ABSTRACT
The ability of bacterial pathogens to acquire essential micronutrients is critical for their survival in the host environment. Manganese plays a complex role in the virulence of a variety of pathogens due to its function as an antioxidant and enzymatic cofactor. Therefore, host cells deprive pathogens of manganese to prevent or attenuate infection. Here, we show that evolution of the human-restricted pathogen Bordetella pertussis has selected for an inhibitory duplication within a manganese exporter of the calcium:cation antiporter superfamily. Intriguingly, upon exposure to toxic levels of manganese, the nonfunctional exporter becomes operative in resister cells due to a unique reverse adaptation mechanism. However, compared with wild-type (wt) cells, the resisters carrying a functional copy of the exporter displayed strongly reduced intracellular levels of manganese and impaired growth under oxidative stress. Apparently, inactivation of the manganese exporter and the resulting accumulation of manganese in the cytosol benefited the pathogen by improving its survival under stress conditions. The inhibitory duplication within the exporter gene is highly conserved among B. pertussis strains, absent from all other Bordetella species and from a vast majority of organisms across all kingdoms of life. Therefore, we conclude that inactivation of the exporter gene represents an exceptional example of a flexible genome decay strategy employed by a human pathogen to adapt to its exclusive host.
KEYWORDS: Bordetella, pathogen adaptation, genome decay, manganese, oxidative stress, Bordetella pertussis
INTRODUCTION
Upon infection, bacterial pathogens enter hostile and variable environments containing limited amounts of essential nutrients and, consequently, face several stresses. One of the major challenges is to sustain metal homeostasis and adapt to nutritional and oxidative stresses. Thus, as an important part of the response to infection, host organisms employ several mechanisms and strategies to unbalance the pathogen metabolism and metal homeostasis (1). Given the transition metal essentiality on one side and their metal-linked toxicity on the other, the host cells exploit in principle two opposite antimicrobial strategies. Either they intoxicate the cells by pumping metal into the pathogen milieu by dedicated transporters (2) or, as a part of so-called nutritional immunity, they deprive the pathogen of the essential metal by means of metal-sequestering proteins (3, 4) and efflux pumps that transport metals out of the bacterium-containing phagosomes.
Manganese (Mn) is an important transition metal and essential micronutrient for bacterial pathogens (5). It is an efficient catalyst in redox transactions and a cofactor in various enzymes (for a review, see references 6 and 7). Importantly, there is growing evidence that bacterial pathogens use Mn as an important element in the defense against oxidative stress (8, 9). Indeed, mutants defective in Mn import displayed increased sensitivity to reactive oxygen species (10, 11). Therefore, to prevent or attenuate the infection, host cells deprive pathogens of extracellular Mn through different mechanisms. Neutrophils and macrophages produce calprotectin, an S100 protein family member consisting of S100A8 and S100A9 subunits (12), which efficiently scavenges Mn and zinc from the host environment and sensitize the pathogen toward oxidative stress effects (13). Furthermore, phagocytic cells can withhold the Mn from internalized bacteria by another mechanism based on the divalent metal exporter NRAMP1 (14, 15). While the procedures of Mn deprivation are well characterized, the molecular mechanisms of host-mediated intoxication of invading cells are poorly understood. Mn exporters have been discovered recently in several pathogenic bacteria, and mutants lacking these exporters displayed attenuated virulence and reduced ability to survive within the host environments (16–19). However, it remains to be clarified whether these effects resulted from Mn intoxication.
Given the role of Mn in redox and metal homeostasis, its transport is tightly regulated (5). Import of Mn into the bacterial cell is facilitated by bacterial homologues of the NRAMP1 transporter family (e.g., MntH [20]) or by ATP-binding cassette (ABC)-type transporters (e.g., MntABC [21]), while the export is accomplished by various efflux pumps belonging to several protein superfamilies, such as lysine exporters (e.g., MntP), cation diffusion facilitators (e.g., MntE), and P-type ATPases (e.g., CtpC) (16, 22–24). Mn trafficking is controlled by the cognate transcriptional regulators MntR (21, 25, 26) and Mur (27, 28) as well as by global regulators involved in metal homeostasis and oxidative stress response, such as Fur and OxyR (23, 29, 30). Furthermore, regulatory RNAs such as riboswitches have been shown to play essential roles in balancing the Mn homeostasis. Riboswitches are highly structured cis-acting groups of noncoding RNAs positioned within the 5′-untranslated regions of, and are cotranscribed with, the genes they regulate (31). Among other metabolites, riboswitch aptamers also recognize and bind metals and, consequently, riboswitches were found to control expression of metal transporters and to contribute to regulation of metal homeostasis (32). Recently, an Mn-responsive riboswitch was shown to control expression of an Mn exporter in Escherichia coli (33) and Lactococcus lactis (34).
Bordetella pertussis is a strictly human reemerging pathogen and causative agent of whooping cough (35). B. pertussis has evolved from a B. bronchiseptica-like ancestor (36), and as a part of the ongoing adaptation to its host, the B. pertussis genome was continuously rearranged, reduced, and decayed (37–39). Thus, the B. pertussis genome contains hundreds of pseudogenes, i.e., nonfunctional genes inactivated mostly by an insertion sequence element transposition and inversion or by deletion/insertion of one or multiple nucleotides (37). Our analysis of the primary transcriptome of B. pertussis revealed hundreds of noncoding transcripts, including several riboswitches (40). Here, we describe an Mn-sensing riboswitch that controls the expression of the downstream BP3410 gene encoding a nonfunctional Mn exporter. Intriguingly, although the functionality of the exporter gene was fully recovered in resister cells formed in the presence of toxic concentrations of Mn, the wild-type (wt) strain carrying the decayed copy of the exporter displayed improved growth under oxidative stress compared to resister cells. We propose that during infection, when access to Mn is limited, the nonfunctional Mn exporter is beneficial to B. pertussis survival.
RESULTS
Expression of the BP3410 gene is controlled by an Mn2+-responsive riboswitch.
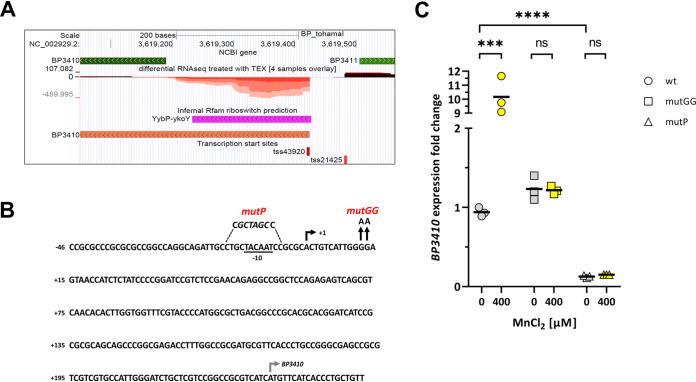
Our recent analysis of the primary transcriptome of Bordetella pertussis revealed a large variety of noncoding transcripts, including several riboswitch structures (40). One of the riboswitches was identified in the 5′-untranslated region (UTR) of the BP3410 gene, which was annotated as a calcium:cation antiporter (Fig. 1A). Curiously, the riboswitch region overlapped a previously identified small RNA, BprK, that initially had been predicted by several bioinformatics tools and then confirmed by Northern blotting to be an approximately 200-nucleotide (nt) transcript (41). However, a search against the Rfam database confirmed that BprK constitutes an orphan yybP-ykoY riboswitch (E value of 8.5 × 10−15). These riboswitches are frequently found upstream of genes encoding transmembrane proteins involved in the transport of metals, and some of them are Mn responsive (33, 34). The Stainer-Scholte (SS) medium, used in our experiments to grow B. pertussis, is not supplemented with Mn (42), and therefore the presumed sRNA most likely represents a BP3410-specific transcript that terminates prematurely within the riboswitch structure in the absence of Mn. In support of this, our primary transcriptome analysis showed that the BP3410 gene is transcribed exclusively from a single promoter upstream of the riboswitch and that this transcript originates from the negative strand as the A residue at position 3,619,424 (TSS43920 in Fig. 1A) within the B. pertussis genome (37, 40). At an appropriate distance from the transcriptional start site, we identified a plausible −10 promoter sequence (TACAAT) (Fig. 1B). Collectively, these data revealed that the BP3410 gene is transcribed from the promoter upstream of the riboswitch; however, in the absence of Mn, the RNA polymerase stops within the riboswitch region.
FIG 1.
Mn-responsive riboswitch yybP-ykoY affects the expression of BP3140 gene. (A) A detailed view of the genome browser (http://bit.ly/2Euo5HZ) (40) showing the dRNA-seq normalized data set of the region between genes BP3410 and BP3411 (green arrows). The plot shows the sequencing depth of expression of the positive-strand (depth above the zero line, dark red) and the negative-strand (depth below the zero line, light red) expression, with different color intensities representing the different library replicates. The transcription start sites of the BP3410 (TSS43920) and BP3411 (TSS21425) genes and the predicted riboswitch region (purple arrows) are also shown. (B) Nucleotide sequence map of the region upstream of the BP3410 gene encompassing the riboswitch region. The transcription start site identified by dRNA-seq analysis (black rectangular arrow, +1), the plausible −10 promoter sequence (underlined), and the start codon of the BP3410 gene (gray rectangular arrow) are shown. The mutations introduced into the riboswitch are indicated above the sequence. To construct the BP3410mutP strain, the sequence around the −10 site was altered to prevent transcription of the BP3410 gene, and the introduced NheI restriction site is shown in italics. The BP3410mutGG mutant was created by replacing two adjacent G residues with A residues (depicted by vertical arrows). (C) RT-qPCR analysis was performed with total RNA isolated from three biological replicates of wt, BP3410mutP, and BP3410mutGG cells induced with 400 μM Mn (yellow symbols) for 60 min or left untreated (gray symbols, controls). The dot plot shows individual data points for all strains; the horizontal black lines represent mean values. Statistical analysis was performed using an unpaired two-tailed t test; ns, P > 0.05; ***, P < 0.0005; ****, P < 0.00005. Expression of BP3410 in the first biological replicate of the wt strain grown in the absence of Mn was arbitrarily set to 1, and all other values are expressed relative to this value.
To test this assumption experimentally, we analyzed the link between the riboswitch, Mn, and BP3410 expression by quantitative PCR (qPCR) and constructed two mutants for this purpose. First, we mutated the plausible −10 region upstream of the riboswitch (BP3410mutP strain), and second, we mutated two conserved G residues within the riboswitch aptamer (BP3410mutGG strain) that were documented to be critical for Mn binding (33, 34) (Fig. 1B). Overnight cultures of wt, BP3410mutP, and BP3410mutGG strains were divided and further incubated either in the absence (controls) or presence of 400 μM Mn for 60 min. BP3410 expression was then examined by reverse transcription-qPCR (RT-qPCR). As demonstrated in Fig. 1C, the addition of Mn induced the expression of the BP3410 gene in the wt strain, while this effect was absent from both mutants. Notably, under noninducing conditions, BP3410 expression was significantly reduced in the BP3410mutP strain compared with the wt and BP3410mutGG strains. This observation suggested that the inhibitory structure of the riboswitch does not completely disrupt transcription driven by the BP3410 promoter even in the absence of Mn. It should be noted that the addition of different concentrations of calcium had no effect on BP3410 gene expression (data not shown). Collectively, our results proved that the BP3410 gene is transcribed from a single promoter upstream of the riboswitch and that binding of Mn to the riboswitch is required for induction of BP3410 expression.
The wild-type strain forms survivor colonies in the presence of toxic manganese concentrations.
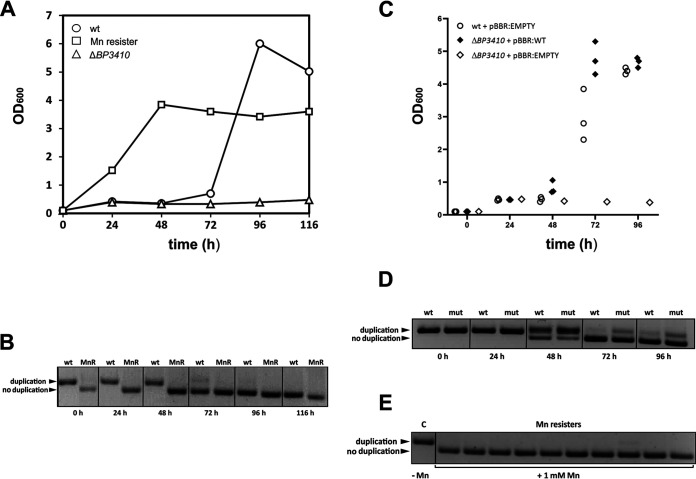
The inducibility of BP3410 expression by Mn, as well as the high similarity between the 5′-UTR of the BP3410 gene and yybP-ykoY riboswitch, suggested that the BP3410 protein constitutes an Mn exporter despite its annotated function. To experimentally confirm this hypothesis, we constructed a markerless in-frame ΔBP3410 deletion mutant of B. pertussis with the rationale that the mutant strain should exhibit a growth defect in the presence of toxic Mn concentrations. We used a disc diffusion assay to compare the sensitivity of the wt and ΔBP3410 strains toward the gradient of Mn concentrations. Surprisingly, both strains exhibited similarly sized zones of inhibition around discs soaked with 10 μl of 100 mM Mn. However, in contrast to the plates inoculated with the ΔBP3410 strain, the plates inoculated with the wt strain revealed distinct colonies within the zone of inhibition (Fig. 2A). To corroborate this observation, we plated both wt and ΔBP3410 cells on agar plates supplemented with 1 mM Mn. While we did not obtain any colonies of the mutant, plating out the wt strain yielded dozens of colonies (Fig. 2B) and allowed us to estimate that these survivors were formed at an approximate frequency of 1.06 × 10−6 ± 0.62 × 10−6.
FIG 2.
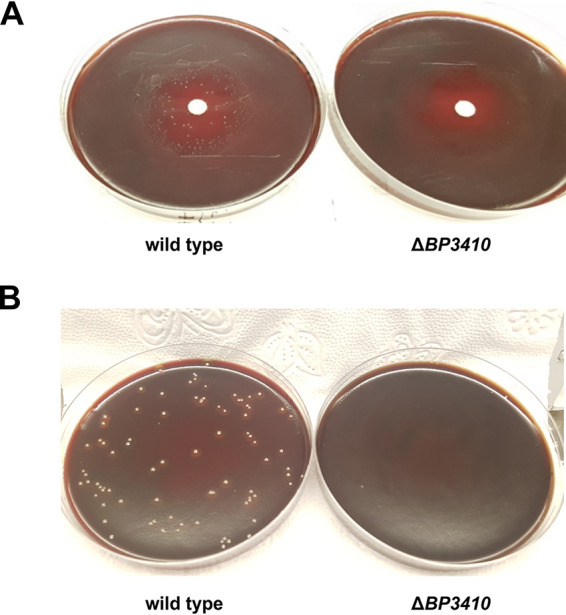
Growth of B. pertussis in the presence of toxic concentrations of manganese. (A) B. pertussis wt and ΔBP3410 cells were grown in SS medium to late exponential phase, and then 100 μl of cell suspension (OD600 of suspension was set to 2) was plated on BG agar. Sensitivity to Mn was tested with a disc diffusion assay in which 10 μl of 100 mM MnCl2 was pipetted onto the disc. (B) B. pertussis wt and ΔBP3410 cells were grown in SS medium to an OD600 of ≈2.0, and then 100 μl of each culture was plated on BG agar supplemented with 1 mM MnCl2.
The Mn resisters lack the unique duplication within the BP3410 gene that is highly conserved in B. pertussis.
Our results indicated that B. pertussis cells can adapt to toxic concentrations of Mn. To clarify the genetic background of this adaptation, the BP3410 locus was sequenced in several survivor cells and compared with the wt strain. Interestingly, all the survivors carried a 30-nt deletion within the coding region of the BP3410 gene, resulting in a deletion of 10 amino acids. Closer inspection of this region revealed that this region is duplicated in the wt strain, whereas all survivors, here referred to as Mn resisters, contained only one copy of this 30-nt segment (Fig. 3). Furthermore, a comparison of the corresponding region in all fully sequenced B. pertussis strains deposited in GenBank showed that this duplication is present in almost 98 % of the strains. The calcium:cation antiporters represent a ubiquitous protein family found across all kingdoms of life. Nevertheless, as shown in Fig. 3, the observed duplication is highly specific only to B. pertussis, with the exception of three recent isolates of Aeromonas veronii (43).
FIG 3.
Conservation of duplication within the BP3410 exporter in other organisms. The amino acid sequences of the BP3410 exporter of B. pertussis and its selected homologues found in different organisms were aligned with the MUSCLE algorithm built in the MEGA software using the neighbor-joining clustering method. The black asterisk indicates a highly conserved negatively charged residue essential for cation transport. Aligned residues that can be considered similar based on their physicochemical distance at a given position are shown in red. Similar residues found in at least 70 % of the aligned sequences are enclosed in blue frames. The triangles mark the duplicated amino acid sequence present only in B. pertussis and three recent isolates of A. veronii (red triangles) and the sequence present in all homologous proteins (blue triangles). The amino acid residues are numbered according to the B. pertussis BP3410 protein. The complete alignment is presented in Fig. S1.
Complete alignment of BP3410 exporter homologues in other organisms. The amino acid sequences of the BP3410 exporter of B. pertussis and its selected homologues found in different organisms were aligned with the MUSCLE algorithm built in the MEGA software using the neighbor joining clustering method. The black asterisk indicates a highly conserved negatively charged residue essential for cation transport. Aligned residues that can be considered similar based on their physicochemical distance at a given position are shown in red. Similar residues found in at least 70 % of the aligned sequences are enclosed in blue frames. The triangles mark the duplicated amino acid sequence present only in B. pertussis and three recent isolates of A. veronii (red triangles), and the sequence present in all homologous proteins (blue triangles). The amino acid residues are numbered according to the B. pertussis BP3410 protein. Download FIG S1, PDF file, 0.03 MB (33KB, pdf) .
Copyright © 2021 Čapek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Duplication in BP3410 is lost in cells growing in the presence of toxic Mn concentrations.
To gain better insight into the dynamics of duplication loss, we monitored the growth characteristics of the wt, ΔBP3410, and Mn resister strains in SS medium supplemented with 200 μM Mn. In parallel with the optical density measurements, we collected samples for isolation of genomic DNA and subsequent amplification of the region around the duplication by PCR. The ΔBP3410 mutant reached a very low OD600 (≈0.3) and then ceased growth, whereas the Mn resister exhibited a standard growth curve (Fig. 4A). Interestingly, the wt strain grew similarly to the ΔBP3410 mutant for the first 3 days, but after this rather long lag time of 72 h, it resumed growth and reached optical densities comparable to those of the Mn resister. PCR analysis of samples collected during incubation clearly showed that by the third day of cultivation, the wt cells carrying the 30-bp duplication were eliminated and replaced by the Mn resisters without the duplication (Fig. 4B). Apparently, growth of B. pertussis in the presence of toxic amounts of Mn did not resume until the duplication in the BP3410 gene was lost. Nevertheless, to rule out the possibility that an additional mutation(s) was responsible for improved growth of the Mn resister, site-directed mutagenesis was used to construct a mutant strain lacking duplication in the BP3410 gene. Compared with the spontaneously evolved resister, this mutant, here referred to as the constructed Mn resister (Mn resisterC), displayed a nearly identical phenotype in the presence of toxic amounts of Mn (data not shown). To further corroborate our results, we tested whether ectopic expression of the BP3410 gene would remedy the growth defect of the ΔBP3410 strain observed in the presence of an inhibitory Mn concentration. Therefore, the ΔBP3410 strain was complemented with the plasmid carrying the wt copy of the BP3410 gene including the native promoter and the Mn-responsive riboswitch. As controls, wt and ΔBP3410 strains carrying an empty vector were grown together with the complemented strain in SS medium supplemented with 200 μM Mn. Again, in parallel with the optical density measurements, we determined the presence of the duplication within the BP3410 gene by PCR. As shown in Fig. 4C, the wt strain (carrying a single copy of the gene) and the complemented strain (carrying multiple copies of the gene) resumed growth after 48 h of incubation, but the complemented strain reached stationary phase earlier than the wt strain. Both strains began to lose the duplication after 48 h, and after 72 h of incubation, they lost the duplication almost completely (Fig. 4D). We wondered whether the presence of the larger product in PCR samples of the complemented mutant indicates that some resister cells carry a mixed population of plasmid-borne copies of the BP3410 gene that either have or have not resolved the duplication. Therefore, we plated the complemented mutant in three biological replicates on agar plates containing 1 mM Mn, and we randomly selected 30 resister colonies (10 clones per replicate) for PCR analysis. All resister clones contained only the functional copy of the BP3410 gene (Fig. 4E). We also plated both the wt strain carrying empty plasmid and the complemented ΔBP3410 strain in biological triplicates on agar plates containing 1 mM Mn and enumerated the colonies to determine the rate of resister formation. The complemented strain formed resister cells at more than 50-fold higher frequency (8.60 ± 1.85 × 10−5) than the wt strain (1.45 ± 0.58 × 10−6). These experiments demonstrated that ectopic expression of the BP3410 gene complements the growth defect of the ΔBP3410 mutant and contributes to more rapid adaptation to toxic Mn concentrations.
FIG 4.
B. pertussis forms resister cells during growth in the presence of toxic concentrations of manganese. (A) B. pertussis wt, ΔBP3410, and Mn resister cells were grown in SS medium in the presence of 200 μM Mn. Optical density (OD600) was measured every 24 h, and samples were collected in parallel for PCR analysis. (B) Genomic DNA was isolated from wt and Mn resister (MnR) cells harvested in parallel with optical density measurements and used as a template for amplification of the region encompassing the duplication within the BP3410 gene. PCR products that either contain or do not contain the duplication are indicated by arrowheads. (C) B. pertussis wt cells carrying an empty plasmid (white circles) and complemented ΔBP3410 cells carrying a copy of the BP3410 gene on the plasmid (black diamonds) were grown in triplicate cultures in SS medium supplemented with 200 μM Mn. As a control, a single culture of ΔBP3410 cells carrying empty plasmid (white diamonds) was grown under the same conditions. The plot shows individual optical density values measured every 24 h. (D) Genomic DNA was isolated from the wt strain carrying an empty plasmid (wt) and the ΔBP3410 mutant carrying plasmid-borne BP3410 gene (mut) harvested in parallel with the optical density measurements (C) and used as a template for amplification of the region encompassing the duplication within the BP3410 gene. PCR products that either contain or do not contain the duplication are indicated by arrowheads. (E) Genomic DNA was isolated from ΔBP3410 cells carrying plasmid-borne BP3410 gene harvested from plates without (control, C) or with 1 mM Mn (Mn resisters). PCR products either containing the duplication or not are indicated by arrowheads. The experiment was performed in three biological replicates. In each experiment, 10 resister clones were analyzed. A representative experiment is shown.
Despite its homology with calcium:cation antiporters, the BP3410 protein exports manganese.
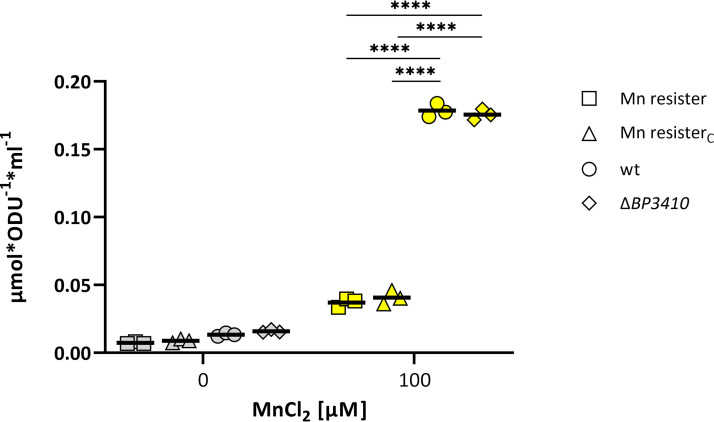
Based on our results, we concluded that B. pertussis produces a nonfunctional Mn exporter, BP3410, that carries a deleterious duplication, and that this alteration prevents growth of the pathogen in the presence of toxic Mn concentrations. Consistent with the annotation of BP3410, the scan of the exporter sequence against the Pfam database identified two domains (E value of 4.4 × 10−13 for the N-terminal domain and E value of 2 × 10−8 for the C-terminal domain) characteristic of calcium:cation antiporters. In addition, searching the PDB database unveiled that the closest crystallized homolog of BP3410 (38.3% amino acid sequence similarity) is the calcium:cation antiporter of Methanocaldococcus jannaschii (44). We hypothesized that, similar to the M. jannaschii antiporter, the B. pertussis protein contains 10 transmembrane helices evenly distributed between two domains and that each domain contains a conservative α-repeat region, which, in complex of both domains, governs the passage of ions. When we mapped the position of the duplicated 10-amino-acid sequence in the M. jannaschii protein structure, we found that this duplication is located directly in the α-repeat region (data not shown) and therefore would drastically disrupt domain symmetry and most likely impair exporter function. Although these in silico analyses suggested that BP3410 shares significant homology with the calcium:cation antiporter superfamily, our experimental data suggested that expression and functionality of the BP3410 gene are tied to the presence of Mn. To prove the affinity of the BP3410 exporter for Mn, we used inductively coupled plasma tandem mass spectrometry (ICP-MS/MS). We reasoned that in contrast to Mn resisters (both evolved and constructed), the strains carrying a nonfunctional copy of the BP3410 gene (wt strain and ΔBP3410 mutant) should accumulate Mn in the presence of this metal. Strains were grown overnight in the absence of Mn and diluted to an OD600 of 0.2, and then the cultures were divided and grown for 8 h in either the absence or presence of 100 μM Mn (a concentration that did not inhibit the growth of strains lacking the functional BP3410 exporter during the 8-h cultivation). Consistent with our assumption, ICP-MS/MS analysis revealed that wt and ΔBP3410 cells contained approximately 5-fold more Mn than either type of Mn resister (Fig. 5).
FIG 5.
Cells producing functional BP3410 protein export manganese. Three biological replicates of wt, ΔBP3410, Mn resister, and Mn resisterC strains were cultured either in the absence (gray symbols) or in the presence (yellow symbols) of 100 μM Mn for 8 h. The amount of intracellular Mn was measured by ICP–MS/MS. The dot plot shows individual data points for all strains, and the horizontal black lines represent mean values. Statistical analysis was performed using an unpaired one-tailed t test; ****, P < 0.00005. ODU, optical density units.
Compared with wt cells producing the nonfunctional exporter, Mn resisterC cells display impaired growth under oxidative stress.
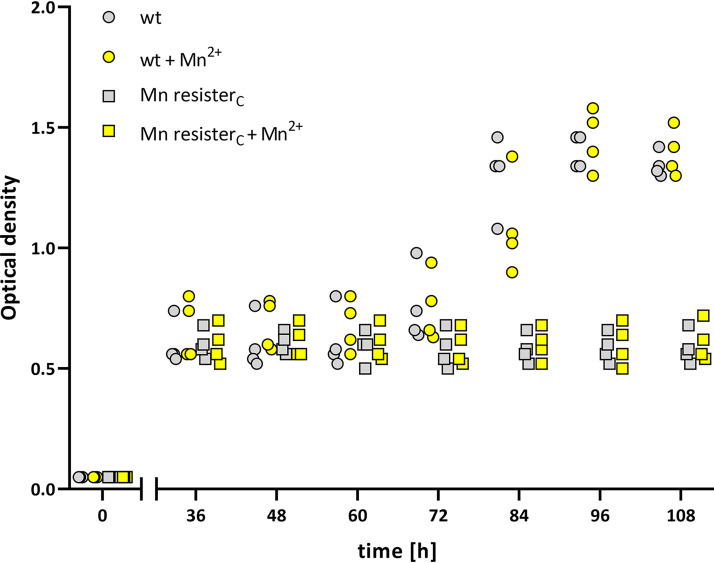
Given the important role of Mn in the adaptation of bacterial pathogens to the host environment, we wondered whether the deleterious duplication that renders the Mn exporter BP3410 nonfunctional confers an advantage to B. pertussis cells. We speculated that the decay of the exporter and the resulting accumulation of Mn within the cytosol favor the survival of the pathogen under oxidative stress. Therefore, to prove the concept, we tested the growth characteristics of B. pertussis strains cultured in the presence of paraquat, a known inducer of oxidative stress (45). In addition, we used iron-limited conditions because the iron-dependent repressor Fur inhibits Mn import (29), and iron deficiency mimics the conditions that prevail during infection in vivo. Therefore, cultures of wt and Mn resisterC strains were passaged several times in the iron-limited SS medium and then incubated in the presence or absence of 100 nM Mn (a concentration that is in the range of Mn concentrations in human blood [46]) for 8 h to allow Mn accumulation. Cultures were then washed with PBS containing 10 mM EDTA and diluted in SS medium supplemented with paraquat and the iron chelator dipyridyl. Growth of all cultures was monitored for 108 h, but optical density was not measured for the first 36 h, as all strains exhibited similar growth during this period (data not shown). As shown in Fig. 6, the optical density of both strains did not change substantially until 60 h of cultivation. Around this time point, however, the wt strain resumed growth, in contrast to the resister strain. Surprisingly, the resumption of growth of the wt strain was not dependent on prior exposure to Mn.
FIG 6.
Mn resister cells do not resist oxidative stress. Four biological replicates of wt (circles) and Mn resisterC strains were passaged 5 times in low-iron SS medium and then pregrown for 8 h in low-iron SS medium in the absence (gray symbols) or presence (yellow symbols) of 100 nM Mn. Cultures were then treated with 70 μM dipyridyl and 0.9 mM paraquat, and after an initial period of 36 h, growth was monitored every 12 h by OD600 measurements. The dot plot shows the individual data points for all strains.
DISCUSSION
B. pertussis represents a respiratory pathogen of humans that is continuously adapting to its exclusive host (36, 37, 47). During this process, B. pertussis underwent a global expansion of insertion elements that resulted in large genome rearrangements (change in gene order), genome reduction (loss of genes), and genome decay (loss of gene functionality) (37–39, 48). Such an evident and substantial genome reorganization process is usually associated with an evolutionarily recent change in the niche and resulting host restriction (49). Gene inactivation, primarily by insertion of massively expanded IS481 elements, resulted in the formation of hundreds of pseudogenes within the B. pertussis genome (37). To date, the inactivation of B. pertussis genes has resulted in permanent loss of their function. Here, we report the first example of a decayed gene whose functionality can be fully restored. Our data demonstrate that the Mn exporter gene BP3410, which is controlled by an Mn-sensing riboswitch, is inactivated in wt cells by a 30-bp tandem duplication and thereby encodes a defective protein. However, exposure of wt cells to toxic levels of Mn resulted in formation of resister cells lacking the deleterious duplication. Loss of the duplication restored the functionality of the BP3410 exporter, as evidenced by significantly reduced intracellular Mn levels in resister cells compared with wt cells.
Our results proved that resumption of growth of the wt strain and the complemented mutant in the presence of toxic Mn concentrations correlated with the loss of duplication of the chromosome and the plasmid-borne gene, respectively. Notably, ectopic expression of the BP3410 gene led to increased frequency of resister formation, indicating that adaptation to Mn stress is a stochastic and gene dose-dependent event. However, PCR analysis of resisters formed from complemented ΔBP3410 strains indicated that all plasmid-borne copies of the BP3410 gene resolved duplication, proposing that loss of duplication is not purely of a stochastic nature. The exact nature of this unique process that ensures precise excision of the duplication region is unclear, but the length, proximity, and 100% homology of the two tandem repeats imply that this process is RecA independent (50).
It remains to be determined whether the loss-of-duplication/gain-of-function process is reversible in an Mn-restrictive environment. We tested dozens of Mn resister clones isolated from human THP-1 macrophages (Mn-limited environment), but none of them carried the inhibitory duplication (data not shown). Therefore, we speculate that loss of duplication is not reversible under in vitro conditions and represents a dead-end state for resister cells. Interestingly, a sequence similarity search revealed that approximately 2 % of B. pertussis isolates carry a functional copy of the BP3410 gene (data not shown) and that these strains are randomly distributed across the phylogenetic tree reconstructed by Bart et al. (51). This observation suggests that strains resistant to toxic Mn concentration have reemerged during the evolution of B. pertussis and that the gain of BP3410 gene function, observed in our experimental setup, represents a biologically relevant backup mechanism. The genus Bordetella appears to be of an environmental origin (52), and species such as B. bronchiseptica and B. petrii can survive outside the host and, thus, may encounter environments with elevated Mn content. Moreover, most environmental isolates of Bordetella sp. were collected from soil (52), and some of the isolates exhibited substantial resistance to heavy metals (53, 54).
Given the uniqueness of the duplication in B. pertussis and the general importance of Mn for virulence, we hypothesized that inactivation of the Mn exporter specifically contributes to the survival of B. pertussis within the human host. Similarly, a homology search against the GenBank database showed that human B. parapertussis isolates carry a frameshift mutation in the BPP3560 gene, a BP3410 homolog. This observation supports our hypothesis that the pseudogenization of Mn exporters arose from adaptation to humans. Moreover, nearly 70 % of Neisseria gonorrhea strains carry a frameshift mutation within the Mn exporter MntX (17). It has been reported that this pathogen uses Mn as a quenching substance to deal with oxidative stress (55). By analogy, we reasoned that the nonfunctional Mn exporter might also help B. pertussis cells to retain the acquired Mn within the cytosol to cope with oxidative stress. Indeed, our experiments demonstrated that compared with the wt strain, the resister cells carrying a functional exporter displayed impaired growth under oxidative stress in vitro. Intriguingly, pretreatment with Mn had no effect on the growth of paraquat-treated wt cells. We presume that the growth medium contained trace amounts of Mn, present as an impurity in other components of the medium, which allowed B. pertussis to resist oxidative stress in vitro independent of additional Mn supplement. Notably, compared with the BP3410mutP strain, BP3410 expression was significantly increased in the wt strain in the absence of Mn supplement. This finding indicated that the exporter gene is expressed to some extent even in the absence of Mn and, thus, would dictate unfavorable Mn export even under Mn-limited conditions during infection. We speculate, in the context of nutritional immunity, that “leaky” expression exposed the exporter to selection pressure that led to its decay.
Notably, we have discovered other links that document the importance of adequate Mn homeostasis for the physiological fitness of B. pertussis. One of the steps in the biosynthesis of heme, the oxidative decarboxylation of coproporphyrinogen III to protoporphyrinogen IX, is catalyzed by two isozymes, HemN and HemF (56). In contrast to HemN, HemF requires Mn as a cofactor, and hemF expression is induced upon oxidative stress in E. coli (57). We noticed that the hemN gene is nonfunctional in B. pertussis, as its coding sequence is interrupted by an IS481 element, making heme biosynthesis in this pathogen most likely Mn dependent. Our ongoing experiments (B. Vecerek, unpublished data) also have shown that intracellular B. pertussis cells strongly upregulate the ATP-binding cassette importer (encoded by the BP3080 to BP3082 genes) during infection of human THP-1 macrophages. This transporter shares substantial homology with the Mn importer SitABCD of Salmonella enterica serovar Typhimurium (58).
Collectively, our results demonstrate that speciation of B. pertussis was accompanied by inhibitory duplication within the Mn exporter and indicate that the inactivation of the exporter represents an adaptive mechanism that maintains sufficient levels of cytosolic Mn, thereby assisting the pathogen in resistance to stress and survival within the human host.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bordetella pertussis Tohama I strain (59) and its derivatives (see Table S1 in the supplemental material) were grown on Bordet-Gengou agar (BGA) plates supplemented with 15% sheep blood for 3 days at 37°C prior to inoculation into liquid medium. For liquid cultures, bacteria were grown in Stainer-Scholte (SS) medium (42) supplemented with 0.1% cyclodextrin (Sigma-Aldrich) and 0.5% Casamino Acids (Difco) in an Innova 43 orbital incubator (Eppendorf) at 37°C and 160 rpm. When required, SS medium was supplemented with various concentrations of manganese chloride (Sigma-Aldrich) as specified in figure legends. Strains bearing the pBBRMCS1 vector were grown on BGA plates supplemented with chloramphenicol (10 μg ml−1; Sigma-Aldrich).
Strains and plasmids used in the study. Download Table S1, PDF file, 0.3 MB (330.6KB, pdf) .
Copyright © 2021 Čapek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of the mutants.
The deletions or point mutations were introduced into B. pertussis Tohama I strain chromosome as already described (60). To construct the ΔBP3410 deletion mutant, two DNA fragments of approximately 750 bp corresponding to the upstream region (ending with the ATG codon of the BP3410 gene and NheI site) and the downstream regions (starting with the NheI site and TGA stop codon of the BP3410 gene) flanking the gene. Obtained PCR products were ligated via an NheI site, and the ligation mixture was used as a template to create an approximately 1.5-kb PCR product containing the intended deletion. In the resulting product, the start and stop codons of BP3410 separated by the NheI restriction site created a markerless in-frame deletion. A similar approach was applied to construct the BP3410mutP strain, carrying the mutated −10 region within the BP3410 promoter. DNA fragments of approximately 750 bp adjacent to the mutated site were amplified using mutagenic primers containing the NheI site and then ligated via the NheI site. The ligation mixture was used to amplify the 1.5-kb product, in which the sequence TGCTACAAT was replaced by the NheI restriction site GCTAGC. To construct the BP3410mutGG strain, where two G residues within the riboswitch were replaced by A residues, the regions upstream and downstream of the mutation site were amplified using primers carrying the mutagenic sequence. Resulting PCR products were joined by an overlap PCR, yielding an ≈1.5-kb PCR product containing the planned mutation. To construct the Mn resisterc strain, the BP3410 gene lacking the 30-nt duplication was directly amplified using the DNA of the evolved Mn resister strain as a template. In all cases, final PCR products were ligated into the allelic exchange plasmid pSS4245 and cloned in the E. coli XL1-Blue strain. Finally, the resulting recombinant plasmid was transformed into the E. coli SM10 strain (donor strain) and transferred to B. pertussis Tohama I (recipient strain) by conjugation, as described elsewhere (61). After two recombination events, the strain carrying the desired mutation was obtained. All deletions and point mutations were confirmed by DNA sequencing.
To complement the deletion of the exporter gene, the plasmid carrying the BP3410 gene, including its promoter region, was constructed as follows. The gene was amplified using forward and reverse primers containing SacI and XhoI restriction sites, respectively. The PCR product was then cleaved with SacI and XhoI enzymes and inserted into the corresponding sites of the plasmid pBBRMCS1. Resulting plasmid was transferred to B. pertussis ΔBP3410 recipient strain by conjugation. The plasmids used in this study and primers used for the construction of the mutants and quantitative PCR are listed in the Tables S1 and S2, respectively.
Determination of manganese toxicity by disc diffusion assays.
Strains were inoculated from BGA plates into SS medium and grown overnight for ≈20 h, until the cells reached late exponential phase (optical density at 600 nm [OD600], ≈1.5 to 2). The next day, the OD600 value of bacterial cultures was adjusted to 2, and 100 μl of the suspension was spread on BGA plates. Next, 6-mm filter paper discs (Oxoid) were placed into the center of the dish and immediately soaked with 10 μl of 100 mM MnCl2. Plates were incubated for 4 to 5 days until the clear inhibition zones were formed.
Determination of frequency of resister formation.
Strains were inoculated from BGA plates into SS medium and grown overnight for ≈20 h until the cells reached late exponential phase (OD600, ≈1.5 to 2). Strains bearing pBBRMCS1 derivatives were inoculated from BGA plates supplemented with chloramphenicol (10 μg ml−1). The next day, the bacterial cultures were either appropriately diluted and spread on BGA plates without Mn or spread undiluted on BGA plates supplemented with 1 mM Mn. CFU were enumerated from both types of plates, and frequency of resister formations for each strain was calculated as a ratio between CFU of formed resisters recovered from plates with Mn and the total number of CFU counted on plates without Mn. Frequencies were calculated as means and standard deviations from at least four independent experiments.
Monitoring of Mn resister formation in liquid cultures.
Strains were grown in SS medium supplemented with 200 μM MnCl2. In parallel to optical density measurements, samples for isolation of the genomic DNA were taken every 24 h. Isolated genomic DNA, serving as a template, and primer pair flanking the 30-nt duplication within the BP3410 gene (Table S2) were used to monitor the loss of the duplication. PCR was performed with Herculase II fusion DNA polymerase (Agilent Technologies) in 20-μl reaction mixtures containing 0.1 μl of the polymerase, 5 nmol each deoxynucleoside triphosphate, 4% dimethyl sulfoxide (Sigma-Aldrich), and 5 pmol each forward and reverse primer. Each reaction consisted of an initial step at 98°C for 2 min, followed by 35 cycles of 98°C for 20 s, 62°C for 30 s, and 72°C for 30 s and finishing with an amplification step at 72°C for 5 min. The amplicons were visualized using Midori Green stain (Nippon Genetics) on 3% agarose gel. The PCR products of 329 bp and 299 bp indicated the presence and absence of the duplication, respectively.
Primers used in this study. Download Table S2, PDF file, 0.3 MB (283.8KB, pdf) .
Copyright © 2021 Čapek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RT-qPCR analysis.
Cells were grown in biological triplicates to mid-exponential phase and then incubated for 60 min in the presence or absence of 400 μM MnCl2. Next, cells were mixed with stop solution (95% ethanol, 5% phenol at 4:1 ratio), pelleted, and stored at −80°C. RT-qPCR analysis of BP3410 transcript levels was performed as described earlier (60). Briefly, bacterial pellets were suspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) containing 1 mg ml−1 lysozyme (Sigma-Aldrich), and total RNA was isolated from lysed cells using TRI reagent (Sigma-Aldrich) according to the manufacturer’s protocol. Removal of DNA was achieved by treatment of samples with DNase I (Sigma-Aldrich) for 15 min at room temperature. After organic extraction, the RNA was precipitated with 75% ethanol and dissolved in RNase-free water. RNA concentration was measured for each sample in technical triplicates using spectrophotometer DS-11 (DeNovix). Isolated RNA (600 ng) was reverse transcribed into cDNA using random hexamers and M-MLV reverse transcriptase (Promega) at 37°C in a 25-μl reaction mix according to the manufacturer’s instructions. RT-qPCRs were performed in at least three technical replicates per sample on a Bio-Rad CFX96 instrument using SYBR green JumpStart Taq ReadyMix (Sigma-Aldrich), 4 pmol of each primer, and 2 μl of 10× diluted cDNA in a 20-μl reaction. Each reaction consisted of an initial step at 95°C for 2 min, 40 cycles of 95°C for 15 s, 65°C for 30 s, and 72°C for 30 s, followed by melting curve recording. The rpoB gene was used as the reference gene and relative gene expression was determined using delta-delta threshold cycle method (62).
ICP-MS/MS analysis.
Throughout this experiment, B. pertussis strains were cultured in SS medium that was not supplemented with iron. Biological triplicates of overnight (≈18 h) cultures of each strain were diluted the next day to an OD600 of 0.2 in 20 ml of the fresh medium and split into two flasks, each containing 10 ml of culture. To the first culture, MnCl2 was added to a final concentration of 100 μM, while the second culture, lacking Mn, served as a control. All cultures were then grown in an orbital incubator for 8 h to allow for Mn accumulation. The OD600 was measured in all cultures to verify that manganese did not impose toxic effect on bacterial growth (i.e., OD600 of Mn-treated cultures was similar to that of control cultures). Next, cells were washed twice in phosphate-buffered saline (PBS; pH 7.4) containing 10 mM EDTA and once with PBS only. Finally, cells were suspended in PBS and normalized to the OD600 so that after centrifugation, each pellet contained cells corresponding to 2 OD600 units. Bacterial pellets were decomposed in 1 ml of 65% HNO3 (Honeywell) for 15 min at 80°C. An aliquot of 0.9 ml was then 10× diluted using 8.1 ml of deionized water. Determination of the intracellular Mn levels by ICP-MS/MS with triple quadrupole was carried out using an Agilent 8900 spectrometer equipped with an SPS 4 autosampler and ISIS3 sample introduction system. Mn was detected at m/z 55 in single-quad He mode (He, 4.5 ml min−1) and at m/z 55→55 and 55→71 in MS/MS O2 reaction mode (O2 40% gas flow rate) to ensure interference-free analysis. Yttrium (m/z 89 and 89→103) was used as an internal standard to correct for sensitivity drifts. Quantification was performed using six-point external calibration (0.125 to 4.0 μg liter−1 Mn). Calibration standards were prepared from Mn standard solution (1 g liter−1 Mn; Sigma-Aldrich) in 10× diluted HNO3. Results were processed using Agilent MassHunter 4.5 and Microsoft Excel software, and the result for each biological replicate is an average from two parallel ICP-MS/MS measurements at three tune modes. Three blank samples without bacteria were processed simultaneously with the experimental samples to determine background concentrations of Mn, which then were subtracted from values obtained with experimental samples.
Multiple-sequence alignment of calcium:cation antiporters.
The alignment of BP3410 exporter with homologous proteins was performed using complete BP3410 sequence as a query in a gapped BLAST search, which is integrated as a first step of PSI-BLAST (63); no further iterations were necessary. When needed, BLOSUM matrix was reduced to 45 in order to detect distantly related proteins. Representative sequence of each species with the highest score was included in the alignment. Additionally, the presence of two calcium:cation antiporter domains was verified by Pfam database search. NCBI accession numbers for each sequence were WP_010931461.1 (B. pertussis), WP_003820973.1 (B. bronchiseptica), WP_115522187.1 (A. veronii AG_5.28.6), WP_115522187.1 (A. veronii VCK_1), WP_115522187.1 (A. veronii BIOO50A), NEY27906.1 (E. coli), WP_164818558.1 (C. perfringens), WP_010869583.1 (M. jannaschii), PNW75261.1 (C. reinhardtii), NP_010155.1 (S. cerevisiae), OAO96401.1 (A. thaliana), NP_001104467.2 (D. melanogaster), XP_637590.1 (D. discoideum), EDL28124.1 (M. musculus), XP_018108579.1 (X. laevis), NP_499146.2 (C. elegans), AAS76476.1 (D. rerio), XP_025006903.1 (G. gallus), and AAM76071.1 (H. sapiens). Sequences were aligned by MUSCLE algorithm (64) built in MEGA 7.0.21 software (65) using the neighbor-joining clustering method, and the resulting alignment was refined by ESPript 3.0 (66). An exception to this procedure was the transporter Vcx1 from Saccharomyces cerevisiae, which has been included based on its known tertiary structure, as a gapped BLAST search did not return a positive result (67). The similarity between BP3410 and M. jannaschii exporter was calculated using the Needleman–Wunsch algorithm (68) using the BLOSUM62 scoring matrix.
Sensitivity to oxidative stress.
In the sensitivity to oxidative stress experiment, B. pertussis cells were cultivated in low-iron SS medium (i.e., medium was not supplemented with 36 μM FeSO4). To reduce the cytosolic content of manganese and iron in cultivated cells, cultures were passaged every 12 h (five passages in total). For the last passage, each culture was diluted to an OD600 of 0.2 into 20 ml of the fresh medium and split into two flasks, each containing 10 ml of culture. To the first culture, MnCl2 was added to a final concentration of 100 nM, while the second culture, lacking Mn, served as a control. All cultures were then grown for an additional 8 h to allow the cells to accumulate manganese. The cells were washed twice with PBS (pH 7.4) containing 10 mM EDTA and once with fresh low-iron SS medium. Finally, cells were resuspended in 1 ml of fresh SS medium and diluted to an OD600 of 0.05 in 8 ml of fresh low-iron SS medium containing 70 μM dipyridyl (Sigma-Aldrich) to chelate the remaining iron and 0.9 mM paraquat (Sigma-Aldrich) to induce the oxidative stress. Cultures were grown for 36 h, and then the optical density of cultures was monitored every 12 h.
Data availability.
All study data are included in the article and/or supporting information.
ACKNOWLEDGMENTS
This work was supported by grant 19-12338S (to B.V.) from the Czech Science Foundation (www.gacr.cz) and by funding from RVO61388971 and RVO68081715. We thank Václava Hejnarová from the Institute of Microbiology, Prague, for help with construction of the mutants and Rudy Antoine (Institute Pasteur, Lille) for helpful discussions.
J.C. performed bacterial growth experiments, site-directed mutagenesis, manganese resistance, and oxidative stress assays, with supervision from B.V. T.M. performed and analyzed ICP-MS/MS measurements with J.C.’s assistance. I.P. and J.C. constructed the mutants and performed and evaluated RT-qPCR assays with supervision from B.V. J.C. and I.P. performed bioinformatics analyses. B.V., D.H., and J.C. designed the study. B.V. and J.C. wrote the original draft of the manuscript. All authors contributed to writing the manuscript.
Footnotes
Citation Čapek J, Procházková I, Matoušek T, Hot D, Večerek B. 2021. A unique reverse adaptation mechanism assists Bordetella pertussis in resistance to both scarcity and toxicity of manganese. mBio 12:e01902-21. https://doi.org/10.1128/mBio.01902-21.
Contributor Information
Branislav Večerek, Email: vecerek@biomed.cas.cz.
Julian Parkhill, Department of Veterinary Medicine.
REFERENCES
- 1.Palmer LD, Skaar EP. 2016. Transition metals and virulence in bacteria. Annu Rev Genet 50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botella H, Stadthagen G, Lugo-Villarino G, de Chastellier C, Neyrolles O. 2012. Metallobiology of host-pathogen interactions: an intoxicating new insight. Trends Microbiol 20:106–112. doi: 10.1016/j.tim.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Weinberg ED. 1975. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA 231:39–41. doi: 10.1001/jama.1975.03240130021018. [DOI] [PubMed] [Google Scholar]
- 4.Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kehres DG, Maguire ME. 2003. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol Rev 27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 6.Lisher JP, Giedroc DP. 2013. Manganese acquisition and homeostasis at the host-pathogen interface. Front Cell Infect Microbiol 3:91. doi: 10.3389/fcimb.2013.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juttukonda LJ, Skaar EP. 2015. Manganese homeostasis and utilization in pathogenic bacteria. Mol Microbiol 97:216–228. doi: 10.1111/mmi.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnese K, Gralla EB, Valentine JS, Cabelli DE. 2012. Biologically relevant mechanism for catalytic superoxide removal by simple manganese compounds. Proc Natl Acad Sci USA 109:6892–6897. doi: 10.1073/pnas.1203051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inaoka T, Matsumura Y, Tsuchido T. 1999. SodA and manganese are essential for resistance to oxidative stress in growing and sporulating cells of Bacillus subtilis. J Bacteriol 181:1939–1943. doi: 10.1128/JB.181.6.1939-1943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng HJ, Srikhanta Y, McEwan AG, Jennings MP. 2001. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol Microbiol 40:1175–1186. doi: 10.1046/j.1365-2958.2001.02460.x. [DOI] [PubMed] [Google Scholar]
- 11.Tseng HJ, McEwan AG, Paton JC, Jennings MP. 2002. Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect Immun 70:1635–1639. doi: 10.1128/IAI.70.3.1635-1639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 13.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. 2011. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. 1993. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell 73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 15.Jabado N, Jankowski A, Dougaparsad S, Picard V, Grinstein S, Gros P. 2000. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J Exp Med 192:1237–1248. doi: 10.1084/jem.192.9.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosch JW, Gao G, Ridout G, Wang YD, Tuomanen EI. 2009. Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol Microbiol 72:12–25. doi: 10.1111/j.1365-2958.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veyrier FJ, Boneca IG, Cellier MF, Taha MK. 2011. A novel metal transporter mediating manganese export (MntX) regulates the Mn to Fe intracellular ratio and Neisseria meningitidis virulence. PLoS Pathog 7:e1002261. doi: 10.1371/journal.ppat.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner AG, Ong CL, Gillen CM, Davies MR, West NP, McEwan AG, Walker MJ. 2015. Manganese homeostasis in group A Streptococcus is critical for resistance to oxidative stress and virulence. mBio 6:e00278-15. doi: 10.1128/mBio.00278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grunenwald CM, Choby JE, Juttukonda LJ, Beavers WN, Weiss A, Torres VJ, Skaar EP. 2019. Manganese detoxification by MntE is critical for resistance to oxidative stress and virulence of Staphylococcus aureus. mBio 10:e02915-18. doi: 10.1128/mBio.02915-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kehres DG, Zaharik ML, Finlay BB, Maguire ME. 2000. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol 36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- 21.Que Q, Helmann JD. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 22.Zeinert R, Martinez E, Schmitz J, Senn K, Usman B, Anantharaman V, Aravind L, Waters LS. 2018. Structure-function analysis of manganese exporter proteins across bacteria. J Biol Chem 293:5715–5730. doi: 10.1074/jbc.M117.790717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waters LS, Sandoval M, Storz G. 2011. The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J Bacteriol 193:5887–5897. doi: 10.1128/JB.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padilla-Benavides T, Long JE, Raimunda D, Sassetti CM, Arguello JM. 2013. A novel P(1B)-type Mn2+-transporting ATPase is required for secreted protein metallation in mycobacteria. J Biol Chem 288:11334–11347. doi: 10.1074/jbc.M112.448175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, Foster SJ. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol 44:1269–1286. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- 26.Huang X, Shin JH, Pinochet-Barros A, Su TT, Helmann JD. 2017. Bacillus subtilis MntR coordinates the transcriptional regulation of manganese uptake and efflux systems. Mol Microbiol 103:253–268. doi: 10.1111/mmi.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, Johnston AWB. 2004. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn(2+)-responsive transcriptional regulator. Microbiology (Reading) 150:1447–1456. doi: 10.1099/mic.0.26961-0. [DOI] [PubMed] [Google Scholar]
- 28.Menscher EA, Caswell CC, Anderson ES, Roop RM, Jr.. 2012. Mur regulates the gene encoding the manganese transporter MntH in Brucella abortus 2308. J Bacteriol 194:561–566. doi: 10.1128/JB.05296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kehres DG, Janakiraman A, Slauch JM, Maguire ME. 2002. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H(2)O(2), Fe(2+), and Mn(2+). J Bacteriol 184:3151–3158. doi: 10.1128/JB.184.12.3151-3158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anjem A, Varghese S, Imlay JA. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol 72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wedekind JE, Dutta D, Belashov IA, Jenkins JL. 2017. Metalloriboswitches: RNA-based inorganic ion sensors that regulate genes. J Biol Chem 292:9441–9450. doi: 10.1074/jbc.R117.787713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dambach M, Sandoval M, Updegrove TB, Anantharaman V, Aravind L, Waters LS, Storz G. 2015. The ubiquitous yybP-ykoY riboswitch is a manganese-responsive regulatory element. Mol Cell 57:1099–1109. doi: 10.1016/j.molcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price IR, Gaballa A, Ding F, Helmann JD, Ke A. 2015. Mn(2+)-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol Cell 57:1110–1123. doi: 10.1016/j.molcel.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams MM, Sen K, Weigand MR, Skoff TH, Cunningham VA, Halse TA, Tondella ML, CDC Pertussis Working Group . 2016. Bordetella pertussis strain lacking pertactin and pertussis toxin. Emerg Infect Dis 22:319–322. doi: 10.3201/eid2202.151332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diavatopoulos DA, Cummings CA, Schouls LM, Brinig MM, Relman DA, Mooi FR. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog 1:e45. doi: 10.1371/journal.ppat.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeno-Tarraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, et al. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 38.Cummings CA, Brinig MM, Lepp PW, van de Pas S, Relman DA. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J Bacteriol 186:1484–1492. doi: 10.1128/JB.186.5.1484-1492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weigand MR, Peng Y, Loparev V, Batra D, Bowden KE, Burroughs M, Cassiday PK, Davis JK, Johnson T, Juieng P, Knipe K, Mathis MH, Pruitt AM, Rowe L, Sheth M, Tondella ML, Williams MM. 2017. The history of Bordetella pertussis genome evolution includes structural rearrangement. J Bacteriol 199:e00806-16. doi: 10.1128/JB.00806-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amman F, D'Halluin A, Antoine R, Huot L, Bibova I, Keidel K, Slupek S, Bouquet P, Coutte L, Caboche S, Locht C, Vecerek B, Hot D. 2018. Primary transcriptome analysis reveals importance of IS elements for the shaping of the transcriptional landscape of Bordetella pertussis. RNA Biol doi: 10.1080/15476286.2018.1462655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hot D, Slupek S, Wulbrecht B, D'Hondt A, Hubans C, Antoine R, Locht C, Lemoine Y. 2011. Detection of small RNAs in Bordetella pertussis and identification of a novel repeated genetic element. BMC Genomics 12:207. doi: 10.1186/1471-2164-12-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stainer DW, Scholte MJ. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol 63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 43.Smyrli M, Triga A, Dourala N, Varvarigos P, Pavlidis M, Quoc VH, Katharios P. 2019. Comparative study on a novel pathogen of European seabass. Diversity of Aeromonas veronii in the Aegean Sea. Microorganisms 7:504. doi: 10.3390/microorganisms7110504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao J, Li H, Zeng W, Sauer DB, Belmares R, Jiang Y. 2012. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science 335:686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 45.Carr RJ, Bilton RF, Atkinson T. 1986. Toxicity of paraquat to microorganisms. Appl Environ Microbiol 52:1112–1116. doi: 10.1128/aem.52.5.1112-1116.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aschner JL, Aschner M. 2005. Nutritional aspects of manganese homeostasis. Mol Aspects Med 26:353–362. doi: 10.1016/j.mam.2005.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linz B, Ivanov YV, Preston A, Brinkac L, Parkhill J, Kim M, Harris SR, Goodfield LL, Fry NK, Gorringe AR, Nicholson TL, Register KB, Losada L, Harvill ET. 2016. Acquisition and loss of virulence-associated factors during genome evolution and speciation in three clades of Bordetella species. BMC Genomics 17:767. doi: 10.1186/s12864-016-3112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seydlova G, Beranova J, Bibova I, Dienstbier A, Drzmisek J, Masin J, Fiser R, Konopasek I, Vecerek B. 2017. The extent of the temperature-induced membrane remodeling in two closely related Bordetella species reflects their adaptation to diverse environmental niches. J Biol Chem 292:8048–8058. doi: 10.1074/jbc.M117.781559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev 14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Bzymek M, Lovett ST. 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci USA 98:8319–8325. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bart MJ, Harris SR, Advani A, Arakawa Y, Bottero D, Bouchez V, Cassiday PK, Chiang C-S, Dalby T, Fry NK, Gaillard ME, van Gent M, Guiso N, Hallander HO, Harvill ET, He Q, van der Heide HGJ, Heuvelman K, Hozbor DF, Kamachi K, Karataev GI, Lan R, Lutyńska A, Maharjan RP, Mertsola J, Miyamura T, Octavia S, Preston A, Quail MA, Sintchenko V, Stefanelli P, Tondella ML, Tsang RSW, Xu Y, Yao S-M, Zhang S, Parkhill J, Mooi FR. 2014. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. mBio 5:e01074. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamidou Soumana I, Linz B, Harvill ET. 2017. Environmental origin of the genus Bordetella. Front Microbiol 8:28. doi: 10.3389/fmicb.2017.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cavalca L, Zanchi R, Corsini A, Colombo M, Romagnoli C, Canzi E, Andreoni V. 2010. Arsenic-resistant bacteria associated with roots of the wild Cirsium arvense (L.) plant from an arsenic polluted soil, and screening of potential plant growth-promoting characteristics. Syst Appl Microbiol 33:154–164. doi: 10.1016/j.syapm.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Bachate SP, Khapare RM, Kodam KM. 2012. Oxidation of arsenite by two beta-proteobacteria isolated from soil. Appl Microbiol Biotechnol 93:2135–2145. doi: 10.1007/s00253-011-3606-7. [DOI] [PubMed] [Google Scholar]
- 55.Seib KL, Tseng HJ, McEwan AG, Apicella MA, Jennings MP. 2004. Defenses against oxidative stress in Neisseria gonorrhoeae and Neisseria meningitidis: distinctive systems for different lifestyles. J Infect Dis 190:136–147. doi: 10.1086/421299. [DOI] [PubMed] [Google Scholar]
- 56.Choby JE, Skaar EP. 2016. Heme synthesis and acquisition in bacterial pathogens. J Mol Biol 428:3408–3428. doi: 10.1016/j.jmb.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mancini S, Imlay JA. 2015. The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress. Mol Microbiol 96:744–763. doi: 10.1111/mmi.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou D, Hardt WD, Galan JE. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect Immun 67:1974–1981. doi: 10.1128/IAI.67.4.1974-1981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kasuga T, Nakase Y, Ukishima K, Takatsu K. 1954. Studies on Haemophilis pertussis. III. Some properties of each phase of H. pertussis. Kitasato Arch Exp Med 27:37–47. [PubMed] [Google Scholar]
- 60.Bibova I, Skopova K, Masin J, Cerny O, Hot D, Sebo P, Vecerek B. 2013. The RNA chaperone Hfq is required for virulence of Bordetella pertussis. Infect Immun 81:4081–4090. doi: 10.1128/IAI.00345-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inatsuka CS, Xu Q, Vujkovic-Cvijin I, Wong S, Stibitz S, Miller JF, Cotter PA. 2010. Pertactin is required for Bordetella species to resist neutrophil-mediated clearance. Infect Immun 78:2901–2909. doi: 10.1128/IAI.00188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waight AB, Pedersen BP, Schlessinger A, Bonomi M, Chau BH, Roe-Zurz Z, Risenmay AJ, Sali A, Stroud RM. 2013. Structural basis for alternating access of a eukaryotic calcium/proton exchanger. Nature 499:107–110. doi: 10.1038/nature12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete alignment of BP3410 exporter homologues in other organisms. The amino acid sequences of the BP3410 exporter of B. pertussis and its selected homologues found in different organisms were aligned with the MUSCLE algorithm built in the MEGA software using the neighbor joining clustering method. The black asterisk indicates a highly conserved negatively charged residue essential for cation transport. Aligned residues that can be considered similar based on their physicochemical distance at a given position are shown in red. Similar residues found in at least 70 % of the aligned sequences are enclosed in blue frames. The triangles mark the duplicated amino acid sequence present only in B. pertussis and three recent isolates of A. veronii (red triangles), and the sequence present in all homologous proteins (blue triangles). The amino acid residues are numbered according to the B. pertussis BP3410 protein. Download FIG S1, PDF file, 0.03 MB (33KB, pdf) .
Copyright © 2021 Čapek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in the study. Download Table S1, PDF file, 0.3 MB (330.6KB, pdf) .
Copyright © 2021 Čapek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, PDF file, 0.3 MB (283.8KB, pdf) .
Copyright © 2021 Čapek et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All study data are included in the article and/or supporting information.