Abstract
Ivermectin (IVM) is an FDA approved macrocyclic lactone compound traditionally used to treat parasitic infestations and has shown to have antiviral potential from previous in-vitro studies. Currently, IVM is commercially available as a veterinary drug but have also been applied in humans to treat onchocerciasis (river blindness - a parasitic worm infection) and strongyloidiasis (a roundworm/nematode infection). In light of the recent pandemic, the repurposing of IVM to combat SARS-CoV-2 has acquired significant attention. Recently, IVM has been proven effective in numerous in-silico and molecular biology experiments against the infection in mammalian cells and human cohort studies. One promising study had reported a marked reduction of 93% of released virion and 99.98% unreleased virion levels upon administration of IVM to Vero-hSLAM cells. IVM's mode of action centres around the inhibition of the cytoplasmic-nuclear shuttling of viral proteins by disrupting the Importin heterodimer complex (IMPα/β1) and downregulating STAT3, thereby effectively reducing the cytokine storm. Furthermore, the ability of IVM to block the active sites of viral 3CLpro and S protein, disrupts important machinery such as viral replication and attachment. This review compiles all the molecular evidence to date, in review of the antiviral characteristics exhibited by IVM. Thereafter, we discuss IVM's mechanism and highlight the clinical advantages that could potentially contribute towards disabling the viral replication of SARS-CoV-2. In summary, the collective review of recent efforts suggests that IVM has a prophylactic effect and would be a strong candidate for clinical trials to treat SARS-CoV-2.
Keywords: Antiviral, Treatment, Inhibition, Drug repurposing, Importin heterodimer complex, STAT3, Cytokine storm, Viral 3CLpro, Streptomyces avermitilis
1. Introduction
The recent pandemic of Covid-19 has been acknowledged as a global health crisis that has threatened public health and safety. This phenomenon has raised awareness and conflict towards the healthcare sector for their negligence and absence of alternative measures that would have prevented or successfully treated novel causative viral agents. Approximately 233,000,000 individuals have been affected during this pandemic, and 4,750,000 associated deaths were correlated worldwide. Currently, the cases of Covid-19 are still growing rapidly on a global scale [1]. The etiological agent that contributed to this disastrous episode is known to be the SARS Coronavirus – 2 (SARS-CoV-2). To date, the origin of SARS-CoV-2 are still debated though evidence suggests bats as the source of the virus, akin to the SARS-CoV outbreak identified during 2002 [2]. Although SARS-CoV-2 carries a 3.4% mortality rate, which is significantly lower than its predecessors (SARS-CoV and MERS-CoV), it is more infectious. Of note, up to 15% of SARS-CoV-2 cases led to several critical complications such as pneumonia, heart arrhythmia, septic shock, multiple organ failure, and the more pronounced acute respiratory distress syndrome (ARDS) [3]. Thus, emphasizing the need for effective antiviral therapy. In this urgent time of need, drug repositioning attempts are crucial to address the immediate demand for a fast and effective antiviral therapy against COVID-19.
Drug repositioning, also known as drug repurposing or drug recycling, is an alternative approach to finding new use of an established drug to treat other diseases aside from the intended ones. A repositioned drug has often gone through all the rigorous safety and pharmacokinetic profiling studies with well-established ADMET (absorption, distribution, metabolism, excretion, toxicity) data [4]. The benefits of utilizing repositioned drugs are the omission of critical and time-consuming drug development stages, which significantly reduces the time needed to produce an effective antiviral drug. In this paradigm, we discuss the numerous drug candidates already proposed for SARS-CoV-2 treatment, such as Remdesivir, Lopinavir-Ritonavir, Oseltamivir, Saquinavir, Tenofovir, and ciclesonide [5].
Firstly, Lopinavir-Ritonavir and Saquinavir has been proven effective in inhibiting the replication of SARS-CoV-2 via forming a stable complex with its main protease (Mpro or 3CLpro), which oversees the regulation of replicase polyprotein proteolytic activity and viral replication [6], [7]. Conversely, tenofovir as a nucleotide analogue for HIV treatment was revealed to interact with papain-like protease (PLpro) and Mpro. This interferes with the binding of SARS-CoV-2 S protein to the host ACE2 receptor, subsequently reducing the production of non-structural protein (NSP) required for viral replication [8]. Additionally, tenofovir also binds to SARS-CoV-2 RNA dependent RNA polymerase (RdRp), an important replicase for viral replication, effectively bringing down the viral load [8]. Ciclesonide is a corticosteroid was reported to have targeted the nsp15 or also known as the nidoviral RNA endoribonuclease uridylate-specific (NendoU) enzyme in SARS-CoV-2. This protein interferes with human innate immune response, contributing to its immune-evasive properties in COVID-19 patients [9].
Most recently, the anti-parasitic drug Ivermectin (IVM) has also gained an interest as a newly repositioned drug against Covid-19 disease. IVM is one of the best known antiparasitic drug discoveries for human and veterinary medicine by microbiologist, Satoshi Ōmura at a Japanese golf course in the 1970s [10]. The active components were identified via NRRL 8165 culture of gram-positive Streptomyces avermitilis, which had revealed a 16-membered macrocyclic lactones, named avermectins (AVMs). These AVMs are responsible for the nematicidal, acaricidal and insecticidal activities [10], [11]. This drug was further investigated by William C. Campbell et al., at Merck, Sharpe and Dohme (MSD), which had led to discovery of IVM variants B1a and B1b that were shown to possess the highest activity and were later selected for commercial development under the name, Ivermectin (IVM) [12], [13]. IVM was first introduced to the commercial market by Merck & Co. in 1981, as a veterinary drug [14]. In 1987, IVM was registered by Merck for the onchocerciasis (river blindness – a parasitic worm infection) in humans under the Mectizan Donation Program (MDP) with a mass treatment in 1988 [15]. As of June 2021, IVM is the only macrocyclic lactone drug approved for onchocerciasis and strongyloidiasis (a roundworm/nematode infection) in humans by the US Food and Drug Administration (FDA) [16]. This incredible discovery and deployment of IVM by Satoshi Ōmura, and William C. Campbell had won them the 2015 Nobel Prize in Physiology or Medicine [17].
Apart from the broad anti-parasitic implications such as onchocerciasis, elephantiasis, scabies and strongyloidiasis, IVM has been found to inhibit several cancer cells through regulation of multiple signalling pathways [17]. For instance, it was discovered that IVM decreased miR-21 levels via inhibition of DDX23 activity, effectively blocking cell proliferation in cancer cells [18]. IVM was also found to bind and activate Farnesoid X receptor (FXR), effectively reducing hepatic lipid accumulation. Subsequently, improving insulin sensitivity in non-alcoholic fatty liver disease as indicated by decreased serum cholesterol and glucose levels [19]. Interestingly, IVM was reported to have exhibited antiviral properties against Flaviviruses and Lentiviruses. It had shown promising results in inhibiting replication of flavivirus by targeting the NS3 helicase [20]. IVM also inhibits the nuclear transport of viral proteins via α/β-mediated nuclear transport, exerting an antiviral effect against HIV-1 and dengue viruses [21]. Recently, IVM has been investigated for its antiviral properties against SARS-CoV-2, which reported a 5000-fold reduction in SARS-CoV-2 RNA levels in an in-vitro study [22].
The deployment of IVM against SARS-CoV-2 may provide an effective antiviral treatment in this urgent time of need. Henceforth, this manuscript aims at highlighting all known molecular mechanisms of IVM and its effective usage in combatting SARS-CoV-2 replication thereby reducing the severity of Covid-19 related pathogenesis in patients.
2. Antiparasitic action of Ivermectin
As aforementioned, IVM was chemically derived from AVMs, a group of 16-membered macrocyclic lactone compounds. IVM possesses two variants as A or B, which are differentiated by the presence of methoxy or hydroxyl groups at C5, respectively. AVMs naturally occur as mixtures of eight compounds A1a, A1b, A2a, A2b, B1a, B1b, B2a and B2b, from which, B1 is administered orally while B2 is parenterally administered [23]. The subscript ‘1’ following the variants indicates the presence of double bonds between C22 and C23 whereas, the subscript of ‘2’ describes the presence of hydrogen and hydroxyl groups at C22 and C23, respectively [23]. As such, IVM is a semi-synthetic derivative of Avermectin B1 where it consists of two homologues, 22, 23-dihydro-avermectin B1a and 22, 23-dihydro-avermectin B1b in the ratio of 80:20 [24]. IVM is capable of affecting the motility, feeding and reproduction of parasites through a high-affinity binding to the γ-aminobutyric acid (GABA)-regulated or glutamate-gated chloride channels. In response, the activities of these channels are enhanced, leading to hyperpolarization of the cell membrane and influx of chloride ions. Subsequently, this inhibits the regulatory light chain of myosin II phosphorylation via p21 activated kinase 1 (PAK1), causing muscle paralysis and eventually, parasite death [25].
Apart from its original function, IVM has been proven effective in numerous antiviral treatments via the inhibition of the nuclear import of viral nucleoproteins. This encompasses HIV-1, West Nile Virus (WNV), tick-borne encephalitis, Zika Virus (ZKV), Venezuelan equine encephalitis virus, Chikungunya virus, Pseudorabies virus, Adenovirus, Influenza virus, SARS-CoV-1, and most recently in SARS-CoV-2 [26], [27], [28]. The mode of inhibition of the aforementioned viruses were described in detail below.
3. Antiviral action of Ivermectin
Several studies in the past have revealed the possible role of SARS-CoV-1 ORF6 interacting with the Karyopherin-α2 (KPNA2), retaining the IMPα/β1 of the Golgi membrane. Thereafter, inhibiting the STAT1 nuclear transport, antagonizing antiviral activity and downplaying the host's antiviral response [28], [29], [30]. Given the role of importin in many viruses, especially SARS-CoV-1, it is of great interest to explore the mechanism of action of IVM for its potential in viral inhibition. We briefly describe the antiviral action of IVM for each of the viruses listed above and postulate its role in the SARS-CoV-2 infection cycle.
3.1. Flavivirus (Zika/West-Nile virus)
The Flavivirus genus spans over numerous types of viruses such as ZKV, WNV and many more. ZKV is an enveloped single-stranded positive-sense RNA virus, akin to the SARS-CoV-2 [31]. Differing from SARS-CoV-2, ZIKV is mosquito-borne via Aedes mosquitoes carrying the possibility of non-mosquito transmissions, such as pregnancy (mother-foetus) and sexual transmission [32]. ZKV increases the risk of neurological complications such as Guillain-Barré syndrome, neuropathy, and myelitis [33]. An in-vitro study conducted by Barrow and colleagues has discovered the antiviral effects of IVM on ZKV, particularly on the ZKV MEX_I_7 strain [32]. In this study, IVM showed the strongest inhibition of ZKV at concentrations of 10 μM and 16 μM in human liver cells (HuH-7) and human amnion epithelial cells (HAECs), respectively. Apart from this, another study has also shown IVM to act as an inhibitor of importin α/β that blocks non-structural protein 5 (NS5) interaction with IMPα/β transporter in ZKV, leading to 60% NS5 reduction in nucleus post-7 h IVM treatment [34]. NS5 is an important protein for the methyltransferase and RNA-dependent RNA polymerase activities in ZKV, which is vital for viral replication [34].
As a member of the Flavivirus genus, the WNV is similar to ZKV, as an enveloped single-stranded positive-sense RNA virus [35]. It is also mosquito-borne, but via Culex mosquitoes [35]. IVM was also found to exhibit an inhibitory effect in WNV through an effective concentration of 50 (EC50) of 4 μM, inhibiting the NS3 helicase. The NS3 helicase is an important protein that mediates the RNA binding and unwinding in WNV, thereby leading towards reduction of viral load [36]. Hence, implicating the huge potential for IVM in antiviral therapeutics.
3.2. Influenza A virus
Influenza A virus (IAV) is an RNA virus that is made up of eight different negative-sense RNA segments which encode for 12 different proteins [37]. To achieve a new IAV lineage in the human population, the IAV needs to overcome MxA and nuclear viral ribonucleoprotein (vRNP) nuclear import restrictions [38]. MxA antiviral proteins are produced by Myxovirus resistance gene1 (MX1) upon induction of the type-I (α/β) or type-III (λ) Interferons (IFN) during the early stages of IAV infection [38]. Interestingly, the determining factor for the strength of MxA inhibition relies upon the viral nucleoprotein (NP) [38]. It was hypothesized that the MxA inhibits IAV in two different pathways. Firstly, it interferes with the transcription of viral RNA via retention of incoming vRNPs in the cytoplasm [39]. Secondly, MxA inhibits the amplification of viral RNA (vRNA) via cytoplasmic sequestering of newly synthesized NP and PB2 [40]. IVM as a importin α/β complex inhibitor shows a complete suspension of the nuclear import for all vRNP complexes in both wild-type and antiviral MxA escape IAV mutants at 10 μM concentration. Therefore, it effectively inhibits the viral replication [41]. These findings provide a clear potential for IVM as an antiviral medication against IAV.
3.3. HIV-1 virus
Human immunodeficiency virus type 1 or HIV-1 is a single-stranded, positive-sense, enveloped RNA virus under the genus Lentivirus. Upon entry into the cell, the vRNA genome of HIV-1 is transcribed into dsDNA (cDNA) via reverse transcriptase [42]. This is followed by the import of dsDNA into the cell nucleus and integration into the cellular DNA by the viral encoded enzyme Integrase (IN), allowing for immune evasion and subsequent vRNA replication. HIV-1 depend heavily on IN for efficient viral production and replication [42]. IVM has been documented to show a reduction of IN nuclear accumulation in HIV-1 infected cells, with a half-maximal inhibitory concentration (IC50) of 2 μM [43]. Another study showed that treatment of 25 μM IVM in HIV-1 infected HeLa cells for 2 h, significantly reduced viral replication with no observable cell toxicity induced [21]. These results illustrates that IVM can inhibit IN nuclear import by reducing the viral protein binding by the importin heterodimer complex (IMPα/β1), halting further HIV-1 replication.
4. General morphology of SARS-CoV-2
The general morphology of SARS-CoV-2 closely resembles other beta-coronaviruses, such as the SARS-CoV and MERS-CoV [44]. The SARS-CoV-2 is a non-segmented positive-sense RNA virus with a diameter of 65-125 nm [45]. The typical layout for SARS-CoV-2 genome is denoted as follows [5′‑leader-UTR-replicase-S-E-M-N-3′-UTR-poly (A) tail], with accessory genes scattered between the structural genes (S-E-M-N) at the 3′ end (38×). Under the envelope, it consists of a ~29.9 kilobase (kb) RNA genome with 2/3rd of the genome containing the main open reading frame 1a and 1b (ORF1ab) replicase gene from the 5′-end, encoding for the non-structural proteins (NSP 1–16) while the remaining 1/3rd genome encodes for the structural proteins (spike protein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N)) [46]. The NSPs play an important role for viral replication in infected host cells. NSP 1 and 3 are known to inhibit IFN signalling, interrupting the translation of RNA and innate immune responses [47]. NSP 3 and 5 promote cytokine expression and viral protein cleavage [47]. NSP-12 is an RNA-dependent RNA polymerase (RdRp) and has been shown to be inhibited by IVM in studies on SARS-CoV and MERS-CoV [48], [49]. This discovery discloses strong possibilities for SARS-CoV-2. RdRp, also known as RNA replicase, is a vital enzyme in the life cycle for RNA viruses since it primarily functions to catalyse the replication of RNA from an existing RNA template in the virus thereby initiating viral replication.
5. The molecular action of Ivermectin on SARS-CoV-2
The SARS-CoV accessory protein ORF6 has been shown to sequester IMPα/β1 on the rough endoplasmic reticulum which antagonizes the STAT1 transcription factor, resulting in an antiviral potential [50]. The genomic similarity between SARS-CoV-2 and the previous SARS-CoV may reveal the role of the importin heterodimer complex (IMPα/β1) for viral protein (NSP12-RdRp) shuttling between the nucleus and cytoplasm upon infection. Currently, there is only one RdRp inhibitor approved by the FDA for Covid-19, namely Remdesivir [51]. However, a recent discovery from Monash University, Australia reported that IVM could inhibit SARS-CoV-2 within a 48 h post-infection, drawing much attention worldwide [22].
IVM, a non-specific inhibitor of IMPα/β1-dependent nuclear import, now shows great potential in reducing SARS-CoV-2 viral replication via different modes. Apart from disrupting the importin heterodimer complex IMPα/β1, IVM also prevents cytokine storm via STAT3 regulation. Besides this, IVM also inhibits the viral entry via the ACE2 receptor and is capable of disrupting the viral 3-chymotrypsin-like enzyme in SARS-CoV-2 [52], [53]. We describe each of the proposed molecular mechanisms below.
5.1. The disruption of Importin heterodimer complex (IMPα/β1)
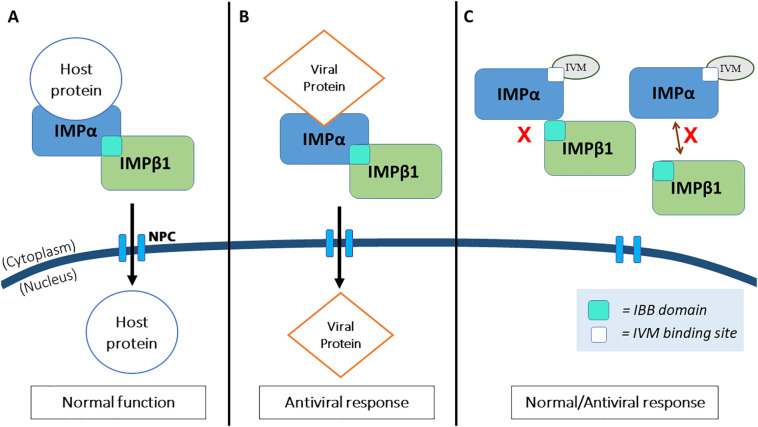
Transporting host proteins, such as STAT or NF-κB transcription factor families, in-and-out of the nucleus is a crucial function for normal nuclear activity in all eukaryotic cells. The signal for transport is mediated by the members of the IMP superfamily transporters, α- and β- types [54]. The import of host protein, such as STAT protein (>45 kDa) into the nucleus requires nuclear localisation signal (NLS) to be recognised by either the IMPα from the importin heterodimer complex (IMPα/β1), IMPβ1 alone or the homologues thereof. From there, the cargo protein interacts with the IMPβ1 and is translocated via the nuclear pore complex (NPC) embedded in the nuclear envelope [55]. This nuclear transport complex has been documented to have been hijacked by many viruses, such as Influenza (NP), Dengue Virus (NS5), HIV-1 (Integrase), SARS-CoV (ORF6), and the most recently by SARS-CoV-2 (NSP12-RdRp) to gain access into the nucleus and to facilitate infection (42×,44×,48×). Theoretically, the IVM binds to the IMPα, dissociating the IMPα from the heterodimer IMPα/β1 complex, thus preventing nuclear import of viral proteins via the NPC, halting the IMPα/β1 dependent nuclear import activities of viral SARS-CoV-2 proteins (NS12-RdRp) [56] (Fig. 1 ).
Fig. 1.
A brief layout illustrating the roles of IMPα/β1 in nuclear transport during normal function and upon SARS-CoV-2 infection with respect to the inhibitory activity by IVM. A) Transporting host proteins (e.g.: STAT) requires interaction with the Importin heterodimer complex (IMPα/β1) via binding at the IMPα, which subsequently brings the whole complex (IMPα/β1) to the nucleus via the nuclear pore complex (NPC), thus establishing normal transcriptional activities. B) However, this process is postulated to be hijacked by the viral protein NS12. C) IVM targets the binding site at the IMPα or binding domain at IMPβ1, leading to dissociation of the complex from the latter, halting the hijacking mechanism mentioned in B.
In the laboratory setting, numerous in-vitro studies on IVM have been carried out recently. An in-vitro infection study on a SARS-CoV-2 isolate, Australia/VIC01/2020 showed a 93% reduction in viral RNA at 24 h and a 99.98% reduction (~5000-fold) at 48 h upon administration of 5 μM IVM [22]. The same study also found no cytotoxicity from administration of IVM, making it a safe and efficacious candidate for further clinical studies [22]. Furthermore, a clinical study (NCT04422561) showed that two doses of IVM at 72 h apart greatly reduced the symptoms of SARS-CoV-2 infection by 7.4% as compared to the control group at 58.4%. The authors conclude the unravelling of a huge potential of IVM as a prophylactic agent [57]. On top of that, 19 ongoing randomized controlled trials on IVM for Covid-19 have shown significantly reduced mortality, improved clinical recovery time, and viral clearance [58]. Meanwhile, an in-silico study revealed that IVM is capable of binding to the RdRp complex at the active residues (Ser759 and Asp760), further insinuating the potential of IVM for inhibiting SARS-CoV-2 viral replication [59].
5.2. The inhibition of cytokine storm in Covid-19
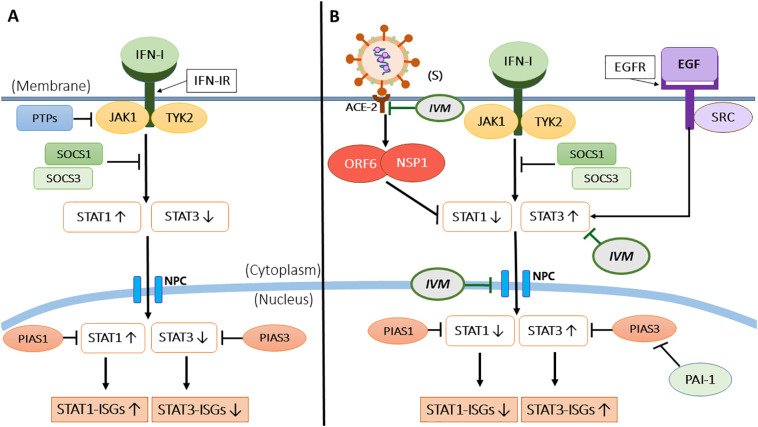
Apart from disrupting the IMPα/β1 complex, SARS-CoV-2 proteins have been postulated to antagonize the antiviral interferon (IFNs) and the downstream JAK-STAT signalling pathway, primarily through IFN and STAT1, resulting in a cytokine storm [60]. Cells recognise foreign viruses via various endosomal or cytosolic pattern recognition receptors (PRRs), such as Toll-like receptors 3, 4, 7 and 8 (TLRs), and retinoic acid-inducible gene I-like receptors (RLRs) (RIG-1, MDA5) [60], [61]. Upon recognition, nuclear factor-κΒ (NF-κB) stimulates the production of pro-inflammatory cytokines (IL-1, IL-6, TNF-α). The interferon regulatory factor 3 and 7 (IRFs) stimulate the production of type I (IFN-α, IFN-β, IFN-ε, IFN-κ, IFN-ω) and type III (IFN-λ) IFNs, respectively [61]. Then, the type I and III IFNs recognised by the JAK proteins (JAK1 and TYK2) and STAT proteins (STAT 1 and 2) stimulate the formation of transcription factor complex ISGF3 (containing STAT1/STAT2/IRF9) to translocate into the nucleus and upregulate interferon-stimulated genes (ISGs) to interfere with the viral replication. Additionally, Karyopherin-α1 (KPNA1) is essential for the nuclear transport of ISGF3 via interaction with STAT1 [62]. The STAT proteins are also involved in the signal transduction of downstream cytokines such as TGF-β, IL-2, IL-4, IL-12, and inflammatory IL-6 [62] (Fig. 2).
Fig. 2.
A schematic layout on STAT dependent IFN-1 signalling, a key contributor to the cytokine storm in SARS-CoV-2 infected individuals and the action of IVM. A) The IFN-I signalling pathway showing JAK1 and TYK2 being activated upon binding of Interferon-1 (IFN-I) at the IFN-1 receptor (IFN-IR). In a normal scenario, STAT1 is predominantly activated to induce interferon-stimulated genes (ISGs)-(STAT1-ISGs) via (STAT1/STAT2/IRF9) complex, known as ISGF3 (not shown here). STAT3 activation is also present here albeit small. B) Upon SARS-CoV-2 infection the activity of STAT1 is inhibited by SARS-CoV-2 proteins (NSP1 and ORF6), leading to the upregulation of STAT3 activity that subsequently induces STAT3-ISGs. Following this, the PIAS1 and PIAS3 regulate the binding activity of STAT1 and STAT3 to the DNA respectively, via negative feedback. However, upon infection, the hyperactivation of STAT3 represses the miR-34a, an inhibitor for plasminogen activator inhibitor-1 (PAI-1), inadvertently enhancing the level of PAI-1, which in turn inhibits the activity of PIAS3. To further stress, the epidermal growth factor receptor (EGFR) produced from the reduced STAT1 activity also activates STAT3, leading to enhanced production of cytokine and chemokines. Here, IVM inhibits STAT3 activity, subsequently reducing inflammatory IL-6 cytokine production, preventing cytokine storm and ADRS. IVM also binds to the viral S protein, thereby prohibiting the attachment of S protein to the host ACE2 receptors for viral entry and reducing the viral load.
SARS-CoV-2 and SARS-CoV share the same cellular signalling pathways and therefore, a similar disruption of viral replication [60]. Upon SARS-CoV-2 infection, the activity of STAT1 is inhibited by the SARS-CoV-2 NSP1 and ORF6 proteins. The reduced STAT1 level is compensated by an enhanced STAT3 activity, subsequently inducing STAT3-ISGs [60]. The reduction of STAT1 or acute lung injury also upregulates epidermal growth factor receptor (EGFR), further enhancing STAT3 activity [63], [64]. Furthermore, the upregulation of EGFR could also impair IFN-1 signalling, affecting its downstream regulation of STAT1 [60]. The hyperactivation of STAT3 also represses miR-34a, an inhibitor for the plasminogen activator inhibitor-1 (PAI-1). PAI-1 is a crucial inhibitor for the regulation of the protein inhibitor for activated STAT3 (PIAS3) activity for negative feedback, an enhanced PAI-1 would lead to further inhibition of PIAS3, causing for unregulated STAT3 levels in the nucleus [60].
In short, the activity of STAT3 is upregulated in many ways, and the activity of STAT1-mediated IFN-I response is downregulated upon SARS-CoV-2 infection. The hyperactivation of STAT3 upon SARS-CoV-2 infection leads to a cascade of deleterious events. Notably, STAT3 causes significant inflammatory cytokine (IL-6) production via increased macrophage activity in Covid-19 patients. Cytokine production is also enhanced, in parallel to PAI-1 that binds to TLR4, leading to the activation of NF-κB which produces a large production of IL-6 and other chemokines [65]. Aside from that, STAT3 can also activate integrin αvβ6 and thrombospondin at the lung extracellular matrix (ECM), contributing to TGF-β activation and pulmonary fibrosis [66]. The TGF-β activation upregulates PAI-1, facilitating the continuous production of IL-6 [66]. Lastly, STAT3 and PAI-1 increase the production of Hyaluronan (HA), leading to severe acute respiratory distress syndrome (ARDS) in Covid-19 patients, as a result of diffuse alveolar damage (DAD) [67].
IVM may be a good therapeutic tool to inhibit the cytokine storm and prevent ADRS in the following pathways. IVM blocks the IMPα/β1-dependent nuclear import of viral proteins (NS12-RdRp) as mentioned prior [56]. IVM also inhibits the activity of STAT3, subsequently reducing inflammatory IL-6 cytokine production. Besides that, IVM promotes the ubiquitination-mediated degradation of p21 activated kinase 1 (PAK1), a key protein that binds to STAT3 for IL-6 gene transcription, disrupting IL-6 production [60], [68]. The reduction of STAT3 levels by IVM could also bring down HA levels, preventing ADRS in Covid-19 patients [67].
5.3. Inhibition of viral entry via ACE2 receptor
Much like the previous SARS-CoV, SARS-CoV-2 also requires the binding of spike (S) protein to the host angiotensin-converting enzyme 2 (ACE2) receptors to infiltrate the host cells [69]. There are two regions in the receptor-binding motif on the S protein that forms an interface between S protein and ACE2. Key residues from the S protein, namely L455 and Q493, possess a high affinity for residues K31 and E35 on the ACE2 via hydrogen bonding, thus encouraging the binding of S protein to the ACE2 receptor [70]. It was evident that the overexpression of ACE2 was highly associated to SARS-CoV-2 replication, from which the concentration of ACE2 remained at a high level at 72 h post-infection [71]. The overexpression of ACE2 can be induced by the interferons and cytokines associated with ARDS, regulated by the JAK-STAT signalling pathways. In conjunction, the ACE2 is also associated with pathways involving immunological activation such as the chemokine signalling pathway and cytokine-cytokine receptor interactions. Several studies have since demonstrated the association between ACE2 expression and the levels of signalling cascade components (IL-7R, JAK1, JAK2, JAK3, TYK2, STAT1, STAT3, STAT4, STAT5) [71], [72]. Given this close association, it can be postulated that the cytokines and interferons produced from the activation of several signalling pathways after SARS-CoV-2 infection may lead to an overexpression of ACE2, further facilitating viral replication and potentially worsened prognosis. This highlights the need for the development of therapies that would inhibit viral entry to prevent the activation of signalling cascades that ultimately leads to further activation of endogenous ACE2 expression.
IVM may achieve this by targeting the interaction between S protein and host ACE2, allowing a conformational change of the S protein and guiding the virus towards an abortive infection due to its inability to infiltrate the host cells [73]. While the exact mechanism behind IVM has yet to be determined, docking studies have demonstrated the ability of IVM to attach to leucine 91 in S protein and histidine 378 of the SARS-CoV-2-ACE2 receptor complex [74]. Other docking complex has also revealed key hydrogen bonding between IVM and Asn487 in the receptor-binding domain (RBD) of the spike (S) protein [75]. In addition, five polar interaction residues (Arg389, Gln484, Gln479, Gly482 and Lys403), and five non-polar interaction residues (Leu441, Tyr491, Tyr481, Tyr439 and Phe483) had took place between IVM and RBD-S protein [75]. Likewise, IVM can also bind with several other residues (LEU492, GLN493, GLY496, TRY505) in S protein via hydrogen bonding. Moreover, there is an interaction between the alkyl group from IVM and aromatic rings of S protein residues (TYR449, TYR489, PHE456, LEU455, PHE490). The LEU455 and GLN493 possesses a high binding affinity with ACE2, showing that IVM binding to these residues will block the attachment of S protein to host ACE2, subsequently obstructing viral entry, and effectively reducing the viral load [52].
5.4. The disruption of 3-chymotrypsin-like enzyme in virus
As described previously, the envelope of SARS-CoV-2 is a ~29.9 kilobases (kb) RNA genome exists containing the ORF-1a and 1b genes to encode NSPs and the structural protein (S, E, M, N) [46]. Upon infection, the SARS-CoV-2 binds to the ACE2 receptor via its S protein, subsequently hijacking the host ribosome and reprogrammes it to translate the viral RNA to large polypeptides [53]. These polypeptides must be auto-cleaved by papain-like proteases (PLpro) and 3-chymotrypsin like protease (3CLpro) to generate the NSP required for viral replication, such as the RdRp enzyme produced by the NSP12 [53]. 3CLpro or Mpro is characterized as a three-domain protein that is highly conserved among coronaviruses, in which mutation of 3CLpro sequences has shown to be fatal for many viruses. Thus, the risk of drug resistance from virus evolution is significantly reduced [76]. Shown here, the 3CLpro shares a high sequence identity of ≥95% between SARS-CoV and SARS-CoV-2, highlighting the potential of 3CLpro as a target for drug repurposing [77].
Recently, the X-ray crystallography revealed that the 3CLpro of SARS-CoV-2 consists of two monomers, which are arranged perpendicularly to form a homodimer via interaction between the N-terminus of one monomer (subunit 1) and the domain II (Glu166) of another monomer (subunit 2) via hydrogen bonding, which led to the formation of the active site [76]. Interestingly, the SARS-CoV adopts the same configuration with only a difference of 12 amino acids [78]. Each monomer is comprised of three domains, namely Domain I (residues 8–101), II (residues 102–184) and III (residues 201–303). Domain I and II are made up of six antiparallel β-barrels whereas domain III is made up of a globular cluster of five α-helices connecting the domain II and III via a long loop region. Notably, there is a catalytic dyad (His41-Cys145) and substrate-binding site (S1′, S1, S2 and S4) in the cleft between domain I and II which corresponds to the amino acid residues; usually numbered as (P4-P3-P2-P1↓P1′-P2′) from the N terminus to C terminus, responsible for the regulation of proteolytic activity of 3CLpro [79]. The His41 acts as a proton acceptor while the Cys145 is primarily targeted by the carbonyl carbon of the substrates. As discussed by Dai et al., the inhibitory activity of drug candidates for 3CLpro confers within its ability to form covalent bonds with Cys145, disrupting dimerization [80], [81], [82]. From which, several key residues (H41, C145, Q192, T190, A191, D187, Q189, M165, S144, D187, M49, G143 and N142) that highly contributed to the binding affinity of 3CLpro inhibitors have been revealed, as H41 and C145 were the most conserved residues among coronaviruses [77]. In particular, perampanel, praziquantel, and nelfinavir have demonstrated better inhibitory activity for 3CLpro in SARS-CoV-2 due to its ability to bind with the abovementioned key residues [7], [77].
In further note, a Michael acceptor-based peptidomimetic inhibitor, N3 has shown great inhibitory activity against SARS-CoV-2 3CLpro over some competitive drug such as lopinavir, and ritonavir which generally requires a high concentration for a significant effect [83], [84], [85]. The N3 functions via covalent binding between the vinyl group of N3 and the aforementioned C145 of the catalytic dyad in 3CLpro. Moreover, N3 possesses several features (lactam ring, aliphatic isobutyl group and methyl group) which corresponds to its side chain (P1, P2 and P4), allowing it to fit tightly to the subsites of S1, S2 and S4 via multiple hydrogen bonds, to effectively hinder the activity of 3CLpro [79], [86]. Similar to the N3 inhibitor molecule, an in-silico study showed that IVM exhibits an inhibitory effect (>85%) against the 3CLpro in SARS-CoV-2 at 50 μM concentration, positioning it as another promising antiviral candidate [53]. It was later found that the carbonyl group in IVM also forms hydrogen bonds with the active site residues (Cys145 and His41) of 3CLpro monomer, destabilising the complex, leading to the loss of function and subsequently reducing its protease activity [53], [87], [88]. Besides that, IVM was also found to have interact with 3CLpro via ten polar residues (Arg188, Asp187, Asn142, His41, His164, Ser46, Glu166, Gly170, Gln189, and Thr169), and seven non-polar residues (Cys145, Leu27, Leu50, Leu167, Met49, Met165, and Pro168) of subunit 1 [75]. The interaction between the two polar residues (Gln306 and Ser1), and two non-polar residues (Phe305 and Val303) of subunit 2 were discovered as well. The stable hydrogen bond interactions were also illustrated by the docking simulation, in which IVM complexes with the subunit 2 of 3CLpro at Thr304, and glu166 residues, forming the greatest binding energy (T). Lastly, the inhibition of the 3CLpro complex has been proven a success in HIV-1 and SARS-CoV previously, making 3CLpro a viable target for IVM [89].
6. Clinical efficacies of IVM
Numerous clinical studies are underway to study the efficacy of IVM. Ivermectin has illustrated great potency towards asymptomatic SARS-CoV-2-positive subjects in a randomized trial in Lebanon [90]. In that study, the IVM group (n = 50) has an increased cycle-threshold (Ct) value from 15.13 to 30.14 whereas the control group (n = 50) has Ct values from 14.20 to 18.96 at 72 h [90]. A higher Ct-value denotes for an insignificant viral remnant or non-viable virus, from which the values of 30 and above are to be considered as negative [91]. Additionally, the subjects from the IVM group developed fewer symptoms compared to the non-IVM group from the reported incidence of fever (2% vs 22%), ageusia (6% vs 24%), anosmia (6% vs 32%), and myalgia (0% vs 18%) [90].
Other notable studies on the positive effects of IVM on COVID-19 patients are as follows. In a study conducted by Elgazzar et al., a randomized 200 healthcare and household contacts with COVID-19 patients consisting of two groups of 100 patients were each given a high dose of 0.4 mg/kg IVM on day 1 followed by a subsequent dose on day 7, or without treatment. The study had reported a significant reduction in PCR contacts testing at 2% on IVM compared to 10% on the non-treatment group [92], [93]. Besides that, Elgazzar et al. had also shown a remarkable outcome of IVM in a randomized controlled trial among 400 hospitalized patients. Their study consisted of four groups with a sample size of 100 patients each. Group 1 was treated with a single IVM dose of 0.4 mg/kg + standard-of-care (SOC); Group 2 was treated with 400 mg of hydroxychloroquine twice during the 1st day followed by a twice daily dose of 200 mg for the 5 consecutive days + SOC; Group 3 and Group 4 obeyed the same treatment plan from Group 1, and Group 2 respectively, with the specific exception of severely ill patients [92]. The outcomes were statistically significant as a lower rate of progression in the Ivermectin Group 1 and 3 vs hydroxychloroquine Group 2 and 4 (1% and 4% vs 22% and 30%), respectively [92]. These results were accompanied by 0 death count and 2% mortality in IVM Group 1 and 3 respectively, followed by 4 death count and 20% mortality for the Group 2 and 4 hydroxychloroquine settings, respectively [92].
Apart from that, IVM also showed remarkable protection against healthcare workers through a prophylactic mechanism. In a randomized trial conducted by Carvallo et al., 131 subjects were administered orally with 0.2 mg of ivermectin drops 5 times per day, and 98 subjects served as the control group. Interestingly, none of those whom received the ivermectin tested positive for SARS-COV-2 compared to the 11.2% positive cases in the control group over the span of 28 days [94]. In addition, Carvallo's follow-up study had also recorded no infections among the 788 healthcare workers who were prescribed 12 mg of Ivermectin weekly compared to the 58% positive cases among the 407 workers in the control group who did not receive IVM treatment [94].
7. Contradictions
Despite numerous positive outcomes for IVM in SARS-CoV-2 treatment, there are several contradictions. For instance, a study has reviewed that the standard dosage of single-dose IVM (200μg/kg) showed no significant clinical and microbiological outcomes compared to the patients that did not receive any IVM [95]. Albeit there was no significant difference, a smaller proportion of the patients from the IVM group required intensive care as compared to the non-IVM group (69% vs 38%), postulating the need for a higher effective dose [95]. Another study pointed no differences in the viral load, outcomes of adverse events, and the laboratory parameters between the IVM treated group (30 participants) and control group (15 participants) from the baseline until day 5 [96]. However, there were positive correlations where the higher IVM plasma concentrations resulted in greater reduction of viral load in nasopharyngeal secretions and viral decay rate, also suggesting the need for a higher dose [96]. A double-blind randomized trial conducted in Cali, Columbia has also shown similar results where the duration of symptoms and time to resolution in 400 patients (200 with IVM, and 200 controls) showed no statistical difference over a 5 day observation [97]. Besides that, another study suggests larger dose (150–200 μg/kg) of IVM may trigger several side effects ranging across symptoms such as rashes, headaches, nausea, ataxia, sweating, tremors, and more severe tachycardia, coma, and death. Thus, implying the need for more controlled trials and safety and efficacies studies for the mass public [98]. Another report showed that IVM alone has a low probability of success in treating COVID-19 as the approved dose or dose of 10× higher than the approved is unlikely to reach the concentration needed for 50% inhibition (IC50) in the lungs after single-dose oral administration, making it less ideal for COVID-19 treatment [99].
8. Conclusion
The rapid emergence of SARS-CoV-2 has put drug repositioning in a critically important position. IVM, an FDA approved antiparasitic drug for onchocerciasis and strongyloidiasis, now sees great opportunity as an interim antiviral drug for SARS-CoV-2. IVM has already been proven effective in numerous in-vitro viral studies such as HIV-1, Flaviviruses, Influenza A virus and many more. The main mode of antiviral action for IVM revolves around the disruption of the Importin heterodimer complex (IMPα/β1), which is a vital complex that shuttles the viral protein (NS12) into the nucleus from the cytoplasm via the NPC upon receiving NLS. In addition, IVM sees a huge potential in inhibiting cytokine storm in Covid-19 patients. This is due to IVM downregulating STAT3, a key protein in the JAK-STAT signalling pathway that contributes to enhanced inflammatory IL-6 production in infected patients. Furthermore, IVM can bind to several residues in the S protein of SARS-CoV-2, concomitantly hindering the attachment of S protein to host ACE-2 receptors for viral entry, effectively reducing the viral load. Finally, IVM binds on the active sites of 3CLpro, a vital protease, for the production of functional NSPs for viral replication. Owing to its multifarious antiviral machinery described above, IVM could be warranted as an interim solution to delay the rapid progression of SARS-CoV-2 in infected patients while significantly reducing the severity of disease by bringing down the viral load and downregulating the post-Covid-19 cytokine storm in recovering patients. There is an urgent demand for more relevant and comprehensive clinical data to accurately define the pharmacokinetic profile of IVM thereby paving the path for its use against Covid-19.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Worldometers Coronavirus Update (Live) COVID-19 Virus Pandemic. https://www.worldometers.info/coronavirus/
- 2.Andersen K., Rambaut A., Lipkin W., Holmes E., Garry R. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehbe Z., Hammoud S., Soudani N., Zaraket H., El-Yazbi A., Eid A. Molecular insights into SARS COV-2 interaction with cardiovascular disease: role of RAAS and MAPK signaling. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Low Z., Farouk I., Lal S. Drug repositioning: new approaches and future prospects for life-debilitating diseases and the COVID-19 pandemic outbreak. Viruses. 2020;12:1058. doi: 10.3390/v12091058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma A., Ahmad Farouk I., Lal S. COVID-19: a review on the novel coronavirus disease evolution, transmission, detectionControl and Prevention. 2021;13:202. doi: 10.3390/v13020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bello M. Prediction of potential inhibitors of the dimeric SARS-CoV2 main proteinase through the MM/GBSA approach. J. Mol. Graph. Model. 2020;101 doi: 10.1016/j.jmgm.2020.107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bello M., Martínez-Muñoz A., Balbuena-Rebolledo I. Identification of saquinavir as a potent inhibitor of dimeric SARS-CoV2 main protease through MM/GBSA. J. Mol. Model. 2020;26 doi: 10.1007/s00894-020-04600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanella I., Zizioli D., Castelli F., Quiros-Roldan E. Tenofovir, another inexpensive, well-known and widely available old drug repurposed for SARS-COV-2 infection. Pharmaceuticals. 2021;14:454. doi: 10.3390/ph14050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeon S., Ko M., Lee J., Choi I., Byun S., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laing R., Gillan V., Devaney E. Ivermectin – old drug, new Tricks? Trends Parasitol. 2017;33:463–472. doi: 10.1016/j.pt.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Õmura S. Ivermectin: 25 years and still going strong. Int. J. Antimicrob. Agents. 2008;31:91–98. doi: 10.1016/j.ijantimicag.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 12.Campbell W., Fisher M., Stapley E., Albers-Schonberg G., Jacob T. Ivermectin: a potent new antiparasitic agent. Science. 1983;221:823–828. doi: 10.1126/science.6308762. [DOI] [PubMed] [Google Scholar]
- 13.Õmura S., Crump A. The life and times of ivermectin — a success story. Nat. Rev. Microbiol. 2004;2:984–989. doi: 10.1038/nrmicro1048. [DOI] [PubMed] [Google Scholar]
- 14.Ashour D. Ivermectin: from theory to clinical application. Int. J. Antimicrob. Agents. 2019;54:134–142. doi: 10.1016/j.ijantimicag.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Burnham G., Mebrahtu T. Review: the delivery of ivermectin (MectizanR) Trop. Med. Int. Health. 2004;9:A26–A44. doi: 10.1111/j.1365-3156.2004.01211.x. [DOI] [PubMed] [Google Scholar]
- 16.Muñoz-Muñoz L., Shoen C., Sweet G., Vitoria A., Bull T., Cynamon M., Thompson C., Ramón-García S. Repurposing avermectins and milbemycins against mycobacteroides abscessus and other nontuberculous mycobacteria. Antibiotics. 2021;10:381. doi: 10.3390/antibiotics10040381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang M., Hu X., Wang Y., Yao X., Zhang W., Yu C., Cheng F., Li J., Fang Q. Ivermectin, a potential anticancer drug derived from an antiparasitic drug. Pharmacol. Res. 2021;163 doi: 10.1016/j.phrs.2020.105207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin J., Park G., Lee J., Choi E., Park J., Kim T., Park N., Jin X., Jung J., Shin D., Hong J., Kim H., Yoo H., Lee S., Kim Y., Park J., Kim J. DEAD-box RNA helicase DDX23 modulates glioma malignancy via elevating miR-21 biogenesis. Brain. 2015;138:2553–2570. doi: 10.1093/brain/awv167. [DOI] [PubMed] [Google Scholar]
- 19.Jin L., Wang R., Zhu Y., Zheng W., Han Y., Guo F., Ye F., Li Y. Selective targeting of nuclear receptor FXR by avermectin analogues with therapeutic effects on nonalcoholic fatty liver disease. Sci. Rep. 2015;5 doi: 10.1038/srep17288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mastrangelo E., Pezzullo M., De Burghgraeve T., Kaptein S., Pastorino B., Dallmeier K., de Lamballerie X., Neyts J., Hanson A., Frick D., Bolognesi M., Milani M. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J. Antimicrob. Chemother. 2012;67:1884–1894. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagstaff K., Sivakumaran H., Heaton S., Harrich D., Jans D. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caly L., Druce J., Catton M., Jans D., Wagstaff K. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wehbe Z., Wehbe M., Iratni R., Pintus G., Zaraket H., Yassine H., Eid A. Repurposing ivermectin for COVID-19: molecular aspects and therapeutic possibilities. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.663586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell W. An introduction to the avermectins. N. Z. Vet. J. 1981;29:174–178. doi: 10.1080/00480169.1981.34836. [DOI] [PubMed] [Google Scholar]
- 25.Kaur H., Shekhar N., Sharma S., Sarma P., Prakash A., Medhi B. Ivermectin as a potential drug for treatment of COVID-19: an in-sync review with clinical and computational attributes. Pharmacol. Rep. 2021;73:736–749. doi: 10.1007/s43440-020-00195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jans D., Wagstaff K. Ivermectin as a broad-Spectrum host-directed antiviral: the real deal? Cells. 2020;9:2100. doi: 10.3390/cells9092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaccour C., Hammann F., Ramón-García S., Rabinovich N. Ivermectin and COVID-19: keeping rigor in times of urgency. Am. J. Trop. Med. Hyg. 2020;102:1156–1157. doi: 10.4269/ajtmh.20-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaddoura M., AlIbrahim M., Hijazi G., Soudani N., Audi A., Alkalamouni H., Haddad S., Eid A., Zaraket H. COVID-19 therapeutic options under investigation. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frieman M., Yount B., Heise M., Kopecky-Bromberg S., Palese P., Baric R. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/golgimembrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wulan W., Heydet D., Walker E., Gahan M., Ghildyal R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrows N., Campos R., Powell S., Prasanth K., Schott-Lerner G., Soto-Acosta R., Galarza-Muñoz G., McGrath E., Urrabaz-Garza R., Gao J., Wu P., Menon R., Saade G., Fernandez-Salas I., Rossi S., Vasilakis N., Routh A., Bradrick S., Garcia-Blanco M. A screen of FDA-approved drugs for inhibitors of zika virus infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen L., Jamieson D., Powers A., Honein M. Zika virus. N. Engl. J. Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 33.Zika Virus. https://www.who.int/news-room/fact-sheets/detail/zika-virus
- 34.Ji W., Luo G. Zika virus NS5 nuclear accumulation is protective of protein degradation and is required for viral RNA replication. Virology. 2020;541:124–135. doi: 10.1016/j.virol.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Colpitts T., Conway M., Montgomery R., Fikrig E. West Nile virus: biology, transmission, and human infection. Clin. Microbiol. Rev. 2012;25:635–648. doi: 10.1128/CMR.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mastrangelo E., Pezzullo M., De Burghgraeve T., Kaptein S., Pastorino B., Dallmeier K., de Lamballerie X., Neyts J., Hanson A., Frick D., Bolognesi M., Milani M. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J. Antimicrob. Chemother. 2012;67:1884–1894. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salomon R., Webster R. The influenza virus enigma. Cell. 2009;136:402–410. doi: 10.1016/j.cell.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Götz V., Magar L., Dornfeld D., Giese S., Pohlmann A., Höper D., Kong B., Jans D., Beer M., Haller O., Schwemmle M. Influenza a viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep. 2016;6 doi: 10.1038/srep23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao H., Killip M., Staeheli P., Randall R., Jackson D. The human interferon-induced MxA protein inhibits early stages of influenza a virus infection by retaining the incoming viral genome in the cytoplasm. J. Virol. 2013;87:13053–13058. doi: 10.1128/JVI.02220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmermann P., Mänz B., Haller O., Schwemmle M., Kochs G. The viral nucleoprotein determines mx sensitivity of influenza a viruses. J. Virol. 2011;85:8133–8140. doi: 10.1128/JVI.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yip T., Selim A., Lian I., Lee S. Advancements in host-based interventions for influenza treatment. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith J., Daniel R. Following the path of the virus: the exploitation of host DNA repair mechanisms by retroviruses. ACS Chem. Biol. 2006;1:217–226. doi: 10.1021/cb600131q. [DOI] [PubMed] [Google Scholar]
- 43.Yang S., Atkinson S., Wang C., Lee A., Bogoyevitch M., Borg N., Jans D. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antivir. Res. 2020;177 doi: 10.1016/j.antiviral.2020.104760. [DOI] [PubMed] [Google Scholar]
- 44.Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W., Si H., Zhu Y., Li B., Huang C., Chen H., Chen J., Luo Y., Guo H., Jiang R., Liu M., Chen Y., Shen X., Wang X., Zheng X., Zhao K., Chen Q., Deng F., Liu L., Yan B., Zhan F., Wang Y., Xiao G., Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shereen M., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: emergence, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naqvi A., Fatima K., Mohammad T., Fatima U., Singh I., Singh A., Atif S., Hariprasad G., Hasan G., Hassan M. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fehr A., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses. 2015:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuman B., Adair B., Yoshioka C., Quispe J., Orca G., Kuhn P., Milligan R., Yeager M., Buchmeier M. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sen Gupta P., Biswal S., Panda S., Ray A., Rana M. Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1839564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frieman M., Yount B., Heise M., Kopecky-Bromberg S., Palese P., Baric R. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic Reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian L., Qiang T., Liang C., Ren X., Jia M., Zhang J., Li J., Wan M., YuWen X., Li H., Cao W., Liu H. RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021;213 doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saha J.K., Raihan M.J. The binding mechanism of ivermectin and levosalbutamol with spike protein of SARS-CoV-2. Struct. Chem. 2021:1–8. doi: 10.1007/s11224-021-01776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mody V., Ho J., Wills S., Mawri A., Lawson L., Ebert M., Fortin G., Rayalam S., Taval S. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun. Biol. 2021;4 doi: 10.1038/s42003-020-01577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jans D., Martin A., Wagstaff K. Inhibitors of nuclear transport. Curr. Opin. Cell Biol. 2019;58:50–60. doi: 10.1016/j.ceb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 55.Caly L., Wagstaff K., Jans D. Nuclear trafficking of proteins from RNA viruses: potential target for antivirals? Antivir. Res. 2012;95:202–206. doi: 10.1016/j.antiviral.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Yang S., Atkinson S., Fraser J., Wang C., Maher B., Roman N., Forwood J., Wagstaff K., Borg N., Jans D. Novel flavivirus antiviral that targets the host nuclear transport importin α/β1 heterodimer. Cells. 2019;8:281. doi: 10.3390/cells8030281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waheed Shouman . Prophylactic Ivermectin in COVID-19 Contacts - Study Results - ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/results/NCT04422561 (Accessed Jun 10, 2021).
- 58.Hariyanto T., Halim D., Rosalind J., Gunawan C., Kurniawan A. Ivermectin and outcomes from Covid-19 pneumonia: a systematic review and meta-analysis of randomized clinical trial studies. Rev. Med. Virol. 2021 doi: 10.1002/rmv.2265. [DOI] [Google Scholar]
- 59.Swargiary A. 2020. Ivermectin as a promising RNA-dependent RNA polymerase inhibitor and a therapeutic drug against SARS-CoV2: Evidence from in silico studies. [Google Scholar]
- 60.Matsuyama T., Kubli S., Yoshinaga S., Pfeffer K., Mak T. An aberrant STAT pathway is central to COVID-19. Cell Death Differ. 2020;27:3209–3225. doi: 10.1038/s41418-020-00633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park A., Iwasaki A. Type I and type III interferons – induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang C., Sun M., Yuan X., Ji L., Jin Y., Cardona C., Xing Z. Enterovirus 71 suppresses interferon responses by blocking Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling through inducing karyopherin-α1 degradation. J. Biol. Chem. 2017;292:10262–10274. doi: 10.1074/jbc.M116.745729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H., Yuan M., Wang S., Zhang L., Zhang R., Zou X., Wang X., Chen D., Wu Z. STAT3 regulates the type I IFN-mediated antiviral response by interfering with the nuclear entry of STAT1. Int. J. Mol. Sci. 2019;20:4870. doi: 10.3390/ijms20194870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finigan J., Downey G., Kern J. Human epidermal growth factor receptor signaling in acute lung injury. Am. J. Respir. Cell Mol. Biol. 2012;47:395–404. doi: 10.1165/rcmb.2012-0100TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta K., Xu Z., Castellino F., Ploplis V. Plasminogen activator inhibitor-1 stimulates macrophage activation through toll-like Receptor-4. Biochem. Biophys. Res. Commun. 2016;477:503–508. doi: 10.1016/j.bbrc.2016.06.065. [DOI] [PubMed] [Google Scholar]
- 66.Aluwihare P., Munger J. What the lung has taught us about latent TGF-β activation. Am. J. Respir. Cell Mol. Biol. 2008;39:499–502. doi: 10.1165/rcmb.2008-0003ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han S., Mallampalli R. The acute respiratory distress syndrome: from mechanism to translation. J. Immunol. 2015;194:855–860. doi: 10.4049/jimmunol.1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dou Q., Chen H., Wang K., Yuan K., Lei Y., Li K., Lan J., Chen Y., Huang Z., Xie N., Zhang L., Xiang R., Nice E., Wei Y., Huang C. Ivermectin induces cytostatic autophagy by blocking the PAK1/Akt Axis in breast cancer. Cancer Res. 2016;76:4457–4469. doi: 10.1158/0008-5472.CAN-15-2887. [DOI] [PubMed] [Google Scholar]
- 69.Choi H., Moon S., Yang H., Kim K. Understanding viral infection mechanisms and patient symptoms for the development of COVID-19 therapeutics. Int. J. Mol. Sci. 2021;22:1737. doi: 10.3390/ijms22041737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yi C., Sun X., Ye J., Ding L., Liu M., Yang Z., Lu X., Zhang Y., Ma L., Gu W., Qu A., Xu J., Shi Z., Ling Z., Sun B. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020;17:621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo J., Lu S., Yu M., Zhu L., Zhu C., Li C., Fang J., Zhu X., Wang X. The potential involvement of JAK-STAT signalling pathway in the COVID-19 infection assisted by ACE2. Gene. 2021;768 doi: 10.1016/j.gene.2020.145325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hennighausen L., Lee H. 2020. Activation of the SARS-CoV-2 receptorAce2by cytokines through pan JAK-STAT enhancers. [Google Scholar]
- 73.Choudhury A., Das N., Patra R., Bhattacharya M., Ghosh P., Patra B., Mukherjee S. Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in-silico approach. Futur. Virol. 2021;16:277–291. [Google Scholar]
- 74.Lehrer S., Rheinstein P. Ivermectin docks to the SARS-CoV-2 spike receptor-binding domain attached to ACE2. In Vivo. 2020;34:3023–3026. doi: 10.21873/invivo.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bello M. Elucidation of the inhibitory activity of ivermectin with host nuclear importin α and several SARS-CoV-2 targets. J. Biomol. Struct. Dyn. 2021:1–9. doi: 10.1080/07391102.2021.1911857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silvestrini L., Belhaj N., Comez L., Gerelli Y., Lauria A., Libera V., Mariani P., Marzullo P., Ortore M., Palumbo Piccionello A., Petrillo C., Savini L., Paciaroni A., Spinozzi F. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-88630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bello M. Prediction of potential inhibitors of the dimeric SARS-CoV2 main proteinase through the MM/GBSA approach. J. Mol. Graph. Model. 2020;101 doi: 10.1016/j.jmgm.2020.107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 80.Kneller D., Phillips G., Kovalevsky A., Coates L. Room-temperature neutron and X-ray data collection of 3CL mprofrom SARS-CoV-2. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2020;76:483–487. doi: 10.1107/S2053230X20011814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dai W., Zhang B., Jiang X., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nukoolkarn V., Lee V., Malaisree M., Aruksakulwong O., Hannongbua S. Molecular dynamic simulations analysis of ritronavir and lopinavir as SARS-CoV 3CLpro inhibitors. J. Theor. Biol. 2008;254:861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choy K., Wong A., Kaewpreedee P., Sia S., Chen D., Hui K., Chu D., Chan M., Cheung P., Huang X., Peiris M., Yen H. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F., Horby P., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cui W., Yang K., Yang H. Recent Progress in the drug development targeting SARS-CoV-2 Main protease as treatment for COVID-19. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.616341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anand K. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 88.Nukoolkarn V., Lee V., Malaisree M., Aruksakulwong O., Hannongbua S. Molecular dynamic simulations analysis of ritronavir and lopinavir as SARS-CoV 3CLpro inhibitors. J. Theor. Biol. 2008;254:861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jamalipour Soufi G., Iravani S. Potential inhibitors of SARS-CoV-2: recent advances. J. Drug Target. 2020;29:349–364. doi: 10.1080/1061186X.2020.1853736. [DOI] [PubMed] [Google Scholar]
- 90.Samaha A., Mouawia H., Fawaz M., Hassan H., Salami A., Bazzal A., Saab H., Al-Wakeel M., Alsaabi A., Chouman M., Moussawi M., Ayoub H., Raad A., Hajjeh O., Eid A., Raad H. Effects of a single dose of ivermectin on viral and clinical outcomes in asymptomatic SARS-CoV-2 infected subjects: a pilot clinical trial in Lebanon. Viruses. 2021;13:989. doi: 10.3390/v13060989. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Kampf G., Lemmen S., Suchomel M. Ct values and infectivity of SARS-CoV-2 on surfaces. Lancet Infect. Dis. 2021;21 doi: 10.1016/S1473-3099(20)30883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kory P., Meduri G., Varon J., Iglesias J., Marik P. Review of the emerging evidence demonstrating the efficacy of ivermectin in the prophylaxis and treatment of COVID-19. Am. J. Ther. 2021;28:e299–e318. doi: 10.1097/MJT.0000000000001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elgazzar A., Eltaweel A., Youssef S., Hany B., Hafez M., Moussa H. 2020. Efficacy and Safety of Ivermectin for Treatment and Prophylaxis of COVID-19 Pandemic. [Google Scholar]
- 94.Héctor C., Roberto H., Psaltis A., Veronica C. Study of the efficacy and safety of topical ivermectin + iota-carrageenan in the prophylaxis against COVID-19 in health personnel. J. Biomed. Res. Clin. Investig. 2020;2 [Google Scholar]
- 95.Camprubí D., Almuedo-Riera A., Martí-Soler H., Soriano A., Hurtado J., Subirà C., Grau-Pujol B., Krolewiecki A., Muñoz J. Lack of efficacy of standard doses of ivermectin in severe COVID-19 patients. PLOS ONE. 2020;15 doi: 10.1371/journal.pone.0242184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krolewiecki A., Lifschitz A., Moragas M., Travacio M., Valentini R., Alonso D., Solari R., Tinelli M., Cimino R., Álvarez L., Fleitas P., Ceballos L., Golemba M., Fernández F., Fernández de Oliveira D., Astudillo G., Baeck I., Farina J., Cardama G., Mangano A., Spitzer E., Gold S., Lanusse C. Antiviral effect of high-dose ivermectin in adults with COVID-19: a proof-of-concept randomized trial. EClinicalMedicine. 2021;37 doi: 10.1016/j.eclinm.2021.100959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.López-Medina E., López P., Hurtado I., Dávalos D., Ramirez O., Martínez E., Díazgranados J., Oñate J., Chavarriaga H., Herrera S., Parra B., Libreros G., Jaramillo R., Avendaño A., Toro D., Torres M., Lesmes M., Rios C., Caicedo I. Effect of ivermectin on time to resolution of symptoms among adults with mild COVID-19. JAMA. 2021;325:1426. doi: 10.1001/jama.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramírez C., Herrera-Paz E., Peralta G., Rodríguez G., Durón R. Is ivermectin ready to be part of a public health policy for COVID-19 prophylaxis? EClinicalMedicine. 2021;32 doi: 10.1016/j.eclinm.2021.100744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmith V., Zhou J., Lohmer L. The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin. Pharmacol. Ther. 2020;108:762–765. doi: 10.1002/cpt.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]