Abstract
Outcomes of anticancer therapy vary dramatically among patients due to diverse genetic and molecular backgrounds, highlighting extensive inter-tumoral heterogeneity. The fundamental tenet of precision oncology defines molecular characterization of tumors to guide optimal patient-tailored therapy. Towards this goal, we have established a compilation of pharmacological landscape of 462 patient-derived tumor cells (PDCs) across 14 cancer types, together with genomic and transcriptomic profiling in 385 of these tumors. Compared to the traditional long-term cultured cancer cell-line models, PDCs recapitulate the molecular properties and biology of the diseases more precisely. In present study, we provide unprecedented insights into dynamic pharmacogenomic associations, including molecular determinants that elicit therapeutic resistance to EGFR inhibitors and potential repurposing of ibrutinib (currently being used in hematological malignancies) for EGFR-specific therapy in gliomas. Lastly, we present potential implementation of PDC-derived drug sensitivities for prediction of clinical response to targeted therapeutics using retrospective clinical studies.
Genomic and molecular tumor profiling enables identification of effective drugs tailored to cancer patients1–9. However, predicting successful anti-cancer therapy remains extremely challenging10–12, largely due to extensive inter and intra-tumoral heterogeneity13–15. Recent efforts have established a framework for genetic predictions of anticancer drug responses using standard in vitro cancer cell-line models16–23. In particular, large-scale drug screening systems, using conventional cancer cell-lines have provided reference points for gene-drug associations, enabling discovery of molecular markers that may predict therapeutic response16,17. However, there are several challenges that hamper broad clinical utility of the current gene-drug association map in the oncology clinic. First, as most solid cancers harbor multiple molecular aberrations, predicting therapeutic efficacy of a targeted agent based on genomic profiling alone can be a complicated process. Second, prediction of treatment outcome extrapolated from conventional cancer cell-lines may not recapitulate each cancer patient’s tumor. To address these challenges, we present a comprehensive integrated approach using genomic analysis of the patient tumor, ex vivo assessment of drug effect on patient tumor derivatives, and in vivo validation of the selected compounds’ therapeutic efficacies.
While patient-derived xenograft (PDX) systems both respect inter-patient genomic diversity and intra-tumor microenvironment24, generation of PDX models exhibit relatively lower tumor formation rate, and require a longer establishment period compared to patient-derived tumor cells (PDCs). In glioblastoma, it takes approximately 2 to 3 weeks to establish neurosphere-like PDCs, whereas well-defined tumor masses in vivo are generally observed after 6 to 7 weeks of patient tumorsphere transplantation20,25,26. More importantly, chemical screening system using PDCs presents significant advantage over PDXs as to substantially enlarge the number of chemicals that could be employed in multiple doses, necessary to generate reliable drug response parameters. PDCs represent unique biology of each corresponding tumor and provide an accurate model system for assessing drug response. We and others previously showed that PDCs and PDX tumors retain the genomic and biological characteristics of tumors in situ26–29. Drug sensitivity screening of PDCs, however, remains at the level of proof-of-concept22,23,30, as large-scale studies have yet to reveal statistically robust connections between pharmacology and genomics across multiple cancer types. Moreover, the number of clinical studies guided by PDC-based drug screening is insufficient to draw concrete conclusions. In present study, we report a unique resource presenting pharmacological landscape of 462 PDCs in 14 cancer types, based on treatment with 60 anti-cancer agents. We show that PDCs faithfully recapitulate the molecular profiles (RNA expression, mutations) of the original primary tumors. Using a combination of machine learning and statistical methods, we discovered lineage-specific drug sensitivities and molecular correlates of drug sensitivity and resistance. Finally, we demonstrate high clinical concordance rate between clinical response and drug susceptibility prediction in retrospective studies.
RESULTS
Genetic and transcriptomic similarity between primary tumors and patient-derived tumor cells
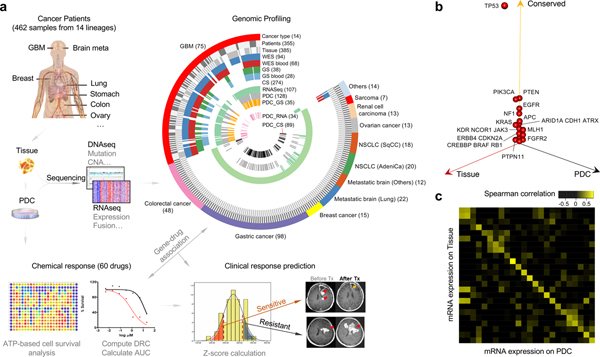
To establish a large-scale PDC library, we derived 462 short-term tumor cell cultures from surgically resected tumor specimens or ascites-derived tumor cells that were isolated from 14 different cancer types, including malignant gliomas, metastatic brain tumors, gastric (GC), colorectal (CRC), lung, breast, ovary cancers, renal cell carcinoma and sarcomas (Fig. 1a and Supplementary Table 1). To identify somatic mutations and copy number alterations (CNAs), exome sequencing was performed on 368 tumor specimens and matched normal specimens. Among them, 94 samples were subjected to whole exome sequencing (WES), while 274 tissue specimens were sequenced for full coding exons of 80 commonly mutated cancer genes (CancerSCAN™, Supplementary Tables 1 and 2)31–34. Forty-one specimens from malignant gliomas were analyzed with a massive parallel targeted sequencing platform, covering exons of cancer-driven and/or glioma-associated genes (GliomSCAN™, Supplementary Tables 1 and 3)1,9,35. We used SAVI2 to identify somatic single-nucleotide variants, as well as short insertions and deletions36. Somatic variants with a mutant allele frequency > 5% were considered. To explore copy number variations (CNVs), EXCAVATOR37 was applied on the samples with available WES data (Supplementary Fig. 1 and Supplementary Table 4). RNA sequencing analysis was performed in 107 tumors to identify structure variations and gene expression profiles. Tumor cell isolates were cultured under serum-free conditions27,28. Patient-tumor derived short-term cultures were further subjected to targeted exome sequencing (GliomaSCAN™ or CancerSCAN™, n=122) and/or RNA-sequencing to determine whether the PDCs retained the spectrum of genomic alterations found in the patient specimen (Fig. 1a and Supplementary Table 1). We observed that somatic variations in major cancer-driven genes including TP53, PTEN, PIK3CA, EGFR, NF1, APC, KRAS, and ATRX were well preserved from the parental tumors to PDCs across major cancer types including gliomas, metastatic brain tumors, GC, CRC and lung adenocarcinomas (n=122; Fig. 1b, Supplementary Fig. 2a and 2b). Next, we measured transcriptomic similarity between the parent tumors and PDCs and discovered a strong positive correlation (Fig. 1c and Supplementary Fig. 3). Gene fusions were identified by chimerascan and characterized by Pegasus38. We found the expression levels of gene fusions were strongly correlated between tumors and PDCs (Supplementary Fig. 2c). In addition, we observed that the mutation and gene expression profiles of our PDC panel were notably similar to tumor tissues (The Cancer Genome Atlas (TCGA) dataset) compared to the previously established conventional cell-lines (Genomics of Drug Sensitivity in Cancer (GDSC) and Cancer Cell Line Encyclopedia (CCLE) datasets), underscoring significant use of patient-derived cells over traditional cancer cell-line models (Supplementary Fig. 4)1,16,17,23. Taken together, our results suggest that PDCs provide genomic/transcriptomic proxies for primary tumors, and are less divergent from the patient tumors than conventional cancer cell-lines.
Figure 1. Patient tumor and derived cell resources for pharmacogenomics analysis.
(a) Overview of the procedure for pharmacogenomics analysis in patient tumor-derived cells (PDCs). A total of 462 PDCs from 14 cancer types were isolated. Genomic contexts were analyzed to identify somatic variants and/or gene expression profiles. Comprehensive genomic profiling from 14 cancer lineages are summarized in a circos plot, demonstrating detailed data structure and size for each type of available molecular data. Short-term cultured PDCs have undergone drug sensitivity screening to 60 molecular targeted agents. Clinical feasibility of PDC screening-guided precision therapy was evaluated. (b) Three-dimensional bubble plot showing the frequency of somatic nonsynonymous mutations exclusively in tissue (red; left axis), exclusively in PDC (black; right axis), and in common to the two (yellow; upper axis). 122 samples with DNA sequencing data from tumor tissue and PDC samples were considered in this analysis. (c) Comparison of mRNA expression profiles between primary tissue and PDC on 24 paired samples with matched RNASeq data. Spearman correlations of mRNA expression between tissue and PDC are shown as a heat map. Paired samples are located along the diagonal.
A 60-drug panel was used for chemical screening of PDCs across multiple tumor lineages. Our panel included well-known receptor tyrosine kinases (RTKs) inhibitors, including epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR)/vascular endothelial growth factor receptor (VEGFR), and phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) (PAM pathway); as well as multi-target drugs and inhibitors for proteasome, poly (ADP-ribose) polymerase (PARP), and histone deacetylase (HDAC) (Supplementary Tables 5 and 6)39–52.
Cell viability was determined after six or seven days of drug treatment using multi-parameter drug sensitivity analyses, including half-maximal inhibitory concentration (IC50) and area under curve (AUC) of the dose-response curve (DRC; Fig. 1a, Supplementary Tables 7 and 8)53–55. The drug panel sensitivity profiles were highly concordant among technical replicates (Supplementary Fig. 5a). To evaluate whether tumor cell expansion affected biological properties, including drug sensitivity, we measured post-drug treatment cell viability using four different sets of the matched PDCs across various in vitro passages or expansion stages (Supplementary Fig. 5b). Drug sensitivities of biological replicates from the same patient-derived tumor cells at different passages showed significant correlations with minimal variations.
Pan-cancer pharmacological profiling reveals lineage-specific drug sensitivity
We next established a pharmacological landscape of 462 PDCs, from 14 different tumor lineages, based on anticancer drug response to 60 compounds. A total of 27,720 drug-PDC combinations were evaluated and analyzed (Supplementary Fig 6). The median IC50 values varied from 0.003–53.22 μM, where 1 μM is a commonly applied threshold for pharmacological relevance (Supplementary Fig. 6a). A subset of compounds, including AUY922 (HSP), BEZ235, PKI-587 (PI3K/mTOR), bortezomib, carfilzomib (proteasome), neratinib (EGFR), panobinostat (HDAC), and trametinib (MEK) showed notably high drug activities. In contrast, several agents such as dabrafenib (BRAF), olaparib (PARP), sotrastaurin (PKC), vismodegib (hedgehog), and XL147 (PI3K) exhibited low anti-tumor activities, each with median IC50 values of >10 μM. RTK blockers, targeting EGFR, MET, FGFR, PDGFR and PAM, showed a wide spectrum of drug sensitivities, suggesting a strong association between pharmacological drug response and genomic aberration (Supplementary Fig. 6b).
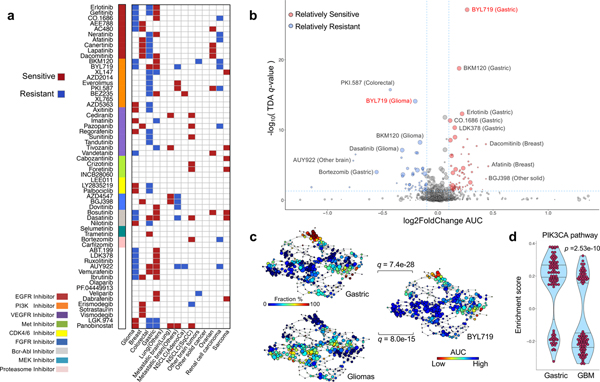
As tumor lineage portrays diverse drug responses16,17, we first analyzed lineage-specific drug sensitivity within our cohort (Fig. 2a). Overall, we discovered robust lineage-specific drug sensitivities across multiple cancer types, among which a significant diversity was observed in glioma, breast, CRC, and GC. Hierarchical clustering of drug sensitivities (normalized AUCs) identified three distinct clusters, enriched with GC, glioma and CRC, suggesting lineage-specific drug response patterns. (Supplementary Fig. 6c). Of note, EGFR inhibitors (erlotinib, dacomitinib, afatinib) showed significantly high activity in both GC and breast cancer models, while demonstrating minimal responses in malignant gliomas and CRC (Fig. 2a and Supplementary Table 9). Interestingly, PDCs from malignant gliomas demonstrated considerable resistance to erlotinib, gefitinib (EGFR) and BKM120 (PI3K), while CDK4/6 inhibitors, LY2835219 and palbociclib, were significantly more sensitive in glioma PDCs compared to the rest (Fig. 2a and Supplementary Fig. 6d)1,56–58. To explore dynamic pharmacological associations among different drug profiles, we performed topological data analysis (TDA, using Mapper, see Supplementary methods), a computational technique to reduce the dimensionality of large data sets while retaining local high dimensional structure13,59–61 of lineage-specific drug associations (Fig. 2b and Supplementary Table 10). PDCs isolated from CRCs were relatively more resistant to multiple PAM inhibitors such as BEZ235, PKI-587, AZD2014, everolimus and BYL719 (Fig. 2b). Additionally, GC PDCs were highly sensitivity to PI3K inhibitors, including BYL719 and BKM12062, which were comparatively less potent in malignant gliomas (Fig. 2b, 2c, Supplementary Fig. 7). Consistently, PIK3CA pathways were found to be more activated in GC PDCs, compared to GBM PDCs (Fig. 2d)63–65. These observations suggest higher likelihood of anti-tumor activity of PAM inhibitors in GC patients when compared with GBM patients.
Figure 2. Therapeutic landscape of PDCs and lineage-specific responses.
(a) Tumor lineage-specific drug association identified using 60 compounds (n=462 biologically independent samples). Two-sided wilcox rank sum test was applied to determine the relative differences of drug sensitivity between specific tumor type and the rest. Only significant associations are marked (q-value < 0.05). Drugs are ordered based on their known targets. (b) A volcano plot representation of TDA analysis showing the magnitude (Fold change, x-axis) and significance (TDA q value, y-axis) of all tumor-drug associations (n=462 biologically independent samples). Each circle represents a single tumor-drug interaction and the size is proportional to the cohort size of that tumor. (c) Distribution of gastric, glioma PDCs and BYL719 drug AUC profile over the topological representation of PDCs (n=462 biologically independent samples). Each node represents a set of PDCs with similar AUC profiles. A PDC can appear in several nodes, and two nodes are connected by an edge if they have at least one PDC in common. The P values were calculated using the pearson correlation test between the fraction distribution of gastric or glioma cell lines and mean AUC values of BYL719 drug over the nodes and they were adjusted using BH method. (d) Violin plots measures the activity level of PI3K–AKT–mTOR pathway on gastric and GBMs using TCGA RNASeq datasets (n=100 biologically independent samples for each group). We adopt the enrichment score derived from ssGSEA analysis as assessment. The P value is calculated from two-sided wilcox rank-sum test. Horizontal lines within the violin plot represent 0.25, 0.50, and 0.75 quantiles.
Genomic predictors of drug sensitivity/resistance based on PDC genome-drug mapping
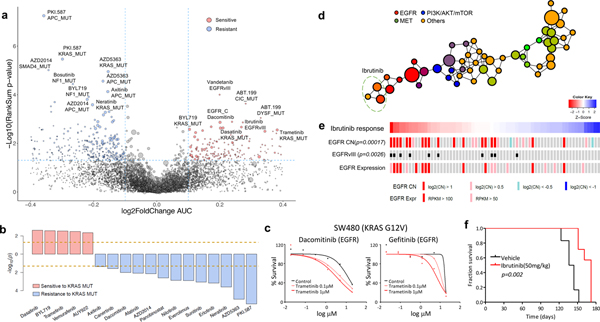
Genomic variations are increasingly being utilized as reliable biomarkers for predicting clinical response to cancer therapy4,5,10,11,16,17. To identify significant genomic correlates of pharmacological sensitivity in PDC models, we assessed individual drug profiles across 462 tumor cells based on single genomic alterations (Fig. 3a and Supplementary Table 11). EGFR somatic alterations, including single-nucleotide variation, copy number amplification and structure variations, were mostly found in GBM tissues and conferred increased sensitivity to relevant EGFR inhibitors, such as dacomitinib, and interestingly to vandetanib (VEGFR) and ibrutinib (BTK)66–68. Previous studies have demonstrated strong association between KRAS mutation and therapeutic resistance to PAM pathway inhibitors, including PKI-587, AZD5363, AZD2014, and multiple EGFR inhibitors, such as neratinib, erlotinib, afatinib, dacomitinib, and canertinib69–72. On the contrary, we discovered that KRAS mutation conferred increased sensitivity to a specific group of drugs, including dasatinib (SRC, BCR-ABL), BYL719 (PI3K) and trametinib (MEK) (Fig 3b)73–76. Although KRAS mutation drive tumor progression through constitutive activation of mitogen activated protein kinase (MAPK) signaling pathway, conflicting evidences have been reported for the efficacy of MEK target therapy in KRAS mutant cancers77–80. We showed that treatment with MEK inhibitors increased the efficacy of EGFR family blockers (dacomitinib and gefitinib) in a KRAS mutant (G12V) CRC cell-line, SW48080, suggesting that simultaneous targeting of EGFR and MEK signaling pathways could potentially treat KRAS-mutant CRC patients (Fig. 3c)19,81,82. Furthermore, mutation in the adenomatous polyposis coli (APC) gene was significantly correlated with minimal responses of multiple drugs, including PKI-587 (PI3K/mTOR), AZD5363 (AKT), AZD2014 (mTOR) (Fig. 3a and Supplementary Fig. 8), consistent with previous observations that the activation of β-catenin signaling pathway may elicit multi-drug resistance across various cancer types83–85.
Figure 3. Pharmacogenomic interactions in PDCs.
(a) A volcano plot representation of correlation analysis showing the magnitude and significance of gene-drug associations (n=360 biologically independent samples). (b) Waterfall plot enumerating significant associations between KRAS mutation and drug sensitivity (n=360 biologically independent samples). Two horizontal dashed lines indicate statistical significance. (c) SW480 was treated with DMSO (control) or trametinib (0.1 or 1 mM), followed by incubation with two EGFR inhibitors, dacomitinib (left) and gefitinib (right). Cell viability for each dose was normalized to DMSO or trametinib (0.1 or 1 mM) treatment only cells. (d) Probability distribution of drug-target families over the topological network. Each node represents a set of drugs with similar AUC profiles. A drug can appear in several nodes, and two nodes are connected by an edge if they have at least one drug in common. Colors of the nodes correspond to mean RGB values of drug families. Ibrutinib belongs to three nodes on the network encompassed with an oval. (e) Drug sensitivities to ibrutinib in 67 PDCs. The red color in the heat map represents sensitivity, while the blue color indicates resistance. EGFR alterations including genomic amplification, vIII, and expression are shown. (f) Kaplan-Meier survival plots for P2.T (EGFR amp/vIII) orthotopic mice model. Once intracranial model was established, Vehicle (0.5% methylcellulose) or iburitnib (50mg/kg/day) was administrated orally (PO) for 5 consecutive days and 2 days of resting period per each cycle (n=8 per group). P-values: a,b,e, two-sided wilcox rank sum test. P-values: f, two-sided Log-rank test.
Activation of various RTKs is a key molecular feature of GBM. As RTK-targeting approaches against GBM have been widely pursued, we examined pharmacological sensitivities of various RTK inhibitors in GBM PDCs via drug-centric TDA (see Supplementary Methods) (Fig. 3d). Interestingly, we have discovered several compounds that are clustered with different class of agents, including erlotinib with PI3K, foretinib and BGJ398 with VEGFR, cediranib with FGFR and ibrutinib with EGFR compounds (Fig. 3d and Supplementary Fig. 9). Ibrutinib-EGFR TDA result was consistent with previous genomic and pharmacological correlate analysis in Fig. 3a. In preclinical and clinical studies on hematologic malignancies, ibrutinib has been reported to promote cellular arrest via inhibition of Bruton’s tyrosine kinase (BTK) phosphorylation68,86–88. Although, a potent anti-tumor effect of ibrutinib was previously demonstrated in non-small-cell lung cancer cells67, it was exclusive to tumors harboring EGFR T790M mutation, which is an erlotinib-resistance–associated event. Notably, we showed that EGFRvIII and EGFR amplification were significantly linked to increased sensitivity to ibrutinib (Fig. 3e). To determine whether the sensitivity to ibrutinib was exclusively associated with EGFR aberration, we surveyed the transcriptome levels of BLK, BMX, and BTK, the proposed targets of ibrutinib. Expression levels of these genes were generally low in GBMs, and more importantly, there was no significant correlation between their transcriptional levels and ibrutinib activity (Supplementary Fig. 10a). In contrast, ibrutinib response significantly correlated with EGFR mRNA expression level (Supplementary Fig. 10b). Such observation was significantly enhanced when only the EGFRvIII-positive tumors were selected.
To further validate and explore ibrutinib-EGFR association and its biological effects in GBM, we performed a series of functional in vitro and in vivo experiments. Consistent with previous findings, Ibrutinib significantly decreased clonogenic growth of EGFR amplification/vIII harboring GBM PDCs compared to those with EGFR wild-type (Supplementary Fig. 11a)89. Immunoblot analyses of EGFR and its downstream effectors, pSTAT3, pERK, pAKT and pS6K, revealed robust inhibition in the presence of ibrutinib treatment (Supplementary Fig. 11b). These activities were further confirmed using different Ibrutinib compounds that were obtained from five different vendors (Supplementary Fig. 11c). Furthermore, in vivo assessment of ibrutinib (50 mg/kg p.o. q.d. for 5 days) demonstrated prolonged survival and reduced tumor volume in PDX models (Fig. 5f and Supplementary Fig. 11d). Histological and immunocytochemical analyses revealed attenuated EGFR expression and its downstream molecules (Supplementary Fig. 11d and 11e). Collectively, our results indicate that EGFR amplification/vIII could be a potential biomarker for sensitivity to ibrutinib in GBM.
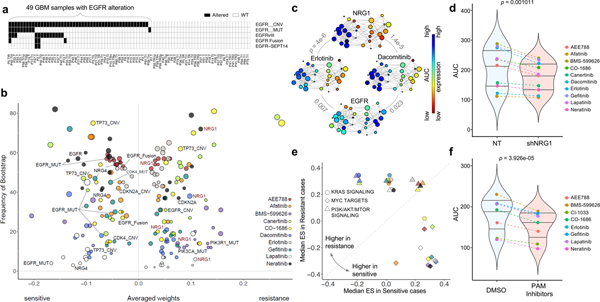
Figure 5. Predictive biomarkers for response to EGFR inhibitors in EGFR altered GBM PDCs.
(a) Mutational landscape of EGFR alterations in GBM cohort. (b) For the 10 EGFR inhibitors, top drug–feature associations identified by dNetFS are plotted for their frequency and effect size (n=49 biologically independent samples in (a). Node size is proportional to the single drug–feature linear correlation. (c) Gene expression profiles of EGFR and NRG1, and AUC drug response profiles of erlotinib and dacomitinib, over the topological representation (n=44 biologically independent samples). (d) Drug response assessment of EGFR inhibitors with shRNA-mediated knockdown of NRG1 or NT (non-target). Cell viability for each dose was normalized to shNRG1 or NT transduced cells only. (n=10 independent experiments with 3 technical replicates) (e) Comparisons of cancer pathway activities between two EGFR-altered GBM subgroups that were most sensitive and most resistance. We adopt the enrichment score derived from ssGSEA analysis as assessment. (f) Drug response assessment of EGFR inhibitors with PI3K-AKT-mTOR (PAM) inhibitors or DMSO. Cell viability for each dose was normalized to PAM or DMSO treated cells only. Mean AUC value for 4 PAM inhibitors (BYL719, BKM120, BEZ235, and AZD2014) was plotted (n=8 independent experiments with 3 technical replicates). P-values: c, pearson correlation test. P-values: d,f, two-sided wilcox rank sum test. Horizontal lines within the violin plot represent 0.25, 0.50, and 0.75 quantiles.
Integrative multi-layer data analysis reveals molecular determinants that dictate drug sensitivity to panobinostat
Prediction of drug response is a complex process which often depends on multiple variables including genetic and transcriptomic profiling of tumor 16,23,48. Given such extensive diversity, we next applied dNetFS (Diffusion kernel based Network method for Feature Selection of drug sensitivity), a regression model-based analysis to uncover cooperative interactions among multiple layers of variables90. dNetFS integrates protein-protein interaction network, and prior knowledge of drug-targets interaction to prioritize genetic and gene expression features that are potentially associated with drug response (Figs. 4a and Supplementary Fig. 12a). Using dNetFS, we detected previously known associations between EGFR, FGFR, CDK4/6, KIT and MET alterations and drugs that directly target these molecules (Supplementary Fig. 12b). In addition, we also found that PDCs harboring EPHB4 mutation showed substantial sensitivity to EGFR inhibitors including Afatinib, Dacomitinib and AEE788. EPHB4 gene encodes for receptor tyrosine kinase, EphB4, which governs tumor progression and portrays dismal prognosis across various cancer types91–95. Our result suggests that EPHB4 mutation could also confer increased sensitivity to EGFR target therapies, for which its underlying mechanism needs to be further evaluated.
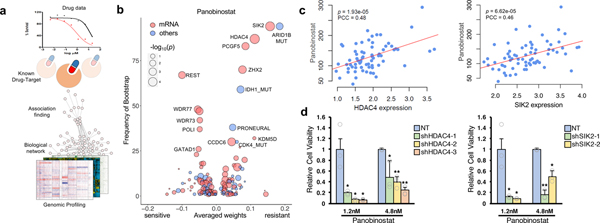
Figure 4. Genomic and transcriptomic correlates of panobinostat sensitivity.
(a) Schematic overview for dNetFS (Diffusion kernel based Network method for Feature Selection of drug sensitivity). In brief, dNetFS integrates genomic/pharmaceutical data, protein-protein interaction network, and prior knowledge of drug-targets interaction to prioritize genetic and gene expression features of PDCs that predict drug response. (b) Predictive features of panobinostat response identified by the dNetFS are plotted for their frequency and effect size. Associations are colored in red for expression features and blue for others. Node size is proportional to the single drug–feature linear correlation. (c) Scatter plot showing linear correlation between panobinostat AUC and HDAC4 expression (left panel) and SIK2 expression (right panel) (n=69 biologically independent samples). The correlation coefficient and the P values were obtained using pearson correlation test. (d) Drug response assessment of panobinostat (1.2nM or 4.8nM) with shRNA-mediated knockdown of HDAC4 or NT (non-target) (left panel) and SIK2 or NT (right panel). Cell viability for each dose was normalized to sole transduced cells only. Data are mean ± s.d. of n=3 technical replicates. Experiments were repeated three times with similar results. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, two-tailed t-test.
We further discovered a group of molecular predictors to panobinostat (HDAC), a drug that has rarely been analyzed in pharmacogenomics studies (Fig. 4b and Supplementary Fig. 12c)96–98. We identified high expression of RE1-Silencing Transcription factor (REST) confers cellular vulnerability to panobinostat, consistent with previous notion that HDAC inhibitors could be potentially employed as therapeutic agents against REST-positive tumors99. Furthermore, mutations on IDH1 and ARID1B, as well as high expression of Zinc finger and homeobox 2 (ZHX2: a transcriptional repressor), polycomb group ring finger 5 (PCGF5: a chromatin modifier), HDAC4 (a transcriptional repressor and class IIa HDAC) and salt inducible kinase 2 (SIK2: an upstream regulator of class IIa HDACs) were significantly associated with resistance to panobinostat (Fig. 4b, Supplementary Fig. 12c and Supplementary Table 12)100–103. Interestingly, although panobinostat is a non-selective inhibitor for HDAC, transcriptome expression of HDAC4 and SIK2, class IIa HDACs and upstream regulator of class IIa HDACs, respectively, were highly correlated with its resistance (Fig. 4c). Consistently, shRNA-mediated knockdown of HDAC4 or SIK2 conferred increased sensitivity to panobinostat (Fig. 4d and Supplementary Fig. 12d). Panobinostat was reported to confine HDAC4 at the cytoplasm104. Cytoplasmic HDAC4, which also could be induced by SIK2 activation, may attenuate the sensitivity to panobinostat by previously proposed mechanisms, such as activation of HDAC-MAPK-activator protein-1 (AP-1) signaling axis or hypoxia inducible factor 1-alpha (HIF-1α) functions100,103–106. These associations represent candidate genomic markers of drug sensitivity which may be useful for future biomarker-adaptive clinical trial design.
Genomic correlates of EGFR inhibitor sensitivity in EGFR-altered subgroup
Clinical response to molecular targeted drugs is likely to depend on the multiplicity of biological processes in tumor cells, even in subpopulations harboring alterations on the corresponding target molecules107–109. Genomic aberrations of EGFR, including copy number amplification, mutation, and structure variations, largely contribute to glioma malignancy in over 50% of the patients1,110. However, targeting EGFR has shown disappointing clinical outcomes due to various resistance-associated biological mechanisms48,111–114. Therefore, we analyzed pharmacological response of 10 EGFR inhibitors on 49 glioma PDCs harboring EGFR alterations, including copy number amplification (n=48), mutations (n=10), vIII (n=13) and fusions including EGFR-SEPT14 (n=4) (Fig. 5a). All 10 EGFR inhibitors showed similar activities across GBM PDCs: A subset of tumor cells that are sensitive to a particular EGFR compound would more likely to respond to other EGFR inhibitors, and vice versa (Supplementary Fig. 10b). EGFR-altered tumors clustered into two major groups based on cellular response to EGFR inhibitors. We next applied dNetFS and TDA to investigate the molecular correlates of drug sensitivity to EGFR inhibitors (Figs. 5b, 5c and Supplementary Fig. 13). As expected, EGFR transcriptional expression, mutations, fusions conferred sensitivity to multiple EGFR inhibitors (Supplementary Table 13). Strikingly, neuregulin 1 (NRG1) transcriptome expression emerged as a robust hit against cellular resistance to a variety of EGFR inhibitors. We further observed similar unresponsiveness behavior to multiple EGFR inhibitors via topological data analysis (Fig 5c and Supplementary Fig. 13). To determine whether NRG1 promotes cellular resistance to EGFR inhibitors, we evaluated proliferative kinetics of EGFR-amplified PDCs in response to 50 ng/ml exogenous NRG1 protein. As suspected, NRG1 treatment induced drug resistance to EGFR inhibitors, compared to the vehicle-treated cells (Supplementary Fig. 14a). Furthermore, silencing of NRG1 through either shRNA-mediated knockdown or neutralizing antibody demonstrated sensitizing effects to EGFR inhibitors, further supporting our observation that NRG1 expression potentially dictates cellular response to EGFR inhibitors (Fig. 5D and Supplementary Fig. 14b and 14c). NRG1 has been reported to enhance survival and proliferation of cancer cells including GBM115,116, and elevated expression of NRG1 was associated with increased sensitivity to EGFR inhibitors through activation of HER3 kinases in HER2 non-amplified cancer cells117. Our results, however, support that NRG1 promotes cellular resistance to EGFR target therapy in a subpopulation of EGFR-altered tumors, potentially due to triggering hetero-dimerization of EGFR and HER3, as previously reported118,119.
To identify cellular signaling pathways that influence pharmacological sensitivity/resistance to EGFR target therapy, we performed ssGSEA analysis between previously identified two major EGFR-altered subgroups (Supplementary Fig. 10b). Interestingly, activation of Myc signaling pathways conferred superior sensitivity to all 10 EGFR inhibitors, while upregulation of KRAS and/or PI3K/AKT/mTOR signaling pathways was associated with resistance to a subset of EGFR-targeting drugs (Fig. 5e). Our results provide a potential drug evasion process where sustainable activation of RAS-MEK or PI3K-AKT signaling molecules could prevent cell death from EGFR inhibition in EGFR-driven tumors108,120. Therefore, we conducted two-drug combinational strategy of applying PI3K-AKT-mTOR (PAM) inhibitors with EGFR compounds, in EGFR-altered tumors. Notably, combining PAM inhibitors significantly increased sensitizing effects of EGFR inhibitors (Fig. 5f and Supplementary Fig. 14d), highlighting potential combinatorial implementation of PAM inhibitors to overcome EGFR-mediated therapeutic resistance 121–124.
Clinical feasibility of patient-centric drug screening-guided therapy
The pharmacogenomic mapping using PDCs have identified several genetic aberrations which may potentially guide in biomarker-driven clinical trials. As demonstrated in recent studies, most refractory cancers have multiple genomic aberrations125, rending optimization of genome-based treatment difficult for each patient. To translate our ex vivo analysis into a clinical framework, we compared PDC-based drug sensitivities with clinical response in retrospective clinical studies.
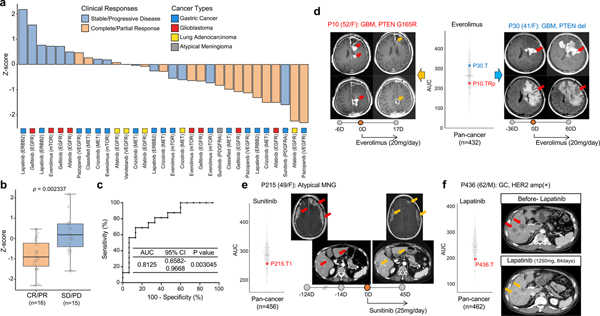
To this end, we explored whether normalized AUC (z-score in pan-cancer AUC dataset, n=462) of drugs targeting EGFR, VEGFR, PI3K/mTOR, MET, or PDGFRA could be used to predict clinical response of 31 patients across 4 major cancer types, including GC (n=17), GBM (n=8), lung adenocarcinoma (n=5), and atypical meningioma (n=1). We discovered that majority of the PDCs with a z-score of less than −0.5, indicating membership in the top ~30% sensitivity to a particular drug, predicted partial or complete response with average duration of 6 months (+/− 3 months) (Fig. 6a)126. In concordance to the clinical outcome, the z-scores for drugs derived from drug sensitivity analysis using patients’ progeny PDCs demonstrated considerably lower values when compared to the values in clinically resistant tumors (Fig. 6b). Next, we evaluated the accuracy of the z-score based drug sensitivity profile as a biomarker for predicting clinical responses by applying receiver-operating characteristic (ROC) curve analysis127. When we integrated z-score and clinical responses, our in vitro drug sensitivity of PDCs (n=31) demonstrated high concordance rate to clinical response (AUC=0.8125, p=0.003045; where AUC=0.5 means the chance discrimination as a diagnostic accuracy)128.
Figure 6. Clinical feasibility of PDC drug screening-guided precision oncology.
(a) Bar graph represents normalized AUCs (Z-scores from pan-cancer dataset) of indicated drugs (n=31 biologically independent samples). Clinical responses were determined according to RECIST. Multiple-target drugs are classified based on corresponding representative genomic targets. (b) Representative box plot of (a). CR (complete response); PR (partial response); SD (stable disease); PD (progressive disease) per RECIST criteria. Box plot spans from the first to third quartiles and the whiskers represent the 1.5 interquartile range. (c) Receiver operating characteristic (ROC) curve was plotted by the sensitivity (%) and 100-specificity (%) values for predicting clinical response rate using z-scores in (a). (d-f) T1-weighted contrast enhanced (T1CE), T2-flare magnetic resonance images (MRI) or computed tomography (CT) images for indicated patients before and after drug treatment were demonstrated. Circles on the bar represent days obtaining presented images, where 0D refers the day starting drug treatment. Negative (−) or positive (+) D represents days before or after treatment, respectively. Red arrows indicate measurable or progressed tumors, and orange arrows represent partial response. Vertical scattered plot for AUCs of indicated drugs in pan-cancer AUC reference dataset for the indicated drug. AUCs of PDCs isolated from the illustrated patients were highlighted. P-value: b, two-sided wilcox rank sum test. P-values: c, two-sided binomial exact test.
Here, we demonstrated examples for clinical concordance of drug response prediction using the progeny PDCs’ drug sensitivity. Two GBM patients had been treated with gefitnib, EGFR inhibitor, based on presence of EGFR amplification with concomitant point mutations, as well as PTEN deletion (Supplementary Fig. 15a and 15b)129. Despite such promising genomic indications, neither patient responded to the drug treatment and their clinical outcomes were consistent with our patient-centric drug sensitivity analysis. Furthermore, multi-centric GBM patient revealed notable concordance between clinical outcome and drug screening results13,130,131. EGFR amplification with concurrent EGFR mutations including vIII was only found in right frontal tumor, and showed partial response on the treatment of afatinib (EGFR), while left frontal tumor did not respond to this therapy (Supplementary Fig. 15c)13,113,132. These clinical results were well correlated with the drug sensitivity profiles of the corresponding PDCs in response to afatinib. Another instance for drug-screening correlation with clinical sensitivities was demonstrated in GBM patient (P10), harboring PTEN mutation, who showed short term partial response to everolimus (mTOR) treatment, while another GBM patient (P30), with PTEN deletion, did not respond to such therapeutic approach (Fig. 6d and Supplementary Fig. 15d and 15e)133. The AUC of P10-derived tumor cells on the treatment of everolimus was notably lower compared to that of P30-derived tumor cells, highlighting accuracy of our drug screening results in relevance to clinical responses. P215, patient who has been diagnosed with atypical meningioma (MNG), showed remarkable in vitro sensitivity to treatment of sunitinib (VEGFR, PDGFR, etc), and accordingly presented partial response on both meningeal and abdominal metastatic lesions (Fig. 6e and Supplementary Fig. 15f)134. HER2 amplification is found in 13~23% of GCs and phase III trials with anti-HER2 targeted agents have shown controversial results135–137. PDC progeny from a GC patient (P436) with HER2 amplification showed profound in vitro sensitivities to lapatinib (HER2, EGFR), which was consistent to clinical partial response (Fig. 6f). Although larger scale clinical study is essential to determine clinical utility of PDCs in predicting clinical response, our findings suggest that integrative analysis of genome and PDC-based drug screening be a potential tool for patient enrichment trial.
These accumulating evidences collectively propose clinical feasibility of combined patient-centric drug sensitivity screening with genomic profiling to facilitate prediction of clinical response to targeted agents.
DISCUSSION
Precision oncology aims to provide personalized treatment option through identifying and targeting genomic and molecular aberration of the individual patient tumor. Such approach holds potential to greatly improve clinical outcomes3. Rapidly accumulating genomic data, spearheaded by TCGA and multiple global collaborative efforts, including the International Cancer Genome Consortium (ICGC), have painted a comprehensive portrait of tumor genome atlas1,8,138. Large-scale drug screening efforts using human cancer cell-line models have begun to establish a collection of gene-drug associations and uncovered potential molecular markers, predictive of therapeutic response16,17,20,21,23. However, conventional cell-line models failed to demonstrate accurate genomic representations of the parental tumors, which rarely lead to successful clinical implementation. Therefore, systematic evaluation and clinical application of patient-derived resources to assess genetic variations that underlie pharmacological drug response would make breakthrough points for precision treatment in cancer patients.
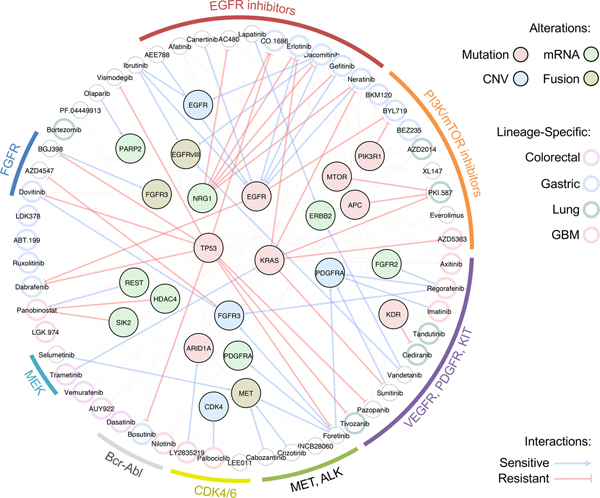
In present study, we demonstrated feasibility and clinical relevance of PDC-based drug screening system for pharmacogenomics analyses. We reported therapeutic landscape of 60 molecular-targeted compounds on 462 PDCs across 14 cancer types, revealing lineage-specific drug associations, such as GCs and PI3K inhibitors. We also integrated cancer-driven genomic variations including somatic mutations, copy number alterations and/or transcriptome expressions. Through large-scale pharmacogenomics analyses, we suggested therapeutic options for KRAS-mutant tumors and identified molecular determinants, such as HDAC4 and SIK2, that dictate panobinostat sensitivity. HDAC4, class IIa HDACs, is preferentially expressed in heart, brain and muscles, inferring a potential role on brain pathogenesis including brain tumor and acquired/intrinsic resistance to chemotherapies100. We proposed potential functional role of HDAC4 and its regulator SIK2 on inducing resistance to the HDAC inhibitor, of which the underlying mechanisms should be further evaluated. Furthermore, we identified molecular predictors of intrinsic resistance to EGFR inhibitors, such as NRG1, in EGFR-altered tumors. These results propose potential combinatorial strategy of combining anti-NRG1 therapy to overcome unresponsiveness to EGFR targeted treatment 139. Moreover, we suggested EGFR alterations, including genomic amplification and structure variation, vIII, as a novel genomic biomarker for a BTK inhibitor, ibrutinib, in GBM patients. Ibrutinib previously showed notable sensitivity to EGFR mutant tumors, such as L858R, Del19 and T790M in NSCLC cells67,140, which are intracellular domain events and critically different from the EGFR alterations that are often found in GBM. As ibrutinib has been suggested to be able to penetrate the blood-brain barrier141, our results provide an important groundwork for clinical feasibility of repurposing ibrutinib for EGFR-altered GBM patients. Collectively, our findings demonstrate significant lineage-specific or molecular correlates of diverse drug agents (Fig. 7).
Figure 7. Schematic illustration of major lineage-specific and genomic associated drug interactions.
Molecular targeting agents are clustered based on drug family classification and connected to various genomic alterations including mutation, mRNA expression, copy number variation (CNV), and fusion. Edges represent sensitive or resistant gene-drug interactions. Lineage-specific drug associations are highlighted in each drug node based on specific lineage type.
Lastly, we demonstrated clinical concordance between PDC-based drug sensitivity and clinical responses, highlighting promising opportunity for drug screening-guided personalized treatment. Although the evaluation of clinical sensitivity was based on early radiological tumor responses, initial response rate has largely been accepted to be significantly correlated with patient survivals142,143. Furthermore, as prior clinical studies have demonstrated, predicting therapeutic efficacy of a targeted compound based on genomic profiling alone is a difficult task as tumors often harbor multiple genetic aberrations144. Therefore, we provide an alternative avenue for predicting clinical response via integrative pharmacological drug response with genomic characterization. As prospective validation, we are currently enrolling GC patients using PDC models to screen for drug sensitivity and to facilitate optimized clinical trials based on integrative genome-drug mapping analysis (clinicaltrials.gov NCT#03170180).
Collectively, our systematic method, using a unique drug sensitivity dataset of molecularly annotated patient-derivatives represents a significant conceptual advance toward precision oncology treatment for cancer patients. In addition, we have developed an interactive, publicly available web resource (cDx, Cancer-Drug eXplorer; cancerdrugexplorer.org, temporary user and password: cdx) for exploration of our pharmacogenomic dataset.
ONLINE METHODS
Tumor specimens and their derivative cells
After receiving informed consent, tumor specimens or malignant ascites with corresponding clinical records were obtained from patients undergoing surgery or paracentesis at Samsung Medical Center (SMC) in accordance with its Institutional Review Board (IRB file #201004004, #201107089, #201310017 and #201310072). This work was performed in compliance with all relevant ethical regulations for research using human specimens. Cells from malignant effusions were collected by centrifugation at 300g for 10 min, followed by washing with Dulbecco’s phosphate-buffered saline (DPBS, Thermo Scientific). Portions of the surgical samples were enzymatically dissociated using Liberase TM (Roche). Patient-derived tumor cells (PDCs) were cultured in neurobasal or DMEM/F12 medium with N2 and B27 supplements (0.5× each; Thermo Scientific) and growth factors based on human recombinant basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF; 20 ng/ml each; R&D Systems)25,27. Short tandem repeat (STR) analysis (AmpFlSTR Identifiler, Applied Biosystems) was performed to verify corresponding normal blood. Universal Mycoplasma Detection Kit 9ATCC 30–1012K) was used to confirm absence of mycoplasma contamination.
Whole- or targeted exome sequencing (WES)
An Agilent SureSelect kit was used to capture the exonic DNA fragments. An Illumina HiSeq 2000 instrument was used for sequencing and generation of 2 × 100-bp paired-end reads. FASTQ files were aligned to the human genome assembly (hg19) using Burrows- Wheeler Aligner (version 0.6.2)145. Before further analysis, the initially aligned BAM files were subjected to preprocessing that sorted, removed duplicated reads, locally realigned reads around potential small indels, and recalibrated base quality scores using SAMtools, Picard (version 1) and Genome Analysis ToolKit (GATK, version 2.5.2)146. In addition to Whole-exome sequencing, we also performed targeted sequencing of full coding exons of 80 commonly mutated cancer genes (CancerSCAN™, Supplementary Tables 1 and 2)31,33. Tissues or PDCs from malignant gliomas were analyzed with massive parallel targeted sequencing platform, covering exons of cancer-driven and/or glioma-associated genes (GliomaSCAN™, Supplementary Tables 1 and 3)1,9,35. Read alignments and conventional preprocessing were similarly conducted as the WES data.
Mutation calling:
To identify somatic mutations from whole-exome sequencing data for tumor samples with matched blood control, we applied variance calling software SAVI2 (statistical algorithm for variant frequency identification), based on the empirical Bayesian method36. Somatic mutations are identified based on the report of SAVI2, and following four additional criteria: 1) Not annotated as synonymous variant, intragenic variant, intron variant; 2) Not annotated as common SNP; 3) Mutation allele frequency of >5% in the tumor sample; 4) Altered read depth < 2 in the matched normal. For targeted DNA sequencing, we only focused on variant callings within the genomic region covered by the protocol. For samples without matching normal control, only the variants with cosmic id were considered.
Copy number alteration:
Excavator was used to generate estimated copy number alterations in a tumor specimen in comparison with its matching blood. For each gene, we calculated copy number as 2x+1, where x is the segmentation mean, which is the log2 (fold change) value for each tumor divided by the normal control. The gene was labeled as ‘amplified’ when the x value is above 0 and ‘deleted’ when it was below 0. The copy number estimation was only conducted on Whole-exome sequencing data.
RNA sequencing
Reads were aligned to the human reference using STAR 2.4.0b147. The genome index was generated using annotation file of the human genome (GRCh37), version 19 (Ensembl 74) from GENCODE. For gene expression analysis, featureCounts from package ‘Subread’ was adopted to calculate RPKM values, followed by log2 transformation, and quantile normalization. Genes with low expression level across the cohort are removed.
Exon skipping & gene fusion analysis:
Chimerascan was used to generate a list of candidate gene fusions using RNASeq data 148. To reduce the false positive rate and nominate potential driver events, we applied the Pegasus annotation and prediction pipeline38. Only previously reported in-frame, highly expressed fusions were reported in this manuscript. To examine rearrangement of EGFR, we applied prada-guess-if from the PRADA package. PRADA is an RNA-seq analysis pipeline developed at MD Anderson149. The transcribed allelic fraction of EGFRvIII was defined as the fraction of junction reads joining exon 1 to exon 8.
A list of major alterations including copy number changes, point mutations and fusion genes was provided in Supplementary Table 2.
PDC-based chemical screening and analysis
Tumor sphere forming PDCs, cultured in serum-free medium, were dissociated into single cells and seeded into 384-well plates (500 cells/well) with technical duplicates. PDCs were treated with 60-drug library, targeting major oncogenic signaling molecules (SelleckChem), in a fourfold and seven-point serial dilution series from 4.88 nM to 20 μM. Two same PDCs and two same cell-lines have been screened every month to confirm preservation of chemical activities of our drug library. After 6 days of incubation at 37°C in a 5% CO2 humidified incubator, cell viability was accessed using adenosine triphosphate (ATP) monitoring system based on firefly luciferase (ATPLite™ 1step, PerkinElmer) and estimated by EnVision Multilabel Reader (PerkinElmer)13. Relative cell viability for each dose was obtained by normalization with dimethyl sulfoxide (DMSO) per each plate. Screening plates were subjected to quality control measurement using Z-factor score, comparing both negative and positive control wells150. Dose response curves (DRCs) were fitted using GraphPad Prism 6 (GraphPad): Best-fit lines and the resulting IC50s were calculated using GraphPad: [log(inhibitor) vs. response – variable slope (four parameters)]. Y=Bottom + (Top-Bottom) / (1+10^((LogIC50-X)*HillSlope)). The area under curve (AUC) for each DRC was calculated using trapezoidal method, ignoring regions defined by fewer than two peaks55. The AUC values from non-convergent or ambiguous DRC were excluded in all analysis.
Transcriptome expression comparison between primary tumors and patient-derived cells
We selected 24 GBM samples with available RNA sequencing on both primary tissues and matching PDCs. In addition to the normalization described above, we further calculate z score for each gene within the tissue/PDC cohort to eliminate batch effects. Then, for each gene, we applied spearman correlation as a score to evaluate its expression similarity. Based on its cutoffs, we generate different lists of genes for comparison (Supplementary Fig. 4). Finally, we calculated pairwise spearman correlation coefficient between the normalized expression values obtained for primary tissues and PDCs.
Molecular profile comparisons among PDC, tumor tissue and conventional cancer cell-lines on glioma samples
To evaluate whether PDC could provide a better representation of primary tumor tissue, we compared recurrent mutation and gene expression profiles of our PDC system, TCGA and two publicly available pharmacogenomics resources using traditional cancer cell-line models: The Cancer Cell Line Encyclopedia (CCLE) and the Genomics of Drug Sensitivity in Cancer (GDSC) datasets. The comparison was conducted on GBM samples only.
For somatic mutation profiles, 33 GBM PDCs with available GliomaScan data and normal controls were selected to call somatic mutations. Mutation reports on TCGA GBM cohort (n=287) were downloaded from cBioPortal. For SGDC, mutation information on GBM cell lines (n=34) was obtained from the supplementary table of Iorio et al., 2016. Binary calls for mutation data was downloaded from CCLE website, and 68 GBM cell lines were extracted. We selected a list of 27 marker genes, which has mutation occurrence > 5 patients on TCGA’s report, and also covered by the protocol of GliomaScan. Next, we calculated and compared the mutation frequency of selected genes on each cohort.
To compare the transcriptome similarities, 24 GBM PDCs with available RNA sequencing were selected. For GBM samples from TCGA, we used the pan-cancer normalized gene expression data (n=172) downloaded from UCSC Xena. RMA normalized expression data of 35 GBM cell lines were downloaded from the Sanger’s website. For CCLE, expression data of 69 GBM cell lines was downloaded from the CCLE website. Then, expression matrixes of 4 cohorts were merged on overlapped genes, and values were quantile normalized on sample level. Next, to evaluate the transcriptome similarities of two different cohorts, we calculated the spearman correlations of mRNA expression for every pair of samples coming from the two different cohorts. Lastly, p-value was generated from Kruskal-Wallis test to estimate the statistical differences of the correlation distributions among multiple comparisons.
Lineage-specific drug identification
Drug response profiles were directly compared between a given tissue type and all other PDCs. Wilcox rank sum test was adopted to show the statistical significance, followed by Benjamini-Hochberg procedure for multiple tests correction. We also applied Topological data analysis for the identification of tissue-specific drug sensitivity.
Topological data analysis (TDA)
Topological representations were constructed using ‘Mapper’ algorithm, as implemented by Ayasdi Inc. Open-source implementations of this algorithm are also available. The output of ‘Mapper’ is a low-dimensional network representation of the data, where nodes represent sets of drugs or PDCs. In the manuscript, we have constructed three topological networks:
The TDA network of 462 PDCs was constructed using Pearson correlation as a similarity measure between AUC profiles across 60 drugs. As ‘Mapper’ parameters, we used two-dimensional MDS projections as lenses with resolution 30 and gain 3 (equalized). Each node of the network represents a set of PDCs with similar AUC profiles. A PDC can appear in several nodes, and two nodes are connected by an edge if they have at least one PDC in common. We then computed mean adjusted AUCs for each node, for each drug: AUCs larger than 300 for a PDC were taken to be 300, and AUCs smaller than 200 were taken to be 0 to emphasize strong drug sensitivity. For each node, we also computed the fraction of PDCs derived from a particular cancer type. The mean adjusted AUC and cancer type fraction were then used both to color nodes and to compute Pearson correlation between drug effect and cancer type. P values of these correlations are used to identify significant relationships between cancer type and drug effect.
The TDA network of 44 EGFR-altered PDCs was constructed based on Euclidean (L2) metric as a similarity measure between gene expression profiles. MDS projection lenses with resolution 12 and gain 4 (equalized) were used as ‘Mapper’ parameters. Each node represents a set of EGFR-altered PDCs with similar gene expression profiles. A PDC can appear in several nodes, and two nodes are connected by an edge if they have at least one PDC in common. We then computed mean AUCs for each drug and mean gene expression for every gene in every node of the network. The mean AUC and gene expression were then used both to color nodes and to compute Pearson correlation between drug effect and gene expression. P values of these correlations are used to identify significant relationships between gene expression and drug effect.
The TDA network of 60 drugs is constructed based on Pearson correlation as a similarity measure between their AUC profiles across 462 PDCs. 2-dimensional MDS projection lenses with resolution 15 and gain 3 (equalized) were used as ‘Mapper’ algorithm parameters. Each node represents a set of drugs with similar AUC profiles. A drug can appear in several nodes, and two nodes are connected by an edge if they have at least one drug in common. Every drug family is assigned a specific color. Mean RGB values are computed for every node to identify the drug family that dominates the node.
Oncogenic signaling pathways activation assessment
To determine the relative activity of cancer pathways between two cohorts, ssGSEA (version gsea2–2.2.1) was applied using patients’ gene expression profiles. To eliminate batch effects, we normalized gene expression by calculating Z score within each cohort. Then, for each sample, we ranked all genes on the basis of their expression values to create a .rnk file as input for the software GseaPreranked. The enrichment score was computed for oncogenic pathways as the assessment. Cancer-related pathway signatures were downloaded from MSigDB. PIK3CA pathway was compared between gastric and glioblastoma of TCGA patients, using the pan-cancer normalized gene expression data downloaded from UCSC Xena. We randomly select 100 TCGA patients from each cohort for ssGSEA analysis. We also compared the activation of signaling pathways between two EGFR-altered GBM subgroups, which demonstrated two drastic responses to EGFR therapy. We first selected 49 GBM PDCs with EGFR alterations. Then, for each EGFR inhibitor, we ranked the drug AUC values for these 49 PDCs, and selected top/bottom 10 ranked PDCs as the resistance/sensitivity subgroups for comparison.
Pan-cancer analyses uncover statistically significant associations between drug sensitivity and genomic alteration.
A pre-selected list of major cancer-driver alterations, including copy number changes, somatic mutations and gene fusions, were considered to identify drug response interaction (Supplementary Table 2). For each drug, sensitivity data were compared between pan-cancer subgroups based on presence or absence of the selected genomic alteration using Wilcoxon rank-sum test. Samples with unknown status of given alteration were excluded from the analysis.
Pharmacogenomic Modeling of Drug Sensitivity.
To predict drug sensitivity profiles based on cooperative interactions among multiple layers of variables, we applied dNetFS (Diffusion kernel based Network method for Feature Selection of drug sensitivity), a regression model-based algorithm (Supplementary Fig. 14) 90. dNetFS integrate protein-protein interaction network, and prior knowledge of drug-targets interaction to prioritize genetic and gene expression features of PDCs that predict drug response.
Generally, our dNetFS pipeline consists of two parts: 1) network propagation to select potential predictive features with a strong network connection to known drug targets. 2) Fitting an elastic-net model to generate a robust list of molecular predictor.
The input variables of the dNetFS includes the pre-selected list of cancer-driver alterations (mutation, CNV, fusion), tumor subtyping and mRNA expression. Here, we only employed expression of 7262 genes, which showed high consistency between primary tissue and matched PDC with a correlation of larger than 0.4 (Figures S5). But still, this number is much larger than the total feature number of driver alterations, leading to an unbalanced feature pools. Also, it falls into the standard sparsely selection problem, where the number of features is far greater than the number of observations. Thus, before any regression model based shrinkage, dNetFS performed a pre-selection on the expression features based on a protein-protein interaction network. We adopted the STRING human network (version 10.0) and only kept the most confident 10% of interactions 151. For a given drug, we first obtained the known drug targeted genes from DGIdb, which is a comprehensive collection of available information on the “actionable” genome 152. Then, a diffusion kernel was applied as a measure to capture information flow on the network to evaluate the closeness between known drug targets and other proteins 153.
It represents the continuous time limit of a lazy random walk, where H is negative Laplacian matrix defined on the adjacency matrix from STRING network. We only selected the top 500 closest genes as candidate features. And the hypothesis is that, correlation-based sensitivity features might be close to the known targets in terms of sharing a common pathway or carrying out the same function.
Notably, one parameter γ was involved in the above diffusion kernel process. Next, we applied an elastic net regression algorithm 154, using only mRNA expression of genes filtered by the network propagation. Then, 10-fold cross validations were used to optimize parameter γ. The final list of gene expression features was determined by diffusion kernel process with optimized γ. γ was screened using 30 values of γ = 0.75^n with n = 1,2,…,30.
Next, we trained the standard elastic net regression (R package: glmnet_2.0–5), combing all alterations, tumor subtyping, and pre-selected mRNA expression as the input features. Given one drug and N PDCs, we used y = (y1, y2, … yn) represents drug sensitivity of n PDCs, and xi = (xi1,..., xip), (i = 1,…, p) indicates p features of ith PDC, the elastic net regression is to solve the following optimization problem:
α controls the relative balance of the L1 and L2 penalty terms, while λ controls the overall penalty level of the regularized term. Similar with γ, α was optimized by 10-fold cross validations, but using all features. α was screened using 50 values with α ⊰ [0,1] equally spaced. For each model fitting, we used the function cv.glmnet, with its optimized λ value provided from the function.
After parameter optimization, we adopt bootstrapping strategy for 100 times to obtain a robust evaluation of the predictive power of features. During each bootstrapping, we randomly select 80% of PDCs with 80% of the features to fit the elastic net with above optimized α. For each feature, the time of appearances (non-zero fitting coefficient) out of the 100 bootstrappings, together with the average of its non-zero weights are used as its final assessment of predictive ability.
Limiting dilution assay
Single-cell suspensions were plated into 96-well plates at 1–250 cells per well. Cells were incubated at 37°C for one to two weeks under serum-free conditions supplemented with EGF and basic FGF (20 ng/ml each), with or without drug treatment. Each well was examined for formation of tumor spheres. Statistical significance was accessed using Extreme Limiting Dilution Analysis (ELDA; Walter+Eliza Hall Bioinformatics)89
shRNA-mediated knockdown
shRNA lentiviral clones for SIK2 (TRCN0000037495 and TRCN0000037497 for clone#1 and #2, respectively), HDAC4 (TRCN0000004832, TRCN0000314667 and TRCN0000314665for clone #1, #2 and #3, respectively) and NRG1 (TRCN0000058303) were purchased from Sigma-Aldrich. Lentiviruses were produced from 293FT cells with packaging mix (ViraPower Lentiviral Expression Systems, Thermofisher) and concentrated by ultracentrifugation. Lentiviral particles were transduced into corresponding tumor cells and subjected for purmycin selection.
Patient-derived xenograft (PDX) animal models and drug treatment
All animal experiments were approved by the Institutional Review Board of the SMC and performed according to the guidelines of the Animal Use and Care Committees at SMC. A GBM orthotopic xenograft model was established as reported previously26. In brief, patient GBM cells (N464; 2X105/mouse) were dissociated, resuspended in 5 μl of Hank’s balanced salt solution (HBSS, Thermo Scientific), and stereotactically (2 mm left and 1 mm anterior of the bregma, 2 mm deep from the dura) injected into the brains of Balb/c nude mice (6~8 week-old female, Orient Bio Inc.). After randomization, vehicle (0.5% methyl cellulose, SigmaAldrich) or Ibrutinib (50 mg/kg/day; ChemieTek) were treated orally for 5 consecutive days per cycle155. Kaplan-Meier survival and immunohistochemical analysis were performed under blinded inspection.
Immunohistochemistry
Paraformaldehyde-fixed tissues were embedded in paraffin and sectioned to a thickness of 4 μm. Sections from the brains of xenograft tumor-bearing mice were stained with hematoxylin and eosin (H&E; Sigma-Aldrich). Paraffin-embedded slides were subjected to antigen retrieval in 10 mM sodium citrate buffer. For immunohistochemistry, tissue sections were incubated with primary antibodies against pEGFR (#2234), pAKT (#4060), and pERK (#9102, all from Cell Signaling Technology), followed by the appropriate biotinylated secondary antibody. Sections were then stained with 3,3-diaminobenzidine substrate and counterstained with H&E solution.
Radiological determination of tumor responses (RESCIST criteria)
All complete and partial responses were confirmed by repeated radiological evaluation including computed tomography (CT) scan or magnetic resonance imaging (MRI) per Response Evaluation Criteria in solid Tumors (RECIST) guideline version 1.1126.
Statistical analysis.
Statistical analyses were conducted by either pearson correlation coefficient test, t-test (two-tailed), wilcox rank sum test (two-sided), or binomial exact test (two-sided). Survival curves were estimated with the Kaplan-Meier method. For in vitro results, two to three independent experiments were conducted. Results are expressed as mean ± s.d. for the indicated number of observations. All statistical analyses were conducted and obtained using the GraphPad Prism software 6 or the R software (https://www.r-project.org).
Reporting Summary.
Further information is available in the Life Sciences Reporting Summary linked to this article.
URLs.
Picard, http://picard.sourceforge.net; cBioPortal, http://www.cbioportal.org/study?id=gbm_tcga#summary; CCLE, https://portals.broadinstitute.org/ccle/data/browseData?conversationPropagation=begin; UCSC Xena, https://xenabrowser.net/datapages/; Sanger, http://www.cancerrxgene.org/gdsc1000/GDSC1000_WebResources/Home.html; Ayasdi Inc., http://danifold.net/mapper, http://github.com/MLWave/kepler-mapper; MSigDB, http://software.broadinstitute.org/gsea/msigdb;
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant of the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3418). This work has been funded by NIH grants (R01 CA185486, R01 CA179044, U54 CA193313 and U54 209997) and NSF/SU2C/V-Foundation Ideas Lab Multidisciplinary Team (PHY-1545805) and Hong Kong RGC grants (N_HKUST601/17 and C6002–17G). The biospecimens for this study were provided by Samsung Medical Center BioBank.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no competing financial interests.
Data availability
All sequenced data have been deposited in the European Genome-phenome Archive (EGA) with accession code EGAS00001002407. Processed data and basic association analysis are publicly available through an interactive webportal (cDx, the Cancer Drug eXplorer, at cancerdrugexplorer.org).
REFERENCES
- 1.Brennan CW et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research, N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45, 1113–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamburg MA & Collins FS The path to personalized medicine. N Engl J Med 363, 301–4 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344, 783–92 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Chapman PB et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364, 2507–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Brien SG et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 348, 994–1004 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Loeb LA Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer 11, 450–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceccarelli M et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 164, 550–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubio-Perez C et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 27, 382–96 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Altman RB Predicting cancer drug response: advancing the DREAM. Cancer Discov 5, 237–8 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Geeleher P, Cox NJ & Huang RS Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol 15, R47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JK et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet 49, 594–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrell RA, McGranahan N, Bartek J & Swanton C The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–45 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Yates LR et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21, 751–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnett MJ et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barretina J et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoemaker RH The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 6, 813–23 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Basu A et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holbeck SL, Collins JM & Doroshow JH Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol Cancer Ther 9, 1451–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garnett MJ & McDermott U The evolving role of cancer cell line-based screens to define the impact of cancer genomes on drug response. Curr Opin Genet Dev 24, 114–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van de Wetering M et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio F et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 166, 740–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao H et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 21, 1318–25 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Galli R et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64, 7011–21 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Joo KM et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep 3, 260–73 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Lee J et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Lee JY et al. Patient-derived cell models as preclinical tools for genome-directed targeted therapy. Oncotarget 6, 25619–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie Y et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine 2, 1351–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanabur P et al. Patient-derived glioblastoma stem cells respond differentially to targeted therapies. Oncotarget 7, 86406–86419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park YH et al. Role of HER2 mutations in refractory metastatic breast cancers: targeted sequencing results in patients with refractory breast cancer. Oncotarget 6, 32027–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim SH et al. The implication of FLT3 amplification for FLT targeted therapeutics in solid tumors. Oncotarget 8, 3237–3245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo KH et al. Genomic Alterations in Biliary Tract Cancer Using Targeted Sequencing. Transl Oncol 9, 173–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song HN et al. Molecular characterization of colorectal cancer patients and concomitant patient-derived tumor cell establishment. Oncotarget 7, 19610–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki H et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47, 458–68 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Trifonov V, Pasqualucci L, Tiacci E, Falini B & Rabadan R SAVI: a statistical algorithm for variant frequency identification. BMC Syst Biol 7 Suppl 2, S2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magi A et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol 14, R120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abate F et al. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol 8, 97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemmon MA & Schlessinger J Cell signaling by receptor tyrosine kinases. Cell 141, 1117–34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gschwind A, Fischer OM & Ullrich A The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer 4, 361–70 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Nakada M et al. Aberrant signaling pathways in glioma. Cancers (Basel) 3, 3242–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joo KM et al. MET signaling regulates glioblastoma stem cells. Cancer Res 72, 3828–38 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Wen PY, Lee EQ, Reardon DA, Ligon KL & Alfred Yung WK Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro Oncol 14, 819–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filbin MG et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med 19, 1518–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen PY & Kesari S Malignant gliomas in adults. N Engl J Med 359, 492–507 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Ohka F, Natsume A & Wakabayashi T Current trends in targeted therapies for glioblastoma multiforme. Neurol Res Int 2012, 878425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puputti M et al. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in gliomas. Mol Cancer Res 4, 927–34 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Taylor TE, Furnari FB & Cavenee WK Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr Cancer Drug Targets 12, 197–209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snuderl M et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20, 810–7 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Szerlip NJ et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A 109, 3041–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cloughesy TF, Cavenee WK & Mischel PS Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol 9, 1–25 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Mellinghoff IK et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 353, 2012–24 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Fallahi-Sichani M, Honarnejad S, Heiser LM, Gray JW & Sorger PK Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat Chem Biol 9, 708–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jang IS, Neto EC, Guinney J, Friend SH & Margolin AA Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data. Pac Symp Biocomput, 63–74 (2014). [PMC free article] [PubMed] [Google Scholar]
- 55.Huang S & Pang L Comparing statistical methods for quantifying drug sensitivity based on in vitro dose-response assays. Assay Drug Dev Technol 10, 88–96 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Raub TJ et al. Brain Exposure of Two Selective Dual CDK4 and CDK6 Inhibitors and the Antitumor Activity of CDK4 and CDK6 Inhibition in Combination with Temozolomide in an Intracranial Glioblastoma Xenograft. Drug Metab Dispos 43, 1360–71 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Cen L et al. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro Oncol 14, 870–81 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroder LB & McDonald KL CDK4/6 Inhibitor PD0332991 in Glioblastoma Treatment: Does It Have a Future? Front Oncol 5, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicolau M, Levine AJ & Carlsson G Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc Natl Acad Sci U S A 108, 7265–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camara PG, Rosenbloom DI, Emmett KJ, Levine AJ & Rabadan R Topological Data Analysis Generates High-Resolution, Genome-wide Maps of Human Recombination. Cell Syst 3, 83–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rizvi AH et al. Applied Topology Delineates Developmental Progression with Single-Cell Resolution Nat Biotech In press(2017). [Google Scholar]
- 62.Bhattacharya B et al. Pharmacologic synergy between dual phosphoinositide-3-kinase and mammalian target of rapamycin inhibition and 5-fluorouracil in PIK3CA mutant gastric cancer cells. Cancer Biol Ther 13, 34–42 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Tapia O et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch 465, 25–33 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Ying J et al. The expression of the PI3K/AKT/mTOR pathway in gastric cancer and its role in gastric cancer prognosis. Onco Targets Ther 8, 2427–33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.TCGA. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yiin JJ et al. ZD6474, a multitargeted inhibitor for receptor tyrosine kinases, suppresses growth of gliomas expressing an epidermal growth factor receptor mutant, EGFRvIII, in the brain. Mol Cancer Ther 9, 929–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao W et al. Selective antitumor activity of ibrutinib in EGFR-mutant non-small cell lung cancer cells. J Natl Cancer Inst 106(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Byrd JC et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med 369, 32–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Misale S et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garassino MC et al. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann Oncol 22, 235–7 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Lievre A et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 66, 3992–5 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Belmont PJ et al. Resistance to dual blockade of the kinases PI3K and mTOR in KRAS-mutant colorectal cancer models results in combined sensitivity to inhibition of the receptor tyrosine kinase EGFR. Sci Signal 7, ra107 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Hutchinson L Targeted therapies: dasatinib sensitizes KRAS-mutant colorectal cancer tumors to cetuximab. Nat Rev Clin Oncol 8, 193 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Ku BM et al. BYL719, a selective inhibitor of phosphoinositide 3-Kinase alpha, enhances the effect of selumetinib (AZD6244, ARRY-142886) in KRAS-mutant non-small cell lung cancer. Invest New Drugs 33, 12–21 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Jing J et al. Comprehensive predictive biomarker analysis for MEK inhibitor GSK1120212. Mol Cancer Ther 11, 720–9 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Infante JR et al. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol 13, 773–81 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Hatzivassiliou G et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature 501, 232–6 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Blumenschein GR Jr. et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)dagger. Ann Oncol 26, 894–901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manchado E et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534, 647–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yeh JJ et al. KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Mol Cancer Ther 8, 834–43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun C et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep 7, 86–93 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Heinemann V, Stintzing S, Kirchner T, Boeck S & Jung A Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev 35, 262–71 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Cui J, Jiang W, Wang S, Wang L & Xie K Role of Wnt/beta-catenin signaling in drug resistance of pancreatic cancer. Curr Pharm Des 18, 2464–71 (2012). [DOI] [PubMed] [Google Scholar]
- 84.Yeung J et al. beta-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell 18, 606–18 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Nagaraj AB et al. Critical role of Wnt/beta-catenin signaling in driving epithelial ovarian cancer platinum resistance. Oncotarget 6, 23720–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ivanescu AM, Oprea M, Turbatu A, Colita A & Lupu AR Ibrutinib, a novel agent in relapsed or refractory chronic lymphocytic leukemia. Maedica (Buchar) 9, 217–8 (2014). [PMC free article] [PubMed] [Google Scholar]
- 87.Rushworth SA, MacEwan DJ & Bowles KM Ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med 369, 1277–8 (2013). [DOI] [PubMed] [Google Scholar]
- 88.Wang ML et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med 369, 507–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu Y & Smyth GK ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347, 70–8 (2009). [DOI] [PubMed] [Google Scholar]
- 90.Wang J, Kribelbauer J & Rabadan R Network propagation reveals novel genetic features predicting drug response of cancer cell lines. Current Bioinformatics 11, 8 (2016). [Google Scholar]
- 91.Munarini N et al. Altered mammary epithelial development, pattern formation and involution in transgenic mice expressing the EphB4 receptor tyrosine kinase. J Cell Sci 115, 25–37 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Kumar SR et al. Receptor tyrosine kinase EphB4 is a survival factor in breast cancer. Am J Pathol 169, 279–93 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang NY, Pasquale EB, Owen LB & Ethell IM The EphB4 receptor-tyrosine kinase promotes the migration of melanoma cells through Rho-mediated actin cytoskeleton reorganization. J Biol Chem 281, 32574–86 (2006). [DOI] [PubMed] [Google Scholar]
- 94.Ferguson BD et al. The EphB4 receptor tyrosine kinase promotes lung cancer growth: a potential novel therapeutic target. PLoS One 8, e67668 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pasquale EB Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer 10, 165–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai Y, Yan X, Zhang G, Zhao W & Jiao S The predictive value of ERCC1 and p53 for the effect of panobinostat and cisplatin combination treatment in NSCLC. Oncotarget 6, 18997–9005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee EQ et al. Phase II study of panobinostat in combination with bevacizumab for recurrent glioblastoma and anaplastic glioma. Neuro Oncol 17, 862–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grasso CS et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 21, 827 (2015). [DOI] [PubMed] [Google Scholar]
- 99.Taylor P et al. REST is a novel prognostic factor and therapeutic target for medulloblastoma. Mol Cancer Ther 11, 1713–23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Z, Qin G & Zhao TC HDAC4: mechanism of regulation and biological functions. Epigenomics 6, 139–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Plass C et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 14, 765–80 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Kawata H et al. Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem J 373, 747–57 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Walkinshaw DR et al. The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J Biol Chem 288, 9345–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Geng L et al. Histone deacetylase (HDAC) inhibitor LBH589 increases duration of gamma-H2AX foci and confines HDAC4 to the cytoplasm in irradiated non-small cell lung cancer. Cancer Res 66, 11298–304 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Geng H et al. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem 286, 38095–102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Choi MC et al. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell 47, 122–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ellis LM & Hicklin DJ Resistance to Targeted Therapies: Refining Anticancer Therapy in the Era of Molecular Oncology. Clin Cancer Res 15, 7471–7478 (2009). [DOI] [PubMed] [Google Scholar]
- 108.Hopper-Borge EA et al. Mechanisms of tumor resistance to EGFR-targeted therapies. Expert Opin Ther Targets 13, 339–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spaans JN & Goss GD Drug resistance to molecular targeted therapy and its consequences for treatment decisions in non-small-cell lung cancer. Front Oncol 4, 190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fan QW et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 24, 438–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nathanson DA et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343, 72–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thiessen B et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol 65, 353–61 (2010). [DOI] [PubMed] [Google Scholar]
- 113.Reardon DA et al. Phase I/randomized phase II study of afatinib, an irreversible ErbB family blocker, with or without protracted temozolomide in adults with recurrent glioblastoma. Neuro Oncol 17, 430–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Uhm JH et al. Phase II evaluation of gefitinib in patients with newly diagnosed Grade 4 astrocytoma: Mayo/North Central Cancer Treatment Group Study N0074. Int J Radiat Oncol Biol Phys 80, 347–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ritch PS, Carroll SL & Sontheimer H Neuregulin-1 enhances survival of human astrocytic glioma cells. Glia 51, 217–28 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sheng Q et al. An activated ErbB3/NRG1 autocrine loop supports in vivo proliferation in ovarian cancer cells. Cancer Cell 17, 298–310 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilson TR, Lee DY, Berry L, Shames DS & Settleman J Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell 20, 158–72 (2011). [DOI] [PubMed] [Google Scholar]
- 118.Dong X, Fernandez-Salas E, Li E & Wang S Elucidation of Resistance Mechanisms to Second-Generation ALK Inhibitors Alectinib and Ceritinib in Non-Small Cell Lung Cancer Cells. Neoplasia 18, 162–71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dhomen NS, Mariadason J, Tebbutt N & Scott AM Therapeutic targeting of the epidermal growth factor receptor in human cancer. Crit Rev Oncog 17, 31–50 (2012). [DOI] [PubMed] [Google Scholar]
- 120.Dempke WC & Heinemann V Ras mutational status is a biomarker for resistance to EGFR inhibitors in colorectal carcinoma. Anticancer Res 30, 4673–7 (2010). [PubMed] [Google Scholar]
- 121.Fan QW et al. A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res 67, 7960–5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yi YW et al. Inhibition of the PI3K/AKT pathway potentiates cytotoxicity of EGFR kinase inhibitors in triple-negative breast cancer cells. J Cell Mol Med 17, 648–56 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tricker EM et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer Discov 5, 960–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang MH et al. MEK inhibitors reverse resistance in epidermal growth factor receptor mutation lung cancer cells with acquired resistance to gefitinib. Mol Oncol 7, 112–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zehir A et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eisenhauer EA et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45, 228–47 (2009). [DOI] [PubMed] [Google Scholar]
- 127.Soreide K Receiver-operating characteristic curve analysis in diagnostic, prognostic and predictive biomarker research. J Clin Pathol 62, 1–5 (2009). [DOI] [PubMed] [Google Scholar]
- 128.Hajian-Tilaki K Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Caspian J Intern Med 4, 627–35 (2013). [PMC free article] [PubMed] [Google Scholar]
- 129.Rich JN et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol 22, 133–42 (2004). [DOI] [PubMed] [Google Scholar]
- 130.Lasocki A, Gaillard F, Tacey M, Drummond K & Stuckey S Multifocal and multicentric glioblastoma: Improved characterisation with FLAIR imaging and prognostic implications. J Clin Neurosci 31, 92–8 (2016). [DOI] [PubMed] [Google Scholar]
- 131.Liu Q et al. Genetic, epigenetic, and molecular landscapes of multifocal and multicentric glioblastoma. Acta Neuropathol 130, 587–97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alshami J et al. Afatinib, an irreversible ErbB family blocker, with protracted temozolomide in recurrent glioblastoma: a case report. Oncotarget 6, 34030–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ma DJ et al. A phase II trial of everolimus, temozolomide, and radiotherapy in patients with newly diagnosed glioblastoma: NCCTG N057K. Neuro Oncol 17, 1261–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaley TJ et al. Phase II trial of sunitinib for recurrent and progressive atypical and anaplastic meningioma. Neuro Oncol 17, 116–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bang YJ et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376, 687–97 (2010). [DOI] [PubMed] [Google Scholar]
- 136.Hecht JR et al. Lapatinib in Combination With Capecitabine Plus Oxaliplatin in Human Epidermal Growth Factor Receptor 2-Positive Advanced or Metastatic Gastric, Esophageal, or Gastroesophageal Adenocarcinoma: TRIO-013/LOGiC--A Randomized Phase III Trial. J Clin Oncol 34, 443–51 (2016). [DOI] [PubMed] [Google Scholar]
- 137.Kim ST et al. Impact of genomic alterations on lapatinib treatment outcome and cell-free genomic landscape during HER2 therapy in HER2-positive gastric cancer patients. Ann Oncol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fernandez-Cuesta L & Thomas RK Molecular Pathways: Targeting NRG1 Fusions in Lung Cancer. Clin Cancer Res 21, 1989–94 (2015). [DOI] [PubMed] [Google Scholar]
- 140.Wu H et al. Ibrutinib selectively and irreversibly targets EGFR (L858R, Del19) mutant but is moderately resistant to EGFR (T790M) mutant NSCLC Cells. Oncotarget 6, 31313–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bernard S et al. Activity of ibrutinib in mantle cell lymphoma patients with central nervous system relapse. Blood 126, 1695–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jain P et al. Early responses predict better outcomes in patients with newly diagnosed chronic myeloid leukemia: results with four tyrosine kinase inhibitor modalities. Blood 121, 4867–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Louvet C et al. Correlation between progression free survival and response rate in patients with metastatic colorectal carcinoma. Cancer 91, 2033–8 (2001). [PubMed] [Google Scholar]
- 144.Tsimberidou AM & Kurzrock R Precision medicine: lessons learned from the SHIVA trial. Lancet Oncol 16, e579–80 (2015). [DOI] [PubMed] [Google Scholar]
- 145.Baras A, Yu Y, Filtz M, Kim B & Moskaluk CA Combined genomic and gene expression microarray profiling identifies ECOP as an upregulated gene in squamous cell carcinomas independent of DNA amplification. Oncogene 28, 2919–24 (2009). [DOI] [PubMed] [Google Scholar]
- 146.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Iyer MK, Chinnaiyan AM & Maher CA ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 27, 2903–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Torres-Garcia W et al. PRADA: pipeline for RNA sequencing data analysis. Bioinformatics 30, 2224–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang JH, Chung TD & Oldenburg KR A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 151.Franceschini A et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41, D808–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Griffith M et al. DGIdb: mining the druggable genome. Nat Methods 10, 1209–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kondor RI & Lafferty J Diffusion kernels on graphs and other discrete structures. in Proceedings of the 19th International Conference on Machine Learning 8 (Morgan Kaufmann, 2002). [Google Scholar]
- 154.Zou H & Hastie T Regularization and Variable Selection via the Elastic Net. J. R. Statist. Soc. B 67, 20 (2005). [Google Scholar]
- 155.Honigberg LA et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A 107, 13075–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequenced data have been deposited in the European Genome-phenome Archive (EGA) with accession code EGAS00001002407. Processed data and basic association analysis are publicly available through an interactive webportal (cDx, the Cancer Drug eXplorer, at cancerdrugexplorer.org).