Abstract
The intestine is fundamental in controlling human health. Intestinal epithelial and immune cells are continuously exposed to millions of microbes that greatly impact on intestinal epithelial barrier and immune function. This microbial community, known as gut microbiota, is now recognized as an important partner of the human being that actively contribute to essential functions of the intestine but also of distal organs. In the gut ecosystem, bidirectional microbiota‐host communication does not involve direct cell contacts. Both microbiota and host‐derived extracellular vesicles (EVs) are key players of such interkingdom crosstalk. There is now accumulating body of evidence that bacterial secreted vesicles mediate microbiota functions by transporting and delivering into host cells effector molecules that modulate host signalling pathways and cell processes. Consequently, vesicles released by the gut microbiota may have great influence on health and disease. Here we review current knowledge on microbiota EVs and specifically highlight their role in controlling host metabolism, intestinal barrier integrity and immune training.
Keywords: bacterial membrane vesicles, gut microbiota, gut permeability, immune regulation, intestinal homeostasis, probiotics
Abbreviations
- BEVs
bacterial extracellular vesicles
- CD
Crohn's disease
- CMV
cytoplasmic membrane vesicles
- DCs
dendritic cells
- EVs
extracellular vesicles
- IBD
inflammatory bowel disease
- IECs
intestinal epithelial cells
- InsP6
inositol polyphosphates
- LPS
lipopolysaccharide
- MAMPs
microbial associated molecular patterns
- miRNAs
microRNAs
- MMP‐9
matrix metalloprotease 9;
- NOD
nucleotide‐binding oligomerization domain
- O‐IMVs
outer‐inner membrane vesicles
- OMVs
outer membrane vesicles
- PRRs
pattern recognition receptors
- PSA
polysaccharide A
- SCFAs
short‐chain fatty acids
- TFF3
trefoil factor 3
- TLRs
toll‐like receptors
- UC
ulcerative colitis
- ZO
zonula occludens
1. INTRODUCTION
The skin and mucosal surfaces of the human body are colonized by a huge number of microbes, collectively called the microbiome (“The Integrative Human Microbiome Project.,” 2019). Colonization is especially relevant in the intestinal tract, where the microbial population reaches numbers that exceed 1011 organisms per gram of wet weight of faecal content in the colon. The gut microbial community, known as the gut microbiota, mainly consist of bacteria, but also archaea, fungi and viruses. Collectively, the gut microbiota contributes with more than three million genes, which far surpass and complement the genetic information encoded by the human genome. In fact, the gut microbiota is considered a “hidden organ” since microbiota‐encoded products actively contribute to numerous essential host functions. In addition to its role in nutrition, metabolism and energy production, the gut microbiota regulates immune homeostasis and inflammatory responses against pathogens and other injuries (Jandhyala et al., 2015). The pivotal role of the gut microbiota in the development of the gastrointestinal immune system has been evidenced in germ‐free mice models. Among other alterations, gnotobiotic animals exhibit impaired immune function with atrophy of lymphoid organs and decreased production of immunoglobulin A (IgA). They are more susceptible to infections and allergies (Fiebiger et al., 2016; Kennedy et al., 2018).
The gut microbiota establishes a complex, dynamic, bidirectional relation with host cells of the intestinal mucosa (Kaparakis‐Liaskos & Ferrero, 2015). Due to the anatomical structure of the gut barrier, interkingdom interactions do not involve direct cell contacts. The mucus layer that covers the intestinal epithelium creates a physical separation that prevents luminal bacteria from accessing the underlining epithelial cells (Donaldson et al., 2016). In the colon, the secreted mucus gel‐forming mucin 2 creates a well‐defined layer, stratified in two sections. The inner layer is highly compact and physically excludes luminal bacteria. In addition, the inner mucus layer contains antimicrobial peptides and immune effectors secreted by epithelial and immune cells that target luminal bacteria. This contributes to spatial host–bacterial segregation (Tropini et al., 2017). The outer mucus layer is loose and serves as an adhesion site for a great number of bacteria that colonize the intestinal tract. This external layer is degraded by specific bacteria of the gut microbiota. Therefore, it needs to be constantly renewed to preserve the inner mucus layer. Production of mucin 2 by goblet cells is upregulated by toll‐like receptor (TLR) signalling in response to its degradation by commensals or mechanical sources.
Since access of the microbiota to the intestinal epithelium is restricted physically and chemically, communication with the host mainly depends on microbiota‐secreted factors such as metabolites, proteins, and extracellular vesicles (EVs) that can go through the mucin layer and reach host cells at the intestinal mucosal surface. There is mounting evidence that interkingdom crosstalk is principally mediated by EVs released by gut microbiota or by host intestinal cells (Diaz‐Garrido et al., 2021). This review is specifically focused on microbiota‐secreted EVs and their role as key mediators of bacteria–host interplay in the gut. In the following sections, we discuss current knowledge of their contribution as signals that modulate functions that are crucial for intestinal homeostasis and human health.
2. BACTERIAL EXTRACELLULAR MEMBRANE VESICLES
The production of EVs by bacteria was discovered in 1967 in Vibrio cholerae. Electron microscopy analyses of logarithmic growing bacterial cells showed the release of spherical membrane‐bound structures produced by pinching off the bacterial outer membrane. These structures were postulated to serve as a mechanism for toxin secretion (Chatterjee & Das, 1967). Initially, these vesicle‐type particles were considered needless, artefactual structures. However, as research advanced, they gained recognition as an active, regulated essential mechanism for bacterial cell physiology and communication. Due to their origin, these vesicles were named as membrane vesicles. In recent decades, intensive research has been carried out to establish their mechanisms of biogenesis, composition and functions (Bitto & Kaparakis‐Liaskos, 2017; Haurat et al., 2015; Schwechheimer & Kuehn, 2015).
It is now well known that all bacteria release EVs as a mechanism of communication between species. Bacterial extracellular vesicles (BEVs) are spherical lipid bilayer nanostructures, ranging from 20 to 300 nm in size. They are produced by Gram‐negative and Gram‐positive bacteria (Kim et al., 2015; Kulp & Kuehn, 2010). They are naturals carriers of bacterial molecules that include lipopolysaccharides (LPS), peptidoglycans, lipids, proteins, nucleic acids and in the case of pathogens, toxins and virulence factors. They act as a secretion mechanism that allows long‐distance delivery of bacterial active compounds in a protected environment, avoiding direct intercellular contact (Gill et al., 2019). The composition and content of BEVs originate in the producer strain and differ depending on the growth phase and conditions (Dauros Singorenko et al., 2017). Currently, it is assumed that the vesicle cargo defines their function.
2.1. Nomenclature and types of BEVs
Studies performed with large number of bacterial species have shown that Gram‐negative bacteria generate different kinds of EVs (Avila‐Calderón et al., 2015; Jan, 2017; Kulp & Kuehn, 2010). The main type derives from the outer membrane and are referred to as “outer membrane vesicles” (OMVs) (Jan, 2017; Schwechheimer & Kuehn, 2015). OMVs are spherical particles consisting of an outer leaflet of LPS and an inner leaflet of phospholipids. They originate by blebbing of the outer membrane and are therefore enriched in outer‐membrane and periplasmic biomolecules (Kulkarni & Jagannadham, 2014; Volgers et al., 2018). In addition, vesicles known as “outer‐inner membrane vesicles” (O‐IMVs) have been observed in Gram‐negative bacteria (Pérez‐Cruz et al., 2013). These vesicles display both the outer and the cytoplasmic membrane. In addition to periplasmic components, they include inner membrane proteins and cytosolic molecules (Perez‐Cruz et al., 2015). It is assumed that most Gram‐negative bacteria produce both kinds of vesicles, OMVs and O‐IMVs. Recent studies have described a new type of Gram‐negative bacteria derived vesicles called “explosive outer‐membrane vesicles” (E‐OMVs), in accordance with the biogenesis mechanism (Toyofuku et al., 2019). This model, which is based on cell lysis induced by phage‐encoded endolysin, could explain why OMVs also contain DNA and cytosolic proteins.
In contrast to Gram‐negative bacteria, the cell wall of Gram‐positive bacteria consists of a thick, rigid external layer of peptidoglycan, which was initially thought to prevent the release of EVs (Gill et al., 2019). For many years, the study of EVs from Gram‐positive bacteria was overlooked. This area has only received more attention in the last decade. Research developed in multiple Gram‐positive species such as Staphylococcus aureus, Bacillus anthracis, Listeria monocytogenes, Clostridium perfringens, or Bacillus subtilis has confirmed that Gram‐positive‐derived EVs are also spherical membrane particles of 20–100 nm in diameter (Brown et al., 2015; Kim et al., 2015; Lee et al., 2009; Palacios et al., 2020), although some species such as Bacillus sp., and C. perfringens produce larger vesicles of up to 400 nm (Brown et al., 2014; Jiang et al., 2014). Given the lack of an outer membrane, the name “cytoplasmic membrane vesicles” (CMVs) has been suggested for Gram‐positive EVs (Toyofuku et al., 2019). EVs from Gram‐positive bacteria lack LPS and periplasmic components but carry similar types of cargo molecules as Gram‐negative EVs, including peptidoglycan, lipids, proteins, and nucleic acids (Brown et al., 2015).
To simplify nomenclature, we use the term “bacterial extracellular vesicle” (BEV) to refer to all types bacteria‐derived vesicles, except when the information that applies to a specific BEV subset (e.g., OMV or CMV) is emphasized. BEV is consistent with the MISEV 2018 guidelines that recommend “extracellular vesicle” (EV) as the generic term for particles naturally released from the cell that are delimited by a lipid bilayer and cannot replicate (Théry et al., 2018).
2.2. Biogenesis of BEVs
Despite the significant biological implication of BEVs, the mechanisms of biogenesis are still poorly understood. Several models have been proposed and reformulated in the light of technological advances and new findings (Schwechheimer & Kuehn, 2015; Zingl et al., 2020).
For Gram‐negative bacteria, initial reports suggested that BEVs are produced under natural conditions as side products of cellular processes related with cell wall turnover. During cell wall recycling, loss of interaction between the outer membrane and peptidoglycan leads to outer membrane budding and the subsequent release of OMVs (Deatherage et al., 2009; Hoekstra et al., 1976; Wensink & Witholt, 1981). BEVs are also shed in response to stress conditions. One model is based on the physical stress caused by accumulation of peptidoglycan fragments or misfolded proteins in the periplasm, which results in increased turgor pressure that leads to the outer membrane bulging out (McBroom et al., 2006; McBroom & Kuehn, 2007). This model supports the idea that OMVs formation could be a helpful waste mechanism to get rid of an excess of harmful proteins (Schwechheimer et al., 2013; Schwechheimer & Kuehn, 2013). Moreover, stress induced in Stenotrophomonas maltophilia by antibiotic ciprofloxacin promotes increased release of a heterogeneous population of BEVs, including OMVs and O‐IMV (Devos et al., 2017). A third model involves structural changes in LPS in response to external factors (Schwechheimer & Kuehn, 2015). Changes in the concentration of cations that cross‐bridge the highly electronegative LPS may lead to accumulation of local negative charges and subsequent repulsion between LPS molecules, which results in local deformation of the bacterial cell membrane and vesicle shedding (Kadurugamuwa & Beveridge, 1995; Mashburn & Whiteley, 2005). Insertion of the Pseudomonas quinolone signal (PQS) into the outer leaflet of the outer membrane increases membrane curvature and OMVs formation through this mechanism. This quorum sensing molecule sequestrates positive charged ions, triggering destabilization of Mg+2 and Ca+2 salt bridges that leads to anionic repulsion between LPS molecules (Mashburn‐Warren et al., 2009; Schertzer & Whiteley, 2012). Other factors that influence LPS remodelling also result in modifications in the outer membrane curvature and OMVs formation (Volgers et al., 2018). LPS is anchored to the outer leaflet of the outer membrane through an acylated disaccharide named lipid A. In Salmonella enterica serovar thyphymurium, deacylation of lipid A results in hypervesiculation and OMVs production. In contrast, pagL mutants, which are defective in the enzyme that catalyses Lipid A deacylation, show significant reduction in OMV release relative to the wild‐type strain (Elhenawy et al., 2016). Recent studies suggest that asymmetric distribution of phospholipids between the inner and outer leaflets of the outer membrane represent a general mechanism for OMV formation under all growth conditions (Roier, Zingl, Cakar, & Schild, 2016; Roier, Zingl, Cakar, Durakovic, et al., 2016). In fact, mutants of the phospholipid transporter VacJ/Yrb show an hypervesiculation phenotype. This is due to the accumulation of phospholipids in the outer leaflet of the outer membrane, which triggers asymmetric expansion, membrane bulging, and the subsequent OMVs release (Henderson et al., 2016; Roier, Zingl, Cakar, Durakovic, et al., 2016). This mechanism involving the VacJ/Yrb ABC transport system is regulated by iron availability in Gram‐negative bacteria (Roier, Zingl, Cakar, Durakovic, et al., 2016). As stated above (Section 2.1), a new mechanism for OMVs formation based on explosive cell lysis has been described in Pseudomonas aeruginosa (Turnbull et al., 2016). In this model, cell lysis is triggered by a phage‐encoded endolysin that degrades the peptidoglycan cell wall. Once the peptidoglycan is degraded, the cells round up and explode. The shattered membrane fragments round up and self‐assemble into E‐OMVs (Turnbull et al., 2016).
The origin of Gram‐positive BEVs remains unclear and is still not fully understood. Earlier studies had suggested that vesicles may be forced through pores in the cell wall by turgor pressure after budding from the cell membrane (Brown et al., 2015). However, the current model involves peptidoglycan‐degrading enzymes in a mechanism that resembles the explosive cell lysis described in P. aeruginosa (Brown et al., 2015; Toyofuku et al., 2017; Wang et al., 2018). This mechanism was evidenced in B. subtilis treated with mitomycin C. This drug induces genotoxic stress that leads to activation of endolysins encoded by prophages integrated in the bacterial chromosome. The subsequent degradation of peptidoglycan causes cells to protrude cytoplasmic material through holes in the peptidoglycan layer, allowing CMV release (Toyofuku et al., 2017). Whereas P. aeruginosa cells round up and explode, B subtilis derived CMVs are released by pores formed in the wall. Additionally, recent studies in S. aureus have indicated that this bacterium produces proteins such as phenol‐soluble modulins and autolysins that increase cell membrane fluidity, facilitating CMV formation (Schlatterer et al., 2018). Likewise, oxidative stress induced by antibiotics in S. aureus triggers CMVs production by increasing the permeability of the peptidoglycan layer in a phage‐independent manner (Andreoni et al., 2019). In addition to enzymatic control of CMV production in Gram‐positive bacteria, genetic regulation of vesiculation has been investigated. Disruption of genes encoding factor σB (sigB) in L. monocytogenes or the two component system CovRS in Streptococcus pyogenes resulted in altered CMV production, thus pointing to a regulatory role in vesicle biogenesis (Lee et al., 2013; Resch et al., 2016).
2.3. Composition of BEVs
Once released, BEVs mediate cell‐to‐cell communication. The effects exerted by BEVs depend on their cargo. Cargo composition is influenced by the producer bacteria, growth and environmental conditions, and the mechanism of biogenesis. Diversity in cargo molecules confer BEVs crucial roles in bacteria–bacteria and bacteria–host interactions. Many studies have revealed that certain proteins and lipids are selectively incorporated into BEVs. However, how these molecules are picked up remains largely unknown. In this context, mechanisms based on preformed outer membrane microdomains could be proposed to explain the differential enrichment of certain proteins in BEVs (Bonnington & Kuehn, 2014). In this section, we recapitulate the general information about the composition of BEVs in terms of proteins, lipids, nucleic acids and small molecules.
Proteins . The protein content of BEVs is not universal and depends on the taxonomic group of the parental bacteria (genus, species, strain) and specific features such as whether they are symbiotic or pathogenic. Studies based on proteomics approaches have identified proteins connected with many biological processes that fit in six categories according to their function as structural proteins, porins, ion channels, transporters, enzymes and proteins related to stress responses (Lee et al., 2008). From these studies we have learned that BEVs released by Gram‐negative bacteria are enriched in outer membrane proteins such as OmpA, OmpC and OmpF, and periplasmic proteins such as AcrA and alkaline phosphatase. Proteomic analysis of BEVs from diverse Gram‐negative pathogens revealed the presence of virulence factors and a large number of proteins involved in biofilm formation, pathogenicity and host–cell interaction (Altindis et al., 2014; Choi et al., 2011; Jang et al., 2014). Proteins involved in vesicle biogenesis or in the modulation of vesicle size, yield and cargo have also been identified in S. enterica Serovar Typhi OMVs (Nevermann et al., 2019). Proteomic studies of vesicles released by Gram‐negative probiotic and commensal strains have shown that they contain proteins that contribute to colonization, competition and bacterial survival in the intestinal tract, and modulation of host immune and defence responses. Some of the identified proteins are strain‐specific (Aguilera et al., 2014; Elhenawy et al., 2014; Hong et al., 2019; Lee et al., 2007; Zakharzhevskaya, Vanyushkina, et al., 2017). Interestingly, the proteomic analysis of BEVs released by the probiotic Escherichia coli Nissle 1917 (EcN) identified a set of proteins that are common to BEVs secreted by Gram‐negative pathogens. These are moonlighting proteins that play different functions depending on the cell location. This group of bacterial proteins is mainly composed by chaperones (GroEL, DnaK) and metabolic enzymes (glyceraldehyde‐3‐phospate dehydrogenase, enolase, phosphoglycerate kinase). Besides their intracellular activity, these proteins have functions related with host interaction and tissue colonization when located extracellularly (Jeffery, 2018). In addition, the study revealed proteins that are encoded by strain‐specific genes. Most of them are adhesins and fitness factors that help bacterial growth and survival in the harmful gut environment (Aguilera et al., 2014; Hong et al., 2019). Reviews with specific information on proteomic analyses of BEVs are available (Kroniger et al., 2018; Lee et al., 2016). Regarding BEVs from Gram‐positive bacteria, proteomic analyses in different species (S. aureus, S. pyogenes, S. pneumoniae, and S. agalactiae, B. anthracis) identified membrane and cytoplasmic proteins related with several biological processes. As in Gram‐negative bacteria, pathogen‐derived BEVs are loaded with virulence factors and toxins that contribute to pathogenicity (Dean et al., 2019; Dean et al., 2020; Jeon et al., 2016; Lee et al., 2009; Olaya‐Abril et al., 2014; Resch et al., 2016; Rivera et al., 2010; Surve et al., 2016; West et al., 2020). Recent research shows that CMVs isolated from several Gram‐positive probiotics transport proteins that are responsible for their beneficial effects in the context of immune modulation and host interaction (Behzadi et al., 2017; Domínguez Rubio et al., 2017; Lee et al., 2016; Li et al., 2017).
Lipids . Lipids are important structural components of the bacterial cell envelop, and the corresponding BEVs from Gram‐negative and Gram‐positive bacteria; the OMVs and CMVs, respectively. Consistently, outer membrane phospholipids are present in OMVs. However, there is evidence that OMVs contain some lipids that are not found in the bacterial outer membrane (Chowdhury & Jagannadham, 2013; Kulkarni & Jagannadham, 2014; Tashiro et al., 2011). Studies in E. coli have revealed that OMVs comprise lipids involved in membrane curvature, such as glycerophospholipids, phosphatidylglycerol, phosphatidylethanolamine and cardiolipin (Barák & Muchová, 2013; Horstman & Kuehn, 2000). The structures of the fatty acid moieties affect the fluidity and rigidity of the lipid membranes. In P. aeruginosa OMVs, the relative amount of saturated fatty acids was around 1.5‐fold higher than in the cellular outer membrane, a fact that correlates with membrane rigidity (Li et al., 2018; Tashiro et al., 2012). In contrast, in Gram‐positive group A Streptococcus strains, overall levels of saturated fatty acids (C16 and C19) were not significantly different between BEVs and bacterial cell membrane. However, distinctive differences in their distribution in cardiolipin and phosphatidylglycerol were apparent (Resch et al., 2016). LPS is a general component of bacterial OMVs, although differences in LPS composition may exist between vesicles and the bacterial outer membrane. In this context, it has been reported that Porphyromonas gingivalis OMVs carry LPS molecules with long sugar chains and deacylated lipid A compared to the cell wall (Haurat et al., 2011). Likewise, deacylated lipid A accumulates in Salmonella OMVs (Elhenawy et al., 2014). In Gram‐positive bacteria, lipidomic studies have revealed that CMVs and the plasma membrane conserve essential aspects but show some differences in certain lipids and lipoproteins. In B. anthracis and S. pneumoniae, vesicles are enriched in C12‐C16 chain saturated fatty acids (Olaya‐Abril et al., 2014; Rivera et al., 2010). Conversely, lipids containing unsaturated fatty acids are more abundant in L. monocytogenes‐ derived vesicles than in bacterial cells. In addition, CMVs from this pathogen are enriched in phosphatidylethanolamine, sphingolipids and triacylglycerols, but glycoglycerolipids are underrepresented (Coelho et al., 2019). Overall, these studies show that the lipid composition of CMVs may vary with the bacterial species. These differences may be related with bacterial adaptation and survival in certain ecological niches (Coelho et al., 2019).
Nucleic acids . Many studies show that Gram‐negative and Gram‐positive‐derived BEVs are loaded with nucleic acids, including DNA and RNA. Therefore, BEVs allow delivery of DNA into other bacterial cells, which mediates horizontal gene transfer (Domingues & Nielsen, 2017). In addition, BEVs facilitate the interaction of transported nucleic acids with their specific intracellular receptors in host cells, which triggers modulation of host immune responses (Gilmore et al., 2021). BEVs contain luminal and surface associated DNA (Bitto & Kaparakis‐Liaskos, 2017; Jiang et al., 2014; Liao et al., 2014; Renelli et al., 2004). In BEVs from P. aeruginosa, most of the luminal DNA correspond to genes involved in specific functions related with antibiotic resistance, virulence and stress response (Bitto & Kaparakis‐Liaskos, 2017). In the Gram‐positive C. perfringens, BEVs carry genes encoding bacterial toxins such as α‐toxin (plc) and perfringolysin O (pfoA) (Jiang et al., 2014). External DNA in Streptococcus mutans‐derived BEVs was mainly related with biofilm formation and maturation. These are essential processes during bacterial adhesion (Liao et al., 2014).
Besides DNA, some studies have shown the presence of RNA in BEVs, with important implications for the regulation of the host immune system and other cellular processes (Lee, 2019). RNA associated with BEVs is protected from degradation, which facilitates its delivery to eukaryotic target cells. A compilation of RNAs associated with BEVs has been published (Dauros‐Singorenko et al., 2018). These studies revealed that the RNA included in BEVs is enriched in small non‐coding RNAs, which are mainly involved in the regulation of gene expression post‐transcriptionally. In some cases, the RNA cargo was shown to induce phenotypic changes in the target cells. In certain pathogenic bacteria, small RNAs secreted through BEVs mediate dysregulation of host immune responses (Choi et al., 2017; Koeppen et al., 2016). Moreover, many sequences of the transported small non‐coding RNAs found in E. coli BEVs align to human genome regions involved in the regulation of gene expression associated with epigenetic mechanisms (chromatin remodelling, histone modifications) or cell‐specific transcriptional control (Celluzzi & Masotti, 2016). It has been suggested that BEV‐associated small RNAs have a function similar to the regulatory role of eukaryotic miRNAs (Dauros‐Singorenko et al., 2018).
Small molecules. BEVs are also loaded with metabolites and effector molecules that modulate functions of target cells. Metabolomic approaches have revealed that BEVs contain metabolites, which were selectively packaged depending on the producer strains. In B. thetaiotaomicron, the varying metabolite content between BEVs isolated from pathogenic or commensal strains was consistent with the ability of each strain to colonize and survive in the intestinal tract (Bryant et al., 2017; Zakharzhevskaya, Vanyushkina, et al., 2017). Other effectors packaged into BEVs also act as fitness factors, helping bacterial survival in specific niches. For instance, BEVs from B. fragilis are loaded with the antimicrobial peptide BSAP, which displays inhibitory activity against other Bacteroidales of the human gut (Chatzidaki‐Livanis et al., 2014). BEVs also serve as vehicles to secrete hydrophobic quorum sensing molecules that mediate communication in bacterial communities, controlling important processes such as virulence or biofilm formation (Brameyer et al., 2018).
2.4. Functions of BEVs
Secretion of BEVs is a costly process that requires energy expenditure. This secretion mechanism was selected by evolution because BEVs play essential roles that give bacteria self‐protection or promote interaction and communication with other members of the microbial community or the host. BEVs are involved in numerous biological functions, which may differ depending on their specific cargo. Functions of BEVs can be classified into two groups: those linked to bacteria–bacteria interaction and those related with bacteria–host communication. Specific reviews are available on this topic (Caruana & Walper, 2020; Jan, 2017; Schwechheimer & Kuehn, 2015).
Regarding bacteria–bacteria interactions, BEVs allow producer bacteria to persist in the ecological niche by helping, competing with, or killing other bacteria. In this context, BEVs may serve as a microbial defence mechanism by providing protective functions against harmful agents that include phages, antibiotics, reactive oxygen species and antimicrobial peptides. Released BEVs can sequester phages, so that they do not directly interact with bacterial cells, preventing deleterious effects (Manning & Kuehn, 2011). BEVs can also serve as decoys for contaminants or antibiotics that target the bacterial membrane (Manning & Kuehn, 2011). Besides their passive role as antibiotic targets, BEVs also release enzymes that confer resistance to antibiotics, which benefits the producer strain and other susceptible bacteria in the microbial community. In this context, beta‐lactamases have been found in BEVs from S. aureus and Bacteroides sp (Lee et al., 2013; Régis Stentz et al., 2015) and enzymes that hydrolyse carbapenem in Acinetobacter baumannii BEVs (Liao et al., 2015). Genes encoding β‐lactamases have been found in BEVs. In this case, BEVs allow intra‐ and interspecies horizontal transfer of genes encoding antibiotic resistance (Chatterjee et al., 2017). In addition, BEVs can confer metabolic advantages. The presence of transport systems for siderophores, amino acids or fatty acids facilitates nutrient acquisition and bacterial survival and may serve as a competing mechanism against other bacteria. BEVs can also harbour hydrolase‐type enzymes that catalyse degradation of the proteins and complex polysaccharides present in the environment. This may assist the microbial community in nutrient acquisition (Elhenawy et al., 2014).
Besides beneficial effects for the bacterial community, BEVs can mediate offensive functions. In this sense, they carry degradative enzymes such as murein hydrolases, peptidoglycan hydrolases or endopeptidases for killing competing bacteria (Jan, 2017; Lee et al., 2008). As an example, BEVs from some Myxococcus sp. species have been shown to contain factors with antimicrobial activity and a great variety of hydrolytic enzymes that cause cell lysis of target bacteria, thus conferring predatory activity (Evans et al., 2012; Marshall & Whitworth, 2019).
In microbial communities, BEVs serve as key players in cell‐to‐cell communication. As stated above, hydrophobic quorum sensing molecules that coordinate bacterial growth and behaviour depending on population density are secreted into BEVs. Quorum sensing molecules also drive the microbial community to grow in biofilms. There is evidence that BEVs are important components of the biofilm matrix, which consists of polysaccharides, lipids, nucleic acids and proteins. Formation of biofilms protects multispecies bacterial communities and has benefits for nutrient acquisition and survival. In this context, BEVs can contribute to these functions (Flemming et al., 2016).
In the context of bacteria–host interactions, BEVs are well adapted and become essential for interkingdom communication. There is evidence that they control host cell pathways and processes by delivering bacterial effectors into target cells. BEVs are equipped with adhesin‐type molecules that help their adhesion to host cells, and microbial molecules recognized by immune receptors involved in the activation of inflammatory and defence responses (see Section 3). In pathogens, BEVs carry virulence factors, toxins and immunomodulatory effectors that help bacteria to attack host cells and subvert the host machinery to escape the immune system (Jan, 2017; Kaparakis‐Liaskos & Ferrero, 2015; Lee et al., 2008). For strains that establish a symbiotic relation with the host, released BEVs serve as a mechanism for the delivery of bacterial mediators that modulate important host functions (Badia & Baldomà, 2020).
In addition, based on their properties as long‐distance delivery vehicles and their intrinsic versatility and biocompatibility, BEVs are being explored as promising novel therapies. Engineered BEVs are being developed as new vaccines and adjuvants or specialized drug delivery vehicles for the treatment of emerging and new diseases. Specific reviews are available on this topic (Bitto & Kaparakis‐Liaskos, 2017; Cai et al., 2018).
3. INTERACTION OF BEVS WITH MAMMALIAN HOST CELLS
3.1. Internalization of BEVs by host cells
BEVs from Gram‐negative and Gram‐positive bacteria can deliver their content into the host cell cytoplasm. The uptake of BEVs by mammalian host cells is mainly mediated by endocytosis. This process occurs through different pathways depending on the surface and cargo of the vesicles (O'Donoghue et al., 2017; O'Donoghue & Krachler, 2016). Phagocytosis is the main route used by phagocytic cells of the immune system (neutrophils, macrophages, dendritic cells) to internalize BEVs. In non‐phagocytic cells, such as epithelial cells, other endocytic routes allow the entry of BEVs. These include macropinocytosis, clathrin‐mediated endocytosis and lipid raft‐mediated processes that may depend on caveolin. Each pathway allows internalized vesicle cargo to be sorted to specific subcellular locations. Actin‐dependent macropinocytosis is driven by the polymerization of an actin ring below the cell membrane. This causes a circular lump that closes at the top to encircle a portion of the extracellular space. The process mostly allows internalization of very large cargos, up to 1 μm. Clathrin‐mediated endocytosis is triggered by ligand binding to a cell surface receptor located in clathrin‐coated pits, which mature into vesicles inside the cells in a process that requires the GTPase dynamin. During intracellular trafficking, bacterial vesicles are sorted to endosomes and lysosomal compartments. Lipid rafts are dynamic membrane microdomains rich in cholesterol, sphingolipids and certain proteins that are associated with distinct endocytic pathways. One such pathway is caveolin‐mediated endocytosis, which involves membrane lipid raft domains that are enriched in caveolin, a protein that triggers membrane invaginations called caveolae and the subsequent formation of endocytic vesicles. Fission from the plasma membrane also requires dynamin, although trafficking of caveolar vesicles does not involve the same endosomal compartments as clathrin‐mediated endocytosis. Finally, BEVs can enter host cells through lipid‐raft domains in a caveolin‐independent manner. These routes are not mutually exclusive, since certain BEVs have been shown to use more than one pathway to enter epithelial cells depending on their size and cargo (O'Donoghue & Krachler, 2016). More details about this topic can be found in specific reviews (Anand & Chaudhuri, 2016; Bitto & Kaparakis‐Liaskos, 2017; Kaparakis‐Liaskos & Ferrero, 2015; O'Donoghue & Krachler, 2016).
Most of the available information on the mechanisms for BEV uptake in mammalian host cells comes from studies dealing with Gram‐negative pathogens. The first study on microbiota‐derived BEVs focused on the probiotic E. coli Nissle 1917 (EcN) and commensal E. coli strains (Cañas et al., 2016). The authors showed that BEVs from beneficial gut resident E. coli strains are internalized by several epithelial cell lines mainly by clathrin‐mediated endocytosis. Consistently, internalized BEVs colocalized with clathrin and specific markers of endosomes and lysosomes. Disruption of lipid rafts and caveolae domains with specific inhibitors did not affect vesicle uptake (Cañas et al., 2016). Furthermore, cryo‐transmission electron microscopy analyses of BEVs isolated from an hypervesiculating EcN tolR mutant evidenced highly structural heterogeneity in the isolated vesicles, with the presence of abundant aberrant structures that did not correspond with common OMVs or O‐IMVs (Pérez‐Cruz et al., 2016). This study revealed that tolR mutant‐derived BEVs displayed diminished cell binding capacity, which in turn, negatively influence the uptake levels in Caco‐2 cells. This indicates that only conventional BEVs efficiently enter epithelial cells through clathrin‐mediated endocytosis (Pérez‐Cruz et al., 2016). A recent study revealed the mechanism of uptake and traffic of BEVs from the resident gut microbiota B. thetaiotaomicron (Jones et al., 2020). By means of in vitro cultures, authors showed that B. thetaiotaomicron BEVs are internalized by intestinal epithelial cells (IECs) principally by dynamin‐dependent endocytosis and are sorted to endo‐lysosomal vesicles. Moreover, in vivo imaging studies showed that a proportion of B. thetaiotaomicron BEVs cross the intestinal epithelium by paracellular transmigration and reach systemic circulation. This demonstrates that BEVs mediate long‐distance communication with extra‐intestinal tissues (Jones et al., 2020).
There are few studies on the internalization pathways used by BEVs from Gram‐positives. In the probiotic Lactiplantibacillus plantarum BGAN8, time‐course internalization experiments performed in the presence of specific inhibitors of endocytosis pathways evidenced that the main entry route for secreted vesicles in IECs is the clathrin‐mediated pathway, although the uptake kinetics was slower than that reported for BEVs of Gram‐negative probiotics. Whereas internalized E. coli BEVs were apparent at 15–30 min incubation, more than 1 h was required to detect intracellular L. plantarum BEVs (Bajic et al., 2020). Authors suggested that the abundant exopolysaccharide material secreted by this probiotic that accompanied isolated BEVs could cause a trapping effect and delay the interaction of BEVs with epithelial cell membrane structures needed for endocytosis. Other internalization mechanisms may operate in parallel, depending on the vesicle size. The exopolysaccharide network that entraps BEVs creates a large volume coating the BEVs isolated from cultures of this probiotic that could be taken up by micropinocytosis (Bajic et al., 2020).
3.2. Recognition of BEVs by immune receptors
BEVs contain biological components produced by the parental strain. In this context, surface cargo molecules that mediate adhesion to host extracellular proteins or specific cell receptors are determinant for the primary interaction with target cells, and consequently with the derived effects (Kulp & Kuehn, 2010). BEVs also carry a set of molecules known as microbe‐associated molecular patterns (MAMPs) that are recognized by specific receptors expressed by host epithelial and immune cells. These pattern recognition receptors (PRRs) are key components of innate immunity as they sense gut microbes and mediate appropriate responses (Figure 1). Mammalian cells express different types of PRRs according to their location and association with cell membranes (Mu et al., 2015; Zhou et al., 2020). TLRs are transmembrane proteins that can be located at the plasma membrane (TLR2, TLR4, TLR5) or at intracellular endosomal membranes (TLR3, TLR7, TLR9). MAMPS present in the surface of BEVs (extracellular polysaccharides, LPS and lipoproteins) can bind extracellular TLRs to activate cell signalling pathways involved in the regulation of immune and defence responses. In addition, BEVs are internalized by mammalian host cells through endocytic pathways. By this mechanism, BEVs allow intracellular delivery of other enclosed MAMPs such as DNA or RNA that are recognized by specific TLR receptors in the membrane of endocytic compartments. At intracellular level, there are cytosolic immune receptors (not associated with organelle membranes), such as nucleotide‐binding oligomerization domain protein 1 (NOD1) and NOD2 that are activated by bacterial peptidoglycan fragments. In non‐invasive Gram‐negative pathogens, internalized BEVs are a mechanism for the delivery of peptidoglycan inside the host cell and subsequent activation of NOD receptors (Chatterjee & Chaudhuri, 2013; Kaparakis et al., 2010; Thay et al., 2014). Studies performed with H. pylori revealed that interaction of NOD1 with peptidoglycan included in the endocytosed vesicles takes place at early endosomes, where NOD1 is recruited during intracellular trafficking of BEVs (Irving et al., 2014). Following peptidoglycan binding, these receptors initiate the inflammatory response through activation of nuclear factor‐kB (NF‐kB) or the mitogen‐activated protein kinase (MAPK) pathways, which ultimately upregulate transcription of inflammatory genes (Figure 1). This mechanism is also activated by BEVs released by microbiota E. coli strains (Section 4.3.1).
FIGURE 1.
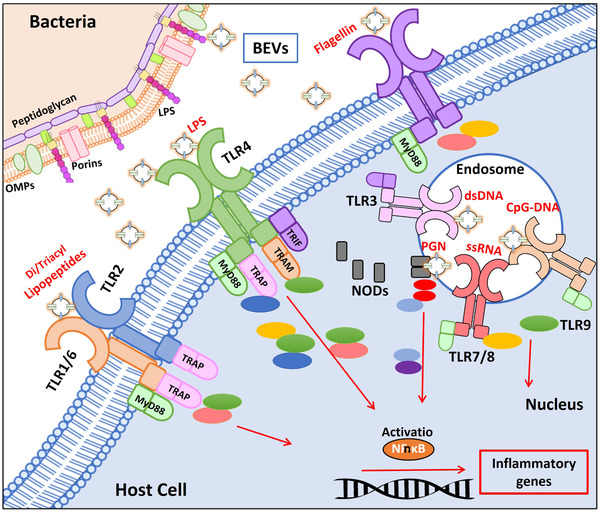
Recognition of BEV‐associated molecular patterns by host immune receptors. The drawing schematically shows toll‐like receptors (TLRs) located at the host cell membrane (TLR1/6, TLR2, TLR4, and TLR5), TLRs located at the endosomal membranes (TLR3, TLR7/8, TLR9) and cytosolic NODs (NOD1/NOD2), and the downstream signalling pathways that lead to activation of NF‐kB. Proteins of the different signalling pathways are drawn as coloured ellipses. The molecular associated pattern that specifically interacts with each receptor is indicated in red. LPS: lipopolysaccharide, PGN: peptidoglycan
4. CONTRIBUTION OF MICROBIOTA BEVS TO INTESTINAL HOMEOSTASIS
In the last 20 years, numerous studies have focused on BEVs from Gram‐negative pathogens, due to their role as vehicles for the delivery of virulence factors and molecules that inhibit and escape host immune responses (Kaparakis‐Liaskos & Ferrero, 2015). The study of probiotic and gut microbiota‐derived vesicles started in the early 2010s (Lopez et al., 2012; Shen et al., 2012). Since then, most studies have focused on Gram‐negative commensals, specifically Bacteroides fragilis, B. thetaiotaomicron, Akkermansia muciniphila, and intestinal E. coli isolates, including the probiotic EcN. Studies on BEVs from Gram‐positive gut microbes started later and were mostly focused on probiotics of the genus Lactobacillus and Bifidobacterium (Liu et al., 2018). In this section, we review current knowledge of the role of BEVs as important players in the gut microbiota effects on intestinal homeostasis. We focus on how microbiota BEVs control food metabolism, epithelial barrier integrity and immune responses. The information provided in this section is summarized in Table 1.
TABLE 1.
Studies evaluating the role of microbiota derived BEVs as modulators of intestinal homeostasis‐related processes
| Bacteria | Mechanism | Experimental approach | References |
|---|---|---|---|
| Gut ecology and food metabolsim | |||
|
Bacteriodes fragilis B. thetaiotaomicron |
Metabolism of complex carbohydrates to produce SFCAs: Expression of glycosyl‐hydrolases, sulfatases, proteases Cholesterol uptake: upregulation NPC1L1 receptor Metabolites in BEVs that facilitate intestinal colonization |
Proteomics of BEVs by mass spectrometry In vitro Caco‐2 cell culture In silico, proteomic and metabolomic analysis |
Valguarnera et al. (2018) Elhenawy et al. (2014) Ahmadi Badi et al. (2020) Bryant et al. (2017) Zakharzhevskaya, Tsvetkov, et al. (2017) Zakharzhevskaya, Vanyushkina, et al. (2017) |
| B. thetaiotaomicron |
Assimilation of dietary Insitol‐P Macrophage internalization (Sulfatases) |
Biochemical characterization of InsP6‐phosphatase Experimental model of colitis using genetically modified mice |
Stentz et al. (2014) Hickey et al. (2015) |
| Bacteroides fragilis | Antibiotic resistance (β‐lactamases) | Knockout mutant of putative β‐lactamase gene | Stentz et al. (2015) |
| Epithelial barrier integrity | |||
|
E. coli Nissle 1917 ECOR63 strain |
Upregulation of TJ proteins ZO‐1 and claudin‐14, downregulation of claudin‐2 Protection against EPEC‐induced damage: preservation of occludin and claudin‐14 mRNA levels, redistribution of ZO1, amelioration of F‐actin disorganization |
In vitro Caco‐2 and T‐84 cell cultures: RT‐qPCR, confocal microscopy In vitro Caco‐2 and T‐84 cell cultures infected with EPEC: RT‐qPCR, confocal microscopy, paracellular permeability assays |
Alvarez et al. (2016) Alvarez et al. (2019) |
| E. coli Nissle 1917 | Upregulation TFF3 and MMP‐9 mRNA | In vivo mice model of DSS‐induced colitis | Fabrega et al. (2017) |
| Akkermansia muciniphila |
Upregulation of ZO‐1, claudin 5 Upregulation of ZO‐1, occludin, claudin‐1 Upregulation of occludin, ZO‐1/2, claudin‐4 |
In vivo high‐fat diet (HFD)‐induced diabetic mice model, and Caco‐2 cell culture In vivo HFD‐induced obesity mice model In vitro Caco‐2 cells challenged with LPS |
Chelakkot et al. (2018) Ashrafian, Shahriary, et al. (2019) Ashrafian, Behrouzi, et al. (2019) |
| Gut immunity: modulation of inflammatory responses through the intestinal epithelium | |||
|
E. coli Nissle 1917 ECOR12 strain |
Upregulation of IL‐6, IL‐8, TNF‐α, IL‐10, MIP1α Upregulation of IL‐22 and β‐defensin Downregulation of IL‐12 Activation of NOD‐1 / NF‐κB pathway |
In vitro Caco‐2/PBMCs cell coculture model Ex vivo model of colonic explants Caco‐2 cells: NOD1 silencing ‐ RIP2 kinase inhibition |
Fabrega et al. (2016) Fabrega et al. (2016) Cañas et al. (2018) |
| E. coli Nissle 1917 | Upregulation of IL‐10; downregulation of IL‐1β, TNF‐α, IL‐6, IL‐12, IL‐17, iNOS and COX‐2 in colonic tissue | In vivo mice model of DSS‐induced colitis | Fabrega et al. (2017) |
|
Lactobacillus kefir L. kefiranofaciens L. kefirgranum |
Downregulation of IL‐8 Counteract oxidative stress by decreasing myeloperoxidase serum levels |
Caco‐2 cells challenged with TNF‐α In vivo mice model of TNBS‐induced IBD |
Seo et al. (2018) |
| Gut immunity: modulation of DCs and derived T cell responses | |||
| Bacteroides fragilis |
Induction Treg cells (CD4+CD25+FOXP3+) and IL‐10 production through a mechanism that involves TLR2 Activation of autophagy. Induction of Treg cells and IL‐10 production depends on functional ATG16L1 and NOD2. |
In vivo mice model of TNBs‐induced colitis In vitro bone marrow‐derived DCs culture BEVs from wild‐type and PSA deficient strains Bone‐marrow derived DCs from wild type, ATG16L1‐ and NOD2 deficient mice. In vitro cocultures of BMDCs with CD4+T cells In vivo mice model DNBS‐induced colitis |
Shen et al. (2012) Chu et al. (2016) |
| Bacteroides vulgatus mpk |
Induction of DC tolerance via TLR2 and TLR4 Upregulation of co‐stimulatory molecules including MHC‐II, CD40, CD80 and CD86 in CD11c+ cells |
In vitro bone marrow‐derived DCs culture TLR4/TLR2 knockout mice model |
Maerz et al. (2018) |
| Lactobacillus rhamnosus JB‐1 |
Increased production of IL‐10 and regulatory (CD4+CD25+FOXP3+) T cells |
In vitro bone marrow‐derived DCs culture In vivo mice model |
Al‐Nedawi et al. (2015) |
| Lactobacillus sakei | Enhance IgA expression | Ex vivo model of murine Peyer's patches | Yamasaki‐Yashiki et al. (2019) |
| Bifidobacterium bifidum LMG13195 | Promote differentiation to regulatory T cells (CD4+CD25+FOXP3+) and IL‐10 secretion | In vitro model of monocyte‐derived DCs co‐cultivated with CD4+ T cells | Lopez et al. (2012) |
| Bifidobacterium longum | Apoptosis of bone‐marrow‐derived mast cells through ESBP vesicular protein | In vivo mouse model of allergen‐induced food allergy | Kim et al. (2016) |
|
E. coli Nissle 1917 Commensal E. coli |
Upregulation of driver Th cytokines by DCs in a strain‐specific manner Differential induction of Th1, Th2, Th17/Th22 and T regulatory responses Regulation of key miRNAs in immunity (miR‐155, miR‐146a/b and miR‐let7i) Differential modulation of miRNAs involved in tolerogenic responses (miR‐125a/99b/let7e, miR‐125b, miR‐24) |
In vitro model of monocyte‐derived DCs co‐cultivated with CD4+ T cells In vitro model of monocyte‐derived DCs: RNA seq approaches to identify differential expressed miRNAs |
Diaz‐Garrido et al. (2019) Diaz‐Garrido et al. (2020) |
4.1. Gut ecology and food metabolism
The gut microbiota maintains a symbiotic association with the host at the intestinal mucosa and provides important metabolic, immunological and gut protective functions in the healthy individual. One important function of the microbiota is to generate metabolites that influence both microbes and host. One of the most abundant phyla present in the intestinal microbiota is Bacteroides spp, which exploit diverse biochemical mechanisms to colonize the harsh environment of the intestine. These commensal bacteria are well studied for their role in carbohydrate metabolism and production of short‐chain fatty acids (SCFAs) that provide daily energy requirements for the host (Wexler & Goodman, 2017; Wexler, 2007). In particular, B. thetaiotaomicron is a good model for investigating commensal bacteria–host interaction, since it has a well‐defined role in complex carbohydrate metabolism and its genome is fully sequenced (Eckburg et al., 2005). In this context, B. thetaiotaomicron codifies several mucin‐degrading enzymes such as glycosyl hydrolases and sulfatases that allow assimilation of mucin glycans and dietary nutrients, thus conferring an adaptive advantage over other bacteria. Some of the breakdown products can be used as a source of nutrients by other components of the gut microbiota. This favours the establishment of a balanced microbial symbiotic community (Benjdia et al., 2011; Martens et al., 2009). Importantly, bacterial metabolic activity on complex carbohydrates generates a pool of SCFAs that are reabsorbed through the large intestine and used by host as an energy source, which provides a significant amount of daily energy requirements (Singh, 2019).
BEVs produced by Bacteroides species are a source of such enzymatic activities and have a great impact on host metabolism (Figure 2). Most studies have been carried out on B. thetaiotaomicron and B. fragilis (Ahmadi Badi et al., 2020). B. fragilis is a good colonizer of the intestinal tract. In fact, it is commonly isolated from healthy human stools (Huang et al., 2011). B. thetaiotaomicron‐derived BEVs contain enzymes that help polysaccharide digestion in the gut. Dietary inositol polyphosphates (InsP6) present in vegetables interfere with polysaccharide digestion by chelating divalent cations. Since human IECs do not express enzymes that can dephosphorylate InsP6, this activity depends on enzymes encoded by gut microbiota. In this context, B. thetaiotaomicron‐derived BEVs are a luminal source of InsP6 phosphatases in the gut (Stentz et al., 2014). In addition, diverse hydrolytic enzymes such as glycosidases, sulfatases and proteases have been found as cargo in Bacteroides‐derived BEVs that participate in the breakdown of host‐indigestible glycans and host mucins (Elhenawy et al., 2014; Valguarnera et al., 2018). There is evidence that B. thetaiotaomicron BEVs are internalized in mucosa resident macrophages in a sulfatase‐dependent manner. By this mechanism, BEVs could cross the mucosal barrier and facilitate the delivery of bacterial antigens and factors to immune cells (Hickey et al., 2015).
FIGURE 2.

Metabolic activities associated with Bacteroides‐derived BEVs known to contribute to gut ecology and food metabolism. (a) The enzymes included as cargo in B. thetaiotaomicron and B. fragilis BEVs responsible for metabolic effects are indicated. (b) The drawing shows the intestinal epithelial barrier and the underlying immune system. The mucin layer (in green) maintains segregation between luminal microbes and the intestinal epithelium. Blue boxes connect the vesicular enzyme (in red) with each specific function. SCFAs: short‐chain fatty acids, InsP6: inositol phosphates
Various studies focus on the association between Bacteroides BEVs and cholesterol metabolism (Figure 2). Production of BEVs from B. thetaiotaomicron and B. fragilis is stimulated by saturated fatty acids, such as palmitate (Mirjafari Tafti et al., 2019). Moreover, BEVs from both bacteria upregulate expression of the apical surface‐expressed transporter Niemann‐Pick C1‐like (NPC1L1) that mediates cholesterol uptake by enterocytes (Ahmadi Badi et al., 2020).
Metabolomic analyses has revealed that BEVs from Bacteroides species are loaded with metabolites with a relevant role in interaction with the host (Bryant et al., 2017; Zakharzhevskaya, Vanyushkina, et al., 2017). Concerning B. thetaiotaomicron BEVs, the putative role of the metabolites was predicted by in silico analysis. This showed that the vesicles are enriched with metabolites known to facilitate intestinal colonization and metabolites that could be incorporated into mouse metabolic pathways (Bryant et al., 2017). Studies performed in B. fragilis revealed significant differences in the metabolomic and proteomic content of BEVs depending on the producer strain (Zakharzhevskaya, Vanyushkina, et al., 2017). Strains of B. fragilis can be distinguished by their ability to produce a zinc‐dependent metalloprotease toxin (BTF). They are classified accordingly as nontoxigenic B. fragilis (NTBF) and enterotoxigenic B. fragilis (ETBF). ETBF strains cause intestinal inflammation and, in some cases, can contribute to colon cancer (Sears et al., 2014). The toxin BTF is secreted through BEVs as a surface exposed protein. In this location, BTF interacts with phospholipids and polysaccharides of the host cell membrane through hydrophobic and electrostatic interactions (Zakharzhevskaya, Tsvetkov, et al., 2017). Besides virulence factors, proteomic and metabolomic characterization of BEVs from NTBF and ETBF strains showed that BEVs from the non‐pathogenic strain are enriched in enzymes required for polysaccharide utilization, a metabolic activity that positively contributes to intestinal ecology. Conversely, BEVs from ETBF contain a high number of catabolic enzymes that belong to energy‐producing pathways, which allow long persistence in the gut. Fluxomic experiments with isotope‐labelled glucose confirmed that these pathways are active in pathogen‐derived BEVs (Zakharzhevskaya, Vanyushkina, et al., 2017).
4.2. Modulation of intestinal epithelial barrier integrity
In the gut, besides host‐derived EVs (Diaz‐Garrido et al., 2021), microbiota BEVs act as regulators of epithelial barrier integrity. This regulation is critical to gut homeostasis, as disruption of this barrier leads to enhanced intestinal permeability, a condition that causes inflammatory, allergic and metabolic diseases (Suzuki, 2013).
The intestinal epithelium serves as a physical barrier that restricts translocation of luminal microbes and antigens into host tissues without interfering with the passage of ions through the paracellular space. An important mechanism governing epithelial barrier function is the formation of tight junctions (TJs) between adjacent IECs. TJs establish intercellular connections in the apical part of the polarized epithelium through transmembrane proteins that include occludin, claudins, tricellulin and junctional adhesion molecules. These proteins are anchored to the actin cytoskeleton by the cytosolic proteins zonula occludens (ZO)‐1 and ZO‐2. TJs are dynamic structures that modulate epithelial barrier integrity and paracellular permeability. Therefore, the expression and subcellular location of TJ proteins are controlled by signalling pathways and mechanisms in response to external stimuli (Shen, 2012). Enteric pathogens cause TJ disruption as part of their infective mechanisms, whereas certain microbiota strains reinforce TJ structures as a mechanism to maintain gut barrier integrity (Hiippala et al., 2018). Besides TJs, other structures maintain connectivity between IECs, namely adherens junctions, gap junctions and desmosomes (Luissint et al., 2016).
Several studies in gut microbiota species show the role of microbiota BEVs as modulators of epithelial barrier integrity (Badia & Baldomà, 2020). Concerning intestinal E. coli isolates, the probiotic EcN has been extensively studied due to its recognized immunomodulatory and anti‐inflammatory properties that positively contribute to microbiota balance (Sonnenborn, 2016; Sonnenborn & Schulze, 2009; Toloza et al., 2015). The probiotic EcN reinforces the epithelial barrier through regulation of TJ proteins ZO‐1 (Ukena et al., 2007), ZO‐2 (Zyrek et al., 2007), claudin‐14 (Hering et al., 2014) and the desmosome protein pinin (Veltman et al., 2012). Hering and co‐workers showed that the probiotic factor that triggers upregulation of claudin‐14 is the secreted protein TcpC, which activates the ERK1/2 signalling pathway (Hering et al., 2014). Further studies in our group with EcN cell‐free supernatants revealed that in addition to TcpC, secreted BEVs have a strengthening barrier function (Alvarez et al., 2016). Our studies included other E. coli strains of intestinal origin, such as ECOR12 (group A; TcpC negative) and ECOR63 (group B2; TcpC positive). We have shown that BEVs released by E. coli strains modulate epithelial barrier integrity in a strain‐specific manner through direct and indirect mechanisms. In monolayers of colonic cell lines, BEVs from EcN and ECOR63 reinforce the epithelial barrier through upregulation of TJ proteins ZO‐1 and claudin‐14. Moreover, BEVs from both strains modulate barrier permeability through downregulation of claudin‐2, a leaky protein that facilitates barrier permeability (Figure 3). These regulatory activities do not depend on TcpC, which is secreted as free soluble protein. Conversely, ECOR12 BEVs have no effect on epithelial barrier integrity (Alvarez et al., 2016). The protective role of EcN and ECOR63 BEVs was studied in a cellular model of epithelial barrier dysfunction induced by infection with enteropathogenic E. coli (EPEC) (Alvarez et al., 2019). In this model, BEVs from both E. coli strains were able to protect the epithelial barrier integrity against EPEC infection by activating compensatory regulatory mechanisms at transcriptional and post‐transcriptional levels that affect TJ proteins. Specifically, BEVs counteracted altered mRNA expression of occludin and claudin‐14, retained ZO‐1 and occludin at TJs in the cell boundaries and ameliorated F‐actin cytoskeleton disorganization (Alvarez et al., 2019). In the human gut, crosstalk between epithelial and immune cells is crucial for maintaining intestinal homeostasis. In this context, we have shown that BEVs released by intestinal E. coli strains activate other mechanisms in the gut mucosa that can synergistically reinforce the epithelial barrier (Figure 3). In human colonic explants, EcN BEVs activate IL‐22 expression. This cytokine helps to preserve the integrity of the epithelial barrier by inducing mucin production by goblet cells (Fabrega et al., 2016). In the experimental mouse model of colitis, oral administration of EcN BEVs preserves the colonic cytoarchitecture of the mucosa, an effect that may be attributed to upregulation of trefoil factor 3 (TFF3), a bioactive peptide involved in epithelial repair. Since TFF3 promotes redistribution of ZO‐1 from the cytoplasmic compartment to the intercellular junctions in IEC monolayers, activation of TFF3 by EcN BEVs indirectly reinforces TJs in colitic mice and counteracts increased barrier permeability (Fabrega et al., 2017). Another barrier protective mechanism exerted by EcN BEVs is downregulation of matrix metalloprotease‐9 (MMP‐9) expression, an enzyme that contributes to intestinal inflammation by disrupting TJs between IECs, which in turn leads to increased intestinal permeability and the subsequent pro‐inflammatory effects (Fabrega et al., 2017).
FIGURE 3.
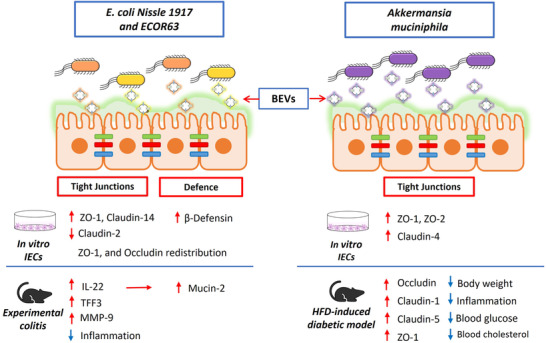
Modulation of the gut epithelial barrier by microbiota BEVs. Schematic representation of the intestinal epithelium, where tight junction (TJ) proteins are indicated by coloured bars connecting adjacent intestinal epithelial cells (IECs). BEVs released by E. coli Nissle 1917 (EcN) and ECOR63 (left panel), or A. muciniphila (right panel) migrate through the inner mucus layer and reach the epithelium. Regulatory mechanisms influencing the integrity of the intestinal barrier known to be activated by BEVs are indicated below the drawing, specifying whether evidences were obtained from in vitro assays (culture of IECs) or in vivo models of increased intestinal permeability (experimental colitis and high fat diet (HFD)‐induced diabetic model). Upregulation/downregulation of gene transcription is indicated by red arrows. For each experimental model, the beneficial effects of BEVs counteracting disease alterations are shown by blue arrows. Overall, the regulatory effects mediated by microbiota BEVs result in gut epithelial barrier reinforcement and the subsequent reduction of intestinal permeability
Reduced microbiota diversity, and especially low numbers of the Gram‐negative bacterium A. muciniphila, has been linked with the development of inflammatory bowel disease (IBD) (Png et al., 2010; Vigsnæs et al., 2012), and metabolic alterations such as diabetes and obesity (Zhou & Zhang, 2019). This microorganism is an abundant inhabitant of the human intestinal tract that provides metabolic benefits to the host by degrading intestinal mucin into SCFAs (de Vos, 2017). Butyrate is used as an energy source by colonic epithelial cells and propionate can signal to the host through GPR41 and GPR43 receptors (Clausen & Mortensen, 1995; Le Poul et al., 2003). Interestingly, SCFAs are also important molecules involved in appetite control and food intake (Flint et al., 2012). In addition, A. muciniphila has other beneficial properties associated with gut barrier strengthening and immune regulation (Ottman et al., 2017). Consistently, pretreatment of mice with A. muciniphila prior to exposure to dextran sulphate sodium (DSS) resulted in a reduction of colitis severity (Bian et al., 2019; Gobert et al., 2016; Zhai et al., 2019). Studies based on the same experimental model of colitis revealed that BEVs released by A. muciniphila mediate the protective effects observed. Oral administration of isolated BEVs ameliorated colitis symptoms and disease scores by reducing infiltration of immune cells in the colonic tissue and production of pro‐inflammatory cytokines (Kang et al., 2013).
As stated above, the increased gut permeability that follows epithelial barrier disruption allows the passage of endotoxins and luminal antigens into the intestinal lamina propria. This trafficking initiates a mucosal immune response that causes chronic low‐grade inflammation, prompting metabolic disorders characterized by non‐alcoholic fatty liver disease and insulin resistance that lead to type 2 diabetes and eventually obesity. Metagenomic analyses revealed diminished abundance of A. muciniphila in patients with obesity and type 2 diabetes, and their stool samples also contain fewer A. muciniphila‐derived MVs than healthy controls (Chelakkot et al., 2018).
The role of A. muciniphila BEVs in the modulation of gut permeability was studied in a high‐fat diet (HFD)‐induced diabetic mice model compared with control mice fed a normal diet (Chelakkot et al., 2018). Oral administration of A. muciniphila BEVs improved all the alterations caused by HFD, affecting gut permeability and metabolic function (Figure 3). Specifically, BEVs restored the intestinal barrier, reduced the subsequent recruitment of immune cells, and upregulated the altered TJ‐proteins occludin, ZO‐1 and claudin‐5. Transcriptional activation of occludin by A. muciniphila BEVs was verified by immunoblotting in Caco‐2 cell monolayers challenged with LPS (Chelakkot et al., 2018). Regarding metabolic effects, the treated mice had decreased body weight and improved glucose tolerance. These metabolic outcomes were attributed to restored barrier integrity, since at six hours post‐administration, BEVs were found in colon, but not in liver, muscle, fat tissue, pancreas or spleen (Chelakkot et al., 2018). Other studies also evidenced the beneficial effects of A. muciniphila‐derived BEVs when given to mice fed an HFD. They could reduce weight gain, adipose tissue inflammation, blood glucose and cholesterol levels (Ashrafian, Shahriary, et al., 2019). Moreover, animals that received A. muciniphila BEVs showed upregulated expression of ZO‐1, occludin and claudin‐1. Interestingly, A. muciniphila BEVs elicited stronger effects than live bacteria (Ashrafian, Shahriary, et al., 2019). Authors confirmed upregulation of ZO‐1, ZO‐2 and claudin‐4 by A. muciniphila BEVs in the human epithelial colorectal adenocarcinoma cell line Caco‐2 (Ashrafian, Behrouzi, et al., 2019). Overall, these studies point to the therapeutic potential of A. muciniphila BEVs against inflammatory diseases, acting as positive regulators of intestinal barrier integrity and thus reducing inflammation (Ashrafian, Behrouzi, et al., 2019).
4.3. Modulation of gut immunity
In this section, we discuss the role of commensal and probiotic‐derived BEVs on the regulation of host immune responses. We consider mechanisms that involve indirect activation of immune cells through the intestinal epithelium and mechanisms that result from direct interaction of microbiota BEVs with immune cells. The main mechanisms described in this section are summarized in Figure 4.
FIGURE 4.
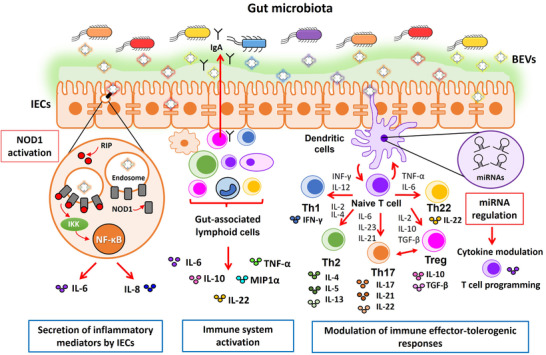
Schematic picture summarizing the immunomodulatory effects elicited by microbiota‐derived BEVs in the gut. The drawing shows the intestinal epithelium covered with the mucin layer that prevents access of luminal microbes while allowing passage of BEVs. Immune cells (lymphocytes, macrophages and dendritic cells) in the lamina propria are shown below the epithelial monolayer. Microbiota derived BEVs exert immune modulation by two main mechanisms. (i) Undirect activation of immune cells through the intestinal epithelium (left scheme). Internalized EVs by intestinal epithelial cells (IECs) activate the cytosolic receptor NOD1 that triggers secretion of immune effectors, which in turn stimulate gut‐associated lymphoid cells to produce a wide range of cytokines. Activation of the NOD1 signalling pathway by microbiota BEVs is shown encircled in more detail. BEVs are internalized through clathrin‐mediated endocytosis and recruit NOD1 (grey cylinders) to early endosomes. Activated NOD1 interacts with the specific kinase RIP2 (red circles), which leads to NF‐kB activation and the subsequent upregulation of host genes involved in the inflammatory response (IL‐6, IL‐8). (ii) Direct activation of gut resident immune cells by microbiota BEVs that leads to secretion of immune mediators and secretory IgA (middle scheme). In addition to direct interaction with microbiota BEVs that reach the gut‐associated lymphoid tissue via transcytosis across M cells, dendritic cells (DCs) also interact with luminal BEVs by extending pseudopodia across the epithelial cell layer (right scheme). Studies with several gut microbiota species revealed that BEVs activate DCs in a strain‐specific manner. Differential regulation of miRNAs in DCs is one of the regulatory mechanisms involved in the specific immunomodulatory effects of BEVs isolated from probiotic and commensal E. coli is (highlighted in the adjacent violet circle). In conclusion, DCs integrate incoming signals delivered by microbiota BEVs and set up specific programs that promote differentiation of naïve T cells into effector T cells (Th1, Th2, Th17, Th22) or regulatory T cells (Treg), thus allowing coordination of suitable T cell responses
4.3.1. Activation of immune responses by microbiota BEVs through the intestinal epithelium
Modulation of the immune system by commensal bacteria has been investigated in in vitro cellular models designed to mimic what occurs in vivo in the intestine. These models aimed to reproduce the crosstalk between the microbiota, the intestinal epithelium and the immune system, and consisted of co‐cultures of epithelial and immune cells in Transwell devices. A monolayer of human IECs is grown in the upper chamber simulating the intestinal epithelial barrier, and immature dendritic cells (or other immune cells) are grown in the basolateral chamber mimicking the underlying immune system of the lamina propria (Bermudez‐Brito et al., 2013). Several studies to evaluate the probiotic properties of Lactobacillus spp strains were performed in this model following apical stimulation with live bacteria (Bermudez‐Brito et al., 2012, 2014).
Subsequently, similar models were used to study microbiota BEVs as modulators of immune responses. In this context, our group investigated the immunomodulatory role of BEVs from the probiotic EcN and commensal E. coli strains following apical stimulation of Caco‐2/ peripheral blood mononuclear cells (PBMCs) co‐cultures as a model to mimic intact intestinal mucosa. For comparison, direct stimulation of PBMCs was approached as a model of intestinal barrier disruption. Secretion of several cytokines and chemokines was analysed in the basolateral compartment, and their expression was assessed in Caco‐2 cells and PBMCs. Whereas both BEVs and bacterial lysates activated the expression of IL‐6, IL‐8, TNF‐ α, IL‐10, and MIP1α in PBMCs treated following the barrier disruption model, only BEVs elicited immunomodulatory effects in the co‐culture model (Fabrega et al., 2016). This proved that the epithelial barrier formed by Caco‐2 cells prevented direct access of bacterial effector molecules to underlying PBMCs, and that PBMCs were activated by factors secreted by the epithelium in response to BEVs. This study showed the ability of microbiota‐derived BEVs to trigger signalling events through the intestinal epithelial barrier (Fabrega et al., 2016). Upregulation of these cytokines by BEVs was verified in human colonic explants. Expression analysis of other immune mediators was also approached in this ex vivo model. As stated above (Section 4.2), BEVs from EcN and the commensal ECOR12 activated the expression of intestinal barrier effectors such as IL‐22, and the antimicrobial peptide β‐defensin‐2 in this ex vivo model. In contrast, BEVs from these strains triggered downregulation of TGF‐β and the pro‐inflammatory cytokine IL‐12 (Fabrega et al., 2016). TGF‐β is a pleiotropic cytokine with relevant regulatory roles in inflammation. This cytokine triggers differentiation of Treg cells with anti‐inflammatory properties. However, in the presence of IL‐6, TGF‐β promotes differentiation of pro‐inflammatory T helper 17 (Th17) cells. The balanced activity of Treg and Th17 cells in the intestinal mucosal surface is important to preserve intestinal homeostasis (Cheng et al., 2019). Imbalance in these phenotypes towards excessive production of Th17 cells can lead to development of inflammatory pathologies including IBD. In this context, reduction in TGF‐β levels caused by BEVs from these microbiota strains may help restore the Th17/Treg balance under inflammatory conditions. Th17 cells also influence production of mucin‐1 by colonic cells. We showed that BEVs from EcN and ECOR12 triggered downregulation of this membrane‐associated mucin in colonic explants. Mucin‐1 and TGF‐β are overexpressed in several cancer types. Regulation of these markers by certain gut beneficial bacteria may reflect the influence of microbiota BEVs on cancer progression or treatment effectiveness (Gopalakrishnan et al., 2018). Overall, these findings support the role of microbiota‐derived BEVs in the activation of immune and defence responses through the intestinal epithelial barrier. This proves that BEVs are an effective strategy used by gut bacteria to communicate with intestinal mucosa cells and regulate intestinal homeostasis.
In addition to in vitro models, the beneficial properties of the probiotic EcN have been shown in mouse models of colitis (Rodríguez‐Nogales et al., 2018; Souza et al., 2016). Oral administration of EcN BEVs to DSS‐treated mice ameliorated colitis symptoms, histological scores, intestinal permeability and inflammatory responses in the same way as the live probiotic (Fabrega et al., 2017). Besides counteracting the altered expression of tissue repair and wound healing markers (Section 4.2), EcN MVs mediated anti‐inflammatory effects. Treatment with BEVs reduced the increased expression of pro‐inflammatory cytokines IL‐1β, TNF‐α, IL‐6, IL‐12, IL‐17, and INF‐γ and increased expression of the anti‐inflammatory cytokine IL‐10. The beneficial effects were also associated with downregulation of well‐known enzymes involved in injury and inflammation, such as cyclooxygenase‐2 and inducible nitric oxide synthase. These outcomes correlated with lesser infiltration of inflammatory cells in EV‐treated mice (Fabrega et al., 2017). Like A. muciniphila BEVs, vesicles from the probiotic EcN could be a safe probiotic‐derived strategy free of bacteria to preserve gastrointestinal health by targeting intestinal inflammatory processes.
There is evidence that BEVs from Gram‐positive bacteria elicit immunomodulatory effects in IECs. Kefir‐derived strains Lactobacillus kefir, Lactobacillus kefiranofaciens, and Lactobacillus kefirgranum alleviate the production of inflammatory cytokines under in vitro and in vivo inflammatory conditions (Seo et al., 2018). Treatment of TNF‐α stimulated Caco‐2 cells with BEVs isolated from these strains significantly diminished production of the proinflammatory cytokine IL‐8 at mRNA and protein‐secreted levels. The mechanism involved inhibition of TNF‐α signalling by reducing phosphorylation of the NF‐kB p65 subunit (Seo et al., 2018). In the mouse model of experimental colitis, administration of kefir Lactobacillus‐derived BEVs alleviated disease symptoms and tissue damage scores. These vesicles also reduced the oxidative stress associated with inflammatory diseases, since serum levels of myeloperoxidase were significantly reduced in mice treated with BEVs (Seo et al., 2018).
Activation of NOD signalling pathways in IECs
There is evidence on the involvement of NOD receptors in the molecular mechanisms and signalling pathways activated by microbiota BEVs in IECs that initiate the innate immune response. These cytosolic receptors are crucial for maintaining intestinal homeostasis. They play relevant roles in host defence responses against bacterial pathogens and regulation of the inflammatory response to microbiota (Caruso et al., 2014; Kaparakis‐Liaskos & Ferrero, 2015). NOD1 detects D‐glutamyl‐meso‐diaminopimelic acid that is mainly present in the peptidoglycan of Gram‐negative bacteria (Chamaillard et al., 2003; Girardin et al., 2003). NOD2 detects muramyl dipeptide that is ubiquitously present in all bacterial peptidoglycan molecules (Girardin et al., 2003). Considering that gut microbiota is mainly composed of non‐invasive bacteria, several pathways have been proposed to mediate intracellular delivery of peptidoglycan into IECs. Peptidoglycan fragments released into the gut lumen during bacterial division can be internalized in IECs through endocytosis or by specific membrane‐associated transporters (Philpott et al., 2014; Swaan et al., 2008). The other pathway, mostly studied in Gram‐negative pathogens, involves peptidoglycan delivery through BEVs (Section 3.2).
The activation of NOD receptors by BEVs from gut microbiota has been studied in probiotic and commensal E. coli strains (Cañas et al., 2018). This study showed that BEVs isolated from the probiotic EcN and the commensal ECOR12 activate NOD1 signalling pathways in IECs. Several experimental approaches were used to confirm involvement of this cytosolic receptor in the cell response to internalized BEVs. These included NOD1 silencing with specific siRNA and the use of inhibitors of the specific kinase of the NOD1 pathway. Both strategies significantly abolished vesicle‐mediated activation of NF‐κB and consequently reduced IL‐6 and IL‐8 expression. Furthermore, BEVs from both microbiota strains colocalized with NOD1 in early endosomes. Interaction with NOD1 led to the formation of NOD aggregates in the cell cytoplasm and the subsequent recruitment of RIP2 kinase that initiates the phosphorylation cascade, resulting in NF‐κB activation. This transcription factor activates genes involved in the inflammatory response that is transmitted to gut resident immune cells through the release of chemokines and other immune effectors (Cañas et al., 2018). This study proved that EVs from beneficial gut bacteria serve as a mechanism for peptidoglycan delivery into the host cytosol. In IECs, continuous stimulation of NOD receptors by microbiota BEVs steadily primes the innate immune system, leading to controlled inflammatory responses that assist pathogen eradication and intestinal homeostasis.
4.3.2. Direct activation of immune cells by microbiota BEVs
To maintain intestinal balance, the immune system must respond to external antigens and trigger immune responses to protect the host against pathogens or injuries. The gut microbiota and the host mucosal immune system interact strongly, with important implications for human health. As stated above, the intestinal epithelium acts as an indirect transmitter of microbiota signals to immune cells. However, specialized cells of the gut‐associated lymphoid tissue such as dendritic cells (DCs) can directly interact with gut microbes and direct appropriate immune responses. DCs detect bacteria or derived BEVs that reach the immune cells underneath the intestinal epithelium by transcytosis across M cells. However, DCs can also sample gut luminal content by extending pseudopodia across the epithelial cell layer (Rescigno et al., 2001). On recognition of MAMPs by PRRs, DCs activate the expression of cytokines and specific surface molecules that promote differentiation of T cells to specific functional subsets (Hooper et al., 2012). Therefore, DCs play a relevant role in connecting innate and adaptive responses, ensuring intestinal homeostasis by orchestrating the host immune responses to gut microbes.
Bacteroides sp . As mentioned in Section 4.1, B. fragilis is a gut resident bacterium with well‐known beneficial properties in food metabolism and gut ecology. The immunomodulatory effects of this commensal have been proved in experimental models of colitis. Administration of B. fragilis protected against colitis development by suppressing activity of inflammatory molecules and inducing production of anti‐inflammatory cytokines (Chang et al., 2017; Chiu et al., 2014; Mazmanian et al., 2008; Round & Mazmanian, 2010).
The first study proving the beneficial effects of BEVs from a commensal bacterium was focused on B. fragilis (Shen et al., 2012). Authors showed that BEVs from B. fragilis contain capsular polysaccharide A (PSA), an immunomodulatory molecule that induces regulatory T cells (Treg) and mucosal tolerance (Mazmanian et al., 2005). In a mouse colitis model, animals treated with BEVs from wild type B. fragilis showed improved disease scores and symptoms compared to animals treated with BEVs derived from a PSA‐deficient mutant (Shen et al., 2012). Regarding molecular mechanisms, there was a strong association between the PSA content in BEVs and the activation of Treg cells. This was confirmed in an in vitro model of bone marrow‐derived DCs pulsed with BEVs from B. fragilis and then co‐cultured with naïve T cells. Increased numbers of CD4+ CD25+ FOXP3+ T cells and IL‐10 production were induced by DCs stimulated with wild‐type BEVs but not by DCs treated with BEVs from the ∆PSA mutant. These immunomodulatory effects were mediated by activation of the TLR2 pathway by PSA in DCs. A microarray analysis comparing mRNA levels induced by wild‐type BEVs or ΔPSA BEVs in DCs allowed identification of genes regulated by PSA in a TLR2‐dependent manner (Shen et al., 2012). Another study showed that immune receptors other than TLR2 contributed to the signalling events activated by B. fragilis BEVs in DCs. This study focused on risk genes proposed for Crohn's disease (CD) development, particularly ATG16L1 and the immune receptor NOD2. These proteins are involved in autophagy regulation and there is evidence that immune cells impaired in autophagy elicit exacerbated inflammatory responses (Chu et al., 2016). Results from co‐cultures of CD4+ T cells with bone marrow‐derived DCs differentiated from wild type, ATG16L1‐deficient or NOD2‐/‐ knockout mice stimulated with B. fragilis BEVs revealed that the immunomodulatory effects of BEVs depend on functional ATG16L1 and NOD2 to induce a non‐canonical autophagy pathway that is essential for colitis protection. In fact, ATG16L1‐deficient DCs did not induce Tregs in response to BEVs from this commensal. Further analysis revealed that B. fragilis BEVs use the noncanonical autophagy pathway LAP (LC3‐associated phagocytosis) to mediate tolerogenic responses (Chu et al., 2016). Likewise, NOD2 knockout bone marrow‐derived DCs failed to induce Treg responses with reduced IL‐10 production, which in turn compromises suppression of mucosal inflammation. This study also showed that immune cells from human subjects with the T300A variant of ATG16L1 linked with a high risk of developing CD are defective in Treg responses to B. fragilis BEVs (Chu et al., 2016).
The immunomodulatory properties of B. thetaiotaomicron have been proved in preclinical models of CD treated with live bacterial suspensions (Delday et al., 2019). The role of BEVs in mediating immunomodulatory effects has been recently evidenced in a study performed in DCs derived from healthy individuals and from patients with CD or ulcerative colitis (UC) stimulated with B. thetaiotaomicron BEVs (Durant et al., 2020). BEVs from this commensal induced IL‐10 expression from healthy colonic DCs, which is an indicator of health state. The most abundant population were myeloid CD11+ DCs. In DCs derived from PBMCs isolated from healthy donors, BEVs also activated expression of the co‐stimulatory marker CD80 and IL‐6. In contrast, B. thetaiotaomicron BEVs failed to induce IL‐10 in colonic DCs from IBD patients. This correlated with reduced numbers of CD103+ DCs, a population involved in T regulatory responses and immune balance (Durant et al., 2020). The activity of disease also influences the regulatory responses in IBD. In this context, this study showed that blood‐derived DCs from patients with inactive and active UC showed variations in IL‐10 levels. DCs from patients with inactive disease had decreased levels of IL‐10. Conversely, DCs from patients with active disease showed IL‐10 levels comparable to those of healthy controls. This suggests that intrinsic alterations in the immune responses to this commensal are important in IBD (Durant et al., 2020). This study indicates that BEVs from B. thetaiotaomicron could drive a balanced immune response to microbiota in healthy individuals. However, this response is altered in IBD states, which contributes to an inflammatory environment in the intestine.
B. vulgatus mpk is another gut symbiont of the Bacteroides family with relevant features that confer immunomodulatory properties. In fact, this commensal ameliorates colitis development in several IBD mouse models (Müller et al., 2008; Perez‐Lopez et al., 2016). The mechanism by which B. vulgatus mpk elicits beneficial immunomodulatory effects is not well‐established but a recent study has shown that secreted BEVs induce a tolerant semi‐mature phenotype in bone marrow‐derived DCs (Maerz et al., 2018). Notably, B. vulgatus BEVs elicited the same immunomodulatory response as the live commensal on CD11c+ DCs, since no differences in the expression levels of MHC‐II, CD40, CD80 and CD86 markers were observed between DCs stimulated with either B. vulgatus mpk or isolated BEVs. To decipher the mechanisms involved in the actions mediated by BEVs, DCs derived from mice deficient in toll‐like receptor 2 (Tlr2 –/–), TLR‐4 (Tlr4 –/–) or both receptors (Tlr2 –/– and Tlr4 –/–) were used as a model. The absence of both receptors impaired vesicle antigen recognition and the subsequent conversion of immature DCs into a semi‐mature state, which correlated with reduced expression of MHC‐II and TNF‐α. The authors conclude that the presence of TLR‐2 and TLR‐4‐ligands in B. vulgatus mpk BEVs could facilitate induction of tolerance in DCs by desensitizing the host to subsequent stimulation with endotoxin. However, the nature of the TLR2 ligand included in BEVs from this commensal is still unknown (Maerz et al., 2018).
Escherichia coli . As mentioned, the probiotic EcN has many beneficial effects that contribute to intestinal homeostasis and microbiota balance. Many studies have been conducted to elucidate the mechanisms and molecular factors that mediate the immunomodulatory and barrier strengthening properties of this probiotic. In the preceding sections, we reviewed the mechanisms activated by EcN BEVs that confer protection against IBD and highlighted their role as regulators of intestinal barrier integrity and immune responses in IECs. Here, we describe current knowledge on the immunomodulatory effects elicited by EcN and other gut resident E. coli on DCs and derived immune responses.
Our group showed that BEVs from EcN and commensal E. coli strains activate DCs to promote differentiation of T cell subsets in a strain‐specific manner. Immature DCs were derived from CD14+ monocytes isolated from buffy coats collected from healthy donors and stimulated with BEVs from EcN and ECOR strains of intestinal origin (Diaz‐Garrido et al., 2019). Maturation of DCs was assessed by increased expression of CD83 and reduced expression of CD209 (DC‐SIGN) markers compared to untreated DCs. For all strains, BEVs significantly increased secretion of the cytokines that drive the Th responses: INF‐γ, IL‐12, IL‐18 (Th1), IL‐6, IL‐23, TNF‐α, and IL‐1β (Th17/Th22) and IL‐10 and TGF‐β (Treg). Vesicles from EcN and ECOR63 (both belonging to the phylogenetic group B2) elicited greater secretion of Th1 polarizing cytokines than BEVs from ECOR12 (phylogenetic group A) or the cytotoxic ECOR53 strain. Conversely, ECOR12 BEVs induced high levels of Treg‐related cytokines (Diaz‐Garrido et al., 2019). The ability of activated‐DCs to differentially modulate activation of CD4+ T cells towards specific effector subsets was evaluated in the in vitro co‐culture DCs/CD4+ T cell model. Cytokine quantification and cytometry analyses of specific T cell markers evidenced that BEVs from all strains activated the Th2 response with release of IL‐4, IL‐5, and IL‐13. BEVs from ECOR12 elicited a strong Treg response with higher levels of TGF‐β and lower expression of proinflammatory cytokines than BEVs from the other strains. Accordingly, the highest number of CD4+ CD25high+ FOXP3+ T cells were obtained in the presence of DCs stimulated with ECOR12 BEVs, and in a smaller proportion with EcN BEVs (Diaz‐Garrido et al., 2019). This study indicated that in the gut, BEVs released by microbiota strains steadily prime the innate immune system through specific activation of DCs, which in turn orchestrate balanced immune tolerance and immune inflammatory responses that are essential to preserve intestinal homeostasis. Consistent with the beneficial effects of EcN, BEVs from this probiotic drive complex Th responses: Th2 (immunity to extracellular parasites and allergic inflammatory responses), Th22 (tissue protection), Th1 (pro‐inflammatory, protective immunity against pathogens), Th17 (pro‐inflam matory, antimicrobial response) and Treg (immune tolerance). Other commensal B2 E. coli strains trigger similar Th responses but with a stronger pro‐inflammatory profile. In contrast, ECOR12 BEVs mainly promote host tolerance against gut microbiota by increasing the Treg/Th17 balance and they protect gut integrity by activating the Th22 response. However, they fail to trigger proper Th1 responses against pathogens (Diaz‐Garrido et al., 2019).
The regulatory mechanisms exploited by microbiota‐derived BEVs to specifically activate DCs to prime appropriate T cell responses are complex and not fully elucidated. Several molecules linked to the signalling pathways activated by BEVs are involved in modulating cytokine and co‐stimulatory molecules that lead to DCs with different activation profiles. In this context, microRNAs (miRNAs) have a relevant role as post‐transcriptional regulators of the immune system that allow fine‐tuning of signalling pathways. We have recently reported that differential immunomodulatory effects mediated by BEVs from probiotic and commensal E.coli in DCs are in part mediated through regulation of miRNAs (Díaz‐Garrido et al., 2020). In this study, deep RNA sequencing was approached to identify miRNAs that are differentially expressed in DCs challenged with BEVs from EcN or ECOR12. This study revealed a common set of miRNAs modulated by BEVs from both strains. In addition, BEVs elicited differential expression of specific miRNAs depending on the producer strain (21 downregulated and 21 upregulated by EcN BEVs; 45 downregulated and 41 upregulated by ECOR12 BEVs). The differential expression of the miRNAs and their targets was consistent with the effects triggered by these BEVs on DCs. These effects are compatible with the ability of ECOR12 BEVs to drive DCs that orchestrate T regulatory responses that are critical for immune tolerance in the gut, and why EcN BEVs programme DCs to mount the Th1 responses required for pathogen eradication (Díaz‐Garrido et al., 2020). Hence, differential regulation of miRNAs by microbiota BEVs exerts immune training to preserve intestinal homeostasis.
Gram‐positive beneficial bacteria: Lactobacillus sp and Bifidobacterium sp . Most microbes documented to date as probiotics are Gram‐positive bacteria. Among them, the genera Lactobacillus and Bifidobacterium are the most widely used and studied due to their health‐promoting and immunomodulatory effects (Grimm et al., 2014; Wells, 2011). The effects of live probiotic suspensions on the modulation of immune responses have been extensively studied. However, few studies have been focused on their BEVs.
The immunomodulatory effects of BEVs from Lactobacillus rhamnosus JB‐1 have been studied in mice models. Besides immunomodulatory effects, this probiotic exerts functional effects on peristalsis by activating the host enteric nervous system. Oral administered BEVs from L. rhamnosus JB‐1 were shown to cross the epithelial layer and reach DCs located at Peyer's patches at early times post‐treatment. After three days, the phenotype of DCs changed to a regulatory profile with increased levels of IL‐10 production and polarization towards functional regulatory T cells CD4+ CD25+ FOXP3+ (Al‐Nedawi et al., 2015). This study evidenced that L. rhamnosus JB‐1‐derived BEVs reproduced the same immunomodulatory and neuron‐stimulating effects as the live probiotic. Moreover, ex vivo experiments revealed that modulation of the neuronal response depended on signals released by the gut epithelium in response to BEVs. These results confirm the key role of IECs as mediators of indirect communication between microbiota and cells of the intestinal submucosa (Al‐Nedawi et al., 2015).
Secretion of IgA by gut mucosal tissue is known to play an important role in modulating microbiota composition and preventing invasion of enteric pathogens. Production of IgA by gut immune cells is activated by probiotic Lactobacillus strains such as L. sakei subsp. sakei NBRC15893. Experiments performed in cells isolated from murine Peyer's patches proved that the probiotic factor that mediates this effect was associated with purified BEVs (Yamasaki‐Yashiki et al., 2019). Thus, by enhancing IgA expression, BEVs also serve as a mechanism for gut beneficial bacteria to reinforce the intestinal barrier and modulate gut microbiome composition.
There is evidence that in addition to protective properties against colitis, probiotics of the genus Bifidobacterium can alleviate allergic responses. Activation of DCs by strains of the genus Bifidobacterium and the subsequent polarization of naïve CD4+ T cells into effector responses differs between strains. Specifically, the probiotic Bifidobacterium bifidum LMG13195 triggers a tolerogenic response (López et al., 2011). Analysis of several probiotic subcellular fractions on DCs proved that the effects were specifically mediated by BEVs. Similar to live Bifidobacterium bifidum LMG13195, BEVs activated DCs to promote differentiation of Treg cells (FOXP3+) and induce higher levels of IL‐10 than Th1/Th17 proinflammatory cytokines. Based on the suppressor effects, the authors suggested a potential application of BEVs from this probiotic as adjuvants for allergen‐specific immunotherapy (Lopez et al., 2012).
Food allergy results from an abnormal immune response to food components, and some probiotics can alleviate symptoms and associated inflammation. In this context, the healing potential of Bifidobacterium longum was assessed in a mouse model of allergen‐induced food allergy with acute diarrhoea (Kim et al., 2016). This probiotic ameliorated inflammation by decreasing the number of mast cells in the small intestine, without counteracting the Th2 response. Authors showed that this activity was mediated by B. longum‐derived BEVs, and specifically by the vesicular protein ESBP (family 5 extracellular solute‐binding protein), which triggered apoptosis of bone marrow‐derived mast cells (Kim et al., 2016).
5. TRANSMIGRATION OF BEVS THROUGH THE GUT EPITHELIUM
There is considerable evidence that gut microbiota‐derived compounds are found in the bloodstream and can be distributed through the body, reaching distal organs and tissues (Arentsen et al., 2017; Cani et al., 2007; Zhan et al., 2016). BEVs are one of the factors released by resident gut microbiota. These vesicles can cross the epithelial barrier to gain access to the underlying submucosa and interact with immune cells such as DCs and macrophages (Kaparakis‐Liaskos & Ferrero, 2015). Besides, microbiota BEVs can be disseminated through the body by systemic or lymphatic circulation, reaching extraintestinal tissues (Stentz et al., 2018). The question of how these gut microbiota‐derived BEVs can reach distant organs remains poorly explored. However, this is now an emerging research area in the field of the gut microbiome and its impact on health and disease.
The main theory about how BEVs reach systemic circulation is based on the leaky gut condition. Many factors that disturb the intestinal environment trigger gut microbiota imbalance. This condition, known as gut dysbiosis, leads to epithelial barrier disruption and a subsequent increase in paracellular permeability that allows translocation of BEVs into the bloodstream (Figure 5) (Chakaroun et al., 2020). Tulkens et al. investigated the presence of BEVs from gut microbes in the systemic circulation of patients with intestinal barrier dysfunction (Tulkens, Vergauwen, et al., 2020). They analysed plasma samples from healthy donors without barrier dysfunction and patients with well‐defined intestinal barrier dysfunction (diagnosed with IBD, HIV and intestinal mucositis). After plasma fractionation, functional assays were performed to quantify BEV‐associated LPS. The results showed increased levels of LPS‐positive BEVs in plasma from patients suffering IBD, HIV and intestinal mucositis. Patients also presented higher plasma levels of zonulin, a marker of impaired barrier integrity. This protein causes TJ disassembly by phosphorylating ZO proteins (Tulkens, Vergauwen, et al., 2020). In this study, the paracellular translocation of BEVs was also confirmed in the in vitro model of Caco‐2 cells challenged with DSS, which mimics barrier disruption. The same group developed an exhaustive protocol to separate BEVs from eukaryotic‐derived EVs in stool and blood samples consisting of orthogonal separation that includes size‐exclusion chromatography and density gradient‐centrifugation. Specific features of BEVs, such as their ultrastructure, proteomic content, and presence of MAMPs were analysed in the isolated samples. This purification procedure preserves the integrity of BEVs and is suitable for further proteomic and biochemical characterization. However, specific settings should be considered to standardize protocols for isolation and handling according to each body fluid. Therefore, its application would be of great help to study BEVs in body fluids and their use as diagnostic tools (Tulkens, De Wever, et al., 2020). The existence of circulating BEVs can explain the presence of bacterial genetic material in body fluids. Several studies have reported the presence of genomic DNA in blood samples from healthy donors (Païssé et al., 2016; Whittle et al., 2019). It is now accepted that blood is not a sterile fluid, and there is a human blood microbiome that is altered under pathological conditions (Castillo et al., 2019). In fact, analysis of the blood microbiome has been suggested as a potential method for cancer diagnosis (Poore et al., 2020). Nevertheless, when sequence‐based techniques are used to study the microbiome in samples with low biomass, it is essential to avoid contamination during sample processing (Salter et al., 2014).
FIGURE 5.

Mechanisms that mediate translocation of microbiota‐derived BEVs across the intestinal epithelium. The processes are numbered and indicated in red. BEVs released by gut microbes are taken up by intestinal epithelial cells (IECs) via endocytosis (1). Internalized BEVs can reach the basolateral membrane of IECs and be translocated into the lamina propria by transcytosis (2). In addition, the paracellular route (3) also allows passage of luminal BEVs to the underlying submucosa. This route is favoured under conditions of gut dysbiosis. Once in the lamina propria, translocated BEVs directly interact with resident gut immune cells, triggering suitable immune responses. Direct sampling and phagocytosis of luminal BEVs by DCs also mediate passage of luminal BEVs into the internal milieu (4). There is evidence that BEVs cross endothelial barriers and reach blood vessels, thus being distributed to distal organs
The idea that translocation of BEVs occurs only and exclusively when barrier disruption exists has been rejected. Recent evidence indicates that BEVs from the commensal bacterium B. thetaiotaomicron migrate through the intestinal epithelium via a paracellular route and enter the circulatory or lymphatic system in healthy conditions (Jones et al., 2020). Following oral administration of B. thetaiotaomicron EVs, in vivo imaging proved that small amounts of BEVs reach distal organs such as the liver, where vesicle passage could be through the portal vein. BEVs were also found in lungs and heart, which suggests that they can cross several cellular barriers, enter the bloodstream, and spread through the body (Jones et al., 2020). The way BEVs cross healthy epithelial barriers to reach the bloodstream remains unknown. However, the most plausible route involves active transcellular transport mediated by either active transporters or endocytosis‐transcytosis‐exocytosis processes (Figure 5). In fact, there is evidence that eukaryotic EVs called exosomes can enter the bloodstream and migrate through the blood‐brain barrier by transcellular transport (Chen et al., 2016). Likewise, tumour‐derived exosomes can cross the blood‐brain barrier during the course of brain metastasis via transcytosis (Morad et al., 2019). Thus, it can be speculated that a similar process occurs with BEVs. In addition, other mechanisms that involve phagocytic cells of the intestinal mucosa facilitate the entry of gut BEVs into the circulation. Specifically, intestinal DCs are specialized in sampling and transporting luminal antigens and BEVs across the gut epithelium (Figure 5). The phagocytic epithelial M cells also contribute to this function. Once activated, DCs from the mucosal surfaces migrate to lymphoid organs to induce coordinated effector T cell responses. From these sites, DCs can spread through distal locations.
There is now a large body of evidence that BEVs cross cellular barriers and reach every tissue in the body (Figure 5). As an example, BEVs from gut‐associated bacteria have been identified in the human milk of healthy lactating mothers (Kim & Yi, 2020). Microbiota BEVs can also reach the brain. This finding could explain the presence of bacterial DNA in the brain, despite the blood‐brain barrier's strict permeability (Zhan et al., 2016). Recent studies focused on the microbiota‐brain axis indicate that BEVs are involved in several brain disorders such as depression, anxiety, stress, autism and Alzheimer's (Haas‐Neill & Forsythe, 2020). In an Alzheimer's disease mouse model, 16S rRNA metagenomic analyses of BEVs in blood allowed identification of more than 3000 taxonomic units that correlated with the gut microbiota profile. Interestingly, important alterations in certain taxonomic phyla were found in mutant mice with an Alzheimer disease phenotype compared with control wild type mice (Park et al., 2017). Based on significant statistical variations between experimental groups, the authors suggested that metagenomic analyses of blood BEVs could be used to identify gut microbiota imbalance associated with specific diseases (Park et al., 2017). Similarly, in humans with autism disorder, great differences in the genomic profile of BEVs isolated from urine samples have been found compared to healthy controls. These differences also reflected the changes in gut microbiome previously described in autism, thus supporting the potential use of BEVs in body fluids for diagnostic purposes (Lee & Kim, 2017).
6. CONCLUDING REMARKS
The relevance of microbiota research is emphasised by the large number of publications, specific meetings and forums dedicated to this topic. Despite intense research in this area, there is still much to learn about the intrinsic mechanisms of microbiota action, especially those concerning the delivery of effectors to target cells. This knowledge is essential to understanding disease and developing translation strategies of microbiota or microbiota‐derived products to human health. In this context, the study of microbiota BEVs is currently receiving growing interest. Evidence from preclinical assays in animal models point to potential applications of microbiota BEVs as therapeutic/nutritional strategies to prevent intestinal infections or inflammatory and immune‐related disorders. Administration of isolated BEVs would overcome the potential risk of the use of live probiotics in immunocompromised individuals.
Imbalances in gut microbiota composition and diversity have been associated with a great variety of diseases including cancer, neurological, metabolic and immune disorders. In this context, gut microbiome profiling is envisaged as a potentially useful tool for diagnosis. The discovery of gut microbiota‐derived BEVs in blood and urine samples of patients and healthy individuals with a profile that highly correlates with the gut microbiome opens novel strategies for diagnostic purposes.
The study of microbiota‐derived EVs is quickly evolving. In the near future, it is expected that research efforts in this area will contribute to understanding the complex microbiota‐host communication network, which is essential to preserve human health, and provide solid basis for propelling new therapeutic strategies based on EVs from gut beneficial bacteria as postbiotics.
FUNDING
This work was supported by Ministerio de Ciencia e Innovación, Gobierno de España, under grant PID2019‐107327RB‐100.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Conceptualization, J.B and L.B; investigation, N.D.G, writing—original draft preparation, N.D.G.; writing—review and editing, L.B and J.B; supervision L.B; project administration, J.B; funding acquisition, L.B and J.B.
ACKNOWLEDGEMENTS
We acknowledge Lucille Banham for assistance in preparing the English manuscript. N. Diaz‐Garrido acknowledges her doctoral fellowship from Agencia Nacional de Investigación y Desarrollo (ANID) / Scholarship Program/DOCTORADO BECAS CHILE/2017—72180379.
Díaz‐Garrido, N. , Badia, J. , & Baldomà, L. (2021). Microbiota‐derived extracellular vesicles in interkingdom communication in the gut. Journal of Extracellular Vesicles, 10, e12161. 10.1002/jev2.12161
Josefa Badia and Laura Baldomà authors contributed equally to the supervision of the study
REFERENCES
- Aguilera, L. , Toloza, L. , Gimenez, R. , Odena, A. , Oliveira, E. , Aguilar, J. , Badia, J. , & Baldoma, L. (2014). Proteomic analysis of outer membrane vesicles from the probiotic strain Escherichia coli Nissle 1917. Proteomics, 14(2–3), 222–229. 10.1002/pmic.201300328 [DOI] [PubMed] [Google Scholar]
- Ahmadi Badi, S. , Moshiri, A. , Ettehad Marvasti, F., Mojtahedzadeh, M. , Kazemi, V. , & Siadat, S. D. (2020). Extraction and evaluation of outer membrane vesicles from two important gut microbiota members, Bacteroides fragilis and Bacteroides thetaiotaomicron . Cell Journal, 22(3), 344–349. 10.22074/cellj.2020.6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Nedawi, K. , Mian, M. F. , Hossain, N. , Karimi, K. , Mao, Y.‐K. , Forsythe, P. , Min, K. K. , Stanisz, A. M. , Kunze, W. A. , & Bienenstock, J. (2015). Gut commensal microvesicles reproduce parent bacterial signals to host immune and enteric nervous systems. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 29(2), 684–695. 10.1096/fj.14-259721 [DOI] [PubMed] [Google Scholar]
- Altindis, E. , Fu, Y. , & Mekalanos, J. J. (2014). Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proceedings of the National Academy of Sciences of the United States of America, 111(15), E1548‐56. 10.1073/pnas.1403683111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, C.‐S. , Giménez, R. , Cañas, M.‐A. , Vera, R. , Díaz‐Garrido, N. , Badia, J. , & Baldomà, L. (2019). Extracellular vesicles and soluble factors secreted by Escherichia coli Nissle 1917 and ECOR63 protect against enteropathogenic E. coli‐induced intestinal epithelial barrier dysfunction. BMC Microbiology, 19(1). 10.1186/s12866-019-1534-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, C. S. , Badia, J. , Bosch, M. , Gimenez, R. , & Baldoma, L. (2016). Outer membrane vesicles and soluble factors released by probiotic Escherichia coli Nissle 1917 and commensal ECOR63 enhance barrier function by regulating expression of tight junction proteins in intestinal epithelial cells. Frontiers in Microbiology, 7, 1981. 10.3389/fmicb.2016.01981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, D. , & Chaudhuri, A. (2016). Bacterial outer membrane vesicles: New insights and applications. Molecular Membrane Biology, 33(6–8), 125–137. 10.1080/09687688.2017.1400602 [DOI] [PubMed] [Google Scholar]
- Andreoni, F. , Toyofuku, M. , Menzi, C. , Kalawong, R. , Mairpady Shambat, S. , François, P. , Zinkernagel, A. S. , & Eberl, L. (2019). Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage‐dependent and ‐independent fashions and via different routes. Antimicrobial Agents and Chemotherapy, 63(2). 10.1128/aac.01439-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentsen, T. , Qian, Y. , Gkotzis, S. , Femenia, T. , Wang, T. , Udekwu, K. , Forssberg, H. , & Diaz Heijtz, R. (2017). The bacterial peptidoglycan‐sensing molecule Pglyrp2 modulates brain development and behavior. Molecular Psychiatry, 22(2), 257–266. 10.1038/mp.2016.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafian, F. , Behrouzi, A. , Shahriary, A. , Ahmadi Badi, S. , Davari, M. , Khatami, S. , Rahimi Jamnani, F. , Fateh, A. , Vaziri, F. , & Siadat, S. D. (2019). Comparative study of effect of Akkermansia muciniphila and its extracellular vesicles on toll‐like receptors and tight junction. Gastroenterology and Hepatology from Bed to Bench, 12(2), 163–168. [PMC free article] [PubMed] [Google Scholar]
- Ashrafian, F. , Shahriary, A. , Behrouzi, A. , Moradi, H. R. , Keshavarz Azizi Raftar, S. , Lari, A. , Hadifar, S. , Yaghoubfar, R. , Ahmadi Badi, S. , Khatami, S. , Vaziri, F. , & Siadat, S. D. (2019). Akkermansia muciniphila‐derived extracellular vesicles as a mucosal delivery vector for amelioration of obesity in mice. Frontiers in microbiology, 10, 2155. 10.3389/fmicb.2019.02155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila‐Calderón, E. D. , Araiza‐Villanueva, M. G. , Cancino‐Diaz, J. C. , López‐Villegas, E. O. , Sriranganathan, N. , Boyle, S. M. , & Contreras‐Rodríguez, A. (2015). Roles of bacterial membrane vesicles. Archives of Microbiology, 197(1), 1–10. 10.1007/s00203-014-1042-7 [DOI] [PubMed] [Google Scholar]
- Badia, J. , & Baldomà, L. (2020). Membrane vesicles from the gut microbiota and their interactions with the host. Bacterial membrane vesicles: Biogenesis, functions and applications (Kaparakis‐Liaskos M. & Kufer T. A. (eds.); pp. 189–217). Springer International Publishing. 10.1007/978-3-030-36331-4_9 [DOI] [Google Scholar]
- Bajic, S. S. , Cañas, M.‐A. , Tolinacki, M. , Badia, J. , Sánchez, B. , Golic, N. , Margolles, A. , Baldomá, L. , & Ruas‐Madiedo, P. (2020). Proteomic profile of extracellular vesicles released by Lactiplantibacillus plantarum BGAN8 and their internalization by non‐polarized HT29 cell line. Scientific Reports, 10(1), 21829. 10.1038/s41598-020-78920-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barák, I. , & Muchová, K. (2013). The role of lipid domains in bacterial cell processes. International Journal of Molecular Sciences, 14(2), 4050–4065. 10.3390/ijms14024050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi, E. , Mahmoodzadeh Hosseini, H. , & Imani Fooladi, A. A. (2017). The inhibitory impacts of Lactobacillus rhamnosus GG‐derived extracellular vesicles on the growth of hepatic cancer cells. Microbial Pathogenesis, 110, 1–6. 10.1016/j.micpath.2017.06.016 [DOI] [PubMed] [Google Scholar]
- Benjdia, A. , Martens, E. C. , Gordon, J. I. , & Berteau, O. (2011). Sulfatases and a radical S‐adenosyl‐L‐methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron . Journal of Biological Chemistry, 286(29), 25973–25982. 10.1074/jbc.M111.228841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez‐Brito, M. , Muñoz‐Quezada, S. , Gomez‐Llorente, C. , Matencio, E. , Bernal, M. J. , Romero, F. , & Gil, A. (2012). Human intestinal dendritic cells decrease cytokine release against Salmonella infection in the presence of Lactobacillus paracasei upon TLR activation. Plos One, 7(8), e43197. e4197. 10.1371/journal.pone.0043197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez‐Brito, M. , Muñoz‐Quezada, S. , Gomez‐Llorente, C. , Romero, F. , & Gil, A. (2014). Lactobacillus rhamnosus and its cell‐free culture supernatant differentially modulate inflammatory biomarkers in Escherichia coli‐challenged human dendritic cells. British Journal of Nutrition, 111(10), 1727–1737. 10.1017/s0007114513004303 [DOI] [PubMed] [Google Scholar]
- Bermudez‐Brito, M. , Plaza‐Díaz, J. , Fontana, L. , Muñoz‐Quezada, S. , & Gil, A. (2013). In vitro cell and tissue models for studying host‐microbe interactions: A review. British Journal of Nutrition, 109 Suppl, S27‐S34. 10.1017/s0007114512004023 [DOI] [PubMed] [Google Scholar]
- Bian, X. , Wu, W. , Yang, L. , Lv, L. , Wang, Q. , Li, Y. , Ye, J. , Fang, D. , Wu, J. , Jiang, X. , Shi, D. , & Li, L. (2019). Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium‐induced ulcerative colitis in mice. Frontiers in microbiology, 10. 10.3389/fmicb.2019.02259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto, N. J. , & Kaparakis‐Liaskos, M. (2017). The therapeutic benefit of bacterial membrane vesicles. International Journal of Molecular Sciences, 18(6). 10.3390/ijms18061287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington, K. E. , & Kuehn, M. J. (2014). Protein selection and export via outer membrane vesicles. Biochimica et Biophysica Acta, 1843(8), 1612–1619. 10.1016/j.bbamcr.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameyer, S. , Plener, L. , Müller, A. , Klingl, A. , Wanner, G. , & Jung, K. (2018). Outer membrane vesicles facilitate trafficking of the hydrophobic signaling molecule CAI‐1 between Vibrio harveyi cells. Journal of Bacteriology, 200(15). 10.1128/JB.00740-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, L. , Kessler, A. , Cabezas‐Sanchez, P. , Luque‐Garcia, J. L. , & Casadevall, A. (2014). Extracellular vesicles produced by the Gram‐positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Molecular Microbiology, 93(1), 183–198. 10.1111/mmi.12650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, L. , Wolf, J. M. , Prados‐Rosales, R. , & Casadevall, A. (2015). Through the wall: Extracellular vesicles in Gram‐positive bacteria, mycobacteria and fungi. Nature Reviews Microbiology, 13(10), 620–630. 10.1038/nrmicro3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, W. A. , Stentz, R. , Le Gall, G. , Sternberg, M. J. E. , Carding, S. R. , & Wilhelm, T. (2017). In silico analysis of the small molecule content of outer membrane vesicles produced by Bacteroides thetaiotaomicron indicates an extensive metabolic link between microbe and host. Frontiers in microbiology, 8, 2440. 10.3389/fmicb.2017.02440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, W. , Kesavan, D. K. , Wan, J. , Abdelaziz, M. H. , Su, Z. , & Xu, H. (2018). Bacterial outer membrane vesicles, a potential vaccine candidate in interactions with host cells based. Diagnostic pathology, 13(1), 95. 10.1186/s13000-018-0768-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas, M. A. , Fabrega, M. J. , Gimenez, R. , Badia, J. , & Baldoma, L. (2018). Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1‐mediated immune responses in intestinal epithelial cells. Frontiers in microbiology, 9, 498. 10.3389/fmicb.2018.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas, M. A. , Gimenez, R. , Fabrega, M. J. , Toloza, L. , Baldoma, L. , & Badia, J. (2016). Outer membrane vesicles from the probiotic Escherichia coli Nissle 1917 and the commensal ECOR12 enter intestinal epithelial cells via clathrin‐dependent endocytosis and elicit differential effects on DNA damage. Plos One, 11(8), e0160374. 10.1371/journal.pone.0160374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani, P. D. , Amar, J. , Iglesias, M. A. , Poggi, M. , Knauf, C. , Bastelica, D. , Neyrinck, A. M. , Fava, F. , Tuohy, K. M. , Chabo, C. , Waget, A. , Delmée, E. , Cousin, B. , Sulpice, T. , Chamontin, B. , Ferrières, J. , Tanti, J.‐F. , Gibson, G. R. , Casteilla, L. , … Burcelin, R. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes, 56(7), 1761–1772. 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- Caruana, J. C. , & Walper, S. A. (2020). Bacterial membrane vesicles as mediators of microbe‐microbe and microbe‐host community interactions. Frontiers in Microbiology, 11. 10.3389/fmicb.2020.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, R. , Warner, N. , Inohara, N. , & Núñez, G. (2014). NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity, 41(6), 898–908. 10.1016/j.immuni.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo, D. J. , Rifkin, R. F. , Cowan, D. A. , & Potgieter, M. (2019). The healthy human blood microbiome: fact or fiction? Frontiers in Cellular and Infection Microbiology, 9, 148. 10.3389/fcimb.2019.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celluzzi, A. , & Masotti, A. (2016). How our other genome controls our epi‐genome. Trends in Microbiology, 24(10), 777–787. 10.1016/j.tim.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Chakaroun, R. M. , Massier, L. , & Kovacs, P. (2020). Gut microbiome, intestinal permeability, and tissue bacteria in metabolic disease: Perpetrators or bystanders? Nutrients, 12(4). 10.3390/nu12041082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard, M. , Hashimoto, M. , Horie, Y. , Masumoto, J. , Qiu, S. , Saab, L. , Ogura, Y. , Kawasaki, A. , Fukase, K. , Kusumoto, S. , Valvano, M. A. , Foster, S. J. , Mak, T. W. , Nuñez, G. , & Inohara, N. (2003). An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nature Immunology, 4(7), 702–707. 10.1038/ni945 [DOI] [PubMed] [Google Scholar]
- Chang, Y.‐C. , Ching, Y.‐H. , Chiu, C.‐C. , Liu, J.‐Y. , Hung, S.‐W. , Huang, W.‐C. , Huang, Y.‐T. , & Chuang, H.‐L. (2017). TLR2 and interleukin‐10 are involved in Bacteroides fragilis‐mediated prevention of DSS‐induced colitis in gnotobiotic mice. Plos One, 12(7), e0180025. 10.1371/journal.pone.0180025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, D. , & Chaudhuri, K. (2013). Vibrio cholerae O395 outer membrane vesicles modulate intestinal epithelial cells in a NOD1 protein‐dependent manner and induce dendritic cell‐mediated Th2/Th17 cell responses. Journal of Biological Chemistry, 288(6), 4299–4309. 10.1074/jbc.M112.408302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. , Mondal, A. , Mitra, S. , & Basu, S. (2017). Acinetobacter baumannii transfers the blaNDM‐1 gene via outer membrane vesicles. The Journal of Antimicrobial Chemotherapy, 72(8), 2201–2207. 10.1093/jac/dkx131 [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. N. , & Das, J. (1967). Electron microscopic observations on the excretion of cell‐wall material by Vibrio cholerae . Journal of General Microbiology, 49(1), 1–11. 10.1099/00221287-49-1-1 [DOI] [PubMed] [Google Scholar]
- Chatzidaki‐Livanis, M. , Coyne, M. J. , & Comstock, L. E. (2014). An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Molecular Microbiology, 94(6), 1361–1374. 10.1111/mmi.12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelakkot, C. , Choi, Y. , Kim, D. K. , Park, H. T. , Ghim, J. , Kwon, Y. , Jeon, J. , Kim, M. S. , Jee, Y. K. , Gho, Y. S. , Park, H. S. , Kim, Y. K. , & Ryu, S. H. (2018). Akkermansia muciniphila‐derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Experimental & Molecular Medicine, 50(2), e450. 10.1038/emm.2017.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. C. , Liu, L. , Ma, F. , Wong, C. W. , Guo, X. E. , Chacko, J. V , Farhoodi, H. P. , Zhang, S. X. , Zimak, J. , Ségaliny, A. , Riazifar, M. , Pham, V. , Digman, M. A. , Pone, E. J. , & Zhao, W. (2016). Elucidation of exosome migration across the blood‐brain barrier model in vitro. Cellular and Molecular Bioengineering, 9(4), 509–529. 10.1007/s12195-016-0458-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H. , Guan, X. , Chen, D. , & Ma, W. (2019). The Th17/Treg cell balance: A gut microbiota‐modulated story. Microorganisms, 7(12). 10.3390/microorganisms7120583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, C.‐C. , Ching, Y.‐H. , Wang, Y.‐C. , Liu, J.‐Y. , Li, Y.‐P. , Huang, Y.‐T. , & Chuang, H.‐L. (2014). Monocolonization of germ‐free mice with Bacteroides fragilis protects against dextran sulfate sodium‐induced acute colitis. BioMed Research International, 2014, 675786. 10.1155/2014/675786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D. S. , Kim, D. K. , Choi, S. J. , Lee, J. , Choi, J. P. , Rho, S. , Park, S. H. , Kim, Y. K. , Hwang, D. , & Gho, Y. S. (2011). Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa . Proteomics, 11(16), 3424–3429. 10.1002/pmic.201000212 [DOI] [PubMed] [Google Scholar]
- Choi, J. W. , Um, J. H. , Cho, J. H. , & Lee, H. J. (2017). Tiny RNAs and their voyage via extracellular vesicles: Secretion of bacterial small RNA and eukaryotic microRNA. Experimental Biology and Medicine (Maywood, N.J.), 242(15), 1475–1481. 10.1177/1535370217723166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, C. , & Jagannadham, M. V. (2013). Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochimica Et Biophysica Acta, 1834(1), 231–239. 10.1016/j.bbapap.2012.09.015 [DOI] [PubMed] [Google Scholar]
- Chu, H. , Khosravi, A. , Kusumawardhani, I. P. , Kwon, A. H. , Vasconcelos, A. C. , Cunha, L. D. , Mayer, A. E. , Shen, Y. , Wu, W. L. , Kambal, A. , Targan, S. R. , Xavier, R. J. , Ernst, P. B. , Green, D. R. , McGovern, D. P. , Virgin, H. W. , & Mazmanian, S. K. (2016). Gene‐microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science, 352(6289), 1116–1120. 10.1126/science.aad9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, M. R. , & Mortensen, P. B. (1995). Kinetic studies on colonocyte metabolism of short chain fatty acids and glucose in ulcerative colitis. Gut, 37(5), 684–689. 10.1136/gut.37.5.684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, C. , Brown, L. , Maryam, M. , Vij, R. , Smith, D. F. Q. , Burnet, M. C. , Kyle, J. E. , Heyman, H. M. , Ramirez, J. , Prados‐Rosales, R. , Lauvau, G. , Nakayasu, E. S. , Brady, N. R. , Hamacher‐Brady, A. , Coppens, I. , & Casadevall, A. (2019). Listeria monocytogenes virulence factors, including listeriolysin O, are secreted in biologically active extracellular vesicles. Journal of Biological Chemistry, 294(4), 1202–1217. 10.1074/jbc.RA118.006472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauros‐Singorenko, P. , Blenkiron, C. , Phillips, A. , & Swift, S. (2018). The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiology Letters, 365(5). 10.1093/femsle/fny023 [DOI] [PubMed] [Google Scholar]
- Dauros Singorenko, P. , Chang, V. , Whitcombe, A. , Simonov, D. , Hong, J. , Phillips, A. , Swift, S. , & Blenkiron, C. (2017). Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity. Journal of Extracellular Vesicles, 6(1), 1324731. 10.1080/20013078.2017.1324731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos, W. M. (2017). Microbe profile: Akkermansia muciniphila: a conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology (Reading, England), 163(5), 646–648. 10.1099/mic.0.000444 [DOI] [PubMed] [Google Scholar]
- Dean, S. N. , Leary, D. H. , Sullivan, C. J. , Oh, E. , & Walper, S. A. (2019). Isolation and characterization of Lactobacillus‐derived membrane vesicles. Scientific Reports, 9(1), 877. 10.1038/s41598-018-37120-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, S. N. , Rimmer, M. A. , Turner, K. B. , Phillips, D. A. , Caruana, J. C. , Hervey, W. J. , Leary, D. H. , & Walper, S. A. (2020). Lactobacillus acidophilus membrane vesicles as a vehicle of bacteriocin delivery. Frontiers in Microbiology, 11, 710. https://www.frontiersin.org/article/10.3389/fmicb.2020.00710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage, B. L. , Lara, J. C. , Bergsbaken, T. , Rassoulian Barrett, S. L. , Lara, S. , & Cookson, B. T. (2009). Biogenesis of bacterial membrane vesicles. Molecular Microbiology, 72(6), 1395–1407. 10.1111/j.1365-2958.2009.06731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delday, M. , Mulder, I. , Logan, E. T. , & Grant, G. (2019). Bacteroides thetaiotaomicron ameliorates colon inflammation in preclinical models of crohn's disease. Inflammatory Bowel Diseases, 25(1), 85–96. 10.1093/ibd/izy281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, S. , Van Putte, W. , Vitse, J. , Van Driessche, G. , Stremersch, S. , Van Den Broek, W. , Raemdonck, K. , Braeckmans, K. , Stahlberg, H. , Kudryashev, M. , Savvides, S. N. , & Devreese, B. (2017). Membrane vesicle secretion and prophage induction in multidrug‐resistant Stenotrophomonas maltophilia in response to ciprofloxacin stress. Environmental Microbiology, 19(10), 3930–3937. 10.1111/1462-2920.13793 [DOI] [PubMed] [Google Scholar]
- Díaz‐Garrido, N. , Bonnin, S. , Riera, M. , Gíménez, R. , Badia, J. , & Baldomà, L. (2020). Transcriptomic microRNA profiling of dendritic cells in response to gut microbiota‐secreted vesicles. Cells, 9(6). 10.3390/cells9061534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz‐Garrido, N. , Fábrega, M.‐J. , Vera, R. , Giménez, R. , Badia, J. , & Baldomà, L. (2019). Membrane vesicles from the probiotic Nissle 1917 and gut resident Escherichia coli strains distinctly modulate human dendritic cells and subsequent T cell responses. Journal of Functional Foods, 61. 103495. 10.1016/j.jff.2019.103495 [DOI] [Google Scholar]
- Diaz‐Garrido, N. , Cordero, C. , Olivo‐Martinez, Y. , Badia, J. , & Baldomà, L. (2021). Cell‐to‐cell communication by host‐released extracellular vesicles in the gut: implications in health and disease. International Journal of Molecular Sciences, 22, 2213. 10.3390/ijms22042213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues, S. , & Nielsen, K. M. (2017). Membrane vesicles and horizontal gene transfer in prokaryotes. Current Opinion in Microbiology, 38, 16–21. 10.1016/j.mib.2017.03.012 [DOI] [PubMed] [Google Scholar]
- Domínguez Rubio, A. P. , Martínez, J. H. , Martínez Casillas, D. C. , Coluccio Leskow, F. , Piuri, M ., & Pérez, O. E. (2017). Lactobacillus casei BL23 produces microvesicles carrying proteins that have been associated with its probiotic effect. Frontiers in microbiology, 8, 1783. 10.3389/fmicb.2017.01783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, G. P. , Lee, S. M. , & Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nature Reviews Microbiology, 14(1), 20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant, L. , Stentz, R. , Noble, A. , Brooks, J. , Gicheva, N. , Reddi, D. , O'Connor, M. J. , Hoyles, L. , McCartney, A. L. , Man, R. , Pring, E. T. , Dilke, S. , Hendy, P. , Segal, J. P. , Lim, D. N. F. , Misra, R. , Hart, A. L. , Arebi, N. , Carding, S. R. , & Knight, S. C. (2020). Bacteroides thetaiotaomicron‐derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome, 8(1), 88. 10.1186/s40168-020-00868-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg, P. B. , Bik, E. M. , Bernstein, C. N. , Purdom, E. , Dethlefsen, L. , Sargent, M. , Gill, S. R. , Nelson, K. E. , & Relman, D. A. (2005). Diversity of the human intestinal microbial flora. Science, 308(5728), 1635–1638. 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy, W. , Bording‐Jorgensen, M. , Valguarnera, E. , Haurat, M. F. , Wine, E. , & Feldman, M. F. (2016). LPS remodeling triggers formation of outer membrane vesicles in Salmonella . MBio, 7(4). 10.1128/mBio.00940-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy, W. , Debelyy, M. O. , & Feldman, M. F. (2014). Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. MBio, 5(2), e00909‐14. 10.1128/mBio.00909-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, A. G. L. , Davey, H. M. , Cookson, A. , Currinn, H. , Cooke‐Fox, G. , Stanczyk, P. J. , & Whitworth, D. E. (2012). Predatory activity of Myxococcus xanthus outer‐membrane vesicles and properties of their hydrolase cargo. Microbiology (Reading, England), 158(Pt 11), 2742–2752. 10.1099/mic.0.060343-0 [DOI] [PubMed] [Google Scholar]
- Fabrega, M. J. , Aguilera, L. , Gimenez, R. , Varela, E. , Alexandra Cañas, M. , Antolin, M. , Badia, J ., & Baldoma, L. (2016). Activation of immune and defense responses in the intestinal mucosa by outer membrane vesicles of commensal and probiotic Escherichia coli strains. Frontiers in microbiology, 7, 705. 10.3389/fmicb.2016.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega, M. J. , Rodriguez‐Nogales, A. , Garrido‐Mesa, J. , Algieri, F. , Badia, J. , Gimenez, R. , Galvez, J. , & Baldoma, L. (2017). Intestinal anti‐inflammatory effects of outer membrane vesicles from Escherichia coli Nissle 1917 in DSS‐experimental colitis in mice. Frontiers in microbiology, 8, 1274. 10.3389/fmicb.2017.01274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebiger, U. , Bereswill, S. , & Heimesaat, M. M. (2016). Dissecting the interplay between intestinal microbiota and host immunity in health and disease: Lessons learned from germfree and gnotobiotic animal models. European Journal of Microbiology & Immunology, 6(4), 253–271. 10.1556/1886.2016.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming, H.‐C. , Wingender, J. , Szewzyk, U. , Steinberg, P. , Rice, S. A. , & Kjelleberg, S. (2016). Biofilms: An emergent form of bacterial life. Nature Reviews. Microbiology, 14(9), 563–575. 10.1038/nrmicro.2016.94 [DOI] [PubMed] [Google Scholar]
- Flint, H. J. , Scott, K. P. , Louis, P. , & Duncan, S. H. (2012). The role of the gut microbiota in nutrition and health. Nature Reviews. Gastroenterology & Hepatology, 9(10), 577–589. 10.1038/nrgastro.2012.156 [DOI] [PubMed] [Google Scholar]
- Gill, S. , Catchpole, R. , & Forterre, P. (2019). Extracellular membrane vesicles in the three domains of life and beyond. Fems Microbiology Reviews, 43(3), 273–303. 10.1093/femsre/fuy042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, W. J. , Johnston, E. L. , Zavan, L. , Bitto, N. J. , & Kaparakis‐Liaskos, M. (2021). Immunomodulatory roles and novel applications of bacterial membrane vesicles. Molecular Immunology, 134, 72–85. 10.1016/j.molimm.2021.02.027 [DOI] [PubMed] [Google Scholar]
- Girardin, S. E. , Boneca, I. G. , Viala, J. , Chamaillard, M. , Labigne, A. , Thomas, G. , Philpott, D. J. , & Sansonetti, P. J. (2003). Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. The Journal of Biological Chemistry, 278(11), 8869–8872. 10.1074/jbc.C200651200 [DOI] [PubMed] [Google Scholar]
- Gobert, A. P. , Sagrestani, G. , Delmas, E. , Wilson, K. T. , Verriere, T. G. , Dapoigny, M. , Del'homme, C. , & Bernalier‐Donadille, A. (2016). The human intestinal microbiota of constipated‐predominant irritable bowel syndrome patients exhibits anti‐inflammatory properties. Scientific Reports, 6, 39399. 10.1038/srep39399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan, V. , Helmink, B. A. , Spencer, C. N. , Reuben, A. , & Wargo, J. A. (2018). The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell, 33(4), 570–580. 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, V. , Westermann, C. , & Riedel, C. U. (2014). Bifidobacteria‐host interactions—An update on colonisation factors. BioMed Research International, 2014, 960826. 10.1155/2014/960826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas‐Neill, S. , & Forsythe, P. (2020). A budding relationship: Bacterial extracellular vesicles in the microbiota‐gut‐brain axis. International Journal of Molecular Sciences, 21(23). 10.3390/ijms21238899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurat, M. F. , Aduse‐Opoku, J. , Rangarajan, M. , Dorobantu, L. , Gray, M. R. , Curtis, M. A. , & Feldman, M. F. (2011). Selective sorting of cargo proteins into bacterial membrane vesicles. Journal of Biological Chemistry, 286(2), 1269–1276. 10.1074/jbc.M110.185744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurat, M. F. , Elhenawy, W. , & Feldman, M. F. (2015). Prokaryotic membrane vesicles: New insights on biogenesis and biological roles. Biological Chemistry, 396(2), 95–109. 10.1515/hsz-2014-0183 [DOI] [PubMed] [Google Scholar]
- Henderson, J. C. , Zimmerman, S. M. , Crofts, A. A. , Boll, J. M. , Kuhns, L. G. , Herrera, C. M. , & Trent, M. S. (2016). The power of asymmetry: Architecture and assembly of the gram‐negative outer membrane lipid bilayer. Annual Review of Microbiology, 70, 255–278. 10.1146/annurev-micro-102215-095308 [DOI] [PubMed] [Google Scholar]
- Hering, N. A. , Richter, J. F. , Fromm, A. , Wieser, A. , Hartmann, S. , Günzel, D. , Bücker, R. , Fromm, M. , Schulzke, J. D. , & Troeger, H. (2014). TcpC protein from E. coli Nissle improves epithelial barrier function involving PKCζ and ERK1/2 signaling in HT‐29/B6 cells. Mucosal Immunology, 7(2), 369–378. 10.1038/mi.2013.55 [DOI] [PubMed] [Google Scholar]
- Hickey, C. A. , Kuhn, K. A. , Donermeyer, D. L. , Porter, N. T. , Jin, C. , Cameron, E. A. , Jung, H. , Kaiko, G. E. , Wegorzewska, M. , Malvin, N. P. , Glowacki, R. W. , Hansson, G. C. , Allen, P. M. , Martens, E. C. , & Stappenbeck, T. S. (2015). Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase‐dependent manner via outer membrane vesicles. Cell Host Microbe, 17(5), 672–680. 10.1016/j.chom.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiippala, K. , Jouhten, H. , Ronkainen, A. , Hartikainen, A. , Kainulainen, V. , Jalanka, J. , & Satokari, R. (2018). The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients, 10(8). 10.3390/nu10080988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra, D. , van der Laan, J. W. , de Leij, L. , & Witholt, B. (1976). Release of outer membrane fragments from normally growing Escherichia coli . Biochimica Et Biophysica Acta, 455(3), 889–899. 10.1016/0005-2736(76)90058-4 [DOI] [PubMed] [Google Scholar]
- Hong, J. , Dauros‐Singorenko, P. , Whitcombe, A. , Payne, L. , Blenkiron, C. , Phillips, A. , & Swift, S. (2019). Analysis of the Escherichia coli extracellular vesicle proteome identifies markers of purity and culture conditions. Journal of extracellular vesicles, 8(1), 1632099. 10.1080/20013078.2019.1632099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, L. V , Littman, D. R. , & Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science, 336(6086), 1268–1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman, A. L. , & Kuehn, M. J. (2000). Enterotoxigenic Escherichia coli secretes active heat‐labile enterotoxin via outer membrane vesicles. Journal of Biological Chemistry, 275(17), 12489–12496. 10.1074/jbc.275.17.12489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. Y. , Lee, S. M. , & Mazmanian, S. K. (2011). The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe, 17(4), 137–141. 10.1016/j.anaerobe.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving, A. T. , Mimuro, H. , Kufer, T. A. , Lo, C. , Wheeler, R. , Turner, L. J. , Thomas, B. J. , Malosse, C. , Gantier, M. P. , Casillas, L. N. , Votta, B. J. , Bertin, J. , Boneca, I. G. , Sasakawa, C. , Philpott, D. J. , Ferrero, R. L. , & Kaparakis‐Liaskos, M. (2014). The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host & Microbe, 15(5), 623–635. 10.1016/j.chom.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Jan, A. T. (2017). Outer Membrane Vesicles (OMVs) of gram‐negative bacteria: A perspective update. Frontiers in microbiology, 8, 1053. 10.3389/fmicb.2017.01053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala, S. M. , Talukdar, R. , Subramanyam, C. , Vuyyuru, H. , Sasikala, M. , & Reddy, D. N. (2015). Role of the normal gut microbiota. World Journal of Gastroenterology, 21(29), 8787–8803. 10.3748/wjg.v21.i29.8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, K. S. , Sweredoski, M. J. , Graham, R. L. , Hess, S. , & Clemons Jr., W. M. (2014). Comprehensive proteomic profiling of outer membrane vesicles from Campylobacter jejuni . Journal of Proteomics, 98, 90–98. 10.1016/j.jprot.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery, C. J. (2018). Protein moonlighting: What is it, and why is it important? Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 373(1738). 10.1098/rstb.2016.0523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, H. , Oh, M. H. , Jun, S. H. , Kim, S. I. , Choi, C. W. , Kwon, H. I. , Na, S. H. , Kim, Y. J. , Nicholas, A. , Selasi, G. N. , & Lee, J. C. (2016). Variation among Staphylococcus aureus membrane vesicle proteomes affects cytotoxicity of host cells. Microbial Pathogenesis, 93, 185–193. 10.1016/j.micpath.2016.02.014 [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Kong, Q. , Roland, K. L. , & Curtiss, R. (2014). Membrane vesicles of Clostridium perfringens Type A strains induce innate and adaptive immunity. International Journal of Medical Microbiology, 304(0), 431–443. 10.1016/j.ijmm.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E. J. , Booth, C. , Fonseca, S. , Parker, A. , Cross, K. , Miquel‐Clopés, A. , Hautefort, I. , Mayer, U. , Wileman, T. , Stentz, R. , & Carding, S. R. (2020). The uptake, trafficking, and biodistribution of Bacteroides thetaiotaomicron generated outer membrane vesicles. Frontiers in microbiology, 11, 57. 10.3389/fmicb.2020.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa, J. L. , & Beveridge, T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: A novel mechanism of enzyme secretion. Journal of Bacteriology, 177(14), 3998–4008. 10.1128/jb.177.14.3998-4008.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, C. S. , Ban, M. , Choi, E. J. , Moon, H. G. , Jeon, J. S. , Kim, D. K. , Park, S. K. , Jeon, S. G. , Roh, T. Y. , Myung, S. J. , Gho, Y. S. , Kim, J. G. , & Kim, Y. K. (2013). Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium‐induced colitis. Plos One, 8(10), e76520. 10.1371/journal.pone.0076520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaparakis‐Liaskos, M , & Ferrero, R. L. (2015). Immune modulation by bacterial outer membrane vesicles. Nature Reviews Immunology, 15(6), 375–387. 10.1038/nri3837 [DOI] [PubMed] [Google Scholar]
- Kaparakis, M. , Turnbull, L. , Carneiro, L. , Firth, S. , Coleman, H. A. , Parkington, H. C. , Le Bourhis, L. , Karrar, A. , Viala, J. , Mak, J. , Hutton, M. L. , Davies, J. K. , Crack, P. J. , Hertzog, P. J. , Philpott, D. J. , Girardin, S. E. , Whitchurch, C. B. , & Ferrero, R. L. (2010). Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cellular Microbiology, 12(3), 372–385. 10.1111/j.1462-5822.2009.01404.x [DOI] [PubMed] [Google Scholar]
- Kennedy, E. A. , King, K. Y. , & Baldridge, M. T. (2018). Mouse microbiota models: Comparing germ‐free mice and antibiotics treatment as tools for modifying gut bacteria. Frontiers in physiology, 9. 10.3389/fphys.2018.01534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. H. , Jeun, E. J. , Hong, C. P. , Kim, S. H. , Jang, M. S. , Lee, E. J. , Moon, S. J. , Yun, C. H. , Im, S. H. , Jeong, S. G. , Park, B. Y. , Kim, K. T. , Seoh, J. Y. , Kim, Y. K. , Oh, S. J. , Ham, J. S. , Yang, B. G. , & Jang, M. H. (2016). Extracellular vesicle‐derived protein from Bifidobacterium longum alleviates food allergy through mast cell suppression. Journal of Allergy and Clinical Immunology, 137(2), 507–516.e8. 10.1016/j.jaci.2015.08.016 [DOI] [PubMed] [Google Scholar]
- Kim, J. H. , Lee, J. , Park, J. , & Gho, Y. S. (2015). Gram‐negative and gram‐positive bacterial extracellular vesicles. Seminars in Cell & Developmental Biology, 40, 97–104. 10.1016/j.semcdb.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Kim, S. Y. , & Yi, D. Y. (2020). Analysis of the human breast milk microbiome and bacterial extracellular vesicles in healthy mothers. Experimental & Molecular Medicine, 52(8), 1288–1297. 10.1038/s12276-020-0470-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen, K. , Hampton, T. H. , Jarek, M. , Scharfe, M. , Gerber, S. A. , Mielcarz, D. W. , Demers, E. G. , Dolben, E. L. , Hammond, J. H. , Hogan, D. A. , & Stanton, B. A. (2016). A novel mechanism of host‐pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathogens, 12(6), e1005672. 10.1371/journal.ppat.1005672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroniger, T. , Otto, A. , & Becher, D. (2018). Proteomic analysis of bacterial (outer) membrane vesicles: Progress and clinical potential. Expert Review of Proteomics, 15(8), 623–626. 10.1080/14789450.2018.1505509 [DOI] [PubMed] [Google Scholar]
- Kulkarni, H. M. , & Jagannadham, M. V. (2014). Biogenesis and multifaceted roles of outer membrane vesicles from Gram‐negative bacteria. Microbiology (Reading, England), 160(Pt 10), 2109–2121. 10.1099/mic.0.079400-0 [DOI] [PubMed] [Google Scholar]
- Kulp, A. , & Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual Review of Microbiology, 64, 163–184. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poul, E. , Loison, C. , Struyf, S. , Springael, J.‐Y. , Lannoy, V. , Decobecq, M.‐E. , Brezillon, S. , Dupriez, V. , Vassart, G. , Van Damme, J. , Parmentier, M. , & Detheux, M. (2003). Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. The Journal of Biological Chemistry, 278(28), 25481–25489. 10.1074/jbc.M301403200 [DOI] [PubMed] [Google Scholar]
- Lee, E.‐Y. , Bang, J. Y. , Park, G. W. , Choi, D.‐S. , Kang, J. S. , Kim, H.‐J. , Park, K.‐S. , Lee, J.‐O. , Kim, Y.‐K. , Kwon, K.‐H. , Kim, K.‐P. , & Gho, Y. S. (2007). Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli . Proteomics, 7(17), 3143–3153. 10.1002/pmic.200700196 [DOI] [PubMed] [Google Scholar]
- Lee, E. Y. , Choi, D. S. , Kim, K. P. , & Gho, Y. S. (2008). Proteomics in gram‐negative bacterial outer membrane vesicles. Mass Spectrometry Reviews, 27(6), 535–555. 10.1002/mas.20175 [DOI] [PubMed] [Google Scholar]
- Lee, E. Y. , Choi, D. Y. , Kim, D. K. , Kim, J. W. , Park, J. O. , Kim, S. , Kim, S. H. , Desiderio, D. M. , Kim, Y. K. , Kim, K. P. , & Gho, Y. S. (2009). Gram‐positive bacteria produce membrane vesicles: proteomics‐based characterization of Staphylococcus aureus‐derived membrane vesicles. Proteomics, 9(24), 5425–5436. 10.1002/pmic.200900338 [DOI] [PubMed] [Google Scholar]
- Lee, H. J. (2019). Microbe‐host communication by small RNAs in extracellular vesicles: Vehicles for transkingdom rna transportation. International Journal of Molecular Sciences, 20(6). 10.3390/ijms20061487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. H. , Choi, C. W. , Lee, T. , Kim, S. I. , Lee, J. C. , & Shin, J. H. (2013). Transcription factor σB plays an important role in the production of extracellular membrane‐derived vesicles in Listeria monocytogenes . Plos One, 8(8), e73196. 10.1371/journal.pone.0073196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Kim, O. Y. , & Gho, Y. S. (2016). Proteomic profiling of Gram‐negative bacterial outer membrane vesicles: Current perspectives. Proteomics: Clinical Applications , 10(9–10), 897–909. 10.1002/prca.201600032 [DOI] [PubMed] [Google Scholar]
- Lee, J. Y. , & Kim, H. S. (2017). Extracellular vesicles in neurodegenerative diseases: A double‐edged sword. Tissue Engineering and Regenerative Medicine , 14(6), 667–678. 10.1007/s13770-017-0090-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A. , Schertzer, J. W. , & Yong, X. (2018). Molecular dynamics modeling of Pseudomonas aeruginosa outer membranes. Physical Chemistry Chemical Physics, 20(36), 23635–23648. 10.1039/c8cp04278k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Lee, K. , Hsu, M. , Nau, G. , Mylonakis, E. , & Ramratnam, B. (2017). Lactobacillus‐derived extracellular vesicles enhance host immune responses against vancomycin‐resistant enterococci. BMC Microbiology [Electronic Resource], 17. 10.1186/s12866-017-0977-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, S. , Klein, M. I. , Heim, K. P. , Fan, Y. , Bitoun, J. P. , Ahn, S. J. , Burne, R. A. , Koo, H. , Brady, L. J. , & Wen, Z. T. (2014). Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. Journal of Bacteriology, 196(13), 2355–2366. 10.1128/jb.01493-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y.‐T. , Kuo, S.‐C. , Chiang, M.‐H. , Lee, Y.‐T. , Sung, W.‐C. , Chen, Y.‐H. , Chen, T.‐L. , & Fung, C.‐P. (2015). Acinetobacter baumannii extracellular OXA‐58 is primarily and selectively released via outer membrane vesicles after sec‐dependent periplasmic translocation. Antimicrobial Agents and Chemotherapy, 59(12), 7346–7354. 10.1128/AAC.01343-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Defourny, K. A. Y. , Smid, E. J. , & Abee, T. (2018). Gram‐positive bacterial extracellular vesicles and their impact on health and disease. Frontiers in Microbiology, 9, 1502. 10.3389/fmicb.2018.01502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López, P. , González‐Rodríguez, I. , Gueimonde, M. , Margolles, A. , & Suárez, A. (2011). Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. Plos One, 6(9), e24776. 10.1371/journal.pone.0024776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, P. , Gonzalez‐Rodriguez, I. , Sanchez, B. , Gueimonde, M. , Margolles, A. , & Suarez, A. (2012). Treg‐inducing membrane vesicles from Bifidobacterium bifidum LMG13195 as potential adjuvants in immunotherapy. Vaccine, 30(5), 825–829. 10.1016/j.vaccine.2011.11.115 [DOI] [PubMed] [Google Scholar]
- Luissint, A.‐C. , Parkos, C. A. , & Nusrat, A. (2016). Inflammation and the intestinal barrier: Leukocyte‐epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology, 151(4), 616–632. 10.1053/j.gastro.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerz, J. K. , Steimle, A. , Lange, A. , Bender, A. , Fehrenbacher, B. , & Frick, J.‐S. (2018). Outer membrane vesicles blebbing contributes to B. vulgatus mpk‐mediated immune response silencing. Gut Microbes, 9(1), 1–12. 10.1080/19490976.2017.1344810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, A. J. , & Kuehn, M. J. (2011). Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiology, 11, 258. 10.1186/1471-2180-11-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, R. C. , & Whitworth, D. E. (2019). Is “Wolf‐Pack” predation by antimicrobial bacteria cooperative? Cell behaviour and predatory mechanisms indicate profound selfishness, even when working alongside kin. BioEssays : News and Reviews in Molecular, Cellular and Developmental Biology, 41(4), e1800247. 10.1002/bies.201800247 [DOI] [PubMed] [Google Scholar]
- Martens, E. C. , Roth, R. , Heuser, J. E. , & Gordon, J. I. (2009). Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. Journal of Biological Chemistry, 284(27), 18445–18457. 10.1074/jbc.M109.008094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn‐Warren, L. , Howe, J. , Brandenburg, K. , & Whiteley, M. (2009). Structural requirements of the Pseudomonas quinolone signal for membrane vesicle stimulation. Journal of Bacteriology, 191(10), 3411–3414. 10.1128/jb.00052-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn, L. M. , & Whiteley, M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature, 437(7057), 422–425. 10.1038/nature03925 [DOI] [PubMed] [Google Scholar]
- Mazmanian, S. K. , Liu, C. H. , Tzianabos, A. O. , & Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell, 122(1), 107–118. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Mazmanian, S. K. , Round, J. L. , & Kasper, D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature, 453(7195), 620–625. 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- McBroom, A. J. , Johnson, A. P. , Vemulapalli, S. , & Kuehn, M. J. (2006). Outer membrane vesicle production by Escherichia coli is independent of membrane instability. Journal of Bacteriology, 188(15), 5385–5392. 10.1128/jb.00498-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom, A. J. , & Kuehn, M. J. (2007). Release of outer membrane vesicles by Gram‐negative bacteria is a novel envelope stress response. Molecular Microbiology, 63(2), 545–558. 10.1111/j.1365-2958.2006.05522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirjafari Tafti, Z. S. , Moshiri, A. , Ettehad Marvasti, F. , Tarashi, S. , Sadati Khalili, S. F. , Motahhary, A. , Fateh, A. , Vaziri, F. , Ahmadi Badi, S. , & Siadat, S. D. (2019). The effect of saturated and unsaturated fatty acids on the production of outer membrane vesicles from Bacteroides fragilis and Bacteroides thetaiotaomicron . Gastroenterology and Hepatology from Bed to Bench, 12(2), 155–162. [PMC free article] [PubMed] [Google Scholar]
- Morad, G. , Carman, C. V , Hagedorn, E. J. , Perlin, J. R. , Zon, L. I. , Mustafaoglu, N. , Park, T.‐E. , Ingber, D. E. , Daisy, C. C. , & Moses, M. A. (2019). Tumor‐derived extracellular vesicles breach the intact blood‐brain barrier via transcytosis. ACS Nano, 13(12), 13853–13865. 10.1021/acsnano.9b04397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, C. , Yang, Y. , & Zhu, W. (2015). Crosstalk between the immune receptors and gut microbiota. Current Protein & Peptide Science, 16(7), 622–631. 10.2174/1389203716666150630134356 [DOI] [PubMed] [Google Scholar]
- Müller, M. , Fink, K. , Geisel, J. , Kahl, F. , Jilge, B. , Reimann, J. , Mach, N. , Autenrieth, I. B. , & Frick, J. S. (2008). Intestinal colonization of IL‐2 deficient mice with non‐colitogenic B. vulgatus prevents DC maturation and T‐cell polarization. Plos One, 3(6), e2376. 10.1371/journal.pone.0002376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevermann, J. , Silva, A. , Otero, C. , Oyarzún, D. P. , Barrera, B. , Gil, F. , Calderón, I. L. , & Fuentes, J. A. (2019). Identification of genes involved in biogenesis of outer membrane vesicles (OMVs) in Salmonella enterica Serovar Typhi. Frontiers in microbiology, 10, 104. 10.3389/fmicb.2019.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue, E. J. , & Krachler, A. M. (2016). Mechanisms of outer membrane vesicle entry into host cells. Cellular Microbiology, 18(11), 1508–1517. 10.1111/cmi.12655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue, E. J. , Sirisaengtaksin, N. , Browning, D. F. , Bielska, E. , Hadis, M. , Fernandez‐Trillo, F. , Alderwick, L. , Jabbari, S. , & Krachler, A. M. (2017). Lipopolysaccharide structure impacts the entry kinetics of bacterial outer membrane vesicles into host cells. Plos Pathogens, 13(11), e1006760. 10.1371/journal.ppat.1006760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya‐Abril, A. , Prados‐Rosales, R. , McConnell, M. J. , Martín‐Peña, R. , González‐Reyes, J. A. , Jiménez‐Munguía, I. , Gómez‐Gascón, L. , Fernández, J. , Luque‐García, J. L. , García‐Lidón, C. , Estévez, H. , Pachón, J. , Obando, I. , Casadevall, A. , Pirofski, L. A. , & Rodríguez‐Ortega, M. J. (2014). Characterization of protective extracellular membrane‐derived vesicles produced by Streptococcus pneumoniae . Journal of Proteomics, 106, 46–60. 10.1016/j.jprot.2014.04.023 [DOI] [PubMed] [Google Scholar]
- Ottman, N. , Reunanen, J. , Meijerink, M. , Pietilä, T. E. , Kainulainen, V. , Klievink, J. , Huuskonen, L. , Aalvink, S. , Skurnik, M. , Boeren, S. , Satokari, R. , Mercenier, A. , Palva, A. , Smidt, H. , de Vos, W. M. , & Belzer, C. (2017). Pili‐like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. Plos One, 12(3), e0173004. 10.1371/journal.pone.0173004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Païssé, S. , Valle, C. , Servant, F. , Courtney, M. , Burcelin, R. , Amar, J. , & Lelouvier, B. (2016). Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion, 56(5), 1138–1147. 10.1111/trf.13477 [DOI] [PubMed] [Google Scholar]
- Palacios, A. , Coelho, C. , Maryam, M. , Luque‐García, J. L. , Casadevall, A. , & Prados‐Rosales, R. (2020). Biogenesis and function of extracellular vesicles in gram‐positive bacteria, mycobacteria, and fungi. In Kaparakis‐Liaskos M. & Kufer T. A. (Eds.), Bacterial Membrane Vesicles: Biogenesis, Functions and Applications (pp. 47–74). Springer International Publishing. 10.1007/978-3-030-36331-4_3 [DOI] [Google Scholar]
- Park, J.‐Y. , Choi, J. , Lee, Y. , Lee, J.‐E. , Lee, E.‐H. , Kwon, H.‐J. , Yang, J. , Jeong, B.‐R. , Kim, Y.‐K. , & Han, P.‐L. (2017). Metagenome analysis of bodily microbiota in a mouse model of alzheimer disease using bacteria‐derived membrane vesicles in blood. Experimental Neurobiology, 26(6), 369–379. 10.5607/en.2017.26.6.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Cruz, C. , Cañas, M. A. , Giménez, R. , Badia, J. , Mercade, E. , Baldomà, L. , & Aguilera, L. (2016). Membrane vesicles released by a hypervesiculating Escherichia coli Nissle 1917 tolR mutant are highly heterogeneous and show reduced capacity for epithelial cell interaction and entry. Plos One, 11(12), e0169186. 10.1371/journal.pone.0169186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Cruz, C. , Carrión, O. , Delgado, L. , Martinez, G. , López‐Iglesias, C. , & Mercade, E. (2013). New type of outer membrane vesicle produced by the Gram‐negative bacterium Shewanella vesiculosa M7T: Implications for DNA content. Applied and Environmental Microbiology, 79(6), 1874–1881. 10.1128/aem.03657-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Cruz, C. , Delgado, L. , Lopez‐Iglesias, C. , & Mercade, E. (2015). Outer‐inner membrane vesicles naturally secreted by gram‐negative pathogenic bacteria. Plos One, 10(1), e0116896. 10.1371/journal.pone.0116896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Lopez, A. , Behnsen, J. , Nuccio, S.‐P. , & Raffatellu, M. (2016). Mucosal immunity to pathogenic intestinal bacteria. Nature Reviews. Immunology, 16(3), 135–148. 10.1038/nri.2015.17 [DOI] [PubMed] [Google Scholar]
- Philpott, D. J. , Sorbara, M. T. , Robertson, S. J. , Croitoru, K. , & Girardin, S. E. (2014). NOD proteins: Regulators of inflammation in health and disease. Nature Reviews. Immunology, 14(1), 9–23. 10.1038/nri3565 [DOI] [PubMed] [Google Scholar]
- Png, C. W. , Lindén, S. K. , Gilshenan, K. S. , Zoetendal, E. G. , McSweeney, C. S. , Sly, L. I. , McGuckin, M. A. , & Florin, T. H. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. American Journal of Gastroenterology, 105(11), 2420–2428. 10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- Poore, G. D. , Kopylova, E. , Zhu, Q. , Carpenter, C. , Fraraccio, S. , Wandro, S. , Kosciolek, T. , Janssen, S. , Metcalf, J. , Song, S. J. , Kanbar, J. , Miller‐Montgomery, S. , Heaton, R. , Mckay, R. , Patel, S. P. , Swafford, A. D. , & Knight, R. (2020). Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature, 579(7800), 567–574. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Renelli, M. , Matias, V. , Lo, R. Y. , & Beveridge, T. J. (2004). DNA‐containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology (Reading, England), 150(Pt 7), 2161–2169. 10.1099/mic.0.26841-0 [DOI] [PubMed] [Google Scholar]
- Resch, U. , Tsatsaronis, J. A. , Le Rhun, A. , Stübiger, G. , Rohde, M. , Kasvandik, S. , Holzmeister, S. , Tinnefeld, P. , Wai, S. N. , & Charpentier, E. (2016). A two‐component regulatory system impacts extracellular membrane‐derived vesicle production in group A Streptococcus. MBio, 7(6). 10.1128/mBio.00207-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno, M. , Urbano, M. , Valzasina, B. , Francolini, M. , Rotta, G. , Bonasio, R. , Granucci, F. , Kraehenbuhl, J. P. , & Ricciardi‐Castagnoli, P. (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nature Immunology, 2(4), 361–367. 10.1038/86373 [DOI] [PubMed] [Google Scholar]
- Rivera, J. , Cordero, R. J. , Nakouzi, A. S. , Frases, S. , Nicola, A. , & Casadevall, A. (2010). Bacillus anthracis produces membrane‐derived vesicles containing biologically active toxins. Proceedings of the National Academy of Sciences of the United States of America, 107(44), 19002–19007. 10.1073/pnas.1008843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Nogales, A. , Algieri, F. , Garrido‐Mesa, J. , Vezza, T. , Utrilla, M. P. , Chueca, N. , Fernández‐Caballero, J. A. , García, F. , Rodríguez‐Cabezas, M. E. , & Gálvez, J. (2018). The administration of Escherichia coli Nissle 1917 ameliorates development of DSS‐induced colitis in mice. Frontiers in Pharmacology, 9, 468. 10.3389/fphar.2018.00468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier, S. , Zingl, F. G. , Cakar, F. , Durakovic, S. , Kohl, P. , Eichmann, T. O. , Klug, L. , Gadermaier, B. , Weinzerl, K. , Prassl, R. , Lass, A. , Daum, G. , Reidl, J. , Feldman, M. F. , & Schild, S. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in Gram‐negative bacteria. Nature communications, 7, 10515. 10.1038/ncomms10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roier, S. , Zingl, F. G. , Cakar, F. , & Schild, S. (2016). Bacterial outer membrane vesicle biogenesis: A new mechanism and its implications. Microbial Cell, 3(6), 257–259. 10.15698/mic2016.06.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round, J. L. , & Mazmanian, S. K. (2010). Inducible Foxp3+ regulatory T‐cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America, 107(27), 12204–12209. 10.1073/pnas.0909122107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter, S. J. , Cox, M. J. , Turek, E. M. , Calus, S. T. , Cookson, W. O. , Moffatt, M. F. , Turner, P. , Parkhill, J. , Loman, N. J. , & Walker, A. W. (2014). Reagent and laboratory contamination can critically impact sequence‐based microbiome analyses. BMC Biology, 12, 87. 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer, J. W. , & Whiteley, M. (2012). A bilayer‐couple model of bacterial outer membrane vesicle biogenesis. MBio, 3(2). 10.1128/mBio.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatterer, K. , Beck, C. , Hanzelmann, D. , Lebtig, M. , Fehrenbacher, B. , Schaller, M. , Ebner, P. , Nega, M. , Otto, M. , Kretschmer, D. , & Peschel, A. (2018). The mechanism behind bacterial lipoprotein release: Phenol‐soluble modulins mediate toll‐like receptor 2 activation via extracellular vesicle release from Staphylococcus aureus . MBio, 9(6). 10.1128/mBio.01851-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C. , & Kuehn, M. J. (2013). Synthetic effect between envelope stress and lack of outer membrane vesicle production in Escherichia coli . Journal of Bacteriology, 195(18), 4161–4173. 10.1128/jb.02192-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C. , & Kuehn, M. J. (2015). Outer‐membrane vesicles from Gram‐negative bacteria: Biogenesis and functions. Nature Reviews Microbiology, 13(10), 605–619. 10.1038/nrmicro3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C. , Sullivan, C. J. , & Kuehn, M. J. (2013). Envelope control of outer membrane vesicle production in Gram‐negative bacteria. Biochemistry, 52(18), 3031–3040. 10.1021/bi400164t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, C. L. , Geis, A. L. , & Housseau, F. (2014). Bacteroides fragilis subverts mucosal biology: From symbiont to colon carcinogenesis. Journal of Clinical Investigation, 124(10), 4166–4172. 10.1172/jci72334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, M. K. , Park, E. J. , Ko, S. Y. , Choi, E. W. , & Kim, S. (2018). Therapeutic effects of kefir grain Lactobacillus‐derived extracellular vesicles in mice with 2,4,6‐trinitrobenzene sulfonic acid‐induced inflammatory bowel disease. Journal of Dairy Science, 101(10), 8662–8671. 10.3168/jds.2018-15014 [DOI] [PubMed] [Google Scholar]
- Shen, L. (2012). Tight junctions on the move: Molecular mechanisms for epithelial barrier regulation. Annals of the New York Academy of Sciences, 1258, 9–18. 10.1111/j.1749-6632.2012.06613.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. , Giardino Torchia, M. L. , Lawson, G. W. , Karp, C. L. , Ashwell, J. D. , & Mazmanian, S. K. (2012). Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe, 12(4), 509–520. 10.1016/j.chom.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. P. (2019). Glycan utilisation system in Bacteroides and Bifidobacteria and their roles in gut stability and health. Applied Microbiology and Biotechnology, 103(18), 7287–7315. 10.1007/s00253-019-10012-z [DOI] [PubMed] [Google Scholar]
- Sonnenborn, U. (2016). Escherichia coli strain Nissle 1917‐from bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. Fems Microbiology Letters, 363(19). 10.1093/femsle/fnw212 [DOI] [PubMed] [Google Scholar]
- Sonnenborn, U. , & Schulze, J. (2009). The non‐pathogenic Escherichia coli strain Nissle 1917 – features of a versatile probiotic. Microbial Ecology in Health and Disease, 21(3–4), 122–158. 10.3109/08910600903444267 [DOI] [Google Scholar]
- Souza, É. L. , Elian, S. D. , Paula, L. M. , Garcia, C. C. , Vieira, A. T. , Teixeira, M. M. , Arantes, R. M. , Nicoli, J. R. , & Martins, F. S. (2016). Escherichia coli strain Nissle 1917 ameliorates experimental colitis by modulating intestinal permeability, the inflammatory response and clinical signs in a faecal transplantation model. Journal of Medical Microbiology, 65(3), 201–210. 10.1099/jmm.0.000222 [DOI] [PubMed] [Google Scholar]
- Stentz, R , Osborne, S. , Horn, N. , Li, A. W. , Hautefort, I. , Bongaerts, R. , Rouyer, M. , Bailey, P. , Shears, S. B. , Hemmings, A. M. , Brearley, C. A. , & Carding, S. R. (2014). A bacterial homolog of a eukaryotic inositol phosphate signaling enzyme mediates cross‐kingdom dialog in the mammalian gut. Cell reports, 6(4), 646–656. 10.1016/j.celrep.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentz, R. , Carvalho, A. L. , Jones, E. J. , & Carding, S. R. (2018). Fantastic voyage: The journey of intestinal microbiota‐derived microvesicles through the body. Biochemical Society Transactions, 46(5), 1021–1027. 10.1042/BST20180114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentz, R. , Horn, N. , Cross, K. , Salt, L. , Brearley, C. , Livermore, D. M. , & Carding, S. R. (2015). Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β‐lactam antibiotics. The Journal of Antimicrobial Chemotherapy, 70(3), 701–709. 10.1093/jac/dku466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surve, M. V , Anil, A. , Kamath, K. G. , Bhutda, S. , Sthanam, L. K. , Pradhan, A. , Srivastava, R. , Basu, B. , Dutta, S. , Sen, S. , Modi, D. , & Banerjee, A. (2016). Membrane vesicles of group B Streptococcus disrupt feto‐maternal barrier leading to preterm birth. Plos Pathogens, 12(9), e1005816. 10.1371/journal.ppat.1005816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T. (2013). Regulation of intestinal epithelial permeability by tight junctions. Cellular and Molecular Life Sciences : CMLS, 70(4), 631–659. 10.1007/s00018-012-1070-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaan, P. W. , Bensman, T. , Bahadduri, P. M. , Hall, M. W. , Sarkar, A. , Bao, S. , Khantwal, C. M. , Ekins, S. , & Knoell, D. L. (2008). Bacterial peptide recognition and immune activation facilitated by human peptide transporter PEPT2. American Journal of Respiratory Cell and Molecular Biology, 39(5), 536–542. 10.1165/rcmb.2008-0059OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro, Y. , Inagaki, A. , Shimizu, M. , Ichikawa, S. , Takaya, N. , Nakajima‐Kambe, T. , Uchiyama, H. , & Nomura, N. (2011). Characterization of phospholipids in membrane vesicles derived from Pseudomonas aeruginosa . Bioscience, Biotechnology, and Biochemistry, 75(3), 605–607. 10.1271/bbb.100754 [DOI] [PubMed] [Google Scholar]
- Tashiro, Y. , Uchiyama, H. , & Nomura, N. (2012). Multifunctional membrane vesicles in Pseudomonas aeruginosa . Environmental Microbiology, 14(6), 1349–1362. 10.1111/j.1462-2920.2011.02632.x [DOI] [PubMed] [Google Scholar]
- Thay, B. , Damm, A. , Kufer, T. A. , Wai, S. N. , & Oscarsson, J. (2014). Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1‐ and NOD2‐dependent NF‐κB activation. Infection and Immunity, 82(10), 4034–4046. 10.1128/IAI.01980-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Integrative Human Microbiome Project . (2019). Nature, 569(7758), 641–648. 10.1038/s41586-019-1238-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J , Anderson, J. D , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of extracellular vesicles, 7(1), 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toloza, L. , Giménez, R. , Fábrega, M. J. , Alvarez, C. S. , Aguilera, L. , Cañas, M. A. , Martín‐Venegas, R. , Badia, J. , & Baldomà, L. (2015). The secreted autotransporter toxin (Sat) does not act as a virulence factor in the probiotic Escherichia coli strain Nissle 1917. Bmc Microbiology [Electronic Resource], 15, 250. 10.1186/s12866-015-0591-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku, M. , Cárcamo‐Oyarce, G. , Yamamoto, T. , Eisenstein, F. , Hsiao, C. C. , Kurosawa, M. , Gademann, K. , Pilhofer, M. , Nomura, N. , & Eberl, L. (2017). Prophage‐triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis . Nature communications, 8(1), 481. 10.1038/s41467-017-00492-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku, M. , Nomura, N. , & Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nature Reviews Microbiology, 17(1), 13–24. 10.1038/s41579-018-0112-2 [DOI] [PubMed] [Google Scholar]
- Tropini, C. , Earle, K. A. , Huang, K. C. , & Sonnenburg, J. L. (2017). The gut microbiome: Connecting spatial organization to function. Cell Host Microbe, 21(4), 433–442. 10.1016/j.chom.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulkens, J. , De Wever, O. , & Hendrix, A. (2020). Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nature Protocols, 15(1), 40–67. 10.1038/s41596-019-0236-5 [DOI] [PubMed] [Google Scholar]
- Tulkens, J. , Vergauwen, G. , Van Deun, J. , Geeurickx, E. , Dhondt, B. , Lippens, L. , De Scheerder, M.‐A. , Miinalainen, I. , Rappu, P. , De Geest, B. G. , Vandecasteele, K. , Laukens, D. , Vandekerckhove, L. , Denys, H. , Vandesompele, J. , De Wever, O. , & Hendrix, A. (2020). Increased levels of systemic LPS‐positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. In Gut (Vol. 69, Issue 1, pp. 191–193). 10.1136/gutjnl-2018-317726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull, L. , Toyofuku, M. , Hynen, A. L. , Kurosawa, M. , Pessi, G. , Petty, N. K. , Osvath, S. R. , Cárcamo‐Oyarce, G. , Gloag, E. S. , Shimoni, R. , Omasits, U. , Ito, S. , Yap, X. , Monahan, L. G. , Cavaliere, R. , Ahrens, C. H. , Charles, I. G. , Nomura, N. , Eberl, L. , & Whitchurch, C. B. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nature communications, 7, 11220. 10.1038/ncomms11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukena, S. N. , Singh, A. , Dringenberg, U. , Engelhardt, R. , Seidler, U. , Hansen, W. , Bleich, A. , Bruder, D. , Franzke, A. , Rogler, G. , Suerbaum, S. , Buer, J. , Gunzer, F. , & Westendorf, A. M. (2007). Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. Plos One, 2(12), e1308. 10.1371/journal.pone.0001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valguarnera, E. , Scott, N. E. , Azimzadeh, P. , & Feldman, M. F. (2018). Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in Bacteroides . MSphere, 3(6). 10.1128/mSphere.00559-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman, K. , Hummel, S. , Cichon, C. , Sonnenborn, U. , & Schmidt, M. A. (2012). Identification of specific miRNAs targeting proteins of the apical junctional complex that simulate the probiotic effect of E. coli Nissle 1917 on T84 epithelial cells. International Journal of Biochemistry & Cell Biology, 44(2), 341–349. 10.1016/j.biocel.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Vigsnæs, L. K. , Brynskov, J. , Steenholdt, C. , Wilcks, A. , & Licht, T. R. (2012). Gram‐negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Beneficial Microbes, 3(4), 287–297. 10.3920/bm2012.0018 [DOI] [PubMed] [Google Scholar]
- Volgers, C. , Savelkoul, P. H. M. , & Stassen, F. R. M. (2018). Gram‐negative bacterial membrane vesicle release in response to the host‐environment: different threats, same trick? Critical Reviews in Microbiology, 44(3), 258–273. 10.1080/1040841x.2017.1353949 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Thompson, C. D. , Weidenmaier, C. , & Lee, J. C. (2018). Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nature communications, 9(1), 1379. 10.1038/s41467-018-03847-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, J. M. (2011). Immunomodulatory mechanisms of lactobacilli. Microbial Cell Factories, 10(1), S17. 10.1186/1475-2859-10-S1-S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink, J. , & Witholt, B. (1981). Outer‐membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. European Journal of Biochemistry Febs, 116(2), 331–335. 10.1111/j.1432-1033.1981.tb05338.x [DOI] [PubMed] [Google Scholar]
- West, C. L. , Stanisz, A. M. , Mao, Y.‐K. , Champagne‐Jorgensen, K. , Bienenstock, J. , & Kunze, W. A. (2020). Microvesicles from Lactobacillus reuteri (DSM‐17938) completely reproduce modulation of gut motility by bacteria in mice. Plos One, 15(1), e0225481. 10.1371/journal.pone.0225481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler, A. G. , & Goodman, A. L. (2017). An insider's perspective: Bacteroides as a window into the microbiome. Nature Microbiology, 2, 17026. 10.1038/nmicrobiol.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler, H. M. (2007). Bacteroides: The good, the bad, and the nitty‐gritty. Clinical Microbiology Reviews, 20(4), 593–621. 10.1128/cmr.00008-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle, E. , Leonard, M. O. , Harrison, R. , Gant, T. W. , & Tonge, D. P. (2019). Multi‐method characterization of the human circulating microbiome . In Frontiers in Microbiology, 9, 3266. 10.3389/fmicb.2018.03266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki‐Yashiki, S. , Miyoshi, Y. , Nakayama, T. , Kunisawa, J. , & Katakura, Y. (2019). IgA‐enhancing effects of membrane vesicles derived from Lactobacillus sakei subsp. sakei NBRC15893. Bioscience of Microbiota, Food and Health, 38(1), 23–29. 10.12938/bmfh.18-015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharzhevskaya, N. B , Tsvetkov, V. B. , Vanyushkina, A. A. , Varizhuk, A. M. , Rakitina, D. V , Podgorsky, V. V , Vishnyakov, I. E. , Kharlampieva, D. D. , Manuvera, V. A. , Lisitsyn, F. V , Gushina, E. A. , Lazarev, V. N. , & Govorun, V. M. (2017). Interaction of Bacteroides fragilis toxin with outer membrane vesicles reveals new mechanism of its secretion and delivery. Frontiers in Cellular and Infection Microbiology, 7. 2. 10.3389/fcimb.2017.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharzhevskaya, N. B , Vanyushkina, A. A. , Altukhov, I. A. , Shavarda, A. L. , Butenko, I. O. , Rakitina, D. V , Nikitina, A. S. , Manolov, A. I. , Egorova, A. N. , Kulikov, E. E. , Vishnyakov, I. E. , Fisunov, G. Y. , & Govorun, V. M. (2017). Outer membrane vesicles secreted by pathogenic and nonpathogenic Bacteroides fragilis represent different metabolic activities. Scientific Reports, 7(1), 5008. 10.1038/s41598-017-05264-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, R. , Xue, X. , Zhang, L. , Yang, X. , Zhao, L. , & Zhang, C. (2019). Strain‐specific anti‐inflammatory properties of two Akkermansia muciniphila strains on chronic colitis in mice. Frontiers in Cellular and Infection Microbiology, 9, 239. 10.3389/fcimb.2019.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, X. , Stamova, B. , Jin, L.‐W. , DeCarli, C. , Phinney, B. , & Sharp, F. R. (2016). Gram‐negative bacterial molecules associate with Alzheimer disease pathology. Neurology, 87(22), 2324–2332. 10.1212/WNL.0000000000003391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B. , Yuan, Y. , Zhang, S. , Guo, C. , Li, X. , Li, G. , Xiong, W. , & Zeng, Z. (2020). Intestinal flora and disease mutually shape the regional immune system in the intestinal tract. Frontiers in Immunology, 11, 575. 10.3389/fimmu.2020.00575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.‐C. , & Zhang, X.‐W. (2019). Akkermansia muciniphila: A promising target for the therapy of metabolic syndrome and related diseases. Chinese Journal of Natural Medicines, 17(11), 835–841. 10.1016/S1875-5364(19)30101-3 [DOI] [PubMed] [Google Scholar]
- Zingl, F. G. , Leitner, D. R. , & Schild, S. (2020). Biogenesis of Gram‐Negative OMVs. In Kaparakis‐Liaskos Maria & Kufer T. A. (Eds.), Bacterial membrane vesicles: Biogenesis, functions and applications (pp. 23–46). Springer International Publishing. 10.1007/978-3-030-36331-4_2 [DOI] [Google Scholar]
- Zyrek, A. A. , Cichon, C. , Helms, S. , Enders, C. , Sonnenborn, U. , & Schmidt, M. A. (2007). Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO‐2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cellular Microbiology, 9(3), 804–816. 10.1111/j.1462-5822.2006.00836.x [DOI] [PubMed] [Google Scholar]


