Abstract
The discovery of insulin 100 years ago and its application to the treatment of human disease in the years since have marked a major turning point in the history of medicine. The availability of purified insulin allowed for the establishment of its physiological role in the regulation of blood glucose and ketones, the determination of its amino acid sequence, and the solving of its structure. Over the last 50 years, the function of insulin has been applied into the discovery of the insulin receptor and its signaling cascade to reveal the role of impaired insulin signaling—or resistance—in the progression of type 2 diabetes. It has also become clear that insulin signaling can impact not only classical insulin-sensitive tissues, but all tissues of the body, and that in many of these tissues the insulin signaling cascade regulates unexpected physiological functions. Despite these remarkable advances, much remains to be learned about both insulin signaling and how to use this molecular knowledge to advance the treatment of type 2 diabetes and other insulin-resistant states.
Keywords: Insulin, Insulin receptor, Insulin signal transduction, Insulin resistance
Highlights
-
•
Insulin receptor signaling involves a complex network that includes tyrosine and serine/threonine phosphorylation events.
-
•
The first intracellular nodes of insulin action are the IRS proteins, which also serve as critical nodes in feedback and the heterologous regulation that promotes insulin resistance.
-
•
Insulin signaling is important for insulin action in both classical (liver, muscle and adipose) and unexpected (pancreatic β-cell, brain and vascular endothelium) tissues.
-
•
Insulin resistance in diabetes and metabolic syndrome is driven by many extrinsic factors and new cell-intrinsic factors that could be new therapeutic targets.
Abbreviations
- AC
adenylate cyclase
- ACADL
acyl-CoA dehydrogenase long chain
- ACACA
acetyl-CoA carboxylase
- ACh
acetylcholine
- AKT
V-Akt Murine thymoma viral oncogene homolog
- AMPK
adenosine monophosphate (AMP) activated kinase
- aPKC
atypical protein kinase C
- ARHGEF
Rho Guanine Nucleotide Exchange Factor
- AS160
Akt substrate of 160 kDa (also called TBC1D4)
- ATGL
adipose triglyceride lipase
- CaN
Calcineurin
- CAP
Cbl-associated protein
- CBL
Cas-Br-M (murine) ectopic retroviral transforming sequence
- ChREBP
carbohydrate response element binding protein
- CREB
cyclic AMP (cAMP) response element binding protein
- CRL7
Cullin–RING-type E3 ligase 7
- CRTC2
CREB regulated transcription coactivator 2
- CUL5
Cullin 5
- CUL7
Cullin 7
- DAG
diacylglycerol
- ELOB
Elongin B
- ELOC
Elongin C
- ERK1/2
Mitogen-activated protein kinase ½
- FA
fatty acid
- FBW8
F-Box and WD repeat domain containing 8
- FOXA2
hepatocyte nuclear factor 3-beta
- FOXK
Forkhead box protein K1
- FOXK2
Forkhead box protein K2
- FOXO1
Forkhead box protein O1
- G3P
glycerol-3-phosphate
- G6P
Glucose-6-Phosphate
- G6Pase
glucose-6-phosphatase
- GLP1
glucagon-like peptide 1
- GLP1R
glucagon like peptide 1 receptor
- GLUT2
glucose transporter 2
- GLUT4
glucose transporter 4
- Gq/G11/Ga
G-proteins
- GRB2
Growth factor receptor bound protein 2
- GSK3β
glycogen synthase kinase 3 beta
- HDAC
Histone deacetylase
- HNF1B
hepatocyte nuclear factor 1-beta
- HSL
hormone sensitive lipase
- IKKα
Inhibitor of nuclear factor kappa B kinase subunit alpha
- IKKβ
Inhibitor of nuclear factor kappa B kinase subunit beta
- IL6
Interleukin 6
- INFγ
Interferon Gamma
- INPPL1
Inositol Polyphosphate Phosphatase Like 1 (SHIP2)
- JAK
Janus kinase
- JNK
C-Jun N-Terminal Kinase 1
- KDM
Lysine demethylase
- KAT
Lysine acetyltransferase
- M3R
muscarinic 3 receptor
- MAPK
mitogen-activated protein kinases
- MEF2C
Myocyte enhancer factor 2C
- MEK1
Mitogen-activated protein kinase kinase 1
- mTORC1
mammalian target of rapamycin complex 1
- mTORC2
mammalian target of rapamycin complex 2
- NAD+
oxidized nicotinamide adenine dinucleotide
- NEMO
Inhibitor of nuclear factor kappa B kinase regulatory subunit
- NFAT
Nuclear factor of activated T-cells
- NGN3
neurogenin 3
- PC1/3
Prohormone convertase 1/3
- PDE3B
phosphodiesterase 3B
- PP2A
PP1, pTEN, C1-TEN and SHIP2. I
- PDHA1
Pyruvate Dehydrogenase E1 Subunit Alpha 1
- PDK1
phosphoinositide-dependent kinase-1
- PDX1
pancreatic and duodenal homeobox 1
- PEPCK
phosphoenolpyruvate carboxykinase
- PI(3,4,5)P3
phosphatidylinositol (3,4,5)-trisphosphate
- PI3K
phosphoinositide 3-kinase
- PKA
protein kinase A
- PKC
protein kinase C
- PLCβ
phospholipase C
- PPP1CA (PP1)
Protein Phosphatase 1 Catalytic Subunit Alpha
- PPP2CA (PP2A)
Protein Phosphatase 2 Catalytic Subunit Alpha
- PPP1R12A
Myosin phosphatase-targeting subunit 1 (MYPT1)
- PTEN
phosphatase and tensin homolog
- PTPN1 (PTP1B)
Protein Tyrosine Phosphatase Non-Receptor Type 1
- PTPN2 (TCPTP)
Protein Tyrosine Phosphatase Non-Receptor Type 2
- RAF
Raf proto-oncogene, serine/threonine kinase
- RBX1
Ring-Box 1
- RHEB
Ras homolog, mTORC1 binding
- RIP1
Receptor interacting serine/threonine kinase 1
- RNU1-1
RNA U1 Small Nuclear 1 (U1)
- RNU2-1
RNA U2 Small Nuclear 1 (U2)
- SETD
SET domain containing histone lysine methyltransferase
- SHC
SH3-containing protein
- SOS1
son of sevenless Ras/Rac guanine nucleotide exchange factor 1
- SR
Serine and arginine rich splicing factor
- SREBF1
sterol responsive element binding factor 1
- TAK1
Transforming growth factor-beta-activated kinase 1
- TBC1D1
TBC1 (Tre-2/USP6, BUB2, Cdc16) Domain Family, Member 1
- TC10
Ras Homolog Gene Family, Member Q
- TG
triglyceride
- TNFα
Tumor necrosis factor alpha
- TRADD
Tumor necrosis factor receptor type 1-associated DEATH domain protein
- TRAF
TNF receptor associated factor 1
- TSC1
Tuberous Sclerosis 1 Protein (Hamartin)
- TSC2
Tuberous Sclerosis 2 Protein (Tuberin)
1. Introduction
The discovery of insulin 100 years ago and its application to the treatment of human disease in the years since have marked a major turning point in the history of medicine [1]. Amazingly, insulin was not initially recognized as a peptide hormone, and virtually nothing was known about its mechanism of action. This all dramatically changed 50 years ago with the identification of the insulin receptor, initially through binding studies [2] and then, 10 years later, by the recognition that the receptor was a tyrosine kinase [3]. We now understand the importance of insulin and insulin-like growth factor signaling systems as integrators of metabolism, growth, and lifespan in species from C. elegans to Homo sapiens. In this brief review, we summarize the nature of this insulin signaling system and how knowing this system has led to better understanding of insulin-resistant states and diabetes. It is also important to realize that despite huge advances, much remains to be learned about both insulin signaling and how to use this knowledge to advance the treatment of type 2 diabetes and other insulin-resistant states.
2. Insulin receptor tyrosine kinase
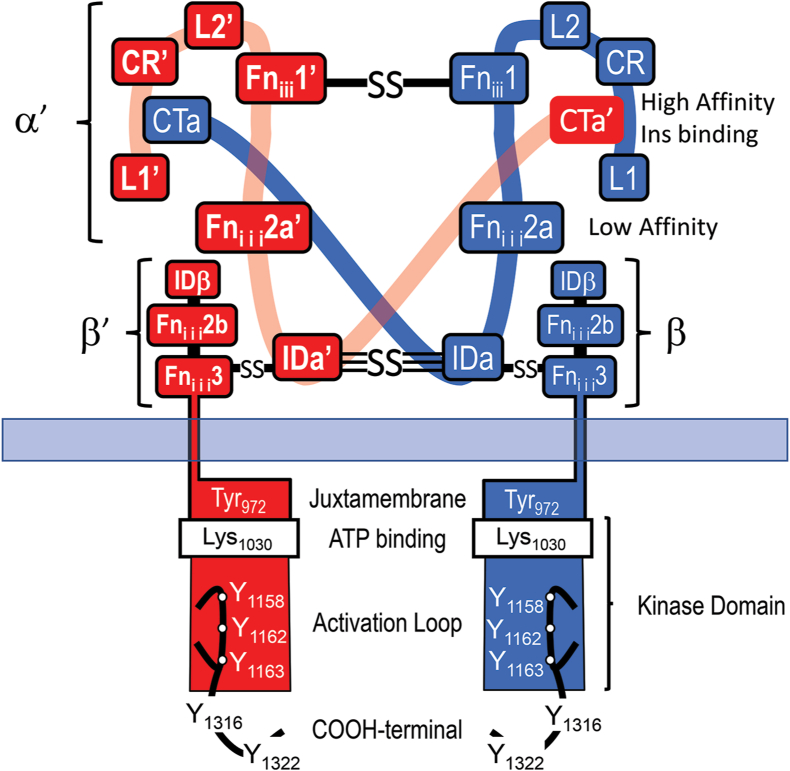
At the cellular level, insulin initiates action by binding to its membrane receptor. The insulin receptor (InsR) is encoded by a 150-kb gene composed of 22 exons on human chromosome 19p13.3-p13.2. During synthesis, a single-chain InsR proreceptor is formed, glycosylated, linked into a dimer by disulfide bonds, and cleaved to generate the α- and β-subunits that create the α2β2 tetramer (Figure 1) [4,5]. The extracellular domain is composed of the entire α-subunit and the NH2-terminal portion of the β-subunit. The α-subunit contains two leucine-rich (L1 and L2) repeats flanking a cysteine-rich region (CR). These are followed by three fibronectin-III (Fniii) motifs, which are interrupted by a 120 amino acid insert domain (ID) that contains the furin cleavage site allowing separation of the α- and β-subunits (Figure 1).
Figure 1.
Schematic diagram of the mature insulin receptor, composed of two extracellular α-subunits (red and blue) and two covalently linked transmembrane β-subunits. Contiguous modules of the α-subunits are labeled with the relative location of disulfide bonds (S–S) between the α- and β-subunits. The high-affinity insulin binding site is created from the L1-CR and CTa’ or L1′-CR′ and CTa domains of the disulfide-linked α- and α′-subunits. The β-subunit is formed upon furin-mediated cleavage of the ID region into IDα and IDβ. The COOH terminus of the Fniii2 and Fniii3 domains forms after the furin cleavage site is separated from the intracellular juxtamembrane region by the hydrophobic transmembrane domain. The tyrosine kinase catalytic domain, including the canonical ATP binding site (Lys1030) and the activation loop with three tyrosine phosphorylation sites, follows immediately after the juxtamembrane region. The β-subunit ends with two tyrosine phosphorylation sites in the COOH terminus.
One of the remarkable insights of the past few years has been visualization of the receptor in its three-dimensional structure. X-ray crystallography and cryo-EM studies reveal that the unoccupied extracellular domain folds into an inverted V, with the apex formed by the L2 and Fniii1 of each α-subunit (Figure 1) [6]. Mutagenesis and affinity-labeling studies predict two insulin binding sites in the α-subunit: a high-affinity site-1, created by the L1 domain from one α-subunit and the C-terminus of the other α-subunit, and site-2, located near the Fniii1→Fniii2 interface (Figure 1) [[6], [7], [8], [9], [10]]. Exon-11 of the InsR gene is alternatively spliced to produce two isoforms (IR-A, which omits exon-11, and IR-B, which includes exon-11), adding 12 amino acids at the C-terminus of the α-subunit [11]. This constitutes part of the insulin binding domain, thus increasing the affinity for insulin [12,13]. These two forms of InsR and the homologous IGF1R can form heterodimers, allowing a total of five receptor “subtypes,” which interact with insulin, IGF-1, and IGF-2 with differing affinities.
Even before its cloning, studies using patient-derived InsR autoantibodies revealed that the InsR β-subunit was a tyrosine kinase [3]. This was a remarkable observation, since at that time, only the epidermal growth factor receptor was known to have this enzymatic activity. Two groups sequenced the InsR cDNA four years later to confirm this discovery [14,15]. Structurally, the intracellular portion of the β-subunit is composed of three regions, each containing clusters of tyrosine autophosphorylation sites: Y965 and Y972 (numbered as in IR-B) in the juxtamembrane domain (JMD), Y1158, Y1162 and Y1163 in the activation loop of the kinase domain, and Y1328 and Y1334 in the carboxy-terminus [16,17] (Figure 1). Insulin binding to sites-1 and -2 converts the ECD into a T-conformation that promotes a convergence of the intracellular domains, thus facilitating full activation of the receptor kinase toward exogenous substrates by tyrosine transphosphorylation [ [18] and article by Lawrence in this issue].
3. The proximal insulin signaling cascade
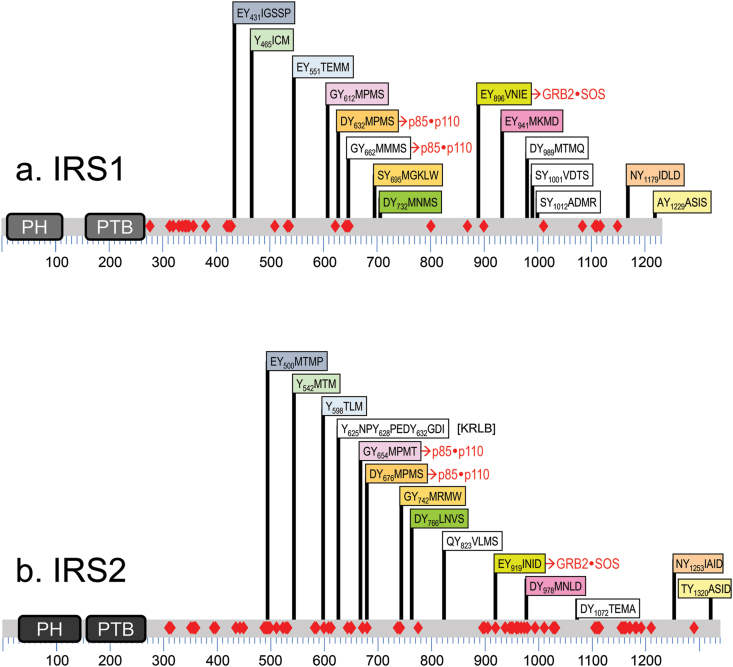
Evidence for an insulin receptor substrate came from anti-phosphotyrosine antibody immunoprecipitates of insulin-stimulated hepatoma cells [19]. Purification and cloning of this protein revealed IRS1, the first member of the insulin receptor substrate family [20]. Two other homologous IRS proteins are present in humans (IRS2 and IRS4), and a fourth (IRS3) in rodents [21]. IRS1 and IRS2 are broadly expressed, whereas IRS3 and IRS4 are tissue-restricted [22]. All IRS proteins have tandem PH (pleckstrin homology) and PTB (phosphotyrosine binding) domains, important for membrane and receptor association, followed by a long unstructured tail containing 14 tyrosine phosphorylation sites (Figure 2). IRS proteins also contain >50 serine/threonine phosphorylation sites (shown as red diamonds in Figure 2), which modulate stability and tyrosine phosphorylation for feedback and heterologous regulation [23]. Upon insulin stimulation, the IRS proteins are recruited to a phosphorylated NPEpY972 motif in the juxtamembrane region of the InsR [24], which facilitates phosphorylation of the tyrosine residues in the IRS tail. These in turn bind to the SH2 domains in various downstream signaling proteins, the most metabolically important of which is the p85 regulatory subunit of the class 1A PI3K (phosphatidylinositol 3-kinase) (Figure 3) [25]. Tyrosine phosphorylation of other sites in IRS-1 (or the alternative substrate SHC) recruit the Grb2•SOS complex, which activates the RAS→MAP kinase cascade, a second major branch of insulin signaling (Figure 3). The recruitment of IRS to the activated InsR/IGF1R is highly regulated and adds an essential level of signaling specificity [24,26,27].
Figure 2.
Comparison of (a) IRS1 and (b) IRS2. Alignments of IRS1 and IRS2 tyrosine phosphorylation sites relative to the amino-terminal pleckstrin homology (PH) and phosphotyrosine binding (PTB) domains. Conserved tyrosine phosphorylation motifs-including their number in the human protein and the surrounding amino acid sequences-are color-coded to highlight alignments between the isoforms: white boxes indicate unique sites in IRS1 or IRS2, including the KRLB (kinase regulatory loop binding) domain in IRS2 located around Y624 in IRS2. The relative position of Ser/Thr phosphorylation sites in IRS1 or IRS2 revealed by MS/MS are indicated with red diamonds.
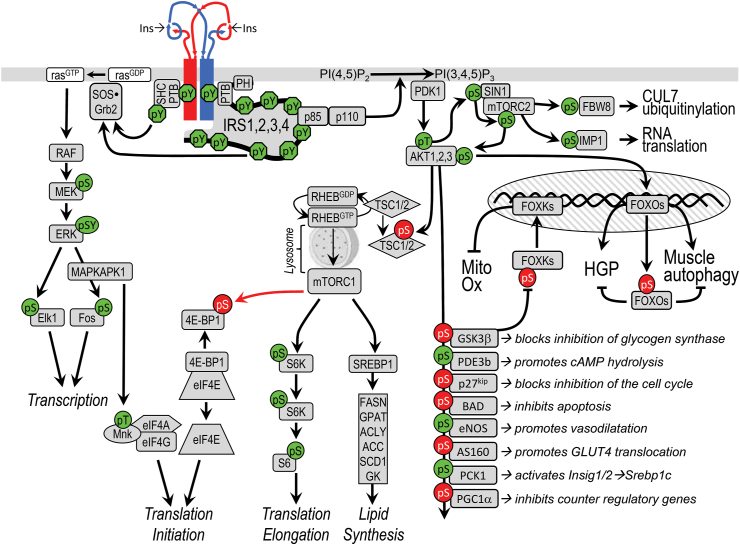
Figure 3.
A canonical insulin/IGF signaling cascade. The InsR subunits are illustrated at the top in red and blue. InsR signals begin with tyrosine phosphorylation of the IRS or SHC. The IRS protein binds and activates the PI3K, which generates PI3,4P2 and PI3,4,5P3 that recruit PDK1, SIN1, and AKT to the plasma membrane. AKT is activated upon phosphorylation at T308 by PDK1 and at S473 by the SIN1•mTORC2 complex. mTORC1 is activated by RhebGTP, which accumulates upon inhibition of the GAP activity of the TSC1•TSC2 complex following AKT-mediated phosphorylation of TSC2. mTORC1-mediates phosphorylation of S6K and SREBP1, which promote protein and lipid synthesis, respectively. AKT phosphorylates many cellular proteins, inactivating PGC1α, p21kip, GSK3β, BAD, and AS160 and activating PDE3b, PCK1, and eNOS. AKT-mediated phosphorylation of FOXO1 and FOXK causes their sequestration in the cytoplasm, which inhibits their influence upon transcriptional activity. GRB2•SOS can bind to IRS or SHC. The Grb2/SOS complex promotes GDP/GTP exchange on p21ras, which activates the ras→raf→MEK→ERK1/2 cascade. Activated ERK stimulates transcriptional activity by direct phosphorylation of ELK1 (ETS domain-containing protein) and by indirect phosphorylation of cFOS through MAPKAPK1 (MAPK-activated protein kinase-1). MAPKAPK1 also phosphorylates other proteins, including S6 (ribosomal protein S6), NFκB, PP1, and MYT1. Insulin stimulates protein synthesis by altering the intrinsic activity or binding properties of key translation initiation and elongation factors (eIFs and eEFs, respectively) as well as critical ribosomal proteins. mTORC1-mediated phosphorylation of 4E-BP1 and S6K plays an important role in stimulating translation initiation and elongation [270]. Stimulatory phosphorylation sites are highlighted in green, and inhibitory sites are highlighted in red.
4. The PI3K-AKT signaling cascade
The PI3K cascade begins when insulin stimulates the tyrosyl phosphorylation of two YMPM-motifs in IRS proteins, which bind and activate the PI3K (Figure 3) [28]. PI3K is composed of a catalytic and a regulatory subunit, each occurring in multiple isoforms encoded by multiple genes. The catalytic subunit—including p110α, p110β, p110δ, and p110γ—is stabilized and inhibited by its association with one regulatory subunit—including p85α or an alternatively spliced isoforms (p55α, p50α), p85β, or p55γ. All the regulatory subunits contain two SH2 domains that bind the phosphorylated YMPM-motifs in IRS1 or IRS2. This binding disinhibits the catalytic subunit's production of PIP3 (phosphatidylinositol 3,4,5-trisphosphate) [[29], [30], [31]]. Chemical or genetic inhibition of PIP3 production blocks almost all metabolic responses stimulated by insulin—including glucose uptake, glycogen and lipid synthesis, and adipocyte differentiation—confirming that the PI3K is a critical node in insulin's action [32,33].
PIP3, in turn, activates PDK1 and SIN1 (also called MAPKAP1). The former phosphorylates AKT at Thr308, while the latter complexes with mTORC2 to phosphorylate AKT at Ser473 [34]. This activates AKT, which phosphorylates consensus RXRXX (pS/pT)Ψ motifs in more than 100 substrates (Figure 3) [35]. AKT1 mainly regulates growth, development, and survival, whereas AKT2 regulates metabolism via GLUT4 translocation and glucose and lipid metabolism [36]. Humans with a dominant negative mutation in AKT2 display features of T2DM [37].
Insulin also activates mTOR (mechanistic target of rapamycin). This is a Ser/Thr kinase that forms two functionally-distinct protein complexes, mTORC1 and mTORC2, both controlled by insulin/IGF1 and other growth factors [38]. In the absence of stimulation, mTORC1 is inhibited through complexing with the TSC1•TSC2 (hamartin-tuberin) complex until AKT phosphorylates and inhibits the GTPase activity of TSC2, which activates RHEB (Figure 3). FKBP8 (FK506 binding protein 8) also inhibits mTORC1 until RHEB•GTP promotes its dissociation from mTORC1 [39]. Regulation by TSC1•TSC2 activation of RHEB is also modulated by AKT-mediated phosphorylation of AKT1S1 (PRAS40), which promotes its dissociation from RAPTOR and thus activates mTORC1, potentiating the role of insulin in growth and proliferation (Figure 3) [40]. mTORC2, on the other hand, in addition to taking part in activating AKT, plays a role in mRNA processing by phosphorylation of IMP1 (Figure 3) [41].
Another important target of AKT is the Forkhead box O family of transcription factors (FOXO1, FOXO3a, FOXO4, and FOXO6). FOXO nuclear localization is regulated by multiple posttranslational modifications, but especially AKT-mediated phosphorylation, which leads to nuclear exclusion of FOXOs following insulin stimulation, turning off their gene targets [42,43]. This leads to decreased glucose production in the liver, decreased autophagy and protein degradation in the muscle, increased adipose differentiation [[44], [45], [46], [47]], and decreased hepatic IGFBP1 expression to increase circulating IGF1 bioavailability and somatic growth [48].
5. Mutations in the insulin signaling system
Because of the critical nature of insulin signaling, mutations in these key proteins are rare. Regardless, the investigation of syndromes of insulin resistance have been extremely informative. Firstly, some patients have insulin resistance due to autoantibodies to the receptor, which provided the first tool for identifying receptor autophosphorylation even before its cloning [3]. Secondly, other patients with severe insulin resistance revealed the first naturally occurring mutations in the insulin receptor. These exhibit several clinical subtypes. Those with mutations in the kinase domain usually present as type A syndrome of insulin resistance and acanthosis nigricans, whereas those with mutations in the extracellular domain can have more severe phenotypes, exemplified by Donohue and Rabson-Mendenhall syndromes [49,50]. While InsR mutations can cause severe insulin resistance, leading to high insulin requirements (>10,000 U/day), some individuals remain near normoglycemic due to massive elevations of endogenous insulin secretion [49]. Loss-of-function mutations in AKT2 are extremely rare, but can also result in severe forms of insulin resistance [[51], [52], [53], [54], [55], [56]]. A natural human variant in IRS1 with a Gly972Arg substitution that reduces insulin-stimulated PI3K signaling is relatively common [57]; however, this polymorphism plays only a minor role in T2DM risk [[51], [52], [53], [54], [55], [56]].
6. Dissecting the insulin signaling system in mice by tissue-specific gene inactivation
6.1. Skeletal muscle
Using genetic approaches in mice, we and others have dissected the InsR and IGF1R signaling pathways in vivo in virtually every tissue in the body. Despite the essential role of muscle as a site for glucose uptake, as evidenced by marked hyperglycemia in muscle GLUT4 knockout mice [58,59], deletion of InsR in skeletal muscle in MIRKO mice results in only mild obesity and elevated circulating FFA and triglycerides, but without elevated glucose and insulin [53,60]. Indeed, even combined deletions of InsR and IGF1R in MIGIRKO mice results in a major reduction in muscle mass, while glucose and insulin tolerance remain normal (Figure 4A). The latter is due at least in part to increased basal glucose uptake, indicating alternative pathways for activation of glucose transport [61]. Likewise, mice with muscle-specific Irs1/Irs2 double knockout (MDKO) also fail to develop hyperglycemia, despite progressive and severe loss of skeletal and cardiac muscle due to unrestrained autophagy [62]. Again, isolated skeletal muscles from MDKO mice show elevated basal glucose uptake, elevated AMP/ATP ratio, and increased AMPK (AMP-activated protein kinase) activity (Figure 4A) [58,59,62,63]. The marked muscle atrophy in the absence of InsR and IGF1R is due to unrestrained activity of FOXOs promoting autophagy (Figure 4A). Thus, deletion of FOXO1, FOXO3, and FOXO4 can prevent muscle loss in MIGIRKO mice [64] and also prevents the muscle atrophy observed in insulin deficient diabetes [46].
Figure 4.
Tissue-specific insulin signaling. The insulin receptor is autophosphorylated on multiple tyrosine residues, allowing the docking and activation of multiple signaling molecules, most notably insulin receptor substrate (IRS) proteins. This in turn activates phosphatidylinositol-3-kinase (PI3K) and Akt to mediate the increases in glucose uptake and metabolism as well as changes in protein and lipid metabolism. While the general pathway is similar in all tissues, the final biological effects are specialized to the roles of insulin in muscle (a), liver (b), and adipose tissue (c).
6.2. Liver
Liver-specific InsR knockout (LIRKO) mice, on the other hand, display moderately elevated fasting and postprandial glucose levels and severe hyperinsulinemia [65]. The latter is due to a combination of increased insulin secretion and reduced hepatic insulin degradation [66]. LIRKO mice also display reduced levels of circulating free fatty acids (FFA) and triglycerides [53,65], and on an atherogenic diet develop dyslipidemia that can progress to atherosclerosis [67]. Insulin receptor deletion dysregulates hundreds of hepatic genes, including reduced GCK (glucokinase) and elevated PCK1 (phosphoenolpyruvate carboxykinase 1), G6PC (glucose-6-phosphatase, catalytic subunit), and PK1 (pyruvate kinase) (Figure 4B) [53,65]. The chronic hyperinsulinemia in LIRKO mice also leads to insulin resistance in other tissues. Thus, somewhat paradoxically, streptozotocin treatment to reduce insulin secretion improves peripheral insulin sensitivity [56,[68], [69], [70]]. Genetic inactivation of Irs1 and Irs2 or Akt1 and Akt2 in the liver in mice [56,[71], [72], [73]] resembles the LIRKO mouse with unsuppressed HGP, hyperinsulinemia, glucose intolerance, and diabetes, consistent with the genes’ roles as critical nodes in insulin action [36,66,[71], [72], [73]]. As with muscle, hepatic inactivation of FOXO1 in either LIRKO or liver IRS1/2 or Akt1/2 double-knockout (DKO) mice reverses dysregulated hepatic gene expression and restores metabolic health, despite lack of upstream insulin signaling (Figure 4B) [36,56,[74], [75], [76]]. At least part of this improvement is due to reversing the increased levels of the FOXO1-dependent hepatokine, follistatin (Fst), which promotes WAT lipolysis and thus propagates systemic metabolic disease during hepatic insulin resistance [56].
6.3. Adipose tissue
Genetic inactivation of insulin/IGF1 signaling in adipose tissue produces different phenotypes depending on the specific promoter used to drive adipocyte Cre expression. The initial fat insulin receptor knockout mice (FIRKO) created using the aP2-Cre transgene displayed partial lipodystrophy and increased longevity [77,78], whereas FIRKO mice created using the more potent adiponectin-Cre displayed more severe lipodystrophy and NAFLD that progressed to NASH and liver dysplasia [79]. Perhaps the most interesting of these models are mice with an inducible fat-specific knockout of IR or IR/IGF1R created using the tamoxifen-regulated adiponectin-Cre [80]. Following induction of recombination, these mice rapidly develop severe lipodystrophy and systemic insulin resistance with β-cell hyperplasia; however, as new adipocytes develop from preadipocytes that have not undergone gene inactivation, this syndrome totally reverses. Finally, in recent unpublished work, Homan et al. have produced mice with a combined knockout of the IR and IGF1R as well as all three FOXO proteins expressed in fat (Foxo1, 3, and 4). As in liver and muscle, deletion of FOXOs rescues much of the phenotype created by IR/IGF1R KO, but in this case the extent of rescue depends on the adipose depot, with complete rescue of brown adipose tissue mass, partial rescue of subcutaneous adipose mass, and no rescue of perigonadal adipose mass indicating differential roles of InsR, IGF1R, and FOXO proteins in adipocyte subtypes (Figure 4C).
7. Insulin action in non-classical target tissues as revealed by gene knockout
7.1. Cardiovascular system
Genetic tools for tissue-specific knockout have provided unique insights into the role of insulin action in tissues not recognized as classical insulin targets. Genetic deletion of insulin and IGF-1 receptors in the heart reveals the essential role of InsR/IGF1R in cardiac development and function [81]; however, absence of InsR alone only leads to changes in potassium channel expression and ventricular repolarization [82]. Deletion of myocardial InsR also decreases VEGF expression, impairing reactive angiogenesis following ischemia reperfusion damage or myocardial infarction [83]. Similarly, muscle deletion of Irs and Irs2 causes severe left ventricular failure in mice between 3 and 4 weeks of age. At the same time, retention of a single allele of Irs1 or Irs2 can prevent sudden death, suggesting that either can mediate enough IR/IGF1R signaling to prevent excess autophagy and fatal cardiac muscle loss [62]; however, Irs1 and Irs2 play different roles. Thus in conditions of pressure overload modeled in mice, cardiac deletion of Irs1 attenuates Akt1 activation, preventing LV remodeling and heart failure. By contrast, deletion of cardiac Irs2 leads to severe LV dysfunction that can be prevented by haploinsufficiency of Akt1 [83,84]. Therefore, InsR, Irs1, and Akt1 are critical signaling nodes required for normal LV remodeling in both mice and humans [84].
Cardiovascular disease and myocardial infarction are major causes of morbidity associated with insulin resistance and T2DM [85]. The progression of atherogenesis can involve dysregulated crosstalk between immune cells, endothelial cells (ECs), and vascular smooth muscle cells (VSMCs) [86]. Deletion of InsR or IGF1R in VSMC shows that the InsR (but not IGF1R) mediates VSMC proliferation and intimal hyperplasia following intimal injury [87]. In contrast, in ApoE-deficient mice bred with mice lacking endothelial InsR, atherosclerotic lesion size increases more than 2-fold [88]. Likewise, ApoE KO mice bred with whole-body IRS2 heterozygous KO mice have increased aortic intima thickness [89]. Endothelial cell-specific IRS2 KO also impairs eNOS phosphorylation, muscle insulin delivery, and glucose uptake [54,90], indicating a special role for IRS2 signaling in cardiovascular complications of diabetes.
7.2. Pancreatic β-cells
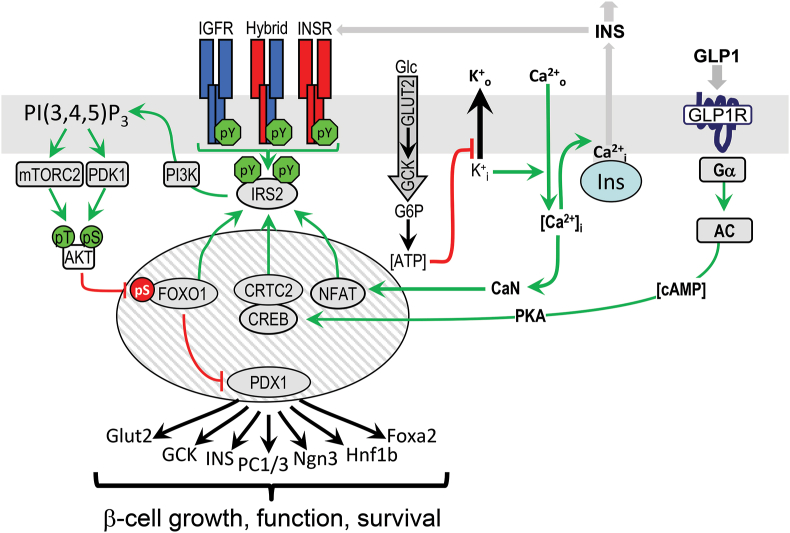
The capacity of pancreatic β-cells to maintain glucose homeostasis during physiologic stress is important for metabolic homeostasis. Since β-cells were thought to always be exposed to high insulin levels, β-cells were initially assumed to be non–insulin responsive, or if they did respond, this was considered part of a negative feedback loop to suppress insulin secretion [91]. However, knockout of InsR in β-cells revealed important roles of insulin signaling in both β-cell growth and glucose sensing [92], which appears to be regulated through IRS2 as mice with IRS2 KO develop β-cell failure (Figure 5) [93]. These results should still be interpreted cautiously, as these experiments were conducted with Rip-Cre mice (B6.Cg-Tg (Ins2-cre)25Mgn/J; JAX Stock No: 003573), which can express Cre at low levels in the hypothalamus [94]. Regardless, β-cell IRS2 expression is known to be regulated through the actions of FOXO1/3, NFAT, and the CREB•CRTC2 [91]. Glucose-stimulated Ca2+ release activates calcineurin, which dephosphorylates NFAT to facilitate its entry into the nucleus, where it induces expression of IRS2 and other genes [95]. Glucose, GLP-1 (glucagon-like peptide-1), and other GPCR agonists also promote IRS2 transcription via increased cAMP [96,97]. IRS2 expression through these mechanisms maintains PDX1 action, which is essential for β-cell growth, function, and survival (Figure 5) [98]. Compounds that augment IRS2 expression may be useful for treatment of β-cell failure during insulin resistance and the progression of T2DM [99].
Figure 5.
The integrative role of IR/IGF1R→IRS2 signaling in pancreatic β-cell function. The diagram shows the relation between the IRS2-branch of the insulin signaling pathway and upstream and downstream mechanisms regulating β-cell growth and function. Since IR and IGF1R are constitutively active in β-cells, activation of GLP1→cAMP→PKA→CREB, glucose→Ca2+→CRTC2, and calcineurin→NFAT induce IRS2 expression to regulate PI3K→AKT cascade, which places β-cell growth, function, and survival under the control of glucose and incretins.
7.3. Brain
Insulin resistance, obesity, and T2DM are associated with age-related cognitive decline and ADRD (Alzheimer's Disease and Related Dementia) [[100], [101], [102]]. How insulin resistance promotes cognitive decline, however, is still only partially understood. In the brain, insulin receptor substrates IRS1, IRS2, and IRS4 link IR-A, IGF1R, and their hybrids to downstream pathways. The neurotrophin receptors TrkA and TrkB, which regulate synaptic strength and plasticity, also engage IRSs in neurons [103,104]. Insulin action in the central nervous system and its effects on mood, behavior, and cognition are apparent in mice with brain knockout of InsR [105] or localized knockout of both insulin and IGF-1 receptors in the hippocampus and amygdala [106]. Alterations in insulin signaling proteins have also been observed in brain samples from humans with Alzheimer's Disease [101,107,108] and mouse models of AD [109], suggesting that enhancing central insulin action could be therapeutic in AD [100]. Mice with brain KO of InsR show increased tau phosphorylation [110]. Brains of AD patients show increased Ser/Thr phosphorylation of IRS1 [107,109,111]. On the other hand, Irs2 deficiency in the brain appears to increase lifespan and improve memory formation in mice [[112], [113], [114], [115]], despite producing obesity, peripheral insulin resistance, and hyperinsulinemia [105,112,[116], [117], [118], [119]]. Similarly, decreasing Irs2 improves motor performance and extends lifespan in a mouse model of Huntington's disease (R6/2), and this is associated with reduced neuronal oxidative stress and enhanced autophagy and mitochondrial function [120]. Lower Irs2 also increases nuclear FOXO1 in R6/2-mice [120], which contribute to prevention of age-related axonal degeneration [121].
8. Regulation of proximal insulin signals
Genetic experiments in mice have established that changes in a broad array of insulin signaling components, nutrient sensors, and their downstream effectors can have profound effects upon insulin sensitivity, β-cell function, and nutrient homeostasis. Indeed, a 50 % reduction in the concentration of IR and IRS1 will cause diabetes in mice [122].
8.1. Transcriptional regulation of IRS1 and IRS2
Physiologically, IRS1 expression is regulated by transcriptional repressors (including the transcription factor AP2β) or the p160 family of nuclear receptor coactivators p/CIP (p300/CBP/cointegrator-associated protein) and SRC1 (steroid receptor coactivator-1) [123,124]. Interestingly, GWAS reveals AP2β as a potential root of obesity and T2DM [125]. In contrast, p/CIP and SRC1 serve as transcriptional coactivators, and increased IRS1 expression following inactivation of p/CIP and SRC1 in mice results in increased glucose uptake and enhanced insulin sensitivity in WAT and skeletal muscle. Finally, muscle-specific knockout mice of the TAZ transcription factor display decreased IRS1 expression and insulin sensitivity [126]. Statins reduce TAZ levels, which may contribute to the insulin resistance observed in some patients on statins.
Expression of IRS2 is regulated by multiple factors, many of which respond to nutrients and energy, such as CREB (cAMP response element binding protein) and its coactivator CRTC2 [127,128]. Regulation of IRS2 by FOXO creates a direct feedback loop to augment insulin signaling during fasting. Fasting and exercise also induce the CREB•CRTC2 transcriptional complex through glucagon signaling; this increases expression of gluconeogenic genes as well as IRS2 [129]. Increases in active hepatic SREBP1c in situations of nutrient excess or chronic insulin stimulation decrease IRS2 expression, creating insulin resistance while promoting lipogenesis [130,131]. Thus, IRS2 expression is regulated through multiple metabolic sensors that modulate insulin sensitivity through feedback and heterologous mechanisms to maintain metabolic homeostasis.
8.2. Regulation of IRS degradation
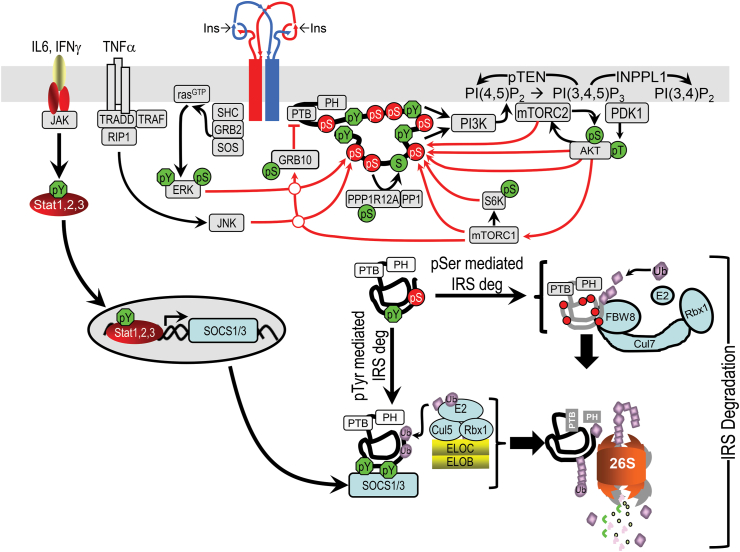
IRS1 and IRS2 can also be regulated by degradation. Both proteins undergo poly-ubiquitinylation during inflammation, chronic nutrient excess, or hyperinsulinemia through various mechanisms, including SOCS1/3 and ubiquitin-mediated degradation (Figure 6) [132,133]. IRS Ser/Thr phosphorylation (see below) also coordinates ubiquitin-mediated degradation [134,135]. Caloric excess induces CBL-Proto-Oncogene B expression, which drives insulin resistance through the polyubiquitinylation and degradation of IRS1 [136].
Figure 6.
Schematic diagram of feedback and heterologous regulation of the insulin signaling cascade. Various kinases in the insulin signaling cascade mediate feedback of Ser/Thr phosphorylation of IRS1/2—including AKT, mTOR, S6K, ERK, and AKT [137]. Other kinases activated by heterologous signals, including IL6, INFγ, and TNFα are also illustrated. Serine phosphorylation of IRS1 can recruit CRL7, which can promote ubiquitination and degradation of IRS1 through the 26S proteasome. Many proinflammatory cytokines cause insulin resistance through SOCS1 or SOCS3, targeting phosphotyrosine-containing IRS1 or IRS2 for ubiquitination by a BC-containing ubiquitin ligase (E3) and degradation [271,272].
8.3. Regulation of IRS signaling by Ser/Thr phosphorylation
Perhaps more important than changes in protein levels, regulation of IRS1 and IRS2 occurs through a complex mechanism involving phosphorylation of more than 50 serine/threonine (Ser/Thr) phosphorylation sites, located mainly in the long unstructured tail of the molecule (See Figure 2, Figure 6). Understanding how phospho-Ser/Thr regulates insulin signaling, however, has been challenging because a multitude of sites and mechanisms can be involved [23]. Proinflammatory cytokines, excess free fatty acids, ceramides, amino acids and glucose, and endoplasmic reticulum stress have all been implicated in increased IRS1/2 Ser/Thr phosphorylation and reduced insulin-stimulated tyrosine phosphorylation [23]. Most IRS1/2 Ser/Thr phosphorylation is stimulated by the PI3K→Akt→mTOR cascade during insulin stimulation [23,137], suggesting that IRS1/2 phospho-Ser/Thr is likely a feedback mechanism that develops during chronic insulin stimulation and can be co-opted by metabolic stress to inhibit insulin signaling (Figure 6) [23,137]. This may also be a link between hyperinsulinemia and insulin resistance.
In mice, phosphorylation of Ser307 (equivalent to human IRS-1 Ser312) is often used as a barometer of insulin resistance [23]. Insulin can promote IRS1-Ser307 phosphorylation through a pathway involving PI3K→AKT→mTORC1→S6K1 (Figure 6) [137]. This phosphorylation is also stimulated by free fatty acids via activation of JNK1 (c-Jun N-terminal kinase) or mTORC1 [23]. Surprisingly, however, knock-in of alanine to replace Ser307 and eliminate the phosphorylation site increases insulin resistance, rather than decreasing it [138]. Likewise, alanine substitution at Ser302, a prime target of mTORC1→S6k mediated phosphorylation, does not prevent insulin resistance [139], indicating that other factors must be involved [[132], [133], [134], [135], [136]].
8.4. Modulation of insulin signaling by protein and lipid phosphatases
Phosphatases modulate insulin signaling by dephosphorylating key proteins or lipids in the signaling cascade. Numerous phosphatases are involved, including PTPN1 (PTP1B), PTPN2 (TCPTP), PP1, PP2A, pTEN, C1-TEN, and SHIP2. Inactivation of PTP1B, a phosphotyrosine phosphatase that can dephosphorylate InsR, increases insulin sensitivity in mice [140]. Inactivation of PTP1B maintains β-cell mass in mice lacking IRS2, preventing an early onset of diabetes [141]. Inhibition of specific hypothalamic neurons or PTP1B or TCPTP in the brain promotes insulin and leptin signaling and prevents diet-induced obesity, T2DM, and fatty liver disease [142]. Thus far, targeting PTP1B and TCPTP for treatment has been problematic due to challenges in drug development and the potential for cancer risk brought about by increasing the activity of other tyrosine kinases; however, intranasal targeting of PTP1B and TCPTP can increase leptin and insulin sensitivity and promotes weight loss by repressing feeding and increasing energy expenditure [143].
PTEN is a lipid phosphatase and a potent negative regulator of insulin action, which attenuates insulin signaling by dephosphorylating PIP3 at the 3-position, thus reducing the activation of PDK1, AKT, and other downstream molecules (Figure 6). PTEN heterozygous knockout can increase peripheral insulin sensitivity in IRS2 KO mice and normalize glucose tolerance [144]. SHIP2 (Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase) attenuates insulin signaling by dephosphorylating the 5′-position of PIP3 (Figure 6). Several genetic studies link SHIP2 to metabolic disorders, and metformin's ability to increase insulin sensitivity is at least in part through inhibition of SHIP2 [145]. Phosphoserine-directed phosphatases, including PP1 and PP2A, have complex effects depending on substrate targeting. PP1 complexed with MYPT1 that can be activated by GPCR-mediated phosphorylation removes phosphates from Ser/Thr sites in IRS1, promoting tyrosine phosphorylation to enhance insulin/IGF signaling by heterologous agonists [146].
8.5. miRNA-mediated posttranscriptional regulation
miRNAs (microRNAs) are short non-coding RNA molecules that negatively modulate gene expression through their specific binding within the 3′UTR sequence of mRNA, inhibiting translation or destabilizing the target mRNA. Most of the components of insulin signaling can be regulated in a tissue-specific way by miRNAs [147]. LET7 miRNA interferes with many proximal insulin signaling components, including IGF1R, INSR, IRS2, PIK3IP1, AKT2, TSC1, and RICTOR [148]. Other miRNAs exhibit specificity against proximal signaling molecules. miR-424–5p, miR-15b, miR-195, and miR-96 increase in mouse livers during the high-fat diet, which associates with less InsR expression [[149], [150], [151], [152]]. IRS1 and IRS2 are targeted by miR-222 in liver and adipose [153]; miR-145 in liver [154]; and miR-29a and miR-29c in muscle [155]. Likewise, IRS2 expression can be suppressed by miR-126 [156], miR-33b, and miR-135a [157].
9. Heterologous systemic mechanisms of insulin resistance
9.1. Introduction
Insulin resistance refers to any state in which the response to insulin (exogenous or endogenous) is lower than normal. Multiple pathologic mechanisms associated with over-nutrition and inactivity promote insulin resistance and type 2 diabetes [60,62,[158], [159], [160], [161], [162]]. In addition to hyperinsulinemia itself, insulin resistance is influenced by many factors, including age, weight, ethnicity, body fat, physical activity, dietary intake, gut microbiota, and medications [[163], [164], [165]]. Elevated circulating insulin concentrations can downregulate insulin receptors and desensitize post-receptor pathways [166], including reducing IRS2 expression in liver [167]. In addition, while elevated insulin works to promote glucose utilization and storage to defend against hyperglycemia, it also increases hepatic lipogenesis, leading to hyperlipidemia, WAT expansion, and hepatic steatosis [164]. This is associated with the accumulation of nutrient-derived toxic metabolites, including NEFA, DAG (diacylglycerol), and ceramides. These can activate novel PKCs (protein kinase C) to mediate Ser/Thr phosphorylation of IRS1 and InsR and thereby inhibit early steps in insulin signaling [165]. The important role of hyperinsulinemia in promoting insulin resistance is substantiated by better glucose tolerances in mice with genetically-attenuated hyperinsulinemia subjected to high-fat diets [168,169]. Nutrient excess can also increase circulating branched-chain amino acid levels, which stimulate mTORC1 that can further inhibit IR→IRS1/IRS2 signaling [170,171]. Chronic mTorc1 activity also exacerbates the ER (endoplasmic reticulum) stress response by activating an UPR (unfolded protein response) that can cause insulin resistance [[172], [173], [174], [175]].
9.2. Inflammation: IL6 and TNFα
Chronic inflammation is an important cause of systemic insulin resistance. It occurs in adipose tissue, liver, pancreatic islets, vasculature, and other tissues during obesity and T2DM [163,172,176]. Expanding visceral adiposity creates a microenvironment conducive to inflammation owing to hypoxia, adipose cell death, and dysregulated adipokines, including decreased adiponectin and increased leptin, resistin, and RBP4 [177,178] as well as accumulation of proinflammatory M1-like adipose tissue macrophages (ATMs) [179]. This contributes to increased local and circulating concentrations of proinflammatory cytokines, including MCP1 (monocyte chemoattractant protein 1), IL6 (interleukin 6), and TNFα (tumor necrosis factor-α) [180,181]. These cytokines contribute to insulin resistance by stimulating Ser/Thr phosphorylation of IRS proteins and increasing levels of SOCS proteins, which can inhibit InsR signaling [182] both directly [132,183] and through the action of JNK1 and IKKβ (Figure 6) [163,165]. Despite extensive evidence for this in murine models, clinical trials of TNFα→JNK inhibition in humans have been disappointing [165].
Activation of the innate immune response during obesity also increases the production of so-called inflammasomes and is associated with significant insulin resistance. This is mediated in part by elevated levels of fatty acids acting on Toll-like receptors, especially TLR2 and TLR4. In mice, activation of TLRs in cells results in insulin resistance, whereas genetic disruption of the TLR4 receptor in mice instead protects against fatty acid–induced insulin resistance [184].
9.3. ER stress and the unfolded protein response
The ER (endoplasmic reticulum) is a network of interconnected membrane-enclosed tubes that are continuous with the outer membrane of the nuclear envelope. The unfolded protein response (UPR) is activated when circumstances disrupt protein folding—including glucose and energy deprivation, cholesterol accumulation, viral infection, and other factors that dysregulate protein synthesis [185,186]. Hyperactivation of mTORC1 secondary to nutrient excess and chronic hyperinsulinemia promotes ER stress through increased flux of newly-synthesized proteins through the ER lumen. Three distinct branches of the UPR are initiated by two type-I transmembrane kinases, PERK (PKR-like endoplasmic reticulum kinase) and IRE1 (inositol requiring enzyme-1), and by type-II transmembrane transcription factor ATF6 [185,186]. PERK-mediated phosphorylation of eIF2 promotes lipid accumulation in the liver, which can contribute to insulin resistance [187]. IRE1 signaling activates JNK in the liver and adipose tissue to increase Ser/Thr phosphorylation of IRS1 [188]. ATF6 upregulates expression of the transcription factor XBP1, which helps resolve ER stress in obesity and insulin resistance [188]. ATF6→XBP1 also interacts with FOXO1 to direct it toward proteasome-mediated degradation, which can contribute to systemic insulin sensitivity [189]. In addition to proteotoxic stress activation, the ATF6 detects sphingolipids and ceramides to link UPR to dysregulated lipid homeostasis [190]. Attenuation of ER stress in obese and diabetic mice by chemical chaperones attenuates insulin resistance and improves glucose tolerance [191,192]. The presence of ER stress in the liver and adipose tissues of obese patients suggests that this system plays a role in the development of obesity-linked insulin resistance [193].
9.4. Adipose tissues and insulin resistance
The association of obesity with T2DM has been recognized for decades. Central (visceral) adiposity, compared to total obesity, is more strongly linked to insulin resistance and metabolic abnormalities, including elevated plasma glucose, insulin, cholesterol, and triglyceride concentrations and decreased plasma HDL cholesterol [194]. By contrast, higher levels of subcutaneous fat may be protective against insulin resistance [195,196]. The reason for the tight link between intra-abdominal fat and metabolism is multifactorial. Abdominal fat is more lipolyticly active than subcutaneous fat, releasing more FFA into circulation [197,198]. Intra-abdominal fat also has higher levels of HSD11B1 (11β-hydroxysteroid dehydrogenase type 1), which enhances conversion of inactive cortisone to active cortisol, promoting insulin resistance. Conversely, subcutaneous fat makes and releases more adiponectin, an insulin-sensitizing adipokine.
Independent of this, when nutrient intake exceeds energy expenditure, the excess calories are stored as adipose or ectopic lipid in myocytes, hepatocytes, vascular cells, and β-cells where it can produce toxic metabolites, including DAG and ceramides. This in turn can trigger activation of PKC isoforms that promote insulin resistance [199,200]. The adipocyte itself can be adversely affected by accumulation of excess nutrients, leading to events that can have adverse consequences on the body, including increased expression of leptin, IL-6, IL-8, MCP-1 (monocyte chemoattractant protein-1), and GCSF (granulocyte colony-stimulating factor). These and other cytokines attract proinflammatory M1-macrophages, which release factors such as TNFα that may have local and systemic inflammatory effects to induce insulin resistance [201].
In addition to energy-storing WAT, humans and most other mammals have energy-burning BAT (brown adipose tissue). The content of BAT in humans is negatively correlated with age, obesity, and insulin resistance [202]. Rodents and humans also have beige or brite (brown in white) adipocytes, which appear mixed with WAT following cold or hormonal stimuli [203]. Like BAT, beige adipocytes express UCP1 (uncoupling protein 1). Higher levels of both brown and beige fat are associated with improved metabolic homeostasis and lower insulin resistance [204]. Whether this is through improved insulin-independent glucose utilization or some direct effect on insulin action remains to be determined.
9.5. Ectopic lipid accumulation
When the storage capacity of adipose tissue is exceeded, lipids accumulate in tissues such as muscle and liver, leading to insulin resistance and metabolic dysfunction. First-degree relatives of T2DM patients have an increase in intramyocellular fat, which correlates with insulin resistance [205]. This accumulation of triglyceride in muscle of obese and insulin-resistant persons is likely related to a mismatch between fatty acid uptake and oxidation. The increased lipolysis associated with obesity increases fatty acid delivery to muscle, which can activate PKC isoforms that inhibit insulin signaling [206,207]. However, increased muscle triglyceride is not always linked to insulin resistance. Indeed, exercise training, which increases insulin sensitivity, is also associated with increased muscle triglycerides [208] and increased fatty acid oxidation [[209], [210], [211], [212], [213]]. The reason for this dissociation is not completely understood, but may be related to differences in perilipin proteins associated with lipid droplets in muscle of obese and trained subjects [214].
Lipid accumulation in liver is a common feature of insulin resistance and T2DM, and when clinically significant is referred to as NAFLD (non-alcoholic fatty liver disease) [215]. However, excess lipid intake is not the only way to develop NAFLD. Feeding mice excess glucose or fructose induces metabolic pathways in liver that lead to NAFLD [216,217], indicating that a combination of excess macronutrients and decreased adipose tissue storage promotes lipid accumulation in the liver, which associates with insulin resistance.
9.6. Mitochondrial abnormalities
A decrease in oxidative capacity is seen in both humans and animals with insulin resistance, obesity, and T2DM [218,219]. Increases in intramyocellular fat content in skeletal muscle, associated with insulin resistance, may be caused by alterations in mitochondrial mass. Expression of nuclear-encoded genes that regulate mitochondrial biogenesis and electron transport chain activity—such as PGC1α and PGC1β—is downregulated in obese patients with impaired glucose tolerance and T2DM [[220], [221], [222]]. The cause–effect relation between alterations in mitochondrial mass/function and skeletal muscle insulin resistance remains debated; however, an impairment of β-oxidation of fatty acids that increases even-chained acyl-carnitine levels in plasma is a marker of insulin resistance [223]. Post-translational modification of mitochondrial proteins by acetylation, succinylation, or malonylation also provides a potential mechanism for control of mitochondrial flux and insulin resistance [224]. This is controlled by levels of substrate and the sirtuin family of dacetylases. Sirt3 is the primary mitochondrial deacetylase that is activated by NAD+ and can deacetylate critical metabolic enzymes, including ACADL (Acyl-CoA Dehydrogenase Long Chain) in liver and the pyruvate dehydrogenase complex in muscle [225,226]. Sirt3 knockout mice exhibit decreased oxygen consumption and develop oxidative stress in skeletal muscle, leading to JNK activation and impaired insulin signaling [226]. Similarly, the NAD+-dependent Sirt5 leads to desuccinylation and demalonylation of mitochondrial enzymes, altering their activity [227,228].
9.7. Skeletal muscle
The primary site of glucose disposal after a meal is skeletal muscle, and the primary mechanism of glucose storage is through its conversion to glycogen [229]. Studies using the hyperinsulinemic-euglycemic clamp technique have demonstrated that insulin-resistant people, with or without T2DM, have a deficiency in the nonoxidative disposal of glucose. This is related primarily to a defect in glycogen synthesis, which itself is secondary to a decrease in insulin-stimulated glucose uptake [230,231]. A major question is to what extent extrinsic factors versus intrinsic factors lead to insulin resistance.
Increased fatty acid flux into skeletal muscle, related to increased visceral lipolysis, has been implicated as one of the extrinsic factors in the inhibition of muscle glucose uptake. More recent studies in humans suggest that the primary effect of fatty acids, at least in the presence of high insulin levels, is a decrease in glucose transport, as measured in vivo using 13C- and 31P nuclear magnetic resonance (NMR) spectroscopy [232,233]. These studies also found increased activity of novel isoforms of protein kinase C, including PKCθ and PKCδ, that might mediate the effect of elevated fatty acids to inhibit PI3K activity [206,207]. PKC-mediated serine phosphorylation of the IKKβ subunit, leading to its degradation and the unregulated translocation of NF-κB into the nucleus, may also be important to fatty acid–induced insulin resistance [234]. Disruption of the IKKβ inflammatory pathway by high-dose salicylate therapy improved insulin sensitivity in a small human trial [235].
Additionally, skeletal muscle insulin resistance may relate to changes in fatty acid and triglyceride (TG) metabolism [236]. Malonyl-CoA is an allosteric inhibitor of CPT1, the enzyme that controls the transfer of long-chain fatty acyl-CoAs into the mitochondria [[237], [238], [239]]. In the presence of elevated glucose and insulin levels, the TCA cycle is activated, resulting in an increase in citrate in the cytoplasm through increased mitochondrial malate cycling. The increased citrate is converted to acetyl-CoA through citrate lyase and thus provides an indirect substrate for ACC (acetyl-CoA carboxylase). Even during insulin-resistant states and T2DM, glucose uptake into skeletal muscle is higher than normal due to elevated circulating glucose and more GLUT4 at the plasma membrane owing to rising AMPK activity [240,241]. This glucose is shunted toward the glycolytic pathway, generating acetyl-CoA that can be converted to malonyl-CoA in the cytoplasm by the action of the highly-regulated enzyme ACACA [242]. This results in a buildup of long-chain acyl-CoAs and diacylglycerols, which can activate one or more PKC isoforms, leading to insulin resistance [236].
9.8. The gut microbiome and metabolome
There is growing evidence that the gut microbiome—the bacteria that reside in the gastrointestinal tract—can be a major mediator of the effect of diet in obesity, diabetes, and metabolic syndrome and can contribute to the development of insulin resistance in these disorders [[243], [244], [245], [246], [247], [248], [249], [250], [251], [252]]. In mice, it has been shown that administration of low dose antibiotics early in life may predispose to obesity and glucose intolerance by perturbing the development of a normal microbiome [253]. The mechanisms by which gut microbiota affect pathogenesis of diabetes, obesity, and insulin resistance are complex. Gut microbiota have major effects on intestinal barrier function, breakdown of otherwise indigestible dietary components, modification of bile acids and other substances, development of the gut, and education of the immune system [243,254]. These effects can lead to a release of bacterial proteins, endotoxins, and cytokines into the bloodstream [255,256] as well as produce changes in hundreds of metabolic products, including bile acids, short-chained fatty acids, amino acids, and many other classes of molecules [246,247,[257], [258], [259], [260]]. Together, these lead to tissue-specific metabolic dysregulation and immune activation, leading to insulin resistance and progression of diabetes pathogenesis. A number of metabolites have been shown to correlate with insulin resistance in both mice [261] and humans, and some, such as 2-aminoadipate, alpha-hydroxybutyrate, and N-acetylglycine, are suggested to be biomarkers of T2DM or insulin resistance [[261], [262], [263], [264]]. Further studies are needed to determine exactly how gut microbiota affect insulin sensitivity and diabetes progression and whether therapies that change gut microbiota can be used to treat or prevent type 2 diabetes.
9.9. Intrinsic factors and cell-autonomous insulin resistance
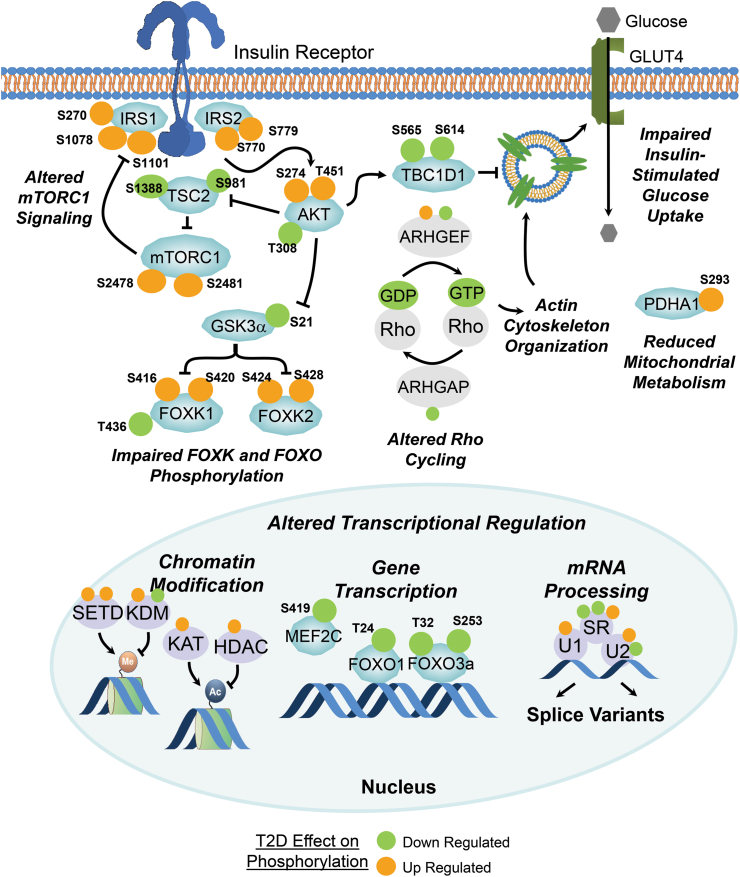
It is well recognized that insulin resistance precedes and predicts type 2 diabetes, even when none of the known extrinsic factors leading to insulin resistance are present [265]. A major challenge has been identifying the intrinsic cellular defects which underlie insulin resistance and might be more intricately linked to the genetic determinants of disease. Muscle biopsies and primary cultured myoblasts derived from people with T2DM show insulin resistance, including impaired insulin signaling at the level of IRS1-associated PI3K and AKT activity and decreased glucose uptake and glycogen synthesis [222,229,[266], [267], [268]]. Recently, Batista et al. have used iPSC (induced pluripotential stem cell) differentiated into muscle (iMyos) to study ex vivo signaling defects in T2DM subjects’ skeletal muscle in the absence of extrinsic stimuli [269]. They showed that iMyos from individuals with T2DM shows defects in insulin signaling at the level of AKT/GSK3/FOXO1 phosphorylation, decreased insulin-stimulated glucose uptake, and altered mitochondrial respiration similar to the defects observed in muscle in T2DM in vivo. Global phosphoproteomics revealed that these defects are part of a much larger multi-dimensional network of signaling changes involving over 1000 Ser/Thr phosphorylation sites on more than 700 different proteins. Only a small proportion of these abnormalities are in classical insulin-regulated phosphorylation that defines critical nodes in insulin action [100]. Indeed, the largest number of perturbations occurred in pathways outside of the canonical insulin signaling pathway and not (acutely) regulated by insulin. These include up- and downregulation of phosphorylation in proteins involved in cytoskeleton remodeling, vesicle trafficking, and RHO GTPase activity and nuclear proteins involved in transcription, mRNA splicing, and chromatin remodeling (Figure 7). These findings indicate that there is a primary cellular defect underlying the insulin resistance of T2DM, and that defining what drives these defects at a molecular and cellular level will not only help in understanding the pathogenesis of type 2 diabetes but open avenues for new treatments.
Figure 7.
Schematic diagram showing some of the changes in phosphorylation observed in iPS cell–derived myoblasts from control and T2DM patients. The sites highlighted in orange are increased in either basal or stimulated phosphorylation in cells of the T2DM patients, whereas those highlighted in green are decreased in their phosphorylation. Note that many of the altered phosphorylations occur in pathways outside the pathways considered canonical insulin signaling. Figure was adapted from the data of Batista et al. [269].
10. Conclusions
Insulin and IGF-1 signaling are present in virtually every cell of the body and play a central role in the control of metabolism, growth, and differentiation. Since the discovery of the insulin receptor 50 years ago, major progress has been made in dissecting these pathways and understanding some of the many drivers of insulin resistance in T2DM, obesity, and the metabolic syndrome. These include a range of both cell-extrinsic and cell-intrinsic factors. It is clear, however, that more remains to be learned about the integration of this systemic regulatory system, translating our understanding of these pathways into new therapies for insulin resistance–associated diseases in an important challenge for the next decade.
Conflict of interests
M.F.W. is an advisory board member of Housey Pharma (https://www.housey.com/). C.R.K. is on the scientific advisory board or serves as a consultant for Kaleido Biosciences, CohBar, ERX Therapeutics, and Cellarity.
Acknowledgements
This work was supported by National Institutes of Health grants DK098655, AG067913, and DK094721 (to M.F.W.) and DK031036, DK05545, and DK121967 (to C.R.K.).
Contributor Information
Morris F. White, Email: morris.white@childrens.harvard.edu.
C. Ronald Kahn, Email: C.Ronald.Kahn@joslin.harvard.edu.
References
- 1.Flier J.S., Kahn C.R. 2021. Insulin: A pacesetter for the shape of modern biomedical science and the nobel prize. Mol metab in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freychet P., Roth J., Neville D.M., Jr. Insulin receptors in the liver: specific binding of ( 125 I)insulin to the plasma membrane and its relation to insulin bioactivity. Proceedings of the National Academy of Sciences of the U S A. 1971;68(8):1833–1837. doi: 10.1073/pnas.68.8.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasuga M., Karlsson F.A., Kahn C.R. Insulin stimulates the phosphorylation of the 95,000-dalton subunit of its own receptor. Science. 1982;215(4529):185–187. doi: 10.1126/science.7031900. [DOI] [PubMed] [Google Scholar]
- 4.Bravo D.A., Gleason J.B., Sanchez R.I., Roth R.A., Fuller R.S. Accurate and efficient cleavage of the human insulin proreceptor by the human proprotein-processing protease furin. Characterization and kinetic parameters using the purified, secreted soluble protease expressed by a recombinant baculovirus. Journal of Biological Chemistry. 1994;269(41):25830–25837. [PubMed] [Google Scholar]
- 5.De Meyts P. Insulin/receptor binding: the last piece of the puzzle? What recent progress on the structure of the insulin/receptor complex tells us (or not) about negative cooperativity and activation. BioEssays. 2015;37(4):389–397. doi: 10.1002/bies.201400190. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker J. In: Post T.W., editor. 2020. Structure and function of the insulin receptor. [Uptodate. Uptodate: Waltham MA] [Google Scholar]
- 7.Gutmann T., Kim K.H., Grzybek M., Walz T., Coskun U. Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. The Journal of Cell Biology. 2018;217(5):1643–1649. doi: 10.1083/jcb.201711047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scapin G., Dandey V.P., Zhang Z., Prosise W., Hruza A., Kelly T. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature. 2018;556(7699):122–125. doi: 10.1038/nature26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson K.M., Hu C., Lemmon M.A. Insulin and epidermal growth factor receptor family members share parallel activation mechanisms. Protein Science. 2020;29(6):1331–1344. doi: 10.1002/pro.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchikawa E., Choi E., Shang G., Yu H., Bai X.C. 2019. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Meyts P. The insulin receptor: a prototype for dimeric, allosteric membrane receptors? Trends in Biochemical Sciences. 2008;33(8):376–384. doi: 10.1016/j.tibs.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Belfiore A., Malaguarnera R., Vella V., Lawrence M.C., Sciacca L., Frasca F. Insulin receptor isoforms in physiology and disease: an updated view. Endocrine Reviews. 2017;38(5):379–431. doi: 10.1210/er.2017-00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y., Kong G.K., Menting J.G., Margetts M.B., Delaine C.A., Jenkin L.M. How ligand binds to the type 1 insulin-like growth factor receptor. Nature Communications. 2018;9(1):821. doi: 10.1038/s41467-018-03219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ullrich A., Bell J.R., Chen E.Y., Herrera R., Petruzzelli L.M., Dull T.J. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature. 1985;313(6005):756–761. doi: 10.1038/313756a0. [DOI] [PubMed] [Google Scholar]
- 15.Ebina Y., Ellis L., Jarnagin K., Edery M., Graf L., Clauser E. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell. 1985;40(4):747–758. doi: 10.1016/0092-8674(85)90334-4. [DOI] [PubMed] [Google Scholar]
- 16.White M.F., Shoelson S.E., Keutmann H., Kahn C.R. A cascade of tyrosine autophosphorylation in the beta-subunit activates the phosphotransferase of the insulin receptor. Journal of Biological Chemistry. 1988;263(6):2969–2980. [PubMed] [Google Scholar]
- 17.Rajagopalan M., Neidigh J.L., McClain D.A. Amino acid sequences Gly-Pro-Leu-Tyr and Asn-Pro-Glu-Tyr in the submembranous domain of the insulin receptor are required for normal endocytosis. Journal of Biological Chemistry. 1991;266(34):23068–23073. [PubMed] [Google Scholar]
- 18.Gutmann T., Schafer I.B., Poojari C., Brankatschk B., Vattulainen I., Strauss M. Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain. Journal of Cell Biology. 2020;219(1) doi: 10.1083/jcb.201907210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White M.F., Maron R., Kahn C.R. Insulin rapidly stimulates tyrosine phosphorylation of a Mr-185,000 protein in intact cells. Nature. 1985;318(6042):183–186. doi: 10.1038/318183a0. [DOI] [PubMed] [Google Scholar]
- 20.Sun X.J., Rothenberg P., Kahn C.R., Backer J.M., Araki E., Wilden P.A. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352(6330):73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 21.Bjornholm M., He A.R., Attersand A., Lake S., Liu S.C., Lienhard G.E. Absence of functional insulin receptor substrate-3 (IRS-3) gene in humans. Diabetologia. 2002;45(12):1697–1702. doi: 10.1007/s00125-002-0945-z. [DOI] [PubMed] [Google Scholar]
- 22.Sadagurski M., Dong X.C., Myers M.G., Jr., White M.F. Irs2 and Irs4 synergize in non-LepRb neurons to control energy balance and glucose homeostasis. Molecular Metabolism. 2014;3(1):55–63. doi: 10.1016/j.molmet.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Copps K.D., White M.F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55(10):2565–2582. doi: 10.1007/s00125-012-2644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White M.F., Livingston J.N., Backer J.M., Lauris V., Dull T.J., Ullrich A. Mutation of the insulin receptor at tyrosine 960 inhibits signal transmission but does not affect its tyrosine kinase activity. Cell. 1988;54(5):641–649. doi: 10.1016/s0092-8674(88)80008-4. [DOI] [PubMed] [Google Scholar]
- 25.Backer J.M., Myers M.G., Jr., Shoelson S.E., Chin D.J., Sun X.J., Miralpeix M. Phosphatidylinositol 3'-kinase is activated by association with IRS-1 during insulin stimulation. The EMBO Journal. 1992;11(9):3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhe-Paganon S., Ottinger E.A., Nolte R.T., Eck M.J., Shoelson S.E. Crystal structure of the pleckstrin homology-phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1. Proceedings of the National Academy of Sciences of the U S A. 1999;96(15):8378–8383. doi: 10.1073/pnas.96.15.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai W., Sakaguchi M., Kleinridders A., Gonzalez-Del Pino G., Dreyfuss J.M., O'Neill B.T. Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nature Communications. 2017;8:14892. doi: 10.1038/ncomms14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorpe L.M., Yuzugullu H., Zhao J.J. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nature Reviews Cancer. 2015;15(1):7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 30.Vanhaesebroeck B., Stephens L., Hawkins P. PI3K signalling: the path to discovery and understanding. Nature Reviews Molecular Cell Biology. 2012;13(3):195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 31.Xu F., Na L., Li Y., Chen L. Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell & Bioscience. 2020;10:54. doi: 10.1186/s13578-020-00416-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Cheatham B., Vlahos C.J., Cheatham L., Wang L., Blenis J., Kahn C.R. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Molecular and Cellular Biology. 1994;14(7):4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hopkins B.D., Pauli C., Du X., Wang D.G., Li X., Wu D. Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. Nature. 2018;560(7719):499–503. doi: 10.1038/s41586-018-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang G., Murashige D.S., Humphrey S.J., James D.E. A positive feedback loop between Akt and mTORC2 via SIN1 phosphorylation. Cell Reports. 2015;12(6):937–943. doi: 10.1016/j.celrep.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Hers I., Vincent E.E., Tavare J.M. Akt signalling in health and disease. Cellular Signalling. 2011;23(10):1515–1527. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Lu M., Wan M., Leavens K.F., Chu Q., Monks B.R., Fernandez S. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nature Medicine. 2012;18(3):388–395. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George S., Rochford J.J., Wolfrum C., Gray S.L., Schinner S., Wilson J.C. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science. 2004;304(5675):1325–1328. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proud C.G. Cell signaling. mTOR, unleashed. Science. 2007;318(5852):926–927. doi: 10.1126/science.1150653. [DOI] [PubMed] [Google Scholar]
- 40.Xie J., Proud C.G. Crosstalk between mTOR complexes. Nature Cell Biology. 2013;15(11):1263–1265. doi: 10.1038/ncb2877. [DOI] [PubMed] [Google Scholar]
- 41.Dai N., Christiansen J., Nielsen F.C., Avruch J. mTOR complex 2 phosphorylates IMP1 cotranslationally to promote IGF2 production and the proliferation of mouse embryonic fibroblasts. Genes & Development. 2013;27(3):301–312. doi: 10.1101/gad.209130.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K., Guo X., Yan H., Wu Y., Pan Q., Shen J.Z. Phosphorylation of Forkhead protein FoxO1 at S253 regulates glucose homeostasis in mice. Endocrinology. 2019;160(5):1333–1347. doi: 10.1210/en.2018-00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown A.K., Webb A.E. Regulation of FOXO factors in mammalian cells. Current Topics in Developmental Biology. 2018;127:165–192. doi: 10.1016/bs.ctdb.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S., Dong H.H. FoxO integration of insulin signaling with glucose and lipid metabolism. Journal of Endocrinology. 2017;233(2):R67–R79. doi: 10.1530/JOE-17-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundell L.S., Massart J., Altintas A., Krook A., Zierath J.R. Regulation of glucose uptake and inflammation markers by FOXO1 and FOXO3 in skeletal muscle. Molecular Metabolism. 2019;20:79–88. doi: 10.1016/j.molmet.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Neill B.T., Bhardwaj G., Penniman C.M., Krumpoch M.T., Suarez Beltran P.A., Klaus K. FoxO transcription factors are critical regulators of diabetes-related muscle atrophy. Diabetes. 2019;68(3):556–570. doi: 10.2337/db18-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samuel V.T., Shulman G.I. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148(5):852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barthel A., Schmoll D., Unterman T.G. FoxO proteins in insulin action and metabolism. Trends in Endocrinology and Metabolism. 2005;16(4):183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Kahn C.R., Flier J.S., Bar R.S., Archer J.A., Gorden P., Martin M.M. The syndromes of insulin resistance and acanthosis nigricans. Insulin-receptor disorders in man. New England Journal of Medicine. 1976;294(14):739–745. doi: 10.1056/NEJM197604012941401. [DOI] [PubMed] [Google Scholar]
- 50.Hosoe J., Kadowaki H., Miya F., Aizu K., Kawamura T., Miyata I. Structural basis and genotype-phenotype correlations of INSR mutations causing severe insulin resistance. Diabetes. 2017;66(10):2713–2723. doi: 10.2337/db17-0301. [DOI] [PubMed] [Google Scholar]
- 51.Semple R.K., Sleigh A., Murgatroyd P.R., Adams C.A., Bluck L., Jackson S. Postreceptor insulin resistance contributes to human dyslipidemia and hepatic steatosis. Journal of Clinical Investigation. 2009;119(2):315–322. doi: 10.1172/JCI37432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature Genetics. 2012;44(9):981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biddinger S.B., Kahn C.R. From mice to men: insights into the insulin resistance syndromes. Annual Review of Physiology. 2006;68:123–158. doi: 10.1146/annurev.physiol.68.040104.124723. [DOI] [PubMed] [Google Scholar]
- 54.Kubota T., Kubota N., Kadowaki T. Imbalanced insulin actions in obesity and type 2 diabetes: key mouse models of insulin signaling pathway. Cell Metabolism. 2017;25(4):797–810. doi: 10.1016/j.cmet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Stefan N., Haring H.U. The role of hepatokines in metabolism. Nature Reviews Endocrinology. 2013;9(3):144–152. doi: 10.1038/nrendo.2012.258. [DOI] [PubMed] [Google Scholar]
- 56.Tao R., Wang C., Stohr O., Qiu W., Hu Y., Miao J. Inactivating hepatic follistatin alleviates hyperglycemia. Nature Medicine. 2018;24(7):1058–1069. doi: 10.1038/s41591-018-0048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almind K., Inoue G., Pedersen O., Kahn C.R. A common amino acid polymorphism in insulin receptor substrate-1 causes impaired insulin signaling. Evidence from transfection studies. Journal of Clinical Investigation. 1996;97(11):2569–2575. doi: 10.1172/JCI118705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zisman A., Peroni O.D., Abel E.D., Michael M.D., Mauvais-Jarvis F., Lowell B.B. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nature Medicine. 2000;6(8):924–928. doi: 10.1038/78693. [DOI] [PubMed] [Google Scholar]
- 59.Herman M.A., Kahn B.B. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. Journal of Clinical Investigation. 2006;116(7):1767–1775. doi: 10.1172/JCI29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruning J.C., Michael M.D., Winnay J.N., Hayashi T., Horsch D., Accili D. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Molecular Cell. 1998;2(5):559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 61.O'Neill B.T., Lauritzen H.P., Hirshman M.F., Smyth G., Goodyear L.J., Kahn C.R. Differential role of insulin/IGF-1 receptor signaling in muscle growth and glucose homeostasis. Cell Reports. 2015;11(8):1220–1235. doi: 10.1016/j.celrep.2015.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Long Y.C., Cheng Z., Copps K.D., White M.F. Insulin receptor substrates Irs1 and Irs2 coordinate skeletal muscle growth and metabolism via the Akt and AMPK pathways. Molecular and Cellular Biology. 2011;31(3):430–441. doi: 10.1128/MCB.00983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bozadjieva N., Heppner K.M., Seeley R.J. Targeting FXR and FGF19 to treat metabolic diseases-lessons learned from bariatric surgery. Diabetes. 2018;67(9):1720–1728. doi: 10.2337/dbi17-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Neill B.T., Lee K.Y., Klaus K., Softic S., Krumpoch M.T., Fentz J. Insulin and IGF-1 receptors regulate FoxO-mediated signaling in muscle proteostasis. Journal of Clinical Investigation. 2016;126(9):3433–3446. doi: 10.1172/JCI86522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michael M.D., Kulkarni R.N., Postic C., Previs S.F., Shulman G.I., Magnuson M.A. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Molecular Cell. 2000;6(1):87–97. [PubMed] [Google Scholar]
- 66.Titchenell P.M., Lazar M.A., Birnbaum M.J. Unraveling the regulation of hepatic metabolism by insulin. Trends in Endocrinology and Metabolism. 2017;28(7):497–505. doi: 10.1016/j.tem.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biddinger S.B., Hernandez-Ono A., Rask-Madsen C., Haas J.T., Aleman J.O., Suzuki R. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metabolism. 2008;7(2):125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fisher S.J., Kahn C.R. Insulin signaling is required for insulin's direct and indirect action on hepatic glucose production. Journal of Clinical Investigation. 2003;111(4):463–468. doi: 10.1172/JCI16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu C., Hoene M., Plomgaard P., Hansen J.S., Zhao X., Li J. Muscle-liver substrate fluxes in exercising humans and potential effects on hepatic metabolism. Journal of Clinical Endocrinology Metabolism. 2020;105(4) doi: 10.1210/clinem/dgz266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Staiger H., Keuper M., Berti L., Hrabe de Angelis M., Haring H.U. Fibroblast growth factor 21-metabolic role in mice and men. Endocrine Reviews. 2017;38(5):468–488. doi: 10.1210/er.2017-00016. [DOI] [PubMed] [Google Scholar]
- 71.Dong X.C., Copps K.D., Guo S., Li Y., Kollipara R., DePinho R.A. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metabolism. 2008;8(1):65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng Z., Guo S., Copps K., Dong X., Kollipara R., Rodgers J.T. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nature Medicine. 2009;15(11):1307–1311. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo S., Copps K.D., Dong X., Park S., Cheng Z., Pocai A. The Irs1 branch of the insulin signaling cascade plays a dominant role in hepatic nutrient homeostasis. Molecular and Cellular Biology. 2009;29(18):5070–5083. doi: 10.1128/MCB.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.I O.S., Zhang W., Wasserman D.H., Liew C.W., Liu J., Paik J. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nature Communications. 2015;6:7079. doi: 10.1038/ncomms8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Titchenell P.M., Quinn W.J., Lu M., Chu Q., Lu W., Li C. Direct hepatocyte insulin signaling is required for lipogenesis but is dispensable for the suppression of glucose production. Cell Metabolism. 2016;23(6):1154–1166. doi: 10.1016/j.cmet.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perry R.J., Camporez J.G., Kursawe R., Titchenell P.M., Zhang D., Perry C.J. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell. 2015;160(4):745–758. doi: 10.1016/j.cell.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bluher M., Michael M.D., Peroni O.D., Ueki K., Carter N., Kahn B.B. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Developmental Cell. 2002;3(1):25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 78.Boucher J., Softic S., El Ouaamari A., Krumpoch M.T., Kleinridders A., Kulkarni R.N. Differential roles of insulin and IGF-1 receptors in adipose tissue development and function. Diabetes. 2016;65(8):2201–2213. doi: 10.2337/db16-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Softic S., Boucher J., Solheim M.H., Fujisaka S., Haering M.F., Homan E.P. Lipodystrophy due to adipose tissue-specific insulin receptor knockout results in progressive NAFLD. Diabetes. 2016;65(8):2187–2200. doi: 10.2337/db16-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakaguchi M., Fujisaka S., Cai W., Winnay J.N., Konishi M., O'Neill B.T. Adipocyte dynamics and reversible metabolic syndrome in mice with an inducible adipocyte-specific deletion of the insulin receptor. Cell Metabolism. 2017;25(2):448–462. doi: 10.1016/j.cmet.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laustsen P.G., Russell S.J., Cui L., Entingh-Pearsall A., Holzenberger M., Liao R. Essential role of insulin and insulin-like growth factor 1 receptor signaling in cardiac development and function. Molecular and Cellular Biology. 2007;27(5):1649–1664. doi: 10.1128/MCB.01110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lopez-Izquierdo A., Pereira R.O., Wende A.R., Punske B.B., Abel E.D., Tristani-Firouzi M. The absence of insulin signaling in the heart induces changes in potassium channel expression and ventricular repolarization. American Journal of Physiology - Heart and Circulatory Physiology. 2014;306(5):H747–H754. doi: 10.1152/ajpheart.00849.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He Z., Opland D.M., Way K.J., Ueki K., Bodyak N., Kang P.M. Regulation of vascular endothelial growth factor expression and vascularization in the myocardium by insulin receptor and PI3K/Akt pathways in insulin resistance and ischemia. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26(4):787–793. doi: 10.1161/01.ATV.0000209500.15801.4e. [DOI] [PubMed] [Google Scholar]
- 84.Riehle C., Weatherford E.T., Wende A.R., Jaishy B.P., Seei A.W., McCarty N.S. Insulin receptor substrates differentially exacerbate insulin-mediated left ventricular remodeling. JCI Insight. 2020;5(6) doi: 10.1172/jci.insight.134920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Riehle C., Abel E.D. Insulin signaling and heart failure. Circulation Research. 2016;118(7):1151–1169. doi: 10.1161/CIRCRESAHA.116.306206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu D., Yin C., Luo S., Habenicht A.J.R., Mohanta S.K. Vascular smooth muscle cells contribute to atherosclerosis immunity. Frontiers in Immunology. 2019;10:1101. doi: 10.3389/fimmu.2019.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Q., Fu J., Xia Y., Qi W., Ishikado A., Park K. Homozygous receptors for insulin and not IGF-1 accelerate intimal hyperplasia in insulin resistance and diabetes. Nature Communications. 2019;10(1):4427. doi: 10.1038/s41467-019-12368-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rask-Madsen C., Li Q., Freund B., Feather D., Abramov R., Wu I.H. Loss of insulin signaling in vascular endothelial cells accelerates atherosclerosis in apolipoprotein E null mice. Cell Metabolism. 2010;11(5):379–389. doi: 10.1016/j.cmet.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clough M.H., Schneider D.J., Sobel B.E., White M.F., Wadsworth M.P., Taatjes D.J. Attenuation of accumulation of neointimal lipid by pioglitazone in mice genetically deficient in insulin receptor substrate-2 and apolipoprotein E. Journal of Histochemistry and Cytochemistry. 2005;53(5):603–610. doi: 10.1369/jhc.4A6590.2005. [DOI] [PubMed] [Google Scholar]
- 90.Kubota T., Kubota N., Kumagai H., Yamaguchi S., Kozono H., Takahashi T. Impaired insulin signaling in endothelial cells reduces insulin-induced glucose uptake by skeletal muscle. Cell Metabolism. 2011;13(3):294–307. doi: 10.1016/j.cmet.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 91.Rhodes C.J., White M.F., Leahy J.L., Kahn S.E. Direct autocrine action of insulin on beta-cells: does it make physiological sense? Diabetes. 2013;62(7):2157–2163. doi: 10.2337/db13-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kulkarni R.N., Bruning J.C., Winnay J.N., Postic C., Magnuson M.A., Kahn C.R. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96(3):329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 93.Withers D.J., Burks D.J., Towery H.H., Altamuro S.L., Flint C.L., White M.F. Irs-2 coordinates Igf-1 receptor-mediated beta-cell development and peripheral insulin signalling. Nature Genetics. 1999;23(1):32–40. doi: 10.1038/12631. [DOI] [PubMed] [Google Scholar]
- 94.Lee J.Y., Ristow M., Lin X., White M.F., Magnuson M.A., Hennighausen L. RIP-Cre revisited, evidence for impairments of pancreatic beta-cell function. Journal of Biological Chemistry. 2006;281(5):2649–2653. doi: 10.1074/jbc.M512373200. [DOI] [PubMed] [Google Scholar]
- 95.Demozay D., Tsunekawa S., Briaud I., Shah R., Rhodes C.J. Specific glucose-induced control of insulin receptor substrate-2 expression is mediated via Ca2+-dependent calcineurin/NFAT signaling in primary pancreatic islet beta-cells. Diabetes. 2011;60(11):2892–2902. doi: 10.2337/db11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Assmann A., Ueki K., Winnay J.N., Kadowaki T., Kulkarni R.N. Glucose effects on beta-cell growth and survival require activation of insulin receptors and insulin receptor substrate 2. Molecular and Cellular Biology. 2009;29(11):3219–3228. doi: 10.1128/MCB.01489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park S., Dong X., Fisher T.L., Dunn S., Omer A.K., Weir G. Exendin-4 uses Irs2 signaling to mediate pancreatic beta cell growth and function. Journal of Biological Chemistry. 2006;281(2):1159–1168. doi: 10.1074/jbc.M508307200. [DOI] [PubMed] [Google Scholar]
- 98.Kushner J.A., Flint C.L., Dow M.A., Dutta S., Schubert M., Montminy M.R. Insulin receptor substrate-2 and PDX-1 act convergently to regulate beta cell mass in vivo. Diabetes. 2001;50:A338. A338. [Google Scholar]
- 99.Kuznetsova A., Yu Y., Hollister-Lock J., Opare-Addo L., Rozzo A., Sadagurski M. Trimeprazine increases IRS2 in human islets and promotes pancreatic beta cell growth and function in mice. JCI Insight. 2016;1(3) doi: 10.1172/jci.insight.80749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kullmann S., Heni M., Hallschmid M., Fritsche A., Preissl H., Haring H.U. Brain insulin resistance at the crossroads of metabolic and cognitive disorders in humans. Physiological Reviews. 2016;96(4):1169–1209. doi: 10.1152/physrev.00032.2015. [DOI] [PubMed] [Google Scholar]
- 101.Arnold S.E., Arvanitakis Z., Macauley-Rambach S.L., Koenig A.M., Wang H.Y., Ahima R.S. Brain insulin resistance in type 2 diabetes and Alzheimer disease: concepts and conundrums. Nature Reviews Neurology. 2018;14(3):168–181. doi: 10.1038/nrneurol.2017.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bischof G.N., Park D.C. Obesity and aging: consequences for cognition, brain structure, and brain function. Psychosomatic Medicine. 2015;77(6):697–709. doi: 10.1097/PSY.0000000000000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Geetha T., Rege S.D., Mathews S.E., Meakin S.O., White M.F., Babu J.R. Nerve growth factor receptor TrkA, a new receptor in insulin signaling pathway in PC12 cells. Journal of Biological Chemistry. 2013;288(33):23807–23813. doi: 10.1074/jbc.M112.436279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang E.J., Reichardt L.F. Trk receptors: roles in neuronal signal transduction. Annual Review of Biochemistry. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 105.Kleinridders A., Ferris H.A., Cai W., Kahn C.R. Insulin action in brain regulates systemic metabolism and brain function. Diabetes. 2014;63(7):2232–2243. doi: 10.2337/db14-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soto M., Cai W., Konishi M., Kahn C.R. Insulin signaling in the hippocampus and amygdala regulates metabolism and neurobehavior. Proceedings of the National Academy of Sciences of the U S A. 2019;116(13):6379–6384. doi: 10.1073/pnas.1817391116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Talbot K., Wang H.Y., Kazi H., Han L.Y., Bakshi K.P., Stucky A. Demonstrated brain insulin resistance in Alzheimer's disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. Journal of Clinical Investigation. 2012;122(4):1316–1338. doi: 10.1172/JCI59903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moloney A.M., Griffin R.J., Timmons S., O'Connor R., Ravid R., O'Neill C. Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer's disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiology of Aging. 2010;31(2):224–243. doi: 10.1016/j.neurobiolaging.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 109.Bomfim T.R., Forny-Germano L., Sathler L.B., Brito-Moreira J., Houzel J.C., Decker H. An anti-diabetes agent protects the mouse brain from defective insulin signaling caused by Alzheimer's disease- associated Abeta oligomers. Journal of Clinical Investigation. 2012;122(4):1339–1353. doi: 10.1172/JCI57256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schubert M., Gautam D., Surjo D., Ueki K., Baudler S., Schubert D. Role for neuronal insulin resistance in neurodegenerative diseases. Proceedings of the National Academy of Sciences of the U S A. 2004;101(9):3100–3105. doi: 10.1073/pnas.0308724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lourenco M.V., Clarke J.R., Frozza R.L., Bomfim T.R., Forny-Germano L., Batista A.F. TNF-alpha mediates PKR-dependent memory impairment and brain IRS-1 inhibition induced by alzheimer's beta-amyloid oligomers in mice and monkeys. Cell Metabolism. 2013;18(6):831–843. doi: 10.1016/j.cmet.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 112.Irvine E.E., Drinkwater L., Radwanska K., Al-Qassab H., Smith M.A., O'Brien M. Insulin receptor substrate 2 is a negative regulator of memory formation. Learning & Memory. 2011;18(6):375–383. doi: 10.1101/lm.2111311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Freude S., Hettich M.M., Schumann C., Stohr O., Koch L., Kohler C. Neuronal IGF-1 resistance reduces Abeta accumulation and protects against premature death in a model of Alzheimer's disease. The FASEB Journal. 2009;23(10):3315–3324. doi: 10.1096/fj.09-132043. [DOI] [PubMed] [Google Scholar]
- 114.Killick R., Scales G., Leroy K., Causevic M., Hooper C., Irvine E.E. Deletion of Irs2 reduces amyloid deposition and rescues behavioural deficits in APP transgenic mice. Biochemical and Biophysical Research Communications. 2009;386(1):257–262. doi: 10.1016/j.bbrc.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cohen E., Paulsson J.F., Blinder P., Burstyn-Cohen T., Du D., Estepa G. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139(6):1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Withers D.J., Gutierrez J.S., Towery H., Burks D.J., Ren J.M., Previs S. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391(6670):900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 117.Taguchi A., Wartschow L.M., White M.F. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317(5836):369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 118.Sadagurski M., White M.F. Integrating metabolism and longevity through insulin and IGF1 signaling. Endocrinology and Metabolism Clinics of North America. 2013;42(1):127–148. doi: 10.1016/j.ecl.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.White M.F. IRS2 integrates insulin/IGF1 signalling with metabolism, neurodegeneration and longevity. Diabetes Obesity Metabolism. 2014;1(16 Suppl):4–15. doi: 10.1111/dom.12347. [DOI] [PubMed] [Google Scholar]
- 120.Sadagurski M., Cheng Z., Rozzo A., Palazzolo I., Kelley G.R., Dong X. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. Journal of Clinical Investigation. 2011;121(10):4070–4081. doi: 10.1172/JCI46305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hwang I., Oh H., Santo E., Kim D.Y., Chen J.W., Bronson R.T. FOXO protects against age-progressive axonal degeneration. Aging Cell. 2018;17(1) doi: 10.1111/acel.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bruning J.C., Winnay J., Bonner-Weir S., Taylor S.I., Accili D., Kahn C.R. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997;88(4):561–572. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- 123.Meng X., Kondo M., Morino K., Fuke T., Obata T., Yoshizaki T. Transcription factor AP-2beta: a negative regulator of IRS-1 gene expression. Biochemical and Biophysical Research Communications. 2010;392(4):526–532. doi: 10.1016/j.bbrc.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 124.Wang Z., Shah O.J., Hunter T. The transcriptional coactivators p/CIP and SRC-1 control insulin resistance through IRS1 in obesity models. PloS One. 2012;7(7):e36961. doi: 10.1371/journal.pone.0036961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maeda S., Tsukada S., Kanazawa A., Sekine A., Tsunoda T., Koya D. Genetic variations in the gene encoding TFAP2B are associated with type 2 diabetes mellitus. Journal of Human Genetics. 2005;50(6):283–292. doi: 10.1007/s10038-005-0253-9. [DOI] [PubMed] [Google Scholar]
- 126.Hwang J.H., Kim A.R., Kim K.M., Il Park J., Oh H.T., Moon S.A. TAZ couples Hippo/Wnt signalling and insulin sensitivity through Irs1 expression. Nature Communications. 2019;10(1):421. doi: 10.1038/s41467-019-08287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jhala U.S., Canettieri G., Screaton R.A., Kulkarni R.N., Krajewski S., Reed J. cAMP promotes pancreatic beta-cell survival via CREB-mediated induction of IRS2. Genes & Development. 2003;17(13):1575–1580. doi: 10.1101/gad.1097103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Besse-Patin A., Jeromson S., Levesque-Damphousse P., Secco B., Laplante M., Estall J.L. PGC1A regulates the IRS1:IRS2 ratio during fasting to influence hepatic metabolism downstream of insulin. Proceedings of the National Academy of Sciences of the U S A. 2019;116(10):4285–4290. doi: 10.1073/pnas.1815150116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Canettieri G., Koo S.H., Berdeaux R., Heredia J., Hedrick S., Zhang X. Dual role of the coactivator TORC2 in modulating hepatic glucose output and insulin signaling. Cell Metabolism. 2005;2(5):331–338. doi: 10.1016/j.cmet.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 130.Shimomura I., Bashmakov Y., Ikemoto S., Horton J.D., Brown M.S., Goldstein J.L. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proceedings of the National Academy of Sciences of the U S A. 1999;96(24):13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ide T., Shimano H., Yahagi N., Matsuzaka T., Nakakuki M., Yamamoto T. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nature Cell Biology. 2004;6(4):351–357. doi: 10.1038/ncb1111. [DOI] [PubMed] [Google Scholar]
- 132.Rui L., Yuan M., Frantz D., Shoelson S., White M.F. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. Journal of Biological Chemistry. 2002;277(44):42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 133.Liu F., Yang T., Wang B., Zhang M., Gu N., Qiu J. Resistin induces insulin resistance, but does not affect glucose output in rat-derived hepatocytes. Acta Pharmacologica Sinica. 2008;29(1):98–104. doi: 10.1111/j.1745-7254.2008.00709.x. [DOI] [PubMed] [Google Scholar]
- 134.Xu X., Sarikas A., Dias-Santagata D.C., Dolios G., Lafontant P.J., Tsai S.C. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Molecular Cell. 2008;30(4):403–414. doi: 10.1016/j.molcel.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu X., Keshwani M., Meyer K., Sarikas A., Taylor S., Pan Z.Q. Identification of the degradation determinants of insulin receptor substrate 1 for signaling cullin-RING E3 ubiquitin ligase 7-mediated ubiquitination. Journal of Biological Chemistry. 2012;287(48):40758–40766. doi: 10.1074/jbc.M112.405209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bonala S., Lokireddy S., McFarlane C., Patnam S., Sharma M., Kambadur R. Myostatin induces insulin resistance via Casitas B-lineage lymphoma b (Cblb)-mediated degradation of insulin receptor substrate 1 (IRS1) protein in response to high calorie diet intake. Journal of Biological Chemistry. 2014;289(11):7654–7670. doi: 10.1074/jbc.M113.529925. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 137.Hancer N.J., Qiu W., Cherella C., Li Y., Copps K.D., White M.F. Insulin and metabolic stress stimulate multisite serine/threonine phosphorylation of insulin receptor substrate 1 and inhibit tyrosine phosphorylation. Journal of Biological Chemistry. 2014;289(18):12467–12484. doi: 10.1074/jbc.M114.554162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Copps K.D., Hancer N.J., Opare-Ado L., Qiu W., Walsh C., White M.F. Irs1 serine 307 promotes insulin sensitivity in mice. Cell Metabolism. 2010;11(1):84–92. doi: 10.1016/j.cmet.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Copps K.D., Hancer N.J., Qiu W., White M.F. Serine 302 phosphorylation of mouse insulin receptor substrate 1 (IRS1) is dispensable for normal insulin signaling and feedback regulation by hepatic S6 kinase. Journal of Biological Chemistry. 2016;291(16):8602–8617. doi: 10.1074/jbc.M116.714915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tiganis T. PTP1B and TCPTP–nonredundant phosphatases in insulin signaling and glucose homeostasis. FEBS Journal. 2013;280(2):445–458. doi: 10.1111/j.1742-4658.2012.08563.x. [DOI] [PubMed] [Google Scholar]
- 141.Kushner J.A., Haj F.G., Klaman L.D., Dow M.A., Kahn B.B., Neel B.G. Islet-sparing effects of protein tyrosine phosphatase-1b deficiency delays onset of diabetes in IRS2 knockout mice. Diabetes. 2004;53(1):61–66. doi: 10.2337/diabetes.53.1.61. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Z.Y., Dodd G.T., Tiganis T. Protein tyrosine phosphatases in hypothalamic insulin and leptin signaling. Trends in Pharmacological Sciences. 2015;36(10):661–674. doi: 10.1016/j.tips.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dodd G.T., Xirouchaki C.E., Eramo M., Mitchell C.A., Andrews Z.B., Henry B.A. Intranasal targeting of hypothalamic PTP1B and TCPTP reinstates leptin and insulin sensitivity and promotes weight loss in obesity. Cell Reports. 2019;28(11):2905–2922. doi: 10.1016/j.celrep.2019.08.019. e2905. [DOI] [PubMed] [Google Scholar]
- 144.Parsons R. Human cancer, PTEN and the PI-3 kinase pathway. Seminars in Cell & Developmental Biology. 2004;15(2):171–176. doi: 10.1016/j.semcdb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 145.Polianskyte-Prause Z., Tolvanen T.A., Lindfors S., Dumont V., Van M., Wang H. Metformin increases glucose uptake and acts renoprotectively by reducing SHIP2 activity. The FASEB Journal. 2019;33(2):2858–2869. doi: 10.1096/fj.201800529RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Carlson C.J., White M.F., Rondinone C.M. Mammalian target of rapamycin regulates IRS-1 serine 307 phosphorylation. Biochemical and Biophysical Research Communications. 2004;316(2):533–539. doi: 10.1016/j.bbrc.2004.02.082. [DOI] [PubMed] [Google Scholar]
- 147.Nigi L., Grieco G.E., Ventriglia G., Brusco N., Mancarella F., Formichi C. MicroRNAs as regulators of insulin signaling: research updates and potential therapeutic perspectives in type 2 diabetes. International Journal of Molecular Science. 2018;19(12) doi: 10.3390/ijms19123705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhu H., Shyh-Chang N., Segre A.V., Shinoda G., Shah S.P., Einhorn W.S. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Min K.H., Yang W.M., Lee W. Saturated fatty acids-induced miR-424-5p aggravates insulin resistance via targeting insulin receptor in hepatocytes. Biochemical and Biophysical Research Communications. 2018;503(3):1587–1593. doi: 10.1016/j.bbrc.2018.07.084. [DOI] [PubMed] [Google Scholar]
- 150.Yang W.M., Jeong H.J., Park S.W., Lee W. Obesity-induced miR-15b is linked causally to the development of insulin resistance through the repression of the insulin receptor in hepatocytes. Molecular Nutrition & Food Research. 2015;59(11):2303–2314. doi: 10.1002/mnfr.201500107. [DOI] [PubMed] [Google Scholar]
- 151.Yang W.M., Jeong H.J., Park S.Y., Lee W. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Letters. 2014;588(21):3939–3946. doi: 10.1016/j.febslet.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 152.Yang W.M., Min K.H., Lee W. Induction of miR-96 by Dietary Saturated Fatty Acids Exacerbates Hepatic Insulin Resistance through the Suppression of INSR and IRS-1. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0169039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ono K., Igata M., Kondo T., Kitano S., Takaki Y., Hanatani S. Identification of microRNA that represses IRS-1 expression in liver. PLoS One. 2018;13(1) doi: 10.1371/journal.pone.0191553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wen F., Yang Y., Jin D., Sun J., Yu X., Yang Z. MiRNA-145 is involved in the development of resistin-induced insulin resistance in HepG2 cells. Biochemical and Biophysical Research Communications. 2014;445(2):517–523. doi: 10.1016/j.bbrc.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 155.Massart J., Sjogren R.J.O., Lundell L.S., Mudry J.M., Franck N., O'Gorman D.J. Altered miR-29 expression in type 2 diabetes influences glucose and lipid metabolism in skeletal muscle. Diabetes. 2017;66(7):1807–1818. doi: 10.2337/db17-0141. [DOI] [PubMed] [Google Scholar]
- 156.Tao H., Wang M.M., Zhang M., Zhang S.P., Wang C.H., Yuan W.J. MiR-126 Suppresses the Glucose-Stimulated Proliferation via IRS-2 in INS-1 beta Cells. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0149954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Agarwal P., Srivastava R., Srivastava A.K., Ali S., Datta M. miR-135a targets IRS2 and regulates insulin signaling and glucose uptake in the diabetic gastrocnemius skeletal muscle. Biochimica et Biophysica Acta. 2013;1832(8):1294–1303. doi: 10.1016/j.bbadis.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 158.Paolisso G., Tagliamonte M.R., Rizzo M.R., Giugliano D. Advancing age and insulin resistance: new facts about an ancient history. European Journal of Clinical Investigation. 1999;29(9):758–769. doi: 10.1046/j.1365-2362.1999.00522.x. [DOI] [PubMed] [Google Scholar]
- 159.Warram J.H., Martin B.C., Krolewski A.S., Soeldner J.S., Kahn C.R. Slow glucose removal rate and hyperinsulinemia precede the development of type II diabetes in the offspring of diabetic parents. Annals of Internal Medicine. 1990;113(12):909–915. doi: 10.7326/0003-4819-113-12-909. [DOI] [PubMed] [Google Scholar]
- 160.Lillioja S., Mott D.M., Howard B.V., Bennett P.H., Yki-Jarvinen H., Freymond D. Impaired glucose tolerance as a disorder of insulin action. Longitudinal and cross-sectional studies in Pima Indians. New England Journal of Medicine. 1988;318(19):1217–1225. doi: 10.1056/NEJM198805123181901. [DOI] [PubMed] [Google Scholar]
- 161.Haffner S.M., Stern M.P., Dunn J., Mobley M., Blackwell J., Bergman R.N. Diminished insulin sensitivity and increased insulin response in nonobese, nondiabetic Mexican Americans. Metabolism. 1990;39(8):842–847. doi: 10.1016/0026-0495(90)90130-5. [DOI] [PubMed] [Google Scholar]
- 162.Reaven G.M., Bernstein R., Davis B., Olefsky J.M. Nonketotic diabetes mellitus: insulin deficiency or insulin resistance? Americas Journal of Medicine. 1976;60(1):80–88. doi: 10.1016/0002-9343(76)90536-2. [DOI] [PubMed] [Google Scholar]
- 163.Donath M.Y., Shoelson S.E. Type 2 diabetes as an inflammatory disease. Nature Reviews Immunology. 2011;11(2):98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- 164.Roden M., Shulman G.I. The integrative biology of type 2 diabetes. Nature. 2019;576(7785):51–60. doi: 10.1038/s41586-019-1797-8. [DOI] [PubMed] [Google Scholar]
- 165.Petersen M.C., Shulman G.I. Mechanisms of insulin action and insulin resistance. Physiological Reviews. 2018;98(4):2133–2223. doi: 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gavin J.R., 3rd, Roth J., Neville D.M., Jr., de Meyts P., Buell D.N. Insulin-dependent regulation of insulin receptor concentrations: a direct demonstration in cell culture. Proceedings of the National Academy of Sciences of the U S A. 1974;71(1):84–88. doi: 10.1073/pnas.71.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kubota N., Kubota T., Itoh S., Kumagai H., Kozono H., Takamoto I. Dynamic functional relay between insulin receptor substrate 1 and 2 in hepatic insulin signaling during fasting and feeding. Cell Metabolism. 2008;8(1):49–64. doi: 10.1016/j.cmet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 168.Mehran A.E., Templeman N.M., Brigidi G.S., Lim G.E., Chu K.Y., Hu X. Hyperinsulinemia drives diet-induced obesity independently of brain insulin production. Cell Metabolism. 2012;16(6):723–737. doi: 10.1016/j.cmet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 169.Page M.M., Skovso S., Cen H., Chiu A.P., Dionne D.A., Hutchinson D.F. Reducing insulin via conditional partial gene ablation in adults reverses diet-induced weight gain. The FASEB Journal. 2018;32(3):1196–1206. doi: 10.1096/fj.201700518R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Newgard C.B., An J., Bain J.R., Muehlbauer M.J., Stevens R.D., Lien L.F. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yu Y., Yoon S.O., Poulogiannis G., Yang Q., Ma X.M., Villen J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332(6035):1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Samuel V.T., Shulman G.I. Nonalcoholic fatty liver disease, insulin resistance, and ceramides. New England Journal of Medicine. 2019;381(19):1866–1869. doi: 10.1056/NEJMcibr1910023. [DOI] [PubMed] [Google Scholar]
- 173.Lo K.A., Labadorf A., Kennedy N.J., Han M.S., Yap Y.S., Matthews B. Analysis of in vitro insulin-resistance models and their physiological relevance to in vivo diet-induced adipose insulin resistance. Cell Reports. 2013;5(1):259–270. doi: 10.1016/j.celrep.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Holland W.L., Miller R.A., Wang Z.V., Sun K., Barth B.M., Bui H.H. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nature Medicine. 2011;17(1):55–63. doi: 10.1038/nm.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Levy J.R., Campbell K.P., Glass D.J. MG53's new identity. Skeletal Muscle. 2013;3(1):25. doi: 10.1186/2044-5040-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Reilly S.M., Saltiel A.R. Obesity: a complex role for adipose tissue macrophages. Nature Reviews Endocrinology. 2014;10(4):193–194. doi: 10.1038/nrendo.2014.12. [DOI] [PubMed] [Google Scholar]
- 177.Wedell-Neergaard A.S., Lang Lehrskov L., Christensen R.H., Legaard G.E., Dorph E., Larsen M.K. Exercise-induced changes in visceral adipose tissue mass are regulated by IL-6 signaling: a randomized controlled trial. Cell Metabolism. 2019;29(4):844–855. doi: 10.1016/j.cmet.2018.12.007. e843. [DOI] [PubMed] [Google Scholar]
- 178.Saltiel A.R. Insulin resistance in the defense against obesity. Cell Metabolism. 2012;15(6):798–804. doi: 10.1016/j.cmet.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 179.Lumeng C.N., Bodzin J.L., Saltiel A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. Journal of Clinical Investigation. 2007;117(1):175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shoelson S.E., Herrero L., Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132(6):2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- 181.Benatti F.B., Pedersen B.K. Exercise as an anti-inflammatory therapy for rheumatic diseases-myokine regulation. Nature Reviews Rheumatology. 2015;11(2):86–97. doi: 10.1038/nrrheum.2014.193. [DOI] [PubMed] [Google Scholar]
- 182.Sadagurski M., Norquay L., Farhang J., D'Aquino K., Copps K., White M.F. Human IL6 enhances leptin action in mice. Diabetologia. 2010;53(3):525–535. doi: 10.1007/s00125-009-1580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ueki K., Kondo T., Kahn C.R. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Molecular and Cellular Biology. 2004;24(12):5434–5446. doi: 10.1128/MCB.24.12.5434-5446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shi H., Kokoeva M.V., Inouye K., Tzameli I., Yin H., Flier J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. Journal of Clinical Investigation. 2006;116(11):3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Marciniak S.J., Ron D. Endoplasmic reticulum stress signaling in disease. Physiological Reviews. 2006;86(4):1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 186.Schroder M., Kaufman R.J. The mammalian unfolded protein response. Annual Review of Biochemistry. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 187.Oyadomari S., Harding H.P., Zhang Y., Oyadomari M., Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metabolism. 2008;7(6):520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ozcan U., Cao Q., Yilmaz E., Lee A.H., Iwakoshi N.N., Ozdelen E. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306(5695):457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 189.Zhou Y., Lee J., Reno C.M., Sun C., Park S.W., Chung J. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nature Medicine. 2011;17(3):356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tam A.B., Roberts L.S., Chandra V., Rivera I.G., Nomura D.K., Forbes D.J. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Developmental Cell. 2018;46(3):327–343. doi: 10.1016/j.devcel.2018.04.023. e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu J., Lee J., Salazar Hernandez M.A., Mazitschek R., Ozcan U. Treatment of obesity with celastrol. Cell. 2015;161(5):999–1011. doi: 10.1016/j.cell.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R.O. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313(5790):1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sharma N.K., Das S.K., Mondal A.K., Hackney O.G., Chu W.S., Kern P.A. Endoplasmic reticulum stress markers are associated with obesity in nondiabetic subjects. Journal of Clinical Endocrinology & Metabolism. 2008;93(11):4532–4541. doi: 10.1210/jc.2008-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cefalu W.T., Werbel S., Bell-Farrow A.D., Terry J.G., Wang Z.Q., Opara E.C. Insulin resistance and fat patterning with aging: relationship to metabolic risk factors for cardiovascular disease. Metabolism. 1998;47(4):401–408. doi: 10.1016/s0026-0495(98)90050-6. [DOI] [PubMed] [Google Scholar]
- 195.Tran T.T., Yamamoto Y., Gesta S., Kahn C.R. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metabolism. 2008;7(5):410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McLaughlin T., Lamendola C., Liu A., Abbasi F. Preferential fat deposition in subcutaneous versus visceral depots is associated with insulin sensitivity. Journal of Clinical Endocrinology & Metabolism. 2011;96(11):E1756–E1760. doi: 10.1210/jc.2011-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Arner P., Hellstrom L., Wahrenberg H., Bronnegard M. Beta-adrenoceptor expression in human fat cells from different regions. Journal of Clinical Investigation. 1990;86(5):1595–1600. doi: 10.1172/JCI114880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nicklas B.J., Rogus E.M., Colman E.G., Goldberg A.P. Visceral adiposity, increased adipocyte lipolysis, and metabolic dysfunction in obese postmenopausal women. American Journal of Physiology. 1996;270(1 Pt 1):E72–E78. doi: 10.1152/ajpendo.1996.270.1.E72. [DOI] [PubMed] [Google Scholar]
- 199.Samuel V.T., Shulman G.I. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. Journal of Clinical Investigation. 2016;126(1):12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the metabolic syndrome. Endocrinology Nutrition. 2013;1(60 Suppl):39–43. doi: 10.1016/s1575-0922(13)70026-3. [DOI] [PubMed] [Google Scholar]
- 201.Karastergiou K., Mohamed-Ali V. The autocrine and paracrine roles of adipokines. Molecular and Cellular Endocrinology. 2010;318(1–2):69–78. doi: 10.1016/j.mce.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 202.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. New England Journal of Medicine. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dempersmier J., Sul H.S. Shades of brown: a model for thermogenic fat. Frontiers in Endocrinology. 2015;6:71. doi: 10.3389/fendo.2015.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Raiko J., Orava J., Savisto N., Virtanen K.A. High Brown fat activity correlates with cardiovascular risk factor levels cross-sectionally and subclinical atherosclerosis at 5-year follow-up. Arteriosclerosis, Thrombosis, and Vascular Biology. 2020;40(5):1289–1295. doi: 10.1161/ATVBAHA.119.313806. [DOI] [PubMed] [Google Scholar]
- 205.Perseghin G., Scifo P., De Cobelli F., Pagliato E., Battezzati A., Arcelloni C. Intramyocellular triglyceride content is a determinant of in vivo insulin resistance in humans: a 1H-13C nuclear magnetic resonance spectroscopy assessment in offspring of type 2 diabetic parents. Diabetes. 1999;48(8):1600–1606. doi: 10.2337/diabetes.48.8.1600. [DOI] [PubMed] [Google Scholar]
- 206.Griffin M.E., Marcucci M.J., Cline G.W., Bell K., Barucci N., Lee D. Free fatty acid-induced insulin resistance is associated with activation of protein kinase C theta and alterations in the insulin signaling cascade. Diabetes. 1999;48(6):1270–1274. doi: 10.2337/diabetes.48.6.1270. [DOI] [PubMed] [Google Scholar]
- 207.Itani S.I., Pories W.J., Macdonald K.G., Dohm G.L. Increased protein kinase C theta in skeletal muscle of diabetic patients. Metabolism. 2001;50(5):553–557. doi: 10.1053/meta.2001.22512. [DOI] [PubMed] [Google Scholar]
- 208.Carlson L.A., Ekelund L.G., Froberg S.O. Concentration of triglycerides, phospholipids and glycogen in skeletal muscle and of free fatty acids and beta-hydroxybutyric acid in blood in man in response to exercise. European Journal of Clinical Investigation. 1971;1(4):248–254. doi: 10.1111/eci.1971.1.4.248. [DOI] [PubMed] [Google Scholar]
- 209.Laws A., Reaven G.M. Effect of physical activity on age-related glucose intolerance. Clinics in Geriatric Medicine. 1990;6(4):849–863. [PubMed] [Google Scholar]
- 210.Gollnick P.D., Saltin B. Significance of skeletal muscle oxidative enzyme enhancement with endurance training. Clinical Physiology. 1982;2(1):1–12. doi: 10.1111/j.1475-097x.1982.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 211.Turcotte L.P., Richter E.A., Kiens B. Increased plasma FFA uptake and oxidation during prolonged exercise in trained vs. untrained humans. American Journal of Physiology. 1992;262(6 Pt 1):E791–E799. doi: 10.1152/ajpendo.1992.262.6.E791. [DOI] [PubMed] [Google Scholar]
- 212.Romijn J.A., Klein S., Coyle E.F., Sidossis L.S., Wolfe R.R. 1993. Strenuous endurance training increases lipolysis and triglyceride-fatty acid cycling at rest. Journal of Applied Physiology. 1985;75(1):108–113. doi: 10.1152/jappl.1993.75.1.108. [DOI] [PubMed] [Google Scholar]
- 213.Phillips S.M., Green H.J., Tarnopolsky M.A., Heigenhauser G.F., Hill R.E., Grant S.M. 1996. Effects of training duration on substrate turnover and oxidation during exercise. Journal of Applied Physiology. 1985;81(5):2182–2191. doi: 10.1152/jappl.1996.81.5.2182. [DOI] [PubMed] [Google Scholar]
- 214.Shepherd S.O., Strauss J.A., Wang Q., Dube J.J., Goodpaster B., Mashek D.G. Training alters the distribution of perilipin proteins in muscle following acute free fatty acid exposure. Journal of Physiology. 2017;595(16):5587–5601. doi: 10.1113/JP274374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Loomba R., Sanyal A.J. The global NAFLD epidemic. Nature Reviews Gastroenterology & Hepatology. 2013;10(11):686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 216.Softic S., Gupta M.K., Wang G.X., Fujisaka S., O'Neill B.T., Rao T.N. Divergent effects of glucose and fructose on hepatic lipogenesis and insulin signaling. Journal of Clinical Investigation. 2017;127(11):4059–4074. doi: 10.1172/JCI94585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Softic S., Cohen D.E., Kahn C.R. Role of dietary fructose and hepatic de novo lipogenesis in fatty liver disease. Digestive Diseases and Sciences. 2016;61(5):1282–1293. doi: 10.1007/s10620-016-4054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Schrauwen P., Hesselink M.K. Oxidative capacity, lipotoxicity, and mitochondrial damage in type 2 diabetes. Diabetes. 2004;53(6):1412–1417. doi: 10.2337/diabetes.53.6.1412. [DOI] [PubMed] [Google Scholar]
- 219.Boirie Y. Insulin regulation of mitochondrial proteins and oxidative phosphorylation in human muscle. Trends in Endocrinology and Metabolism. 2003;14(9):393–394. doi: 10.1016/j.tem.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 220.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 221.Patti M.E., Butte A.J., Crunkhorn S., Cusi K., Berria R., Kashyap S. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proceedings of the National Academy of Sciences of the U S A. 2003;100(14):8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kelley D.E., He J., Menshikova E.V., Ritov V.B. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51(10):2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 223.Schooneman M.G., Vaz F.M., Houten S.M., Soeters M.R. Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes. 2013;62(1):1–8. doi: 10.2337/db12-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.LaBarge S., Migdal C., Schenk S. Is acetylation a metabolic rheostat that regulates skeletal muscle insulin action? Molecular Cell. 2015;38(4):297–303. doi: 10.14348/molcells.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hirschey M.D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D.B. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jing E., O'Neill B.T., Rardin M.J., Kleinridders A., Ilkeyeva O.R., Ussar S. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62(10):3404–3417. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.He W., Newman J.C., Wang M.Z., Ho L., Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends in Endocrinology and Metabolism. 2012;23(9):467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 228.Newman J.C., He W., Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. Journal of Biological Chemistry. 2012;287(51):42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Marette A., Liu Y., Sweeney G. Skeletal muscle glucose metabolism and inflammation in the development of the metabolic syndrome. Reviews in Endocrine & Metabolic Disorders. 2014;15(4):299–305. doi: 10.1007/s11154-014-9296-6. [DOI] [PubMed] [Google Scholar]
- 230.Del Prato S., Bonadonna R.C., Bonora E., Gulli G., Solini A., Shank M. Characterization of cellular defects of insulin action in type 2 (non-insulin-dependent) diabetes mellitus. Journal of Clinical Investigation. 1993;91(2):484–494. doi: 10.1172/JCI116226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Freymond D., Bogardus C., Okubo M., Stone K., Mott D. Impaired insulin-stimulated muscle glycogen synthase activation in vivo in man is related to low fasting glycogen synthase phosphatase activity. Journal of Clinical Investigation. 1988;82(5):1503–1509. doi: 10.1172/JCI113758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Roden M., Price T.B., Perseghin G., Petersen K.F., Rothman D.L., Cline G.W. Mechanism of free fatty acid-induced insulin resistance in humans. Journal of Clinical Investigation. 1996;97(12):2859–2865. doi: 10.1172/JCI118742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Jucker B.M., Rennings A.J., Cline G.W., Shulman G.I. 13C and 31P NMR studies on the effects of increased plasma free fatty acids on intramuscular glucose metabolism in the awake rat. Journal of Biological Chemistry. 1997;272(16):10464–10473. doi: 10.1074/jbc.272.16.10464. [DOI] [PubMed] [Google Scholar]
- 234.Hundal R.S., Petersen K.F., Mayerson A.B., Randhawa P.S., Inzucchi S., Shoelson S.E. Mechanism by which high-dose aspirin improves glucose metabolism in type 2 diabetes. Journal of Clinical Investigation. 2002;109(10):1321–1326. doi: 10.1172/JCI14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Goldfine A.B., Fonseca V., Jablonski K.A., Pyle L., Staten M.A., Shoelson S.E. The effects of salsalate on glycemic control in patients with type 2 diabetes: a randomized trial. Annals of Internal Medicine. 2010;152(6):346–357. doi: 10.1059/0003-4819-152-6-201003160-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ruderman N.B., Saha A.K., Vavvas D., Witters L.A. Malonyl-CoA, fuel sensing, and insulin resistance. American Journal of Physiology. 1999;276(1):E1–E18. doi: 10.1152/ajpendo.1999.276.1.E1. [DOI] [PubMed] [Google Scholar]
- 237.McGarry J.D. Glucose-fatty acid interactions in health and disease. American Journal of Clinical Nutrition. 1998;67(3 Suppl):500S–504S. doi: 10.1093/ajcn/67.3.500S. [DOI] [PubMed] [Google Scholar]
- 238.McGarry J.D. Malonyl-CoA and satiety? Food for thought. Trends in Endocrinology and Metabolism. 2000;11(10):399–400. doi: 10.1016/s1043-2760(00)00331-3. [DOI] [PubMed] [Google Scholar]
- 239.Swanson S.T., Foster D.W., McGarry J.D., Brown N.F. Roles of the N- and C-terminal domains of carnitine palmitoyltransferase I isoforms in malonyl-CoA sensitivity of the enzymes: insights from expression of chimaeric proteins and mutation of conserved histidine residues. Biochemical Journal. 1998;335(Pt 3):513–519. doi: 10.1042/bj3350513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Kelley D.E., Mandarino L.J. Hyperglycemia normalizes insulin-stimulated skeletal muscle glucose oxidation and storage in noninsulin-dependent diabetes mellitus. Journal of Clinical Investigation. 1990;86(6):1999–2007. doi: 10.1172/JCI114935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kelley D.E., Simoneau J.A. Impaired free fatty acid utilization by skeletal muscle in non-insulin-dependent diabetes mellitus. Journal of Clinical Investigation. 1994;94(6):2349–2356. doi: 10.1172/JCI117600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bavenholm P.N., Pigon J., Saha A.K., Ruderman N.B., Efendic S. Fatty acid oxidation and the regulation of malonyl-CoA in human muscle. Diabetes. 2000;49(7):1078–1083. doi: 10.2337/diabetes.49.7.1078. [DOI] [PubMed] [Google Scholar]
- 243.Shreiner A.B., Kao J.Y., Young V.B. The gut microbiome in health and in disease. Current Opinion in Gastroenterology. 2015;31(1):69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ussar S., Griffin N.W., Bezy O., Fujisaka S., Vienberg S., Softic S. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metabolism. 2015;22(3):516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tilg H., Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J.Clin. Invest. 2011;121(6):2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Fujisaka S., Ussar S., Clish C., Devkota S., Dreyfuss J.M., Sakaguchi M. Antibiotic effects on gut microbiota and metabolism are host dependent. Journal of Clinical Investigation. 2016;126(12):4430–4443. doi: 10.1172/JCI86674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 248.Tilg H., Adolph T.E. Influence of the human intestinal microbiome on obesity and metabolic dysfunction. Current Opinion in Pediatrics. 2015;27(4):496–501. doi: 10.1097/MOP.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 249.Perry R.J., Peng L., Barry N.A., Cline G.W., Zhang D., Cardone R.L. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534(7606):213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Nieuwdorp M., Gilijamse P.W., Pai N., Kaplan L.M. Role of the microbiome in energy regulation and metabolism. Gastroenterology. 2014;146(6):1525–1533. doi: 10.1053/j.gastro.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 251.Ussar S., Fujisaka S., Kahn C.R. Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome. Mol Metab. 2016;5(9):795–803. doi: 10.1016/j.molmet.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tremaroli V., Karlsson F., Werling M., Stahlman M., Kovatcheva-Datchary P., Olbers T. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metabolism. 2015;22(2):228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Blaser M.J. The microbiome revolution. Journal of Clinical Investigation. 2014;124(10):4162–4165. doi: 10.1172/JCI78366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nature Reviews Immunology. 2016;16(6):341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167(4):1125–1136. doi: 10.1016/j.cell.2016.10.020. e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Burcelin R., Serino M., Chabo C., Blasco-Baque V., Amar J. Gut microbiota and diabetes: from pathogenesis to therapeutic perspective. Acta Diabetologica. 2011;48(4):257–273. doi: 10.1007/s00592-011-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Joyce S.A., MacSharry J., Casey P.G., Kinsella M., Murphy E.F., Shanahan F. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proceedings of the National Academy of Sciences of the U S A. 2014;111(20):7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Allayee H., Hazen S.L. Contribution of gut bacteria to lipid levels: another metabolic role for microbes? Circulation Research. 2015;117(9):750–754. doi: 10.1161/CIRCRESAHA.115.307409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Vinje S., Stroes E., Nieuwdorp M., Hazen S.L. The gut microbiome as novel cardio-metabolic target: the time has come! European Heart Journal. 2014;35(14):883–887. doi: 10.1093/eurheartj/eht467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Fujisaka S., Avila-Pacheco J., Soto M., Kostic A., Dreyfuss J.M., Pan H. Diet, genetics, and the gut microbiome drive dynamic changes in plasma metabolites. Cell Reports. 2018;22(11):3072–3086. doi: 10.1016/j.celrep.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Newgard C.B. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metabolism. 2012;15(5):606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Gall W.E., Beebe K., Lawton K.A., Adam K.P., Mitchell M.W., Nakhle P.J. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PloS One. 2010;5(5):e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Menni C., Fauman E., Erte I., Perry J.R., Kastenmuller G., Shin S.Y. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62(12):4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Martin B.C., Warram J.H., Krolewski A.S., Bergman R.N., Soeldner J.S., Kahn C.R. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet. 1992;340(8825):925–929. doi: 10.1016/0140-6736(92)92814-v. [DOI] [PubMed] [Google Scholar]
- 266.Czech M.P. Insulin action and resistance in obesity and type 2 diabetes. Nature Medicine. 2017;23(7):804–814. doi: 10.1038/nm.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Cusi K., Maezono K., Osman A., Pendergrass M., Patti M.E., Pratipanawatr T. Insulin resistance differentially affects the PI 3-kinase- and MAP kinase-mediated signaling in human muscle. Journal of Clinical Investigation. 2000;105(3):311–320. doi: 10.1172/JCI7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Mashili F., Chibalin A.V., Krook A., Zierath J.R. Constitutive STAT3 phosphorylation contributes to skeletal muscle insulin resistance in type 2 diabetes. Diabetes. 2013;62(2):457–465. doi: 10.2337/db12-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Batista T.M., Jayavelu A.K., Wewer Albrechtsen N.J., Iovino S., Lebastchi J., Pan H. A cell-autonomous signature of dysregulated protein phosphorylation underlies muscle insulin resistance in type 2 diabetes. Cell Metabolism. 2020;32(5):844–859. doi: 10.1016/j.cmet.2020.08.007. e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Rhoads R.E. Signal transduction pathways that regulate eukaryotic protein synthesis. Journal of Biological Chemistry. 1999;274(43):30337–30340. doi: 10.1074/jbc.274.43.30337. [DOI] [PubMed] [Google Scholar]
- 271.Kamura T., Sato S., Haque D., Liu L., Kaelin W.G., Jr., Conaway R.C. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes & Development. 1998;12(24):3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zhang Z., Elly C., Qiu L., Altman A., Liu Y.C. A direct interaction between the adaptor protein Cbl-b and the kinase zap-70 induces a positive signal in T cells. Current Biology. 1999;9(4):203–206. doi: 10.1016/s0960-9822(99)80090-6. [DOI] [PubMed] [Google Scholar]