Abstract
Background and Aims
Understanding the population genetics and evolutionary history of endangered species is urgently needed in an era of accelerated biodiversity loss. This knowledge is most important for regions with high endemism that are ecologically vulnerable, such as the Qinghai–Tibet Plateau (QTP).
Methods
The genetic variation of 84 juniper trees from six populations of Juniperus microsperma and one population of Juniperus erectopatens, two narrow-endemic junipers from the QTP that are sister to each other, was surveyed using RNA-sequencing data. Coalescent-based analyses were used to test speciation, migration and demographic scenarios. Furthermore, positively selected and climate-associated genes were identified, and the genetic load was assessed for both species.
Key Results
Analyses of 149 052 single nucleotide polymorphisms showed that the two species are well differentiated and monophyletic. They diverged around the late Pliocene, but interspecific gene flow continued until the Last Glacial Maximum. Demographic reconstruction by Stairway Plot detected two severe bottlenecks for J. microsperma but only one for J. erectopatens. The identified positively selected genes and climate-associated genes revealed habitat adaptation of the two species. Furthermore, although J. microsperma had a much wider geographical distribution than J. erectopatens, the former possesses lower genetic diversity and a higher genetic load than the latter.
Conclusions
This study sheds light on the evolution of two endemic juniper species from the QTP and their responses to Quaternary climate fluctuations. Our findings emphasize the importance of speciation and demographic history reconstructions in understanding the current distribution pattern and genetic diversity of threatened species in mountainous regions.
Keywords: Bottleneck event, conservation genomics, demographic history, effective population size, positively selected genes, habitat loss, Juniperus microsperma, Juniperus erectopatens, speciation history
INTRODUCTION
As a result of global climate change and the anthropogenic alteration of natural habitats (Dirzo et al., 2014; Ceballos et al., 2015, 2017; Cronk, 2016; Miraldo et al., 2016), numerous endangered species have experienced population declines (Allendorf et al., 2010; Wang et al., 2018) leading to higher than expected extinction rates (Barnosky et al., 2011).Therefore, conservation genetic and ecological studies that aim to minimize the loss of rare species and maximize conservation efforts are urgently required (Segelbacher et al., 2010; Casas-Marce et al., 2013; Harrisson et al., 2014; Garner et al., 2016; Hamabata et al., 2019). Habitat loss is often the primary factor that causes fragmentation of rare species, resulting in small and isolated populations over short evolutionary time scales (Frankham, 2005; Hung et al., 2014; Rogers and Slatkin, 2017). This may lead to reduced genetic diversity due to the more pronounced effects of genetic drift in small and isolated populations (Keller and Waller, 2002; Spielman et al., 2004; Jacquemyn et al., 2009). In general, threatened species tend to have low genetic diversity (Spielman et al., 2004; Allendorf et al., 2010), which increases the extinction risk (Saccheri et al., 1998; Frankham, 2005). Therefore, it is crucial to understand how past events have shaped the demographic history of a species if we want to make predictions about how populations may respond to future challenges (Ellegren and Galtier, 2016; Fan et al., 2018). A key event in demographic histories is the fast reduction and subsequent increase of the effective population size (Ne), usually known as a genetic bottleneck (Tajima, 1989a). This often negatively impacts the viability of a species through the loss of genetic diversity and various stochastic demographic processes (Frankham et al., 1999; Frankham, 2005; Lima et al., 2017). Therefore, knowledge of bottleneck events in the evolutionary history of a species, and of current levels of genetic diversity, are of critical importance when designing conservation and management strategies.
The Qinghai–Tibet Plateau (QTP) sensu lato, especially its eastern and southern regions, holds tremendous biodiversity due to past geological and climatic changes (Wang et al., 2007; Favre et al., 2015; Fu et al., 2020; Spicer et al., 2021). It is one of the most important alpine biodiversity hotspots in the world and a natural laboratory for evolutionary studies (Wen et al., 2014; Huang et al., 2018). Diverse habitats and ecological niches generated by the uplift of mountains (Rahbek et al., 2019; Spicer et al., 2021) have promoted speciation (Liu et al., 2013; Rahbek et al., 2019) via allopatric and ecological speciation. Furthermore, the extensive oscillations of the climate on the QTP during the Quaternary led to repeated cycles of climate-driven habitat change, which promoted hybrid speciation as well as extinction over a short period of time (Fjeldså, 1994; Mosbrugger et al., 2018; Nevado et al., 2018; Muellner-Riehl, 2019; Rahbek et al., 2019). The QTP contains about 9000-12 000 species of plants (in about 1500 genera), of which more than 20 % are endemic (Liu et al., 2014; Wen et al., 2014; Zhang et al., 2016). Many of these endemic plant species are listed as rare and/or endangered (Duan and Liu, 2006; Liu et al., 2011, 2013; Wang and Li, 2016; Fu et al., 2019), and face a high risk of extinction (López-Pujol et al., 2011; Liu et al., 2014) due to global warming and increased human activities on the QTP (He et al., 2005; Diffenbaugh and Giorgi, 2012).
Conservation genomics has been applied to a number of endangered species, and provided insight into the genetic diversity and evolutionary history of these species, leading to informed and effective conservation and management plans (O’Brien, 1994; Pautasso, 2009; Allendorf et al., 2010; Ouborg et al., 2010; Segelbacher et al., 2010). Unfortunately, such studies are lacking for endangered conifers on the QTP, despite their ecological importance. Here, we aim to unravel the evolutionary history of two rare endemic conifers, Juniperus microsperma and Juniperus erectopatens, in the eastern QTP (Adams and Schwarzbach, 2013; Adams, 2014; Shang et al., 2015; Xu et al., 2019). Juniperus microsperma and J. erectopatens have slender leaves and irregular globose glaucous seed cones of similar size but differ in the number of seeds per cone [(1-)2-seeded vs. 1-seeded, respectively] and branchlet morphology (ascending vs. largely pendulous, respectively) (Adams, 2014). Although J. microsperma is the closest relative of J. erectopatens (Adams and Schwarzbach, 2013), they occur in areas ~800 km apart (Adams, 2014; Xu et al., 2019). According to our field surveys, these two species are very rare. Juniperus microsperma is currently found only in the Parlung Zangbo Valley, Bomi County, south-eastern QTP (Adams, 2014; Shang et al., 2015), and J. erectopatens occurs only in one wooded area of ~2 km2 at Anhong, Songpan, Sichuan, China (Adams, 2014; Xu et al., 2019). Previous studies of these two species were limited to a small number of single nucleotide polymorphisms (SNPs) or indels from chloroplast and nuclear DNA markers (Shang et al., 2015; Xu et al., 2019), which might not reflect the genome-wide genetic variation. Due to recent advances in molecular biology technologies, genome-wide data can be generated rapidly by high-throughput sequencing (Ellegren, 2014; Goodwin et al., 2016; Todd et al., 2016; Fuentes-Pardo and Ruzzante, 2017) and analysed using various bioinformatic tools (Ouborg et al., 2010; Hamabata et al., 2019).
Here, we employed high-throughput sequencing to detect the evolutionary history and its influence on the genetic diversity of two rare and closely related juniper trees, J. microsperma and J. erectopatens. Specifically, we aimed to address the following questions: (1) What is the pattern of genetic diversity and genetic load in J. microsperma and J. erectopatens? (2) When did these two species diverge from their most recent ancestor and what biogeographical history did they experience? (3) Which model of demographic history best explains the current genetic diversity? (4) How did orogenic events and climatic changes affect the evolutionary history of J. microsperma and J. erectopatens? (5) Did habitat adaptation play a role during the evolution of the two species? Answering these questions will shed light on the conservation genomics and evolutionary history of QTP endemics as well as threatened plant species that occur in other mountainous regions worldwide.
MATERIALS AND METHODS
Sampling and RNA sequencing
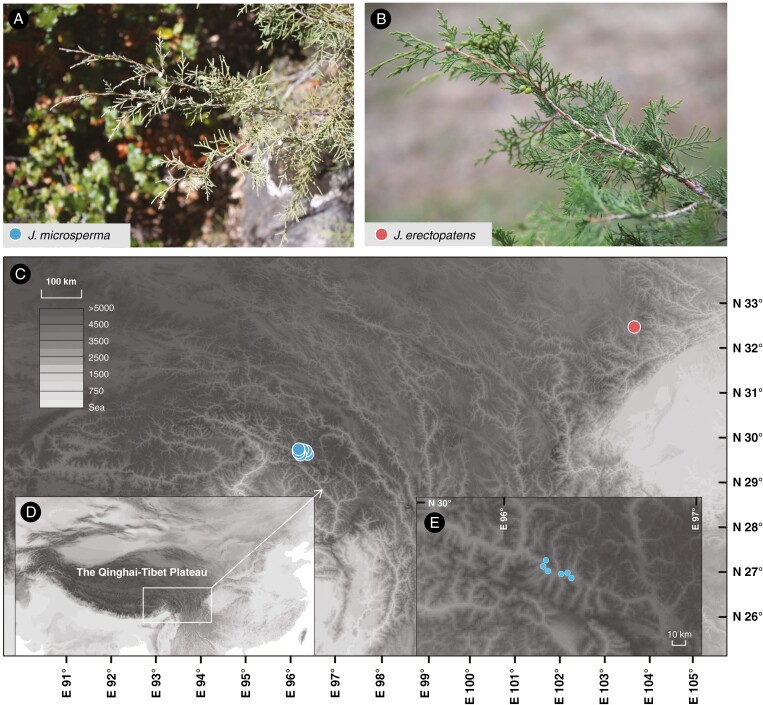
Leaf samples were collected from 84 mature individuals comprising 56 from Juniperus microsperma (Cheng & L. K. Fu) R. P. Adams and 28 from Juniperus erectopatens (Cheng & L. K. Fu) R. P. Adams. The distance between sampled trees was more than 50 m (Table 1; Supplementary Data Table S1; Fig. 1C). In addition, 15 trees of J. sabina L. and five trees of J. saltuaria Rehder & E.H. Wilson were sampled as outgroups for further analysis. Fresh leaves were frozen in liquid nitrogen immediately after picking and kept at −80 °C prior to RNA extraction. Total RNA was extracted using the RNeasy Kit (Qiagen, Germany) and TRIzol reagent (Invitrogen, CA, USA). RNA purity was checked using a NanoPhotometer spectrophotometer (IMPLEN, CA, USA) and the RNA concentration was measured using a Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Sequencing libraries were prepared using a NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, MA, USA) following standard RNA-sequencing (RNA-seq) methodology. After the above steps, 150-bp paired-end raw reads were generated on an Illumina Hiseq PE150 platform (Novogene, China).
Table 1.
Location details of the sampled populations of Juniperus microsperma and J. erectopatens
| ID | Species | n a | Location | Latitude N | Longitude E | Altitude (m) |
|---|---|---|---|---|---|---|
| 1 | J. microsperma | 8 | Bomi, Xizang, China | 29°36.98′ | 96°19.92′ | 3325 |
| 2 | J. microsperma | 12 | Bomi, Xizang, China | 29°37.12′ | 96°18.98′ | 3253 |
| 3 | J. microsperma | 5 | Bomi, Xizang, China | 29°37.29′ | 96°18.05′ | 3221 |
| 4 | J. microsperma | 5 | Bomi, Xizang, China | 29°38.94′ | 96°13.13′ | 3171 |
| 5 | J. microsperma | 17 | Bomi, Xizang, China | 29°39.94′ | 96°12.34′ | 3167 |
| 6 | J. microsperma | 9 | Bomi, Xizang, China | 29°40.78′ | 96°12.56′ | 3202 |
| 7 | J. erectopatens | 28 | Songpan, Sichuan, China | 32°27.94′ | 103°40.06′ | 2714 |
aThe number of samples.
Fig. 1.
(A) Photograph of J. microsperma. (B) Photograph of J. erectopatens. (C) Location of sampled populations of J. microsperma and J. erectopatens. See also Supplementary Data Table S1. (D) Location of the Qinghai–Tibet Plateau. (E) Sampling sites of J. microsperma along the Parlong Zangpo river valley.
Read trimming and transcriptome de novo assembly
We used Trimmomatic ver.0.36 (Bolger et al., 2014) to trim and filter Illumina raw reads, during which adapter sequences, and poly-N and low-quality reads (Q < 30) were discarded. Filtered reads from one J. microsperma individual were assembled into contigs using Trinity ver.2.8.4 (Grabherr et al., 2011) with default parameters. To obtain high-quality contigs, sequences were aligned to the microbial genome database (MBGD: http://mbgd.genome.ad.jp/; Uchiyama et al., 2010) via BLASTN ver.2.7.1 (Altschul et al., 1997) and sequences with more than 90 % similarity were discarded. CD-HIT ver.4.6.8 (Li and Godzik, 2006) was used to remove redundant sequences of assembled contigs with a threshold value of 0.99. Transcripts of <200 bp were removed and the longest transcripts were selected in case of alternative splicing. Finally, 61 700 contiguous expressed sequences were obtained as the reference transcriptome.
Read mapping and variant calling
High-quality reads of J. microsperma and J. erectopatens individuals were aligned to the reference transcriptome using BWA-MEM ver.0.7.17 (Li and Durbin, 2009) with default parameters. To identify the ancestral state of characters for these two species, and for the phylogenetic analysis, we mapped the reads of one J. oxycedrus L., one J. phoenicea L., 15 J. sabina and five J. saltuaria samples to the reference transcriptome. Transcriptome sequence data of J. oxycedrus and J. phoenicea were taken from Mao et al. (2019). SAMTOOLS ver.1.9 (Li et al., 2009) was used to convert Sequence Alignment/Map (SAM) files to Binary Alignment/Map (BAM) files, followed by sorting of the BAM files. Duplicate reads were marked and excluded from further analysis using PICARDTOOLS ver.2.18.11 (https://github.com/broadinstitute/picard/). The local regions around indels were realigned using the RealignerTargetCreator and IndelRealigner tools in GATK ver.3.8.1 (DePristo et al., 2011). Variant calling was conducted using the ‘mpileup’ command in SAMTOOLS ver.1.9 (Li et al., 2009) with parameters ‘-t AD,ADF,ADR,DP,SP -Q 20 -q 20’.
To obtain a high-quality SNP set, we filtered those sites with mapping quality <30 and removed indels within a 5-bp window. Sites with coverage depth (DP) <10 were considered as missing for each sample and SNPs with >50 % of missing bases within either species were excluded. In addition, we used VCFTOOLS ver. 0.1.15 (Danecek et al., 2011) to remove variant sites which differed significantly from Hardy–Weinberg equilibrium (P < 0.001) or which had a minimum allele frequency of <0.05 to reduce the false discovery rate. In addition, we calculated the number of shared and species-specific SNPs of J. microsperma and J. erectopatens.
The SNP dataset was used for population genetic and phylogenetic analyses, demographic reconstruction, gene flow estimation, and the detection of environment-associated loci. In parallel, we generated a gene sequence dataset to identify positively selected genes and the Gene Ontology (GO) annotation.
Population genetic and phylogenetic analysis
The genetic structure of the two species was examined using a Bayesian clustering (ADMIXTURE) and principal component analysis (PCA). The SNP variant calling format was converted into binary ped format for downstream analysis using VCFTOOLS ver.0.1.15. We used PLINK ver.1.90 (Purcell et al., 2007) to remove sites in linkage disequilibrium (LD) with parameters ‘--indep-pairwise 50 10 0.4’. The software ADMIXTURE ver.1.3.0 (Alexander and Lange, 2011) was used to estimate the most likely number of distinct genetic clusters (K) by varying K from 1 to 10 and by computing the parameters’ maximum-likelihood (ML) estimates. Twenty independent replicates were run for each K to calculate the cross-validation errors. The optimal K was indicated by the lowest cross-validation error. In addition, PCA was performed via the SMARTPCA module in the software EIGENSOFT ver.7.2.1 (Price et al., 2006).
For phylogenetic analysis, we used a Perl script (Ru et al., 2018) to convert filtered SNPs to concatenated sequences for each individual. An ML phylogenetic tree for J. microsperma and J. erectopatens was reconstructed by RAxML ver.8.2.11 (Stamatakis, 2014) under the GTRGAMMA model using J. oxycedrus and J. phoenicea as outgroups. We ran 200 replicates to calculate bootstrap support values. The final phylogenetic tree was visualized using Figtree ver.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
To investigate the genetic differentiation between the two species, we calculated FST (Weir and Cockerham, 1984) using VCFTOOLS. The transcriptome-wide distribution of nucleotide diversity, θπ (based on pairwise differences between sequences; Nei and Li, 1979) and θw (based on number of segregating sites between sequences; Watterson, 1975) were estimated using VCFTOOLS and the equations of Watterson (1975), respectively. Observed heterozygosity (Ho) and expected heterozygosity (He) were calculated for each population averaging across the individuals’ total heterozygosity using an unpublished in-house Python script. The estimation of θπ, θw, Ho and He was performed on a subsample of seven individuals in Populations 1, 2, 5, 6 and 7, and five individuals in Populations 3 and 4 as there were only five individuals in these two populations (Table 1; Supplementary Data Table S1). Also, Tajima’s D (Tajima, 1989b) was calculated with VCFTOOLS for each of the two studied species. The mean value of genome-wide FST, (negative FST value for any locus was set to zero when calculating) and the average number of nucleotide substitutions dXY (Foote et al., 2016) per locus was calculated using a Perl script (Ru et al., 2018).
Demographic history and gene flow
Gene flow between J. microsperma and J. erectopatens was estimated via the detection of shared haplotypes between individuals of both species using a refined identity-by-descent (IBD) blocks analysis which was performed in Refined IBD with the parameters ‘window = 0.001 lod = 3.0 length = 0.0001 trim = 0.00001’ (Browning and Browning, 2013).
We used the maximum-composite-likelihood approach based on the joint site frequency spectrum (SFS) implemented in fastsimcoal2 (Excoffier et al., 2013) to assess the fit of various demographic models and to infer the final optimal demographic scenario for J. microsperma and J. erectopatens. First, to minimize the effects of natural selection on demographic inference, we extracted a total of 1 050 559 four-fold degenerate synonymous (4DTV) sites from the SNP dataset, which contained no missing bases across all individuals. A folded two-dimensional site frequency spectrum (2D-SFS) for J. microsperma and J. erectopatens was inferred based on 4DTV sites, and the joint SFS of each species was also inferred. The scripts from Ru et al. (2018) were used for extracting 4DTV sites and generating 2D-SFS, which was subsequently used to infer divergence time, bottleneck size and gene flow size between the two studied species. The mutation rate per generation was set to 9.7 × 10−9 and the generation time to 50 years following the parameters for Cupressaceae species in Li et al. (2012). Global ML estimates were derived from 50 independent runs, with 100 000 coalescent simulations and 40 likelihood maximization algorithm cycles. The Akaike information criterion (AIC; Akaike, 1974) was used to assess the relative fit of each model and the best fit with the lowest AIC was chosen. Ninety-five per cent non-parametric bootstrap confidence intervals (CIs) were constructed by sampling the 2D-SFS based on parameters of the preferred demographic scenario (Excoffier et al., 2013).
In addition, we used Stairway Plot (Liu and Fu, 2015) to investigate the changes in effective population size over time for J. microsperma and J. erectopatens based on the joint SFS of each species. Two hundred subsamples of 67 % of all sites were created, the median value of the estimation was used as a final output and the 95 % CI of each value was produced (Liu and Fu, 2015).
Detection of positive selection and GO annotation
Coding and peptide sequences in the form of open reading frames (ORFs) were predicted for the reference transcriptome using TRANSDECODER ver.5.5 (http://github.com/TransDecoder/TransDecoder/wiki/). To establish homology, the peptide sequences in the ORF were blasted to the Swiss-Prot protein sequences database (Bairoch and Apweiler, 2000) and the NR plant database (https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/) using BLASTP ver.2.7.1 (Altschul et al., 1997). We also used a Python script and idmapping_selected.tab.gz (https://ftp.uniprot.org/pub/databases/uniprot/knowledgebase/idmapping) to determine gene functions according to the GO terms (Ashburner et al., 2000). Then we merged the results of the Swiss-Prot database and the NR plant database as the final annotation.
The population branch statistic (PBS) analysis (Yi et al., 2010) and the Hudson–Kreitman–Aguadé (HKA) test were applied to detect candidate genes under positive selection within each of the two target species, J. microsperma and J. erectopatens. Fifteen individuals of J. sabina were used as the control group, and five individuals of J. saltuaria served as the outgroup. We calculated the population branch statistic for two triplets, J. microsperma–J. sabina–J. saltuaria and J. erectopatens–J. sabina–J. saltuaria, using the Perl script ‘PBS_test.pl’ from Ma et al. (2019) (https://github.com/mayz11/cypress). The FST value was calculated between the following population pairs: (1) the target population and the control population, (2) the target population and outgroup, and (3) the control population and outgroup.
We also used the custom Perl script ‘HKA_test.pl’ from Ma et al. (2019) (https://github.com/mayz11/cypress) to carry out the HKA tests (Hudson et al., 1987). We considered the number of polymorphic sites in the target population (J. microsperma or J. erectopatens) as A, and the number of fixed differences between the target populations and both the control group (J. sabina) and the outgroup (J. saltuaria) as B. The ratio of A/B for each unigene was compared to the transcriptome-wide average, and the null hypothesis A(unigene)/B(unigene) = A (transcriptome-wide)/B(transcriptome-wide) was tested using Pearson’s chi-square test for the 2 × 2 contingency table.
Unigenes with the highest 10 % of the population branch statistic with a significant P-value (<0.05) for the HKA test were considered as candidate genes under positive selection in J. microsperma or J. erectopatens. GO enrichment analysis was conducted using TBtools (Chen et al., 2020). Fisher’s exact test was used to examine the significance of the GO enrichment in which the corrected P-values <0.05 were considered significant.
Detection of environment-associated loci
Latent factor mixed models (LFMMs) (Frichot et al., 2013) were used to measure the associations between SNPs and climatic gradients while accounting for underlying population structure. This method estimates allele–environment correlations between each SNP and each variable at a time. In LFMMs, environmental variables are tested separately and introduced into each model as fixed effects. The number of latent factors (K) is included in the model as a covariate and the environmental gradients were not considered in the analysis (Frichot et al., 2013).
A total of 19 climate variables (Supplementary Data Table S2; 1970–2000) were retrieved from WorldClim (https://www.worldclim.org/; Hijmans et al., 2005). We extracted the climate variables for each population location using the R package DISMO (Hijmans et al., 2015). To avoid multicollinearity, we discarded variables that were highly correlated (Pearson’s r > 0.8). The remaining subset of four uncorrelated BIOCLIM variables, BIO3, BIO10, BIO17, and BIO19, was retained for further analysis. For each of the four uncorrelated BIOCLIM variables, we ran five independent runs to simulate the correlation between SNPs and climate factors, using 100 000 iterations and 50 000 burn-in with a latent factor of K = 2, according to the results of ADMIXTURE, in the R package LEA (Frichot and François, 2015). The five separate runs had nearly similar |z|-scores. Second, we calculated the mean |z|-scores and considered a false discovery rate (FDR) of 0.05 to be significant. Finally, we conducted a GO enrichment analysis.
Assessment of genetic load
Nucleotide diversity (θ π) of zero-fold and four-fold degenerate sites, as well as the ratio between the two indices, were calculated within coding regions based on the annotation of J. microsperma (Petit et al., 2008; Marsden et al., 2016; Wang et al., 2021). The zero-fold and four-fold degenerate sites were identified by iterating across all four possible bases at each site along a transcript and recording the changes in the resulting amino acid using a Perl script (https://github.com/wk8910/bio_tools/blob/master/00.scripts/get_0fold-4fold_sites.pl). Sites were classified as zero-fold degenerate when the four different bases resulted in four different amino acids. However, sites were considered as four-fold degenerate when no changes in amino acids were detected. We also estimated the θπ(0-fold)/θπ(4-fold) ratios for each species to test the accumulation of missense mutations (Wang et al., 2021).
RESULTS
Reference transcriptome assembly and SNP calling
The assembled reference transcriptome of J. microsperma contained 61 700 unigenes with an average length of 907 bp and a contig N50 of 1793 bp. After removing low-quality Illumina sequences, 3.6 Gb of reads remained for the assayed 84 samples (56 from J. microsperma and 28 from J. erectopatens). Following the alignment of the transcriptome reads from all individuals to the reference transcriptome and undertaking stringent quality filtering with 50 % missing data (SNP) per species, we identified 149 052 high-quality SNPs across all studied individuals. A total of 81 775 SNPs were shared between J. microsperma and J. erectopatens, and the number of species-specific SNPs were 20 758 and 46 519, respectively (Supplementary Data Fig. S1).
Population genetic structure and genetic diversity
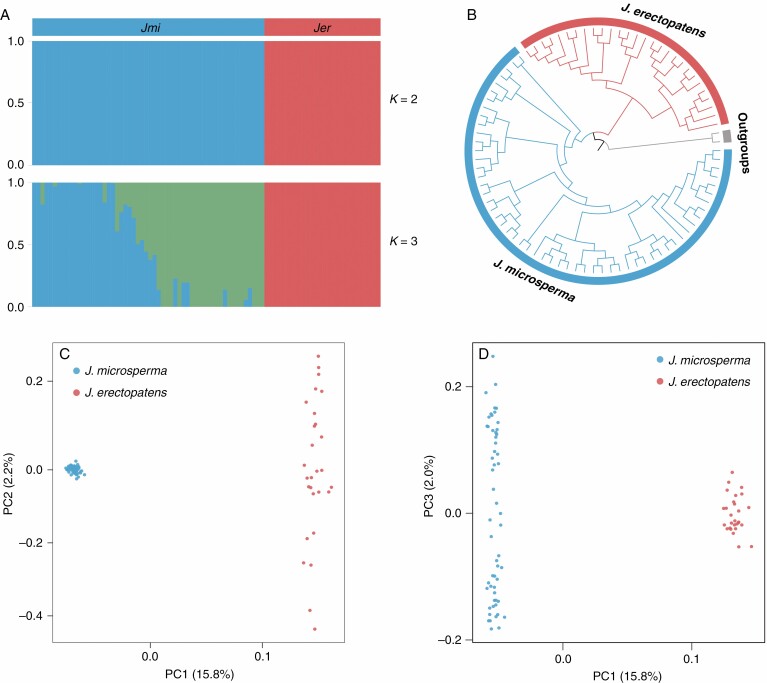
Our phylogenetic analysis based on 149 052 SNPs showed that samples belonging to each species, J. microsperma and J. erectopatens, were clustered into a separate monophyletic clade (Fig. 2B). PCA based on SNPs also distinguished the two species along PC1 (variance explained = 15.8 %; Fig. 2C, D). Furthermore, analysis of population genetic structure clearly assigned the individuals of J. microsperma and J. erectopatens into a species-specific group when K = 2, which was estimated to be the optimal value of K (Fig. 1C; Supplementary Data Figs S2 and S3). In addition, the two species were well differentiated based on FST (0.233) and dXY (0.319) values (Table 2; Fig. S4).
Fig. 2.
Population genetic and phylogenetic analyses of J. microsperma and J. erectopatens based on 149 052 SNPs. (A) ADMIXTURE plots for K = 2 and 3. The x-axis shows the individuals of J. microsperma (Jmi) and J. erectopatens (Jer) with each vertical bar representating an individual; the y-axis quantifies the proportion of an individual’s inferred ancestry. See also Supplementary Data Figs S2 and S3. (B) Maximum-likelihood phylogenetic tree. (C, D) Principal component analysis (PCA) plots of the two studied species showing PC1 vs. PC2 and PC1 vs. PC3, respectively.
Table 2.
Population genetic statistics of J. microsperma and J. erectopatens
| Population | θ π a | θ w b | H o c | H e d | Tajima’s D | F ST e | d XY f |
|---|---|---|---|---|---|---|---|
| J. erectopatens | J. erectopatens | ||||||
| J. microsperma | 0.00129 | 0.00099 | 0.19768 | 0.16934 | 0.68151 | 0.23363 | 0.31924 |
| J. erectopatens | 0.00222 | 0.00139 | 0.36917 | 0.28872 | 1.50340 | - | - |
aNucleotide diversity calculated by pairwise differences between sequences.
bNucleotide diversity calculated by numbers of segregating sites between sequences.
cObserved heterozygosity.
dExpected heterozygosity.
ePairwise mean relative divergence.
fPairwise mean absolute sequence divergence.
The transcriptome-wide average values for the two indicators of nucleotide diversity, θπ and θw, were higher for J. erectopatens (θπ = 0.00222, θw = 0.00139) than for J. microsperma (θπ = 0.00129, θw = 0.00099) (Table 2). Observed (Ho) and expected (He) heterozygosity were lower in J. microsperma (Ho = 0.198, He = 0.169) compared to J. erectopatens (Ho = 0.369, He = 0.289; Table 2). To avoid a potential bias due to different sample numbers per population, we selected a small subset of individuals (seven from five populations, and five from two populations) for measurements of nucleotide diversity parameters. All six populations of J. micropserma presented values of θπ, θw, Ho and He lower than for the single population of J. erectopatens (Supplementary Data Table S3). Tajima’s D indicated that a large number of alleles with medium frequency were retained in J. microsperma (D = 0.68151) and J. erectopatens (D = 1.50340, Table 2), suggesting that both species may have experienced population bottleneck(s) and/or balanced selection in their population evolutionary history.
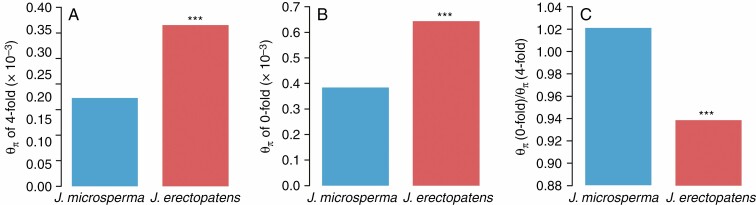
Juniperus microsperma had relatively lower θπ compared with J. erectopatens in both zero- and four-fold sites (Fig. 3). The θπ (0-fold degeneration variants)/θπ (4-fold degeneration variants) ratio for each species was higher for J. microsperma than for J. erectopatens (Fig. 3C), suggesting that J. microsperma has accumulated more missense mutations and possesses a higher genetic load.
Fig. 3.
Population genetics statistics. (A) Nucleotide diversity (θ π) on four-fold degeneration sites. (B) Nucleotide diversity (θ π) on zero-fold degeneration sites. (C) Nucleotide diversity (θ π) of zero-fold degeneration sites over nucleotide diversity (θ π) of four-fold degeneration sites. For each statistic, Juniperus erectopatens populations were compared with the J. microsperma population. ***Non-adjusted P < 0.001.
Gene flow and demographic history
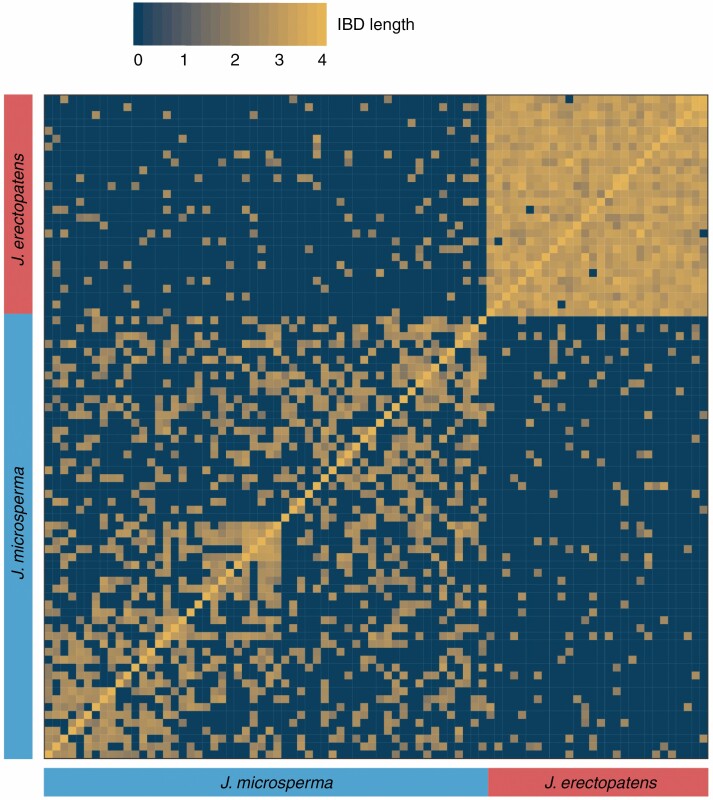
To detect whether gene flow existed between J. microsperma and J. erectopatens, we identified shared haplotypes between these two species using the refined IBD approach (Browning and Browning, 2013). Pairwise comparisons between individuals showed that haplotype sharing was common between J. microsperma and J. erectopatens (131 out of 1568 pairwise comparisons indicated haplotype sharing; Fig. 4). All shared haplotypes were short in length, due to the relatively short length of unigenes in the transcriptome dataset. However, they were of sufficient length to reveal intra- and inter-specific gene flow (Ma et al., 2019). In summary, these results indicated that historical gene flow has occurred between J. microsperma and J. erectopatens.
Fig. 4.
Estimation of shared haplotypes between individuals of J. microsperma and J. erectopatens. Heatmap colours represent the local length of IBD blocks for each pairwise comparison.
The timing of speciation, population contraction(s) and expansion(s), and levels of gene flow between the two species were estimated using coalescent-based simulations in fastsimcoal2 (Excoffier et al., 2013). First, we established six demographic models for J. microsperma and J. erectopatens covering a large range of demographic possibilities (Supplementary Data Fig. S5, Model-A to Model-F). These six demographic models were identified as representative scenarios based on the current population size and distribution of J. microsperma and J. erectopatens. These models were also representative of the demographic histories of QTP’s rare conifers in previous studies (Shang et al., 2015; Xu et al., 2019). AIC results favoured for J. microsperma the model where two sequential bottleneck events occurred, whereas for J. erectopatens, they favoured the model in which one bottleneck event took place (Supplementary Data Fig. S5). Based on these preferred models, we merged the best suggested demographic scenarios to form the optimal model for the demographic relationship between the two species. Because the bottleneck events detected in the two species may have occurred in their common ancestor, we included in the models further bottleneck events and divergence times. After comparing four models (Fig. S5, Model-1 to Model-4), Model-3 was shown to be the optimal one. This model suggested that J. microsperma and J. erectopatens diverged from their common ancestor after the first bottleneck event (Fig. S5).
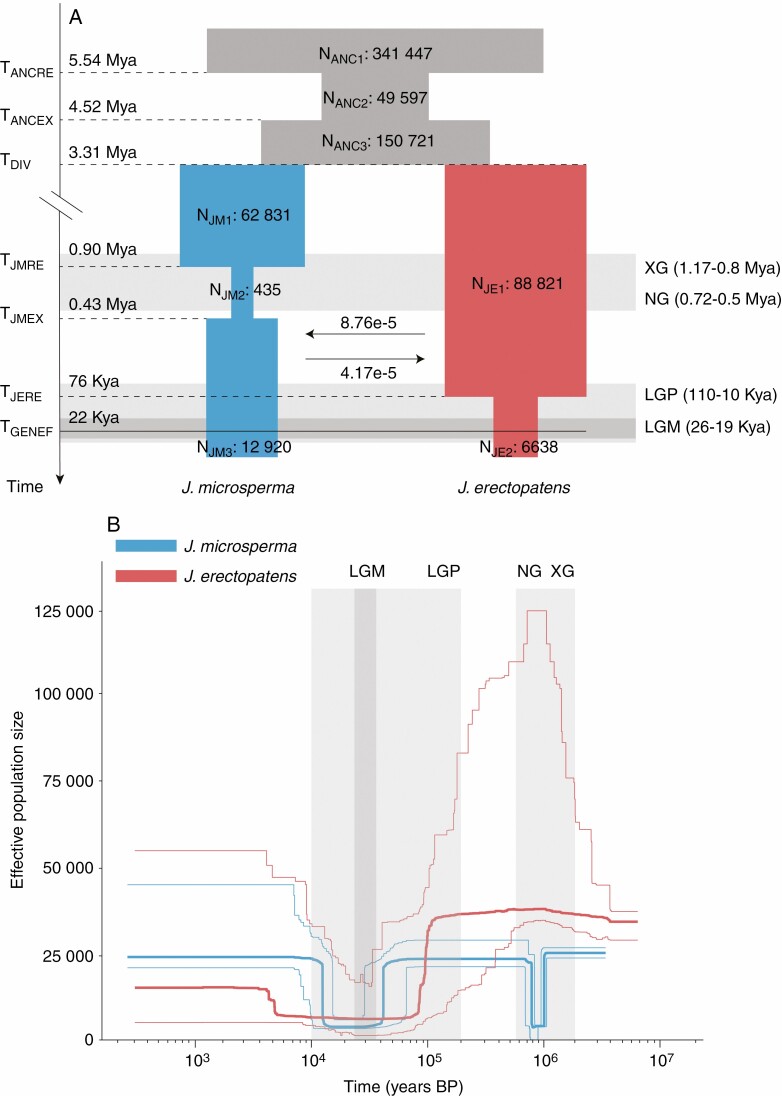
The best-fitting model (with minimum Akaike’s weight value) containing 17 parameters (Supplementary Data Tables S4 and S5; Fig. 5A) indicated that J. microsperma and J. erectopatens diverged from each other ~3.31 million years ago (Mya) (95 % CI: 2.62–3.74 Mya; Table S5; Fig. 5A). After divergence, J. microsperma experienced one bottleneck event between 0.90 Mya (CI: 0.67–1.34 Mya) and 0.43 Mya (CI: 0.31–0.48 Mya) with a decrease in effective population size (Ne) from 62 831 (CI: 33 345–85 507) to 435 (CI: 124–2533). Later, the population size of J. microsperma increased to 12 920 (CI: 12 157–16 570) but failed to recover to the pre-bottleneck size (Fig. 5A; Table S5). In contrast, J. erectopatens only experienced one population contraction at ~76 kya (CI: 51–124 kya), with a decrease in Ne from 88 821 (CI: 63 250–95 861) to 6638 (CI: 6207–9243) (Table S5; Fig. 5A). After divergence, the gene flow between the two species persisted until ~22 kya (CI: 16–35 kya) and was slightly higher from J. erectopatens to J. microsperma than in the opposite direction (8.76 × 10−5 vs. 4.17 × 10−5) (Table S5; Fig. 5A).
Fig. 5.
Demographic history of J. microsperma and J. erectopatens. The light grey areas represent different glaciation events during the Pleistocene (XG, Xixiabangma Glaciation; NG, Naynayxungla Glaciation; LGP, Last Glaciation Period; LGM, Last Glaciation Maximum). (A) Schematic illustration of the best demographic scenario using fastsimcoal2. Estimated effective population sizes (Ne), divergence time, bottleneck time and phased gene flow are indicated. The number next to the horizontal arrows represents the per-generation migration rate between populations. The full line indicates the time when gene flow stopped. See also Supplementary Data Table S5. (B) Changes in Ne over time in J. microsperma and J. erectopatens inferred by the Stairway Plot method. Thick and thin light lines represent the median and the 95 % pseudo-CI defined by the 2.5 % and 97.5 % estimations, respectively, using the site frequency spectrum analysis.
To obtain more robust interpretations of the demographic history of these two species, we used a Stairway Plot analysis. This analysis showed that the effective population size of J. microsperma declined sharply at ~1.0 Mya, and then quickly arose about 0.8 Mya (Fig. 5B). After this, Ne declined again around 40 kya and increased to its current level at about 13 kya (Fig. 5B), suggesting that J. microsperma experienced two severe population bottleneck events. The Ne of J. erectopatens displayed a slow decline around ~70–100 kya and then expanded rapidly around 3–5 kya (Fig. 5B). This indicates that J. erectopatens experienced a single bottleneck event that lasted longer, yet was less severe and less recovered than the (roughly) contemporary bottleneck experienced by J. microsperma.
Detection of positive selection genes
We found a total of 24 847 coding peptide sequences from the reference transcriptome. Of these, 17 883 and 18 056 peptide sequences were annotated using the Swiss-prot and the NR plant database, respectively. After merging the outputs from both databases, a total of 19 786 peptide sequences were annotated.
To explore the genetic basis of possible adaptation in J. microsperma and J. erectopatens, PBS and HKA tests were conducted to identify genes under positive selection within the two species. In total, 183 and 85 positively selected candidate genes were identified within J. microsperma and J. erectopatens, respectively. The over-represented GO terms identified by GO enrichment analysis for positively selected candidate genes within J. microsperma included aerenchyma formation, positive regulation of salicylic acid-mediated signalling pathway, response to decreased oxygen levels, and the terpenoid biosynthetic process (Supplementary Data Table S6). In contrast, no significant GO enrichment category was detected for the positively selected candidate genes within J. erectopatens (Table S7). Nevertheless, four marginally significant GO enrichment categories were detected, including glycosyl and organonitrogen compound biosynthetic process, antiporter and secondary active transmembrane transporter activity (0.07 < P < 0.08; Table S7).
Detection of environment-associated loci
We found that 18 723, 7883, 10 118 and 9799 SNPs were associated with the climate variables BIO3, BIO10, BIO17 and BIO19 (described above), respectively (FDR-corrected P < 0.05). From the 2127 SNPs associated with all four variables, we identified 49 categories of genes of various function detailed in Supplementary Data Table S8. Furthermore, six of these gene categories are involved in response to abiotic and biotic stimuli such as the immune system process, ethylene biosynthetic process, response to oxygen-containing compounds, defence response to insect, response to stress and response to hormone (Table 3).
Table 3.
GO enrichment of environment-associated genes (LFMM) within J. microsperma and J. erectopatens
| GO.ID | Term | Annotated | Significant | Expected | P-value |
|---|---|---|---|---|---|
| GO:0002376 | Immune system process | 39 | 4 | 1.33 | 4.10E-06 |
| GO:0009693 | Ethylene biosynthetic process | 26 | 3 | 0.88 | 5.00E-05 |
| GO:1901700 | Response to oxygen-containing compound | 99 | 2 | 3.37 | 0.03501 |
| GO:0002213 | Defence response to insect | 14 | 1 | 0.48 | 0.03798 |
| GO:0006950 | Response to stress | 291 | 6 | 9.9 | 0.04289 |
| GO:0009725 | Response to hormone | 17 | 1 | 0.58 | 0.04593 |
P-value was calculated by Classic Fisher.
DISCUSSION
Genetic differentiation and diversity of J. microsperma and J. erectopatens
All our population genomic analyses suggested that J. microsperma and J. erectopatens are well differentiated (Fig. 2A, C, D). Similarly, the phylogenetic tree showed that all individuals of J. microsperma and J. erectopatens formed a monophyletic clade (Fig. 2B). This is consistent with the most recent phylogenetic study for Juniperus based on ITS and four chloroplast genes (Adams and Schwarzbach, 2013). Furthermore, the pairwise FST and dXY values indicated moderate to high levels of genetic differentiation between these two species (Table 2; Supplementary Data Fig. S4).
In general, species that occupy a larger distribution range tend to possess higher levels of genetic variation than those with a smaller range and population size (O’Brien, 1994; Frankham, 1997). However, we detected a contrasting pattern for these two rare juniper species, because J. erectopatens occurs in a much smaller geographical area than J. microsperma (Fig. 1) and has twice the number of species-specific SNPs compared to J. microsperma (Supplementary Data Fig. S1). Furthermore, the nucleotide (θ) and gene diversity (He) of J. erectopatens were higher than those of J. microsperma (Table 2; Table S3), suggesting higher levels of diversity in J. erectopatens. Interestingly, the nucleotide diversities of both J. microsperma (θπ = 0.00129, θw = 0.00099) and J. erectopatens (θπ = 0.00222, θw = 0.00139) found in this study were lower than those previously published for other conifer species: for example, θπ = 0.0029 for Cupressus gigantea W.C.Cheng & L.K.Fu., θπ = 0.0031 for C. duclouxiana Hickel (Ma et al., 2019), θπ = 0.00770 for C. chengiana S.Y.Hu (Li et al., 2020b), θπ = 0.00411 for Picea likiangensis (Franch) E.Pritz., θπ = 0.00392 for P. purpurea Mast. and θπ = 0.00392 for P. wilsonii Mast. (Ru et al., 2018), whereas θw = 0.0144 for C. gigantea and θw = 0.0151 for C. duclouxiana (Ma et al., 2019). Different factors may have contributed to the relatively low levels of genetic diversity of J. microsperma and J. erectopatens, such as the evolutionary history and the current small population sizes as indicated by the narrow geographical distribution of the two species. This is supported by Tajima’s D < 0 (Table 2), which suggests that the two junipers have experienced bottlenecks or balancing selection in their demographic history. Rare alleles are lost during such events, leading to reduced diversity (Frankham et al., 1999; Spielman et al., 2004; Ellegren and Galtier, 2016). Furthermore, the loss of rare alleles due to genetic drift is more pronounced when Ne is small, further decreasing genetic diversity (Stoffel et al., 2018; Hahn, 2019; Freeland, 2020). Hence, the relatively small population sizes in both J. microsperma and J. erectopatens would also have contributed to the low genetic diversity within these two species.
Contrasting range and diversity pattern reflects different demographic history
Our demographic models using fastsimcoal2 and Stairway Plot indicated that the two species went through bottlenecks in the past. However, the results for each species were somewhat contradictory between the two approaches as J. microsperma experienced one bottleneck according to the fastsimcoal2 analysis (Supplementary Data Table S5; Fig. 5A) and two according to Stairway Plot (Fig. 5B). Similarly, although J. erectopatens had one bottleneck event in both simulations, a modest population expansion was detected using Stairway Plot which was not apparent using fastsimcoal2 (Fig. 5B). Both methods rely on the (SNP) SFS to infer the demographic history of species, although the internal algorithm of each software is different (Excoffier et al., 2013; Liu and Fu, 2015). Fastsimcoal2 uses a series of pre-defined demographic models and an ML approach to estimate the best-fitting model (Excoffier et al., 2013), whereas Stairway Plot is a model-independent method that uses the expected composite likelihood method (Liu and Fu, 2015). In our analyses, both methods detected demographic events around or before the Last Glacial Period (LGP), yet Stairway Plot detected recent demographic events that fastsimcoal2 did not (Fig. 5). Hence, the resolution of fastsimcoal2 to recent demographic events may be less sensitive compared to Stairway Plot. However, it has been shown that despite these shortcomings a combination of the two approaches can reveal deeper insights into the demographic history of a species (Hansen et al., 2018).
The key points which emerge from the demographic analyses are that during the Quaternary J. microsperma experienced considerable population bottleneck(s), which probably caused the current low levels of genetic diversity. One bottleneck occurred between 0.90 and 0.43 Mya (Supplementary Data Table S5; Fig. 5A), which corresponds to two of the most intense periods of glaciation on the QTP, namely the Xixiabangma Glaciation (1.2–0.8 Mya) and the Naynayxungla Glaciation (0.72–0.5 Mya) (Zheng et al., 2002; Zhou et al., 2011). During this bottleneck event, the Ne of J. microsperma shrank severely, from 62 831 to 435 and only recovered to about one-fifth of the pre-bottleneck size (12 920; Table S5; Fig. 5A), which emphasizes the severity of this event. Another bottleneck might have occurred more recently between 13 and 40 kya (Table S5; Fig. 5B) which coincides with the Last Glacial Maximum (LGM; Yokoyama et al., 2000; Clark et al., 2009), in which the population shrank and then expanded rapidly to nearly 100 % of pre-bottleneck levels. These periods coincided with the Xixiabangma Glaciation and the LGM (Yokoyama et al., 2000; Clark et al., 2009). The effective population size Ne of J. microsperma was reduced by 80–99 % but recovered substantially after the event(s) to somewhere between 25 % and nearly 100 % of pre-bottleneck levels, depending on the demographic approach (Fig. 5). Despite the substantially different estimates of the effective population sizes between the two approaches, the results are consistent in that J. microsperma experienced major population losses but was able to increase population size significantly after the events. This is also consistent with previous studies which showed that the periods with the most extensive glaciation and the LGM have caused significant changes to the distribution and Ne of plants on the QTP (Sun et al., 2014; Ren et al., 2017; Chen et al., 2019; Ma et al., 2019; Feng et al., 2020; Li et al., 2020a, b).
Unlike for J. microsperma, both methods indicated only a single population contraction event for J. erectopatens. This occurred around 100–70 kya (depending on the method used) with a reduction in Ne of between 81% and 93 % (Supplementary Data Table S5; Fig. 5A). This event coincided with the beginning of the LGP around 110 kya (Thompson et al., 1997) and is consistent with previous studies that also indicated that this glacial period affected the distribution of plants on the QTP (Wang et al., 2009; Shimono et al., 2010). After a considerable period of population size depression, J. erectopatens may have experienced a population expansion around 5–3 kya when the climate became more stable (Yu et al., 1997; Hou et al., 2017) but this is only evident from the Stairway Plot analysis (Fig. 5B).
One unexpected result of this study was the lower level of (whole transcriptome-wide) genetic diversity in J. microsperma compared to J. erectopatens, despite the currently larger population size and distribution range of J. microsperma. The results from both demographic analyses suggested that J. microsperma had about a 30 % smaller ancestral effective population size (NJM1, Fig. 5) than J. erectopatens (NJE1, Fig. 5). Therefore, it is likely that this also corresponds to an initially lower level of genetic diversity, which was further reduced during one or even two bottleneck events (Fig. 5).
The contrasting demographic history of the two junipers may also be a reflection of the heterogeneity of changes in the local climate on the eastern QTP. Previous studies have shown that climate changes on the QTP affected the distribution and effective population size of plants (Ru et al., 2016; Chen et al., 2019; Ma et al., 2019; Zhang et al., 2019; Li et al., 2020b). For example, species which occur in different regions on the QTP may have experienced a distinct demographic history due to different local climates and geographical factors (Owen and Dortch, 2014). Juniperus microsperma occurs in the deep river valley of the Parlung Zangpo around 3150–3350 m (Adams, 2014; Shang et al., 2015), which is surrounded by numerous mountain peaks (>5000 m) and modern glaciers (Zheng and Rutter, 1998; Yang et al., 2010). In contrast, J. erectopatens occurs in the upper Minjiang river valley (Adams, 2014), where the local terrain is less rugged and is covered by fewer glaciers (Wang et al., 2017). Hence, the expansion of local glaciers during colder periods (Zhou et al., 2010) might have affected J. microsperma more severely, reducing its population size but permitting rapid expansion during warmer periods. This could be the reason why Stairway Plot identified two bottleneck events for J. microsperma, which coincide with two glacial periods on the QTP. Our results are in agreement with the geological evidence suggesting that the palaeoclimatic changes in different regions on the QTP and the Himalayas may be driven by different factors. Indeed, while glaciers in some regions are likely to respond to the climate oscillations of the Northern Hemisphere, other regions may be more influenced by the southern Asian monsoon (Owen and Dortch, 2014).
Speciation history of the two junipers
The divergence time of J. microsperma and J. erectopatens was estimated to have occurred during the Pliocene, at around 3.13 Mya (CI: 2.62–3.74 Mya) (Supplementary Data Table S5; Fig. 5A). A possible trigger for this speciation might have been the uplift events on the eastern QTP, although the timing of these events is controversial (Wang et al., 2008; Deng and Ding, 2015; Renner, 2016; Su et al., 2019). The QTP has experienced phased uplifts since the Eocene (Wang et al., 2014; Favre et al., 2015), but these events were heterogeneous both spatially and temporally. Indeed, different parts of the QTP have different uplift histories as well as climatic histories (Mulch and Chamberlain, 2006; Deng and Ding, 2015; Favre et al., 2015; Muellner-Riehl, 2019). By the late Pliocene, mountain uplifts on the eastern QTP (including the Hengduan Mountains, HDM) were completed (~3.6 Mya; Sun et al., 2011; Xing and Ree, 2017), and vegetation reconstruction based on macrofossil floras suggested that the south-eastern QTP had probably reached roughly its current elevation by around the same period (Sun et al., 2011). The rugged terrain of the HDM may have acted as a natural geographical barrier to pollen flow and seed dispersal for many taxa in this region (Feng et al., 2016; Shahzad et al., 2017; Ru et al., 2018; Zhang et al., 2018; Li et al., 2020b). The fastsimcoal2 analysis suggested that the common ancestor of J. microsperma and J. erectopatens experienced a population expansion about 4.52 Mya (Fig. 5B). The geological changes during the late Pliocene with the uplift of the HDM might have fragmented these populations which ultimately diverged under the influence of genetic drift and adapted to different habitats giving rise to the lineages of J. microsperma and J. erectopatens. The divergence of several conifer lineages which are endemic to the QTP and the HDM have also been associated with the uplift of the HDM in the late Pliocene (Sun et al., 2018; Ma et al., 2019; Li et al., 2020b).
Adaptation of the two junipers to different habitats
Elucidating the genetic basis of local adaptation to environmental factors is crucial for our understanding of plant evolution. In the long term, local adaptation helps plants to face selection pressures due to climate change and environmental heterogeneity. We found genetic evidence that J. microsperma and J. erectopatens may have adapted to different environmental stressors. A total of 183 and 85 loci were identified as positively selected candidate genes in J. microsperma and J. erectopatens, respectively, but only in J. microsperma were significant GO enrichment categories detected (Supplementary Data Tables S6 and S7). Enriched categories included response to decreased oxygen levels, aerenchyma formation and response to external stimulus (Table S6). Aerenchyma is a cavity-filled parenchymatous tissue that can be found in the roots, stems and leaves, especially from plants growing in oxygen-deficient environments (Schussler and Longstreth, 1996; Watkin et al., 1998). The presence of J. microsperma at high altitudes (3150–3350 m), which are characterized by lower oxygen levels and stronger UV radiation compared to lower altitudes, might have contributed to the positive selection of the related genes to adapt to these environmental stressors. Furthermore, genes related to the regulation of the salicylic acid biosynthetic process, the positive regulation of the salicylic acid-mediated signalling pathway, and the response to trehalose were also enriched (Table S6). Salicylic acid, a phenolic compound, and trehalose, a disaccharide, play a role in the resistance of plants to stress (Ding et al., 2002; Kaplan et al., 2004). These positively selected genes related to stress resistance might have resulted from the climate oscillation and constant environmental changes that occurred during the evolutionary history of J. microsperma.
Although no significant GO enrichment categories were detected in J. erectopatens, four were close to being significant (0.07 < P < 0.08), including the glycosyl and organonitrogen compound biosynthetic process as well as the antiporter and secondary active transmembrane transporter activity (Supplementary Data Table S7). The GO enrichment categories found in J. erectopatens do not overlap with those detected in J. microsperma, which suggests that the evolution of the species pair may have responded to different environmental stressors.
Furthermore, our results suggest that based on the local climate data for the period between 1970 and 2000, thousands of SNPs are associated with the climate variables BIO3, BIO10, BIO17 and BIO19, of which 2127 SNPs were shared between all four variables. In addition, we identified 49 categories of genes associated with the local environment (Supplementary Data Table S8). Six of these gene categories are involved in the response to abiotic and biotic stimuli, such as the immune system process, the ethylene biosynthetic process, the response to oxygen-containing compounds, the defence against insects, the response to stress and the response to hormones (Table 3). We also found several gene categories related to habitat adaptation, such as the carbohydrate metabolic process, embryo development and reproduction (Table S8). There was also an overlap between significantly enriched gene categories of positively selected genes and climate-associated SNPs (Tables S6 and S8). This indicates that the two studied juniper species have the capacity to adapt to climatic and environmental fluctuations, suggesting that this ability might have been an important factor in their survival and adaptation to historical abiotic and biotic stressors.
Conservation implications
Narrow endemics such as J. microsperma and J. erectopatens are very vulnerable to extinction, especially in a rapidly changing climate and during increased habitat fragmentation. However, many of the remaining trees in fragmented populations may serve as reproductively viable individuals and would therefore be very important for the long-term recovery of populations and genetic conservation programmes (Ralls et al., 2020). Thus, studying the genetic diversity and demographic history of threatened species may inform conservation programmes of these species.
Based on our recent field surveys, it is likely that the two studied juniper species would be evaluated as ‘Endangered’ according to IUCN Red List criteria (IUCN, 2012) (see also Shang et al., 2015; Tso et al., 2019; Xu et al., 2019). Therefore, conservation measures should be taken to minimize the risk of extinction. Based on the relatively low genetic diversity and high θπ(0-fold)/θπ(4-fold) ratio of J. microsperma compared to J. erectopatens, this species is likely to have a higher genetic load than J. erectopatens. This suggests lower fitness at the population level, which might increase the probability of extinction (Klekowski, 1988; Glémin et al., 2003; Stewart et al., 2017). Moreover, our field surveys from 2011 to 2018 indicated that the area of occupancy and habitat quality of this rare juniper is declining due to human activities (Shang et al., 2015; Tso et al., 2019). It is therefore important to raise awareness in the local community of the importance of protecting J. microsperma.
Although J. erectopatens has higher genetic diversity and probably a lower genetic load than J. microsperma, only about 100 mature individuals occur in an area of around 2 km2 at Anhong, Songpan, Sichuan (Xu et al., 2019). No natural regeneration of J. erectopatens was evident during our field surveys, which might indicate that this species has a higher risk of extinction compared to J. microsperma. In this case, seed collection for ex situ conservation and artificial breeding programmes using seeds and cuttings should be considered as top priority conservation actions. Educating the local community on the ecological importance of J. erectopatens is an equally important measure to maintain and improve the long-term chances of its survival.
CONCLUSION
Here we have presented evidence that uplift of the eastern QTP triggered the speciation of two well-differentiated, narrow-endemic juniper species, and that climatic changes in the last million years have shaped their demographic histories differently. Our study highlights the importance of speciation and demographic history reconstruction to understand the current distribution pattern and genetic diversity level of threatened species and to help implement conservation and management strategies.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1. Location information for sampled individual of J. microsperma and J. erectopatens. Table S2. Environmental variables used in this study. Table S3. Genetic diversity of J. microsperma and J. erectopatens populations based on seven randomly selected individuals per population apart from populations 3 and 4 where only five individuals per population were available. Table S4. Summary of parameters for the five final candidate models tested using fastsimcoal2. Table S5. Inferred parameters and confidence intervals for the best-fitting demographic model presented in Fig. 5A. Table S6. GO enrichment of positively selected genes within J. microsperma. Table S7. GO enrichment of positively selected genes within J. erectopatens. Table S8. GO enrichment of environment-associated genes (LFMM) within J. microsperma and J. erectopatens. Figure S1. Number of shared and species-specific SNPs in J. microsperma and J. erectopatens. Figure S2. The cross-validation (CV) error for each K value that was estimated using ADMIXTURE. Figure S3. Population structure plots for K = 2 to K = 4. Figure S4. Genetic differentiation between J. microsperma and J. erectopatens. Figure S5. Schematic illustration of candidate demographic models that were simulated and tested in fastsimcoal2; the value below each model is the AIC result of that model.
AUTHOR CONTRIBUTIONS
K.M. designed and supervised this study. J.M., W.W., J.L. and J.X. managed fieldwork and collected the materials. J.M., J.L. and H.Y. analysed the data. J.M. and K.M. wrote the manuscript. P.F., R.M., M.R., G.M. and L.O. revised the manuscript. J.M., P.F., W.W., M.R., H.Y. and K.M. finalized the manuscript.
FUNDING
This work was supported by the National Natural Science Foundation of China (grant numbers U20A2080, 31622015), the National Basic Research Program of China (grant number 2014CB954100) and Sichuan University (Fundamental Research Funds for the Central Universities, SCU2021D006, SCU2020D003). The Royal Botanic Garden Edinburgh is supported by the Scottish Government’s Rural and Environment Science and Analytical Services Division.
LITERATURE CITED
- Adams RP. 2014. Junipers of the world: the genus Juniperus, 4th edn. Bloomington: Trafford Publishing. [Google Scholar]
- Adams RP, Schwarzbach AE. 2013. Phylogeny of Juniperus using nrDNA and four cpDNA regions. Phytologia 95: 179–187. [Google Scholar]
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- Alexander DH, Lange K. 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf FW, Hohenlohe PA, Luikart G. 2010. Genomics and the future of conservation genetics. Nature Reviews Genetics 11: 697–709. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research 28: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnosky AD, Matzke N, Tomiya S, et al. 2011. Has the Earth’s sixth mass extinction already arrived? Nature 471: 51–57. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. 2013. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Marce M, Soriano L, López-Bao JV, Godoy JA. 2013. Genetics at the verge of extinction: insights from the Iberian lynx. Molecular Ecology 22: 5503–5515. [DOI] [PubMed] [Google Scholar]
- Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. 2015. Accelerated modern human-induced species losses: entering the sixth mass extinction. Science Advances 1: e1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos G, Ehrlich PR, Dirzo R. 2017. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proceedings of the National Academy of Sciences of the United States of America 114: E6089–E6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, et al. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant 13: 1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen JH, Huang Y, Brachi B, et al. 2019. Genome-wide analysis of Cushion willow provides insights into alpine plant divergence in a biodiversity hotspot. Nature Communications 10: 5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, et al. 2009. The last glacial maximum. Science (New York, N.Y.) 325: 710–714. [DOI] [PubMed] [Google Scholar]
- Cronk Q. 2016. ECOLOGY. Plant extinctions take time. Science 353: 446–447. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. ; 1000 Genomes Project Analysis Group . 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Ding L. 2015. Paleoaltimetry reconstructions of the Tibetan Plateau: progress and contradictions. National Science Review 2: 417–437. [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffenbaugh NS, Giorgi F. 2012. Climate change hotspots in the CMIP5 global climate model ensemble. Climatic Change 114: 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding CK, Wang CY, Gross KC, Smith DL. 2002. Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta 214: 895–901. [DOI] [PubMed] [Google Scholar]
- Dirzo R, Young HS, Galetti M, Ceballos G, Isaac NJ, Collen B. 2014. Defaunation in the Anthropocene. Science 345: 401–406. [DOI] [PubMed] [Google Scholar]
- Duan Y-W, Liu J-Q. 2006. Pollinator shift and reproductive performance of the Qinghai-Tibetan Plateau endemic and endangered Swertia przewalskii (Gentianaceae). Biodiversity and Conservation 16: 1839–1850. [Google Scholar]
- Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends in Ecology & Evolution 29: 51–63. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Galtier N. 2016. Determinants of genetic diversity. Nature Reviews Genetics 17: 422–433. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. 2013. Robust demographic inference from genomic and SNP data. PLoS Genetics 9: e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Zheng H, Milne RI, Zhang L, Mao K. 2018. Strong population bottleneck and repeated demographic expansions of Populus adenopoda (Salicaceae) in subtropical China. Annals of Botany 121: 665–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre A, Päckert M, Pauls SU, et al. 2015. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biological Reviews of the Cambridge Philosophical Society 90: 236–253. [DOI] [PubMed] [Google Scholar]
- Feng L, Ruhsam M, Wang YH, Li ZH, Wang XM. 2020. Using demographic model selection to untangle allopatric divergence and diversification mechanisms in the Rheum palmatum complex in the Eastern Asiatic Region. Molecular Ecology 29: 1791–1805. [DOI] [PubMed] [Google Scholar]
- Feng L, Zheng QJ, Qian ZQ, et al. 2016. Genetic structure and evolutionary history of three Alpine sclerophyllous oaks in East Himalaya-Hengduan mountains and adjacent regions. Frontiers in Plant Science 7: 1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjeldså J. 1994. Geographical patterns for relict and young species of birds in Africa and South America and implications for conservation priorities. Biodiversity and Conservation 3: 207–226. [Google Scholar]
- Foote AD, Vijay N, Ávila-Arcos MC, et al. 2016. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nature Communications 7: 11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. 1997. Do island populations have less genetic variation than mainland populations? Heredity 78: 311–327. [DOI] [PubMed] [Google Scholar]
- Frankham R. 2005. Genetics and extinction. Biological Conservation 126: 131–140. [Google Scholar]
- Frankham R, Lees K, Montgomery ME, England PR, Lowe EH, Briscoe DA. 1999. Do population size bottlenecks reduce evolutionary potential? Animal Conservation 2: 255–260. [Google Scholar]
- Freeland JR. 2020. Molecular ecology. Oxford: John Wiley & Sons Ltd. [Google Scholar]
- Frichot E, François O. 2015. LEA: an R package for landscape and ecological association studies. Methods in Ecology and Evolution 6: 925–929. [Google Scholar]
- Frichot E, Schoville SD, Bouchard G, François O. 2013. Testing for associations between loci and environmental gradients using latent factor mixed models. Molecular Biology and Evolution 30: 1687–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li S, Guo Q, Zheng W, Yang R, Li H. 2019. Genetic diversity and population structure of two endemic Cupressus (Cupressaceae) species on the Qinghai-Tibetan plateau. Journal of Genetics 98: 14. [PubMed] [Google Scholar]
- Fu PC, Sun SS, Khan G, et al. 2020. Population subdivision and hybridization in a species complex of Gentiana in the Qinghai-Tibetan Plateau. Annals of Botany 125: 677–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Pardo AP, Ruzzante DE. 2017. Whole-genome sequencing approaches for conservation biology: advantages, limitations and practical recommendations. Molecular Ecology 26: 5369–5406. [DOI] [PubMed] [Google Scholar]
- Garner BA, Hand BK, Amish SJ, et al. 2016. Genomics in conservation: case studies and bridging the gap between data and application. Trends in Ecology & Evolution 31: 81–83. [DOI] [PubMed] [Google Scholar]
- Glémin S. 2003. How are deleterious mutations purged? Drift versus nonrandom mating. Evolution 57: 2678–2687. [DOI] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR. 2016. Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics 17: 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. 2019. Molecular population genetics. New York: Sinauer Associates. [Google Scholar]
- Hamabata T, Kinoshita G, Kurita K, et al. 2019. Endangered island endemic plants have vulnerable genomes. Communications Biology 2: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CCR, Hvilsom C, Schmidt NM, et al. 2018. The muskox lost a substantial part of its genetic diversity on its long road to Greenland. Current Biology 28: 4022–4028. e5. [DOI] [PubMed] [Google Scholar]
- Harrisson KA, Pavlova A, Telonis-Scott M, Sunnucks P. 2014. Using genomics to characterize evolutionary potential for conservation of wild populations. Evolutionary Applications 7: 1008–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Lu A, Zhang Z, Pang H, Zhao J. 2005. Seasonal variation in the regional structure of warming across China in the past half century. Climate Research 28: 213–219. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology: A Journal of the Royal Meteorological Society 25: 1965–1978. [Google Scholar]
- Hijmans RJ, Phillips S, Leathwick J, Elith J. 2015. R package dismo: species distribution modeling, version 1.3-3. http://cran.r-project.org/web/packages/dismo/index.html (17 September 2021).
- Hou J, D’Andrea WJ, Wang M, He Y, Liang J. 2017. Influence of the Indian monsoon and the subtropical jet on climate change on the Tibetan Plateau since the late Pleistocene. Quaternary Science Reviews 163: 84–94. [Google Scholar]
- Huang J, Yang LQ, Yu Y, et al. 2018. Molecular phylogenetics and historical biogeography of the tribe Lilieae (Liliaceae): bi-directional dispersal between biodiversity hotspots in Eurasia. Annals of Botany 122: 1245–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CM, Shaner PJ, Zink RM, et al. 2014. Drastic population fluctuations explain the rapid extinction of the passenger pigeon. Proceedings of the National Academy of Sciences of the United States of America 111: 10636–10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN. 2012. IUCN red list categories and criteria: version 3.1. Gland and Cambridge: IUCN. [Google Scholar]
- Jacquemyn H, Vandepitte K, Roldán-Ruiz I, Honnay O. 2009. Rapid loss of genetic variation in a founding population of Primula elatior (Primulaceae) after colonization. Annals of Botany 103: 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, et al. 2004. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiology 136: 4159–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller LF, Waller DM. 2002. Inbreeding effects in wild populations. Trends in Ecology and Evolution 17: 230–241. [Google Scholar]
- Klekowski EJ. 1988. Genetic load and its causes in long-lived plants. Trees-Structure and Function 2: 195–203. [Google Scholar]
- López-Pujol J, Zhang FM, Sun HQ, Ying TS, Ge S. 2011. Centres of plant endemism in China: places for survival or for speciation? Journal of Biogeography 38: 1267–1280. [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Markus R, Wang Y-H, Li Z-H, Wang X-M. 2020a. Using demographic model selection to untangle allopatric divergence and diversification mechanisms in the Rheum palmatum complex in the Eastern Asiatic Region. Molecular Ecology 29: 1791–1805. [DOI] [PubMed] [Google Scholar]
- Li J, Milne RI, Ru D, et al. 2020. b. Allopatric divergence and hybridization within Cupressus chengiana (Cupressaceae), a threatened conifer in the northern Hengduan Mountains of western China. Molecular Ecology 29: 1250–1266. [DOI] [PubMed] [Google Scholar]
- Li Z, Zou J, Mao K, et al. 2012. Population genetic evidence for complex evolutionary histories of four high altitude juniper species in the Qinghai-Tibetan Plateau. Evolution 66: 831–845. [DOI] [PubMed] [Google Scholar]
- Lima JS, Telles MP, Chaves LJ, Lima-Ribeiro MS, Collevatti RG. 2017. Demographic stability and high historical connectivity explain the diversity of a savanna tree species in the Quaternary. Annals of Botany 119: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JQ, Duan YW, Hao G, Ge XJ, Sun H. 2014. Evolutionary history and underlying adaptation of alpine plants on the Qinghai-Tibet Plateau. Journal of Systematics and Evolution 52: 241–249. [Google Scholar]
- Liu X, Fu YX. 2015. Exploring population size changes using SNP frequency spectra. Nature Genetics 47: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Möller M, Provan J, Gao LM, Poudel RC, Li DZ. 2013. Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. The New Phytologist 199: 1093–1108. [DOI] [PubMed] [Google Scholar]
- Liu X-L, Qian Z-G, Liu F-H, Yang Y-W, Pu C-X. 2011. Genetic diversity within and among populations of Neopicrorhiza scrophulariiflora (Scrophulariaceae) in China, an endangered medicinal plant. Biochemical Systematics and Ecology 39: 297–301. [Google Scholar]
- Ma Y, Wang J, Hu Q, et al. 2019. Ancient introgression drives adaptation to cooler and drier mountain habitats in a cypress species complex. Communications Biology 2: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Ruhsam M, Ma Y, et al. 2019. A transcriptome-based resolution for a key taxonomic controversy in Cupressaceae. Annals of Botany 123: 153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD, Ortega-Del Vecchyo D, O’Brien DP, et al. 2016. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proceedings of the National Academy of Sciences of the United States of America 113: 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraldo A, Li S, Borregaard MK, et al. 2016. An Anthropocene map of genetic diversity. Science 353: 1532–1535. [DOI] [PubMed] [Google Scholar]
- Mosbrugger V, Favre A, Muellner-Riehl AN, Päckert M, Mulch A. 2018. Cenozoic evolution of geo-biodiversity in the Tibeto-Himalayan region. Mountains, climate, and biodiversity. Hoboken: John Wiley & Sons. [Google Scholar]
- Muellner-Riehl AN. 2019. Mountains as evolutionary arenas: patterns, emerging approaches, paradigm shifts, and their implications for plant phylogeographic research in the Tibeto-Himalayan Region. Frontiers in Plant Science 10: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulch A, Chamberlain CP. 2006. Earth science: the rise and growth of Tibet. Nature 439: 670–671. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the United States of America 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevado B, Contreras-Ortiz N, Hughes C, Filatov DA. 2018. Pleistocene glacial cycles drive isolation, gene flow and speciation in the high-elevation Andes. The New Phytologist 219: 779–793. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ. 1994. A role for molecular genetics in biological conservation. Proceedings of the National Academy of Sciences of the United States of America 91: 5748–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouborg NJ, Pertoldi C, Loeschcke V, Bijlsma RK, Hedrick PW. 2010. Conservation genetics in transition to conservation genomics. Trends in Genetics 26: 177–187. [DOI] [PubMed] [Google Scholar]
- Owen LA, Dortch JM. 2014. Nature and timing of Quaternary glaciation in the Himalayan-Tibetan orogen. Quaternary Science Reviews 88: 14–54. [Google Scholar]
- Pautasso M. 2009. Geographical genetics and the conservation of forest trees. Perspectives in Plant Ecology, Evolution and Systematics 11: 157–189. [Google Scholar]
- Petit RJ, Hu FS, Dick CW. 2008. Forests of the past: a window to future changes. Science 320: 1450–1452. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbek C, Borregaard MK, Antonelli A, et al. 2019. Building mountain biodiversity: geological and evolutionary processes. Science 365: 1114–1119. [DOI] [PubMed] [Google Scholar]
- Ralls K, Sunnucks P, Lacy RC, Frankham R. 2020. Genetic rescue: a critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biological Conservation 251: 108784. [Google Scholar]
- Ren G, Mateo RG, Liu J, et al. 2017. Genetic consequences of Quaternary climatic oscillations in the Himalayas: Primula tibetica as a case study based on restriction site-associated DNA sequencing. The New Phytologist 213: 1500–1512. [DOI] [PubMed] [Google Scholar]
- Renner SS. 2016. Available data point to a 4-km-high Tibetan Plateau by 40 Ma, but 100 molecular-clock papers have linked supposed recent uplift to young node ages. Journal of Biogeography 43: 1479–1487. [Google Scholar]
- Rogers RL, Slatkin M. 2017. Excess of genomic defects in a woolly mammoth on Wrangel island. PLoS Genetics 13: e1006601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru D, Mao K, Zhang L, Wang X, Lu Z, Sun Y. 2016. Genomic evidence for polyphyletic origins and interlineage gene flow within complex taxa: a case study of Picea brachytyla in the Qinghai-Tibet Plateau. Molecular Ecology 25: 2373–2386. [DOI] [PubMed] [Google Scholar]
- Ru D, Sun Y, Wang D, et al. 2018. Population genomic analysis reveals that homoploid hybrid speciation can be a lengthy process. Molecular Ecology 27: 4875–4887. [DOI] [PubMed] [Google Scholar]
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. 1998. Inbreeding and extinction in a butterfly metapopulation. Nature 392: 491–494. [Google Scholar]
- Schussler EE, Longstreth DJ. 1996. Aerenchyma develops by cell lysis in roots and cell separation in leaf petioles in Sagittaria lancifolia (Alismataceae). American Journal of Botany 83: 1266–1273. [Google Scholar]
- Segelbacher G, Cushman SA, Epperson BK, et al. 2010. Applications of landscape genetics in conservation biology: concepts and challenges. Conservation Genetics 11: 375–385. [Google Scholar]
- Shahzad K, Jia Y, Chen FL, Zeb U, Li ZH. 2017. Effects of mountain uplift and climatic oscillations on phylogeography and species divergence in four endangered Notopterygium herbs. Frontiers in Plant Science 8: 1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang HY, Li ZH, Dong M, et al. 2015. Evolutionary origin and demographic history of an ancient conifer (Juniperus microsperma) in the Qinghai-Tibetan Plateau. Scientific Reports 5: 10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono A, Ueno S, Gu S, Zhao X, Tsumura Y, Tang Y. 2010. Range shifts of Potentilla fruticosa on the Qinghai-Tibetan Plateau during glacial and interglacial periods revealed by chloroplast DNA sequence variation. Heredity 104: 534–542. [DOI] [PubMed] [Google Scholar]
- Spielman D, Brook BW, Frankham R. 2004. Most species are not driven to extinction before genetic factors impact them. Proceedings of the National Academy of Sciences of the United States of America 101: 15261–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer R, Su T, Valdes P, et al. 2021. Why ‘the uplift of the Tibetan Plateau’ is a myth? National Science Review 8: nwaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel MA, Humble E, Paijmans AJ, et al. 2018. Demographic histories and genetic diversity across pinnipeds are shaped by human exploitation, ecology and life-history. Nature Communications 9: 4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Morris MR, Genis AB, et al. 2017. The power of evolutionary rescue is constrained by genetic load. Evolutionary Applications 10: 731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Farnsworth A, Spicer R, et al. 2019. No high Tibetan plateau until the Neogene. Science Advances 5: eaav2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Abbott RJ, Li L, Li L, Zou J, Liu J. 2014. Evolutionary history of Purple cone spruce (Picea purpurea) in the Qinghai-Tibet Plateau: homoploid hybrid origin and Pleistocene expansion. Molecular Ecology 23: 343–359. [DOI] [PubMed] [Google Scholar]
- Sun Y, Abbott RJ, Lu Z, et al. 2018. Reticulate evolution within a spruce (Picea) species complex revealed by population genomic analysis. Evolution; International Journal of Organic Evolution 72: 2669–2681. [DOI] [PubMed] [Google Scholar]
- Sun B-N, Wu J-Y, Liu Y-SC, et al. 2011. Reconstructing Neogene vegetation and climates to infer tectonic uplift in western Yunnan, China. Palaeogeography, Palaeoclimatology, Palaeoecology 304: 328–336. [Google Scholar]
- Tajima F. 1989a. The effect of change in population size on DNA polymorphism. Genetics 123: 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989b. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson Lo, Yao T, Davis M, et al. 1997. Tropical climate instability: the last glacial cycle from a Qinghai-Tibetan ice core. Science 276: 1821–1825. [Google Scholar]
- Todd EV, Black MA, Gemmell NJ. 2016. The power and promise of RNA-seq in ecology and evolution. Molecular Ecology 25: 1224–1241. [DOI] [PubMed] [Google Scholar]
- Tso S, Li J, Xie S, et al. 2019. Characterization of the complete chloroplast genome of Juniperus microsperma (Cupressaceae), a rare endemic from the Qinghai-Tibet Plateau. Conservation Genetics Resources 11: 325–328. [Google Scholar]
- Uchiyama I, Higuchi T, Kawai M. 2010. MBGD update 2010: toward a comprehensive resource for exploring microbial genome diversity. Nucleic Acids Research 38: D361–D365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Abbott RJ, Zheng W, Chen P, Wang Y, Liu J. 2009. History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Molecular Ecology 18: 709–721. [DOI] [PubMed] [Google Scholar]
- Wang P, Burley JT, Liu Y, et al. 2021. Genomic consequences of long-term population decline in brown eared pheasant. Molecular Biology and Evolution 38: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chai K, Liu S, Wei J, Jiang Z, Liu Q. 2017. Changes of glaciers and glacial lakes implying corridor–barrier effects and climate change in the Hengduan Shan, southeastern Tibetan Plateau. Journal of Glaciology 63: 535–542. [Google Scholar]
- Wang C, Dai J, Zhao X, et al. 2014. Outward–growth of the Tibetan Plateau during the Cenozoic: a review. Tectonophysics 621: 1–43. [Google Scholar]
- Wang A, Li W. 2016. Genetic diversity of Rheum tanguticum (Polygonaceae), an endangered species on Qinghai–Tibetan Plateau. Biochemical Systematics and Ecology 69: 132–137. [Google Scholar]
- Wang Y, Liang Q, Hao G, Chen C, Liu J. 2018. Population genetic analyses of the endangered alpine Sinadoxa corydalifolia (Adoxaceae) provide insights into future conservation. Biodiversity and Conservation 27: 2275–2291. [Google Scholar]
- Wang YJ, Liu JQ, Miehe G. 2007. Phylogenetic origins of the Himalayan endemic Dolomiaea, Diplazoptilon and Xanthopappus (Asteraceae: Cardueae) based on three DNA regions. Annals of Botany 99: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhao X, Liu Z, et al. 2008. Constraints on the early uplift history of the Tibetan Plateau. Proceedings of the National Academy of Sciences of the United States of America 105: 4987–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkin EL, Thomson CJ, Greenway H. 1998. Root development and aerenchyma formation in two wheat cultivars and one triticale cultivar grown in stagnant agar and aerated nutrient solution. Annals of Botany 81: 349–354. [Google Scholar]
- Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wen J, Zhang JQ, Nie ZL, Zhong Y, Sun H. 2014. Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Frontiers in Genetics 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Ree RH. 2017. Uplift–driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proceedings of the National Academy of Sciences of the United States of America 114: E3444–E3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Song X, Ruhsam M, et al. 2019. Distinctiveness, speciation and demographic history of the rare endemic conifer Juniperus erectopatens in the eastern Qinghai–Tibet Plateau. Conservation Genetics 20: 1289–1301. [Google Scholar]
- Yang W, Yao T, Xu B, Ma L, Wang Z, Wan M. 2010. Characteristics of recent temperate glacier fluctuations in the Parlung Zangbo River basin, southeast Tibetan Plateau. Chinese Science Bulletin 55: 2097–2102. [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y, Lambeck K, De Deckker P, Johnston P, Fifield LK. 2000. Timing of the last Glacial maximum from observed sea-level minima. Nature 406: 713–716. [DOI] [PubMed] [Google Scholar]
- Yu Z, McAndrews JH, Eicher U. 1997. Middle Holocene dry climate caused by change in atmospheric circulation patterns: evidence from lake levels and stable isotopes. Geology 25: 251–254. [Google Scholar]
- Zhang D, Hao GQ, Guo XY, Hu QJ, Liu JQ. 2019. Genomic insight into ‘sky island’ species diversification in a mountainous biodiversity hotspot. Journal of Systematics and Evolution 57: 633–645. [Google Scholar]
- Zhang JM, López-Pujol J, Gong X, Wang HF, Vilatersana R, Zhou SL. 2018. Population genetic dynamics of Himalayan-Hengduan tree peonies, Paeonia subsect. Delavayanae. Molecular Phylogenetics and Evolution 125: 62–77. [DOI] [PubMed] [Google Scholar]
- Zhang DC, Ye JX, Sun H. 2016. Quantitative approaches to identify floristic units and centres of species endemism in the Qinghai–Tibetan Plateau, south-western China. Journal of Biogeography 43: 2465–2476. [Google Scholar]
- Zheng B, Rutter N. 1998. On the problem of Quaternary glaciations, and the extent and patterns of Pleistocene ice cover in the Qinghai–Xizang (Tibet) Plateau. Quaternary International 45: 109–122. [Google Scholar]
- Zheng B, Xu Q, Shen Y. 2002. The relationship between climate change and Quaternary glacial cycles on the Qinghai–Tibetan Plateau: review and speculation. Quaternary International 97: 93–101. [Google Scholar]
- Zhou S, Li J, Zhao J, Wang J, Zheng J. 2011. Quaternary glaciations: extent and chronology in China. In: Ehlers J, Gibbard PL, Hughes PD, eds. Quaternary glaciations – extent and chronology: a closer look, Developments in Quaternary Sciences. Amsterdam: Elsevier, 981–1002. [Google Scholar]
- Zhou S, Wang J, Xu L, Wang X, Colgan PM, Mickelson DM. 2010. Glacial advances in southeastern Tibet during late Quaternary and their implications for climatic changes. Quaternary International 218: 58–66. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.