Abstract
Biomolecular condensates are found throughout eukaryotic cells, in the nucleus, cytoplasm and on various membranes. They are also found across the biological spectrum, organizing molecules that act in processes ranging from RNA metabolism to signaling to gene regulation. Early work in the field focused on identifying condensates, understanding their physical properties and regulation, and how these arise from their molecular constituents. Recent years have brought a focus on understanding condensate functions. Studies have revealed functions that span from molecular scales, modulating the rates of chemical reactions; to the mesoscale, organizing large structures within cells; to cellular scales, facilitating localization of cellular materials and homeostatic responses. In this Roadmap, we discuss examples from the recent literature and organize condensate functions into a series of non-exclusive classes across these length scales. Beyond these few examples discussed in detail, we provide a more comprehensive listing of condensates that play roles in a large variety of biological processes and which classes of functional mechanisms have been implicated. We conclude with a discussion of areas of current interest and challenges in the field, and thoughts about how progress may be made.
Introduction
Over the last decade, it has become broadly appreciated that many cellular structures consist of membraneless assemblies of proteins and/or nucleic acids. These structures, termed biomolecular condensates due to their ability to selectively concentrate molecules in defined foci, are pervasive through biology and provide a fundamental mechanism of cellular organization.
A large number of studies using both natural and engineered proteins designed to form condensates, as well as theoretical and experimental results from polymer chemistry and physics, have provided a solid foundation explaining how condensates form in regulated fashion. This work has shown that many condensates form via liquid-liquid phase separation (LLPS), a thermodynamic process by which above a threshold macromolecule concentration separate, coexisting liquid phases form to minimize free energy. It is important to note, however, that alternate mechanisms of condensate formation can also exist (Box 1). Material properties of condensates are distinct from their surroundings, including higher viscosity and internal structure that can exclude molecules in a size-dependent manner1. LLPS results in interfacial tension between the dense and dilute phases, resulting in spherical dense-phase droplets that relax upon fusion. In addition to forming three-dimensional droplets, condensates may also form on membranes2.
Box 1 | Separate liquid phases or macromolecular assemblies?
As interest in LLPS as a mechanism for organizing macromolecules in cells has increased, so have criticisms of the quality of evidence for LLPS in vivo137,167. Other mechanisms for generating biomolecular condensates—defined as concentrated foci lacking a surrounding membrane—have been proposed and the full range of mechanisms should be considered when interpreting experimental results.
Condensates can form via either active processes, which consume energy to organize molecules, or passive thermodynamic mechanisms, which do not. Both mechanisms may contribute to one condensate, which appears to occur for the nucleolus168. Active processes take many forms24, for example deposition of molecules at a defined location by motors or constraint of molecules by applied forces, and we will not consider them further here.
For passive thermodynamic mechanisms, important considerations include assembly cooperativity, size scaling as a function of macromolecule concentration, stoichiometry of components within the condensate, and condensate material properties. LLPS lies at one end of the spectrum as the transition between one-phase and two-phase regimes is infinitely cooperative, the dense phase volume grows without bounds as macromolecule concentration increases before ultimately re-entering the one-phase dense regime, and (for multicomponent systems) dense phases can form with a wide range of subcomponent stoichiometries169. Distinct material properties arise from LLPS, most prominently interfacial tension between the two phases.
At the other end of the spectrum, condensates may form as molecules bind to static cellular structures, for example RNA aggregates or genomic DNA. Formation of such condensates would be conceptually equivalent to ligands binding to a receptor with multiple binding sites, and may or may not exhibit cooperativity. Condensate size would be limited by the number of binding sites on the underlying structure. Likewise, the stoichiometry of components within the condensate would be dictated by binding site numbers, interaction affinities, and total concentrations for each component. Importantly, assembly behavior and composition of such condensates may resemble LLPS in certain affinity, cooperativity, and concentration regimes. Further challenges arise from the thermodynamics of small-number systems, where stochastic assembly state fluctuations become significant, as described elsewhere in this review145,146. Experimental findings such as rapid recovery of fluorescent signal after photobleaching are not sufficient to conclude a condensate forms via LLPS, and single-molecule imaging can provide more detailed information137. Similarly, the absence of a single fixed threshold concentration for condensate formation is insufficient to exclude LLPS if the system consists of more than one component125. Researchers working to define condensate assembly mechanisms should therefore focus on the key factors of assembly cooperativity, size scaling, stoichiometry of subcomponents, and material properties of apparent dilute-dense phase interfaces, particularly the dynamics of molecules at such boundaries.
Finally, as this review aims, in part, to describe the cellular functions of biomolecular condensates, it is important to consider how proposed functions depend on specific condensate assembly mechanisms. For example, models invoking condensation to buffer gene expression noise35 require formation via LLPS, but biochemical reaction rates could be enhanced in clusters of molecules bound to a static scaffold. Thus, researchers should consider whether functions ascribed to condensates require formation via a specific mechanism and whether a given formation mechanism excludes or indicates certain functional properties.
Studies have revealed the central role of multivalent interactions in driving condensate formation3,4. Many instantiations of multivalency have been described in condensates, involving folded protein domains, intrinsically disordered regions (IDR)3,5, nucleic acids3, and chromatin6. For IDRs, a ‘molecular grammar’ model has emerged, wherein the abundance and patterning of certain amino acids within the sequence influences both the drive to form and the physical properties of the condensates7–10. Analyses of nucleic acids have revealed the importance of base pairing and secondary structure in promoting assembly and specificity11,12. Further studies have demonstrated how the formation and composition of condensates can be regulated by post-translational modification13,14, binding interactions15–18 and environmental conditions19–21. A great deal of work has also demonstrated the relationship of aberrant condensates to diseases including neurodegeneration and cancer 22,23. As mechanisms of condensate formation have become better understood and broadly appreciated, attention has begun to shift toward condensate functions. This has brought new challenges to the field and has required development of new technologies and experimental systems. Here we focus on condensate functions in normal physiology, and direct readers to excellent reviews on the physicochemical underpinnings of condensate formation24–26 and the role of condensates in human disease22,23,26.
In this Roadmap, we highlight several examples from the recent literature and organize these findings into a framework that allows the role of condensates in disparate biological processes to be understood from a common conceptual viewpoint. We emphasize the importance of the length scale(s) on which functional mechanisms operate, which span from molecular to cellular (Figure 1). As we focus on illustrating classes of functional mechanisms within this framework with a few examples, we are unable to describe in detail all the biological processes where roles for condensates have been identified by this rapidly growing field; we thus provide a more comprehensive list of processes, organized by the class of functional mechanism by which condensates are proposed to function (Table 1). Finally, we provide suggestions for interesting avenues of future research, including the need for new technologies to address challenges in developing and confirming functional models.
Figure 1 |. Overview of biomolecular condensate functions across scales.
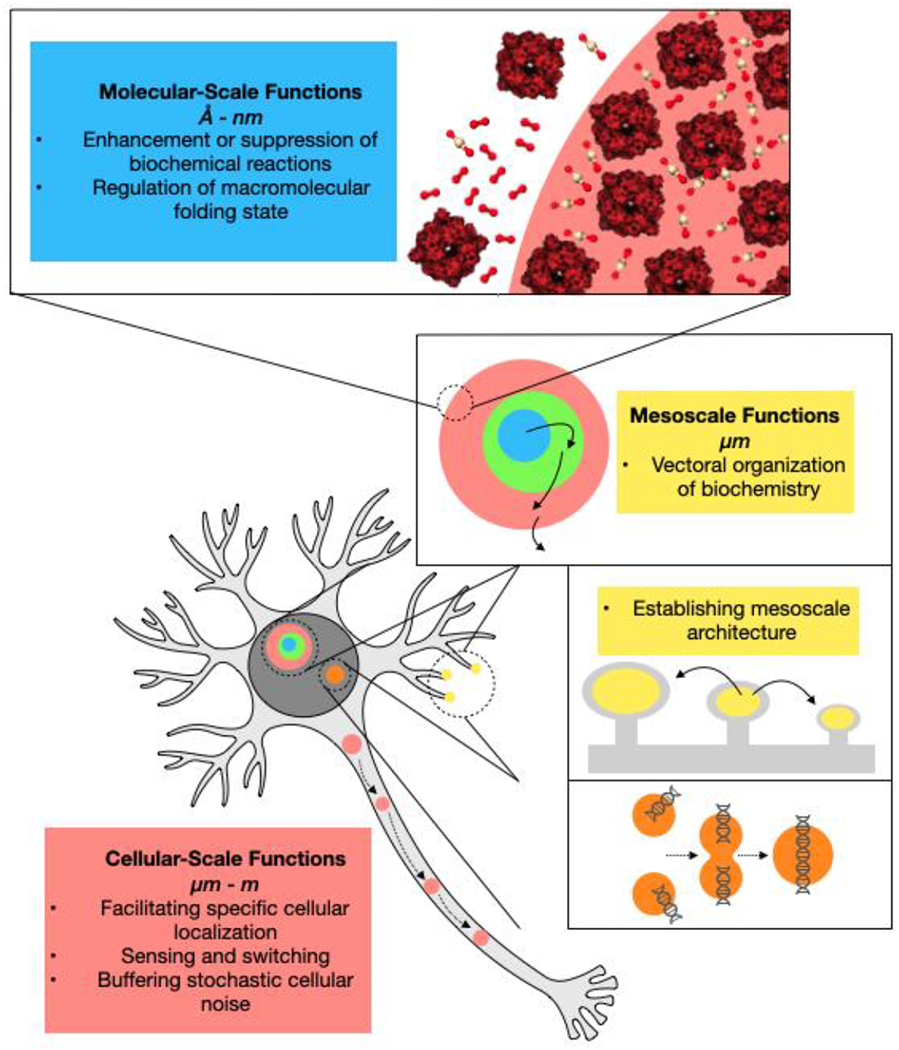
The functions of biomolecular condensates operate on multiple length scales, ranging from atomic or molecular-level enhancement or suppression or biochemical reaction rates to cellular localization, which in principle operates on meter length scales in cells such as mammalian neurons. A condensate may have functions on more than one length scale, for instance participating in mesoscale vectoral organization of biochemistry while also enhancing reaction rates via mass action due to increased concentration of substrates and/or enzymes within condensates.
Table 1 |.
Examples of the functional mechanisms of biomolecular condensates in biological processes
| Functional mechanism | Biological process | In vitro | In vivo |
|---|---|---|---|
| Enhancement or suppression of biochemical reactions | |||
| Enhanced activity through concentrating enzymes and substrates | Carbon fixation | 38,40 | 41 |
| Innate immune signaling | 47 | 47 | |
| RNA interference | 50 | ||
| Microtubule nucleation/spindle assembly | 54–57,199 | 54,199 | |
| Cell-cell adhesion | 59 | 58,59 | |
| Autophagy | 106 | 106 | |
| RNA degradation | 200 | 200 | |
| RNA splicing | 201 | 201 | |
| Pre-mRNA processing | 202,203 | ||
| Inhibition of activity through sequestration | Transcriptional regulation | 60,61 | |
| Cell fate determination under stress | 204 | ||
| Modulation of activity through exclusion of factors | Adaptive immune signaling | 63 | 63 |
| Synaptic transmission | 65 | ||
| Factors beyond concentration that modulate enzyme activity | Signal transduction | 69 | |
| Actin cytoskeleton | 68 | 68 | |
| Post-transcriptional regulation | 70 | ||
| Bacterial cell polarization | 205 | ||
| Membrane protein trafficking | 72 | ||
| Modulation of macromolecule folding | Nuclear protein homeostasis | 27 | |
| RNA homeostasis | 28,29,76 | 28,29,76,77 | |
| Vectoral organization of biochemistry | Ribosome biogenesis | 31,81 | 30,80 |
| Transgenerational RNAi inheritance | 82 | ||
| Establishing mesoscale architecture | Synaptic transmission | 65,84,85,185 | 85 |
| DNA damage response | 32,92,96,97 | 32,92,96,97 | |
| DNA replication | 98 | ||
| Autophagy | 102,104–106 | 102,104–106 | |
| Centriole biogenesis | 206 | 206 | |
| Facilitating specific cellular localization | Localized neuronal translation | 34 | 34 |
| Heterochromatin maintenance | 120 | 120 | |
| Germ cell fate maintenance | 207 | 33 | |
| Buffering stochastic cellular noise | Signal transduction | 35 | |
| Sensing and switching | Heat shock response | 19,128 | 19,128 |
| Nutrient stress response | 20,130,131 | 20,130,131 | |
| Protein homeostasis | 208 | ||
| Unknown or debated functions | Transcription | 149,150,152,156,209,210 | 149–156,209,210 |
| Chromatin | 6,117,119,188,211 | 119,188,211 | |
| Viral replication/evasion of host innate immunity | 212–214 | ||
| Protein degradation | 215 | 215 | |
| Cell motility | 216 | ||
| Sorting RNA to distinct granules | 12 | 12 | |
| Nucleocytoplasmic transport | 217 |
A typology of condensate functions
Contributions from a large number of researchers have demonstrated a range of mechanisms by which condensates contribute to biological processes26. Here, we organize these findings into a framework of functional classes, with an eye toward identifying mechanistic commonalities between condensates that play roles in different biological processes. We emphasize that these mechanistic classes are not intended to be mutually exclusive; as our framework spans a broad range of length scales (Figure 1), it is likely that more than one principle operates simultaneously for a given condensate. Further, we feel that blurring of boundaries between mechanistic classes will serve as useful focal points for future experimental efforts. The functional classes are below, listed by length scale from short to long. In addition to detailed discussion of several examples below, we provide a more comprehensive list of biological processes where condensate functions have been characterized, including some cases where functions are unclear or under debate (Table 1).
Enhancement or Suppression of Biochemical Reactions.
At the shortest length scale, condensate formation may enhance reaction rates due to high local concentration (mass action), which can lead to specificity and feedback. Condensates may also enhance or suppress reactions through mechanisms beyond mass action, such as by controlling the structural organization or dynamics of molecules (Figure 2A–C). These functions have been described in diverse systems.
Figure 2 |. Molecular scale functions of biomolecular condensates.
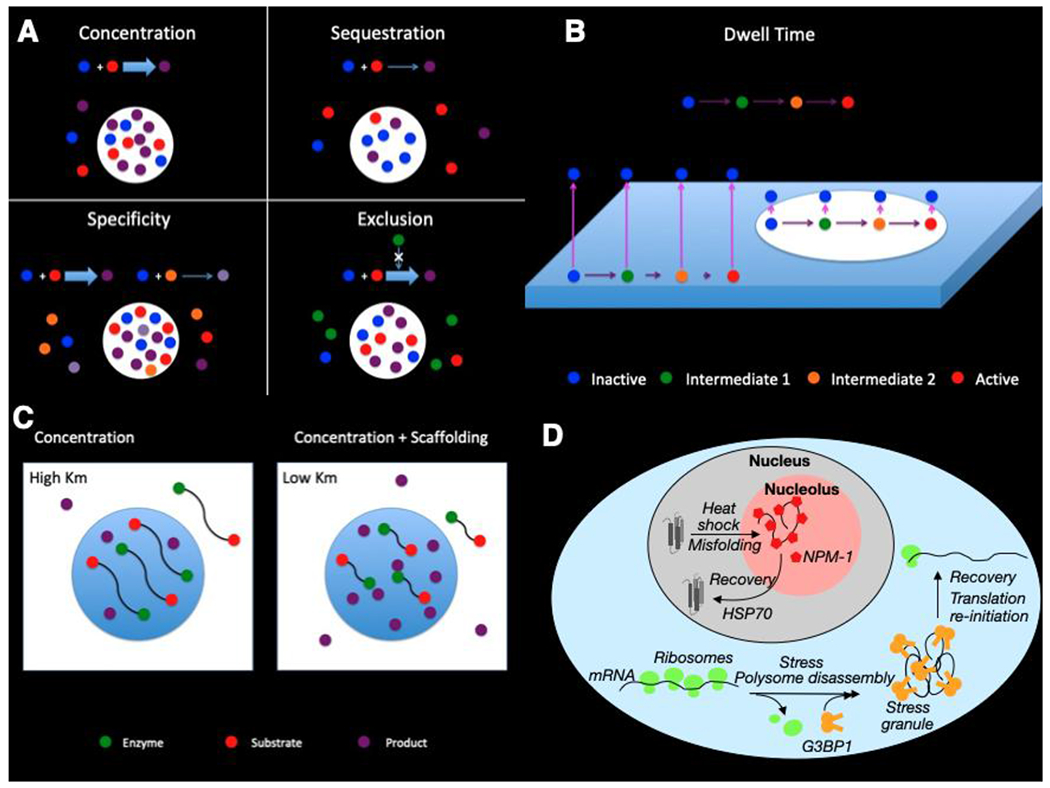
A. Concentration of enzyme (E) and substrate (S) can be enhancing or inhibiting depending upon which are enriched in the condensates. Enrichment of both enzyme (blue) and substrate (red) into condensates results in a substantial increase in product (purple) within the condensate, with unchanged or decreased activity in the surrounding bulk. Enrichment of only enzyme (blue), for example, will result in higher condensate activity but lower bulk activity due to depletion. The overall activity decreases due to the relatively small condensate volume being unable to compensate for the loss of activity everywhere else. If there is more than one substrate competing for the same enzyme, selective enrichment of one substrate in the condensate promotes reaction specificity. In this scenario, substrate 1 (red) is enriched while substrate 2 (orange) is not, resulting in preferential activity toward the former. Product 1 (purple) is higher in the condensate and lower in the bulk, as in A, whereas product 2 (light purple) is lower everywhere. If there is an inhibitor (green) that is excluded, concentration of enzyme (blue) and substrate (red) within condensates gives a bigger increase than in A due to the combined effects of higher enzyme and substrate and decreased inhibitor.
B. Concentration-independent mechanisms such as dwell time and scaffolding can also enhance reaction rates. For slow reactions, decreasing the rate at which enzyme (red) diffuses away from the membrane increases the probability of a productive reaction (yellow). Concentration-independent mechanisms such as dwell time and scaffolding can also enhance reaction rates. For slow reactions, decreasing the rate at which enzyme (red) diffuses away from the membrane increases the probability of a productive reaction (yellow). Pink arrows represent diffusion off the membrane, and purple arrows represent reaction flux to the next step
C. Molecular organization can increase product (yellow) by closely tethering enzyme (red) and substrate (green). If tethering is sufficiently close, this can result in an apparent decrease in Km, accelerating the reaction under otherwise equivalent conditions.
D. Nuclear proteins unfold upon heat shock and are recruited into the granular component of the nucleolus, where interactions with NPM-1 (red pentagons) maintain misfolded proteins in a state where refolding can occur aided by molecular chaperones including HSP7027. In the cytoplasm, stress causes polysome disassembly, leading to formation of stress granules by G3BP1 and other proteins, preventing base-pairing and aberrant RNA aggregation.
Regulation of Macromolecular Folding State.
The chemically and physically distinct nature of condensates compared with bulk cytoplasm may facilitate changes in the folding state of macromolecules upon partitioning into the condensate. Similarly, interactions between unfolded macromolecules and condensate-forming proteins can suppress aggregation, facilitating refolding after stress has subsided. This function appears to operate in the nucleolus and stress granules under stress conditions (Figure 2D)27–29.
Vectoral Organization of Biochemistry.
Many biochemical processes occur via multi-step cascades of substrate modification, including production of ribosomal RNAs in the nucleolus, a condensate with multiple subphases30,31. In such systems, enzymatic modification can be vectorially coupled with net transport from one subphase to the next, potentially enhancing efficiency of these pathways (Figure 3A).
Figure 3 |. Mesoscale functions of biomolecular condensates.
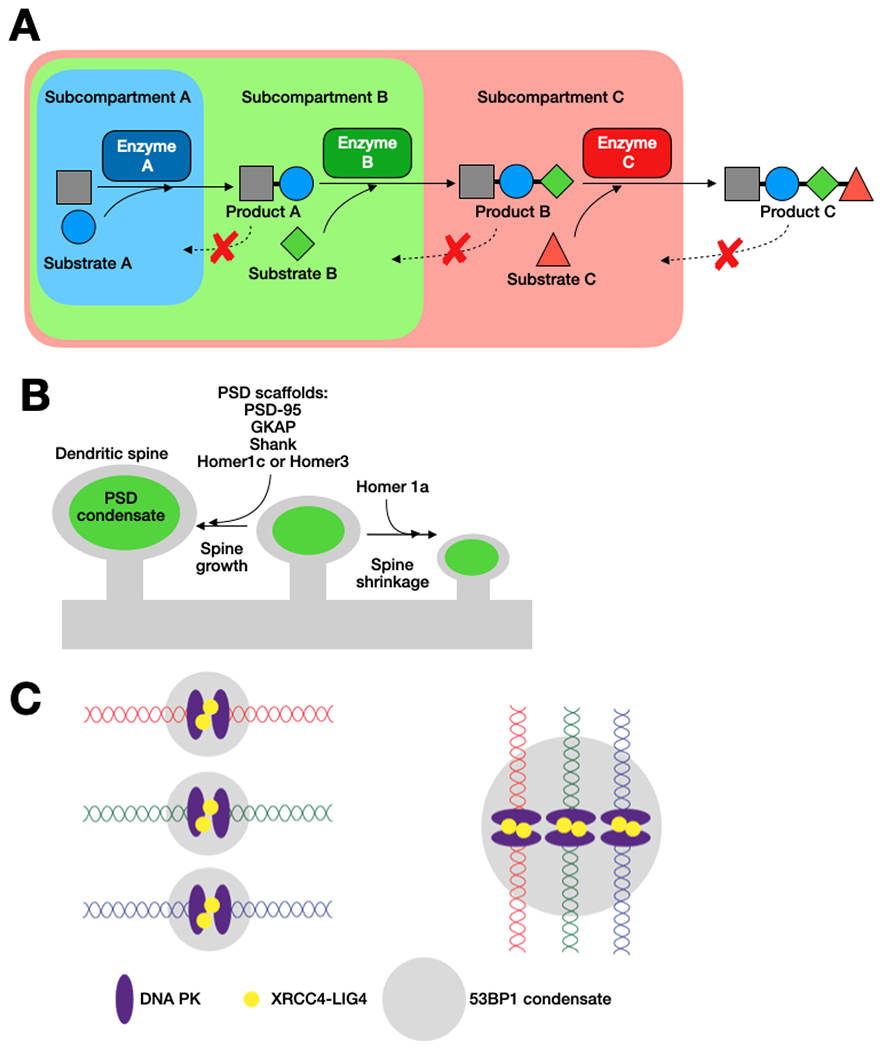
A. In condensates with more than one subcompartment, differential enrichment of enzymes in different subcompartments can lead to vectoral modification of substrates if substrate molecules are enriched in a given phase, but reaction products are excluded from the phase30,31.
B. Size-scaling of dendritic spines dictated by growth of the PSD condensate, composed of PSD-95, GKAP, Shank, and Homer1c or Homer 3. Homer1a causes dispersal of the PSD condensate, potentially causing shrinkage of the dendritic spine.
C. Representation of architectural role of 53BP1 condensate in DNA damage repair by non-homologous end joining, an example of how condensate formation and fusion can shape genomic architecture to facilitate joining of potentially distant DNA ends. 53BP1 promotes stabilization of the break and facilitates recruitment of repair components. Binding of the DNA ends by DNA PK recruits the XRCC4/LIG4 complex promote religation96,198.
Establishing Mesoscale Architecture.
Operating on micron length scales, condensates may serve an architectural role via their material properties, for example bridging distant DNA double-stranded breaks via condensate fusion to facilitate repair (Figure 3B)32. These functions may use material properties to organize large objects in space, and simultaneously modulate reaction rates within the condensate, highlighting that our functional typologies are not mutually exclusive and depend on the length scale under examination (Figure 1).
Facilitating Specific Cellular Localization.
Cellular localization is one of the best characterized functions of condensates in vivo, beginning with foundational work on P-granules in C. elegans embryos33. Cells can localize molecules via specific dissolution and assembly of condensates, or by coupling condensates to motor proteins for transport (Figure 4A)34. Efficiency of motor-driven transport is greatly enhanced by packaging many molecules into a condensate that can be trafficked as a discrete structure. This mechanism operates on cellular length scales, and can extend over meters in mammalian neurons.
Figure 4 |. Cellular scale mechanisms of biomolecular condensate functions.
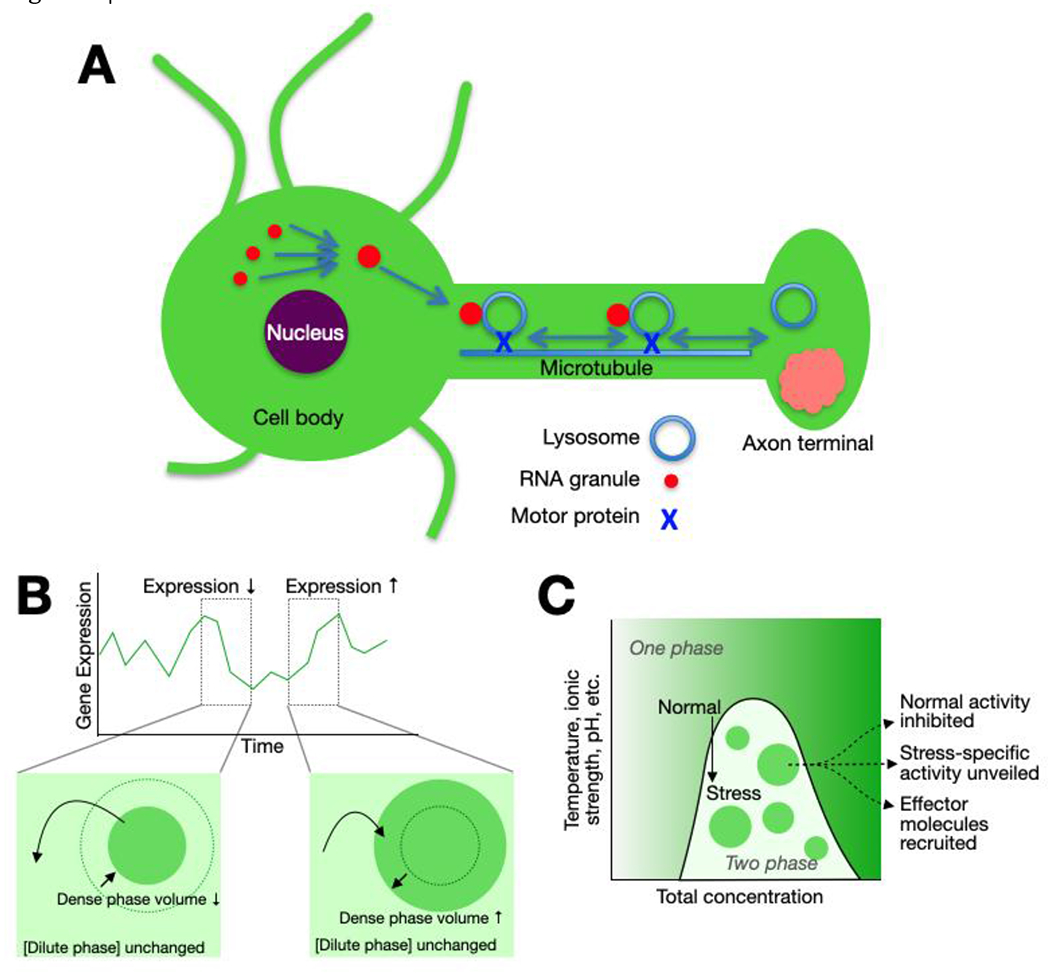
A. Localization of neuronal RNA granules via hitchhiking on lysosomal traffic. RNA granules formed in the cell body following transcription are tethered to a lysosome via the protein ANXA11, facilitating transport along microtubules to the site of translation in the axon (or dendrite), where RNAs are released for subsequent translation34.
B. Condensates formed via LLPS can integrate information about macromolecule concentration over the entire cellular volume to buffer stochastic fluctuations in gene expression. In the two-phase regime, transient fluctuations in protein concentration are buffered by transfer of molecules into or out of the dense phase, changing the dense phase volume while leaving the dilute phase concentration unchanged35.
C. Condensate formation integrates information at a cellular scale by coupling changes in the environment to the transition from the one-phase to two-phase regimes and can mediate activation of appropriate homeostatic responses19,20.
Buffering Stochastic Cellular Noise.
Single-component systems undergoing LLPS exhibit a threshold concentration above which two coexisting liquid phases form. In this two-phase regime, the dilute and dense phases maintain constant concentrations, with any fluctuation in total concentration (e.g. from the stochastic nature of gene expression35) manifesting as changes in the relative volumes of the two phases (Figure 4B). By integrating concentration information over the entire cell volume, this mechanism operates at large length scales and could potentially be coupled with other mechanisms that operate at shorter length scales in the condensate
Sensing and switching.
Phase-separated systems exhibit rapid transitions between the one- and two-phase regimes upon changes in the chemical or physical environment. For example, changes in temperature and/or pH during heat stress drive rapid condensation of Pab1 in budding yeast19. Cells may use these transitions to sense environmental stresses and activate homeostatic responses (Figure 4C). Again, this mechanism integrates information about the cellular environment over large length scales and is likely coupled to modulation of biochemistry at short length scales (Figure 1).
In the sections that follow, we review illustrative evidence for the existence of these classes of functional mechanisms from the recent literature, highlighting particularly compelling pieces of evidence, noting some limitations of existing studies, and suggesting particularly fruitful lines of future inquiry.
Molecular-scale functions
Enhancement or suppression of biochemical reaction rates
One of the best-studied and most intuitive examples of short length scale condensate function is modulation of biochemical activity. Since condensates concentrate certain molecules, and can potentially exclude others, rates of chemical reactions (binding, catalysis, etc.) should be different inside and outside of the structures according to the law of mass action. Recent data have borne out this prediction in a variety of biological systems, and shown different modes of regulation depending on which molecules are concentrated (sections A-C below). Physical mechanisms beyond simple mass action have also been demonstrated to control chemistry within condensates (section D). We describe recent, illustrative examples of these mechanisms below.
A. Enhanced activity through concentrating enzymes and substrates.
A variety of studies in diverse biological systems have shown enhanced catalytic activity within condensates due to co-concentration of enzymes and substrates (Figure 2A). One such example has been proposed to accelerate the rate-limiting step in carbon fixation, carboxylation of ribulose bisphosphate (RuBP) by the enzyme ribulose bisphosphate carboxylase/oxygenase (Rubisco)36. Rubisco is a very slow enzyme and has a nonproductive side reaction, involving oxygenation of RuBP instead of carboxylation. Prokaryotic and eukaryotic photosynthetic organisms have independently solved these problems by evolving distinct multivalent ligands that interact with the octameric rubisco to drive LLPS37–41. These condensates each concentrate rubisco and CO2, greatly enhancing carboxylation activity42,43. Mutations disrupting Rubisco condensation or CO2 enrichment produce dramatic growth defects, strongly suggesting that concentrating Rubisco is required for full activity38,44–46. These data explain how photosynthetic organisms overcome Rubisco’s shortcomings through compartmentalization. Condensation thus enhances both concentration and specificity as activity increases due to enzyme and substrate concentration and also preferential concentration of CO2 over the competing O2 substrate (Figure 2A). For these and other condensates for which function arises at least partly from increased concentration of small-molecule substrates, it is important to understand the nature of the interactions between the condensate and small molecules. For example, certain IDR sequence features may promote interactions with hydrophobic or charged small molecules. Metabolomics experiments, coupled with computational studies, may help discern whether certain classes of small molecules are preferentially enriched in condensates and identify molecular mechanisms of enrichment.
A second example of this phenomenon can be found in the innate immune system DNA sensor, cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) synthase, (cGAS). Both in vitro and in cells, cGAS phase separates upon binding long nucleosome-free double stranded DNA47. Structural studies indicate that DNA also induces activating conformational changes in the cGAS catalytic site48. Together, concentration and allostery dramatically increase the enzymatic activity of cGAS in vitro and lead to increased cGAMP levels in cells49. Mutations in cGAS that abrogate condensation have large effects on activity47. Deconvolving the roles of condensation and allosteric activation remains challenging, though both are likely necessary for maximal activation. These examples demonstrate the ability of condensates to accelerate reactions through co-enrichment of enzymes and substrates.
A third example occurs in the RNAi pathway, where mRNA deadenylation is thought to occur in cytoplasmic condensates (RNA granules) containing Ago2. It was shown in biochemical reconstitutions that Ago2 makes multivalent interactions with its RNA granule ligand, GW182, which drive LLPS50. The resulting droplets concentrate the deadenylation enzyme complex, CCR4-NOT, and target mRNA substrates, accelerating the deadenylation reaction. Acceleration occurs either when both Ago2 and GW182 are above the LLPS threshold concentration, or at sub-threshold concentrations when LLPS is induced by crowding agents, suggesting that deadenylation is increased specifically via condensation. The role of condensation in vivo remains unclear, as previous work demonstrated very little effect on RNA silencing upon condensate disruption51,52. A possible explanation for this finding is that silencing may occur in condensates that are too small to observe using standard microscopy methods, a question discussed in general terms below. This possibility could be addressed through super-resolution imaging studies if activity-dependent fluorescent probes were developed to allow simultaneous analysis of condensate formation, structure, and enzymatic activity.
Nucleation-dependent processes are particularly sensitive to changes in concentration, and thus should be especially susceptible to regulation by condensates. This has been demonstrated in control of microtubule polymerization, a process that is rate-limited by nucleation and only occurs stably above a threshold concentration of the αβ-tubulin dimer53. Recent work biochemically reconstituted centrosomes, condensates that nucleate microtubules in vivo, through self-assembly of the key scaffold protein, SPD-5, along with other centrosome proteins PLK-1, SPD-2, ZYG-9, and TPXL-154. When maintained in a liquid-like state, these condensates recruit and concentrate αβ-tubulin, accelerating microtubule nucleation. Similarly, it was shown that TPX2, a protein that enhances nucleation of new microtubules that branch from the sides of existing microtubules, undergoes LLPS with αβ-tubulin55. These condensates interact with the microtubule lattice and accelerate branching nucleation in cell extracts. Nucleation enhancement correlates with αβ-tubulin enrichment, suggesting the condensate functions by increasing concentration to increase assembly rates. Similar mechanisms may be involved in the function of the mitotic spindle protein BugZ, which undergoes LLPS in vitro and recruits αβ-tubulin and the spindle assembly factor Aurora A kinase56,57. Together these studies illustrate the ways condensates may enhance formation of nucleated assemblies, which may occur in other biological contexts including cell-cell adhesion58,59.
When examining rate enhancement via simple mass action as described in the examples above, it is important to consider how enhancement is constrained by the properties of specific enzymes and substrates, as recent experiments have demonstrated (Figure 2A). In a biochemical reconstitution of the SUMOylation cascade in engineered condensates, when two substrates are present only the one recruited into the condensates was SUMOylated efficiently (W.P., M.K.R., unpublished). However, the relative change in reaction rate due to condensate formation depends strongly on the KM for each individual substrate. Substrates present at concentrations far above their KM exhibit little change in reaction rate upon condensate formation as the enzyme’s catalytic rate is already saturated even in the absence of condensates. On the other hand, when substrates are present at concentrations well below their KM, formation of condensates results in strong reaction rate enhancement as the enzyme’s reaction rate is far from the saturation regime. Thus, reaction rates for specific substrates can be enhanced upon condensate formation while others change only marginally, dictated by their concentrations and KM. This may play an important role in pathways such as RNAi, where hundreds of thousands of different substrates can compete for the same enzyme.
B. Inhibition of activity through sequestration
In addition to enhancing activity, condensates may act to decrease activity by sequestering molecules away from their sites of action and/or substrates (Figure 2A). As detailed below, this activity requires substantial depletion of molecules from the cytoplasm/nucleoplasm, which demands very large condensates and/or very high degrees of concentration in them. Nevertheless, one system that appears to act by sequestration is the nuclear condensate, paraspeckles. The primary scaffold for paraspeckles is the long non-coding RNA, NEAT1, which recruits the transcriptional regulator protein SFPQ. It has been shown that proteasome inhibition or viral infection upregulate NEAT1 transcription and enlarge paraspeckles, thereby depleting SFPQ by up to 50% from the nucleoplasm60. This depletion then leads to repression of SFPQ target transcripts, providing an example of how sequestration of proteins within condensates can exert regulatory functions.
A second interesting example of sequestration comes from plants, in which the auxin response factor (ARF) proteins, ARF7 and ARF19, are localized primarily within cytoplasmic condensates. Cytosolic sequestration of these transcription factors inhibits their nuclear translocation, blocking their gene regulatory activities61. Cell-to-cell differences in ARF7/19 nuclear localization give rise to varying auxin responsiveness, such that actively growing cells respond quite differently from quiescent cells even in the presence of the same hormone signal. Sequestration in cytoplasmic condensates thus represents a short length-scale biochemical inhibition that is coupled to cellular-scale effects on nucleocytoplasmic transport and response to signaling molecules, demonstrating how condensates can have inter-related functions at different length scales.
Together, these examples illustrate how condensates can also inhibit activity through sequestration of transcription factors away from their sites of action, an effect that can be readily tuned and reversed upon environmental changes or signaling.
C. Modulation of activity through exclusion of factors
In addition to recruiting components, condensates can also enhance activity by excluding a negative regulator (Figure 2A). Two recent studies have demonstrated this effect in vitro. The first involves T-cell receptor (TCR) signaling clusters, which form in T cells upon antigen recognition62–64. In this system, TCR activation causes phosphorylation of the scaffold protein, LAT, which undergoes LLPS with a group of adaptor proteins. LAT phosphorylation is antagonized by the phosphatase, CD45. In a biochemical reconstitution, LAT clusters partially exclude CD45 due to charge-mediated repulsion, thereby stabilizing the clusters against dephosphorylation and dissolution63. The authors speculate that in cells, CD45 exclusion could provide positive feedback in LAT phosphorylation, prolonging TCR signaling and increasing sensitivity to antigen stimulation.
The second study investigated condensation of components of the postsynaptic density (PSD), a protein dense structure that clusters postsynaptic ion channels for activation in response to neurotransmitter release65,66. Synapses can be excitatory or inhibitory, and their developmental pathways are mutually exclusive, suggesting competition between the two outcomes. Gephyrin is an inhibitory postsynaptic scaffold protein. The authors find that gephyrin is partially excluded from biochemically reconstituted PSD condensates by an unknown mechanism not based on molecular size. This exclusion could have ramifications for establishing either an excitatory or inhibitory synapse during development. These two examples illustrate how exclusion of molecules from condensates can have both short length-scale effects via changing biochemistry, and large length-scale effects on synaptic function.
In general, the physical mechanisms of exclusion from condensates are unclear. Condensates are composed of macromolecular networks, which result in variously sized pores that can impose diffusion barriers. With improved physical models of condensates, it may be possible to determine constraints on charge density required for charge repulsion-mediated exclusion, and to discover sequence features that dictate effective pore volume of condensates and eventually whether evolution has acted on condensate porosity to favor recruitment or exclusion of molecules.
D. Factors beyond concentration that modulate enzyme activity
Do condensates modulate activity solely by concentrating selected groups of molecules, or do they also operate through other mechanisms related to their internal structure, material properties, or other features? Recent studies have shown that indeed, condensates can act beyond mass action to control internal reactions through changes in membrane dwell time of constituent proteins (Figure 2B), the effects of multiple phases, and molecular organization (Figure 2C).
It was recently shown that in systems where phase separation occurs on lipid bilayers, membrane dwell time of constituent proteins is substantially higher within the condensate than on surrounding regions of the membrane. It was further suggested that this could lead to enhanced signaling when reactions are slow, multi-step, and driven out of equilibrium, through a process akin to kinetic proofreading67. Classically, kinetic proofreading occurs in multistep reaction pathways when desired and undesired substrates exhibit different rates of dissociation from an enzyme, resulting in enhanced specificity. In such systems flux through the pathway is correlated with dissociation rate(s) of key steps. In the context of membrane-associated condensates, the analog of dissociation from an enzyme is dwell time on the membrane. Thus, flux increases within the condensate because dwell time is longer there. (Figure 2B). These predictions were borne out in two recent studies. The first study examined the nucleation of actin filaments by membrane-associated signaling clusters, a process that meets the criteria for kinetic proofreading68. It was found that the specific activity (filament nucleation activity per molecule) of the cluster components N-WASP and Arp2/3 complex, were strongly correlated with dwell time. A parallel study examined the activation of the Ras GTPase by distinct, but analogous signaling clusters69. This system also meets the criteria for kinetic proofreading, and Ras activation was dependent on the membrane dwell time of the guanine nucleotide exchange factor, SOS. These observations demonstrate the importance of physical properties (which are determined by interaction kinetics and network structure), beyond simply concentration, in determining how condensates affect molecular activities. Both studies also reported that since dwell time is related to network connectivity, which is determined by relative stoichiometry of condensate components, stoichiometry can control activities of molecules within the condensate. This mechanism represents a new means of regulation that is unique to the stoichiometrically variable nature of phase-separated structures.
In addition to dwell time, it has also been shown that the molecular organization imparted by condensate scaffolds can also enhance activity (W.P. and M.K.R., unpublished). When the SUMOylation cascade is recruited into condensates, close spatial tethering of enzyme and substrate upon binding the scaffolds decreases the apparent KM of the reaction. Thus the SUMOylation rate is higher in the condensate than when enzyme and substrate are at equivalent concentrations but are not recruited into condensates. Increasing the length of the scaffold, which increases the distance between enzyme and substrate, eliminates this effect on KM, and condensate enhancement becomes purely concentration-based (Figure 2C).
Thus, both temporal and spatial mechanisms can produce activity beyond that predicted by mass action alone. This indicates that condensates have generic concentration-based effects but also pathway/reaction-specific effects, such that maximal activity is achieved only when condensate properties are tuned for a particular reaction.
Aside from dwell time and molecular organization, the ability of condensates to form multiple phases can also significantly impact activity70. A system consisting of the RNA-binding proteins, FMRP and CAPRIN1, and RNA, can form either single-phase or two-phase condensates depending on which protein is phosphorylated. Single-phase condensates formed by phosphoFMRP and CAPRIN1 homogeneously recruit RNA and the deadenylase, CNOT7. Two-phase condensates formed by FMRP and phosphoCAPRIN1 concentrate RNA and CNOT7 into distinct inner and outer phases, respectively. Despite spatial segregation, deadenylation activity is higher in the two-phase system than the one-phase system, suggesting that co-enrichment does not always lead to higher activity. In fact, CNOT7 activity toward RNA is the same in the presence or absence of the single-phase condensates. A similar lack of enhancement was also observed in ubiquitination reactions occurring within condensates of SPOP and DAXX 71. In both systems, further quantitative studies could reveal whether the lack of enhancement is due to inhibitory activities within the condensate that counter the effects of concentration, or simply to saturation of the enzyme systems at the concentrations employed. As an example of the converse situation, TIS granules, endoplasmic reticulum-associated condensates formed by the RNA-binding protein TIS11B, are necessary for assembly of complexes of membrane proteins and the protein SET, despite SET being present at higher concentration outside the condensates72. This indicates mechanisms beyond mass action are at play in enhancing the assembly reaction within TIS granules. Together, these data suggest that localization both to and within condensates can have significant, and sometimes non-intuitive, effects on function.
These examples demonstrate that condensates can modulate enzyme reactions through both mass action and other physical mechanisms. The frequent observation of such modulation in mechanistic studies suggests that control of function on short, biochemical length scales is likely to be pervasive in condensate biology.
Regulation of macromolecule folding state
Organisms have evolved elaborate mechanisms for preventing aggregation of biomolecules during heat shock or other stresses, and the unique properties of condensates relative to bulk cytoplasm or nucleoplasm may facilitate these processes. A recent study on protein misfolding in the nucleus suggests that the nucleolar granular component (GC), a phase-separated condensate enriched in nucleophosmin 1 (NPM1), exhibits non-enzymatic chaperone activity in concert with the ATPase chaperone Hsp7027 (Figure 4D). Upon heat stress, a thermally unstable reporter enzyme and several native proteins migrate into the GC (Figure 2D). After recovery from heat stress, misfolded proteins exit the GC in an Hsp70-dependent manner, with the reporter enzyme recovering lost enzymatic activity. Dissolving the nucleolus abolishes the chaperone activity of the GC, resulting in amyloid-like aggregation of nuclear proteins. RNA can also form aberrant aggregated structures, which have been implicated in neurodegenerative disease11 and may form during stress-induced translational arrest when mRNA is released from polysomes73. Upon insults including heat, osmotic, or oxidative stresses, cytoplasmic condensates termed stress granules (SGs) form dependent on the RNA-binding protein G3BP174–77. G3BP1 forms condensates with long unstructured RNA, thereby inhibiting formation of entangled, base-paired RNA species29. This may facilitate recovery of mRNA and re-initiation of translation after stress subsides. DEAD-box RNA helicases may play similar roles in other RNA granules, including processing bodies, as they form condensates with RNA that dissolve upon stimulation of their ATPase activity28.
These studies demonstrate how the properties of condensates, acting on short length scales, can prevent aggregation of both proteins and RNA. For G3BP1 in stress granules, co-condensation with unstructured RNA appears to sterically occlude potential base-pairing interactions. The mechanism by which the nucleolus aids refolding remains unclear, though a conceptually similar mechanism may operate, with aggregation-prone unfolded protein regions bound by NPM-1. Alternatively, the material properties of the condensate itself may favor certain conformations of macromolecular chains or assemblies in a manner that suppresses aggregation or favors folding. Condensates formed by the IDR of Ddx4 have been shown to destabilize duplex DNA and favor compact single-stranded oligonucleotide conformations, as free energy is minimized by oligonucleotide conformations that minimally distort the mesh-like structure of the condensate interior78. The chemical environment within condensates may also be distinct, with measured polarities approximately 50-70% that of water13,79. NMR studies may allow these mechanisms to be distinguished by interrogating conformations, interactions, and dynamics of misfolded macromolecules within condensate dense phases. Additionally, FRET reporters for the conformation of misfolded proteins could be developed to enable in vivo interrogation of chaperone activity.
Mesoscale functions
Vectoral Organization of Biochemistry
Efficient production of biomolecules requires high catalytic efficiency without product inhibition, competing back-reactions, and formation of dead-end products by non-specific activity. Two properties of condensates formed via LLPS provide potential solutions to these constraints. First, solutions containing multiple macromolecules can produce multiple dense phases, allowing different enzymes within a cascade to be concentrated in different compartments. Second, condensates can concentrate or exclude molecules to varying degrees depending on the physical properties of the molecules, so condensates may recruit substrates then exclude them following enzymatic modification. These properties combine to yield micrometer-scale organization where a molecule can be vectorially transferred from one dense phase to another in a manner dependent on enzymatic modification in each phase (Figure 3A). The best-studied example of vectoral organization of biochemistry via biomolecular condensation is production of ribosomes in the nucleolus, where pre-ribosomal RNA is transcribed in the innermost phase and is processed and assembled with ribosomal proteins as it transits through the outer phases 30,31,80,81. As this process is reviewed elsewhere in this issue, we will briefly discuss another, recently discovered condensate with properties that may also facilitate vectoral biochemistry.
A genetic screen in C. elegans identified a requirement for the proteins ZNFX-1 and the Argonaute homolog WAGO-4 in transgenerational inheritance of RNA interference82. In early germline progenitor cells, ZNFX-1 and WAGO-4 colocalized in P-granules, phase-separated ribonucleoprotein condensates where small RNA biogenesis and post-transcriptional regulation occur33. Later in development, ZNFX-1 and WAGO-4 transition to perinuclear puncta termed Z-granules, which are found immediately adjacent to P-granules and exhibit properties consistent with formation via LLPS. Remarkably, Z-granules further associate with yet another punctate body, Mutator foci, that contains proteins involved in siRNA amplification and RNA silencing83. Thus, Z-granules can form a bridge connecting P-granules with Mutator foci.
While mechanistic details have yet to be uncovered, the intimate association of three condensates with roles in RNAi provides a compelling reason for further investigation with the vectoral processing framework in mind. Studies of nucleolar biochemistry provide an excellent path toward addressing this possibility. For vectoral organization in general, the field would benefit greatly from new reporters for enzyme activity that can be read out spatially via fluorescence microscopy (Box 2).
Box 2 | Technological Advances.
Scientific advances are often driven by technological advances. As the great molecular biologist Sidney Brenner once said “Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.” Below we list some of the questions currently of interest in the field, why they are important, and technological advances that would foster their answers.
Q: What are the compositions of proteins, RNA and small molecules in condensates, and how do they arise from molecular properties and environmental conditions? Relatedly, how are molecules (both macromolecules and small molecules) excluded from certain condensates?
I: Composition is a primary determinant of condensate physical properties and functions.
Combined development of high-throughput cell engineering, quantitative imaging and mass spectrometry, as well as improved probes, particularly for small molecules.
Development of new theories to integrate experimental data and understand how multi-valent, multi-component physical interactions drive partitioning into condensates.
Q: What functions arise specifically from higher-order assembly of individual molecules?
I: This issue is at the center of condensate biology, and its answer explains why condensates exist and have come about through evolution.
Methods to precisely and quantitatively enhance or inhibit higher-order assembly (e.g. phase separation) without altering activities of individual molecules. Current methods to achieve this are generally either non-specific (e.g. hexane 1,6 diol, solution conditions) or perturb interactions known to be functionally important.
Improved understanding of how surface properties of folded protein domains, sequence patterns of disordered proteins and sequence/structural features of RNA lead to assembly and/or phase separation (to design perturbations).
Spatially and temporally precise perturbations, e.g. using optogenetics.
Q: How can biochemical activities be measured inside condensates in cells?
I: Understanding the biochemical consequences of condensate formation in vivo remains a major challenge in the field.
Novel fluorescence techniques for rapid, live cell imaging at super-resolution and/or single molecule sensitivity.
High resolution imaging mass spectrometry to quantify small molecules in cellular condensates (see 170 for a beautiful recent example).
Novel probes for real time, spatially localized imaging of small molecule enzyme substrates and products, and activity-specific protein/RNA probes, such as a recently developed fluorescent probe for mRNA decapping171.
Q: What is the internal structure of condensates, and how does it relate to composition, dynamics and function?
I: Increasing data show that many condensates are not homogeneous, but are composed of subcompartments and structured elements. Structure-function relations are central in structural biology as they explain how activity arises from atomic organization. The issue is similar in condensates, but at larger length scales.
Multi-color super resolution imaging of fixed and live cells, single-molecule imaging
Correlative cryo-super resolution fluorescence imaging and cryo-electron tomography
Means of perturbing internal structure in cells and creating it biochemically, to observe effects on molecular behaviors and functions
Improved structure-specific probes (e.g. for amyloid fibers) for cellular analyses
Q: What is the range of mechanisms by which condensates form in cells, and how can these be clearly distinguished?
I: Formation of condensates is closely tied to their regulation and sometimes to their functions.
Advanced single molecule and super-resolution imaging, quantitative imaging in general
High-throughput cell engineering to observe and manipulate multiple molecules simultaneously
Improved theoretical descriptions of condensate formation and behaviors, particularly for small molecule numbers
Q: How do macroscopic material properties contribute to condensate function?
I: Some condensates sense and transduce forces, and it is unknown how the material properties of such condensates affect these activities. Material properties can also be regulated and are defective in neurodegenerative diseases.
Improved methods to exert quantitatively defined forces on condensates and monitor their functional responses in vivo, including optical trapping172, generation of intracellular flows173, and others.
Approaches to specifically modulate condensate material properties in cells--genetically, chemically, optically30—combined with measurements of function.
Methods to disrupt force transduction by condensates in vitro and in cells, e.g. laser ablation.
Q: Is the subcellular spatial organization of condensates functionally important?
I: Many condensates are not evenly distributed throughout the cytoplasm and nucleoplasm, but, for many, the reason for this is unknown.
Establishing mesoscale architecture
The previous sections describe mechanisms that modulate or organize biochemical reactions occurring at sub-micrometer length scales. How might condensates contribute to cellular function at larger scales?
The elaborate cellular morphologies of neurons provide two such examples. Both pre-synaptic axon terminals and post-synaptic dendrites contain local zones of electron-dense material. The mesoscale architectures of both the pre-synaptic active zones (AZ) and the post-synaptic density (PSD, see above) may be organized by condensates formed by LLPS65,84,85. The AZ and PSD play conceptually similar roles in the spatial organization of the pre- and post-synapse66,86. While the AZ clusters synaptic vesicles in proximity to voltage-gated calcium channels (VGCCs), the PSD clusters excitatory glutamate receptors. Two components of the AZ, RIM1α and RIM-BP, undergo LLPS in vitro84. Importantly, RIMα-RIM-BP condensates recruit VGCCs at a density similar to estimates from EM studies of native synapses, consistent with a role for LLPS in organizing the AZ. On the post-synaptic side, major proteins of the PSD—PSD-95, GKAP, Shank3, and Homer1c or Homer3—also undergo LLPS and form condensates anchored to membranes via interaction with the C-terminal tail of the NMDA receptor, thereby clustering and dramatically concentrating the receptor tail65,85. Homer1a, a splice isoform of Homer1, disperses PSD condensates, potentially providing a mechanism to control the size of dendritic spines and thus synapse strength (Figure 3B). Shank3 directly interacts with cortactin, an activator of the actin-nucleating Arp2/3 complex, providing a connection between the PSD and the actin cytoskeleton. These examples demonstrate how condensates can provide a physical means of maintaining cellular structures in proximity, such as neurotransmitter vesicles and VGCCs in the AZ condensate, as well as a mechanism for controlling the size of cellular structures, as postulated for Homer1a (Figure 3B). Condensates may play roles in the mesoscale organization of other membrane-bound structures, including clustering of mitochondria in dormant oocytes by the Balbiani body87, and potentially in organization of the Golgi apparatus88. These examples suggest organization of membrane-bound organelles by membraneless condensates may be an important means of generating cell organization.
A second recent example where condensates play an important architectural role, generating mesoscale organization, is in DNA damage repair (DDR) (Figure 3C). DNA double strand breaks (DSBs) are toxic to cells, and can lead to chromosomal translocations if misrepaired. DDR poses a spatial challenge for the enzymes involved, as DNA ends can diffuse away from each other. It has long been known that condensates containing various DNA repair factors form rapidly at sites of DNA damage repair89,90. Phase separating proteins targeted to specific genomic loci can mechanically exclude chromatin while preferentially incorporating distant targeted loci via coalescence of multiple condensates, indicating the material properties of nuclear condensates, including potentially DDR foci, can reshape the mesoscale architecture of the genome91.
Shortly after induction of DNA damage, FUS, a protein known to phase separate and form amyloid fibers, and polyADP-ribose (PAR) polymerase I (PARI) localize to DSBs92,93. Condensates form via interaction between FUS and PAR generated by PARI at the DSB. PAR removal by PAR glycosylase results in rapid dissolution of the FUS foci92. Once FUS foci are dispersed, they can be replaced by p53 binding protein 1 (53BP1) foci94,95, liquid-like condensates that form via interactions between 53BP1 and long non-coding RNA transcribed near the DSB96,97, committing the break to repair by non-homologous end joining. Together, FUS and 53BP1 condensates can reshape the mesoscale architecture of the nucleus in a manner that facilitates repair of DNA damage, with accompanying biochemical effects likely occurring at shorter length scales. A further example occurs in budding yeast, where DNA damage foci not only form and fuse but are physical translocated by nuclear microtubules to the nuclear periphery where repair takes place32. Condensates formed by the metazoan DNA replication machinery may serve analogous architectural roles by bringing distant replication origins into close proximity in replication factories, the spatial organization of which is believed to coordinate firing between different origins98–100.
Formation of condensates has also been implicated in autophagy, wherein a double-membrane structure termed the autophagosome is synthesized around cytoplasmic components, facilitating their degradation by fusion with a lysosome101. Polyubiquitinated proteins targeted for autophagy form condensates via multivalent interactions with p62102, which forms filaments and acts as a cargo receptor by interacting with Atg8/LC3 on the autophagosome inner membrane 103,104. Thus, condensate formation is a mechanism to generate discrete mesoscale structures that can be specifically targeted for autophagic degradation. Similar results have been obtained for the protein Ape1 in yeast, which forms condensates that recruit Atg19 at the interface between dense and dilute phases, which acts as a cargo receptor for the autophagosome105. In addition to these examples, the pre-autophagosomal structure (PAS), which forms on the yeast vacuole under starvation conditions, is a condensate formed by the complex composed of Atg1, Atg13, Atg17, Atg29, and Atg31106. This condensate is specifically targeted to vacuoles by the membrane protein Vac8, again facilitating formation of discrete mesoscale structures that are then engulfed by the autophagosome. The kinase activity of Atg1 is enhanced within the PAS condensate and must be balanced by phosphatase activity to prevent condensate dissolution, highlighting the fact that short-length scale biochemical functions of the condensate operate at the same time as its mesoscale functions.
Given the relatively simple reconstitution experiments performed thus far it is unclear whether establishing mesoscale architecture through the physical properties of a condensate91 versus controlling molecular scale biochemical activities is most important in cells. The answer will likely vary from system to system. To address this issue further, experiments that break the large scale organization without disrupting the smaller scale biochemistry are necessary. In the yeast DDR example, this could be done through microtubule disruption, but would be difficult in the mammalian DDR, AZ, or PSD systems. This might require optical trapping and chemo- or optogenetic techniques to prevent fusion of multiple clusters or change their material properties. Finally, for the neuronal systems, improved live-cell and intravital imaging studies are also needed to better characterize the structure and function of AZ and PSD condensates in vivo.
Cellular-Scale Functions
Facilitating Specific Subcellular Localization
Cells are highly spatially organized at multiple levels, from the micron-scale architectures of endomembrane systems through the potentially meter-scale organization of mammalian neurons. Such large-scale organization means proteins, RNAs, or vesicles may be produced far from the site of their function. Cells have thus evolved active mechanisms by which to transport these structures to specific locations and retain them there107. Recent data have shown that biomolecular condensates have important functions in controlling subcellular localization of specific molecules and processes.
Membrane-bound organelles, such as endosomes and mitochondria, can traffic along microtubules through direct binding to microtubule motor proteins (reviewed in 108). While occurring in most cells, the need for microtubule based transport is particularly acute in neurons, where molecules and assemblies must be transported from the cell body to the ends of axons and dendritic arbors, which can be microns to meters away109. Protein translation in neurons often occurs locally at these distant sites, necessitating transport of numerous mRNA molecules and ribosomes large distances110. Transport efficiency is greatly enhanced by packaging mRNA into RNA transport granules, which are liquid-like protein-RNA condensates akin to stress granules and P-bodies in non-neuronal cells111. These condensates form in the cell body, are trafficked by motors along axons and deposit their cargo at the axon terminal or growth cone111,112 (Figure 4A). A recent study reported that neuronal RNA granules are coupled indirectly to motors through tethering to lysosomes, which are directly coupled to motor proteins34. The authors identify the protein, annexin A11 (ANXA11) as a key tethering factor. ANXA11 can form condensates and colocalize with RNA granules, and also binds to lysosomes in a PIP3 and Ca2+-dependent manner. These properties allow it to attach granules to lysosomes. ALS-associated ANXA11 mutations alter the dynamics of RNA granules and impair their interactions with lysosomes, causing defects in granule trafficking and mRNA delivery to growth cones, highlighting the importance of these condensates and their ability to efficiently translocate mRNA via organelle hitchhiking34.
Chromatin in the eukaryotic nucleus is highly heterogeneous and organized across a range of length scales into compositionally and functionally distinct domains113–116. One high level of organization involves the division into transcriptionally active euchromatin and transcriptionally repressed heterochromatin. A major question in chromosome biology is how this organization is maintained despite profound short- and long-range reorganization during DNA replication and cell division. Heterochromatin is defined in part through trimethylation of histone 3 at lysine 9 (H3K9me3), a modification recognized by heterochromatin protein 1 (HP1α). Phase separation of HP1α is thought to play an important role in compaction and biochemical identification of heterochromatin117–119. A condensate was recently found to play a role in maintaining heterochromatin in dividing neural precursors (NPs) and initiating the establishment of heterochromatin during differentiation120. Phase separation of the Prospero/Prox1 transcription factor is responsible for its retention at pericentromeric heterochromatin throughout mitosis, when many gene regulatory proteins, including HP1, leave chromatin and become diffuse121,122. This localization of Prospero allows rapid recruitment of HP1 and the H3K9 methyltransferase following mitosis, leading to heterochromatin compaction and spreading. The inability of Prospero to phase separate impairs heterochromatin maintenance and can lead to dedifferentiation, highlighting the importance of maintaining heterochromatin throughout rounds of cell division and in differentiation. Thus, like RNA granules, condensation of Prospero into a discrete body allows it to maintain localization at a specific nuclear location despite dramatic reorganization of the genome over the course of mitosis.
Buffering Stochastic Cellular Noise
Due to the stochastic nature of gene expression, cells exhibit fluctuations in protein concentration over time, a phenomenon known as gene expression noise123. Despite noise, biological processes are generally robust and precise in space and time. This robustness has been attributed to noise-resistant signal transduction network architectures, analogous to active noise-filtering concepts in engineering124. LLPS of individual proteins has been proposed to afford passive noise filtering, since in the two-phase regime, fluctuations in total concentration alter the volume of the condensed phase, but concentrations in both dilute and condensed phases are held constant (Figure 4B). Recent work has experimentally demonstrated the feasibility of such passive noise filtering through LLPS.
A theoretical model of LLPS in the presence of stochastic concentration fluctuations predicts a sharp decrease in dilute phase protein concentration noise at the phase separation threshold concentration, with the ultimate reduction in noise depending on protein lifetime and rate of diffusion between dense and dilute phases35. When transfer of protein molecules between phases is much faster than protein synthesis and degradation, the minimum noise approaches the theoretical lower limit. Phase separation of both a synthetic protein and the endogenously expressed nucleolar component nucleophosmin results in reduced concentration fluctuations, consistent with the model.
While a convincing proof-of-concept, it remains to be seen whether native systems exploit LLPS to buffer noise in a functionally significant manner. As noise buffering via this model absolutely depends on condensate formation via LLPS, any native condensates proposed to serve a buffering role must be conclusively shown to form via LLPS. Further theoretical developments are also necessary to encompass any noise buffering effects of multicomponent phase separating systems, which are more prevalent in vivo, where the saturation concentration depends on the total concentration of each component125 (see below).
Sensing and switching
Given the extraordinary sensitivity of phase transitions to solution conditions including temperature, pH, and ionic strength, biological systems may have evolved to exploit them as a mechanism to sense potentially deleterious environmental conditions and induce appropriate homeostatic responses126 (Figure 4C). A compelling example of such a mechanism occurs in budding yeast, where the poly-A binding protein Pab1 undergoes rapid condensation upon heat shock19. Pab1 condensation is exquisitely sensitive to temperature, with a 10 °C increase in temperature accelerating the rate of condensation by more than 300-fold (versus 2–4-fold in other biological systems), with mutations perturbing temperature sensitivity reducing cellular fitness. Pab1 releases bound RNA upon heat shock and condensation, suggesting that translation of mRNAs with A-rich 5ʹ untranslated regions, which includes many heat shock protein mRNAs127, will be selectively enhanced under conditions that favor Pab1 condensation. A parallel pathway operates by very similar means via heat-induced condensation of the translation initiation factor Ded1, an RNA helicase that facilitates ribosomal start site scanning by resolving secondary structure in mRNA 5ʹ untranslated regions (UTRs) 128. Under heat stress, transcripts with complex 5ʹ UTRs are preferentially downregulated, favoring expression of mRNAs with simple UTRs, which includes the heat-shock transcripts with A-rich UTRs described above.
Sensing functions such as this, and similarly for Sup35 in budding yeast20, can be considered to operate at multiple length scales. As temperature, pH, and ionic strength affect physiology at the level of cells or even whole organisms, such stress-triggered condensation serves to integrate information at long length scales. At the same time, the biochemical function of the condensing molecule changes, with RNA binding, translation initiation, and translation termination activities inhibited for Pab1, Ded1, and Sup35, respectively19,20. Delineating the distinctions between sensing large-scale environmental conditions and short-length scale biochemical interactions is likely to be a fruitful avenue for future experiments. Characterizing the effects of other cellular-level changes in the cytoplasmic environment is also warranted, as nutrient availability decreases the crowdedness of the cytoplasm via the mechanistic target of rapamycin complex (mTORC) pathway downregulating ribosome abundance129. As condensate formation typically decreases as solution crowding decreases, this could be a potential mechanism by which nutrient sensation by mTORC is coupled to functions of condensate-forming proteins.
Formation of prions can also be considered a condensate-based switching behavior that operates on transgenerational timescales, rather than the transient responses described above. This is particularly intriguing given the frequently observed time-dependent maturation of liquid-like condensates to solid-like structures with amyloid features, as well as the well-known transition of Sup35 to the [PSI+] prion. Supporting this idea, the non-amyloid [SMAUG+] prion, which is a non-dynamic condensate composed of the RNA-binding protein Vts1, is thought to be induced by enhanced expression and self-assembly during nutrient replenishment after starvation130,131. The [SMAUG+] prion state of Vts1 enhances its mRNA degradation activity, creating a heritable post-transcriptional regulatory program that confers growth advantages in nutrient-limited conditions. Thus, it will be important to explore the timescales on which condensate-based switching behaviors occur, and how such behaviors are altered by transitions to amyloid- or prion-like states.
Challenges and opportunities for the future
Great progress has been made over the past several years in understanding the functions of biomolecular condensates across the biological spectrum. This work has revealed functions that span from molecular to cellular scales, and enabled generalities to be discerned. Nevertheless, interesting challenges and exciting opportunities in understanding the roles of biomolecular condensates remain. Below we describe some of the areas that we are enthusiastic about for the future and ideas about how progress may be made.
Understanding the Complexity of Natural Condensates
Most mechanistic studies of condensate function to date have been reductionist by necessity, both to reduce technical complexity and also because the basic principles were poorly understood. With technology advancing and basic principles increasingly clear, it will be important to understand how those principles are modulated by the complexity of native cellular condensates.
Natural condensates contain many molecular species with heterogeneous distributions of physical properties. Proteomic and imaging data have shown that tens to hundreds of molecules localize to individual condensates132–136. Concentrations of species within a condensate can vary from < 1 µM to > 10 µM132, with partition coefficients ranging from ~1 to > 10029,76,77,132. Some molecules have been shown to be excluded from condensates (partition coefficient < 1) in vitro63,65 and in cells137,138. Different species can have widely variable dynamic properties; some exchange with the surroundings in seconds and others exchange only over minutes or longer132.
In more collective aspects of complexity, molecules can contribute very differently to the formation and composition of condensates. An early model based largely on simplified synthetic systems classified molecules into scaffolds, whose assembly drives formation of the compartments, and clients, which are recruited into condensates through interactions with scaffolds14. It has become clear over time that in native condensates, the behavior of molecules spans a continuum from absolute requirement for condensate formation, to recruitment without altering condensate properties76. Although there tend to be few absolute scaffolds in any condensate and many clients, there can also be a number of species with intermediate behavior, whose deletion will shift the saturation concentration of other components (for systems that form by LLPS) by greater or lesser degrees. It is important to note that with even two species phase separating together, the simple features of single-component phase separation often invoked in the literature4,137,139–142 are not observed14,125,143. There is no longer a single phase separation threshold concentration, but rather the threshold of each species differs depending on the concentrations of others. Moreover, the concentrations in the condensate and surroundings are no longer invariant, but shift as individual components change, generating cell-to-cell and perhaps even subcellular variability. Mapping of genetic, proteomic and biochemical data onto known protein-protein and protein-RNA interaction maps has suggested that molecules with higher connectivity to other molecules in the condensate, and thus located more centrally in the condensate interaction network, are likely to behave more scaffold-like, while those with lower connectivity are likely to behave more client-like76,132. Effects on the threshold concentration of a phase separating condensate are also likely coupled to effects on composition, since highly connected molecules will recruit more species into the structure than those with only few interaction partners. So deletion of a more central molecule in the condensate interaction network is likely to strongly impact both formation of the structure and the collection of molecules contained within it.
These features have important implications for the behaviors and functions of natural condensates. Different components will have different physical signatures depending on how they contribute to a condensate (measurable by various microscopies (Box 2)). Scaffold-like components will show behaviors expected for a phase separated molecule, with a threshold concentration for assembly, and slow exchange dynamics that contribute strongly to macroscopic viscosity and surface tension. Client-like components will behave more akin to molecules binding a porous, static scaffold144, with Michaelis-like recruitment into the condensate, slow and fast dynamic components representing bound and free species, and less contribution to macroscopic properties. It remains unclear how coupled the physical properties of individual molecules are to one another, and thus how the properties of a condensate arise from its component collection. Thus, the extent to which alterations of some molecules (e.g. those with more scaffold like-or client-like behaviors) by natural regulatory factors or experimental perturbations will affect others is largely unknown. Importantly, it is also unclear how those physical properties, both the dynamics of individual molecules and material properties of the whole structure, will impact functions on different length scales.
Thus, to fully understand the formation and activity of a condensate, it is necessary to understand these features in detail. What is the collection of components in a condensate, and what are their concentrations and dynamics inside and outside of the structure? What is the interaction network among the species, and which molecules are more central or peripheral? Thus, which species are more likely to have scaffold-like and client-like behaviors? Obtaining this knowledge will require combining proteomics analyses with quantitative biochemical and biophysical studies to develop a quantitative understanding of composition and connectivity. The more complete this information, the better the physiology of a condensate can be captured in a biochemical reconstitution, and the more mechanistically one can understand the cellular behaviors of the compartment.
In summary, these considerations speak to the need for detailed, quantitative, in vivo studies of the physical properties of condensate components. In order to properly interpret these data, they must be further combined with an understanding of condensate complexity—the connectivity and binding affinities of the species—and physical theories that relate the properties of individual condensate species to the properties of the collective. Together, this information will allow us to understand how molecular scale features produce the cellular scale behaviors and functions of condensates.
The question of size
Some condensates are macroscopic in size, being much larger than the diffraction limit of light in standard microscopes and containing tens of thousands of molecules. These include compartments like the nucleolus and stress granules, and virtually all phase separated liquid droplets studied in vitro. However, in vivo many condensates are appreciably smaller, being <100-300 nm in diameter, and sometimes containing only tens to hundreds of individual molecular species. These include transcriptional foci, some signaling puncta and sub-diffraction RNP assemblies. It is unclear how the properties of such small particle number biological systems will compare to those that are much larger, and in general how function scales with size.
This is an interesting conceptual question but also one of practical importance, in correlating properties and functions of large phase separated droplets generated in vitro with their much smaller counterparts in cells. Relatedly, in cases where function has been ascribed to the higher order assembly of macromolecules, it is unclear at what size (molecule number) this new function arises. This latter question is important both in basic biophysics/biology, and also in development of therapeutic agents designed to disrupt condensates, a topic of considerable current interest (Box 3). That is, if functionality is manifest in small clusters, a potential drug cannot merely eliminate the macroscopic, readily observable condensate, but must reduce the system below the functional size threshold, which could be much smaller.
Box 3 | Condensates as Therapeutic Targets.
Our increasing understanding of condensate function provides opportunities to target the structures in treatment of disease. Defects in condensates have been implicated in numerous diseases. Most notably, defects in stress granules due to mutations in RNA binding proteins hnRNPA1, hnRNPA2, FUS, TDP-43 and others, as well as RNA regulatory proteins, have been associated with the neurodegenerative diseases amyotrophic lateral sclerosis, frontotemporal dementia and multisystem proteinopathy22,174. Dipeptide repeat proteins expressed from the c9Orf72 gene cause defects in multiple RNA-based condensates and the nuclear pore complex, and are also associated with ALS and FTD22. Aberrant nuclear condensates produced by repetitive RNA sequences can produce different degenerative diseases11. Defective condensates have also been implicated in various cancers, which are driven by fusions of self-associating sequences from proteins such as EWS, FUS and EML4 to transcription factors or kinases175–178. As described above, condensates also play roles in many biological processes relevant to disease, including innate and adaptive immune signaling47,63, DNA repair32,93, gene regulation149,150,153, cell adhesion179 and synaptic transmission65,84.
Thus, modulation of condensates has promise for broad impact in disease treatment. Many mechanisms can be envisioned for such modulation. Therapeutics could act directly on molecules within a condensate to disrupt their interactions and dissolve the structure. The small hydrophobic molecule lipoic acid has recently been reported to have such activity in vitro and in cells180. The formation and dissolution of many condensates are naturally regulated by covalent modification of components by enzymes141,181 such as kinases3,63,182–186, poly(ADP-ribose) polymerases187, methyl transferases13,16,188,189, ubiquitin transferases102,190 and acetyl transferases6,191, which could themselves be targeted therapeutically. Similarly, protein and RNA disaggregases control the turnover and dynamics of condensates28,192, and could be targeted as well. These strategies could be used to both destroy or enhance the formation of condensates, both of which could be valuable depending on the disease mechanism. In CAR T-cell therapy, increased phase separation through increased numbers of tyrosine phosphorylation sites could be used to increase the efficiency of signaling63,193. In neurodegenerative diseases, aberrant condensates appear to have slowed dynamics of their constituents, behaving more like solids than their natural liquids, and this change in material properties may contribute to pathogenesis22. In such cases, using sub-threshold amounts of disrupting agents, or distinct means of altering material properties, could reverse solidification and perhaps restore normal function. Capping amyloid fiber formation could be a mechanism to achieve this activity194–197. Because molecules act cooperatively to form condensates and define their material properties, it may be possible to target a disease-causing mutant protein indirectly, by altering the existence or activity of its neighbors in the structure. Finally, with a better understanding of the physical factors that control partitioning of small molecules into condensates, it may be possible to design inhibitors that selectively concentrate with their condensate-resident targets. Alternatively such compounds could be used to direct other inhibitory or activating molecules to such targets. Both approaches could enhancing potency or specificity of target modulation. Together, these strategies could provide novel means of treating disease based on the unique properties of biomolecular condensates. If these strategies could be generalized, they would also provide powerful tools to advance our basic understanding of condensate functions in biology. Such work thus presents exciting opportunities for future investigation, both practical and fundamental.
While we are not aware of studies of macromolecules that address these issues, some information can be inferred from theory, computer simulations, and experimental studies of colloidal and viral particles. These have shown that, unlike large-number systems, small systems do not show sharp phase transitions between discrete states145. Rather, transitions are broader, and intermediate states with intermediate compositions and densities exist. Stochastic fluctuations in assembly, existence and size are also significant146–148. Thus, very small condensates need not be long-lived and homogeneous, nor form in switch like fashion. These considerations may be relevant to resolving current disagreements in the transcription field, where some groups have argued that transcriptional foci form through LLPS based on correlations between in vitro phase separation of the transcriptional machinery and cellular behaviors of these factors149–152, while others have argued that the physical properties of transcriptional foci in cells (e.g. stochastic fluctuations, lack of a single threshold concentration) are inconsistent with a LLPS mechanism of formation137,153–156.
In functional considerations, an important parameter in small systems is surface tension, which can produce different structures/organizations of molecules in the center of an aggregate than at the periphery, which in turn can alter function. In simulations of assembly of small numbers (≤100) of water molecules or non-adhesive particles, surface tension increases as clusters grow, reaching macroscopic values at ~50-100 molecules/particles157,158. Analogous behavior has been observed experimentally in studies of weakly adhesive colloidal particles, with quantitative differences depending on the magnitude of inter-particle forces159. So for very small biological condensates, depending on the molecular interaction strengths, surface tension effects might be substantial. It has been proposed that the balance of interfacial tensions between phase separated droplets and membranes and between droplets and cytosol can be used to do mechanical work on the membrane160. Thus, since surface tension increases with the size of small droplets, the amount of work (and thus deformation of a membrane) could increase as droplets grow in such systems. CryoEM studies of phase separation of rod-like viral particles show that the transition from surface-like to interior-like structure can occur gradually over multiple particle layers161. Thus, surface tension may not manifest structurally only at the immediate periphery of a condensate, but can propagate internally as well. In very small condensates, more molecules may be more surface-like than interior-like in their organization and activities. Thus, studies of macroscopic droplets in vitro may not capture the functional nuances or features of their much smaller counterparts in cells. In general, physical studies, coupled with theory and simulation, focused on the scale dependence of the organization and activities of biomolecular condensates represents an interesting area of future investigation.
In addition to physical considerations of small condensates, it is important to note the biochemical implications of molecules forming small condensates inside cells132,137. Where quantified, condensates only take up a small fraction of the cell volume. Yeast P bodies, for example, are on average only ~1% of cytoplasm; PML NBs are only 0.2 – 2 % of the nuclear volume in PML−/− HeLa cells re-expressing PML (Allyson Rice, unpublished). Moreover, where quantified, most proteins are only concentrated 2-150-fold in cellular condensates, with most falling in the 2-20-fold range (it is possible that RNA molecules may partition more strongly due to kinetic trapping)29,76,77,132. Taken together with small size, this means that most molecules of a given condensate component are not located within the compartment, but rather are in surrounding cytoplasm or nucleoplasm. In yeast P bodies, for example, even when correcting for potential undercounting due to sub-diffraction assemblies, only four components of 31 studied were found to have > 50% of molecules in the condensate, while 25 had < 20% there132. Models based on inhibitory sequestration by condensates must account for such quantitative information. Similarly, models invoking enhanced activity within a condensate must also contend with the fact that much of the reaction can also take place through molecules outside the compartment. Because of these considerations, condensates may be most important biochemically in organizing cascades of reactions, where enhancements of reactivity and/or specificity in individual steps will be magnified when all components are concentrated together. In such cases, overall flux through a pathway could be vastly higher than in the surroundings, and condensation could provide substantial increases in overall activity or specificity. This may explain why molecules involved in common processes are often co-concentrated into condensates. These arguments point to the importance of quantitative analyses of condensate properties in order to accurately define their cellular functions.
Adaptation, evolution, and fitness value of biomolecular condensates
Several aspects of evolution have been mostly unexplored in regard to biomolecular condensates, and addressing some of these questions could enhance understanding of condensate functions. First, as has been proposed for other forms of colocalization, condensates may act as ‘crucibles’ for evolutionary adaptation, as client molecule colocalization at high concentrations may allow for novel interactions to arise upon a single point mutation. These interactions, if adaptive, could then be refined through further mutation and selection162. Second, comparative studies of homologous condensate-forming molecules in related organisms adapted to life in different climates could reveal how condensate formation and function have evolved to compensate for environmental difference. Finally, several studies have demonstrated impacts on organismal fitness upon disruption of condensates. Fitness analysis, more broadly applied, may contribute to a better understanding of the adaptive value of condensates.
How do functionally important protein-protein interactions evolve? A single point mutation is likely to produce only a very small increase in affinity for a non-specific interaction partner. When both molecules are free in solution, this change will increase the population of their complex only infinitesimally, producing no selective advantage. However, if molecules are colocalized, the system is tuned differently, and small increases in affinity can manifest as large differences in population of the complex. If these new interactions have adaptive value, for example by allosterically modulating the activity of one or both molecules, over time these mutations may become fixed in the population, allowing for further adaptation to favor the novel function162. In this sense, the high concentration of molecules within condensates may have adaptive value simply in producing colocalization, and thus enabling generation of novel functions through mutation and selection. Bioinformatic analyses of proteins containing amino acid homorepeat segments, which include many proteins that form condensates, have identified more rapid divergence at putative sites of protein-protein interaction than in proteins lacking homorepeats, consistent with the idea that condensates may facilitate evolutionary re-wiring of protein interaction networks163. The experimental route for examining such possibilities is challenging, but a potential proof-of-concept could lie in synthetic systems that recruit two or more client molecules that do not otherwise interact, such as a split enzyme system bearing mutations that prevent fragment complementation. Carefully designed selection schemes and experimental evolution164 could favor mutations allowing formation of the functional enzyme, with co-localization in the condensate allowing evolution of a functional enzyme in fewer generations than in populations lacking condensates.
Condensate formation via LLPS depends strongly on temperature and ionic strength, and should also be sensitive to pressure. Related organisms that live at, for instance, high versus low latitudes or are surface-dwelling versus deep-sea must have acquired adaptive mutations that favor condensate formation in the appropriate environmental conditions, as was recently demonstrated for the yeast protein Ded1p128. Little is known about how condensate-forming molecules change to compensate for life in different environments. Researchers should examine factors such as regulatory changes that favor higher or lower expression depending on environment, as well as sequence variation that could modify the saturation concentration, such as by altering disordered regions, changing oligomerization state, or acquiring or losing protein-protein interaction domains. Identifying such changes would provide powerful evidence of selection for, and adaptive value of, condensate formation. Characterizing how condensate properties vary in different environments could also provide hints regarding their functions. Aggregation-prone proteins and proteins with amino acid homorepeats are, on average, expressed at lower rates and turned over more rapidly, likely to keep total concentrations below the threshold for aggregation to prevent any deleterious effects 163,165. Thus, for functional condensates to evolve, both genomic, mRNA, and protein-level regulatory mechanisms and condensate-favoring, protein-intrinsic properties must evolve simultaneously. Identifying sets of homologous proteins where, under endogenous conditions, some are expressed at levels exceeding the threshold concentration and form condensates while others are not may help reveal how evolutionary processes act in concert to tune expression level and select for protein-intrinsic condensation behavior.
As described above, Pab1 shows signs of selection for condensation upon heat shock, and indeed mutants that impair condensation are significantly less fit for growth under heat stress conditions19. Such population-level analyses of fitness under varying environmental conditions may reveal other ways that condensate formation has adaptive value for populations of cells. For example, proteins that condense into static, inactive aggregates of varying size can increase the phenotypic diversity among a population of cells by impacting the concentration of active protein166. Higher phenotypic diversity can allow some individuals to survive a period of environmental insult, while a more homogeneous population might uniformly die off. Given that many condensates formed via LLPS exhibit time-dependent changes leading to a static state, an intriguing possibility is that cells might regulate these liquid-solid transitions as a means of tuning population-level variability.
In summary, we see collaboration between biochemists, cell biologists, experimental evolution specialists, and evolutionary biologists and theorists as a particularly fruitful future development for the field.
Conclusions
Biomolecular condensates function in diverse cellular processes spanning molecular to cellular scales. We have proposed a framework for understanding condensate functions based on the length scale on which they operate, with interrelated functions possibly occurring at different length scales. As examples, at the shortest scales, condensates can selectively concentrate macromolecules; at intermediate scales, they can spatially organize large structures; and at cellular scales they can sense environmental changes. We also recognize that additional functional classes may be discovered over time. It is our hope that this framework will provide a useful point of embarkation for studies of novel condensates, and spur new thinking about molecular mechanisms, experimental approaches, technological advances, biological processes and therapeutic strategies.
Acknowledgments
Research on biomolecular condensates is supported in the Rosen lab by the Howard Hughes Medical Institute, a Paul G. Allen Frontiers Distinguished Investigator Award to M.K.R. and grants from the Welch Foundation (I-1544 to M.K.R.) and NIH (F32 GM136058 to A.S.L.).
Glossary
- multivalent interactions
Interactions occurring between macromolecules with multiple sites of interaction, such that each molecule can interact with multiple binding partners
- intrinsically disordered region (IDR)
A protein region that does not adopt any stable ordered three-dimensional structure
- ribulose bisphosphate carboxylase/oxygenase (Rubisco)
Enzyme acting in carbon fixation in photosynthetic organisms, catalyzing reaction between ribulose bisphosphate and atmospheric carbon dioxide
- cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) synthase (cGAS)
Innate immune signaling enzyme that senses cytosolic DNA, a pathogen associated molecular pattern, and produces cGAMP, which activates the Stimulator of Interferon Genes (STING) protein to induce pro-inflammatory transcriptional responses
- allostery
Regulation of enzyme activity via binding by a second molecule at a site other than the enzyme’s active site, often by inducing a conformational change
- scaffold and client molecules
In simple cases, scaffolds are macromolecules that are required for condensate formation, while client molecules bind to and selectively partition into condensates without affecting condensate formation. In more complicated situations, as is typically the case in cells, macromolecules can have varying impacts on condensate formation, such as by modulating saturation concentration or significantly altering condensate composition
- KM
Parameter of the Michaelis-Menten model of enzyme kinetics, describing the concentration of substrate molecule at which the rate of product formation reaches half the maximum possible rate under a given set of conditions. If the rate of enzyme-substrate binding is rapid relative to catalysis, KM is approximates the dissociation constant for of the enzyme-substrate complex
- kinetic proofreading
A biochemical error-correction mechanism favoring reaction pathways that lead to correct over incorrect products, wherein an irreversible step that leads to exit of reaction intermediates from the pathway is more likely to occur for incorrect intermediates
- transgenerational epigenetic inheritance
Biological processes that allow transmission of epigenetic regulatory molecules or modifications, such as RNA interference factors or DNA methylation, from parent to offspring without altering DNA sequences
- P-granules
Biomolecular condensates formed by LLPS in C. elegans composed of RNA and proteins involved in maintenance of germ cell fate via post-transcriptional regulation and small RNA biogenesis
- Z-granules
Biomolecular condensates in C. elegans containing the proteins ZNFX-1 and WAGO-4 required for transgenerational epigenetic inheritance of RNA interference. Associates with both P-granules and Mutator foci, forming a bridge between the two condensates
- Mutator foci
Biomolecular condensate in C. elegans consisting of proteins encoded by mutator class genes, originally discovered in genetic screens for activation of transposons in the germline. Functions in siRNA amplification and RNA silencing
- voltage-gated calcium channels (VGCC)
Membrane protein channels that allow ingress of calcium into the cell at pre-synaptic terminals of neurons when activated by membrane depolarization. Calcium activates exocytosis of neurotransmitter vesicles
- N-methyl-D-aspartate (NMDA) receptor
Postsynaptic membrane protein channel activated by the excitatory neurotransmitter glutamate, allowing ingress of cations to depolarize the postsynaptic neuron
- dendritic spines
Small protrusions on postsynaptic dendrites that are sites of excitatory signaling by glutamate neurotransmitter receptors
- partition coefficient
The ratio of molecular concentration within a biomolecular condensate relative to the concentration in the surrounding solution
- Michaelis-like recruitment
For the binding of a molecule to some structure, a non-linear, saturable relationship between molecular concentration and fraction bound described by the rectangular hyperbola of the Michaelis-Menten model of enzyme kinetics
- surface or interfacial tension
For separate liquid phases in contact with each other, the work required to increase the surface area of contact between the two phases. In the absence of external forces, surface tension causes phase separated liquids to form spherical droplets as spheres have minimal surface area for a given volume
- optogenetics
A class of experimental techniques using light-responsive proteins or engineered protein domain fusions to acutely modulate cellular or protein activities by illuminating cells or biochemical reactions
Footnotes
Competing interests:
MKR is a co-founder of the biotechnology company Faze Medicines.
References
- 1.Elbaum-Garfinkle S et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences of the United States of America 112, 7189–7194, doi: 10.1073/pnas.1504822112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banjade S & Rosen MK Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li P et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340, doi: 10.1038/nature10879 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298, doi: 10.1038/nrm.2017.7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato M et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell 149, 753–767, doi: 10.1016/j.cell.2012.04.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson BA et al. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 179, 470–484 e421, doi: 10.1016/j.cell.2019.08.037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699. e616, doi: 10.1016/j.cell.2018.06.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin EW et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699, doi: 10.1126/science.aaw8653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin YH, Forman-Kay JD & Chan HS Theories for Sequence-Dependent Phase Behaviors of Biomolecular Condensates. Biochemistry 57, 2499–2508, doi: 10.1021/acs.biochem.8b00058 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Vernon RM et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486, doi: 10.7554/eLife.31486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain A & Vale RD RNA phase transitions in repeat expansion disorders. Nature 546, 243–247, doi: 10.1038/nature22386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langdon EM et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927, doi: 10.1126/science.aar7432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nott TJ et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Molecular cell 57, 936–947, doi: 10.1016/j.molcel.2015.01.013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banani SF et al. Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663, doi: 10.1016/j.cell.2016.06.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshizawa T et al. Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 173, 693–705 e622, doi: 10.1016/j.cell.2018.03.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qamar S et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 173, 720–734 e715, doi: 10.1016/j.cell.2018.03.056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L et al. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 173, 677–692. e620, doi: 10.1016/j.cell.2018.03.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofweber M et al. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 173, 706–719. e713, doi: 10.1016/j.cell.2018.03.004 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Riback JA et al. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 168, 1028–1040. e1019, doi: 10.1016/j.cell.2017.02.027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzmann TM et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654, doi: 10.1126/science.aao5654 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Kato M et al. Redox State Controls Phase Separation of the Yeast Ataxin-2 Protein via Reversible Oxidation of Its Methionine-Rich Low-Complexity Domain. Cell 177, 711–721. e718, doi: 10.1016/j.cell.2019.02.044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nedelsky NB & Taylor JP Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nature Reviews Neurology 15, 272–286, doi: 10.1038/s41582-019-0157-5 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Alberti S & Dormann D Liquid–Liquid Phase Separation in Disease. Annual Review of Genetics 53, 171–194, doi: 10.1146/annurev-genet-112618-043527 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Berry J, Brangwynne CP & Haataja M Physical principles of intracellular organization via active and passive phase transitions. Reports on Progress in Physics 81, 046601, doi: 10.1088/1361-6633/AAA61E (2018). [DOI] [PubMed] [Google Scholar]
- 25.Brangwynne CP, Tompa P & Pappu RV Polymer physics of intracellular phase transitions. Nat Phys 11, 899–904, doi: 10.1038/nphys3532 (2015). [DOI] [Google Scholar]
- 26.Shin Y & Brangwynne CP Liquid phase condensation in cell physiology and disease. Science 357, doi: 10.1126/science.aaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Frottin F et al. The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342–347, doi: 10.1126/science.aaw9157 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Hondele M et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148, doi: 10.1038/s41586-019-1502-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guillen-Boixet J et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361 e317, doi: 10.1016/j.cell.2020.03.049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu L et al. Controlling the material properties and rRNA processing function of the nucleolus using light. Proceedings of the National Academy of Sciences of the United States of America 116, 17330–17335, doi: 10.1073/pnas.1903870116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feric M et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 165, 1686–1697, doi: 10.1016/j.cell.2016.04.047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oshidari R et al. DNA repair by Rad52 liquid droplets. Nature communications 11, 695, doi: 10.1038/s41467-020-14546-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brangwynne CP et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732, doi: 10.1126/science.1172046 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Liao YC et al. RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell 179, 147–164. e120, doi: 10.1016/j.cell.2019.08.050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klosin A et al. Phase separation provides a mechanism to reduce noise in cells. Science 367, 464–468, doi: 10.1126/science.aav6691 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Andersson I & Backlund A Structure and function of Rubisco. Plant Physiol Biochem 46, 275–291, doi: 10.1016/j.plaphy.2008.01.001 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Wunder T, Oh ZG & Mueller-Cajar O CO2 -fixing liquid droplets: Towards a dissection of the microalgal pyrenoid. Traffic 20, 380–389, doi: 10.1111/tra.12650 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Wang H et al. Rubisco condensate formation by CcmM in beta-carboxysome biogenesis. Nature 566, 131–135, doi: 10.1038/s41586-019-0880-5 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Oltrogge LM et al. Multivalent interactions between CsoS2 and Rubisco mediate alpha-carboxysome formation. Nat Struct Mol Biol 27, 281–287, doi: 10.1038/s41594-020-0387-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wunder T, Cheng SLH, Lai S-K, Li H-Y & Mueller-Cajar O The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nature Communications 9, 5076, doi: 10.1038/s41467-018-07624-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freeman Rosenzweig ES et al. The Eukaryotic CO2-Concentrating Organelle Is Liquid-like and Exhibits Dynamic Reorganization. Cell 171, 148–162. e119, doi: 10.1016/j.cell.2017.08.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giordano M, Beardall J & Raven JA CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol 56, 99–131, doi: 10.1146/annurev.arplant.56.032604.144052 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Kaplan A & Reinhold L Co2 Concentrating Mechanisms in Photosynthetic Microorganisms. Annu Rev Plant Physiol Plant Mol Biol 50, 539–570, doi: 10.1146/annurev.arplant.50.1.539 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Nishimura T, Yamaguchi O, Takatani N, Maeda S & Omata T In vitro and in vivo analyses of the role of the carboxysomal beta-type carbonic anhydrase of the cyanobacterium Synechococcus elongatus in carboxylation of ribulose-1,5-bisphosphate. Photosynth Res 121, 151–157, doi: 10.1007/s11120-014-9986-7 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Dou Z et al. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem 283, 10377–10384, doi: 10.1074/jbc.M709285200 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Varesio LM, Willett JW, Fiebig A & Crosson S A Carbonic Anhydrase Pseudogene Sensitizes Select Brucella Lineages to Low CO2 Tension. J Bacteriol 201, doi: 10.1128/JB.00509-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du M & Chen ZJ DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709, doi: 10.1126/science.aat1022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep 6, 421–430, doi: 10.1016/j.celrep.2014.01.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830, doi: 10.1126/science.1229963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheu-Gruttadauria J & MacRae IJ Phase Transitions in the Assembly and Function of Human miRISC. Cell 173, 946–957 e916, doi: 10.1016/j.cell.2018.02.051 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu CY & Rana TM Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4, e210, doi: 10.1371/journal.pbio.0040210 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eulalio A, Behm-Ansmant I, Schweizer D & Izaurralde E P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27, 3970–3981, doi: 10.1128/MCB.00128-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brouhard GJ & Rice LM Microtubule dynamics: an interplay of biochemistry and mechanics. Nature Reviews Molecular Cell Biology 19, 451–463, doi: 10.1038/s41580-018-0009-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodruff JB et al. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169, 1066–1077 e1010, doi: 10.1016/j.cell.2017.05.028 (2017). [DOI] [PubMed] [Google Scholar]
- 55.King MR & Petry S Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat Commun 11, 270, doi: 10.1038/s41467-019-14087-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang H et al. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 163, 108–122, doi: 10.1016/j.cell.2015.08.010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y et al. Aurora A activation in mitosis promoted by BuGZ. J Cell Biol 217, 107–116, doi: 10.1083/jcb.201706103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwayer C et al. Mechanosensation of Tight Junctions Depends on ZO-1 Phase Separation and Flow. Cell 179, 937–952. e918, doi: 10.1016/j.cell.2019.10.006 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Beutel O, Maraspini R, Pombo-García K, Martin-Lemaitre C & Honigmann A Phase Separation of Zonula Occludens Proteins Drives Formation of Tight Junctions. Cell 179, 923–936. e911, doi: 10.1016/j.cell.2019.10.011 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Hirose T et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 25, 169–183, doi: 10.1091/mbc.E13-09-0558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Powers SK et al. Nucleo-cytoplasmic Partitioning of ARF Proteins Controls Auxin Responses in Arabidopsis thaliana. Mol Cell 76, 177–190 e175, doi: 10.1016/j.molcel.2019.06.044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakraborty AK & Weiss A Insights into the initiation of TCR signaling. Nature Immunology 15, 798–807, doi: 10.1038/ni.2940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su X et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science, doi: 10.1126/science.aad9964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Douglass AD & Vale RD Single-Molecule Microscopy Reveals Plasma Membrane Microdomains Created by Protein-Protein Networks that Exclude or Trap Signaling Molecules in T Cells. Cell 121, 937–950, doi: 10.1016/j.cell.2005.04.009 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng M et al. Reconstituted Postsynaptic Density as a Molecular Platform for Understanding Synapse Formation and Plasticity. Cell 174, 1172–1187. e1116, doi: 10.1016/j.cell.2018.06.047 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Zhu J, Shang Y & Zhang M Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nature Reviews Neuroscience 17, 209–223, doi: 10.1038/nrn.2016.18 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Huang WYC et al. Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proceedings of the National Academy of Sciences 113, 8218–8223, doi: 10.1073/pnas.1602602113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Case LB, Zhang X, Ditlev JA & Rosen MK Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097, doi: 10.1126/science.aau6313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang WYC et al. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103, doi: 10.1126/science.aau5721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim TH et al. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829, doi: 10.1126/science.aax4240 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Bouchard JJ et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol Cell 72, 19–36 e18, doi: 10.1016/j.molcel.2018.08.027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma W & Mayr C A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3′UTR-Mediated Protein-Protein Interactions. Cell 175, 1492–1506. e1419, doi: 10.1016/j.cell.2018.10.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kedersha N & Anderson P in Progress in Molecular Biology and Translational Science Vol. 90 155–185 (Academic Press, 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kedersha N et al. G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212, doi: 10.1083/jcb.201508028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsuki H et al. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes to Cells 18, 135–146, doi: 10.1111/gtc.12023 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Yang P et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 181, 325–345 e328, doi: 10.1016/j.cell.2020.03.046 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanders DW et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324 e328, doi: 10.1016/j.cell.2020.03.050 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nott TJ, Craggs TD & Baldwin AJ Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nature chemistry 8, 569–575, doi: 10.1038/nchem.2519 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Küffner AM et al. Acceleration of an Enzymatic Reaction in Liquid Phase Separated Compartments Based on Intrinsically Disordered Protein Domains. ChemSystemsChem 2, e2000001, doi: 10.1002/syst.202000001 (2020). [DOI] [Google Scholar]
- 80.Yao R-W et al. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Molecular cell 76, 767–783. e711, doi: 10.1016/j.molcel.2019.08.014 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Mitrea DM et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nature Communications 9, 842, doi: 10.1038/s41467-018-03255-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wan G et al. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679–683, doi: 10.1038/s41586-018-0132-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Phillips CM, Montgomery TA, Breen PC & Ruvkun G MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes and Development 26, 1433–1444, doi: 10.1101/gad.193904.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu X et al. RIM and RIM-BP Form Presynaptic Active-Zone-like Condensates via Phase Separation. Molecular cell 73, 971–984. e975, doi: 10.1016/j.molcel.2018.12.007 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Zeng M et al. Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 166, 1163–1175. e1112, doi: 10.1016/j.cell.2016.07.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Südhof TC The presynaptic active zone. Neuron 75, 11–25, doi: 10.1016/j.neuron.2012.06.012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boke E et al. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 166, 637–650, doi: 10.1016/j.cell.2016.06.051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rebane AA et al. Liquid–liquid phase separation of the Golgi matrix protein GM130. FEBS Letters 594, 1132–1144, doi: 10.1002/1873-3468.13715 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maser RS, Monsen KJ, Nelms BE & Petrini JH hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol 17, 6087–6096, doi: 10.1128/mcb.17.10.6087 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haaf T, Golub EI, Reddy G, Radding CM & Ward DC Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci U S A 92, 2298–2302, doi: 10.1073/pnas.92.6.2298 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shin Y et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 175, 1481–1491 e1413, doi: 10.1016/j.cell.2018.10.057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singatulina AS et al. PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell reports 27, 1809–1821. e1805, doi: 10.1016/j.celrep.2019.04.031 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Patel A et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077, doi: 10.1016/j.cell.2015.07.047 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Altmeyer M et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 6, 8088, doi: 10.1038/ncomms9088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bunting SF et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254, doi: 10.1016/j.cell.2010.03.012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kilic S et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. Embo J 38, e101379, doi: 10.15252/embj.2018101379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pessina F et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nature Cell Biology 21, 1286–1299, doi: 10.1038/s41556-019-0392-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parker MW et al. A new class of disordered elements controls DNA replication through initiator self-assembly. eLife 8, e48562, doi: 10.7554/eLife.48562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xiang W et al. Correlative live and super-resolution imaging reveals the dynamic structure of replication domains. J Cell Biol 217, 1973–1984, doi: 10.1083/jcb.201709074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cook PR The organization of replication and transcription. Science 284, 1790–1795, doi: 10.1126/science.284.5421.1790 (1999). [DOI] [PubMed] [Google Scholar]
- 101.Wang Z & Zhang H Phase Separation, Transition, and Autophagic Degradation of Proteins in Development and Pathogenesis. Trends in Cell Biology 29, 417–427, doi: 10.1016/j.tcb.2019.01.008 (2019). [DOI] [PubMed] [Google Scholar]
- 102.Sun D, Wu R, Zheng J, Li P & Yu L Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res 28, 405–415, doi: 10.1038/s41422-018-0017-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ciuffa R et al. The Selective Autophagy Receptor p62 Forms a Flexible Filamentous Helical Scaffold. Cell reports 11, 748–758, doi: 10.1016/j.celrep.2015.03.062 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Zaffagnini G et al. p62 filaments capture and present ubiquitinated cargos for autophagy. The EMBO journal 37, e98308, doi: 10.15252/embj.201798308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamasaki A et al. Liquidity Is a Critical Determinant for Selective Autophagy of Protein Condensates. Molecular cell 77, 1163–1175. e1169, doi: 10.1016/j.molcel.2019.12.026 (2020). [DOI] [PubMed] [Google Scholar]
- 106.Fujioka Y et al. Phase separation organizes the site of autophagosome formation. Nature 578, 301–305, doi: 10.1038/s41586-020-1977-6 (2020). [DOI] [PubMed] [Google Scholar]
- 107.Weatheritt RJ, Gibson TJ & Babu MM Asymmetric mRNA localization contributes to fidelity and sensitivity of spatially localized systems. Nature structural & molecular biology 21, 833–839, doi: 10.1038/nsmb.2876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maday S, Twelvetrees AE, Moughamian AJ & Holzbaur EL Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84, 292–309, doi: 10.1016/j.neuron.2014.10.019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ishizuka N, Cowan WM & Amaral DG A quantitative analysis of the dendritic organization of pyramidal cells in the rat hippocampus. J Comp Neurol 362, 17–45, doi: 10.1002/cne.903620103 (1995). [DOI] [PubMed] [Google Scholar]
- 110.Donnelly CJ, Fainzilber M & Twiss JL Subcellular communication through RNA transport and localized protein synthesis. Traffic 11, 1498–1505, doi: 10.1111/j.1600-0854.2010.01118.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gopal PP, Nirschl JJ, Klinman E & Holzbaur EL Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc Natl Acad Sci U S A 114, E2466–E2475, doi: 10.1073/pnas.1614462114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Holt CE & Schuman EM The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80, 648–657, doi: 10.1016/j.neuron.2013.10.036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meaburn KJ & Misteli T Cell biology: chromosome territories. Nature 445, 379–781, doi: 10.1038/445379a (2007). [DOI] [PubMed] [Google Scholar]
- 114.Bintu B et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, doi: 10.1126/science.aau1783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lieberman-Aiden E et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293, doi: 10.1126/science.1181369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rao SSP et al. Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320 e324, doi: 10.1016/j.cell.2017.09.026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Larson AG et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240, doi: 10.1038/nature22822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanulli S et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394, doi: 10.1038/s41586-019-1669-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Strom AR et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245, doi: 10.1038/nature22989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu X et al. Mitotic Implantation of the Transcription Factor Prospero via Phase Separation Drives Terminal Neuronal Differentiation. Dev Cell 52, 277–293 e278, doi: 10.1016/j.devcel.2019.11.019 (2020). [DOI] [PubMed] [Google Scholar]
- 121.Zaidi SK et al. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet 11, 583–589, doi: 10.1038/nrg2827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Palozola KC, Lerner J & Zaret KS A changing paradigm of transcriptional memory propagation through mitosis. Nat Rev Mol Cell Biol 20, 55–64, doi: 10.1038/s41580-018-0077-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eldar A & Elowitz MB Functional roles for noise in genetic circuits. Nature 467, 167–173, doi: 10.1038/nature09326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stoeger T, Battich N & Pelkmans L Passive Noise Filtering by Cellular Compartmentalization. Cell 164, 1151–1161, doi: 10.1016/j.cell.2016.02.005 (2016). [DOI] [PubMed] [Google Scholar]
- 125.Riback JA et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214, doi: 10.1038/s41586-020-2256-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yoo H, Triandafillou C & Drummond DA Cellular sensing by phase separation: Using the process, not just the products. Journal of Biological Chemistry 294, 7151–7159, doi: 10.1074/jbc.TM118.001191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lindquist S & Petersen R Selective Translation and Degradation of Heat-Shock Messenger RNAs in Drosophila. Enzyme 44, 147–166 (1990). [DOI] [PubMed] [Google Scholar]
- 128.Iserman C et al. Condensation of Ded1p Promotes a Translational Switch from Housekeeping to Stress Protein Production. Cell 181, 818–831. e819, doi: 10.1016/j.cell.2020.04.009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Delarue M et al. mTORC1 Controls Phase Separation and the Biophysical Properties of the Cytoplasm by Tuning Crowding. Cell, doi: 10.1016/j.cell.2018.05.042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chakravarty AK, Smejkal T, Itakura AK, Garcia DM & Jarosz DF A Non-amyloid Prion Particle that Activates a Heritable Gene Expression Program. Molecular cell 77, 251–265. e259, doi: 10.1016/j.molcel.2019.10.028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Itakura AK, Chakravarty AK, Jakobson CM & Jarosz DF Widespread Prion-Based Control of Growth and Differentiation Strategies in Saccharomyces cerevisiae. Molecular cell 77, 266–278. e266, doi: 10.1016/j.molcel.2019.10.027 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xing W, Muhlrad D, Parker R & Rosen MK A quantitative inventory of yeast P body proteins reveals principles of compositional specificity. BioRxiv 489658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jain S et al. in Cell Vol. 164 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Markmiller S et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604 e513, doi: 10.1016/j.cell.2017.12.032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Youn JY et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell 69, 517–532 e511, doi: 10.1016/j.molcel.2017.12.020 (2018). [DOI] [PubMed] [Google Scholar]
- 136.Hubstenberger A et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell 68, 144–157 e145, doi: 10.1016/j.molcel.2017.09.003 (2017). [DOI] [PubMed] [Google Scholar]
- 137.McSwiggen DT, Mir M, Darzacq X & Tjian R Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes & development 33, 1619–1634, doi: 10.1101/gad.331520.119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M & Parker R Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382, doi: 10.1261/rna.7258505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wheeler RJ & Hyman AA Controlling compartmentalization by non-membrane-bound organelles. Philos Trans R Soc Lond B Biol Sci 373, doi: 10.1098/rstb.2017.0193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Alberti S, Gladfelter A & Mittag T Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434, doi: 10.1016/j.cell.2018.12.035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Soding J, Zwicker D, Sohrabi-Jahromi S, Boehning M & Kirschbaum J Mechanisms for Active Regulation of Biomolecular Condensates. Trends Cell Biol 30, 4–14, doi: 10.1016/j.tcb.2019.10.006 (2020). [DOI] [PubMed] [Google Scholar]
- 142.Bratek-Skicki A, Pancsa R, Meszaros B, Van Lindt J & Tompa P A guide to regulation of the formation of biomolecular condensates. FEBS J 287, 1924–1935, doi: 10.1111/febs.15254 (2020). [DOI] [PubMed] [Google Scholar]
- 143.Choi JM, Dar F & Pappu RV LASSI: A lattice model for simulating phase transitions of multivalent proteins. PLoS Comput Biol 15, e1007028, doi: 10.1371/journal.pcbi.1007028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cai LH, Panyukov S & Rubinstein M Mobility of Nonsticky Nanoparticles in Polymer Liquids. Macromolecules 44, 7853–7863, doi: 10.1021/ma201583q (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hill TL Thermodynamics of Small Systems, Parts 1 & 2. (Dover Publications, 2013). [Google Scholar]
- 146.Puglisi A, Sarracino A & Vulpiani A Thermodynamics and Statistical Mechanics of Small Systems. (published by the authors, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Elowitz MB, Levine AJ, Siggia ED & Swain PS Stochastic gene expression in a single cell. Science 297, 1183–1186, doi: 10.1126/science.1070919 (2002). [DOI] [PubMed] [Google Scholar]
- 148.Ozbudak EM, Thattai M, Kurtser I, Grossman AD & van Oudenaarden A Regulation of noise in the expression of a single gene. Nat Genet 31, 69–73, doi: 10.1038/ng869 (2002). [DOI] [PubMed] [Google Scholar]
- 149.Boija A et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855 e1816, doi: 10.1016/j.cell.2018.10.042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sabari BR et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, doi: 10.1126/science.aar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cho WK et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415, doi: 10.1126/science.aar4199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lu H et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323, doi: 10.1038/s41586-018-0174-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chong S et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, doi: 10.1126/science.aar2555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mir M et al. Dense Bicoid hubs accentuate binding along the morphogen gradient. Genes Dev 31, 1784–1794, doi: 10.1101/gad.305078.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mir M et al. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 7, doi: 10.7554/eLife.40497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Boehning M et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol 25, 833–840, doi: 10.1038/s41594-018-0112-y (2018). [DOI] [PubMed] [Google Scholar]
- 157.Julin J, Napari I, Merikanto J & Vehkamaki H A thermodynamically consistent determination of surface tension of small Lennard-Jones clusters from simulation and theory. J Chem Phys 133, 044704, doi: 10.1063/1.3456184 (2010). [DOI] [PubMed] [Google Scholar]
- 158.Lau GV, Hunt PA, Muller EA, Jackson G & Ford IJ Water droplet excess free energy determined by cluster mitosis using guided molecular dynamics. J Chem Phys 143, 244709, doi: 10.1063/1.4935198 (2015). [DOI] [PubMed] [Google Scholar]
- 159.Nguyen VD, Schoemaker FC, Blokhuis EM & Schall P Measurement of the Curvature-Dependent Surface Tension in Nucleating Colloidal Liquids. Phys Rev Lett 121, 246102, doi: 10.1103/PhysRevLett.121.246102 (2018). [DOI] [PubMed] [Google Scholar]
- 160.Bergeron-Sandoval L-P et al. Endocytosis caused by liquid-liquid phase separation of proteins. bioRxiv, 145664, doi: 10.1101/145664 (2018). [DOI] [Google Scholar]
- 161.Gibaud T et al. Reconfigurable self-assembly through chiral control of interfacial tension. Nature 481, 348–351, doi: 10.1038/nature10769 (2012). [DOI] [PubMed] [Google Scholar]
- 162.Kuriyan J & Eisenberg D The origin of protein interactions and allostery in colocalization. Nature 450, 983–990, doi: 10.1038/nature06524 (2007). [DOI] [PubMed] [Google Scholar]
- 163.Chavali S et al. Constraints and consequences of the emergence of amino acid repeats in eukaryotic proteins. Nature structural & molecular biology 24, 765–777, doi: 10.1038/nsmb.3441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kawecki TJ et al. Experimental evolution. Trends in Ecology and Evolution 27, 547–560, doi: 10.1016/j.tree.2012.06.001 (2012). [DOI] [PubMed] [Google Scholar]
- 165.Gsponer J & Babu MM Cellular Strategies for Regulating Functional and Nonfunctional Protein Aggregation. Cell reports 2, 1425–1437, doi: 10.1016/j.celrep.2012.09.036 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sanchez de Groot N et al. The fitness cost and benefit of phase-separated protein deposits. Molecular Systems Biology 15, e8075, doi: 10.15252/msb.20178075 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.A P & Weber SC. Evidence for and against Liquid-Liquid Phase Separation in the Nucleus. Non-Coding RNA 5, 50, doi: 10.3390/ncrna5040050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Falahati H & Wieschaus E Independent active and thermodynamic processes govern the nucleolus assembly in vivo. Proceedings of the National Academy of Sciences of the United States of America 114, 1335–1340, doi: 10.1073/pnas.1615395114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dill KA & Bromberg S Molecular Driving Forces. Statistical Thermodynamics in Biology, Chemistry, Physics, and Nanoscience. 2 edn, (Garland Science, 2010). [Google Scholar]
- 170.Pareek V, Tian H, Winograd N & Benkovic SJ Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science 368, 283–290, doi: 10.1126/science.aaz6465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tibble RW, Depaix A, Kowalska J, Jemielity J & Gross JD Biomolecular condensates amplify mRNA decapping by coupling protein interactions with conformational changes in Dcp1/Dcp2. bioRxiv, 2020.2007.2009.195057, doi: 10.1101/2020.07.09.195057 (2020). [DOI] [Google Scholar]
- 172.Itia AF-B, Alexander BS, Ethan KS & Halina R-D Optical trapping in vivo: theory, practice, and applications. Nanophotonics 8, 1023–1040, doi: 10.1515/nanoph-2019-0055 (2019). [DOI] [Google Scholar]
- 173.Mittasch M et al. Non-invasive perturbations of intracellular flow reveal physical principles of cell organization. Nature Cell Biology 20, 344–351, doi: 10.1038/s41556-017-0032-9 (2018). [DOI] [PubMed] [Google Scholar]
- 174.Taylor JP, Brown RH Jr. & Cleveland DW Decoding ALS: from genes to mechanism. Nature 539, 197–206, doi: 10.1038/nature20413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Boulay G et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 171, 163–178 e119, doi: 10.1016/j.cell.2017.07.036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tulpule A et al. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. BioRxiv 704312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Takeuchi K Discovery Stories of RET Fusions in Lung Cancer: A Mini-Review. Front Physiol 10, 216, doi: 10.3389/fphys.2019.00216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Grunewald TGP et al. Ewing sarcoma. Nat Rev Dis Primers 4, 5, doi: 10.1038/s41572-018-0003-x (2018). [DOI] [PubMed] [Google Scholar]
- 179.Beutel O, Maraspini R, Pombo-Garcia K, Martin-Lemaitre C & Honigmann A Phase Separation of Zonula Occludens Proteins Drives Formation of Tight Junctions. Cell 179, 923–936 e911, doi: 10.1016/j.cell.2019.10.011 (2019). [DOI] [PubMed] [Google Scholar]
- 180.Wheeler RJ et al. Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. BioRxiv 721001 (2020). [Google Scholar]
- 181.Berchtold D, Battich N & Pelkmans L A Systems-Level Study Reveals Regulators of Membrane-less Organelles in Human Cells. Mol Cell 72, 1035–1049 e1035, doi: 10.1016/j.molcel.2018.10.036 (2018). [DOI] [PubMed] [Google Scholar]
- 182.Rai AK, Chen JX, Selbach M & Pelkmans L Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216, doi: 10.1038/s41586-018-0279-8 (2018). [DOI] [PubMed] [Google Scholar]
- 183.Monahan Z et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. Embo J 36, 2951–2967, doi: 10.15252/embj.201696394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang JT et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3, e04591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Milovanovic D, Wu Y, Bian X & De Camilli P A liquid phase of synapsin and lipid vesicles. Science 361, 604–607, doi: 10.1126/science.aat5671 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang G, Wang Z, Du Z & Zhang H mTOR Regulates Phase Separation of PGL Granules to Modulate Their Autophagic Degradation. Cell 174, 1492–1506 e1422, doi: 10.1016/j.cell.2018.08.006 (2018). [DOI] [PubMed] [Google Scholar]
- 187.Altmeyer M et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 6, 8088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang L et al. Histone Modifications Regulate Chromatin Compartmentalization by Contributing to a Phase Separation Mechanism. Mol Cell 76, 646–659 e646, doi: 10.1016/j.molcel.2019.08.019 (2019). [DOI] [PubMed] [Google Scholar]
- 189.Guccione E & Richard S The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol 20, 642–657, doi: 10.1038/s41580-019-0155-x (2019). [DOI] [PubMed] [Google Scholar]
- 190.Dao TP et al. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol Cell 69, 965–978 e966, doi: 10.1016/j.molcel.2018.02.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Saito M et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat Chem Biol 15, 51–61, doi: 10.1038/s41589-018-0180-7 (2019). [DOI] [PubMed] [Google Scholar]
- 192.Buchan JR, Kolaitis RM, Taylor JP & Parker R Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474, doi: 10.1016/j.cell.2013.05.037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.James JR Tuning ITAM multiplicity on T cell receptors can control potency and selectivity to ligand density. Science Signaling 11, eaan1088, doi: 10.1126/scisignal.aan1088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cao Q, Boyer DR, Sawaya MR, Ge P & Eisenberg DS Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils. Nature structural & molecular biology 27, 653–659, doi: 10.1038/s41594-020-0435-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Griner SL et al. Structure-based inhibitors of amyloid beta core suggest a common interface with tau. eLife 8, e46924, doi: 10.7554/eLife.46924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lu J et al. Structure-Based Peptide Inhibitor Design of Amyloid-β Aggregation. Frontiers in Molecular Neuroscience 12, 54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sangwan S et al. Inhibition of synucleinopathic seeding by rationally designed inhibitors. eLife 9, e46775, doi: 10.7554/eLife.46775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pessina F et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol 21, 1286–1299, doi: 10.1038/s41556-019-0392-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.So C et al. A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science 364, doi: 10.1126/science.aat9557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Al-Husini N, Tomares DT, Bitar O, Childers WS & Schrader JM α-Proteobacterial RNA Degradosomes Assemble Liquid-Liquid Phase-Separated RNP Bodies. Molecular cell 71, 1027–1039. e1014, doi: 10.1016/j.molcel.2018.08.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ying Y et al. Splicing Activation by Rbfox Requires Self-Aggregation through Its Tyrosine-Rich Domain. Cell 170, 312–323. e310, doi: 10.1016/j.cell.2017.06.022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tatomer DC et al. Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J Cell Biol 213, 557–570, doi: 10.1083/jcb.201504043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Novotný I, Blažíková M, Staněk D, Herman P & Malinsky J In vivo kinetics of U4/U6·U5 tri-snRNP formation in Cajal bodies. Molecular biology of the cell 22, 513–523, doi: 10.1091/mbc.e10-07-0560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Samir P et al. DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594, doi: 10.1038/s41586-019-1551-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lasker K et al. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nature Microbiology 5, 418–429, doi: 10.1038/s41564-019-0647-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Park J-E et al. Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nature Communications 10, 4959, doi: 10.1038/s41467-019-12619-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Saha S et al. Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell 166, 1572–1584. e1516, doi: 10.1016/j.cell.2016.08.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gaglia G et al. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nature Cell Biology 22, 151–158, doi: 10.1038/s41556-019-0458-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Guo YE et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548, doi: 10.1038/s41586-019-1464-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zamudio AV et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Molecular cell 76, 753–766. e756, doi: 10.1016/j.molcel.2019.08.016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Erdel F et al. Mouse Heterochromatin Adopts Digital Compaction States without Showing Hallmarks of HP1-Driven Liquid-Liquid Phase Separation. Molecular cell 78, 236–249. e237, doi: 10.1016/j.molcel.2020.02.005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Davis D et al. Human Antiviral Protein MxA Forms Novel Metastable Membraneless Cytoplasmic Condensates Exhibiting Rapid Reversible Tonicity-Driven Phase Transitions. J Virol 93, e01014–01019, doi: 10.1128/jvi.01014-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Heinrich BS, Maliga Z, Stein DA, Hyman AA & Whelan SPJ Phase Transitions Drive the Formation of Vesicular Stomatitis Virus Replication Compartments. mBio 9, e02290–02217, doi: 10.1128/mBio.02290-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhou Y, Su JM, Samuel CE & Ma D Measles Virus Forms Inclusion Bodies with Properties of Liquid Organelles. J Virol 93, e00948–00919, doi: 10.1128/JVI.00948-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yasuda S et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300, doi: 10.1038/s41586-020-1982-9 (2020). [DOI] [PubMed] [Google Scholar]
- 216.Sala K et al. The ERC1 scaffold protein implicated in cell motility drives the assembly of a liquid phase. Scientific reports 9, 13530, doi: 10.1038/s41598-019-49630-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Celetti G et al. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J Cell Biol 219, doi: 10.1083/jcb.201907157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


