Abstract
Transcription factor Mrr1, best known for its regulation of Candida azole resistance genes such as MDR1, regulates other genes that are poorly characterized. Among the other Mrr1-regulated genes are putative methylglyoxal reductases. Methylglyoxal (MG) is a toxic metabolite that is elevated in diabetes, uremia, and sepsis, which are diseases that increase the risk for candidiasis, and MG serves as a regulatory signal in diverse organisms. Our studies in Clavispora lusitaniae, also known as Candida lusitaniae, showed that Mrr1 regulates expression of two paralogous MG reductases, MGD1 and MGD2, and that both participate in MG resistance and MG catabolism. Exogenous MG increased Mrr1-dependent expression of MGD1 and MGD2 as well as expression of MDR1, which encodes an efflux pump that exports fluconazole. MG improved growth in the presence of fluconazole and this was largely Mrr1-dependent with contributions from a secondary transcription factor, Cap1. Increased fluconazole resistance was also observed in mutants lacking Glo1, a Mrr1-indedependent MG catabolic enzyme. Isolates from other Candida species displayed heterogeneity in MG resistance and MG stimulation of azole resistance. We propose endogenous and host-derived MG can induce MDR1 and other Mrr1-regulated genes causing increased drug resistance, which may contribute to some instances of fungal treatment failure.
Keywords: Candida, Fluconazole, Pyruvaldehyde, Candida lusitaniae
Summary
While it is clear that constitutively active variants of the transcription factor Mrr1 confer resistance to the antifungal fluconazole in Candida species, the natural roles of Mrr1 remain poorly understood. Here, we report that in Clavispora lusitaniae, also known as Candida lusitaniae, two of the genes regulated by Mrr1 encode enzymes that detoxify methylglyoxal, a toxic metabolite and regulatory signal in diverse species. Serum methylglyoxal is elevated in conditions that are also associated with increased risk of infection by Candida species, such as diabetes and kidney failure. We discovered that methylglyoxal increased Mrr1-dependent expression of the methylglyoxal metabolic enzymes as well as MDR1, a drug efflux pump. Furthermore, methylglyoxal enhanced growth with azole antifungals in strains of C. lusitaniae and other Candida species. We posit that the induction of azole resistance in response to methylglyoxal may contribute to some instances of fungal treatment failure.
Introduction
Candida species are among the most prominent fungal pathogens, with mortality rates for candidemia ranging from 28 to 72% depending on geographic location (reviewed in (Lamoth et al., 2018)), and recent decades have seen a worldwide increase in the overall incidence of candidemia (Arendrup, 2010). Treatment failure of invasive fungal infections remains an important clinical issue (Nucci & Perfect, 2008) due to long-term complications, high mortality rates, and elevated healthcare costs. Perplexingly, treatment may fail even in cases where isolates from a patient have tested as susceptible to a certain antifungal in vitro, suggesting that cryptic factors which are not present during in vitro testing may influence the outcome of antifungal therapy in vivo.
In Candida species, one mechanism of azole resistance is overexpression of the gene MDR1 (Hiller et al., 2006, Jin et al., 2018, Wirsching et al., 2001, Demers et al., 2018), which encodes an efflux pump. Overexpression of MDR1 is usually caused by gain-of-function mutations in the gene encoding the zinc-cluster transcription factor Mrr1 (Dunkel et al., 2008, Schubert et al., 2008, Morschhauser et al., 2007, Demers et al., 2018). Many studies have focused on the relationship between Candida Mrr1 and resistance against clinical, host, and microbially-produced antifungal compounds (Dunkel et al., 2008, Liu & Myers, 2017, Mogavero et al., 2011, Morschhauser et al., 2007, Schubert et al., 2011, Hampe et al., 2017, Demers et al., 2018). However, little is known about other genes that Mrr1 regulates and thus, the natural role of Mrr1 beyond its involvement in drug resistance is not well understood. By studying the biological functions of Mrr1-regulated genes, it is possible to gain insight into important questions such as the evolutionary purpose of Mrr1, drivers of selection for gain-of-function mutations in Mrr1, and other consequences of high Mrr1 activity aside from drug resistance. Independent studies in C. albicans (Karababa et al., 2004, Schubert et al., 2011, Hoehamer et al., 2009, Rogers & Barker, 2003, Morschhauser et al., 2007), Candida parapsilosis (Silva et al., 2011), and Clavispora (Candida) lusitaniae (Demers et al., 2018, Kannan et al., 2019) have revealed genes that appear coordinately upregulated in fluconazole (FLZ)-resistant isolates with gain-of-function mutations in MRR1.
Previously, we demonstrated a link between FLZ resistance and specific single nucleotide polymorphisms in the MRR1 locus (CLUG_00542) among twenty clinical C. lusitaniae isolates from a single patient with cystic fibrosis (Demers et al., 2018). We identified multiple MRR1 alleles containing gain-of-function mutations that correlated with elevated FLZ resistance, though the presence of MRR1 alleles conferring high FLZ resistance within this population was unexpected, as the patient had no prior history of antifungal use. Thus, we became interested in other potential factors that could have selected for gain-of-function mutations in MRR1. An RNA-Seq analysis comparing several isolates with high- or low-activity Mrr1 variants identified nineteen genes that may be regulated by Mrr1 in C. lusitaniae, including two genes that encoded putative methylglyoxal (MG) reductases (Demers et al., 2018). Although homologs of CaGRP2/MGD1 were known to be more highly expressed in FLZ-resistant Candida strains with high Mrr1 activity across multiple species (Karababa et al., 2004, Schubert et al., 2011, Hoehamer et al., 2009, Rogers & Barker, 2003, Silva et al., 2011, Demers et al., 2018, Kannan et al., 2019), the relationship between Mrr1 and MG has not been described. Recently, genome analyses by Kannan, Sanglard, and colleagues (Kannan et al., 2019) found a possible expansion of putative aldehyde reductases including MG reductases in the C. lusitaniae genome.
MG is a reactive compound that forms spontaneously during multiple metabolic processes in all known organisms (Fig. 1). Because it is a highly reactive electrophile, MG can irreversibly modify proteins, lipids, and nucleic acids in a nonenzymatic reaction known as glycation, resulting in cellular damage and stress (Zuin et al., 2005, Takatsume et al., 2006). Serum levels of MG are elevated in patients with diabetes (Lu et al., 2011, Wang et al., 2019, McLellan et al., 1994), sepsis (Brenner et al., 2014), and uremia (Mukhopadhyay et al., 2008, Lapolla et al., 2005, Karg et al., 2009, Odani et al., 1998) relative to healthy controls. Additionally, evidence suggests that MG is generated during inflammation as part of the neutrophil respiratory burst (Zhang et al., 2016). In fungi, MG can be formed during metabolism, for example, in Saccharomyces cerevisiae, a positive correlation has been shown between rate of glycolysis and MG levels (Stewart et al., 2013). Catabolism of MG can occur through a glutathione-dependent glyoxalase system, Glo1 and Glo2 (Thornalley, 1990), or through NADH- or NADPH-dependent MG reductases (Ray & Ray, 1984) (Fig. 1). MG reductases have been characterized in S. cerevisiae, Gre2 (Chen et al., 2003), and C. albicans, Grp2 (Kwak et al., 2018).
Fig. 1. Schematic of Methylglyoxal (MG) metabolism and catabolism.
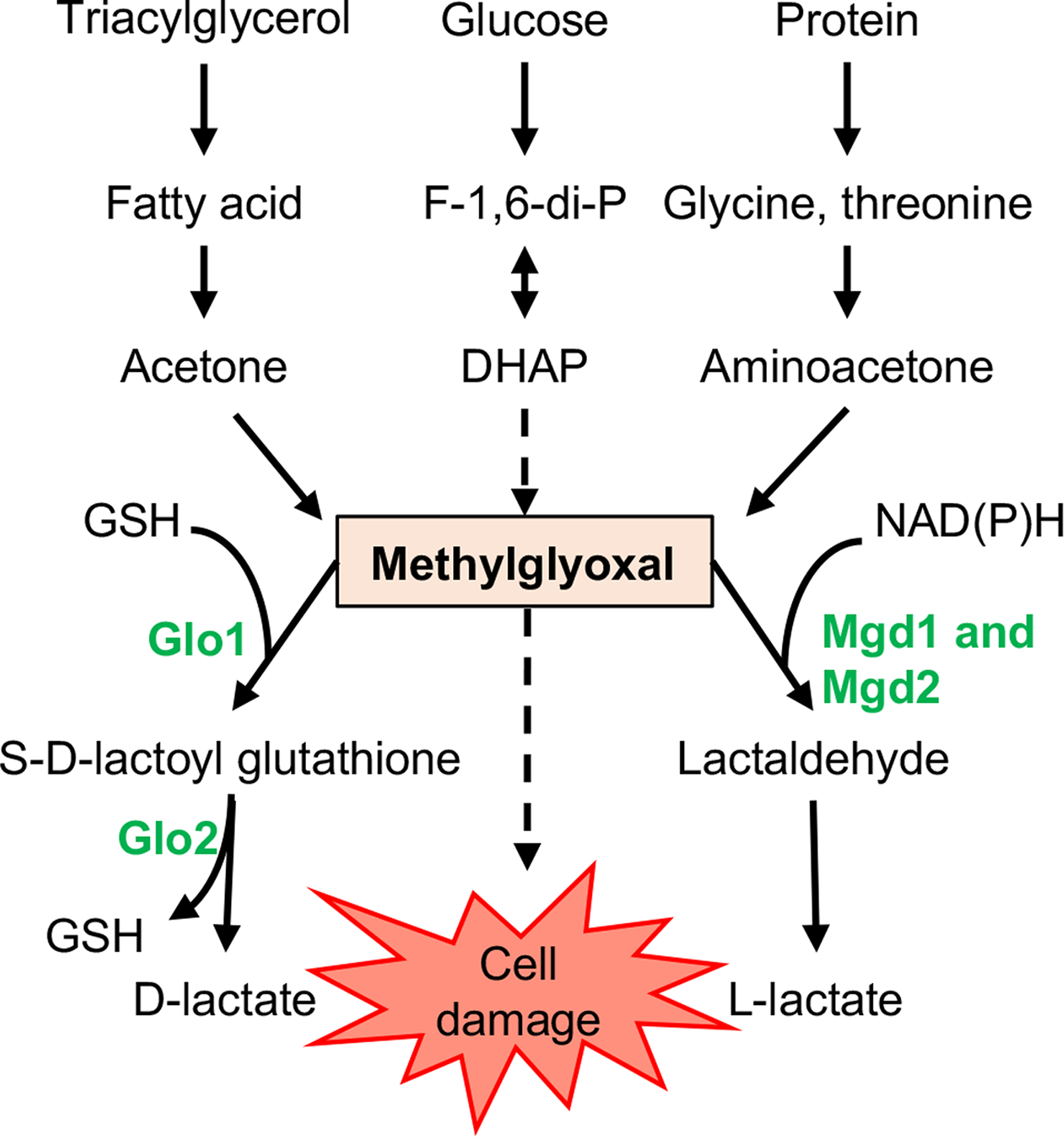
MG is a highly reactive, toxic product that forms spontaneously during the catabolism of sugars, fatty acids, and proteins. It can be detoxified to D-lactate via the GSH-dependent glyoxalase system, consisting of Glo1 and Glo2, or to lactaldehyde through NAD(P)H-dependent MG reductases such as Mgd1 and Mgd2, which are homologs of C. albicans Grp2. F-1,6-di-P, fructose 1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GSH, reduced glutathione. Solid arrows represent enzymatic processes; dashed arrows represent nonenzymatic processes.
In the present study, we demonstrated that in C. lusitaniae, Mrr1 regulates MGD1 (CLUG_01281) and MGD2 (CLUG_04991), both of which encode proteins important for the detoxification and metabolism of MG. Deletion of one or both genes led to increased sensitivity to high concentrations of exogenous MG and decreased ability to use MG as a sole carbon source. In addition, we demonstrated that MG can induce Mrr1-dependent expression of MGD1 and MGD2, as well as expression of MDR1 in a partially Mrr1-dependent manner. MG increased growth in FLZ, and this response was largely dependent on MRR1 and MDR1. Furthermore, deletion of GLO1 increased FLZ resistance, likely due to elevated endogenous levels of MG. Finally, we showed that though MG sensitivity varies across Candida species, stimulation of azole resistance by MG is not exclusive to C. lusitaniae. Together, these data demonstrate a broader role for Mrr1 in a metabolic process and describe a mechanism by which host or microbial metabolism could increase resistance to azoles in vivo.
Results
C. lusitaniae MGD1 and MGD2 contribute to the detoxification and metabolism of MG
In our previous work, an RNA-seq analysis of clinical C. lusitaniae isolates from a chronic lung infection showed that two genes with high sequence identity to each other, CLUG_01281 and CLUG_04991, were significantly upregulated in isolates with gain-of-function mutations in MRR1 (Demers et al., 2018). The protein sequences encoded by CLUG_01281 and CLUG_04991 are 88% percent identical to each other, and both have 59% and 58% identity to C. albicans Grp2 and S. cerevisiae Gre2, respectively (Kwak et al., 2018, Chen et al., 2003) (Fig. 2A). Based on sequence homology to previously characterized MG reductases and the experimental data shown below, from here forward CLUG_01281 and CLUG_04991 are referred to as MGD1 and MGD2, respectively. We further analyzed the relationships between MGD1, MGD2, and other putative MG reductases with homology to C. albicans GRP2 in select Candida spp. using FungiDB (Stajich et al., 2012, Basenko et al., 2018) (Fig. 2A). An interesting phylogeny emerged among the homologs with at least 50% amino acid identity to C. albicans Grp2. C. lusitaniae MGD1 and MGD2 were more similar to each other than to homologs in other Candida species, and other Candida species, including Candida auris, Candida parapsilosis, and Candida tropicalis also had at least one set of highly similar paralogous putative MG reductases (Fig. 2A). Candida glabrata has a pair of related putative MG reductases that are homologous to S. cerevisiae Gre2 (Fig. 2A). The phylogeny of Grp2 homologs suggests that a duplication of MG reductase genes has occurred in many Candida species, indicating that this function may be biologically important within the natural niches of Candida.
Fig. 2. MGD1 and MGD2 are required for fitness in the presence of high MG.
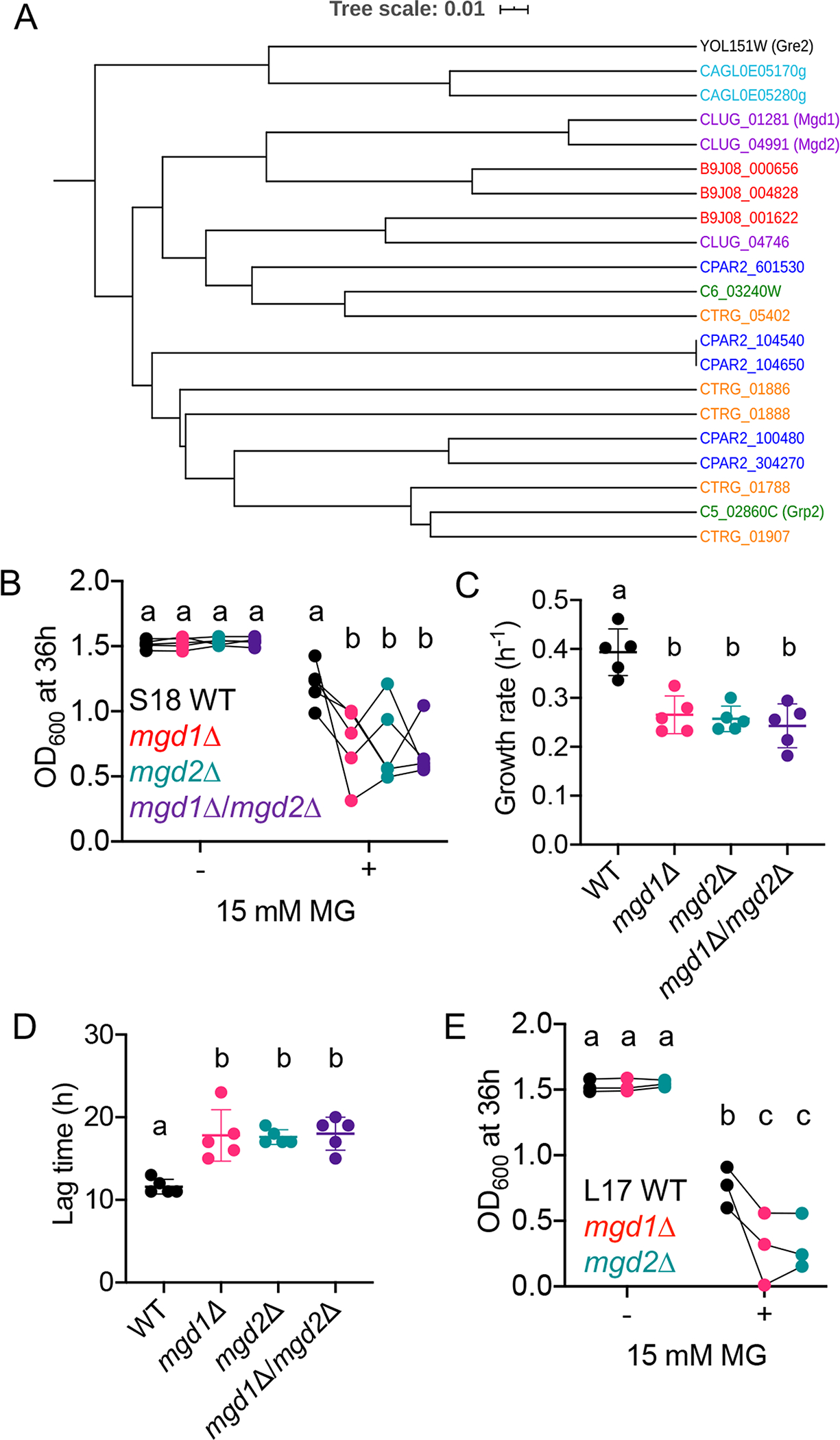
(A) Phylogeny of known and putative MG reductase based on amino acid sequences with homology to C. albicans Grp2, S. cerevisiae Gre2, and C. lusitaniae Mgd1 and Mgd2. Candida species is denoted by color: C. lusitaniae (purple); C. auris (red); C. tropicalis (orange); C. parapsilosis (blue); C. glabrata (teal) and C. albicans (green). (B-D) Growth of C. lusitaniae S18 WT (black), mgd1Δ (red), mgd2Δ (teal), and mgd1Δ/mgd2Δ (purple) strains in YPD with or without 15 mM MG in terms of OD600 after 36 h (B), exponential growth rate (C), and lag time (D). Data shown represent the mean ± SD from five independent experiments. (E) OD600 after 36 h of strain L17 WT (black), mgd1Δ (red), and mgd2Δ (teal) in YPD with or without 15 mM MG. Data shown represent the mean ± SD from three independent experiments. Ordinary two-way ANOVA with Tukey’s multiple comparison test was used for statistical evaluation in (B) and (E); a-b, a-c, b-c, p < 0.05. Data points connected by line in (B) and (E) are from the same experiment. Ordinary one-way ANOVA with Tukey’s multiple comparison test was used for statistical evaluation in (C) and (D); a-b, p < 0.01.
To determine if MGD1 and MGD2 were involved in MG resistance and gain more insight into the respective roles of these two similar genes, we knocked out each gene independently and in combination in the previously characterized C. lusitaniae clinical isolate S18, which contains a constitutively active Mrr1 variant, H467L (referred to as H4) (Demers et al., 2018). We found that although the mgd1Δ, mgd2Δ, and mgd1Δ/mgd2Δ mutants grew similarly to the S18 parental strain in the absence of MG, they grew significantly worse in the presence of 15 mM MG, with a lower OD600 after 36 hours, slower growth rate, and longer lag time (Fig. 2B, 2C and 2D; see Fig. S1A and S1C for representative growth curves). To our surprise, the double mutant did not exhibit a more severe phenotype than either single mutant, suggesting that these genes are not redundant and that both enzymes are required for full function of the cell’s NADPH- or NADH-dependent MG reductase machinery. We confirmed these phenotypes in the L17 isolate, which is closely related to S18 and shares the constitutively active Mrr1-H4 variant, and similarly found that the mgd1Δ and mgd2Δ mutants were more sensitive to MG than the parental strains (Fig. 2E; see Fig. S1D and Fig. S1B for representative growth curves). We were unable to generate an mgd1Δ/mgd2Δ double mutant in the L17 background for reasons that are not yet known but do not appear to relate to the selectable markers used as each selectable marker can be used singly.
To determine if MGD1 and MGD2 also contributed to MG metabolism, we tested whether the mgd1Δ, mgd2Δ, or mgd1Δ/mgd2Δ mutants were deficient in utilizing MG as a sole carbon source. In minimal YNB medium with 5 mM glucose, none of the mutants displayed a significant difference in OD600 at 36 h relative to the WT (Fig. S2A). With 5 mM MG as the sole carbon source, neither single mutants exhibited a significant defect in growth, but the mgd1Δ/mgd2Δ displayed a 26.8% reduction in yield (p < 0.05) relative to the WT (Fig. S2B). Together, these data suggest that both MGD1 and MGD2 play a role in the detoxification and metabolism of MG.
Mrr1 strongly regulates expression of MGD1, but MGD2 is not highly expressed under standard conditions
We have previously reported (Demers et al., 2018) an RNA-seq analysis that showed that clinical isolates with constitutive Mrr1 activity had higher levels of MGD1 and MGD2 expression than strains with low basal Mrr1 activity. Furthermore, analyses of C. albicans, C. parapsilosis, and an independent collection of clinical C. lusitaniae isolates also found that gene expression for MGD1 and MGD2 and their was elevated in azole-resistant strains with gain-of-function mutations in Mrr1 (Karababa et al., 2004, Schubert et al., 2011, Hoehamer et al., 2009, Rogers & Barker, 2003, Silva et al., 2011, Kannan et al., 2019). In both the S18 and L17 isolate backgrounds, the mrr1Δ mutant was significantly more sensitive to 15 mM MG than the WT despite no difference in growth between the isogenic parental and mrr1Δ strains in control conditions (Fig. 3A and 3B). Furthermore, we found that S18 mrr1Δ had a 32% lower yield relative to the parental strain in MG as a sole carbon source, with no defects in growth on glucose, and that S18 mrr1Δ phenocopied the mgd1Δ/mgd2Δ mutant in this assay (Fig. S2).
Fig. 3. Mrr1 regulates MG resistance and basal expression of MGD1 but not MGD2.
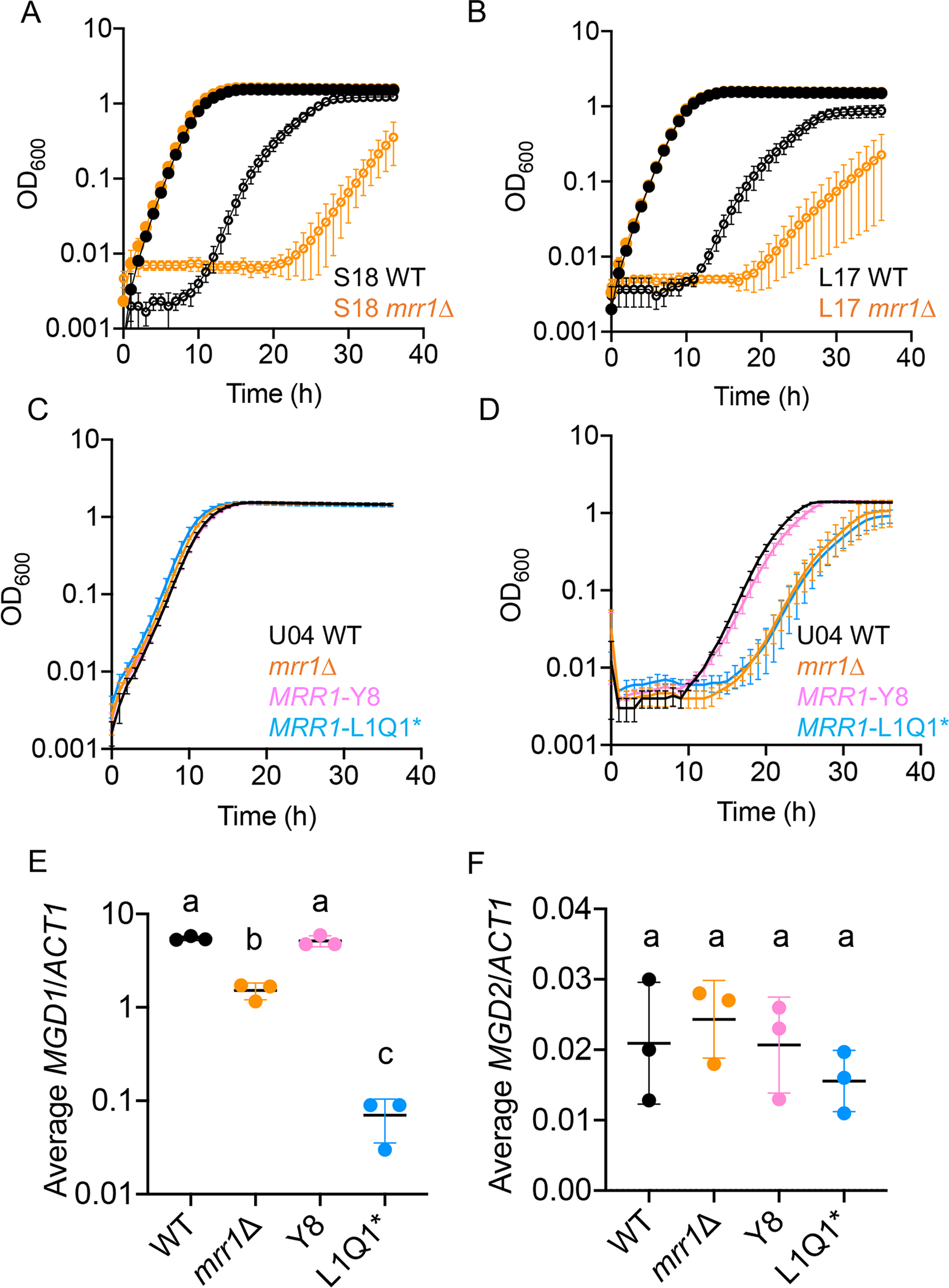
(A-B) Growth curves of C. lusitaniae S18 (A) and L17 (B) wild type (black) and mrr1Δ (orange) in YPD alone (closed circles) or with 15 mM MG (open circles). One representative experiment out of three independent experiments is shown; error bars represent the standard deviation of technical replicates within the experiment. (C-D) Growth curves of C. lusitaniae U04 (black), U04 mrr1Δ (orange), U04 mrr1Δ + MRR1Y8 (pink) and U04 mrr1Δ + MRR1L1Q1* (light blue) in YPD alone (C) or with 15 mM MG (D). One representative experiment out of three independent experiments is shown; error bars represent the standard deviation of technical replicates within the experiment. (E-F) Expression of MGD1 (E) and MGD2 (F) in C. lusitaniae U04 WT (black), U04 mrr1Δ (orange), U04 mrr1Δ + MRR1Y8 (pink) and U04 mrr1Δ + MRR1L1Q1* (light blue). Data shown represent the mean ± SD from three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparison test was used for statistical evaluation in (E-F); a-b and a-c, p < 0.0001; b-c, p < 0.01.
To directly assess whether Mrr1 regulates expression of MGD1 and MGD2, we developed a set of isogenic strains that differed only by which MRR1 allele was present at the native locus. These MRR1 alleles were complemented into the mrr1Δ derivative of U04, which we have previously described (Demers et al., 2018). The naturally-occurring MRR1 alleles in this set included a high activity variant (Mrr1-Y813C, referred to as Y8) and a low activity variant (Mrr1-L1191H + Q1197* referred to as L1Q1*); the strain with the high activity Mrr1-Y8 variant had a FLZ minimum inhibitory concentration (MIC) that was 64–128-fold higher than the strain with the low activity Mrr1-L1Q1* variant (Table 1). The mrr1Δ derivative of U04 had an 8-fold higher FLZ MIC than the strain with a low activity allele, though the mechanism for this is not known (Table 1). As expected, based on results shown in Fig. 2, we found that strains with high Mrr1 activity grew better in medium with MG compared to strains with low or no Mrr1 activity; no growth differences were observed between strains in control conditions (Fig. 3C and 3D). We found that the mrr1Δ mutant had significantly lower levels of basal expression of MGD1 relative to the relative to WT and Y8 revertant, and the strain with low activity Mrr1 variant had even lower MGD1 expression (Fig. 3E). MGD2 levels were 10–100-fold lower than MGD1, as judged using a standard curve of input DNA with primer sets for MGD1, MGD2, and ACT1 (see Methods). Surprisingly, MGD2 levels were not different across the U04 strains with different Mrr1 variants in YPD medium without MG (Fig. 3F).
Table 1.
FLZ MIC and relative Mrr1 activity of C. lusitaniae strains used in this paper
| Strain | FLZ MIC (μg mL−1) | Relative Mrr1 activity |
|---|---|---|
| S18 | 8 | High |
| S18 mgd1Δ | 8 | High |
| S18 mgd2Δ | 8 | High |
| S18 mgd1Δ/mgd2Δ | 8 | High |
| S18 mrr1Δ | 4 | N/A |
| S18 cap1Δ | 8 | High |
| S18 mrr1Δ/cap1Δ | 4 | N/A |
| S18 mdr1Δ | 2 | High |
| S18 glo1Δ | 8 | High |
| L17 | 8 | High |
| L17 mgd1Δ | 8 | High |
| L17 mgd2Δ | 8 | High |
| L17 mrr1Δ | 4 | N/A |
| L17 cap1Δ | 8 | High |
| L17 mdr1Δ | 2 | High |
| U04 | 32 | High |
| U04 mrr1Δ | 4–8 | N/A |
| U04 mrr1Δ + MRR1-Y8 | 32 | High |
| U04 mrr1Δ + MRR1-L1Q1* | 0.25 – 0.5 | Low |
| U05 | 0.5 – 1 | Low |
| L14 | 0.5 – 1 | Low |
Exogenous MG induces Mrr1-regulated genes through Mrr1 with contributions from Cap1
Because of our observations that MGD1 and MGD2 are involved in detoxification and metabolism of MG (Fig. 2 and S2B), we tested whether MG induced their expression through Mrr1 in the S18 background. As shown in Fig. 4A and 4B, 5 mM MG significantly induced expression of MGD1 by 2-fold at 15 minutes and MGD2 by 16-fold at 30 minutes in the unaltered S18 isolate. Expression of both genes remained elevated after 60 minutes of MG exposure, although they appeared to be trending downward and the difference at 60 min relative to basal expression only reached statistical significance for MGD1. MG also induced expression of another Mrr1-regulated gene, MDR1, by 6-fold at 15 and 30 minutes, but as with MGD1 and MGD2, relative MDR1 levels began trending downward by 60 minutes (Fig. 4C).
Fig. 4. Levels of MGD1, MGD2, and MDR1 transcripts were increased in response to MG in a partially Mrr1- and Cap1-dependent manner.
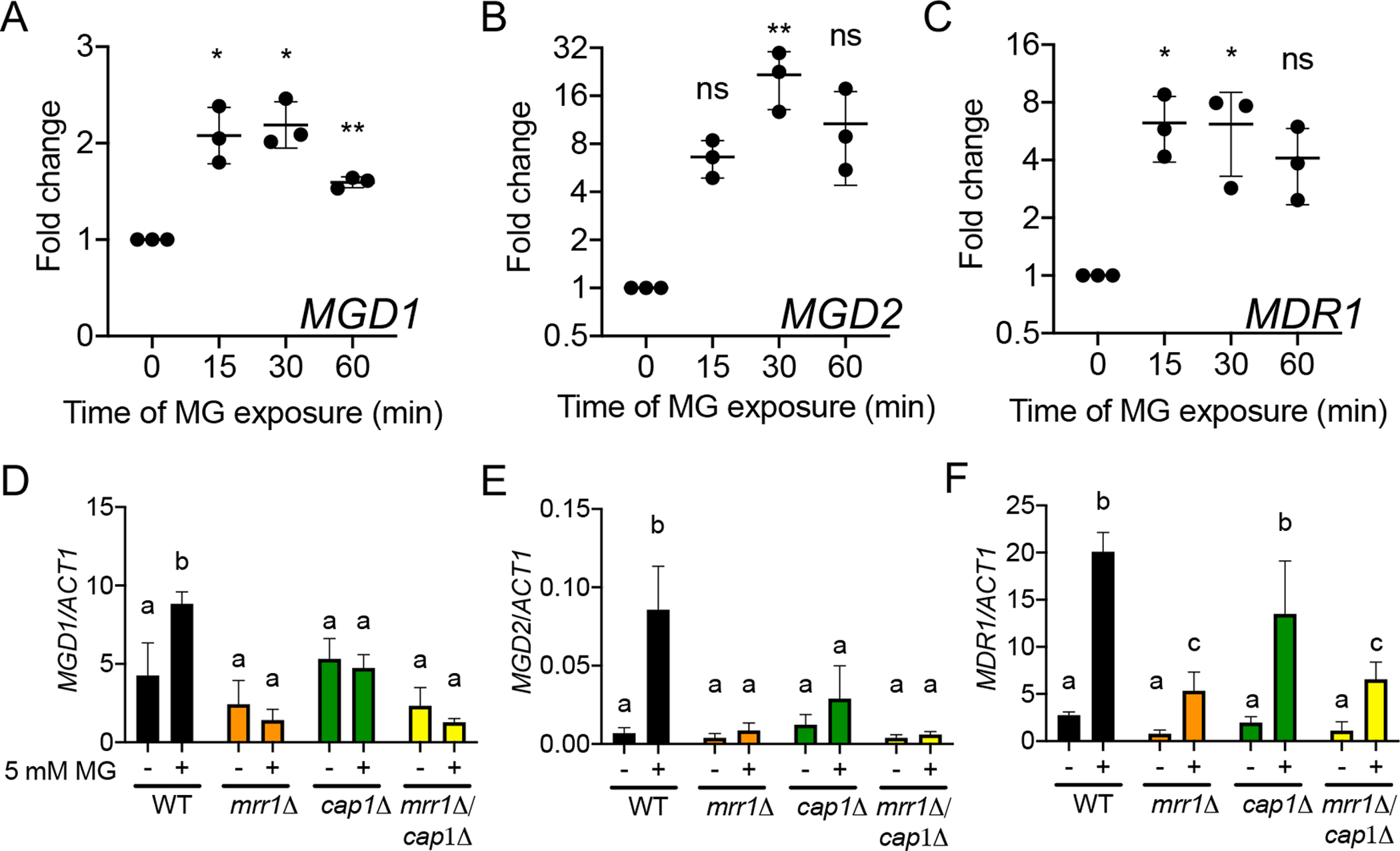
(A-C) C. lusitaniae isolate S18 was grown to exponential phase at 30°C and treated with 5 mM MG for the time indicated prior to analysis of MGD1 (A), MGD2 (B), and MDR1 (C) transcript levels by qRT-PCR. Transcript levels are normalized to levels of ACT1 and presented as ratio at each time point relative to 0 min for three independent experiments. Error bars represent the standard deviation across the three independent experiments. Ordinary one-way ANOVA with Dunnett’s multiple comparison test was used for statistical evaluation of each time point compared to t=0; * p < 0.05, ** p < 0.01, ns not significant. (D-F) C. lusitaniae S18 wild type (black) and mrr1Δ (orange), cap1Δ (green), and mrr1Δ/cap1Δ (yellow) mutants were grown to exponential phase at 30°C and treated with 5 mM MG for 15 minutes prior to analysis of MGD1 (D), MGD2 (E), and MDR1 (F) transcript levels by qRT-PCR. Transcript levels are normalized to ACT1. Data shown represent the mean ± SD for three independent experiments. Ordinary two-way ANOVA with Tukey’s multiple comparison test was used for statistical evaluation; a-b, a-c, and b-c p < 0.05.
As C. albicans Mrr1 induces MDR1 in response to benomyl and hydrogen peroxide (H2O2) in conjunction with another transcription factor, Cap1 (Schubert et al., 2011), we hypothesized that Cap1 may similarly contribute to Mrr1 induction of MDR1 in C. lusitaniae. Furthermore, in S. cerevisiae, MG directly modifies the Cap1 ortholog Yap1 by reversibly oxidizing cysteines, thereby inducing nuclear translocation (Maeta et al., 2004). To determine whether C. lusitaniae MRR1 and/or CAP1 (CLUG_02670) were required for the transcriptional response observed in Fig. 4A–C, we used isogenic mrr1Δ, cap1Δ, and mrr1Δ/cap1Δ mutants in the S18 background. Consistent with the results in Fig. 4A and 4B, MG induced expression of MGD1 (Fig. 4D) and MGD2 (Fig. 4E) by two-fold and 12-fold, respectively in the S18 parental strain, while the mrr1Δ, cap1Δ, or mrr1Δ/cap1Δ derivatives of S18 did not exhibit a significant change in expression of either gene in response to 5 mM MG (Fig. 4D and 4E). These results support the hypothesis that both Mrr1 and Cap1 are necessary for induction of MGD1 and MGD2 expression in response to MG. Additionally, the S18 cap1Δ mutant was also defective in growth in YPD + 15 mM MG (Fig. S3) providing further evidence that Cap1 plays an important role the upregulation of genes involved in MG detoxification.
Consistent with the transcriptomics evidence that Mrr1 coregulates MGD1 and MGD2 with MDR1(Demers et al., 2018), that all three genes are induced by MG (Fig. 4A–C), and that MG induction of MGD1 and MGD2 depended on Mrr1, we found that Mrr1 also played a role in MG induction of MDR1. While there were no differences in MDR1 levels among the WT, mrr1Δ, cap1Δ and mrr1Δ/cap1Δ in control conditions, the S18 mrr1Δ and the S18 mrr1Δ/cap1Δ had significantly lower MDR1 levels than the WT and cap1Δ in medium with MG (Fig. 4F). To confirm these results in strain L17, we repeated our analysis of MDR1 expression in the original isolate and its mrr1Δ and cap1Δ derivatives. In agreement with the results in Fig. 4C and 4F, the parental L17 exhibited a significant increase in MDR1 expression when exposed to MG, and knocking out MRR1 reduced MDR1 levels in medium with MG. (Fig S4). In L17, the cap1Δ also had significantly lower MDR1 levels when compared to the WT. Together, it appears that Mrr1 and Cap1 each play a role in MG-dependent MDR1 induction, though the effects of loss of Cap1 were only significant in strain L17. The weak stimulation of MDR1 by MG in the S18 mrr1Δ/cap1Δ background leads us to suggest that there are other factors may also influence the levels of MDR1 in response to MG as we discuss below.
MG stimulates growth in FLZ in an Mrr1- and Mdr1-dependent manner
Due to the induction of MDR1 expression by MG (Fig. 4F), we hypothesized that MG could increase MDR1-dependent FLZ resistance in C. lusitaniae. To test this, we used FLZ at a concentration equal to the MIC (Table 1) and 5 mM MG. While MG alone did not alter the growth of S18 WT (Fig. S5A), it drastically improved growth in the presence of FLZ (Fig. 5A), resulting in a OD600 at 16 h that was, on average, 5.2-fold higher than in FLZ alone (Fig. 5). The S18 mrr1Δ and mrr1Δ/cap1Δ mutants exhibited a significantly lower fold increase in yield at 16 h in FLZ upon amendment of the medium with MG compared to the S18 parental strain, 2.4- and 1.8-fold, respectively and S18 mdr1Δ was similar to the mrr1Δ and mrr1Δ/cap1Δ mutants (Fig. 5B). The cap1Δ mutant exhibited on average a 4.6-fold increase in growth in FLZ with MG which was not significantly different from the parental S18 in these analyses, but trended lower (Fig. 5B).
Fig. 5. MG increases FLZ resistance via MRR1 and MDR1.
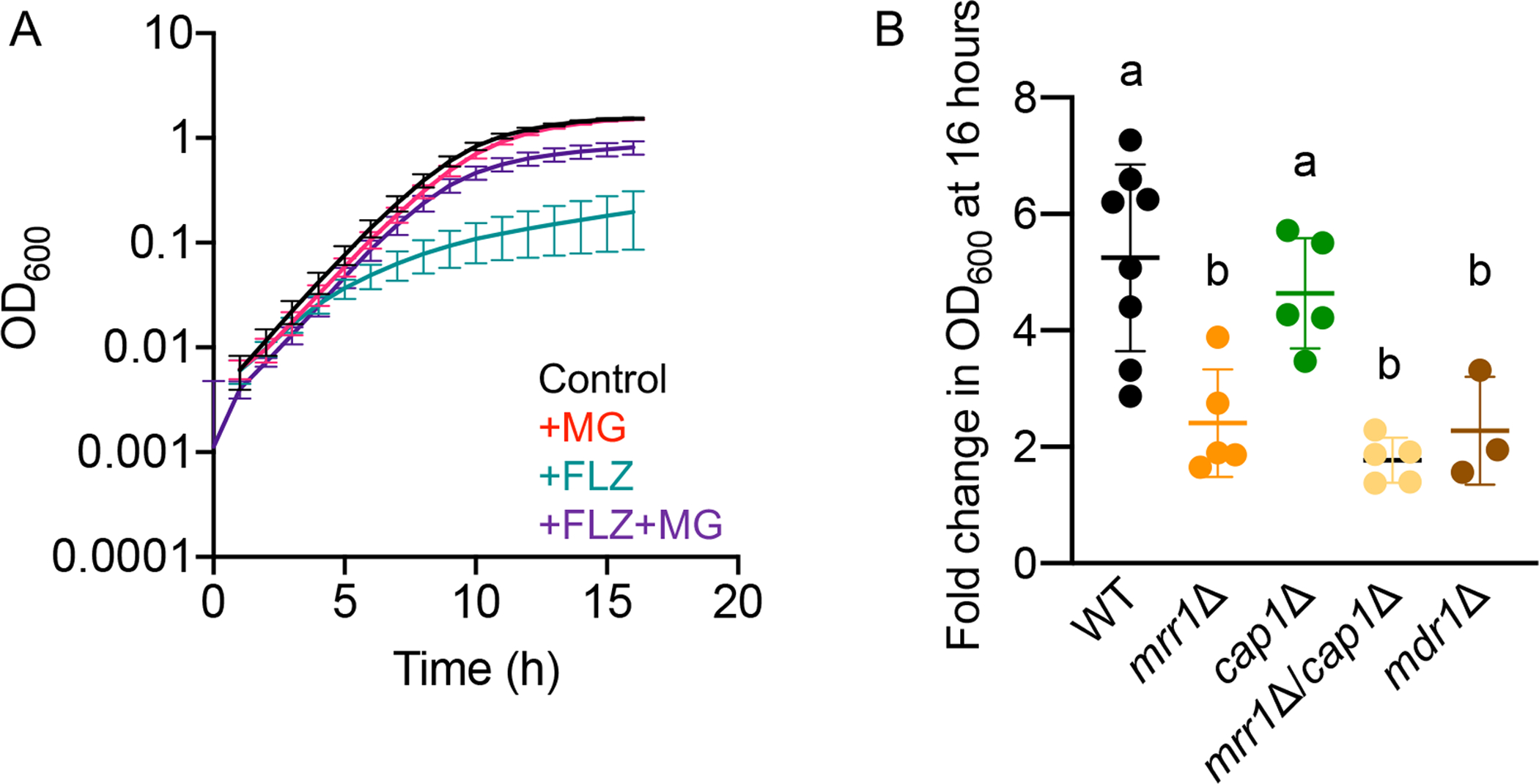
(A) C. lusitaniae isolate S18 was grown at 37°C in YPD alone (black), or with 5 mM MG (red), FLZ (equal to the MIC) (teal), or FLZ + 5 mM MG (purple). Data shown represent the mean ± SD for eight independent experiments. (B) Fold change in OD600 after 16 hours of growth for each indicated strain at 37°C in FLZ versus FLZ + 5 mM MG. Data shown represent the mean ± SD from at least three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparison test was used for statistical evaluation; a-b, p < 0.05.
We repeated these growth assays in the L17 background with strains lacking MRR1, CAP1, or MDR1. Again, MG did not alter growth for any of the strains relative to the YPD control (Fig. S5B), but it did lead to a robust stimulation of growth in FLZ, with an average fold change in OD600 of 8.5 (Fig. S5 C and D). The stimulation of growth in FLZ by MG was partially dependent on Mrr1 as the mrr1Δ mutant exhibited a fold change in OD600 of 4.2 which was significantly lower than the S18 WT (Fig. S5D). Similar to the S18 background, the L17 mdr1Δ mutant exhibited a fold change in OD600 at 16h that was significantly lower than the parental isolate (2.7-fold). Consistent with the MDR1 expression analysis of L17 strains that found that both Mrr1 and Cap1 contributed to the induction of MDR1 (Fig. S4), both Mrr1 and Cap1 contributed to increased FLZ resistance in the presence of MG (Fig. S5D). The differences between the S18 and L17 backgrounds in the robustness of the cap1Δ mutant phenotype, with Cap1 appearing to play a greater role in MDR1 regulation in L17, suggest that strain-dependent variables may influence the relative importance of the two transcription factors in the MG response.
Strains with constitutively active Mrr1 variants exhibit greater growth with MG in FLZ than strains with low activity Mrr1 variants
Given our discovery of repeated selection for Mrr1 variants with constitutive activity within a chronic C. lusitaniae lung infection (Demers et al., 2018), we sought to determine if higher basal Mrr1 activity effected the magnitude of stimulation of FLZ resistance by MG. We compared the effects of a sub-inhibitory concentration of MG on growth in the presence of inhibitory concentrations of FLZ for C. lusitaniae strains S18 and L17, which both express the constitutively active Mrr1-H4 variant, to previously published strains U05 and L14, which express the low activity Mrr1-L1Q1* variant. While there were no differences in growth among strains in reference conditions, the combination of FLZ and MG significantly increased the growth of isolates with high Mrr1 activity (S18 and L17) by ~6 fold relative to growth with FLZ alone and isolates with low Mrr1 activity (U05 and L14) showed similar trends, though the differences were not significant (Fig. 6B). These results show strains with highly active Mrr1 variants were able to reach more robust levels of FLZ resistance in response to MG than strains with low Mrr1 activity.
Fig. 6. Strains with a constitutively active Mrr1 variant show a greater increase in growth with FLZ by MG than strains with low activity Mrr1 variants.
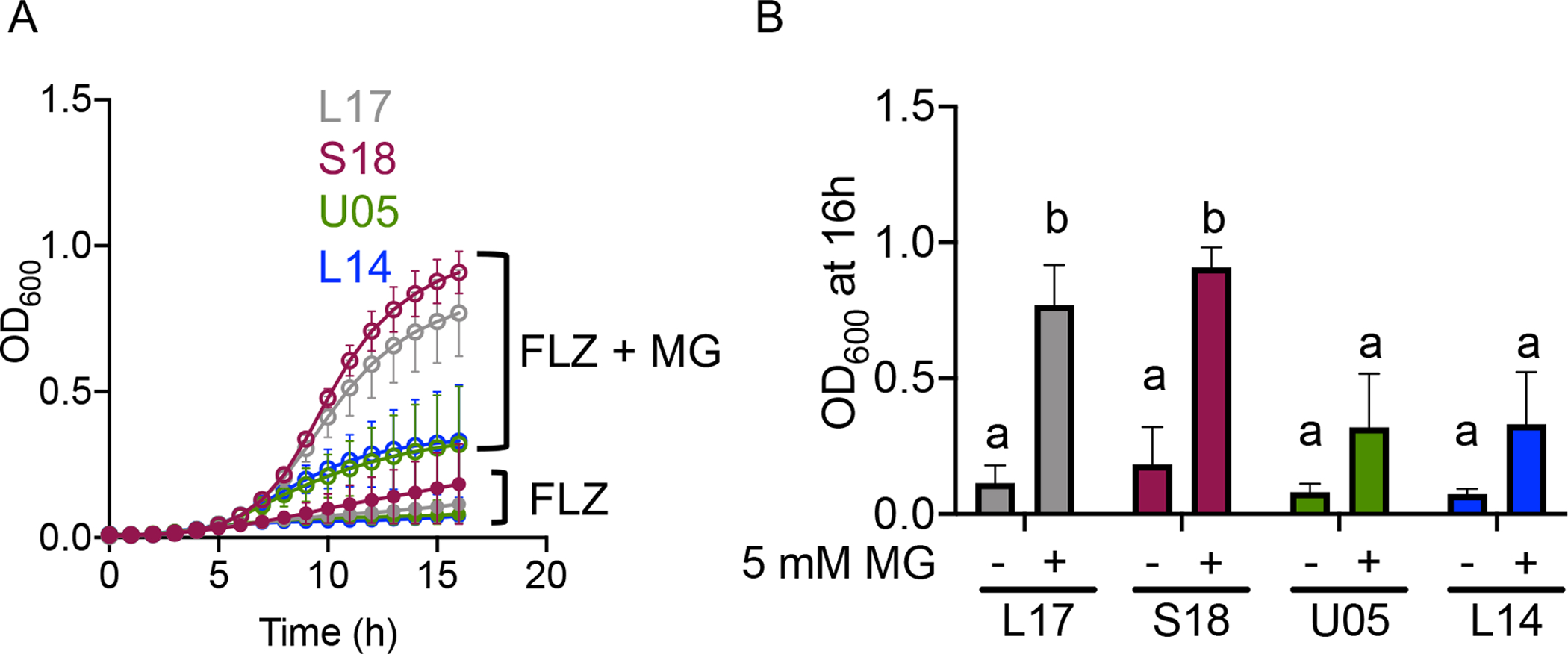
C. lusitaniae isolates with low activity Mrr1 variants, U05 (green) and L14 (blue), or with constitutively active Mrr1 variants, L17 (grey) and S18 (dark red), were grown at 37°C in YPD with FLZ in the presence or absence of 5 mM MG. (A) Growth kinetics of isolates U05, L14, S18, and L17 in FLZ with (open circles) or without (closed circles) 5 mM MG. Data shown represent the mean ± SD from three independent experiments. (B) OD600 after 16 hours of growth for each indicated strain at 37°C in FLZ with or without 5 mM MG as indicated. Data shown represent the mean ± SD from three independent experiments. Ordinary two-way ANOVA Tukey’s multiple comparison test was used for statistical analysis; a-b, p < 0.05.
Absence of GLO1 causes increased sensitivity to MG and increased resistance to FLZ
The experiments above focused on the effects of exogenous MG, but endogenously generated MG is also an important signal that modulates cell behavior (Moraru et al., 2018, Irshad et al., 2019, Nokin et al., 2019, Antognelli et al., 2019). Disruption of the glyoxalase pathway in S. cerevisiae has been shown to cause an accumulation of intracellular MG (Maeta et al., 2004, Penninckx et al., 1983) and render cells highly sensitive to exogenous MG (Inoue & Kimura, 1996). The glyoxalase pathway, which consists of the glutathione-dependent enzymes Glo1 and Glo2, is widely recognized as a major mechanism for MG catabolism in eukaryotic cells (see Fig. 1) (Thornalley, 1996). Thus, we were interested in whether the S18 glo1Δ mutant (lacking CLUG_04105) was more resistant to FLZ than its parent in the absence of exogenously added MG. We found that S18 glo1Δ was highly sensitive to 15 mM MG (Fig. 7A), even more so than the S18 mgd1Δ, mgd2Δ, and mgd1Δ/mgd2Δ mutants (Fig. S2C). Although S18 glo1Δ had similar growth kinetics in YPD as the S18 WT (Fig. 7A), the glo1Δ mutant grew substantially better in FLZ compared to its parent strain (Fig 7B). These data lead us to speculate that the absence of GLO1 in C. lusitaniae leads to an accumulation of intracellular MG, which may influence the activity of Mrr1, causing an increase in FLZ resistance.
Fig 7. The absence of GLO1, which encodes a MG catabolizing enzyme, leads to increased sensitivity to MG and increased resistance to FLZ.
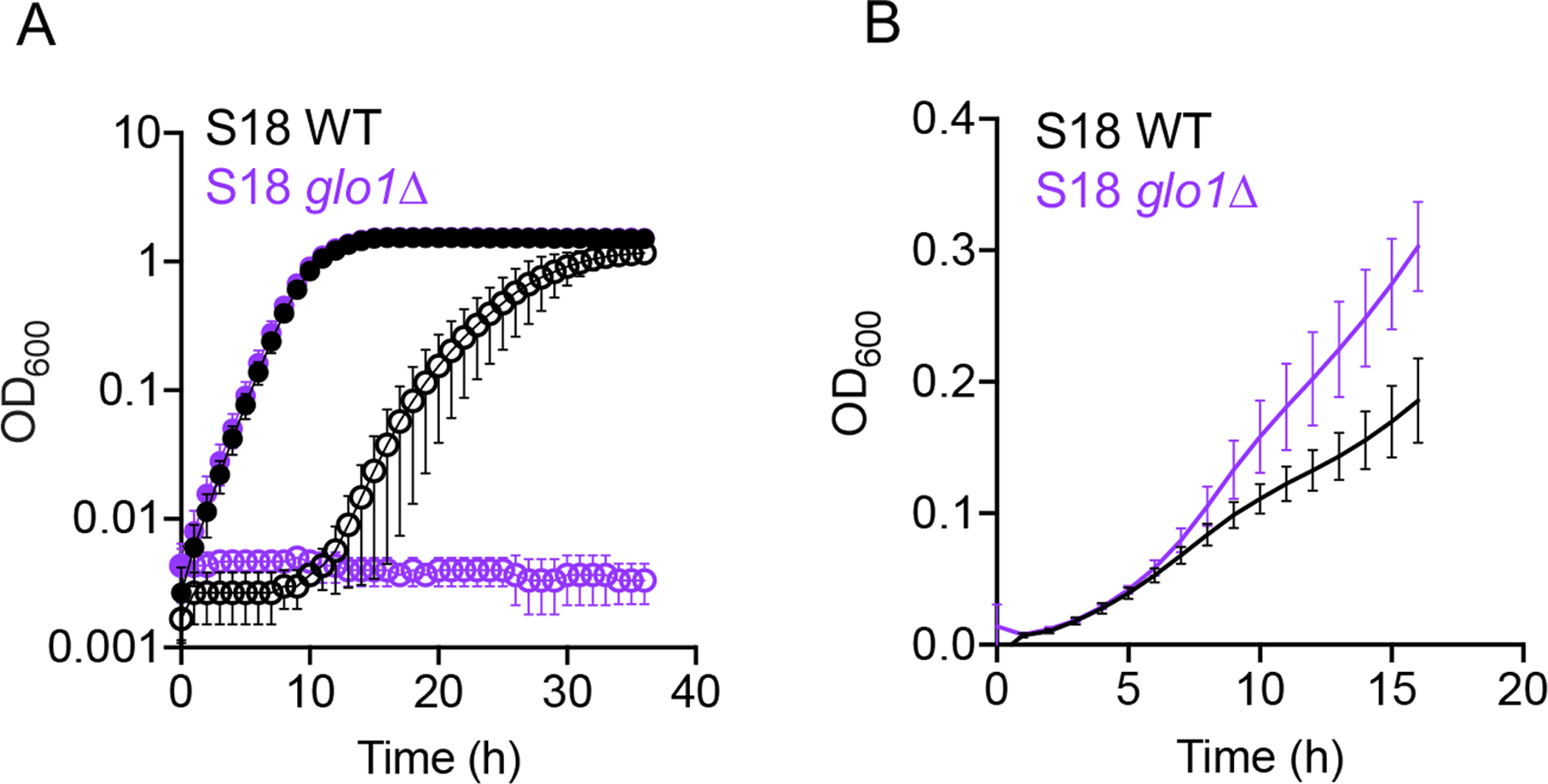
(A) C. lusitaniae S18 wild type (WT, black) and glo1Δ derivative (purple) were grown in YPD with (open circles) or without (closed circles) 15 mM MG. (B) Growth of S18 wild type (WT, black) and glo1Δ (purple) derivative in YPD with 8 μg mL−1 FLZ. Data shown represent the mean ± SD from three independent experiments.
C. lusitaniae is more resistant to MG than many other Candida species, and some strains of other species exhibit induction of azole resistance by MG
To assess intrinsic MG resistance across multiple Candida species, we assessed growth for a panel of isolates representing seven Candida species on YPD agar plates in the presence and absence of 15 mM MG. As controls, we included the C. lusitaniae S18 isolate and S18 glo1Δ, shown above to be highly sensitive to MG (Fig. 7). We found that C. lusitaniae and Candida dubliniensis strains were only minimally inhibited by 15 mM MG on plates. There was, however, heterogeneity in growth on MG among C. auris and C. albicans strains, and the tested Candida guilliermondii, C. glabrata, and C. parapsilosis strains were highly sensitive to MG (Fig. 8A). Overall, the results in Fig. 8A, using a limited number of strains, suggest that intrinsic MG resistance varies between Candida species and strains.
Fig 8. MG sensitivity and MG stimulation of azole resistance varies among Candida species and strains.
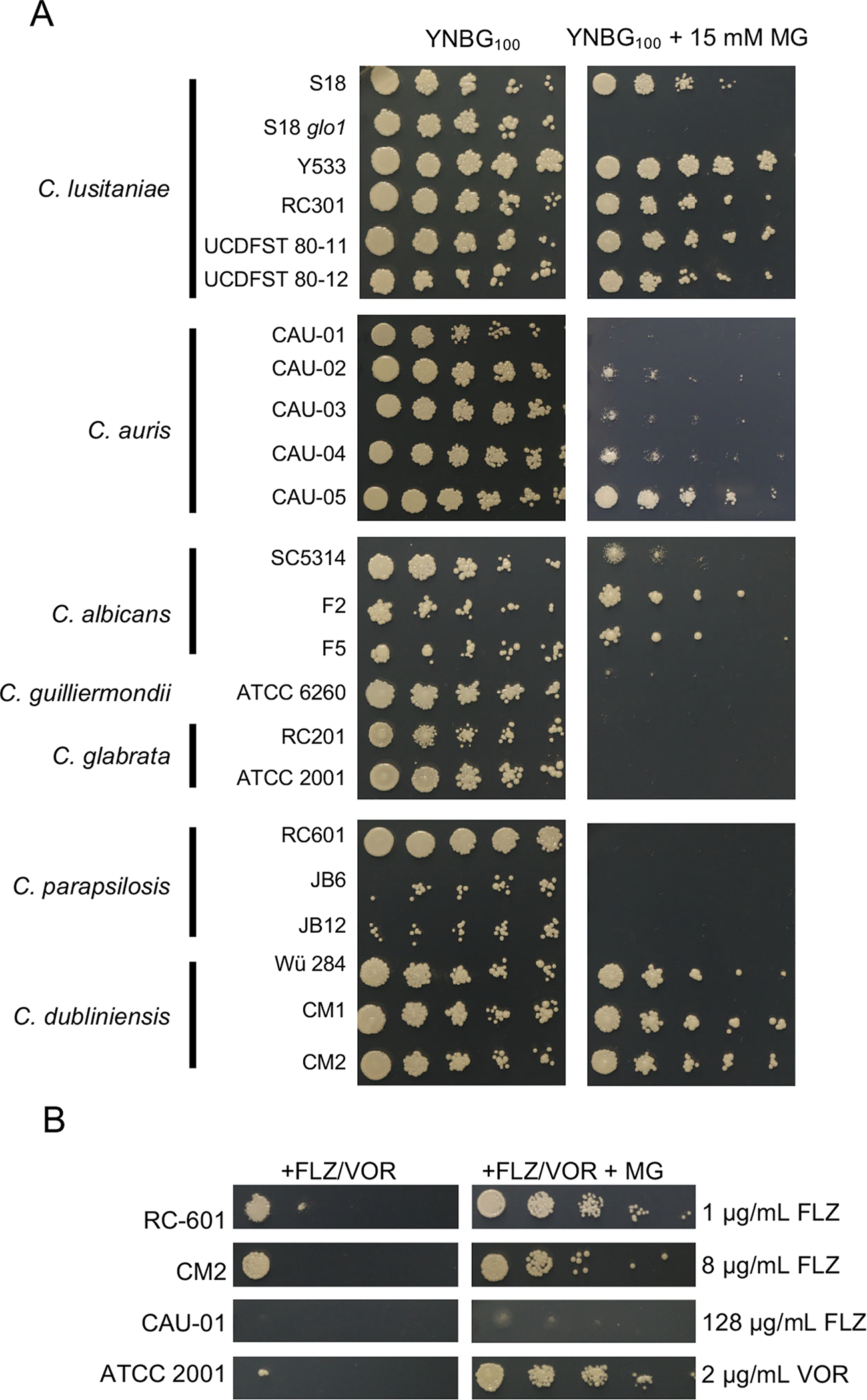
(A) Serial 1:10 dilutions of each Candida strain were spotted onto YNBG100 plates without or with 15 mM MG, then grown at 37°C for two days. One representative out of three independent experiments is shown. (B) Serial 1:10 dilutions of Candida strains were spotted onto YNBG100 plates containing the indicated concentration of FLZ or VOR without or with 3 mM MG. Only strains that demonstrated improved growth with the presence of 3 mM MG are shown, the other strains are shown in Fig. S6. One representative experiment out of two independent experiments is shown.
We used the same strains as in Fig. 8A to determine if the increase in FLZ resistance in the presence of MG was conserved across Candida species. Using 3 mM MG, a lower concentration of MG than in Fig. 8A because of high MG sensitivity of some species, we determined resistance to increasing concentrations of either FLZ or voriconazole (VOR) depending on the species. As shown in Fig. 8B, C. parapsilosis RC-601 and C. dubliniensis CM2 displayed a striking increase of growth on FLZ with MG and C. glabrata ATCC 2001 exhibited a striking increase of growth on VOR with MG. C. auris CAU-01 demonstrated a more subtle increase in growth with MG (Fig. 8B). Strains that did not demonstrate visible stimulation of growth on FLZ or VOR by MG under the tested conditions are shown in Fig S6. These results suggest that MG stimulation of azole resistance is not exclusive to C. lusitaniae, but not every strain within a species can be stimulated under the conditions tested. Future studies are required to determine what factors determine whether a strain is or is not capable of being induced by MG to have higher azole resistance.
Discussion
The findings from this study show that Mrr1 plays an important role in regulating genes other than MDR1 in ways that impact growth and fitness, thereby adding to the growing appreciation of MG as an important biological signal across the tree of life. Although the serum concentrations of MG reported in humans are lower than those used in vitro for this study (Beisswenger et al., 1999, Ogasawara et al., 2016), local MG levels at sites of infection are hard to measure as MG is highly reactive. At the site of a chronic infection, it is likely that microbes are exposed to MG from a variety of exogenous and endogenous sources including the host immune system, other microbes, and the pathogen’s own metabolic activity (see Fig. 1 and reviewed in (Allaman et al., 2015)). Evidence for the generation of MG in vivo comes from the fact that group A Streptococci require glyoxalase I for resistance to neutrophil killing, suggesting that neutrophils may be a source of MG in vivo (Zhang et al., 2016). In addition, CaGRP2, along with other stress-response genes, was upregulated in C. albicans cells grown in the murine cecum (Rosenbach et al., 2010). Even low levels of exogenous MG may stimulate a transcriptional response if endogenous MG is already high due to basal metabolism or depletion of the reducing agents required for MG detoxification. Production of MG can be affected by the local environment with low carbon or phosphate increasing MG production in mammalian and bacterial cells, respectively (Liu et al., 2011, Masterjohn et al., 2013, Ferguson et al., 1998). In addition, MG reaction with arginine, lysine, and cysteine residues on proteins forms both reversible and irreversible adducts, and thus some effects of MG on transcriptional activation may increase over time upon low level exposure (Zuin et al., 2005, Takatsume et al., 2006). Our demonstration of the induction of azole resistance by MG could be an important step toward understanding and preventing treatment failure in populations who are susceptible to Candida infection.
Previous studies of Mrr1 in multiple Candida species have focused on the regulation and biological significance of only a small number of Mrr1-regulated genes, primarily the two efflux pumps encoded by MDR1 (Hiller et al., 2006, Jin et al., 2018, Wirsching et al., 2001, Demers et al., 2018) and FLU1 (Calabrese et al., 2000, Li et al., 2013, Hampe et al., 2017). Here, we show that isogenic C. lusitaniae strains with gain-of-function mutations in Mrr1 led to higher levels of MGD1 and MGD2 transcripts, and higher resistance to exogenous MG (Fig. 3D) than strains with low Mrr1 activity. Furthermore, we showed that MG induced Mrr1 activity to increase the expression of not just MGD1 and MGD2, but also MDR1. The co-regulation of genes involved in the detoxification of metabolic by-products with efflux pumps may highlight a broad coordination of a stress response that could be important in vivo. Future studies will determine whether MG enhances FLZ resistance in vivo and if MG exposure can contribute to the selection for high activity Mrr1 variants.
While multiple chemical inducers of Mrr1 activity have been described, including methotrexate, 4-nitroquinoline-N-oxide, o-phenanthroline, benomyl, diethyl maleate, diamide, and H2O2, (Harry et al., 2005, Mogavero et al., 2011, Schubert et al., 2011), little is known about why or how these inducers activate Mrr1. It has been postulated that many of these compounds many directly or indirectly induce oxidative stress, which then activates Mrr1. MG is especially interesting as a natural inducer of Mrr1 activity because i) it is produced by cells during metabolism and in vivo as a antimicrobial agent, ii) Mrr1 regulates enzymes that specifically metabolize and detoxify this compound, and iii) it has similarly been documented to cause oxidative stress like other known inducers of Mrr1 activity. Though the mechanism by which MG activates transcription in C. lusitaniae will be the subject of future work, in S. cerevisiae MG has been shown to activate the Cap1 homolog Yap1 by reversibly modifying cysteine residues (Maeta et al., 2004). Multiple studies have established that the transcription factors Mrr1 and Cap1, a regulator of oxidative stress, can cooperate to regulate the expression of MDR1 in C. albicans (Schubert et al., 2011, Mogavero et al., 2011) and we found evidence that this can be the case in C. lusitaniae. We do not yet know if Mrr1 or Cap1 is directly modified by MG. C. lusitaniae Mrr1 contains many cysteine residues near the C-terminal portion that could be react with MG in a manner similar to S. cerevisiae Yap1. Furthermore, the observation that MG slightly induced MDR1 even in the absence of both MRR1 and CAP1 (Fig. 4F) suggests that other transcription regulators may play a role in MDR1 induction in response to MG. Other known regulators of MDR1 expression in C. albicans include the transcription factors Mcm1, which is required for induction of MDR1 by benomyl and by hyperactive Mrr1, but not induction by H2O2 (Mogavero et al., 2011), and Upc2 (Schubert et al., 2011, Znaidi et al., 2008), as well as the Swi/Snf chromatin remodeling complex (Liu & Myers, 2017).
As MG is elevated in many diseases associated with Candida infections, we were struck by the implications of subinhibitory levels of exogenous MG inducing Mrr1 activity and by extension FLZ treatment outcomes. Diabetes (reviewed in (Rodrigues et al., 2019)) and uremia (Pyrgos et al., 2009, Jawale et al., 2018) are considered risk factors for infection by a variety of Candida species, and both are associated with higher levels of MG. Our studies with the C. lusitaniae glo1Δ mutant suggest that intracellular MG can also influence FLZ resistance (Penninckx et al., 1983, Maeta et al., 2004). The glyoxalase system, utilizing Glo1 and Glo2, requires reduced glutathione (GSH) to function (Fig. 1), so it is possible that oxidants encountered in vivo could deplete GSH and cause increased intracellular MG. In fact, GSH levels are lower in chronic infections, such as those associated with cystic fibrosis (Kettle et al., 2014, Dickerhof et al., 2017). It is also worth noting that diethyl maleate, a compound shown to induce MDR1 expression in C. albicans (Harry et al., 2005), is commonly used in laboratory studies to deplete GSH (Yamauchi et al., 2011, Urban et al., 2017, Enkvetchakul & Bottje, 1995, Mitchell et al., 1983, Zheng et al., 2018).
Importantly, we found that MG induction of azole resistance was not specific to C. lusitaniae but more broadly applicable to other Candida species though with clear strain-to-strain differences in MG sensitivity (Fig. 8B). Interestingly, several species of bacteria exhibit an increase of drug resistance-related genes in response to MG; for example, MG induces expression of the MexEF-OprN multidrug efflux system in Pseudomonas aerguinosa (Juarez et al., 2017), and derepresses Escherichia coli TetR family repressor NemR (Lee et al., 2013). Clearly, MG is an important stimulus and stressor that many microbes encounter and understanding how MG affects microbial physiology and drug resistance can open doors to novel means of modulating pathogenic and/or commensal microbes for better health outcomes. For example, it would be interesting to investigate whether supplementation with carnosine, a known scavenger of MG (Hipkiss & Chana, 1998) that is readily available as a dietary supplement, could improve the efficacy when treating infection by Candida species, particularly in patients who are predisposed to elevated serum MG.
Methods
Generation of MG reductase phylogenetic tree
Orthologs of CaGrp2 from S. cerevisiae and multiple Candida species were identified in FungiDB (https://fungidb.org) (Stajich et al., 2012, Basenko et al., 2018) and selected for a protein Clustal Omega multiple sequence alignment (Sievers et al., 2011). The resulting alignment was then used to generate a phylogenetic tree using the Interactive Tree of Life (ITOL) tool (https://itol.embl.de) (Letunic & Bork, 2007).
Strains, media, and growth conditions
The sources of all strains used in this study are listed in Table S1. All strains were stored long term in a final concentration of 25% glycerol at −80°C and freshly streaked onto yeast extract peptone dextrose (YPD) agar (10 g/L yeast extract, 20 g/L peptone, 2% glucose, 1.5% agar) once every seven days and maintained at room temperature. Cells were grown in YPD, yeast nitrogen base (YNB) (0.67% yeast nitrogen base medium with ammonium sulfate (RPI Corp)) supplemented with either 5 mM dextrose or 5 mM MG (Sigma-Aldrich, 5.55 M), or RPMI-1640 (Sigma, containing L-glutamine, 165 mM MOPS, 2% glucose at pH 7) liquid as noted. Media was supplemented with FLZ (Sigma-Aldrich, stock 4 mg mL−1 in DMSO) or 3 mM, 5 mM or 15 mM MG as noted. Unless otherwise noted, all overnight cultures were grown in 5 mL YPD liquid medium (10 g/L yeast extract, 20 g/L peptone, 2% glucose) on a rotary wheel at 30°C. E. coli strains were grown in LB with either 100 μg mL−1 carbenicillin (carb) or 15 μg mL−1 gentamycin (gent) as necessary.
Plasmids for complementation of MRR1
We amplified i) the MRR1 gene and terminator with ~1150 bp upstream for homology from the appropriate strain’s genomic DNA, ii) the selective marker, HygB from pYM70 (Basso et al., 2010), and iii) ~950 bp downstream of MRR1 for homology from genomic U05 (identical sequence for all relevant strains using primers listed in Table S2. PCR products were cleaned up using the Zymo DNA Clean & Concentrator kit (Zymo Research) and assembled using the S. cerevisiae recombination technique previously described (Shanks et al., 2006). Plasmids created in S. cerevisiae were isolated using a yeast plasmid miniprep kit (Zymo Research) and transformed into High Efficiency NEB®5-alpha competent E. coli (New England BioLabs). E. coli containing pMQ30 derived plasmids were selected for on LB containing 15 μg mL−1 gentamycin. Plasmids from E. coli were isolated using a Zyppy Plasmid Miniprep kit (Zymo Research) and subsequently verified by Sanger sequencing with the Dartmouth College Genomics and Molecular Biology Shared Resources Core. MRR1 complementation plasmids were linearized with Not1-HF (New England BioLabs), cleaned up the Zymo DNA Clean & Concentrator kit (Zymo Research) and eluted in molecular biology grade water (Corning) before transformation of 2 μg into C. lusitaniae strain U04 mrr1Δ as described below.
All plasmids for complementing MRR1 were constructed using the S. cerevisiae recombination technique previously described (Shanks et al., 2006) and primers listed in Table S2. To create the precursor plasmid pMQ30MRR1-L1191H+Q1197*-URA3, MRR1 with ~1150 bp upstream of MRR1 and separately ~950 bp downstream of MRR1 were amplified from genomic U05 (containing MRR1L1191H+Q1197*) DNA and C. albicans URA3 under the controls of a TEF1 promoter was amplified from pTEF1-URA3. This construct did not restore growth of 5-FOA resistant C. lusitaniae strains on uracil deplete medium, so we replaced URA3 with a different selectable marker. Linearized pMQ30MRR1-L1191H+Q1197*-URA3 (using XbaI) and PCR amplified HygB, the hygromycin B resistance gene from pYM70 (Basso et al., 2010), were combined using the S. cerevisiae recombination technique to create pMQ30MRR1-L1191H+Q1197*-HygB. To create the pMQ30MRR1-Y813C-HygB plasmid, pMQ30MRR1-L1191H+Q1197*-HygB was linearized with XbaI and NotI to remove MRR1 and the upstream sequence. Replacement sequence including MRR1 with ~1150 bp upstream of MRR1 were amplified from U04 (MRR1Y813C) gDNA.
Plasmids created in S. cerevisiae were isolated using a yeast plasmid miniprep kit (Zymo Research) and transformed into High Efficiency NEB®5-alpha competent E. coli (New England BioLabs). E. coli containing pMQ30 derived plasmids were selected for on LB containing 15 μg mL−1 gentamycin. Plasmids from E. coli were isolated using a Zyppy Plasmid Miniprep kit (Zymo Research) and subsequently verified by Sanger sequencing with the Dartmouth College Genomics and Molecular Biology Shared Resources Core. All restriction enzymes were purchased from New England BioLabs and used as recommended by the manufacturer.
Mutant construction
Mutants were generated using an expression-free CRISPR-Cas9 method, as previously described (Grahl et al., 2017), with the exception of the mgd1Δ/mgd2Δ double mutant, as detailed below. In brief, cultures were grown to exponential phase in 50 mL YPD on a shaker at 150 rpm, then washed and incubated in TE buffer and 0.1 M lithium acetate at 30°C for one hour. Dithiothreitol was added to a final concentration of 100 mM and cultures were incubated for an additional 30 minutes at 30°C. Cells were washed and resuspended in 1 M sorbitol before being transferred to electroporation cuvettes. To each cuvette was added 1.5 μg of DNA for the knockout or MRR1 complementation construct and Cas9 ribonucleoprotein containing crRNA specific to the target gene. Following electroporation, cells were allowed to recover in YPD at 30°C for four to six hours. Cells were then plated on YPD agar supplemented with 200 μg mL−1 nourseothricin (NAT) or 600 μg mL−1 hygromycin B (HYG) and incubated at 30°C for two days. The mgd1Δ/mgd2Δ double mutant was generated from the S18 mgd1Δ single mutant using the microhomology repair method (Al Abdallah et al., 2017). In brief, the knockout construct containing 50 bp homology to the flanking regions of MGD2 was transformed alongside Cas9 complexed with two crRNA, targeting the 5’ and 3’ region immediately adjacent to MGD2. PCR with primers inside the NAT1 or HygB cassette and in the flanking regions of the genes outside of each construct were used to confirm all mutants. Primers (IDT) used to create knockout constructs and verify mutants are listed in Table S2.
Minimum Inhibitory Concentration (MIC) Assay
MIC assays for FLZ were performed as described in (Demers et al., 2018) using the broth microdilution method. In brief, overnight cultures were diluted to an OD600 of 0.1 in 200 μL dH2O and 60 μL of each dilution were added to 5 mL RPMI-1640 medium. FLZ was serially diluted across a clear, flat-bottom 96-well plate (Falcon) from 128 μg mL−1 down to 0.25 μg mL−1 in RPMI-1640. To each well was added 100 μL of cell suspension in RPMI-1640. Upon addition of cells, the final concentration of FLZ ranged from 64 μg mL−1 to 0.125 μg mL−1. Plates were incubated at 35°C and scored for growth at 24 hours; the results are summarized in Table 1. The MIC was defined as the drug concentration that abolished visible growth compared to a drug-free control.
Growth Kinetics
C. lusitaniae cultures were grown overnight, diluted 1:50 into 5 mL fresh YPD, and grown for four to six hours at 30°C. After washing, the cultures were diluted to OD600 of 1 in 200 μL dH2O. Each inoculum was prepared by pipetting 60 μL of the OD600 of 1 suspension into 5 mL YPD. Clear 96-well flat-bottom plates (Falcon) were prepared by adding 100 μl per well YPD or YPD with MG and/or FLZ at twice the desired final concentrations. 100 μL of inoculum was added to each row of the plate. Each plate was set up in technical triplicate for each strain and condition. The plates were incubated in a Synergy Neo2 Microplate Reader (BioTek, USA) to generate a kinetic curve. The plate reader protocol was as follows: heat to 37°C, start kinetic, read OD600 every 60 minutes for 16 or 36 hours, end kinetic.
Spot Assays
Candida cultures were grown overnight, diluted 1:50 into 5 mL fresh YPD, and grown for four to six hours at 30°C. Cultures were diluted to OD600 of 1 in 200 μL dH2O. Each strain was then serially diluted by 1:10 down to an OD600 of approximately 1 × 10−6. 5 μL of each dilution was spotted onto YPD alone or YPD containing the specified concentrations of MG, FLZ or VOR (Cayman Chemical Company, stock 1 mg mL−1 in DMSO). Plates were incubated at 37°C for two days before imaging.
Quantitative Real-Time PCR
C. lusitaniae cultures were grown overnight, diluted 1:50 into 5 mL fresh YPD, and grown for four hours at 30°C. Control cultures were harvested at this point and MG was added to a final concentration of 5 mM to all other cultures, which were returned to 30°C on a roller drum. Cultures were then harvested after 15, 30, or 60 minutes. To harvest, 2 mL of culture was spun in a tabletop centrifuge at 13.2 × g for 5 min and supernatant was discarded. RNA isolation, gDNA removal, cDNA synthesis, and quantitative real-time PCR were performed as previously described (Demers et al., 2018). Transcripts were normalized to ACT1 expression. Primers are listed in Table S2.
Statistical Analysis and Figure preparation
All graphs were prepared with GraphPad Prism 8.3.0 (GraphPad Software). One- and two-way analysis of variance (ANOVA) tests were performed in Prism; details on each test are described in the corresponding figure legends. All p values were two-tailed and p < 0.05 were considered to be significant for all analyses performed and are indicated with asterisks or letters in the text: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. The graphical abstract was prepared using BioRender (biorender.com).
Supplementary Material
Acknowledgements
We thank Theodore White, Richard Calderone, Lawrence Myers, Joachim Morschhäuser, Isabel Miranda, Kyria Boundy-Mills and the FDA-CDC Antimicrobial Resistance Isolate Bank for providing strains. We thank Judith Berman for the pGEM-URA3 plasmid.
Funding.
This study was supported by grants R01 5R01 AI127548 to DAH and AI133956 to EGD. Core services were provided by STANTO19R0 to CFF RDP, P30-DK117469 to DartCF, and P20-GM113132 to BioMT. Sequencing services and specialized equipment were provided by the Genomics and Molecular Biology Shared Resource Core at Dartmouth, NCI Cancer Center Support Grant 5P30 CA023108-41. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing interests. The authors have declared that no competing interests exist.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
- Al Abdallah Q, Ge W, and Fortwendel JR (2017) A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro-Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates. mSphere 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaman I, Belanger M, and Magistretti PJ (2015) Methylglyoxal, the dark side of glycolysis. Front Neurosci 9: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antognelli C, Moretti S, Frosini R, Puxeddu E, Sidoni A, and Talesa VN (2019) Methylglyoxal Acts as a Tumor-Promoting Factor in Anaplastic Thyroid Cancer. Cells-Basel 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup MC (2010) Epidemiology of invasive candidiasis. Curr Opin Crit Care 16: 445–452. [DOI] [PubMed] [Google Scholar]
- Basenko EY, Pulman JA, Shanmugasundram A, Harb OS, Crouch K, Starns D, Warrenfeltz S, Aurrecoechea C, Stoeckert CJ Jr., Kissinger JC, Roos DS, and Hertz-Fowler C (2018) FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J Fungi (Basel) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso LR Jr., Bartiss A, Mao Y, Gast CE, Coelho PS, Snyder M, and Wong B (2010) Transformation of Candida albicans with a synthetic hygromycin B resistance gene. Yeast 27: 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisswenger PJ, Howell SK, Touchette AD, Lal S, and Szwergold BS (1999) Metformin reduces systemic methylglyoxal levels in type 2 diabetes. Diabetes 48: 198–202. [DOI] [PubMed] [Google Scholar]
- Brenner T, Fleming T, Uhle F, Silaff S, Schmitt F, Salgado E, Ulrich A, Zimmermann S, Bruckner T, Martin E, Bierhaus A, Nawroth PP, Weigand MA, and Hofer S (2014) Methylglyoxal as a new biomarker in patients with septic shock: an observational clinical study. Crit Care 18: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese D, Bille J, and Sanglard D (2000) A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 146 (Pt 11): 2743–2754. [DOI] [PubMed] [Google Scholar]
- Chen CN, Porubleva L, Shearer G, Svrakic M, Holden LG, Dover JL, Johnston M, Chitnis PR, and Kohl DH (2003) Associating protein activities with their genes: rapid identification of a gene encoding a methylglyoxal reductase in the yeast Saccharomyces cerevisiae. Yeast 20: 545–554. [DOI] [PubMed] [Google Scholar]
- Demers EG, Biermann AR, Masonjones S, Crocker AW, Ashare A, Stajich JE, and Hogan DA (2018) Evolution of drug resistance in an antifungal-naive chronic Candida lusitaniae infection. Proc Natl Acad Sci U S A 115: 12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerhof N, Pearson JF, Hoskin TS, Berry LJ, Turner R, Sly PD, Kettle AJ, and Arest CF (2017) Oxidative stress in early cystic fibrosis lung disease is exacerbated by airway glutathione deficiency. Free radical biology & medicine 113: 236–243. [DOI] [PubMed] [Google Scholar]
- Dunkel N, Blass J, Rogers PD, and Morschhauser J (2008) Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol 69: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkvetchakul B, and Bottje WG (1995) Influence of diethyl maleate and cysteine on tissue glutathione and growth in broiler chickens. Poult Sci 74: 864–873. [DOI] [PubMed] [Google Scholar]
- Ferguson GP, Totemeyer S, MacLean MJ, and Booth IR (1998) Methylglyoxal production in bacteria: suicide or survival? Archives of microbiology 170: 209–218. [DOI] [PubMed] [Google Scholar]
- Grahl N, Demers EG, Crocker AW, and Hogan DA (2017) Use of RNA-Protein Complexes for Genome Editing in Non-albicans Candida Species. mSphere 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe IAI, Friedman J, Edgerton M, and Morschhauser J (2017) An acquired mechanism of antifungal drug resistance simultaneously enables Candida albicans to escape from intrinsic host defenses. PLoS Pathog 13: e1006655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry JB, Oliver BG, Song JL, Silver PM, Little JT, Choiniere J, and White TC (2005) Drug-induced regulation of the MDR1 promoter in Candida albicans. Antimicrob Agents Chemother 49: 2785–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller D, Sanglard D, and Morschhauser J (2006) Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob Agents Chemother 50: 1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipkiss AR, and Chana H (1998) Carnosine protects proteins against methylglyoxal-mediated modifications. Biochem Biophys Res Commun 248: 28–32. [DOI] [PubMed] [Google Scholar]
- Hoehamer CF, Cummings ED, Hilliard GM, Morschhauser J, and Rogers PD (2009) Proteomic analysis of Mrr1p- and Tac1p-associated differential protein expression in azole-resistant clinical isolates of Candida albicans. Proteomics Clin Appl 3: 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, and Kimura A (1996) Identification of the structural gene for glyoxalase I from Saccharomyces cerevisiae. J Biol Chem 271: 25958–25965. [PubMed] [Google Scholar]
- Irshad Z, Xue M, Ashour A, Larkin JR, Thornalley PJ, and Rabbani N (2019) Activation of the unfolded protein response in high glucose treated endothelial cells is mediated by methylglyoxal. Sci Rep 9: 7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawale C, Ramani K, and Biswas PS (2018) Defect in neutrophil function accounts for impaired anti-fungal immunity in kidney dysfunction. J Immunol 200. [Google Scholar]
- Jin L, Cao Z, Wang Q, Wang Y, Wang X, Chen H, and Wang H (2018) MDR1 overexpression combined with ERG11 mutations induce high-level fluconazole resistance in Candida tropicalis clinical isolates. BMC Infect Dis 18: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez P, Jeannot K, Plesiat P, and Llanes C (2017) Toxic Electrophiles Induce Expression of the Multidrug Efflux Pump MexEF-OprN in Pseudomonas aeruginosa through a Novel Transcriptional Regulator, CmrA. Antimicrob Agents Chemother 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan A, Asner SA, Trachsel E, Kelly S, Parker J, and Sanglard D (2019) Comparative genomics for the elucidation of multidrug resistance in Candida lusitaniae. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karababa M, Coste AT, Rognon B, Bille J, and Sanglard D (2004) Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Chemother 48: 3064–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg E, Papp F, Tassi N, Janaky T, Wittmann G, and Turi S (2009) Enhanced methylglyoxal formation in the erythrocytes of hemodialyzed patients. Metabolism 58: 976–982. [DOI] [PubMed] [Google Scholar]
- Kettle AJ, Turner R, Gangell CL, Harwood DT, Khalilova IS, Chapman AL, Winterbourn CC, Sly PD, and Arest CF (2014) Oxidation contributes to low glutathione in the airways of children with cystic fibrosis. The European respiratory journal 44: 122–129. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Ku M, and Kang SO (2018) Inducible NAD(H)-linked methylglyoxal oxidoreductase regulates cellular methylglyoxal and pyruvate through enhanced activities of alcohol dehydrogenase and methylglyoxal-oxidizing enzymes in glutathione-depleted Candida albicans. Biochim Biophys Acta Gen Subj 1862: 18–39. [DOI] [PubMed] [Google Scholar]
- Lamoth F, Lockhart SR, Berkow EL, and Calandra T (2018) Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother 73: i4–i13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapolla A, Flamini R, Lupo A, Arico NC, Rugiu C, Reitano R, Tubaro M, Ragazzi E, Seraglia R, and Traldi P (2005) Evaluation of glyoxal and methylglyoxal levels in uremic patients under peritoneal dialysis. Ann N Y Acad Sci 1043: 217–224. [DOI] [PubMed] [Google Scholar]
- Lee C, Shin J, and Park C (2013) Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol Microbiol 88: 395–412. [DOI] [PubMed] [Google Scholar]
- Letunic I, and Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128. [DOI] [PubMed] [Google Scholar]
- Li R, Kumar R, Tati S, Puri S, and Edgerton M (2013) Candida albicans Flu1-mediated efflux of salivary histatin 5 reduces its cytosolic concentration and fungicidal activity. Antimicrob Agents Chemother 57: 1832–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang R, Desai K, and Wu L (2011) Upregulation of aldolase B and overproduction of methylglyoxal in vascular tissues from rats with metabolic syndrome. Cardiovasc Res 92: 494–503. [DOI] [PubMed] [Google Scholar]
- Liu Z, and Myers LC (2017) Candida albicans Swi/Snf and Mediator complexes differentially regulate Mrr1-induced MDR1 expression and fluconazole resistance. Antimicrob Agents Chemother 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Randell E, Han Y, Adeli K, Krahn J, and Meng QH (2011) Increased plasma methylglyoxal level, inflammation, and vascular endothelial dysfunction in diabetic nephropathy. Clinical biochemistry 44: 307–311. [DOI] [PubMed] [Google Scholar]
- Maeta K, Izawa S, Okazaki S, Kuge S, and Inoue Y (2004) Activity of the Yap1 transcription factor in Saccharomyces cerevisiae is modulated by methylglyoxal, a metabolite derived from glycolysis. Mol Cell Biol 24: 8753–8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterjohn C, Park Y, Lee J, Noh SK, Koo SI, and Bruno RS (2013) Dietary fructose feeding increases adipose methylglyoxal accumulation in rats in association with low expression and activity of glyoxalase-2. Nutrients 5: 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan AC, Thornalley PJ, Benn J, and Sonksen PH (1994) Glyoxalase system in clinical diabetes mellitus and correlation with diabetic complications. Clin Sci (Lond) 87: 21–29. [DOI] [PubMed] [Google Scholar]
- Mitchell JB, Russo A, Biaglow JE, and Mcpherson SJ (1983) Cellular glutathione depletion by diethyl maleate or buthionine sulfoximine and its effects on the oxygen enhancement ratio. Radiat Res 94: 612–612. [PubMed] [Google Scholar]
- Mogavero S, Tavanti A, Senesi S, Rogers PD, and Morschhauser J (2011) Differential requirement of the transcription factor Mcm1 for activation of the Candida albicans multidrug efflux pump MDR1 by its regulators Mrr1 and Cap1. Antimicrob Agents Chemother 55: 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraru A, Wiederstein J, Pfaff D, Fleming T, Miller AK, Nawroth P, and Teleman AA (2018) Elevated levels of the reactive metabolite methylglyoxal recapitulate progression of type 2 diabetes. Cell Metab 27: 926–934 e928. [DOI] [PubMed] [Google Scholar]
- Morschhauser J, Barker KS, Liu TT, Bla BWJ, Homayouni R, and Rogers PD (2007) The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog 3: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Ghosh A, and Kar M (2008) Methylglyoxal increase in uremia with special reference to snakebite-mediated acute renal failure. Clin Chim Acta 391: 13–17. [DOI] [PubMed] [Google Scholar]
- Nokin MJ, Bellier J, Durieux F, Peulen O, Rademaker G, Gabriel M, Monseur C, Charloteaux B, Verbeke L, van Laere S, Roncarati P, Herfs M, Lambert C, Scheijen J, Schalkwijk C, Colige A, Caers J, Delvenne P, Turtoi A, Castronovo V, and Bellahcene A (2019) Methylglyoxal, a glycolysis metabolite, triggers metastasis through MEK/ERK/SMAD1 pathway activation in breast cancer. Breast Cancer Res 21: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucci M, and Perfect JR (2008) When primary antifungal therapy fails. Clin Infect Dis 46: 1426–1433. [DOI] [PubMed] [Google Scholar]
- Odani H, Shinzato T, Usami J, Matsumoto Y, Brinkmann Frye E, Baynes JW, and Maeda K (1998) Imidazolium crosslinks derived from reaction of lysine with glyoxal and methylglyoxal are increased in serum proteins of uremic patients: evidence for increased oxidative stress in uremia. FEBS Lett 427: 381–385. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Tanaka R, Koike S, Horiuchi Y, Miyashita M, and Arai M (2016) Determination of methylglyoxal in human blood plasma using fluorescence high performance liquid chromatography after derivatization with 1,2-diamino-4,5-methylenedioxybenzene. J Chromatogr B Analyt Technol Biomed Life Sci 1029–1030: 102–105. [DOI] [PubMed] [Google Scholar]
- Penninckx MJ, Jaspers CJ, and Legrain MJ (1983) The glutathione-dependent glyoxalase pathway in the yeast Saccharomyces cerevisiae. J Biol Chem 258: 6030–6036. [PubMed] [Google Scholar]
- Pyrgos V, Ratanavanich K, Donegan N, Veis J, Walsh TJ, and Shoham S (2009) Candida bloodstream infections in hemodialysis recipients. Med Mycol 47: 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M, and Ray S (1984) Purification and partial characterization of a methylglyoxal reductase from goat liver. Biochimica et biophysica acta 802: 119–127. [DOI] [PubMed] [Google Scholar]
- Rodrigues CF, Rodrigues ME, and Henriques M (2019) Candida sp. infections in patients with diabetes mellitus. J Clin Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PD, and Barker KS (2003) Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob Agents Chemother 47: 1220–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbach A, Dignard D, Pierce JV, Whiteway M, and Kumamoto CA (2010) Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell 9: 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, Dunkel N, Aid M, Boucher G, Rogers PD, Raymond M, and Morschhauser J (2011) Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob Agents Chemother 55: 2212–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S, Rogers PD, and Morschhauser J (2008) Gain-of-function mutations in the transcription factor MRR1 are responsible for overexpression of the MDR1 efflux pump in fluconazole-resistant Candida dubliniensis strains. Antimicrob Agents Chemother 52: 4274–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, and O’Toole GA (2006) Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72: 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, and Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AP, Miranda IM, Guida A, Synnott J, Rocha R, Silva R, Amorim A, Pina-Vaz C, Butler G, and Rodrigues AG (2011) Transcriptional profiling of azole-resistant Candida parapsilosis strains. Antimicrob Agents Chemother 55: 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ Jr., and Roos DS (2012) FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40: D675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BJ, Navid A, Kulp KS, Knaack JLS, and Bench G (2013) D-Lactate production as a function of glucose metabolism in Saccharomyces cerevisiae. Yeast 30: 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsume Y, Izawa S, and Inoue Y (2006) Methylglyoxal as a signal initiator for activation of the stress-activated protein kinase cascade in the fission yeast Schizosaccharomyces pombe. The Journal of biological chemistry 281: 9086–9092. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ (1990) The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. The Biochemical journal 269: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley PJ (1996) Pharmacology of methylglyoxal: formation, modification of proteins and nucleic acids, and enzymatic detoxification--a role in pathogenesis and antiproliferative chemotherapy. Gen Pharmacol 27: 565–573. [DOI] [PubMed] [Google Scholar]
- Urban N, Tsitsipatis D, Hausig F, Kreuzer K, Erler K, Stein V, Ristow M, Steinbrenner H, and Klotz LO (2017) Non-linear impact of glutathione depletion on C. elegans life span and stress resistance. Redox Biol 11: 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Ma SB, Liu ZF, Li H, and Gao WY (2019) Elevated levels of alpha-dicarbonyl compounds in the plasma of type II diabetics and their relevance with diabetic nephropathy. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences 1106–1107: 19–25. [DOI] [PubMed] [Google Scholar]
- Wirsching S, Moran GP, Sullivan DJ, Coleman DC, and Morschhauser J (2001) MDR1-mediated drug resistance in Candida dubliniensis. Antimicrob Agents Chemother 45: 3416–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi S, Kiyosawa N, Ando Y, Watanabe K, Niino N, Ito K, Yamoto T, Manabe S, and Sanbuissho A (2011) Hepatic transcriptome and proteome responses against diethyl maleate-induced glutathione depletion in the rat. Arch Toxicol 85: 1045–1056. [DOI] [PubMed] [Google Scholar]
- Zhang MM, Ong CL, Walker MJ, and McEwan AG (2016) Defence against methylglyoxal in Group A Streptococcus: a role for Glyoxylase I in bacterial virulence and survival in neutrophils? Pathog Dis 74. [DOI] [PubMed] [Google Scholar]
- Zheng J, Hu CL, Shanley KL, and Bizzozero OA (2018) Mechanism of protein carbonylation in glutathione-depleted rat brain slices. Neurochem Res 43: 609–618. [DOI] [PubMed] [Google Scholar]
- Znaidi S, Weber S, Al-Abdin OZ, Bomme P, Saidane S, Drouin S, Lemieux S, De Deken X, Robert F, and Raymond M (2008) Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot Cell 7: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin A, Vivancos AP, Sanso M, Takatsume Y, Ayte J, Inoue Y, and Hidalgo E (2005) The glycolytic metabolite methylglyoxal activates Pap1 and Sty1 stress responses in Schizosaccharomyces pombe. The Journal of biological chemistry 280: 36708–36713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


