ABSTRACT
To develop an effective Pseudomonas aeruginosa outer-membrane-vesicle (OMV) vaccine, we eliminated multiple virulence factors from a wild-type (WT) P. aeruginosa strain, PA103, to generate a recombinant strain, PA-m14. Strain PA-m14 was tailored with a pSMV83 plasmid carrying the pcrV-hitAT fusion gene to produce OMVs. The recombinant OMVs (termed OMV-PH) enclosed increased amounts of the PcrV-HitAT bivalent antigen (PH) and exhibited lower toxicity than did the OMVs from PA103. Intramuscular vaccination with OMV-PH from PA-m14(pSMV83) afforded 70% protection against intranasal challenge with 6.5 × 106 CFU (∼30 50% lethal doses [LD50]) of PA103, while immunization using OMVs without the PH antigen (termed OMV-NA) or the PH antigen alone failed to offer effective protection against the same challenge. Further immune analysis showed that OMV-PH immunization significantly stimulated potent antigen-specific humoral and T-cell (Th1/Th17) responses over those with PH or OMV-NA immunization in mice and that these more-potent responses can effectively hinder P. aeruginosa infection. Undiluted antisera from OMV-PH-immunized mice displayed significantly more opsonophagocytic killing of WT PA103 than antisera from PH antigen- or OMV-NA-immunized mice. Moreover, OMV-PH immunization afforded significant antibody-independent cross-protection to mice against PAO1 and the AMC-PA10 clinical isolate. Taking our findings together, the recombinant P. aeruginosa OMV delivering the bivalent PH antigen exhibits high immunogenicity and may be a promising next-generation vaccine candidate against P. aeruginosa infection.
KEYWORDS: P. aeruginosa, outer membrane vesicles, nanoparticle, vaccine, protective immunity
INTRODUCTION
Pseudomonas aeruginosa, a Gram-negative bacterium, is one of the major opportunistic bacterial pathogens in health care settings (1). P. aeruginosa is listed as one of the leading nosocomial pathogens responsible for life-threatening pneumonia, surgical infection, and bacteremia (2), especially among immunocompromised individuals with underlying diseases such as cancer, AIDS (3), or cystic fibrosis (CF) (4) and among patients in intensive care units (5). P. aeruginosa has a complex gene regulation network including hundreds of genes that enable the bacterium to adapt rapidly to many different environments (6), resulting in its intrinsic resistance to treatment with antibiotics. Recently, the resistance rates of P. aeruginosa have been increasing in many parts of the world. Multidrug-resistant (MDR) and extensively drug-resistant (XDR) high-risk strains are widespread in health care settings (7). Therefore, the treatment of P. aeruginosa infections is becoming extremely challenging, and development of an effective vaccine for active and/or passive immunization is imperative to prevent P. aeruginosa infection and reduce the spread of MDR and XDR P. aeruginosa strains. In the past several decades, vigorous efforts have been aimed at developing an effective P. aeruginosa vaccine (2). Although several P. aeruginosa vaccines have been assessed in clinical trials, no licensed vaccines are available for humans yet (8).
A growing body of evidence has shown that mice immunized with outer membrane vesicles (OMVs) packaging homologous or heterologous antigens can prime significant protective responses counteracting the pathogens from which these homologous or heterologous antigens originated (9). OMVs from Neisseria meningitidis as a component of the vaccine against N. meningitidis serogroup B have been licensed (10), highlighting the potential of OMV-based vaccines to prevent infection by drug-resistant bacteria. P. aeruginosa OMVs are involved in pathogenesis by delivering numerous virulence factors to distant locations (11–13) but also contain abundant OM proteins, such as porins OprF and OprH/OprG and flagellin (14), which are potential protective antigens (15). Protection against P. aeruginosa infection by immunization with OMVs directly purified from wild-type (WT) P. aeruginosa has been observed (16, 17), but OMV toxicity, a major obstacle to OMV vaccines, was not mentioned in those studies. A range of bacteria are being engineered to generate safe and immunogenic OMV vaccines (18), but the use of genetically modified P. aeruginosa strains for making OMV vaccines is largely unexplored.
P. aeruginosa PcrV is located at the tip of its type III secretion system (T3SS) needle complex, which is required for translocation of the effectors (19), and is critical for pathogenicity (20). Studies have demonstrated that immunization with either PcrV alone or PcrV fusion antigens protects against pulmonary and burn infections by P. aeruginosa (21–24). Also, PcrV-specific antibodies are effective in counteracting P. aeruginosa infection in different animal models (25) and can reduce inflammation and damage of the airways of CF patients (26). Thus, PcrV seems to be an ideal antigen. However, PcrV as a vaccine component has not been evaluated in human clinical trials thus far, probably due in part to difficulties in the production of high-quality PcrV (23). In addition, the iron acquisition systems play an important role in the virulence of P. aeruginosa (27, 28). Among them, the ferric iron-binding periplasmic proteins HitA (PA4687) and HitB (PA4688) are involved in iron transportation (29) and are associated with bacterial virulence (30), rendering them potential vaccine candidates. HitA immunization offers protection against systemic infection with P. aeruginosa in the murine model (31). Moreover, protein alignment shows that both PcrV and HitA have 98% to 100% amino acid identity among different clinical isolates. Our previous study demonstrated that immunization with OMVs carrying a vector that oversynthesized the LcrV antigen of Yersinia pestis afforded enhanced protection against pneumonic plague (32). Thus, immunization with OMVs containing increased amounts of the PcrV and HitA antigens might potentiate protective immunity against P. aeruginosa infection. In this study, we genetically manipulated P. aeruginosa PA103, a serotype O11 strain that is prevalent in hospital settings (33), to eliminate an array of virulence factors. The mutant strain was tailored with a plasmid to oversynthesize the PcrV-HitA fusion antigen (PH) and produce immunogenic self-adjuvanting OMVs with diminished toxicity. Immunization with OMVs enclosing PH offered significant protection against lethal pneumonic infection with PA103, stimulated potent humoral and cellular immune responses, and provided broad protection against P. aeruginosa strains of different serotypes.
RESULTS
Trimming P. aeruginosa to mitigate the toxicity of outer membrane vesicles.
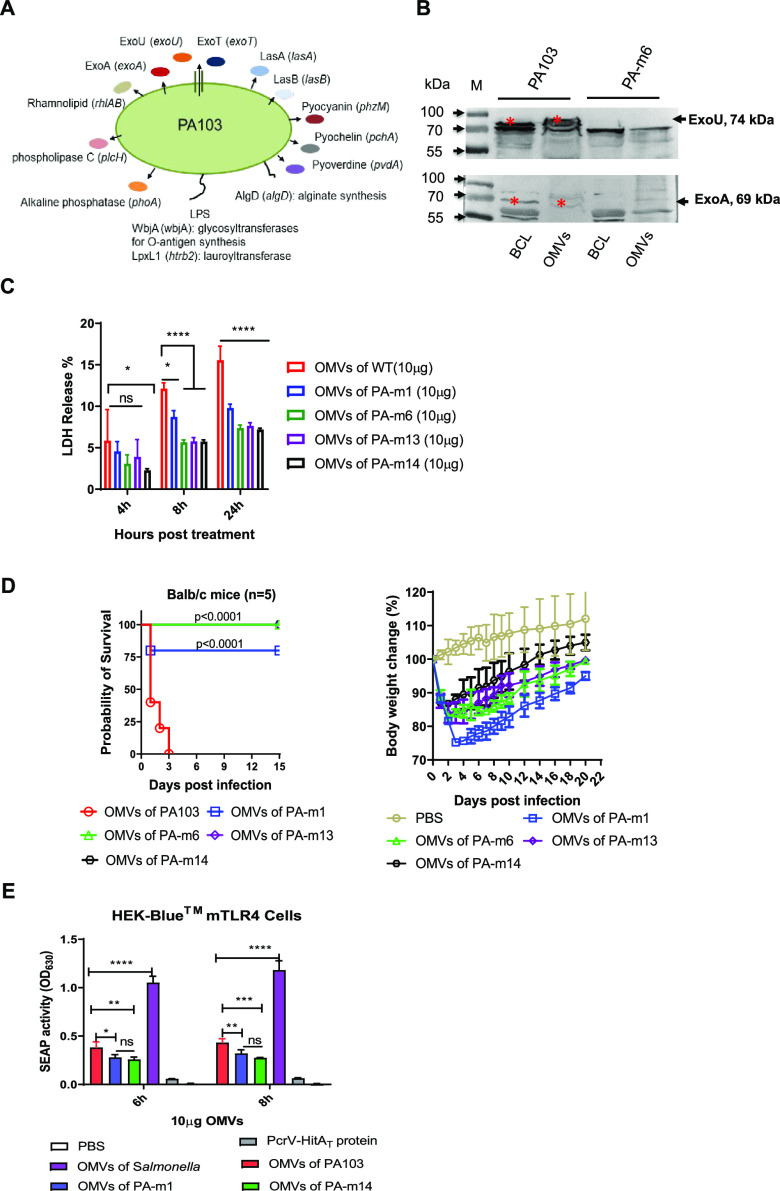
A multitude of virulence factors (Fig. 1A) produced by P. aeruginosa are involved in acute and chronic infections (34). Studies have illustrated that OMVs from WT P. aeruginosa can package numerous virulence factors, such as toxic effectors of the type III secretion system (T3SS), among other toxins, and deliver them into host cells, leading to cytotoxicity and impairment of host defense (11–13). The toxins (ExoU, ExoT, or ExoS) secreted by the T3SS enable P. aeruginosa to breach the epithelial barrier by antagonizing wound healing during colonization and to promote cell injury, ultimately causing pneumonia (35). Additionally, several toxic effectors (exotoxin A, LasA, and LasB) of the type II secretion system (T2SS) contribute to bacterial pathogenicity (36, 37). As shown in Fig. 1B, considerable amounts of known toxins (ExoA and ExoU) were present in OMVs isolated from WT PA103 but absent from OMVs from strain PA-m6, with deletions of multiple toxin genes. The O-antigen moiety of lipopolysaccharides (LPS) is one of the immunogenic antigens in P. aeruginosa. O-antigen immunization confers high levels of protection against the homologous strain but is largely inefficient against different serotypes (38, 39). WbjA (encoded by wbjA), a glycosyltransferase, adds glucose to complete the O-antigen trisaccharide repeating unit of the LPS of PA103 (40). Thus, mutants (from PA-m6 to PA-m14) (Table 1) carrying the wbjA mutation are devoid of full-length O antigen (see Fig. S1A in the supplemental material), eliminating the immune response to the specific O antigen. Besides the toxic factors mentioned above, alginate or elastases can induce high levels of antibodies during P. aeruginosa infection. However, these antibodies have poor opsonic activities, especially in CF patients (41), fail to clear the infection effectively (42), and even exacerbate the lung infection (43). Siderophores (pyochelin and pyoverdine), rhamnolipids, and alkaline phosphatases can promote P. aeruginosa pathogenicity and infection (34, 44). To further mitigate the potential toxicity of OMVs, we consecutively deleted genes encoding different virulence factors (Fig. 1A) to generate a PA-m13 mutant (Table 1).
FIG 1.
Analysis of outer membrane vesicles (OMVs) from the genetically manipulated P. aeruginosa strain PA103. (A) Schematic diagram of genes and their encoding proteins. The 14 genes were deleted constitutively to generate the final strain, PA-m14, producing OMVs of low toxicity. (B) Determining the presence of the ExoU and ExoA toxins in a bacterial cell lysate (BCL) and in OMVs from wild-type PA103 or the PA-m6 mutant strain by Western blotting. (C) Quantification of LDH release into culture supernatants of human THP-1 cells treated with 10 μg/ml of OMVs from WT PA103, PA-m1, PA-m6, PA-m13, or PA-m14 for 4, 8, and 24 h (3 replications). PBS was used for a control group. (D) Toxicities of different OMVs from wild-type PA103 or its derived mutants in BALB/c mice. BALB/c mice (n = 5) were injected intramuscularly with 50 μg of OMVs from either wild-type PA103, PA-m1, PA-m6, PA-m11, or PA-m14. Mouse body weight changes after intramuscular injection with OMVs isolated from different strains were measured. Mice were monitored daily for 2 weeks. Statistical significance was analyzed by the log rank (Mantel-Cox) test. (E) TLR4 activation of OMVs in vitro. Secreted embryonic alkaline phosphatase (SEAP) activities in HEK-Blue cells with murine TLR4 were compared. HEK-Blue mTLR4 cells (InvivoGen) were cocultured with 10 μg/ml OMVs from WT PA103, PA103 ΔlpxL1, or PA-m14 for 6 or 8 h. OMVs from Salmonella Typhimurium were used as a positive control, and 10 μg/ml of purified PcrV-HitAT protein or PBS was used as a negative control. The statistical significance of differences among the groups was analyzed by two-way multivariant ANOVA with a Tukey post hoc test (ns, no significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristics | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| Top10 | F– mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| χ6212 | F– λ– ϕ80 Δ(lacZYA-argF) endA1 recA1 hsdR17 deoR thi-1 glnV44 gyrA96 relA1 ΔasdA4 | 85 |
| SM10(λpir) | Kmr; thi-1 thr-1 leuB26 tonA21 lacY1 supE44 recA integrated RP4-2 Tcr::Mu aphA+ (RP4-2 is RP4 ΔTn1) | 86 |
| RHO3 | Kms; SM10(λpir) Δasd::FRT ΔaphA::FRT | 87 |
| P. aeruginosa | ||
| PA103 | Wild-type strain | Joanna B. Goldberg |
| PAO1 | Wild-type strain | Shouguang Jin |
| AMC-PA10 | Clinical isolate from a patient sputum sample; resistant to piperacillin-tazobactam, ceftazidime, cefepime, aztreonam, ciprofloxacin, amikacin | Albany Medical Center |
| PA103 ΔexoU | ΔexoU | 88 |
| PA-m1 | ΔlpxL1 | This study |
| PA-m2 | ΔexoU ΔexoA | This study |
| PA-m3 | ΔexoU ΔexoA ΔexoT | This study |
| PA-m4 | ΔexoU ΔexoA ΔexoT ΔlasA | This study |
| PA-m5 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB | This study |
| PA-m6 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA | This study |
| PA-m7 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA ΔpchA | This study |
| PA-m8 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA ΔpchA ΔphzM | This study |
| PA-m9 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA ΔpchA ΔphzM Δalg | This study |
| PA-m10 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA ΔpchA ΔphzM Δalg ΔrhlAB | This study |
| PA-m11 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA ΔpchA ΔphzM Δalg ΔrhlAB ΔpvdA | This study |
| PA-m12 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA ΔpchA ΔphzM Δalg ΔrhlAB ΔpvdA ΔplcH | This study |
| PA-m13 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA ΔpchA ΔphzM Δalg ΔrhlAB ΔpvdA ΔplcH ΔphoA | This study |
| PA-m14 | ΔexoU ΔexoA ΔexoT ΔlasA ΔlasB ΔwbjA ΔpchA ΔphzM Δalg ΔrhlAB ΔpvdA ΔplcH ΔphoA ΔlpxL | This study |
| Plasmids | ||
| pYA3342 | Asd+ vector, Ptrc, pBR ori | 85 |
| pYA3493 | Asd+ vector with β-lactamase N-terminal signal sequence, Ptrc, pBR ori | 85 |
| pDMS197 | Suicide vector; Tetr; mob (RP4) R6K ori, sacB | 82 |
| pUCP20 | E. coli-Pseudomonas shuttle vector; Apr Cbr | 89 |
| pSMV81 | The pcrV-hitAT DNA fragment was cloned into EcoRI and HindIII sites in pYA3494 | This study |
| pSMV82 | The pcrV-hitAT-6×His fragment was cloned into NcoI and HindIII sites in pYA3342 | This study |
| pSMV83 | The Ptrc-bla ss-pcrV-hitAT DNA fragment from pSMV81 was cloned into pUCP20 | This study |
The lipid A moiety of LPS in Gram-negative bacteria is another major contributor to toxicity (45). The presence of two acyltransferase HtrB (LpxL) homologs, PA0011 (HtrB1) and PA3242 (HtrB2), in strain PAO1 might modify lipid A via the addition of 2-hydroxylaurate at the C-2 and C-2′ positions, respectively (46). In silico analysis demonstrated that PA103 also has two LpxL homologs, PA103_1714 (99.038% identity to PA3242; designated LpxL1) and PA103_4391 (100% identity to PA0011; designated LpxL2). The lpxL1 deletion was successful in PA103, but not the lpxL2 deletion (lab observation). Thus, adding the lpxL1 mutation to strain PA-m13 so as to generate strain PA-m14 may further reduce bacterial OMV toxicity. Lipid analysis indicated that OMVs from WT PA103 contained both hexa-acylated and hepta-acylated lipid A species, as characterized in P. aeruginosa isolates from cystic fibrosis patients (47, 48), whereas OMVs from PA-m14 completely lost hepta-acylated lipid A species and contained mainly hexa-acylated lipid A species (Fig. S1B and C). Therefore, disruption of LpxL1 led to the loss of a secondary laurate acyl chain. However, the predicted hexa-acylated lipid A species in our OMVs were not present in the study of Ernst and colleagues (46), which reported only penta- or tetra-acylated lipid A species. Additionally, the transmission electron microscopy (TEM) images showed that the morphology of PA-m14 was slightly altered from that of PA103 but that the OMVs from PA-m14 were much smaller than those from WT PA103 (Fig. S2B).
To evaluate the toxicity of P. aeruginosa OMVs, lactate dehydrogenase (LDH) release from human THP-1 cells was measured as described in Materials and Methods below. The results showed that OMVs from WT PA103, PA-m1, PA-m6, and PA-m13 caused comparable cytotoxicity, while the OMVs of PA103 caused significantly more cytotoxicity than the OMVs of PA-m14, after 4 h of treatment (Fig. 1C). The cytotoxicity profiles were similar after 8 and 24 h of treatment. The highest cytotoxicity was observed in cells treated with OMVs from PA103. OMVs from PA-m1, with the elimination of one fatty acid chain of lipid A, presented significantly lower cytotoxicity than OMVs from PA103 but still retained slightly higher cytotoxicity than OMVs from PA-m6, PA-m13, or PA-m14 (Fig. 1C). Subsequently, in vivo toxicity testing of different OMVs showed that mice injected intramuscularly (i.m.) with 50 μg OMVs from WT PA103 succumbed within 3 days, while 80% of mice survived i.m. injection with 50 μg OMVs from strain PA-m1 (with a single lpxL1 mutation), and i.m. injection with 50 μg OMVs from PA-m6, PA-m13, or PA-m14 did not cause any death in mice (Fig. 1D). Injection with OMVs from either PA-m6 or PA-m13 caused mice to gain weight more slowly than with injection with OMVs from PA-m14 over a 20-day observation period (Fig. 1D). The results implied that the deletion of multiple virulence factors and the elimination of one fatty acid chain of lipid A significantly diminished the toxicity of P. aeruginosa OMVs. Further, we compared the secreted embryonic alkaline phosphatase (SEAP) activities of HEK-Blue murine Toll-like receptor 4 (mTLR4) cells cultured with different OMVs. The TLR4-stimulatory activities of OMVs from P. aeruginosa were all dramatically lower than those of OMVs from the Salmonella enterica serovar Typhimurium strain UK1 (a positive control) but significantly higher than those of the purified PcrV-HitAT fusion protein and phosphate-buffered saline (PBS) controls (Fig. 1E). The SEAP activities of OMVs from the P. aeruginosa ΔlpxL1 and PA-m14 strains were comparable but were significantly lower than those of OMVs from the WT P. aeruginosa strain (Fig. 1E). Taken together, these results indicate that PA-m14 OMVs are less toxic than other OMVs. Therefore, we chose to use strain PA-m14 for generating OMVs in this study as a vaccine candidate for a proof of concept.
Increasing the amounts of the PcrV-HitAT fusion antigen enclosed by P. aeruginosa OMVs.
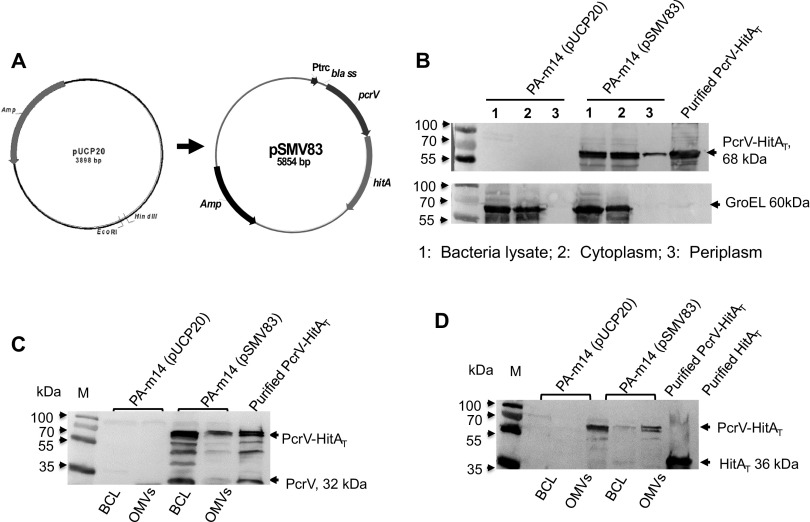
As mentioned above, conservative PcrV and HitA antigens have been evaluated as vaccine candidates (21, 22, 31). P. aeruginosa OMVs contained an array of conservative protein antigens (14), but the amounts of protective antigens (PcrV and HitA) enclosed in the OMVs directly isolated from the mutant strain PA-m14 were marginal (Fig. 2), which could limit OMV immunogenicity. Our previous study indicated that immunization with OMVs carrying increased amounts of the Y. pestis LcrV antigen offered greater protection against plague challenge than immunization with OMVs direct from WT Y. pestis, containing very small amounts of LcrV (32). Thus, we sought to increase the amounts of PcrV and HitA enclosed in OMVs by oversynthesizing a fusion antigen designated PH (68 kDa), which is composed of both truncated PcrV (E28 to I294, with the signal peptide removed) and HitA (D28 to N355) from PA103. Antigens guided by the T2SS into the bacterial periplasm space could increase the antigen amounts in the lumina of OMVs, significantly increasing protective immunity (32, 49, 50). Therefore, we constructed the pSMV83 plasmid (Table 1 and Fig. 2A), in which the bla ss-pcrV-hitAT fragment, encoding an N-terminal β-lactamase signal peptide to facilitate secretion of the PH fusion antigen into the periplasm of P. aeruginosa, was driven by a strong Ptrc promoter. Subsequently, the pSMV83 plasmid was introduced into strain PA-m14 to determine the synthesis of the PH antigen in bacteria and their fractions (cytoplasm, periplasm, and OMVs). The results showed that the cytoplasmic fraction of the PA-m14(pSMV83) strain contained larger amounts of PH than the periplasmic fraction (Fig. 2B), and OMVs from this strain carried significant amounts of the PH antigen (Fig. 2C and D). No PH was detected in those fractions in PA-m14 harboring the empty plasmid pUCP20 (Fig. 2B, C, and D).
FIG 2.
Enhancement of the PcrV-HitAT fusion antigen in P. aeruginosa OMVs. (A) Construction of the pSMV83 plasmid, containing a fusion gene encoding the PH fusion antigen. The Ptrc-bla ss-pcrV-hitAT gene fragment was inserted into pUCP20. (B) Comparison of PH amounts in different cell fractions. The total-cell lysates and subcellular fractions, including the cytoplasmic and periplasmic fractions, were prepared from the PA-m14(pUCP20) and PA-m14(pSMV83) strains. The cells were grown in LB broth at 37°C for 16 h, as described in the Materials and Methods section of the supplemental material. Fractions with 25-μl volumes from cultures grown to an OD600 of 0.8 were evaluated by immunoblotting with a PcrV-specific polyclonal mouse antibody. GroEL was used as a cytoplasmic marker for fractionation. Five micrograms of purified PH protein was used as a loading control. (C) The PH fusion antigen in the BCL (6 × 108 CFU bacterial lysate) and 33 μg OMVs from wild-type PA103 or mutant PA-m14 was detected by Western blotting using mouse primary anti-PcrV antibodies. PH protein (3.5 μg) was used as a loading control. (D) The PH fusion antigen in the BCL (6 × 108 CFU bacterial lysate) and 33 μg OMVs from wild-type PA103 or the PA-m14 mutant was detected by Western blotting using mouse primary anti-HitAT antibodies. PH protein (3.5 μg) was used as a loading control.
Immunization with recombinant P. aeruginosa OMVs induces protection against P. aeruginosa infection.
Before the challenge study, we determined that the LD50 (50% of the lethal dose) of WT PA103 in BALB/c mice by intranasal (i.n.) administration was 2 × 105 CFU (Fig. 3A). Meanwhile, groups of mice (n = 10; 5 males and 5 females) were i.m. immunized with 50 μg of OMVs purified from PA-m14(pSMV83) (referred to as OMV-PH) in 100 μl PBS, which contained ∼2 μg PH, and were then boosted 21 days after prime immunization (Fig. 3B). Compared to the others, immunization with 50 μg of either OMV-PH or OMVs from PA-m14(pUCP20) (termed OMV-NA) affected mouse weight gain (Fig. 3C) and led to moderate swelling at the injection site 1 week after injection (observation data) but did not cause observable disease symptoms in mice. Immunization with 50 μg of OMV-NA, PH (10 μg)-Alhydrogel, or PBS-Alhydrogel (referred to as PBS) was used as an experimental control. On day 42 after the initial vaccination, mice were challenged with P. aeruginosa by the i.n. route. Vaccination with OMV-PH afforded 70% protection for mice infected with 6.5 × 106 CFU (∼30 LD50) of PA103, but only 20% of mice immunized with PH or OMV-NA survived the same challenge (Fig. 3D). None of the PBS-immunized mice survived the challenge (Fig. 3D). Das et al. have reported that vaccination with a PcrV-PopB fusion protein adjuvanted with dmLT reduced P. aeruginosa lung burden (51), so we attempted to evaluate the immune protection of PH plus the dmLT adjuvant in mice. However, no significant differences in antibody titers or protective efficacy between PH-Alhydrogel and PH-dmLT immunization were observed in our study (Fig. S3A).
FIG 3.
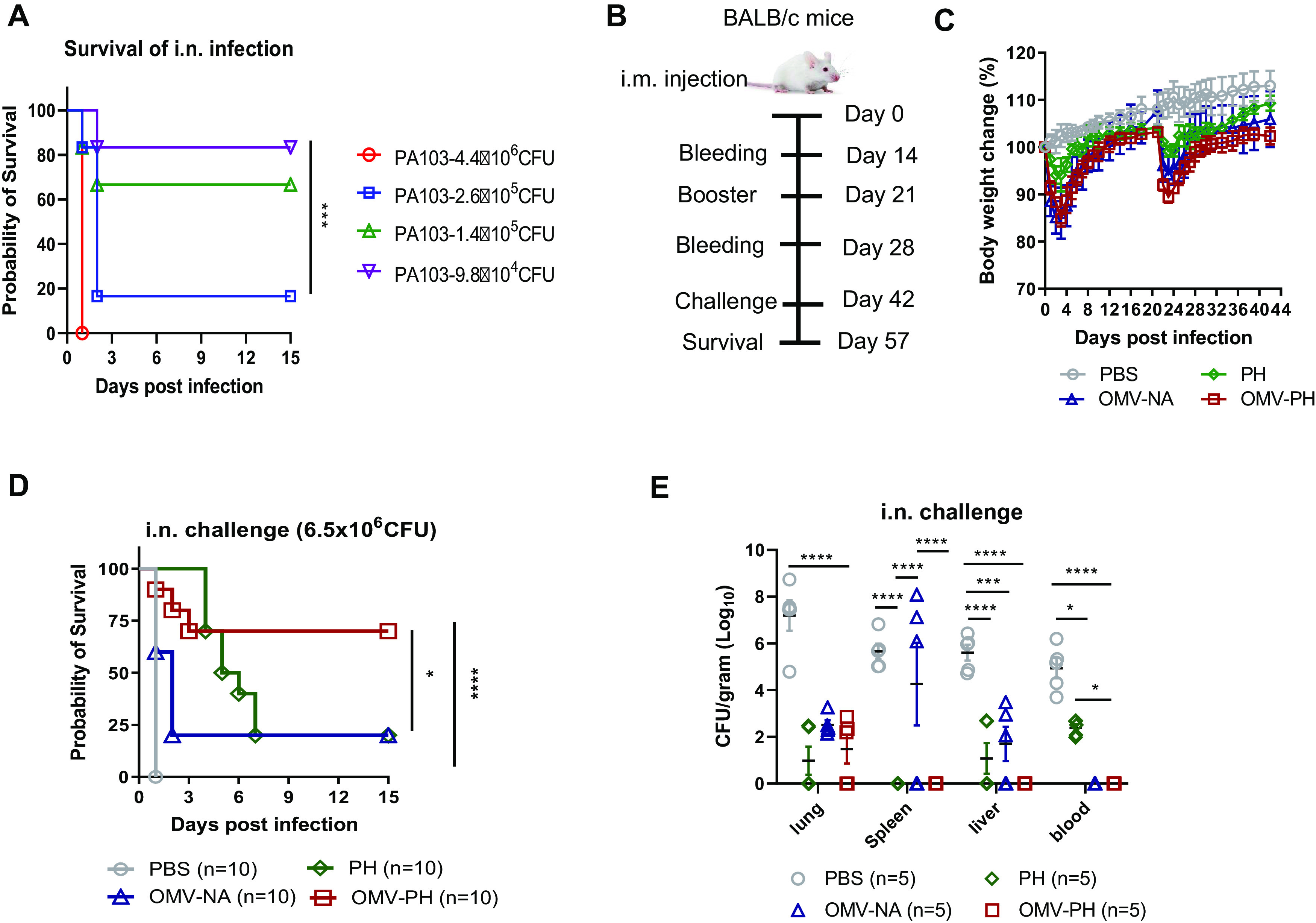
Protective efficacy of P. aeruginosa OMVs against acute pneumonic P. aeruginosa infection. (A) LD50 of intranasal (i.n.) administration. BALB/c mice (n = 10; equal numbers of males and females) were challenged with wild-type PA103 at 4.4 × 106, 2.6 × 105, 1.4 × 105, or 9.8 × 104 CFU/mouse by i.n. administration, and animal survival was recorded for 15 days. (B) Immunization regimen used for the mouse study. (C) BALB/c mice (n = 10) were immunized with either PBS-Alhydrogel, 10 μg of PH-Alhydrogel, 50 μg of OMV-NA, or 50 μg of OMV-PH by i.m. injection and were boosted 21 days after the prime immunization. Mouse weight was monitored and recorded for 6 weeks. (D) On day 42 after the initial immunization, mice were challenged with 6.5 × 106 CFU of wild-type PA103 (∼30 LD50) by i.n. administration, and animal survival was recorded for 15 days. The experiments were performed twice, and data were combined for analysis. Statistical significance was analyzed by the log rank (Mantel-Cox) test. (E) On day 42 after the initial immunization, BALB/c mice (n = 5) were infected i.n. with a sublethal dose (5 × 105 CFU) of PA103. On day 2 postchallenge, different tissues (lung, liver, spleen, and blood) were collected from euthanized mice. Data are shown as means ± SD. The experiments were performed twice, and data were combined for analysis. The statistical significance of differences among the groups was analyzed by two-way multivariant ANOVA with a Tukey post hoc test (ns, no significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Further, groups of immunized mice (n = 5) were challenged with a lethal dose of PA103 (5 × 105 CFU) to determine bacterial burdens in major organs. At 2 days postinfection (dpi), PBS-immunized mice had substantially higher P. aeruginosa titers in lungs (mean, 7.2 log10 CFU/g tissue), spleens (mean, 5.7 log10 CFU/g tissue), livers (mean, 5.6 log10 CFU/g tissue), and blood (mean, 5.2 log10 CFU/g tissue) than PH-, OMV-NA-, or OMV-PH-immunized mice. In PH-immunized mice, bacteria reached moderate levels in livers (mean, 1.2 log10 CFU/g tissue) and blood (mean, 2.5 log10 CFU/g tissue), but no bacteria were detected in spleens (Fig. 3E). In OMV-NA-immunized mice, bacteria reached moderate levels in spleens (mean, 4.3 log10 CFU/g tissue) and livers (mean, 1.2 log10 CFU/g tissue); however, no bacteria were detected in blood (Fig. 3E). No P. aeruginosa was detected in the spleens, livers, and blood of OMV-PH-immunized mice (Fig. 3E). In addition, all OMV-immunized mice survived subcutaneous challenge with 7.4 × 107 CFU (10 LD50) of PA103, while 40% of PH-immunized mice survived the same challenge, and PBS-immunized mice succumbed to the challenge within 4 days (Fig. S3B).
Serum antibody responses and the microbial killing capacity in vitro.
Antibody measurement showed that the highest anti-PH IgG titers among all immunized groups were mounted by the PH-immunized mice at week 2, 4, or 6 postimmunization. OMV-PH immunization stimulated significantly higher anti-PH IgG titers than OMV-NA immunization, but lower anti-PH IgG titers than PH immunization, at week 2, 4, or 6 postimmunization (Fig. 4A). Anti-PH IgG titers from both the OMV-PH- and PH-immunized groups were significantly boosted at week 4 and were maintained at week 6 (Fig. 4A). To distinguish between Th1/Th2 responses in immunized mice (52), analysis of IgG subclasses in response to PH showed that the IgG2a/IgG1 ratios in OMV-immunized mice were close to 1 at weeks 4 and 6 postimmunization, while the IgG2a/IgG1 ratios in PH-immunized mice were less than 0.7 at different points (Fig. 4B). Also, measurement of IgG titers in response to a P. aeruginosa cell lysate (PCL) showed that higher anti-PCL IgG titers were mounted in both OMV-PH- and OMV-NA-immunized mice at 2 weeks postimmunization and that these titers were significantly boosted at week 4 and maintained at week 6. However, low anti-PCL IgG titers were maintained in PH-immunized mice even after a booster (Fig. 4C). Analysis of IgG subclasses in response to PCL showed that the IgG2a/IgG1 ratios in mice immunized with either OMV-PH or OMV-NA were ≥1 at weeks 4 and 6 postimmunization, while the IgG2a/IgG1 ratios were much less than 1 (∼0.6) in mice immunized with PH throughout the entire period (Fig. 4D). Collectively, the OMV-immunized mice generated broader antibody responses against multiple antigens and more-balanced Th1/Th2 responses than the PH-immunized mice.
FIG 4.
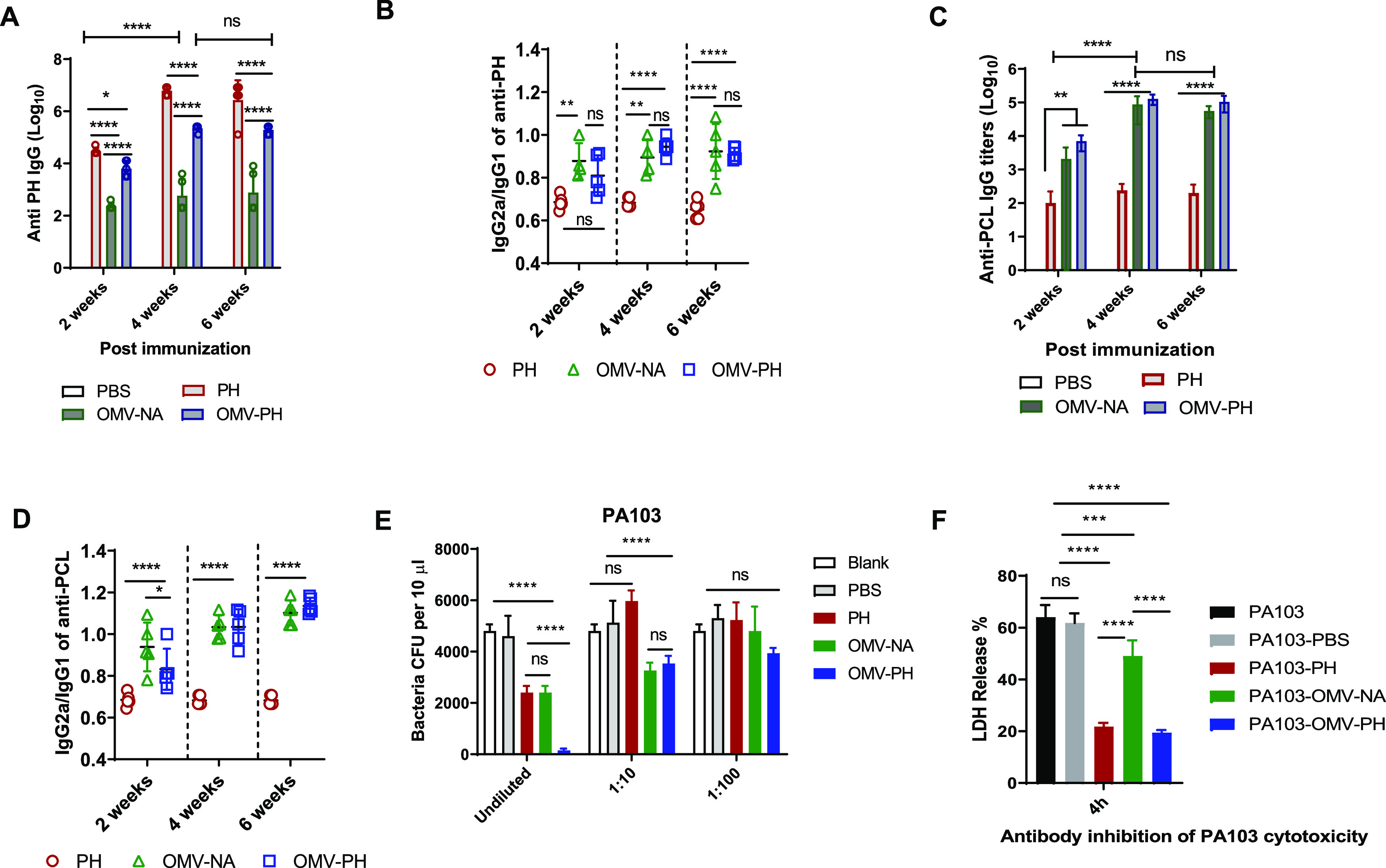
Antibody responses to the PH fusion antigen in immunized mice and antibody opsonophagocytic killing capacity. BALB/c mice were immunized with either PBS-Alhydrogel, 10 μg of PH-Alhydrogel, 50 μg of OMV-NA, or 50 μg of OMV-PH by i.m. administration and were then boosted on day 21 after prime immunization. Blood was collected on days 14, 28, and 42, and antigen-specific antibodies were determined by ELISA. Data represent 5 mice per group. (A) Total anti-PH IgG titers at days 14, 28, and 42 in differently immunized mice. (B) IgG2a/IgG1 ratios in response to the PH fusion antigen at days 14, 28, and 42. (C) Total anti-PCL IgG titers at days 14, 28, and 42 in differently immunized mice. (D) IgG2a/IgG1 ratios in response to a PCL at days 14, 28, and 42. (E) Comparative analysis of opsonophagocytic killing activity against PA103 using antisera from differently immunized mice. (F) Assay of antibody inhibition of PA103 cytotoxicity to HeLa cells. Sera collected from different immunized mice were used for this assay (see Materials and Methods). Data are shown as means ± SD. The statistical significance of differences among groups was analyzed by two-way multivariant ANOVA with a Tukey post hoc test (ns, no significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (F) Inhibition of PA103 cytotoxicity by sera from immunized mice.
Since an opsonophagocytic killing (OPK) assay has already been established to evaluate the correlation of functional antibody levels in serum samples with protection (53, 54), we used it to determine whether the P. aeruginosa-specific antibodies were protective. Undiluted sera from OMV-PH-immunized mice exhibited the highest killing activity (∼97% of PA103 organisms were killed), and undiluted sera from OMV-NA- or PH-immunized mice also exhibited significantly higher opsonophagocytic activity (∼50%) for PA103 than sera from PBS-immunized mice (Fig. 4E). The OPK activity of 10-fold-diluted sera from OMV-PH- or OMV-NA-immunized mice decreased to around 35% but was still substantially higher than that from PBS- or PH-immunized mice, while sera from PH-immunized mice completely lost OPK activities after 10-fold dilution (Fig. 4E). There were no significant differences in serum OPK activity after 100-fold dilution (Fig. 4E). The results suggested that antibodies from OMV-PH- or OMV-NA-immunized mice exhibited significant OPK activity in a concentration-dependent manner. Surprisingly, sera from all the immunized mice described above failed to show significant OPK activity for PAO1 (serotype O5) or a clinical isolate from patient sputum, AMC-PA10 (Table 1; also Fig. S3C and D). Since P. aeruginosa PA103 is a cytotoxic strain (55), we determined whether sera generated from immunized mice could block the cytotoxicity of PA103 for HeLa cells. The results showed that sera from OMV-NA-immunized mice afforded moderate protection against PA103 cytotoxicity in comparison to the PBS control but significantly less protection than sera from either PH- or OMV-PH-immunized mice (Fig. 4F). This indicates that the PH-specific antibody is the major contributor to mitigating P. aeruginosa cytotoxicity, while blocking P. aeruginosa cytotoxicity alone is not sufficient to prevent infection.
Cell-mediated immune responses induced by OMV-PH immunization.
After 48 h of in vitro induction with the PH fusion antigen, lung and spleen cells were stained and were analyzed using flow cytometry. Lung CD4+ T cells from OMV-PH-immunized mice displayed dramatically higher production of gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin 17A (IL-17A) than those from OMV-NA-, PH-, or PBS-immunized mice (Fig. 5). The numbers of CD4+ IFN-γ-producing cells in the lungs from PH- or OMV-NA-immunized mice were comparable but were significantly higher than those from control mice (Fig. 5, right). After PH stimulation, the largest amounts of TNF-α and IFN-γ were produced in lung CD8+ T cells from OMV-PH-immunized mice. The amounts of TNF-α and IFN-γ produced by lung CD8+ T cells from OMV-NA- or PH-immunized mice were comparable but higher than those from control animals (Fig. S4). There were no significant differences in lung CD8+ T cells producing IL-17A among OMV-PH-, OMV-NA-, and PH-immunized mice (Fig. S4).
FIG 5.
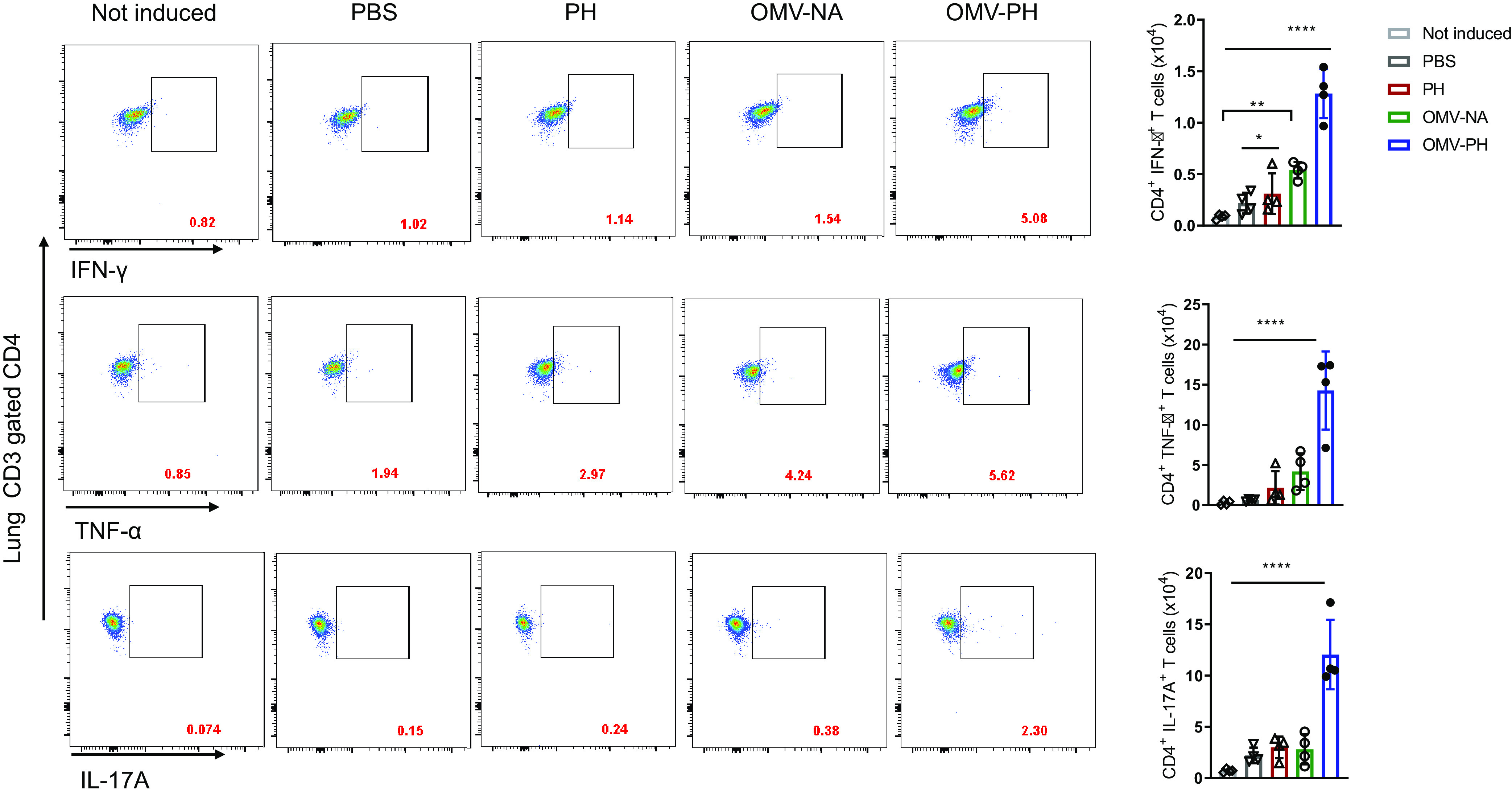
Analysis of antigen-specific lung CD4+ T-cell responses in immunized mice. BALB/c mice (n = 4) were immunized with PBS-Alhydrogel, 10 μg of PH-Alhydrogel, 50 μg of OMV-NA, or 50 μg of OMV-PH by i.m. administration. On day 42 after the initial immunization, lymphocytes from the lungs were aseptically isolated from mice and were stimulated in vitro with 10 μg/ml of purified recombinant PcrV-HitAT fusion protein (PH) for 48 h to detect specific CD4+ T cells producing IFN-γ, TNF-α, or IL-17A. PBS-immunized mouse lung cells were used as controls. (Left) Representative flow cytometry profiles of lung CD4+ T cells producing IFN-γ, TNF-α, or IL-17A from differently immunized mice. (Right) Quantification of CD4+ IFN-γ+, CD4+ TNF-α+, and CD4+ IL-17A+ cells. Each symbol represents a data point obtained from an individual mouse. Bars represent means; error bars, SD. The experiments were performed twice, and data were combined for analysis. The statistical significance of differences among the groups was analyzed by two-way multivariant ANOVA with a Tukey post hoc test (ns, no significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
The numbers of spleen CD4+ T cells producing IFN-γ were comparable for OMV-PH- and OMV-NA-immunized mice but were significantly higher than those from PH-immunized and PBS-immunized mice (Fig. 6). The numbers of spleen CD4+ T cells producing TNF-α and IL-17A from OMV-PH-immunized mice were dramatically higher than those from OMV-NA-, PH-, and PBS-immunized mice (Fig. 6). Spleen CD8+ T cells from mice immunized with either type of OMV produced higher levels of IFN-γ and TNF-α than cells from PH- or PBS-immunized mice (Fig. S5). Similarly, there were no significant differences in spleen CD8+ T cells producing IL-17A among OMV-PH-, OMV-NA-, and PH-immunized mice (Fig. S5). Taking these findings together, the OMV-PH vaccination elicited more-potent antigen-specific Th1 and Th17 responses in the lungs and spleens of mice than the other vaccinations.
FIG 6.
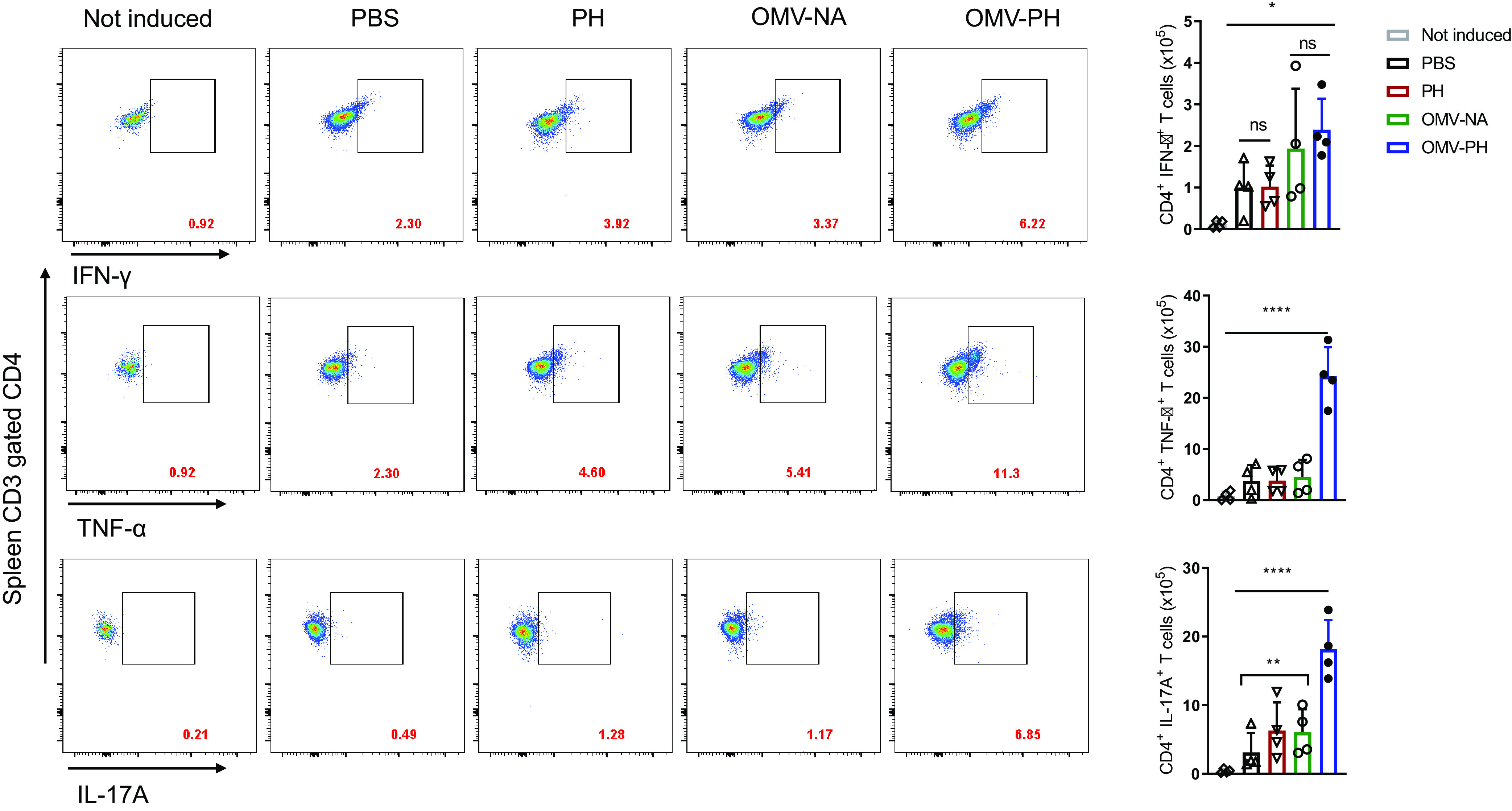
Analysis of antigen-specific spleen CD4+ T-cell responses in immunized mice. BALB/c mice (n = 4) were immunized with either PBS-Alhydrogel, 10 μg of PH-Alhydrogel, 50 μg of OMV-NA, or 50 μg of OMV-PH by i.m. administration. On day 42 after the initial immunization, lymphocytes from the spleen were aseptically isolated from mice and were stimulated in vitro with 10 μg/ml of PH for 48 h to detect specific CD4+ T cells producing IFN-γ, TNF-α, or IL-17A. PBS-immunized mouse lung cells were used as controls. (Left) Representative flow cytometry profiles of spleen CD4+ T cells producing IFN-γ, TNF-α, or IL-17A from differently immunized mice. (Right) Quantification of CD4+ IFN-γ+, CD4+ TNF-α+, and CD4+ IL-17A+ cells. Each symbol represents a data point obtained from an individual mouse. Bars represent means; error bars, SD. The experiments were performed twice, and data were combined for analysis. The statistical significance of differences among the groups was analyzed by two-way multivariant ANOVA with a Tukey post hoc test (ns, no significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
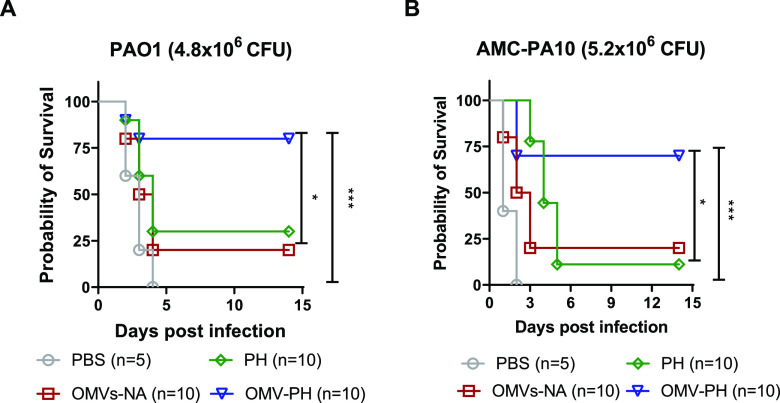
OMV-PH vaccination offers protection against P. aeruginosa strains of different serotypes in murine pneumonia models.
Further, we investigated whether OMV-PH immunization could offer broad protection. At 42 days after the initial immunization, OMV-PH-, OMV-NA-, PH-, or PBS-immunized mice were challenged with the most commonly used laboratory-adapted strain, PAO1 (serotype O5), or the clinical isolate AMC-PA10. OMV-PH immunization was able to provide 60% protection against i.n. challenge with 4.8 × 106 CFU of PAO1 and 4.8 × 106 CFU of AMC-PA10 (Fig. 7). Low percentages of PH- or OMV-NA-immunized mice survived the same challenge, and no PBS-immunized mice survived this challenge (Fig. 7).
FIG 7.
Evaluation of broad protection against pulmonary infection with different P. aeruginosa strains in immunized mice. BALB/c mice (n, 5 to 10; mixed males and females) were immunized with either PBS-Alhydrogel, 10 μg of PH-Alhydrogel, 50 μg of OMV-NA, or 50 μg of OMV-PH by i.m. administration and were then boosted on day 21 after prime immunization. (A) Survival of mice challenged intranasally with a lethal dose (4.8 × 106 CFU) of PAO1 on day 42 after the initial immunization. (B) Survival of mice challenged intranasally with a lethal dose (5.2 × 106 CFU) of AMC-PA10 on day 42 after the initial immunization. The experiments were performed twice, and data were combined for analysis. Statistical significance was analyzed by the log rank (Mantel-Cox) test (ns, no significance; *, P < 0.05; ***, P < 0.001).
DISCUSSION
The biogenesis of OMVs from pathogenic Gram-negative bacteria is associated with numerous cellular behaviors, such as interbacterial communication, threat avoidance, virulence, and modulation of the host immune response (56). OMVs from Gram-negative bacteria intrinsically contain different pathogen-associated molecular patterns (PAMPs) and an array of potential antigens that can activate innate and adaptive immune responses (57); thus, they possess high potential as vaccines. The goal of this study was to build a proof of concept for developing recombinant P. aeruginosa OMVs as vaccines to prevent surges of drug-resistant P. aeruginosa in health care settings.
Rational elimination of multiple known toxins (ExoU, ExoT, and ExoA) and other virulence factors significantly decreased the toxicity of P. aeruginosa OMVs (Fig. 1C). In addition, OMVs isolated from strain PAO1 induce potent detrimental inflammation reactions in the lung via TLR2 and TLR4 pathways in vivo (58). Lipid A, one of the moieties of endotoxin (LPS) sensed by the TLR4 complex, can lead to toxicity and even septic shock (59). Tetra- and penta-acylated lipid A species in P. aeruginosa lack immunostimulatory activity and cause fewer neutrophil respiratory bursts than several hexa- and hepta-acylated lipid A species (60, 61). However, the single lpxL1 mutation, removing a secondary laurate acyl chain in the lipid A species of PA-m1 and PA-m14 OMVs, significantly reduced toxicity (Fig. 1C and D) and TLR4 activation from those for OMVs from WT PA103 (Fig. 1E). So far, OMVs from PA-m14 still contained abundant hexa-acylated lipid A species (see Fig. S1B in the supplemental material) and uncharacterized TLR2 agonists that may contribute to the remaining toxicity. We noticed that the lipid A species in OMVs from WT PA103 and from PA-m14 with the lpxL1 mutation (Fig. S1B) were not completely consistent with those in previous studies (46–48). These inconsistencies may be due to different P. aeruginosa strains, culture conditions, or sample preparations. Also, the disruption of LpxL2 that seemed to mediate the addition of the C-2 position 2-hydroxylaurate in P. aeruginosa (46) was not achievable in strain PA103 (lab observations). Regarding the above discrepancies, we speculate that the presence of high phenotypic heterogeneity among different P. aeruginosa species (62) might be one of the reasons. Moreover, the presence of PagP, a lipid A palmitoyltransferase for the addition of a palmitate (C16:0) acyl chain (47), and PagL, a lipid A deacetylase for removing the C10 acyl chain at position 3 (63–65), in P. aeruginosa may impact lipid A fatty acid acylation. Further studies on lipid A synthesis in strain PA103 will be pursued to mitigate the toxicity of P. aeruginosa OMVs. Regarding the fact that PA-m14, with multiple mutations, produces OMVs smaller than those of the WT (Fig. S1D), we speculate that deletions of O antigen (wbjA), exopolysaccharide (algD), or quorum-sensing signaling systems (lasAB and rhlAB) might have led to this occurrence.
Antigens guided by the T2SS into the periplasmic space of P. aeruginosa could increase the antigen amounts in the lumina of OMVs (Fig. 2B, C, and D) and enhance protective immunity (Fig. 3D). However, the amounts of PH antigen encased in OMVs were still relatively low. The reason might be that the Bla SS fragment leading fusion antigen secretion by the T2SS, which originated from Escherichia coli, may not be fully compatible with the T2SS of P. aeruginosa. The N-terminal amino acid residues of P. aeruginosa exotoxin A (residues 1 to 120) are sufficient to direct β-lactamase secretion (66), implying that N-terminal signal peptides of P. aeruginosa T2SS substrates fused with homologous or heterologous proteins might enhance the secretion of these fusion proteins into the periplasm of P. aeruginosa. Alternatively, increased bacterial membrane curvature favors OMV biogenesis (67), which may promote antigen enclosure in OMVs. Disruption of tolR in E. coli (68) and Salmonella enterica (69) resulted in high levels of OMV formation without significantly compromising the cell envelope and growth. Protein alignments showed that PA103_1767 (39% amino acid identity) in strain PA103 was homologous to E. coli TolR. However, the tolR mutation in PA103 was unsuccessful (lab observation). It is not clear whether the TolR in PA103 has other functions essential for bacterial replication. In addition, the crude OMVs used in the current study may contain protein aggregates, bacterial debris, and OMVs of mixed sizes (70, 71), which may distract immune responses after immunization. Also, several disadvantages of density gradient ultracentrifugation, such as lower volume processability, high equipment requirements, time-consuming and labor-intensive processes, and low portability, limit the quantitative yield of OMVs. Since the current yield of OMV-PH was relatively low, we used the crude OMVs without further purification via the density gradient ultracentrifugation in this study. In future studies, we will seek to greatly increase P. aeruginosa OMV production by disrupting genes associated with membrane curvature and to prepare purified OMVs for immunization.
PH immunization generated higher PH-specific antibody titers than OMV-PH or OMV-NA immunization (Fig. 4A), and OMV-NA immunization induced levels of anti-PCL titers comparable to those with OMV-PH immunization (Fig. 4C). However, OMV-NA or PH immunization failed to offer good protection against pulmonary challenge with PA103 (Fig. 3D) and did not effectively prevent bacterial persistence in the lungs or dissemination to livers and spleens (Fig. 3E) compared to OMV-PH immunization. Also, sera from PH-immunized mice could not effectively kill P. aeruginosa in the in vitro OPK assay compared to sera from OMV-PH- or OMV-NA-immunized mice (Fig. 4E). Our results were inconsistent with those of several previous studies, in which anti-PcrVNH sera from PcrVNH-immunized mice (24) or POH-specific antibodies from mice vaccinated with the trivalent subunit PcrV-OprI-Hcp1 (POH) (23) exhibited significant OPK activity against P. aeruginosa. One possible explanation is that different P. aeruginosa strains used in the OPK assay or sera from mice immunized with different antigen combinations caused this inconsistency. Another explanation is that OMVs enrich bacterial outer membrane and periplasmic components. Thus, high anti-PCL antibody titers raised from mice immunized with OMVs, instead of PH antigen (Fig. 4C), may target many P. aeruginosa factors. Intriguingly, undiluted sera from OMV-PH-, OMV-NA-. or PH-immunized mice had marginal opsonic killing activity against PAO1 and AMC-PA10 in vitro (Fig. S3B and C). Strain PA-m14, with a wbjA mutation, lacked full-length O antigen attached to the LPS core (Fig. S1A). Thus, OMV-PH immunization is supposed to generate antibodies in mice to an array of conserved antigens of P. aeruginosa, but not to the O antigen of PA103. Currently, the reason why the OMV-PH immunization sera did not have OPK activity against strains PAO1 and AMC-PA10 in vitro is unclear. A study reported that the combination of flagellin-, OprI-, and OprF-specific IgG antibodies triggered the highest level of C3-deposition-mediated opsonic killing activity (72). Strain PA103 did not synthesize flagella composed of flagellin units (73). Thus, the lack of flagellin-specific IgG antibodies, and the low levels of OprI/OprF-specific IgG antibodies. induced by the OMV-PH immunization may lead to the deficiency of broad OPK activity against different types of P. aeruginosa strains. In addition, the quality of antibodies generated in PH-, OMV-PH-, or OMV-NA-immunized mice may not be optimal.
Unlike OMV-NA or PH immunization, OMV-PH immunization induced high PH-specific antibody titers, but also balanced Th1/Th2 or Th1-biased immune responses (Fig. 4B and D). Moreover, lung and spleen CD4+ T cells from OMV-PH-immunized mice produced significant levels of Th1/Th17 cytokines (IFN-γ, IL-17A, or TNF-α) after in vitro PH stimulation in comparison to cells from OMV-NA- or PH-immunized mice (Fig. 5 and 6). Growing clinical and experimental evidence suggests that an excellent P. aeruginosa vaccine must stimulate antibodies and Th1/Th17-type T-cell responses to provide effective protection against pulmonary and systemic infection with P. aeruginosa (23, 74, 75). The PH-specific antibodies from PH or OMV-PH immunization both significantly inhibited cytotoxicity caused by PA103 infection (Fig. 4F), but in vivo protection against PA103 infection differed substantially between PH and OMV-PH immunization (Fig. 3D; also Fig. S3B), further suggesting that antibody alone is not sufficient to prevent P. aeruginosa infection. Thus, both potent antigen-specific antibody and T-cell responses to OMV-PH immunization can explain why only the OMV-PH immunization could afford significant broad protection against different P. aeruginosa strains (Fig. 3 and 7). The detailed underlying mechanisms for protection will be interrogated further.
Chronic lung infection with P. aeruginosa accounts for most of the morbidity and mortality in CF patients (76). Studies have shown that high levels of antibodies against alginate or elastases were induced upon P. aeruginosa infection, but these antibodies had poor opsonic activities, especially in CF individuals (41), where they failed to clear the infection effectively (42, 77) and could even exacerbate lung infection (43). Increasing numbers of studies have demonstrated that humoral and cellular immune responses play synergistic roles in protection against P. aeruginosa infection (8, 78). Th17-mediated protection against P. aeruginosa in mice is antibody independent (79). The absence of alginate or elastases in OMV-PH might eliminate the potential adverse effects of immunization on CF individuals. Moreover, immunization with OM-PH induced potent antigen-specific Th1 and Th17 responses that facilitated P. aeruginosa clearance in the respiratory tract and reduced mortality (Fig. 3, 5, and 6). Although OMV-PH exhibits higher potential than other formulations in this study, more efforts are needed to improve P. aeruginosa OMV vaccines further. Ultimately, the concept in this work will be valuable in the development of OMV-based vaccines against other drug-resistant pathogens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacterial growth conditions are described in the supplemental material.
Constructions of plasmids and PA103 mutant strain.
PA103 mutant strains and plasmids are listed in Table 1, and the DNA primers used in this study are listed in Table S1 in the supplemental material. For the detailed procedure for constructing each plasmid, see the supplemental material. The procedure for PA103 mutant construction using the sacB-based sucrose counterselectable suicide vectors was similar to those in previous reports, with minor modifications (80, 81). Briefly, a ∼1-kb flanking region of each gene was assembled by overlapping PCR using the corresponding primers listed in Table S1 and was individually cloned into the XbaI and SacI sites of pDMS197 (Tetr) (82) to generate the corresponding gene deletion suicide vector, which was conjugated into the P. aeruginosa strain by allelic exchange. The resulting mutant was confirmed by PCR.
OMV isolation, quantification, and cytotoxicity measurement.
OMVs were isolated from P. aeruginosa strains as described previously, with minor modifications (32). The detailed procedure is described in the supplemental material. OMV cytotoxicity was assayed in vitro as reported previously (83), with minor modifications. Human THP-1 cells were seeded at a density of 2.5 × 105 per well in a 48-well plate and were treated with 10 μg of different OMVs. PBS was used as a control. The release of lactate dehydrogenase (LDH) in the supernatants of OMV-treated cells was determined at 4, 8, and 24 h posttreatment using a CytoTox 96 Non-Radioactive Cytotoxicity Assay kit (Promega). Cell death was expressed as a percentage of maximum LDH release. The percentage of cytotoxicity was calculated as follows: (optical density at 490 nm [OD490] of treated cells – OD490 of untreated cells)/(OD490 of lysed untreated cells – OD490 of untreated cells) × 100%.
Stimulation assay in cell lines.
To determine the stimulatory activity of OMVs via Toll-like receptor 4 (TLR4), HEK-Blue human TLR4 cells (InvivoGen, San Diego, CA, USA) were maintained at 37°C under 5% CO2 in Dulbecco’s modified Eagle medium (DMEM; Gibco BRL, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS) supplemented with 100 μg/ml penicillin, 100 μg/ml streptomycin, and 100 μg/ml Normocin. Cells were seeded at a density of 5 × 104 per well in 96-well tissue culture plates (Costar, Washington, DC) and were stimulated with 20 μl OMVs isolated from different strains (final concentration, 10 μg/ml) for 8 h. Purified protein and PBS were used as negative controls. Relative NF-κB activity was determined by measuring the secreted embryonic alkaline phosphatase (SEAP) activity in the culture supernatant according to the manufacturer’s instructions (InvivoGen).
Animal studies.
Animal care and experimental protocols were conducted according to the NIH Guide for the Care and Use of Laboratory Animals (84) and were approved by the Institutional Animal Care and Use Committee at Albany Medical College (IACUC protocol 20-02001). Six-week-old male and female BALB/c mice were purchased from Taconic (Germantown, NY) and were acclimated for 1 week after arrival. The groups of mice were intramuscularly (i.m.) immunized with 50 μg OMVs in 100 μl PBS buffer, 10 μg PcrV-HitAT–Alhydrogel in a 100-μl mixture as a subunit vaccine control, or 100 μl PBS-Alhydrogel as a negative control. Booster vaccinations were then administered 3 weeks after the initial vaccination. Blood samples were collected via submandibular veins at intervals of 2 weeks in order to harvest sera for antibody analysis. At 42 days after the initial vaccination, the animals were anesthetized with a 1:5 xylazine-ketamine mixture and were intranasally challenged with PA103 in 40 μl PBS to mimic pneumonic infection (23). All infected animals were observed over 15 days. The number of bacterial CFU was determined by plating serial dilutions of the inoculum onto LB agar plates.
For determination of the bacterial burden, animals were euthanized with an overdose of sodium pentobarbital at 36 h postinfection. Lungs, livers, and spleens were removed and homogenized in ice-cold PBS (pH 7.4) using a bullet blender (Bullet Blender Blue; Next Advance, Inc., Troy, NY, USA) at power 7 for 2 min. Serial dilutions of each organ homogenate were plated onto LB agar, and each count was confirmed with duplicate plates to determine the titers of bacteria per gram of tissue. The experiments were performed twice, and the data were combined for analysis.
Antibody responses, opsonophagocytic killing assay, and inhibition of P. aeruginosa cytotoxicity assay.
Antibody titers were measured using an enzyme-linked immunosorbent assay (ELISA) as described in the supplemental material. The opsonophagocytic killing assay was carried out as described previously (23). Briefly, HL-60 cells (ATCC; CCL-240) were differentiated into granulocyte-like cells in a growth medium containing 100 mM N′,N-dimethylformamide (Sigma) for 5 days. Serum samples from immunized mice containing opsonic antibodies were heat inactivated (56°C, 30 min) and serially diluted with opsonization buffer (a mixture of 80 ml of sterile water, 10 ml of 10× Hanks’ balanced salt solution, 10 ml of 1% gelatin, and 5.3 ml of fetal bovine serum). We added the following components to each well in a 96-well plate: 40 μl of 4 × 105 HL60 cells, 103 CFU of PA103 in 10 μl of opsonophagocytic buffer, 20 μl of serum, and 10 μl of 1% infant rabbit serum as a complement source (Sigma). Blank wells with the same system in the absence of mouse serum were used as negative controls. After a 2-h incubation, 10 μl of each sample was plated onto LB agar medium. Each sample was performed in triplicate. The opsonophagocytic killing ability was defined as a reduction in CFU compared with the CFU in the sera from unimmunized mice. The assay of inhibition of P. aeruginosa cytotoxicity is described in the supplemental material.
Analysis of T-cell responses.
Lungs and spleens were obtained aseptically from euthanized animals and were dissociated with 70-μm strainers to obtain single cells. The-individual cell populations (2 × 106) derived from the lysis of red blood cells (RBC) were seeded in 12-well cell culture plates and were stimulated in vitro for 48 h with 10 μg/ml of recombinant PcrV-HitAT (rPcrV-HitAT). Four hours before the collection of cells, the culture medium in each well was supplemented with brefeldin A and a monensin cocktail (1:1 ratio) to block Golgi apparatus-mediated cytokine secretion. For the flow cytometric analysis of the T-cell populations and their corresponding cytokines, the induced cells were harvested and resuspended in a fluorescence-activated cell sorter (FACS) staining buffer containing CD16/32 antibodies (1:200) for 10 min on ice. The T-cell-specific markers were stained using anti-mouse CD3 (with fluorescein isothiocyanate [FITC]), CD4 (with phycoerythrin [PE]), and CD8 (with allophycocyanin [APC]) antibodies (BioLegend, CA), followed by intracellular cytokine staining (for IFN-γ, peridinin chlorophyll protein [PerCP] Cy5.5; for TNF-α, BV510; for IL17A, APC-Cy7) according to the manufacturer’s protocol. The events (50,000 cells) were acquired on BD flow cytometers (LSR II) and were analyzed using FlowJo, v.10.
Statistical analysis.
The statistical analyses of the data and comparisons among the groups were performed by one-way analysis of variance (ANOVA)/univariate or two-way ANOVA with Tukey post hoc tests. The log rank (Mantel-Cox) test was used for survival analysis. All data were analyzed using GraphPad Prism software (version 8.0). The data are represented as means ± standard deviations (SD), and levels of significance are indicated as follows: ns, no significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
ACKNOWLEDGMENTS
We thank Herbert P Schweizer for sharing the RHO3 strain, Roy Curtiss III for sharing the χ6212 strain and pYA plasmids, Shouguang Jin for sharing the pUCP20 plasmid and PAO1 strains, Joanna B. Goldberg for sharing the PA103 strain, and Alan R. Hauser for sharing the PA103 ΔexoU strain and antibodies against ExoU. Also, we thank Ravindra Thakkar at the Nanotechnology Innovation Center of Kansas State University for obtaining the OMV images via electron microscopy.
This work was supported by the Albany Medical College startup fund and was also partially supported by National Institutes of Health grant R21AI139703 to W.S.
Experiments were conceived and designed by W.S., P.L., and X.W. and were performed by P.L., X.W., and X.S. Data were analyzed by P.L., X.W., Z.G., and W.S. The manuscript was written by P.L., X.W., Z.G., and W.S. and was edited by J.C., Z.G., and W.S.
All authors declare that they have no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Wei Sun, Email: sunw@amc.edu.
Igor E. Brodsky, University of Pennsylvania
REFERENCES
- 1.Wolfgang MC, Kulasekara BR, Liang XY, Boyd D, Wu K, Yang Q, Miyada CG, Lory S. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 100:8484–8489. 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grimwood K, Kyd JM, Owen SJ, Massa HM, Cripps AW. 2015. Vaccination against respiratory Pseudomonas aeruginosa infection. Hum Vaccin Immunother 11:14–20. 10.4161/hv.34296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolston KV. 2017. Infections in cancer patients with solid tumors: a review. Infect Dis Ther 6:69–83. 10.1007/s40121-017-0146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LiPuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church D, Elsayed S, Reid O, Winston B, Lindsay R. 2006. Burn wound infections. Clin Microbiol Rev 19:403–434. 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahajan-Miklos S, Rahme LG, Ausubel FM. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol Microbiol 37:981–988. 10.1046/j.1365-2958.2000.02056.x. [DOI] [PubMed] [Google Scholar]
- 7.Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, Benito N, Grau S. 2019. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev 32:e00031-19. 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Priebe GP, Goldberg JB. 2014. Vaccines for Pseudomonas aeruginosa: a long and winding road. Expert Rev Vaccines 13:507–519. 10.1586/14760584.2014.890053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Pol L, Stork M, van der Ley P. 2015. Outer membrane vesicles as platform vaccine technology. Biotechnol J 10:1689–1706. 10.1002/biot.201400395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holst J, Martin D, Arnold R, Huergo CC, Oster P, O’Hallahan J, Rosenqvist E. 2009. Properties and clinical performance of vaccines containing outer membrane vesicles from Neisseria meningitidis. Vaccine 27(Suppl 2):B3–B12. 10.1016/j.vaccine.2009.04.071. [DOI] [PubMed] [Google Scholar]
- 11.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5:e1000382. 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadurugamuwa JL, Beveridge TJ. 1997. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J Antimicrob Chemother 40:615–621. 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- 13.Ballok AE, Filkins LM, Bomberger JM, Stanton BA, O’Toole GA. 2014. Epoxide-mediated differential packaging of Cif and other virulence factors into outer membrane vesicles. J Bacteriol 196:3633–3642. 10.1128/JB.01760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS. 2011. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11:3424–3429. 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- 15.Sharma A, Krause A, Worgall S. 2011. Recent developments for Pseudomonas vaccines. Hum Vaccin 7:999–1011. 10.4161/hv.7.10.16369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Yang F, Zou J, Wu W, Jing H, Gou Q, Li H, Gu J, Zou Q, Zhang J. 2018. Immunization with Pseudomonas aeruginosa outer membrane vesicles stimulates protective immunity in mice. Vaccine 36:1047–1054. 10.1016/j.vaccine.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 17.Wang SH, Gao J, Li M, Wang LG, Wang ZJ. 2018. A facile approach for development of a vaccine made of bacterial double-layered membrane vesicles (DMVs). Biomaterials 187:28–38. 10.1016/j.biomaterials.2018.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerritzen MJH, Martens DE, Wijffels RH, van der Pol L, Stork M. 2017. Bioengineering bacterial outer membrane vesicles as vaccine platform. Biotechnol Adv 35:565–574. 10.1016/j.biotechadv.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Goure J, Pastor A, Faudry E, Chabert J, Dessen A, Attree I. 2004. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect Immun 72:4741–4750. 10.1128/IAI.72.8.4741-4750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee PC, Stopford CM, Svenson AG, Rietsch A. 2010. Control of effector export by the Pseudomonas aeruginosa type III secretion proteins PcrG and PcrV. Mol Microbiol 75:924–941. 10.1111/j.1365-2958.2009.07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawa T, Yahr TL, Ohara M, Kurahashi K, Gropper MA, Wiener-Kronish JP, Frank DW. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat Med 5:392–398. 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 22.Holder IA, Neely AN, Frank DW. 2001. PcrV immunization enhances survival of burned Pseudomonas aeruginosa-infected mice. Infect Immun 69:5908–5910. 10.1128/IAI.69.9.5908-5910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang F, Gu J, Yang L, Gao C, Jing H, Wang Y, Zeng H, Zou Q, Lv F, Zhang J. 2017. Protective efficacy of the trivalent Pseudomonas aeruginosa vaccine candidate PcrV-OprI-Hcp1 in murine pneumonia and burn models. Sci Rep 7:3957. 10.1038/s41598-017-04029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan C, Zhang J, Zhao LQ, Cheng X, Gao C, Wang Y, Xu WT, Zou QM, Gu J. 2019. Rational design of a chimeric derivative of PcrV as a subunit vaccine against Pseudomonas aeruginosa. Front Immunol 10:781. 10.3389/fimmu.2019.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawa T, Ito E, Nguyen VH, Haight M. 2014. Anti-PcrV antibody strategies against virulent Pseudomonas aeruginosa. Hum Vaccin Immunother 10:2843–2852. 10.4161/21645515.2014.971641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.François B, Luyt CE, Dugard A, Wolff M, Diehl JL, Jaber S, Forel JM, Garot D, Kipnis E, Mebazaa A, Misset B, Andremont A, Ploy MC, Jacobs A, Yarranton G, Pearce T, Fagon JY, Chastre J. 2012. Safety and pharmacokinetics of an anti-PcrV PEGylated monoclonal antibody fragment in mechanically ventilated patients colonized with Pseudomonas aeruginosa: a randomized, double-blind, placebo-controlled trial. Crit Care Med 40:2320–2326. 10.1097/CCM.0b013e31825334f6. [DOI] [PubMed] [Google Scholar]
- 27.Vasil ML, Ochsner UA. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol Microbiol 34:399–413. 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 28.Visca P, Leoni L, Wilson MJ, Lamont IL. 2002. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol Microbiol 45:1177–1190. 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- 29.Li KW, Xu C, Jin YX, Sun ZY, Liu C, Shi J, Chen GK, Chen RH, Jin SG, Wu WH. 2013. SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio 4:e00419-13. 10.1128/mBio.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nde CW, Jang HJ, Toghrol F, Bentley WE. 2008. Toxicogenomic response of Pseudomonas aeruginosa to ortho-phenylphenol. BMC Genomics 9:473. 10.1186/1471-2164-9-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elhosary MA, Bahey-El-Din M, AbdelBary A, El Guink N, Aboushleib HM. 2019. Immunization with the ferric iron-binding periplasmic protein HitA provides protection against Pseudomonas aeruginosa in the murine infection model. Microb Pathog 131:181–185. 10.1016/j.micpath.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Singh AK, Zhang X, Sun W. 2020. Induction of protective antiplague immune responses by self-adjuvanting bionanoparticles derived from engineered Yersinia pestis. Infect Immun 88:e00081-20. 10.1128/IAI.00081-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thrane SW, Taylor VL, Freschi L, Kukavica-Ibrulj I, Boyle B, Laroche J, Pirnay JP, Lévesque RC, Lam JS, Jelsbak L. 2015. The widespread multidrug-resistant serotype O12 Pseudomonas aeruginosa clone emerged through concomitant horizontal transfer of serotype antigen and antibiotic resistance gene clusters. mBio 6:e01396-15. 10.1128/mBio.01396-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gellatly SL, Hancock REW. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 35.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz MJ, Speelman P, Zaat SA, Hack CE, van Deventer SJ, van der Poll T. 2000. The effect of Pseudomonas exotoxin A on cytokine production in whole blood exposed to Pseudomonas aeruginosa. FEMS Immunol Med Microbiol 29:227–232. 10.1111/j.1574-695X.2000.tb01527.x. [DOI] [PubMed] [Google Scholar]
- 37.Mariencheck WI, Alcorn JF, Palmer SM, Wright JR. 2003. Pseudomonas aeruginosa elastase degrades surfactant proteins A and D. Am J Respir Cell Mol Biol 28:528–537. 10.1165/rcmb.2002-0141OC. [DOI] [PubMed] [Google Scholar]
- 38.Haghbin M, Armstrong D, Murphy ML. 1973. Controlled prospective trial of Pseudomonas aeruginosa vaccine in children with acute leukemia. Cancer 32:761–766. . [DOI] [PubMed] [Google Scholar]
- 39.Pennington JE, Reynolds HY, Wood RE, Robinson RA, Levine AS. 1975. Use of a Pseudomonas aeruginosa vaccine in patients with acute leukemia and cystic fibrosis. Am J Med 58:629–636. 10.1016/0002-9343(75)90498-2. [DOI] [PubMed] [Google Scholar]
- 40.Dean CR, Datta A, Carlson RW, Goldberg JB. 2002. WbjA adds glucose to complete the O-antigen trisaccharide repeating unit of the lipopolysaccharide of Pseudomonas aeruginosa serogroup O11. J Bacteriol 184:323–326. 10.1128/JB.184.1.323-326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fick RB. 1989. Lung humoral response to Pseudomonas species. Eur J Clin Microbiol Infect Dis 8:29–34. 10.1007/BF01964117. [DOI] [PubMed] [Google Scholar]
- 42.Ciofu O, Petersen TD, Jensen P, Høiby N. 1999. Avidity of anti-P aeruginosa antibodies during chronic infection in patients with cystic fibrosis. Thorax 54:141–144. 10.1136/thx.54.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Høiby N. 1993. Antibiotic therapy for chronic infection of Pseudomonas in the lung. Annu Rev Med 44:1–10. 10.1146/annurev.me.44.020193.000245. [DOI] [PubMed] [Google Scholar]
- 44.Lau GW, Ran HM, Kong FS, Hassett DJ, Mavrodi D. 2004. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun 72:4275–4278. 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hittle LE, Powell DA, Jones JW, Tofigh M, Goodlett DR, Moskowitz SM, Ernst RK. 2015. Site-specific activity of the acyltransferases HtrB1 and HtrB2 in Pseudomonas aeruginosa lipid A biosynthesis. Pathog Dis 73:ftv053. 10.1093/femspd/ftv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thaipisuttikul I, Hittle LE, Chandra R, Zangari D, Dixon CL, Garrett TA, Rasko DA, Dasgupta N, Moskowitz SM, Malmström L, Goodlett DR, Miller SI, Bishop RE, Ernst RK. 2014. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol Microbiol 91:158–174. 10.1111/mmi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ernst RK, Moskowitz SM, Emerson JC, Kraig GM, Adams KN, Harvey MD, Ramsey B, Speert DP, Burns JL, Miller SI. 2007. Unique lipid A modifications in Pseudomonas aeruginosa isolated from the airways of patients with cystic fibrosis. J Infect Dis 196:1088–1092. 10.1086/521367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muralinath M, Kuehn MJ, Roland KL, Curtiss R, III.. 2011. Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect Immun 79:887–894. 10.1128/IAI.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elluri S, Enow C, Vdovikova S, Rompikuntal PK, Dongre M, Carlsson S, Pal A, Uhlin BE, Wai SN. 2014. Outer membrane vesicles mediate transport of biologically active Vibrio cholerae cytolysin (VCC) from V. cholerae strains. PLoS One 9:e106731. 10.1371/journal.pone.0106731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das S, Howlader DR, Zheng Q, Ratnakaram SSK, Whittier SK, Lu T, Keith JD, Picking WD, Birket SE, Picking WL. 2020. Development of a broadly protective, self-adjuvanting subunit vaccine to prevent infections by Pseudomonas aeruginosa. Front Immunol 11:583008. 10.3389/fimmu.2020.583008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Germann T, Bongartz M, Dlugonska H, Hess H, Schmitt E, Kolbe L, Kölsch E, Podlaski FJ, Gately MK, Rüde E. 1995. Interleukin-12 profoundly up-regulates the synthesis of antigen-specific complement-fixing IgG2a, IgG2b and IgG3 antibody subclasses in vivo. Eur J Immunol 25:823–829. 10.1002/eji.1830250329. [DOI] [PubMed] [Google Scholar]
- 53.Tsoi SK, Smeesters PR, Frost HR, Licciardi P, Steer AC. 2015. Correlates of protection for M protein-based vaccines against group A Streptococcus. J Immunol Res 2015:167089. 10.1155/2015/167089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamei A, Coutinho-Sledge YS, Goldberg JB, Priebe GP, Pier GB. 2011. Mucosal vaccination with a multivalent, live-attenuated vaccine induces multifactorial immunity against Pseudomonas aeruginosa acute lung infection. Infect Immun 79:1289–1299. 10.1128/IAI.01139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kudoh I, Wiener-Kronish JP, Hashimoto S, Pittet JF, Frank D. 1994. Exoproduct secretions of Pseudomonas aeruginosa strains influence severity of alveolar epithelial injury. Am J Physiol 267:L551–L556. 10.1152/ajplung.1994.267.5.L551. [DOI] [PubMed] [Google Scholar]
- 56.Anand D, Chaudhuri A. 2016. Bacterial outer membrane vesicles: new insights and applications. Mol Membr Biol 33:125–137. 10.1080/09687688.2017.1400602. [DOI] [PubMed] [Google Scholar]
- 57.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park KS, Lee J, Jang SC, Kim SR, Jang MH, Lötvall J, Kim YK, Gho YS. 2013. Pulmonary inflammation induced by bacteria-free outer membrane vesicles from Pseudomonas aeruginosa. Am J Respir Cell Mol Biol 49:637–645. 10.1165/rcmb.2012-0370OC. [DOI] [PubMed] [Google Scholar]
- 59.Opal SM. 2007. The host response to endotoxin, antilipopolysaccharide strategies, and the management of severe sepsis. Int J Med Microbiol 297:365–377. 10.1016/j.ijmm.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 60.SenGupta S, Hittle LE, Ernst RK, Uriarte SM, Mitchell TC. 2016. A Pseudomonas aeruginosa hepta-acylated lipid A variant associated with cystic fibrosis selectively activates human neutrophils. J Leukoc Biol 100:1047–1059. 10.1189/jlb.4VMA0316-101R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korneev KV, Arbatsky NP, Molinaro A, Palmigiano A, Shaikhutdinova RZ, Shneider MM, Pier GB, Kondakova AN, Sviriaeva EN, Sturiale L, Garozzo D, Kruglov AA, Nedospasov SA, Drutskaya MS, Knirel YA, Kuprash DV. 2015. Structural relationship of the lipid A acyl groups to activation of murine Toll-like receptor 4 by lipopolysaccharides from pathogenic strains of Burkholderia mallei, Acinetobacter baumannii, and Pseudomonas aeruginosa. Front Immunol 6:595. 10.3389/fimmu.2015.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kordes A, Preusse M, Willger SD, Braubach P, Jonigk D, Haverich A, Warnecke G, Häussler S. 2019. Genetically diverse Pseudomonas aeruginosa populations display similar transcriptomic profiles in a cystic fibrosis explanted lung. Nat Commun 10:3397. 10.1038/s41467-019-11414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561–1565. 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 64.Ernst RK, Adams KN, Moskowitz SM, Kraig GM, Kawasaki K, Stead CM, Trent MS, Miller SI. 2006. The Pseudomonas aeruginosa lipid A deacylase: selection for expression and loss within the cystic fibrosis airway. J Bacteriol 188:191–201. 10.1128/JB.188.1.191-201.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trent MS, Pabich W, Raetz CR, Miller SI. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem 276:9083–9092. 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 66.Lu HM, Lory S. 1996. A specific targeting domain in mature exotoxin A is required for its extracellular secretion from Pseudomonas aeruginosa. EMBO J 15:429–436. 10.1002/j.1460-2075.1996.tb00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pérez-Cruz C, Cañas MA, Giménez R, Badia J, Mercade E, Baldomà L, Aguilera L. 2016. Membrane vesicles released by a hypervesiculating Escherichia coli Nissle 1917 tolR mutant are highly heterogeneous and show reduced capacity for epithelial cell interaction and entry. PLoS One 11:e0169186. 10.1371/journal.pone.0169186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nevermann J, Silva A, Otero C, Oyarzún DP, Barrera B, Gil F, Calderón IL, Fuentes JA. 2019. Identification of genes involved in biogenesis of outer membrane vesicles (OMVs) in Salmonella enterica serovar Typhi. Front Microbiol 10:104. 10.3389/fmicb.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang D, Zhang W, Zhang H, Zhang F, Chen L, Ma L, Larcher LM, Chen S, Liu N, Zhao Q, Tran PHL, Chen C, Veedu RN, Wang T. 2020. Progress, opportunity, and perspective on exosome isolation — efforts for efficient exosome-based theranostics. Theranostics 10:3684–3707. 10.7150/thno.41580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qing G, Gong N, Chen X, Chen J, Zhang H, Wang Y, Wang R, Zhang S, Zhang Z, Zhao X, Luo Y, Liang X-J. 2019. Natural and engineered bacterial outer membrane vesicles. Biophys Rep 5:184–198. 10.1007/s41048-019-00095-6. [DOI] [Google Scholar]
- 72.Weimer ET, Lu HP, Kock ND, Wozniak DJ, Mizel SB. 2009. A fusion protein vaccine containing OprF epitope 8, OprI, and type A and B flagellins promotes enhanced clearance of nonmucoid Pseudomonas aeruginosa. Infect Immun 77:2356–2366. 10.1128/IAI.00054-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jain R, Kazmierczak BI. 2014. A conservative amino acid mutation in the master regulator FleQ renders Pseudomonas aeruginosa aflagellate. PLoS One 9:e97439. 10.1371/journal.pone.0097439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu W, Huang J, Duan B, Traficante DC, Hong H, Risech M, Lory S, Priebe GP. 2012. Th17-stimulating protein vaccines confer protection against Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 186:420–427. 10.1164/rccm.201202-0182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bayes HK, Bicknell S, MacGregor G, Evans TJ. 2014. T helper cell subsets specific for Pseudomonas aeruginosa in healthy individuals and patients with cystic fibrosis. PLoS One 9:e90263. 10.1371/journal.pone.0090263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med 168:918–951. 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 77.Bruderer U, Cryz SJ, Schaad UB, Deusinger M, Que JU, Lang AB. 1992. Affinity constants of naturally acquired and vaccine-induced anti-Pseudomonas aeruginosa antibodies in healthy adults and cystic fibrosis patients. J Infect Dis 166:344–349. 10.1093/infdis/166.2.344. [DOI] [PubMed] [Google Scholar]
- 78.Worgall S. 2012. 40 years on: have we finally got a vaccine for Pseudomonas aeruginosa? Future Microbiol 7:1333–1335. 10.2217/fmb.12.106. [DOI] [PubMed] [Google Scholar]
- 79.Priebe GP, Walsh RL, Cederroth TA, Kamei A, Coutinho-Sledge YS, Goldberg JB, Pier GB. 2008. IL-17 is a critical component of vaccine-induced protection against lung infection by lipopolysaccharide-heterologous strains of Pseudomonas aeruginosa. J Immunol 181:4965–4975. 10.4049/jimmunol.181.7.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun W, Sanapala S, Henderson JC, Sam S, Olinzock J, Trent MS, Curtiss R, III.. 2014. LcrV delivered via type III secretion system of live attenuated Yersinia pseudotuberculosis enhances immunogenicity against pneumonic plague. Infect Immun 82:4390–4404. 10.1128/IAI.02173-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schweizer HP. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol 6:1195–1204. 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 82.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 83.Vrla GD, Esposito M, Zhang C, Kang Y, Seyedsayamdost MR, Gitai Z. 2020. Cytotoxic alkyl-quinolones mediate surface-induced virulence in Pseudomonas aeruginosa. PLoS Pathog 16:e1008867. 10.1371/journal.ppat.1008867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 85.Wang S, Li Y, Shi H, Sun W, Roland KL, Curtiss R, III.. 2011. Comparison of a regulated delayed antigen synthesis system with in vivo-inducible promoters for antigen delivery by live attenuated Salmonella vaccines. Infect Immun 79:937–949. 10.1128/IAI.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.López CM, Rholl DA, Trunck LA, Schweizer HP. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol 75:6496–6503. 10.1128/AEM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rabin SD, Hauser AR. 2005. Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infect Immun 73:573–582. 10.1128/IAI.73.1.573-582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86. 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI.00396-21-s0001.pdf, PDF file, 0.2 MB (210.5KB, pdf)
Supplemental material. Download IAI.00396-21-s0002.pdf, PDF file, 0.5 MB (543.1KB, pdf)