREP1Scd2, for overexpression of Scd2, and pAAUCM (B), pREP1 and pAAUCMShk1R1, for overexpression of the Shk1 R1 subdomain (C), pREP1Scd2 and pAAUCMShk1R1, for overexpression of Scd2 and the Shk1 R1 subdomain (D), pREP1 and pAAUCMShk1R3, for overexpression of the Shk1 R3 subdomain (E), and pREP1Scd2 and pAAUCMShk1R3, for overexpression of Scd2 and the Shk1 R3 subdomain (F). Note large size and gross morphological defects of cells overexpressing the Shk1R1 subdomain (C). This phenotype is suppressed by overexpression of Scd2 (D). Bars correspond to 10 μm. (G) Growth curves for CHP428 cells cotransformed with pREP1 and pAAUCM ( ), pREP1Scd2 and pAAUCM (
), pREP1Scd2 and pAAUCM ( ), pREP1 and pAAUCMShk1R1 (●), and pREP1Scd2 and pAAUCMShk1R1 (■).
), pREP1 and pAAUCMShk1R1 (●), and pREP1Scd2 and pAAUCMShk1R1 (■).
The R1 domain of Shk1 and the second SH3 domain of Scd2 are both necessary for the native proteins to be fully functional.
We took a genetic approach to investigate whether the R1 domain and the second SH3 domain are important for the function of Shk1 and Scd2, respectively. Overexpression of Shk1 caused wild-type fission yeast cells to exhibit a slower growth than cells transformed with a control plasmid (Fig. 4, top). By contrast, overexpression of Shk1ΔN118, which lacks most of the R1 subdomain to which Scd2 binds but retains catalytic function in vivo (Fig. 5C), was not inhibitory to the growth of wild-type fission yeast cells (Fig. 4, bottom). Thus, Shk1 overexpression-induced toxicity in fission yeast is dependent on the Scd2-interacting domain (R1) of Shk1.
FIG. 4.
Overexpression of full-length Shk1, but not Shk1ΔN118, is inhibitory to growth of wild-type fission yeast cells. CHP428 cells were transformed with pART1CMShk1, for overexpression of full-length Shk1, pART1CMShk1ΔN118, for overexpression of a truncated Shk1 protein lacking most of the R1 subdomain, or the control plasmid pART1CM. Fresh transformants were streaked onto EMM and grown for 3 to 4 days at 30°C prior to photographing of the plates. The degree of growth inhibition resulting from Shk1 overexpression was markedly reduced as cultures aged and/or underwent subculturing, and this reduced toxicity was correlated with a reduction in Shk1 protein expression (data not shown).
FIG. 5.
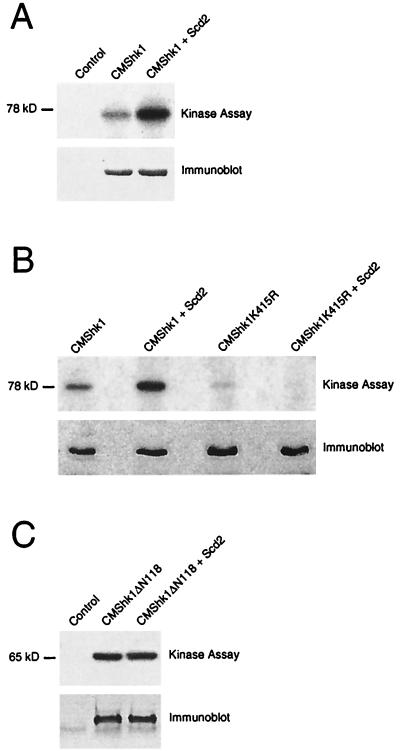
Scd2 stimulates Shk1 autophosphorylation activity in vivo. (A) CHP428 cells were cotransformed with control plasmids pAAUCM and pREP1 (Control), pAAUCMShk1 and pREP1 (CMShk1), or pAAUCMShk1 and pREP1Scd2 (CMShk1+Scd2). Transformed cells were lysed, and CMShk1 immune complexes were isolated and either assayed for protein kinase activity and resolved by autoradiography after SDS-PAGE (top) or subjected to immunoblot analysis using c-Myc antibody 9E10 (bottom) (Materials and Methods). (B) CHP428 cells were cotransformed with pAAUCMShk1 and pREP1 (CMShk1), pAAUCMShk1 and pREP1Scd2 (CMShk1+Scd2), pAAUCMShk1K415R, for overexpression of kinase defective Shk1, and pREP1 (CMShk1K415R), or pAAUCMShk1K415R and pREP1Scd2 (CMShk1K415R+Scd2). Cultures were lysed, and CMShk1 or CMShk1K415R immune complexes were then isolated and assayed for protein kinase activity (top) or subjected to immunoblot analysis (bottom) as for panel A. (C) Shk1ΔN118 is not stimulated by Scd2 in vivo. CHP428 cells were cotransformed with the control plasmids pAAUCM and pREP1 (Control), pAAUCMShk1ΔN118 and pREP1 (CMShk1ΔN118), or pAAUCMShk1ΔN118 and pREP1Scd2 (CMShk1ΔN118+Scd2). Cells were lysed, and CMShk1ΔN118 immune complexes were isolated and assayed for protein kinase activity (top) or subjected to immunoblot analysis (bottom) as for panel A.
Scd2, like Shk1, is required both for maintenance of normal cell morphology and for mating in fission yeast (7). To examine the importance of the SH3 domains of Scd2, we tested whether overexpressing full-length or truncated Scd2 proteins (Fig. 1) could rescue the sterility of an scd2 mutant strain. As shown in Table 2, while overexpression of full-length Scd2 effectively rescued the sterility of the scd2 mutant, Scd2 proteins that lack either one of the SH3 domains (Scd2ΔN93 and Scd2Δ114-177) were much less capable of doing so. The Scd2 mutant protein lacking both SH3 domains (Scd2ΔN184) showed no ability to restore mating to the scd2 mutant. These results demonstrate that both SH3 domains of Scd2 are required for it to function effectively for mating.
TABLE 2.
Suppression of the mating defect of the scd2-1 mutant by various forms of Scd2
scd2 sequences were expressed from plasmid pART1CM, which allows for the expression of c-Myc epitope-tagged proteins under the control of the adh1 promoter (see Materials and Methods).
S. pombe SPM2 (7) was used for the mating assays (Materials and Methods).
Scd2 stimulates Shk1 autophosphorylation activity in fission yeast.
We next addressed whether Scd2 affects the catalytic function of Shk1 in fission yeast. We overexpressed wild-type Shk1, a kinase-defective Shk1 mutant protein (Shk1K415R), and the non-Scd2-interacting N-terminally truncated Shk1 protein Shk1ΔN118 as c-Myc epitope-tagged proteins (CMShk1, CMShk1K415R, and CMShk1ΔN118, respectively), either alone or in combination with overexpressed Scd2 in fission yeast cells. CMShk1 proteins were then immunoprecipitated from cell lysates and assayed for kinase activity in vitro, as measured by autophosphorylation of CMShk1. As shown in Fig. 5A, autophosphorylation of CMShk1 was detected in this kinase assay and the level of autophosphorylation was significantly greater for CMShk1 isolated from cells that overexpressed Scd2 than for cells that did not overexpress Scd2. CMShk1K415R was also weakly phosphorylated in this assay, but the level of phosphorylation was not affected by Scd2 overexpression (Fig. 5B). The weak phosphorylation of CMShk1K415R detected in this assay was probably the result of either residual kinase activity for the mutant protein (Shk1 actually has two consecutive lysines in this region, and only one was mutated) and/or the association of endogenous wild-type Shk1 protein or another protein kinase in the immune complex. Significantly, we also observed that the autophosphorylation activity of CMShk1ΔN118 was unaffected by overexpression of Scd2 (Fig. 5C). These results suggest that Scd2 stimulates the autophosphorylation activity of Shk1 in fission yeast and, consistent with the two-hybrid data described above, that this effect is dependent on the R1 subdomain of Shk1.
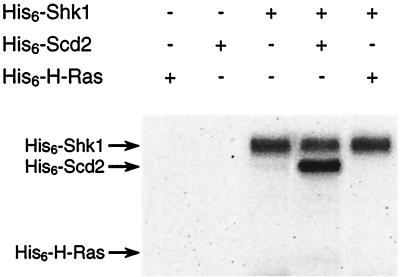
Having determined that Scd2 stimulates Shk1 autophosphorylation activity in fission yeast, we tested whether Scd2 could directly stimulate Shk1 autophosphorylation in vitro by using recombinant His6-tagged proteins purified from bacterial cells. We determined that His6-Scd2 did not stimulate autophosphorylation of His6-Shk1 in vitro (Fig. 6), despite the fact that the two proteins bind in vitro. This result suggests that Scd2 by itself cannot activate Shk1 and that its in vivo stimulation of Shk1 autophosphorylation probably involves other factors. Interestingly, we observed that His6-Shk1 phosphorylated His6-Scd2 protein in vitro (Fig. 6). This result raises the possibility that Scd2 may be both a regulator and a substrate of Shk1 (see Discussion).
FIG. 6.
Shk1 directly phosphorylates Scd2 in vitro. Approximately 80 ng of His6-Shk1, 250 ng of His6-Scd2, and 500 ng of His6-H-Ras were subjected to protein kinase assays either separately or in combination as indicated. Samples were resolved by autoradiography after SDS-PAGE. The positions of His-tagged Shk1, Scd2, and H-Ras are marked by arrows.
Scd2 positively modulates interaction between Cdc42 and Shk1.
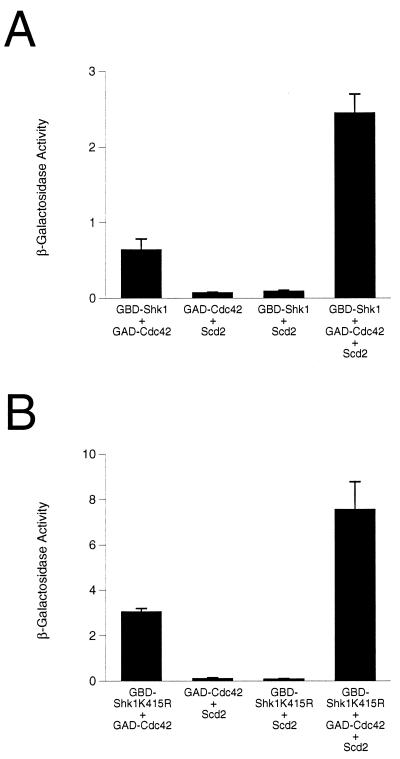
The molecular mechanisms by which Shk1 can be activated remain largely unknown. However, Tu and Wigler (37) have used the two-hybrid system to provide evidence that Shk1 is regulated by autoinhibition and that Cdc42 relieves Shk1 from the presumptive autoinhibitory configuration by inhibiting Shk1-Shk1 homomeric interaction. We used the two-hybrid system to examine the possible role of Scd2 in regulating Shk1-Cdc42 interaction. We determined that overexpression of Scd2 from a third plasmid can enhance the two-hybrid interaction between Cdc42 and Shk1 (Fig. 7A). This effect of Scd2 is not dependent on Shk1 catalytic function, as Scd2 also increased the interaction between Cdc42 and the kinase-defective Shk1K415R mutant protein (Fig. 7B). These results suggest that Scd2 may positively affect Shk1 function by promoting interaction between Cdc42 and Shk1.
FIG. 7.
Scd2 positively affects Cdc42-Shk1 (A) and Cdc42-Shk1K415R (B) interactions in the two-hybrid system. The proteins tested in reporter strain SFY526 are indicated below each bar graph. The plasmids used for expressing various proteins were pGBDShk1, for expression of GBD-Shk1, pGBDShk1K415R, for expression of GBD-Shk1K415R, pGADCdc42, for expression of GAD-Cdc42, and pAAScd2, for expression of Scd2. Values on the y axis represent Miller units for β-galactosidase (Materials and Methods), calculated from the average of at least four determinations using at least two independent transformants.
DISCUSSION
In this report, we have provided evidence for both in vivo and direct in vitro interaction between the fission yeast PAK Shk1 and the dual-SH3-domain-containing protein Scd2. Scd2 binds via the second of its two SH3 domains to the R1 regulatory subdomain of Shk1. Our data suggest that while the second SH3 domain of Scd2 is both necessary and sufficient for interaction with Shk1, the non-SH3 C-terminal domain may confer specificity in binding domain recognition by Scd2. We have shown previously that Scd2 binds directly to the putative Cdc42 GEF Scd1, and genetic epistasis analyses suggested that Scd2 acts upstream of Scd1 (7). We speculated from these data that Scd2 might influence the action of components that function downstream of Cdc42. Consistent with this notion, we demonstrated in this report that Scd2 stimulates the autophosphorylation activity of wild-type Shk1 in vivo but does not affect the activity of a truncated Shk1 protein that lacks the R1 subdomain with which Scd2 interacts. In addition, we showed genetically that Scd2 and Shk1 mutant proteins that lack the domains needed for them to interact appear to be less functional than their wild-type counterparts. Even though Scd2 can stimulate Shk1 autophosphorylation activity in vivo, purified recombinant Scd2 does not stimulate Shk1 autophosphorylation in vitro. Thus, it is likely that the in vivo stimulation of Shk1 autophosphorylation by Scd2 involves other factors.
What is the molecular function of Scd2 with respect to its stimulation of Shk1 kinase activity? Results presented in this report, combined with previously published findings by us and other investigators, may provide at least a partial answer. Tu and Wigler (37) recently used the two-hybrid system to provide evidence that the Cdc42 binding domain of Shk1 can form a complex with the Shk1 catalytic domain. Shk1 in such a closed conformation would presumably be held in a catalytically inactive state due to blocking of the catalytic domain. Tu and Wigler provided genetic evidence supporting this idea (37). They also showed that the binding of Cdc42 to the CRIB site of Shk1 can inhibit interaction between the Shk1 autoinhibitory and catalytic domains. Results of our two-hybrid experiments suggest that Scd2 positively modulates the interaction between Cdc42 and Shk1. The contribution of Scd2 to Shk1 regulation is further underscored by the fact that Scd2 can induce interaction between Cdc42 and its presumptive GEF Scd1 in the two-hybrid system (7), which suggests that Scd2 also plays a pivotal role in Cdc42 activation. The non-SH3 C-terminal domain of Scd2 is responsible for interaction with both Scd1 and Cdc42 (7), while Scd2’s interaction with Shk1 is mediated by the second of its two N-terminal SH3 domains (this study). Thus, Scd2 binds to Shk1 and to Cdc42 and Scd1 via separate domains. Cumulatively, these various observations lead us to propose a model in which Scd2 functions as an organizing center, or scaffold, for assembly of the Cdc42 signaling module consisting of Cdc42, Scd1, and Shk1. Scd2 may function both to positively affect Cdc42 activity by modulating the interaction between Scd1 and Cdc42, as we have shown in a previous study (7), and to regulate Shk1 function by recruiting it to the activated Cdc42 complex. Both Scd2 (7) and Shk1 (27) are required for normal cytoskeletal regulation in fission yeast, and Scd2 has been shown to localize to areas in the cell where active remodeling of the cytoskeleton occurs (32). It is enticing to speculate that Scd2 may function to recruit Shk1 to sites in the cell where the cytoskeleton is undergoing reorganization.
Is the interaction between Scd2 and Shk1 reflective of a general mechanism of regulation for members of the PAK family of protein kinases? We believe this is likely to be the case. As already noted, recent studies have provided evidence that the mammalian SH3-domain-containing proteins NCK and PIX function to recruit PAKs to Cdc42 and/or Rac complexes in mammalian cells (3, 23). In addition, homologs of Shk1 and Scd2, Ste20 and Bem1, respectively, have been shown to form a complex in the evolutionarily distant budding yeast S. cerevisiae (20). Although it has not been demonstrated whether the interaction between Ste20 and Bem1 is direct or whether Bem1 is involved in Ste20 activation, the protein domains involved in complex formation between these two proteins are similar to those required for interaction between Shk1 and Scd2 (20). For example, Bem1 forms a complex with the N-terminal regulatory domain of Ste20, and this interaction is dependent on the second SH3 domain of Bem1. In addition, the non-SH3 C termini of both Scd2 and Bem1 are required for the two proteins to properly interact with their partners, Shk1 and Ste20. These various observations from yeast and mammalian systems suggest that regulation by SH3 domain proteins is, indeed, a highly conserved mechanism of PAK regulation.
Interestingly, we found that Scd2 is an in vitro substrate for Shk1 (this study) and have also obtained evidence that Scd2 is a phosphoprotein in fission yeast (27a). It is noteworthy that mammalian Pak1/α-Pak can also phosphorylate Nck, although the physiological significance of this phosphorylation is unknown (4). The physiological significance of Scd2 phosphorylation and possible regulation by Shk1 is currently under investigation.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Sherry Yen and Leonille Douglas for technical assistance and members of the Chang and Marcus labs for critical comments on the manuscript.
This work was supported by a Whitehead Fellowship from New York University (E.C.), American Cancer Society grant RPG 97-137-01-MGO (E.C.), and National Institutes of Health grant R01GM53239 (S.M.).
REFERENCES
- 1.Abo A, Qu J, Cammarano M S, Dan C, Fritsch A, Baud V, Belisle B, Minden A. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 3.Bagrodia S, Taylor S J, Jordon K A, Van Aelst L, Cerione R A. A novel regulator of p21-activated kinases. J Biol Chem. 1998;273:23633–23636. doi: 10.1074/jbc.273.37.23633. [DOI] [PubMed] [Google Scholar]
- 4.Bokoch G M, Wang Y, Bohl B P, Sells M A, Quilliam L A, Knaus U G. Interaction of the Nck adapter protein with p21-activated kinase (Pak1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 5.Brzeska H, Young R, Knaus U, Korn E D. Myosin I heavy chain kinase: cloning of the full-length gene and acidic lipid-dependent activation by Rac and Cdc42. Proc Natl Acad Sci USA. 1999;96:394–399. doi: 10.1073/pnas.96.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbelo P D, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 7.Chang E C, Barr M, Wang Y, Jung V, Xu H P, Wigler M H. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen C R, Li Y C, Chen J, Hou M C, Papadaki P, Chang E C. Moe1, a conserved protein in Schizosaccharomyces pombe, interacts with a Ras effector, Scd1, to affect proper spindle formation. Proc Natl Acad Sci USA. 1999;96:517–522. doi: 10.1073/pnas.96.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cvrckova F, De Virgilio C, Manser E, Pringle J R, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 10.Daniels R H, Zenke F T, Bokoch G M. alpha-Pix stimulates p21-activated kinase activity through exchange factor-dependent and -independent mechanisms. J Biol Chem. 1999;274:6047–6050. doi: 10.1074/jbc.274.10.6047. [DOI] [PubMed] [Google Scholar]
- 11.Faure S, Vigneron S, Doree M, Morin N. A member of the Ste20/PAK family of protein kinases is involved in both arrest of Xenopus oocytes at G2/prophase of the first meiotic cell cycle and in prevention of apoptosis. EMBO J. 1997;16:5550–5561. doi: 10.1093/emboj/16.18.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faure S, Vigneron S, Galas S, Brassac T, Delsert C, Morin N. Control of G2/M transition in Xenopus by a member of the p21-activated kinase (PAK) family: a link between protein kinase A and PAK signaling pathways? J Biol Chem. 1999;274:3573–3579. doi: 10.1074/jbc.274.6.3573. [DOI] [PubMed] [Google Scholar]
- 13.Frost J A, Khokhlatchev A, Stippec S, White M A, Cobb M H. Differential effects of PAK1-activating mutations reveal activity-dependent and -independent effects on cytoskeletal regulation. J Biol Chem. 1998;273:28191–28198. doi: 10.1074/jbc.273.43.28191. [DOI] [PubMed] [Google Scholar]
- 14.Frost J A, Xu S, Hutchison M R, Marcus S, Cobb M H. Actions of Rho family small G proteins and p21-activated protein kinases on mitogen-activated protein kinase family members. Mol Cell Biol. 1996;16:3707–3713. doi: 10.1128/mcb.16.7.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukui Y, Yamamoto M. Isolation and characterization of Schizosaccharomyces pombe mutants phenotypically similar to ras1. Mol Gen Genet. 1988;215:26–31. doi: 10.1007/BF00331298. [DOI] [PubMed] [Google Scholar]
- 16.Gilbreth M, Yang P, Bartholomeusz G, Pimental R A, Kansra S, Gadiraju R, Marcus S. Negative regulation of mitosis in fission yeast by the Shk1-interacting protein Skb1 and its human homolog, Skb1Hs. Proc Natl Acad Sci USA. 1998;95:14781–14786. doi: 10.1073/pnas.95.25.14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb M H, Marcus S. The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc Natl Acad Sci USA. 1996;93:13802–13807. doi: 10.1073/pnas.93.24.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes D A, Ashworth A, Marshall C J. Complementation of byr1 in fission yeast by mammalian MAP kinase kinase requires coexpression of Raf kinase. Nature. 1993;364:349–352. doi: 10.1038/364349a0. [DOI] [PubMed] [Google Scholar]
- 19.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas D Y, Leberer E. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- 21.Manser E, Huang H Y, Loo T H, Chen X Q, Dong J M, Leung T, Lim L. Expression of constitutively active α-Pak reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manser E, Leung T, Salihuddin H, Zhao Z S, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 23.Manser E, Loo T H, Koh C G, Zhao Z S, Chen X Q, Tan L, Tan I, Leung T, Lim L. Pak kinases are directly coupled to the Pix family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 24.Marcus S, Polverino A, Chang E, Robbins D, Cobb M H, Wigler M H. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 26.Miller P J, Johnson D I. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ottilie S, Miller P J, Johnson D I, Creasy C L, Sells M A, Bagrodia S, Forsburg S L, Chernoff J. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 1995;14:5908–5919. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Pimental, R., and S. Marcus. Unpublished results.
- 28.Polverino A, Frost J, Yang P, Hutchison M, Neiman A M, Cobb M H, Marcus S. Activation of mitogen-activated protein kinase cascades by p21-activated protein kinases in cell-free extracts of Xenopus oocytes. J Biol Chem. 1995;270:26067–26070. doi: 10.1074/jbc.270.44.26067. [DOI] [PubMed] [Google Scholar]
- 29.Ramer S W, Davis R W. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 31.Rudel T, Bokoch G M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1578. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 32.Sawin K E, Nurse P. Regulation of cell polarity by microtubules in fission yeast. J Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sells M A, Barratt J T, Caviston J, Ottilie S, Leberer E, Chernoff J. Characterization of Pak2p, a pleckstrin homology domain-containing, p21-activated protein kinase from fission yeast. J Biol Chem. 1998;273:18490–18498. doi: 10.1074/jbc.273.29.18490. [DOI] [PubMed] [Google Scholar]
- 34.Sells M A, Chernoff J. Emerging from the Pak—the p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 35.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 36.Shibuya E K, Polverino A J, Chang E, Wigler M, Ruderman J V. Oncogenic ras triggers the activation of 42-kDa mitogen-activated protein kinase in extracts of quiescent Xenopus oocytes. Proc Natl Acad Sci USA. 1992;89:9831–9835. doi: 10.1073/pnas.89.20.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu H, Wigler M. Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol Cell Biol. 1999;19:602–611. doi: 10.1128/mcb.19.1.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verde F, Wiley D J, Nurse P. Fission yeast Orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci USA. 1998;95:7526–7531. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Xu H P, Riggs M, Rodgers L, Wigler M. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol Cell Biol. 1991;11:3554–3563. doi: 10.1128/mcb.11.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang P, Kansra S, Pimental R A, Gilbreth M, Marcus S. Cloning and characterization of shk2, a gene encoding a novel p21-activated protein kinase from fission yeast. J Biol Chem. 1998;273:18481–18489. doi: 10.1074/jbc.273.29.18481. [DOI] [PubMed] [Google Scholar]
- 43.Zhang S, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]