Abstract
Correct removal of RNA primers of Okazaki fragments during lagging-strand DNA synthesis is a critical process for the maintenance of genome integrity. Disturbance of this process has severe mutagenic consequences and could contribute to the development of cancer. The role of the mammalian nucleases RNase HI and FEN-1 in RNA primer removal has been substantiated by several studies. Recently, RNase H(35), the Saccharomyces cerevisiae homologue of mammalian RNase HI, was identified and its possible role in DNA replication was proposed (P. Frank, C. Braunshofer-Reiter, and U. Wintersberger, FEBS Lett. 421:23–26, 1998). This led to the possibility of moving to the genetically powerful yeast system for studying the homologues of RNase HI and FEN-1, i.e., RNase H(35) and Rad27p, respectively. In this study, we have biochemically defined the substrate specificities and the cooperative as well as independent cleavage mechanisms of S. cerevisiae RNase H(35) and Rad27 nuclease by using Okazaki fragment model substrates. We have also determined the additive and compensatory pathological effects of gene deletion and overexpression of these two enzymes. Furthermore, the mutagenic consequences of the nuclease deficiencies have been analyzed. Based on our findings, we suggest that three alternative RNA primer removal pathways of different efficiencies involve RNase H(35) and Rad27 nucleases in yeast.
Replication of double-stranded DNA is an asymmetric process. While leading-strand synthesis proceeds continuously, lagging-strand synthesis takes place by synthesis, processing, and ligation of Okazaki fragments (39). These fragments, measuring about 200 nucleotides (nt) in eukaryotes, are primed by DNA polymerase alpha/primase with a short oligoribonucleotide of 7 to 14 residues. Before the nascent Okazaki fragments are ligated to form a continuous lagging strand, the short RNA primers must be removed by an enzyme exhibiting RNase H activity. Although several such enzymes from eukaryotes are known, the process of RNA primer hydrolysis is as yet not fully understood.
An RNase H was first detected in calf thymus extracts (57). Subsequently, RNase H enzymatic activity was detected in all prokaryotes and eukaryotes examined as well as in a bacteriophage and in retroviruses as a part of reverse transcriptases (for reviews, see references 12 and 66). Generally, RNases H are defined as ribonucleotide-specific endonucleases, cleaving the RNA portion of RNA-DNA/DNA or RNA/DNA duplexes. Several RNases H implicated in RNA primer removal have been purified and/or cloned from diverse organisms ranging from bacteriophages to human cells (see, e.g., references 8, 9, 13, 14, 24, 26, 37, 51, and 52). Nevertheless, conclusive evidence of the involvement of these enzymes in primer removal is still lacking. In the budding yeast, Saccharomyces cerevisiae, three different RNases H were identified and partially characterized as RNase H(70), RNase H1, and RNase H(35) (17, 32, 34). These enzymes are evolutionarily related to prokaryotic as well as to mammalian counterparts, as summarized in Table 1. The amino acid sequences are highly conserved within each RNase H family (18); however, sequence similarity between families is very low except at acidic residues of putative active sites. Mammalian RNase HI and HII proteins can be distinguished from each other based on size, charge, metal cation requirements, and serological properties (62). Moreover, evidence from the bovine system suggests that the activity of the large RNase HI correlates with DNA replication whereas that of the small RNase HII correlates with transcription (7). RNA-DNA junction-specific RNase activity of bovine RNase HI was recently demonstrated by using a model Okazaki fragment as substrate (45), which indicates the participation of this enzyme in the removal of RNA primers during lagging-strand DNA synthesis. Upon purification of calf thymus RNase HI, two polypeptides with molecular masses of 32 and 21 kDa were obtained (8, 18). Recent studies by Frank et al. (18) demonstrated that (i) a 33.4-kDa human polypeptide, the equivalent of the above-mentioned enzymatically active protein, is the large subunit of RNase HI, and (ii) this subunit is evolutionarily conserved in various organisms including S. cerevisiae and Escherichia coli.
TABLE 1.
Evolutionary relationship of RNase H
| RNase H in:
|
References | |||
|---|---|---|---|---|
| Retroviruses | E. coli | S. cerevisiae | Mammalian cells (e.g., bovine and human) | |
| Reverse transcriptase (RNase H domain) | RNase HI | RNase H1 | RNase HII | 12, 32, 66 |
| —a | RNase HII | RNase H(35) | RNase HI (large subunit) | 8, 17, 31, 45 |
| — | — | RNase H(70) | GOR antigen | 19 |
—, not detected.
The flap endonucleases 1 represent another family of nucleases involved in RNA primer removal. This evolutionarily conserved family includes mammalian FEN-1 nucleases, S. cerevisiae Rad27p, archaebacterial FEN-1 nucleases, 5′-nuclease domains of bacterial DNA polymerases I, and viral 5′-exonucleases. Mammalian FEN-1, a Mg2+-dependent metallonuclease, possesses both 5′→3′ exonuclease and flap endonuclease activities (3, 11, 40, 54). As an exonuclease, the enzyme recognizes and cleaves the 5′ phosphodiester bond within a 3′ overhang of a double-stranded nucleic acid substrate. The exonuclease activity is stimulated by an upstream primer and is most efficient for nicked duplex substrates. As a flap endonuclease, the enzyme recognizes branched nucleic acid structures containing a single-stranded 5′ flap. It cleaves at junctions where the two strands of duplex DNA adjoin a single-stranded arm without the need for any accessory proteins (23, 25, 30, 41, 44, 47, 67). The enzyme is unable to cleave bubble substrates, 3′ single-stranded flaps, heterologous loops, and Holliday junctions (40). Crystal structures of several FEN-1 nuclease homologues have been determined, facilitating the elucidation of molecular mechanisms and structure-function relationships of this structure-specific nuclease family (10, 27, 29, 36, 43).
Rad27p is the yeast homologue of mammalian FEN-1, and therefore it is reasonable to assume that it might have analogous functions in this lower eukaryote. Several distinct phenotypes have been identified in the yeast null mutant, including conditional lethality, elevated spontaneous recombination rate, chromosomal instability, and high sensitivity to methyl methanesulfonate and UV radiation (46, 50). These identified phenotypes indicate the in vivo functions of Rad27 nuclease in DNA replication, repair (1, 35, 38), and prevention of trinucleotide repeat expansion and contraction (20, 22). In vitro reconstitution experiments demonstrated that mammalian FEN-1 together with RNase HI is required for RNA primer removal (23, 28, 30, 45, 61, 63, 64). It has been proposed that in this process FEN-1 participates in RNA primer removal by either of two pathways: endonucleolytically, by cleaving the 5′ flap structure resulting from a DNA polymerase-displaced 5′ end of the downstream Okazaki fragment, or exonucleolytically, by cleaving the last ribonucleotide adjacent to the deoxyribonucleotide portion of the Okazaki fragment after RNase H had degraded the rest of the RNA primer (3).
Recently, RNase H(35), the S. cerevisiae homologue of mammalian RNase HI, was identified and its possible role in DNA replication was proposed (17). This led to the possibility of moving from the mammalian system to the genetically powerful yeast system for studying the homologues of RNase HI and FEN-1 at the same time. In this study, we have biochemically determined the substrate specificity and the cooperative as well as independent cleavage mechanisms of S. cerevisiae RNase H(35) and Rad27 nucleases by using Okazaki fragment model substrates. We have examined the additive and compensatory pathological effects of these two enzymes by gene deletion and overexpression. Furthermore, the mutagenic consequences of the nuclease deficiency have been analyzed. Based on the above findings, we have suggested three alternative RNA primer removal pathways involving RNase H(35) and Rad27 nucleases in yeast.
MATERIALS AND METHODS
Biochemical reagents and yeast media.
Oligonucleotides used for amplifying genes and constructing null mutant strains were synthesized in the City of Hope Cancer Center core facility. The vector pET-28b was from Novagen (Madison, Wis.), and E. coli XL2-Blue and pCR-Script vector were from Stratagene (La Jolla, Calif.). Restriction enzymes and T4 polynucleotide kinase were obtained from New England Biolabs (Beverly, Mass.). [γ-32P]ATP, [α-32P]dGTP, and cordycepin-5′-[α-32P]triphosphate were purchased from NEN (Boston, Mass.). Pre-packed FPLC Ni2+ chelating columns and desalting columns were purchased from Pharmacia Biotech (Piscataway, N.J.). The protein assay kit was from Bio-Rad (Hercules, Calif.). Yeast culture media including yeast extract-peptone-dextrose (YPD), synthetic complete (SC), minimal-sporulation, and synthetic dextrose minimal (SD) media were prepared by the method of Sherman et al. (55). Amino acids and all other medium components and chemicals were purchased from Sigma (St. Louis, Mo.). PCR reagents were purchased from Promega (Madison, Wis.).
Plasmids.
The marker genes TRP1, HIS3, and URA3 were used to disrupt the RNH1, RNH35, and RNH70 genes, respectively. pRUT7, the plasmid harboring the marker gene TRP1, was used for deletion of the RNH1 gene. It is a hybrid of the 4,887-bp EcoRI-SalI fragment of YIp5 and the 2,108-bp fragment produced by partial digestion of YRp7 with EcoRI and SalI (58) and was constructed by E. Heidenreich (see below for more details). The plasmid used for RNH35 deletion was pJJ215 (marker HIS3), a gift from L. Prakash’s laboratory (33). RNH70 was deleted by using pYEUra3 (marker URA3; Clontech).
The yeast expression plasmid for RNase H(35) was constructed with pDB 20, a URA3- (selection marker) and ADH1 (promoter)-based yeast expression vector (16). The open reading frame (ORF) of the RNH35 gene was PCR amplified with a pair of primers, RNHF (5′-AGTGAAAGCTTCATATGGTACCCCCCACGGTAG-3′) and RNHR (5′-CACAGAATTCACTCGAGCCGGTACCAATTATCTAGG-3′). The amplified DNA fragment (blunt ended) was then inserted into the pCR-Script Amp SK(+) cloning vector (Stratagene). The plasmid with the insertion of the RNH35 ORF in the forward orientation was digested with HindIII. The fragment was then inserted into pDB 20 at the HindIII cloning site. The orientation of the resulting plasmid, pDB-RNH35, was determined. The plasmid was sequenced to confirm the integrity of the coding sequence and transformed into yeast cells. (iii) Plasmids for His6-tagged RNase H(35) and Rad27 protein overexpression in E. coli were constructed as follows. The coding sequence of the S. cerevisiae RNH35 gene was cloned into pET-28b vector (Novagen) with PCR primers RNHF (5′-AGTGAAAGCTTCATATGGTACCCCCCACGGTAG-3′) and RNHR (5′-CACAGAATTCACTCGAGCCGGTACCAATTATCTAGG-3′) containing NdeI and XhoI sites, respectively (underlined); and purified total S. cerevisiae genomic DNA as a template. The resulting plasmid, pET-RNH35, was sequenced, and no mutation was found. To overexpress Rad27 protein in E. coli, the RAD27 ORF was PCR amplified with primers RAD-OV1 (5′-ACAGCAGAAGCTTCCATGGGTATTAAAGGTTTGA-3′) (the NcoI restriction site is underlined) and RAD-OV2 (5′-ACCTAGGAAGCTTACTCGAGTCTTCTTCCCTTTGTGACT-3′) (the XhoI restriction site is underlined). The amplified DNA fragment was then cloned into pET28b at the NcoI and XhoI sites and sequenced to confirm its correct nucleotide sequence. The resulting plasmid was named pET-RAD27.
Overexpression and purification of RNase H(35).
To overexpress the RNase H(35) protein, pET-28b and pET-RNH were transformed into GJ1158, an E. coli strain harboring a chromosomal T7 RNA polymerase gene inducible by high salt concentrations (4). Colonies were inoculated into 1 liter of LBON (NaCl omitted from Luria broth) broth supplemented with 30 μg of kanamycin per ml. Cultures were grown at 37°C to an optical density (OD) of 0.6 and then subjected to induction at 30°C with 0.3 M NaCl for 3 h. The cells were harvested and stored at −80°C until use. RNase H(35) protein was highly expressed, but a major portion of the protein remained in inclusion bodies.
The protein was purified from inclusion bodies by using a built-in C-terminal His tag as follows. All of the purification steps were performed at 4°C. The stored cells were thawed in a mixture of ice and water and resuspended in 80 ml of lysis solution (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl [pH 8.0], 0.5 mg of lysozyme powder per ml). The solution was incubated at room temperature for 1 h and then was centrifuged at 10,000 × g for 20 min. The pellet was resuspended in 40 ml of ice-cold lysis solution (100 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, 50 mM Tris-HCl [pH 8.0]) and incubated for 10 min on ice with occasional mixing. The lysate was pushed through an 18-gauge needle 10 times and centrifuged at 23,000 × g for 20 min to reduce the viscosity. The pellet was washed by being resuspended in 40 ml of wash buffer (1% Nonidet P-40, 100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl [pH 8.0]) and in 40 ml of wash buffer without Nonidet P-40. After centrifugation at 23,000 × g for 20 min, the pellet was dissolved in 25 ml of buffer A (10 mM Tris-HCl, 0.5 N NaCl, 5 mM imidazole, 6 M urea, [pH 7.9]) and incubated on ice for 1 h to dissolve the inclusion bodies. Then the sample was centrifuged at 23,000 × g for 45 min to remove the debris. A 5-ml HiTrap chelating Ni2+ column (Pharmacia) was equilibrated with buffer A by fast protein liquid chromatography (FPLC) (Pharmacia). After loading, the column was washed with 25 ml of buffer A and 25 ml of buffer A with 30 mM imidazole and eluted with a 50-ml linear gradient from 30 to 300 mM imidazole in buffer A at 2 ml/min. An aliquot of each fraction was run on a sodium dodecyl sulfate (SDS)–10%-polyacrylamide gel and stained with Coomassie brilliant blue R-250 to inspect the purity. Protein from the appropriate pooled fractions was renatured by exchanging the buffer with decreasing concentrations of urea. Finally the protein fractions were desalted with a desalting buffer (10 mM Tris-HCl, 150 mM NaCl [pH 8.0]) on a HiTrap desalting column (Pharmacia). Protein concentrations were determined by the Bio-Rad protein assay.
An attempt was also made to purify the RNase H(35) from the soluble fraction of transformed E. coli cell crude extract by following the protocol recommended by Novagen and using nondenaturing conditions. Multiple proteins were obtained with this preparation, including a dominant band migrating at the same molecular weight as the purified and denatured RNase H(35). This partially purified enzyme was used in an RNase H activity assay with substrate 4 and the conditions described below. The first five amino acid residues at the N- terminus of the RNase H(35) proteins purified under both conditions and excised from the SDS-gel were sequenced in the City of Hope Comprehensive Cancer Center core facility. This was done to confirm that the desired protein was used in all subsequent experiments. The RNase H activity of the purified and renatured enzyme has been compared to the partially purified native protein to confirm the restoration of the enzyme activity. Total intensities of the products produced within 10 min at 37°C were normalized with the total protein(s) added to the reaction mixture (see Fig. 1C).
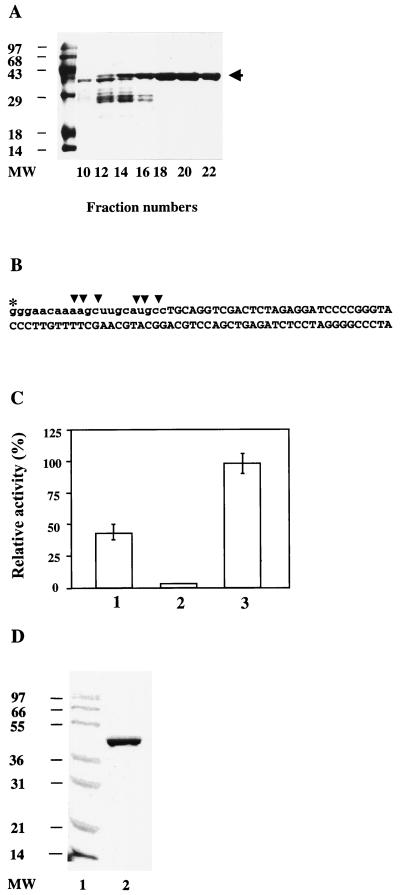
FIG. 1.
Purification of recombinant S. cerevisiae RNase H(35) and Rad27p. (A) The ORF of the RNH35 gene was subcloned into the pET28b vector for addition of a His6 tag and expression in E. coli GJ1153 with a salt-inducible promoter for T7 RNA polymerase (4). As described in detail in Materials and Methods, inclusion bodies were isolated from the transformed cells and dissolved with 6 M urea. RNase H(35) protein was purified on a Ni2+ column with an FPLC system, and fractions were analyzed by SDS-polyacrylamide gel electrophoresis. Indicated is the Coomassie brilliant blue staining of FPLC gradient fractions 10, 12, 14, 16, 18, 20, and 22 and the molecular weight markers (in thousands) (MW). The arrow on the right indicates the desired band of RNase H(35). (B) The RNase H substrate used to quantify the relative activities in panel C. The asterisk indicates the radioactively labeled site. The vertical arrowheads indicate the major cleavage sites. Lowercase letters indicate the RNA portion of the substrate. (C) RNase H activity restoration by renaturation of the RNase H(35) protein purified under denaturing conditions. Bar 1, Activity of partially purified protein from the supernatant of E. coli crude extract overexpressing RNase H(35). The protein was eluted from a chelating Ni2+ column under nondenaturing conditions. Multiple bands were observed on a SDS gel of eluates due to low concentration of the desired protein in the soluble fraction. Bar 2, Activity of RNase H(35) purified by FPLC under denaturing conditions. Bar 3, Activity of renatured RNase H(35). Approximately 100 ng of protein was used for each reaction with the substrate illustrated in panel B under standard conditions described in Materials and Methods. The reaction time is 10 min. (D) The coding sequence of Rad27p was subcloned into the vector pET28b and transformed into E. coli B121(DE3). After induction for 3 h with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), cells were harvested and Rad27p was again purified on a Ni2+ column with an FPLC system. The pooled fractions containing Rad27p (lane 2) were, together with protein size markers (lane 1), subjected to SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue.
RNA/DNA hybrid substrate preparation.
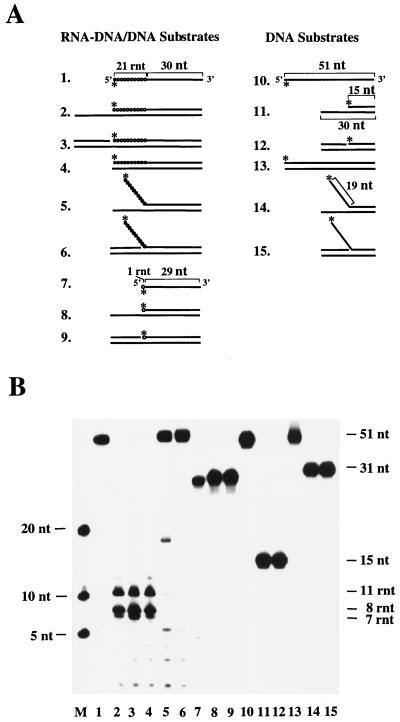
Nine RNA/DNA hybrid substrates were constructed for the assays described below. These substrates are schematically depicted in Fig. 2A and were designed to mimic Okazaki fragments in different dynamic situations. The activities of purified recombinant RNase H(35) and Rad27 nuclease were tested on these substrates. Substrates were labeled on the 5′ or 3′ end by using T4 polynucleotide kinase (NEB) or terminal transferase (Gibco-BRL) with [γ-32P]ATP or cordycepin-5′-[α-32P]triphosphate, respectively. The labeled 21-ribonucleotide–30-deoxyribonucleotide junction oligonucleotide (5′-gggaacaaaagcuugcaugccTGCAGGTCGACTCTAGAGGATCCCCGGGTA-3′) was used as a single-stranded RNA-DNA substrate (substrate 1) (RNA is in lowercase letters, and DNA is in capital letters). This oligonucleotide was also annealed to the 72-mer DNA template (5′-TACCCGGGGATCCTCT AGAGTCGACCTGCAGGCATGCAAGCTTTTGTTCCCCATTACGGCTCT CCGAGTTAT-3′) to obtain a 3′ overhang (substrate 2); annealing substrate 1 and the oligonucleotide 5′-ATAACTCGGAGAGCCGTAATG-3′ produced a nicked duplex, substrate 3. The 51-mer RNA-DNA oligonucleotide formed a complete duplex when it was annealed to a complementary DNA strand (substrate 4). The labeled substrate 1 was also annealed to a partially complementary 51-mer oligodeoxynucleotide (5′-TACCCGGGGATCCTCTAGAGTCGACCTGCAGTAGACGTCTGACACAGCCGT-3′) to form a pseudo-Y substrate (substrate 5) or a flap substrate (substrate 6) when an upstream primer (5′-ACGGCTGTGTCAGACGTCTA-3′) was included. The single-stranded 1-ribonucleotide–30-deoxyribonucleotide junction oligonucleotide (5′-cTGCAGGTCGACTCTAGAGGATCCCCGGGTA-3′) served as substrate 7. It was annealed to a 51-mer complementary DNA strand to form a 3′ overhang (substrate 8) or a nicked duplex (substrate 9) when an upstream primer (5′-ACGGCTGTGTCAGACGTCTA-3′) was included. Substrates 10 to 15 are control DNA substrates analogous to substrates 1 to 6.
FIG. 2.
Substrate specificity of RNase H(35) nuclease as determined with Okazaki fragment-mimicking substrates and DNA substrates as controls. (A) Schematic representation of the substrates, with a stretch of circles indicating the RNA portion and lines representing the DNA portion of the RNA-DNA junction-containing substrates (substrates 1 to 9). An asterisk marks the positions of the 32P label. Substrates 10 to 15 are “DNA-only” substrates with similar configurations corresponding to substrates 1 to 9 as controls. (B) Products obtained from incubation with RNase H(35) were separated on a sequencing gel. Size markers (lane M) are shown as numbers of nucleotides on the left, while the sizes of the substrates and products are given on the right as number of nucleotides (nt) or ribonucleotides (rnt).
RNase activity assays.
For RNase H assays, standard reaction mixtures contained 0.8 pmol of γ-32P-labeled substrate, 50 mM Tris (pH 8.0), 10 mM MgCl2, and 100 ng of enzyme in 13 μl and were incubated at 37°C for 10 min. An equal volume of stop solution (USB) was added to stop the reactions. The samples were mixed, boiled for 3 min, and cooled in ice. A 3-μl volume of each reaction product was run on a 15% denaturing polyacrylamide gel and exposed to Kodak X-ray film.
Construction of RNH single null mutants and the RNH35/RAD27 double mutant.
All of the S. cerevisiae strains used in this study are listed in Table 2. The deletion of RNH35 (strain BFRH35a) is described elsewhere (17). Deletion of the RNH1 gene (4a) to generate BC39a (AK310a rnh1::TRP1) was done by PCR with genomic yeast DNA (Promega) as the template and primers rnh1/d5 (5′- CGTAGAGGTACCAAGCGGTTGAT-CTTGGCTGTAGCACTTATAC), which contains a KpnI site, and rnh1/d4 (5′-GATGTCCTGCAGGAAGTACAAGTAGATGATCTTGCT-GAACGT), which contains a PstI site. The 1,711-bp product was then inserted in pUC18. The 1,049-bp XbaI-ClaI fragment, including almost the entire ORF, was replaced by the TRP1 gene, amplified by PCR with pRUT7 as the template and two trp1-specific primers, trp/1 (5′-GATGTCATCGATAATTCGGTCGAAAAAAGAAAAGGAGAGGGC) and trp/2 (5′-GATGCTACTAGTGAGAAAAGGCTAGCAAGAATCGGGTCATTG), and digested with ClaI and SpeI. The 1,741-bp EcoRI-SphI fragment of pUC18 with the above RNH1-TRP1 insert was then ligated into pRUT7. To integrate the plasmid at the RNH1 locus, the plasmid was linearized with MamI and introduced into AK310 by pop-in/pop-out replacement (53). Yeast cells were transformed by the lithium acetate method (21) and plated on selective medium.
TABLE 2.
S. cerevisiae strains used in this study
| Straina | Deleted gene(s) | Genotype | Sources |
|---|---|---|---|
| W1021-7c | Wild type | a ade2-1 can1-100 his3-11,17 leu2-3 ura3-1 | R. Rothstein |
| W1089-6c | Wild type | α ade2-1 can1-100 his3-11,17 trp1-1 ura3-1 | R. Rothstein |
| *IC2-1 | RAD27 | α rad27::LEU2 | This work |
| *R27-H35 | RAD27 and RNH35 | a rad27::LEU2 rnh35::HIS3 | This work |
| AK310 | Wild type | a ade2-1 can1-100 his3-11,15 leu2-3,112 ura3 ssd1 | U. Wintersberger |
| ΔBFRH35a | RNH35 | a rnh35::HIS3 | P. Frank |
| ΔBFRH35b | RNH35 | α rnh35::HIS3 | P. Frank |
| ΔBC39a | RNH1 | a rnh1::TRP1 | P. Frank |
| ΔBC39b | RNH1 | α rnh1::TRP1 | P. Frank |
| ΔBC70a | RNH70 | a rnh70::URA3 | P. Frank |
| ΔBC70b | RNH70 | α rnh70::URA3 | P. Frank |
| RKY2672 | Wild type | a ade2-1 ade8 hom3-10 his3Δ200,17 leu2Δ1 lys2ΔBgl trp1Δ63 ura3-52 | R. D. Kolodner |
| · RKY2608 | RAD27 | a rad27::hisG-URA3-hisG | R. D. Kolodner |
| · MEXO1a | EXO1 | a exo1::LEU2 | This work |
| · MEXO1b | EXO1 | a exo1::URA3 | This work |
| · MRAD27a | RAD27 | a rad27::LEU2 | This work |
| · MRAD27b | RAD27 | a rad27::URA3 | This work |
| · MMSH2a | MSH2 | a msh2::TRP1 | This work |
| · MMSH2b | MSH2 | a msh2::hisG-URA3-hisG | This work |
| · MBFRH35a | RNH35 | a rnh35::HIS3 | This work |
| · MBFRH35b | RNH35 | α rnh35::HIS3 | This work |
| · MR27H35 | RAD27 and RNH35 | — rad27::URA3 rnh35::HIS3b | This work |
| · MBC39a | RNH1 | a rnh1::TRP1 | This work |
| · MBC39b | RNH1 | α rnh1::TRP1 | This work |
| · MBC70a | RNH70 | a rnh70::URA3 | This work |
| · MBC70b | RNH70 | α rnh70::URA3 | This work |
*, derivatives of W1021-7c and W1089-6c; Δ, derivatives of AK310; · , derivatives of RKY2672. The genotype of all the derivatives from the same parental strains is indicated only in their differences from the parental strains.
—, mating type not determined.
To amplify the RNase H(70) coding sequence (yeast ORF YGR276c) for the production of a deletion allele of the RNH70 gene, PCRs were performed with yeast genomic DNA (Promega) as a template. The following primers were used: forward primer 5′-TGTGACTGAATTCTAATGTACTTGGAGGAAGGGAGGAAGACG-3′ and reverse primer rh70d4/HindIII (5′-CGGATGTAAGCTTTCTACAGAGGGAGAGCTGTGAGGATATATT-3′). The 2,273-bp PCR product was cloned into pUC18. An SspBI-SmaI fragment of 1,549 bp including nearly the complete ORF was replaced by an 1,125 bp fragment carrying the URA3 gene. The isolated and gel-purified deletion allele was used for integrative transformation (53), and Ura+ transformants were isolated. They were checked by specific PCR for successful gene replacement and analyzed for phenotypes (19).
For construction of the double null mutant, a rad27 null mutant (IC2-1) (16) was crossed with a rnh35 null mutant (BFRH35a) to produce heterozygous diploids. The diploid cells were then sporulated and dissected. Spores from microdissection were incubated at 30°C for 2 days, and colonies were replica plated onto different SD media with one or two amino acids eliminated to determine their genotypes. The rad27 single null mutant grows on SD-Leu medium, and the rnh35 single null mutant grows on SD-His medium. The double null mutant (R27-H35) can grow on both SD-His and SD-Leu media. Deletions of the genes were also confirmed by PCR analysis.
Cell proliferation of yeast mutant strains.
Different sizes of colonies descending from wild-type and mutant spores were observed and photographed. To further determine growth rates, cells were grown in YPD liquid medium. The initial concentration was standardized. The cells were grown at 30 or 37°C, and OD values were measured. The experiments were repeated six times, and the growth curves show mean OD values and standard deviation.
RNH35 overexpression in yeast.
To determine a possible suppression effect of overexpression in different genetic backgrounds, the pDB20 expression vector with or without insertion of RNH35 was transformed into rnh35 and rad27 single null mutants, a rad27/rnh35 double null mutant, and a wild-type strain of S. cerevisiae. For the observation of cell proliferation, the transformants were cultured in SD-Ura liquid medium at 30 or 37°C. OD values were then measured and growth curves were determined as described above.
Mutation frequency assay.
To test the possible role of RNase H(35) and RNase H(35)/Rad 27p in mutation avoidance, the mutator assays based on the CAN1, HOM3, and LYS2 genes (42, 48, 60, 65) were used. A single-knockout RNH35 mutant (BFRH35b) was crossed with RKY2672, which contains three assay genes, CAN1, HOM3, and LYS2, on the chromosome. The diploids were sporulated, and tetrads were dissected. Colonies with single disruption of the RNH35 gene were selected for all four genetic markers: HIS3, CAN1, HOM3, and LYS2. The resulting mutant strain was named MBFRH35b. The rad27 null mutant in an RKY2672 background used in this study was RKY2608 (a gift from R. Kolodner, University of California, San Diego, Calif.). The rnh35 null mutant BFRH35b was also crossed with RKY2608 to produce a rnh35/rad27 double-deletion strain. After sporulation and dissection, the resulting double mutant was named MR27H35, representing a rnh35/rad27 double null mutant with three assay genes. The RNH1 and RNH70 mutant versions in RKY2672 were constructed in a similar way (all S. cerevisiae mutants used in this study are listed in Table 2). For the mutation assay, cells were grown to saturation. Then 100 μl of cultured cells was plated onto the corresponding selection medium, i.e., medium SD with 60 mg of canavanine per liter and all the required amino acids for the Canr assay, SD medium without threonine for the HOM3 gene assay, and SD medium without lysine for the LYS2 gene assay. In addition, 10,000-fold dilutions of each culture were plated onto YPD medium for counting viable cells. Finally, colonies from selection and YPD plates were counted. The mutation frequency was calculated by dividing the colony number on the selection medium by that on YPD medium. For each assay gene, three independent experiments were performed and values from the three experiments were averaged.
RESULTS
Purification of RNase H(35) and Rad27 recombinant proteins.
To obtain a highly purified enzyme, we PCR cloned the RNase H(35) coding region from S. cerevisiae genomic DNA and expressed it as a His6-tagged protein by using the T7 RNA polymerase system in E. coli. RNase H(35)-His6 was expressed to a high level (∼40% of total E. coli cellular proteins). However, more than 90% of the recombinant protein was found in insoluble inclusion bodies. The inclusion bodies were purified from the cell extract, and the recombinant RNase H was purified by FPLC under denaturing conditions. Figure 1A shows the FPLC fractions which contained recombinant protein. These fractions were pooled, and the protein was renatured by step-gradient dialysis to gradually remove the urea. To confirm the restoration of the enzyme activity, the relative RNase H activities were determined by using native (partially purified from the crude extracts of transformed E. coli), denatured, and renatured RNase H(35) proteins (Fig. 1B and C). In contrast to the protein purified from inclusion bodies, multiple protein bands were observed on the SDS gel with the nondenaturing protein preparation from the whole-cell extract. This was probably due to the low concentration of RNase H(35) protein in the soluble fraction of the E. coli crude extract. Since the RNase H activities were normalized by the total protein added to the reaction mixture, the purified and renatured protein had twice the specific activity as the protein purified under nondenaturing conditions. The highly purified and renatured enzyme was used for all subsequent experiments described below.
Recombinant Rad27 nuclease was cloned and overexpressed by a method similar to that for the human FEN-1 protein (47). It was purified from the E. coli crude extract by using the built-in His tag and FPLC under nondenaturing conditions (Fig. 1D). When assayed with DNA substrates, the enzyme showed structure-specific flap endonuclease and nick-specific exonuclease activities resembling those of the human FEN-1 enzyme (see below).
Biochemical properties of the purified RNase H(35) enzyme.
Analysis of the effects of salt concentration, pH, and divalent-metal-ion concentration on the RNase H activity were performed under standard conditions as described in Materials and Methods. RNase H(35) was found to be optimally active at pH 7.7, although it tolerated a wide range of pH (from 5.5 to 10.3). Maximal activity required the presence of divalent cations (Mg2+ or Mn2+), although some activity was observed in their absence. Stimulation by Mg2+ displayed an optimum at 4 mM, while the activity was completely inhibited at 80 mM. With Mn2+, optimal activity was found at 40 mM. Both metal ions were able to activate the enzyme to a similar extent. Salt (KCl) stimulated RNase H(35) activity approximately 1.5-fold, with a broad optimum around 80 mM.
Substrate specificity.
To obtain information about a possible role of RNase H(35) in RNA primer removal, it is critical to characterize its substrate specificity. Experiments with RNase H(35) were conducted in parallel with Rad27p, using a set of nine RNA/DNA hybrid substrates and six DNA substrates as controls. These substrates were designed to mimic Okazaki fragments in different dynamic situations. Essentially, four groups of substrates were made as outlined in Fig. 2A. The first group was derived from a 21-RNA–30-DNA oligonucleotide which was annealed to a DNA oligonucleotide to form a 3′ overhang, a nicked double-stranded duplex, or a blunt-ended duplex. The second group was constructed to form the pseudo-Y and the flap structure by annealing the same oligonucleotide to a DNA oligonucleotide. However, in this group the DNA oligonucleotide was only complementary to the DNA portion of the 21-RNA–30-DNA oligonucleotide. The third group was similar to the first except that the RNA-DNA hybrid had only one ribonucleotide attached to the DNA portion; it tested whether the enzyme was able to remove the last ribonucleotide of an Okazaki fragment. The last group of substrates, serving as control, had the same configurations as the other groups but contained only DNA.
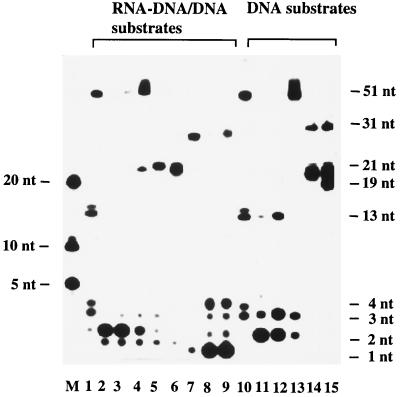
Figure 2B shows that RNase H(35) is highly specific in its substrate requirement. The nuclease removed only RNA portions (7, 8, and 11 ribonucleotides in length) from the RNA-DNA/DNA duplexes and did not discriminate between substrates with a single-stranded 3′ overhang (Fig. 2B, lane 2), an upstream DNA double strand (lane 3), or a blunt-ended duplex (lane 4). Moreover, the enzyme did not recognize the single-stranded RNA-DNA substrate (lane 1), in contrast to the previously published result obtained with calf enzyme (45). It had very low activity on a flap or a pseudo-Y substrate containing single-stranded RNA (lanes 5 and 6) and on the last ribonucleotide adjacent to the DNA (lane 8 and 9). When the Rad27 nuclease was subjected to the same set of substrates, a different cleavage pattern was obtained (Fig. 3). Rad27p cleaved dinucleotides from the RNA and DNA of both 3′ overhang and nicked duplex substrates in a processive manner, but its activity was very much reduced when the substrate was a blunt-ended duplex (Fig. 3, lanes 2 to 4 and 11 to 13). It digested both RNA and DNA single-stranded portions from pseudo-Y and flap substrates as a flap endonuclease (lanes 5 and 6 and lanes 14 and 15). In contrast to the RNase H(35), Rad27p had strong hydrolytic activity against the last ribonucleotide attached to a DNA strand of double-stranded DNA or of a nicked DNA · RNA-DNA/DNA duplex ( · indicates the nick, - indicates the RNA-DNA junction, and / indicates the complementary nucleic acid strand) (lanes 8 and 9). Rad27 nuclease also cleaved longer single-stranded RNA-DNA or DNA substrates (lanes 1 and 10), in contrast to previous observations with other FEN-1 family members (3, 40, 54).
FIG. 3.
Substrate specificity of Rad27 nuclease as determined with Okazaki fragment-mimicking substrates and DNA substrates as a control. The same set of substrates as in Fig. 2A was used in this experiment. The size markers and labeling designations of the lanes are the same as in Fig. 2B. The product spectrum produced by Rad27 nuclease is indicated on the right.
Cooperative removal of RNA primers by RNase H(35) and Rad27 nucleases.
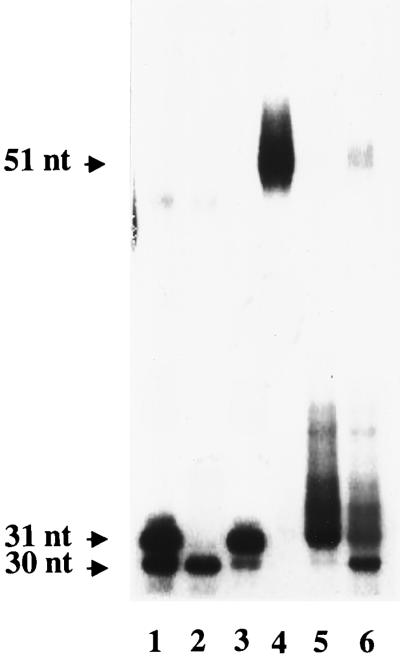
To evaluate the fate of the 3′ portion of the segment after RNase H(35) cleavage and release of 7 to 11 nucleotides, we labeled the RNA-DNA junction strand of substrate 3 at the 3′ terminus. When the reaction was solely driven by RNase H(35), the products separated on the gel were smeared, probably due to cleavage at multiple sites. The smeared product pattern changed depending on the incubation time. When the incubation time reached 20 min, the major product observed was 31 nt, with very little product of 30 nt (Fig. 4, lane 5). This indicates that RNase H(35) made a first cut at the 7th, 8th, or 11th phosphodiester bond from the 5′ end and then a second cut at 1, 4, and 5 nt upstream of the RNA-DNA junction, as shown in Fig. 1B, when substrate 4 was used in the experiment. Furthermore, a possible cooperative RNA primer removal mechanism involving RNase H(35) and Rad27p was explored. When the two enzymes were mixed and added to the above reaction, more than 50% of the 31-nt and longer products were converted to a 30-nt product within 10 min (Fig. 4, lane 6). This probably indicates the complete removal of the RNA primer by cooperative action of the two enzymes.
FIG. 4.
Combined action of RNase H(35) and Rad27p. Substrate 3 in Fig. 2A was labeled at the 3′ end of the DNA part of the RNA-DNA junction-containing strand (lane 4). The reaction mixture was incubated with RNase H(35) (lane 5) or with RNase H(35) plus Rad27p (lane 6) for 10 min under the standard conditions described in Materials and Methods. The sizes of the reaction products in nucleotides are indicated on the left. Lanes 1 to 3 are size markers (30 and 31 nt).
Cells lacking both RNH35 and RAD27 are viable but proliferate very slowly.
Up to now, three yeast RNases H have been described: RNase H(70), RNase H1, and RNase H(35). Since RNase H(35) is the yeast homologue of mammalian RNase HI enzyme and since it was able to cooperate in vitro with Rad27p in hydrolyzing the RNA part of a model Okazaki fragment, we wanted to observe the in vivo effects of deleting the genes for both of these enzymes in one cell. Deletion of the gene for RNase H(35) alone is known to reduce the RNase H activity considerably, as measured in vitro with cell extracts but has only a very small effect on viability and proliferation rate of the deletion mutant (17). On the other hand, deletion of the RAD27 gene has severe consequences, one of which is temperature-sensitive lethality (50, 56). The previously reported mutant strain had been shown to grow slowly at 30°C whereas it was unable to form colonies at 37°C. However, the rad27 mutant strain derived from parental strains W1021-7c and W1089-6c in our laboratory showed very small and heterogeneous colonies on plates and delayed proliferation in liquid medium at 37°C. The arrested cells were twice as big as the wild-type cells, which indicates a deficiency in progression of DNA replication (data not shown).
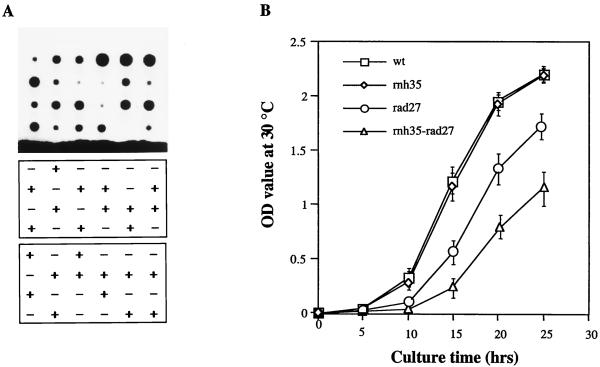
When we crossed an rnh35 deletion mutant with a rad27 deletion mutant, we were surprised that after sporulation we obtained tetrads comprising four viable spore colonies. However, as shown in Fig. 5A, these colonies grown at 30°C differed in their sizes (top panel) in a pattern reflecting their genotypes (lower panels): spores possessing both wild-type genes formed the largest colonies, whereas those carrying a single deletion were slightly smaller in the case of the rnh35 mutant and much smaller in that of the rad27 mutant. The double-deletion mutants formed the smallest colonies. Figure 5B shows growth curves of haploid strains from one of the tetrads depicted in Fig. 5A. The data indicate that the double mutant grew even slower than the rad27 single mutant at 30°C (Fig. 5B) and 37°C (data not shown). The finding that the double mutant survived at all (probably only under laboratory conditions) indicates that another nuclease might partially replace the functions of RNase H(35) and Rad27 nucleases (see Discussion).
FIG. 5.
Double-deletion mutants of RNH35 and RAD27 are extremely retarded in growth. (A) Genetic analysis of spore clones of tetrads descending from a cross of an rnh35 deletion mutant with a rad27 deletion mutant. (Top) Sizes of haploid colonies (incubated at 30°C). (Middle and bottom) Genotypes of the spore clones as determined by growth on selective media lacking either histidine (middle) or leucine (bottom). Growth on His dropout medium (+) indicates deletion of RNH35, and growth on Leu dropout medium (+) indicates deletion of RAD27. Double-deletion mutants grow on both selective media. (B) Growth curves of the four isogenic strains (fifth column from the left of the top panel in panel A) at 30°C.
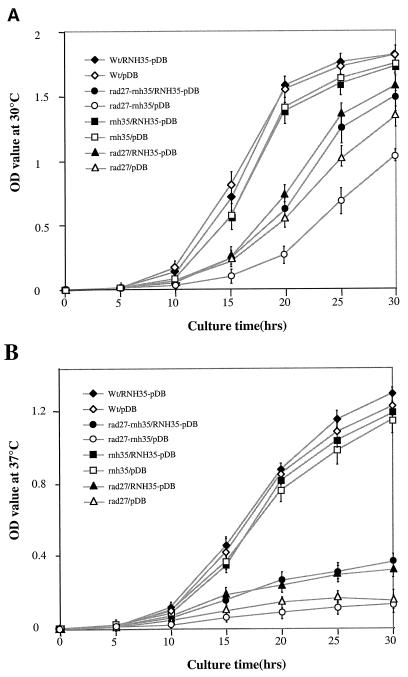
Overexpression of RNH35 in RNH35/RAD27-proficient cells and in rnh35, rad27, and rnh35/rad27 mutant backgrounds.
If there actually exists an additional pathway for removing the RNA parts of Okazaki fragments, which requires only RNase H(35) but not Rad27p, one would expect that overexpression of RNH35 in the rad27 background might partially rescue the rad27 growth defect. Therefore, the RNH35 coding region was subcloned into pDB20, a URA3 and ADH1 promoter-based yeast expression vector. Plasmids with or without the RNH35 insert were transformed into four different S. cerevisiae strains: a RNH35/RAD27-proficient strain, a RNH35 deletion mutant, a RAD27 deletion mutant, and a RNH35/RAD27 double-deletion mutant. Growth curves of the above eight strains were measured at both 30°C (Fig. 6A) and 37°C (Fig. 6B). Based on their growth rates, the eight strains can be divided into three groups: (i) strains not affected by transformation with the plasmid, regardless of whether it carried the RNH35 gene or not, were the RNH35/RAD27 proficient strain and the rnh35 single-deletion mutant; (ii) the poor growth of deletion mutants rad27 and rnh35/rad27 was not influenced by their transformation with the plasmid alone; however, (iii) the poor growth was ameliorated by their transformation with the plasmid overexpressing the RNH35 gene. Northern analysis confirmed that expression of the plasmid-borne RNH35 gene was elevated 15-fold in all strains relative to endogenous RNase H(35) mRNA in the RNH35/RAD27 strain (data not shown).
FIG. 6.
Overexpression of RNH35 with the pDB 20 yeast expression vector partially suppresses the growth defect of rad27 and rnh35/rad27 mutants. (A) The growth curves of four different S. cerevisiae strains with or without RNH35 expression were determined at 30°C each with six independent cell cultures, and mean values were plotted. (B) Growth curves were determined at 37°C.
Mutagenic consequences in rnh35 and rnh35/rad27 mutants.
Defects in the removal of RNA primers lead to mutations (mainly duplications [60]) in the genome. We therefore determined the reversion frequencies of nuclease single-deletion and double-deletion mutants containing the frameshift mutations hom3-10, a +1 insertion in a stretch of 6 T’s of the HOM3 gene, or lys2-Bgl, a 4-base insertion in the LYS2 gene. In addition, we used the forward-mutation assay, which measures the frequency of canavanine-resistant (Canr) mutants. The results are summarized in Table 3. Both rnh35 and rnh1 single deletions caused increased frequencies of mutations leading to canavanine resistance, a phenotype which can arise by many different changes affecting the CAN gene (see Discussion for further considerations). Deletion of the RNH35 gene also led to an increase of Lys+ but not of Hom+ revertants. Reversions of the frameshift in the HOM3 gene occur by single-nucleotide deletions which are typical for mismatch repair defects, as shown by our result with the mismatch repair mutant msh2 and by previously published data (42, 60). Cells lacking RNH70 did not exhibit enhanced mutation frequencies to Canr or to Hom+. A strain harboring a deletion of another recently identified nuclease gene, EXO1, produced around sixfold-higher frequencies of Canr mutants and Hom+ revertants. In accordance with earlier studies (60), disruption of the RAD27 gene increased the mutation frequencies in all three tested genes to a larger extent than did the single disruptions of the other nucleases (Canr, 58-fold; frameshifts to HOM3, 21-fold; and Lys+, 48-fold). The mutation frequencies occurring in the double-deletion mutant rnh35/rad27 were higher than those calculated from addition but lower than expected from multiplication of the single-deletion frequencies, indicating that the two gene products may participate in two different pathways to the same end product.
TABLE 3.
Mutation frequencies of S. cerevisiae strains with deletions of the nuclease genes
| Strain | Mutation frequency (fold)
|
||
|---|---|---|---|
| Canr | Hom+ | Lys+ | |
| Wild type | 3.1 × 10−7 (1) | 1.0 × 10−8 (1) | 1.3 × 10−8 (1) |
| rnh35 | 1.9 × 10−6 (6.1) | 9.0 × 10−9 (0.9) | 6.6 × 10−8 (5.07) |
| rnh1 | 1.0 × 10−6 (3.2) | 3.0 × 10−9 (0.3) | NDa |
| rnh70 | 3.2 × 10−7 (1) | 3.8 × 10−9 (0.38) | ND |
| rad27 | 1.8 × 10−5 (58) | 2.1 × 10−7 (21) | 6.1 × 10−7 (48) |
| rad27-rnh35 | 3.2 × 10−5 (103) | 4.6 × 10−7 (46) | 1.5 × 10−6 (115) |
| exo1 | 2.2 × 10−6 (6.5) | 5.6 × 10−8 (5.6) | 3.5 × 10−8 (2.7) |
| msh2 | 2.9 × 10−6 (9.3) | 5.0 × 10−6 (500) | 8.1 × 10−7 (58) |
ND, not determined.
DISCUSSION
Fidelity of DNA polymerases, proofreading of 3′ exonucleases, and postreplication repair represent mechanisms for maintenance of genome stability. Proper and prompt removal of RNA primers, particularly during lagging-strand DNA synthesis, is another vital process, which has not yet been intensively studied in eukaryotic cells. Here we took advantage of the availability of highly purified recombinant enzymes and budding yeast as a powerful genetic tool to study this mechanism and the mutagenic consequences of failing to remove RNA primers. S. cerevisiae RNase H(35) was identified recently by Frank et al. (17). For in vitro biochemical studies, we exploited the feasibility of its purification from inclusion bodies and subsequent renaturation and successfully attained the active recombinant enzyme of RNase H(35). The purification of active recombinant Rad27 enzyme has also been described in this work. The budding yeast, S. cerevisiae, was chosen because it serves as an ideal model organism for studying biological processes of eukaryotes. Frequently, the yeast system allows more efficient analysis of the properties and functions of genes and proteins homologous to those originally discovered in a multicellular organism. Thus, establishment of the evolutionary and functional relationship between S. cerevisiae RNase H(35) and mammalian RNase HI made it possible to do biochemical and genetic experiments with the simple microbe (18).
Here we have reported a series of in vitro experiments to elucidate the mechanism of action of RNase H(35), as well as genetic tests examining the in vivo consequences of deletion or overexpression of the RNH35 gene. Furthermore, we studied possible cooperative actions of RNase H(35) and Rad27 nucleases in the context of their possible involvement in RNA primer removal from Okazaki fragments during lagging-strand DNA synthesis and in mutation avoidance.
Substrate specificities and cooperative action of recombinant RNase H(35) and Rad27 nucleases.
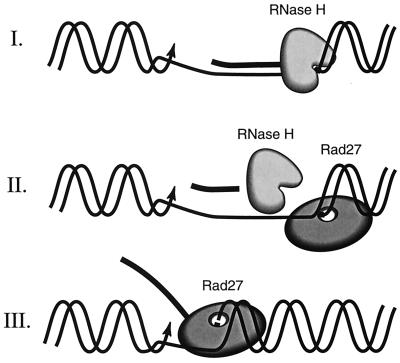
By using 15 substrates mimicking Okazaki fragments in different dynamic situations, the specificities of RNase H(35) and Rad27 nuclease were determined and compared. As expected from its close evolutionary relationship to the FEN-1 family of nucleases, Rad27p was found to cleave single-stranded RNA and DNA flaps protruding from nucleic acid double strands. In addition, Rad27p hydrolyzed single-stranded nucleic acids per se. This stands in marked contrast to RNase H(35), whose activity was absolutely restricted to RNA/DNA hybrid duplex substrates. The yeast RNase H(35) readily removed the RNA portion from these substrates, except for the last ribonucleotide adjacent to the DNA part of an RNA-DNA/DNA duplex. RNase H(35) may be inefficient in hydrolysis of the final ribonucleotide adjoining the DNA, since it has difficulty in recognizing the phosphodiester bond between a ribonucleotide and deoxyribonucleotide. However, from time to time, we did observe monoribonucleotide products after incubating RNase H(35) with substrates 8 and 9 and exposing the gel to the film for an extended time. In contrast, our in vitro results indicate that Rad27p is predisposed for this reaction. We considered it very likely that the two enzymes might cooperate in RNA primer removal from Okazaki fragments in an in vivo process (shown as major pathway II in Fig. 7. An additional in vitro experiment corroborated this idea of cooperation: when substrate 4 (mimicking an Okazaki fragment with its RNA primer still attached) was incubated with both enzymes, the RNA primer was perfectly removed. These results are consistent with in vitro reconstitution studies involving the simian virus 40 replication system in mammalian cells (61, 63, 64). However, a recent paper by Murante et al. (45) indicates that the calf RNase HI is able to make a junction-specific cleavage on a single stranded RNA-DNA substrate. Failure of RNase H(35) to recognize and cleave the single stranded RNA-DNA substrate in vitro may be due to the absence of an interacting polypeptide(s).
FIG. 7.
A working model of RNA primer removal during S. cerevisiae DNA replication involving RNase H(35) and Rad27 nucleases and three possible pathways. (I) A minor pathway. RNase H(35) alone removes 7 to 11 nt endonucleotically as well as the last ribonucleotide adjacent to the DNA. (II) A major pathway. The combined action of RNase H and Rad27 nucleases completely removes the RNA primer. The RNase H(35) removes the main part of the RNA primer, while the last ribonucleotide is removed by Rad27 nuclease. (III) A major alternative pathway. In the absence of RNase H(35), Rad27p as a FEN removes the displaced single-stranded RNA primer adjacent to the newly synthesized DNA.
Deletion of both RNH35 and RAD27 genes severely impairs vitality of yeast cells.
Earlier it was observed that cells survive quite well without RNH35 under laboratory conditions, whereas the absence of Rad27p results in death at elevated temperature. The arrest of rad27 mutant cells in S phase at high temperature indicates failure of an essential step in DNA replication. It has been suggested that this step is probably RNA primer removal from Okazaki fragments (50, 56). Molecular mechanisms and roles of nucleases in RNA primer removal were previously proposed but based solely on studies involving in vitro reconstitution of DNA replication (3, 61, 63, 64) (illustrated as major [II] and alternative major [III] pathways in Fig. 7). Since RNase H(35) has recently been identified as the homolog of the large subunit of mammalian RNase HI and was recognized as the main RNase H activity in cell extracts, we were in a position to perform direct genetic studies to determine if RNase H and Rad27p might both be involved in RNA primer removal. We found that the double-deletion mutant was further impaired in its growth relative to the rad27 single-deletion mutant and that overexpression of RNase H(35) partially rescued growth of the double-deletion mutant. Taken together, these results are consistent with RNase H(35) contributing to a minor pathway (I in Fig. 7) of Okazaki fragment processing and being partially responsible for the survival of rad27 deletion mutants. Unexpectedly, we found that spores lacking both functions of Rad27p and RNase H(35) were still able to germinate and to form small colonies at 30°C. This observation leads us to conclude that another RNase H, one that may or may not be already identified, could act as an inefficient substitute of RNase H(35). Experimental evidence also suggests that exonuclease 1, an enzyme recently identified in several eukaryotic organisms (15, 59), is involved in RNA primer removal (49).
Mutagenic consequences of deficiencies for various nucleases.
Our main interest was to determine the roles of RAD27 and RNH35 and their cooperation in mutation avoidance. For comparison, we also analyzed the roles of the two other known RNH genes as well as of EXO1 and the mismatch repair gene MSH2 (Table 3). For all single-nuclease mutants except the msh2 mutant, the enhancement of the frequency of forward mutation to Canr was greater than the enhancement of the reversion frequency of the hom3-10 and lys2-Bgl frameshift mutations. The enhanced forward mutation to Canr is not unexpected, since any sequence change that knocks out the function of arginine permease coded for by the 1.8-kb CAN1 gene will lead to canavanine resistance. Such changes can even include large insertions or duplications and other drastic sequence alterations. Remarkably, deletion of the RNH70 gene did not enhance any of the observed mutation frequencies, including that of the Canr marker. Hence, RNase H(70) might not play a role in mutation avoidance. In contrast, both rnh35 and rnh1 single-deletion mutants generated slightly enhanced numbers of Canr progeny and the rnh35 deletion mutant also exhibited a somewhat higher reversion frequency of the lys2-Bgl frameshift mutation. Such reversions can only arise from frameshift mutations within a 134-bp region bounded by the upstream and downstream termination codons flanking the lys2-Bgl 4-base insertion. The assay using the hom3-10 marker is specific for a deletion of a single T in a stretch of 6 T’s; hence, it is most suitable for detecting mismatch repair gene deficiencies, as verified by our experiment with the msh2 strain. Given our findings, we can assert that the absence of active RNase H(35) leads to a weak mutator phenotype, caused most probably by a slight defect in RNA primer removal. Our data with the rad27 single-deletion mutant are consistent with those obtained earlier by Tishkoff et al. (60). Determination of the exact sequence changes generated by each of the gene deletions is one of our future aims.
In this study, we focused mainly on the effects caused by loss of both RNase H(35) and Rad27 nuclease functions. The mutation frequencies of such a double mutant were found to be more enhanced than would be expected if one of the two functions were strictly epistatic to the other. However, the observed frequencies are lower than expected if the two proteins were participants in two completely distinct pathways. Therefore, we believe that the additional mutations in the rad27/rnh35 strain, compared with those already caused by the absence of Rad27p alone (and thus erasing pathways II and III) result from the failure of pathway I.
Working hypothesis for RNA primer removal from Okazaki fragments in S. cerevisiae.
As indicated above, two major (II and III) pathways of RNA primer removal involving nucleases have been proposed previously, based mainly on in vitro reconstitution experiments (references 3, 61, 63, and 64 and references therein). Our in vivo data further corroborate the previous proposal. In addition, we propose a minor pathway (I) as well as a very inefficient compensatory fourth mechanism for RNA primer hydrolysis. The most efficient pathway, pathway II, requires RNase H(35) to endonucleolytically cleave the initiator RNA as an intact fragment up to 1 nt upstream of the RNA-DNA junction; the remaining ribonucleotide is then eliminated by Rad27p. In pathway III, which is as efficient as pathway II under laboratory conditions, the FEN activity of Rad27 nuclease is capable of bypassing the need for RNase H(35) in Okazaki fragment processing and of cleaving the entire RNA primer. However, the FEN activity of Rad27p requires separation of strands, which can be achieved by two alternative ways. In one, the RNA primer might be displaced by upstream DNA synthesis during DNA replication. Alternatively, strand separation could be performed by a DNA/RNA helicase. In S. cerevisiae, the Dna2 helicase is a good candidate for this process. The enzyme was found to interact physically with Rad27 nuclease, and high expression of Rad27p complements a temperature-sensitive dna2 mutation and vice versa (5, 6). More recently, it was shown that the Dna2p has a flap endonuclease activity (2). Further experiments are needed to test whether Dna2p is involved in RNA primer removal (described above). If damaged cells must resort to pathway I as the sole mechanism of RNA primer removal, RNase H(35) will slowly hydrolyze the remaining ribonucleotide after removing the main body of the primer as in pathway II. Remarkably, this mechanism is unable to work at an elevated temperature, and cells dependent exclusively on RNase H(35) exhibit a hyperrecombination phenotype. Such a phenotype is symptomatic of a defect in Okazaki fragment processing that produces long-lived single-stranded DNA regions in the chromosomes of affected cells. In such a scenario, when all three pathways fail, a fourth and extremely inefficient mechanism must be postulated. We propose that exonuclease 1 may represent the last alternative for avoiding the life-threatening circumstance of RNA primer persistence during DNA replication (49).
ACKNOWLEDGMENTS
J.Q. and Y.Q. contributed to this work equally.
We thank Yehua Weng and Alexandra Bogusch for technical assistance in the RNase H substrate preparation and activity assays. We also thank R. Kolodner and R. Rothstein for generously providing the yeast strains listed in Table 2 and Arthur Partikian and Susan Kane for critical reading of the manuscript.
The work was supported by NIH grants CA 85344 CA 73764 to B.H.S.
REFERENCES
- 1.Alleva J L, Doetsch P W. Characterization of Schizosacharomyces pombe Rad2 protein, a FEN-1 homolog. Nucleic Acids Res. 1998;26:3645–3650. doi: 10.1093/nar/26.16.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae S-H, Choi E, Lee K-H, Park J S, Lee S-H, Seo Y-S. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J Biol Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 3.Bambara R A, Murante R S, Henricksen L A. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 4.Bhandari P, Gowrishankar J. An Escherichia coli host strain useful for efficient overproduction of cloned gene products with NaCl as the inducer. J Bacteriol. 1997;179:4403–4406. doi: 10.1128/jb.179.13.4403-4406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Braunshofer-Reiter C. Ph.D. thesis. Vienna, Austria: University of Agricultural Sciences; 1999. [Google Scholar]
- 5.Budd M E, Choe W-C, Campbell J L. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26722–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 6.Budd M E, Campbell J L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busen W, Peters J H, Hausen P. Ribonuclease H levels during the response of bovine lymphocytes to concanavalin A. Eur J Biochem. 1977;74:203–208. doi: 10.1111/j.1432-1033.1977.tb11382.x. [DOI] [PubMed] [Google Scholar]
- 8.Busen W. Purification, subunit structure, and serological analysis of calf thymus ribonuclease HI. J Biol Chem. 1980;255:9434–9443. [PubMed] [Google Scholar]
- 9.Cathala G, Rech J, Huet J, Jeanteur P. Isolation and characterization of two types of ribonucleases H in Krebs II ascites cells. J Biol Chem. 1979;254:7353–7361. [PubMed] [Google Scholar]
- 10.Ceska T A, Sayers J R, Stier G, Suck D. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature. 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 11.Ceska T A, Sayers J R. Structure-specific DNA cleavage by 5′ nucleases. Trends Biochem Sci. 1998;23:331–336. doi: 10.1016/s0968-0004(98)01259-6. [DOI] [PubMed] [Google Scholar]
- 12.Crouch R J, Toulme J J, editors. Ribonucleases H. 1998. Editions INSERM, Paris, France. [Google Scholar]
- 13.DeFrancesco R A, Lehman I R. Interaction of ribonuclease H from D. melanogaster embryos with DNA polymerase-primase. J Biol Chem. 1985;260:14764–14770. [PubMed] [Google Scholar]
- 14.Eder P S, Walder J A. Ribonuclease H from K562 human erythroleukemia cells. J Biol Chem. 1991;266:6472–6479. [PubMed] [Google Scholar]
- 15.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank G, Qiu J, Somsouk M, Weng Y, Somsouk L, Nolan J P, Shen B. Partial functional deficiency of E160D flap endonuclease-1 mutant in vitro and in vivo is due to defective cleavage of DNA substrates. J Biol Chem. 1998;273:33064–33072. doi: 10.1074/jbc.273.49.33064. [DOI] [PubMed] [Google Scholar]
- 17.Frank P, Braunshofer-Reiter C, Wintersberger U. Yeast RNase H(35) is the counterpart of the mammalian RNase HI and is evolutionarily related to prokaryotic RNase HII. FEBS Lett. 1998;421:23–26. doi: 10.1016/s0014-5793(97)01528-7. [DOI] [PubMed] [Google Scholar]
- 18.Frank P, Braunshofer-Reiter C, Wintersberger U, Grimm R, Büsen W. Cloning of the cDNA encoding the large subunit of human RNase HI, a homologue of the prokaryotic RNase HII. Proc Natl Acad Sci USA. 1998;95:12872–12877. doi: 10.1073/pnas.95.22.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank P, Braunshofer-Reiter C, Karwan A, Grimm R, Wintersberger U. Purification of S. cerevisiae RNase H(70) and identification of the corresponding gene. FEBS Lett. 1999;450:251–256. doi: 10.1016/s0014-5793(99)00512-8. [DOI] [PubMed] [Google Scholar]
- 20.Freudenreich C H, Kantrow S M, Zakian V A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 21.Gietz R D, Schiestl R H. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 22.Gordenin D A, Kunkel T A, Resnick M A. Repeat expansion—all in a flap? Nat Genet. 1998;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 23.Goulian M, Richards S H, Heard C J, Bigsby B M. Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem. 1990;265:18461–18471. [PubMed] [Google Scholar]
- 24.Hagemeier A, Grosse F. A distinct form of ribonuclease H from calf thymus stimulates its homologous DNA-polymerase-α/primase complex. Eur J Biochem. 1989;185:621–628. doi: 10.1111/j.1432-1033.1989.tb15158.x. [DOI] [PubMed] [Google Scholar]
- 25.Harrington J J, Lieber M R. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollingsworth H C, Nossal N G. Bacteriophage T4 encodes an RNase H which removes RNA primers made by the T4 DNA replication system in vitro. J Biol Chem. 1991;266:1888–1897. [PubMed] [Google Scholar]
- 27.Hosfield D J, Mol C D, Shen B, Tainer J A. Crystal structure of the DNA repair and replication exo- and endonuclease FEN-1: implications for coupling DNA and PCNA binding to enzymatic activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 28.Huang L, Kim Y, Turchi J J, Bambara R A. Structure-specific cleavage of the RNA primer from Okazaki fragments by calf thymus RNase H. J Biol Chem. 1994;269:25922–25927. [PubMed] [Google Scholar]
- 29.Hwang K Y, Baek K, Kim H-Y, Cho Y. Crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat Struct Biol. 1998;5:707–713. doi: 10.1038/1406. [DOI] [PubMed] [Google Scholar]
- 30.Ishimi Y, Claude A, Bullok P, Hurwitz J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J Biol Chem. 1988;263:19723–19733. [PubMed] [Google Scholar]
- 31.Itaya M. Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc Natl Acad Sci USA. 1990;87:8587–8591. doi: 10.1073/pnas.87.21.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itaya M, McKelvin D, Chatterjie S K, Crouch R J. Selective cloning of genes encoding RNase H from Salmonella typhimurium, Saccharomyces cerevisiae and Escherichia coli rnh mutant. Mol Gen Genet. 1991;227:438–445. doi: 10.1007/BF00273935. [DOI] [PubMed] [Google Scholar]
- 33.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 34.Karwan R, Blutsch H, Wintersberger U. Physical association of DNA polymerase stimulating activity with a RNase H purified from yeast. Biochemistry. 1983;22:5500–5502. [Google Scholar]
- 35.Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8848. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y, Eom S H, Wang J, Lee D S, Suh S W, Steitz T A. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 37.Kitahara N, Sawai Y, Tsukada K. Purification and properties of magnesium- and manganese-dependent ribonucleases H from chick embryo. J Biochem. 1982;92:855–864. doi: 10.1093/oxfordjournals.jbchem.a133999. [DOI] [PubMed] [Google Scholar]
- 38.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kornberg A, Baker T A. DNA replication. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 40.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination, and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 41.Lyamichev V, Brow M A D, Dahlberg J E. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993;193:963–968. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 42.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 43.Mueser T C, Nossal N G, Hyde C C. Structure of bacteriophage T4 RNase II, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 44.Murante R S, Rust L, Bambara R A. Calf 5′ to 3′ exonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 45.Murante R S, Henricksen L A, Bambara R A. Junction ribonuclease: an activity in Okazaki fragment processing. Proc Natl Acad Sci USA. 1998;95:2244–2249. doi: 10.1073/pnas.95.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray J M, Tavassoli M, Al-Harithy R, Sheldrick K S, Lehmann A R, Carr A R, Watts F Z. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol Cell Biol. 1994;14:4878–4888. doi: 10.1128/mcb.14.7.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolan J P, Shen B, Park M S, Sklar L A. Kinetic analysis of the human flap endonuclease-1 by flow cytometry. Biochemistry. 1996;35:11668–11676. doi: 10.1021/bi952840+. [DOI] [PubMed] [Google Scholar]
- 48.Prolla T A, Christie D-M, Liskay R M. A requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu J, Qian Y, Chen V, Guan M, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 50.Reagan M S, Pittenberger C, Siede W, Friedberg E C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roewekamp W, Sekeris C E. Purification and characteristics of hybridase (ribonuclease H) from rat liver cytosol. Eur J Biochem. 1974;43:405–413. doi: 10.1111/j.1432-1033.1974.tb03426.x. [DOI] [PubMed] [Google Scholar]
- 52.Rong Y W, Carl P L. On the molecular weight and subunit composition of calf thymus ribonuclease H1. Biochemistry. 1990;29:3838–389. doi: 10.1021/bi00454a012. [DOI] [PubMed] [Google Scholar]
- 53.Rothstein R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:281–301. [PubMed] [Google Scholar]
- 54.Shen B, Qiu J, Hosfield D J, Tainer J A. Flap endonuclease homologues identified in archaebacteria exist as independent proteins. Trends Biochem Sci. 1998;23:171–173. doi: 10.1016/s0968-0004(98)01199-2. [DOI] [PubMed] [Google Scholar]
- 55.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 56.Sommers C H, Miller E J, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 57.Stein H, Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA hybrids: effect on DNA-dependent RNA polymerase. Science. 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- 58.Struhl K, Stinchcomb D T, Scherer S, Davis R W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szankasi P, Smith G R. A role of Exo 1 from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1168. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 60.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:263–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 61.Turchi J J, Huang L, Murante R S, Kim Y, Bambara R A. Enzymatic completion of mammalian lagging-strand DNA replication. Proc Natl Acad Sci USA. 1994;91:9803–9807. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vonwirth H, Frank P, Busen W. Serological analysis and characterization of calf thymus ribonuclease H IIb. Eur J Biochem. 1989;184:321–329. doi: 10.1111/j.1432-1033.1989.tb15022.x. [DOI] [PubMed] [Google Scholar]
- 63.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 64.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 65.Williamson M S, Game J C, Fogel S. Meiotic gene conversion mutants in Saccharomyces cerevisiae strains that are isogenic to S288c. Yeast. 1985;11:53–55. [Google Scholar]
- 66.Wintersberger U. Ribonucleases H of retroviral and cellular origin. Pharmacol Ther. 1990;48:259–280. doi: 10.1016/0163-7258(90)90083-e. [DOI] [PubMed] [Google Scholar]
- 67.Zhu F X, Biswas E E, Biswas S B. Purification and characterization of the DNA polymerase α associated exonuclease: the RTH1 gene product. Biochemistry. 1997;36:5947–5954. doi: 10.1021/bi962889v. [DOI] [PubMed] [Google Scholar]