Main text
(iScience 23, 101068-1–101068-11; May 22, 2020)
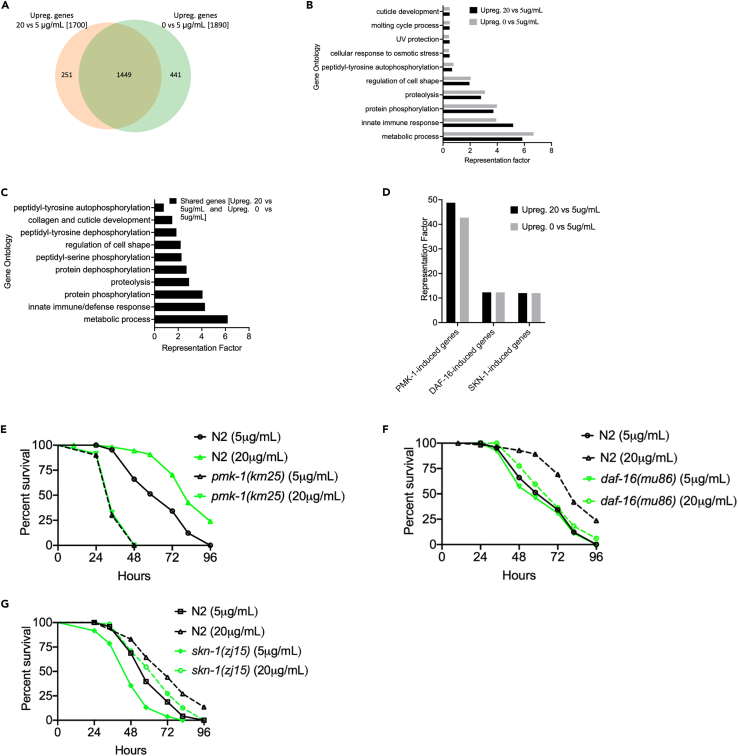
In the originally published article, the authors made a mistake in analyzing RNA sequencing (RNA-seq) data corresponding to 5 μg/ml vs. 0 cholesterol presented. Therefore, incorrect annotations were placed in the data corresponding to 5 μg/ml cholesterol as shown in Figures 2A–2D and S4 and their corresponding legends, Tables S1, S2, S3, S4, and S5, and associated text throughout the manuscript. Values were mistakenly presented in the reversed order (i.e., the reported genes were annotated as downregulated instead of upregulated in the absence of cholesterol). These are vital considerations necessary for reporting and reproducibility of data, and the authors apologize for this error. They further confirm that this does not change the scientific message of the paper.
Figure 2. Cholesterol-Mediated Immunity Primarily Acts through a p38/PMK-1 MAPK Pathway
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103361.
Supplemental information
The RNA-seq raw data were submitted and assigned Gene Expression Omnibus (GEO) accession numbers: GSE136881 and GSE137058, Related to Figure 2
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The RNA-seq raw data were submitted and assigned Gene Expression Omnibus (GEO) accession numbers: GSE136881 and GSE137058, Related to Figure 2