Abstract
The coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is currently a global pandemic. Extensive investigations have been performed to study the clinical and cellular effects of SARS-CoV-2 infection. Mass spectrometry-based proteomics studies have revealed the cellular changes due to the infection and identified a plethora of interactors for all SARS-CoV-2 components, except for the longest non-structural protein 3 (NSP3). Here, we expressed the full-length NSP3 proteins of SARS-CoV and SARS-CoV-2 to investigate their unique and shared functions using multi-omics methods. We conducted interactome, phosphoproteome, ubiquitylome, transcriptome, and proteome analyses of NSP3-expressing cells. We found that NSP3 plays essential roles in cellular functions such as RNA metabolism and immune response (e.g., NF-κB signal transduction). Interestingly, we showed that SARS-CoV-2 NSP3 has both endoplasmic reticulum and mitochondrial localizations. In addition, SARS-CoV-2 NSP3 is more closely related to mitochondrial ribosomal proteins, whereas SARS-CoV NSP3 is related to the cytosolic ribosomal proteins. In summary, our integrative multi-omics study of NSP3 improves the understanding of the functions of NSP3 and offers potential targets for the development of anti-SARS strategies.
Keywords: NSP3, SARS-CoV-2, SARS-CoV, Multi-omics, Mitochondria
Introduction
A few clusters of severe pneumonia from unknown sources were reported from Wuhan at the end of 2019 [1]. The causative factor of this pneumonia was soon isolated, sequenced, and designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the disease was named coronavirus disease 2019 (COVID-19) [1], [2], [3], [4]. Although approximately 80% of SARS-CoV-2-infected patients have mild to no symptoms, 15% of patients could develop pneumonia and dyspnea, whereas the remaining 5% of patients, especially those with chronic disease, including lung diseases or asthma, are at high risk of developing severe illness with high rates of mortality and morbidity [5], [6]. As SARS-CoV-2 is easily transmitted via respiratory droplets, COVID-19 has spread worldwide very rapidly and became a pandemic by March 2020 [7]. As of Aug 25th, 2021, more than 210 million infections and 4.4 million deaths were reported according to the Center for Systems Science and Engineering at the Johns Hopkins University (https://coronavirus.jhu.edu/). Encouragingly, the research society has made tremendous progress in understanding of the molecular mechanisms behind the viral infection, as well as in the development of antiviral compounds and vaccines to treat COVID-19 [8], [9].
SARS-CoV-2 is a beta coronavirus and possesses a ∼ 30 kb positive, single-strand genome. Phylogenetic sequence analysis revealed that the SARS-CoV-2 is closely related to two human transmissible beta coronaviruses, SARS-CoV (80% gene sequence similarity) and MERS-CoV (50% gene sequence similarity) [6], [10]. Bioinformatic predictions suggest that SARS-CoV-2 encodes four structural proteins, nine accessory proteins, and two long polypeptides (pp1a and pp1ab) [4], [11]. These two long polypeptides are translated from ORF1a and ORF1ab. ORF1ab is produced via a −1 ribosomal frameshift from the stop codon of ORF1a. These two polypeptides together produce a total of 15 non-structural proteins (NSPs) via proteolysis. The processing of pp1a and pp1ab requires the protease activity of NSP3 (papain-like protease, PLpro) and NSP5 (chymotrypsin-like protease, 3CLPro) [11].
Previous studies on SARS-CoV revealed that NSPs are the primary constituents for the assembly of the replication and transcription complex (RTC), where viral genome RNA synthesis occurs, and double-strand RNA is abundantly expressed [12]. Co-expression of NSP3, NSP4, and NSP6 induces the formation of double-membrane vesicles (DMVs), which are continuous with the endoplasmic reticulum (ER) membrane. These DMVs have similar morphology to the organelle-like structures induced by SARS-CoV infection [12], [13]. This multiple protein- and membrane-containing organelle-like RTC facilitates the synthesis of positive-strand RNA viruses where no interference from the host cell innate immune system occurs [14]. The NSP3 of SARS-CoV is the largest and most essential component of the RTC complex. It has 1922 amino acids and multiple functional domains with single-strand RNA (ssRNA) binding, deMARylation, G-quadruplex binding, and cysteine protease activities [15], [16]. Therefore, it is not surprising that NSP3 plays pivotal roles in the viral life cycle, and the inhibition of NSP3 PLpro activity prevents SARS-CoV replication [17]. NSP3 of SARS-CoV-2 is a polypeptide of 1945 amino acids that has conserved functional domains with an overall 86% protein sequence similarity to that of SARS-CoV. Recent studies have demonstrated the deMARylation, deubiquitination, and deISGylation functions of the Macro and PLpro domains of NSP3 from SARS-CoV-2 [16], [18], [19], [20]. More importantly, two studies showed that PLpro inhibition results in excellent anti-SARS-CoV-2 activity [18], [19]. However, despite years of extensive research into the functions of NSP3 from SARS-CoV and recently into SARS-CoV-2, the functions of full-length NSP3 are still only partially understood. We only found two reports that successfully expressed full-length NSP3, one for SARS-CoV, and one for SARS-CoV-2 [12], [19]. In addition, three very recent interaction studies were performed for SARS-CoV-2 NSP3, with two studies using deletion mutants, whereas the third one failed to detect the expression of NSP3 [21], [22], [23].
In this study, we successfully cloned and expressed full-length NSP3 proteins of SARS-CoV and SARS-CoV-2. Using these plasmids, we presented the transcriptomic and proteomic (interactome, proteome, phosphoproteome, and ubiquitylome) landscapes of NSP3. These parallel studies were used to investigate the similarities and differences between NSP3 proteins. Specifically, we investigated how NSP3 protein interacts with host cells. Furthermore, we assessed the signaling pathways that are regulated by the NSP3 protein. Lastly, we also aimed to offer a list of compounds that could potentially be repurposed for further investigations of COVID-19 treatment.
Results
Expression of the full-length NSP3 proteins of SARS-CoV and SARS-CoV-2
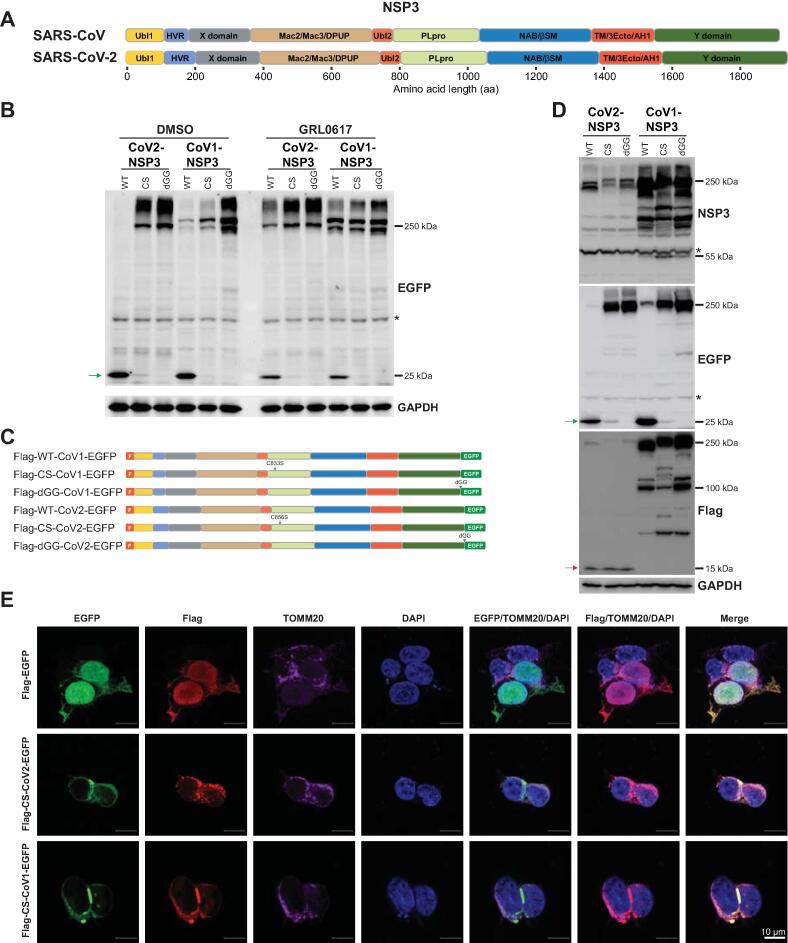
Current studies into the functions of NSP3 proteins (Figure 1A) of both SARS-CoVs (hereafter referred to as CoV1-NSP3 and CoV2-NSP3) are mainly limited to their truncated deletions. We successfully cloned the wild-type (WT), catalytically inactive (CS-mutated), or dGG-mutated (last two glycine residues deleted, resistant to PLpro cleavage) full-length NSP3 proteins of both SARS-CoVs (without codon optimization) into a lentivirus vector containing a C-terminal Enhanced Green Fluorescent Protein (EGFP) tag, as illustrated in Figure S1A. We expressed all six plasmids in HEK293T cells and detected their expression using anti-EGFP antibody. As shown in Figure S1B, the expression of CS- and dGG-mutated NSP3 proteins was detected using anti-EGFP antibody (∼ 250 kDa), whereas no obvious expression of WT-NSP3 proteins was detected. This is due to the self-cleavage activity of PLpro toward the Leu-Lys-Gly-Gly of its C-terminus, as dGG-NSP3 is expressed well [24]. We verified this hypothesis by treating cells with the PLpro inhibitor GRL0617 [17], [18]. As shown in Figure 1B, anti-EGFP antibody could detect the WT-NSP3 proteins in GRL0617-treated cells. This observation also explains why a previous study failed to detect the expression of NSP3 using a C-terminal tag [22].
Figure 1.
Expression and subcellular localization ofNSP3 proteins ofSARS-CoVand SARS-CoV-2
A. Schematic illustration of NSP3 proteins of SARS-CoV and SARS-CoV-2. Domains of NSP3 proteins of SARS-CoV and SARS-CoV-2 are illustrated according to Lei et al. [15] and Alhammad et al. [16]. B. GRL0617 inhibits the release of C-terminal EGFP from NSP3-EGFP proteins. Cells transfected with indicated plasmids (Figure S1A) were treated with DMSO or GRL0617 for 24 h. Cells were lysed and anti-EGFP antibody was used for immunoblotting. Green arrow indicates the released EGFP protein, and asterisk indicates non-specific signal. GAPDH was used to serve as a loading control. C. Schematic illustration of plasmids used in (D). D. CoV2-NSP3 is processed at the N-terminus. Green arrow indicates the released EGFP, and red arrow around 15 kDa indicates the processed N-terminus of CoV2-NSP3. Asterisks indicate non-specific signals. E. N-terminus of CoV2-NSP3 prefers to co-localize with the mitochondrial marker TOMM20. HEK293T cells transfected with Flag-CoV1/2-NSP3-EGFP plasmids were fixed 48 h post-transfection. Anti-Flag and anti-TOMM20 antibodies were used to visualize the co-localization. Scale bar, 10 μm. NSP3, non-structural protein 3; SARS-CoV, severe acute respiratory syndrome coronavirus; Ubl1, ubiquitin like domain 1; HVR, hyper variable region; X/Mac1, macrodomain I; Mac2/Mac3/DPUP, macrodomain II, macrodomain III, and domain preceding Ubl2 and PLpro; Ubl2, ubiquitin like domain 2; PLpro, Papain-like protease; NAB/βSM, nucleic acid-binding and beta-coronavirus-specific marker domain; TM/3Ecto/AH1, transmembrane region, 3 ectodomain, and amphipathic helix1; Y, a domain with unknown functions; EGFP, Enhanced Green Fluorescent Protein; DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CoV1-NSP3, NSP3 of SARS-CoV; CoV2-NSP3, NSP3 of SARS-CoV-2; WT, wild-type NSP3; CS, catalytic inactivated NSP3; dGG, GlyGly deficient NSP3; F, Flag; DAPI, 4′,6-diamidino-2-phenylindole.
Next, we performed immunoblotting using an antibody specific for CoV1-NSP3. The specificity of this anti-CoV1-NSP3 antibody to CoV2-NSP3 was verified by immunoblotting and immunostaining (Figure S1B and C; hereafter referred to as anti-NSP3 antibody). Interestingly, we noticed that CoV2-NSP3 had a lower molecular weight than CoV1-NSP3, with both anti-EGFP and anti-NSP3 antibodies (Figure 1B, Figure S1B). This implies that either the amino acid composition of NSP3 protein affects its molecular weight or that CoV2-NSP3 undergoes extra processing at its N-terminus. We therefore subcloned NSP3 into a vector with N-terminal Flag and C-terminal EGFP tags (Figure 1C). Interestingly, we observed a very specific processed band of ~ 15 kDa for all three CoV2-NSP3 proteins (Figure 1D, anti-Flag antibody). In addition, anti-Flag antibody detected most of CoV1-NSP3 proteins at the correct position, thought multiple smaller signals were also observed, suggesting the existence of extra processing (Figure 1D).
NSP3 proteins of SARS-CoV and SARS-CoV-2 have different cellular localization
We sought to determine the cellular localization of the NSP3 protein by immunostaining. As shown in Figure S1C, EGFP and NSP3 signals were well-localized, especially in CS- and dGG-NSP3-expressing cells. Interestingly, cells expressing CS-CoV1-NSP3 showed very large vesicle-like structures, which were not observed in cells expressing WT and dGG-mutated NSP3 proteins, as well as CS-CoV2-NSP3. Next, we stained NSP3-expressing cells with ER, autophagosome, endosome, and mitochondrial makers, as all four organelles are potential origins of DMVs [25], [26], [27], [28]. Both CoV1-NSP3 and CoV2-NSP3 proteins showed no obvious co-localization with the endosome marker RAB7 or autophagosome marker LC3 (data not shown). As shown in Figure S2A, we found that CS-CoV1-NSP3 co-localized with the ER marker Calnexin (CANX), particularly in the large vesicle-like structures, but not with mitochondrial marker TOMM20 (Figure S2B). In contrast, CS-CoV2-NSP3 protein showed partial co-localization with both TOMM20 and CANX (Figure S2A and B). We next investigated whether the extra N-terminal processed band might somehow affect the localization of CoV2-NSP3. As shown in Figure S2C, anti-NSP3 antibody showed less co-localization signals with Flag-CS-CoV2-NSP3, compared to the co-localization signals of anti-NSP3 and anti-EGFP antibodies. In addition, anti-Flag antibody also detected vesicle-like structures, though smaller in size than those in CS-CoV1-NSP3-expressing cells (Figure S2C). These vesicle-like structures showed strong co-localization with TOMM20, but not with CANX (Figure 1E, Figure S2D). Interestingly, the EGFP signals of CS-CoV2-NSP3 still co-localized with CANX (Figure S2D). These observations indicate that there are two cellular localizations of CoV2-NSP3, influenced by the extra processing at its N-terminus. In contrast, Flag staining of CS-CoV1-NSP3 showed the same localization pattern with CANX as with EGFP (Figure S2D). Finally, extracting data from interactome studies verified the localization preference of different NSP3 proteins (Figure S2E).
The N-terminus determines the localization of CoV2-NSP3 to mitochondria
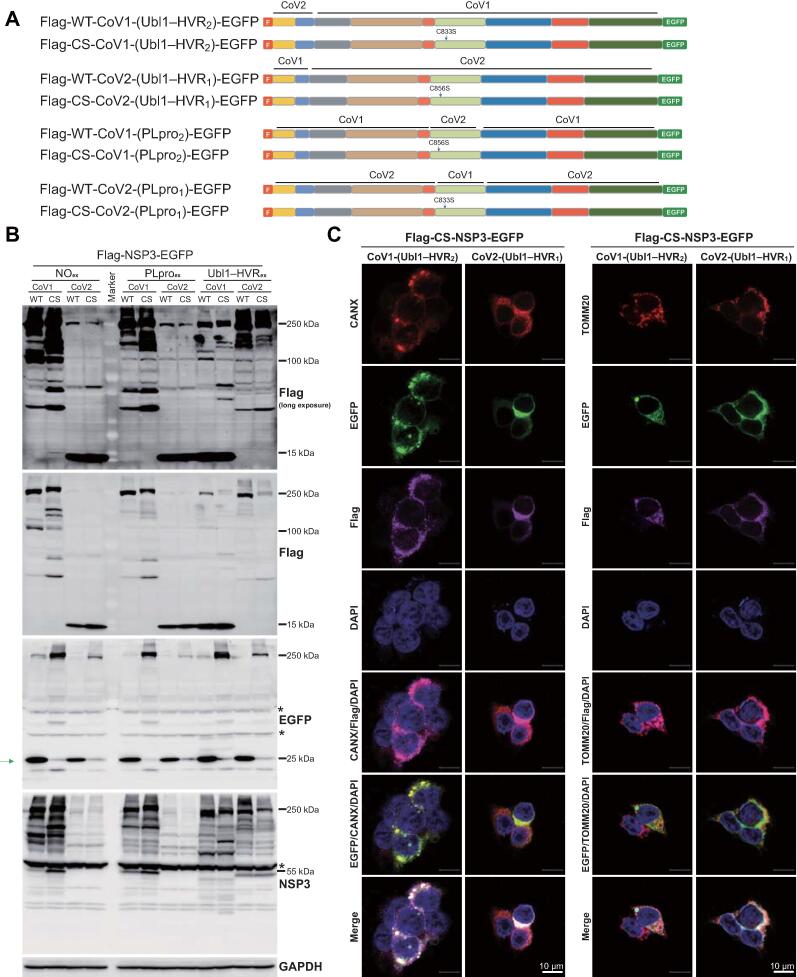
To further investigate the cellular localization of CoV1-NSP3 and CoV2-NSP3, we exchanged their Ubl1 and HVR domains reciprocally (Figure 2A). We also exchanged their PLpro domains, as the CS-CoV1-NSP3 mutant showed large vesicle-like structures. As shown in Figure 2B, when CoV2-NSP3 was equipped with the Ubl1 and HVR domains of CoV1-NSP3, the N-terminal extra band disappeared. In contrast, a 15-kDa band was observed when CoV1-NSP3 was equipped with the Ubl1 and HVR domains of CoV2-NSP3. In addition, we showed that the PLpro domain has no obvious influence on processing of the CoV2-NSP3 protein (Figure 2B).
Figure 2.
N-terminusdetermines the extra processing and mitochondriallocalization of NSP3
A. Schematic illustration of chimeric plasmids used in this figure. B. The processing site of CoV2-NSP3 is localized at its N-terminus. Green arrow indicates the released EGFP. NOex indicates no exchange, PLproex indicates the exchange of PLpro domain, whereas Ubl1–HVRex indicates the exchange of Ubl1 and HVR domains. Asterisks indicate non-specific signals. C. The N-terminus of CoV2-NSP3 re-locates CoV1-NSP3 to mitochondria. Scale bar, 10 μm. CANX, Calnexin (an ER marker); ER, endoplasmic reticulum.
Next, we used immunostaining to study the localization of these fused proteins (Figure 2C). As expected, CS-CoV1-NSP3, detected with anti-Flag antibody, co-localized with TOMM20 when its N-terminus was replaced to that of CoV2-NSP3. In contrast, CS-CoV2-NSP3 fused with the N-terminus of CoV1-NSP3 was predominantly co-localized with CANX. However, the EGFP signals of both fused proteins were still co-localized with CANX. Therefore, we concluded that the C-terminus of both NSP3 proteins co-localizes with ER, whereas the N-terminus of CoV2-NSP3 prefers to bind to mitochondria. Furthermore, we showed that the exchange of the PLpro domain has no obvious influence on NSP3 protein localization (Figure S3).
Interactome analysis of NSP3 proteins
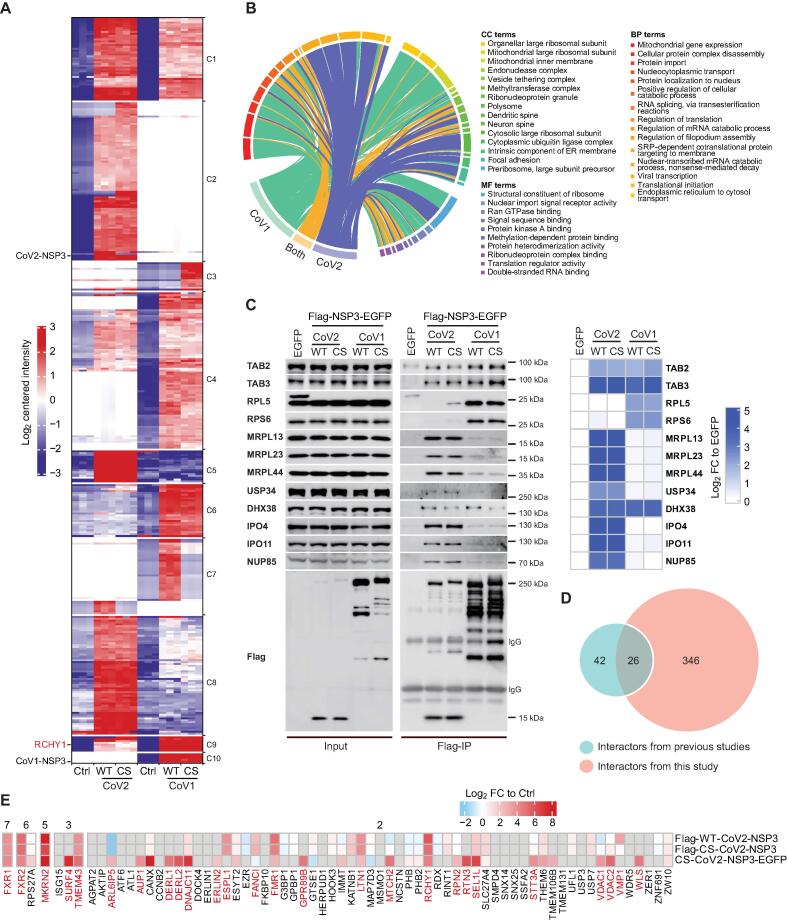
To study how NSP3 protein contributes to cellular function regulation during infection, we conducted a multi-omics study (interactome, phosphoproteome, ubiquitylome, proteome, and transcriptome) on NSP3-expressing cells (Figure S4A). We performed all experiments in biological triplicates, and we showed that the distribution of total intensity was consistent within every experimental condition (Figure S4B). We first analyzed the interactome of NSP3 using Flag-immunoprecipitation coupled with mass spectrometry (Flag-IP/MS) identification. As shown in Figure S5A, principal component analysis (PCA) of the top 1000 variable proteins showed a strong correlation within the triplicate experiments for NSP3 proteins. In addition, the interactomes of the WT and CS-mutated NSP3 proteins showed a good correlation, implying that the catalytic activity has minor effects on the NSP3 interactome. In total, 211 and 226 of significant interactors were enriched for CoV1-NSP3 and CoV2-NSP3, respectively, using thresholds based on the fold change (FC) > 4 and adjusted P < 0.001 (Table S1). Next, we compared these two NSP3 interactomes side by side to study their shared and specific interactions (Figure 3A; Table S1). We noticed that RCHY1, a known interactor of the macrodomain and PLpro of NSP3, was enriched by both NSP3 proteins [29]. However, the overall similarities between the two NSP3 interactomes was low (55 out of 382 interactors; Figure 3A, Figure S5B). Gene Ontology (GO) and Reactome [30] analyses of significant interactors highlighted the roles of both NSP3 proteins in RNA splicing, NF-κB signaling, and the regulation of translation (Figure 3B, Figure S5C and D; Table S1). The most striking difference between the two interactomes was that CoV2-NSP3 was linked to mitochondrial translation, whereas CoV1-NSP3 was associated with cytosolic translation. We verified a few specific and shared interactions using Flag-IP followed by immunoblotting (Figure 3C). For example, TAB2 and TAB3 showed similar binding ability to both NSP3 proteins, whereas cytosolic ribosomal proteins and mitochondrial ribosomal proteins preferred to bind to CoV1-NSP3 and CoV2-NSP3, respectively.
Figure 3.
Interactome analysis of NSP3 proteins and host proteins
A. Clustering of significant interactors of NSP3 proteins. Significant interactors of NSP3 were analyzed to show the shared and unique interactions (Table S1). Red indicates enrichment, while blue indicates lack of enrichment. Interactors that do not present in either NSP3 interactome are left empty. RCHY1 and NSP3 proteins are indicated on the left. C1–C10 on the right indicate Clusters 1–10. B. Circos diagram showing representative enriched GO items of identified interactors. Full list of GO terms is shown in Table S1. C. Validation of interactome result. HEK293T cells transfected with Flag-NSP3-EGFP plasmids were harvest for Flag-IP assay. Indicated antibodies were used to detect the enrichment. LFQ intensity of each protein was extracted from IP-MS result, log2-transformed, and then substracted its intensity identified in EGFP condition. Black asterisk indicates non-specific signal, whereas green asterisk indicates leaked EGFP signal. D. Venn diagram showing the overlap of CoV2-NSP3 interactors identified in our study and interactors obtained from BioGRID database [32] and Almasy and his colleagues [23]. 68 interactors identified in at least two individual studies were compared with interactors in this study. E. Heatmap showing the enrichment (log2 FC) of 68 previously reported CoV2-NSP3 interactors in this study. Number on top of the heatmap indicates the frequency of previously reported interactors identified in BioGRID database [32] and Almasy and his colleagues [23]. The 26 reported interactors also significantly enriched (log2 FC > 2, FDR < 0.05) in our IP assay were labeled by red. Detailed information about the comparison is available in Table S3. GO, Gene Ontology; CC, cellular component; BP, biological process; MF, molecular function; FC, fold change; IP, immunoprecipitation; LFQ, Label Free Quantification.
To avoid missing of transient protein–protein interactions (PPIs) and to identify a core PPI complex, we repeated our interactome study using a crosslinking method called rapid immunoprecipitation mass spectrometry of endogenous protein (RIME) [31] (Figure S6A–D; Table S2). Approximately 30% and 55% of high confident interactors (Figure S6B) from crosslinking experiments were found in non-crosslinking experiments for CoV2-NSP3 and CoV1-NSP3, respectively. Strikingly, most of the interactors of CoV2-NSP3 were mitochondrial proteins including mitochondrial ribosomal protein large subunits (MRPLs), mitochondrial ribosomal protein small subunits (MRPSs), and coenzyme Q proteins (COQs) (Figure S6C). In addition, GO biological process (BP) analysis showed that proteins that related to the mitochondrial gene expression, quinone biosynthetic process, and cellular respiration were significantly enriched with CoV2-NSP3 (Figure S6D; Table S2), further validating the intimate interaction between CoV2-NSP3 and mitochondria.
Because the extra processing of CoV2-NSP3 at its N-terminus, it was therefore necessary to study the interactome with a EGFP tag that localized to the C-terminus of the NSP3 protein (Figure S7A–D; Table S3). As shown in Figure S7B, RCHY1 was again enriched with both NSP3 proteins again. The overall similarities between the two EGFP interactomes was a slightly higher than those of Flag-IP (27% vs. 15%; Figures S5B and S7C). Consistent with immunostaining results using anti-EGFP antibody, interactors of both NSP3 proteins were related to the ER (Figure S7D; Table S3). In addition, shared interactors were enriched in IL-1 signaling and regulation of RNA metabolism.
To study the differences between the NSP3 interactomes obtained with full-length protein and deletions. We compared our result with previously reported NSP3 interactomes that based on deletions. Here, we combined reported NSP3 interaction data from BioGRID database [32] and from a recently published study using three deletions to cover the NSP3 protein [23]. Among the 68 NSP3-interacting proteins that were found in at least two previous studies, 26 were identified in our studies (Figure 3D; Table S3). Importantly, interactors that have higher identification frequency in previous studies had a better chance to be identified in our study (Figure 3E), indicating that interactome based on full-length NSP3 is more reliable. Therefore, 346 specific interactors for full-length NSP3 in our study are novel candidates for further investigation.
Viral protein–host protein interaction network
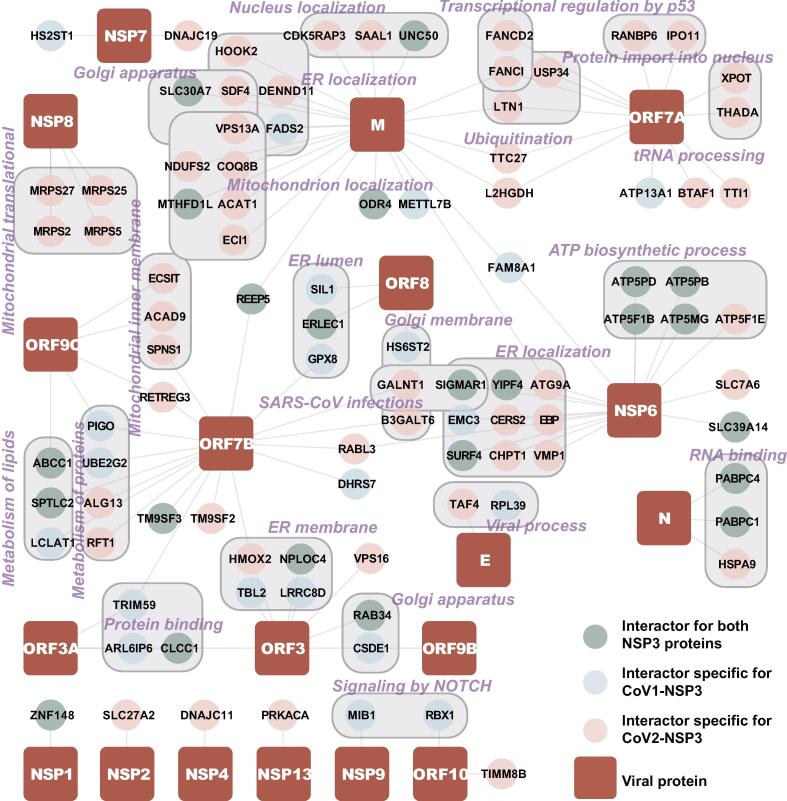
To study how the NSP3 protein is connected to other SARS-CoV-2 proteins and host proteins, we mined our data with reported SARS-CoV-2 interactors [29], [33]. Here, we combined all our identified NSP3 interactors with reported interactors for all other SARS-CoV-2 proteins. As shown in Figure 4 and Table S4, NSP3 was linked to multiple viral proteins via its interacting partners, suggesting that they work coordinately to regulate host cell functions. We found that NSP3, ORF3, and NSP6 shared multiple ER-localized interactors, which is consistent with the observation of ER-localization of both NSP3 proteins. Furthermore, both NSP3 proteins were linked to NSP6 via ATP metabolism-related proteins. Interestingly, only CoV2-NSP3 was associated with NSP8 and M proteins via mitochondrial-related proteins. One exception is that MTHFD1L also interacts with CoV1-NSP3. In addition, ORF7A almost exclusively interacted with CoV2-NSP3, but not with CoV1-NSP3. These results indicate that NSP3 coordinates with other viral proteins to regulate cellular functions.
Figure 4.
Viral protein–host protein interaction network
The interactors of NSP3 proteins were combined with reported interactors of all other SARS-CoV-2 proteins to create viral protein–host protein interaction network (Table S4). Interactors specific for CoV1-NSP3 and CoV2-NSP3 are displayed as different colors. Cytoscape was used to create the connection map of NSP3, interactors, and other SARS-CoV-2 proteins as described in Material and methods.
Taken together, our interactome results indicate the following: 1) regulation of RNA metabolism could be the main function of NSP3; 2) both NSP3 proteins are associated with protein translation but employ different translation machinery; 3) the N-terminus of CoV2-NSP3 determines the interaction with mitochondria; and 4) NSP3 functions together with other SARS-CoV-2 proteins to control host cell functions.
Phosphoproteome analysis of NSP3-expressing cells
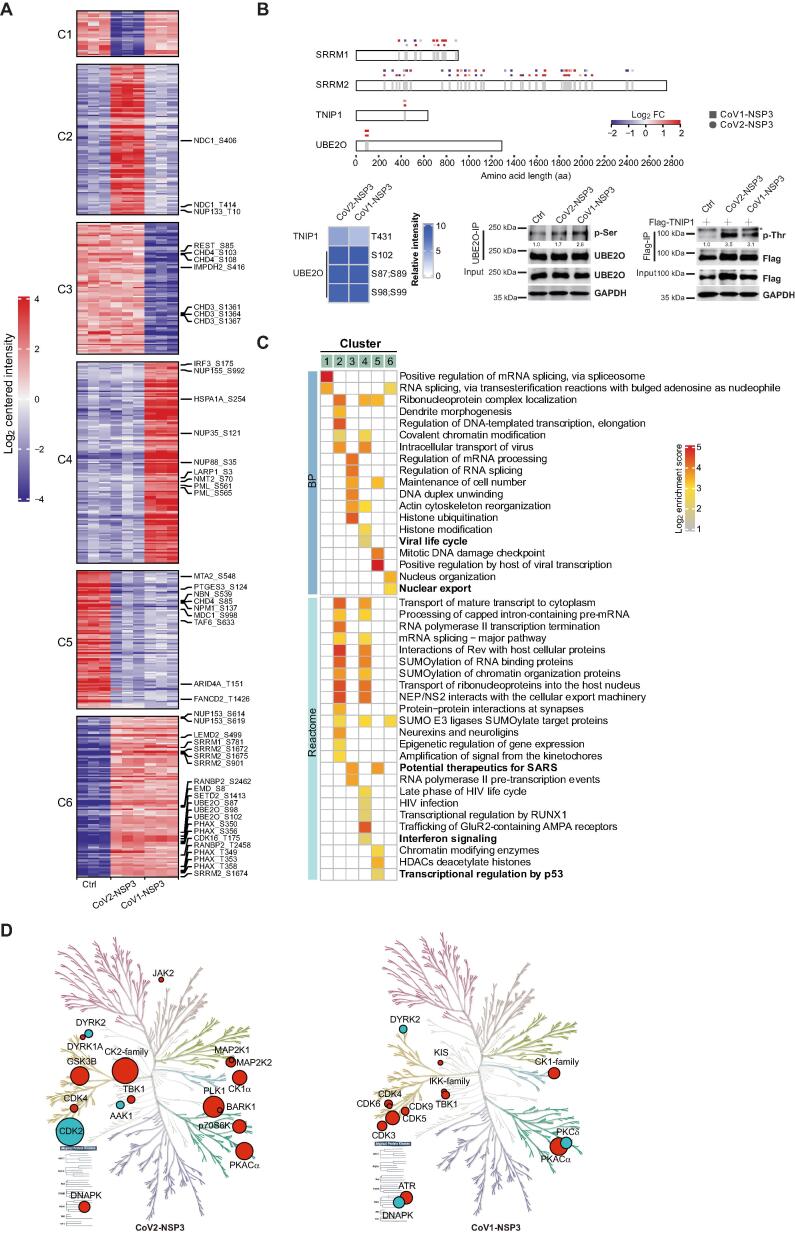
We next explored the effects of NSP3 on the phosphoproteome of host cells using a highly sensitive phosphoproteome preparation and identification workflow (EasyPhos) [34]. Differentially enriched phosphorylated peptides (DEPPs) were identified if they appeared at least three times in one condition based on an ANOVA test, with thresholds of a Benjamini–Hochberg (BH) adjusted P < 0.01, and |log2 FC| > 2. For all analyses, we chose 0.7 as the minimum cutoff for the localization probability (Table S5). In total, 659 DEPPs were identified for NSP3-expressing cell lines. Moreover, some of the DEPPs showed specificity in either NSP3-expressing cell line (Figure 5A, Figure S8A; Table S5). We validated the serine and threonine phosphorylation of UBE2O and TNIP1 using IP followed by immunoblotting (Figure 5B). BP and Reactome analyses of NSP3-shared DEPPs (Clusters 5 and 6; Figure 5A and C) showed that terms related RNA metabolism, the regulation of viral transcription, and potential therapeutics for SARS were enriched for both NSP3 proteins. Interestingly, similar BP and Reactome terms were found for DEPPs in Cluster 2 and Cluster 4, although they were specific DEPPs for CoV2-NSP2 and CoV1-NSP3, respectively. To dissect the contributions of NSP3 to the host cell phosphoproteome during viral infection, we compared our results with recently reported SARS-CoV-2 phosphoproteome datasets [35], [36]. In total, 90 DEPPs in our dataset were matched to previous reports, and a large proportion of them showed similar regulation trends (Figure S8B). For example, DEPPs with similar regulatory trends were found for SRRM2, UBE2O, SRSF3, and SIPA1L1, emphasizing the contributions of NSP3 to these phosphorylation events during viral infection.
Figure 5.
Phosphoproteome analysis of NSP3-expressing cells
A. Clustering of DEPPs in NSP3-expressing cells. DEPPs showing significantly differences in CoV1-NSP3- and CoV2-NSP3-expressing cells compared to control cells were clustered together. C1–C6 on the left indicate Clusters 1–6. Full list of DEPPs is available in Table S5. Representative DEPPs with highlighted GO in (C) were shown on the right. B. Validation of phosphoproteome studies. Top panel: schematic representation of phosphorylated sites of indicated proteins. Square indicates a DEPP with significant change in CoV1-NSP3-expressing cells, whereas circle indicates a DEPP with significant change in CoV2-NSP3-expressing cells. Bottom panels: HEK293T cells transfected with indicated plasmids were subjected to IP assays (anti-Flag-resin or anti-UBE2O antibody with protein A/G), following immunoblotting with phosphorylated serine/threonine antibodies. ImageJ was used to quantify the relative expression of each protein to its matching loading control. C. Representative enriched GO BP and Reactome terms. Labels on top of the heatmap correspond to the clusters in (A). Full list of the GO and Reactome terms is shown in Table S5. Bold terms containing DEPPs that are highlighted on the right of (A). D. KinMaps showing altered kinases whose activities mapped to DEPPs in NSP3-expressing cells. Significantly enriched DEPPs in CoV1-NSP3- and CoV2-NSP3-expressing cells were matched to kinases. Kinome profiling was performed using KinMap interface. Red indicates up-regulation, whereas cyan indicates down-regulation. Size of the circles reflects the number of DEPPs. DEPP, differentially enriched phosphorylated peptide.
We then mapped our DEPPs to kinases that regulate these phosphorylation sites and visualized the data using KinMap [37] (Figure 5D; Table S5). We found that the altered activities of CDK4, CK1α, TBK1, and DYRK2 were conserved in both NSP3-expressing cell lines. However, the activities of multiple CDKs were up-regulated in CoV1-NSP3-expressing cells, whereas only CDK4 activity was up-regulated in CoV2-NSP3-expressing cells. The decrease of CDK2 activity in CoV2-NSP3-expressing cells is consistent with the results obtained from virus-infected cells [36]. As CDKs play an important role in cell cycle progression, we showed that the expression of both NSP3 proteins caused a prolonged G2 phase compared to that of control cells (Figure S8C). We also mapped inhibitors to kinases for which the activity was significantly changed in NSP3-expressing cells (Figure S8D and E). We retrieved 12 kinases and 18 kinase inhibitors from our phosphoproteome data of CoV2-NSP3-expressing cells, whereas 7 kinases and 13 kinase inhibitors were found for CoV1-NSP3-expressing cells. Further, one of the inhibitors, fostamatinib, a treatment for chronic immune thrombocytopenia targeting multiple mapped kinase pathways [38], has been investigated as a potential treatment for acute lung injury for COVID-19 patients [39]. Interestingly, we noticed that the interferon activators, IKK-family and TBK1 kinases, were significantly present in CoV1-NSP3-expressing cells (Figure S8E and F), indicating the different phosphorylation regulatory effects of NSP3 proteins.
Ubiquitylome analysis of NSP3-expressing cells
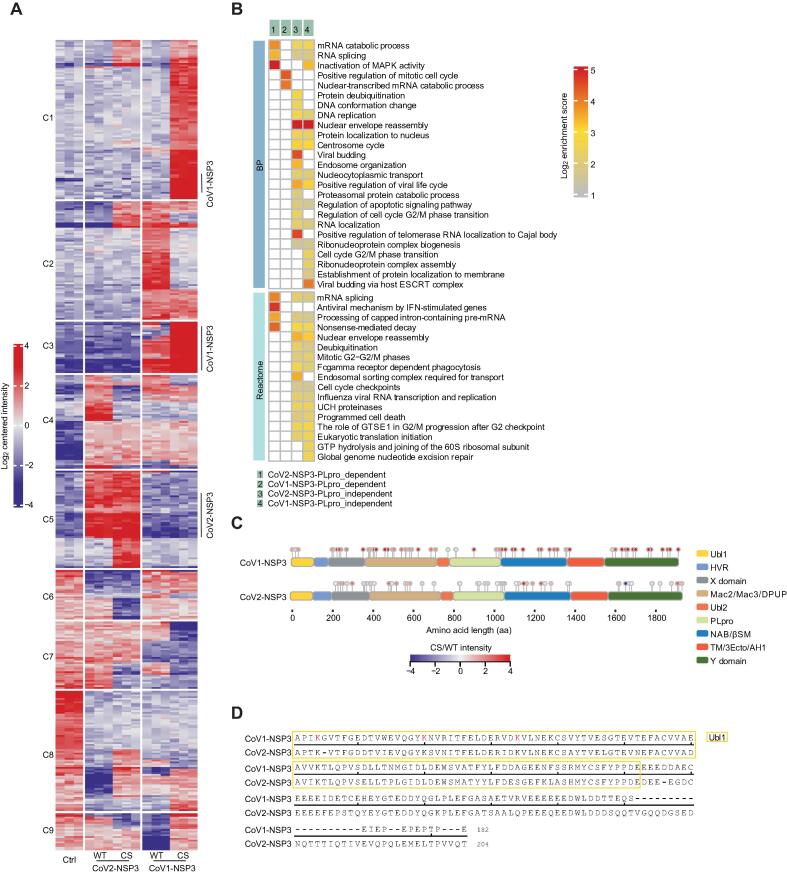
Ubiquitination is used by both viruses and host cells to combat each other [40]. For SARS-CoVs, they both have a PLpro protease domain within NSP3 proteins that is involved in deubiquitination and deISGylation activities [18]. We used the antibody-based K-ε-GG peptide enrichment method to study the effects of WT and CS-mutated NSP3 proteins on the ubiquitylome of the host cells. Because the levels of ISGylation and neddylation are relatively low in cells, we referred to the enriched peptides as ubiquitinated peptides (ubiquitylome) hereafter.
In total, 449 differentially enriched ubiquitinated peptides (DEUPs) showed significant differences with thresholds of localization probability > 0.7, |log2 FC| > 2, and adjusted P < 0.001 (Figure 6A, Figure S9A; Table S6). We noticed that most of the ubiquitination events, except some in Clusters 1, 4, and 8, were not consistent in NSP3-expressing cells. However, GO analysis showed similar enriched terms, such as positive regulation of viral life cycle, mRNA splicing, cell cycle regulation, and translation initiation, for both NSP3 proteins (Figure 6B). These results indicate that two NSP3 proteins have similar cellular functions, although they might function through different substrates. In addition, only 22 DEUPs and 26 DEUPs showed catalytic activity-dependent regulation in CoV2-NSP3- and CoV1-NSP3-expressing cells, respectively (Figure S9B). This observation suggests that the marked effects of NSP3 on the cellular ubiquitylome are catalytic activity-independent, which has also been observed for other deubiquitinases [41].
Figure 6.
Ubiquitylome analysis of NSP3-expressing cells
A. Clustering of DEUPs in NSP3-expressing cells. DEUPs showing significantly differences in NSP3-expressing cells compared to control cells were clustered together. C1–C9 on the left indicate Clusters 1–9. Full list of DEUPs is available in Table S6. B. GO BP and Reactome analyses of catalytic activity-dependent and -independent DEUPs. Full list of enriched terms is available in Table S6. C. Schematic representation of identified ubiquitination sites of NSP3 proteins. The intensity of each ubiquitinated site was log2-transformed and compared between CS-mutated and WT NSP3-expressing cells. D. Alignment of the Ubl1 and HVR domains of NSP3 proteins. Red-labeled K indicates identified ubiquitinated lysine residue of CoV1-NSP3. DEUP, differentially enriched ubiquitinated peptide.
We next mapped all ubiquitinate sites of NSP3 proteins to their functional domains and found that the overall ubiquitination sites were conserved in both NSP3 proteins (Table S6). The three domains, HVR, Ubl2, and TM/3Ecto/AH1, had almost no ubiquitinated sites, explained by the fact that these domains only have a few lysine residues. Interestingly, we noticed that the N-terminus of CoV1-NSP3 had three ubiquitinated sites, whereas no such ubiquitination was observed in CoV2-NSP3 (Figure 6C). However, these three lysine amino acids were also conserved in CoV2-NSP3 (Figure 6D), raising questions about why the N-termini of NSP3 proteins are differently ubiquitinated, as well as the mechanisms underlying and consequences of these differences.
Transcriptome and proteome changes in NSP3-expressing cells
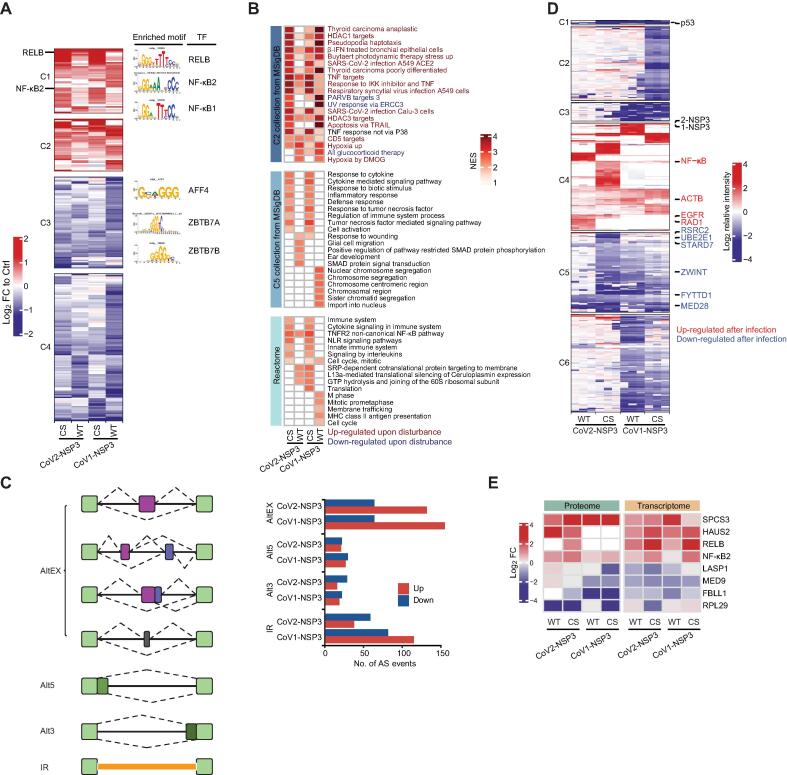
We also measured the transcriptome and proteome of control and NSP3-expressing cells using deep-sequencing and filter-aided sample preparation (FASP)-based proteomics analyses. As shown in Figure 7A, differentially expressed genes (DEGs) were calculated using thresholds of false discovery rate (FDR) < 0.05 and |log2 FC| > 1 compared with the expression in control cells (Table S7). Using motif enrichment analysis, we found that RELB-, NF-κB1-, and NF-κB2-binding motifs were highly represented in the up-regulated DEGs, whereas ZBTB7A-, ZBTB7B- and AFF4-binding motifs were enriched in the down-regulated DEGs (Figure 7A; Table S7). We then matched our gene lists with C2 collection (curated gene sets; containg two sub-collections: chemical and genetic perturbations and canonical pathways) and C5 collection (ontology gene sets; containing two sub-collections: GO and Human Phenotype Ontology) from the Molecular Signatures Database (MsigDB) by gene set enrichment analysis (GSEA) [42], [43]. As shown in Figure 7B and Table S7, our up-regulated genes were correlated with multiple reported gene sets, and a large portion of them was related to perturbations in NF-κB signaling. More importantly, our gene sets had a significant correlation with that of SARS-CoV-2 infection of Calu-3 and A549 cells and respiratory syncytial virus infection of A549 cells.
Figure 7.
Transcriptome and proteome analyses of NSP3-expressing cells
A. Motif enrichment analysis of DEGs in NSP3-expressing cells. DEGs showing significantly differences in NSP3-expressing cells compared to control cells were clustered together. C1–C4 on the left indicate Clusters 1–4. RELB and NF-κB were up-regulated in NSP3-expressing cells. Enriched motifs are shown on the right of the heatmap. Full list of the DEGs and genes with enriched motifs is available in Table S7. B. GSEA of C2 collection (curated gene sets; containg two sub-collections: chemical and genetic perturbations and canonical pathways) and C5 collection (ontology gene sets; containing two sub-cellections: GO and Human Phenotype Ontology) from MSigDB v7.2 and pathway gene sets from Reactome pathway database. Full list of the GO and GSEA is available in Table S7. C. AS events in NSP3-expressing cells. Schematic illustration of AS events identified in NSP3-expressing cells (left panel). AS events was identified from the RNA-seq data of NSP3-expressing cells compared to control cells. Full list of the AS events is available in Table S7. D. Clustering of DEPs in NSP3-expressing cells. C1–C6 on the left indicate Clusters 1–6. Proteins showing consistent regulation in viral infection are labeled on the right of the heatmap [45]. Full list of DEPs is available in Table S8. E. Clustering of genes showing consistent expression trends in both transcriptome and proteome data. DEG, differentially expressed gene; TF, transcription factor; GSEA, gene set enrichment analysis; MSigDB, Molecular Signatures Database; NES, normalized enrichment score; AS, alternative splicing; AltEX, alternative exon; Alt5, alternative 5′ UTR; Alt3, alternative 3′ UTR; IR, intron retention; DEP, differentially expressed protein.
Our proteomics (interactome, phosphoproteome, and ubiquitylome) analysis identified the significant enrichment of proteins related to mRNA splicing. Therefore, we performed the alternative splicing analyses of our transcriptome results. We observed 381 and 514 alternative splicing (AS) events in CoV2-NSP3- and CoV1-NSP3-expressing cells, respectively (Figure 7C; Table S7). Among them, a large portion of AS events were detected on the exon cassettes and intron retention (IR) regions, indicating that NSP3 proteins alter host cell RNA processing.
Further, we performed FASP-based proteome analysis of NSP3-expressing cells in parallel. Differentially expressed proteins (DEPs) were calculated with thresholds of FDR < 0.05, and |log2 FC| > 1 (Figure 7D; Table S8). Among them, we found that a known target of CoV1-NSP3, p53, was down-regulated by both NSP3 proteins, and this down-regulation was not CS-dependent [44]. Unfortunately, no significant GO terms or pathways were enriched for these DEPs. We compared our results with the proteome data after SARS-CoV-2 infection reported by Bojkova and colleagues [45] and highlighted overlapping proteins on the left of the heatmap (Figure 7D; Table S8). A comparison of transcriptome and proteome data revealed that NF-κB2 was up-regulated at both levels, indicating a positive feedback regulation, and highlighting the pivotal roles of NSP3 in NF-κB signaling (Figure 7E).
Multi-omics reveals cellular events regulated by NSP3 and the potential repurposing of drugs
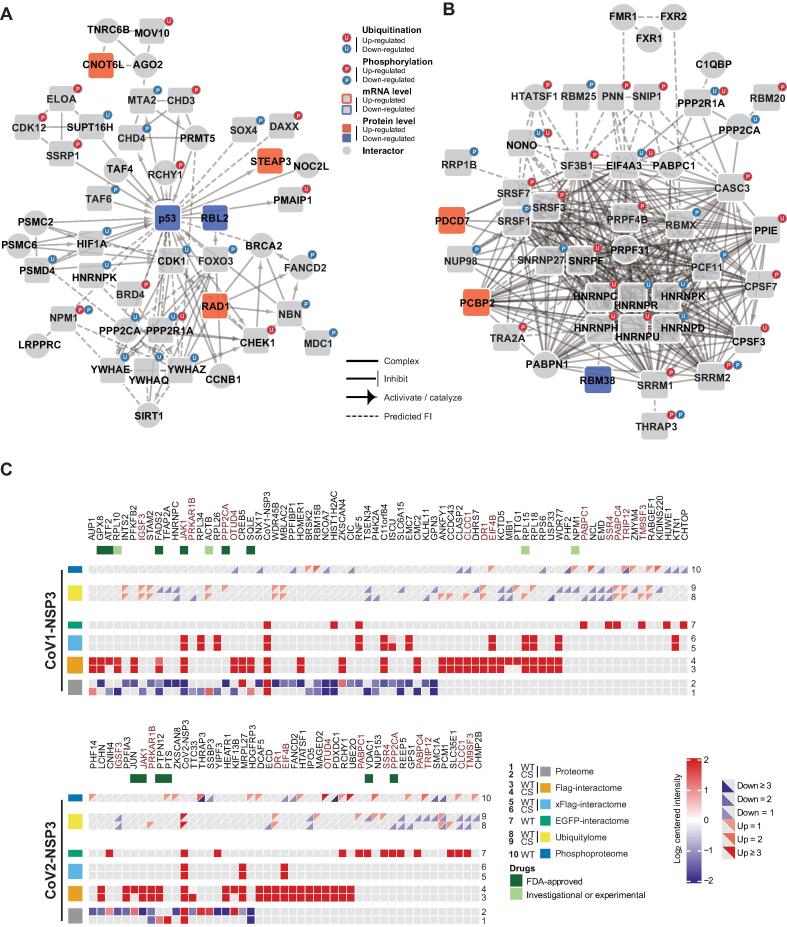
We combined all our significant candidates to systematically analyze CoV2-NSP3-regulated cellular events. We included known PPIs and regulation events to expand the connections of our candidates. Among the many cellular events we found, p53 was one of the proteins that were extensively regulated by CoV2-NSP3 (Figure 8A). We found interactors, activators, and inhibitors of p53 in our multi-omics studies. In addition, these regulators were associated with other candidates, further expanding the diffusion network. Therefore, we concluded that CoV2-NSP3 attacks p53 at multiple levels to circumvent the antiviral defense effects of p53 [44], [46]. Furthermore, the innate immune response, a surveillance system that SARS-CoV-2 needs to evade [47], was determined to be regulated by CoV2-NSP3 expression (Figure S10). Besides regulating antiviral effects, CoV2-NSP3 contributes to SARS-CoV-2-induced RNA AS [35]. We observed that proteins related to RNA splicing are regulated by CoV2-NSP3 at post-translational levels (Figure 8B). For example, the phosphorylation of splicing factors and ubiquitination of heterogeneous nuclear ribonucleoproteins were changed in CoV2-NSP3-expressing cells. Collectively, these data imply that CoV2-NSP3 has diverse cellular effects in SARS-CoV-2 infection.
Figure 8.
Regulation of cellular events by NSP3 and potential repurposing drugs for COVID-19
A. and B. Regulation of p53 (A) and proteins related to RNA AS (B) by CoV2-NSP3. Multi-omics data were combined. Network was created using Cytoscape with ReactomeFI v7.2.3. C. Candidate proteins for drug repurposing. Candidate proteins that are regulated by either CoV1-NSP3 or CoV2-NSP3 in at least two omics studies were shown. Candidate proteins regulated by both NSP3 proteins were labeled in red. The rectangular is divided into two triangles to indicate up-regulated or down-regulated ubiquitination or phosphorylation events. Proteins with FDA-approved or investigational drugs are labeled with green square (Table S9). “x” in xFlag-interactome represnts crosslinking. FI, functional interaction; FDA, Food and Drug Administration.
Finally, we analyzed our data to identify proteins that were significantly enriched in at least two omics studies of NSP3-expressing cells (Figure 8C; Table S9). In the meantime, we searched these candidate proteins against the drug database drugbank (https://go.drugbank.com/). In total, 13 proteins were regulated by both NSP3 proteins, and two (JAK1 and PPP2CA) out of them had 9 Food and Drug Administration (FDA)-approved drugs (Table S9). Importantly, drugs that target JAK1 are under investigation for COVID-19 [48], [49], [50]. Therefore, it would be interesting to investigate the antiviral effects of these candidate proteins with FDA-approved drugs.
Discussion
As the largest polypeptide of SARS-CoV, NSP3 interacts with host cellular functions in a versatilely manner and plays a pivotal role in the viral life cycle. To meaningfully study the functions of NSP3, it needs to be expressed in its full-length form in mammalian cells. Recent reports demonstrated that PLpro alone is not sufficient to cleave the NSP1-NSP2 polypeptide, and the PLpro domain has a very similar interactome with NSP3 without the transmembrane region, suggesting that the localization and integrity of NSP3 are important for its function and interaction with host cells [18], [21]. A previous study using deletions showed that CoV2-NSP3 has a general cytosolic localization, which is contradictory to our results [51]. We showed that CoV2-NSP3 has both ER and mitochondrial localizations, whereas CoV1-NSP3 localizes to ER as reported [15]. By comparing our results with recently published interactome studies using NSP3 deletions, we found that approximately 40% of interactors from at least two reported studies were found in our study. Importantly, reported interactors with high identification frequency are prone to be identified in our study. These results indicate that the expression of full-length NSP3 protein with proper cellular localization is important for its interaction with host cell proteins. For instance, we observed a strong association between NSP3 proteins and ribosomal proteins in our study, which is lacking in the previous studies.
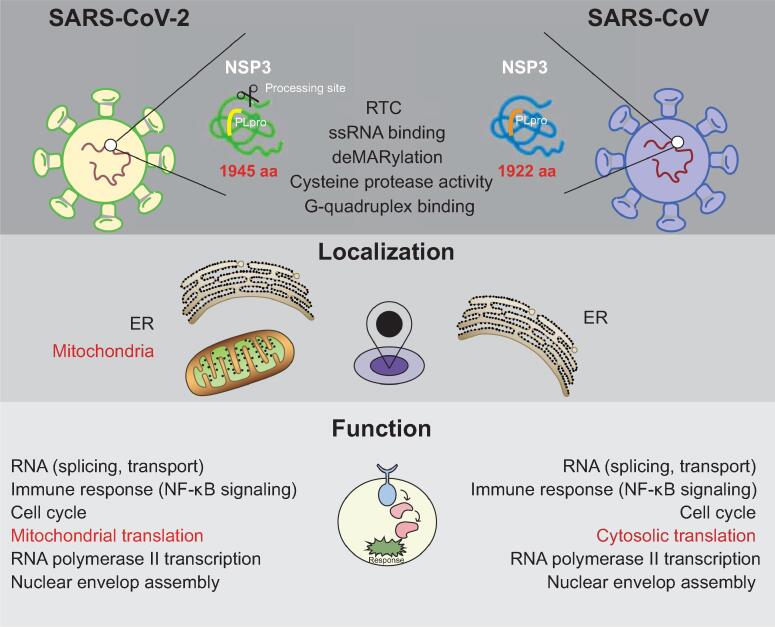
Understanding the contributions of each SARS-CoV-2 protein to the functions of host cells is important to follow the pathogenesis of viral infection, and eventually develop antiviral compounds. Multi-omics methods, such as proteomics-based interaction and proteome studies have been proven to be useful tools for studying virus–host cell interactions [29], [45]. In this study, we combined the interactome, phosphoproteome, transcriptome, proteome, and ubiquitylome to provide an integrative landscape of unique and shared functions of the NSP3 proteins. These parallel multi-omics datasets provide a rich resource for future mechanistic studies on how NSP3 regulates host cell functions in detail. Overall, we showed that NSP3 proteins are connected to multiple fundamental cellular functions, including RNA metabolism, immune response, cell cycle, translation, transcription, and others (Figure 9). These host cellular functions and signaling pathways are also largely affected by SARS-CoV-2 infection [29], [45], indicating that NSP3 is one of the key polypeptides of SARS-CoVs in attacking host cell functions. Interestingly, multi-omics results showed that although NSP3 proteins regulate similar cellular functions, they utilize different proteins to achieve these regulatory functions. For example, CoV2-NSP3 prefers to associate with the mitochondrial translational machinery, whereas CoV1-NSP3 is linked to the cytosolic translational machinery. These results suggest a conserved cellular effect of SARS-CoV NSP3 proteins during the evolution.
Figure 9.
Multi-omics analysis of NSP3 functions
Schematic overview of NSP3 regulated cellular functions. Red indicates difference of two NSP3 proteins. RTC, replication and transcription complex; ssRNA, single-strand RNA.
Previous studies on SARS-CoV have suggested that the combination of three transmembrane-localized NSPs (NSP3, NSP4, and NSP6) induces DMV formation, which together with other NSPs and host cell proteins, forms the RTC. Therefore, the membrane localization of NSP3 might be a key factor that the virus uses to select the organelles for RTC formation. In the present study, we found that unlike CoV1-NSP3, CoV2-NSP3 has two cellular localizations, N-terminal mitochondrial and C-terminal ER localizations. In addition, we showed that the N-terminal enrichment of CoV2-NSP3 extensively results in the IP of mitochondrial proteins. Furthermore, Wu and colleagues [52] showed that the SARS-CoV-2 RNA genome and small-guide RNAs (sgRNAs) are enriched in the host mitochondrial matrix using computational modeling. These results suggest that during SARS-CoV-2 infection, NSP3 might exploit both ER and mitochondria to initiate the RTC formation. In addition, we showed that proteins related to mRNA exporting and translation are enriched for both NSP3 proteins. In combination with the fact that the RTC is the location for RNA synthesis, we speculate that NSP3 couples RNA transport and translation together to rapidly synthesize viral proteins. However, we should note that the interaction between ribosomal proteins and NSP3 is not mediated by RNA, as we introduced universal nucleases to our cell lysis processes.
In summary, we expressed and studied the localization of NSP3 proteins. We identified a rich candidate resource for studying the functions of NSP3 proteins and provided a systematic analysis of how NSP3 contributes to the regulation of cellular functions. Importantly, we showed that although both NSP3 proteins shared extensive similar functions, SARS-CoV-2 NSP3 is different from SARS-CoV NSP3 in its connection with mitochondrial proteins. Moreover, we provided a list of FDA-approved compounds that could serve as a starting point for drug repurposing with some already under investigation.
Materials and methods
Plasmids, cell culture, transfection, immunoblotting, and IP
Full-length coding sequences of SARS-CoV NSP3 (GenBank: FJ882960) and SARS-CoV-2 NSP3 (GenBank: MN988669) with earlier inserted stop codon but without codon optimization were synthesized from IGEbio, Guangzhou, China. Polymerase Chain Reaction (PCR) amplified products were subcloned into homemade lentivirus vector either with a C-terminal EGFP tag or N-terminal Flag and C-terminal EGFP tags. Mutagenesis PCR was performed to mutate the earlier stop codon to generate WT NSP3 using Phanta Max Super-Fidelity DNA Polymerase (Catalog No. P505-D2, Vazyme, Nanjing, China). The same method was used to create catalytic inactivated (CS-mutated), GlyGly deficient (dGG-mutated), and domain exchanged NSP3 plasmids. UBE2O plasmid was described before [53] and TNIP cDNA was amplified from HeLa cDNA and cloned in to pCS2 vector with Flag tag. All batches of purified plasmids (Catalog No. 12165, Qiagen, Hilden, Germany) were confirmed by DNA sequencing (IGEbio).
HEK293T cells from American Type Culture Collection (ATCC) were cultured in Dulbecco’s Modified Eagle’s Medium (Catalog No. C11995500BT, ThermoFisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Catalog No. SV30160.03, Cytiva, Boston, MA) and 1× penicillin/streptomycin (Catalog No. SV30010-10, Cytiva). HEK293T cells were tested mycoplasma free using MycoAlert Detection Kit (Catalog No. LT07-318, Lonza, Basel, Switzerland) before the whole experiments.
HEK293T cells were transfected using Polyethylenimine (PEI; Catalog No. 24765-2, Polysciences, Warrington, PA) with a ratio of plasmid:PEI = 1:3. Cells were harvest using whole cell lysate buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris at pH 8.0, and 10% glycerol) with 1× cOmplete™ Protease Inhibitor Cocktail (Catalog No. 11873580001, Sigma, St Louis, MO) 48 h post-transfection, following with a maximum speed centrifugation at 4 °C. Supernatant was collected and protein concentration was measured using BCA assay kit (Catalog No. 23225, ThermoFisher Scientific, Rochford, IL). Equivalent amounts of proteins were boiled with 1× LDS loading buffer (Catalog No. NP0008, ThermoFisher Scientific) for 5 min at 95 °C and resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting. For Flag-IP assays, anti-Flag-M2 agarose beads (Catalog No. A2220, Sigma) were used to enrich NSP3 and interactors. For phosphorylation validation, 1× PhosSTOP (Catalog No. 4906837001, Sigma) was added to lysis buffer, and IP assays were performed either with anti-Flag agarose beads for 2 h or with 1 μg anti-UBE2O antibody (Catalog No. A301-873A, Bethyl, Montgomery, AL) overnight followed by 1.5 h incubation with 15 μl prewashed protein A/G beads (1:1 mixture; Catalog Nos. 1614013 and 1614023, Bio-Rad, Hercules, CA). Bound proteins were boiled with 1× LDS loading buffer for 5 min at 95 °C after 3 washes with lysis buffer. Antibodies used in these experiments included anti-Flag (Catalog No. F1804, Sigma), anti-EGFP (Catalog No. 50430-2-AP, Proteintech, Rosemont, IL), anti-NSP3 (Catalog No. ab181620, Abcam, Cambridge, UK), anti-UBE2O (Catalog No. A301-873A, Bethyl), pSerine (Catalog No. gtx26639, GeneTex, Irvine, CA), pThreonine (Catalog No. 9386, Cell Signaling, Beverly, CA), anti-TAB2 (Catalog No. A302-759A, Bethyl), anti-TAB3 (Catalog No. A302-208A, Bethyl), anti-RPL5 (Catalog No. A303-933A, Bethyl), anti-RPS6 (Catalog No. A300-556A, Bethyl), anti-MRPL13 (Catalog No. 16241-1-AP, Proteintech), anti-MRPL23 (Catalog No. 11706-1-AP, Proteintech), anti-MRPL44 (Catalog No. 16394-1-AP, Proteintech), anti-USP34 (Catalog No. A300-824A, Bethyl), anti-DHX38 (Catalog No. A300-858A-T, Bethyl), anti-IPO4 (Catalog No. 11679-1-AP, Proteintech), anti-IPO11 (Catalog No. 14403-1-AP, Proteintech), anti-NUP85 (Catalog No. 19370-1-AP, Proteintech), and GAPDH-HRP (Catalog No. KC-5G4, KANGCHENG, Shanghai, China).
Immunofluorescence
Cells grown on 0.1% gelatin coated coverslips were transfected with indicated plasmids for 36 h. 4% paraformaldehyde (Catalog No. D60714, Deweibio, Guangzhou, China) was used to fix the cells at room temperature for 30 min followed by quenching with 2 mg/ml glycine. Cells were then permeabilized with 0.2% Triton X-100 (Catalog No. X-100, Sigma) and blocked with 10% FBS. Indicated first antibody and second antibody were added for overnight at 4 °C and 1 h at room temperature, respectively. Cells were mounted with ProLong Gold Antifade Reagent with DAPI (Catalog No. 8961S, Cell Signaling). Images were taken and processed using LSM800 and LSM900 (Zeiss, Oberkochen, Germany). Antibodies used for the experiments are: anti-CANX (Catalog No. 2679, Cell Signaling), anti-TOMM20 (Catalog No. ab56783, Abcam), anti-Flag (Catalog Nos. F1804 and F7425, Sigma), and anti-NSP3 (Catalog No. ab181620, Abcam).
Fluorescence-activated cell sorting
Cell cycle and mitochondrial potential experiments were performed using Fluorescence-activated cell sorting (FACS). For cell cycle analysis, approximately 1 ml of 1 × 106/ml cells transfected with indicated plasmids were harvest and fixed with 70% ethanol (Catalog No. 1.00983.1011, Sigma) overnight. 0.1% Triton X-100 was used to permeabilize cells before the staining with propidium iodide (Catalog No. P4864, Sigma) for 15 min at room temperature. Cells were analyzed with phycoerythrin (PE) channel using Beckman CytoFLEX.
Interactome sample preparation
HEK293T cells transfected with indicated plasmids in biological triplicates were lysed using whole cell lysis buffer [50 mM Tris at pH 8.0 (Catalog No. 15504020, ThermoFisher Scientific), 150 mM NaCl (Catalog No. 106443, Sigma), 10% glycerol (Catalog No. 104032, Sigma), and 0.5% NP-40 (Catalog No. I8896, Sigma)] with fresh added 1× Complete Protease inhibitors and Universal Nuclease (1:5000; Catalog No. 88702, ThermoFisher Scientific). For crosslinking IP-MS with RIME method, transfected cells were incubated with 2 mM discuccinimidyl glutarate (Catalog No. sc-285455A, Santa Cruz, Dallas, TX) for 20 min at room temperature first, then replaced with 1% formaldehyde (Catalog No. F8775, Sigma) for 10 min at room temperature, followed by quenching with 0.1 M glycine for another 10 min at room temperature. Cells were incubated for 2 h at 4 °C on a rotation wheel. Soluble cell lysates were collected after maximum speed centrifugation at 4 °C for 15 min. 1 mg of cell lysates were incubated with anti-Flag-M2 (Catalog No. M8823, Sigma) or GFP (Catalog No. gtma, Chromotek, Planegg-Martinsried, Germany) magnetic agarose for 1.5 h. On-bead digestion was performed to digest immunoprecipitated protein to peptides for MS measurements [54]. In short, beads after incubation were washed 3 times with cell lysis buffer and 1 time with phosphate-buffered saline (PBS; Catalog No. C10010500BT, ThermoFisher Scientific). After complete removal of PBS, beads were incubated with 100 μl of elution buffer [2 M urea (Catalog No. U5378, Sigma), 10 mM dithiothreitol (DTT; Catalog No. D9779, Sigma), and 50 mM Tris at pH 8.5] for 20 min. Afterwards, iodoacetamide (IAA; Catalog No. I1149, Sigma) was added to a final concentration of 50 mM for 10 min in the dark, followed by partial digestion with 250 ng of trypsin (Catalog No. V5280, Promega, Madison, WI) for 2 h. After incubation, the supernatant was collected into a separate tube. The beads were then incubated with 100 μl of elution buffer for another 5 min, and the supernatant was collected in the same tube. All these steps were performed at room temperature in a thermoshaker C at 1500 r/min. Combined elutes were digested with additional 200 ng of trypsin overnight at room temperature. Finally, tryptic peptides were acidified to pH < 2 by adding 10 μl of 10% trifluoroacetic acid solution (TFA; Catalog No. 1002641000, Sigma).
Phosphoproteome sample preparation
HEK293T cells transfected with indicated plasmids in biological triplicates were lysed using sodium deoxycholate (SDC) buffer [4% SDC (Catalog No. D6750, Sigma), 100 mM Tris at pH 8.5], and phosphoproteome sample preparation was performed according to the previous report [34]. In brief, 200 μg of cell lysates were reduced and alkylated with Tris (2-carboxyethyl) phosphine hydrochloride (TCEP; Catalog No. 75268, Sigma) and 2-chloroacetamide (CAA; Catalog No. C0267, Sigma) at 45 °C for 5 min, following with overnight digestion with Lys-C (Catalog No. 129-02541, Wako Chemicals, Osaka, Japan) and trypsin (Catalog No. T6567, Sigma). TiO2 beads (Catalog No. 5010-21315, GL Sciences, Tokyo, Japan) were used to enrich phosphorylated peptides, and homemade C8 StageTip (Catalog No. 66882-U, Sigma) was used to trap the TiO2 beads for washes. Eluted phosphorylated peptides were loaded into homemade SDB-RPS StageTip (Catalog No. 66886-U, Sigma) for desalting.
Ubiquitylome sample preparation
HEK293T cells transfected with indicated plasmids in biological triplicates were lysed using buffer containing 8 M urea, 50 mM Tris-HCl at pH 8, 150 mM NaCl, 1 mM EDTA (Catalog No. EDS, Sigma), 1 mM CAA, and 1× Complete Protease inhibitors. K-ε-GG Ubiquitin Remnant Motif Enrichment Kit (Catalog No. 5562, Cell Signaling) was used to enrich K-ε-GG peptides. The whole process was performed according to the manufacturer’s instruction except in the steps of digestion and peptide desalting. We used Lys-C to digest 3 mg of proteins in 8 M urea buffer for 3 h at 37 °C first, and then diluted the buffer to 2 M urea and performed trypsinization overnight at 37 °C. For peptide desalting, we used the Blond Elute LRC-C18 200 mg column (Catalog No. 12113024, Agilent, Santa Clara, CA), followed by lyophilization for 2 days.
Proteome sample preparation
HEK293T cells transfected with indicated plasmids in biological triplicates were lysed with 8 M urea with 10 mM DTT and 0.1 M Tris (pH 8.5). Soluble cell lysates were collected after 10 cycles of sonication for 30 s followed by 30 s at 4 °C using a Biorupter Pico Sonicator (Diagenode, Liége, Belgium), followed by maximum speed centrifugation at room temperature for 15 min. Whole proteome peptides were prepared using the FASP protocol as described before [54]. In brief, 50 μg of soluble lysates were added onto 30 kDa cutoff filter (MRCF0R030, Sigma) and centrifuged at 11,000 r/min at 20 °C for 15 min. 50 mM IAA in urea buffer was used to alkylate proteins at 20 °C for 15 min. After 3 washes with urea lysis buffer and 3 washes with 50 mM ammonium biocarbonate (ABC; Catalog No. 09830, Sigma) buffer, 500 ng of trypsin in 50 μl of 50 mM ABC buffer was used to digest proteins in a wet chamber overnight at 37 °C. Peptides were extracted by 50 mM ABC buffer and acidified to pH < 2 by adding 10 μl of 10% TFA.
RNA-seq
HEK293T cells transfected with indicated plasmids in biological triplicates were harvested, and total RNA was isolated using ipureTRizol kit (Catalog No. K417, IGEbio) according to manufacturer’s instruction. Sequencing libraries were generated using Next UltraTM RNA Library Prep Kit for Illumina (Catalog No. E7760, NEB, Ipswich, UK) following manufacturer’s recommendations. In brief, mRNA was captured using mRNA capture beads, followed by fragmentation and cDNA synthesis using random hexamers. DNA clean beads were used to purified dsDNA after the second strand synthesis, followed by end repairing, A tailing, adapter ligation, PCR amplification, and library purification. Sequencing was done by Illumina NovaSeq platform (IGEbio).
MS measurements
Desalted peptides were separated and analyzed with an Easy-nLC 1200 (ThermoFisher Scientific) connected online to Fusion Lumos or Fusion Eclipse (crosslinking experiment, ThermoFisher Scientific) mass spectrometer equipped with FAIMS pro (high-Field Asymmetric waveform Ion Mobility Spectrometry, ThermoFisher Scientific) using different gradient of buffer B [80% acetonitrile (Catalog No. 1000041001, Sigma) and 0.1% formic acid (Catalog No. 1002641000, Sigma)]. For interactome study, a gradient of total 140 min of buffer B (2%–22%, 100 min; 22%–28%, 20 min; 28%–36%, 12 min; 100%, 8 min) was used; for proteome and ubiquitylome analyses, a gradient of total 240 min of buffer B (2%, 1 min; 2%–7%, 10 min; 7%–28%, 200 min; 28%–36%, 15 min; 36%–60%, 5 min; 95%, 7 min) was used; for phosphoproteome analysis, a gradient of total 120 min of buffer B (2%–22%, 80 min; 22%–28%, 20 min; 28%–40%, 12 min; 95%, 8 min) was used. Data dependent analysis was used as data acquisition mode. Detailed information about the gradient and the setting of the MS can be found in the raw files (ProteomeXchange: PXD023927). Raw data were first transformed using FAIMS MzXML Generator and then analyzed using MaxQuant version 1.6.17.0 [55] search against Human Fasta database with CoV1-NSP3 and CoV2-NSP3 protein sequences. Label-free quantification and match between run functions were enabled for all analyses. For phosphoproteome and ubiquitylome analyses, pSTY (phospho-Serine, Threonine, Tyrosine) and Gly-Gly (K, not C-term) in variable modifications were added.
PCA
PCA for all omics studies were performed using top 1000 variable events with maximum standard deviation of NSP3-expressing cells. PC1–PC2 scatterplot exhibits the magnitude of the difference between samples.
Proteome and interactome data analysis
The MaxQuant output files “proteinGroups” were used for the subsequent analysis of proteome and interactome. We filtered out the protein that labeled as reverse or potential contaminant, and analyzed the non-crosslinking interactome and proteome data using DEPs [56]. The significant thresholds were log2 FC > 2, FDR < 0.001 for interactome analyses, and |log2 FC| > 1, FDR < 0.05 for proteome analyses. For crosslinking interactome analysis, we used an alternative method described before, which used a variable filter combining log2 FC (enrichment) and adjusted P value [57]. Briefly, to determine enriched interactors, we used a cutoff line with the function y > c/(x − x0) on scatters where x is the log2 FC and y is adjusted P value (c = curvature, x0 = minimum FC). The distribution of log2 FC between NSP3 overexpression and EGFP control was fitted to a Gaussian curve using least squares fitting (excluding outliers) to determine the standard deviation σ. We set a stringent FC cutoff x0 to 2σ, while selected a relatively loose c to get a higher positive coverage rate. Then we verified the overlap between crosslinking IP-MS and non-crosslinking IP-MS under selected curvature c.
PPI network and pathway annotation of interactors were constructed by Cytoscape plugin ReactomeFI v7.2.3 [58]. Interactors of NSP3 were divided into 3 groups (CoV1-NSP3-specific, CoV2-NSP3-specific, and both) and enriched GO terms were shown using clusterProfiler package [59]. Circos plots were performed using circlize package [60].
Phosphoproteome data analysis
The MaxQuant output files designated “Phospho(STY)sites” were used for testing intensity differences of phosphosites between control and NSP3-expressing cells. We performed Student’s t-test as two-sample-test and one-way analysis of variance (ANOVA) test following Tukey’s significant honest difference post-hoc test (THSD) as multiple-sample-test. P values were adjusted by BH method. We defined peptides with adjusted P < 0.01, |log2 FC| > 2, and localization possibility > 0.7 as significant enriched peptides.
To study change of kinase activities, we ranked phosphosites by log2 FC and performed Kinase-Substrate Enrichment Analysis (KSEA) using KSEAapp package [61] in combination with a reported kinase-substrate relationship [62]. We filtered out the kinases of which matching sites were less than 3 or Z-score was lower than 1 and created human kinome tree illustration using KinMap [37]. The targeting drugs that regulate activity-changed kinases were extracted from drugbank (https://go.drugbank.com/).
Ubiquitylome data analysis
The ubiquitylome data were analyzed as described for phosphoproteome. Here, we defined peptides with adjusted P < 0.001, |log2 FC| > 2, and localization possibility > 0.7 as significant enriched peptides.
Transcriptome data analysis
Raw reads (GSA: HRA000634) were aligned to the human genome (hg38) using HISAT2 [63]. Next, we assembled reads to transcripts and quantified the read counts of each gene utilizing StringTie [64]. Lowly expressed genes were filtered out using a Counts per million (CPM) threshold. We kept genes with CPM greater than 1 in at least 2 samples, and analyzed theDEGs using edgeR [65]. Top 5000 up-regulated and 5000 down-regulated genes were extracted and ordered by log2 FC as the gene list input of GSEA. DEGs between control and NSP3-expressing cells were characterized for each sample with thresholds of |log2 FC| > 1 and FDR < 0.05.
Motif enrichment
The DEGs were divided into 2 gene sets according to their expression change statuses (Clusters 1 and 2 for up-regulated genes and Clusters 3 and 4 for down-regulated genes) as represented in Figure 7A. Then, the putative transcription factor (TF)-targeting regulons of these 2 gene sets were identified by employing RcisTarget [66]. We choose human motif collection v9 as TF annotations and the motif-rankings in 10 kb around transcription start sites (TSSs) of hg38 genome as region databases. The motifs with most target genes as well as a normalized enrichment score (NES) lager than 3 were determined as significant enriched regulons.
AS analysis
We performed AS analysis based on transcription raw reads with vast-tools V2.5.1 [67] in combination with vastdb hs2.23.06.20. To determine significant AS events, we kept the AS events which have more than 10 reads in at less two samples and applied a further filter by setting a noB3 parameter to alternative exon (AltEx) and a 0.05 binomial-test p_IR parameter to IR in vast-tools’ diff module. Significant AS events were defined according to the following requirements: change was greater than 10 delta Percent-Spliced-In (dPSI)/delta Percent-Intron-Retention (dPIR), and minimal difference value (where P > 0.95) was greater than 0.
GO and GSEA
GO and GSEA were implemented based on clusterProfiler package. C2 (chemical and genetic perturbation sub-collection) and C5 (GO sub-collection) from MSigDB v7.2 [42] and the pathway gene sets from Reactome Pathway Database [30] were chosen for GSEA. Significant terms were chosen by the threshold that P < 0.05, FDR < 0.25, and |NES| > 1. Most significant terms were selected to plot heatmap.
Both GO and Reactome pathway enrichments were applied using the Over Representation Analysis (ORA) based on the hypergeometric distribution and P value of each term was adjusted by BH method. GO analyses of BP, cellular component (CC), molecular function (MF) were performed using enrichR in clusterProfiler package. Reactome pathway analysis was performed using enrichPathway function in ReactomePA packages [8]. Gene sets large than 500 or less than 15 genes were excluded from ORA. Significant GO terms were enriched by a threshold of adjusted P value < 0.05 and q-value < 0.25.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository (ProteomeXchange: PXD023927) [68], and are publicly accessible at http://proteomecentral.proteomexchange.org. The raw sequence data are deposited in the Genome Sequence Archive [69] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: HRA000634), and are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human/.
CRediT author statement
Ruona Shi: Conceptualization, Investigation, Validation, Visualization. Zhenhuan Feng: Conceptualization, Formal analysis, Data curation, Visualization. Xiaofei Zhang: Conceptualization, Supervision, Writing - original draft, Writing - review & editing, Funding acquisition. All authors have read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
Acknowledgments
We like to thank Bioland Laboratory, Guangzhou, China for the generous startup package. Work in Zhang lab is supported by the Guangdong Science and Technology Project, China (Grant Nos. 2018B030306047 and 2020B1212060052), the National Natural Science Foundation of China (Grant Nos. 31770889 and 31801180), the Guangzhou Science and Technology Project, China (Grant No. 201904010469), and the Guangzhou Regenerative Medicine and Health Guangdong Laboratory project, China (Grant No. 2018GZR110104003). We would like to apologize to all researchers whose work were unable to be cited and discussed due to space constraints. We thank all researchers whose excellent work helped us to understand and tackle the COVID-19. We would like to thank Prof. Duanqing Pei from Westlake University, China for valuable suggestions. We would like to thank all members in Zhang lab for fruitful discussions, Liman Guo from Proteomics & Metabolomics Centre of Bioland Laboratory for excellent mass-spectrometry measurement assistance, Yu Fu from Microscope & Imaging Centre of Bioland Laboratory for confocal imaging, and Chan Rong from Center for Cell Lineage and Atlas of Bioland Laboratory for FACS assistance.
Handled by Yu Xue
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2021.09.007.
Supplementary material
The following are the Supplementary data to this article:
Expression and subcellular localization of NSP3 proteins with C-terminal tag A. Schematic illustration of plasmids used in Figures 1B, S1B, and S1C. B. NSP3 cleaves itself at the C-terminus. HEK293T cells transfected with CoVs-NSP3-EGFP plasmids were lysed and EGFP and NSP3 antibodies were used after immunoblotting. EGFP was released in WT, but not in CS- or dGG-mutated NSP3 samples. Green arrow indicates the released EGFP, whereas asterisk indicates non-specific signal. C. Validation of the specificity of CoV-1 NSP3 antibody. HEK293T cells transfected with indicated plasmids were fixed 48 h post-transfection. EGFP and NSP3 antibodies were used to detect the localization of NSP3 proteins.
NSP3 proteins have preferable cellular localization A and B. Cellular localization of NSP3 protein detected by EGFP antibody. HEK293T cells transfected with indicated plasmids were fixed 48 h post-transfection. EGFP fluorescence was used to detect the localization of CS-NSP3-EGFP protein. CANX and TOMM20 antibodies (red) were used to serve as markers for the ER (A) and mitochondria (B). Red arrow indicates that the large vesicle-like structure of CoV2-NSP3 was not co-localized with TOMM20. C. CoV2-NSP3 has two cellular localizations. Immunostaining was performed as in Figure S2A. Merged signals show that GFP and NSP3, but not NSP3 and Flag, are co-localized well. D. Flag-CoV1-NSP3 prefers to co-localize with CANX. Immunostaining was performed as in Figure S2A. Flag and CANX antibodies were used to visualize the co-localization between CoV1-NSP3 and the ER marker CANX. E. Interaction preference of NSP3 protein and CANX/TOMM20. Interactome studies of NSP3 based on Flag (Table S1) and EGFP (Table S3) IP-MS were analyzed to show the binding preference between NSP3 proteins and CANX/TOMM20. ER, endoplasmic reticulum; IP-MS, Immunoprecipitation-Mass Spectrometry.
PLpro domain of CoV1-NSP3 is responsible for the formation of the large vesicle-like structures CoV2-NSP3 fused with PLpro domain of CoV1-NSP3 induces large-vesicle structures. Immunostaining was performed as in Figure S2A.
Workflow and quality control of multi-omics study A. Schematic overview of the workflow to study the function of NSP3. B. Boxplots of log2 imputed intensity for all proteomics studies. FASP, Filter Aided Sample Preparation; xIP-MS, crosslinking Immunoprecipitation-Mass Spectrometry.
Protein–protein interaction network of NSP3 interactomes A. PCA analysis of top 1000 variable proteins of NSP3 interactomes. Top 1000 variable proteins with highest standard deviation (Table S1) were used as input for PCA analysis. B. Upset and Venn diagrams showing the overlap of NSP3 interactors. Significant interactors for WT and CS of NSP3 proteins were used as input. Data used for the analysis is available in Table S1. C and D. Protein-protein interaction network of CoV2-NSP3 (C) and CoV1-NSP3 (D) interactomes. Significant interactors of NSP3 were clustered by gene annotation and protein–protein interaction network was constructed using Cytoscape with ReactomeFI v7.2.3. PCA, principal component analysis; PC, principal component.
Crosslinking IP-MS to identify NSP3 interactome A. PCA analysis of top 1000 variable proteins of NSP3 interactomes. Top 1000 variable proteins of each condition (Table S2) were used as input for PCA analysis. B. Venn diagram showing the overlap of NSP3 interactors identified in crosslinking and non-crosslinking experiments. C. Clustering of significant interactors of NSP3 proteins using crosslinking IP-MS. Analysis and color as Figure 3A. Full list of interactors is available in Table S2. D. Representative enriched GO of identified interactors in crosslinking IP-MS. Analysis as Figure 3B. Full list of GO terms is available in Table S2.
NSP3 interactome identified by EGFP IP-MS. A. PCA analysis of top 1000 variable proteins of NSP3-EGFP interactomes. Top 1000 variable proteins with highest standard deviation (Table S3) were used as input for PCA analysis. B. Clustering of significant interactors of NSP3 proteins. The relative intensity to control samples of significant interactors was compared between CoV1-NSP3 and CoV2-NSP3. Full list of interactors is available in Table S3. C. Venn diagram showing the overlap of CoV1-NSP3 and CoV2-NSP3 interactomes. D. Representative enriched GO of identified interactors. Analysis as Figure 3B. Full list of GO terms is available in Table S3.
Phosphoproteome related analyses A. PCA analysis of top 1000 variable phosphorylated peptides of NSP3 expressing cells. Top 1000 variable phosphorylated peptides with standard deviation (Table S5) were used as input for PCA analysis. B. Clustering of DEPPs which were also found in SARS-CoV-2 infection studies. Two reported phosphoproteome data were used as inputs to identify consistent phosphorylated peptides. Analysis and color as Figure 3A. Full list of DEPPs is available in Table S5. C. Cell cycle analysis of NSP3 expressing cells. Cells transfected with indicated plasmids were harvest for FACS analysis using propidium iodide. Significant differences compared to control cells were calculated using Student’s t-tests. The graph showed mean ± SD, n = 3, * indicates P < 0.05, ** indicates P < 0.01. One representative experiment was shown. D. and E. Kinases inhibitors mapped to kinases whose activity mapped to DEPPs in CoV2-NSP3 (D) and CoV1-NSP3 (E) expressing cells. Red indicates upregulated activity, whereas blue indicates downregulated activity. F. Clustering of DEPPs that regulated by IKK-family and TBK1. Analysis and color as Figure 3A. FACS, Fluorescence-activated Cell Sorting; SD, Standard Deviation; pSTY, phospho-Serine, Threonine, Tyrosine; G0, G0 phase; G1, G1 phase; G2, G2 phase; S, S phase; M, M phase.
Ubiquitylome related analyses A. PCA analysis of top 1000 variable ubiquitinated peptides of NSP3 expressing cells. Top 1000 variable ubiquitinated peptides with highest standard deviation (Table S6) were used as input for PCA analysis. B. Deubiquitinase activity dependent ubiquitylome of NSP3 protein. Analysis and color as in Figure 3A. Full list of DEUPs is available in Table S6.
Regulation of innate response by NSP3. Multi-omics data were combined to model the regulation innate response by NSP3. Network was created using Cytoscape with ReactomeFI v7.2.3.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., et al. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm Sin B. 2021;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Chin J Epidemiol. 2020;41:145–151. (in Chinese with an English abstract) [Google Scholar]
- 6.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharjee A., Saha M., Halder A., Debnath A., Mukherjee O. Therapeutics and vaccines: strengthening our fight against the global pandemic COVID-19. Curr Microbiol. 2021;78:435–448. doi: 10.1007/s00284-020-02310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan J.W., Kok K.H., Zhu Z., Chu H., To K.W., Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J., Moscona A. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4:e00524–13. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoops K., Kikkert M., van der Worm S.H.E., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belov G.A., van Kuppeveld F.J.M. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr Opin Virol. 2012;2:740–747. doi: 10.1016/j.coviro.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alhammad Y.M.O., Kashipathy M.M., Roy A., Gagné J.P., McDonald P., Gao P., et al. The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. J Virol. 2021;95:e01969–20. doi: 10.1128/JVI.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39:e106275. doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claverie J.M. A putative role of de-mono-ADP-ribosylation of STAT1 by the SARS-CoV-2 Nsp3 protein in the cytokine storm syndrome of COVID-19. Viruses. 2020;12:646. doi: 10.3390/v12060646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armstrong LA, Lange SM, dee Cesare V, Matthews SP, Nirujogi RS, Cole I, et al. Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its inhibition by nanobodies. PLoS One 2021;16:e0253364. [DOI] [PMC free article] [PubMed]
- 22.Chen Z., Wang C., Feng X., Nie L., Tang M., Zhang H., et al. Interactomes of SARS-CoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. EMBO J. 2021;40:e107776. doi: 10.15252/embj.2021107776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almasy K.M., Davies J.P., Plate L. Comparative host interactomes of the SARS-CoV-2 nonstructural protein 3 and human coronavirus homologs. Mol Cell Proteomics. 2021;20:100120. doi: 10.1016/j.mcpro.2021.100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Meer Y., Snijder E.J., Dobbe J.C., Schleich S., Denison M.R., Spaan W.J., et al. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clementz M.A., Kanjanahaluethai A., O'Brien T.E., Baker S.C. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology. 2008;375:118–129. doi: 10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verheije M.H., Raaben M., Mari M., te Lintelo E.G., Reggiori F., van Kuppeveld F.J.M., et al. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2020;48:D498–503. [DOI] [PMC free article] [PubMed]
- 31.Mohammed H., Taylor C., Brown G.D., Papachristou E.K., Carroll J.S., D'Santos C.S. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11:316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 32.Oughtred R., Rust J., Chang C., Breitkreutz B.J., Stark C., Willems A., et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- 34.Humphrey S.J., Karayel O., James D.E., Mann M. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat Protoc. 2018;13:1897–1916. doi: 10.1038/s41596-018-0014-9. [DOI] [PubMed] [Google Scholar]
- 35.Hekman R.M., Hume A.J., Goel R.K., Abo K.M., Huang J., Blum B.C., et al. Actionable cytopathogenic host responses of human alveolar type 2 cells to SARS-CoV-2. Mol Cell. 2020;80:1104–1122. doi: 10.1016/j.molcel.2020.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eid S., Turk S., Volkamer A., Rippmann F., Fulle S. KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinformatics. 2017;18:16. doi: 10.1186/s12859-016-1433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connell N.T., Berliner N. Fostamatinib for the treatment of chronic immune thrombocytopenia. Blood. 2019;133:2027–2030. doi: 10.1182/blood-2018-11-852491. [DOI] [PubMed] [Google Scholar]
- 39.Kost-Alimova M., Sidhom E.H., Satyam A., Chamberlain B.T., Dvela-Levitt M., Melanson M., et al. A high-content screen for mucin-1-reducing compounds identifies fostamatinib as a candidate for rapid repurposing for acute lung injury. Cell Rep Med. 2020;1:100137. doi: 10.1016/j.xcrm.2020.100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu H., Fada B.J. Specificity in ubiquitination triggered by virus infection. Int J Mol Sci. 2020;21:4088. doi: 10.3390/ijms21114088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Q, Wu Y, Qin Y, Hu J, Xie W, Qin FX, et al. Broad and diverse mechanisms used by deubiquitinase family members in regulating the type I interferon signaling pathway during antiviral responses. Sci Adv 2018;4:eaar2824. [DOI] [PMC free article] [PubMed]
- 42.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma-Lauer Y., Carbajo-Lozoya J., Hein M.Y., Müller M.A., Deng W., Lei J., et al. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc Natl Acad Sci U S A. 2016;113:E5192–E5201. doi: 10.1073/pnas.1603435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cardozo C.M., Hainaut P. Viral strategies for circumventing p53: the case of severe acute respiratory syndrome coronavirus. Curr Opin Oncol. 2021;33:149–158. doi: 10.1097/CCO.0000000000000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amor S., Blanco L.F., Baker D. Innate immunity during SARS-CoV-2: evasion strategies and activation trigger hypoxia and vascular damage. Clin Exp Immunol. 2020;202:193–209. doi: 10.1111/cei.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan B, Freiwald T, Chauss D, Wang L, West E, Bibby J, et al. SARS-CoV2 drives JAK1/2-dependent local and systemic complement hyper-activation. Sci Immunol 2021;6:eabg0833.
- 49.Bagca B.G., Avci C.B. The potential of JAK/STAT pathway inhibition by ruxolitinib in the treatment of COVID-19. Cytokine Growth Factor Rev. 2020;54:51–61. doi: 10.1016/j.cytogfr.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stebbing J, Nievas GS, Falcone M, Youhanna S, Richardson P, Ottaviani S, et al. JAK inhibition reduces SARS-CoV-2 liver infectivity and modulates inflammatory responses to reduce morbidity and mortality. Sci Adv 2020;7:eabe4724. [DOI] [PMC free article] [PubMed]
- 51.Zhang J., Cruz-cosme R., Zhuang M.W., Liu D., Liu Y., Teng S., et al. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct Target Ther. 2020;5:269. doi: 10.1038/s41392-020-00372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu K.E., Fazal F.M., Parker K.R., Zou J., Chang H.Y. RNA-GPS predicts SARS-CoV-2 RNA residency to host mitochondria and nucleolus. Cell Syst. 2020;11:102–108. doi: 10.1016/j.cels.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X., Zhang J., Bauer A., Zhang L., Selinger D.W., Lu C.X., et al. Fine-tuning BMP7 signalling in adipogenesis by UBE2O/E2-230K-mediated monoubiquitination of SMAD6. EMBO J. 2013;32:996–1007. doi: 10.1038/emboj.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X., Smits A.H., van Tilburg G.B.A., Jansen P.W.T.C., Makowski M.M., Ovaa H., et al. An interaction landscape of ubiquitin signaling. Mol Cell. 2017;65:941–955. doi: 10.1016/j.molcel.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 55.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X., Smits A.H., van Tilburg G.B., Ovaa H., Huber W., Vermeulen M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat Protoc. 2018;13:530–550. doi: 10.1038/nprot.2017.147. [DOI] [PubMed] [Google Scholar]
- 57.Keilhauer E.C., Hein M.Y., Mann M. Accurate protein complex retrieval by affinity enrichment mass spectrometry (AE-MS) rather than affinity purification mass spectrometry (AP-MS) Mol Cell Proteomics. 2015;14:120–135. doi: 10.1074/mcp.M114.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Res 2014;3:146. [DOI] [PMC free article] [PubMed]
- 59.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize implements and enhances circular visualization in R. Bioinformatics 2014;30:2811–2. [DOI] [PubMed]
- 61.Wiredja DD, Koyuturk M, Chance MR. The KSEA App: a web-based tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics 2017;33:3489–91. [DOI] [PMC free article] [PubMed]
- 62.Wang C., Xu H., Lin S., Deng W., Zhou J., Zhang Y., et al. GPS 5.0: an update on the prediction of kinase-specific phosphorylation sites in proteins. Genomics Proteomics Bioinformatics. 2020;18:72–80. doi: 10.1016/j.gpb.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tapial J., Ha K.C.H., Sterne-Weiler T., Gohr A., Braunschweig U., Hermoso-Pulido A., et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017;27:1759–1768. doi: 10.1101/gr.220962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma J, Chen T, Wu S, Yang C, Bai M, Shu K, et al. iProX: an integrated proteome resource. Nucleic Acids Res 2019;47:D1211–7. [DOI] [PMC free article] [PubMed]
- 69.Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., et al. The Genome Sequence Archive Family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021;19:578–583. doi: 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression and subcellular localization of NSP3 proteins with C-terminal tag A. Schematic illustration of plasmids used in Figures 1B, S1B, and S1C. B. NSP3 cleaves itself at the C-terminus. HEK293T cells transfected with CoVs-NSP3-EGFP plasmids were lysed and EGFP and NSP3 antibodies were used after immunoblotting. EGFP was released in WT, but not in CS- or dGG-mutated NSP3 samples. Green arrow indicates the released EGFP, whereas asterisk indicates non-specific signal. C. Validation of the specificity of CoV-1 NSP3 antibody. HEK293T cells transfected with indicated plasmids were fixed 48 h post-transfection. EGFP and NSP3 antibodies were used to detect the localization of NSP3 proteins.
NSP3 proteins have preferable cellular localization A and B. Cellular localization of NSP3 protein detected by EGFP antibody. HEK293T cells transfected with indicated plasmids were fixed 48 h post-transfection. EGFP fluorescence was used to detect the localization of CS-NSP3-EGFP protein. CANX and TOMM20 antibodies (red) were used to serve as markers for the ER (A) and mitochondria (B). Red arrow indicates that the large vesicle-like structure of CoV2-NSP3 was not co-localized with TOMM20. C. CoV2-NSP3 has two cellular localizations. Immunostaining was performed as in Figure S2A. Merged signals show that GFP and NSP3, but not NSP3 and Flag, are co-localized well. D. Flag-CoV1-NSP3 prefers to co-localize with CANX. Immunostaining was performed as in Figure S2A. Flag and CANX antibodies were used to visualize the co-localization between CoV1-NSP3 and the ER marker CANX. E. Interaction preference of NSP3 protein and CANX/TOMM20. Interactome studies of NSP3 based on Flag (Table S1) and EGFP (Table S3) IP-MS were analyzed to show the binding preference between NSP3 proteins and CANX/TOMM20. ER, endoplasmic reticulum; IP-MS, Immunoprecipitation-Mass Spectrometry.
PLpro domain of CoV1-NSP3 is responsible for the formation of the large vesicle-like structures CoV2-NSP3 fused with PLpro domain of CoV1-NSP3 induces large-vesicle structures. Immunostaining was performed as in Figure S2A.
Workflow and quality control of multi-omics study A. Schematic overview of the workflow to study the function of NSP3. B. Boxplots of log2 imputed intensity for all proteomics studies. FASP, Filter Aided Sample Preparation; xIP-MS, crosslinking Immunoprecipitation-Mass Spectrometry.
Protein–protein interaction network of NSP3 interactomes A. PCA analysis of top 1000 variable proteins of NSP3 interactomes. Top 1000 variable proteins with highest standard deviation (Table S1) were used as input for PCA analysis. B. Upset and Venn diagrams showing the overlap of NSP3 interactors. Significant interactors for WT and CS of NSP3 proteins were used as input. Data used for the analysis is available in Table S1. C and D. Protein-protein interaction network of CoV2-NSP3 (C) and CoV1-NSP3 (D) interactomes. Significant interactors of NSP3 were clustered by gene annotation and protein–protein interaction network was constructed using Cytoscape with ReactomeFI v7.2.3. PCA, principal component analysis; PC, principal component.
Crosslinking IP-MS to identify NSP3 interactome A. PCA analysis of top 1000 variable proteins of NSP3 interactomes. Top 1000 variable proteins of each condition (Table S2) were used as input for PCA analysis. B. Venn diagram showing the overlap of NSP3 interactors identified in crosslinking and non-crosslinking experiments. C. Clustering of significant interactors of NSP3 proteins using crosslinking IP-MS. Analysis and color as Figure 3A. Full list of interactors is available in Table S2. D. Representative enriched GO of identified interactors in crosslinking IP-MS. Analysis as Figure 3B. Full list of GO terms is available in Table S2.
NSP3 interactome identified by EGFP IP-MS. A. PCA analysis of top 1000 variable proteins of NSP3-EGFP interactomes. Top 1000 variable proteins with highest standard deviation (Table S3) were used as input for PCA analysis. B. Clustering of significant interactors of NSP3 proteins. The relative intensity to control samples of significant interactors was compared between CoV1-NSP3 and CoV2-NSP3. Full list of interactors is available in Table S3. C. Venn diagram showing the overlap of CoV1-NSP3 and CoV2-NSP3 interactomes. D. Representative enriched GO of identified interactors. Analysis as Figure 3B. Full list of GO terms is available in Table S3.
Phosphoproteome related analyses A. PCA analysis of top 1000 variable phosphorylated peptides of NSP3 expressing cells. Top 1000 variable phosphorylated peptides with standard deviation (Table S5) were used as input for PCA analysis. B. Clustering of DEPPs which were also found in SARS-CoV-2 infection studies. Two reported phosphoproteome data were used as inputs to identify consistent phosphorylated peptides. Analysis and color as Figure 3A. Full list of DEPPs is available in Table S5. C. Cell cycle analysis of NSP3 expressing cells. Cells transfected with indicated plasmids were harvest for FACS analysis using propidium iodide. Significant differences compared to control cells were calculated using Student’s t-tests. The graph showed mean ± SD, n = 3, * indicates P < 0.05, ** indicates P < 0.01. One representative experiment was shown. D. and E. Kinases inhibitors mapped to kinases whose activity mapped to DEPPs in CoV2-NSP3 (D) and CoV1-NSP3 (E) expressing cells. Red indicates upregulated activity, whereas blue indicates downregulated activity. F. Clustering of DEPPs that regulated by IKK-family and TBK1. Analysis and color as Figure 3A. FACS, Fluorescence-activated Cell Sorting; SD, Standard Deviation; pSTY, phospho-Serine, Threonine, Tyrosine; G0, G0 phase; G1, G1 phase; G2, G2 phase; S, S phase; M, M phase.
Ubiquitylome related analyses A. PCA analysis of top 1000 variable ubiquitinated peptides of NSP3 expressing cells. Top 1000 variable ubiquitinated peptides with highest standard deviation (Table S6) were used as input for PCA analysis. B. Deubiquitinase activity dependent ubiquitylome of NSP3 protein. Analysis and color as in Figure 3A. Full list of DEUPs is available in Table S6.
Regulation of innate response by NSP3. Multi-omics data were combined to model the regulation innate response by NSP3. Network was created using Cytoscape with ReactomeFI v7.2.3.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository (ProteomeXchange: PXD023927) [68], and are publicly accessible at http://proteomecentral.proteomexchange.org. The raw sequence data are deposited in the Genome Sequence Archive [69] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: HRA000634), and are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human/.