Abstract
It has been shown that gut microbiota dysbiosis leads to physiological changes and links to a number of diseases, including cancers. Thus, many cancer categories and treatment regimens should be investigated in the context of the microbiome. Owing to the availability of metagenome sequencing and multiomics studies, analyses of species characterization, host genetic changes, and metabolic profile of gut microbiota have become feasible, which has facilitated an exponential knowledge gain about microbiota composition, taxonomic alterations, and host interactions during tumorigenesis. However, the complexity of the gut microbiota, with a plethora of uncharacterized host‐microbe, microbe‐microbe, and environmental interactions, still contributes to the challenge of advancing our knowledge of the microbiota‐cancer interactions. These interactions manifest in signaling relay, metabolism, immunity, tumor development, genetic instability, sensitivity to cancer chemotherapy and immunotherapy. This review summarizes current studies/molecular mechanisms regarding the association between the gut microbiota and the development of cancers, which provides insights into the therapeutic strategies that could be harnessed for cancer diagnosis, treatment, or prevention.
Keywords: cancer biomarkers, chemotherapy, fecal microbiota transplantation, gut microbiome, immunotherapy, microbiota, probiotics
This review summarizes current studies/molecular mechanisms regarding the association between the gut microbiota and the development of cancers, which provides insights into the therapeutic strategies that could be harnessed for cancer diagnosis, treatment or prevention.
Abbreviations
- 3‐oxoLCA
3 oxolithocholic acid
- AhR
aryl hydrocarbon receptor
- AhR
aryl hydrocarbon receptor
- AOM
axozymethane
- AOM
azoxymethane
- C. rodentium
Citrobacter rodentium
- CDK
Cyclin‐dependent kinase
- CS
cesarean section
- CSGG
cell surface β‐glucan/galactan
- CTLA‐4
cytotoxic T‐lymphocyte‐associated antigen 4
- DC
dendritic cell
- DCA
deoxycholic acid
- DP IEL
double‐positive intraepithelial lymphocyte
- DSB
double‐strand break
- DSS
dextran sulfate sodium
- E. coli
Escherichia coli
- EAC
esophageal adenocarcinoma
- EGFR
epidermal growth factor receptor
- ENO1
enolase 1
- ESCC
esophageal squamous cell carcinoma
- F. nucleatum
Fusobacterium nucleatum
- FMT
Fecal microbiota transplantation
- GBC
gallbladder cancer
- H. pylori
Helicobacter pylori
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- ICI
immune checkpoint inhibitor
- IFN‐γ
interferon gamma
- KRT7
Keratin7
- KRT7‐AS
Keratin7‐antisense
- L. reuteri
Lactobacillus reuteri
- lncRNA
long non‐coding RNA
- MAMP
microbe‐associated molecular pattern
- MDSC
myeloid‐derived suppressor cell
- METTL3
methyltransferase‐like 3
- NAD
nicotinamide adenine dinucleotide
- NADP
nicotinamide adenine dinucleotide phosphate
- NEIL2
Nei‐like DNA glycosylase 2
- NF‐κB
nuclear factor‐κB
- OSCC
oral squamous cell carcinoma
- PCWBR2
putative cell wall binding repeat 2
- PD‐1
programmed cell death protein 1
- PDAC
pancreatic ductal adenocarcinoma
- PD‐L1
programmed death‐ligand 1
- PMN
polymorphonuclear
- PTEN
phosphatase and tensin homolog deleted on chromosome 10
- SCFA
short‐chain fatty acid
- SGOC
ser‐gly one‐carbon
- sIgA
secretory IgA
- T2D
type 2 diabete
- TAMO
trimethylamine‐N‐oxide
- TCGA
The Cancer Genome Atlas
- Th17
T helper 17
- TLR
Toll‐like receptor
- TMA
trimethylamine
- TMAO
trimethylamine N‐Oxide
- Treg
T regulatory cells
- Trp
tryptophan
- WSD
western style diet
- WT
Wild type
1. BACKGROUND
The gut microbiota, containing at least 38 trillion bacteria, is pivotal for maintaining homeostasis and health [1, 2]. Gut microbiota is appreciated as a microbial human organ, which has 100‐fold of human genome [3], involved in immunity regulation, metabolic functions, and others. Many studies have indicated that many diseases, including cancers, may result from microbiota dysbiosis [4]. Importantly, microbial pathogens cause tumorigenesis in high percentage (∼20%) of cancers [5]. Thus, it is important to investigate the involvement of these microbial pathogens during tumorigenesis.
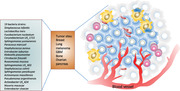
Progress in metagenome‐wide association studies of fecal samples, such as from colorectal cancer (CRC) patients, has characterized some important microbial markers of cancers [3, 6]. However, some of the causal effects of bacteria on cancer promotion remain to be further recognized [7]. In addition to fecal microbiota, a recent study has shown that distinct tumor‐associated microbiome had been characterized in several cancers including breast, lung, ovarian, pancreatic, melanoma, bone, and brain tumors [8]. Particularly, 19 prevalent bacteria were characterized [8]. The intratumor bacteria are present in cancer or immune cells (Figure 1). Little is known about the impact of these tumor‐residing bacteria on tumors in different body sites. The challenge is that tumor‐residing bacteria are very difficult to be manipulated for further studies because of low biomass.
FIGURE 1.
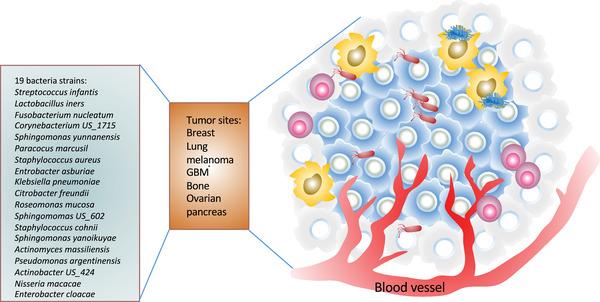
Bacteria species reside in different types of tumors. The 19 indicated bacteria strains reside in 7 indicated types of tumors [8]. The intratumor bacteria are present in cancer or immune cells such as macrophages (in yellow). These tumor‐associated bacteria may participate in cancer signaling to promote cancer growth. GBM, glioblastoma multiforme
Importantly, how microbiota may increase or suppress host's cancer susceptibility is a critical question. Characterizing the causal roles of specific microbes and microbiota, analyzing the host‐microbiota interactions during carcinogenesis, and harnessing this knowledge for cancer diagnosis and therapeutic design are of great interest to physicians and scientists. In general, when dysbiosis of microbiota occurs in several organs, it may contribute to epithelial barrier breach as well as reprogram immune and metabolic signaling, including affecting other hallmarks of cancer [9], such as causing inflammation, regulating cellular proliferation and apoptosis, instigating genome instability, promoting angiogenesis, and influencing cell stemness. However detailed mechanistic regulations of these phenomena remain to be determined.
Cancers are generally caused by host genetic alterations and environmental factors, but microorganisms are involved in human cancers as well [10], reiterating the importance of understanding the roles of microbes and microbiota during tumorigenesis. Microbiome may be influenced by environmental factors such as infection, diet and lifestyle, and these factors can lead to microbiome dysbiosis and promote diseases and cancers [3]. Therefore, characterizing the impacts of these factors and the causal roles of microbiota during tumorigenesis, understanding host‐microbiota interactions in carcinogenesis, and exploiting the knowledge in cancer diagnosis, treatment, and prevention are of great importance in future cancer research endeavor [11].
2. CANCER‐PROMOTING BACTERIA
Based on examined whole‐genome and whole‐transcriptome sequencing studies in The Cancer Genome Atlas (TCGA) for microbial reads, unique microbial signatures in tissue and blood were observed in many types of cancer [12]. Thus, microbiome‐based oncology diagnostic tool seems to be established. It is then clear that there are associations between diverse types of cancer and specific microbiota. Also, it is important to point out that using plasma‐derived, cell‐free microbial nucleic acid examination allows discriminating among healthy, cancer‐free individuals from patients with multiple types of cancer [12, 13] if potential contamination problem is carefully addressed.
As mentioned earlier, a recent comprehensive investigation of microbiomes across seven cancer types indicated that intracellular bacteria are widespread in tumors [8], including genera of staphylococcus and fusobacterium (Figure 1). It is not clear whether the enriched bacterial species or genera can be further identified in other types of cancer to facilitate the microenvironment to boost cancer growth. It is possible that these species may impose immune inflammatory and metabolic burden, and further studies are warranted. These studies suggest that cancer‐specific microbial taxa may serve as sensitive and specific clinical diagnostic markers. It is possible that targeting these bacterial strains may be effective and beneficial to cancer patients. Correct microbial assessment will aid in the detection and treatment of cancer in the future.
There are three major mechanisms by which microbiome can initiate cancer growth and development: (1) causing DNA damage and instigating mutagenesis; (2) triggering oncogenic signals; and (3) disturbing the immune response system. Here, oncobacterial signals and important factors involved in tumorigenesis are highlighted.
2.1. Oncobacteria examples
2.1.1. Helicobacter pylori (H. pylori)
Chronic H. pylori infection appears to be the strongest risk factor associated with gastric cancer [14]. Chronic H. pylori infection results in reduced acid secretion, thereby leading to the growth of a different gastric bacterial community [15]. This change in the bacterial composition may cause increased aggression to the gastric mucosa and lead to malignancy.
H. pylori infection leads to the suppression of Nei‐like DNA glycosylase 2 (NEIL2), which is a mammalian DNA glycosylase that specifically removes oxidized bases, thereby increasing the accumulation of DNA damage during the tumorigenesis of gastric cancer [16]. H. pylori infection in gastric organoids leads to the production of various inflammatory cytokines. Notably, the H. pylori‐infected Neil2‐knockout murine gastric organoid exhibited more DNA damage and manifested greater inflammation and more epithelial cell damage than wild type (WT), suggesting that NEIL2 down‐regulation caused by H. pylori infection dampens DNA damage repair and amplifies the inflammatory response to promote cancer formation [16].
H. pylori increased epidermal growth factor receptor (EGFR) phosphorylation during carcinogenesis [17]. Gefitinib, a specific EGFR inhibitor, can reduce C‐X‐C motif chemokine ligand Cxcl1 and Cxcl2 expression in gastric epithelial cells, decrease myeloperoxidase‐positive inflammatory cells in the mucosa, and quench epithelial DNA damage [17]. H. pylori infection instigates DNA damage via suppression of Rad51 expression through inhibition of autophagy and accumulation of p62 during gastric carcinogenesis [18]. H. pylori infection leads to upregulation of long non‐coding RNA (lncRNA) SNHG17, which increases levels of double‐strand breaks [19]. In addition, SNHG17 is associated with polycomb repressive complex 2 and is involved in epigenetic repression of cyclin‐dependent kinase (CDK) inhibitors, including p15 and p57, thereby promoting cell cycle progression and proliferation [20]. H. pylori contain genes encoding a secreted effector protein (chronic active gastritis A, CagA) and components of a type IV secretion system (Cag T4SS) [21], which forms needle‐like pili to bind the integrin‐β1 receptor and results in injection of the CagA oncoprotein. CagA can activate the p70 S6 kinase pathway and promotes programmed death‐ligand 1 (PD‐L1) expression.
Together, H. pylori releases virulence factors, such as CagA, and activates several pathways, including the EGFR pathway, the S6 kinase pathway, and the cell cycle progression pathway, thereby promoting cancer development [22].
2.1.2. Fusobacteria
Fusobacterium nucleatum (F. nucleatum) is an oral bacterium and functions as an “oncobacterium” [23, 24, 25] due to its capability to promote cancer growth. F. nucleatum triggers the Wnt signaling activity to promote CRC growth [26]. Basically, the virulence factor FadA from F. nucleatum can signal through E‐cadherin and increase expression of annexin A1 [22]. Importantly, FadA activates Wnt/β‐catenin signaling, thereby upregulating c‐Myc and cyclin D1 [23]. Further, other virulence factors of F. nucleatum, including Fap2, lipopolysaccharide (LPS) and cell wall extracts, can affect the shift of normal epithelial cells into tumor cells [27].
Furthermore, F. nucleatum mitigates T cell‐mediated immune responses of CRC [28]. The outer membrane protein Fap2 interacts with inhibitory T cell immunoreceptor with Ig and ITIM domains (TIGIT) receptor on natural killer (NK) cells and T cells, thereby facilitating cancer immune evasion. It also activates Toll‐like receptors (TLR)4, which leads to activation of nuclear factor‐κB (NF‐κB) and subsequent expression of the oncogenic microRNA21 (miR21) [29]. miR21 reduced levels of the RAS GTPase and was overexpressed in CRC [29]. F. nucleatum also activates TLR4 and myeloid differentiation primary response protein 88 (MyD88) immune signaling to induce specific microRNAs (miRNA18a and miRNA4802) to activate the autophagy, thereby affecting CRC chemotherapeutic response [30]. Animal experiments also show that Apc Min/+ mice fed with F. nucleatum developed more colorectal and small intestinal tumors when compared with sham‐fed controls. Importantly, the NF‐κB pathway is induced, leading to the expression of several pro‐inflammatory cytokines, such as tumor necrosis factor (TNF), interleukin IL‐6, IL‐8, and IL‐1β. Recent studies indicate that F. nucleatum significantly upregulated the expression of lncRNA Keratin7‐antisense (KRT7‐AS) and Keratin7 (KRT7) in CRC cells, thus promoting cell migration and metastasis [31]. F. nucleatum‐infected cells’ exosomes can deliver miR‐1246/92b‐3p/27a‐3p and C‐X‐C motif chemokine ligand CXCL16/RhoA/IL‐8 into non‐infected cells to increase cell migration ability and promote tumor metastasis [32]. F. nucleatum is involved in glucose metabolism by regulating enolase 1 (ENO1) through upregulating an lncRNA (ENO1‐IT1), leading to high glucose metabolism and poor prognosis in CRC patients [33]. Measurement of ENO1‐IT1, ENO1 and F. nucleatum levels may serve as prognosis markers [33].
Due to its oncogenic role, the fecal abundance of F. nucleatum may serve as a much needed biomarker for non‐invasive screening of CRC. Also, detection of IgA or IgG antibodies against F. nucleatum in the serum may provide a diagnostic strategy.
2.1.3. Streptococcus gallolyticus/peptostreptococcus anaerobius
CRC‐specific conditions can promote Streptococcus gallolyticus gut colonization [34]. Streptococcus gallolyticus subsp. gallolyticus (SGG) is associated with the occurrence of CRC [34]. It strongly activates Wnt pathway, thereby promoting signaling alterations in CRC. Basically, SGG induces IL‐1, cyclooxygenase‐2 (COX‐2), and IL‐8, β‐catenin oncogenic downstream targets (such as c‐Myc and cyclin D) to increase cell proliferation. Peptostreptococcus anaerobius can adhere to the CRC mucosa and facilitate CRC growth in ApcMin/+ mouse cancer model [35]. Its surface protein, putative cell wall binding repeat 2 (PCWBR2), interacts with α2/β1 integrin to induce the activation of the Phosphoinositide 3‐kinase (PI3K)‐Akt pathway, thereby causing cell proliferation and NF‐κB activation. Blockade of integrin α2/β1 by RGDS peptide leads to abolishing peptostreptococcus anaerobius‐mediated oncogenic activity [35].
Together, these observations highlight the rationale of targeting “oncobacteria” for certain cancer therapy. Further efforts should be the identification of more tumor‐promoting bacteria in cancer patients and the translation to novel, bacteria‐directed therapies.
2.2. Mechanisms of oncobacteria in instigating tumorigenesis
Gut microbiota can impact oncogenesis by a variety of molecular mechanisms including aberrant signal transduction, epigenetic regulation, immunoregulation, mucosa deregulation, p53 regulation, and metabolite contribution.
2.2.1. Aberrant signal transducer/epigenetic regulation
The stool from patients with CRC fed into axozymethane (AOM)‐treated or germ‐free mice leads to increased number of polyps or high levels of intestinal dysplasia and proliferation [36, 37] (Figure 2). In AOM experiment, mice administrated with stool from CRC patients demonstrate markers of inflammation and high T helper 1 (Th1) and Th17 cells in the colon when compared with stool from healthy person without CRC [37]. It was shown that stool from CRC patients induces cell proliferation and activates β‐catenin as well as other oncogenic factors such as Aurora Kinase A in the germ‐free mouse experimental group [37]. This study highlights the interactions among host immune system/oncogenic signaling, microbiota, and intestinal tumor formation. How the microbial community from CRC patients in promoting carcinogenesis remains to be addressed as it is not clear whether this change in cancer phenotype is due to an increase in tumor‐promoting bacteria or a decrease in anti‐tumorigenic bacteria.
FIGURE 2.
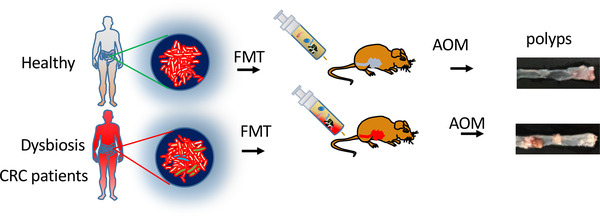
Microbial community from colorectal cancer patients promotes carcinogenesis. The stool from patients with colorectal cancer fed into axozymethane (AOM)‐treated mice leads to increased number of polyps. Stool from healthy person is used as a control
It was shown that microbiota can induced epigenetic programming based on whole‐genome bisulfite sequencing of conventionally raised, germ‐free mice [38]. Basically, commensal microbiota can induce local DNA methylation changes at regulatory elements through TET2/3 regulation (hydroxylate 5mC) [38]. Indeed, the microbiota can induce changes in gene expression through profound DNA methylation and chromatin accessibility changes at regulatory elements. Another example is that Escherichia coli (E. coli) can produce colibactin, which can alkylate DNA through electrophilic cyclopropane [39]. Obviously, DNA methylation will result in alterations in gene expression programs, thereby affecting diseases or cancer formation. Collectively, a deeper understanding of the cancer in microbiome environment and the early aberrant signals in cancer cells associated with microbiome might guide the therapeutic targeting strategy.
2.2.2. Deregulated immune modulation
The immune system is a dominant force in controlling cancer growth. Attenuation of immunity leads to carcinogenesis, cancer progression, and ill responses to cancer therapy [40]. The gut microbiota plays a critical role in immune functions. On the other hand, the host immune system developed multiple ways to maintain its functional relationship with the microbiota [41].
Gut microbiota acts through the function of metabolites, including short‐chain fatty acids (SCFA), bile acids, and tryptophan metabolites, to regulate the multiple processes in immune cells [42, 43, 44]. For example, the aryl hydrocarbon receptor (AhR) is involved in the regulation of intestinal immunity by bacterial tryptophan (Trp) metabolites (indole, indolic acid, skatole, and tryptamine) to regulate intestinal immunity [45]. Also, microbiota‐derived Toll‐like and nucleotide‐binding oligomerization domain receptor (TLR) ligands can impact on local intestinal cells and also penetrate beyond the mucosa into the circulation system to affect immune cells [41]. TLR1, 2, 4, 5, 6, 7, and 9 contribute to the recognition of various bacterial components [46]. Gut microbiota also activates NLRP6 inflammasome through LPS and/or bile‐acid conjugate taurine, which leads to the production of epithelial IL‐18 and antimicrobial peptides [47].
Gut microbiota can modulate systemic immune responses. Basically, microbial metabolites are able to penetrate the epithelial barrier, thereby entering the host circulatory system where they are sensed and responded by immune cells. For example, bacteria‐produced metabolite tryptophol impacts on interferon (IFN)‐γ production [48]. This microbiome‐cytokine interaction can be characterized, and certain bacterial taxa can be predicted to impact cytokine production. Also other bacterial metabolites can activate local dendritic cells, which in turn activate naïve T cells to effector T cells, T regulatory cells (T reg), or Th17 cells. For example, T reg can secret IL‐10 to regulate local anti‐inflammatory cytokines. Th17 cells produce IL‐17 to increase Paneth cell‐mediated production of antimicrobial peptides [40]. It is important to point out that Th17 cells are important lymphocytes connecting host microbiota and cancer [49]. Lung cancer can be caused by chronic inflammation. Local lung microbiota elicits inflammation associated with lung adenocarcinoma through activating lung‐resident γδ T cells [50]. In an animal study, germ‐free or antibiotic‐treated mice are resistant to lung cancer development induced by Kras mutation and p53 loss [50], suggesting the microbiota‐lung tumor development link. This process involves bacteria‐mediated inflammation [50].
Because of the critical relationship between the microbiome and immune regulation, numerous evidence suggests that modulating the gut microbiome can impact responses to cancer immunotherapy. For examples, several ways, including life style change (diet, exercise, psychosocial condition), prebiotics, probiotics, and Fecal microbiota transplantation (FMT), have been exploited to modify the gut microbiome (Figure 3) to enhance therapeutic responses to cancer immunotherapy such as immune checkpoint inhibitors (ICI) [40].
FIGURE 3.
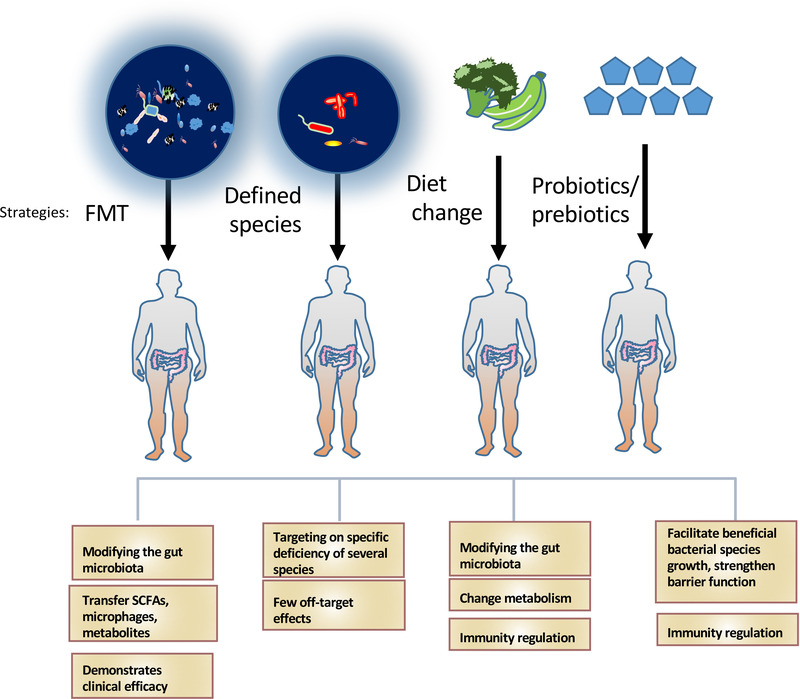
Impacts of modifying gut microbiota for improving health. Techniques to modulate the gut microbiota: FMT, the administration of defined bacterial species, diet change, the use of probiotics or prebiotics. Effects of each technique in improving health are listed
How and why the gut microbiome can influence the therapeutic response to ICI in various cancers remain hot topics in cancer research field [40]. Characterizing the intricate regulation among cancer, immunosurveillance, host immunity, metabolism, and the gut microbiome becomes very critical in exploiting strategies for cancer therapy [40]. The effectiveness of approved ICIs shown to be dependent on the composition of gut microbiota[22] has led to increased great interest in identifying more bacteria strains that could promote reinvigoration of anticancer immune responses.
2.2.3. Mucosa deregulation
It was shown that diets lacking dietary fiber can cause degrading of host glycans of the intestinal mucus layer by gut microbial species [51, 52]. Basically, fiber deficiency leads to erosion of mucus barrier and allows gut bacteria to use mucus glycoproteins as a nutrient source [53]. As a result, a defective mucus layer increases the infection by citrobacter rodentium as mucus layer is a defense against many pathogens. Interestingly, the presence of Bifidobacterium longum can revert such an impact [54]. Likewise, Western style diet (WSD) can alter the gut microbiota composition in colonic lumen and mucus layer [54]. Especially, Bifidobacterium is reduced in mucus layer because of WSD. Thus, dietary fiber is known to affect the gut microbiota composition, thereby influencing the colonic mucus barrier, which is a primary defense against enteric pathogens. When dietary fiber is deficient, the gut microbiota resorts to secreted mucus glycoproteins as a nutrient source, causing erosion of the colonic mucus barrier integrity. This forms a diet‐microbiota‐intestinal barrier integrity axis, which is important for health and cancer prevention [55]. This information is useful for elaborating strategies to strengthen mucus layer.
2.2.4. p53 regulation/DNA damage
p53 is a tumor suppressor and is the most frequently mutated or deleted gene in many types of tumors [56, 57]. Surprisingly, p53 mutant can avoid activating Wnt signaling, thereby inhibiting tumorigenesis. However, this phenomenon is reversed by the gut commensal metabolite gallic acid [58, 59]. Thus, the impact of microbiome creates a microenvironment to impact a cancer gene's functional activity although the detailed mechanism required further studies.
Many bacteria demonstrated mechanisms to cause DNA damage, so it can kill other competitors and survive among bacteria species. In the meanwhile, these bacterial defensive systems can promote mutations that lead to carcinogenesis. For examples, colibactin from Escherichia coli, Bacteroides fragilis toxin from enterotoxigenic Bacteroides fragilis, and cytolethal distending toxin from ε‐ and γ‐proteobacteria are either causing DNA damage directly or eliciting high production of ROS that leads to DNA damage [11].
2.2.5. Contributions of metabolites in promoting/regulating cancers
Microbiota can generate several small molecules and metabolites to promote tumorigenesis and to affect therapeutic responses through both local and systemic impacts [6, 60, 61]. Metabolites of microbiota are the functional output of host‐microorganism interactions, contributing to physiological regulations of host [62, 63]. They are known to demonstrate direct and indirect genotoxic activity. Products from protein (including H2S, p‐cresol) and secondary bile acids (e.g., deoxycholic acid [DCA]) plus products of the breakdown of liver‐detoxified xenobiotics contribute to cancer initiation and progression [64]. Several metabolites, such as butyrate, DCA, trimethylamine‐N‐oxide (TMAO), and others known to regulate the development of cancer, are discussed (Figure 4).
FIGURE 4.
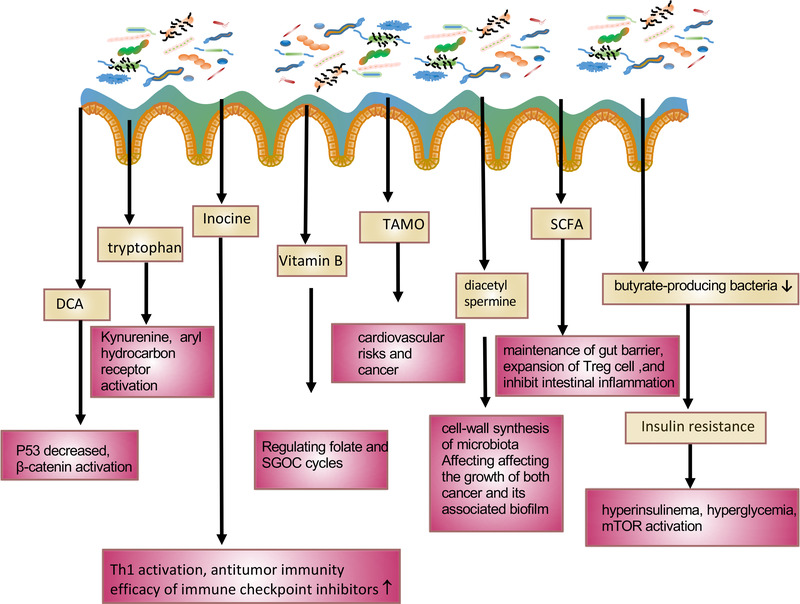
Impacts of bacteria‐derived metabolites on host. Listed bacteria‐derived metabolites and their impacts on affecting host/cancer cell signaling. DCA, deoxycholic acid; SGOC, ser gly one carbon pathway; TAMO, trimethylamine N‐oxide; SCFA, short chain fatty acid; mTOR, the mammalian target of rapamycin; T reg, T regulatory cell; Th1, T helper 1 cell
2.2.5.1. SCFA/DCA
SCFAs play pivotal roles in many host physiological and biochemical functions, such as maintenance of gut barrier function, gut motility, secretion of serotonin/5‐hydroxytryptamine, gut hormones, gastric inhibitory peptide and glucagon‐like peptide 1, chromatin regulation, the gut‐brain axis, immunological function, and others [43, 62]. For example, dietary fiber undergoes anaerobic fermentation in the presence of the commensal microbiota, such as Clostridia spp., to generate SCFAs, which stimulate expansion of Treg cells and inhibit intestinal inflammation. Also, butyrate serves as metabolic energy source for colonocytes but becomes detrimental to stem cells [47].
Bile acid is synthesized by the liver, stored in the gallbladder, and processed by intestinal bacteria, such as Clostridium hylemonae and Clostridium hiranonis [65], to produce DCA. High‐fat diet elevates the hepatic synthesis of bile acids. Excess levels of secreted bile acids can enter the colon, facilitating increased conversion of primary to secondary bile acids by colonic bacteria through 7α‐dehydroxylation to generate high levels of tumor‐promoting DCA. The genus Clostridium was characterized to have the enzymatic activities (7α‐dehydroxylase) to perform 7α‐dehydroxylation, thereby producing secondary bile acid DCA [65]. DCA plays critical roles in different tumors by mediating various signaling pathways, including regulating microRNA, enhancing EGFR‐MAPK signaling, decreasing p53 levels, and increasing β‐catenin activation [66]. For example, in a high‐fat diet mouse model, DCA supplementation leads to increased hepatocellular carcinoma development. On the other hand, using antibiotics to eradicate DCA‐producing bacteria can reverse this phenomenon [67]. Also, DCA can antagonize Farnsoid X receptor to induce DNA damage in Lgr5 + cells, which leads to CRC progression [68]. Bile acids are converted by bacteria to generate many bioactive molecules. The 3 oxolithocholic acid (3‐oxoLCA) and isoalloLCA are identified as important T cell regulators [69]. 3‐oxoLCA can inhibit the differentiation of Th17 cells while isoalloLCA promotes the differentiation of T reg cells. It is then conceivable that microbiota dysbiosis will disturb the modulation of the balance of Th17 and T reg cells, which are critical in tumor immune surveillance [69]. CRC patients have increased amounts of DCA in serum, bile, and stool [66]. Bile acid can be conjugated by glycine or taurine. Taurine‐conjugated bile acids can be metabolized by gut microbes to generate hydrogen sulfide and DCA, which are genotoxic, thereby promoting cancer growth [66]. Further, bile acid pool in gut can affect the population of colonic FOXP3+ Treg cells, suggesting a role in regulating immunity [70].
As discussed, DCA seems to promote cancer growth. However, a recent study shows that DCA is downregulated in gallbladder cancer (GBC), and reduced DCA is correlated with patient poor survival [71]. Surprisingly, DCA treatment suppressed tumor growth by inhibiting cell proliferation. Mechanistically, DCA reduced miR‐92b‐3p expression through N6‐methyladenosine‐dependent posttranscriptional regulation by facilitating dissociation of methyltransferase‐like 3 (METTL3) from METTL3‐METTL14‐WTAP complex, thereby increasing phosphatase and tensin homolog deleted on chromosome 10 (PTEN) tumor suppressor expression [71]. miR‐92b‐3p can inhibit the expression of PTEN. The studies reveal that DCA impacts on PTEN expression to suppress GBC growth. Thus, DCA may serve as a tumor suppressive agent for GBC. The discrepancy regarding DCA's role in different cancers might, in part, be explained by the heterogeneity of different cancers.
2.2.5.2. Trimethylamine N‐oxide (TMAO)
Choline, carnitine, creatinine, betaine or lecithin metabolized by host gut microbes to synthesize trimethylamine (TMA). TMA can be absorbed through the intestinal wall and transported to the liver. TMA then would be metabolized into TAMO through oxygenation by hepatic flavin‐containing monooxygenase 3 [72]. Thus, TAMO is a gut microbiota‐dependent metabolite from fat and dietary meat. TAMO has been shown to be involved in cardiovascular risks (myocardial infarction, or stroke) [73, 74]. Omnivorous human subjects produce more TMAO when compared with vegans or vegetarians as L‐carnitine from red meat is processed by gut microbiota to subsequently produce TAMO [75]. Also, circulating TMAO is particularly elevated during the aging process and may play a role in the pathogenesis of Alzheimer's disease [76]. Importantly, TMAO level is associated with cancer risk, such as CRC [77]. TMAO levels are correlated with CRC risks [77]. It is implied that TAMO‐induced inflammation could be culprit for cancer formation, but other mechanisms exist. Also, reticulum stress kinase PERK can function as a receptor for TMAO. TAMO can activate the PERK‐mediated unfolded protein response, thereby inducing the transcription factor Forkhead box protein O (FoxO)1, which is a key regulator of metabolism [72]. Thus, the compositional changes of intestinal microbiota and TMAO are linked to cancer risk. TMA and TMAO production is an important factor that links diet, intestinal microbiota and cancer. Further understanding the role of TAMO in cancer pathogenesis will help to determine how to cope with diet, microbiota, and TAMO signaling in the control and prevention of cancers.
2.2.5.3. Tryptophan
Colon cancer cells can uptake and process tryptophan more than normal colonic cells. Tryptophan can be metabolized through the kynurenine pathway, the serotonin pathway, protein synthesis, or transformation to various compounds [62]. Oncogenic Myc promotes the expression of solute carrier family (SLC)1A5, SLC7A5, and the tryptophan‐metabolizing enzyme arylformamidase, thereby generating kynurenine to promote cell proliferation [62]. Kynurenine can enhance spheroid growth and increase invasive potential of pancreatic cancer cell lines [78]. Kynurenine is an oncometabolite that can promote nuclear translocation of the transcription factor AhR, a transcription factor for inflammation and immunity [79]. Interestingly, various bacterial enzymes are homologous to the enzymes of human kynurenine pathways. It remains to be determined if gut microbiota is involved in Myc‐regulated kynurenine‐AhR axis for promoting cancer growth, as tryptophan can be converted into various catabolites by the gut microbiota. For example, Lactobacilli can convert tryptophan into indole‐3‐aldehyde, which functions as an AhR agonist. AhR activation will enhance gene expression of IL‐22 microbicidal factors and increasing Th17 cell activity [80].
2.2.5.4. Insulin resistance
In a Swedish study, gut microbiota composition is changed in people with impaired glucose tolerance or combined glucose intolerance and type 2 diabetes (T2D) [81]. Interestingly, the abundance of several butyrate‐producing bacteria are reduced both in prediabetes and T2D patients [81, 82]. It was then concluded that insulin resistance is strongly associated with microbial dysbiosis [81]. In another study, T2D‐associated bacteria produce imidazole propionate metabolite from histidine, which can impair glucose tolerance and insulin signaling [83]. Thus, imidazole propionate level is high in T2D patients [84, 85]. Insulin resistance leads to a potential to promote cancer growth: impacts include causing hyperinsulinema, hyperglycemia, and mTOR activation [86, 87]. Indeed, imidazole propionate can activate mTOR [83]. Insulin resistance results in metabolic alterations, which are important in supporting the uncontrolled growth of tumor cells [86, 88, 89]. Therefore, it is conceivable that impacts of gut microbiota dysbiosis on causing insulin resistance might be involved in promoting cancer growth. Modifying gut microbiota may be a feasible way for the prevention and/or delay of T2D onset or cancer formation.
2.2.5.5. Inosine
Three bacterial species, Bifidobacterium pseudolongum (B. pseudolongum), Lactobacillus johnsonii, and Olsenella, have been identified to positively impact the efficacy of immune checkpoint inhibitors in mouse cancer models [90]. Significantly, B. pseudolongum can enhance immunotherapy efficacy by producing the metabolite inosine. Indeed, inosine is the immunotherapy‐promoting metabolite and experimentally demonstrates its impact in intestinal cancer, bladder cancer, and melanoma [90]. Basically, inosine mechanistically promotes Th1 activation and antitumor immunity, and Th1 immunity is beneficial for most antitumor responses. Inosine regulates T cell‐specific A2AR signaling to promote Th1 cell activation. It is then possible to develop inosine‐based adjuvant therapies as inosine boost the efficacy of ICI. Further development of metabolite biomarkers in refining the efficacy of ICI therapies and deeper understanding the mechanisms behind will help determine the ICI treatment strategies.
2.2.5.6. Niacin
Niacin functions as a precursor of nicotinamide adenine dinucleotide (NAD) and nicotinamide adenine dinucleotide phosphate (NADP), and is a cofactor of many enzymes. NAD plays critical roles in redox reactions, in which NAD and NADH are interconverted. NAD also functions as a substrate for sirtuins, NAD‐dependent protein deacetylases that connects transcriptional regulation to cellular energetics [91]. Dietary fiber and carbohydrates from host mucins undergo fermentative pathways to generate monosaccharides by bacterial polysaccharidases and/or glycosidases. These monosaccharides are catabolized further through either the pentose phosphate pathway or the Embden‐Meyerhof‐Parnas pathway to generate pyruvate or NADH [62]. G protein‐coupled receptor (GPR)109A, a tumor suppressor, is a receptor for both butyrate and niacin in the colon [92]. Niacin is a pharmacological GPR109A agonist and can inhibit colon cancer growth in a GPR109A‐dependent manner [93]. Interestingly, niacin demonstrated beneficial effects on DSS‐induced colitis in mice by enhancing the production of prostaglandin D2 [94].
2.2.5.7. Vitamin B
B vitamins are critical for human health, acting as enzyme cofactors involved in important functions such as energy metabolism, DNA and protein synthesis. For example, B vitamins are critical in regulating ser‐gly one‐carbon (SGOC) metabolism [62]. Indeed, they are substrates or cofactors in the folate and SGOC cycles. Gut bacteria species are able to synthesize 8 B vitamins (vitamins B1, B2, B3, B5, B6, B7, B9 and B12) and are critical for human health as human is unable to synthesize enough B vitamins [62]. Since SGOC pathway is frequently deregulated in cancers [95, 96], it is conceivable that composition changes in Vitamin B‐producing bacterial species will impact on tumorigenesis.
2.2.5.8. Diacetyl spermine/urolithin/oncotoxins
Bacterial biofilms, contributing to polyamine pool, are very important in altering the host tissue microenvironment [97]. There is an upregulation of polyamine metabolites in tissues from cancer patients [98, 99]. Importantly, antibiotic treatment can clear bacterial film, thereby decreasing N(1),N(12)‐diacetylspermine [100], a polyamine metabolite affecting the growth of both cancer and biofilm. Mechanistically, increased polyamine concentrations correlated with eukaryotic proliferation and cell‐wall synthesis of microbiota. Thus, the upregulation of polyamine metabolism will facilitate cancer growth. It will then be interesting to explore inhibiting polyamine metabolism for suppressing cancer growth. Eggethellaceae family bacteria were identified to generate urolithin metabolites [101]. Microbial metabolite urolithin A is derived from polyphenol of several fruits and have anti‐oxidative, anti‐inflammatory, and anti‐ageing activities. It activates AhR to upregulate tight junction proteins [102]. It is conceivable that these activities may be involved in regulating tumorigenesis.
In addition, the carcinogenic versions of the bacterial species Escherichia coli and Bacteroides fragilis can generate secreted oncotoxins to cause cancer in Familial adenomatous polyposis (FAP) patients [103]. Cytolethal‐distending toxin produced by enteric pathogens (Escherichia and Campylobacter spp.) can cause double‐strand DNA breaks and is carcinogenic [104]. Colibactin is produced by members of the Enterobacteriaceae family to induce DNA damages [105]. Thus, microbiome produces certain oncogenic toxins and/or metabolites to influence cancer prognosis. Modulating the microbiome and its products can be useful for cancer treatments [106]. Together, the above mentioned information might be useful for elaborating strategies to overcome cancer development caused by certain “oncobacteria”.
3. CANCER‐PROTECTING BACTERIA
The important advances in microbiome studies have led to the appreciation of the critical protecting role of intestinal microbes, such as probiotics, in human diseases including cancers. Probiotic bacteria impact on physiological and immunological mechanisms; therefore, they may have antitumor activity. Several mechanisms have been described: altering the intestinal microflora, inactivating carcinogenic compounds, competing with pathogenic microbiota, improving immune system, regulating apoptosis and cell differentiation, producing healthy metabolites, maintaining barrier integrity of gut mucosa [107] (Figure 3). Many research findings regarding the protective role of probiotics/prebiotics in diseases/cancers are very encouraging [108, 109, 110, 111]. For example, probiotics and prebiotics can modulate the gut microbiota and is beneficial to improve human health [112, 113]. Lactobacillus, Bifidobacterium, and Saccharomyces are safe and effective probiotics. Others, such as Roseburia spp. [114], Akkermansia spp. [115, 116, 117], Propionibacterium spp. [118, 119], have potentials to be characterized as probiotics. Nonetheless, clinical trials are still required to examine the safety and effectiveness of probiotics for cancer therapy.
3.1. Beneficial probiotics
3.1.1. Lactobacillus
Lactobacillus spp., is a common probiotic used in dietary supplement. Its cancer protection role has been examined in mouse cancer model. The effect of Lactobacillus fermentum, Lactobacillus acidophilus and Lactobacillus rhamnosus on cancer growth is demonstrated in azoxymethane/dextran sulfate sodium (AOM/DSS)‐induced colitis‐associated cancer model [120]. Lactobacillus fermentum can inhibit colonic tumor formation and mitigate pro‐inflammatory cytokine production. Further it can alter the composition of gut microbiota by reducing the presence of Bacteroides [120]. Thus, Lactobacillus probiotics is beneficial in alleviating colon cancer progression.
It was shown that Lactobacillus reuteri (L. reuteri) can maintain the cell number of Lgr5+ cells and stimulate intestinal epithelial growth to repair epithelial damage, thereby protecting the intestinal mucosal barrier integrity [121]. Further, L. reuteri can decrease Citrobacter rodentium (C. rodentium) colonization, thereby ameliorating intestinal inflammation in mice. It is a promising therapeutic agent for intestinal inflammation. L. reuteri is also critical to drive maturation and function of the immune system. For example, L. reuteri can induce CD4+CD8αα+ double‐positive intraepithelial lymphocytes (IELs) by activating AHR [122]. Basically, L. reuteri can metabolize dietary tryptophan to indole derivatives to activate AHR, which in turn downregulates Thpok (Th‐inducing BTB/POZ‐Kruppel‐like factor) transcription factor to reprogram CD4+ IELs into double‐positive IELs. These cells may be critical for preventing pathogen infection and protecting epithelial barrier in the gut. These results highlight potential therapeutic use of Lactobacillus in tumors.
3.1.2. Bifidobacterium
Bifidobacterium can modulate cancer immunotherapeutic efficacy. Oral administration of Bifidobacterium in a melanoma mouse model can improve tumor control as efficient as PD‐L1 antibody therapy (checkpoint blockade) [123]. Importantly, Bifidobacterium and anti‐PD‐L1 combination treatment is very efficient in suppressing tumor outgrowth [123]. Interestingly, mice fed with a WSD have an altered gut microbiota that instigates increased penetrability and decreased growth rate of the inner mucus layer. Importantly, Bifidobacterium longum can restore mucus growth in WSD‐fed mice, thereby protecting mucus function [124]. Cell surface β‐glucan/galactan (CSGG) polysaccharides of Bifidobacterium bifidum is critical for Treg induction [125]. CSGG activates regulatory dendritic cells through TLR 2, which in turn leads to immune suppressive activity. Dysregulation of intestinal microflora, such as Bifidobacterium bifidum, may cause inflammatory disorders due to compromised immunosuppressive functions of Foxp3+ Treg cells [125]. These studies highlight the therapeutic potentials of Bifidobacterium in cancer treatment or prevention as it impacts on immune regulation and mucus protection.
3.1.3. Faecalibaculum rodentium
Faecalibaculum rodentium of the mouse microbiota and its human homologue, Holdemanella biformis, are under‐represented or lost bacteria during tumourigenesis [126]. Both Faecalibaculum rodentium and Hemicrepidius biformis have generated SCFA metabolites (mainly butyrate) that inhibit calcineurin and nuclear factor of activated T‐cells (NFAT)c3 activation to control protein acetylation and tumor cell proliferation [126]. Administration of F. rodentium in ApcMin/+, which harbors a mutation in the Adenomatous polyposis coli (APC) gene mutated in more than 80% of sporadic CRC, or azoxymethane‐ and dextran sulfate sodium‐treated mice can mitigate tumor growth. Similarly, Holdemanella biformis seems to behave like Faecalibaculum rodentium in inhibiting cancer cell growth in ApcMin/+ model through the action of butyrate, an Histone deacetylase (HDAC) inhibitor. Thus, these anti‐tumorigenic bacterial strains may be useful for cancer therapeutic design (Figure 3). Nevertheless, the impact of Holdemanella biformis on human tumor growth remains to be evaluated. If successful, it may also serve as a cancer biomarker for early detection of cancers.
3.1.4. Streptococcus thermophilus
Streptococcus thermophilus is one of the many lactic acid bacteria and is a powerful probiotic found in the colon and has shown digestive, immunity, and other health benefits. Importantly, Streptococcus thermophilus was found to be depleted in CRC patients [127]. Consistently, Streptococcus thermophilus has tumor‐suppressive activity. This was demonstrated in mouse models: Apcmin/+ and azoxymethane‐injected mice [127]. Oral gavage of Streptococcus thermophilus leads to significantly reduced tumor formation in these two mouse models. Mass spectrometry studies clearly demonstrate that a protein called β‐galactosidase secreted from Streptococcus thermophilus has tumor‐suppressive impact on cancer growth [127]. β‐galactosidase secreted by Streptococcus thermophilus reduced cell proliferation, inhibited colony formation, caused cell cycle arrest, and led to apoptosis of CRC cells and impeded the tumor growth in mouse CRC xenograft study [127]. Impressively, β‐galactosidase secreted from Streptococcus thermophilus can boost the abundance of two well‐known probiotics, such as Bifidobacterium and Lactobacillus, suggesting a collaborative effect between probiotics [127]. Mechanistically, β‐Galactosidase‐dependent production of galactose reprogrammed energy homeostasis via changing oxidative phosphorylation and hampered the Hippo pathway kinases, thereby mediating the tumor‐suppressive effects of S. thermophilus [127]. However, the detailed mechanism of β‐galactosidase on human tumor growth required further studies.
Streptococcus thermophilus strains can also produce and release folate during growth [111]. Folate serves as an important factor in diet and is critical in cell metabolism, including DNA replication, repair, methylation, and synthesis of nucleotides. Studies indicate that folate deficiency is quite common among people [128]. It is possible that folate released from Streptococcus thermophilus may play roles in tumor‐suppressive effects of Streptococcus thermophilus. Also, Streptococcus thermophilus have impacts on the severity of colitis, lymphocyte profile, and regulatory T‐cell response [129], which are critical in ameliorating symptoms by modulating the immune response under the dextran sulfate sodium (DSS)‐induced colitis condition. In summary, impact of β‐galactosidase and folate from Streptococcus thermophilus on human tumor growth needs further studies, and the applications in tumor diagnosis/treatment could be explored through clinical trials.
3.2. Mechanisms for cancer‐protecting bacteria
3.2.1. Immunity boosting
The gut microbiota plays a pivotal role in regulating the innate and adaptive immune systems [130, 131]. The impact of the microbiota on cancer development relies on the intricate interaction between the microbiota, the tumor, and the immune system [60]. Gut bacterial species can boost immune cells to target cancer and/or protect against pathogen infection. Eleven bacterial strains that were isolated from healthy human stool samples were verified to induce IFN‐γ‐producing CD8 T cells in the intestine [132] (Figure 5). Importantly, the 11 strains augment host resistance against pathogen Listeria monocytogenes infection. Further, they boost the therapeutic efficacy of immune checkpoint inhibitor anti‐programmed cell death protein 1 (anti‐PD‐1) in mouse cancer models [132]. In both cases, the 11 bacterial strains caused an increase in CD8+ IFN‐γ+ T cells. There is a great potential of using these 11 strains for effective biotherapeutics (Figure 5). Also, SCFA‐producing bacteria can bind to GPR41, GPR43, and GPR109A located on the surface of epithelial cells and immune cells. SCFA promotes the production of mucus from goblet cells, hindering the activity of NF‐κB, eliciting signaling of inflammasomes and production of IL‐18, facilitating the secretion of secretory IgA (sIgA) from B cells, and increasing the function of colonic Treg cells [97, 133]. A recent study shows that microbiota can activate protective immunity against colitis and CRC [134]. Odoribacter splanchnicus leads to Th17 cell development and allows the host to be resistant to colitis and CRC [134]. Therefore, an important role of gut microbiota is probably to aid the activity of immune system.
FIGURE 5.
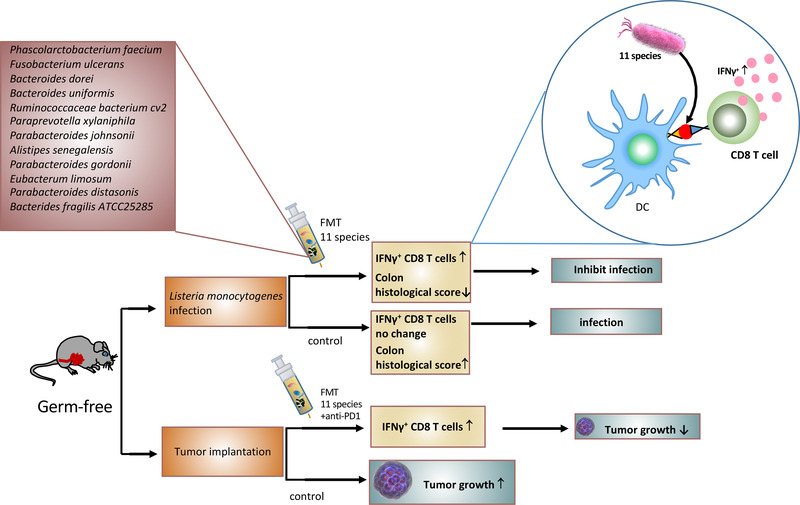
Bacterial strains boost immune responses to fight pathogen infection and enhance anti‐PD‐1 treatment efficacy in cancer. Eleven indicated bacterial strains [132] isolated from healthy human stool samples can induce interferon‐γ‐producing CD8 T cells in the intestine. In mouse models, the 11 strains delivered through FMT can increase interferon‐γ‐producing CD8 T cells, thereby augmenting host resistance against pathogen Listeria monocytogenes infection. Also in tumor implantation study, they caused an increase in CD8+ IFN‐γ+ T cells to boost the therapeutic efficacy of immune checkpoint inhibitor anti‐PD‐1, thereby inhibiting cancer growth. PD‐1, the programmed cell death protein 1
3.2.2. Metabolite regulation
About 50% of metabolites in the human plasma are from bacterial origin. For example, all SCFAs and secondary bile acids are synthesized by gut microbiome. These metabolites have impacts on cancer development and affect the efficacy of cancer therapies.
It was demonstrated that mice fed with a high‐fiber diet may have an elevated level of SCFA [135]. SCFAs produced from commensal bacteria stimulate generation of Treg cells, which is critical in limiting inflammatory responses in the intestine [133]. Importantly, butyrate's HDAC inhibitory activity leads to reduced proinflammatory cytokine expression [133]. SCFAs can induce macrophage differentiation and enhance antimicrobial function of macrophage. It inhibits HDAC3 activity to impact glycolysis, mTOR activity, and autophagy [43]. SCFAs can augment immunity via IgA, which blocks bacterial adherence to epithelial cells. IgA can cause agglutination, entrapment, and clearance and has impacts on bacterial virulence [40].
Butyrate and niacin are bacterial products due to the fermentation of dietary fiber in the colon. They bind to Niacr1, a receptor for butyrate and niacin, to suppress intestinal inflammation [93], thus mediating the beneficial effects of gut microbiota. Chromatin is a signal integrator within cells, and it takes environmental cues from small‐molecule metabolites to reprogram gene expression (chromatin modification) in response to various stimuli. For example, methyl donor S‐adenosyl methionine and acetyl‐Coenzyme A can regulate the activity of modifying enzymes that add and remove chromatin modifications. Also microbial‐derived butyrate is known to acetylate histones and regulate target gene expression. However, there is controversial evidence that butyrate can promotes cancer [136], suggesting that detailed mechanistic regulation by butyrate needs to be further characterized. Maintaining intestinal homeostasis requires cross‐talk between host and microbes. There is a link between butyrate, macrophage differentiation and antimicrobial activity. Schulthess et al. [137] showed that butyrate‐induced antimicrobial activity is linked to a shift in macrophage metabolism, decreasing mTOR kinase activity, an increase in microtubule‐associated protein 1 light chain (LC3)‐associated host defense, and anti‐microbial peptide production. Basically, butyrate‐mediated histone deacetylase 3 inhibition can drive macrophage differentiation. Thus, butyrate induced the antimicrobial activity of intestinal macrophages. The role of butyrate as a differentiation factor for monocyte‐derived macrophages can be further exploited for disease treatment or cancer therapy.
Systemic inflammation, bacterial dissemination, butyrate depletion, and mortality were observed in mice subjected to taurocholate‐induced necrotizing pancreatitis under a Western style diet [138]. Significantly, butyrate supplementation can decrease mortality, bacterial dissemination, and reverse the microbiota alterations. Microbiota analysis demonstrated that patients with acute pancreatitis have an increase in Proteobacteria and a decrease of butyrate producers [138]. Pancreatitis is linked to pancreatic cancer; it is then conceivable that butyrate producers can be designed for pancreatic cancer prevention. Thus, these studies underscore the possibility of using butyrate or butyrate‐producing bacteria for cancer prevention and/or therapy. More studies should focus on fecal metabolomics profiling as fecal metabolomics is the important readout of microbial metabolism and annotates microbial interaction with host environment [139]. Fecal metabolomics profiling can provide insight into the relationship between fecal metabolites and cancer genetics.
Collectively, gut microbiota can generate/lose metabolites in the intestinal environment to promote genetic and epigenetic changes that lead to cancer. It is then critical to combine metabolites and microbiome analyses to illustrate interactions between gut microbiota, metabolism, and the host in terms of understanding microbiota‐regulated tumorigenesis.
4. NEW DIAGNOSTIC APPROACHES USING MICROBIAL MARKERS
Since gut microbiota are involved in the development of cancer, it is possible to exploit the microbiota identified by metagenome sequencing analysis for cancer diagnosis. If successfully executed, microbiota biomarkers can be explored as a potential screen for early‐stage cancers. For example, traces of microbes’ DNA and RNA are found in various tissues, blood, and tumors, and they can be employed as a signature of cancer patients or healthy individuals [12]. It has been demonstrated that plasma‐derived, cell‐free microbial nucleic acids from patients can be used for diagnosis of several types of cancer, including prostate cancer, lung cancer, and melanoma based on artificial‐intelligence programs [12, 140, 141]. Thus, microbiome‐based diagnostic tool for cancers can be established in near future to serve as biomarkers. However, it will be critical to obtain insights into the distribution (location) and function of these microbial signatures. Also, the mechanisms by which microbes enter and reside in cancerous tissues remain to be determined. Further, mechanistic insights into how to target these microbes for cancer treatment and prevention is required.
4.1. CRC microbiota
The colon is a location with the largest number of gut microbes, which links to CRC [142, 143, 144]. Enterococcus faecalis, Shigella, Escherichia coli NC101, Bacteroides fragilis, Streptococcus bovis, H. pylori, F. nucleatum are known to promote CRC cancer growth, while Bifidobacterium, Eubacterium rectale, Faecalibacterium prausnitzii, Lactobacillus may reduce the growth of CRC [6]. Bacteroides dorei, Bacteroides vulgatus, Bacteroides massiliensis, and E coli, are involved in systemic inflammation and in promoting CRC tumor growth [145]. F. nucleatum is involved in recruiting tumor‐infiltrating myeloid cells to establish a proinflammatory microenvironment to promote tumorigenesis [146]. Fusobacterium spp. were abundant in tumors when compared with adjacent healthy tissues [147]. Microbial biomarkers, including fusobacteria, porphyromonas [147], improve the accuracy of predictive models for adenoma and carcinoma groups. F. nucleatum, Bacteroides clarus, Roseburia intestinalis, Clostridium hathewayi were identified in fecal samples of CRC patients, and thus can serve as diagnosis biomarkers of CRC [148]. It has been shown that combination of three or four markers can improve the diagnostic ability for CRC.
Peptostreptococcus anaerobius was identified as one of the microbial species associated with CRC [35]. In animal studies, its oncogenic role was confirmed to facilitate CRC development in Apc Min/+ mice [35]. Its PCWBR2 surface protein is critical in binding to integrin α2/β1 to activate Akt and NF‐κB pathway, thereby promoting cell proliferation and pro‐inflammatory immune response [35]. Through its adherence and colonization in guts, it was able to promote tumor development [34. However, it remains to be determined if this is the only mechanism to initiate tumorigenesis although eliciting pro‐inflammation response is also observed. Moreover, several other bacterial species have been described in promoting CRC development, such as Enterococcus faecalis, Alistipes spp., Bifidobacterium spp., and Bacteroides thetaiotamicron [9]; they are close to the epithelium and are involved in mucosal immune system. Further progress in understanding more molecular mechanisms underlying dysbiosis of microbiota in colon cancer could lead to novel therapeutic strategies to control the growth of CRC.
4.2. Gastric microbiota
In a gastric cancer study, analysis of microbiota dysbiosis has the capacity to differentiate between gastritis and gastric carcinoma [15]. Importantly, functional analysis of the gastric cancer microbiota identified nitrosating microbial community in gastric carcinoma [15]. Also, it has been shown that Proteobacteria, Firmicutes, Actinobacteria and Fusobacteria phyla are frequently detected in gastric biopsies [15]. Noticeably, H. pylori infection as mentioned above appeared as a major risk factor for gastric cancer (GC). However, only 3% of H. pylori infection accounts for gastric cancer, suggesting that other bacteria strains are involved in gastric tumorigenesis [149]. Indeed, five bacteria strains Peptostreptococcus stomatis, Streptococcus anginosus, Parvimonas micra, Slackia exigua and Dialister pneumosintes were characterized to be enriched in gastric cancer [150]. Future studies to understand the pathological impacts from these bacterial strains are warranted. Thus, they could serve as non‐invasive diagnosis markers for gastric cancer. Further, these five bacterial strains are important members of the human oral microbiome, suggesting that oral hygiene is critical in preventing gastric cancer. Clostridium colicanis and F. nucleatum are also enriched in gastric cancer and demonstrate an excellent predictive ability for prognosis [151].
4.3. Esophageal microbiota
The microbiome is less well characterized in the context of esophageal squamous cell carcinoma (ESCC). Only a small number of studies characterized the human esophageal microbiota in health and disease [152, 153, 154]. A recent study demonstrated significant enrichments of Treponema amylovorum, Streptococcus infantis, Prevotella nigrescens, Porphyromonas endodontalis, Veillonella dispar, Aggregatibacter segnis, Prevotella melaninogenica, Prevotella intermedia, Prevotella tannerae, Prevotella nanceiensis and Streptococcus anginosus in ESCC [155]. These 10 bacterial strains are involved in oral health, suggesting that oral hygiene plays important roles in tumor development of ESCC [155]. Consistent with these data, it was demonstrated that tooth losses can potentially increase the risk of ESCC development [156] and that ESCC with high burden of F. nucleatum, which inhabits the oral cavity and causes periodontal disease, correlates with poor recurrence‐free survival (RFS) [157]. In addition, Porphyromonas gingivalis, a Gram‐negative bacterial species plays a critical role in periodontal diseases, is a pivotal biomarker of ESCC [158]. Studies reveal that one of the top‐ranked Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in these 10 ESCC‐associated bacterial strains is nitrate reductase function [155]. Nitrate is important inorganic nitrogen sources for microbes, and many bacteria express assimilatory nitrate reductase to catalyze the rate‐limiting reduction of nitrate to nitrite [159]. Nitrate reduction plays a pivotal role in pathogenic/neoplastic progression [160]. The observation that deregulation of nitrate reductase functions in the microbiota of ESCC may impose pathogenic effects during ESCC tumorigenesis [155]. It is then possible that targeting those bacteria strains involved in nitrate regulation may be feasible in treating ESCC.
In esophageal adenocarcinoma (EAC), lipopolysaccharides, a major structure of the outer membrane in gram‐negative bacteria, upregulate gene expression of proinflammatory cytokines via activation of the Toll‐like receptor 4 and NF‐κB pathway and promote the occurrence of Barrett esophagus and EAC [161, 162]. The periodontal pathogen Tannerella forsythia is associated with higher risk of EAC while the periodontal pathogen Porphyromonas gingivalis, a Gram‐negative bacterial species involved in periodontal diseases, is associated with higher risk of ESCC [158]. Together, these studies foster an interest in characterizing several oral bacteria strains as a cancer biomarker and promoting strategies designed to modulate these bacteria strains for efficient therapeutic applications.
4.4. Pancreatic cancer microbiota
Pancreatic cancer is one of the most lethal cancers, with a 5‐year survival rate of less than 5%. Pushalkar et al. [163] have detected specific gut and tumor microbiome in mouse models of pancreatic cancer, indicating bacterial translocation from the gut into the tumor. However, the composition of the human pancreatic ductal adenocarcinoma (PDAC) microbiome that leads to pancreatic cancer growth remains to be further studied. Recently, several bacteria species, such as Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans, are associated with increased risk of pancreatic cancer [164]. Further, it is demonstrated that PDAC patients with long‐term survival have an intra‐tumoral microbiome signature (Pseudoxanthomonas‐Streptomyces‐Saccharopolyspora‐Bacillus clausii) when compared with patients with short‐term survival [165], suggesting that the gut microbiome can influence tumor microbiome and tumor growth [166]. Indeed, PDAC microbiota compositions are significantly different between long‐term survival and short‐term survival. Also, it has been shown that the pancreas is colonized by Malassezia [167, 168]. It has been shown that Malassezia fungi can migrate from the gut lumen to the pancreas to accelerate the oncogenesis of PDAC [168]. Fungal ablation studies indicate that repopulating with Malassezia globosa is sufficient to accelerate the tumorigenesis of PDAC [168]. Further delineation of the possible cancer‐promoting mechanisms of these bacteria or fungi remains to be demonstrated although inflammatory process/complement cascade is proposed.
The gut microbiota causes infections in necrotizing pancreatitis, which in turn might have impact on tumor development. For example, in an animal model with taurocholate‐induced necrotizing pancreatitis, systemic inflammation and bacterial dissemination were elevated in mice fed with Western style diet [138]. Gut microbiota analysis and metabolism profiling demonstrated a loss of diversity and enrichment of Escherichia coli, and butyrate depletion. Interestingly, butyrate supplementation can reduce bacterial dissemination, and reverse the microbiota alterations.
Not just limited to gut microbiota, salivary microbiota analysis reveals a difference in salivary microflora between pancreatic cancer and healthy control. Two bacterial candidates (Neisseria elongata and Streptococcus mitis) were identified [169]. In addition, these two bacteria candidates demonstrated variation between chronic pancreatitis samples and controls [169]. These observations suggest that these two candidates can serve as non‐invasive biomarkers of pancreatic cancer and chronic pancreatitis.
4.5. Breast cancer microbiota
The microbiome linked to breast tissue and breast diseases is poorly understood although studies have shown that a distinct microbiome with certain species enriched in the breast cancer tissue itself [8, 106, 170, 171]. Recently, breast cancer animal model studies have revealed that microbiota dysbiosis facilitates circulating of tumor cells, thereby enhancing dissemination of cancer cells to the lymph nodes and lungs [172], suggesting that the gut microbiome is critical in breast cancer metastasis. The underlying mechanism is that microbiota dysbiosis promotes inflammation and enhances fibrosis and collagen deposition plus myeloid recruitment in tumor microenvironment. Certainly, further studies are required to exploit this finding in human cancer for interventions or diagnosis to improve treatment outcome. In breast cancer patients, several species were particularly enriched in postmenopausal patients, such as Escherichia coli, Klebsiella sp_1_1_55, Prevotella amnii, Enterococcus gallinarum, Actinomyces sp. HPA0247, Shewanella putrefaciens, and Erwinia amylovora [173]. Further understanding the cause and effect of these bacterial strains may help diagnosis or treatment. Also, studies have shown that a distinct microbiome with particular species is enriched in breast tissues, the nipple aspirate, and the gut of breast cancer patients [171]. Together, the breast cancer microbiomes play a critical role in therapeutic response, diagnosis, and staging.
4.6. Melanoma microbiota
Approximately 70% of melanoma patients are resistant to ICI therapy [174]. Evidence has suggested that the gut microbiome is critical in determining the treatment efficacy of ICI therapy, including anti‐PD‐1, ‐PD‐L1, ‐cytotoxic T‐lymphocyte‐associated antigen 4 (CTLA‐4), or combination immunotherapy. Indeed, there is a fecal microbiome signature inherent across ICI responders [175]. Differences of microbial compositions between ICI responders and nonresponders have been demonstrated. For example, analysis identified important bacteria strains enriched in responders, such as Faecalibacterium and Barnesiella intestinihominis [175, 176]. These microbiome signatures of ICI responders can be exploited for diagnosis and therapeutic designs. Also, resistance may be caused by particular bacterial metabolites, such as SCFA, which could impact the efficacy of ICI. The microbiota has obviously opened up a new avenue for predicting the efficacy of immunotherapy. Combining microbiome composition with tumor genomics and metabolomics will be more accurately in predicting the efficacy of ICI treatments.
4.7. Liver cancer microbiota
Evidence demonstrates the important role of the gut microbiota in promoting the carcinogenesis of hepatocellular carcinoma [177, 178]. Basically, gut microbiome dysbiosis and barrier damage‐mediated leaky gut can facilitate the progression of liver diseases and cancer. For example, the impact of DCA metabolite from gut microbiome can promote cancer and senescence; lipopolysaccharide can activate microbe‐associated molecular patterns (MAMPs) through Toll‐like receptor to elicit inflammation, fibrosis, proliferation, and anti‐apoptotic signals [62].
Alterations of the human gut microbiome have been characterized in liver cirrhosis [179]. Streptococcus spp. and Veillonella spp. are particularly abundant in liver cirrhosis patients, implying that these two genera might play a critical role during liver cirrhosis [179] although the molecular mechanism of promoting liver cirrhosis remains to be investigated. Primary sclerosing cholangitis or colitis are two risk factors for cholangiocarcinoma as they facilitate tumor development by causing an accumulation of C‐X‐C chemokine receptor type 2(CXCR)2+ polymorphonuclear myeloid‐derived suppressor cells (PMN‐MDSC) that protect tumors from elimination by immune cells. Importantly, gut barrier dysfunction allowed gut bacteria and lipopolysaccharide to appear in the liver, thereby inducing CXCL1 expression in hepatocytes to form an immunosuppressive environment by increasing PMN‐MDSC to promote liver cancer [180]. Together, it can be emphasized that gut‐liver axis is critical in promoting liver‐related diseases. Interrupting DCA signaling, regulating MAMP, or strengthening barrier integrity could be considered to design novel therapeutic strategies to control the development of liver cancer/diseases.
4.8. Oral microbiota and cancer
There are about 700 bacterial species that reside in the oral cavity [181]. Oral microbiome is associated with oral cancers such as oral squamous cell carcinoma (OSCC) [182]. Well‐known causative factors of oral cancer such as tobacco, alcohol, and betel nut may change the oral microbiome composition, thereby affecting oral cancer development. The change of oral microorganisms may alter the inflammatory microenvironment and deregulate host signaling pathways that are critical in controlling, inflammation, cell viability, proliferation, or differentiation. For example, colonization by Porphyromonas gingivalis and Streptococcus gordonii can cause impaired innate host defense and trigger inflammation [182]. Porphyromonas gingivalis, which is a critical pathogen in chronic periodontitis, can antagonize chemically induced apoptosis [183]. Porphyromonas gingivalis results in activation of Jak1/Akt/Stat3 signaling pathway that regulates intrinsic mitochondrial apoptosis pathway [184].
Porphyromonas gingivalis expresses surface molecules that can activate the Toll‐like receptor 2‐TLR1 complex and secretes arginine‐specific cysteine proteinases (HRgpA and RgpB gingipains enzymes) that act on the complement component C5 to generate C5a, a ligand of complement C5a receptor 1 C5aR1 [185]. Therefore, Porphyromonas gingivalis can activate both C5aR1 and TLR2 in phagocytic cells to promote the expansion of inflammophiles and subsequent dysbiosis and disease‐provoking state. Porphyromonas gingivalis can also inhibit the phagocytosis of OSCC cells by macrophages, thereby promoting immunoevasion of oral cancer by protecting cancer from the attack of macrophages [186]. F. nucleatum is an oral bacterium. As mentioned earlier, it can promote CRC chemoresistance through regulating TLR4‐MYD88 pathway and function as an oncobacterium [23, 30]. The role of F. nucleatum in oral cancer has not been well‐documented, but it is linked to head and neck cancer [187], acute appendicitis [188], inflammatory bowel disease (IBD) [189], metastasis [32], esophageal cancer [155], breast cancer/pancreatic cancer [8]. It is conceivable that it may have a role in the development of oral cancer. It is worthwhile to point out that the impact of the oral microbiome extends beyond the oral cavity and oral cancer. Oral microbes can affect coronary artery disease [190, 191], preterm delivery of low‐birthweight neonates [192], Alzheimer's disease [193], and rheumatoid arthritis [182].
Thus, oral microbiome is associated with various cancers/diseases through direct toxic impacts of bacteria and their products and/or through indirect impacts of inflammatory pathology and signal transductions. Pathological effects due to oral microbial dysbiosis warrants further investigations.
5. GUT MICROBIOTA AND CANCER THERAPY EFFICACY
The crosstalk between microbes and host cells is critical for health and regulation of many physiological functions both locally and systemically [194]. For example, gut microbiota can affect locally regarding the nutrient absorption, vitamin B synthesis [195], bile acid metabolisms [196], carbohydrate fermentation [197], maintenance of barrier integrity [55], and regulating mucosa immunity [198] (Figure 6). Importantly, the gut microbiota can also systemically impact the host's metabolism, behavioral and cognitive activities [199], cardiovascular functions [200], hematopoiesis functions [201], ageing [202], inflammation and immunity [203, 204], and circadian rhythm [205, 206] (Figure 6), thereby affecting cancer development and cancer therapeutic efficacy and toxicity [140]. Also, tumor‐associated microbiota (Figure 1) may involve in cancer initiation, progression, and responses to cancer therapies [207]. Of note, microbiome regulates complex cellular networks involved in enhancing or attenuating the formation of cancer (Figure 7). Therefore, it is critical to investigate how the gut microbiome changes during the development of cancers and whether these changes can contribute to drug resistance during chemotherapy or other types of treatment. Investigating the role of gut microbiome/tumor‐associated microbiota will provide promising insights into diagnostic tools, biomarkers, and therapeutic intervention strategies for cancers.
FIGURE 6.
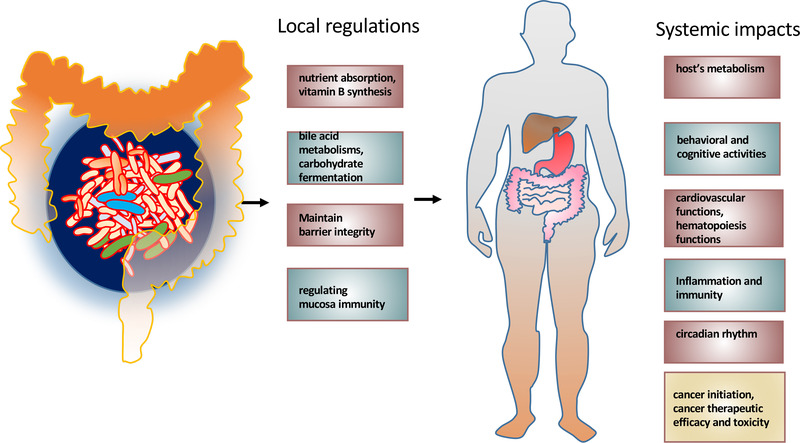
Gut microbiota's local and systemic impacts. Gut microbiota affects locally in listed functions and mucosa immunity. The gut microbiota can also systemically impact the host's many listed physiological functions. Both can possibly influence cancer development and cancer therapeutic efficacy and toxicity [210]
FIGURE 7.
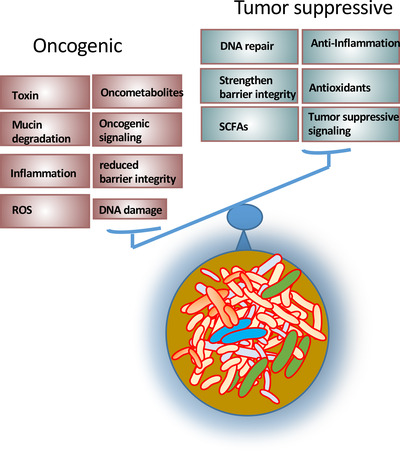
Mechanistic roles of microbiota in oncogenesis and tumor suppression. Roles of microbiota are pivotal in regulating oncogenesis or tumor suppression. Microbiota can contribute to oncogenesis or tumor suppression via the listed metabolites or functions. Microbiota dysbiosis may tilt the balance toward oncogenesis
Because the gut bacteria have the modifiable nature, gut microbiome can be modified for the purpose of cancer therapy via FMT, the administration of probiotics or certain bacterial species, altering lifestyle or changing diet [208], and using well‐designed antibiotic or tailored bacteriophages (Figure 3). Use of techniques in modulating the gut microbiota for cancer therapy will be discussed. Studies to unravel the relationship between microbiota and cancer drug resistance may have potential implications for overcoming drug resistance in clinical situation.
5.1. Microbiota impacts chemotherapeutic efficacy
The drug resistance to chemotherapy is a difficult challenge for treating cancers. Cancer cells have intrinsic genetic mechanisms that mediate chemo resistance, but little is known if other non‐cancer cells may impose an impact on drug resistance. Until recently, growing evidence suggests that microbes have impacts on the chemotherapeutic drug efficacy of cancer therapies [209]. Further, several microbial species are characterized to modulate radiotherapy and immunotherapy [210]. For example, higher F. nucleatum burden leads to poor response to neoadjuvant chemotherapy [157, 211]. It is then important to understand the mechanism of action from these microbial species, so improving anticancer efficacy is possible.
It was shown that glycolysis generates ATP and NAD+ that are used by poly(ADP‐ribose) polymerase (PARP) to repair DNA damage and provide energy for Multidrug Resistance Efflux pumps on cellular membranes to discard toxic chemotherapy agents. Thus, enhanced glycolysis can have impact on drug resistance. It is known that dietary fiber by increasing the abundance of Prevotella can improve glucose metabolism [212]. Barley Kernel supplements or high fiber leads to increased Prevotella in gut microbiota [212]. Prevotella subsequently protects against Bacteroides‐induced glucose intolerance [212]. Glucose intolerance can lead to accelerated tumor growth [86, 213]. A recent study indicates that ARAF mutations conferred resistance to RAF inhibitor [214]. CRC with Kras or RAF mutations often upregulates glucose transporter 1 (GLUT1), a gene encoding glucose transporter‐1 involved in glycolysis, to reprogram cancer energy metabolism [215], implying that the glycolysis deregulation is critical for cancer development and drug resistance. It is conceivable that microbes can affect drug efficacy through moderating glucose intolerance.
5.1.1. Gemcitabine
Also, the microbiota is involved in direct metabolic processes of drugs, including reduction, hydrolysis, dehydroxylation, and dealkylation [209], so the microbiota impacts drug pharmacokinetics, anticancer activity and toxicity. Thus, it is possible that controlling gut microbiota may be a useful strategy in reducing drug resistance. For example, it was shown that bacteria can metabolize gemcitabine (2',2'‐difluorodeoxycytidine) into its inactive form, 2',2'‐difluorodeoxyuridine [216]. The metabolic process is dependent on bacterial enzyme cytidine deaminase found in intratumoral Gammaproteobacteria. Gemcitabine is commonly used in treating PDAC, and many PDAC patients are positive for Gammaproteobacteria, implying that many of them may be resistant to Gemcitabine treatment. Targeting Gammaproteobacteria may have potential implications for overcoming gemcitabine resistance in pancreatic cancer.
5.1.2. Erlotinib
Specific taxa related to different cancer treatment responses in multiple cancer types are identified [217]. Functional profiles of intestinal microbiota of cancer patients were assessed. On the basis of this information, microbiota composition and functionality can predict the response to cancer treatments including cytotoxic or targeted chemotherapy, immunotherapy, or a combination [217]. Bacteroides ovatus and Bacteroides xylanisolvens were then identified in cancer treatment responders, and thus were positively correlated with treatment outcomes [217]. Significantly, administration of these two responder bacteria strains can boost the treatment efficacy of erlotinib in mouse lung cancer model [217]. They are able to increase the expression of the chemokine (C‐X‐C motif) ligand 9, CXCL10, and IFN‐γ in the tumors of erlotinib‐treated mice, implying their impact on cytokine expression [217]. This microbiota signature to strengthen the treatment efficacy may be applied to various cancer therapeutic designs as the signature is independent of cancer types. The potential of combining this bacterial signature with various types of cancer therapies should be addressed soon in clinical trials involving patients with tumors. Similarly, it was shown that combination of L. acidophilus and Bifidobacterium bifidum can reduce intestinal toxicity in cancer patients treated with both radiotherapy and cisplatin [218]. Further, they actually enhance anti‐tumor effect of cisplatin. Therefore, specific gut microbes can be characterized to significantly increase the effect of chemotherapy.
Also prebiotics (such as inulin or oligofructose) and postbiotics (such as butyrate) have the potential of preventing cancer [112, 219, 220]. Lactobacillus and Bifidobacterium are two genera of probiotics. They are efficient in decomposing sugars and producing lactic acid [220]. It is known that probiotics are beneficial to human health [220]. Probiotics will have impacts on the microbiota by inhibiting colonization of pathogenic bacteria, strengthen gut barrier, reduce colonic immunity or enhance anti‐cancer immunity, influence neuromuscular function, and drive the microbiota‐gut‐brain axis [220, 221] (Figure 3). It is then conceivable that administrations of beneficial bacteria (such as Bacteroides ovatus and Bacteroides xylanisolvens and other probiotics) together with prebiotics may be employed as anticancer adjuvant agents in enhancing cancer therapeutic regimens.
5.1.3. Oxaliplatin
Efficacy of oxaliplatin was diminished due to reduced intratumoral ROS production in germ‐free mice model, suggesting the therapeutic effects of oxaliplatin depend on the presence of microbiota [222]. Thus, these data support that the microorganisms play a critical role in determining the efficacy and toxicity of chemotherapeutic agents. However, the complexity remains to be characterized to understand the host‐microbial interactions in modulating the human clinical outcomes of drugs.
Collectively, the dysbiosis of microbiota could be the culprit of drug resistance, and a strategic intervention against the specific bacterium associated with drug resistance may significantly improve therapeutic response in patients with cancer. Gut microbiota modification could be a strategy to boost the efficacy and/or reduce the potential toxicity and adverse effects of cancer therapeutic treatment [223].
5.2. Microbiota affecting immunity/immunotherapy
Microbial products cause MAMPs. MMAPs and damage‐associated molecular patterns can interact with membrane‐bound and cytoplasmic innate immune receptors to regulate metabolism, inflammation, innate and adaptive immunity [210]. Metabolites of microbiota or bacteria themselves are involved in activating local dendritic cells to activate naive T cells to effector T cells, such as Tregs or Th17 cells. Tregs can secret IL‐10 to establish a local anti‐inflammatory cytokine milieu, while Th17 cells can produce IL‐17 to increase Paneth cell production of antimicrobial peptides [40]. It is interesting to point out that in gut microbiome of corona virus disease 2019 (COVID‐19) patients’ immunomodulatory potential bacterial strains, such as Faecalibacterium prausnitzii, Eubacterium rectale and Bifidobacteria, were particularly depleted, suggesting the link of immunity, virus infection, and gut microbiome [224]. Strong evidence has indicated a critical role of microbiome in regulating human immunity as broad‐spectrum antibiotics treatment can affect the immune response to vaccination by affecting inflammatory signatures, metabolism, and inflammasome activation [225].
Several cancer therapies, including the trendy immunotherapy, have improved the clinical outcomes for many cancer patients, but sometimes the outcomes are heterogeneous and required further insights to understand the discrepancy [226, 227, 228, 229, 230]. The predictors of response to cancer therapy have been proposed to be the microbiome, as the microbiome is critical in influencing host immunity, which is involved in impacting responses to cancer treatment [231]. Indeed, it was shown that in anti‐PD‐1‐based cancer immunotherapy of melanoma patients, Bifidobacterium longum, Collinsella aerofacieus, and Enterococcus faecium were more abundant in responders. Also, anti‐PD‐1 cancer immunotherapy efficacy is correlated with fecal SCFA concentrations [232]. Basically, high concentrations of SCFAs, including acetic acid, propionic acid, butyric acid, valeric acid, and isovaleric acid, are associated with longer progression‐free survival [232]. Thus, SCFAs act as the effectors between the gut microbiota and anti‐PD‐1 cancer immunotherapy efficacy. Because fecal examinations are completely noninvasive, they may be applicable for routine monitoring of patients. It is then critical to identify the effectors affecting the gut microbiota's role in therapy in order to design strategies to modify the microbiome to boost cancer therapeutic responses [40, 233].
The gut microbiota impacts systemic immune function through local interactions within the gut mucosa and gut‐associated lymphoid tissue. Several microbial taxa are enriched in response to cancer immunotherapy agents (such as anti‐CTLA‐4 and/or anti‐PD‐1). On the other hand, several microbial taxa are enriched in nonresponders. Whether these associations are correlative in nature or indeed manifest causality remains to be established. Recently, it has been shown that efficacy of anti‐CTLA‐4 and anti‐PD‐L1 therapies require Bacteroides enrichment and Bifidobacterium abundance respectively [234], illustrating that gut microbiota is a critical modulator of the immune system. Together, the interaction between microorganisms and the host immune regulation is frequently observed [235]. Of note, gut microbiota has impacts in regulating inflammation [236], a well‐characterized hallmark of cancers, thereby furthering the development of cancers [237, 238]. This characteristic also participates in deciding the immunotherapy efficacy.
5.3. FMT applications in clinical medicine
FMT was first used in patients roughly 1700 years ago by a Chinese physician/chemist named Ge Hong [239]. He orally administered ‘Huang Long Tang’, a stool liquid from a healthy person, to cure patients of severe diarrhea. FMT was used as a treatment for Clostridium difficile infection in 1958 [240]. Recently, there are several clinical trials using FMT to treat colon cancer, breast cancer, melanoma, renal cell carcinoma, liver cancer, gynecological cancer, head‐and‐neck cancer, and lung cancer [241]. Therefore, the use of FMT in cancer patients for cancer therapy purpose is emerging [242].
5.3.1. Successful FMT treatment in diseases
Dysbiosis of microbiota leads to a plethora of disease conditions, such as skin, inflammatory, metabolic, neurological disorders, and cancers. Understanding host‐microbe interaction is critical for diagnosis and critical therapeutic design for these ailments. Indeed, several studies have demonstrated that FMT is effective in treating ulcerative colitis, irritable bowel syndrome (IBS), and hepatic encephalopathy [243, 244, 245]. Some of these applications serve as the important examples to follow for cancer therapy.
5.3.1.1. Depression, autism, aging, birth
FMT from depression patients into germ‐free mice leads to increased immobility times in the forced swimming and tail suspension tasks, suggesting that gut microbial dysbiosis is involved in stress‐related behavior in human depression [246]. Gut microbiota dysbiosis is involved in the pathophysiology of autism as gut microbiota dysbiosis and barrier dysfunction can cause behavioral changes as well as neurodevelopment disorder [243]. Children with autism often suffer gastrointestinal problems, suggesting that microbiome correlates with autism [247]. Indeed, microbiome links to abnormal metabolites and behavior of children with autism. Significantly, FMT can alleviate behavioral symptoms in children with autism. The engraftment of donor microbiota and changes in the gut environment lead to the improvement. Several taxa including Bifidobacterium, Prevotella, and Desulfovibrio are increased after FMT, suggesting that they may play important roles in autism [247].
In a mouse model study, FMT from aged donor mice resulted in compromised spatial learning and memory in young recipients [248]. This phenomenon is due to altered expression of proteins involved in synaptic plasticity and neurotransmission in the hippocampus [248]. Cesarean section (CS) infants lack for the mother‐to‐neonate transmission of maternal fecal microbes. Intriguingly, gut microbiota of CS‐born infants can be restored by maternal FMT [249]. Not only do these FMT applications lead to impressive results in animal experiments, but results in human clinical applications have substantial therapeutic potential, thus highlighting the importance of developing FMT for these diseases or conditions.
5.3.1.2. Obesity, IBS, clostridium difficile infection
It has been shown that the intestinal microbiome is also correlated with several diseases including obesity, IBS and Clostridium difficile infection [250]. Characterizing the relationships and understanding the roles of microbial strains involved in these diseases will allow future diagnosis and treatment [250]. For obesity, it was shown that FMT from lean donors can improve insulin sensitivity in obese recipients with metabolic syndrome [251]. IBS is a common disorder with altered gut function. IBS FMT was administrated to healthy mice, and these mice demonstrated faster gastrointestinal transit, barrier integrity dysfunction, innate immune system activation, and even behavior change [252]. Thus, microbiome has impact on IBS progression. FMT was applied in the treatment of Clostridium difficile infection [253]. FMT for Clostridium difficile infection has been remarkably successful [254]. The mechanistic impacts from FMT include competition with Clostridium difficile, rebuilding secondary bile acid metabolism, and restoring gut barrier integrity. Although FMT has successfully treated relapsed or refractory Clostridium difficile infection, the recent incident of bacteremia caused by extended‐spectrum beta‐lactamase‐producing Escherichia coli is alarming [255]. It is important to point out that enhanced donor screening is required to mitigate the transmission of bacteria that could cause adverse infectious events.
Together, it has been shown that the intestinal microbiome is correlated with several diseases including obesity, IBS, and CDI, and that FMT showcases its potential in clinical applications. Characterizing the relationships and understanding the roles of microbial strains involved in these diseases will allow more future use of these bacterial strains in diagnosis and in improving treatment [250].
5.3.2. Promising FMT in cancer therapy
As for the cancers, they are described below.
5.3.2.1. ICI therapy
Recently, ICI immunotherapy has made a big progress. Many factors are deciding the immunotherapy efficacy, including mutational load, cancer metabolism, and microbiota [40]. Evidence demonstrates that the composition of microbiota impacts responses to anticancer immunotherapies and can be a novel biomarker of response assessment [256, 257]. It was demonstrated that the presence of specific immune‐potentiating bacteria, including Akkermansia muciniphila, Faecalibacterium prausnitzii, Bifidobacterium spp., and Bacteroides fragilis, is critical for ICI therapy. Therefore, modulation of the microbiota composition appears to be a critical strategy for future treatment [258].
Although ICI therapy targets CTLA‐4, PD‐1 and PD‐L1 to increase T cell activation for effective anti‐cancer immune responses, treatments often lead to immune‐related adverse effects, such as ICI‐associated colitis. There is an urgent need to deal with this issue. Significantly, modulation of the gut microbiome with FMT seems to be effective in abrogating ICI‐associated colitis [259]. These results demonstrate that therapeutic responses of ICI have specific dependencies on bacterial composition; in the clinic, the adverse effects of ICI‐related colitis in ICI treatment could be ameliorated by FMT.
5.3.2.2. Melanoma, gynecology cancer, and FMT
Several strategies can be employed for modifying the gut microbiota of cancer patients, including FMT from specific donors, administration of single bacterial species or mixed several bacterial species, using antibiotics to reduce “bad” bacterial species, using prebiotics to facilitate beneficial bacterial species growth or direct probiotics to strengthen the microenvironment (Figure 3) [258]. The major indirect impacts of FMT include immunotherapy efficacy modulation, regulation of bile acid metabolism/SCFA metabolism, and restoration of intestinal microbiota diversity. Direct impacts can be obtained from bacteriophages and metabolites transferred [243] (Figure 3).
Several years ago, it was documented that the gut microbiome plays an important role to affect the response of anti‐PD‐1 immunotherapy in mouse models and patient cohorts [226, 227, 228]. Basically, these investigations serve as a proof of concept. But FMT has not been exploited in cancer patients to investigate whether modulation of gut microbiota can really increase the efficacy of anti‐PD‐1 immunotherapy until recently. Phase I clinical trials indicate that patients with anti‐PD‐1‐refractory metastatic melanoma had their responses to anti‐PD‐1 immunotherapy improved after FMT from anti‐PD‐1 responders [260, 261, 262], suggesting a big step in improving cancer immunotherapy. Further, in human clinical trials from Davar et al. [262] showed that FMT increased abundance of taxa of Bifidobacteriacae, Coriobacteriacae, Ruminococcaceae, Lachnospiraceae in patients responded to anti‐PD‐1 immunotherapy.
FMT contains the microbiome, which is critical in maintaining homeostasis and health. Microbiota dysbiosis is also responsible for gynecological cancer [263, 264, 265]. Epidemiological studies indicate that the urogenital microbiota dysbiosis is implied in gynecological cancers, particularly cervical cancer [263, 264, 266]. Probiotics or microbiota transplant seems to be effective in improving responsiveness to treatment of gynecological cancer [263, 267, 268]. In the presence of damaged barrier integrity, the microbiota may cause carcinogenesis by impacting host cell proliferation and death, disturbing the regulation of immune system, and affecting metabolism of host. Gut microbiota modulation, such as FMT, to correct microbial dysbiosis is a rational therapeutic strategy for prevention and treatment of cancer.
Collectively, clinical trials evaluating the efficacy of FMT in non‐responder of melanoma patients regarding ICI treatment efficacy are quite promising. These inspirational clinical trials will enable more development of effective new immunotherapies combined with FMT in many types of cancer.
5.3.2.3. FMT in clinical trials
FMT is very effective in treating Clostridium difficile infection. Clinical trials have demonstrated that oral capsules or colonoscopy delivery is equally effective [269]. This resolves the concern regarding how to deliver microbiota much more efficiently. The clinical trials showed high efficacy of FMT for Clostridium difficile infection patients. Short‐term follow‐up demonstrated a very good safety profile [270]. However, continuing long‐term follow‐up will be required for establishing the full safety profile of FMT.
There are more than 200 studies about FMT in clinical applications listed in ClinicalTrials.gov, suggesting the enthusiasm from the medical field [243, 271]. However, microbiota‐targeting of certain bacterial strains remains to be further established. Certainly, FMT standardized treatment approaches to a particular disorder or cancer will need further refinement. Long‐term safety of FMT needs to be established and monitored, such as side effect in inflammation or autoimmune reactions. Further, quality control, central registration of stool banks, and clinical outcome with careful follow‐up are required to be established. The rational design of novel FMT‐based combinations with chemotherapy or immunotherapy for use in cancer patients will need a well‐established framework to select the most appropriate therapeutic agents. FMT carries tremendous promise to improve the treatment and survival of cancers, and further investigations/clinical trials will ultimately improve the lives of cancer patients.
5.3.2.4. FMT and future applications
FMT is indeed effective for a variety of human illnesses. However, for microbiota‐based therapeutic strategy to be more practical in clinic applications, we still have a long way to go. FMT may have advantages, as this process can transfer microbiota, SCFAs, other metabolites, and microphages, which will have uncharacterized impacts in providing advantage in therapeutic efficacy (Figure 3). However, the use of defined bacterial species will probably increase the robustness of efficacy and decrease the risks associated with FMT from unknown bacterial composition. Promising evidence indicates that we are making progress in harnessing the power of microbiota in fighting diseases and cancers [260, 272].
The caveats of FMT are that live microorganisms pose safety concerns, especially when they are modified genetically. But it is possible to use phage, small molecules or macromolecules that can modify the natural gut microbiota or provide microbiota's beneficial effects to overcome potential concerns. The availability of stool banks makes FMT a useful new treatment option for many other disorders, including cancers, possible in addition to Clostridium difficile infection. FMT has been investigated in several diseases, such as IBD, IBS, metabolic syndromes, autism, neurodevelopmental disorders, and cancer diseases [243]. Although FMT study is still in its infancy, we expect to see more applications in the future. It is a very exciting field, and well‐executed clinical trials will be furthering the applications in many microbiota‐related conditions.
6. CONCLUSIONS
Although cancer is generally considered a disease of the gene deregulation, more studies have demonstrated that the microbiome contributes to the tumorigenesis of cancers. Contributions of the fecal microbiome to gastrointestinal cancers are evident, and this phenomenon has extended to other types of cancer. Future applications in terms of diagnostics based on the microbial signatures have great potentials. Microbiome investigation has led to promising development of cancer care during the past few years. The relationship characterization between the microbiome and cancer biology has emerged. The critical cancer gene–microbiome environmental interactions in cancer have been demonstrated. The impact of diet on the microbiome composition/metabolism, which in turn affects the cancer risk, has been illustrated. Modifying microbiota to affect the efficacy of chemotherapies and immunotherapies has been successful. Thus, the gut microbiome has a strong potential to be exploited as both a diagnostic tool and a therapeutic strategy for anticancer therapy.
Nonetheless, our current understanding of how human microbiota can confer susceptibility to certain cancers is far from complete; for example, there are still extensive gaps in understanding the compositions of microbiota as many of the bacterial species are not easy to be cultured; the crosstalk between the gut microbiota and the physiological systems remains to be characterized; beneficial impacts of prebiotic molecules or probiotic strains remain to be further investigated in human clinical studies; the roles of phages, archaebacteria, fungus in regulating cancer growth remain to be explored; direct effect of local presence within the tumor microenvironment or regulating the systemic impact of distant microbiota or physiology warrants further investigation. This field is still young, and we are left with many challenges described above. It is important to characterize the mechanisms of action from bacterial species and to determine the bacterial species that can be important in mediating anticancer effects.
The gut microbiome has become an efficient target for making cancer chemotherapy/immunotherapy safer or efficient, which will further improve cancer survival rates in the future. Certainly, multifaceted strategies will need to be devised to monitor and to modulate the impact of microbiota, such as FMT applications. The field is extraordinarily promising and rapidly expanding. It is expected that more clinical trials will be employed to provide important insights into the application potential of the gut microbiota in cancer treatment. We can imagine that there is a humongous opportunity in microbiota research, from basic and translational to clinical applications and epidemiological analyses, to advance our knowledge of this complicated biological system created by microbiota.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
The author declares that they have no competing interests.
FUNDING
This study was supported in part by National key research and development program (2020YFA0803300 and 2018YFC0910300) and National Natural Science Foundation of China (81630072), and Shenzhen Municipal Government of China (KQTD20170810160226082).
AUTHORS' CONTRIBUTIONS
M.H.L. contributed substantially to the conception of the study, drafted the article, and provided final approval of the version to publish.
ACKNOWLEDGEMENTS
Not applicable.
Lee M‐H. Harness the functions of gut microbiome in tumorigenesis for cancer treatment. Cancer Commun. 2021;41:937–967. 10.1002/cac2.12200
REFERENCES
- 1. Fischbach MA. Microbiome: Focus on Causation and Mechanism. Cell. 2018;174(4):785‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14(8):e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rajagopala SV, Vashee S, Oldfield LM, Suzuki Y, Venter JC, Telenti A, et al. The Human Microbiome and Cancer. Cancer Prev Res (Phila). 2017;10(4):226‐34. [DOI] [PubMed] [Google Scholar]
- 5. Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin. 2017;67(4):326‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zou S, Fang L, Lee MH. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol Rep (Oxf). 2018;6(1):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170(3):548‐63 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type‐specific intracellular bacteria. Science. 2020;368(6494):973‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fulbright LE, Ellermann M, Arthur JC. The microbiome and the hallmarks of cancer. PLoS Pathog. 2017;13(9):e1006480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13(6):607‐15. [DOI] [PubMed] [Google Scholar]
- 11. Garrett WS. Cancer and the microbiota. Science. 2015;348(6230):80‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579(7800):567‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dohlman AB, Arguijo Mendoza D, Ding S, Gao M, Dressman H, Iliev ID, et al. The cancer microbiome atlas: a pan‐cancer comparative analysis to distinguish tissue‐resident microbiota from contaminants. Cell Host Microbe. 2021;29(2):281‐98.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stewart OA, Wu F, Chen Y. The role of gastric microbiota in gastric cancer. Gut Microbes. 2020;11(5):1220‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferreira RM, Pereira‐Marques J, Pinto‐Ribeiro I, Costa JL, Carneiro F, Machado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer‐associated microbiota. Gut. 2018;67(2):226‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sayed IM, Sahan AZ, Venkova T, Chakraborty A, Mukhopadhyay D, Bimczok D, et al. Helicobacter pylori infection downregulates the DNA glycosylase NEIL2, resulting in increased genome damage and inflammation in gastric epithelial cells. J Biol Chem. 2020;295(32):11082‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sierra JC, Asim M, Verriere TG, Piazuelo MB, Suarez G, Romero‐Gallo J, et al. Epidermal growth factor receptor inhibition downregulates Helicobacter pylori‐induced epithelial inflammatory responses, DNA damage and gastric carcinogenesis. Gut. 2018;67(7):1247‐1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie C, Li N, Wang H, He C, Hu Y, Peng C, et al. Inhibition of autophagy aggravates DNA damage response and gastric tumorigenesis via Rad51 ubiquitination in response to H. pylori infection. Gut Microbes. 2020;11(6):1567‐1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han T, Jing X, Bao J, Zhao L, Zhang A, Miao R, et al. H. pylori infection alters repair of DNA double‐strand breaks via SNHG17. J Clin Invest. 2020;130(7):3901‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang G, Xu Y, Wang S, Gong Z, Zou C, Zhang H, et al. LncRNA SNHG17 promotes gastric cancer progression by epigenetically silencing of p15 and p57. J Cell Physiol. 2019;234(4):5163‐74. [DOI] [PubMed] [Google Scholar]
- 21. Tegtmeyer N, Wessler S, Necchi V, Rohde M, Harrer A, Rau TT, et al. Helicobacter pylori Employs a Unique Basolateral Type IV Secretion Mechanism for CagA Delivery. Cell Host Microbe. 2017;22(4):552‐60.e5. [DOI] [PubMed] [Google Scholar]
- 22. Matson V, Chervin CS, Gajewski TF. Cancer and the Microbiome: Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology. 2021;160(2):600–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brennan CA, Garrett WS. Fusobacterium nucleatum ‐ symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leung A, Tsoi H, Yu J. Fusobacterium and Escherichia: models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev Gastroenterol Hepatol. 2015;9(5):651‐7. [DOI] [PubMed] [Google Scholar]
- 25. Hashemi Goradel N, Heidarzadeh S, Jahangiri S, Farhood B, Mortezaee K, Khanlarkhani N, et al. Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J Cell Physiol. 2019;234(3):2337‐44. [DOI] [PubMed] [Google Scholar]
- 26. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gholizadeh P, Eslami H, Kafil HS. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed Pharmacother. 2017;89:918‐25. [DOI] [PubMed] [Google Scholar]
- 28. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1(5):653‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll‐Like Receptor 4 Signaling to Nuclear Factor‐κB, and Up‐regulating Expression of MicroRNA‐21. Gastroenterology. 2017;152(4):851‐866.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170(3):548‐63.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen S, Su T, Zhang Y, Lee A, He J, Ge Q, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7‐AS/KRT7. Gut Microbes. 2020;11(3):511‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo S, Chen J, Chen F, Zeng Q, Liu WL, Zhang G. Exosomes derived from Fusobacterium nucleatum‐infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR‐1246/92b‐3p/27a‐3p and CXCL16. Gut. 2020. [DOI] [PubMed] [Google Scholar]
- 33. Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang X, et al. F. nucleatum targets lncRNA ENO1‐IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2020. [DOI] [PubMed] [Google Scholar]
- 34. Aymeric L, Donnadieu F, Mulet C, du Merle L, Nigro G, Saffarian A, et al. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc Natl Acad Sci U S A. 2018;115(2):E283‐E291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4(12):2319‐30. [DOI] [PubMed] [Google Scholar]
- 36. Rackaityte E, Lynch SV. The human microbiome in the 21(st) century. Nat Commun. 2020;11(1):5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ‐Free and Conventional Mice. Gastroenterology. 2017;153(6):1621‐1633.e6. [DOI] [PubMed] [Google Scholar]
- 38. Ansari I, Raddatz G, Gutekunst J, Ridnik M, Cohen D, Abu‐Remaileh M, et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat Microbiol. 2020;5(4):610‐9. [DOI] [PubMed] [Google Scholar]
- 39. Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 2019;363(6428). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33(4):570‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Belkaid Y, Harrison OJ. Homeostatic Immunity and the Microbiota. Immunity. 2017;46(4):562‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Michaudel C, Sokol H. The Gut Microbiota at the Service of Immunometabolism. Cell Metab. 2020;32(4):514‐23. [DOI] [PubMed] [Google Scholar]
- 43. Schulthess J, Pandey S, Capitani M, Rue‐Albrecht KC, Arnold I, Franchini F, et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity. 2019;50(2):432‐45 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He Y, Fu L, Li Y, Wang W, Gong M, Zhang J, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. 2021;33(5):988‐1000.e7. [DOI] [PubMed] [Google Scholar]
- 45. Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, et al. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front Cell Infect Microbiol. 2018;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30(1):16‐34. [DOI] [PubMed] [Google Scholar]
- 47. Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol. 2017;18(8):851‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167(4):1125‐36.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bellone M, Brevi A, Huber S. Microbiota‐Propelled T Helper 17 Cells in Inflammatory Diseases and Cancer. Microbiol Mol Biol Rev. 2020;84(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell. 2019;176(5):998‐1013.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (Oxf). 2019;7(1):3‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Belzer C, Chia LW, Aalvink S, Chamlagain B, Piironen V, Knol J, et al. Microbial Metabolic Networks at the Mucus Layer Lead to Diet‐Independent Butyrate and Vitamin B(12) Production by Intestinal Symbionts. mBio. 2017;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A Dietary Fiber‐Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167(5):1339‐53 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, et al. Bifidobacteria or Fiber Protects against Diet‐Induced Microbiota‐Mediated Colonic Mucus Deterioration. Cell Host Microbe. 2018;23(1):27‐40 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A Dietary Fiber‐Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167(5):1339‐53.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee MH, Lozano G. Regulation of the p53‐MDM2 pathway by 14‐3‐3 sigma and other proteins. Semin Cancer Biol. 2006;16(3):225‐34. [DOI] [PubMed] [Google Scholar]
- 57. Yang HY, Wen YY, Chen CH, Lozano G, Lee MH. 14‐3‐3 sigma positively regulates p53 and suppresses tumor growth. Mol Cell Biol. 2003;23(20):7096‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kadosh E, Snir‐Alkalay I, Venkatachalam A, May S, Lasry A, Elyada E, et al. The gut microbiome switches mutant p53 from tumour‐suppressive to oncogenic. Nature. 2020;586(7827):133‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lau HCH, Yu J. Gut microbiome alters functions of mutant p53 to promote tumorigenesis. Signal Transduct Target Ther. 2020;5(1):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Elinav E, Garrett WS, Trinchieri G, Wargo J. The cancer microbiome. Nat Rev Cancer. 2019;19(7):371‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schroeder BO, Backhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22(10):1079‐89. [DOI] [PubMed] [Google Scholar]
- 62. Krautkramer KA, Fan J, Bäckhed F. Gut microbial metabolites as multi‐kingdom intermediates. Nat Rev Microbiol. 2020. [DOI] [PubMed] [Google Scholar]
- 63. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661‐72. [DOI] [PubMed] [Google Scholar]
- 65. Ridlon JM, Devendran S, Alves JM, Doden H, Wolf PG, Pereira GV, et al. The ‘in vivo lifestyle’ of bile acid 7α‐dehydroxylating bacteria: comparative genomics, metatranscriptomic, and bile acid metabolomics analysis of a defined microbial community in gnotobiotic mice. Gut Microbes. 2020;11(3):381‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ridlon JM, Wolf PG, Gaskins HR. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes. 2016;7(3):201‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity‐induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97‐101. [DOI] [PubMed] [Google Scholar]
- 68. Fu T, Coulter S, Yoshihara E, Oh TG, Fang S, Cayabyab F, et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell. 2019;176(5):1098‐112 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. 2019;576(7785):143‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature. 2020;577(7790):410‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lin R, Zhan M, Yang L, Wang H, Shen H, Huang S, et al. Deoxycholic acid modulates the progression of gallbladder cancer through N(6)‐methyladenosine‐dependent microRNA maturation. Oncogene. 2020;39(26):4983‐5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen S, Henderson A, Petriello MC, Romano KA, Gearing M, Miao J, et al. Trimethylamine N‐Oxide Binds and Activates PERK to Promote Metabolic Dysfunction. Cell Metab. 2019;30(6):1141‐51.e5. [DOI] [PubMed] [Google Scholar]
- 73. Xu R, Wang Q, Li L. A genome‐wide systems analysis reveals strong link between colorectal cancer and trimethylamine N‐oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics. 2015;16(7):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, et al. Non‐lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell. 2015;163(7):1585‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L‐carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gao Q, Wang Y, Wang X, Fu S, Zhang X, Wang RT, et al. Decreased levels of circulating trimethylamine N‐oxide alleviate cognitive and pathological deterioration in transgenic mice: a potential therapeutic approach for Alzheimer's disease. Aging (Albany NY). 2019;11(19):8642‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chan CWH, Law BMH, Waye MMY, Chan JYW, So WKW, Chow KM. Trimethylamine‐N‐oxide as One Hypothetical Link for the Relationship between Intestinal Microbiota and Cancer ‐ Where We Are and Where Shall We Go? J Cancer. 2019;10(23):5874‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang L, Tang W, Yang S, He P, Wang J, Gaedcke J, et al. NO(•) /RUNX3/kynurenine metabolic signaling enhances disease aggressiveness in pancreatic cancer. Int J Cancer. 2020;146(11):3160‐3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Venkateswaran N, Lafita‐Navarro MC, Hao YH, Kilgore JA, Perez‐Castro L, Braverman J, et al. MYC promotes tryptophan uptake and metabolism by the kynurenine pathway in colon cancer. Genes Dev. 2019;33(17‐18):1236‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “western‐lifestyle” inflammatory diseases. Immunity. 2014;40(6):833‐42. [DOI] [PubMed] [Google Scholar]
- 81. Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Krämer M, et al. The Gut Microbiota in Prediabetes and Diabetes: A Population‐Based Cross‐Sectional Study. Cell Metab. 2020;32(3):379‐90.e3. [DOI] [PubMed] [Google Scholar]
- 82. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås‐Holm L, et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell. 2018;175(4):947‐61.e17. [DOI] [PubMed] [Google Scholar]
- 84. Molinaro A, Bel Lassen P, Henricsson M, Wu H, Adriouch S, Belda E, et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat Commun. 2020;11(1):5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koh A, Mannerås‐Holm L, Yunn NO, Nilsson PM, Ryu SH, Molinaro A, et al. Microbial Imidazole Propionate Affects Responses to Metformin through p38γ‐Dependent Inhibitory AMPK Phosphorylation. Cell Metab. 2020;32(4):643‐653.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Velazquez‐Torres G, Fuentes‐Mattei E, Choi HH, Yeung SJ, Meng X, Lee MH. Diabetes mellitus type 2 drives metabolic reprogramming to promote pancreatic cancer growth. Gastroenterol Rep (Oxf). 2020;8(4):261‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fuentes‐Mattei E, Velazquez‐Torres G, Phan L, Zhang F, Chou PC, Shin JH, et al. Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor‐positive breast cancer. J Natl Cancer Inst. 2014;106(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. He X, Esteva FJ, Ensor J, Hortobagyi GN, Lee MH, Yeung SC. Metformin and thiazolidinediones are associated with improved breast cancer‐specific survival of diabetic women with HER2+ breast cancer. Ann Oncol. 2012;23(7):1771‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. He XX, Tu SM, Lee MH, Yeung SC. Thiazolidinediones and metformin associated with improved survival of diabetic prostate cancer patients. Ann Oncol. 2011;22(12):2640‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome‐derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369(6510):1481‐9. [DOI] [PubMed] [Google Scholar]
- 91. Peterson CT, Rodionov DA, Osterman AL, Peterson SN. B Vitamins and Their Role in Immune Regulation and Cancer. Nutrients. 2020;12(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, et al. GPR109A is a G‐protein‐coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69(7):2826‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li J, Kong D, Wang Q, Wu W, Tang Y, Bai T, et al. Niacin ameliorates ulcerative colitis via prostaglandin D(2)‐mediated D prostanoid receptor 1 activation. EMBO Mol Med. 2017;9(5):571‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li K, Wu JL, Qin B, Fan Z, Tang Q, Lu W, et al. ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer. Cell Res. 2020;30(2):163‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Choi HH, Zou S, Wu JL, Wang H, Phan L, Li K, et al. EGF Relays Signals to COP1 and Facilitates FOXO4 Degradation to Promote Tumorigenesis. Adv Sci (Weinh). 2020;7(20):2000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hosseinkhani F, Heinken A, Thiele I, Lindenburg PW, Harms AC, Hankemeier T. The contribution of gut bacterial metabolites in the human immune signaling pathway of non‐communicable diseases. Gut Microbes. 2021;13(1):1‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li J, Meng Y, Wu X, Sun Y. Polyamines and related signaling pathways in cancer. Cancer Cell Int. 2020;20(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li L, Mao Y, Zhao L, Li L, Wu J, Zhao M, et al. p53 regulation of ammonia metabolism through urea cycle controls polyamine biosynthesis. Nature. 2019;567(7747):253‐6. [DOI] [PubMed] [Google Scholar]
- 100. Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21(6):891‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Selma MV, Beltran D, Luna MC, Romo‐Vaquero M, Garcia‐Villalba R, Mira A, et al. Isolation of Human Intestinal Bacteria Capable of Producing the Bioactive Metabolite Isourolithin A from Ellagic Acid. Front Microbiol. 2017;8:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Singh R, Chandrashekharappa S, Bodduluri SR, Baby BV, Hegde B, Kotla NG, et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019;68(2):289‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille‐Michaud A, et al. High prevalence of mucosa‐associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One. 2013;8(2):e56964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15(8):465‐78. [DOI] [PubMed] [Google Scholar]
- 107. Plaza‐Diaz J, Ruiz‐Ojeda FJ, Gil‐Campos M, Gil A. Mechanisms of Action of Probiotics. Adv Nutr. 2019;10(1):S49‐s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Motevaseli E, Dianatpour A, Ghafouri‐Fard S. The Role of Probiotics in Cancer Treatment: Emphasis on their In Vivo and In Vitro Anti‐metastatic Effects. Int J Mol Cell Med. 2017;6(2):66‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Banna GL, Torino F, Marletta F, Santagati M, Salemi R, Cannarozzo E, et al. Lactobacillus rhamnosus GG: An Overview to Explore the Rationale of Its Use in Cancer. Front Pharmacol. 2017;8:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wei D, Heus P, van de Wetering FT, van Tienhoven G, Verleye L, Scholten RJ. Probiotics for the prevention or treatment of chemotherapy‐ or radiotherapy‐related diarrhoea in people with cancer. Cochrane Database Syst Rev. 2018;8(8):Cd008831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tarrah A, de Castilhos J, Rossi RC, Duarte VDS, Ziegler DR, Corich V, et al. In vitro Probiotic Potential and Anti‐cancer Activity of Newly Isolated Folate‐Producing Streptococcus thermophilus Strains. Front Microbiol. 2018;9:2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16(10):605‐16. [DOI] [PubMed] [Google Scholar]
- 113. Britton RA, Hoffmann DE, Khoruts A. Probiotics and the Microbiome‐How Can We Help Patients Make Sense of Probiotics? Gastroenterology. 2021;160(2):614‐23. [DOI] [PubMed] [Google Scholar]
- 114.La Rosa SL, Leth ML, Michalak L, Hansen ME, Pudlo NA, Glowacki R, et al. The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary β‐mannans. Nat Commun. 2019;10(1):905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019;364(6446):1179‐1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107‐13. [DOI] [PubMed] [Google Scholar]
- 117. Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8(+) T cells in mice. Gut. 2020;69(11):1988‐1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xie C, Coda R, Chamlagain B, Varmanen P, Piironen V, Katina K. Co‐fermentation of Propionibacterium freudenreichii and Lactobacillus brevis in Wheat Bran for in situ Production of Vitamin B12. Front Microbiol. 2019;10:1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yeom J, Ma S, Lim YH. Probiotic Propionibacterium freudenreichii MJ2 Enhances Osteoblast Differentiation and Mineralization by Increasing the OPG/RANKL Ratio. Microorganisms. 2021;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Chou YC, Ho PY, Chen WJ, Wu SH, Pan MH. Lactobacillus fermentum V3 ameliorates colitis‐associated tumorigenesis by modulating the gut microbiome. Am J Cancer Res. 2020;10(4):1170‐81. [PMC free article] [PubMed] [Google Scholar]
- 121. Wu H, Xie S, Miao J, Li Y, Wang Z, Wang M, et al. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut Microbes. 2020;11(4):997‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cervantes‐Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8αα(+) T cells. Science. 2017;357(6353):806‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sivan A, Corrales L, Hubert N, Williams JB, Aquino‐Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti‐PD‐L1 efficacy. Science. 2015;350(6264):1084‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Schroeder BO, Birchenough GMH, Ståhlman M, Arike L, Johansson MEV, Hansson GC, et al. Bifidobacteria or Fiber Protects against Diet‐Induced Microbiota‐Mediated Colonic Mucus Deterioration. Cell Host Microbe. 2018;23(1):27‐40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Verma R, Lee C, Jeun EJ, Yi J, Kim KS, Ghosh A, et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci Immunol. 2018;3(28). [DOI] [PubMed] [Google Scholar]
- 126. Zagato E, Pozzi C, Bertocchi A, Schioppa T, Saccheri F, Guglietta S, et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat Microbiol. 2020;5(3):511‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Li Q, Hu W, Liu WX, Zhao LY, Huang D, Liu XD, et al. Streptococcus thermophilus Inhibits Colorectal Tumorigenesis Through Secreting β‐Galactosidase. Gastroenterology. 2021;160(4):1179‐1193. [DOI] [PubMed] [Google Scholar]
- 128. Konings EJ, Roomans HH, Dorant E, Goldbohm RA, Saris WH, van den Brandt PA. Folate intake of the Dutch population according to newly established liquid chromatography data for foods. Am J Clin Nutr. 2001;73(4):765‐76. [DOI] [PubMed] [Google Scholar]
- 129. Wasilewska E, Zlotkowska D, Wroblewska B. Yogurt starter cultures of Streptococcus thermophilus and Lactobacillus bulgaricus ameliorate symptoms and modulate the immune response in a mouse model of dextran sulfate sodium‐induced colitis. J Dairy Sci. 2019;102(1):37‐53. [DOI] [PubMed] [Google Scholar]
- 130. Chen F, Stappenbeck TS. Microbiome control of innate reactivity. Curr Opin Immunol. 2019;56:107‐13. [DOI] [PubMed] [Google Scholar]
- 131. Jain T, Sharma P, Are AC, Vickers SM, Dudeja V. New Insights Into the Cancer‐Microbiome‐Immune Axis: Decrypting a Decade of Discoveries. Front Immunol. 2021;12:622064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti‐cancer immunity. Nature. 2019;565(7741):600‐5. [DOI] [PubMed] [Google Scholar]
- 133. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature. 2013;504(7480):451‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Xing C, Wang M, Ajibade AA, Tan P, Fu C, Chen L, et al. Microbiota regulate innate immune signaling and protective immunity against cancer. Cell Host Microbe. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom‐Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159‐66. [DOI] [PubMed] [Google Scholar]
- 136. Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr. 2004;134(2):479‐82. [DOI] [PubMed] [Google Scholar]
- 137. Schulthess J, Pandey S, Capitani M, Rue‐Albrecht KC, Arnold I, Franchini F, et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity. 2019;50(2):432‐45.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. van den Berg FF, van Dalen D, Hyoju SK, van Santvoort HC, Besselink MG, Wiersinga WJ, et al. Western‐type diet influences mortality from necrotising pancreatitis and demonstrates a central role for butyrate. Gut. 2021;70(5):915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zierer J, Jackson MA, Kastenmüller G, Mangino M, Long T, Telenti A, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. 2018;50(6):790‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Rajpoot M, Sharma AK, Sharma A, Gupta GK. Understanding the microbiome: Emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin Cancer Biol. 2018;52(Pt 1):1‐8. [DOI] [PubMed] [Google Scholar]
- 141. Hamada T, Nowak JA, Milner DA, Jr. , Song M, Ogino S. Integration of microbiology, molecular pathology, and epidemiology: a new paradigm to explore the pathogenesis of microbiome‐driven neoplasms. J Pathol. 2019;247(5):615‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15(3):317‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690‐704. [DOI] [PubMed] [Google Scholar]
- 144. Yang J, Yu J. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell. 2018;9(5):474‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma‐carcinoma sequence. Nat Commun. 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 146. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host Microbe. 2013;14(2):207‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zackular JP, Rogers MA, MTt Ruffin, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7(11):1112‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res. 2017;23(8):2061‐70. [DOI] [PubMed] [Google Scholar]
- 149. Peek RM, Jr. , Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208(2):233‐48. [DOI] [PubMed] [Google Scholar]
- 150. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67(6):1024‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, et al. Increased Abundance of Clostridium and Fusobacterium in Gastric Microbiota of Patients with Gastric Cancer in Taiwan. Sci Rep. 2018;8(1):158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Di Pilato V, Freschi G, Ringressi MN, Pallecchi L, Rossolini GM, Bechi P. The esophageal microbiota in health and disease. Ann N Y Acad Sci. 2016;1381(1):21‐33. [DOI] [PubMed] [Google Scholar]
- 153. Baba Y, Iwatsuki M, Yoshida N, Watanabe M, Baba H. Review of the gut microbiome and esophageal cancer: Pathogenesis and potential clinical implications. Ann Gastroenterol Surg. 2017;1(2):99‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Nardone G, Compare D, Rocco A. A microbiota‐centric view of diseases of the upper gastrointestinal tract. Lancet Gastroenterol Hepatol. 2017;2(4):298‐312. [DOI] [PubMed] [Google Scholar]
- 155. Yang W, Chen C‐H, Jia M, Xing X, Gao L, Tsai H‐T, et al. Tumor‐Associated Microbiota in Esophageal Squamous Cell Carcinoma. Frontiers in Cell and Developmental Biology. 2021;9:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Chen X, Yuan Z, Lu M, Zhang Y, Jin L, Ye W. Poor oral health is associated with an increased risk of esophageal squamous cell carcinoma ‐ a population‐based case‐control study in China. Int J Cancer. 2017;140(3):626‐35. [DOI] [PubMed] [Google Scholar]
- 157. Yamamura K, Izumi D, Kandimalla R, Sonohara F, Baba Y, Yoshida N, et al. Intratumoral Fusobacterium Nucleatum Levels Predict Therapeutic Response to Neoadjuvant Chemotherapy in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2019;25(20):6170‐6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Malinowski B, Węsierska A, Zalewska K, Sokołowska MM, Bursiewicz W, Socha M, et al. The role of Tannerella forsythia and Porphyromonas gingivalis in pathogenesis of esophageal cancer. Infect Agent Cancer. 2019;14:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Tan W, Liao TH, Wang J, Ye Y, Wei YC, Zhou HK, et al. A recently evolved diflavin‐containing monomeric nitrate reductase is responsible for highly efficient bacterial nitrate assimilation. J Biol Chem. 2020;295(15):5051‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wang Z, Gao X, Zeng R, Wu Q, Sun H, Wu W, et al. Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis. Front Microbiol. 2020;11:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yang L, Francois F, Pei Z. Molecular pathways: pathogenesis and clinical implications of microbiome alteration in esophagitis and Barrett esophagus. Clin Cancer Res. 2012;18(8):2138‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Zhang G, Meredith TC, Kahne D. On the essentiality of lipopolysaccharide to Gram‐negative bacteria. Curr Opin Microbiol. 2013;16(6):779‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018;8(4):403‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population‐based nested case‐control study. Gut. 2018;67(1):120‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell. 2019;178(4):795‐806.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. McAllister F, Khan MAW, Helmink B, Wargo JA. The Tumor Microbiome in Pancreatic Cancer: Bacteria and Beyond. Cancer Cell. 2019;36(6):577‐9. [DOI] [PubMed] [Google Scholar]
- 167. Kiss B, Mikó E, Sebő É, Toth J, Ujlaki G, Szabó J, et al. Oncobiosis and Microbial Metabolite Signaling in Pancreatic Adenocarcinoma. Cancers (Basel). 2020;12(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ingman WV. The Gut Microbiome: A New Player in Breast Cancer Metastasis. Cancer Res. 2019;79(14):3539‐41. [DOI] [PubMed] [Google Scholar]
- 171. Chen J, Douglass J, Prasath V, Neace M, Atrchian S, Manjili MH, et al. The microbiome and breast cancer: a review. Breast Cancer Res Treat. 2019;178(3):493‐6. [DOI] [PubMed] [Google Scholar]
- 172. Buchta Rosean C, Bostic RR, Ferey JCM, Feng TY, Azar FN, Tung KS, et al. Preexisting Commensal Dysbiosis Is a Host‐Intrinsic Regulator of Tissue Inflammation and Tumor Cell Dissemination in Hormone Receptor‐Positive Breast Cancer. Cancer Res. 2019;79(14):3662‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zhu J, Liao M, Yao Z, Liang W, Li Q, Liu J, et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome. 2018;6(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Puglisi R, Bellenghi M, Pontecorvi G, Pallante G, Carè A, Mattia G. Biomarkers for Diagnosis, Prognosis and Response to Immunotherapy in Melanoma. Cancers (Basel). 2021;13(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Limeta A, Ji B, Levin M, Gatto F, Nielsen J. Meta‐analysis of the gut microbiota in predicting response to cancer immunotherapy in metastatic melanoma. JCI Insight. 2020;5(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Tomela K, Pietrzak B, Schmidt M, Mackiewicz A. The Tumor and Host Immune Signature, and the Gut Microbiota as Predictive Biomarkers for Immune Checkpoint Inhibitor Response in Melanoma Patients. Life (Basel). 2020;10(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Zhou A, Tang L, Zeng S, Lei Y, Yang S, Tang B. Gut microbiota: A new piece in understanding hepatocarcinogenesis. Cancer Lett. 2020;474:15‐22. [DOI] [PubMed] [Google Scholar]
- 178. Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14(9):527‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59‐64. [DOI] [PubMed] [Google Scholar]
- 180. Zhang Q, Ma C, Duan Y, Heinrich B, Rosato U, Diggs LP, et al. Gut microbiome directs hepatocytes to recruit MDSC and promote cholangiocarcinoma. Cancer Discov. 2021;11(5):1248–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Kolenbrander PE, Palmer RJ, Jr. , Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell‐cell distance. Nat Rev Microbiol. 2010;8(7):471‐80. [DOI] [PubMed] [Google Scholar]
- 182. Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16(12):745‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9(8):1997‐2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Yilmaz O, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3‐kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72(7):3743‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K, et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. 2014;15(6):768‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Liu S, Zhou X, Peng X, Li M, Ren B, Cheng G, et al. Porphyromonas gingivalis Promotes Immunoevasion of Oral Cancer by Protecting Cancer from Macrophage Attack. J Immunol. 2020;205(1):282‐9. [DOI] [PubMed] [Google Scholar]
- 187. Chen Z, Wong PY, Ng CWK, Lan L, Fung S, Li JW, et al. The Intersection between Oral Microbiota, Host Gene Methylation and Patient Outcomes in Head and Neck Squamous Cell Carcinoma. Cancers (Basel). 2020;12(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Swidsinski A, Dörffel Y, Loening‐Baucke V, Theissig F, Rückert JC, Ismail M, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60(1):34‐40. [DOI] [PubMed] [Google Scholar]
- 189. Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, et al. Invasive potential of gut mucosa‐derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17(9):1971‐8. [DOI] [PubMed] [Google Scholar]
- 190. Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15(1):30‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Han YW, Houcken W, Loos BG, Schenkein HA, Tezal M. Periodontal disease, atherosclerosis, adverse pregnancy outcomes, and head‐and‐neck cancer. Adv Dent Res. 2014;26(1):47‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Ye C, Xia Z, Tang J, Khemwong T, Kapila Y, Kuraji R, et al. Unculturable and culturable periodontal‐related bacteria are associated with periodontal inflammation during pregnancy and with preterm low birth weight delivery. Sci Rep. 2020;10(1):15807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, et al. Porphyromonas gingivalis in Alzheimer's disease brains: Evidence for disease causation and treatment with small‐molecule inhibitors. Sci Adv. 2019;5(1):eaau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Dzutsev A, Badger JH, Perez‐Chanona E, Roy S, Salcedo R, Smith CK, et al. Microbes and Cancer. Annu Rev Immunol. 2017;35:199‐228. [DOI] [PubMed] [Google Scholar]
- 195. Li Y, Luo ZY, Hu YY, Bi YW, Yang JM, Zou WJ, et al. The gut microbiota regulates autism‐like behavior by mediating vitamin B(6) homeostasis in EphB6‐deficient mice. Microbiome. 2020;8(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome‐mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360(6391). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Patnode ML, Beller ZW, Han ND, Cheng J, Peters SL, Terrapon N, et al. Interspecies Competition Impacts Targeted Manipulation of Human Gut Bacteria by Fiber‐Derived Glycans. Cell. 2019;179(1):59‐73.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Bergstrom K, Shan X, Casero D, Batushansky A, Lagishetty V, Jacobs JP, et al. Proximal colon‐derived O‐glycosylated mucus encapsulates and modulates the microbiota. Science. 2020;370(6515):467‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. O'Donnell MP, Fox BW, Chao PH, Schroeder FC, Sengupta P. A neurotransmitter produced by gut bacteria modulates host sensory behaviour. Nature. 2020;583(7816):415‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Gil‐Cruz C, Perez‐Shibayama C, De Martin A, Ronchi F, van der Borght K, Niederer R, et al. Microbiota‐derived peptide mimics drive lethal inflammatory cardiomyopathy. Science. 2019;366(6467):881‐6. [DOI] [PubMed] [Google Scholar]
- 201. Vicente‐Dueñas C, Janssen S, Oldenburg M, Auer F, González‐Herrero I, Casado‐García A, et al. An intact gut microbiome protects genetically predisposed mice against leukemia. Blood. 2020;136(18):2003‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M, et al. Regulation of life span by the gut microbiota in the short‐lived African turquoise killifish. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167(7):1897. [DOI] [PubMed] [Google Scholar]
- 204. Guedj A, Volman Y, Geiger‐Maor A, Bolik J, Schumacher N, Künzel S, et al. Gut microbiota shape ‘inflamm‐ageing’ cytokines and account for age‐dependent decline in DNA damage repair. Gut. 2020;69(6):1064‐1075. [DOI] [PubMed] [Google Scholar]
- 205. Parkar SG, Kalsbeek A, Cheeseman JF. Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health. Microorganisms. 2019;7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science. 2019;365(6460):1428‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Wong‐Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. 2021;12(5):426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362(6416):776‐80. [DOI] [PubMed] [Google Scholar]
- 209. Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14(6):356‐65. [DOI] [PubMed] [Google Scholar]
- 210. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17(5):271‐85. [DOI] [PubMed] [Google Scholar]
- 211. Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin Cancer Res. 2016;22(22):5574‐81. [DOI] [PubMed] [Google Scholar]
- 212. Kovatcheva‐Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary Fiber‐Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015;22(6):971‐82. [DOI] [PubMed] [Google Scholar]
- 213. Chou PC, Choi HH, Huang Y, Fuentes‐Mattei E, Velazquez‐Torres G, Zhang F, et al. Impact of diabetes on promoting the growth of breast cancer. Cancer Commun (Lond). 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Yen I, Shanahan F, Lee J, Hong YS, Shin SJ, Moore AR, et al. ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma. Nature. 2021. [DOI] [PubMed] [Google Scholar]
- 215. Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325(5947):1555‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Geller LT, Barzily‐Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Heshiki Y, Vazquez‐Uribe R, Li J, Ni Y, Quainoo S, Imamovic L, et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome. 2020;8(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Chitapanarux I, Chitapanarux T, Traisathit P, Kudumpee S, Tharavichitkul E, Lorvidhaya V. Randomized controlled trial of live lactobacillus acidophilus plus bifidobacterium bifidum in prophylaxis of diarrhea during radiotherapy in cervical cancer patients. Radiat Oncol. 2010;5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Quigley EMM. Prebiotics and Probiotics in Digestive Health. Clin Gastroenterol Hepatol. 2019;17(2):333‐44. [DOI] [PubMed] [Google Scholar]
- 220. Eslami M, Yousefi B, Kokhaei P, Hemati M, Nejad ZR, Arabkari V, et al. Importance of probiotics in the prevention and treatment of colorectal cancer. J Cell Physiol. 2019;234(10):17127‐43. [DOI] [PubMed] [Google Scholar]
- 221. Marinelli L, Tenore GC, Novellino E. Probiotic species in the modulation of the anticancer immune response. Semin Cancer Biol. 2017;46:182‐90. [DOI] [PubMed] [Google Scholar]
- 222. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Ma W, Mao Q, Xia W, Dong G, Yu C, Jiang F. Gut Microbiota Shapes the Efficiency of Cancer Therapy. Front Microbiol. 2019;10:1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID‐19. Gut. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, et al. Antibiotics‐Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell. 2019;178(6):1313‐28.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science. 2018;359(6371):91‐7. [DOI] [PubMed] [Google Scholar]
- 227. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti‐PD‐1 immunotherapy in melanoma patients. Science. 2018;359(6371):97‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti‐PD‐1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Elkrief A, Derosa L, Zitvogel L, Kroemer G, Routy B. The intimate relationship between gut microbiota and cancer immunotherapy. Gut Microbes. 2019;10(3):424‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Cogdill AP, Gaudreau PO, Arora R, Gopalakrishnan V, Wargo JA. The Impact of Intratumoral and Gastrointestinal Microbiota on Systemic Cancer Therapy. Trends Immunol. 2018;39(11):900‐20. [DOI] [PubMed] [Google Scholar]
- 231. McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019;20(2):e77‐e91. [DOI] [PubMed] [Google Scholar]
- 232. Nomura M, Nagatomo R, Doi K, Shimizu J, Baba K, Saito T, et al. Association of Short‐Chain Fatty Acids in the Gut Microbiome With Clinical Response to Treatment With Nivolumab or Pembrolizumab in Patients With Solid Cancer Tumors. JAMA Netw Open. 2020;3(4):e202895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Azizian K, Pustokhina I, Ghanavati R, Hamblin MR, Amini A, Kouhsari E. The potential use of theranostic bacteria in cancer. J Cell Physiol. 2021;236(6):4184–4194. [DOI] [PubMed] [Google Scholar]
- 234. Longhi G, van Sinderen D, Ventura M, Turroni F. Microbiota and Cancer: The Emerging Beneficial Role of Bifidobacteria in Cancer Immunotherapy. Front Microbiol. 2020;11:575072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Todoric J, Antonucci L, Karin M. Targeting Inflammation in Cancer Prevention and Therapy. Cancer Prev Res (Phila). 2016;9(12):895‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, et al. Inflammation and colorectal cancer, when microbiota‐host mutualism breaks. World J Gastroenterol. 2014;20(4):908‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700‐year‐old fecal microbiota transplantation? Am J Gastroenterol. 2012;107(11):1755; author reply p.‐6. [DOI] [PubMed] [Google Scholar]
- 240. Eiseman B, Silen W, Bascom GS, Kauvar AJ. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958;44(5):854‐9. [PubMed] [Google Scholar]
- 241. Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25(3):377‐88. [DOI] [PubMed] [Google Scholar]
- 242. Vivarelli S, Salemi R, Candido S, Falzone L, Santagati M, Stefani S, et al. Gut Microbiota and Cancer: From Pathogenesis to Therapy. Cancers (Basel). 2019;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Ooijevaar RE, Terveer EM, Verspaget HW, Kuijper EJ, Keller JJ. Clinical Application and Potential of Fecal Microbiota Transplantation. Annu Rev Med. 2019;70:335‐51. [DOI] [PubMed] [Google Scholar]
- 244. Wang Z, Hua W, Li C, Chang H, Liu R, Ni Y, et al. Protective Role of Fecal Microbiota Transplantation on Colitis and Colitis‐Associated Colon Cancer in Mice Is Associated With Treg Cells. Front Microbiol. 2019;10:2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, et al. Effect of Fecal Microbiota Transplantation on 8‐Week Remission in Patients With Ulcerative Colitis: A Randomized Clinical Trial. Jama. 2019;321(2):156‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Vuong HE, Yano JM, Fung TC, Hsiao EY. The Microbiome and Host Behavior. Annu Rev Neurosci. 2017;40:21‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open‐label study. Microbiome. 2017;5(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. D'Amato A, Di Cesare Mannelli L, Lucarini E, Man AL, Le Gall G, Branca JJV, et al. Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity‐ and neurotransmission‐related proteins in young recipients. Microbiome. 2020;8(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Korpela K, Helve O, Kolho KL, Saisto T, Skogberg K, Dikareva E, et al. Maternal Fecal Microbiota Transplantation in Cesarean‐Born Infants Rapidly Restores Normal Gut Microbial Development: A Proof‐of‐Concept Study. Cell. 2020;183(2):324‐34.e5. [DOI] [PubMed] [Google Scholar]
- 250. Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology. 2013;145(5):946‐53. [DOI] [PubMed] [Google Scholar]
- 251. Aron‐Wisnewsky J, Clément K, Nieuwdorp M. Fecal Microbiota Transplantation: a Future Therapeutic Option for Obesity/Diabetes? Curr Diab Rep. 2019;19(8):51. [DOI] [PubMed] [Google Scholar]
- 252. De Palma G, Lynch MD, Lu J, Dang VT, Deng Y, Jury J, et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci Transl Med. 2017;9(379). [DOI] [PubMed] [Google Scholar]
- 253. Borody TJ, Eslick GD, Clancy RL. Fecal microbiota transplantation as a new therapy: from Clostridioides difficile infection to inflammatory bowel disease, irritable bowel syndrome, and colon cancer. Curr Opin Pharmacol. 2019;49:43‐51. [DOI] [PubMed] [Google Scholar]
- 254. Khoruts A, Staley C, Sadowsky MJ. Faecal microbiota transplantation for Clostridioides difficile: mechanisms and pharmacology. Nat Rev Gastroenterol Hepatol. 2020. [DOI] [PubMed] [Google Scholar]
- 255. DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, et al. Drug‐Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N Engl J Med. 2019;381(21):2043‐2050. [DOI] [PubMed] [Google Scholar]
- 256. Christofi T, Baritaki S, Falzone L, Libra M, Zaravinos A. Current Perspectives in Cancer Immunotherapy. Cancers (Basel). 2019;11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Choudhry H. The Microbiome and Its Implications in Cancer Immunotherapy. Molecules. 2021;26(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382‐96. [DOI] [PubMed] [Google Scholar]
- 259. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor‐associated colitis. Nat Med. 2018;24(12):1804‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Baruch EN, Youngster I, Ben‐Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy‐refractory melanoma patients. Science. 2021;371(6529):602–609. [DOI] [PubMed] [Google Scholar]
- 261. McQuade JL, Ologun GO, Arora R, Wargo JA. Gut Microbiome Modulation Via Fecal Microbiota Transplant to Augment Immunotherapy in Patients with Melanoma or Other Cancers. Curr Oncol Rep. 2020;22(7):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti‐PD‐1 therapy in melanoma patients. Science. 2021;371(6529):595‐602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Łaniewski P, Ilhan ZE, Herbst‐Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol. 2020;17(4):232‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Han Y, Liu Z, Chen T. Role of Vaginal Microbiota Dysbiosis in Gynecological Diseases and the Potential Interventions. Front Microbiol. 2021;12:643422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Borella F, Carosso AR, Cosma S, Preti M, Collemi G, Cassoni P, et al. Gut Microbiota and Gynecological Cancers: A Summary of Pathogenetic Mechanisms and Future Directions. ACS Infect Dis. 2021;7(5):987‐1009. [DOI] [PubMed] [Google Scholar]
- 266. Sims TT, El Alam MB, Karpinets TV, Dorta‐Estremera S, Hegde VL, Nookala S, et al. Gut microbiome diversity is an independent predictor of survival in cervical cancer patients receiving chemoradiation. Commun Biol. 2021;4(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Lev‐Sagie A, Goldman‐Wohl D, Cohen Y, Dori‐Bachash M, Leshem A, Mor U, et al. Vaginal microbiome transplantation in women with intractable bacterial vaginosis. Nat Med. 2019;25(10):1500‐4. [DOI] [PubMed] [Google Scholar]
- 268. DeLong K, Bensouda S, Zulfiqar F, Zierden HC, Hoang TM, Abraham AG, et al. Conceptual Design of a Universal Donor Screening Approach for Vaginal Microbiota Transplant. Front Cell Infect Microbiol. 2019;9:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, et al. Effect of Oral Capsule‐ vs Colonoscopy‐Delivered Fecal Microbiota Transplantation on Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. Jama. 2017;318(20):1985‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Kelly CR, Yen EF, Grinspan AM, Kahn SA, Atreja A, Lewis JD, et al. Fecal Microbiota Transplantation Is Highly Effective in Real‐World Practice: Initial Results From the FMT National Registry. Gastroenterology. 2021;160(1):183‐92.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Dronkers TMG, Ouwehand AC, Rijkers GT. Global analysis of clinical trials with probiotics. Heliyon. 2020;6(7):e04467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Dedrick RM, Guerrero‐Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug‐resistant Mycobacterium abscessus. Nat Med. 2019;25(5):730‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


