Abstract
Initiation of DNA replication in eukaryotes requires the assembly of prereplication complexes (pre-Rcs) at the origins of replication. The assembly and function of the pre-Rcs appear to be controlled by phosphorylation events. In this study we report the detailed characterization of the cell cycle phosphorylation of one component of the Xenopus pre-Rcs, the Mcm protein complex. We show that individual Mcm subunits are differentially phosphorylated during the cell cycle. During mitosis, the Mcm4 subunit is hyperphosphorylated, while the other subunits are not actively phosphorylated. The mitotic phosphorylation of Mcm4 requires Cdc2-cyclin B and other unknown kinases. Following exit from mitosis, the Mcm4 subunit of the cytosolic interphase complex undergoes dephosphorylation, and the Mcm2, Mcm3, or Mcm6 subunits are then actively phosphorylated by kinase(s) other than cyclin-dependent kinases (Cdks) or Cdc7. The association of the Mcm complex with the pre-Rcs correlates with the formation of a transient interphase complex. This complex contains an intermediately phosphorylated Mcm4 subunit and is produced by partial dephosphorylation of the mitotic hyperphosphorylated Mcm4 protein. Complete dephosphorylation of the Mcm4 subunit inactivates the Mcm complex and prevents its binding to the chromatin. Once the Mcm complex is assembled on the chromatin the Mcm4 and the Mcm2 proteins are the only subunits phosphorylated during the activation of the pre-Rcs. These chromatin-associated phosphorylations require nuclear transport and are independent of Cdk2-cyclin E. These results suggest that the changes in Mcm4 phosphorylation regulate pre-Rc assembly and the function of the pre-Rcs on the chromatin.
Initiation of DNA replication in eukaryotes is a two-step process (reviewed in references 12 and 39). First, the DNA is licensed through the establishment of prereplication complexes (pre-Rcs) at the replication origins. Second, the pre-Rcs are activated, resulting in the unwinding of the origins and loading of the replication machinery. Studies using Saccharomyces cerevisiae, Xenopus laevis, and cultured mammalian cells as model systems indicate that these two steps are highly conserved among eukaryotes and tightly regulated during the cell cycle. The formation of the pre-Rcs involves the sequential loading onto the chromatin of the origin recognition complex (ORC), followed by Cdc6 and the minichromosome maintenance proteins (6, 10). All of these proteins are essential for the initiation of replication and homologs have been identified in yeast, frogs, flies, and mammals. The ORC in budding yeast is a multisubunit complex that binds to the origin of replication located within the autonomous replicating sequence (2, 15). The observations that the ORC does not have DNA unwinding activity and can be removed from the chromatin before initiation indicate that its role is more likely to target other components of the pre-Rcs to the chromatin (10, 18). The Cdc6 protein is a single polypeptide, and its recruitment to the pre-Rcs, through its interaction with ORC, is required for the loading of the Mcm proteins (6). The fact that the Cdc6 protein shares sequence similarity with the eukaryotic and prokaryotic clamp loader suggests that Cdc6 is the loading factor that clamps the Mcm proteins around the DNA duplex (30, 33). The Mcm proteins form a heterohexameric complex that is composed of different subcomplexes. Mcm4, Mcm6, and Mcm7 form a tightly interacting core complex that interacts with Mcm2 and also with a dimer formed between Mcm3 and Mcm5 (7, 37). The relationship between these subcomplexes and the biochemical properties of the various Mcm proteins is currently unknown. Several lines of evidence suggest that the Mcm proteins contain helicase activity. For example, they have a hexameric structure and an ATPase motif common to several known helicases (21). Further, the Mcm proteins are physically associated with the moving replication fork in vivo (1). Finally, they exhibit helicase activity in vitro (20). However, the lack of processivity of this in vitro Mcm helicase activity raises questions about its physiological relevance.
After assembly, the activation of the pre-Rcs requires the action of at least two distinct kinases, the S-phase cyclin-dependent kinase (Cdk) and the Cdc7-Dbf4 kinase (reviewed in reference 23). While it is likely that the activation of the pre-Rcs leads to origin unwinding and priming of replication, the molecular events of activation are yet to be determined. In particular, the role of these two kinase complexes relative to each other and their in vivo targets are presently unknown. Recent work using S. cerevisiae and X. laevis indicates that the S-phase Cdk regulates, directly or indirectly, the association between the chromatin and the Cdc45 protein, an essential initiation factor (27, 41). On the other hand, the Cdc7-Dbf4 kinase is required throughout S-phase for initiation at both early- and late-firing origins (3, 9). Genetic analysis in S. cerevisiae has shown that both Cdc7 and Dbf4 interact with the ORC and Mcm proteins (11, 16). Furthermore, yeast and human homologs of the Cdc7-Dbf4 kinase can phosphorylate some members of the Mcm protein family in vitro (5, 24, 36). This evidence raises the possibility that the Cdc7-Dbf4 kinase regulates the function of the Mcm proteins. In addition to their positive effects on the initiation of replication, S-phase Cdks also exert a negative effect on the assembly of the pre-Rcs and thereby provide a mechanism to prevent rereplication during S and G2 phases. While we do not know exactly how this inhibitory effect is achieved, there is some evidence that it targets the ability of the Cdc6 protein to load the Mcm proteins (reviewed in reference 32).
It is clear that phosphorylation regulates the assembly and disassembly and the function of the pre-Rcs. While the phosphorylation targets are unknown, some likely candidates are components of the pre-Rcs. In this report we provide a detailed characterization of the cell cycle phosphorylation of one component of the Xenopus pre-Rcs, the Mcm proteins. We show that individual subunits of the Xenopus Mcm complex are differentially phosphorylated during the cell cycle. The changes in phosphorylation are most pronounced for the Mcm4 subunit and correlate with the ability of the Mcm complexes to bind to the pre-Rcs.
MATERIALS AND METHODS
Extract preparation.
Interphase and metaphase arrested Xenopus extracts were prepared according to the method of Murray (29), with the exception that interphase extract buffer was supplemented with 0.25 mg of cycloheximide per ml. Interphase high-speed supernatant extract (HSS) was prepared by additional centrifugation of the interphase extract at 100,000 × g for 50 min at 4°C. Membrane fractions were isolated according to the method of Smythe and Newport (38). All extracts were supplemented with 3% glycerol, aliquoted, and stored at −80°C.
Chromatin isolation.
Demembranated Xenopus sperm nuclei (25) were incubated in Xenopus extract for the indicated times at 23°C with 3,500 sperm heads/μl of extract. To prepare chromatin, samples were diluted 10-fold into an ice-cold nuclei buffer containing 20 mM HEPES (pH 7.4), 50 mM sucrose, 50 mM KCl, 5 mM MgCl2, and 0.1% NP-40. The chromatin was then isolated by centrifugation through a 15% sucrose cushion prepared in the same nuclei buffer. The pelleted material was either resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for Western blot analysis or in kinase buffer containing 1000 U of DNase I per ml for immunoprecipitation experiments.
Antibodies.
W. Dunphy and H. Masai kindly provided Xenopus Cdc6 and Cdc7 cDNAs, respectively. His-tagged recombinant proteins were produced in Escherichia coli, isolated from the inclusion bodies and purified by SDS-PAGE. Proteins were electroeluted from the gels, mixed with adjuvant, and injected into New Zealand White rabbits. Antibodies were purified by affinity chromatography as described previously (8). Both antibodies specifically recognize either the Xenopus Cdc6 or the Cdc7 protein in the extracts or in vitro-translated protein (TNT Kit; Promega). A fraction of the purified anti-Cdc7 antibodies were biotinylated using the EZ-link Sulfo-NHS-LC-LC-Biotin reagent (Pierce). These modified antibodies were used for Western blot analyzes to detect the Cdc7 protein in immunoprecipitated samples. The biotin-strepavidin detection system minimized the background due to the immunoglobulin G molecules that migrated closely to the Cdc7 protein. Antibodies against the other Mcm subunits were generous gifts of R. Laskey (Xenopus Mcm3), H. Takisawa (Xenopus Mcm6 and Mcm7), I. Todorov (human Mcm2), and T. T. Su (Drosophila Mcm5).
Immunodepletion and isolation of Mcm complexes.
For immunodepletion, 100 μl of extract was incubated with 20 to 100 μg of purified antibodies bound to 50 μl of protein A-Sepharose (Pharmacia). After a 1-h incubation at 4°C with constant rotation, the Sepharose was harvested by low-speed centrifugation. The beads were then washed two to three times with extract buffer (29) or kinase buffer (20 mM HEPES, pH 7.4; 50 mM sucrose; 50 mM KCl; 10 mM MgCl2; 100 μM ATP; 10 mM NaF; 80 mM β-glycerophosphate; 0.1% NP-40) before phosphorylation experiments or Western blot analyses. Interphase extract immunodepleted using anti-Mcm-4 or anti-Cdc7 antibodies was unable to replicate double-stranded DNA (data not shown).
32P radiolabeling of Mcm proteins.
Radiolabeling of proteins in the extracts was done by adding 1 μCi of [γ-32P]ATP per μl of extract. After a 1-h incubation at 23°C, the Mcm complexes were isolated from the extract as described above. Labeled subunits of the Mcm complex were then separated by SDS-PAGE and analyzed by Western blotting and autoradiography. Phosphorylation of Mcm proteins bound to chromatin was analyzed under the same conditions in an extract containing 3,000 demembranated sperm heads/μl. At the indicated times chromatin was isolated, and the phosphorylated Mcm proteins associated with the chromatin were immunoprecipitated using anti-Mcm4 antibodies. In vitro phosphorylation of Mcm complexes by its associated kinases was accomplished by incubating the Mcm complexes attached to Sepharose beads in kinase buffer containing 0.5 μCi of [γ-32P]ATP/μl of reaction. In vitro phosphorylation of the Mcm4 protein by purified Cdc2-cyclin B kinase or by mitotic Xenopus extract was performed as follows. In vitro-translated Mcm4 proteins were first immunoprecipitated with anti-Mcm4 antibodies, followed by incubation in kinase buffer containing 0.5 μCi of [γ-32P]ATP/μl of reaction volume plus either 50 U of purified Cdc2-cyclin B (New England Biolabs) or 10 μl of mitotic extract. Similar experiments were performed using immunopurified Mcm interphase complexes as substrate containing about 300 ng of each Mcm protein.
Identification of phosphorylated Mcm proteins.
Different criteria were applied for the identification of the phosphorylated Mcm proteins (Mcm2, Mcm4, Mcm3, or Mcm6). First, for each 32P-labeled band, we performed an alignment between the autoradiogram and both the Western blot and Ponceau patterns. Second, for the identification of phosphorylated Mcm3 and Mcm2, we treated Mcm complexes (which had been immunoprecipitated with anti-Mcm4 antibodies) with 1 M NaCl. This treatment resulted in partial dissociation of the complex and the release of Mcm2 and Mcm3 proteins from the core complex containing Mcm4, Mcm6, and Mcm7. Dissociated Mcm2 and Mcm3 were then immunoprecipitated with specific antibodies and unambiguously identified as 32P labeled. However, because the salt dissociation of the complex was not total and some Mcm3 protein remained in the complex, it was difficult to determine if the remaining 32P-labeling was associated with Mcm3 protein or the Mcm6 protein that comigrates with Mcm3. Thus, we do not differentiate between the Mcm3 phosphoisoform and the potential Mcm6 phosphoisoform. Verification of the identity of each Mcm4 phosphoisoform was obtained by their ability to undergo a mobility shift resulting from phosphorylation and/or dephosphorylation.
Phosphopeptide map analysis.
Following 32P labeling in Xenopus extracts or in vitro, the Mcm4 protein was retrieved by immunoprecipitation, separated by SDS-PAGE, and transferred to nitrocellulose. The blot was then exposed to autoradiography film. Alignment of the blot and the autoradiograph allowed excision of the Mcm4 band. The sample was then digested with trypsin. Digested tryptic peptides were separated on thin-layer chromatography plates by electrophoresis in pH 1.9 buffer in the first dimension using an HTLE-7000 apparatus and ascending chromatography in the second dimension (4). The phosphorylated peptides were visualized using a PhosphorImager (Molecular Dynamics).
RESULTS
Mitotic and interphase Xenopus Mcm complexes contain different phosphoisoforms of the Mcm4 protein.
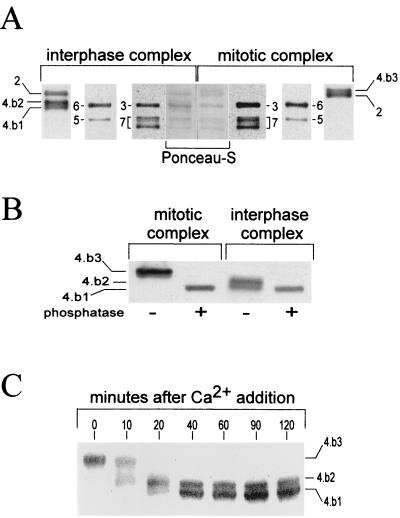
We have previously reported that the Xenopus Mcm4 protein is hyperphosphorylated during mitosis and hypophosphorylated in interphase cytosol (8). These two phosphoisoforms were identified in gels as they exhibit distinct electrophoretic mobilities. Additional work has revealed that interphase extracts contain not one but two electrophoretically distinct isoforms of Mcm4. To better understand the nature of each of these Mcm4 isoforms, we carefully reexamined which Mcm4 isoforms were associated with mitotic and interphase Mcm complexes. We also used this opportunity to look for potential phosphorylation of the other Mcm subunits in these complexes. After immunoprecipitation of the complexes from Xenopus egg extracts the proteins were separated by SDS-PAGE under conditions which allowed the maximum separation between the different isoforms and then subjected to Western blot analyses using antibodies against each individual Mcm subunit. Among all of the Mcm proteins, the Mcm4 subunit was the only one found to exhibit different electrophoretic mobilities between the mitotic and the interphase complexes (Fig. 1A). One slowly migrating Mcm4 isoform (Mcm4:band-3) was associated with mitotic complexes, and two faster-migrating bands (Mcm4:band-2 and Mcm4:band-1) were found in interphase complexes. Phosphatase treatment of the mitotic and interphase complexes confirmed that these three Mcm4 gel bands corresponded to different phosphoisoforms of the Mcm4 protein (Fig. 1B). The Mcm4:band-3 is the hyperphosphorylated mitotic form that we had previously identified (8). The Mcm4:band-1 comigrated with the alkaline phosphatase dephosphorylated form and is indeed the unphosphorylated form of the Mcm4 protein (data not shown). The Mcm4:band-2 appeared to be an intermediately phosphorylated form of the Mcm4 protein. Densitometric analysis of the Ponceau staining of the Mcm proteins in complexes (Fig. 1A, middle lanes) suggested that the majority of the mitotic and interphase complexes have a heterohexameric nature. Therefore, two different Mcm complexes seemed to be present during interphase; one contained the Mcm4:band-2 subunit, and the other contained the Mcm4:band-1 subunit. Since we have previously shown that the Mcm4 protein undergoes dephosphorylation at the mitosis-interphase transition (8), we wondered if the Mcm4:band-2 was a transient form in the dephosphorylation pathway between the mitotic Mcm4:band-3 and the fully dephosphorylated Mcm4:band-1. When a mitotic extract was induced to enter interphase by Ca2+ treatment, the mitotic Mcm4:band-3 was rapidly converted into the Mcm4:band-2, which in turn was more slowly converted into the Mcm4:band-1 (Fig. 1C). This result indicated that the Mcm4:band-2 is an intermediate of dephosphorylation and that the ratio of the two types of Mcm interphase complexes containing different Mcm4 phosphoisoforms varies with time after exit from mitosis.
FIG. 1.
Mitotic and interphase Mcm complexes contain different Mcm4 phosphoisoforms. (A) Western blot analyses of the Mcm subunits composing the interphase and mitotic complexes. Mcm complexes were obtained by immunoprecipitation using anti-Mcm4 antibodies. Each Mcm subunit is referred to by its corresponding number. The three Mcm4 phosphoisoforms are the Mcm4:band-1 (4.b1) and the Mcm4:band-2 (4.b2), present in interphase complexes, and the Mcm4:band-3 (4.b3) present in the mitotic complex. In order to obtain the best resolution between the different Mcm4 isoforms, the Mcm complexes were separated by SDS–7.5% PAGE, run until the 68-kDa molecular mass marker reached the bottom of the gel. (B) Phosphatase treatment of mitotic and interphase extracts. Mcm complexes were incubated with 2 U of alkaline phosphatase for 20 min at 30°C. (C) Kinetics of Mcm4 dephosphorylation following the addition of 0.4 mM CaCl2 to a mitotic extract.
The Mcm4 protein is the major phosphorylated subunit of the mitotic Xenopus Mcm complex.
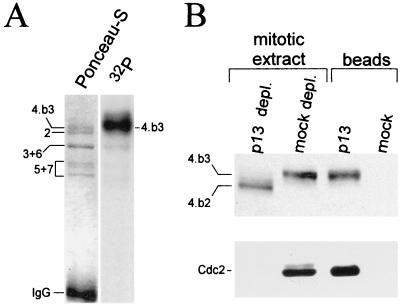
While we had clearly established the hyperphosphorylation of the Mcm4 protein during mitosis, we had not determined the phosphorylation state of the other Mcm proteins since they displayed no obvious differences in their electrophoretic mobilities. To directly test if other Mcm subunits could be phosphorylated during mitosis, we incubated a mitotic extract in the presence of [γ-32P]ATP and immunoprecipitated the Mcm complex with an anti-Mcm4 antibody. Figure 2A shows that the Mcm4:band-3 isoform was the major phosphorylated protein in the complex, while other Mcm subunits were not strongly labeled. Based on the fact that the hyperphosphorylation of Mcm4 proteins correlates with high levels of Cdc2-cyclin B kinase activity in the mitotic extract, we had previously speculated that this kinase may be involved in phosphorylating Mcm4 during mitosis. To further examine this possibility, we first tested whether the mitotic Mcm complex and the Cdc2-cyclin B protein kinase directly interact. Immunoprecipitation experiments using anti-Mcm4 or anti-Cdc2 antibodies did not reveal any interaction between these proteins (data not shown). However, when a mitotic Xenopus egg extract was depleted of all Cdk proteins (including Cdc2) using p13-Suc1 beads, about 15% of the total Mcm4 proteins and other Mcm subunits were also depleted (Fig. 2B). Interestingly, while the Mcm4 protein associated with the beads remained hyperphosphorylated, the Mcm4 protein left in the depleted extract underwent dephosphorylation. All together these results indicate that if any interaction between the Mcm and Cdc2 proteins exists it is not strong enough to be detected using the coimmunoprecipitation procedure. Further, the proteins associated with the p13-Suc1 beads seem to be involved, directly or indirectly, in the mitotic phosphorylation of the Mcm4 proteins. Thus, among these proteins, the Cdc2-cyclin B kinase remains a candidate for phosphorylating Mcm4.
FIG. 2.
Phosphorylation of the Mcm4 subunit of the mitotic Mcm complex is Cdk dependent. (A) The Mcm complex was radiolabeled with 32P in a mitotic extract and then isolated by immunoprecipitation using anti-Mcm4 antibodies. The subunits of the complex were resolved by SDS–7.5% PAGE and transferred to nitrocellulose, and the radiolabeled proteins were visualized with a PhosphorImager. (B) Depletion of Cdks (and their associated proteins) from a mitotic extract was achieved by incubating the extract with an equal volume of p13-Suc1 beads. The beads were recovered by low-speed centrifugation and washed several times with extract buffer containing 0.1% NP-40. The beads and depleted extracts were analyzed for Mcm4 and Cdc2 content by Western blotting using rabbit polyclonal Mcm4 antibodies an a monoclonal anti-Cdc2 antibody (Sc-54; Santa Cruz).
Mitotic phosphorylation of the Mcm4 protein requires the Cdc2-cyclin B and other kinases.
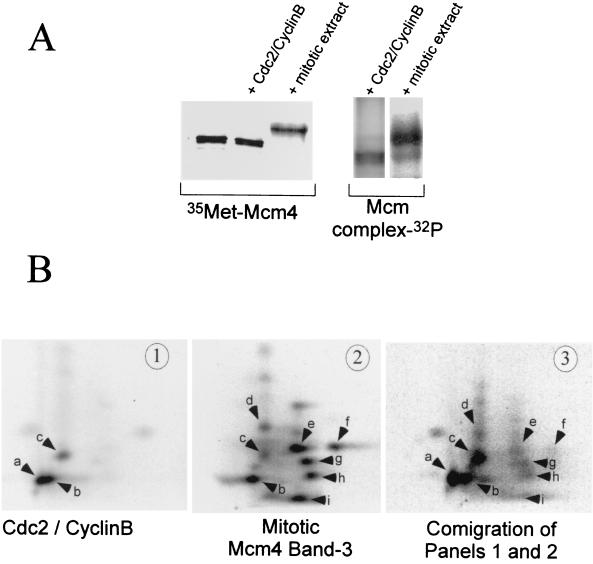
We further tested the ability of the Cdc2-cyclin B kinase to phosphorylate the Mcm4 protein. Mcm4 protein prepared by in vitro translation or as part of an immunoprecipitated interphase Mcm complex was incubated in the presence of [γ-32P]ATP with either purified Cdc2-cyclin B kinase or a small amount of mitotic Xenopus egg extract. In both cases phosphorylation of the Mcm4 protein was observed (Fig. 3A). However, the mobility shift corresponding to the mitotic hyperphosphorylation of the Mcm4 protein was only observed in the presence of mitotic extract and not with purified Cdc2-cyclin B kinase. Since the amount of purified Cdc2-cyclin B we used had >5 times the H1 kinase activity than the added mitotic extract, we do not believe that the inability of the Cdc2-cyclin B kinase to hyperphosphorylate Mcm4 was due to insufficient kinase activity. Several of our experiments also indicated that the Mcm4 antibodies used to immunoprecipitate the protein prior to phosphorylation did not interfere with the ability of Cdc2-cyclin B to fully hyperphosphorylate the Mcm4 protein. As shown in Fig. 3A, the ability of the Mcm4 subunit of the interphase complex to be fully phosphorylated by a small amount of mitotic extract and not by purified Cdc2-cyclin B indicates that the anti-Mcm4 antibodies do not interfere with Mcm4 phosphorylation. This conclusion was not only supported by the mobility shift analysis (Fig. 3A) but also by phosphopeptide map analysis (Fig. 3B, panel 2, and data not shown). The noninterference of the anti-Mcm4 antibodies was also demonstrated by the observation that the same phosphopeptide map was obtained for the in vitro-translated Mcm4 protein phosphorylated by Cdc2-cyclin B either in the presence or absence of these antibodies (Fig. 3B, panel 1, and data not shown).
FIG. 3.
Mitotic hyperphosphorylation of the Mcm4 protein requires Cdc2-cyclin B and other kinases. (A) In vitro phosphorylation of Mcm4 by purified Cdc2-cyclin B or a mitotic extract. Phosphorylation reactions were run in kinases buffer containing either 50 U of purified Cdc2-cyclin B or 10 μl of mitotic extract. The substrate was 4 μl of an in vitro-translated [35S]methionine labeled Mcm4 protein or Mcm4 present in an interphase Mcm complex. After a 1-h incubation at room temperature the Mcm proteins were immunoprecipitated with an anti-Mcm4 antibody and separated by SDS-PAGE. The mobility shift of the Mcm4 protein was detected by visualizing both the [35S]methionine label and the incorporated 32P by using a PhosphorImager. The mobility shift of the Mcm4 protein was also confirmed by Western blot analyses (data not shown). (B) Two-dimensional tryptic phosphopeptide mapping was performed on the Mcm4 protein phosphorylated in vitro by Cdc2-cyclin B (panel 1) or in a mitotic extract (panel 2). In panel 3 the two-dimensional phosphopeptide map of a mixture containing samples 1 and 2 is shown. Electrophoretic separation was performed along the horizontal axis, and ascending chromatography was done along the vertical axis.
To determine if the in vitro phosphorylation of the Mcm4 protein by the Cdc2-cyclin B kinase occurred at physiologically relevant sites, we performed tryptic phosphopeptide analyses. We compared the phosphopeptide map of Mcm4 phosphorylated in vitro by Cdc2-cyclin B or in a mitotic Xenopus egg extract (Fig. 3B). Again, we used either an in vitro-translated Mcm4 protein or Mcm4 protein from an interphase complex as the phosphorylation substrates. The Cdc2-cyclin B kinase phosphorylated both substrates on only a subset of the mitotic peptides phosphorylated in the extract (Fig. 3B, peptides b and c). Weak phosphorylation of peptide g could also be observed when we used the Mcm complex as substrate (data not shown). One possibility is that peptide g is more accessible for phosphorylation by Cdc2-cyclin B when the Mcm4 protein is within the Mcm complex. Cdc2-cyclin B also phosphorylated Mcm4 in vitro on an additional site that is not present in the hyperphosphorylated mitotic form of the protein (peptide a). Based on its migration, peptide a seemed to be a more acidic version of peptide b which could originate from an extra phosphorylation site in the peptide or a different partial tryptic digestion. Phosphoamino acid analysis of all the peptides phosphorylated by Cdc2-cyclin B in vitro (peptides a, b, c, and g) revealed that phosphorylation occurs only on threonine residues (data not shown). Possible residues are Thr-7, Thr-102, or Thr-110 since they are positioned in the three best Cdk phosphorylation consensus sequences found within the Mcm4 protein. On the other hand, phosphoamino acid analysis of the Mcm4:band-3 isoform showed that in a mitotic extract the Mcm4 protein is phosphorylated on both threonine and serine residues (data not shown). Collectively, our results demonstrate that Cdc2-cyclin B could partially phosphorylate Mcm4 during mitosis but that full hyperphosphorylation of the protein requires an additional kinase(s).
Phosphorylation of Mcm subunits within the interphase complex does not involve Cdc7.
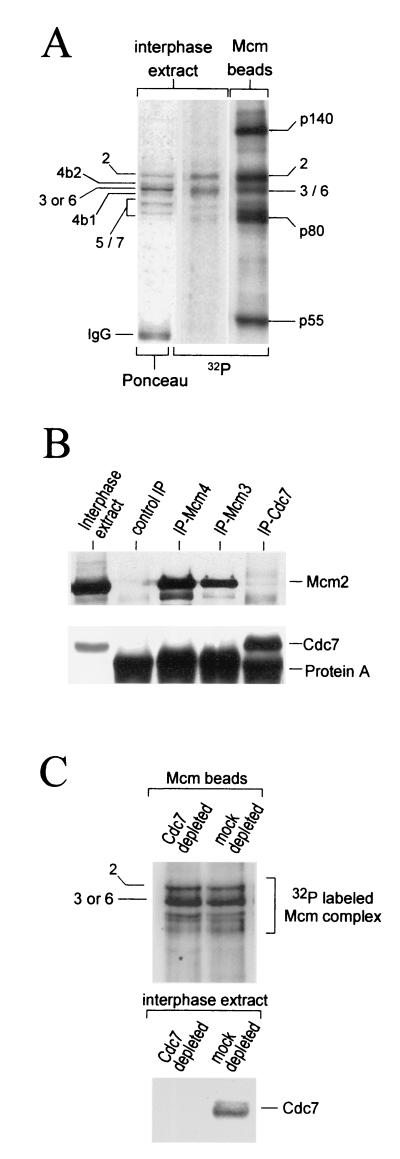
While we had established that the Mcm4 protein within the Mcm complex is actively dephosphorylated in Xenopus interphase cytosol, we wanted to examine the phosphorylation state of the other Mcm subunits. Labeling of the interphase extracts in the presence of [γ-32P]ATP followed by immunoprecipitation of the Mcm complex indicated that the Mcm2 protein and the Mcm3 or Mcm6 protein were phosphorylated in the interphase complex (Fig. 4A). Since Mcm3 and Mcm6 always comigrated on gels, we could not distinguish between different phosphorylated isoforms, if they exist. As expected, we never observed 32P incorporation into the Mcm4:band-2 or the Mcm4:band-1 (see Fig. 4A and Fig. 6B for better resolution). No obvious phosphorylation of the Mcm5 or Mcm7 proteins was observed in interphase complexes. The phosphorylation of Mcm2, Mcm3, or Mcm6 was insensitive to the presence of p21-Cip1 in the extract, indicating that the Cdk2-cyclin E kinase was not involved (data not shown). Since our interphase extract, prepared in the presence of cycloheximide (a protein synthesis inhibitor), did not contain any cyclin A or cyclin B, we can also exclude their associated kinases as potential Mcm interphase kinases. To determine if kinase(s) were physically associated with the interphase Mcm complexes, we tested the kinase activity of the immunoprecipitated complexes. As in the interphase extract, we detected strong phosphorylation of the Mcm2 protein and the Mcm3 or Mcm6 proteins, as well as some other non-Mcm proteins. Three phosphorylated Mcm associated proteins were identified based on their apparent molecular weights as p140, p80, and p55. In-gel kinase assays using the Mcm interphase complexes revealed that polypeptides comigrating with p80 and p55 were autophosphorylated and therefore could be the kinases responsible for phosphorylating the Mcm interphase complexes (data not shown). In particular, the Cdc7 kinase has a molecular mass of about 55 kDa and has been reported, in yeast, to phosphorylate itself and some of the Mcm proteins in vitro (5). We therefore tested the possibility that Cdc7 was associated with Mcm complexes during interphase. Using a rabbit polyclonal antibody directed against the Xenopus Cdc7 protein we could not detect any association between the Cdc7 protein kinase and the Mcm interphase complexes immunoprecipitated with either anti-Mcm4 or anti-Mcm3 antibodies (Fig. 4B). Reciprocally, anti-Cdc7 antibodies did not coimmunoprecipitate the Mcm interphase complexes. Finally, depletion of the Cdc7 protein kinase from an interphase extract did not significantly affect the phosphorylation of the Mcm2, Mcm3, or Mcm6 subunits in the isolated complex (Fig. 4C). All together, these data indicated that the Cdc7 kinase and the Mcm interphase complexes do not strongly interact in interphase extracts and that the Cdc7 kinase is unlikely to be the kinase associated with the interphase Mcm complex.
FIG. 4.
Phosphorylation of the Mcm interphase complex is independent of Cdc7 kinase activity. (A) The Mcm interphase complex was phosphorylated in the extract (lanes 1 and 2) or in vitro by its associated kinase (lane 3). The phosphorylated proteins associated with the immunoprecipitated Mcm complex were separated by SDS-PAGE, transferred to nitrocellulose, and visualized by Ponceau-S staining and autoradiography. (B) Interphase extracts were subjected to immunoprecipitation with antibodies against the Xenopus Mcm3, Mcm4, and Cdc7 proteins, as well as preimmune control antibodies. The immunoprecipitates were resolved by SDS-PAGE and Western blotting using anti-Mcm2 and biotinylated anti-Xenopus Cdc7 antibodies. (C) Interphase extracts were depleted with preimmune (mock) or with anti-Xenopus Cdc7 antibodies bound to protein A-Sepharose beads. Mcm complexes immunoprecipitated from the mock and Cdc7 extracts were then incubated in kinase buffer containing [γ-32P]ATP. After a 1-h incubation at room temperature, the phosphorylated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and visualized by autoradiography.
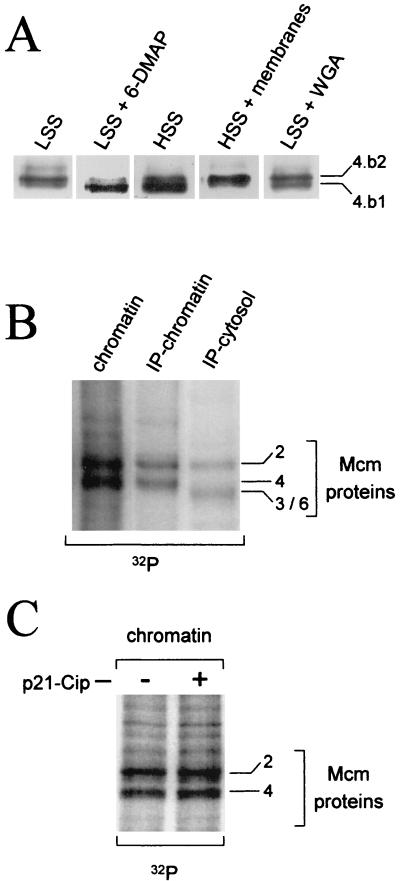
FIG. 6.
Phosphorylation of the Mcm4 protein on the replicative chromatin. (A) Demembranated sperm nuclei were incubated in a cytosolic high-speed extract (HSS) or interphase extracts (LSS) containing 50 μg of aphidicolin per ml. When indicated, the cytosolic extract was supplemented with a 1/10 volume of a membrane fraction, and the interphase extract was supplemented with either 3 mM DMAP or 0.5 mg of wheat germ agglutinin (WGA) per ml. After a 90-min incubation at room temperature, the chromatin was isolated and the Mcm4 isoform associated with the chromatin was identified by Western blot analysis. (B) Demembranated sperm nuclei were incubated in interphase extracts containing 50 μg of aphidicolin per ml and 1 μCi of [γ-32P]ATP per μl of extract. After a 1-h incubation at room temperature, the chromatin was separated from the cytosol. Mcm proteins were immunoprecipitated from the cytosolic and from the chromatin fractions using anti-Mcm4 antibodies. Western blot analysis and autoradiography identified the phosphorylated Mcm subunits. Total chromatin was also analyzed. (C) Interphase extract was incubated with or without 0.5 μM p21-Cip recombinant protein (40) for 10 min at room temperature. Sperm nuclei, aphidicholin, and [γ-32P]ATP were then added to the extract and incubated for an additional hour. Phosphorylated Mcm proteins on the chromatin were purified as in panel B.
Mcm interphase complexes containing Mcm4:band-2 but not Mcm4:band-1 bind to pre-Rcs and displace Cdc6.
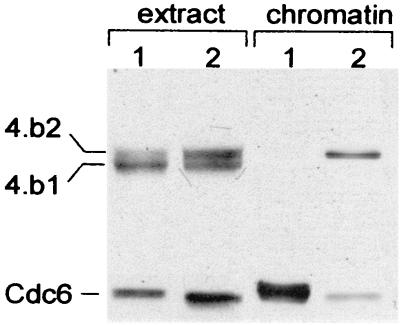
The existence of two different soluble interphase Mcm complexes, containing either the Mcm4:band-1 or the Mcm4:band-2 isoform raises the possibility that these two complexes differ in their ability to bind chromatin. To test this possibility, sperm nuclei were incubated in interphase extracts and the binding of the Mcm4 protein to chromatin was determined after a 15-min incubation. This short incubation time did not allow complete nuclear reconstitution or active phosphorylation of the Mcm4 protein while it was chromatin bound (see next section for a description of chromatin-associated Mcm4 phosphorylation). When the interphase extract contained an equal amount of the Mcm4:band-2 and the Mcm4:band-1 isoforms, we only observed binding of Mcm4:band-2 to chromatin (Fig. 5, lane 2). Furthermore, when the extract contained primarily Mcm4:band-1 at the time of sperm addition, no Mcm4 or any other Mcm protein binding to chromatin was observed, regardless of the incubation time (Fig. 5, lane 1). Because the loading of Mcms on the chromatin depends on Cdc6, we checked if the failure of Mcm4 to bind resulted from failure of Cdc6 to bind as well. As shown in Fig. 5, the amount of Cdc6 bound to the chromatin was significantly higher in the absence of Mcm binding. Such large amounts of Cdc6 bound to chromatin were only observed when sperm were incubated in Mcm-depleted interphase extracts (data not shown). However, in a control extract the release of the Cdc6 protein bound to chromatin occurred early and correlated with the binding of the Mcm complex containing the Mcm4:band-2 isoform. The results from these experiments suggest that the interphase Mcm complexes containing the Mcm4:band-2 protein are the active complexes that associate with the pre-Rcs and lead to the displacement of Cdc6 from the chromatin. Further dephosphorylation of the Mcm4:band-2 subunit in these complexes, generating Mcm4:band-1, appears to inactivate the ability of the Mcm complexes to bind the pre-Rcs and displace Cdc6.
FIG. 5.
Dephosphorylation of the Mcm4 subunit prevents the binding of the Mcm complex to pre-Rcs. Chromatin binding assays were performed by incubating sperm nuclei (3,000 sperm heads/μl of extract) in interphase Xenopus extract enriched in the dephosphorylated Mcm4:band-1 isoform or in extract containing equivalent amounts of Mcm4:band-2 and Mcm4:band-1. After 15 min of incubation at room temperature, the chromatin was isolated and the presence of Mcm4 and Cdc6 proteins on the chromatin was detected by Western blot analysis.
Phosphorylation of the Mcm4 protein bound to chromatin during S phase is independent of Cdks but requires nuclear transport.
Our previous work showed that the only Mcm4 isoform found in association with chromatin during S phase was hypophosphorylated and migrated upon SDS-PAGE between the hyperphosphorylated mitotic form (Mcm4:band-3) and the dephosphorylated form (Mcm4:band-1). We have now established that the chromatin-bound form of Mcm4 comigrates with the Mcm4:band-2 phosphoisoform that is the dephosphorylation intermediate found in the interphase cytosol (Fig. 6A, lane 1). This finding raised two possibilities: (i) the Mcm4:band-2 binds to the chromatin and, as a result, is protected against dephosphorylation that is otherwise observed in the interphase cytosol; or (ii) the Mcm4 protein is actively phosphorylated on the chromatin. To distinguish between these possibilities, we first looked at the effect of 6-DMAP, a serine/threonine kinase inhibitor, on the phosphorylation state of the chromatin-bound Mcm4 protein. In the next series of experiments, aphidicolin was also added to the interphase extract in order to block the progress of the elongation fork and thereby prevent the displacement of the Mcm proteins associated with the chromatin. When 6-DMAP and sperm nuclei were added simultaneously to an interphase extract, we first observed the binding of the Mcm4:band-2 isoform and then its dephosphorylation on the chromatin. After 90 min only the dephosphorylated Mcm4:band-1 was found on the chromatin (Fig. 6A, lane 2). In the control extract we observed the intermediately phosphorylated isoform we previously described (Fig. 6A, lane 1). This result suggested an active phosphorylation of the Mcm4 protein on chromatin. Confirmation of this phosphorylation was obtained by 32P labeling of the chromatin-bound proteins. Sperm nuclei were incubated in an interphase extract containing aphidicolin and [γ-32P]ATP. After 60 min the chromatin fraction was isolated and the phosphorylation of the Mcm proteins associated with chromatin was examined directly or after immunoprecipitation of the Mcm complex (Fig. 6B). The Mcm2 and Mcm4 subunits were two of the most intensely radiolabeled proteins bound to the chromatin. After immunoprecipitation of the Mcm complex no significant phosphorylation of other Mcm subunits was detected. In the case of the Mcm2 protein our data did not allow us to discriminate between the phosphorylation of the protein in the interphase cytosolic complex or on the chromatin after binding (compare chromatin and cytosol in Fig. 6B). However, we can conclude that while the Mcm4 complex subunit is dephosphorylated in the cytosol, it is actively phosphorylated on the chromatin by a serine/threonine kinase. We next examined the requirements for the phosphorylation of the Mcm4 subunit on the chromatin. As shown in Fig. 6A, this phosphorylation required the formation of an intact nuclear membrane and functioning nuclear transport. Rather than being phosphorylated, the Mcm4 protein was dephosphorylated on chromatin assembled in a high-speed interphase extract incapable of nuclear reconstitution (Fig. 6A, lane 3). The addition of a nuclear membrane fraction to such a high-speed extract restored Mcm4 phosphorylation on the chromatin (Fig. 6A, lane 4). Wheat germ agglutinin, an inhibitor of nuclear transport, also prevented chromatin-bound Mcm phosphorylation (Fig. 6A, lane 5). Since Cdk2-cyclin E kinase is required for the initiation of DNA replication and has been shown to be actively transported into the nucleus at the beginning of S phase (19), we sought to determine if this kinase was involved in the phosphorylation of Mcm4 on the chromatin. The addition of p21-Cip, a specific inhibitor of the Cdk2-cyclin E kinase, to the interphase extract did not affect Mcm4 phosphorylation but did completely block DNA synthesis (Fig. 6C and data not shown). Therefore, we conclude that Cdk2-cyclin E is not directly or indirectly responsible for the phosphorylation of the Mcm4 protein while it is bound to chromatin. Again, the absence of cyclin A or cyclin B in the extract rules out the possibility that their associated Cdks are involved in the phosphorylation of Mcm4 on the chromatin.
The chromatin-bound Mcm4 and cytosolic Mcm4:band-2 are distinct phosphoisoforms.
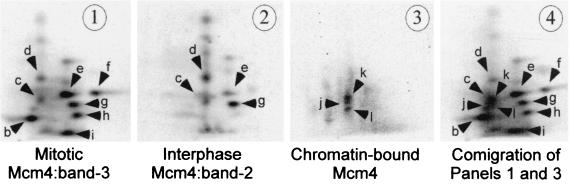
While the Mcm4:band-2 of the interphase cytosolic complex and the Mcm4 chromatin-bound isoform have the same electrophoretic mobilities, we have shown that they are generated differently through dephosphorylation of Mcm4:band-3 and active phosphorylation on the chromatin, respectively. However, the possibility remained that the two isoforms were phosphorylated on the same residues. To address this issue, we compared the phosphopeptide map of the mitotic hyperphosphorylated Mcm4:band-3, the interphase cytosolic Mcm4:band-2, and the chromatin-bound Mcm4 phosphoisoforms. Each form was labeled with 32P in extracts and then immunoprecipitated using an anti-Mcm4 antibody followed by phosphopeptide map analysis. To obtain labeled Mcm4:band-2, we first labeled Mcm4:band-3 in a mitotic extract and then Ca2+ activated the extract to induce the dephosphorylation of Mcm4:band-3 into Mcm4:band-2. Figure 7 shows that only four of the eight mitotic phosphopeptides were conserved upon dephosphorylation of the mitotic form of Mcm4 (Mcm4:band-3) into the Mcm4:band-2 of the interphase cytosol. Three phosphopeptides were detected from the Mcm4 chromatin-bound form, and none of these peptides were common to either the interphase Mcm4:band-2 or to the mitotic Mcm4:band-3 derived peptides. Phosphoamino acid analysis of the peptides from the chromatin-bound Mcm4 isoform indicated that they were only phosphorylated on serine residues, while the mitotic peptides showing the closest migration (peptides b and c) contained only phosphothreonine residues (data not shown). All together, these data indicate that the chromatin-bound and the transient interphase Mcm4:band-2 are distinct Mcm4 phosphoisoforms and are therefore likely to be phosphorylated by different kinases.
FIG. 7.
Tryptic phosphopeptide maps of the chromatin-bound Mcm4 and cytosolic Mcm4:band-2 isoforms. The phosphorylation of the Mcm protein on the chromatin and of the mitotic Mcm4:band-3 was performed as described in Fig. 6B and 3B, respectively. The 32P-labeled Mcm4:band-2 isoform was obtained by partial dephosphorylation of the mitotic 32P-labeled Mcm4:band-3 following the addition of 0.4 mM CaCl2 to a mitotic extract. Two-dimensional tryptic phosphopeptide mapping of each of the three Mcm4 phosphoisoforms are shown in panel 1 (Mcm4:band-3), panel 2 (Mcm4:band-2), and panel 3 (chromatin-bound Mcm4). Electrophoretic separation was performed along the horizontal axis, and ascending chromatography was done along the vertical axis.
DISCUSSION
In this study we have analyzed the cell cycle phosphorylation of one component of the pre-Rcs, the Mcm proteins. We report the first characterization of the phosphorylation state of the different subunits of the Mcm complex found in mitotic and interphase Xenopus extracts or associated with the replicative chromatin. We present evidence that individual Mcm subunits are differentially phosphorylated during the cell cycle and that the Mcm4 protein is the subunit that exhibits the most dramatic changes in its phosphorylation state. Some of these changes in Mcm4 phosphorylation correlate with the ability of the Mcm complex to bind pre-Rcs. In particular, we show that the active Mcm complex involved in the formation of the pre-Rcs is a transient complex containing an intermediately phosphorylated subunit (Mcm4:band-2) that is produced by partial dephosphorylation of the mitotic hyperphosphorylated Mcm4 protein (Mcm4:band-3). In addition, complete dephosphorylation of the Mcm4 subunit inactivates the Mcm complex and prevents its binding to the chromatin. Once the Mcm complex is assembled on the chromatin we also find that the Mcm4 subunit is phosphorylated during the activation of the pre-Rcs. We propose that phosphorylation of Mcm4 on the chromatin regulates the activity of the Mcm complex. Finally, we provide information pertinent to the identification of the kinases involved in the cell cycle phosphorylation of the Mcm complex.
We have shown that during mitosis the Mcm4 protein is the only subunit of the Mcm complex that is highly and actively phosphorylated. While we did not observe significant 32P incorporation into other Mcm subunits, we do believe that the mitotic Mcm complex contains a phosphorylated Mcm2 isoform. In fact, our observations indicate that the Mcm2 subunit is always phosphorylated and exhibits a fast electrophoretic mobility compared to its dephosphorylated forms (our unpublished data). Therefore, the very low phosphate turnover in the Mcm2 subunit indicates that somehow the conformation of the protein in the mitotic complex makes it rather inaccessible to phosphatases. This possibility is further strengthened by the fact that we found the Mcm2 subunit much more resistant than the Mcm4 subunit to in vitro phosphatase treatment of the Mcm mitotic complex (unpublished data). Our results indicate that the mitotic hyperphosphorylation of the Xenopus Mcm4 subunit requires different kinases. One of these appears to be the Cdc2-cyclin B kinase since it is able to phosphorylate in vitro a subset of the mitotic Mcm4 physiological phosphorylation sites. However, other unidentified kinases are also required for the full mitotic hyperphosphorylation of the protein. Our findings do not totally agree with the study of Hendrickson et al. (17) that reported Cdc2-cyclin B as the only kinase phosphorylating the Xenopus Mcm4 protein during mitosis. Concerned with this discrepancy, we repeated the in vitro phosphorylation experiment using different preparations and amounts of Cdc2-cyclin B kinase as well as different Mcm4 substrates. In each of these experiments we were never able to obtain full mitotic phosphorylation of the Mcm4 protein in the presence of purified Cdc2-cyclin B kinase. While we do not know the precise reason for the difference between the findings of Hendrickson et al. and this study, it could be related to the purity of the Mcm proteins used for the 32P-labeling experiments. Finally, additional support for the existence of other mitotic kinase(s) phosphorylating Mcm4 come from our finding that the mitotic phosphorylation of the Mcm4 protein occurs on serine and threonine residues, while the Cdc2-cyclin B phosphorylates Mcm4 on threonine residues only, as predicted by the presence of only threonine Cdk consensus phosphorylation sites in the Mcm4 protein. A similar conclusion implicating the Cdc2-cyclin B and other unknown kinase(s) in the phosphorylation of Mcm4 has also been reached based on experiments with the human Mcm4 protein at the G2/M transition (14). In the case of the human mitotic Mcm complex, other subunits beside the Mcm4 protein appear to be actively phosphorylated as well (including Mcm2 and Mcm3). The identity of the other mitotic kinase(s) is presently unknown. However, the fact that depletion of Cdks from a mitotic extract induces the dephosphorylation of the Mcm4 protein left in the depleted extract allows us to envision three hypotheses concerning the identity of the unknown mitotic kinase(s). First, beside Cdc2-cyclin B, the two other Cdks present in mitotic Xenopus extracts (i.e., Cdc2-cyclin A and Cdk2-cyclin E) are involved in Mcm4 hyperphosphorylation. Second, the unknown kinase(s) are somehow associated with Cdks and are simultaneously depleted by p13-Suc1 beads. Third, the unknown kinase(s) are not associated with the Cdks but are directly or indirectly under their control. While we think the first hypothesis is unlikely, we cannot distinguish yet between the last two hypotheses. Several reasons lead us to believe that Cdc2-cyclin A and Cdk2-cyclin E kinases are unlikely to be involved in the mitotic phosphorylation of Mcm4. Previous reports indicate that Cdk2-cyclin E cannot phosphorylate the Xenopus Mcm4 protein in vitro or any other subunit of the Mcm complex (13, 17). In agreement with these finding we found that high concentrations of Cdk2-cyclin E in the S-phase nucleus is not responsible for the phosphorylation of Mcm4 on the chromatin or in the nucleoplasm (Fig. 6C and unpublished data). Thus, there seems to be no evidence that the Mcm4 protein (and probably other Mcm subunits as well) is a substrate for the Cdk2-cyclin E kinase. On the other hand, the Cdc2-cyclin A kinase has recently been reported to phosphorylate the Xenopus Mcm4 protein in vitro and apparently on the same sites as the Cdc2-cyclin B kinase (13). Although the phosphopeptide map was not included in this report, we anticipate that the targeted peptides are peptides a, b, c, and possibly g as described in Fig. 3B. Considering that the Cdc2-cyclin B and Cdc2-cyclin A use the same Mcm4 phosphorylation sites and the kinase activity of Cdc2-cyclin A is rather low in mitotic extracts (28, 34) we believe that Cdc2-cyclin B is more likely to play a role in the mitotic hyperphosphorylation of the Mcm4 protein. Finally, our previous report that the addition of a constitutively active cyclin B mutant (Δ90) to an interphase extract lacking any endogenous cyclin A or cyclin B induces the mitotic hyperphosphorylation and mobility shift of the Mcm4 protein certainly supports the idea that Cdc2-cyclin A is not required for the hyperphosphorylation of Mcm4 (8). The results from this experiment also support the idea that the activity of the unknown Mcm4 mitotic kinase(s) is Cdc2-cyclin B dependent.
The observation that the Mcm4 subunit of the mitotic Mcm complex is highly phosphorylated raises the question of functional significance. Because the mitotic phosphorylation of the Xenopus or human Mcm proteins correlates with the proteins not being bound to the chromatin, it has been proposed that such phosphorylation regulates the chromatin binding of the Mcm proteins. In the Xenopus system the experimental data accumulated so far are conflicting. While addition of Cdc2-cyclin B has been reported to release Mcm from the chromatin in one study (17), the addition of MPF activity or purified Cdc2-cyclin B to prebound Mcm chromatin did not displace the Mcm protein in other studies (13, 22). Furthermore, it has been shown that mitotic Xenopus extract contains an active RLFM fraction (i.e., Mcm proteins fraction) and that the binding activity of this fraction is regulated by the presence of an inhibitor and not necessarily by the phosphorylation of the Mcm4 protein (26). Based on the data reported here, we believe that the phosphorylation of the Mcm4 protein at mitosis could be a preactivation step for the binding of the Mcm complex to the pre-Rcs. As a preactivation step, the mitotic phosphorylation of Mcm4 would ensure that a few key sites would remain phosphorylated in the active form of the interphase Mcm complexes that bind chromatin. Our finding that the partial dephosphorylation of the Mcm4 protein at the exit from mitosis creates a transient active Mcm complex containing the Mcm4:band-2 isoform supports this hypothesis. Unraveling the role of Mcm4 mitotic phosphorylation will require the identification and mutation of the phosphorylation sites involved. Generation of mutant proteins which are nonphosphorylatable or that mimic constitutive phosphorylation should definitively establish if the mitotic Mcm4 hyperphosphorylation is either a preactivation or an inactivation step regulating the binding of the Mcm complex to the chromatin. The inactivation of the Mcm interphase complex resulting from total dephosphorylation of the Mcm4 subunit could explain why the licensing activity of an interphase extract decreases with time after Ca2+ activation (26). While the physiological relevance of the cytoplasmic inactivation of the Mcm proteins during S phase is unknown, we speculate that it may be important in the prevention of rereplication events. This cytoplasmic mechanism would coexist with other nuclear mechanisms that also prevent pre-Rc assembly. Inactivation of the cytoplasmic Mcm complex might be important for the early embryonic Xenopus cell cycle since the amount of Mcm proteins in the nucleus during S phase represents only a few percent of the total Mcm protein accumulated during oogenesis. It remains to be seen if such an inactivation mechanism is also present in other organisms. Finally, it is interesting to note that while the initial binding of the Mcm complex is inhibited by the dephosphorylation of the Mcm4 subunit, the release of the Mcm complex from the chromatin is not observed when dephosphorylation of the Mcm4 subunit occurs on the chromatin. Assuming that the Mcm complex acts as a DNA clamp, we can envision that the dephosphorylation of the complex prevents the opening of the clamp and thereby prevents the loading and unloading of the Mcm complex from the chromatin. Another way in which phosphorylation and dephosphorylation could regulate loading and unloading of Mcm proteins is by affecting the interaction of Mcm proteins with other factors, such as loading (e.g., Cdc6) or unloading factors.
While the Mcm4 subunit undergoes dephosphorylation in the interphase cytosol, the Mcm2 and the Mcm3 or Mcm6 subunits are actively phosphorylated. The phosphorylation of these proteins might be required for the formation of an active Mcm complex, but it is certainly not sufficient since these proteins are also phosphorylated in the inactive complex containing the Mcm4:band-1 dephosphorylated isoform. The kinases responsible for the phosphorylation of these subunits during interphase are unknown but are not Cdk2-cyclin E or the Cdc7 kinase. So far we have not been able to show interaction between the soluble interphase Mcm complex and the Cdc7 kinase which has been reported to phosphorylate in vitro some of the Mcm proteins. Using antibodies specific for Mcm4, Mcm2, Mcm3, or Cdc7, we have not been able to reproduce the coimmunoprecipitation data reported recently by Roberts et al. (35). Furthermore, we observed that depletion of Cdc7 protein from an interphase extract does not affect the phosphorylation of Mcm2, Mcm3, or Mcm6 subunits in the isolated Mcm complex. A recent study reported that in S. cerevisiae the chromatin association of the Dbf4 protein, the Cdc7 regulatory subunit, is dependent on ORC but not on Cdc6 or the Mcm proteins (31). This raises the possibility that an interaction between the Xenopus Cdc7 kinase and Mcm proteins might only occur on the chromatin and not in the interphase cytosol. Although this possibility needs to be further investigated, it would be consistent with the presumptive role of the Cdc7 kinase in the activation of the pre-Rcs and not in their assembly.
Interestingly, the phosphorylation state of the Mcm proteins also varies during S phase, based on their association with the chromatin. We clearly show that the Mcm4 subunit is dephosphorylated in the interphase cytoplasm but becomes phosphorylated on the chromatin during S phase. By contrast, the Mcm3 or Mcm6 proteins are phosphorylated in the soluble interphase complex and dephosphorylated on the replicative chromatin. We have also shown that the Mcm2 protein bound to chromatin is phosphorylated but our data cannot distinguish between an active phosphorylation on the chromatin or the binding and stabilization of an already phosphorylated protein that was initially phosphorylated in the cytosol. Finally, our data indicate that the phosphorylation of Mcm4 on the chromatin occurs after establishment of the pre-Rcs. It requires the assembly of a nuclear membrane and probably the transport of a critical factor into the nucleus. It is independent of Cdk2-cyclin E kinase, or any cyclin A- and cyclin B-associated kinase activities. It is also unaffected by the presence of aphidicolin, an inhibitor of DNA polymerases, in the extract. Therefore, phosphorylation of Mcm4 on the chromatin seems to occur during the activation step of initiation and to precede the elongation step of DNA replication. Furthermore, it seems to occur independently of Cdc45 binding to the chromatin, an event which is Cdk2-cyclin E dependent. The role of Mcm4 and perhaps Mcm2 phosphorylation, while they are bound to chromatin, is presently unclear. Among different possibilities, it could facilitate the binding of other proteins involved in downstream initiation events or the disassembly of the pre-Rcs during S phase. Further experiments are required to explore these possibilities. Another point of interest is the identification of the responsible kinase. According to our results this kinase is a nuclear serine/threonine kinase that phosphorylates the Mcm4 and possibly Mcm2 subunits of the Mcm complex during the activation step of the pre-Rcs. Among the two identified kinases required for pre-Rcs activation, we showed that Cdk2-cyclin E is not responsible for the phosphorylation of the Mcm proteins on the chromatin. Therefore, it is tempting to postulate that the other pre-Rc activating kinase, Cdc7-Dbf4, is the relevant kinase. This possibility is supported by genetic evidence indicating a close relationship between the Cdc7-Dbf4 kinase and the Mcm proteins and also by biochemical evidence showing that the Cdc7-Dbf4 kinase is able to phosphorylate several Mcm proteins in vitro. We are actively investigating this possibility. Overall, our results indicate that the Mcm4 subunit of the Xenopus Mcm complex is the major target for phosphorylation during the cell cycle. These phosphorylation changes of the Mcm4 protein regulate pre-Rc assembly and probably their function on the chromatin.
ACKNOWLEDGMENTS
This work was supported by grants from the American Heart Association, Texas Affiliate, and the South Plains Foundation to M.C.
We thank W. Dunphy, T. Hunt, R. Laskey, H. Masai, H. Takisawa, I. Todorov, and T. T. Su for reagents and C. M. Pfarr for critical reading of the manuscript.
REFERENCES
- 1.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 2.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 3.Bousset K, Diffley J F. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. . (Erratum, 12:1072.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 5.Brown G W, Kelly T J. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- 6.Coleman T R, Carpenter P B, Dunphy W G. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 7.Coue M, Amariglio F, Maiorano D, Bocquet S, Mechali M. Evidence for different MCM subcomplexes with differential binding to chromatin in Xenopus. Exp Cell Res. 1998;245:282–289. doi: 10.1006/excr.1998.4271. [DOI] [PubMed] [Google Scholar]
- 8.Coue M, Kearsey S E, Mechali M. Chromotin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J. 1996;15:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson A D, Fangman W L, Brewer B J. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donovan S, Harwood J, Drury L S, Diffley J F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowell S J, Romanowski P, Diffley J F. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- 12.Dutta A, Bell S P. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 13.Findeisen M, El-Denary M, Kapitza T, Graf R, Strausfeld U. Cyclin A-dependent kinase activity affects chromatin binding of ORC, cdc6, and MCM in egg extracts of Xenopus laevis. Eur J Biochem. 1999;264:415–426. doi: 10.1046/j.1432-1327.1999.00613.x. [DOI] [PubMed] [Google Scholar]
- 14.Fujita M, Yamada C, Tsurumi T, Hanaoka F, Matsuzawa K, Inagaki M. Cell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinase. J Biol Chem. 1998;273:17095–17101. doi: 10.1074/jbc.273.27.17095. [DOI] [PubMed] [Google Scholar]
- 15.Gavin K A, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 16.Hardy C F, Dryga O, Seematter S, Pahl P M, Sclafani R A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson M, Madine M, Dalton S, Gautier J. Phosphorylation of MCM4 by cdc2 protein kinase inhibits the activity of the minichromosome maintenance complex. Proc Natl Acad Sci USA. 1996;93:12223–12228. doi: 10.1073/pnas.93.22.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua X H, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X H, Yan H, Newport J. A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J Cell Biol. 1997;137:183–192. doi: 10.1083/jcb.137.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. . (Erratum, 273:23616, 1998.) [DOI] [PubMed] [Google Scholar]
- 21.Koonin E V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- 24.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohka M J, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- 26.Mahbubani H M, Chong J P, Chevalier S, Thommes P, Blow J J. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minshull J, Golsteyn R, Hill C S, Hunt T. The A- and B-type cyclin-associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle. EMBO J. 1990;9:2865–2875. doi: 10.1002/j.1460-2075.1990.tb07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray A W. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 30.Neuwald A F, Aravind L, Spouge J L, Koonin E V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 31.Pasero P, Duncker B P, Schwob E, Gasser S M. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 1999;13:2159–2176. doi: 10.1101/gad.13.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasero P, Gasser S M. New systems for replicating DNA in vitro. Curr Opin Cell Biol. 1998;10:304–310. doi: 10.1016/s0955-0674(98)80004-5. [DOI] [PubMed] [Google Scholar]
- 33.Perkins G, Diffley J F. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 34.Rempel R E, Sleight S B, Maller J L. Maternal Xenopus Cdk2-cyclin E complexes function during meiotic and early embryonic cell cycles that lack a G1 phase. J Biol Chem. 1995;270:6843–6855. doi: 10.1074/jbc.270.12.6843. [DOI] [PubMed] [Google Scholar]
- 35.Roberts B T, Ying C Y, Gautier J, Maller J L. DNA replication in vertebrates requires a homolog of the Cdc7 protein kinase. Proc Natl Acad Sci USA. 1999;96:2800–2804. doi: 10.1073/pnas.96.6.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato N, Arai K, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman D A, Pasion S G, Forsburg S L. Multiple domains of fission yeast Cdc19p (MCM2) are required for its association with the core MCM complex. Mol Biol Cell. 1998;9:1833–1845. doi: 10.1091/mbc.9.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smythe C, Newport J W. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- 39.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1564. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 40.Strausfeld U P, Howell M, Rempel R, Maller J L, Hunt T, Blow J J. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr Biol. 1994;4:876–883. doi: 10.1016/s0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 41.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]