Abstract
In Saccharomyces cerevisiae, RAD1 and RAD52 are required for alternate pathways of mitotic recombination. Double-mutant strains exhibit a synergistic interaction that decreases direct repeat recombination rates dramatically. A mutation in RFA1, the largest subunit of a single-stranded DNA-binding protein complex (RP-A), suppresses the recombination deficiency of rad1 rad52 strains (J. Smith and R. Rothstein, Mol. Cell. Biol. 15:1632–1641, 1995). Previously, we hypothesized that this mutation, rfa1-D228Y, causes an increase in recombinogenic lesions as well as the activation of a RAD52-independent recombination pathway. To identify gene(s) acting in this pathway, temperature-sensitive (ts) mutations were screened for those that decrease recombination levels in a rad1 rad52 rfa1-D228Y strain. Three mutants were isolated. Each segregates as a single recessive gene. Two are allelic to RSP5, which encodes an essential ubiquitin-protein ligase. One allele, rsp5-25, contains two mutations within its open reading frame. The first mutation does not alter the amino acid sequence of Rsp5, but it decreases the amount of full-length protein in vivo. The second mutation results in the substitution of a tryptophan with a leucine residue in the ubiquitination domain. In rsp5-25 mutants, the UV sensitivity of rfa1-D228Y is suppressed to the same level as in strains overexpressing Rfa1-D228Y. Measurement of the relative rate of protein turnover demonstrated that the half-life of Rfa1-D228Y in rsp5-25 mutants was extended to 65 min compared to a 35-min half-life in wild-type strains. We propose that Rsp5 is involved in the degradation of Rfa1 linking ubiquitination with the replication-recombination machinery.
RAD1 and RAD52 are involved in different pathways of mitotic recombination in yeast (19, 39, 56). Although loss of function of neither RAD1 nor RAD52 alone has a significant effect on most mitotic direct repeat recombination assays, rad1 rad52 double mutants exhibit a synergistic interaction that decreases the recombination rates dramatically (19, 39, 56). This result has led to the hypothesis that RAD1 and RAD52 act in two alternate recombination pathways.
Strains that are mutated for RAD1 are highly sensitive to UV light and are completely defective in the incision step of excision repair of damaged DNA (12, 35). In vivo Rad1 and Rad10 form a stable complex that exhibits a single-stranded DNA endonuclease activity (57). The function of this complex is to remove the 3′ nonhomologous regions of single-strand DNA that interfere with annealing and/or strand invasion during mitotic recombination. Consequently, rad1 cells cannot efficiently complete recombination when the ends of the double-strand break contain approximately 60 bp of nonhomology (9). Efficient recombination is restored when the ends of the break are homologous to the donor sequences or less than 40 bp (30).
RAD52 was identified as a mutation that confers extreme sensitivity to gamma irradiation and to methyl methanesulfonate (33). Genetic analysis has revealed that wild-type RAD52 function is required to repair double-strand breaks (23, 29, 34). More importantly, Rad52 is a conserved DNA binding protein and promotes DNA strand annealing (26, 52). It interacts physically with Rad51, a RecA homolog, that has been shown to catalyze strand exchange (25, 42, 51, 53). Recently, it was demonstrated that Rad52 stimulates Rad51-dependent strand exchange reactions and that the binding of Rad52 to Rad51 is necessary for this stimulatory effect (3, 27, 43).
A classical genetic approach was taken to study rad1 rad52 double mutants (7). A suppressor mutation, rfa1-D228Y, was identified that restores wild-type levels of direct repeat recombination in rad1 rad52 strains (45). The wild-type RFA1 gene encodes the largest component of an essential three-subunit complex, replication factor A (4). Its human homolog is required for the initiation and elongation steps of in vitro simian virus 40 replication, as well as for excision repair (1, 5, 8). The rfa1-D228Y allele on its own causes a 15-fold increase in direct repeat recombination levels. This hyper-recombination phenotype is RAD52 independent and only partially dependent on the RAD1 gene product. In addition, the mutant strains display increased UV sensitivity and slow growth. Overexpression of the mutant protein in rfa1-D228Y strains results in partial suppression of the recombination defect and complete suppression of the UV sensitivity phenotype. Interestingly, the amount of RP-A complex present in extracts from rfa1-D228Y cells is reduced twofold compared to the amount present in wild-type cells. Taken together, these results suggest that the mutant RP-A complex is unstable and that this instability can be compensated for by overexpression of mutant Rfa1 (45).
To define further the rfa1-D228Y-dependent recombination pathway, rad1 rad52 rfa1-D228Y triple-mutant strains were screened for new mutations that decrease recombination. Here, we report that three temperature-sensitive (ts) mutations were isolated. Cloning and sequencing analyses showed that two (rsp5-25 and rsp5-26) are allelic to RSP5, a ubiquitin-protein ligase. RSP5, repressor of spt3 phenotype, was first isolated as a suppressor of Spt3, a subunit of the SAGA complex (36). Subsequently, rsp5 mutations have been observed to affect many diverse cellular processes. These include stability of both the largest subunit of RNA polymerase II, Rpb1, and the uracil permease, Fur4 (11, 18). Also, it was shown that both the localization of Mod5 (a tRNA modifier) and the mitogen-activated protein kinase cascade are affected by rsp5 mutations (59, 62). One of the alleles described here, rsp5-25, contains two changes in its open reading frame (ORF). The first changes a tyrosine codon to an ochre codon, resulting in the truncation of the full-length protein. In the genetic background used for the screen, the ochre mutation is partially suppressed by the tyrosine-inserting tRNA suppressor, SUP4-o. This mutation does not create a change in the amino acid sequence of Rsp5; however, it causes a decrease in the level of full-length protein in vivo. The second mutation results in the substitution of a tryptophan residue with a leucine residue in the ubiquitination domain. Biochemical and genetic studies show that Rfa1-D228Y is stabilized in rsp5-25 mutant strains. Thus, the increased amount of Rfa1-D228Y protein causes a decrease in the recombination levels of rad1 rad52 rfa1-D228Y strains. We hypothesize that Rsp5 is involved in the degradation of Rfa1, thereby linking ubiquitin-dependent protein degradation to the replication-recombination machinery.
MATERIALS AND METHODS
Media, strains, and genetic methods.
All media were prepared as described previously (24, 45). Standard genetic techniques were employed (41). All yeast strains are derivatives of a RAD5 W303-1B (46, 55) unless otherwise noted and are listed in Table 1. W303 derivatives containing the SUP4 duplication, leu2 direct repeats, and rad1::HIS5, rad52::HIS5, rfa1-D228Y, and can1-100,x mutations were described earlier (24, 45, 46). A modified version of the SUP4 construct was used in this study (U928). The URA3 gene flanked by the SUP4 repeats was disrupted by the HIS3 gene. Lastly, cim3 cim5 strains (the 26S proteasome mutants) and their congenic wild-type strain were gifts from C. Mann.
TABLE 1.
Strains used in this study
| Straina | Genotype | Source |
|---|---|---|
| W1588-4A | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 | Rothstein lab |
| W1100-3B | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 SUP4-o::CAN1-URA3::sup4+ | Rothstein lab |
| W1088-1A | MATα ade2-1 can1-100 his5Δ leu2-3,112 trp1-1 ura3-1 | Rothstein lab |
| W1088-10D | MATα ade2-1 can1-100 his5Δ leu2-3,112 trp1-1 ura3-1 | Rothstein lab |
| W1698-11C | MATα ade2-1 can1-100,x his3-11,15 leu2-3, 112 trp1-1 ura3-1 RAD5 rad1::HIS5 rad52::HIS5 rfa1-D228Y SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| J709 | MATα ade2-1 can1-100,x his3-11,15 leu2-3, 112 trp1-1 ura3-1 RAD5 rad1::HIS5 rad52::HIS5 rfa1-D228Y rsp5-25 SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| J714 | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rad1::HIS5 rad52::HIS5 rfa1-D228Y rsp5-26 SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| W1713-1B | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rsp5-25 SUP4-o::CAN1-HIS3::sup4+ | This study |
| W1713-22D | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rad1::HIS5 rad52::HIS5 rfa1-D228Y rsp5-25 SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| U1025 | MATa/α ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rsp5Δ SUP4-o::CAN1-HIS3::sup4+ | This study |
| W1713-1A | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rfa1-D228Y SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| W2013-4B | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rsp5-Y647och SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| W2012-2D | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rsp5-W650L SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| W1713-22A | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rfa1-D228Y rsp5-25 SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| W2013-11C | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rfa1-D228Y rsp5-Y647ochSUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| W2012-15C | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rfa1-D228Y rsp5-W650L SUP4-o::CAN1-HIS3::sup4+ leu2ΔEcoRI::URA3::leu2BstEII | This study |
| W2015-1A | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rfa1Δ rsp5-25 SUP4-o::CAN1-HIS3::sup4+ pWJ709-GAL1-rfa1-D228Y | This study |
| W2015-4A | MATα ade2-1 can1-100,x his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 rfa1Δ SUP4-o::CAN1-HIS3::sup4+ pWJ709-GAL1-rfa1-D228Y | This study |
| W2016-1B | MATα ade2-1 can1-100,x his3-11, 15 leu2-3, 112 trp1-1 ura3-1 RAD5 rfa1Δ SUP4-o::CAN1-HIS3::sup4+ pWJ708-GAL1-RFA1 | This study |
| CMY763 | MATα cim3-1 ura3-52 leu2Δ1 | Carl Mann |
| CMY765 | MATα cim5-1 ura3-52 leu2Δ1 his3Δ-200 | Carl Mann |
| CMY826 | MATa ura3-52 leu2Δ1 his3Δ-200 trp1Δ63 lys2-801 ade2-101 bar1::HIS3 | Carl Mann |
All strains used are derivatives of W303-1B (55). The genotypes are identical to that of W303-1B except where noted. In most instances several strains with the same genotype (except the mating type) were used. However, only one strain is noted. In addition, can1-100,x is an allele of can1-100 in which an unidentified secondary mutation prevents suppression by SUP4-o.
The rfa1-D228Y mutation creates a novel AccI restriction site, which permits the segregation of this allele to be monitored by PCR analysis by using the RFA1-2A (5′-CAGAGCATCCAAATGAAACC-3′) and the RFA1-3B (5′-TTTGGATAATACCGAGGACG-3′) primers as described earlier (45). The presence of rad1::HIS5 and rad52::HIS5 alleles were identified by complementation test for their respective UV and X-ray sensitivity phenotypes.
In this study, a novel allele of RSP5 was isolated and named rsp5-25. The rsp5-25 allele was scored in two ways: the presence of the mutation was confirmed by crosses to a rad1 rad52 rfa1-D228Y strain and determining whether lower recombination frequency and a ts phenotype is observed in half of the rad1 rad52 rfa1-D228Y segregants. Later, we found that the rsp5-25 allele contains an additional MunI restriction enzyme site not present in the wild-type RSP5 sequence. Therefore, it is easily detected by colony PCR by using RSP5-G (5′-GAAGGTGTTGACGCAGAGGTG-3′) and RSP5-H (5′-CCGCAATACCACCGATCAATAG-3′) primers and a diagnostic MunI restriction enzyme digestion. To perform linkage analysis of RSP5 and mrr1, a 5.9-kb SalI fragment of the complementing library insert was cloned into an integrating vector, YIp5. The plasmid was linearized by BglII digestion, which cuts uniquely in the SalI fragment, and transformed into a wild-type strain (W1588-4C). As a result, the 5.9-kb region was duplicated and marked with the URA3 gene in the genome (U957).
Strains containing only the ochre mutation (rsp5-Y647och) or only the missense mutation (rsp5-W650L) of rsp5-25 were constructed by using a PCR-based allele replacement method as described previously (20). The primers used for the construction of rsp5-Y647och strain were Stop1 (5′-GGTAATAAGAAAGAATATGTCGAATTATAAACCCAATGGAGAATTGTTGATAGAGTTCAAGTTTTCCCAGTCACG-3′) and Stop2 (5′-TTGAACTCTATCAACAATTCTCCATTGGGTTTATAATTCGACATATTCTTTCTTATTACCGGATCCTCTAGAGTCG-3′). Similarly, the primers for creating the rsp5-W650L allele were WtoL1 (5′-AGAAAGAATATGTCGAATTATATACCCAATTGAGAATTGTTGATAGAGTTCAAGAACAATGTTTTCCCAGTCACG-3′) and WtoL2 (5′-AATGTTCTTGAACTCTATCAACAATTCTCAATTGGGTATATAATTCGACATATTCTTTCTGGATCCTCTAGAGTCG-3′). Each pair of 75-mer oligonucleotides were constructed so that the last 15 nucleotides of the oligonucleotides (italics) were used to amplify URA3 from pUC18-URA3. The remaining nucleotides of both primers are complementary to each other and contain the desired mutation (underlined letter) in the middle of 60 nucleotides of the RSP5 gene. After PCR amplification, a linear fragment was transformed into a diploid strain that contains the SUP4 construct. The Ura+ transformants were screened for the desired integration events by PCR with an internal URA3 primer (5′-CAACACTACATATGCG-3′) and the RSP5-H primer (5′-CCGCAATACCACCGATCAATAG-3′), which only give a product if the construct integrates at the RSP5 locus. Transformants integrated into the RSP5 locus for rsp5-Y647och and for rsp5-W650L mutations were identified (J717 and J718, respectively). After transformation with a plasmid that contains the wild-type RSP5 gene, these diploid strains were sporulated and dissected. The Ura+ segregants were plated on 5-fluoro-orotic acid medium to select for direct repeat recombinants that “pop-out” the URA3 gene. The rsp5-Y647och strains were verified by their inability to lose the SUP4-o allele, since the ochre mutation causes a truncation in an essential protein and requires SUP4-o for viability. The presence of the rsp5-W650L allele was identified by PCR analysis for a MunI restriction polymorphism as described above.
The RSP5 gene disruption was constructed in a diploid strain (W1588) by a PCR-based gene disruption method utilizing TRP1 as the selective marker (2). The following primers were used for disruption: RSP5-T1 (5′-ATGCCTTCAT CCATATCCGTCAAGTTAGTGGCTGCAGAGTCATTATATAAGAGGGA CGTATTTCCCAGTCACGACG-3′) and RSP5-T2 (5′-TCATTCTTGACCAAACCCTATGGTTTCTTCCACGGCCAATGTTAGCTTCTGTTTCATGCTGCAAGTGCACAAACAAT-3′). The italicized sequences are those that are required to amplify the TRP1 gene. Three Trp+ transformants were obtained after the transformation. Disruption of the RSP5 locus was confirmed by genomic blot analysis.
Mutagenesis.
A rad1 rad52 rfa1-D228Y strain with SUP4 and leu2 direct repeat constructs (W1698-11C) was mutagenized with 0.3% ethyl methanesulfonate to 10% survival as described previously (45). Next, 200,000 mutagenized colonies were grown at 23°C and were first screened for temperature sensitivity at 37°C. A total of 2,347 ts mutants were further tested for decreased recombination by replica plating onto canavanine-containing medium at 30°C. Recombination frequencies of mutants were measured by both SUP4 and leu2 direct repeats constructs as described previously (45, 46).
Plasmid constructions.
The plasmid pWJ611 containing RAD1, RAD52, and RFA1 used for complementation was created in three steps. First, a 2.5-kb PstI-HindIII fragment of RFA1 was end filled by Klenow polymerase, and BamHI linkers were added. This fragment was cloned into the BamHI site of pRS414 (44) to create pWJ609. A 3.3-kb SalI fragment that contains full-length RAD52 was cloned into the SalI site of pWJ609 to generate pWJ610. Lastly, a 5.9-kb Klenow end-filled SalI fragment of RAD1 was cloned into the SmaI site of pWJ610 to create pWJ611.
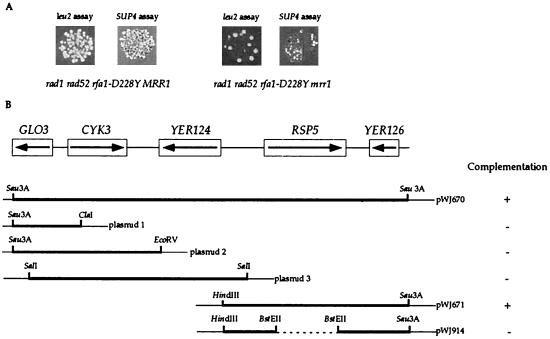
The rsp5-25 complementing clone (pWJ670) was isolated from a library of Sau3A-digested yeast genomic DNA fragments cloned into YCp50 (38) and was shown by DNA sequence and restriction digest analyses to contain five ORFs (GLO3, YCK3, YER124, RSP5, and YER126). For linkage analysis, a 5.9-kb SalI fragment containing part of GLO3, as well as full-length YCK3 and YER124, was inserted into the SalI site of an integrating vector, YIp5 (48). As shown in Fig. 2B, three ClaI-ClaI fragments of 1.3, 2.5, and 4.9 kb were deleted to remove YCK3, YER124, RSP5, and YER126 (plasmid 1). To delete YER124, RSP5, and YER126, pWJ670 was also digested with EcoRV and religated (plasmid 2). The 5.9-kb SalI fragment from pWJ670 was ligated into the corresponding site in YCp50 (plasmid 3). In pWJ671, a 6.0-kb HindIII-HindIII fragment was deleted from pWJ670 to remove GLO3, YCK3, and YER124. In pWJ914, a 2.0-kb BstEII-BstEII fragment is deleted from pWJ671 so that most of the RSP5 ORF is also removed.
FIG. 2.
Identification of mrr mutants and cloning of the mrr1 mutation. (A) Papillation phenotype of the mrr1 mutation in rad1 rad52 rfa1-D228Y after replica plating to the selective media for SUP4 and leu2 direct repeat recombination assays. (B) mrr1 is allelic to RSP5. The five ORFs present in pWJ670 are illustrated, as well as the five subclones constructed from this plasmid by using the indicated restriction enzymes. Each subclone was used to determine which of the five ORFs is MRR1. The Sau3A sites represent the junction of the genomic yeast sequences and the plasmid sequences. The thin lines depict the plasmid sequences. The plasmid, pWJ914, was derived from pWJ671 and carries a deletion of an internal BstEII fragment indicated as the dashed line. Each plasmid was tested for complementation of both the ts and the reduced recombination phenotypes in rad1 rad52 rfa1-D228Y mrr1 strains, and the results are shown adjacent to each plasmid.
pWJ708 and pWJ709 were constructed by cloning the wild-type RFA1 and rfa1-D228Y alleles, respectively, into pYX243 plasmid (kindly provided by Morten Dunø). The RFA1 alleles were amplified by PCR by using RFA1BH5 (5′-CGAGGATCCTATGAGCAGTGTTCAACTTTC-3′) and RFA1BH3 (5′-CGAGGATCCGCTAACAAAGCC-3′). BamHI sites were introduced by PCR primers to permit the in-frame fusion of hemagglutinin (HA) tags at the C termini of RFA1 and rfa1-D228Y ORFs to aid in detection of Rfa1 protein by the HA antibody during protein blot analysis.
Mapping and sequencing of rsp5-25.
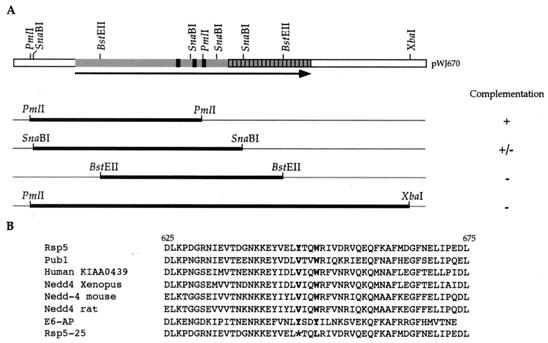
The mutations in the rsp5-25 allele were localized by a gap repair experiment (28). pWJ671 was digested with various combinations of the following enzymes: PmlI, SnaBI, BstEII, and XbaI to create several overlapping gaps in the RSP5 ORF (the thick lines in Fig. 3A). These linear plasmid fragments were transformed into W1713-1B, an rsp5-25 strain, and repaired circular plasmids were selected as Ura+ transformants. After rescue in Escherichia coli, several gap-repaired plasmids were retransformed into W1713-22D, a rad1 rad52 rfa1-D228Y rsp5-25 strain, to test for complementation. A 395-bp region between the SnaBI and BstEII sites in RSP5 was identified that contains the rsp5-25 mutation. Both mutant and wild-type plasmids were sequenced by using RSP5-G and RSP5-H primers.
FIG. 3.
Localization of the mutations in rsp5-25. (A) The rsp5-25 mutation was mapped by using a plasmid that contains the wild-type RSP5 allele, digested with indicated restriction enzymes for use in plasmid gap repair experiments. The gapped regions are indicated as thick lines. After transformation into an rsp5-25 strain, plasmid gap repair results in the copying of the corresponding genomic region onto the plasmid. The results of complementation experiments in rad1 rad52 rfa1-D228Y rsp5-25 are depicted. The black boxes on the RSP5 ORF depict the WW domains, and the hatched box represents the ubiquitination domain of the protein. (B) Localization of the two rsp5-25 mutations in the ubiquitination domain. The mutated tryptophan residue is indicated as a boldface letter, and the ochre codon is indicated by an asterisk. The protein sequences of several Rsp5 homologs are also shown to indicate the conservation near the mutated residues.
Analysis of UV sensitivity.
For any given genotype, UV sensitivity was determined by analyzing three segregants three times as described previously (45).
Analysis of degradation kinetics of Rfa1-D228Y and immunoprecipitations.
Both RSP5 (W2015-1A) and rsp5-25 (W2015-4A) strains with the plasmid that contains the rfa1-D228Y gene under the control of GAL1 promoter were grown to 1 × 107 to 2 × 107 cells/ml in galactose liquid medium lacking leucine. Next, the cells were washed and resuspended in rich liquid medium (YPD liquid). For the S-phase experiments, 100 mM hydroxyurea (HU) was added to an early-log-phase culture for 6 h at 30°C. Next, the cells were washed and resuspended in YPD liquid medium with 100 mM HU. For both HU-arrested and nonarrested cultures, samples were taken every 15 and 30 min, respectively, for 3 h at 30°C.
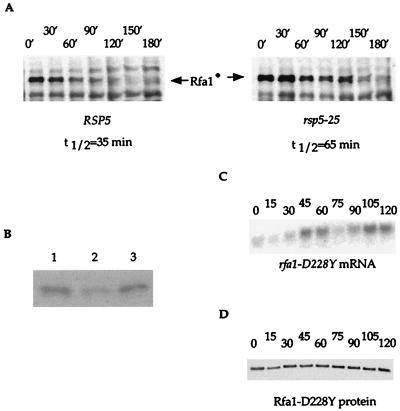
Total protein extract was prepared from each sample by using glass bead disruption as described earlier (14). Then, 15 μg of total protein (determined by using the Bio-Rad Protein Concentration Assay) were separated by electrophoresis with sodium dodecyl sulfate (SDS)–10% polyacrylamide gels (Bio-Rad). The extracts were transferred to polyvinylidene difluoride membranes, and protein blot analysis was performed as described previously (45). The resulting bands on Kodak X-Omat films were quantitated by using a computing densitometer (model 300A; Molecular Dynamics). The exponential decay of concentration with time exhibited first-order reaction kinetics and was used to calculate the relative half-lives. The relative half-life of each genotype was determined by averaging the results from 9 to 12 trials. One representative of each is shown in Fig. 6A. It is important to note that the observed decay is a function of protein half-life, as well as mRNA half-life.
FIG. 6.
Protein stability and mRNA levels of rfa1-D228Y. (A) A protein blot analysis of the stability of Rfa1-D228Y in the RSP5 and rsp5-25 mutant strains was performed. Rfa1-D228Y protein is tagged with HA and is indicated on the blots as Rfa1•. Other bands are due to background obtained with the HA antibody. The half-lives were calculated as described in Materials and Methods. (B) Protein levels of Rfa1-D228Y in rfa1-D228Y strains overexpressing rfa1-D228Y (lane 1) and rfa1-D228Y rsp5-25 strains (lane 3) were compared by a protein blot analysis. rfa1-D228Y strains (lane 2) were included as a control. (C and D) Measurement of mRNA and protein levels in rfa1-D228Y strains during the cell cycle. To measure mRNA and protein levels of rfa1-D228Y during the cell cycle, cells were released from alpha-factor arrest, samples were taken every 15 min, total mRNA was extracted, and total yeast protein extracts were prepared as described in Materials and Methods. (C) RNA blots were performed by using an internal fragment of RFA1 as a probe and URA3 as a loading control. The mRNA levels for URA3 were unchanged (data not shown). (D) Protein blot analysis was performed by using Rfa1 antibody. As a control, the same blot was reprobed with Rsp5 antibody. No variation in Rsp5 protein levels was detected (data not shown). After release from arrest, cells with small buds started to appear at 30 min and peaked at 45 min for the first cell cycle. The peak for cells with small buds for the second cell cycle occurs at 105 min.
The levels of Rsp5 protein present in different RSP5 backgrounds were determined by protein blot analysis as described above. The blots were incubated with monoclonal antibody raised against Rsp5, a kind gift from J. Huibregste.
cim3 and cim5 mutant strains that overexpress rfa1-D228Y under the GAL1 promoter were used for the pulse-chase experiments as described previously (13). Immunoprecipitations were performed as described previously (4). After separation of the immunoprecipitates on SDS–10% polyacrylamide gels, the gels were processed with the Entensify Kit (DuPont) according to the manufacturer's instructions.
RESULTS
Isolation of mutations that reduce the recombination levels of rad1 rad52 rfa1-D228Y strains.
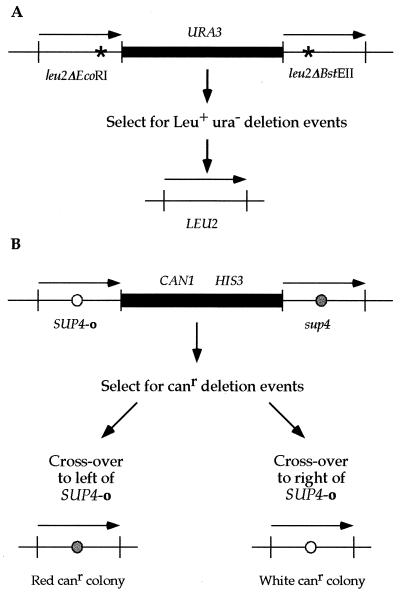
To identify mutations that decrease the recombination levels of rad1 rad52 rfa1-D228Y strains, two direct repeat recombination constructs were employed. In each assay, decreased recombination is indicated by fewer papillae in the selective medium. Each assay was chosen to utilize different metabolic pathways to avoid specifically mutations that affect a metabolic pathway. The first construct is a nontandem leu2 direct repeat consisting of two leu2 mutant alleles, leu2-ΔEcoRI and leu2-ΔBstEII, that are separated from each other by E. coli plasmid sequences and a yeast selectable marker, URA3 (Fig. 1A). Direct repeat recombination events that generate a single, wild-type LEU2 allele and delete the intervening sequences are selected first on medium lacking leucine and screened for uracil auxotrophy.
FIG. 1.
The direct repeat recombination constructs for leu2 and SUP4 are depicted. Both assays utilize a direct repeat of two alleles that are separated by plasmid and selectable marker sequences. Recombination events between the repeats that result in the retention of one allele and deletion of the intervening sequences can be selected. The leu2 and SUP4 direct repeats are 2.4 kb and are not drawn to scale. (A) In the leu2 assay (46), both alleles contain a frameshift mutation created by filling in a restriction enzyme site (EcoRI or BstEII). Leu+ recombinants are first selected for on medium lacking leucine and further identified as Ura− colonies after replica plating on medium lacking uracil. (B) The SUP4 assay (24) was modified by disrupting URA3 with HIS3 as described in Materials and Methods. The alleles differ by a single nucleotide change in the anticodon. Canr recombinants are selected on canavanine-containing medium. The strain contains the ochre suppressible ade2-1 allele, which enables colony color to designate which SUP4 allele is retained in the genome after direct repeat recombination: sup4 colonies are red, while SUP4-o colonies remain white. Deletions are confirmed by their failure to grow on histidine-less medium.
The second construct utilizes direct repeats of a yeast tyrosine tRNA gene where one copy is wild type (sup4) and the other copy is an ochre suppressor (SUP4-o), as depicted in Fig. 1B (24, 37). These two alleles differ at a single position in the anticodon. The repeats are separated from each other by plasmid sequences and two yeast selectable markers, CAN1 and HIS3. The CAN1 gene encodes the arginine permease, which allows the uptake of canavanine, an arginine analog and a cell poison (16). Cells that undergo a deletion event between the SUP4 alleles become Canr and His− and are selected on canavanine-containing medium.
For both the leu2 and the SUP4 constructs, several possible recombination mechanisms can be envisioned to lead to the retention of a single repeat. Such mechanisms include intrachromosomal recombination, unequal sister chromatid exchange, unequal sister chromatid gene conversion, replication slippage, and single-strand annealing (6, 21, 22, 24, 31, 47, 54). Neither of the two constructs allows these mechanisms to be distinguished from one another, and thus all recombination events are collectively referred to as plasmid loss. However, both constructs permit the visualization of recombination levels simply by replica plating yeast strains onto selective media. In rad1 rad52 rfa1-D228Y strains, typically 20 to 30 papillae are observed after the recombination event between the direct repeats (Fig. 2A). However, in rad1 rad52 strains, the number of papillae is lowered at least 10-fold (data not shown).
We mutagenized a rad1 rad52 rfa1-D228Y strain to 10% survival with ethyl methanesulfonate and screened for ts mutations among 200,000 colonies, generating a collection of 2,345 ts strains. Next, these ts strains were screened for their recombination phenotype at 30°C, a semipermissive temperature. A decreased number of papillae on selective medium with both recombination constructs reflects a decrease in recombination (Fig. 2A). Three mutants consistently displayed this phenotype and were provisionally named mrr (mutated for rfa1-stimulated recombination). None of the three exhibited a specific cell cycle arrest phenotype.
Each mutant was crossed to a wild-type strain, and random spore analyses were performed. For each mutation, half of the rad1 rad52 rfa1-D228Y segregants were both ts and recombination-deficient, indicating that a mutation in a single gene is likely responsible for both traits and that it is unlinked to RAD1, RAD52, or RFA1 (data not shown). Crosses to rad1 rad52 rfa1-D228Y MRR strains revealed that all three mutations are recessive for temperature sensitivity since the diploids are temperature resistant. Similarly, after the three mutant strains were crossed to each other, the resulting diploids were no longer ts, indicating that the mrr mutations define three recessive complementation groups. Each mutant displays a 5- to 10-fold reduction with the leu2 construct and at least 40-fold reduction with the SUP4 construct in the levels of recombination compared to the parental MRR strain.
To investigate the phenotype of mrr single mutants, a plasmid that complements the rad1, rad52, and rfa1-D228Y mutations, pWJ611, was transformed into each strain. After transformation, mrr2 strains remain ts, indicating that the mrr2 mutation on its own causes temperature sensitivity. In contrast, pWJ611-containing mrr1 and mrr3 strains are no longer ts. Results from genetic crosses indicated that mrr1 strains are ts only in the absence of a functional RAD52 gene. On the other hand, mrr3 is ts with any combination of the rad1, rad52, or rfa1-D228Y mutations. Additionally, mrr3 mutants are ts in a rad51 mutant background. We focused on characterizing mrr1 and mrr3 since these two mutations display conditional lethal interactions in conjunction with other recombination mutations.
mrr1 and mrr3 are alleles of RSP5, an essential ubiquitin-protein ligase.
A centromere-based yeast library (38) was transformed into a rad1 rad52 rfa1-D228Y mrr1 strain to identify a wild-type clone by complementation. Among 5,000 transformants screened for temperature resistance and subsequently for complementation of decreased recombination, one complementing clone was identified. To determine whether this clone encodes MRR1, we first demonstrated that sequences from the library clone were genetically linked to mrr1. Part of the insert was cloned into YIp5 as described in Materials and Methods and integrated into a wild-type strain by homologous recombination to mark the genomic integration site with URA3. This Ura+ strain was crossed to a rad1 rad52 rfa1-D228Y mrr1 strain. The resulting diploid was sporulated and dissected. Eight rad1 rad52 rfa1-D228Y Ura− segregants were tested, and each displayed a ts and a low recombination phenotype, indicating the presence of mrr1. In contrast, eight Ura+ rad1 rad52 rfa1-D228Y segregants were temperature resistant and had high levels of recombination. This demonstrates that the complementing clone is genetically linked to the wild-type MRR1 gene.
The complementing clone contains five ORFs; therefore, a series of subclones were constructed to determine which ORF corresponds to MRR1. Only constructs containing the full-length RSP5 gene complement both the ts and recombination phenotype of rad1 rad52 rfa1-D228Y mrr1 strains (pWJ670 and pWJ671 in Fig. 2B). Next, we showed that disruption of the plasmid-based RSP5 gene eliminated complementation, confirming that mrr1 is an allele of RSP5 (pWJ914 in Fig. 2B). Hereafter, the mrr1 mutation is referred to as rsp5-25.
Similarly, mrr3 was cloned by complementing its recessive ts phenotype. One of 10,000 transformants displayed temperature resistance and exhibited the recombination levels of rad1 rad52 rfa1-D228Y strains. DNA sequence analysis revealed that the complementing plasmid also contained the RSP5 ORF and surrounding region. By using the same strategy described for the mrr1 above, mrr3 was shown to be an allele of RSP5 and was renamed rsp5-26. Thus, mrr1 and mrr3 are alleles of the same gene and display intragenic complementation. This is consistent with Rsp5 functioning as a multimer (61). Since both mrr mutations were allelic to each other and display a very similar phenotype, we only characterized the rsp5-25 allele in more detail.
The defect in rsp5-25 is due to two mutations localized in its ubiquitination domain.
Plasmid gap repair was used to localize the genetic alteration in the rsp5-25 mutant allele (28). By this analysis, a 395-bp region between the SnaBI and BstEII enzyme sites was shown to contain the mutation(s) (Fig. 3A). DNA sequence analysis of this region revealed two changes. The first, a T-to-A transversion at position 1941 in the RSP5 ORF, results in an ochre codon in place of a tyrosine codon (Fig. 3B) and truncates the protein from 803 to 647 amino acids. The second mutation is a T-to-G transversion at position 1950, resulting in a leucine-to-tryptophan substitution at residue 650 (Fig. 3B).
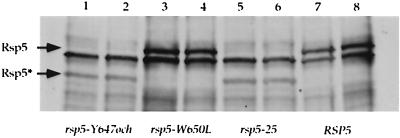
Genetic crosses reveal that, in the absence of SUP4-o, the rsp5-25 mutation is lethal (see Materials and Methods). Such a mutation could be isolated since the parental strain contains the SUP4-o suppressor, which recognizes the ochre codon to produce full-length Rsp5. However, since suppression is only partial, an approximately 10-fold decrease in the level of full-length Rsp5 protein is observed by protein blot analysis (Fig. 4). Interestingly, ochre suppression does not create an amino acid change, since both wild-type and SUP4-o-suppressed proteins contain a tyrosine residue in this position (32).
FIG. 4.
Protein blot analysis of different RSP5 alleles. Extracts from two rsp5-25, rsp5-Y647och, rsp5-W650L, and RSP5 strains were prepared, and the total amount of Rsp5 protein was detected by protein blot analysis by using Rsp5 antibody. The full-length Rsp5 proteins (92 kDa) is indicated as Rsp5. In rsp5-25 and rsp5-Y647och strains, an additional truncated form of Rsp5 (72 kDa) was observed and is indicated as Rsp5*. This is due to the ochre codon present at the position of residue 647. The SUP4-o suppressor also present in these strains recognizes the ochre codon to produce full-length Rsp5. However, since SUP4-o suppression is only partial, both full-length and truncated forms of Rsp5 are observed. The other bands are background obtained with the Rsp5 antibody.
The second mutation, the tryptophan-to-leucine substitution at position 650, is in the ubiquitination domain of Rsp5 (reference 17 and Fig. 3B). To establish whether the effect of rsp5-25 is due to the lowered amount of Rsp5 and/or the missense mutation, we separated the two mutations from each other by a PCR-based allele transfer method (20). The ochre mutation was named rsp5-Y647och, and the second mutation was named rsp5-W650L. Since both rsp5-Y647och and rsp5-25 mutations are lethal in the sup4 background, all tests of their effects were performed in a SUP4-o background. These results will be discussed below.
Both mutations in rsp5-25 are required to suppresses rfa1-D228Y.
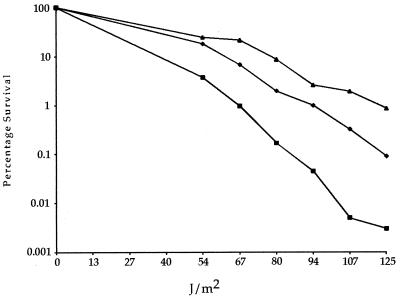
Since rsp5-25 was isolated as a suppressor of rfa1-D228Y-stimulated direct repeat recombination, we next asked whether it suppresses the UV sensitivity exhibited by rfa1-D228Y strains. As a control, we showed that the rsp5-25 mutant strain itself is not sensitive to UV damage (data not shown). Figure 5 shows that the UV sensitivity of rfa1-D228Y is partially suppressed in an rsp5-25 rfa1-D228Y double mutant.
FIG. 5.
UV survival curves. Wild-type (▴), rfa1-D228Y (■), and rfa1-D228Y rsp5-25 (⧫) strains were exposed to increasing amounts of UV irradiation, and their survival was plotted. Both rsp5-25 strains and rfa1-D228Y strains overexpressing rfa1-D228Y exhibit identical survival to wild-type strains (reference 45 and data not shown).
Next, we determined the extent to which each separate alteration in rsp5-25 contributes to its phenotype. All tests were performed in the presence of SUP4-o. Neither rsp5-Y647och nor rsp5-W650L on its own reduces direct repeat recombination levels in a rad1 rad52 rfa1-D228Y background (data not shown). Similarly, neither allele displays temperature sensitivity in a rad52 mutant background nor can either one suppress the UV sensitivity of rfa1-D228Y (data not shown). Thus, both the decreased level of full-length protein caused by rsp5-Y647och and the rsp5-W650L missense mutation in the ubiquitination domain are necessary to observe the rsp5-25 phenotype, indicating a synergistic interaction between the two mutations.
The stability of Rfa1-D228Y is increased in rsp5-25 strains.
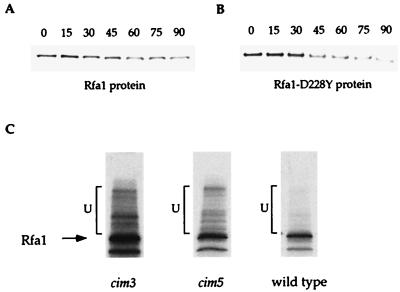
The relative stability of Rfa1-D228Y was measured and compared in rsp5-25 and wild-type strains. A plasmid containing a functional HA-tagged rfa1-D228Y allele under the control of the GAL1 promoter was introduced into both rsp5-25 rfa1Δ and RSP5 rfa1Δ strains. These strains were grown in the presence of galactose until early log phase and then shifted to glucose-containing medium to repress the GAL1 promoter and to turn off the production of Rfa1-D228Y. Samples were taken at 30-min intervals for 3 h, and the Rfa1-D228Y protein levels were evaluated by protein blot analysis. As shown in Fig. 6A, under these conditions the half-life of Rfa1-D228Y in the rsp5-25 strain is 65 min compared to 35 min in the RSP5 strain. Furthermore, Fig. 6B shows that the same steady-state levels of Rfa1-D228Y protein is seen in rfa1-D228Y rsp5-25 strains and rfa1-D228Y strains where rfa1-D228Y is overexpressed.
Next, the protein levels of Rfa1-D228Y were measured during the cell cycle, where RFA1 mRNA levels have been shown to increase fourfold at the beginning of S phase (reference 5 and Fig. 6C). Surprisingly, Rfa1-D228Y protein levels remain constant throughout the cell cycle (Fig. 6D). Similar results have been observed in RFA1 wild-type strains (reference 5 and data not shown). This suggests that either regulation at the level of translation and/or a faster rate of degradation of Rfa1 during S phase may account for the constant steady-state protein levels. We tested the second possibility by measuring the degradation kinetics of both wild-type Rfa1 and Rfa1-D228Y proteins in S-phase-arrested cells. Cells were arrested by incubation with HU for 6 h in the presence of inducer (galactose). Next, they were switched to glucose medium, still in the presence of HU, and the half-lives of the Rfa1 proteins were determined by protein blot analysis. In HU-arrested cells, wild-type and mutant Rfa1 proteins were shown to have approximately 65- and 35-min half-lives, respectively (Fig. 7A and B), which is not different from the findings with unarrested cells. Thus, it is more likely that the difference in mRNA and protein levels during S phase is due to regulation at the level of translation.
FIG. 7.
Degradation of Rfa1 during S phase and its in vivo ubiquitination. (A and B) Protein blot analyses of the stability of Rfa1 and Rfa1-D228Y during S phase. RFA1 and rfa1-D228Y cells were arrested with HU at S phase, and samples were taken every 15 min. Total yeast protein extracts were prepared and subjected to protein blot analysis by using the Rfa1 antibody. The half-lives of each were identical to that determined for logarithmically growing cells (see Fig. 6A). (C) In vivo ubiquitination of Rfa1. Wild-type, cim3, and cim5 mutant strains that overexpress rfa1-D228Y under the GAL1 promoter were pulse labeled with [35S]methionine. After incubation at 37°C, the nonpermissive temperature for the cim mutants, total yeast protein extract was prepared, and immunoprecipitations were performed with Rfa1 antibody. By immunoprecipitation, putative ubiquitinated forms of Rfa1 were visualized in cim3, cim5, and wild-type strains respectively. The bands corresponding to the nonubiquitinated forms of the Rfa1 are indicated. The higher-molecular-weight bands, indicated by brackets, represent the putative ubiquitinated forms.
Rfa1 may be ubiquitinated in vivo.
The observation that a mutation in Rsp5 destabilizes Rfa1 suggests that its turnover may be controlled by ubiquitination. Therefore, we examined Rfa1 for ubiquitin conjugates in vivo. Total Rfa1 protein was immunoprecipitated with Rfa1 antibodies from a strain containing cim3 and cim5 ts mutations that affect 26S proteasome function (13). In these strains, degradation of ubiquitinated proteins in the 26S proteasome is impaired and, at the restrictive temperature, substrates accumulate with branched ubiquitin chains. To visualize potential ubiquitinated forms of Rfa1, both cim3 and cim5 strains expressing wild-type Rfa1 endogenously and Rfa1-D228Y from a plasmid were labeled with [35S]methionine in a pulse-chase experiment. After the shift to the restrictive temperature, total Rfa1 was immunoprecipitated. Upon gel electrophoresis, a ladder of higher molecular weight forms of Rfa1 protein was detected, a finding that is consistent with heterogeneous ubiquitination (Fig. 7C).
DISCUSSION
Ubiquitin-dependent proteolysis is a major pathway for protein degradation. It functions to regulate many cellular processes by modulating the availability of proteins in the cell (reviewed in reference 15). Ubiquitin, a very conserved 76-amino-acid peptide, marks proteins for degradation when it is covalently attached to them. Degradation occurs mainly in the 26S proteasome but also occasionally in the vacuole. A key group of enzymes of this pathway are the ubiquitin-protein ligases. Their role is to recognize substrates and to promote the transfer of ubiquitin, directly or indirectly, onto the substrates, thus targeting them for degradation (15).
In our search for mutations that reduce recombination levels in rad1 rad52 rfa1-D228Y strains, we isolated two alleles of RSP5, a gene encoding an essential protein ubiquitin ligase in S. cerevisiae (17, 58). In the present study we extensively characterized one of these alleles, rsp5-25. We observed that many aspects of the rsp5 rfa1-D228Y phenotype could be mimicked by overexpressing rfa1-D228Y. First, the fivefold reduction in leu2 direct repeat recombination observed in rad1 rad52 rfa1-D228Y rsp5-25 strains is similar to the 4.2-fold reduction seen when the Rfa1-D228Y protein is overexpressed in rad1 rad52 rfa1-D228Y strains (unpublished data). Similarly, the level of suppression of UV sensitivity in rfa1-D228Y rsp5-25 strains is the same as that seen when Rfa1-D228Y is overproduced in an rfa1-D228Y strain (45; Fig. 5). Third, the protein levels of Rfa1-D228Y in those same strains is increased to approximately the same levels (Fig. 6B). Furthermore, the half-life of Rfa1-D228Y is doubled in rsp5-25 strains (Fig. 6A). Finally, the ubiquitination and degradation of Rfa1 likely occurs in the 26S proteasome in vivo (Fig. 7C). Taken together, these results implicate Rsp5 in the degradation of Rfa1, a protein involved in DNA repair, recombination, and replication.
Molecular characterization revealed that the rsp5-25 allele contains two nucleotide changes. The first mutation causes a tyrosine-to-ochre codon change at position 647, thus truncating the protein in a sup4 background. In contrast, in a SUP4-o tyrosine ochre suppressor background, tyrosine is inserted as in the wild-type protein. However, suppression by SUP4-o is only partial; thus, the amount of the full-length Rsp5 protein is reduced approximately 10-fold. The second mutation substitutes a leucine residue for a conserved tryptophan residue at position 650. The two mutations were separated from each other, and both mutations are required in order to observe the rsp5-25 phenotype. Thus, the decreased level of full-length protein as well as the missense mutation in the conserved domain are necessary to alter the degradation kinetics of Rfa1-D228Y.
Although the consequences of rsp5 mutations on a variety of cellular processes are dramatic, their effects on the protein stability of known substrates (Fur4, Rfa1, and Rbp1) are only increased two- to fivefold (11, 18; this study). We offer several hypotheses to explain this observation. First, an even greater increase in stability may be observed in some alleles; however, such mutants may be lethal and thus could not be recovered. Second, cells may not be able to tolerate an increase in the stability of certain proteins, especially if these proteins are found in complexes. One example is the RP-A complex itself, where overexpression of only one RP-A subunit causes slow growth (5), which is likely due to a disruption in the equilibrium of the subunit concentrations. Third, Rsp5 may only recognize a specific form of its substrate, e.g., a phosphorylated form. Therefore, when the amount of the substrate form is low compared to the total amount of that protein, the consequent stabilization of only the minor form in an rsp5 mutant will not significantly affect the half-life measurement of that protein. Finally, Rsp5 may ubiquitinate a substrate at a specific point of the cell cycle. Thus, measurement of total protein turnover in an asynchronous population would obscure cell-cycle-specific changes for a particular protein. Similar observations have been made for some cyclin proteins (40). In fact, we observed that while the transcripts levels of RFA1 increase during S phase, Rfa1 protein levels remain constant throughout the cell cycle (Fig. 6C and D). We also found that the degradation kinetics of Rfa1 protein does not differ significantly between S-phase-arrested and nonarrested cells (data not shown), suggesting that regulation is at the level of translation. However, we cannot exclude the possibility, as discussed above, that a critical subpopulation of the total Rfa1 protein is degraded in a cell-cycle-dependent manner, since we can only measure the total amount of the Rfa1 protein in our experiments.
Studies on the molecular genetics and biochemistry of both RP-A and Rad52 may help explain the ts phenotype associated with the two rsp5 alleles in various genetic backgrounds. Recent work has shown that excess RP-A can actually inhibit DNA annealing in vitro (49, 52) and that Rad52 protein, which stimulates DNA annealing reactions (26, 52), can overcome this inhibition (49). Thus, rad52 rsp5 strains may be ts due to an increased level of Rfa1 that dramatically inhibits spontaneous annealing reactions. In support of this, we also observed that rad52 rfa1-D228Y strains that overexpress rfa1-D228Y display a similar ts phenotype (unpublished data). For the rsp5-26 allele, synthetic lethal interactions occur with any combination of rad1, rad52, rfa1-D228Y, or rad51. Although there are many possible explanations for this phenotype, perhaps an additional protein(s) is stabilized in the rsp5-26 strain that inhibits recombination in the presence of these mutations. Alternatively, the stabilized protein(s) may lead to an increase in recombinogenic lesions, which would be detrimental in the absence of efficient DNA repair.
The spontaneous direct repeat recombination events assayed here in rad1 rad52 rfa1-D228Y strains likely proceed via single-strand annealing according to the following scenario (45). The first step is a double-strand break in the plasmid sequence, after which degradation of the 5′ ends leads to the formation of 3′ single-stranded DNA tails (46, 50, 60). These single-stranded tails participate in the homology search and subsequent annealing to promote the recombination event (10). In rad52 strains, where annealing is greatly reduced, more-extensive degradation results in longer single-stranded 3′ DNA (60). It was demonstrated recently that the rfa1-D228Y mutation suppresses the formation of the long single-stranded 3′ DNA tracts normally found in rad52 mutants (46). Therefore, we suggest that, in a rad1 rad52 rfa1-D228Y rsp5 quadruple mutant, as a consequence of the prolonged half-life of Rfa1, the concentration of RP-A increases, allowing it to bind more extensively to single-stranded DNA. Consequently, this may reduce the ability of these DNA molecules to participate in the homology search and/or annealing reactions. This effectively reverses the recombination phenotype of the triple mutant (i.e., the increased recombination observed in rad1 rad52 rfa1-D228Y is reduced by the addition of an rsp5 mutation).
At the onset of this study, we mutagenized a rad1 rad52 rfa1-D228Y strain to help define the “pathway” responsible for the observed increased recombination. For example, if the rfa1-D228Y mutation specifically activated an alternative recombination pathway, we expected to find mutations that subsequently reduced various steps. Surprisingly, none were found. Instead, our extensive screen to identify genes in this putative pathway resulted in the isolation of a single ubiquitination gene twice. Both rsp5 alleles display synthetic lethality with rad52 at a restrictive temperature and, furthermore, turnover of the Rfa1-D228Y protein is slowed in the rsp5-25 background. These results argue that the rsp5 mutations affect recombination by altering the cellular pathway involved in protein stability.
ACKNOWLEDGMENTS
We thank Uffe Mortensen, Steve Sturley, Xiaolan Zhao, and especially Marcel Wehrli for comments on the manuscript and helpful discussions. We also thank Jon Huibregtse, Carl Mann, Bruce Stillman, and Steve Brill for the kind gifts of strains, plasmids, and antibodies. Finally, we thank Adam Bailis, Serge Gangloff, John McDonald, Julie Smith, and Hui Zou for encouragement and discussion throughout the course of this work.
This work was supported by National Institutes of Health grant GM50237 (R.R.).
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson F E, Baumann P, West S C. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 4.Brill S J, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 5.Brill S J, Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 6.Broker T, Lehman I. Branched DNA molecules: intermediates in T4 recombination. J Mol Biol. 1971;60:131–149. doi: 10.1016/0022-2836(71)90453-0. [DOI] [PubMed] [Google Scholar]
- 7.Clark A J. Toward a metabolic interpretation of genetic recombination of Escherichia coli and its phages. Annu Rev Microbiol. 1971;25:438–464. doi: 10.1146/annurev.mi.25.100171.002253. [DOI] [PubMed] [Google Scholar]
- 8.Fairman M P, Prelich G, Tsurimoto T, Stillman B. Replication of SV40 in vitro using proteins derived from a human cell extract. J Cell Sci. 1989;12:161–169. doi: 10.1242/jcs.1989.supplement_12.14. [DOI] [PubMed] [Google Scholar]
- 9.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 10.Fishman-Lobell J, Rudin N, Haber J E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan J M, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 12.Game J C, Cox B S. Allelism tests of mutants affecting sensitivity to radiation in yeast and a proposed nomenclature. Mutat Res. 1971;6:37–55. [Google Scholar]
- 13.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 14.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 15.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann W. Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae. A transmembrane protein without N-terminal hydrophobic signal sequence. J Biol Chem. 1985;260:11831–11837. [PubMed] [Google Scholar]
- 17.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huibregtse J M, Yang J C, Beaudenon S L. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein H L. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics. 1988;120:367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langle-Rouault F, Jacobs E. A method for performing precise alterations in the yeast genome using a recycable selectable marker. Nucleic Acids Res. 1995;23:3079–3081. doi: 10.1093/nar/23.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larionov V L, Grishin A V, Smirnov M N. 3 μm DNA—an extrachromosomal ribosomal DNA in the yeast Saccharomyces cerevisiae. Gene. 1980;12:41–49. doi: 10.1016/0378-1119(80)90014-1. [DOI] [PubMed] [Google Scholar]
- 22.Lin F L, Sperle K, Sternberg N. Homologous recombination in mouse L cells. Cold Spring Harbor Symp Quant Biol. 1984;49:139–149. doi: 10.1101/sqb.1984.049.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Malone R E, Esposito R E. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci USA. 1980;77:503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald J P, Rothstein R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milne G T, Weaver D T. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 1993;7:1755–1765. doi: 10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- 26.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 28.Orr-Weaver T L, Szostak J W, Rothstein R J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 29.Orr-Weaver T L, Szostak J W, Rothstein R J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petes T D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980;19:765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- 32.Piper P W, Wasserstein M, Engbaek F, Kaltoft K, Celis J E, Zeuthen J, Liebman S, Sherman F. Nonsense suppressors of Saccharomyces cerevisiae can be generated by mutation of the tyrosine tRNA anticodon. Nature. 1976;262:757–761. doi: 10.1038/262757a0. [DOI] [PubMed] [Google Scholar]
- 33.Resnick M A. Genetic control of radiation sensitivity in Saccharomyces cerevisiae. Genetics. 1969;62:519–531. doi: 10.1093/genetics/62.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resnick M A, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds R J, Love J D, Friedberg E C. Molecular mechanisms of pyrimidine dimer excision in Saccharomyces cerevisiae: excision of dimers in cell extracts. J Bacteriol. 1981;147:705–708. doi: 10.1128/jb.147.2.705-708.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronne H, Rothstein R. Mitotic sectored colonies: evidence of heteroduplex DNA formation during direct repeat recombination. Proc Natl Acad Sci USA. 1988;85:2696–2700. doi: 10.1073/pnas.85.8.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 39.Schiestl R H, Prakash S. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol Cell Biol. 1988;8:3619–3626. doi: 10.1128/mcb.8.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 41.Sherman F. Getting started with yeast. Vol. 194. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 42.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 43.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 44.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith J, Rothstein R. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith J, Rothstein R. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics. 1999;151:447–458. doi: 10.1093/genetics/151.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Struhl K, Stinchcomb D T, Scherer S, Davis R W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiyama T, New J H, Kowalczykowski S C. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 51.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 52.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 53.Sung P, Robberson D L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 54.Szostak J W, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 55.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 56.Thomas B J, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomkinson A E, Bardwell A J, Bardwell L, Tappe N J, Friedberg E C. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature. 1993;362:860–862. doi: 10.1038/362860a0. [DOI] [PubMed] [Google Scholar]
- 58.Wang G, Yang J, Huibregtse J M. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yashiroda H, Oguchi T, Yasuda Y, Toh E A, Kikuchi Y. Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3255–3263. doi: 10.1128/mcb.16.7.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zolladek T, Tobiasz A, Vaduva G, Boguta M, Martin N C, Hopper A K. MDP1, a Saccharomyces cerevisiae gene involved in mitochondrial/cytoplasmic protein distribution, is identical to the ubiquitin-protein ligase gene RSP5. Genetics. 1997;145:595–603. doi: 10.1093/genetics/145.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]