Abstract
The alkaline comet assay, or single cell gel electrophoresis, is one of the most popular methods for assessing DNA damage in human population. One of the open issues concerning this assay is the identification of those factors that can explain the large inter-individual and inter-laboratory variation. International collaborative initiatives such as the hCOMET project - a COST Action launched in 2016 - represent a valuable tool to meet this challenge. The aims of hCOMET were to establish reference values for the level of DNA damage in humans, to investigate the effect of host factors, lifestyle and exposure to genotoxic agents, and to compare different sources of assay variability. A database of 19,320 subjects was generated, pooling data from 105 studies run by 44 laboratories in 26 countries between 1999 and 2019. A mixed random effect log-linear model, in parallel with a classic meta-analysis, was applied to take into account the extensive heterogeneity of data, due to descriptor, specimen and protocol variability. As a result of this analysis interquartile intervals of DNA strand breaks (which includes alkali-labile sites) were reported for tail intensity, tail length, and tail moment (comet assay descriptors). A small variation by age was reported in some datasets, suggesting higher DNA damage in oldest age-classes, while no effect could be shown for sex or smoking habit, although the lack of data on heavy smokers has still to be considered. Finally, highly significant differences in DNA damage were found for most exposures investigated in specific studies. In conclusion, these data, which confirm that DNA damage measured by the comet assay is an excellent biomarker of exposure in several conditions, may contribute to improving the quality of study design and to the standardization of results of the comet assay in human populations.
Keywords: Comet assay, DNA damage, Pooled analysis, Human biomonitoring, Biomarkers
1. Introduction
The alkaline comet assay, or single cell gel electrophoresis, is one of the most popular methods for the assessment of DNA damage in human populations [1–5]. The test is quick, simple, and sensitive, qualities that make the comet assay very well suited to study the effect of exposure to occupational or environmental mutagens, to quantify the influence of lifestyle, diet and dietary supplementation on genetic stability, and to check the levels of DNA damage in relation to various diseases [6], in fresh or frozen samples of human cells. The presence of DNA damage, or its inadequate repair is a well-known factor in the etiology of cancer and other non-communicable diseases, and therefore the comet assay can in theory be used to detect early genetic damages that indicate the risk of future disease [7]. One of the open issues concerning this assay is the identification of those factors that can explain the large inter-individual and inter-laboratory variation in results. Several potential sources of variability of the assay have been identified in human biomonitoring studies, including technical heterogeneity, lifestyle, and individual characteristics. Differences in protocols and interpretation of the outcomes contribute to multiply the effect of biological variability, making results of multi-centre biomonitoring studies with the comet assay difficult to interpret, and greatly limiting the potential of this approach. To warrant widespread acceptance and credibility in human population studies, the comet assay requires standardization of the protocol and a better knowledge of critical features affecting the assay outcomes, and the role of host factors. These challenges can be met by international collaborative initiatives, involving a large network of laboratories [8]. Here we are interested specifically in levels of background DNA damage, i.e. DNA strand breaks (and alkali-labile sites) present in the cells (whether from control or exposed individuals), measured with the comet assay, without any treatment of cells ex vivo. The large pooled database resulting from such efforts represents a valuable tool to evaluate the assay’s performance and to study the causes and consequences of DNA damage. Initiatives have been proposed by international bodies and individual research groups, especially within the European Comet Assay Validation Group (ECVAG) [9–16]. An international collaborative study on the Comet assay, originally known by the name of ComNet, was launched during the International Comet Assay Workshop meeting held in Kusadasi, Turkey (September 13–16, 2011) with the aim of investigating whether the comet assay is a reliable and validated biomarker assay for use in human biomonitoring [17]. Building on these efforts, the hCOMET COST (European Cooperation in Science and Technology) Action (CA15132) has assembled a pooled data set of nearly 20,000 individuals, collecting data from existing studies, and protocol details from most laboratories in the field.
The aims of this first paper analysing the large hCOMET dataset are: 1) to establish reference values for background levels of DNA damage, 2) to investigate the effect of host factors, lifestyle and exposure to genotoxic agents on the level of DNA damage measured by the comet assay, and 3) to model and compare different sources of the assay’s variability.
2. Materials and methods
2.1. The hCOMET database
More detail about the creation of the hCOMET databases can be found in the previous publications of the ComNet consortium, which indicated the strategy later followed by hCOMET [6.17]. In brief, 44 laboratories contributed data referring to 19,320 subjects within the framework of the hCOMET EU COST Action CA15132. Data included in the database were generated by 105 studies published between 1999 and 2019. Studies using the comet assay in whole blood (i.e., white blood cells), peripheral blood mononuclear cells (commonly referred to as lymphocytes), or in other tissues in human populations, with an epidemiological design, the presence of a control group, and with an adequate description of the protocol(s) used, were included in the analysis. Whenever available in the original set of data, detailed information was collected on those parameters that according to published literature were potentially relevant for the technical, biological or epidemiological features of the comet assay, namely demographic parameters, lifestyle, occupational exposure, smoking habit, diet-related factors, genetic profile, and diagnoses of chronic disease. The large heterogeneity among the laboratories contributing data in terms of quality and quantity of information collected did not allow fine-tuning in the analysis of parameters such as smoking and occupational exposures to genotoxic agents. For the latter, subjects were broadly classified as exposed individuals or unexposed controls according to criteria used to define exposure in the original studies. The list of types of exposure is heterogeneous and includes occupational, environmental, and lifestyle exposures to genotoxic agents (e.g., antineoplastic drugs, anesthetics, dust, ceramic material, styrene, air pollution, formaldehyde, metal and metalloids, polycyclic aromatic hydrocarbons, dyes, pesticides, tobacco farming and manufacturing, ionizing and non-ionizing radiation, diet-related factors, alcohol, and smoking habit). In some studies, the presence of diseases or disease-associated conditions were considered (e.g., cancer, cardiovascular diseases, diabetes, obesity, kidney failure, hemodialysis, renal transplant, inflammatory bowel disease). This approach has given the possibility to identify a large group of presumably unexposed subjects representing a robust reference group for several analyses. The 133 papers (referring to 105 studies) from which data were included in the hCOMET dataset - as communicated by individual contributors - are listed in the supplementary Table 1. Data gathering was coordinated by the IRCCS San Raffaele Pisana, Rome, Italy, and the Institute for Medical Research and Occupational Health, Zagreb, Croatia. The pooled analysis of data was approved by the ethics committee of the IRCCS San Raffaele Pisana, Rome, Italy (12 December 2015, Prot. N. 10/15), i.e., the centre coordinating data collection and running the statistical analysis of data. Each study had already received ethical approval from local ethical committees for the collection and analysis of individual coded data in accordance with the Declaration of Helsinki, and all the measures of General Data Protection Regulation (EU) 2016/679 (GDPR) were respected. The list of laboratories contributing data used for this paper with the corresponding codes used for statistical analysis is given in the supplementary Table 2, while their geographical distribution is mapped in the Fig. 1. The identification of technical issues that most affect the assay descriptors will be evaluated in another publication, while results involving the enzyme-modified comet assay (testing for damage such as oxidised bases) were too sparse for a proper statistical analysis. Therefore, this paper will present only data referring to the frequency of DNA strand breaks (including alkali-labile sites) as evaluated by the classic alkaline version of the assay.
Fig. 1.
Geographical distribution of the 26 countries contributing data to the hCOMET dataset (dark grey).
2.2. Laboratory methods
An extensive questionnaire collecting technical detail of protocols used was filled in by participating laboratories, but to characterize the dataset and describe critical sources of technical variability, only main differences between laboratories were evaluated - namely 1) use of fresh (57.5 % of subjects) or frozen specimens (41.6 %), 2) use of standard [1] (46.0 %) or modified protocols, e.g., with the addition of 10 % dimethyl sulfoxide (DMSO) in the lysis step (44.0 %), 3) use of different staining methods, of which the most common were DAPI (40,6-diamidino-2-phenylindole (10.1 %), SYBR1 Gold Nucleic Acid Gel Stain (34 %), and ethidium bromide (30.1 %). Minor differences were found for the concentration of the (low melting) agarose layer with the cells (from 0.5 to 0.8 %), duration of the electrophoresis step (21.8 % lasted more than 20 min), and voltage used during the electrophoresis (from 20 to 30 V using electrophoresis tanks of different lengths, resulting in a wide range of potential gradients (V/cm)). The number of cells scored was variable, although a minimum of 100 cells were scored for 92.8 % of specimens. Finally, in 23.1 % of subjects the analyzed specimen was whole blood rather than isolated lymphocytes. Given the fragmented distribution of these parameters among laboratories, only the major discrepancies (in terms of numbers and biological significance) were included in the multivariate modelling, i.e., cell type (isolated lymphocytes or whole blood) and sample processing (fresh/frozen sample).
2.3. Statistical analysis
The most commonly used descriptors of the comet assay were considered for statistical analysis, i.e., tail length (TL), % tail DNA (% T), tail moment (TM), and visual scoring (VS). Descriptive statistics referring to background level of DNA damage reported both mean and median as a measure of central tendency, and standard deviation and the interquartile ranges as measures of variability. The frequency of these descriptors is reported 1) for each laboratory included in the study, 2) for the total of the hCOMET dataset, and 3) for the subgroup of subjects classified as controls in the original studies. As regards VS, most laboratories used 5 classes of cell damage accounting for a maximum possible value of 400, while others used 3 or fewer classes. This difference in scoring contributed to generate the great deal of heterogeneity observed for this descriptor, and therefore no reference values or pooled estimates of effect by age or sex have been reported for VS. Studies using tissues different from whole blood or isolated lymphocytes, mostly exfoliated epithelial cells, were not included in the statistical analyses. The dataset was searched for subjects with repeated values (such as in before-and-after studies or in extended surveillance programs), and only baseline measures were left in the dataset. These restrictions brought the number of subjects to 15,421. A further screening identified outliers, technical errors, subjects without valid measures, and a group of newborns, whose values were hardly comparable with adult groups. After removing these cases, the final valid number of subjects included in the statistical analyses was 13,553. Univariate parametric tests were used on the original or log-transformed data for all analyses according to Lowell and Omori [18]. Since data from each individual study can be considered as a cluster of correlated observations, all regression models considered the within- and between-studies variance components. A mixed random effect log-linear model (REM) including the fixed effects of age, sex, smoking habit, cell populations (isolated lymphocytes or whole blood), and cell processing (fresh or frozen samples), and the random effects of laboratory, study and subjects, was fitted to the whole dataset, providing a suitable range of expected values [19,20]. The use of random effect modelling (REM) allowed estimation of the adjusted mean ratio (MR) i.e. the ratio between the average values of DNA damage in the categories to be compared. REM allowed us also to estimate the Variance Partition Coefficient (VPC), which is a summary measure expressing the relative contribution of each component to the total variance of data. To better explore the relationship between the descriptor of the comet assay and age, the restricted cubic spline technique was applied from 0 to 70 years of age [21]. To allow a more straightforward and reliable interpretation of results, estimates provided by REM are accompanied in each dataset, when suitable, by a classic meta-analysis. The forest plot, together with meta-estimates, has been reported for data on sex (females vs. males) and genotoxic exposure (subjects reported as exposed in original studies vs. non-exposed subjects). This approach was not used to study the effect of age (because of the non-linearity of the relationship with DNA damage), or the effect of smoking habit (given the inter-study heterogeneity in the quality of data). Age was included in multivariate models as a continuous variable, while smoking could be evaluated only by comparing subjects reporting to have ever smoked (ever-smokers) with those that declared to have never smoked (non-smokers).
The MR for the variables included in the meta-analyses was computed for each study, adjusting for the other confounding factors. Subsequently, a pooled value was estimated as suggested by DerSimonian and Laird [22]. The main difference between these approaches is that a meta-analysis takes into account only the between-study heterogeneity, while multilevel modelling quantifies variance components at each level. SPSS 26 and STATA 14.2 statistical software was used for all analyses.
3. Results
3.1. Background frequency of DNA damage
To identify the range of values, which is likely to represent the background frequency of DNA damage, we investigated a dataset of 13,553 subjects with valid results from at least one of the descriptors of the comet assay. Descriptive statistics concerning the descriptors considered are reported in Table 1(a–d). The most popular descriptor was the %T, with a total number of 8293 subjects screened for this parameter, and measured by 28 out of 40 laboratories contributing valid data to hCOMET (70.0 %). The other three descriptors (TL, TM, and VS) were evaluated in 3002, 2793, and 4513 subjects, respectively. The numbers of laboratories using these descriptors were 15 for TL, 14 for TM, and 13 for VS, corresponding to 37.5, 35.0, and 32.5 % of the total, respectively. A graphical summary of values of each descriptor per laboratory using box-and-whiskers plot is reported in Fig. 2 (statistics refer to unexposed cases only). Model estimates of the coefficient of variation (CV) based on the expected values, i.e., taking into account the effect of laboratory, study, and repeated measures, showed CVs of 42.9 %, 92.6 %, and 305 %, for TL, %T, and TM, respectively (data not shown). As expected, a positively skewed distribution was generally observed for all descriptors, particularly severe for the TM, due to the frequent presence of extreme values. In most cases, due to departure from normality, the median value is the most reliable index of central tendency, and the interquartile interval of each descriptor provides the most reliable range of frequent results. However, to provide a complete view of descriptive statistics also the mean, the standard deviation (SD), and the corresponding confidence interval are reported. According to these results, a typical biomonitoring study, using %T as the descriptor of DNA damage should estimate in most cases an overall median value falling between 1.6 and 9.9 %. This interval does not change markedly when restricted to the group of subjects designated as controls (4625 subjects) and in this case the interquartile interval falls between 1.1 and 6.7 %. A summary report for reference values for different descriptors of the comet assay is reported in Table 2together with the results of expected values from multivariate modelling. Measures of central tendency and variability intervals are reported for the overall dataset and for the subgroup of subjects classified as controls in the original studies. The expected distribution of TL, %T, and TM estimated by the mixed model did not differ from univariate estimates, but should be preferred since main confounders are taken into account and the influence of extreme values is smoothed.
Table 1.
Spontaneous frequency of DNA damage in the individual datasets contributed by participating laboratories. 1a) tail length; 1b) % tail DNA; 1c) tail moment, and 1d) arbitrary units.
|
1a) Tail Length (μm) | ||||
|---|---|---|---|---|
| Laboratory | N | Mean (SD) | Median | 25°−75° Percentile |
| AS4 | 77 | 49.91 (11.7) | 48.9 | 41.1‒59.9 |
| CSA5 | 138 | 39.9 (22.7) | 42.5 | 17.6‒54.1 |
| CSA6 | 129 | 27.3 (6.6) | 26.6 | 22.9‒30.1 |
| EU12 | 42 | 10.1 (3.3) | 9.6 | 7.9‒13.2 |
| EU15 | 101 | 28.3 (18.1) | 21.3 | 15.0‒37.3 |
| EU17 | 18 | 48.2 (4.2) | 47.5 | 45.9‒51.5 |
| EU25 | 76 | 3.1 (1.1) | 3.1 | 2.4‒3.6 |
| EU26 | 973 | 36.2 (19.0) | 28.2 | 21.7‒47.1 |
| EU30 | 92 | 1.5 (1.0) | 1.5 | 0.7‒2.0 |
| EU32 | 562 | 41.8 (14.0) | 46.7 | 41.4‒50.6 |
| EU33 | 97 | 30.2 (14.7) | 32.5 | 16.5‒41.9 |
| EU36 | 302 | 45.1 (20.2) | 38.9 | 29.5‒54.1 |
| EU4 | 171 | 13.9 (1.2) | 13.9 | 13.1‒14.6 |
| EU41 | 187 | 6.2 (3.7) | 5.40 | 3.5‒8.5 |
| EU50 | 37 | 21.8 (5.9) | 21.4 | 18.1‒23.7 |
| TOTAL | 3,002 | 32.3 (20.4) | 28.6 | 16.6‒47.1 |
| Controls only | 1,228 | 28.6 (17.5) | 26.6 | 14.2‒43.6 |
|
1b) Tail Intensity (%) | ||||
| Laboratory | N | Mean (SD) | Median | 25°−75° Percentile |
|
| ||||
| AS4 | 77 | 4.5 (1.4) | 4.2 | 3.6‒5.0 |
| EU12 | 42 | 0.01 (0.01) | 0.02 | 0.01‒0.02 |
| EU15 | 101 | 24.0 (9.1) | 23.6 | 18.8‒31.1 |
| EU17 | 115 | 7.4 (4.6) | 5.4 | 4.6‒9.1 |
| EU18 | 57 | 6.7 (2.1) | 6.0 | 5.5‒7.0 |
| EU22 | 752 | 8.1 (7.3) | 7.2 | 1.1‒14.6 |
| EU23 | 86 | 1.2 (1.2) | 0.8 | 0.4‒1.7 |
| EU24 | 92 | 13.8 (9.0) | 13.3 | 5.2‒21.1 |
| EU25 | 76 | 3.1 (1.0) | 3.0 | 2.4‒3.3 |
| EU26 | 1,076 | 9.5 (7.3) | 7.2 | 4.3‒12.2 |
| EU27 | 17 | 2.9 (0.7) | 3.2 | 2.5‒3.4 |
| EU28 | 32 | 6.5 (2.8) | 6.5 | 4.0‒8.7 |
| EU3 | 1,637 | 2.7 (2.3) | 2.0 | 1.1‒3.6 |
| EU30 | 92 | 0.9 (0.9) | 0.6 | 0.3‒1.2 |
| EU31 | 142 | 30.0 (16.0) | 29.0 | 15.5‒39.0 |
| EU32 | 494 | 0.7 (1.0) | 0.2 | 0.1‒0.8 |
| EU33 | 1,061 | 16.5 (12.2) | 12.9 | 7.0‒21.3 |
| EU34 | 170 | 6.4 (3.1) | 6.4 | 3.8‒8.2 |
| EU36 | 441 | 6.7 (2.5) | 6.6 | 5.0‒8.3 |
| EU37 | 287 | 10.7 (2.4) | 10.4 | 9.2‒12.6 |
| EU4 | 111 | 1.1 (0.6) | 1.1 | 0.7‒1.6 |
| EU41 | 187 | 7.0 (3.6) | 6.4 | 4.3‒9.2 |
| EU43 | 162 | 2.7 (2.1) | 2.1 | 1.2‒3.6 |
| EU45 | 93 | 1.4 (4.3) | 0.4 | 0.1‒1.0 |
| EU46 | 329 | 3.2 (1.9) | 2.8 | 1.8‒4.0 |
| EU50 | 37 | 6.4 (1.5) | 6.2 | 5.8‒7.1 |
| EU6 | 325 | 1.1 (1.0) | .9 | 0.3‒1.6 |
| OCNA1 | 202 | 9.5 (5.6) | 7.7 | 6.2‒11.0 |
| TOTAL | 8,293 | 7.4 (8.8) | 4.5 | 1.6‒9.9 |
| Controls only | 4,625 | 4.8 (5.5) | 2.9 | 1.1‒6.7 |
|
1c) Tail Moment (arbitrary units) | ||||
| Laboratory | N | Mean (SD) | Median | 25°−75° Percentile |
|
| ||||
| AS4 | 77 | 1.6 (0.4) | 1.5 | 1.3‒1.8 |
| CSA5 | 35 | 2.0 (1.3) | 1.4 | 1.2‒2.4 |
| CSA6 | 129 | 1.3 (1.0) | 1.1 | 0.5‒1.7 |
| EU12 | 221 | 0.4 (0.4) | 0.4 | 0.1‒0.7 |
| EU15 | 173 | 27.3 (15.7) | 25.2 | 16.0‒34.8 |
| EU21 | 151 | 1.8 (1.7) | 1.4 | 0.7‒2.4 |
| EU23 | 86 | 0.6 (0.9) | 0.3 | 0.1‒0.9 |
| EU25 | 76 | 0.3 (0.2) | 0.2 | 0.1‒0.3 |
| EU26 | 824 | 2.1 (2.7) | 1.2 | 0.6‒2.6 |
| EU30 | 92 | 0.5 (1.3) | 0.01 | 0.01‒0.2 |
| EU33 | 273 | 10.2 (13.5) | 3.0 | 1.3‒18.4 |
| EU36 | 302 | 1.4 (0.7) | 1.4 | 0.9‒1.8 |
| EU4 | 111 | 0.1 (0.1) | 0.1 | 0.1‒0.2 |
| EU8 | 41 | 0.3 (0.2) | 0.3 | 0.2‒0.4 |
| OCNA1 | 202 | 1.5 (1.9) | 0.9 | 0.6‒1.6 |
| TOTAL | 2,793 | 3.96 (8.9) | 1.1 | 0.5‒2.4 |
| Controls only | 1,181 | 2.8 (6.9) | 0.9 | 0.4‒1.8 |
|
1d) Visual scoring (arbitrary units) | ||||
| Laboratory | N | Mean (SD) | Median | 25°−75° Percentile |
|
| ||||
| CSA2 | 71 | 114.2 (31.0) | 118.0 | 89.0‒138.0 |
| CSA3 | 56 | 8.8 (5.3) | 7.8 | 5.1‒10.5 |
| CSA5 | 55 | 204.8 (53.6) | 217.0 | 159.0‒254.0 |
| CSA6 | 76 | 74.1 (7.9) | 73.5 | 70.0‒78.5 |
| CSA7 | 1,206 | 21.6 (21.7) | 15.0 | 6.0‒30.0 |
| EU14 | 103 | 198.1 (29.3) | 202.5 | 190.0‒207 |
| EU19 | 89 | 288.7 (36.2) | 284.0 | 264.5‒315.8 |
| EU30 | 41 | 28.5 (20.5) | 24.0 | 12.5‒42.5 |
| EU31 | 1,468 | 78.9 (59.0) | 59.1 | 41.0‒95.3 |
| EU36 | 55 | 116.5 (7.6) | 118.0 | 110.0‒122.0 |
| EU37 | 238 | 17.9 (16.1) | 13.0 | 7.0‒24.0 |
| EU42 | 49 | 23.5 (14.0) | 21.8 | 12.4‒32.3 |
| EU7 | 1,006 | 26.0 (16.0) | 23.0 | 18.0‒31.0 |
| TOTAL | 4,513 | – | – | – |
Fig. 2.
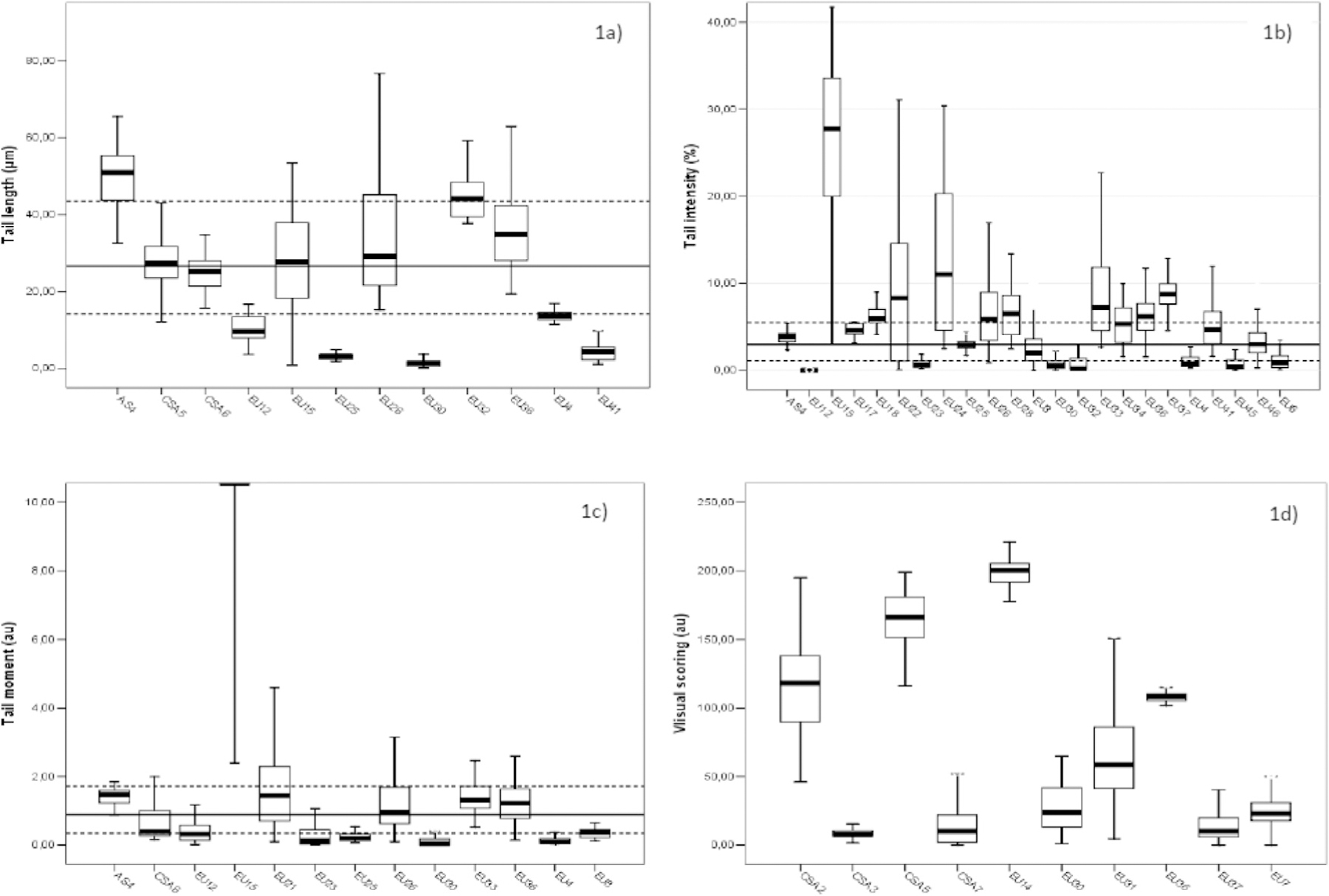
Frequency (median and interquartile distance) of DNA damage measured in the whole dataset for all endpoints. 1a) Tail length; 1b) Tail intensity; 1c) Tail moment; 1d) Visual Scoring. Reference lines corresponds to the overall median values for the specific endpoint. Overall median and interquartile interval is reported with whole and dotted lines, respectively in Figure 1a, 1b, and 1c. All statistics shown in this table refers to unexposed cases only.
Table 2.
Reference values for selected descriptors of the comet assay in the whole dataset and in the subset of control individuals.
| Descriptor | N | Observed data |
Modelled data |
|||
|---|---|---|---|---|---|---|
| Mean (SD) | Median | 25°−75° percentile | Expected Mean*(SD) | Expected Median*(25°−75°) | ||
| Tail length (μm) Overall | 3,002 | 32.3 (20.4) | 28.6 | 16.6‒47.1 | 22.4 (9.6) | 23.3 (14.8‒26.9) |
| Controls | 1,228 | 28.6 (17.5) | 26.6 | 14.2‒43.6 | 21.1 (8.6) | 22.4 (14.3‒25.6) |
| Tail Intensity (%) Overall | 8,293 | 7.4 (8.8) | 4.5 | 1.6‒9.9 | 5.4 (5.0) | 2.8 (1.3‒5.4) |
| Controls | 4,625 | 4.8 (5.5) | 2.9 | 1.1–6.7 | 3.9 (3.5) | 3.9 (1.8‒7.7) |
| Tail Moment (au) Overall | 2,793 | 3.96 (8.9) | 1.1 | 0.5‒2.4 | 2.0 (6.1) | 1.1 (0.5‒2.1) |
| Controls | 1,181 | 2.8 (6.9) | 0.9 | 0.4‒1.8 | 2.0 (9.0) | 0.9 (0.5‒1.6) |
Predicted values from a mixed random effect log-linear model including the fixed effects of age, sex, smoking habit, exposure to genotoxic agents, cell type (isolated lymphocytes or whole blood), sample processing (fresh/frozen) and the random effects of laboratory, study and subject; 5°–95° correspond to the 25th-75th percentiles. Standard deviation (SD), arbitrary units(au)..
3.2. Distribution of DNA damage by age and sex
A summary view of the level of DNA damage by age-class is shown in Table 3, where absolute number, mean and SD, and medians are shown according to age-classes and sex. The univariate analysis of data shows an increasing trend for mean values of TL and especially for %T (p < 0.01), while unexpectedly, TM showed the opposite trend. The multivariate analysis of the pooled dataset based on a mixed random effect log-linear model and taking into account sex, smoking habit, exposure to genotoxic agents, cell population and sample processing did not show any significant association between DNA damage and age, with a progressive non-significant increase of 4, 2, and 6% of TL by age-class of 19‒40 y, 41‒60 y, and 61+ years, respectively, when compared to the youngest class. A cubic spline analysis applied to these data suggested a potential increase of DNA damage in the older age-classes. Tail intensity rises steeply up to 20 years; after that it seems to stabilize, with modest fluctuations, but returns to rise at about 60 years of age (Supplementary Fig. 1). Due to the small numbers and the large variability of the oldest age-classes, this analysis has been truncated to 70 years of age. The other descriptors evaluated with this approach did not show any significant variations by age class (data not shown).
Table 3.
Distribution of selected descriptors of the comet assay by age class and sex (numbers in each cell represent: absolute frequency; mean (SD), and median). All values refer to total hCOMET population.
| Age-Class | Tail Length (μm) | Tail Intensity (%)* | Tail Moment (au) |
|---|---|---|---|
| 0‒18 years | 138 | 1,226 | 166 |
| 16.6 (19.4) | 3.2 (3.1) | 7.0 (12.8) | |
| 12.9 | 2.1 | 0.2 | |
| 19‒40 years | 1,437 | 2,575 | 1,217 |
| 33.9 (18.1) | 7.2 (7.5) | 4.0 (8.9) | |
| 32.9 | 5.3 | 1.2 | |
| 41‒60 years | 1,098 | 2,657 | 903 |
| 31.2 (20.4) | 7.6 (9.9) | 4.2 (9.9) | |
| 26.4 | 4.0 | 1.1 | |
| 61+ years | 312 | 1,329 | 261 |
| 35.1 (26.7) | 10.5 (11.2) | 3.0 (3.8) | |
| 30.7 | 6.2 | 1.6 | |
| Sex | Tail Length (μm) | Tail Intensity (%)** | Tail Moment (au)** |
|
| |||
| Females | 1,330 | 3,598 | 1,217 |
| 31.7 (20.3) | 7.1 (8.0) | 3.6 (7.9) | |
| 27.5 | 4.4 | 1.2 | |
| Males | 1,671 | 4,489 | 1,332 |
| 32.8 (20.4) | 7.6 (9.5) | 4.7 (10.3) | |
| 29.6 | 4.3 | 1.2 | |
=Test for linear trend p < 0.01.
Student’s t-test p < 0.01; Arbitrary units (au).
The level of DNA damage according to sex was initially evaluated with univariate analysis as shown in Table 3. Males showed higher mean rates of DNA damage, significant for %T and TM, while median values did not differ between sexes. However, the observed differences were not homogeneous, and therefore the effect of this parameter was further estimated in each laboratory through meta-analysis, and described with a forest plot. All descriptors yielded similar results and therefore only the forest plot of %T was shown (Fig. 3). The ratio between the mean of %T in males to females is reported for each study, together with the 95 % confidence interval and the proportional weight of each study. The level of heterogeneity (I2 < 5%) was very low, and only one study found a borderline statistically significant difference between sexes, i.e., 6% higher %T in males. The overall estimate of the meta-MR did not show any effects of sex, i.e., 1.01. 95 % CI 0.98‒1.03. The estimates from random effect modelling confirmed the results of meta-analysis and failed to find significant differences between males and females in the level of DNA damage measured with the comet assay (MR females vs males for %T = 1.00, 95 % CI = 0.91–1.11).
Fig. 3.
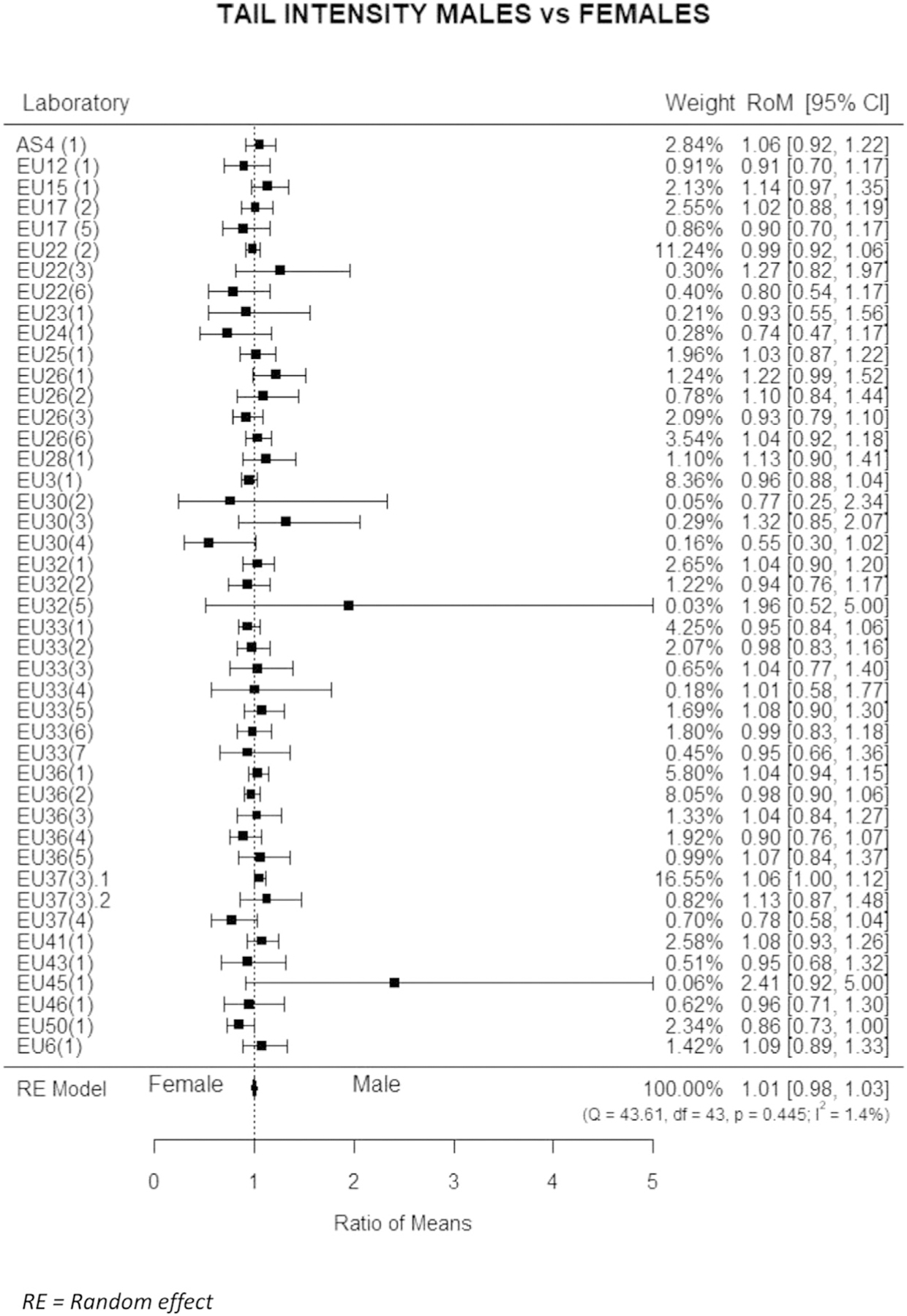
Estimated variation of Tail intensity by sex. Mean ratios (MR), i.e., the ratio of mean DNA values of males to the mean values of females, with 95 % confidence intervals (95 % CI) are reported for individual studies and for the whole dataset.
3.3. Other major confounding factors
As discussed in the methods section, results concerning smoking habit suffer from a great deal of heterogeneity in the quantity and quality of information retrieved, limiting the comparability of results among laboratories. The evaluation of MRs for ever-smokers vs non-smokers in each study allowed a standardized evaluation of this parameter at study level. Results did not show any increases in ever-smokers, which for %T showed higher levels of DNA damage in the group of smokers only in 15 studies out of 36 (41.6 %), and never significantly. The results of REM did not find significant differences in the level of DNA damage between ever-smokers and non-smokers (MR = 0.98; 95 % CI 0.93–1.02). All descriptors showed results similar to %T.
The only covariate which showed constantly a significant effect reported by most studies in the meta-analysis was the exposure studied in each specific dataset (a list of most common exposures is reported in the methods section). As shown by the forest plot of Fig. 4 referring to %T, the large majority of studies, independently of the exposure investigated, showed significant differences in DNA damage when compared with unexposed controls. The meta-estimate confirms the excess of DNA damage in the group of exposed subjects (MR = 1.32; 95 % CI 1.20–1.45), in agreement with the mixed random effect model, which estimated a significant increase of DNA damage in exposed subjects for all descriptors, i.e, MR = 1.46 (95 % CI = 1.19–1.79) for %T, 1.16 (1.13–1.19) for TL, 1.55 (1.42–1.69) for TM.
Fig. 4.
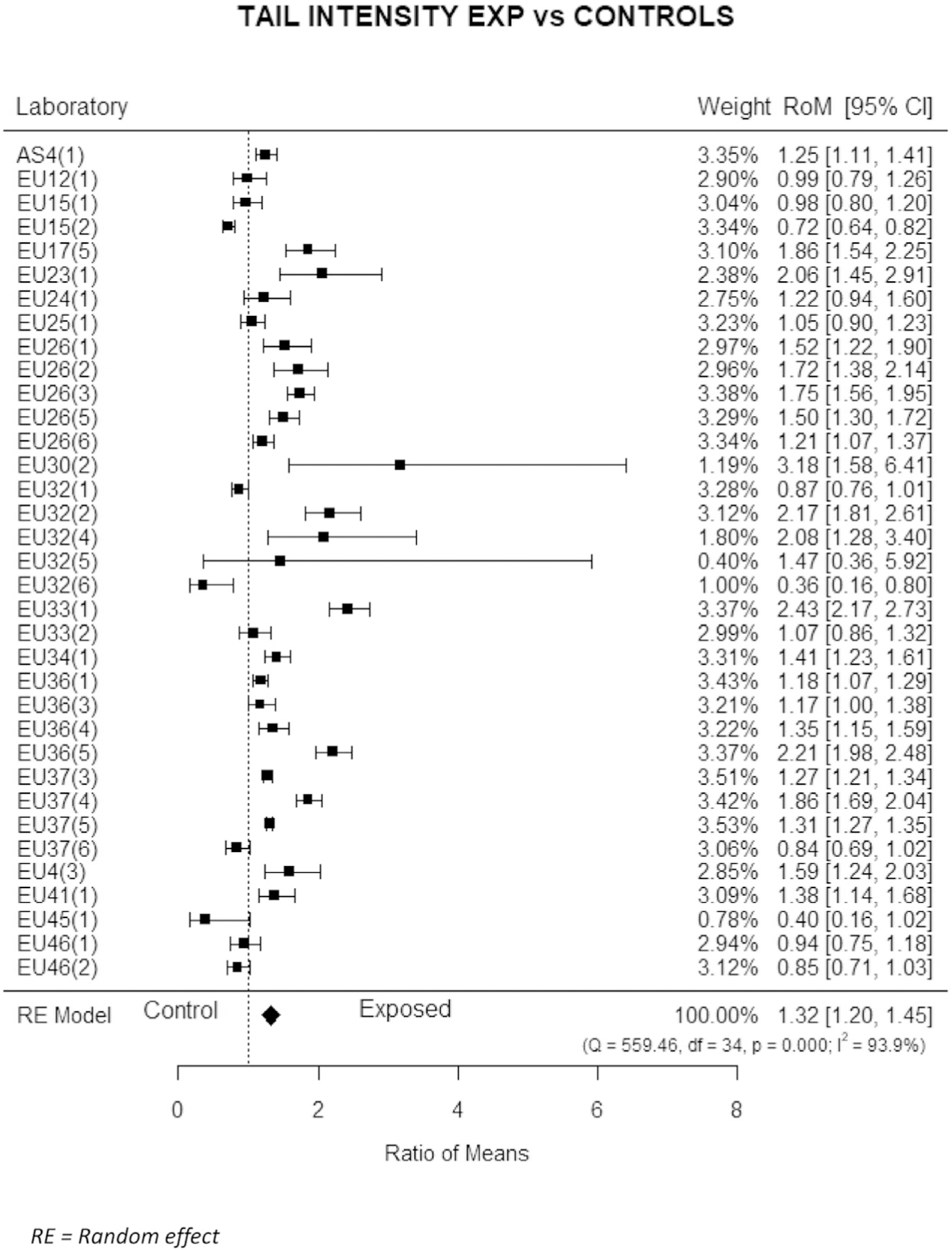
Estimated variation of Tail intensity for study groups as compared to unexposed controls. Estimates for individual study and for the whole dataset. Mean ratios (MR), i.e., the ratio of mean DNA values of subjects exposed to the mean values of unexposed controls, with 95 % confidence intervals (95 % CI) are reported for individual studies and for the whole dataset.
Among protocol features that were used as potential confounder in these models, but that will require a dedicated analysis, assays using whole blood showed for all descriptors a much lower frequency of DNA damage than assays using isolated lymphocytes, e.g., for %T the MR was 0.35 (95 % CI 0.18‒0.71).
Figures reported in the supplementary Table 3 show that while most variability of TL can be explained with the heterogeneity of estimates between laboratories, studies and subjects, i.e., 91.0 %, for the other descriptors higher proportions of residual, unexplained variability were found, i.e., 42.6 % for %T, 23.4 % for TM, and 55.0 % for VS. For all descriptors the most important component of variance is the laboratory, even after removing the heterogeneity between single studies.
The correlation between descriptors was evaluated in those datasets where more than one descriptor was simultaneously evaluated. Correlation coefficients were highly significant for the pairs %T-TL (r = 0.25; p < 0.001), %T-TM (r = 0.41; p < 0.001), and TL-TM (r = 0.25; p < 0.001). Positive correlations were found for %T and TL with VS, but the small numbers prevented any reliable evaluation. A detailed table with correlation coefficients for all descriptors and all laboratories is reported in the Supplementary table 4. An example of the strength of correlation between laboratories scoring %T and TL is showed in the Supplementary Fig. 2.
4. Discussion
Only validated biomarkers can be efficiently used in molecular epidemiological studies. Validation includes a proper understanding of technical and host factors that contribute to the large extent of variability which characterizes these studies. To achieve validation, biomarkers of genotoxicity, such as the comet assay, need joint efforts to reach sufficient numbers of individuals/observations to do reliable analyses. Indeed, critical improvements in the quality and reliability of other popular biomarkers of DNA damage and genetic instability results have been achieved as a consequence of large international collaborative initiatives [23–26]. The availability of the pooled database assembled within the hCOMET international collaborative project offered a unique opportunity to obtain comprehensive information about the comet assay. This information includes the background level of DNA damage in the general population, the role of host factors such as sex and age on the level of DNA damage, the role of occupational and environmental exposure to DNA-damaging agents, and the comparison of the four different descriptors most commonly used over the 20-year span during which these data were obtained.
When the ComNet project was launched in 2011 [17] and taken to the next level by the hCOMET COST Action since 2016, all these points were planned with the purpose of improving our knowledge of the comet assay and making DNA damage measures more comparable between different laboratories. The levels of DNA damage estimated by the comet assay are rather heterogeneous, mostly because of the large number of technical protocols available, which leads to large inter-laboratory variation [27]. A detailed analysis of different protocols, and their influence on the final estimates of DNA damage is beyond the scope of this paper. Nevertheless, the role of technical variability was evaluated as a whole, considering the variability between laboratories, studies, and subjects. The importance of this variability has been confirmed by the present study, which gathered together a large number of laboratories and researchers, revealing differences in technical protocols and contributing altogether individual data of thousands of subjects. On the other hand, this variability reflects the sensitivity of the assay, which is well suited to detect differences, even small, in populations exposed to DNA-damaging agents, as shown by the analysis of individual studies. The high level of heterogeneity concerning background level of DNA damage in the individual datasets was consistent for all four descriptors evaluated. The source of this heterogeneity has been evaluated within multilevel modelling by the Variance Partition Coefficient, which showed a high proportion of unexplained variance for %T and VS (42.6 and 55.0 %, respectively), while TL variation is mostly explained by the variance associated with the heterogeneity between laboratories (76.2 %) and between single studies (14.8 %). An additional explanation for the large inter-laboratory variability of descriptors based on metric measures such as TL (76.2 %) is the lack of calibration. The larger number of studies reporting %T as the favourite descriptor reflects the orientation of the literature, and the recommendation of international collaborative consortia [5,6], while the large number of subjects screened using the VS reflects the lower cost of this approach, and therefore its greater accessibility. It should be noted that the high level of correlation between the descriptors evaluated shows clearly that they are all well suited to efficiently measure DNA strand breaks. It is also worth pointing out that these are not independent measures; TM is essentially the product of %T and TL.
The identification of a range of values, reflecting the ‘background’ interval for the level of DNA damage for the different descriptors of the comet assay will help new laboratories in the field to establish a range of referent values, and all laboratories to revisit and revalidate their results. Furthermore these data will contribute to standardization of results of the comet assay in human populations. In spite of the fact that some laboratories generated data that were inconsistent with other datasets, our analysis suggests a range of credible values, reflecting the typical values scored by the majority of laboratories in the field. In addition, the use of multilevel modeling allowed us to take into account the variability generated by protocol and specimen heterogeneity, providing a range that can be applied to different conditions. The substantial consistency between the estimates of background %T frequency provided by the hCOMET pooled reanalysis i.e., 1.6‒9.9 %, and those reported by a classic review paper which evaluated 125 published papers, i.e., 7–11 %, adds reliability to these estimates [9]. Estimates of baseline values from another ECVAG initiative suffered from unexpected internal variability, with participating laboratories clustering around lower (1.8–2.4%) or higher values (13–21%) [15].
A general limitation of the comet assay is that the TL, TM, %T and VS are primary descriptors of DNA migration in the agarose gel and information on actual levels of DNA damage can only be obtained by indirect calibration using ionizing radiation [28]. Calibration curves are prepared by exposing e.g. lymphocytes to ionizing irradiation, which produces a fairly linear relationship between the dose and any primary comet assay descriptor. As the yield of DNA strand breaks is known for ionizing radiation, one can obtain a comet assay calibration curve - essentially the linear relationship between the primary comet assay descriptor and the number of breaks in the DNA, see [29]). This method has been used in the European Standards Committee on Oxidative DNA Damage (ESCODD) ring-trials, where baseline levels of DNA damage measured by the comet assay did not differ from the alkaline elution and alkaline unwinding methods that also use independent calibration with ionizing radiation to estimate absolute levels of DNA damage [30–32]. The ECVAG ring-trials used calibration curves to standardize results between laboratories because it was noted from the results obtained by ECSODD that differences in DNA damage levels in lymphocytes from human donors strongly correlated with DNA damage levels in reference standards used by different labs [10]. Importantly, it is mainly the slope of the calibration curves that differs between labs, indicating that inter-lab (or inter-study) differences in DNA damage levels increase proportionally to the level of DNA damage (see e.g [12].). As background DNA damage levels in lymphocytes from human donors are relatively low, the inter-lab difference in comet assay protocols is not a major issue when interpreted in terms of actual numbers of DNA lesions. The median in our pooled analysis (percent tail DNA = 2.9 %) corresponds to 0.08 lesions/106 base pairs or 475 lesions per cell, using the calibration curve from ECVAG, i.e. one % tail DNA unit = 0.0273 lesions/106 base pairs and the fact that the human diploid cell contains 6 × 109 base pairs [12]. In comparison, a previous pooled analysis of leukocyte polycyclic aromatic hydrocarbon (PAH)-DNA adducts data from several laboratories actually reported lower levels of lesions in current smokers compared to never smokers (0.67 versus 0.88 lesions/108 nucleotides; corresponding to 80 and 105 lesions per cell, respectively) [33]. The actual number of DNA lesions in our pooled analysis of DNA strand breaks in lymphocytes (i.e. 475 lesions per cell) is somewhat higher than PAH-DNA adducts (80–105 lesions per cell) [33], although it is much lower than the median level of oxidative stress generated 8-oxo-7,8-dihydro-2ʹdeoxyguanosine lesions measured by chromatographic assays in the ESCODD ring-trial on DNA damage levels in human lymphocytes (i.e. 4.4 lesions per million deoxyguanosine or 15,840 lesions per cell, assuming that 30 % of the genome is guanine nucleobases) [32].
As regards the potential role of host factors as confounders or effect modifiers, a minimal effect on DNA damage was found, and this result is consistent in most studies included in the meta-analysis. The lack of association between DNA damage and sex shown by multivariate models apparently contrasts with the higher levels reported in the univariate analysis in males, but is fully justified by the presence of confounding due to a higher prevalence of males in the group of exposed subjects (p < 0.001). More complex is the role of ageing, particularly as far as the increase observed in the oldest age-classes is concerned. Several interpretations can be proposed for this pattern, but due to the limited robustness of this finding (only a limited number of laboratories evaluated subjects over 65 years of age), further evidence to establish the reliability of this association is required. Nevertheless, this observation is in keeping with the age-dependent increase in DNA damage reported in a meta-analysis that used standardized results of different comet assay descriptors [34]. The effect of smoking habit could not be properly evaluated because of the lack of very informative details such as the number of cigarettes smoked per day, the history of smoking habit, or the source of information about smoking habit, which were not available for most studies. The extreme heterogeneity of available data, which in different studies ranged from extensively detailed (e.g., exposomics) to basic “yes/no” classifications, did not allow us to take into account specific features of smoking habit. Therefore, a simple comparison between those who declared to have ever smoked and those that declared to have never smoked was the only suitable option for assessing confounding from smoking. A lack of association between cigarette smoking and DNA damage has been previously reported for the comet assay [35] and for other DNA damage biomarkers [36]. In particular Hoffmann and colleagues [35] showed that the lower quality of exposure assessment may dilute the estimates of the genotoxic effect of smoking in human studies. Despite the contrasting evidence from our study on the genotoxic effect of cigarette smoking, this exposure should always be considered in the design and statistical analysis of biomonitoring studies. The experience with the micronucleus assay (MN), where an increase was observed only in heavy smokers, suggests that taking into account the number of cigarettes smoked is always recommended [36]. The presence of an adaptive response, as proposed for the MN results, could not be excluded in the present analysis. In conclusion, the results concerning host factors and lifestyle are in keeping with similar findings reported in a literature review published by Møller [9], including the uncertainty concerning the effect of ageing. As regards exposure to genotoxic agents, the level of DNA damage measured by the comet assay in different datasets confirmed for most genotoxic agents the sensitivity already described by inter-laboratory validation trials [11,12]. The fragmentation of the database as a whole prevented the generation of meaningful statistics for specific exposures or diseases. The need to take into consideration several technical and biological confounding factors considerably restricted the numbers of observations available for each analysis, and estimates of exposure- or disease-specific risk could not be reliably estimated. The most critical advantage of pooled analyses in this connection is to produce risk estimates which are stable and thus overcome the variability of individual studies, but since this was not possible here, we decided to show only analyses referring to the group of exposed as a whole. A collaborative study published by a large group of leading scientists in the field of the comet assay [14] estimated that 79.2 % of total variability was attributable to inter-laboratory variation, while the contribution of intra-laboratory and inter-subject variability was minimal (0.3 and 0.5 %. respectively). Actually, in our study, inter-study variability explained a notable proportion of variability, as high as 22.6 % for the TM. The size of human biomonitoring studies using the comet assay is typically around a hundred subjects, but often as few as a dozen, and so a study of several thousand subjects is an outstanding opportunity to substantiate or dismiss many common beliefs about this assay [8,37]. A further advantage is the achievement – in contrast with validation studies designed on purpose in small study groups and with fixed conditions - of a real-life picture of the assay, based on the experience of several laboratories, allowing consideration of all technical and epidemiological features. The final advantage is the creation of a network of laboratories, which can contribute to the definition of new standards, validation exercises, and ad hoc studies, etc.
The major limitation of this pooled analysis is the limited control of inter-laboratory heterogeneity. The overall number of 19,320 subjects, when stratified by cell type and divided by descriptor, fell into much smaller groups, with major discrepancies, such as the difference between fresh/frozen tissue and all the other covariates. The use of powerful statistical tools such as random effect modeling, which allows joint evaluation of the main components of the variability, and the simple but effective use of techniques from meta-analysis allowed us to provide reliable summary estimates of effect, although the whole size of the dataset could not be exploited. From a more general perspective, the role of DNA repair, which may reverse the individual results considered in the pooled analysis, is still to be defined at a population level. The level of DNA damage could be affected by a modified efficiency of DNA repair in specific groups, such as older age-classes, subjects repeatedly exposed to DNA damaging agents, smokers, etc. To address these issues an ad hoc study has been launched involving those hCOMET laboratories with data on DNA repair.
5. Conclusions
In conclusion, a number of valuable results have been produced by the pooled analysis of the hCOMET dataset for the validation of the comet assay in human biomonitoring studies; namely 1) a range of measures to be considered as the baseline level of DNA damage in non-exposed individuals has been provided for %T, TL, and TM; 2) studies measuring two or more descriptors showed a high correlation between descriptors; 3) sex is not consistently associated with any of the descriptors of the comet assay evaluated in this analysis; 4) Age showed contrasting results, with univariate analyses suggesting a moderate and constant increase of DNA damage for all the descriptors evaluated, and meta-analysis showing no age effect in the large majority of studies. The results from multivariate random effects modeling which suggest a higher level of DNA damage in oldest age-classes provide a reasonable interpretation of these results; 5) no association between smoking habit and all descriptors of the comet assay was evident for all approaches, although the lack of data on heavy smokers has still to be considered; 6) DNA damage measured by the comet assay was confirmed as a sensitive biomarker of exposure in several conditions, showing significant association in almost all exposures considered in the hCOMET dataset.
Supplementary Material
Acknowledgements
We would like to thank all those researchers who have expressed their interest in the hCOMET project and who have contributed their data, their ideas, and their enthusiasm to this consortium. We thank also all volunteers who agreed to participate to our studies, and generously donated biological samples and a little of their time.
Funding
This article is based upon work from COST Action hCOMET CA15132, supported by COST (European Cooperation in Science and Technology www.cost.eu) - STSM fellowships for Mirta Milić (IMROH, EU 19); IMROH, Zagreb, Croatia, Institute for Medical Research and Occupational Health (IMROH), Zagreb, Croatia, and the Ministry of Science, Education and Sports of the Republic of Croatia (Grant No. 022-0222148-2125) (EU4); Cancer Plan for PestiBG; Grant number: no ENV201401(EU 8, EU9); Italian Ministry of Education, University and Research PRIN 2005, prot. 2005058197 and Cariplo Foundation (Milan, Italy), Rif. Pratica 2007-5810 and Rif. Pratica 2010.2303 (EU 18); Associazione Italiana per la Ricerca sul Cancro (AIRC) (IG 2015/17564). (EU19); European Union Integrated Projects New Generis, 6th Framework Programme, Priority 5: Food Quality and Safety; Newborns and Genotoxic Exposure Risks, FOOD-CT-2005-016320 (EU22); ACT project No. 036APy/09 and No. 005DBB/12 (EU 24); FCT-SFRH/BPD/96196/2013, SFRH/BPD/100948/2014, Portugal (EU 26); MZ 2012/8-UKBA-8; VEGA 1/0703/13, APVV 15-0063 (EU30); Xunta de Galicia (XUGA 10605B98; INCITE08PXIB106155PR; ED481B2016/190-0; Grants ED431B2019/02), Spain (EU 32); Grant 01 173034, Ministry of Education, Science and Technological Development of the Republic of Serbia (EU 42); The Centre for Industrial and Technological Development within National Strategic Consortia for Techical Research (Industrial Research diets and food with specific characteristics for elderly, SENIFOOD); University of Navarra LE/97; Physiopathology of Obesity and Nutrition (CIBER Obn); Carlos III Health Research Institute (CB12/03/30002); Ministerio de Economia y Compatitividad (‘Ramón y Cajal’ Programme, RYC-2013-14370) of the Spanish Government for personal support (EU 45); the Ministry of Education, Youth and Sports of the Czech Republic project Healthy Aging in Industrial Environment HAIE (CZ.02.1.01/0.0/0.0/16_019/0000798) which is co-financed by the European Union (European Structural and Investment funds; Operation Programme Research, Development and Education); MYES LO 1508 (EU 46); MICRODIAB Study; ClinicalTrials.org (#NCT02231736) (EU 52); The study was funded by the Italian Ministry for Education, University and Scientific Research (MIUR) - Research No. 2005-062547 (EU14, EU53); Projects financed from Serbian Ministry of Education, Science and Technological Development #11146002, #175035, #173034 (EU 54); Mehr foundation organisation, UK (EU 55); MCTI/CNPQ No. 01/2016-Universal; FAPESC No. 09/2015; MEC/MCTI/CAPES/CNPQ/FAPS/No. 09/2014, Brazil (CSA 6); the National Nuclear Energy Agency of Indonesia (Badan Tenaga Nuklir Nasional) with contract number 080.01.06 3447.001 001.052.A (AS4); Slovak Grant Agency (APVT-21 013202, APVT-21- 017704); Ministry of Health, Slovak Republic (2005/43-SZU-21, 2006/07- SZU-02 MZ SR, <GN21>2005/42-SZU-20</GN21), New Generis, 6th Framework Programme QLK4-CT-1999-01629 and ERBICI 15-CT96-1012, CIPA-CT94-0129. (EU31); R.G. and S.L. thank the European Network of Excellence (NoE) ‘Environmental Cancer risk, Nutrition and Individual Susceptibility’ (ECNIS), sixth Framework programme (FP6), FOOD-CT-2005-513943 for financial support (EU21).
Footnotes
Declaration of Competing Interest
The authors report no declarations of interest.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.mrrev.2021.108371.
References
- [1].Collins AR, The comet assay for DNA damage and repair: principles, applications, and limitations, Mol. Biotechnol 26 (2004) 249–261 [DOI] [PubMed] [Google Scholar]
- [2].Azqueta A, Collins AR, The essential comet assay: a comprehensive guide to measuring DNA damage and repair, Arch. Toxicol 87 (2013) 949–968. [DOI] [PubMed] [Google Scholar]
- [3].Møller P, The comet assay: ready for 30 more years, Mutagenesis 33 (2018) 1–7. [DOI] [PubMed] [Google Scholar]
- [4].Neri M, Milazzo D, Ugolini D, Milic M, Campolongo A, Pasqualetti P, Bonassi S, Worldwide interest in the comet assay: a bibliometric study, Mutagenesis 30 (2015) 155–163. [DOI] [PubMed] [Google Scholar]
- [5].Azqueta A, Ladeira C, Giovannelli L, Boutet-Robinet E, Bonassi S, Neri M, Gajskii G, Duthie S, Del Bo C, Riso P, Koppen G, Basaran N, Collins A, Møller P, Application of the comet assay in human biomonitoring:an hCOMET, Mutat. Res 783 (2020) 82–88. [DOI] [PubMed] [Google Scholar]
- [6].Collins A, Koppen G, Valdiglesias V, Dusinska M, Kruszewski M, Møller P, Rojas E, Dhawan A, Benzie I, Coskun E, Moretti M, Speit G, Bonassi S for the ComNet project, The comet assay as a tool for human biomonitoring studies: the ComNet project, Rev. Mutat. Res 759 (2014) 27–39. [DOI] [PubMed] [Google Scholar]
- [7].Milic M, Frustaci A, Del Bufalo A, Sánchez-Alarcón J, Valencia-Quintana R, Russo P, Bonassi S, DNA damage in non-communicable diseases: a clinical and epidemiological perspective, Mutat. Res 776 (2015) 118–127. [DOI] [PubMed] [Google Scholar]
- [8].Taioli E, Bonassi S, Pooled analysis of epidemiological studies involving biological markers, Int. J. Hyg. Environ. Health 206 (2003) 109–115. [DOI] [PubMed] [Google Scholar]
- [9].Møller P, Assessment of reference values for DNA damage detected by the comet assay in human blood cell DNA, Mutat. Res 612 (2006) 84–104. [DOI] [PubMed] [Google Scholar]
- [10].Møller P, Mőller L, Godschalk RWL, Jones GDD, Assessment and reduction of comet assay variation in relation to DNA damage: studies from the European Comet Assay Validation Group, Mutagenesis 25 (2010) 109–111. [DOI] [PubMed] [Google Scholar]
- [11].Johansson C, Møller P, Forchhammer L, Loft S, Godschalk RWL, Langie SAS, Lumeij S, Jones GDD, Kwok RWL, Azqueta A, Phillips DH, Sozeri O, Routledge MN, Charlton AJ, Riso P, Porrini M, Allione A, Matullo G, Palus J, Stepnik M, Collins AR, Möller L, An ECVAG trial on assessment of oxidative damage to DNA measured by the comet assay, Mutagenesis 25 (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Forchhammer L, Johansson C, Loft S, Möller L, Godschalk RWL, Langie SAS, Jones GDD, Kwok RWL, Collins AR, Azqueta A, Phillips DH, Sozeri O, Stepnik M, Palus J, Vogel U, Wallin H, Routledge MN, Handforth C, Allione A, Matullo G, Teixeira JP, Costa S, Riso P, Porrini M, Møller P, Variation in the measurement of DNA damage by comet assay measured by the ECVAG inter-laboratory validation trial, Mutagenesis 25 (2010) 113–123. [DOI] [PubMed] [Google Scholar]
- [13].Forchhammer L, Ersson C, Loft S, Möller L, Godschalk RWL, van Schooten FJ, Jones GDD, Higgins JA, Cooke M, Mistry V, Karbaschi M, Collins AR, Azqueta A, Phillips DH, Sozeri O, Routledge MN, Nelson-Smith K, Riso P, Porrini M, Matullo G, Allione A, Stepnik M, Komorowska M, Teixeira JP, Costa S, Corcuera LA, López de Cerain A, Laffon B, Valdiglesias V, Møller P, Inter-laboratory variation in DNA damage using a standard comet assay protocol, Mutagenesis 27 (2012) 665–672. [DOI] [PubMed] [Google Scholar]
- [14].Ersson C, Møller P, Forchhammer L, Loft S, Azqueta A, Godschalk RWL, van Schooten FJ, Jones GDD, Higgins JA, Cooke MS, Mistry V, Karbaschi M, Phillips DH, Sozeri O, Routledge MN, Nelson-Smith K, Riso P, Porrini M, Matullo G, Allione A, Stepnik M, Ferlińska M, Teixeira JP, Costa S, Corcuera LA, López A, Laffon de Cerain B, Valdiglesias V, Collins AR, Möller L, An ECVAG inter-laboratory validation study of the comet assay: inter-laboratory and intra-laboratory variations of DNA strand breaks and FPG-sensitive sites in human mononuclear cells, Mutagenesis 28 (2013) 279–286. [DOI] [PubMed] [Google Scholar]
- [15].Godschalk RWL, Ersson C, Stepnik M, Ferlińska M, Palus J, Teixeira JP, Costa S, Jones GDD, Higgins JA, Kain J, Möller L, Forchhammer L, Loft S, Lorenzo Y, Collins AR, van Schooten FJ, Laffon B, Valdiglesias V, Cooke M, Mistry V, Karbaschi M, Phillips DH, Sozeri O, Routledge MN, Nelson-Smith K, Riso P, Porrini M, López A, de Cerain A, Azqueta A, Matullo G, Allione A, Møller P, Variation of DNA damage levels in peripheral blood mononuclear cells isolated in different laboratories, Mutagenesis 29 (2014) 241–249. [DOI] [PubMed] [Google Scholar]
- [16].Zainol M, Stoute J, Almeida GM, Rapp A, Bowman KJ, Jones GDD, ECVAG. Introducing a true internal standard for the Comet assay to minimize intra- and inter-experiment variability in measures of DNA damage and repair, Nucleic Acids Res 37 (2009) e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Collins AR, Anderson D, Coskun E, Dhawan A, Dusinska M, Koppen G, Kruszewski M, Moretti M, Rojas E, Speit G, Valverde M, Bonassi S, Launch of the ComNet (comet network) project on the comet assay in human population studies during the International Comet Assay Workshop meeting in Kusadasi, Turkey (September 13–16, 2011), Mutagenesis 27 (2012) 385–866. [DOI] [PubMed] [Google Scholar]
- [18].Lovell DP, Omori T, Statistical issues in the use of the comet assay, Mutagenesis 23 (2008) 171–182. [DOI] [PubMed] [Google Scholar]
- [19].Goldstein H, Multilevel Statistical Models, third ed., Oxford University Press Inc., New York, 2003. [Google Scholar]
- [20].Harrell FE Jr., Regression Modeling Strategies: with Application to Linear Model, Logistic Regression, and Survival Analysis, Springer, New York, 2001. [Google Scholar]
- [21].Merlo J, Chaix B, Ohlsson H, Beckman A, Johnell K, Hjerpe P, Råstam L, Larsen K, A brief conceptual tutorial of multilevel analysis in social epidemiology: using measures of clustering in multilevel logistic regression to investigate contextual phenomena, J. Epidemiol. Community Health 60 (2006) 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Der Simonian R, Laird N, Meta-analysis in clinical trials, Control. Clin. Trials 7 (1986) 177–188. [DOI] [PubMed] [Google Scholar]
- [23].Brøgger A, Hagmar L, Hansteen IL, Heim S, Högstedt B, Knudsen L, Lambert B, Linnainmaa K, Mitelman F, Nordenson I, et al. , An inter-Nordic prospective study on cytogenetic endpoints and cancer risk. Nordic Study Group on the Health Risk of Chromosome Damage, Cancer Genet. Cytogenet 45 (1990) 85–92. [DOI] [PubMed] [Google Scholar]
- [24].Bolognesi C, Abbondandolo A, Barale R, Casalone R, Dalpra L, De Ferrari M, Degrassi F, Forni A, Lamberti L, Lando C, Migliore L, Padovani P, Pasquini R, Puntoni R, Sbrana I, Stella M, Bonassi S, Age-related increase of baseline frequencies of sister chromatid exchanges, chromosome aberrations, and micronuclei in human lymphocytes, Cancer Epidemiol. Biomarkers Prev 6 (1997) 249–256. [PubMed] [Google Scholar]
- [25].Bonassi S, Fenech M, Lando C, Lin YP, Ceppi M, Chang WP, Holland N, Kirsch-Volders M, Zeiger E, Ban S, Barale R, Bigatti MP, Bolognesi C, Jia C, Di Giorgio M, Ferguson LR, Fucic A, Lima OG, Hrelia P, Krishnaja AP, Lee TK, Migliore L, Mikhalevich L, Mirkova E, Mosesso P, Müller WU, Odagiri Y, Scarfi MR, Szabova E, Vorobtsova I, Vral A, Zijno A, HUman MicroNucleus project: international database comparison for results with the cytokinesis-block micronucleus assay in human lymphocytes: I. effect of laboratory protocol, scoring criteria, and host factors on the frequency of micronuclei, Environ. Mol. Mutagen 37 (2001) 31–45. [PubMed] [Google Scholar]
- [26].Bonassi S, Coskun E, Ceppi M, Lando C, Bolognesi C, Burgaz S, Holland N, Kirsh-Volders M, Knasmueller S, Zeiger E, Carnesoltas D, Cavallo D, da Silva J, de Andrade VM, Demircigil GC, Domínguez Odio A, Donmez-Altuntas H, Gattas G, Giri A, Giri S, Gómez-Meda B, Gómez-Arroyo S, Hadjidekova V, Haveric A, Kamboj M, Kurteshi K, Martino-Roth MG, Montero Montoya R, Nersesyan A, Pastor-Benito S, Favero Salvadori DM, Shaposhnikova A, Stopper H, Thomas P, Torres-Bugarín O, Yadav AS, Zúñiga González G, Fenech M, The HUman MicroNucleus project on eXfoLiated buccal cells (HUMN (XL)): the role of lifestyle, host factors, occupational exposures, health status, and assay protocol, Mutat. Res 728 (2011) 88–97. [DOI] [PubMed] [Google Scholar]
- [27].Azqueta A, Muruzabal D, Boutet-Robinet E, Milic M, Dusinska M, Brunborg G, Møller P, Collins AR, Technical recommendations to perform the alkaline standard and enzyme-modified comet assay in human biomonitoring studies, Mutat. Res 843 (2019) 24–32. [DOI] [PubMed] [Google Scholar]
- [28].Møller P, Loft S, Ersson C, Koppen G, Dusinska M, Collins A, On the search for an intelligible comet assay descriptor, Front. Genet 5 (2014) 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Møller P, Cooke MS, Collins A, Olinski R, Rozalski R, Loft S, Harmonising measurements of 8-oxo-7,8-dihydro-2’-deoxyguanosine in cellular DNA and urine, Free Radic. Res 46 (2012) 541–553. [DOI] [PubMed] [Google Scholar]
- [30].ESCODD (European Standards Committee on Oxidative DNA Damage), Comparative analysis of baseline 8-oxo-7,8-dihydroguanine in mammalian cell DNA, by different methods in different laboratories: an approach to consensus, Carcinogenesis 23 (2002) 2129–2133. [DOI] [PubMed] [Google Scholar]
- [31].European Standards Committee on Oxidative DNA Damage (ESCODD), Measurement of DNA oxidation in human cells by chromatographic and enzymic methods, Free Radic. Biol. Med 34 (2003) 1089–1099. [DOI] [PubMed] [Google Scholar]
- [32].ESCODD (European Standards Committee on Oxidative DNA Damage), Gedik CM, Collins A, Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study, FASEB J 19 (2005) 82–84. [DOI] [PubMed] [Google Scholar]
- [33].Ricceri F, Godschalk RW, Peluso M, Phillips DH, Agudo A, Georgiadis P, Loft S, Tjonneland A, Raaschou-Nielsen O, Palli D, Perera F, Vermeulen R, Taioli E, Sram RJ, Munnia A, Rosa F, Allione A, Matullo G, Vineis P, Bulky DNA adducts in white blood cells: a pooled analysis of 3,600 subjects, Cancer Epidemiol. Biomarkers Prev 19 (2010) 3174–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Møller P, Effect of age and sex on the level of DNA strand breaks and oxidatively damaged DNA in human blood cells, Mutat. Res 838 (2019) 16–21. [DOI] [PubMed] [Google Scholar]
- [35].Hoffmann H, Högel J, Speit G, The effect of smoking on DNA effects in the comet assay: a meta-analysis, Mutagenesis 20 (2005) 455–466. [DOI] [PubMed] [Google Scholar]
- [36].Bonassi S, Neri M, Lando C, Ceppi M, Lin YP, Chang WP, Holland N, Kirsch-Volders M, Zeiger E, Fenech M, For the HUMN collaborative group, effect of smoking habit on the frequency of micronuclei in human lymphocytes: results from the Human Micro Nucleus project, Mutat. Res 543 (2003) 155–166. [DOI] [PubMed] [Google Scholar]
- [37].Key TJ, Appleby PN, Allen NE, Reeves GK, Pooling biomarker data from different studies of disease risk, with a focus on endogenous hormones, Cancer Epidemiol. Biomarkers Prev 19 (2010) 960–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


