Abstract
Nanoscale electrophoresis allows for unique separations of single molecules, such as DNA/RNA nucleobases, and thus has the potential to be used as single molecular sensors for exonuclease sequencing. For this to be envisioned, label-free detection of the nucleotides to determine their electrophoretic mobility (i.e., time-of-flight, TOF) for highly accurate identification must be realized. Here, we for the first time show a novel nanosensor that allows discriminating four 2-deoxyribonucleoside 5’-monophosphates, dNMPs, molecules in a label free manner by nanoscale electrophoresis. This was made possible by positioning two sub-10 nm in-plane pores at both ends of a nanochannel column used for nanoscale electrophoresis and measuring the longitudinal transient current during translocation of the molecules. The dual nanopore TOF sensor with 0.5, 1, and 5 μm long nanochannel column lengths discriminated different dNMPs with a mean accuracy of 55, 66, and 94%, respectively. This nanosensor format could broadly be applicable to label-free detection and discrimination of other single molecules, vesicles, and particles by changing the dimensions of the nanochannel column and in-plane nanopores and integrating different pre- and post-processing units to the nanosensor. This is simple to accomplish because the nanosensor is contained within a fluidic network made in a plastic via replication.
Keywords: nanoscale electrophoresis, polymer nanofluidic biosensors, in-plane nanopore sensing, molecular time-of-flight, identification of mononucleotides, nanoimprint lithography
Graphical Abstract
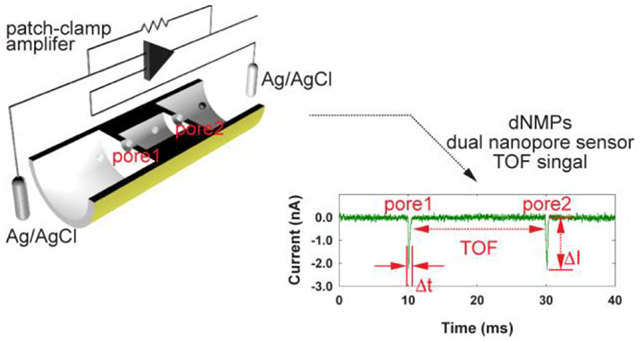
A dual-nanopore time-of-flight (TOF) sensor, which consists of two in-plane nanopores formed at both ends of a nanochannel column, is designed and fabricated to determine the mobility of single molecules in nanoscale electrophoresis in a label-free manner. This novel nanosensor with a 5 μm long nanochannel column demonstrates discrimination of four dNMPs molecules with a mean identification accuracy of 94%.
INTRODUCTION
Single-molecule sequencing has attracted significant interest in the area of diagnostics as their development paradigm moves towards precision medicine that requires production and handling of a large amount of genomic data from small amounts of input material.[1, 2] While the sequencing-by-synthesis technology dominates currently, this technology requires significant sample preparation and amplification steps, the use of costly biological reagents such as fluorescently labeled molecules, and expensive imaging and data-handling instruments.[3-7] Single-molecule sequencing, which does not require amplification and labeling steps, would simplify the entire sequencing process significantly reducing costs and time of acquiring sequencing data, and is thus more adaptable to clinical translation to enable precision medicine even from mass-limited samples such as those provided by liquid biopsies.[8]
Nanopores offer a fast and low-cost sequencing platform that does not require labeling and sample amplification.[6, 7, 9-13] The basic principle of nanopore sequencing is to electrically drive charged single molecules, either an intact DNA molecule in strand sequencing or individual nucleotides cleaved from the DNA in exonuclease sequencing,[10, 13-15] through a nanopore and determine the identity of each constituent nucleotide by monitoring small changes in the ionic current flowing through the pore while individual nucleotides temporarily reside within the pore (i.e., resistive pulse sensing, RPS).[13, 16] Strand nanopore sequencing has demonstrated whole-genome sequencing[14] with long read lengths to 134 kb.[15] Compared to strand nanopore sequencing,[9, 12-14, 17, 18] exonuclease sequencing, a sequencing-by-subtraction technology, has far less been explored despite its potential of long reads and the fact that only a single nucleotide occupies the pore at any time that can improve read accuracy. A major huddle is that diffusion of cleaved nucleotides makes many bases escape prior to being shuttled into the detection element of the nanopore.[13, 17, 19] An increase in driving voltage may reduce diffusional artifacts, but at the same time it lowers the accuracy of discriminating molecules due to fast translocation, thus limiting the use of nanopore sensors for exonuclease sequencing.[19] This limitation is associated with the nature of current nanopore sensing, where both sensing and discriminating of nucleotides are done simultaneously using current transient amplitudes. A nanosensor that decouples sensing and discriminating functions with no unfavorable effects arising from increasing driving voltage would assist in using nanopore sensors for exonuclease sequencing.
In the present work, we demonstrate a novel nanosensor format, which combines nanoscale electrophoresis with nanopore sensing that can identify single nucleotides in a label free manner with high accuracy. Nanoscale electrophoresis was used as the discrimination mechanism of different nucleotides while nanopore sensing was used to detect the passage time of a single molecule between both ends of a nanochannel column to determine the electrophoretic-dependent time-of-flight (TOF). Recently, near baseline separation of four fluorescently-labeled deoxynucleotide monophosphates (dNMPs) has been demonstrated using nanoscale electrophoresis in thermoplastic nanochannel columns built in poly(methyl methacrylate), PMMA.[20] Herein we report an identification accuracy of four dNMPs of 94% with the nanosensor using a 5 μm nanochannel column length and this identification accuracy was found to be nanochannel column length dependent. Our nanosensor, when combined with exonuclease clipping of DNA/RNA biopolymers prior to nanosensor readout,[21] will allow realization of an exonuclease sequencing platform with high single read accuracy.
RESULTS AND DISCUSSION
Design and fabrication of dual-nanopore TOF sensor.
The dual-nanopore TOF sensor consists of a pair of in-plane nanopores poised on either end of a nanochannel used for the electrophoresis, which was made in a polymer substrate in a single step via nanoimprint lithography, NIL (Figure 1). When a molecule was electrokinetically driven through the first in-plane nanopore, the nanochannel column, and the second in-plane nanopore sequentially, two consecutive current transient signals were produced from which the molecular TOF was read in a label-free manner (Figure 1a). There are two important design requirements for the nanosensor. First, the nanochannel width and depth should be less than 100 nm to allow for molecular-dependent transport time determined by nanoscale electrophoresis, which uses a combination of electrophoretic and chromatographic effects for the dNMPs.[20] A series of nanopores have been used to improve detection capabilities of RPS and to measure the electrophoretic mobilities of virus capsids[22-26] and DNAs.[27, 28] While the use of a series of nanopores reduced the standard deviation of the pulse amplitude distribution in determining the sizes of virus capsids[25, 26] and molecular noise and trans-pore linearization in nanopore sensing of DNA,[28] the electrophoretic mobilities did not differentiate particles of different sizes.[23, 27] Unique to our nanosensor design is the use of a sub-100 nm column for electrophoresis instead of hundreds of nanometers- or micron-scale columns in the previous examples,[22-27] which provides a significant longitudinal electric field drop between the two in-plane pores to drive electrophoresis (Figure S1 and Figure S2, COMSOL simulation results in Supporting Information). Second, the two in-plane nanopores should be small enough to create current transient signals with sufficient signal-to-noise ratios to detect the TOF of the molecules being interrogated. For detecting the passage of nucleotides, the equivalent diameter of the in-plane nanopores after cover plate bonding should be sub-10 nm to ensure sufficient signal-to-noise ratios, which will be discussed later.
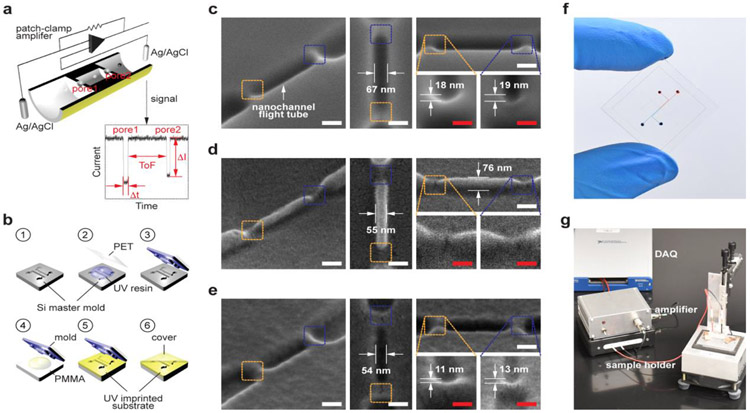
Figure 1.
Dual-nanopore time-of-flight (TOF) sensor. (a) Schematics of the dual-nanopore TOF sensor, which consists of a pair of in-plane nanopores poised on either side of a nanochannel used as the nanochannel column for nanoscale electrophoresis. Upon translocation of a dNMP molecule through the first in-plane nanopore, the nanochannel column, and the second in-plane nanopore sequentially, two consecutive current transient signals are generated. The time interval between the two current pulses corresponds to the TOF of the dNMP molecule through the nanochannel column. (b) Schematics of the device fabrication process: fabrication of Si master mold (1); fabrication of a UV resin mold via replication from the Si mater (2-3); UV-NIL into a PEGDA substrate from the UV resin mold (4-5); and simultaneous thermal bonding with a PEGDA cover plate resulting in pore reduction (6). Top and tilted view SEM images of (c) Si master mold, (d) the UV resin mold, and (e) the UV-imprinted PEGDA substrate. Scale bars, 100 nm in white and 50 nm in red. (f) A photograph of the TOF nanopore sensor fabricated by NIL. (g) TOF measurement system used included a sample holder, a custom current amplifier, and a data acquisition (DAQ) system. The current amplifier consists of a standard topology transimpedance amplifier (TIA) that is battery powered with a gain of 100 nA·V−1 and a single-pole −3dB bandwidth of 10 kHz. The output of the TIA is digitized by a National Instruments, Inc. model NI PXIe-6341 DAQ system at a sampling rate of 250 ksps.
The dual-nanopore TOF sensor was fabricated using a single step NIL process following a procedure (Figure 1b) that has been developed previously.[29] Figure 1c-e shows scanning electron microscopy (SEM) images of the Si master, the cured UV-resin mold, and the UV-imprinted PEGDA substrate, which clearly shows two in-plane pores at both ends of the nanochannel column. Different from the circular pore shape in conventional nanopore membranes, the in-plane nanopores in our sensor are a shallow V-shape attributed to the nature of the FIB milling used for fabricating the Si master. For the imprinted PEGDA devices, the depth of the in-plane nanopores was 12 ± 2 nm (Figure 1e) and the width and depth of the nanochannel column were 54 ± 3 nm and 75 ± 3 nm, respectively.
The final, but critical step in the device fabrication was thermal fusion bonding of a cover plate to the imprinted PEGDA substrate, and this was done at 1 MPa and 70 °C for 15 min to form an enclosed nanofluidic device. The size of the enclosed in-plane nanopores is determined during this step because the bonding process inevitably involves mixing of polymer chains at the interface resulting in a reduction in the pore size. Because it was difficult to determine the exact pore size and shape after the cover plate bonding using microscopy, we estimated the pore size by measuring the conductance through the pores after filling the device with a 1 m KCl containing 1× TE (pH 8.0) and comparing the measured conductance with the simulated conductance (Figure S3, Supporting Information).[30] Only devices showing an equivalent diameter <10 nm were used for dNMP translocation experiments. With the imprinting and bonding parameters determined, the process yield to produce nanopores with diameters less than 10 nm was ~ 90%. However, it is difficult to control the pore size within the sub-10 nm range with the current fabrication protocol of passively controlling imprinting and bonding conditions. Controlling the pore size within the sub-10 nm regime may be possible if the pore size can be controlled actively in a closed loop with the measured transient currents as a control input while applying bonding force. A similar concept has been demonstrated,[31, 32] where the nanochannel size made in PMDS was actively controlled by applied external force.
Label-free detection of dNMPs.
The ability of the dual-nanopore TOF sensor to obtain molecular TOFs was examined by translocating unlabeled dNMPs through the sensor electrokinetically and measuring the longitudinal transient currents through the sensor. All experiments were carried out with a 1 m KCl solution containing 0.5× TBE at room temperature. The dNMP concentration was 0.01 ng·μL−1. Figure 2a shows a typical transient current (I-t) trace obtained for dCMP at pH 10.0 in the nanosensor using a 0.5 μm-long nanochannel column at a driving voltage of 3 V. An open-pore current of 11.3 nA corresponds to an equivalent pore diameter of ~3 nm assuming that the nanopore length is 10 nm as determined by the COMSOL open-pore current simulation. At pH 10.0, the transient current peaks were detected when molecules were driven to the cathode (in the direction of electroosmotic flow, EOF)[33] while the molecular translocation to the anode (in the direction of electrophoresis, EP) was observed at a pH value of 8.0.
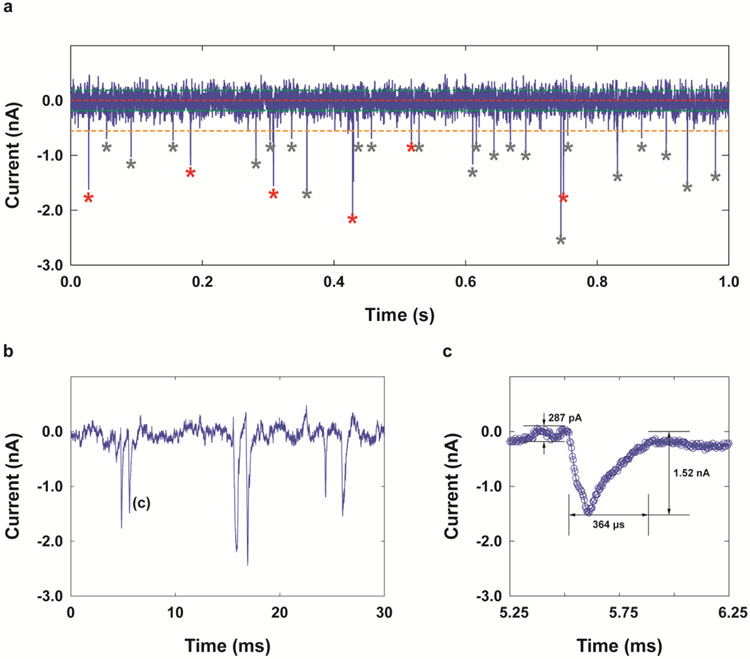
Figure 2.
Determination of TOF from current-time (I-t) traces obtained for dNMPs using a dual-nanopore TOF sensor. (a) An example overview plot of transient current measured when dCMP molecules were introduced into the dual-nanopore TOF sensor with a 0.5 μm-long nanochannel column and sub-10 nm dual-nanopores under a driving voltage of 3 V with a bandwidth of 10 kHz and a digital sampling rate of 250 ksps allowing for a time resolution of 35 μs. The baseline or open current of ~11.3 nA was subtracted from the measured current. Only 1 s of the trace is displayed for clarity; the entire data set spans over 600 s. The red, green, and orange horizontal lines indicate the overall baseline, the RMS noise level, and the detection threshold (e.g. 3 times the RMS noise of the baseline current trace), respectively. The red asterisks indicate paired peaks that were selected using the selection criteria described in the Supporting Information. The grey asterisks show the peaks whose amplitudes met the detection threshold but that did not meet the maximum TOF criterion, and thus were not selected as paired. (b) Example peak pairs. (c) A magnified transient current peak shown in (b). The peak amplitude was in the range of 0.8-2.5 nA. The dwell time at individual pores ranged from 0.18-0.58 ms.
An example peak pair and a magnified transient current peak are shown in Figure 2b and Figure 2c. The peak amplitude and dwell time were in the range of 0.8-2.5 nA and 0.18-0.58 ms, respectively. The peak amplitudes measured were comparable to that obtained by static COMSOL simulations with/without the presence of a 1 nm particle (Figure S3, Supporting Information). The detection of mononucleotides with 3-10 nm pores is unprecedented, which is partly due to the in-plane design with sequential nanostructures associated with our nanosensor design. To date, the detection and differentiation of dNMPs has only been reported via transient current amplitudes with biological protein nanopores[13, 17, 34] and synthetic nanopores with a diameter of ~2.3 nm formed in a monolayer MoS2 membrane[35] and a 1.8 nm pore formed in a Si3N4 membrane.[36] The transient current signals of the dNMPs seen for our in-plane nanopores are attributed to the combined effects of ion concentration gradients present at both sides of the in-plane nanopores[37] and the compartmental limitation due to ionic flow induced by the presence of the nanochannel column and inlet/exit nanochannels.[38] Lee et al. showed that a 500 nm long guide nanochannel with a diameter of 150 nm built prior to a 5 nm Si3N4 pore resulted in a 2.5-fold increase in the peak amplitudes and a 20% reduction in the translocation velocities for detection of DNA compared to those without the guide nanochannel.[38] The conical shape of the in-plane nanopores also contributes to the enhanced signal.[39]
The dwell time of individual dNMPs through in-plane nanopores was significantly longer compared to the speed of DNA translocation of ~1 base/μs through nanopore membranes without an adaptor modification.[40] The slow translocation time was also associated with the presence of multiple nanostructures around the in-plane nanopores.[38] The COMSOL simulation results from Lee et al. showed that the slow translocation of DNA is associated with a strong EOF drag in the guide nanochannel, where the maximum difference would be in the order of 100.[38] Thus, the measurement of small currents with our nanosensor does not seem to require high bandwidth in the MHz range even for relatively high applied voltages. Also, a sampling rate of 250 kHz used in our experiments was sufficient to detect the passage of molecules using our dual-nanopore TOF sensor platform without signal aliasing.
Among the peaks that were observed in Figure 2a, most were paired, as marked by red asterisks. The first and second peaks of each pair were assigned to the passage of a dCMP molecule through the first and second in-plane pores, respectively, and the time interval between the two peaks corresponded to the TOF of the molecule through the nanochannel column. We developed a selection criteria to determine peak pairs,[27] the details of which are provided in the Supporting Information (Figure S4). In previous work by Langecker et al.[27] where double-stranded (ds)-DNA was detected by stacked nanopores with diameters of 23 and 28 nm, 94% of detected peaks could unambiguously be assigned to the translocating dsDNA. The reasons for unpaired peaks found in this work are mainly two folds; first, one of the two translocation events from a single molecule is undetectable (missing peaks) and, second, more than one molecule could be located in the flight column (molecule overlapping). The former case (missing peaks) may be attributed to the relatively large size (3-10 nm in equivalent diameter)[41] and the shallow V-shape of the in-plane nanopores,[42] so that molecules translocate at different locations within the nanopore leading to variations in transient current signals giving rise to missing peaks. Another potential reason for missing peaks would be fast translocation of molecules, which cannot be detected with the bandwidth (10 kHz) of the current amplifier used for our experiments. Higher bandwidth current amplifiers would enable overcoming this limitation.[43, 44] On the other hand, the latter case (molecule overlapping) can be reduced simply by reducing the dNMP concentration. More importantly, molecule overlapping may be avoided in real exonuclease sequencing process with our dual-nanopore TOF sensor by using enzymes whose clipping rates are lower than TOFs of the clipped mononucleotides in the flight column. For example, the clipping rates of dNMPs and rNMPs digested from λ-DNA and RNA by λ-Exonuclease and XRN1 immobilized on polymer substrates are 0.9 ms·nt−1 (1100 ± 100 nt·s−1)[21] and 38 ms·nt−1 (26 ± 5 nt·s−1)[45], respectively. The clipping rate of dNMPs is comparable to the measured TOF of this work and thus to reduce the molecule overlapping it may be necessary to modify electrokinetic conditions to slow down the enzyme function. On the other hand, no molecule overlapping is expected with sequencing RNA because the clipping rate of rNMPs is significantly lower than the measured TOF.
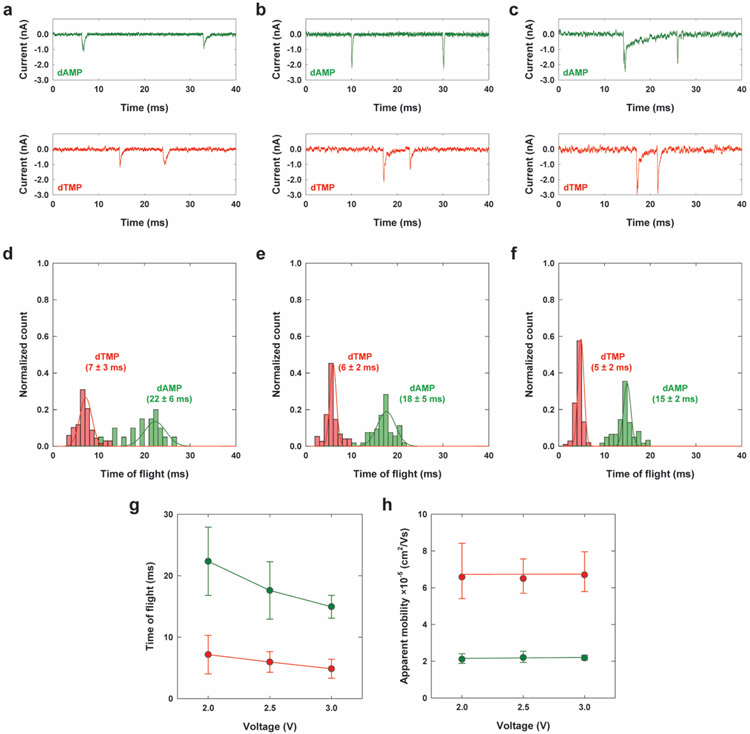
Figure 3 shows TOF of unlabeled dAMP and dCMP versus driving voltage measured with the dual-nanopore TOF sensor with a 5 μm long nanochannel column. Increasing the electric field strength in the nanochannel decreased the TOF while the molecular electrophoretic mobilities were unchanged. Interestingly, increasing the applied voltage also increased the signal-to-noise ratio of transient current peaks and reduced the standard deviation of the measured TOF values. Similar behavior was observed for translocation of dsDNA through a nano-slit, where the standard deviation of the measured mobility decreased with increasing electric field strength.[46] This attribute is important in nanoscale electrophoresis as the mechanism of differentiating dNMP molecules for exonuclease sequencing. Different from nanopore sensing,[17, 19] the use of a high driving voltage simultaneously reduces diffusional misordering of dNMP molecules after cleavage from the intact DNA strand and also increases the identification accuracy of dNMPs via nanoscale electrophoresis.[20] The contribution of the molecular Brownian motion during translocation to the TOF spread was negligible (See the calculation in the Supporting Information).
Figure 3.
TOF of dAMP and dTMP versus driving voltage using a dual-nanopore TOF sensor. (a-c) Current traces of example dual peaks for dAMP and dTMP obtained from the nanosensor with a 5 μm-long nanochannel column under driving voltages of 2, 2.5, and 3 V, respectively. (d-f) Histograms of normalized counts for TOF for dAMP and dTMP shown in (a-c), respectively. The corresponding Gaussian fits were also included. (g-h) TOF and the corresponding apparent mobilities versus driving voltage.
Label-free identification of dNMPs using nanoscale electrophoresis.
Nanoscale electrophoresis has recently been demonstrated for the identification of fluorescently-labeled single molecules, such as dNMPs,[20] rNMPs,[47] and oligonucleotides of different lengths (35-, 50-, and 70-mer)[48] via their TOF through a 100 nm plastic nanochannel column made via NIL. Our dual-nanopore TOF sensor was designed to extend the TOF differentiation of single molecules to a label-free approach. Figure 4 shows histograms of the TOFs for four different dNMPs that were obtained using our dual-nanopore TOF sensors with three different nanochannel column lengths of 0.5, 1, and 5 μm (Figure 4a-c) under a driving voltage of 3 V in 1 m KCl containing 0.5× TBE (pH 10.0) at room temperature. As the nanochannel length increased, the TOF increased, but the order of TOF values for the dNMPs remained unchanged with an order of dTMP < dCMP < dAMP < dGMP. This order of the TOFs was the same as that seen for the fluorescently labeled dNMPs identified by nanoscale electrophoresis through a nanochannel column.[20]
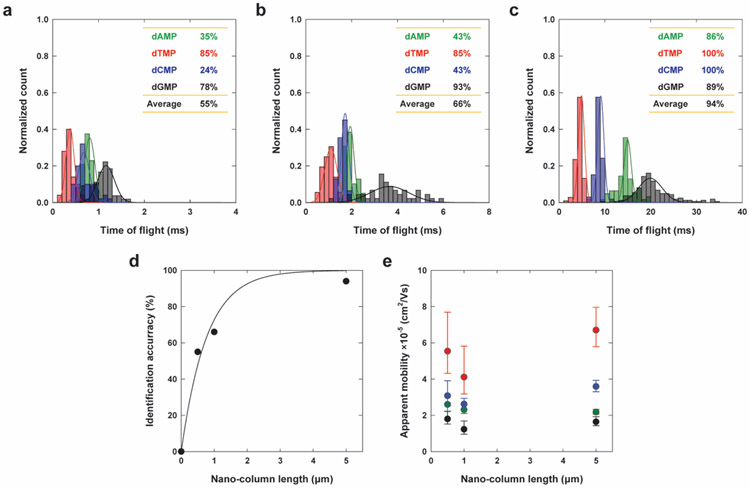
Figure 4.
Label-free identification of dNMPs using the dual-nanopore TOF sensor. (a-c) Histograms for the normalized counts of TOF obtained with four dNMPs using a nanosensor with 0.5, 1, and 5 μm-long nanochannel column, respectively, and the corresponding Gaussian fits. The insets show the identification accuracies of dNMPs, where each value was obtained by the ratio of the non-overlapped area to the entire area of the Gaussian curve fit for a histogram. (d) Average identification accuracy. The fitting curve with IA (%) = 100[1 − exp(−1.2998 · l)] was included, where IA (%) is the identification accuracy in % and l is the nanochannel column length. (e) Apparent mobility versus the length of the nanochannel column. Data were acquired with a 1 m KCl solution containing 0.5× TBE, pH 10.0 under a driving voltage of 3 V. The nucleotide concentration in cis-compartment was 0.01 ng·μL−1.
Nanoscale electrophoresis occurs by a combined action of differences in the electrophoretic mobility of the dNMPs and the stick-slip process of the molecules at the nanochannel wall,[46, 49-51] which results in a mobility difference that is the combined action of electrophoresis and chromatography.[49, 52, 53] The separation is dependent on the identity of the molecule, such as chemical structure (charges and functional groups), size, interactions between the molecules and counterions surrounding the molecules, and the surface of the nanochannel column.[52, 53] Overall, the larger molecules with the purine base (dAMP and dGMP) showed longer TOFs (slower translocation) compared to those with the pyrimidine base (dCMP and dTMP).[54, 55] This result is in good agreement with the MD simulations of dNMP translocation through a nano-slit made in PMMA, which showed that the larger dAMP and dGMP pair (purines) are slowed down more compared to dCMP and dTMP (pyrimidines) when adsorbed to the channel wall due to their larger contact area with the surface.[53]
The chromatographic feature of nanoscale electrophoresis in our dual-nanopore TOF sensor is confirmed by examining the effect of nanochannel column length on the identification accuracy (Figure 4d). The identification accuracy of each dNMP molecule was determined by the non-overlapped area to the total area of the Gaussian curve fit of the histogram. As the length of the nanochannel column increased from 0.5, 1, and to 5 μm, the separation of the histograms of the four dNMP molecules was more obvious and the average identification accuracy calculated increased from 55%, 66%, and 94%. Even with the limited number of data points, the curve fitted reasonably well to IA (%) = 100[1 − exp(−1.2998 · l)] , where IA (%) is the identification accuracy in % and l is the nanochannel column length.
The identification accuracy of dNMPs by nanoscale electrophoresis was also influenced by electrokinetic conditions such as pH, which would result in a change in the charge states of dNMPs as well as the nanochannel surface. The TOF histograms of dNMPs obtained from the translocation experiments at pH 8.0 were significantly overlapped and thus resulted in poor base identification efficiencies (Figure S5, Supporting Information), in agreement with the work by O’Neil et al. on nanoscale electrophoresis with fluorescently-labeled dNMPs.[20] The results also support that EOF-dominant electrokinetic conditions in conjunction with chromatography are in effect for the nanoscale electrophoresis of the dNMPs.
To improve the identification accuracy, an active control method of driving molecules may be adopted, as demonstrated by Liu et al. where an automated “flossing” control logic was used to move a DNA molecule back-and-force in a dual nanopore device enabling multiread coverage data.[12] Similarly, a device with more than two in-plane nanopores formed in series would produce more than two sets of TOF data from a single molecule translocation event.[11] These methods will increase the number of reads, sharpen the TOF histograms, and thus improve the identification accuracy.
Exonuclease sequencing and beyond.
This work focuses on a nanosensor, which enabled the label-free identification with high accuracy of dNMPs via their TOFs using nanoscale electrophoresis. To realize exonuclease sequencing, single nucleotides disassembled from a DNA/RNA molecule by an enzymatic reaction need to be driven sequentially to the dual-nanopore TOF sensor without misordering. The in-plane design of our sensor allows easy addition of nanofluidic structures immediately in front of the sensor so that the enzymatic reactions can occur with nanoscale confinement under high electric field strength, significantly minimizing molecular misordering. Recently, Oliver-Calixte et al. demonstrated that the catalytic efficiency of double-stranded DNA digestion by λ-exonuclease was similar when the enzyme was attached to a solid support compared to the free solution digestion, but the processivity was significantly improved.[21] Compared to enzymatic digestion of a DNA molecule in solution, disassembled nucleotides by the enzymatic digestion at a solid phase bioreactor will enter into the nanosensor in an ordered fashion, allowing for significantly reduced misordering. Increasing the driving voltage to reduce diffusional misordering will also help increase the identification accuracy determined by nanoscale electrophoresis with our sensor. The length of the nanochannel column needs to be optimized via a trade-off between the increased identification accuracy and using exonuclease enzymes that provide clipping rates that minimize the possibility of more than one molecule residing in the nanochannel column at a given time leading to difficulty in identifying peaks during signal processing.
Due to the decoupling of sensing and identification of molecules using our nanosensor, nanopores are used only to detect the passage of molecules, the resolution and sensitivity needed to discriminate nucleotides by transient current amplitudes are not a requirement, which presents a tremendous advantage in the design and fabrication of the nanosensor. However, transient current amplitudes could be used as an additional molecular signature to multiplex with TOF discrimination, which could further improve the identification accuracy (Figure S6, Supporting Information).
While we could detect single-molecule passage with the in-plane nanopores, the transient current background signal showed drift over a 3-5 h operational time, which could have arisen from the use of the hydrogel PEGDA as the substrate and its low Young’s modulus and susceptibility to swelling when in contact with aqueous electrolytes. Therefore, instead of PEGDA use of a hard plastic such as PMMA or cyclic olefin copolymer (COC), which allows achieving both high replication fidelity in the fabrication process using thermal NIL or injection molding and long-term stability against solvents and electrolytes, is desirable. In this case, controllability of the pore size during bonding may be different and thus optimal bonding conditions needs to be determined. There may also be a need to develop a functionalization protocol to achieve non-fouling surfaces of the hard plastics, easy filling of electrolytes into the nanofluidic structures, and the desired EP or EOF force(s) to drive the molecules through the nanochannel column and in-plane pores.
While this study demonstrated the ability of the dual-nanopore TOF sensor to detect and efficiently identify dNMPs, the sensor could provide molecular information from other unamplified targets (DNA, RNA, proteins, exosomes, other single molecules/nanoparticles) by tailoring the dimensions and shape of the pores and the column to produce a molecular-specific current transient signal and TOF. Also, multiple pores can be configured in series to improve the precision in determining the electrophoretic mobility (TOF) of single molecules.
CONCLUSIONS
In conclusion, we have demonstrated single nucleotide identification via molecular-dependent TOF measurements using nanoscale electrophoresis. Flanking the nanochannel column with two sub-10 nm in-plane nanopores allowed identification of the molecular-dependent TOF in a label-free manner. The identification efficiencies of the four dNMP molecules showed a strong dependence on the length of the nanochannel column, reaching 94% when a 5 μm column length was used confirming that the separation was a combination of chromatography through molecule/nanochannel column wall interactions as well as electrokinetic effects. The sensor was formed in a polymer substrate by a simple, cost-effective, and high throughput NIL technique, which is easily expandable to a production format. By combining with a nanoscale reactor for exonuclease digestion of a DNA or RNA strand, the dual-nanopore TOF sensor has the potential to be an essential component to realize exonuclease sequencing.
METHODS
Chemicals and Materials.
Chemicals and materials were obtained from the following sources and used without further purification: 2'-Deoxyadenosine-5'-monophosphate (Alfa Aesar); Thymidine-5'-monophosphate disodium salt (Alfa Aesar); 2'-Deoxycytidine-5'-monophosphate (Alfa Aesar); 2'-Deoxyguanosine-5'-monophosphate disodium salt hydrate (Alfa Aesar); potassium chloride (1 and 3 m, Sigma-Aldrich); Tris-EDTA (100×, Sigma-Aldrich); Tris-Borate-EDTA (10×, Sigma-Aldrich); silver (Ag) wires (>99%, Sigma-Aldrich); S1813 photoresist (MicroChem); MF319 developer (MicroChem); potassium hydroxide (KOH) pellets (Fisher Scientific); hydrofluoric acid (HF) (Sigma-Aldrich); Fluorolink MD 700 (Solvay); 2-hydroxy-2-methylpropiophenone (Sigma-Aldrich); NOA72 (Norland Products); poly(ethylene glycol) diacrylate (PEGDA) (Mn = 250, Sigma-Aldrich); 2,2-dimethoxy-2-phenylacetophenone (Sigma-Aldrich); Si wafers (P/B, resistivity 5-10 Ωcm, orientation of (100), and 525 ± 25 μm thickness) (WaferPro); polyethylene terephthalate (PET) with 250 μm thickness (Goodfellow); polymethylmethacrylate (PMMA) with a thickness of 250 μm (Goodfellow); polymethylmethacrylate (PMMA) of 1 mm thickness (ePlastics).
Silicon (Si) Mater Mold Fabrication.
A Si master mold was prepared by using a combination of photolithography, wet-chemical etching, and focused ion beam (FIB) milling. Si wafers with a 100 nm thick silicon nitride (Si3N4) layer on each side were used for fabricating the master mold. 10 μm deep microchannels were fabricated using a combination of photolithography and wet-chemical etching. Details on the microchannel fabrication can be found in our previous work.[14] Then, the nanochannel flight tube combined with in-plane nanopores was fabricated using FIB milling (Quanta 3D Dual Beam system, FEI). The milling was performed at a beam voltage and current of 30 kV and 10 pA, respectively, in a bitmap mode.
Polymer Device Fabrication.
The Si master mold was used to produce a resin mold by using a UV resin (Fluorolink MD 700) solution containing a UV initiator (2-hydroxy-2-methylpropiophenone) (1 wt%). Details on the UV resin fabrication can be found in our previous work.[42, 56] PEGDA containing the UV initiator (2,2-dimethoxy-2-phenylacetophenone) (1 wt%) was used as the polymer device material. Drops of the UV resin were distributed on the UV resin mold. A PMMA substrate (1 mm thick) was then pressed against the drop and used as a supporting substrate. In order to cure, PEGDA was exposed to flash-type UV light (250-400 nm) for 1 min at an intensity of ~1.8 Wcm−2 by using a nanoimprinter (Eitre6, Obducat). For bonding, a thin PEGDA layer on a thin PMMA sheet (250 μm thick) were used as the cover plate. The cover plate was placed over the imprinted substrate and bonding was performed at 70 °C at 1 MPa for 15 min. Details on the device fabrication can be found in our previous work.[29, 42]
dNMP Translocation Experiments and Data Analysis.
Prior to TOF measurements with dNMPs, the nanosensor was filled with a KCl solution (1 m) containing 1× TE at pH 8.0 and the conductance was measured between two electrodes that were placed across the nanosensor to determine the size of the in-plane nanopores found within the enclosed nanosensor (Figure S3, Supporting Information). For dNMP translocation experiments, the solution in the nanosensor was replaced by a KCl solution (1 m) containing 0.5× TBE and then the dNMP solution with a concentration of 0.01 ng·μL−1 was introduced into the cis-compartment of the nanosensor. The current trace recordings were carried out with a custom current amplifier (UNC CRITICL, https://chem.unc.edu/critcl-main/). The current amplifier consists of a standard topology transimpedance amplifier (TIA) that is battery powered with a gain of 100 nA·V−1 and a single-pole −3dB bandwidth of 10 kHz (Figure S7, Supporting Information). The output of the TIA was digitized by a National Instruments, Inc. model NI PXIe-6341 DAQ system at a sample rate of 250 ksps by running WinEDR software (Strathclyde Electrophysiology Software, http://spider.science.strath.ac.uk/sipbs/software_ses.htm).
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the National Institutes of Health for funding of this work (NIBIB: P41 EB020594). The authors also thank the LSU Shared Instrumentation Facilities (SIF) and Center for Advanced Materials and Devices (CAMD) for assistance in performing focused ion beam milling and lithography, respectively, used in this work.
Footnotes
The authors declare no competing financial interest.
Supporting Information The supporting information is available free of charge at https://pubs.acs.org/
Additional figures and text (PDF)
REFERENCES
- 1.Hamburg MA; Collins FS, New England Journal of Medicine 2010, 363 (4), 301–304. DOI 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 2.Whirl-Carrillo M; McDonagh EM; Hebert JM; Gong L; Sangkuhl K; Thorn CF; Altman RB; Klein TE, Clinical Pharmacology & Therapeutics 2012, 92 (4), 414–417. DOI 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodwin S; McPherson JD; McCombie WR, Nature Reviews Genetics 2016, 17 (6), 333–351. DOI 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang B; Lee JH; Bang D, Experimental & Molecular Medicine 2018, 50 (8), 1–14. DOI 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardis ER, Nature Protocols 2017, 12 (2), 213–218. DOI 10.1038/nprot.2016.182. [DOI] [PubMed] [Google Scholar]
- 6.Slatko BE; Gardner AF; Ausubel FM, Current Protocols in Molecular Biology 2018, 122 (1), e59. DOI 10.1002/cpmb.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reuter JA; Spacek DV; Snyder MP, Molecular Cell 2015, 58 (4), 586–97. DOI 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos CDM; Jackson JM; Witek MA; Soper SA, The Cancer Journal 2018, 24 (2), 93–103. DOI 10.1097/ppo.0000000000000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee M; Burns MA, Trends Biotechnol 2006, 24 (12), 580–6. DOI 10.1016/j.tibtech.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Laver T; Harrison J; O’Neill PA; Moore K; Farbos A; Paszkiewicz K; Studholme DJ, Biomolecular Detection and Quantification 2015, 3, 1–8. DOI 10.1016/j.bdq.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y; Yang Q; Wang Z, Frontiers in Genetics 2015, 5 (449). DOI 10.3389/fgene.2014.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaulaurier J; Schadt EE; Fang G, Nature Reviews Genetics 2019, 20 (3), 157–172. DOI 10.1038/s41576-018-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke J; Wu H-C; Jayasinghe L; Patel A; Reid S; Bayley H, Nature Nanotechnology 2009, 4 (4), 265–270. DOI 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 14.Bowden R; Davies RW; Heger A; Pagnamenta AT; de Cesare M; Oikkonen LE; Parkes D; Freeman C; Dhalla F; Patel SY; Popitsch N; Ip CLC; Roberts HE; Salatino S; Lockstone H; Lunter G; Taylor JC; Buck D; Simpson MA; Donnelly P, Nature Communications 2019, 10 (1), 1869. DOI 10.1038/s41467-019-09637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyson JR; O'Neil NJ; Jain M; Olsen HE; Hieter P; Snutch TP, Genome Research 2018, 28 (2), 266–274. DOI 10.1101/gr.221184.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasianowicz JJ; Brandin E; Branton D; Deamer DW, Proceedings of the National Academy of Sciences 1996, 93 (24), 13770–13773. DOI 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branton D; Deamer DW; Marziali A; Bayley H; Benner SA; Butler T; Di Ventra M; Garaj S; Hibbs A; Huang X; Jovanovich SB; Krstic PS; Lindsay S; Ling XS; Mastrangelo CH; Meller A; Oliver JS; Pershin YV; Ramsey JM; Riehn R; Soni GV; Tabard-Cossa V; Wanunu M; Wiggin M; Schloss JA, Nature Biotechnology 2008, 26 (10), 1146–1153. DOI 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatesan BM; Bashir R, Nature Nanotechnology 2011, 6 (10), 615–624. DOI 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 19.Reiner JE; Balijepalli A; Robertson JWF; Drown BS; Burden DL; Kasianowicz JJ, The Journal of Chemical Physics 2012, 137 (21), 214903. DOI 10.1063/1.4766363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Neil C; Amarasekara CA; Weerakoon-Ratnayake KM; Gross B; Jia Z; Singh V; Park S; Soper SA, Analytica Chimica Acta 2018, 1027, 67–75. DOI 10.1016/j.aca.2018.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliver-Calixte NJ; Uba FI; Battle KN; Weerakoon-Ratnayake KM; Soper SA, Analytical Chemistry 2014, 86 (9), 4447–4454. DOI 10.1021/ac5002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balakrishnan KR; Anwar G; Chapman MR; Nguyen T; Kesavaraju A; Sohn LL, Lab on a Chip 2013, 13 (7), 1302–1307. DOI 10.1039/C3LC41286E. [DOI] [PubMed] [Google Scholar]
- 23.Harms ZD; Haywood DG; Kneller AR; Selzer L; Zlotnick A; Jacobson SC, Analytical Chemistry 2015, 87 (1), 699–705. DOI 10.1021/ac503527d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondylis P; Schlicksup CJ; Katen SP; Lee LS; Zlotnick A; Jacobson SC, ACS Infectious Diseases 2019, 5 (5), 769–777. DOI 10.1021/acsinfecdis.8b00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondylis P; Zhou J; Harms ZD; Kneller AR; Lee LS; Zlotnick A; Jacobson SC, Analytical Chemistry 2017, 89 (9), 4855–4862. DOI 10.1021/acs.analchem.6b04491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J; Kondylis P; Haywood DG; Harms ZD; Lee LS; Zlotnick A; Jacobson SC, Analytical Chemistry 2018, 90 (12), 7267–7274. DOI 10.1021/acs.analchem.8b00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langecker M; Pedone D; Simmel FC; Rant U, Nano Letters 2011, 11 (11), 5002–5007. DOI 10.1021/nl2030079. [DOI] [PubMed] [Google Scholar]
- 28.Liu X; Zimny P; Zhang Y; Rana A; Nagel R; Reisner W; Dunbar WB, Small 2020, 16 (3), 1905379. DOI 10.1002/smll.201905379. [DOI] [PubMed] [Google Scholar]
- 29.Wu J; Chantiwas R; Amirsadeghi A; Soper SA; Park S, Lab on a Chip 2011, 11 (17), 2984–2989. DOI 10.1039/C1LC20294D. [DOI] [PubMed] [Google Scholar]
- 30.Choi J; Lee CC; Park S, Microsystems & Nanoengineering 2019, 5 (1), 12. DOI 10.1038/s41378-019-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huh D; Mills KL; Zhu X; Burns MA; Thouless MD; Takayama S, Nature Materials 2007, 6 (6), 424–428. DOI 10.1038/nmat1907. [DOI] [PubMed] [Google Scholar]
- 32.Fanzio P; Manneschi C; Angeli E; Mussi V; Firpo G; Ceseracciu L; Repetto L; Valbusa U, Sci. Rep 2012, 2 (1), 791. DOI 10.1038/srep00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Firnkes M; Pedone D; Knezevic J; Döblinger M; Rant U, Nano Letters 2010, 10 (6), 2162–2167. DOI 10.1021/nl100861c. [DOI] [PubMed] [Google Scholar]
- 34.Astier Y; Braha O; Bayley H, Journal of the American Chemical Society 2006, 128 (5), 1705–1710. DOI 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 35.Feng J; Liu K; Bulushev RD; Khlybov S; Dumcenco D; Kis A; Radenovic A, Nature Nanotechnology 2015, 10 (12), 1070–1076. DOI 10.1038/nnano.2015.219. [DOI] [PubMed] [Google Scholar]
- 36.Yang H; Li Z; Si W; Lin K; Ma J; Li K; Sun L; Sha J; Chen Y, The Journal of Physical Chemistry B 2018, 122 (32), 7929–7935. DOI 10.1021/acs.jpcb.8b06056. [DOI] [PubMed] [Google Scholar]
- 37.Sha J; Shi H; Zhang Y; Chen C; Liu L; Chen Y, ACS Sensors 2017, 2 (4), 506–512. DOI 10.1021/acssensors.6b00718. [DOI] [PubMed] [Google Scholar]
- 38.Lee K; Lee H; Lee S-H; Kim H-M; Kim K-B; Kim SJ, Nanoscale 2017, 9 (45), 18012–18021. DOI 10.1039/C7NR05840C. [DOI] [PubMed] [Google Scholar]
- 39.Tu B; Bai S; Lu B; Fang Q, Scientific Reports 2018, 8 (1), 9097. DOI 10.1038/s41598-018-27517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farimani AB; Min K; Aluru NR, ACS Nano 2014, 8 (8), 7914–7922. DOI 10.1021/nn5029295. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y; Zhao J; Si W; Kan Y; Xu Z; Sha J; Chen Y, Small Methods 2020, 4 (11), 1900893. DOI 10.1002/smtd.201900893. [DOI] [Google Scholar]
- 42.Jia Z; Choi J; Park S, ACS Applied Materials & Interfaces 2018, 10 (47), 40927–40937. DOI 10.1021/acsami.8b14423. [DOI] [PubMed] [Google Scholar]
- 43.Hartel AJW; Shekar S; Ong P; Schroeder I; Thiel G; Shepard KL, Analytica Chimica Acta 2019, 1061, 13–27. DOI 10.1016/j.aca.2019.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenstein JK; Wanunu M; Merchant CA; Drndic M; Shepard KL, Nature Methods 2012, 9 (5), 487–492. DOI 10.1038/nmeth.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Athapattu US; Amarasekara CA; Immel JR; Bloom S; Barany F; Nagel AC; Soper SA, Nucleic Acids Research 2021, 49 (7), e41–e41. DOI 10.1093/nar/gkab001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chantiwas R; Hupert ML; Pullagurla SR; Balamurugan S; Tamarit-López J; Park S; Datta P; Goettert J; Cho Y-K; Soper SA, Lab on a Chip 2010, 10 (23), 3255–3264. DOI 10.1039/C0LC00096E. [DOI] [PubMed] [Google Scholar]
- 47.Amarasekara CA; Rathnayaka C; Athapattu US; Zhang L; Choi J; Park S; Nagel AC; Soper SA, Journal of Chromatography A 2021, 1638, 461892. DOI 10.1016/j.chroma.2021.461892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amarasekara CA; Athapattu US; Rathnayaka C; Choi J; Park S; Soper SA, ELECTROPHORESIS 2020, 41 (18-19), 1627–1640. DOI 10.1002/elps.202000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell LC; Wilkinson MJ; Manz A; Camilleri P; Humphreys CJ, Lab on a Chip 2004, 4 (3), 225–229. DOI 10.1039/B312592K. [DOI] [PubMed] [Google Scholar]
- 50.Castillo-Fernandez O; Salieb-Beugelaar GB; van Nieuwkasteele JW; Bomer JG; Arundell M; Samitier J; van den Berg A; Eijkel JCT, ELECTROPHORESIS 2011, 32 (18), 2402–2409. DOI 10.1002/elps.201100278. [DOI] [PubMed] [Google Scholar]
- 51.Salieb-Beugelaar GB; Teapal J; Nieuwkasteele J. v.; Wijnperlé D; Tegenfeldt JO; Lisdat F; van den Berg A; Eijkel JCT, Nano Letters 2008, 8 (7), 1785–1790. DOI 10.1021/nl080300v. [DOI] [PubMed] [Google Scholar]
- 52.Novak BR; Moldovan D; Nikitopoulos DE; Soper SA, The Journal of Physical Chemistry B 2013, 117 (12), 3271–3279. DOI 10.1021/jp309486c. [DOI] [PubMed] [Google Scholar]
- 53.Xia K; Novak BR; Weerakoon-Ratnayake KM; Soper SA; Nikitopoulos DE; Moldovan D, The Journal of Physical Chemistry B 2015, 119 (35), 11443–11458. DOI 10.1021/acs.jpcb.5b02798. [DOI] [PubMed] [Google Scholar]
- 54.Zwolak M; Di Ventra M, Reviews of Modern Physics 2008, 80 (1), 141–165. DOI 10.1103/RevModPhys.80.141. [DOI] [Google Scholar]
- 55.Zwolak M; Di Ventra M, Nano Letters 2005, 5 (3), 421–424. DOI 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 56.Choi J; Jia Z; Park S, Microelectronic Engineering 2018, 199, 101–105. DOI 10.1016/j.mee.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.