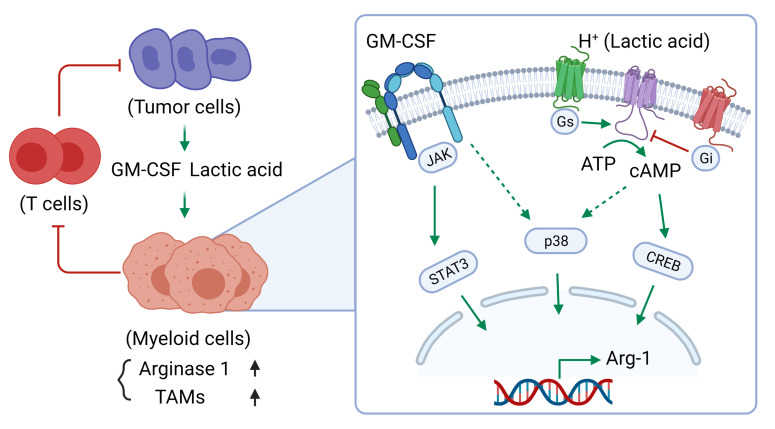
Abstract
Tumor-infiltrating myeloid cells contribute to the development of the immunosuppressive tumor microenvironment. Myeloid cell expression of arginase 1 (ARG1) promotes a protumor phenotype by inhibiting T cell function and depleting extracellular l-arginine, but the mechanism underlying this expression, especially in breast cancer, is poorly understood. In breast cancer clinical samples and in our mouse models, we identified tumor-derived GM-CSF as the primary regulator of myeloid cell ARG1 expression and local immune suppression through a gene-KO screen of breast tumor cell–produced factors. The induction of myeloid cell ARG1 required GM-CSF and a low pH environment. GM-CSF signaling through STAT3 and p38 MAPK and acid signaling through cAMP were required to activate myeloid cell ARG1 expression in a STAT6-independent manner. Importantly, breast tumor cell–derived GM-CSF promoted tumor progression by inhibiting host antitumor immunity, driving a significant accumulation of ARG1-expressing myeloid cells compared with lung and melanoma tumors with minimal GM-CSF expression. Blockade of tumoral GM-CSF enhanced the efficacy of tumor-specific adoptive T cell therapy and immune checkpoint blockade. Taken together, we show that breast tumor cell–derived GM-CSF contributes to the development of the immunosuppressive breast cancer microenvironment by regulating myeloid cell ARG1 expression and can be targeted to enhance breast cancer immunotherapy.
Keywords: Immunology, Oncology
Keywords: Breast cancer, Cancer immunotherapy, Macrophages
Introduction
Immunotherapy is heralded for its promise in the treatment of cancer (1). Emerging clinical evidence suggests that the immunosuppressive tumor microenvironment (TME) represents a major obstacle for treatment success against several tumor types, including breast cancer (1–3). Higher numbers of tumor-infiltrating lymphocytes (TILs) are associated with an increased durable response and better survival (1, 4–6) and have shown promise as indicators of a positive response to immune checkpoint blockade (ICB) treatment (1, 7). However, the accumulation of inhibitory cells, such as myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs), as well as metabolic constraints inherent to the TME are important contributors to T cell dysfunction (2, 3, 8, 9). Thus far, no robust predictive biomarkers for ICB have been established in breast cancer (1).
With tumor progression, a supportive and immunosuppressive TME develops, comprising immune cells, cancer-associated fibroblasts (CAFs), endothelial cells, and stromal cells, all of which are embedded within an extracellular matrix (ECM) (3, 10). Heterogeneous populations of tumor-infiltrating myeloid cells (TIMs) are the most abundant immune-related cells in the TME and include monocytes, tumor-associated neutrophils, tumor-associated DCs, MDSCs, and TAMs (2, 8, 9). In the TME, TIMs exhibit a diverse range of functional phenotypes from antitumor to protumor (2, 8, 9). TAMs, for example, are often broadly divided into 1 of 2 categories: (a) antitumorigenic or M1 TAMs, which express high levels of TNF, inducible NOS (iNOS, or NOS2), and MHC class II molecules; and (b) protumorigenic or M2 TAMs, which express high levels of arginase 1 (ARG1), IL-10, and CD206 (2). In many cancer types, including breast cancer, TIMs often exhibit a protumor phenotype in the TME, where they promote tumor growth and metastasis and are associated with a poor prognosis for patients with cancer (2, 11–13). Indeed, protumor TIMs, especially M2 TAM subsets, can directly inhibit cytotoxic T lymphocyte (CTL) function through at least 3 distinct mechanisms, including direct cell-cell contact inhibition through checkpoint-inhibitory molecules such as programmed death ligand 1 (PD-L1), production of inhibitory cytokines such as IL-10 and TGF-β, and modulation of the metabolic environment through depletion of l-arginine and production of ROS (2, 3).
Breast cancer tissue is usually infiltrated with a high number of myeloid cells (2, 13), and the presence of abundant CD68+ myeloid cells with low CD8+ CTL infiltration is negatively correlated with patient survival (13). Breast tumor cell–derived factors, especially members of the CSF superfamily such as macrophage CSF (M-CSF or CSF1), granulocyte CSF (G-CSF or CSF3), and granulocyte macrophage CSF (GM-CSF or CSF2), can directly influence myeloid cell populations, regulating their recruitment, proliferation, and function (2, 9, 11, 12). GM-CSF is expressed by nearly 60% of patients’ breast cancer tumors (14) and has been shown to activate plasmacytoid pre-DCs (pDCs) to promote a regulatory Th2 response and to induce the production of immunosuppressive myeloid cells (14–17).
ARG1 is a well-characterized marker of immunosuppressive myeloid cells, and myeloid cell arginine metabolism regulates both innate and adaptive immunity (18). Compared with NOS2 or ARG2, ARG1 expression in macrophages plays a larger role in regulating extracellular l-arginine levels (19). TIM-derived ARG1 can directly suppress T cell function by depleting l-arginine, inhibiting T cell receptor (TCR) expression and responses (2, 3, 9, 20). Inhibition of ARG1 reduces the growth of tumors only in immunocompetent mice (20), and macrophage-specific deletion of Arg1 in mice improved responses to adoptive cell transfer therapy (21). However, in breast cancer, the role and mechanism of ARG1 expression are not fully understood.
In this study, we found increased numbers of ARG1+ myeloid cells in breast cancer preclinical models and in breast cancer tumors from patients compared with lung cancer and melanoma tumors. Using CRISPR/Cas9-mediated gene-KO screening in breast cancer cells, we identified GM-CSF as a critical cytokine regulating myeloid cell ARG1 expression in the breast cancer TME. We show that GM-CSF promoted primary and metastatic tumor growth through these effects on myeloid cell ARG1 expression. GM-CSF and the acidic TME were required for TIM ARG1 expression through JAK/STAT3 and cAMP pathways, respectively. Finally, disruption of tumoral GM-CSF reversed resistance to T cell therapy and immune checkpoint inhibitors. Our data suggest that breast tumor cell–derived GM-CSF contributes to the development of the immunosuppressive breast cancer TME and can be rationally targeted to enhance breast cancer immune therapy.
Results
ARG1-expressing myeloid cells accumulate in the breast TME.
In patients with breast cancer, an abundance of myeloid cells and low T cell infiltration in the tumor tissue are associated with poor outcomes (13). To evaluate myeloid cell and T cell dynamics in the breast cancer TME, we performed FACS analysis of immune cell populations in PyMT-BO1-GFP-Luc (BO1) syngeneic C57BL/6J mice, an orthotopic model of murine estrogen receptor–positive (ER+) luminal B breast cancer (22), at several time points during tumor progression. We found that the percentage of CD4+ and CD8+ T cells among total tumor-infiltrating leukocytes decreased over time, while CD11b+ myeloid cell percentages increased. Moreover, the majority of infiltrated TAMs were of the CD206-expressing, protumorigenic M2 phenotype (Figure 1, A and B). The murine ER–, progesterone receptor–negative, human epidermal growth factor receptor 2–negative (ER–PR–HER2–) triple-negative 4T1 breast tumor cell line has also been shown to recruit high levels of M2 polarized macrophages and exhibits reduced levels of infiltrating T cells in syngeneic BALB/c mice (23).
Figure 1. Immunosuppressive ARG1-expressing myeloid cells accumulate in the breast TME.
(A) PyMT-BO1 breast tumor cells (1 × 105) were injected into the MFP tissue of C57BL/6J female mice, and tumor growth was measured by digital calipers. (B) Single-cell suspensions from whole-tumor tissue were analyzed by FACS on days 6, 10, and 15 of tumor growth. The TIM and T cell populations are shown. TAM markers are CD45+, CD11b+, Ly6C–, Ly6G–, and F4/80+. M1 TAMs are MHC-II+, and M2 TAMs are CD206+. (C) Weights of PyMT-BO1 orthotopic tumors from YARG mice. (D) Percentage of ARG1-expressing myeloid cells in CD45+CD11b+ TIMs. (E) Percentage of TIM subpopulations in total CD45+CD11b+ myeloid cells from day-10 tumors. The ARG1– cells (white) and ARG1+ cells (blue) are labeled for each subpopulation. (F) ARG1-expressing myeloid cells in CCR2hi and CCR2loCD45+CD11b+ myeloid cells. FSC-W, forward scatter width. (G) ARG1-expressing CD45+CD11b+ myeloid cells in PyMT-BO1 breast cancer MFP tumor tissue, B16F10 melanoma, and LLC lung cancer subcutaneous tumor tissue. (H) Immunofluorescence staining of paraffin-embedded BO1 MFP tissue. Scale bars: 1 mm (whole-tumor tissue); 25 μm (enlarged images).
ARG1 is an established biomarker for protumor M2-polarized macrophages (2). To track ARG1-expressing macrophages during tumor development in vivo, we established orthotopic BO1 tumors in ARG1 reporter mice (YARG), in which an enhanced yellow fluorescent protein (EYFP) is expressed under the control of the ARG1 promoter without disruption of normal ARG1 expression (24). We found that the majority of ARG1-expressing cells in the breast TME were myeloid cells, and that the tumor-infiltrating CD45+CD11b+ myeloid cell population consistently represented approximately half of the cells expressing ARG1 (Figure 1, C and D). Importantly, we observed ARG1+ cells in all myeloid cell subpopulations studied (Figure 1E). Further, we found that ARG1-expressing TIMs were mostly CCR2+, pointing toward a bone marrow (BM) origin (refs. 25, 26 and Figure 1F). However, we did not observe ARG1 induction in the spleen or in BM-resident myeloid cells from tumor-bearing mice, suggesting that ARG1 induction in TIMs primarily occurred within the breast TME (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI145296DS1).
Comparing BO1 breast tumors with subcutaneous B16F10 melanoma or LLC lung tumor tissue in C57BL/6J mice, we found that the number of ARG1+ myeloid cells was exceptionally high in murine breast tumor tissue (Figure 1G). IHC staining confirmed high numbers of ARG1-expressing cells in the murine BO1 and 4T1 breast tumor tissues (Supplemental Figure 2). By multicolor immunofluorescence, we found that most ARG1-expressing myeloid cells were located in the tumor core (Figure 1H).
We next evaluated breast cancer, melanoma, and lung cancer tissues from patients for myeloid cell ARG1 expression by immunofluorescence staining (Figure 2F). All cancer types showed infiltrating CD68+ myeloid cells, but the breast cancer tissues had a significantly higher number of CD68+ infiltrating cells per tissue area (Figure 2A). Moreover, less than 10% of CD68+ cells in the melanoma and lung cancer tissues expressed ARG1, whereas over 45% of CD68+ cells were ARG1+ in the breast cancer tissue (Figure 2B). Evaluation of myeloid subsets in the most common breast cancer subtypes (ER+, HER2+, and triple-negative breast cancer [TNBC]) showed higher levels of infiltrating CD68+ myeloid cells compared with levels in normal breast tissue (Figure 2C). In contrast to TNBC, the ER+ and HER2+ breast cancer subsets had similar ratios of CD68+ cells expressing ARG1 (Figure 2D). When comparing the ARG1+CD68+ cell numbers based on tumor stage, we found that the number of ARG1+CD68+ myeloid cells was increased in later-stage cancer (Figure 2E).
Figure 2. ARG1-expressing myeloid cells accumulate in human breast cancer tissue.
Lung cancer (n = 10), melanoma (n = 10), and breast cancer (n = 60) tissue microarrays (TMAs) were used for immunofluorescence staining. (A) CD68+ myeloid cell number per mm2 tissue area, and (B) percentage of ARG1+ cells in total CD68+ cells from lung cancer, melanoma, and breast cancer tissues. (C) CD68+ myeloid cell number per mm2 tissue area, and (D) percentage of ARG1+ cells in total CD68+ cells from breast cancer (BC) subtypes. (E) ARG1+CD68+ myeloid cell number per mm2 tissue area from breast cancer based on cancer stage. (F) Representative immunofluorescence images of stained paraffin-embedded human cancer tissue. Scale bars: 500 μm (whole-tissue images); 100 μm (enlarged images). Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-tailed, unpaired Student’s t test with Welch’s correction.
These data demonstrate that ARG1-expressing myeloid cells accumulated at high levels in the breast TME in the 3 major breast cancer subtypes. Our data also suggest that the breast cancer TME has unique properties that drive TIM ARG1 expression compared with lung or melanoma tumors.
Tumor cell–produced GM-CSF is necessary to induce myeloid cell ARG1 expression.
To test whether tumor cells can directly induce myeloid cell ARG1 expression, we treated bone marrow macrophages (BMMs) with conditioned media (CM) collected from tumor cells. We used BMM because the majority of TIMs expressed high levels of CCR2, suggestive of an origin in the BM; additionally, they represent an abundant and reliable source of myeloid cells for mechanistic biochemical studies. We found that both BO1 and 4T1 breast tumor cell–derived CM strongly induced ARG1 expression in BMMs, whereas CM collected from Lewis lung carcinoma (LLC) or B16F10 melanoma cells did not (Figure 3A). Tumor cell–derived CM had no direct inhibitory effect on T cell proliferation (Figure 3B), prompting us to evaluate the effect of CM from ARG1+ macrophages. BMMs were exposed to tumor cell–derived CM for 24 hours and sorted into ARG1+ and ARG1–CD11b+ cell populations by FACS; CM from these sorted cell populations were then harvested and added to the T cell proliferation assay. We found that only CM from ARG1+ myeloid cells had an inhibitory effect on activated T cell proliferation (Figure 3B and Supplemental Figure 3).
Figure 3. Tumor cell–produced GM-CSF is necessary to induce myeloid cell ARG1 expression.
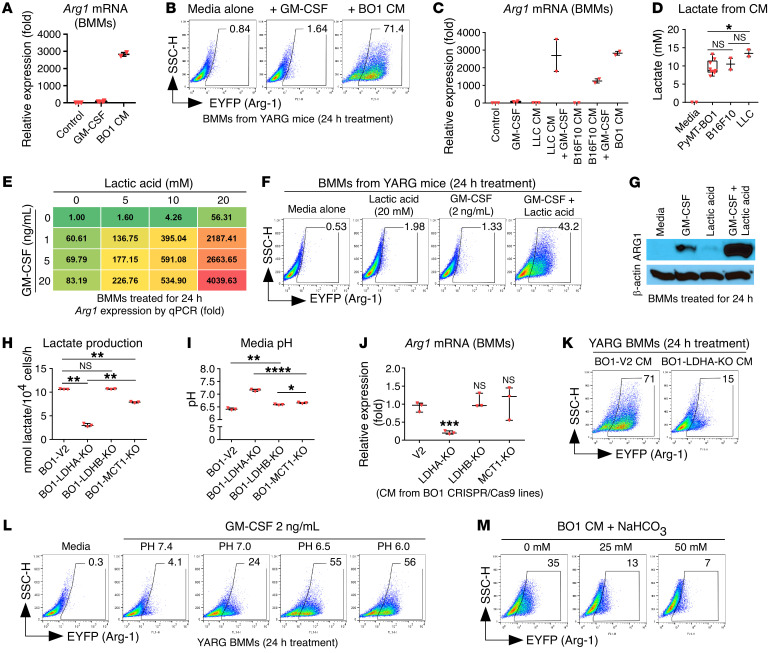
(A) Arg1 mRNA expression in BMMs treated with LLC, B16F10, PyMT-BO1, and 4T1 tumor cell CM (n = 2–3). (B) CFSE-labeled whole splenic cells stimulated (stim) with plate-banded anti-CD3E antibody and soluble anti-CD28 antibody, cocultured with 1:1 diluted CM from PyMT-BO1 or FACS-sorted ARG1+ or ARG1– BMMs pretreated with PyMT-BO1 CM. T cell proliferation was measured from the quantification of CFSE dilution in gated CD4+ T cells by FACS (n = 3). (C) Microarray analysis of gene expression in breast tumor cells and CD206+ TAMs sorted by FACS from the same tumor tissue. (D) Arg1 mRNA expression in BMMs treated with tumor cell CM from PyMT-BO1 or CRISPR/Cas9-mediated gene-KO PyMT-BO1 tumor cells (n = 2). (E) Western blot of ARG1 in tumor cell CM–treated BMMs. (F) Quantification of GM-CSF levels in tumor cell CM by ELISA (n 2–4). (G and H) Arg1 mRNA expression in BMMs treated with tumor cell CM that included anti-CSF2 (αCSF2) antibody or CSF2 (n = 2). (I) ARG1 expression quantified as EYFP expression by FACS of YARG BMMs treated as indicated. SSC, side scatter. (J) BO1-WT (vector control) or BO1-CSF2–KO breast tumor cells (1 × 105) were injected into MFP tissues of 8-week-old female YARG mice. On day 10, single-cell suspensions from whole-tumor tissue were analyzed by FACS. ARG1 expression in TIMs was quantified as EYFP expression. In E and I, data are representative of 3 independent experiments. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-tailed, unpaired Student’s t test with Welch’s correction. iso-Ab, isotype control antibody.
To identify candidate tumor-derived secreted factors that could promote protumor myeloid cell phenotypes in the TME, we performed gene expression analysis of FACS-sorted tumor cells (GFP+) and TAMs (CD206+) from BO1 mammary fat pad (MFP) tumors. We found that breast tumor cells expressed many cytokines and growth factor genes and that TAMs from the breast cancer microenvironment expressed their reciprocal receptors (Figure 3C). We performed a KO screen of candidate secreted factors in BO1 and 4T1 breast cancer cells using the CRISPR/Cas9 lentiviral vector system. We validated targeted gene KO and evaluated tumor cell CM from these KO lines for the ability to induce BMM ARG1 expression, identifying GM-CSF as a prime candidate (Figure 3, D and E, and Supplemental Figure 4). We confirmed that GM-CSF was present in the CM from WT BO1 and 4T1 cell line cultures, but absent in CM from the KO cell line cultures, as measured by ELISA. Interestingly, neither B16F10 melanoma cells nor LLC lung cancer cells exhibited detectable GM-CSF in their CM (Figure 3F).
Consistent with KO studies, pharmacologic blockade of the GM-CSF present in breast cancer cell CM using neutralizing antibodies resulted in decreased Arg1 mRNA expression in BMMs; likewise, exogenous GM-CSF added to CM from CSF2-KO breast cancer cells rescued Arg1 mRNA expression in BMMs (Figure 3, G and H). We further confirmed the effect of GM-CSF depletion or rescue on ARG1 expression by FACS using BMMs from YARG mice (Figure 3I).
To evaluate whether GM-CSF can regulate ARG1 expression in vivo, we injected BO1-WT or BO1-CSF2–KO breast tumor cells into MFP tissue of YARG mice and evaluated ARG1-expressing myeloid cells by FACS. We detected a significantly lower number of ARG1-expressing myeloid cells in the BO1-CSF2–KO tumors (Figure 3J).
To evaluate the role of GM-CSF in myeloid cell ARG1 expression in other tumor types, we directly injected GM-CSF into established B16F10 tumors and found that the number of ARG1-expressing myeloid cells was significantly increased 24 hours after GM-CSF injection (Supplemental Figure 5).
To further confirm these results, we engineered B16 melanoma and LLC lung cancer cells to overexpress GM-CSF. CM from GM-CSF–expressing B16 and LLC cells induced ARG1 expression in BMMs. In vivo, we found that enforced GM-CSF expression yielded cell line–derived tumors with significantly more ARG1-expressing TIMs (Supplemental Figure 6).
Together, these results demonstrate that tumor cell–produced GM-CSF is necessary to induce myeloid cell ARG1 expression.
GM-CSF and lactic acid synergistically induce myeloid cell ARG1 expression.
GM-CSF can regulate ARG1 expression in BMMs, but the mechanism is not clear (27). When BMMs were treated with GM-CSF alone in naive media, the induced Arg1 mRNA level was surprisingly low compared with the response observed following treatment with BO1 CM (Figure 4A). A similar experiment with YARG BMMs demonstrated minimal Arg1 reporter gene expression after GM-CSF alone treatment (Figure 4B). By contrast, the addition of GM-CSF to B16F10 or LLC CM, neither of which is sufficient to induce ARG1, induced Arg1 mRNA expression in BMMs to a level comparable to that seen in BO1 CM (Figure 4C). These data indicate that GM-CSF requires a tumor-derived secreted cofactor to induce myeloid cell ARG1 expression.
Figure 4. GM-CSF and LA synergistically induce myeloid cell ARG1 expression.
(A) Arg1 mRNA expression in BMMs treated with recombinant GM-CSF or PyMT-BO1 tumor cell CM (n = 2–3). (B) FACS quantification of ARG1+ cells from YARG BMMs treated with recombinant GM-CSF or PyMT-BO1 tumor cell CM. (C) Arg1 mRNA expression in BMMs treated with tumor cell CM plus recombinant GM-CSF (n = 2–3). (D) Quantification of lactate from tumor cell CM. (E) Arg1 mRNA expression in BMMs treated with recombinant GM-CSF and LA. (F) ARG1+ cells quantified by FACS. (G) ARG1 expression in BMMs was detected by Western blotting after GM-CSF and LA treatment. (H) Lactate production from tumor cell CM (n = 3). (I) Tumor cell CM pH measurement (n = 3). (J) Arg1 mRNA expression in BMMs (n = 3). (K) ARG1+ cells quantified by FACS. (L) YARG BMMs were treated with GM-CSF. The media pH was adjusted with hydrochloric acid. (M) BO1 tumor CM were premixed with NaHCO3 at the indicated concentrations before being added to YARG BMMs. In B, F, G, and K–M, data are representative of 3 independent experiments. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-tailed, unpaired Student’s t test with Welch’s correction. SSC-H, side scatter height.
Our top candidate for such a cofactor was lactic acid (LA), a common component of the TME in multiple cancer types (28) and previously shown to upregulate macrophage ARG1 expression (29). LA dissociates into lactate and free hydrogen ions, actively acidifying the TME, and the anion lactate can be measured as a surrogate of tumor cell LA production. CM from BO1, B16F10, and LLC tumor cells had similar concentrations of lactate (10–15 mM; Figure 4D). As with GM-CSF alone in cell culture media, we found that LA alone induced Arg1 mRNA expression in BMMs, without significant upregulation at the protein level. Testing multiple combinations of GM-CSF and LA at various concentrations, we found that they synergized to generate high-level Arg1 mRNA expression from BMMs in a dose-dependent manner (Figure 4E). We confirmed the induction of protein expression using this combination both by FACS of BMMs from YARG reporter mice and by Western blotting (Figure 4, F and G).
Lactate dehydrogenase A (LDHA), lactate dehydrogenase B (LDHB), and monocarboxylate transporter 1 (MCT1) are important for LA production in tumor cells (28). We genetically knocked out these genes in BO1 cells to quantify their lactate production and the media pH and found that Ldha-KO cells had decreased lactate production and increased media pH (Figure 4, H and I). Compared with CM from WT cancer cells, CM from Ldha-KO cells resulted in diminished induction of ARG1 expression in BMMs (Figure 4, J and K).
Next, we asked whether the molecule lactate, independent of H+, could work with GM-CSF to induce myeloid cell ARG1 expression. Surprisingly, the combination of sodium lactate and GM-CSF did not induce ARG1 expression in BMMs (Supplemental Figure 7), suggesting that acidified culture conditions may play a role in the induction of ARG1 expression. We tested the addition of GM-CSF to media across pH values ranging from 6 to 7.4 for induction of ARG1 expression. We found that GM-CSF only induced myeloid cell ARG1 expression in acidified media (Figure 4L), and that neutralization of acidic tumor CM with sodium bicarbonate (NaHCO3) could block the induction of ARG1 expression (Figure 4M).
Together, these data indicate that breast tumor–derived GM-CSF is necessary, but not sufficient, to induce myeloid cell ARG1 expression. LA acidification may be functioning as a cofactor that synergistically induces myeloid cell ARG1 expression along with GM-CSF.
Tumor-derived GM-CSF drives ARG1 expression through noncanonical signaling pathways.
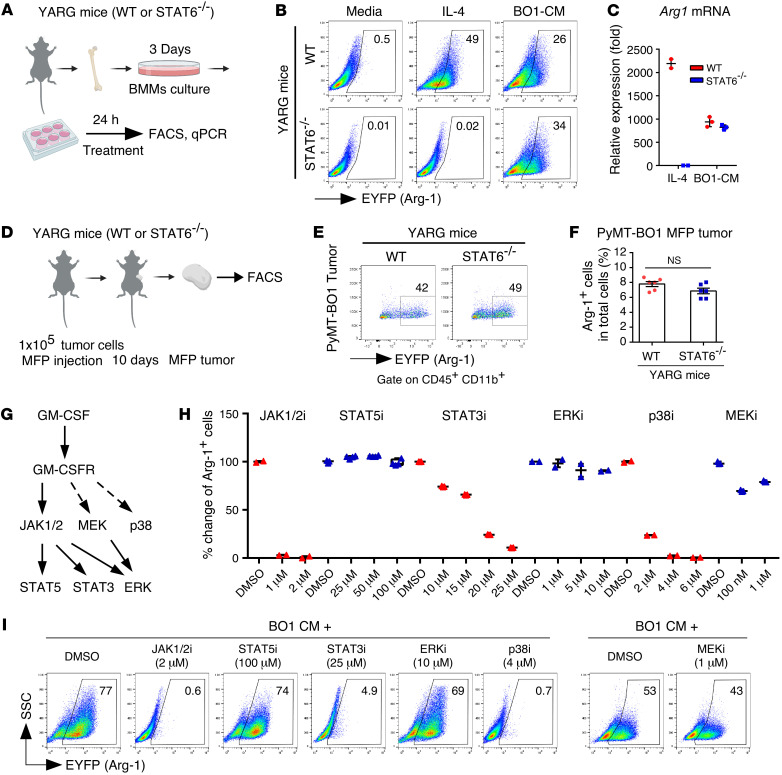
STAT6 signaling is required for macrophage ARG1 expression induced by IL-4 (12). To test the role of STAT6 signaling in breast tumor–induced macrophage ARG1 expression, we treated BMMs from WT YARG mice or Stat6–/– YARG mice with either IL-4 or BO1 CM and detected reporter gene EYFP expression by FACS. We found that IL-4 induced ARG1 expression in WT BMMs, but not in Stat6–/– BMMs, whereas BO1 CM induced ARG1 expression in both WT and Stat6–/– BMMs (Figure 5, A and B). We obtained the same results by quantitative PCR (qPCR) for Arg1 mRNA expression in a parallel experiment (Figure 5C). In vivo, we found that BO1 orthotopic MFP tumors had induced expression of ARG1 by TIMs in both WT and Stat6–/– mice (Figure 5, D–F). These data suggest that STAT6 signaling is dispensable in breast tumor–induced myeloid cell ARG1 expression.
Figure 5. GM-CSF signaling regulates myeloid cell ARG1 expression through noncanonical pathways.
(A) Experimental scheme. (B) WT or Stat6–/– YARG BMMs were treated with recombinant IL-4 or PyMT-BO1 tumor cell CM for 24 hours, and ARG1+ cells were quantified by FACS. (C) Arg1 mRNA expression in WT and Stat6–/– BMMs treated with IL-4 or PyMT-BO1 tumor cell CM (n = 2–3). (D) Experimental scheme. (E and F) PyMT-BO1 tumor cells (1 × 105) were injected into MFP tissue of WT or Stat6–/– YARG mice. After tumors reached 500 mm3 in size, ARG1+ cells from whole-tumor tissue single-cell suspensions were quantified by FACS (n = 6). (G) Working model of GM-CSF receptor signaling. (H and I) ARG1+ cells quantified by FACS were from YARG BMMs pretreated with DMSO, the JAK1/2 inhibitor (JAK1/2i) ruxolitinib, the STAT3 inhibitor C188-9, the STAT5 inhibitor CAS 285986-31-4, the ERK1/2 inhibitor ulixertinib, the p38 inhibitor SB203580, or the MEK inhibitor trametinib for 1 hour, followed by treatment with BO1 tumor cell CM for 24 hours (n = 2–5). Data are shown as the mean ± SEM. Two-tailed, unpaired Student’s t test with Welch’s correction.
Classical GM-CSF signaling proceeds through JAK2/STAT5; however, studies have shown that GM-CSF can also activate STAT3, p38 MAPK, MEK, and ERK1/2 signaling pathways (30–32). To evaluate the downstream signaling pathways essential to breast tumor–induced ARG1 expression in myeloid cells, we pretreated BMMs from YARG mice with inhibitors of JAK1/2, STAT3, STAT5, p38, MEK, and ERK1/2 and then added BO1 CM for 24 hours and analyzed ARG1 expression by FACS. We found that the clinical JAK1/2 inhibitor ruxolitinib completely blocked BO1 CM–induced ARG1 expression; a JAK2 downstream STAT5 inhibitor, meanwhile, had no effect. Inhibition of STAT3 and p38 reduced BO1 CM–induced ARG1 expression in a dose-dependent manner. The clinically used MEK inhibitor trabectedin partially inhibited ARG1 expression, while ERK1/2 inhibition had no effect (Figure 5, G–I, and Supplemental Figure 8).
These results suggest that tumor-derived GM-CSF drives breast tumor cell–induced myeloid cell ARG1 expression through noncanonical JAK/STAT3 and p38 MAPK signaling pathways.
Tumor-derived GM-CSF requires cAMP signaling to drive ARG1 expression.
Cells can respond to changes in environmental pH through pH-sensing GPCRs (33). GPCR downstream signaling proceeds through the G α subunit, which negatively or positively regulates adenylyl cyclase (AC) activity through inhibitory G protein (Gi) or stimulatory G protein (Gs), respectively (refs. 34, 35 and Figure 6A). We first used the Gi inhibitor pertussis toxin (PTX) to pretreat BMMs from YARG mice for 1 hour, and then added breast tumor CM for a further 24 hours before analyzing ARG1 expression by FACS. We found that PTX dose-dependently enhanced ARG1 expression (Figure 6B). Forskolin, an AC activator, similarly enhanced breast tumor CM–induced BMM ARG1 expression (Figure 6C). When combined with GM-CSF, we found that forskolin was sufficient to induce ARG1 expression in BMMs (Figure 6D), suggesting that acidified tumor CM or LA may signal through the cAMP pathway to induce ARG1 expression in myeloid cells.
Figure 6. Tumor cell–derived, GM-CSF–induced myeloid cell ARG1 expression requires cAMP signaling.
(A) Working model illustrating that GPCR-associated subunits Gs and Gi regulate cAMP levels. (B) BMMs (5 × 105) from ARG1-YFP mice were seeded in a 6-well plate overnight. BMMs were pretreated with PTX for 2 hours before addition of tumor cell CM. (C and D) Forskolin (Fors) was added to the BMMs at the same time as the indicated treatments. (E and F) Inhibitors were added 1 hour before CM or 2 ng/mL GM-CSF plus 20 mM LA (CSF2/LA) treatment. All BMMs were treated for 24 hours with CM or CSF2/LA before FACS analysis. In B–F, data are representative of 3 independent experiments. (G) Working model showing that GM-CSF (CSF2) and cAMP signaling combine to induce myeloid cell ARG1 expression.
AC activation increases intracellular cAMP concentrations and regulates many cellular functions through cAMP response element–binding protein (CREB) (35). To test the necessity of cAMP for promoting ARG1 expression, we pretreated BMMs with either MDL-12, an AC inhibitor, or KG-501, an inhibitor of CREB binding, and then added breast tumor CM or GM-CSF in combination with LA to the BMMs. In both conditions, we observed a dose-dependent inhibition of ARG1 expression (Figure 6, E and F).
Taken together, these results suggest that tumor cell–derived GM-CSF–induced myeloid cell ARG1 expression requires both cAMP signaling and the JAK/STAT3 signaling pathway (see proposed model in Figure 6G).
Breast tumor–derived GM-CSF promotes tumor growth through the modulation of host immune cells.
To evaluate the functional role of tumor-derived GM-CSF in breast tumor growth in vivo, we inoculated 4T1-WT or 4T1-CSF2–KO (GM-CSF–KO) cells into the MFP tissue of BALB/c mice and measured tumor growth by digital calipers. We found that GM-CSF–KO tumor cells had significantly decreased tumor growth (Figure 7A) in immunocompetent mice. We performed the same experiment using BO1-WT or BO1-CSF2–KO breast cancer cell injection into the MFP tissue of C57BL/6J mice and obtained the same result (Figure 7B). To rule out the effect of genetic differences between CRISPR-modified cell lines, we established BO1 orthotopic MFP tumors and pharmacologically inhibited GM-CSF in the TME using a neutralizing anti–GM-CSF antibody. Compared with isotype control antibody treatment, WT C57BL/6J mice that received anti–GM-CSF antibody had significantly decreased tumor growth (Figure 7C). Importantly, pharmacological or genetic disruption of tumor-derived GM-CSF did not affect tumor growth in immunocompromised NOD-SCID gamma (NSG) mice (Figure 7, D–F). These results indicate that tumor-derived GM-CSF promotes tumor growth by modulating host immune responses.
Figure 7. Breast tumor–derived GM-CSF promotes tumor growth through the modulation of host immune cells.
(A) 4T1-WT (vector control) or 4T1-CSF2–KO breast tumor cells (1 × 105) were injected into MFP tissue of 8-week-old female BALB/C mice (n = 7–8). (B) BO1-WT (vector control) or BO1-CSF2–KO breast tumor cells (1 × 105) were injected into MFP tissue of 8-week-old female C57BL/6J mice (n = 9–11). (C) BO1-GFP-Luc breast tumor cells (1 × 105) were mixed with Matrigel plus an isotype antibody (control) or anti-CSF2 antibody before injection into MFP tissue of 8-week-old female C57BL/6J mice (n = 9). (D–F) The same experiments in A–C were performed in NSG mice (n = 4–8). (G) The same experiment in B was performed in Arg1fl/fl LySM-Cre+/– mice (n = 8–10). For all of the above experiments, tumor growth was measured by digital calipers. On day 24, MFP tumors were dissected and weighed. (H–J) Single-cell suspensions from day-10 BO1-WT or BO1-CSF2–KO whole-tumor tissue (C57BL/6J mice) were analyzed by FACS. TIM and T cell populations are shown (n = 7). Gran, granulocytes; Mono, monocytes; MoDC, monocyte-derived DCs; Tex, exhausted T cells. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-way, repeated-measures ANOVA or 2-tailed, unpaired Student’s t test with Welch’s correction between groups for column data.
We also tested the role of GM-CSF in a spontaneous tumorigenesis model by crossing MMTV-PyMT mice with GM-CSF–KO (Csf2–/–) mice to generate Csf2–/– MMTV-PyMT mice and evaluated tumor development. We found that the median time to tumor appearance in WT MMTV-PyMT mice was 96 days versus 130.5 days in Csf2–/– MMTV-PyMT mice. We performed immune cell profiling of tumors from these mice (harvested at the same tumor size) and found that tumors from Csf2–/– MMTV-PyMT mice had decreased ARG1+CD11b+ myeloid cells and CD4+ T cell infiltration, while CD8+ T cells numbers were increased (Supplemental Figure 9). These data show that GM-CSF can modulate tumor growth and alter tumor immune profiles in a spontaneous breast cancer model.
It is known that myeloid cells can inhibit T cell function through ARG1 expression (2, 3, 9, 20), and our in vitro data indicated that BMMs exposed to breast tumor–secreted GM-CSF expressed ARG1 and inhibited T cell function (Figure 3). This prompted us to evaluate WT and GM-CSF–KO tumor cell growth in myeloid cell–specific ARG1-KO mice (Arg1fl/fl LysM-Cre+/–). Interestingly, as with our studies in immunocompromised NSG mice, we found no significant differences in tumor growth in mice lacking ARG1 in myeloid cells (Figure 7G), suggesting that ARG1 protein expression in myeloid cells is particularly important for GM-CSF–induced enhancement of tumor growth.
We next evaluated the immune profile of BO1-WT and BO1-CSF2–KO tumors in WT mice. We found that in the myeloid cell compartment, total CD45+CD11b+ myeloid cell numbers were decreased in BO1-CSF2–KO tumors. Among all myeloid cell populations, the number of monocytes, granulocytes, and monocyte-derived DCs was not significantly different between WT and GM-CSF–KO BO1 tumors; however, the number of TAMs was decreased in BO1-CSF2–KO tumors (Figure 7H and Supplemental Figure 10). Compared with WT BO1 tumors, TAMs in the BO1-CSF2–KO tumors were also less polarized toward an M2 phenotype (Figure 7I). Evaluation of the entire population of tumor-infiltrating CD45+ cells revealed that the percentages of CD4+Foxp3– conventional T (Tconv) cells and CD8+ T cells in BO1-WT versus BO1-CSF2–KO tumors were not significantly different. However, BO1-CSF2–KO tumors exhibited higher ratios of memory Tconv (CD62L+CD44+) and memory CD8+ T (CD3+CD8+CD62L+CD44+) cells, with reduced percentages of PD-1+TIM3+CD8+ T cells compared with BO1-WT tumors (Figure 7J and Supplemental Figure 11). These data suggest that loss of tumoral GM-CSF may provide a favorable environment for T cells in breast cancer.
To determine whether ARG1 and GM-CSF mRNA expression relates to T cell phenotypes in human breast cancer samples, we queried the Gene Expression of Normal and Tumor tissues 2 (GENT2) database (36) for correlations between T cell markers and ARG1 or CSF2. Across all breast cancer subtypes, we identified a significant (P < 0.05) positive correlation between ARG1 expression and CSF2 expression. In some breast cancer subtypes, especially in luminal B cancers, ARG1 or CSF2 expression negatively correlated with CD3E expression (Supplemental Figure 12). We also evaluated T cell subset markers (CD4 and CD8A) and markers that reflect T cell function, including LAG3, HAVCR2, PDCD1, and FOXP3. We found that expression of the Treg marker FOXP3 significantly (P < 0.05) correlated with ARG1 and CSF2 expression across all breast cancer subtypes. In some breast cancer subtypes, we also observed a correlation between ARG1 or CSF2 expression and putative markers of T cell exhaustion, but this signal was not robust across all breast cancer subtypes from the current data set (Supplemental Tables 3 and 4). These data suggest that CSF2 expression correlates with ARG1 expression in human breast cancer tissue and that expression of CSF2 or ARG1 may reflect an unfavorable environment for T cell function.
Taken together, tumor-derived GM-CSF modulates tumor-infiltrating host immune cells in a myeloid ARG1 protein expression–dependent manner, resulting in enhanced tumor growth. Interruption of tumor cell–derived GM-CSF resulted in decreased TAM infiltration and diminished myeloid cell ARG1 expression and may enhance the antitumor function of infiltrating T cells.
Disruption of tumor cell–produced GM-CSF enhances the efficacy of immune therapy.
Because myeloid cells exposed to breast tumor cell–produced GM-CSF suppressed T cell function in vitro and enhanced tumor growth in immunocompetent mice, we hypothesized that blockade of GM-CSF production in breast tumor cells would increase the efficacy of T cell–targeted cancer immune therapy.
We first evaluated the effect of the disruption of tumor-derived GM-CSF on the efficacy of adoptive T cell transfer therapy. We used CD45.1+ OT-1 TCR-transgenic T cells that are specific for the chicken OVA epitope SIINFEKL (OVA254–267) peptide bound to H-2Kb as the source of antitumor-specific T cells (37). We genetically modified tumor cell lines to express OVA254–267 by a retroviral vector (38) and identified them as B16-OVA, BO1-OVA, BO1-WT-OVA, and BO1-CSF2–KO-OVA cells, respectively. The expression levels of SIINFEKL-bound H-2Kb and MHC-I on B16-OVA and BO1-OVA cells were similar (Supplemental Figure 13).
BO1 and mCherry-expressing BO1-OVA cells were cultured together at a 1:1 ratio and exposed to in vitro–expanded OT-1 T cells. After 16 hours, BO1-OVA cells without OT-1 T cell treatment retained their population, while in the treatment group, OT-1 T cells effectively killed OVA254–267 expressing BO1-OVA cells (Figure 8A). In vivo, adoptively transferred CD45.1+ OT-1 T cells effectively inhibited melanoma B16-OVA subcutaneous tumor growth (Figure 8B) but had no effect on breast cancer BO1-WT-OVA MFP tumor growth (Figure 8C). In contrast, OT-1 T cells attenuated the growth of BO1-CSF2–KO-OVA MFP tumors lacking GM-CSF gene expression (Figure 8D). These results show that, in comparison with B16 melanoma cells, BO1 breast cancer cells were resistant to adoptive T cell transfer therapy and that this resistance could be overcome by disrupting tumor cell–produced GM-CSF.
Figure 8. Disruption of tumor cell–produced GM-CSF enhances breast cancer immune therapy.
(A) BO1 cells (GFP+) were cocultured with BO1-OVA cells (GFP+ mCherry+) at a 1:1 ratio, and then OT-1 T cells were added for 16 hours and analyzed by FACS. (B) B16F10-OVA cells (1 × 106) were subcutaneously injected into C57BL/6J mice. On day 5, one group of mice was treated with 5 × 106 in vitro–expanded OT-1 T cells via intravenous injection. Tumor size was measured by digital calipers. (C and D) PyMT-BO1-WT-OVA or PyMT-BO1-CSF2–KO-OVA breast tumor cells (1 × 105) were inoculated with PBS and injected into MFP tissue. On day 7, OT-1 T cells (5 × 106) were intravenously injected. (E) PyMT-BO1-WT-OVA or PyMT-BO1-CSF2–KO-OVA breast tumor cells (1 × 105) were intracardially injected into 6-week-old female C57BL/6J mice (n = 5–6). On day 5, OT-1 T cells (5 × 106) were intravenously injected. Representative BLI images on day 12 are shown. (F) PyMT-BO1-OVA breast tumor cells (1 × 105) were intracardially injected into 6-week-old female C57BL/6J mice (n = 5–6). On days 5, 7, and 11, anti-CSF2 antibodies were intravenously injected into mice in the antibody treatment groups. On day 5, OT-1 T cells (5 × 106) were intravenously injected. Representative BLI images on day 12 are shown. (G) PyMT-BO1-V2 breast tumor cells (1 × 105) were injected into MFP tissue of 8-week-old female C57BL/6J mice. On days 6, 8, and 10, anti–PD-1 and anti-CTLA4 antibodies (2.5 mg/kg) were intravenously injected. (H) Primary tumor mastectomies were performed when the tumor size reached 1200 mm3. The primary tumor weight after mastectomy is shown. (I) Four weeks after primary tumor mastectomy, distant metastasis was detected by BLI. The rate of metastatic events and representative BLI images are shown. (J–L) The same experiments were performed using PyMT-BO1-CSF2KO breast tumor cells. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-way ANOVA.
As bone metastasis is common and often challenging to treat in patients with breast cancer (39), we asked whether disruption of GM-CSF production could enhance adoptive T cell therapy in a murine bone metastasis model. We intracardially injected BO1-WT-OVA and BO1-CSF2–KO-OVA tumor cells to establish tumor infiltration into bones and visceral organs in WT C57BL/6J mice and injected OT-1 T cells on day 5, monitoring tumor burden by bioluminescence imaging (BLI). We found that without OT-1 T cell treatment, there was no significant difference in bone tumor burden between WT and GM-CSF–KO tumors; however, with OT-1 T cell treatment, the bone tumor burden in GM-CSF–KO bone metastases was significantly lower than in WT mice (Figure 8E). We further evaluated whether pharmacological blockade of GM-CSF with a neutralizing antibody could enhance the efficacy of OT-1 T cell treatment on bone metastases. We found that anti–GM-CSF antibody treatment alone did not significantly decrease bone tumor burden in our experimental metastasis model; however, when combined with OT-1 T cell treatment, neutralization of GM-CSF significantly reduced the bone tumor burden (Figure 8F). These data suggest that genetic or pharmacologic disruption of tumor cell–derived GM-CSF in preclinical models of metastasis can enhance the efficacy of tumor-specific adoptive T cell therapy.
Next, we tested this hypothesis in a model of ICB therapy. The most common ICB targets on T cells are cytotoxic T lymphocyte antigen 4 (CTLA4) and programmed cell death 1 (PD-1) signaling (40). We administered anti–PD-1 antibody and anti-CTLA4 antibody as a combined neoadjuvant therapy in an orthotopic spontaneous metastatic breast cancer model. We established BO1-WT MFP tumors in C57BL/6J mice and intravenously injected anti–PD-1 and anti-CTLA4 antibodies on days 6, 8, and 10, starting when T cells and myeloid cells are present within the TME. When the primary tumor size reached approximately 1000 mm3, we surgically removed the tumor and monitored the mice by BLI for evidence of spontaneous metastases for 4 weeks. We found that ICB treatment did not significantly change BO1-WT primary tumor growth or the rate of metastasis (Figure 8, G–I). However, the same experiment performed with BO1-CSF2–KO tumors showed that ICB treatment decreased GM-CSF–KO primary tumor growth in WT mice. Notably, after resection of tumors that had reached approximately 1000 mm3 in size, the metastasis rate in the control group was approximately 40%, whereas no metastases were observed in the ICB-treated group (Figure 8, J–L). These data suggest that disruption of tumor cell–derived GM-CSF can enhance the efficacy of neoadjuvant ICB treatment in both primary and metastatic settings.
Overall, our results implicate GM-CSF as a contributor to the development of immunosuppression in the TME and suggest that targeting GM-CSF could enhance the efficacy of immune therapy.
Discussion
ARG1 expression and its effect on myeloid cells have been well reported, but not much is known about the role and regulation of ARG1 in breast cancer. Here, we identify GM-CSF produced by breast cancer cells as a critical regulator of the immune-suppressive TME through effects on ARG1 expression in protumor immune-suppressive myeloid cells. We show that the effect of GM-CSF on ARG1 expression in myeloid cells was triggered by the acidic TME and required the JAK/STAT3 and p38 MAPK signaling pathways. Moreover, breast tumor cell–derived GM-CSF promoted tumor progression through inhibition of host antitumor immunity in preclinical mouse models of primary and metastatic tumor growth. We also show that blockade of tumoral GM-CSF enhanced the efficacy of tumor-specific adoptive T cell therapy and ICB. Therefore, targeting GM-CSF or downstream pathways of GM-CSF could be an alternate way to inhibit myeloid cell ARG1 expression, reduce immunosuppression in the breast cancer TME, and enhance immunotherapy efficacy.
GM-CSF is largely redundant for steady-state myelopoiesis and virtually undetectable in circulation (41) but can be administered systemically to promote neutrophil recovery after cytotoxic chemotherapy (42). Physiologically, GM-CSF exerts most of its effects at the local level during immune responses and inflammation (30). GM-CSF can enhance DC function to prime T cells (30, 43) and has been used as an immune adjuvant in cancer vaccines (44). In contrast to the antitumor effects noted above, several studies have reported that tumor cell–derived GM-CSF can suppress immune responses, enhancing tumor-infiltrating immunosuppressive cells, including TAMs, granulocytic MDSCs, Tregs, and plasmacytoid pDCs (14, 45–48), although the mechanism is not fully understood. Importantly, in human breast cancer, higher levels of GM-CSF expression have been correlated with increased metastasis and reduced survival (14–17).
Here, we show that the protumor and immune-suppressive effects of breast tumor–derived GM-CSF require the induction of ARG1 in TIMs that are primarily recruited from the BM. Genetic disruption of GM-CSF in both TNBC and luminal B subtype breast cancer cell lines (4T1 and PyMT-BO1) substantially decreased the percentage of ARG1+ myeloid cells in primary tumor tissue but did not completely inhibit ARG1 expression, suggesting that host-derived GM-CSF could also contribute to myeloid cell ARG1 expression in the TME. Multiple cellular sources of GM-CSF have been described, including epithelial cells, endothelial cells, fibroblasts, stromal cells, and hematopoietic cells (30, 31, 49), and future studies using fresh cancer tissue are aimed at delineating sources of local GM-CSF in the breast cancer TME that may contribute to myeloid ARG1 induction and subsequent immune suppression.
ARG1 expression is an established marker of protumor myeloid cells, which have been associated with immune suppression and enhanced tumor growth (18–20). ARG1 metabolizes the semi-essential amino acid l-arginine into urea and l-ornithine and is required for the maintenance of normal cell growth, collagen synthesis, and neuronal development, as well as tissue repair from injury (19). In the TME, infiltrating ARG1-expressing myeloid cells drive immunosuppression by depleting extracellular l-arginine, resulting in blockade of TCR ζ chain synthesis and inhibition of T cell proliferation (20). Culturing of T cells in media with reduced l-arginine levels markedly impairs T cell function (20, 50), whereas the culturing of T cells in high levels of l-arginine enhances T cell antitumor activity (51). In murine models, treatment of tumor-bearing mice with either l-arginine or ARG1 inhibitors or KO of ARG1 in myeloid cells decreases tumor growth and metastasis and relieves myeloid cell–mediated immune suppression (29, 52–54). We found that the significant difference in growth between WT and GM-CSF–KO breast tumors was eliminated in Arg1fl/fl LysM-Cre+/– mice, suggesting that the effect of breast tumor–derived GM-CSF on the immune-suppressive TME specifically requires ARG1 expression by myeloid cells.
Overexpression of ARG1 mRNA is a poor prognostic factor in many cancer types, including neuroblastoma (55), acute myeloid leukemia (AML) (56), ovarian carcinoma (57), and colorectal cancer (58). In patients with breast cancer, increased arginase activity in both tumor tissue and blood was reported, although its prognostic utility has not been established (59–61). We found that in both patient samples and preclinical cancer models, melanoma and lung tumors had significantly lower numbers of infiltrating ARG1+ myeloid cells compared with breast tumors.
We used a gene expression screen of breast tumor–secreted factors to identify GM-CSF as a critical factor to promote immune-suppressive macrophage polarization. We identified LA as a factor that could combine with GM-CSF to induce myeloid ARG1 levels in a dose-dependent manner. Tumor cell metabolic reprogramming can generate high levels of LA in the TME, secreted in the form of lactate and H+ ions by the MCT. Lactate concentration is therefore often used as a surrogate measure of metabolic acidosis (2, 28). Neither high levels of LA nor GM-CSF alone could induce high expression levels of ARG1 in myeloid cells (Figure 4), suggesting that induction of ARG1 expression in TIMs might be heavily dependent on local, overlapping gradients of GM-CSF and LA, in addition to other factors yet to be identified. Our screens focused on secreted factors and did not evaluate the impact of direct cell-cell interactions, which probably also play a critical role in sculpting immune-suppressive TMEs. The significance of this highly local, context-dependent ARG1 expression by TIMs for a response to immune therapy in human breast cancer demands further study.
Myeloid cell ARG1 expression has been shown to be regulated by IL-4 or IL-13 signaling through STAT6 (2, 59), but the STAT6 pathway was dispensable for both breast tumor CM and GM-CSF– and LA-induced ARG1 expression (Figure 5). A well-known signaling pathway downstream of GM-CSF is JAK2/STAT5. Inhibition of JAK1/2 by ruxolitinib completely blocked tumoral GM-CSF–induced myeloid cell ARG1 expression; however, the STAT5 inhibitor CAS 285986-31-4 had no effect. Accordingly, we turned to other pathways known to regulate immunosuppressive MDSCs, such as STAT3 (62–64), a previously identified target of GM-CSF signaling in human neutrophils (65, 66). Studies have also showed that both STAT3 and p38 MAPK signaling can be regulated by JAK2 (67–69). Indeed, we found that blockade of STAT3 or p38 MAPK signaling abrogated ARG1 expression in myeloid cells (Figure 5). Further, we found that acid signaling through the GPCR/cAMP pathway was also required for robust ARG1 induction in combination with GM-CSF signaling (Figure 6). Therefore, our data suggest that GM-CSF signals through the STAT3 and p38 MAPK pathways within an acidified environment, cooperating with signaling through GPPCRs/cAMP to trigger myeloid cell ARG1 expression.
The immune profile of GM-CSF–KO tumors in this study revealed 2 significant differences: a decreased percentage of ARG1+ myeloid cells and decreased total TAM infiltration (Figure 7). Either or both phenotypes could contribute to the reduced tumor growth phenotype observed in GM-CSF–KO breast tumors. Importantly, GM-CSF disruption restored sensitivity to ICB and antigen-specific CD8+ T cell therapy. Thus, tumor cell–produced GM-CSF modulated TIMs and contributed to an immunosuppressive TME.
Enhanced antitumor T cell activity is critical in current immune checkpoint therapy (1, 70, 71). The majority of breast cancer clinical trials are focused on TNBC, because this subtype has higher numbers of TILs (1). However, the overall response rates to checkpoint treatment in breast cancer are much lower compared with other cancer types like melanoma or lung cancer. Numerous mechanisms of immune suppression have been identified in the breast TME, a better understanding of which is clearly required to design rational combination treatments for patients. One major potential reason for the failure of such therapeutic interventions is that tumor cell metabolism can impact the TME to limit immune responses and present barriers to cancer therapy (2, 10, 70, 71). l-Arginine is one of the essential nutrients required for proper T cell function that can be depleted by ARG1-expressing myeloid cells in the TME (19). Because of in vivo stability, bioavailability, and safety issues, there are currently no arginase inhibitors available for clinical use (59, 72). Here, we found that GM-CSF, a cytokine that normally promotes immunity, could induce the immune-inhibitory enzyme ARG1 in acidic microenvironments common across a variety of cancers. We described the mechanisms and highlighted 2 signaling pathways, GM-CSF and cAMP, which are critical for myeloid ARG1 induction. We also tested several key molecules — JAK1/2, STAT3, p38, AC, and CREB — that can be targeted by inhibitors to block ARG1 expression in vitro. Several clinical inhibitors targeting JAK2, STAT3, and p38 MAPK pathways for cancer treatment are either FDA approved or currently in clinical trials. Future studies are underway to evaluate the inhibition of ARG1 expression using these inhibitors prior to checkpoint therapy in preclinical models.
Together, our data indicate that targeting of tumor cell–produced GM-CSF to alter the myeloid cell phenotype may alleviate immunosuppression and improve antitumor immunity in breast cancer. This finding extends our understanding of the immune-suppressive TME and provides a new strategy for circumventing microenvironment-mediated resistance when designing immunotherapy.
Methods
Additional details on methods are provided in the Supplemental Methods.
Mice.
WT mice (JAX 000664); LysM-Cre mice (JAX 004781; ref. 73); ARG1 reporter mice (YARG, JAX 015857; ref. 24); Arg1fl/fl mice (JAX 008817; ref. 74); GM-CSF–KO (Csf2–/–) mice (JAX 026812; ref. 75); OT-1 mice (JAX 003831; ref. 37); CD45.1 mice (JAX 002014); Stat6–/– mice (JAX 005977; ref. 76); and MMTV-PyMT mice (JAX 022974; ref. 77) were all on a C57BL/6J background and obtained from The Jackson Laboratory. BALB/c mice (JAX 000651) and NSG mice (JAX 005557) were also from purchased from The Jackson Laboratory. Arg1fl/fl mice were crossed with LysM-Cre mice to obtain ARG1 myeloid cell conditional-KO mice (Arg1fl/fl LysM-Cre+/–). YARG mice were crossed with Stat6–/– mice to obtain Stat6–/– YARG mice. OT-1 mice were crossed with CD45.1 mice to obtain OT-1-CD45.1 mice. For in vivo experiments, 8- to 12-week-old mice were used. Mice used for in vitro experiments were 6–12 weeks of age. All mice were housed under pathogen-free conditions.
Cell lines and constructs.
The BALB/c background 4T1-FL-GFP murine mammary tumor cell line was originally from David Piwnica-Worms (The University of Texas, Houston, Texas, USA) as previously described (78). The C57BL/6J background PyMT-BO1-GFP-Luc murine mammary tumor cell line was previously described (22). Both murine breast tumor cell lines had been previously modified to express firefly luciferase and GFP. The B16F10-Luc murine melanoma and LLC-Luc murine lung cancer cell lines were provided by K. Hodivala-Dilke (Barts Cancer Institute, Queen Mary University of London, London, United Kingdom) (79). Nonlabeled B16 and LLC cell lines were from the American Type Culture Collection (ATCC). Gene-KO cell lines were made using the lentiCRISPR version 2 vector system (Addgene, 52961), as described previously (80). All guide RNA (gRNA) sequences used for CRISPR-KO experiments are listed in Supplemental Table 1. Virus was packaged using 293T cells and the helper plasmids pCMV-DR8.2 and pCMV-VSVG. Tumor cell lines were transduced with viral supernatant for 12 hours at 37°C in 6-well tissue culture plates and selected with 5–10 μg/mL puromycin for 3 days. All CRISPR-KO cell lines were validated by ELISA or Western blotting.
GM-CSF–overexpressing B16 and LLC cell lines were established by transfection of plasmid DNA with the murine Csf2 vector pCR3.1-mGM-CSF (Addgene, 74465; ref. 81) using Lipofectamine 3000 (Thermo Fisher Scientific) and selected with 1 mg/mL G418 for 1 week. GM-CSF expression from established cell lines (B16-GM and LLC-GM) was confirmed by ELISA.
OVA257–264–expressing cell lines were established as described previously (38). Briefly, HEK293T Phoenix-Ampho cells were transfected with a PresentER-SIINFEKL (mCherry) vector (Addgene, 102945). After 24 hours, cell culture viral supernatant was harvested every 12 hours. Tumor cell lines were transduced with viral supernatant for 12 hours at 37°C in 6-well tissue culture plates. All cell lines were selected with puromycin and purified by FACS according to mCherry expression.
All cell lines were maintained in DMEM (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS (MilliporeSigma) and penicillin/streptomycin (Gibco, Thermo Fisher Scientific). All cell lines tested negative for mycoplasma.
Mouse tumor models.
4T1 cells were implanted into BALB/C mice, and PyMT-BO1 cells were implanted into C57BL/6J mice. NSG mice received cell lines from mice on both backgrounds. In vivo orthotopic breast tumor models were established by injection of 1 × 105 tumor cells, mixed with BD Matrigel (BD Biosciences) or PBS in a total of 40 μL, into the fourth MFP tissue of 8-week-old female mice.
For subcutaneous injections, 1 × 106 tumor cells in 200 μL PBS were injected into the flank of mice. Tumor growth was monitored and measured with digital calipers.
Breast tumor bone metastasis models were established by intracardiac injection of 1 × 105 tumor cells in 50 μL PBS into 6-week-old mice, as previously described (82). BLI was used to quantify tumor growth after injection. For antibody treatment, mice were given anti–PD-1 and anti-CTLA4 antibodies (2.5 mg/kg, Bio X Cell) by intravenous injection at the indicated time points. For localized anti–GM-CSF antibody treatment, 1 × 105 BO1-GFP-Luc breast tumor cells were mixed with Matrigel plus either an isotype antibody (control) or anti–GM-CSF antibody before injection into the MFP tissue of 8-week-old female C57BL/6J mice. In the metastasis models, anti–GM-CSF antibody (2.5 mg/kg, Bio X Cell) was administered by intravenous injection at the indicated time points.
For adoptive T cell treatment experiments, OT-1 T cells were expanded in vitro before intravenous injection. Briefly, splenic cells from OT-1-CD45.1 mice were harvested and stimulated with 0.5 μg/mL OVA peptide and 10 ng/mL IL-2 in a T-75 flask, and the media were refreshed every day with IL-2 for 3 days. On day 4, T cells were harvested and CD8+ T cells were purified with MACS CD8α magnetic beads and counted, followed by resuspension in PBS before injection.
In vivo BLI.
For BLI of live animals, as previously described (82), mice were injected intraperitoneally with 150 μg/g d-luciferin (Biosynth) in PBS, anesthetized with 2.5% isoflurane, and imaged with a charge-coupled device (CCD) camera–based BLI system (IVIS 100, Caliper Life Sciences; exposure time 1–60 seconds, binning 8, field of view 12, f/stop 1, open filter, anterior side). Signal was displayed as photons/s/cm2/sr. Regions of interest (ROI) were defined manually around the legs using Living Image and Igor Pro Software (version 2.50, WaveMetrics).
BMM culture and treatment.
To generate primary BMMs, whole BM was extracted from the femurs and tibias of mice, plated in Petri dishes in DMEM containing 10% FBS and 50 ng/mL M-CSF, and cultured in a 37°C, 5% CO2 incubator. Day-3 cultured BMMs were plated at 5 × 105 cells per well in 6-well cell culture plates and treated with GM-CSF (2 ng/mL), IL-4 (2 ng/mL), or LA (0–20 mM) for 24 hours before analysis. The following pathway agonists and inhibitors were used: JAK1/2 inhibitor (ruxolitinib, MilliporeSigma); STAT3 inhibitor (C188-9, MilliporeSigma); STAT5 inhibitor (CAS 285986-31-4, MilliporeSigma); MEK inhibitor (trametinib, Cell Signaling Technology); ERK inhibitor (ulixertinib, Chemie Tek); P38 MAPK inhibitor (SB203580, AdipoGen); PTX (MilliporeSigma); forskolin (MilliporeSigma); AC inhibitor (MDL-12, MilliporeSigma); and CREB inhibitor (KG-501, MilliporeSigma). All inhibitors were used as a pretreatment 1–2 hours before administration of cytokines or tumor CM. Tumor cell CM were collected from 24-hour-cultured tumor cells with a cell density of 5 × 105 cells/1 mL DMEM. Tumor cell CM were diluted at 1:1 or 1:2 with fresh media before BMM treatment. After 24 hours or the indicated treatment duration, cells were detached with Dulbecco’s PBS (DPBS) plus 5 mM EDTA and directly used for FACS analysis, lysed for Western blotting, or harvested for qPCR.
Microarray analysis.
Microarray analysis was performed at the Genome Technology Access Center of the Washington University School of Medicine. The microarray data from this study are available in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE75882) (22). PyMT-BO1-GFP-Luc cells and CD206hi TAMs from day-10 MFP tumor tissue were sorted by FACS. RNA was isolated from tumor cells and CD206hi TAMs using a Nucleospin RNA II Kit (Clontech). Data were analyzed as described previously (22).
Lactate and media pH measurements.
Lactate concentrations in cell culture CM were measured with an l-Lactate Assay Kit I (120001, Eton Bioscience) according to the manufacturer’s protocol. Cell culture CM pH was measured by a digital pH meter.
Flow cytometry.
Tumor tissues were prepared in single-cell suspensions for FACS analysis. Briefly, tumor tissue was manually minced using a scalpel, followed by enzymatic digestion with 1 mg/mL collagenase A (Roche) and DNase I (MilliporeSigma) for 30 minutes at 37°C with constant stirring. Cells were filtered through 100 μm nylon strainers (Thermo Fisher Scientific), and then washed twice in 2% FBS PBS. After counting, 1 × 106 total cells were placed in 200 μL buffer (PBS plus 2% FBS) and incubated for 20 minutes with fluorophore-conjugated anti-mouse antibodies using the manufacturers’ recommended concentrations. All antibodies used are listed in Supplemental Table 2. Data acquisition was performed on an LSR-II or X20 system (BD Biosciences), and FlowJo software (version 10, Tree Star) was used for analysis.
Statistics.
GraphPad Prism, version 7 (GraphPad Software), was used for statistical analyses. Differences between groups were evaluated by unpaired 2-tailed Student’s t test or 2-way ANOVA for repeated measures. Data are presented as the mean ± SEM. P values of less than 0.05 were considered statistically significant.
Study approval.
All animal studies were performed according to the guidelines established by the IACUC of the Washington University School of Medicine. Mice were analyzed using approved protocols and were provided appropriate care while undergoing research that complied with the standards set forth in the Guide for the Use and Care of Laboratory Animals (National Academies Press, 2011) and the Animal Welfare Act.
Author contributions
XS supervised the project, designed and performed experiments, and wrote the manuscript. XS, YX, GCF, JX, KAK, JIB, WCL, TK, HMT, JS, and MAM conducted experiments. XS, YX, GCF, JX, JLD, JIB, WHW, FF, LFHA, and DJV analyzed data. LFHA, SJB, SAS, CE, PK, DJV, DGD, GML, KAK, and SA reviewed and edited the manuscript. KNW supervised the project, designed experiments, and reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
This study was supported by the following grants: National Cancer Institute (NCI), NIH, P01 CA100730; NCI R01 CA216840; NIH U54CA199092; US Department of Defense (DoD) BCRP W81XWH-16-1-0286; National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), NIH, R21 AR073507; NIAMS R01 AR070030; and NCI R01CA177670. The authors thank Kendall Blumer, Vivek Arora, Brian Van Tine, and Christopher Maher for their valuable expert suggestions and criticism, and Crystal Idleburg, Lynne Collins, and Julie Prior for their expert technical assistance. The authors also thank the Musculoskeletal Research Center for histology (NIH P30-AR057235); the Molecular Imaging Center at Washington University (NIH, NCI P50-CA09056); the DDRCC Morphology core (grant P30 DK52574); The Pat Burkhart Breast Cancer Fund; The Barnes-Jewish Foundation; The St. Louis Men’s Group Against Cancer; Washington University MSTP grant (GM07200); and the Hope Center Alafi Neuroimaging Lab (NIH Shared Instrumentation grant S10 RR027552).
Version 1. 09/14/2021
In-Press Preview
Version 2. 10/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(20):e145296.https://doi.org/10.1172/JCI145296.
Contributor Information
Xinming Su, Email: xinmingsu@wustl.edu.
Yalin Xu, Email: xuyalin@wustl.edu.
Gregory C. Fox, Email: foxg@wustl.edu.
Jingyu Xiang, Email: xiangjingyu@wustl.edu.
Kristin A. Kwakwa, Email: kristin.kwakwa@wustl.edu.
Jennifer L. Davis, Email: jldavis@wustl.edu.
Jad I. Belle, Email: j.belle@wustl.edu.
Wen-Chih Lee, Email: mosquito1213@gmail.com.
Wing H. Wong, Email: winghing@wustl.edu.
Francesca Fontana, Email: francesca.fontana@wustl.edu.
Takayuki Kobayashi, Email: tkobayashi.g@gmail.com.
Helen M. Tomasson, Email: htomasson@uchicago.edu.
Junyi Su, Email: jsu@wustl.edu.
Suzanne J. Bakewell, Email: suzanne@beestonbiotech.com.
Sheila A. Stewart, Email: Sheila.stewart@wustl.edu.
Christopher Egbulefu, Email: cegbulefu@wustl.edu.
Partha Karmakar, Email: karmakarp@wustl.edu.
Deborah J. Veis, Email: dveis@wustl.edu.
David G. DeNardo, Email: ddenardo@wustl.edu.
Gregory M. Lanza, Email: greg.lanza@me.com.
Samuel Achilefu, Email: achilefu@wustl.edu.
Katherine N. Weilbaecher, Email: kweilbae@wustl.edu.
References
- 1.Adams S, et al. Current landscape of immunotherapy in breast cancer: a review. JAMA Oncol. 2019;5(8):1205–1214. doi: 10.1001/jamaoncol.2018.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19(6):369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson KG, et al. Obstacles posed by the tumor microenvironment to T cell activity: a case for synergistic therapies. Cancer Cell. 2017;31(3):311–325. doi: 10.1016/j.ccell.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corredor G, et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2019;25(5):1526–1534. doi: 10.1158/1078-0432.CCR-18-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Q, et al. Prognostic value of tumor-infiltrating lymphocytes in melanoma: a systematic review and meta-analysis. Oncoimmunology. 2019;8(7):1593806. doi: 10.1080/2162402X.2019.1593806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao G, et al. Prognostic value of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: a systematic review and meta-analysis. BMC Cancer. 2020;20(1):179. doi: 10.1186/s12885-020-6668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havel JJ, et al. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awad RM, et al. Turn back the TIMe: targeting tumor infiltrating myeloid cells to revert cancer progression. Front Immunol. 2018;9:1977. doi: 10.3389/fimmu.2018.01977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engblom C, et al. The role of myeloid cells in cancer therapies. Nat Rev Cancer. 2016;16(7):447–462. doi: 10.1038/nrc.2016.54. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11(11):750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 13.DeNardo DG, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1(1):54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghirelli C, et al. Breast cancer cell-derived GM-CSF licenses regulatory Th2 induction by plasmacytoid predendritic cells in aggressive disease subtypes. Cancer Res. 2015;75(14):2775–2787. doi: 10.1158/0008-5472.CAN-14-2386. [DOI] [PubMed] [Google Scholar]
- 15.Burga RA, et al. Liver myeloid-derived suppressor cells expand in response to liver metastases in mice and inhibit the anti-tumor efficacy of anti-CEA CAR-T. Cancer Immunol Immunother. 2015;64(7):817–829. doi: 10.1007/s00262-015-1692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohanbash G, et al. GM-CSF promotes the immunosuppressive activity of glioma-infiltrating myeloid cells through interleukin-4 receptor-α. Cancer Res. 2013;73(21):6413–6423. doi: 10.1158/0008-5472.CAN-12-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su S, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25(5):605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez PC, et al. Arginine metabolism in myeloid cells shapes innate and adaptive immunity. Front Immunol. 2017;8:93. doi: 10.3389/fimmu.2017.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldwell RW, et al. Arginase: a multifaceted enzyme important in health and disease. Physiol Rev. 2018;98(2):641–665. doi: 10.1152/physrev.00037.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez PC, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64(16):5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 21.Marigo I, et al. T cell cancer therapy requires CD40-CD40L activation of tumor necrosis factor and inducible nitric-oxide-synthase-producing dendritic cells. Cancer Cell. 2016;30(3):377–390. doi: 10.1016/j.ccell.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su X, et al. Antagonizing integrin β3 increases immunosuppression in cancer. Cancer Res. 2016;76(12):3484–3495. doi: 10.1158/0008-5472.CAN-15-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen BM, et al. Systemic dysfunction and plasticity of the immune macroenvironment in cancer models. Nat Med. 2020;26(7):1125–1134. doi: 10.1038/s41591-020-0892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reese TA, et al. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. 2007;447(7140):92–96. doi: 10.1038/nature05746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsou CL, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest. 2007;117(4):902–909. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesokhin AM, et al. Monocytic CCR2(+) myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res. 2012;72(4):876–886. doi: 10.1158/0008-5472.CAN-11-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jost MM, et al. Divergent effects of GM-CSF and TGFbeta1 on bone marrow-derived macrophage arginase-1 activity, MCP-1 expression, and matrix metalloproteinase-12: a potential role during arteriogenesis. FASEB J. 2003;17(15):2281–2283. doi: 10.1096/fj.03-0071fje. [DOI] [PubMed] [Google Scholar]
- 28.Corbet C, Feron O. Tumour acidosis: from the passenger to the driver’s seat. Nat Rev Cancer. 2017;17(10):577–593. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 29.Colegio OR, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513(7519):559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becher B, et al. GM-CSF: from growth factor to central mediator of tissue inflammation. Immunity. 2016;45(5):963–973. doi: 10.1016/j.immuni.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 31.Dougan M, et al. GM-CSF, IL-3, and IL-5 family of cytokines: regulators of inflammation. Immunity. 2019;50(4):796–811. doi: 10.1016/j.immuni.2019.03.022. [DOI] [PubMed] [Google Scholar]
- 32.Hansen G, et al. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134(3):496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig MG, et al. Proton-sensing G-protein-coupled receptors. Nature. 2003;425(6953):93–98. doi: 10.1038/nature01905. [DOI] [PubMed] [Google Scholar]
- 34.Rosenbaum DM, et al. The structure and function of G-protein-coupled receptors. Nature. 2009;459(7245):356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell AP, Smrcka AV. Targeting G protein-coupled receptor signalling by blocking G proteins. Nat Rev Drug Discov. 2018;17(11):789–803. doi: 10.1038/nrd.2018.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SJ, et al. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics. 2019;12(suppl 5):101. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76(1):17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 38.Gejman RS, et al. Rejection of immunogenic tumor clones is limited by clonal fraction. Elife. 2018;7:e41090. doi: 10.7554/eLife.41090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weilbaecher KN, et al. Cancer to bone: a fatal attraction. Nat Rev Cancer. 2011;11(6):411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Topalian SL, et al. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton JA, Anderson GP. GM-CSF biology. Growth Factors. 2004;22(4):225–231. doi: 10.1080/08977190412331279881. [DOI] [PubMed] [Google Scholar]
- 42.Mehta HM, et al. G-CSF and GM-CSF in neutropenia. J Immunol. 2015;195(4):1341–1349. doi: 10.4049/jimmunol.1500861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greter M, et al. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 2012;36(6):1031–1046. doi: 10.1016/j.immuni.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emens LA, et al. Timed sequential treatment with cyclophosphamide, doxorubicin, and an allogeneic granulocyte-macrophage colony-stimulating factor-secreting breast tumor vaccine: a chemotherapy dose-ranging factorial study of safety and immune activation. J Clin Oncol. 2009;27(35):5911–5918. doi: 10.1200/JCO.2009.23.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayne LJ, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21(6):822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marigo I, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010;32(6):790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Quail DF, et al. Obesity alters the lung myeloid cell landscape to enhance breast cancer metastasis through IL5 and GM-CSF. Nat Cell Biol. 2017;19(8):974–987. doi: 10.1038/ncb3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reggiani F, et al. Adipose progenitor cell secretion of GM-CSF and MMP9 promotes a stromal and immunological microenvironment that supports breast cancer progression. Cancer Res. 2017;77(18):5169–5182. doi: 10.1158/0008-5472.CAN-17-0914. [DOI] [PubMed] [Google Scholar]
- 49.Hamilton JA. GM-CSF in inflammation. J Exp Med. 2020;217(1):e20190945. doi: 10.1084/jem.20190945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5(8):641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 51.Geiger R, et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167(3):829–842. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miret JJ, et al. Suppression of myeloid cell arginase activity leads to therapeutic response in a NSCLC mouse model by activating anti-tumor immunity. J Immunother Cancer. 2019;7(1):32. doi: 10.1186/s40425-019-0504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Secondini C, et al. Arginase inhibition suppresses lung metastasis in the 4T1 breast cancer model independently of the immunomodulatory and anti-metastatic effects of VEGFR-2 blockade. Oncoimmunology. 2017;6(6):e1316437. doi: 10.1080/2162402X.2017.1316437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steggerda SM, et al. Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J Immunother Cancer. 2017;5(1):101. doi: 10.1186/s40425-017-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mussai F, et al. Neuroblastoma arginase activity creates an immunosuppressive microenvironment that impairs autologous and engineered immunity. Cancer Res. 2015;75(15):3043–3053. doi: 10.1158/0008-5472.CAN-14-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mussai F, et al. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood. 2013;122(5):749–758. doi: 10.1182/blood-2013-01-480129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Czystowska-Kuzmicz M, et al. Small extracellular vesicles containing arginase-1 suppress T-cell responses and promote tumor growth in ovarian carcinoma. Nat Commun. 2019;10(1):3000. doi: 10.1038/s41467-019-10979-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma Z, et al. Overexpression of arginase-1 is an indicator of poor prognosis in patients with colorectal cancer. Pathol Res Pract. 2019;215(6):152383. doi: 10.1016/j.prp.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 59.Grzywa TM, et al. Myeloid cell-derived arginase in cancer immune response. Front Immunol. 2020;11:938. doi: 10.3389/fimmu.2020.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Boniface J, et al. Expression patterns of the immunomodulatory enzyme arginase 1 in blood, lymph nodes and tumor tissue of early-stage breast cancer patients. Oncoimmunology. 2012;1(8):1305–1312. doi: 10.4161/onci.21678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez G, et al. Arginase activity in patients with breast cancer: an analysis of plasma, tumors, and its relationship with the presence of the estrogen receptor. Onkologie. 2012;35(10):570–574. doi: 10.1159/000343005. [DOI] [PubMed] [Google Scholar]
- 62.Chalmin F, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120(2):457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poschke I, et al. Immature immunosuppressive CD14+HLA–DR–/low cells in melanoma patients are Stat3hi and overexpress CD80, CD83, and DC-sign. Cancer Res. 2010;70(11):4335–4345. doi: 10.1158/0008-5472.CAN-09-3767. [DOI] [PubMed] [Google Scholar]
- 64.Vasquez-Dunddel D, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123(4):1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Shami A, et al. Granulocyte-macrophage colony-stimulating factor-activated signaling pathways in human neutrophils. Selective activation of Jak2, Stat3, and Stat5b. J Biol Chem. 1998;273(2):1058–1063. doi: 10.1074/jbc.273.2.1058. [DOI] [PubMed] [Google Scholar]
- 66.Brizzi MF, et al. Granulocyte-macrophage colony-stimulating factor stimulates JAK2 signaling pathway and rapidly activates p93fes, STAT1 p91, and STAT3 p92 in polymorphonuclear leukocytes. J Biol Chem. 1996;271(7):3562–3567. doi: 10.1074/jbc.271.7.3562. [DOI] [PubMed] [Google Scholar]
- 67.Rawlings JS, et al. The JAK/STAT signaling pathway. J Cell Sci. 2004;117(pt 8):1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 68.Rebe C, et al. STAT3 activation: a key factor in tumor immunoescape. JAKSTAT. 2013;2(1):e23010. doi: 10.4161/jkst.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu T, Lobie PE. Janus kinase 2-dependent activation of p38 mitogen-activated protein kinase by growth hormone. Resultant transcriptional activation of ATF-2 and CHOP, cytoskeletal re-organization and mitogenesis. J Biol Chem. 2000;275(3):2103–2114. doi: 10.1074/jbc.275.3.2103. [DOI] [PubMed] [Google Scholar]
- 70.Colligan SH, et al. Myeloid-driven mechanisms as barriers to antitumor CD8+ T cell activity. Mol Immunol. 2020;118:165–173. doi: 10.1016/j.molimm.2019.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bader JE, et al. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell. 2020;78(6):1019–1033. doi: 10.1016/j.molcel.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clemente GS, et al. Arginase as a potential biomarker of disease progression: a molecular imaging perspective. Int J Mol Sci. 2020;21(15):5291. doi: 10.3390/ijms21155291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clausen BE, et al. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 74.El Kasmi KC, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9(12):1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dranoff G, et al. Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science. 1994;264(5159):713–716. doi: 10.1126/science.8171324. [DOI] [PubMed] [Google Scholar]
- 76.Kaplan MH, et al. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4(3):313–319. doi: 10.1016/S1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 77.Davie SA, et al. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res. 2007;16(2):193–201. doi: 10.1007/s11248-006-9056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith MC, et al. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64(23):8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 79.Baker M, et al. Stromal Claudin14-heterozygosity, but not deletion, increases tumour blood leakage without affecting tumour growth. PLoS One. 2013;8(5):e62516. doi: 10.1371/journal.pone.0062516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanjana NE, et al. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heller L, et al. In vivo electroporation of plasmids encoding GM-CSF or interleukin-2 into existing B16 melanomas combined with electrochemotherapy induces long-term antitumour immunity. Melanoma Res. 2000;10(6):577–583. doi: 10.1097/00008390-200012000-00010. [DOI] [PubMed] [Google Scholar]
- 82.Su X, et al. The ADP receptor P2RY12 regulates osteoclast function and pathologic bone remodeling. J Clin Invest. 2012;122(10):3579–3592. doi: 10.1172/JCI38576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.